-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Ethanolamine Signaling Promotes Niche Recognition and Adaptation during Infection
Chemical signaling underlies all cellular processes. Bacteria rely on chemical signaling to gain information about the local environment and precisely regulate gene expression. Ethanolamine is an abundant molecule within mammalian hosts that plays an important role in mammalian physiology and also serves as a carbon and nitrogen source for bacteria. Here we show that the foodborne pathogen Salmonella enterica exploits ethanolamine as a signal of distinct host environments to coordinate metabolism and virulence, which enhances disease progression during infection. The ability to sense ethanolamine is conserved in diverse bacteria; thus, these studies reveal that ethanolamine signaling may be important for bacterial adaptation to the mammalian host.
Published in the journal: . PLoS Pathog 11(11): e32767. doi:10.1371/journal.ppat.1005278
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1005278Summary
Chemical signaling underlies all cellular processes. Bacteria rely on chemical signaling to gain information about the local environment and precisely regulate gene expression. Ethanolamine is an abundant molecule within mammalian hosts that plays an important role in mammalian physiology and also serves as a carbon and nitrogen source for bacteria. Here we show that the foodborne pathogen Salmonella enterica exploits ethanolamine as a signal of distinct host environments to coordinate metabolism and virulence, which enhances disease progression during infection. The ability to sense ethanolamine is conserved in diverse bacteria; thus, these studies reveal that ethanolamine signaling may be important for bacterial adaptation to the mammalian host.
Introduction
Chemical and nutrient signaling mediate diverse biological processes, and underlie interactions among the mammalian host, the resident microbiota, and invading pathogens [1]. Ethanolamine is abundant in cell membranes, as a component of phosphatidylethanolamine as well as in modified lipid molecules such as N-acylethanolamines [2]. These ethanolamine-containing compounds play important roles in mammalian cell signaling and influence diverse physiological effects, including cytokinesis, immunomodulation, food intake and energy balance [2–4]. Ethanolamine is abundant in the intestinal tract due to the turnover and exfoliation of enterocytes and bacterial cells [5,6], and intracellular pools of ethanolamine are maintained by low and high affinity uptake systems as well as through internal recycling of phosphatidylethanolamine [7–10].
Bacterial pathogens compete for nutrients with the resident microbiota and rely on environmental cues to control virulence gene expression. Salmonella enterica serovar Typhimurium (S. Typhimurium) is a facultative intracellular pathogen and a leading cause of acute gastroenteritis, which can progress to systemic infection in susceptible individuals [11]. S. Typhimurium encodes two type three secretion systems (T3SSs) that are important for pathogenesis. S. Typhimurium uses the T3SS encoded within the Salmonella pathogenicity island (SPI)-1 to invade intestinal epithelial cells and penetrate to the lamina propria [12]. There, S. Typhimurium is taken up by macrophages, where it survives and replicates. Intracellular survival is mediated by the T3SS and effectors encoded in SPI-2 [13–15]. Ethanolamine can serve as a carbon and/or nitrogen source for bacteria in the intestine as well as within epithelial cells [16,17]. The aim of this work was to determine whether S. Typhimurium relies on ethanolamine as a signal to coordinate gene expression and augment virulence in vivo. Here, we show that the intramacrophage environment promotes expression of the ethanolamine utilization transcription factor EutR, which directly activates SPI-2. Moreover, we demonstrate that EutR signaling during systemic infection is specific to the intracellular environment and is important for robust S. Typhimurium dissemination. Altogether, our findings suggest that ethanolamine, an intrinsic component of bacterial and mammalian cell membranes, functions as a signal to modulate metabolism and virulence and suggest a new layer of complexity in chemical signaling that underlies pathogenicity.
Results and Discussion
EutR contributes to dissemination in vivo
Genes encoding for ethanolamine metabolism are clustered in the eut operon [18] (Fig 1A). In the Enterobacteriaceae, expression of this operon is regulated by the eut-encoded transcription factor EutR. EutR is constitutively expressed at low levels from its own promoter and binds to the promoter region immediately upstream of eutS. In the presence of ethanolamine and vitamin B12, EutR activates transcription of this operon [19,20]. In enterohemorrhagic Escherichia coli (EHEC), EutR senses ethanolamine to activate virulence gene expression in vitro, independently of ethanolamine metabolism [19,21,22]. To determine whether EutR influences S. Typhimurium disease progression during infection, we generated an eutR deletion strain (ΔeutR) that cannot sense ethanolamine as well as an eutB deletion strain (ΔeutB) that lacks the large subunit of the ethanolamine ammonia lyase, and thus is unable to catabolize ethanolamine. The eutR and eutB mutations did not result in a general loss of fitness, as the ΔeutR and ΔeutB strains exhibited no measurable growth defects in vitro (Fig 1B). Importantly, the eutB mutation is nonpolar as this mutant can respond to ethanolamine (Fig 1C). Subsequently, we performed competitive infections in which streptomycin-treated mice were orally infected with an equal mixture of wild type (WT) and ΔeutB (ΔeutB::CmR) strains or the WT and ΔeutR (ΔeutR::CmR) strains. S. Typhimurium infection presents as intestinal outgrowth, invasion of epithelial cells, and subsequent uptake by macrophages and dissemination to secondary lymphoid tissue. Therefore, to monitor the course of S. Typhimurium infection, we analyzed the number of recovered bacteria from the intestinal contents, the colon, and the spleen. At 2 and 4 days post infection (dpi), the ΔeutR and ΔeutB strains were significantly outcompeted by the WT strain in intestinal contents (Fig 1D and 1E). These data underscore the importance of ethanolamine metabolism in S. Typhimurium colonization of the intestinal tract, and these findings are consistent with previous work by Thiennimitr et al., who showed that ethanolamine metabolism provides a growth advantage to S. Typhimurium during intestinal colonization [17].
Fig. 1. EutR in pathogen-microbiota-host interactions. 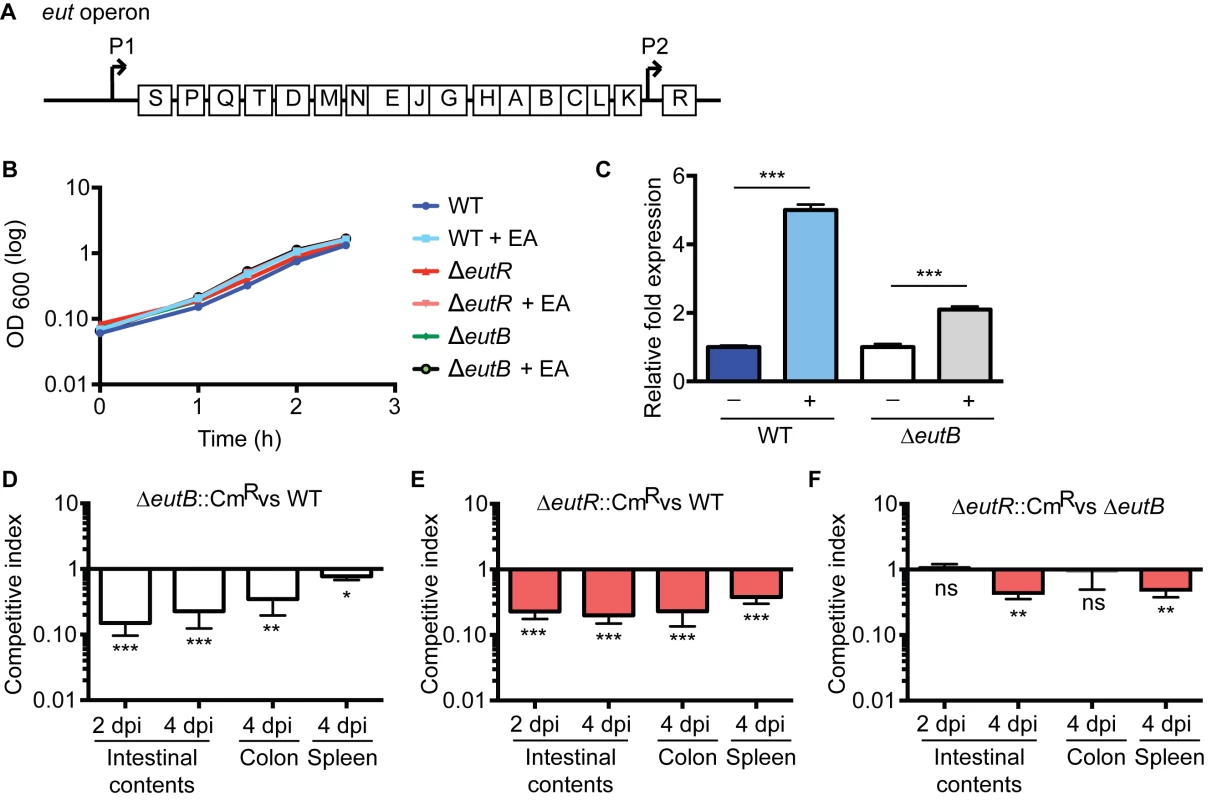
(A) Schematic of the eut operon. (B) In vitro growth curve of S. Typhimurium WT (SL1344), ΔeutR (CJA009), or ΔeutB (CJA020) strains in LB without or with supplementation of 5 mM ethanolamine (EA). Each data point shows the average of three independent experiments. (C) qRT-PCR of eutR in WT or the ΔeutB (CJA020) S. Typhimurium strains grown in Dulbecco’s Modified Eagle Medium (DMEM) or DMEM supplemented with 5 mM EA. n = 3; error bars represent the geometric mean ± standard deviation (SD); strB was used as the endogenous control. (D-F) Competition assays between (D) ΔeutB::CmR (CJA018) and WT strains; (E) ΔeutR::CmR (CJA007) and WT strains; or (F) ΔeutR::CmR (CJA007) and ΔeutB (CJA020) strains. Mice were orogastrically inoculated with 1:1 mixtures of indicated strains. Colony forming units (cfu) were determined at indicated time points. Each bar represents a competition index (CI). Horizontal lines represent the geometric mean value ± standard error (SE) for each group (n = 2 litters (6–8 animals)). *, P ≤0.05; **, P ≤ 0.005; ***, P ≤0.0005; P > 0.05 = ns. Defects in ethanolamine metabolism have been reported to result in mild or no attenuation during S. Typhimurium systemic infection [23,24]; however the role of EutR, specifically, in contributing to dissemination has not been investigated. Therefore, to assess this, we harvested the colons and the spleens of infected mice at 4 dpi. The ΔeutR and ΔeutB strains were both recovered at significantly lower numbers than WT from the colon and spleen; however, the competition indices measured from the spleen from the ΔeutR/WT infections were significantly greater than between WT and the ΔeutB strain (P = 0.002). These findings led us to hypothesize that EutR plays a more extensive role in S. Typhimurium pathogenesis that is distinct from its function to promote ethanolamine metabolism.
