-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Viral Enhancer Mimicry of Host Innate-Immune Promoters
article has not abstract
Published in the journal: . PLoS Pathog 10(2): e32767. doi:10.1371/journal.ppat.1003804
Category: Opinion
doi: https://doi.org/10.1371/journal.ppat.1003804Summary
article has not abstract
The inflammatory milieu is the natural habitat for a pathogenic infection, characterised by activity of pro-inflammatory signalling pathways and inflammatory cytokines. Viral entry rapidly activates a range of innate-immune signalling events such as the activation of Pattern Recognition Receptors (PRRs) [1]–[5]. A virus must therefore counteract intrinsic cellular and innate-immune responses to successfully complete the replication cycle. Frequently this is accomplished by encoding viral effector molecules that block these cellular responses by working as either structural or functional mimics of host target proteins [6]–[11]. Nuclear DNA viruses are dependent on the host transcriptional machinery to express the first viral genes; for example the immediate-early (IE) control elements of DNA viruses are by definition absolutely dependent on host transcription factors (TF) [12]. Therefore, these viruses are particularly hostage to their host transcriptional environment [13], [14]. Here we propose that mimicry of regulatory DNA sequences by viral regulatory regions may also provide an additional strategy to counteract at IE times of infection the innate-immune response. In this context, viral IE control elements might functionally mimic innate-immune enhancers, taking advantage of the activated immune signalling TFs for promoting viral IE gene expression.
In other words: “If you can't beat ‘em. Join ‘em.”
In exploring this possibility, we present a synopsis of the promoter-regulatory elements from seven extensively studied mammalian viruses with a DNA stage, and seven promoters representing prototypical cellular innate-immune genes. These are the SV-40 early enhancer, the E1A enhancer of HAdV5, the long terminal repeat (LTR) of HIV-1, the E6/7 long control region (LCR) of both HPV-16 and HPV-18, the major IE (MIE) enhancer of HCMV, and the enhancer-1 (Eh-1) regulatory region of HBV for viral sequences, and the enhancer regions of human IFNB1, IFNG, TNF, IRF1, IL8, IL12B, and IL1B for host sequences. First, we consider similarities between the primary sequence structures of the enhancers. Second, we present arguments for convergent evolution and structural flexibility inherent to enhancer sequences. Third, we discuss functional features and regulatory hallmarks that may be used to define viral enhancer mimicry of cellular immune enhancers.
Do Viral and Cellular Enhancers Display Any Primary Sequence Similarity?
To investigate if there is any similarity of primary sequences and therefore structural mimicry between the selected viral and cellular enhancers, we used the BLAST tool to compare the sequences against each other (Table 1) and applied an exhaustive pairwise multi-way alignment (CloneManager suite 7.0) to search for similarities in this group of sequences (Figure 1A). While multi-way alignment of the various selected viral and cellular promoter-regulatory regions (Figure 1A, top panel) reveals a lack of extended primary sequence homology, the pairwise BLAST comparison showed that small islands of sequence identity or high similarity are present (Table 1). We randomly compared some of these short sequence motifs with the JASPAR CORE (Vertebrae) database [15] and found that all checked motifs have similarities with consensus binding motifs for TFs (e.g., AP1, SP1, YY1, or RelA with relative scores of >0.8). This finding raises the question of whether there might be functional similarity. We therefore consider in the next section how convergent evolution of viral enhancers may have resulted in functional mimicry of the transcription control elements of innate-immune genes, providing a co-opting strategy for immune evasion.
