-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Symbionts Commonly Provide Broad Spectrum Resistance to Viruses in Insects: A Comparative Analysis of Strains
In recent years it has been discovered that many organisms are infected with bacterial symbionts that protect them against pathogens. Wolbachia is a bacterial symbiont that is found in many species of insects, and several strains are known to protect the insects against viral infection. We took 19 strains of Wolbachia from different species of Drosophila fruit flies, transferred them into Drosophila simulans, and then infected these flies with two different viruses. We found that about half of the strains slowed the death of flies after viral infection. Given that 40% of terrestrial arthropods may be infected with Wolbachia, this suggests that many species may benefit from this protection. These increases in survival were tightly linked to reductions in the levels of the virus in the insect, suggesting that Wolbachia is reducing the viruses' ability to replicate. Despite the two viruses we used being very different, the level of protection that a Wolbachia strain provided against the two viruses tended to be very similar, suggesting that a single general mechanism underlies the antiviral effects. The extent to which a Wolbachia strain provides protection against viral infection depends largely on the bacterial density — the more Wolbachia, the greater the protection.
Published in the journal: . PLoS Pathog 10(9): e32767. doi:10.1371/journal.ppat.1004369
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1004369Summary
In recent years it has been discovered that many organisms are infected with bacterial symbionts that protect them against pathogens. Wolbachia is a bacterial symbiont that is found in many species of insects, and several strains are known to protect the insects against viral infection. We took 19 strains of Wolbachia from different species of Drosophila fruit flies, transferred them into Drosophila simulans, and then infected these flies with two different viruses. We found that about half of the strains slowed the death of flies after viral infection. Given that 40% of terrestrial arthropods may be infected with Wolbachia, this suggests that many species may benefit from this protection. These increases in survival were tightly linked to reductions in the levels of the virus in the insect, suggesting that Wolbachia is reducing the viruses' ability to replicate. Despite the two viruses we used being very different, the level of protection that a Wolbachia strain provided against the two viruses tended to be very similar, suggesting that a single general mechanism underlies the antiviral effects. The extent to which a Wolbachia strain provides protection against viral infection depends largely on the bacterial density — the more Wolbachia, the greater the protection.
Introduction
Heritable symbionts are major players in arthropod evolution owing to their high incidence and the diversity of effects they have on their host's phenotype [1]–[3]. Primary (obligate) symbionts are mutualists that play some essential role — typically synthesizing nutrients missing from the insect's diet — and they often form stable associations with their hosts that can last for many millions of years [4]–[6]. Secondary (facultative) symbionts have more diverse effects, which range from parasitism to mutualism [3], [7]. The parasites mostly manipulate their host's reproduction to enhance their transmission to the next generation, for example by distorting the sex ratio towards females (the sex that transmits the bacteria). The mutualists can supply nutrients, or protect against environmental stresses [8] or natural enemies [9]–[11]. Furthermore, symbionts can combine several strategies at once, with some ‘Jekyll and Hyde’ strains simultaneously exhibiting mutualistic and parasitic phenotypes [12].
As secondary symbionts occasionally jump between different host species [13]–[16], they can result in rapid evolutionary change in their hosts. This process may be quite different to selection acting on the host genome, as when a host acquires a novel symbiont it can instantly acquire a complex adaptation encoded by many genes. Striking examples of rapid evolution resulting from the spread of symbionts include a Rickettsia bacterium infecting whiteflies which rapidly spread through US populations by causing sex ratio distortion as well as increased fecundity and survival [17], and a Spiroplasma bacterium that spread through populations of Drosophila neotestacea, protecting the hosts against a parasitic nematode [10].
Wolbachia is a maternally-transmitted alphaproteobacterium that is estimated to infect around 40% of terrestrial arthropods [18] and can act as both a parasite and a mutualist. Until recently it was viewed primarily as a parasite that manipulates host reproduction, most commonly by inducing cytoplasmic incompatibility (CI) [19]–[23]. CI allows Wolbachia to invade populations by causing embryonic mortality when uninfected females mate with infected males, thus conferring a selective advantage to infected females [24]. Recently it was discovered that Wolbachia can also protect Drosophila melanogaster against several RNA viruses [25], [26], and subsequently similar antiviral effects have been reported in other Drosophila species [27], [28], as well as in mosquitoes [29]–[32]. In most cases, Wolbachia has been shown to be associated with a decrease in viral titer [26], [27], [29]. However, Wolbachia increased the survival of the flies but had no effect on viral titer in D. melanogaster infected with Flock House virus (FHV) [26] as well as in one case in D. simulans infected with Drosophila C virus (DCV) [27], suggesting that Wolbachia might also allow its host to tolerate viral infections without affecting the pathogen load. Wolbachia has also been associated with protection against filarial nematodes, Plasmodium parasites and pathogenic bacteria in mosquitoes [29], [33]–[36]. However, it is not known whether the mechanisms of protection acting against these parasites are the same that are involved in protection against viruses.
Antiviral protection by Wolbachia could potentially be used to control vector-borne diseases such as dengue fever [37], [38]. When artificially introduced into Aedes aegypti, the main vector of dengue virus, Wolbachia was shown to limit the replication of dengue virus as well as chikungunya, yellow fever and West Nile viruses [29], [39], [40]. Furthermore, when Wolbachia infected mosquitoes were released into the wild, the bacterium spread through the mosquito populations due to the induction of CI [41], [42].
In both Drosophila and mosquitoes, different Wolbachia strains are associated with different levels of antiviral protection [27], [32], [39], [43], [44], even among very closely related strains [45]. The causes of this variation are not entirely clear, as relatively few Wolbachia strains have been characterized for their level of protection, not all studies control the host genetic background, and none have controlled for the confounding effects of the bacterial phylogeny. Nonetheless, several studies found that the Wolbachia strains with the highest density within the host provide the strongest protection against viruses, and tissue tropism may also play a role [27], [30], [44]–[47]. Overall, little is known about how commonly Wolbachia protects insects against viral infection, how this trait is distributed across the Wolbachia phylogeny, and therefore to what extent it has contributed to the evolutionary success of Wolbachia.
The mechanisms by which Wolbachia protects hosts against viruses remain to be elucidated. The protection could be caused by direct interactions between Wolbachia and viruses, competition for shared resources, or indirectly through the regulation of host gene expression [26]. In particular, it was speculated that Wolbachia infection may up-regulate the host immune system. While this was shown to occur after transinfection of Wolbachia from Drosophila into Ae. aegypti and Anopheles gambiae [33], [48]–[50], such an effect was not observed in D. melanogaster naturally infected with Wolbachia [50]–[55] or transinfected with a non-native strain [44]. Similarly, the small interfering RNA (siRNA) pathway, which provides broad-spectrum antiviral defense in insects, is not required for Wolbachia to confer antiviral protection in flies [56]. Recent results suggest that, in Ae. aegypti, Wolbachia has an indirect effect on viral replication through the manipulation of host microRNAs [57], [58]. In this species Wolbachia suppresses the expression of AaDnmt2, a methyltransferase gene, by up-regulating the microRNA aae-miR-2940 [57]. Overexpression of AaDnmt2 decreases Wolbachia density and increases the titer of Dengue virus, suggesting a causal link between Wolbachia and viral replication. However, Dnmt2, the homolog of AaDnmt2 found in D. melanogaster, was shown to have an antiviral effect against Drosophila C virus [59], contradicting the effect observed in case of dengue infection.
To overcome the lack of experimental tools available for Wolbachia — the bacterium cannot be cultured outside of insect cells and cannot be genetically manipulated or cloned — we have taken a comparative approach, looking for genetic correlations between levels of antiviral protection and potential causes such as changes in gene expression. To allow us to do this, we compared the level of protection of 19 Wolbachia strains from a diverse range of Drosophila species that we transferred into a common D. simulans genetic background. We used Drosophila C virus and Flock House virus, which are both RNA viruses with positive-sense single-stranded genomes. Drosophila C virus belongs to the family Dicitroviridae [60] and is naturally found in D. melanogaster and D. simulans [61]–[63]. FHV belongs to the family Nodaviridae and was initially isolated from a beetle [64].
Using this comparative approach, we show that Wolbachia strains vary considerably in the extent to which they increase the survival of flies after viral infection. There is little specificity, with strong genetic correlations between protection against FHV and DCV, despite these viruses being distantly related. The increases in survival can largely be explained by Wolbachia reducing viral titer. The variation in antiviral protection is largely explained by differences in the density of Wolbachia in host tissues. However, there is no evidence that either activation of the humoral immune response or up-regulation of the methyltransferase gene Dnmt2 play any role in antiviral protection.
Results
Diversity of Wolbachia in the genus Drosophila
We assembled a panel of 19 Wolbachia strains that naturally infect 16 different species of Drosophila (Table 1). We reconstructed the phylogeny of these strains using sequences from eight multilocus sequence typing (MLST) genes and a Bayesian method that accounts for recombination between strains [65]. The phylogeny reveals that 18 of the strains clustered in what is commonly regarded as the supergroup A [66], with wMa being the only strain from the supergroup B (Figure 1A). Many of the strains are very closely related.
