-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Regulation of p53 and Rb Links the Alternative NF-κB Pathway to EZH2 Expression and Cell Senescence
Although the classical NF-κB pathway is frequently associated with the induction of cellular senescence and the senescence associated secretory phenotype (SASP), the role of the alternative NF-κB pathway, which is frequently activated in hematological malignancies as well as some solid tumors, has not been defined. We therefore investigated the role of the alternative NF-κB pathway in this process. Here we report that NF-κB2 and RelB, the effectors of the alternative NF-κB pathway, suppress senescence through inhibition of p53 activity. Using primary human fibroblasts, we demonstrate that this is accomplished through NF-κB2/RelB dependent control of a previously unknown pathway, incorporating regulation of CDK4 and 6 expression as well as regulators of p21WAF1 and p53 protein stability. Loss of NF-κB2/RelB results in suppression of retinoblastoma (Rb) tumour suppressor phosphorylation, which in turn leads to inhibition of EZH2 expression and de-repression of p53 activity. Interestingly, we find that CD40 ligand stimulation of cells from Chronic Lymphocytic Leukemia patients, which strongly induces the alternative NF-κB pathway, also induces EZH2 expression. We propose that the alternative NF-κB pathway can promote tumorigenesis through suppression of p53 dependent senescence, a process that may have relevance to cancer cells retaining wild type p53.
Published in the journal: . PLoS Genet 10(9): e32767. doi:10.1371/journal.pgen.1004642
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004642Summary
Although the classical NF-κB pathway is frequently associated with the induction of cellular senescence and the senescence associated secretory phenotype (SASP), the role of the alternative NF-κB pathway, which is frequently activated in hematological malignancies as well as some solid tumors, has not been defined. We therefore investigated the role of the alternative NF-κB pathway in this process. Here we report that NF-κB2 and RelB, the effectors of the alternative NF-κB pathway, suppress senescence through inhibition of p53 activity. Using primary human fibroblasts, we demonstrate that this is accomplished through NF-κB2/RelB dependent control of a previously unknown pathway, incorporating regulation of CDK4 and 6 expression as well as regulators of p21WAF1 and p53 protein stability. Loss of NF-κB2/RelB results in suppression of retinoblastoma (Rb) tumour suppressor phosphorylation, which in turn leads to inhibition of EZH2 expression and de-repression of p53 activity. Interestingly, we find that CD40 ligand stimulation of cells from Chronic Lymphocytic Leukemia patients, which strongly induces the alternative NF-κB pathway, also induces EZH2 expression. We propose that the alternative NF-κB pathway can promote tumorigenesis through suppression of p53 dependent senescence, a process that may have relevance to cancer cells retaining wild type p53.
Introduction
In mammalian cells the NF-κB family of transcription factors consists of five subunits, RelA (p65), c-Rel, RelB, NF-κB1 (p105/p50) and NF-κB2 (p100/p52), which form a wide variety of homodimeric and heterodimeric complexes [1], [2]. In most normal, unstimulated cells, NF-κB complexes are held in an inactive form, bound to one of a family of inhibitory proteins, termed IκBs. The precursor proteins p100 and p105 can also function as IκBs, prior to their processing to p52 and p50, which function as nuclear regulatory subunits. The classical (or canonical) NF-κB pathway typically leads to the induction of RelA or c-Rel containing complexes and involves the degradation of IκBα in a manner dependent on IκB kinase (IKK) β and the IKK regulatory subunit NEMO (IKKγ). The alternative (or non-canonical) pathway, involves the inducible processing of p100 to p52, leading to the induction of p52/RelB containing complexes, and is dependent on IKKα and NF-κB inducing kinase (NIK).
Aberrantly active NF-κB is associated with many diseases, including cancer [3]. The ability to both respond to and induce inflammatory stimuli is an important component of NF-κB's role in disease [3]. NF-κB also has other functions and can contribute to tumorigenesis through inducing proliferation, metastasis as well as resistance to apoptosis [4]. However, NF-κB can also exhibit apparently contradictory functions, more akin to those of a tumor suppressor. These include pro-apoptotic activity in response to some stimuli and induction of cellular senescence [4], [5]. NF-κB is also associated with the senescence associated secretory phenotype (SASP), which can exhibit tumor promoting properties but also contribute to the effectiveness of cancer therapy [6], [7]. Other studies have suggested a role for NF-κB in protecting against senescence [8]. One common feature of these studies has been a focus on the classical branch of the NF-κB pathway and any role for the alternative NF-κB pathway has not generally been considered or functionally analyzed.
A possible explanation for apparently contradictory functions of NF-κB lies in the ‘tumor suppressor status’ of the cell. A number of studies have shown that tumor suppressors can modulate NF-κB activity and function [4]. The most studied example of tumor suppressor crosstalk with NF-κB involves p53 [2], [4], [9]–[11]. NF-κB and p53 can both be activated by many of the same stimuli with a common link frequently being DNA damaging agents, which include reactive oxygen species (ROS) [2], [9]–[12]. Crosstalk between these factors can take many forms, with reports indicating both antagonistic and co-operative behavior. Although often appearing contradictory, one conclusion of these studies is that p53 and NF-κB can modulate each-others activity, and consequently cell survival, but that the exact outcome is dependent upon the cell context. As many of these studies have been performed in cancer cell lines exhibiting a variety of genetic backgrounds, with a range of different stimuli, variability in the nature and outcome of any crosstalk might be expected. We have previously identified crosstalk between the alternative NF-κB pathway subunit p52 and p53, involving p53 modulation of p52 homodimer transcriptional activity, by inducing a change from p52/Bcl3 to p52/HDAC complexes, in addition to direct recruitment of p52 to p53 target gene promoters [13], [14]. However, there remain many unanswered questions, including how the effectors of the alternative NF-κB pathway, p52 and RelB can affect p53 dependent senescence. Moreover, whether there also exists crosstalk between these NF-κB proteins and another important tumor suppressor, the retinoblastoma gene product, Rb, is largely unexplored.
Genes whose promoters and enhancers are regulated by both p53 and NF-κB have the potential to act as ‘nodes of integration’ between these pathways. That is, they form a route through which both factors come together to influence cell fate. A number of such genes have been identified, including DR5 and Caspase 10 [15], [16]. We were interested in identifying genes encoding chromatin remodellers and transcriptional co-regulators that exhibited co-operative or antagonistic regulation by NF-κB and p53 as these have the potential to re-program the transcriptional ‘landscape’ of the cell [17]. A candidate gene that fitted this category was the Polycomb protein enhancer of zeste homolog 2 (EZH2), a histone H3 K27 methylase and component of the PRC2 complex, which previously has been shown to be repressed by p53 [18]. p53 repression of EZH2 expression is thought to be indirect, resulting from transcriptional upregulation of p21WAF1 expression [18]. This in turn leads to Rb mediated repression of E2F activity, a key transcription factor driving EZH2 expression [19].
EZH2 is a tumor promoter and is found over-expressed or mutated in many solid tumors and hematological malignancies [20]. A component of its ability to drive tumorigenesis derives from its ability to suppress cellular senescence [21]–[23]. This is achieved, in part, through EZH2 repression of the CDKN2A locus [21], [24], encoding the CDK inhibitor p16Ink4a and the tumor suppressor p14ARF, which can induce p53 activity through binding its inhibitor Mdm2. p16Ink4a and p14ARF are both important regulators of cell senescence [25]. EZH2 has also been previously linked to NF-κB activity, where in ER negative breast cancer it can function as a coactivator, independent of its methylase activity, for both RelA and RelB [26].
In this report, we define a regulatory network through which the alternative NF-κB pathway regulates Rb activity and consequently EZH2 expression. We demonstrate that EZH2 functions as a critical ‘node’ of crosstalk between NF-κB and p53 that controls a gene regulatory network through which p52 and RelB act to suppress p53 and Rb mediated cellular senescence.
Results
Human dermal fibroblasts grown under normoxic conditions contain a basal level of NF-κB2 and p53 activity
To investigate the function of the alternative NF-κB pathway in untransformed cells, we analyzed its expression in non-immortalized, normal human dermal (NHD) fibroblasts, cultured for a limited number of passages. This revealed constitutive processing of the p100 NF-κB subunit to p52 as well as a basal level of the p53 tumor suppressor (Fig. 1A). To determine if these resulted from oxidative stress due to normoxic culture conditions, the NHD fibroblasts were grown under low oxygen tension (3% O2 or treated with the antioxidant epigallocatechin-3-gallate (ECGC). This resulted in loss of p53 while processing of p100 to p52 was unaffected, suggesting the latter effect results from factors present in the media (Fig. 1A & S1A). The Ataxia Telangiectasia Mutated (ATM) kinase can be activated by ROS, independently of DNA damage [27] and consistent with this we found that the basal levels of p53 seen in this experiment as well as inducible levels seen in later experiments were reduced by treatment with an ATM kinase inhibitor (Fig. S1B). Interestingly we observed that 7 days after treatment of these cells with hydrogen peroxide (H2O2) to induce cellular senescence, a significant reduction in the processing of p100 to p52 occurred, concomitant with activation of p53 suggesting a possible antagonistic relationship between these factors in these cells (Figs. S1C&D).
Fig. 1. EZH2 is an NF-κB regulated target gene. 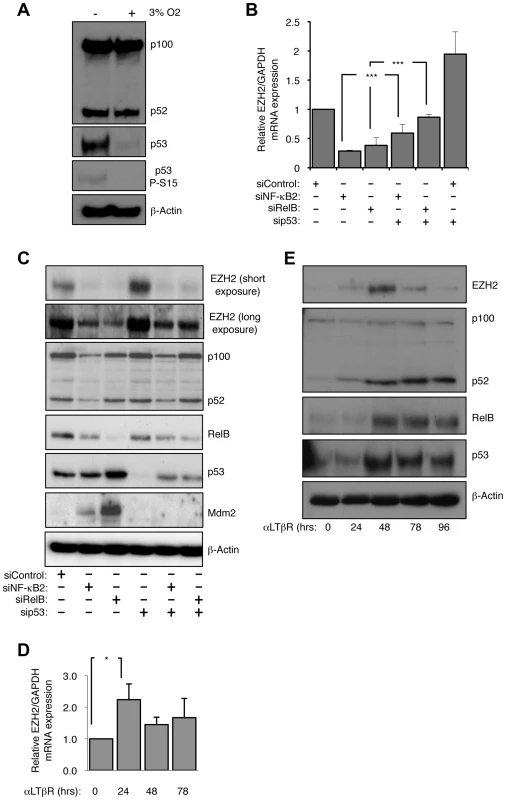
(A) The basal level of p53 protein in NHD fibroblasts is ROS dependent. NHD fibroblasts were grown under normoxia at 3% O2 for 7 days before western blot analysis. (B & C) siRNA mediated knock-down of NF-κB2 and RelB leads to a reduction in EZH2 mRNA and protein levels. RNA (B) or protein (C) was prepared from NHD fibroblasts treated with the indicated siRNAs 48 hours after transfection and Q-PCR or western blot analysis was performed to determine EZH2 expression. *** P≤0.001. (D & E) Lymphotoxin β receptor stimulation leads to induction of EZH2 expression. NHD fibroblasts were treated with LTβR agonist antibody for the times indicated and either Q-PCR (D) or western blot analysis (E) was performed to determine the expression of EZH2 (D) or EZH2, p52/p100, RelB and p53 (E). * P≤0.05. Depletion of NF-κB2 or RelB results in repression of EZH2 expression
We decided to exploit the observation that NHD fibroblasts exhibited activation of both the alternative NF-κB pathway and p53 to determine how these pathways might be integrated in a non-cancerous, non-immortalized cellular context. Consequently, we used siRNAs to deplete p53 and the alternative NF-κB pathway subunits p52/p100 (encoded by the NFKB2 gene) and RelB in the NHD fibroblasts. siRNA knockdowns of p52/p100 expression will henceforth be referred to as NF-κB2, while analysis of the individual proteins will refer to either p52 or p100.
As described above, a candidate target for NF-κB/p53 crosstalk was EZH2, a histone H3 K27 methylase and component of the PRC2 complex, previously shown to be repressed by p53 [18]. As expected siRNA knockdown of p53 resulted in an increase in EZH2 RNA and protein levels 48 hours after transfection (Fig. 1B&C). However, siRNA depletion of both alternative NF-κB pathway subunits and the p52 coactivator Bcl3 had the opposite effect, leading to almost complete loss of EZH2 expression (Fig. 1B&C, Fig. S1E–G). Knockdown of NF-κB2/RelB or Bcl3 with p53 resulted in a partial rescue of EZH2 protein levels. These effects were also seen with an EZH2 promoter luciferase construct, where loss of NF-κB2/RelB resulted in less promoter activity, while depletion of p53 had a strong stimulatory effect (Fig. S1H). As reported previously [18], depletion of the p53 target, the CDK inhibitor p21WAF1, also induced EZH2 promoter activity (Fig. S1H). These data suggested that the alternative NF-κB pathway and p53 antagonistically regulate EZH2 expression and that this is mediated, at least in part, through direct effects on EZH2 transcription driven by its promoter.
