-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Evidence for a Transketolase-Mediated Metabolic Checkpoint Governing Biotrophic Growth in Rice Cells by the Blast Fungus
The blast fungus Magnaporthe oryzae destroys rice and wheat harvests and could compromise global food security. Following penetration into the rice cell, M. oryzae elaborates bulbous invasive hyphae that grow in living rice cells for most of the infection cycle without causing disease symptoms. Little is known about the physiological processes governing this important biotrophic stage of fungal growth. Here, we used gene functional analysis to show how the primary metabolic enzyme transketolase is essential for hyphal growth in rice cells. Loss of transketolase did not affect the ability of the fungus to gain entry into rice cells, but invasive hyphal growth was curtailed in transketolase null mutants. Biotrophic growth was restored in transketolase mutants by the addition of exogenous ATP. We conclude that M. oryzae metabolism is dedicated to metabolizing glucose through transketolase in planta in order to provide ATP as a trigger for biotrophic growth and infection. This work is significant because it reveals important—but previously unknown—metabolic strategies employed by M. oryzae to facilitate rice infection. These strategies might be open to abrogation by chemical or biological means and are likely relevant to other rapidly proliferating intracellular pathogens.
Published in the journal: . PLoS Pathog 10(9): e32767. doi:10.1371/journal.ppat.1004354
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1004354Summary
The blast fungus Magnaporthe oryzae destroys rice and wheat harvests and could compromise global food security. Following penetration into the rice cell, M. oryzae elaborates bulbous invasive hyphae that grow in living rice cells for most of the infection cycle without causing disease symptoms. Little is known about the physiological processes governing this important biotrophic stage of fungal growth. Here, we used gene functional analysis to show how the primary metabolic enzyme transketolase is essential for hyphal growth in rice cells. Loss of transketolase did not affect the ability of the fungus to gain entry into rice cells, but invasive hyphal growth was curtailed in transketolase null mutants. Biotrophic growth was restored in transketolase mutants by the addition of exogenous ATP. We conclude that M. oryzae metabolism is dedicated to metabolizing glucose through transketolase in planta in order to provide ATP as a trigger for biotrophic growth and infection. This work is significant because it reveals important—but previously unknown—metabolic strategies employed by M. oryzae to facilitate rice infection. These strategies might be open to abrogation by chemical or biological means and are likely relevant to other rapidly proliferating intracellular pathogens.
Introduction
The rice blast fungus Magnaporthe oryzae causes the most serious disease of cultivated rice [1], [2] and is a significant challenge to global food security [3], [4]. Infection involves the elaboration of a specialized structure, the appressorium, from a germinating three-celled conidium (spore) on the surface of the leaf [1], [5], [6]. Each cell of the conidium contains a nucleus, the most apical of which migrates into the germ tube during appressorium development where it undergoes closed mitosis [7]. One of the resulting daughter nuclei returns to the conidium to be degraded during autophagic cell-death [8]; the other enters the incipient appressorium [7] to become the source of M. oryzae genetic material during host infection. Disrupting autophagy or blocking the cell cycle at three checkpoints (S-phase, mitosis and exit from mitosis) abolishes infection [8], [9]. At maturation, enormous turgor is generated in the appressorium through the accumulation of glycerol and the ingress of water that becomes trapped due to a layer of melanin deposited on the wall of the appressorium [10], [11]. This hydrostatic pressure acts on a penetration peg [5], [6], forcing it through the leaf cuticle. In the cell lumen, the penetration peg becomes a thin primary hypha that invaginates the host plasma membrane before differentiating into bulbous invasive hyphae (IH) that are sealed in the plant-derived extra-invasive hyphal membrane (EIHM) compartment [12]. IH moves into adjacent cells by crossing the cell wall at plasmodesmata, and the biotrophic invasion process is repeated in successive rice cells [12]. After four to five days of biotrophic growth in susceptible cultivars, host cells die and M. oryzae enters its necrotrophic growth phase, causing spreading necrotic lesions on the leaf surface from which conidia are produced on aerial hyphae [1].
During appressorium development, lipid and glycogen stores in the spore are mobilized and transported to the appressorium in a manner dependent on the MAP kinase - and cAMP-dependent protein kinase A-signaling pathways [13], [14]. Following lipolysis in the incipient appressorium [15], a large body of evidence indicates that organelles and biochemical pathways that metabolize fatty acids, such as mitochondrial and peroxisomal β-oxidation and the glyoxylate cycle, are essential for the development of infection-competent appressoria [15]–[20]. Thus, fatty acid β-oxidation and the distribution of the resulting acetyl-CoA subunits into cellular processes such as cell wall biosynthesis [17] and pyruvate formation [19] are dominant biochemical pathways during appressorial development.
In contrast to appressorial function (which requires lipid catabolism through β-oxidation and the glyoxylate cycle), post-penetrative host infection is dependent on glucose 6-phosphate (G6P) sensing by trehalose-6-phosphate synthase 1 (Tps1) and the concomitant production of NADPH through the pentose phosphate pathway [2], [21]–[24]. The binding of G6P by Tps1 leads, via an NADPH-dependent signaling pathway, to the induced expression of genes encoding NADPH-requiring enzymes [22], [24] and the repression of genes required for utilizing alternative sources of carbon [23]. At least two NADPH-requiring processes under Tps1 control, the glutathione and thioredoxin antioxidation systems, are glucose-responsive and required for biotrophic growth in rice cells [24]. Taken together, we have proposed that these observations are consistent with the occurrence of a metabolic shift from lipid metabolism by M. oryzae on the host surface to glucose metabolism in the host cell [25]. This metabolic reprogramming hypothesis is supported by the observations that the glyoxylate enzyme isocitrate lyase is required for appressorium function but is not required for growth in planta [16]. However, which glucose metabolizing pathways are active during M. oryzae growth in planta, and whether glucose metabolism contributes to appressorial development, is not known.
Here, we sought to bolster our understanding of the metabolic strategies governing M. oryzae in planta growth by focusing on how glucose-metabolizing pathways contribute to rice blast infection. We used gene functional analysis and live-cell imaging to show how glucose-metabolizing pathways are not required for appressorium formation and function. Instead, glucose metabolism via transketolase (encoded by TKL1) is essential for the post-invasive colonization of rice cells by M. oryzae. The loss of transketolase function resulted in Δtkl1 mutant strains that could form appressoria, penetrate host cells and elaborate IH, but were attenuated for growth and cell cycle progression in planta. Δtkl1 strains were depleted for ATP, and IH growth was restored in planta following the treatment of Δtkl1 strains with exogenous ATP, suggesting ATP acts downstream of transketolase to signal growth [26], [27]. Moreover, evidence is provided to suggest ATP acts on the TOR signaling pathway and together our data indicate a functional connection between TKL1, ATP and the TOR signaling pathway. These results are consistent with a novel role for transketolase in mediating an in planta-specific metabolic checkpoint that regulates the cell cycle, via TOR activation, in order to permit rice cell colonization by M. oryzae. This work thus gives fresh insights into the metabolic demands of biotrophy and points to novel signaling roles for transketolase that will be important for other areas of biology.
Results
Early glycolysis, late gluconeogenesis and the pentose phosphate pathway are not required for appressorium formation
To develop a more robust understanding of the biochemical processes underlying plant infection by M. oryzae, we first sought to determine what glucose-metabolizing pathways might be important for rice infection. Following Berg and associates, [28], we considered four metabolic destinations for G6P in M. oryzae: into glycolysis via the action of phosphoglucose isomerase 1 (Pgi1) if more ATP than NADPH or ribose 5-phosphate is needed (Mode 1 in Figure 1); into early glycolysis and the non-oxidative pentose phosphate pathway if ribose 5-phosphate production for nucleotide biosynthetic purposes is predominant (Mode 2 in Figure 1); into the pentose phosphate pathway (PPP) and recycling via late gluconeogenesis until G6P is fully oxidized to CO2 if NADPH production is paramount (Mode 3 in Figure 1); into the PPP and glycolysis to form pyruvate if NADPH and ATP production is required (Mode 4 in Figure 1). Searching the M. oryzae genome [29], we found single orthologues of FBP1 (MGG_08895) encoding fructose 1,6-bisphosphatase in the gluconeogenic pathway, PGI1 (MGG_12822) involved in both glycolysis and gluconeogenesis, and TKL1 (MGG_02471) encoding transketolase, a non-oxidative PPP enzyme linking the PPP and glycolysis. Using our high-throughput, PCR-based split marker method for gene deletion [22], we replaced part of the coding regions of FBP1 and PGI1 with the ILV1 gene conferring sulphonyl urea resistance, and the coding region of TKL1 with the Bar gene conferring bialaphos resistance. Figure 2A shows that the resulting three mutant strains - Δfbp1, Δpgi1 and Δtkl1 - grew without impediment on glucose-rich complete media (CM) compared to wild type Guy11 strains. Despite these similarities in growth, Figure 2B shows that both PGI1 and TKL1, but not FBP1, contributed significantly (Student's t-test p≤0.05) to sporulation rates on CM media. This indicates nucleotide biosynthesis via Mode 2 (Figure 1) might be important for sporulation, perhaps due to the nucleotide demands of DNA replication during spore production.
