-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaTranscription Factor SomA Is Required for Adhesion, Development and Virulence of the Human Pathogen
Invasive fungal infections affecting immunocompromised patients are emerging worldwide. Among various human fungal pathogens, Aspergillus fumigatus is one of the most common molds causing severe invasive aspergillosis in immunocompromised patients. The conidia, which can evade from innate immunity and adhere to epithelial cells of alveoli in human lungs will start to germinate and cause the disease. Currently, the understanding of the molecular mechanisms of adherence of fungal cells to hosts is scarce. The transcription factor Flo8 controls adhesion to biotic or abiotic surfaces and morphological development in baker’s yeast. Flo8 homologues in the dimorphic human pathogenic yeast Candida albicans or the filamentous plant pathogen Magnaporthe oryzae are required for development and virulence. We found in this study that the Flo8 homologue SomA of A. fumigatus is required for adhesion and conidiation. Two independent invasive aspergillosis assays using chicken eggs or mouse demonstrated that deletion of the corresponding gene resulted in attenuated virulence. SomA represents an important fungal transcription factor at the interface between adherence, asexual spore formation and pathogenicity in an important opportunistic human pathogen.
Published in the journal: . PLoS Pathog 11(11): e32767. doi:10.1371/journal.ppat.1005205
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1005205Summary
Invasive fungal infections affecting immunocompromised patients are emerging worldwide. Among various human fungal pathogens, Aspergillus fumigatus is one of the most common molds causing severe invasive aspergillosis in immunocompromised patients. The conidia, which can evade from innate immunity and adhere to epithelial cells of alveoli in human lungs will start to germinate and cause the disease. Currently, the understanding of the molecular mechanisms of adherence of fungal cells to hosts is scarce. The transcription factor Flo8 controls adhesion to biotic or abiotic surfaces and morphological development in baker’s yeast. Flo8 homologues in the dimorphic human pathogenic yeast Candida albicans or the filamentous plant pathogen Magnaporthe oryzae are required for development and virulence. We found in this study that the Flo8 homologue SomA of A. fumigatus is required for adhesion and conidiation. Two independent invasive aspergillosis assays using chicken eggs or mouse demonstrated that deletion of the corresponding gene resulted in attenuated virulence. SomA represents an important fungal transcription factor at the interface between adherence, asexual spore formation and pathogenicity in an important opportunistic human pathogen.
Introduction
Adherence to host cells represents a key step for pathogenesis of bacterial or fungal microorganisms. Prerequisites at the molecular level include cell wall adhesive or hydrophobic proteins, carbohydrate components of the cell wall and the extracellular matrix. Gene families responsible for adherence comprise the FLO adhesins (flocculins) of Saccharomyces cerevisiae or the ALS agglutinins (agglutinins like sequence) of Candida albicans [1, 2]. Conidial adherence of the opportunistic human pathogen Aspergillus fumigatus requires the hydrophobin RodA, the laminin-binding protein AspF2 or glycans as important constituents of the cell wall [3–5].
Adherence is triggered by different environmental stimuli which are sensed by receptors and which induce various signaling pathways [2, 6]. A prominent example is the cyclic adenosine monophosphate (cAMP) dependent signaling pathway, which is highly conserved from bacteria to mammals. In eukaryotic cells, the cAMP dependent protein kinase A (PKA) signaling pathway is activated by the G protein-coupled receptors [7]. The corresponding Gα subunit activates adenylate cyclases, which convert ATP to cAMP. This secondary messenger binds to the regulatory subunits of PKA. The catalytic subunits of the enzyme are released and activate downstream transcription factors by phosphorylation [7]. The cAMP/PKA pathway plays a crucial role in development and pathogenesis in animal or plant pathogenic fungi such as C. albicans, Cryptococcus neoformans, Magnaporthe oryzae and Ustilago maydis [8–12]. This link between development, virulence and the cAMP/PKA pathway is conserved in the filamentous fungus and opportunistic pathogen A. fumigatus. Components of the A. fumigatus cAMP/PKA pathway include the GpaA and GpaB Gα subunits of the heterotrimertic G protein, the AcyA adenylate cyclase, the PkaR regulatory and the PkaC1/PkaC2 catalytic subunits of PKA. Deletion of gpaB or acyA results in reduced conidiation and a reduced growth rate in the ΔacyA strain [13]. The regulatory PkaR and the catalytic PkaC1 proteins are required to promote germination, growth and accurate conidiation [14–16]. Null mutants of the previous described genes showed attenuated virulence and indicate a role of the cAMP/PKA pathway in the pathogenicity process. This process needs to be further elucidated because of the importance of the A. fumigatus opportunistic pathogen, which can cause invasive aspergillosis in immunocompromised individuals with mortality rates of more than 60% [17–19].
The cAMP/PKA pathway activates several downstream factors like the S. cerevisiae transcription factor Flo8, which controls adhesive and filamentous growth. It induces the expression of the FLO11 gene for an adhesin, which is required for flocculation or the establishment of biofilms in haploid and pseudohyphae formation in diploid yeast strains [2]. The Flo8 counterpart of the dimorphic yeast C. albicans regulates hyphal development and virulence factors [20]. In both yeasts, Flo8 interacts with additional co-activators as for example Mfg1, which is also required for invasive and hyphal growth [21, 22]. The current knowledge about the transcription factors and their corresponding genes which represent the homologues of the FLO8 gene are limited. Among filamentous fungi, only the gene for MoSom1 corresponding to Flo8 in the plant pathogenic filamentous fungus M. oryzae has been examined. The Flo8 counterpart of the dimorphic yeast C. albicans is the only analyzed protein in a human pathogen. Representatives of constitutively filamentous fungi of human pathogens have not yet been studied. MoSom1 carries like the corresponding yeast protein the N-terminal LUFS (LUG/LUH-Flo8-single-stranded DNA binding) domain and can complement adhesive growth in a Δflo8 yeast mutant strain. MoSom1 controls the gene for the hydrophobin MoMpg1, which is required for fungal attachment to plant leaves during infection. Deletion of the Mosom1 gene results in loss of asexual or sexual development and impairs pathogenicity [23].
The development of Aspergilli has been primarily analyzed in the model fungus Aspergillus nidulans [24, 25]. The C2H2 zinc finger transcription factor BrlA represents a central regulator of asexual development in A. nidulans as well as in A. fumigatus and controls the formation of vesicles, which are required for conidiation at the top of aerial hyphae. BrlA induces the expression of the downstream abaA and wetA regulatory genes, which induce differentiation of phialides as spore forming cells and the subsequent maturation of conidia, which represent the asexual spores [26]. The flbB, flbC and flbD regulatory genes are genetically located upstream the expression of brlA [25]. MedA and the APSES (Asm1, Phd1, Sok2, Efg1, and StuA) protein StuA regulate transcription of the brlA gene in A. nidulans where they are required for metulae cell formation from vesicles followed by phialide cell formation [27]. A. fumigatus forms only phialides as asexual spore forming cells but does not produce an additional layer of metulae cells. Though, lack of either MedA or StuA also impairs conidiation in A. fumigatus where their exact molecular function is yet unknown [28, 29]. Additionally, MedA and StuA control adhesion and virulence in A. fumigatus by regulating the gene uge3 encoding uridine diphosphate (UDP)-glucose-epimerase, which is essential for adherence through mediating the synthesis of galactosaminogalactan [30].
The main objective of this study was to examine the function of A. fumigatus SomA and putative interaction partners. SomA corresponds to the Flo8/Som1 regulator described in other fungi. Our data show that SomA in collaboration with its co-regulator PtaB plays a key role in a transcriptional network controlling conidiation and adhesion and that SomA is required for virulence of filamentous pathogens A. fumigatus.
Results
SomA of A. fumigatus complements the defects of S. cerevisiae flo8 strains in haploid adhesive and diploid pseudohyphal growth
Flo8 is a regulator of S. cerevisiae dimorphism and its counterpart Som1 in the filamentous fungus M. oryzae is required for plant pathogenicity in rice [20, 23, 31]. These proteins share with the A. fumigatus protein SomA (AFUA_7G02260) the LUFS domain, which contains a LisH (Lis homology) motif for protein dimerization and tetramerization at the N-terminus (Fig 1A). SomA shows identities of 15.7% and 20.5% to the Flo8 proteins of S. cerevisiae and C. albicans, respectively, and 39% to Som1 of M. oryzae. In addition, there is a conserved nuclear localization signal (NLS) PSPSKRPRLE in filamentous fungi. These data suggest that the proteins derived from all homologous genes have a nuclear function. Exons of somA were identified by comparing the DNA sequence of the genomic locus with cDNAs, which were amplified from total mRNA. Sequencing of the resulting plasmid revealed that somA carries five exons of a size of 486 bp, 152 bp, 1279 bp, 267 bp and 171 bp (pME4192) resulting in a deduced protein of 784 amino acids with a molecular weight of 84.59 kDa. An additional splice variant (pME4193) was found (Fig 1A) Both splice variants of somA could be identified in the ΔakuA strain after 20 h vegetative growth in liquid minimal medium (MM).
Fig. 1. The SomA corresponds to Flo8 and complements developmental phenotypes in S. cerevisiae flo8 mutant strains. 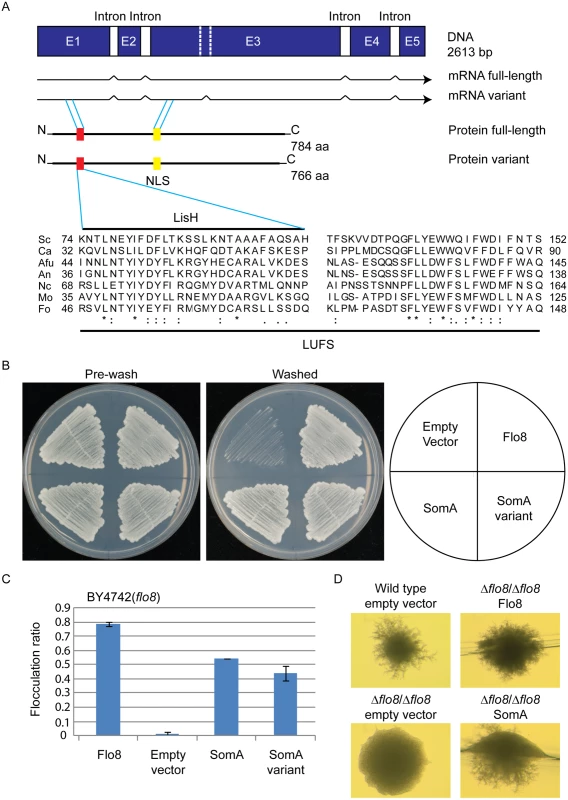
(A) The somA gene locus results in two mRNA splice variants and two deduced proteins including the LisH motif (red) and a predicted nuclear localization signal (NLS, yellow). The sequence alignments of the LUFS domain including the LisH motif of A. fumigatus SomA and the corresponding proteins of other fungi. Asterisks indicate identical residues; highly (colon) or modestly (period) similar residues are marked. Abbreviation: Sc, S. cerevisiae; Ca, C. albicans; Afu, A. fumigatus; An, A. nidulans; Nc, Neurospora crassa; Mo, M. oryzae; Fo, F. oxysporum. (B) Haploid invasive growth of S. cerevisiae strain BY4742 (flo8) expressing the indicated proteins or empty vector as negative control. Strains were grown on SC-Ura plates for 3 days at 30°C, and the plates were photographed before and after washing under water stream. (C) Flocculation of yeast strain BY4742 (flo8) expressing the indicated proteins or empty vector as negative control. Strains were grown in 10 mL SC-Ura medium for one day at 30°C. Graph indicates mean ± standard error of triplicate measurements. (D) Diploid pseudohyphal growth of yeast strain RH2660 (Δflo8/Δflo8) expressing the indicated proteins or empty vector as negative control. The wild type (RH2656) carrying empty vector was used as positive control. Strains were grown on SLAD for 6 days at 30°C and photographed. Cross-species complementation of the somA gene was performed in yeast flo8 mutant strains to verify whether both variants share similar functions with Flo8. Expression of either somA or its splice variant (pME4194 and pME4195) under the MET25 yeast promoter could rescue invasive growth (cell-surface adhesion) in the flo8 (truncated FLO8) haploid mutant (BY4742) on solid agar (Fig 1B). Flocculation (cell-cell adhesion) in liquid medium was complemented similar to Flo8 (pME4197) (Fig 1C). In addition, expression of somA in Δflo8 diploid strain (RH2660) restored pseudohyphal growth (Fig 1D). These data support that SomA and Flo8 can fulfill similar cellular functions in yeast.
SomA and Flo8 act through similar promoter sites on FLO11 expression
Flo8 is a transcription factor, which binds and promotes transcription of the FLO11 gene encoding the flocculin Flo11 [32, 33], which is a key determinant for adhesion in yeast [2]. We performed β-galactosidase assays with the 3 kb FLO11 promoter fused to the bacterial LacZ reporter gene to examine whether SomA complements the adhesive phenotypes in flo8 or Δflo8 yeasts (Fig 1B and 1D) by activating FLO11 gene expression. As shown in Fig 2A, both SomA and its splice variant showed significantly increased FLO11 promoter driven LacZ activity in comparison to the mutant strain transformed with the empty plasmid.
