-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Microbial Pathogens Trigger Host DNA Double-Strand Breaks Whose Abundance Is Reduced by Plant Defense Responses
Multicellular organisms are continuously exposed to microbes and have developed sophisticated defense mechanisms to counter attack by microbial pathogens. Organisms also encounter many types of DNA damage and have evolved multiple mechanisms to maintain their genomic integrity. Even though these two fundamental responses have been characterized extensively, the relationship between them remains largely unclear. Our study demonstrates that microbial plant pathogens with diverse life styles, including bacteria, oomycete and fungal pathogens, induce double-strand breaks (DSBs) in the genomes of infected host plant cells. DSB induction is apparently a common feature during plant-pathogen interactions. DSBs are the most deleterious form of DNA damage and can lead to chromosomal aberrations and gene mutations. In response to pathogen infection, plant immune responses are activated and contribute to suppressing pathogen-induced DSBs, thereby maintaining better genome integrity and stability. The findings identify important ways that the plant immune and DNA damage repair responses are interconnected. Awareness of the above phenomena may foster future development of disease management approaches that improve crop productivity under biotic stress.
Published in the journal: . PLoS Pathog 10(4): e32767. doi:10.1371/journal.ppat.1004030
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1004030Summary
Multicellular organisms are continuously exposed to microbes and have developed sophisticated defense mechanisms to counter attack by microbial pathogens. Organisms also encounter many types of DNA damage and have evolved multiple mechanisms to maintain their genomic integrity. Even though these two fundamental responses have been characterized extensively, the relationship between them remains largely unclear. Our study demonstrates that microbial plant pathogens with diverse life styles, including bacteria, oomycete and fungal pathogens, induce double-strand breaks (DSBs) in the genomes of infected host plant cells. DSB induction is apparently a common feature during plant-pathogen interactions. DSBs are the most deleterious form of DNA damage and can lead to chromosomal aberrations and gene mutations. In response to pathogen infection, plant immune responses are activated and contribute to suppressing pathogen-induced DSBs, thereby maintaining better genome integrity and stability. The findings identify important ways that the plant immune and DNA damage repair responses are interconnected. Awareness of the above phenomena may foster future development of disease management approaches that improve crop productivity under biotic stress.
Introduction
Organisms continuously encounter many types of DNA damage and have evolved elegant mechanisms to maintain their genomic integrity [1], [2]. DNA damage can be induced by a variety of exogenous stresses such as ultraviolet light or genotoxic chemicals, and by endogenous insults such as reactive oxygen species and DNA replication errors [1]–[3]. DNA double-strand breaks (DSBs) can trigger cell cycle arrest and programmed cell death, and are among the most serious types of DNA damage. Surveillance for DSBs and signaling in response to DSBs are therefore critical for cells to orchestrate DNA repair pathways not only in the germ line but also in somatic tissues, to sustain genome stability and survival of the organism [1], [2].
Pathogen management of their own (microbial) DNA integrity has a long history of study [4], as does the study of interactions between viruses and host DNA damage repair processes [5]. There have been far fewer reports or studies of damage to host DNA caused by microbial pathogens. However, it has recently been established that microbial pathogens of animals can induce host DNA damage [6]–[12].
Multicellular organisms are continuously exposed to microbes and have developed effective immune systems to resist attacks by pathogens [13], [14]. Organisms are challenged to balance the health-promoting impacts of antimicrobial responses and the potential toxic effects on surrounding tissue caused by excessive or chronic inflammation. In animal pathogenesis studies, carcinogenic effects of innate immune responses mediated by Toll-like receptors have been reported [15]. An oxidative burst is a common element of plant and animal antimicrobial responses [14], [16], [17], but reactive oxygen species (ROS) also have well-known DNA damaging activities [3], [18]. There is evidence that a significant component of the host genotoxicity of certain microbial infections in animals is attributable to host-generated ROS [8]–[11]. In plants, the relative contribution of the defense-associated ROS burst to pathogen restriction as opposed to genotoxicity (DNA damage) remains to be explored.
Plant DNA damage repair pathways have received extensive study [19], [20]. Although the tie-ins of plant DNA damage to other aspects of organismal physiology are often plant-specific, many elements of the plant DNA damage repair pathways resemble those of animals due to conservation of core DNA damage repair mechanisms [19], [20]. A hallmark DNA damage response conserved across multicellular organisms is the rapid phosphorylation of histone variant H2AX in the chromatin that flanks break sites, forming γ-H2AX [21], [22]. The phosphatidylinositol 3-kinase-like kinases ATM (ataxia telangiectasia mutated) and ATR (ataxia telangiectasia mutated and Rad3-related) are central mediators of these and other cellular responses to DSBs [23]–[25]. γ-H2AX is one of the most sensitive indicators of DNA DSBs [26].
Associations between DNA damage and plant immune responses have been identified. Exposure of plants to the salicylic acid analogs BTH or INA, or inoculation with the oomycete pathogen H. arabidopsidis, was shown to increase the frequency of somatic homologous recombination [27]. Infection of tobacco and Arabidopsis leaves with either tobacco masaic virus or oilseed rape mosaic virus (ORMV) results in both local and systemic increases in homologous recombination frequency, and ORMV inoculations elicited DNA damage [28], [29]. DNA damaging agents induce pathogenesis-related gene expression [30], [31], and the DNA damage repair proteins RAD51D, BRCA2 and SSN2 are now known to be involved in regulation of gene expression during plant immune responses [32]–[34]. Poly(ADP-ribosyl)ation, a process frequently associated with DNA damage repair, has been shown to impact plant responses to microbial pathogens [35], [36]. Lastly, during the programmed cell death of the hypersensitive response to pathogens expressing effectors (Avr gene products) recognized by a corresponding plant R gene product, increased signal in TUNEL (terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling) assays can be observed, as is also common in animal cell apoptosis [37], [38]. In spite of the above, DNA damage during plant interactions with virulent pathogens is largely undescribed, and whether DNA damage arises during responses activated by core plant defense mediators such as salicylic acid, jasmonic acid or activated microbe-associated molecular pattern (MAMP) receptors also is not known.
Here, we demonstrate that diverse microbial pathogens induce DNA double-strand breaks in host plant genomes. Surprisingly (in light of the mutagenic nature of ROS in many settings), infection-associated AtrbohD - and AtrbohF-dependent ROS production is not required for pathogen-induced elevation of γ-H2AX. Instead, we find that plant antimicrobial defense mechanisms contribute to suppressed formation and/or rapid repair of γ-H2AX-associated lesions. DNA DSB damage is apparently a common aspect of plant pathogenesis by virulent microbial pathogens, and protection against DNA damage is an important feature of effective plant disease resistance.
Results
Induction of DNA double-strand breaks by bacterial pathogens
To investigate interactions between pathogen infection and genome stress, we used γ-H2AX [21], [26] to monitor the extent of DNA damage in response to bacterial pathogens in Arabidopsis. Wild-type Arabidopsis Col-0 plants were challenged with virulent Pseudomonas syringae pv. tomato (Pst) strain DC3000 and levels of γ-H2AX at various time points after infection were determined using an anti-γ-H2AX antibody. Accumulation of γ-H2AX was readily detected as early as 2 h after infiltration, with a progressive increase at the indicated time points after infiltration (Figures 1A and 1B). No γ-H2AX accumulation was observed after mock treatment with 10 mM MgCl2, suggesting that the elevated levels of DNA damage were triggered by the pathogen rather than by any physical perturbations associated with plant inoculation.
Fig. 1. Host DNA damage by Pseudomonas syringae pv. tomato (Pst). 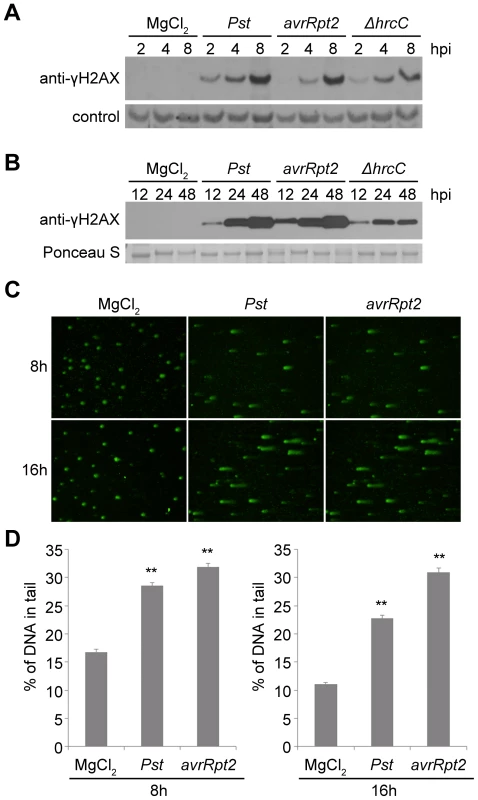
(A–B) Accumulation of γ-H2AX during infection. Wild-type Arabidopsis Col-0 plants were vacuum-inoculated with (left to right) 10 mM MgCl2, Pst DC3000, Pst DC3000(avrRpt2) or Pst DC3000(ΔhrcC) at 1×107 cfu/ml. The level of γ-H2AX was monitored at (A) 2, 4, 8 h, or (B) 12, 24, 48 h after inoculation, by immunoblot using anti-γ-H2AX antibody. Controls for equivalent loading included a non-specific band detected by the antibody (control) or Ponceau S staining of the same blot. Similar results were obtained in at least three separate experiments. (C) Representative Pst-induced DNA damage detected by comet assay. Wild-type Col-0 plants were inoculated with 10 mM MgCl2, or with Pst DC3000 or Pst DC3000(avrRpt2) at 1×107 cfu/ml. Tissues were collected 8 or 16 h after inoculation and nuclei were subjected to comet assays. (D) Comet assay data presented as mean ± SE from at least 200 randomly selected nuclei for each treatment; data for 8 and 16 h are from separate experiments. **: significantly different from MgCl2-treated control (ANOVA P<0.01). We also measured the phosphorylation of H2AX in response to Pst DC3000(avrRpt2), a strain that is isogenic with Pst DC3000 except for its expression of the effector AvrRpt2. AvrRpt2 induces a strong host resistance response (an R gene-mediated incompatible interaction) in plants that express the resistance gene RPS2 [39], [40]. Across four independent experiments, the induction of γ-H2AX levels between 2 and 48 h after inoculation was relatively similar between Pst DC3000(avrRpt2) and Pst DC3000. At the early 2, 4 and 8 h time points, minor differences between the two strains in the γ-H2AX levels induced at the same time after inoculation were not reproducible across experiments. To determine whether the high γ-H2AX accumulation after infection is related to the suite of virulence-promoting bacterial effectors delivered via type III secretion, Arabidopsis Col-0 plants were infected with Pst DC3000(ΔhrcC) that carries a deletion in the hrcC gene that encodes a key component of the type III secretion system [41]. Decreased γ-H2AX accumulation was observed with Pst DC3000(ΔhrcC) 4 to 8 h after infiltration compared with virulent Pst DC3000 treatment, and the difference became statistically significant at 24 and 48 h after infiltration (Figures 1A, 1B and S1). As points of reference, our group and other laboratories have previously found that the onset of Pst DC3000-induced plant cell death is not observed until ∼16–24 h after infection, and Pst DC3000(avrRpt2)-induced hypersensitive response cell death also occurs relatively late, with an onset ∼14–18 h after inoculation [e.g.], [ 42], [43]–[45]. Electrolyte leakage (an early sign of the resistance response) is first detected 5–6 h after inoculation of resistant Arabidopsis with Pst DC3000(avrRpt2) [46], and with Pst DC3000, cytosolic Ca2+ increases are not observed for at least 150 min after inoculation [47], well after the 2 h time point at which γ-H2AX accumulation is first detected (Figure 1A).
