-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
BMPs Regulate Gene Expression in the Dorsal Neuroectoderm of and Vertebrates by Distinct Mechanisms
The trunk nervous system of both vertebrates and invertebrates develops from three primary rows of neural stem cells whose fate is determined by neural identity genes expressed in an evolutionarily conserved dorso-ventral pattern. Establishment of this pattern requires a shared signaling pathway in both groups of animals. Previous studies suggested that a shared signaling pathway functions in opposite ways in vertebrates and invertebrates, despite the final patterning outcomes having remained the same. Here, we employ bioinformatics, biochemistry, and transgenic animal technology to elucidate the genetic mechanism by which this pathway can engage the same components to generate opposite instructions and yet arrive at similar outcomes in patterning of the nervous system. Our findings highlight how natural selection can act to conserve a particular output pattern despite changes during evolution in the genetic mechanisms underlying the formation of this pattern.
Published in the journal: . PLoS Genet 10(9): e32767. doi:10.1371/journal.pgen.1004625
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004625Summary
The trunk nervous system of both vertebrates and invertebrates develops from three primary rows of neural stem cells whose fate is determined by neural identity genes expressed in an evolutionarily conserved dorso-ventral pattern. Establishment of this pattern requires a shared signaling pathway in both groups of animals. Previous studies suggested that a shared signaling pathway functions in opposite ways in vertebrates and invertebrates, despite the final patterning outcomes having remained the same. Here, we employ bioinformatics, biochemistry, and transgenic animal technology to elucidate the genetic mechanism by which this pathway can engage the same components to generate opposite instructions and yet arrive at similar outcomes in patterning of the nervous system. Our findings highlight how natural selection can act to conserve a particular output pattern despite changes during evolution in the genetic mechanisms underlying the formation of this pattern.
Introduction
In both Drosophila melanogaster and vertebrates, Bone Morphogenetic Proteins (BMPs) are expressed in the epidermal ectoderm abutting the dorsal border of the neuroectoderm [1]. The genetic network that underlies formation of a centralized nervous system consisting of segregated motor and sensory centers appears to have been conserved across bilaterians (animals with right-left symmetry) [2]. BMPs are thought to exert a common function in the early epidermal ectoderm during neural induction (i.e., suppressing expression of neural genes in epidermal regions that experience peak BMP levels). BMP signaling also acts subsequently in a dose dependent fashion to pattern dorsal versus medial regions of the neuroectoderm. For example, the trunk Central Nervous System (CNS) of both invertebrates and vertebrates consists of three primary rows of neuroblasts that are determined by the expression of three conserved transcription factors. In metazoan species spanning all three primary branches (e.g., Ecdysozoa -Drosophila, lophotrochozoa – annelids, and deuterostomes - vertebrates) “neural identity” genes (vnd/nkx2.2, ind/gsh and msh/msx) are expressed in the same relative order and orientation with respect to the dorsal-ventral axis and an epidermal BMP source. Moreover, in a broad range of organisms, BMPs and opposing antagonists have been found to play a key role in patterning the ectoderm and establishing neuronal fates. These commonalities suggest an ancestral origin for the CNS among bilateria [1]–[4] and raise the possibility that BMPs play a conserved role in patterning the CNS axis.
Despite their consistent role in promoting epidermal over neuronal cells fates in diverse species, BMPs and other extracellular factors are deployed in diverse patterns and may act by distinct mechanisms to achieve D/V patterning [5]. For instance, in Drosophila, BMPs originating in the presumptive epidermis act to repress expression of neural genes during both neural induction [6] and subsequent neuroectodermal patterning [3], [7]. In vertebrates, however, the prevailing view is that BMPs act as they do in flies to repress expression of neural genes within epidermal regions early during neural induction [1], [8] but switch function later to activate expression of orthologous neural identity genes in dorsal regions of the neural tube (e.g., the msh orthologs Msx1/2) [9]. Thus, in mice, ectopic BMP signaling leads to ventral expansion of msx expression in the neural tube [10]. In contrast, in Drosophila, the absence of BMPs leads to msh expanding dorsally into non-neural domains [11]. In zebrafish, there is evidence that BMPs act in a bimodal fashion where intermediate BMP levels are necessary for activating Msx genes, while both low and high levels of BMPs repress or fail to activate these target genes [12]. Similarly, in amphioxus, a basal chordate, msx is expressed more broadly but at reduced levels in response to ectopic BMP signaling [13]. In Echinoderms, where BMPs and chordin are co-expressed in the ventral ectoderm that gives rise to neural tissue [14], msx is expressed dorsally and is activated by peak levels of BMPs that diffuse dorsally from their ventral source into non-neural regions while Chordin remains restricted to ventral regions where it blocks the BMP response in neural cells [15]. While these conserved suites of gene expression strongly suggest a common ancestral origin for BMPs in axial patterning, it is unclear whether the regulatory mechanisms establishing these patterns have been similarly conserved during evolution.
BMPs signal via hetero-tetrameric receptor complexes consisting of two type-I and two type-II subunits, which in turn phosphorylate the cytoplasmic transducing-SMAD proteins (Mothers Against Dpp (Mad) in Drosophila, SMAD1/5/8 in vertebrates). Once phosphorylated, pMad/pSMAD1/5/8 translocates into the nucleus in a complex with Medea/Smad4 whereupon they act as transcription factors to regulate expression of BMP target genes (reviewed in [16]). Mad and Medea (Med) bind DNA as a heteromeric complex consisting of two Mad subunits and one Med subunit to regulate genes through interactions with binding sites composed by a Mad (GC-rich) site separated, by a variable length spacer, from a Med (Smad Binding Element or SBE) site. One of the best characterized such sites in Drosophila is the brinker (brk) Silencer Element (SE) which has a spacer length of 5 nucleotides [17]–[20]. Brk encodes a transcriptional repressor protein and the brk gene itself is repressed by Dpp (the Drosophila BMP4 homologue) signaling. Repression of brk through its SEs requires the presence of the zinc-finger protein Schnurri (Shn) [21]–[23], which is provided maternally and is also expressed zygotically in dorsal epidermal regions of the early embryo. Hence, in Drosophila, genes that are repressed by BMPs have been found to have binding sites for pMad/Med/Shn (henceforth, pMMS) complexes in their cis-Regulatory Modules (CRMs) while genes that are directly activated by BMPs, such as the inhibitory SMAD daughters-against-dpp (dad), contain activating elements (AE) in their CRMs [24]. These AE elements also share a bipartite configuration (GC-rich/spacer/SBE), but have configurations (spacing and sequence constraints) that do not allow for Shn binding and lead instead to the recruitment of activating transcriptional co-factors.
Here, we compare BMP-mediated regulation of CRMs controlling the expression of the Drosophila msh and zebrafish and mouse msx genes in the early dorsal nerve chord. We identify zebrafish and mouse msx neuroectodermal CRMs that drive expression in the dorsal neuroectoderm. We find that both Drosophila msh and zebrafish msxB CRM-reporter transgenes respond to BMPs and characterize BMP responsive sites within these elements. Consistent with prior genetic studies [7], the Drosophila msh CRM contains Shn-dependent SE sites that are required for BMP repression. Surprisingly, it also harbors sites that resemble known BMP-responsive activation sites, which, however, do not bind to pMad/Medea (pMM) complexes in vitro, but are nonetheless required for msh expression. In addition, we characterize a single SMAD binding site with a novel spacing of SMAD1/5/8 and SMAD4 binding motifs in a minimal zebrafish msxB CRM that is required for dorsal neuroectodermal expression. This comparison suggests that while overall gene expression patterns have been conserved between flies and zebrafish and are both regulated by BMP signaling, distinct mechanisms have evolved to generate the shared output patterns in these two widely separated metazoan lineages.
Results
The Drosophila msh CRM responds to BMPs
A 700 bp msh CRM (henceforth referred to as ME for Msh Element) has been identified that is directly repressed by Ind [25]. The response of the ME to BMP-mediated regulation has not yet been investigated, however. As is the case for the endogenous msh gene (Fig. 1A), the expression of a ME-lacZ construct expands throughout the dorsal region of the embryo in dpp - mutants (Fig. 1B). In order to determine whether Dpp regulates msh directly or indirectly, we analyzed BMP regulation of the ME element. Consistent with a direct role of BMP signaling on this CRM, genome wide chromatin immune precipitation (ChIP) data [26], [27] revealed DNA binding sites for the BMP effectors Mad, Medea and Shn within the ME region in blastoderm stage embryos (available on the UCSC genome browser - http://genome.ucsc.edu/ or the Berkeley Drosophila Transcription network Project - http://bdtnp.lbl.gov/Fly-Net/chipchip.jsp) (Fig. 1D). We confirmed the involvement of Shn in regulating msh within the neuroectoderm by examining homozygous zygotic shn - mutant embryos, which exhibit a partial dorsal expansion of msh expression (Fig. 1C).
