-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Chitin Synthases from Are Involved in Tip Growth and Represent a Potential Target for Anti-Oomycete Drugs
Oomycetes represent some of the most devastating plant and animal pathogens. Typical examples are Phytophthora infestans, which causes potato and tomato late blight, and Saprolegnia parasitica, responsible for fish diseases. Despite the economical and environmental importance of oomycete diseases, their control is difficult, particularly in the aquaculture industry. Carbohydrate synthases are vital for hyphal growth and represent interesting targets for tackling the pathogens. The existence of 2 different chitin synthase genes (SmChs1 and SmChs2) in Saprolegnia monoica was demonstrated using bioinformatics and molecular biology approaches. The function of SmCHS2 was unequivocally demonstrated by showing its catalytic activity in vitro after expression in Pichia pastoris. The recombinant SmCHS1 protein did not exhibit any activity in vitro, suggesting that it requires other partners or effectors to be active, or that it is involved in a different process than chitin biosynthesis. Both proteins contained N-terminal Microtubule Interacting and Trafficking domains, which have never been reported in any other known carbohydrate synthases. These domains are involved in protein recycling by endocytosis. Enzyme kinetics revealed that Saprolegnia chitin synthases are competitively inhibited by nikkomycin Z and quantitative PCR showed that their expression is higher in presence of the inhibitor. The use of nikkomycin Z combined with microscopy showed that chitin synthases are active essentially at the hyphal tips, which burst in the presence of the inhibitor, leading to cell death. S. parasitica was more sensitive to nikkomycin Z than S. monoica. In conclusion, chitin synthases with species-specific characteristics are involved in tip growth in Saprolegnia species and chitin is vital for the micro-organisms despite its very low abundance in the cell walls. Chitin is most likely synthesized transiently at the apex of the cells before cellulose, the major cell wall component in oomycetes. Our results provide important fundamental information on cell wall biogenesis in economically important species, and demonstrate the potential of targeting oomycete chitin synthases for disease control.
Published in the journal: . PLoS Pathog 6(8): e32767. doi:10.1371/journal.ppat.1001070
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1001070Summary
Oomycetes represent some of the most devastating plant and animal pathogens. Typical examples are Phytophthora infestans, which causes potato and tomato late blight, and Saprolegnia parasitica, responsible for fish diseases. Despite the economical and environmental importance of oomycete diseases, their control is difficult, particularly in the aquaculture industry. Carbohydrate synthases are vital for hyphal growth and represent interesting targets for tackling the pathogens. The existence of 2 different chitin synthase genes (SmChs1 and SmChs2) in Saprolegnia monoica was demonstrated using bioinformatics and molecular biology approaches. The function of SmCHS2 was unequivocally demonstrated by showing its catalytic activity in vitro after expression in Pichia pastoris. The recombinant SmCHS1 protein did not exhibit any activity in vitro, suggesting that it requires other partners or effectors to be active, or that it is involved in a different process than chitin biosynthesis. Both proteins contained N-terminal Microtubule Interacting and Trafficking domains, which have never been reported in any other known carbohydrate synthases. These domains are involved in protein recycling by endocytosis. Enzyme kinetics revealed that Saprolegnia chitin synthases are competitively inhibited by nikkomycin Z and quantitative PCR showed that their expression is higher in presence of the inhibitor. The use of nikkomycin Z combined with microscopy showed that chitin synthases are active essentially at the hyphal tips, which burst in the presence of the inhibitor, leading to cell death. S. parasitica was more sensitive to nikkomycin Z than S. monoica. In conclusion, chitin synthases with species-specific characteristics are involved in tip growth in Saprolegnia species and chitin is vital for the micro-organisms despite its very low abundance in the cell walls. Chitin is most likely synthesized transiently at the apex of the cells before cellulose, the major cell wall component in oomycetes. Our results provide important fundamental information on cell wall biogenesis in economically important species, and demonstrate the potential of targeting oomycete chitin synthases for disease control.
Introduction
Saprolegnia parasitica is the most important oomycete fish pathogen [1]. This micro-organism is ubiquitous in fresh water and infects salmonids by acting as a primary pathogen [2]–[4]. The group of diseases caused by S. parasitica and other members of the order Saprolegniales is termed Saprolegniosis. These diseases are of great concern for the aquaculture industry and represent a serious threat to fish populations in natural habitats [1]. Multiple strains of S. parasitica that cause more than 90% fish mortality have been isolated [2], [5]. Due to the lack of efficient methods to control pathogenic Saprolegniales, there is a dramatic re-emergence of Saprolegniosis in aquaculture [1], [3]. This problem represents important economical loss and a serious threat to the activities of fish farmers worldwide [1], [3]. Thus, there is an important need for developing efficient and sustainable methods to stop the spread of these pathogens and their virulence. The enzymes involved in cell integrity, such as those responsible for cell wall formation, represent interesting potential targets of anti-oomycete drugs.
The architecture and physical properties of oomycete cell walls are governed by a network of cellulose microfibrils that play a similar role as chitin in the walls of fungi [6], [7]. By forming a scaffold for other abundant β-glucans, such as β-(1→3) and β-(1→6) glucans, cellulose microfibrils provide structural support to the hyphae [6]. While chitin represents the most important ultrastructural compound of fungal walls [7], a crystalline form of this polymer has been reported in a few oomycete species only, in which it never accounts for more than 0.5% of the total cell wall content [8]–[11]. Other oomycetes like Aphanomyces euteiches are devoid of chitin, but their cell walls contain about 10% of other GlcNAc-based carbohydrates [12]. Unlike chitin, these compounds are amorphous and soluble in water, possibly due to the occurrence of unidentified sugars different from GlcNAc in their structure [12]. As opposed to the soluble GlcNAc-based carbohydrates which contributeto cell integrity in A. euteiches [12], the role played by chitin in the walls of the oomycete species in which it has been detected is unknown.
Although chitin is not a typical cell wall carbohydrate of oomycetes [8]–[11], genes that encode putative chitin synthases are present in species from different genera, including species in which chitin has never been detected. For instance, analysis of the full genome of the plant pathogen Phytophthora infestans [13] has revealed the existence of a putative chitin synthase gene in this species, while its cell wall seems to be devoid of chitin. Similarly, partial sequences of putative chitin synthases have been isolated from Plasmopara viticola [14], Phytophthora capsici [15] and Achlya ambisexualis [15]. Despite their structural difference with chitin, it was proposed that the cell wall GlcNAc-based carbohydrates from A. euteiches are synthesized by the product of one or both putative chitin synthase genes identified in this species, but this has not been demonstrated [12]. In fact, to date there is not a single example of an oomycete “chitin synthase” gene product for which the enzymatic activity has been experimentally demonstrated. This is essentially due to the difficulty in expressing these polytopic transmembrane proteins in heterologous systems. In all cases, the function of the products of the identified genes has been assumed on the sole basis of sequence similarities with fungal or yeast chitin synthases. Chitin synthase 2 from Saccharomyces cerevisiae represents the only example for which the capacity to catalyze the formation of β-(1→4)-N-acetylglucosaminyl linkages has been demonstrated in vitro [16].
Saprolegnia monoica is the only oomycete in which both crystalline chitin and putative chitin synthase sequences have been identified [11], [15]. Isolated membranes and detergent extracts of plasma membranes from this species exhibit chitin synthase activity in vitro [11], [17] but there is no evidence that the detected activity corresponds to the sequences identified [15]. Preliminary work in S. monoica using the chitin synthase inhibitor polyoxin D suggested that chitin may be involved in hyphal growth [11]. Thus, chitin synthases may represent interesting targets for growth inhibitors of Saprolegnia species and possibly other oomycetes that contain chitin in their walls. The potential use of drugs that interfere with chitin biosynthesis is particularly attractive since higher plants and all vertebrates that are infected by pathogenic oomycetes are devoid of chitin synthase activity.
We describe here the isolation and functional characterization of 2 chitin synthase genes from S. monoica. For the first time, we have successfully expressed an oomycete chitin synthase that is catalytically active in vitro. The recombinant enzyme was used to confirm directly the inhibitory activity of nikkomycin Z on oomycete chitin synthases. In addition, we demonstrate that chitin biosynthesis is involved in tip growth in Saprolegnia and that chitin synthase activities are vital for the micro-organism. Interestingly, the economically important pathogen S. parasitica was shown to be more sensitive to nikkomycin Z than S. monoica. These findings pave the way for the establishment of sustainable methods for controlling the fish diseases caused by Saprolegniales.
Results
Isolation of the SmChs genes
Two Chs genes, named SmChs1 and SmChs2, were identified in S. monoica in 1997 [15]. While the complete sequence of SmChs2 was obtained (GenBank accession number U19946), only a short segment of 600 bp corresponding to SmChs1 could be amplified by PCR and sequenced [15]. Here, we have isolated the full-length sequence of SmChs1 and re-analyzed the sequence of SmChs2 by cloning and sequencing the genes from mycelial cDNA (sequences submitted to the GenBank database under accession numbers GQ252794 and GQ252795, respectively). Surprisingly the SmChs2 sequence determined earlier from genomic DNA [15] differed in multiple places compared to our new sequence, which caused a shift in the reading frame. Consequently, the previously reported amino acid sequence [15] was incorrect at the N-terminus of the protein; in addition, the stop codon had been wrongly predicted.