Fig. 1. Comparison of host innate-immune and viral regulatory regions. 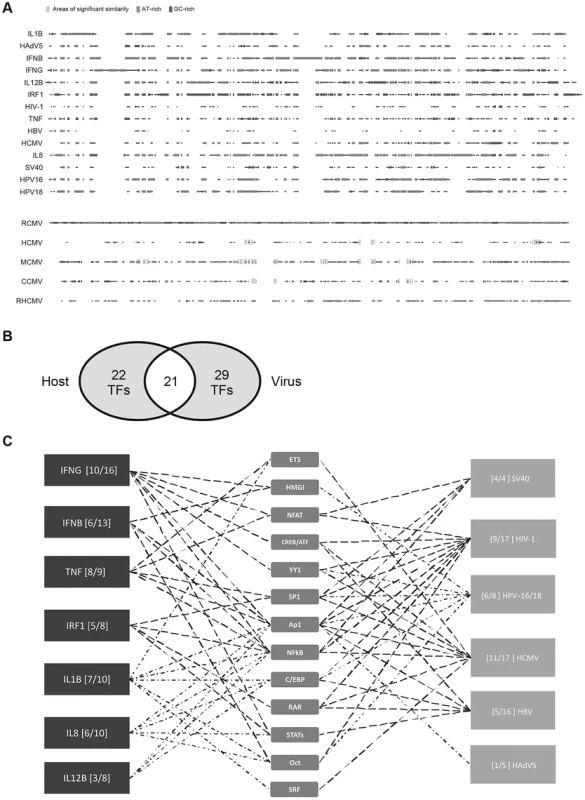
A) Multi-way alignment of analysed enhancer sequences shows no sequence similarity. Narrow grey boxes mark AT-rich stretches and dark grey boxes mark GC-rich stretches. Overall, sequence similarity was too low to produce a phylogenetic tree. To analyse sequence similarity within one family of viruses, we compared the major immediate-early enhancer region of rat-CMV (RCMV) with those of human (HCMV), murine (MCMV), chimpanzee (CCMV) and rhesus (RHCMV) cytomegalovirus. Small stretches of sequences similarity to the RCMV sequence are indicated by wide light grey boxes (similarity >80%). B) Venn diagram of 72 TFs identified in our literature search to interact with the analysed regulatory sequences. Detailed SBGN diagrams of all elements and interactions can be found at [46]–[52] except for TNF [57]. C) Simplified summary of transcription factor families shared between analysed innate-immune regulatory regions and viral control elements. For simplification interactions with members belonging to a family of TFs are represented by only one symbol (e.g., p50, p65, and RelA interactions are all represented by the “NFkB” symbol). The summary was produced in the “MSc by research in genomics and pathway biology” project by literature review. Digits in brackets indicate the number of shared interactions (left of dash) and total number of interactions for the specific enhancer (right of dash). TFs that are more highly connected with viral and host elements were placed toward the centre. Tab. 1. Summary of pairwise sequence comparison. 
Pairwise comparison of analysed enhancer sequences and number of similarity islands identified by BLAST (blastn) alignment. Each regulatory region was used as a reference sequence and compared to all other sequence elements. The number of sequence motifs with high similarity produced from this analysis are given in parentheses; comparisons that produced no significant similarities are not shown. Could Viral Regulatory Regions Evolve as Functional Mimics of Innate-Immune Enhancers without Extensive Sequence Similarity?
There are two principal genetic mechanisms that could lead to viral mimicry of host enhancers, horizontal transfer of cellular sequences to viral genomes or genetic drift of viral sequences. The first possibility, acquisition of cellular sequences through horizontal sequence transfer, could arise through illegitimate recombination with host DNA, for example by retro-transposition of non-coding RNA transcripts, resulting in the virus hijacking host transcription control sequences. If this were the general case, we would, however, expect significant structural similarity, which we did not find in our analysis. Alternatively, but not mutually exclusive from horizontal transfer, viral enhancer mimics could arise through neutral evolution and genetic drift by sequence duplication or accumulation of point mutations. Duplicated sequence features are hallmarks for many viral and cellular enhancers [16]–[24]. For instance, deletion or loss of enhancer sequences in SV40 and JC polyomavirus promotes restoration of enhancer function through duplication of flanking sequences [25]–[28]. A third possibility is the accumulation of point mutations in enhancer sequences and subsequent fixation [29]. It has recently been described for a wide range of species that evolution of host-cell transcriptional control can occur in relatively short time spans and is mainly driven by the rapid and flexible emergence or loss of binding motifs rather than by evolution of the TF proteins themselves [30]–[36]. The described mechanisms of rapid enhancer evolution argue that viral enhancers could acquire functionality that mimics innate-immune enhancers without any extensive sequence homology, and this is consistent with the comparison of cellular and viral enhancers shown in Figure 1A. This possibility is underscored by the fact that promoter sequences seem to be poorly conserved even among members within a virus-family yet share many of the same regulatory elements [37]. For example the MIE enhancers of cytomegaloviruses show low levels of primary sequence similarity between the different species strains (Figure 1A, lower panel). Despite these differences, functionality of the enhancers is conserved between hosts for different CMV species strains, e.g., the human CMV enhancer can functionally complement deletion of the murine CMV enhancer [38] and human CMV enhancer sequences recapitulate in vivo biological sites of infection in species from mice to zebra fish [39]–[41].
What Features Would Classify a Viral Enhancer as an Innate-Immune Enhancer Mimic?
Since our work and that of others discussed so far indicates that viral enhancers are functional rather than structural mimics of host innate-immune enhancers, we suggest four principal hallmarks of functional enhancer mimicry. These are: 1) shared TF interactions independent of sequence structure, 2) similar kinetics of gene induction between cellular innate-immune and viral IE genes, 3) positive responsiveness to immune-stimulatory ligands, and 4) susceptibility to inhibition of inflammatory signalling. In the following section we briefly discuss these hallmarks.