Fig. 1. The msh CRM responds to BMPs. 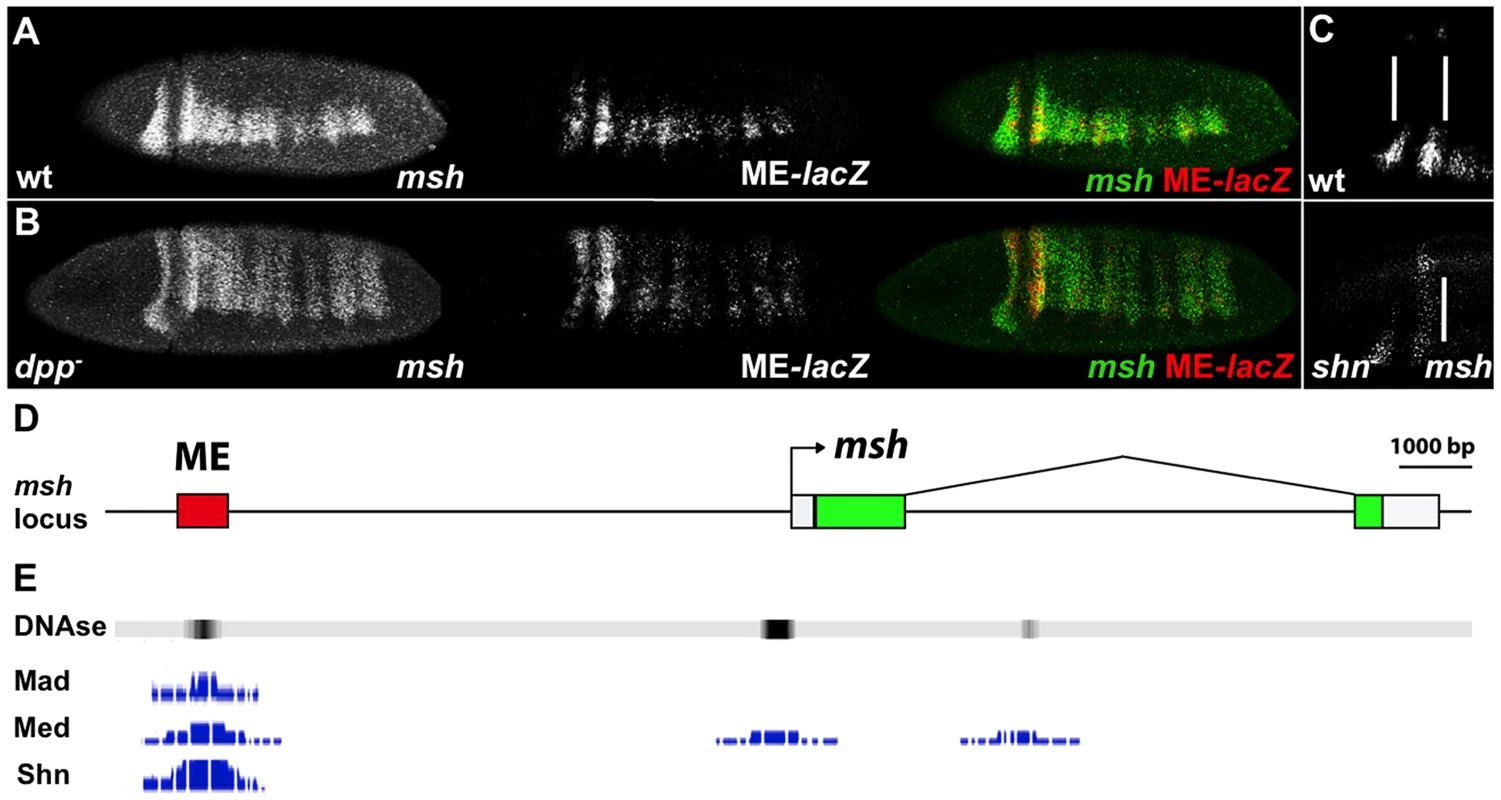
Lateral and dorso-lateral views (anterior to the left) of blastoderm stage Drosophila embryos showing msh, shn and ME-lacZ reporter construct expression detected by in situ hybridization, and schematic overview of the msh locus. (A) Transgenic embryos carrying the lacZ reporter gene under control of the ME were hybridized with probes against msh (green) and ME-lacZ (red). (B) The ME-lacZ construct was crossed into a dpp null background. Expression of both msh and ME-lacZ expands dorsally in dpp- homozygous mutant embryos. (C) In shn- homozygous zygotic mutant embryos, anterior msh expression expands towards the dorsal regions of the embryo compared to wild-type. A maternal contribution of shn remains intact in these embryos. (D) The msh locus depicting the location of the ME CRM. (E) Genome wide ChIP data representation depicting peaks of Mad, Med and Shn binding signal as well as DNase hypersensitivity sites reflecting regions of open chromatin in the ME CRM region. The msh CRM contains direct BMP-responsive sites involved in repression
To identify BMP responsive sites within the ME, we first scanned this element for known consensus binding sites for Mad, Med, Shn, and Brk. The two best characterized BMP responsive elements are the Silencer Element (SE), which binds a trimeric complex comprised of pMMS (GNCGNC(N)5GNCTG), and the activator element (AE), which binds pMM heteromers (GGCGCCA(N)4GNCV). Brk binding sites ((T)GGCGYY) overlap with a subset of AE elements [24]. Although there are no perfect consensus SE, AE, or Brk sites within the ME, we identified several candidate sites with either single base-pair mismatches to the SE or AE elements or variable spacer length (N)5–6. We defined three such candidate SE sites (SE1, SE2 and SE3) with a single nucleotide mismatch and two conserved candidate AE sites with a spacer of 6 nucleotides (conforming to the expanded consensus: GNCGNC(N)6GNCV) and tested each of these sites for direct DNA binding of pMM or pMMS complexes in vitro using Electrophoretic Mobility Shift Assays (EMSAs).
The SE1 and SE2 candidate silencer sites (Fig. 2A) both conform to the relaxed consensus of GNYGNC(N)5GNCTG (where Y can be either C or T). EMSA experiments using DNA oligonucleotide probes reveal that pMM and pMMS complexes assembled on the SE1 and SE2 sites in a BMP dependent fashion (Fig. 2B) but not on the SE3 site (Fig. S1C). As expected, mutation of the Med (SBE) motif within the SE1 (SE1SBE) or SE2 (SE2SBE) sites abolished binding of all BMP responsive complexes in vitro. In contrast, none of the candidate AE or Brk sites bound pMM, pMMS, or Brk complexes (Fig. S1C) (see below however, regarding effects of mutating or deleting the candidate AE sites).
Fig. 2. The msh CRM contains BMP Silencer Elements contributing to dorsal repression. 
(A) Diagram of ME showing the location of Silencer Elements SE1 and SE2 within highly conserved regions. Conservation score (Cons) is based on alignment of the ME region in 12 Drosophila species [61]. (B) Autoradiograph of electrophoretic mobility shift assay gels with radiolabeled oligonucleotide probes incubated with extracts from S2 cells over-expressing activated Tkv (to induce BMP signaling), Med and Mad in the presence or absence of over-expressed C-terminal Schnurri (Shn). EMSA probe sequences depict the location of the GC-rich (red) and SBE (blue) motifs as well as the ind SE site [20]. Mutations relative to wild type sequences are indicated by lower case. SE1 and SE2 containing probes show higher molecular weight retardation when Shn is present versus Med and Mad alone (lanes 1–3 and 6–8). When the SBE motif of either SE1 or SE2 is mutated, probe retardation is no longer observed (lanes 4/5 and 9/10). When SE2 is mutated to conform to the canonical SE consensus, SE2*, the amount of probe retained appears higher compared to wild-type SE2 (lanes 11–13, see Fig. S1D for further verification of this effect in a competition assay). (C) Lateral and dorsal views (anterior to the left) of in situ hybridization detection of lacZ expression in ME-lacZ embryos demonstrating the in vivo effects of mutating SE1 and SE2 SBE motifs. ME: wild type embryos containing the intact ME-lacZ construct; dpp: dpp- mutant embryos show complete dorsal expansion of lacZ expression; D1: deletion of the first 100 bp of ME spanning the SE1 SE2 region leads to partial dorsal expansion accompanied by reduced levels of expression; SE1: mutating SE1 leads to modest dorsal expansion; SE2: mutating SE2 leads to significant dorsal expansion; SE1 SE2: mutating both SE1 and SE2 results in yet more pronounced dorsal expansion although not as extensive as observed for the wt-ME in a dpp- mutant background; SE2*: Converting SE2 to an optimal (ind-like) SE site results in reduced lacZ expression; SE2* dpp-: SE2* in a dpp- background shows complete dorsal expansion of lacZ. In order to test the in vivo roles of the SE sites, we mutated each site (i.e., using the same SBE mutations that abolished all BMP responsive DNA binding in vitro described above) and generated a series of small deletions spanning virtually the entire ME (i.e., all but 36 bp). These mutant constructs were inserted into the same chromosomal integration site as the reference ME construct using the PhiC31 transgenesis system [28]. Deletion of the 5′ most 100 bp of ME, which contains both SE sites, led to dorsal expansion of reporter gene expression (Fig. 2C). Transgene expression, however, was also weaker within its normal neuroectodermal domain, suggesting that contributing activation sites are also present within this region. Targeted mutation of the individual SE1 and SE2 sites also led to discernable dorsal expansion of reporter gene expression, which was more pronounced for the SE2 mutant. Mutating both SE sites in combination (SE1, SE2 double mutant) resulted in more prominent dorsal expansion than observed for either mutant alone, but still less than that observed for the wild-type ME (or the endogenous msh gene) crossed into a dpp - mutant background. We conclude that SE elements mediate direct BMP-dependent repression of the ME and that additional direct or indirect BMP-dependent inputs also contribute to negatively regulating this CRM.
Differential affinities of pMMS complexes for SE sites in the ind and msh CRMs may contribute to threshold-dependent repression of these genes
Our prior genetic studies revealed that BMP signaling is more effective in repressing expression of ind than msh [7]. One possible explanation for this differential response is that the ind CRM might contain higher affinity SE sites than those in the msh CRM. Indeed, a single perfect consensus matching SE site in the ind CRM (Fig. 2B) has been shown to be required for repression of this element dorsally [20], [29]. In line with the possibility that SE sites in the ind and msh CRMs have differing affinities for binding pMMS complexes, modifying the SE2 site by one base-pair to adhere to the optimal SE consensus resulted in greater pMMS binding (Fig. 2B - SE2*), which was most evident in competition experiments (Fig. S1D). We tested whether the optimized ind-like SE2* site would result in repression of msh CRM activity in vivo. In support of this site being more effective at recruiting repressive pMMS complexes, reporter gene expression driven by the SE2* ME was greatly reduced relative to that of the wild-type ME. This reduced expression was BMP-dependent since SE2*ME-driven reporter gene expression was restored and expanded throughout the dorsal region in a dpp - mutant background to a degree comparable to that observed for the intact ME (Fig. 2C). Taken together, these results suggest that differential affinities of pMMS complexes for SE sites in the ind and msh CRMs contribute to the mechanism by which silencer elements mediate graded BMP responses of these two genes in the Drosophila neuroectoderm.
