-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
DNA Methylation and Histone Modifications Regulate Shoot Regeneration in by Modulating Expression and Auxin Signaling
Plants have a profound capacity to regenerate organs from differentiated somatic tissues, based on which propagating plants in vitro was made possible. Beside its use in biotechnology, in vitro shoot regeneration is also an important system to study de novo organogenesis. Phytohormones and transcription factor WUSCHEL (WUS) play critical roles in this process but whether and how epigenetic modifications are involved is unknown. Here, we report that epigenetic marks of DNA methylation and histone modifications regulate de novo shoot regeneration of Arabidopsis through modulating WUS expression and auxin signaling. First, functional loss of key epigenetic genes—including METHYLTRANSFERASE1 (MET1) encoding for DNA methyltransferase, KRYPTONITE (KYP) for the histone 3 lysine 9 (H3K9) methyltransferase, JMJ14 for the histone 3 lysine 4 (H3K4) demethylase, and HAC1 for the histone acetyltransferase—resulted in altered WUS expression and developmental rates of regenerated shoots in vitro. Second, we showed that regulatory regions of WUS were developmentally regulated by both DNA methylation and histone modifications through bisulfite sequencing and chromatin immunoprecipitation. Third, DNA methylation in the regulatory regions of WUS was lost in the met1 mutant, thus leading to increased WUS expression and its localization. Fourth, we did a genome-wide transcriptional analysis and found out that some of differentially expressed genes between wild type and met1 were involved in signal transduction of the phytohormone auxin. We verified that the increased expression of AUXIN RESPONSE FACTOR3 (ARF3) in met1 indeed was due to DNA demethylation, suggesting DNA methylation regulates de novo shoot regeneration by modulating auxin signaling. We propose that DNA methylation and histone modifications regulate de novo shoot regeneration by modulating WUS expression and auxin signaling. The study demonstrates that, although molecular components involved in organogenesis are divergently evolved in plants and animals, epigenetic modifications play an evolutionarily convergent role in this process.
Published in the journal: . PLoS Genet 7(8): e32767. doi:10.1371/journal.pgen.1002243
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002243Summary
Plants have a profound capacity to regenerate organs from differentiated somatic tissues, based on which propagating plants in vitro was made possible. Beside its use in biotechnology, in vitro shoot regeneration is also an important system to study de novo organogenesis. Phytohormones and transcription factor WUSCHEL (WUS) play critical roles in this process but whether and how epigenetic modifications are involved is unknown. Here, we report that epigenetic marks of DNA methylation and histone modifications regulate de novo shoot regeneration of Arabidopsis through modulating WUS expression and auxin signaling. First, functional loss of key epigenetic genes—including METHYLTRANSFERASE1 (MET1) encoding for DNA methyltransferase, KRYPTONITE (KYP) for the histone 3 lysine 9 (H3K9) methyltransferase, JMJ14 for the histone 3 lysine 4 (H3K4) demethylase, and HAC1 for the histone acetyltransferase—resulted in altered WUS expression and developmental rates of regenerated shoots in vitro. Second, we showed that regulatory regions of WUS were developmentally regulated by both DNA methylation and histone modifications through bisulfite sequencing and chromatin immunoprecipitation. Third, DNA methylation in the regulatory regions of WUS was lost in the met1 mutant, thus leading to increased WUS expression and its localization. Fourth, we did a genome-wide transcriptional analysis and found out that some of differentially expressed genes between wild type and met1 were involved in signal transduction of the phytohormone auxin. We verified that the increased expression of AUXIN RESPONSE FACTOR3 (ARF3) in met1 indeed was due to DNA demethylation, suggesting DNA methylation regulates de novo shoot regeneration by modulating auxin signaling. We propose that DNA methylation and histone modifications regulate de novo shoot regeneration by modulating WUS expression and auxin signaling. The study demonstrates that, although molecular components involved in organogenesis are divergently evolved in plants and animals, epigenetic modifications play an evolutionarily convergent role in this process.
Introduction
Differentiated somatic tissues of plants can be reprogrammed to generate various organs, a process called de novo organogenesis. This feature is not only critical for in vitro plant propagation and application of biotechnology, but also provides a good experimental system for understanding regulatory mechanisms underlying organogenesis.
Recent studies have revealed some molecular mechanisms underlying de novo shoot regeneration in Arabidopsis [1]–[4], in which WUS, a transcription factor, plays a key role [5], [6]. WUS is a master regulator of stem cell fate determination in shoot apical meristem (SAM), on which many signaling pathways converge [7]. It turned out to be also critical for de novo shoot regeneration. During de novo shoot regeneration in Arabidopsis, expression of WUS is sufficient to specify the organizing center, which is required for stem cell induction and subsequent shoot regeneration [5], [6], [8]. WUS induction is also essential for shoot formation during de novo somatic embryogenesis [9]. Induction of the WUS expression during de novo shoot regeneration was regulated by the master phytohormone auxin [2], [5]. Recently, WUS expression in the organizing center of the Arabidopsis plant SAM was shown to be regulated by epigenetic modifications [10].
Epigenetic modifications, including DNA methylation and histone modifications, occur extensively during cellular differentiation and development in mammals [11]–[13]. In mammals, the patterns of DNA methylation are established by de novo DNA methyltransferase 3 (DNMT3) family and maintained by methyltransferase DNMT1 [14]. DNMT1 plays a vital role in controlling the self-renewal and differentiation of stem cells during hematopoiesis and leukemogenesis and is critical for progenitor maintenance and self-renewal in mammalian somatic tissues [15], [16]. DNA methylation and histone modifications regulate gene expression through changing chromatin structure and transcriptional activities [17]–[19]. For instance, transcriptional repression is associated with hypermethylation of DNA, histone deacetylation and histone H3K9 methylation, whereas active chromatin is linked with hypomethylation of DNA, histone acetylation and histone H3K4 methylation [17], [20].
In plants, pattern changes of DNA methylation and histone modifications leading to changes in chromatin state occur in plant cells undergoing dedifferentiation [21]–[24]. Furthermore, DNA methylation at some promoters is critical for establishing or maintaining the undifferentiated cell state in plants [25]. However, whether and how epigenetic modifications are involved in cell differentiation during de novo shoot regeneration is unknown. Here we showed that mutations of key epigenetic genes altered WUS expression and developmental rates of regenerated shoots in vitro. In addition, epigenetic marks of DNA methylation and histone modifications in the regions of WUS underwent dynamic changes during de novo shoot regeneration, correlating with dynamic WUS expression levels. Genome-wide transcriptional analysis indicated that some genes involved in auxin signaling and meristem development were methylated within the callus, but were demethylated following an induction treatment. Based on these results, we propose that dynamic DNA methylation and histone modifications mediate de novo shoot regeneration in Arabidopsis through WUS and auxin signaling.
Results/Discussion
Mutations interfering with epigenetic modifications changed developmental rates of de novo shoot regeneration
To find out whether DNA methylation and histone modifications played roles in de novo shoot regeneration, we first compared the capacity and rates of shoot regeneration between wild type and various epigenetic mutants after calli being transferred onto a shoot induction medium (SIM) from a callus induction medium (CIM) [26]. Arabidopsis METHYLTRANSFERASE1 (MET1), KRYPTONITE (KYP), JMJ14 and HISTONE ACETYLTRANSFERASE1 (HAC1), among diverse genes involved in epigenetic modifications, have been well characterized [27]–[31]. MET1 is an ortholog of DNMT1, which maintains DNA methylation directly at CpG motif and indirectly at non-CG motif [27], [32], [33]. Functional loss of MET1 resulted in delayed transition from vegetative phase to reproductive phase [32]. KYP encodes histone H3K9 methyltransferase, and mutation of which resulted in abnormal number of floral organs [28]. JMJ14 encodes histone H3K4 demethylase that inhibited flowering under long-day condition [29], [34]. HAC1 encodes histone acetyltransferase, regulating flowering time through histone acetylation [31], [35]. We used the final percentage of shoot primordia on SIM to reflect the capacity of de novo shoot regeneration, whereas the timely appearance of shoot primordia to reflect their developmental rates.