We next determined if induction of the alternative NF-κB pathway by a physiological stimulus would also regulate EZH2 expression. To achieve this NHD fibroblasts were treated with a lymphotoxin β receptor (LTβR) agonist antibody. Consistent with the results obtained with basal level activity of the alternative pathway, LTβR activation resulted in increased levels of EZH2 protein and mRNA (Fig. 1E & F).
In many experiments, with different siRNAs, we noted a partial depletion of RelB levels seen upon NF-κB2 knockdown. However, as there is no effect of NF-κB2 siRNAs on RelB mRNA levels and vice versa (Fig. S1F) this probably represents RelB protein instability due to an inability to form homodimers, with the remaining RelB dimerized to p50 with which it forms an active complex [28].
EZH2 expression is induced upon CD40L stimulation of primary B-cell chronic lymphocytic leukemia (CLL) cells
To extend our observation that LTβR stimulation induced EZH2 expression, we were interested in whether this pathway was also associated with activation of the alternative NF-κB pathway in a pathological setting. Primary chronic lymphocytic leukemia (CLL) cells from patients can be cultured in vitro and induced to proliferate when stimulated with CD40 ligand (CD40L), which induces both the classical and alternative NF-κB pathways [29], [30] (Fig. S2A). Significantly, we observed a CD40L dependent increase in EZH2 mRNA and protein levels that was seen up to 7 days after plating (Fig. 2A&B). The reproducibility of this effect between patients was confirmed with analysis of 4 different isolates (Fig. S2B). Western blot analysis confirmed activation of the alternative NF-κB pathway, with both an increase in nuclear and overall levels of NF-κB2 and RelB being observed (Fig. 2B & S2B). The latter likely results from activation of the classical pathway by CD40L, which can ‘prime’ the alternative NF-κB pathway through inducing NF-κB2 and RelB expression levels [1]. We exploited this characteristic to confirm the role of NF-κB in EZH2 induction in CLL cells: treatment with the IKKβ inhibitor TPCA-1 efficiently blocked the induction of NF-κB2/RelB protein and mRNA levels and also abolished induction of EZH2 (Fig. 2C & D).
Fig. 2. CD40 stimulation leads to NF-κB activation and CLL induction in Chronic Lymphocytic Leukemia cells. 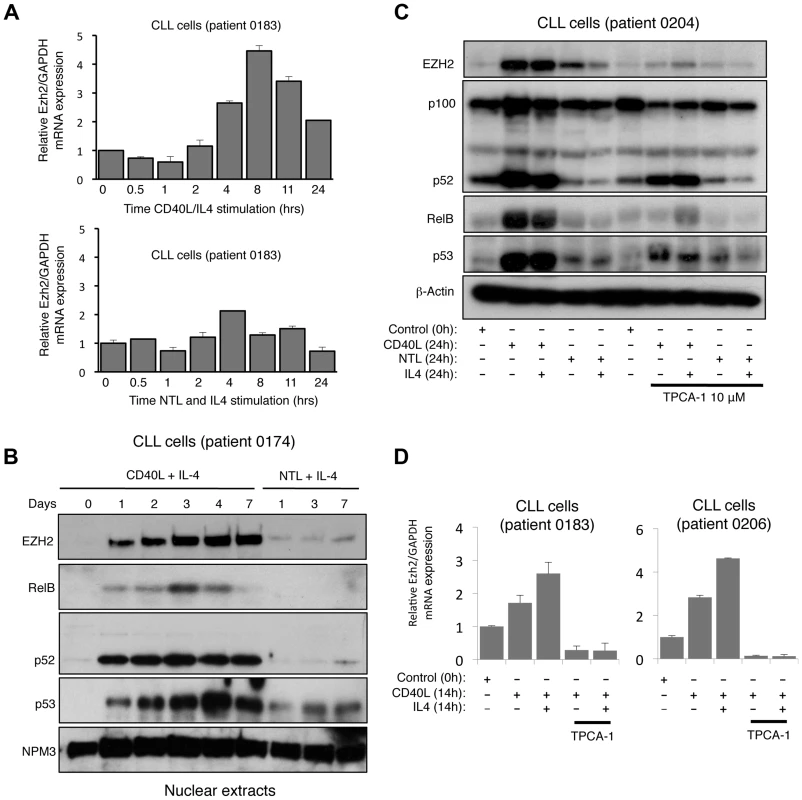
(A) Analysis of EZH2 mRNA expression in CLL cells. RNA was prepared from CLL cells stimulated with CD40L/IL4 expressing mouse fibroblasts or with untransfected fibroblasts (NTL) and IL4 for the indicated times and Q-PCR analysis of EZH2 expression was performed. (B) Analysis of EZH2 protein level in CLL cells. Western blot analysis of nuclear extracts from CLL cells stimulated with CD40L/IL4 or untransfected fibroblasts (NTL) and IL4 for the indicated times. (C & D) EZH2 protein and RNA levels in CLL cells is NF-κB dependent. Whole cell protein lysates (C) and RNA (D) were prepared from CLL cells stimulated for 24 hours with CD40L/IL4 and treated with the IKKβ inhibitor TPCA-1 where indicated. In addition to further confirmation of EZH2 regulation by a physiological inducer of the alternative NF-κB pathway this data also demonstrated that this pathway is not restricted to fibroblasts. Interestingly, we also observed that in most patient cells (except 0205 where p53 appears to be mutant), CD40L stimulation also induced p53 protein levels (Fig. 2A & B, S2B), an effect also seen with LTβR stimulation (Fig. 1E). As these CLL cells are being induced to proliferate (Fig. S2A) this suggests ongoing suppression/modulation of p53 activity and function.
NF-κB2 and RelB suppress p53 dependent senescence
When analyzing siRNA depletion of NF-κB2 and RelB in NHD fibroblasts, we also observed that cells ceased to proliferate, changed morphology and after 7 days in culture, using the acidic β galactosidase assay, were found to enter a senescent state (Fig. 3A&B). This effect was confirmed with different NF-κB2 and RelB siRNAs (Fig. S3A). These effects were also associated with induction of reaction oxygen species (ROS) and loss of Lamin B1, both markers of senescence [12], [31] (Fig. 3C, Fig. S3B). Treatment of cells with the antioxidant N-acetyl cysteine (NAC) prevented senescence induced upon NF-κB2 and RelB depletion, an effect also seen with an ATM inhibitor, or ECGC (Fig. 3A, Fig. S3C–E). By contrast, depletion of the NF-κB1 (p50/p105) subunit had no detectable effect on senescence in this system (Fig. S3F). Importantly induction of senescence and ROS upon depletion of NF-κB2, RelB and Bcl-3 was p53 dependent (Fig. 3D, S3G). p53 dependent senescence was also observed upon depletion of the p52 coactivator Bcl3 (Fig. 3D). Activation of the alternative NF-κB pathway through LTβR stimulation also inhibited the basal level of senescence in NHD fibroblasts (Fig. 3E). Therefore we concluded that in NHD fibroblasts, the basal level of p53 activity induced by oxidative stress is suppressed by the alternative NF-κB pathway. Depletion of either of 3 components of this pathway, results in loss of this suppression leading to p53 dependent production of ROS and senescence.
Fig. 3. The alternative NF-κB pathway suppresses p53 mediated senescence in primary fibroblasts. 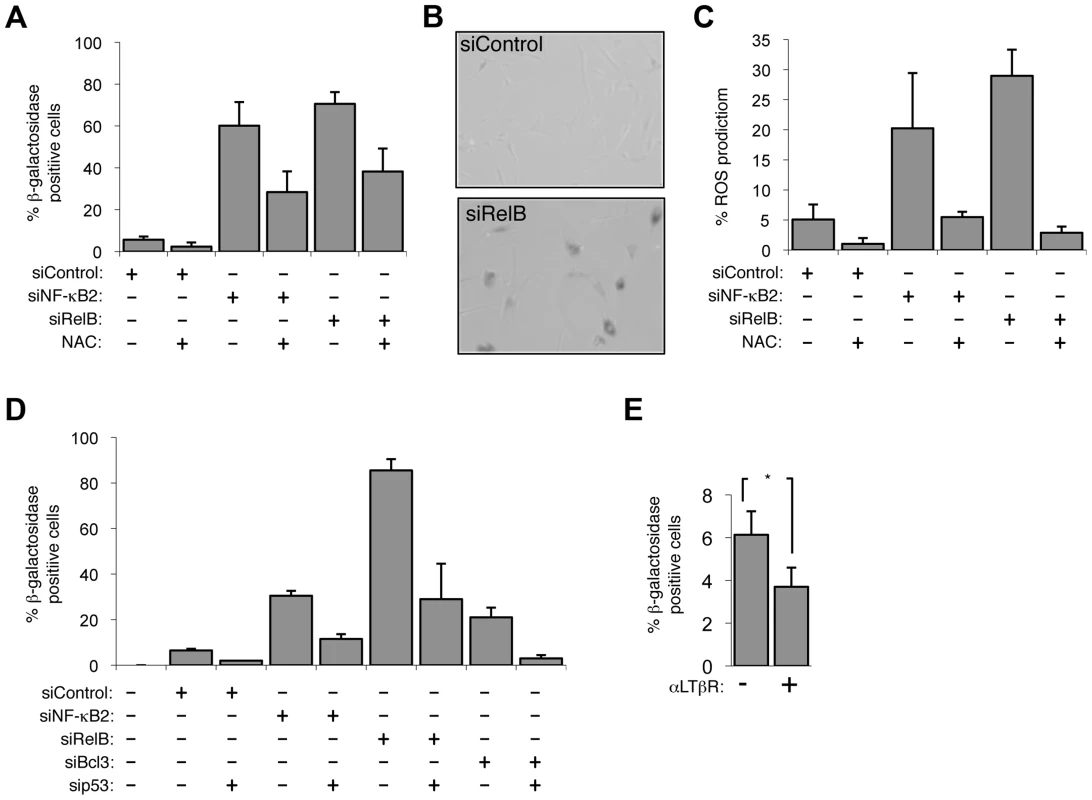
(A & B) Senescence induced by siRNA knock down of NF-κB2 and RelB is ROS dependent. NHD fibroblasts were transfected with the siRNAs shown and treated, where indicated, 2 days later with the anti-oxidant N-Acetyl cysteine (NAC). 7 days after transfection cells analyzed for senescence by acidic β-galactosidase staining (A). An image of RelB siRNA transfected cells after staining is shown (B). (C) siRNA mediated knock down of NF-κB2 and RelB induces ROS production. NHD fibroblasts were transfected with the siRNAs shown and treated, where indicated, 2 days later with NAC. After 7 days they were incubated for 30 minutes with 5 mM DCF-DA and analyzed by FACs. The percentage of cells with higher than baseline ROS levels are shown. (D) siRNA knock down of NF-κB2, RelB and Bcl3 induce cellular senescence in a p53 dependent manner. NHD fibroblasts were transfected with the listed siRNAs and analyzed for senescence by β-galactosidase staining after 7 days. (E) Lymphotoxin β receptor stimulation represses basal level senescence in fibroblasts. NHD fibroblasts were treated with LTβR agonist antibody and after 7 days analyzed for senescence by β-galactosidase staining. * P≤0.05. EZH2 is a major effector of alternative NF-κB pathway transcriptional effects
We next examined the effect of depleting EZH2 itself. Since EZH2 can act to suppress senescence [21]–[23] we were interested in whether regulation of its expression provided a mechanism through which the alternative NF-κB pathway and p53 could exert antagonistic effects on senescence. Consistent with this hypothesis we observed that cells treated with an EZH2 siRNA became senescent and also showed elevated levels of ROS (Fig. 4A & B, S4A). Co-depletion of EZH2 with NF-κB2, RelB, Bcl-3 and p53 revealed that while EZH2 associated ROS production and senescence is p53 dependent, no additional effects were seen with the NF-κB subunits (Fig. 4A & B). This is consistent with EZH2 being a downstream ‘effector’ and master regulator of NF-κB2/RelB's ability to suppress p53 dependent senescence.
Fig. 4. EZH2 siRNA associated senescence and ROS production is p53 dependent. 