The msh CRM contains essential activation sites that resemble BMP responsive sequences
As mentioned above, in our initial search for BMP-responsive sites in the ME we identified two sites that were similar to activation elements (AE) but that did not bind pMM complexes in EMSA assays (Fig. S1C). We nonetheless tested for potential roles of these sites by deleting them or creating a point mutation in one of them (AE2). Deletions encompassing either AE1 or AE2 or the AE2 point mutation greatly reduced ME-lacZ expression (Fig. 3B,C,E), while deletion of the 3′ most region containing a previously reported Ind site [25] resulted in ventral expansion of reporter gene expression as expected. We tested the possibility that activation of the ME via AE2 might be balanced against repression mediated by the SE1 and SE2 sites by constructing a triple mutant in which all three sites were eliminated. We reasoned that if the AE2 site, which acts as a bonafide activation site, functions in a BMP independent manner, combining it with the double SE site mutant might result in loss of expression (the AE mutant phenotype). On the other hand, if the AE2 site were providing an important activation function in the neuroectoderm via BMP signaling, the triple mutant should at least show ectopic expression dorsally (e.g., if this was a BMP-dependent activation site, relieving repression would give rise to normalized expression since we would be removing both activating and repressing components). We found loss of expression in this triple mutant comparable to that of the AE2 single mutant (Fig. 3F), suggesting that activation via the AE2 site is BMP-independent.
Fig. 3. The msh CRM contains AE-resembling sites required for expression. 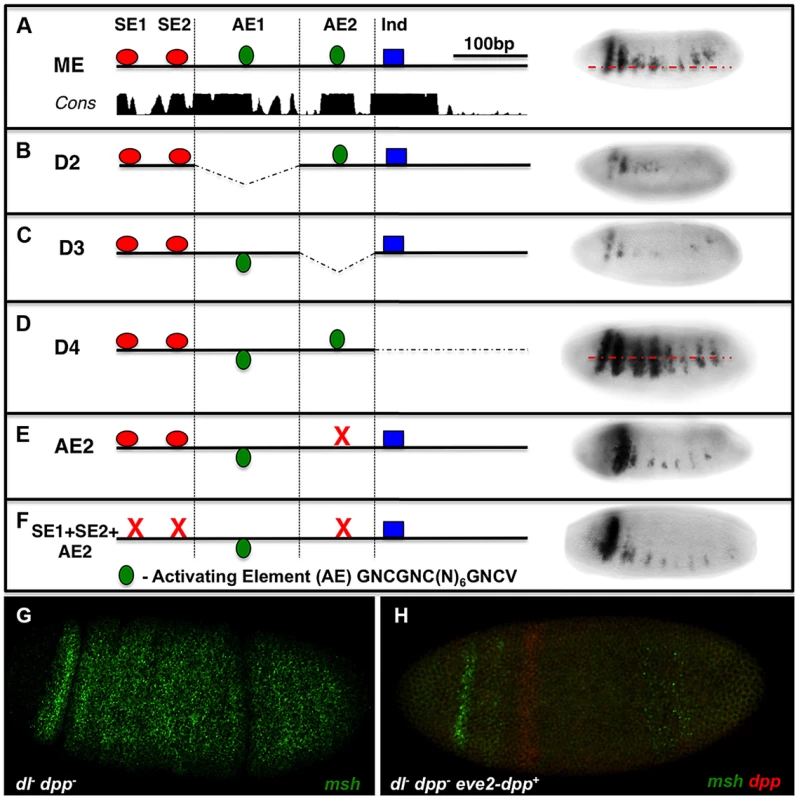
Embryos are viewed laterally (anterior to the left). ME deletions spanning 4 sequential blocks (D1-D4) contain distinct islands of conserved sequences on sequence conservation and binding site clusters. lacZ-reporter constructs carrying these deletions were then tested in vivo in transgenic embryos to identify additional potential BMP responsive sequences. (A) Wild-type ME depicting the location of putative AE sites in relation to SE sites and an Ind binding site. (B) Deletion of D2 where AE1 is located results in reduced lacZ reporter expression. (C) Deletion of D3, which contains the AE2 region results in severely reduced lacZ reporter expression. (D) Deletion of D4 results in ventrally expanded lacZ reporter expression consistent with an essential Ind repressor binding site being present [7]. (E) Site-directed mutagenesis of AE2 results in similarly reduced reporter expression as observed for the D3 deletion, demonstrating an essential role of AE2 site as an activation sequence. (F) A triple mutant comprised of point mutations in the SE1, SE2 and AE2 sites results in reduced reporter expression comparable to that observed in the AE2 mutant alone suggesting that this site does not mediate BMP-dependent activation. The AE consensus shown (GNCGNC(N)6GNCV) is an expanded version based on the standard consensus indicated in the text (GGCGCCA(N)4GNCV) and on our hand curation from the literature. In addition, sensitized embryos were tested for potential msh activation by particular doses of Dpp. (G) msh expression in an embryo lacking maternal Dorsal and lacking zygotic Dpp (dl- dpp-), msh (green) is ubiquitously expressed as previously reported [11]. (H) The addition of a single copy of a wild-type dpp gene under the control of the even-skipped stripe 2 CRM, which creates a Dpp gradient emanating from the zone of expression (red) [7] in dl- dpp- embryos abolishes most msh expression (green) throughout the embryo. These results suggest that Dpp does not have a positive role in msh regulation in the absence of Dorsal signaling in Drosophila at any dose. Although the above analysis suggests that the AE2 site acts in a BMP-independent fashion, we further examined the possibility that BMPs might play an activating as well as repressive role in regulating msh expression. Embryos that are dorsal - (maternal); dpp - (zygotic) double mutants express msh ubiquitously [11]. To test whether there might be a threshold at which Dpp enhances rather than suppress msh expression, we attempted to augment msh expression locally by generating embryos that lack Dorsal and whose only source of Dpp is one copy of dpp driven in a narrow stripe by the eve 2 CRM (Fig. 3G,H) or by, adding progressive amounts of Dpp (by varying copy number of the dpp locus – Fig. S2A,B). In both cases, we observed only a diminution in msh expression, further arguing against any activating role for Dpp. Finally, we considered the possibility that BMPs might act indirectly to regulate msh expression via non-canonical mechanisms (e.g. via ETS or the HMG-box Cic transcription factors) by altering EGF-R signaling. We found no evidence, however, for a role of EGFR signaling in influencing the position of the dorsal border of msh expression (Fig. S2C-E). In aggregate, our experiments suggest that BMP-dependent regulation of the ME is mediated by SE sites and by additional inhibitory inputs, which may act either directly or indirectly.
Identification of early embryonic vertebrate msx CRMs
The above analysis of the msh CRM in Drosophila is consistent with genetic data indicating that BMPs act by dosage sensitive repression of neural identity gene expression [7]. To determine the mechanism by which BMPs regulate expression of orthologous vertebrate Msx genes we sought to identify the zebrafish (Danio rerio) msxB CRM using the powerful tol2 transgenesis system [30]. We choose to focus on regulation of the msxB gene among the zebrafish Msx paralogs as this gene has the earliest onset and most specific pattern of expression in the dorsal neuroectoderm [31]. We identified a 2.4 Kb region of DNA immediately upstream of the zebrafish msxB coding region that drives faithful reporter gene expression in the dorsal neuroectoderm in both neural plate and early neural tube stages (i.e., 3–6 somite stage embryos) of a stable transformant line (Fig. 4A,B). This fragment has two peaks of strong sequence conservation among vertebrates, which overlap regions of predicted open chromatin [32] (Fig. 4A). Later during neural tube stages, the early neural plate expression pattern fuses into a single dorsal zone (e.g., top panels in Fig. 4D).
Fig. 4. Identification of vertebrate msx CRMs. 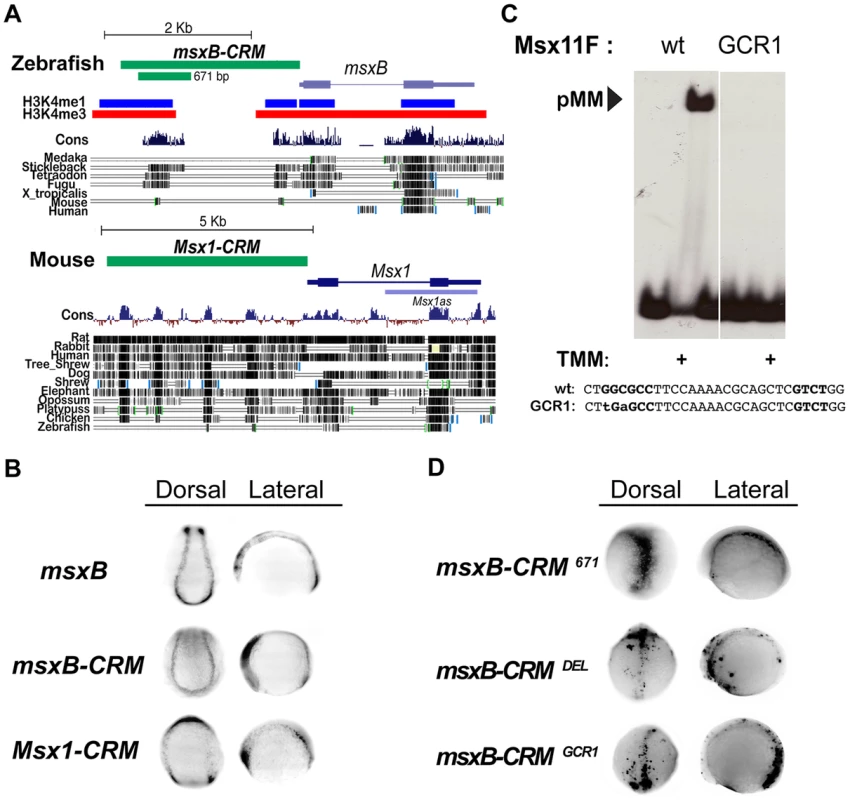
(A) Representation of the zebrafish msxB and mouse Msx1 loci depicting the location of the CRMs and vertebrate sequence conservation (Cons) (http://genome.ucsc.edu/). For zebrafish, histone 3 lysine 4 single and triple methylation patterns indicative of open chromatin are also indicated. Block conservation tracks for select species are represented for both loci. (B) In situ expression driven by msx CRMs. Dorsal (anterior to the top) and lateral (anterior to the left) views of transgenic zebrafish embryos at the open neural plate stages (3–6 somites). Embryos were injected with either msxB-CRM or Msx1-CRM constructs driving gfp and stable transgenic lines were subsequently bred. Stable transgenic embryos were stained for msxB and gfp expression, which was detected by in situ hybridization. Both CRMs drive patterns resembling the endogenous msxB pattern. The zebrafish DNA isolated contains sufficient information to drive a pattern resembling the endogenous msxB expression pattern. The cloned mouse CRM also is capable of responding to regulatory cues in the zebrafish embryo to drive expression resembling that of the zebrafish msxB gene (as well as the endogenous Msx1 gene in mice [62]). Note that this embryo is tilted in a slightly more rostral direction than the other embryos shown from the dorsal perspective, which results in bands of anterior expression coming into view. (C) EMSA experiment in which a radiolabeled oligonucleotide probe carrying the zAE element (Msx11F in Fig. S3) was incubated with extracts from S2 cells over-expressing activated Tkv (to induce BMP signaling), Med and Mad. When the GC-rich region of the mad1 binding site is mutated (GCR1), pMM biding is abolished (the same loss of binding was also observed for the mutation in the mad2 site, GCR2 - see Fig. S3C). (D) Mutation of the pMM zAE site greatly reduces specific expression driven by the 671 bp msxB CRM. Dorsal (anterior to the top) and lateral (anterior to the left) views of injected zebrafish embryos (6–8 somites). Embryos were injected with GFP-reporter constructs under the control of the intact 671 bp msxB-CRM, a 30 bp mutant deleting the zAE (msxB-CRMDEL), or a point mutant version of the zAE that abolishes pMM binding – see panel C (msxB-CRMGCR1). Both zAE mutant constructs show greatly reduced reporter expression. Transient gfp mRNA expression was detected by in situ hybridization. We also tested a 5 Kb genomic fragment upstream of the mouse (Mus musculus) Msx1 gene, which like the zebrafish msxB CRM carries sequences lying immediately upstream of the transcriptional start site (Fig. 4A). When the mouse CRM-GFP construct was introduced into zebrafish embryos, it drove expression in a pattern (Fig. 4B) very similar to that of the fish msxB gene as well as that observed endogenously in mice. These results suggest that both the zebrafish and mouse CRMs contain sufficient information to correctly direct expression to the dorsal ectoderm despite the fact that they show only limited sequence conservation. These observations provide another clear example of the highly conserved function of vertebrate CRMs from lineages that diverged over 400 MYA in the absence of obvious sequence conservation in these non-coding regions [33], [34].