Fig. 2. SomA and Flo8 activate FLO11 expression and act through similar regions of the FLO11 promoter. 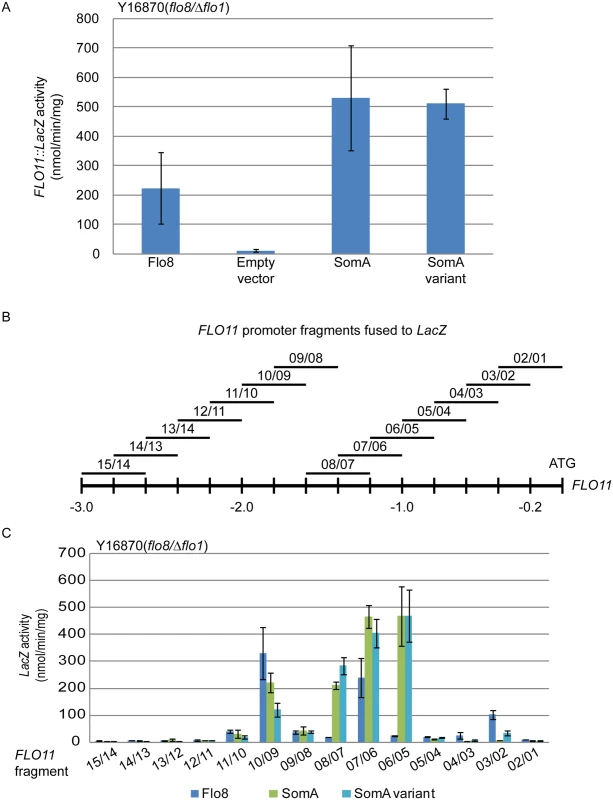
(A) Expression of FLO11::LacZ was determined in haploid Y16870 strain expressing the indicated proteins or empty vector as negative control. (B) Schematic overview of 14 different 400 bp constructs of the FLO11 promoter region fused to CYC1::LacZ reporter [31]. (C) Expression of LacZ gene fused to different FLO11 promoter fragments in Y16870 strain expressing the indicated proteins. In all experiments, strains were grown on 10 mL SC-Ura-Leu medium as pre-culture, then 1 mL of samples were inoculated into SC-Ura-Leu-Met medium as main culture for 6 h before the β-galactosidase activities were determined. Graph indicates mean ± standard error of triplicate measurements. We took a more detailed look at the FLO11 promoter to determine whether SomA and Flo8 bind to similar regions of the promoter. A set of 14 reporter constructs containing 400 bp FLO11 promoter fragments that overlap by 200 bp [34] (Fig 2B) was analyzed in the flo8/Δflo1 yeast strain (Y16870). As shown in Fig 2C, two promoter regions were affected by both Flo8 and SomA. Comparison of Fig 2B and 2C indicated that these two regions are located at 1.8 kb and 1.2 kb upstream of the start codon of FLO11. SomA presumably recognizes two additional regions located at 1.4 kb and 1 kb upstream of the FLO11 open reading frame. These data indicate that SomA and Flo8 share molecular functions in recognizing and controlling similar regions of the FLO11 promoter and hence complemented adhesion and filamentous growth in flo8 and Δflo8 yeast strains.
SomA physically interacts with PtaB
Yeast Flo8 is part of a protein complex required for regulating cellular development, and Mfg1 represents another subunit of this complex [21, 22]. We analyzed whether the similarity of SomA to Flo8 and the similar function of both proteins are reflected by similar interaction partners of SomA in A. fumigatus. A GFP tagged somA gene was constructed (AfGB75) to identify interaction partners of SomA. A GFP-Trap was performed and the recruited proteins were analyzed by LC/MS. Proteins identified in the GFP control strain were considered as unspecific background identifications (LC/MS raw data in S1 Table). Apart from SomA itself, the PtaB protein (AFUA_2G12910), a homologue of yeast Mfg1, was identified by LC/MS. This protein was absent in GFP control strain (Fig 3A). The detailed LC/MS data are shown in S2 Table.
Fig. 3. SomA interacts with PtaB in A. fumigatus. 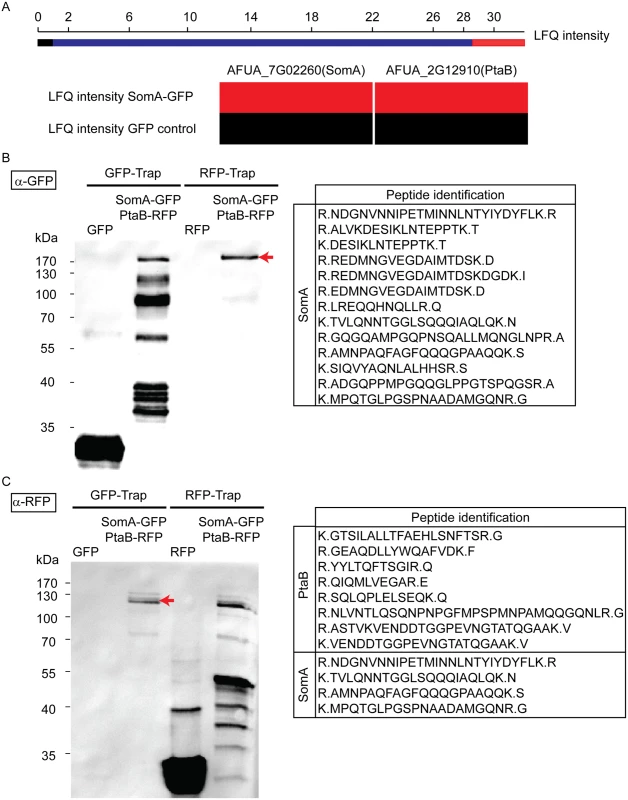
(A) The abundance of SomA and PtaB was measured by LC/MS and estimated based on MaxQuant’s logarithmized label free quantification (log2 LFQ) intensities. High and intermediate LFQ intensities are shown in red and blue. Absence of peptides and low LFQ intensities are presented in black. (B) Western hybridization of GFP-Trap and RFP-Trap enrichments with α-GFP antibody. The single band in the RFP-Trap indicated by an arrow was identified as SomA-GFP by LC/MS with the given peptides. (C) Reciprocal western to (B) but an α-RFP antibody instead of the α-GFP antibody as a different probe. The double band (arrow) correspond to PtaB and SomA by LC/MS. For all experiments, the strains were grown in MM medium for 24 h at 37°C. Protein extracts were performed with either GFP-Trap or RFP-Trap beads and the eluted proteins were separated by 12% SDS-PAGE. We further performed a co-immunoprecipitation to verify whether PtaB and SomA are interaction partners (Fig 3B and 3C). A strain expressing SomA-GFP and PtaB-RFP fusion proteins was constructed (AfGB117). Application of the α-GFP antibody recognized GFP in the trap enrichment of the GFP control strain (Fig 3B). Several signals in the GFP-Trap of the strain expressing both fusions (somA-gfp/ptaB-rfp) presumably represent SomA-GFP and its degradation products. Only a single signal in the RFP-Trap was detected by the α-GFP antibody. The single signal (arrow, Fig 3B) at approximately 170 kDa was verified by LC/MS as SomA-GFP. This suggests that the SomA-GFP protein has been recruited through the RFP-Trap by PtaB-RFP. The reciprocal experiment using an α-RFP antibody and the same trap enrichments resulted in the recognition of RFP in the RFP control strain and several signals in the RFP-Trap presumably representing PtaB-RFP and its derivatives. Two signals at 120 kDa in the GFP trap enrichment (arrow, Fig 3C) were identified by the α-RFP antibody and were determined as the PtaB-RFP protein with a calculated size of 106 kDa by LC/MS. In addition, several SomA-GFP peptides were identified by LC/MS which presumably correspond to the SomA-GFP degradation products which are visible in Fig 3B (GFP-Trap lane). Taken together, these data suggest that SomA and PtaB physically interact in A. fumigatus similar to their counterparts Flo8 and Mfg1 in the two yeasts S. cerevisiae and C. albicans [22].
Deletion of the somA gene blocks A. fumigatus asexual development at aerial hyphae
The somA gene was deleted in a ΔakuA background strain (AfS35) to analyze the function of this gene in correlation with growth, adhesion and development. Cultivation on solid MM plates revealed slow growth (2.8 mm/day) of the ΔsomA strain (AfGB77) in comparison to ΔakuA strain (6.1 mm/day). This ΔsomA growth phenotype was verified by complementation with the respective wild type gene. The complemented strain (AfGB105) showed improved growth rate (5.1 mm/day), indistinguishable from ΔakuA strain (Fig 4A). We also analyzed PtaB as the physical interaction partner by genetic analysis. The deletion of ptaB (AfGB115) resulted also in a reduced growth rate phenotype (4.6 mm/day) which was less pronounced in comparison to the ΔsomA phenotype. In addition, a delayed conidiation was observed in the ptaB null mutant (arrow, S1 Fig). The growth defect and the delayed asexual development of the ΔptaB strain were restored by complementation with a ptaB-rfp fusion (S1 Fig).
Fig. 4. SomA promotes growth and conidia formation of A. fumigatus. 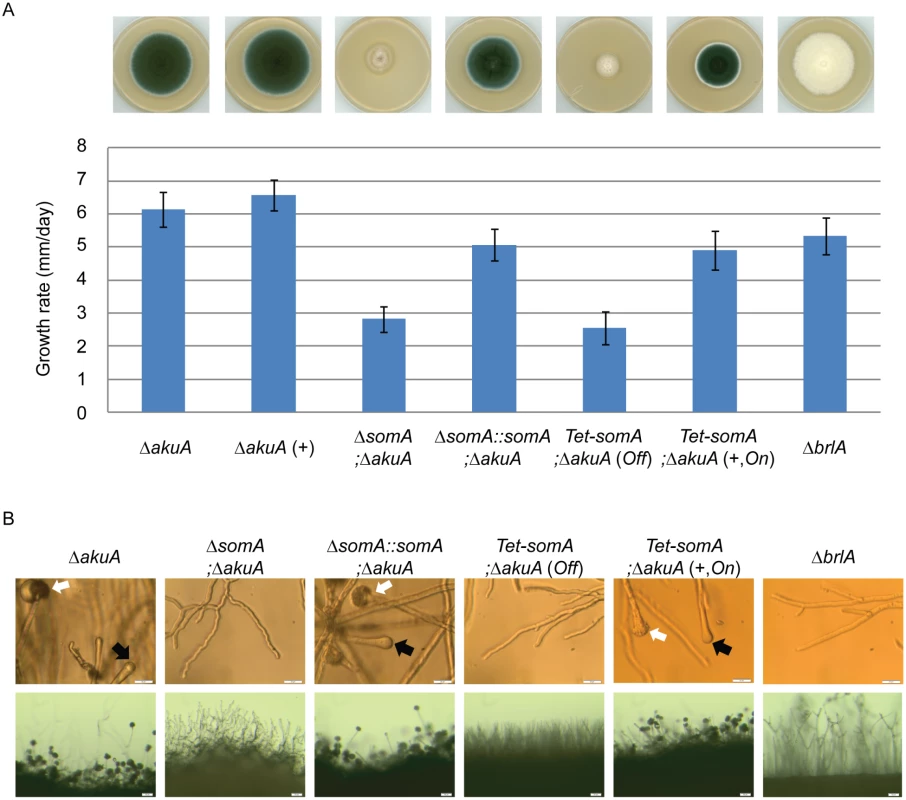
(A) Colony morphology and growth rate of the indicated strains. All strains were grown on either MM plate or MM plate with 5 mg/L doxycycline indicated as (+) for 5 days at 37°C. Values in the graph are indicated as means ± standard error. (B) Morphology of conidiation in the indicated strains. Upper panel: Strains were grown on MM or MM with 5 mg/L doxycycline agar-coated slides for 28 h at 37°C. The open arrows indicate conidiophores and filled arrows represent the vesicles for sporulation. Lower panel: Strains were grown on MM or MM with 5 mg/L doxycycline agar-slides for 28 h at 37°C. Scale bars represent 20 μm (upper panel) and 50 μm (lower panel). Strains grow on MM agar with doxycycline indicated as (+). For all experiments, the Tet-somA was induced (On state) at the present of doxycycline. Asexual spores are normally produced at conidiophores consisting of aerial hyphae with a vesicle on top where the conidia are pinched off [27]. The ΔsomA strain formed exclusively aerial hyphae and was incapable of forming conidiophores. To have a detailed look on conidiation, the strains were inoculated on MM-agar coated microscope slides or MM agar on microscope slides and incubated for 28 h at 37°C. As shown in Fig 4B, the ΔsomA mutant showed no mature conidiophores. In contrast, the ΔakuA strain and the somA complemented strain revealed conidiophores (white arrow) and vesicle formation (black arrow) on top of the aerial hyphae. Furthermore, macroscopic inspection indicated that the ΔakuA strain produces aerial hyphae and conidiophores similar to the somA complemented strain. In contrast, the ΔsomA mutant showed only aerial hyphae (Fig 4B). The defect in vesicle formation of the ΔsomA mutant was similar to the defect in a ΔbrlA strain except of the growth retardation (Fig 4A) [35]. We analyzed the SomA dependent step in asexual development in more detail. A Tet-somA strain (AfGB74) was constructed by replacing the promoter region with the inducible Tet-On system [36] which could conditionally express the somA gene upon addition of doxycycline to the medium.
The growth and conidiation phenotype of the ΔakuA strain were not affected by the presence of doxycycline (Fig 4A). The Tet-somA strain grew as slowly as the ΔsomA mutant and had severely impaired sporulation when doxycycline was absent (Off state). In contrast, these impaired phenotypes were complemented when the promoter was induced by doxycycline (Fig 4A). Further observation showed that the Tet-somA strain revealed conidiophores (white arrow) and vesicle formation (black arrow) on top of the aerial hyphae as the ΔakuA strain only under inducing conditions (Fig 4B). Taken together, these results support a function of SomA in asexual development and fungal growth.
SomA and PtaB are required for biofilm formation
Flo8 is required for adherence of S. cerevisiae by regulating FLO gene expression [2]. Therefore, the impact of the loss of the somA gene on the adherence to plastic or fibronectin were examined. Due to the fact that the ΔsomA mutant has a defect in asexual development, we used the Tet-somA strain to perform the adherence assay and the ΔakuA strain was used as control. As a pilot test the adherence of germlings was tested. Germlings of the ΔakuA strain and Tet-somA mutant (On state) displayed 25% adherence to polystyrene plates and fibronectin-coated plates. In contrast, the Tet-somA germlings (Off state) showed only 5% adherence to both surfaces (Fig 5A).
Fig. 5. SomA and PtaB are involved in biofilm formation in A. fumigatus. 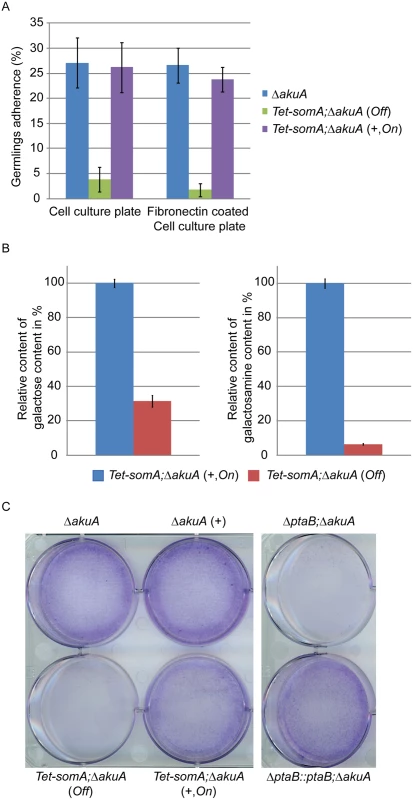
(A) Germlings adherence of the indicated strains to plastic surfaces and fibronectin coated wells. Addition of 5 mg/L doxycycline is indicated with (+). Values in the graph are indicated as means ± standard error with triplicate determinations. (B) Galactosaminogalactan (GAG) assay for the Tet-somA strain. Results amounts of galactose and galactosamine are shown. Levels for the Tet-somA On state were set to 100%. (C) Biofilm formation of the indicated strains inoculated on polystyrene plates for 24 h. The wells were washed with PBS and stained with crystal violet. Addition of 5 mg/L doxycycline is indicated with (+). For all experiments, the Tet-somA was induced (On state) at the present of doxycycline. The polysaccharide galactosaminogalactan (GAG) from the fungal cell wall is composed of α1,4-linked galactose and N-acetylgalactosamine and plays a role in fungal adherence [30]. We tested whether the loss of germling adhesion in the Tet-somA mutant (Off state) is due to reduced GAG production. We cultivated the strain under inducing and non-inducing conditions and after precipitation and hydrolysis of GAG, the amounts of galactose and galactosamine were measured by GC-MS [30]. The amount of galactose was reduced to 31% and the amount of galactosamine was reduced to 6% in the Tet-somA strain (Off) compared to the Tet-somA strain (On), respectively (Fig 5B).
