-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaPost-Replication Repair Suppresses Duplication-Mediated Genome Instability
RAD6 is known to suppress duplication-mediated gross chromosomal rearrangements (GCRs) but not single-copy sequence mediated GCRs. Here, we found that the RAD6 - and RAD18-dependent post-replication repair (PRR) and the RAD5-, MMS2-, UBC13-dependent error-free PRR branch acted in concert with the replication stress checkpoint to suppress duplication-mediated GCRs formed by homologous recombination (HR). The Rad5 helicase activity, but not its RING finger, was required to prevent duplication-mediated GCRs, although the function of Rad5 remained dependent upon modification of PCNA at Lys164. The SRS2, SGS1, and HCS1 encoded helicases appeared to interact with Rad5, and epistasis analysis suggested that Srs2 and Hcs1 act upstream of Rad5. In contrast, Sgs1 likely functions downstream of Rad5, potentially by resolving DNA structures formed by Rad5. Our analysis is consistent with models in which PRR prevents replication damage from becoming double strand breaks (DSBs) and/or regulates the activity of HR on DSBs.
Published in the journal: . PLoS Genet 6(5): e32767. doi:10.1371/journal.pgen.1000933
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1000933Summary
RAD6 is known to suppress duplication-mediated gross chromosomal rearrangements (GCRs) but not single-copy sequence mediated GCRs. Here, we found that the RAD6 - and RAD18-dependent post-replication repair (PRR) and the RAD5-, MMS2-, UBC13-dependent error-free PRR branch acted in concert with the replication stress checkpoint to suppress duplication-mediated GCRs formed by homologous recombination (HR). The Rad5 helicase activity, but not its RING finger, was required to prevent duplication-mediated GCRs, although the function of Rad5 remained dependent upon modification of PCNA at Lys164. The SRS2, SGS1, and HCS1 encoded helicases appeared to interact with Rad5, and epistasis analysis suggested that Srs2 and Hcs1 act upstream of Rad5. In contrast, Sgs1 likely functions downstream of Rad5, potentially by resolving DNA structures formed by Rad5. Our analysis is consistent with models in which PRR prevents replication damage from becoming double strand breaks (DSBs) and/or regulates the activity of HR on DSBs.
Introduction
Post-replication repair (PRR) was first identified in bacteria as a pathway for the repair of single-stranded gaps in DNA produced during the replication of DNA that had been damaged by exposure to ultraviolet light resulting in replication blocking lesions [1], [2]. PRR was also identified in the eukaryote Saccharomyces cerevisiae and found to be dependent on RAD6 and RAD18 [3]. PRR in both bacteria and eukaryotes is thought to not directly repair the replication-blocking lesions, but rather allows the replication machinery to bypass lesions. In eukaryotes, PRR has at least two downstream branches (reviewed in [4]). One branch extends nascent strands that are blocked by replication stalling lesions on the template strand using translesion or “error-prone” DNA polymerases, including DNA polymerase eta (Rev3-Rev7) and zeta (Rad30), which contribute to DNA damage-induced mutagenesis. The other “error-free” branch depends on RAD5, MMS2, and UBC13 that is believed to allow extension by transiently pairing the blocked nascent strand and the other newly synthesized strand (“template-switching”). Template switching may occur by isomerization of the replication fork by Rad5 as demonstrated in vitro [5] and as first proposed thirty years ago [6], [7]. Alternatively, template switching might be mediated by a cross-fork template-switching mechanism proposed based on genetic similarities between E. coli dnaK mutants and S. cerevisiae rad5 mutants and as suggested by the formation RAD18-, RAD5-, and RAD51-dependent double Holliday junctions in sgs1Δ mutants [8]–[10]. Importantly, these two models for template switching may not be mutually exclusive.
Many of the eukaryotic PRR genes encode proteins mediating protein ubiquitination [11], [12]. Rad6 is an E2 ubiquitin conjugase that is covalently linked by a thioester bond to the C-terminus of ubiquitin and transfers ubiquitin to targets recruited by the E3 ubiquitin ligases Bre1, Rad18 and Ubr1. Rad6 and Rad18 are required for PRR whereas Rad6-Bre1 mediates ubiquitination of histone H2B leading to transcriptional and checkpoint signaling [13]–[15] and Rad6-Ubr1 targets N-end rule substrates for degradation [16]. The Rad6-Rad18 complex monoubiquitinates PCNA at Lys164 [17] and the Rad17 subunit of the PCNA-like 9-1-1 checkpoint clamp at Lys197 [18]. Monoubiquitinated PCNA has been implicated in recruiting translesion polymerases [19], [20] as well as serving as substrate for synthesis of a Lys63-linked polyubiquitin chain by Mms2-Ubc13 E2 ubiquitin conjugase in conjunction with the Rad5 E3 ubiqutin ligase/DNA helicase [21], [22]. How the activities of Mms2 and Ubc13, and PCNA polyubiqutination channel DNA damage to error-free repair remains unclear [5]–[7].
In addition to roles in mediating tolerance to replication blocking DNA lesions, PRR genes have complex roles in maintaining genome stability. Both rad5Δ and rad18Δ mutants have elevated levels of spontaneous recombination [23] and rapid expansion of trinucleotide repeats [24]. Deletions of PRR genes appear to generally suppress gross chromosomal rearrangements (GCRs) mediated by single-copy sequences; rad6Δ suppresses the increased GCR rates caused by the pif1-m2 allele [25] and deletion of RAD5, RAD6, RAD18, UBC13 and MMS2 similarly suppress the increased GCR rates caused by an asf1 mutation [26]. In contrast, we found that deletion of RAD6 dramatically increases the rate of GCRs mediated by homologous recombination (HR) between imperfect duplications resulting in increased accumulation of GCRs [27]. These differences likely reflect the fact that HR suppresses single copy sequence-mediated GCRs whereas HR produces duplication-mediated GCRs. Here we have sought to understand how the RAD6 pathways function to specifically suppress duplication-mediated GCRs and to use the sensitivity of the duplication-mediated GCR assay to defects in RAD6 to analyze interactions between components of RAD6-dependent pathways.
Results
RAD6 suppression of duplication-mediated GCRs was epistatic to the replication stress checkpoint
Deletion of RAD6 was previously found to specifically increase the spontaneous rate of duplication-mediated GCRs by comparing the rates of loss of a CAN1/URA3 cassette on chromosome V in the yel068c::CAN1/URA3 GCR assay, which lacks a duplication in the breakpoint region, with the yel072w::CAN1/URA3 GCR assay, which contains the DSF1-HXT13 duplicated region in the breakpoint region (Table 1; Figure 1a)[27]. The DSF1-HXT13 region shares ∼4.2 kb of homology with chromosome XIV and ∼1.7 kb of homology with highly similar regions of chromosomes IV and X, and consequently most of the duplication-mediated GCRs are translocations between the DSF1 HXT13 region on chromosome V and the homology regions on chromosomes XIV, IV and X. We analyzed the GCRs obtained in the yel072w::CAN1/URA3 assay in the rad6Δ background and observed that the increased rates of forming homology-mediated t(V;XIV) and t(V;IV or X) translocations were responsible for most of the rate increases (Figure 1b). The majority of both homology and non-homology-mediated GCRs lost the telomeric end of chromosome V as determined by the loss of the telomeric hygromycin resistance marker (Table S1).