To test this, we performed competition infections between the ΔeutB and ΔeutR (ΔeutR::CmR) strains. At 2 days post infection (dpi), the ΔeutR and ΔeutB strains were recovered at similar numbers from intestinal contents (Fig 1F), indicating that at this initial stage of colonization, EutR functions to drive ethanolamine metabolism. However, at 4 dpi, which is a time point consistent with the progression to systemic infection [25], the ΔeutR strain was significantly outcompeted by the ΔeutB strain (Fig 1F). Significantly, although equal numbers of the ΔeutR and ΔeutB strains were recovered from the colon, the ΔeutR strain was significantly outcompeted by the ΔeutB strain in the spleen (Fig 1F). These data suggest that EutR, independent of its role in ethanolamine metabolism, is important to S. Typhimurium dissemination during infection.
EutR does not influence invasion of epithelial cells
To further explore how ethanolamine signaling contributes to S. Typhimurium dissemination, we examined S. Typhimurium virulence gene expression in vitro. To examine ethanolamine-mediated expression of SPI-1, we measured expression of sipC, a SPI-1 encoded translocase that plays a role in invasion of epithelial cells [26]. For this, we grew S. Typhimurium in LB, which induces SPI-1 expression [27] as well as in DMEM used for cell culture assays. In both cases, expression of sipC was slightly decreased when ethanolamine was included in the culture medium (Fig 2A and 2B and S1 Fig). To determine whether the ethanolamine-dependent decrease in sipC expression impacted S. Typhimurium invasion of epithelial cells, we infected HeLa cells with either WT S. Typhimurium or the ΔeutR strain. WT S. Typhimurium and ΔeutR invaded HeLa cells at nearly equivalent levels in the presence or absence of ethanolamine (Fig 2C and 2D). Because we did not observe EutR-dependent effects on epithelial invasion under the specified conditions, the influence of ethanolamine at this stage in S. Typhimurium dissemination was not pursued further.
Fig. 2. Effect of ethanolamine and EutR on SPI-1. 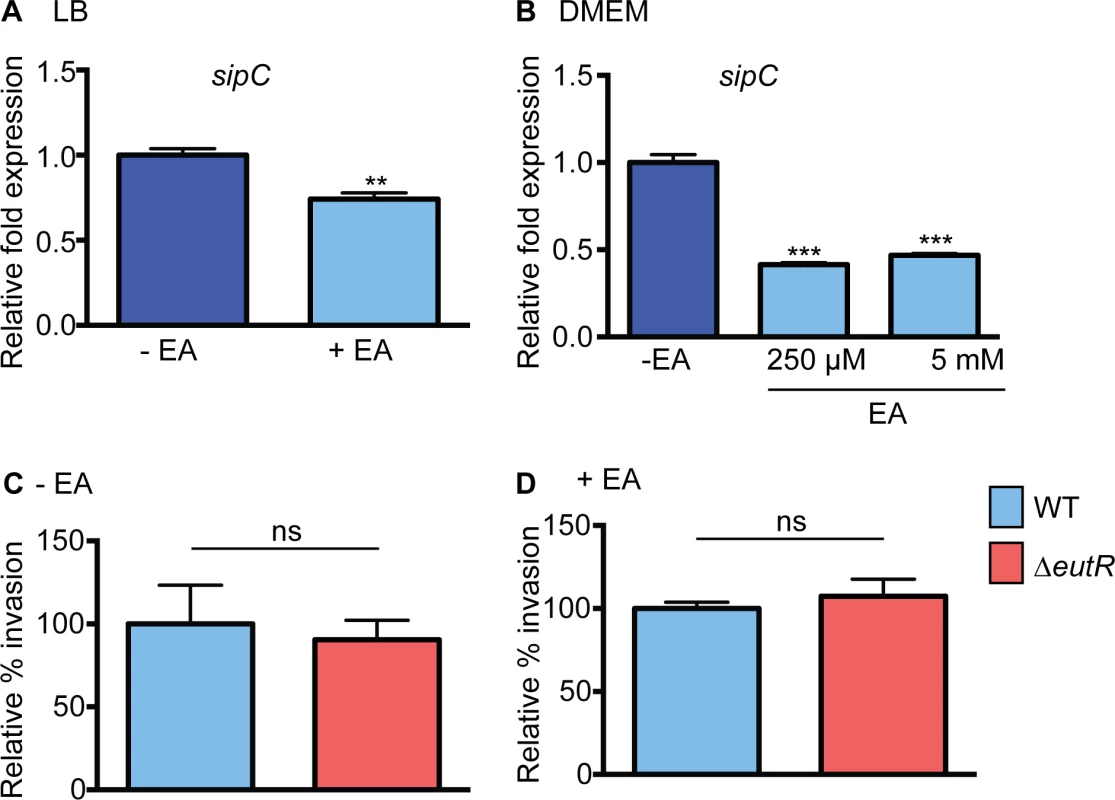
(A) qRT-PCR of sipC from WT S. Typhimurium (SL1344) grown in LB or LB supplemented with 5 mM ethanolamine (EA). (B) qRT-PCR of sipC from WT S. Typhimurium (SL1344) grown in DMEM or DMEM supplemented with ethanolamine (EA) as indicated. For (A) and (B), n = 3; error bars represent the geometric mean ± SD. Statistical significance is shown relative to cells grown without EA supplementation; strB was used as the endogenous control. (C) Invasion of HeLa cells by WT (SL1344) and the ΔeutR (CJA009) strains. Mean ± SE of nine independent experiments. (D) Invasion of HeLa cells by WT (SL1344) and the ΔeutR (CJA009) strains. Mean ± SE of six independent experiments with supplementation of 5 mM EA. **, P ≤ 0.005; P > 0.05 = ns. Ethanolamine influences SPI-2 expression
Next, we investigated whether ethanolamine impacted SPI-2 expression. SsrB is a SPI-2 encoded transcriptional regulator that is required for expression of all the SPI-2-encoded genes, as well as for expression of effectors and virulence genes encoded outside of SPI-2 [28–31]. To test the influence of ethanolamine, we measured expression of ssrB in low magnesium, minimal medium, a condition that induces SPI-2 expression [32] (S2 Fig) without supplementation or with supplementation of 250 μM or 5 mM ethanolamine. These concentrations were used because 250 μM ethanolamine was the lowest concentration with which we could readily detect EutR expression (S3 Fig), whereas 5 mM is similar to ethanolamine concentrations in the gastrointestinal tract [33]. When 250 μM ethanolamine was added to the SPI-2 inducing medium, expression of ssrB was significantly increased compared to medium without supplementation, but unchanged when 5 mM ethanolamine was added (Fig 3A). These data suggest that ethanolamine may enhance the response of S. Typhimurium in adapting to the intramacrophage environment. Expression of SPI-2 is tightly regulated and is induced specifically in the intracellular environment [34], or in conditions that mimic the intracellular environment. In accordance, ssrB expression was not induced in DMEM or LB when ethanolamine was supplemented to the medium (Fig 3B and S4 Fig), indicating that ethanolamine in and of itself does not override additional regulatory factors that direct ssrB expression. However, ssrB expression was decreased in DMEM with the addition of 5 mM ethanolamine (Fig 3B). Altogether these data raised the possibility that ethanolamine signaling enhances niche recognition.