Independent evidence that P. syringae pv. tomato infection increases DSBs in Arabidopsis was obtained in comet assays (Figure 1C and 1D). Comet assays measure DNA damage by directly monitoring the increased capacity of fractured DNA to electrophoretically migrate out of isolated nuclei [48], [49]. The roughly comparable levels of DNA damage caused by Pst DC3000(avrRpt2) and Pst DC3000 may be a result of infiltration directly into the leaf interior of the relatively high (1×107 cfu/ml) and equivalent populations of the two pathogen strains. However, these initial results also suggested that stress may be imposed on plant DNA by plant defense responses.
A number of non-pathogenic bacteria were then tested for their ability to induce DNA DSBs in the host plant. When wild-type Arabidopsis Col-0 plants were infiltrated with E. coli strain DH5α at a dose similar to the above Pst inoculations, no γ-H2AX was observed (Figure 2A). Three plant-associated bacterial strains that are not pathogenic on Arabidopsis were also tested: P. syringae pv. glycinea (Psg) is a soybean pathogen that multiplies and produces few visible symptoms when introduced into Arabidopsis leaf mesophyll [50], [51], Psg (avrRpt2) expresses the effector AvrRpt2 and induces R gene-mediated defenses in Arabidopsis Col-0 despite the low virulence of the parent strain [44] and P. fluorescens WCS417r is a biological control strain that has been widely used to trigger induced systemic resistance (ISR) in plants [52], [53]. Similar to E. coli strain DH5α, P. fluorescens WCS417r did not induce detectable levels of γ-H2AX. However, Psg and Psg (avrRpt2) caused elevated phosphorylation of H2AX (Figure 2B).
Fig. 2. Accumulation of γ-H2AX induced by non-pathogenic pathogens and abiotic stresses. 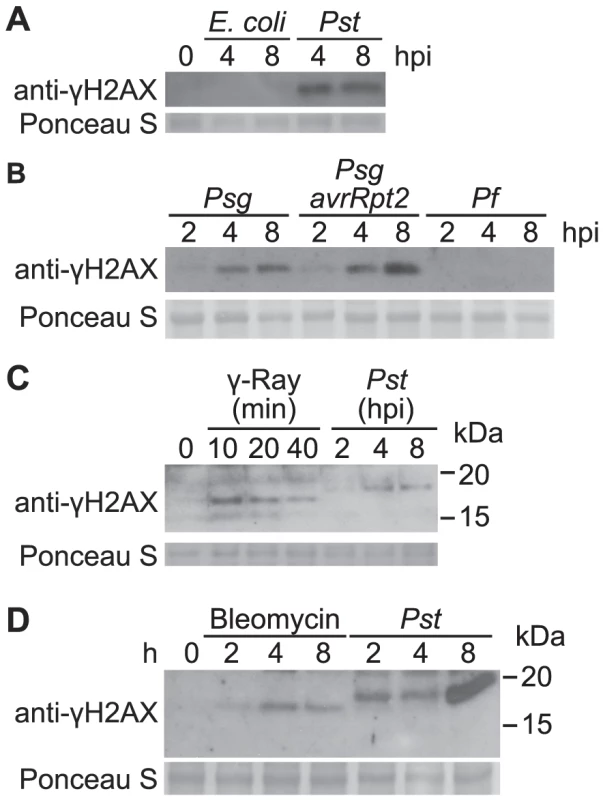
(A) Arabidopsis Col plants were vacuum-infiltrated with an E. coli DH5α strain or Pst DC3000 at a concentration of 1×107 cfu/ml. The level of γ-H2AX was assessed at 0, 4 and 8 h postinoculation by immunoblot using anti-γ-H2AX antibody. (B) Arabidopsis Col plants were vacuum-infiltrated with Psg, Psg (avrRpt2) or P. fluorescens WCS417r at a concentration of 1×107 cfu/ml. The level of γ-H2AX was assessed at 2, 4 and 8 h postinoculation by immunoblot using anti-γ-H2AX antibody. (C) Arabidopsis Col plants were irradiated with 100 Gy of gamma-rays and harvested at 10, 20 and 40 min, or vacuum-infiltrated with Pst DC3000 at a concentration of 1×107 cfu/ml and harvested at 2, 4 and 8 h postinoculation. The level of γ-H2AX was assessed by immunoblot using anti-γ-H2AX antibody. (D) Arabidopsis Col plants were treated with 2.5 µg/ml of bleomycin or vacuum-infiltrated with Pst DC3000 at a concentration of 1×107 cfu/ml. The level of γ-H2AX was assessed at 0, 2, 4 and 8 h post treatment by immunoblot using anti-γ-H2AX antibody. Equivalent loading of lanes was verified using Ponceau S stain. Similar results were obtained in separate replicate experiments. To compare the extent of Pst-induced DSB damage to other known stress conditions, Col-0 plants were irradiated with gamma-rays. Phosphorylation of H2AX was readily detected after exposure to 100 Gy of ionizing gamma irradiation (Figure 2C). Interestingly, the γ-H2AX band induced by Pst migrated slightly slower in SDS-PAGE than that induced by gamma-rays, suggesting that additional sites may be phosphorylated upon pathogen infection other than the highly conserved serine at the C-terminus of H2AX protein (Figure 2C). Similarly, when plants were treated with bleomycin, a DNA damage agent that generates DNA DSBs, accumulation of γ-H2AX was induced and a small size difference was observed when compared with the γ-H2AX triggered by Pst (Figure 2D).
Oomycete and fungal pathogens of multiple plant species induce DSBs
To investigate whether DSBs are induced by pathogens other than bacteria, and in other plant species, we examined the level of γ-H2AX in potato and tomato in response to strains of the oomycete pathogen Phytophthora infestans. Katahdin, a potato variety susceptible to late blight disease, was challenged with a US23 isolate of P. infestans. Figure 3 shows that accumulation of γ-H2AX was induced 3 days after inoculation and significantly increased between 5–7 days after inoculation, a time coincident with visible lesion formation. Extensive γ-H2AX accumulation was similarly detected in tomato variety Bonny Best after inoculation with a US22 isolate of P. infestans that is virulent on Bonny Best (Figure 3). At late time points after compatible interactions of potato or tomato with P. infestans (i.e., 7 days post-infection) an additional more slowly migrating band was also detected with the anti-γ-H2AX antibody (Figures 3A and 3B). We speculate that H2AX may by that point carry other post-translational modifications [54], [55]. For example, ionizing radiation can induce formation of a ubiquitinated H2AX that migrates at a higher molecular weight and is detected using anti-γ-H2AX antibodies [56].
Fig. 3. Accumulation of γ-H2AX induced by oomycete and fungal pathogens but not paraquat. 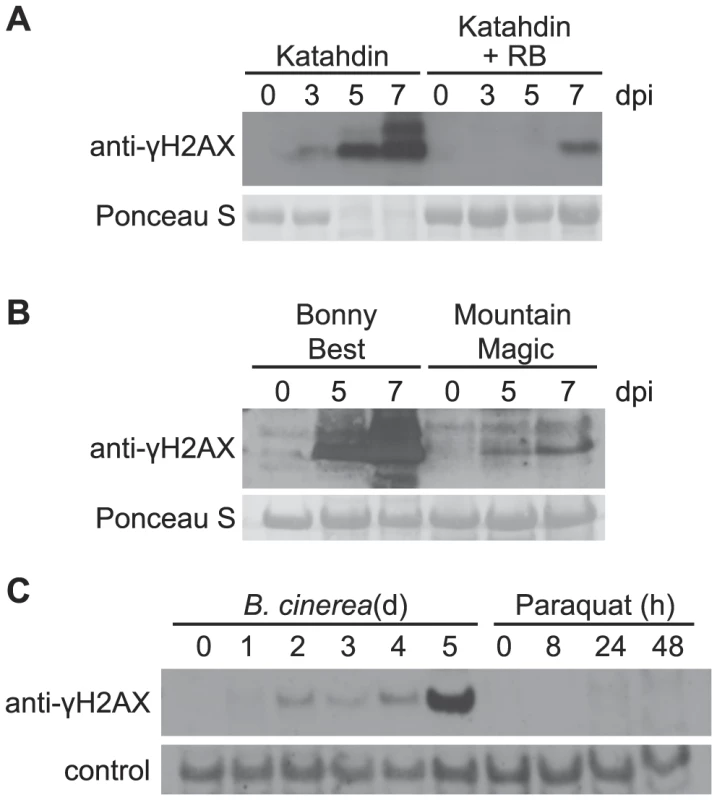
The level of γ-H2AX was assessed at indicated times by immunoblot using anti-γ-H2AX antibody. (A) Katahdin and transgenic Katahdin potato plants carrying a single copy of the RB gene were spray-inoculated with 5×104 sporangia/ml of a US23 isolate of P. infestans. dpi: days post-inoculation. (B) Two tomato varieties, Bonny Best (susceptible) and Mountain Magic (resistant), were spray-inoculated with 5×104 sporangia/ml of a US22 isolate of P. infestans. (C) Wild-type Arabidopsis Col-0 plants were spray-treated with either a Botrytis cinerea spore suspension (1×105 spores/ml) or 50 µM paraquat, and leaf samples were removed for analysis at the indicated days (d) or hours (h) after treatment. Equivalent loading of lanes was verified using Ponceau S stain or a non-specific band detected by the antibody (control). Incompatible interactions were also tested with these pathogens, to determine if a net increase or decrease in DNA damage is observed relative to compatible interactions in which R gene-mediated defenses are not prominent. US23 isolates of P. infestans are recognized by the product of the RB resistance gene, and RB mediates a mild hypersensitive response in potato [57], [58]. Much less accumulation of γ-H2AX was detected when the US23 isolate infected the resistant Katahdin SP951 transgenic potato line (Figure 3A), relative to Katahdin lines that lack the single transgene copy of RB. In tomato as well (Figure 3B), significantly less γ-H2AX accumulation was observed when the above-noted US22 P. infestans isolate was sprayed onto the tomato variety Mountain Magic that carries the Ph-2 and Ph-3 loci that confer resistance to P. infestans [59]. This is consistent with the finding that TMV triggered systemic activation of homologous recombination is blocked when resistance gene N is absent [28].
