-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
An Anti-Checkpoint Activity for Rif1
article has not abstract
Published in the journal: . PLoS Genet 7(12): e32767. doi:10.1371/journal.pgen.1002421
Category: Perspective
doi: https://doi.org/10.1371/journal.pgen.1002421Summary
article has not abstract
Chromosomal double-strand breaks (DSBs) are among the most severe lesions a cell has to deal with: if left unrepaired, they may lead to cell death or cancer. Thus, efficient mechanisms have evolved that respond to the presence of DSBs. These are collectively called the “DNA damage response” (DDR), or the “DNA damage checkpoint”. As a result of intensive studies by many research groups in several model organisms, the basic mechanisms that respond to DNA damage have been delineated: following the formation of DSBs, the broken ends are resected, exposing single-stranded DNA (ssDNA) which gets covered by Replication Protein A (RPA), eliciting cell cycle arrest through a complex cascade of protein recruitment and phosphorylation in which several kinases take part (reviewed in [1]). The ends of linear eukaryotic chromosomes, called telomeres, resemble DSBs; however, they do not normally elicit the checkpoint: the DNA ends are somehow “hidden” from the checkpoint-activating mechanisms. This is a very important feature, as it prevents continuous cell cycle arrests or inappropriate (and undesirable) repair of the natural chromosome ends. However, the precise mechanism(s) by which telomeres avoid checkpoint activation have remained elusive. In the accompanying paper, Xue et al. [2] identify Saccharomyces cerevisiae Rif1 as an important telomeric factor with an anti-checkpoint role.
Yeast telomeres maintain their integrity by the action of three different protein complexes: the CST (Cdc13-Stn1-Ten1) complex, which resembles RPA and binds to the telomeric G-rich single-stranded 3’ end; the Yku70/80 heterodimer, which blocks single-stranded DNA formation specifically in G1 [3]; and the Rap1 protein, which binds the TG-rich telomeric dsDNA and recruits two additional proteins, Rif1 and Rif2, via its C-terminus [4]. The Rif1 and Rif2 proteins seem to have important, yet different, roles in determining the integrity and length of telomeres [4]–[6]. Xue and co-workers [2] have studied the recruitment of several proteins to the telomeres in a strain carrying the temperature-sensitive cdc13-1 allele. In such strains, upon transfer of the cells to the restrictive temperature (e.g., 36°C) telomeres become uncapped and DNA resecting factors such as Sgs1 and Exo1 are recruited, generating ssDNA [7]. The authors followed the recruitment of the various factors, as well as the binding of checkpoint proteins, by chromatin immuno-precipitation (ChIP) at telomeric, subtelomeric, and unrelated sequences after transfer of the cells to the restrictive temperature.
As expected, once resection by Sgs1 and Exo1 started, the amount of Rap1 bound to the telomeric sequences diminished (as Rap1 binds dsDNA); however, surprisingly, Rif1 accumulated with the same pattern as that of the DNA processing enzymes. This was true even in strains in which the C-terminus of Rif1 (thought to be essential for its recruitment) was deleted. Thus, Rif1 can associate to resected telomeres independently of Rap1.
The presence of Rif1 had a negative effect on the recruitment of the checkpoint sensors RPA, Ddc2ATRIP, Ddc1RAD9, and Rad953BP1: a much higher recruitment of these proteins was seen in strains lacking Rif1 than in the wild type. Moreover, with time after temperature shift, the negative effect of Rif1 was stronger at proximal sites than at the subtelomeric sequences, suggesting that the Rif1 protein itself moves; these effects were not caused by increased ssDNA levels or by changes in the dynamics of resection. Thus, it appears that Rif1 travels with the resection machinery at telomeres, preventing the local activation of the checkpoint by interfering with the recruitment of RPA and checkpoint sensors (Figure 1). Rif1 seems to act by de-sensitizing cells to the presence of ssDNA: whereas cdc13-1 RIF1+/RIF1-CΔ cells respond to the presence of ssDNA when its level reaches 6%–10% (at 27°C) but not at low ssDNA levels (e.g., at 25°C), cdc13-1 rif1Δ cells already arrest in the cell cycle in the presence of only 2% ssDNA (at 25°C).
