-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Mutation Disrupts Synaptonemal Complex Formation, Recombination, and Chromosome Segregation in Mammalian Meiosis
Meiosis is unique to germ cells and essential for reproduction. During the first meiotic division, homologous chromosomes pair, recombine, and form chiasmata. The homologues connect via axial elements and numerous transverse filaments to form the synaptonemal complex. The synaptonemal complex is a critical component for chromosome pairing, segregation, and recombination. We previously identified a novel germ cell–specific HORMA domain encoding gene, Hormad1, a member of the synaptonemal complex and a mammalian counterpart to the yeast meiotic HORMA domain protein Hop1. Hormad1 is essential for mammalian gametogenesis as knockout male and female mice are infertile. Hormad1 deficient (Hormad1−/−) testes exhibit meiotic arrest in the early pachytene stage, and synaptonemal complexes cannot be visualized by electron microscopy. Hormad1 deficiency does not affect localization of other synaptonemal complex proteins, SYCP2 and SYCP3, but disrupts homologous chromosome pairing. Double stranded break formation and early recombination events are disrupted in Hormad1−/− testes and ovaries as shown by the drastic decrease in the γH2AX, DMC1, RAD51, and RPA foci. HORMAD1 co-localizes with γH2AX to the sex body during pachytene. BRCA1, ATR, and γH2AX co-localize to the sex body and participate in meiotic sex chromosome inactivation and transcriptional silencing. Hormad1 deficiency abolishes γH2AX, ATR, and BRCA1 localization to the sex chromosomes and causes transcriptional de-repression on the X chromosome. Unlike testes, Hormad1−/− ovaries have seemingly normal ovarian folliculogenesis after puberty. However, embryos generated from Hormad1−/− oocytes are hyper - and hypodiploid at the 2 cell and 8 cell stage, and they arrest at the blastocyst stage. HORMAD1 is therefore a critical component of the synaptonemal complex that affects synapsis, recombination, and meiotic sex chromosome inactivation and transcriptional silencing.
Published in the journal: . PLoS Genet 6(11): e32767. doi:10.1371/journal.pgen.1001190
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1001190Summary
Meiosis is unique to germ cells and essential for reproduction. During the first meiotic division, homologous chromosomes pair, recombine, and form chiasmata. The homologues connect via axial elements and numerous transverse filaments to form the synaptonemal complex. The synaptonemal complex is a critical component for chromosome pairing, segregation, and recombination. We previously identified a novel germ cell–specific HORMA domain encoding gene, Hormad1, a member of the synaptonemal complex and a mammalian counterpart to the yeast meiotic HORMA domain protein Hop1. Hormad1 is essential for mammalian gametogenesis as knockout male and female mice are infertile. Hormad1 deficient (Hormad1−/−) testes exhibit meiotic arrest in the early pachytene stage, and synaptonemal complexes cannot be visualized by electron microscopy. Hormad1 deficiency does not affect localization of other synaptonemal complex proteins, SYCP2 and SYCP3, but disrupts homologous chromosome pairing. Double stranded break formation and early recombination events are disrupted in Hormad1−/− testes and ovaries as shown by the drastic decrease in the γH2AX, DMC1, RAD51, and RPA foci. HORMAD1 co-localizes with γH2AX to the sex body during pachytene. BRCA1, ATR, and γH2AX co-localize to the sex body and participate in meiotic sex chromosome inactivation and transcriptional silencing. Hormad1 deficiency abolishes γH2AX, ATR, and BRCA1 localization to the sex chromosomes and causes transcriptional de-repression on the X chromosome. Unlike testes, Hormad1−/− ovaries have seemingly normal ovarian folliculogenesis after puberty. However, embryos generated from Hormad1−/− oocytes are hyper - and hypodiploid at the 2 cell and 8 cell stage, and they arrest at the blastocyst stage. HORMAD1 is therefore a critical component of the synaptonemal complex that affects synapsis, recombination, and meiotic sex chromosome inactivation and transcriptional silencing.
Introduction
Mammalian meiosis is unique to germ cells and a critical step in sexual reproduction. Meiosis reduces the chromosome complement to haploidy in preparation for fertilization. The first meiotic division is unique in pairing of homologous chromosomes, homologous recombination, and formation of chiasmata. The reduction in chromosome numbers happens when homologous chromosomes segregate to opposite poles during the first meiotic division. Proper disjunction (separation) requires crossovers (manifested cytologically as chiasmata). The sister chromatids organize along structures called axial elements (AEs) and transverse elements connect AEs to form the synaptonemal complex (SC) [1]. SC is a proteinaceous structure that connects paired homologous chromosomes during prophase I of meiosis, and SC is critical for wild-type levels of crossovers to occur during meiosis. AEs are critical part of the SCs and mutations in proteins that form AEs disrupt sister chromatid cohesion, recombination, and chromosome segregation [2]–[4]. Proteins with HORMA domain are critical components of the axial elements [5]. HORMA domain proteins are predicted to form globular structure that may sense specialized chromatin states, such as those associated with double strand breaks (DSBs) or other forms of DNA damage [5]. Several mammalian proteins that contain HORMA domain, such as mitotic arrest deficient protein 2, MAD2, are essential for mitosis [6]–[7]. Mice lacking MAD2 unsurprisingly die during early embryogenesis [7]. In lower organisms, several meiotic specific HORMA proteins are known and all are critical for meiosis. These HORMA proteins are: Hop1 [8] and Red1 [9] in yeast; Him-3 [10] in nematodes; and Asy1 [11] in plants. Him-3 localizes to the axial cores of both synapsed and unsynapsed chromosomes. C. elegans Him-3 mutants are deficient in chromosome pairing, synapsis, and the regulation of double strand break repair [10], [12]–[13]. Synapsis in both male and female Asy1 mutants is disrupted [14]–[15]. In yeast, plants, and nematodes, HORMA domain proteins are critical components of the synaptonemal complex and essential for meiosis I. Others and we identified a previously uncharacterized gene that we named Nohma, later re-named to Hormad1 [16]–[18]. Hormad1 encodes a protein that contains a HORMA domain, and unlike Mad2, Hormad1 expression is germ cell–specific [16]. Mouse and human HORMAD1 are highly conserved and share 77% amino acid identity overall, and share 89% amino acid identity in the HORMA domain. Moreover, mouse and human HORMAD1 HORMA domains share 28% amino acid identity with Hop1 HORMA domain. Hop1 in yeast appears to bind near or at the sites of DSB formation and may modulate the initial DSB cleavage [19]. Hop1 mutants in yeast have reduced number of DSBs [20], and Hop1 may participate in recruiting DMC1, RAD51 and other proteins that are required for DNA repair during meiotic synapsis and recombination [19]–[20]. Phosphorylation of Hop1 by Mec1/Tel1 yeast kinases is important for interhomologue recombination and prevents DMC1-independent repair of meiotic DSBs [21]. Here we report that HORMAD1 is likely the mammalian counterpart of Hop1, and that HORMAD1 deficiency disrupts mammalian synaptonemal complex formation, meiotic recombination, and chromosome segregation.
Results
HORMAD1 is essential for spermatogenesis
We previously showed that Hormad1 RNA expression in testes began at postnatal day 10, with little expression detected at birth or postnatal day 5 [16]. Hormad1 RNA expression pattern coincided with the onset of meiosis, and appearance of primary spermatocytes in the developing testes. In situ hybridization with anti-sense Hormad1 riboprobe revealed that Hormad1 expression was confined to germ cells, and specifically spermatocytes, with no signal detected in spermatogonia or sertoli cells [16]. We generated antibodies against HORMAD1 and studied its protein localization pattern in testes. HORMAD1 localized exclusively in germ cells, specifically in zygotene, and early pachytene spermatocytes as previously described for the RNA expression [16].
Since HORMAD1 protein showed localization consistent with its potential role in meiosis I and contains the HORMA domain, we disrupted the Hormad1 gene to examine its requirement for germ cell development and meiosis in mouse. Hormad1 is located on chromosome 3 and composed of sixteen exons. We deleted exons 4 and 5 (Figure S1A), and this mutation is predicted to remove 33 amino acids from the highly conserved HORMA domain and to cause a frame shift mutation. Small amounts of truncated Hormad1 RNA transcripts were detectable on RT-PCR, and Western blots on testes extracts showed absence of HORMAD1 protein in knockout mice as expected (Figure S1B and S1C).
Female and male heterozygote matings produced expected Mendelian ratios, averaged 8.1±2 pups per litter (n = 20 breeding pairs) over a 6-month period, and remained fertile for at least 9 months. The litter size was statistically not significantly different from the wild-type average (8.4±2 pups per litter). Male and female mice heterozygous for the mutation (Hormad1+/−) were fertile with grossly normal male and female gonadal morphology and histology. However, both Hormad1−/− males and females were infertile with no pups produced over a period of 6 months from mating with wild-type female and male mice, respectively.
While ovaries showed no gross morphologic differences between the knockout and wild-type mice, Hormad1 knockout adult testes were significantly smaller than the wild-type testes (Figure S2A). Testes in the 7-day-old Hormad1−/− mice were grossly normal and weighed 8.0±2.0 mg/pair and did not significantly differ from the wild-type, 9.0±3.0 mg/pair of testes. By 4 weeks of postnatal life, the knockout testes (47±6 mg/pair) were 50% of the wild-type weight (94±1.7 mg/pair), and by 8 weeks the knockout testes (60±7.7 mg/pair) were 27% of the wild-type weight (225±2.7 mg/pair) (Figure S2B).
Histology at 6 weeks showed hypocellular seminiferous tubules with clumps of sertoli cells in the lumen. We observed spermatogonia and early spermatocytes, but no post-meiotic germ cells such as spermatids or spermatozoa (Figure 1A–1H). We therefore carefully examined spermatogenesis in Hormad1−/− mice. Spermatogenesis is a complex process that involves differentiating and proliferating self-renewing spermatogonia that differentiate into spermatozoa. Type A spermatogonia self-renew and can initiate differentiation into Type B spermatogonia which in turn differentiate into primary spermatocytes. Primary spermatocytes undergo meiosis I to form secondary spermatocytes. Secondary spermatocytes enter meiosis II and divide to produce haploid spermatids. We examined Hormad1 knockout testes histology during gonadal development to determine the stage at which spermatogenesis is disrupted. Identical testes weights at postnatal day 7, and similar histology between Hormad1−/− and wild-type testes argue that pre-spermatogonia in Hormad1−/− testes proliferate into Type A spermatogonia without major disruption. Immunohistochemistry with antibodies directed against PLZF and SOHLH1, markers that identify self-renewing (PLZF) and differentiating spermatogonia (SOHLH1), showed the presence of both proteins in the wild-type as well as the knockout animals, confirming that spermatogonia are unaffected (Figure 1J, 1K, 1N, 1O). At postnatal day 10, testes contain preleptotene/leptotene primary spermatocytes, and there was no gross difference between wild-type and Hormad1−/− testes. At 14 days, testes contain pachytene spermatocytes, and there was a significant difference between the wild-type and Hormad1−/− testes, with many apoptotic cells and few pachytene spermatocytes in Hormad1−/− testes (), and rising apoptotic index with age in the knockout as compared to the wild-type (Figure S3K). We counted leptotene, zygotene and pachytene spermatocytes in 6 week old wild-type and Hormad1−/− testes. Hormad1−/− testes showed declining number of spermatocytes beginning in stages II-III with 28±8 spermatocytes as compared to 52±12 in the wild-type (Figure 1A and 1E). No spermatocytes were noted in stages IV-IX in the Hormad1−/− testes (Figure 1F and 1G), and no significant difference was noted in the spermatocyte number in stages X-XII between the wild-type and knockout testes (Figure 1D and 1H). These results indicate that Hormad1 deficiency in the male gonad caused meiotic arrest at the pachytene stage.
Fig. 1. HORMAD1 is required for spermatogenesis. 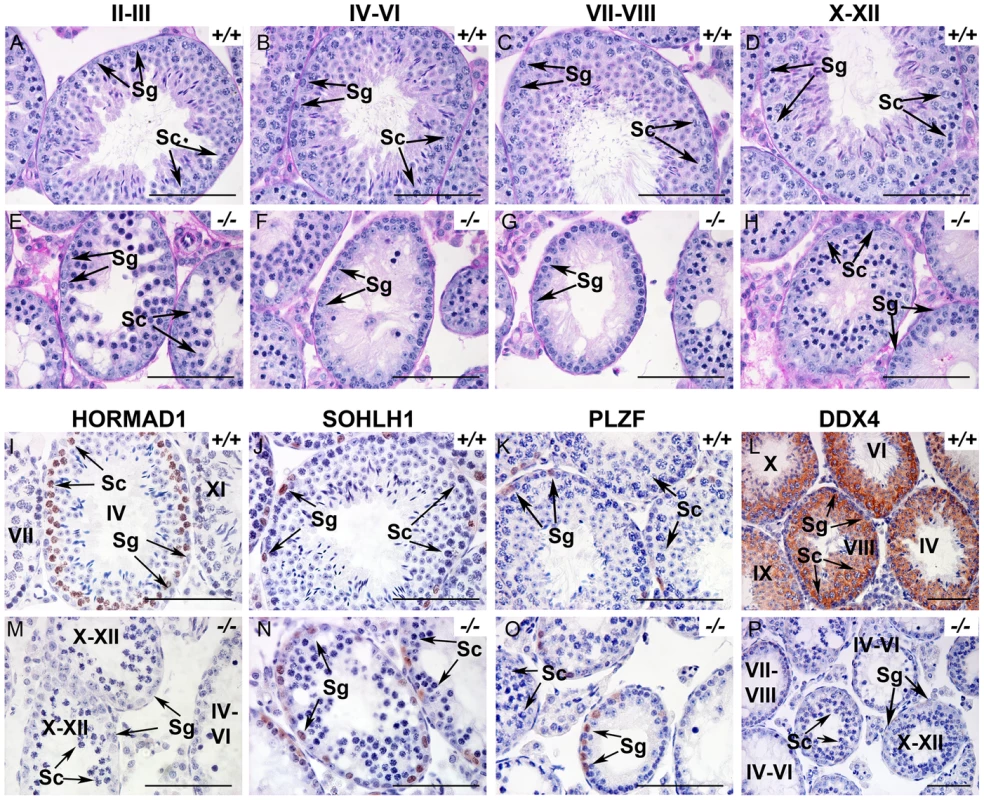
(A–H). Periodic acid/Schiff reagent (PAS) stained cross sections from 6 weeks old wild-type testes (A-D) and Hormad1 −/− (E-H) testes. Seminiferous tubule stages are shown above the panels. Lack of mature sperm and arrest in spermatogenesis is shown at different tubular stages. (I-P). Immunohistochemistry (IHC) of 6 weeks old wild type and Hormad1−/− testes with anti HORMAD1, SOHLH1, PLZF and DDX4 antibody. IHC with anti-HORMAD1 antibody shows HORMAD1 expression (brown) in wild-type stage IV pachytene, and stage XI zygotene spermatocytes (I), while no expression was observed in the knockout (M). SOHLH1 is expressed in differentiating spermatogonia and is present in wild-type and Hormad1−/− testes as shown by arrows (J and N). PLZF identifies differentiating and self-renewing spermatogonia and is expressed in both wild-type and Hormad1−/− testes as shown by arrows (K and O). DDX4 is an RNA binding protein specific for germ cells, and is under-expressed in Hormad1−/− spermatocytes (L and P). Arrows point to Spermatogonia (Sg) and Spermatocytes 1(Sc). Scale bars: 50 µm. HORMAD1 is critical for chromosome synapsis
Previous studies on HORMA domain proteins indicate their specific involvement in cell division. MAD2 is a ubiquitously expressed mammalian HORMA domain protein involved in both meiosis and mitosis [7], while yeast HOP1, RED1, nematode HIM3 and plant ASY1 genes are specifically involved in meiotic segregation, synapsis and recombination [2], [10]–[11], [13], [15], [22]. No mammalian counterparts to Hop1, Red1, Him3 and Asy1 have been functionally evaluated up to date. We previously hypothesized that HORMAD1 is a functional counterpart to Hop1, Him3 and Asy1 [16]. Critical components of the synaptonemal complex include meiosis specific SYCP1, SYCP2 and SYCP3 proteins. SYCP1 is a major component of the transverse filaments, while both SYCP2 and SYCP3 are components of the axial lateral elements [23]–[26]. To determine HORMAD1 localization during meiosis, and whether HORMAD1 localizes to the axial elements, or transverse filaments, we used antibodies against SYCP1, SYCP2 and SYCP3 to study their respective co-localization with HORMAD1. HORMAD1 co-localized with SYCP3 and SYCP2 but did not co-localize with SYCP1, which indicates that HORMAD1 is located along the axial elements (Figure 2A, 2C, and 2E). Recent studies also show that HORMAD1 localizes to the axial elements [17]–[18]. We also studied whether absence of SYCP2 affected HORMAD1 localization along the chromosomes. HORMAD1 localization is independent of major germ cell–specific components of the axial elements of the synaptonemal complex because neither Sycp2 (Figure 3A–3F) nor Sycp3 mutation affected HORMAD1 localization to the axial elements [18].