The zebrafish msxB CRM contains a BMP responsive site mediating activation
We pared down the zebrafish msxB CRM in transient transformant embryos and identified a minimal 671 bp fragment containing the most conserved island that also faithfully recapitulates msxB expression in dorsal neuroectodermal/neural crest progenitor cells (Fig. 4D). Paralleling our approach in Drosophila, we searched for BMP responsive sites within the minimal msxB CRM by first scanning bioinformatically for candidate SE or AE sites using the SMAD1/5/8 consensus GNCKNC and SMAD4 consensus GNC(T/V) with relaxed spacing constraints, and then testing by EMSA whether oligonucleotides containing these sites could indeed assemble Drosophila pMM and/or pMMS complexes in response to BMP signaling in vitro (Fig. S3). This analysis identified a single highly conserved site (zAE) to which BMP signal-dependent pMM (but no pMMS) DNA binding was observed. The zAE contains candidate SMAD1/5/8 and SMAD4 binding sites separated by an unusually long 16 bp spacer (Fig. S3A,B). These sites are also present in mouse albeit with different spacing (12 bp). Further analysis of this binding motif revealed that the SMAD1/5/8 and SMAD4 sites are each required, suggesting that the functional zAE includes both sites (Fig. S3C). Changing the sequence or length of the spacer DNA linking the two sites did not affect the ability to form pMM complexes in vitro indicating that the exceptional length of the zAE spacer is not required for SMAD complex formation in vitro. Interestingly, however, changing the linker length to 5 bp allowed the formation of trimeric pMMS complexes (Fig. S3D).
We generated a 36 base pair deletion spanning the zAE (and both the SMAD1/5/8 and SMAD4 candidate binding sites – DEL mutant) in the context of the 671 bp msxB CRM and observed that GFP reporter gene expression was lost in transient transformant embryos (Fig. 4D). Similarly, mutation of two core base pairs in the GC-rich region of the zAE (GCR1 mutant), which abolished pMM binding in vitro (Fig. 4C; Fig. S3C), also reduced reporter expression in vivo in transient transformant embryos (Fig. 4D). These results indicate that a single BMP responsive site within the 671 bp zebrafish msxB CRM is required for mediating reporter gene activation by this element in vivo.
Fly and vertebrate msx CRMs respond oppositely to BMPs
The above dissection of BMP-responsive sequences within the Drosophila and zebrafish msh/msx CRMs suggests that they are under opposing forms of BMP regulation: repression in Drosophila versus activation in zebrafish. To test this hypothesis further, we compared the response of these CRMs to alterations in BMP signaling in vivo (Fig. 5). In Drosophila, we examined msh and ME reporter gene expression in both a dpp - mutant background and in embryos ectopically expressing dpp in the dorsal epidermis. As mentioned above, msh and ME-reporter gene expression both expand dorsally in dpp - mutant (Fig. 1B; Fig. 5A). Conversely, ectopic dpp expressed from a Heat Shock-dpp construct (HS-dpp) resulted in loss of msh expression within its normal domain (Fig. 5C). In zebrafish, a stable transgenic line carrying the 2.4 kb msxB-GFP reporter construct was crossed to lines carrying either a Heat Shock-chordin (HS-CHD) or a Heat Shock-BMP (HS-BMP) construct. When the BMP antagonist Chordin was induced by heat treatment (Fig. 5G), msxB-GFP reporter expression was strongly suppressed, as was endogenous msxB expression (Fig. 5D). The opposite effect was observed in HS-BMP embryos, however, where expression of endogenous (Fig. 5F) and reporter (Fig. 5I) genes was broadened compared to control embryos (Fig. 5E and 5H, respectively) that were subjected to the same conditions. Thus, consistent with the inverse effects of mutagenizing BMP-responsive sites in the Drosophila msh and zebrafish msxB CRMs, these two elements respond in an opposing fashion to equivalent manipulations of BMP signaling in vivo. Our analysis strongly suggests that BMPs pattern the neuroectoderm primarily via repression in Drosophila, while in zebrafish, BMPs function, at least in part, to activate the orthologous msxB gene.
Fig. 5. Manipulating BMP signaling elicits opposite responses from msh and msxB in Drosophila and zebrafish embryos. 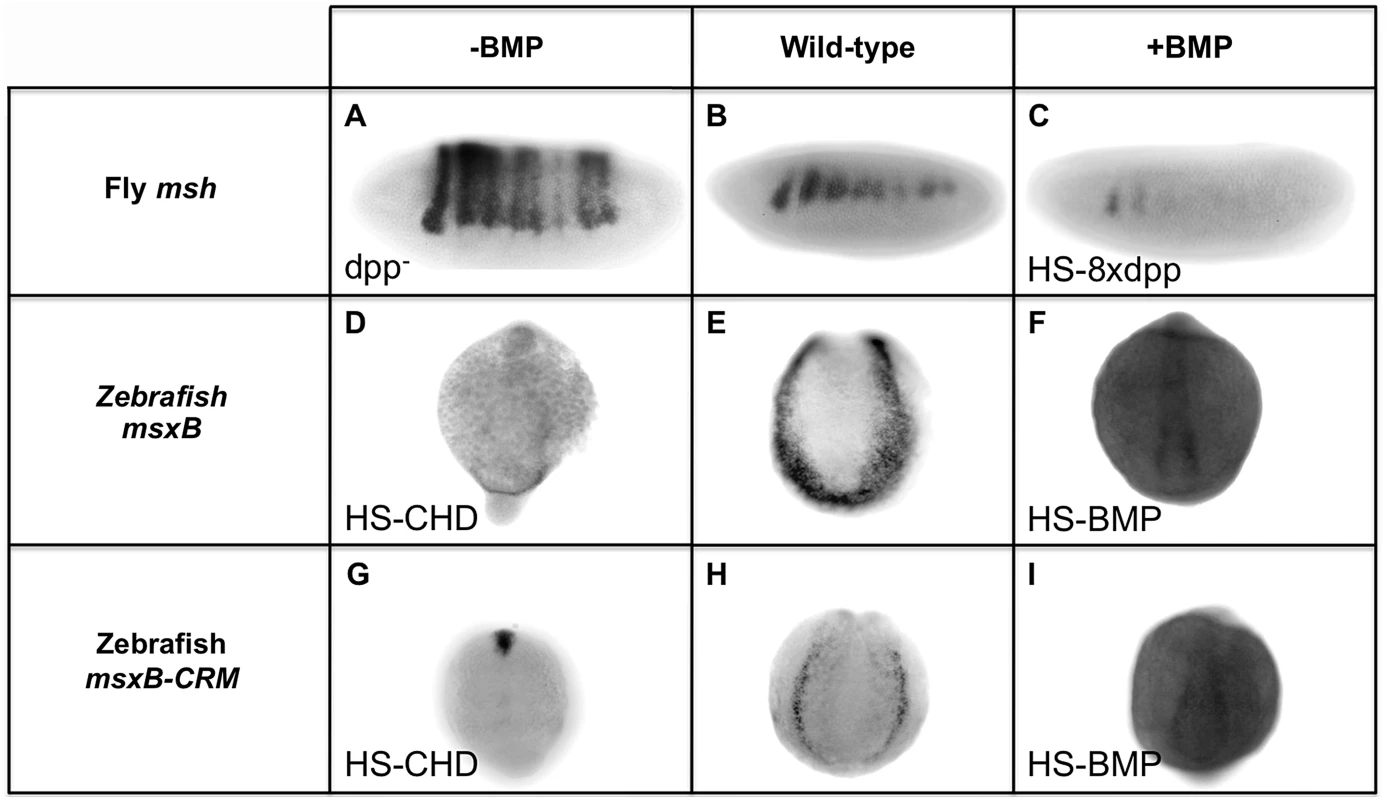
Comparison of equivalent BMP manipulations in Drosophila (Dmel) and zebrafish (Drer) embryos and their effects on msh, msxB and msxB-CRM driving GFP assayed by in situ hybridization. All embryos are oriented with dorsal at the top. Drosophila embryos are oriented with anterior to the left, while zebrafish embryos are view from a posterior perspective. Loss of BMP signaling is Drosophila was examined in dpp null mutant (dpp−) embryos (A) and ectopic BMP signaling was generated by heat induction of transgenic embryos carrying eight copies of a heat-shock dpp construct (HS-8xdpp) (C). In zebrafish, BMP signaling was reduced by heat induction of transgenic embryos carrying a Heat Shock Chordin construct (HS-CHD) (D, G), while BMP over-expression was accomplished by induction of transgenic embryos carrying a Heat Shock BMP2 construct (HS-BMP) (F,I). In dpp− embryos, msh expression expands dorsally as shown also in Fig. 1B. In, contrast, in HS-CHD embryos, expression is weakened relative to the wild-type pattern for both the endogenous msxB gene (E) and msxB-CRM-gfp reporter construct (H). Drosophila HS-8xdpp embryos show reduced msh expression compared to wild type embryos (B) consistent with Dpp having a repressive action on msh expression, while zebrafish HS-BMP embryos exhibit ectopic expression of both the endogenous msxB gene and the GFP reporter gene driven by the msxB-CRM when compared to wild-type. Discussion
BMPs play a highly conserved role in neural induction and also contribute to establishment of dorsal-ventral polarity within the CNS. In the latter case, however, it has not been established whether they act through common or distinct mechanisms to effect dose-dependent patterning of neural identity genes. Since BMPs regulate expression of highly conserved members of the ancient msh/msx family in the dorsal neuroectoderm of both Drosophila and vertebrate embryos, comparing cis-regulation of these genes by BMPs provides an excellent paradigm for addressing whether cis-regulatory processes are maintained across distant taxa. Our analysis of the Drosophila msh embryonic CRM suggests that BMPs act in part through two SE-type binding sites that mediate repression, while activation sites do not appear to mediate responses to BMP signaling. In contrast, we identified a single SMAD binding site within an embryonic zebrafish msxB CRM that is required for BMP-dependent activation. These findings suggest that BMPs act on msh/msx CRMs by opposite mechanisms in these two lineages, while nonetheless driving similar output expression patterns in the dorsal neuroectoderm.