In order to determine the total number of Chs genes in S. monoica, Southern blot analyses were performed using 3 different restriction enzymes (SacI, ClaI and PstI) and 2 different probes (see Materials and Methods and Table S1 in Supplementary Material). The blots performed on the enzymatic digests obtained with SacI using either of the 2 probes revealed 3 bands of different apparent molecular weights (Figure S1 in Supplementary Material). Two of the bands (ca 1 and 3 kb) systematically exhibited a higher intensity and were easily detectable, while the third band (ca 1.2 kb) was much fainter, even when using alternative sensitive detection methods (32P-labelled probes and autoradiography, or biotinylated probes and chemiluminescent detection) (Figure S1). This third band was never visible on any of the blots performed on the ClaI and PstI digests (not shown), suggesting that its occurrence in the blots performed on the SacI digests may have arisen from nonspecific hybridization. This is further supported by the fact that all PCR approaches used to isolate the Chs genes from S. monoica systematically generated fragments corresponding to either SmChs1 or SmChs2. In addition, previous work based on chromosomal localization and Southern blot analyses suggested the occurrence of 2 Chs genes only in S. monoica [15]. Altogether, our data and earlier investigations [15] point toward the existence of a total of 2 Chs genes in S. monoica.
Sequence analysis of SmCHS1 and SmCHS2
SmChs1 codes for a polypeptide of 910 amino acids with a theoretical mass and pI of 103,100.95 Da and 8.41, respectively, while SmChs2 encodes a protein of 962 amino acids, corresponding to a theoretical mass of 107,220.75 Da and a pI of 6.33. Detailed analysis of the SmCHS1 and SmCHS2 sequences revealed that both proteins contain the yeast and fungal conserved CHS motifs (motifs a to h, Figure S2 in Supplementary Material), including the catalytic pfam 03142 subdomain [18], [19]. The pentapeptide forming the g motif (Q(R/G)RRW) is a signature of all known N-acetylglucosaminyltransferases with processive activity [18], [19] and it also occurs more generally in the form QXXRW in most processive glycosyltransferases from family 2 [20]. Motifs d and e are similar to those found in fungal CHS proteins from division 1 [18], which further supports the phylogenetic relationship between oomycete and fungal chitin synthases from division 1 [12], [18]. The full-length amino acid sequences of SmCHS1 and SmCHS2 (Figure S2) share 42% similarity with each other, while 52% amino acid identity was observed when comparing the conserved segments between motifs a and h. These regions exhibited 43–66% identity with the predicted sequences from other oomycete CHS proteins (Figure 1A). The alignments revealed that SmCHS2 exhibits greater similarity with AeCHS2 than with SmCHS1, and SmCHS1 is more similar to its orthologue in A. euteiches than to SmCHS2 (Figure 1A). Thus, as for fungi, oomycete orthologous CHS proteins exhibit a higher similarity with each other than with their paralogues [12]. Phylogenetic analysis showed that SmCHS1 and SmCHS2 group into the 2 clusters designated previously as “CHS1” and “CHS2” [12]. However, the P. capsici protein groups together with SmCHS1 and not with the P. infestans CHS (Figure 1B), providing further evidence that oomycetes CHSs share a common ancestor that duplicated before the segregation of the orders Saprolegniales and Peronosporales.
Fig. 1. Sequence identities and phylogenetic analysis of oomycete CHS proteins. 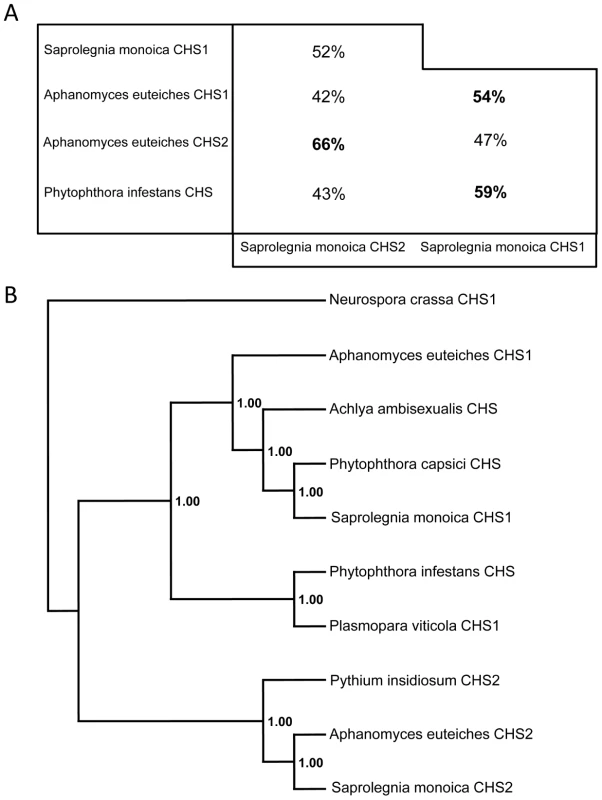
(A) Amino acid identity between the sequences of the conserved domains of SmCHS1 and SmCHS2, and the similar segments from other putative oomycete CHS proteins. The percentage of amino acid identity was calculated following alignments of the 245–744 amino acid portion of SmCHS1 and the 291–794 amino acid segment of SmCHS2. (B) Majority rule consensus tree of oomycete CHS proteins rooted with the sequence of CHS1 from Neurospora crassa. Numbers refer to posterior probabilities. Accession numbers are listed in Materials and Methods. Interestingly, the N-terminal ends of SmCHS1 and SmCHS2 exhibit high scores with the pfam 04212 domain, which corresponds to a so-called microtubule interacting and trafficking (MIT) domain [21] (Figure S2). MIT domains occur in numerous proteins having a wide range of functions [22], but their presence in carbohydrate synthases has never been described. Secondary structure prediction suggests an organization of the SmCHS1 and SmCHS2 MIT domains in 3 consecutive α-helices (Figure S3 in Supplementary Material), consistent with the typical asymmetric three-helix bundle structure experimentally determined for other MIT domains [23]. The overall topologies of SmCHS1 and SmCHS2 were predicted from hydropathy analyses (Figure S3). Both proteins exhibit a hydrophilic N-terminal end followed by a neutral segment containing the CHS consensus motifs a–h, the D,D,D,QXXRW signature of most processive glysosyltransferases [20], and the MIT domain (Figure S3). The proteins are predicted to be anchored in the plasma membrane through 7 C-terminal transmembrane domains. Searches for post-translational modifications revealed the presence of multiple potential phosphorylation sites in both proteins as well as 2 potential N-glycosylation sites for CHS1 but none for CHS2 (Figure S2 in Supplementary Material).
Functional characterization of the chitin synthase genes
In order to confirm that SmChs1 and SmChs2 encode chitin synthases, both proteins were expressed in Pichia pastoris with the eGFP marker at their C-termini. Chitin synthase activity was measured in vitro using CHAPS extracts of membranes from the recombinant zeocin-resistant yeast cells as a source of enzyme. CHAPS-solubilized microsomal fractions from the cells expressing SmCHS2 showed a 4-fold increased CHS activity compared to the control prepared from wild-type cells (Figure 2A). The lower activity observed with the control sample is attributable to chitin synthase from Pichia. The detergent extracts containing the recombinant SmCHS2 protein synthesized crystallites of chitin in vitro (Figure 2B) that were not detected when using similar preparations from the control wild-type strain (not shown). The radioactive products and the chitin crystallites synthesized in vitro by the detergent extracts containing SmCHS2 (Figure 2) were sensitive to chitinase digestion, thereby confirming the function of the gene and the catalytic activity of the corresponding protein. CHAPS-extracts of membranes isolated from the cells expressing SmCHS1 did not exhibit a higher level of activity compared to the control, which suggests that SmCHS1 is not responsible for the polymerization of chitin chains, as opposed to SmCHS2 (Figure 2A).