The yeast Flo8-Mfg1 complex is required for biofilm formation [22]. We analyzed whether SomA and PtaB play a similar role in the A. fumigatus life style. As shown in Fig 5C, the hyphae of the Tet-somA strain (On) formed biofilm when the promoter was induced by doxycycline (+). The ΔakuA strain with (+) or without the drug showed similar biofilm formation (Fig 5C). In contrast, the complete mycelium was washed off when the Tet-somA strain was at Off state. A similar phenotype was detected for PtaB. The ΔptaB mutant strain resulted in a defect of biofilm formation and this phenotype could be rescued by re-introducing the ptaB-rfp fused gene in the deletion strain (Fig 5C).
Taken together, these data show a common function of SomA and PtaB in biofilm formation. Furthermore, SomA is required for germling adherence to plastic surfaces or fibronectin and GAG production.
SomA controls the expression of genes related to the process of conidiation and adherence in A. fumigatus
The cellular function of SomA as a transcription factor involved in asexual development and adherence was examined by quantitative transcript analysis of putative target genes. We could show that the Tet-somA strain has a similar asexual development as the ΔsomA mutant when doxycycline is absent (Fig 4). In addition, we showed that SomA plays an important role in adherence and GAG production using the Tet-somA strain (Fig 5). Hence, we used the Tet-somA strain to test the role of SomA in gene regulation. The ΔakuA mutant and the Tet-somA strain were incubated in liquid minimal medium (MM) for 18 h. Afterwards, the mycelium was shifted to liquid MM for 8 h and solid MM plate for 24 h with or without doxycycline. The drug had no effect on gene expressions in the ΔakuA mutant (S2 Fig). Transcript analysis revealed that the Tet-somA strain (Off) abolished brlA expression in contrast to the On state of the Tet-somA strain (Fig 6). In A. fumigatus, FlbB is necessary for flbD expression and FlbD might be essential for expression of brlA [25]. The expression of flbBCD genes in the Tet-somA strain was decreased in the Off state in comparison to the On state (Fig 6). The velvet domain protein family and LaeA also control fungal development and secondary metabolism in filamentous fungi including conidiation [24, 37, 38]. The expression of members of the velvet domain family was not significantly affected except for transcription of the velC gene, which was impaired by the Tet-somA Off state (Fig 6). These results suggest a broader role of SomA in fungal development.
Fig. 6. SomA regulates genes for conidiation and adhesion. 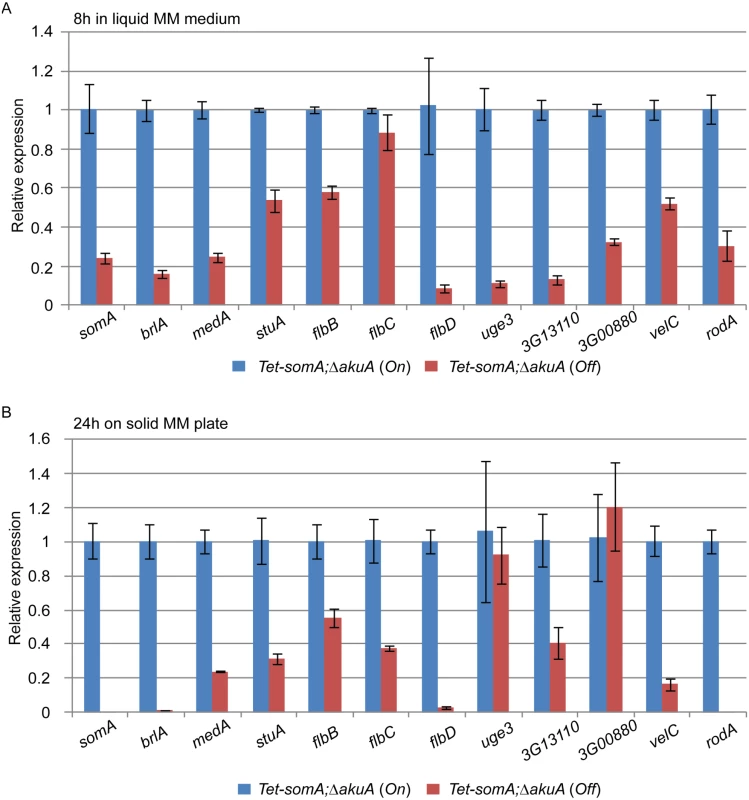
Relative expression of genes encoding proteins that regulate conidiation and adhesion in the Tet-somA strain. The Tet-somA strain was cultivated in liquid MM medium for 18 h at 37°C and shifted to (A) liquid MM medium for 8 h at 37°C and (B) solid MM plate for 24 h at 37°C. Addition of 5 mg/L doxycycline is indicated as (On). Levels for the Tet-somA On state were set to 1. Graph indicates mean ± standard errors from two independent experiments. Gravelat et al (2013) showed that medA and stuA genes are required for adhesion and regulate some putative adhesins [28, 30] and the transcript levels of medA and stuA were significantly reduced in the Tet-somA Off state (Fig 6). Possible adherence genes located downstream of the medA and stuA genes were further evaluated. Three genes (AFUA_3G13110, AFUA_3G00880 and uge3) encoding possible adherence-associated proteins with high scores in bioinformatic prediction [39] were analyzed. The transcript levels of all three genes are reduced in the absence of somA (Off state) (Fig 6). A similar transcript analysis was also observed in the ΔsomA mutant in comparison to the ΔakuA background strain and the somA complemented strain (S3 Fig).
Deletion of the SomA interaction partner PtaB also resulted in a delayed conidiation (S1 Fig) and, a defect of biofilm formation (Fig 5C). This suggests that PtaB might also contribute to the SomA control of gene transcriptions. The transcript levels showed that the ΔptaB mutant strain had a significant effect on the expression of the development and adherence related genes which are also controlled by SomA (S4 Fig).
SomA, FlbB, MedA and StuA represent fungal transcription factors controlling a complex developmental regulatory transcriptional network. To identify the interaction between SomA and these three transcription factors, an epistatic analysis was performed. The overexpression of somA did not change the phenotype of either ΔakuA background, ΔflbB, ΔmedA or ΔstuA mutant strains and had also no significant effect on colony growth (Fig 7A). Double deletion strains revealed a different picture. An additional somA deletion in the ΔflbB background resulted no more in a ΔflbB but in a ΔsomA colony phenotype including the reduced growth rate of the colony. The same phenotype indistinguishable from the ΔsomA mutant was observed in the double mutant strain with ΔmedA (Fig 7B). The ΔstuA ΔsomA double mutant (AfGB114) showed a more complex phenotype which does not completely correspond to the ΔsomA deletion. More aerial hyphae on the surface and a lighter color on the back are visible compared to ΔsomA single or the other double deletion strains.
Fig. 7. SomA acts upstream of flbB, medA and stuA genes. 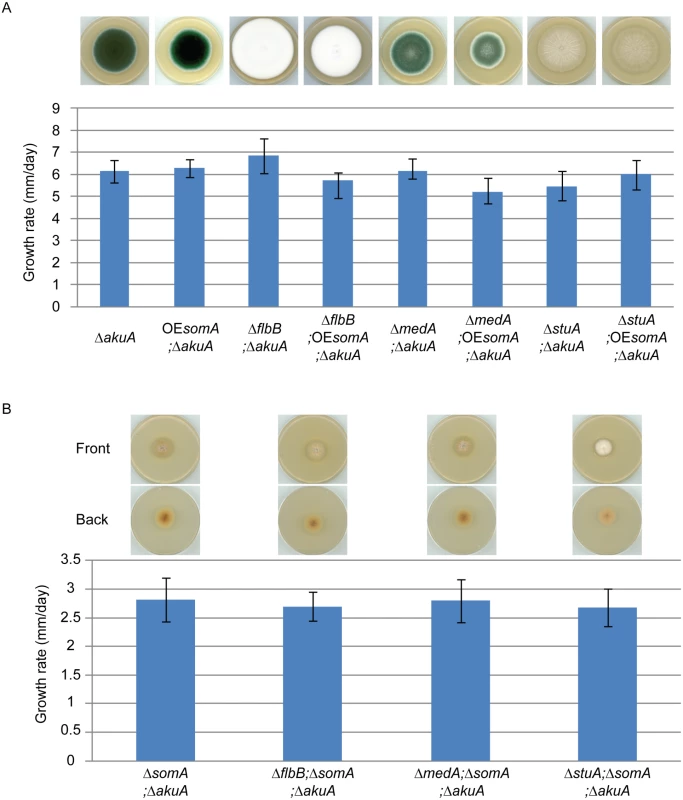
(A) Colony morphology and growth rate of the indicated strains. (B) Colony morphology and growth rate of the corresponding strains. All strains were grown on MM plate for 5 days at 37°C. Values in the graph are indicated as means ± standard error. Taken together, our combined genetic and transcriptional analysis supports that SomA regulates asexual development regulatory genes flbB and flbD and, through this pathway, affects the brlA master gene of conidiation. There is presumably an additional combinatory effect between SomA and the regulator StuA. The network of SomA, PtaB, StuA and MedA finally results in a SomA-mediated control of various adhesins encoding genes in A. fumigatus.
SomA is required for virulence in an egg and a mouse infection model
SomA is presumably required for adhesion by affecting medA expression and a ΔmedA deletion results in reduced virulence in a mice model [28]. Hence, we addressed whether SomA plays a role in virulence in animals. We established the Tet-On system in an egg infection model as a pilot study to carry out the virulence experiments. This model mimics the pulmonary invasive aspergillosis model in mice by infecting the chorioallantoic membrane in eggs [40].
In an egg infection model, the ΔsomA mutant was not included due to the severely impaired conidiation. The eggs infected with the inactive Tet-somA strain without doxycycline (Off) had no significant difference in mortality of infected eggs compared to the PBS control (p = 0.58; log-rank test). The Tet-somA (Off) showed attenuated virulence compared to the ΔakuA strain, the somA complemented and the Tet-somA (On) (p<0.05). In contrast, the On state of the Tet-somA strain which was induced by doxycycline showed similar virulence to the ΔakuA strain or the somA complemented strain (Fig 8A).
Fig. 8. SomA is required for virulence in an egg and a mouse model of invasive aspergillosis. 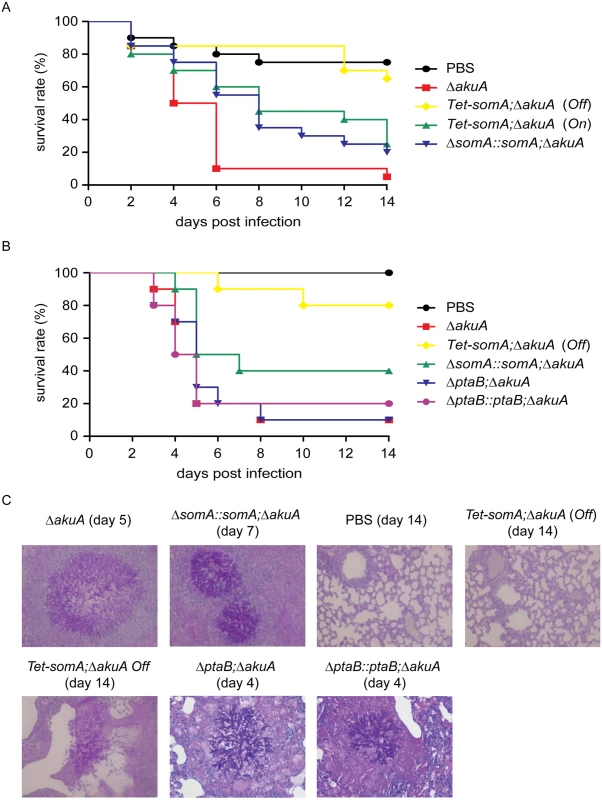
(A) Survival rate of eggs inoculated with the indicated strains. Addition of doxycycline was not performed in the somA complemented mutant and Tet-somA Off state. (B) Survival rate of mice infected with the indicated strains. (C) Histological pictures of mouse lungs obtained from indicated dates after infection with the corresponding strains or PBS. We verified the egg infection experiments by virulence assays of the mutant compared to the ΔakuA background strain in a mouse infection model for pulmonary aspergillosis [41]. As in the egg model, the ΔsomA mutant was not included due to the severely impaired conidiation. Addition of doxycycline in mouse model was not performed due to the fact that the somA complemented strain and the Tet-somA (On) strain show the virulence in the egg infection (Fig 8A). In this model, the Tet-somA mutant (Off) displayed attenuation in virulence, which was statistically significant compared to the ΔakuA background and the somA complemented strain (p<0.05) (Fig 8B). In neutropenic mice, a cellular immune response is severely restricted and development of invasive aspergillosis is characterized by massive invasive growth of the fungus. Accordingly, the presence of invasive mycelia was confirmed by histopathology in mice infected with the ΔakuA background and the complemented conidia. Even the two mice that died after infection with Tet-somA (Off) showed fungal growth within the lung tissue. However, no mycelium could be found in the majority of mice that survived the infection with Tet-somA conidia (Off) (Fig 8C). We had shown that the ΔptaB mutant had a defect of biofilm formation (Fig 5C) and PtaB interacts with SomA (Fig 3). This suggests that PtaB might also contribute to the SomA control of pathogenesis. However, a mice infection model showed that the ΔptaB mutant strain had normal virulence and invasive mycelia as the ΔakuA background strain (Fig 8B and 8C).