Comparable maximal percentages of shoot primordia were reached after 18 days of incubation on SIM for both wild type and all tested mutants, including met1, kyp, jmj14 and hac1 (Figure 1A–1C), indicating that there was no significant difference in the capacities of de novo shoot regeneration. However, it took different induction time for the wild-type calli and the mutant calli to reach half of the maxima (Figure 1A–1C). Specifically, the mutants whose epigenetic changes were associated with more active transcription, such as met1, kyp, jmj14 [27]–[29], took significantly less time to reach half of the maxima as compared to the wild type (Figure 1A–1C). In contrast, the mutant associated with more repressed transcription such as hac1 took significantly more time to reach half of the maxima (Figure 1C). We obtained similar results indicating precocious or delayed initiation of shoots in these mutants using either pistils or roots as explants (Figure 1A–1C, Figure S1). Interestingly, calli of met1 cultured on SIM develop differently from those of the wild type (Figure 1D). At 4 days on SIM, around 70% met1 calli contained green regions from which the shoots would differentiate, but these green regions could not be identified in the wild-type calli. At 6 to 14 days on SIM, more shoots emerged from the met1 calli than those from the wild-type calli (Figure 1D). At 18 days on SIM, the shoots from the met1 calli were much precocious compared with those from the wild-type calli although the percentages of shoots from both the wild-type and the met1 calli were similar (Figure 1A). We also obtained similar results with roots as explants (Figure S2). Thus, these results indicated that epigenetic modifications, including DNA methylation and histone modifications, played roles in mediating developmental rate of de novo shoot regeneration.
Fig. 1. Mutation in key epigenetic genes alters the rate of Arabidopsis shoot regeneration in vitro. 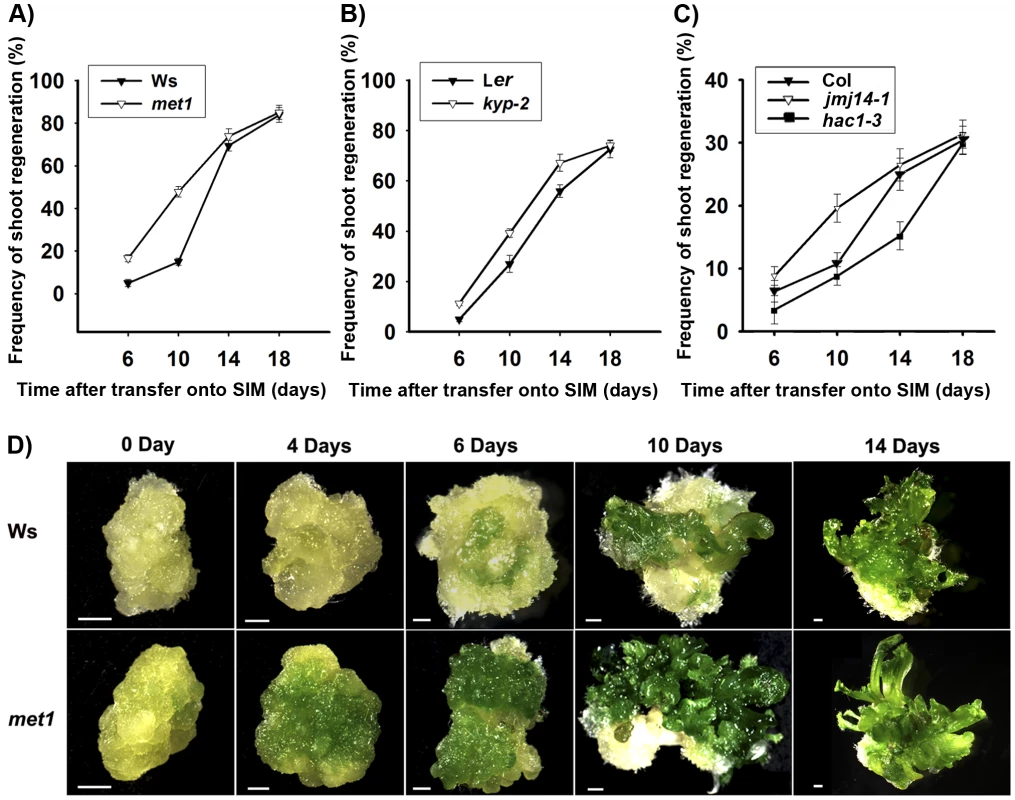
A) Frequency of shoot regeneration from calli of the wild type (Ws) and the mutant met1. B) Frequency of shoot regeneration from calli of the wild type (Ler) and the mutant kyp-2. C) Frequency of shoot regeneration from calli of the wild type (Col) and the mutants jmj14-1 and hac1-3. Calli were induced from pistils on CIM, and were then transferred onto SIM for shoot induction. Data are presented as mean values from three sets of biological replicates. In each replicate, at least 60 calli were examined. D) Calli of the wild type (Ws) and the mutant met1 cultured on SIM for 0 day, 4 days, 6 days, 10 days and 14 days. Scale bars, 1 mm. Regulation of WUS expression during de novo shoot regeneration may have resulted from dynamic DNA methylation
It was well established that WUS expression is critical for stem cell formation during de novo shoot regeneration [5], [6]. Here, we showed that induction of wild-type calli on SIM for 4 days (S4) and 6 days (S6) was accompanied by a significant increase of WUS level through qRT-PCR analysis (Figure 2A). In contrast, WUS transcripts were in a low level in wild-type calli on CIM for 16 days (C16) and 20 days (S0, non-induced calli), and similar results were obtained in the prolonged time, such as calli on CIM for 24 days (C24) and 26 days (C26). We further determined the expression patterns of WUS by pWUS::GUS reporter and in situ hybridization, and the results demonstrated that local distribution of WUS transcripts occurred in wild-type calli on SIM (Figure 3, Figure S3). Because it was shown previously that WUS expression was mediated by epigenetic factors [10], we were tempted to hypothesize that the regulation of WUS expression during de novo shoot regeneration might have resulted from reduced DNA methylation.
