-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
The Transcription Factor Rbf1 Is the Master Regulator for -Mating Type Controlled Pathogenic Development in
In the phytopathogenic basidiomycete Ustilago maydis, sexual and pathogenic development are tightly connected and controlled by the heterodimeric bE/bW transcription factor complex encoded by the b-mating type locus. The formation of the active bE/bW heterodimer leads to the formation of filaments, induces a G2 cell cycle arrest, and triggers pathogenicity. Here, we identify a set of 345 bE/bW responsive genes which show altered expression during these developmental changes; several of these genes are associated with cell cycle coordination, morphogenesis and pathogenicity. 90% of the genes that show altered expression upon bE/bW-activation require the zinc finger transcription factor Rbf1, one of the few factors directly regulated by the bE/bW heterodimer. Rbf1 is a novel master regulator in a multilayered network of transcription factors that facilitates the complex regulatory traits of sexual and pathogenic development.
Published in the journal: . PLoS Pathog 6(8): e32767. doi:10.1371/journal.ppat.1001035
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1001035Summary
In the phytopathogenic basidiomycete Ustilago maydis, sexual and pathogenic development are tightly connected and controlled by the heterodimeric bE/bW transcription factor complex encoded by the b-mating type locus. The formation of the active bE/bW heterodimer leads to the formation of filaments, induces a G2 cell cycle arrest, and triggers pathogenicity. Here, we identify a set of 345 bE/bW responsive genes which show altered expression during these developmental changes; several of these genes are associated with cell cycle coordination, morphogenesis and pathogenicity. 90% of the genes that show altered expression upon bE/bW-activation require the zinc finger transcription factor Rbf1, one of the few factors directly regulated by the bE/bW heterodimer. Rbf1 is a novel master regulator in a multilayered network of transcription factors that facilitates the complex regulatory traits of sexual and pathogenic development.
Introduction
In a wide range of fungi, complex developmental traits such as cell identity, morphogenesis and sexual development are controlled by mating type loci [1], [2], [3]. In the smut fungi, a group of plant pathogens, these traits also include the ability to infect their host plants. In Ustilago maydis, a smut fungus that infects maize, it is the b-mating type locus that is critical for both sexual as well as for pathogenic development. Similar to other smuts, U. maydis exhibits a dimorphic life cycle. The haploid, cigar-shaped cells, called sporidia, multiply by yeast-like budding, and the dikaryon, which is formed upon the fusion of two compatible sporidia, grows as a filament. This switch in cell morphology is accompanied by an alteration of the life-style. While the sporidia are apathogenic and grow strictly saprophytic, the filament is biotrophic, i.e. it depends on the living tissue of its host plant maize for further development. Initially, the dikaryotic hypha consists of a long tip cell with the accumulated cytoplasm; the succeeding, older parts consist of “empty” cells that are separated by regularly spaced septae. Cell division is stalled until the hypha has penetrated the cuticula of a corn plant, and only then a “true” filament with multiple septated compartments is formed. Upon plant invasion, hyphae traverse the plant without harming the cells and without an apparent host defense response. After several days, the fungus induces plant tumors, coinciding with a massive proliferation of fungal hyphae [for review, see 4].
In order to fuse and to form the pathogenic filament, the two sporidia must carry different alleles both of the biallelic a - and of the multiallelic b-mating type locus. The a-locus encodes a pheromone/receptor system required for cell sensing, initiation of filamentous conjugation tubes, and cell fusion. After fusion, the crucial step for the initiation of the pathogenic phase is the formation of a heterodimeric complex of two homeodomain proteins, bE and bW, which are encoded by the b-mating type. This bE/bW complex is formed only when the two proteins are derived from different b-alleles, and is sufficient to initiate the switch from budding to filamentous growth. Concomitantly, activation of b leads to a cell cycle arrest that is only released after host plant infection. It has been shown conclusively that the bE/bW complex is sufficient to initiate the pathogenic development, as exemplified by haploid “solopathogenic” strains that harbor different alleles of bE and bW and that are capable to infect plants without a mating partner [5]. Thus, it is conceivable that genes regulated by the bE/bW heterodimer are involved in (1) the establishment of the biotrophic phase, (2) cell cycle regulation and (3) the dimorphic transition from budding to the polarized growth of the filament. However, until now, only four b-regulated genes have been identified with impact on these processes, three of which are required during the very early infection stages. biz1 encodes a zinc finger transcription factor that is involved in the G2 cell cycle arrest preceding plant penetration as well as in the induction of appressoria, specific infection structures at the tip of penetrating hyphae [6]. The mitogen-activated protein (MAP) kinase Kpp6 is required for the subsequent step: U. maydis strains harboring a non-activatable kpp6 allele still form appressoria, but are defective in the penetration of the plant cuticula [7]. After plant penetration, the clp1 gene is required for further proliferation of dikaryotic filaments in planta. clp1 mutant strains still penetrate the plant cuticula, but development is stalled prior the first mitotic division; in addition, mutant strains do not form clamps, a structure that ensures the proper distribution of nuclei in the dikaryotic hyphae [8]. Interestingly, the induced expression of clp1 strongly interferes with the b-dependent induction of several of the genes regulated by the bE/bW-heterodimer, indicating that Clp1 may modulate the activity of the bE/bW complex. And finally, the b-dependently expressed cyclin Pcl12 is involved in the polarized growth of the b-dependent filament, but is dispensable for pathogenic development [9].
The bE/bW heterodimer binds to a conserved sequence motif, the b-binding sequence (bbs) that has been identified in the b-dependently induced lga2-gene [10]. Out of the 20 b-dependent genes identified so far, only two additional genes were found to harbor the bbs-motif: the above mentioned clp1 gene, and frb52, a gene with unknown function [11]. As the majority of b-controlled genes is obviously not directly regulated by bE/bW, it appears likely that the bE/bW heterodimer triggers a regulatory cascade with a limited number of direct targets genes. Thus, these “class I” genes should encompass regulators that trigger the regulation of the larger number of indirect, “class II” b targets. It was proposed that these regulators play pivotal roles either in all (as master regulator) or distinct (as relay) b-dependent processes.
Here, we employed U. maydis strains that harbor inducible combinations of the bE and bW genes [11] and DNA array technology to investigate the b-dependent processes in a time-resolved manner. Our analysis provides insight in the complex interconnection of cell cycle regulation during the dimorphic switch and highlights the specific characteristics of the “pathogenic” hyphae. Most important, we identify the zinc-finger transcription factor Rbf1 as a novel master regulator that is required for all b-dependent processes.
Results
Genes regulated by the bE/bW heterodimer are involved in cell cycle regulation, cell wall remodeling and secretion of effector candidates
In order to identify genes regulated by the bE/bW heterodimer, we performed microarray experiments with custom Affymetrix arrays (MPIUstilagoA) covering 5823 of the predicted 6786 U. maydis genes. Changes in gene expression were monitored during a 12-h time course (with samples taken at 1h, 2h, 3h, 5h, 12h) using the haploid U. maydis strains AB31 and AB33 that harbor the bE1 and bW2 genes under the control of the arabinose-inducible crg1 promoter and the nitrate-inducible nar1 promoter, respectively [11]. Induction of bE1/bW2 in these strains results in a filament that resembles the infectious hypha formed after fusion of compatible sporidia [11]. Strains AB32 and AB34, which harbor the incompatible bE2 and bW2 combination, were used as controls. Expression of bE and bW genes was induced by a shift from glucose - to arabinose (AB31 and AB32) or from glutamine - to nitrate containing media (AB33 and AB34). The expression profiles after b-induction in AB31 and AB33 were similar, but not identical. Firstly, the use of different media had an effect on gene expression, and, secondly, the use of the crg1 promoter resulted in gene expression values that were two - to fivefold higher when compared with nar1-driven gene expression (Suppl. Fig. S1). To account for expression changes caused by the medium shift, we considered changes only as relevant when the expression for a particular gene was altered significantly in both AB31 and AB33 in at least one time point (change in expression ≥2, adjusted p-value ≤0.01). Using these criteria, 206 genes were induced and 139 were repressed in response to b-induction (Fig. 1; Suppl. Table S1). Within this list, all genes with a significant b-dependent regulation identified in previous studies were present, emphasizing the validity of the global approach and the quality of our data set (Suppl. Table S2).
Fig. 1. b-dependent gene expression. 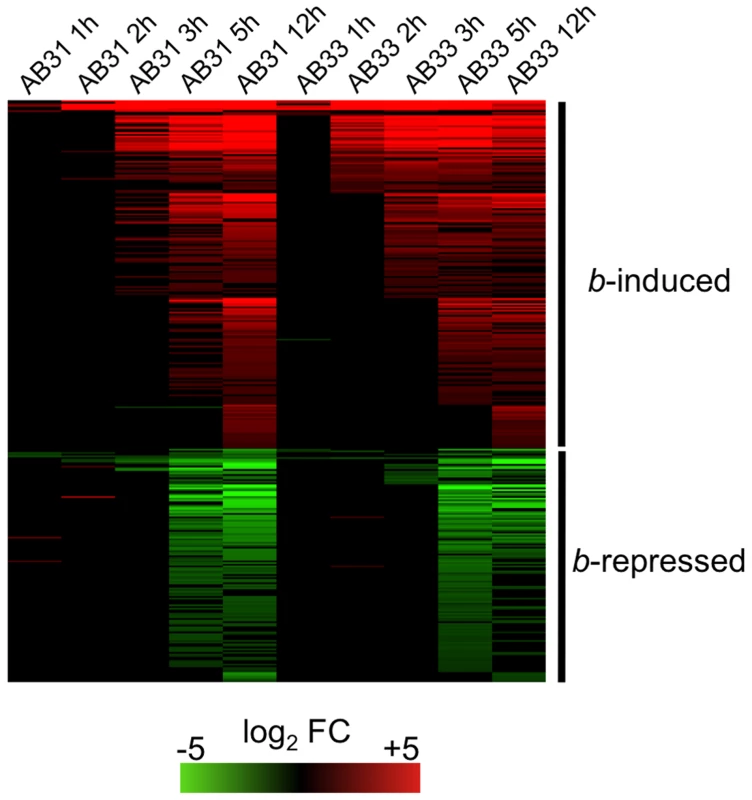
In U. maydis strains AB31 and AB33, expression of the bE1 and bW2 genes is controlled via the arabinose-inducible crg1-promoter and the nitrate-inducible nar1-promoter, respectively. Strains AB32 and AB34 are isogenic to AB31 and AB33, respectively, but harbor the incompatible bE2 and bW2 genes as control. bE/bW genes were induced by shifting the cells to array medium containing arabinose (AB31 and AB32) or nitrate (AB33 and AB34). Cells were harvested at the given time points, RNA was extracted and used for microarry-analysis (Affymetrix MPIUstilagoA gene chip). The heat-map indicates the significant changes (adjusted P<0.01; change >2) in gene expression for AB31 and AB33 at a given time point relative to the control strains AB32 and AB34. Order of genes from top to bottom and expression changes are given in Suppl. Table S1. Details for the statistics and filters applied are given in “Material and Methods”. From the 345 b-regulated genes, a total of 239 were functionally classified using the Blast2Go tool [12]. Using enrichment analysis, we did not observe a significant over-representation of b-induced genes in any of the Gene Ontology (GO) categories (http://www.geneontology.org). However, for the b-down-regulated genes, we observed a significant enrichment of the GO categories “Cell Cycle” (GO:0007049; 29 genes), “Chromosome” (GO:0005694; 25 genes) and “DNA metabolic process” (GO:0006259; 19 genes), “Cytoskeleton” (GO0005856; 16 genes) and “Microtuble cytoskeleton” (GO:0015630; 9 genes) (Suppl. Table S3).