Shared Transcription Factor Interactions
The human genome encodes an estimated 1,700 to 1,900 TFs, with 1,391 representing high-confidence candidates [42]. These proteins represent an ample resource for viruses to harness. To probe, in more detail, the TF usage of the 14 viral and innate-immune enhancers selected (Table 1), we constructed unambiguous diagrams [43]–[45] of known TF interactions—available as an online resource on Figshare [46]–[52]. Using this approach we identified 72 interactions (Table 2) between the selected host and viral regulatory regions and host TFs. Of the 72 interactions identified, 43 were described for cellular enhancers and 50 for viral enhancers and 21 interactions (49% and 42% respectively) are shared among innate-immune enhancers and viral enhancers (Figure 1B). Annotation of this dataset using the BioMART tool (v0.7, ENSEMBL release v72) identified 31 TFs associated with “regulation of immune processes” [GO:0002376] in our 72 identified interactions. Notably, the extent to which the distinct viral enhancers share factorswith the innate-immune genes varies (Figure 1C). This may be explained by the different physiological roles of the innate-immune genes and lifestyles of the selected viruses. Among the viruses, HCMV and HIV-1 enhancers show the largest TF overlap in total numbers of interactions with the innate-immune genes. In summary, we identified a substantial overlap in TF interactions between host and viral regulatory regions.
Tab. 2. List of identified interactions for the selected viral and host enhancers. 
Comparable Expression Kinetics
It is noteworthy that host immediate-early response genes and viral immediate-early genes are, by definition, identified by the same criterion, namely that their expression is independent of newly synthesised proteins [12], [53], [54]. Upon infection of permissive cells, viral promoters are activated within the first hour of infection. This follows a typical expression profile with a peak between 2–6 h followed by reduced expression levels. This expression pattern has parallels with the temporal expression of host innate-immune genes, e.g., IFNB1, IL6, or TNF that are rapidly induced after PRR activation [55]–[58]. Most notably, it has recently been demonstrated in a genome-wide transcriptome study with murine CMV that the mRNA synthesis rate of viral IE transcripts is rapidly induced and subsequently strongly downregulated, following the expression kinetic profile for many innate immune genes in this dataset [59].
Response to Immune-Stimulatory Ligands
A corollary of viral enhancer mimicry of innate-immune regulatory functions is that the viral promoters/enhancers should be activated by the same signalling events as innate-immune genes. This implies that events during the infection process that trigger “antiviral” signalling cascades actually facilitate the initial viral transcription. In this context, it has been shown that activation of TLRs by LPS and CpG [60], [61] increases activity in isolated HCMV-enhancer and HIV-LTR–driven reporter constructs [62]–[64]. This also seems to apply in the context of viral infection since cytokine signalling stimulates HBV gene expression [65] and HIV needs TLR-8 signalling in specific cell types for replication [66]. It is also notable that all of the viral control regions examined here have been shown to interact with AP-1 (Figure 1C). While AP-1 is not exclusively associated with innate-immune signalling, it can be activated by TLR signalling via MAPK-activation or by cAMP-related signalling during infection [67], [68] and subsequently also binds to innate-immune enhancers. Taken together, these examples indicate that so called “antiviral” processes have the potential to facilitate viral IE gene expression and replication. In the future, their importance and potentially proviral role should be examined in viral infection models.
Responsiveness to Negative Feedback Control
Immune signalling pathways are tightly regulated by negative feedback with the inhibitors of signalling activity acting in a matter of minutes to hours [69], [70]. Thus, innate-immune negative feedback loops should also inhibit viral gene expression and may play a role in viral latency. This hallmark of viral enhancer mimicry might prove the most challenging for scientific investigation. Interference with negative feedback regulators before infection may lead to an exacerbated immune response, either inducing an elevated antiviral state in the cell before the experimental infection or driving it into apoptosis. Still, proving that this hallmark is applicable to viral infections might provide new drug targets to inhibit viral infections. While, to our knowledge, no direct effects of negative regulators of inflammatory signalling on viral gene expression have been reported so far, it has been shown that anti-inflammatory drugs and chemical inhibitors of pro-inflammatory signalling, expected to increase viral replication, actually can inhibit viral gene expression and replication of HCMV, HBV, and HIV-1 [67], [68], [71]–[74].
Concluding Remarks
TFs activating innate-immune genes are regulated by PRR signalling that cannot be efficiently inhibited by viruses as their activation occurs during the viral entry process. Mimicking an innate-immune enhancer therefore has the advantage that TFs, already activated by the viral entry process, can be directly utilised in a time restricted manner to ensure viral gene expression at IE times. We hope this opinion opens debate and provides new insights for either reexamination or future-based investigations toward understanding viral gene activation and latency. Indeed we believe that the principle of viruses co-opting host-innate regulatory signals has broad implications toward understanding the biological role of viral enhancers, in acute and latent viral infections, and prospective host-directed antiviral therapeutic and vaccine strategies.
Zdroje
1. TabetaK, GeorgelP, JanssenE, DuX, HoebeK, et al. (2004) Toll-like receptors 9 and 3 as essential components of innate immune defense against mouse cytomegalovirus infection. Proc Natl Acad Sci U S A 101 : 3516–3521.