If Rif1 sets the threshold for the DDR, then overexpression of the protein might elevate the threshold: indeed, cdc13-1 cells overexpressing Rif1 were able to grow at 29°C, an effect similar to the one obtained by deleting checkpoint components such as RAD24RAD17 and RAD17RAD9 [8]. Thus, Rif1 over-expression has the same effect as a checkpoint knockout, abrogating cell cycle arrest. Moreover, increasing expression of Rif1 in cdc13-1 cells already arrested at the restrictive temperature allowed them to exit the cell cycle arrest, demonstrating that Rif1 can out-compete the checkpoint proteins already present at the telomeres and extinguish an ongoing checkpoint response. Interestingly, this effect was telomere specific, as no anti-checkpoint effect could be seen associated with non-telomeric-induced DSBs.
Some time ago Weinert and colleagues [9] showed that the presence of a telomeric tract close to an artificial DSB gradually turned off the DDR elicited by the DSB. The molecular nature of this anti-checkpoint effect was not clear at the time, but the Rif1 protein seems to fit all the requirements for such an anti-checkpoint factor: it is specific for telomeres, acts in cis, and does not affect the resection or the repair of the broken ends.
The identification of Rif1 as an anti-checkpoint factor is a huge step forward; however, many questions remain: If Rif1 activity is independent of Rap1, what is the mechanism of its recruitment? Unlike its vertebrate ortholog, the yeast Rif1 lacks a C-terminal DNA-binding domain [10]. Does yeast Rif1 require an additional factor for binding? Does it move with the DNA-resection machinery by being somehow linked to it? Interestingly, the vertebrate Rif1 protein was shown to interact with DNA and with the BLM protein (the ortholog of yeast’s Sgs1) [10]. An intriguing hypothesis is that Rif1 may be bound to Rap1 at normal telomeres; when telomeres become uncapped, the resection machinery may advance along the chromosome, dislodging Rap1 and concomitantly recruiting Rif1. What then is the role of Rif2? Genetic analysis has shown that its role is independent of Rif1 in determining telomere length [4]. Finally, what is the mechanism by which Rif1 can turn off an ongoing checkpoint response? An attractive idea proposed by Xue et al. [2] is that Rif1 may help recruit phosphatases to de-phosphorylate the central checkpoint kinases.
Interestingly, mammalian Rif1 was thought to function differently from yeast Rif1, as it can be found at non-telomeric locations and does not co-localize with Rap1 at normal telomeres [11]. The data presented here, however, suggest that Rif1 activity in yeast is independent of Rap1 and that yeast and mammalian proteins may share more features than originally thought. Remarkably, Rif1 expression is elevated in human breast tumors, and its expression status is also positively correlated with differentiation degrees of invasive ductal carcinoma of the breast [12]. If the anti-checkpoint role of Rif1 is conserved in mammalian cells, the increased levels of Rif1 may artificially increase the threshold for ssDNA recognition, allowing cells to continue their proliferation in the presence of unrepaired DNA damage without eliciting the DDR.