Fig. 1. Potential metabolic destinations for G6P. 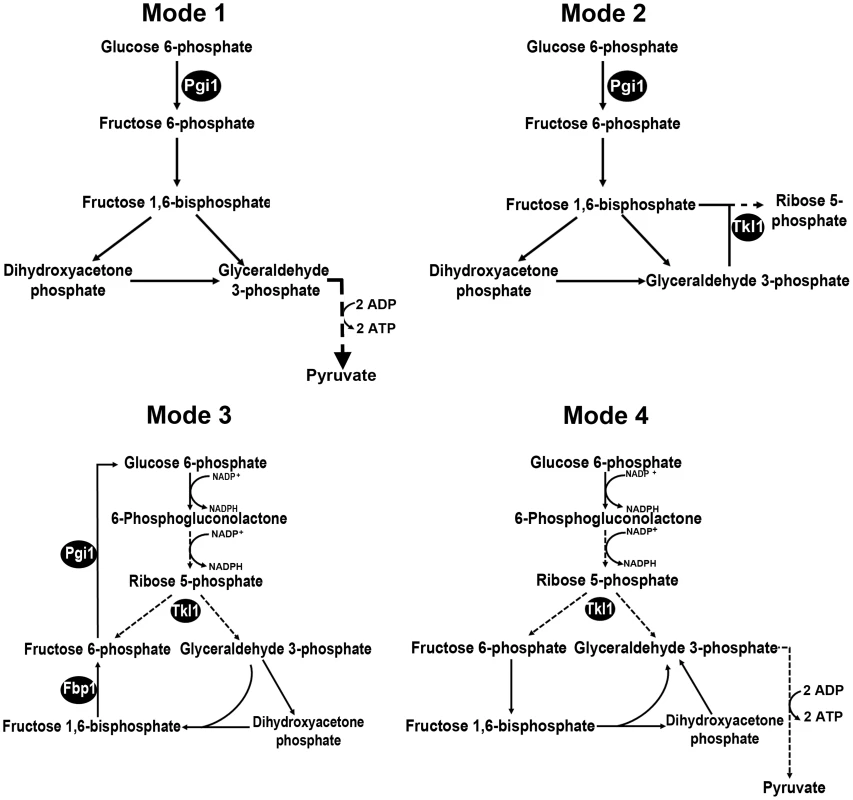
Following Berg and colleagues, [28], we considered four scenarios for the fate of glucose 6-phosphate (G6P) during infection-related processes. Mode 1: More ATP than NADPH is required. G6P enters glycolysis through its conversion to fructose 6-phosphate by phosphoglucose isomerase 1 (Pgi1). Mode 2: M. oryzae cells require more ribose 5-phosphate for biosynthetic purposes than NADPH. Fructose 6-phosphate and glyceraldehyde 3-phosphate in the glycolytic pathway are converted to ribose 5-phosphate in the non-oxidative pentose phosphate pathway (PPP) by the action of transaldolase and transketolase (Tkl1). Mode 3: more NADPH than ribose 5-phosphate is required. G6P fuels NADPH production by G6P dehydrogenase (G6PDH) in the oxidative PPP and is resynthesized in the gluconeogenic pathway by fructose 1,6-bisphosphatase (Fbp1) and Pgi1. G6P is completely oxidized to CO2. Mode 4: NADPH and ATP are required. G6P fluxes through the PPP and into glycolysis via Tkl1 resulting in the production of NADPH and ATP. G6P is converted into pyruvate. Dashed lines indicate steps that have been omitted for clarity. Products of genes functionally characterized in the text are indicated by black spheres. Fig. 2. Dissection of G6P-utilizing pathways by gene functional analysis. 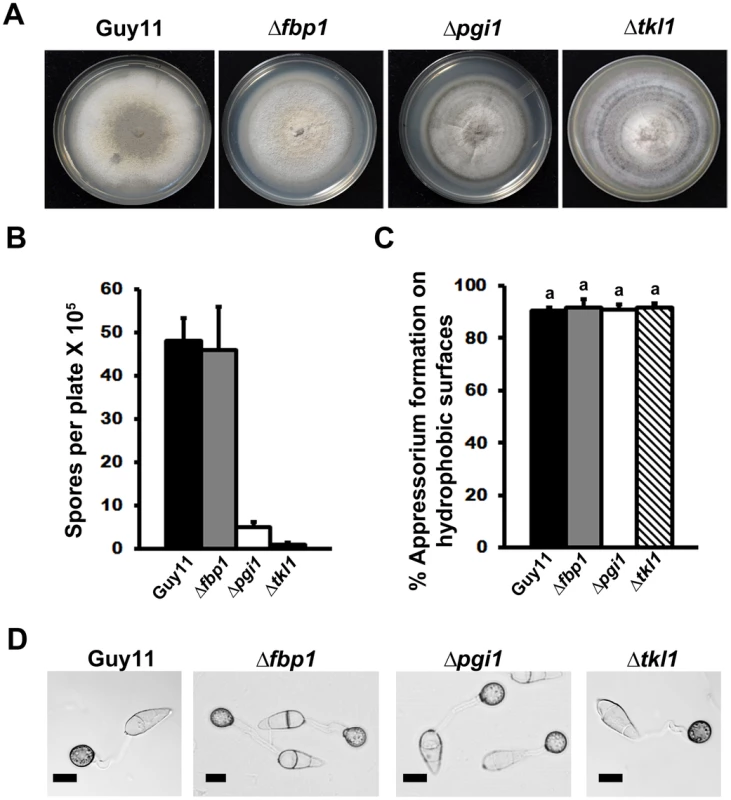
(A) Radial growth on complete media (CM) was not impaired in mutant strains carrying gene deletions in FBP1, PGI1 or TKL1. Guy11 is the wild type isolate used in this study. (B) Sporulation was impaired in strains lacking functional PGI1 and TKL1 genes, but not FBP1, during growth on CM. Results are the mean of three or more independent measurements. Error bars denote SD. Bars with the same letters are not significantly different (Student's t-test p≤0.05). (C) Spores of Guy11, Δpgi1, Δfbp1 and Δtkl1 were applied to artificial hydrophobic surfaces. At 24 hr post inoculation (hpi), the rate of appressorial formation by either gene deletion strain was not significantly different to Guy11. Error bars are SD. Bars with the same letter are not significantly different (Student's t-test p≤0.05). (D) Loss of PGI1, FBP1 or TKL1 did not impair the formation of melanized appressoria on artificial hydrophobic surfaces by 24 hpi. Scale bar is 10 µm. Interestingly, all the mutant strains in this study formed melanized appressoria on hydrophobic surfaces (Figure 2C and 2D) at rates that were indistinguishable from Guy11 (Figure 2C, Student's t-test p>0.05). This indicates that the glucose utilization pathways shown in Figure 1 are not required for appressorium formation and is consistent with previous work emphasizing the requirement for lipid metabolism and the glyoxylate cycle, rather than sugar metabolism, during appressorium formation [15]–[20]. Taken together, G6P metabolism via Mode 2 contributes to sporulation, but neither Mode described in Figure 1 is necessary for appressorial formation.
Transketolase is essential for invasive hyphal growth in host rice cells
Although all the mutant strains were able to form appressoria on artificial hydrophobic surfaces, Figure 3A shows that when spores were applied to whole plants, only the Δtkl1 mutant strain was unable to cause disease. TKL1 is thus revealed here as an essential and previously unknown determinant of pathogenicity by the rice blast fungus.
Fig. 3. The transketolase-encoding gene TKL1 is required for in planta growth. 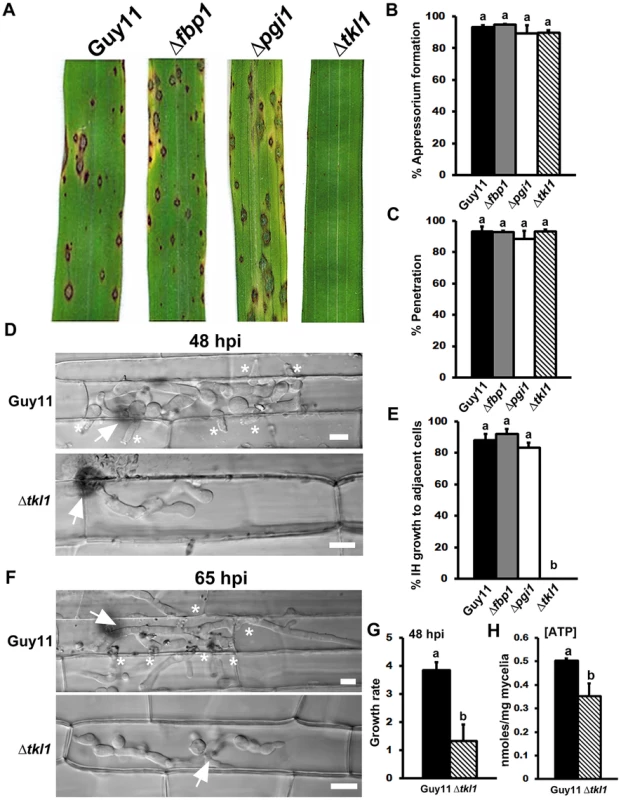
(A) TKL1, but not PGI1 or FBP1, is essential for pathogenicity. Spore suspensions of each strain were applied to susceptible CO-39 plants at a rate of 1×105 spores per mL. (B) Despite being non-pathogenic, the rate of appressorial formation by Δtkl1 strains was not significantly different to Guy11 on rice leaf surfaces at 24 hpi and (C) the rate of rice leaf penetration by Δtkl1 strains, determined at 30 hpi, was equivalent to Guy11. Values are the mean of three independent replicates. Error bars denote SD. Bars with the same letters are not significantly different (Student's t-test p≤0.05). (D) Live-cell imaging at 48 hpi of Guy11 and Δtkl1 strains infecting susceptible CO-39 rice leaf sheaths showed Δtkl1 was impaired in IH growth compared to Guy11. Scale bar is 5 µm. Arrows indicate appressoria on the surface of the leaf corresponding to the point of penetration. Asterisks indicate IH movement from primary infected cells to adjacent cells. (E) IH movement to cells adjacent to the point of penetration was impaired in Δtkl1 strains compared to Guy11, Δpgi and Δfbp1 strains. Values are the percentage of IH in primary infected cells that have spread to neighboring cells at 48 hpi and represent the mean of three independent replicates. Error bars denote SD. Bars with the same letters are not significantly different (Student's t-test p≤0.05). (F) Live-cell imaging at 65 hpi of Guy11 and Δtkl1 strains infecting susceptible CO-39 rice leaf sheaths showed Δtkl1 was impaired in IH growth compared to Guy11. Scale bar is 5 µm. Arrows indicate appressoria on the surface of the leaf corresponding to the point of penetration. Asterisks indicate IH movement from primary infected cells to adjacent cells. (G) The in planta growth rate of Δtkl1 IH, determined at 48 hpi, was significantly reduced (Student's t-test p≤0.05) compared to Guy11. The growth rate of IH was measured by using the 1–4 scale described in [59]. Values are the mean of three independent replicates. Error bars denote SD. Bars with the same letters are not significantly different (Student's t-test p≤0.05). (H) Intracellular ATP levels were reduced in mycelia of Δtkl1 strains compared to Guy11 following growth in minimal media (MM). [ATP] was determined by LC-MS/MS, as described in Materials and Methods. Metabolite extractions were performed in triplicate for each strain. Error bars denote SD. Bars with the same letters are not significantly different (Student's t-test p≤0.05). The inability of Δtkl1 strains to infect rice leaves was not due to defective appressorium formation or function on host surfaces because, like Δpgi and Δfbp1 mutant strains, Δtkl1 formed appressoria on rice leaf surfaces at the same rates as Guy11 (Figure 3B; Student's t-test p = 0.64). Also, the rates of cuticle penetration on detached rice leaf sheaths were not significantly different (Student's t-test p = 0.37) for Guy11 and any of the mutant strains generated for this study, including Δtkl1 (Figure 3C).
If appressorium formation and function did not depend on a functional Tkl1 enzyme, how, then, does TKL1 contribute to pathogenicity? To address this question, we continued to characterize the role of theTKL1 gene in pathogenicity using live-cell imaging of detached rice leaf sheaths colonized with either Δtkl1 or Guy11 strains. At 48 hr post inoculation (hpi), Δtkl1 mutant strains were found to elaborate bulbous IH from primary hyphae, but IH growth was severely restricted to the first infected cell compared to Guy11 (Figure 3D). At 48 hpi, more than 80% of Guy11, Δpgi and Δfbp1 primary infection sites had resulted in IH spreading to adjacent cells, but IH movement to adjacent cells by Δtkl1 mutant strains was not observed (Figure 3E). Δtkl1 strains had also not spread to adjacent cells by 65 hpi (Figure 3F). Growth rates in rice cells at 48 hpi was scored for 50 infected cells/strain (repeated in triplicate) using our 4 point scale (where 1 = IH length shorter than 10 µm with no branching; 2 = IH length is 10–20 µm with 0–2 branches; 3 = IH length is longer than 20 µm and/or with more than 2 branches within one cell; 4 = IH has spread to adjacent cells). The average growth rate score for Δtkl1 strains in rice cells at 48 hpi was significantly reduced compared to Guy11 (Figure 3G; Student's t-test p = 0.0078). Therefore, TKL1 is not required for appressorium formation or function, or for IH elaboration, but is essential for strong biotrophic growth in rice cells and the movement of IH to neighboring cells.
Glucose is metabolized via transketolase during rice infection
Figure 3A and 3E shows that early glycolytic and late gluconeogenic steps are dispensable for biotrophic growth in rice cells and disease development, whereas the non-oxidative PPP enzyme Tkl1 is essential. This indicates that G6P metabolism via Mode 4 (Figure 1) is likely the dominant pathway for glucose utilization by M. oryzae during host infection. Further experimental support for Mode 4—which would involve Tkl1 connecting the PPP and later glycolytic steps to yield NADPH and ATP from glucose [28]—is shown in Figure 3G where, following axenic growth in minimal media (MM), Δtkl1 mycelia was found by LC-MS/MS analysis to contain significantly less ATP than the mycelia of Guy11 strains (Figure 3H). Therefore, ATP production is disrupted in Δtkl1 strains.
Δtkl1 mutant strains are downregulated for genes associated with translation initiation and protein biosynthesis in planta
We next sought to determine the molecular basis for the observed attenuated growth of Δtkl1 mutant strains in planta (Figure 3). First, we wished to ascertain if Δtkl1 mutants were senescent or killed in host cells after a short period of initial IH growth. We used quantitative real time PCR (qPCR) to measure M. oryzae actin (MoACT1) gene expression in Guy11 and Δtkl1 mutant strains, normalized against rice actin (OsACT2) gene expression, during the infection of rice epidermal cells at 48 hpi. The ratio of MoACT1 to OsACT2 gene expression was significantly higher in rice cells infected with Guy11 than Δtkl1 (Figure 4A). This was to be expected considering the greater growth rate of Guy11 in rice cells (Figure 3G). Nonetheless, MoACT1 gene expression was detectable in rice cells infected with Δtkl1 mutant strains. CT values for MoACT1 expression, before rice actin normalization, were 28.5±0.2 for rice cells infected with Δtkl1 mutants compared to 23.7±0.1 for those infected with Guy11. These results indicate Δtkl1 mutant strains were alive in rice cells at 48 hpi and undertaking some gene transcription despite the reduced growth rate of these strains (Figure 3G).