Fig. 2. DNA methylation and histone modifications regulate WUS transcript levels. 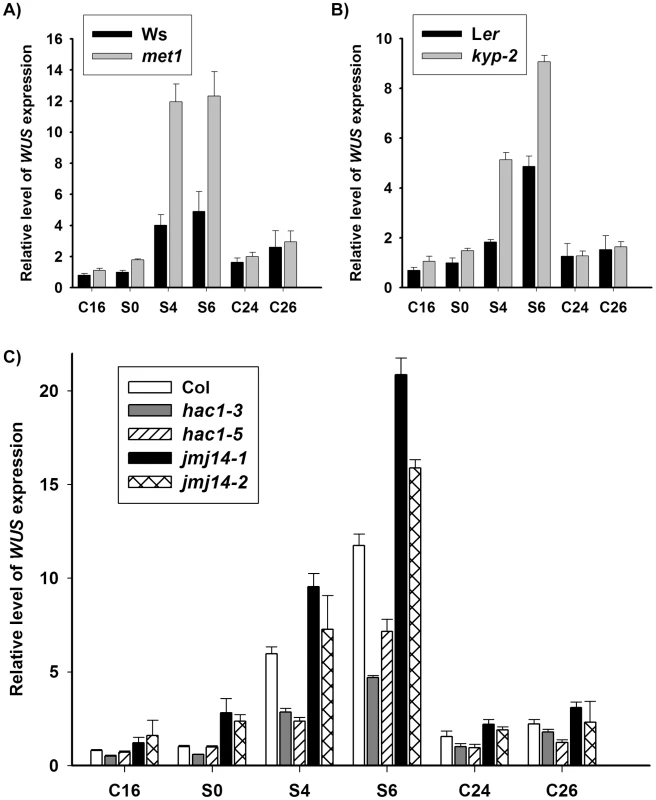
A) Transcript levels of WUS in calli of the wild type (Ws) and the mutant met1. B) Transcript levels of WUS in calli of the wild type (Ler) and the mutant kyp-2. C) Transcript levels of WUS in calli of the wild type (Col) and the mutants, hac1-3, hac1-5, jmj14-1 and jmj14-2. Total RNAs were isolated from calli of wild type (Ws, Ler and Col) and various mutants (met1, kyp-2, jmj14-1, jmj14-2, hac1-3 and hac1-5) cultured on SIM at the indicated time points, respectively. WUS transcript levels were quantified by qRT-PCR. The results are shown as mean values of three biological replicates with standard errors. The relative expression level of WUS gene, corresponding to the expression level of TUBULIN2, was calculated using the comparative C(T) method. After incubating on CIM for 20 days (S0), some of the calli were transferred onto SIM for further induction for 4 days (S4) and 6 days (S6), other calli were still cultured on CIM as controls (C24, C26). C16, C24, C26 indicated that pistils as explants were cultured on CIM for 16 days, 24 days and 26 days, respectively. Fig. 3. Regulation of WUS expression in met1 mutant. 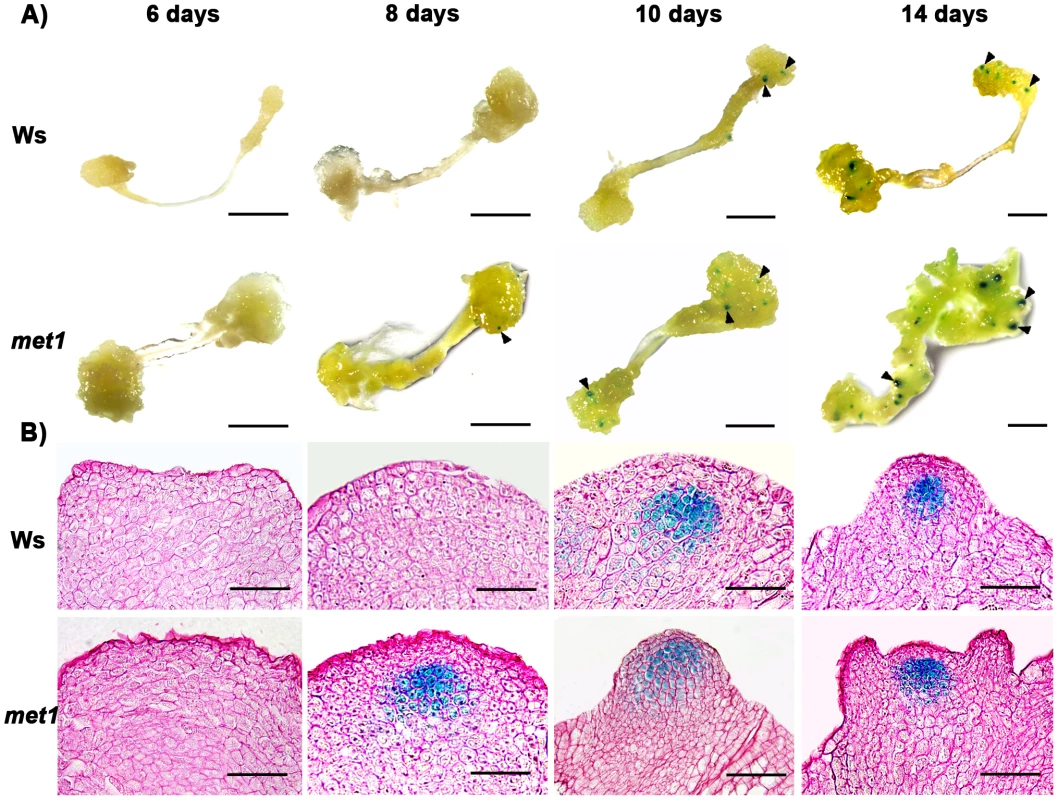
A) By roots as explants, pWUS::GUS transgenic calli in the wild type transferred onto SIM for 6 days, 8 days, 10 days and 14 days, and pWUS::GUS transgenic calli in the met1 mutant transferred onto SIM for 6 days, 8 days, 10 days and 14 days. Arrowheads indicate pWUS::GUS signals. Scale bars, 1 mm. B) Longitudinal sections of pWUS::GUS transgenic calli in both the wild type and the met1 mutant transferred onto SIM for 6 days, 8 days, 10 days and 14 days, respectively. Scale bars, 50 µm. To test this possibility, we first compared DNA methylation of the ∼10 kb WUS genomic sequences between the calli of wild type on CIM (C16 and S0) and those on SIM (S6) by bisulfite genomic sequencing. Three regions within the WUS genomic sequences were hyper-methylated in S0 calli but substantially decreased in S6 calli (Figure 4A and 4B). Among the three regions, region I was previously proposed to regulate WUS expression [36]. Both CpG dinucleotide motifs and non-CG motifs in the three regions of the WUS genomic sequences showed induced demethylation upon induction on SIM (Figure 4B). These results showed that de novo shoot regeneration was accompanied with demethylation on methylated WUS genomic sequences. That could partially contribute to the regulation of WUS expression during de novo shoot regeneration.
Fig. 4. Analysis of WUS methylation through bisulfite genomic sequencing. 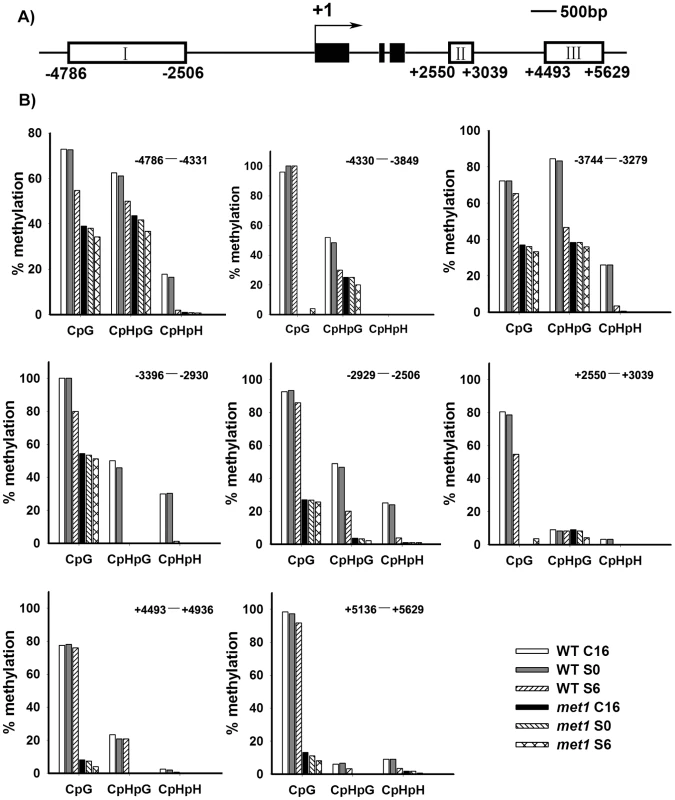
A) A diagram of WUS structure, with +1 as the transcription start site and rectangles representing the methylated region I, II and III. B) Cytosine methylation at region I, II and III of WUS was determined by bisulfite genomic sequencing. Genomic DNA methylation status of WUS is shown in calli of the wild type on CIM for 16 days (WT, C16) and for 20 days (WT, S0), and on SIM for 6 days (WT, S6). Calli of met1 are incubated on CIM for 16 days (met1, C16) and for 20 days (met1, S0), and on SIM for 6 days (met1, S6). H represents A, T or C. Demethylation and regulation of WUS expression in met1 mutant
Because DNA methylation was significantly reduced in met1 mutant [27], we wondered whether DNA methylation in the WUS genomic sequences would be affected in met1 mutant. To find out, we used two approaches. First, we compared the expression patterns of WUS in wild-type calli and met1 calli at different induction points. Indeed, the met1 mutant showed much higher WUS level than that in the wild type at each time point by qRT-PCR (Figure 2A). Then, in situ hybridization analysis demonstrated that localization of WUS in the met1 calli on SIM was earlier than that in the wild-type calli on SIM (Figure S3A–S3F, Table S1). GUS staining confirmed that the pattern of WUS expression is similar to that in situ hybridization (Figure 3), and the number of GUS signal distribution in both the met1 calli and the wild-type calli on SIM is consistent to percentages of shoot primordia on SIM at different induction points (Figure 3, Figure S3, Table S2). Thus, the results indicated that WUS expression and corresponding developmental rate of de novo shoot regeneration were mediated by reduced DNA methylation.
