-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
HIF-1 Regulates Iron Homeostasis in by Activation and Inhibition of Genes Involved in Iron Uptake and Storage
Caenorhabditis elegans ftn-1 and ftn-2, which encode the iron-storage protein ferritin, are transcriptionally inhibited during iron deficiency in intestine. Intestinal specific transcription is dependent on binding of ELT-2 to GATA binding sites in an iron-dependent enhancer (IDE) located in ftn-1 and ftn-2 promoters, but the mechanism for iron regulation is unknown. Here, we identify HIF-1 (hypoxia-inducible factor -1) as a negative regulator of ferritin transcription. HIF-1 binds to hypoxia-response elements (HREs) in the IDE in vitro and in vivo. Depletion of hif-1 by RNA interference blocks transcriptional inhibition of ftn-1 and ftn-2 reporters, and ftn-1 and ftn-2 mRNAs are not regulated in a hif-1 null strain during iron deficiency. An IDE is also present in smf-3 encoding a protein homologous to mammalian divalent metal transporter-1. Unlike the ftn-1 IDE, the smf-3 IDE is required for HIF-1–dependent transcriptional activation of smf-3 during iron deficiency. We show that hif-1 null worms grown under iron limiting conditions are developmentally delayed and that depletion of FTN-1 and FTN-2 rescues this phenotype. These data show that HIF-1 regulates intestinal iron homeostasis during iron deficiency by activating and inhibiting genes involved in iron uptake and storage.
Published in the journal: . PLoS Genet 7(12): e32767. doi:10.1371/journal.pgen.1002394
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002394Summary
Caenorhabditis elegans ftn-1 and ftn-2, which encode the iron-storage protein ferritin, are transcriptionally inhibited during iron deficiency in intestine. Intestinal specific transcription is dependent on binding of ELT-2 to GATA binding sites in an iron-dependent enhancer (IDE) located in ftn-1 and ftn-2 promoters, but the mechanism for iron regulation is unknown. Here, we identify HIF-1 (hypoxia-inducible factor -1) as a negative regulator of ferritin transcription. HIF-1 binds to hypoxia-response elements (HREs) in the IDE in vitro and in vivo. Depletion of hif-1 by RNA interference blocks transcriptional inhibition of ftn-1 and ftn-2 reporters, and ftn-1 and ftn-2 mRNAs are not regulated in a hif-1 null strain during iron deficiency. An IDE is also present in smf-3 encoding a protein homologous to mammalian divalent metal transporter-1. Unlike the ftn-1 IDE, the smf-3 IDE is required for HIF-1–dependent transcriptional activation of smf-3 during iron deficiency. We show that hif-1 null worms grown under iron limiting conditions are developmentally delayed and that depletion of FTN-1 and FTN-2 rescues this phenotype. These data show that HIF-1 regulates intestinal iron homeostasis during iron deficiency by activating and inhibiting genes involved in iron uptake and storage.
Introduction
Iron is essential due to its presence in proteins involved in DNA synthesis, mitochondrial respiration and oxygen transport. Regulation of cellular iron content is crucial: excess cellular iron catalyzes the generation of reactive oxygen species that damage DNA and proteins, while cellular iron deficiency causes cell cycle arrest and cell death. Dysregulation of iron homeostasis caused by iron deficiency or iron excess leads to hematological, neurodegenerative and metabolic diseases in humans. Iron must therefore be maintained within a narrow range to avoid the adverse consequences of iron depletion or excess.
Maintaining iron content within this physiological range requires precise mechanisms for regulating its uptake, storage and export (for reviews, see [1], [2]). In mammals, dietary non-heme Fe3+ is reduced by membrane bound ferric reductases (e.g. duodenal cytochrome B or DCYTB) before transport across the enterocyte apical membrane by divalent metal transporter-1 (DMT1, also known as NRAMP2, SLC11a2 and DCT1) [3]. Cytosolic iron is either transported across the basolateral membrane into the circulation by ferroportin or sequestered in ferritin in a form unable to catalyze free radical formation [4], [5]. Iron export by ferroportin is dependent on oxidation to Fe3+ by membrane and soluble multicopper oxidases where it is incorporated into transferrin for delivery to tissues. When body iron stores are high, cytosolic iron is not exported into blood, and is instead sequestered into ferritin [5], . Iron in ferritin is lost by sloughing of enterocytes into the intestinal lumen.
Mammalian intestinal iron transport increases during iron deficiency due to hypoxia-inducible factor-2α (HIF-2α) mediated expression of DMT1 and DCYTB [7], [8]. HIFs (HIF-1 and HIF-2) are key regulators of cellular and systemic oxygen homeostasis (for reviews, see [9], [10]). HIF transcription factors consist of an oxygen-regulated α subunit (HIF-1α, HIF-2α and a constitutively expressed β subunit (HIF-1β, also known as aryl hydrocarbon nuclear translocator or ARNT). In the presence of iron and oxygen, HIF-α subunits are hydroxylated by iron - and oxygen-dependent prolyl hydroxylases (PHDs) and are targeted for proteasomal degradation by the von Hippel-Lindau (VHL) E3 ubiquitin ligase. During hypoxia or iron deficiency, PHDs are inactivated, allowing HIF-1α /HIF-2β to accumulate. HIF-1α/HIF-2α dimerizes with HIF-β and binds to HREs in target genes to increase transcription. HIF regulates genes in diverse pathways including erythropoiesis, iron homeostasis, glucose metabolism, angiogenesis and cell survival (for reviews, see [9]–[11] ).
Oxygen and iron homeostasis pathways are conserved in Caenorhabditis elegans. The HIF-1 pathway in C. elegans consists of hif-1, aha-1, vhl-1 and egl-9, which are orthologous to genes encoding mammalian HIF-1α, HIF-1β, VHL and PHD [12]–[14]. C. elegans express a single hif gene, which encodes a protein homologous to vertebrate HIF-1α and HIF-2α [12]. C. elegans HIF-1 regulates target genes involved in metabolism, extracellular remodeling [15], nervous system development [16], oxygen-dependent behavior [17] and modulation of life span [18]. hif-1 mutant animals display increased embryonic and larval lethality in oxygen concentrations less than 1%, demonstrating the importance of HIF-1 for survival during hypoxia [13], [19].
C. elegans express genes homologous to ferritin (ftn-1 and ftn-2), DMT1 (smf-1, smf-2 and smf-3) and ferroportin (fpn-1.1, fpn-1.2 and fpn-1.3). Vertebrate ferritin is a mixture of 24 light-(L) and heavy-(H) subunits that form a shell that can accommodate up to 4500 iron atoms. The H-subunits exhibit ferroxidase activity and facilitate the oxidation of iron, whereas the L-subunits function with the H-subunits in iron nucleation [20], [21]. C. elegans FTN-1 and FTN-2 display greater homology to the human H-subunit (55% and 60%) than to the L-subunit (46% and 50%), and notably both proteins contain ferroxidase active-site residues. ftn-1 and ftn-2 genes are transcriptionally repressed during iron deficiency, which is dependent on an iron-dependent enhancer (IDE) located in the ftn-1 and ftn-2 promoters [22]–[24]. The IDE contains two GATA binding sites for the intestinal specific ELT2 transcription factor that regulates basal ftn-1 and ftn-2 transcription, but the mechanism regulating iron-dependent transcriptional repression is unknown. Unlike ftn-1 and ftn-2, vertebrate ferritin-H and -L subunit mRNAs are translationally repressed by iron-regulatory proteins 1 and 2 (IRP1 and IRP2) during iron deficiency (for reviews, see [25]–[27]). SMF-1, SMF-2 and SMF-3 display 55–58% amino acid identity with mammalian DMT1 and are involved in Mn2+ uptake and sensing, but the role of these transporters in iron uptake is not well understood [28]–[30]. The function of ferroportin homologs FPN1.1, FPN-1.2 and FPN-1.3 in iron homeostasis has not been reported.
Here, we show that HIF-1 activates smf-3 transcription and inhibits ftn-1 and ftn-2 transcription during iron deficiency. Transcriptional activation of smf-3 and repression of ftn-1 and ftn-2 is dependent on IDEs in their promoters that are similar but not identical. These studies show that HIF-1 is a key regulator of intestinal iron uptake and storage during iron deficiency in C. elegans.
Results
HIF-1 inhibits iron-dependent transcription of ftn-1 and ftn-2
Previous studies showed that ftn-1 and ftn-2 transcription is activated by iron and inhibited by iron chelators [22]–[24]. Transcriptional regulation is mediated by the IDE located in the promoters of ftn-1 and ftn-2 genes (Figure 1A). The IDE contains two WGATAR sequences that are binding sites for the intestinal specific ELT2 GATA transcription factor. Mutation of either of the WGATAR sequences abolished expression of an ftn-1::GFP-his reporter showing that ELT-2 is required for ftn-1 transcription under iron sufficient conditions [24]. The IDE also contains three canonical HREs (TACGTG) in the reverse orientation that have been identified in hif-1 target genes [15], [31], suggesting a role for HIF-1 in iron-dependent ftn-1 and ftn-2 regulation.