The induction of the active bE1/bW2-heterodimer leads to a G2 cell cycle arrest, and in accordance with this observation we found cln1, clb1 and clb2, which encode a G1-type cyclin and two B-type cyclins [13], [14] among the down-regulated genes (−29.9-fold, −7.7-fold and −2.6-fold, respectively; Suppl. Table S1). cln1 and clb1 are involved in G1 to S transition, clb1 and clb2 in the G2 to M transition; thus, it is expected that these genes are poorly expressed in cells that are arrested in G2. For clb1 it has been shown previously that the repression leads to a G2 cell cycle arrest [14]; thus, the observed low expression of this cyclin may trigger the b-induced G2 arrest. Additionally, we find um03928, encoding a homologue of the S. pombe Cdr2 protein, as 40.7 fold down-regulated. In S. pombe, Cdr2 functions as a mitotic inducer via the Wee1 kinase and is required for G2/M transition [15]. In U. maydis, Wee1 has been shown to be a central regulator for G2/M transition [16]; however, a function for the Cdr2 homologue um03928 has not been assigned yet. Another level of complexity may be achieved via the up-regulation of the Cdk5 associated Pho80 Cyclin Like protein Pcl12 (49.1-fold, Suppl. Table S1). Induced expression of pcl12 leads to a G2 cell cycle arrest, and promotes filamentous growth [C. Pothiratana and J. Kämper, unpublished; 9]. Thus, the b-induced cell cycle arrest may be realized via the synchronized regulation of independent pathways. In line with the cell cycle arrest, we observe the repression of genes involved in DNA-replication and nucleotide metabolism, as, for example, um01008, encoding the catalytic subunit of DNA polymerase epsilon (3.6-fold down-regulated at 12h), or um06402, encoding a DNA replication licensing factor (3,2-fold down-regulated at 12 h; Suppl. Table S1, FunCat DNA).
Several of the b-regulated genes can be attributed to the morphological switch from budding - to filamentous growth. A total of 20 genes with a potential function in cell wall synthesis or modification was found to be induced, starting 3 h after b-induction, which coincides with the onset of filamentation; five additional genes were repressed (Suppl. Table S1, FunCat: CW). These genes encode for chitin synthases as well as for exochitinases, chitin deacetylases, and exo - and endoglucanases, indicating that the cell wall composition is altered during the switch from sporidia to hyphae.
The largest “functional” group (74 genes) encodes for potentially secreted proteins. 34 of them have no ascribed function, and of these 15 are specific for U. maydis. Such secreted proteins are candidates for effectors that may play a role in the establishment of the biotrophic interaction (Suppl. Table S1, secreted).
The bE/bW heterodimer regulates the expression of transcription factors required for pathogenic development
To identify b-dependent genes important for pathogenic development, we focused initially on genes whose expression was “strictly” dependent on the presence of the bE/bW heterodimer, i.e. genes that showed only basal expression levels in strains AB32 and AB34 and showed a more than 10-fold induction upon expression of an active bE1/bW2-heterodimer. None of the 53 genes that fulfilled these criteria showed a significant similarity to known pathogenicity factors. Potential exceptions were dik6 and dkh6, which encode two related seven trans-membrane (7TM) domain proteins. 7TM proteins represent an extended protein family in M. grisea that is discussed to function in plant/pathogen interactions [17]. However, neither the single, nor the double deletion of the two genes had an impact on pathogenic development (Suppl. Table S1, G. Weinzierl and J. Kämper, unpublished). In total, we deleted 30 of the 53 strictly b-dependent genes in the haploid, solopathogenic strain SG200; in addition, nine genes have been analyzed in the course of previous studies. 35 of the 39 deletion strains did not show altered virulence when assayed in plant infection assays.
However, the individual deletion of each of the five genes encoding proteins with potential regulatory functions affected pathogenic development or filamentous growth (Suppl. Table S1). Among these genes was clp1 (um02438), which has been identified in the course of this study and has been shown to be required for pathogenic development and in planta proliferation [8]. The biz1 gene (um02549) encodes a C2H2 zinc finger transcription factor that is required for pathogenic development and efficient appressoria formation [6]. In addition, we could show that the deletion of two genes encoding potential homeodomain transcription factors (um12024 and um04928, termed hdp1 and hdp2) impaired filamentous growth or led to loss of pathogenicity, respectively; the detailed characterization of these two genes will be published elsewhere. Here we will focus on the analysis of um03172, encoding a potential C2H2 zinc finger transcription factor.
The b-dependently induced rbf1 gene encodes a zinc-finger transcription factor
Due to the initially observed phenotype (see below), the U. maydis gene um03172 was termed rbf1 (regulator of b-filament). According to our microarray analysis, rbf1 expression was strongly induced early after b-induction (Fig. 2A). Significant expression was detected already 1h after b-induction in AB33 (13.6-fold induction), and expression peaked at 2h to 3 h (176.3-fold in AB33 at 2h and 297.4-fold in AB31 at 3 h, respectively; Fig. 2A). In the control strains AB32 and AB34 rbf1 expression was not detectable. The b-dependent expression of rbf1 was confirmed by qRT-PCR using strains AB31 and AB32 (Fig. 2B). Within the rbf1 promoter, we identified three motifs with similarities to the previously identified b-binding sequences (bbs) (Fig. 2C). We used an AB31 derivative expressing the bE1 protein fused to a triple HA-tag (AB31bE1 : 3xHA) for quantitative chromatin immunoprecipitation analysis (qChIP). Induction of bE1 : 3xHA/bW2 genes in this strain led to filamentous growth (see below), demonstrating that the bE1 : 3HA protein is functional (data not shown). In a qChIP analysis with bE1 : 3xHA, a significant enrichment (P = 5.7 10−5, Students t-test) was observed for the PCR amplicon covering the bbs-motif located at position −1377, when compared to a amplicon covering a region further upstream in the rbf1 promoter (Fig. 2D). The bbs−1377-motif shares also the highest sequence similarity with the previously described bbs-motifs (Fig. 2C). When the rbf1 gene with a promoter fragment deleted for bbs−1377 was used for transformation of a strain deleted for rbf1, the rbf1 deletion phenotype could not be complemented (Fig. 3D, Table 1, see below), demonstrating that bbs−1377 is required for expression of rbf1. The early induction of rbf1 upon b-activation, the presence of a conserved bbs-motif which is bound by the bE/bW heterodimer in vivo, and the requirement of this bbs-motif for rbf1-function strongly suggest that rbf1 is a direct target of the bE/bW heterodimer.