Fig. 2. HORMAD1 co-localizes with SYCP2 and SYCP3, but not with SYCP1. 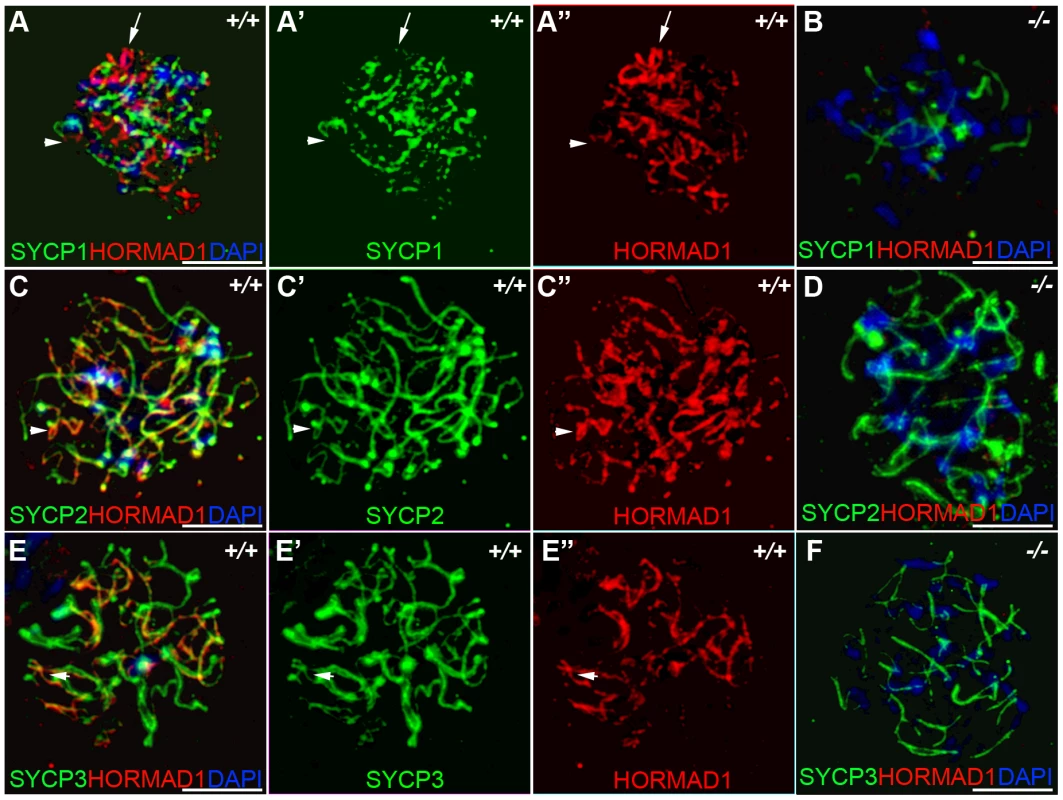
Chromosomal spread assay in wild-type (+/+) and Hormad1−/− (−/−) zygotene stage spermatocytes. (A–B) Immunofluorescence staining with anti-SYCP1 (Green) and anti-HORMAD1 (Red) antibody. HORMAD1 preferentially localizes to unsynapsed regions of chromosome axes (arrow head) and sex body (arrow) but does not co-localize with SYCP1 (A-A”). Hormad1 deficiency does not affect SYCP1 localization to the axes (B). HORMAD1 co-localizes with SYCP2 on the unsynapsed chromosome axes (C-C”), but HORMAD1 deficiency does not affect SYCP2 localization to the axes (D). SYCP3, another integral and critical component of the synaptonemal complex co-localizes with HORMAD1 on unsynapsed axes (E-E”). Similar to SYCP1 and SYCP2, SYCP3 localizes to the axes despite HORMAD1 deficiency (F). DNA was stained with DAPI. Scale bars: 10 µm. Fig. 3. HORMAD1 localization in Sycp2 mutant spermatocytes. 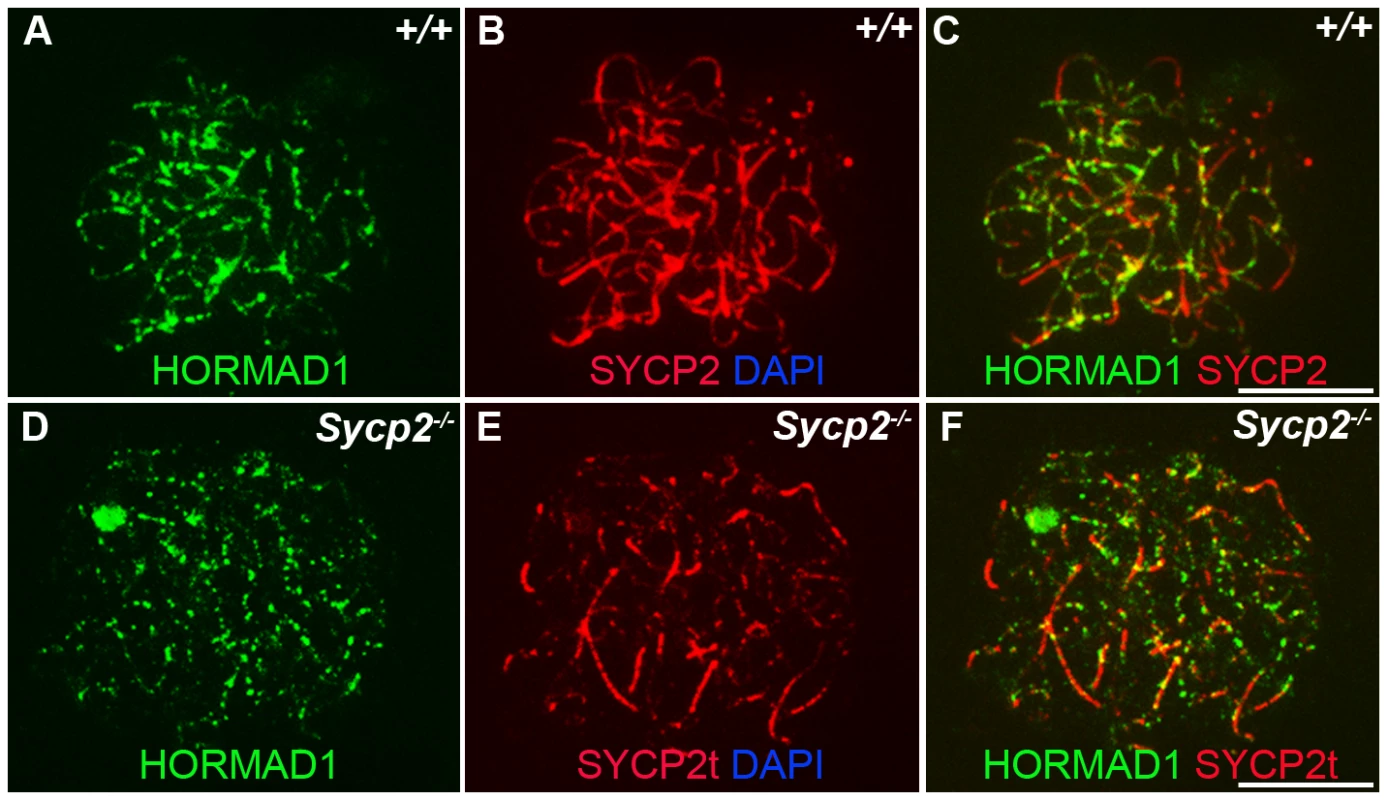
Chromosome spread assay was performed on the wild-type (+/+) (A–C) and Sycp2−/− (D–F) zygotene spermatocytes with anti-HORMAD1 (green) and anti-SYCP2 (red) antibodies. The truncated SYCP2 protein (SYCP2t) in Sycp2−/− mice is still made and localizes to axial chromosomal cores[48]. SYCP2t lacks the domain necessary to bind SYCP3. Scale bars: 10 µm. We examined localization of germ cell–specific synaptonemal complex proteins SYCP1, SYCP2 and SYCP3 in Hormad1−/− testes. SYCP1 is a major component of the transverse filaments and is known to form fibrillar structures in Sycp3 mutant spermatocytes [27]. However, the fibers are truncated, contain axial gaps, and do not associate with the centromeres of the meiotic chromosomes. We also observed truncated fibers with anti-SYCP1 antibodies in Hormad1−/− mice (Figure 2B). HORMAD1 is therefore not necessary for SYCP1 binding to the chromatin. We also examined localization of SYCP2 and SYCP3 proteins in Hormad1−/− mice. The localization of SYCP2 and SYCP3 to the chromatin was not significantly affected by the lack of HORMAD1 (Figure 2D and 2F). These results indicate that HORMAD1 is not necessary for SYCP2 and SYCP3 localization to the axial elements.
The deficiency in synaptonemal complex proteins such as SYCP3 is known to affect chromosome synapsis [27]. In order to determine the effect of HORMAD1 deficiency on chromosome synapsis during meiosis I, we utilized CREST sera. CREST sera labels centromeres and allows the determination of the pairing status during meiosis [28]. In the wild type spermatocytes, prior to the synapsis, 40 centromeres are usually observed in the leptotene stage. The number of visible centromeres become reduced as the synapsis of homologues progresses. At the completion of the synapsis in pachytene, 20 centromeric foci are usually observed corresponding to 19 autosomal homologues and partially paired X-Y chromosomes. We examined CREST foci formation in Hormad1−/− spermatocytes. Examination of over 100 Hormad1−/− spermatocytes and oocytes in meiosis I, revealed greater than 20 centromeric foci in both male and female germ cells, most containing 40 CREST foci (Figure 4A, 4D, and data not shown). These results indicate that Hormad1 deficient germ cells cannot complete homologous chromosome pairing, and Hormad1 is therefore critical for chromosome synapsis during meiosis.