Fig. 1. HIF-1 is required for ftn-1 and ftn-2 transcriptional repression during iron deficiency. 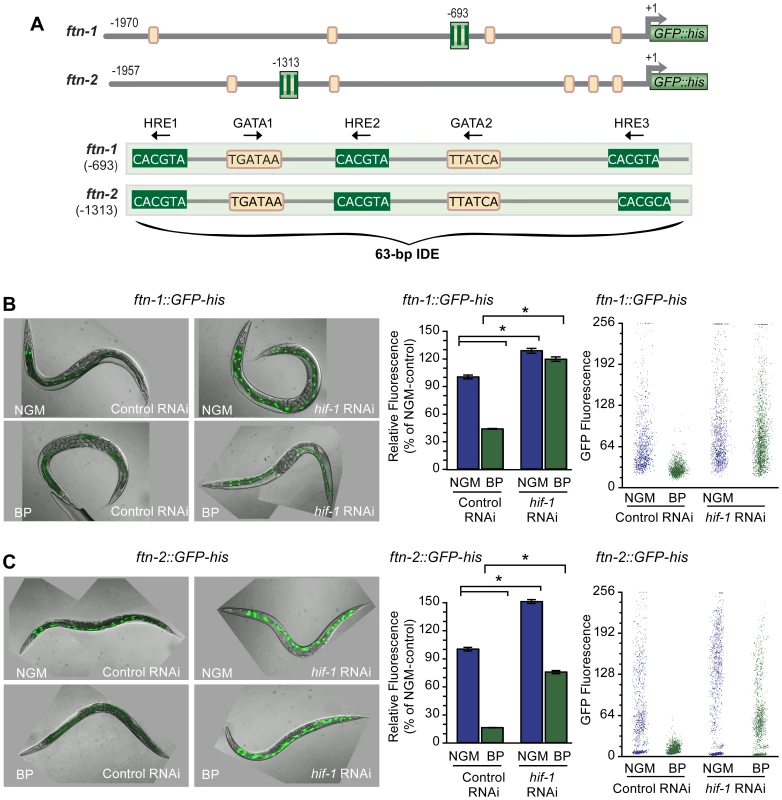
(A) Conserved sequences in the 5′ promoter regions of ftn-1 and ftn-2. Large stripped boxes, IDE; small boxes, GATA binding motifs. The IDE is enlarged showing HRE1, HRE2 and HRE3 and GATA1 and GATA2 sequences. Arrows indicate orientation of motifs. (B and C) Expression of ftn-1::GFP-his and ftn-2::GFP-his reporters in L4 larvae fed control (empty vector L4440) RNAi or hif-1 RNAi after culture on NGM or NGM-BP for 16 h. Quantification of GFP expression in (B and C) was carried out using the COPAS Biosort. Dot plots of sorted worms are shown where each dot represents one worm. Data are mean GFP fluorescence ± SEM reported as a percentage of worms fed control RNAi on NGM (n = 1000 worms per treatment), *p<0.0001. To test this model, hif-1 RNAi was used to deplete HIF-1 in worms carrying an ftn-1::GFP-his or an ftn-2::GFP-his reporter. GFP expression was quantified using the COPAS Biosort after growth in NGM or NGM supplemented with the membrane permeable Fe2+ chelator, 2,2′-dipyridyl (NGM-BP) [32]. These reporters contain 1.9 kb of ftn-1 or ftn-2 promoter sequences, including the IDE, fused to the initiator ATG of nuclear-localized GFP-histone [24]. BP reduces expression of ftn-1::GFP-his and ftn-2::GFP-his in worms fed control (empty vector L4400) RNAi by 60% and 80%, respectively, compared to worms grown on NGM (Figure 1B and 1C). By contrast, the BP - induced reduction in GFP expression is blocked by hif-1 RNAi. Furthermore, hif-1 RNAi increases GFP expression in worms grown on NGM, indicating that HIF-1 is expressed under normal growth conditions, and is capable of inhibiting ftn-1 and ftn-2 transcription.
We next determined whether endogenous ftn-1 and ftn-2 mRNAs are regulated by HIF-1. ftn-1 and ftn-2 mRNAs were measured in N2 wildtype animals cultured on NGM or NGM-BP and in strains carrying the loss-of function mutations in hif-1(ia04) and vhl-1(ok161). vhl-1(ok161) mutant animals lack VHL required for HIF-1 ubiquitination and proteasomal degradation, leading to constitutive expression of HIF-1 [14], [31], [33]. Western blots confirm the absence of HIF-1 in hif-1(ia04) mutant animals and increased HIF-1 levels in vhl-1(ok161) mutant and in N2 wildtype animals cultured in NGM-BP (Figure 2A). BP reduces ftn-1 and ftn-2 mRNA levels 75% and 20%, respectively, compared to untreated N2 wildtype animals. This is consistent with our previous studies showing that ftn-1 is more sensitive to iron chelators as compared to ftn-2 [22], [24]. By contrast, ftn-1 and ftn-2 mRNA levels are not reduced by BP in hif-1(ia04) mutant animals, and notably hif-1(ia04) animals cultured in NGM express higher amounts of ftn-1 mRNA compared to N2 wildtype animals (Figure 2B). In vhl-1(ok161) mutant animals, ftn-1 and ftn-2 mRNAs are reduced to levels found in N2 wildtype animals cultured in NGM-BP. Taken together, these data show that ftn-1 and ftn-2 are transcriptionally inhibited by HIF-1 during iron deficiency.
Fig. 2. Endogenous ftn-1 and ftn-2 mRNA expression is regulated by HIF-1. 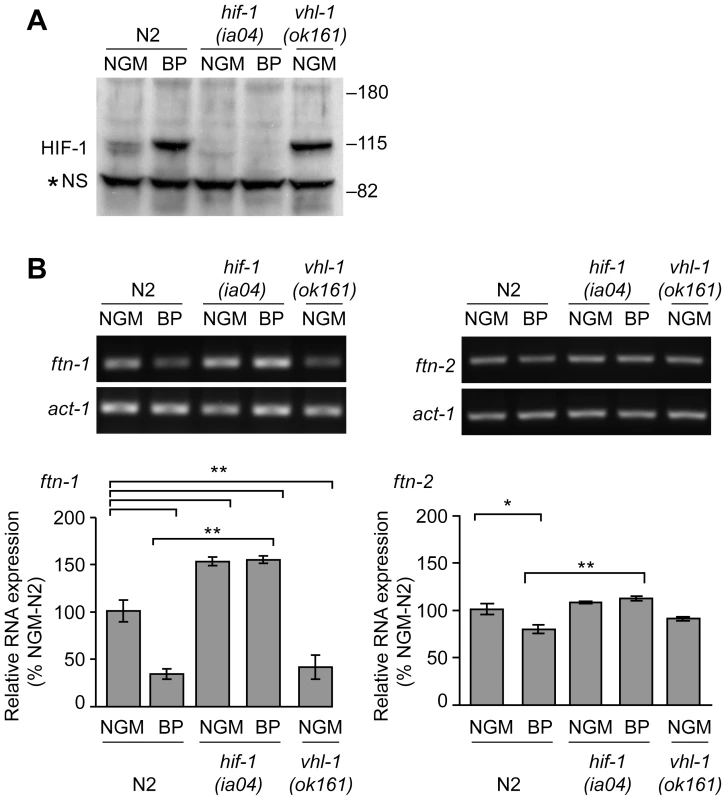
(A) N2 wildtype, hif-1(ia04) and vhl-1(ok161) strains were cultured on NGM or NGM-BP, and HIF-1 was detected in extracts by western blots using CeHIF-1 antibodies. *NS, non-specific band. (B) RNA was isolated from the strains in (A) and ftn-1, ftn-2 and actin (act-1) mRNAs were quantified by RT-PCR. Relative changes in ftn-1 and ftn-2 mRNAs are expressed as a percentage of N2 wildtype animals on NGM after normalization to act-1 mRNA. Five (ftn-1) and three (ftn-2) independent experiments were performed and the data are reported as means ± SEM, *p<0.05, **p<0.01. HIF-1 binds to the IDE in vitro and in vivo
Electrophoretic mobility shift assays were used to determine whether HIF-1 binds to the HREs in the IDE. Radiolabeled wildtype IDE or an IDE containing mutations in the three HRE sites (HRE3m) was incubated with reticulocyte lysate-synthesized HIF-1 and AHA-1 (Figure 3A). Complex formation is only observed when HIF-1 and AHA-1 are present together in the reaction with wildtype IDE (Figure 3A). Addition of HIF-1 antibody to the reaction led to the formation of a slower migrating HIF-1/AHA-IDE complex (Figure 3A, lane 5). Formation of HIF-1/AHA-1-IDE complexes is competed away by unlabeled wildtype IDE but not by unlabeled HRE3m IDE, showing that HIF-1 specifically binds to the HREs (Figure 3B).