The resistance of oxidative stress and cell wall integrity are important virulent factors in A. fumigatus infection [42, 43]. Due to the fact that loss of somA was avirulent whereas the ΔptaB strain had normal virulence (Fig 8B), we performed a stress test to study the function of these two genes in stress response. As shown in S5 Fig, the ptaB null strain was resistant to H2O2 (3 mM) in comparison to the ΔakuA background strain. In contrast to this result the loss of somA does not increase resistance to H2O2.
Taken together, these data show that the Tet-On system is functional in the egg infection assay of A. fumigatus. The egg as well as the mouse model as established infection assays support that SomA is contributing to virulence of the opportunistic fungal pathogen A. fumigatus.
Discussion
The current understanding of Flo8/Som1 homologues and their role in adhesion and virulence is primarily based on yeasts with their dimorphic life style switching between a single cell yeast growth form and a pseudohyphal or hyphal growth mode [20, 31]. In addition, Som1 had been analyzed in plant pathogenic and saprophytic filamentous fungi [23, 44]. Here, we show that similar to the Flo8-Mfg1 complex in yeast the corresponding pair SomA-PtaB is required for biofilm formation in the opportunistic human pathogenic filamentous fungus A. fumigatus. Application of the Tet-On system revealed that the Flo8/Som1 counterpart SomA of this fungus functions in development and virulence in embryonated hen egg as well as a mouse infection model.
The mechanism of adherence has most extensively been studied in the yeast S. cerevisiae. Since adhesion is highly correlated with pathogenicity in fungi, S. cerevisiae has been used as a model for detecting adhesins encoding genes and identifying control genes of adhesion from C. albicans [45] and the filamentous fungus Verticillium longisporum [46]. Flo8 is one of the most prominent yeast regulators of adhesion and it had been demonstrated that Flo8 functions downstream of the cAMP/PKA pathway [47]. The binding of Flo8 to target promoters is regulated in budding yeast by Tpk2, which is one of the catalytic subunits of PKA, and loss of either Flo8 or Tpk2 blocks pseudohyphal growth [48]. We showed that heterologous SomA protein complements the defects of flo8 yeast in haploid adhesive (cell-cell and cell-surface) and diploid pseudohyphal filamentous growth, which require the expression of FLO11 or FLO1 [2, 32]. Furthermore, the expression of FLO11 could be activated by SomA in flo8 yeast. These results suggest that SomA might be activated by Tpk2 in S. cerevisiae and it can activate downstream genes as FLO11.
Higher expression levels of FLO11::LacZ reporter were measured by heterologous SomA compared to the Flo8. Further analysis of FLO11 promoter regions indicated that SomA binds to two similar promoter regions as Flo8 and two additional regions. All these four regions contain the Flo8 consensus binding sequence TTTGC [49]. The higher promoter binding activity of SomA might be due to a higher overall binding affinity to the FLO11 promoter compared to Flo8. Flocculation requires both adhesins Flo1 as well as Flo11 [33, 50]. The observed decreased flocculation by SomA suggests a poor binding activity to the promoter of FLO1. The fact that heterologous SomA activates the expression of FLO11 by binding to similar regions of the promoter as Flo8 supports an evolutionary conserved strategy of gene activation in yeast and filamentous fungi.
Flo8/Som1 is a transcription factor that regulates downstream targets together with other interaction partners [22, 23]. Here, we showed that SomA interacts with PtaB, the A. fumigatus homologue of yeast Mfg1. PtaB and Mfg1 proteins are members of the LIM-domain binding protein family, which play a role in development in eukaryotic cells [51, 52]. In S. cerevisiae, the Mfg1 protein forms even a ternary complex with Mss11 and Flo8 leading to efficient FLO11 expression and hence mediating invasive growth and pseudohyphal formation [22]. Deletion of either partner gene leads to the loss of these growth forms. In C. albicans, this heterotrimeric complex is also required for hyphal growth [21], but overexpression of FLO8 complemented the defect of Δmss11 in hyphal growth [53]. This indicated that Flo8 and Mss11 might share functions in development. The closest relative of an Mss11 encoding gene in A. fumigatus is somA with no other putative paralogue present in the genome. One possible explanation is that the yeast Flo8-Mss11-Mfg1 complex might correspond to a SomA-SomA-PtaB complex in A. fumigatus. The MSS11 gene in yeasts might be either a product of gene duplication in yeasts or might have been lost in filamentous fungi as the Aspergilli.
Increased protein sizes of SomA-GFP and PtaB-RFP were detected with the respective antibodies in Western experiments (Fig 3B and 3C). These results imply that SomA-GFP and PtaB-RFP is modified post-translationally. Bioinformatic prediction tools [54] suggest 10 putative ubiquitination sites (medium confidence) in SomA and three additional sites in the deduced PtaB primary amino acid sequence. Consistently, SomA-GFP is simultaneously recognized with α-Ubi and α-GFP antibodies (S6 Fig).
Asexual development in Aspergilli is a morphological change, which is reminiscent to the dimorphic life style of yeasts. Aerial hyphae are formed which can differentiate into conidiophores, containing many single cell conidia with a single nucleus per cell. These asexual spores are released into the air for dispersal of the fungus [25, 27]. The combined data of the deletion analysis and the regulated promoter suggest that SomA is a regulator of asexual spore formation and is controlling developmental steps after the formation of aerial hyphae during the asexual cycle (Fig 4B). The similar developmental phenotypes in vesicle formation between the ΔsomA (AfGB77) and the ΔbrlA (A1176) strain and the fact that SomA regulates the expression of flbB, flbD and brlA genes suggest that SomA and BrlA might be part of the same regulatory pathway. This is consistent with earlier findings where lack of conidiation was also observed in M. oryzae and A. nidulans when Som1 is inactivated [23, 44].
The connection between Flo8/Som1 and the PKA pathway seems to be conserved between yeasts and filamentous fungi. The ΔsomA strain in A. fumigatus was reduced in its growth rate in comparison to the wild type and resembles the ΔacyA mutant phenotype, which is deficient in the adenylate cyclase producing cAMP where growth was reduced and nearly no conidiation was observed [13]. MoSom1 interacts with the CpkA catalytic subunit of protein kinase A [23]. A. fumigatus pkaC1 and pkaC2 encode two cAMP dependent PKA catalytic subunits. PkaC1 belongs to the class I PKAs similar to Tpk proteins of S. cerevisiae whereas PkaC2 is dispensable for conidiation. In contrast, PkaC1 is responsible for conidiation and vegetative growth [15]. This suggests that the protein kinase PkaC1 may regulate activation of SomA and subsequently control asexual development.
SomA controls conidiation and adhesion primarily by affecting the expression of the three regulatory genes flbB, stuA and medA (Fig 9). FlbB is a bZIP transcription factor which controls together with the cMyb transcription factor FlbD the expression of the major regulatory gene brlA. The resulting protein BrlA is a C2H2 zinc finger transcription factor, which plays a key role in asexual development in the pathogen A. fumigatus and the model fungus A. nidulans [35, 55]. Deletion of either flbB or flbD results in fluffy phenotypes resembling the ΔbrlA mutant strain in A. nidulans. The FlbB impact on conidiation is similar in A. fumigatus, but the FlbD impact is less pronounced. The flbB deletion abolishes flbD expression and delays brlA expression [56]. In A. fumigatus, expression of the flbD gene requires in addition to FlbB also FlbE as further developmental regulator. Consequently, conidiation is delayed and reduced in a ΔflbB mutant [25, 56–58].
Fig. 9. Model of the SomA and SomA-PtaB genetic network in A. fumigatus. 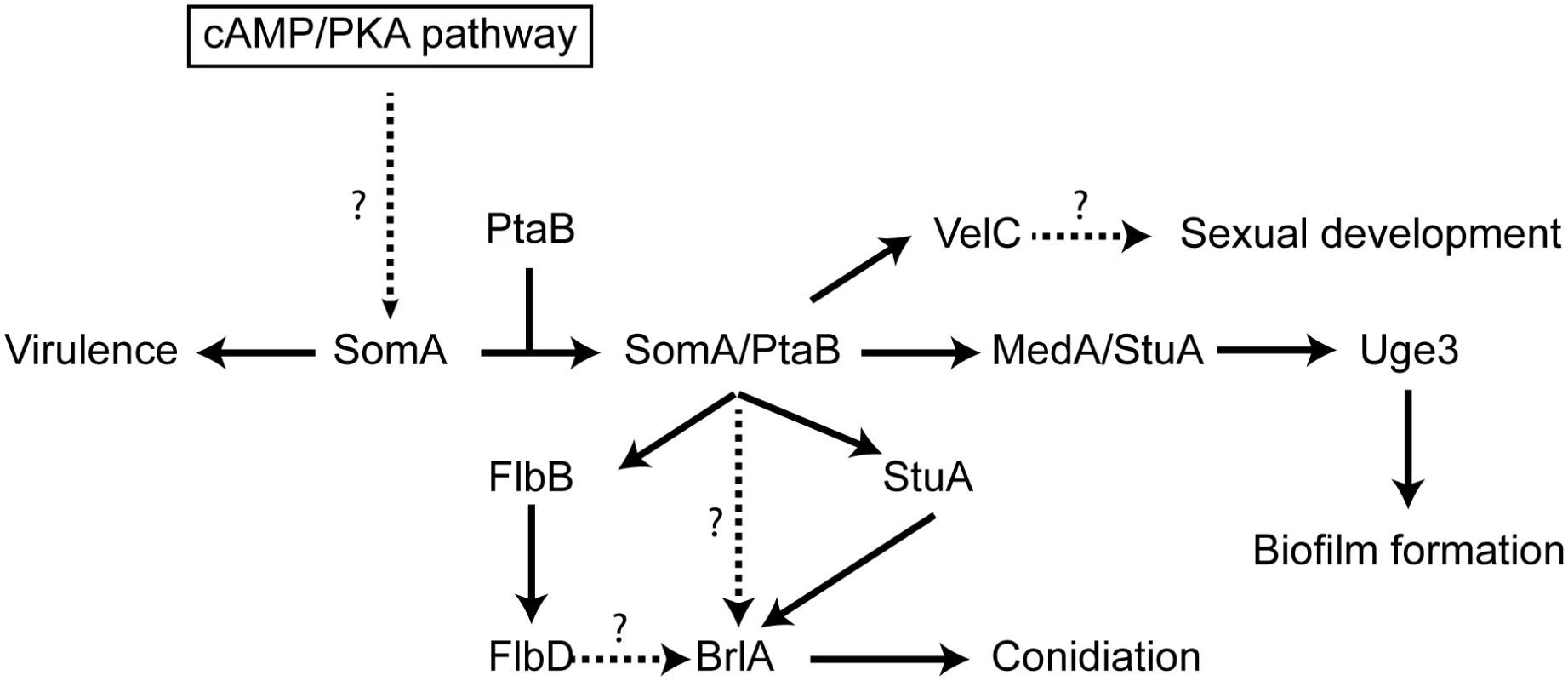
The solid arrows indicate the presented data and the results of previous studies [25, 28–30, 56]. We showed that SomA/PtaB complex is a transcriptional activator for downstream targets (Fig 6 and S3 Fig) and these proteins had different cellular functions in A. fumigatus (See Discussion). SomA controls stuA and medA expressions in the correlation between conidiation and adherence in A. fumigatus. StuA and MedA contribute to accurate spatial and temporal expression of brlA in A. nidulans [59]. Consistently, disruption of stuA and medA results in abnormal conidiophores and reduced conidiation in A. fumigatus [27–29]. The severe impairment of conidiation in the somA deletion mutant can be attributed to both StuA and MedA, which are also required for activating expression of genes for adherence. Uge3 is an UDP-glucose epimerase that interconverts UDP-glucose and UDP-galactose and mediates formation of galactosaminogalactan (GAG) [30]. This compound is part of the extracellular matrix and is required for biofilm formation as well as adherence and therefore plays a prominent role in pathogenesis of A. fumigatus [60]. The StuA binding sites (A/T)CGCG(T/A)N(A/C) had been defined [61] and are also present in the promoter regions, of the uge3 (position -1651 and -1108 bp) or AFUA_3G00880 (position -3627 bp) genes for adhesion and as well in the brlA promoter region (position -507, -753 and -3276 bp) for asexual development (S7 Fig). This indicates that StuA has a dual role in directly activating the transcription of genes for adhesion and conidiation by binding to the corresponding promoters (Fig 9). SomA is required for the expression of stuA and medA and thereby plays an important role for conidiation as well as adhesion.
In this study, an attenuated virulence of the Tet-somA mutant (Off state) resulting in significantly reduced cellular SomA protein levels was observed in infection models with embryonated egg or with mice (Fig 8). An interesting question is which of the phenotypes which are observed in the absence of sufficient amounts of SomA protein is causing the reduction in virulence. SomA is required for the normal fungal growth rate, for asexual spore formation and for adherence. The ΔstuA mutant which is located downstream of the somA gene (Fig 9) showed abnormal impaired conidiation with a nonsignificant reduced virulence [29]. Loss of the gpaB gene, which is the Gα subunit of a heterotrimeric G proteins upstream of somA resulted in no growth retardation but attenuated virulence in a mouse model [16]. Loss of somA gene resulted in abolished biofilm formation and adherence to various surfaces as well as decreased uge3 expression and GAG production (Figs 5 and 6). Gravelat et al. showed that adhesion is an important factor for full virulence in mice model. The deletion of uge3 resulted in a normal growth and morphology together with an impaired adherence and reduced virulence in mice model [30]. In contrast, the ΔptaB mutant showed normal virulence in mice model incorporate with reduced expression of uge3 gene and resistance to oxidative stress (Fig 5C and S4 and S5 Figs). These data support a complex interplay of SomA with several genetic networks which ultimately has an important impact on fungal virulence in host cells (Fig 9).
The Flo8/Som1 protein family functions downstream of the cAMP/PKA signaling pathway and this pathway regulates asexual development in both A. nidulans and A. fumigatus [25]. We showed that SomA is upstream of flbB, medA and stuA genes (Fig 7). Overexpression of the somA gene in the ΔstuA mutant had no significant phenotype compared to the ΔstuA mutant. In contrast, the ΔstuA ΔsomA double mutant strain resulted in a complex phenotype with more aerial hyphae and different lighter color in the back as a hint of changes in secondary metabolite production. This distinct phenotype of the ΔstuA ΔsomA double deletion strain could reflect the activation of additional signal pathways. Previous studies indicated that StuA is required for conidiation and the regulation of secondary metabolite clusters [29, 35]. These studies suggested that the difference of color on the back might be due to the absence of StuA protein in the double deletion mutant. The stuA gene was expressed, but was reduced, in the ΔsomA mutant (Fig 6 and S3 Fig). Macheleidt et al. showed that expression of stuA is regulated by the cAMP/PKA pathway [62]. SomA might affect the expression of the stuA gene but the interplay between both gene products is presumably more complex and might rather resemble a genetic network than a single signal transduction pathway (Fig 9).