2. KumarH, KawaiT, AkiraS (2011) Pathogen recognition by the innate immune system. Int Rev Immunol 30 : 16–34.
3. KawaiT, AkiraS (2006) TLR signaling. Cell Death Differ 13 : 816–825.
4. ZhongB, TienP, ShuHB (2006) Innate immune responses: crosstalk of signaling and regulation of gene transcription. Virology 352 : 14–21.
5. O'NeillLAJ, GolenbockD, BowieAG (2013) The history of Toll-like receptors - redefining innate immunity. Nat Rev Immunol 13 : 453–460.
6. EldeNC, MalikHS (2009) The evolutionary conundrum of pathogen mimicry. Nat Rev Micro 7 : 787–797.
7. WuB, HurS (2013) Viral counterattack against the host innate immune system. Cell Res 23 : 735–736.
8. DraymanN, GlickY, Ben-nun-ShaulO, ZerH, ZlotnickA, et al. (2013) Pathogens use structural mimicry of native host ligands as a mechanism for host receptor engagement. Cell Host Microbe 14 : 63–73.
9. SlobedmanB, BarryPA, SpencerJV, AvdicS, AbendrothA (2009) Virus-encoded homologs of cellular interleukin-10 and their control of host immune function. J Virol 83 : 9618–9629.
10. AlcamiA (2003) Viral mimicry of cytokines, chemokines and their receptors. Nat Rev Immunol 3 : 36–50.
11. Engel P, Angulo A (2012) Viral immunomodulatory proteins: usurping host genes as a survival strategy. In: López-Larrea C, editors. Self and nonself. Springer US. pp.256–276.
12. HonessRW, RoizmanB (1974) Regulation of herpesvirus macromolecular synthesis I. Cascade regulation of the synthesis of three groups of viral proteins. J Virol 14 : 8–19.
13. GhazalP, GonzalezAJC, Garcia-RamirezJJ, KurzS, AnguloA (2000) Viruses: hostages to the cell. Virology 275 : 233–237.
14. GhazalP, Garcia-RamirezJJ, GonzalezAJC, KurzS, AnguloA (2000) Principles of homeostasis in governing virus activation and latency. Immunol Res 21 : 219–223.
15. BryneJC, ValenE, TangMHE, MarstrandT, WintherO, et al. (2008) JASPAR, the open access database of transcription factor-binding profiles: new content and tools in the 2008 update. Nucl Acids Res 36: D102–D106.
16. BoshartM, WeberF, JahnG, Dorsch-HaslerK, FleckensteinB, et al. (1985) A very strong enhancer is located upstream of an immediate early gene of human cytomegalovirus. Cell 41 : 521–530.
17. HarlanSM, ReiterRS, SigmundCD, LinJL-C, LinJJ-C (2008) Requirement of TCTG(G/C) direct repeats and overlapping GATA site for maintaining the cardiac-specific expression of cardiac troponin T in developing and adult mice. Anat Rec 291 : 1574–1586.
18. GhazalP, LubonH, Reynolds-KohlerC, HennighausenL, NelsonJA (1990) Interactions between cellular regulatory proteins and a unique sequence region in the human cytomegalovirus major immediate-early promoter. Virology 174 : 18–25.
19. LeeW, HaslingerA, KarinM, TjianR (1987) Activation of transcription by two factors that bind promoter and enhancer sequences of the human metallothionein gene and SV40. Nature 325 : 368–372.
20. KroppKA, SimonCO, FinkA, RenzahoA, KühnapfelB, et al. (2009) Synergism between the components of the bipartite major immediate-early transcriptional enhancer of murine cytomegalovirus does not accelerate virus replication in cell culture and host tissues. J Gen Virol 90 : 2395–2401.
21. HaslingerA, KarinM (1985) Upstream promoter element of the human metallothionein-IIA gene can act like an enhancer element. Proc Natl Acad Sci U S A 82 : 8572.
22. UchiumiF, MiyazakiS, TanumaSi (2011) The possible functions of duplicated ets (GGAA) motifs located near transcription start sites of various human genes. Cell Mol Life Sci 68 : 2039–2051.
23. KimuraA, IsraëlA, Le BailO, KourilskyP (1986) Detailed analysis of the mouse H-2Kb promoter: enhancer-like sequences and their role in the regulation of class I gene expression. Cell 44 : 261–272.
24. HonkakoskiP, MooreR, WashburnKA, NegishiM (1998) Activation by diverse xenochemicals of the 51-base pair phenobarbital-responsive enhancer module in the CYP2B10 gene. Mol Pharmacol 53 : 597–601.
25. WeberF, de VilliersJ, SchaffnerW (1984) An SV40 “enhancer trap” incorporates exogenous enhancers or generates enhancers from its own sequences. Cell 36 : 983–992.