Fig. 3. HIF-1 binds to the ftn-1 IDE in vitro and in vivo. 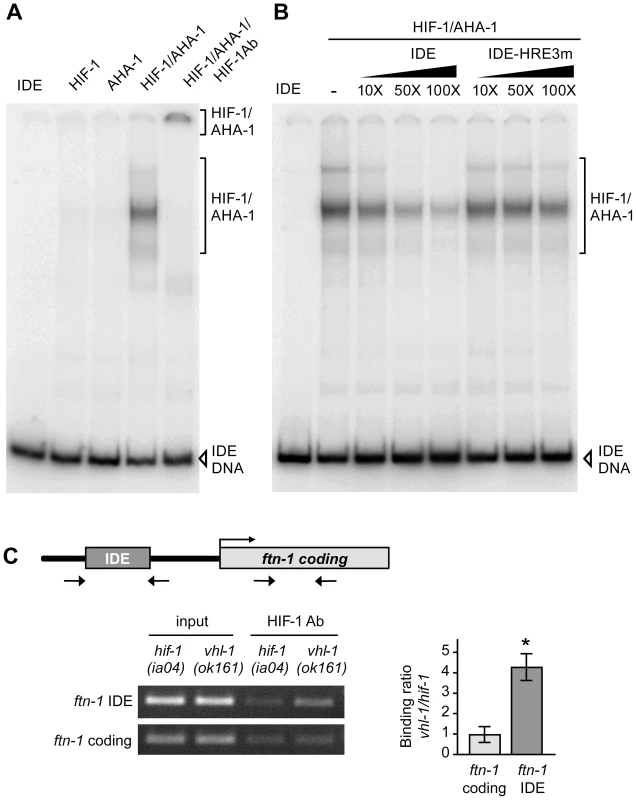
(A) HIF-1 and AHA-1 synthesized in reticulocyte lysates were incubated singly or together with a 32P-labeled ftn-1 IDE probe. HIF-1/AHA-1/IDE complexes were analyzed by non-denaturing polyacrylamide gels. Free IDE probe and HIF-1/AHA-1/IDE complexes are indicated. HIF-1 antibody was added to the reaction (lane 5). (B) 32P-labeled IDE probe was incubated with HIF-1/AHA-1 with increasing molar amounts of unlabeled wildtype IDE or mutant IDE-HRE3m. (C) Diagram of the location of the IDE in the ftn-1 promoter and the location of the primers used for amplification. ChIP was performed with chromatin isolated from a mixed-staged population of vhl-1(ok161) animals constitutively expressing HIF-1 or hif-1(iao4) animals. Chromatin was immunoprecipitated with CeHIF-1 antibody and bound DNA sequences were analyzed by qPCR using IDE or coding region primers. Chromatin extract (input) was amplified to determine the total amount of genomic DNA prior to immunoprecipitation. The binding of HIF-1 to the IDE is expressed as a ratio of binding in vhl-1(ok161) versus hif-1(ia04) ± SEM, *p = 0.019. A representative gel of three independent experiments is shown. To determine whether the IDE is a direct HIF-1 target in vivo, ChIP was performed on chromatin isolated from vhl-1(ok161) and hif-1(ia04) mutant animals. The binding of HIF-1 to the IDE or to the ftn-1 coding region used as a negative control was determined by ChIP using HIF-1 antibody. IDE DNA was enriched 4-fold in vhl-1(ok161) immunoprecipitates as compared to hif-1(ia04) immunoprecipitates (Figure 3C). These studies indicate that HIF-1 binds to HREs in vitro and occupies the ftn-1 IDE in vivo.
The DMT1 ortholog SMF-3 is regulated by HIF-1 during iron deficiency
The activation of DMT1 by HIF-2α in iron-deficient mice increases intestinal iron uptake [7], [8]. C. elegans express three DMT1 homologs, SMF-1, SMF-2 and SMF-3. SMF-1 and SMF-3 are expressed in the apical membrane in intestinal cells and are involved in Mn2+ uptake [28], [30], whereas SMF-2 is mainly expressed in pharyngeal epithelial cells [28]–[30]. To determine whether HIF-1 regulates smf-1, smf-2 or smf-3 expression during iron deficiency, their mRNA levels were quantified in N2 wildtype and hif-1(ia04) mutant animals cultured in NGM or NGM-BP. BP increases smf-3 mRNA levels 2-fold as compared to untreated N2 wildtype animals, but has no effect on smf-1 or smf-2 mRNA levels (Figure 4A and data not shown). In hif-1(ia04) mutant animals, smf-3 mRNA levels are reduced by 50% as compared to N2 wildtype animals, and are not increased by BP (Figure 4A).
Fig. 4. Activation of smf-3 during iron deficiency is dependent on an IDE in the smf-3 promoter. 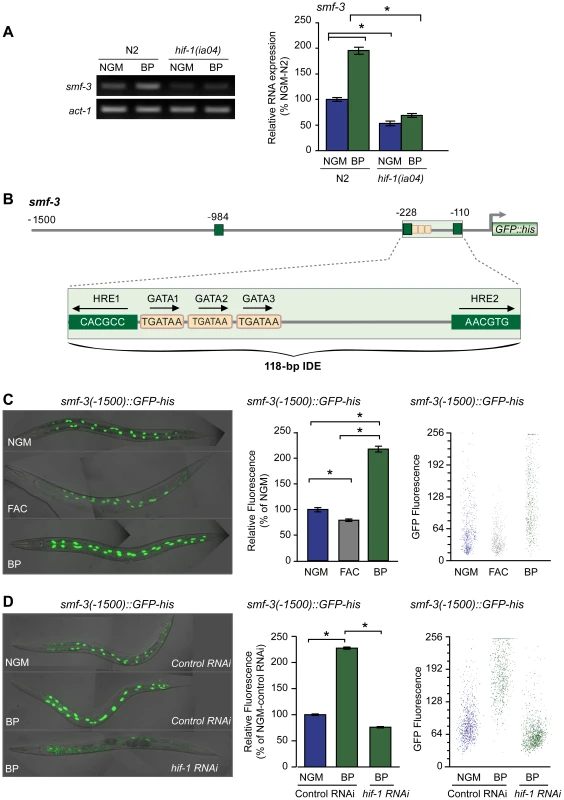
(A) smf-3 mRNA was quantified by RT-PCR in RNA isolated from N2 wildtype and hif-1(ia04) mutant strains cultured in NGM or NGM-BP for 16 h. Relative changes in smf-3 mRNA are expressed as a percentage of N2 wildtype animals cultured on NGM after normalization to act-1 mRNA. Five independent experiments were performed and the data are reported as means ± SEM, *p<0.0015. (B) Schematic diagram of the 5′ promoter region of the smf-3 gene. Small boxes, GATA binding motifs; large boxes, HREs. The IDE is enlarged showing HRE1, HRE2 and HRE3 and GATA1 and GATA2 sequences. Arrows indicate motif orientation. (C) Expression smf-3(-1500)::GFP-his containing 1.5 kb of promoter sequences in worms after culture in NGM, NGM-BP or NGM supplemented with FAC for 18 h. (D) Expression of smf-3(-1500)::GFP-his reporter in L4 animals fed control RNAi or hif-1 RNAi after culture in NGM or NGM-BP for 16 h. GFP fluorescence was quantified in (C and D) using the COPAS Biosort as described in Figure 1. Data are mean GFP fluorescence ±SEM as a percentage of untreated worms grown in NGM for (C) or as a percentage of worms fed control RNAi on NGM for (D), n = 1000 worms per treatment, *p<0.001. Inspection of the 5′ upstream regulatory region of smf-3 reveals a 118-nt element harboring three tandem GATA binding sites flanked by two HREs (Figure 4B). To determine whether this element functions as an iron enhancer, transgenic strains carrying a transcriptional reporter containing 1500 nt of smf-3 promoter sequences fused to GFP-his were generated. GFP expression was quantified in these strains after growth on NGM, NGM-ferric ammonium citrate (NGM-FAC) or NGM-BP. FAC reduces GFP expression, whereas BP increases GFP expression as compared to untreated worms (Figure 4C). To show that the BP-induced increase in GFP expression is due to iron chelation, GFP expression was quantified after culture of worms on NGM-BP in the presence of an equimolar amount of FAC or MnCl2. BP plus FAC, but not BP plus MnCl2, reduces smf-3(-1500)::GFP-his expression (Figure S1). hif-1 RNAi completely blocks BP-induced increased GFP expression (Figure 4D). Similarly, expression of a smf-3(-1500)::GFP-his reporter containing 250 nt of upstream sequences containing the IDE is increased by BP in control RNAi fed worms and is reduced by BP in hif-1 RNAi fed worms (Figure S2). Taken together, these data indicate that smf-3 is transcriptionally activated by HIF-1 during iron deficiency.
Iron content is reduced in smf-3(ok1035) mutant animals
The localization of SMF-3 to the apical membrane of intestinal cells [28], [30] and its regulation by HIF-1 suggest a role for SMF-3 in intestinal iron uptake. If SMF-3 has a role in iron uptake, iron content might be expected to be reduced in smf-3(ok1035) mutant animals. We found that ftn-1 mRNA levels, which are positively correlated with cellular iron levels (Figure 2), are reduced in smf-3(ok1035) mutants as compared to N2 wildtype animals, but not in smf-1(ok1748) or smf-2(gk133) mutant animals (Figure 5A). Quantification of metal content by inductively-coupled plasma spectroscopy (ICP) shows that total iron content in smf-3(ok1035) mutant animals is 45% of N2 wildtype animals consistent with reduced ftn-1 mRNA levels in these animals (Figure 5B). The total iron content in smf-1(ok1748) and smf-2(gk133) mutant animals is not significantly different as compared to N2 wildtype animals. The total Mn content in smf-3(ok1035) mutant animals is 60% of N2 wildtype controls (Figure 5B) in agreement with SMF-3 as a regulator of Mn uptake [28], [30]. The Mn content in the smf-1(ok1748) and smf-2(gk133) strains tended to be lower as compared to N2 wildtype controls, but did not reach significance. These results are consistent with a role for SMF-3 in intestinal iron transport.
Fig. 5. Total Fe content is reduced in smf-3(1035) mutant animals. 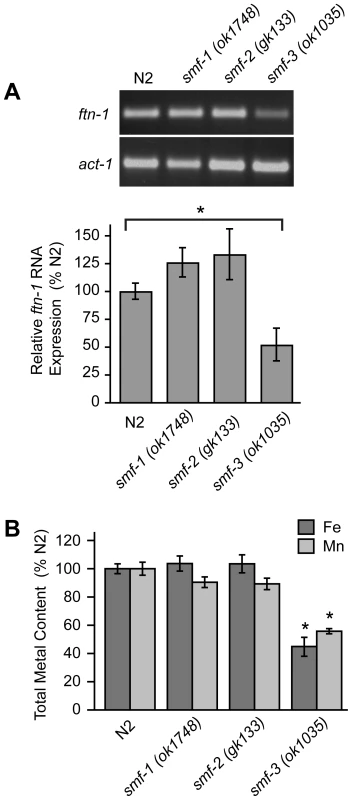
(A) ftn-1 mRNA was quantified by RT-PCR from RNA isolated from N2 wildtype and smf-1(ok1748), smf-2(gk133) and smf-3(ok1035) strains. Relative changes in ftn-1 mRNA are expressed as a percentage of N2 wildtype animals after normalization to act-1 mRNA. Five independent experiments were performed and data are reported as means ± SEM, * p<0.05. (B) Total content of Fe and Mn in N2 wildtype and smf mutant animals determined by ICP spectroscopy is expressed as a percentage of N2 wildtype animals for each metal. At least three independent experiments were performed and the data are reported as means ± SEM, *p<0.01. Reduced viability of hif-1(ia04) mutant animals by iron deficiency is rescued by ftn-1 and ftn-2 RNAi
hif-1(ia04) mutant animals have no overt phenotype in normoxia, but display reduced viability in oxygen concentrations less than 1% [13], [34]. Similarly, we find that hif-1(ia04) mutant animals develop normally under normoxic conditions, but are developmentally delayed when cultured under iron deficient normoxic conditions (NGM plus 20 µM BP) as compared to N2 wildtype animals (Figure 6A). As total iron content is reduced in smf-3(ok1035) mutant animals and smf-3 expression is reduced in hif-1(ia04) animals, these data suggest that total iron content might also be reduced in hif-1(ia04) animals. ICP analyses show that the total iron content in hif-1(ia04) mutant animals is 60% of N2 wildtype animals (Figure 6B). The total Mn content is also reduced in hif-1(ia04) mutant animals.
