-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Heritable Epigenetic Variation among Maize Inbreds
Epigenetic variation describes heritable differences that are not attributable to changes in DNA sequence. There is the potential for pure epigenetic variation that occurs in the absence of any genetic change or for more complex situations that involve both genetic and epigenetic differences. Methylation of cytosine residues provides one mechanism for the inheritance of epigenetic information. A genome-wide profiling of DNA methylation in two different genotypes of Zea mays (ssp. mays), an organism with a complex genome of interspersed genes and repetitive elements, allowed the identification and characterization of examples of natural epigenetic variation. The distribution of DNA methylation was profiled using immunoprecipitation of methylated DNA followed by hybridization to a high-density tiling microarray. The comparison of the DNA methylation levels in the two genotypes, B73 and Mo17, allowed for the identification of approximately 700 differentially methylated regions (DMRs). Several of these DMRs occur in genomic regions that are apparently identical by descent in B73 and Mo17 suggesting that they may be examples of pure epigenetic variation. The methylation levels of the DMRs were further studied in a panel of near-isogenic lines to evaluate the stable inheritance of the methylation levels and to assess the contribution of cis - and trans - acting information to natural epigenetic variation. The majority of DMRs that occur in genomic regions without genetic variation are controlled by cis-acting differences and exhibit relatively stable inheritance. This study provides evidence for naturally occurring epigenetic variation in maize, including examples of pure epigenetic variation that is not conditioned by genetic differences. The epigenetic differences are variable within maize populations and exhibit relatively stable trans-generational inheritance. The detected examples of epigenetic variation, including some without tightly linked genetic variation, may contribute to complex trait variation.
Published in the journal: . PLoS Genet 7(11): e32767. doi:10.1371/journal.pgen.1002372
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002372Summary
Epigenetic variation describes heritable differences that are not attributable to changes in DNA sequence. There is the potential for pure epigenetic variation that occurs in the absence of any genetic change or for more complex situations that involve both genetic and epigenetic differences. Methylation of cytosine residues provides one mechanism for the inheritance of epigenetic information. A genome-wide profiling of DNA methylation in two different genotypes of Zea mays (ssp. mays), an organism with a complex genome of interspersed genes and repetitive elements, allowed the identification and characterization of examples of natural epigenetic variation. The distribution of DNA methylation was profiled using immunoprecipitation of methylated DNA followed by hybridization to a high-density tiling microarray. The comparison of the DNA methylation levels in the two genotypes, B73 and Mo17, allowed for the identification of approximately 700 differentially methylated regions (DMRs). Several of these DMRs occur in genomic regions that are apparently identical by descent in B73 and Mo17 suggesting that they may be examples of pure epigenetic variation. The methylation levels of the DMRs were further studied in a panel of near-isogenic lines to evaluate the stable inheritance of the methylation levels and to assess the contribution of cis - and trans - acting information to natural epigenetic variation. The majority of DMRs that occur in genomic regions without genetic variation are controlled by cis-acting differences and exhibit relatively stable inheritance. This study provides evidence for naturally occurring epigenetic variation in maize, including examples of pure epigenetic variation that is not conditioned by genetic differences. The epigenetic differences are variable within maize populations and exhibit relatively stable trans-generational inheritance. The detected examples of epigenetic variation, including some without tightly linked genetic variation, may contribute to complex trait variation.
Introduction
Much of the heritable variation within a species is a consequence of differences in the primary DNA sequence of different individuals. However, there is growing evidence for heritable variation in the absence of DNA sequence polymorphisms, termed epigenetic variation [1]. Cytosine methylation is one of the molecular mechanisms that can contribute to epigenetic variation and often acts to suppress the activity of transposable elements, repetitive sequences, pseudogenes, and in some cases otherwise active genes [2], [3]. There is evidence that epigenetic changes can lead to stable phenotypic variation in plant and animal species [4]–[10]. However, the abundance and role of epigenetic, relative to genetic, variation has not been well characterized. Maize (Zea mays) provides a useful model to study the role of epigenetic variation. Genetically, maize is a highly diverse species [11], [12] with a large, complex genome with many interspersed genic and repetitive regions [13], [14]. While in the past this complex genomic structure has complicated the ability to perform genome-wide analyses it also is likely to contribute to higher levels of epigenetic variation relative to less complex genomes such as Arabidopsis [3], [15]. In addition, there are outstanding resources for the analysis of quantitative trait variation in maize [16], [17] that may allow for a better understanding of the relative roles of genetic and epigenetic variation in controlling quantitative trait variation.
In plants, the majority of genome-wide methylation studies have been conducted in Arabidopsis [18]–[20], [8]. In these studies DNA methylation was frequently associated with heterochromatic regions, transposable elements, and repetitive DNA [18]. In general, lower levels of methylation occur within gene promoter sequences; however when present, promoter methylation shows a negative correlation with gene expression [19]. Within gene bodies, regions of DNA methylation have been observed uniquely in the CG context, but no major impact on gene expression is associated with this modification [20]. The exact role of gene body methylation is unclear, but it may preferentially affect moderately-transcribed genes [19], and be under different regulatory control than that of transposable element methylation [21]. Similar genome-wide patterns of DNA methylation have also been observed in rice and poplar [22]. A recent analysis of DNA methylation in maize used a 0.3× coverage sequencing of McrBC digested DNA to show evidence for mutually exclusive patterns of DNA methylation and H3K27me3 near genes with low, or no expression [23].
DNA methylation has been proposed to play a role in generating variation that could provide adaptation to environmental stresses [5], [24]–[27]. Two groups have recently developed epiRIL populations in which epigenetic states were altered by passage through DNA methylation mutants [28]–[31]]. The existence of quantitative trait variation in these populations suggests that alteration of DNA methylation patterns can induce phenotypic change although it is difficult to rule out the potential for primary sequence changes due to activated transposition. These studies have been very useful for documenting an important role for DNA methylation in regulating complex traits but do not provide information on natural variation for epigenetics states. There is evidence that DNA methylation patterns at specific loci can vary within Arabidopsis ecotypes [8], [32]–[35] and there are several specific examples of epigenetic variation that result in phenotypic variation in a variety of species [36]–[44]. However, there are limited analyses of genome-wide methylation variation conducted in plant species. A detailed contrast of chromosome 4 methylation patterns in Arabidopsis thaliana ecotypes shows very similar targeting of transposable elements and repetitive sequences, yet genic partial methylation states were highly polymorphic across ecotypes [8]. A study in maize found evidence for variable effects of CHG methylation on transcription patterns in different inbreds of maize [45].
Despite the evidence for variation in DNA methylation patterns among individuals of the same species relatively little is known about the nature of the inheritance of these methylation differences. One study in Arabidopsis found that gene body methylation was only partially heritable and was lost at a relatively high frequency [8]. Richards [5] provided a description of how methylation variation may be dependent upon, conditioned by, or independent of DNA sequence change and termed these as examples of obligatory, facilitated or pure epialleles, respectively. Obligatory epialleles exhibit different levels of DNA methylation but are entirely dependent upon DNA sequence changes at linked or unlinked sites. In contrast, facilitated and pure epialleles exhibit stochastic variation that can be conditioned with or without genetic differences, respectively. In addition, there is evidence that genetic variation at unlinked genomic regions can direct epigenetic changes elsewhere in the genome, presumably through an RNA-direct DNA methylation (RdDM) pathway [36], [46], [47].
We sought to characterize the variation in DNA methylation patterns in two maize inbred genotypes, B73 and Mo17. Genome-wide profiling of DNA methylation patterns was used to assess the relationship of methylation to chromosomal and gene structures. Although the majority of the genome shows highly similar methylation patterns in both inbreds there are several hundred differentially methylation regions (DMRs) found throughout the maize genome. The analyses of several identical-by-descent regions of the B73 and Mo17 genomes provides evidence that epigenetic variation can occur in the absence of nearby genetic polymorphisms. A population of near-isogenic lines was used to further characterize the heritable behavior of the DMRs and to assess the genomic regions that controlled the differential methylation.
Results
An array platform containing 2.1 million long oligonucleotide probes was designed to profile genomic DNA methylation patterns in low-copy sequences throughout the maize genome (Table S1; Methods). Methylated DNA immunoprecipitation (meDIP) was performed on fragmented genomic DNA using a 5-methylcytosine antibody. This approach is very useful for providing cost-effective quantitative estimates of DNA methylation density [19]. This method can detect substantial differences in the proportion of methylated cytosines in genomic regions but cannot accurately assess individual bases or differentiate the different types of DNA methylation such as CG, CHG, CHH. The enrichment of methylated DNA was confirmed (Figure S1A) by assessing the enrichment for a region known to be methylated and lack of enrichment for a region known to lack significant DNA methylation [48]. meDIP was performed on three biological replicates of leaf blade tissue isolated from the third expanded leaf of greenhouse-grown B73 and Mo17 seedlings; the resulting enriched fractions were labeled and hybridized to the array along with un-enriched control input DNA. Linear model ANOVA was used to estimate values for input DNA, B73 methylation, Mo17 methylation and relative methylation in B73 and Mo17 (Figure 1).