Fig. 1. PRR defects result in increased rates of duplication-mediated translocations. 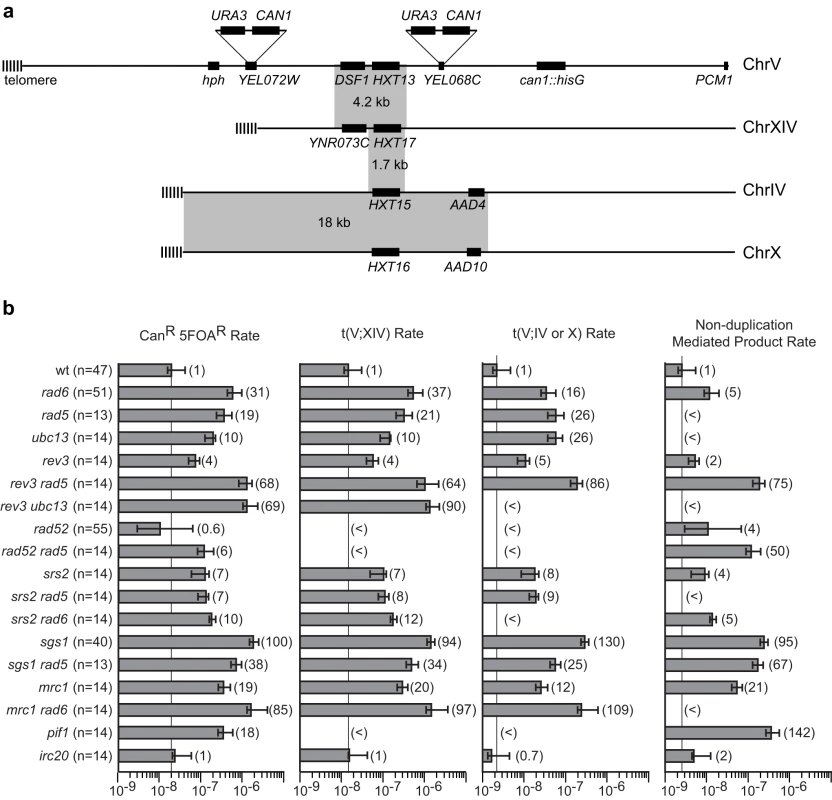
A. The pre-duplication (yel068c::CAN1/URA3) and post-duplication (yel072w::CAN1/URA3) assays differ by whether or not they include the DSF1-HXT13 homology in the breakpoint region (the left arm of chromosome V between the CAN1/URA3 cassette and the most telomeric essential gene, PCM1). The hygromycin resistance marker is indicated by hph. Grey boxes indicate regions of homologies between the chromosomes. B. The rates of the total CanR 5FOAR product and the rates of t(V;XIV) and t(V;IV or X) translocations, and non-duplication-mediated GCR products in the yel072w::CAN1/URA3 assay are depicted in a bar graph. Error bars indicate 95% confidence intervals and the fold increase for each rate is displayed in parentheses, (<) indicates that no isolates of that class were identified. The number of isolates analyzed is shown in parentheses after the genotype. The numerical GCR rates are presented in Tables 1, 2, 4 and 5. Tab. 1. Effects of combining RAD6 and checkpoint gene mutations on duplication-mediated GCRs. 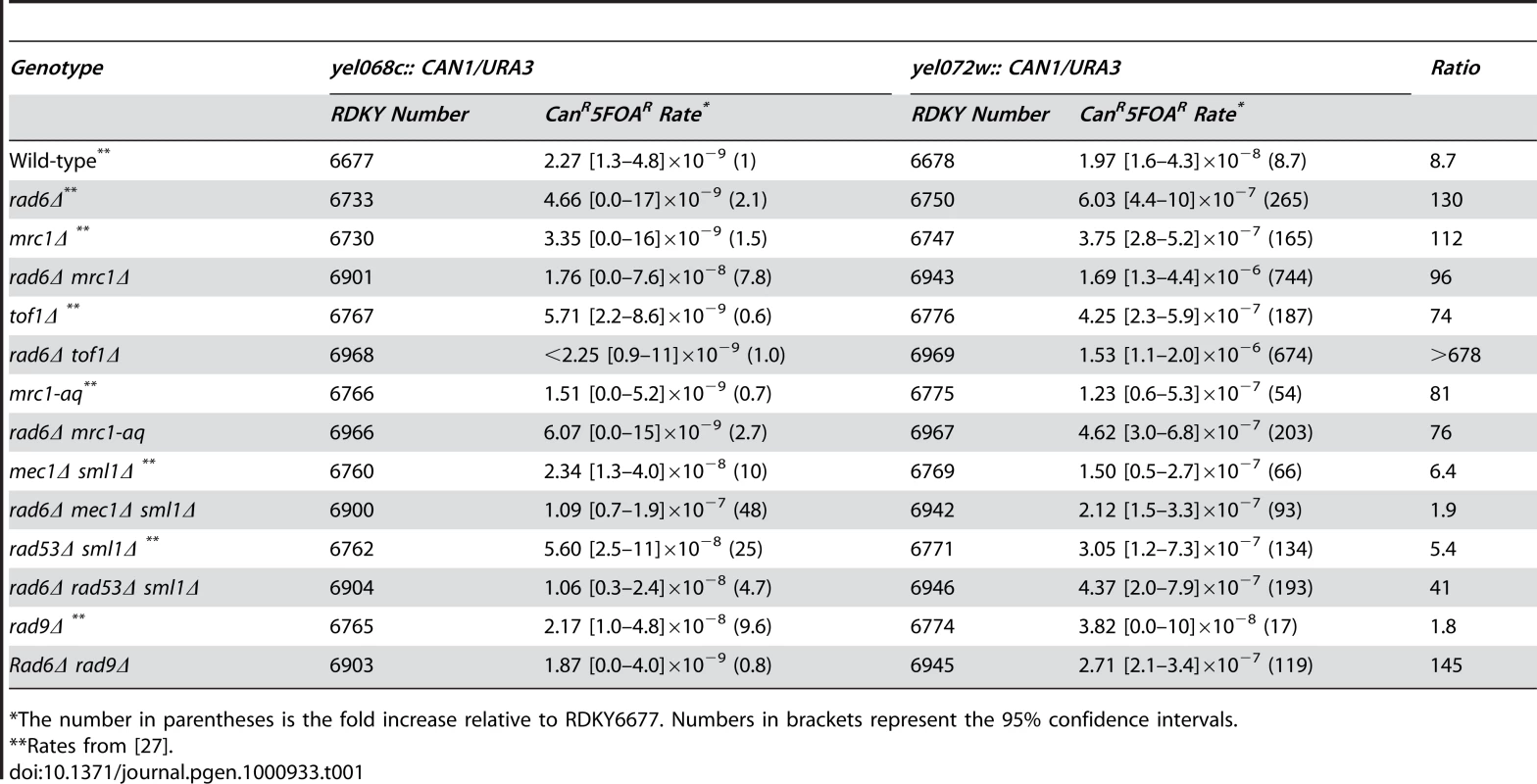
*The number in parentheses is the fold increase relative to RDKY6677. Numbers in brackets represent the 95% confidence intervals. Like RAD6, components of the replication stress checkpoint also has roles in specifically suppressing duplication-mediated GCRs [27]. To investigate the possibility that RAD6 and the replication stress checkpoints function in the same pathway, we constructed double mutants containing a rad6Δ mutation along with different checkpoint defective mutations (Table 1). Remarkably, rad6Δ caused a synergistic increase in GCR rate that was statistically significant (p<0.0001 for the difference being due to chance) when combined with deletions of MRC1 or TOF1, which encode signaling components of the replication fork that also have roles in sister chromatid cohesion [28], [29]. Comparison of the GCR products isolated from the mrc1Δ single mutant strain and rad6Δ mrc1Δ double mutant strain revealed that the increase in rate was primarily due to the formation of homology-mediated rearrangements (Figure 1b). Unlike mrc1Δ, however, rad6Δ appeared to be epistatic to the mrc1-aq allele (p = 0.1), which is specifically defective in the MRC1 checkpoint function but not the replication function [30]. Similar to mrc1-aq, deletion of MEC1 or RAD53, which encode protein kinases involved in the checkpoint response [31], appeared to be epistatic with rad6Δ (p = 0.09 and p = 0.4, respectively). In contrast, deletion of RAD9, which specifically impairs the DNA damage checkpoint but not the replication stress checkpoint [32], suppressed the rate of a rad6Δ mutation (p = 0.002), although the rad6Δ rad9Δ double mutant had a significantly higher duplication-specific GCR rate than the rad9Δ single mutant (p = 0.01). Taken together, these data suggest that RAD6 functions in a pathway channeling replication damage away from duplication-mediated GCR formation in concert with replication stress checkpoint signaling and that deletion of MRC1 and TOF1 either causes increased replication errors that lead to GCRs or allows HR to target homology regions at dispersed chromosomal locations due to defects in sister chromatid cohesion that might restrict HR to sister chromatids.
PRR is the major RAD6-dependent pathway that suppresses HR-dependent GCRs
To identify the RAD6-dependent pathways that suppress GCRs, each gene encoding a Rad6-associated E3 ubiquitin ligase was deleted in both the yel068c::CAN1/URA3 and the yel072w::CAN1/URA3 strain backgrounds. The ubr1Δ and bre1Δ mutations did not cause increased GCR rates in the yel068c::CAN1/URA3 assay lacking a duplication (Table 2), consistent with previous results [25], [26]. Both mutations caused a small rate increase in the yel072w::CAN1/URA3 duplication-mediated GCR assay, but these rates were substantially lower than that caused by deletion of RAD6 (p<0.0001 for both). Deletion of LGE1, which encodes a protein that may function with Bre1 [33], was not distinguishable from deletion of BRE1 in both GCR assays (p = 0.4). In contrast, the rad18Δ mutation, like a rad6Δ mutation caused little increase in the GCR rate in the yel068c::CAN1/URA3 GCR assay but caused a large increase in the GCR rate in the yel072w::CAN1/URA3 duplication-mediated GCR assay (Table 2). Thus, the Rad6-Rad18-dependent PRR branch appears to be the major pathway that functions in the RAD6-dependent suppression of duplication-mediated GCR formation.
Tab. 2. Effects of mutations in PRR subpathways on duplication-mediated GCRs. 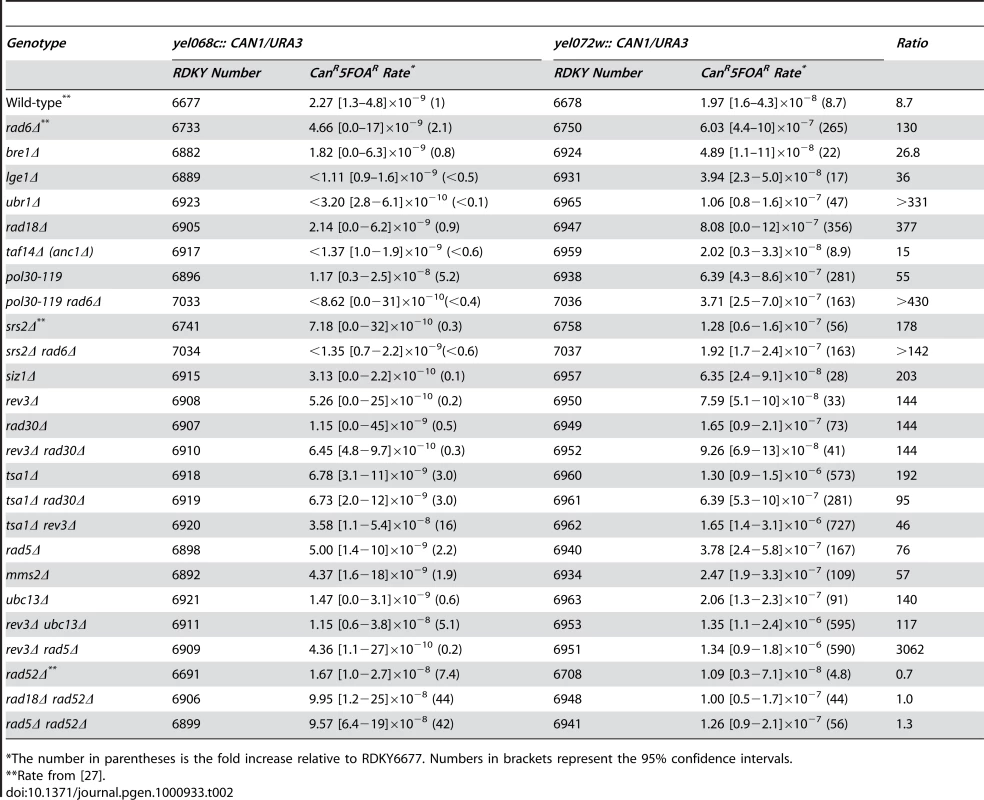
*The number in parentheses is the fold increase relative to RDKY6677. Numbers in brackets represent the 95% confidence intervals. Monoubiquitination of PCNA by Rad6-Rad18 is an early event in PRR [17]. We therefore tested the pol30-119 allele, which encodes a Lys164Arg mutant PCNA that lacks the PCNA ubiquitination site [17], and found that pol30-119 caused essentially the same increase in the rate of duplication-mediated GCRs as caused by both rad6Δ and rad18Δ mutations (Table 2; p>0.01 and overlapping 95% confidence intervals for all pairwise comparisons). As the pol30-119 allele also eliminates a major sumoylation site on PCNA [17], we also tested the effects of deleting SIZ1, which encodes a PCNA-modifying SUMO ligase, and SRS2, which encodes a helicase recruited to sumoylated PCNA [34], [35] that is also epistatic to PRR [36]. Neither of these deletions affected the rate of GCRs mediated by single copy sequences, consistent with previous data [25], [26]. Both siz1Δ and srs2Δ mutations caused a moderate increase in the rate of duplication-mediated GCRs, though the effect was significantly less than that caused by the rad6Δ, rad18Δ or pol30-119 (p≤0.0001 for all pairwise comparisons). Consistent with this, the increased rates of duplication-mediated GCR products in the srs2Δ mutant were lower than that seen in the rad6Δ mutant (Figure 1b). Thus, the primary defect of the pol30-119 allele in the suppression of duplication-mediated GCRs appears to be due to defects in PRR-mediated ubiquitination and rather than sumoylation. To confirm this, we analyzed pol30-119 rad6Δ double mutants and observed that the increased GCR rate seen in the yel072w::CAN1/URA3 assay was indistinguishable from that caused by pol30-119 and rad6Δ single mutations (Table 2; p>0.01 and overlapping 95% confidence intervals for all pairwise comparisons). We also analyzed the srs2Δ rad6Δ double mutants and observed a slight, but significant suppression of the rad6Δ duplication-mediated GCR rate (Table 2; p<0.0001), consistent with partial, but incomplete, epistasis of RAD6 to SRS2 for suppression of duplication-mediated GCRs.