Fig. 4. In planta gene expression in Δtkl1 strains suggests attenuated growth. 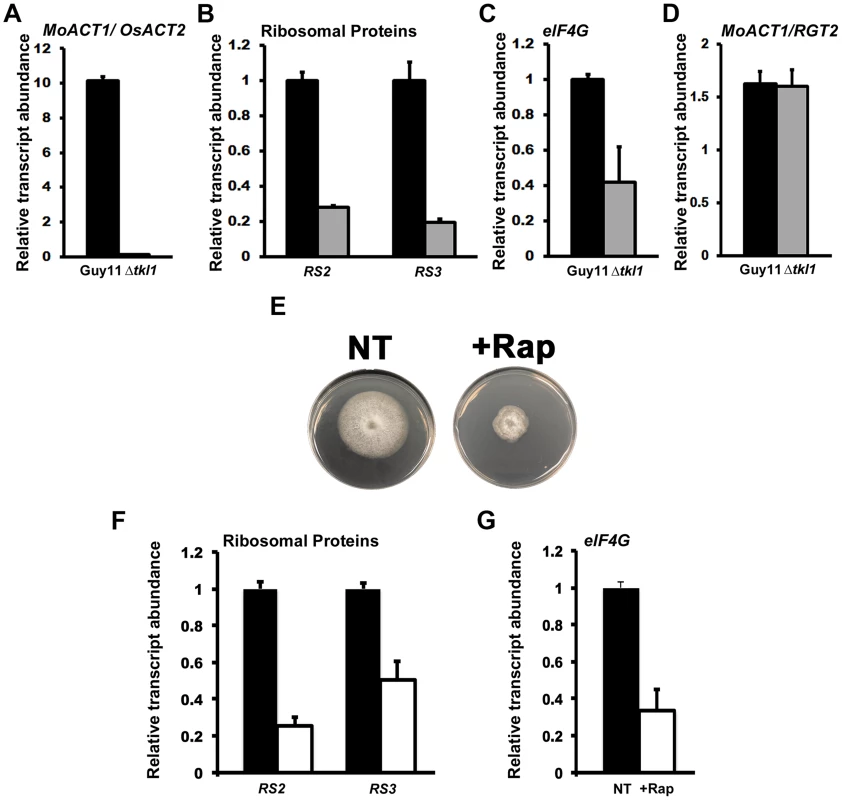
(A–D) In planta gene expression was analyzed by qPCR using cDNAs generated from infected rice leaf sheaths at 48 hpi. Values are the mean of three independent replicates. Error bars are SD. Closed bars are Guy11 expression levels; open bars are Δtkl1 expression levels. (A) Guy11 was 10 fold more abundant than Δtkl1 strains in rice cells at 48 hpi, as determined from the expression of MoACT1 normalized against rice actin (OsACT2) gene expression. Values are given as fold differences between each strain. Closed bar is the ratio of the mean normalized MoACT1 Ct values for Guy11: mean normalized MoACT1 Ct values for Δtkl1; open bar is the ratio of the mean normalized MoACT1 Ct values for Δtkl1: mean normalized MoACT1 Ct values for Guy11. (B) The expression of the M. oryzae ribosomal protein genes RS2 and RS3 was reduced in planta in Δtkl1 strains compared to Guy11 when normalized against MoACT1 expression. Expression changes in Δtkl1 strains are shown relative to Guy11. (C) eIF4G expression was downregulated in Δtkl1 strains compared to Guy11 in planta following normalization against MoACT1 expression. Expression changes are relative to Guy11. (D) Expression levels of MoAct1, relative to a second constitutively expressed gene RGT2, were not different between Guy11 and Δtkl1 strains in planta at 48 hpi. (E) The radial growth of Guy11 on minimal media (MM) containing the preferred carbon source glucose and the preferred nitrogen source ammonium was compromised when 50 nM (final concentration) of rapamycin, the specific TOR kinase inhibitor, was added to the media. + Rap = 50 nM rapamycin was added to the media. NT = no rapamycin treatment. (F) The gene expression levels of RS2, RS3 and (G) eIF4G were reduced in Guy11 following growth in MM containing glucose and ammonium in the presence of 50 nM rapamycin. Values are the mean of three independent replicates. Error bars are SD. Closed bars are gene expression levels following growth in media without rapamycin treatment; open bars are gene expression levels following growth in media with 50 nM rapamycin treatment. Expression levels were normalized against TUB2 gene expression and are given relative to Guy11 without rapamycin treatment. Because Δtkl1 mutant strains were alive in rice cells, we next asked what molecular mechanism might account for their attenuated growth in planta. Quiescence is associated with arrested growth and has been described as either an extended G1 stage or a distinct G0 state [26], [27]. Severely reduced growth states such as those exhibited by Δtkl1 strains also exhibit some features of quiescence [26], [27], [30] and it is not known if quiescence is a distinct state from slow growth [31]. Cellular quiescence occurs in yeast and mammals via the inactivation of the TOR signaling pathway in response to nutrient depletion. Conversely, the activated TOR signaling pathway promotes growth under favorable nutrient conditions. TOR inactivation results in, amongst other outcomes, cell cycle arrest [32] and translation suppression [33], [34]. Translation suppression occurs at the level of ribosome biogenesis, including TOR-dependent downregulation of ribosomal gene expression [35], [34]. To determine if the TOR signaling pathway might play a role in the poor growth of Δtkl1 strains in rice cells, we first examined the expression of two ribosomal genes, RS2 and RS3 (encoding ribosomal protein S2-like and 40S ribosomal protein S3, respectively), and the translation initiation factor eIF4G, in Guy11 and Δtkl1 mutant strains in planta at 48 hpi. eIF4G expression is reduced when mammalian mTOR is inhibited in an immortalized breast epithelial cell, and eIF4G depletion results in impaired cell proliferation and increased autophagy [36]. Compared to Guy11, and following normalization against MoACT1, both ribosomal protein genes (Figure 4B) and eIF4G (Figure 4C) were downregulated in Δtkl1 mutant strains in planta.
To ensure that the observed downregulation of gene expression in Δtkl1 mutants in planta was not attributable to a general decrease in MoACT1 expression in Δtkl1 strains compared to Guy11, we analyzed the expression of a putative glucose transporter encoded by RGT2, previously shown to be highly and constitutively expressed during rice infection [37]. Figure 4D shows that the ratio of MoACT1 gene expression to RGT2 gene expression, in planta, is the same for both Guy11 and Δtkl1, indicating MoACT1 gene expression was not reduced in Δtkl1 strains compared to Guy11.
We next sought to determine if RS2, RS3 and eIF4G gene expression was under TOR signaling control in M. oryzae. We studied the expression levels of these genes under optimal growth conditions (ie. minimal media containing the preferred carbon and nitrogen sources glucose and ammonium, respectively [2], [23]), and following growth on the same media containing the specific TOR kinase inhibitor rapamycin [33], [35]. Figure 4E shows that Guy11 grew poorly on preferred media when 50 nM rapamycin was added, confirming that optimal growth requires an activated TOR signaling pathway. Figure 4F and 4G shows, consistent with observations in yeast and mammalian cells, that poor growth in media containing rapamycin was associated with RS2, RS3 and eIF4G downregulation. Taken together, Figure 4 suggests Δtkl1 mutant strains might be downregulated - in a TOR-dependent manner - for translation initiation and protein biosynthesis during growth in rice cells.
Mitosis is delayed in Δtkl1 mutant strains in planta
In response to nutrient deprivation, TOR inactivation can arrest yeast cells in G1 to induce quiescence [33]. In addition, TOR inactivation prolongs the G2/M transition in yeast [33]. Thus, cell growth and cell cycle progression are linked by common signaling pathways, including TOR [33]. In order to continue investigating the role of TKL1 during in planta growth, we next turned our attention to the cell nucleus. We performed live-cell imaging of M. oryzae IH using a Guy11 strain expressing a histone H1 protein fused to the tdTomato variant [38] of red fluorescent protein (H1:RFP) - described in [9] - and the same strain lacking a functional copy of TKL1 due to homologous recombination with the ILV1 gene conferring sulphonyl urea resistance (Δtkl1 H1:RFP). The individual nuclei of these strains were visualized by epifluorescence microscopy, and Figure 5A shows that at 32 hpi, when both strains had elaborated IH in rice cells and growth differences between the two strains were not as pronounced as at later timepoints, the IH of Guy11 H1:RFP contained significantly more (Student's t-test p≤0.05) nuclei, per infected rice cell, than the IH of Δtkl1 H1:RFP strains (quantified below). We also quantified the rate of mitosis in both strains at 32 hpi by measuring the number of nuclei per unit length of mycelia. Figure 5B shows that 10 µm lengths of Δtkl1 H1:RFP IH carried significantly less nuclei (Student's t-test p≤0.05) than 10 µm lengths of Guy11 H1:RFP IH. Thus, attenuated in planta growth by Δtkl1 strains is accompanied by delayed (but not arrested) mitotic progression. Interestingly, no significant differences in nuclei number (Student's t-test p>0.05) were observed in Δtkl1 H1:RFP vegetative hyphae compared to Guy11 H1:RFP following the growth of both strains in glucose rich CM (Figure S1A and B) or in defined glucose minimal media (GMM) with varying concentrations of glucose (Figure S2). This is consistent with Figure 2A that showsTKL1 is not required for optimal radial growth on CM.
Fig. 5. Nuclear division is attenuated in Δtkl1 mutant strains in planta. 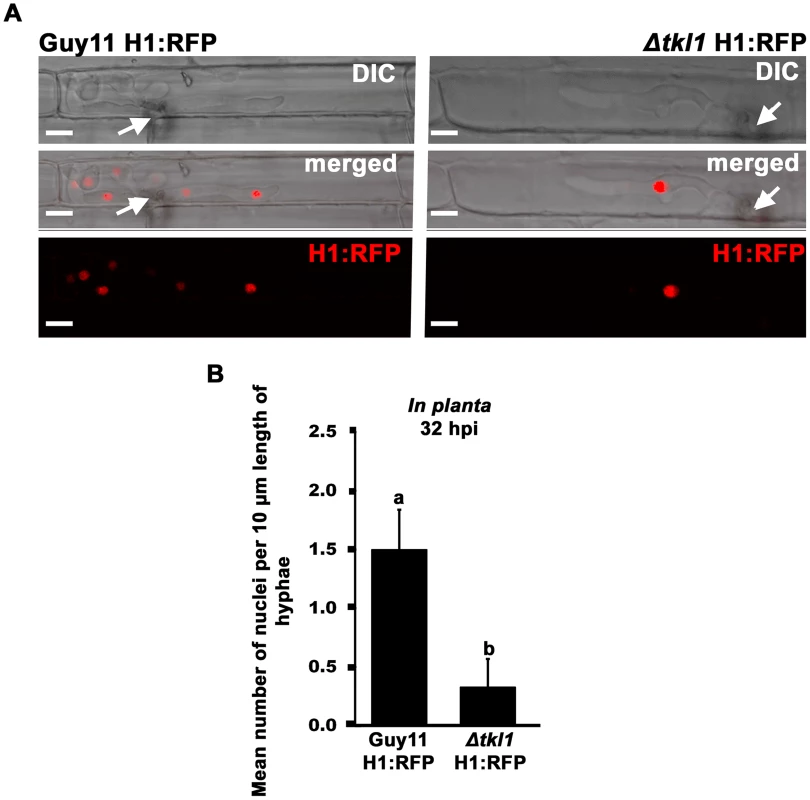
(A) Live-cell imaging at 32 hpi of Guy11 H1:RFP and Δtkl1 H1:RFP strains infecting susceptible CO-39 rice leaf sheaths showed Δtkl1 H1:RFP strains were impaired in nuclear proliferation in epidermal cells compared to Guy11 H1:RFP. Arrows indicate point of penetration. Scale bar is 5 µm. (B) The mean number of nuclei in 10 µm lengths of IH was calculated, using ImageJ, for each strain at 32 hpi, indicating Δtkl1 H1:RFP strains were delayed in mitosis. Values are the mean of at least six independent replications. Error bars denote SD. Bars with the same letters are not significantly different (Student's t-test p≤0.05). Together with the transcript data presented in Figure 4, and the physiological data in Figure 3, the results presented in Figure 5 support the notion that a functional TKL1 gene is required for cell cycle progression and growth during the biotrophic colonization of host rice cells.
Mitotic delay in Δtkl1 mutant strains is obviated by treatment with ATP
Why does loss of TKL1 result in mitotic delay and growth attenuation? We speculated that, considering the Tkl1 enzyme is involved in glucose metabolism, the lack of a metabolite(s) or metabolic pathway(s) downstream of Tkl1 might impact, directly or indirectly, cell cycle progression. From Figure 3H we knew that Δtkl1 strains were significantly reduced in ATP production compared to Guy11, such as would be predicted from perturbations to Mode 4 (Figure 1). In the absence of evidence for other metabolic changes, we hypothesized, then, that treatment of Δtkl1 mutant strains with exogenous ATP might remediate IH growth and affect mitotic rates upon host infection.