(A & B) NHD fibroblasts were transfected with the siRNAs shown and analyzed for senescence (A) and ROS production (B) after 7 days. EZH2 represses expression from the CDKN2A locus encoding the CDK inhibitor p16Ink4a and the tumor suppressor p14ARF, both of which are important regulators of senescence [21], [24]. Interestingly, 48 hours after siRNA transfection, when we analyzed effects on gene expression, no changes in p16Ink4a or p14ARF protein or mRNA levels could be observed (Fig. S4B & C). However, 7 days after transfection, when cells begin to senesce, this had changed with induction of the mRNAs for both factors being seen (Fig. S4C). Moreover, siRNA depletion of p14ARF activity confirmed its requirement for NF-κB2, RelB and EZH2 induced senescence (Fig. S4D). Therefore, while these proteins are required, as expected, for eventual induction of the senescent phenotype, they did not seem to be directly involved in the early regulatory events linking NF-κB2, RelB to EZH2 and p53 function that are the focus of this study.
To further investigate the significance of this regulatory pathway and gain insights into the early mechanisms through which these effects were achieved prior to induction of p16Ink4a or p14ARF, gene expression profiling was performed on cells 48 hours after transfection with siRNAs targeting NF-κB2, RelB, EZH2 and p53 (accession number for microarray data is E-MTAB-1593). Significantly, a cluster of genes co-regulated by EZH2, NF-κB2 and RelB were also antagonistically regulated by p53, and co-depletion of p53 generally abolished these effects (Fig. 5A&B, see also Figure S5A and Table S1). More detailed analysis of the 975 genes significantly (>1.5 fold) affected by p53 depletion revealed a group of genes normally repressed by p53 (which are therefore induced upon p53 siRNA treatment), antagonistically regulated by EZH2, NF-κB2 and RelB (Fig. 5C, full gene list in Table S2). The potential importance of EZH2 regulation as an ‘effector’ of antagonistic crosstalk between NF-κB and p53 was demonstrated by analysis of the genes where NF-κB and p53 depletions had directly opposing effects: of the 142 RelB/p53 genes in this category, 93 were also regulated by EZH2, while of the 82 genes similarly regulated by NF-κB2/p53, 60 genes were also regulated by EZH2 (Table S2). Interestingly, of the smaller subset of genes regulated in the same manner by NF-κB and p53, that is where both either induce or repress, EZH2 depletion was found to have generally minimal effects (7/54 for RelB/p53 and 1/17 for NF-κB2/p53) (Table S2). The overlap between NF-κB2 and RelB regulated genes was less than expected, given that these factors are often depicted as being favored dimer partners. This most probably results from circumstances where the effect of one dimer partner fell below the 1.5× effect cut off used in this analysis. However, it may also result from compensation by other NF-κB dimer complexes in which these proteins participate, such as p52 homodimer/Bcl3 or p50/RelB complexes.
Fig. 5. EZH2 is a critical effector of an antagonistic cross-talk between NF-κB and p53. 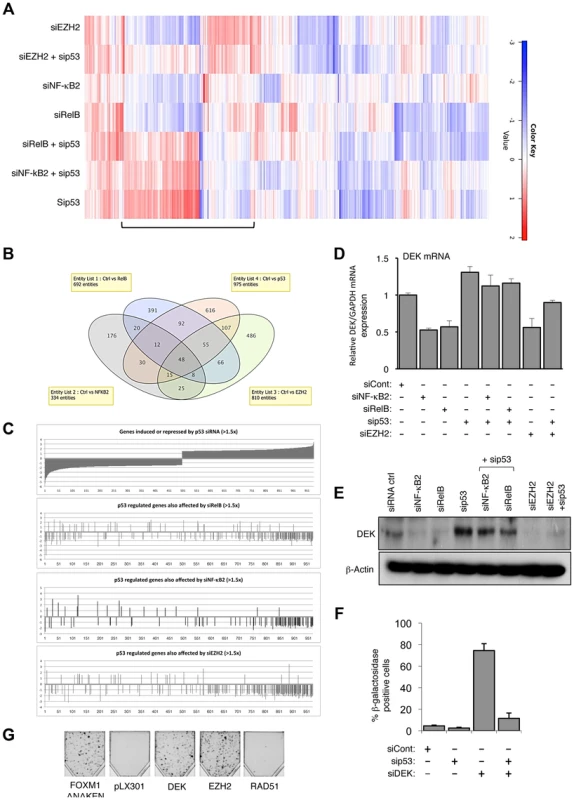
(A) Heatmap showing effects on gene expression of NHD fibroblasts depleted for EZH2, NF-κB2, RelB and p53. NHD fibroblasts were transfected in triplicate with the listed siRNAs. After 48 hours RNA was extracted for microarray analysis. Shown with a bar is the group of genes where EZH2, NF-κB2 and RelB antagonize p53 dependent gene expression. (B) A subset of genes is co-regulated by NF-κB2, RelB, EZH2 and p53 in NHD fibroblasts. (C) Graphical representation of the 975 p53 regulated genes whose expression changes >1.5 fold that are also regulated (>1.5×) by NF-κB2, RelB and Ezh2. (D–E) NF-κB2, RelB, EZH2 regulate DEK expression in NHD fibroblasts. RNA (D) and whole cell protein lysates (E) was prepared from NHD fibroblasts treated with the indicated siRNAs and Q-PCR or western blot analysis of DEK expression was performed. Note that (E) is a reprobing of blots used in Fig. 1C and the β-actin blot shown here is the same as in that figure. (F) siRNA mediated knock down of DEK induces cellular senescence. NHD fibroblasts were transfected with the siRNAs shown and analyzed for senescence after 7 days. (G) EZH2 and DEK alone can rescue senescence. Fibroblasts conditionally immortalized with temperature sensitive T antigen (described in Rovillain et al.) were shifted to the non-permissive temperature and subjected to a clonogenic assay with and without expression of the indicated genes. The panel shows an image of cells upon completion of the assay. Genes regulated by NF-κB/EZH2/p53 are part of a senescence associated gene signature
An NF-κB associated gene signature has been previously described in a study of senescence induced in conditionally immortalized human fibroblasts upon activation of the p16-pRB and p53-p21 tumor suppressor pathways [32]. We were therefore interested in any similarities between the gene signature we had identified and that seen by Rovillain et al., especially as EZH2 expression was also downregulated in this study [32]. Therefore, we integrated our gene expression signature with that of Rovillain et al., to produce a combined heat map (Fig. S5B). This exercise confirmed that a significant proportion of the genes we had previously identified as belonging to the NF-κB2/RelB/EZH2 regulatory network also formed part of the previously identified NF-κB dependent senescence gene signature. Importantly, the heat map reveals that genes co-regulated by EZH2, NF-κB2 and RelB were also antagonistically regulated by p53, and that co-depletion of p53 generally abolished these effects.
To confirm that genes identified as being regulated by the NF-κB/EZH2 pathway did contribute towards suppression of p53 mediated cell senescence, we analyzed the DEK oncogene and histone chaperone that also been described as an inhibitor of senescence [33], [34]. Our microarray analysis revealed its expression to be down-regulated upon NF-κB2, RelB or EZH2 depletion but induced upon treatment with the p53 siRNA, which we confirmed by Q-PCR and western blot analysis (Fig. 5D&E). siRNA depletion of DEK resulted in a striking, p53 dependent induction of senescence (Fig. 5F) but did not affect EZH2 or p53 mRNA levels, consistent with it being a downstream effector of this regulatory pathway (Fig. S5C–E). By contrast, as a control, siRNA depletion of tp53INP1, which is induced upon NF-κB2, RelB or EZH2 depletion but down-regulated upon treatment with the p53 siRNA did not induce senescence or affect senescence induced upon NF-κB2 or RelB depletion (Fig. S5F&G).
The importance of both EZH2 and DEK as regulators of senescence was confirmed by performing a reconstitution experiment, using the conditionally immortalized fibroblast cells described in [32]. Lentiviral gene transfer was used to express DEK and EZH2 in cells at 34°C, before being shifted to 38°C to induce Rb and p53 dependent cellular senescence. A constitutively active FOXM1ΔNΔKEN mutant was included as a positive control [32]. Expression of both EZH2 and DEK proteins was found to suppress p53 and Rb induced senescence, as seen by the formation of colonies in the clonogenic assay (Fig. 5G).
NF-κB2/RelB/EZH2 regulation of ROS production requires RAC1 and CDC42 activity
As described above, p53 dependent generation of ROS is a requirement for senescence induced upon depletion of NF-κB2, RelB and EZH2. We therefore investigated if any of the genes regulated by this pathway could account for these effects. Although NF-κB activity has previously been associated with regulation of ROS levels, we did not find any well-known target genes in our list, such as Manganese Superoxide Dismutase (MnSOD, also known as SOD2). Further analysis using Q-PCR and western blot confirmed that its levels were not significantly changing upon the treatments used in this study (Fig. S6A&B). However, analysis of the genes in Table S2 revealed that expression of Rac GTPase Activating Protein 1 (RACGAP1) fell into the category of genes whose expression was downregulated upon depletion of NF-κB2/RelB/EZH2 and were antagonistically regulated by p53 (Fig. 6A), a result confirmed by Q-PCR (Fig. 6B). Moreover, analysis of the senescence gene expression signature of obtained by Rovillain et al., also showed that RACGAP1 expression was down-regulated upon senescence arrest and reversed upon senescence bypass but was not commented on or further analyzed in that study [32]. RACGAP1 regulates the activity of RAC1 (ras-related C3 botulinum toxin substrate 1) and CDC42 (Cell Division Cycle 42), both members of the RHO family of small GTP binding proteins [35]. Importantly, in the context of this study, both RAC1 and CDC42 can regulate the NADPH oxidase and thereby ROS production [36], [37]. Consistent with the hypothesis that down regulation of RACGAP1 could account, at least in part, for the effects seen on ROS levels, its siRNA depletion resulted in an increase in ROS, while MnSOD did not (Fig. 6C). Moreover, depletion of either RAC1 or CDC42 both inhibited the increases in ROS levels seen upon down regulation of NF-κB2 or RelB, to a similar level seen with the p53 siRNA (Fig. 6D). As a control, no effect was seen with PUMA, a downstream p53 target previously linked to ROS production (Fig. 6D) [12].
Fig. 6. The alternative NF-κB pathway suppresses ROS production through the regulation of RacGAP. 
(A) Table summarizing the fold effect on RACGAP1 expression induced by transfection of the listed siRNAs in the microarray analysis. (B) NF-κB2, RelB and EZH2 regulate RACGAP1 expression in NHD fibroblasts. RNA was prepared from NHD fibroblasts treated with the indicated siRNAs and Q-PCR analysis of RACGAP1 expression was performed. (C) siRNA mediated knock-down of RACGAP1 induces ROS production. NHD fibroblasts were transfected with the siRNAs shown and analyzed for ROS production after 4 days. (D) ROS production induced by siRNA mediated knock down of NF-κB2 and RelB is dependent upon Rac1 and Cdc42. NHD fibroblasts were transfected with the siRNAs shown and analyzed for ROS production after 4 days. Taking these results together with those from Figures 1–5, this demonstrated that regulation of EZH2 by the alternative NF-κB pathway provides a mechanism to control p53 dependent cellular senescence. We next investigated the mechanisms through which regulation of EZH2 by NF-κB is achieved.
p52 and RelB regulate EZH2 expression through modulation of Rb activity
EZH2 expression is regulated by Rb/E2F signaling [19]. We therefore investigated the effect of NF-κB2 and RelB siRNA depletion, and found strong inhibition of Rb phosphorylation, as well as reductions in the levels of Rb family members p107 and p130, (Fig. 7A, Fig. S7A). Moreover, LTβR stimulation also induced Rb phosphorylation (Fig. S7B), consistent with the induction of EZH2 seen before (Fig. 1D & E). Confirming the importance of this pathway, co-depletion of Rb rescued loss of EZH2 expression after treatment with both NF-κB2 and RelB siRNAs (Fig. 7A). Therefore NF-κB2 and RelB regulation of EZH2 is Rb dependent.