Next, we tested whether MET1 loss of function affected the methylation status of WUS genomic region by bisulfite genomic sequencing. We found that the calli of met1 mutant on CIM (C16 and S0) and on SIM (S6) showed much lower level of DNA methylation in the WUS genomic region than those of wild type under the same condition (Figure 4B). WUS expression was detected in met1 calli earlier than in wild type based on in situ hybridization and GUS reporter analysis (Figure 3 and Figure S3). In addition, met1 contained more WUS-expressing regions than wild type, indicating that increased WUS expression level contributed to elevated the number of organizing centers (Figure 3 and Figure S3). These results suggested that the regulation of WUS expression in met1 mutant during de novo shoot regeneration could at least partially be contributed by DNA demethylation on methylated WUS genomic sequences.
Dynamic changes of histone modifications at the genomic regions of WUS during de novo shoot regeneration
Higher WUS level in the met1 mutant suggested the involvement of MET1-mediated DNA methylation in the regulation of WUS expression. However, the expression of WUS still responded to the induction by incubation on SIM in met1 mutant (Figure 2A), indicating additional pathways that regulated the dynamic expression of WUS. Because we showed that histone modifications were also important for de novo shoot regeneration (Figure 2B and 2C), we next tested whether histone modifications played a role in mediating WUS expression during de novo shoot regeneration.
We analyzed several histone modifications for the WUS genomic sequences using chromatin immunoprecipitation at two developmental stages: S0 and S6. Methylation at histone H3 at lysine 4 (H3K4me3) was shown to occur in euchromatin undergoing active transcription [37]. Whereas methylation at histone H3 at lysine 9 (H3K9me2) was shown to inhibit transcription [38]. Additionally, acetylation at histone H3 at lysine 9 (H3K9ac) is one of the most characterized epigenetic marks invariably associated with active transcription in all species investigated so far [18]. It also plays a crucial role in plant development [39].
Our results showed that these three histone modifications were dynamically regulated at the WUS genomic sequences during de novo shoot regeneration. Compared with S0, S6 showed an increase in the levels of H3K4me3 at region a and d, but not at b and c (Figure 5A and 5B). H3K4me3 occurred in euchromatin undergoing active transcription [37], therefore increased H3K4me3 levels were consistent with WUS induction during de novo shoot regeneration (Figure 1C, Figure 2C). A mark for chromatin acetylation, H3K9ac, also showed increased levels at all four regions during induction (Figure 5C). In contrast to these epigenetic marks associated with active transcription, H3K9me2, which is associated with transcription suppression [37] were reduced during de novo shoot regeneration in all four regions (Figure 5B). The changes at these epigenetic marks around WUS genomic region explained the active state of WUS chromatin structure, and might well contribute to the regulation of WUS expression during de novo shoot regeneration.
Fig. 5. ChIP assays of calli of wild type on SIM at the WUS locus. 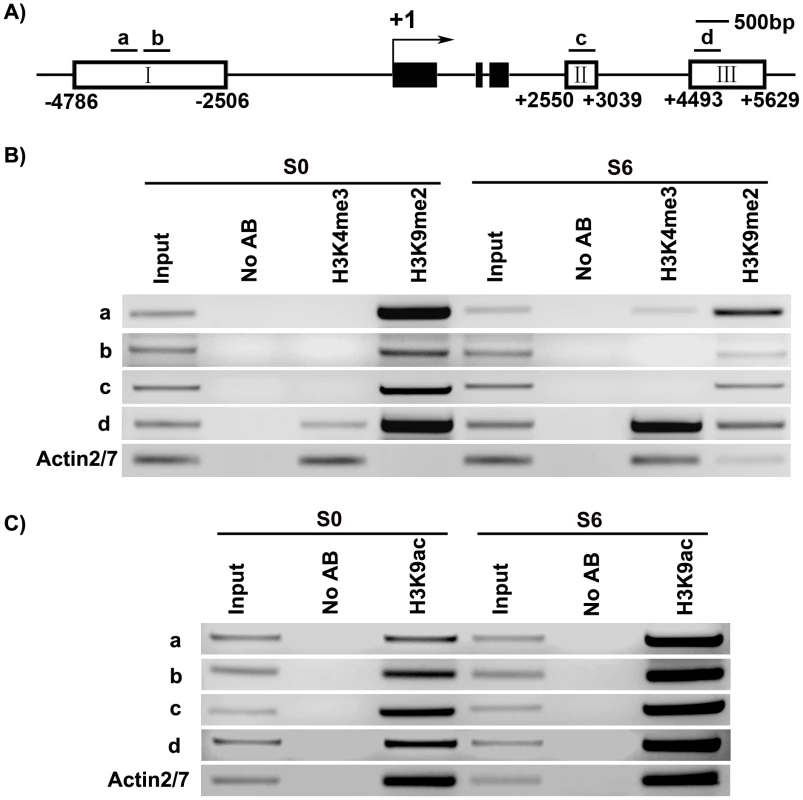
A) A diagram of WUS structure, with +1 as the transcription start site, and bars representing the regions examined by ChIP. B) ChIP analysis using antibodies against trimethyl H3K4 and dimethyl H3K9 at 5′ and 3′ regions of WUS in calli of wild type for 20 days on CIM (S0) and 6 days on SIM (S6). C) ChIP analysis using antibodies against H3 acetyl Lys 9 at 5′ and 3′ regions of WUS in calli of wild type (S0, S6). ACTIN was used as a control. The input was chromatin before immunoprecipitation. ‘No AB’ corresponds to chromatin treated with normal mouse IgG as the negative control. Three biological replicates were analyzed and each was tested by three technical replicates, and similar results were obtained. Representative data were shown. WUS expression was changed in mutants defective in histone modifications
Dynamic histone modifications at the genomic regions of WUS during de novo shoot regeneration indicated that histone modifications contributed to regulation of WUS expression during de novo shoot regeneration. To provide further evidence that histone modifications regulated WUS expression in this process, we examined transcript level of WUS in mutants that were defective in histone modifications by qRT-PCR. As stated before, KYP, JMJ14 and HAC1 encoded enzymes for histone modification, mutations of which affected the developmental rate of de novo shoot regeneration (Figure 1B and 1C, Figure S1). Comparing with the wild-type calli, levels of WUS expression in the calli of the mutant kyp-2 were significantly enhanced compared to those of wild type for 6 days on SIM (Figure 2B). Similar results were obtained for the mutants jmj14-1 and jmj14-2 (Figure 2C). Contrast to the mutants kyp and jmj14, the levels of WUS transcripts in two different allelic hac1 mutants were reduced compared to that of wild type (Figure 2C).
Then, we used kyp-2 calli on SIM (S0, S4, and S6) to do in situ hybridization analysis. The results showed that localization of WUS signals in kyp-2 calli on SIM occurred early comparing to that in wild-type calli on SIM (Figure S3G–S3L). Also, the number of localized WUS signals in kyp-2 calli on SIM (S4 and S6) was more than that in wild-type calli at the same time points (Table S1). Similar to the case of met1, expression of WUS appeared earlier in kyp-2 calli than in wild type (Figure S3). Thus, changes of WUS expression in these mutants correlated with their different developmental rates of de novo shoot regeneration, suggesting that WUS expression was regulated by histone modifications.