Fig. 1. Synopsis of chromosome-level methylation and variation. 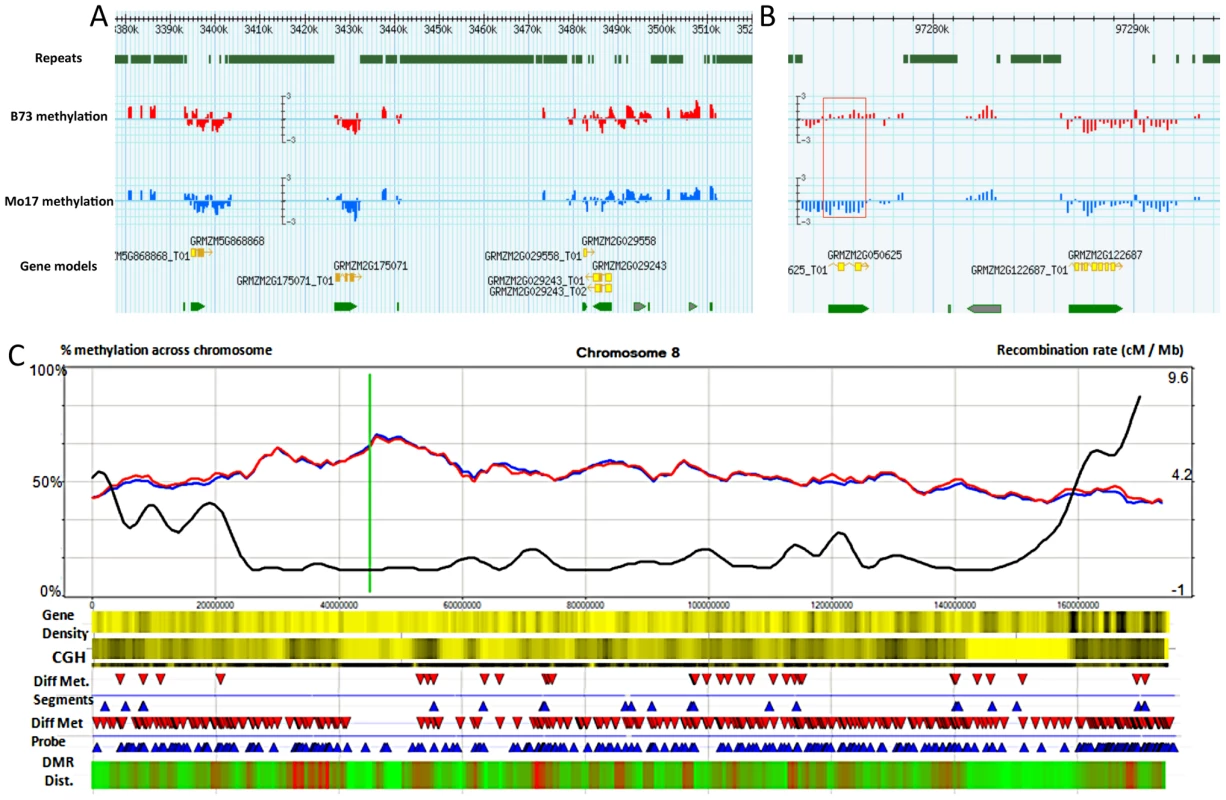
(A–B) A Gbrowse view is presented for 140 kb of chromosome 8 (3,380 kb–3,520 kb) and a closer view of 22 kb (97,273–97,294K) that includes a differentially methylated region (red box). The top track shows the regions that are annotated as repetitive sequences using the MIPS/Recat repeat catalog. The next two tracks show the B73 (red) and Mo17 (blue) relative methylation levels for each of the probes within these regions. Methylation levels are defined as the normalized log2 ratio of IP enriched sample to un-enriched genomic DNA on a scale of −3 to 3 indicating methylation enrichment (>0) and depletion (<0) respectively. The bottom tracks illustrate the gene models. The gray arrows indicate gene models that were rejected from the FGS. (C) Shows a chromosomal view of methylation levels in B73 and Mo17. The percentage of methylation is plotted as a 5 Mb window sliding 1 Mb downstream across the chromosome Blue and red lines indicate percent methylation of all probes for B73 and Mo17 respectively. The green line indicates centromere position. The black line shows the cm/Mb across the chromosome. The first heatmap provides a visualization of gene density. Yellow and black values indicate lower and higher relative gene density values respectively. The second heatmap provides visualization for the genomic structural variation between B73 and Mo17 using Comparative Genomic Hybridization (CGH) values. A 5 Mb sliding window across the chromosome indicates regions of high diversity (black) to low diversity (yellow). The differential methylation regions (DMRs) are shown using red (Mo17 hypermethylation) and blue (B73 hypermethylation) arrows. The next track shows the location of individual probes that have significant (q<0.001) methylation variation between B73 and Mo17. The final heat map indicates the relative enrichment for differentially methylation probes across the chromosome with enriched regions indicated by red and regions with depleted levels of methylation variation in green. The probe sequences were designed based on the sequence of the B73 reference genome, but previous studies have documented abundant DNA sequence polymorphisms [49] and structural variants between B73 and Mo17 [50], [51]. We investigated the methylation levels at sequences that exhibit structural genomic variation such as copy number variation (CNV) and presence-absence variation (PAV). Comparative genomic hybridization (CGH) data were obtained from the hybridization of input B73 and Mo17 genomic DNA. DNAcopy [52] was performed, followed by expectation maximization [53] model analysis to identify segments that exhibit significantly more copies in Mo17 than in B73 (M>B CNV) and to identify segments that exhibit significant fewer or no copies in Mo17 relative to B73 (M<B CNV and PAV) (Table S2). While there are examples of CNV or PAV that show high levels of methylation, there is little evidence for substantial differences in the overall methylation levels of sequences that exhibit structural genome variation relative to sequences that do not show structural variation (Figure S2, Figure S3A–S3C). Even so, for subsequent analyses, we focused on regions that do not have any evidence for CNV/PAV based on the CGH data. The values from the remaining probes were adjusted using the B73-input vs. Mo17-input hybridization coefficient to control for differential hybridization efficiency while still estimating methylation differences.
Microarray probes were selected to target low-copy regions of the maize genome by using repeat-masked sequences (provided by J. Stein and D. Ware). This repeat masking does not, however, remove all multi-copy sequences. The number of exact (100% identity and coverage) or close matches (>90% identity and coverage) was determined for each probe (Table S3). Slightly over half (58%) of the probes present on the array have only a single perfect match in the B73 reference genome and no other close matches. As the numbers of perfect or close matches for probes increase there is a significant increase in the levels of methylation they detect (Figure S3D–S3E). This copy-number dependent increase in methylation levels is observed in both B73 and Mo17 (data not shown). The subsequent genome-wide analyses of DNA methylation are confined to the subset of probes that are present as a single copy within the B73 genome (Table 1). The genome-wide analysis of methylation levels in Mo17 is further restricted to those probes that do not exhibit evidence for substantial differences in CGH values in the two inbreds (Table 1). By focusing on these subsets of probes the effects of probe copy number and genomic polymorphism on the detected methylation levels are minimized.
Tab. 1. Methylation levels in subsets of probes. 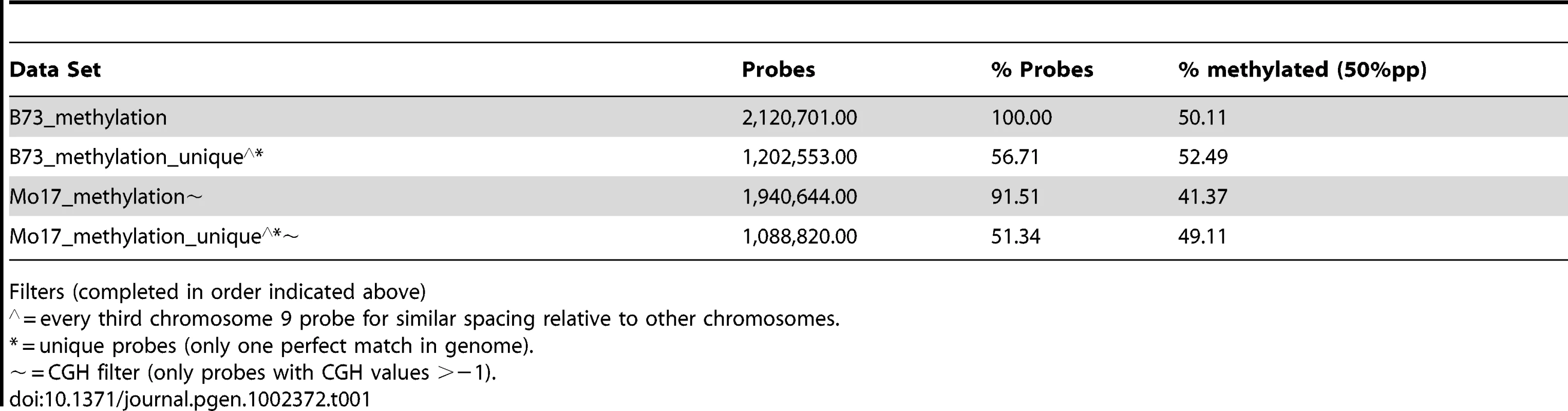
Filters (completed in order indicated above) The distribution of per-probe methylation estimates provides evidence for a bi-modal distribution (Figure S1B) with the two distributions accounting for methylated and un-methylated genomic regions. Application of expectation maximization allows classification of the methylation status of each probe (Table 1). The genomic distribution of DNA methylation patterns was visualized across each of the maize chromosomes (Figure 1C and Figure S4). Similar to other species, methylation levels are higher in the pericentromic regions of maize chromosomes than at the ends of chromosomal arms. There are several regions of higher methylation throughout the chromosome that do not correlate with the centromeric position and do not correlate well with cytologically visible features such as knobs or rDNA sites. In general, the relative levels of DNA methylation are inversely correlated with gene density. The relative levels of DNA methylation in parental lines also show a negative correlation with recombination rates measured in a set of intermated B73×Mo17 RILs [54]. However, the exact parents for this population may have slight differences relative to the B73 and Mo17 profiled in this study and we have not measured actual DNA methylation profiles in any specific RIL genotype.
Comparative genomic analysis of DNA methylation dynamics for maize genes
The location of each probe was determined relative to the gene models of annotation 5a.59 (www.maizesequence.org). Version 5a.59 of the maize working gene set contains 104,369 annotated genes which include 39,384 genes models that are part of the high-confidence filtered gene set (FGS) and another 64,985 genes that were rejected from the FGS. In both B73 and Mo17 the FGS genes show substantially lower methylation levels within and surrounding the genes relative to the rejected genes (Figure 2A). The reasons for rejecting genes from the FGS include low confidence FGENESH models, probable transposons and probable pseudogenes. Rejected genes that fall into these classes exhibit significantly higher methylation than the genes in the FGS (Figure S5). The methylation pattern for FGS genes has reduced methylation in the 300 bp upstream of the transcription start site (TSS) then has a short “peak” of methylation in the very beginning of the gene which drops off quickly in the 3′ direction. There is a region of low methylation at the 3′ ends of FGS genes, but methylation returns to genome-wide average levels within 500 bp of the transcription termination site. This distribution of methylation levels, particularly the increased methylation at the 5′ ends of genes, is distinct from patterns observed in other species [55], [22] but is consistent with a previous report from maize [23]. The methylation pattern observed across the gene body is related to the distribution of CpG dinucleotides (Figure S6A–S6B). However, the analysis of the region immediately upstream of the transcription site reveals that this region with increased CpG content does not show increased DNA methylation levels which confirms hypomethylation of these promoter regions and provides evidence that observed methylation levels are not strictly driven by CpG content (Figure S6C). In addition to the dynamics of methylation along the length of the genes, there are also significant differences in the methylation levels of exons, introns and UTRs relative to intergenic probes (Figure S6D). Introns show relatively low methylation levels throughout the gene body while exon sequences exhibit relatively high methylation in the 5′ end and low methylation in the 3′ end of the gene (Figure S6E). Many of these differences reflect the relatively high CpG content of the first exons of maize genes.