Fig. 7. NF-κB2 and RelB regulate EZH2 in an Rb/E2F dependent manner. 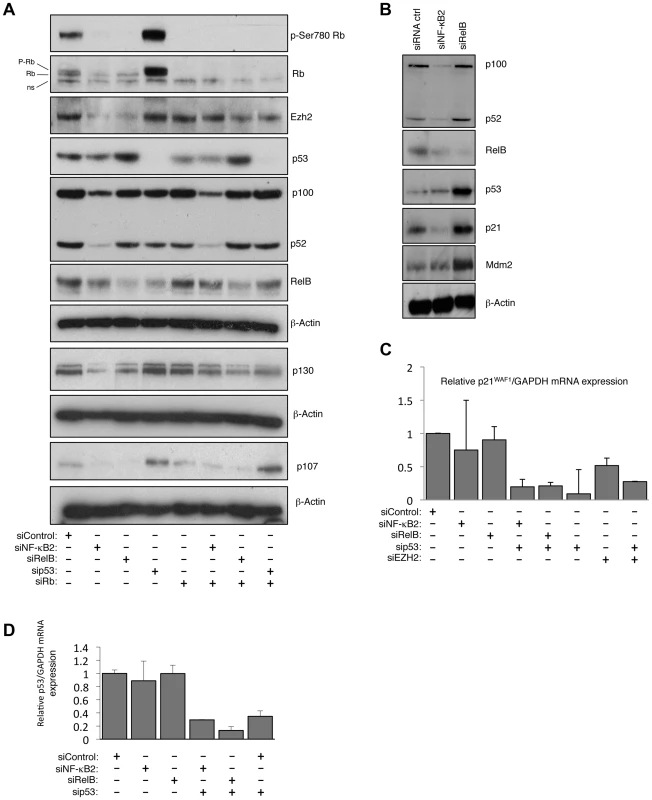
(A) siRNA mediated knock-down of NF-κB2 and RelB leads to a reduction of Rb- phosphorylation. Western blot analysis of whole cell lysates prepared from NHD fibroblasts 48 hours after transfection with the indicated siRNAs. (B) siRNA mediated knock down of RelB leads to accumulation of p53 and p21WAF1 protein level. Western blot analysis of NHD fibroblasts treated with the indicated siRNAs. Whole cell lysates were prepared 48 hours after transfection. (C & D) NF-κB2 and RelB depletion does not affect p21WAF1 or p53 mRNA levels. RNA was prepared from NHD fibroblasts treated with the indicated siRNAs and Q-PCR analysis of p21WAF1 (C) or p53 (D) expression was performed. RelB regulates p21WAF1 and p53 protein stability
Further investigation revealed unexpected differences between the pathways controlled by RelB and NF-κB2. Interestingly, loss of RelB but not NF-κB2 resulted in a strong induction of p21WAF1 and p53 protein (Fig. 7B but see also Fig. 1C). Additional investigation revealed that this was a consequence of an effect of RelB on p21WAF1 and p53 protein stability rather than gene transcription: Q-PCR analysis showed that RelB depletion did not significantly affect p21WAF1 (CDKN1A) or p53 mRNA levels (Fig. 7C&D, see also Table S3), despite the increase in p53 protein levels. However depletion of p53 in these cells did reduce overall p21WAF1 expression (Fig. 7C). This suggested two effects. Firstly that the basal level of p53 in these cells is required for the basal level of p21WAF1 expression, while secondly RelB acts to suppress p53 and p21WAF1 protein stability, thereby permitting cell proliferation. In this model loss of RelB leads to stabilization of p21WAF1 protein, thus inhibiting Rb phosphorylation by Cyclin/CDK complexes, which in turn suppresses E2F induction of EZH2 expression.
NF-κB2 regulates CDK4 and CDK6 expression
Since depletion of NF-κB2 did not lead to induction of p53 or p21WAF1 (Fig. 7B) we investigated if there was an alternative explanation for its effect on Rb phosphorylation. Analysis of our microarray data confirmed that there were no effects on the CDK inhibitors analyzed (Table S3). However, effects on other cell cycle regulatory proteins were seen (Table S4), and in particular we observed NF-κB2 specific downregulation of CDK4 and CDK6, both of which are known to directly phosphorylate Rb [38]. Q-PCR and western blot analysis confirmed that CDK4 and CDK6 expression is selectively lost upon NF-κB2 depletion in NHD fibroblasts, with no effect being seen with the RelB siRNA (Fig. 8A–C). Interestingly, in U2OS cells, we have also observed that CDK4 expression is lost upon siRNA depletion of NF-κB2 [39]. CDK4's key regulatory role as an effector of the effects seen upon depletion of NF-κB2 was confirmed when its siRNA depletion resulted in loss of Rb phosphorylation, reduction in EZH2 expression and induction of senescence (Fig. 8D&E, S8A & B). Furthermore, CDK4 re-expression partially recovered the loss of RB phosphorylation and downregulation of EZH2 expression seen upon depletion of NF-κB2 (Fig. 8F). This was not seen with RelB siRNA treatment.
Fig. 8. NF-κB2 controls Rb phosphorylation, EZH2 expression and senescence through CDK4 and CDK6 regulation. 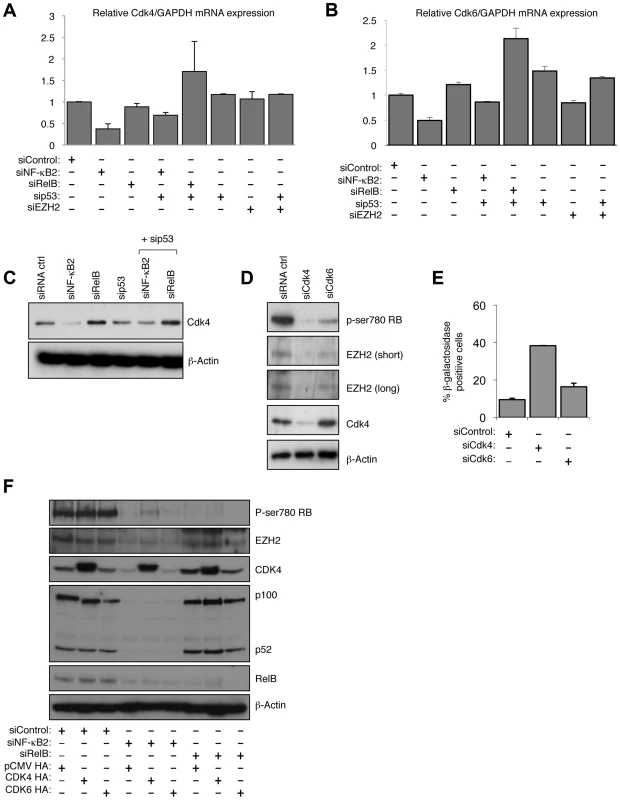
(A & B) NF-κB2 regulates CDK4 & 6 expression. RNA was prepared from NHD fibroblasts treated with the indicated siRNAs and Q-PCR analysis of CDK4 (A) and CDK6 (B) expression was performed. (C) NF-κB2 regulates CDK4 expression. Western blot analysis of NHD fibroblasts treated with the indicated siRNAs. Note that this is a reprobing of blots used in Fig. 2A and the β-actin blot shown here is the same as in that figure. (D) siRNA mediated knock down of CDK4 and CDK6 results in loss of Rb phosphorylation and EZH2 expression. Western blot analysis of NHD fibroblasts treated with the indicated siRNAs. (E) siRNA mediated knock down of CDK4 induces cellular senescence. NHD fibroblasts were transfected with the listed siRNAs and analyzed for senescence by β-galactosidase staining after 7 days. (F) Re-expression of CDK partially recovers the effects of NF-κB2 siRNA depletion. 96 hours after the transfection of NHD fibroblasts treated with the indicated siRNAs, cells were further transfected with CDK4 and CDK6 expression plasmids. After an additional 24 hours, protein extracts were prepared and western blot analysis performed. Chromatin Immunoprecipitation (ChIP) analysis confirmed that both CDK4 and CDK6 are direct p52 target genes in NHD fibroblasts (Fig. S8C&D). Furthermore, demonstrating the generality of this effect, data extracted from a ChIP-Seq analysis of the EBV-transformed human lymphoblastoid B-cell line (LCL) GM12878 [40] confirmed both CDK4 and CDK6 as NF-κB regulated genes (Figs. S8E&F). In GM12878, the EBV-encoded membrane protein LMP1 mimics activated CD40 to stimulate canonical and non-canonical NF-κB pathway activity [40]. Interestingly, this ChIP-Seq analysis revealed that although multiple NF-κB subunits bind the promoters of these genes, including p52, RelB was not found to significantly bind the CDK4 promoter, while p50 was not seen at the CDK6 promoter (Fig. S8E&F). Such differential subunit binding may explain some of the differential effects seen with NF-κB2 and RelB siRNAs.
RelB regulation of PSMA5 regulates p21WAF1 protein stability
A potential explanation for the effects of RelB on p53 protein stability could result from regulation of Mdm2 levels. Mdm2 is a ubiquitin ligase that induces degradation of p53 and has also been shown to be an NF-κB target gene [41]. However, RelB depletion was seen to consistently induce Mdm2 protein levels (Fig. 1C, 7B), suggesting it is not acting as a RelB effector in this case. We therefore again analyzed our microarray data for RelB regulated genes, whose products are known to have effects on protein stability (Table S5). A number of such genes were identified and a mini-siRNA screen was performed in NHD fibroblasts to ascertain if any had the potential to regulate p53 and p21WAF1 stability (Fig. 9A and S9A–C). Particularly striking effects were observed with siRNAs targeting PSMA5 (proteasome (prosome, macropain) subunit, α type, 5) and ANAPC1 (APC1, Anaphase Promoting Complex Subunit 1). Notably, knock down of PSMA5 was the only one to significantly stabilize p21WAF1 levels, with the effect of ANAPC1 depletion specifically affecting p53 protein levels (Fig. 9A and S9A). PSMA5 has been previously shown to be transcriptionally upregulated by the antioxidative nuclear Factor E2-related factor 2 (Nrf2) [42] but has not, to the best of our knowledge, been linked to p21WAF1 or p53 protein stability, EZH2 or senescence. The Anaphase Promoting Complex (APC/C), of which ANAPC1 is a component, is a cell cycle-regulated E3 ubiquitin ligase. It controls progression through the G1 and M phases of the cell cycle and has been previously associated with senescence induced upon acute loss of the tumour suppressor PTEN [43].
Fig. 9. RelB controls Rb phosphorylation, EZH2 expression and senescence through PSMA5 induced regulation of p21WAF1 and p53 protein stability. 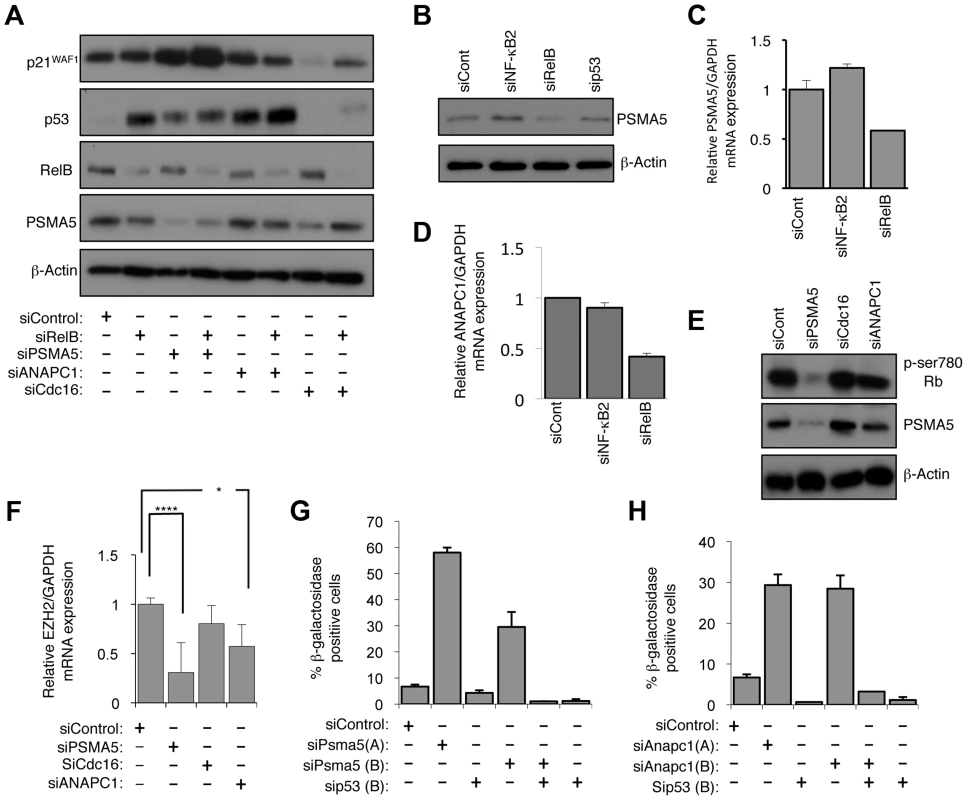
(A) PSMA5 and ANAPC1 regulate p21WAF1 and p53 protein stability. Western blot analysis of NHD fibroblasts treated with the indicated siRNAs. (B & C) RelB regulates PSMA5 expression. Whole cell protein lysates (B) or RNA (C) was prepared from NHD fibroblasts treated with the indicated siRNAs and western blot or Q-PCR analysis of PSMA5 expression was performed. (D) RelB regulates ANAPC1 expression. RNA was prepared from NHD fibroblasts treated with the indicated siRNAs and Q-PCR analysis of ANAPC1 was performed. (E) siRNA mediated knock down of PSMA5 results in loss of Rb phosphorylation. Western blot analysis of NHD fibroblasts treated with the indicated siRNAs. (F) siRNA mediated knock down of PSMA5 results in loss of EZH2 expression. RNA was prepared from NHD fibroblasts treated with the indicated siRNAs and Q-PCR analysis of EZH2 was performed. Psma5: (*** p≤0.001) Anapc1: (* p≤0.05). (G & H) siRNA mediated knock down of PSMA5 (G) or ANAPC1 (H) induces p53 dependent cellular senescence. NHD fibroblasts were transfected with the listed siRNAs and analyzed for senescence by β-galactosidase staining after 7 days. Further analysis confirmed that depletion of RelB but not NF-κB2 resulted in reduced levels of PSMA5 and ANAPC1 (Fig. 9B–D). Interestingly, PSMA5 siRNA depletion resulted in almost complete loss of Rb phosphorylation and a significant reduction in EZH2 mRNA levels (Fig. 9E&F, S9C). By contrast, depletion of ANAPC1 and another control siRNA from our initial screen, CDC16, had no effect on Rb phosphorylation (Fig. 9E) although the former did also affect EZH2 expression (Fig. 9F). This implies that the increase in p53 levels alone seen upon depleting ANAPC1 may also repress EZH2 in an Rb independent manner. Consistent with these effects, siRNA depletion of both PSMA5 and ANAPC1 resulted in significant p53 dependent senescence (Fig. 9G&H).