The inducibility of DSBs by microbial plant pathogens was further examined in Arabidopsis infected by the necrotrophic fungal pathogen Botrytis cinerea. γ-H2AX was induced and its presence sustained between 2 and 4 days after inoculation, and then γ-H2AX increased significantly on the 5th day after inoculation (Figure 3C). Host cell death and leaf collapse also became prevalent on the 5th day after inoculation.
Paraquat-associated plant cell death does not include strong γ-H2AX induction
Virulent P. syringae, P. infestans and B. cinerea all eventually cause tissue necrosis and plant cell death. To investigate the hypothesis that the elevation of DSBs in plants infected with these virulent pathogens is an event common to any dying plant cells, experiments were conducted with paraquat (methyl viologen). Paraquat is an herbicide that blocks photosynthetic electron transport and causes excess superoxide generation leading to plant cell death [60], but we found no evidence of strong DSB induction by paraquat. Paraquat was applied to Arabidopsis in the same experiment described above in which Botrytis cinerea induced γ-H2AX accumulation prior to the appearance of necrotic lesions. With application of 50 µM paraquat, leaves started to wilt 8 h after spraying and developed extensive necrotic lesions by 24 h, but only minimal increases in γ-H2AX abundance were observed (Figure 3C). When 5 µM paraquat was misted onto Arabidopsis leaves in a separate experiment, multiple isolated necrotic lesions formed over the next few days but no elevation of γ-H2AX was observed (in contrast to the Pst-inoculated positive control; Figure S2).
Pathogen-triggered ROS are not a primary cause of pathogen-induced DSBs
Elevated ROS are a primary feature of plant defense responses and a possible source of the host DNA damage associated with pathogen infections [3], [17], [18], [61], [62]. Virulent P. syringae elicit a rapid but transient accumulation of ROS in plants over approximately the first half hour after infection, while P. syringae expressing a recognized avirulence gene induce the first wave as well as a second wave of elevated ROS that is more massive and prolonged [16]. However, ROS production induced by bacterial and oomycete pathogens is nearly eliminated in Arabidopsis atrbohD single and atrbohDF double mutants with disruptions in the corresponding NADPH oxidase catalytic subunits [17], [63]. We examined the γ-H2AX level in response to pathogen infections in the atrbohD and atrbohDF mutant plants. There was no obvious reduction of γ-H2AX (Figure 4), in response to either virulent Pst DC3000 or avirulent Pst DC3000(avrRpt2), indicating that pathogen-triggered NADPH-derived ROS production is not the primary cause or a required component of the formation of pathogen-induced DSBs.
Fig. 4. Pst-induced γ-H2AX accumulation is independent of Pst-triggered ROS production. 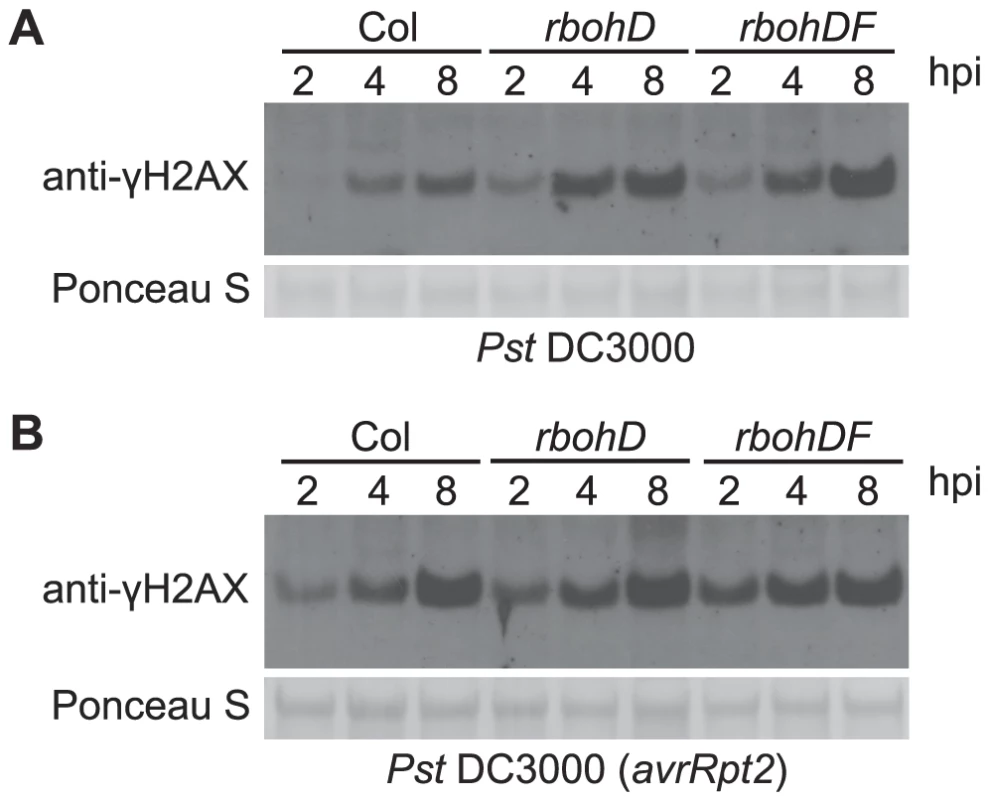
Four-week old wild-type Arabidopsis Col, atrbohD and atrbohDF plants were vacuum-infiltrated with (A) Pst DC3000 or (B) Pst DC3000(avrRpt2) at a concentration of 1×107 cfu/ml. The level of γ-H2AX was assessed at 2, 4 and 8 h postinoculation by immunoblot using anti-γ-H2AX antibody. Equivalent loading of lanes was verified using Ponceau S stain. Salicylate-mediated defenses reduce pathogen-induced DSBs
Salicylic acid (SA) is a key signaling molecule that activates defense responses against pathogens in plants, including cellular redox shifts and other physiological responses that could lead to DNA damage [64]–[66]. We investigated if SA induces γ-H2AX accumulation. After wild-type Arabidopsis plants were sprayed with 1 mM SA (a defense-inducing level [67]), no γ-H2AX accumulation was detected at time points up to 48 h after SA treatment (Figure S3). To test if SA-mediated defenses reduce pathogen-induced DNA damage, plants were treated with 1 mM SA for 1 day to induce systemic acquired resistance (SAR), then vacuum-inoculated with virulent Pst DC3000. Pretreatment with SA strongly reduced the γ-H2AX accumulation caused by Pst DC3000, compared with H2O-pretreated controls (Figures 5A and S4A). The finding that SAR reduces Pst-induced DNA damage prompted us to investigate the accumulation of γ-H2AX in the SAR-deficient Arabidopsis mutant npr1. We detected increased γ-H2AX accumulation induced by Pst DC3000 in the npr1mutant (Figures 5B and S4B). These data indicate that, rather than causing greater DNA damage, SA-mediated signaling reduces Pst-induced damage to host DNA.
Fig. 5. Pst-induced γ-H2AX accumulation during salicylic acid signaling and perception. 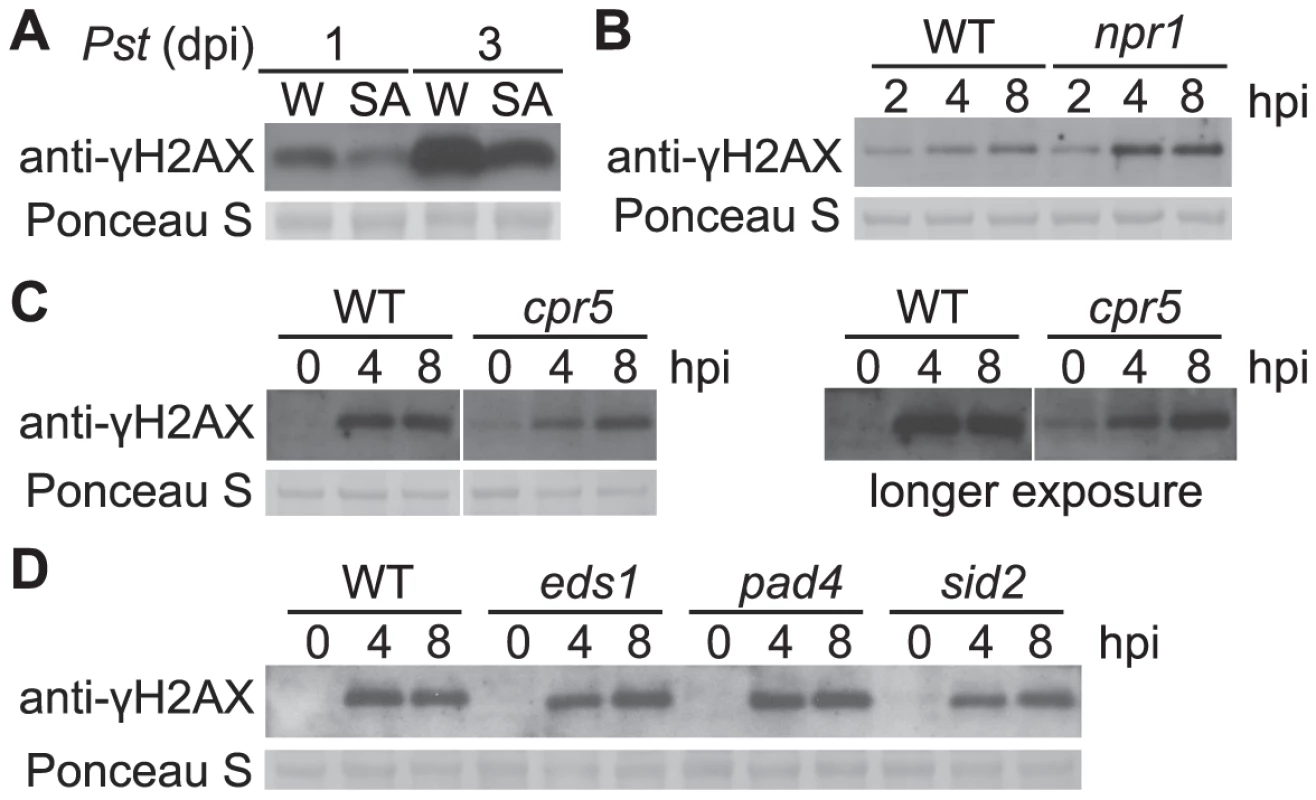
(A) Wild-type Arabidopsis Col-0 plants were pretreated with H2O (W) or 1 mM SA for 1 day and then vacuum-inoculated with Pst DC3000 at a concentration of 1×106 cfu/ml. The level of γ-H2AX was assessed at 1 and 3 days postinoculation by immunoblot using anti-γ-H2AX antibody. (B) Wild-type Col-0 and Col-0 npr1 mutant plants were spray-inoculated with Pst DC3000 at a concentration of 1×107 cfu/ml. The level of γ-H2AX was assessed at 2, 4 and 8 h postinoculation. (C) Arabidopsis Col-0 and Col-0 cpr5 mutant plants were vacuum-inoculated with Pst DC3000 at a concentration of 1×107 cfu/ml. The level of γ-H2AX was assessed at 0, 4 and 8 h postinoculation by immunoblot using anti-γ-H2AX antibody. Results with both shorter and longer exposure times were shown. (D) Arabidopsis Col-0, eds1, pad4 and sid2 plants were vacuum-inoculated with Pst DC3000 at a concentration of 1×107 cfu/ml. The level of γ-H2AX was assessed at 0, 4 and 8 h postinoculation by immunoblot using anti-γ-H2AX antibody. Equivalent loading of lanes was verified using Ponceau S stain. Similar results were obtained in two additional experiments. Other defense mutants that exhibit altered SA perception and/or signaling were also examined. Arabidopsis cpr5 plants exhibit constitutive defense responses such as PR gene expression, and constitutively elevated levels of SA [68], [69]. A low but constitutive presence of γ-H2AX was detected in the cpr5 mutant prior to pathogen infection (Figures 5C and S4C). In the Arabidopsis SA signaling mutants eds1 and pad4, and in the SA synthesis mutant sid2, no detectable changes of γ-H2AX accumulation after Pst infection were observed (Figures 5D and S4D).