Fig. 2. In vitro synthesis of chitin by SmCHS1 and SmCHS2 expressed in Pichia pastoris. 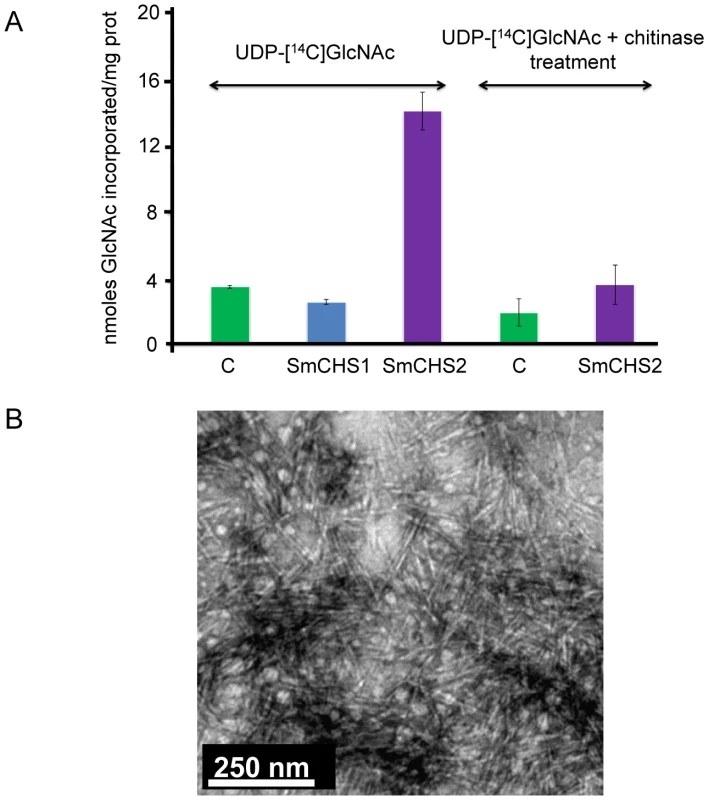
(A) Chitin synthase activities in detergent extracts of membranes from P. pastoris expressing SmCHS1 and SmCHS2. Chitin synthase activity was assayed in the presence of 14C-labelled UDP-GlcNAc, using CHAPS extracts prepared from the membrane fractions of yeast cells expressing SmCHS1 (blue bar) or SmCHS2 (purple bars) as a source of enzyme. A control was performed in the same conditions using a CHAPS extract from untransformed P. pastoris cells (green bars). The radioactive polymer synthesized in vitro was shown to correspond to chitin by measuring its sensitivity to the action of a specific chitinase from S. marcescens. Standard deviations from 3 independent experiments in which each assay was performed in triplicate are shown in the Figure. (B) Electron micrograph showing crystallites of chitin synthesized in vitro by the recombinant SmCHS2 solubilized with CHAPS (negative staining using 2% uranyl acetate). A control performed in the same conditions using a CHAPS extract of membranes from untransformed Pichia cells revealed the absence of the type of crystallites visible in the micrograph. Effect of nikkomycin Z on chitin synthase activity, mycelial growth and chitin content, and expression of SmChs genes
Nikkomycin Z is a well-known inhibitor of yeast and fungal chitin synthases [24], but its effect on enzymes and mycelial growth in oomycete has not been investigated. Here, we have first tested the effect of nikkomycin Z in vitro, using total membranes of S. monoica mycelium or a CHAPS extract from Pichia cells expressing SmCHS2 as a source of enzyme. In both cases, the drug caused a decrease in chitin synthase activity in a concentration-dependent manner, and enzyme kinetics revealed a competitive type of inhibition, with an apparent inhibition constant (Kiapp) of 4.6+0.4 µM (Figure S4 in Supplementary Material). The apparent Km with respect to UDP-N-acetylglucosamine was 150+12 µM.
Cultures of S. monoica hyphae in the presence of 50 µM nikkomycin Z showed that about 50% of the cells exhibited abnormal morphologies within 6 h of growth. The effect was more pronounced when using 200 µM of the inhibitor, with up to 80% abnormal cells after 6 h of culture. The morphological abnormalities consisted essentially of swollen and/or distorted hyphae (Figure S5 in Supplementary Material), reflecting an alteration of the growth most likely due to the perturbation of cell wall biosynthesis. About 50% of the cells survived after 5 days of culture in the presence of 50 µM nikkomycin Z, but all exhibited morphological abnormalities. In agreement with this rate of survival, 50% less biomass was recovered than in the absence of the drug. The cells that survived contained a similar amount of chitin in their walls as the control grown in the absence of nikkomycin Z (not shown), indicating that they were able to compensate for the inhibition of chitin synthase by the drug. Above 500 µM, nikkomycin Z provoked a dramatic decrease of the cell density (Figure 3A and B) and the systematic occurrence of numerous abnormalities along every individual hyphae. In addition, scanning electron microscopy revealed that none of the cells grew within the agar medium containing nikkomycin Z, as opposed to the control performed in the absence of inhibitor (panel C in Figure S5). Instead, all cells were crawling along the surface of the solid medium, changing their direction of growth multiple times, as if they were trying to avoid contact with the culture medium containing nikkomycin Z. Similar experiments conducted on S. parasitica revealed a significantly higher sensitivity of the mycelium to nikkomycin Z compared to S. monoica, with hardly any growth observed after several days at concentrations as low as 200 µM inhibitor (not shown).
Fig. 3. Effect of nikkomycin Z on the growth of S. monoica mycelium. 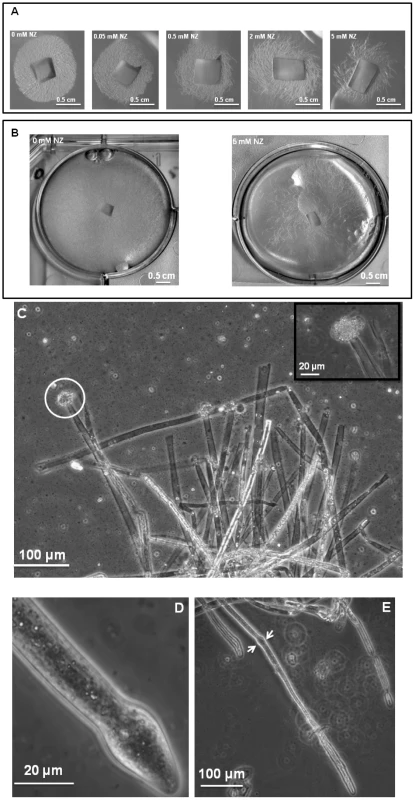
(A) Colony growth at different concentrations of nikkomycin Z (NZ) after 2 days of culture on PDA medium. (B) As in (A) but after 5 days of culture. (C) Microscopic observations of S. monoica mycelium grown in the presence of 50 µM nikkomycin Z showing cell death by tip bursting (the insert shows a magnification of a bursting tip). (D) Close view of a swollen tip. (E) Observation of the same cell as in (D) but after one additional hour of growth. A deformation of the cell wall resulting from the initial tip swelling remained in this surviving cell (arrows). In order to investigate the underlying compensation mechanism in the cells that survived the presence of 50 µM nikkomycin Z, the expression of SmChs1 and SmChs2 was analyzed by quantitative RT-PCR using mycelium grown in the presence and absence of inhibitor (50 µM) for up to 5 days. In the absence of inhibitor, the level of expression of SmChs2 was more than 100,000 times higher than that of SmChs1 (Figure 4). This important difference suggests that SmChs2 plays a major role in chitin biosynthesis in the hyphae. The presence of nikkomycin Z in the culture medium provoked an increase by 70% of the expression of SmChs2 (Figure 4). Even though the overall expression of SmChs1 was negligible compared to that of SmChs2, the expression of SmChs1 also increased significantly (300%) in the presence of nikkomycin Z (Figure 4). These data suggest that the cells that survived the presence of nikkomycin Z did so by compensating the inhibitory effect of the drug by up-regulating the expression of the chitin synthase genes.