Fig. 2. Gene body methylation and expression levels in maize genes. 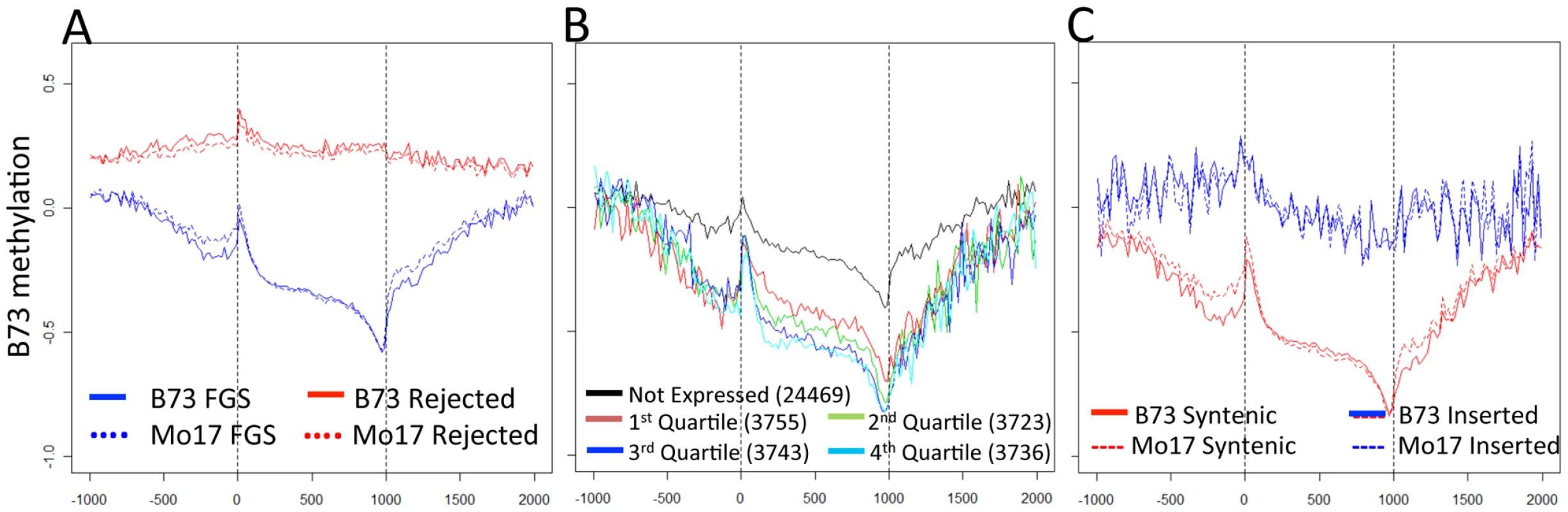
(A) The relative methylation levels (log2(IP/input)) were assessed for probes within 1000 bp of the transcription start and termination site. The relative position for probes within the gene was normalized to a scale of 1000. The genes in the filtered gene set (FGS) exhibit a lower methylation and a more dynamic pattern across the length of the gene than the rejected genes. The vertical dashed lines indicated the beginning and end of transcription for each gene. (B) The relative expression level for all FGS genes was assessed using published RNAseq data from B73 leaf tissue [56] and genes were assigned as not expressed or quartile 1–4 based on their expression level. In general, the genes show similar patterns of methylation but the higher expressed genes exhibit lower levels of methylation within and around the gene. (C) Each of the FGS genes was also classified according to whether it was located in a syntenic position relative to sorghum and/or rice or in a non-syntenic position. The syntenic genes exhibit much lower levels of methylation than the non-syntenic genes. This difference between syntenic and non-syntenic genes can also be seen in the regions immediately surrounding the gene. To determine the relationship between DNA methylation and gene expression, the relative expression levels of FGS genes in B73 leaf tissue were used to divide genes into five categories: non-expressed; and four quartiles based on RNAseq data from Li et al. [56]. As expected, highly expressed genes show the lowest levels of methylation. There are significant differences in DNA methylation values among all quartiles of genes except between the two quartiles containing highly expressed genes (Figure S7A). Genes that are not expressed have higher levels of methylation in nearby regions as well as within the gene body (Figure 2B). We proceeded to assess methylation levels of FGS genes in a comparative genomics context. Schnable et al. [57] used comparative genomic approaches to identify homoeologous regions of the maize genome derived from a whole genome duplication event and to then assign them to sub-genome 1 and sub-genome 2 based on the level of fractionation observed. Sub-genome 1 has retained a larger proportion of the ancestral genes and generally exhibits higher mRNA expression levels as compared to sub-genome 2. Despite the trend for lower expression levels for genes in sub-genome 2 [57], there was no evidence for differences in methylation levels in genes present in sub-genome 1 relative to sub-genome 2 (Figure S7B). However, there was evidence for substantial differences in the methylation levels of genes in the FGS that are in syntenic positions relative to sorghum and/or rice relative to FGS genes that are located in non-syntenic positions (Figure 2C). The non-syntenic genes are enriched (chi-square p value<0.001) for genes that are not expressed or are in the lowest quartile of expressed genes based on the data of Li and coworkers [56].
Variability for B73-Mo17 methylation
A visual analysis of the B73 and Mo17 methylation patterns revealed that while the majority of loci exhibit very similar patterns, there are examples of altered methylation levels between the two genotypes (Figure 1). Two different approaches were used to discover differentially methylated regions (DMRs) in B73 and Mo17. One approach identified individual probes that exhibit significant (q<0.001) differences in the contrast of B73 and Mo17 methylation (Table 2). A second approach implemented the DNAcopy segmentation algorithm on the relative methylation values followed by expectation maximization to identify segments of at least three probes that exhibit altered methylation (Figure 3A; Table 3). The per-probe analysis is capable of identifying small regions with altered methylation whereas the analysis of segments defined by adjacent probes will identify larger, high-confidence DMRs.
Fig. 3. DMRs in B73 and Mo17. 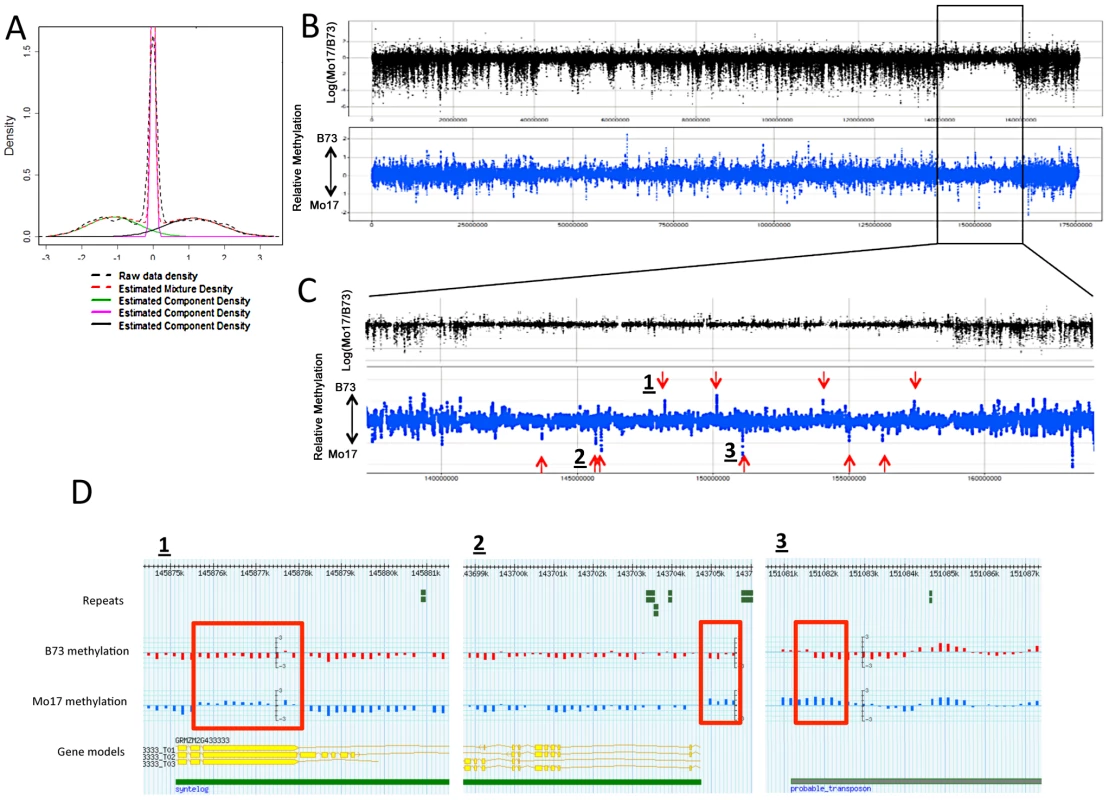
(A) The relative methylation levels for all probes were used to perform DNAcopy segmentation followed by expectation maximization. The black dashed lines show the observed distribution of the segment means. This distribution can be approximated (red dashed line) by a model that is derived from three normal distributions including B73 hypermethylation (right peak), Mo17 hypermethylation (left peak) and unchanged regions (middle peak). (B) The structural variation across chromosome 8 is shown in the plot with black spots. The blue spots show relative methylation in B73 and Mo17. The region of low structural diversity (boxed region) is magnified in (C). (D) Gbrowse views for three DMRs located within this region are shown. The data tracks show the position of repeats, genes, B73 methylation, Mo17 methylation and relative methylation. Each bar showing methylation values represents an individual probe. The actual DMRs are shown by the red boxes. Tab. 2. Probes with variable DNA methylation in B73 and Mo17. 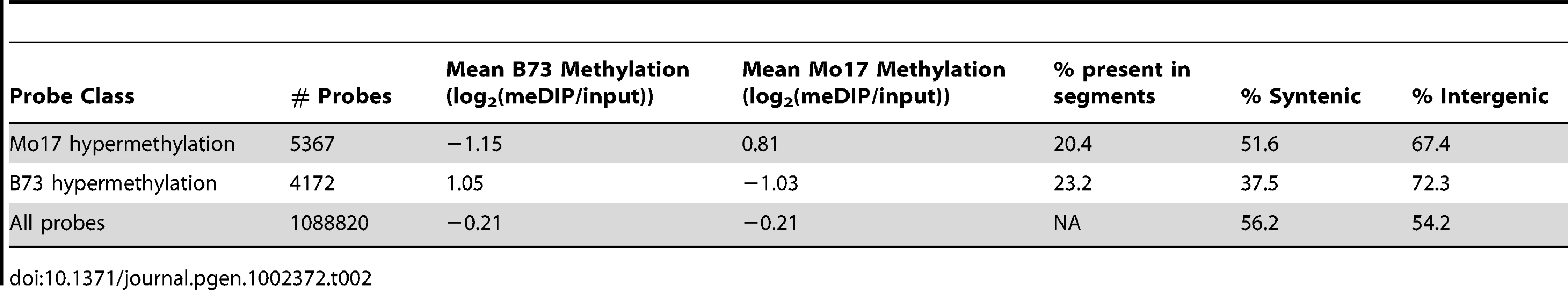