Silencer sites mediate graded BMP responses in the Drosophila neuroectoderm
Mutational analysis of the Drosophila msh CRM in this study supports a direct role for BMP repression acting via the SE1 and SE2 sites to suppress activity of this element in the dorsal ectoderm where there are likely to be moderate levels of BMP signaling. Mutation of either of these sites results in modest dorsal expansion of reporter gene expression while elimination of both sites by point mutations or a deletion spanning both sites causes more pronounced ectopic dorsal expression. The dorsal expansion in SE1, SE2 double mutants is less complete, however, than that observed when the intact ME is crossed into a dpp - background, indicating that additional inputs are also involved in repressing the activity of this element dorsally. These additional BMP-dependent inputs might act either directly or indirectly. Since each of the three deletions spanning the remaining portions of the CRM (i.e. sequences outside of the deletion covering the SE1 and SE2 sites) result in reduced CRM activity it is possible that the effects of such hypothetical additional BMP responsive sites are canceled out by the deletion of necessary adjacent activation sites (e.g., deletion of the A2 site in the D3 region, Fig. 3C). If these hypothetical repressor sites act directly on the msh CRM they would presumably bind Mad, Medea and Schnurri, MAPK pathway transcriptional effectors, or possibly yet unknown BMP mediators alone or in conjunction with other transacting factors. Our detailed bioinformatic analysis and systematic experimental EMSA surveys have failed to identify any such sites, however. It is also possible that part of the BMP response of the msh CRM is mediated indirectly. For example, we have previously reported that localized overexpression of Brk can de-repress msh expression dorsally [7], yet there are no consensus Brk sites in the ME and we were unable to detect Brk protein binding to any closely related candidate Brk sites by EMSA (Fig. S1). Thus, Brk may act via regulating expression of other components required for BMP signaling such as the BMP type-1 receptor Thick veins [35]. Alternatively, activators of the ME may be under negative BMP/Brk regulation.
The SE1 and SE2 sites that play a role in repressing ME activity dorsally are imperfect matches to the consensus SE sites determined by Pyrowolakis and colleagues [36]. The ind CRM, however, which according to genetic data is more sensitive to BMP repression than msh [7], contains a perfect SE site required for repressing activity of this element dorsally [20]. When the SE2 site in the ME was mutated to similarly match the ideal SE consensus sequence (SE2*) it repressed ME expression in its normal dorsal ectodermal domain in a dpp-dependent fashion (Fig. 2C). In addition, competition experiments indicate that Mad/Schnurri/Medea bind to the ind-like SE element with higher affinity than the msh SE2 element (Fig. S1D). These combined findings suggest that differences in affinity of SE sites for forming Mad/Med/Shn complexes contribute to the distinct responses of the two CRMs to BMP-mediated threshold-dependent repression.
A single BMP responsive site is required for activity of the msxB CRM
Using a combination of bioinformatics and efficient transgenesis in zebrafish we identified genomic fragments upstream of the zebrafish msxB and mouse Msx1 genes that drive neuroectodermal GFP-reporter gene expression at the open neural plate stage in zebrafish embryos. Further analysis of a minimal 671 bp zebrafish CRM identified a single conserved SMAD binding site that is required for activity of this element. An novel feature of this BMP-activation site is that the SMAD1/5/8 and SMAD4 binding site motifs are separated by a 16 bp spacer, which interposes approximately one and half turns of the DNA helix between these two sites, thus differing from other characterized vertebrate BMP activation sites in which these SMAD binding sites are closer [37]. Interestingly, deletion of 11 bp (about one turn of the helix) endows this modified site with the ability to bind the pMMS repressor complex in vitro. Whether this unique architecture of the msxB BMP activation site is relevant to activity within the neuroectoderm remains to be explored.
We also examined the in vivo response of the endogenous zebrafish msxB gene and the msxB-CRM to inhibition of BMP signaling or ectopic expression of BMPs and compared these responses to equivalent manipulations of BMP signaling in Drosophila. In Drosophila, msh or ME-lacZ expression expands dorsally in a dpp - mutant while msh expression is repressed within its normal dorsal neuroectodermal domain by ectopic dpp expression. In contrast, expression of the zebrafish msxB gene, which is mirrored by activity of the msxB-CRM, is lost upon inhibition of BMP signaling and expanded or elevated in response to ectopic BMPs. Thus, both mutational analysis and in vivo testing suggest opposing mechanisms for BMP-dependent regulation of the msh and msxB genes in the early neuroectoderm.
Given the opposing mechanisms by which the msh and msxB CRMs respond to BMPs, it is intriguing that a site within the msh CRM closely resembling an activation site (AE2) is required for activation of this CRM. Also, another AE-like site (AE1) lies within a region which when deleted greatly reduces ME driven reporter gene expression, although the role of that AE1 site remains to be examined. These AE-like sites, while having only single mismatches to consensus Mad-Medea binding sites, did not bind Mad-Medea complexes in vitro, indicating that they are most likely not involved in mediating a BMP response. Additionally, experiments designed to identify potential positive roles of BMP signaling in regulating ME activity provided no evidence for such an effect. Given the known role of AE sites in other genes to BMP-dependent activation and the evidence that BMPs can act positively to promote msx gene expression in vertebrates, it is tempting to speculate that these sites could once have been BMP responsive activation sites and were subsequently co-opted by different transcription factors (possibly a TAGteam motif [38] binding protein) in the course of evolution to maintain msh expression in a BMP-independent fashion. Identifying such transcriptional activators is an interesting goal for future experiments.
Evolution of conserved gene expression patterns
In Drosophila, Evo/Devo studies of the even-skipped stripe 2 CRM have suggested that regulatory mechanisms that lead to a particular gene expression pattern are extremely flexible, i.e., the same pattern can be achieved in multiple ways [39]. Accordingly, in the current case of BMP-dependent regulation of msh/msxB expression, natural selection may have operated similarly to maintain relevant gene expression patterns that fulfill a particular function (i.e. dorsal neuroectodermal expression) while allowing the upstream mechanisms generating that pattern to change over time.
As summarized above, our analysis strongly suggests that BMPs pattern the neuroectoderm primarily via repression in Drosophila, while in zebrafish, BMPs function, at least in part, to activate the orthologous msxB gene. Genetic studies and exogenous BMP treatment in zebrafish suggest that msx gene expression may also be repressed by high levels of BMP signaling. Whether the BMP-responsive site in the 671 bp msxB CRM together with other potential BMP-responsive elements mediate such a biphasic response will be interesting to address in future experiments. In the future, it will also be important to determine whether expression of other msx paralogs in the dorsal CNS of zebrafish (e.g., msxC,E [31]) or msx genes in other vertebrates (e.g., the murine Msx1 neuroectodermal CRM identified here) are similarly regulated by BMPs. Analysis of these additional vertebrate msx CRMs should reveal whether distinct evolutionary trajectories have shaped the BMP responsiveness of these elements. Such comparative studies may also shed light on whether there is a single or multiple independent origin(s) of BMP regulation of vertebrate msx genes. Furthermore, analysis of the CRM driving BMP-dependent expression of an echinoderm Msx homolog in regions of peak BMP activity [14] will be informative since this gene is expressed in the non-neural ectoderm. In this case, one might predict finding only positively acting AE-like BMP-responsive sites.