Defects in translesion polymerase-dependent PRR branch cause only moderate increases in HR-dependent GCRs
To understand which PRR branch suppresses duplication-mediated GCRs, we first analyzed the role of the translesion polymerases. Deletion of REV3, encoding the catalytic subunit of DNA polymerase zeta, and RAD30, encoding DNA polymerase eta, caused very small increases in the rate of single copy sequence-mediated GCRs (Table 2), consistent with previous results [25], [26], [37], but both caused moderate increases in the rate of duplication-mediated GCRs, although the rates were not increased to the extent seen for rad6Δ or rad18Δ mutations (Table 2). Moreover, the rev3Δ mutation caused roughly equivalent fold increases in the rates of forming both duplication-mediated and non-duplication-mediated GCR products (Figure 1b). The rev3Δ rad30Δ double mutant also had a moderate increase in the rate of duplication-mediated GCRs that was indistinguishable by 95% confidence intervals to the rate caused by the rev3Δ and rad30Δ single mutants, suggesting involvement in a single genetic pathway consistent with biochemical experiments demonstrating that DNA polymerases eta and zeta function sequentially to bypass specific lesions [38]. Deletion of TSA1, which encodes a thioredoxin peroxidase that suppresses oxidative damage of DNA in S. cerevisiae [37], [39] caused over a 65-fold increase in the rate of duplication-mediated GCRs (Table 2). Surprisingly, the rev3Δ and rad30Δ mutations did not cause synergistic interactions in either GCR assay when combined with a tsa1Δ mutation. This lack of a synergistic interaction is consistent with previous results obtained for GCR rates in single-copy sequences [37]. This suggests that the translesion polymerases either do not repair or bypass the oxidative damage that leads to duplication-mediated GCRs in tsa1Δ mutants or that other repair pathways, such as base-excision repair, nucleotide-excision repair or mismatch repair [39], [40], can efficiently repair tsa1Δ-mediated damage in the absence of REV3 or RAD30.
Defects in the error-free PRR branch caused large increases in HR-dependent GCRs
Deletion of RAD5, MMS2 or UBC13 that function in the error-free PRR branch as well as deletion of RAD6 and RAD18 did not cause a substantial increase in the rate of single copy sequence-mediated GCRs (Table 2). These results were consistent with those of one previous study [26], but the results for rad5Δ and rad18Δ were inconsistent with the results of another study that reported that rad5Δ and rad18Δ mutations caused an increase in the rate of single copy sequence-mediated GCRs [25]. Despite this, all of these deletions caused a significant increase in the rate of duplication-mediated GCRs (Table 2), which suggests that the error-free branch, and not the translesion polymerase branch, is the major PRR pathway that suppresses duplication-mediated GCRs. The rate increases caused by rad5Δ and ubc13Δ mutations in the yel072w::CAN1/URA3 assay were due to increased rates of formation of the t(V;XIV) and t(V;IV or X) non-reciprocal translocations (Figure 1b). Deletion of TAF14 (ANC1), which has been reported to be epistatic to RAD5 in the repair of alkylation damage [41] and which encodes a protein involved in the RNA polymerase II-associated complexes TFIID, TFIIF, RSC, SWI/SNF, INO80, NuA3, and Mediator [42], had no effect on the GCR rate in either the single copy sequence - or duplication-mediated GCR assays (Table 2). Double mutants including the rev3Δ ubc13Δ and rev3Δ rad5Δ double mutants in which both the translesion polymerase and error-free PRR branches were defective had low GCR rates in the yel068c::CAN1/URA3 assay. In contrast, these double mutants had increased rates of duplication-mediated GCRs that were significantly higher than seen in either single mutant individually but were not significantly different (by their 95% confidence intervals) from the rates of duplication-mediated GCRs seen in the rad6Δ and rad18Δ single mutants (Table 2). The t(V;XIV) duplication-mediated GCR product dominated the GCR products obtained in the yel072w::CAN1/URA3 assay in the rev3Δ ubc13Δ and rev3Δ rad5Δ mutants; however, increases in the rates of t(V;IV or X) translocations and non-duplication-mediated GCRs were also observed in the rev3Δ rad5Δ double mutant (Figure 1b).
A rad52Δ mutation eliminated the increased rate of duplication-mediated GCRs rate due to the DSF1-HXT13 duplication in the yel072w::CAN1/URA3 assay caused by the rad18Δ and rad5Δ mutations (Table 2), consistent with the previously determined role of RAD52 in the formation of duplication-mediated GCRs [27]. Similarly, no homology-mediated translocations were observed among the GCRs identified in the yel072w::CAN1/URA3 GCR assay in the rad52Δ single or rad5Δ rad52Δ double mutants (Figure 1b). Remarkably, the rad18Δ rad52Δ and rad5Δ rad52Δ double mutants showed a synergistic increase in the rate of single copy sequence-mediated GCRs in the yel068c::CAN1/URA3 assay (Table 2; p = 0.005 and p<0.0001, respectively), suggesting that PRR and HR are redundant in suppressing single-copy sequence-mediated GCRs such as de novo telomere additions and chromosome fusions that occur in the absence of extensive homology targets [43], [44]; consistent with this, the rate of non-duplication-mediated GCRs in the yel072w::CAN1/URA3 assay was increased 50-fold in the rad5Δ rad52Δ double mutant compared to 5-fold and <11-fold increases in the rad52Δ and rad5Δ single mutants, respectively (Figure 1b).
The Rad5 helicase activity, but not its RING-finger domain, suppresses duplication-mediated GCRs
Rad5 is a DNA helicase as well as an E3 ubiquitin ligase; both activities are required for the function of Rad5 in repair of UV damage by PRR [45]. Thus, we tested the ability of RAD5 plasmids containing different rad5 mutations to complement the defects in suppressing duplication-mediated GCRs caused by a rad5Δ mutation (Table 3). In contrast to the effects of rad5 mutations on UV damage, we found that a rad5 mutant plasmid containing RING finger mutations (C914A C917A) was able to significantly complement the rad5Δ mutant (p<0.0001). However, the GCR rate seen with the RING finger plasmid was 2-fold higher than that seen with a plasmid bearing a wild-type RAD5 gene, which is a small but significant increase (p = 0.009). In contrast, a rad5 mutant plasmid with defects in the Walker B motif of the helicase domain (D681A E682A) resulted in an increase in the rate of duplication-mediated GCRs similar to that of the empty vector control (p = 0.4). These results indicate that the helicase activity of Rad5, in contrast to its RING finger-dependent E3 ubiquitin ligase activity, is the most important Rad5 activity required for the RAD5-dependent suppression of duplication-mediated rearrangements.