Fig. 1. Phylogeny of Wolbachia strains and respective level of protection and within-host density. 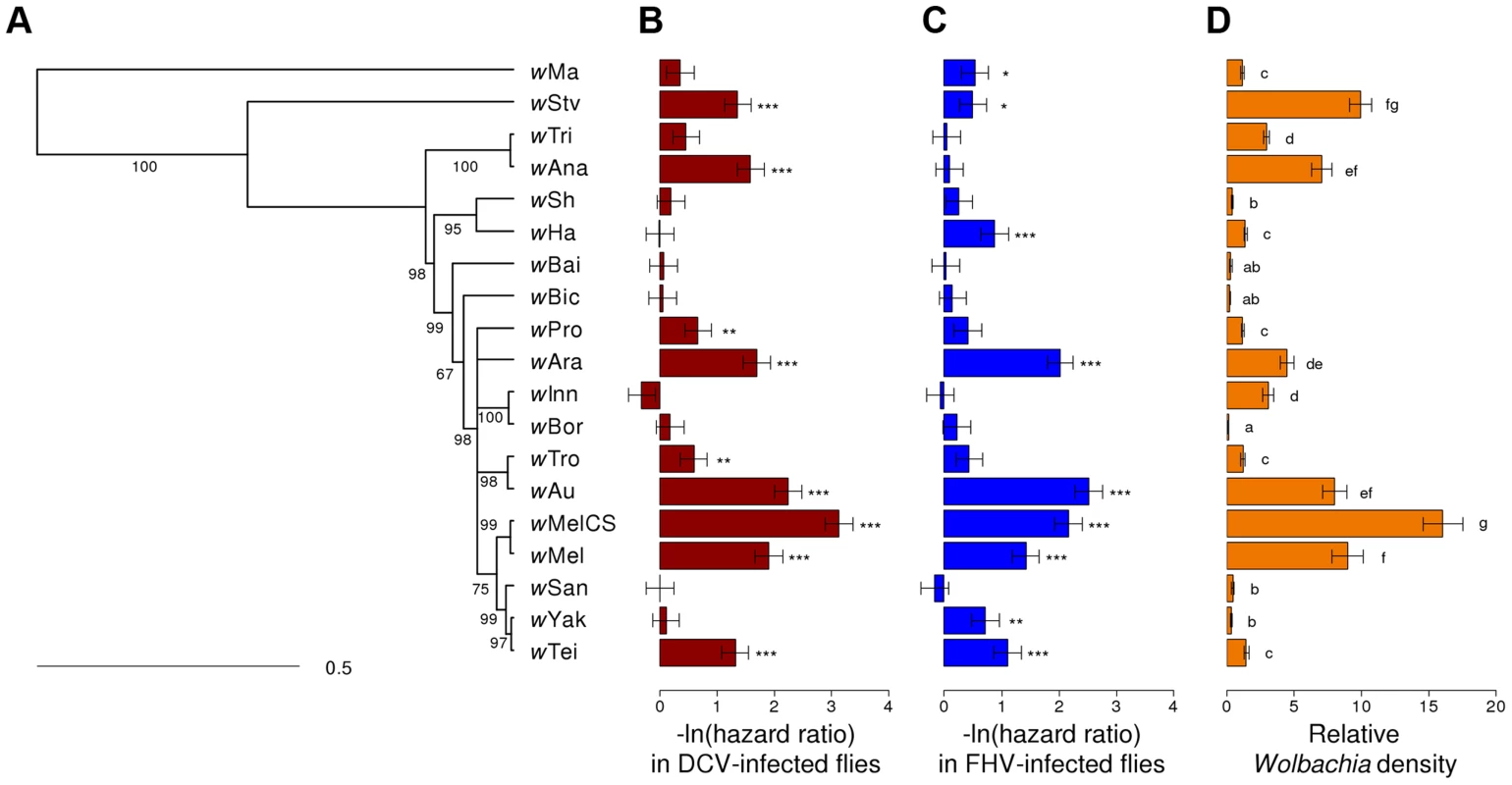
(A) The phylogeny is based on the sequence of the MLST genes 16S rRNA, aspC, atpD, ftsZ, sucB, groEL, coxA and fbpA. Branch labels represent posterior support values. Nodes with less than 50% support were collapsed. The scale bar indicates time in coalescent units. (B–C) Flies were either infected with (B) DCV or (C) FHV. Survival is expressed as the negative natural log of the hazard ratio compared to Wolbachia-free flies, as estimated from a Cox's mixed-effect model. Error bars are standard errors. Symbols above the bars give the significance relative to the Wolbachia-free controls (*: P<0.05; **: P<0.01; ***: P<0.001). (D) Wolbachia density is expressed as the ratio of Wolbachia genomic DNA to Drosophila genomic DNA, as estimated by quantitative PCR. Different letters indicate significant differences based on a Tukey's honest significance test on ln-transformed data. Tab. 1. Wolbachia strains used in this study. 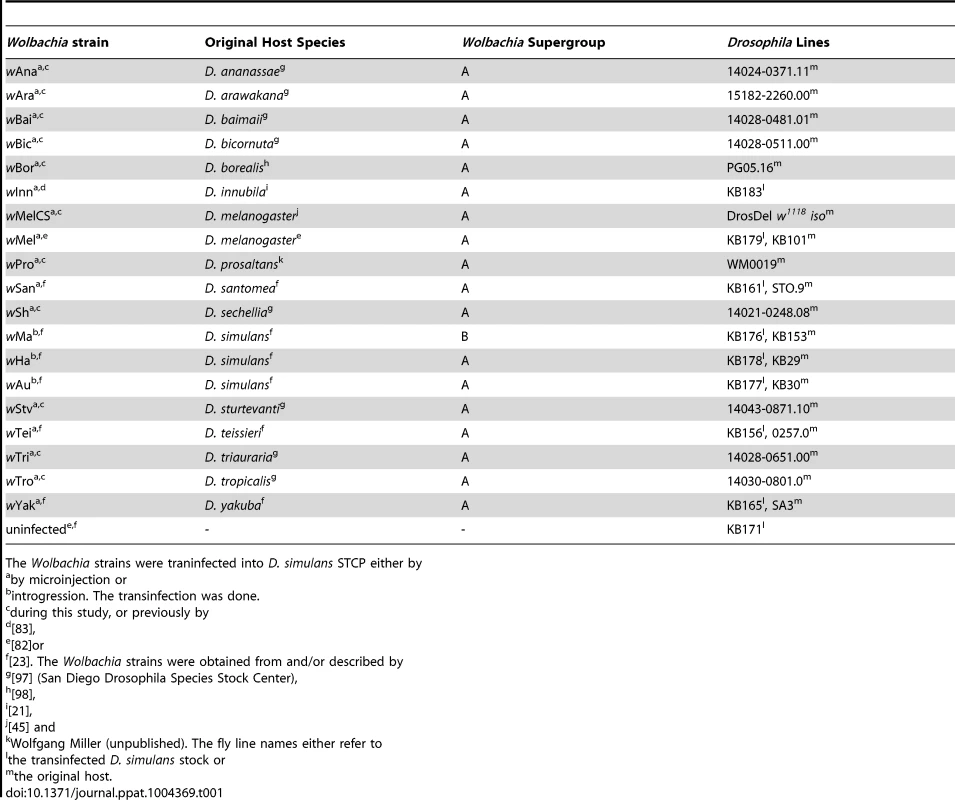
The Wolbachia strains were traninfected into D. simulans STCP either by To allow us to compare the level of antiviral protection that the different Wolbachia strains provide to their hosts, we transferred them into the same inbred D. simulans genetic background. Eleven of the strains were transferred as part of this study, and the remaining eight have been reported before (Table 1).
Wolbachia strains vary in the extent to which they increase survival after viral infection
Flies from the 19 D. simulans lines carrying the different Wolbachia strains, together with a Wolbachia-free control, were stabbed with a needle that had been dipped in DCV, FHV or Ringer's solution (2528, 2527 and 2492 flies were stabbed for each treatment respectively). We then followed their survival for 25 days (Figure 2A–C). In the mock-infected flies (Ringer), there was significant heterogeneity among the 20 fly lines (Cox's mixed-effect model, χ2 = 47, d.f. = 19, P = 0.0004; Figure S1), possibly reflecting either intrinsic effects of Wolbachia on survival or some other difference between the lines, such as remaining differences in the host genetic background. The overall survival of the mock-infected flies was low, likely due to this being a weak inbred stock (Figure 2C).
Fig. 2. Survival of flies carrying different Wolbachia strains or being Wolbachia-free. 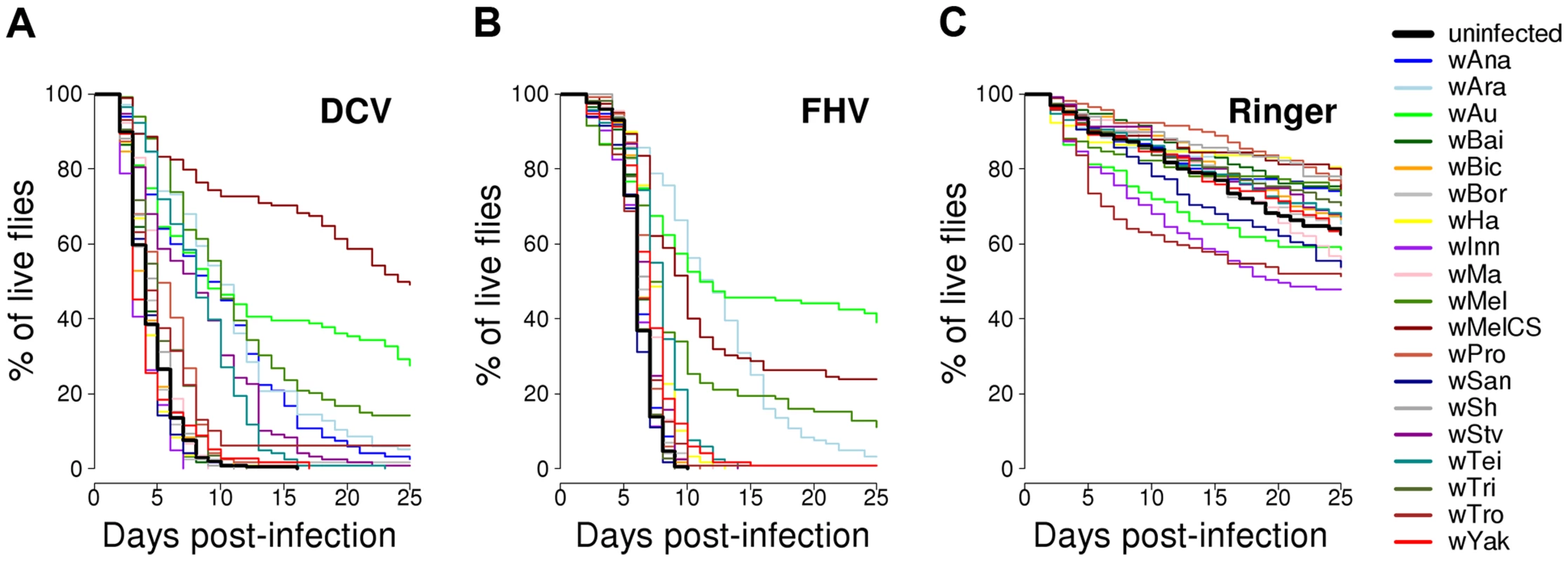
Flies were either infected with (A) DCV, (B) FHV or (C) mock-infected with Ringer's solution. There was a substantial variation among Wolbachia strains in the degree to which they protect their hosts against viral infection (Figure 2A & B). Twelve of the Wolbachia-infected lines showed significantly reduced mortality relative to the Wolbachia-free flies within either the DCV or FHV treatments (Figure 1B & 1C). To account for the slight variation in the survival of the mock-infected controls, we tested whether survival of the 20 fly lines was affected by a statistical interaction between the Wolbachia strain and infection (whether the flies were infected with a virus or mock-infected). There was a highly significant interaction for both DCV and FHV (Cox's mixed-effect models; DCV: χ2 = 127.4, d.f. = 19, P<10−15; FHV: χ2 = 107.6, d.f. = 19, P<10−14). Using this more conservative approach of testing for an interaction of Wolbachia and infection treatment, nine of the 19 Wolbachia strains provided a significant level of antiviral protection, with six protecting against both viruses, one protecting against just FHV and two protecting against just DCV (Table S1). This protective phenotype is widespread across the Wolbachia phylogeny and is not restricted to particular clades (Figure 1A–C).