Tab. 3. Segments with variable DNA methylation levels in B73 and Mo17. 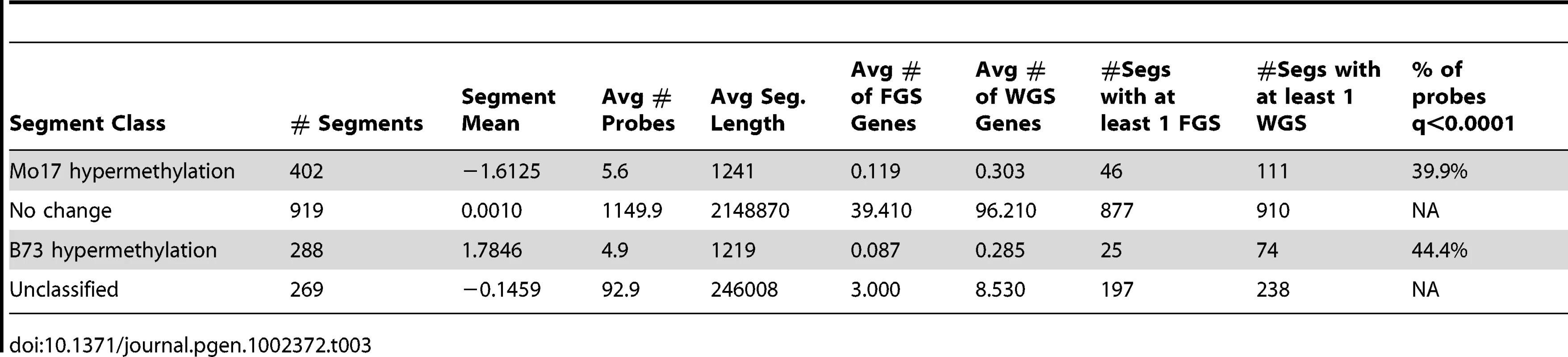
The DMRs identified by segmentation were further characterized as they have evidence for altered methylation from multiple adjacent probes encompassing a region of at least several hundred base pairs. There are nearly 700 DMR segments each that exhibit either B73 or Mo17 hypermethylation (Table 3; full list in table S4). The Mo17 hypermethylation segments include a total of 500 kb of DNA while the B73 hypermethylation segments include a total of 350 kb of DNA. The majority of the DMRs (674/690) are less than 5 kb in length and only one segment is over 10 kb (Figure S8A). The majority of the DMRs occurred in intergenic regions and relatively few even overlap with a FGS gene or a member of the WGS (Table 3). Those genes that were contained within DMRs were enriched for non-syntenic genes, inclusion in sub-genome 1, and for those that are not expressed in leaf tissue (Figure S8B–S8C). A genomic visualization of probes and/or segments of differential methylation (Figure 1; Figure S4) revealed a non-uniform genomic distribution.
The DMRs may be conditioned by local sequence differences in B73 and Mo17 or may be the result in stochastic epigenetic differences that are not directly attributable to genetic differences. We focused on genomic regions of low diversity to identify potential examples of epigenetic differences that are not directly attributable to local sequence changes. As previously reported [50] there are several large seemingly non-polymorphic regions in B73 relative to Mo17. These are likely identical-by-descent (IBD) regions that represent shared inheritance of the same haplotype block in these two different inbred genotypes from a common parent [50]. We analyzed 10 putative IBD regions in the B73-Mo17 genome that are at least 2 Mb in length, have no evidence for structural variation, and have extremely low SNP densities (Table 4). The SNP rates in these regions (1 every 44.2 kb) are below the levels of sequence error rates reported for the B73 reference genome [14]. Despite the near-absence of genetic variation within these regions there are 52 differentially methylated probes and 9 DMRs within these regions. The large low diversity region on chromosome 8 provides several examples of altered methylation levels within a large region that lacks sequence differences (Figure 3B and 3C). We used the Mo17 whole-genome shotgun sequences and targeted PCR to confirm that absence of any InDels within 2 kb of each of the nine DMRs located within the IBD regions. The majority (8/9) DMRs in IBD regions did not have any InDels. Only one of these DMRs in an IBD region exhibit sequence polymorphism. At this DMR there is evidence for a recent insertion of a repetitive element in the B73 allele and B73 is more highly methylated than Mo17.
Tab. 4. IBD regions in the B73-Mo17 genome. 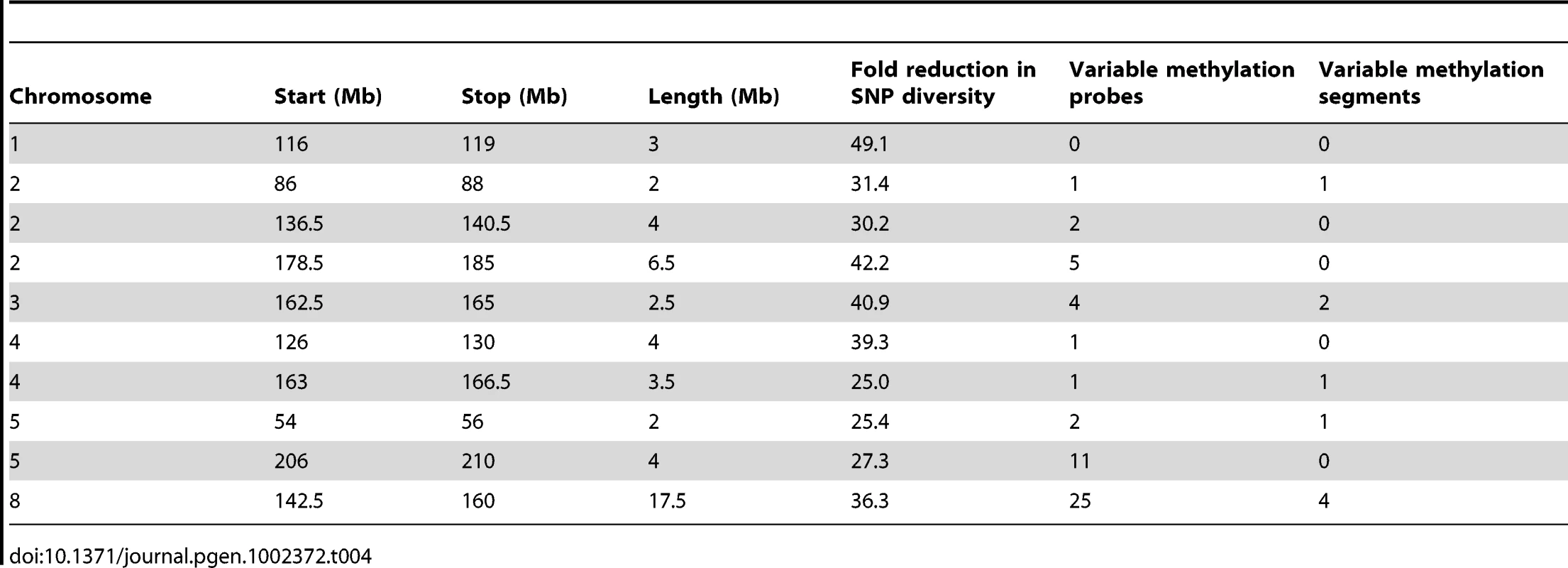
Characterization of inheritance for differential methylation
A set of 33 DMR regions consisting of 14 regions of B73 hypermethylation and 19 regions of Mo17 hypermethylation was selected for further characterization (Table 5). These included eight DMRs that were present in the IBD regions. Quantitative PCR-based assays were developed to assess relative methylation levels following digestion with the methylation dependent enzymes MspJI and FspEI. The methylation differences observed in the full-genome profiling were confirmed for 28/33 of these regions in independent biological samples of B73 and Mo17 DNA (Table 5). The differential methylation was also assessed using the methylation-sensitive enzymes HpaII and/or PstI for ten of these same DMRs (Table S5) including the three that had not been supported by methylation-dependent digests. All DMRs (4/4) that include a PstI (CHG sensitive) site were validated and 8 of the 10 DMRs that had a HpaII site were validated (Table S5). Two of the three regions that were not conclusively validated by the methylation-dependent enzyme digests did exhibit differential methylation when tested with HpaII. The other DMR was not supported by assays with either methylation-dependent or –sensitive enzymes. The classification of differential methylation was also supported by an analysis of read counts from methyl-sensitive and insensitive sequencing libraries from Gore et al [47].