Fig. 6. Delayed growth of hif-1(ia04) mutant animals during iron deficiency is rescued by ftn-1 and ftn-2 RNAi. 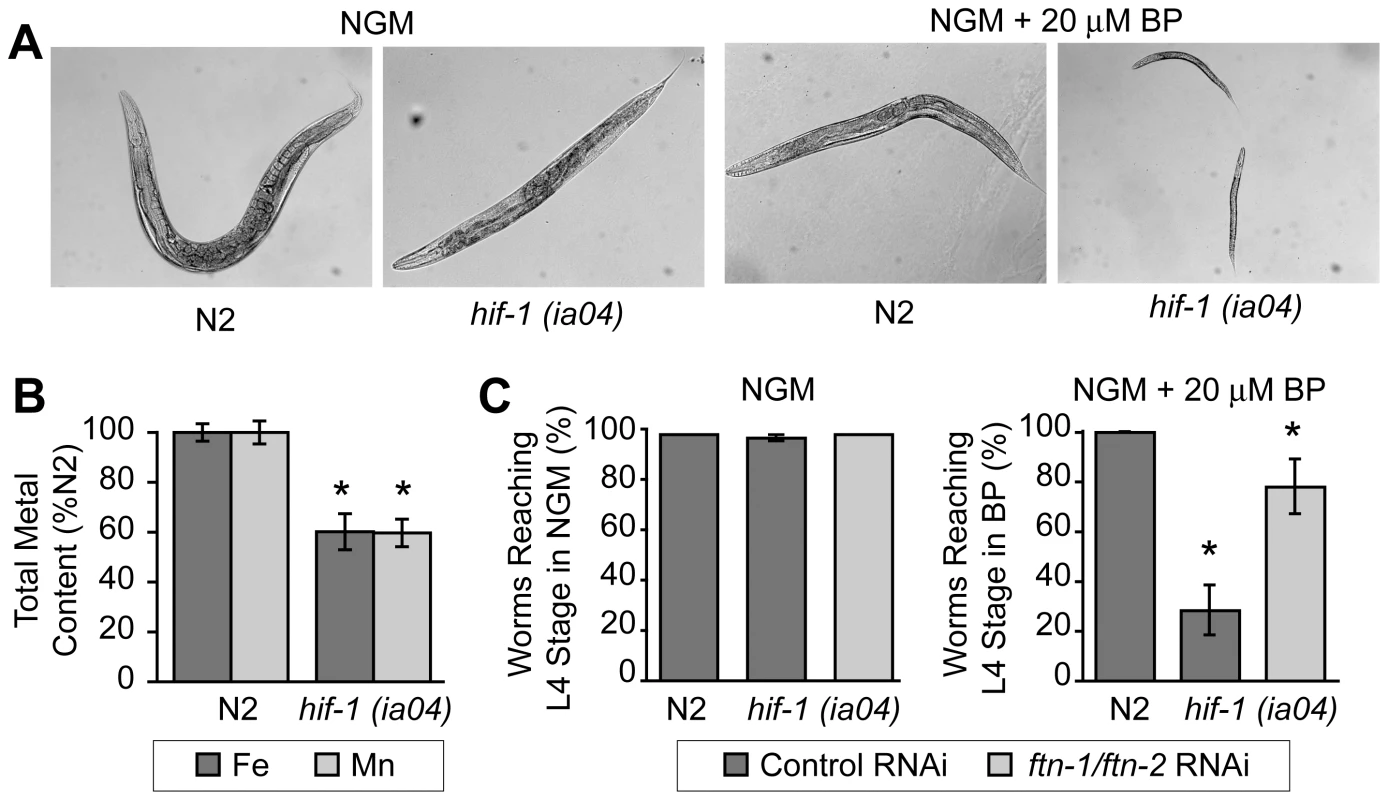
(A) N2 wildtype and hif-1(ia04) mutant L1 stage animals were grown on NGM with or without 20 µM BP for 48 h. hif-1(ia04) mutant animals are developmentally delayed when grown on NGM-20 µM BP as compared to growth on NGM. (B) Total iron content in N2 wildtype and hif-1(ia04) mutant animals determined by ICP is expressed as a percentage of N2 wildtype animals. Four independent experiments were performed and the data are reported as means ± SEM, *p<0.02. (C) N2 wildtype and hif-1(ia04) animals were fed control RNAi or ftn-1 and ftn-2 RNAi and cultured on NGM or NGM+20 µM BP. The data are expressed as the number of animals reaching L4 stage as a percentage of N2 wildtype animals. Three independent experiments were performed and the data are reported as means ± SEM, *p<0.05, hif-1(ia04) (control RNAi) versus hif-1(ia04) (ftn-1/ftn-2 RNAi). We next questioned whether the developmental delay observed in hif-1(ia04) mutant animals cultured in BP can be rescued by reducing FTN-1 and FTN-2 expression, which would lead to an increase in the cellular labile iron pool. This pool contains chelatable redox-active iron that constitutes less than 5% of total cellular iron [32], [35]. The modulation of ferritin levels is one mechanism for regulating the labile iron pool in mammalian cells: ferritin overexpression reduces the iron pool, while ferritin depletion increases this pool [36]–[38]. We depleted ftn-1 and ftn-2 by RNAi in N2 wildtype and in hif-1(ia04) mutant animals cultured on NGM or NGM plus 20 µM BP, and the number of worms reaching L4 stage was measured. ftn-1/ftn-2 RNAi increases the number of hif-1(ia04) mutant animals reaching L4 stage from 28% in untreated animals to 78% in ftn-1/ftn-2 RNAi-fed animals (Figure 6C). These data indicate that the developmental delay observed in hif-1(ia04) mutant animals during iron deficiency can be partially rescued by reducing FTN-1 and FTN-2 levels.
Discussion
Previous studies showed that ftn-1 and ftn-2 transcription in intestine is activated by iron and inhibited by iron deficiency [22]–[24]. Transcriptional regulation is dependent on an IDE containing HIF-1 and GATA binding sites. The GATA factor ELT2 regulates basal intestinal ftn-1 and ftn-2 transcription during iron sufficiency, but how transcription is repressed by iron deficiency was not understood. Here, we identify HIF-1 as a negative regulator of ftn-1 and ftn-2 transcription during iron deficiency. We also show that smf-3 is regulated by HIF-1 during iron deficiency, but unlike ftn-1 and ftn-2, HIF-1 activates smf-3 transcription. The activation of smf-3 and inhibition of ftn-1 and ftn-2 by HIF-1 provides a mechanism to increase iron uptake and decrease iron storage during iron deficiency.
In mice, increased iron uptake by DMT1 during iron deficiency is mediated by HIF-2α [7], [8]. In contrast to the transcriptional regulation of ftn-1 and ftn-2 in C. elegans, vertebrate ferritin H - and -L subunit mRNAs are translationally repressed during iron deficiency by the binding of IRP1 and IRP2 to an iron-responsive element (IRE) in ferritin mRNAs [26]. When cellular iron levels increase, IRP1 is converted into a cytosolic aconitase concomitant with loss of RNA-binding activity and IRP2 is degraded, which leads to increased ferritin translation. Although C. elegans express an IRP1 homolog (ACO-1), it lacks RNA-binding activity and functions solely as a cytosolic aconitase [22]. The regulation of iron homeostasis by ftn-1and ftn-2 is essential as depletion of FTN-1 and FTN-2 rescues the growth delay observed in hif-1 null worms grown under iron limiting conditions. Recent studies show that constitutive expression of ferritin-H and -L subunits in mice with intestinal specific deletion of IRP1 and IRP2 have reduced survival rates [39]. Depletion of the intestinal ferritin-H gene in mice leads to reduced iron sequestration in intestine and increased body iron stores [40]. Taken together, these studies show that the precise regulation of intestinal ferritin expression is essential for appropriate control of iron absorption.
The transcriptional inhibition of ftn-1and ftn-2 genes by HIF-1 during iron deficiency was unexpected. HIF-1 is a potent transcriptional activator of hundreds of HRE target genes [15], [41], [42]. HIF has also been reported to function as a repressor, but the mechanism of transcriptional repression is not fully understood [43]–[48]. For some HIF-negatively regulated genes, transcriptional repression may be an indirect effect due to HIF activation of a transcriptional repressor [41], [42]. HIF-1 has also been shown to upregulate microRNA mir-120, which represses gene expression [49]. For other genes, direct binding of HIF to HREs in the promoters of negatively-regulated genes has been demonstrated using ChIP analysis [45]–[47]. These studies led to models whereby HIF-1 negatively regulates transcription by the recruitment of corepressors and histone modifying complexes or by competing with transcriptional activators for HRE binding.