There are two possible explanations for distinct mechanisms of BMP-regulation of msh/msxB expression in flies versus fish. One is that these genes independently evolved BMP responsiveness. Alternatively, BMP-dependent regulation may be an ancestral trait dating back to the first bilaterians with a condensed CNS. We favor the latter alternative for the following reasons. First, the co-linearity of msh-msx, ind-gsh/pax, and vnd-Nkx2.2 genes relative to the source of BMPs and the BMP responsiveness of these genes in species from all three primary branches of bilateria - flies (ecdysozoa), vertebrates (deuterostome chordates), and annelid worms [40] (lophotrochozoa) - provides a compelling argument for this arrangement reflecting the ancestral state. Second, a polarized source of BMPs was present in diploblasts (e.g., corals [41], [42], jellyfish [43], and the sea anemone [44], [45]) and therefore preceded evolution of bilaterian triploblasts and a condensed CNS. Thus, it is plausible that a single species evolved a condensed CNS which deployed neural identity genes along the DV axis in much the same way that Hox genes are expressed in sequential order along the AP axis. Finally, if one looks more broadly among the 30 bilaterian phyla, a striking trend is that at least some clades within most of these phyla have a condensed CNS with three primary axon bundles [46], suggestive of an ancestral tripartite subdivision of the CNS. It is true that there are also many examples of species scattered among these phyla that either secondarily lost a condensed polarized CNS or retained a prior ancestral state in which there was only a distributed nervous system. Echinoderms in which Msx genes are expressed in the non-neural ectoderm (see above) or the hemichordate Saccoglossus kowalevskii which has lost bilateral symmetry to become radially organized [47] may be examples of such derived simplifications of the nervous system. Thus, in our view, the most likely scenario is that the ancestral bilaterian CNS was a condensed nervous system partitioned into at least three DV domains and that loss of centralization has occurred numerous times in different lineages undergoing morphological simplification.
If one assumes a common ancestral origin for BMP-regulation of msx genes, one can imagine various scenarios under which BMP-mediated regulation of msh/msx genes could have switched its effect during evolution. In vertebrates, BMP targets frequently contain Drosophila SEs that activate rather than repress transcription. This might be due to Shn proteins losing their repressive activity through changes in the Shn amino acid sequence and/or the lack of components required for repression downstream of Shn. The molecular relatedness of SEs and AEs raises the possibility that ancestral SE-mediated repressive effects on msh/Msx expression may have been relatively easy to convert into activating effects in the vertebrate lineage by the loss of the Shn repressor function. Consequently, the increased linker length of zAE could be accounted for by the lack of evolutionary pressure on the SE to meet the sequence requirements for Shn recruitment.
Since the Drosophila msh gene is weakly repressed by BMPs (e.g., relative to ind and other neural genes such as AS-C, scrt or sna [6]), while vertebrate msx genes are weakly activated by BMPs (i.e., high neuroectodermal levels of BMPs are required to activate msx genes) an intermediate CRM state may have existed in which BMPs both weakly activated msx gene expression within the neuroectoderm at moderate levels while repressing gene expression at the peak BMP levels present in the adjacent epidermis. Indeed the zebrafish msxB gene may represent such a bifunctional intermediate condition since in vivo studies indicate that high levels of BMPs can inhibit msxB expression [12]. It remains to be determined whether such proposed positive and negative inputs are mediated by a single or multiple independent CRM(s). Within different evolutionary lineages such biphasic responses could have then been rendered monophasic in opposing directions to account for the observed differences in the Drosophila versus vertebrate or echinoderm Msx CRMs. In vertebrates, one potential driving force for reducing the effect of BMP-mediated inhibition may have been the incorporation of BMP expression within the dorsal neural tube itself since this would be expected to generate much higher BMP levels than would result from BMPs diffusing in from the adjacent epidermal ectoderm (e.g., as is the case in Drosophila).
In future analyses it will also be important to examine BMP-mediated regulation of additional neural identity genes expressed along the dorsal-ventral axis including the Gsh ≈ ind and Nxk2.2 ≈ vnd genes as CRMs controlling expression of each of these genes will have undergone independent evolutionary trajectories. Since there is evidence that laterally and ventrally expressed genes in vertebrates are inhibited by BMPs [48]–[52], and because the more ventrally expressed ind gene in Drosophila is more sensitive to BMP-mediated repression than msh [7], one might expect to find similar, and perhaps conserved ancestral modes, of BMP-mediated repression of these genes across bilateria.
It will also be interesting to understand how flexible the ancestral metazoan state was by investigating the relationship between BMPs and msx genes in basal metazoans such as jellyfish. In these diploblastic animals, although the BMP-msx relationship has not been tested, BMP2/4 [53] and msx [54] homologues are expressed in adjacent regions during development, as is the case in the majority of triploblastic animals.
Materials and Methods
Bioinformatics
We identified candidate SE and AE sites in the msh, msxB and msx1 CRMs using binding site consensus sequences curated from the literature referenced and used Gene Palette [55]. For this analysis, we used the consensus sequence GNCGNC(N)5GNCTG to identify candidate Silencer Elements (SE) and the consensus GGCGCCA(N)4GNCV for Activator elements (AE) allowing for single base-pair mismatches to these consensus sequences. We identified candidate zebrafish msxB and mouse Msx1 CRMs by using genome wide alignments for multiple vertebrate species, which indicates regions of high sequence conservation as provided by the UCSC genome browser (http://genome.ucsc.edu).
CRM-reporter construction and analysis
The 700 bp msh CRM is described in Von Ohlen et al., 2009 [25]. All primers used in this study and the corresponding constructs generated can be found in Table S1. The various Drosophila msh-CRM constructs were subcloned in pCR-TOPO vectors (Invitrogen) and subsequently cloned into the [P]acman vector [28] as NotI and KpnI restriction fragments. Site-directed mutagenesis PCR methods were adapted from [56]. The primers used to isolate the zebrafish msxB CRMs and the mouse msx1 CRM can be found in Table S1. Zebrafish constructs were cloned into pENTR-TOPO (Invitrogen), transferred to pTol2 by Gateway Recombination and injected in zebrafish embryos as previously described [30].
Genetic strains and crossing schemes
The Drosophila dpph46 null allele used in this study is Flybase stock number 2061. The 8x HS-dpp stock and its use are described in Biehs et al 1996 [6]. The schnurri mutant allele is shn04738. To generate the dl dpp st2-dpp+ embryos, females that are Dpdpp/+; dl1 cn1 sca1/dpph46 wgsp dl1 were crossed to yw/Y; dpph46 wgsp st2-dpp+,w+/CyO males. The fly strain used to inject all constructs has genotype PBac{yellow[+]-attP-3BVK00002 and injections were outsourced to BestGeneInc (http://www.thebestgene.com/).
The zebrafish strains containing the hsp70:bmp2b [57] and hsp70:chd [58] transgenes were crossed to stable transgenic lines containing the msxB-CRM construct. Embryos at the sphere stage (4hpf) were subjected to heat shock at 37°C for 1 hour and then returned to normal temperature of 28.5°C until they were fixed at the bud – 6 somite (10–11hpf) stage as necessary.
Electrophoretic mobility shift assays
For electrophoretic mobility shift assays (EMSA), Drosophila S2 cells were co-transfected with 50 ng TkvQD and 175 ng Mad - and Med-expression plasmids or with 400 ng of a ShnCT-expression plasmid. Cells were harvested 72 hr after transfection and lysed for 10 min at 4°C in 100 µl of 100 mM Tris (pH 7.5), 1 mM DTT, 0.5% TritonX100 and 1%NP40. Radioactively labeled probes were generated by annealing and filling in partially overlapping oligonucleotides in the presence of [P]-32 ATP. Binding reactions were carried out in a total volume of 25 µl containing 12.5 µl 2x binding buffer (200 mM KCl, 40 mM HEPES (pH 7.9), 40% glycerol, 2 mM DTT, 0.6% BSA and 0.02% NP40), 10000 cpm of radioactively-labeled probe, 1 µl poly dIdC (1 mM) and 7 µl of cleared S2 cell extracts. After incubation for 30 min at 4°C, the reactions were analyzed by non-denaturing 4%polyacrylamide gel electrophoresis followed by autoradiography.
In situ hybridization
Fluorescent in situ hybridization methods used were performed according to [59] in Drosophila embryos and adapted to zebrafish embryos by increasing the hybridization temperature: 55°C in Drosophila to 65°C in zebrafish embryos. Antibodies used: dpERK (Cell Signaling Technology #5683), anti-digoxigenin (Roche), anti-biotin (Roche), Alexa fluor 488, 594, 647 (Invitrogen). We also used colorimetric staining methods performed according to O'Neill and Bier [60]. The DNA template used to generate the msxB probe was a generous gift from the Riley lab. Histochemical stain images were acquired using a Nikon optical microscope and fluorescent stain images were collected using a LEICA SP2 confocal microscope. Images were adjusted for color, brightness and contrast using Adobe Photoshop software.
Supporting Information
Zdroje
1. BierE (1997) Anti-neural-inhibition: a conserved mechanism for neural induction. Cell 89 : 681–684.
2. ArendtD, DenesAS, JékelyG, Tessmar-RaibleK (2008) The evolution of nervous system centralization. Philos Trans R Soc Lond, B, Biol Sci 363 : 1523–1528.
3. MizutaniCM, BierE (2008) EvoD/Vo: the origins of BMP signalling in the neuroectoderm. Nat Rev Genet 9(9): 663–77.
4. RustenTE, CanteraR, KafatosFC, BarrioR (2002) The role of TGF beta signaling in the formation of the dorsal nervous system is conserved between Drosophila and chordates. Development 129 : 3575–3584.
5. BierE (2011) Evolution of development: diversified dorsoventral patterning. Curr Biol 21: R591–594.
6. BiehsB, FrançoisV, BierE (1996) The Drosophila short gastrulation gene prevents Dpp from autoactivating and suppressing neurogenesis in the neuroectoderm. Genes & Development 10 : 2922–2934.
7. MizutaniCM, MeyerN, RoelinkH, BierE (2006) Threshold-dependent BMP-mediated repression: a model for a conserved mechanism that patterns the neuroectoderm. PLoS Biol 4: e313.
8. De RobertisEM, LarrainJ, OelgeschlagerM, WesselyO (2000) The establishment of Spemann's organizer and patterning of the vertebrate embryo. Nat Rev Genet 1 : 171–181.
9. LeeKJ, JessellTM (1999) The specification of dorsal cell fates in the vertebrate central nervous system. Annu Rev Neurosci 22 : 261–294.
10. HuQ, UenoN, BehringerRR (2004) Restriction of BMP4 activity domains in the developing neural tube of the mouse embryo. EMBO Rep 5 : 734–739.
11. Von OhlenT (2000) Convergence of Dorsal, Dpp, and Egfr Signaling Pathways Subdivides the Drosophila Neuroectoderm into Three Dorsal-Ventral Columns. Developmental Biology 224 : 362–372.
12. TribuloC, AybarMJ, NguyenVH, MullinsMC, MayorR (2003) Regulation of Msx genes by a BMP gradient is essential for neural crest specification. Development 130 : 6441–6452.
13. YuJ-K, MeulemansD, MckeownSJ, Bronner-FraserM (2008) Insights from the amphioxus genome on the origin of vertebrate neural crest. Genome Research 18 : 1127–1132.
14. LaprazF, BesnardeauL, LepageT (2009) Patterning of the dorsal-ventral axis in echinoderms: insights into the evolution of the BMP-chordin signaling network. Plos Biol 7: e1000248.
15. SaudemontA, HaillotE, MekpohF, BessodesN, QuirinM, et al. (2010) Ancestral regulatory circuits governing ectoderm patterning downstream of Nodal and BMP2/4 revealed by gene regulatory network analysis in an echinoderm. PLoS Genet 6: e1001259.
16. RossS, HillCS (2008) How the Smads regulate transcription. Int J Biochem Cell Biol 40 : 383–408.
17. PyrowolakisG, HartmannB, MullerB, BaslerK, AffolterM (2004) A simple molecular complex mediates widespread BMP-induced repression during Drosophila development. Dev Cell 7 : 229–240.
18. GaoS, SteffenJ, LaughonA (2005) Dpp-responsive silencers are bound by a trimeric Mad-Medea complex. J Biol Chem 280 : 36158–36164.
19. CrockerJ, ErivesA (2013) A Schnurri/Mad/Medea complex attenuates the dorsal-twist gradient readout at vnd. Dev Biol 378 : 64–72.
20. GarciaM, StathopoulosA (2011) Lateral gene expression in Drosophila early embryos is supported by grainyhead-mediated activation and tiers of dorsally-localized repression. PLoS ONE 6: e29172.
21. DaiH (2000) The Zinc Finger Protein Schnurri Acts as a Smad Partner in Mediating the Transcriptional Response to Decapentaplegic. Developmental Biology 227 : 373–387.
22. MartyT, MullerB, BaslerK, AffolterM (2000) Schnurri mediates Dpp-dependent repression of brinker transcription. Nat Cell Biol 2 : 745–749.
23. MüllerB, HartmannB, PyrowolakisG, AffolterM, BaslerK (2003) Conversion of an extracellular Dpp/BMP morphogen gradient into an inverse transcriptional gradient. Cell 113 : 221–233.
24. WeissA, CharbonnierE, EllertsdóttirE, TsirigosA, WolfC, et al. (2010) A conserved activation element in BMP signaling during Drosophila development. Nat Struct Mol Biol 17 : 69–76.
25. Von OhlenT, MosesC, PoulsonW (2009) Ind represses msh expression in the intermediate column of the Drosophila neuroectoderm, through direct interaction with upstream regulatory DNA. Dev Dyn 238 : 2735–2744.
26. LiXY, MacArthurS, BourgonR, NixD, PollardDA, et al. (2008) Transcription factors bind thousands of active and inactive regions in the Drosophila blastoderm. PLoS Biol 6: e27.
27. MacArthurS, LiXY, LiJ, BrownJB, ChuHC, et al. (2009) Developmental roles of 21 Drosophila transcription factors are determined by quantitative differences in binding to an overlapping set of thousands of genomic regions. Genome Biol 10: R80.
28. VenkenKJT, HeY, HoskinsRA, BellenHJ (2006) P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science 314 : 1747–1751.
29. StathopoulosA, LevineM (2005) Localized repressors delineate the neurogenic ectoderm in the early Drosophila embryo. Dev Biol 280 : 482–493.
30. FisherS, GriceEA, VintonRM, BesslingSL, UrasakiA, et al. (2006) Evaluating the biological relevance of putative enhancers using Tol2 transposon-mediated transgenesis in zebrafish. Nat Protoc 1 : 1297–1305.
31. PhillipsBT, KwonHJ, MeltonC, HoughtalingP, FritzA, et al. (2006) Zebrafish msxB, msxC and msxE function together to refine the neural-nonneural border and regulate cranial placodes and neural crest development. Dev Biol 294 : 376–390.
32. AdayAW, ZhuLJ, LakshmananA, WangJ, LawsonND (2011) Identification of cis regulatory features in the embryonic zebrafish genome through large-scale profiling of H3K4me1 and H3K4me3 binding sites. Dev Biol 357 : 450–462.
33. FisherS, GriceEA, VintonRM, BesslingSL, McCallionAS (2006) Conservation of RET regulatory function from human to zebrafish without sequence similarity. Science 312 : 276–279.
34. KagueE, BesslingSL, LeeJ, HuG, Passos-BuenoMR, et al. (2010) Functionally conserved cis-regulatory elements of COL18A1 identified through zebrafish transgenesis. Dev Biol 337 : 496–505.
35. JaźwińskaA, RushlowC, RothS (1999) The role of brinker in mediating the graded response to Dpp in early Drosophila embryos. Development 126 : 3323–3334.
36. PyrowolakisG, HartmannB, MüllerB, BaslerK, AffolterM (2004) A simple molecular complex mediates widespread BMP-induced repression during Drosophila development. Dev Cell 7 : 229–240.
37. YaoL-C, BlitzIL, PeifferDA, PhinS, WangY, et al. (2006) Schnurri transcription factors from Drosophila and vertebrates can mediate Bmp signaling through a phylogenetically conserved mechanism. Development 133 : 4025–4034.
38. SatijaR, BradleyRK (2012) The TAGteam motif facilitates binding of 21 sequence-specific transcription factors in the Drosophila embryo. Genome Research 22 : 656–665.
39. ArnostiDN, BaroloS, LevineM, SmallS (1996) The eve stripe 2 enhancer employs multiple modes of transcriptional synergy. Development 122 : 205–214.
40. DenesAS, JekelyG, SteinmetzPR, RaibleF, SnymanH, et al. (2007) Molecular architecture of annelid nerve cord supports common origin of nervous system centralization in bilateria. Cell 129 : 277–288.
41. HaywardDC, SamuelG, PontynenPC, CatmullJ, SaintR, et al. (2002) Localized expression of a dpp/BMP2/4 ortholog in a coral embryo. Proc Natl Acad Sci U S A 99 : 8106–8111.
42. SamuelG, MillerD, SaintR (2001) Conservation of a DPP/BMP signaling pathway in the nonbilateral cnidarian Acropora millepora. Evol Dev 3 : 241–250.
43. Reber-MullerS, Streitwolf-EngelR, YanzeN, SchmidV, StierwaldM, et al. (2006) BMP2/4 and BMP5–8 in jellyfish development and transdifferentiation. Int J Dev Biol 50 : 377–384.
44. RentzschF, AntonR, SainaM, HammerschmidtM, HolsteinTW, et al. (2006) Asymmetric expression of the BMP antagonists chordin and gremlin in the sea anemone Nematostella vectensis: implications for the evolution of axial patterning. Dev Biol 296 : 375–387.
45. FinnertyJR, PangK, BurtonP, PaulsonD, MartindaleMQ (2004) Origins of bilateral symmetry: Hox and dpp expression in a sea anemone. Science 304 : 1335–1337.
46. Valentine JW (2004) On The Origin of Phyla. Chicago: University of Chicago Press. 614 p.
47. LoweCJ, TerasakiM, WuM, FreemanRMJr, RunftL, et al. (2006) Dorsoventral patterning in hemichordates: insights into early chordate evolution. PLoS Biol 4: e291.
48. FurutaY, PistonDW, HoganBL (1997) Bone morphogenetic proteins (BMPs) as regulators of dorsal forebrain development. Development 124 : 2203–2212.
49. GoldenJA, BracilovicA, McFaddenKA, BeesleyJS, RubensteinJL, et al. (1999) Ectopic bone morphogenetic proteins 5 and 4 in the chicken forebrain lead to cyclopia and holoprosencephaly. Proc Natl Acad Sci U S A 96 : 2439–2444.
50. HartleyKO, HardcastleZ, FridayRV, AmayaE, PapalopuluN (2001) Transgenic Xenopus embryos reveal that anterior neural development requires continued suppression of BMP signaling after gastrulation. Dev Biol 238 : 168–184.
51. LiemKFJr, JessellTM, BriscoeJ (2000) Regulation of the neural patterning activity of sonic hedgehog by secreted BMP inhibitors expressed by notochord and somites. Development 127 : 4855–4866.
52. PieraniA, Brenner-MortonS, ChiangC, JessellTM (1999) A sonic hedgehog-independent, retinoid-activated pathway of neurogenesis in the ventral spinal cord. Cell 97 : 903–915.
53. Reber-MüllerS, Streitwolf-EngelR, YanzeN, SchmidV, StierwaldM, et al. (2006) BMP2/4 and BMP5–8 in jellyfish development and transdifferentiation. Int J Dev Biol 50 : 377–384.
54. TakahashiH, KamiyaA, IshiguroA, SuzukiAC, SaitouN, et al. (2008) Conservation and diversification of Msx protein in metazoan evolution. Mol Biol Evol 25 : 69–82.
55. RebeizM, PosakonyJW (2004) GenePalette: a universal software tool for genome sequence visualization and analysis. Dev Biol 271 : 431–438.
56. HanssonMD, RzeznickaK, RosenbäckM, HanssonM, SirijovskiN (2008) PCR-mediated deletion of plasmid DNA. Anal Biochem 375 : 373–375.
57. ChocronS, VerhoevenMC, RentzschF, HammerschmidtM, BakkersJ (2007) Zebrafish Bmp4 regulates left-right asymmetry at two distinct developmental time points. Dev Biol 305 : 577–588.
58. HashiguchiM, MullinsMC (2013) Anteroposterior and dorsoventral patterning are coordinated by an identical patterning clock. Development 140 : 1970–1980.
59. KosmanD, MizutaniC, LemonsD, CoxG, McGinnisW, et al. (2004) Multiplex detection of RNA expression in Drosophila embryos. Science 305 : 846.