SIM-induced as well as MET1-dependent transcriptional changes during de novo shoot regeneration
Our results showed that DNA methylation and histone modifications regulated WUS expression during de novo shoot regeneration. To get a whole picture of epigenetic modifications during this process, we decided to do a genome-wide expression profiling using the Affymetrix ATH1 full genome array. We analyzed the transcriptomes of wild-type calli being transferred to CIM for 20 days (S0) and to SIM for 6 days (S6). Because met1 calli showed significantly different developmental rate from wild-type calli, we also analyzed transcriptomes of met1 calli being transferred to CIM for 20 days (M0) for comparison. Significance Analysis of Microarrays software package analysis was conducted for three biological samples replicates between the Ws and met1. The q value≤0.05 and fold change ≥2 were used as the threshold for candidate gene selection (Figure 6A). This criterion gave 1334 upregulated genes, and 501 downregulated genes by induction on SIM (S6 versus S0) (Table S3). 768 candidate genes showed over 2 fold difference between M0 and S0, suggesting that they might be regulated by MET1-dependent DNA methylation (Table S4). 308 candidate genes showed over 2 fold difference both between S6 versus S0 and between M0 versus S0, suggesting that they might be induced on SIM and be regulated by MET1-dependent DNA methylation (Table S5). By qRT-PCR analysis, we confirmed the microarray data (Figure S4).
Fig. 6. Identification of the candidate genes regulated by DNA methylation. 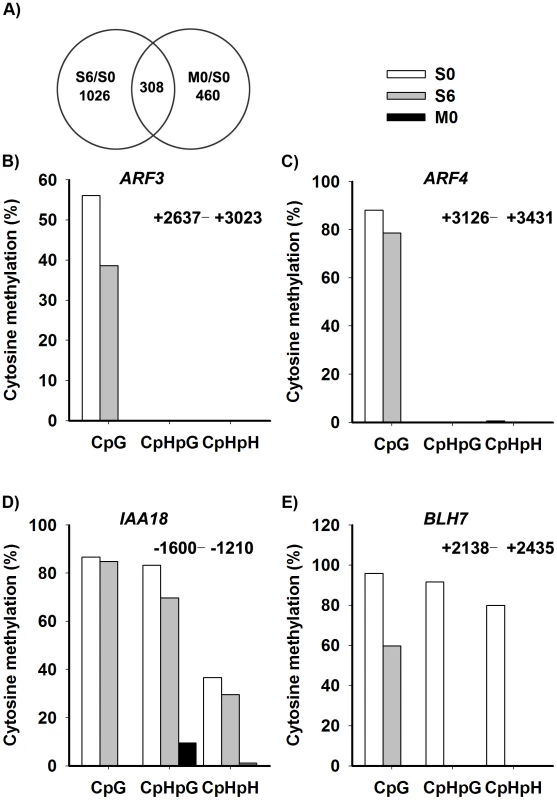
A) The overlap between differentially-expressed genes of S6 versus S0 (Table S3) and M0 versus S0 (Table S4) were identified as candidate genes, and were listed in Table S5. A two-fold difference in the expression level of genes with a q value≤0.05 between S6 versus S0 and M0 versus S0 was set as the threshold for the selection of differentially-expressed genes. B)–E) Cytosine methylation levels of ARF3, ARF4, IAA18 and BLH7 genes in calli of wild type (S0, S6), and calli of met1 (M0) were determined by bisulfite genomic sequencing. H represents A, T or C. Because auxin and cytokinin are essential for de novo shoot regeneration [2], [5], we selected genes involved in cytokinin and auxin signaling for bisulfite sequencing analysis. Indeed, some displayed differential methylation patterns during de novo shoot regeneration, such as AUXIN RESPONSE FACTOR3 (ARF3), AUXIN RESPONSE FACTOR4 (ARF4), INDOLE-3-ACETIC ACID INDUCIBLE18 (IAA18) and BELL1-LIKE HOMEODOMAIN7 (BLH7) (Figure 6B–6E). A loss of DNA methylation occurred in these genes, along with increased levels of their transcription in induced wild-type calli (Figure S4). Their expression levels were also higher in met1 than those in the wild type, suggesting that the expression of these genes might be regulated by a MET1-dependent dynamic DNA methylation during shoot regeneration. On the other hand, some candidate genes selected from SIM-induced and MET1-dependent pathways displayed no methylation, such as ASMMETRIC LEAVES1 (AS1), ARABIDOPSIS RESPONSE REGULATOR15 (ARR15), CYTOKININ OXIDASE/DEHYDROGENASE1 (CKX1), INDOLE-3-ACETIC ACID27 (IAA27) and PINOID2 (PID2), but they displayed great changes in their transcriptional levels upon SIM-induction, implying that those genes might not be directly regulated by MET1 (Table S5).
Epigenetic modifications: evolutionary recurring themes for reprogramming
DNA methylation and histone modifications are critical epigenetic processes that control chromatin structure and gene expression during development and differentiation [17], [18], and there are likely complicated interactions between these processes [20], [40]. In human, a crosstalk between DNA methylation and histone modifications has been proposed to regulate gene transcription in tumors [20]. Similarly, DNA methylation controls histone H3K9 methylation and further affect heterochromatin assembly in Arabidopsis [41]. Recent study has indicated that chromatin status facilitates the accessibility of transcription factor to FLOWERING LOCUS T (FT) in Arabidopsis, and distant regulatory regions are required for FT transcription [42]. WUS transcription is regulated through a fairly complicated chromatin remodeling mechanism in the SAM of the Arabidopsis plant [43]. It was shown that WUS expression was positively correlated with FASCIATA1 (FAS1)/FAS2, subunits of ASSEMBLY FACTOR-1 (CAF-1), and BRUSHY1 (BRU1), both of which regulate post-replicative stabilization of chromatin structure [44], [45]. Another study showed that the chromatin remodeling factor SPLAYED (SYD) directly regulated WUS to maintain proper WUS transcript levels in its spatial expression domain [46]. It has been demonstrated that at least 3.5 kb fragment upstream of WUS is required for its spatiotemporal expression during plant development [36]. Here, we showed that the 5′ and 3′ regions of WUS were regulated by SIM-induced changes of DNA methylation and histone modifications. Because the met1-3 kyp-7 double mutant displayed more severe phenotypes than each single mutant [19], we propose that regulation of WUS by DNA methylation and histone modifications may function in a partially redundant manner during de novo shoot regeneration. To understand mechanism of the in vitro organogeneis mediated by the factors involved in both DNA methylation and histone modifications, knocking out both DNA methylation and histone modifications remains to be investigated in the future.
It has long been thought that animal cells, once committed to a specific lineage, can no longer change their fate. However, recent studies suggested that differentiated animal cells do maintain plasticity and can be induced to undergo reprogramming [47], [48]. Further studies have shown that differentiated cells in mouse can be reprogrammed to pluripotent stem cells by introducing four transcription factors [49]. Plant cells can easily regenerate organs from the differentiated tissues under proper cultured conditions [1]. Previously, we used Arabidopsis ptstils as explants on CIM to obtain the callus, a mass of pluripotent cells [26], and by transferring calli onto SIM, the expression of WUS was induced in a group of cells termed the organizing center as a self-renewing source of stem cells within calli. The induced organizing center and stem cells were responsible for subsequent shoot regeneration. Here, we showed that expression of many genes was induced by SIM-induction (Figure 6A). Those genes were divided into either MET1-dependent or MET1-independent. Among MET1-dependent genes, WUS is a key transcription factor to regulate shoot regeneration [1]. ARF3 was required for shoot induction (Cheng et al., unpublished data). Previous study showed that ARF3 and ARF4 act redundantly to establish the abaxial cell fate of the Arabidopsis leaves [50]. Thus, ARF3 and ARF4 may function on de novo meristem formation mediated by epigenetic modifications. MET1-independent genes might also be involved in the process of shoot induction. Our results suggested that pluripotent cells of the callus can be reprogrammed to stem cells and subsequent, shoot formation through the regulation of both MET1-dependent genes, such as WUS and ARFs, and some MET1-independent genes.