Fig. 4. Expression analysis of SmChs1 and SmChs2 in S. monoica mycelium grown in the absence and presence of 50 µM nikkomycin Z (NZ). 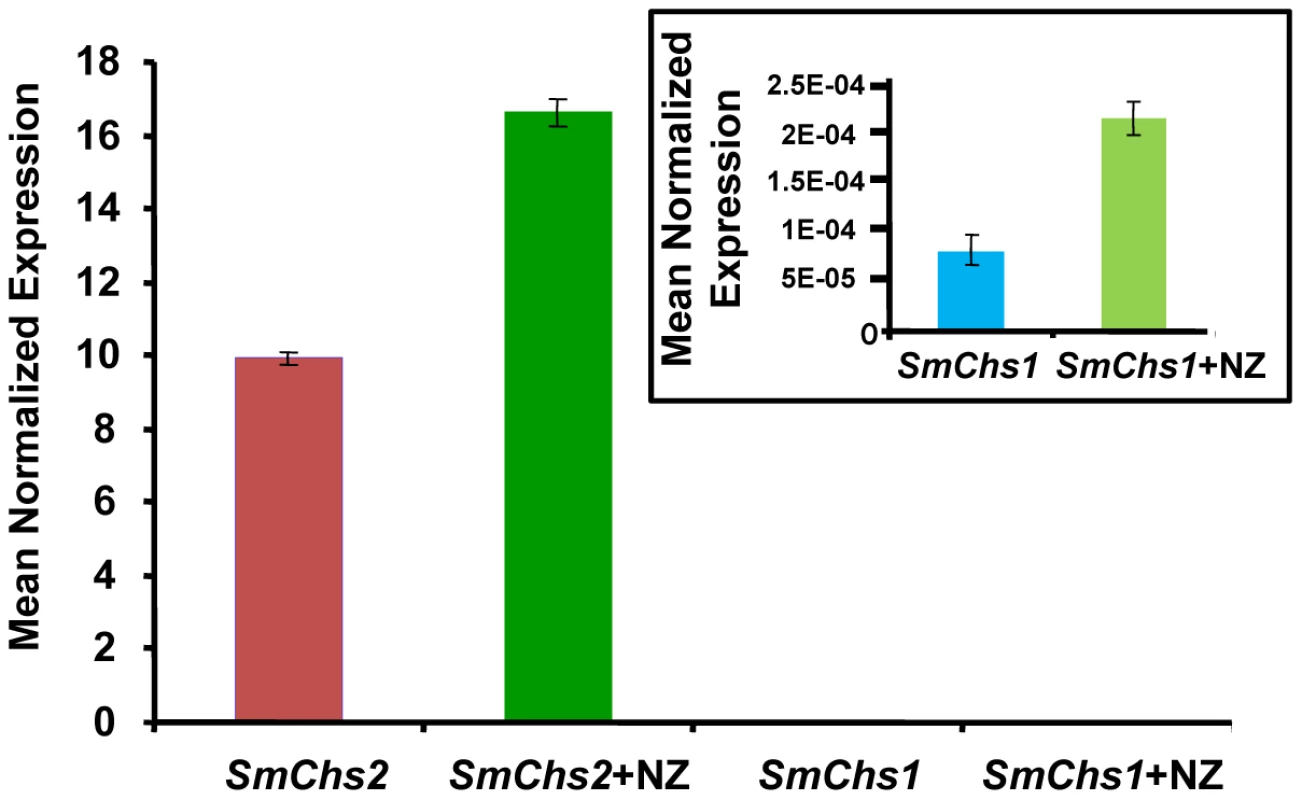
Quantitative RT-PCR experiments were performed on 3-day-old mycelium. The insert shows the expression levels of SmChs1 with a different scale on the y axis. Levels of expression were normalized with the 18S rRNA housekeeping gene and expressed as Mean Normalized Expression. Standard deviations from 3 independent sets of experiments are shown. The mode of action of nikkomycin Z on cell death was determined by continuously observing individual cells in the presence of 50 µM inhibitor during a period of 1 h. Within a few minutes, the tip of about 50% of the cells swelled and burst, leading to cell death through the release of the cellular content in the extracellular medium (Figure 3C). This indicates that the cell wall was more fragile at the tip of these hyphae than in the absence of nikkomycin Z, most likely due to a lack or lower amount of chitin. The data suggest that chitin, which represents less than 0.5% of the total cell wall carbohydrates [11] is primarily synthesized at the tip of the hyphae where the nikkomycin-Z-sensitive chitin synthase is most active. The tips of the cells that survived also swelled and the change of shape of the cell wall remained as the hyphae elongated (Figure 3, D and E). In these surviving cells, the compensation of the chitin default occurred through overexpression of chitin synthase, as suggested above with the demonstration of the overexpression of SmChs1 and SmChs2 in the presence of nikkomycin Z.
Discussion
In our early work on oomycete chitin synthases, we preliminarily reported that the chitin synthase inhibitor polyoxin D has a negative effect on the growth of S. monoica [11]. Thus, enzymes involved in the biosynthesis of minor cell wall carbohydrates such as chitin synthases may play important roles in cell integrity and represent potential targets for growth inhibitors of oomycetes, in addition to enzymes that form major cell wall components. We have now demonstrated this hypothesis by using S. monoica as a model species and showing that the more economically important pathogen S. parasitica exhibits an even higher sensitivity to the chitin synthase inhibitor nikkomycin Z. We also show for the first time that the Saprolegnia chitin synthases are primarily located at the apex of the mycelium and involved in tip growth.
Several oomycete genes are considered to encode chitin synthases owing to their sequence similarity with fungal and yeast chitin synthase genes [13]–[15]. However, their function has never been experimentally demonstrated. Here we provide the first evidence that the protein SmCHS2 from S. monoica indeed catalyses the formation of chitin. The protein expressed in P. pastoris as a GFP fusion was correctly targeted to the plasma membrane (data not shown) and was isolated in a catalytically active form after solubilization from the yeast membranes. The extracted protein was able to incorporate radioactive GlcNAc from 14C––labeled UDP-GlcNAc into chitin crystallites that were sensitive to the action of a specific chitinase. There is so far only one example where the heterologous expression of a chitin synthase has been reported, but the data were obtained in a non-oomycete species, the yeast Saccharomyces cerevisiae [16]. In general, such experiments have proven to be extremely challenging due to the occurrence of multiple transmembrane domains in chitin synthases. In addition to providing the first direct evidence for the catalytic activity of an oomycete chitin synthase, we confirm that the enzyme is inhibited by nikkomycin Z in a competitive manner. The Kiapp value is in the same range as previously reported for different yeast and fungal crude chitin synthase preparations while the apparent Km is 3–10 times lower [25]–[27].
Despite the occurrence in SmCHS1 of the same typical conserved chitin synthase motifs as in SmCHS2 and other CHS from division I [12], [19], no activity was measured in vitro when the protein was expressed in the same yeast system as SmCHS2. PCR experiments and observation by confocal microscopy of the recombinant GFP fusion protein confirmed that it was expressed and correctly targeted to the plasma membrane (not shown). If we assume that the enzyme was correctly folded, these data suggest that, as opposed to SmCHS2, SmCHS1 is not involved in the polymerization of insoluble chains of chitin. Interestingly A. euteiches has been shown to synthesize rather abundant (10% of cell wall content) GlcNAc-based carbohydrates that are water-soluble and exposed at the surface of the cells [12]. These carbohydrates were clearly different from chitin and, due to their solubility in water, it was concluded that they correspond to heterosaccharides that are possibly branched [12]. Since the cell wall of A. euteiches is devoid of chitin, it was proposed that the AeChs1 and AeChs2 genes encode proteins that do not form chitin but that are involved in the formation of the soluble GlcNAc-based heterosaccharides [12]. However, the actual N-acetylglucosaminyltransferase activity of AeCHS1 and AeCHS2, if any, remains to be demonstrated. By analogy with the Aphanomyces proteins, it is possible that SmCHS1 is involved in the biosynthesis of GlcNAc-based carbohydrates different from chitin. This hypothesis would concur with the inability of the recombinant protein to form chitin in vitro, as shown here. Alternatively, SmCHS1 may be involved in chitin biosynthesis during non-mycelial developmental stages, which would be consistent with the observed low level of expression in the mycelium. It is noteworthy that the assay used here was optimized for the measurement of GlcNAc incorporation into insoluble chitin polymers or into chito-oligosaccharides of a degree of polymerization higher than 6 [11], [17]. Thus, another possibility explaining the absence of in vitro activity with the recombinant SmCHS1 is that the protein may form chito-oligosaccharides of a low degree of polymerization that could possibly be involved in priming chitin biosynthesis.
Our data based on the use of nikkomycin Z strongly suggest that chitin synthase activity is essentially located at the hyphal tip in Saprolegnia. Indeed, inhibition of chitin biosynthesis led to the formation of a weaker wall at the apex of the hyphae, which was accompanied by a swelling of the cell primarily at its tip, followed by cell lysis. From these observations, and similar to the endosome-to-plasma membrane pathway described in yeast for membrane proteins [28], it is tempting to speculate that the MIT domains of SmCHS1 and SmCHS2 are involved in the apical delivery of chitin synthase via a direct or indirect interaction with the cytosleleton, and/or in the recycling of the enzyme through endosomal membranes. Membrane microdomains similar to lipid rafts have been shown to be involved in cell polarization in fungi [29]. This is consistent with our recent finding that the Saprolegnia chitin synthase activity is associated to such membrane structures [30], and with the present observation that chitin synthase is primarily located at the apex of the cells.
To date the CHS proteins from S. monoica represent the only examples of processive carbohydrate synthases that contain MIT domains. Similarly to SmCHS1 and SmCHS2, some of the Saprolegnia [31] and Phytophthora [32] cellulose synthases contain domains that seem to be specific to the oomycete enzymes. These domains belong to the Pleckstrin Homology (PH) family of domains [31], [32]. PH domains occur in a wide range of proteins involved in intracellular signaling or as constituents of the cytoskeleton [33]. They have been associated to different functions, especially the targeting of proteins to appropriate cellular compartment [33]. Altogether, these observations suggest that oomycete carbohydrate synthases have evolved by acquiring specific domains at their N-terminal ends that guide their cellular trafficking and membrane targeting.