60. O'Neill JW, Bier E (1994) Double-label in situ hybridization using biotin and digoxigenin-tagged RNA probes. Biotechniques 17: : 870, 874–875.
61. BlanchetteM, LabourierE, GreenRE, BrennerSE, RioDC (2004) Genome-wide analysis reveals an unexpected function for the Drosophila splicing factor U2AF50 in the nuclear export of intronless mRNAs. Mol Cell 14 : 775–786.
62. HouzelsteinD, Auda-BoucherG, CheraudY, RouaudT, BlancI, et al. (1999) The homeobox gene Msx1 is expressed in a subset of somites, and in muscle progenitor cells migrating into the forelimb. Development 126 : 2689–2701.
63. AjuriaL, NievaC, WinklerC, KuoD, SamperN, et al. (2011) Capicua DNA-binding sites are general response elements for RTK signaling in Drosophila. Development 138 : 915–924.
Štítky
Genetika Reprodukční medicína
Článek An Evolutionarily Conserved Role for the Aryl Hydrocarbon Receptor in the Regulation of MovementČlánek Requirement for Drosophila SNMP1 for Rapid Activation and Termination of Pheromone-Induced ActivityČlánek Co-regulated Transcripts Associated to Cooperating eSNPs Define Bi-fan Motifs in Human Gene NetworksČlánek Identification of a Regulatory Variant That Binds FOXA1 and FOXA2 at the Type 2 Diabetes GWAS LocusČlánek tRNA Modifying Enzymes, NSUN2 and METTL1, Determine Sensitivity to 5-Fluorouracil in HeLa CellsČlánek Derlin-1 Regulates Mutant VCP-Linked Pathogenesis and Endoplasmic Reticulum Stress-Induced ApoptosisČlánek A Genetic Assay for Transcription Errors Reveals Multilayer Control of RNA Polymerase II FidelityČlánek The Proprotein Convertase KPC-1/Furin Controls Branching and Self-avoidance of Sensory Dendrites inČlánek Regulation of p53 and Rb Links the Alternative NF-κB Pathway to EZH2 Expression and Cell SenescenceČlánek Unkempt Is Negatively Regulated by mTOR and Uncouples Neuronal Differentiation from Growth Control
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 9- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
-
Všechny články tohoto čísla
- Translational Regulation of the Post-Translational Circadian Mechanism
- An Evolutionarily Conserved Role for the Aryl Hydrocarbon Receptor in the Regulation of Movement
- Eliminating Both Canonical and Short-Patch Mismatch Repair in Suggests a New Meiotic Recombination Model
- Requirement for Drosophila SNMP1 for Rapid Activation and Termination of Pheromone-Induced Activity
- Co-regulated Transcripts Associated to Cooperating eSNPs Define Bi-fan Motifs in Human Gene Networks
- Targeted H3R26 Deimination Specifically Facilitates Estrogen Receptor Binding by Modifying Nucleosome Structure
- Role for Circadian Clock Genes in Seasonal Timing: Testing the Bünning Hypothesis
- The Tandem Repeats Enabling Reversible Switching between the Two Phases of β-Lactamase Substrate Spectrum
- The Association of the Vanin-1 N131S Variant with Blood Pressure Is Mediated by Endoplasmic Reticulum-Associated Degradation and Loss of Function
- Identification of a Regulatory Variant That Binds FOXA1 and FOXA2 at the Type 2 Diabetes GWAS Locus
- Regulation of Flowering by the Histone Mark Readers MRG1/2 via Interaction with CONSTANS to Modulate Expression
- The Actomyosin Machinery Is Required for Retinal Lumen Formation
- Plays a Conserved Role in Assembly of the Ciliary Motile Apparatus
- Hidden Diversity in Honey Bee Gut Symbionts Detected by Single-Cell Genomics
- Ribosome Rescue and Translation Termination at Non-Standard Stop Codons by ICT1 in Mammalian Mitochondria
- tRNA Modifying Enzymes, NSUN2 and METTL1, Determine Sensitivity to 5-Fluorouracil in HeLa Cells
- Causal Variation in Yeast Sporulation Tends to Reside in a Pathway Bottleneck
- Tissue-Specific RNA Expression Marks Distant-Acting Developmental Enhancers
- WC-1 Recruits SWI/SNF to Remodel and Initiate a Circadian Cycle
- Clonal Expansion of Early to Mid-Life Mitochondrial DNA Point Mutations Drives Mitochondrial Dysfunction during Human Ageing
- Methylation QTLs Are Associated with Coordinated Changes in Transcription Factor Binding, Histone Modifications, and Gene Expression Levels
- Differential Management of the Replication Terminus Regions of the Two Chromosomes during Cell Division
- Obesity-Linked Homologues and Establish Meal Frequency in
- Derlin-1 Regulates Mutant VCP-Linked Pathogenesis and Endoplasmic Reticulum Stress-Induced Apoptosis
- Stress-Induced Nuclear RNA Degradation Pathways Regulate Yeast Bromodomain Factor 2 to Promote Cell Survival
- The MAPK p38c Regulates Oxidative Stress and Lipid Homeostasis in the Intestine
- Widespread Genome Reorganization of an Obligate Virus Mutualist
- Trans-kingdom Cross-Talk: Small RNAs on the Move
- The Vip1 Inositol Polyphosphate Kinase Family Regulates Polarized Growth and Modulates the Microtubule Cytoskeleton in Fungi
- Myosin Vb Mediated Plasma Membrane Homeostasis Regulates Peridermal Cell Size and Maintains Tissue Homeostasis in the Zebrafish Epidermis
- GLD-4-Mediated Translational Activation Regulates the Size of the Proliferative Germ Cell Pool in the Adult Germ Line
- Genome Wide Association Studies Using a New Nonparametric Model Reveal the Genetic Architecture of 17 Agronomic Traits in an Enlarged Maize Association Panel
- Translational Regulation of the DOUBLETIME/CKIδ/ε Kinase by LARK Contributes to Circadian Period Modulation
- Positive Selection and Multiple Losses of the LINE-1-Derived Gene in Mammals Suggest a Dual Role in Genome Defense and Pluripotency
- Out of Balance: R-loops in Human Disease
- A Genetic Assay for Transcription Errors Reveals Multilayer Control of RNA Polymerase II Fidelity
- Altered Behavioral Performance and Live Imaging of Circuit-Specific Neural Deficiencies in a Zebrafish Model for Psychomotor Retardation
- Nipbl and Mediator Cooperatively Regulate Gene Expression to Control Limb Development
- Meta-analysis of Mutations in Autism Spectrum Disorders: A Gradient of Severity in Cognitive Impairments
- The Proprotein Convertase KPC-1/Furin Controls Branching and Self-avoidance of Sensory Dendrites in
- Hydroxymethylated Cytosines Are Associated with Elevated C to G Transversion Rates
- Memory and Fitness Optimization of Bacteria under Fluctuating Environments
- Regulation of p53 and Rb Links the Alternative NF-κB Pathway to EZH2 Expression and Cell Senescence
- Interspecific Tests of Allelism Reveal the Evolutionary Timing and Pattern of Accumulation of Reproductive Isolation Mutations
- PRO40 Is a Scaffold Protein of the Cell Wall Integrity Pathway, Linking the MAP Kinase Module to the Upstream Activator Protein Kinase C
- Low Levels of p53 Protein and Chromatin Silencing of p53 Target Genes Repress Apoptosis in Endocycling Cells
- SPDEF Inhibits Prostate Carcinogenesis by Disrupting a Positive Feedback Loop in Regulation of the Foxm1 Oncogene
- RRP6L1 and RRP6L2 Function in Silencing Regulation of Antisense RNA Synthesis
- BMPs Regulate Gene Expression in the Dorsal Neuroectoderm of and Vertebrates by Distinct Mechanisms
- Unkempt Is Negatively Regulated by mTOR and Uncouples Neuronal Differentiation from Growth Control
- Atkinesin-13A Modulates Cell-Wall Synthesis and Cell Expansion in via the THESEUS1 Pathway
- Dopamine Signaling Leads to Loss of Polycomb Repression and Aberrant Gene Activation in Experimental Parkinsonism
- Histone Methyltransferase MMSET/NSD2 Alters EZH2 Binding and Reprograms the Myeloma Epigenome through Global and Focal Changes in H3K36 and H3K27 Methylation
- Bipartite Recognition of DNA by TCF/Pangolin Is Remarkably Flexible and Contributes to Transcriptional Responsiveness and Tissue Specificity of Wingless Signaling
- The Olfactory Transcriptomes of Mice
- Muscular Dystrophy-Associated and Variants Disrupt Nuclear-Cytoskeletal Connections and Myonuclear Organization
- Interplay of dFOXO and Two ETS-Family Transcription Factors Determines Lifespan in
- Evidence for Widespread Positive and Negative Selection in Coding and Conserved Noncoding Regions of
- Genome-Wide Association Meta-analysis of Neuropathologic Features of Alzheimer's Disease and Related Dementias
- Rejuvenation of Meiotic Cohesion in Oocytes during Prophase I Is Required for Chiasma Maintenance and Accurate Chromosome Segregation
- Admixture in Latin America: Geographic Structure, Phenotypic Diversity and Self-Perception of Ancestry Based on 7,342 Individuals
- Local Effect of Enhancer of Zeste-Like Reveals Cooperation of Epigenetic and -Acting Determinants for Zygotic Genome Rearrangements
- Differential Responses to Wnt and PCP Disruption Predict Expression and Developmental Function of Conserved and Novel Genes in a Cnidarian
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Admixture in Latin America: Geographic Structure, Phenotypic Diversity and Self-Perception of Ancestry Based on 7,342 Individuals
- Nipbl and Mediator Cooperatively Regulate Gene Expression to Control Limb Development
- Genome Wide Association Studies Using a New Nonparametric Model Reveal the Genetic Architecture of 17 Agronomic Traits in an Enlarged Maize Association Panel
- Histone Methyltransferase MMSET/NSD2 Alters EZH2 Binding and Reprograms the Myeloma Epigenome through Global and Focal Changes in H3K36 and H3K27 Methylation
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