There is an important interplay between conidiation and cell-cell adhesion. The formation of aerial hyphae results in vesicles, which further differentiate into spore forming cells by a polar budding process reminiscent to the yeast single cell growth mode [27, 63]. In A. nidulans, this results primarily in the formation of metulae cells, which are absent in A. fumigatus where directly the spore forming phialides are formed. Phialides are the cells, which produce the small hydrophobic non-motile conidia in a process similar to pseudohyphae formation where at the beginning the spores are attached to each other. Pseudohyphae formation in yeast requires adhesins such as Flo11, which mediate cell-cell adhesion [2]. StuA controls metulae and phialide differentiation in A. nidulans and consistently, Phd1 as homologue of StuA, governs pseudohyphal growth in S. cerevisiae [63]. Asexual spore formation also requires AbaA, which is located downstream of BrlA in the developmental cascade. Consistently, the AbaA corresponding yeast protein Tec1 is required for pseudohyphae formation. Tec1 can be replaced by A. nidulans AbaA to repair the defect of a Δtec1 S. cerevisiae mutant strain [64].
Several proteins providing adherence in filamentous fungi have been identified. SomA affects several adhesion related genes (Fig 6), which results in the SomA mediated plastic adherence observed in this study. Hydrophobins also play a specific role in adhesion and are amphiphilic proteins involved in aerial hyphae formation and conidiation [65]. Hydrophobins as Mhp1 of the plant pathogen Magnaporthe grisea or Mpg1 of M. oryzae are responsible for appressorium development and subsequent entry into the plant host [66, 67]. RodA is a spore hydrophobin of A. fumigatus which prevents immune recognition and provides adherence of conidia to collagen or albumin [3]. The expression of the rodA gene depends on regulators as BrlA and AbaA [25] and the rodA expression is affected by SomA (Fig 6). Recently, galactosaminogalactan was found to be an adhesive compound produced by the A. fumigatus Uge3 epimerase, which is required for virulence [30]. We showed that SomA and PtaB regulate uge3 expression as well as stuA and medA (Fig 6 and S4 Fig), which have been shown to control transcription of uge3. In addition, loss of somA showed reduced galactose and galactosamine production which are required for galactosaminogalactan formation (Fig 5B). Interestingly, the homologues of these genes are also present in the non-pathogenic A. nidulans. StuA, MedA and Som1 are required for normal conidiation in A. nidulans [27, 44] and StuA binding sequence is present in the promoter of the uge3 homologue. This indicated that SomA might regulate other unknown genes required for adhesion or pathogenicity.
Most of the filamentous pathogenic fungi show host specificity and can only infect and cause disease in either plants or animals. Only a limited number of fungal pathogens such as Aspergillus flavus or Fusarium oxysporum can cause infections in both kingdoms. Plants and animals possess distinct protective systems to prevent fungal infections. This includes the plant cuticle as barrier or increased temperatures of 37°C and higher in humans [68]. Genes which are required for cuticle degradation or appressorium formation are specific for plant pathogens [69]. The set of genes from filamentous fungal pathogens, which is important for plant as well as human pathogenic fungi is limited and includes siderophores for the uptake of iron ions and adhesins as hydrophobins [70, 71]. The transcription factor Flo8/Som1 is an interesting control gene which is required for adherence, development and pathogenicity of pathogens which include filamentous fungal plant pathogens as M. oryzae as well as dimorphic and constitutively filamentous fungal human pathogens as C. albicans and A. fumigatus, respectively.
Materials and Methods
Strains and growth conditions
The fungal strains used in this study are listed in Table 1. The AfS35 (ΔakuA) was used as the background strain [72]. A. fumigatus was grown at 37°C in minimal medium (MM) as previously described [73, 74]. 1% D-glucose as carbon source and 10 mM ammonium tartrate as nitrogen source were supplemented. 2% agar and 1 mg/L pyrithiamine were used for solid medium and selection, respectively. For pyrithiamine marker recycling, 0.5% xylose was supplemented. Escherichia coli strain DH5α was used for construction of plasmid and was propagated in LB medium (0.5% yeast extract, 1% bacto-tryptone and 1% NaCl) at 37°C. S. cerevisiae strains BY4742, Y16870, RH2656 and RH2660 (Table 1) were used for cross-species complementation. The BY4742 (flo8) is derived from S288c carrying truncated FLO8 gene [31] and RH2660 (Δflo8) is derived from Σ1278b with deletion of FLO8 gene [75, 76]. S. cerevisiae was cultivated at 30°C in either non-selective YEPD medium (1% yeast extract, 2% peptone and 2% glucose) or in SC-3 medium (0.15% yeast nitrogen base without amino acid and ammonium sulfate, 0.5% (NH4)2SO4, 2% glucose and 0.2% amino acid mixture lacking uracil, L-methionine and L-leucine). Appropriate amino acids were supplemented as required for adhesive assay and flocculation assay. For solid medium, 2% agar was added.
Tab. 1. Fungal strains used in this study. 
To determine the growth rate of the strains, 500 conidia or a portion of mycelia of the strains were inoculated in the middle of MM plates for 5 days. The colony diameters were measured every day. Doxycycline (5 mg/L) was supplemented as required.
Nucleic acid isolation and manipulations
Recombinant DNA technologies were performed according to standard methods [77]. DNA fragments for plasmid construction, hybridization probes or sequencing were amplified by Phusion polymerase (Thermo Fisher Scientific GmbH). Primers used for plasmid construction are listed in S3 Table. Isolation of genomic DNA from A. fumigatus was performed as previously described [78]. Isolation of plasmid DNA and RNA were performed using either QIAprep Miniprep Kit (Qiagen) or RNeasy Plant Mini Kit (Qiagen) referring to user’s manual, respectively. The cDNA of A. fumigatus was generated from total RNA using the QuantiTect Reverse Transcription Kit (Qiagen) following the user’s manual.
Deletion of somA
The 5’ and 3’ UTR regions were amplified with the corresponding primers HO499/500 or HO501/502. These two PCR products were fused by amplifying with the primer pair HO499/502 to yield a fragment which contains a restriction site for SfiI in the middle and a restriction site for HindIII at both ends. Then it was cloned into pJET1.2 Blunt cloning vector (Fermentas GmbH). The self excising marker system, which harbors a xylose driven β-recombinase, a pyrithiamine resistance cassette and two flanking binding sites (six), was isolated from pSK485 [79] with SfiI restriction enzyme. This recyclable marker fragment was cloned into the SfiI restriction site in the previous plasmid containing fused 5’ and 3’ UTR regions to generate pME4188.
Transformations were performed as previously described to construct A. fumigatus mutants [80]. The ΔakuA strain (AfS35) was transformed with the deletion fragment isolated from pME4188 using HindIII restriction enzyme and was selected with pyrithiamine. The positive mutant (ΔsomA::ptrA) was streaked out on MM plates containing 0.5% xylose and glucose to remove the pyrithiamine resistance and resulted in ΔsomA mutant (AfGB77).
Construction of the somA complemented strain
For complementation, the fragment containing 5’ UTR and somA gene was amplified with the primer pair HO603/601 and cloned into SmaI digested pUC19 (Fermentas GmbH) using the In-fusion HD Cloning Kit (Takara BioEurope/Clontech) to generate pME4189. Linear pME4189 amplified with primers HO711/611 was fused with the recyclable marker fragment and the 3’ UTR of somA which was amplified with the primer pair HO677/501 to yield pME4190. The complement fragment isolated from pME4190 by HindIII digestion was transformed into ΔsomA mutant and the pyrithiamine resistance was removed to generate the complemented mutant (AfGB105).
Construction of the Tet-somA strain
To overcome the defect of conidiation in the somA deletion mutant, a strain containing the conditional expression of somA gene was generated. First, the pyrithiamine resistance cassette and the Tet-On system were amplified with the primer pair HO116/675 using pCH008 [36] as template. This fragment was cloned into the linear pME4189 amplified with primers HO710/676 to yield plasmid pME4191. The Tet-On system fragment replaced 602 bp of the 5’ UTR region (position -602~-1) of somA gene. The fragment isolated from pME4191 using HindIII restriction enzyme was transformed into the ΔakuA strain to generate the Tet-somA mutant (AfGB74).
Deletion of ptaB
The deletion of the ptaB gene was carried out as followed. The 5’ UTR was amplified via PCR using the primer pair PtaB-1/PtaB-2. For the 3’ UTR the primers PtaB-3/PtaB-4 were used. As selection marker the flipper cassette published by [79] was used. The cassette was received by digestion of the plasmid pSK485 with SfiI. The three received fragments were integrated in the pBluescript II KS+ via an EcoRV restriction site. Therefore, the GeneArt Seamless Cloning and Assembly Kit (Invitrogen) was used according to user’s manual. The generated plasmid pME4361 was digested with PmeI and the received knock out fragment was integrated in the ΔakuA strain. Transformants were selected on pyrithiamine containing media and checked for correctness via Southern hybridization. To recycle the resistance cassette clones were streaked out on minimal medium containing 0.5% glucose and 0.5% xylose. The loss of the resistance cassette was checked by Southern hybridization to generate the ptaB knock out strain without pyrithiamine resistance (AfGB115).
Construction of the ptaB complemented strain
To complement the ptaB deletion mutant, a fragment containing the ptaB gene with 5’ and 3’ UTR region was amplified with primers HO885/701 and was cloned into pUC19. The rfp gene and the trpC terminator were amplified in accordance with the primer pair HO872/873 or HO874/890 and fused by amplifying with the primer pair HO872/890 to yield a 1.5 kb fragment. This fragment was cloned into the plasmid, which contains 5’UTR-ptaB-3’UTR in pUC19, amplified with primers HO887/888 to yield plasmid contains ptaB-rfp fused gene. Afterwards, the recyclable marker was cloned into the SfiI site in previous plasmid containing 5’UTR-ptaB-rfp-trpCt-3’UTR to generate pME4362. For complementation, the ΔptaB mutant was transformed with the fragment isolated from pME4362 with HindIII restriction enzyme to generate the complemented strain (AfGB116).
Construction of ΔmedA, ΔstuA and ΔflbB mutants and derivatives
To construct plasmid pME4359, the 5’ and 3’ UTR regions of the medA gene were amplified with the corresponding primers HO852/881 or HO854/855 and fused by HO852/855. The fused fragment was further cloned into pUC19. The recyclable marker was cloned into the plasmid, which contains used 5’ and 3’ UTR of medA, amplified with primers HO854/881 to generate pME4359. The construction of plasmids pME4360 and pME4358 was similar to pME4359. The 5’ and 3’ UTR regions of stuA were amplified with primer pairs HO848/880 and HO850/851. These two PCR products were fused with primer HO848/851, cloned into pUC19 and linked with recyclable marker to yield pME4360. For pME4358, two UTR regions of flbB were amplified with primers HO844/879 and HO846/847 and fused by HO 844/847. The fused PCR was cloned into pUC19. The recyclable marker was cloned into the plasmid, which contains fused UTRs of flbB, amplified with primers HO882/846 to generate pME4358. The ΔakuA strain was transformed with the deletion fragment isolated from pME4359, pME4360 or pME4358 using HindIII and ApaLI restriction enzymes to obtain ΔmedA; ΔakuA (AfGB107), ΔstuA; ΔakuA (AfGB108) or ΔflbB; ΔakuA (AfGB106) mutants.
For construction of the somA overexpression plasmid, the somA gene was amplified with primer pair HO531/532 and cloned into PmeI site in pSK379 to yield pME4363. Plasmid pME4363 was transformed into the ΔakuA background, ΔmedA, ΔstuA or ΔflbB mutants to generate the OEsomA (AfGB119), ΔmedA;OEsomA (AfGB110), ΔstuA;OEsomA (AfGB111) or ΔflbB;OEsomA (AfGB109) strains, respectively. The somA gene is ectopical overexpression. Double deletion strains ΔmedA; ΔsomA (AfGB113), ΔstuA; ΔsomA (AfGB114) or ΔflbB; ΔsomA (AfGB112) were constructed by transformation of the deletion fragment isolated from pME4188 using HindIII restriction enzyme into the corresponding strain ΔmedA, ΔstuA or ΔflbB mutant.
Southern hybridization
All mutants were confirmed by Southern hybridization which was performed as described previously [81]. Preparation of probes was carried out using AlkPhos Direct Labelling Reagents Kit (GE Healthcare) according to user’s manual. Detection of probes was performed with the CDP-Star Detection reagent (GE Healthcare) following user’s manual.
Computational analysis
Blast searches and protein conserved domain identification were conducted at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov). Protein alignments were made by Clustal Omega at European Molecular Biology Laboratory–European Bioinformatics Institute (http://www.ebi.ac.uk). Nuclear localization signal (NLS) was predicted at cNLS mapper (http://nls-mapper.iab.keio.ac.jp/). The protein name and gene number of A. fumigatus are according to the AspGD (http://www.aspergillusgenomes.org) [82]. Protein and DNA sequence analysis was performed using Lasergene software (Dna Star Inc., Madison, WI, USA).
Yeast complementation
To complement flo8-deficient yeasts, two SomA cDNA variants were amplified with primers HO441/HO442, digested with SpeI/HindIII and cloned into pME2786 and pME2787 to generate plasmids pME4192, pME4193, pME4194 and pME4195, respectively (Table 2). For positive control, S. cerevisiae Flo8 (FLO8) was amplified with primers HO446/447 and cloned into SmaI digested pME2786 and pME2787 to generate plasmids pME4196 and pME4197, respectively. Empty plasmids pME2786 and pME2787 were used as negative control. Transformation of S. cerevisiae was performed as described previously [83]. The pME2786 based plasmids were transformed into haploid strain BY4742 and selected on SC-Leu medium; meanwhile, the pME2787 based plasmids were transformed into haploid strain BY4742 and diploid strain RH2656 and RH2660 using selection medium SC-Ura.
Tab. 2. Plasmids used in this study. 
