-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Inferring Influenza Infection Attack Rate from Seroprevalence Data
Seroprevalence studies have been regarded as the most practical method for accurately estimating the number of infections in influenza epidemics and pandemics. However, methods for inferring the number of infections from seroprevalence data in previous studies have mostly been based on conventional practice instead of standardized criteria. Specifically, there are no systematic criteria on how to select the seropositivity threshold and adjust for the proportion of infections that become seropositive. Here, we showed that under the conventional criteria, the number of 2009 pandemic influenza A/H1N1 infections had been substantially underestimated in Hong Kong as well as other countries, mostly due to overestimation of the proportion of infections that became seropositive. Our results highlighted the need to reexamine the widely accepted practice in interpreting seroprevalence data, especially in the context of pandemics when little is known but robust and comparable estimates of the number of infections and severity are most needed for informing situational awareness and guiding control policies.
Published in the journal: . PLoS Pathog 10(4): e32767. doi:10.1371/journal.ppat.1004054
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1004054Summary
Seroprevalence studies have been regarded as the most practical method for accurately estimating the number of infections in influenza epidemics and pandemics. However, methods for inferring the number of infections from seroprevalence data in previous studies have mostly been based on conventional practice instead of standardized criteria. Specifically, there are no systematic criteria on how to select the seropositivity threshold and adjust for the proportion of infections that become seropositive. Here, we showed that under the conventional criteria, the number of 2009 pandemic influenza A/H1N1 infections had been substantially underestimated in Hong Kong as well as other countries, mostly due to overestimation of the proportion of infections that became seropositive. Our results highlighted the need to reexamine the widely accepted practice in interpreting seroprevalence data, especially in the context of pandemics when little is known but robust and comparable estimates of the number of infections and severity are most needed for informing situational awareness and guiding control policies.
Introduction
Severity of influenza infection is defined as the probability of severe complications (e.g. hospitalization or death) if infected [1]. Timely and accurate estimates of severity are extremely valuable for informing decisions about the scale and targeting of response to an emerging pandemic [2]. In 2011, the International Health Regulations Review Committee highlighted the lack of “a consistent, measurable and understandable depiction of severity” as a major shortcoming of global response to the 2009 influenza pandemic [3]. Real-time serial cross-sectional or longitudinal seroprevalence studies can address this shortcoming in future pandemics by providing direct estimates of infection attack rate (IAR) as the denominator for severity [4].
In serial cross-sectional seroprevalence studies, with the absence of vaccination, IARs are estimated from seroprevalence rise (ΔS). These studies typically entail selecting an arbitrary titer threshold for seropositivity. Although many influenza seroprevalence studies have been conducted, there is no consensus on how to select seropositivity thresholds and adjust for the proportion of infections that became seropositive (infection-seropositivity probability, ISP). Haemagglutinin-inhibition (HI) titer 1∶40 and microneutralization (MN) titer 1∶40 have been commonly used as seropositivity thresholds [5]; ISP has either been ignored (IAR≈ΔS, e.g. [6]–[11]) or assumed to be similar to the proportion of clinical cases that became seropositive during convalescence (IAR≈ΔS/(proportion of clinical cases seropositive), e.g. [12]–[15]). Historically, seropositivity thresholds were often chosen by conventions instead of systematic evaluation and ISP was rarely included or discussed [4]. Previous studies have noted the arbitrariness associated with predefined seropositivity thresholds and proposed to circumvent such arbitrariness by fitting the cross-sectional titer distribution to a mixture of probability distributions for estimating IAR [16]. A simple example of these so-called mixture models is the superposition of two lognormal distributions which correspond to the titer distributions of the uninfected and infected populations [17]. In this study, we incorporated such mixture model structure into a transmission model to show that conventional seropositivity thresholds and ISP adjustments had probably led to underestimation of IARs in many seroprevalence studies of 2009 pandemic influenza A/H1N1 (pdmH1N1). Our results thus resonate with these earlier studies regarding the lack of robustness in conventional practice for inferring IAR from seroprevalence data, not only for influenza but also other infectious diseases [18], [19]. Our results highlighted the need to reexamine the widely accepted practice in interpreting seroprevalence data, especially in the context of pandemics when little is known but robust and comparable estimates of the number of infections and severity are most needed for informing situational awareness and guiding control policies.
Results
Seroprevalence data
During the 2009 influenza pandemic in Hong Kong, we conducted a large serial cross-sectional seroprevalence study with ∼14,800 serum samples from individuals aged 3–59 years, the details of which have been previously documented [11], [13]. Briefly, for samples collected before or in July 2009, we tested whether they were seropositive with respect to MN titer 1∶10, 1∶20, 1∶40, 1∶80, 1∶160, 1∶320, 1∶640, 1∶1280, and 1∶2560 (Figure 1A). Due to logistical constraints, for samples collected after July 2009, we only tested whether they were MN1∶20 and MN1∶40 seropositive, e.g. if a sample was MN1∶80 seropositive, we would only know that it was MN1∶20 and MN1∶40 seropositive. We denoted the seroprevalence, seroprevalence rise and infection-seropositivity probability for MN1:X by SX, ΔSX and ISPX, respectively.
Fig. 1. Prepandemic seroprevalence and the epidemic curve of pdmH1N1 in Hong Kong. 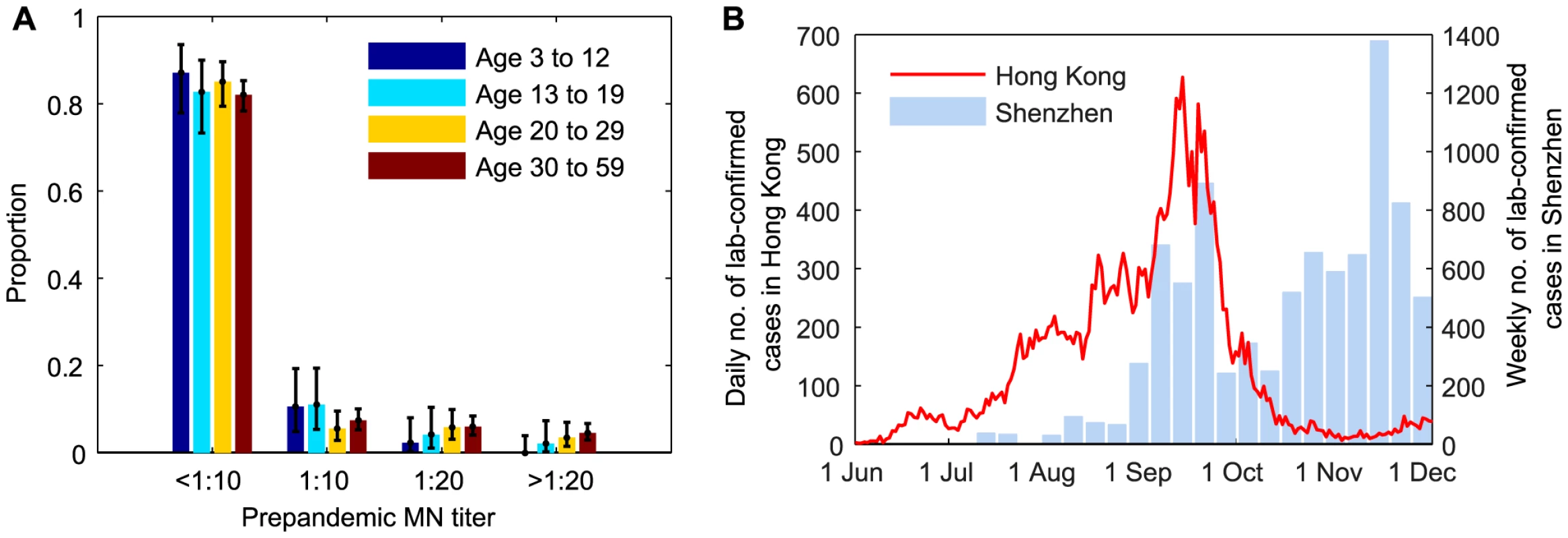
A Age-stratified pre-pandemic MN titer distributions which were estimated from serum samples collected in June and early-July 2009. For samples collected after July 2009, we only tested whether they were MN1∶20 and MN1:40 seropositive because of logistical constraints. B Epidemic curves of pdmH1N1 in Hong Kong and Shenzhen. Estimated weekly numbers of lab-confirmed cases in Shenzhen were extracted from [38]. Hospitalization data
The bulk of the first wave of pdmH1N1 in Hong Kong occurred between 1 June and 30 November 2009 (Figure 1B). Age-stratified daily number of pdmH1N1 hospitalizations during this period was provided by the Hong Kong Hospital Authority [20], [21]. Since May 2009, patients admitted with acute respiratory illnesses routinely underwent laboratory testing for pdmH1N1 [22]. Due to containment efforts enforced until June 29, all lab-confirmed pdmH1N1 cases before that date were hospitalized for isolation regardless of their clinical conditions. Therefore, our analysis only used hospitalization data from June 30 onwards during which only those required hospital care were admitted.