As mentioned above, our data concur with a localization of chitin synthase activity at the apex of Saprolegnia hyphae, which implies a role in tip growth. This represents the first evidence of the biological function of chitin synthases in oomycetes. The data suggest that chitin is essentially synthesized at the tip of the hyphae, consistent with the previous observation that chitin is a minor cell wall component of S. monoica mycelial walls (<0.5% of the total cell wall carbohydrates) [11]. The chitin granules that have been shown to occur in all parts of the mycelial cell wall [11] most likely arise primarily from the apical site of synthesis and, as the cells grow, get distributed along the older parts of the hyphal walls. Interestingly, ultrastructural studies of the cell wall of the closely related species Achlya bisexualis have shown that cellulose is mainly located in the subapical and older parts of the hyphae, while the tip of the cells are devoid of cellulose and consist essentially of β-(1→3)-glucan [34], [35]. Although no similar studies have been performed in Saprolegnia, Saprolegnia cells most likely exhibit a similar distribution of cellulose and β-(1→3)-glucan in their walls as Achlya. In this case, chitin would substitute for cellulose at the tip of the hyphae to maintain a sufficient mechanical support to the apical wall. Indeed, the type of crystalline chitin that occurs in Saprolegnia is similar to fungal chitin and exhibits comparable physical and mechanical properties as cellulose [11]. A matrix that would consist essentially of amorphous β-(1→3)-glucan is not expected to be able to provide sufficient mechanical support to the cell and needs to be reinforced by crystalline polymers such as chitin or cellulose [6], [7]. From these observations, we reasoned that despite its low abundance in Saprolegnia cell walls, chitin plays an important role in tip growth and stabilization of the apical wall, and thus represents an interesting target of anti-oomycete drugs. This was verified by confirming that nikkomycin Z was indeed able to inhibit the mycelial chitin synthase activity and provoked cell lysis through tip bursting. Interestingly, the mycelium from S. parasitica showed an increased sensitivity to the inhibitor compared to S. monoica, which confirms the potential of using chitin synthase as a target for controlling the diseases provoked by devastating pathogenic oomycetes. The nikkomycin Z concentrations required for inhibiting the growth of S. parasitica were in the range 200 µM (data not shown). The mycelium from S. monoica could overcome such concentrations, with about 50% cell death, but all surviving cells grew slower and exhibited multiple abnormalities compared to the untreated control. The morphological alterations were similar to those reported earlier when growing the mycelium in the presence of Congo Red [36], which interferes with both chitin and cellulose formation. The cells surviving the nikkomycin Z treatment exhibited a higher level of expression of SmChs1 and SmChs2, most likely to compensate for the inhibitory effect of the drug.
In conclusion, due to the rather high concentrations of nikkomycin required to provoke cell lysis, or at least inhibit the growth of the mycelium, this drug may not be usable in practice to control Saprolegnia infections of fish. It remains however that chitin synthase represents one of the potential targets of anti-Saprolegnia drugs. The development of drugs with a higher affinity for chitin synthases than nikkomycin Z should allow the use of lower concentrations of inhibitors and be thus friendlier to the environment. Combining such chitin synthase inhibitors with compounds that are directed toward other cell wall synthesizing enzymes or other key biochemical pathways represents an interesting strategy to control the diseases caused by Saprolegnia.
Materials and Methods
Strain and culture conditions
The strains S. monoica Pringsheim 53–967 Dick and S. parasitica Coker were obtained from the Centraal Bureau voor Schimmel Culture (CBS, Baarn, The Netherlands) and maintained on Potato Dextrose Agar (PDA). The mycelium used for all experiments was grown in the liquid medium of Machlis [37] for 3–5 days at 25°C, as previously described [11].
RNA extraction, RACE and qRT-PCR
Total RNA was extracted and purified from 100 mg of 3-days old S. monoica mycelium, using the RNeasy Plant Mini Kit (Qiagen) and an on-column DNaseI digestion. Reverse transcription experiments were performed with 4.5 µg total RNA using the Superscript III First Strand cDNA Synthesis kit (Invitrogen) following the manufacturer's instructions. For the PCR amplification of SmChs2, 1 µl of the synthesized cDNA was used with primers (Chs2Fwd and Chs2Rev, Table S1 in Supplementary Material) designed from the putative SmChs2 gene sequence isolated previously (GenBank accession number U19946). Similarly, Chs1Fwd and Chs1Rev primers (Table S1) were designed from the partial sequence of the putative Chs1 gene from P. capsici (GenBank accession number U42304) and used for the amplification of a conserved region of Chs1. All PCR reactions were performed using Phusion High-Fidelity DNA Polymerase (Finnzymes), according to the manufacturer's instructions. An RNA ligase-mediated RACE kit (RLM-RACE kit, Ambion) was used to isolate the 5′ and 3′ ends of SmChs1 and amplify the sequence of the 3′ end of SmChs2 for verifying its sequence. The following sets of nested reverse primers were used to obtain the 5′ end of SmChs1: Chs1Rev1 and Chs1Rev2 (Table S1). The 3′ end of the gene was isolated using the nested forward primers Chs1Fwd1 and Chs1Fwd2 (Table S1), while the 3′ end of SmChs2 was amplified with the primers Chs2Fwd1 and Chs2Fwd2 (Table S1). The full-length SmChs1 and SmChs2 genes were amplified from mycelial cDNA to confirm their complete sequences. The following primers were used for this purpose: Chs1FLFwd and Chs1FLRev for the amplification of SmChs1, and Chs2Fwd and Chs2FLRev for the amplification of SmChs2 (Table S1). The full-length genes were cloned into the pENTR-D-TOPO vector (Invitrogen) using the Gateway technology, according to the manufacturer's instructions. The constructs were transformed into One Shot TOP10 Escherichia coli cells that were chemically competent (Invitrogen) and their sequenced confirmed (MWG, Germany).
Quantitative Real-Time PCR (qRT-PCR) analyses were normalized against the 18S rRNA [31], ubiquitin and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) genes from S. monoica. Since the sequences of the latter 2 genes were unknown, we first amplified them by designing primers from the S. parasitica ATCC90214 mycelial sequence library (www.oomycete.org). UbFwd and UbRev were used to amplify the ubiquitin gene, while GAPDHFwd and GAPDHRev were the primers used for isolating the GAPDH gene (Table S1). The PCR products were cloned into pUC19 using the SmaI site and the constructs were transformed into E. coli competent cells as above. For qRT-PCR experiments, total RNA was extracted from the mycelium grown in the absence or in the presence of 50 µM nikkomycin Z for either 3 days or 5 days. One µg RNA was converted into cDNA using the SuperScript III First-Strand Synthesis SuperMix kit for qRT-PCR (Invitrogen). The cDNA was diluted to 2.4 ng/µl, and 2.5 µl of the diluted cDNA was used as a template for qRT-PCR analysis. The sequences of the primers are presented in Table S2 (Supplementary Material). The cDNA was amplified using the iQ SYBR Green Supermix (Bio-Rad) on a Bio-Rad iCycler. The reactions were performed in triplicates and repeated on two independent biological samples. The PCR conditions consisted of an initial denaturation at 95° for 3 minutes, followed by 60 cycles of denaturation at 95° for 30 seconds, annealing at 58° for 30 seconds and elongation at 72° for 30 seconds. A dissociation kinetics analysis was performed at the end of the experiment to check the specificity of the annealing. Results were analysed with the Q-gene software [38]. Identical results were obtained with all 3 housekeeping genes used. Only the normalized results obtained with the 18S rRNA gene are presented.
Southern blot analysis
DNA from S. monoica mycelium grown for 3 days in the liquid culture of Machlis [37] was extracted in the presence of cetyltrimethylammonium bromide [39]. The genomic DNA was resuspended in TE buffer (10 mM Tris-HCl pH 8.5 containing 0.1 mM EDTA) and 100 µg was digested with SacI, ClaI or PstI (Fermentas) at 37°C overnight. Aliquots of the digests containing the restriction fragments were separated by electrophoresis on a 0.7% (w/v) agarose gel. After depurination and alkaline denaturation, the DNA fragments were transferred to a positive nylon membrane (Hybond N+, Amersham) and subjected to Southern hybridization [40]. The probes corresponding to conserved amino acid sequences were prepared using the NEBlot Phototope kit (New England BioLabs) after PCR amplification of a plasmid DNA template containing SmChs2, using the primer pairs Chs2Probe1Fwd/Chs2Probe1Rev and Chs2Probe2Fwd/Chs2Probe2Rev (Table S1). The probes corresponding to the 1404–1704 nucleotide segment of SmChs2 (Probe 1) and 876–1233 nucleotide sequence of SmChs2 (Probe 2) were amplified using the Phusion High-Fidelity DNA Polymerase from Finnzymes. They were labeled with biotin and 32P in the presence of [α-32P]dCTP (Perkin Elmer, Boston, MA; Megaprime DNA Labeling Systems, Amersham Biosciences, Uppsala, Sweden) according to the manufacturer's instructions. Hybridization was performed overnight at 65°C in the presence of 20 ng/ml of each probe. The detection of the signals was performed using the chemiluminescent Phototope Star detection kit following the manufacturer's instructions (New England BioLabs), and a Biorad CCD camera.