There is a strong genetic correlation in the level of protection against DCV and FHV
To examine the extent to which the effects of Wolbachia are specific to different viruses, we estimated the genetic correlation between protection to DCV and FHV (the proportion of the genetic variance shared by the two traits). The genetic correlation between protection to DCV and FHV was high (Model 1: rg = 0.81, 95% CI: 0.55,0.97; Figure 3A), indicating that most of the genetic variance in antiviral protection affects both DCV and FHV. There was no evidence of a genetic correlation between the survival of virus-infected and mock-infected flies (Model 1, DCV-Ringer: rg = 0.31, 95% CI = −0.37,0.87; Model 1, FHV-Ringer: rg = 0.61, 95% CI = −0.14,0.99).
Fig. 3. Correlation between protection, viral titers and Wolbachia density. 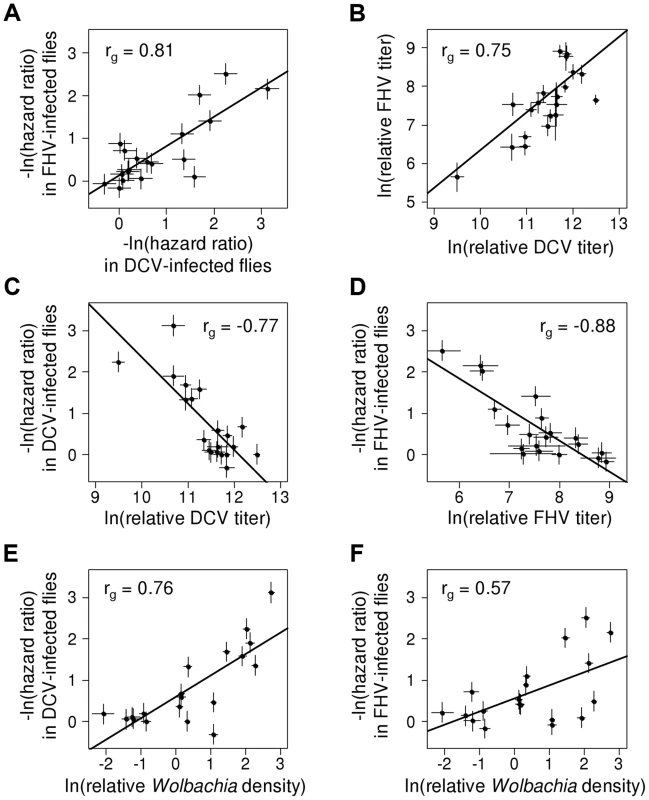
Dots indicate mean value of the traits for each Wolbachia strain. Error bars are standard errors. Solid lines show predicted values from linear regressions for illustrative purposes. rg is the genetic correlation between traits. (A) Correlation of survival between DCV- and FHV-infected flies (negative natural log of hazard ratios). (B) Correlation between DCV and FHV titers. (C–D) Correlation between viral titer and survival following (C) DCV infection or (D) FHV infection. Viral titers were estimated as viral RNA concentrations relative to the Drosophila gene EF1α100E. (E–F) Relationship between Wolbachia density and survival in (E) DCV- and (F) FHV-infected flies. Wolbachia density was estimated as the ratio between copy numbers ofthe Wolbachia gene atpD and the Drosophila gene Actin 5C. This genetic correlation between viruses could arise either because there is a causal relationship between DCV and FHV protection, or as a consequence of common ancestry (phylogenetic non-independence). We used a phylogenetic mixed model to partition the variance in the two traits into a component that can be explained by correlations across the Wolbachia phylogeny and a strain specific component that is independent of phylogeny. If there is a causal link between the traits, then the strength of their association will be the same for the phylogenetic and strain components. There was no significant difference in the strength of the genetic correlation for the phylogenetic and strain-specific components, consistent with a causal link between the traits. As we have limited power to separate these effects in a single model, we also fitted a model with just the phylogenetic effect. This model again produced a very similar correlation between viruses (rg = 0.95, 95% CI: 0.84,0.99), and the convergence of parameter estimates was improved.
Increased survival is genetically correlated to reduced viral titers
To investigate the effect of Wolbachia on viral titers, flies were stabbed with DCV or FHV, and relative viral RNA levels measured at two days post infection (dpi) by reverse transcription-quantitative PCR (RT-qPCR). For both DCV and FHV, viral titer was affected by the Wolbachia-infection status of flies (ANOVA on ln(viral titer); DCV: F19,165 = 23.4, P<10−16; FHV: F19,166 = 12.1, P<10−16; Figure 4A & 4B). Wolbachia strains tended to have similar effects on the two viruses, with a strong positive genetic correlation across the Wolbachia strains between titers of DCV and FHV (Model 2: rg = 0.75, 95% CI = 0.48,0.94; Figure 3B).
Fig. 4. Effect of Wolbachia strains on viral titers. 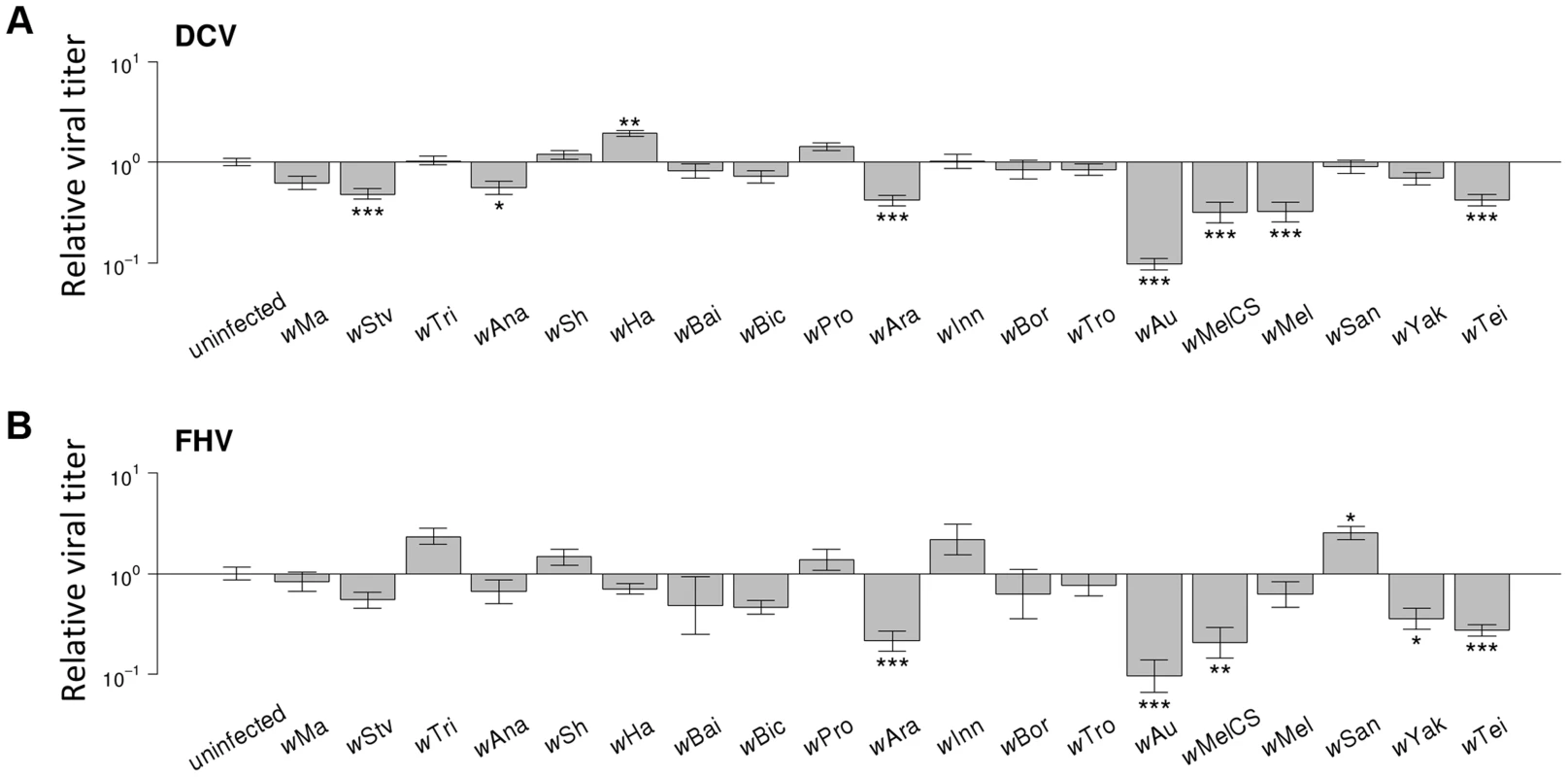
(A–B) Relative viral titer in (A) DCV- and (B) FHV-infected flies. Relative titers are normalised by the mean titer of Wolbachia-free controls (uninfected). Error bars are standard errors. Symbols above the bars give the significance relative to the Wolbachia-free controls based on a Dunnett's test (*: P<0.05; **: P<0.01; ***: P<0.001). Wolbachia could increase the survival of Drosophila after infection either by reducing viral titers (increasing resistance), or by allowing flies to better cope with infection damage (increasing tolerance). To test whether Wolbachia provides resistance to infection, we compared the survival of flies and viral titers across the 19 Wolbachia strains. Relative to the Wolbachia-free control, seven of the strains were individually associated with significantly reduced DCV titers, and five with reduced FHV titers (Figure 4A & 4B). Interestingly, for the strains wHa in DCV - and wSan in FHV-infected flies, there was a significant increase in viral RNA levels (Figure 4A & 4B) and this result was replicable by repeating the experiment on these lines (J. Martinez, personal observation). Overall, we found that the titer of both DCV and FHV was negatively genetically correlated to the survival of DCV and FHV infected flies respectively (Model 3, DCV: rg = −0.77, 95% CI = −0.95,−0.49; Model 4, FHV: rg = −0.88, 95% CI = −1.00,−0.64; Figure 3C & 3D). Therefore, resistance could be the primary explanation for Wolbachia-induced protection.