To test this hypothesis, we first needed to determine if M. oryzae was capable of acquiring or metabolizing ATP from the external milieu. In a previous study, we had shown how an adenine-requiring mutant, Δade1, could not grow on minimal media unless supplemented with adenosine or adenine [37]. Figure 6A shows how this same Δade1 mutant strain was remediated for growth on minimal media in the presence of ATP at a final concentration of 5 mM. This suggests that M. oryzae is capable of acquiring exogenous ATP directly from the media and, in adenine-requiring Δade1 mutant strains, this remediates axenic growth by converting ATP to adenine through the purine salvage pathway [37]. In addition, because Δade1 mutant strains can penetrate host cells but fail to establish infection, the growth tests in Figure 6A indicate plant sources of ATP, like adenine and adenosine [37], are not available to M. oryzae during growth in rice cells.
Fig. 6. Exogenous ATP promotes biotrophic growth and cell-to-cell movement by Δtkl1 mutant strains. 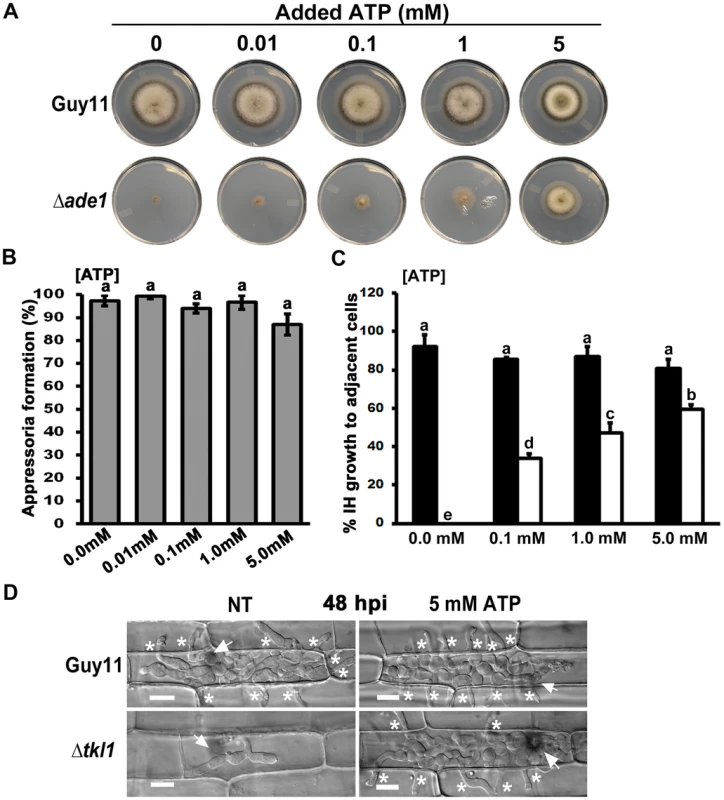
(A) Exogenous ATP can remediate growth of the adenine requiring mutant Δade1 on MM at the final concentrations indicated. Plates were imaged after 10 days of growth. (B) Spores of Δtkl1 were treated with the indicated concentrations of ATP and applied to rice leaf sheaths. Appressorium formation rates were determined at 24 hpi by counting the number of appressoria that had elaborated from 50 spores/leaf sheath for each treatment. Values are the mean of three independent replicates. Error bars denote SD. Bars with the same letters are not significantly different (Student's t-test p≤0.05). (C) Spores of Guy11 (closed bar) or Δtkl1 (open bar) were treated with ATP at the indicated final concentrations before applying to detached leaf surfaces. The rate of cell-to-cell movement was determined by counting how many of 50 primary infected cells had colonized adjacent cells by 48 hpi. Values are the mean of three independent replicates. Error bars denote SD. Bars with the same letters are not significantly different (Student's t-test p≤0.05). (D) Compared to untreated controls, the treatment of Δtkl1 spores with 5 mM ATP resulted in increased cell-to-cell movement in detached leaf sheaths at 48 hpi. NT = no treatment. Scale bar is 5 µm. Arrows indicate appressoria on the surface of the leaf corresponding to the point of penetration. Asterisks indicate IH movement from primary infected cells to adjacent cells. To determine whether exogenous ATP might interfere with the cAMP-signaling pathway controlling appressorium development [1], we added increasing amounts of ATP to Guy11 spores and applied them to leaf surfaces. No significant (Student's t-test p≤0.05) inhibition of appressorium formation was observed using final concentrations up to and including 5 mM ATP (Figure 6B). Δtkl1 spores treated with the same concentrations of ATP were then applied to detached leaf sheaths. Live-cell imaging showed cell-to-cell movement was improved in Δtkl1 mutant strains following treatment with ATP in a dose-dependent manner (Figure 6C and 6D).
We next added 5 mM ATP to spores of our RFP labeled histone H1 strains and applied them to detached rice leaf sheaths. Figure 7A and 7B shows how the addition of exogenous ATP increased the total number of nuclei in the IH of Δtkl1 H1:RFP, in rice cells at 32 hpi, to levels comparable to those observed in the IH of Guy11 H1:RFP. The rate of mitosis was also remediated in Δtkl1 H1:RFP strains by ATP treatment (Figure 7C). Similarly, at 48 hpi, the mitotic rate of untreated Δtkl1 H1:RFP was significantly less (Student's t-test p≤0.05) than Guy11 H1:RFP but was restored by ATP treatment (Figure S3A). However, at 65 hpi, there was no significant difference (Student's t-test p≤0.05) in the rates of mitosis between Δtkl1 H1:RFP and Guy11 H1:RFP strains in untreated samples (Figure S3B), suggesting mitotic delay is surmounted in Δtkl1 strains at later timepoints, perhaps in response to other metabolites.
Fig. 7. Mitotic delay is remediated in planta by treating Δtkl1 H1:RFP mutant strains with exogenous ATP. 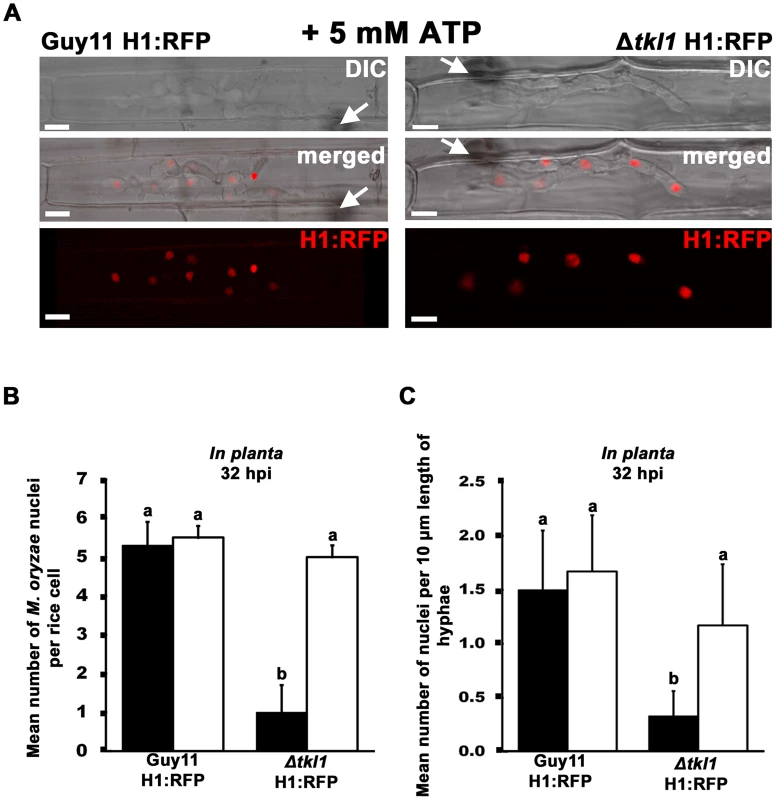
(A) Spores of Guy11 H1:RFP and Δtkl1 H1:RFP strains were treated with 5 mM ATP and applied to rice leaf sheaths. Live-cell imaging at 32 hpi shows ATP treated Δtkl1 H1:RFP strains were restored for nuclear proliferation in rice epidermal cells compared to Guy11 H1:RFP. Arrows indicate point of penetration. Scale bar is 5 µm. (B) Spores of Guy11 H1:RFP and Δtkl1 H1:RFP strains were either untreated (closed bar) or treated with 5 mM ATP (open bar) and applied to rice leaf sheaths. The mean total number of nuclei produced by Guy11 H1:RFP and Δtkl1 H1:RFP in planta at 32 hpi was calculated for each infected cell. Nuclei were counted in 50 infected cells per leaf sheath. Values are the mean of three independent replicates. Error bars denote SD. Bars with the same letters are not significantly different (Student's t-test p≤0.05). (C) The mean number of nuclei in 10 µm lengths of IH was calculated, using ImageJ, at 32 hpi for each strain. Closed bars are untreated controls, open bars are strains treated with 5 mM ATP. Values are the mean of at least six independent replications. Error bars denote SD. Bars with the same letters are not significantly different (Student's t-test p≤0.05). The remediating effect of ATP on Δtkl1 H1:RFP IH nuclear proliferation at 32 hpi was also observed when Δtkl1 H1:RFP spores were treated with the related compound adenosine (possibly due to its conversion to ATP through the purine salvage pathway [37]), but not when treated with 0.4 mM of the unrelated ROS quencher diphehyleneiodonium chloride (data not shown). Thus, we conclude exogenous sources of ATP can stimulate cell cycle progression in Δtkl1 mutant strains.
Δtkl1 mutant strains are not impaired in mitosis or autophagy during appressorium development
The preceding results demonstrated that following host penetration, nuclear division rates in the IH of Δtkl1 H1:RFP mutant strains were reduced compared to those of Guy11 H1:RFP, but this mitotic delay could be avoided by treatment with ATP prior to infection. However, untreated Δtkl1 mutant strains could still form infection-competent appressoria on the host surface (Figure 3B and C). This was interesting considering appressorial developmental involves, amongst other processes, cAMP signaling [1] and an active cell cycle [7], [9]. How can these observations be reconciled? To address this question, we applied spores of Guy11 H1:RFP and Δtkl1 H1:RFP strains to detached leaf surfaces and observed nuclei behaviour during appressorium development and early infection. Figure S4 shows that, up to 21 hpi, there was little difference in the behaviour of the nuclei of either strain, with each strain demonstrating three conidial nuclei at 13 hpi, and a punctate, appressorial nucleus at 18 hpi. Figure 8A and Figure S4 shows that by 21 hpi, no nuclei remained in the conidium of either strain, indicating that appressorial development and autophagic cell death of the conidium and its contents [8] had progressed normally in both strains. The timing of these events on rice cuticles is consistent with what has previously been demonstrated during appressorial formation on artificial hydrophobic surfaces [9]. However, at 21 hpi, whereas H1:RFP was still localized to a single, punctate appressorial nucleus in Guy11 H1:RFP strains, H1:RFP was diffused throughout the appressoria of Δtkl1 H1:RFP strains (Figure 8A and Figure S4). Differences were also seen at 24 hpi when, by this time, the single nucleus of each Guy11 H1:RFP appressorium had migrated from the leaf surface to primary hyphae in the host cell (Figure 8A), but no nuclear migration was evident for Δtkl1 H1:RFP (Figure 8A). However, Δtkl1 H1:RFP strains treated with ATP displayed a single, punctate appressorial nucleus at 21 hpi which had migrated into primary hyphae by 24 hpi (Figure 8A). These results suggest TKL1 is not required for the cell cycle events that give rise to the single nucleus of the appressorium, or for autophagic cell death and degradation of the remaining nuclei in the conidium. Instead, the results presented here indicate TKL1 acts after autophagy, via ATP, to maintain a punctate appressorial nucleus and control its migration into primary hyphae in the host cell.