Fig. 2. Structure and expression of Rbf1. 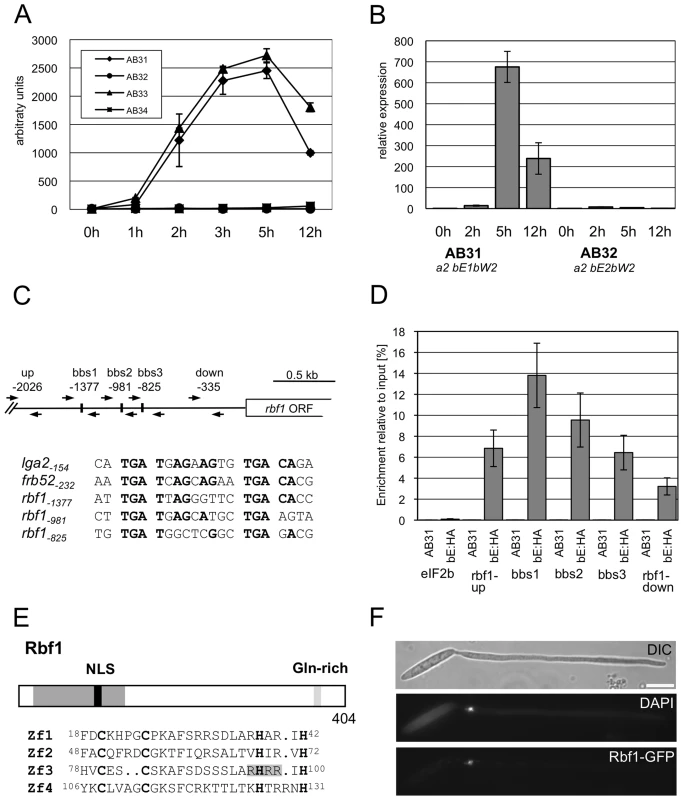
(A) Microarray analysis of b-dependent rbf1 expression after induction of compatible (AB31 and AB33) and incompatible (AB32 and AB34) combinations of bE and bW. Shown are the mean expression values of two biological replicates and the standard deviation (SD) (B) qRT-PCR analysis of rbf1 expression after induction of compatible (AB31) and incompatible (AB32) combinations of bE and bW. Samples were taken at the time-points indicated. qRT-PCR analysis was performed using the constitutively expressed ppi gene (um03726) for normalization. Expression was calculated relative to the lowest expression value. Shown are mean values of two technical replicates. (C) Overview of primer binding sites in the rbf1-promoter used for qChIP experiments and alignment of three putative b-binding sites (bbs) in the rbf1 promoter region to the bbs of lga2 and frb52 [10], [11]. Nucleotide positions indicated are relative to the start codon. Nucleotides identical to the bbs in lga2 [10] and frb52 [11] are in bold. (D) qChIP analysis of bE1 binding to the rbf1-promoter in strains AB31 and AB31bE1:3xHA 5h after induction of the bE1/bW2-heterodimer. AB31bE1:3xHA harbours a HA-tagged bE1 protein used for immunoprecipitation with anti-HA-antibody. Numbers give the enrichment in % of the input-DNA of the PCR amplicons in DNA co-immunoprecipitated with HA-antibody. No significant enrichment was observed in control strain AB31. In AB31bE1:3xHA, the PCR-amplicon spanning bbs1 (bbs−1377) is significantly enriched (t-test) when compared to the amplicon spanning a control region (−2026) (p = 5.71 10−5). As additional control, a region from the eIF2b gene (um04869) was used. Given are the mean values of three technical replicates of three independent experiments each, and the standard deviation (SD). (E) Structure of the Rbf1 protein. The potential C2H2 zinc finger domain (aa 18 to 131) and a putative NLS (RHRR) (aa 95 to 98) within this domain are marked in dark grey and black, a glutamine rich sequence (aa 365 to 373) is marked in grey. The alignment shows the four C2H2 zinc finger domains; the conserved cysteine and histidine residues are in bold. (F) Subcellular localization of the Rbf1-3xeGFP fusion protein. Strain AB31rbf1:3eGFP (UMS63) was induced in CM medium supplemented with 1% arabinose (CMA) for eight hours. The functional Rbf1-3xeGFP fusion protein localizes to the nucleus. Cells were stained with DAPI to visualize nuclei. Scale bar corresponds to 10 µm. Fig. 3. Rbf1 is required for b-dependent filament formation. 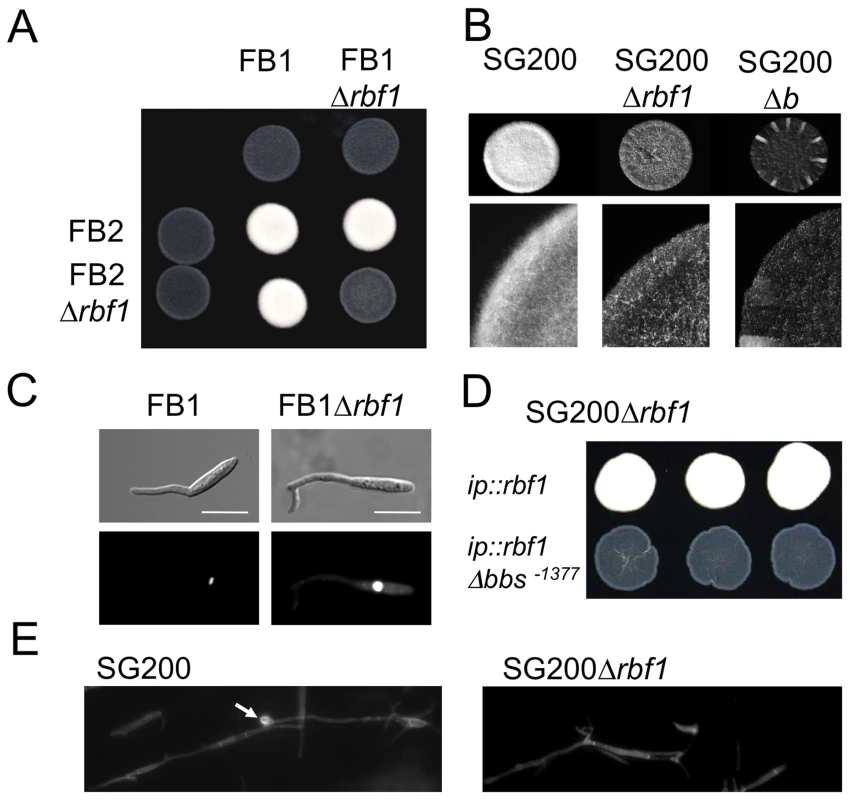
(A) Mixtures of wild-type U. maydis strains FB1 (a1b1) and FB2 (a2b2) and of the respective Δrbf1 strains and (B) of the solopathogenic strain SG200 (a1mfa2 bE1bW2) and its Δrbf1 and Δb derivatives were spotted on PD charcoal plates to induce filament formation. Filamentation is drastically reduced in all Δrbf1-strains. (C) FB1 and FB1Δrbf1 cells grown in CM medium supplemented with 1% glucose (CMD) (OD600 = 0.2) were treated with synthetic a2 pheromone (2.5 µg/ml) for six hours; nuclei were visualized by DAPI staining. Rbf1 is not required for the pheromone-induced conjugation tube formation and cell cycle arrest. Scale bar = 10 µm. (D) Transformants of SG200Δrbf1 obtained by integration into the ip-locus of (1) the rbf1 open reading frame with the wild-type 3 kb 5′-upstream region (ip::rbf1), or (2) the rbf1 open reading frame with a 3 kb 5′-fragment harbouring a deletion of the bbs-motif at position −1377 (ip::rbf1Δbbs−1377). Shown are three independent transformants each spotted on PD charcoal plates. While the rbf1 gene with the wt-promoter fragment is capable to complement the rbf1 deletion, restoring filamentous growth, the deletion of bbs−1377 renders the transforming fragment inactive, indicating that binding of the bE1/bW2-heterodimer to bbs−1377 is required for rbf1 expression. (E) Calcofluor staining of fungal hyphae on the leaf surface. 16 hours post inoculation, both U. maydis SG200 and SG200Δrbf1 have formed filaments; no appressoria were found in SG200Δrbf1. The arrow marks the appressorium. Tab. 1. Pathogenicity of rbf1 deletion strains. 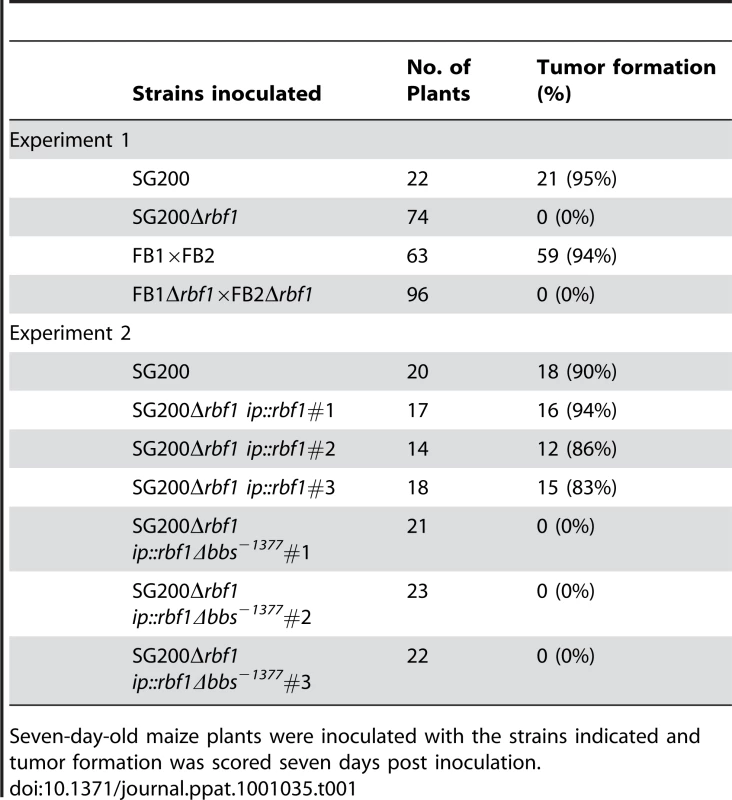
Seven-day-old maize plants were inoculated with the strains indicated and tumor formation was scored seven days post inoculation. The cDNA-copy of rbf1 was obtained by RACE and revealed four introns when compared to the genomic locus. The predicted open reading frame encodes a protein of 404 amino acids (aa) with an N-terminal C2H2 zinc finger domain (aa 18 to 131), a putative nuclear localization sequence (RHRR, aa 95 to 98) within the zinc finger domain and a C-terminal glutamine-rich sequence (aa 365 to 373) (Fig. 2E). To determine the localization of Rbf1, the open reading frame was fused to a triple eGFP gene and integrated into strain AB31 via homologous recombination, thereby replacing the native rbf1 gene. Fluorescence microscopy of the resulting strain AB31rbf1 : 3eGFP (UMS63) revealed a nuclear localization of the functional Rbf1-3xGFP fusion protein upon induction of the bE/bW heterodimer (Fig. 2F), fostering the assumption that rbf1 encodes a C2H2 zinc finger transcription factor.
Rbf1 is required for b-dependent filament formation and pathogenicity
To investigate the biological function of rbf1, the gene was deleted in the haploid solopathogenic strain SG200 (a1mfa2bE1bW2) and in the haploid U. maydis wild-type strains FB1 (a1b1) and FB2 (a2b2), producing strains SG200Δrbf1 (UMS20), FB1Δrbf1 (UMS49) and FB2Δrbf1 (UMS51), respectively. In all strains, the deletion of rbf1 did not cause any obvious phenotype in haploid sporidia growing in axenic culture, and the growth rate was not altered in different minimal or complete media (data not shown). However, when the compatible strains FB1Δrbf1 and FB2Δrbf1 were crossed on charcoal containing plates, only very short filaments were observed at the edge of the forming colonies, while the cross of FB1 or FB2 resulted in fuzzy white colonies indicative for the formation of the filamentous dikaryon (Fig. 3A). Similarly, only scarce filament formation was observed in SG200Δrbf1 (Fig. 3B). Since SG200 cells undergo the dimorphic switch without the need of a mating partner on charcoal containing media, we can exclude that the drastically reduced filamentation is caused by a defect in cell-cell fusion. Treatment of FB1Δrbf1 cells with synthetic a2 pheromone resulted in the formation of conjugation tubes which were indistinguishable from those produced by wild-type FB1 cells, indicating that deletion of rbf1 does not affect polarized growth per se (Fig. 3C). Transformation of SG200Δrbf1 with plasmid pRbf1 harboring the rbf1 gene and 3kb of 5′sequence restored the fuzzy colony appearance; three independent transformants (SG200Δrbf1 ip::rbf1) were indistinguishable from the SG200 wild type strain (Fig. 3 D). However, the rbf1 deletion phenotype was not complemented when plasmid pRbf1Δbbs−1377, in which the bbs-motif at position −1377 in the rbf1 promoter was deleted, was used for transformation (SG200Δrbf1 ip::rbf1Δbbs−1377) (Fig. 3 D).