Tab. 3. Complementation of <i>rad5Δ</i> in the <i>yel072w::CAN1/URA3</i> assay. 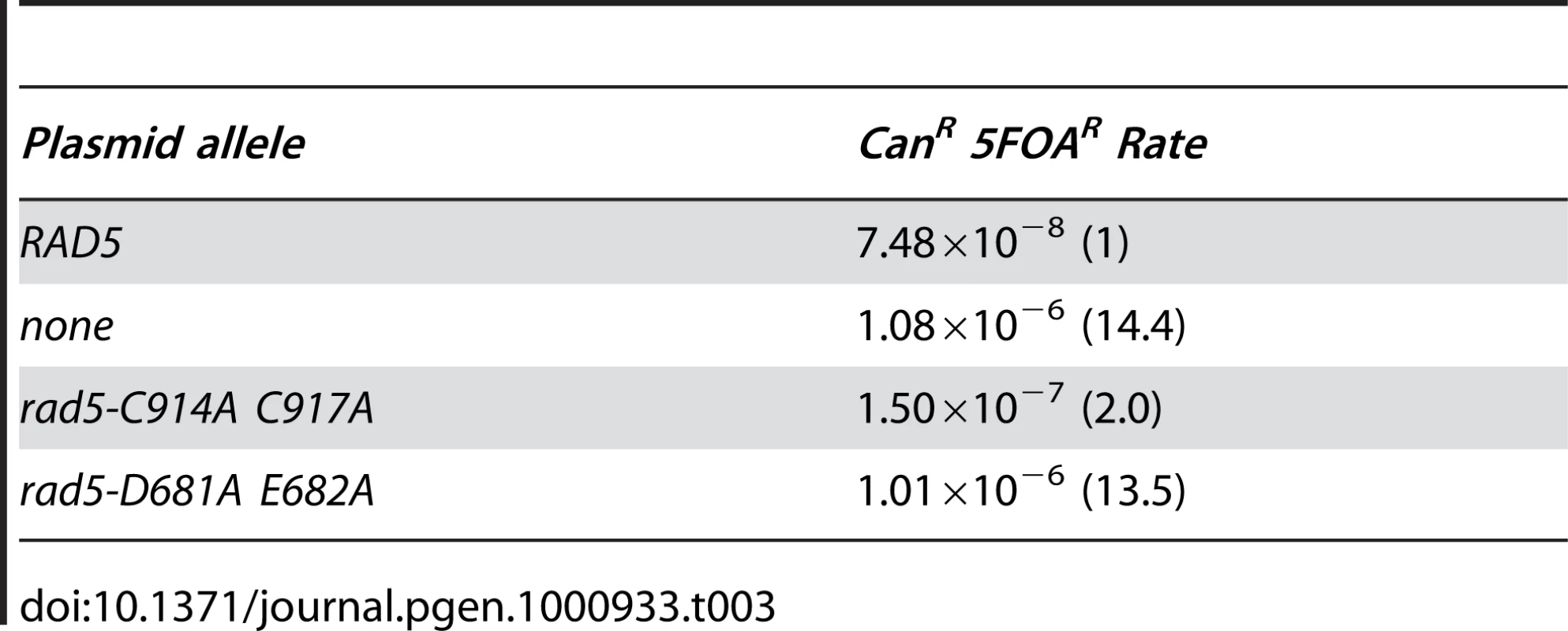
RAD5 has RAD6 - and UBC13-independent but PCNA Lys164-dependent roles in suppressing duplication-mediated GCRs
RAD5 has complex genetic relationships with other PRR genes. RAD5 has MMS2 - and UBC13-independent roles in the repair of UV damage that is processed through the action of translesion polymerases [45], [46]. rad5Δ single mutants also have higher UV sensitivity than ubc13Δ and mms2Δ single mutants [45], and the rad5Δ mms2Δ double mutant is more sensitive to DNA damaging agents than either single mutant [47]. We therefore tested the rad5Δ ubc13Δ double mutant and found that like the single mutants, the double mutant did not have an increased rate of single copy sequence-mediated GCRs in the yel068c::CAN1/URA3 assay (Table 4). In contrast, the double mutant had an increased rate of accumulating duplication-mediated GCRs in the yel072w::CAN1/URA3 assay compared to the respective single mutants (p = 0.0004 relative to rad5Δ and p<0.0001 relative to ubc13Δ), although this increased GCR rate could not be distinguished from that of rad6Δ and rad18Δ single mutants or rev3Δ rad5Δ and rev3Δ ubc13Δ double mutants (Table 2; p = 0.2 and p = 0.4 respectively). These results suggest that RAD5 and UBC13 may have some independent roles in suppressing duplication-mediated GCRs.
Tab. 4. Effects of combining rad5Δ with mutations in RAD6-pathway genes on duplication-mediated GCRs. 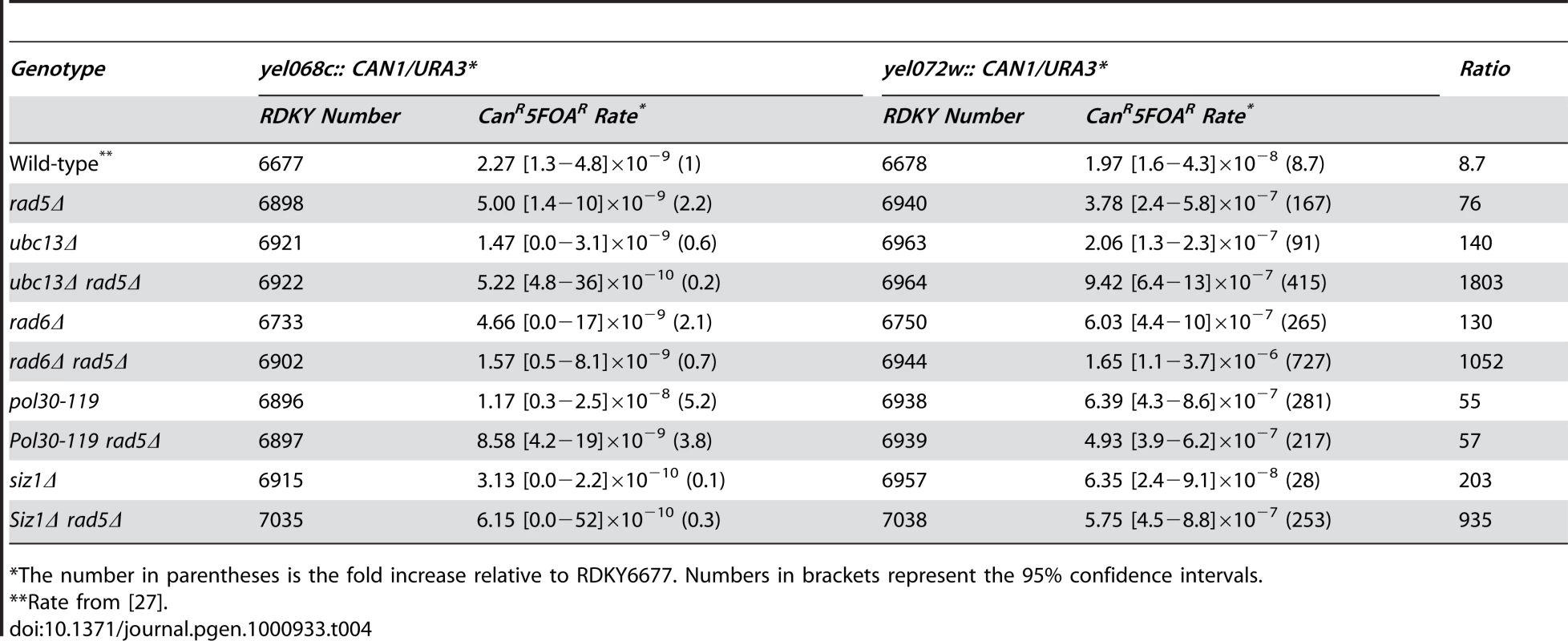
*The number in parentheses is the fold increase relative to RDKY6677. Numbers in brackets represent the 95% confidence intervals. To investigate the possibility that RAD5 has some functions in suppressing duplication-mediated genome rearrangements that are independent of PRR, we tested the rad5Δ rad6Δ and rad5Δ pol30-119 double mutants. The rad5Δ rad6Δ double mutant had an increased rate of accumulating duplication-mediated GCRs relative to that of the individual single mutants (Table 4; p<0.0001). In contrast, the rad5Δ pol30-119 double mutant had an increased rate of accumulating duplication-mediated GCRs that was indistinguishable from that of the respective single mutants (p = 0.04 for rad5Δ and p = 0.08 for pol30-119), suggesting epistasis of RAD5 to post-translational modifications of PCNA at Lys164. Remarkably, the rad5Δ siz1Δ double mutant and the rad5Δ single mutant had indistinguishable rates of accumulating duplication-mediated GCRs, suggesting that sumoylation of PCNA at Lys164 was of limited importance for the suppression of duplication-mediated GCRs (Table 4).
SRS2 is epistatic to RAD5, and deletion of RAD5 partially suppresses an SGS1 deletion
Both replication fork regression [5] and cross-fork [8]–[10] template-switching mechanisms involving Rad5 helicase action would result in branched DNA structures requiring additional processing. We therefore used the sensitivity of the duplication-mediated GCR assay to deletion of RAD5 to screen for helicases that might act in concert with RAD5.
Deletion of MGS1, MPH1, RRM3, HRQ1 or IRC20 did not increase the rate of single copy sequence mediated GCRs in the yel068c::CAN1/URA3 assay and caused small, up to 6-fold, increases in the rate of duplication-mediated GCRs in the yel072w::CAN1/URA3 assay (Table 5). Deletion of IRC20 had no effect on the overall duplication-mediated GCR rate or the rate of any specific class of GCR (Figure 1b). The mgs1Δ rad5Δ and mph1Δ rad5Δ double mutants did not have increased rates of single copy sequence-mediated GCRs but had significantly increased rates of duplication-mediated GCRs relative to the single mutants (p = 0.0001) that were greater than additive (mgs1Δ rad5Δ) or at least as high as additive (mph1Δ rad5Δ), suggesting that Mgs1 and Mph1 may have roles in suppressing duplication mediated GCRs that are independent of Rad5. The rrm3Δ rad5Δ, hrq1Δ rad5Δ and irc20Δ rad5Δ double mutants did not have increased rates of single copy sequence-mediated GCRs and had increased rates of duplication-mediated GCRs that were the same as that of the rad5Δ single mutant and which could not be distinguished from additivity; in our view this latter double mutant analysis provided no strong evidence for epistasis because of the very small affect of rrm3Δ, hrq1Δ and irc20Δ single mutations on duplication-mediated GCR rates.