We also tested the plant defense signaling molecule jasmonic acid [66]. Similar to SA, jasmonic acid did not induce γ-H2AX accumulation (Figure S3).
MAMPs do not induce detectable DSBs
We then investigated whether pathogen-free activation of MAMP-induced defense signaling induces γ-H2AX accumulation. The response was monitored from 15 min. to 2 h, which is beyond the half-hour time period when the ROS burst, MAP kinase activation, ethylene synthesis, changes in gene expression and the other primary responses to MAMPs arise [70]. When wild-type Arabidopsis Col-0 seedlings were exposed to 0.1 µM of the bacterial EF-Tu epitope elf18 (a dose sufficient to saturate induction of most elf18-induced plant defense responses [71]), no γ-H2AX accumulation was detected (Figure S5A). The ability of MAMPs to induce γ-H2AX accumulation was also investigated using the flagellin epitope flg22. Again, no γ-H2AX accumulation was observed after a high-dose 1 µM flg22 treatment (Figure S5B), suggesting that typical MAMP-induced plant defense responses do not in general induce sufficient DNA DSBs to cause detectable phosphorylation of H2AX.
Minimal impact of sni1, ssn2 and rad51D mutants on Pst-induced DNA damage
The DNA damage (DNA protection) proteins SNI1, SSN2, and RAD51D have been shown to play roles in both homologous recombination and defense gene transcription [32], [34]. To test for a possible contribution of these proteins to prevention or reduction of Pst-induced DNA damage, the level of γ-H2AX in response to Pst was examined in the respective Arabidopsis Col-0 mutants. SNI1 is a subunit of the Structural Maintenance of Chromosome (SMC) 5/6 complex involved in DNA damage response [72] and functions as a negative regulator of some plant defense responses [32]–[34]. The sni1 single mutant was recently reported to exhibit a constitutive DNA damage response [72]. Consistent with this, we observed phosphorylation of H2AX in -sni1 plants in the absence of pathogen infection (Figures 6 and S6A). However, no reproducibly significant changes in the time course of Pst-induced γ-H2AX were observed. SSN2 is a SWIM-domain containing protein that acts at early steps of homologous recombination, and the ssn2 mutation partially suppresses sni1 [34], [73]. Rad51D complexes with SSN2 and SNI1 during homologous recombination and the rad51d mutation also suppresses sni1 [32]. With rad51d and ssn2 single mutants, we found that the time course of Pst-induced γ-H2AX again was comparable to that in wild-type plants (Figures 6 and S6A). When the rad51d mutant was grown under short-day conditions, it displayed a spontaneous lesion phenotype and exhibited elevated levels of γ-H2AX without pathogen infection (Figures 6 and S6B).
Fig. 6. Pst-induced γ-H2AX accumulation in mutants involved in homologous-recombination pathway. 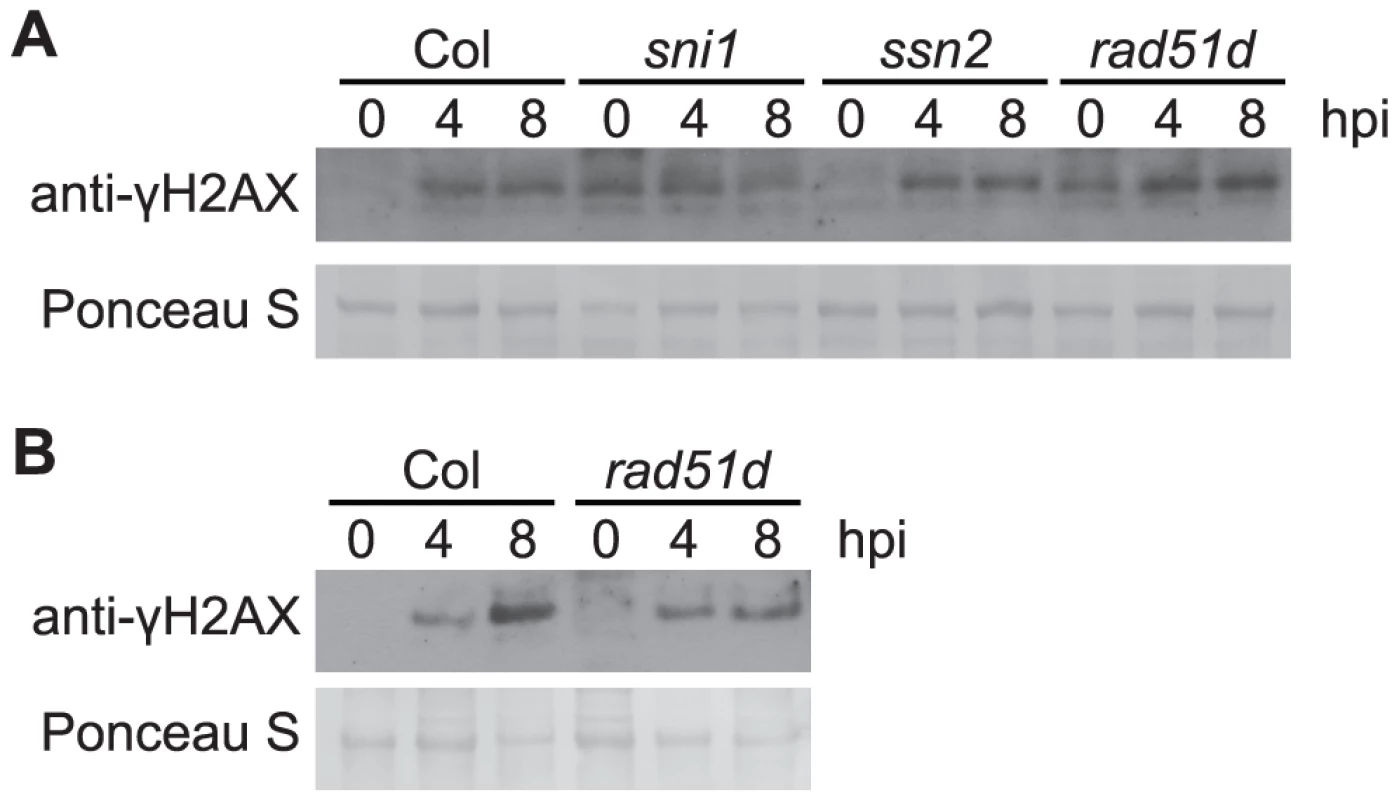
Wide-type Arabidopsis Col, sni1, ssn2, rad51d plants grown under short-day conditions (A), or wild-type Arabidopsis Col and rad51d plants grown under long-day conditions (B), were vacuum-inoculated with Pst DC3000 at a concentration of 1×107 cfu/ml. The level of γ-H2AX was assessed at 0, 4 and 8 h after inoculation by immunoblot using anti-γ-H2AX antibody. Equivalent loading of lanes was verified using Ponceau S stain. Similar results were obtained in two additional experiments. ATR and ATM contribute to restriction of virulent and avirulent Pst bacteria but are not required for pathogen-induced H2AX phosphorylation
The accumulation of γ-H2AX after exposure to ionizing radiation is largely dependent on ATM in Arabidopsis, although ATR can contribute to formation of DSBs to a lesser extent [25]. To determine whether Arabidopsis ATR or ATM is required for the phosphorylation of H2AX in response to pathogen infection, we examined γ-H2AX levels in atr and atm single mutants and in atr atm double mutant plants. We used SALK T-DNA insertion lines atr-2 and atm-2 in the Col-0 background, that carry an insertion in exon 10 of ATR and intron 64 of ATM respectively, and have been characterized previously [74], [75]. Similar levels of γ-H2AX were induced by virulent Pst DC3000 in the atr-2 and atm-2 single mutants and the atr-2 atm-2 double mutants compared with wild-type Col-0 (Figure 7A). To verify this result we also tested atr-3 and atm-1, which carry in the Ws genetic background a T-DNA insertion in the highly conserved C-terminal kinase domain (atr-3) or in the 3′ region of the gene (atm-1), and both likely act as null alleles [74]. γ-H2AX induction after Pst infection was detected in atr-3 and atm-1 single mutants and in atr-3 atm-1 double mutants at comparable levels to wild-type (Figure S7). Similar results were obtained in two independent experiments with the Col atr and atm mutants, and in three independent experiments with the Ws atr and atm mutants. These experiments are consistent with the slightly larger γ-H2AX band observed after Pst treatment as opposed to gamma-rays or bleomycin (Figure 2), and suggest that protein kinases other than ATR and ATM are engaged to mediate pathogen-induced γ-H2AX formation.