To assess the role of rbf1 during pathogenic development, seven days old maize plants were inoculated with SG200Δrbf1, or with a mixture of FB1Δrbf1 and FB2Δrbf1, and scored for tumor formation. Seven days post inoculation (dpi) 95% and 94% of the plants inoculated with SG200 and a mixture of FB1 and FB2, respectively, had developed tumors. In contrast, inoculation with the respective Δrbf1 mutants resulted in the complete absence of infection symptoms (Table 1). As expected, transformation of SG200Δrbf1 with pRbf1 (SG200Δrbf1 ip::rbf1) restored pathogenicity, while transformation with pRbf1Δbbs−1377 (SG200Δrbf1 ip::rbf1Δbbs−1377) did not (Table 1). To determine at which stage of pathogenic development the rbf1 mutant strains were blocked, fungal hyphae were stained with calcofluor at 2 dpi. Microscopic observation revealed that the Δrbf1-strains formed filaments on the leaf surface (Fig. 3E), however, we did not observe any hyphae within plant cells. To assess whether SG200Δrbf1 was able to form appressoria, we co-inoculated plants with a mixture of SG200 and SG200Δrbf1 strains, each expressing either cytoplasmatically localized CFP or YFP to distinguish the strains. In the combinations SG200-CFP/SG200Δrbf1-YFP and SG200-YFP/SG200Δrbf1-CFP, we counted 57 and 60 appressoria for the SG200 strains on the leaf surface, respectively. By contrast, we were unable to detect any appressoria formation for the SG200Δrbf1 strains in the same surface areas. Thus, the observed pathogenicity defect of Δrbf1 strains results from the inability to form appressoria and to penetrate the plant cuticle.
Rbf1 is required and sufficient to induce filamentous growth and a G2 cell-cycle arrest
To get a more detailed view on the role of rbf1 during b-dependent filament formation, we deleted the gene in strain AB31. More than 90% of the cells had switched to filamentous growth 12h after b-gene induction in AB31, while in AB31Δrbf1 (UMS25) no filament formation was observed (Fig. 4A). Upon induction of bE1/bW2 in AB31 the cells stop to divide; in contrast, in AB31Δrbf1 cells continued to grow by budding (Fig. 4A and 4C), indicating that rbf1 is required for both filamentous growth as well as for the b-dependent cell cycle arrest. FACS analysis of AB31 cells revealed an accumulation of cells containing a 2C DNA content upon b-induction, indicative for the b-induced G2-cell cycle arrest. In AB31Δrbf1, however, the distribution of cells with 1C and 2C DNA content was comparable to the wild-type strain FB2, corroborating the requirement of rbf1 for the b-induced cell cycle arrest (Fig. 4B).
Fig. 4. rbf1 is required and sufficient for b-dependent filamentation and G2 cell cycle arrest. 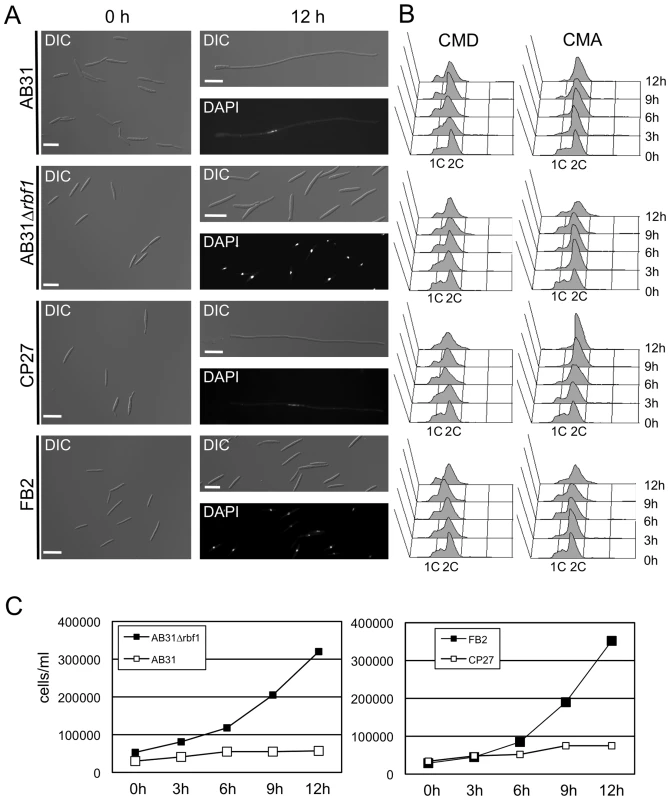
(A) Microscopic analysis of AB31, AB31Δrbf1, CP27 (FB2Δb::Pcrg1:rbf1) and FB2 after induction of the active bE1/bW2-heterodimer or Rbf1, respectively. Filament formation was observed 12 hours after b-induction in strain AB31 and rbf1-induction in strain CP27 (FB2Δb::Pcrg1:rbf1). In contrast, strain AB31Δrbf1 did not switch to filamentous growth and continued to grow by budding, similar to the FB2 strain that was used as negative control. Scale bar = 10 µm. (B) FACS analysis of AB31, AB31Δrbf1, CP27 (FB2Δb::Pcrg1:rbf1) and FB2 under non inducing conditions (CMD) or inducing conditions (CMA). Induction of the active bE1/bW2 combination in AB31 or induction of rbf1 in strain CP27 resulted in an enrichment of cells with a 2C content, indicative of a G2 cell cycle arrest. In AB31Δrbf1 no enrichment of cells with 2C was observed, similar to control strain FB2. (C) Measurement of cell numbers/ml in strains AB31, AB31Δrbf1, CP27 (FB2Δb::Pcrg1:rbf1) and FB2 grown in CMA for the time indicated. Strains AB31Δrbf1 and FB2 show a typical exponential growth curve, whereas strains AB31 and CP27 stopped to grow after induction of the bE1/bW2-heterodimer or rbf1, respectively, indicating a cell cycle arrest. To dissect b-dependent and rbf1-dependent processes, we constructed strain CP27 (a2Δb::Pcrg1:rbf1), an FB2 derivative in which the b-locus was replaced by a copy of rbf1 under control of the arabinose-inducible crg1 promoter. Induction of rbf1 in CP27 resulted in the formation of filamentous cells that were indistinguishable from b-induced filaments: the cells contained single nuclei (Fig. 4A) and stopped to divide (Fig. 4C). FACS analysis revealed that rbf1 induction in CP27 leads to a G2 cell cycle arrest (Fig. 4B) analogous to that observed after b-induction. In summary, our results demonstrate that rbf1 is required for b-dependent filament formation and G2 cell cycle arrest and, in addition, sufficient to induce these developmental steps in the absence of an active bE/bW-heterodimer.
Rbf1 is a master regulator for b-dependent processes
To analyze the connection between b - and rbf1-mediated gene-regulation in more detail, we performed DNA-array analysis. b-dependent genes for which rbf1 is required for expression were identified by comparing the transcriptional profile of strains AB31Δrbf1 and AB31 at 3h, 5h and 12h after b-induction. Induced expression (5h) of rbf1 in strain CP27 (a2Δb::Pcrg1:rbf1) was used to identify genes for which rbf1 is sufficient for regulation. 189 (91.7%) out of the 206 previously identified b-induced genes showed no significant changes in expression after b-induction in strain AB31Δrbf1 (Fig. 5; Suppl. Table S4). From the remaining 17 genes, 11 showed comparable expression levels after b-induction in AB31 and AB31Δrbf1, and 6 genes showed significant, but reduced expression levels in AB31Δrbf1. The 11 genes that showed no altered b-dependent expression upon rbf1 deletion did not respond to rbf1 induction in strain CP27; we consider these genes to be regulated only by b, and not by Rbf1 (“only b”, Fig. 5, Suppl. Table S4). With the exception of um00027, all these genes harbor sequence motifs with similarities to the b-binding site within their promoter sequences. In addition, they are all up-regulated early upon b-induction, indicating that these genes are most likely direct targets of the bE/bW heterodimer. The six genes that show a significant, but reduced b-responsive expression in AB31Δrbf1 all respond to rbf1 induction in CP27. Four of the genes harbor b-binding sites in their promoter regions; apparently, these genes may be regulated directly via b and, in addition, independently via rbf1 (“rbf1 OR b sufficient”, Fig. 5, Suppl. Table S4).