Tab. 5. Effects of combining defects in RAD5 with other helicase-encoding genes on duplication-mediated GCRs. 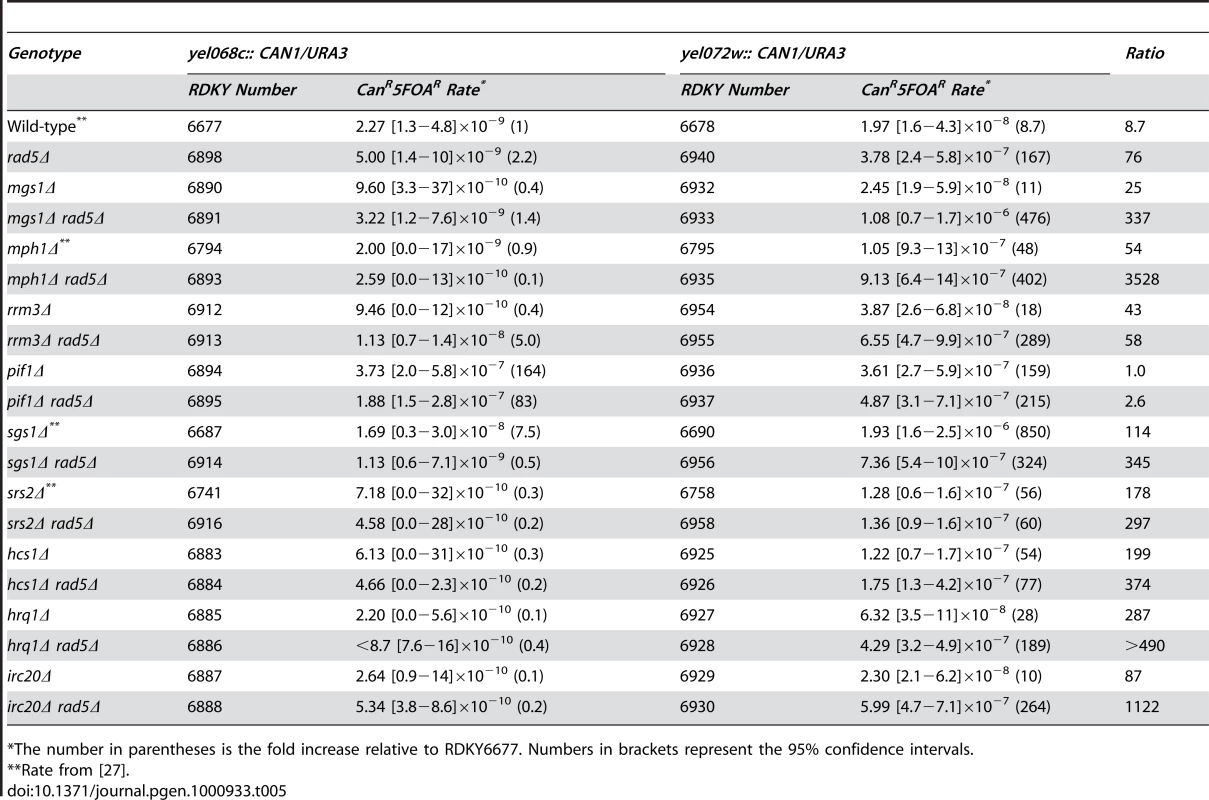
*The number in parentheses is the fold increase relative to RDKY6677. Numbers in brackets represent the 95% confidence intervals. A pif1Δ mutation caused a similar increase in the rate of GCRs in both the yel068c::CAN1/URA3 and yel072w::CAN1/URA3 GCR assays (Table 5); the lack of a duplication-specific increase in the GCR rate is consistent with the fact that Pif1 functions to suppress the healing of broken chromosomes by de novo telomere addition [48] and the fact that none of the isolates from the yel072w::CAN1/URA3 GCR assay were homology-mediated translocations (Figure 1b). The rad5Δ pif1Δ double mutant had a modest decrease in the rate of single-copy sequence mediated GCRs compared to the pif1Δ mutant consistent with published results [49], whereas the rad5Δ pif1Δ double mutant had that same rate of duplication-mediated GCRs as both the pif1Δ and rad5Δ single mutants (p = 0.2). This latter result could be explained by pif1Δ and rad5Δ affecting the same pathway or by GCR-producing pathways activated by pif1Δ and rad5Δ mutations competing for the same source of broken chromosomes with one pathway being dominant.
An srs2Δ mutation had no affect on the rate of single copy sequence-mediated GCRs and caused an increased in the rate of duplication-mediated GCRs that was less than that caused by a rad5Δ mutation. The srs2Δ rad5Δ double mutant did not have an increased rate of single copy sequence-mediated GCRs (Table 5) but had an increased rate of duplication mediated GCRs that was the same as that of the srs2Δ single mutant (p = 0.6) and less than that of the rad5Δ single mutant (Table 5; p<0.0001). These results are consistent with previously observed epistasis between srs2Δ and rad5Δ for spontaneous recombination, triplet-repeat expansion and UV sensitivity [23], [24], [50], [51], and the partial epistasis for srs2Δ and rad6Δ observed above (Table 4). Thus it seems likely that RAD5 might function in an SRS2-dependent pathway. Remarkably, mutations in HCS1, which encodes a DNA polymerase alpha-associated helicase [52], behaved exactly the same as srs2Δ mutations suggesting that an hcs1Δ mutation might also be epistatic with or slightly suppress a rad5Δ mutation in the duplication-mediated GCR assay.
The rad5Δ sgs1Δ double mutant had a lower rate of accumulating duplication-mediated GCRs than an sgs1Δ single mutant (p<0.0001), and the double mutant rate was similar to but somewhat higher than that of a rad5Δ single mutant (Table 5; p = 0.002). This partial epistasis of sgs1Δ to rad5Δ is consistent with a role for SGS1 downstream of RAD5. The partial nature of the epistasis, however, suggests that SGS1 also has RAD5-independent roles as well. Despite the indication that RAD5 might function upstream of SGS1, deletion of RAD5 did not suppress the synthetic lethality and growth defects observed between an sgs1Δ mutation and srs2Δ, rrm3Δ, mus81Δ, slx1Δ, slx5Δ or slx8Δ mutations nor did deletion of both RAD5 and RAD52 suppress the lethality between an sgs1Δ mutation and slx4Δ or slx8Δ mutations (not shown).
Discussion
In the present study, we have demonstrated that suppression of duplication-mediated GCRs by RAD6 is epistatic to the replication stress checkpoint and that the RAD18-, RAD5-, UBC13-, and MMS2-dependent error-free PRR pathway is the RAD6-dependent pathway that is primarily responsible for suppressing duplication-mediated GCRs. The translesion polymerase-dependent pathways for PRR and the BRE1 - and UBR1-dependent RAD6 pathways played small roles in suppressing duplication-mediated GCRs. In addition, genes that are not typically considered as encoding components of the PRR pathways, but which have been implicated in PRR by a few genetic studies, including RAD9 [53] and TAF14 (ANC1) [41], as well as the Shu complex genes, PSY3 and CSM2, implicated as acting downstream of RAD5 [54], did not appear to play significant roles in suppressing duplication-mediated GCRs (Tables 1 & 2, and not shown). The suppression of duplication-mediated GCRs exhibited remarkably complex genetic interactions between downstream PRR components (Figure 2a), involved the helicase and not the RING-finger functions of Rad5, and required Sgs1 for processing of repair intermediates. Our analysis using the sensitive duplication-mediated GCR assay revealed a number of surprising results that appear paradoxical in the context of commonly accepted models for PRR [4], but fit with a growing body of evidence that indicate that the in vivo pathways are more complicated than can be accounted for by present models [26], [45]–[47], [55]–[57].
Fig. 2. Models for the suppression of duplication-mediated GCRs by PRR. 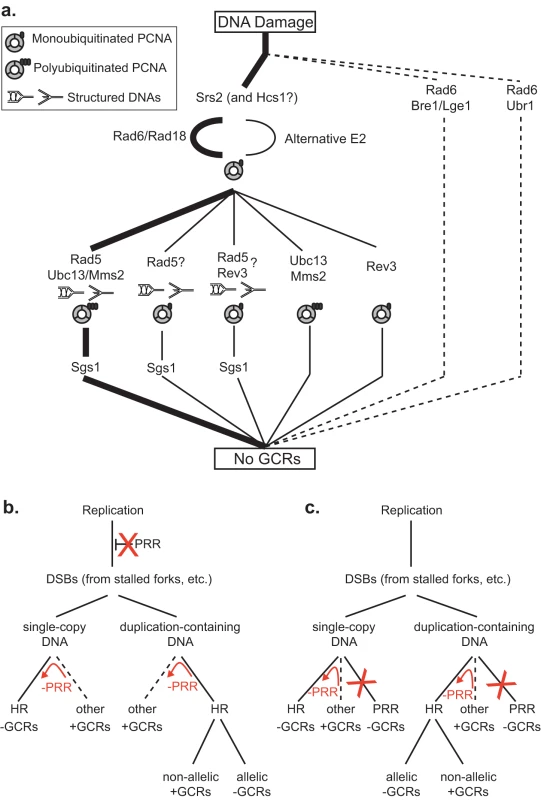
A. The most important RAD6-dependent pathway that suppresses duplication-mediated GCRs (thick lines) corresponds to the “error-free” PRR branch, which is downstream of Srs2. Other Rad6- and Rad18-dependent branches are less important (thin lines). The presence of specific PCNA and DNA states are inferred based on the genes involved in the pathway. Sgs1 appears to act downstream of the Rad5-dependent branches. The existence of Rad5 branches that are independent of Ubc13 and Rev3 that could be dependent upon Rad6 and Rad18 or independent of Rad6 is inferred by the observation of synergistic interactions between mutations in RAD5 and mutations in RAD6, UBC13 and REV3. Our data do not directly address the previously identified Rad5- and Rev3-dependent branch [46]. B. PRR could potentially suppress duplication-mediated GCRs by preventing replication damage from being converted into DSBs and other HR substrates. Suppression of single-copy GCRs also requires that PRR promotes other GCR forming pathways (such as NHEJ and de novo telomere addition) or requires PRR-dependent suppression of HR. C. PRR could potentially suppress duplication-mediated GCRs by functioning as an alternative to HR. Suppression of single-copy GCRs also requires that PRR promotes other GCR forming pathways (such as NHEJ and de novo telomere addition) or requires PRR-dependent suppression of HR. The red arrows and Xs in B and C indicate the consequences of PRR defects. The first surprising result is the lack of epistasis between RAD5 and UBC13 in the duplication-mediated GCR assay, as the Ubc13-Mms2 synthesis of Lys63-linked polyubiquitin chains on PCNA is dependent upon the E3 function of Rad5 [21], [57]. This lack of epistasis is consistent with a function of Rad5 that is independent of PCNA polyubiquitination, consistent with our observation of weak defects caused by mutations affecting the Rad5 RING finger function but large defects caused by mutations affecting the Rad5 helicase function, and consistent with a role of Rad5 in some translesion polymerase-dependent events [45], [46] and the ability of Rad5 to recognize and bind PCNA with a similar affinity regardless of its ubiquitination status [57]. This lack of epistasis also argues for a role of Ubc13 independent of Rad5 that would not have been predicted by the lesser sensitivity of UBC13 and MMS2 mutants to DNA damaging agents than seen with rad5 mutants [45], [47], and may reconcile the weak effect of the Rad5 RING finger mutation in the duplication-mediated GCR assay with the stronger effect of the ubc13Δ mutation. Together with the observation that a rev3Δ mutation shows synergistic interactions with both rad5Δ and ubc13Δ mutations in the duplication-mediated GCR assay, this supports the idea that there are individual pathways that repair spontaneous damage that are solely dependent upon REV3, RAD5 or UBC13 in addition to pathways that are dependent upon combinations of these genes (Figure 2a); in the context of this model, the rates of duplication-mediated GCRs seen in different mutants suggest that UBC13 and RAD5 function in the same pathway more frequently than other combinations in accord with more simple models of “error-free” and “error-prone” branches [4].