Fig. 7. Pst-induced γ-H2AX accumulation is independent of ATR and ATM but atr atm double mutants are more susceptible to growth of Pst bacteria. 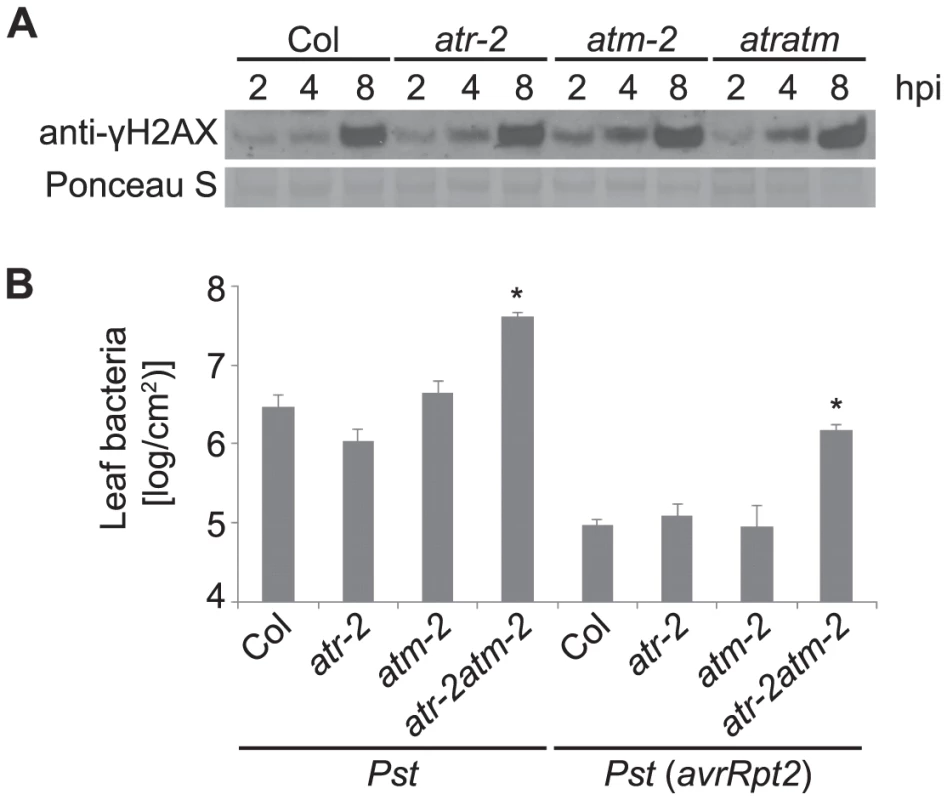
(A) Wild-type Arabidopsis Col, Col atr-2 or Col atm-2 single mutants, or Col atr-2 atm-2 double mutant plants were vacuum-inoculated with Pst DC3000 at a concentration of 1×107 cfu/ml. The level of γ-H2AX was assessed at 2, 4 and 8 h after inoculation by immunoblot using anti-γ-H2AX antibody. Equivalent loading of lanes was verified using Ponceau S stain. Similar results were obtained in three additional experiments. (B) Growth of Pst within leaves. Plants were infiltrated with Pst DC3000 or Pst DC3000(avrRpt2) at a concentration of 1×105 cfu/ml. Error bars are SEM for four replicates for each sample within the experiment. * indicates significant difference from Col-0 (ANOVA, Tukey pairwise comparisons, P<0.05). Similar results were obtained in two additional experiments. The response of atr-2 and atm-2 mutants to pathogen infection was examined using Pst DC3000. The atr-2 atm-2 double mutants were more susceptible to infection than wild type plants whereas the atr-2 and atm-2 single mutants were similar to wild type (Figure 7B), indicating that ATR and ATM play overlapping roles in basal defense. To test whether ATR and ATM are required for effector triggered immunity, atr-2 and atm-2 lines were inoculated with the avirulent pathogen Pst DC3000(avrRpt2). As shown in Figure 7B, the atr-2 atm-2 double mutants exhibited enhanced susceptibility. Taken together with previously published findings [74]–[78], these bacterial growth data provide another example that Arabidopsis ATR and ATM can play broad roles in plant development, DNA damage repair, and now, plant immunity.
Discussion
The present study discovered that host DNA damage is induced, both in the model organism Arabidopsis thaliana and in tomato and potato crop plants, in response to plant pathogens with diverse life styles including a hemibiotrophic bacterial species, an oomycete and a necrotrophic fungus. Similar or reduced levels of DNA DSBs were induced during incompatible interactions when compared with compatible interactions. Plant defense mediators such as ROS, jasmonic acid and MAMP receptors did not on their own increase DSBs, and SA-mediated defenses reduced rather than elevated pathogen-induced DNA damage. These findings provide a new type of evidence of links between the plant immune and DNA damage responses. Prevention and repair of DNA damage is needed, to a greater extent than was previously understood, as an element of the plant defense response.
Previously discovered associations between DNA damage and plant immune responses were noted in the introduction [27]–[38]. In addition, Yan et al. very recently reported that salicylic acid activates DNA damage responses as part of the plant immune response [72]. Similar associations have been established in animal systems. For example, DNA damage can regulate human inflammatory responses through activation of the tumor suppressor p53 and elevated expression of Toll-like receptors [79], and the DNA damage response induces expression of innate immune system ligands of the NKG2D receptor [80]. The fact that DNA damage induces immune responses suggests that multicellular organisms associate DNA damage with, among other things, microbial infections. The present work and [29] provide experimental evidence for a key part of this arrangement, by showing that diverse plant pathogens elicit plant DNA damage.
One point of note is the short time after Pst infection at which γ-H2AX becomes apparent. The flagellin or EF-Tu MAMPs flg22 or elf18 did not elicit detectable DNA DSBs, but γ-H2AX was reproducibly present within 2 h after Pst infection. The pathbreaking findings of [27], [28] indicate that pathogens do not even need to be physically present at a cell for that cell to experience pathogenesis-associated genome stress. They measured homologous recombination rather than directly monitoring DNA damage, but as one example, those researchers reported increased homologous recombination in non-inoculated leaves as early as 8 h after inoculation of tobacco with Tobacco mosaic virus [28]. This is faster than the virus itself moves. The SA analogs BTH or INA induced a 1.5 to 7 fold increase of homologous recombination frequency 14 days after chemical treatment [28], while we did not observe increases in DNA DSBs after treatment with SA or JA. However, we examined the level of γ-H2AX at much earlier times points 12, 24 and 48 h after SA or JA application. Yan et al. reported, from comet assays, that SA can induce DNA damage in wild-type and npr1 mutant plants [72]. This discrepancy between the two studies may have been caused by the use of different DNA damage detection methods or by differences in plant growth and treatment conditions. The main conclusion of Yan et al. [72], that SA activates DNA damage responses to potentiate plant immunity, is highly consistent with our main finding that pathogenic microorganisms can induce plant DNA damage and that plant defense mechanisms help to suppress rather than promote this damage.
Production of ROS is one of the earliest cellular responses of plants to pathogens and is also a common response to pathogens in animals [14], [16], [17]. The genotoxicity of certain microbial infections in animals has been attributed to host-generated ROS [8]–[11]. We found that Arabidopsis rbohD and rbohDF mutants that are defective in pathogenesis-induced ROS burst still produced extensive pathogenesis-induced DSBs. In addition, the strong ROS inducer paraquat failed to induce extensive DSBs. Furthermore, despite the contrasting induction of a less strong early ROS burst in response to virulent pathogens vs. the combined early ROS burst and a stronger and more prolonged later ROS burst in response to avirulent pathogens [16], both types of Pst DC3000 pathogens induced similar formation of DSBs. Recognition of diverse MAMPs including both flg22 and elf18 also triggers an oxidative burst [70] but failed to induce the generation of DSBs. Collectively, these findings indicate that ROS are not key mediators required for pathogen induction of DSBs in the plant pathosystems that we analyzed. A recent paper has analogously suggested that the host ROS triggered by H. pylori infection is not required for DSB formation in animals [7].
Because in our studies the MAMPs flg22 and elf18, the signaling molecules salicylic acid and jasmonic acid, and ROS-generating paraquat each failed to induce detectable level of DSBs, we postulate that direct interaction with one or more pathogen-derived effectors, toxins, or other molecules is required for pathogen induction of DNA damage. Type-III secretion-defective Pst DC3000 ΔhrcC induced fewer DSBs, suggesting a contribution of one or more Type III-secreted effectors to bacteria-induced plant DNA damage. However, that contribution may be direct, or indirect through effector elicitation of specific host responses, or indirect due to general enhancement of pathogen population sizes. The more significant result of the DC3000 ΔhrcC experiments may be that substantial DSB induction was evident even when the Type-III secretion system was disabled.
In the present study, host DNA DSB induction was observed following infection by plant pathogens but not after introduction of non-pathogenic bacteria. Prior work provides some context for this result. For example, the non-pathogenic bacteria E. coli DH5α and P. fluorescens Pf101 elicit defense transcript accumulation and phytoalexin biosynthesis in bean [81]. Inoculation of Arabidopsis roots with P. fluorescens strain WCS417r (the strain used in this study) activates induced systemic resistance, which is independent of SA accumulation and pathogenesis-related (PR) gene activation but primes plants to respond faster or stronger to pathogen attack [82], [83]. Induction of those responses, then, is not likely to elicit DNA DSBs, although the failure of those non-pathogenic bacteria to induce DNA DSBs may alternatively be attributable to the weak defense response they trigger. In contrast to P. fluorescens WCS417r, the soybean pathogen Psg was previously shown to trigger a marked systemic increase of SA level and PR gene expression in Arabidopsis, leading to elevated systemic resistance to secondary infection, although to a lesser extent compared to those induced by virulent Pst and avirulent Pst avrRpm1 [50]. So although we do not yet know the mechanism of DNA DSB induction by plant pathogens, and did not observed its induction by ROS, SA, JA, or MAMPs, strength of host defense induction is a feature that correlates with the DNA DSB-inducing behavior of the strains we studied. Recent work with Psg on Arabidopsis reminds us that molecular mechanisms of non-host resistance against different plant pathogens can be distinct. The Arabidopsis non-host resistance gene PSS1 confers a new form of non-host resistance against both a hemibiotrophic oomycete pathogen, P. sojae and a necrotrophic fungal pathogen, F. virguliforme, but not the bacterial pathogen Psg [84].
ATM and ATR are the two primary known signal transducers of DNA breakage, and they initiate a phosphorylation-mediated signal transduction cascade that leads to cell-cycle arrest and repair of DSBs [23]–[25]. In addition to ATM and ATR, a related mammalian enzyme, the DNA-dependent protein kinase (DNA-PK), is capable of phosphorylating H2AX in response to DSBs [85]. The relative roles of ATM and ATR in DSB-dependent γ-H2AX induction have been debated, and apparently vary depending on the biological context. For example, Kuhne et al. provided evidence that ATM contributes to ionizing radiation-induced γ-H2AX formation in mouse fibroblasts [86], whereas a separate report suggested that ATM was not required or played minor role in ionizing radiation-dependent γ-H2AX accumulation [87]. In Arabidopsis the accumulation of γ-H2AX in response to ionizing radiation-induced DSBs is dependent on both ATM and ATR, with a predominant role for ATM [25]. We found that pathogen-induced γ-H2AX accumulation was not reduced in two different Arabidopsis atr atm double mutant lines. Our finding unsettles the concept that only ATR and ATM carry out this process in plants. Because no obvious homologs of DNA-PK are present in nonvertebrates, our experiments suggest that plants have one or more kinases other than ATM, ATR or DNA-PK that can phosphorylate H2AX, and which do so in response to pathogen infection. This finding of pathogen-induced γ-H2AX accumulation in Arabidopsis atr atm double mutants is supported by a recent discovery made using primary cultures of human renal proximal tubule epithelial cells, where knockdown of all three major phosphatidylinositol 3-kinase-like kinases (ATM, ATR, and DNA-PKcs) did not abolish the activation of γ-H2AX during viral infection by BKPyV [88]. Viral infection by BKPyV did cause severe DNA damage in the absence of ATM or ATR [88], and as previously noted, numerous animal and plant studies have shown that ATM and ATR play central roles in DNA damage repair [19], [24], [25]. Hence it is not overly surprising that Arabidopsis atr atm double mutant plants exhibit heightened disease susceptibility, even though ATM and ATR are not the sole means through which plant H2AX can be phosphorylated after pathogen infection.