Fig. 5. Rbf1 is required and sufficient for expression of b-dependent genes. 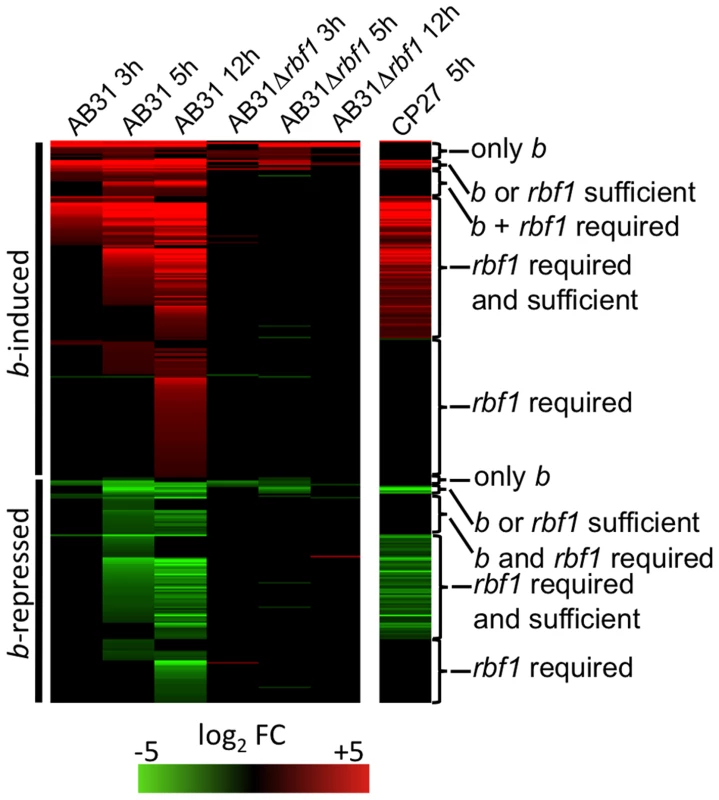
The bE1 and bW2 genes were induced in strains AB31 or AB31Δrbf1; as a control for both strains, the incompatible bE2 and bW2 genes were induced in AB32. Strain CP27 (a2 Δb:: Pcrg1:rbf1) was used for induced expression of rbf1, with strain JB2 (a2Δb) as control. Induction was performed by shifting the cells from array medium containing 1% glucose to array medium containing 1% arabinose. Cells were harvested at the given time points, RNA was extracted and used for microarry-analysis (Affymetrix MPIUstilagoA gene chip). The heat-map indicates the significant changes (adjusted P<0.01; change >2) in gene expression for all b-regulated genes depicted in Fig. 1 at the given time points relative to the control strains. Most genes do not show altered expression values in response to b-induction in AB31Δrbf1, indicating that rbf1 is required for the expression of the majority of b-dependent genes. In strain CP27, the induced expression of rbf1 causes the induction or repression of a large group of the b-responsive genes, indicating that Rbf1 is sufficient for the regulation of these genes. Order of genes and expression changes are given in Suppl. Table S4. Details for the statistics and filters applied are given in “Material and Methods”. For a large fraction (46%) of the b-dependent genes rbf1 is both sufficient as well as required for expression; for these genes, deletion of rbf1 abolishes the b-dependent induction, and they respond to rbf1-induction in CP27. It is likely that the regulation of these genes occurs by a b-mediated regulatory cascade via Rbf1 as a central regulator (“rbf1 required AND sufficient”, Fig. 5, Suppl. Table S4). Expression of the remaining 102 genes was dependent on rbf1, however, no significant induction was detected 5h after rbf1 induction in CP27. Notably, 63 of these genes were late b-induced (12h after b-induction in AB31), and additional 22 genes were only weakly induced (less than 3-fold), or only transiently induced 5h after b-induction in AB31. It is well possible that these genes respond to rbf1 only after prolonged rbf1 induction (>5h). For 16 genes, we observed a significant b-dependent induction, no induction in AB31Δrbf1, and no rbf1-dependent induction in CP27. Thus, we have to assume that for the regulation of these genes the action of both b and rbf1 is required (“rbf1 AND b required”, Fig. 5, Suppl. Table S4).
An analogous scenario was found for the b-dependently repressed genes: of the 139 b-dependently repressed genes, the repression was abrogated for 129 (92.8%) genes in AB31Δrbf1. For a total of 69 genes rbf1 was both required and sufficient for repression. Formally, the b-repressed genes can be grouped equivalent to the b-induced genes (b only; rbf1 AND b; rbf1 OR b; only rbf1; Fig. 5, Suppl. Table S4).
To assess whether the rbf1-dependent gene expression involves the binding of Rbf1, we dissected the promoter of dik6, one of the rbf1 responsive genes, by means of qChIP analysis. We used an AB31 derivative where the rbf1 gene was replaced by a rbf1-3xHA fusion (AB31rbf1 : 3xHA). Induction of bE1/bW2 in this strain triggers the expression of the Rbf1-3xHA fusion protein, which results in filamentous growth, demonstrating that the Rbf1-3xHA fusion protein is functional (Data not shown). qChIP analysis was performed via a set of 9 overlapping amplicons spanning 930 bp of the dik6 promoter (Fig. 6A); as controls, we used an amplicon upstream of the potential promoter (−1703 to −1829 with respect to the ATG) and an amplicon within the dik6 ORF (Fig. 6A). With the exception of an amplicon spanning the region from −9 to −157, all amplicons within the promoter showed significant (P<0.001) differences in enrichment when compared to the control amplicon located within the ORF. The amplicons with the highest enrichment were found to span the region from −825 to −422 (Fig. 6B) The functional analysis of the dik6 promoter by means of promoter-GFP fusions revealed that Rbf1-induced GFP expression levels declined when the dik6 promoter was truncated from 816 to 638 bp, while a 298 bp fragment was not sufficient to mediate expression (Fig. 6A,C). Internal promoter deletions corresponding to the amplicons used for the qChip analysis revealed that deletion of the dik6 promoter region from −825 to −680 (corresponding to amplicon 3) led to reduced Rbf1-dependent induction, while the deletion of the promoter region from −601 to −500 (corresponding to amplicon 5) abolished expression completely (Fig. 6A,C). In summary, our data indicate that the dik6 promoter has at least one binding site for Rbf1 that is required for Rbf1-mediated dik6-expression.
Fig. 6. Rbf1 binds to the promoter of the Rbf1-dependently expressed dik6 gene. 
(A) Overview of the dik6 promoter region investigated by qChIP in strain AB31 rbf1:3xHA. Shown are the positions of the amplicons used for qChIP (numbered from 1 to 10) and the promoter truncations and internal deletions assayed in the GFP-reporter assay. (B) qChIP analysis of Rbf1-binding to the dik6-promoter in strains AB31 and AB31rbf1:3xHA 5h after induction of the bE1/bW2-heterodimer in CMA. AB31rbf1:3xHA expresses an HA-tagged Rbf1 protein used for immunoprecipitation with anti-HA-antibody. Numbers give the enrichment in % of the input-DNA of the PCR amplicons in DNA co-immunoprecipitated with HA-antibody Start and end of the amplicons are given in nucleotides (nt) relative to the start codon of dik6. The relative positions of the amplicons are given in (A). As additional control for qChIP, a region from the eIF2b gene (um04869) was used. Given are the mean values of two technical replicates of three independent experiments each, and the standard deviation (SD). Significance of the difference to values obtained for the control-amplicon located within the dik6 open reading frame (amplicon 11) was calculated using Students t-test; the respective p-values are given for AB31rbf1:3xHA. Highest enrichment was observed for amplicons 3, 4, 5 and 6. No significant enrichment was observed in control strain AB31. (C) The dik6 promoter fragments outlined in (A) were fused to GFP as a reporter and integrated in single copy into the ip-locus of U. maydis strain CP27 (a2 Δb::Pcrg1:rbf1). GFP-expression was visualized microscopically 5 hours after induction of rbf1 expression in CMA medium. GFP expression declined when the promoter was truncated from 816 bp to 638 bp, and was abolished when a 298 bp promoter fragment was used. Similarly, the internal deletion Δ3 led to reduced GFP expression, while no GFP signal was detectable in the Δ 5 deletion. Scale bar = 20 µm. Previously, it was shown that rbf1 is induced when haploid cells are treated with compatible pheromone [18]. Since both bE as well as bW are also induced upon pheromone treatment [19], we asked whether Rbf1 might be required for pheromone-dependent expression of the b genes. However, real time qRT-PCR analysis revealed no difference in the abundance of bE and bW transcripts in the strains FB1 and FB1Δrbf1 (UMS49; a1b1Δrbf1) upon treatment (75 min) with synthetic a2 pheromone (see Suppl. Fig. S2). Thus, we can exclude the possibility that Rbf1 is required for the pheromone-dependent b-induction.
In summary, our data identify Rbf1 as the central regulatory switch within the b-dependent regulatory cascade, which is not only required for the regulation of the majority of the b-dependent genes, but also indispensable for all b-mediated developmental processes.
Discussion
The switch from saprophytic to biotrophic growth of U. maydis requires a meticulous coordination of different processes, such as cell cycle control, the change to polarized growth, and, most interestingly, the onset of a program facilitating plant invasion and colonization. The top-most control instance for these processes is the b-mating type locus; it has been conclusively shown that compatible b-alleles are both required and sufficient for pathogenic development [5]. The bE/bW-heterodimer also controls polarized cell growth and induces a G2 cell cycle arrest, but not exclusively, since both can be triggered as well via the pheromone/receptor system encoded by the a-mating type locus [20]. Necessarily, the a and b loci are cross-controlled: activation of the a-pathway leads to induction of the bE and bW genes via Prf1, and the formation of an active bE/bW-heterodimer after cell fusion leads to a down-regulation of the a-pathway [19], [21].
Since a direct binding of the bE/bW heterodimer to promoters of the plethora of genes associated with b-dependent processes appeared unlikely, we have favored a model that places b on top of regulatory proteins (relays) mediating the regulation of further downstream targets. We have now identified the C2H2 zinc finger transcription factor Rbf1 as a central key player within this regulatory network.
The fast induction of rbf1 upon b-activation, the binding of the bE/bW-heterodimer to a defined b-binding site in the rbf1-promoter region as well as the requirement of this binding site for rbf1 function define rbf1 as a direct target of the bE/bW-heterodimer. Deletion of rbf1 abolishes all b-mediated processes, and induction of rbf1 leads to filamentation and a G2 cell cycle arrest analogous to that observed upon b-induction. In addition, we could show that rbf1 is required for regulation of the far majority of b-regulated genes, and, for a large fraction, also sufficient. Thus, we consider Rbf1 as a key master regulator whose action is sufficient to induce an entire complex developmental pathway. Despite of the essential function of Rbf1 within the b-regulatory cascade, we consider it unlikely that rbf1 alone is sufficient to trigger pathogenic development of U. maydis, because clp1, which was shown to be required for pathogenicity [8], is induced directly by b and independently from rbf1. We were not able to address this question experimentally, since transformants with a constitutively expressed rbf1 gene were not viable, most probably as a result of the rbf1 induced cell cycle arrest.
In fungi, only few master regulators of pathogenic development have been identified yet. In Candida albicans, WOR1 is the master regulator of white to opaque switching [22], and the C. neoformans Gat201 [23] is a key regulator of melanin production and capsule formation. The C. neoformans Cir1 transcriptional regulator integrates the sensing of iron with the expression of virulence factors, with signalling pathways influencing virulence, and with growth at elevated temperature [24], [25]. WOR1 and Gat201 are required (and sufficient) for the initiation of specific programs that are tightly linked to fungal pathogenesis. In contrast, nearly all of the genes regulated by b require rbf1 for their expression, and it is not possible to assign specific, common functions to the rbf1-regulated genes, or to the few genes that are not regulated by Rbf1. Thus, different from WOR1 and Gat201, Rbf1 regulates not the genes of a specific, defined pathway, but is required for the regulation of all b-dependent processes. Similarly, the C. neoformans Cir1 regulator is involved in the regulation of all major virulence traits [24], [25].