Similar to our previous results with CDK4/6, ChIP analysis of NHD fibroblasts and analysis of ChIP-Seq data from the EBV-transformed human lymphoblastoid B-cell line (LCL) GM12878 [40] demonstrated that PSMA5 and ANAPC1 are direct NF-κB target genes (Fig. S10A–D).
These results indicate that RelB can regulate numerous genes associated with protein stability. Of these PSMA5 is a key effector of RelB regulation of p21WAF1 protein stability, Rb phosphorylation and EZH2 levels, while ANAPC1 contributes to p53 stability and EZH2 repression through an independent pathway.
EZH2 is a direct NF-κB target gene
Although these results provided an explanation for the Rb dependent regulation of EZH2 expression by the alternative NF-κB pathway, we also investigated whether EZH2 is also be a direct target for p52 and RelB. ChIP analysis of the EZH2 promoter in NHD fibroblasts confirmed binding by E2F and Rb, as previously reported [19] (Fig. 10A&B). Moreover, this analysis also revealed recruitment of p52 and RelB. This result was confirmed by mining of ChIP-Seq data from GM12878 B-cells [40], where binding of all NF-κB subunits to the EZH2 promoter was seen (Fig. 10C). Taken together these results indicate that p52 and RelB induce EZH2 expression both through regulation of Rb/E2F activity and also directly, through binding the EZH2 promoter.
Fig. 10. NF-κB subunits bind the EZH2 promoter in fibroblasts and B-cells. 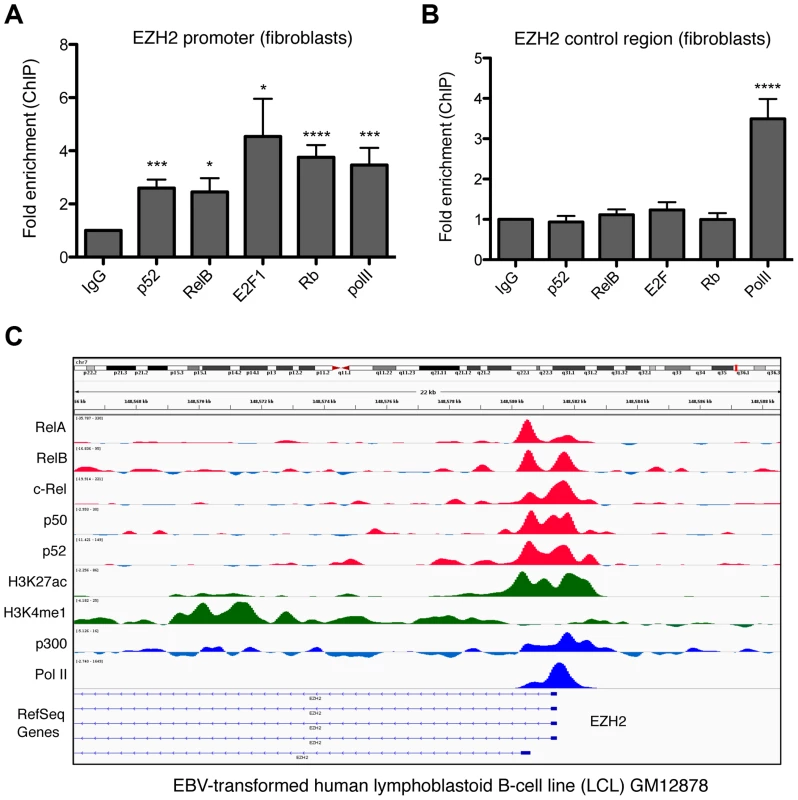
(A & B) ChIP analysis of the EZH2 promoter was performed in NHD fibroblast cells using primers close to the core promoter of the EZH2 gene (A) (+596/+888) or an upstream control region (B) (−2802/−2600). Results shown are representative of a minimum of 3 separate experiments. * P≤0.05, ** P≤0.01, *** P≤0.001, **** P≤0.0001. (C) ChIP Seq data showing NF-κB subunit binding in the region of the EZH2 gene in the human EBV-transformed lymphoblastoid B-cell line (LCL) GM12878. Discussion
NF-κB and senescence
NF-κB activation has previously been associated with induction of senescent cells and the senescence associated secretory phenotype [5]–[7]. However, these studies have focused on the canonical NF-κB pathway and have generally been performed in immortalized or transformed cells. Here we have described a previously unknown pathway through which the alternative NF-κB pathway can suppress cellular senescence. We show that both NF-κB2 and RelB regulate pathways leading to control of Rb phosphorylation and hence determine the level of EZH2 expression (summarized in Fig. 11). Notably, this is achieved through separate but complementary routes, with NF-κB2 regulating expression of CDK4 and CDK6 and RelB regulating the stability of p53 and p21WAF1 protein. We demonstrate that this latter effect is achieved through RelB specific regulation of PSMA5 and ANAPC1. It is possible that other NF-κB2 and RelB regulated genes contribute to this process. We then demonstrate that EZH2 antagonizes a subset of p53-regulated genes associated with cell senescence. This included RACGAP1, which through regulating the activity of Rac1 and CDC42, mediated induction of ROS, required for the senescent phenotype. Moreover, through regulating EZH2 expression, this pathway represents the major route of crosstalk between the alternative NF-κB pathway and p53 under these experimental conditions.
Fig. 11. Summary of the results identified in this manuscript through which NF-κB2 and RelB regulate p53 dependent cellular senescence in primary human NHD fibroblasts. 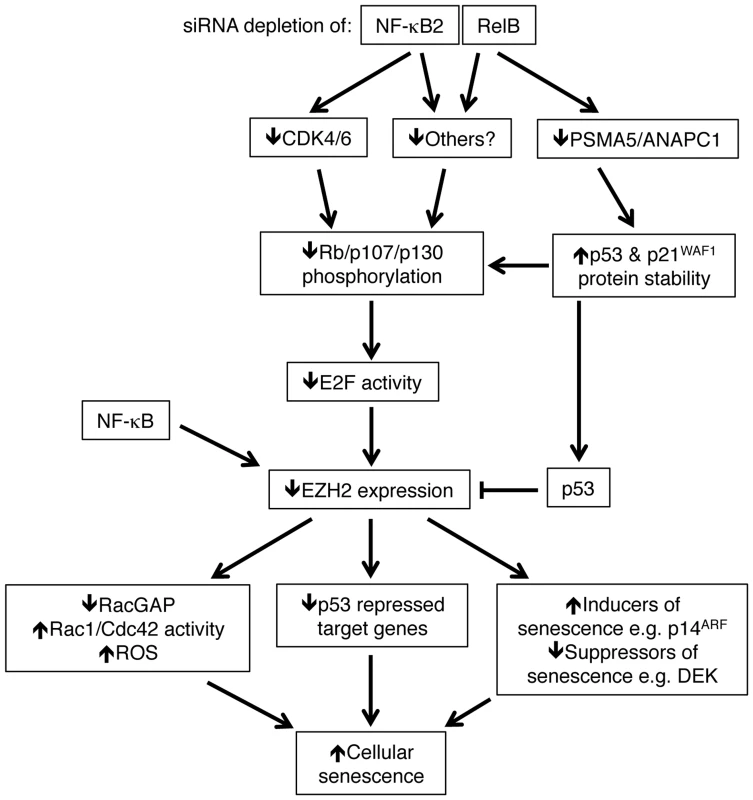
Depletion of NF-κB2 and RelB leads to a decrease in Rb phosphorylation. Unphosphorylated Rb represses E2F transcriptional activity and consequently inhibits EZH2 expression (which is negatively regulated by p53). However, this occurs through distinct pathways. Depletion of NF-κB2 leads to down regulation of CDK4 and CDK6, which are known to phosphorylate Rb directly. RelB depletion leads to an increase in p53 and p21 protein stability as a consequence of loss of expression of genes such as PSMA5 and ANAPC1. Other gene targets may be involved in these processes. NF-κB subunits also bind directly to the EZH2 promoter and this may also contribute towards its regulation. As a consequence of this pathway, down regulation of EZH2 results in numerous changes in gene expression, including p53 dependent repression of a number of gene targets. These include inhibition of RACGAP1 expression, resulting in Rac1/Cdc42 dependent induction of reactive oxygen species (ROS). Together with other changes, such as suppression of DEK and ultimately induction of p14ARF and p16INK4a, the ultimate consequence of NF-κB2 and RelB siRNA depletion and subsequent loss of EZH2 expression is p53 dependent cell senescence. Note, RACGAP1 activity as well as various inducers/suppressors or senescence may also be regulated by p53. Many of these genes may also be direct targets of NF-κB. Not shown is that oxidative stress is required to drive the basal level p53 activity seen in these cells. Both ChIP analysis of NHD fibroblasts as well as mining of ChIP-Seq data from GM12878 B-cells [40] revealed that CDK4, CDK6, PSMA5 and ANAPC1, together with EZH2 itself, are direct NF-κB target genes. However, it is not clear if this recruitment is a result of direct binding to κB elements or occurs through ‘piggy-backing’ on other transcription factors, such as E2F. The presence of multiple NF-κB subunits on these promoters may also explain why we see differential effects upon knockdown of NF-κB2 and RelB, as there is the potential for other subunits to compensate in ways that may be promoter specific. Moreover, ChIP-Seq analysis showed a lack of recruitment of RelB to the CDK4 promoter (Fig. S8F) in B-cells that may account for why this gene was not seen to be RelB regulated in our experiments. In addition, although p52 and RelB are frequently found in the same NF-κB complex, removal of either subunit will not have the same effect. For example, loss of RelB can still leave p52 homodimers or alternative p52 heterodimers active in the cell. By contrast, RelB does not homodimerise [44] and so loss of p52 can only be potentially compensated for by the activity of p50/RelB heterodimers or other NF-κB complexes.
Crosstalk between NF-κB and p53
Under normal circumstances, cross regulation between NF-κB and p53, leading to modulation of activity and transcriptional output, also has the potential to influence physiological responses to stress and determine cell fate or behavior under circumstances where both pathways are active. There are a number of situations where such simultaneous activation of both pathways may occur, including many types of DNA damage and oncogene activation. Furthermore, where NF-κB is activated during chronic inflammation, a process shown to promote tumorigenesis, p53 will likely also be induced due to production of ROS [2], [9]–[11]. Indeed we see induction of p53 by both LTβR activation and CD40L stimulation, both inducers of the NF-κB pathway (Fig. 1E, 2B&C, S2B). The consequences of such dual activation may depend on the relative levels of activity of both pathways. In this study, we focused our analysis on the ability of NF-κB to modulate basal level of p53 activity, induced by chronic oxidative stress. Under these conditions, NF-κB activity appears dominant and acts to promote proliferation and suppress senescence. However, under circumstances where p53 is induced to a high level, either by acute administration of a DNA damaging agent or activation of potent oncogene, we propose that the balance would shift and that p53 activity would dominate. Indeed, we have previously observed such an effect where artificial induction of p53 or treatment with ultraviolet light can induce a switch from p52/Bcl-3 complexes to p52/HDAC1 complexes, resulting in a change from activation of Cyclin D1 expression to repression [13]. Both aspects of NF-κB/p53 behavior have physiological relevance and reflect the complex nature of crosstalk between these pathways.