Fig. 4. HORMAD1 is required for chromosome synapsis during male meiosis. 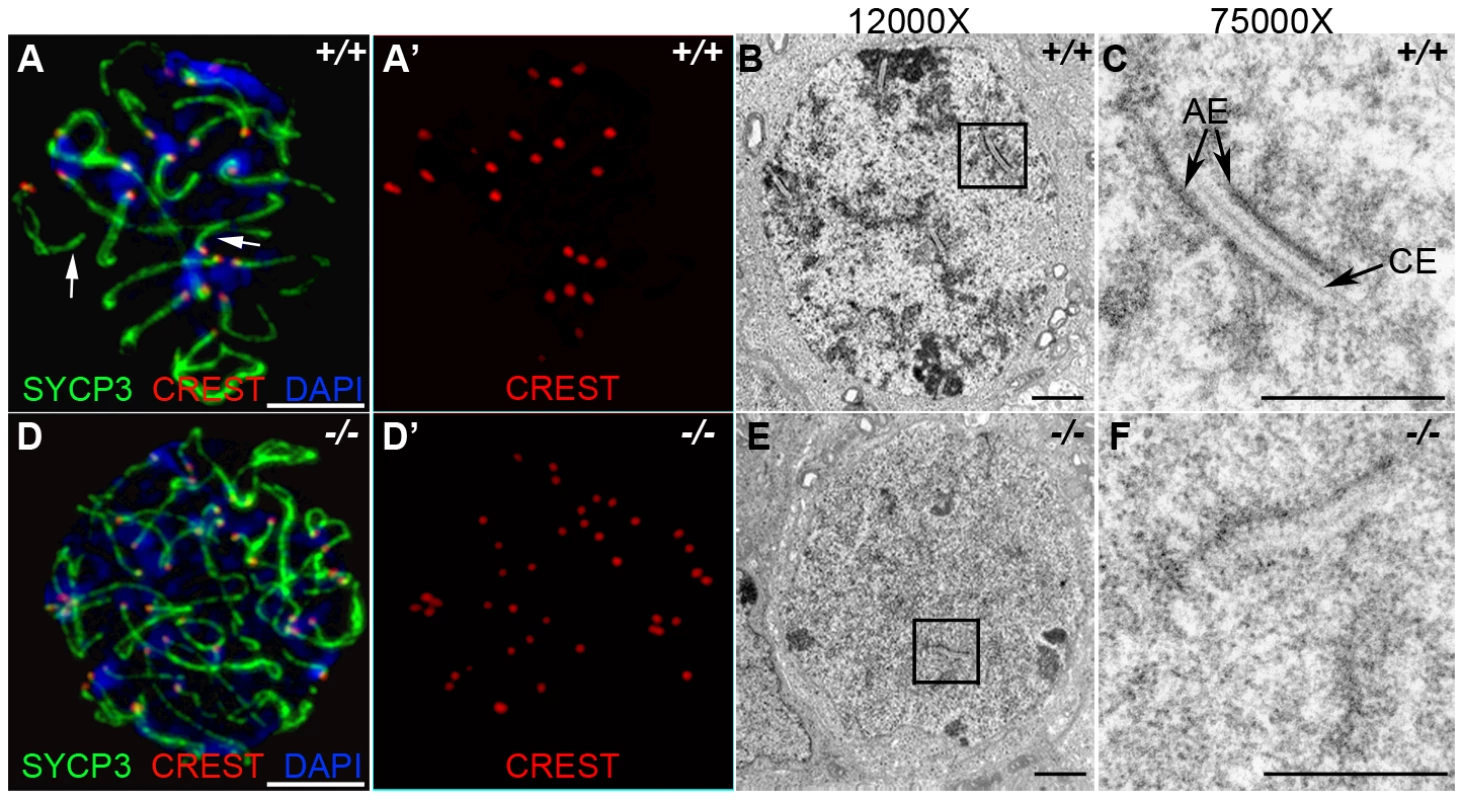
(A and D) Immunofluorescence with CREST (red) and anti-SYCP3 (green) antibodies. Anti-CREST antibody recognizes chromosome centromeres. Synapsed wild-type zygotene spermatocytes contain about 20 CREST foci (n = 50) (A-A'). However, Hormad1−/− spermatocytes contain approximately 40 CREST foci (n = 50), an indication that synapsis is disrupted (D-D'). Arrow indicates chromosome synapses in the wild-type. Scale bars: 10 µm. (B–C, E–F) Electron microscopy analysis of synaptonemal complexes in 2 week old wild-type and Hormad1−/− spermatocyte at different magnifications. The typical, tripartite structure of the synaptonemal complex consists of one central element (CE) that connects with two axial elements (AE) (C). We did not identify normal tripartite synaptonemal complex structure in Hormad1−/− spermatocytes. Scale bars: 1 µm. Our experimental evidence strongly suggests that HORMAD1 localizes to the axial core and is yet another critical component of the synaptonemal complex. To determine the effect of Hormad1 deficiency on the structure of the synaptonemal complex, we visualized synaptonemal complexes during meiosis I in wild-type and Hormad1−/− spermatocytes using electron microscopy. In the wild-type, synaptonemal complexes were well visualized during the pachytene stage of meiosis (Figure 4B and 4C). Electron microscopy examination of one hundred and ten Hormad1−/− spermatocytes from three independent experiments revealed the lack of the typical tripartite synaptonemal complex structure (Figure 4E and 4F). Persistence of pre-synaptic number of centromeric foci (CREST staining) in Hormad1−/− spermatocytes, as well as non-visualization of the tripartite synaptonemal complex structure by electron microscopy, demonstrate that HORMAD1 is essential for chromosomal synapsis.
Hormad1 deficiency disrupts localization of proteins important in early recombination
Previous studies have indicated that Sycp3 deficiency has subtle effects on meiotic recombination [27]. Early recombination events do not seem to be disrupted in Sycp3, as similar number of DMC1 foci are present in Sycp3 mutant and wild-type meiosis [29]. DMC1 is a meiotic specific recombinase that together with ubiquitously expressed RAD51 catalyzes homologous pairing and DNA strand exchange [30]–[31]. These early steps in recombination are critical for establishing the physical connections between homologous chromosomes during meiosis. Hop1 has been implicated in modulating the formation and processing of double stranded breaks [19]. We examined formation of DMC1, RAD51, and RPA foci in zygotene stage Hormad1−/− spermatocytes. There is a dramatic decrease in the number of DMC1 foci as compared to the wild-type (Figure 5A and 5B). We counted a total of 98.9±28.2 DMC1 foci in the wild-type spermatocytes (n = 50) and 9.28±3.9 DMC1 foci in the Hormad1−/− spermatocytes (n = 45). The number of RAD51 foci was also decreased from 189.3±31.8 in the wild-type spermatocytes (n = 50), to 69.3±34.5 in the Hormad1−/− spermatocytes (n = 40) (Figure 5C and 5D).
Fig. 5. Hormad −/− spermatocytes are defective in early recombination. 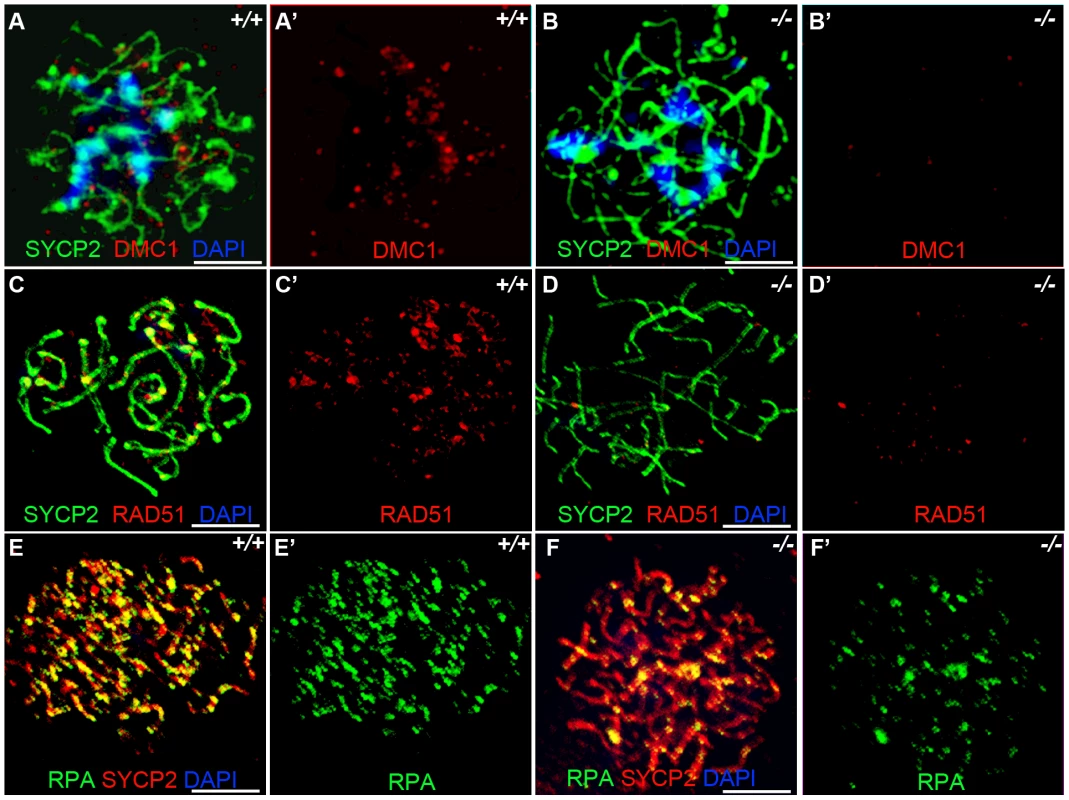
(A-B') Immunofluorescence assay with anti-SYCP2 (Green) and anti-DMC1 (Red) antibody in zygotene spermatocytes. (C-D') Immunofluorescence assay with anti-RAD51 (Red) and anti-SYCP2 (Green) antibody in zygotene spermatocytes. (E-F') Immunofluorescence assay with anti-RPA (Green) and anti-SYCP2 (Red) antibody in zygotene spermatocytes. DMC1 catalyzes DNA strand exchange during recombination, and DMC1, RAD51 and RPA foci mark early recombination events. DMC1, RAD51 and RPA foci are drastically decreased in Hormad1−/− spermatocytes. Scale bars: 10 µm. Following double-strand breaks formation by SPO11, RPA is recruited together with RAD51 to the single stranded DNA regions [32]. We counted a total of 194.5±64.5 RPA foci in the wild-type spermatocytes (n = 30) as compared to 70.1±38.5 foci in the Hormad1−/− spermatocytes (n = 20) (Figure 5E and 5F). These results indicate that early meiotic recombinational events were disrupted and not surprisingly, MLH1, a protein that forms foci in later stages of recombination and required for the formation of most of the crossovers (chiasmata) observed in mice [33], was dramatically reduced in Hormad1−/− spermatocytes (data not shown).
We also examined DMC1, RAD51 and RPA foci formation in female meiocytes at embryonic day 15.5 (E15.5). Embryonic ovaries contain zygotene to early pachytene oocytes at E15.5 [34]. We counted a total of 208.7±117.1 DMC1 foci in the wild-type E15.5 oocytes (n = 50), and 79.1±81.5 foci in the Hormad1−/− oocytes (n = 30) (Figure 6A and 6B), a total of 197.9±46.0 RAD51 foci in the wild-type oocytes (n = 50) versus 85.1±37.6 foci in the Hormad1−/− oocytes (n = 40) (Figure 6C and 6D) and a total of 317.16±135.3 RPA in the wild-type oocytes (n = 50) and 51.7±48.8 in the Hormad1−/− oocytes (n = 50) (Figure 6E and 6F). DMC1, RAD51 and RPA foci are therefore, similar to our observations in spermatocytes, significantly decreased in Hormad1 deficient female meiocytes.
Fig. 6. Hormad −/− fetal oocytes are defective in early recombination. 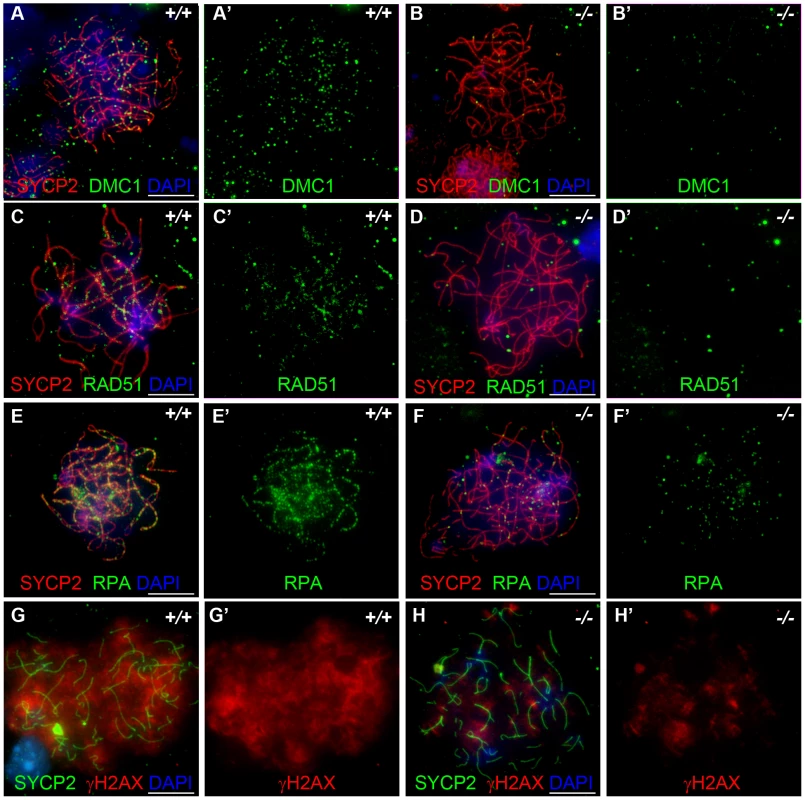
Chromosome spread assay in the wild-type and Hormad1−/− E15.5 fetal ovary. A-E represent zygotene stage, G and H represent leptotene stage. (A-B') Immunofluorescence assay with anti-SYCP2 (Red) and anti-DMC1 (Green) antibody. (C-D') Immunofluorescence assay with anti-SYCP2 (Red) and anti-RAD51 (Green) antibody. (E-F') Immunofluorescence assay with anti-SYCP2 (Red) and anti-RPA (Green) antibody. (G-H') Immunofluorescence assay with anti-SYCP2 (Green) and anti-γH2AX (Red) antibody. DMC1, RAD51, RPA and γH2AX signals were significantly decreased in Hormad1−/− fetal oocyte. DNA was stained with DAPI (Blue). Scale bars: 10 µm. These results indicate that homologous recombination is significantly affected in Hormad1−/− mammalian germ cells, as previously reported for HOP1 [35]. We also observed effects of Hormad1 deficiency on γH2AX staining (a phosphorylated form of histone H2AX), a well known surrogate marker for DSB formation [36]. In the leptotene stage, phosphorylation of H2AX is induced by SPO11 catalyzed DSBs in meiotic DNA, and γH2AX appears as large, cloud-like patterns thatdisappearat the pachytene stage [37]. At the leptotene stage, γH2AX staining in Hormad1−/− spermatocytes was significantly decreased (76% decrease in signal intensity) as compared to the wild-type (Figure 7A–7D). γH2AX staining was also significantly decreased in Hormad1−/− fetal oocytes (71% decrease in signal intensity) (Figure 6G and 6H). These results suggest that similar to Hop1 mutants, DSBs do not efficiently form in Hormad1 mutants.