Fig. 1. Rif1 works as an anti-checkpoint protein. 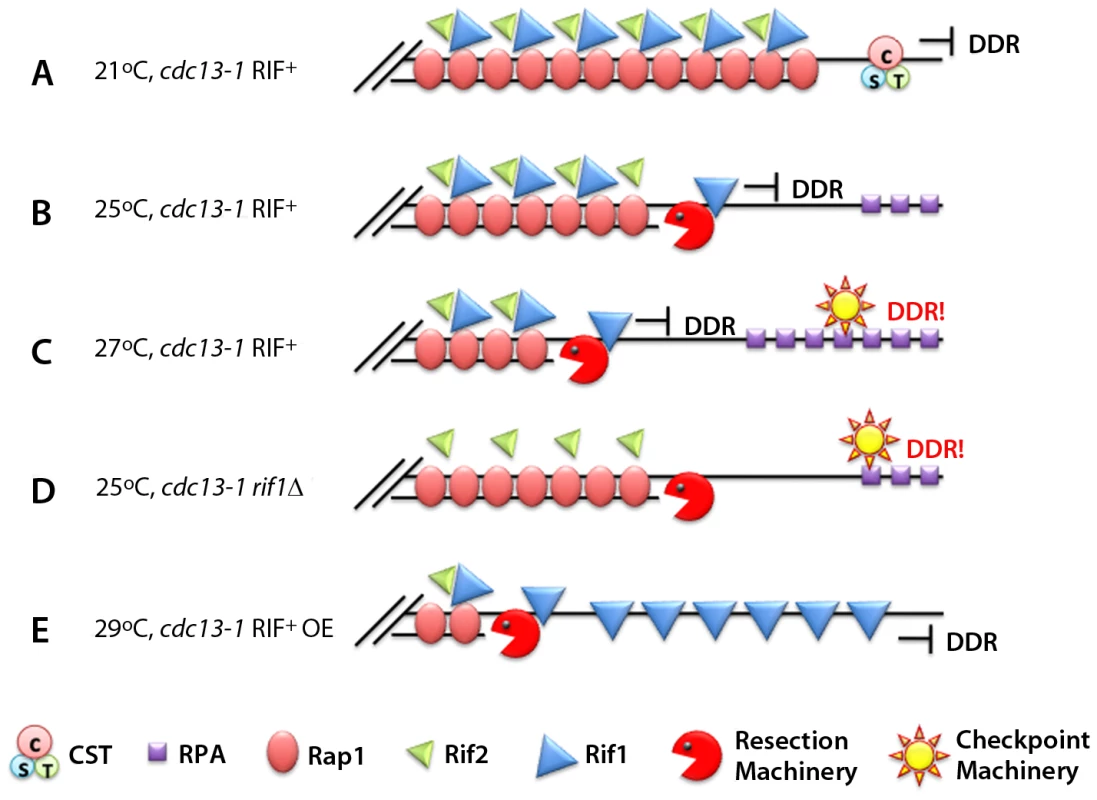
(A) At 21°C the CST complex in a cdc13-1 RIF1+ strain is still functional and “caps” the telomeres, preventing the DNA damage response (DDR). (B) At 25°C the CST is not entirely functional. The resection machinery (Sgs1, Exo1, etc.) creates ssDNA. The presence of Rif1 prevents DDR activation. (C) At 27°C the CST becomes non-functional, and the amount of Rif1 available cannot prevent binding of RPA and additional checkpoint proteins. (D) In the absence of Rif1, the checkpoint is elicited even at 25°C. (E) Over-expressing Rif1 allows the cells to grow at 29°C without eliciting the DDR.
Zdroje
1. WarmerdamDOKanaarR 2010 Dealing with DNA damage: relationships between checkpoint and repair pathways. Mutat Res 704 2 11
2. XueYRushtonMDMaringeleL 2011 A novel checkpoint and RPA inhibitory pathway regulated by Rif1. PLoS Genet 7 e1002417 doi:10.1371/journal.pgen.1002417
3. FrankCJHydeMGreiderCW 2006 Regulation of telomere elongation by the cyclin-dependent kinase CDK1. Mol Cell 24 423 432
4. WottonDShoreD 1997 A novel Rap1p-interacting factor, Rif2p, cooperates with Rif1p to regulate telomere length in Saccharomyces cerevisiae. Gene Dev 11 748 760
5. BournsBDAlexanderMKSmithAMZakianVA 1998 Sir proteins, Rif proteins, and Cdc13p bind Saccharomyces telomeres in vivo. Mol Cell Biol 18 5600 5608
6. AnbalaganSBonettiDLucchiniGLongheseMP 2011 Rif1 supports the function of the CST complex in yeast telomere capping. PLoS Genet 7 e1002024 doi:10.1371/journal.pgen.1002024
7. BonettiDMartinaMClericiMLucchiniGLongheseMP 2009 Multiple pathways regulate 3' overhang generation at S. cerevisiae telomeres. Mol Cell 35 70 81
8. MaringeleLLydallD 2004 Telomerase - and recombination-independent immortalization of budding yeast. Gene Dev 18 2663 2675
9. MichelsonRJRosensteinSWeinertT 2005 A telomeric repeat sequence adjacent to a DNA double-stranded break produces an anticheckpoint. Gene Dev 19 2546 2559
10. XuDMuniandyPLeoEYinJThangavelS 2010 Rif1 provides a new DNA-binding interface for the Bloom syndrome complex to maintain normal replication. EMBO J 29 3140 3155
11. SilvermanJTakaiHBuonomoSBEisenhaberFde LangeT 2004 Human Rif1, ortholog of a yeast telomeric protein, is regulated by ATM and 53BP1 and functions in the S-phase checkpoint. Gene Dev 18 2108 2119
12. WangHZhaoAChenLZhongXLiaoJ 2009 Human RIF1 encodes an anti-apoptotic factor required for DNA repair. Carcinogenesis 30 1314 1319
Štítky
Genetika Reprodukční medicína
Článek A Complex Genomic Rearrangement Involving the Locus Causes Dermal Hyperpigmentation in the ChickenČlánek Genome Instability and Transcription Elongation Impairment in Human Cells Depleted of THO/TREXČlánek A Population Genetics-Phylogenetics Approach to Inferring Natural Selection in Coding SequencesČlánek Interspecific Sex in Grass Smuts and the Genetic Diversity of Their Pheromone-Receptor SystemČlánek Genomic Distribution and Inter-Sample Variation of Non-CpG Methylation across Human Cell Types