Data in this report also underlines the impact that the activity of tumor suppressors can have on NF-κB dependent gene expression. For example, the effects we see in NHD fibroblasts are dependent upon Rb expression (Fig. 7A) and would not be seen in an Rb null tumor cell line. Similarly, the transcriptional consequences of activation of the alternative NF-κB pathway will differ in cells either with mutant or absent p53. That much research on NF-κB activity in cancer occurs in such cell lines, or is not taken into consideration in model systems, might account for some of the apparently contradictory effects reported in the literature.
The alternative NF-κB pathway and cancer
The alternative NF-κB pathway can become deregulated in hematological malignancies such as multiple myeloma, through mutation of upstream regulators such as NF-κB inducing kinase (NIK) [45]–[47]. Indeed the NF-κB2 gene itself is subject to translocation, leading to truncation and constitutive processing to p52, in a subset of B and T cell lymphomas [48], [49]. Moreover, the tumor microenvironment can induce alternative NF-κB pathway activity through, for example, the CD40 receptor [45], [46], [50]. Although this branch of NF-κB signalling is associated with the adaptive immune response, the mechanisms through which it can promote tumorigenesis have received little attention and remain poorly defined, compared to the parallel IKKβ/RelA dependent classical pathway. However, due to feed forward mechanisms, in which the classical pathway can prime the activity of the alternative pathway through inducing the expression of the NF-κB2 and RelB genes [4], it is possible that some effects have been assigned to the former pathway but in fact result from the latter.
This pathway provides a mechanism through which deregulated NF-κB2/RelB activity can promote tumorigenesis in cancer cells that retain wild type p53. Consistent with this hypothesis, we show that in CLL, where only 10–15% of tumors at diagnosis contain mutated p53 [51], stimulation of B-CLL cells by CD40L receptor, which induces the alternative NF-κB pathway, results in induction of EZH2 expression (Fig. 2). Moreover, mining of data from ChIP-Seq analysis of the EBV-transformed human lymphoblastoid B-cell line (LCL) GM12878 [40] confirmed that EZH2 is an NF-κB target in this cell type (Fig. 10B). In diffuse large B-cell lymphoma (DLBCL), where activating mutations in EZH2 occur, EZH2 function is essential during B-cell activation and clonal expansion in the germinal center. Recent evidence demonstrated that small molecule inhibition of EZH2 significantly reduced growth of germinal center-derived DLBCL cells, and conditional expression of an EZH2 mutant lymphoma allele was shown to drive lymphomagenesis [52]. These studies highlight that EZH2 has a key role during B-cell activation and together with our data, present the possibility of interplay between NF-κB signaling and EZH2 to enhance survival and proliferation of tumor cells. We propose that this pathway provides a mechanism where activation of NF-κB, either as consequence of the tumor microenvironment or through mutation of upstream signaling pathways (such as occurs in a number of hematological malignancies [53]–[55]), can promote tumorigenesis in cells retaining wild type p53 by suppressing the consequences of p53 activation and providing a window during which further mutagenesis can occur. These effects need not be limited to effects on senescence but may also include suppression of apoptosis, cell cycle arrest and metabolic effects of p53 activity. Our data suggests that inhibition of the alternative pathway, through for example inhibitors of the NIK or IKKα kinases, could have the potential to treat select hematological malignancies that retain wild type p53 and Rb.
Materials and Methods
Cells
Primary normal human juvenile dermal fibroblasts were purchased from Promocell (c-12300) and maintained in Fibroblast growth media supplemented with 2% supplement mix (Promocell c-23010) and 1% Pen/Strep/Fungizone Solution (Promocell c-42020). Cells were cultured from passage 2 to passage 10 before being discarded.
CLL cells were cultured with 10 ng/ml IL-4 (R&D Systems, Abingdon, UK). CD40L cell stimulation of CLL cells was performed essentially as described (Pepper et al., 2011). Untransfected L-cells (NTL) and CD40L-expressing mouse fibroblast L-cells were cultured in in RPMI-1640 medium supplemented with 10% foetal bovine serum, 50 units/ml penicillin and 50 mg/ml streptomycin and seeded into 12-well plates (0.6×106/well) and irradiated with 75 Gy. L cells were left to attach for at least 4 h prior to the addition of the CLL cells. The study was approved by the UK NHS Research Ethics Service, and samples were obtained from the Newcastle Haematology Biobank (http://www.ncl.ac.uk/nbb/collections/nhb). Following written informed consent, patients provided peripheral blood samples, from which CLL cells were isolated using Lymphoprep (Axis Shield, Cambridgeshire, UK).
Inhibitors and treatments
Hydrogen peroxide (H2O2) was purchased from Sigma (H1009). 3-Deazaneplanocin-A (DZNep) was purchased from Cayman Chemicals (13828). ATM inhibitor was purchased from Tocris (KU55933), Epigallocatechin-Gallate (ECGC) was purchased from Calbiochem (324880), N-acetyl-L-Cysteine (NAC) and IKK-β inhibitor (TPCA-1) were purchased from Sigma (A9165-5gr and T1452).
Concentrations used were: H2O2 (100 & 200 µM), DZNep: (0.5 µM), KU55933: (10 µM), ECGC: (10 µM), NAC: (5 mM), TPCA-1: (10 µM).
Lymphotoxin β receptor agonist antibody was used at a final concentration used of 2 µG/mL.
Microarray analysis
Cells were separately transfected in triplicate with siRNAs to generate biological replicates. After 48 hours, RNA was extracted using a PeqLab gold total RNA extraction kit (12-6634-02). Q-PCR and subsequent Principal Components analysis confirmed consistent levels of depletion in all biological replicates, apart from one double EZH2/p53 knockdown, where problems were encountered due to the strong induction of EZH2 levels upon loss of p53 (Table S1). This sample was not included in subsequent analysis. Microarray analysis was performed by Cambridge Genomic Services.
Bioinformatics analysis
The Illumina Human HT12v4 Expression BeadChip data was background corrected in Illumina Beadstudio, subsequent analysis proceeded using the lumi and limma packages in R (Bioconductor) [56]–[58]. Variant Stabilisation Transform and Robust Spline Normalisation were applied in lumi. Differential expression was detected using linear models and empirical Bayes statistics in limma. A list of genes for each comparison was generated using a Benjamini Hochberg false discovery rate correct p-value of 0.05 and a fold change of 1.5 as cut - offs. Gene lists were integrated with data from Rovillain et al. [32] by comparison across gene names. Genes found in all experiments were retained for inclusion in the integrated heat map.
Flow cytometric analysis of ROS production
Cells were incubated for 30 minutes with 5 mM 2′,7′ –dichlorofluorescin diacetate (D399 Invitrogen). Cells were then washed twice in phosphate buffered saline (PBS) and resuspended in 200 µl PBS. Samples were analyzed using a FacsCanto flow cytometer (excitation 488 nm, emission 530 nm). Data shown in figures is the average derived from three separate experiments.
β galactosidase staining
β galactosidase staining was performed according to [59]. Images of senescent stained cells were taken using a Canon power shot A640 camera. The proportion of cells positive for β-galactosidase activity was determined by counting the number of blue cells in the total population, 20 hours after staining. Results shown are averages derived from three separate experiments and error bars indicate the standard deviation.
CFSE staining
CLL cells were stained with carboxyfluorescein diacetate succinimidyl ester (CFSE) (Life Technologies). These cells were co-cultured, with 10 ng/mL Interleukin 4, on CD40L-expressing fibroblast cells (or on non-CD40L-expressing control (NTL) cells) that had been growth-arrested (with 75 Gy ionising radiation). Quantification of CFSE in CD19+ve cells by flow cytometry [60], was used to show CLL cell proliferation (seen by sub-peaks of CFSE fluorescence).
Clonogenic assays
Clonogenic assays to measure recovery from senescence were performed essentially as described [32]. Briefly, lentiviral gene transfer was used to express constitutively active FOXM1ΔNΔKEN mutant, DEK, and EZH2 in conditionally immortalized fibroblast cells at 34°C. These cells were then shifted to 38°C, which in control transfected cells induces cellular senescence, causing no colonies to appear in this assay. Colonies were stained with methylene blue.
Quantitative PCR analysis
Total RNA was extracted with PeqLab gold total RNA extraction kit (12-6634-02), according to the manufacturer's directions. For reverse transcriptase PCR (RT-PCR), 1 µg RNA sample were transcribed with Quantitect Reverse Transcription Kit (QIAgen; 205313). The cDNA stock was diluted by 200 and 5 µl was used for PCR with GoTaq flexi DNA-polymerase (Promega; M8305).
Quantitative PCR data was generated on a Rotor-Gene Q (Qiagen) using the following experimental settings: Hold 50°C for 3 min; Hold 95°C 10 min; Cycling (95°C for 20 sec; 58°C for 20 sec; 72°C for 20 sec with fluorescence measurement)×45; Melting Curve 50–99°C with a heating rate of 1°C every 5 sec. All values were calculated relative to untreated levels and normalized to GAPDH levels using the Pfaffl method [61]. Each RNA sample was assayed in triplicate and the results shown are averages derived from three separate experiments with error bars indicating the standard deviation.
Luciferase assay
Cells were transfected with siRNAs. 24 hours later, they were transfected with 0.8 µg of pGL3 luciferase reporter vector containing the EZH2 promoter region (Tang et al., 2004) (kind gift of Dr. Tomer Cooks, Weizmann Institute, Israel). After 48 hours, cells were lysed in 100 µl Passive lysis buffer and luciferase was performed using a Dual - Luciferase Reporter Assay System kit (Promega E1910). Luciferase activity was read in a luminometer (Lumat LB9507, Berthold technologies) and normalized to protein content. Results shown are averages derived from five separate experiments and error bars indicate the standard deviation.
Chromatin Immunoprecipitation (ChIP)
NHD fibroblasts cells, either grown to 70% confluency or analysed 48 hours after transfection, were cross-linked with 1% formaldehyde at room temperature for 10 min. Cells were washed once with cold glycine and then scraped into 0.5 mL of RIPA buffer (0.1% SDS, 1% Triton, 0.5% deoxycholate, 0.5% NP40, 50 mM Tris-HCl pH 7.5, 150 mM NaCl, 50 µg/ml PMSF, 1 µg/ml leupeptin, 1 µg/ml aprotinin, 1 µg/ml pepstatin, Na3VO4, 50 µg/ml & 50 µg/ml NaF) and left on ice for 10 minutes. Samples were then sonicated on ice nine times. Each sonication was for 30 seconds with a 30 seconds gap between each sonication. Supernatants were recovered by centrifugation at 12,000 rpm in an eppendorf microfuge for 10 min at 4°C before being diluted 1∶1 in dilution buffer (1% Triton, 2 mM EDTA, 20 mM Tris-HCl pH 8.1, 150 mM NaCl supplemented with 0.1% NP40, protease and phosphatase inhibitors). Samples were then precleared for 2 hours at 4°C with sheared salmon sperm DNA (1 µg/ml) and 20 µl of protein A and G-agarose beads. At this stage, 20 µl of the material was kept as Input material. Immunoprecipitations were performed overnight with specific antibodies (2 µg). The immune complexes were captured by incubation with 20 µl of protein A and G-agarose beads and salmon sperm DNA (1 µg/ml) for 1 hour at 4°C. The immunoprecipitates were washed sequentially for 5 minutes each at 4°C in TSE 1(0.1% SDS, 1% Triton, 2 mM EDTA, 20 mM Tris-HCl pH 8.1,150 mM NaCl), TSE 2 (0.1% SDS, 1% Triton, 2 mM EDTA, 20 mM Tris-HCl pH 8.1,500 mM NaCl), Buffer 3 (250 mM LiCl, 1% NP40, 1% deoxycholate, 1 mM EDTA, 10 mM Tris-HCl pH 8.1 and TE buffer (1 mM EDTA, 10 mM Tris-HCl pH = 8.1). Beads were then eluted with 500 µl of Elution Buffer (1% SDS, 100 mM NaHCO3).