26. HerrW, ClarkeJ (1986) The SV40 enhancer is composed of multiple functional elements that can compensate for one another. Cell 45 : 461–470.
27. NakamichiK, KishidaS, TanakaK, SuganumaA, SanoY, et al. (2013) Sequential changes in the non-coding control region sequences of JC polyomaviruses from the cerebrospinal fluid of patients with progressive multifocal leukoencephalopathy. Arch Virol 158 : 639–650.
28. MarkowitzRB, TolbertS, DynanWS (1990) Promoter evolution in BK virus: functional elements are created at sequence junctions. J Virol 64 : 2411–2415.
29. SchmidtD, WilsonMD, BallesterB, SchwaliePC, BrownGD, et al. (2010) Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science 328 : 1036–1040.
30. HareEE, PetersonBK, IyerVN, MeierR, EisenMB (2008) Sepsid even-skipped enhancers are functionally conserved in Drosophila despite lack of sequence conservation. PLoS Genet 4: e1000106.
31. ArnostiDN, KulkarniMM (2005) Transcriptional enhancers: intelligent enhanceosomes or flexible billboards? J Cell Biochem 94 : 890–898.
32. KuoD, LiconK, BandyopadhyayS, ChuangR, LuoC, et al. (2010) Coevolution within a transcriptional network by compensatory trans and cis mutations. PCR Methods Appl 20 : 1672–1678.
33. MacArthurS, BrookfieldJFY (2004) Expected rates and modes of evolution of enhancer sequences. Mol Biol Evol 21 : 1064–1073.
34. VincesMD, LegendreM, CaldaraM, HagiharaM, VerstrepenKJ (2009) Unstable tandem repeats in promoters confer transcriptional evolvability. Science 324 : 1213–1216.
35. ArbizaL, GronauI, AksoyBA, HubiszMJ, GulkoB, et al. (2013) Genome-wide inference of natural selection on human transcription factor binding sites. Nat Genet 45 : 723–729.
36. WeirauchMT, HughesTR (2010) Conserved expression without conserved regulatory sequence: the more things change, the more they stay the same. Trends Genet 26 : 66–74.
37. StinskiMF, IsomuraH (2008) Role of the cytomegalovirus major immediate early enhancer in acute infection and reactivation from latency. Med Microbiol Immunol 197 : 223–231.
38. AnguloA, MesserleM, KoszinowskiUH, GhazalP (1998) Enhancer requirement for murine cytomegalovirus growth and genetic complementation by the human cytomegalovirus enhancer. J Virol 72 : 8502–8509.
39. Mella-AlvaradoV, GautierA, Le GacF, LareyreJJ (2013) Tissue and cell-specific transcriptional activity of the human cytomegalovirus immediate early gene promoter (UL123) in zebrafish. Gene Expr Patterns 13 : 91–103.
40. KoedoodM, FichtelA, MeierP, MitchellPJ (1995) Human cytomegalovirus (HCMV) immediate-early enhancer/promoter specificity during embryogenesis defines target tissues of congenital HCMV infection. J Virol 69 : 2194–2207.
41. BaskarJF, SmithPP, CimentGS, HoffmannS, TuckerC, et al. (1996) Developmental analysis of the cytomegalovirus enhancer in transgenic animals. J Virol 70 : 3215–3226.
42. VaquerizasJM, KummerfeldSK, TeichmannSA, LuscombeNM (2009) A census of human transcription factors: function, expression and evolution. Nat Rev Genet 10 : 252–263.
43. NovereNL, HuckaM, MiH, MoodieS, SchreiberF, et al. (2009) The systems biology graphical notation. Nat Biotech 27 : 735–741.
44. WattersonS, MarshallS, GhazalP (2008) Logic models of pathway biology. Drug Discov Today 13 : 447–456.
45. WattersonS, GhazalP (2010) Use of logic theory in understanding regulatory pathway signaling in response to infection. Future Microbiol 5 : 163–176.
46. Yadaf A, Kropp KA, Mazein A, Watterson S, Roy D, et al. (2013) Exploring possible mimicry of host immune genes by viruses. Available: http://dx.doi.org/10.6084/m9.figshare.703130. Accessed 5 June 2013.
47. Ming-Yuan Huang J, Kropp KA, Mazein A, Watterson S, Roy D, et al. (2013) Transcriptional regulation of viral and innate immune genes - a comparison of IFNβ and HCMV MIE genes. Available: http://dx.doi.org/10.6084/m9.figshare.703089. Accessed 5 June 2013.
48. Ba Abdullah MM, Kropp KA, Mazein A, Watterson S, Roy D, et al. (2013) Logic based mapping of the promoter enhancer regions of HPV16 and Interferon-γ: a search for similarity. Available: http://dx.doi.org/10.6084/m9.figshare.753326. Accessed 5 June 2013.