Fig. 3. The impact of ethanolamine on SPI-2 expression in vitro. 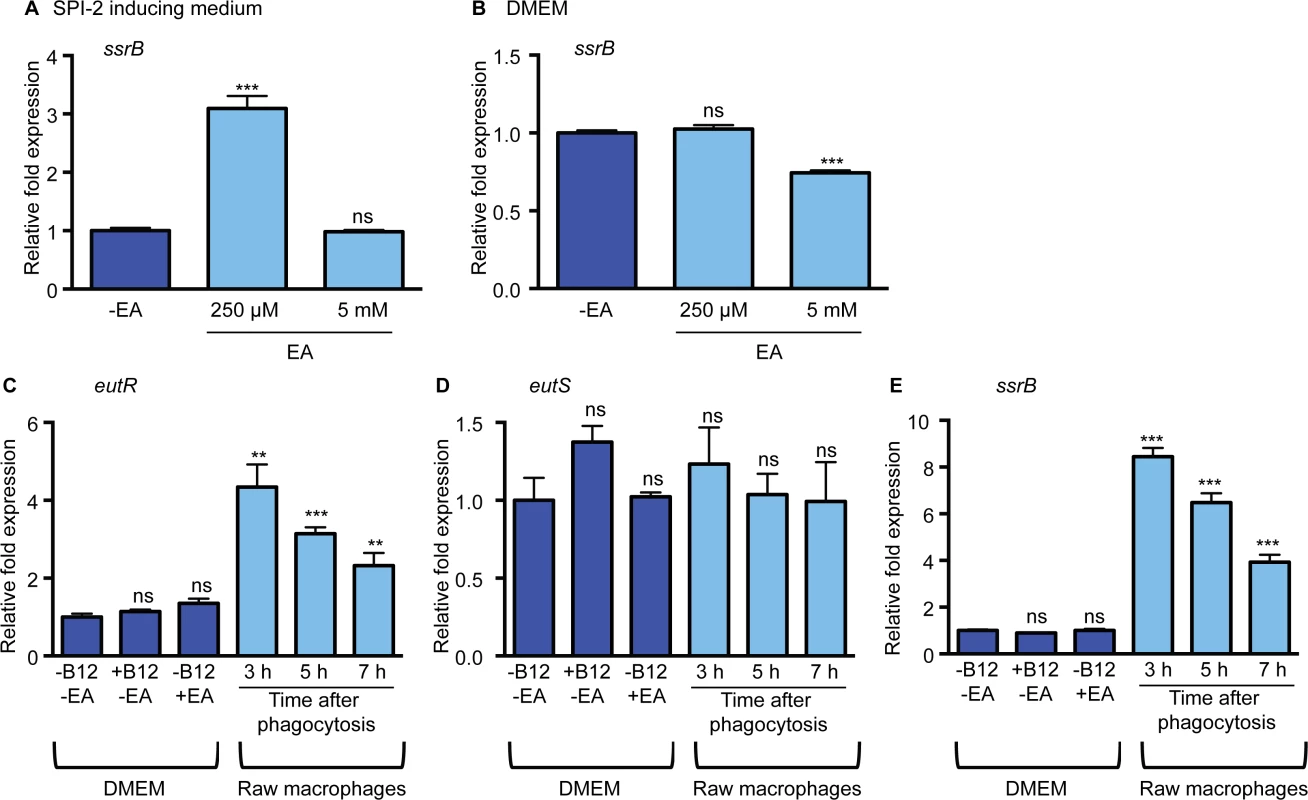
(A) qRT-PCR of ssrB from RNA isolated from the S. Typhimurium (SL1344) grown in SPI-2 inducing medium with ethanolamine (EA) supplementation as indicated. Statistical significance is shown relative to cells grown without EA supplementation. (B) qRT-PCR of ssrB from RNA isolated from the S. Typhimurium (SL1344) grown in DMEM with EA supplementation as indicated. Statistical significance is shown relative to cells grown without EA supplementation. (C) qRT-PCR of eutR from RNA isolated from S. Typhimurium (AJK61) grown in DMEM with supplementation as indicated or after phagocytosis in RAW macrophages. Statistical significance relative to cells grown in DMEM is indicated. (D) qRT-PCR of eutS from RNA isolated from S. Typhimurium (AJK61) grown in DMEM with supplementation as indicated or after phagocytosis in RAW macrophages. Statistical significance relative to cells grown in DMEM is indicated. (E) qRT-PCR of ssrB from RNA isolated from the S. Typhimurium strain (AJK61) grown in DMEM with supplementation as indicated or after phagocytosis in RAW macrophages. Statistical significance relative to cells grown in DMEM is indicated. For all, n = 3; error bars represent the geometric mean ± SD; strB was used as the endogenous control. *, P ≤ 0.05; **, P ≤ 0.005; ***, P ≤0.0005; P > 0.05 = ns. Macrophages play a significant role in the pathogenesis of S. Typhimurium infection by providing protected sites for intracellular replication and a means of dissemination [35]. Robust expression of EutR requires ethanolamine as well as the cofactor vitamin B12 [20]. S. Typhimurium synthesizes vitamin B12 under anaerobic conditions [36]; however, S. Typhimurium must acquire ethanolamine from the environment [18]. Therefore, we investigated whether the intracellular environment induces eutR expression. For this, we infected macrophages with S. Typhimurium in the absence of any exogenous ethanolamine or vitamin B12. Subsequently, RNA was extracted from internalized S. Typhimurium at 3, 5, and 7 h post phagocytosis, and eutR transcript levels were analyzed and compared to eutR transcript levels from S. Typhimurium grown in the absence of macrophages. Expression of eutR was significantly increased in phagocytized S. Typhimurium throughout infection compared to cells grown in the absence of macrophages (Fig 3C and S5 Fig). Moreover, neither vitamin B12 or ethanolamine alone activated eutR expression in tissue culture medium, indicating that the intramacrophage environment is conducive to EutR-dependent signaling.
The addition of ethanolamine and vitamin B12 to SPI-2 inducing medium or DMEM resulted in an increase in expression of the eut operon (as indicated by eutS expression (Fig 1A)) that corresponded with an increase in eutR expression (S6 Fig). Notably, the eut operon was not induced within macrophages (Fig 3D). These findings indicate that the ethanolamine metabolic genes are not activated within the intramacrophage environment. Interestingly, the expression pattern of eutR in internalized S. Typhimurium was similar to ssrB expression (Fig 3E); therefore, we hypothesized that EutR regulates SPI-2 expression.
EutR directly regulates SPI-2 expression
SPI-2 contains four major operons that encode a T3SS, chaperone and effector proteins, as well as the transcriptional regulator SsrB (Fig 4A). To test our hypothesis, we examined transcription of ssrB and one gene from each of the other major operons encoded in SPI-2 using RNA harvested from phagocytized WT or ΔeutR S. Typhimurium strains. Transcription of ssrB was significantly decreased in the ΔeutR strain compared to WT (Fig 4B and S7 Fig), and we measured a concomitant decrease in expression of all the SPI-2 operons, as well as the SPI-2-associated effector sifA (Fig 4B and S8 Fig). Expression of SPI-2 encoded and associated factors enhances the intrinsic ability of S. Typhimurium to withstand and disrupt host defense mechanisms [37,38], and these data revealed that EutR influences this critical aspect of S. Typhimurium virulence.
Fig. 4. EutR regulates SPI-2 expression. 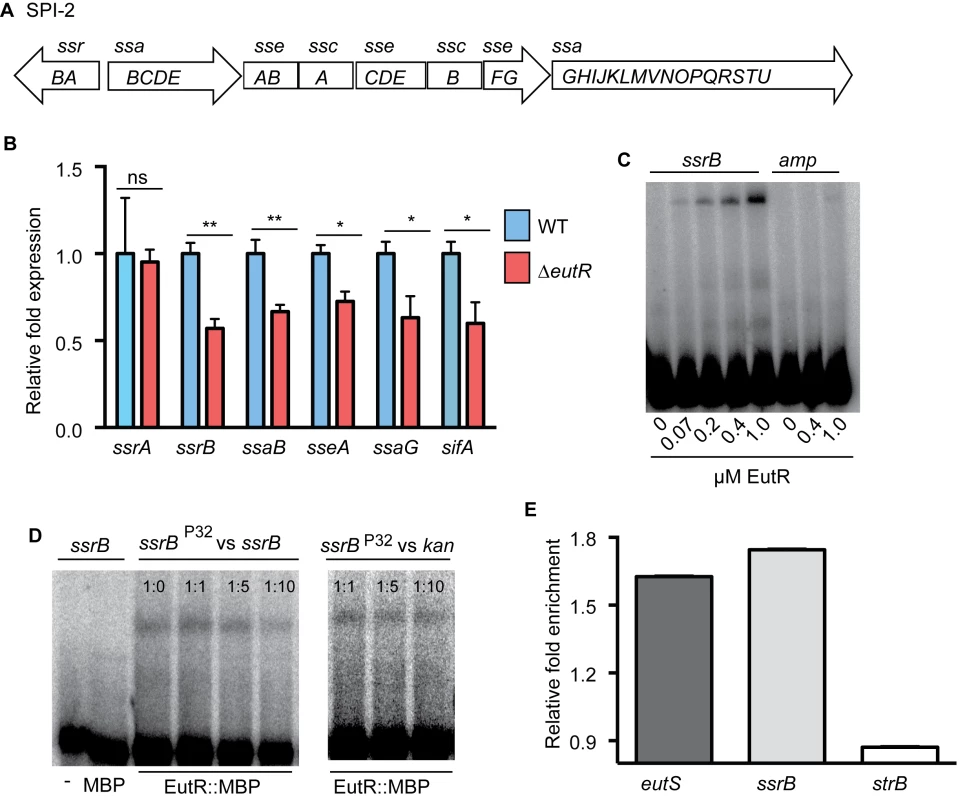
(A), Schematic of SPI-2. (B) qRT-PCR analysis of SPI-2-encoded and associated (sifA) genes from RNA isolated from S. Typhimurium (AJK61) or the ΔeutR (CJA023) strains after 5 h phagocytosis in RAW macrophages. n = 3; error bars represent the geometric mean ± SD; strB was used as the endogenous control. (C) EMSAs of ssrB and amp (ampicillin) with EutR::MBP. (D) EMSAs of ssrB with MBP or EutR::MBP. Also, competition EMSAs with EutR::MBP. The assay was performed with increasing amounts of unlabeled ssrB promoter probe. A competition assay was also performed using the kan promoter as a negative control. The ratios represent hot:cold probe. (E) qPCR showing enrichment of eutS, ssrB, and strB from in vivo ChIP of EutR::MBP (n = 2). *, P ≤ 0.05; **, P ≤ 0.005; ***, P ≤0.0005; P > 0.05 = ns. SsrB is a response regulator that comprises a two component system with the sensor kinase SsrA (also referred to as SpiR) [13,15]. SsrA autophosphorylates in response to the acidic environment of the Salmonella-containing vacuole (SCV) within host cells [39,40], which initiates a signaling cascade that promotes SsrB activity as well as ssrAB expression [41]. Importantly, the ssrB gene contains its own promoter [41]. The genetic data indicated that EutR influenced expression of ssrB and downstream targets, but that EutR did not impact ssrA expression (Fig 4B and S8 Fig), indicating that EutR may regulate SPI-2 expression by binding the ssrB promoter. To examine this, we purified an EutR::MBP fusion protein. Electrophoretic mobility shift assays (EMSAs) indicated that EutR directly binds the ssrB promoter to activate expression of SPI-2 (Fig 4C). To confirm specificity of binding, EMSAs with purified MBP alone as well as competitions assays with unlabeled probes were performed. MBP alone did not bind the ssrB promoter (Fig 4D). Furthermore, EutR binding was outcompeted by the addition of unlabeled ssrB probe; however, the addition of unlabeled kan probe, as a negative control reaction, showed no competition (Fig 4D).
Consistent with these results, there was a significant enrichment of the ssrB promoter when EutR-DNA interactions were analyzed using in vivo chromatin immunoprecipitation followed by qPCR (Fig 4E). As a positive control, we also measured enrichment of the eutS promoter, an established binding target of EutR [19], and observed similar enrichment of both targets (Fig 4E); strB DNA was used as a negative control. Control of ssrB expression is complex and also includes activation by additional TCS PhoP/PhoQ and EnvZ/OmpR, which respond to signals within the Salmonella containing vesicle (SCV) [41,42]. Our findings suggest that EutR-dependent activation of ssrB enables S. Typhimurium to integrate intrinsic information regarding the host cell through ethanolamine signaling with SCV-specific signals to coordinate efficient spatiotemporal expression of SPI-2 and SPI-2 associated effectors.
EutR enhances intramacrophage survival
Next, we tested the consequences of EutR-dependent activation of SPI-2 on S. Typhimurium fitness during macrophage infection. Following infection of RAW or peritoneal exudate macrophages (PEMs), the ΔeutR strain was recovered at significantly lower numbers compared to the WT strain (Fig 5A–5C and S9 Fig). Additionally, complementation of the ΔeutR strain with eutR expressed from the native promoter (eutR+) restored intracellular survival to WT levels during primary macrophage infection (Fig 5B).