Fig. 7. HORMAD1 disrupts γH2AX, BRCA1, and ATR localization to the XY chromosomes. 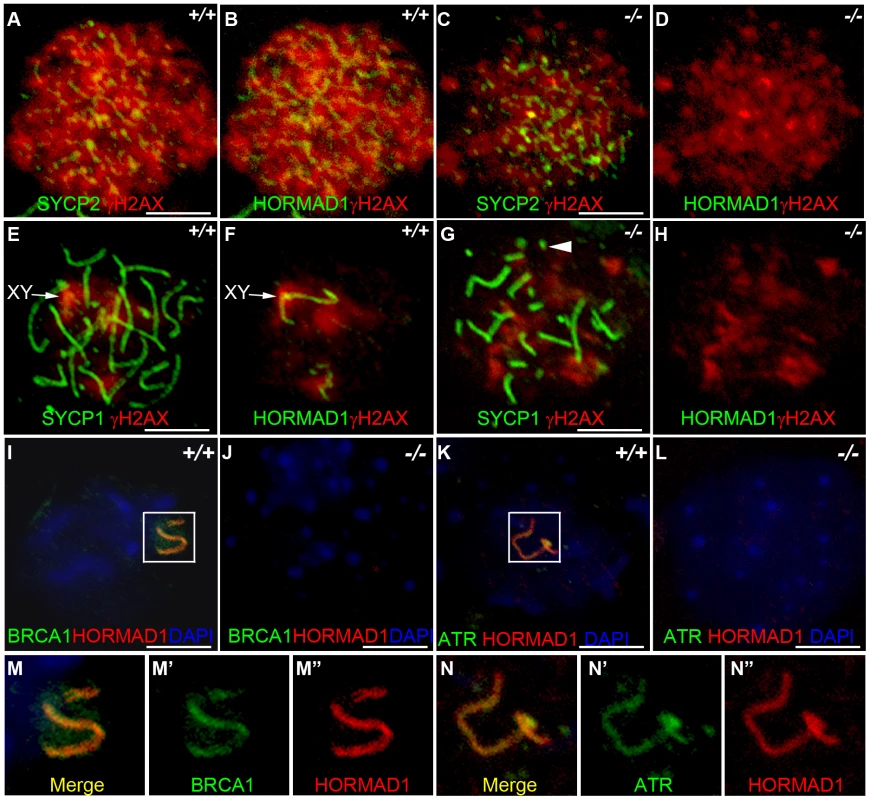
Chromosome spread assay was performed on wild-type and Hormad1−/− leptotene (A-D), zygotene (E-H) and pachytene (I-L) spermatocytes to determine effects of HORMAD1 deficiency on γH2AX, BRCA1 and ATR localization to the sex chromosomes. (A-D) Immunofluorescence assay with anti-SYCP2 (Green) or anti-HORMAD1 (Green) and anti-γH2AX (Red). (E-H) Immunofluorescence assay with anti-SYCP1 (Green) or anti-HORMAD1 (Green) and anti-γH2AX (Red) antibody. (C and D) At the leptotene stage, γH2AX and SYCP2 localization to chromatin can be detected in Hormad1−/− spermatocytes, however, γH2AX is substantially reduced. (F and H) γH2AX staining localizes preferentially to the sex chromosome in the wild-type pachytene stage spermatocytes but no preferential localization to the sex chromosomes was observed in the Hormad1−/− spermatocytes, which is not surprising since sex body does not form in Hormad1 mutants. (G) Arrow head indicates truncated axial fiber. (I, M-M”) BRCA1 and HORMAD1 co-localize to the sex chromosomes, but Hormad1 deficiency disrupts BRCA1 localization (J). Similarly, ATR co-localizes with HORMAD1 to the XY chromosomes (K, N-N”), and Hormad1 deficiency disrupts ATR localization (L). These results indicate that HORMAD1 is upstream of the currently known critical components of the meiotic sex chromosome inactivation complex, γH2AX, BRCA1 and ATR. DNA was stained with DAPI (Blue), and arrows indicate sex chromosomes. Scale bars: 10 µm. HORMAD1 is a germ cell–specific component of the meiotic sex chromosome inactivation complex
HORMAD1 protein localization along autosomes was previously shown to be transient [17] (Figure 7B and 7F). HORMAD1 staining was highest along unsynapsed chromosome axis in the zygotene to pachytene stage and diminished significantly along autosomes in the pachytene [17]–[18] (Figure 7F and 7I). Interestingly, HORMAD1 localized strongly along desynapsed autosomes in diplotene meiocytes [17]. During the pachytene stage, HORMAD1 is faintly visible along the autosomes, but co-localizes strongly with γH2AX on the XY chromosomes, and specifically along the axial elements [17]–[18]. γH2AX is a phosphorylated form of histone H2AX, and a marker for DSBs [36], [38]. H2AX is phosphorylated throughout the chromatin in leptotene spermatocytes and by the pachytene stage [17]–[18], γH2AX staining is undetectable on autosomes and restricted to the sex body (Figure 7E and 7F) [39]. H2ax knockout shows the essential role for H2AX in sex body formation and meiotic sex chromosome inactivation [39]. Meiotic sex chromosome inactivation also involves ATR and BRCA1 dependent phosphorylation of H2AX [18], [40]. Interestingly, Hormad1 deficiency, similar to H2ax deficiency, abolishes the formation of the sex body (Figure 7G and 7H, and data not shown). The lack of sex body formation is most likely due to the disruption of Hormad1−/− spermatocytes prior to the pachytene. We examined the γH2AX localization in the wild-type and Hormad1−/− spermatocytes. Chromatin in Hormad1−/− spermatocytes stained with anti-γH2AXantibodies, but no preferential localization to the sex chromosomes was observed (Figure 7G and 7H). BRCA1 and HORMAD1 have recently been shown to co-localize in the sex body [18]. We determined whether BRCA1 localization is dependent on HORMAD1. BRCA1 protein could not be localized in Hormad1−/− spermatocytes (Figure 7I–7J, 7M). This finding is unlike H2ax knockout, where BRCA1 is still detected on the sex chromosomes despite the lack of the sex body [40]. We also examined ATR localization in wild-type and Hormad1−/− testes. In wild-type testes, HORMAD1 and ATR co-localized in the sex body, but we could not detect ATR in Hormad1−/− spermatocytes (Figure 7K–7L, 7N). Above data suggest that HORMAD1 may be involved in the recruitment of BRCA1, ATR and γH2AX to the sex chromosome.
Since ATR, BRCA1 and γH2AX are involved in the transcriptional silencing of sex chromosomes, we examined whether Hormad1 deficiency affects transcriptional repression. Previous studies have shown that H2ax and Brca1 deficiencies individually, lead to the over-expression of genes exclusively expressed from the X or Y chromosome [40]. We examined whether X-linked germ cell–specific genes were over-expressed in Hormad1−/− testes compared to wild-type testes. We performed quantitative real-time PCR on select autosomal genes (Hormad2, Rnh2 and Mov10l1) as well as X-chromosome derived genes (Usp26, Fthl17, Pramel3, Tex11, and Tex13) on wild-type and Hormad1−/− testes (Figure 8A–8H). Rnh2 and Mov10l1 are germ cell–specific transcripts, derived from autosomes, and were not differentially expressed between wild-type and Hormad1−/− testes (Figure 8B and 8C). In contrast, all of the germ cell–specific transcripts transcribed from the X chromosome were significantly elevated in Hormad1−/− testes over the wild-type. These include Usp26 (4.5 fold increase), Fthl17 (6.5 fold increase), Pramel3 (3.5 fold increase), Tex11 (2.2 fold increase), and Tex13 (2.8 fold increase) (Figure 8D–8H). Moreover, RNA expression microarray analyses comparing two week old Hormad1 deficient testes with corresponding wild-type, indicate that almost 20% of the up-regulated genes derive from the X chromosome (Figure 8I). Our results are remarkably similar to transcriptional de-repression observed in H2ax and Brca1 mutants [39]–[40], and indicate that HORMAD1 is a germ cell and meiosis specific factor critical in meiotic sex chromosome inactivation and transcriptional silencing.
Fig. 8. Sex chromosome expressed transcripts escape meiotic sex chromosome inactivation in Hormad −/− spermatocytes. 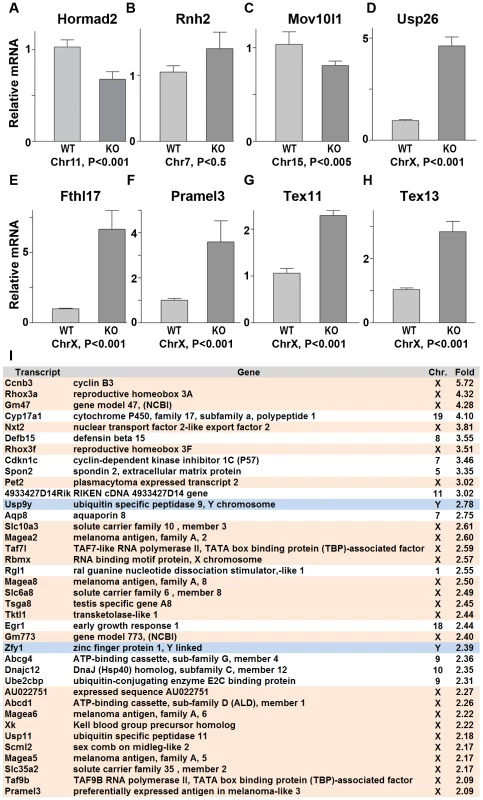
(A–H) Quantitative real time PCR analyses show that expression of testes specific genes derived from the autosome, Hormad2, Rnh2, and Mov10l1, did not significantly differ between the wild-type and the Hormad1−/− testes (A-C). However, X chromosome linked, testis specific genes, Usp26, Fthl17, Pramel3, Tex11 and Tex13 transcript were 2–6 fold increased (D-H). (I) Microarray analysis shows preponderance of X and Y derived transcripts among the top 38 up-regulated genes in Hormad −/− testes. Data are normalized to β-actin expression and presented as the mean relative quantity (compared to wild-type) with error bars representing the standard error of mean. Student's t test was used to calculate the P values. Hormad1 deficiency abrogates ATM autophosphorylation
HORMAD1 is a likely mammalian counterpart to the yeast HORMA domain meiotic protein, Hop1. Hop1 is phosphorylated by Mec1/Tel1, the budding yeast homologue to the mammalian ATR and ATM kinases, and this phosphorylation is thought to play an important role in inter-homologous recombination [21]. We therefore examined HORMAD1 expression in Atm deficient mice as well as ATM expression in Hormad1−/− animals. ATM is a serine/threonine-specific protein kinase that has been associated with cell cycle regulation, apoptosis, and response to DNA damage repair. ATM kinase activation is associated with increased auto-phosphorylation of ATM at multiple sites including serine 1981 [41]. We examined HORMAD1 protein expression in testes between postnatal days 5–21 (Figure 9A). Eight to ten day old testes contain spermatogonial cells as well as preleptotene/leptotene spermatocytes [34]. At postnatal day 14–18, testes contain pachytene spermatocytes and visible sex bodies [34]. HORMAD1 is known to be phosphorylated [17] (Figure 9B). Western blot analysis on testes extracts detected phospho-HORMAD1 beginning at postnatal day 14 (Figure 9A). Phospho-HORMAD1 protein was decreased in postnatal day 18 and 21 wild-type testes (Figure 9A). Phospho-HORMAD1 appearance correlates temporally with sex body formation. HORMAD1 phosphorylation was not affected by Atm deficiency (Figure 9A). These results indicate that ATM is not responsible for HORMAD1 phosphorylation. We also examined HORMAD1 localization in Atm−/− spermatocytes. We observed HORMAD1 localization to chromosomal axes in Atm deficient spermatocytes, as previously described by others [17] (Figure 9C and 9D). Since synaptonemal complexes do not form in Atm mutants, HORMAD1 association with unsynapsed chromosomes does not require ATM. The anti-ATM phospho-S1981 antibody did not detect phosphorylated ATM in Hormad1−/− testes (Figure 9E and 9F). These results suggest that HORMAD1 is upstream of ATM auto-phosphorylation, and therefore likely upstream of ATM kinase activation.
Fig. 9. HORMAD1 phosphorylation is unaffected by Atm deficiency while Hormad1 deficiency disrupts ATM autophosphorylation. 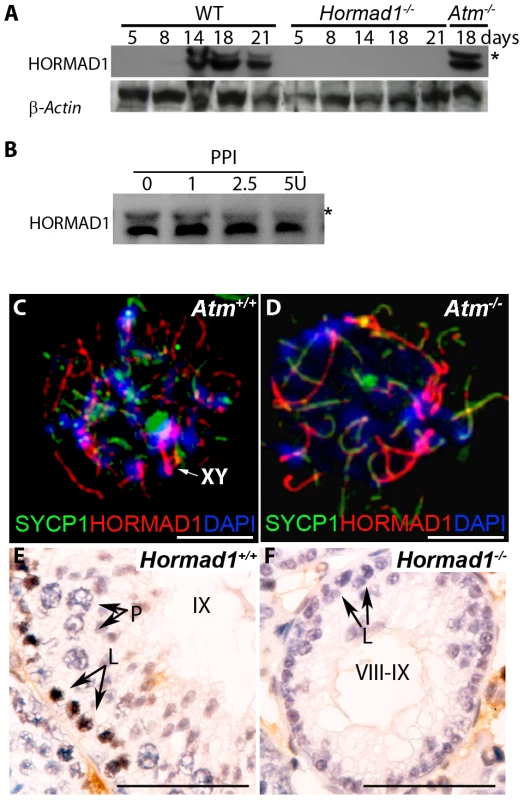
(A) Western blots analyses with anti-HORMAD1 specific antibodies show that HORMAD1 protein and its phosphorylated form (*) appear circa post-natal day 14. Atm1 deficient testes (Atm−/−) express both forms of HORMAD1 and HORMAD1 is therefore unlikely to be ATM1 substrate. (B) The presumed phosphorylated form of HORMAD1 (higher molecular weight band indicated by asterisk) decreases in intensity after protein phosphatase I (PPI) treatment. (C and D) Chromosome spread assay in wild-type (Atm+/+), and Atm deficient spermatocytes (Atm−/−). HORMAD1 preferentially localizes to unsynapsed regions of chromosome axes (C), and HORMAD1 antibody stains unsynapsed chromosome axes in Atm−/− spermatocytes (D) more intensely than in the wild-type spermatocytes. Scale bars: 10 µm. (E and F) Immunohistochemistry analysis in 6 week old wild-type and Hormad1−/− testes with anti-phospho ATM-S1981 antibody. Phospho-ATM S1981 was detected in wild-type zygotene, but not detected in Hormad1−/− zygotene spermatocytes. Scale bars: 50 µm. Hormad1 deficiency does not affect gross ovarian development
We have previously shown that Hormad1 RNA expression in the ovary was confined to the germ cell [16]. Meiosis I in the female gonad commences circa E13.5 and most oocytes arrest at the dictyate stage by the time of birth. Antibodies against HORMAD1 recognized HORMAD1 protein at E14.5 (leptotene) and E18.5 (arrest in diplotene) oocytes (Figure 10A and 10B), but little HORMAD1 protein was detected in the newborn ovary oocytes (Figure 10C), at the time when oocytes are arrested in diplotene. Deficiency in genes critical in meiosis can disrupt early ovarian development, as is the case for Dmc1, Msh5, Spo11 and Atm [42]–[44].