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2011 Číslo 12- Akutní intermitentní porfyrie
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Růst a vývoj dětí narozených pomocí IVF
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
-
Všechny články tohoto čísla
- The Connection between Space and Thinking: An Interview with Rafael Viñoly
- An Assessment of the Individual and Collective Effects of Variants on Height Using Twins and a Developmentally Informative Study Design
- Widespread Cotranslational Formation of Protein Complexes
- Genomes Reveal Transition of Bacteria from Aquatic to Terrestrial Environments
- A Complex Genomic Rearrangement Involving the Locus Causes Dermal Hyperpigmentation in the Chicken
- Plasticity of BRCA2 Function in Homologous Recombination: Genetic Interactions of the PALB2 and DNA Binding Domains
- Transcription Is Required to Establish Maternal Imprinting at the Prader-Willi Syndrome and Angelman Syndrome Locus
- Substitutions in the Amino-Terminal Tail of Neurospora Histone H3 Have Varied Effects on DNA Methylation
- MAPK/ERK Signaling Regulates Insulin Sensitivity to Control Glucose Metabolism in
- A Comprehensive Analysis of Shared Loci between Systemic Lupus Erythematosus (SLE) and Sixteen Autoimmune Diseases Reveals Limited Genetic Overlap
- Genome Instability and Transcription Elongation Impairment in Human Cells Depleted of THO/TREX
- Genome-Wide Meta-Analysis of Five Asian Cohorts Identifies as a Susceptibility Locus for Corneal Astigmatism
- A Population Genetics-Phylogenetics Approach to Inferring Natural Selection in Coding Sequences
- HIF-1 Regulates Iron Homeostasis in by Activation and Inhibition of Genes Involved in Iron Uptake and Storage
- Ror2 Enhances Polarity and Directional Migration of Primordial Germ Cells
- DNA Methylation of the Gonadal Aromatase () Promoter Is Involved in Temperature-Dependent Sex Ratio Shifts in the European Sea Bass
- A Genetic Screening Strategy Identifies Novel Regulators of the Proteostasis Network
- Interspecific Sex in Grass Smuts and the Genetic Diversity of Their Pheromone-Receptor System
- The Synthetic Multivulva Genes Prevent Ras Pathway Activation by Tightly Repressing Global Ectopic Expression of EGF
- Mining the Allelic Spectrum Reveals the Contribution of Rare and Common Regulatory Variants to HDL Cholesterol
- Identification of a Genomic Reservoir for New Genes in Primate Genomes
- Genomic Distribution and Inter-Sample Variation of Non-CpG Methylation across Human Cell Types
- Identification of Evolutionarily Conserved Exons as Regulated Targets for the Splicing Activator Tra2β in Development
- Acute Multiple Organ Failure in Adult Mice Deleted for the Developmental Regulator Wt1
- Age-Related Neuronal Degeneration: Complementary Roles of Nucleotide Excision Repair and Transcription-Coupled Repair in Preventing Neuropathology
- Target Site Recognition by a Diversity-Generating Retroelement
- Ancestral Components of Admixed Genomes in a Mexican Cohort