Tab. 5. Characterization of variable methylation segments. 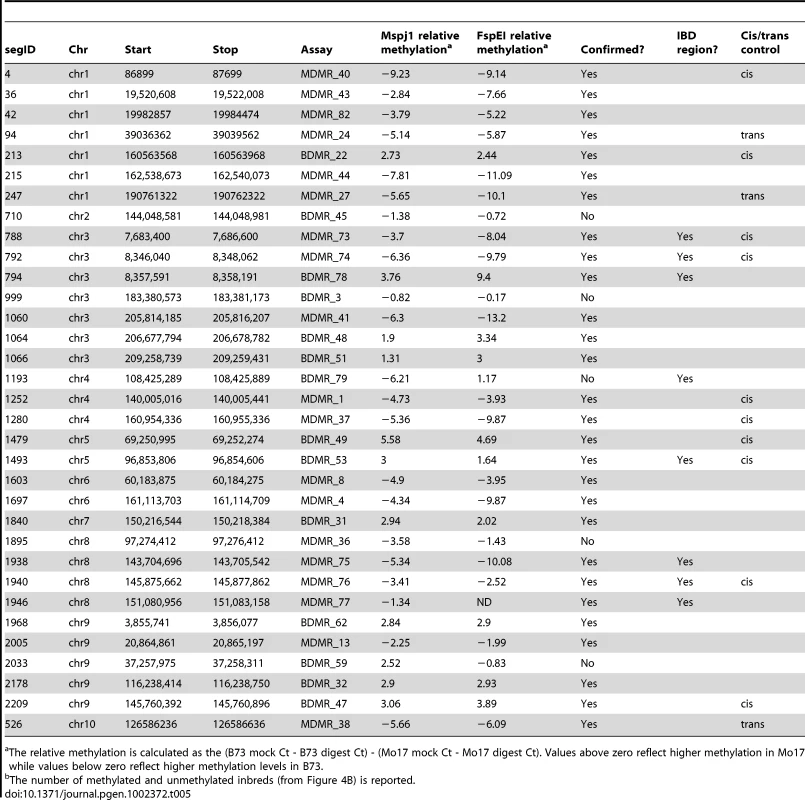
The relative methylation is calculated as the (B73 mock Ct - B73 digest Ct) - (Mo17 mock Ct - Mo17 digest Ct). Values above zero reflect higher methylation in Mo17 while values below zero reflect higher methylation levels in B73. We assessed relative methylation levels for 13 of the DMRs in selected genotypes from a population of near-isogenic lines (NILs) derived from B73 and Mo17 [58]. The levels of methylation in NILs can be used to evaluate the relative contribution of linked and unlinked genomic regions and to test for paramutation-like transfer of information between alleles. The expected results for each of these potential scenarios are shown in Figure 4A. The genotype for the chromosomal region containing the DMR is expected to predict the methylation state in the NIL if the methylation change is purely epigenetic or if linked sequence polymorphisms regulate methylation levels. Alternatively, if unlinked genomic regions are directing the methylation levels at DMRs introgressed into a NIL then the DMR is expected to exhibit methylation levels similar to the recurrent parent. For each of the DMRs we selected several genotypes that provided an introgression of the locus into either a B73 or Mo17 genomic background. In addition, as a control we monitored DNA methylation levels in several NIL genotypes that did not have an introgression at the locus of the DMR. In general, the control assays show a high stability of DNA methylation levels at these DMRs. The analysis of the NILs with introgressions at the DMR loci reveal that 10/13 have methylation levels that can be predicted by the haplotype of the region surrounding the DMR. This could reflect stable inheritance of epigenetic variation or cis-linked genetic changes that are directing the methylation difference. The other three DMRs that were mapped have DNA methylation patterns that are influenced by genomic regions that are unlinked to the DMR locus, suggesting that the methylation levels of these loci are controlled by trans-acting loci. Four (of the 13) DMRs that were mapped are located within the IBD regions and each of these exhibited methylation patterns that were controlled by cis-linked regions despite the absence of closely linked genetic variation within these regions.
Fig. 4. Variable DNA methylation patterns in near-isogenic lines and diverse inbreds. 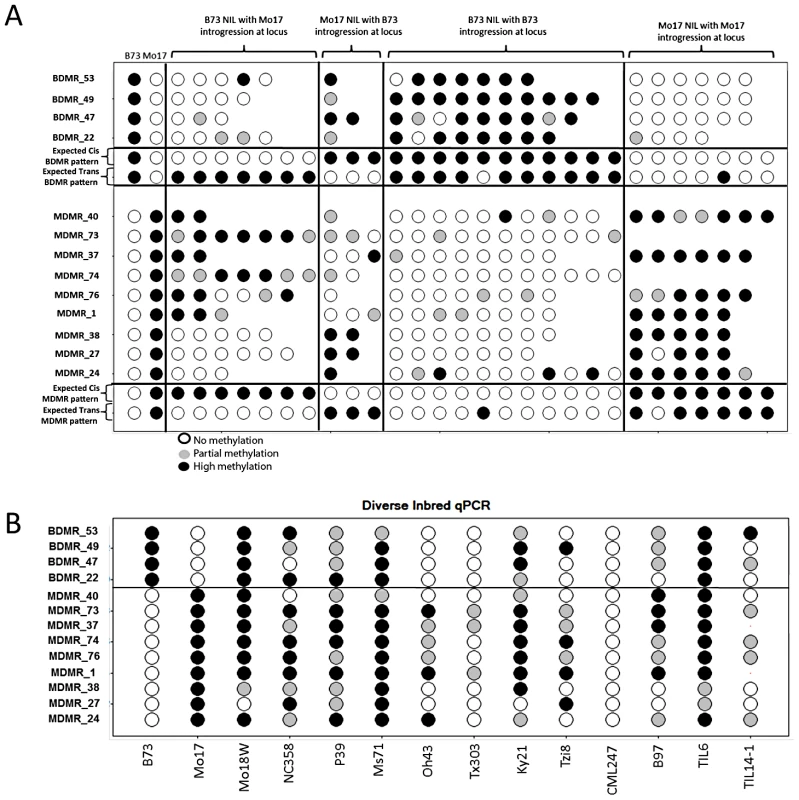
(A) The relative DNA methylation levels in selected near-isogenic lines was tested by digestion with the methylation dependent restriction enzyme MspjI followed by qPCR. Different subsets of NILs were selected and analyzed for each of 13 DMRs. The first two columns show the data from B73 and Mo17. Open circles reflect low methylation levels and black circles indicate high methylation levels. Intermediate methylation levels are indicated by gray color. The next group of 2–7 genotypes show the data from NILs that have B73 as the recurrent parent (>95% of the genome) and have introgression of the Mo17 haplotype in the region containing the DMR. The variable number of genotypes tested reflects the fact that some DMR loci are have more NILs with an introgression than others. The next group of 1–3 genotypes are NILs that are primarily Mo17 but have B73 introgressed at the DMR. The next two groups provide “control” genotypes of B73-like or Mo17-like NILs that do not have an introgression at the DMR. The expected patterns for cis (local) inheritance of DNA methylation or trans (unlinked) control of DNA methylation are shown. Note that the expected pattern for trans control would include a small number of genotypes with the methylation pattern from the introgressed genotype in cases where the trans-acting locus is introgressed. (B) The same type of assays were performed on a panel of 12 diverse inbred genotypes, including two inbred teosinte lines, to monitor the frequency for the hyper- and hypo-methylated states. The relative DNA methylation patterns for these 13 DMRs were also assessed in a panel of 10 other inbred lines of maize and two teosinte inbred lines (TILs) (Figure 4B). Each of these DMRs exhibits at least one other genotype with high or low levels of methylation indicating that the B73-Mo17 states are not unique within maize.
Discussion
Maize has a rich history of serving as a model for epigenetic studies. The first examples of imprinting and paramutation were discovered in maize [59], [60] and there have been a number of pioneering studies on the epigenetic regulation of transposable element in maize [61], [62]. While these discoveries have been enabled by the ease of genetic studies in maize it is also likely that the complex organization of the maize genome with many interspersed transposons and genes has led to numerous examples of epigenetic regulation. In this study we have performed a genome-wide characterization of DNA methylation levels in two inbred lines of maize and found hundreds of loci with differences in DNA methylation levels. This study of natural epigenetic variation also demonstrates the utility of near-isogenic lines for characterizing epialleles.
Methylation dynamics along maize chromosomes and genes
Our data provide evidence for higher levels of DNA methylation in the pericentromeric regions of maize chromosomes. This is quite similar to observations in Arabidopsis [18]–[20]. Although there is a general negative correlation between methylation density and recombination, methylation density does not exactly mirror recombination rates. Recombination happens much more frequently near the ends of the chromosomes and quickly drops to a lower level internally [54]. The DNA methylation patterns exhibit a much more gradual change along the length of the chromosomes, potentially suggesting that the differences in recombination rate along the length of a chromosome are not directly related to DNA methylation levels. In general, the methylation patterns within maize gene bodies are similar to the density of CG sites. However, it is clear that the methylation levels in regions immediately upstream and downstream of gene bodies are not reflective of CG density. Interestingly, while short maize genes have elevated CG content throughout the gene body the longer maize genes have elevated CG content only in the first 500–1000 bp.
We also noted very distinct patterns of DNA methylation in maize genes. In general, methylation is lower in high-confidence genes (i.e., members of the FGS), highly expressed genes and genes in syntenic positions relative to other grass species. Putative genes that were rejected during stringent genome annotation (often partial length sequences or putative transposons) are more highly methylated. The lack of detectable methylation differences between genes located in the maize1 and maize2 sub-genomes was surprising because there is evidence that maize1 genes are generally more highly expressed than maize2 genes [57]. Therefore, DNA methylation is unlikely to provide the mechanism for controlling the expression differences between the two sub-genomes of maize. The significant difference in average methylation levels for syntenic and non-syntenic genes is correlated with lower expression levels for non-syntenic genes. The non-syntenic genes that are conserved among the grasses but exhibit different genomic positions are the result of gene movement, perhaps mediated by transposons. Approximately 1/3 of the maize FGS genes are not in syntenic positions relative to other grasses, yet nearly all genes investigated by classical genetics to date belong to the fraction of the genome located in syntenic positions [63]. Their higher levels of methylation may reflect the presence of transposon sequences near these genes or may result from the insertion of a gene into new chromosomal environments that are lacking some of their ancestral regulatory sequences.