To understand how Wolbachia affects the dynamics of infection, we followed DCV and FHV titers for five days in Wolbachia-free flies and flies infected with the protective strain wAu that conferred strong protection against both viruses, the non-protective strain wSh and wAna, which protected against DCV but not FHV. In all treatments, including in the presence of the strongly protective strain wAu, DCV and FHV were able to replicate within the flies (Figure S2). For example, at 2 dpi, DCV titres had increased ∼18.000-fold in wAu-infected flies compared to ∼290.000-fold in Wolbachia-free flies. At 2 dpi, the timepoint chosen in the previous experiment, viruses were still in their growth phase, with a plateau of DCV titres being reached at around 3 dpi (Figure S2). Resistance conferred by wAu seemed to occur earlier against FHV (1 dpi at start growth phase) than against DCV (2 dpi at end growth phase) and viral titers were reduced from those points on, including at the end of the growth phase.
Wolbachia density is positively genetically correlated to antiviral protection
There was significant variation in the relative density of the different Wolbachia strains (ANOVA on ln(relative density): F18,150 = 115.6, P<10−16; Figure 1D) in 3–6 day old virus-free flies (the same age as the flies that were infected to estimate survival and viral titer). The highest density strain, wMelCS, had 114 fold higher density than the lowest density strain, wBor.
Flies infected with high density Wolbachia strains tended to live longer after viral infection (Figure 3E & F) and had lower viral titers. This is reflected in strong genetic correlations between bacterial density and survival after both DCV and FHV infection (Model 1; DCV: rg = 0.76, 95% CI: 0.49,0.93; FHV: rg = 0.57, 95% CI: 0.20,0.86). This appears to be a specific effect of Wolbachia on survival after viral infection as there is no support for a correlation between Wolbachia density and the survival of the mock-infected flies in this assay (Model 1; Ringer: rg = 0.18, 95% CI: −0.41,0.77). Similar to the survival analysis, bacterial density is negatively genetically correlated with DCV titers (Model 2: rg = −0.53, 95% CI: −0.82,−0.13). However, there is no support for a correlation between FHV titer and Wolbachia density (Model 2: rg = −0.29, 95% CI: −0.65,0.18).
To examine whether there is likely to be a causal relationship between Wolbachia density and survival after viral infection, we partitioned the variance in these traits into components that are dependent and independent of the Wolbachia phylogeny. The regression coefficient of survival against bacterial density was not significantly different for the phylogenetic and strain components for the two viruses. We are therefore unable to find evidence to suggest that this correlation is an artefact of phylogenetic relatedness, although we would caution that this analysis has very limited statistical power (non-significant phylogenetic component). Given this limited power to partition the variance across the two components, we also fitted a model with phylogenetic component only, and again the correlations between protection and Wolbachia density were similar to the model without the phylogeny (DCV: rg = 0.71, 95% CI: 0.32,0.92; FHV: rg = 0.63, 95% CI: 0.24,0.90).
Antiviral protection is not correlated with immune or methyltransferase gene expression
We finally investigated if antiviral protection could be explained by an effect of Wolbachia on host gene expression. We first tested the hypothesis that Wolbachia prime the immune system of flies, by measuring the expression of Drosomycin and Diptericin as reporters of Toll and IMD pathway activation respectively. We did not detect a significant genetic correlation between expression of either immune genes and DCV protection (Model 5; Drosomycin: rg = 0.26, 95% CI: −0.37,0.83; Diptericin: rg = −0.05, 95% CI: −0.79,0.66; Figure S3A & S3C) or FHV protection (Model 6; Drosomycin: rg = 0.25, 95% CI: −0.29,0.72; Diptericin: rg = 0.28, 95% CI: −0.37,0.95; Figure S3B & S3D). Finally, the expression of a putative candidate for protection, the methyltransferase gene Dnmt2, was not significantly affected by the Wolbachia-infection status (ANOVA on ln(expression level); DCV: F16,122 = 1.56, 0.09; FHV: F16,122 = 0.85, P = 0.63; Figure S3E & S3F) and did not show any correlation with level of protection in DCV-infected (Model 5; Dnmt2: rg = 0.12, 95% CI: −0.59,0.90) or FHV-infected flies (Model 6; Dnmt2: rg = −0.35, 95% CI: −0.98,0.49).
Discussion
Protective symbionts can be an important component of an organism's defenses against infection, in some cases even being the primary mode of defense [67]. Despite their importance being increasingly recognized, studying these symbionts remains challenging. Many cannot survive outside of host cells, be genetically manipulated or cloned. Our approach to circumvent these problems has been to assemble a large panel of different Wolbachia strains in a common host genetic background. This allows us to detect genetic correlations between traits, and infer whether these traits are causally linked.
Our results suggest that symbionts may play a role in protecting a substantial proportion of insect species against viral infection. Wolbachia is probably the most widespread symbiont in arthropods [68] and its wide distribution is partly attributable to its ability to manipulate host reproduction as well as its tendency to be horizontally-acquired between different host species over evolutionary time scales [2]. Recently, it has been shown to confer protection against natural enemies, in particular against RNA viruses [25], [26], [29]. By assessing the level of protection among several Wolbachia strains, we showed that, far from being an exception, Wolbachia-mediated protection is a common phenomenon, which could potentially have contributed to its evolutionary success.
Among the tested strains, about half were able to confer some level of protection in D. simulans. Assuming that Wolbachia is found in 40% of arthropod species [18], our results suggest that 20% of arthropods may benefit from such a protection. This extrapolation relies on strains retaining their ability to protect their original host, but the host species could also influence the expression of the protective phenotype. It was previously shown that protective strains native to D. melanogaster also protect mosquito hosts after artificial transfer. In contrast, the strain wInn, which did not confer protection in this study, was previously found to protect against FHV in its original host D. innubila [28]. Host genotype effects on the Wolbachia density have previously been found [69], [70]. Given the correlation between protection and density, the expression of protection is likely to be under the control of both the Wolbachia strain and the host genotype.
Wolbachia has previously been shown to protect insects against a remarkably taxonomically diverse array of RNA viruses [25], [26], [29], [31], [40]; and this could either reflect a broad-spectrum antiviral mechanism or Wolbachia may have independently evolved different ways of targeting different viruses. Previously it has been observed that strains that protect strongly against one virus tend to protect against other viruses, suggesting the former explanation is true [27], [44]–[45]. We found that an estimated 81% (rg = 0.81) of the genetic variation among strains in DCV and FHV protection is common to the two viruses. Furthermore, this pattern appears to be independent of the bacterial phylogeny, indicating that the same genes underlie the level of protection to the two viruses tested. This supports the hypothesis that Wolbachia has a single broad-spectrum mechanism of antiviral protection.
The increased survival of Wolbachia-infected flies after viral infection could result from the symbiont increasing either resistance or tolerance to infection [71], [72]. Resistance occurs where increases in survival are caused by reductions in viral titers, while tolerance describes the situation where hosts are better able to survive a given viral load [71]. Both of these effects have been ascribed to the antiviral properties of Wolbachia in the past [26], [27], [29], [31], [40], [44], [45]. Our analysis allows us to test the effect of resistance by estimating the proportion of the variation in survival that can be explained by differences in viral titer. The genetic correlation between titer and survival was very high for both viruses, so in this instance it seems likely that the between-strain variation in survival is mainly due to resistance to virus infection. Our data cannot exclude a role for tolerance, as Wolbachia may be altering the amount of harm that a given viral titre causes. However, were this to be the case, then it is likely to be a common underlying link, such as both traits relying on Wolbachia density or the same mechanism.
In some studies it was shown that Wolbachia infection can lead to higher viral titers or virus-induced mortality [73], [74]. Interestingly, in our experiment two Wolbachia strains were associated with an increase in viral titer, although not with increased mortality. This is a tantalizing result, which would suggest care should be taken when introducing Wolbachia into disease vector populations. However, we would caution that this result needs to be investigated in more detail – we measured many traits across many strains, so rare outliers could be an artefact of confounding factors like remaining differences in the genetic background of the strains.
The density of Wolbachia plays a key role in determining the level of antiviral protection it provides to its host. This has been previously demonstrated experimentally by manipulating Wolbachia density using antibiotics, and by comparisons of high and low density strains [27], [44], [45], [46]. Our results strengthen this conclusion, as we show that the relationship of density and survival is strong and highly significant across a large panel of strains. Furthermore, this association does not appear to be a consequence of phylogenetic relatedness, suggesting that higher Wolbachia density is causing higher levels of resistance to viruses.
Do any factors other than Wolbachia density cause between-strain variation in the level of resistance to viruses? Our analysis provides only weak support for other factors being important, as while the genetic correlation (the proportion of genetic variance shared) between Wolbachia density and survival ranges from 0.57–0.76, the upper confidence intervals for all estimates are greater than 0.86. Therefore, while our data suggests other factors are important, the evidence is not strong. Any of these Wolbachia strains may have the intrinsic ability to provide resistance to viruses – they simply need to be present at a sufficiently high density. If true, it is tempting to speculate what this might imply about the underlying mechanism of resistance. It seems more compatible with a mechanism whereby the presence of Wolbachia per se makes cells or the host less hospitable to viruses, such as through competition for resources [75] or remodeling of the cellular environment. In contrast, if Wolbachia was expressing specific antiviral factors, then these might be easily gained or lost through evolution, breaking the genetic correlation of resistance and Wolbachia density. It is likely that various mechanisms can lead to variation in bacterial density and thus affect within-host density. For example, it was found recently that a ∼21 kb region encoding eight genes is amplified three to seven times in different wMelPop isolates relative to wMelCS [45], [76], and is associated with much higher density and stronger protection against viruses [45]. However, copy number variation of this region does not explain differences in density or protection between wMelCS and wMel-like strains [45]. It is therefore tempting to speculate that genomic differences between Wolbachia strains that confer differential protection to viruses will only reveal different ways of varying the bacterial density rather than the actual antiviral mechanisms.
One way of rendering the host less hospitable for viruses is through the regulation of host genes. It was argued in the past that Wolbachia infection may lead to the activation of immune pathways that in turn could limit the multiplication of other parasites. Previous studies in mosquitoes showed that even if Wolbachia can prime the host immune system and increase antiviral resistance, such an effect is absent in D. melanogaster [44], [50], [51], [55], and flies deficient in both the Toll and IMD pathways still display Wolbachia-mediated resistance [54]. In agreement with previous studies, our results support the conclusion that Toll and IMD pathways are not required for antiviral protection since both Drosomycin and Diptericin expression level (reporters of Toll and IMD pathways respectively) were uncorrelated to the survival of virus-infected flies. However, other immune pathways and restriction factors could still be involved.