The discovery of pathogen-induced plant DNA damage [present study and 29] opens intriguing avenues for future study. For example, research to pinpoint pathogenesis-induced DSB sites may reveal if preferential sites exist. Investigation of the pathogen factors or pathogen-induced plant factors that lead to infection-associated DNA damage will be a priority, and this may lend insight into disease management mechanisms that can protect the genome from the damage induced by pathogens. The subject has clear implications for improved crop productivity under conditions of biotic stress [19], [20].
Materials and Methods
Plant treatments
Arabidopsis plants were grown at 22°C under 9-h light/15-h cycles in Fison's Sunshine Mix #1. The rad51d mutant line was also grown at 16-h light/8-h dark cycles. Five-week-old Arabidopsis plants were typically used in various treatments unless otherwise indicated. Pseudomonas syringae pv. tomato bacterial strains used in this study were Pst DC3000, Pst DC3000(avrRpt2) [39] and Pst DC3000(ΔhrcC) [41]. Nonpathogenic bacteria strains included in this study were E. coli DH5α grown on LB plates, or P. syringae pv. glycinea (Psg), Psg (avrRpt2) [44] or P. fluorescens WCS417r [52] grown on NYGA plates. For DNA damage experiments, above-soil portions of intact plants were briefly inverted into a bacterial solution at 1×107 cfu/ml in 10 mM MgCl2, bacteria were introduced into leaf mesophyll by vacuum infiltration [89], plants were returned to their normal growth environment, and samples were collected at the specified time points. For SAR induction, plants were pretreated with 10 mM MgCl2 or 1 mM SA for 1 day followed by vacuum-infiltration with virulent Pst DC3000 at 1×106 cfu/ml. For ionizing radiation, Arabidopsis Col-0 plants were irradiated at 100 Gy with a 137Cs source and collected at indicated times. For bleomycin treatment, Arabidopsis Col-0 plants were incubated with 2.5 µg/ml of bleomycin for indicated times. For Botrytis cinerea infection [90], plants were sprayed with 1×105 spores/ml and samples were collected 1 to 5 days post inoculation. Inoculation of Phytophthora infestans was carried out by spraying plants with sporangial suspensions according to previously described procedures [57]. Potato and tomato plants were infected with US23 or US22 P. infestans isolates, respectively, provided courtesy of Amilcar Sánchez-Pérez and Dennis Halterman. Treatment with paraquat (methyl viologen; Sigma-Aldrich, St. Louis, MO) was performed on Arabidopsis plants by spraying an aqueous suspension to run-off at concentrations of 5 or 50 µM. To obtain atr atm double mutants, progeny plants from self-fertilized atr-3/atr-3,ATM/atm-1 or atr-2/atr-2,ATM/atm-2 lines [74], [75] were genotyped by PCR for presence of the relevant T-DNA insertion and separately for absence of the wild-type allele using the methods of http://signal.salk.edu/tdnaprimers.2.html. The resulting double mutant lines were observed to be sterile.
Bacterial growth assay
Five-week-old Arabidopsis seedlings were inoculated with Pst DC3000 or Pst DC3000(avrRpt2) at 1×105 cfu/ml by infiltration of leaf mesophyll using a 1 cc plastic syringe with no needle or vacuum infiltration of immersed rosette leaves. After 3 days, leaf discs were taken from eight inoculated fully expanded rosette leaves and samples from two leaves were combined to form a single replicate and macerated in 10 mM MgCl2. The samples were then diluted serially, plated on NYGA plates and colony counts were recorded two days after incubation at 28°C.
Histone preparations and immunoblotting
Histones were extracted from plant leaf tissue nuclear preparations as previously described [25], [91]. Protein samples were subjected to SDS-PAGE, blotted and immunodetected with rabbit anti-human γ-H2AX antibody at 1∶5000 dilution (Sigma-Aldrich, St. Louis, MO). Band intensity on immunoblots was quantified using Image Studio Lite software version3.1 (LI-COR Biosciences, Lincoln, Nebraska) and statistical tests of significance were performed on the resulting data as described.
Comet assay
Comet assays [48], [49] were performed using the CometAssay kit from Trevigen (Gaithersburg, MD) with minor modifications. Leaf tissues were cut into pieces with a razor blade in 500 µl 1× PBS buffer supplemented with 20 mM EDTA on ice. Nuclei suspension was filtered into an Eppendorf tube through 50 µm nylon mesh, combined with Comet low-melting-point agarose at a ratio of 1∶ 10 and pipetted onto CometSlides. After incubation in lysis solution for 1 h at 4°C, the slides were placed in 1× Tris-Acetate electrophoresis buffer for 30 min prior to electrophoresis in the same buffer for 10 min at 4°C. Nuclei were stained with SYBR green. Images were captured and quantified with CometScore software (Tritek Co., Sumerduck, VA). At least 200 nuclei were scored per slide.
Arabidopsis gene numbers
H2AXa: AT1G08880; H2AXb: AT1G54690; NPR1: AT1G64280; CPR5: AT5G64930; EDS1: AT3G48090; PAD4: AT3G52430; SID2: AT1G74710; FLS2: AT5G46330; EFR: AT5G20480; SNI1: AT4G18470; SSN2: AT4G33925; RAD51D: AT1G07745; ATR: AT5G40820; ATM: AT3G48190.
Supporting Information
Zdroje
1. JacksonSP, BartekJ (2009) The DNA-damage response in human biology and disease. Nature 461 : 1071–1078.
2. CicciaA, ElledgeSJ (2010) The DNA damage response: making it safe to play with knives. Mol Cell 40 : 179–204.
3. SedelnikovaOA, RedonCE, DickeyJS, NakamuraAJ, GeorgakilasAG, et al. (2010) Role of oxidatively induced DNA lesions in human pathogenesis. Mutat Res 704 : 152–159.
4. AmburOH, DavidsenT, FryeSA, BalasinghamSV, LagesenK, et al. (2009) Genome dynamics in major bacterial pathogens. FEMS Microbiol Rev 33 : 453–470.
5. WeitzmanMD, LilleyCE, ChaurushiyaMS (2010) Genomes in conflict: maintaining genome integrity during virus infection. Annu Rev Microbiol 64 : 61–81.
6. NougayredeJP, HomburgS, TaiebF, BouryM, BrzuszkiewiczE, et al. (2006) Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science 313 : 848–851.
7. TollerIM, NeelsenKJ, StegerM, HartungML, HottigerMO, et al. (2011) Carcinogenic bacterial pathogen Helicobacter pylori triggers DNA double-strand breaks and a DNA damage response in its host cells. Proc Natl Acad Sci U S A 108 : 14944–14949.
8. ChakrabortySP, Kar MahapatraS, SahuSK, DasS, TripathyS, et al. (2011) Internalization of Staphylococcus aureus in lymphocytes induces oxidative stress and DNA fragmentation: possible ameliorative role of nanoconjugated vancomycin. Oxid Med Cell Longev 2011 : 942123.
9. RakkestadKE, SkaarI, AnsteinssonVE, SolhaugA, HolmeJA, et al. (2010) DNA damage and DNA damage responses in THP-1 monocytes after exposure to spores of either Stachybotrys chartarum or Aspergillus versicolor or to T-2 toxin. Toxicol Sci 115 : 140–155.
10. TouatiE (2010) When bacteria become mutagenic and carcinogenic: lessons from H. pylori. Mutat Res 703 : 66–70.
11. MangerichA, KnutsonCG, ParryNM, MuthupalaniS, YeW, et al. (2012) Infection-induced colitis in mice causes dynamic and tissue-specific changes in stress response and DNA damage leading to colon cancer. Proc Natl Acad Sci U S A 109: E1820–1829.
12. ArthurJC, Perez-ChanonaE, MuhlbauerM, TomkovichS, UronisJM, et al. (2012) Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 338 : 120–123.
13. DoddsPN, RathjenJP (2010) Plant immunity: towards an integrated view of plant-pathogen interactions. Nat Rev Genet 11 : 539–548.
14. Murphy K, Travers P, Walport M, Janeway C (2012) Janeway's immunobiology. New York: Garland Science. xix, 868 p. p.
15. Rakoff-NahoumS, MedzhitovR (2009) Toll-like receptors and cancer. Nat Rev Cancer 9 : 57–63.
16. LambC, DixonRA (1997) The Oxidative Burst in Plant Disease Resistance. Annu Rev Plant Physiol Plant Mol Biol 48 : 251–275.
17. O'BrienJA, DaudiA, ButtVS, BolwellGP (2012) Reactive oxygen species and their role in plant defence and cell wall metabolism. Planta 236 : 765–779.
18. Roldan-ArjonaT, ArizaRR (2009) Repair and tolerance of oxidative DNA damage in plants. Mutat Res 681 : 169–179.
19. WaterworthWM, DruryGE, BrayCM, WestCE (2011) Repairing breaks in the plant genome: the importance of keeping it together. New Phytol 192 : 805–822.
20. BalestrazziA, ConfalonieriM, MacoveiA, DonaM, CarboneraD (2011) Genotoxic stress and DNA repair in plants: emerging functions and tools for improving crop productivity. Plant Cell Rep 30 : 287–295.
21. RogakouEP, PilchDR, OrrAH, IvanovaVS, BonnerWM (1998) DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem 273 : 5858–5868.
22. IacovoniJS, CaronP, LassadiI, NicolasE, MassipL, et al. (2010) High-resolution profiling of gammaH2AX around DNA double strand breaks in the mammalian genome. EMBO J 29 : 1446–1457.
23. AbrahamRT (2001) Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev 15 : 2177–2196.
24. SperkaT, WangJ, RudolphKL (2012) DNA damage checkpoints in stem cells, ageing and cancer. Nat Rev Mol Cell Biol 13 : 579–590.
25. FriesnerJD, LiuB, CulliganK, BrittAB (2005) Ionizing radiation-dependent gamma-H2AX focus formation requires ataxia telangiectasia mutated and ataxia telangiectasia mutated and Rad3-related. Mol Biol Cell 16 : 2566–2576.
26. KinnerA, WuW, StaudtC, IliakisG (2008) Gamma-H2AX in recognition and signaling of DNA double-strand breaks in the context of chromatin. Nucleic Acids Res 36 : 5678–5694.
27. LuchtJM, Mauch-ManiB, SteinerHY, MetrauxJP, RyalsJ, et al. (2002) Pathogen stress increases somatic recombination frequency in Arabidopsis. Nat Genet 30 : 311–314.