Fig. 10. HORMAD1 is expressed in embryonic but not post-natal oocytes. 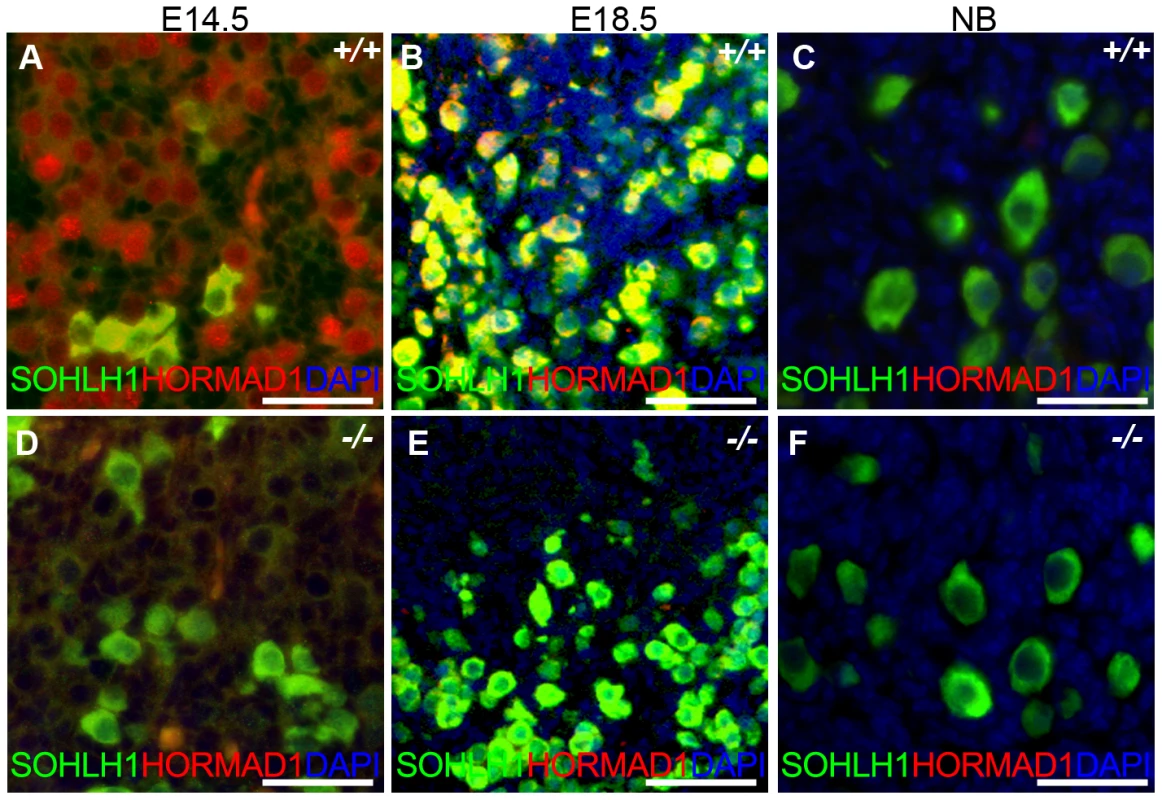
(A–C) HORMAD1 protein was detected by immunofluorescence in the embryonic ovary at E14.5 (A), E18.5 (B), but not in the newborn ovary (C). (D–F) Oocytes in Hormad1−/− ovary (−/−) were stained with antibodies against the germ cell–specific transcriptional regulator SOHLH1 at E14.5 (D), E18.5 (E), and newborn ovary (F). DNA was stained with DAPI (Blue). Scale bars: 20 µm. We therefore examined ovarian development in Hormad1−/− females. Antibodies against germ cell–specific transcriptional regulator SOHLH1 [45] stained wild-type and knockout oocytes throughout embryonic gonadal development with no significant differences noted (Figure 10A–10F). Moreover, the numbers of primordial, primary and secondary follicle counts did not significantly differ between the mutant and wild-type ovaries at post-natal day 8 (Figure S4). These results indicate that Hormad1 deficiency does not affect embryonic ovarian development, germ cell cyst breakdown, and primordial follicle formation. We also examined the histology of wild-type and Hormad1−/− ovaries between 2 and 30 weeks of life. Mice reach sexual maturity around 6 weeks of life, and mouse ovaries at this time consist of all of the follicular types including corpora lutea, an indication that the ovaries are ovulating. Postnatal Hormad1−/− ovaries were grossly indistinguishable from wild-type mice between 2 and 30 weeks of life, with abundant corpora lutea in the Hormad1−/− ovaries indicating that the normal process of oocyte maturation was not disrupted, and ovulation has occurred (Figure 11A–11H). We induced superovulation in knockout and wild-type mice with exogenous gonadotropins to determine whether subtler ovarian defects contributed to infertility in Hormad1−/− mice. Hormad1−/− females super-ovulated 28±11 eggs (n = 22), while wild-type animals superovulated 29±14 eggs (n = 13). We therefore did not observe significant difference between the number of eggs superovulated from wild-type versus knockout mice. These results indicate that ovarian development is grossly normal in Hormad1−/− mice, and that ovarian defects are unlikely to account for observed infertility.
Fig. 11. Histological analysis of wild-type and Hormad1–/– ovaries. 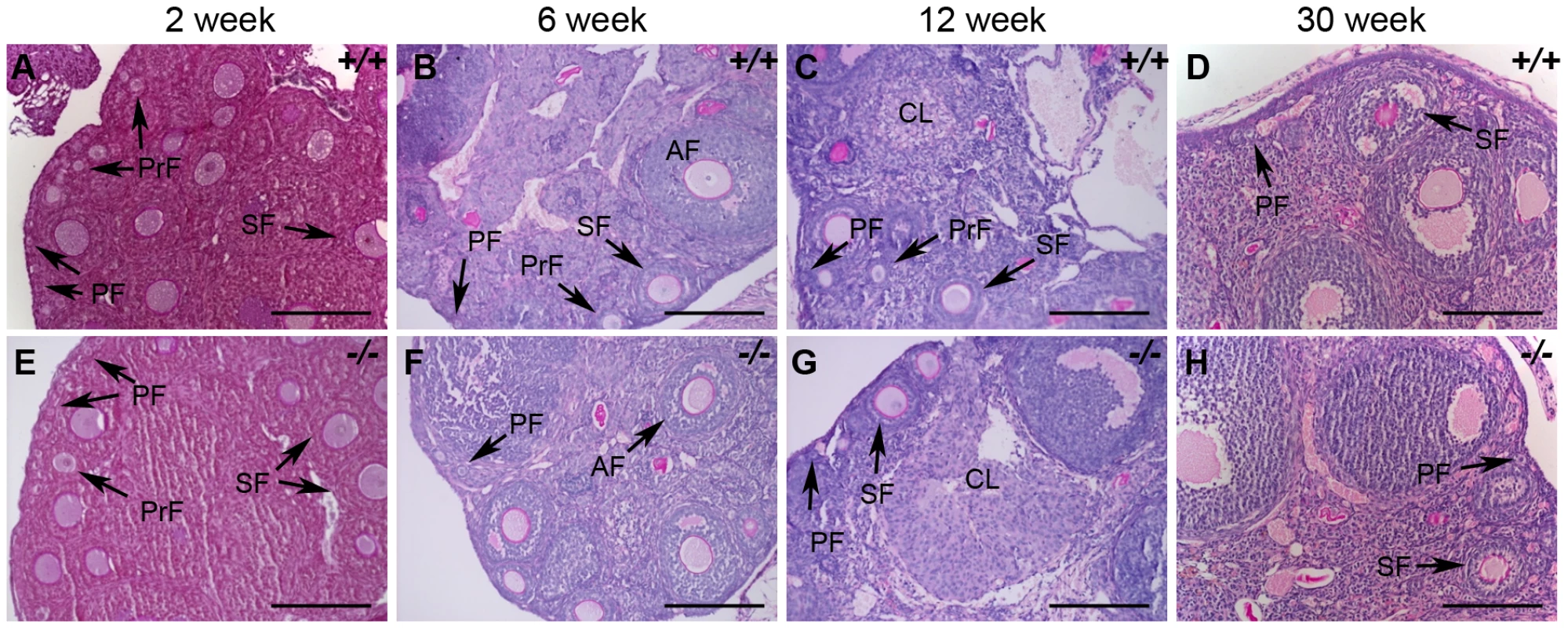
(A–H) PAS staining of wild-type (+/+) and Hormad1−/− (−/−) ovaries does not show gross histological differences. Wild-type and Hormad1−/− ovaries are shown at different post-natal ages that show the full range of ovarian follicles: primordial follicles (PF), primary follicles (PrF), secondary follicles (SF), antral follicles (AF) and corpus luteum (CL). Scale bars: 50 µm. Hormad1 deficient eggs fertilize and embryonic development arrests at blastocyst stage due to aneuploidy
We studied early embryonic development of fertilized Hormad1−/− oocytes because ovarian defects were unlikely to explain observed infertility. We recovered embryos from wild-type male matings with Hormad1−/− females at E0.5, E1.5, E2.5, and E3.5. Comparable numbers of morphologically indistinct 1-cell zygotes were recovered from oviducts of control and mutant female mice at E0.5 (Figure 12A–12C), and little difference was noted at the 2-cell stage, except for an increased number of 1-cell embryos in the knockouts, indicating a lag in the progression from the 1-cell to 2 - cell stage (Figure 12D–12F). By E2.5, the number of normal appearing 8-cell stage embryos was significantly less in Hormad1−/− fertilized eggs as opposed to the wild-type (Figure 12G–12I), and no morphologically normal blastocysts were observed in the Hormad1−/− fertilized eggs at E3.5. It is interesting to note that at E3.5, a significant number of 4 and 8 cell stage embryos were observed in the knockout while only blastocysts were observed in the wild-type E3.5 embryos (Figure 12J–12L). We also tested, using Chicago Sky Blue 6B dye (Sigma, MO, USA) injection into the tail vein [46], whether blastocysts derived from Hormad1−/− fertilized eggs could implant. We did not detect implantation of Hormad1−/− fertilized eggs (Figure 12M). These results indicate that a defect in early embryogenesis led to premature loss of embryos.
Fig. 12. Hormad1–/– oocyte derived embryos arrest at blastocyst stage. 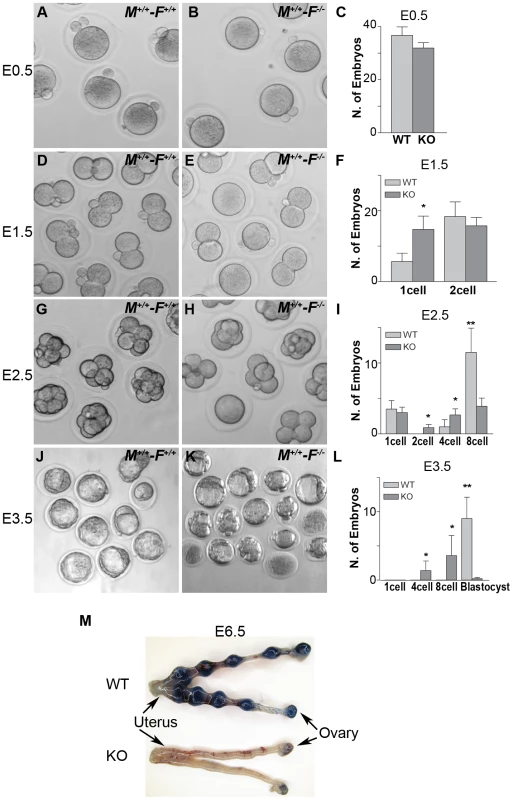
Embryos derived from wild-type matings (M+/+-F+/+) and matings involving Hormad1−/− oocytes fertilized by wild-type males (M+/+-F−/−), were collected at E0.5, E1.5, E2.5 and E3.5 for the analysis of the early embryonic development. (A-C) No significant differences were noted between wild-type and Hormad1−/− at E 0.5, with normal appearing one cell embryos and polar bodies visible in both. (D-F) At E1.5, there is an excess of 1 cell embryos in Hormad1−/− fertilized oocytes, indicating a lag in embryo development at this stage. (G-I) At E2.5 there is a significantly less number of eight cell embryos formed in Hormad1−/− fertilized oocytes as opposed to the wild-type. A large proportion of 8-cell embryos appear disorganized and apoptotic. (J-L) Blastocysts form at E3.5 in wild-type but very few normal blastocysts are visible in Hormad1−/− embryos. The count only includes morphologically normal blastocysts (L). Error bars represent standard error of the mean. Student's t test was used to calculate P values (*:P<0.5, **:P<0.001). (M) Embryos derived from Hormad1−/− fertilized oocytes can not implant, as visualized by Sky Blue dye injected into the tail vein of pregnant females at E6.5 gestational age. Hormad1 deficiency causes aneuploidy and early embryonic demise
We hypothesized that aneuploidy is the major cause of embryo wastage in Hormad1−/− females. Unlike meiosis in testes, errors during oocyte meiosis have milder effect on oocyte loss and apoptosis, as has been observed in Sycp3 and Smc1β knockouts [27], [29], [47]. The presence of growing oocytes in the Hormad1−/− ovaries, as opposed to early germ cell loss observed in testes during pachytene stage of meiosis, indicates greater tolerance of Hormad1 deficiency in the oocytes as compared to spermatocytes. The whole range of ovarian follicle types are present in the Hormad1−/− ovaries, including primordial follicles, primary, secondary and antral follicles. In contrast to other critical genes during meiosis, such as Dmc1, Msh5, Spo11 and Atm that cause early ovarian failure due to rapid loss of oocytes following disruption of meiosis I [42]–[44], Hormad1 deficiency does not activate apoptotic pathways and does not lead to gross premature loss of oocytes. We used chromosome specific mouse BAC (bacterial artificial chromosome) clones to perform fluorescent in situ hybridization (FISH) on germinal vesicle (GV) oocytes, 2 cell and 8 cell stage embryos to determine whether aneuploidy is significantly higher in Hormad1−/− embryos. Wild-type GV oocytes are arrested in meiosis I and contain bivalent chromosomes that consist of four chromatids. Examination of 58 GV oocytes from three independent knockout animals and 112 GV oocytes from three wild-type animals with BAC probes specific for chromosomes 19, 18 and X, revealed no significant differences, with presence of four chromatids for each of the chromosome examined as expected (Figure 13A–13C). Following fertilization and completion of meiosis II, each cell in the developing embryo should contain two chromosomes except for the sex chromosomes. We examined by FISH, mouse chromosomes 6 and X in the 2-cell wild-type and Hormad1−/− embryos. Sixty three cells examined for chromosome 6 in the Hormad1−/− 2-cell embryos revealed that 22% of the cells had 4 signals corresponding to chromosome 6, 35% had 3 signals corresponding to chromosome 6, 24% had 1 signal, and 5% had no signal. Wild-type 2-cell embryos were also examined by chromosome specific FISH, and 44 cells examined out of 46 showed only 2 signals, as expected. In 2-cell Hormad1−/− embryos, out of sixty three cells examined for chromosome X, 44% showed four, three or no signal. Wild-type 2-cell stage embryos, as expected, showed only 2 or 1 signal, consistent with either XX or XY sex of the cell. These results indicate widespread hypo and hyperdiploidy in 2-cell embryos (Figure 13D–13F).