- Targeted Proteolysis of Plectin Isoform 1a Accounts for Hemidesmosome Dysfunction in Mice Mimicking the Dominant Skin Blistering Disease EBS-Ogna
- Autosomal Recessive Dilated Cardiomyopathy due to Mutations Results from Abnormal Dystroglycan O-Mannosylation
- SREBP Coordinates Iron and Ergosterol Homeostasis to Mediate Triazole Drug and Hypoxia Responses in the Human Fungal Pathogen
- The RNA Silencing Enzyme RNA Polymerase V Is Required for Plant Immunity
- An Anti-Checkpoint Activity for Rif1
- The FGFR4-G388R Polymorphism Promotes Mitochondrial STAT3 Serine Phosphorylation to Facilitate Pituitary Growth Hormone Cell Tumorigenesis
- Common Variants Show Predicted Polygenic Effects on Height in the Tails of the Distribution, Except in Extremely Short Individuals
- The Fission Yeast Stress-Responsive MAPK Pathway Promotes Meiosis via the Phosphorylation of Pol II CTD in Response to Environmental and Feedback Cues
- Integrating Genome-Wide Genetic Variations and Monocyte Expression Data Reveals -Regulated Gene Modules in Humans
- Repetitive Elements May Comprise Over Two-Thirds of the Human Genome
- A Novel Checkpoint and RPA Inhibitory Pathway Regulated by Rif1
- Hierarchical Generalized Linear Models for Multiple Groups of Rare and Common Variants: Jointly Estimating Group and Individual-Variant Effects
- The Major Roles of DNA Polymerases Epsilon and Delta at the Eukaryotic Replication Fork Are Evolutionarily Conserved
- A High-Resolution Whole-Genome Map of Key Chromatin Modifications in the Adult
- A Densely Interconnected Genome-Wide Network of MicroRNAs and Oncogenic Pathways Revealed Using Gene Expression Signatures
- A Functional Phylogenomic View of the Seed Plants
- Histone H3K9 Trimethylase Eggless Controls Germline Stem Cell Maintenance and Differentiation
- Ribosomal Protein Mutants Control Tissue Growth Non-Autonomously via Effects on the Prothoracic Gland and Ecdysone
- , , and Are Required to Activate or Delimit the Spread of the Transcriptional Response to Epidermal Wounds in
- Mechanisms Establishing TLR4-Responsive Activation States of Inflammatory Response Genes
- Candidate Gene Screen in the Red Flour Beetle Reveals as Ancient Regulator of Anterior Median Head and Central Complex Development
- Charcot-Marie-Tooth–Linked Mutant GARS Is Toxic to Peripheral Neurons Independent of Wild-Type GARS Levels
- The RNA–Methyltransferase Misu (NSun2) Poises Epidermal Stem Cells to Differentiate
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Targeted Proteolysis of Plectin Isoform 1a Accounts for Hemidesmosome Dysfunction in Mice Mimicking the Dominant Skin Blistering Disease EBS-Ogna
- The RNA Silencing Enzyme RNA Polymerase V Is Required for Plant Immunity
- The FGFR4-G388R Polymorphism Promotes Mitochondrial STAT3 Serine Phosphorylation to Facilitate Pituitary Growth Hormone Cell Tumorigenesis
- Target Site Recognition by a Diversity-Generating Retroelement
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