Preliminary analysis
In our previous IAR estimates, we (i) adopted the conventional MN1∶40 seropositivity threshold because the proportion of pdmH1N1 clinical cases who became MN1∶20 and MN1∶40 seropositive during convalescence were ∼100% and 90%, respectively [23], [24]; and (ii) assumed that ISP of all pdmH1N1 cases (i.e. including mild and asymptomatic infections) were similar to the proportion of clinical cases that became seropositive, i.e., ISP20≈1 and ISP40≈0.9–1. Because IAR≈ΔSX/ISPX, it follows that ΔS40/ΔS20≈ISP40/ISP20. The assumption ISP20≈1 and ISP40≈0.9–1 thus implied ΔS40/ΔS20>0.9. However, this contradicted our serial cross-sectional seroprevalence data which suggested that ΔS40/ΔS20 was consistently much smaller than 0.9 in all cross-sections throughout the first wave for all age groups, especially among older adults (Figure 2). The contribution of seasonal influenza to ΔS20 was small because (i) <34% of influenza A isolates during the first wave were seasonal influenza (http://www.chp.gov.hk/en/epidemiology/304/518/519.html); and (ii) in a Hong Kong study of within-household influenza transmission [25], only a small percentage of subjects infected with seasonal influenza became MN1∶20 seropositive against pdmH1N1 (unpublished data, BJ Cowling). Thus, given that pdmH1N1 vaccination was absent during the study period, ΔS20 could only be attributed to pdmH1N1 infections. This preliminary analysis strongly suggested that a substantial proportion of pdmH1N1 infections (e.g. mild and asymptomatic infections) did not become MN1∶40 seropositive. To substantiate this hypothesis, we developed a mathematical model to fully characterize the transmission dynamics and seroprevalence rises of pdmH1N1 during its first wave in Hong Kong.
Fig. 2. Age-specific ΔS40/ΔS20 during the first wave of pdmH1N1 in Hong Kong. 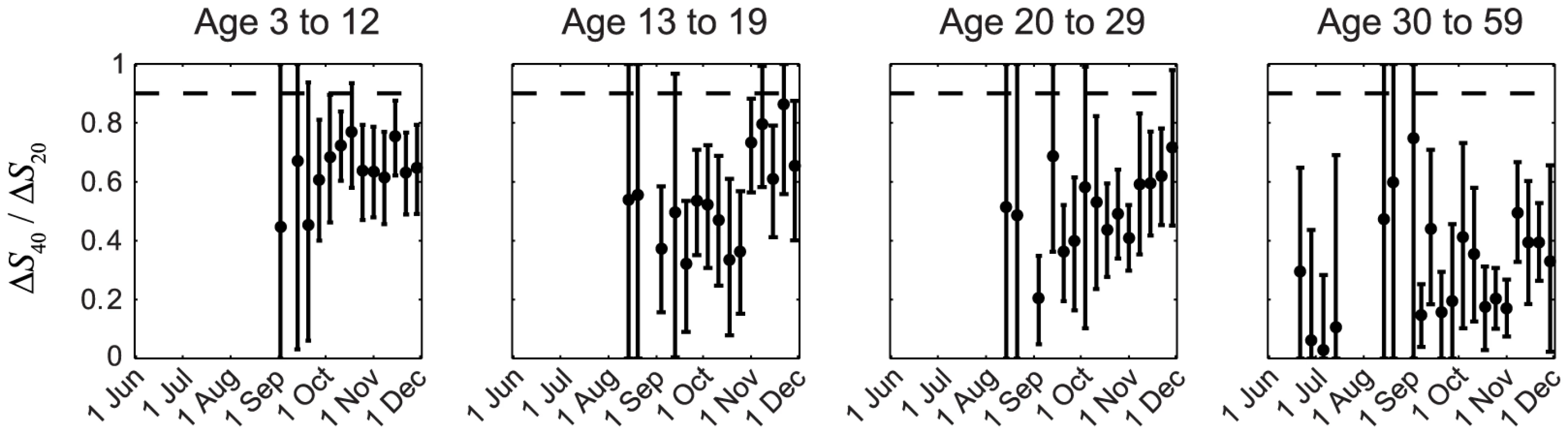
ΔS40 and ΔS20 at each cross-section were estimated using the method described in our previous work [11]. If ISP20 and ISP40 (among all pdmH1N1 infections) were the same as the proportions of clinical cases that became MN1:20 and MN1∶40 seropositive (i.e. around 100% and 90%, respectively [23], [24]), ΔS40/ΔS20 should have remained close to 0.9–1 (the horizontal dashed line) throughout the first wave, which was not the case in reality as shown here. Transmission dynamics and ISP estimates
We used an age-structured Susceptible-Exposed-Infected-Recovered (SEIR) model with 4 age groups (age 3–12 y, 13–19 y, 20–29 y and 30–59 y) to simulate pdmH1N1 transmission between 1 June and 30 November 2009. The 0–2 and ≥60 age groups were omitted because (i) reliable serologic data from them were not available and (ii) they only represented 2% of all lab-confirmed pdmH1N1 cases and 5% of all pdmH1N1 hospitalizations and thus likely to have small contribution in pdmH1N1 transmission. In our sensitivity analysis, we showed that our results remained almost unchanged if we included these age groups in disease transmission. We used the POLYMOD matrices constructed for European countries (8 matrices and their average PAVG) as the contact matrix C because analogous data was unavailable from Hong Kong [26] and most of our results were insensitive to the choice of contact matrix. We included the effect of infection importations from Shenzhen, a large city adjacent to Hong Kong with a population of 13 million (Figure 1B).
We fitted the transmission model to the seroprevalence and hospitalization data by estimating the parameters listed in Table 1. All parameters were identifiable (Figure 3 and Table 1) and the fitted model was congruent with the data (Figure 4). Parameter estimates were very similar across all nine contact matrices except for age-specific susceptibility (see below for details). Partial rank correlation coefficient (PRCC) analysis did not indicate any unexpected confounding effects (see Text S1).