In the mosquito Ae. aegypti, the methyltransferase AaDnmt2, whose homolog in Drosophila methylates transfer RNAs and other nucleic acids, has been proposed as a potential candidate to explain the antiviral effect of Wolbachia [57]. Wolbachia was shown to decrease the expression of AaDnmt2 through the induction of the expression of aae-miR-2940 microRNA. Conversely, the overexpression of AaDnmt2 led to a decrease in Wolbachia density and an increase in the titer of dengue virus. However, it was recently shown that the Drosophila homolog Dnmt2 has an antiviral effect against DCV and Nora virus, the opposite to the pattern seen in mosquito cells infected with dengue virus [59]. We found that Wolbachia has no consistent effect on Dnmt2 expression in D. simulans, and variation in Dnmt2 expression does not explain any of the variation in survival after infection. This suggests changes to Dnmt2 expression are not a general explanation of the antiviral effects of Wolbachia. It is possible that a different mechanism of resistance applies to mosquitoes and dengue virus. However, we would argue that the critical experiment to reach this conclusion would be to show that the antiviral effects of Wolbachia on dengue virus require AaDnmt2 or aae-miR-2940, and this experiment has yet to be performed.
Together with previous studies, our results show that antiviral protection is very common among Wolbachia strains. As such, it has to be taken into account if we are to draw a complete picture of Wolbachia ecology and evolution. For example, protection may favor the rapid sweeps of Wolbachia observed in natural populations [77], [78] and explain why strains such as wMel and wAu, that induce weak or no CI, can be maintained in natural populations [79], [80]. Owing to the high incidence of Wolbachia and the broad spectrum of viruses affected by the protection, it is likely that Wolbachia-mediated protection has substantially contributed to the evolution of arthropods. By protecting against infection, symbiont-based immunity may in turn influence the evolution of the host immune system. Although the mechanisms remain to be elucidated, protection is tightly linked to the bacterial density. Therefore, variation in the selective pressure exerted by viruses could partly explain why Wolbachia strains vary so much in density, and why some are found in somatic tissues whereas other are restricted to the germ cells [81]. From an applied perspective, our study extends the panel of Wolbachia strains that could be introduced into mosquito populations to limit the spread of arboviruses. However, for successful introduction, the choice of a strain should not only be based on the level of protection but also consider costs on host fitness and strength of CI that will affect the invasive potential of Wolbachia.
Materials and Methods
Wolbachia strains, Drosophila lines and fly rearing
The origin of the 19 Wolbachia strains used in this study and their original host line are listed in Error! Reference source not found. To control for host genetic effects, all Wolbachia strains were transferred into the D. simulans STCP genetic background. This line was previously obtained through six generations of brother-sister crossing [23], [82], [83]. Eight of the strains were transferred into the STCP background in previous studies (Table 1; [23], [82], [83]). Of these, the three strains naturally-infecting D. simulans were generated by six generations of backcrossing Wolbachia-infected females to STCP males, and the remaining five were transferred by microinjection (Table 1). We microinjected eleven more strains into the STCP line (Table 1). Microinjections were performed as previously described using a microcapillary needle to transfer cytoplasm of infected embryos into uninfected STCP embryos [82]. All microinjected lines were maintained in the lab for at least 10 generations before the beginning of the experiments.
Two generations before the beginning of the experiments, the Wolbachia infection status of the STCP lines was checked by PCR using the diagnostic primers wsp81F and wsp691R [84], and the PCR products were sequenced. For strains microinjected in this study, vertical transmission was also assessed with PCR by testing 48 offspring per strain originating from Wolbachia-infected mothers (data not shown). Three fly stocks, transinfected with the strains wBai, wBic and wBor, showed imperfect vertical transmission (54%, 91% and 62% respectively). For those three strains, the presence of Wolbachia was checked by PCR one generation before each experiment and only offspring from infected mothers were used in the experiments. Additionally, in qPCR assays (see below), flies of those three strains that were used in the experiments were first isolated individually, their Wolbachia infection status was confirmed and only Wolbachia-infected individuals were kept and pooled in groups of 4–6 flies.
For all the experiments, flies were maintained on a cornmeal diet (agar: 1%, dextrose: 8.75%, maize: 8.75%, yeast: 2%, nipagin: 3%) at a constant temperature of 25°C with a 12-hour light/dark cycle at 70% relative humidity.
Inference of the Wolbachia phylogeny
The phylogeny of the 19 Wolbachia strains was inferred from the partial sequences of the eight genes 16S rRNA, aspC, atpD, ftsZ, sucB, groEL, coxA and fbpA previously used in Multilocus Sequence Typing studies [66], [85]. The gene sequences were either obtained from GenBank, or were sequenced using the protocol described in Paraskevopoulos et al. (2006). Accession numbers and the origins of the sequences are described in Table S2. Each gene was individually aligned using Mauve v2.3.1 [86] and the phylogeny was inferred using ClonalFrame v1.2 to take into account recombination between strains [65]. To check for convergence, 9 independent runs were done with 100,000 MCMC iterations after 100,000 burn-in iterations with parameter recording every 100 iterations. For the first 8 runs, a uniformly chosen coalescent tree was used as the initial tree, and for the 9th run, a UPGMA tree was used. The UPGMA starting tree was compared to the eight other trees and showed a good convergence with seven of them based on the tree comparison tool implemented in ClonalFrame v1.2 [65]. Parameter estimates for the UPGMA starting tree also showed a good convergence based on the Gelman and Rubin test [87] in ClonalFrame v1.2. A consensus tree with branch support values was built from the posterior sample of the UPGMA starting tree at 50% majority-rule in MEGA 5.2 [88] and visualized in R using the ape package [89]. The consensus tree was visually compared with a tree inferred from the concatenated sequence of the eight genes using PhyML v3.1 with 500 bootstrap replicates [90] to assess the effect of recombination on the phylogenetic signal. All clades inferred from ClonalFrame were retrieved in the maximum likelihood tree. Therefore, the ClonalFrame tree with the UPGMA starting tree was used in further analyses.
Viral isolates
Viruses were produced and titrated as in [26], with minor changes. DCV was produced and titrated in Schneider's Line 2 cells (SL-2), while FHV was titrated in Schneider Drosophila line 2 cells (DL2). For each infection assay, one viral aliquot was defrosted on the day of infection and diluted in Ringer's solution [91] to reach a viral concentration of 5×108 TCID50/mL for DCV and 3.38×108 TCID50/mL for FHV.
Survival assay
For each fly line, 3–6 day-old female flies were collected. After being anaesthetized with CO2, flies were either infected with DCV, FHV, or mock-infected with Ringer's solution [91]. The inoculum was administered by stabbing flies into the left pleural suture on the thorax with a 0.15 mm diameter anodized steel needle (Austerlitz Insect Pins) bent ∼0.25 mm from the end (∼half of the dorsal width of the thorax), dipped into viral or Ringer's solution as in [92]. Twenty stabbed flies were placed in a vial of fly cornmeal medium and dead flies were recorded every day for 25 days after infection. Flies were transferred into fresh vials of food every 3 days.
The survival assay was replicated on six consecutive days. On each day two vials of flies from each Wolbachia strain were assigned to two of the three treatments. The same was done for the Wolbachia-free flies, except that the number of replicates was doubled to increase statistical power. The stabbing order of the fly lines as well as the sequence of treatments were randomized each day. Mortality that occurred on the day following infection was attributed to stabbing injuries and was discarded from the analyses.
Quantitative PCR
The Wolbachia density, DCV and FHV titers as well as the expression of the three host genes Drosomycin, Diptericin and Dnmt2 were measured by qPCR on a BioRad iQ5 thermocycler using primers, probes and cycle conditions listed in Table S3. Wolbachia density was measured on pools of 10 virus-free 3–6 day-old female flies (n = 10 pools) from which DNA was extracted using the Gentra Puregene kit (Qiagen). For viral titers and host gene expression, 3–6 day-old flies were first infected with DCV or FHV, as described above, and, 2 days after infection, 10 flies were pooled (n = 10 pools per virus), homogenized in TRIzol Reagent (Ambion) and frozen at −80°C. Total RNA was extracted using the Direct-zol-96 RNA kit (Zymo Research) by following the manufacturer's instructions, including a 15 min DNase I digestion step.
For host gene expression, total RNA was reverse-transcribed using the GoScript Reverse Transcription System (Promega) with random primers. Host gene expression and the Wolbachia density were measured relative to the endogenous control gene actin 5C (Table S3) using the SensiFAST SYBR & Fluorescein kit (Bioline).
The copy-number of viral genomic RNA was measured relative to the control gene EF1α100E (Table S3) in a one-step RT-qPCR reaction using the QuantiTect Virus kit (Qiagen). For each virus, both viral and fly cDNAs were amplified in a duplex reaction using virus and fly primers in association with dual-labeled (hydrolysis) fluorescent probes (Sigma) (Table S3). For each sample, two RT-qPCR reactions were carried out and the mean of these two technical replicates was used as the relative viral titer in the statistical analysis.
The efficiency of the PCR amplication was checked using a dilution series for each set of primers. The relative Wolbachia density and viral titers were calculated as follows: , where Ct is the cycle threshold and .
Because qPCR efficiencies tended to be different between the control gene actin 5C and both the immune and the methyltransferase genes, we use the Pfaffl method to take into account those differences [93]. As dilution series analysis shows the qPCR efficiency for actin 5C to be 100%, the relative efficiency E for the gene of interest can be estimated from the experimental data as . Following [93], Ct values for the gene of interest were corrected for differences in qPCR efficiency as . Levels of gene expression were then estimated as follow: , where . We also normalized the results across 96 well plates (sets of samples were kept in plates for both RNA extraction and qPCR). Thus, expression level for a given sample j was normalized by the mean, , and standard deviation of the corresponding plate i for each gene of interest as follow: . The strains wBai, wBic and wBor were not included in the analysis of host gene expression.
Time-course analysis of viral infection
In addition to the single timepoint analysis of viral titers, variation of titers was measured in another infection experiment for Wolbachia-free flies and for the Wolbachia strains wAu, wSh and wAna over a 5 day period. Flies were infected with DCV or FHV and maintained in the same conditions as for the other infection experiments. Live flies were frozen everyday from the day of infection until 5 dpi. For each day and each strain, the RNA was extracted from two pools of ten flies and viral titers measured as explained above except that the RT-qPCR was run on a StepOnePlus thermocycler (Applied Biosystems).