Fig. 8. TKL1 is required for the migration of appressorial nuclei into primary hyphae in host cells. 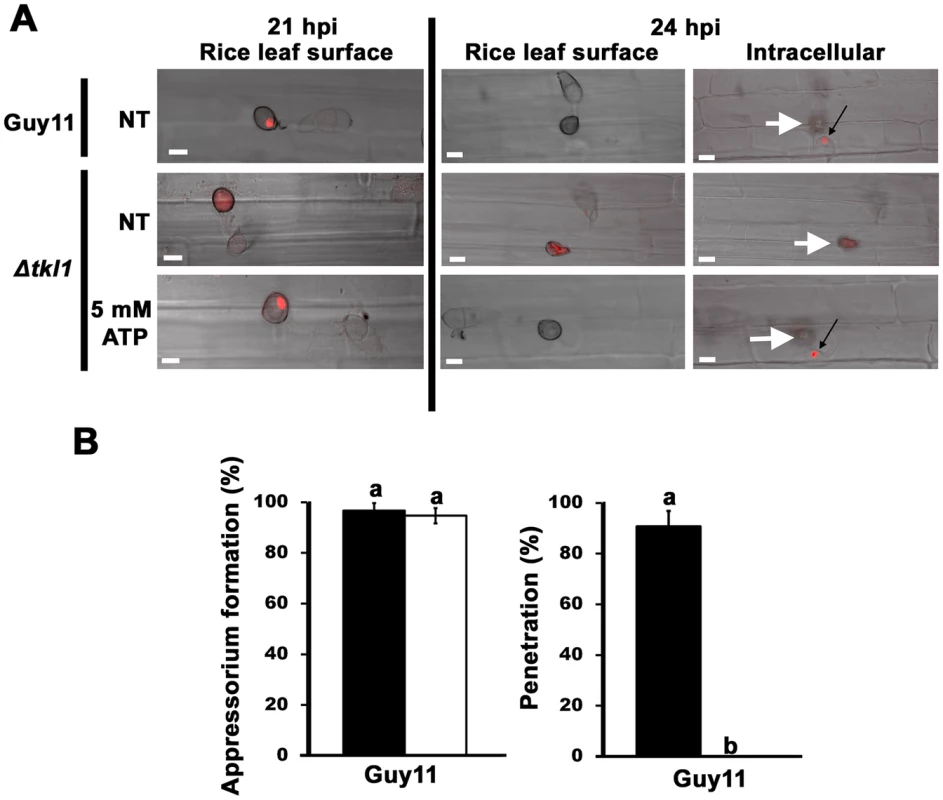
(A) Spores of Guy11 H1:RFP and Δtkl1 H1:RFP strains were either not treated (NT) or treated with 5 mM ATP and applied to rice leaf sheaths. At 21 hpi, untreated Guy11 H1:RFP and Δtkl1 H1:RFP strains treated with 5 mM ATP displayed a single punctate nucleus in the appressorium which had migrated into primary hyphae by 24 hpi (black arrows). In contrast, untreated Δtkl1 H1:RFP strains lacked a punctate nuclear structure at 21 hpi, and no punctate nucleus was evident in primary hyphae by 24 hpi. All panels are merged DIC and fluorescence images. Scale bar is 10 µm. White arrows indicate appressoria on the surface of the leaf corresponding to the point of penetration. (B) Spores of Guy11 were either untreated (closed bars) or treated (open bars) with 5 mM AMP-PNP, a non-hydrolysable analogue of ATP. Appressorium formation on rice leaf sheaths was unaffected by AMP-PNP treatment, but penetration was abolished. For each treatment, appressorium formation rates were determined at 24 hpi by counting the number of appressoria that had elaborated from 50 spores/leaf sheath for each treatment. Penetration rates were determined at 24 hpi by counting the number of successful cuticle breaches made by 50 appressoria/leaf sheath. Values are the mean of three independent replicates. Error bars denote SD. Bars with the same letters are not significantly different (Student's t-test p≤0.05). Additional evidence that ATP acts after appressorial formation comes from the use of the non-hydrolysable analogue of ATP, adenosine 5′-adenylyl imidodiphosphate (AMP-PNP). Treating Guy11 spores with AMP-PNP did not prevent appressorium formation on detached leaf sheaths but did abolish appressorium penetration of host surfaces (Figure 8B). This could indicate AMP-PNP is not taken up from the environment until after the appressorium has formed, but this is not likely considering cAMP is readily taken up into the cell prior to appressorial development [25]. Rather, the results with AMP-PNP are consistent with TKL1 and ATP being necessary for cuticle penetration and post-penetration development but not appressorial development and autophagy.
The M. oryzae TOR signaling pathway is activated downstream of TKL1 in response to ATP
The results presented above suggest TKL1 temporally controls the migration of appressorial nuclei into primary hyphae, and is required for the subsequent establishment of IH biotrophic growth, using a mechanism that involves ATP. Because TOR inactivation slows growth and extends mitosis [33], [34], we considered that a functional relationship between TKL1, ATP and the TOR signaling pathway might exist to promote biotrophic growth in planta.
To tests this hypothesis and thus gain more mechanistic insights into the regulation of M. oryzae gene expression during biotrophic growth, we studied the transcript levels for a number of known TOR read-out genes [34], [35], [39] in Guy11 and Δtkl1 mutant strains in planta at 48 hpi. These included M. oryzae genes shown in a previous study [39], or known from yeasts studies [35], to be positively regulated by the activated TOR pathway, such as those involved in nitrogen uptake and utilization (Nii1 encoding nitrate reductase, an aspartate semi-aldehyde dehygrogenase-encoding gene and GAP1 encoding the general amino acid permease [39]), a laccase putatively involved in cell wall modifications [39], and the single M. oryzae TOR-encoding gene TOR1 [35]. We also studied the expression of the ATG8 gene involved in autophagy [8], a process upregulated when TOR signaling is inactivated [34]. Figure 9A shows that all TOR readout genes were downregulated in expression in Δtkl1 strains, in planta, compared to Guy11 except ATG8, which was more highly expressed in Δtkl1 strains. These expression patterns are consistent with reduced TOR activity in Δtkl1 strains compared to Guy11 during in planta biotrophic growth at 48 hpi. Gene expression patterns were restored in Δtkl1 mutant strains when the infection was repeated using Guy11 and Δtkl1 spores treated with 5 mM ATP (Figure 9B). These results provide evidence that TKL1 activates the TOR signaling pathway via ATP (or an ATP derivative) in order to regulate gene expression in planta, promote biotrophic growth and prevent mitotic delay.
Fig. 9. TKL1 regulates the expression of TOR pathway read-out genes, via ATP, in planta. 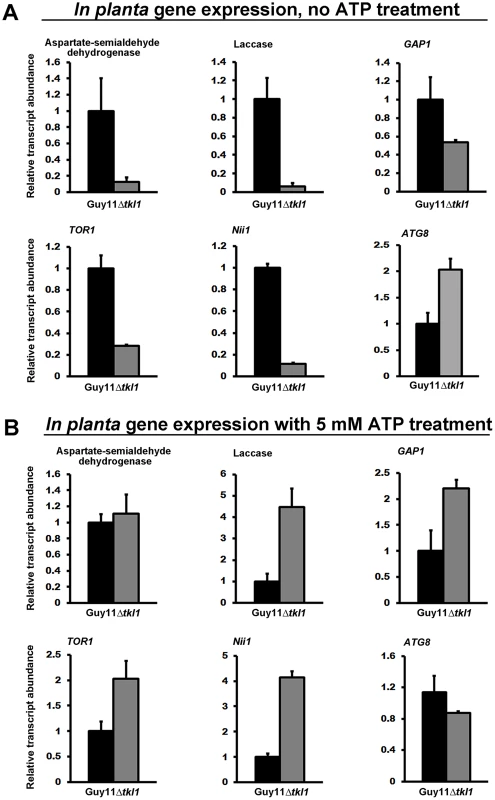
M. oryzae in planta gene expression was analyzed by qPCR using cDNAs generated from infected rice leaf sheaths at 48 hpi. Transcript abundance was normalized against MoACT1 gene expression. Expression changes in Δtkl1 strains are shown relative to Guy11. Values are the mean of three independent replicates. Error bars are SD. Closed bars are Guy11 expression levels; open bars are Δtkl1 expression levels. (A) Without ATP treatment, genes under positive TOR pathway control were downregulated in Δtkl1 strains compared to Guy11. By contrast, the ATG8 gene was upregulated in Δtkl1 strains compared to Guy11. (B) In planta Δtkl1 gene expression patterns were reversed when spores were treated with 5 mM ATP before applying to the host surface. Further evidence for a connection between ATP and the TOR signaling pathway in M. oryzae is shown in Figure 10. Compared to axenic growth in liquid minimal media with glucose and ammonium (preferred carbon and nitrogen sources for M. oryzae [2], [23]), growth on the same media with 50 nM rapamycin [33], [35] resulted in elevated expression of the autophagy gene ATG8. This observation is consistent with studies in yeast, which showed that rapamycin inhibits the TOR signaling pathway and activates autophagy [33]–[35]. However, rapamycin induction of ATG8 expression in M. oryzae was not observed when exogenous ATP was also present (Figure 10). Thus in M. oryzae, at least under some growth conditions, exogenous ATP can positively influence the TOR signaling pathway and override negative-acting signals such as those deriving from rapamycin treatment.
Fig. 10. Exogenous ATP activates the TOR signaling pathway. 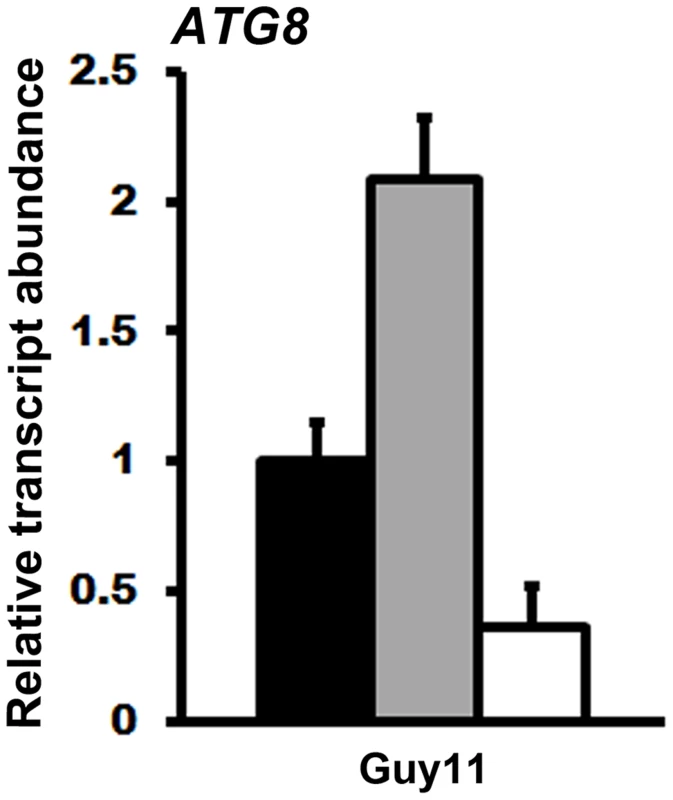
cDNA for qPCR analysis of ATG8 expression was obtained from the mycelia of Guy11 grown for 16 hr in liquid MM with 1% glucose and 10 mM ammonium (GMM) (closed bar), GMM with 50 nM rapamycin (grey bar), and GMM containing both 50 nM rapamycin and 5 mM ATP (open bar). Transcript abundance was normalized against TUB2 expression and gene expression changes are shown relative to ATG8 expression levels in GMM. Values are the mean of three independent replicates. Error bars are SD. In summary, the results presented above suggest that TKL1 and ATP influence the expression of at least some known TOR read-out genes during biotrophic growth. How ATP affects the TOR signalling pathway, and the implications for biotrophy, require future elucidation.
Discussion
M. oryzae is a serious threat to world rice harvests and spends most of its infection cycle as a symptomless biotroph growing cell-to-cell in rice leaves before the onset of necrosis. Compared to appressorial development on the host surface, little is known about the metabolic and physiological demands of the fungus during in planta growth [25]. Improving our basic understanding of the biology of biotrophy might contribute to devising effective, long-term resistance strategies using novel chemical or biological interventions. Here, we aimed to add to our knowledge of M. oryzae by describing how some glucose-metabolizing pathways contribute to rice blast disease. In doing so, we revealed that TKL1, encoding transketolase, is essential for pathogenicity and controls biotrophic growth in rice cells via a mechanism involving ATP, TOR and the regulation of cell cycle progression.
This work started with the hypothesis that M. oryzae undergoes metabolic reprogramming during infection, switching from lipid metabolism on the surface of the leaf during appressorium formation to glucose metabolism in host cells. On the leaf surface, β-oxidation and the glyoxylate cycle are required for developing infection-competent appressoria [16], [17], but at least the glyoxylate enzyme isocitrate lyase is not required for growth in planta [16]. In contrast, our genetic dissections of glucose metabolism in M. oryzae show that early glycolysis, late gluconeogenesis and the PPP are not required for appressorium formation or function on the rice leaf surface (Figure 3B and C). Rather, glucose utilization through transketolase (but not early glycolysis or late gluconeogenesis) is essential for growth in host cells (Figure 3A and D). Glucose utilization through transketolase likely occurs in the direction of glycolysis because ATP levels are reduced in the mycelia of Δtkl1 strains compared to Guy11 (Figure 3H). Together, the genetic and biochemical evidence point to transketolase being important in connecting the PPP with glycolysis during in planta growth (Mode 4, Figure 1).
