Unkempt Is Negatively Regulated by mTOR and Uncouples Neuronal Differentiation from Growth Control
The development of a functional nervous system requires that nerve cells are generated at exactly the right time and place to be correctly integrated. Defects in the timing at which nerve cells are generated, or ‘differentiate’, lead to neurological disorders such as epilepsy and autism. However, very little is known about the identity of the genes that control the timing of nerve cell differentiation. Using developing photoreceptor nerves in the eye of the fruit fly, Drosophila, as a model, we showed previously that a molecular pathway known as ‘mTOR signalling’ is a key regulator of the timing of differentiation. In this study we have identified two new genes, unkempt and headcase, which control the timing of photoreceptor differentiation in Drosophila. The activity of unkempt and headcase is controlled by mTOR signalling and it is through these genes that mTOR is able to control nerve cell differentiation. The proteins encoded by unkempt and headcase form a complex and act synergistically to control the development of Drosophila photoreceptors. mTOR signalling controls a number of important cellular processes, but unkempt and headcase are the first components of this pathway to be identified that control nerve cell differentiation.
Published in the journal:
. PLoS Genet 10(9): e32767. doi:10.1371/journal.pgen.1004624
Category:
Research Article
doi:
https://doi.org/10.1371/journal.pgen.1004624
Summary
The development of a functional nervous system requires that nerve cells are generated at exactly the right time and place to be correctly integrated. Defects in the timing at which nerve cells are generated, or ‘differentiate’, lead to neurological disorders such as epilepsy and autism. However, very little is known about the identity of the genes that control the timing of nerve cell differentiation. Using developing photoreceptor nerves in the eye of the fruit fly, Drosophila, as a model, we showed previously that a molecular pathway known as ‘mTOR signalling’ is a key regulator of the timing of differentiation. In this study we have identified two new genes, unkempt and headcase, which control the timing of photoreceptor differentiation in Drosophila. The activity of unkempt and headcase is controlled by mTOR signalling and it is through these genes that mTOR is able to control nerve cell differentiation. The proteins encoded by unkempt and headcase form a complex and act synergistically to control the development of Drosophila photoreceptors. mTOR signalling controls a number of important cellular processes, but unkempt and headcase are the first components of this pathway to be identified that control nerve cell differentiation.
Introduction
Neural progenitors in the developing human brain generate up to 250,000 neurons per minute. After differentiating from these neural progenitors, neurons migrate and are then integrated into neural circuits. Temporal control of neurogenesis is therefore critical to produce a complete and fully functional nervous system. Loss of the precise temporal control of neuronal cell fate can lead to defects in cognitive development and to neurodevelopmental disorders such as epilepsy and autism.
Mechanistic target of rapamycin (mTOR) signalling has recently emerged as a key regulator of neurogenesis [1]. mTOR is a large serine/threonine kinase that forms two complexes, known as mTORC1 and mTORC2 [2]. mTORC1 is rapamycin sensitive and is regulated upstream by mitogen signalling, such as the insulin receptor (InR)/insulin like growth factor (IGF) pathway, amino acids, hypoxia, cellular stress and energy levels [3]. mTORC1 positively regulates a large number of cellular processes including growth, autophagy, mitochondrial biogenesis and lipid biosynthesis and activation of mTOR has been linked to cancer. Hyperactivation of mTOR signalling in neurological disease is best understood in the dominant genetic disorder tuberous sclerosis complex (TSC), which causes epilepsy and autism [4]. mTOR signalling has also been shown to be activated in animal models of epilepsy and in human cortical dysplasia [5]–[7].
The control of neurogenesis by the InR/mTOR pathway was first discovered in the developing Drosophila melanogaster retina, where activation of the pathway caused precocious differentiation of photoreceptor neurons and inhibition caused delayed differentiation [8]–[10]. Subsequent in vitro studies demonstrated that insulin induces neurogenesis of neonatal telencephalonic neural precursor cells in an mTOR dependent manner and that Pten negatively regulates neuronal differentiation of embryonic olfactory bulb precursor cells [11], [12]. More recently, in vivo studies have shown that inhibition of mTOR suppresses neuronal differentiation in the developing neural tube [13]. Furthermore, knock-down of the mTOR pathway negative regulator RTP801/REDD1 causes precocious differentiation of neural progenitors in the mouse embryonic subventricular zone (SVZ), while overexpression of RTP801/REDD1 delays neuronal differentiation [14]. Loss of Pten, Tsc1, or overexpression of an activated form of Rheb, also cause premature differentiation of neurons in the SVZ [15]–[17]. These studies have demonstrated that InR/mTOR signalling plays a conserved role in regulating neurogenesis in several different neural tissues. However, the downstream effectors of InR/mTOR signalling in neurogenesis are completely unknown.
To identify neurogenic downstream regulators of InR/mTOR signalling we screened genes that were previously shown to be transcriptionally regulated by mTOR in tissue culture cells [18], for in vivo neurogenic phenotypes in the developing Drosophila retina. From this screen we identified the zinc finger/RING domain protein Unkempt (Unk) as a negative regulator of photoreceptor differentiation. Loss of unk phenocopies the differentiation phenotype of InR/mTOR pathway activation and Unk expression is negatively regulated by InR/mTOR signalling. Importantly, unk does not regulate cell proliferation or cell size and so uncouples the function of InR/mTOR signalling in growth from its role in neurogenesis. We also identified the evolutionarily conserved basic protein Headcase (Hdc) [19], as a physical interactor of Unk and show that loss of hdc causes precocious differentiation of photoreceptors. Hdc expression is regulated by the InR/mTOR pathway and by unk, demonstrating that Hdc and Unk work together downstream of InR/mTOR signalling in neurogenesis. Unk also regulates the expression of and interacts with D-Pax2, suggesting a model for the regulation of neurogenesis by the InR/mTOR pathway. We also show that one of the mammalian homologs of Unk, Unkempt-like, is expressed in the developing mouse retina and in the early postnatal brain. We have thus identified the Unk/Hdc complex as the first component of the InR/mTOR pathway that regulates the timing of neuronal differentiation.
Results
unkempt negatively regulates the timing of photoreceptor differentiation
The eight photoreceptors (R1-R8) that constitute each ommatidium (facet) of the Drosophila compound eye (Figure S1A) differentiate in a stereotypical sequence that is initiated by R8 in response to signalling events around the morphogenetic furrow (MF). The MF is a physical indentation that traverses the eye imaginal disc from posterior to anterior during the final 48 hours of larval development and the early pupal stage (Figure 1A) [20]. Photoreceptors differentiate posterior to the MF in rows that are aligned along a differentiation front (Figure 1A, dotted line), with each new row forming every two hours. Adult ommatidia are organised in rows forming a mirror image about the equator (Figure S1A, dotted line). We have previously shown that the timing of differentiation of R1/6/7 and cone cells, but not R3/4 and R2/5, is regulated by InR/mTOR signalling [8], [10]. In clones that are mutant for Tsc1, in which mTOR signalling is activated, R1/6 differentiate two to three rows ahead of the differentiation front (Figure 1B, arrows), whereas differentiation is severely delayed in clones that are mutant for Rheb (Figure 1J), in which the pathway is inhibited. To identify novel factors that regulate photoreceptor differentiation downstream of mTOR, we screened genes that are transcriptionally regulated by mTOR in Drosophila S2 cells [18] for differentiation phenotypes in vivo by RNAi. Flp-out clones expressing dsRNAs against 28 mTOR regulated genes (Table S1), were generated to test for photoreceptor differentiation phenotypes. Using this approach we identified unkempt (unk) as a potential negative regulator and three molecular chaperone encoding genes (Hsc70Cb, Hsp60 and Hsp83) as potential positive regulators of R1/6 differentiation (Table S1 and Figure S1B). Hsp83 mutant clones are cell lethal, suggesting that the delayed differentiation phenotype caused by RNAi of Hsp83 observed in the screen, and potentially also Hsc70Cb and Hsp60, may be due to cell death rather than a genuine delay in differentiation. These chaperones were not studied further and we focused on unk as a potential negative regulator of photoreceptor differentiation.