The cell cycle block of the b-induced filaments is only released upon plant penetration. Our data reveal a complex contribution of different key players to control the cell cycle. At least four different transcription factors, namely bE/bW, Rbf1, and the two Rbf1-dependent factors Biz1 and Hdp1 are involved in cell cycle regulation. The ectopic expression of any of these factors leads to the formation of G2 arrested hyphae [6], [8, C. Pothiratana and J. Kämper, unpublished], which argues for a complex transcriptional network with different levels of relays that allow the integration of various stimuli, as for example, the unknown signal that leads to the release of the cell cycle after penetration of the host plant. The regulatory control achieved via b, Rbf1, Biz1 and Hdp1 may funnel into the transcriptional regulation of different key factors for cell cycle control, as we observe the transcriptional regulation of different cyclins (cln1, clb1, clb2 and pcl12) and of the potential Wee1 kinase Um03928. The Um03928 homologue in S. pombe, Cdr2, is required for the proper formation of septae, and functions as mitotic inducer via the negative regulation of the central cell cycle regulator Wee1 [15], [26]; the U. maydis Wee1 was shown to trigger filamentous growth and a G2 arrest [9], [16]. Obviously, the b-induced G2 cell cycle arrest is controlled by several independent regulatory pathways.
The induction of b leads to the formation of polar growing hyphae, and several of the b-dependently regulated genes reflect this morphological change and the altered requirements of the cell for e.g. long distance transport or cell wall remodeling. However, the most interesting trait by which the b-induced filament differs from other filaments like the pheromone-induced conjugation tube is its ability to infect the host. The exploitation of the b-dependently regulated genes provides for the first time comprehensive insights into the complex developmental processes during morphogenic switching and pathogenic development of U. maydis. The pathogenic potential of the hyphae may for once be marked by an altered cell wall composition, as we observe the differential regulation of several genes involved in cell wall synthesis, including chitin synthases and chitin deacetylases. Rebuilding or masking of the cell wall is a strategy of pathogens to evade perception or to protect themselves from defense responses of the host [27]. However, deletion of either of the two b-regulated chitin deacetyases does not affect virulence in U. maydis (B. Günther, J. Kämper, B. Moerschbacher, unpublished), and neither are the two chitin synthases chs1 and chs4 required for pathogenicity [28], most likely due to overlapping and/or redundant functions.
The other intriguing characteristic of the b-filament is the secretion of various potential effector proteins. Such effectors are thought to be involved in suppression of host defense responses and redirection of nutrient flow during biotrophic growth. The expression of putative effectors prior to the contact with the plant indicates a priming mechanism of the fungal hypha to facilitate rapid suppression of plant defense responses and the fast establishment of the biotrophic interface subsequent to plant penetration. The observation that the temperature-induced inactivation of the bE1/bW2-heterodimer in planta abolishes expression of various additional candidate effector genes [29] that are not identified as b-regulated in this study, implies that the temporal expression of these genes is subject to combinatorial gene regulation involving the bE/bW-heterodimer and other plant-induced factors.
The competence of the b-filaments to penetrate the plant cuticula is reflected by the induction of Biz1 and the MAP kinase Kpp6. Both factors have been shown to be required for efficient formation of appressoria and subsequent penetration. Rbf1 is required for the b-dependent induction of both genes, which explains the absence of appressoria in rbf1 mutant strains.
Rbf1 is required for the induction of most, but not all b-regulated genes. All genes that are exclusively regulated by bE/bW harbor b-binding sites within their promoters, and it is conceivable that these genes are regulated via direct binding of the bE/bW-heterodimer. The majority of the b-regulated genes, however, lack putative b-binding sites, indicating that the b-dependent regulatory circuit involves additional transcription factors.
Similarly, those genes for which rbf1 is required and sufficient for regulation may be directly regulated by Rbf1. Our data indicate that Rbf1 binding to the promoter of the dik6 gene is required for Rbf1-mediated dik6 expression, which emphasizes the function of Rbf1 as a transcription factor. However, the actual Rbf1 binding site has not been determined yet. The in silico analysis of rbf1-regulated genes may be constrained by the fact that Rbf1 triggers the induction of at least three transcription factors, leading to a superimposition of direct and indirect effects. For a small fraction of genes, both b and rbf1 are required for regulation, which can be explained by a combinatorial action of two transcription factors [30]. A substantial number of genes is down-regulated upon b-induction. Intriguingly, two of the very few known transcription factors that can act both as transcriptional activators and repressors, the S. cerevisiae Rme1 protein and the human YY1 protein, are both C2H2 zinc-finger proteins [31], [32]. Thus, it is well possible that the repression of genes is also directly mediated via Rbf1.
The dimorphic switch and the onset of pathogenic development trigger a multilayered regulatory cascade that involves several transcription factors (Fig. 7). Is there a specific reason that the bE/bW-heterodimer regulates only a small number of genes directly and more than 90% in dependence on a second master regulator? For once, additional regulators allow more signals to be integrated into the regulatory circuits, which may help to quickly adapt to changing environmental conditions during biotrophic development, thereby avoiding nutrient stress or plant defense responses. In particular, Rbf1 interconnects the a - and b-dependent regulatory pathways, as both pheromone-response [18] as well as b-induction leads to rbf1 expression. Since we could not determine a specific function for Rbf1 in the pheromone-dependent signalling pathway and deletion of rbf1 is not required for conjugation tube formation and for pheromone-induced G2 cell cycle arrest, we consider it unlikely that rbf1 plays a central role in a-dependent signalling or gene regulation. One possibility is that the pheromone-induced expression of b and rbf1 primes the cells for post-fusion events. The a - and b-dependently induced cell cycle arrest is independently coordinated; thus, the pheromone-induced rbf1 expression facilitates rapid switching of developmental programs thereby minimizing the time preceding plant infection (Fig. 7).
Fig. 7. Key factors of a- and b-dependent development in U. maydis. 
(A) Activation of the pheromone pathway after pheromone (Mfa) binding to the cognate receptor (Pra) triggers a signalling casacde via PKA and MAPK phospho-relays. Differential phosphorylation of the HMG transcription factor Prf1, in turn, leads to expression of the b-mating type locus gene bE and bW. (B) After cell fusion, sexual and pathogenic development is orchestrated by the bE/bW-heterodimer, coordinating the key players of important developmental steps: Clp1 is required for in planta proliferation; Rbf1 is the central transcriptional regulator, controlling appressoria formation and penetration of the host plant via the transcription factor Biz1 and the MAPK Kpp6, as well as filamentous growth and the G2 cell cycle arrest by concerted action of Biz1 and Hdp1. In essence, our study provides fundamental new insights into the complex regulatory traits of sexual, as well as pathogenic development of U. maydis. The identification of key factors points towards an emerging picture that explains how multilayered regulatory pathways can dynamically interact to control complex developmental decisions. We believe that this work is not only relevant for U. maydis but can also serve as a model for other fungi and higher organisms.
Methods
Strains and growth conditions
Escherichia coli strain TOP10 (Invitrogen) was used for cloning purposes. Growth conditions and media for E. coli [33] and U. maydis [8], [34], [35] and the quantification of appressoria formation [6] have been described previously. U. maydis strains with relevance for this study are listed in Suppl. Table S5. U. maydis strains carrying crg1 expression constructs were induced in array medium [8] or CM medium [34] supplemented with 1% arabinose instead of 1% glucose as described in [8]; equivalently, for nar1-induction, array medium supplemented with 3.8g/l KNO3 instead of glutamine (4.38 g/l) as nitrogen source was used. Mating assays and plant infections are described in reference [35]. For pheromone stimulation of U. maydis cells we followed the protocol of [36].
DNA and RNA procedures
Molecular methods followed described protocols [33]. DNA isolation and transformation procedures for U. maydis were carried out as described [37]. For all gene deletions, we used the PCR based approach described in [38]. For the Rbf1-3xeGFP fusion, 1 kb of the 3′end of the rbf1 ORF and 1 kb of the 3′ UTR were PCR-amplified, introducing two SfiI sites and removing the stop-codon of rbf1; both fragments were ligated to an SfiI 3xeGFP-HygR fragment of pUMA647 (K. Zarnack and M. Feldbrügge, unpublished) in pCR2.1 TOPO (Invitrogen) as backbone, yielding pMS85.
To replace the b-mating type locus with the arabinose inducible rbf1 allele, the rbf1-ORF was PCR amplified, creating an NdeI site at the start and a NotI site following the stop codon, and cloned into pCR2.1 TOPO. The NdeI-NotI rbf1-ORF-fragment, a 1.3 kb BstXI(blunt)-NdeI crg1-promotor fragment and a 0.3 kb NotI-EcoRI(blunt) nos-terminator fragment from pRU12 [11] were integrated into the StuI site of pCRΔb [38] to generate pCRΔb-crg:rbf1. For the generation of HA-tagged bE1 - and Rbf1-fusion proteins 1 kb of the 3′end of the ORF and 1 kb of the 3′ UTR were PCR-amplified, introducing two SfiI sites and removing the stop-codon; respective fragments were ligated to an SfiI 3xHA-HygR fragment of pUMA792 (M. Feldbrügge, unpublished) and cloned into pCRII TOPO, yielding plasmids pDS1 and pDS3. After linearization plasmids were integrated into the bE1 and rbf1 loci, respectively, of strain AB31 by homologous recombination.
All PCR amplified fragments were verified by sequencing. For transformation, either linearized plasmid DNA or PCR generated linear DNA was used; homologous integration was verified by Southern blot. For complementation of the rbf1 deletion, a 3kb region upstream of the rbf1 ORF was PCR amplified introducing a 5′-FseI and a 3′-NdeI restriction site and inserted with the NdeI-NotI rbf1-ORF fragment of pCRΔb-crg:rbf1 into pRU11-NotI6474 (a pRU11 [11] derivative in which the NotI site at position 6474 has been removed by a fill up reaction) by three-fragment ligation to generate pRbf1. Generation of pRbf1Δbbs−1377 was performed as described for pRbf1, with the exception that the bbs-motif at position −1377 within the 3kb rbf1 - upstream region was removed by standard PCR techniques [39].