The critical post-penetration, in planta-specific role of TKL1 indicates that once in the host cell, M. oryzae metabolism might be dedicated to NADPH and ATP production from glucose. NADPH is likely important, amongst other processes, for recycling antioxidation systems [40], [41] and maintaining redox balance during biotrophic growth [24]; ATP is likely required for meeting the energetic demands of the fungus in planta. A major and novel finding of this work is that ATP produced via Tkl1 can also act as a signal, likely upstream of the TOR pathway, to promote cell cycle progression and trigger the biotrophic growth of M. oryzae in planta. Delayed mitotic progression was reversed by the addition of exogenous ATP, but not unrelated compounds, to Δtkl1 spores, thus providing a functional connection between TKL1, ATP and TOR. Indeed, exogenous ATP was shown to override the specific TOR kinase inhibitor rapamycin. Although we do not know how ATP acts on TOR, these results are consistent with observations in mammalian cells, where inactivated mTOR promotes cellular quiescence while mTOR activation is dependent on the monitoring of ATP levels, either directly [42] or via the AMP-sensing TOR regulator AMPK [43]. Therefore, one explanation for the results presented here could be that, following the transition of the fungus into the host cell, TKL1 is required to metabolize glucose and provide an ATP signal that mediates a metabolic checkpoint - via TOR - in order to control the cell cycle and promote hyphal growth in planta. Metabolic checkpoints sense metabolites to regulate cellular functions and have recently been implicated in cell cycle quiescence in hematopoietic stem cells [44]. However, although TOR can control quiescence in yeast [33], [34], Δtkl1 strains are likely in a slow growing rather than quiescent state (Figure 3F and 3G). Moreover, in addition to extending G1, inactivating TOR can affect the G2/M transition, resulting in G2-delay in yeast [33], [45]. Therefore, we cannot state at this time where in the cell cycle of M. oryzae the TOR pathway regulates mitosis in response to ATP, except that its abrogation results in mitotic delay.
Additional evidence that TKL1 mediates an in planta-specific metabolic checkpoint in M. oryzae comes from the observations that outside the host plant, TKL1 was not required for cell cycle regulation or autophagic cell death of the spore during appressorium formation and rice leaf penetration. Instead, TKL1 (or exogenous ATP treatment of Δtkl1 strains) was required to maintain a punctate appressorial nucleus and ensure the correct timing of nuclear migration from the appressorium into the primary hyphae in the plant. H1:RFP was not localized to a punctate nuclear structure in Δtkl1 H1:RFP strains following host penetration at 21 hpi and was instead diffused throughout the appressorium. In fibroblasts, different histone H1 subtypes have different cellular localizations during cell division. For example, Histone H1.2 is associated with chromatin during prophase but cytoplasmically localized during metaphase and early anaphase [46]. Histone H1.5 is partitioned between chromatin and cytoplasm during metaphase and early anaphase [46]. In addition, histone H1 diffusion dynamics in HeLa cells are affected by ATP depletion [47]. Thus, H1:RFP might be localized throughout the appressorium in Δtkl1 H1:RFP strains at 21 hpi due to altered cell cycle progression and/or altered histone dynamics in response to ATP (compared to Guy11 H1:RFP at 21 hpi). This hypothesis remains to be tested.
In Δtkl1 strains in the absence exogenous ATP, at least one nucleus has migrated into the IH of Δtkl1 mutant strains by at least 32 hpi (Figure 5), suggesting the TKL1-dependent metabolic checkpoint is reversible and perhaps responds to other metabolites. Reversibility is a hallmark of slow growth and quiescence and distinguishes it from other non-growing cell states such as senescence, apoptosis or terminal differentiation [31].
Taken together, our data suggests the following testable model of infection. Appressorial formation occurs under the nutrient-starvation conditions found on the host surface and requires autophagic cell death of the spore and recycling of the spore contents into the incipient appressorium [8]. β-oxidation and the glyoxylate cycle are the dominant metabolic pathways during this stage of development [25]. A single mitotic cell cycle event occurs in the germ tube and one daughter nucleus migrates into the incipient appressorium [9]. The remaining conidial nuclei are degraded during autophagy [8], resulting in a sole appressorial nucleus. Following host penetration, and perhaps in response to G6P sensing by Tps1 [25], M. oryzae metabolism switches to glucose metabolism through the PPP and transketolase, resulting in NADPH production for redox and ATP production for satisfying the energetic demands of the growing fungus. ATP is also a signal, likely acting via TOR pathway activation, to control the migration of the appressorial nucleus into primary hyphae in the host cell. Once IH have developed in the host cell, transketolase is further required to propagate the ATP signal and, via TOR pathway activation, prevent delayed mitotic progression in order to permit vigorous cell-to-cell growth in planta. Deleting TKL1 or treating spores with the non-hydrolysable ATP analogue AMP-PNP does not impact appressorium development but subsequent infection steps are delayed or abolished.
The work presented here has highlighted mechanisms controlling the transition from appressorial development to biotrophic growth in rice cells, but important questions remain. We wish to understand 1) whether ATP affects TOR signaling due to a direct interaction of ATP and the Tor1 kinase, or indirectly due to an interaction between ATP and additional TOR interacting protein(s) or pathways; 2) how the germ tube mitotic cell cycle event occurs during autophagy when TOR signaling is presumably inactive, whereas nuclear division in IH requires an activated TOR pathway; 3) what is the relationship between autophagy in the spore, G6P sensing by Tps1 in planta, and the TOR signaling pathway; 4) how does G6P sensing by Tps1 switch carbon metabolism from β-oxidation in mitochondria and peroxisomes to glucose metabolism through the PPP in the cytoplasm. Addressing these points will require further, detailed explorations of the biology of M. oryzae biotrophy.
In addition to shedding new light on the metabolic strategies governing rice infection, the characterizations of TKL1 function in M. oryzae might also have important clinical relevance. Metabolizing glucose through the PPP and transketolase to generate NADPH is a metabolic strategy observed in cancer cells [48], activated macrophages [49] and the growth of T cells in response to pathogen challenge [50], [51]. Similar to our observations with M. oryzae, the transition of naive T cells to actively growing T cells is accompanied by a metabolic shift from β-oxidation to glucose utilization via the PPP [50], [51]. Moreover, transketolase, which controls the PPP along with G6PDH [52], is upregulated in certain tumors [53] and has been implicated in metastasis [52]. Transketolase inhibitors have been shown to reduce the rate of proliferation of pancreatic adenocarcinoma cells in culture [54] while, conversely, stimulating transketolase activity in cancer cells using thiamine promoted tumor growth in mice [55]. Thus, switching to glucose utilization via transketolase might be a conserved feature of proliferating cells. Consequently, small-molecule inhibitors of transketolase, such as those developed as anti-cancer therapies [54], [56], could hold promise for combating recalcitrant intracellular fungal pathogens.
The work presented here also contributes to our understanding of metabolic checkpoints by revealing a previously unknown functional relationship between TKL1 and TOR that provides new information on the regulation of the TOR pathway. This connection might have importance beyond the field of plant pathogenicity due to the role activated TOR plays in some cancers and other serious disorders such as cardiovascular disease [57]. Moreover, metabolic checkpoints are required for T cell differentiation and immune responses [51], and this requires signaling through a metabolic checkpoint mediated by the mTOR pathway [51].
In conclusion, this study gives fresh insights into the metabolic strategies controlling, and committing, intracellular pathogens to biotrophic growth. This knowledge is likely applicable to a wide range of important fungal pathogens. Moreover, our characterization of transketolase as a metabolic checkpoint mediator might inform other areas of biology where this enzyme and its downstream targets are important, such as during cancer cell proliferation.
Materials and Methods
Ex planta culture conditions and physiological analyses
All strains used in this study are stored in the Wilson lab. Guy11 was used as the wild-type isolate [58] and all mutant strains described in this study were generated from the Guy11 parental strain (Table S1). M. oryzae was cultured and stored using standard procedures [37]. Strains were grown on complete medium (CM) or Cove's minimal media (MM), as described previously [59]. Glucose was used in MM at a final concentration of 1% w/v. Nitrogen sources were used in MM at a final concentration of 10 mM. Physiological analyses were performed on media as described previously [24]. Plates were imaged with a Sony Cyber-shot digital camera, 14.1 mega pixels, after 10 days of growth. For sporulation rates, strains were grown on at least three independent CM plates for 12 days before the spores were harvested and spore concentrations were measured using a hemocytometer (Corning). Appressorium developmental assays were performed on hydrophobic microscope coverslips (Fisherbrand). Spores were harvested and diluted into 1×105 spores ml−1 in sterile distilled water. 200 µl of spore suspension was placed on three plastic coverslips per strain and placed in a plastic box with a wet paper towel on the bottom simulating a humid chamber. After 24 hr of incubation, the number of appressoria formed from 50 spores was determined for each replicate, and an average percent value generated. The non-hydrolysable ATP analogue adenosine 5′-adenylyl imidodiphosphate (AMP-PNP) was purchased from Sigma and added to Guy11 spores to a final concentration of 5 mM. The NADPH oxidase inhibitor diphehyleneiodonium chloride was purchased from Sigma and added to Guy11 spores to a final concentration of 0.4 mM, following [60].
Homologous gene replacement strategies
Transformation-competent protoplasts were produced as previously described [22]. Guy11 was grown in liquid CM with agitation at 150 rpm for 48 hr. Mycelia was harvested and treated with lytic enzymes (Glucanex, Sigma) for 3 hr at 29°C in order to produce protoplasts. All targeted gene deletions mentioned in this study were generated using the split marker approach in which a selectable marker replaces the native gene of interest in the Guy11 genome (following [22]). PGI1, and FBP1 genes were replaced in Guy11 with the ILV1 gene conferring resistance to sulphonyl urea [22]. TKL1 was replaced in Guy11 with the Bar gene confirming resistance to bialaphos, and replaced in Guy11 H1:RFP with the ILV1 gene conferring resistance to sulphonyl urea [22]. In all cases, primers were designed to amplify a 1 kb sequence upstream and a 1 kb sequence downstream of the gene of interest (Table S2). These primers were used in a first round PCR reaction, which amplified both the right and left flanks of the gene of interest. The thermocycler conditions for the first round were 1 min at 95°C initial denaturation, followed by 34 cycles of 95°C for 30 sec denaturation, 63°C for 30 sec annealing and 68°C for 1 min extension. A second round of PCR was conducted, in which each flanking region of the gene of interest was fused to one of two overlapping pieces of the ILV1 or Bar gene. For the second round PCR, similar thermocycler conditions as first round were used except for a 3 min extension time. The resulting PCR products were transformed directly into protoplasts of M. oryzae. Homologous gene replacements by the ILV1 or Bar resistance markers were initially selected on the basis of sulphonyl urea or bialaphhos resistance, respectively. Strains carrying homologous gene replacement of the gene of interest were identified by PCR as described by Wilson et al. [22] using the oligonucleotide primers shown in Table S2. The conditions were 2 min at 95°C initial denaturation, followed by 35 cycles of 95°C for 30 sec denaturation, 63°C for 1 min annealing and 68°C for 5 min extension. A minimum of two transformants was analyzed per gene of interest. To ensure that the observed phenotype for Δtkl1 mutant strains were solely the result of TKL1 deletion, a TKL1 complementation vector was constructed using the primers in Table S2, following the protocol outlined in [24], and introduced into Δtkl1 mutant strains to restore virulence.
Rice blast pathogenicity assays
Rice blast pathogenicity assays were performed as described previously [37]. Briefly, three to four week old rice seedlings from a susceptible cultivar, CO-39, were spray inoculated with spore suspensions at a rate of 1×105 spores ml−1 in a 0.2% gelatin (Difco) solution. Plants were placed in a growth chamber with 12 hr light/dark periods. After five days, the infected leaves were collected and scored for disease symptoms. Images of the infected leaves were taken by using an Epson Workforce scanner at a resolution of 600 dpi.