Fig. 3. Posterior distributions of parameter estimates. 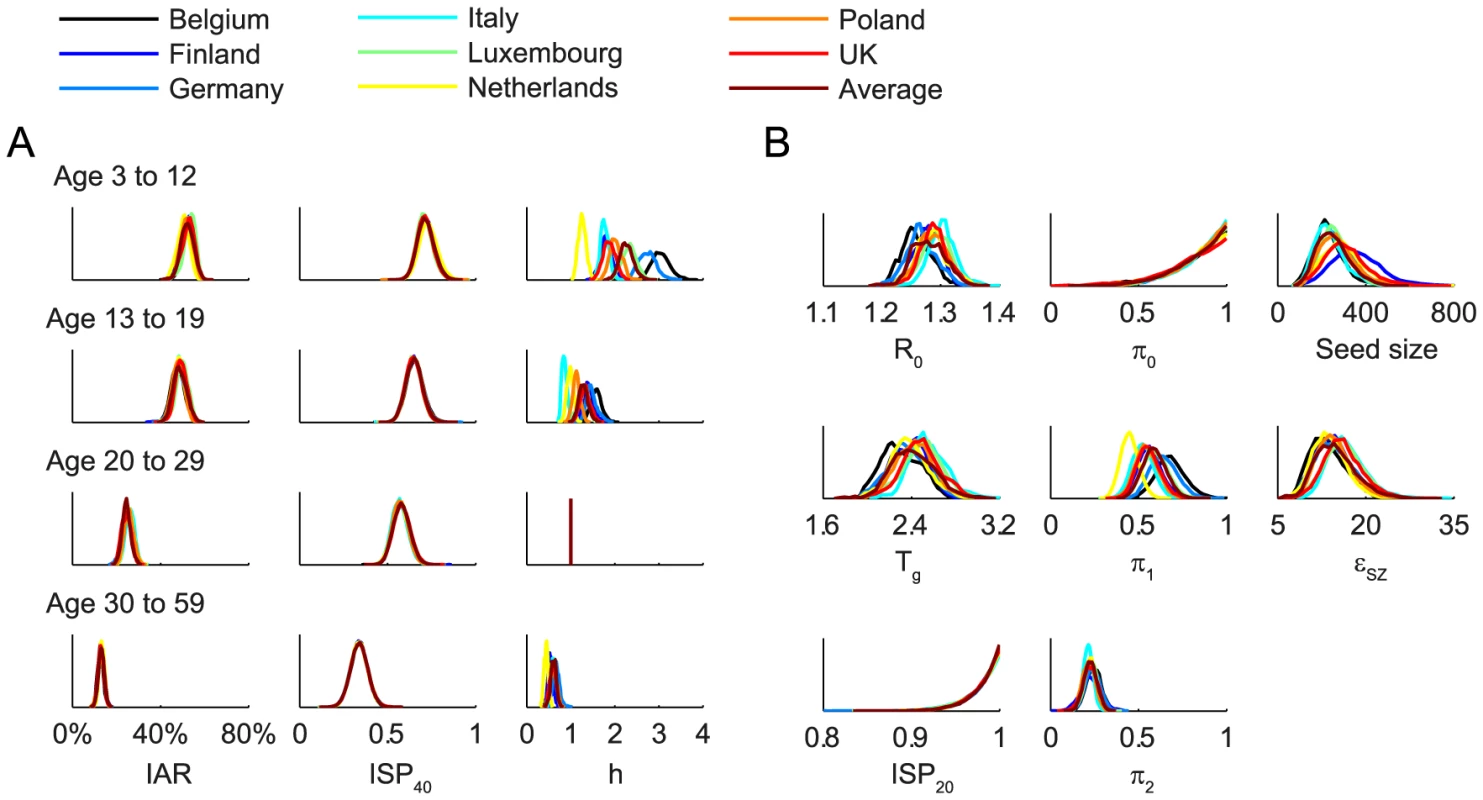
Different colors correspond to different POLYMOD contact matrices. A Age-dependent parameters including IARs (first column), ISP40 (second), and age-specific susceptibility (third). B Other parameters including R(0), Tg, ISP20, reduction in within-age-group mixing due to school closure (π0, π1, π2), seed size, and scaling factor for FOI from Shenzhen (εSZ). Fig. 4. Comparison of the data and the fitted model. 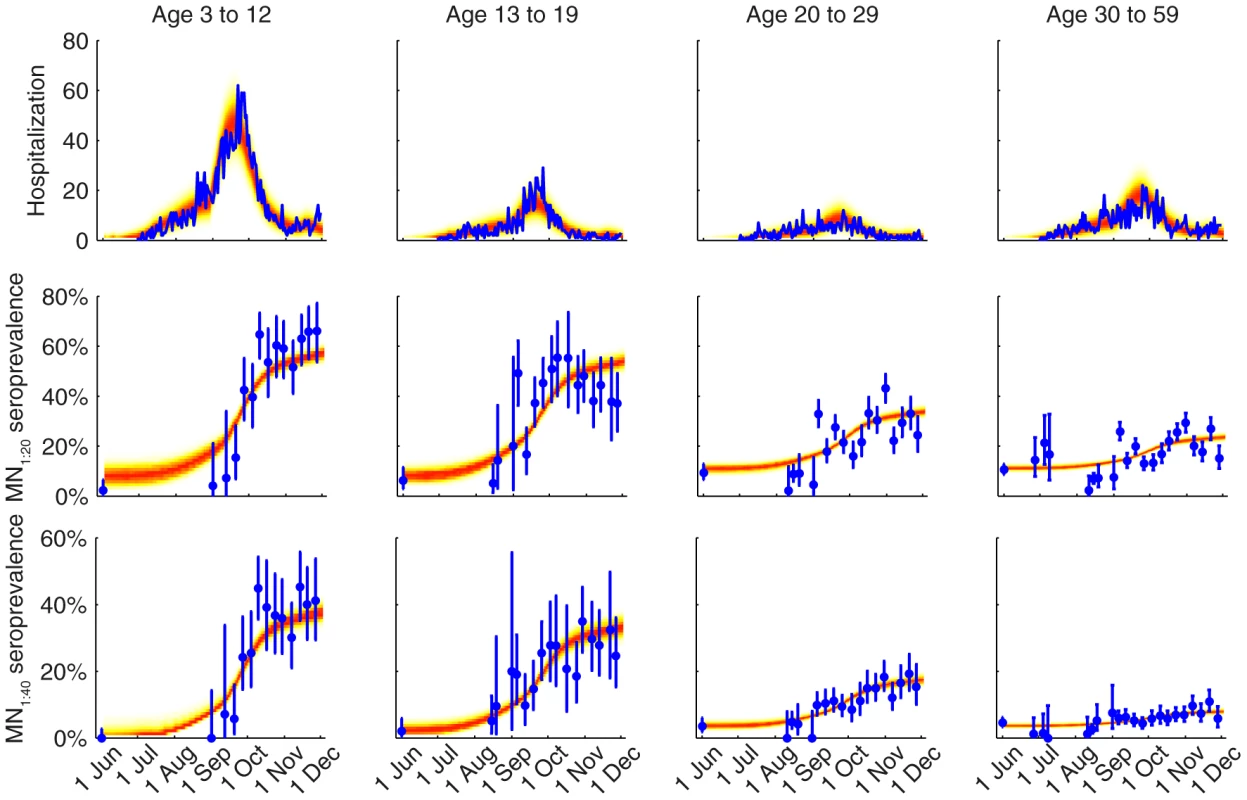
The hospitalization and serial cross-sectional seroprevalence data are shown in blue (vertical bars indicate 95% confidence intervals). Posterior intervals of hospitalizations and seroprevalence in the fitted model are shown as heat shades in which darker colors represent higher probability densities (i.e. highest density in red and zero density in white). Tab. 1. Model parameters and their posterior statistics. 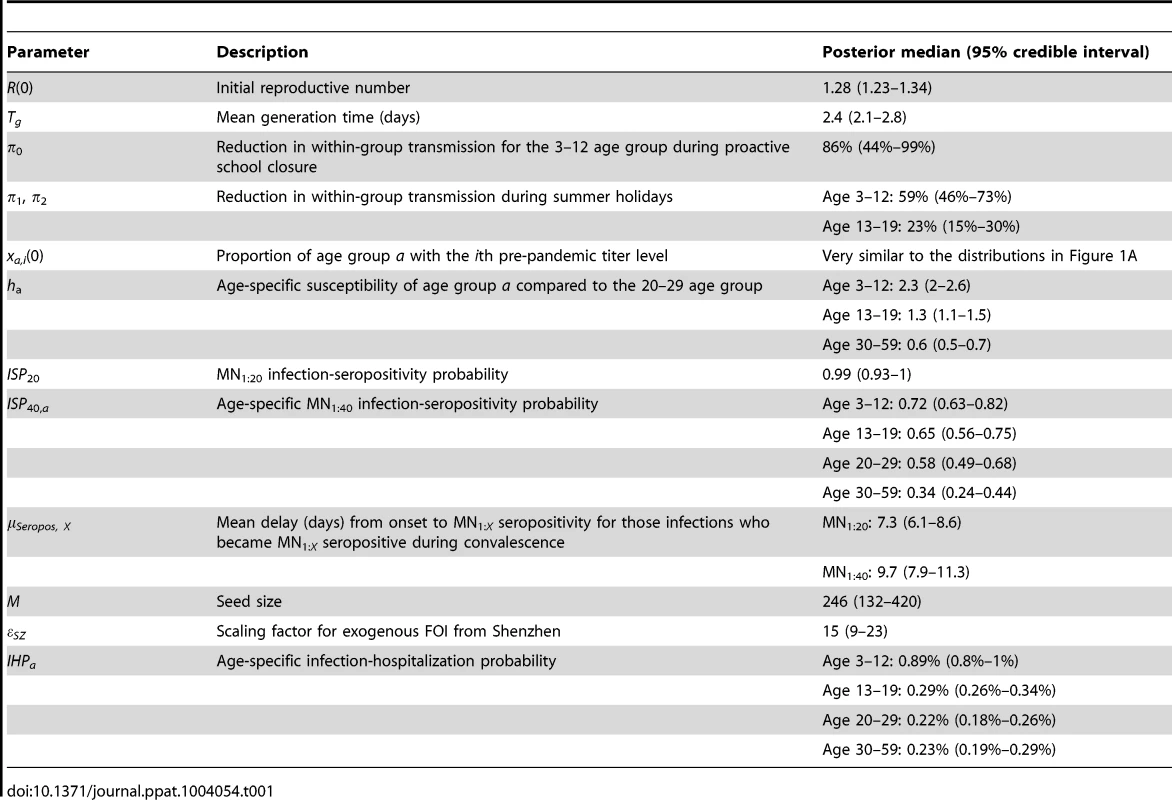
We estimated that the initial reproductive number R(0) was 1.28 (95% credible interval, 1.23–1.34) and mean generation time Tg was 2.4 (2.1–2.8) days, i.e. consistent with estimates of pdmH1N1 transmission parameters in other studies [27]. The scaling factor for the force of infection (FOI) from Shenzhen was εSZ = 15 (9–23), which conformed with the intuition that
(see Text S1).Among infected individuals who were MN1∶20 seronegative before infection, 99% (93%–100%) became MN1∶20 seropositive with a mean delay of 7.3 (6.1–8.6) days after onset. Among infected individuals who were MN1∶40 seronegative before infection, 72% (63%–82%), 65% (56%–75%), 58% (49%–68%) and 34% (24%–44%) among the 3–12, 13–19, 20–29 and 30–59 age group became MN1∶40 seropositive with a mean delay of 9.5 (7.9–11.3) days after onset. Hence, ISP40 decreased with age and was much lower compared to the 90–100% of clinical cases that became MN1∶40 seropositive [23], [24]. Consequently, IAR estimates here were significantly higher than our previous estimates, especially for the 30–59 age group [13]: 52% (46%–58%), 49% (43%–55%), 25% (21%–29%) and 13% (10%–16%) for age 3–12, 13–19, 20–29 and 30–59.
Proactive closure of kindergartens and primary schools reduced mixing among children aged 3–12 by 86% (44%–99%). Summer holidays reduced within-age-group mixing by 59% (46%–73%) and 23% (15%–30%) for age 3–12 and 13–19. A weaker effect of school closing for age 13–19 was plausible because older teenagers were more likely to actively mix with their peers in non-school settings while schools were closed.