Statistical analysis
Survival data were analyzed with a Cox's proportional hazards mixed-effect model using the coxme package in R [89]. The Cox's model estimates hazard ratios, which is the probability of a Wolbachia-infected fly dying at a given time-point divided by the probability of a Wolbachia-free fly dying. The infection treatment (DCV, FHV or mock-infected), the Wolbachia infection status (the 19 strains and no Wolbachia) were treated as fixed effects, and the replicate vial as a random effect. The overall significance of multilevel factors or their interactions was tested using likelihood ratio tests to compare models with or without these terms. Flies that were alive at the end of the experiment were treated as censored data. Variation in Wolbachia density and viral titers was analysed using linear models on ln-transformed data to reach the assumptions of normality and homoscedasticity. Differences in viral titer with the Wolbachia-free control were assessed using Dunnett's tests in order to correct for multiple comparisons.
Genetic correlations between traits were estimated by fitting a series of multi-response mixed models using a Bayesian approach in the R package MCMCglmm [94] as follows:
where ytwi is the response of the ith biological replicate for Wolbachia strain w, for which we have measured trait t. βt is the intercept term for trait t with a level for each trait, and it can be interpreted as the mean trait value across the Wolbachia strains. The Wolbachia strain random effect, us:tw, is the deviation from the expected value for trait t in strain w. Random effects were assumed to be from multivariate normal distributions with zero mean vectors (illustrated for a model with three traits): where σ2s:t1 is the genetic variance for trait t1, and σs:t1,t2 is the genetic covariance between trait t1 and t2. etwi is a residual capturing the between-vial variation for each trait (within-strain effects, environmental effects and experimental error). Residuals were assumed to be normally distributed and a separate variance was estimated for each trait with the following variance-covariance structure (illustrated for a model with three traits): where I is an identity matrix indicating that strain effects within traits are independent of each other since traits were measured on different biological replicates.The traits included in these models included the survival of DCV-, FHV - or mock-infected flies (estimated as a negative ln hazard ratio for each vial of flies), the Wolbachia density, viral titer and gene expression (all estimated as ln ). We fitted six different models with different trait combinations. Model 1 included four traits: survival after DCV-, FHV - and mock-infection, and Wolbachia density. Model 2 included three traits: DCV titer, FHV titer and Wolbachia density. Models 3 and 4 included two traits for each virus respectively: survival after viral infection and viral titer. Models 5 and 6 included four traits for each virus respectively: survival as well as Drosomycin, Diptericin and Dnmt2 expression levels after viral infection.
Genetic correlations between two traits can arise either because the traits are causally related or because of phylogenetic non-independence. To explore these explanations we also fitted a phylogenetic mixed model, which included an additional random effect, up:tw, which is the deviation from the expected value for trait t in strain w due to the phylogeny, i.e. the component of the between-strain variation that is explained by the phylogeny [95], [96]:
In this model the strain random effect us:tw is the variation that is not accounted for by the phylogeny under a Brownian model of evolution [95]. The intercept βt can be interpreted as the trait value in the Wolbachia strain at the root of the phylogeny. For the phylogenetic effect, up:tw, the following variance-covariance structure was assumed (illustrated for a model with three traits): where A is a matrix with elements ajk standing for the proportion of time that strain j and k have had shared ancestry since the root of the phylogeny. σ2p:t1 is the variance of the phylogenetic effect for trait t1, and σp:t1,t2 is the covariance of phylogenetic effects between trait t1 and t2. Under a Brownian model of evolution, the phylogenetic covariance between two Wolbachia strains is inversely proportional to the time since they diverged from their common ancestor. The phylogenetic effects themselves were poorly estimated and are therefore not reported. We also fitted these phylogenetic models without the strain effect as this improved model convergence and statistical power to test for genetic correlations.Independent normal priors with zero mean and large variance (1010) were used for the fixed effects and are virtually non-informative in this context. We used several prior probability distributions for the Vp and Vs covariance matrices to ensure our results were robust to the prior selected. Results presented were obtained using parameter expanded priors, but we also fitted models with inverse-Wishart and flat priors that gave equivalent results. We also repeated the analyses after removing outliers so that the distribution of the residuals was normal. In all cases our conclusions were unaffected by these changes. Finally, models were run after removing strains wBai, wBic and wBor, since those strains showed unstable Wolbachia infection status. Estimates of genetic correlations as well as their statistical significance were very similar to models that include these 3 strains, and are therefore not reported. The models were run for 13,000,000 iterations with a burn-in of 3,000,000. We checked for convergence by visually examining the trace of the posterior sample and ensuring the autocorrelation between successive samples in the MCMC chain was <0.1. Credible intervals (CI) were estimated from the posterior distribution of parameter estimates as the 95% highest posterior density intervals.
Supporting Information
Zdroje
1. MoranNA (2007) Symbiosis as an adaptive process and source of phenotypic complexity. Proc Natl Acad Sci U S A 15 : 8627–8633.
2. WerrenJH, BaldoL, ClarkME (2008) Wolbachia: master manipulators of invertebrate biology. Nature 6 : 741–751.
3. EngelstädterJ, HurstGDD (2009) The Ecology and Evolution of Microbes that Manipulate Host Reproduction. Annu Rev Ecol Evol Syst 40 : 127–149.
4. BaumannP (2005) Biology bacteriocyte-associated endosymbionts of plant sap-sucking insects. Annu Rev Microbiol 59 : 155–189.
5. MoranN, TranP, GerardoN (2005) Symbiosis and insect diversification: an ancient symbiont of sap-feeding insects from the bacterial phylum Bacteroidetes. Appl Environ Microbiol 71 : 8802–8810.
6. DouglasAE (2009) The microbial dimension in insect nutritional ecology. Funct Ecol 23 : 38–47.
7. OliverKM, DegnanPH, BurkeGR, MoranNA (2010) Facultative Symbionts in Aphids and the Horizontal Transfer of Ecologically Important Traits. Annu Rev Entomol 55 : 247–266.
8. RussellJ, MoranN (2006) Costs and benefits of symbiont infection in aphids: variation among symbionts and across temperatures. Proc R Soc B Biol Sci 273 : 603–610.
9. OliverKM, RussellJA, MoranNA, HunterMS (2003) Facultative bacterial symbionts in aphids confer resistance to parasitic wasps. Proc Natl Acad Sci U S A 100 : 1803–1807.
10. JaenikeJ, UncklessR, CockburnSN, BoelioLM, PerlmanSJ (2010) Adaptation via symbiosis: recent spread of a Drosophila defensive symbiont. Science 329 : 212–215.
11. XieJL, VilchezI, MateosM (2010) Spiroplasma Bacteria Enhance Survival of Drosophila hydei Attacked by the Parasitic Wasp Leptopilina heterotoma. PLoS One 5: e12149.
12. JigginsFM, HurstGDD (2011) Rapid insect evolution by symbiont transfer. Science 332 : 185–186.
13. BaldoL, AyoubNA, HayashiCY, RussellJA, StahlhutJK, et al. (2008) Insight into the routes of Wolbachia invasion: high levels of horizontal transfer in the spider genus Agelenopsis revealed by Wolbachia strain and mitochondrial DNA diversity. Mol Ecol 17 : 557–569.
14. SchulerH, BertheauC, EganSP, FederJL, RieglerM, et al. (2013) Evidence for a recent horizontal transmission and spatial spread of Wolbachia from endemic Rhagoletis cerasi (Diptera: Tephritidae) to invasive Rhagoletis cingulata in Europe. Mol Ecol 22 : 4101–4111.
15. StahlhutJK, DesjardinsCA, ClarkME, BaldoL, RussellJA, et al. (2010) The mushroom habitat as an ecological arena for global exchange of Wolbachia. Mol Ecol 19 : 1940–1952.
16. HenryLM, PeccoudJ, SimonJ-C, HadfieldJD, MaidenMJC, et al. (2013) Horizontally Transmitted Symbionts and Host Colonization of Ecological Niches. Curr Biol 23 : 1713–1717.
17. HimlerAG, Adachi-HagimoriT, BergenJE, KozuchA, KellySE, et al. (2011) Rapid spread of a bacterial symbiont in an invasive whitefly is driven by fitness benefits and female bias. Science 332 : 254–256.
18. ZugR, HammersteinP (2012) Still a host of hosts for Wolbachia: analysis of recent data suggests that 40% of terrestrial arthropod species are infected. PLoS One 7: e38544.
19. CharlatS, NirgianakiA, BourtzisK, MerçotH (2002) Evolution of Wolbachia-induced cytoplasmic incompatibility in Drosophila simulans and D. sechellia. Evolution (N Y) 56 : 1735–1742.
20. BourtzisK, NirgianakiA, MarkakisG, SavakisC (1996) Wolbachia Infection and Cytoplasmic Incompatibility in Drosophila Species. Genetics 144 : 1063–1073.
21. DyerKa, JaenikeJ (2004) Evolutionarily stable infection by a male-killing endosymbiont in Drosophila innubila: molecular evidence from the host and parasite genomes. Genetics 168 : 1443–1455.
22. CharlatS, HurstGDD, MerçotH (2003) Evolutionary consequences of Wolbachia infections. 19 : 217–223.
23. ZabalouS, ApostolakiA, PattasS, VenetiZ, ParaskevopoulosC, et al. (2008) Multiple rescue factors within a Wolbachia strain. Genetics 178 : 2145–2160.
24. StouthamerR, BreeuwerJAJ, HurstGDD (1999) Wolbachia Pipientis: Microbial Manipulator of Arthropod Reproduction. Annu Rev Microbiol 53 : 71–102.
25. HedgesL, BrownlieJ, O'NeillS, JohnsonK (2008) Wolbachia and virus protection in insects. Science 322 : 702.
26. TeixeiraL, FerreiraA, AshburnerM (2008) The Bacterial Symbiont Wolbachia Induces Resistance to RNA Viral Infections in Drosophila melanogaster. PLoS Biol 6 : 2753–2763.
27. OsborneSE, LeongYS, O'NeillSL, JohnsonKN (2009) Variation in Antiviral Protection Mediated by Different Wolbachia Strains in Drosophila simulans. PLoS Pathog 5 : 9.
28. UncklessRL, JaenikeJ (2011) Maintenance of a male-killing Wolbachia in Drosophila innubila by male-killing dependent and male-killing independent mechanims. Evolution 66 : 678–689.
29. MoreiraLA, Iturbe-OrmaetxeI, JefferyJA, LuG, PykeAT, et al. (2009) A Wolbachia symbiont in Aedes aegypti limits infection with dengue, Chikungunya, and Plasmodium. Cell 139 : 1268–1278.