To reverse the crosslinks, samples, including ‘Input’, were incubated at 65°C overnight in a waterbath with 0.2M NaCl. DNA was ethanol precipitated following Phenol-Chloroform extraction. For PCR, 5 µl of DNA was used from an 80 µl DNA preparation and subjected to 40 cycles of PCR amplifications.
For all ChIP results shown are averages derived from three separate experiments and error bars indicate the standard deviation.
ChIP-Seq
ChIP Seq data shown here was extracted from a previously published analysis of the EBV-transformed lymphoblastoid B-cell line (LCL) GM12878 using validated anti-RelA, RelB, cRel, p52 and p50 antibodies [40]. GM12878 are one of three ENCODE project Tier 1 cell lines. It is an original HapMap cell line used in many genetic studies including the 1000 Genomes Project and has a relatively normal karyotype. Reads from biological replicate ChIP-seq experiments were mapped to the hg19/GRCh37 build of the human genome using bowtie v0.12.8 [62]. ChIP-Seq binding profiles were visualized by the Integrated Genome Viewer (IGV) [63].
Plasmids
Cdk4-HA (no. 1876), Cdk6-HA (no. 1868) and EZH2-HA (no. 24230) expression plasmids were purchased from Addgene.
Antibodies
Antibodies used were: anti-EZH2 (3147S Cell Signaling), anti-p52/p100 (05-361 Millipore), anti-RelB (4954S Cell Signaling), anti-p53 (DO-1 sc-126 Santa Cruz), anti-β - Actin (A5441, Sigma), anti-p21 (sc-397 Santa Cruz), anti-Rb (sc-50 Santa Cruz), anti - Cdk4 (sc-260 Santa Cruz), anti-DEK (610948 BD transduction Laboratories), anti-Bcl3 (PA1-41087 Pierce), anti-Lamin B1 (sc-374015 Santa Cruz), anti-p50 (3035S Cell Signaling), anti-MDM2 (OP46 Calbiochem), anti-p130 (610261 BD transduction Laboratories), anti-p107 (sc-318 Santa Cruz), anti-PSMA5 (2457S Cell Signaling), anti-p14ARF (14PO2 Calbiochem), anti-p16INK4a (sc-56330 Santa Cruz), anti MnSOD (sc-133134 Santa Cruz). Phospho-antibodies used were S15-p53 (9284S Cell Signaling) and S780 - Rb (8180S Cell Signaling). Lymphotoxin β receptor agonist antibody (anti-HuLTβR:Fc Ab) was a kind gift of Prof. Carl Ware (Sanford/Burnham Medical Research Institute) [64].
Other procedures
Transfections of siRNAs were performed when cells were at low (<50%) confluency, essentially as described previously [14]. Western blots shown are representative of at least 3 separate experiment and were performed as described [14] using 15–25 µg of protein extracts.
Details of oligonucleotides, siRNAs and primer sequences can be found in Supporting information (Text S1).
Accession numbers
Microarray data has been submitted to ArrayExpress with accession number is: E-MTAB-1593.
NF-κB ChIP-seq datasets have been published [40] and are deposited in the gene expression omnibus, accession code GSE55105.
Supporting Information
Zdroje
1. HaydenMS, GhoshS (2008) Shared principles in NF-κB signaling. Cell 132 : 344–362.
2. PerkinsND (2007) Integrating cell-signalling pathways with NF-κB and IKK function. Nat Rev Mol Cell Biol 8 : 49–62.
3. Ben-NeriahY, KarinM (2011) Inflammation meets cancer, with NF-κB as the matchmaker. Nat Immunol 12 : 715–723.
4. PerkinsND (2012) The diverse and complex roles of NF-κB subunits in cancer. Nat Rev Cancer 12 : 121–132.
5. VaughanS, JatPS (2011) Deciphering the role of nuclear factor-κB in cellular senescence. Aging 3 : 913–919.
6. ChienY, ScuoppoC, WangX, FangX, BalgleyB, et al. (2011) Control of the senescence-associated secretory phenotype by NF-κB promotes senescence and enhances chemosensitivity. Genes Dev 25 : 2125–2136.
7. JingH, KaseJ, DorrJR, MilanovicM, LenzeD, et al. (2011) Opposing roles of NF-κB in anti-cancer treatment outcome unveiled by cross-species investigations. Genes Dev
8. SfikasA, BatsiC, TselikouE, VartholomatosG, MonokrousosN, et al. (2012) The canonical NF-κB pathway differentially protects normal and human tumor cells from ROS-induced DNA damage. Cellular Signalling 24 : 2007–2023.
9. DeyA, TergaonkarV, LaneDP (2008) Double-edged swords as cancer therapeutics: simultaneously targeting p53 and NF-κB pathways. Nat Rev Drug Discov 7 : 1031–1040.
10. AkP, LevineAJ (2010) p53 and NF-κB: different strategies for responding to stress lead to a functional antagonism. FASEB J 24 : 3643–3652.
11. SchneiderG, KramerOH (2011) NFκB/p53 crosstalk-a promising new therapeutic target. Biochim Biophys Acta 1815 : 90–103.
12. VigneronA, VousdenKH (2010) p53, ROS and senescence in the control of aging. Aging (Albany NY) 2 : 471–474.
13. RochaS, MartinAM, MeekDW, PerkinsND (2003) p53 represses cyclin D1 transcription through down regulation of Bcl-3 and inducing increased association of the p52 NF-κB subunit with histone deacetylase 1. Mol Cell Biol 23 : 4713–4727.
14. SchummK, RochaS, CaamanoJ, PerkinsND (2006) Regulation of p53 tumour suppressor target gene expression by the p52 NF-κB subunit. EMBO J 25 : 4820–4832.
15. ShettyS, GrahamBA, BrownJG, HuX, Vegh-YaremaN, et al. (2005) Transcription factor NF-κB differentially regulates death receptor 5 expression involving histone deacetylase 1. Mol Cell Biol 25 : 5404–5416.
16. FrankAK, LeuJI, ZhouY, DevarajanK, NedelkoT, et al. (2011) The codon 72 polymorphism of p53 regulates interaction with NF-κB and transactivation of genes involved in immunity and inflammation. Mol Cell Biol 31 : 1201–1213.
17. De SantaF, NarangV, YapZH, TusiBK, BurgoldT, et al. (2009) Jmjd3 contributes to the control of gene expression in LPS-activated macrophages. EMBO J 28 : 3341–3352.
18. TangX, MilyavskyM, ShatsI, ErezN, GoldfingerN, et al. (2004) Activated p53 suppresses the histone methyltransferase EZH2 gene. Oncogene 23 : 5759–5769.
19. BrackenAP, PasiniD, CapraM, ProsperiniE, ColliE, et al. (2003) EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J 22 : 5323–5335.
20. ChaseA, CrossNC (2011) Aberrations of EZH2 in cancer. Clin Cancer Res 17 : 2613–2618.
21. BrackenAP, Kleine-KohlbrecherD, DietrichN, PasiniD, GargiuloG, et al. (2007) The Polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes Dev 21 : 525–530.
22. FanT, JiangS, ChungN, AlikhanA, NiC, et al. (2011) EZH2-dependent suppression of a cellular senescence phenotype in melanoma cells by inhibition of p21/CDKN1A expression. Mol Cancer Res 9 : 418–429.
23. TzatsosA, PaskalevaP, LymperiS, ContinoG, StoykovaS, et al. (2011) Lysine-specific demethylase 2B (KDM2B)-let-7-enhancer of zester homolog 2 (EZH2) pathway regulates cell cycle progression and senescence in primary cells. J Biol Chem 286 : 33061–33069.
24. MargueronR, ReinbergD (2011) The Polycomb complex PRC2 and its mark in life. Nature 469 : 343–349.
25. LaniganF, GeraghtyJG, BrackenAP (2011) Transcriptional regulation of cellular senescence. Oncogene 30 : 2901–2911.
26. LeeST, LiZ, WuZ, AauM, GuanP, et al. (2011) Context-Specific Regulation of NF-κB Target Gene Expression by EZH2 in Breast Cancers. Mol Cell 43 : 798–810.
27. GuoZ, KozlovS, LavinMF, PersonMD, PaullTT (2010) ATM activation by oxidative stress. Science 330 : 517–521.
28. ShihVF, Davis-TurakJ, MacalM, HuangJQ, PonomarenkoJ, et al. (2012) Control of RelB during dendritic cell activation integrates canonical and noncanonical NF-κB pathways. Nat Immunol 13 : 1162–1170.
29. PepperC, MahdiJG, BugginsAG, HewamanaS, WalsbyE, et al. (2011) Two novel aspirin analogues show selective cytotoxicity in primary chronic lymphocytic leukaemia cells that is associated with dual inhibition of Rel A and COX-2. Cell Prolif 44 : 380–390.
30. HostagerBS, BishopGA (2013) CD40-Mediated Activation of the NF-κB2 Pathway. Front Immunol 4 : 376.
31. FreundA, LabergeRM, DemariaM, CampisiJ (2012) Lamin B1 loss is a senescence-associated biomarker. Mol Biol Cell 23 : 2066–2075.
32. RovillainE, MansfieldL, CaetanoC, Alvarez-FernandezM, CaballeroOL, et al. (2011) Activation of Nuclear Factor-κB signalling promotes cellular senescence. Oncogene 30 : 2356–2366.
33. KavanaughGM, Wise-DraperTM, MorrealeRJ, MorrisonMA, GoleB, et al. (2011) The human DEK oncogene regulates DNA damage response signaling and repair. Nucleic Acids Res 39 : 7465–7476.
34. LiuK, FengT, LiuJ, ZhongM, ZhangS (2012) Silencing of the DEK gene induces apoptosis and senescence in CaSki cervical carcinoma cells via the up-regulation of NF-κB p65. Biosci Rep 32 : 323–332.
35. RaptisL, ArulanandamR, GeletuM, TurksonJ (2011) The R(h)oads to Stat3: Stat3 activation by the Rho GTPases. Exp Cell Res 317 : 1787–1795.
36. ChengG, DieboldBA, HughesY, LambethJD (2006) Nox1-dependent reactive oxygen generation is regulated by Rac1. J Biol Chem 281 : 17718–17726.
37. QianY, LiuKJ, ChenY, FlynnDC, CastranovaV, et al. (2005) Cdc42 regulates arsenic-induced NADPH oxidase activation and cell migration through actin filament reorganization. J Biol Chem 280 : 3875–3884.
38. PolagerS, GinsbergD (2008) E2F - at the crossroads of life and death. Trends Cell Biol 18 : 528–535.
39. LedouxAC, SellierH, GilliesK, IannettiA, JamesJ, et al. (2013) NFκB regulates expression of Polo-like kinase 4. Cell Cycle 12 : 3052–3062.
40. ZhaoB, BarreraLA, ErsingI, WilloxB, SchmidtSCS, et al. (2014) The NF-κB Genomic Landscape in Lymphoblastoid B-cells. Cell Reports In Press.
41. TergaonkarV, PandoM, VafaO, WahlG, VermaI (2002) p53 stabilization is decreased upon NFκB activation: a role for NFκB in acquisition of resistance to chemotherapy. Cancer Cell 1 : 493–503.
42. ArltA, SebensS, KrebsS, GeismannC, GrossmannM, et al. (2013) Inhibition of the Nrf2 transcription factor by the alkaloid trigonelline renders pancreatic cancer cells more susceptible to apoptosis through decreased proteasomal gene expression and proteasome activity. Oncogene 32 : 4825–4835.
43. SongMS, CarracedoA, SalmenaL, SongSJ, EgiaA, et al. (2011) Nuclear PTEN regulates the APC-CDH1 tumor-suppressive complex in a phosphatase-independent manner. Cell 144 : 187–199.
44. VuD, HuangDB, VemuA, GhoshG (2013) A structural basis for selective dimerization by NF-κB RelB. J Mol Biol 425 : 1934–1945.
45. AnnunziataCM, DavisRE, DemchenkoY, BellamyW, GabreaA, et al. (2007) Frequent engagement of the classical and alternative NF-κB pathways by diverse genetic abnormalities in multiple myeloma. Cancer Cell 12 : 115–130.
46. DemchenkoYN, GlebovOK, ZingoneA, KeatsJJ, BergsagelPL, et al. (2010) Classical and/or alternative NF-κB pathway activation in multiple myeloma. Blood 115 : 3541–3552.