Fig. 5. EutR enhances S. Typhimurium survival within macrophages. 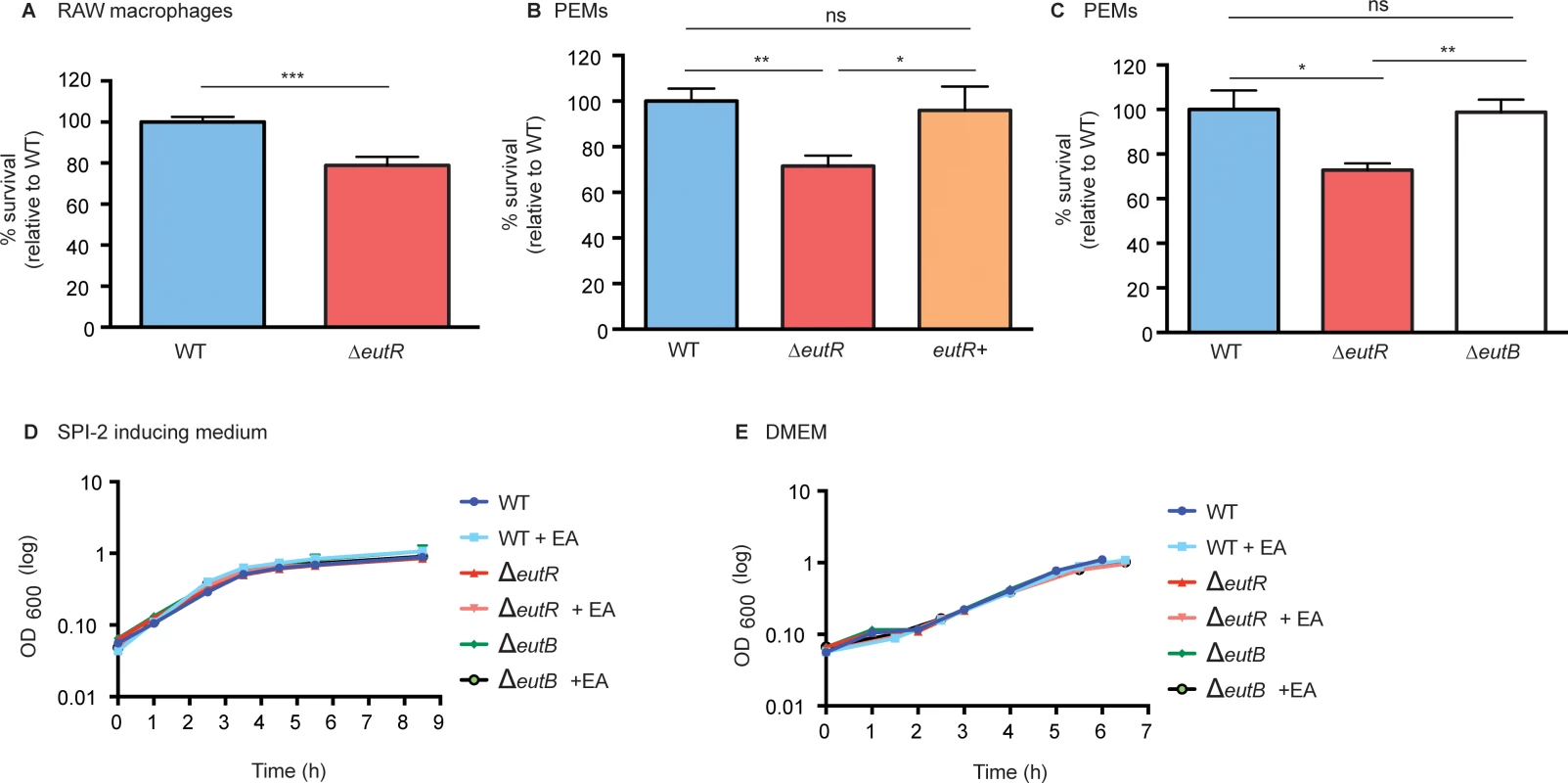
(A) Intramacrophage survival and replication of S. Typhimurium (AJK61) and the ΔeutR (CJA023) strains after 5 h phagocytosis in RAW macrophages (error bars represent the geometric mean value ± SE of 24 independent experiments). (B) Intramacrophage survival and replication of S. Typhimurium (CJA034), ΔeutR (CJA032), and ΔeutR complemented with eutR (eutR+) (CJA033) strains after 5 h phagocytosis in peritoneal exudate macrophages (PEMs) (error bars represent the geometric mean value ± SE of nine independent experiments). (C) Intramacrophage survival and replication of S. Typhimurium (AJK61), ΔeutR (CJA023) and ΔeutB (CJA028) strains after 5 h phagocytosis in PEMs (error bars represent the geometric mean value ± SE of six independent experiments). (D) In vitro growth curve of S. Typhimurium WT (SL1344), ΔeutR (CJA009), or ΔeutB (CJA020) strains in SPI-2 inducing medium without or with supplementation of 5 mM ethanolamine (EA). Each data point shows the average of three independent experiments. (E) In vitro growth curve of S. Typhimurium WT (SL1344), ΔeutR (CJA009), or ΔeutB (CJA020) strains in tissue culture medium without or with supplementation of 5 mM ethanolamine (EA). Each data point shows the average of three independent experiments. *, P ≤ 0.05; **, P ≤ 0.005; ***, P ≤ 0.0005; P > 0.05 = ns. To verify that the defect in the ΔeutR strain was not the result of a defect in ethanolamine metabolism, we assessed survival of the ΔeutB strain within PEMs. The ΔeutB strain was recovered at similar numbers to WT and at significantly higher numbers than the ΔeutR strain (Fig 5C). Importantly, the ΔeutR mutant grows similarly to WT and ΔeutB strains in SPI-2 inducing medium and tissue culture medium with or without the addition of ethanolamine (Fig 5D and 5E), confirming that the decrease in intracellular survival is not a result of a EutR-dependent growth defect. These findings indicate that ethanolamine-associated signaling, but not catabolism, impacts S. Typhimurium survival within macrophages. Moreover, these findings, in conjunction with lack of eut operon induction within macrophages (Fig 3D), reveals that S. Typhimurium relies on EutR to direct gene expression in a manner that is particular to a specific niche.
EutR provides a gauge of the intracellular environment in vivo
Next, we confirmed that EutR mediates dissemination specifically through intracellular survival in vivo. To further discriminate between ethanolamine-associated signaling and ethanolamine metabolism, we infected mice with equal numbers of the ΔeutR::CmR and ΔeutB strains by intraperitoneal injection. At 6 h pi, the ΔeutR strain was recovered at significantly lower numbers compared to the ΔeutB strain from the spleen (Fig 6A). Furthermore, we assessed bacterial burden in the peritoneal cavity. At this site, there were no significant differences between the ΔeutR and ΔeutB strains in the total bacteria recovered (Fig 6B), the majority of which were extracellular (S10 Fig). However, the ΔeutR strain was recovered at significantly lower numbers compared to the ΔeutB strain in the phagocytized population of S. Typhimurium within the peritoneal cavity (Fig 6C). These findings reveal that EutR augments S. Typhimurium fitness during systemic infection. Our findings differ from a previous study that used a genetic screen to identify genes important for systemic virulence [43]. Discrepancies may reflect differences in study design such as the age and genetic background of mice, route of infection, and/or duration of infection. Importantly, using in vitro and in vivo approaches, our data establish a genetic and functional role for EutR in S. Typhimurium systemic disease, and altogether, these results indicate that EutR contributes to the ability of S. Typhimurium to gauge and adapt to the intracellular environment in vivo.
Fig. 6. EutR promotes recognition and adaptation to the intracellular environment. 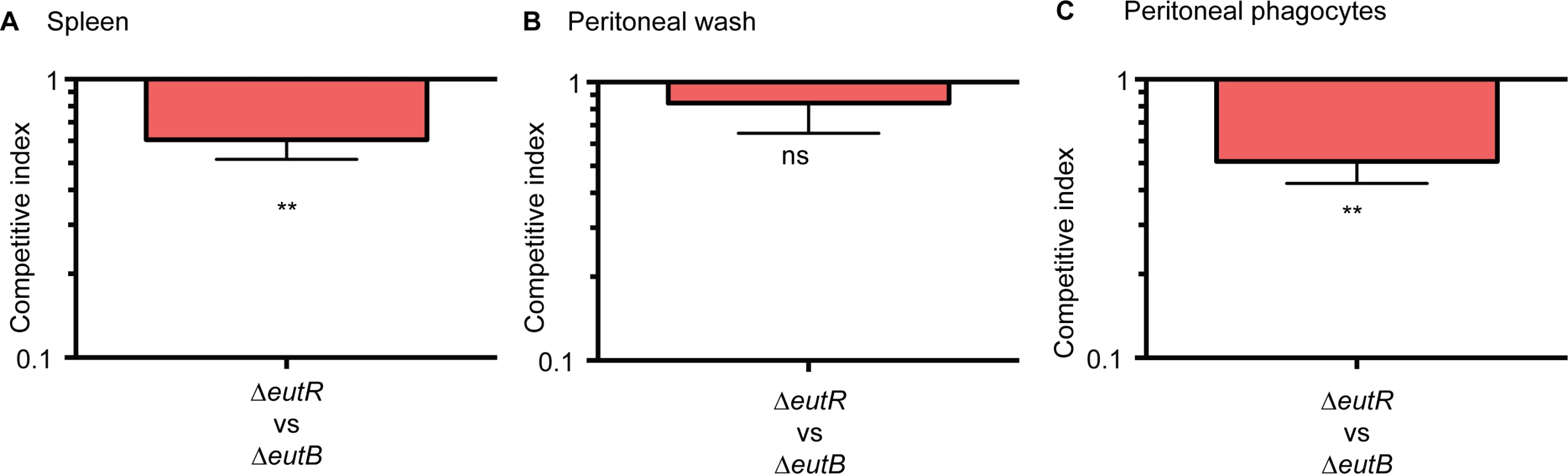
(A-C) Competition assays between ΔeutR::CmR (CJA007) and ΔeutB (CJA020) strains collected from (A) harvested spleens, (B) the peritoneal cavity, or (C) phagocytized cells at 6 h pi. Mice were intraperitoneally infected with 1:1 mixtures of the ΔeutR and ΔeutB strains. Each column represents a CI. Each column shows the geometric mean value ± SE for each group (n = 2 litters (6–8 animals)). *, P ≤ 0.05; **, P ≤ 0.005; ***, P ≤ 0.0005; P > 0.05 = ns. EutR signaling during systemic infection
The in vitro studies identified targets of EutR-dependent gene regulation. To test our findings within the complexities of the in vivo environment, we assessed EutR-dependent regulation of ssrB using single strain infections and purified S. Typhimurium RNA from harvested spleens. Expression of ssrB was significantly decreased in the ΔeutR strain compared to WT (Fig 7A and S11 Fig), which is consistent with the data presented in Fig 4B. Additionally, we measured expression of eutR and eutS in WT S. Typhimurium recovered from the spleen relative to S. Typhimurium grown in vitro. Notably, eutR expression was significantly increased in the spleen, whereas expression of eutS was not detectable (Fig 7B). These data further highlight the dynamic role of EutR in S. Typhimurium pathogenesis from driving ethanolamine metabolism in the intestine to promoting virulence gene expression in later stages of disease.
Fig. 7. EutR-associated signaling in vivo. 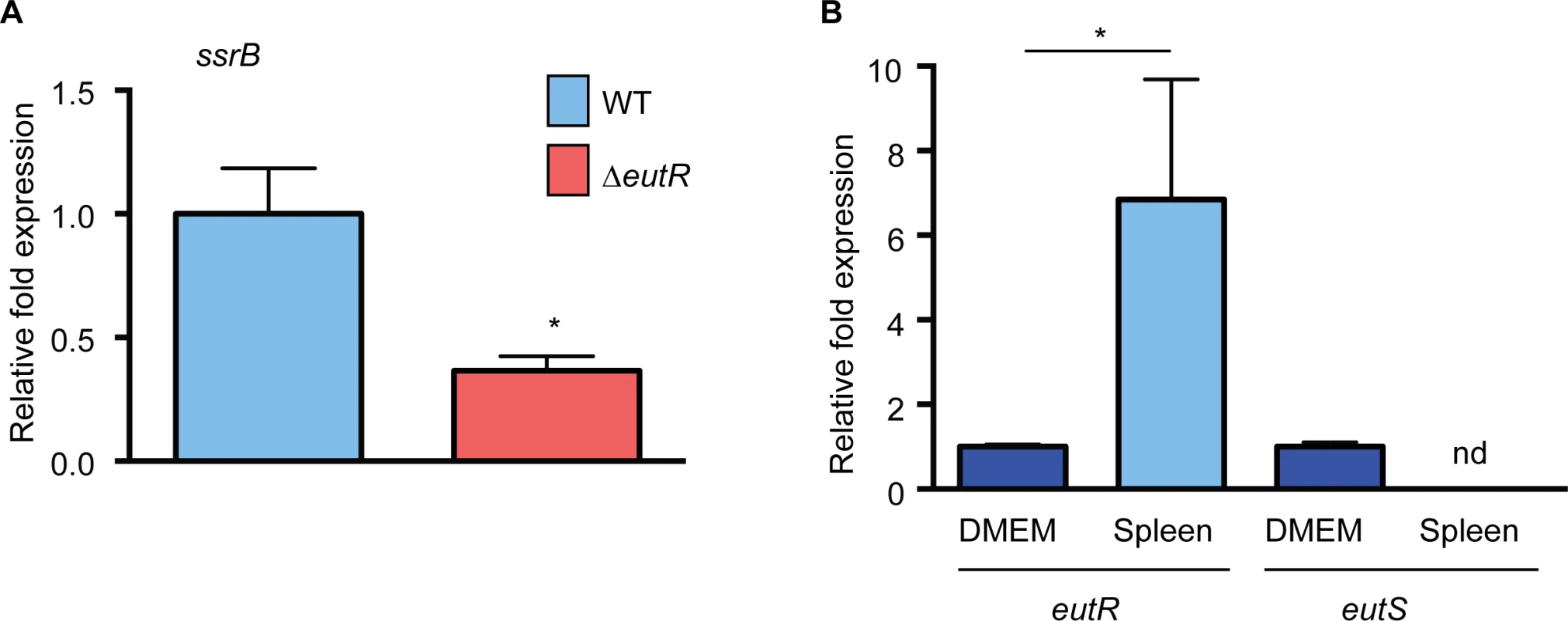
(A) qRT-PCR analysis of ssrB expression in WT S. Typhimurium (SL1344) or the ΔeutR strain (CJA009) harvested from infected spleens. (B) qRT-PCR analysis of eutR or eutS expression in WT S. Typhimurium (SL1344) harvested from infected spleens compared to S. Typhimurium (SL1344) grown in tissue culture medium (DMEM). For (A) and (B), n = 2–3; error bars represent the geometric mean ± SD; strB was used as the endogenous control. *, P ≤ 0.05. nd = not detected. Conclusions
These findings reveal a novel signaling pathway critical for S. Typhimurium to enhance disease progression during infection. We propose a model in which S. Typhimurium relies on ethanolamine signaling through EutR to gauge distinct environments in the host and then modulate expression of genes encoding metabolism and virulence (Fig 8). The resident microbiota do not readily metabolize ethanolamine [33]. Thus, to establish infection, S. Typhimurium sidesteps nutritional competition by respiring ethanolamine in conjunction with tetrathionate, an electron acceptor generated specifically during intestinal inflammation [17,44]. Fermentation of ethanolamine provides very little growth [17]; hence, outside of the intestine, and in the absence of bacterial competition, S. Typhimurium preferentially utilizes alternative nutrients [23]. This enables EutR to direct expression of traits necessary for dissemination and systemic infection. Additional experiments are necessary to determine what factors influence the transition from driving metabolism to influencing virulence in the intestine.
Fig. 8. EutR in S. Typhimurium niche adaptation. 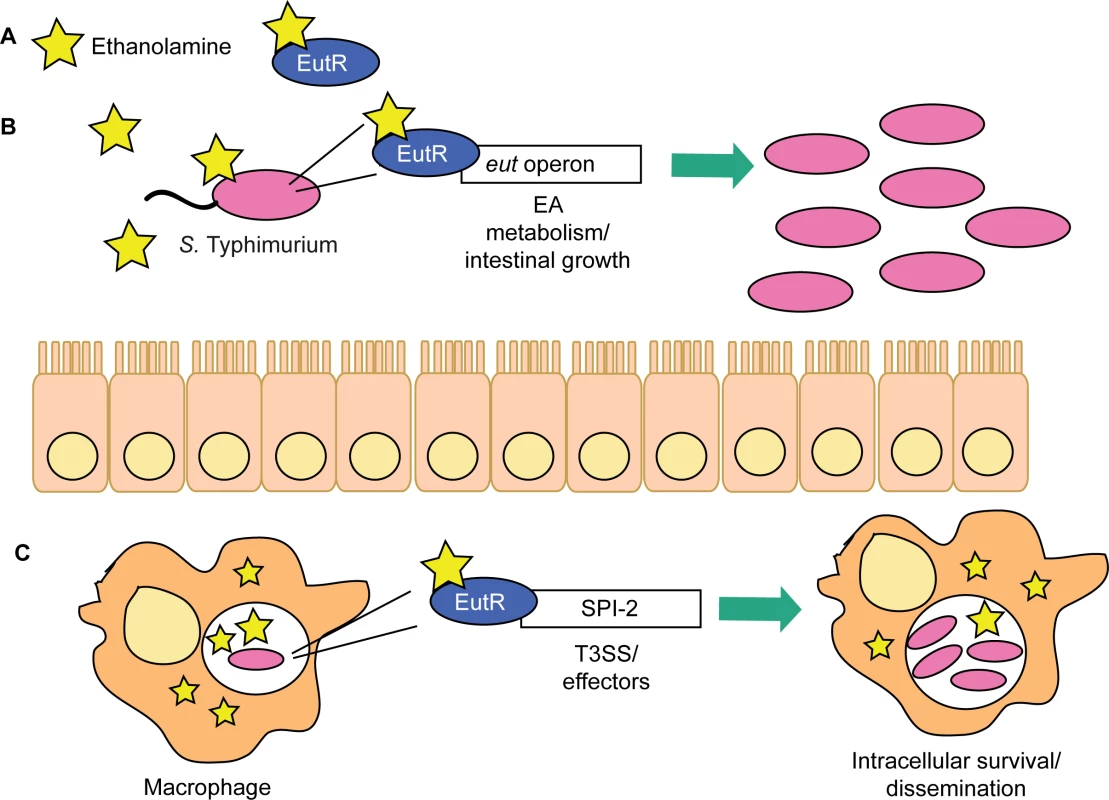
(A) EutR senses ethanolamine to activate transcription. (B) In the intestine, EutR promotes expression of the eut operon that encodes ethanolamine metabolism, thereby enhancing S. Typhimurium growth. (C) EutR expression in macrophages activates expression of genes in SPI-2, which are required for intramacrophage survival and dissemination. Genes encoding ethanolamine utilization are widespread in pathogenic bacteria as well as in members of the resident microbiota [45], and the extracellular pathogen EHEC responds to ethanolamine to regulate virulence gene expression [22]. Therefore, ethanolamine signaling may be a conserved strategy used by diverse pathogens to coordinate metabolism and virulence in response to distinct host environments. Our findings highlight a sophisticated mechanism in which S. Typhimurium exploits an abundant and essential molecule within the host to gain specific information about the localized environment and modulate gene expression to overcome bacterial and host resistance mechanisms.
Materials and Methods
Strains and plasmids
All strains and plasmids used in this study are listed in S1 Table. Luria-Bertani (LB), Dulbecco’s Modified Eagle Medium (DMEM) (Invitrogen), or minimal medium (described below) were used as indicated. Ethanolamine (Sigma) and/or vitamin B12 (Sigma) were supplemented to the media as indicated in the main text. Unless indicated otherwise, 150 nM vitamin B12 was added whenever ethanolamine was added to the growth medium. Antibiotics were used in the following concentrations: ampicillin (100μg/ml), streptomycin (100μg/ml), chloramphenicol (20μg/ml), and kanamycin (50μg/ml). Recombinant DNA and molecular biology techniques were performed as described previously [46].