30. FrentiuFD, RobinsonJ, YoungPR, McGrawEa, O'NeillSL (2010) Wolbachia-mediated resistance to dengue virus infection and death at the cellular level. PLoS One 5: e13398.
31. GlaserRL, MeolaMa (2010) The native Wolbachia endosymbionts of Drosophila melanogaster and Culex quinquefasciatus increase host resistance to West Nile virus infection. PLoS One 5: e11977.
32. BlagroveMSC, Arias-GoetaC, FaillouxA-B, SinkinsSP (2012) Wolbachia strain wMel induces cytoplasmic incompatibility and blocks dengue transmission in Aedes albopictus. Proc Natl Acad Sci U S A 109 : 255–260.
33. KambrisZ, CookP, PhucH, SinkinsS (2009) Immune activation by life-shortening Wolbachia and reduced filarial competence in mosquitoes. Science 326 : 134–136.
34. ZéléF, Nicota, DuronO, Riveroa (2012) Infection with Wolbachia protects mosquitoes against Plasmodium-induced mortality in a natural system. J Evol Biol 25 : 1243–1252.
35. YeYH, WoolfitM, RancèsE, O'NeillSL, McGrawEa (2013) Wolbachia-Associated Bacterial Protection in the Mosquito Aedes aegypti. PLoS Negl Trop Dis 7: e2362.
36. HughesGL, KogaR, XueP, FukatsuT, RasgonJL (2011) Wolbachia infections are virulent and inhibit the human malaria parasite Plasmodium falciparum in Anopheles gambiae. PLoS Pathog 7: e1002043.
37. CookPE, McGrawEa (2010) Wolbachia pipientis: an expanding bag of tricks to explore for disease control. Trends Parasitol 26 : 373–375.
38. VavreF, CharlatS (2012) Making (good) use of Wolbachia: what the models say. Curr Opin Microbiol 15 : 263–268.
39. HussainM, LuG, TorresS, EdmondsJH, KayBH, et al. (2013) Effect of Wolbachia on replication of West Nile virus in a mosquito cell line and adult mosquitoes. J Virol 87 : 851–858.
40. Van den HurkAF, Hall-MendelinS, PykeAT, FrentiuFD, McElroyK, et al. (2012) Impact of Wolbachia on infection with chikungunya and yellow fever viruses in the mosquito vector Aedes aegypti. PLoS Negl Trop Dis 6: e1892.
41. WalkerT, JohnsonPH, MoreiraLa, Iturbe-OrmaetxeI, FrentiuFD, et al. (2011) The wMel Wolbachia strain blocks dengue and invades caged Aedes aegypti populations. Nature 476 : 450–453.
42. HoffmannAA, MontgomeryBL, PopoviciJ, Iturbe-OrmaetxeI, JohnsonPH, et al. (2011) Successful establishment of Wolbachia in Aedes populations to suppress dengue transmission. Nature 476 : 454–457.
43. LongdonB, FabianDK, HurstGD, JigginsFM (2012) Male-killing Wolbachia do not protect Drosophila bifasciata against viral infection. BMC Microbiol 12 Suppl 1: S8.
44. ChrostekE, MarialvaMSP, YamadaR, O'NeillS, et al. (2014) High antiviral protection without immune upregulation after interspecies Wolbachia transfer. PLoS one 9: e99025.
45. ChrostekE, MarialvaMSP, EstevesSS, WeinertLa, MartinezJ, et al. (2013) Wolbachia Variants Induce Differential Protection to Viruses in Drosophila melanogaster: A Phenotypic and Phylogenomic Analysis. PLoS Genet 9: e1003896.
46. OsborneSE, Iturbe-OrmaetxeI, BrownlieJC, O'NeillSL, JohnsonKN (2012) Antiviral protection and the importance of Wolbachia density and tissue tropism in Drosophila simulans. Appl Environ Microbiol 78 : 6922–6929.
47. LuP, BianG, PanX, XiZ (2012) Wolbachia induces density-dependent inhibition to dengue virus in mosquito cells. PLoS Negl Trop Dis 6: e1754.
48. KambrisZ, BlagboroughAM, PintoSB, BlagroveMSC, GodfrayHCJ, et al. (2010) Wolbachia stimulates immune gene expression and inhibits plasmodium development in Anopheles gambiae. PLoS Pathog 6: e1001143.
49. PanX, ZhouG, WuJ, BianG, LuP, et al. (2012) Wolbachia induces reactive oxygen species (ROS)-dependent activation of the Toll pathway to control dengue virus in the mosquito Aedes aegypti. Proc Natl Acad Sci U S A 109: E23–31.
50. RancèsE, YeYH, WoolfitM, McGrawEa, O'NeillSL (2012) The relative importance of innate immune priming in Wolbachia-mediated dengue interference. PLoS Pathog 8: e1002548.
51. BourtzisK, PettigrewMM, O'NeillSL (2000) Wolbachia neither induces nor suppresses transcripts encoding antimicrobial peptides. Insect Mol Biol 9 : 635–639.
52. RottschaeferSM, LazzaroBP (2012) No effect of Wolbachia on resistance to intracellular infection by pathogenic bacteria in Drosophila melanogaster. PLoS One 7: e40500.
53. WongZS, HedgesLM, BrownlieJC, JohnsonKN (2011) Wolbachia-mediated antibacterial protection and immune gene regulation in Drosophila. PLoS One 6: e25430.
54. RancèsE, JohnsonTK, PopoviciJ, Iturbe-OrmaetxeI, ZakirT, et al. (2013) The toll and imd pathways are not required for Wolbachia-mediated dengue virus interference. J Virol 87 : 11945–11949.
55. TeixeiraL (2012) Whole-genome expression profile analysis of Drosophila melanogaster immune responses. Brief Funct Genomics 11 : 375–386.
56. HedgesLM, YamadaR, O'NeillSL, JohnsonKN (2012) The small interfering RNA pathway is not essential for Wolbachia-mediated antiviral protection in Drosophila melanogaster. Appl Environ Microbiol 78 : 6773–6776.
57. ZhangG, HussainM, O'NeillSL, AsgariS (2013) Wolbachia uses a host microRNA to regulate transcripts of a methyltransferase, contributing to dengue virus inhibition in Aedes aegypti. Proc Natl Acad Sci U S A 110 : 10276–10281.
58. HussainM, FrentiuFD, MoreiraLa, O'NeillSL, AsgariS (2011) Wolbachia uses host microRNAs to manipulate host gene expression and facilitate colonization of the dengue vector Aedes aegypti. Proc Natl Acad Sci U S A 108 : 9250–9255.
59. DurdevicZ, HannaK, GoldB, PollexT, CherryS, et al. (2013) Efficient RNA virus control in Drosophila requires the RNA methyltransferase Dnmt2. EMBO Rep 14 : 269–275 Available: http://www.ncbi.nlm.nih.gov/pubmed/23370384.
60. HuszarT, ImlerJ-L (2008) Drosophila viruses and the study of antiviral host-defense. Adv Virus Res 72 : 227–265.
61. PlusN, CroizierG, JoussetF, DavidJ (1975) Picornaviruses of laboratory and wild Drosophila melanogaster: geographical distribution and serotypic composition. Ann Microbiol (Paris) 126 : 107–117.
62. Christian P, Scotti P (1998) Picornalike Viruses of Insects. In: Miller L, Ball LA, editors. The Insect Viruses SE - 10. Springer US. pp. 301–336.
63. ComendadorM, PlusN, LouisC, Lopez-FerberM, KuhlA, et al. (1986) Endemic microorganisms of a Drosophila simulans strain and their relationships with the non-mendelian transmission of a character. Génétique, sélection, évolution 18 : 131–144.
64. ScottiP, DearingS, MossopD (1983) Flock house virus: A Nodavirus isolated from Costelytra zealandica (White)(Coleoptera: Scarabaeida). Arch Virol 75 : 181–189.
65. DidelotX, FalushD (2007) Inference of Bacterial Microevolution Using Multilocus Sequence Data. Genet 175 1251–1266.
66. BaldoL, Dunning HotoppJC, JolleyKa, BordensteinSR, BiberSa, et al. (2006) Multilocus sequence typing system for the endosymbiont Wolbachia pipientis. Appl Environ Microbiol 72 : 7098–7110.
67. OliverKM, MoranNA, HunterMS (2005) Variation in resistance to parasitism in aphids is due to symbionts not host genotype. 102 : 12795–12800.
68. DuronO, BouchonD, BoutinS, BellamyL, ZhouL, et al. (2008) The diversity of reproductive parasites among arthropods: Wolbachia do not walk alone. BMC Biol 6 : 1–12.
69. KondoN, ShimadaM, FukatsuT (2005) Infection density of Wolbachia endosymbiont affected by co-infection and host genotype. Biol Lett 1 : 488–491.
70. MoutonL, HenriH, CharifD, BoulétreauM, VavreF (2007) Interaction between host genotype and environmental conditions affects bacterial density in Wolbachia symbiosis. Biol Lett 3 : 210–213.
71. RåbergL, GrahamAL, ReadAF (2009) Decomposing health: tolerance and resistance to parasites in animals. Philos Trans R Soc Lond B Biol Sci 364 : 37–49.
72. MedzhitovR, SchneiderDS, SoaresMP (2012) Disease tolerance as a defense strategy. Science 335 : 936–941.
73. GrahamR, GrzywaczD, MushoboziWL, WilsonK (2012) Wolbachia in a major African crop pest increases susceptibility to viral disease rather than protects. PLoS Pathog 9 : 993–1000.
74. DodsonB, HughesG, PaulO, MatacchieroAC, KramerLD, RasgonJL (2014) Wolbachia enhances West Nile Virus (WNV) infection in the mosquito Culex tarsalis. PLoS Negl Trop Dis 7: e2965.
75. CaragataEP, RancèsE, HedgesLM, GoftonAW, JohnsonKN, et al. (2013) Dietary cholesterol modulates pathogen blocking by Wolbachia. PLoS Pathog 9: e1003459.
76. WoolfitM, Iturbe-OrmaetxeI, BrownlieJC, WalkerT, RieglerM, et al. (2013) Genomic evolution of the pathogenic Wolbachia strain, wMelPop. Genome Biol Evol 1–61.
77. TurelliM, HoffmannA (1991) Rapid spread of an inherited incompatibility factor in California Drosophila. Nature 353 : 440–442.
78. KriesnerP, HoffmannAA, LeeSF, TurelliM, WeeksAR (2013) Rapid Sequential Spread of Two Wolbachia Variants in Drosophila simulans. PLoS Pathog 9: e1003607.
79. HoffmannA, ClancyD, DuncanJ (1996) Naturally-occurring Wolbachia infection in Drosophila simulans that does not cause cytoplasmic incompatibility. Heredity 76 : 1–8.