For haploid adhesion assays, the transformed strains were grown on corresponding selective plates for 3 days at 30°C, and then those plates were washed with water until the negative control was washed away. The plates were photographed before and after washing. The flocculation assay in haploid yeasts was performed as previously described [33]. Briefly, the transformed BY4742 strains were inoculated in either SC-Leu or SC-Ura liquid medium and incubated for one day at 30°C. Following, 1 mL of EDTA (0.5 M, pH8) was added and the samples were vortexed until flocculation were disrupted. The value of flocculation was determined by F = 1-B/A, where A is OD600 in solution without 0.1% CaCl2 and B is OD600 in the presence of 0.1% CaCl2. For pseudohyphal growth, diploid transformants were grown on SLAD (synthetic low ammonium dextrose medium) for 6 days at 30°C, and then the plates were photographed.
Analysis of FLO11 promoter elements and promoter binding
To verify that SomA could act on the FLO11 promoter, both pME4192 and pME4193 (Table 2) were co-transformed with plasmid pME2167 which contains 3 kb of the FLO11 promoter fused with LacZ reporter gene, in Y16870 strain (flo8/Δflo1) and transformants were selected on SC-Ura-Leu medium. Transformations of plasmid pME2167 with either FLO8 (pME4196) or empty vector (pME2786) in Y16870 strain were used as positive or negative control, respectively. To identify the specific DNA motif that was regulated by SomA and Flo8, 14 reporter constructs containing 400 bp FLO11 promoter fragments which have 200 bp overlaps and were cloned in front of CYC1::LacZ fused gene [34]. These 14 reporter plasmids (pFLO11-2/1 to pFLO11-15/14) were co-transformed with plasmids pME4192, pME4193 or pME4196 in Y16870 and selected with SC-Ura-Leu medium. The regulation of SomA and Flo8 on the FLO11 promoter was determined by β-galactosidase assays.
β-galactosidase activity assays
The assays were performed as previously described [75]. Yeast strains were grown in SC-Ura-Leu liquid medium overnight as pre-culture, 1 mL of the pre-culture was added to 10 ml of SC-Ura-Leu-Met liquid medium as main-culture for 6 h. β-galactosidase activities were calculated by following formula (OD420 × 0.35) / (0.0045 × protein concentration × extract volume × time) [84]. Protein concentrations were determined by OD595 with Bradford assay [85].
Microscopy analysis
To analyze mycelial morphology, strains were grown on agar-coated slides with thin layer of MM agar for 28 h at 37°C and then were observed under the Olympus Axiolab microscope with 400-fold magnification. For aerial hyphae visualization, strains were grown on agar slides with either MM agar or MM agar containing 5 mg/L doxycycline for 28 h at 37°C, and were observed under the microscope as well.
Construction of somA-gfp and somA-gfp/ptaB-rfp strains
To construct SomA-GFP tagged protein, gfp gene was amplified with primers HO210/713 from pME4435. The 3’ UTR region of somA gene was amplified with primer set HO648/677. This fragment and gfp fragment were cloned into linear pME4189 containing 5’UTR-somA gene amplified with primers HO611/712 using In-fusion Kit resulting plasmid carrying somA-gfp fused gene together with 5’ and 3’ UTR regions. Then, the recyclable marker fragment was cloned into previous plasmid amplified with HO697/501 to yield pME4198. The ΔakuA background strain was transformed with somA-gfp fused fragment which was isolated from pME4198 using HindIII restriction enzyme and selected by pyrithiamine to yield somA-gfp strain (AfGB65). The ΔakuA strain was transformed with pME4435 containing overexression gfp for GFP-Trap control. To perform the co-immunoprecipitation experiments, the fragment isolated from pME4362 with HindIII restrict enzyme was transformed into the somA-gfp mutant and resulted in ptaB-rfp/somA-gfp strain (AfGB117).
Immunoprecipitation and Trypsin digestion
A GFP overexpression strain and two independent SomA-GFP strains were grown in 200 mL of MM for 24 h at 37°C. Total proteins were extracted by mixing grinded mycelia with B* buffer (300 mM NaCl, 100mM Tris pH 7.0, 10% glycerol, 2 mM EDTA, 0.02% NP40, 2 mM DTT, 1 mM PMSF, 2 protease inhibitor pills/1 mL (Complete, EDTA-free, Roche)). The GFP trapping was performed by following steps: The crude protein extracts were mixed with 15 μL GFP-Trap beads (ChromoTek) which has been washed with B* buffer and incubated for 2 h on a rotating machine at 4°C. After the incubation, the beads were washed twice with 1.5 mL and 1 mL of B* buffer. After the centrifugation for 1 min at 4500 rpm at 4°C, the supernatant was removed. The beads were resuspended with 40 μL 6 X loading dye (250 mM Tris pH 6.8, 15% β-mercaptoethanol, 30% glycerol, 7% SDS, 0.3% bromophenol blue) and were boiled for 6–8 min at 95°C to separate the proteins from the beads. The eluted protein were applied to 12% SDS-PAGE. Gel pieces were isolated and performed trypsin digestion. Trypsin digestion was performed as previously described [86, 87].
The somA-gfp/ptaB-rfp strain (AfGB117), GFP (AfGB76) or RFP overexpression (AfGB118) strains were grown in 1 L of MM for 24 h at 37°C for co-immunoprecipitation experiment. Protein extraction and immunoprecipitation were performed as previous described. RFP-Trap beads (ChromoTek) was used for the RFP trapping.
Western experiments
20 μg of eluted protein from co-immunoprecipitations were subjected to 12% SDS-PAGE after heating in SDS loading dye at 95°C for 10 min and were transferred to a nitrocellulose membrane (Whatman). The enhanced chemiluminescence method was used for detection as previous described [88]. Signals were recorded with a Fusion-SL7 detection system (Peqlab) and the Bio 1D software (Peqlab). Detection of GFP or RFP fused proteins was carried out using α-GFP (Santa Cruz) or α-RFP antibody (ChromoTek), respectively. UbiA antibody (Genescript) was used for ubiquitin detection. The horseradish peroxidase-coupled rabbit (Invitrogen) or mouse antibody (Jackson ImmunoResearch) were used as secondary antibodies.
Mass spectrometry analysis and database searching
Mass spectrometry analysis was performed as previously described [87, 89]. Orbitrap raw files were analyzed with the Proteome Discoverer 1.4 software (Thermo Scientific, San Jose, Ca, USA) using the Mascot and SequestHT search engines with an A. fumigatus protein database with the following criteria: Peptide mass tolerance 10 ppm; MS/MS ion mass tolerance 0.8 Da, and up to two missed cleavages allowed. The variable modification considered was methionine oxidation, and carbamidomethylation was considered as fixed modification. High peptide confidence and a minimum of two peptides per protein were used as result filters. Heatmaps of MaxQuant results (version 1.4.1.2 v) were made with Perseus (version 1.4.1.3 v) software [90].
Adherence assays
Two adherence assays were performed as previous described [25]. For biofilm formation 1 mL of Sabouraud broth containing 105 conidia of strains were inoculated into 6-well Nunclon Δ surface culture plates (Nunc) at 37°C for 24 h and 5 mg/L doxycycline was supplemented in the medium of indicated strains. Afterwards, the culture medium was removed and the wells were washed three times with PBS. Biofilm was visualized by applying 2 mL of 0.1% crystal violet solution for 5 min. Excess stain was removed and the plates were washed three time with 3 mL of sterile water. The remained stain in biofilm was extracted by adding 1 mL of ethanol. The biofilm density was determined by measuring the absorbance of the destained solution at 570 nm. The biofilm formation assays were performed in triplicate and experiments were repeated 3 times.
To test the adherence of mutants to plastic and fibronectin, 104 conidia in 1 mL of Sabouraud broth were incubated at 37°C for 8 h for germination and 5 mg/L doxycycline were supplemented in the medium of the indicated strains. The 6-well culture plates were untreated or coated with 0.01 mg/mL of fibronectin overnight and were inoculated with 150 germlings of the mutants and incubated at 37°C for 30 min. Afterwards, wells were washed 3 times with 3 mL of PBS and covered with YEPD agar. Fungal colony was quantified after incubation at 37°C. The adherence assay was performed in triplicates and experiments were repeated 3 times.
Quantitative real-time PCR analysis
Quantitative real-time PCR (qRT-PCR) was performed with MESA GREEN qPCR MasterMix Plus for SYBR (Eurogentec) using CFX Connect Real-Time PCR system (Bio-Rad). The ΔakuA strain and mutants were grown in liquid MM medium for 20 h at 37°C and RNA was extracted using RNeasy Plant Mini Kit (Qiagen). DNase digestion and subsequent cDNA synthesis was carried out using 0.8 μg of RNA with the QuantiTect Reverse Transcription Kit (Qiagen). To perform the shift experiments, the indicated strains grown on liquied MM medium for 18 h at 37°C. Afterwards, the mycelium were shifted to liquid MM medium for 8 h or solid MM plate for 24 h. Addition of Doxycycline (5 mg/L) was noted. Each sample was performed in duplicates and the experiment was repeated two times. The histone H2A (3G05360) was used as reference gene for normalization. The relative expression of the gene of interest was calculated using the ΔΔCT method as previously described [91]. All the primers used for quantitative real-time PCR were listed in S3 Table.
Galactosaminogalactan (GAG) assay
The assay was performed as previous described with modification [30, 92]. 5*107 spores of the strain Tet-somA were inoculated in 100 ml modified Brian medium with 50 μg/ml doxycycline and grown for 20 hours at 37°C. The mycelium was shifted to 100 ml modified Brian medium with and without doxycycline. After 24 hours of growth at 37°C, the culture supernatants were precipitated with 2.5 volumes of ethanol. The pellets were washed with 150 mM NaCl and extracted with 8 M urea. The supernatants were dialyzed against water and dried. Polysaccharides were hydrolyzed (4 M TFA, 100°C, 4 h) and the obtained monosaccharides were derivatized with MSTFA and measured in Gas chromatography-mass spectrometry (GC-MS) (Varian CP-Sil8 CB for amines (30 m, 0.25 mm, 0.25 μm)).
Stress test
Fresh harvest spores were counted and adjusted to 106 spores/ml. Approximately 2000 spores were spotted on minimal medium plates and incubated for three days at 37°C. For inducing cell wall stress conditions SDS was used at a final concentration of 0.01%. For oxidative stress H2O2 was used at a final concentration of 2 mM and 3 mM, respectively
Ethics statement
Mice were cared for in accordance with the principles outlined by the European Convention for the Protection of Vertebrate Animals Used for Experimental and Other Scientific Purposes. According to that, chicken embryo is not animal (http://conventions.coe.int/Treaty/en/Treaties/Html/123.htm). All animal experiments were in compliance with the German animal protection law and were approved by the responsible Federal State authority “Thüringer Landesamt für Lebensmittelsicherheit und Verbraucherschutz” and ethics committee “Beratende Komission nach § 15 Abs. 1 Tierschutzgesetz” with the permit Reg.-Nr. 03-001/12. Fertilized chicken eggs were obtained from a local producer and stored at 4°C for maximum of 4 days before incubation. Embryonation was performed by incubation of the fertilized eggs in an incubator with 60% humidity at 37°C in the lab. The embryonated eggs were infected with spores on day 10 of embryonation and the infection experiments was terminated on day 17 of embryonation.
Egg infection model
Egg infection model was performed as described previously [40]. Eggs were incubated in an incubator with 60% humidity for 10 days at 37°C. Each A. fumigatus strain was grown on malt extract agar (Oxoid) with 5 mg/L doxycycline for 7 days and the conidia were harvested freshly on the day for infection. Each egg was inoculated with 1000 conidia in 100 μL PBS with 5 mg/L doxycycline (corresponding to the volume of the egg) and 20 eggs were inoculated for one strain. Doxycycline was not added to the PBS solution in the Tet-somA (Off state) and the complemented strain. The embryonic death was determined by the loss of movement. Survival data were plotted by Kaplan–Meier curves and statistically analyzed by log rank test using Graph Pad Prism 5.00 (GraphPad Software).
Mouse infection model
Virulence of A. fumigatus mutant strains was tested in an established murine model for invasive pulmonary aspergillosis [41]. To induce neutropenia in female CD-1 mice (Charles River), cyclophosphamide (140 mg/kg; Sigma-Aldrich) was injected intraperitoneally on days -4, -1, 2, 5, 8 and 11, with an additional subcutaneous dose of cortisone acetate (200 mg/kg) on day -1. Mice were anaesthetized and intranasally infected with 20 μL of a fresh suspension containing 2 x 105 conidia. A control group was mock-infected with PBS.
The health status was monitored twice a day for 14 days and moribund animals, defined by severe dyspnoea, severe lethargy, or weight loss > 20%, were sacrificed. Infections were performed with a group of 10 mice for each tested strain. A control group of 5 mice was not infected (inhaled PBS). Survival data were plotted by Kaplan–Meier curves and statistically analyzed by log rank test and Gehan-Breslow-Wilcoxon test using Graph Pad Prism 5.00 (GraphPad Software).
Lungs from sacrificed animals were fixed in formalin and paraffin-embedded for histopathological analyses according to standard protocols. 4-μm sections were stained using Periodic acid-Schiff (PAS, hyphae stained pink). Sections were analyzed with the Zeiss Axio Imager.M2 microscope and images were taken with a Zeiss Axiocam 105 color camera and analyzed by Zen 2012 software (Zeiss).
Accession number
S. cerevisiae: Flo8, P40068; Mfg1, Q07684; Mss11, Q03825; Flo11, P08640.
A. fumigatus: SomA, Q4WAR8; PtaB, Q4X0N1; BrlA, Q4WRE4; RodA, P41746; FlbB, Q4X053; FlbC, Q4X0E3; FlbD, Q4WK99; StuA, Q4X228; MedA, Q4X0J5; Uge3, Q4WX18; AfuA_3g13110, Q4WYI0; AfuA_3g00880, Q4WFV6; VelC, Q4WPM3.