Many stable DMRs are found in B73 and Mo17
Several groups have demonstrated that perturbation of epigenetic information can affect quantitative traits [28]–[31]. In addition, the existence of natural epigenetic variation has been demonstrated for individual loci [32]–[44] or chromosomes [8]. A primary objective of this study was to document the prevalence and distribution of variable methylation levels in different maize genotypes. There are hundreds of examples of differential methylation in B73 and Mo17. In general, many of these DMRs are located in intergenic regions and may reflect differences in transposon silencing among the genotypes. However, at least 71 of the 690 variable methylation regions are found within 500 bp of a high-confidence gene (FGS). Following the discovery of these regions we were able to pursue further characterization using a population of NILs. The NILs provide a useful tool for assessing the stability of DNA methylation patterns and for testing whether the epigenetic variation is caused by genetic differences elsewhere in the genome.
The analysis of DNA methylation levels at several DMRs within the NILs addressed the stability of the DNA methylation patterns. The near-isogenic lines were developed by three back-crosses followed by at least four rounds of self-pollination [59]. In an analysis of the control lines (lines without an introgression at the DMR locus for eleven cis-controlled DMRs) there is evidence for stable inheritance as 85% of the assays reveal the expected methylation level. There are a small number of assays (5/150) that exhibit a completely changed methylation state and another 17/150 exhibit a partial gain or loss of DNA methylation. These examples may reflect inaccuracies in our measurements of DNA methylation or actual instability of DNA methylation patterns. In general, we observe relatively stable inheritance with rare examples of both gains and losses of DNA methylation. We did not observe evidence for paramutation-like effects where methylation levels were affected by heterozygosity for the DMRs.
Stable differences in DNA methylation levels between two genotypes can be the result of differences in epigenetic state that are faithfully propagated to offspring. Alternatively, they may be the result of genetic changes elsewhere in the genome that direct epigenetic modifications at an unlinked site. For example, structural rearrangements of the PAI1-PAI4 locus on chromosome 1 of Arabidopsis control the methylation state of the two other PAI loci on chromosomes 1 and 5 [36]. The conditioning of DNA methylation state by either linked or unlinked genomic regions would be examples of obligatory epialleles where a genetic change at one locus programs variable methylation at another locus. We studied the contribution of linked and unlinked genomic regions to the methylation differences between B73 and Mo17 for 13 of the DMRs using a series of near-isogenic lines [58]. Three of the regions exhibit evidence for trans-acting control of DNA methylation patterns. The remaining ten loci have methylation patterns that are either stably inherited or are continuously directly by local sequence changes.
Evidence for pure epialleles in maize
A major unresolved question about epigenetic variation is whether the majority of epigenetic variation exhibits strong linkage disequilibrium with nearby genetic differences [9]. If genetic markers, such as SNPs, are in strong linkage disequilibrium with epigenetic changes then the functional consequences of epigenetic differences would likely be revealed by assays of linked genetic differences. In particular, “obligatory” epialleles are entirely conditioned by nearby genetic changes [5]. Alternatively, “facilitated” epialleles exhibit stochastic variation in epigenetic state with a conditioning genetic change, and “pure” epialleles exhibit stochastic variation in epigenetic state independent of any genetic changes [5]. Both facilitated and pure epialleles will show differences in epigenetic state that are not completely linked to, or predicted, by nearby genetic polymorphisms.
We were interested in whether some of the epigenetic changes between B73 and Mo17 might be due to epigenetic changes that are not directly caused by nearby genetic differences such as transposon insertions or by unlinked rearrangements that might direct methylation in trans via RNA-directed DNA methylation. This led us to focus on the 10 extended B73-Mo17 identical-by-descent regions. These regions are most likely the result of shared inheritance of a chromosomal region from a genotype that was used in the pedigree of both B73 and Mo17. At least one genotype, CI187-2, is present in the pedigree of both B73 and Mo17 [64]. It is therefore possible that these regions could exhibit identity by descent (IBD). It is worth noting that other maize genotypes have alternative haplotypes in these regions so they are not the result of large selective sweeps among all maize genotypes [65]. The absence of detected structural variation and few SNPs within these regions suggest that any observed epigenetic differences between B73 and Mo17 are not the result of nearby genetic polymorphisms. We found 9 DMR (of 690 genome-wide) within these IBD regions and only one of these has evidence for a nearby genetic change. The number of DMRs within the IBD regions (9) is very close to the number we would have expected (13) based on the frequency per Mb within the whole genome. The finding that all four of these regions that were assessed in NILs show stable inheritance provides evidence for heritable epigenetic information in the absence of genetic differences. Our initial focus on IBD regions allowed the discovery of epigenetic variation without nearby genetic changes given the extended regions of identity. However, it is likely that many of the DMRs that are located in non-identical by descent genomic regions may also be the result of purely epigenetic changes. We noted that one-third of the 690 DMRs do not contain any SNPs in B73 relative to Mo17 within 1 kb of the DMR and may represent epigenetic differences that are not conditioned by genetic differences.
This study provides a detailed view of the distribution of cytosine methylation in two maize inbreds. The evidence for faithfully inherited methylation differences, even in the absence of nearby genetic polymorphisms, provide evidence for at least partially stable epigenetic variation in maize that would not be revealed by high-resolution analyses of genetic differences. There are likely functional consequences of the altered methylation levels in B73 and Mo17. There are several examples in which a DMR within an identical by descent region is near the promoter for a FGS gene and several of these genes exhibit differential expression in other tissues of B73 and Mo17 (data not shown). Further characterization of the relationship between expression variation and methylation variation may identify examples of epigenetic variation that affect phenotypic differences among inbred lines. This study, in combination with recent analyses of epiRILs in Arabidopsis [28]–[31], provides evidence for heritable epigenetic information that may contribute to quantitative trait differences within species. Future research is required to uncover evidence for the contribution of the variable methylation we have described in this study to phenotypic differences among maize genotypes.
Materials and Methods
Plant materials and DNA isolation
Three replications of B73 and Mo17 seedlings were grown in a randomized block design. The seeds for each replication came from a unique, single source (ear). For each replication, 10 seedlings were grown in pots (5 seedlings per pot) that were assigned random positions. Seedlings were grown under controlled conditions in a greenhouse at the University of Minnesota (St. Paul, MN) with a light cycle of 15 hours lights on and 9 hours lights off each day. Seedlings were watered daily as needed. After 18 days of growth, the 3rd leaf (L3) of each plant was harvested and pooled with other plants from the same pot/replication and immediately frozen in liquid nitrogen. DNAs were isolated using the CTAB method. Phenol∶chloroform extraction and subsequent precipitation in 0.1× volume Na-Acetate (3 M) and 2× volume 100% EtOH was conducted to purify the DNA samples. 15–30 ug of gDNA in 650–700 uL nuclease-free water was sonicated for five, ten - second pulses as per the methods of [66]. Samples were quantified and run on 1.5% agarose gels to verify that DNAs were fragmented to 200–400 bp.
Array design and annotation
A NimbleGen 2.1 M feature long oligonucleotide array was designed using B73 RefGen2 assembly (provided by the Arizona Genomics Institute). The maize genome exhibits a complex architecture with many repetitive sequences interspersed with low-copy genic sequences [14]. A repeat masked version of the pseudomolecule sequences from RefGenv2 of the B73 genome (provided by J Stein and D Ware) were used to design probes to low-copy regions. Thermally balanced probes were designed every ∼200 bp across the low-copy portion of the maize genome. The actual spacing varies in some cases to allow for ideal probe selection. In addition, a higher density of probes (one probe every ∼56 bp) was used for chromosome 9 to determine whether higher probe density provided increased resolution for methylation detection (Table S1). The probes were each annotated with respect to their copy number in the B73 genome, the number of close matches and their location relative to gene models. Syntenic orthologs of maize genes in sorghum, rice, and brachypodium were identified using the combined synonymous substitution rate of syntenic blocks method described in [57]. A maize gene was considered to be recently inserted if orthologous locations could be identified in rice, sorghum, and brachypodium by the syntenic conservation of up and downstream genes, but no homologous gene nor unannotated homologous sequence was identified in any species at the predicted orthologous location.
Immunoprecipitation of methylated DNA, labeling, and hybridization
Methylated DNA was immunoprecipitated with an anti-5-methylcytosine monoclonal antibody from 400 ng sonicated DNA using the Methylated DNA IP Kit (Zymo Research, Orange, CA; Cat # D5101). For each replication and genotype, whole genome amplification was conducted on 50–100 ng IP DNA and also 50–100 ng of sonicated DNA (input control) using the Whole Genome Amplification kit (Sigma Aldrich, St. Louis, MO, Cat # WGA2-50RXN). For each amplified IP input sample, 3 ug amplified DNA were labeled using the Dual-Color Labeling Kit (Roche NimbleGen, Cat # 05223547001) according to the array manufacturer's protocol (Roche NimbleGen Methylation UserGuide v7.0). Each IP sample was labeled with Cy5 and each input/control sonicated DNA was labeled with Cy3. The no anti-body control (negative control) was also labeled, using the average volumes required for the experimental samples. Samples were hybridized to the custom 2.1 M probe array (GEO Platform GPL13499) for 16–20 hrs at 42°C. Slides were washed and scanned according to NimbleGen's protocol for the GenePix4000B scanner. Images were aligned and quantified using NimbleScan software (Roche NimbleGen) producing raw data reports for each probe on the array.
Normalization and linear modeling
Pair files exported from NimbleScan were imported into the Bioconductor statistical environment (http://bioconductor.org/). Microarray data channels were assigned the following factors: B73, Mo17, B73 input, or Mo17 input depending on sample derivation. Non-maize probes and vendor-supplied process control probes were configured to have analytical weights of zero. Variance-stabilizing normalization was used to account for array-specific effects. Factor-specific hybridization coefficients were estimated by fitting fixed linear model accounting for dye and sample effects to the data using the limma package [67]. To compute biologically relevant information about B73 and Mo17 DNA methylation, the following contrasts were then computed: B73 IP vs B73 input (B73 methylation); Mo17 input vs B73 input (CGH and differential hybridization efficiency); Mo17 IP vs [Mo17 input vs B73 input] (Mo17 methylation corrected for differential hybridization efficiency); B73 IP vs [Mo17 IP vs [Mo17 input vs B73 input]] (differential DNA methylation corrected for differential hybridization efficiency). Moderated t-statistics and the log-odds score for differential MeDIP enrichment were computed by empirical Bayes shrinkage of the standard errors with the False Discovery Rate controlled to 0.05 [67]. Full results are available for download from the following URL: http://genomics.tacc.utexas.edu/data/eichten-plos-genetics-2011-a. Microarray results were deposited with NCBI GEO under accession GSE29099.
Defining CGH copy-number variations
CGH data were obtained from the hybridization of un-enriched B73 and Mo17 genomic DNA (B73 and Mo17 input channels). DNAcopy [52] was performed to identify segments showing similar hybridization patterns based on chromosomal order of probes. Defined segments were analyzed by expectation maximization model analysis [52], [68] to identify segments that fall into three predicted sub-distributions with non-uniform variances. For each segment, the posterior probability that it occurred in each of the three distributions was determined. Segments that had >0.95 probability of falling into either the first or third sub-distributions were defined as containing more copies in Mo17 than in B73 (M>B CNV) or significant fewer to no copies in Mo17 relative to B73 (M<B CNV and PAV) respectively.
Analysis of variable methylation
To define probes with differential methylation between the B73 and Mo17 inbreds, the significance values developed from the B73 vs Mo17 relative methylation linear model probes were used. Probes with a significance value of <0.001 were considered differentially methylated between the two inbreds. The direction of the variation was determined based on the positive or negative value of the B73 methylation minus the Mo17 methylation state. A total of 4172 B73 hypermethylated and 5367 Mo17 hypermethylated probes were classified using this method.
To identify segments showing differential methylation between B73 and Mo17, the DNAcopy algorithm [52] was used on 1,088,820 Mo17 unique probes in the B73 vs. Mo17 relative methylation linear model results. The EM algorithm [53], [68] was used to estimate the mixing proportion, mean, and variance associated with three predicted sub-distributions with non-uniform variances found within the B73 vs Mo17 segments. For each segment, the posterior probability that it occurred in each of the three distributions was determined. Segments that had >0.95 probability of falling into either the first or third sub-distributions were called as Mo17 hypermethylated or B73 hypermethylated segments respectively.
qPCR
Quantitative PCR (qPCR) was performed to evaluate the efficiency of 5-methylcytosine immunoprecipitation using regions of the Mez1 gene known to have methylation (5′region) and non-methylation (Exon 9) [66]. Primers were designed within the methylated region (forward primer 5′ - TTGTGTCGAGGTCTCGAATG-3′, reverse primer 5′ - TGTTGAAGCGCATTAGCACT -3′) and within the non-methylated region (forward primer 5′ - CAACAAAGTGAAAGCTCTTCAACTGCAA-3′, reverse primer 5′-CACAACACTCCCCTAGTCCCTCAAAAGTT-3′). Primer amplification and efficiency were tested in B73 and Mo17 genomic DNA. Three technical replications were included for each of two biological replications of B73 and Mo17 IP and input DNA samples. The relative amount of immunoprecipitated DNA (percentage of the input control DNA for each sample) was calculated (Figure S1). As expected, the IP negative control and qPCR no template controls either did not amplify or amplified approximately 10 cycles after the experimental samples (>1000 fold difference, data not shown). Mez1 qPCR reactions were conducted using 100 ng DNA and Light Cycler480 SYBR Green I Master (Roche, Cat # 04707516001) on the LightCycler480 instrument (Roche) in accordance with Roche's protocol for SYBR Green on the LightCycler480.
Primers were designed for 33 regions within DMRs (Table S6). 1 microgram of genomic DNA was digested for 16 hr with MspJI or FspEI (New England Biolabs). Mock digestions were performed substituting glycerol for restriction enzyme. qPCR reactions were performed using 37 ng DNA and SsoFast EvaGreen Supermix (BioRad) on the Chromo4 instrument (BioRad) in accordance with SsoFast protocol. The difference between digest C(t) and mock C(t) was calculated for each genotype tested. As our selected enzymes target methylated cytosines, higher methylation leads to increased digestion and subsequently longer C(t) times. DMRs in B73 and Mo17 were validated as higher methylation levels for larger differential C(t) value between the inbred lines. NIL and diverse inbred samples were compared across individual primer pairs and methylation state was determined by comparing C(t) difference values.
Supporting Information
Zdroje
1. BirdA 2007 Perceptions of epigenetics. Nature 447 7143 396 398
2. ChanSWHendersonIRJacobsenSE 2005 Gardening the genome: DNA methylation in arabidopsis thaliana. Nat Rev Genet 6 351 360
3. SlotkinRKMartienssenR 2007 Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet 8 272 285 10.1038/nrg2072
4. RasmussonDCPhillipsRL 1997 Plant breeding progress and genetic diversity from de novo variation and elevated epistasis. Crop Sci 37 303 310
5. RichardsEJ 2006 Inherited epigenetic variation–revisiting soft inheritance. Nat Rev Genet 7 395 401
6. FeinbergAP 2007 Phenotypic plasticity and the epigenetics of human disease. Nature 447 7143 433 440
7. JirtleRLSkinnerMK 2007 Environmental epigenomics and disease susceptibility. Nat Rev Genet 8 253 262
8. VaughnMWTanurdzicMLippmanZJiangHCarrasquilloR 2007 Epigenetic natural variation in arabidopsis thaliana. PLoS Biol 5 e174 doi:10.1371/journal.pbio.0050174
9. RichardsEJ 2008 Population epigenetics. Curr Opin Genet Dev 18 221 226
10. JohannesFColotVJansenRC 2008 Epigenome dynamics: A quantitative genetics perspective. Nat Rev Genet 9 883 890
11. BucklerESGautBSMcMullenMD 2006 Molecular and functional diversity of maize. Curr Opin Plant Biol 9 172 176
12. MessingJDoonerHK 2006 Organization and variability of the maize genome. Curr Opin Plant Biol 9 157 163
13. RabinowiczPDBennetzenJL 2006 The maize genome as a model for efficient sequence analysis of large plant genomes. Curr Opin Plant Biol 9 149 156
14. SchnablePSWareDFultonRSSteinJCWeiF 2009 The B73 maize genome: Complexity, diversity, and dynamics. Science 326 5956 1112 1115
15. WeilCMartienssenR 2008 Epigenetic interactions between transposons and genes: Lessons from plants. Curr Opin Genet Dev 18 188 192
16. YuJHollandJBMcMullenMDBucklerES 2008 Genetic design and statistical power of nested association mapping in maize. Genetics 178 539 551
17. McMullenMDKresovichSVilledaHSBradburyPLiH 2009 Genetic properties of the maize nested association mapping population. Science 325 5941 737 740
18. LippmanZGendrelAVBlackMVaughnMWDedhiaN 2004 Role of transposable elements in heterochromatin and epigenetic control. Nature 430 6998 471 476
19. ZhangXYazakiJSundaresanACokusSChanSW 2006 Genome-wide high-resolution mapping and functional analysis of DNA methylation in arabidopsis. Cell 126 1189 1201
20. ZilbermanDGehringMTranRKBallingerTHenikoffS 2007 Genome-wide analysis of arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat Genet 39 61 69
21. MiuraANakamuraMInagakiSKobayashiASazeH 2009 An arabidopsis jmjC domain protein protects transcribed genes from DNA methylation at CHG sites. EMBO J 28 1078 1086
22. FengSCokusSJZhangXChenPYBostickM 2010 Conservation and divergence of methylation patterning in plants and animals. Proc Natl Acad Sci U S A 107 8689 8694
23. WangXEllingAALiXLiNPengZ 2009 Genome-wide and organ-specific landscapes of epigenetic modifications and their relationships to mRNA and small RNA transcriptomes in maize. Plant Cell 21 1053 1069
24. FinneganEJ 2002 Epialleles - a source of random variation in times of stress. Curr Opin Plant Biol 5 101 106
25. KaliszSPuruggananMD 2004 Epialleles via DNA methylation: Consequences for plant evolution. Trends Ecol Evol 19 309 314
26. LukensLNZhanS 2007 The plant genome's methylation status and response to stress: Implications for plant improvement. Curr Opin Plant Biol 10 317 322
27. RichardsEJ 2011 Natural epigenetic variation in plant species: A view from the field. Curr Opin Plant Biol 14 204 209
28. ReindersJWulffBBMirouzeMMari-OrdonezADappM 2009 Compromised stability of DNA methylation and transposon immobilization in mosaic arabidopsis epigenomes. Genes Dev 23 939 950
29. JohannesFPorcherETeixeiraFKSaliba-ColombaniVSimonM 2009 Assessing the impact of transgenerational epigenetic variation on complex traits. PLoS Genet 5 e1000530 doi:10.1371/journal.pgen.1000530
30. RouxFColome-TatcheMEdelistCWardenaarRGuercheP 2011 Genome-wide epigenetic perturbation jump-starts patterns of heritable variation found in nature. Genetics 188 1015 1017