49. MacDonald D, Kropp KA, Mazein A, Watterson S, Roy D, et al. (2013) Common transcription factors between Hepatitis B virus (HBV) and Interleukin-8 (IL8). Available: http://dx.doi.org/10.6084/m9.figshare.776903. Accessed 5 June 2013.
50. Mallikarjun V, Kropp KA, Mazein A, Watterson S, Roy D, et al. (2013) Investigating a noval kind of molecular mimicry using pathway mapping based approached, in particular for IFNγ, SV40 and Adenovirus. Available: http://dx.doi.org/10.6084/m9.figshare.748790. Accessed 5 June 2013.
51. Zakirova Z, Kropp KA, Mazein A, Watterson S, Roy D, et al. (2013) Mapping innate immune host and viral gene interactions in monoctyte/macrophages, in particular for IL1β and the LTR region of HIV-1. Available: http://dx.doi.org/10.6084/m9.figshare.741713. Accessed 5 June 2013.
52. Weber J, Kropp KA, Mazein A, Watterson S, Roy D, et al. (2013) Examining common transcription factors and functional sequences that might indicate viral mimicry between the activation of Human Papilloma Virus-18 proteins E6 and E7 and the immune genes IRF1 and IL12. Available: http://dx.doi.org/10.6084/m9.figshare.753078. Accessed 5 June 2013.
53. GrassoRJ, BuchananJM (1969) Synthesis of early RNA in bacteriophage T4-infected Escherichia coli B. Nature 224 : 882–885.
54. LauLF, NathansD (1987) Expression of a set of growth-related immediate early genes in BALB/c 3T3 cells: coordinate regulation with c-fos or c-myc. Proc Natl Acad Sci U S A 84 : 1182–1186.
55. TakaokaA, YanaiH (2006) Interferon signalling network in innate defence. Cell Microbiol 8 : 907–922.
56. BeutlerB, JiangZ, GeorgelP, CrozatK, CrokerB, et al. (2006) Genetic analysis of host resistence: Toll-like receptor signaling and immunity at large. Annu Rev Immunol 24 : 353–389.
57. FalvoJV, TsytsykovaAV, GoldfeldAE (2010) Transcriptional control of the TNF gene. Curr Dir Autoimmun 11 : 27–60.
58. BauerJ, GanterU, GeigerT, JacobshagenU, HiranoT, et al. (1988) Regulation of interleukin-6 expression in cultured human blood monocytes and monocyte-derived macrophages. Blood 72 : 1134–1140.
59. MarcinowskiL, LidschreiberM, WindhagerL, RiederM, BosseJB, et al. (2012) Real-time transcriptional profiling of cellular and viral gene expression during lytic cytomegalovirus infection. PLoS Pathog 8: e1002908.
60. LeeY, SohnWJ, KimDS, KwonHJ (2004) NF-KB and c-Jun-dependent regulation of human cytomegalovirus immediate-early gene enhancer/promoter in response to lipopolysaccharide and bacterial CpG-oligodeoxynucleotides in macrophage cell line RAW 264.7. Euro J Biochem 271 : 1094–1105.
61. IversenAC, SteinkjerB, NilsenN, BohnhorstJ, MoenSH, et al. (2009) A proviral role for CpG in cytomegalovirus infection. J Immunol 182 : 5672–5681.
62. NetterwaldJ, YangS, WangW, GhannyS, CodyM, et al. (2005) Two gamma interferon-activated site-like elements in the human cytomegalovirus major immediate-early promoter/enhancer are important for viral replication. J Virol 79 : 5035–5046.
63. EquilsO, FaureE, ThomasL, BulutY, TrushinS, et al. (2001) Bacterial lipopolysaccharide activates HIV long terminal repeat through Toll-like receptor 4. J Immunol 166 : 2342–2347.
64. ZimmermannA, TrillingM, WagnerM, WilbornM, BubicI, et al. (2005) A cytomegaloviral protein reveals a dual role for STAT2 in IFN-γ signaling and antiviral responses. J Exp Med 201 : 1543–1553.
65. WarisG, SiddiquiA (2002) Interaction between STAT-3 and HNF-3 leads to the activation of liver-specific hepatitis B virus enhancer 1 function. J Virol 76 : 2721–2729.
66. GringhuisSI, van der VlistM, van den BergLM, den DunnenJ, LitjensM, et al. (2010) HIV-1 exploits innate signaling by TLR8 and DC-SIGN for productive infection of dendritic cells. Nat Immunol 11 : 419–426.
67. MocarskiES (2002) Virus self-improvement through inflammation: no pain, no gain. Proc Natl Acad Sci U S A 99 : 3362–3364.
68. ZhuH, CongJP, YuD, BresnahanWA, ShenkTE (2002) Inhibition of cyclooxygenase 2 blocks human cytomegalovirus replication. Proc Natl Acad Sci U S A 99 : 3932–3937.