Age-specific susceptibilities ha's were sensitive to the choice of contact matrix C because disease transmission was essentially driven by the matrix {Wab = haCab}. For all POLYMOD matrices, adults aged 30–59 were 0.4–0.7 times as susceptible as those aged 20–29. Children aged 3–12 were ∼2–3 times as susceptible as those aged 20–29 except for the Netherlands matrix which gave an estimate of 1.3 (1.1–1.4). Children aged 13–19 were 1.1–1.6 times more susceptible than adults aged 20–29 except for the Italy matrix which gave an estimate of 0.9 (0.8–1). In summary, these age-specific susceptibility estimates were consistent with analogous estimates from studies which showed that susceptibility decreased with age after adjusting for preexisting antibody titers and close contacts [28], [29].
Discussion
We hypothesized that influenza seroprevalence studies might substantially underestimate IARs if ISP is ignored or based on data from patients presenting to healthcare providers with clinically overt disease. We substantiated this hypothesis with pdmH1N1 seroprevalence data from Hong Kong. To further examine the validity of this conjecture, we performed crude analyses of published pdmH1N1 seroprevalence data from other countries to examine the robustness of their IAR estimates across different seropositivity thresholds and ISP adjustments (see Text S1). In a study in Germany with HI1∶40 threshold and no ISP adjustments, ΔSHI 1∶40/ΔS HI 1∶20 was around 0.9, 0.7, and 0.4 among unvaccinated individuals of age 18–32, 33–52 and >52 [8]. Similarly, in a study in New Zealand with HI1∶40 threshold and no ISP adjustments, ΔSHI 1∶40/ΔS HI 1∶20 was around 0.7, 0.9, and 0.6 among individuals of age 1–4, 5–19 and 20–59 [6]. Therefore, IAR have probably been underestimated in these studies, especially among older adults.
To our knowledge, only four pdmH1N1 studies had adjusted for ISP: one from the UK with HI1∶32 threshold [12], two from the US with HI1∶40 threshold [14], [15], and the remaining one by ourselves previously with MN1∶40 threshold [13]. All four assumed that ISP was similar to the proportion of patients with clinical disease presenting to healthcare providers who became seropositive. We have already shown that this assumption was inconsistent with population-level seroprevalence rises in Hong Kong where only 60%–70% and 34% of pdmH1N1 infections among age 3–29 and 30–59 became MN1∶40 seropositive (Table 1). In the UK study [12], [30], the HI1∶8 IAR estimate was 1.2–1.4 times the HI1∶32 estimate for those aged 25–44. Similarly, in the study in Florida [14], the HI1∶20 IAR estimate was around 1.5–1.7 and 1.9–2.1 times the HI1∶40 estimate for those aged 25–49 and 50–64. In the US multi-state study [15], the HI1∶20 IAR estimate was 1.2–1.3 times the HI1∶40 estimate for those aged 25–64. These results support our conjecture that serologic responses of clinical cases are not necessarily representative.