28. KovalchukI, KovalchukO, KalckV, BoykoV, FilkowskiJ, et al. (2003) Pathogen-induced systemic plant signal triggers DNA rearrangements. Nature 423 : 760–762.
29. YaoY, KathiriaP, KovalchukI (2013) A systemic increase in the recombination frequency upon local infection of Arabidopsis thaliana plants with oilseed rape mosaic virus depends on plant age, the initial inoculum concentration and the time for virus replication. Front Plant Sci 4 : 61.
30. ChoiJJ, KlostermanSJ, HadwigerLA (2001) A comparison of the effects of DNA-damaging agents and biotic elicitors on the induction of plant defense genes, nuclear distortion, and cell death. Plant Physiol 125 : 752–762.
31. KunzBA, CahillDM, MohrPG, OsmondMJ, VonarxEJ (2006) Plant responses to UV radiation and links to pathogen resistance. Int Rev Cytol 255 : 1–40.
32. DurrantWE, WangS, DongXN (2007) Arabidopsis SNI1 and RAD51D regulate both gene transcription and DNA recombination during the defense response. Proc Natl Acad Sci U S A 104 : 4223–4227.
33. WangS, DurrantWE, SongJ, SpiveyNW, DongX (2010) Arabidopsis BRCA2 and RAD51 proteins are specifically involved in defense gene transcription during plant immune responses. Proc Natl Acad Sci U S A 107 : 22716–22721.
34. SongJ, DurrantWE, WangS, YanS, TanEH, et al. (2011) DNA repair proteins are directly involved in regulation of gene expression during plant immune response. Cell Host Microbe 9 : 115–124.
35. Adams-PhillipsL, WanJ, TanX, DunningFM, MeyersBC, et al. (2008) Discovery of ADP-ribosylation and other plant defense pathway elements through expression profiling of four different Arabidopsis-Pseudomonas R-avr interactions. Mol Plant Microbe Interact 21 : 646–657.
36. Adams-PhillipsL, BriggsAG, BentAF (2010) Disruption of poly(ADP-ribosyl)ation mechanisms alters responses of Arabidopsis to biotic stress. Plant Physiol 152 : 267–280.
37. MurLA, KentonP, LloydAJ, OughamH, PratsE (2008) The hypersensitive response; the centenary is upon us but how much do we know? J Exp Bot 59 : 501–520.
38. RyersonDE, HeathMC (1996) Cleavage of nuclear DNA into oligonucleosomal fragments during cell death induced by fungal infection or by abiotic treatments. Plant Cell 8 : 393–402.
39. KunkelBN, BentAF, DahlbeckD, InnesRW, StaskawiczBJ (1993) RPS2, an Arabidopsis disease resistance locus specifying recognition of Pseudomonas syringae strains expressing the avirulence gene avrRpt2. Plant Cell 5 : 865–875.
40. YuGL, KatagiriF, AusubelFM (1993) Arabidopsis mutations at the RPS2 locus result in loss of resistance to Pseudomonas syringae strains expressing the avirulence gene avrRpt2. Mol Plant Microbe Interact 6 : 434–443.
41. BochJ, JoardarV, GaoL, RobertsonTL, LimM, et al. (2002) Identification of Pseudomonas syringae pv. tomato genes induced during infection of Arabidopsis thaliana. Mol Microbiol 44 : 73–88.
42. WhalenMC, InnesRW, BentAF, StaskawiczBJ (1991) Identification of Pseudomonas syringae pathogens of Arabidopsis and a bacterial locus determining avirulence on both Arabidopsis and soybean. Plant Cell 3 : 49–59.
43. RitterC, DanglJL (1996) Interference between two specific pathogen recognition events mediated by distinct plant disease resistance genes. Plant Cell 8 : 251–257.
44. YuI-C, ParkerJ, BentAF (1998) Gene-for-gene disease resistance without the hypersensitive response in Arabidopsis dnd1 mutant. Proc Natl Acad Sci USA 95 : 7819–7824.
45. GaoX, ChenX, LinW, ChenS, LuD, et al. (2013) Bifurcation of Arabidopsis NLR immune signaling via Ca(2)(+)-dependent protein kinases. PLoS Pathog 9: e1003127.
46. ZhangC, GutscheAT, ShapiroAD (2004) Feedback control of the Arabidopsis hypersensitive response. Mol Plant Microbe Interact 17 : 357–365.
47. GrantM, BrownI, AdamsS, KnightM, AinslieA, et al. (2000) The RPM1 plant disease resistance gene facilitates a rapid and sustained increase in cytosolic calcium that is necessary for the oxidative burst and hypersensitive cell death. Plant J 23 : 441–450.
48. CollinsAR, AzquetaA (2012) DNA repair as a biomarker in human biomonitoring studies; further applications of the comet assay. Mutat Res 736 : 122–129.
49. DhawanA, BajpayeeM, ParmarD (2009) Comet assay: a reliable tool for the assessment of DNA damage in different models. Cell Biol Toxicol 25 : 5–32.
50. MishinaTE, ZeierJ (2007) Pathogen-associated molecular pattern recognition rather than development of tissue necrosis contributes to bacterial induction of systemic acquired resistance in Arabidopsis. Plant J 50 : 500–513.
51. HuynhTV, DahlbeckD, StaskawiczBJ (1989) Bacterial blight of soybean: Regulation of a pathogen gene determining host cultivar specificity. Science 245 : 1374–1377.
52. PieterseCM, van WeesSC, HofflandE, van PeltJA, van LoonLC (1996) Systemic resistance in Arabidopsis induced by biocontrol bacteria is independent of salicylic acid accumulation and pathogenesis-related gene expression. Plant Cell 8 : 1225–1237.
53. HaasD, DefagoG (2005) Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat Rev Microbiol 3 : 307–319.
54. LukasJ, LukasC, BartekJ (2011) More than just a focus: The chromatin response to DNA damage and its role in genome integrity maintenance. Nat Cell Biol 13 : 1161–1169.
55. SrivastavaN, GochhaitS, de BoerP, BamezaiRN (2009) Role of H2AX in DNA damage response and human cancers. Mutat Res 681 : 180–188.
56. HuenMS, GrantR, MankeI, MinnK, YuX, et al. (2007) RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell 131 : 901–914.
57. SongJ, BradeenJM, NaessSK, RaaschJA, WielgusSM, et al. (2003) Gene RB cloned from Solanum bulbocastanum confers broad spectrum resistance to potato late blight. Proc Natl Acad Sci U S A 100 : 9128–9133.
58. KramerLC, ChoudoirMJ, WielgusSM, BhaskarPB, JiangJ (2009) Correlation between transcript abundance of the RB gene and the level of the RB-mediated late blight resistance in potato. Mol Plant Microbe Interact 22 : 447–455.
59. GardnerRG, PantheeDR (2010) NC 1 CELBR and NC 2 CELBR: Early Blight and Late Blight-resistant Fresh Market Tomato Breeding Lines. Hortscience 45 : 975–976.
60. BowlerC, van MontaguM, InzeD (1992) Superoxide dismutase and stress tolerance. Ann Rev Plant Physiol Plant Mol Biol 43 : 83–116.
61. LevineA, TenhakenR, DixonR, LambCJ (1994) H2O2 from the oxidative burst orchestrates the plant hypersensitive disease resistance response. Cell 79 : 583–593.
62. SuzukiN, MillerG, MoralesJ, ShulaevV, TorresMA, et al. (2011) Respiratory burst oxidases: the engines of ROS signaling. Curr Opin Plant Biol 14 : 691–699.
63. TorresMA, DanglJL, JonesJD (2002) Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc Natl Acad Sci U S A 99 : 517–522.
64. RaoMV, PaliyathG, OrmrodDP, MurrDP, WatkinsCB (1997) Influence of salicylic acid on H2O2 production, oxidative stress, and H2O2-metabolizing enzymes. Salicylic acid-mediated oxidative damage requires H2O2. Plant Physiol 115 : 137–149.
65. DurrantWE, DongX (2004) Systemic acquired resistance. Annu Rev Phytopathol 42 : 185–209.
66. PieterseCM, Van der DoesD, ZamioudisC, Leon-ReyesA, Van WeesSC (2012) Hormonal modulation of plant immunity. Annu Rev Cell Dev Biol 28 : 489–521.
67. CaoH, BowlingSA, GordonS, DongX (1994) Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6 : 1583–1592.
68. KirikV, BouyerD, SchobingerU, BechtoldN, HerzogM, et al. (2001) CPR5 is involved in cell proliferation and cell death control and encodes a novel transmembrane protein. Curr Biol 11 : 1891–1895.
69. BowlingSA, ClarkeJD, LiuY, KlessigDF, DongX (1997) The cpr5 mutant of Arabidopsis expresses both NPR1-dependent and NPR1-independent resistance. Plant Cell 9 : 1573–1584.
70. BollerT, FelixG (2009) A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol 60 : 379–406.
71. KunzeG, ZipfelC, RobatzekS, NiehausK, BollerT, et al. (2004) The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. Plant Cell 16 : 3496–3507.
72. YanS, WangW, MarquesJ, MohanR, SalehA, et al. (2013) Salicylic acid activates DNA damage responses to potentiate plant immunity. Mol Cell 52 : 602–610.
73. MartinV, ChahwanC, GaoH, BlaisV, WohlschlegelJ, et al. (2006) Sws1 is a conserved regulator of homologous recombination in eukaryotic cells. Embo J 25 : 2564–2574.
74. CulliganK, TissierA, BrittA (2004) ATR regulates a G2-phase cell-cycle checkpoint in Arabidopsis thaliana. Plant Cell 16 : 1091–1104.
75. GarciaV, BruchetH, CamescasseD, GranierF, BouchezD, et al. (2003) AtATM is essential for meiosis and the somatic response to DNA damage in plants. Plant Cell 15 : 119–132.
76. VespaL, CouvillionM, SpanglerE, ShippenDE (2005) ATM and ATR make distinct contributions to chromosome end protection and the maintenance of telomeric DNA in Arabidopsis. Genes Dev 19 : 2111–2115.
77. AmiardS, DepeigesA, AllainE, WhiteCI, GallegoME (2011) Arabidopsis ATM and ATR kinases prevent propagation of genome damage caused by telomere dysfunction. Plant Cell 23 : 4254–4265.
78. AdachiS, MinamisawaK, OkushimaY, InagakiS, YoshiyamaK, et al. (2011) Programmed induction of endoreduplication by DNA double-strand breaks in Arabidopsis. Proc Natl Acad Sci U S A 108 : 10004–10009.
79. MenendezD, ShatzM, AzzamK, GarantziotisS, FesslerMB, et al. (2011) The Toll-like receptor gene family is integrated into human DNA damage and p53 networks. PLoS Genet 7: e1001360.
80. GasserS, OrsulicS, BrownEJ, RauletDH (2005) The DNA damage pathway regulates innate immune system ligands of the NKG2D receptor. Nature 436 : 1186–1190.