In conclusion, our results indicate that dynamic DNA methylation and histone modifications contribute to the control of stem-cell formation and subsequent shoot regeneration. These epigenetic modifications regulate WUS and probably hormone-related genes, whose spatiotemporal expression was critical for de novo shoot regeneration. In mammals, epigenetic modifications of transcription factors and of components in hormone signaling pathways also play crucial roles in cell differentiation and organogenesis [51], [52]. Our results thus provide an interesting scenario in which epigenetic modifications were adopted as recurring themes during evolution for de novo organogenesis.
Materials and Methods
Plant materials
The met1 mutant in the Wassilewskija (Ws) background was a kind gift from Dr. J. Bender (The MCB Department of Brown University) [27]. The kyp-2 [28] mutant in the Landsberg (Ler) background, jmj14-1, jmj14-2 [29], hac1-3, and hac1-5 [31] mutants in the Columbia (Col) background were generously provided by Dr. Xiaofeng Cao (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences).
Plant growth and shoot regeneration
Plants were grown as previously described [9]. Arabidopsis seeds were surface sterilized and plated on germination medium [53]. After cold treatment for 2 days at 4°C in the dark, they were transferred to sterile conditions or the growth chamber at 22°C in a 16 h light/8 h dark cycle. Shoot regeneration procedures used in this study were based on the previously described protocols [26], [54]. Pistils were excised from sterile Arabidopsis plants and transferred onto callus induction medium (CIM, MS medium [53] with 0.5 mg/L 2, 4-dichlorophenoxyacetic acid (2, 4-D) and 1.0 mg/L 6-benzylaminopurine (6-BA)). The explants were incubated for 20 days on CIM to induce callus production, and calli were then transferred onto shoot induction medium (SIM, MS medium with 0.01 mg/L indole-3-acetic acid (IAA) and 2 mg/L zeatin (ZT)). Root explants of 5–10 mm length were excised from 7-day-sterile seedlings, then transferred onto callus induction medium (CIM, Gamborg's B5 medium [55] with 0.5 g/L MES, 2% glucose, 0.2 µmol/L kinetin, and 2.2 µmol/L 2,4-dichlorophenoxyacetic acid (2,4-D), 0.8% agar), and incubated for 6 days in continuous light. Finally, explants were transferred onto shoot-inducing medium (SIM, Gamborg's B5 medium with 0.5 g/L MES, 2% glucose, 0.9 µmol/L 3-indoleacetic acid, 0.5 µmol/L 2-isopentenyladenine) and incubated in continuous light.
The morphology of calli was examined and photographed with an Olympus microscope. We defined the number of regenerated shoots as the number of at least 2 mm long shoots on each callus.
In situ hybridization
Probes were labeled using digoxigenin RNA labeling kit (Boehringer Mannheim). An antisense probe from a full-length WUS cDNA clone was generated using T7 RNA polymerase, and a sense probe was synthesized using SP6 RNA polymerase. The detailed protocol was carried out as described previously [56]. Primer sequences used for probes amplification are summarized in Table S6.
β-glucuronidase (GUS) assay
Plant tissues were incubated in GUS assay solution (50 mmol/L Na2HPO4, 50 mmol/L KH2PO4, pH 7.2, 10 mmol/L Na2EDTA, 0.5 mmol/L K3Fe(CN)6, 0.5 mmol/L K4Fe(CN)6, 1% Triton X-100 and 2 mmol/L X-Gluc (Bio. Basic Inc., Canada)) at 37°C for 12 h. To further investigate WUS expression pattern, some GUS-stained tissues were embedded in paraffin (Sigma) and sectioned. To display the outline of cells clearly, ruthenium red (200 mg/L) was used to stain cell walls.
Genomic bisulfite sequencing
DNA methylation assays were performed by bisulfite sequencing as previously described [57]. PCR products were cloned into the pMD19-T Simple Vector (Takara), and 12 clones were sequenced to determine the methylation status of a locus in each genotype. Primer sequences are shown in Table S6. Bisulfite sequencing data were analyzed by the CyMATE software [58]. The results returned by CyMATE were input into SigmaPlot 10.0 to illustrate DNA methylation frequencies at CG, CHG and CHH (where H = A, C or T) at the various cultured stages of each genotype.
Chromatin immunoprecipitation assay
The Arabidopsis calli grown on CIM for 20 days (S0) and on SIM for 6 days (S6) were vacuum-infiltrated with formaldehyde crosslinking solution. Chromatin immunoprecipitation was performed according to manufactures' instructions (Epigentek Group Inc. USA, Catalogno. P-2014). Chromatin samples were immunoprecipitated with antibodies against a negative control normal mouse IgG and H3 dimethyl Lys 9 (both included in EpiQuik™ Plant ChIP Kit), or with antibodies against H3 trimethyl Lys 4 (Abcam USA, Catalogno. ab1012) and H3 acetyl Lys 9 (Abcam USA, Catalogno. ab10812). PCR amplification was performed in 25 µL volumes for 32 to 37 cycles to determine the appropriate conditions for the PCR products of each region. Primer sequences are shown in Table S6. The PCR products were electrophoresed in a 2% agarose gel. Three biological replicates were analyzed and each was tested by three technical replicates.
Total RNA isolation and quantitative real-time PCR analysis
Total RNAs were isolated from callus tissues 2 to 3 mm deep from the surface. Quantitative real-time PCRs (qRT-PCRs) were performed as described previously [9]. To check the specificity of amplification, the melting curve of the PCR products was detected. The expression levels of specific genes were standardized to the housekeeping gene TUBULIN2. Each reaction was carried out in three biological replicates. The relative expression level of each gene, corresponding to the expression level of TUBULIN2, was calculated using the comparative CT method [59]. Primer sequences used for qRT-PCR are summarized in Table S6.
DNA microarray analysis
RNA of three plant samples was prepared from each of the following tissue types: the wild-type calli cultured on CIM for 20 days (S0), and on SIM for 6 days (S6); the met1 mutant calli cultured on CIM for 20 days (M0). RNA purification, probe labeling, chip hybridization, probe array scanning and data pre-processing normalization were performed by the Affymetrix custom service (CapitalBio, Beijing, China). Significance Analysis of Microarrays software package analysis was conducted for three biological samples replicates between the Ws and met1. When all replicates clustered together, further analysis was performed based on mean values. A two-fold change in the gene expression levels between one versus another samples with a q value≤0.05 was set as the threshold for altered gene expression. Microarray data are available in the ArrayExpress database (www.ebi.ac.uk/arrayexpress) under accession number E-MEXP-3120.