The most plausible and straightforward explanation was that mild and asymptomatic cases were less likely to become seropositive compared to clinical cases. Testing this hypothesis would require studying serologic responses of infected cases with different severity which would be feasible only with a large prospective cohort study with intensive monitoring to identify mild and asymptomatic cases. Nonetheless, some data from independent studies support this hypothesis. Hung et al reported that among 881 lab-confirmed pdmH1N1 symptomatic patients in Hong Kong, convalescent MN titer correlated well with initial viral load and was independently associated with severity [23]. Specifically, being afrebile on presentation was associated with poorer MN convalescent response. Among 44 RT-PCR confirmed cases (22 cohort subjects with mild symptoms and 22 hospital patients) in Singapore, 89% and 57% became MN1∶20 and MN1∶40 seropositive [7]. However, there are also published data which contradict the hypothesis. In a study of 24 patients and their 34 household infectees (all RT-PCR confirmed) in Canada, the MN1∶20 and MN1∶40 seropositivity rates were both 83% [31]. Nonetheless, we caution that titer measurements from different studies might not be directly comparable because serologic titer from different laboratories might vary due to differences in serologic assay protocols and endpoint analysis methods [32]. In particular, serologic follow-ups of clinical cases and seroprevalence studies are often conducted by separate groups with different laboratories, thus adding uncertainty to the consistency between ISP adjustments and seroprevalence data. Our serologic methods were the same as that in the serologic follow-up studies in Hung et al and Mak et al [23], [33], so the results therein should be readily comparable with ours. The Consortium for the Standardization of Influenza Seroepidemiology (CONSISE) is a recent global initiative aiming to standardize both laboratory and field investigation protocols for influenza seroepidemiology (http://consise.tghn.org). Our study suggested that collective interpretation of seroprevalence data and convalescent serologic data should also be an essential part of this standardization effort (e.g. real-time assessment of bias in ISP adjustments by evaluating the consistency of IAR across multiple thresholds and with mixture models; see below for more detailed discussions). Robust sero-surveillance requires an integrated understanding and standardization of the field, laboratory and analytical components of seroepidemiology.
Our results indicated that preexisting MN titers and age group mixing alone could not explain the age distribution of infections. The age-specific susceptibility estimates (Table 1) suggested that older individuals were protected from pdmH1N1 infections by some forms of immunity not reflected by pre-existing MN titers (e.g. cell-mediated immunity). Cytotoxic T cells established by prior seasonal influenza infections were demonstrated to cross react with pdmH1N1 viruses and it is conceivable that such cross-reactive T cell immunity increases with age [34]. Furthermore, the substantial proportion of infections that remained MN1∶40 seronegative might have relatively weak and short-lived immunity against pdmH1N1. Waning of such immunity might have subsequently replenished the pool of susceptibles and permitted a second epidemic of pdmH1N1 to occur in Hong Kong in 2011.
Our study has several limitations. First, we assumed that MN titer rises were entirely attributable to pdmH1N1 infection with immunity (lasting until at least 30 November 2009). In theory, it might be possible that individuals could be exposed to pdmH1N1, became MN1∶20 seropositive but MN1∶40 seronegative, and remained susceptible and noninfectious (i.e. weak serologic response without infection and immunity). This could be an alternative explanation for the discrepancy between ΔS40/ΔS20 in seroprevalence data and the ratio of clinical cases that became MN1∶40 and MN1∶20 seropositive in Hong Kong (Figure 2). In this case, ISP20 and ISP40 could remain at 1 and 0.9 (as observed among clinical cases) for all infections and the gap between ΔS40/ISP40 and ΔS20/ISP20 would comprise these exposed but uninfected individuals who became MN1∶20 seropositive but MN1∶40 seronegative. Second, our serologic data were collected via convenience sampling of blood donors, hospital outpatients and participants in community-based studies and hence did not necessarily provide a representative description of pdmH1N1 seroprevalence in the general population. Third, we did not account for any seasonal effects of influenza transmission. The bulk of pdmH1N1 first wave transmission in Hong Kong occurred between 1 September and 30 November 2009, a period during which circulation of seasonal influenza is typically low [35]. As such, the effect of school closure might be stronger than estimated here if seasonality had substantially reduced the transmissibility of pdmH1N1 during September-November 2009. Fourth, we did not consider the potential effect of oseltamivir use on serologic responses. Although oseltamivir use might attenuate serologic response of pdmH1N1 cases [25], treatment coverage was unlikely to be high enough to have a substantial impact on ΔS40/ΔS20. Finally, we did not have local social contact data to parameterize our transmission model and had to resort to uncertainty analysis using the POLYMOD matrices. However, this does not imply that we expect the contact pattern in Hong Kong to be similar to that in the European countries. Instead, we showed that our results were robust against the choice of contact matrix because given any contact matrix, the age-specific susceptibility was adjusted by the Bayesian inference algorithm accordingly to result in similar transmission dynamics and hence goodness-of-fit (Figure S4, S5, S6, S7, S8, S9, S10, S11, Figure S12).
Sero-epidemiologic study is the most practical method for accurately estimating influenza IAR, disease severity and population-level immunity which in turn are used to inform vaccination policies and decisions [5]. Our study emphasizes that IAR estimates in seroprevalence studies are sensitive to not only seropositivity thresholds but also ISP adjustments. Steens et al has made a similar observation when they compared pdmH1N1 IAR estimates obtained from conventional thresholds with that from mixture model [17]. Seropositivity thresholds have been typically chosen based on conventions instead of systematic criteria [4]. ISP adjustments have either been ignored or based on clinical patients whose antibody kinetics might not be representative for all infections in the community. Although we have shown that conventional seropositivity thresholds and ISP adjustments have probably led to underestimation of the incidence of pdmH1N1, such bias associated with conventional practice is not specific to pdmH1N1 or the serial cross-sectional design of sero-epidemiology. The longitudinal (cohort) design relies on the definition of seroconversion and infection-seroconversion probability. A recent study by Cauchemez et al reported that under the conventional criterion of seroconversion, namely 4-fold rise or more in antibody titers, influenza IARs were substantially underestimated when there were a significant proportion of subjects with 2-fold rises not explainable by measurement errors alone [36].
These studies and ours thus indicated the need for reevaluating current methods for analyzing influenza serologic data. For example, our group and Baguelin et al previously considered a method for generating real-time estimates of IAR and disease severity for pandemic influenza from serial cross-sectional seroprevalence and clinical surveillance data [12], [13]. This method requires a priori specifying the seropositivity threshold and ISP. Although basing ISP on antibody kinetics of clinical cases is likely to be the best a priori option in the real-time pandemic setting, the associated bias can and should be assessed by evaluating the consistency of IAR and severity estimates across multiple thresholds and with mixture models. A natural extension of the method is to analyze seroprevalence data at multiple thresholds under a Bayesian framework using ISPs among clinical cases as priors (possibly with the extension of integrating transmission dynamics as done here and in Birrel et al [37]). Within this framework, ISP can be continuously updated by the posteriors to reconcile discrepancies between seroprevalence data and ISP priors (e.g. Figure 2). Although the potential bias in ISP priors may not be completely eliminated in real-time, the resulting IAR and severity estimates will likely remain sufficiently precise for informing situational awareness and pandemic responses. In conclusion, our results indicated the need for reexamining conventional practice in influenza sero-epidemiology to develop standards for analyzing influenza serologic data, especially in the context of pandemics when robustness and comparability of IAR estimates are most needed for informing situational awareness and risk assessment. While these studies were conducted within the context of influenza, these methodological approaches are broadly applicable to other infectious disease outbreaks.