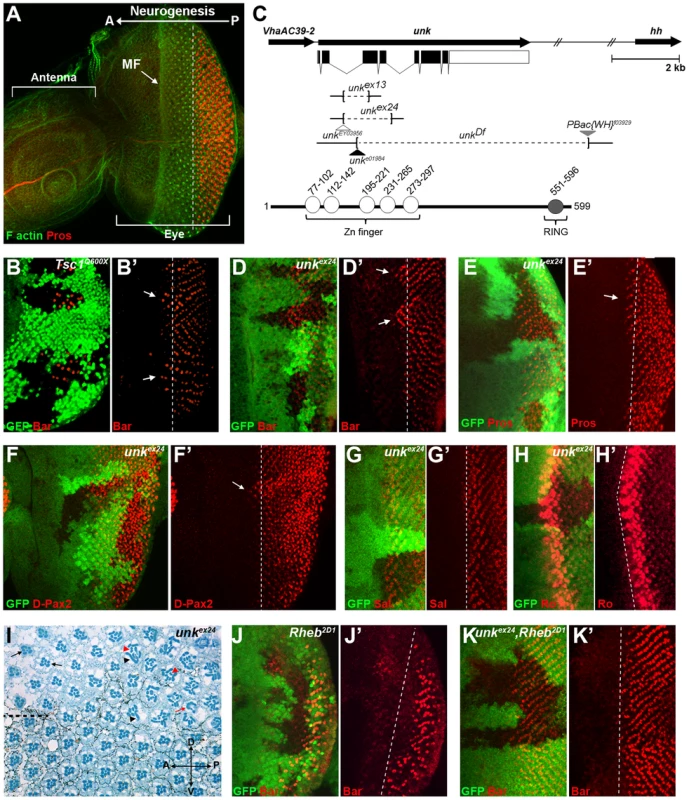
The Unk protein contains an N-terminal zinc finger domain and C-terminal RING domain (Figure 1C). Unk physically interacts with mTOR, Raptor and 4E-BP in Drosophila Kc167 cells and the strength of these interactions is regulated by insulin [21]. Moreover, phosphopeptides corresponding to one of the two mammalian homologs of Unk were identified in mouse and human cells in both of the recent mTOR phosphoproteome studies [22], [23]. Null alleles for unk are lethal at around mid-pupal development, while hypomorphic alleles develop to become adults with an ‘unkempt’ phenotype (small rough eyes, held out wings and crossed scutellar bristles) [24]. However, nothing else is known about the function of unk. The previously isolated unk mutants are no longer extant. Therefore, we generated novel mutations in unk (Figure 1C, see Materials and Methods). Animals homozygous for unkex13, unkex24, unkDf, unke01984 or heteroallelic combinations of these alleles are pupal lethal and mutant clones showed a complete absence of Unk protein expression (Figure S1C), suggesting that they are null alleles.
unk mutant clones cause precocious differentiation of R1/6/7 and cone cells (Figure 1D, E, F, arrows), very similar to the phenotype seen in Tsc1 mutant clones (Figure 1B). Moreover, unk mutant clones have no effect on the differentiation of R3/4 (Figure 1G), or R2/5 (Figure 1H). Co-staining for the pan-neuronal marker Elav, or markers of R3/4 and R8, with either Bar or Prospero also demonstrated that the precocious differentiation in unk mutant clones is not due to ectopic expression of Bar or Prospero (Figure S2 A–C). Thus, loss of unk phenocopies the precocious photoreceptor differentiation phenotype caused by activation of InR/mTOR signalling. This suggests that unk activity is normally repressed by mTOR during photoreceptor differentiation. The R1/6 precocious differentiation phenotype in unk mutant clones was rescued by overexpression of unk cDNA (Figure S1D), demonstrating that the precocious differentiation phenotype is specifically due to loss of unk.
To test whether increased expression of unk is sufficient to delay photoreceptor differentiation, MARCM (mosaic analysis with a repressible cell marker) clones were generated that overexpressed unk. No change in the timing of differentiation of R1/6 was seen in these clones (Figure S1E). Therefore, unk is necessary but not sufficient to regulate the timing of differentiation of R1/6/7 and cone cells.
In the adult eye unk mutant clones had a striking phenotype. Mutant ommatidia in the anterior half of the eye had a normal structure but had rotation defects (Figure 1I, black arrows), similar to Tsc1 and Pten mutant clones [8]. Ommatidia in the posterior half of the eye were missing photoreceptors (red arrow) and contained photoreceptors with elliptical (black arrowheads) and split (red arrowheads) rhabdomeres (Figure 1I). This phenotype is typical of perturbation of the F-actin cytoskeleton and Pten and Tsc1 mutant clones cause similar defects in photoreceptor apical membrane morphogenesis ([25], [26] and Figure S1F). In summary these data demonstrate that unk is necessary for the normal timing of differentiation and morphogenesis of photoreceptors.
Loss of unk suppresses the delay in photoreceptor differentiation caused by inhibition of InR/mTOR signalling
To test whether unk acts genetically downstream of InR/mTOR signalling, we generated double mutant clones that lacked both unk and Rheb. Compared to Rheb clones, which cause a strong delay in the differentiation of R1/6 (Figure 1J), differentiation in unk, Rheb clones appeared normal (Figure 1K). Thus, the strong delay caused by the loss of Rheb was suppressed, but photoreceptors in these double mutant clones did not differentiate precociously as in unk mutant clones (Figure 1D). In the adult eye the elliptical and split rhabdomere phenotype seen in unk mutant clones was suppressed in unk, Rheb mutant clones (Figure S1H). However, both Rheb and unk, Rheb mutant clones contained mis-rotated ommatidia and missing photoreceptors (Figure S1G, H). Therefore, although unk suppresses the delay in photoreceptor differentiation caused by inhibition of InR/mTOR signalling, there may be an additional factor(s) that regulates R1/6/7 and cone cell fate and acts in parallel with unk (see Discussion).
Unk expression is negatively regulated by InR/mTOR signalling in photoreceptor neurons
Unk is a ubiquitously expressed cytoplasmic protein (Figure 2A, B). In the eye disc Unk is expressed in undifferentiated cells anterior to the MF, in photoreceptor precursors posterior to the MF, photoreceptors and cone cells (Figure 2A, B and 3B, C). Moreover, Unk is expressed more strongly posterior to the MF in the apical plane of the disc containing differentiated photoreceptors and cone cells (Figure 2A and S3B). Although localised to the cytoplasm, Unk has a partially punctate distribution (Figure 2B). In accordance with the negative regulation of unk expression by mTOR in S2 cells ([18] and Table S1), Unk expression is reduced in Tsc1 clones posterior to the MF both in differentiated photoreceptors and undifferentiated photoreceptor precursors (Figure 2C, D, arrows). However, Unk expression is unchanged in Tsc1 clones anterior to the MF (Figure 2D, arrowheads). Thus, Unk expression is negatively regulated by the InR/mTOR pathway in differentiating photoreceptor neurons and photoreceptor precursors. Inhibition of InR/mTOR signalling, using Dp110, or Rheb mutant clones, did not cause an increase in Unk expression (Figure S4) and so Unk expression is not positively regulated by inhibition of mTOR signalling.