The second surprising result from our studies is the fact that the increased duplication-mediated GCR rate caused by the rad5Δ mutation was not affected by a deletion of SIZ1 but the rad5Δ mutation was epistatic to both a deletion of SRS2 and the pol30-119 mutation in the duplication-mediated GCR assay. The SRS2 gene was originally identified through the isolation of a mutation that suppressed the trimethoprim - and UV-sensitive phenotypes of rad6Δ and rad18Δ mutants [36] where HR was required for suppression [58]. Epistasis of a rad5Δ mutation with a srs2Δ mutation is consistent with previous observations of a requirement for SRS2 for RAD5-dependent error-free PRR [51], and could be due to direct recruitment of Rad5 to the site of DNA damage by Srs2 or indirect recruitment via a role of Srs2 in suppressing HR [59], [60]. Our results are not consistent, however, with an absolute requirement of Siz1-mediated PCNA sumoylation and subsequent Srs2 recruitment for Srs2 function to suppress duplication-mediated GCRs. For example, a srs2Δ mutation caused a greater GCR rate in the duplication-mediated GCR assay than a siz1Δ mutation and was strongly epistatic to PRR gene deletions, which is consistent with previously published results that an srs2Δ mutation causes a greater suppression of the DNA damaging agent sensitivity caused by a rad6Δ mutation than the level of suppression caused by a siz1Δ mutation [34]. The observation of Cdk1 - and PCNA-independent roles of Srs2 in the completion of synthesis-dependent strand annealing [56] is also consistent with a Siz1-independent role of Srs2. However, this contrasts with suggestions of SIZ1-dependence of PRR based on genetic interactions between siz1Δ and rad18Δ mutations [8], [34].
The third surprising result from our studies is the synergistic interaction between the deletion of RAD6 and the deletion of RAD5 in the duplication-mediated GCR assay, as Rad5 is typically considered to function downstream of Rad6-Rad18-mediated monoubiquitination of PCNA at Lys163 [4], [17]. This result is even more surprising given the equivalent duplication-mediated GCR rates observed in rad6Δ, rad18Δ, pol30-119, and rad6Δ pol30-119 mutants and the apparent epistasis of rad5Δ and pol30-119 mutations in the duplication-mediated GCR assay. The epistasis of pol30-119, but not rad6Δ, to rad5Δ, and the lack of effect of combining siz1Δ and rad5Δ mutations are inconsistent with models suggesting Rad6-dependent monoubiquition of PCNA at Lys164 is absolutely required for Rad5 function. However, these results are consistent with the possibility that ubiquitin ligases other than Rad6 can modify Lys164 of PCNA in vivo, which has been observed to occur at low levels in rad6Δ mutants [26].
Extensive pathway analysis has led to the hypothesis that replication errors are a major form of spontaneous DNA damage giving rise to duplication-mediated GCRs [27]. Thus, the apparent epistasis of RAD6 to components of the replication stress checkpoint suggests that maintaining appropriate DNA structures at the replication checkpoint [61], [62] is important for the PRR pathway to suppress duplication-mediated GCRs, and might be required for PRR to repair replication damage via template-switching pathways [6]–[10], which likely operates in competition with other pathways that might excise such DNA damage [5], [45]. Generation of potential template-switched products by the Rad5 helicase activity would produce molecules requiring further processing. We found that a rad5Δ mutation partially suppressed the defects of the sgs1Δ mutation, potentially suggesting that RAD5-dependent DNA structures that lead to GCRs accumulate in sgs1Δ mutants. This idea is consistent with the observation of HR-dependent DNA intermediates in sgs1Δ strains that accumulate in a RAD5-dependent manner [8] and the observed patterns of sensitivity to DNA damaging agents caused by different combinations of sgs1Δ, mms2Δ, and pol30-119 mutations [8], [54]. This observed partial epistasis is also consistent with the ability of SGS1 and its human homolog BLM to unwind Holliday junctions and other branched DNA structures [63]–[66] and resolve double-Holliday junctions [67]. Interestingly, we also found that srs2Δ and hcs1Δ mutations were epistatic to a rad5Δ mutation suggesting that the Srs2 and Hcs1 helicases may also act in processing stalled replication forks.
Our data suggest how PRR defects cause increased rates of duplication-mediated GCRs, but not single-copy sequence mediated-GCRs, yet suppress the increased rates of single-copy sequence-mediated GCRs caused by mutations in genes such as ASF1 [26], PIF1 ([25], Table 5) and RAD53 (Table 1) (Figures 2b,c). These phenotypes are not simply a matter of PRR mutants having a hyperrecombination phenotype [23], [68], as other hyperrecombination mutants, such as rad27Δ [69], [70] and mre11Δ, rad50Δ and xrs2Δ mutants [71]–[74] have increased rates of both single-copy sequence - and duplication-mediated GCRs [27], [75] and likely have an increased basal level of spontaneous DNA damage. Rather, PRR must function either by preventing damage from becoming HR substrates (Figure 2b) or as an alternative pathway to HR in the processing of damage (Figure 2c). PRR defects would thus increase the potential for HR, increasing the rate of duplication-mediated GCRs resulting from non-allelic HR while having little affect or even suppressing the rate of single-copy sequence-mediated GCRs as increased allelic HR acts on single-copy sequences to suppress GCRs [25], [26]. These models are consistent with the synergisitic effects of deleting RAD5 or RAD18 in conjunction with deleting RAD52 on the rate of single copy sequence-mediated GCRs as well as the decreased rates of duplication-mediated GCRs caused by deleting RAD52 in PRR mutants. Moreover, an additional role of PRR is suggested by the fact that PRR defective mutations also suppress the high rate of single copy sequence-mediated GCRs caused by different mutations. This additional role could be indirectly or directly suppressing HR, such as by controlling the nature of damaged DNA or by the Srs2-mediated suppression of Rad51 filaments [59], [60]. Alternatively, this additional function of PRR could promote the processing of DNA damage by non-HR mediated mechanisms like non-homologous end-joining (NHEJ) or chromosome healing by de novo telomere addition [43], [44]. We note that the generality of PRR defective mutations in suppressing the increased rates of single copy sequence-mediated GCRs caused by different mutations has not yet been broadly established; in addition, RAD5 has been reported to suppress NHEJ [76]. The role of PRR in specifically suppressing duplication-mediated GCRs suggests that PRR plays critical roles in suppressing non-allelic HR in genomes containing high levels of duplicated sequences. In humans, suppression of non-allelic HR is likely important for preventing genome rearrangements from occurring due to the large numbers of duplicated sequences in the human genome [77], [78] and to suppress copy number variations that contribute to human genetic variation and genetic disease [79], [80].
Materials and Methods
Construction and propagation of strains
Synthetic drop-out media for propagation of strains and measuring GCR rates were as described [75]. GCR assays were performed using derivatives of RDKY6678 (yel072w::CAN1/URA3) or RDKY6677 (yel068c::CAN1/URA3) that in addition have the genotype MATa leu2Δ1 his3Δ200 trp1Δ63 lys2ΔBgl hom3-10 ade2Δ1 ade8 ura3-52 can1::hisG iYEL072::hph as previously described as was the analysis of the structure of the resulting GCRs [27]. Mutant derivatives of these strains (Table S2) were constructed using standard PCR-based gene disruption methods as described [75].
Statistical methods
The lower and upper bounds of 95% confidence intervals of the median were calculated as described (http://www.math.unb.ca/~knight/utility/MedInt95.htm). We calculated probabilities for the null model of the observed distributions being generated by the same underlying rate using the two-tailed Mann-Whitney U-test (http://faculty.vassar.edu/~lowry/utest.html). Statistically significant differences in rates were taken to be cases where the probability of the null model was 0.01 or less.