31. JohannesFColome-TatcheM 2011 Quantitative epigenetics through epigenomic perturbation of isogenic lines. Genetics 188 215 227
32. RiddleNCRichardsEJ 2002 The control of natural variation in cytosine methylation in arabidopsis. Genetics 162 355 363
33. RiddleNCRichardsEJ 2005 Genetic variation in epigenetic inheritance of ribosomal RNA gene methylation in arabidopsis. Plant J 41 524 532
34. WooHRPontesOPikaardCSRichardsEJ 2007 VIM1, a methylcytosine-binding protein required for centromeric heterochromatinization. Genes Dev 21 267 277
35. ZhangXShiuSHCalABorevitzJO 2008 Global analysis of genetic, epigenetic and transcriptional polymorphisms in arabidopsis thaliana using whole genome tiling arrays. PL+oS Genet 4 e1000032
36. BenderJFinkGR 1995 Epigenetic control of an endogenous gene family is revealed by a novel blue fluorescent mutant of arabidopsis. Cell 83 725 734
37. JacobsenSEMeyerowitzEM 1997 Hypermethylated SUPERMAN epigenetic alleles in arabidopsis. Science 277 5329 1100 1103
38. CubasPVincentCCoenE 1999 An epigenetic mutation responsible for natural variation in floral symmetry. Nature 401 6749 157 161
39. MorganHDSutherlandHGMartinDIWhitelawE 1999 Epigenetic inheritance at the agouti locus in the mouse. Nat Genet 23 314 318
40. ChandlerVLEgglestonWBDorweilerJE 2000 Paramutation in maize. Plant Mol Biol 43 2–3 121 145
41. StokesTLKunkelBNRichardsEJ 2002 Epigenetic variation in arabidopsis disease resistance. Genes Dev 16 171 182
42. RakyanVKChongSChampMECuthbertPCMorganHD 2003 Transgenerational inheritance of epigenetic states at the murine axin allele occurs after maternal and paternal transmission. Proc Natl Acad Sci U S A 100 2538 2543
43. SuterCMMartinDIWardRL 2004 Germline epimutation of MLH1 in individuals with multiple cancers. Nat Genet 36 497 501
44. ManningKTorMPooleMHongYThompsonAJ 2006 A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat Genet 38 948 952
45. MakarevitchIStuparRMIniguezALHaunWJBarbazukWB 2007 Natural variation for alleles under epigenetic control by the maize chromomethylase zmet2. Genetics 177 749 760
46. SlotkinRKFreelingMLischD 2005 Heritable transposon silencing initiated by a naturally occurring transposon inverted duplication. Nat Genet 37 641 644
47. Arteaga-VazquezMSidorenkoLRabanalFAShrivistavaRNobutaK 2010 RNA-mediated trans-communication can establish paramutation at the b1 locus in maize. Proc Natl Acad Sci U S A 107 12986 12991
48. HaunWJLaoueille-DupratSO'connellMJSpillaneCGrossniklausU 2007 Genomic imprinting, methylation and molecular evolution of maize enhancer of zeste (mez) homologs. Plant J 49 325 337
49. GoreMAChiaJMElshireRJSunQErsozES 2009 A first-generation haplotype map of maize. Science 326 5956 1115 1117
50. SpringerNMYingKFuYJiTYehCT 2009 Maize inbreds exhibit high levels of copy number variation (CNV) and presence/absence variation (PAV) in genome content. PLoS Genet 5 e1000734 doi:10.1371/journal.pgen.1000734
51. BeloABeattyMKHondredDFenglerKALiB 2010 Allelic genome structural variations in maize detected by array comparative genome hybridization. Theor Appl Genet 120 355 367
52. VenkatramanESOlshenAB 2007 A faster circular binary segmentation algorithm for the analysis of array CGH data. Bioinformatics 23 657 663
53. DempsterAPLairdNMRubinDB 1977 Maximum likelihood from incomplete data via the EM algorithm. Journal of the Royal Statistical Society.Series B (Methodological) 39 1 38
54. LiuSYehCTJiTYingKWuH 2009 Mu transposon insertion sites and meiotic recombination events co-localize with epigenetic marks for open chromatin across the maize genome. PLoS Genet 5 e1000733 doi:10.1371/journal.pgen.1000733
55. ListerRO'MalleyRCTonti-FilippiniJGregoryBDBerryCC 2008 Highly integrated single-base resolution maps of the epigenome in arabidopsis. Cell 133 523 536
56. LiPPonnalaLGandotraNWangLSiY 2010 The developmental dynamics of the maize leaf transcriptome. Nat Genet 42 1060 1067
57. SchnableJCSpringerNMFreelingM 2011 Differentiation of the maize subgenomes by genome dominance and both ancient and ongoing gene loss. Proc Natl Acad Sci U S A 108 4069 4074
58. EichtenSRFoersterJMde LeonNKaiYYehCT 2011 B73-Mo17 near-isogenic lines demonstrate dispersed structural variation in maize. Plant Physiol 156 1679 1690
59. ChandlerVLStamM 2004 Chromatin conversations: Mechanisms and implications of paramutation. Nat Rev Genet 5 532 544
60. SpringerNMGutierrez-MarcosJF 2009 Imprinting in maize. Anonymous Handbook of Maize Springer New York 429 440
61. FedoroffNSchlappiMRainaR 1995 Epigenetic regulation of the maize spm transposon. Bioessays 17 291 297
62. LischD 2002 Mutator transposons. Trends Plant Sci 7 498 504
63. SchnableJCFreelingM 2011 Genes identified by visible mutant phenotypes show increased bias toward one of two subgenomes of maize. PLoS ONE 6 e17855 doi:10.1371/journal.pone.0017855
64. HanseyCNJohnsonJMSekhonRSKaepplerSMLeonNd 2011 Genetic diversity of a maize association population with restricted phenology. Crop Sci 51 704 715
65. Swanson-WagnerRAEichtenSRKumariSTiffinPSteinJC 2010 Pervasive gene content variation and copy number variation in maize and its undomesticated progenitor. Genome Res 20 1689 1699
66. HaunWJSpringerNM 2008 Maternal and paternal alleles exhibit differential histone methylation and acetylation at maize imprinted genes. Plant J 56 903 912
67. SmythGK 2004 Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3 Article3
68. FraleyCRafteryAE 2006 MCLUST version 3 for R: Normal mixture modeling and model-based clustering. Department of Statistics: University of Washington
Štítky
Genetika Reprodukční medicína
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2011 Číslo 11- Akutní intermitentní porfyrie
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Intrauterinní inseminace a její úspěšnost
- Růst a vývoj dětí narozených pomocí IVF
- Pánevní endometrióza spojená s volnou tekutinou v peritoneální dutině snižuje úspěšnost otěhotnění po intrauterinní inseminaci
-
Všechny články tohoto čísla
- De Novo Origins of Human Genes
- Duplication Hotspots Are Associated with Late-Replicating Regions of the Genome
- De Novo Origin of Human Protein-Coding Genes
- Cyclin D/CDK4 and Cyclin E/CDK2 Induce Distinct Cell Cycle Re-Entry Programs in Differentiated Muscle Cells
- Short Day–Mediated Cessation of Growth Requires the Downregulation of AINTEGUMENTALIKE1 Transcription Factor in Hybrid Aspen
- Physiological IRE-1-XBP-1 and PEK-1 Signaling in Larval Development and Immunity
- Role of Pirh2 in Mediating the Regulation of p53 and c-Myc
- Signatures of Environmental Genetic Adaptation Pinpoint Pathogens as the Main Selective Pressure through Human Evolution
- FOXO Regulates Organ-Specific Phenotypic Plasticity In
- Heritable Epigenetic Variation among Maize Inbreds
- Foxn1 Regulates Lineage Progression in Cortical and Medullary Thymic Epithelial Cells But Is Dispensable for Medullary Sublineage Divergence
- Attenuation of the Sensing Capabilities of PhoQ in Transition to Obligate Insect–Bacterial Association
- A Novel Protein LZTFL1 Regulates Ciliary Trafficking of the BBSome and Smoothened
- Activation of Bmp2-Smad1 Signal and Its Regulation by Coordinated Alteration of H3K27 Trimethylation in -Induced Senescence
- Histone H3K56 Acetylation, CAF1, and Rtt106 Coordinate Nucleosome Assembly and Stability of Advancing Replication Forks
- The SUN Protein Mps3 Is Required for Spindle Pole Body Insertion into the Nuclear Membrane and Nuclear Envelope Homeostasis
- Evidence-Based Annotation of Gene Function in MR-1 Using Genome-Wide Fitness Profiling across 121 Conditions
- Effect of Host Species on the Distribution of Mutational Fitness Effects for an RNA Virus
- Pch2 Acts through Xrs2 and Tel1/ATM to Modulate Interhomolog Bias and Checkpoint Function during Meiosis
- SOX9 Governs Differentiation Stage-Specific Gene Expression in Growth Plate Chondrocytes via Direct Concomitant Transactivation and Repression
- from the Aphid : A Missing Link from Facultative to Obligate Insect Endosymbiont
- Recessive Antimorphic Alleles Overcome Functionally Redundant Loci to Reveal Function in Flowers and Meristems
- Over-Expression of DSCAM and COL6A2 Cooperatively Generates Congenital Heart Defects
- Consequences of Eukaryotic Enhancer Architecture for Gene Expression Dynamics, Development, and Fitness
- Distinct Genetic Architectures for Male and Female Inflorescence Traits of Maize
- Capture of MicroRNA–Bound mRNAs Identifies the Tumor Suppressor miR-34a as a Regulator of Growth Factor Signaling
- For Male , Sperm Activation Is a “Just-in-Time” Event
- PcG Complexes Set the Stage for Epigenetic Inheritance of Gene Silencing in Early S Phase before Replication
- The Gene Contains Hotspots for L1 Endonuclease-Dependent Insertion
- Relative Burden of Large CNVs on a Range of Neurodevelopmental Phenotypes
- Multiple Means to the Same End: The Genetic Basis of Acquired Stress Resistance in Yeast
- Genome-Wide Crossover Distribution in Meiosis Reveals Sex-Specific Patterns along Chromosomes
- TRY-5 Is a Sperm-Activating Protease in Seminal Fluid
- Homologs of Retinoblastoma-Associated Protein 46/48 Associate with a Histone Deacetylase to Act Redundantly in Chromatin Silencing
- Genetic Interaction Maps in Reveal Functional Crosstalk among Cell Envelope Biogenesis Pathways
- The ERI-6/7 Helicase Acts at the First Stage of an siRNA Amplification Pathway That Targets Recent Gene Duplications
- PBX1 Genomic Pioneer Function Drives ERα Signaling Underlying Progression in Breast Cancer
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Evidence-Based Annotation of Gene Function in MR-1 Using Genome-Wide Fitness Profiling across 121 Conditions
- De Novo Origins of Human Genes
- Capture of MicroRNA–Bound mRNAs Identifies the Tumor Suppressor miR-34a as a Regulator of Growth Factor Signaling
- TRY-5 Is a Sperm-Activating Protease in Seminal Fluid
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