For generation of dik6 promoter-GFP fusion constructs the 2448 bp dik6 promoter fragment was PCR-amplified and integrated into pRU4 [11] digested with HpaI and NdeI. From the resulting plasmid dik6 promoter fragments of 816 bp, 638 bp and 298 bp were recovered as BclI(blunt)/NdeI, MscI(blunt)/NdeI and HindIII(blunt)/NdeI fragments and integrated into pRU4 [11] digested with HpaI and NdeI. Internal deletions in the dik6 promoter were introduced by standard PCR techniques [39]. PCR amplified fragments were integrated into pRU11 via FseI/NdeI restriction sites [11].
RNA extraction and qRT-PCR analysis for rbf1, bW, bE and ppi was performed as described [8]. Full-length rbf1 cDNA was isolated using the GeneRacer Kit (Invitrogen), cloned in pCR2.1 TOPO (Invitrogen) and sequenced. For overview of primers used see Suppl. Table S6.
Quantitative Chromatin Immunoprecipitation (qChIP)
50 ml cultures of U. maydis were grown until OD600 = 0,6–1,0 and cross-linked by addition of formaldehyde (1% final concentration) for 15 min at RT; glycine was added to a final concentration of 0.125 M, cells were harvested by centrifugation and washed three times in TBS (50 mM Tris-HCl, 150 mM NaCl, pH 7.6). The pellet was resupended in 1.5 ml FA lysis buffer (50 mM HEPES-KOH [pH 7.5], 150 mM NaCl, 1 mM EDTA, 1% [v/v] Triton-X-100, 0.1% [w/v] sodium deoxycholate, 0.1% [w/v] sodium dodecyl sulfate [SDS]) supplemented with 2 mM PMSF, 5 mM benzamidine and 1× Complete EDTA-free (Roche). Cells were lysed with a cell mill (Retsch MM200, 25Hz, 5min) in liquid nitrogen pre-cooled grinding jars and the powdery cell extract thawed on ice. 1 ml aliquots of the resulting suspension were sonicated on ice; sonication was set to yield a DNA average size of 400–500 bp. After centrifugation (17000g, 15 min, 4°C) the supernatant was used as the input sample in immunoprecipitation experiments. For each experiment, 400 µl aliquots of the input sample were incubated with 25 µl monoclonal anti-HA-agarose beads (Sigma-Aldrich) over night at 4°C on a rotating wheel.
Washing of beads and recovery of the immunoprecipitated DNA was done according to the ChIP protocol from the Haber Lab (http://www.bio.brandeis.edu/haberlab/jehsite/protocol.html) with the following modifications. All washing steps were carried out at 4°C and repeated one more time, with exception of the TE wash. Proteinase K treatment was done with 50 µl TE containing 3.5 mg/ml Proteinase K without glycogen, and phenol/chlorophorm extraction was done without LiCl. Samples were analysed by qPCR on a Bio-Rad iCycler using the Mesa Green qPCR MasterMix Plus (Eurogentec) with 400 nM Primer (each) and 1 µl immunoprecipitated DNA or 1/100 diluted input DNA, respectively. Amplicons were normalized to input DNA using the Bio-Rad IQ5 software.
DNA array and data analysis
Custom-designed Affymetrix chips were used for DNA-array analysis. Probe sets for the individual genes are visualized at http://mips.helmholtz-muenchen.de/genre/proj/ustilago/Target preparation, hybridization and data analysis was performed essentially as described before [40], with the following alterations: 5 µg RNA were used for first strand cDNA synthesis at 50°C with Superscript II (Invitrogen); for all experiments, an adjusted P-value for the false discovery rate [41] of ≤0.01 and a change in expression of ≥2 was used for filtering. For analysis of b-dependent gene expression strain AB31 (a2 Pcrg1:bE1/bW2) was compared to strain AB32 (a2 Pcrg1:bE2/bW2) and strain AB33 (a2 Pnar1:bE1/bW2) was compared to strain AB34 (a2 Pnar1:bE2/bW2) at the given time points. For analysis of rbf1-dependent gene expression strain AB31 (a2 Pcrg1:bE1/bW2) was compared to AB31Δrbf1 (a2 Pcrg1:bE1/bW2 Δrbf1) and strain CP27 (a2 Δb::Pcrg1:rbf1) was compared to strain JB2 (a2 Δb) at the given time points. Expression values were calculated as mean of two biological replicates. All array data have been submitted to GEO/NCBI (GSE18750, GSE18754 and GSE18756).
De novo promoter motif search was performed using the TAMO framework [42] extended to include AlignAce, Bioprospector, Cismodul, Improbizer, Meme, MDScan and Weeder. Output of each algorithm was collected, converted into a position weight matrix and scored with a hypergeometric test reflecting a random selection null hypothesis [43].
FACS analysis
Flow cytometry measurements were performed as described before [20]. Cell counting was performed with a Neubauer counting chamber.
Microscopy
Microscopic analysis was performed using an Axioimager equipped with an Axiocam MRm camera or a Lumar V12 equipped with an Axiocam HRc (Zeiss, Jena, Germany). Nuclei were stained with DAPI Vectashield H-1200 (Vector Laboratories), fungal cell walls with 2 µg/ml Calcofluor white (Sigma, St. Louis, MO) in PBS. All images were processed with Axiovision (Zeiss, Jena, Germany).
Accession numbers
clp1 (um02438) XP_758585, rbf1 (um03172) XP_759319, hdp1 (um12024) XP_761909.1, hdp2 (um04928) XP_761075, biz1 (um02549) XP_758696, cln1 (um04791) XP_760938, clb1 (um03758) XP_759905, clb2 (um10279) XP_758735, cdr2-like protein (um03928) XP_760075, pcl12 (um10529.2) XP_760585, DNA polymerase epsilon (um01008) XP_757155, DNA replication licensing factor (um06402) XP_762549.
Supporting Information
Zdroje
1. HsuehYP
HeitmanJ
2008 Orchestration of sexual reproduction and virulence by the fungal mating-type locus. Curr Opin Microbiol 11 517 524
2. HiscockSJ
KüesU
1999 Cellular and molecular mechanisms of sexual incompatibility in plants and fungi. Int Rev Cytol 193 165 295
3. MorrowCA
FraserJA
2009 Sexual reproduction and dimorphism in the pathogenic basidiomycetes. FEMS Yeast Res 9 161 177
4. BrefortT
DoehlemannG
Mendoza-MendozaA
ReissmannS
DjameiA
2009 Ustilago maydis as a Pathogen. Annu Rev Phytopathol 47 423 445
5. BölkerM
GeninS
LehmlerC
KahmannR
1995 Genetic regulation of mating, and dimorphism in Ustilago maydis. Can J Bot 73 320 325
6. Flor-ParraI
VranesM
KämperJ
Perez-MartinJ
2006 Biz1, a zinc finger protein required for plant invasion by Ustilago maydis, regulates the levels of a mitotic cyclin. Plant Cell 18 2369 2387
7. BrachmannA
SchirawskiJ
MüllerP
KahmannR
2003 An unusual MAP kinase is required for efficient penetration of the plant surface by Ustilago maydis. EMBO J 22 2199 2210
8. SchererM
HeimelK
StarkeV
KämperJ
2006 The Clp1 protein is required for clamp formation and pathogenic development of Ustilago maydis. Plant Cell 18 2388 2401
9. Flor-ParraI
Castillo-LluvaS
Perez-MartinJ
2007 Polar growth in the infectious hyphae of the phytopathogen Ustilago maydis depends on a virulence-specific cyclin. Plant Cell 19 3280 3296
10. RomeisT
BrachmannA
KahmannR
KämperJ
2000 Identification of a target gene for the bE/bW homeodomain protein complex in Ustilago maydis. Mol Microbiol 37 54 66
11. BrachmannA
WeinzierlG
KämperJ
KahmannR
2001 Identification of genes in the bW/bE regulatory cascade in Ustilago maydis. Mol Microbiol 42 1047 1063
12. ConesaA
GotzS
Garcia-GomezJM
TerolJ
TalonM
2005 Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21 3674 3676
13. Castillo-LluvaS
Perez-MartinJ
2005 The induction of the mating program in the phytopathogen Ustilago maydis is controlled by a G1 cyclin. Plant Cell 17 3544 3560
14. Garcia-MuseT
SteinbergG
Perez-MartinJ
2004 Characterization of B-type cyclins in the smut fungus Ustilago maydis: roles in morphogenesis and pathogenicity. J Cell Sci 117 487 506
15. KanohJ
RussellP
1998 The protein kinase Cdr2, related to Nim1/Cdr1 mitotic inducer, regulates the onset of mitosis in fission yeast. Mol Biol Cell 9 3321 3334
16. SgarlataC
Perez-MartinJ
2005 Inhibitory phosphorylation of a mitotic cyclin-dependent kinase regulates the morphogenesis, cell size and virulence of the smut fungus Ustilago maydis. J Cell Sci 118 3607 3622
17. KulkarniRD
ThonMR
PanH
DeanRA
2005 Novel G-protein-coupled receptor-like proteins in the plant pathogenic fungus Magnaporthe grisea. Genome Biol 6 R24
18. ZarnackK
EichhornH
KahmannR
FeldbruggeM
2008 Pheromone-regulated target genes respond differentially to MAPK phosphorylation of transcription factor Prf1. Mol Microbiol 69 1041 1053
19. UrbanM
KahmannR
BölkerM
1996 Identification of the pheromone response element in Ustilago maydis. Mol Gen Genet 251 31 37
20. Garcia-MuseT
SteinbergG
Perez-MartinJ
2003 Pheromone-induced G2 arrest in the phytopathogenic fungus Ustilago maydis. Eukaryot Cell 2 494 500
21. HartmannHA
KahmannR
BölkerM
1996 The pheromone response factor coordinates filamentous growth and pathogenicity in Ustilago maydis. EMBO J 15 1632 1641
22. HuangG
WangH
ChouS
NieX
ChenJ
2006 Bistable expression of WOR1, a master regulator of white-opaque switching in Candida albicans. Proc Natl Acad Sci U S A 103 12813 12818
23. LiuOW
ChunCD
ChowED
ChenC
MadhaniHD
2008 Systematic genetic analysis of virulence in the human fungal pathogen Cryptococcus neoformans. Cell 135 174 188
24. JungWH
KronstadJW
2008 Iron and fungal pathogenesis: a case study with Cryptococcus neoformans. Cell Microbiol 10 277 284
25. JungWH
ShamA
WhiteR
KronstadJW
2006 Iron regulation of the major virulence factors in the AIDS-associated pathogen Cryptococcus neoformans. PLoS Biol 4 e410
26. BreedingCS
HudsonJ
BalasubramanianMK
HemmingsenSM
YoungPG
1998 The cdr2(+) gene encodes a regulator of G2/M progression and cytokinesis in Schizosaccharomyces pombe. Mol Biol Cell 9 3399 3415
27. El GueddariNE
RauchhausU
MoerschbacherBM
DeisingHB
2002 Developmentally regulated conversion of surface-exposed chitin to chitosan in cell walls of plant pathogenic fungi. New Phytol 156 103 112
28. WeberI
AssmannD
ThinesE
SteinbergG
2006 Polar localizing class V myosin chitin synthases are essential during early plant infection in the plant pathogenic fungus Ustilago maydis. Plant Cell 18 225 242
29. WahlR
WippelK
GoosS
KämperJ
SauerN
2010 A Novel High-Affinity Sucrose Transporter Is Required for Virulence of the Plant Pathogen Ustilago maydis. PLoS Biol 8 e1000303
30. ReedBD
CharosAE
SzekelyAM
WeissmanSM
SnyderM
2008 Genome-wide occupancy of SREBP1 and its partners NFY and SP1 reveals novel functional roles and combinatorial regulation of distinct classes of genes. PLoS Genet 4 e1000133
31. TooneWM
JohnsonAL
BanksGR
ToynJH
StuartD
1995 Rme1, a negative regulator of meiosis, is also a positive activator of G1 cyclin gene expression. EMBO J 14 5824 5832