Fig. 13. Hormad1–/– fertilized oocytes show widespread hypo- and hyperdiploidy. 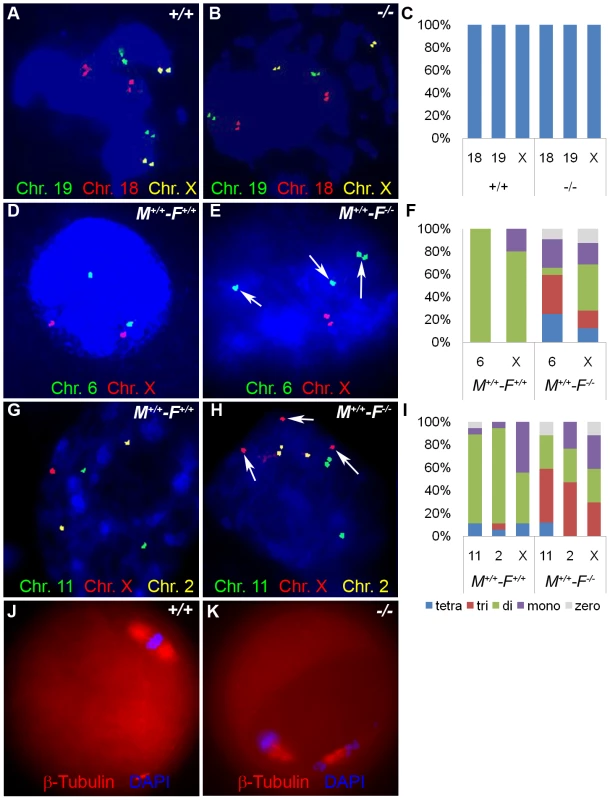
(A-B) In GV stage oocytes, parental chromosomes have replicated to produce identical sister chromatids, which are visualized as closely located double signals for examined chromosomes 18, 19 and X. (C) Histogram of individual chromosomes, does not show significant difference between the wild type and mutants. Fifty eight knockout GV oocytes and 112 wild type GV oocytes were scored. Abbreviations stand for the following: tetra-Tetrasomy (blue), tri-Trisomy (red), di-Disomy (green), mono-Monosomy (purple), zero-Zerosomy (grey). (D-F) E1.5 day (2-cell stage embryo) FISH assay using chromosomes 6 (green) and X-specific DNA probes (red). The wild type embryos were diploid for 6 and X (D). Trisomy for chromosome 6 (arrows) is noted in the knockout (E). In addition, chromosomes 6 and X demonstrate double hybridization signals, the same FISH pattern as observed during the GV stage, and indicative of sister chromatids. (F) Histograms for individual chromosomes show high prevalence of chromosomal aneuploidies in embryos derived from mutant oocytes. We scored 63 knockout and 46 wild type cells. (G-I) E2.5 day (8-cell stage embryo) FISH assay with chromosomes X (red), 11 (red) and 2 (yellow) specific probes. (G) A hybridization pattern of a normal diploid number of chromosomes in the wild type embryo. (H) Trisomy for chromosome X (arrow), as well as double signals likely produced by sister chromatids on chromosome 2 and 11 were observed in embryos derived from knockout eggs. (I) Histogram shows substantial number of trisomies and monosomies in the knockout as compared to the wild type. We scored 84 wild type and 96 knockout cells. (J and K) Immunofluorescence with β-tublin antibody (red) in wild-type and Hormad −/− MII stage oocytes. Hormad −/− oocytes are grossly disrupted with mis-orientation of the chromatid attachment to the spindle. DNA was stained with DAPI (Blue). We also examined cells from the 8-cell stage embryos. A total of 84 cells from the Hormad1−/− embryos were examined with FISH probes specific for chromosomes 11, X and 2. Hormad1−/− cells from the 8-cell stage, hybridized with BAC specific to chromosome 11, showed 4 signals in 10% of the cells, and 3 signals in 55% of the cells. Therefore, a total of 65% of the cells were either monosomic or trisomic for chromosome 11. In the wild-type 8 cell stage embryo, a total of 96 cells were scored for chromosome 11, and only 2 out of 96 (2%) cells showed a single signal, while 94 out of 96 cells (98%) showed two signals as expected. Similar results were obtained for chromosome 2 (Figure 13G–13I). Our results indicate widespread aneuploidy involving different chromosomes in both 2-cell and 8-cell embryos. Such aneuploidy causes early embryo demise and failure of implantation.
We also visualized second metaphase (M2) spindle using anti-β-tubulin antibodies in Hormad1−/− oocytes. During M2, sister chromatids migrate to the opposite pole to form functional, haploid gametes. Sister chromatids are bi-oriented on the spindle and congregate in the center of the spindle prior to separation (Figure 13J). Each sister kinetochore in a pair is attached to the opposite spindle, and such arrangement generates sufficient tension to cause proper segregation. M2 in Hormad1−/− oocytes is grossly disrupted with mis-orientation of chromatid attachment to the spindle observed in all M2 oocytes examined (Figure 13K) (n = 50). These results indicate importance of Hormad1 in proper spindle formation and segregation.
Discussion
We discovered Hormad1 (Nohma), using in silico screen to identify germ cell–specific transcripts critical for gonadal development [16]. We showed that Hormad1 expression was confined to germ cells, and that Hormad1 transcripts were mostly concentrated in the pachytene spermatocytes and early oocytes. Here we show that Hormad1 is essential for both male and female fertility in the mouse. HORMA domain-containing proteins interact with chromatin, particularly chromatin associated with DNA adducts, and are critical mitotic spindle checkpoint proteins [5]. HORMA domain-containing proteins are critical regulatory proteins for mitosis as exemplified by MAD2 protein, and meiosis as exemplified by Hop1p [8], and Red1p [9] in yeast, Him-3 [10] in nematodes, and Asy1 [11] in plants. The 205-aa HORMA domain in the mouse HORMAD1, shares highest similarity to the Saccharomyces cerevisiae Hop1 protein (28% amino acid similarity). Yeast Hop1 mutants have defects in chromosome condensation, synapsis and recombination [4], [8], and Hop1p binds DSBs during meiotic prophase and appears to play an important role in interhomologue recombination. We previously hypothesized, based on homology to Hop1 in yeast, that Hormad1 plays an important role in mammalian meiosis. Our results indicate that HORMAD1 is the mammalian homologue to the Hop1 protein in yeast.
Our current studies, and those of others, have shown that HORMAD1 protein is confined to germ cells and co-localizes with synaptonemal complex proteins SYCP2 and SYCP3 on the chromatin as part of the axial core [27], [48]. Synaptonemal complex is a complex structure composed of multiple germ cell–specific and ubiquitously expressed proteins that connect paired homologous chromosomes. Similar to railroad tracks, the synaptonemal complex axial lateral elements (SYCP2, SYCP3) are connected to each other by proteins known as transverse filaments (SYCP1, TEX12) [1]. The axial/lateral elements play critical roles in chromosome condensation, pairing, and repress recombination pathways that involve sister chromatids [1]. Synaptonemal complex formation is associated with HORMAD1 depletion from the axes [17]–[18]. Our data show that HORMAD1 phosphorylation peaks at the time when HORMAD1 localization shifts to the sex body. ATM and ATR do not appear to be involved in HORMAD1 phosphorylation and binding to chromosomal axes. Previous study showed that Trip13, a homologue of yeast Pch2 kinase, may be involved in HORMAD1 depletion from synapsed chromosome axes [17]. Kinases, such as Mec1/Tel1 in yeast are important to effect inter-homologous recombination [21] by phosphorylating Hop1, but not much is known regarding HORMAD1. Future studies are necessary to understand HORMAD1 phosphorylation sites and responsible kinases.
Although HORMAD1 is not essential for the binding of well-characterized germ cell–specific synaptonemal complex proteins such as SYCP1, SYCP2 and SYCP3, HORMAD1 deficient spermatocytes are defective in synapsis and do not form recognizable synaptonemal complexes. Hormad1 deficiency causes a male meiotic arrest, that is similar to male meiotic arrests observed in other components of the synaptonemal complex and recombination including: Atm, Spo11, Sycp1, Sycp2, Sycp3 and Dmc1 [27], [30]–[31], [48]–[50]. These findings are not surprising as HORMAD1 co-immunoprecipitates with known axial proteins, SYCP2, SYCP3, REC8, and SMC1β [18]. Whether these interactions are biologically significant, and whether HORMAD1 interacts with other members of the synaptonemal complex, including newly discovered HORMAD 2 protein, is unclear. Interestingly, Hop1 interacts with another phosphoprotein, Red1 [35], [51], and whether HORMAD2 is the mammalian counterpart of Red1 remains to be seen.
Like its counterpart in yeast, Hop1, Hormad1 deficiency disrupts early and later stages of recombination as shown by the drastic diminution in the formation of DMC1, RAD51, RPA and MLH1 foci. These findings differ from the Sycp3 mutant, which affects later stages of recombination [29]. DMC1 is a meiotic specific recombinase that together with ubiquitously expressed RAD51, catalyzes homologous pairing and DNA strand exchange, and marks earlier stages of recombination than MLH1 foci [30]. HORMAD1 expression is not significantly affected in Dmc1 mutant spermatocytes [17]. These results indicate that unrepaired DSBs and synaptonemal complexes between non-homologous chromosomes seen in Dmc1 mutants, do not affect HORMAD1 localization. In Hop1 mutants, DMC1 foci are only faintly detected when compared to controls [19]. These findings have led to a suggestion that Hop1 binds at or near the sites of DNA double strand break and modulates the action and perhaps recruitment of recombinases such as DMC1 and RAD51 [19]. Moreover, Hop1 appears to be part of the meiosis specific surveillance system that monitors meiotic double stranded break repair [21]. Our observations support the possibility that HORMAD1 affects early recombination and may therefore perform similar functions in the mammalian meiosis.
During mammalian meiosis, X and Y chromosomes are inactivated and transcriptionally silenced (meiotic sex chromosome inactivation, MSCI) and chromatin condenses to form a sex (XY) body. The epigenetics of MSCI involves γH2AX, BRCA1 and ATR at a minimum. The most intriguing finding regarding HORMAD1 is its co-localization with BRCA1, ATR and γH2AX in the sex chromosomes during pachytene [18]. Hormad1 deficiency, similar to H2ax deficiency, abrogates formation of the sex body, as well as γH2AX localization to the sex chromosomes. Hormad1 deficiency also abrogates BRCA1 and ATR localization to the sex chromosomes. Interestingly, H2ax deficiency does not affect BRCA1 localization to the sex chromosome, despite anomalous sex body, while BRCA1 deficiency does affect both ATR and γH2AX localization to the sex body. These findings indicate that HORMAD1 is involved in the initial recruitment of BRCA1, ATR and γH2AX to the sex body. The mechanism whereby HORMAD1 affects ATR, γH2AX and BRCA1 localization is unclear, but our results are consistent with the conclusion that HORMAD1 is upstream of ATR, γH2AX and BRCA1, and establishes HORMAD1 as an essential component of the MSCI complex. Moreover, we have shown here that Hormad1 deficient testes preferentially over-express X-linked genes as observed in mice deficient for other components of the MSCI complex such as γH2AX and BRCA1 [17]. Almost 20% of genes preferentially up-regulated from the Hormad1−/− testes are derived from the X chromosome. Hormad1 is therefore the first germ cell–specific component known to play a role in MSCI.
Unlike males, meiosis I in females arrests during the embryonic development at the diplotene stage and is completed upon ovulation. The lack of dramatic germ cell apoptosis during ovarian follicle development has also been observed for Sycp2 and Sycp3 deficient mice [48], [52], and reiterates the laxity of oocytes to early meiotic errors, as opposed to spermatogenesis, where meiotic check point causes the dramatic phenotype observed in male gonads. Hormad1 deficiency does not affect folliculogenesis nor ovulation. These findings are unusual, given that Hormad1 affects recombination, and recombination associated proteins DMC1 and MLH1 are important in early oogenesis. Unrepaired recombination intermediates or defects in homologous chromosome pairing and synapsis are likely causes of early oocyte loss [53]. Spo11 deficient animals can ameliorate observed oocyte losses in recombination defective mutants, presumably due to the failure of Spo11−/− deficient mice to form DSBs that initiate meiotic recombination. Spo11 deficiency does not affect HORMAD1 association with the chromosome axes [17], a finding similarly observed for Hop1 localization in Spo11−/− yeast [9]. Moreover, Hop1 mutants are required for full levels of DSBs formation [37], [54]. It is therefore possible that early loss of oocytes in Hormad1 deficient animals does not occur in part because unrepaired DSBs do not form. Reduced staining of γH2AX in Hormad1 mutant leptotene stage oocytes argues that unrepaired DSB formation is indeed reduced. However, unlike Spo11−/− ovaries that have abnormal folliculogenesis, Hormad1−/− ovaries are not defective in folliculogenesis and other mechanisms must protect Hormad1 mutant oocytes from early loss.
Infertility in Hormad1−/− females is due to early embryo demise. This is in stark contrast to males, where spermatocytes are eliminated due to the pachytene defect. The greater ability of oocytes to tolerate meiosis I errors is well known [55], and exemplified by Sycp3 and Smc1β knockouts [52], [56], which have defects in synapsis and recombination. Mechanisms that underlie oocyte's insensitivity to such errors are not well understood [57]. Embryos derived from Hormad1−/− oocytes fertilized with wild-type sperm, cannot implant. Examination of post-fertilization events in fertilized Hormad1−/− oocytes, showed significant abnormalities in early embryo development, including growth lag, and severe disruption in development prior to the blastocyst stage. Very few blastocysts were produced in fertilized Hormad1−/− oocytes, and most were abnormal in morphology. We utilized FISH hybridization against specific chromosomes to show widespread hypo and hyperdiploidy during early embryonic development. These results are reminiscent of Sycp3 and Smc1β knockout embryos. SYCP3 and SMC1β are involved in the synaptonemal complex formation, are germ cell–specific, and implicated in human aneuploidy. Sycp3 deficient females are subfertile, but one third of the embryos die in utero due to aneuploidy [29]. Smc1β encodes a meiosis specific component of the cohesin complex, important in sister chromatid cohesion and recombination [58]. Smc1β deficient females are sterile, like Hormad1−/− females, and meiosis continues until metaphase II [59].
However, unlike Smc1β−/−, Hormad1−/− ovaries do not lose oocytes by 6 months of age, and postnatal oocyte counts do not differ significantly from the wild type. Sycp3−/− ovaries show significant decline in the primordial follicle pool at postnatal day 8, indicating accelerated apoptosis of oocytes as germ cell clusters break down to form primordial follicles [52]. DSBs, as indicated by the assembly of the histone variant γH2AX, form in Smc1β−/− and Sycp3−/− meiocytes and may lead to higher rate of oocyte loss in these mutants [47], [52]. Hormad1 deficient ovaries do not display loss of primordial follicles. Hormad1−/− ovaries may not be losing oocytes at an accelerated rate due to our observations that DSBs, as assessed by γH2AX staining, are substantially reduced in Hormad1 mutants. Similar to Hop1 mutants in yeast, lack of HORMAD1 may derepress Dmc1-independent inter-sister repair pathway, resulting in efficient DNA break repairs [21]. The reduced DSBs in Hormad1−/− meiocytes, and de-repression of Dmc1-independent inter-sister repair pathways, may not affect regular apoptotic pathways that eliminate substantial number of oocytes during germ cell clusters breakdown to form primordial follicles at the time of birth, resulting in no decrease in Hormad1−/− primordial oocyte numbers.