One question is how HIF-1 mediates transcriptional activation and inhibition through the IDE. The ftn-1, ftn-2 and smf-3 IDEs differ in number, spacing and orientation of the GATA and HIF-1 binding sites. It is likely that architecture of the IDE dictates physical interactions of ELT-2, HIF-1, coactivators and other transcription factors to activate or repress transcription. Further studies are needed to determine how the structure of the IDE affects HIF-1 transcriptional responses.
HREs can be flanked by transcription factor binding sites, and it is the cooperation of HIF with these transcription factors that enhance transcription or direct cell specific expression [11]. The GATA binding sites flanking the HREs in ftn-1, ftn-2 and smf-3 are essential for intestinal expression because mutations of these sites or depletion of ELT-2 reduces intestinal expression of ftn-1 and smf-3 GFP transcriptional reporters [24] (preliminary data, SJR and EAL). Computational studies have shown that the majority of intestinal specific genes contain GATA binding sequences, leading to the notion that ELT2 is a global regulator of intestinal gene expression [50], [51]. Several studies have shown that other transcription factors cooperate with ELT-2 to modulate its function in response to nutritional or physiological signals [52]–[56].
Based on our findings, a model for HIF-1 regulation of intestinal iron homeostasis is proposed (Figure 7). During iron sufficiency, ELT-2 binds to GATA binding sites in the IDE to activate intestinal expression of ftn-1 and ftn-2. Both GATA binding sites are required for ftn-1 expression as deletion of either site abolished expression of an ftn-1 GFP transcriptional reporter [24]. In addition, mutation of all three HREs in the ftn-1 IDE abolished GFP reporter expression, suggesting that a transcriptional activator may bind to the HREs enhancing ELT-2 function [24]. This activator may be a member of the basic helix-loop-helix (bHLH/PAS) family or a bHLH transcriptional activator that binds to noncanonical E-box elements [57]–[59]. smf-3 transcription is reduced during iron sufficiency due to decreased HIF-1α levels. Our data also show that a small amount of HIF-1 is expressed during normal growth conditions that can interact with HREs in ftn-1 and smf-3. During iron deficiency, HIF-1 is stabilized and transported to the nucleus where it dimerizes with AHA-1. We propose that HIF-1/AHA-1 competes with a transcriptional activator for binding to the ftn-1 and ftn-2 HREs, inhibiting ftn-1 and ftn-2 transcription. HIF-1/AHA-1 binds to the smf-3 HREs, recruits coactivators and cooperates with ELT-2 to activate transcription.
Fig. 7. Model for HIF-1 iron-dependent activation and inhibition of intestinal iron uptake and storage. 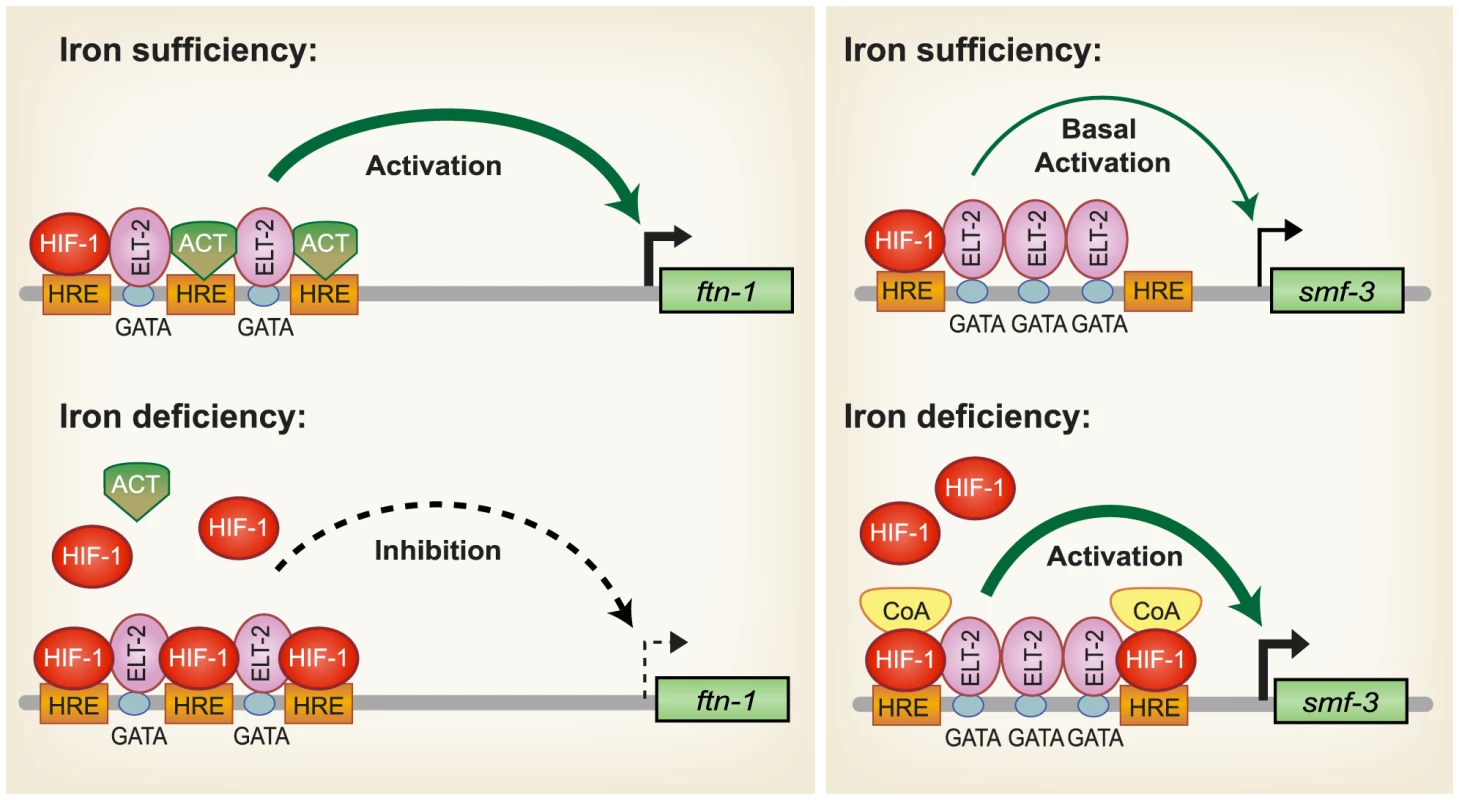
Iron sufficiency: ELT-2 binds to GATA binding sites located in the ftn-1 and ftn-2 IDEs. We propose that ELT- 2 cooperates with an unidentified transcriptional activator (ACT) that binds to the HREs and enhances transcription. smf-3 is transcribed at basal levels during iron sufficiency. HIF-1 is expressed during normal growth conditions, but at low levels. Iron deficiency: HIF-1 is stabilized and dimerizes with AHA-1. HIF-1/AHA-1 (denoted by HIF-1) displaces the unidentified transcriptional activator binding to the ftn-1 and ftn-2 HREs and inhibits transcription. HIF-1/AHA-1 binds to the smf-3 HREs, recruits coactivators and cooperates with ELT-2 to enhance smf-3 transcription. Whether ELT-2 is bound to the ftn-1 GATA sites during iron deficiency and to the smf-3 GATA sites during iron sufficiency has not been determined. SMF-1, SMF-2 and SMF-3 have been characterized with regard to their role in Mn2+ homeostasis and sensitivity [28], [30]. SMF-1 and SMF-3 are localized to the apical intestinal epithelium consistent with a role in metal uptake [28], [30], whereas SMF-2 is primarily expressed in pharyngeal epithelium [28], [29]. SMF-1 and SMF-2 are also expressed in dopaminergic neurons where they mediate sensitivity of neurons to Mn and neurotoxins [29]. The high homology of SMF-3 with mammalian DMT1, its localization in the apical membrane of intestinal cells [28], [30], its regulation by iron and a reduction in total iron content in smf-3(ok1035) mutant animals indicate a role for SMF-3 is intestinal iron transport in C. elegans. The regulation of iron homeostasis by HIF-1 provides a mechanism to ensure that C. elegans maintain sufficient iron stores for growth and survival when iron is limiting.
Materials and Methods
C. elegans strains and culture
Strains were cultivated on nematode growth medium (NGM) agar plates seeded with Escherichia coli OP50 at 20–22°C. For iron chelation experiments, synchronized larvae were grown for 24 h on NGM plates then transferred to NGM plates supplemented with 100 uM 2,2′-dipyridyl (BP) for 18 h unless indicated. For iron experiments, larvae were grown for 18 h on NGM plates supplemented with 6.6 mg/ml ferric ammonium citrate (FAC). The pH of FAC-supplemented NGM agar was adjusted to pH 7.0.
The strains provided by C. elegans Genetics Center are: wild-type Bristol N2, vhl-1 (ok161) [13], hif-1 (ia04) [14], RB1491 smf-1(ok1748), VC171 smf-2(gk133) and RB1074 smf-3(ok1035). XA6900 pha-1(e2123ts) III; qaEx1 [ftn-1::Δpes-10::GFP-his, pha-1+] and XA6901 lin-15(n765ts) X; qaEx2 [ftn-2::Δpes-10::GFP-his, lin-15+] were previously described (Romney et al., 2008). Strains generated in this study are: XA6904 pha-1 (e2123ts) III; qaEx04 [smf-3(-1500)::Δpes-10::GFP-his, pha-1+] and XA6905 pha-1(e2123ts) III; qaEx05 [smf-3(-250)::Δpes-10::GFP-his, pha-1+].