Construction of isogenic mutants
S. Typhimurium SL1344 [47] and its derivatives were used in all experiments. The invG mutant (strain AJK61) was a gift from James Casanova and was constructed as previously described [48]. Nonpolar eutR and eutB deletion strains were generated in WT and ΔinvG backgrounds using λ-red mutagenesis [49]. Briefly, PCR products (obtained with primers listed in S2 Table) were amplified from plasmid pKD3 or pKD4 with flanking regions matching eutR or eutB and then transformed into S. Typhimurium expressing the Red genes from plasmid pKD46. The resistance cassette was resolved with flippase from temperature-sensitive plasmid pCP20, which was then cured through growth at 42°C. Unresolved strains were used in the murine competition assays as indicated in the text and figure legends. All deletions were confirmed by sequencing. The eutR mutant was complemented with pCJA002. Plasmid pCJA002 was constructed by amplifying S. Typhimurium genomic DNA using primers specific to the eutR gene, including 206 nucleotides upstream of the ATG start site (listed in S2 Table). Amplified DNA was digested with HindIII and BamHI and inserted into pGEN-MCS [50] (Addgene MTA). As controls, WT and the ΔeutR strains were transformed with empty pGEN-MCS vectors for use in the complementation experiments. The EutR::Flag strain was generated as described for the deletion strains, using pSUB11 as described [51].
Culture conditions for growth curves and gene expression analyses
All cultures were grown overnight in LB and then diluted 1 : 100 in the indicated medium and grown at 37°C. For RNA expression studies, cultures were grown until mid-logarithmic phase (OD600 = 0.45–0.55). Cultures grown in DMEM were incubated statically under a 5% CO2 atmosphere (to mimic tissue culture conditions). SPI-2 inducing medium was prepared as previously described (100mM Bis/Tris-HCl pH 7.0, 5mM KCl, 7.5mM (NH4)2SO4, 0.5mM K2SO4, 1mM KH2PO4, 38mM glycerol, 0.1% casamino acids, and 8μM MgCl2) [32], and cultures were grown aerobically with agitation [32].
RNA extraction and quantitative reverse transcription polymerase chain reaction (qRT-PCR)
For the in vitro studies, RNA was extracted from S. Typhimurium cells grown in culture medium as described or from phagocytized S. Typhimurium. Cells were resuspended in Trizol (Life Technologies) and RNA was purified using the RiboPure kit (Ambion). For the in vivo studies, spleens were harvested at 2 dpi and homogenized in 1 mL Trizol per 100 mg tissue [52]. RNA was isolated using standard molecular biological procedures. Primer validation and qRT-PCR was performed as described previously [53] using primers listed in S2 Table. Briefly, RNA was extracted from three biological replicates, and qRT-PCR was performed in a one-step reaction using an ABI 7500 sequence detection system (Applied Biosystems). Data were collected using the ABI software Detection 1.2 software (Applied Biosystems). All data were normalized to the endogenous control strB (main text) or to 16S rRNA (RNA was diluted 1 : 1000) as previously performed [54,55]. Controls were used as indicated in figure legends and analyzed using the comparative critical threshold (CT) method. The Student’s unpaired t test was used to determine statistical significance.
Tissue culture
RAW, J774, and HeLa cells were routinely cultured in DMEM supplemented with 10% FBS and 1x penicillin-streptomycin–glutamine; peritoneal exudate macrophages (PEMs) were cultured in RPMI 1640 supplemented with 10% FBS, 20% L-929 conditioned medium, and 1x penicillin-streptomycin–glutamine. PEMs were isolated as described [56]. Antibiotics were omitted during bacterial infections.
For epithelial cell infection bacterial cultures were grown under invasion-inducing conditions [27]. Briefly, overnight cultures were diluted back 1 : 100 and grown without agitation in LB until late logarithmic phase (OD600 of approximately 1.0) at 37°C. Bacterial cells were washed and resuspended in 1x phosphate buffered saline before infection. HeLa cells were placed in DMEM or DMEM supplemented with 5 mM ethanolamine and 150 nM vitamin B12. HeLa cells were infected at a multiplicity of infection (MOI) of 100 for 1 h and either lysed directly or treated with 100 μg/ml gentamicin for 30 min to kill any extracellular bacteria. Percent invasion was calculated as the number of intracellular bacteria as a percent of the directly lysed sample and normalized such that wild type was equal to 100%.
For macrophage assays, we used an invG mutant (deficient in cell invasion) as the WT strain, and we generated corresponding ΔeutRΔinvG and ΔeutBΔinvG strains (described above). These strains were used because invasive S. Typhimurium rapidly kills macrophages [57]. Additionally, expression of invasion-associated genes are down-regulated after entry into host cells; therefore, this strain more closely mimics S. Typhimurium as it is encountered by professional phagocytes after penetration of the epithelial barrier [58].
Gentamicin protection assays were performed according to published methods [14,57,59,60]. S. Typhimurium was grown overnight in LB, washed and re-suspended in PBS before incubation with macrophages (without the addition of ethanolamine or vitamin B12) at an MOI of 50. After 30 min of incubation, extracellular bacteria were killed with 100 μg/ml gentamicin treatment for 30 min, before replacement with media containing 10 μg/ml gentamicin for the remainder of the assay. Cells were lysed at indicated time points in 1% Triton-X and colony forming units (cfu) determined by serial dilutions and plating onto LB agar. After internalization, cells were treated with gentamicin and lysed to enumerate viable intracellular bacteria at time 0 h. Survival was calculated as previously described [14]. Briefly, the viable cfu at the indicated time points were determined as the percentage of this intracellular time 0 h population and normalized such that wild type was equal to 100%. For all assays, the Student t test was used to determine statistical significance.
Mouse studies
All experiments were approved by the Institutional Animal Care and Use Committee at the University of Virginia School of Medicine. For the colitis infections, female C57BL/6 (10–12 week old) mice were given a single dose of 20 mg streptomycin 24 h prior to infection [61]. Mice were infected with an equal mixture of 5 x 108 cfu of the indicated strains. Fresh fecal pellets were collected daily, and mice were euthanized at 4 dpi to assess bacterial burden in the colon and spleen. Tissue samples were weighed, homogenized in 1 ml PBS, and bacterial numbers were quantified by plating serial dilutions of homogenates on MacConkey agar supplemented with streptomycin or with chloramphenicol. The competitive index was calculated as the ratio of ΔeutR to wild type (WT) or ΔeutB strains or the ratio of the ΔeutB to WT recovered normalized to the ratio in the inoculum. Statistical significance was determined by one-sample t test with an expected value of 1. Comparisons between competitive indexes were performed using the Mann-Whitney U test.
For the systemic competition experiments, mice were infected intraperitoneally (i.p.) with 1x105 cfu of the ΔeutR and ΔeutB strains. Spleens were harvested at 6 h and bacterial burden was assessed as described above. Bacterial burden in the peritoneal cavity was assessed as described [62]. Briefly, following euthanasia, 5mL of PBS was injected into the peritoneal cavity, aspirated, and immediately placed on ice. Samples were split into two aliquots, one receiving 100 μg/ml gentamicin treatment. After 30 minutes on ice, samples were washed, lysed and plated as described above. For RNA analyses, mice were infected by i.p. with 1x104 cfu of WT or the ΔeutR strain, and spleens were harvested at 2 dpi.
Electrophoretic mobility shift assays (EMSAs)
Plasmid pDC24 or the empty vector pMAL-c5X was used for the EMSA assays. This plasmid was constructed by amplifying the eutR gene from the S. Typhimurium strain SL1344 with indicated primers (S2 Table). The resulting PCR product was cloned into the Nco1/Sbf1 cloning site of vector pMAL-c5X. EutR was purified under native conditions as described [19]. Briefly, the MBP-tagged EutR protein was purified by growing the E. coli strain NEBexpress cells (NEB) containing pDC24 at 37°C in LB with glucose (0.2% final concentration) and ampicillin (100 μg/ml) to an OD600 of 0.5, at which point IPTG was added to a final concentration of 0.3 mM and allowed to induce overnight at 18°C. Cells were harvested by centrifugation at 4000 x g for 20 min and then resuspended in 25 mL column buffer (20 mM Tris-HCl; 200 mM NaCl; 1 mM EDTA) and lysed by homogenization. The lysed cells were centrifuged, and the lysate was loaded onto a gravity column (Qiagen) with amylose resin. The column was washed with column buffer and then eluted with column buffer containing 10 mM maltose. Fractions containing purified proteins were confirmed by SDS-PAGE and Western analysis, and the protein concentration was determined using a NanoDrop Spectrophotometer. PCR-amplified DNA probes (listed in text and described in S2 Table) were generated as previously described [19,63]. DNA probes were end-labeled with [γ-32P]-ATP (Perkin-Elmer) using T4 polynucleotide kinase (NEB) following standard procedures [64]. End-labeled fragments were run on a 6% polyacrylamide gel, excised, and purified using the Qiagen PCR purification kit.
EMSAs were performed by adding purified EutR-MBP or MBP to labeled DNA in binding buffer (500 μg ml-1 BSA (NEB), 50 ng poly-dIdC, 60 mM HEPES pH 7.5, 5 mM EDTA, 3 mM dithiothreitol (DTT), 300 mM KCl, and 25 mM MgCl2). Ethanolamine (1 mM) and vitamin nM B12 (150 nM) were added to the reactions. Reactions were incubated for 25 minutes at 25°C. Then, a 1% Ficoll solution was added to the reactions immediately before loading the samples on the gel. The reactions were electrophoresed for approximately 6 h at 150 V on a 6% polyacrylamide gel, dried, and imaged with a phosphorimager (Molecular Dynamics).
Chromatin immunoprecipitation (ChIP) and ChIP qPCR
ChIP was performed using an WT S. Typhimurium (untagged EutR) or with the S. Typhimurium eutR mutant transformed with the EutR::MBP plasmid. Strains were grown in DMEM supplemented with 5 mM EA, 150 nM B12, and 0.5 μM IPTG until cells reached an OD600 of approximately 0.8. Cross-linking and ChIP were performed based on established methods [65]. Formaldehyde was added (1% final concentration) for cross-linking, and cells were incubated at room temperature for 20 min. Reactions were quenched with 0.5 M glycine, then samples were pelleted, resuspended in TBS, and washed. Cells were lysed with 2 mg/ml lysozyme and incubated at 37°C for 30 min. Subsequently, samples were placed on ice and sonicated. Insoluble cell debris was removed by centrifugation, and supernatants were saved. Immunoprecipitation was carried out by incubating samples with amylose beads (NEB) in buffer for 2 h at 4°C with gentle mixing. Beads were pelleted and washed. Then the samples were incubated for 10 min at 65°C in elution buffer with occasional gentle mixing. Samples were centrifuged and supernatants were collected. To reverse the cross-link, samples were boiled for 10 min and DNA was purified using the Qiagen PCR purification kit. For ChIP-quantitative PCR (qPCR) experiments, untreated chromatin was de-cross-linked by boiling for 10 min and purified, for use as the “input” control. Primers amplifying the strB gene were used as the negative control. The fold enrichment of each promoter represents the value of the immunoprecipitated DNA divided by the input unprecipitated DNA [66,67]. These values were normalized to the values obtained for each promoter precipitated using untagged EutR in order to account for non-specific enrichment.
Accession numbers for genes/proteins mentioned in text
eutR, NP_461389; eutS, NP_461405; eutB, NP_461393; sipC, NP_461805; ssrA, NP_460357; ssrB, NP_460356; ssaB, NP_460358; sseA, NP_460362; ssaG, NP_460371; sifA, NP_460194.
Supporting Information
Zdroje
1. Hughes DT, Sperandio V (2008) Inter-kingdom signalling: communication between bacteria and their hosts. Nature Rev Microbiol 6 : 111–120.
2. Bakovic M, Fullerton MD, Michel V (2007) Metabolic and moleclar aspects of ethanolamine phospholipid biosynthesis: the role of CTP:phosphoethanolamine cytidylyl-transferase (Pcyt2). Biochem Cell Biol 85 : 283–300. 17612623
3. Meijerink J, Plastina P, Vincken JP, Poland M, Attya M, et al. (2011) The ethanolamide metabolite of DHA, docosahexaenoylethanolamine, shows immunomodulating effects in mouse peritoneal and RAW264.7 macrophages: evidence for a new link between fish oil and inflammation. Br J Nutr 105 : 1798–1807. doi: 10.1017/S0007114510005635 21294934
4. Sugiura T, Kobayashi Y, Oka S, Waku K (2002) Biosynthesis and degradation of anandamide and 2-arachidonoylglycerol and their possible physiological significance. Prostaglandins Leukot Essent Fatty Acids 66 : 173–192. 12052034
5. Cotton PB (1972) Non-dietary lipid in the intestinal lumen. Gut 13 : 675–681. 4639402
6. Garsin DA (2010) Ethanolamine utilization in bacterial pathogens: roles and regulation. Nature Rev Microbiol 8 : 290–295.
7. Lipton BA, Davidson EP, Ginsberg BH, Yorek MA (1990) Ethanolamine metabolism in cultured bovine aortic endothelial cells. J Biol Chem 265 : 7195–7201. 2110161
8. Lipton BA, Yorek MA, Ginsberg BH (1988) Ethanolamine and choline transport in cultured bovine aortic endothelial cells. J Cell Physiol 137 : 571–576. 3192633
9. Nikawa J, Tsukagoshi Y, Yamashita S (1986) Cloning of a gene encoding choline transport in Saccharomyces cerevisiae. J Bacteriol 166 : 328–330. 3514579
10. Sandra A, Cai J (1991) Plasma membrane appearance of phosphatidylethanolamine in stimulated macrophages. J Leukoc Biol 50 : 19–27. 2056244
11. Cohen JI, Bartlett JA, Corey GR (1987) Extra-intestinal manifestations of salmonella infections. Medicine (Baltimore) 66 : 349–388.
12. Galan JE, Curtis R III (1989) Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc Natl Acad Sci 86 : 6383–6387. 2548211
13. Cirillo DM, Valdivia RH, Monack DM, Falkow S (1998) Macrophage-dependent induction of the Salmonella pathogenicity island 2 type III secretion system and its role in intracellular survival. Mol Microbiol 30 : 175–188. 9786194
14. Hensel M, Shea JE, Waterman SR, Mundy R, Nikolaus T, et al. (1998) Genes encoding putative effector proteins of the type III secretion system of Salmonella pathogenicity island 2 are required for bacterial virulence and proliferation in macrophages. Mol Microbiol 30 : 163–174. 9786193
15. Ochman H, Soncini FC, Solomon F, Groisman EA (1996) Identification of a pathogenicity island required for Salmonella survival in host cells. Proc Natl Acad Sci 93 : 7800–7804. 8755556
16. Joseph B, Przybilla K, Stuhler C, Schauer K, Slaghis J, et al. (2006) Identification of Listeria monocytogenes genes contributing to intracellular replication by expression profiling and mutant screening. J Bacteriol 188 : 556–568. 16385046
17. Thiennimitr P, Winter SE, Winter MG, Xavier MN, Tolstikov V, et al. (2011) Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc Natl Acad Sci 108 : 17480–17485. doi: 10.1073/pnas.1107857108 21969563
18. Roof DM, Roth JR (1988) Ethanolamine utilization in Salmonella typhimurium. J Bacteriol 170 : 3855–3863. 3045078
19. Luzader DH, Clark DE, Gonyar LA, Kendall MM (2013) EutR is a direct regulator of genes that contribute to metabolism and virulence in enterohemorrhagic Escherichia coli O157:H7. J Bacteriol 195 : 4947–4953. doi: 10.1128/JB.00937-13 23995630
20. Roof DM, Roth JR (1992) Autogenous regulation of ethanolamine utilization by a transcriptional activator of the eut operon in Salmonella typhimurium. J Bacteriol 174 : 6634–6643. 1328159
21. Gonyar LA, Kendall MM (2014) Ethanolamine and choline promote expression of putative and characterized fimbriae in enterohemorrhagic Escherichia coli O157:H7. Infect Immun 82 : 193–201. doi: 10.1128/IAI.00980-13 24126525
22. Kendall MM, Gruber CC, Parker CT, Sperandio V (2012) Ethanolamine controls expression of genes encoding components involved in interkingdom signaling and virulence in enterohemorrhagic Escherichia coli O157:H7. mBio 3: e00050–00012. doi: 10.1128/mBio.00050-12 22589288
23. Steeb B, Claudi B, Burton NA, Tienz P, Schmidt A, et al. (2013) Parallel exploitation of diverse host nutrients enhances Salmonella virulence. PLoS Pathog 9: e1003301. doi: 10.1371/journal.ppat.1003301 23633950
24. Stojiljkovic I, Bäumler AJ, Heffron F (1995) Ethanolamine utilization in Salmonella typhimurium: nucleotide sequence, protein expression, and mutational analysis of the cchA cchB eutE eutJ eutG eutH gene cluster. J Bacteriol 177 : 1357–1366. 7868611
25. Hapfelmeier S, Hardt W-D (2005) A mouse model for S. typhimurium-induced enterocolits. Trends Microbiol 13 : 497–503. 16140013
26. Lara-Tejero M, Galán JE (2009) Salmonella enterica serovar typhimurium pathogenicity island 1-encoded type III secretion system translocases mediate intimate attachment to nonphagocytic cells. Infect Immun 77 : 2635–2642. doi: 10.1128/IAI.00077-09 19364837
27. Lee CA, Falkow S (1990) The ability of Salmonella to enter mammalian cells is affected by growth state. Proc Natl Acad Sci 87 : 4303–4308.
28. Brown NF, Rogers LD, Sanderson KL, Gouw JW, Hartland EL, et al. (2014) A horizontally acquired transcription factor coordinates Salmonella adaptations to host microenvironments. mBio 5: e01727–01714. doi: 10.1128/mBio.01727-14 25249283
29. Feng X, Walthers D, Oropeza R, Kenney LJ (2004) The response regulator SsrB activates transcription and binds to a region overlapping OmpR binding sites at Salmonella pathogenicity island 2. Mol Microbiol 54 : 823–835. 15491370
30. Walthers D, Carroll RK, Navarre WW, Libby SJ, Fang FC, et al. (2007) The response regulator SsrB activates expression of diverse Salmonella pathogenicity island 2 promoters and counters silencing by the nucleoid-associated protein H-NS. MolMicrobiol 65 : 477–493.
31. Worley MJ, Ching KH, Heffron F (2000) Salmonella SsrB activates a global regulon of horizontally acquired genes. Mol Microbiol 36 : 749–761. 10844662
32. Deiwick J, Nikolaus T, Erdogan S, Hensel M (1999) Environmental regulation of Salmonella pathogenicity island 2 gene expression. Mol Microbiol 31 : 1759–1773. 10209748
33. Bertin Y, Girardeau JP, Chaucheyras-Durand F, Lyan B, Pujos-Guillot E, et al. (2011) Enterohaemorrhagic Escherichia coli gains a competitive advantage by using ethanolamine as a nitrogen source in the bovine intestinal content. Environ Microbiol 13 : 365–377. doi: 10.1111/j.1462-2920.2010.02334.x 20849446
34. Coombes BK, Brown NF, Valdez Y, Brumell JH, Finlay BB (2004) Expression and secretion of Salmonella pathogenicity island-2 virulence genes in response to acidification exhibit differential requirements of a functional type III secretion apparatus and SsaL. J Biol Chem 279 : 49804–49815. 15383528
35. Vazquez-Torres A, Fang FC (2001) Salmonella evasion of the NADPH phagocyte oxidase. Microbes Infect 3 : 1313–1320. 11755420
36. Jeter RM, Olivera B, Roth JR (1984) Salmonella typhimurium synthesizes cobalamin vitamin B12 de novo under anaerobic growth conditions. J Bacteriol 159 : 206–213. 6376471
37. Chakravortty D, Hansen-Wester I, Hensel M (2002) Salmonella pathogenicity island 2 mediates protection of intracellular Salmonella from reactive nitrogen intermediates. J Exp Med 195 : 1155–1166. 11994420
38. Vazquez-Torres A, Xu Y, Jones-Carson J, Holden DW, Lucia SM, et al. (2000) Salmonella pathogenicity island 2-dependent evasion of the phagocyte NADPH oxidase. Science 287 : 1655–1658. 10698741
39. Miao EA, Freeman JA, Miller SI (2002) Transcription of the SsrAB regulon is repressed by alkaline pH and is independent of PhoPQ and magnesium concentration. J Bacteriol 184 : 1493–1497. 11844786
40. Mulder DT, McPhee JB, Savchenko A, Coombes BK (2015) Multiple histidines in the periplasmic domain of the Salmonella enterica sensor kinase SsrA enhance signaling in response to extracellular acidification. Mol Microbiol 95 : 678–691. doi: 10.1111/mmi.12895 25442048
41. Feng X, Oropeza R, Kenney LJ (2003) Dual regulation by phospho-OmpR of ssrA/B gene expression in Salmonella pathogenicity island 2. Mol Microbiol 48 : 1131–1143. 12753201
42. Bijlsma JJE, Groisman EA (2005) The PhoP/PhoQ system controls the intramacrophage type three secretion system of Salmonella enterica. Mol Microbiol 57 : 85–96. 15948951
43. Chaudhuri RR, Morgan E, Peters SE, Pleasance SJ, Hudson DL, et al. (2013) Comprehensive assignment of roles for Salmonella typhimurium genes in intestinal colonization of food-producing animals. PLoS Genet 9: e1003456. doi: 10.1371/journal.pgen.1003456 23637626
44. Winter SE, Thiennimitr P, Winter MG, Butler BP, Huseby DL, et al. (2010) Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 467 : 426–429. doi: 10.1038/nature09415 20864996
45. Garsin DA (2012) Ethanolamine: a signal to commence a host-associated lifestyle? mBio 3: e00172–00112. doi: 10.1128/mBio.00172-12 22761393
46. Alteri CJ, Mobley HLT (2012) Escherichia coli physiology and metabolism dictates adaptation to diverse host microenvironments. Curr Opin Microbiol 15 : 3–9. doi: 10.1016/j.mib.2011.12.004 22204808
47. Hoiseth SK, Stocker BA (1982) Aromatic-dependent Salmonella typhimurium are non-virulenct and effective as live vaccines. Nature 291 : 238–239.