Materials and Methods
Animal breeding
All murine experiments were carried out on the 129S7/SvEvBrd x C57BL/6 hybrid background. Litters were weaned at 3 weeks and breeding pairs set up at 6 weeks of age. One mating pair was placed per cage and inspected daily for presence of litters. All experimental and surgical procedures complied with the Guide for the Care and Use of Laboratory Animals and were approved by the Institutional Agricultural Animal Care and Use of Committee of Baylor College of Medicine and the Institutional Animal Care and Use of Committee of University of Pittsburgh.
Targeting construct, generation of transgenic mice, and genotyping
Targeting construct, electroporation and ES cell selection was done as previously described [60]–[61]. Exons 4 and 5 of the mouse Hormad1 gene were replaced with a neomycin resistance gene flanked by 5.3 kb (5′) and 6.2 kb (3′) homologous sequences (Figure S1A). Targeted ES cells were verified by Southern blotting, and microinjected into the C57BL/6 blastocysts to produce chimeric mice that carried the mutation into the germ line. These mice were mated with C57BL/6 wild-type mice to generate Hormad1 heterozygous animals that were subsequently crossed to produce F2 offspring for functional analysis. PCR genotyping was performed using the following primers: WT-forward (5′ - TCAAGACCAACCTGGGCTAC -3′) and WT-reverse (5′ - CCATGTGGGTTGTAGGGAGT -3′) to amplify a 196-nucleotide wild-type band, and KO - forward (5′ - TCAAGACCAACCTGGGCTAC -3′) and KO - reverse (5′ - GGGGAACTTCCTGACTAGGG -3′) to amplify a 505-nucleotide mutant band (Figure S1A).
Histology and immunohistochemistry analysis
Testes were fixed in Bouin's solution (Sigma-Aldrich, MO, USA) and ovaries were fixed in 10% buffered formalin or 4% paraformaldehyde. Fixed tissues were embedded in paraffin, serially sectioned (5 µm) and stained with hematoxylin and eosin or with Periodic Acid Schiff (PAS). At least five pairs of testes and ovaries of each genotype were subjected to gross and microscopic analyses for each time point. Germ cell cysts, primordial, primary, and secondary follicles were defined as described [45]. We used antibodies against SOHLH1 [45], PLZF ( sc-22839, Santa Cruz, CA,USA), DDX4 (ab13840, Abcam, MA, USA), ATM phospho-S1981 (05-740, Millipore, CA, USA), and HORMAD1 proteins. Affinity purified, anti-HORMAD1 guinea pig and rabbit antibodies, were generated against part of HORMAD1 protein (amino acids 23-373) at Cocalico Biologicals (Lancaster, PA, USA).
Meiotic chromosome analysis and immunostaining
Oocytes and spermatocytes for chromosome analyses were prepared essentially as described previously [62]–[63]. Ovaries and testes were incubated in trypsin-EDTA solution at 37°C for 15 min and washed briefly in PBS. Trypsinized ovaries and testes were pipetted repeatedly and centrifuged, followed by resuspension in PBS. Cell suspensions were placed on poly-L-lysine-coated slides containing 120 mM sucrose solution and 0.05% Triton X-100. The slides were fixed in 2% paraformaldehyde and 0.02% SDS for 1 hour at room temperature, washed in distilled water and air dried, and stored at −80°C before use. Immunostaining was preformed as described [62]–[63]. Slides were incubated with the primary antibody overnight at 4°C, washed with PBS and incubated with Alexa-488 and Alexa-595 (Invitrogen, CA, USA) secondary antibody (1∶300 dilutions) for 1 hour at room temperature. After washing with PBS, slides were mounted using VECTASHIELD medium with 4,6-diamidino-2-phenylindole (DAPI) (Vector Laboratories, CA, USA). SYCP2 rabbit and guinea pig immunoaffinity-purified antibody [48], DMC1 (sc-8973, Santa Cruz, CA, USA), RAD51 (ab1837, Abcam, CA, USA) and RPA (ab87272, Abcam, CA, USA) were used. Dr. Christer Höög (Karolinska Institutet, Sweden) kindly provided SYCP1 and SYCP3 rabbit immunoaffinity-purified antibodies. Dr. William R. Brinkley (Baylor College of Medicine, TX, USA) kindly provided anti-CREST human serum.
Quantification of the immunofluorescence signal
Wild-type and mutant oocyte and spermatocyte spreads were stained at the same time with the same mixture of antibodies. In each experiment, when comparing wild type and mutants, imaging of the cells was performed on the same day with the same microscope and camera settings. PerkinElmer Volocity software 5.3 was used to control for possible changes in illumination during the course of imaging and measurement of the immunofluorescence. We measured total immunofluorescence of γH2AX in identical-sized rectangles that were placed over the cell boundaries. Fifty individual leptotene stage spreads from wild type, mutant spermatocytes and oocytes were subjected to immunofluorescence analysis. A two-tailed non-parametric Wilcoxon–Mann–Whitney two-sample rank-sum test was used for sample comparison.
Electron microscopy analysis
Testes were dissected into ∼0.25 cm pieces and fixed in 1x PBS with 2% formaldehyde and 3% glutaraldehyde (Ted Pella Inc., CA, USA), for 2 hr at room temperature. Samples were treated with 0.5% uranyl acetate and osmium tetraoxide, dehydrated with ethanol, and embedded in LX-112 medium (Ladd Research Industries, VT, USA). The samples were polymerized in a 70°C oven for 2 days. Ultrathin sections (70 –100 nm) were cut in a Leica Ultracut microtome (Leica, IL, USA), stained for 5 min in 1% aqueous uranyl acetate and 2 min in 1% aqueous lead citrate at room temperature in a Leica EM Stainer, and examined by a JEM 1010 transmission electron microscope (JEOL, MA, USA) at an accelerating voltage of 80 kV. Digital images were obtained using AMT Imaging System (Advanced Microscopy Techniques Corp, MA, USA).
Western blot analysis and phosphatase treatment
Western blot analysis was performed essentially as described previously [64]. Postnatal day 5, 8, 14, 18 and 21 testes were collected and lysed in RIPA buffer (50 mM Tris-HCl, pH 7.4, 1% NP-40, 0.25% sodium deoxycholate, 150 mM NaCl, 1 mM EDTA with or without protease inhibitor cocktail (Roche)). Total protein (20 µg) was loaded onto the SDS–PAGE gel, transferred to nitrocellulose membrane, and immunoblotted with HORMAD1 rabbit antibody (1∶5000 dilutions). Varying amounts of protein phosphatase I (Sigma P7937, MO, USA) were incubated with 20 µg of 18 day old wild-type total testes lysates, for 30 min on ice. Phosphatase treated testes lysates were electrophoresed, transferred to nitrocellulose membrane and immunoblotted with anti-HORMAD1 rabbit antibody.
RNA isolation, RT-PCR, quantitative real-time PCR (Q-PCR), and microarray analysis
RT-PCR and quantitative real time PCR were performed as described previously [45], [65]. We used previously published oligonucleotide sequences corresponding to Rnh2, Mov10l1, Pramel3, Usp26, Fthl17, Tex11, Tex13 [66], and Hormad1 (forward 5′ CTGCTGACACCAAGAAAGCA 3′ - and reverse 5′ - CCTGGTGGTTGGTAATCTGG -3′) and Hormad2 (forward 5′ GCTCATCAGGGGCTAGACTG 3′ - and reverse 5′ - TGGTTCGCTGACCTTCTTCT -3′) primers. Q-PCR was performed using the SyberGreen PCR Master Mix (Bio-Red, CA, USA) and probe set specific for each gene under investigation. Each sample was analyzed in triplicate from at least three independent wild type and Hormad1−/− testes cDNA samples. The relative amount of transcripts was calculated by the ΔΔCT method as described by Applied Biosystems, and normalized to β-actin. The average and standard errors were calculated for the triplicate measurements, and the relative amount of target gene expression for each sample was plotted. Student's t test was used to compute the P values. Significance was defined as a P value<0.05.
RNA expression arrays on wild-type and Hormad1−/− 2 week old testes were performed on the Illumina BeadChip MouseWG-6 2.0 arrays and analyzed as previously described (GEO accession numbers, GSE21524) [67]–[68].
Superovulation and oocyte collection in mouse
Mouse oocytes and pre-implantation embryos were collected by using standard protocols for timed mating [69]. Briefly, 4 to 6 weeks female mice were superovulated by injection of 5 IU of pregnant mare serum gonadotropin (PMSG) (Prospec, Rehovot, Israel) and 48 h later 5 IU of human chorionic gonadotropin (hCG) (Sigma, MO, USA). Females were placed individually with 10-week old males immediately after the injection of hCG. Collection of one cell, two cell, four cell, eight cell, and blastocyst stages was performed at 24 h, 42 h, 68 h, and 96 h after hCG injection, by flushing the oviducts and uterine horns with HEPES-buffered media under the microscope. Removal of the cumulus cells was achieved in HEPES-buffered media containing 1 mg/ml hyaluronidase (Sigma, MO, USA).
FISH analysis
Oocytes and embryos were exposed to a hypotonic solution containing 0.1% sodium citrate, 0.1% bovine serum albumin for about 15 min and gently transferred onto the slides. Cells were fixed with fresh methanol: acetic acid (3∶1) solution dispensed carefully over the surface of the slide. The slides were air-dried and dehydrated at room temperature in a series of 70%, 85% and 100% ethanol solutions before FISH analysis.
Twenty BAC (bacterial artificial chromosomes) clones from a mouse RPCI-23 library representing sequences unique to specific chromosomes were selected from the UCSC Genome browser (http://genome.ucsc.edu) and NCBI (http://www.ncbi.nlm.nih.gov) databases (Table S1). Probe DNA extraction was performed according to the standard alkaline lysis method and labeled by standard nick translation with Spectrum Orange - or Spectrum Green-dUTP using a commercially available kit (Abbott/Vysis, IL, USA). Sequential fluorescence in situ hybridization experiments using a mixture of three probes were performed according to manufacturer's instructions (Abbott/Vysis, IL, USA). Each mixture contained a red, green and yellow (combined green/red) fluorescent probes. Probes were applied to slides, hybridized for 20 hr at 37°C, washed with 0.4×SSC/0.3% NP-40 for 2 minutes at 73°C and with 2×SSC/0.1% NP-40 for 1 minute at room temperature, and counterstained with DAPI. Digital FISH images were captured by a Power Macintosh G3 System using MacProbe software version 4.4 (Applied Imaging, CA, USA).