69. SchroderK, HertzogPJ, RavasiT, HumeDA (2004) Interferon-gamma: an overview of signals, mechanisms and functions. J Leukoc Biol 75 : 163–189.
70. LacazeP, RazaS, SingG, PageD, ForsterT, et al. (2009) Combined genome-wide expression profiling and targeted RNA interference in primary mouse macrophages reveals perturbation of transcriptional networks associated with interferon signalling. BMC Genomics 10 : 372.
71. SpeirE, YuZX, FerransVJ, HuangES, EpsteinSE (1998) Aspirin attenuates cytomegalovirus infectivity and gene expression mediated by cyclooxygenase-2 in coronary artery smooth muscle cells. Circ Res 83 : 210–216.
72. FiorinoS, CursaroC, LorenziniS, LoggiE, BrodosiL, et al. (2011) The pharmacology and activity of non-steroidal anti-inflammatory drugs (NSAIDs): a review of their use as an adjuvant treatment in patients with HBV and HCV chronic hepatitis. Ital J Med 5 : 82–89.
73. KoppE, GhoshS (1994) Inhibition of NF-kappa B by sodium salicylate and aspirin. Science 265 : 956–959.
74. DeMerittIB, PodduturiJP, TilleyAM, NogalskiMT, YurochkoAD (2006) Prolonged activation of NF-kappaB by human cytomegalovirus promotes efficient viral replication and late gene expression. Virology 346 : 15–31.
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Structure of the Membrane Anchor of Pestivirus Glycoprotein E, a Long Tilted Amphipathic HelixČlánek Iron Acquisition in : The Roles of IlsA and Bacillibactin in Exogenous Ferritin Iron MobilizationČlánek AvrBsT Acetylates ACIP1, a Protein that Associates with Microtubules and Is Required for ImmunityČlánek Viral MicroRNA Effects on Pathogenesis of Polyomavirus SV40 Infections in Syrian Golden HamstersČlánek Genome-Wide RNAi Screen Identifies Broadly-Acting Host Factors That Inhibit Arbovirus Infection
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2014 Číslo 2- Stillova choroba: vzácné a závažné systémové onemocnění
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Jak souvisí postcovidový syndrom s poškozením mozku?
- Familiární středomořská horečka
-
Všechny články tohoto čísla
- Viral Enhancer Mimicry of Host Innate-Immune Promoters
- The Epstein-Barr Virus-Encoded MicroRNA MiR-BART9 Promotes Tumor Metastasis by Targeting E-Cadherin in Nasopharyngeal Carcinoma
- Implication of PMLIV in Both Intrinsic and Innate Immunity
- The Consequences of Reconfiguring the Ambisense S Genome Segment of Rift Valley Fever Virus on Viral Replication in Mammalian and Mosquito Cells and for Genome Packaging
- Substrate-Induced Unfolding of Protein Disulfide Isomerase Displaces the Cholera Toxin A1 Subunit from Its Holotoxin
- Male-Killing Induces Sex-Specific Cell Death via Host Apoptotic Pathway
- Highly Active Antiretroviral Therapies Are Effective against HIV-1 Cell-to-Cell Transmission
- The microRNAs in an Ancient Protist Repress the Variant-Specific Surface Protein Expression by Targeting the Entire Coding Sequence
- Transmission-Blocking Antibodies against Mosquito C-Type Lectins for Dengue Prevention
- Type III Secretion Protein MxiI Is Recognized by Naip2 to Induce Nlrc4 Inflammasome Activation Independently of Pkcδ
- Lundep, a Sand Fly Salivary Endonuclease Increases Parasite Survival in Neutrophils and Inhibits XIIa Contact Activation in Human Plasma
- Induction of Type I Interferon Signaling Determines the Relative Pathogenicity of Strains
- Structure of the Membrane Anchor of Pestivirus Glycoprotein E, a Long Tilted Amphipathic Helix
- Foxp3 Regulatory T Cells Delay Expulsion of Intestinal Nematodes by Suppression of IL-9-Driven Mast Cell Activation in BALB/c but Not in C57BL/6 Mice
- Iron Acquisition in : The Roles of IlsA and Bacillibactin in Exogenous Ferritin Iron Mobilization
- MicroRNA Editing Facilitates Immune Elimination of HCMV Infected Cells
- Reversible Silencing of Cytomegalovirus Genomes by Type I Interferon Governs Virus Latency
- Identification of Host-Targeted Small Molecules That Restrict Intracellular Growth
- A Cyclophilin Homology Domain-Independent Role for Nup358 in HIV-1 Infection
- Engagement of NKG2D on Bystander Memory CD8 T Cells Promotes Increased Immunopathology following Infection
- Suppression of RNA Silencing by a Plant DNA Virus Satellite Requires a Host Calmodulin-Like Protein to Repress Expression
- CIB1 Synergizes with EphrinA2 to Regulate Kaposi's Sarcoma-Associated Herpesvirus Macropinocytic Entry in Human Microvascular Dermal Endothelial Cells