Supporting Information
Zdroje
1. ChePLallSNettletonDHowellSH 2006 Gene expression programs during shoot, root, and callus development in Arabidopsis tissue culture. Plant Physiol 141 620 637
2. AttaRLaurensLBoucheron-DubuissonEGuivarc'hACarneroE 2009 Pluripotency of Arabidopsis xylem pericycle underlies shoot regeneration from root and hypocotyl explants grown in vitro. Plant J 57 626 644
3. PernisováMKlímaPHorákJVálkováMMalbeckJ 2009 Cytokinins modulate auxin-induced organogenesis in plants via regulation of the auxin efflux. Proc Natl Acad Sci USA 106 3609 3614
4. ChePLallSHowellSH 2007 Developmental steps in acquiring competence for shoot development in Arabidopsis tissue culture. Planta 226 1183 1194
5. GordonSPHeislerMGReddyGVOhnoCDasP 2007 Pattern formation during de novo assembly of the Arabidopsis shoot meristem. Development 134 3539 3548
6. GalloisJLNoraFRMizukamiYSablowskiR 2004 WUSCHEL induces shoot stem cell activity and developmental plasticity in the root meristem. Genes Dev 18 375 380
7. DodsworthS 2009 A diverse and intricate signalling network regulates stem cell fate in the shoot apical meristem. Dev Biol 336 1 9
8. SchoofHLenhardMHaeckerAMayerKFXJürgensG 2000 The stem cell population of Arabidopsis shoot meristems is maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100 635 644
9. SuYHZhaoXYLiuYBZhangCLO'NeillSD 2009 Auxin-induced WUS expression is essential for embryonic stem cell renewal during somatic embryogenesis in Arabidopsis. Plant J 59 448 460
10. ShenWHXuL 2009 Chromatin remodeling in stem cell maintenance in Arabidopsis thaliana. Mol Plant 2 600 609
11. MeissnerAMikkelsenTSGuHWernigMHannaJ 2008 Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 454 766 770
12. WangKChenYChangEAKnottJGCibelliJB 2009 Dynamic epigenetic regulation of the Oct4 and Nanog regulatory regions during neural differentiation in rhesus nuclear transfer embryonic stem cells. Cloning Stem Cells 11 483 496
13. ShuklaVVaissièreTHercegZ 2008 Histone acetylation and chromatin signature in stem cell identity and cancer. Mutat Res 637 1 15
14. GollMGBestorTH 2005 Eukaryotic cytosine methyltransferases. Annu Rev Biochem 74 481 514
15. BröskeAMVockentanzLKharaziSHuskaMRManciniE 2009 DNA methylation protects hematopoietic stem cell multipotency from myeloerythroid restriction. Nat Genet 41 1207 1215
16. SenGLReuterJAWebsterDEZhuLKhavariPA 2010 DNMT1 maintains progenitor function in self-renewing somatic tissue. Nature 463 563 567
17. VaillantIPaszkowskiJ 2007 Role of histone and DNA methylation in gene regulation. Curr Opin Plant Biol 10 528 533
18. KouzaridesT 2007 Chromatin modifications and their function. Cell 128 693 705
19. MathieuOReindersJČaikovskiMSmathajittCPaszkowskiJ 2007 Transgenerational stability of the Arabidopsis epigenome is coordinated by CG methylation. Cell 130 851 862
20. VaissièreTSawanCHercegZ 2008 Epigenetic interplay between histone modifications and DNA methylation in gene silencing. Mutat Res 659 40 48
21. GrafiG 2004 How cells dedifferentiate: a lesson from plants. Dev Biol 268 1 6
22. KoukalovaBFojtovaMLimKYFulnecekJLeitchAR 2005 Dedifferentiation of Tobacco cells is associated with ribosomal RNA gene hypomethylation, increased transcription, and chromatin alterations. Plant Physiol 139 275 286
23. GrafiGBen-MeirHAviviYMosheMDahanY 2007 Histone methylation controls telomerase-independent telomere lengthening in cells undergoing dedifferentiation. Dev Biol 306 838 846
24. GrafiGAviviY 2004 Stem cells: a lesson from dedifferentiation. Trends in Biotechnology 22 388 389
25. BerdascoMAlcázarRGarcía-OrtizMVBallestarEFernándezAF 2008 Promoter DNA hypermethylation and gene repression in undifferentiated Arabidopsis cells. PLoS ONE 3 e3306 doi:10.1371/journal.pone.0003306
26. ChengZJZhuSSGaoXQZhangXS 2010 Cytokinin and auxin regulates WUS induction and inflorescence regeneration in vitro in Arabidopsis. Plant Cell Reports
27. BarteeLBenderJ 2001 Two Arabidopsis methylation-deficiency mutations confer only partial effects on a methylated endogenous gene family. Nucleic Acids Res 29 2127 2134
28. JacksonJPLindrothAMCaoXJacobsenSE 2002 Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature 416 556 560
29. LuFCuiXZhangSLiuCCaoX 2010 JMJ14 is an H3K4 demethylase regulating flowering time in Arabidopsis. Cell Res 20 387 390
30. SearleIRPontesOMelnykCWSmithLMBaulcombeDC 2010 JMJ14, a JmjC domain protein, is required for RNA silencing and cell-to-cell movement of an RNA silencing signal in Arabidopsis. Genes Dev 24 986 991
31. DengWLiuCPeiYDengXNiuL 2007 Involvement of the histone acetyltransferase AtHAC1 in the regulation of flowering time via repression of FLOWERING LOCUS C in Arabidopsis. Plant Physiol 143 1660 1668
32. KankelMWRamseyDEStokesTLFlowersSKHaagJR 2003 Arabidopsis MET1 cytosine methyltransferase mutants. Genetics 163 1109 1122
33. ListerRO'MalleyRCTonti-FilippiniJGregoryBDBerryCC 2008 Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 133 523 536
34. YangWJiangDJiangJHeY 2010 A plant-specific histone H3 lysine 4 demethylase represses the floral transition in Arabidopsis. Plant J 62 663 673
35. EarleyKWShookMSBrower-TolandBHicksLPikaardCS 2007 In vitro specificities of Arabidopsis co-activator histone acetyltransferases: implications for histone hyperacetylation in gene activation. Plant J 52 615 626
36. BäeurleILauxT 2005 Regulation of WUSCHEL transcription in the stem cell niche of the Arabidopsis shoot meristem. Plant Cell 17 2271 2280
37. BarskiACuddapahSCuiKRohTYSchonesDE 2007 High-resolution profiling of histone methylations in the human genome. Cell 129 823 837
38. JacksonJPJohnsonLJasencakovaZZhangXPerezBurgosL 2004 Dimethylation of histone H3 lysine 9 is a critical mark for DNA methylation and gene silencing in Arabidopsis thaliana. Chromosoma 112 308 315
39. ZhouJWangXHeKCharronJBEllingAA 2010 Genome-wide profiling of histone H3 lysine 9 acetylation and dimethylation in Arabidopsis reveals correlation between multiple histone marks and gene expression. Plant Mol Biol 72 585 595
40. DelerisAGreenbergMVAusinILawRWMoissiardG 2010 Involvement of a Jumonji-C domain-containing histone demethylase in DRM2-mediated maintenance of DNA methylation. EMBO Rep 11 950 955
41. SoppeWJJasencakovaZHoubenAKakutaniTMeisterA 2002 DNA methylation controls histone H3 lysine 9 methylation and heterochromatin assembly in Arabidopsis. EMBO J 21 6549 6559
42. AdrianJFarronaSReimerJJAlbaniMCCouplandG 2010 cis-Regulatory elements and chromatin state coordinately control temporal and spatial expression of FLOWERING LOCUS T in Arabidopsis. Plant Cell 22 1425 1440
43. WilliamsLFletcherJC 2005 Stem cell regulation in the Arabidopsis shoot apical meristem. Curr Opin Plant Biol 8 582 586
44. KayaHShibaharaK-iTaokaK-iIwabuchiMStillmanB 2001 FASCIATA genes for Chromatin Assembly Factor-1 in Arabidopsis maintain the cellular organization of apical meristems. Cell 104 131 142
45. TakedaSTadeleZHofmannIProbstAVAngelisKJ 2004 BRU1, a novel link between responses to DNA damage and epigenetic gene silencing in Arabidopsis. Genes Dev 18 782 793
46. KwonCSChenCWagnerD 2005 WUSCHEL is a primary target for transcriptional regulation by SPLAYED in dynamic control of stem cell fate in Arabidopsis. Genes Dev 19 992 1003
47. GrafT 2002 Differentiation plasticity of hematopoietic cells. Blood 99 3089 3101
48. LiuYRaoMS 2003 Transdifferentiation-fact or artifact. J Cell Biochem 88 29 40
49. TakahashiKYamanakaS 2006 Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126 663 676
50. PekkerIAlvarezJPEshedY 2005 Auxin response factors mediate Arabidopsis organ asymmetry via modulation of KANADI activity. Plant Cell 17 2899 2910
51. BirnbaumKDAlvaradoAS 2008 Slicing across kingdoms: regeneration in plants and animals. Cell 132 697 710
52. LiuJCasacciaP 2010 Epigenetic regulation of oligodendrocyte identity. Trends in Neurosciences 33 193 201
53. MurashigeTSkoogF 1962 A revised medium for rapid growth and bioassays with Tobacco tissue cultures. Physiologia Plantarum 15 473 497
54. BuechelSLeibfriedAToJPZhaoZAndersenSU 2010 Role of A-type ARABIDOPSIS RESPONSE REGULATORS in meristem maintenance and regeneration. Eur J Cell Biol 89 279 284
55. GamborgOLMillerRAOjimaK 1968 Nutrient requirements of suspension cultures of soybean root cells. Exp Cell Res 50 151 158
56. ZhaoXYChengZJZhangXS 2006 Overexpression of TaMADS1, a SEPALLATA-like gene in wheat, causes early flowering and the abnormal development of floral organs in Arabidopsis. Planta 223 698 707
57. JacobsenSESakaiHFinneganEJCaoXMeyerowitzEM 2000 Ectopic hypermethylation of flower-specific genes in Arabidopsis. Curr Biol 10 179 186
58. HetzlJFoersterAMRaidlGScheidOM 2007 CyMATE: a new tool for methylation analysis of plant genomic DNA after bisulphite sequencing. Plant J 51 526 536
59. SchmittgenTDLivakKJ 2008 Analyzing real-time PCR data by the comparative CT method. Nat Protocols 3 1101 1108
Štítky
Genetika Reprodukční medicína
Článek The T-Box Factor MLS-1 Requires Groucho Co-Repressor Interaction for Uterine Muscle SpecificationČlánek B Chromosomes Have a Functional Effect on Female Sex Determination in Lake Victoria Cichlid FishesČlánek Distinct Cdk1 Requirements during Single-Strand Annealing, Noncrossover, and Crossover RecombinationČlánek Specification of Corpora Cardiaca Neuroendocrine Cells from Mesoderm Is Regulated by Notch SignalingČlánek Ongoing Phenotypic and Genomic Changes in Experimental Coevolution of RNA Bacteriophage Qβ and
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2011 Číslo 8- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- IVF a rakovina prsu – zvyšují hormony riziko vzniku rakoviny?