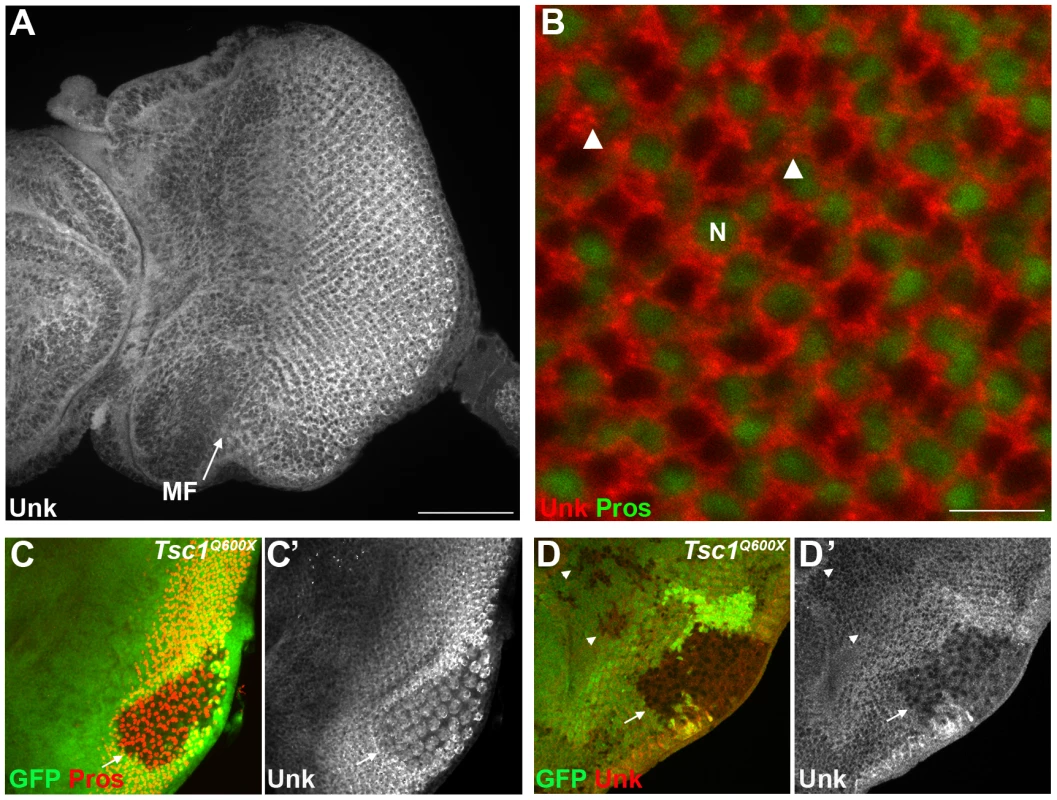
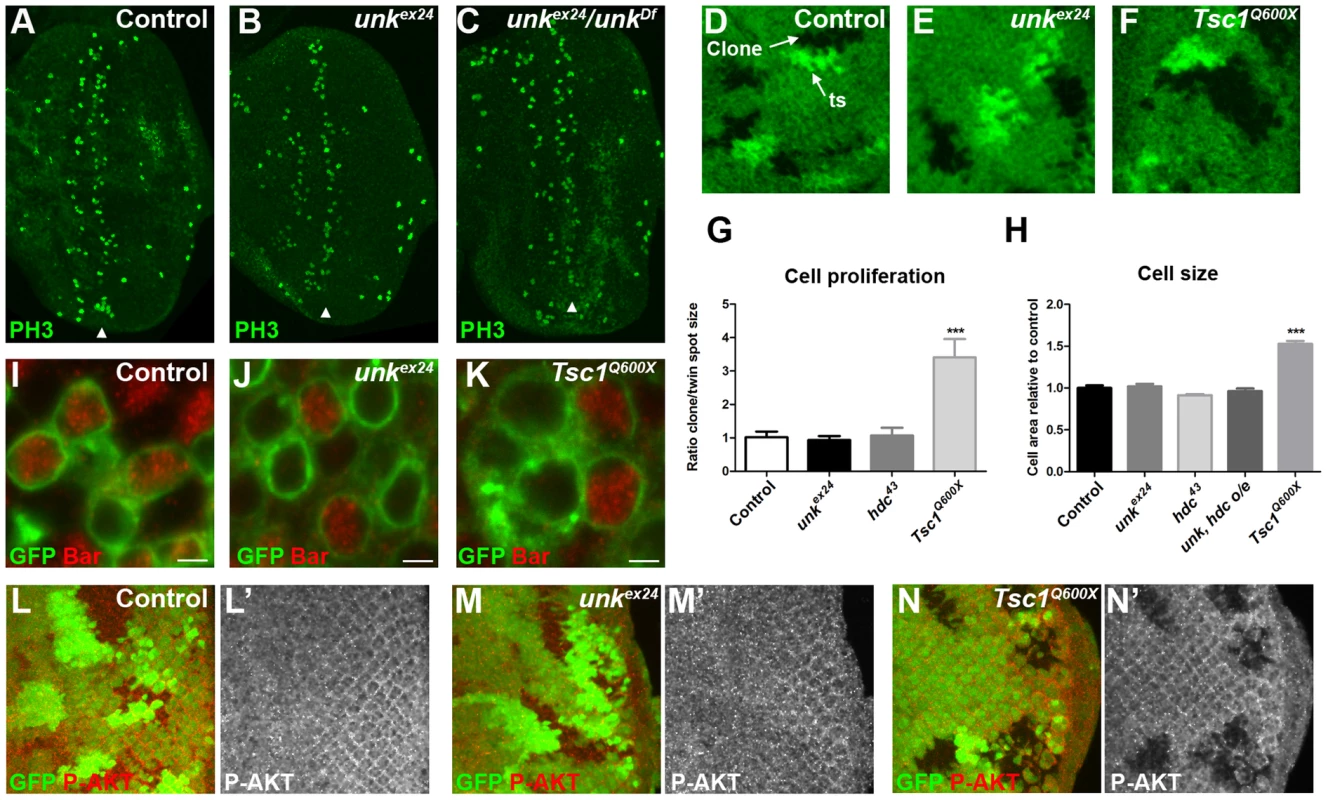
unk does not regulate cell size or proliferation
Several lines of evidence demonstrate that unk does not regulate growth. First, eye discs from unkex24 homozygous larvae, or an unk transheterozygous mutant, had similar levels of phospho-histone H3 expression posterior to the MF (arrowheads) as wild-type discs (Figure 3A–C). Second, unlike Tsc1 mutant clones (Figure 3F, G), unk mutant clones were a similar size to the control (Figure 3D, E, G). Third, unlike cells mutant for Tsc1 (Figure 3K, H), cells mutant for unk in the eye disc were a similar size to controls (Figure 3I, J, H). Phospho-S6K or phospho 4E-BP antibodies did not work as a direct readout of mTOR activity by immunofluorescence. We therefore used phospho-AKT (P-AKT) expression as an indirect readout of mTOR pathway activity through negative feedback regulation via S6K [27]. As expected, expression of P-AKT was decreased in Tsc1 mutant clones (Figure 3N). However, no change in P-AKT expression was observed in unk mutant clones (Figure 3M), suggesting that unk is not required for the canonical regulation of S6K by mTOR. Together these data show that unk plays no role in the regulation of growth. Therefore, unk is the first gene to uncouple the function of InR/mTOR signalling in growth control from its role in regulating the transition of a post-mitotic precursor cell to a neuronal fate.
Headcase interacts with Unk to control photoreceptor differentiation downstream of InR/mTOR signalling
To provide insight into how Unk regulates neurogenesis we interrogated protein interaction maps to identify proteins that might interact with Unk. Headcase (Hdc) was identified as interacting with Unk in two independent protein interaction maps and in a study that characterised the Drosophila InR/mTOR pathway interaction proteome [21], [28], [29]. Hdc is an evolutionarily conserved basic protein with no conserved motifs [19]. hdc mRNA generates two overlapping proteins, a short form (HdcS) and a full-length form (HdcFL), as a result of a novel translational readthrough mechanism [30]. To provide direct evidence for the physical interaction between Unk and Hdc, we expressed epitope-tagged forms of these proteins in S2 cells. Immunoprecipitation assays in both directions showed that Venus-Unk and FLAG-HdcS co-immunoprecipitate (Figure 4A). These assays confirm that Unk and Hdc physically interact in S2 cells.