Supporting Information
Zdroje
1. RuppWD
Howard-FlandersP
1968 Discontinuities in the DNA synthesized in an excision-defective strain of Escherichia coli following ultraviolet irradiation. J Mol Biol 31 291 304
2. RuppWD
WildeCE3rd
RenoDL
Howard-FlandersP
1971 Exchanges between DNA strands in ultraviolet-irradiated Escherichia coli. J Mol Biol 61 25 44
3. di CaprioL
CoxBS
1981 DNA synthesis in UV-irradiated yeast. Mutat Res 82 69 85
4. AndersenPL
XuF
XiaoW
2008 Eukaryotic DNA damage tolerance and translesion synthesis through covalent modifications of PCNA. Cell Res 18 162 173
5. BlastyakA
PinterL
UnkI
PrakashL
PrakashS
2007 Yeast Rad5 protein required for postreplication repair has a DNA helicase activity specific for replication fork regression. Mol Cell 28 167 175
6. HigginsNP
KatoK
StraussB
1976 A model for replication repair in mammalian cells. J Mol Biol 101 417 425
7. FujiwaraY
TatsumiM
1976 Replicative bypass repair of ultraviolet damage to DNA of mammalian cells: caffeine sensitive and caffeine resistant mechanisms. Mutat Res 37 91 110
8. BranzeiD
VanoliF
FoianiM
2008 SUMOylation regulates Rad18-mediated template switch. Nature 456 915 920
9. GoldflessSJ
MoragAS
BelisleKA
SuteraVAJr
LovettST
2006 DNA repeat rearrangements mediated by DnaK-dependent replication fork repair. Mol Cell 21 595 604
10. LiberiG
MaffiolettiG
LuccaC
ChioloI
BaryshnikovaA
2005 Rad51-dependent DNA structures accumulate at damaged replication forks in sgs1 mutants defective in the yeast ortholog of BLM RecQ helicase. Genes Dev 19 339 350
11. BaillyV
LauderS
PrakashS
PrakashL
1997 Yeast DNA repair proteins Rad6 and Rad18 form a heterodimer that has ubiquitin conjugating, DNA binding, and ATP hydrolytic activities. J Biol Chem 272 23360 23365
12. JentschS
McGrathJP
VarshavskyA
1987 The yeast DNA repair gene RAD6 encodes a ubiquitin-conjugating enzyme. Nature 329 131 134
13. GiannattasioM
LazzaroF
PlevaniP
Muzi-FalconiM
2005 The DNA damage checkpoint response requires histone H2B ubiquitination by Rad6-Bre1 and H3 methylation by Dot1. J Biol Chem 280 9879 9886
14. FlemingAB
KaoCF
HillyerC
PikaartM
OsleyMA
2008 H2B ubiquitylation plays a role in nucleosome dynamics during transcription elongation. Mol Cell 31 57 66
15. HwangWW
VenkatasubrahmanyamS
IanculescuAG
TongA
BooneC
2003 A conserved RING finger protein required for histone H2B monoubiquitination and cell size control. Mol Cell 11 261 266
16. DohmenRJ
MaduraK
BartelB
VarshavskyA
1991 The N-end rule is mediated by the UBC2(RAD6) ubiquitin-conjugating enzyme. Proc Natl Acad Sci U S A 88 7351 7355
17. HoegeC
PfanderB
MoldovanGL
PyrowolakisG
JentschS
2002 RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419 135 141
18. FuY
ZhuY
ZhangK
YeungM
DurocherD
2008 Rad6-Rad18 mediates a eukaryotic SOS response by ubiquitinating the 9-1-1 checkpoint clamp. Cell 133 601 611
19. HaracskaL
Torres-RamosCA
JohnsonRE
PrakashS
PrakashL
2004 Opposing effects of ubiquitin conjugation and SUMO modification of PCNA on replicational bypass of DNA lesions in Saccharomyces cerevisiae. Mol Cell Biol 24 4267 4274
20. StelterP
UlrichHD
2003 Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature 425 188 191
21. HofmannRM
PickartCM
1999 Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell 96 645 653
22. UlrichHD
JentschS
2000 Two RING finger proteins mediate cooperation between ubiquitin-conjugating enzymes in DNA repair. Embo J 19 3388 3397
23. LiefshitzB
SteinlaufR
FriedlA
Eckardt-SchuppF
KupiecM
1998 Genetic interactions between mutants of the ‘error-prone’ repair group of Saccharomyces cerevisiae and their effect on recombination and mutagenesis. Mutat Res 407 135 145
24. DaeeDL
MertzT
LahueRS
2007 Postreplication repair inhibits CAG.CTG repeat expansions in Saccharomyces cerevisiae. Mol Cell Biol 27 102 110
25. MotegiA
KuntzK
MajeedA
SmithS
MyungK
2006 Regulation of gross chromosomal rearrangements by ubiquitin and SUMO ligases in Saccharomyces cerevisiae. Mol Cell Biol 26 1424 1433
26. KatsES
EnserinkJM
MartinezS
KolodnerRD
2009 The Saccharomyces cerevisiae Rad6 postreplication repair and Siz1/Srs2 homologous recombination-inhibiting pathways process DNA damage that arises in asf1 mutants. Mol Cell Biol 29 5226 5237
27. PutnamCD
HayesTK
KolodnerRD
2009 Specific pathways prevent duplication-mediated genome rearrangements. Nature 460 984 989
28. CalzadaA
HodgsonB
KanemakiM
BuenoA
LabibK
2005 Molecular anatomy and regulation of a stable replisome at a paused eukaryotic DNA replication fork. Genes Dev 19 1905 1919
29. XuH
BooneC
KleinHL
2004 Mrc1 is required for sister chromatid cohesion to aid in recombination repair of spontaneous damage. Mol Cell Biol 24 7082 7090
30. OsbornAJ
ElledgeSJ
2003 Mrc1 is a replication fork component whose phosphorylation in response to DNA replication stress activates Rad53. Genes Dev 17 1755 1767
31. PutnamCD
JaehnigEJ
KolodnerRD
2009 Perspectives on the DNA damage and replication checkpoint responses in Saccharomyces cerevisiae. DNA Repair (Amst) 8 974 982
32. AlcasabasAA
OsbornAJ
BachantJ
HuF
WerlerPJ
2001 Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat Cell Biol 3 958 965
33. PanX
YeP
YuanDS
WangX
BaderJS
2006 A DNA integrity network in the yeast Saccharomyces cerevisiae. Cell 124 1069 1081
34. PfanderB
MoldovanGL
SacherM
HoegeC
JentschS
2005 SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature 436 428 433
35. PapouliE
ChenS
DaviesAA
HuttnerD
KrejciL
2005 Crosstalk between SUMO and ubiquitin on PCNA is mediated by recruitment of the helicase Srs2p. Mol Cell 19 123 133
36. LawrenceCW
ChristensenRB
1979 Metabolic suppressors of trimethoprim and ultraviolet light sensitivities of Saccharomyces cerevisiae rad6 mutants. J Bacteriol 139 866 876
37. RaguS
FayeG
IraquiI
Masurel-HenemanA
KolodnerRD
2007 Oxygen metabolism and reactive oxygen species cause chromosomal rearrangements and cell death. Proc Natl Acad Sci U S A 104 9747 9752
38. JohnsonRE
WashingtonMT
HaracskaL
PrakashS
PrakashL
2000 Eukaryotic polymerases iota and zeta act sequentially to bypass DNA lesions. Nature 406 1015 1019
39. HuangME
KolodnerRD
2005 A biological network in Saccharomyces cerevisiae prevents the deleterious effects of endogenous oxidative DNA damage. Mol Cell 17 709 720
40. DegtyarevaNP
ChenL
MieczkowskiP
PetesTD
DoetschPW
2008 Chronic oxidative DNA damage due to DNA repair defects causes chromosomal instability in Saccharomyces cerevisiae. Mol Cell Biol 28 5432 5445
41. ErlichRL
FryRC
BegleyTJ
DaeeDL
LahueRS
2008 Anc1, a protein associated with multiple transcription complexes, is involved in postreplication repair pathway in S. cerevisiae. PLoS One 3 e3717
42. KabaniM
MichotK
BoschieroC
WernerM
2005 Anc1 interacts with the catalytic subunits of the general transcription factors TFIID and TFIIF, the chromatin remodeling complexes RSC and INO80, and the histone acetyltransferase complex NuA3. Biochem Biophys Res Commun 332 398 403
43. PutnamCD
PennaneachV
KolodnerRD
2005 Saccharomyces cerevisiae as a model system to define the chromosomal instability phenotype. Mol Cell Biol 25 7226 7238
44. PutnamCD
PennaneachV
KolodnerRD
2004 Chromosome healing through terminal deletions generated by de novo telomere additions in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 101 13262 13267
45. GangavarapuV
HaracskaL
UnkI
JohnsonRE
PrakashS
2006 Mms2-Ubc13-dependent and -independent roles of Rad5 ubiquitin ligase in postreplication repair and translesion DNA synthesis in Saccharomyces cerevisiae. Mol Cell Biol 26 7783 7790
46. PagesV
BressonA
AcharyaN
PrakashS
FuchsRP
2008 Requirement of Rad5 for DNA polymerase zeta-dependent translesion synthesis in Saccharomyces cerevisiae. Genetics 180 73 82
47. XiaoW
ChowBL
BroomfieldS
HannaM
2000 The Saccharomyces cerevisiae RAD6 group is composed of an error-prone and two error-free postreplication repair pathways. Genetics 155 1633 1641
48. SchulzVP
ZakianVA
1994 The saccharomyces PIF1 DNA helicase inhibits telomere elongation and de novo telomere formation. Cell 76 145 155
49. MyungK
ChenC
KolodnerRD
2001 Multiple pathways cooperate in the suppression of genome instability in Saccharomyces cerevisiae. Nature 411 1073 1076
50. FriedlAA
LiefshitzB
SteinlaufR
KupiecM
2001 Deletion of the SRS2 gene suppresses elevated recombination and DNA damage sensitivity in rad5 and rad18 mutants of Saccharomyces cerevisiae. Mutat Res 486 137 146
51. UlrichHD
2001 The srs2 suppressor of UV sensitivity acts specifically on the RAD5 - and MMS2-dependent branch of the RAD6 pathway. Nucleic Acids Res 29 3487 3494
52. BiswasEE
FrickeWM
ChenPH
BiswasSB
1997 Yeast DNA helicase A: cloning, expression, purification, and enzymatic characterization. Biochemistry 36 13277 13284
53. BarbourL
BallLG
ZhangK
XiaoW
2006 DNA damage checkpoints are involved in postreplication repair. Genetics 174 1789 1800
54. BallLG
ZhangK
CobbJA
BooneC
XiaoW
2009 The yeast Shu complex couples error-free post-replication repair to homologous recombination. Mol Microbiol 73 89 102
55. ChenCC
MotegiA
HasegawaY
MyungK
KolodnerR