Materials and Methods
Ethics statement
All study protocols were approved by the Institutional Review Board of The University of Hong Kong/Hospital Authority Hong Kong West Cluster. All adult subjects provided written informed consent, and a parent or guardian of any child participant provided written consent on their behalf.
Transmission modeling
Major modeling assumptions are summarized below (see Text S1 for further technical details):
-
Antibody kinetics and testing. Each infection in age group a became MN1:X seropositive with probability ISPX,a if they were MN1:X seronegative before infection. Because ISP20,a and ISP40,a were not simultaneously identifiable from our data, we assumed that ISP20,a was independent of age. We assumed four pre-pandemic MN titer levels (<1∶10, 1∶10, 1∶20, and ≥1∶40; Figure 1A) and that the ith pre-pandemic titer level reduced susceptibility by 1−gi compared to the lowest level (i.e. g1 = 1). The onset-to-seropositivity duration was estimated using antibody kinetics data from clinical cases in Hong Kong [33]. Sensitivity (specificity) of serologic testing, defined as the probability that the serologic result was positive (negative) if the specimen was truly seropositive (seronegative), was assumed to be 100%. Imperfect sensitivity and specificity had little impact on our conclusions (see Text S1).
-
Age-specific susceptibility. Age group a was ha times as susceptible compared to the 20–29 age group, i.e. h3 = 1. These age-specific susceptibility parameters modeled differential susceptibility not explainable by the contact matrix and pre-pandemic MN titers.
-
School closure. As a proactive mitigation measure, the Hong Kong government closed all kindergartens and primary schools on 11 June 2009 until summer holidays. We assumed that summer holidays and fall semester started on 10 July and 1 September, respectively. Within-age-group mixing was reduced by π0 for age 3–12 during proactive school closure, and by π1 and π2 for age 3–12 and 13–19 during summer holidays.
-
Importation of infections. We seeded the pandemic on 1 June 2009 with M infectious cases. In addition, we assumed that Hong Kong was subject to an exogenous force of infection that was εSZ times the estimated daily number of lab-confirmed cases in Shenzhen [38] because (i) an average of ∼350,000 people crossed the border on a daily basis; and (ii) sustained low levels of transmission in Hong Kong during November 2009 was likely fueled by the Shenzhen epidemic which peaked in that month [38] (Figure 1B).
-
Hospitalization. We assumed that each infection in age group a required hospitalization with probability IHPa (infection-hospitalization probability).
-
Infectiousness and antibody response. We assumed that all infected individuals were equally infectious regardless of their antibody response. In the Text S1, we showed that our results were robust against potential association between infectiousness and antibody response.
Statistical analysis
We fitted the transmission model to the seroprevalence and hospitalization data by estimating the parameters listed in Table 1 using Markov Chain Monte Carlo methods with non-informative flat priors. Because around 85% and 10% of each age group had pre-pandemic MN titer <1∶10 and 1∶10 (Figure 1A), the gi's were not identifiable. As such, we assumed gi = gi which had negligible effect because the small proportion of individuals who had pre-pandemic titer >1∶10 had little impact on transmission dynamics. Partial rank correlation coefficients (PRCC) among estimated parameters were calculated to identify any strong (defined here as |PRCC|>0.5) but unexpected confounding effects.
For uncertainty analysis, we performed statistical inference for g = 0, 0.5 and 1 and each of the nine POLYMOD matrices, i.e. a total of 27 scenarios. Higher g (i.e. preexisting MN titer conferred weaker protection) resulted in slightly higher IARs and lower R(0) and Tg (Figure S4, S5, S6, S7, S8, S9, S10, S11, Figure S12). Otherwise, all combinations of g and C resulted in similar goodness-of-fit and parameter estimates except for age-specific susceptibilities. As such, we describe in the main text the inference results (posterior medians and 95% credible intervals) for g = 0.5 and C = PAVG unless parameter estimates were sensitive to g and C (i.e for age-specific susceptibilities).
Supporting Information
Zdroje
1. Van KerkhoveMD, AsikainenT, BeckerNG, BjorgeS, DesenclosJ-C, et al. (2010) Studies Needed to Address Public Health Challenges of the 2009 H1N1 Influenza Pandemic: Insights from Modeling. PLoS Med 7: e1000275.
2. LipsitchM, FinelliL, HeffernanRT, LeungGM, ReddSC, et al. (2011) Improving the evidence base for decision making during a pandemic: the example of 2009 influenza A/H1N1. Biosecurity and bioterrorism : biodefense strategy, practice, and science 9 : 89–115.
3. Fineberg HV, Aavitsland P, Aditama T, Bino S, Carmo EH, et al.. (2011) Implementation of the international health regulations (2005): report of the Review Committee on the Functioning of the International Health Regulations (2005) and on Pandemic Influenza A(H1N1) 2009. Geneva: World Health Organization.
4. BrobergE, NicollA, Amato-GauciA (2011) Seroprevalence to influenza A(H1N1) 2009 virus–where are we? Clin Vaccine Immunol 18 : 1205–1212.
5. Van KerkhoveMD, HirveS, KoukounariA, MountsAW (2013) Estimating age-specific cumulative incidence for the 2009 influenza pandemic: a meta-analysis of A(H1N1)pdm09 serological studies from 19 countries. Influenza and Other Respiratory Viruses 7 : 872–886.
6. BandaranayakeD, HuangQS, BissieloA, WoodT, MackerethG, et al. (2010) Risk Factors and Immunity in a Nationally Representative Population following the 2009 Influenza A(H1N1) Pandemic. PLoS ONE 5: e13211.
7. ChenMI, BarrIG, KohGCH, LeeVJ, LeeCPS, et al. (2010) Serological Response in RT-PCR Confirmed H1N1-2009 Influenza A by Hemagglutination Inhibition and Virus Neutralization Assays: An Observational Study. PLoS ONE 5: e12474.
8. DudarevaS, SchweigerB, ThammM, HöhleM, StarkK, et al. (2011) Prevalence of Antibodies to 2009 Pandemic Influenza A (H1N1) Virus in German Adult Population in Pre - and Post-Pandemic Period. PLoS ONE 6: e21340.
9. McVernon JLK, NolanT, OwenR, IrvingD, CapperH, et al. (2010) Seroprevalence of 2009 pandemic influenza A(H1N1) virus in Australian blood donors, October - December, 2009. Eurosurveillance 15: pii = 19678.
10. MillerE, HoschlerK, HardelidP, StanfordE, AndrewsN, et al. (2010) Incidence of 2009 pandemic influenza A H1N1 infection in England: a cross-sectional serological study. The Lancet 375 : 1100–1108.
11. WuJT, MaESK, LeeCK, ChuDKW, HoP-L, et al. (2010) The Infection Attack Rate and Severity of 2009 Pandemic H1N1 Influenza in Hong Kong. Clinical Infectious Diseases 51 : 1184–1191.
12. BaguelinM, HoschlerK, StanfordE, WaightP, HardelidP, et al. (2011) Age-Specific Incidence of A/H1N1 2009 Influenza Infection in England from Sequential Antibody Prevalence Data Using Likelihood-Based Estimation. PLoS ONE 6: e17074.