Supporting Information
Zdroje
1. de Groot PW, Bader O, de Boer AD, Weig M, Chauhan N. Adhesins in human fungal pathogens: glue with plenty of stick. Eukaryot Cell. 2013;12(4):470–81. Epub 2013/02/08. doi: 10.1128/EC.00364-12 23397570
2. Brückner S, Mösch HU. Choosing the right lifestyle: adhesion and development in Saccharomyces cerevisiae. FEMS Microbiol Rev. 2012;36(1):25–58. Epub 2011/05/20. doi: 10.1111/j.1574-6976.2011.00275.x 21521246
3. Thau N, Monod M, Crestani B, Rolland C, Tronchin G, Latgé JP, et al. rodletless mutants of Aspergillus fumigatus. Infect Immun. 1994;62(10):4380–8. Epub 1994/10/01. 7927699
4. Sheppard DC. Molecular mechanism of Aspergillus fumigatus adherence to host constituents. Curr Opin Microbiol. 2011;14(4):375–9. Epub 2011/07/23. doi: 10.1016/j.mib.2011.07.006 21784698
5. Aimanianda V, Bayry J, Bozza S, Kniemeyer O, Perruccio K, Elluru SR, et al. Surface hydrophobin prevents immune recognition of airborne fungal spores. Nature. 2009;460(7259):1117–21. doi: 10.1038/nature08264 19713928
6. Mayer FL, Wilson D, Hube B. Candida albicans pathogenicity mechanisms. Virulence. 2013;4(2):119–28. Epub 2013/01/09. doi: 10.4161/viru.22913 23302789
7. Li L, Wright SJ, Krystofova S, Park G, Borkovich KA. Heterotrimeric G protein signaling in filamentous fungi. Annu Rev Microbiol. 2007;61 : 423–52. Epub 2007/05/18. 17506673
8. Mitchell TK, Dean RA. The cAMP-dependent protein kinase catalytic subunit is required for appressorium formation and pathogenesis by the rice blast pathogen Magnaporthe grisea. Plant Cell. 1995;7(11):1869–78. Epub 1995/11/01. 8535140
9. Dürrenberger F, Wong K, Kronstad JW. Identification of a cAMP-dependent protein kinase catalytic subunit required for virulence and morphogenesis in Ustilago maydis. Proc Natl Acad Sci U S A. 1998;95(10):5684–9. Epub 1998/05/20. 9576944
10. Kozubowski L, Lee SC, Heitman J. Signalling pathways in the pathogenesis of Cryptococcus. Cell Microbiol. 2009;11(3):370–80. Epub 2008/12/19. doi: 10.1111/j.1462-5822.2008.01273.x 19170685
11. Hogan DA, Sundstrom P. The Ras/cAMP/PKA signaling pathway and virulence in Candida albicans. Future Microbiol. 2009;4(10):1263–70. Epub 2009/12/10. doi: 10.2217/fmb.09.106 19995187
12. Ramanujam R, Naqvi NI. PdeH, a high-affinity cAMP phosphodiesterase, is a key regulator of asexual and pathogenic differentiation in Magnaporthe oryzae. PLoS Pathog. 2010;6(5):e1000897. Epub 2010/05/14. doi: 10.1371/journal.ppat.1000897 20463817
13. Liebmann B, Gattung S, Jahn B, Brakhage AA. cAMP signaling in Aspergillus fumigatus is involved in the regulation of the virulence gene pksP and in defense against killing by macrophages. Mol Genet Genomics. 2003;269(3):420–35. Epub 2003/05/07. 12734751
14. Zhao W, Panepinto JC, Fortwendel JR, Fox L, Oliver BG, Askew DS, et al. Deletion of the regulatory subunit of protein kinase A in Aspergillus fumigatus alters morphology, sensitivity to oxidative damage, and virulence. Infect Immun. 2006;74(8):4865–74. Epub 2006/07/25. 16861675
15. Fuller KK, Richie DL, Feng X, Krishnan K, Stephens TJ, Wikenheiser-Brokamp KA, et al. Divergent Protein Kinase A isoforms co-ordinately regulate conidial germination, carbohydrate metabolism and virulence in Aspergillus fumigatus. Mol Microbiol. 2011;79(4):1045–62. Epub 2011/01/06. doi: 10.1111/j.1365-2958.2010.07509.x 21210869
16. Liebmann B, Müller M, Braun A, Brakhage AA. The cyclic AMP-dependent protein kinase a network regulates development and virulence in Aspergillus fumigatus. Infect Immun. 2004;72(9):5193–203. Epub 2004/08/24. 15322014
17. Kontoyiannis DP, Marr KA, Park BJ, Alexander BD, Anaissie EJ, Walsh TJ, et al. Prospective surveillance for invasive fungal infections in hematopoietic stem cell transplant recipients, 2001–2006: overview of the Transplant-Associated Infection Surveillance Network (TRANSNET) Database. Clin Infect Dis. 2010;50(8):1091–100. Epub 2010/03/12. doi: 10.1086/651263 20218877
18. Pappas PG, Alexander BD, Andes DR, Hadley S, Kauffman CA, Freifeld A, et al. Invasive fungal infections among organ transplant recipients: results of the Transplant-Associated Infection Surveillance Network (TRANSNET). Clin Infect Dis. 2010;50(8):1101–11. Epub 2010/03/12. doi: 10.1086/651262 20218876
19. Tekaia F, Latgé JP. Aspergillus fumigatus: saprophyte or pathogen? Curr Opin Microbiol. 2005;8(4):385–92. Epub 2005/07/16. 16019255
20. Cao F, Lane S, Raniga PP, Lu Y, Zhou Z, Ramon K, et al. The Flo8 transcription factor is essential for hyphal development and virulence in Candida albicans. Mol Biol Cell. 2006;17(1):295–307. Epub 2005/11/02. 16267276
21. Shapiro RS, Ryan O, Boone C, Cowen LE. Regulatory circuitry governing morphogenesis in Saccharomyces cerevisiae and Candida albicans. Cell Cycle. 2012;11(23):4294–5. Epub 2012/10/24. doi: 10.4161/cc.22608 23095675
22. Ryan O, Shapiro RS, Kurat CF, Mayhew D, Baryshnikova A, Chin B, et al. Global gene deletion analysis exploring yeast filamentous growth. Science. 2012;337(6100):1353–6. Epub 2012/09/18. doi: 10.1126/science.1224339 22984072
23. Yan X, Li Y, Yue X, Wang C, Que Y, Kong D, et al. Two novel transcriptional regulators are essential for infection-related morphogenesis and pathogenicity of the rice blast fungus Magnaporthe oryzae. PLoS Pathog. 2011;7(12):e1002385. Epub 2011/12/01. doi: 10.1371/journal.ppat.1002385 22144889
24. Bayram Ö, Braus GH. Coordination of secondary metabolism and development in fungi: the velvet family of regulatory proteins. FEMS Microbiol Rev. 2012;36(1):1–24. Epub 2011/06/11. doi: 10.1111/j.1574-6976.2011.00285.x 21658084
25. Yu JH. Regulation of Development in Aspergillus nidulans and Aspergillus fumigatus. Mycobiology. 2010;38(4):229–37. Epub 2010/12/31. doi: 10.4489/MYCO.2010.38.4.229 23956662
26. Mah JH, Yu JH. Upstream and downstream regulation of asexual development in Aspergillus fumigatus. Eukaryot Cell. 2006;5(10):1585–95. Epub 2006/10/13. 17030990
27. Adams TH, Wieser JK, Yu JH. Asexual sporulation in Aspergillus nidulans. Microbiol Mol Biol Rev. 1998;62(1):35–54. Epub 1998/04/08. 9529886
28. Gravelat FN, Ejzykowicz DE, Chiang LY, Chabot JC, Urb M, Macdonald KD, et al. Aspergillus fumigatus MedA governs adherence, host cell interactions and virulence. Cell Microbiol. 2010;12(4):473–88. Epub 2009/11/04. doi: 10.1111/j.1462-5822.2009.01408.x 19889083
29. Sheppard DC, Doedt T, Chiang LY, Kim HS, Chen D, Nierman WC, et al. The Aspergillus fumigatus StuA protein governs the up-regulation of a discrete transcriptional program during the acquisition of developmental competence. Mol Biol Cell. 2005;16(12):5866–79. Epub 2005/10/05. 16207816
30. Gravelat FN, Beauvais A, Liu H, Lee MJ, Snarr BD, Chen D, et al. Aspergillus galactosaminogalactan mediates adherence to host constituents and conceals hyphal β-glucan from the immune system. PLoS Pathog. 2013;9(8):e1003575. Epub 2013/08/22. doi: 10.1371/journal.ppat.1003575 23990787
31. Liu H, Styles CA, Fink GR. Saccharomyces cerevisiae S288C has a mutation in FLO8, a gene required for filamentous growth. Genetics. 1996;144(3):967–78. Epub 1996/11/01. 8913742
32. Fichtner L, Schulze F, Braus GH. Differential Flo8p-dependent regulation of FLO1 and FLO11 for cell-cell and cell-substrate adherence of S. cerevisiae S288c. Mol Microbiol. 2007;66(5):1276–89. Epub 2007/11/16. 18001350
33. Bester MC, Pretorius IS, Bauer FF. The regulation of Saccharomyces cerevisiae FLO gene expression and Ca2+ -dependent flocculation by Flo8p and Mss11p. Curr Genet. 2006;49(6):375–83. Epub 2006/03/25. 16568252
34. Rupp S, Summers E, Lo HJ, Madhani H, Fink G. MAP kinase and cAMP filamentation signaling pathways converge on the unusually large promoter of the yeast FLO11 gene. EMBO J. 1999;18(5):1257–69. Epub 1999/03/04. 10064592
35. Twumasi-Boateng K, Yu Y, Chen D, Gravelat FN, Nierman WC, Sheppard DC. Transcriptional profiling identifies a role for BrlA in the response to nitrogen depletion and for StuA in the regulation of secondary metabolite clusters in Aspergillus fumigatus. Eukaryot Cell. 2009;8(1):104–15. Epub 2008/11/21. doi: 10.1128/EC.00265-08 19028996
36. Helmschrott C, Sasse A, Samantaray S, Krappmann S, Wagener J. Upgrading fungal gene expression on demand: improved systems for doxycycline-dependent silencing in Aspergillus fumigatus. Appl Environ Microbiol. 2013;79(5):1751–4. Epub 2012/12/28. doi: 10.1128/AEM.03626-12 23275515
37. Park HS, Bayram Ö, Braus GH, Kim SC, Yu JH. Characterization of the velvet regulators in Aspergillus fumigatus. Mol Microbiol. 2012;86(4):937–53. Epub 2012/09/14. doi: 10.1111/mmi.12032 22970834
38. Ahmed YL, Gerke J, Park HS, Bayram Ö, Neumann P, Ni M, et al. The velvet family of fungal regulators contains a DNA-binding domain structurally similar to NF-κB. PLoS Biol. 2013;11(12):e1001750. Epub 2013/12/31. doi: 10.1371/journal.pbio.1001750 24391470
39. Chaudhuri R, Ansari FA, Raghunandanan MV, Ramachandran S. FungalRV: Adhesin prediction and immunoinformatics portal for human fungal pathogens. BMC Genomics. 2011;12(1):192–205. Epub 2011/04/19.
40. Jacobsen ID, Große K, Slesiona S, Hube B, Berndt A, Brock M. Embryonated eggs as an alternative infection model to investigate Aspergillus fumigatus virulence. Infect Immun. 2010;78(7):2995–3006. Epub 2010/04/26. doi: 10.1128/IAI.00268-10 20421382
41. Kupfahl C, Heinekamp T, Geginat G, Ruppert T, Hartl A, Hof H, et al. Deletion of the gliP gene of Aspergillus fumigatus results in loss of gliotoxin production but has no effect on virulence of the fungus in a low-dose mouse infection model. Mol Microbiol. 2006;62(1):292–302. Epub 2006/08/31. 16956378
42. Ben-Ami R, Lewis RE, Leventakos K, Latge JP, Kontoyiannis DP. Cutaneous model of invasive aspergillosis. Antimicrob Agents Chemother. 2010;54(5):1848–54. Epub 2010/02/11. doi: 10.1128/AAC.01504-09 20145078
43. Wagener J, Echtenacher B, Rohde M, Kotz A, Krappmann S, Heesemann J, et al. The putative alpha-1,2-mannosyltransferase AfMnt1 of the opportunistic fungal pathogen Aspergillus fumigatus is required for cell wall stability and full virulence. Eukaryot Cell. 2008;7(10):1661–73. Epub 2008/08/19. doi: 10.1128/EC.00221-08 18708564
44. Lee BY, Han SY, Choi HG, Kim JH, Han KH, Han DM. Screening of growth - or development-related genes by using genomic library with inducible promoter in Aspergillus nidulans. J Microbiol. 2005;43(6):523–8. Epub 2006/01/18. 16410769
45. Li F, Palecek SP. Identification of Candida albicans genes that induce Saccharomyces cerevisiae cell adhesion and morphogenesis. Biotechnol Prog. 2005;21(6):1601–9. Epub 2005/12/03. 16321041
46. Tran VT, Braus-Stromeyer SA, Kusch H, Reusche M, Kaever A, Kuhn A, et al. Verticillium transcription activator of adhesion Vta2 suppresses microsclerotia formation and is required for systemic infection of plant roots. New Phytol. 2014;202(2):565–81. Epub 2014/01/17. doi: 10.1111/nph.12671 24433459
47. Pan X, Heitman J. Protein kinase A operates a molecular switch that governs yeast pseudohyphal differentiation. Mol Cell Biol. 2002;22(12):3981–93. Epub 2002/05/25. 12024012
48. Pan X, Heitman J. Cyclic AMP-dependent protein kinase regulates pseudohyphal differentiation in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19(7):4874–87. Epub 1999/06/22. 10373537
49. Kim TS, Kim HY, Yoon JH, Kang HS. Recruitment of the Swi/Snf complex by Ste12-Tec1 promotes Flo8-Mss11-mediated activation of STA1 expression. Mol Cell Biol. 2004;24(21):9542–56. Epub 2004/10/16. 15485921
50. Govender P, Domingo JL, Bester MC, Pretorius IS, Bauer FF. Controlled expression of the dominant flocculation genes FLO1, FLO5, and FLO11 in Saccharomyces cerevisiae. Appl Environ Microbiol. 2008;74(19):6041–52. Epub 2008/08/15. doi: 10.1128/AEM.00394-08 18708514
51. Jurata LW, Kenny DA, Gill GN. Nuclear LIM interactor, a rhombotin and LIM homeodomain interacting protein, is expressed early in neuronal development. Proc Natl Acad Sci U S A. 1996;93(21):11693–8. Epub 1996/10/15. 8876198
52. van Meyel DJ, Thomas JB, Agulnick AD. Ssdp proteins bind to LIM-interacting co-factors and regulate the activity of LIM-homeodomain protein complexes in vivo. Development. 2003;130(9):1915–25. Epub 2003/03/19. 12642495
53. Su C, Li Y, Lu Y, Chen J. Mss11, a transcriptional activator, is required for hyphal development in Candida albicans. Eukaryot Cell. 2009;8(11):1780–91. Epub 2009/09/04. doi: 10.1128/EC.00190-09 19734367
54. Radivojac P, Vacic V, Haynes C, Cocklin RR, Mohan A, Heyen JW, et al. Identification, analysis, and prediction of protein ubiquitination sites. Proteins. 2010;78(2):365–80. Epub 2009/09/02. doi: 10.1002/prot.22555 19722269
55. Tao L, Yu JH. AbaA and WetA govern distinct stages of Aspergillus fumigatus development. Microbiology. 2011;157(Pt 2):313–26. Epub 2010/10/21. doi: 10.1099/mic.0.044271-0 20966095
56. Xiao P, Shin KS, Wang T, Yu JH. Aspergillus fumigatus flbB encodes two basic leucine zipper domain (bZIP) proteins required for proper asexual development and gliotoxin production. Eukaryot Cell. 2010;9(11):1711–23. Epub 2010/09/17. doi: 10.1128/EC.00198-10 20852021
57. Garzia A, Etxebeste O, Herrero-Garcia E, Fischer R, Espeso EA, Ugalde U. Aspergillus nidulans FlbE is an upstream developmental activator of conidiation functionally associated with the putative transcription factor FlbB. Mol Microbiol. 2009;71(1):172–84. Epub 2008/11/05. doi: 10.1111/j.1365-2958.2008.06520.x 19007409
58. Garzia A, Etxebeste O, Herrero-Garcia E, Ugalde U, Espeso EA. The concerted action of bZip and cMyb transcription factors FlbB and FlbD induces brlA expression and asexual development in Aspergillus nidulans. Mol Microbiol. 2010;75(5):1314–24. Epub 2010/02/01. doi: 10.1111/j.1365-2958.2010.07063.x 20132447
59. Krijgsheld P, Bleichrodt R, van Veluw GJ, Wang F, Müller WH, Dijksterhuis J, et al. Development in Aspergillus. Stud Mycol. 2013;74(1):1–29. Epub 2013/03/02. doi: 10.3114/sim0006 23450714
60. Loussert C, Schmitt C, Prevost MC, Balloy V, Fadel E, Philippe B, et al. In vivo biofilm composition of Aspergillus fumigatus. Cell Microbiol. 2010;12(3):405–10. Epub 2009/11/04. doi: 10.1111/j.1462-5822.2009.01409.x 19889082
61. Dutton JR, Johns S, Miller BL. StuAp is a sequence-specific transcription factor that regulates developmental complexity in Aspergillus nidulans. EMBO J. 1997;16(18):5710–21. Epub 1997/10/06. 9312029
62. Macheleidt J, Scherlach K, Neuwirth T, Schmidt-Heck W, Straßburger M, Spraker J, et al. Transcriptome analysis of cyclic AMP-dependent protein kinase A-regulated genes reveals the production of the novel natural compound fumipyrrole by Aspergillus fumigatus. Mol Microbiol. 2015;96(1):148–62. Epub 2015/03/11. doi: 10.1111/mmi.12926 25582336
63. Gimeno CJ, Fink GR. Induction of pseudohyphal growth by overexpression of PHD1, a Saccharomyces cerevisiae gene related to transcriptional regulators of fungal development. Mol Cell Biol. 1994;14(3):2100–12. Epub 1994/03/01. 8114741
64. Gavrias V, Andrianopoulos A, Gimeno CJ, Timberlake WE. Saccharomyces cerevisiae TEC1 is required for pseudohyphal growth. Mol Microbiol. 1996;19(6):1255–63. Epub 1996/03/01. 8730867
65. Linder MB, Szilvay GR, Nakari-Setälä T, Penttilä ME. Hydrophobins: the protein-amphiphiles of filamentous fungi. FEMS Microbiol Rev. 2005;29(5):877–96. Epub 2005/02/21. 16219510
66. Kim S, Ahn IP, Rho HS, Lee YH. MHP1, a Magnaporthe grisea hydrophobin gene, is required for fungal development and plant colonization. Mol Microbiol. 2005;57(5):1224–37. Epub 2005/08/17. 16101997
67. Talbot NJ, Ebbole DJ, Hamer JE. Identification and characterization of MPG1, a gene involved in pathogenicity from the rice blast fungus Magnaporthe grisea. Plant Cell. 1993;5(11):1575–90. Epub 1993/11/01. 8312740
68. Gauthier GM, Keller NP. Crossover fungal pathogens: the biology and pathogenesis of fungi capable of crossing kingdoms to infect plants and humans. Fungal Genet Biol. 2013;61 : 146–57. Epub 2013/09/08. doi: 10.1016/j.fgb.2013.08.016 24021881
69. Sexton AC, Howlett BJ. Parallels in fungal pathogenesis on plant and animal hosts. Eukaryot Cell. 2006;5(12):1941–9. Epub 2006/10/13. 17041185
70. Bayry J, Aimanianda V, Guijarro JI, Sunde M, Latgé JP. Hydrophobins—unique fungal proteins. PLoS Pathog. 2012;8(5):e1002700. Epub 2012/05/31. doi: 10.1371/journal.ppat.1002700 22693445
71. Haas H, Eisendle M, Turgeon BG. Siderophores in fungal physiology and virulence. Annu Rev Phytopathol. 2008;46 : 149–87. Epub 2008/08/06. doi: 10.1146/annurev.phyto.45.062806.094338 18680426
72. Krappmann S, Sasse C, Braus GH. Gene targeting in Aspergillus fumigatus by homologous recombination is facilitated in a nonhomologous end - joining-deficient genetic background. Eukaryot Cell. 2006;5(1):212–5. Epub 2006/01/10. 16400185
73. Käfer E. Meiotic and mitotic recombination in Aspergillus and its chromosomal aberrations. Adv Genet. 1977;19 : 33–131. Epub 1977/01/01. 327767
74. Krappmann S, Bayram Ö, Braus GH. Deletion and allelic exchange of the Aspergillus fumigatus veA locus via a novel recyclable marker module. Eukaryot Cell. 2005;4(7):1298–307. Epub 2005/07/09. 16002655
75. Braus GH, Grundmann O, Brückner S, Mösch HU. Amino acid starvation and Gcn4p regulate adhesive growth and FLO11 gene expression in Saccharomyces cerevisiae. Mol Biol Cell. 2003;14(10):4272–84. Epub 2003/06/27. 14517335
76. Guo B, Styles CA, Feng Q, Fink GR. A Saccharomyces gene family involved in invasive growth, cell-cell adhesion, and mating. Proc Natl Acad Sci U S A. 2000;97(22):12158–63. Epub 2000/10/12. 11027318
77. Sambrook J, Fritsch E, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press: New York; 1989.
78. Lee B, Taylor J. Isolation of DNA from fungal mycelia and single spores. Innis M, Gelfand D, Sninsky J, White T, editors. Academic Press Inc: San Diego; 1990. 282–7 p.
79. Hartmann T, Dümig M, Jaber BM, Szewczyk E, Olbermann P, Morschhäuser J, et al. Validation of a self-excising marker in the human pathogen Aspergillus fumigatus by employing the β-rec/six site-specific recombination system. Appl Environ Microbiol. 2010;76(18):6313–7. Epub 2010/07/23. doi: 10.1128/AEM.00882-10 20656854
80. Yelton MM, Hamer JE, Timberlake WE. Transformation of Aspergillus nidulans by using a trpC plasmid. Proc Natl Acad Sci U S A. 1984;81(5):1470–4. Epub 1984/03/01. 6324193
81. Southern EM. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98(3):503–17. Epub 1975/11/05. 1195397
82. Arnaud MB, Cerqueira GC, Inglis DO, Skrzypek MS, Binkley J, Chibucos MC, et al. The Aspergillus Genome Database (AspGD): recent developments in comprehensive multispecies curation, comparative genomics and community resources. Nucleic Acids Res. 2012;40(Database issue):D653–9. Epub 2011/11/12. doi: 10.1093/nar/gkr875 22080559
83. Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153(1):163–8. Epub 1983/01/01. 6336730
84. Rose M, Botstein D. Construction and use of gene fusions to lacZ (β-galactosidase) that are expressed in yeast. Methods Enzymol. 1983;101 : 167–80. Epub 1983/01/01. 6310320
85. Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72 : 248–54. Epub 1976/05/07. 942051
86. Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68(5):850–8. Epub 1996/03/01. 8779443
87. von Zeska Kress MR, Harting R, Bayram Ö, Christmann M, Irmer H, Valerius O, et al. The COP9 signalosome counteracts the accumulation of cullin SCF ubiquitin E3 RING ligases during fungal development. Mol Microbiol. 2012;83(6):1162–77. Epub 2012/02/22. doi: 10.1111/j.1365-2958.2012.07999.x 22329854
88. Tesfaigzi J, Smith-Harrison W, Carlson DM. A simple method for reusing western blots on PVDF membranes. Biotechniques. 1994;17(2):268–9. Epub 1994/08/01. 7980922
89. Harting R, Bayram Ö, Laubinger K, Valerius O, Braus GH. Interplay of the fungal sumoylation network for control of multicellular development. Mol Microbiol. 2013;90(5):1125–45. Epub 2013/11/06. doi: 10.1111/mmi.12421 24279728
90. Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26(12):1367–72. Epub 2008/11/30. doi: 10.1038/nbt.1511 19029910
91. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-ΔΔC(T)) Method. Methods. 2001;25(4):402–8. Epub 2002/02/16. 11846609
92. Fontaine T, Delangle A, Simenel C, Coddeville B, van Vliet SJ, van Kooyk Y, et al. Galactosaminogalactan, a new immunosuppressive polysaccharide of Aspergillus fumigatus. PLoS Pathog. 2011;7(11):e1002372. Epub 2011/11/22. doi: 10.1371/journal.ppat.1002372 22102815
93. Mumberg D, Müller R, Funk M. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 1994;22(25):5767–8. Epub 1994/12/25. 7838736
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2015 Číslo 11- Stillova choroba: vzácné a závažné systémové onemocnění
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Parazitičtí červi v terapii Crohnovy choroby a dalších zánětlivých autoimunitních onemocnění
- Choroby jater v ordinaci praktického lékaře – význam jaterních testů
-
Všechny články tohoto čísla
- Parasite Glycobiology: A Bittersweet Symphony
- On the Discovery of TOR As the Target of Rapamycin
- Broadening of Virus-Specific CD8 T-Cell Responses Is Indicative of Residual Viral Replication in Aviremic SIV Controllers
- PML/TRIM19-Dependent Inhibition of Retroviral Reverse-Transcription by Daxx
- Cleavage of a Neuroinvasive Human Respiratory Virus Spike Glycoprotein by Proprotein Convertases Modulates Neurovirulence and Virus Spread within the Central Nervous System
- Kaposi’s Sarcoma-Associated Herpesvirus (KSHV) Induces the Oncogenic miR-17-92 Cluster and Down-Regulates TGF-β Signaling
- Interferon-α Subtypes in an Model of Acute HIV-1 Infection: Expression, Potency and Effector Mechanisms
- Perivascular Arrest of CD8 T Cells Is a Signature of Experimental Cerebral Malaria
- Targeting HIV Reservoir in Infected CD4 T Cells by Dual-Affinity Re-targeting Molecules (DARTs) that Bind HIV Envelope and Recruit Cytotoxic T Cells
- Evolution and Emergence of Enteroviruses through Intra- and Inter-species Recombination: Plasticity and Phenotypic Impact of Modular Genetic Exchanges in the 5’ Untranslated Region
- Interferon-γ Inhibits Ebola Virus Infection
- Dengue Virus Non-structural Protein 1 Modulates Infectious Particle Production via Interaction with the Structural Proteins
- P-Type Cyclin CYC3 Modulates Endomitotic Growth during Oocyst Development in Mosquitoes
- Diversity of across Evolutionary Scales
- 50 Years of Disease in Humans: The Dramatic Emergence of a Cluster of Novel Fungal Pathogens
- Worse Comes to Worst: Bananas and Panama Disease—When Plant and Pathogen Clones Meet
- Arenavirus Glycan Shield Promotes Neutralizing Antibody Evasion and Protracted Infection
- Infection-Induced Retrotransposon-Derived Noncoding RNAs Enhance Herpesviral Gene Expression via the NF-κB Pathway
- Structural Insight into Archaic and Alternative Chaperone-Usher Pathways Reveals a Novel Mechanism of Pilus Biogenesis
- Increased Susceptibility of Humanized NSG Mice to Panton-Valentine Leukocidin and Skin Infection
- Global Analysis of the Fungal Microbiome in Cystic Fibrosis Patients Reveals Loss of Function of the Transcriptional Repressor Nrg1 as a Mechanism of Pathogen Adaptation
- The Transcription and Translation Landscapes during Human Cytomegalovirus Infection Reveal Novel Host-Pathogen Interactions
- The N-terminal Helical Region of the Hepatitis C Virus p7 Ion Channel Protein Is Critical for Infectious Virus Production
- Activation of Type I and III Interferon Response by Mitochondrial and Peroxisomal MAVS and Inhibition by Hepatitis C Virus
- Hsp70 Isoforms Are Essential for the Formation of Kaposi’s Sarcoma-Associated Herpesvirus Replication and Transcription Compartments
- Distinct Upstream Role of Type I IFN Signaling in Hematopoietic Stem Cell-Derived and Epithelial Resident Cells for Concerted Recruitment of Ly-6C Monocytes and NK Cells via CCL2-CCL3 Cascade
- and Bats: Story of an Emerging Friendship
- Emergence of Pathogenicity in Lagoviruses: Evolution from Pre-existing Nonpathogenic Strains or through a Species Jump?
- Ebolavirus Evolution: Past and Present
- Host and Symbiont Jointly Control Gut Microbiota during Complete Metamorphosis
- Non-Human Primates Harbor Diverse Mammalian and Avian Astroviruses Including Those Associated with Human Infections
- Lactate Dehydrogenase Is Associated with the Parasitophorous Vacuole Membrane and Is a Potential Target for Developing Therapeutics
- Five Questions about Mycoviruses
- Phosphorylation of a Myosin Motor by TgCDPK3 Facilitates Rapid Initiation of Motility during egress
- Ethanolamine Signaling Promotes Niche Recognition and Adaptation during Infection
- Cross-Species Transmission and Differential Fate of an Endogenous Retrovirus in Three Mammal Lineages
- Memory Th1 Cells Are Protective in Invasive Infection
- Transcription Factor SomA Is Required for Adhesion, Development and Virulence of the Human Pathogen
- An -Methyltransferase Is Required for Infection of Tick Cells by
- RNA-seq Brings New Insights to the Intra-Macrophage Transcriptome of Typhimurium
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Dengue Virus Non-structural Protein 1 Modulates Infectious Particle Production via Interaction with the Structural Proteins
- On the Discovery of TOR As the Target of Rapamycin
- Parasite Glycobiology: A Bittersweet Symphony
- Broadening of Virus-Specific CD8 T-Cell Responses Is Indicative of Residual Viral Replication in Aviremic SIV Controllers
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