-
Všechny články tohoto čísla
- Polo, Greatwall, and Protein Phosphatase PP2A Jostle for Pole Position
- Genome-Wide Association Analysis of Incident Coronary Heart Disease (CHD) in African Americans: A Short Report
- The T-Box Factor MLS-1 Requires Groucho Co-Repressor Interaction for Uterine Muscle Specification
- B Chromosomes Have a Functional Effect on Female Sex Determination in Lake Victoria Cichlid Fishes
- Analysis of DNA Methylation in a Three-Generation Family Reveals Widespread Genetic Influence on Epigenetic Regulation
- PP2A-Twins Is Antagonized by Greatwall and Collaborates with Polo for Cell Cycle Progression and Centrosome Attachment to Nuclei in Drosophila Embryos
- Discovery of Sexual Dimorphisms in Metabolic and Genetic Biomarkers
- Pervasive Sharing of Genetic Effects in Autoimmune Disease
- DNA Methylation and Histone Modifications Regulate Shoot Regeneration in by Modulating Expression and Auxin Signaling
- Mutations in and Reveal That Cartilage Matrix Controls Timing of Endochondral Ossification by Inhibiting Chondrocyte Maturation
- Variance of Gene Expression Identifies Altered Network Constraints in Neurological Disease
- Frequent Beneficial Mutations during Single-Colony Serial Transfer of
- Increased Gene Dosage Affects Genomic Stability Potentially Contributing to 17p13.3 Duplication Syndrome
- Distinct Cdk1 Requirements during Single-Strand Annealing, Noncrossover, and Crossover Recombination
- Hunger Artists: Yeast Adapted to Carbon Limitation Show Trade-Offs under Carbon Sufficiency
- Suppression of Scant Identifies Endos as a Substrate of Greatwall Kinase and a Negative Regulator of Protein Phosphatase 2A in Mitosis
- Temporal Dynamics of Host Molecular Responses Differentiate Symptomatic and Asymptomatic Influenza A Infection
- MK2-Dependent p38b Signalling Protects Hindgut Enterocytes against JNK-Induced Apoptosis under Chronic Stress
- Specification of Corpora Cardiaca Neuroendocrine Cells from Mesoderm Is Regulated by Notch Signaling
- Genome-Wide Gene-Environment Study Identifies Glutamate Receptor Gene as a Parkinson's Disease Modifier Gene via Interaction with Coffee
- Identification of Functional Toxin/Immunity Genes Linked to Contact-Dependent Growth Inhibition (CDI) and Rearrangement Hotspot (Rhs) Systems
- Genomic Analysis of the Necrotrophic Fungal Pathogens and
- Celsr3 Is Required for Normal Development of GABA Circuits in the Inner Retina
- Genetic Architecture of Aluminum Tolerance in Rice () Determined through Genome-Wide Association Analysis and QTL Mapping
- Predisposition to Cancer Caused by Genetic and Functional Defects of Mammalian
- Regulation of p53/CEP-1–Dependent Germ Cell Apoptosis by Ras/MAPK Signaling
- and but Not Interact in Genetic Models of Amyotrophic Lateral Sclerosis
- Gamma-Tubulin Is Required for Bipolar Spindle Assembly and for Proper Kinetochore Microtubule Attachments during Prometaphase I in Oocytes
- Ongoing Phenotypic and Genomic Changes in Experimental Coevolution of RNA Bacteriophage Qβ and
- Genetic Architecture of a Reinforced, Postmating, Reproductive Isolation Barrier between Species Indicates Evolution via Natural Selection
- -eQTLs Reveal That Independent Genetic Variants Associated with a Complex Phenotype Converge on Intermediate Genes, with a Major Role for the HLA
- The GATA Factor ELT-1 Works through the Cell Proliferation Regulator BRO-1 and the Fusogen EFF-1 to Maintain the Seam Stem-Like Fate
- and Control Optic Cup Regeneration in a Prototypic Eye
- A Comprehensive Map of Mobile Element Insertion Polymorphisms in Humans
- An EMT–Driven Alternative Splicing Program Occurs in Human Breast Cancer and Modulates Cellular Phenotype
- Evidence for Hitchhiking of Deleterious Mutations within the Human Genome
- A Broad Brush, Global Overview of Bacterial Sexuality
- Global Chromosomal Structural Instability in a Subpopulation of Starving Cells
- A Pre-mRNA–Associating Factor Links Endogenous siRNAs to Chromatin Regulation
- Glutamine Synthetase Is a Genetic Determinant of Cell Type–Specific Glutamine Independence in Breast Epithelia
- The Repertoire of ICE in Prokaryotes Underscores the Unity, Diversity, and Ubiquity of Conjugation
- Genome-Wide Association Analysis of Autoantibody Positivity in Type 1 Diabetes Cases
- Natural Polymorphism in BUL2 Links Cellular Amino Acid Availability with Chronological Aging and Telomere Maintenance in Yeast
- Chromosome Painting Reveals Asynaptic Full Alignment of Homologs and HIM-8–Dependent Remodeling of Chromosome Territories during Meiosis
- Ku Must Load Directly onto the Chromosome End in Order to Mediate Its Telomeric Functions
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- An EMT–Driven Alternative Splicing Program Occurs in Human Breast Cancer and Modulates Cellular Phenotype
- Chromosome Painting Reveals Asynaptic Full Alignment of Homologs and HIM-8–Dependent Remodeling of Chromosome Territories during Meiosis
- Discovery of Sexual Dimorphisms in Metabolic and Genetic Biomarkers
- Regulation of p53/CEP-1–Dependent Germ Cell Apoptosis by Ras/MAPK Signaling
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