13. WuJT, HoA, MaESK, LeeCK, ChuDKW, et al. (2011) Estimating Infection Attack Rates and Severity in Real Time during an Influenza Pandemic: Analysis of Serial Cross-Sectional Serologic Surveillance Data. PLoS Med 8: e1001103.
14. CoxCM, GoodinK, FisherE, DawoodFS, HamiltonJJ, et al. (2011) Prevalence of 2009 pandemic influenza A (H1N1) virus antibodies, Tampa Bay Florida–November-December, 2009. PLoS ONE 6: e29301.
15. ReedC, KatzJM, HancockK, BalishA, FryAM, et al. (2012) Prevalence of Seropositivity to Pandemic Influenza A/H1N1 Virus in the United States following the 2009 Pandemic. PLoS ONE 7: e48187.
16. Hens N, Shkedy Z, Aerts M, Faes C, Van Damme P, et al.. (2012) Modeling Infectious Disease Parameters Based on Serological and Social Contact Data: Springer. 298 p.
17. SteensA, WaaijenborgS, TeunisPFM, ReimerinkJHJ, MeijerA, et al. (2011) Age-Dependent Patterns of Infection and Severity Explaining the Low Impact of 2009 Influenza A (H1N1): Evidence From Serial Serologic Surveys in the Netherlands. American Journal of Epidemiology 174 : 1307–1315.
18. HardelidP, WilliamsD, DezadeuxC, TookeyPA, PeckhamCS, et al. (2008) Analysis of rubella antibody distribution from newborn dried blood spots using finite mixture models. Epidemiology & Infection 136 : 1698–1706.
19. VyseAJ, AndrewsNJ, HeskethLM, PebodyR (2007) The burden of parvovirus B19 infection in women of childbearing age in England and Wales. Epidemiology & Infection 135 : 1354–1362.
20. CowlingBJ, LauMSY, HoL-M, ChuangS-K, TsangT, et al. (2010) The Effective Reproduction Number of Pandemic Influenza: Prospective Estimation. Epidemiology 21 : 842–846 810.1097/EDE.1090b1013e3181f20977.
21. WuJT, CowlingBJ, LauEHY, IpDKM, HoL-M, et al. (2010) School Closure and Mitigation of Pandemic (H1N1) 2009, Hong Kong. Emerging Infectious Diseases 16 : 538–541.
22. WuJT, MaESK, LeeCK, ChuDKW, HoPL, et al. (2010) The infection attack rate and severity of 2009 pandemic influenza (H1N1) in Hong Kong. Clinical Infectious Diseases 51 : 1184–1191.
23. HungIFN, ToKKW, LeeC-K, LinC-K, ChanJFW, et al. (2010) Effect of Clinical and Virological Parameters on the Level of Neutralizing Antibody against Pandemic Influenza A Virus H1N1 2009. Clinical Infectious Diseases 51 : 274–279.
24. VeguillaV, HancockK, SchifferJ, GargiulloP, LuX, et al. (2011) Sensitivity and Specificity of Serologic Assays for Detection of Human Infection with 2009 Pandemic H1N1 Virus in U.S. Populations. Journal of Clinical Microbiology 49 : 2210–2215.
25. CowlingBJ, ChanKH, FangVJ, LauLLH, SoHC, et al. (2010) Comparative Epidemiology of Pandemic and Seasonal Influenza A in Households. New England Journal of Medicine 362 : 2175–2184.
26. MossongJ, HensN, JitM, BeutelsP, AuranenK, et al. (2008) Social Contacts and Mixing Patterns Relevant to the Spread of Infectious Diseases. PLoS Med 5: e74.
27. BoëlleP-Y, AnsartS, CoriA, ValleronA-J (2011) Transmission parameters of the A/H1N1 (2009) influenza virus pandemic: a review. Influenza and Other Respiratory Viruses 5 : 306–316.
28. CauchemezS, DonnellyCA, ReedC, GhaniAC, FraserC, et al. (2009) Household Transmission of 2009 Pandemic Influenza A (H1N1) Virus in the United States. New England Journal of Medicine 361 : 2619–2627.
29. CauchemezS, BhattaraiA, MarchbanksTL, FaganRP, OstroffS, et al. (2011) Role of social networks in shaping disease transmission during a community outbreak of 2009 H1N1 pandemic influenza. Proceedings of the National Academy of Sciences 108 : 2825–2830.
30. HardelidP, AndrewsN, HoschlerK, StanfordE, BaguelinM, et al. (2010) Assessment of baseline age-specific antibody prevalence and incidence of infection to novel influenza A/H1N1. Health Technology Assessment 14 : 115–192.
31. PapenburgJ, BazM, HamelinM-È, RhéaumeC, CarbonneauJ, et al. (2011) Evaluation of Serological Diagnostic Methods for the 2009 Pandemic Influenza A (H1N1) Virus. Clinical and Vaccine Immunology 18 : 520–522.
32. WoodJM, Gaines-DasRE, TaylorJ, ChakravertyP (1994) Comparison of influenza serological techniques by international collaborative study. Vaccine 12 : 167–174.
33. MakGC, ChoyPWW, LeeWY, WongAH, NgKC, et al. (2010) Sero-immunity and serologic response to pandemic influenza A (H1N1) 2009 virus in Hong Kong. Journal of Medical Virology 82 : 1809–1815.
34. TuW, MaoH, ZhengJ, LiuY, ChiuSS, et al. (2010) Cytotoxic T lymphocytes established by seasonal human influenza cross-react against 2009 pandemic H1N1 influenza virus. Journal of virology 84 : 6527–6535.
35. TangJW, NgaiKL, LamWY, ChanPK (2008) Seasonality of influenza A(H3N2) virus: a Hong Kong perspective (1997–2006). PLoS ONE 3: e2768.
36. CauchemezS, HorbyP, FoxA, Mai leQ, Thanh leT, et al. (2012) Influenza infection rates, measurement errors and the interpretation of paired serology. PLoS Pathog 8: e1003061.
37. BirrellPJ, KetsetzisG, GayNJ, CooperBS, PresanisAM, et al. (2011) Bayesian modeling to unmask and predict influenza A/H1N1pdm dynamics in London. Proceedings of the National Academy of Sciences 108 : 18238–18243.
38. XieX, LuSQ, ChengJQ, ChengXW, XuZH, et al. (2012) Estimate of 2009 H1N1 influenza cases in Shenzhen–the biggest migratory city in China. Epidemiol Infect 140 : 788–797.
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Affinity Proteomics Reveals Elevated Muscle Proteins in Plasma of Children with Cerebral MalariaČlánek The Transcriptional Activator LdtR from ‘ Liberibacter asiaticus’ Mediates Osmotic Stress ToleranceČlánek Complement-Related Proteins Control the Flavivirus Infection of by Inducing Antimicrobial PeptidesČlánek Fungal Chitin Dampens Inflammation through IL-10 Induction Mediated by NOD2 and TLR9 ActivationČlánek Parasite Fate and Involvement of Infected Cells in the Induction of CD4 and CD8 T Cell Responses to
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2014 Číslo 4- Stillova choroba: vzácné a závažné systémové onemocnění
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Choroby jater v ordinaci praktického lékaře – význam jaterních testů
- Diagnostický algoritmus při podezření na syndrom periodické horečky
-
Všechny články tohoto čísla
- , , , Genetic Variability: Cryptic Biological Species or Clonal Near-Clades?