Reporter constructs and transgenic lines
smf-3(-1500)::GFP-his and smf-3(-250)::GFP-his were generated by PCR amplification of sequences 1500 nt or 250 nt, respectively, upstream from the initiation ATG of smf-3 (Y69A2AR.4) using primers containing SalI and NheI restriction sites on the 5′ and 3′ termini, respectively. The PCR products were cloned into TOPO Zeroblunt (Invitrogen) followed by digestion and insertion into the SalI and NheI sites of pAP.10. Transgenic strains were made according to standard microinjection procedures. Plasmids B696 (smf-3(-1500)::GFP-his) and B698 (smf-3(-250)::GFP-his) were each co-injected (20 ng/ul) with selection plasmid pBX-1(100 ng/ul). Transgenic worms were recovered after growth at 20–22°C.
RNAi
RNAi clones against ftn-1 (C54F6.14) and ftn-2 (D1037.3) were from the ORFeome-based RNAi library [60] and hif-1 (F38A6.3) was from the Ahringer feeding library [61]. Empty vector (L4400) was used as a control. Worms were grown on RNAi plates (NGM containing 1 mM IPTG and 50 ug/ml ampicillin) seeded with bacteria expressing dsRNA corresponding to ftn-1and ftn-2, hif-1 or L4400.
Rescue
Mixed-stage populations of N2 wildtype and hif-1(ia04) animals containing gravid adults were transferred to RNAi plates seeded with bacteria expressing ftn-1 and ftn-2 dsRNA or empty vector control (L4400) for 24 h at 20–22°C. Synchronized larval stage (L1) worms were obtained by treating RNAi fed gravid adults with alkaline hypochlorite and allowing eggs to hatch overnight in sterile S-basal media. L1 larvae were spotted on ftn-1 and ftn-2 or L4400 RNAi plates supplemented with or without 20 µM BP and incubated at 20–22°C for 48 h prior to scoring for larval stage using a Leica MZ9.5 stereomicroscope.
C. elegans sorting
ftn-1::GFP-his, ftn-2::GFP-his, smf-3(-1500)::GFP-his and smf-3(-250)::GFP-his reporter lines were synchronized by treating gravid adults with alkaline hypochlorite followed by hatching eggs in S-basal medium (0.1 M NaCl, 0.05 M potassium phosphate, pH 6.0, 5 ug/ml cholesterol) overnight. Synchronized L1 worms were grown on control or hif-1 RNAi plates for 32 h. Worms were then rinsed from the plates and washed in M9 buffer (22 mM KH2PO4, 42 mM Na2HPO4, 86 mM NaCl, 1 mM MgSO4) and spotted onto fresh 10 cm control or hif-1 RNAi plates supplemented with 100 µM BP and grown for 18 h. Worms were rinsed from plates and washed with MT buffer (M9 buffer containing 0.1% Triton X-100) by repeated rounds of centrifugation until free of debris. Worms were analyzed using the COPAS Biosort (Union Biometrica, Somerville, MA) as described [24]. Prior to data acquisition, gating parameters were established by visualizing a sorted population by microscopy. The same gating parameters were used for all experimental conditions during GFP fluorescence acquisition and subsequent analysis. Data were analyzed using FCS Express Version 3.0 lite (De Novo Software, Ontario, Canada). Procedures for sorting non-RNAi-treated worms was the same as above except that synchronized L1 stage worms were grown on NGM plates seeded with OP50 for 32 h prior to being transferred to fresh NGM plates and NGM plates supplemented with either 100 uM BP, 100 uM FAC, 100 uM BP plus100 uM MnCl2 or 100 uM BP plus 100 uM FAC. Worms were grown for an additional 18 h prior to data acquisition as performed above.
Microscopy
Images of GFP expression were captured using an Axio Imager (Carl Zeiss MicroImaging, Inc, Thornwood NY) outfitted with the Zeiss filter set 38HE (BP 470/40HE, dichroic FT 495 HE, BP 525/50 HE) and an AxioCam HRm camera (Carl Zeiss MicroImaging, Inc, Thornwood NY) using AxioVision software. Following acquisition, images were rotated, cropped and sized using Adobe Photoshop.
Western blotting
Mixed-stage populations of N2, hif-1(ia04) and vhl-1(ok161) animals were transferred from NGM plates to fresh NGM and 100 uM BP supplemented plates for 16 hr prior to harvesting. Worms were collected, washed with ddH2O and resuspended in 200 µl lysis buffer (20 mM HEPES pH 7.5, 25 mM KCl, 0.5% NP-40). Worms were sonicated twice using a Misonix 3000 (8 pulses, output 3), centrifuged and the protein concentration of the clarified lysate was determined using Coomassie Plus protein assay reagent (Thermo). Samples (20 ug) were fractionated on a NuPAGE 4–12% Bis-Tris gel (Invitrogen) and western blotting was carried out using rabbit CeHIF-1 antibody at (1∶5,000) [14] (a gift from Dr. Peter Ratcliffe, Oxford) followed by incubation with horseradish peroxidase-conjugated secondary antibodies. Proteins were visualized using Western Lighting Chemiluminescence Reagent Plus (PerkinElmer Life Sciences).
RT–PCR
Synchronized L1 larvae were grown on NGM plates seeded with OP50 for 48 h prior to harvest. For iron chelation experiments, synchronized worms were transferred to new NGM plates or NGM plates supplemented with 100 uM BP after 32 h and harvested at 48 h. At harvest, worms were washed from plates and cleaned by repeated rounds of centrifugation and resuspension in ddH2O. Pelleted worms were resuspended in TRIZOL (Invitrogen) and total RNA was purified according to manufacturer's protocol. cDNA was synthesized using 1 ug of purified total RNA using SuperscriptIII Supermix for qRT-PCR (Invitrogen). Semiquantitative RT-PCR (sqRT-PCR) was performed using Recombinant Taq Polymerase (Invitrogen). PCR products were run on ethidium bromide stained 1.2% agarose gels. Images were captured on a FluorChem IS-8900 (Alpha Innotech) and analyzed using ImageQuaNT (Molecular Dynamics) and ImageJ. Primer sequences are shown in Table S1.
Chromatin immunoprecipitation (ChIP)
ChIP assays were performed as previously described [62]. Mixed-stage vhl-1 (ok161) and hif-1 (ia04) worms were grown on two 150 mm NGM plates and harvested. Worm pellets were homogenized in crosslinking buffer (1% formaldehyde in PBS) and incubated for 20 min at room temperature. The reactions were quenched with glycine for 20 min and snap frozen. The frozen pellets were resuspended in HLB buffer (50 mM HEPES-KOH, pH 7.5, 150 mM NaCl, 1 mM EDTA, 0.1% sodium deoxycholate, 1% Triton-X100, 0.1% SDS, 1 mM PMSF) containing protease inhibitor cocktail (Calbiochem), sonicated and the supernatants were precleared using 30 ul salmon sperm DNA/protein A agarose beads. After centrifugation, 10% of each supernatant was kept as input, while the remaining supernatants were incubated with CeHIF antibody (4 ul) overnight at 4°C. DNA-protein antibody complexes were incubated with Protein A agarose (30 ul slurry) for 2 h at 4°C, and the beads were then resuspended in proteinase K buffer (20 mg/ml proteinase K) and incubated at 45°C for 2 h. The samples were further incubated at 65°C overnight to reverse crosslinks. DNA was purified using a QIAquick PCR purification kit (Qiagen) according to manufacturer's protocol. Quantitative (q) PCR was then performed using Sybr Green mix (Invitrogen) run on an ABI 7000 Sequence Detection System and additionally visualized by 2% ethidium bromide stained agarose gels. Primers flanking the ftn-1 IDE and control primers within the ftn-1 coding region are shown in Table S1.
Electrophoretic mobility shift assay (EMSA)
Wildtype IDE and IDE containing mutations in the three HRE binding sites (HRE3m) (CACGTAGC>ACATGCTA) were excised from TopoZero blunt (Invitrogen) with EcoRI and gel purified. Wildtype IDE was radiolabeled with 50 µCi of 32P[α-dATP] using Klenow DNA polymerase. HIF-1 and AHA-1 were synthesized in TNT SP6 Quick Coupled Transcription/Translation system (Promega) using hif-1 (pSP64-HIF-1) and aha-1 (pJ343) [12]. pSP64-HIF-1 was generated by excision of hif-1 cDNA from pR33 [13] and insertion into pSP64 (Promega). EMSA reactions (20 µl) were performed in reaction buffer (10 mM Tris-HCl, pH 7.5, 4% glycerol, 1 mM MgCl2, 1 mM DTT) containing 32P-labeled wildtype or HRE3m probes (0.4–1 ng), poly dI-dC (80 ng) and HIF-1 and/or AHA-1 (1–4 µl) at room temperature for 30 min. CeHIF-1 antibody (2 ul) was added during the last 5 min of the binding reactions. For competition experiments, unlabeled IDE or HRE3m DNA (10–100× molar amounts) was added to the reactions 5 min before addition of 32P-labeled probes. Samples were fractionated on a 5% non-denaturing polyacrylamide gel (37.5∶1 acrylamide∶bis).
Metal content by inductively-coupled plasma-optical emission spectroscopy (ICP)
Synchronized L1 worms were obtained by treating gravid adults from each strain with alkaline hypochlorite followed by hatching eggs in S-basal medium overnight. L1 worms were grown on OP-50 seeded NGM plates. L4 worms were washed extensively with M9 buffer and incubated in M9 buffer for 30 min at room temperature to allow for purging of the gut followed by two rinses with ddH2O. Empty tubes were run in parallel to serve as controls. Samples were pelleted and frozen at −80C°. Samples and controls were brought up in 200 ul of metal free 40% nitric acid (Optima) and heated to 95°C for 2 min in a heating block. Solubilized samples were diluted to a final nitric acid concentration of 10% with ddH2O and measured on an Optima 3000 XL ICP-OES (Perkin Elmer). Serial dilutions of commercially available mixed metal standards were used to calibrate the instrument. Results were normalized to the simultaneously acquired signal for sulfur for each sample [63]. The data are presented as a percentage of N2 wildtype animals. At least three independent biological replicates were performed.