48. Criss AK, Ahlgren DM, Jou TS, McCormick BA, Casanova JE (2001) The GTPase Rac1 selectively regulates Salmonella invasion at the apical plasma membrane of polarized epithelial cells. J Cell Sci 114 : 1331–1341. 11256999
49. Datsenko KA, Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci 97 : 6640–6645. 10829079
50. Lane MC, Alteri CJ, Smith SN, Mobley HLT (2007) Expression of flagella is coincident with uropathogenic Escherichia coli ascension to the upper urinary tract. Proc Natl Acad Sci 104 : 16669–16674. 17925449
51. Uzzau S, Figuerosa-Bossi N, Rubino S, Bossi L (2001) Epitope tagging of chromosomal genes in Salmonella. PNAS 98 : 15264–15269. 11742086
52. Curtis MM, Hu Z, Klimko C, Narayanan S, Deberardinis R, et al. (2014) The gut commensal Bacteroides thetaiotaomicron exacerbates enteric enfection through modification of the metabolic landscape. Cell Host Microbe 16 : 759–769. doi: 10.1016/j.chom.2014.11.005 25498343
53. Kendall MM, Gruber CC, Rasko D, A., Hughes DT, Sperandio V (2011) Hfq virulence regulation in enterohemorrhagic Escherichia coli O157:H7 strain 86–24. J Bacteriol.
54. Lee E-J, Groisman EA (2012) Control of a Salmonella virulence locus by an ATP-sensing leader messenger RNA. Nature 486 : 271–275. doi: 10.1038/nature11090 22699622
55. Shin D, Lee E-J, Huang H, Groisman EA (2006) A positive feedback loop promotes transcription surge that jump-starts Salmonella virulence circui. Science 314 : 1607–1609. 17158330
56. Zhang X, Goncalves R, Mosser DM (2008) The isolation and characterization of murine macrophages. In: Coligan JE, editor. Curr Prot Immunol.
57. Chen LM, Kaniga K, Galan JE (1996) Salmonella spp. are cytotoxic for cultured macrophages. Mol Microbiol 21 : 1101–1115. 8885278
58. Laughlin RC, Knodler LA, Barhoumi R, Payne HR, Wu J, et al. (2014) Spatial segregation of virulence gene expression during acute enteric infection with Salmonella enterica serovar Typhimurium. mBio 5: e00946–00913. doi: 10.1128/mBio.00946-13 24496791
59. Buchmeier NA, Heffron F (1989) Intracellular survival of wild-type Salmonella typhimurium and macrophage-sensitive mutants in diverse populations of macrophages. Infect Immun 57 : 1–7. 2642463
60. Owen K, Meyer CB, Bouton AH, Casanova JE (2014) Activation of focal adhesion kinase by Salmonella suppresses autophagy via an Akt/mTOR signaling pathway and promotes bacterial survival in macrophages. PLoS Pathog 10: e1004159. doi: 10.1371/journal.ppat.1004159 24901456
61. Barthel M, Hapfelmeier S, Quintanilla-Martínez L, Kremer M, Rohde M, et al. (2003) Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect Immun 71 : 2839–2858. 12704158
62. Edwards RA, Schifferli DM, Maloy SR (2000) A role for Salmonella fimbriae in intraperitoneal infections. Proc Natl Acad Sci 97 : 1258–1262. 10655518
63. Kendall MM, Rasko D, A., Sperandio V (2010) The LysR-type regulator QseA regulates both characterized and putative virulence genes in enterohaemorrhagic Escherichia coli O157:H7. Mol Microbiol 76 : 1306–1321. doi: 10.1111/j.1365-2958.2010.07174.x 20444105
64. Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
65. Rhodius VA, Wade JT (2009) Technical considerations in using DNA microarrays to define regulons. Methods 47 : 63–72. doi: 10.1016/j.ymeth.2008.10.017 18955146
66. Kuras L, Struhl K (1999) Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature 399 : 609–613. 10376605
67. Shin D, Groisman EA (2005) Signal-dependent binding of the response regulators PhoP and PmrA to their target promoters in vivo. J Biol Chem 280 : 4089–4094. 15569664
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Increased Susceptibility of Humanized NSG Mice to Panton-Valentine Leukocidin and Skin InfectionČlánek Phosphorylation of a Myosin Motor by TgCDPK3 Facilitates Rapid Initiation of Motility during egress
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2015 Číslo 11- Stillova choroba: vzácné a závažné systémové onemocnění
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Choroby jater v ordinaci praktického lékaře – význam jaterních testů
- Diagnostický algoritmus při podezření na syndrom periodické horečky
-
Všechny články tohoto čísla
- Parasite Glycobiology: A Bittersweet Symphony
- On the Discovery of TOR As the Target of Rapamycin
- Broadening of Virus-Specific CD8 T-Cell Responses Is Indicative of Residual Viral Replication in Aviremic SIV Controllers
- PML/TRIM19-Dependent Inhibition of Retroviral Reverse-Transcription by Daxx
- Cleavage of a Neuroinvasive Human Respiratory Virus Spike Glycoprotein by Proprotein Convertases Modulates Neurovirulence and Virus Spread within the Central Nervous System
- Kaposi’s Sarcoma-Associated Herpesvirus (KSHV) Induces the Oncogenic miR-17-92 Cluster and Down-Regulates TGF-β Signaling
- Interferon-α Subtypes in an Model of Acute HIV-1 Infection: Expression, Potency and Effector Mechanisms
- Perivascular Arrest of CD8 T Cells Is a Signature of Experimental Cerebral Malaria
- Targeting HIV Reservoir in Infected CD4 T Cells by Dual-Affinity Re-targeting Molecules (DARTs) that Bind HIV Envelope and Recruit Cytotoxic T Cells
- Evolution and Emergence of Enteroviruses through Intra- and Inter-species Recombination: Plasticity and Phenotypic Impact of Modular Genetic Exchanges in the 5’ Untranslated Region
- Interferon-γ Inhibits Ebola Virus Infection
- Dengue Virus Non-structural Protein 1 Modulates Infectious Particle Production via Interaction with the Structural Proteins
- P-Type Cyclin CYC3 Modulates Endomitotic Growth during Oocyst Development in Mosquitoes
- Diversity of across Evolutionary Scales
- 50 Years of Disease in Humans: The Dramatic Emergence of a Cluster of Novel Fungal Pathogens
- Worse Comes to Worst: Bananas and Panama Disease—When Plant and Pathogen Clones Meet
- Arenavirus Glycan Shield Promotes Neutralizing Antibody Evasion and Protracted Infection
- Infection-Induced Retrotransposon-Derived Noncoding RNAs Enhance Herpesviral Gene Expression via the NF-κB Pathway
- Structural Insight into Archaic and Alternative Chaperone-Usher Pathways Reveals a Novel Mechanism of Pilus Biogenesis
- Increased Susceptibility of Humanized NSG Mice to Panton-Valentine Leukocidin and Skin Infection
- Global Analysis of the Fungal Microbiome in Cystic Fibrosis Patients Reveals Loss of Function of the Transcriptional Repressor Nrg1 as a Mechanism of Pathogen Adaptation
- The Transcription and Translation Landscapes during Human Cytomegalovirus Infection Reveal Novel Host-Pathogen Interactions
- The N-terminal Helical Region of the Hepatitis C Virus p7 Ion Channel Protein Is Critical for Infectious Virus Production
- Activation of Type I and III Interferon Response by Mitochondrial and Peroxisomal MAVS and Inhibition by Hepatitis C Virus
- Hsp70 Isoforms Are Essential for the Formation of Kaposi’s Sarcoma-Associated Herpesvirus Replication and Transcription Compartments
- Distinct Upstream Role of Type I IFN Signaling in Hematopoietic Stem Cell-Derived and Epithelial Resident Cells for Concerted Recruitment of Ly-6C Monocytes and NK Cells via CCL2-CCL3 Cascade
- and Bats: Story of an Emerging Friendship
- Emergence of Pathogenicity in Lagoviruses: Evolution from Pre-existing Nonpathogenic Strains or through a Species Jump?
- Ebolavirus Evolution: Past and Present
- Host and Symbiont Jointly Control Gut Microbiota during Complete Metamorphosis
- Non-Human Primates Harbor Diverse Mammalian and Avian Astroviruses Including Those Associated with Human Infections
- Lactate Dehydrogenase Is Associated with the Parasitophorous Vacuole Membrane and Is a Potential Target for Developing Therapeutics
- Five Questions about Mycoviruses
- Phosphorylation of a Myosin Motor by TgCDPK3 Facilitates Rapid Initiation of Motility during egress
- Ethanolamine Signaling Promotes Niche Recognition and Adaptation during Infection
- Cross-Species Transmission and Differential Fate of an Endogenous Retrovirus in Three Mammal Lineages
- Memory Th1 Cells Are Protective in Invasive Infection
- Transcription Factor SomA Is Required for Adhesion, Development and Virulence of the Human Pathogen
- An -Methyltransferase Is Required for Infection of Tick Cells by
- RNA-seq Brings New Insights to the Intra-Macrophage Transcriptome of Typhimurium
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Dengue Virus Non-structural Protein 1 Modulates Infectious Particle Production via Interaction with the Structural Proteins
- On the Discovery of TOR As the Target of Rapamycin
- Parasite Glycobiology: A Bittersweet Symphony
- Broadening of Virus-Specific CD8 T-Cell Responses Is Indicative of Residual Viral Replication in Aviremic SIV Controllers
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