Bioinformatics and phylogenetic analyses
All sequences were edited using the software BioEdit [41] and assembled from an initial database of oomycete chitin synthases. Alignments were performed using ClustalW [42]. The SmCHS1 and SmCHS2 full-length sequences as well as the MIT domain sequences alone were used to perform a BLASTp search against non-redundant protein databases from the National Centre for Biotechnology (http://www.ncbi.nlm.nih.gov). The full-length sequences were also used for similarity searches against the P. infestans database at the Broad Institute (http://www.broadinstitute.org/annotation/genome/phytophthora_infestans/MultiHome.html). The resulting oomycete dataset consisted of the putative chitin synthases (accession numbers in parentheses) from P. infestans (PITG_02050.1), A. euteiches (EU522489 and EU447431 for AeCHS1 and AeCHS2 respectively), Plasmopara viticola (AY05254), Pythium insidiosum (DQ116421), P. capsici (U42304), and Achlya ambisexualis (U55044). The phylogenetic tree was rooted with the sequence of CHS1 from Neurospora crassa (accession number M73437). The dataset of MIT-containing proteins was prepared by gathering the MIT domain sequences (accession numbers in parentheses) from Mus musculus (1WFD_A), the Aspergillus oryzae calpain-like protease PalBory (AB020321), the vacuolar sorting protein VPS4 from Entamoeba histolytica (XM_649013), the Sulfolobus solfataricus AAA ATPase family protein (NP_342401), the Homo sapiens sorting nexin 15 (AAH14520), and the Theileria annulata putative AAA family ATPase (XM_949947).
The phylogenetic analyses of the protein datasets were performed with the software MrBayes (v3.1; [43]) using a Bayesian inference [44] under the model suggested by the program ProtTest (http://darwin.uvigo.es/software/prottest_server.html; [45]), which estimates likelihood scores under different models. The WAG+G+F amino acid substitution model was suggested and implemented in the Bayesian inference [46]. The Bayesian analyses consisted of two Markov chains run in parallel. Each of them was flanked to other three simultaneous “heated” chains (Metropolis coupled Markov chain Monte Carlo; [44]) to avoid getting trapped in a local maximum. The chains were run for 500,000 generations, with sampling only once every 100 generations and collecting at the end 5,000 trees. In order to avoid early pre-convergence trees, the first 25% (burning) trees were discarded. The majority rule consensus tree was calculated on the remaining trees, with posterior probability on every node as a measure of confidence of the analyses.
Transmembrane domains were predicted using the TMHMM [47] program at http://www.cbs.dtu.dk/services/TMHMM-2.0/and Kyte and Doolittle hydropathy plots [48] were obtained using the tool available at http://fasta.bioch.virginia.edu/fasta_www2/fasta_www.cgi. Potential phosphorylation and glycosylation sites were identified using the NetPhos 2.0 server (http://www.cbs.dtu.dk/services/NetPhos/) and the NetNGlyc 1.0 server (http://www.cbs.dtu.dk/services/NetNGlyc/), respectively. The search for putative domains in SmCHS1 and SmCHS2 was performed in the Pfam motif database using MotifScan [49] (http://hits.isb-sib.ch/cgi-bin/PFSCAN). The diagrams representing the predicted topology of SmCHS1 and SmCHS2 were drawn using the TOPO2 program at http://www.sacs.ucsf.edu/TOPO-run/wtopo.pl.
Treatment with nikkomycin Z and optical microscopy
The effect of the chitin synthase inhibitor nikkomycin Z on the growth of S. monoica and S. parasitica was tested on cultures of mycelium on PDA plates supplemented with 0, 25, 50, 200, 500, 2000 or 5000 µM nikkomycin Z (Sigma) in either 90-mm Petri plates or 6-well culture plates (Falcon). The microscopic observations were performed on the mycelium grown in 20 ml of liquid medium [37] supplemented with the indicated concentrations of nikkomycin Z. Aliquots of the control and nikkomycin-treated cultures were aseptically removed at different times and mounted on microscope slides. Observations were performed using an Olympus BX-51 optical microscope fitted with a camera.
Field Emission-Scanning Electron Microscopy (FE-SEM)
The mycelium from the control and nikkomycin-treated cultures (5 mM inhibitor) grown on PDA was flash-frozen in liquid nitrogen, freeze-dried and observed by FE-SEM. The samples were mounted onto a metal substrate using carbon tape, and coated with a thin layer of Au–Pd. A Hitachi S-4800 scanning electron microscope operated at 1 kV was used to capture secondary electron images of the surfaces of the samples.
Preparation of microsomal fractions from S. monoica mycelium, chitin synthase assay and enzyme kinetics
S. monoica was grown in 140-mm Petri plates for 3 days in 100 ml liquid medium (Machlis) [37] inoculated with about 30 agar plugs of 5 mm cut from cultures grown on PDA. The mycelium was collected, washed extensively with water and dried under vacuum on filter paper. All the subsequent steps were performed at 4°C. The cells were homogenized in an extraction buffer (Tris-HCl 10 mM pH 7.4) containing a plant protease inhibitor cocktail (Sigma) using a Waring blender. The homogenate was centrifuged at 5,000 g for 10 minutes. The supernatant was filtered through two layers of Miracloth and centrifuged at 100,000 g for 1 hour (Beckman-Coulter Optima L-100 XP ultracentrifuge). The pelleted membranes were resuspended in extraction buffer and used for protein [50] and chitin synthase assays.
Chitin synthase activity was measured as described earlier [11], [17]. Briefly, 50 µl of enzyme preparation was mixed with 150 µl of reaction buffer consisting of 10 mM Tris-HCl pH 7.4, 10 mM MgCl2, 20 mM N-acetyl-d-glucosamine, 1.25 µg trypsin/ml, 0.5 mM UDP-N-acetyl-d-glucosamine and 434 nM UDP-N-acetyl-d-[U-14C]glucosamine (318 mCi/mmol; Amersham). The reaction was incubated for 1 hour at room temperature and stopped with 400 µl of ethanol. After an overnight precipitation at −20°C, insoluble polysaccharides were recovered on Whatman GF/C glass-fiber filters and subsequently washed with water and 70% ethanol. The radioactivity retained on the filters was detected by liquid scintillation (Packard 1500 Tri-Carb). The determination of the apparent Km of chitin synthase was performed in the assay conditions described above, but by using different UDP-GlcNAc concentrations (0.1, 0.5, 1 and 2 mM). The apparent Ki of the enzyme for nikkomycin Z was obtained from Dixon plots by performing kinetics in the presence of varying concentrations of nikkomycin Z (0, 0.2, 1, 5, 15 µM) and UDP-GlcNAc (0.1, 0.5 and 1 mM) over a period of 30 min reaction.
Determination of mycelial chitin content
The chitin content of mycelial cells was measured by assaying the amount of glucosamine released by acid hydrolysis of the wall-bound chitin [51]. Cell wall samples were prepared from about 1 g fresh weight of mycelium by grinding the cells in liquid nitrogen and resuspending the homogenate in 2 ml deionized water. After centrifugation at 13,000 g for 10 minutes at 4°C, the pelleted material was freeze-dried overnight. Known amounts of the samples (2 to 6 mg) were hydrolyzed with 1 ml of 6 M HCl at 100°C for 4 h. After cooling to room temperature, the hydrolysate (0.2 ml) was added to 0.25 ml of 4% acetylacetone in 1.25 M sodium carbonate and heated for 1 h at 90°C. After cooling, 2 mL of ethanol were added under agitation to dissolve the precipitate. Finally, 0.25 ml of Ehrlich reagent (1.6 g of N,N-dimethyl-p-aminobenzaldehyde in 60 mL of a 1∶1 mixture of ethanol and concentrated HCl) was added and the absorbance was measured at 530 nm. The chitin content, expressed as micrograms of glucosamine hydrochloride per mg of cell wall (dry weight) was calculated from a standard curve prepared with known amounts of glucosamine hydrochloride.
Expression of SmCHS1 and SmCHS2 in Pichia pastoris
SmCHS1 and SmCHS2 were expressed as fusion proteins with eGFP at their C-terminus. For this purpose, the corresponding genes were cloned in the pDONR221 vector using the BP clonase kit (Invitrogen) and the primers Chs1PichiaFwd/Chs1PichiaRev and eGfpFwdChs1/eGfpRevChs1 for SmChs1, and Chs2PichiaFwd/Chs2PichiaRev and eGfpFwdChs2/eGfpRevChs2 for SmChs2 (Table S1). The donor vectors were recombined into a Gateway compatible version of the Pichia vector pPICZ using the LR clonase kit (Invitrogen). Zeocin resistant colonies were screened for GFP fluorescence using a confocal microscope (LSM510, Zeiss) and the positive clones exhibiting the highest signal were used for subsequent chitin synthase assays.