32. ThomasMJ
SetoE
1999 Unlocking the mechanisms of transcription factor YY1: are chromatin modifying enzymes the key? Gene 236 197 208
33. SambrookJ
FrischEF
ManiatisT
1989 Molecular Cloning: A Laboratory Manual Cold Spring Harbour, New York Cold Spring Harbour Laboratory Press
34. HollidayR
1974 Ustilago maydis.
KingRC
Handbook of Genetics New York, USA Plenum Press 575 595
35. GillissenB
BergemannJ
SandmannC
SchröerB
BölkerM
1992 A two-component regulatory system for self/non-self recognition in Ustilago maydis. Cell 68 647 657
36. MüllerP
WeinzierlG
BrachmannA
FeldbrüggeM
KahmannR
2003 Mating and pathogenic development of the Smut fungus Ustilago maydis are regulated by one mitogen-activated protein kinase cascade. Eukaryot Cell 2 1187 1199
37. SchulzB
BanuettF
DahlM
SchlesingerR
SchäferW
1990 The b alleles of U. maydis, whose combinations program pathogenic development, code for polypeptides containing a homeodomain-related motif. Cell 60 295 306
38. KämperJ
2004 A PCR-based system for highly efficient generation of gene replacement mutants in Ustilago maydis. Mol Genet Genomics 271 103 110
39. HiguchiR
1990 Recombinant PCR.
InnisMA
GelfandDH
SninskyJJ
WhiteTJ
PCR Protocols: a guide to methods and applications San Diego, USA Academic Press 177 183
40. EichhornH
LessingF
WinterbergB
SchirawskiJ
KämperJ
2006 A ferroxidation/permeation iron uptake system is required for virulence in Ustilago maydis. Plant Cell 18 3332 3345
41. BenjaminiY
HochbergY
1995 Controling the false discovery rate: A practical and powefull approach to multiple testing. J R Stat Soc Ser B 57 289 300
42. GordonDB
NekludovaL
McCallumS
FraenkelE
2005 TAMO: a flexible, object-oriented framework for analyzing transcriptional regulation using DNA-sequence motifs. Bioinformatics 21 3164 3165
43. BarashY
BejeranoG
FriedmanN
2002 A simple hyper-geometric approach for discovering putative transcription factor binding sites. Algorithms in Bioinformatics Berlin/Heidelberg Springer 278 293
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2010 Číslo 8- Stillova choroba: vzácné a závažné systémové onemocnění
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Choroby jater v ordinaci praktického lékaře – význam jaterních testů
- Diagnostický algoritmus při podezření na syndrom periodické horečky
-
Všechny články tohoto čísla
- Dissecting the Genetic Architecture of Host–Pathogen Specificity
- The Battle for Iron between Bacterial Pathogens and Their Vertebrate Hosts
- Global Genotype-Phenotype Correlations in
- Burkholderia Type VI Secretion Systems Have Distinct Roles in Eukaryotic and Bacterial Cell Interactions
- Chitin Synthases from Are Involved in Tip Growth and Represent a Potential Target for Anti-Oomycete Drugs
- Distinct Merkel Cell Polyomavirus Molecular Features in Tumour and Non Tumour Specimens from Patients with Merkel Cell Carcinoma
- Biological and Structural Characterization of a Host-Adapting Amino Acid in Influenza Virus
- Functional Characterisation and Drug Target Validation of a Mitotic Kinesin-13 in
- CTCF Prevents the Epigenetic Drift of EBV Latency Promoter Qp
- The Human Fungal Pathogen Escapes Macrophages by a Phagosome Emptying Mechanism That Is Inhibited by Arp2/3 Complex-Mediated Actin Polymerisation
- Bim Nuclear Translocation and Inactivation by Viral Interferon Regulatory Factor
- Cyst Wall Protein 1 Is a Lectin That Binds to Curled Fibrils of the GalNAc Homopolymer
- Reciprocal Analysis of Infections of a Model Reveal Host-Pathogen Conflicts Mediated by Reactive Oxygen and imd-Regulated Innate Immune Response
- A Subset of Replication Proteins Enhances Origin Recognition and Lytic Replication by the Epstein-Barr Virus ZEBRA Protein
- Damaged Intestinal Epithelial Integrity Linked to Microbial Translocation in Pathogenic Simian Immunodeficiency Virus Infections
- Kaposin-B Enhances the PROX1 mRNA Stability during Lymphatic Reprogramming of Vascular Endothelial Cells by Kaposi's Sarcoma Herpes Virus
- Direct Interaction between Two Viral Proteins, the Nonstructural Protein 2C and the Capsid Protein VP3, Is Required for Enterovirus Morphogenesis
- A Novel CCR5 Mutation Common in Sooty Mangabeys Reveals SIVsmm Infection of CCR5-Null Natural Hosts and Efficient Alternative Coreceptor Use
- Micro RNAs of Epstein-Barr Virus Promote Cell Cycle Progression and Prevent Apoptosis of Primary Human B Cells
- Enterohemorrhagic Requires N-WASP for Efficient Type III Translocation but Not for EspF-Mediated Actin Pedestal Formation
- Host Imprints on Bacterial Genomes—Rapid, Divergent Evolution in Individual Patients
- UNC93B1 Mediates Host Resistance to Infection with
- The Transcription Factor Rbf1 Is the Master Regulator for -Mating Type Controlled Pathogenic Development in
- Protective Efficacy of Cross-Reactive CD8 T Cells Recognising Mutant Viral Epitopes Depends on Peptide-MHC-I Structural Interactions and T Cell Activation Threshold
- Bacteriophage Lysin Mediates the Binding of to Human Platelets through Interaction with Fibrinogen
- Insecticide Control of Vector-Borne Diseases: When Is Insecticide Resistance a Problem?
- Immune Modulation with Sulfasalazine Attenuates Immunopathogenesis but Enhances Macrophage-Mediated Fungal Clearance during Pneumonia
- PKC Signaling Regulates Drug Resistance of the Fungal Pathogen via Circuitry Comprised of Mkc1, Calcineurin, and Hsp90
- A Multi-Step Process of Viral Adaptation to a Mutagenic Nucleoside Analogue by Modulation of Transition Types Leads to Extinction-Escape
- “Everything You Always Wanted to Know about Sex (but Were Afraid to Ask)” in after Two Decades of Laboratory and Field Analyses
- Norovirus Gastroenteritis, Carbohydrate Receptors, and Animal Models
- Variations in TcdB Activity and the Hypervirulence of Emerging Strains of
- SWAN-1 Binds to EGL-9 and Regulates HIF-1-Mediated Resistance to the Bacterial Pathogen PAO1
- Conformational Adaptation of Asian Macaque TRIMCyp Directs Lineage Specific Antiviral Activity
- The Proteasome Active Site Threonine Is Essential for Persistence Yet Dispensable for Replication and Resistance to Nitric Oxide
- Characterization of Oseltamivir-Resistant 2009 H1N1 Pandemic Influenza A Viruses
- The Pneumococcal Serine-Rich Repeat Protein Is an Intra-Species Bacterial Adhesin That Promotes Bacterial Aggregation and in Biofilms
- Regulatory T Cell Suppressive Potency Dictates the Balance between Bacterial Proliferation and Clearance during Persistent Infection
- Structural Alterations in a Component of Cytochrome Oxidase and Molecular Evolution of Pathogenic in Humans
- A Limited Number of Antibody Specificities Mediate Broad and Potent Serum Neutralization in Selected HIV-1 Infected Individuals
- Spliced Leader Trapping Reveals Widespread Alternative Splicing Patterns in the Highly Dynamic Transcriptome of
- Contribution of Coagulases towards Disease and Protective Immunity
- Early Severe Inflammatory Responses to Uropathogenic Predispose to Chronic and Recurrent Urinary Tract Infection
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- The Transcription Factor Rbf1 Is the Master Regulator for -Mating Type Controlled Pathogenic Development in
- PKC Signaling Regulates Drug Resistance of the Fungal Pathogen via Circuitry Comprised of Mkc1, Calcineurin, and Hsp90
- Contribution of Coagulases towards Disease and Protective Immunity
- Early Severe Inflammatory Responses to Uropathogenic Predispose to Chronic and Recurrent Urinary Tract Infection
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