In planta physiological analyses
Live-cell imaging of fungal colonization of rice epidermal cells was achieved using detached rice leaf sheaths from the susceptible cultivar CO-39. Leaf sheaths were inoculated with fungal spores (1×105 spores ml−1 in 0.20% gelatin) in the hollow interior of the sheaths as described previously [37]. Infected sheaths were kept horizontal in a glass container with humid conditions for up to 48 hr. Starting at 24 hpi, the rice sheaths were excised and observed under a light microscope (Zeiss AxioSkop). For each strain, appressorium development from 50 spores was measured on each of three independent leaf surfaces at 24 hpi to obtain an average rate of appressorium formation. Appressorium penetration rates at 30 hpi, and IH growth rates and movement to adjacent cells at 48 hpi, were determined from fifty appressoria per treatment, repeated in triplicate, following [59]. IH growth rates were determined using a 4-point scale where 1 = IH length shorter than 10 µm with no branching; 2 = IH length is 10–20 µm with 0–2 branches; 3 = IH length is longer than 20 µm and/or with more than 2 branches within one cell; 4 = IH has spread to adjacent cells. Images were taken using a Nikon A1 laser scanning confocal mounted on a Nikon 90i compound microscope (software version: NIS Elements 4.13 Build914) at the University of Nebraska-Lincoln Microscopy Center. A 1.5 zoom Z series step (1 µm) was used in the 60× lens. Transmitted light and fluoresce for td tomato were imaged with a 561.5 nm laser. td tomato fluoresce was detected at 570–620 nm. The number of nuclei per 10 µm of IH within infected rice cells was quantified using ImageJ software (rsbweb.nih.gov/ij). For all the images, the scale was set (Analyze-Set scale) and a 10 µm line was drawn inside the hyphae (Analyze-Measure). The number of nuclei along the 10 µm line was counted for each strain at 32, 48 and 65 hpi. For each timepoint, six independent replications were analyzed.
Gene transcript analysis
Total RNA from infected leaf tissue or fungal mycelium was extracted using the RNeasy Plant Mini Kit from Qiagen. For leaf RNA extractions, deteached rice leaf sheaths were inoculated with 1×105 spores ml−1 of the appropriate strain, isolated at 48 hpi, frozen in liquid nitrogen and ground in a mortar with a pestle. For mycelial RNA extraction, strains were grown in CM for 48 hr before switching to Cove's minimal media containing 5 nM rapamycin, 5 mM rapamycin +5 mM ATP, or no treatment, for 16 hr following [23]. Mycelia was harvested, frozen in liquid nitrogen, and lyophilized for 24 hr. A total of 100 mg of each mycelial sample was used to perform RNA extractions. RNA from mycelia or rice leaf tissue was treated with DNase I (Invitrogen) and converted to cDNA using the qScript reagents from Quantas. The cDNA reactions were performed in 20 µl reaction volumes containing 1× of qScrip cDNA Super Mix (5×) and 1–10 µg of total treated RNA. cDNA synthesis conditions were: 5 min at 25°C, 30 min at 42°C and 5 min at 85°C. After completion of cDNA synthesis, the first reaction was diluted 5 fold for PCR amplification. The resulting cDNA was analyzed by quantitative real-time PCR (qPCR) in an Eppendorf Mastercycler ep Realplex real-time PCR system. Reactions were performed in a 25 µl reaction containing 12.5 µl 2× Quantifast SYBR Green PCR Master Mix (Qiagen), 100 nM of oligonucleotide primers and 2.5 µl (≤100 ng) of cDNA. The primer sequences are provided in Table S2. Thermocycler conditions were: 5 min at 95°C initial denaturation, followed by 40 cycles of 95°C for 30 sec denaturation, 63°C for 30 sec annealing and 72°C for 1 min extension. Gene expression of each gene was normalized against the expression levels of either the M. oryzae actin gene (MoACT1) for in planta analysis or the M. oryzae β-tubulin gene (TUB2) for mycelial transcript analysis, as described previously [23]. Results are the average of three technical replications and at least two biological replications.
Nucleotide quantification
The analysis of nucleotides in mycelial samples was performed using LC-MS/MS by separation of the nucleotides using hydrophilic interaction chromatography (HILIC). Samples of ground, lyophilized mycelia were weighed at about 10 mg per sample in seal proof Eppendorf tubes and extracted at 4°C with 0.6 mL of 80% MeOH/20% H2O (both LC-grade, Fisher Scientific). The samples were subjected to extraction at room temperature using a Bullet Blender Homogenizer (Averill Park, NY) after the addition of ∼100 µL of zirconia beads. The samples were then centrifuged for 10 minutes, the supernatants were removed, and a second extraction with 0.4 mL of 50% MeOH/50% H2O was performed by the same procedure. After centrifugation of the samples, aliquots were transferred into plastic LC-vials capped with sealed septums and loaded onto the LC-MS/MS system. The latter consisted on an AbSciex 4000 Qtrap Hybrid LC-MS/MS system (Framingham, MA) operating in triple quad mode using a multiple reaction monitoring (MRM) method. The instrument was interfaced to an Agilent LC1200 which included an autosampler with the samples thermostated at 4°C. The nucleotides were separated using a Phenomenex (Torrance, CA) Luna-NH2 column with dimensions of 2×250 mm; 100 Å pore size and 5 µm particle size. The MRM transitions were monitored in positive mode following [61]. The transitions were optimized with pure nucleotide solutions by infusion of 10 µM solutions directly into the mass spectrometer, followed by final ion source optimization by loop injection of those solutions. Stock solutions of ATP were prepared and quantified by their UV/visible spectra using a Cary-100 spectrophotometer with available extinction coefficients. External calibration curves were generated for 2 transitions of the nucleotide by performing injections of serial dilutions of the stock using the HILIC column interfaced with the LC-MS/MS system. Plots of integrated areas vs. Concentration in the range from 50–1000 pmoles yielded linear fits with correlation coefficients >0.99. The amounts of nucleotide was estimated from the integration of the more predominant MRM transition by comparison with the calibration curve and normalized by the total weight of the samples. Reinjection of the samples after 24 hr yielded the same areas, indicating that no significant hydrolysis of the nucleotides was occurring during LC runs. All solvents were purchased from J.T. Baker and were of LC-MS Grade Purity.
Supporting Information
Zdroje
1. WilsonRA, TalbotNJ (2009) Under pressure: investigating the biology of plant infection by Magnaporthe oryzae. Nat Rev Microbiol 7 : 185–195.
2. FernandezJ, WilsonRA (2012) Why no feeding frenzy? Mechanisms of nutrient acquisition and utilization during Infection by the rice blast fungus Magnaporthe oryzae. Mol Plant-Microbe Interact 25 : 1286–1293.
3. FisherMC, HenkDA, BriggsCJ, BrownsteinJS, MadoffLC, et al. (2012) Emerging fungal threats to animal, plant and ecosystem health. Nature 484 : 186–194.
4. KupferschmidtK (2012) Attack of the clones. Science 337 : 636–638.
5. DagdasYF, YoshinoK, DagdasG, RyderLS, BielskaE, et al. (2012) Septin-mediated plant cell invasion by the rice blast fungus, Magnaporthe oryzae. Science 336 : 1590–1595.
6. RyderLS, DagdasYF, MentlakTA, KershawMJ, ThorntonCR, et al. (2013) NADPH oxidases regulate septin-mediated cytoskeletal remodeling during plant infection by the rice blast fungus. Proc Natl Acad Sci USA 110 : 3179–3184.
7. SaundersDG, DagdasYF, TalbotNJ (2010) Spatial uncoupling of mitosis and cytokinesis during appressorium-mediated plant Infection by the rice blast fungus Magnaporthe oryzae. Plant Cell 22 : 2417–2428.
8. Veneault-FourreyC, BarooahM, EganM, WakleyG, TalbotNJ (2006) Autophagic fungal cell death is necessary for infection by the rice blast fungus. Science 312 : 580–583.
9. SaundersDG, AvesSJ, TalbotNJ (2010) Cell cycle-mediated regulation of plant infection by the rice blast fungus. Plant Cell 22 : 497–507.
10. HowardRJ, ValentB (1996) Breaking and entering: Host penetration by the fungal rice blast pathogen Magnaporthe grisea. Annu Rev Microbiol 50 : 491–512.
11. de JongJC, McCormackBJ, SmirnoffN, TalbotNJ (1997) Glycerol generates turgor in rice blast. Nature 389 : 244–244.
12. KankanalaP, CzymmekK, ValentB (2007) Roles for rice membrane dynamics and plasmodesmata during biotrophic invasion by the blast fungus. Plant Cell 19 : 706–724.
13. ThinesE, WeberRW, TalbotNJ (2000) MAP kinase and protein kinase A-dependent mobilization of triacylglycerol and glycogen during appressorium turgor generation by Magnaporthe grisea. Plant Cell 12 : 1703–1718.
14. WangZY, JenkinsonJM, HolcombeLJ, SoanesDM, Veneault-FourreyC, et al. (2005) The molecular biology of appressorium turgor generation by the rice blast fungus Magnaporthe grisea. Biochem Soc Trans 33 : 384–388.
15. WangZY, SoanesDM, KershawMJ, TalbotNJ (2007) Functional analysis of lipid metabolism in Magnaporthe grisea reveals a requirement for peroxisomal fatty acid beta-oxidation during appressorium-mediated plant infection. Mol Plant-Microbe Interact 20 : 475–491.
16. WangZY, ThorntonCR, KershawMJ, DebaoL, TalbotNJ (2003) The glyoxylate cycle is required for temporal regulation of virulence by the plant pathogenic fungus Magnaporthe grisea. Mol Microbiol 47 : 1601–1612.
17. BhambraGK, WangZY, SoanesDM, WakleyGE, TalbotNJ (2006) Peroxisomal carnitine acetyl transferase is required for elaboration of penetration hyphae during plant infection by Magnaporthe grisea. Mol Microbiol 61 : 46–60.
18. Ramos-PamplonaM, NaqviNI (2006) Host invasion during rice-blast disease requires carnitine-dependent transport of peroxisomal acetyl-CoA. Mol Microbiol 61 : 61–75.
19. BhadauriaV, BannizaS, VandenbergA, SelvarajG, WeiY (2012) Peroxisomal alanine: glyoxylate aminotransferase AGT1 is indispensable for appressorium function of the rice blast pathogen, Magnaporthe oryzae. PLOS One 7: e36266.
20. PatkarRN, Ramos-PamplonaM, GuptaAP, FanY, NaqviNI (2012) Mitochondrial β-oxidation regulates organellar integrity and is necessary for conidial germination and invasive growth in Magnaporthe oryzae. Mol Microbiol 86 : 1345–1363.
21. WilsonRA, JenkinsonJM, GibsonRP, LittlechildJA, WangZY, et al. (2007) Tps1 regulates the pentose phosphate pathway, nitrogen metabolism and fungal virulence. EMBO J 26 : 3673–3685.
22. WilsonRA, GibsonRP, QuispeCF, LittlechildJA, TalbotNJ (2010) An NADPH-dependent genetic switch regulates plant infection by the rice blast fungus. Proc Natl Acad Sci USA 107 : 21902–21907.
23. FernandezJ, WrightJD, HartlineD, QuispeCF, MadayiputhiyaN, et al. (2012) Principles of carbon catabolite repression in the rice blast fungus: Tps1, Nmr1–3, and a MATE–Family Pump regulate glucose metabolism during Infection. PLoS Genet 8: e1002673.
24. FernandezJ, WilsonRA (2014) Characterizing roles for the glutathione reductase, thioredoxin reductase and thioredoxin peroxidase-encoding genes of Magnaporthe oryzae during rice blast disease. PLoS One 9: e87300.
25. FernandezJ, WilsonRA (2014) Cells in cells: morphogenetic and metabolic strategies conditioning rice infection by the blast fungus Magnaporthe oryzae. Protoplasma 251 : 37–47.
26. CollerHA (2011) The essence of quiescence. 334 : 1074–1075.
27. KlosinskaMM, CrutchfieldCA, BradleyPH, RabinowitzJD, BroachJR (2012) Yeast cells can access distinct quiescent states. Genes Dev 25 : 336–349.
28. Berg JM, Tymoczko JL, Stryer L (2011) Biochemistry. Basingstoke, NY: W.H. Freeman & Co Ltd.
29. DeanRA, TalbotNJ, EbboleDJ, FarmanML, MitchellTK, et al. (2005) The genome sequence of the rice blast fungus Magnaporthe grisea. Nature 434 : 980–986.
30. LuC, BrauerMJ, BotsteinD (2009) Slow growth induces heat-shock resistance in normal and respiratory-deficient yeast. Mol Biol Cell 20 : 891–903.
31. CollerHA, SangL, RobertsJM (2006) A new description of cellular quiescence. PLoS Biol 4: e83.
32. BarbetNC, SchneiderU, HelliwellSB, StansfieldI, TuiteMF, et al. (1996) TOR controls translation initiation and early G1 progression in yeast. Mol Biol Cell 7 : 25–42.
33. LoewithR, HallMN (2011) Target of rapamycin (TOR) in nutrient signaling and growth control. Genetics 189 : 1177–1201.
34. De VirgilioC (2012) The essence of yeast quiescence. FEMS Microbiol Rev 36 : 306–339.
35. HardwickJS, KuruvillaFG, TongJK, ShamjiAF, SchreiberSL (1999) Rapamycin-modulated transcription defines the subset of nutrient-sensitive signaling pathways directly controlled by the Tor proteins. Proc Natl Acad Sci U S A 96 : 14866–14870.