81. JakobekJL, LindgrenPB (1993) Generalized Induction of Defense Responses in Bean Is Not Correlated with the Induction of the Hypersensitive Reaction. Plant Cell 5 : 49–56.
82. PieterseCMJ, van WeesSCM, van PeltJA, KnoesterM, LaanR, et al. (1998) A novel signaling pathway controlling induced systemic resistance in Arabidopsis. Plant Cell 10 : 1571–1580.
83. VerhagenBW, GlazebrookJ, ZhuT, ChangHS, van LoonLC, et al. (2004) The transcriptome of rhizobacteria-induced systemic resistance in arabidopsis. Mol Plant Microbe Interact 17 : 895–908.
84. SumitR, SahuBB, XuM, SandhuD, BhattacharyyaMK (2012) Arabidopsis nonhost resistance gene PSS1 confers immunity against an oomycete and a fungal pathogen but not a bacterial pathogen that cause diseases in soybean. BMC Plant Biol 12 : 87.
85. ParkEJ, ChanDW, ParkJH, OettingerMA, KwonJ (2003) DNA-PK is activated by nucleosomes and phosphorylates H2AX within the nucleosomes in an acetylation-dependent manner. Nucleic Acids Res 31 : 6819–6827.
86. KuhneM, RiballoE, RiefN, RothkammK, JeggoPA, et al. (2004) A double-strand break repair defect in ATM-deficient cells contributes to radiosensitivity. Cancer Res 64 : 500–508.
87. KarlssonKH, StenerlowB (2004) Focus formation of DNA repair proteins in normal and repair-deficient cells irradiated with high-LET ions. Radiat Res 161 : 517–527.
88. JiangM, ZhaoL, GamezM, ImperialeMJ (2012) Roles of ATM and ATR-mediated DNA damage responses during lytic BK polyomavirus infection. PLoS Pathog 8: e1002898.
89. KatagiriF, ThilmonyR, HeSY (2002) The Arabidopsis thaliana-pseudomonas syringae interaction. Arabidopsis Book 1: e0039.
90. GengerRK, JurkowskiGI, McDowellJM, LuH, JungHW, et al. (2008) Signaling pathways that regulate the enhanced disease resistance of Arabidopsis “defense, no death” mutants. Mol Plant Microbe Interact 21 : 1285–1296.
91. JacksonJP, JohnsonL, JasencakovaZ, ZhangX, PerezBurgosL, et al. (2004) Dimethylation of histone H3 lysine 9 is a critical mark for DNA methylation and gene silencing in Arabidopsis thaliana. Chromosoma 112 : 308–315.
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Affinity Proteomics Reveals Elevated Muscle Proteins in Plasma of Children with Cerebral MalariaČlánek The Transcriptional Activator LdtR from ‘ Liberibacter asiaticus’ Mediates Osmotic Stress ToleranceČlánek Complement-Related Proteins Control the Flavivirus Infection of by Inducing Antimicrobial PeptidesČlánek Fungal Chitin Dampens Inflammation through IL-10 Induction Mediated by NOD2 and TLR9 ActivationČlánek Parasite Fate and Involvement of Infected Cells in the Induction of CD4 and CD8 T Cell Responses to
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2014 Číslo 4- Stillova choroba: vzácné a závažné systémové onemocnění
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Choroby jater v ordinaci praktického lékaře – význam jaterních testů
- Diagnostický algoritmus při podezření na syndrom periodické horečky
-
Všechny články tohoto čísla
- , , , Genetic Variability: Cryptic Biological Species or Clonal Near-Clades?
- Early Mortality Syndrome Outbreaks: A Microbial Management Issue in Shrimp Farming?
- Wormholes in Host Defense: How Helminths Manipulate Host Tissues to Survive and Reproduce
- Plastic Proteins and Monkey Blocks: How Lentiviruses Evolved to Replicate in the Presence of Primate Restriction Factors
- The 2010 Cholera Outbreak in Haiti: How Science Solved a Controversy
- Affinity Proteomics Reveals Elevated Muscle Proteins in Plasma of Children with Cerebral Malaria
- Noncanonical Role for the Host Vps4 AAA+ ATPase ESCRT Protein in the Formation of Replicase
- Efficient Parvovirus Replication Requires CRL4-Targeted Depletion of p21 to Prevent Its Inhibitory Interaction with PCNA
- Host-to-Pathogen Gene Transfer Facilitated Infection of Insects by a Pathogenic Fungus
- The Transcriptional Activator LdtR from ‘ Liberibacter asiaticus’ Mediates Osmotic Stress Tolerance
- Coxsackievirus B Exits the Host Cell in Shed Microvesicles Displaying Autophagosomal Markers
- TCR Affinity Associated with Functional Differences between Dominant and Subdominant SIV Epitope-Specific CD8 T Cells in Rhesus Monkeys
- Coxsackievirus-Induced miR-21 Disrupts Cardiomyocyte Interactions via the Downregulation of Intercalated Disk Components
- Ligands of MDA5 and RIG-I in Measles Virus-Infected Cells
- Kind Discrimination and Competitive Exclusion Mediated by Contact-Dependent Growth Inhibition Systems Shape Biofilm Community Structure
- Structural Differences Explain Diverse Functions of Actins
- HSCARG Negatively Regulates the Cellular Antiviral RIG-I Like Receptor Signaling Pathway by Inhibiting TRAF3 Ubiquitination Recruiting OTUB1
- Vaginitis: When Opportunism Knocks, the Host Responds
- Complement-Related Proteins Control the Flavivirus Infection of by Inducing Antimicrobial Peptides
- Fungal Chitin Dampens Inflammation through IL-10 Induction Mediated by NOD2 and TLR9 Activation
- Microbial Pathogens Trigger Host DNA Double-Strand Breaks Whose Abundance Is Reduced by Plant Defense Responses
- Alveolar Macrophages Are Essential for Protection from Respiratory Failure and Associated Morbidity following Influenza Virus Infection
- An Interaction between Glutathione and the Capsid Is Required for the Morphogenesis of C-Cluster Enteroviruses
- Concerted Spatio-Temporal Dynamics of Imported DNA and ComE DNA Uptake Protein during Gonococcal Transformation
- Potent Dengue Virus Neutralization by a Therapeutic Antibody with Low Monovalent Affinity Requires Bivalent Engagement
- Regulation of Human T-Lymphotropic Virus Type I Latency and Reactivation by HBZ and Rex
- Functionally Redundant RXLR Effectors from Act at Different Steps to Suppress Early flg22-Triggered Immunity
- The Pathogenic Mechanism of the Virulence Factor, Mycolactone, Depends on Blockade of Protein Translocation into the ER
- Role of Calmodulin-Calmodulin Kinase II, cAMP/Protein Kinase A and ERK 1/2 on -Induced Apoptosis of Head Kidney Macrophages
- An Overview of Respiratory Syncytial Virus
- First Experimental Model of Enhanced Dengue Disease Severity through Maternally Acquired Heterotypic Dengue Antibodies
- Binding of Glutathione to Enterovirus Capsids Is Essential for Virion Morphogenesis
- IFITM3 Restricts Influenza A Virus Entry by Blocking the Formation of Fusion Pores following Virus-Endosome Hemifusion
- Parasite Fate and Involvement of Infected Cells in the Induction of CD4 and CD8 T Cell Responses to
- Deficient IFN Signaling by Myeloid Cells Leads to MAVS-Dependent Virus-Induced Sepsis
- Pernicious Pathogens or Expedient Elements of Inheritance: The Significance of Yeast Prions
- The HMW1C-Like Glycosyltransferases—An Enzyme Family with a Sweet Tooth for Simple Sugars
- The Expanding Functions of Cellular Helicases: The Tombusvirus RNA Replication Enhancer Co-opts the Plant eIF4AIII-Like AtRH2 and the DDX5-Like AtRH5 DEAD-Box RNA Helicases to Promote Viral Asymmetric RNA Replication
- Mining Herbaria for Plant Pathogen Genomes: Back to the Future
- Inferring Influenza Infection Attack Rate from Seroprevalence Data
- A Human Lung Xenograft Mouse Model of Nipah Virus Infection
- Mast Cells Expedite Control of Pulmonary Murine Cytomegalovirus Infection by Enhancing the Recruitment of Protective CD8 T Cells to the Lungs
- Cytosolic Peroxidases Protect the Lysosome of Bloodstream African Trypanosomes from Iron-Mediated Membrane Damage
- Abortive T Follicular Helper Development Is Associated with a Defective Humoral Response in -Infected Macaques
- JC Polyomavirus Infection Is Strongly Controlled by Human Leucocyte Antigen Class II Variants
- Cationic Antimicrobial Peptides Promote Microbial Mutagenesis and Pathoadaptation in Chronic Infections
- Estimating the Fitness Advantage Conferred by Permissive Neuraminidase Mutations in Recent Oseltamivir-Resistant A(H1N1)pdm09 Influenza Viruses
- Progressive Accumulation of Activated ERK2 within Highly Stable ORF45-Containing Nuclear Complexes Promotes Lytic Gammaherpesvirus Infection
- Caspase-1-Like Regulation of the proPO-System and Role of ppA and Caspase-1-Like Cleaved Peptides from proPO in Innate Immunity
- Is Required for High Efficiency Viral Replication
- Modified Vaccinia Virus Ankara Triggers Type I IFN Production in Murine Conventional Dendritic Cells via a cGAS/STING-Mediated Cytosolic DNA-Sensing Pathway
- Evidence That Bank Vole PrP Is a Universal Acceptor for Prions
- Rapid Response to Selection, Competitive Release and Increased Transmission Potential of Artesunate-Selected Malaria Parasites
- Inactivation of Genes for Antigenic Variation in the Relapsing Fever Spirochete Reduces Infectivity in Mice and Transmission by Ticks
- Exposure-Dependent Control of Malaria-Induced Inflammation in Children
- A Neutralizing Anti-gH/gL Monoclonal Antibody Is Protective in the Guinea Pig Model of Congenital CMV Infection
- The Apical Complex Provides a Regulated Gateway for Secretion of Invasion Factors in
- A Highly Conserved Haplotype Directs Resistance to Toxoplasmosis and Its Associated Caspase-1 Dependent Killing of Parasite and Host Macrophage
- A Quantitative High-Resolution Genetic Profile Rapidly Identifies Sequence Determinants of Hepatitis C Viral Fitness and Drug Sensitivity
- Histone Deacetylase Inhibitor Romidepsin Induces HIV Expression in CD4 T Cells from Patients on Suppressive Antiretroviral Therapy at Concentrations Achieved by Clinical Dosing
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- The 2010 Cholera Outbreak in Haiti: How Science Solved a Controversy
- , , , Genetic Variability: Cryptic Biological Species or Clonal Near-Clades?
- Efficient Parvovirus Replication Requires CRL4-Targeted Depletion of p21 to Prevent Its Inhibitory Interaction with PCNA
- An Overview of Respiratory Syncytial Virus
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