47. GilmoreTD (2007) Multiple myeloma: lusting for NF-κB. Cancer Cell 12 : 95–97.
48. MigliazzaA, LombardiL, RocchiM, TreccaD, ChangCC, et al. (1994) Heterogeneous chromosomal aberrations generate 3′ truncations of the NFKB2/lyt-10 gene in lymphoid malignancies. Blood 84 : 3850–3860.
49. ThakurS, LinH, TsengW, KumarS, BravoR, et al. (1994) Rearrangement and altered expression of the NFKB-2 gene in human cutaneous T-lymphoma cells. Oncogene 9 : 2335–2344.
50. Homig-HolzelC, HojerC, RastelliJ, CasolaS, StroblLJ, et al. (2008) Constitutive CD40 signaling in B cells selectively activates the noncanonical NF-κB pathway and promotes lymphomagenesis. J Exp Med 205 : 1317–1329.
51. PospisilovaS, GonzalezD, MalcikovaJ, TrbusekM, RossiD, et al. (2012) ERIC recommendations on TP53 mutation analysis in chronic lymphocytic leukemia. Leukemia 26 : 1458–1461.
52. BeguelinW, PopovicR, TeaterM, JiangY, BuntingKL, et al. (2013) EZH2 Is Required for Germinal Center Formation and Somatic EZH2 Mutations Promote Lymphoid Transformation. Cancer Cell 23 : 677–692.
53. BraunT, CarvalhoG, FabreC, GrosjeanJ, FenauxP, et al. (2006) Targeting NF-κB in hematologic malignancies. Cell Death Differ 13 : 748–758.
54. FuchsO (2010) Transcription factor NF-κB inhibitors as single therapeutic agents or in combination with classical chemotherapeutic agents for the treatment of hematologic malignancies. Curr Mol Pharmacol 3 : 98–122.
55. KeutgensA, RobertI, ViatourP, ChariotA (2006) Deregulated NF-κB activity in haematological malignancies. Biochem Pharmacol 72 : 1069–1080.
56. DuP, KibbeWA, LinSM (2008) lumi: a pipeline for processing Illumina microarray. Bioinformatics 24 : 1547–1548.
57. GentlemanRC, CareyVJ, BatesDM, BolstadB, DettlingM, et al. (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80.
58. Smyth GK (2005) Limma: linear models for microarray data. In: Gentleman RC, Carey VJ, Dudoit S, Irizarry R, Huber W, editors. ‘Bioinformatics and Computational Biology Solutions using R and Bioconductor. New York: Springer. pp. 397–420.
59. DimriGP, LeeX, BasileG, AcostaM, ScottG, et al. (1995) A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci U S A 92 : 9363–9367.
60. LyonsAB, ParishCR (1994) Determination of lymphocyte division by flow cytometry. J Immunol Methods 171 : 131–137.
61. PfafflMW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45.
62. LangmeadB, TrapnellC, PopM, SalzbergSL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25.
63. RobinsonJT, ThorvaldsdottirH, WincklerW, GuttmanM, LanderES, et al. (2011) Integrative genomics viewer. Nat Biotechnol 29 : 24–26.
64. RooneyI, ButrovichK, WareCF (2000) Expression of lymphotoxins and their receptor-Fc fusion proteins by baculovirus. Methods Enzymol 322 : 345–363.
Štítky
Genetika Reprodukční medicína
Článek An Evolutionarily Conserved Role for the Aryl Hydrocarbon Receptor in the Regulation of MovementČlánek Requirement for Drosophila SNMP1 for Rapid Activation and Termination of Pheromone-Induced ActivityČlánek Co-regulated Transcripts Associated to Cooperating eSNPs Define Bi-fan Motifs in Human Gene NetworksČlánek Identification of a Regulatory Variant That Binds FOXA1 and FOXA2 at the Type 2 Diabetes GWAS LocusČlánek tRNA Modifying Enzymes, NSUN2 and METTL1, Determine Sensitivity to 5-Fluorouracil in HeLa CellsČlánek Derlin-1 Regulates Mutant VCP-Linked Pathogenesis and Endoplasmic Reticulum Stress-Induced ApoptosisČlánek A Genetic Assay for Transcription Errors Reveals Multilayer Control of RNA Polymerase II FidelityČlánek The Proprotein Convertase KPC-1/Furin Controls Branching and Self-avoidance of Sensory Dendrites inČlánek BMPs Regulate Gene Expression in the Dorsal Neuroectoderm of and Vertebrates by Distinct MechanismsČlánek Unkempt Is Negatively Regulated by mTOR and Uncouples Neuronal Differentiation from Growth Control
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 9- Růst a vývoj dětí narozených pomocí IVF
- Délka menstruačního cyklu jako marker ženské plodnosti
- Vztah užívání alkoholu a mužské fertility
- Šanci na úspěšný průběh těhotenství snižují nevhodné hladiny progesteronu vznikající při umělém oplodnění
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
-
Všechny články tohoto čísla
- Translational Regulation of the Post-Translational Circadian Mechanism
- An Evolutionarily Conserved Role for the Aryl Hydrocarbon Receptor in the Regulation of Movement
- Eliminating Both Canonical and Short-Patch Mismatch Repair in Suggests a New Meiotic Recombination Model
- Requirement for Drosophila SNMP1 for Rapid Activation and Termination of Pheromone-Induced Activity
- Co-regulated Transcripts Associated to Cooperating eSNPs Define Bi-fan Motifs in Human Gene Networks
- Targeted H3R26 Deimination Specifically Facilitates Estrogen Receptor Binding by Modifying Nucleosome Structure
- Role for Circadian Clock Genes in Seasonal Timing: Testing the Bünning Hypothesis
- The Tandem Repeats Enabling Reversible Switching between the Two Phases of β-Lactamase Substrate Spectrum
- The Association of the Vanin-1 N131S Variant with Blood Pressure Is Mediated by Endoplasmic Reticulum-Associated Degradation and Loss of Function
- Identification of a Regulatory Variant That Binds FOXA1 and FOXA2 at the Type 2 Diabetes GWAS Locus
- Regulation of Flowering by the Histone Mark Readers MRG1/2 via Interaction with CONSTANS to Modulate Expression
- The Actomyosin Machinery Is Required for Retinal Lumen Formation
- Plays a Conserved Role in Assembly of the Ciliary Motile Apparatus
- Hidden Diversity in Honey Bee Gut Symbionts Detected by Single-Cell Genomics
- Ribosome Rescue and Translation Termination at Non-Standard Stop Codons by ICT1 in Mammalian Mitochondria
- tRNA Modifying Enzymes, NSUN2 and METTL1, Determine Sensitivity to 5-Fluorouracil in HeLa Cells
- Causal Variation in Yeast Sporulation Tends to Reside in a Pathway Bottleneck
- Tissue-Specific RNA Expression Marks Distant-Acting Developmental Enhancers
- WC-1 Recruits SWI/SNF to Remodel and Initiate a Circadian Cycle
- Clonal Expansion of Early to Mid-Life Mitochondrial DNA Point Mutations Drives Mitochondrial Dysfunction during Human Ageing
- Methylation QTLs Are Associated with Coordinated Changes in Transcription Factor Binding, Histone Modifications, and Gene Expression Levels
- Differential Management of the Replication Terminus Regions of the Two Chromosomes during Cell Division
- Obesity-Linked Homologues and Establish Meal Frequency in
- Derlin-1 Regulates Mutant VCP-Linked Pathogenesis and Endoplasmic Reticulum Stress-Induced Apoptosis
- Stress-Induced Nuclear RNA Degradation Pathways Regulate Yeast Bromodomain Factor 2 to Promote Cell Survival
- The MAPK p38c Regulates Oxidative Stress and Lipid Homeostasis in the Intestine
- Widespread Genome Reorganization of an Obligate Virus Mutualist
- Trans-kingdom Cross-Talk: Small RNAs on the Move
- The Vip1 Inositol Polyphosphate Kinase Family Regulates Polarized Growth and Modulates the Microtubule Cytoskeleton in Fungi
- Myosin Vb Mediated Plasma Membrane Homeostasis Regulates Peridermal Cell Size and Maintains Tissue Homeostasis in the Zebrafish Epidermis
- GLD-4-Mediated Translational Activation Regulates the Size of the Proliferative Germ Cell Pool in the Adult Germ Line
- Genome Wide Association Studies Using a New Nonparametric Model Reveal the Genetic Architecture of 17 Agronomic Traits in an Enlarged Maize Association Panel
- Translational Regulation of the DOUBLETIME/CKIδ/ε Kinase by LARK Contributes to Circadian Period Modulation
- Positive Selection and Multiple Losses of the LINE-1-Derived Gene in Mammals Suggest a Dual Role in Genome Defense and Pluripotency
- Out of Balance: R-loops in Human Disease
- A Genetic Assay for Transcription Errors Reveals Multilayer Control of RNA Polymerase II Fidelity
- Altered Behavioral Performance and Live Imaging of Circuit-Specific Neural Deficiencies in a Zebrafish Model for Psychomotor Retardation
- Nipbl and Mediator Cooperatively Regulate Gene Expression to Control Limb Development
- Meta-analysis of Mutations in Autism Spectrum Disorders: A Gradient of Severity in Cognitive Impairments
- The Proprotein Convertase KPC-1/Furin Controls Branching and Self-avoidance of Sensory Dendrites in
- Hydroxymethylated Cytosines Are Associated with Elevated C to G Transversion Rates
- Memory and Fitness Optimization of Bacteria under Fluctuating Environments
- Regulation of p53 and Rb Links the Alternative NF-κB Pathway to EZH2 Expression and Cell Senescence
- Interspecific Tests of Allelism Reveal the Evolutionary Timing and Pattern of Accumulation of Reproductive Isolation Mutations
- PRO40 Is a Scaffold Protein of the Cell Wall Integrity Pathway, Linking the MAP Kinase Module to the Upstream Activator Protein Kinase C
- Low Levels of p53 Protein and Chromatin Silencing of p53 Target Genes Repress Apoptosis in Endocycling Cells
- SPDEF Inhibits Prostate Carcinogenesis by Disrupting a Positive Feedback Loop in Regulation of the Foxm1 Oncogene
- RRP6L1 and RRP6L2 Function in Silencing Regulation of Antisense RNA Synthesis
- BMPs Regulate Gene Expression in the Dorsal Neuroectoderm of and Vertebrates by Distinct Mechanisms
- Unkempt Is Negatively Regulated by mTOR and Uncouples Neuronal Differentiation from Growth Control
- Atkinesin-13A Modulates Cell-Wall Synthesis and Cell Expansion in via the THESEUS1 Pathway
- Dopamine Signaling Leads to Loss of Polycomb Repression and Aberrant Gene Activation in Experimental Parkinsonism
- Histone Methyltransferase MMSET/NSD2 Alters EZH2 Binding and Reprograms the Myeloma Epigenome through Global and Focal Changes in H3K36 and H3K27 Methylation
- Bipartite Recognition of DNA by TCF/Pangolin Is Remarkably Flexible and Contributes to Transcriptional Responsiveness and Tissue Specificity of Wingless Signaling
- The Olfactory Transcriptomes of Mice
- Muscular Dystrophy-Associated and Variants Disrupt Nuclear-Cytoskeletal Connections and Myonuclear Organization
- Interplay of dFOXO and Two ETS-Family Transcription Factors Determines Lifespan in
- Evidence for Widespread Positive and Negative Selection in Coding and Conserved Noncoding Regions of
- Genome-Wide Association Meta-analysis of Neuropathologic Features of Alzheimer's Disease and Related Dementias
- Rejuvenation of Meiotic Cohesion in Oocytes during Prophase I Is Required for Chiasma Maintenance and Accurate Chromosome Segregation
- Admixture in Latin America: Geographic Structure, Phenotypic Diversity and Self-Perception of Ancestry Based on 7,342 Individuals
- Local Effect of Enhancer of Zeste-Like Reveals Cooperation of Epigenetic and -Acting Determinants for Zygotic Genome Rearrangements
- Differential Responses to Wnt and PCP Disruption Predict Expression and Developmental Function of Conserved and Novel Genes in a Cnidarian
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Admixture in Latin America: Geographic Structure, Phenotypic Diversity and Self-Perception of Ancestry Based on 7,342 Individuals
- Nipbl and Mediator Cooperatively Regulate Gene Expression to Control Limb Development
- Genome Wide Association Studies Using a New Nonparametric Model Reveal the Genetic Architecture of 17 Agronomic Traits in an Enlarged Maize Association Panel
- Histone Methyltransferase MMSET/NSD2 Alters EZH2 Binding and Reprograms the Myeloma Epigenome through Global and Focal Changes in H3K36 and H3K27 Methylation
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