Chitin synthase assay using detergent extracts of Pichia membranes
The Pichia cells grown in the buffered glycerol complex medium (BMGY: 100 mM potassium phosphate pH 6.0, 1% yeast extract, 2% peptone, 1.34% yeast nitrogen base, 4×10−5% biotin, 1% glycerol) for one day at 30°C were collected by low-speed centrifugation and resuspended in the BMMY medium at a final OD600 of 1 (BMMY medium: as BMGY but with 0.5% methanol instead of glycerol). The cells were induced for one day at 30°C with 0.5% (v/v) methanol, harvested and resuspended in a lysis buffer (Tris-HCl 10 mM pH 7.4) supplemented with a yeast protease inhibitor cocktail (Sigma). The cells were disrupted in an Aminco French pressure cell (10,000 psi), and the microsomal fraction was obtained by differential centrifugation as described above for the isolation of the membrane fraction from the mycelium of S. monoica. The membrane fraction was resuspended in lysis buffer and diluted to a final concentration of 1 mg/ml protein prior to protein extraction in the presence of 0.5% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS) at 4°C under gentle stirring. The preparation was centrifuged at 100,000 g for 1 h (4°C) and the supernatant corresponding to the CHAPS extract was used for chitin synthase assays or enzyme kinetics as described above.
Characterization of the chitin synthesized in vitro and observation by transmission electron microscopy
Product characterization was performed by enzymatic hydrolysis of the radioactive chitin synthesized in vitro in the presence of UDP-N-acetyl-d-[U-14C]glucosamine (see enzyme assay conditions). The ethanol-insoluble polysaccharides were pelleted by a 10 min centrifugation at 10,000 g, washed with 1 ml hydrolysis buffer (50 mM sodium acetate pH 5.0) and pelleted again. The insoluble polysaccharides were resuspended in 200 µl of hydrolysis buffer containing the specific chitinase from Serratia marcescens (Sigma) used at 25 µg/ml (final concentration). Triplicates were run in parallel. The samples were incubated for 48 hours at 25°C and the reaction mixtures were supplemented every 24 hours with freshly prepared chitinase. The non-hydrolyzed polysaccharides were precipitated overnight in ethanol at −20°C and recovered by filtration on glass-fiber filters. The radioactivity retained on the filters was determined by scintillation counting as described above. The extent of hydrolysis was determined by comparison with controls in which the hydrolytic enzyme had been replaced by the corresponding incubation buffer. For electron microscopy observation of the chitin synthesized in vitro by the recombinant chitin synthases expressed in Pichia and solubilized with CHAPS, a drop of the reaction mixture obtained after incubation in the same conditions as for a chitin synthase assay, but in the presence of nonradioactive UDP-GlcNAc, was deposited on carbon-coated grids and observed after negative staining with 2% uranyl acetate as described earlier [17]. Control experiments were performed by using a similar protein fraction prepared from the wild-type Pichia cells.
Supporting Information
Zdroje
1. van WestP
2006 Saprolegnia parasitica, an oomycete pathogen with a fishy appetite: new challenges for an old problem. Mycologist 20 99 104
2. StuelandS
HataiK
SkaarI
2005 Morphological and physiological characteristics of Saprolegnia spp. Strains pathogenic to Atlantic salmon, Salmo salar L. J Fish Dis 28 445 453
3. PhillipsAJ
AndersonVL
RobertsonEJ
SecombesCJ
van WestP
2008 New insights into animal pathogenic oomycetes. Trends Microbiol 16 13 19
4. JohnsonTWJr
SeymourRL
PadgettDE
2002 Biology and Systematics of the Saprolegniaceae. Book published on the World-Wide-Web: http://dl.uncw.edu
5. Diéguez-UribeondoJ
CereniusL
SöderhällK
1996 Physiological characterization of Saprolegnia parasitica isolates from brown trout. Aquaculture 140 247 257
6. Bartnicki-GarciaS
1968 Cell wall chemistry, morphogenesis and taxonomy of fungi. Annu Rev Microbiol 22 87 108
7. WesselsJGH
SietsmaJH
1981 Fungal cell walls: a survey.
TannerW
LoewusFA
Encyclopedia of plant physiology. Plant carbohydrates II Berlin Springer Verlag 352 394
8. LinCC
AronsonJM
1970 Chitin and cellulose in the cell walls of the oomycetes Apodachlya. Arch Mikrobiol 72 111 114
9. AronsonJM
LinCC
1978 Hyphal cell wall chemistry of Leptomitus lacteus. Mycologia 70 363 369
10. Campos-TakakiGM
DietrichSMC
MascarenhasY
1982 Isolation and characterization of chitin from the cell walls of Achlya radiosa. J Gen Microbiol 128 207 209
11. BuloneV
ChanzyH
GayL
GirardV
FèvreM
1992 Characterization of chitin and chitin synthase from the cellulosic cell wall fungus Saprolegnia monoica. Exp Mycol 16 8 21
12. BadreddineI
LafitteC
HeuxL
SkandalisN
SpanouZ
2008 Cell wall chitosaccharides are essential components and exposed patterns of the phytopathogenic oomycete Aphanomyces euteiches. Eukaryot Cell 7 1980 1993
13. HaasBJ
KamounS
ZodyMC
JiangRHY
HandsakerRE
2009 Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature 461 393 398
14. WernerS
SteinerU
BecherR
KortekampK
ZyprianE
2002 Chitin synthesis during in planta growth and asexual propagation of the cellulosic oomycete and obligate biotrophic grapevine pathogen Plasmopara viticola. FEMS Microbiol Lett 208 169 173
15. Mort-BontempsM
GayL
FèvreM
1997 CHS2, a chitin synthase gene from the oomycete Saprolegnia monoica. Microbiology 143 2009 2020
16. Martínez-RucoboFW
Eckhardt-StrelauL
Terwisscha van ScheltingaAC
2009 Yeast chitin synthase 2 activity is modulated by proteolysis and phosphorylation. Biochem J 417 547 554
17. GayL
BuloneV
GirardV
FèvreM
ChanzyH
1992 Chitin pleomorphism in the cellulosic cell wall fungus Saprolegnia. FEMS Microbiol Lett 100 405 410
18. Ruiz-HerreraJ
Gonzalez-PrietoJM
Ruiz-MedranoR
2002 Evolution and phylogenetic relationships of chitin synthases from yeasts and fungi. FEMS Yeast Res 1 247 256
19. ChoquerM
BoccaraM
GonçalvesIR
SouliéMC
Vidal-CrosA
2004 Survey of the Botrytis cinerea chitin synthase multigenic family through the analysis of six euascomycetes genomes. Eur J Biochem 271 2153 2164
20. CampbellJA
DaviesGJ
BuloneV
HenrissatB
1997 A classification of nucleotide-diphospho-sugar glycosyltransferases based on amino-acid sequence similarities. Biochem J 326 929 939
21. CiccarelliFD
ProukakisC
PatelH
CrossH
AzamS
2003 The identification of a conserved domain in both spartin and spastin mutated in hereditary spastic paraplegia. Genomics 81 437 441
22. RigdenDJ
LiuH
HayesSD
UrbéS
ClagueMJ
2009 Ab initio protein modelling reveals novel human MIT domains. FEBS Lett 583 872 878
23. ScottA
GasparJ
Stuchell-BreretonMD
AlamSL
SkalickyJJ
2005 Structure and ESCRT-III protein interactions of the MIT domain of human VPS4A. Proc Natl Acad Sci U S A 102 13813 13818