Supporting Information
Zdroje
1. PageSL
HawleyRS
2004 The genetics and molecular biology of the synaptonemal complex. Annu Rev Cell Dev Biol 20 525 558
2. BailisJM
RoederGS
1998 Synaptonemal complex morphogenesis and sister-chromatid cohesion require Mek1-dependent phosphorylation of a meiotic chromosomal protein. Genes Dev 12 3551 3563
3. KleinF
MahrP
GalovaM
BuonomoSB
MichaelisC
1999 A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell 98 91 103
4. LoidlJ
KleinF
ScherthanH
1994 Homologous pairing is reduced but not abolished in asynaptic mutants of yeast. J Cell Biol 125 1191 1200
5. AravindL
KooninEV
1998 The HORMA domain: a common structural denominator in mitotic checkpoints, chromosome synapsis and DNA repair. Trends Biochem Sci 23 284 286
6. MichelLS
LiberalV
ChatterjeeA
KirchweggerR
PascheB
2001 MAD2 haplo-insufficiency causes premature anaphase and chromosome instability in mammalian cells. Nature 409 355 359
7. DoblesM
LiberalV
ScottML
BenezraR
SorgerPK
2000 Chromosome missegregation and apoptosis in mice lacking the mitotic checkpoint protein Mad2. Cell 101 635 645
8. HollingsworthNM
GoetschL
ByersB
1990 The HOP1 gene encodes a meiosis-specific component of yeast chromosomes. Cell 61 73 84
9. SmithAV
RoederGS
1997 The yeast Red1 protein localizes to the cores of meiotic chromosomes. J Cell Biol 136 957 967
10. ZetkaMC
KawasakiI
StromeS
MullerF
1999 Synapsis and chiasma formation in Caenorhabditis elegans require HIM-3, a meiotic chromosome core component that functions in chromosome segregation. Genes Dev 13 2258 2270
11. ArmstrongSJ
CarylAP
JonesGH
FranklinFC
2002 Asy1, a protein required for meiotic chromosome synapsis, localizes to axis-associated chromatin in Arabidopsis and Brassica. J Cell Sci 115 3645 3655
12. ColaiacovoMP
MacQueenAJ
Martinez-PerezE
McDonaldK
AdamoA
2003 Synaptonemal complex assembly in C. elegans is dispensable for loading strand-exchange proteins but critical for proper completion of recombination. Dev Cell 5 463 474
13. CouteauF
NabeshimaK
VilleneuveA
ZetkaM
2004 A component of C. elegans meiotic chromosome axes at the interface of homolog alignment, synapsis, nuclear reorganization, and recombination. Curr Biol 14 585 592
14. RossKJ
FranszP
ArmstrongSJ
VizirI
MulliganB
1997 Cytological characterization of four meiotic mutants of Arabidopsis isolated from T-DNA-transformed lines. Chromosome Res 5 551 559
15. CarylAP
ArmstrongSJ
JonesGH
FranklinFC
2000 A homologue of the yeast HOP1 gene is inactivated in the Arabidopsis meiotic mutant asy1. Chromosoma 109 62 71
16. PangasSA
YanW
MatzukMM
RajkovicA
2004 Restricted germ cell expression of a gene encoding a novel mammalian HORMA domain-containing protein. Gene Expr Patterns 5 257 263
17. WojtaszL
DanielK
RoigI
Bolcun-FilasE
XuH
2009 Mouse HORMAD1 and HORMAD2, two conserved meiotic chromosomal proteins, are depleted from synapsed chromosome axes with the help of TRIP13 AAA-ATPase. PLoS Genet 5 e1000702 doi:10.1371/journal.pgen.1000702
18. FukudaT
DanielK
WojtaszL
TothA
HoogC
2010 A novel mammalian HORMA domain-containing protein, HORMAD1, preferentially associates with unsynapsed meiotic chromosomes. Exp Cell Res 316 158 171
19. KironmaiKM
MuniyappaK
FriedmanDB
HollingsworthNM
ByersB
1998 DNA-binding activities of Hop1 protein, a synaptonemal complex component from Saccharomyces cerevisiae. Mol Cell Biol 18 1424 1435
20. Mao-DraayerY
GalbraithAM
PittmanDL
CoolM
MaloneRE
1996 Analysis of meiotic recombination pathways in the yeast Saccharomyces cerevisiae. Genetics 144 71 86
21. CarballoJA
JohnsonAL
SedgwickSG
ChaRS
2008 Phosphorylation of the axial element protein Hop1 by Mec1/Tel1 ensures meiotic interhomolog recombination. Cell 132 758 770
22. BailisJM
SmithAV
RoederGS
2000 Bypass of a meiotic checkpoint by overproduction of meiotic chromosomal proteins. Mol Cell Biol 20 4838 4848
23. MeuwissenRL
OffenbergHH
DietrichAJ
RiesewijkA
van IerselM
1992 A coiled-coil related protein specific for synapsed regions of meiotic prophase chromosomes. EMBO J 11 5091 5100
24. DobsonMJ
PearlmanRE
KaraiskakisA
SpyropoulosB
MoensPB
1994 Synaptonemal complex proteins: occurrence, epitope mapping and chromosome disjunction. J Cell Sci 107 Pt 10 2749 2760
25. LammersJH
OffenbergHH
van AalderenM
VinkAC
DietrichAJ
1994 The gene encoding a major component of the lateral elements of synaptonemal complexes of the rat is related to X-linked lymphocyte-regulated genes. Mol Cell Biol 14 1137 1146
26. OffenbergHH
SchalkJA
MeuwissenRL
van AalderenM
KesterHA
1998 SCP2: a major protein component of the axial elements of synaptonemal complexes of the rat. Nucleic Acids Res 26 2572 2579
27. YuanL
LiuJG
ZhaoJ
BrundellE
DaneholtB
2000 The murine SCP3 gene is required for synaptonemal complex assembly, chromosome synapsis, and male fertility. Mol Cell 5 73 83
28. del MazoJ
KremerL
AvilaJ
1987 Centromeric proteins recognized by CREST sera and meiotic chromosome segregation. Chromosoma 96 55 59
29. YuanL
LiuJG
HojaMR
WilbertzJ
NordqvistK
2002 Female germ cell aneuploidy and embryo death in mice lacking the meiosis-specific protein SCP3. Science 296 1115 1118
30. PittmanDL
CobbJ
SchimentiKJ
WilsonLA
CooperDM
1998 Meiotic prophase arrest with failure of chromosome synapsis in mice deficient for Dmc1, a germline-specific RecA homolog. Mol Cell 1 697 705
31. de VriesFA
de BoerE
van den BoschM
BaarendsWM
OomsM
2005 Mouse Sycp1 functions in synaptonemal complex assembly, meiotic recombination, and XY body formation. Genes Dev 19 1376 1389
32. GasiorSL
WongAK
KoraY
ShinoharaA
BishopDK
1998 Rad52 associates with RPA and functions with rad55 and rad57 to assemble meiotic recombination complexes. Genes Dev 12 2208 2221
33. EdelmannW
CohenPE
KaneM
LauK
MorrowB
1996 Meiotic pachytene arrest in MLH1-deficient mice. Cell 85 1125 1134
34. KolasNK
MarconE
CrackowerMA
HoogC
PenningerJM
2005 Mutant meiotic chromosome core components in mice can cause apparent sexual dimorphic endpoints at prophase or X-Y defective male-specific sterility. Chromosoma 114 92 102
35. WolteringD
BaumgartnerB
BagchiS
LarkinB
LoidlJ
2000 Meiotic segregation, synapsis, and recombination checkpoint functions require physical interaction between the chromosomal proteins Red1p and Hop1p. Mol Cell Biol 20 6646 6658
36. WardIM
ChenJ
2001 Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress. J Biol Chem 276 47759 47762
37. PrielerS
PenknerA
BordeV
KleinF
2005 The control of Spo11's interaction with meiotic recombination hotspots. Genes Dev 19 255 269
38. NakamuraTM
DuLL
RedonC
RussellP
2004 Histone H2A phosphorylation controls Crb2 recruitment at DNA breaks, maintains checkpoint arrest, and influences DNA repair in fission yeast. Mol Cell Biol 24 6215 6230
39. Fernandez-CapetilloO
MahadevaiahSK
CelesteA
RomanienkoPJ
Camerini-OteroRD
2003 H2AX is required for chromatin remodeling and inactivation of sex chromosomes in male mouse meiosis. Dev Cell 4 497 508
40. TurnerJM
AprelikovaO
XuX
WangR
KimS
2004 BRCA1, histone H2AX phosphorylation, and male meiotic sex chromosome inactivation. Curr Biol 14 2135 2142
41. KozlovSV
GrahamME
PengC
ChenP
RobinsonPJ
2006 Involvement of novel autophosphorylation sites in ATM activation. EMBO J 25 3504 3514
42. YoshidaK
KondohG
MatsudaY
HabuT
NishimuneY
1998 The mouse RecA-like gene Dmc1 is required for homologous chromosome synapsis during meiosis. Mol Cell 1 707 718
43. EdelmannW
CohenPE
KneitzB
WinandN
LiaM
1999 Mammalian MutS homologue 5 is required for chromosome pairing in meiosis. Nat Genet 21 123 127
44. BarlowC
LiyanageM
MoensPB
TarsounasM
NagashimaK
1998 Atm deficiency results in severe meiotic disruption as early as leptonema of prophase I. Development 125 4007 4017
45. PangasSA
ChoiY
BallowDJ
ZhaoY
WestphalH
2006 Oogenesis requires germ cell-specific transcriptional regulators Sohlh1 and Lhx8. Proc Natl Acad Sci U S A 103 8090 8095
46. DebK
ReeseJ
PariaBC
2006 Methodologies to study implantation in mice. Methods Mol Med 121 9 34
47. RevenkovaE
EijpeM
HeytingC
HodgesCA
HuntPA
2004 Cohesin SMC1 beta is required for meiotic chromosome dynamics, sister chromatid cohesion and DNA recombination. Nat Cell Biol 6 555 562
48. YangF
De La FuenteR
LeuNA
BaumannC
McLaughlinKJ
2006 Mouse SYCP2 is required for synaptonemal complex assembly and chromosomal synapsis during male meiosis. J Cell Biol 173 497 507
49. XuY
AshleyT
BrainerdEE
BronsonRT
MeynMS
1996 Targeted disruption of ATM leads to growth retardation, chromosomal fragmentation during meiosis, immune defects, and thymic lymphoma. Genes Dev 10 2411 2422
50. RomanienkoPJ
Camerini-OteroRD
2000 The mouse Spo11 gene is required for meiotic chromosome synapsis. Mol Cell 6 975 987
51. de los SantosT
HollingsworthNM
1999 Red1p, a MEK1-dependent phosphoprotein that physically interacts with Hop1p during meiosis in yeast. J Biol Chem 274 1783 1790
52. WangH
HoogC
2006 Structural damage to meiotic chromosomes impairs DNA recombination and checkpoint control in mammalian oocytes. J Cell Biol 173 485 495
53. Di GiacomoM
BarchiM
BaudatF
EdelmannW
KeeneyS
2005 Distinct DNA-damage-dependent and -independent responses drive the loss of oocytes in recombination-defective mouse mutants. Proc Natl Acad Sci U S A 102 737 742
54. HollingsworthNM
PonteL
1997 Genetic interactions between HOP1, RED1 and MEK1 suggest that MEK1 regulates assembly of axial element components during meiosis in the yeast Saccharomyces cerevisiae. Genetics 147 33 42
55. HassoldT
HuntP
2001 To err (meiotically) is human: the genesis of human aneuploidy. Nat Rev Genet 2 280 291
56. HodgesCA
RevenkovaE
JessbergerR
HassoldTJ
HuntPA
2005 SMC1beta-deficient female mice provide evidence that cohesins are a missing link in age-related nondisjunction. Nat Genet 37 1351 1355
57. HandelMA
SchimentiJC
2010 Genetics of mammalian meiosis: regulation, dynamics and impact on fertility. Nat Rev Genet 11 124 136
58. RevenkovaE
EijpeM
HeytingC
GrossB
JessbergerR
2001 Novel meiosis-specific isoform of mammalian SMC1. Mol Cell Biol 21 6984 6998
59. GoateA
Chartier-HarlinMC
MullanM
BrownJ
CrawfordF
1991 Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer's disease. Nature 349 704 706
60. WarmingS
CostantinoN
CourtDL
JenkinsNA
CopelandNG
2005 Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res 33 e36
61. RajkovicA
PangasSA
BallowD
SuzumoriN
MatzukMM
2004 NOBOX deficiency disrupts early folliculogenesis and oocyte-specific gene expression. Science 305 1157 1159
62. BaltusAE
MenkeDB
HuYC
GoodheartML
CarpenterAE
2006 In germ cells of mouse embryonic ovaries, the decision to enter meiosis precedes premeiotic DNA replication. Nat Genet 38 1430 1434
63. ChoiY
BallowDJ
XinY
RajkovicA
2008 Lim homeobox gene, lhx8, is essential for mouse oocyte differentiation and survival. Biol Reprod 79 442 449
64. LiM
ShinYH
HouL
HuangX
WeiZ
2008 The adaptor protein of the anaphase promoting complex Cdh1 is essential in maintaining replicative lifespan and in learning and memory. Nat Cell Biol
65. RajkovicA
YanMSC
KlysikM
MatzukM
2001 Discovery of germ cell-specific transcripts by expressed sequence tag database analysis. Fertil Steril 76 550 554
66. WangPJ
McCarreyJR
YangF
PageDC
2001 An abundance of X-linked genes expressed in spermatogonia. Nat Genet 27 422 426
67. MouilletJF
ChuT
NelsonDM
MishimaT
SadovskyY
2010 MiR-205 silences MED1 in hypoxic primary human trophoblasts. FASEB J
68. SmythGK
2004 Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3 Article3
69. SeliE
LaliotiMD
FlahertySM
SakkasD
TerziN
2005 An embryonic poly(A)-binding protein (ePAB) is expressed in mouse oocytes and early preimplantation embryos. Proc Natl Acad Sci U S A 102 367 372
Štítky
Genetika Reprodukční medicína
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2010 Číslo 11- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
-
Všechny články tohoto čísla
- Genome-Wide Association Meta-Analysis of Cortical Bone Mineral Density Unravels Allelic Heterogeneity at the Locus and Potential Pleiotropic Effects on Bone
- Beyond QTL Cloning
- An Evolutionary Framework for Association Testing in Resequencing Studies
- Genome-Wide Association Study Identifies Two Novel Regions at 11p15.5-p13 and 1p31 with Major Impact on Acute-Phase Serum Amyloid A
- The Functional Interplay between Protein Kinase CK2 and CCA1 Transcriptional Activity Is Essential for Clock Temperature Compensation in Arabidopsis
- Endogenous Viral Elements in Animal Genomes
- Analysis of the 10q11 Cancer Risk Locus Implicates and in Human Prostate Tumorigenesis
- DNA Methylation and Normal Chromosome Behavior in Neurospora Depend on Five Components of a Histone Methyltransferase Complex, DCDC
- Sarcomere Formation Occurs by the Assembly of Multiple Latent Protein Complexes
- Genetic Basis of Growth Adaptation of after Deletion of , a Major Metabolic Gene
- Nomadic Enhancers: Tissue-Specific -Regulatory Elements of Have Divergent Genomic Positions among Species
- The Parental Non-Equivalence of Imprinting Control Regions during Mammalian Development and Evolution
- CTCF-Dependent Chromatin Bias Constitutes Transient Epigenetic Memory of the Mother at the Imprinting Control Region in Prospermatogonia
- Systematic Dissection and Trajectory-Scanning Mutagenesis of the Molecular Interface That Ensures Specificity of Two-Component Signaling Pathways
- Nucleolin Is Required for DNA Methylation State and the Expression of rRNA Gene Variants in
- The Complex Genetic Architecture of the Metabolome
- ATM Limits Incorrect End Utilization during Non-Homologous End Joining of Multiple Chromosome Breaks
- Mutation Disrupts Synaptonemal Complex Formation, Recombination, and Chromosome Segregation in Mammalian Meiosis
- Mismatch Repair–Independent Increase in Spontaneous Mutagenesis in Yeast Lacking Non-Essential Subunits of DNA Polymerase ε
- The Kinesin-3 Motor UNC-104/KIF1A Is Degraded upon Loss of Specific Binding to Cargo
- Epigenetic Silencing of Spermatocyte-Specific and Neuronal Genes by SUMO Modification of the Transcription Factor Sp3
- A Coastal Cline in Sodium Accumulation in Is Driven by Natural Variation of the Sodium Transporter AtHKT1;1
- Cyclin B3 Is Required for Multiple Mitotic Processes Including Alleviation of a Spindle Checkpoint–Dependent Block in Anaphase Chromosome Segregation
- Altered DNA Methylation in Leukocytes with Trisomy 21
- Human-Specific Evolution and Adaptation Led to Major Qualitative Differences in the Variable Receptors of Human and Chimpanzee Natural Killer Cells
- Leptotene/Zygotene Chromosome Movement Via the SUN/KASH Protein Bridge in
- RACK-1 Acts with Rac GTPase Signaling and UNC-115/abLIM in Axon Pathfinding and Cell Migration
- Genome-Wide Effects of Long-Term Divergent Selection
- Endless Forms Most Viral
- Conflict between Noise and Plasticity in Yeast
- Essential Functions of the Histone Demethylase Lid
- The Transcriptional Regulator Rok Binds A+T-Rich DNA and Is Involved in Repression of a Mobile Genetic Element in
- The Cellular Robustness by Genetic Redundancy in Budding Yeast
- Localization of a Guanylyl Cyclase to Chemosensory Cilia Requires the Novel Ciliary MYND Domain Protein DAF-25
- A Buoyancy-Based Screen of Drosophila Larvae for Fat-Storage Mutants Reveals a Role for in Coupling Fat Storage to Nutrient Availability
- A Functional Genomics Approach Identifies Candidate Effectors from the Aphid Species (Green Peach Aphid)
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Genome-Wide Association Study Identifies Two Novel Regions at 11p15.5-p13 and 1p31 with Major Impact on Acute-Phase Serum Amyloid A
- Analysis of the 10q11 Cancer Risk Locus Implicates and in Human Prostate Tumorigenesis
- The Parental Non-Equivalence of Imprinting Control Regions during Mammalian Development and Evolution
- Genome-Wide Effects of Long-Term Divergent Selection
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