Statistical analysis
Data are presented as the means ± SEM. Two-tailed unpaired Student's t test were used for statistical analysis. Data are considered statistically significant at p<0.05.
Supporting Information
Zdroje
1. AndrewsNCSchmidtPJ 2007 Iron homeostasis. Annu Rev Physiol 69 69 85
2. ZhangASEnnsCA 2009 Iron homeostasis: recently identified proteins provide insight into novel control mechanisms. J Biol Chem 284 711 715
3. MackenzieBGarrickMD 2005 Iron Imports. II. Iron uptake at the apical membrane in the intestine. Am J Physiol Gastrointest Liver Physiol 289 G981 986
4. HarrisonPMArosioP 1996 The ferritins: molecular properties, iron storage function and cellular regulation. Biochim Biophys Acta 1275 161 203
5. TortiFMTortiSV 2002 Regulation of ferritin genes and protein. Blood 99 3505 3516
6. TheilEC 2009 Mining ferritin iron: 2 pathways. Blood 114 4325 4326
7. ShahYMMatsubaraTItoSYimSHGonzalezFJ 2009 Intestinal hypoxia-inducible transcription factors are essential for iron absorption following iron deficiency. Cell Metab 9 152 164
8. MastrogiannakiMMatakPKeithBSimonMCVaulontS 2009 HIF-2α, but not HIF-1α, promotes iron absorption in mice. J Clin Invest 119 1159 1166
9. SemenzaGL 2007 Life with oxygen. Science 318 62 64
10. KaelinWGJrRatcliffePJ 2008 Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol Cell 30 393 402
11. KaluzSKaluzovaMStanbridgeEJ 2008 Regulation of gene expression by hypoxia: integration of the HIF-transduced hypoxic signal at the hypoxia-responsive element. Clin Chim Acta 395 6 13
12. Powell-CoffmanJABradfieldCAWoodWB 1998 Caenorhabditis elegans orthologs of the aryl hydrocarbon receptor and its heterodimerization partner the aryl hydrocarbon receptor nuclear translocator. Proc Natl Acad Sci U S A 95 2844 2849
13. JiangHGuoRPowell-CoffmanJA 2001 The Caenorhabditis elegans hif-1 gene encodes a bHLH-PAS protein that is required for adaptation to hypoxia. Proc Natl Acad Sci U S A 98 7916 7921
14. EpsteinACGleadleJMMcNeillLAHewitsonKSO'RourkeJ 2001 C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell 107 43 54
15. ShenCNettletonDJiangMKimSKPowell-CoffmanJA 2005 Roles of the HIF-1 hypoxia-inducible factor during hypoxia response in Caenorhabditis elegans. J Biol Chem 280 20580 20588
16. PocockRHobertO 2008 Oxygen levels affect axon guidance and neuronal migration in Caenorhabditis elegans. Nat Neurosci 11 894 900
17. ChangAJBargmannCI 2008 Hypoxia and the HIF-1 transcriptional pathway reorganize a neuronal circuit for oxygen-dependent behavior in Caenorhabditis elegans. Proc Natl Acad Sci U S A 105 7321 7326
18. ZhangYShaoZZhaiZShenCPowell-CoffmanJA 2009 The HIF-1 hypoxia-inducible factor modulates lifespan in C. elegans. PLoS ONE 4 e6348 doi:10.1371/journal.pone.0006348
19. PadillaPARothMB 2001 Oxygen deprivation causes suspended animation in the zebrafish embryo. Proc Natl Acad Sci U S A 98 7331 7335
20. LiuXTheilEC 2005 Ferritins: dynamic management of biological iron and oxygen chemistry. Acc Chem Res 38 167 175
21. Bou-AbdallahF 2010 The iron redox and hydrolysis chemistry of the ferritins. Biochim Biophys Acta 1800 719 731
22. GourleyBLParkerSBJonesBJZumbrennenKBLeiboldEA 2003 Cytosolic aconitase and ferritin are regulated by iron in Caenorhabditis elegans. J Biol Chem 278 3227 3234
23. KimYIChoJHYooOJAhnnJ 2004 Transcriptional regulation and life-span modulation of cytosolic aconitase and ferritin genes in C.elegans. J Mol Biol 342 421 433
24. RomneySJThackerCLeiboldEA 2008 An iron enhancer element in the FTN-1 gene directs iron-dependent expression in Caenorhabditis elegans intestine. J Biol Chem 283 716 725
25. HentzeMWMuckenthalerMUAndrewsNC 2004 Balancing acts: molecular control of mammalian iron metabolism. Cell 117 285 297
26. WallanderMLLeiboldEAEisensteinRS 2006 Molecular control of vertebrate iron homeostasis by iron regulatory proteins. Biochim Biophys Acta 1763 668 689
27. RouaultTA 2006 The role of iron regulatory proteins in mammalian iron homeostasis and disease. Nat Chem Biol 2 406 414
28. AuCBenedettoAAndersonJLabrousseAEriksonK 2009 SMF-1, SMF-2 and SMF-3 DMT1 orthologues regulate and are regulated differentially by manganese levels in C. elegans. PLoS ONE 4 e7792 doi:10.1371/journal.pone.0007792
29. SettivariRLevoraJNassR 2009 The divalent metal transporter homologues SMF-1/2 mediate dopamine neuron sensitivity in Caenorhabditis elegans models of manganism and parkinson disease. J Biol Chem 284 35758 35768
30. BandyopadhyayJSongHOParkBJSingaraveluGSunJL 2009 Functional assessment of Nramp-like metal transporters and manganese in Caenorhabditis elegans. Biochem Biophys Res Commun 390 136 141
31. BishopTLauKWEpsteinACKimSKJiangM 2004 Genetic analysis of pathways regulated by the von Hippel-Lindau tumor suppressor in Caenorhabditis elegans. PLoS Biol 2 e289 doi:10.1371/journal.pbio.0020289
32. BreuerWEpsztejnSCabantchikZI 1995 Iron acquired from transferrin by K562 cells is delivered into a cytoplasmic pool of chelatable iron(II). J Biol Chem 270 24209 24215
33. ShenCShaoZPowell-CoffmanJA 2006 The Caenorhabditis elegans rhy-1 gene inhibits HIF-1 hypoxia-inducible factor activity in a negative feedback loop that does not include vhl-1. Genetics 174 1205 1214
34. PadillaPANystulTGZagerRAJohnsonACRothMB 2002 Dephosphorylation of cell cycle-regulated proteins correlates with anoxia-induced suspended animation in Caenorhabditis elegans. Mol Biol Cell 13 1473 1483
35. KakhlonOCabantchikZI 2002 The labile iron pool: characterization, measurement, and participation in cellular processes(1). Free Radic Biol Med 33 1037 1046
36. PicardVEpsztejnSSantambrogioPCabantchikZIBeaumontC 1998 Role of ferritin in the control of the labile iron pool in murine erythroleukemia cells. J Biol Chem 273 15382 15386
37. EpsztejnSGlicksteinHPicardVSlotkiINBreuerW 1999 H-ferritin subunit overexpression in erythroid cells reduces the oxidative stress response and induces multidrug resistance properties. Blood 94 3593 3603
38. KakhlonOGruenbaumYCabantchikZI 2001 Repression of ferritin expression increases the labile iron pool, oxidative stress, and short-term growth of human erythroleukemia cells. Blood 97 2863 2871
39. GalyBFerring-AppelDKadenSGroneHJHentzeMW 2008 Iron regulatory proteins are essential for intestinal function and control key iron absorption molecules in the duodenum. Cell Metab 7 79 85
40. VanoaicaLDarshanDRichmanLSchumannKKuhnLC 2010 Intestinal ferritin H is required for an accurate control of iron absorption. Cell Metab 12 273 282
41. ManaloDJRowanALavoieTNatarajanLKellyBD 2005 Transcriptional regulation of vascular endothelial cell responses to hypoxia by HIF-1. Blood 105 659 669
42. MoleDRBlancherCCopleyRRPollardPJGleadleJM 2009 Genome-wide association of hypoxia-inducible factor (HIF)-1α and HIF-2α DNA binding with expression profiling of hypoxia-inducible transcripts. J Biol Chem 284 16767 16775
43. NarravulaSColganSP 2001 Hypoxia-inducible factor 1-mediated inhibition of peroxisome proliferator-activated receptor alpha expression during hypoxia. J Immunol 166 7543 7548
44. MazureNMChauvetCBois-JoyeuxBBernardMANacer-CherifH 2002 Repression of alpha-fetoprotein gene expression under hypoxic conditions in human hepatoma cells: characterization of a negative hypoxia response element that mediates opposite effects of hypoxia inducible factor-1 and c-Myc. Cancer Res 62 1158 1165
45. EltzschigHKAbdullaPHoffmanEHamiltonKEDanielsD 2005 HIF-1-dependent repression of equilibrative nucleoside transporter (ENT) in hypoxia. J Exp Med 202 1493 1505
46. ChenKFLaiYYSunHSTsaiSJ 2005 Transcriptional repression of human cad gene by hypoxia inducible factor-1α. Nucleic Acids Res 33 5190 5198
47. IblaJCKhouryJKongTRobinsonAColganSP 2006 Transcriptional repression of Na-K-2Cl cotransporter NKCC1 by hypoxia-inducible factor-1. Am J Physiol Cell Physiol 291 C282 289
48. PeyssonnauxCZinkernagelASSchuepbachRARankinEVaulontS 2007 Regulation of iron homeostasis by the hypoxia-inducible transcription factors (HIFs). J Clin Invest 117 1926 1932
49. HuangXDingLBennewithKLTongRTWelfordSM 2009 Hypoxia-inducible mir-210 regulates normoxic gene expression involved in tumor initiation. Mol Cell 35 856 867
50. McGheeJDSleumerMCBilenkyMWongKMcKaySJ 2007 The ELT-2 GATA-factor and the global regulation of transcription in the C. elegans intestine. Dev Biol 302 627 645
51. PauliFLiuYKimYAChenPJKimSK 2006 Chromosomal clustering and GATA transcriptional regulation of intestine-expressed genes in C. elegans. Development 133 287 295
52. FukushigeTGoszczynskiBYanJMcGheeJD 2005 Transcriptional control and patterning of the pho-1 gene, an essential acid phosphatase expressed in the C. elegans intestine. Dev Biol 279 446 461