36. Ramírez-ValleF, BraunsteinS, ZavadilJ, FormentiSC, SchneiderRJ (2008) eIF4GI links nutrient sensing by mTOR to cell proliferation and inhibition of autophagy. J Cell Biol 181 : 293–307.
37. FernandezJ, YangKT, CornwellKM, WrightJD, WilsonRA (2013) Growth in rice cells requires de novo purine biosynthesis by the blast fungus Magnaporthe oryzae. Sci Rep 3 : 2398.
38. ShanerNC, CampbellRE, SteinbachPA, GiepmansBN, PalmerAE, et al. (2004) Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol 22 : 1567–1572.
39. FranceschettiM, BuenoE, WilsonRA, TuckerSL, Gómez-MenaC, et al. (2011) Fungal virulence and development is regulated by alternative pre-mRNA 3′ end processing in Magnaporthe oryzae. PLoS Pathog 7: e1002441.
40. PollakN, DölleC, ZieglerM (2007) The power to reduce: pyridine nucleotides – small molecules with a multitude of functions. Biochem J 402 : 205–218.
41. YingW (2008) NAD+/NADH and NADP+/NADPH in cellular functions and cell death: regulation and biological consequences. Antioxidants & redox signaling 10 : 179–206.
42. DennisPB, JaeschkeA, SaitohM, FowlerB, KozmaSC, et al. (2001) Mammalian TOR: a homeostatic ATP sensor. Science 294 : 1102–1105.
43. GwinnDM, ShackelfordDB, EganDF, MihaylovaMM, MeryA, et al. (2008) AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell 30 : 214–226.
44. TakuboK, NagamatsuG, KobayashiCI, Nakamura-IshizuA, KobayashiH, et al. (2013) Regulation of glycolysis by Pdk functions as a metabolic checkpoint for cell-cycle quiescence in hematopoietic stem cells. Cell Stem Cell 12 : 49–61.
45. NakashimaA, MarukiY, ImamuraY, KondoC, KawamataT, et al. (2008) The Yeast Tor Signaling Pathway Is Involved in G2/M Transition via Polo - Kinase. PLoS ONE 3 (5) e2223.
46. GréenA, LönnA, PetersonKH, OllingerK, RundquistI (2010) Translocation of histone H1 subtypes between chromatin and cytoplasm during mitosis in normal human fibroblasts. Cytometry A 77 (5) 478–484.
47. BhattacharyaD, MazumderA, MiriamSA, ShivashankarGV (2006) EGFP-tagged core and linker histones diffuse via distinct mechanisms within living cells. Biophys J 91 : 2326–2336.
48. SchulzeA, HarrisAL (2012) How cancer metabolism is tuned for proliferation and vulnerable to disruption. Nature 491 : 364–373.
49. O'NeillLAJ, HardieDG (2013) Metabolism of inflammation limited by AMPK and pseudo-starvation. Nature 493 : 346–355.
50. GerrietsVA, RathmellJC (2012) Metabolic pathways in T cell fate and function. Trends Immunol 33 : 168–173.
51. WangR, GreenDR (2012) Metabolic checkpoints in activated T cells. Nat Immunol 13 : 907–915.
52. Diaz-MoralliS, Tarrado-CastellarnauM, AlendaC, CastellsA, CascanteM (2011) Transketolase-like 1 expression is modulated during colorectal cancer progression and metastasis formation. PLOS One 6: e25323.
53. LangbeinS, ZerilliM, Zur HausenA, StaigerW, Rensch-BoschertK, et al. (2006) Expression of transketolase TKTL1 predicts colon and urothelial cancer patient survival: Warburg effect reinterpreted. Br J Cancer 94 : 578–585.
54. BorosLG, PuigjanerJ, CascanteM, LeeWN, BrandesJL, et al. (1997) Oxythiamine and dehydroepiandrosterone inhibit the nonoxidative synthesis of ribose and tumor cell proliferation. Cancer Res 57 : 4242–4248.
55. Comín-AnduixB, BorenJ, MartinezS, MoroC, CentellesJJ, et al. (2001) The effect of thiamine supplementation on tumour proliferation. A metabolic control analysis study. Eur J Biochem 268 : 4177–4182.
56. DuMX, SimJ, FangL, YinZ, KohS, et al. (2004) Identification of novel small-molecule inhibitors for human transketolase by high-throughput screening with fluorescent intensity (FLINT) assay. J Biomol Screen 9 : 427–433.
57. WullschlegerS, LoewithR, HallMN (2006) TOR signaling in growth and metabolism. Cell 124 : 471–484.
58. TalbotNJ, EbboleDJ, HamerJE (1993) Identification and characterization of MPG1, a gene involved in pathogenicity from the rice blast fungus Magnaporthe grisea. Plant Cell 5 : 1575–1590.
59. WilsonRA, FernandezJ, QuispeCF, GradnigoJ, SengA, et al. (2012) Towards defining nutrient conditions encountered by the rice blast fungus during host infection. PLoS ONE 7: e47392.
60. ChiMH, ParkSY, KimS, LeeYH (2009) A novel pathogenicity gene is required in the rice blast fungus to suppress the basal defenses of the host. PLoS Pathogen 5: e1000401.
61. BajadS, ShulaevV (2011) LC-MS-based metabolomics. Methods Mol Biol 708 : 213–228.
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Out-of-Sequence Signal 3 as a Mechanism for Virus-Induced Immune Suppression of CD8 T Cell ResponsesČlánek RNF26 Temporally Regulates Virus-Triggered Type I Interferon Induction by Two Distinct MechanismsČlánek Mouse, but Not Human, ApoB-100 Lipoprotein Cholesterol Is a Potent Innate Inhibitor of Pneumolysin
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2014 Číslo 9- Stillova choroba: vzácné a závažné systémové onemocnění
- Jak souvisí postcovidový syndrom s poškozením mozku?
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Infekční komplikace virových respiračních infekcí – sekundární bakteriální a aspergilové pneumonie
-
Všechny články tohoto čísla
- Virus Control Goes Epigenetic
- The Role of Iron in Prion Disease and Other Neurodegenerative Diseases
- The Ins and Outs of Rust Haustoria
- Prion Strains and Amyloid Polymorphism Influence Phenotypic Variation
- Teaching Fido New ModiFICation Tricks
- Can Enhance Infection in Mosquitoes: Implications for Malaria Control?
- MIF Contributes to Associated Immunopathogenicity Development
- Persistence of Virus Reservoirs in ART-Treated SHIV-Infected Rhesus Macaques after Autologous Hematopoietic Stem Cell Transplant
- Bacillus Calmette-Guerin Infection in NADPH Oxidase Deficiency: Defective Mycobacterial Sequestration and Granuloma Formation
- EhCoactosin Stabilizes Actin Filaments in the Protist Parasite
- Molecular Insights Into the Evolutionary Pathway of O1 Atypical El Tor Variants
- LprG-Mediated Surface Expression of Lipoarabinomannan Is Essential for Virulence of
- Structural Correlates of Rotavirus Cell Entry
- Multivalent Adhesion Molecule 7 Clusters Act as Signaling Platform for Host Cellular GTPase Activation and Facilitate Epithelial Barrier Dysfunction
- The Effects of Vaccination and Immunity on Bacterial Infection Dynamics
- Myeloid Derived Hypoxia Inducible Factor 1-alpha Is Required for Protection against Pulmonary Infection
- Functional Characterisation of Germinant Receptors in and Presents Novel Insights into Spore Germination Systems
- Global Analysis of Neutrophil Responses to Reveals a Self-Propagating Inflammatory Program
- Host Cell Invasion by Apicomplexan Parasites: The Junction Conundrum
- Comparative Phenotypic Analysis of the Major Fungal Pathogens and
- Unravelling the Multiple Functions of the Architecturally Intricate β-galactosidase, BgaA
- Sialylation of Prion Protein Controls the Rate of Prion Amplification, the Cross-Species Barrier, the Ratio of PrP Glycoform and Prion Infectivity
- Symbionts Commonly Provide Broad Spectrum Resistance to Viruses in Insects: A Comparative Analysis of Strains
- Ontogeny of Recognition Specificity and Functionality for the Broadly Neutralizing Anti-HIV Antibody 4E10
- Identification and Characterisation of a Hyper-Variable Apoplastic Effector Gene Family of the Potato Cyst Nematodes
- Crimean-Congo Hemorrhagic Fever Virus Entry into Host Cells Occurs through the Multivesicular Body and Requires ESCRT Regulators
- Age-Dependent Enterocyte Invasion and Microcolony Formation by
- CD160-Associated CD8 T-Cell Functional Impairment Is Independent of PD-1 Expression
- Functional Fluorescent Protein Insertions in Herpes Simplex Virus gB Report on gB Conformation before and after Execution of Membrane Fusion
- The Tudor Domain Protein Spindlin1 Is Involved in Intrinsic Antiviral Defense against Incoming Hepatitis B Virus and Herpes Simplex Virus Type 1
- Transgenic Analysis of the MAP Kinase MPK10 Reveals an Auto-inhibitory Mechanism Crucial for Stage-Regulated Activity and Parasite Viability
- Evidence for a Transketolase-Mediated Metabolic Checkpoint Governing Biotrophic Growth in Rice Cells by the Blast Fungus
- Incomplete Deletion of IL-4Rα by LysM Reveals Distinct Subsets of M2 Macrophages Controlling Inflammation and Fibrosis in Chronic Schistosomiasis
- Identification and Functional Expression of a Glutamate- and Avermectin-Gated Chloride Channel from , a Southern Hemisphere Sea Louse Affecting Farmed Fish
- Out-of-Sequence Signal 3 as a Mechanism for Virus-Induced Immune Suppression of CD8 T Cell Responses
- Strong Epistatic Selection on the RNA Secondary Structure of HIV
- Hematopoietic but Not Endothelial Cell MyD88 Contributes to Host Defense during Gram-negative Pneumonia Derived Sepsis
- Delineation of Interfaces on Human Alpha-Defensins Critical for Human Adenovirus and Human Papillomavirus Inhibition
- Exploitation of Reporter Strains to Probe the Impact of Vaccination at Sites of Infection
- RNF26 Temporally Regulates Virus-Triggered Type I Interferon Induction by Two Distinct Mechanisms
- Helminth Infections Coincident with Active Pulmonary Tuberculosis Inhibit Mono- and Multifunctional CD4 and CD8 T Cell Responses in a Process Dependent on IL-10
- MHC Class II Restricted Innate-Like Double Negative T Cells Contribute to Optimal Primary and Secondary Immunity to
- Reactive Oxygen Species Regulate Caspase-11 Expression and Activation of the Non-canonical NLRP3 Inflammasome during Enteric Pathogen Infection
- Evolution of Plastic Transmission Strategies in Avian Malaria
- A New Human 3D-Liver Model Unravels the Role of Galectins in Liver Infection by the Parasite
- Translocates into the Myocardium and Forms Unique Microlesions That Disrupt Cardiac Function
- Mouse, but Not Human, ApoB-100 Lipoprotein Cholesterol Is a Potent Innate Inhibitor of Pneumolysin
- The Cofilin Phosphatase Slingshot Homolog 1 (SSH1) Links NOD1 Signaling to Actin Remodeling
- Kaposi's Sarcoma Herpesvirus MicroRNAs Induce Metabolic Transformation of Infected Cells
- Reorganization of the Endosomal System in -Infected Cells: The Ultrastructure of -Induced Tubular Compartments
- Distinct Dictation of Japanese Encephalitis Virus-Induced Neuroinflammation and Lethality via Triggering TLR3 and TLR4 Signal Pathways
- Exploitation of the Complement System by Oncogenic Kaposi's Sarcoma-Associated Herpesvirus for Cell Survival and Persistent Infection
- The Secreted Peptide PIP1 Amplifies Immunity through Receptor-Like Kinase 7
- Structural Insight into Host Recognition by Aggregative Adherence Fimbriae of Enteroaggregative
- The CD14CD16 Inflammatory Monocyte Subset Displays Increased Mitochondrial Activity and Effector Function During Acute Malaria
- Infection Induces Expression of a Mosquito Salivary Protein (Agaphelin) That Targets Neutrophil Function and Inhibits Thrombosis without Impairing Hemostasis
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- The Secreted Peptide PIP1 Amplifies Immunity through Receptor-Like Kinase 7
- Symbionts Commonly Provide Broad Spectrum Resistance to Viruses in Insects: A Comparative Analysis of Strains
- MIF Contributes to Associated Immunopathogenicity Development
- The Ins and Outs of Rust Haustoria
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