- A Gammaherpesvirus Bcl-2 Ortholog Blocks B Cell Receptor-Mediated Apoptosis and Promotes the Survival of Developing B Cells
- Metabolic Reprogramming during Purine Stress in the Protozoan Pathogen
- The Post-transcriptional Regulator / Activates T3SS by Stabilizing the 5′ UTR of , the Master Regulator of Genes, in
- Tailored Immune Responses: Novel Effector Helper T Cell Subsets in Protective Immunity
- AvrBsT Acetylates ACIP1, a Protein that Associates with Microtubules and Is Required for Immunity
- Epstein-Barr Virus Large Tegument Protein BPLF1 Contributes to Innate Immune Evasion through Interference with Toll-Like Receptor Signaling
- The Major Cellular Sterol Regulatory Pathway Is Required for Andes Virus Infection
- Insights into the Initiation of JC Virus DNA Replication Derived from the Crystal Structure of the T-Antigen Origin Binding Domain
- Domain Shuffling in a Sensor Protein Contributed to the Evolution of Insect Pathogenicity in Plant-Beneficial
- Lectin-Like Bacteriocins from spp. Utilise D-Rhamnose Containing Lipopolysaccharide as a Cellular Receptor
- A Compositional Look at the Human Gastrointestinal Microbiome and Immune Activation Parameters in HIV Infected Subjects
- Exploits Asparagine to Assimilate Nitrogen and Resist Acid Stress during Infection
- Interleukin-33 Increases Antibacterial Defense by Activation of Inducible Nitric Oxide Synthase in Skin
- Protective Vaccination against Papillomavirus-Induced Skin Tumors under Immunocompetent and Immunosuppressive Conditions: A Preclinical Study Using a Natural Outbred Animal Model
- Gem-Induced Cytoskeleton Remodeling Increases Cellular Migration of HTLV-1-Infected Cells, Formation of Infected-to-Target T-Cell Conjugates and Viral Transmission
- Viral MicroRNA Effects on Pathogenesis of Polyomavirus SV40 Infections in Syrian Golden Hamsters
- Genome-Wide RNAi Screen Identifies Broadly-Acting Host Factors That Inhibit Arbovirus Infection
- Inflammatory Monocytes Orchestrate Innate Antifungal Immunity in the Lung
- Quantitative and Qualitative Deficits in Neonatal Lung-Migratory Dendritic Cells Impact the Generation of the CD8+ T Cell Response
- Human Genome-Wide RNAi Screen Identifies an Essential Role for Inositol Pyrophosphates in Type-I Interferon Response
- The Master Regulator of the Cellular Stress Response (HSF1) Is Critical for Orthopoxvirus Infection
- Code-Assisted Discovery of TAL Effector Targets in Bacterial Leaf Streak of Rice Reveals Contrast with Bacterial Blight and a Novel Susceptibility Gene
- Competitive and Cooperative Interactions Mediate RNA Transfer from Herpesvirus Saimiri ORF57 to the Mammalian Export Adaptor ALYREF
- The Type III Secretion Chaperone Slc1 Engages Multiple Early Effectors, Including TepP, a Tyrosine-phosphorylated Protein Required for the Recruitment of CrkI-II to Nascent Inclusions and Innate Immune Signaling
- Yeasts: How Many Species Infect Humans and Animals?
- Clustering of Pattern Recognition Receptors for Fungal Detection
- Distinct Antiviral Responses in Pluripotent versus Differentiated Cells
- Igniting the Fire: Virulence Factors in the Pathogenesis of Sepsis
- Inactivation of the Host Lipin Gene Accelerates RNA Virus Replication through Viral Exploitation of the Expanded Endoplasmic Reticulum Membrane
- Inducible Deletion of CD28 Prior to Secondary Infection Impairs Worm Expulsion and Recall of Protective Memory CD4 T Cell Responses
- Clonal Expansion during Infection Dynamics Reveals the Effect of Antibiotic Intervention
- The Secreted Triose Phosphate Isomerase of Is Required to Sustain Microfilaria Production
- Unifying Viral Genetics and Human Transportation Data to Predict the Global Transmission Dynamics of Human Influenza H3N2
- ‘Death and Axes’: Unexpected Ca Entry Phenologs Predict New Anti-schistosomal Agents
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Reversible Silencing of Cytomegalovirus Genomes by Type I Interferon Governs Virus Latency
- Implication of PMLIV in Both Intrinsic and Innate Immunity
- Transmission-Blocking Antibodies against Mosquito C-Type Lectins for Dengue Prevention
- Lundep, a Sand Fly Salivary Endonuclease Increases Parasite Survival in Neutrophils and Inhibits XIIa Contact Activation in Human Plasma
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