53. NevesAEnglishKPriessJR 2007 Notch-GATA synergy promotes endoderm-specific expression of ref-1 in C. elegans. Development 134 4459 4468
54. Anokye-DansoFAnyanfulASakubeYKagawaH 2008 Transcription factors GATA/ELT-2 and forkhead/HNF-3/PHA-4 regulate the tropomyosin gene expression in the pharynx and intestine of Caenorhabditis elegans. J Mol Biol 379 201 211
55. InoueHNishidaE 2010 The DM domain transcription factor MAB-3 regulates male hypersensitivity to oxidative stress in Caenorhabditis elegans. Mol Cell Biol 30 3453 3459
56. SinclairJHamzaI 2010 A novel heme-responsive element mediates transcriptional regulation in Caenorhabditis elegans. J Biol Chem 285 39536 39543
57. WoodsSLWhitelawML 2002 Differential activities of murine single minded 1 (SIM1) and SIM2 on a hypoxic response element. Cross-talk between basic helix-loop-helix/per-Arnt-Sim homology transcription factors. J Biol Chem 277 10236 10243
58. KewleyRJWhitelawMLChapman-SmithA 2004 The mammalian basic helix-loop-helix/PAS family of transcriptional regulators. Int J Biochem Cell Biol 36 189 204
59. FarrallALWhitelawML 2009 The HIF1α-inducible pro-cell death gene BNIP3 is a novel target of SIM2s repression through cross-talk on the hypoxia response element. Oncogene 28 3671 3680
60. RualJFCeronJKorethJHaoTNicotAS 2004 Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res 14 2162 2168
61. KamathRSFraserAGDongYPoulinGDurbinR 2003 Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421 231 237
62. MukhopadhyayADeplanckeBWalhoutAJTissenbaumHA 2008 Chromatin immunoprecipitation (ChIP) coupled to detection by quantitative real-time PCR to study transcription factor binding to DNA in Caenorhabditis elegans. Nat Protoc 3 698 709
63. BlairBGLarsonCASafaeiRHowellSB 2009 Copper transporter 2 regulates the cellular accumulation and cytotoxicity of Cisplatin and Carboplatin. Clin Cancer Res 15 4312 4321
Štítky
Genetika Reprodukční medicína
Článek A Complex Genomic Rearrangement Involving the Locus Causes Dermal Hyperpigmentation in the ChickenČlánek Genome Instability and Transcription Elongation Impairment in Human Cells Depleted of THO/TREXČlánek A Population Genetics-Phylogenetics Approach to Inferring Natural Selection in Coding SequencesČlánek Interspecific Sex in Grass Smuts and the Genetic Diversity of Their Pheromone-Receptor SystemČlánek Genomic Distribution and Inter-Sample Variation of Non-CpG Methylation across Human Cell Types
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2011 Číslo 12- Růst a vývoj dětí narozených pomocí IVF
- Délka menstruačního cyklu jako marker ženské plodnosti
- Vztah užívání alkoholu a mužské fertility
- Šanci na úspěšný průběh těhotenství snižují nevhodné hladiny progesteronu vznikající při umělém oplodnění
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
-
Všechny články tohoto čísla
- The Connection between Space and Thinking: An Interview with Rafael Viñoly
- An Assessment of the Individual and Collective Effects of Variants on Height Using Twins and a Developmentally Informative Study Design
- Widespread Cotranslational Formation of Protein Complexes
- Genomes Reveal Transition of Bacteria from Aquatic to Terrestrial Environments
- A Complex Genomic Rearrangement Involving the Locus Causes Dermal Hyperpigmentation in the Chicken
- Plasticity of BRCA2 Function in Homologous Recombination: Genetic Interactions of the PALB2 and DNA Binding Domains
- Transcription Is Required to Establish Maternal Imprinting at the Prader-Willi Syndrome and Angelman Syndrome Locus
- Substitutions in the Amino-Terminal Tail of Neurospora Histone H3 Have Varied Effects on DNA Methylation
- MAPK/ERK Signaling Regulates Insulin Sensitivity to Control Glucose Metabolism in
- A Comprehensive Analysis of Shared Loci between Systemic Lupus Erythematosus (SLE) and Sixteen Autoimmune Diseases Reveals Limited Genetic Overlap
- Genome Instability and Transcription Elongation Impairment in Human Cells Depleted of THO/TREX
- Genome-Wide Meta-Analysis of Five Asian Cohorts Identifies as a Susceptibility Locus for Corneal Astigmatism
- A Population Genetics-Phylogenetics Approach to Inferring Natural Selection in Coding Sequences
- HIF-1 Regulates Iron Homeostasis in by Activation and Inhibition of Genes Involved in Iron Uptake and Storage
- Ror2 Enhances Polarity and Directional Migration of Primordial Germ Cells
- DNA Methylation of the Gonadal Aromatase () Promoter Is Involved in Temperature-Dependent Sex Ratio Shifts in the European Sea Bass
- A Genetic Screening Strategy Identifies Novel Regulators of the Proteostasis Network
- Interspecific Sex in Grass Smuts and the Genetic Diversity of Their Pheromone-Receptor System
- The Synthetic Multivulva Genes Prevent Ras Pathway Activation by Tightly Repressing Global Ectopic Expression of EGF
- Mining the Allelic Spectrum Reveals the Contribution of Rare and Common Regulatory Variants to HDL Cholesterol
- Identification of a Genomic Reservoir for New Genes in Primate Genomes
- Genomic Distribution and Inter-Sample Variation of Non-CpG Methylation across Human Cell Types
- Identification of Evolutionarily Conserved Exons as Regulated Targets for the Splicing Activator Tra2β in Development
- Acute Multiple Organ Failure in Adult Mice Deleted for the Developmental Regulator Wt1
- Age-Related Neuronal Degeneration: Complementary Roles of Nucleotide Excision Repair and Transcription-Coupled Repair in Preventing Neuropathology
- Target Site Recognition by a Diversity-Generating Retroelement
- Ancestral Components of Admixed Genomes in a Mexican Cohort
- Targeted Proteolysis of Plectin Isoform 1a Accounts for Hemidesmosome Dysfunction in Mice Mimicking the Dominant Skin Blistering Disease EBS-Ogna
- Autosomal Recessive Dilated Cardiomyopathy due to Mutations Results from Abnormal Dystroglycan O-Mannosylation
- SREBP Coordinates Iron and Ergosterol Homeostasis to Mediate Triazole Drug and Hypoxia Responses in the Human Fungal Pathogen
- The RNA Silencing Enzyme RNA Polymerase V Is Required for Plant Immunity
- An Anti-Checkpoint Activity for Rif1
- The FGFR4-G388R Polymorphism Promotes Mitochondrial STAT3 Serine Phosphorylation to Facilitate Pituitary Growth Hormone Cell Tumorigenesis
- Common Variants Show Predicted Polygenic Effects on Height in the Tails of the Distribution, Except in Extremely Short Individuals
- The Fission Yeast Stress-Responsive MAPK Pathway Promotes Meiosis via the Phosphorylation of Pol II CTD in Response to Environmental and Feedback Cues
- Integrating Genome-Wide Genetic Variations and Monocyte Expression Data Reveals -Regulated Gene Modules in Humans
- Repetitive Elements May Comprise Over Two-Thirds of the Human Genome
- A Novel Checkpoint and RPA Inhibitory Pathway Regulated by Rif1
- Hierarchical Generalized Linear Models for Multiple Groups of Rare and Common Variants: Jointly Estimating Group and Individual-Variant Effects
- The Major Roles of DNA Polymerases Epsilon and Delta at the Eukaryotic Replication Fork Are Evolutionarily Conserved
- A High-Resolution Whole-Genome Map of Key Chromatin Modifications in the Adult
- A Densely Interconnected Genome-Wide Network of MicroRNAs and Oncogenic Pathways Revealed Using Gene Expression Signatures
- A Functional Phylogenomic View of the Seed Plants
- Histone H3K9 Trimethylase Eggless Controls Germline Stem Cell Maintenance and Differentiation
- Ribosomal Protein Mutants Control Tissue Growth Non-Autonomously via Effects on the Prothoracic Gland and Ecdysone
- , , and Are Required to Activate or Delimit the Spread of the Transcriptional Response to Epidermal Wounds in
- Mechanisms Establishing TLR4-Responsive Activation States of Inflammatory Response Genes
- Candidate Gene Screen in the Red Flour Beetle Reveals as Ancient Regulator of Anterior Median Head and Central Complex Development
- Charcot-Marie-Tooth–Linked Mutant GARS Is Toxic to Peripheral Neurons Independent of Wild-Type GARS Levels
- The RNA–Methyltransferase Misu (NSun2) Poises Epidermal Stem Cells to Differentiate
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Targeted Proteolysis of Plectin Isoform 1a Accounts for Hemidesmosome Dysfunction in Mice Mimicking the Dominant Skin Blistering Disease EBS-Ogna
- The RNA Silencing Enzyme RNA Polymerase V Is Required for Plant Immunity
- The FGFR4-G388R Polymorphism Promotes Mitochondrial STAT3 Serine Phosphorylation to Facilitate Pituitary Growth Hormone Cell Tumorigenesis
- Target Site Recognition by a Diversity-Generating Retroelement
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