2006 Genetic analysis of ionizing radiation-induced mutagenesis in Saccharomyces cerevisiae reveals TransLesion Synthesis (TLS) independent of PCNA K164 SUMOylation and ubiquitination. DNA Repair (Amst) 5 1475 1488
56. SaponaroM
CallahanD
ZhengX
KrejciL
HaberJE
2010 Cdk1 targets srs2 to complete synthesis-dependent strand annealing and to promote recombinational repair. PLoS Genet 6 e1000858
57. CarlileCM
PickartCM
MatunisMJ
CohenRE
2009 Synthesis of free and proliferating cell nuclear antigen-bound polyubiquitin chains by the RING E3 ubiquitin ligase Rad5. J Biol Chem 284 29326 29334
58. SchiestlRH
PrakashS
PrakashL
1990 The SRS2 suppressor of rad6 mutations of Saccharomyces cerevisiae acts by channeling DNA lesions into the RAD52 DNA repair pathway. Genetics 124 817 831
59. KrejciL
Van KomenS
LiY
VillemainJ
ReddyMS
2003 DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature 423 305 309
60. VeauteX
JeussetJ
SoustelleC
KowalczykowskiSC
Le CamE
2003 The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature 423 309 312
61. LopesM
Cotta-RamusinoC
PellicioliA
LiberiG
PlevaniP
2001 The DNA replication checkpoint response stabilizes stalled replication forks. Nature 412 557 561
62. SogoJM
LopesM
FoianiM
2002 Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science 297 599 602
63. BachratiCZ
HicksonID
2006 Analysis of the DNA unwinding activity of RecQ family helicases. Methods Enzymol 409 86 100
64. BennettRJ
KeckJL
WangJC
1999 Binding specificity determines polarity of DNA unwinding by the Sgs1 protein of S. cerevisiae. J Mol Biol 289 235 248
65. KarowJK
ConstantinouA
LiJL
WestSC
HicksonID
2000 The Bloom's syndrome gene product promotes branch migration of holliday junctions. Proc Natl Acad Sci U S A 97 6504 6508
66. van BrabantAJ
YeT
SanzM
GermanIJ
EllisNA
2000 Binding and melting of D-loops by the Bloom syndrome helicase. Biochemistry 39 14617 14625
67. WuL
HicksonID
2003 The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature 426 870 874
68. RongL
PalladinoF
AguileraA
KleinHL
1991 The hyper-gene conversion hpr5-1 mutation of Saccharomyces cerevisiae is an allele of the SRS2/RADH gene. Genetics 127 75 85
69. SymingtonLS
1998 Homologous recombination is required for the viability of rad27 mutants. Nucleic Acids Res 26 5589 5595
70. TishkoffDX
FilosiN
GaidaGM
KolodnerRD
1997 A novel mutation avoidance mechanism dependent on S. cerevisiae RAD27 is distinct from DNA mismatch repair. Cell 88 253 263
71. AjimuraM
LeemSH
OgawaH
1993 Identification of new genes required for meiotic recombination in Saccharomyces cerevisiae. Genetics 133 51 66
72. IvanovEL
KorolevVG
FabreF
1992 XRS2, a DNA repair gene of Saccharomyces cerevisiae, is needed for meiotic recombination. Genetics 132 651 664
73. MaloneRE
WardT
LinS
WaringJ
1990 The RAD50 gene, a member of the double strand break repair epistasis group, is not required for spontaneous mitotic recombination in yeast. Curr Genet 18 111 116
74. SchiestlRH
ZhuJ
PetesTD
1994 Effect of mutations in genes affecting homologous recombination on restriction enzyme-mediated and illegitimate recombination in Saccharomyces cerevisiae. Mol Cell Biol 14 4493 4500
75. ChenC
KolodnerRD
1999 Gross chromosomal rearrangements in Saccharomyces cerevisiae replication and recombination defective mutants. Nat Genet 23 81 85
76. AhneF
JhaB
Eckardt-SchuppF
1997 The RAD5 gene product is involved in the avoidance of non-homologous end-joining of DNA double strand breaks in the yeast Saccharomyces cerevisiae. Nucleic Acids Res 25 743 749
77. DeiningerPL
BatzerMA
1999 Alu repeats and human disease. Mol Genet Metab 67 183 193
78. GordeninDA
ResnickMA
1998 Yeast ARMs (DNA at-risk motifs) can reveal sources of genome instability. Mutat Res 400 45 58
79. BatzerMA
DeiningerPL
2002 Alu repeats and human genomic diversity. Nat Rev Genet 3 370 379
80. JiY
EichlerEE
SchwartzS
NichollsRD
2000 Structure of chromosomal duplicons and their role in mediating human genomic disorders. Genome Res 10 597 610
Štítky
Genetika Reprodukční medicína
Článek Epistasis of Transcriptomes Reveals Synergism between Transcriptional Activators Hnf1α and Hnf4αČlánek SMA-10/LRIG Is a Conserved Transmembrane Protein that Enhances Bone Morphogenetic Protein SignalingČlánek B1 SOX Coordinate Cell Specification with Patterning and Morphogenesis in the Early Zebrafish EmbryoČlánek Bulk Segregant Analysis by High-Throughput Sequencing Reveals a Novel Xylose Utilization Gene fromČlánek Abundant Quantitative Trait Loci Exist for DNA Methylation and Gene Expression in Human Brain
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2010 Číslo 5- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- IVF a rakovina prsu – zvyšují hormony riziko vzniku rakoviny?
-
Všechny články tohoto čísla
- Digital Quantification of Human Eye Color Highlights Genetic Association of Three New Loci
- Common Genetic Variants near the Brittle Cornea Syndrome Locus Influence the Blinding Disease Risk Factor Central Corneal Thickness
- Age- and Temperature-Dependent Somatic Mutation Accumulation in
- All About Mitochondrial Eve: An Interview with Rebecca Cann
- Aging and Chronic Sun Exposure Cause Distinct Epigenetic Changes in Human Skin
- Transposed Genes in Arabidopsis Are Often Associated with Flanking Repeats
- A Survey of Genomic Traces Reveals a Common Sequencing Error, RNA Editing, and DNA Editing
- Gene Transposition Causing Natural Variation for Growth in
- The Polyproline Site in Hinge 2 Influences the Functional Capacity of Truncated Dystrophins
- Epistasis of Transcriptomes Reveals Synergism between Transcriptional Activators Hnf1α and Hnf4α
- Integration of Light Signals by the Retinoblastoma Pathway in the Control of S Phase Entry in the Picophytoplanktonic Cell
- The Proprotein Convertase Encoded by () Is Required in Corpora Cardiaca Endocrine Cells Producing the Glucose Regulatory Hormone AKH
- Sgs1 and Exo1 Redundantly Inhibit Break-Induced Replication and Telomere Addition at Broken Chromosome Ends
- A MATE-Family Efflux Pump Rescues the 8-Oxoguanine-Repair-Deficient Mutator Phenotype and Protects Against HO Killing
- The Relationship among Gene Expression, the Evolution of Gene Dosage, and the Rate of Protein Evolution
- SMA-10/LRIG Is a Conserved Transmembrane Protein that Enhances Bone Morphogenetic Protein Signaling
- The Nuclear Receptor DHR3 Modulates dS6 Kinase–Dependent Growth in
- Involvement of Global Genome Repair, Transcription Coupled Repair, and Chromatin Remodeling in UV DNA Damage Response Changes during Development
- B1 SOX Coordinate Cell Specification with Patterning and Morphogenesis in the Early Zebrafish Embryo
- Linkage and Association Mapping of Flowering Time in Nature
- Bulk Segregant Analysis by High-Throughput Sequencing Reveals a Novel Xylose Utilization Gene from
- Abundant Quantitative Trait Loci Exist for DNA Methylation and Gene Expression in Human Brain
- Ablation of Whirlin Long Isoform Disrupts the USH2 Protein Complex and Causes Vision and Hearing Loss
- Characterization of Oxidative Guanine Damage and Repair in Mammalian Telomeres
- DNA Adenine Methylation Is Required to Replicate Both Chromosomes Once per Cell Cycle
- Genome-Wide Copy Number Variation in Epilepsy: Novel Susceptibility Loci in Idiopathic Generalized and Focal Epilepsies
- FACT Prevents the Accumulation of Free Histones Evicted from Transcribed Chromatin and a Subsequent Cell Cycle Delay in G1
- GC-Biased Evolution Near Human Accelerated Regions
- Liver and Adipose Expression Associated SNPs Are Enriched for Association to Type 2 Diabetes
- Myeloid Cell-Restricted Insulin Receptor Deficiency Protects Against Obesity-Induced Inflammation and Systemic Insulin Resistance
- The Mating Type Locus () and Sexual Reproduction of : Insights into the Evolution of Sex and Sex-Determining Chromosomal Regions in Fungi
- B-Cyclin/CDKs Regulate Mitotic Spindle Assembly by Phosphorylating Kinesins-5 in Budding Yeast
- Post-Replication Repair Suppresses Duplication-Mediated Genome Instability
- Genome Sequence of the Plant Growth Promoting Endophytic Bacterium sp. 638
- Transcription Factors Mat2 and Znf2 Operate Cellular Circuits Orchestrating Opposite- and Same-Sex Mating in
- The Use of Orthologous Sequences to Predict the Impact of Amino Acid Substitutions on Protein Function
- Mutation in Archain 1, a Subunit of COPI Coatomer Complex, Causes Diluted Coat Color and Purkinje Cell Degeneration
- Shelterin-Like Proteins and Yku Inhibit Nucleolytic Processing of Telomeres
- Affecting Function Causes a Dilated Heart in Adult
- Manipulation of Behavioral Decline in with the Rag GTPase
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Common Genetic Variants near the Brittle Cornea Syndrome Locus Influence the Blinding Disease Risk Factor Central Corneal Thickness
- All About Mitochondrial Eve: An Interview with Rebecca Cann
- The Relationship among Gene Expression, the Evolution of Gene Dosage, and the Rate of Protein Evolution
- SMA-10/LRIG Is a Conserved Transmembrane Protein that Enhances Bone Morphogenetic Protein Signaling
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