24. GoodayGW
1990 Inhibition of chitin metabolism.
KuhnPJ
TrinciAPJ
JungMJ
GoseyMW
CoppingLG
Biochemistry of cell walls and membranes in fungi Berlin Springer Verlag 61 79
25. GowLA
SelitrennikoffCP
1984 Chitin synthetase of Neurospora crassa: Inhibition by nikkomycin, polyoxin B, and UDP. Curr Microbiol 11 211 216
26. GaughranJP
LaiMH
KirschDR
SilvermanSJ
1994 nikkomycin Z is a specific inhibitor of Saccharomyces cerevisiae chitin synthase isozyme Chs3 in vitro and in vivo. J Bacteriol 176 5857 5860
27. DuranA
CabibE
1978 Solubilization and partial purification of yeast chitin synthetase. Confirmation of the zymogenic nature of the enzyme. J Biol Chem 253 4419 4425
28. LuoW
ChangA
2000 An endosome-to-plasma membrane pathway involved in trafficking of a mutant plasma membrane ATPase in yeast. Mol Biol Cell 11 579 592
29. MartinSW
KonopkaJB
2004 Lipid raft polarization contributes to hyphal growth in Candida albicans. Eukaryot Cell 3 675 684
30. BriolayA
BouzenzanaJ
GuichardantM
DeshayesC
SindtN
2009 Cell wall polysaccharide synthases are located in detergent-resistant membrane microdomains in oomycetes. Appl Environ Microbiol 75 1938 1949
31. FugelstadJ
BouzenzanaJ
DjerbiS
GuerrieroG
EzcurraI
2009 Identification of the cellulose synthase genes from the oomycete Saprolegnia monoica and effect of cellulose synthesis inhibitors on gene expression and enzyme activity. Fungal Genet Biol 46 759 767
32. Grenville-BriggsLJ
AndersonVL
FugelstadJ
AvrovaAO
BouzenzanaJ
2008 Cellulose synthesis in Phytophthora infestans is required for normal appressorium formation and successful infection of potato. Plant Cell 20 720 738
33. RebecchiMJ
ScarlataS
1998 Pleckstrin homology domains: a common fold with diverse functions. Annu Rev Biophys Biomol Struct 27 503 528
34. ShapiroA
MullinsJT
2002 Hyphal tip growth in Achlya bisexualis. I. Distribution of 1,3-β-glucans in elongating and non-elongating regions of the wall. Mycologia 94 267 272
35. ShapiroA
MullinsJT
2002 Hyphal tip growth in Achlya bisexualis. II. Distribution of cellulose in elongating and non-elongating regions of the wall. Mycologia 94 273 279
36. NodetP
CapellanoA
FèvreM
1990 Morphogenetic effects of Congo red on hyphal growth and cell wall development of the fungus Saprolegnia monoica. J Gen Microbiol 136 303 310
37. MachlisL
1953 Growth and nutrition of water molds in the subgenus Euallomyces. II. Optimal composition of the minimal medium. Am J Bot 40 449 460
38. MullerPY
JanovjakH
MiserezAR
DobbieZ
2002 Processing of gene expression data generated by quantitative real-time RT-PCR. Biotechniques 32 1372 1378
39. MurrayMG
ThompsonWF
1980 Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8 4321 4325
40. SouthernEM
1975 Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol 98 503 517
41. HallTA
1999 BioEdit: a user-friendly biological sequence alignment editor and analysis. http://www.mbio.ncsu.edu/BioEdit/bioedit.html
42. LarkinMA
BlackshieldsG
BrownNP
ChennaR
McGettiganPA
2007 Clustal W and Clustal X version 2.0. Bioinformatics 23 2947 2948
43. RonquistF
HuelsenbeckJP
van der MarkP
2005 MrBayes 3.1 manual, Draft 5/17/2005 http://mrbayes.csit.fsu.edu/mb3.1_manual.pdf
44. HuelsenbeckJP
RonquistFR
2001 MrBayes: Bayesian inference of phylogeny. Biometrics 17 754 755
45. AbascalF
ZardoyaR
PosadaD
2005 ProtTest: Selection of best-fit models of protein evolution. Bioinformatics 21 2104 2105
46. WhelanS
GoldmanN
2001 A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol 18 691 699
47. KroghA
LarssonB
von HeijneG
SonnhammerELL
2001 Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J Mol Biol 305 567 580
48. KyteJ
DoolittleRF
1982 A simple method for displaying the hydropathic character of a protein. J Mol Biol 157 105 132
49. HuloN
BairochA
BulliardV
CeruttiL
CucheBA
2008 The 20 years of PROSITE. Nucleic Acids Res 36 D245 D249
50. BradfordMM
1976 A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72 248 254
51. ChenGC
JohnsonBR
1983 Improved colorimetric determination of cell wall chitin in wood decay fungi. Appl Environ Microbiol 46 13 16
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2010 Číslo 8- Stillova choroba: vzácné a závažné systémové onemocnění
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Jak souvisí postcovidový syndrom s poškozením mozku?
- Familiární středomořská horečka
-
Všechny články tohoto čísla
- Dissecting the Genetic Architecture of Host–Pathogen Specificity
- The Battle for Iron between Bacterial Pathogens and Their Vertebrate Hosts
- Global Genotype-Phenotype Correlations in
- Burkholderia Type VI Secretion Systems Have Distinct Roles in Eukaryotic and Bacterial Cell Interactions
- Chitin Synthases from Are Involved in Tip Growth and Represent a Potential Target for Anti-Oomycete Drugs
- Distinct Merkel Cell Polyomavirus Molecular Features in Tumour and Non Tumour Specimens from Patients with Merkel Cell Carcinoma
- Biological and Structural Characterization of a Host-Adapting Amino Acid in Influenza Virus
- Functional Characterisation and Drug Target Validation of a Mitotic Kinesin-13 in
- CTCF Prevents the Epigenetic Drift of EBV Latency Promoter Qp
- The Human Fungal Pathogen Escapes Macrophages by a Phagosome Emptying Mechanism That Is Inhibited by Arp2/3 Complex-Mediated Actin Polymerisation
- Bim Nuclear Translocation and Inactivation by Viral Interferon Regulatory Factor
- Cyst Wall Protein 1 Is a Lectin That Binds to Curled Fibrils of the GalNAc Homopolymer
- Reciprocal Analysis of Infections of a Model Reveal Host-Pathogen Conflicts Mediated by Reactive Oxygen and imd-Regulated Innate Immune Response
- A Subset of Replication Proteins Enhances Origin Recognition and Lytic Replication by the Epstein-Barr Virus ZEBRA Protein
- Damaged Intestinal Epithelial Integrity Linked to Microbial Translocation in Pathogenic Simian Immunodeficiency Virus Infections
- Kaposin-B Enhances the PROX1 mRNA Stability during Lymphatic Reprogramming of Vascular Endothelial Cells by Kaposi's Sarcoma Herpes Virus
- Direct Interaction between Two Viral Proteins, the Nonstructural Protein 2C and the Capsid Protein VP3, Is Required for Enterovirus Morphogenesis
- A Novel CCR5 Mutation Common in Sooty Mangabeys Reveals SIVsmm Infection of CCR5-Null Natural Hosts and Efficient Alternative Coreceptor Use
- Micro RNAs of Epstein-Barr Virus Promote Cell Cycle Progression and Prevent Apoptosis of Primary Human B Cells
- Enterohemorrhagic Requires N-WASP for Efficient Type III Translocation but Not for EspF-Mediated Actin Pedestal Formation
- Host Imprints on Bacterial Genomes—Rapid, Divergent Evolution in Individual Patients
- UNC93B1 Mediates Host Resistance to Infection with
- The Transcription Factor Rbf1 Is the Master Regulator for -Mating Type Controlled Pathogenic Development in
- Protective Efficacy of Cross-Reactive CD8 T Cells Recognising Mutant Viral Epitopes Depends on Peptide-MHC-I Structural Interactions and T Cell Activation Threshold
- Bacteriophage Lysin Mediates the Binding of to Human Platelets through Interaction with Fibrinogen
- Insecticide Control of Vector-Borne Diseases: When Is Insecticide Resistance a Problem?
- Immune Modulation with Sulfasalazine Attenuates Immunopathogenesis but Enhances Macrophage-Mediated Fungal Clearance during Pneumonia
- PKC Signaling Regulates Drug Resistance of the Fungal Pathogen via Circuitry Comprised of Mkc1, Calcineurin, and Hsp90
- A Multi-Step Process of Viral Adaptation to a Mutagenic Nucleoside Analogue by Modulation of Transition Types Leads to Extinction-Escape
- “Everything You Always Wanted to Know about Sex (but Were Afraid to Ask)” in after Two Decades of Laboratory and Field Analyses
- Norovirus Gastroenteritis, Carbohydrate Receptors, and Animal Models
- Variations in TcdB Activity and the Hypervirulence of Emerging Strains of
- SWAN-1 Binds to EGL-9 and Regulates HIF-1-Mediated Resistance to the Bacterial Pathogen PAO1
- Conformational Adaptation of Asian Macaque TRIMCyp Directs Lineage Specific Antiviral Activity
- The Proteasome Active Site Threonine Is Essential for Persistence Yet Dispensable for Replication and Resistance to Nitric Oxide
- Characterization of Oseltamivir-Resistant 2009 H1N1 Pandemic Influenza A Viruses
- The Pneumococcal Serine-Rich Repeat Protein Is an Intra-Species Bacterial Adhesin That Promotes Bacterial Aggregation and in Biofilms
- Regulatory T Cell Suppressive Potency Dictates the Balance between Bacterial Proliferation and Clearance during Persistent Infection
- Structural Alterations in a Component of Cytochrome Oxidase and Molecular Evolution of Pathogenic in Humans
- A Limited Number of Antibody Specificities Mediate Broad and Potent Serum Neutralization in Selected HIV-1 Infected Individuals
- Spliced Leader Trapping Reveals Widespread Alternative Splicing Patterns in the Highly Dynamic Transcriptome of
- Contribution of Coagulases towards Disease and Protective Immunity
- Early Severe Inflammatory Responses to Uropathogenic Predispose to Chronic and Recurrent Urinary Tract Infection
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- The Transcription Factor Rbf1 Is the Master Regulator for -Mating Type Controlled Pathogenic Development in
- PKC Signaling Regulates Drug Resistance of the Fungal Pathogen via Circuitry Comprised of Mkc1, Calcineurin, and Hsp90
- Contribution of Coagulases towards Disease and Protective Immunity
- Early Severe Inflammatory Responses to Uropathogenic Predispose to Chronic and Recurrent Urinary Tract Infection
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