80. Hoffmannaa, HercusM, DagherH (1998) Population dynamics of the Wolbachia infection causing cytoplasmic incompatibility in Drosophila melanogaster. Genetics 148 : 221–231.
81. FariaVG, SucenaE (2013) Wolbachia in the Malpighian tubules: evolutionary dead-end or adaptation? J Exp Zool B Mol Dev Evol 320 : 195–199.
82. PoinsotD, BourtzisK, MarkakisG, SavakisC, MerçotH (1998) Wolbachia Transfer from Drosophila melanogaster into D. simulans: Host Effect and Cytoplasmic Incompatibility Relationships. Genetics 150 : 227–237.
83. VenetiZ, ZabalouS, PapafotiouG, ParaskevopoulosC, PattasS, et al. (2012) Loss of reproductive parasitism following transfer of male-killing Wolbachia to Drosophila melanogaster and Drosophila simulans. Heredity 109 : 306–312.
84. ZhouW, RoussetF, O'NeillS (1998) Phylogeny and PCR–based classification of Wolbachia strains using wsp gene sequences. Proc R Soc B-Biological Sci 265 : 509–515.
85. ParaskevopoulosC, BordensteinSR, WernegreenJJ, WerrenJH, BourtzisK (2006) Toward a Wolbachia multilocus sequence typing system: discrimination of Wolbachia strains present in Drosophila species. Curr Microbiol 53 : 388–395.
86. DarlingA, MauB, BlattnerF, PernaN (2004) Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14 : 1394–1403.
87. GelmanA, RubinD (1992) Inference from iterative simulation using multiple sequences. Stat Sci 7 : 457–511.
88. TamuraK, PetersonD, PetersonN, StecherG, NeiM, et al. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28 : 2731–2739.
89. R Core Team (2013) R: A Language and Environment for Statistical Computing.
90. GuindonS, DufayardJ-F, LefortV, AnisimovaM, HordijkW, et al. (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59 : 307–321.
91. Sullivan W, Ashburner M, Hawley R (2000) Drosophila Protocols. New York: Cold Spring Harbor Laboratory Press.
92. LongdonB, CaoC, MartinezJ, JigginsFM (2013) Previous exposure to an RNA virus does not protect against subsequent infection in Drosophila melanogaster. PLoS One 8: e73833.
93. PfafflMW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45.
94. HadfieldJ (2010) MCMC methods for multi-response generalized linear mixed models: the MCMCglmm R package. J Stat Softw 33 : 1–22.
95. HousworthEa, MartinsEP, LynchM (2004) The phylogenetic mixed model. Am Nat 163 : 84–96.
96. HadfieldJD, NakagawaS (2010) General quantitative genetic methods for comparative biology: phylogenies, taxonomies and multi-trait models for continuous and categorical characters. J Evol Biol 23 : 494–508.
97. MateosM, CastrezanaSJ, NankivellBJ, EstesAM, MarkowTA, et al. (2006) Heritable Endosymbionts of Drosophila. Genetics 174 : 363–376.
98. SheeleySL, McAllisterBF (2009) Mobile male-killer: similar Wolbachia strains kill males of divergent Drosophila hosts. Heredity 102 : 286–292.
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Out-of-Sequence Signal 3 as a Mechanism for Virus-Induced Immune Suppression of CD8 T Cell ResponsesČlánek RNF26 Temporally Regulates Virus-Triggered Type I Interferon Induction by Two Distinct MechanismsČlánek Mouse, but Not Human, ApoB-100 Lipoprotein Cholesterol Is a Potent Innate Inhibitor of Pneumolysin
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2014 Číslo 9- Stillova choroba: vzácné a závažné systémové onemocnění
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Jak souvisí postcovidový syndrom s poškozením mozku?
- Infekční komplikace virových respiračních infekcí – sekundární bakteriální a aspergilové pneumonie
-
Všechny články tohoto čísla
- Virus Control Goes Epigenetic
- The Role of Iron in Prion Disease and Other Neurodegenerative Diseases
- The Ins and Outs of Rust Haustoria
- Prion Strains and Amyloid Polymorphism Influence Phenotypic Variation
- Teaching Fido New ModiFICation Tricks
- Can Enhance Infection in Mosquitoes: Implications for Malaria Control?
- MIF Contributes to Associated Immunopathogenicity Development
- Persistence of Virus Reservoirs in ART-Treated SHIV-Infected Rhesus Macaques after Autologous Hematopoietic Stem Cell Transplant
- Bacillus Calmette-Guerin Infection in NADPH Oxidase Deficiency: Defective Mycobacterial Sequestration and Granuloma Formation
- EhCoactosin Stabilizes Actin Filaments in the Protist Parasite
- Molecular Insights Into the Evolutionary Pathway of O1 Atypical El Tor Variants
- LprG-Mediated Surface Expression of Lipoarabinomannan Is Essential for Virulence of
- Structural Correlates of Rotavirus Cell Entry
- Multivalent Adhesion Molecule 7 Clusters Act as Signaling Platform for Host Cellular GTPase Activation and Facilitate Epithelial Barrier Dysfunction
- The Effects of Vaccination and Immunity on Bacterial Infection Dynamics
- Myeloid Derived Hypoxia Inducible Factor 1-alpha Is Required for Protection against Pulmonary Infection
- Functional Characterisation of Germinant Receptors in and Presents Novel Insights into Spore Germination Systems
- Global Analysis of Neutrophil Responses to Reveals a Self-Propagating Inflammatory Program
- Host Cell Invasion by Apicomplexan Parasites: The Junction Conundrum
- Comparative Phenotypic Analysis of the Major Fungal Pathogens and
- Unravelling the Multiple Functions of the Architecturally Intricate β-galactosidase, BgaA
- Sialylation of Prion Protein Controls the Rate of Prion Amplification, the Cross-Species Barrier, the Ratio of PrP Glycoform and Prion Infectivity
- Symbionts Commonly Provide Broad Spectrum Resistance to Viruses in Insects: A Comparative Analysis of Strains
- Ontogeny of Recognition Specificity and Functionality for the Broadly Neutralizing Anti-HIV Antibody 4E10
- Identification and Characterisation of a Hyper-Variable Apoplastic Effector Gene Family of the Potato Cyst Nematodes
- Crimean-Congo Hemorrhagic Fever Virus Entry into Host Cells Occurs through the Multivesicular Body and Requires ESCRT Regulators
- Age-Dependent Enterocyte Invasion and Microcolony Formation by
- CD160-Associated CD8 T-Cell Functional Impairment Is Independent of PD-1 Expression
- Functional Fluorescent Protein Insertions in Herpes Simplex Virus gB Report on gB Conformation before and after Execution of Membrane Fusion
- The Tudor Domain Protein Spindlin1 Is Involved in Intrinsic Antiviral Defense against Incoming Hepatitis B Virus and Herpes Simplex Virus Type 1
- Transgenic Analysis of the MAP Kinase MPK10 Reveals an Auto-inhibitory Mechanism Crucial for Stage-Regulated Activity and Parasite Viability
- Evidence for a Transketolase-Mediated Metabolic Checkpoint Governing Biotrophic Growth in Rice Cells by the Blast Fungus
- Incomplete Deletion of IL-4Rα by LysM Reveals Distinct Subsets of M2 Macrophages Controlling Inflammation and Fibrosis in Chronic Schistosomiasis
- Identification and Functional Expression of a Glutamate- and Avermectin-Gated Chloride Channel from , a Southern Hemisphere Sea Louse Affecting Farmed Fish
- Out-of-Sequence Signal 3 as a Mechanism for Virus-Induced Immune Suppression of CD8 T Cell Responses
- Strong Epistatic Selection on the RNA Secondary Structure of HIV
- Hematopoietic but Not Endothelial Cell MyD88 Contributes to Host Defense during Gram-negative Pneumonia Derived Sepsis
- Delineation of Interfaces on Human Alpha-Defensins Critical for Human Adenovirus and Human Papillomavirus Inhibition
- Exploitation of Reporter Strains to Probe the Impact of Vaccination at Sites of Infection
- RNF26 Temporally Regulates Virus-Triggered Type I Interferon Induction by Two Distinct Mechanisms
- Helminth Infections Coincident with Active Pulmonary Tuberculosis Inhibit Mono- and Multifunctional CD4 and CD8 T Cell Responses in a Process Dependent on IL-10
- MHC Class II Restricted Innate-Like Double Negative T Cells Contribute to Optimal Primary and Secondary Immunity to
- Reactive Oxygen Species Regulate Caspase-11 Expression and Activation of the Non-canonical NLRP3 Inflammasome during Enteric Pathogen Infection
- Evolution of Plastic Transmission Strategies in Avian Malaria
- A New Human 3D-Liver Model Unravels the Role of Galectins in Liver Infection by the Parasite
- Translocates into the Myocardium and Forms Unique Microlesions That Disrupt Cardiac Function
- Mouse, but Not Human, ApoB-100 Lipoprotein Cholesterol Is a Potent Innate Inhibitor of Pneumolysin
- The Cofilin Phosphatase Slingshot Homolog 1 (SSH1) Links NOD1 Signaling to Actin Remodeling
- Kaposi's Sarcoma Herpesvirus MicroRNAs Induce Metabolic Transformation of Infected Cells
- Reorganization of the Endosomal System in -Infected Cells: The Ultrastructure of -Induced Tubular Compartments
- Distinct Dictation of Japanese Encephalitis Virus-Induced Neuroinflammation and Lethality via Triggering TLR3 and TLR4 Signal Pathways
- Exploitation of the Complement System by Oncogenic Kaposi's Sarcoma-Associated Herpesvirus for Cell Survival and Persistent Infection
- The Secreted Peptide PIP1 Amplifies Immunity through Receptor-Like Kinase 7
- Structural Insight into Host Recognition by Aggregative Adherence Fimbriae of Enteroaggregative
- The CD14CD16 Inflammatory Monocyte Subset Displays Increased Mitochondrial Activity and Effector Function During Acute Malaria
- Infection Induces Expression of a Mosquito Salivary Protein (Agaphelin) That Targets Neutrophil Function and Inhibits Thrombosis without Impairing Hemostasis
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- The Secreted Peptide PIP1 Amplifies Immunity through Receptor-Like Kinase 7
- Symbionts Commonly Provide Broad Spectrum Resistance to Viruses in Insects: A Comparative Analysis of Strains
- MIF Contributes to Associated Immunopathogenicity Development
- The Ins and Outs of Rust Haustoria
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání