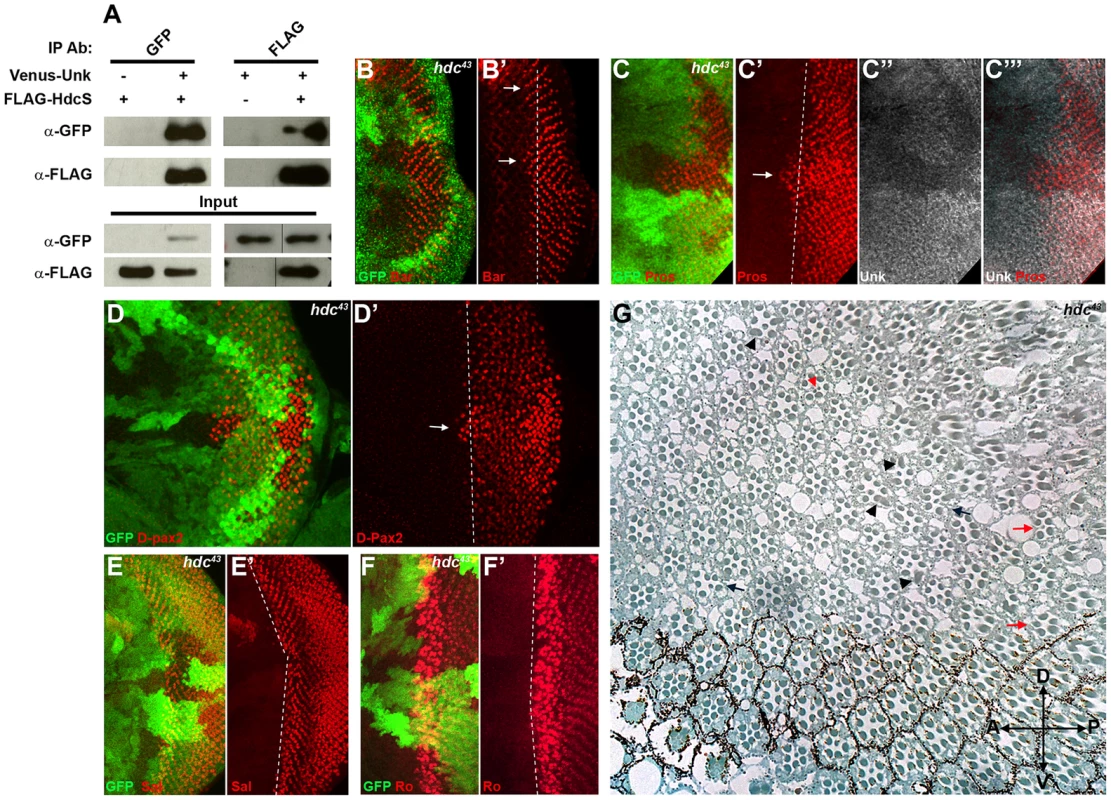
R1/6/7 and cone cells differentiated precociously in clones that were mutant for a null allele of hdc (Figure 4B, C, D, arrows), but the differentiation of R3/4 and R2/5 was not affected (Figure 4E, F). Also there was no ectopic expression of markers for R3/4 or R8 in hdc mutant clones (Figure S2D–F). In the adult hdc mutant ommatidia in the anterior half of the eye had a normal structure but had mis-rotation defects (Figure 4G, black arrows), while ommatidia in the posterior half of the eye had missing photoreceptors (red arrows), elliptical (black arrowheads) and split rhabdomeres (red arrowhead) (Figure 4G). These phenotypes are similar to but weaker than those observed in unk mutant ommatidia (Figure 1I). Loss of hdc also had no effect on photoreceptor proliferation or cell size (Figure 3G, H). Together these data demonstrate that hdc is necessary to regulate the timing of photoreceptor differentiation and photoreceptor morphogenesis.
Hdc is a ubiquitously expressed cytoplasmic protein and in the eye disc, similar to Unk, is expressed more strongly posterior to the MF in the apical plane of the disc containing differentiated photoreceptors and cone cells (Figure 5A and S3B). Hdc is evenly distributed throughout the cytoplasm and does not have any punctate localisation (Figure 5B). In unk mutant clones Hdc expression was increased in a specific spatiotemporal pattern; Hdc expression began to increase two to three rows posterior to the differentiation front for R1/6 (Figure 5C, D, H). This dynamic increase in Hdc expression was also observed in Tsc1 mutant clones (Figure 5E, F, I). Thus, Hdc expression is positively regulated by mTOR signalling and negatively regulated by unk, but only after the R1/6 differentiation front. Hdc expression was also slightly decreased in Rheb mutant clones, in which InR/mTOR signalling is inhibited (Figure 5G, J).
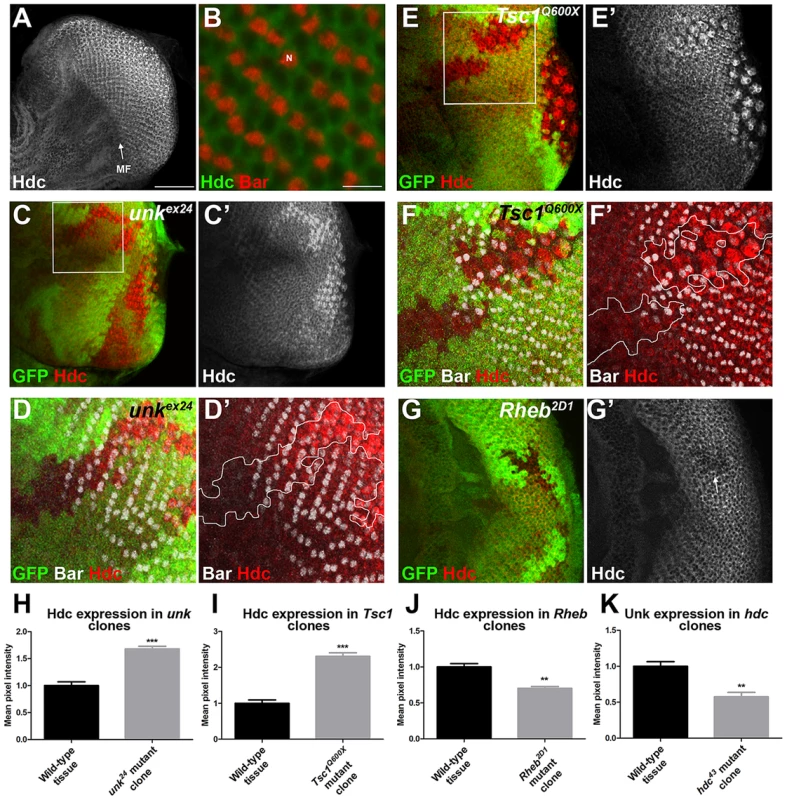
We also tested whether hdc regulates Unk expression. Unk expression was decreased in hdc mutant clones both anterior and posterior to the MF (Figure 4C′′′ and 5K). Therefore, Unk and Hdc are mutually dependent on each other to maintain their expression levels and the precocious differentiation phenotype caused by loss of hdc may be due to its requirement to maintain the expression of Unk.
unk and hdc act together to control the timing of photoreceptor differentiation
To test whether unk and hdc act together double mutant clones were generated. unk, hdc double mutant clones caused precocious differentiation of R1/6 two to three rows ahead of the differentiation front (Figure 6A, arrow), similar to the phenotype of either single mutant. unk, hdc double mutant clones in the adult eye also caused a similar phenotype to unk and hdc mutant clones (Figure 6B). Therefore, unk and hdc act in the same pathway to regulate the timing of photoreceptor differentiation and photoreceptor morphogenesis.
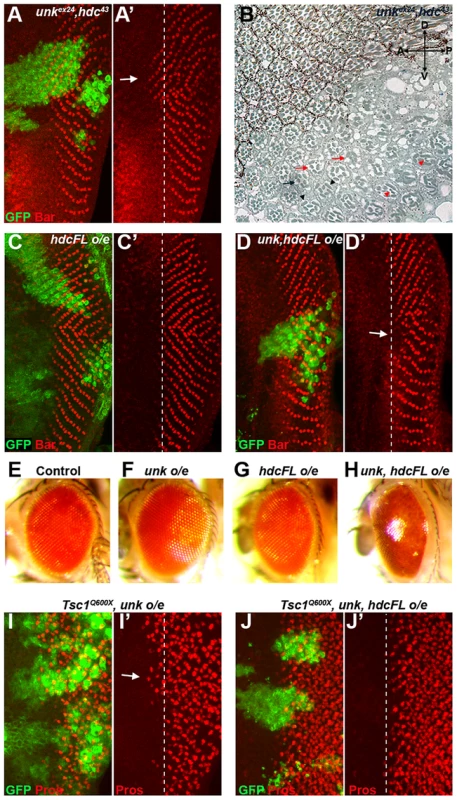
Overexpression of cDNAs for either unk or hdcFL (that expresses both the short and full length forms of hdc) alone in clones did not affect photoreceptor differentiation (Figure S1E and 6C). However, combined overexpression of unk and hdcFL delayed the differentiation of R1/6 (Figure 6D, arrow), but did not delay the differentiation of R3/4 (Figure S5F), or affect cell growth (Figure 3H). Moreover, overexpression of either gene individually in all cells posterior to the MF using GMR-Gal4 had no effect in the adult eye (Figure 6F, G), but combined overexpression of unk and hdcFL caused the eye to have a glassy appearance, indicating a strong defect in eye development (Figure 6H). Neither the delay in differentiation, nor the rough eye phenotype are due to increased apoptosis (Figure S5A−E). Importantly, while overexpression of unk in Tsc1 mutant clones did not affect the precocious differentiation phenotype (Figure 6I), co-overexpression of unk and hdcFL completely suppressed the precocious differentiation of R7 and cone cells caused by loss of Tsc1 (Figure 6J). These data strongly suggest that unk and hdc act together to negatively regulate photoreceptor differentiation downstream of mTOR.
Unk physically interacts with and negatively regulates the expression of D-Pax2
D-Pax2 is a paired domain transcription factor and is the main regulator of cone cell fate in the developing eye [31]. The timing of cone cell differentiation is regulated by InR/mTOR signalling and cone cells precociously differentiate in unk and hdc mutant clones (Figure 1E, F and 4C, D). We noticed that D-Pax2 expression increased in unk, hdc and Tsc1 mutant clones (Figure 1F, 4D and S6A), demonstrating that the Unk/Hdc complex and mTOR signalling regulate D-Pax2 protein level in developing cone cells. Pax8, part of the Pax2/Pax5/Pax8 subgroup of paired domain transcription factors that is homologous to D-Pax2, physically interacts with the human Unk homolog [32]. We therefore tested whether Unk and D-Pax2 physically interact by expressing epitope-tagged forms of these proteins in S2 cells. Immunoprecipitation assays showed that Venus-Unk and FLAG-D-Pax2 co-immunoprecipitate (Figure S6B). Therefore, the regulation of D-Pax2 protein levels by mTOR signalling and Unk/Hdc may contribute to the rate of cone cell differentiation though a direct physical interaction with D-Pax2.
Unkempt-like is expressed in the developing mammalian retina and the brain
There are two homologs of Unk in vertebrates, known as Unkempt (Unk) and Unkempt-like (Unkl). Mouse Unk and Unkl both have a similar degree of identity to Drosophila Unk (45%, Figure S7 and S8). By staining cells overexpressing an HA-tagged version of mouse Unkl we found that an antibody generated against human Unkl also recognises mouse Unkl (Figure 7A). Staining of mouse embryonic day 14.5 (E14.5) tissue with this antibody showed that Unkl is strongly expressed in the developing retina (Figure 7B), suggesting that Unk may have a conserved role in eye development in Drosophila and mammals.
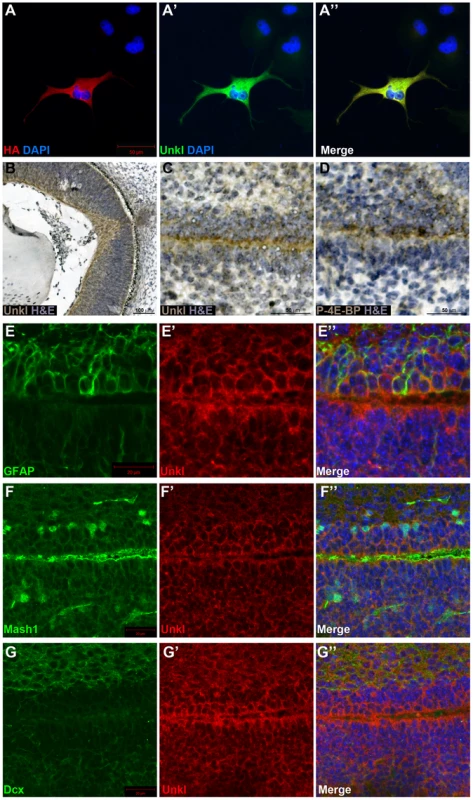
mTOR signalling has recently been shown to be active in the early postnatal mouse subventricular zone (SVZ), where it regulates neural stem cell (NSC) self renewal and differentiation [33]. Staining of the early postnatal brain showed that Unkl is expressed throughout the brain (Figure S9A). Unkl is expressed throughout the SVZ, but is most strongly expressed in the cells close to the ventricle, similar to phosphorylated 4E-BP (P-4E-BP), a marker of mTOR pathway activity (Figure 7C, D). Further analyses showed that glial fibrillary acidic protein (GFAP) positive NSCs, Mash1 positive transit amplifying progenitors (TAPs) and neuroblasts (identified by the expression of doublecortin (Dcx)) all express Unkl (Figure 7E–G and S9B–D). These data suggest that Unkl may play a role in mTOR-dependent neural stem/progenitor cell differentiation in the mammalian CNS. Thus, Unk may act downstream of InR/mTOR signalling to regulate neuronal differentiation in both Drosophila and mammals.
Discussion
Several lines of evidence together demonstrate that unk and hdc act downstream of InR/mTOR signalling to negatively regulate the timing of photoreceptor cell fate. First, loss of either unk or hdc causes precocious differentiation of the same cells and to the same degree as activation of InR/mTOR signalling. Second, the expression of both Unk and Hdc are regulated by InR/mTOR signalling. Third, loss of unk suppresses the strong delay in photoreceptor differentiation caused by inhibition of the InR/mTOR pathway and combined overexpression of unk and hdc suppresses the precocious photoreceptor differentiation caused by loss of Tsc1. Fourth, although Unk has been shown to physically interact with mTOR [21], neither unk nor hdc regulate cell or tissue growth. Taken together these data show that unk and hdc are novel downstream components of the InR/mTOR pathway that regulate the timing of neuronal differentiation (Figure 8).
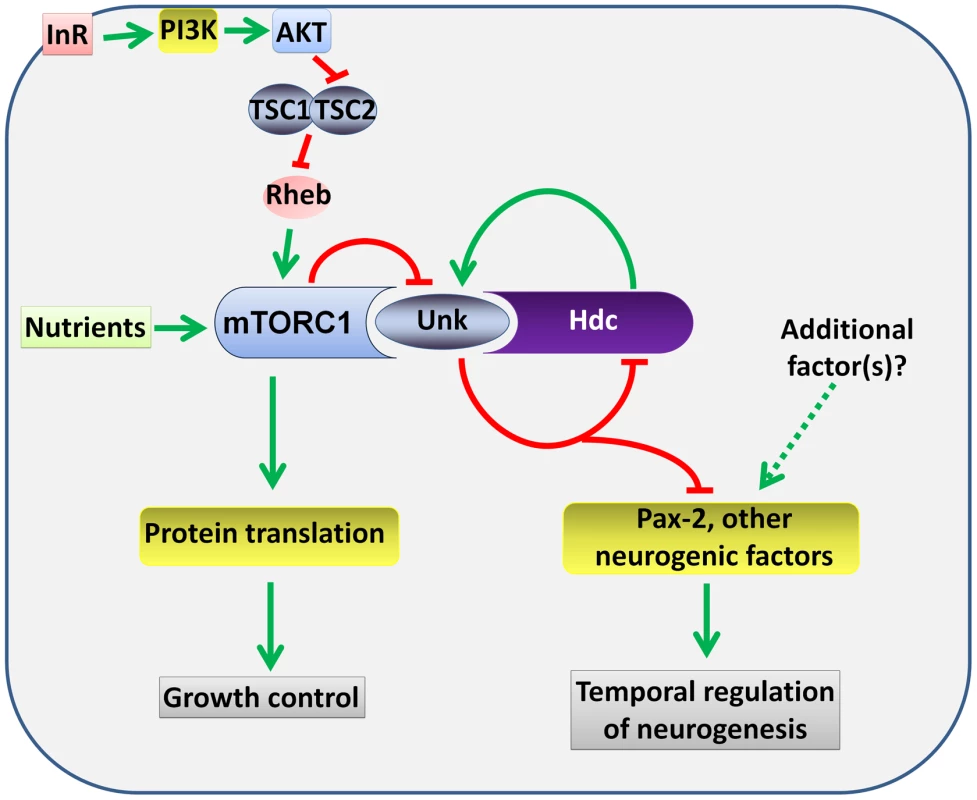
InR/mTOR signalling is a major regulator of cell growth. In Drosophila activation of InR/mTOR signalling by loss of either Tsc1, Tsc2, Pten, or overexpression of Rheb causes increased cell size and proliferation [34]–[37]. In the genetic disease TSC, which is caused by mutations in Tsc1 or Tsc2, patients develop benign tumours in multiple organs including the brain [4]. The previously identified components of the InR/mTOR pathway regulate both growth and neurogenesis in Drosophila and vertebrate model systems [8], [10], [13], [14], [16], [17], [38]. unk and hdc therefore represent a branchpoint in the pathway where its function in neurogenesis bifurcates from that in growth control (Figure 8). Moreover, our analysis of unk and hdc demonstrates that regulation of cell growth can be uncoupled from and is not required for the function of InR/mTOR signalling in the temporal control of neuronal differentiation.
At the protein level we show that Unk and Hdc physically interact in S2 cells. Although this interaction remains to be demonstrated in vivo, the additional observations that they both regulate each other's expression and act synergistically in vivo strongly support the model that they physically interact (Figure 8). Moreover, Unk and Hdc have also previously been shown to physically interact by yeast-2-hybrid and co-immunoprecipitation [21], [28], [29]. Unk and Hdc are both expressed in all developing photoreceptors and so we hypothesise that they control the timing of differentiation through the regulation of neurogenic factors whose expression is restricted to R1/6/7 and cone cells (Figure 8). Loss of unk causes increased expression of D-Pax2, the main regulator of cone cell differentiation. hdc and Tsc1 mutant clones also cause a similar increase in D-Pax2 expression. Overexpression of D-Pax2 alone is insufficient to induce cone cell differentiation, which requires overexpression of both D-Pax2 and Tramtrack88 (TTK88) [39]. Thus, regulation of D-Pax2 expression by mTOR signalling may contribute to the rate of cone cell differentiation, while overall control would require the regulation of additional factors such as TTK88 (Figure 8). Pax8, part of the Pax2/Pax5/Pax8 paired domain transcription factor subgroup that is homologous to D-Pax2, has been shown to physically interact with one of the two human homologs of Unkempt [32]. We find that Drosophila Unk physically interacts with D-Pax2, demonstrating that the physical interaction between Unk and this group of transcription factors is conserved. We suggest that D-Pax2 may be one of several neurogenic factors regulated by InR/mTOR signalling, through a physical interaction with the Unk/Hdc complex, to control the timing of R1/6/7 and cone cell fate (Figure 8).
Unk has been shown to physically interact with mTOR and the strength of this interaction is regulated by insulin [21]. This suggests the intriguing possibility that the inhibition of Unk activity by InR/mTOR signalling is dependent on the strength of the physical interaction between Unk and the mTORC1 complex. Unk was also identified as part of the mTOR-regulated phosphoproteome in both human and murine cells [22], [23]. Thus, Unk may potentially be regulated by mTOR through phosphorylation. Future studies will fully characterise the mechanism by which mTORC1 regulates Unk activity.
Our study represents the first demonstration of a role for unk in specific developmental processes. By contrast, hdc has previously been shown to regulate dendritic pruning during metamorphosis and to act as a branching inhibitor during tracheal development [30], [40]. A screen for genes affecting tracheal tube morphogenesis and branching recently identified Tsc1 [41], suggesting that InR/mTOR also regulates tracheal development. Thus, hdc and unk may act repeatedly as downstream effectors of the InR/mTOR pathway during Drosophila development.
The one previous study of either of the mammalian Unk homologs showed that Unkl binds specifically to an activated form of the Rac1 GTPase [42]. If this function is conserved in Drosophila then the defects in photoreceptor apical membrane morphogenesis caused by activation of mTOR signalling or loss of unk/hdc may be mediated through Rac1.
The function of the two unk homologs, unk and unkl, in mammalian development is not known, but unk has been shown to be expressed in the mouse early postnatal mouse retina [43]. We find that Unkl is also expressed in the developing mouse retina, suggesting that Unk may play a conserved role in eye development in both flies and mammals. InR/mTOR signalling acts as a pro-survival pathway preventing retinal degeneration [44], but its role in mammalian eye development has not been characterised. By contrast InR/mTOR signalling has a well characterised role in NSC self-renewal and differentiation in the mouse SVZ. Loss of Tsc1 or expression of a constitutively active form of Rheb in neural progenitor cells in the postnatal mouse SVZ causes the formation of heterotopias, ectopic neurons and olfactory micronodules [15], [16]. Furthermore, individuals with TSC, which results in activated mTOR signalling, have aberrant cortical neurogenesis and develop benign cortical tumours during foetal development and throughout childhood [4], [45]. mTOR signalling has been shown to be active in proliferative NSCs and TAPs in the neonatal SVZ and inhibition of mTOR signalling prevents NSC differentiation [33]. We find that Unkl is expressed in both NSCs and TAPs in the early postnatal SVZ. Thus, Unkl may regulate NSC differentiation downstream of mTOR signalling in the mammalian brain. Unkempt may therefore play a conserved role in regulating the timing of neural cell fate downstream of mTOR signalling in both flies and mammals.
Materials and Methods
Fly strains and genetic crosses
Flies were maintained on standard yeast, glucose, cornmeal, agar food at 25°C unless stated otherwise. Fly stocks were FRT82B, Dp110A [46], FRT82B, Tsc1Q600X [34], FRT82B, Tsc1Q87X [37], FRT82B, Rheb2D1 [36], FRT82B, hdc43 [19] and UAS-hdcFL [30] and UAS-unk (this study). For clonal experiments the stocks used were y, w, hs-flp; FRT82B, ubi-GFP, y, w, hs-flp; FRT82B, M[95A]Rps63, ubi-GFP and y, w, hs-flp;tub-Gal4, UAS-mCD8GFP;FRT82B, tub-Gal80 (MARCM stock). RNAi lines were obtained from the Vienna Drosophila Resource Centre. All other stocks were obtained from the Bloomington Stock Centre. For mosaic analysis mutant clones were generated by Flp/FRT mediated recombination using heat-shock-flp or by MARCM [47]. For clonal analysis in eye discs larvae were heat-shocked for 1-1.5 hours at 37°C 24 hours after egg laying (AEL). For adult clones larvae were heat-shocked for 30 minutes at 37°C 24 hours and again at 48 hours AEL. Adult eye sections were prepared as described previously [48].
The line P[EPgy2]unkEY03956 was used to generate mutants unkex13 and unkex24 through imprecise P-element excision [49]. The deleted region in both unkex13 and unkex24 begins 557 bp into the second intron and for unkex13 ends 219 bp into the third exon and for unkex24 ends 150 bp into the fourth intron. The PiggyBac lines, PBac[RB]unke01984 and PBac[WH]f03929 were used to generate unkDf using a Flp/FRT –based precise excision strategy [50]. Mutation break points and deleted regions were confirmed or determined by PCR and sequencing.
Generation of the Unk antibody, staining and immunofluorescence
The Unk antibody was generated using a GST-fusion protein as described in Text S1. The antibodies used were: rat anti-Unk3 (1∶500, this study), mouse anti-Hdc (a gift from Robert White, 1∶5) [19], rat anti-Bar (1∶500) [10], rat anti-Elav (DSHB, 1∶100), mouse anti-Prospero (DSHB, 1∶100), mouse anti-Rough (DSHB, 1∶10), rabbit anti-Spalt (a gift from R. Barrio, 1∶500), guinea pig anti-senseless (a gift from Hugo Bellen, 1∶500), rabbit anti-cleaved caspase 3 (Cell Signalling, 1∶100), rabbit anti-D-Pax2 (a gift from M. Noll, 1∶20) [31], rabbit anti-P-AKT (Cell Signalling, 4045S, 1∶200), rabbit or mouse anti-GFP (Life Technologies, 1∶1000), rabbit anti-PH3 (Upstate, 1∶50), rabbit anti RING finger protein unkempt like (Abcam ab155197, 1∶300), rat anti HA (Roche), mouse anti-Mash1 (a gift from Francois Guillemot, 1∶20), mouse anti GFAP (Sigma, G3893, 1∶1000) and goat anti Dcx (Santa Cruz Sc8066, 1∶200).
For immunofluorescence, dissected third instar larvae were fixed for 30 minutes in 4% paraformaldehyde, then washed five times for 10 minutes in PBS/0.1% triton (PBST), before blocking in PBST/1% normal goat serum (PBST-NGS) for an additional hour. Eye discs were incubated with primary antibody overnight in PBST-NGS at 4°C. After five to six washes of 10 minutes in PBST discs were incubated for two hours with secondary antibody at room temperature, washed five to six times in PBST and then mounted in Vectashield (Vectalabs). Phospho-AKT staining was performed exactly as described [51].
P0 mouse CNS tissue was fixed overnight in 4% paraformaldehyde then embedded in paraffin and 6 µm sections were cut. To generate primary neural progenitor monolayers, subventricular zone fragments from P1 mice were triturated with 2 ml of HBSS containing 0.25% trypsin (Gibco) and 40 µl of DNAse l (1 mg/ml, Worthington) and incubated at 37°C for 2 minutes. The trypsin was inactivated with 5 ml of DMEM (Gibco) containing 10% FCS and the solution was centrifuged at 1,500 rpm for 5 minutes. After another two washes with DMEM/10% FCS to remove any traces of trypsin, the pellet was re-suspended in pre-equilibrated (at 37°C/5% CO2) Neurobasal complete medium (Gibco) containing B27 supplement, 2 mM L-glutamine (Invitrogen) and 0.6% glucose (Sigma). Cells were plated onto 24 well plates (2.5×106 cells/well) on glass coated with polyornithine (0.5 mg/ml, Sigma). Cells were maintained in Neurobasal complete medium at 37°C/5% CO2 for 24 hours then fixed and stained as for COS-7 cells (below).
COS-7 cells were fixed for 30 minutes in 4% paraformaldehyde, washed several times in PBS, blocked for one hour in PBS/1% bovine serum albumen/0.2% triton/0.02% sodium azide, then incubated overnight at 4°C in the primary antibody diluted in blocking buffer. Secondary antibodies were FITC donkey anti-mouse, Cy3 donkey anti-rat, Cy5 donkey anti-rat and Cy5 donkey anti-mouse (Jackson Immunolabs); Alexa488 anti-rabbit, Alexa594 anti-rabbit and Alexa594 anti-mouse (Life Technologies). Images were acquired on a Zeiss LSM710 confocal microscope and processed in Photoshop CS4 (Adobe). The differentiation front was marked with a dotted line positioned just ahead of the most anterior row of photoreceptors.
Cloning of unk, hdc and D-Pax2
Details of the cloning of unk, hdc and D-Pax2 are described in Text S1. UAS-unk transgenic lines were generated by germline transformation (BestGene Inc.). Overexpression of Unk using these lines was confirmed by immunostaining.
Cell culture and immunoprecipitation
For immunoprecipitation (IP) experiments Venus-unk, FLAG-hdcS and FLAG-D-Pax2 were expressed in S2 cells (DGRC) cultured in SF9-S2 medium (PAA laboratories). For IP experiments, cells were seeded in six well plates at a density of around 1.5 million cells per well. Cells were then transfected with 4.5 µg of Venus-unk, with or without 0.5 µg FLAG-hdcS or 0.5 µg FLAG-D-Pax2 using transfectin (Biorad) according to the manufacturer's instructions. The IP protocol was adapted from [52]. For each condition after 48 hours cells from two wells were manually detached, washed in cold PBS and lysed for 1.5 hours in lysis buffer (25 mM Tris pH 8, 150 mM NaCl, 5% glycerol, 1% triton, 1 mM PMSF, 1× protease inhibitor cocktail (Roche)) at 4°C. 800 µg of protein was then used for IPs. After clearing, lysates were incubated overnight with the antibody at 4°C. Venus-Unk was immunoprecipitated with 4 µg of rabbit anti-GFP antibody (Life Technologies). FLAG-HdcS and FLAG-D-Pax2 were immunoprecipitated with 3 µg of rabbit anti-FLAG (Fisher). The complexes were then immunoprecipitated for 3 hours using Protein G agarose beads (Thermo Scientific Pierce). After washes in 25 mM Tris pH 8, 150 mM NaCl, immunoprecipitated proteins were recovered by boiling the beads in 2× SDS-PAGE loading buffer and then subjected to SDS-PAGE followed by immunoblot with rabbit anti-GFP (1∶1000) and mouse monoclonal anti-FLAG M2 (1∶500, Agilent).
For overexpression of HA-Unkl COS-7 cells were grown in Dulbecco's modified Eagle's medium (Life Technologies) supplemented with 10% fetal bovine serum (Life Technologies), in a humidified atmosphere of 5% CO2 at 37°C. For transfection COS-7 cells were seeded at a density of 2.5×105 cells per well in Opti-MEM Reduced Serum Medium (Life Technologies) and transfected with 5 µg of pcDNA3-HA-unkl [42] using Lipofectamine 2000 (Life Technologies) according to the manufacturer's instructions.
Quantification and statistical analysis
For quantification of photoreceptor cell areas confocal images of MARCM clones stained with Bar were used. CD8-GFP expression was used to identify individual cell membranes, which were manually outlined at the level of R1/6 nuclei (identified by Bar staining) and cell areas were calculated using ImageJ. Clone and twin spot areas were manually outlined and the areas calculated using ImageJ. The numbers of active caspase 3 positive cells in eye discs were quantified using the quantification tool in Volocity (Perkin Elmer) from three dimensional confocal images of the whole disc with scans every 1 µm. Expression levels were determined in ImageJ using the Measure tool and four mutant clones/four adjacent areas of wild-type tissue were quantified for each genotype. Statistical analysis was performed in Graphpad Prism 5. Statistical significance was determined using an unpaired Student's t test for pairwise comparisons, or one way analysis of variance (ANOVA) with Dunnett's multiple comparison post hoc test for multiple comparisons to the control.
Supporting Information
Zdroje
1. RussellRC, FangC, GuanKL (2011) An emerging role for TOR signaling in mammalian tissue and stem cell physiology. Development 138: 3343–3356.
2. WullschlegerS, LoewithR, HallMN (2006) TOR signaling in growth and metabolism. Cell 124: 471–484.
3. ZoncuR, EfeyanA, SabatiniDM (2011) mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol 12: 21–35.
4. OrlovaKA, CrinoPB (2010) The tuberous sclerosis complex. Ann N Y Acad Sci 1184: 87–105.
5. LjungbergMC, BhattacharjeeMB, LuY, ArmstrongDL, YoshorD, et al. (2006) Activation of mammalian target of rapamycin in cytomegalic neurons of human cortical dysplasia. Ann Neurol 60: 420–429.
6. RussoE, CitraroR, ConstantiA, De SarroG (2012) The mTOR signaling pathway in the brain: focus on epilepsy and epileptogenesis. Mol Neurobiol 46: 662–681.
7. ZengLH, RensingNR, WongM (2009) The mammalian target of rapamycin signaling pathway mediates epileptogenesis in a model of temporal lobe epilepsy. J Neurosci 29: 6964–6972.
8. BatemanJM, McNeillH (2004) Temporal control of differentiation by the insulin receptor/tor pathway in Drosophila. Cell 119: 87–96.
9. BatemanJM, McNeillH (2006) Insulin/IGF signalling in neurogenesis. Cell Mol Life Sci 63: 1701–1705.
10. McNeillH, CraigGM, BatemanJM (2008) Regulation of neurogenesis and epidermal growth factor receptor signaling by the insulin receptor/target of rapamycin pathway in Drosophila. Genetics 179: 843–853.
11. HanJ, WangB, XiaoZ, GaoY, ZhaoY, et al. (2008) Mammalian target of rapamycin (mTOR) is involved in the neuronal differentiation of neural progenitors induced by insulin. Mol Cell Neurosci 39: 118–124.
12. OtaegiG, Yusta-BoyoMJ, Vergano-VeraE, Mendez-GomezHR, CarreraAC, et al. (2006) Modulation of the PI 3-kinase-Akt signalling pathway by IGF-I and PTEN regulates the differentiation of neural stem/precursor cells. J Cell Sci 119: 2739–2748.
13. FishwickKJ, LiRA, HalleyP, DengP, StoreyKG (2010) Initiation of neuronal differentiation requires PI3-kinase/TOR signalling in the vertebrate neural tube. Dev Biol 338: 215–225.
14. MalageladaC, Lopez-ToledanoMA, WillettRT, JinZH, ShelanskiML, et al. (2011) RTP801/REDD1 regulates the timing of cortical neurogenesis and neuron migration. J Neurosci 31: 3186–3196.
15. FelicianoDM, QuonJL, SuT, TaylorMM, BordeyA (2012) Postnatal neurogenesis generates heterotopias, olfactory micronodules and cortical infiltration following single-cell Tsc1 deletion. Hum Mol Genet 21: 799–810.
16. LafourcadeCA, LinTV, FelicianoDM, ZhangL, HsiehLS, et al. (2013) Rheb activation in subventricular zone progenitors leads to heterotopia, ectopic neuronal differentiation, and rapamycin-sensitive olfactory micronodules and dendrite hypertrophy of newborn neurons. J Neurosci 33: 2419–2431.
17. ZhuG, ChowLM, BayazitovIT, TongY, GilbertsonRJ, et al. (2012) Pten deletion causes mTorc1-dependent ectopic neuroblast differentiation without causing uniform migration defects. Development 139: 3422–3431.
18. GuertinDA, GunturKV, BellGW, ThoreenCC, SabatiniDM (2006) Functional genomics identifies TOR-regulated genes that control growth and division. Curr Biol 16: 958–970.
19. WeaverTA, WhiteRA (1995) headcase, an imaginal specific gene required for adult morphogenesis in Drosophila melanogaster. Development 121: 4149–4160.
20. Wolff T, Ready D (1993) Pattern Formation in the Drosophila Retina. In: M B, A M-A, editors. The Development of Drosophila melanogaster: CSHL Press. pp.1277–1326.
21. GlatterT, SchittenhelmRB, RinnerO, RoguskaK, WepfA, et al. (2011) Modularity and hormone sensitivity of the Drosophila melanogaster insulin receptor/target of rapamycin interaction proteome. Mol Syst Biol 7: 547.
22. HsuPP, KangSA, RamesederJ, ZhangY, OttinaKA, et al. (2011) The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science 332: 1317–1322.
23. YuY, YoonSO, PoulogiannisG, YangQ, MaXM, et al. (2011) Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science 332: 1322–1326.
24. MohlerJ, WeissN, MurliS, MohammadiS, VaniK, et al. (1992) The embryonically active gene, unkempt, of Drosophila encodes a Cys3His finger protein. Genetics 131: 377–388.
25. GoberdhanDC, ParicioN, GoodmanEC, MlodzikM, WilsonC (1999) Drosophila tumor suppressor PTEN controls cell size and number by antagonizing the Chico/PI3-kinase signaling pathway. Genes Dev 13: 3244–3258.
26. PinalN, GoberdhanDC, CollinsonL, FujitaY, CoxIM, et al. (2006) Regulated and polarized PtdIns(3, 4, 5)P3 accumulation is essential for apical membrane morphogenesis in photoreceptor epithelial cells. Curr Biol 16: 140–149.
27. HarringtonLS, FindlayGM, GrayA, TolkachevaT, WigfieldS, et al. (2004) The TSC1-2 tumor suppressor controls insulin-PI3K signaling via regulation of IRS proteins. J Cell Biol 166: 213–223.
28. GiotL, BaderJS, BrouwerC, ChaudhuriA, KuangB, et al. (2003) A protein interaction map of Drosophila melanogaster. Science 302: 1727–1736.
29. VeraksaA, BauerA, Artavanis-TsakonasS (2005) Analyzing protein complexes in Drosophila with tandem affinity purification-mass spectrometry. Dev Dyn 232: 827–834.
30. StenebergP, SamakovlisC (2001) A novel stop codon readthrough mechanism produces functional Headcase protein in Drosophila trachea. EMBO Rep 2: 593–597.
31. FuW, NollM (1997) The Pax2 homolog sparkling is required for development of cone and pigment cells in the Drosophila eye. Genes Dev 11: 2066–2078.
32. Miyamoto-SatoE, FujimoriS, IshizakaM, HiraiN, MasuokaK, et al. (2010) A comprehensive resource of interacting protein regions for refining human transcription factor networks. PLoS One 5: e9289.
33. HartmanNW, LinTV, ZhangL, PaqueletGE, FelicianoDM, et al. (2013) mTORC1 targets the translational repressor 4E-BP2, but not S6 kinase 1/2, to regulate neural stem cell self-renewal in vivo. Cell Rep 5: 433–444.
34. GaoX, PanD (2001) TSC1 and TSC2 tumor suppressors antagonize insulin signaling in cell growth. Genes Dev 15: 1383–1392.
35. PotterCJ, HuangH, XuT (2001) Drosophila Tsc1 functions with Tsc2 to antagonize insulin signaling in regulating cell growth, cell proliferation, and organ size. Cell 105: 357–368.
36. StockerH, RadimerskiT, SchindelholzB, WittwerF, BelawatP, et al. (2003) Rheb is an essential regulator of S6K in controlling cell growth in Drosophila. Nat Cell Biol 5: 559–565.
37. TaponN, ItoN, DicksonBJ, TreismanJE, HariharanIK (2001) The Drosophila tuberous sclerosis complex gene homologs restrict cell growth and cell proliferation. Cell 105: 345–355.
38. FelicianoDM, SuT, LopezJ, PlatelJC, BordeyA (2011) Single-cell Tsc1 knockout during corticogenesis generates tuber-like lesions and reduces seizure threshold in mice. J Clin Invest 121: 1596–1607.
39. ShiY, NollM (2009) Determination of cell fates in the R7 equivalence group of the Drosophila eye by the concerted regulation of D-Pax2 and TTK88. Dev Biol 331: 68–77.
40. LoncleN, WilliamsDW (2012) An interaction screen identifies headcase as a regulator of large-scale pruning. J Neurosci 32: 17086–17096.
41. GhabrialAS, LeviBP, KrasnowMA (2011) A systematic screen for tube morphogenesis and branching genes in the Drosophila tracheal system. PLoS Genet 7: e1002087.
42. LoresP, VisvikisO, LunaR, LemichezE, GaconG (2010) The SWI/SNF protein BAF60b is ubiquitinated through a signalling process involving Rac GTPase and the RING finger protein Unkempt. FEBS J 277: 1453–1464.
43. BlackshawS, HarpavatS, TrimarchiJ, CaiL, HuangH, et al. (2004) Genomic analysis of mouse retinal development. PLoS Biol 2: E247.
44. PunzoC, KornackerK, CepkoCL (2009) Stimulation of the insulin/mTOR pathway delays cone death in a mouse model of retinitis pigmentosa. Nat Neurosci 12: 44–52.
45. WeiJ, LiP, ChiribogaL, MizuguchiM, YeeH, et al. (2002) Tuberous sclerosis in a 19-week fetus: immunohistochemical and molecular study of hamartin and tuberin. Pediatr Dev Pathol 5: 448–464.
46. WeinkoveD, NeufeldTP, TwardzikT, WaterfieldMD, LeeversSJ (1999) Regulation of imaginal disc cell size, cell number and organ size by Drosophila class I(A) phosphoinositide 3-kinase and its adaptor. Curr Biol 9: 1019–1029.
47. LeeT, LuoL (1999) Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron 22: 451–461.
48. YangCH, SimonMA, McNeillH (1999) mirror controls planar polarity and equator formation through repression of fringe expression and through control of cell affinities. Development 126: 5857–5866.
49. RobertsonHM, PrestonCR, PhillisRW, Johnson-SchlitzDM, BenzWK, et al. (1988) A stable genomic source of P element transposase in Drosophila melanogaster. Genetics 118: 461–470.
50. ThibaultST, SingerMA, MiyazakiWY, MilashB, DompeNA, et al. (2004) A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat Genet 36: 283–287.
51. KockelL, KerrKS, MelnickM, BrucknerK, HebrokM, et al. (2010) Dynamic switch of negative feedback regulation in Drosophila Akt-TOR signaling. PLoS Genet 6: e1000990.
52. MachadoP, RostaingP, GuigonisJM, RennerM, DumoulinA, et al. (2011) Heat shock cognate protein 70 regulates gephyrin clustering. J Neurosci 31: 3–14.
Štítky
Genetika Reprodukční medicínaČlánek vyšel v časopise
PLOS Genetics
2014 Číslo 9
- Srdeční frekvence embrya může být faktorem užitečným v předpovídání výsledku IVF
- Souvislost haplotypu M2 genu pro annexin A5 s opakovanými reprodukčními ztrátami
- Primární hyperoxalurie – aktuální možnosti diagnostiky a léčby
- Příjem alkoholu a menstruační cyklus
- Mateřský haplotyp KIR ovlivňuje porodnost živých dětí po transferu dvou embryí v rámci fertilizace in vitro u pacientek s opakujícími se samovolnými potraty nebo poruchami implantace
Nejčtenější v tomto čísle
- Admixture in Latin America: Geographic Structure, Phenotypic Diversity and Self-Perception of Ancestry Based on 7,342 Individuals
- Nipbl and Mediator Cooperatively Regulate Gene Expression to Control Limb Development
- Genome Wide Association Studies Using a New Nonparametric Model Reveal the Genetic Architecture of 17 Agronomic Traits in an Enlarged Maize Association Panel
- Histone Methyltransferase MMSET/NSD2 Alters EZH2 Binding and Reprograms the Myeloma Epigenome through Global and Focal Changes in H3K36 and H3K27 Methylation