- Early Mortality Syndrome Outbreaks: A Microbial Management Issue in Shrimp Farming?
- Wormholes in Host Defense: How Helminths Manipulate Host Tissues to Survive and Reproduce
- Plastic Proteins and Monkey Blocks: How Lentiviruses Evolved to Replicate in the Presence of Primate Restriction Factors
- The 2010 Cholera Outbreak in Haiti: How Science Solved a Controversy
- Affinity Proteomics Reveals Elevated Muscle Proteins in Plasma of Children with Cerebral Malaria
- Noncanonical Role for the Host Vps4 AAA+ ATPase ESCRT Protein in the Formation of Replicase
- Efficient Parvovirus Replication Requires CRL4-Targeted Depletion of p21 to Prevent Its Inhibitory Interaction with PCNA
- Host-to-Pathogen Gene Transfer Facilitated Infection of Insects by a Pathogenic Fungus
- The Transcriptional Activator LdtR from ‘ Liberibacter asiaticus’ Mediates Osmotic Stress Tolerance
- Coxsackievirus B Exits the Host Cell in Shed Microvesicles Displaying Autophagosomal Markers
- TCR Affinity Associated with Functional Differences between Dominant and Subdominant SIV Epitope-Specific CD8 T Cells in Rhesus Monkeys
- Coxsackievirus-Induced miR-21 Disrupts Cardiomyocyte Interactions via the Downregulation of Intercalated Disk Components
- Ligands of MDA5 and RIG-I in Measles Virus-Infected Cells
- Kind Discrimination and Competitive Exclusion Mediated by Contact-Dependent Growth Inhibition Systems Shape Biofilm Community Structure
- Structural Differences Explain Diverse Functions of Actins
- HSCARG Negatively Regulates the Cellular Antiviral RIG-I Like Receptor Signaling Pathway by Inhibiting TRAF3 Ubiquitination Recruiting OTUB1
- Vaginitis: When Opportunism Knocks, the Host Responds
- Complement-Related Proteins Control the Flavivirus Infection of by Inducing Antimicrobial Peptides
- Fungal Chitin Dampens Inflammation through IL-10 Induction Mediated by NOD2 and TLR9 Activation
- Microbial Pathogens Trigger Host DNA Double-Strand Breaks Whose Abundance Is Reduced by Plant Defense Responses
- Alveolar Macrophages Are Essential for Protection from Respiratory Failure and Associated Morbidity following Influenza Virus Infection
- An Interaction between Glutathione and the Capsid Is Required for the Morphogenesis of C-Cluster Enteroviruses
- Concerted Spatio-Temporal Dynamics of Imported DNA and ComE DNA Uptake Protein during Gonococcal Transformation
- Potent Dengue Virus Neutralization by a Therapeutic Antibody with Low Monovalent Affinity Requires Bivalent Engagement
- Regulation of Human T-Lymphotropic Virus Type I Latency and Reactivation by HBZ and Rex
- Functionally Redundant RXLR Effectors from Act at Different Steps to Suppress Early flg22-Triggered Immunity
- The Pathogenic Mechanism of the Virulence Factor, Mycolactone, Depends on Blockade of Protein Translocation into the ER
- Role of Calmodulin-Calmodulin Kinase II, cAMP/Protein Kinase A and ERK 1/2 on -Induced Apoptosis of Head Kidney Macrophages
- An Overview of Respiratory Syncytial Virus
- First Experimental Model of Enhanced Dengue Disease Severity through Maternally Acquired Heterotypic Dengue Antibodies
- Binding of Glutathione to Enterovirus Capsids Is Essential for Virion Morphogenesis
- IFITM3 Restricts Influenza A Virus Entry by Blocking the Formation of Fusion Pores following Virus-Endosome Hemifusion
- Parasite Fate and Involvement of Infected Cells in the Induction of CD4 and CD8 T Cell Responses to
- Deficient IFN Signaling by Myeloid Cells Leads to MAVS-Dependent Virus-Induced Sepsis
- Pernicious Pathogens or Expedient Elements of Inheritance: The Significance of Yeast Prions
- The HMW1C-Like Glycosyltransferases—An Enzyme Family with a Sweet Tooth for Simple Sugars
- The Expanding Functions of Cellular Helicases: The Tombusvirus RNA Replication Enhancer Co-opts the Plant eIF4AIII-Like AtRH2 and the DDX5-Like AtRH5 DEAD-Box RNA Helicases to Promote Viral Asymmetric RNA Replication
- Mining Herbaria for Plant Pathogen Genomes: Back to the Future
- Inferring Influenza Infection Attack Rate from Seroprevalence Data
- A Human Lung Xenograft Mouse Model of Nipah Virus Infection
- Mast Cells Expedite Control of Pulmonary Murine Cytomegalovirus Infection by Enhancing the Recruitment of Protective CD8 T Cells to the Lungs
- Cytosolic Peroxidases Protect the Lysosome of Bloodstream African Trypanosomes from Iron-Mediated Membrane Damage
- Abortive T Follicular Helper Development Is Associated with a Defective Humoral Response in -Infected Macaques
- JC Polyomavirus Infection Is Strongly Controlled by Human Leucocyte Antigen Class II Variants
- Cationic Antimicrobial Peptides Promote Microbial Mutagenesis and Pathoadaptation in Chronic Infections
- Estimating the Fitness Advantage Conferred by Permissive Neuraminidase Mutations in Recent Oseltamivir-Resistant A(H1N1)pdm09 Influenza Viruses
- Progressive Accumulation of Activated ERK2 within Highly Stable ORF45-Containing Nuclear Complexes Promotes Lytic Gammaherpesvirus Infection
- Caspase-1-Like Regulation of the proPO-System and Role of ppA and Caspase-1-Like Cleaved Peptides from proPO in Innate Immunity
- Is Required for High Efficiency Viral Replication
- Modified Vaccinia Virus Ankara Triggers Type I IFN Production in Murine Conventional Dendritic Cells via a cGAS/STING-Mediated Cytosolic DNA-Sensing Pathway
- Evidence That Bank Vole PrP Is a Universal Acceptor for Prions
- Rapid Response to Selection, Competitive Release and Increased Transmission Potential of Artesunate-Selected Malaria Parasites
- Inactivation of Genes for Antigenic Variation in the Relapsing Fever Spirochete Reduces Infectivity in Mice and Transmission by Ticks
- Exposure-Dependent Control of Malaria-Induced Inflammation in Children
- A Neutralizing Anti-gH/gL Monoclonal Antibody Is Protective in the Guinea Pig Model of Congenital CMV Infection
- The Apical Complex Provides a Regulated Gateway for Secretion of Invasion Factors in
- A Highly Conserved Haplotype Directs Resistance to Toxoplasmosis and Its Associated Caspase-1 Dependent Killing of Parasite and Host Macrophage
- A Quantitative High-Resolution Genetic Profile Rapidly Identifies Sequence Determinants of Hepatitis C Viral Fitness and Drug Sensitivity
- Histone Deacetylase Inhibitor Romidepsin Induces HIV Expression in CD4 T Cells from Patients on Suppressive Antiretroviral Therapy at Concentrations Achieved by Clinical Dosing
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- The 2010 Cholera Outbreak in Haiti: How Science Solved a Controversy
- , , , Genetic Variability: Cryptic Biological Species or Clonal Near-Clades?
- Efficient Parvovirus Replication Requires CRL4-Targeted Depletion of p21 to Prevent Its Inhibitory Interaction with PCNA
- An Overview of Respiratory Syncytial Virus
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání




