-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Bulk Segregant Analysis by High-Throughput Sequencing Reveals a Novel Xylose Utilization Gene from
Fermentation of xylose is a fundamental requirement for the efficient production of ethanol from lignocellulosic biomass sources. Although they aggressively ferment hexoses, it has long been thought that native Saccharomyces cerevisiae strains cannot grow fermentatively or non-fermentatively on xylose. Population surveys have uncovered a few naturally occurring strains that are weakly xylose-positive, and some S. cerevisiae have been genetically engineered to ferment xylose, but no strain, either natural or engineered, has yet been reported to ferment xylose as efficiently as glucose. Here, we used a medium-throughput screen to identify Saccharomyces strains that can increase in optical density when xylose is presented as the sole carbon source. We identified 38 strains that have this xylose utilization phenotype, including strains of S. cerevisiae, other sensu stricto members, and hybrids between them. All the S. cerevisiae xylose-utilizing strains we identified are wine yeasts, and for those that could produce meiotic progeny, the xylose phenotype segregates as a single gene trait. We mapped this gene by Bulk Segregant Analysis (BSA) using tiling microarrays and high-throughput sequencing. The gene is a putative xylitol dehydrogenase, which we name XDH1, and is located in the subtelomeric region of the right end of chromosome XV in a region not present in the S288c reference genome. We further characterized the xylose phenotype by performing gene expression microarrays and by genetically dissecting the endogenous Saccharomyces xylose pathway. We have demonstrated that natural S. cerevisiae yeasts are capable of utilizing xylose as the sole carbon source, characterized the genetic basis for this trait as well as the endogenous xylose utilization pathway, and demonstrated the feasibility of BSA using high-throughput sequencing.
Published in the journal: . PLoS Genet 6(5): e32767. doi:10.1371/journal.pgen.1000942
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1000942Summary
Fermentation of xylose is a fundamental requirement for the efficient production of ethanol from lignocellulosic biomass sources. Although they aggressively ferment hexoses, it has long been thought that native Saccharomyces cerevisiae strains cannot grow fermentatively or non-fermentatively on xylose. Population surveys have uncovered a few naturally occurring strains that are weakly xylose-positive, and some S. cerevisiae have been genetically engineered to ferment xylose, but no strain, either natural or engineered, has yet been reported to ferment xylose as efficiently as glucose. Here, we used a medium-throughput screen to identify Saccharomyces strains that can increase in optical density when xylose is presented as the sole carbon source. We identified 38 strains that have this xylose utilization phenotype, including strains of S. cerevisiae, other sensu stricto members, and hybrids between them. All the S. cerevisiae xylose-utilizing strains we identified are wine yeasts, and for those that could produce meiotic progeny, the xylose phenotype segregates as a single gene trait. We mapped this gene by Bulk Segregant Analysis (BSA) using tiling microarrays and high-throughput sequencing. The gene is a putative xylitol dehydrogenase, which we name XDH1, and is located in the subtelomeric region of the right end of chromosome XV in a region not present in the S288c reference genome. We further characterized the xylose phenotype by performing gene expression microarrays and by genetically dissecting the endogenous Saccharomyces xylose pathway. We have demonstrated that natural S. cerevisiae yeasts are capable of utilizing xylose as the sole carbon source, characterized the genetic basis for this trait as well as the endogenous xylose utilization pathway, and demonstrated the feasibility of BSA using high-throughput sequencing.
Introduction
It is clear that society has a responsibility to address the anthropogenic causes of climate change. Current estimates indicate that about 95% of the world's energy comes from burning fossil fuels [1], which is the leading contributor of carbon dioxide emissions. Combustion of liquid fossil fuels for transportation is responsible for a large fraction of these carbon dioxide emissions in the United States, second only to electricity generation (U.S. Environmental Protection Agency). For these reasons, creating “carbon neutral” liquid transportation fuels should be an important part of global efforts to reduce carbon emissions.
One solution already in widespread use is bioethanol fermented from sugar cane (Brazil) or cornstarch (U.S.) by various strains of Saccharomyces cerevisiae [2], which is used as a major component or additive to liquid transportation fuels. For bioethanol to become a sustainable, economically viable commodity, and not to compete with food sources, it is necessary to move away from sugar cane or corn biomass toward lignocellulosic biomass sources such as corn stover or other agricultural wastes, wood byproducts, or dedicated fuel crops such as Miscanthus or switchgrass [3]–[5]. However, there are technical challenges that must be overcome before this is possible. For sugar cane and corn biomass, the predominant sugars are glucose and/or fructose, both of which are readily fermented to ethanol by various S. cerevisiae yeast strains, usually wild isolates that are particularly suited for large-scale fermentations [6], [7]. However, in lignocellulosic biomass sources, the second most abundant carbohydrate after glucose is xylose, the major pentose of hemicellulose. There is as yet no known strain of Saccharomyces that is able to convert xylose to ethanol as efficiently as glucose. Because the mass proportion of hemicellulose ranges from 20–50% in common agricultural lignocellulosic biomasses, finding both a cost-effective and energy-efficient conversion of xylose to ethanol is a critical hurdle [8].
The budding yeast Saccharomyces cerevisiae is the microorganism of choice for industrial fermentations for a variety of reasons, mainly due to its high ethanol productivity both aerobically and anaerobically, its high ethanol and low pH tolerance, and its resistance to many of the harmful compounds in a typical biomass hydrolysate. Despite recent evidence that some natural S. cerevisiae can grow, albeit poorly, on xylose [9], it has generally been reported that both natural and laboratory S. cerevisiae strains do not ferment xylose [10]–[12] leading to the assumption that they cannot, without recourse to genetic engineering, be utilized for efficient conversion of lignocellulose to ethanol. While S. cerevisiae strains were shown to be able to ferment the xylose isomer xylulose and to possess genes putatively encoding enzymes capable of xylose reduction (GRE3, GCY1, YPR1, YDL124W, YJR096W), xylitol oxidation (XYL2, SOR1, SOR2), and xylulose phosphorylation (XKS1) (Figure 1), there have been a number of experimental observations indicating that S. cerevisiae could not ferment xylose [13]–[15]. Such observations include low levels of gene expression of the endogenous enzymes, poor transport of xylose, redox cofactor imbalances, and insufficient flux through the pentose phosphate shunt [16], [17]. Despite these issues being well characterized in laboratory strains of S. cerevisiae, little is known about natural variation within Saccharomyces yeasts as it relates to xylose utilization which, as has already been shown [9], is likely to be relevant to this phenotype.
Fig. 1. Endogenous xylose pathway. 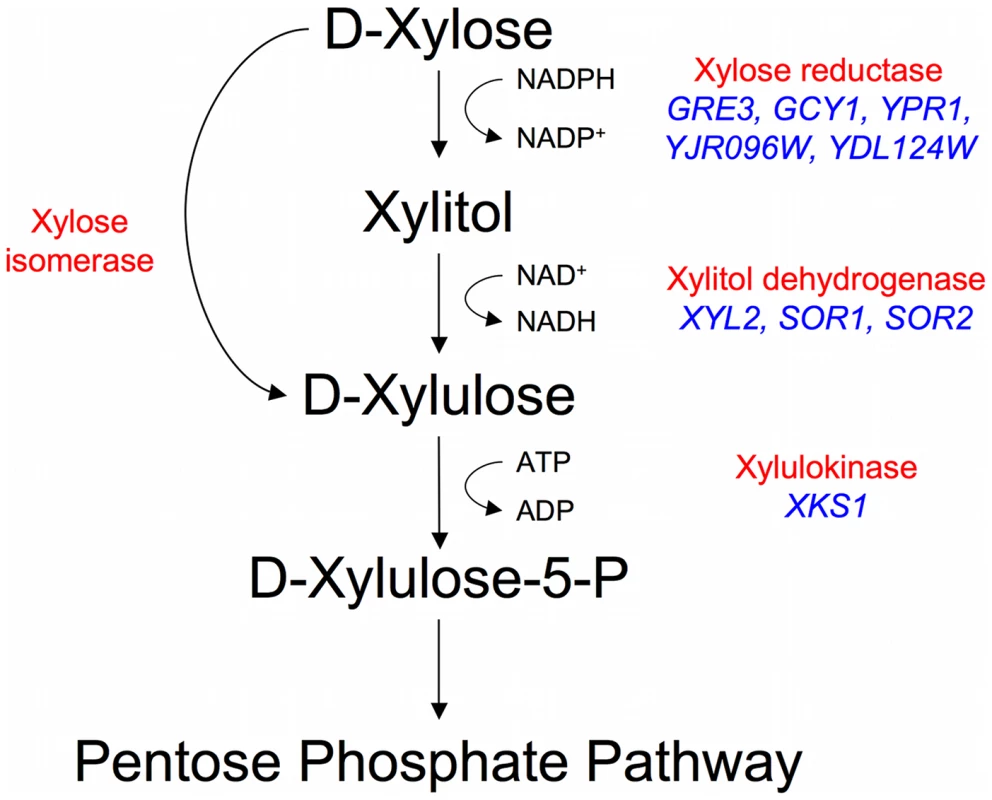
The canonical reduction-oxidation (fungi) and isomerization (bacteria and fungi) pathways with biochemical activities labeled in red. The putative Saccharomyces cerevisiae enzymes are in blue (there is no known xylose isomerase in S. cerevisiae). A significant amount of progress has been made over the last 30 years toward solving these problems, with much of the work focused on introducing foreign xylose pathway enzymes into S. cerevisiae: either the genes that code for xylose reductase [XR], xylitol dehydrogenase [XDH], or xylulokinase [XK] from the xylose-utilizing fungus Pichia stipitis [18]–[22], or genes coding for a xylose isomerase [XI] from other fungi and bacteria [23]–[28]. There have also been efforts to increase or adjust xylose pathway enzyme activities (XR, XK, XDH) [15], [29]–[33] and pentose phosphate flux [34], [35], reduce redox imbalances [36]–[41], and use directed evolution or random mutagenesis to increase xylose utilization [42]–[45]. Despite this large body of work, the fermentation of xylose to ethanol in these strains is still much slower than that of glucose, and there is still significant room for improvement in xylose fermentation, as well as co-fermentation of xylose and glucose, by S. cerevisiae for industrial scale applications.
As mentioned above, it has been determined that some natural strains of Saccharomyces cerevisiae are capable of growing on xylose, contrary to the notion that S. cerevisiae does not recognize this pentose as a usable carbon source [9]. It is also well characterized that there is abundant natural genetic and phenotypic variation within S. cerevisiae and closely related species [46]–[51]. In this work, we have screened a large number of wild, industrial and laboratory yeast strains to determine if other xylose-utilizing strains of Saccharomyces already exist in nature, and if so, to determine the genetic basis or bases for the phenotype. We screened 647 strains, and found a number of different Saccharomyces yeasts, predominantly wine yeasts, which are capable of utilizing xylose, albeit modestly. Through the application of high-throughput sequencing to Bulk Segregant Analysis [BSA] [52], we were able to identify the gene responsible for xylose utilization in a wine strain of S. cerevisiae, which encodes a novel putative xylitol dehydrogenase that we named XDH1. We observed that this gene is present in many different wine strains and is responsible for xylose utilization in these strains, however we have identified other strains in our screen that appear to have an independent genetic basis for their xylose utilization. We also carried out transcriptional profiling to characterize gene expression patterns during xylose utilization in wine strain derivatives and determined the contribution of native S. cerevisiae xylose pathway enzymes to the phenotype we observed. These data suggest that the putative enzyme encoded by XDH1 works in combination with the native xylose pathway to permit natural S. cerevisiae strains to recognize and utilize xylose.
Results
Screen for Xylose Utilization
To identify natural Saccharomyces species/strains that are able to utilize xylose, we screened each strain in our yeast collection for the ability, when placed in liquid medium with xylose as the sole carbon source, to increase in optical density [OD] after several days of incubation at 25°C. We measured the OD of 647 strains (Table S1) in a sealed 96-well plate format with constant, orbital shaking (see Materials and Methods). The collection largely comprises S. cerevisiae strains from various sources, including wine, brewing, baking, laboratory and clinical isolates, but it also contains other Saccharomyces sensu stricto yeasts and various hybrids between them. Of the 647 strains tested, we identified 38 strains that had some observable increase in OD (Table 1). These “xylose-positive” strains were predominantly (29/38) S. cerevisiae wine yeasts (although not all wine yeasts were xylose-positive), with the remainder being interspecific hybrids within the sensu stricto group. These xylose-positive hybrid strains generally reached higher OD in xylose media compared to the S. cerevisiae wine strains. Figure 2A shows a typical S. cerevisiae wine strain profile as well as the profile from one of the best hybrids, comparing growth in a xylose-containing medium to the same medium with no carbon source. While increase in OD does not provide evidence for fermentation of xylose to ethanol, or even of cell division, these data do show that there are natural Saccharomyces yeasts capable of utilizing xylose to accumulate biomass.
Fig. 2. Wine strains display a xylose-utilization phenotype controlled by a single gene. 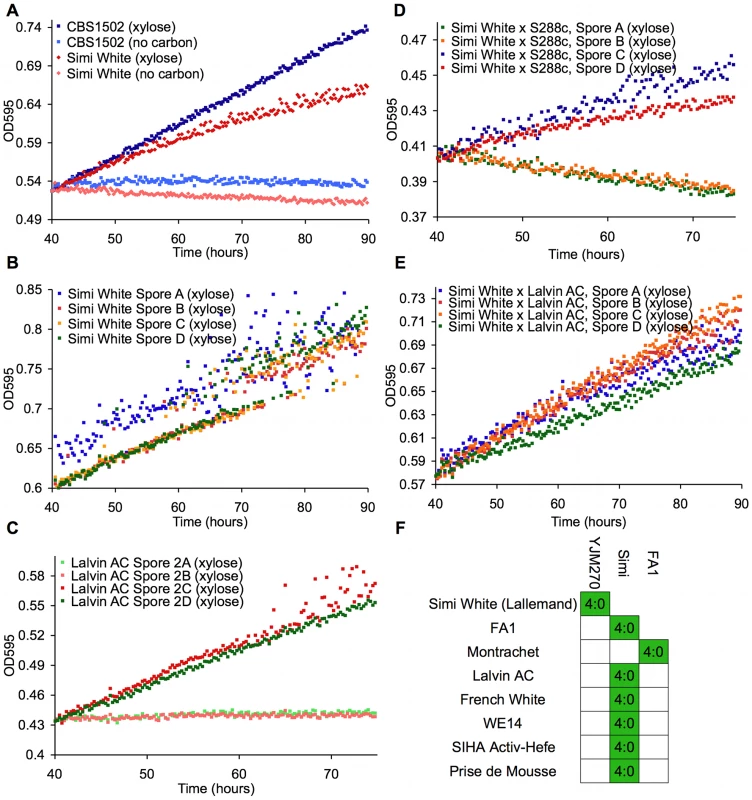
These panels show growth curves measured in the TECAN. Curves are normalized to the first time point, and the initial growth phase due to trehalose (present in YP) was removed from the analysis in these and all other growth curves shown. (A) S. cerevisiae (Simi White) and hybrid (CBS1502) grown in YP media. (B–E) Complete tetrads of Simi White; Lalvin AC; Simi White×S288c; and Simi White×Lalvin AC. (F) This table represents all the wine strains that were able to be interbred, and all show a 4∶0 segregation of xylose utilizing∶xylose non-utilizing. Tab. 1. “Xylose positive” strains. 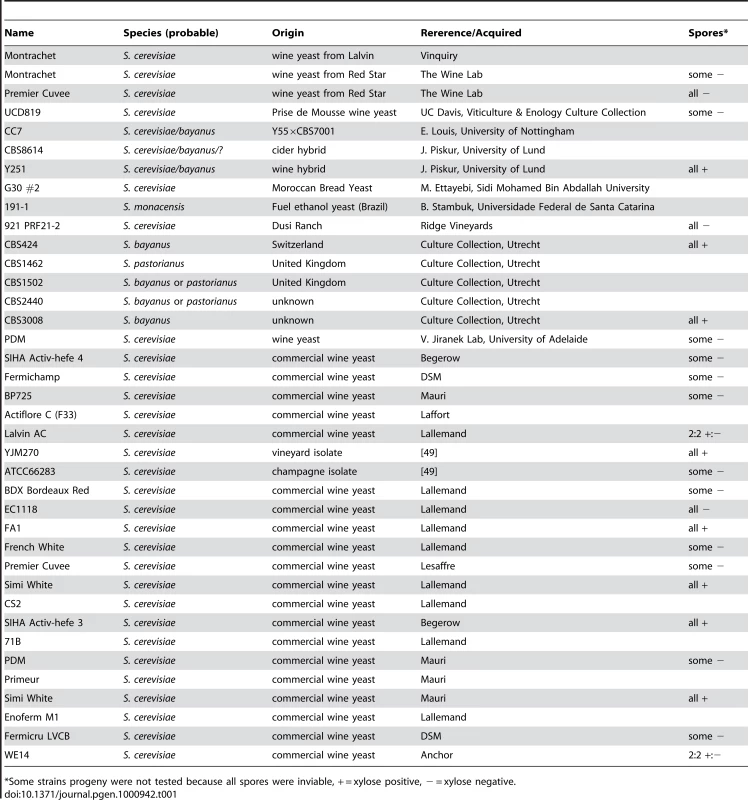
*Some strains progeny were not tested because all spores were inviable, + = xylose positive, − = xylose negative. To understand the genetic basis of this xylose utilization we chose to focus on the wine strains because many could be sporulated and crossed to a laboratory strain of S. cerevisiae, and we could thus determine the segregation pattern of the phenotype. Twenty-five of these xylose-positive S. cerevisiae strains could be sporulated and tetrads dissected (Table 1). Note that because the strains have a wild-type HO gene, the spore products obtained after tetrad dissection are actually fully homozygous diploids, due to self-mating of the haploid spore during its growth on the dissection plate. In 8/25 of the xylose-positive S. cerevisiae strains, all of the spore products were xylose-positive (e.g. Simi White, Figure 2B), while in 2/25 the trait segregated 2 xylose-positive: 2 xylose-negative (e.g. Lalvin AC, Figure 2C). In the remaining 15 strains, including three strains from which no xylose-positive spores were recovered, spore viability was so poor that no complete tetrads were obtained, and thus the segregation pattern(s) could not be identified. We then took xylose-positive spore products from all of the strains from which such spores could be obtained, and crossed them (see Materials and Methods) to a laboratory S. cerevisiae strain, S288c. We observed that all of the resulting diploids were xylose-positive, indicating that the phenotype is dominant (data not shown). The resulting strains were then sporulated, and in those strains where a segregation pattern could be established, the xylose-positive trait segregated to produce two positive and two negative spores, suggesting that a single gene was responsible for the xylose-positive trait (e.g. Simi White, Figure 2D). To determine if the same locus is responsible for xylose utilization in these various wine strains, we crossed xylose-positive spores between the various wine strains and determined the segregation pattern of the xylose phenotype in the progeny of these crosses. In all of the crosses that were performed, the xylose-positive phenotype segregated 4∶0 in six tetrads (Figure 2E); this defines a cohort of at least 9 wine strains containing a single complementation group (locus) responsible for the phenotype (Figure 2F). These data indicate that a single, dominant locus is responsible for permitting xylose utilization in these S. cerevisiae strains and suggest that this mechanism of xylose utilization is common to all of the xylose-positive wine yeasts that we identified. These data also suggest that this locus may be identical by descent, consistent with evidence that wine strains are very closely related and have probably only diverged a few thousand years ago [46], [49], [50].
Identification of the Responsible Gene by Bulk Segregant Analysis
To determine the genomic location of the gene that permits xylose utilization we conducted BSA [53] using Affymetrix yeast tiling arrays. BSA works by taking advantage of DNA sequence polymorphisms between different strains and of the fact that it is relatively easy to pool large numbers of meiotic spore products (segregants) in yeast. Pooling segregants based on their phenotype allows the region of the genome responsible for the phenotype to be detected because DNA polymorphisms in regions unlinked to the responsible locus will segregate randomly and be “evened” out, while sequences or polymorphisms either directly responsible for the trait, or very closely linked to it, will be present in all positive segregants and absent in all negative segregants. In our case, the Simi White wine strain carrying the locus responsible for xylose utilization was crossed to a laboratory strain; the wine strain was previously estimated to carry DNA polymorphisms relative to the laboratory strain at a level of approximately .5% [54]. Spores from the Simi White/S288c diploid were screened for the xylose utilization phenotype and 39 positive spores were combined into one pool and 39 negative spores into another pool, and genomic DNA [gDNA] was isolated from each pool. We then hybridized the positive and negative gDNA pools to tiling microarrays (based on the S288c reference genome) with the expectation that regions of the genome derived from Simi White will hybridize less robustly to the array because of the DNA polymorphisms between Simi White and S288c. Log2 ratios of probe intensities were calculated (negative/positive), and a peak was evident by visual inspection in the chromosome XV right subtelomeric region that corresponds to less robust hybridization to the microarray of the positive pool gDNA (Figure 3). We confirmed the localization of the xylose-positive trait to this region by linkage analysis using strains from the yeast deletion collection, showing that the xylose-positive trait co-segregated meiotically with PHR1 (YOR386W), YOR378W, and YOR365C (2). We cloned a 10 kilobase [kb] region of the genome distal to PHR1 (containing YOR389W, YOR390W, HSP33, YOR392W, ERR1, and PAU21) from haploid, xylose-positive segregants of Simi White (GSY2469) and Lalvin AC (GSY1362) and independently transformed an S288c-based laboratory strain (FY2) with the constructs, but neither the Simi White nor the Lalvin AC derived constructs conferred a xylose-positive phenotype (data not shown), suggesting that the responsible gene was not within this 10kb region. Because yeast telomeric regions are susceptible to amplifications, insertions and translocations [55], we instead considered the possibility that the trait of interest may lie in an insertion distal to the subtelomeric sequences present in the S288c reference genome.
Fig. 3. Bulk Segregant Analysis by Affymetrix Yeast tiling microarrays. 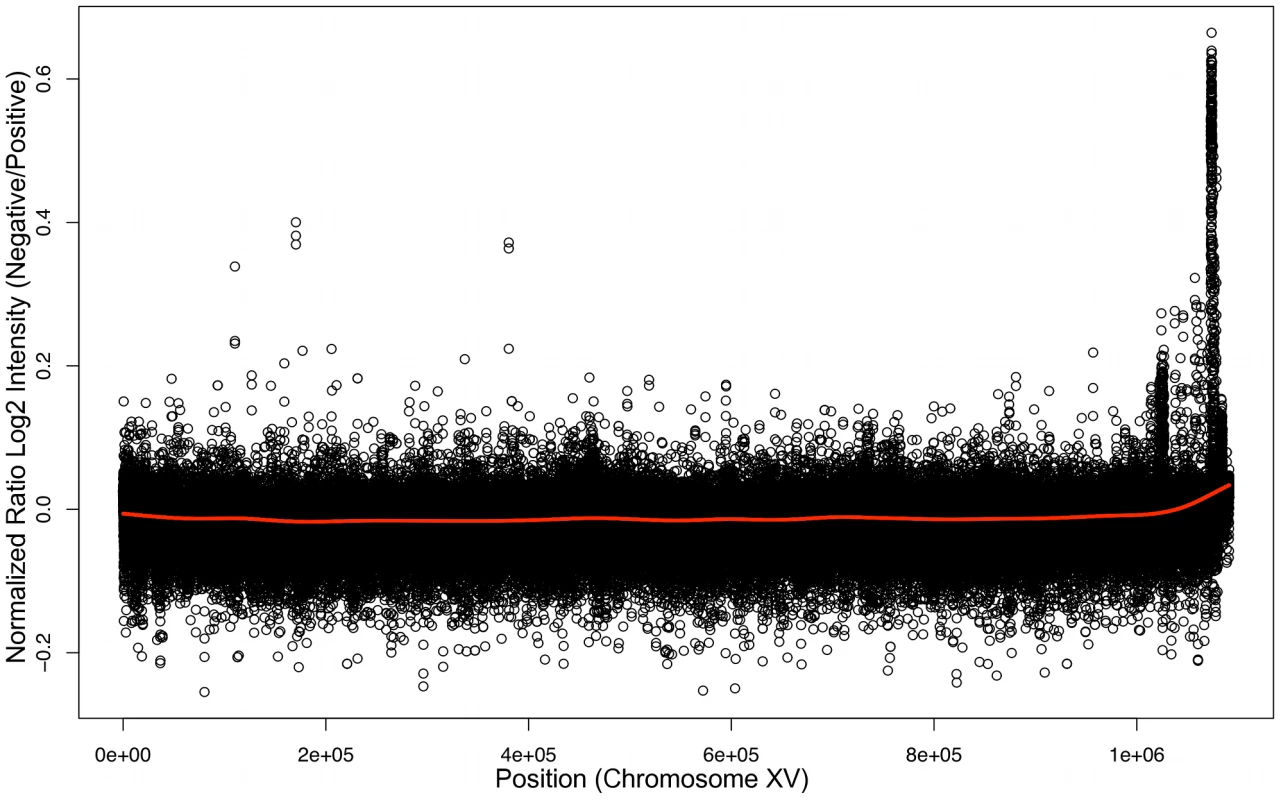
Genomic DNA from pools of xylose utilizing or non-utilizing segregants were hybridized independently to Affymetrix tiling microarrays. Plotted here is a ratio of the log2 intensities of the xylose non-utilizing versus xylose utilizing microarray experiments along chromosome XV. To identify whether there is an insertion on chromosome XV that contains the gene responsible for the xylose utilization phenotype, we repeated BSA using Illumina high-throughput sequencing on the same Simi White gDNA pools, as well as four additional pools, containing 19 positives and 16 negatives derived from a Lalvin AC/S288c cross and 16 positives and 16 negatives from a SIHA Activ-Hefe 4/S288c cross. We chose BSA over sequencing individual isolates to enrich for sequences responsible for (or tightly linked to) the xylose-positive phenotype, as there are likely to be many other novel sequences in the wine strains that are not present in the S228c genome but are unrelated to the xylose phenotype. Simi White positive and negative pools were sequenced to approximately 50× coverage of the S288c genome (∼17M mapped reads per pool), and the Lalvin AC and SIHA pools were sequenced to ∼25× coverage (∼8M mapped reads per pool) (Table S3). The 36 base pair sequence reads were aligned to the S288c reference genome using the software program MAQ [56].
To determine if any sequences were present in the positive pool that were not present in the negative pool, we performed de novo assembly of the reads that did not map to the S288c reference genome. Because de novo assembly with short sequence reads is challenging, it is important to have deep coverage and include only high quality sequence reads. To achieve this coverage and quality, we compiled all of the high-quality unmapped reads (where “high quality” reads were defined as those that did not contain any uncalled bases) from all three positive gDNA pools and used the software program Velvet [57] to perform the assembly. We then used MAQ to independently align the unmapped reads from all six gDNA pools (positive and negative) to the Velvet contigs created from the positive pools. We identified 9 individual contigs with a combined length of approximately 55kb that had no or very few reads map to them from the three negative pools. We designed primers that would amplify each of these 9 contigs and performed linkage analysis to confirm that these contigs are linked to the xylose-positive trait and yor365cΔ (Table S4). We then determined that there were approximately 28 open reading frames [ORFs] (>100 amino acids) within these 9 contigs and that a number of the ORFs are homologous to sugar metabolism genes, including a xylitol/sorbitol dehydrogenase homolog (Figure 4). The presence of a large insertion relative to the S288c reference genome containing these ORFs within the right sub-telomeric region of chromosome XV has independently been recently observed in the EC1118 wine strain genome sequence [54], [58]. The total size of the insertion is 65kb, indicating that de novo assembly identified most of the region. These data, combined with our observation that none of the previously annotated genes in the S288c reference genome distal to PHR1 were able to confer the xylose phenotype, strongly suggested that the xylose utilization trait resided in this telomeric insertion.
Fig. 4. 65kb insertion in subtelomeric region of chromosome XV. 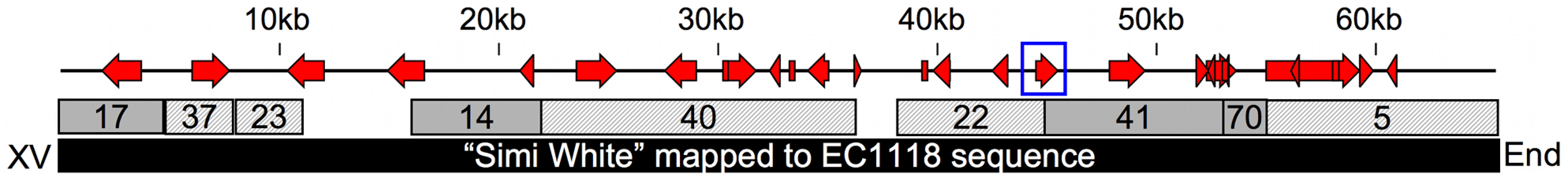
This map shows the positions of open reading frames within a novel chromosome XV subtelomeric region common amongst some wine strains. The blue box denotes the position of the putative xylitol dehydrogenase homolog. Numbered boxes represent Velvet contigs created from de novo assembly of the filtered (solid = Watson strand, hashed = Crick strand), unmapped reads compiled from the three positive pools. The black box represents the 65kb region identified by the EC1118 wine yeast genome sequence used to map the Simi White unmapped reads and find all the open reading frames in the region. Necessity and Sufficiency of Novel XDH Homolog
Of the ORFs within the chromosome XV insertion, the putative xylitol/sorbitol dehydrogenase was particularly interesting to us because it has homology to xylitol dehydrogenases from S. cerevisiae and other species (Figure S1), and we hypothesized that this gene was a likely candidate for the xylose utilization trait. We amplified this gene from both Simi White and Lalvin AC, along with approximately 400 bases of upstream and downstream sequences, and cloned it into the CEN/ARS vector pRS316 [59] to create pGS104 and pGS105. When either of these constructs were transformed into S288c, they were sufficient to permit xylose utilization in this previously non-xylose-utilizing laboratory strain (Figure 5A and data not shown). The phenotype is dependent on the presence of the plasmid containing the gene, as the xylose phenotype was lost when the transformants lost the plasmid (Figure 5A). These data show that this gene, which we have named XDH1, is sufficient to permit xylose utilization in an otherwise wild type, but xylose-negative strain.
Fig. 5. Novel XDH homolog is sufficient and necessary for xylose utilization. 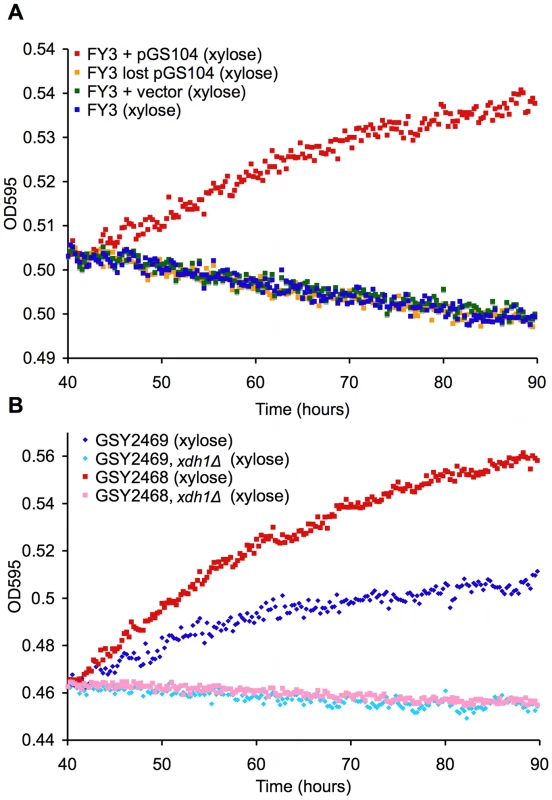
TECAN growth curves in YP with 2% xylose of (A) xylose-positive strain, laboratory strain transformed with pGS104 (pRS316::XDH1), laboratory strain transformed with pRS316 alone, and laboratory strain transformed with pGS104 but allowed to lose the plasmid and (B) two independent xylose-positive strains with an xdh1Δ::KanMX disruption and their parents. To show necessity of XDH1 for the phenotype, we created a deletion strain (xdh1Δ) and measured xylose utilization as before. Two Simi White derivatives (GSY2468/9) were transformed with a KanMX deletion cassette containing sequences (∼400 bases) immediately up and downstream of XDH1. The deletion strains (GSY2472/1) were confirmed by PCR. Deletion of XDH1 completely abrogated the phenotype (Figure 5B). We crossed the deletion strain to another haploid derivative of Simi White and confirmed that the deletion always segregates in opposition to the xylose-positive phenotype in 9 tetrads tested (data not shown). These data prove that XDH1 is not only sufficient but also necessary for xylose utilization.
Having shown that XDH1 is responsible for xylose utilization in at least two S. cerevisiae wine strains (Simi White and Lalvin AC), and also considering our observation that all the other wine strains we were able to test appeared to be in the same complementation group, we sought to determine whether XDH1 is present in all of the xylose-positive S. cerevisiae strains and other Saccharomyces hybrids that we initially identified in our screen. To test for the presence of XDH1 in those strains, we performed colony PCR on all of the xylose-positive strains that were identified in the screen (Table 2). In 33/38 xylose-positive isolates, XDH1 was present. Interestingly, the 5 xylose-positive strains from which we could not amplify XDH1 were all recorded as being either S. bayanus or hybrids between S. bayanus and S. cerevisiae.
Tab. 2. Presence of XDH1 in xylose positive or negative strains. 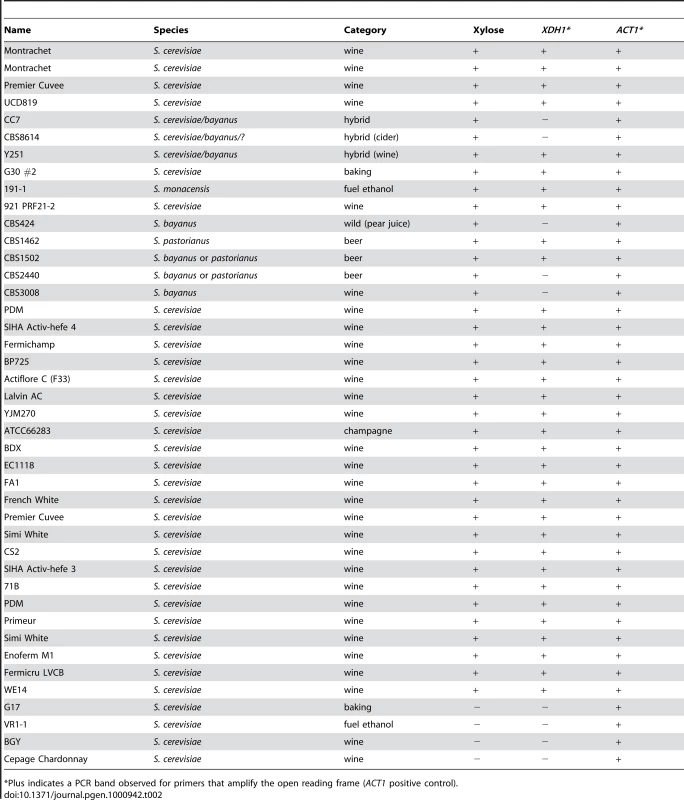
*Plus indicates a PCR band observed for primers that amplify the open reading frame (ACT1 positive control). Some of the positive strains from our screen were heterozygous for xylose utilization, because when sporulated, the trait segregated to produce two positive and two negative spores (or some number of each type in cases where there were not enough viable spores to determine a distinct segregation pattern) (Table 1). We performed colony PCR on some of these spores to test for the presence of XDH1 (Table 3). Among the meiotic progeny of these heterozygotes, every xylose-positive segregant contained this gene. In four cases (Lalvin AC, PDM, SIHA Activ-hefe 4, and WE14) the presence of XDH1 segregated with the xylose-positive spores, while the negative spores did not contain XDH1. Surprisingly, we found instances where some negative spores did contain the XDH1 gene. In one instance, one of the two negative spores contained XDH1, while the other negative spore did not (ATCC66283, note that the four spores not from the same tetrad). In the four other cases (Montrachet, BDX, Fermichamp, French White), all the negative spores tested positive by PCR for XDH1. We sequenced XDH1 and approximately 200 bases up and downstream of the ORF from all spores of two of these heterozygous tetrads (Fermichamp tetrad 1A–D, BDX tetrad 1A–D) and did not observe any DNA sequence polymorphisms between the xylose-positive and negative spores (data not shown). This suggests that there may be another locus that is epistatic to XDH1 in these strains. Overall, the ubiquity of XDH1 in the xylose-positive strains is consistent with the hypothesis that this gene is necessary for xylose utilization in natural S. cerevisiae strains.
Tab. 3. Presence of XDH1 in xylose positive progeny. 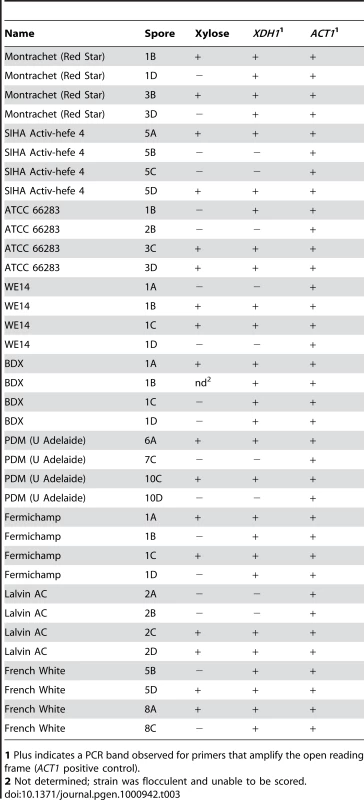
1 Plus indicates a PCR band observed for primers that amplify the open reading frame (ACT1 positive control). Genetic Dissection of Endogenous Xylose Pathway
As described above, there are genes encoding putative xylose pathway enzymes in the S288c reference genome, and it has previously been suggested that the major XR contributors are GRE3, YPR1, and YJR096W [14]. It has also been observed that co-over-expression of GRE3 and XYL2, which encodes a putative XDH, can confer a xylose-positive phenotype [14], [15]. To assess the contribution of these and the other endogenous xylose genes to our xylose phenotype, we deleted either singly or in various combinations these genes from a haploid, xylose-positive Simi White derivative (GSY2469) and assessed the growth phenotypes of the various deletion mutants (Figure 6).
Fig. 6. Genetic dissection of the endogenous xylose pathway. 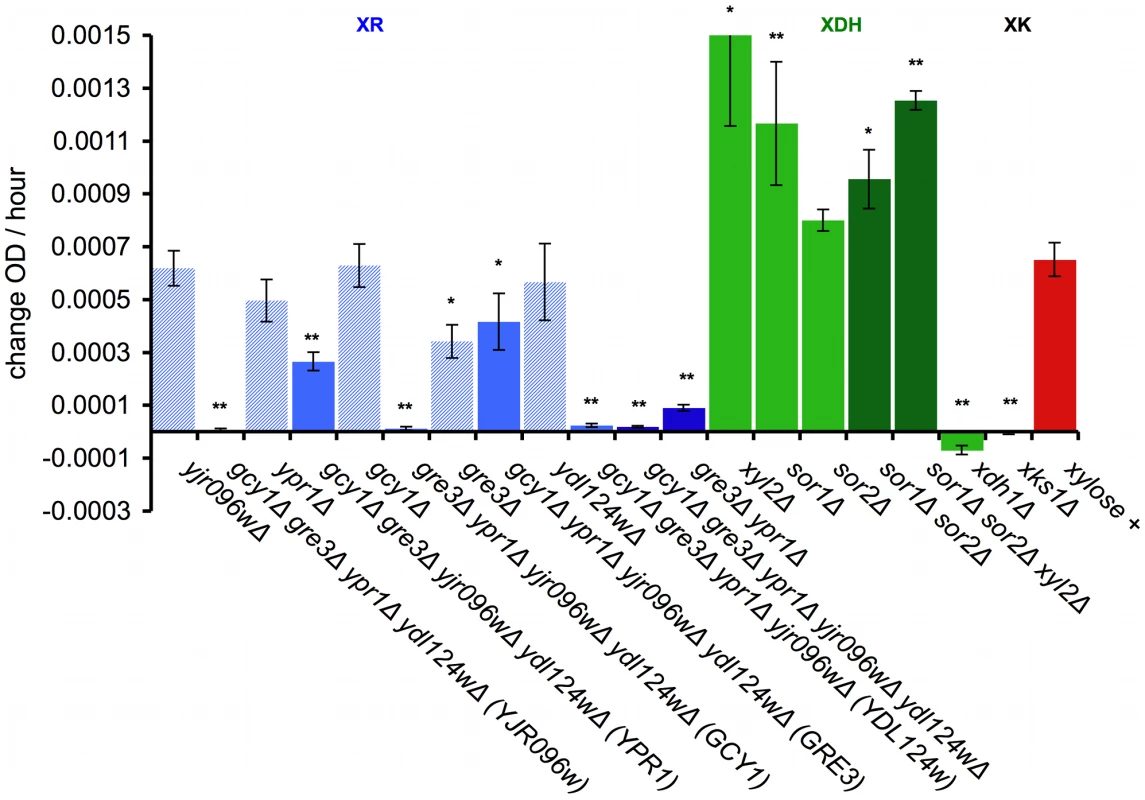
Quantification of increase in OD over time for the indicated deletions that were crossed into the Simi White haploid derivative background (GSY2469). Growth was measured in the TECAN plate reader in minimal media. OD increase calculated from slope of xylose – no carbon subtraction. * = p<.05 and ** = p<.01 in two sample t-test compared to GSY2469 (xylose +). XR = xylose reductase; XDH = xylitol dehydrogenase; XK = xylulokinase. Error bars show the standard error of the mean. To test the contribution of each of the five putative xylose reductase genes, we introduced deletions of each of them individually in the XDH1 background. Only GRE3 significantly affected the phenotype, and none of the xylose reductase genes, when deleted individually, completely abrogated the phenotype (Figure 6, XR). We also tested sufficiency for each of the reductases by creating quadruple deletion mutants, leaving only one putative reductase gene intact (Figure 6, XR). The only two putative xylose reductases that alone contributed significantly to the ability to utilize xylose in our background were GRE3 and YPR1. The other three putative xylose reductases are insufficient by themselves to allow xylose utilization (YDL124W, GCY1, YJR096w). We also created a gre3Δ ypr1Δ double deletion in which the phenotype is almost completely removed (Figure 6, XR), though these data are not inconsistent with the other three putative xylose reductases contributing some residual XR activity. These data together suggest that both GRE3 and YPR1 are the major contributors to XR activity in a natural S. cerevisiae derivative.
Next, we tested the contribution of three putative xylitol dehydrogenases to the observed phenotype (Figure 6, XDH). Interestingly, when each potential XDH was deleted individually in the XDH1 background (sor1Δ, sor2Δ, xyl2Δ), the deletion mutants showed an improved xylose utilization phenotype relative to the positive control. Furthermore, when all three were deleted together (sor1Δ sor2Δ xyl2Δ), the phenotype was further enhanced (Figure 6, XDH). These data suggest that these putative xylitol dehydrogenases may actually be hampering the ability of this strain (and possibly all non-xylose utilizing S. cerevisiae strains) to utilize xylose, and thus are consistent with our newly identified Xdh1 protein being responsible for the presumptive xylitol dehydrogenase step of the canonical xylose utilization pathway.
Finally, we introduced an xks1Δ deletion into the XDH1 background, which encodes the putative xylulokinase, which is responsible for the phosphorylation of the fermentable metabolite xylulose to xylulose-5-phosphate [60], [61]. Deletion of XKS1 completely removed the ability of this strain to utilize xylose (Figure 6, XK), suggesting that the canonical pathway in this strain is responsible for metabolizing xylose and that XKS1 encodes the sole xylulokinase necessary for the xylose utilization phenotype we observe.
Transcriptional Profiling during Xylose Utilization
In addition to understanding how the endogenous xylose pathway genes contribute to the xylose phenotype, we sought to characterize how the presence or absence of xylose in the growth medium affected the S. cerevisiae transcriptional program over time, within the genomic context of presence or absence of the XDH1 gene. To do so, we measured mRNA levels in three pairs of sister spores from a Simi White strain that was backcrossed twice to S288c. Each pair of spores was from an independent tetrad, and contained one XDH1-containing spore (“positive”, GSY2465, 2466, 2469) and one spore that does not contain the XDH1 gene (“negative”, GSY2464, 2467, 2470). We pre-grew each of the six spores in YPD and used these cultures to inoculate minimal medium with or without 2% xylose as the sole carbon source (where the absence of xylose is the “no carbon” condition). Samples were taken from these cultures beginning immediately after inoculation (t = 0) and continuing every 8 hours for 72 hours. We then assayed relative RNA abundance versus a pooled reference, containing equimolar amounts of each sample, using Agilent yeast catalog arrays. The gene expression measurements (Log2(sample/reference)) were averaged among the three positive spores and the three negative spores at each time point.
To determine if the endogenous xylose pathway responds to the presence of xylose in the xylose-positive strain, we qualitatively compared the expression levels of all the putative xylose-pathway genes that are present in the S. cerevisiae S288c genome (Figure 7). In positive spores the putative xylose reductase genes are up-regulated compared to the reference only in the presence of xylose, while in the negative spores the xylose reductase genes are repressed under all conditions; the only exception is YDL124W, which appears to be up-regulated vs. the reference in all spore types and all growth conditions. The pattern of expression for the putative XDH XYL2 is similar to that of the xylose reductase genes; it is highly expressed across the time course in the positive strain in the presence of xylose, but is repressed over the time course in the positive strain in the no carbon medium and in both the xylose and no carbon media in the negative strain.
Fig. 7. Endogenous xylose pathway gene expression. 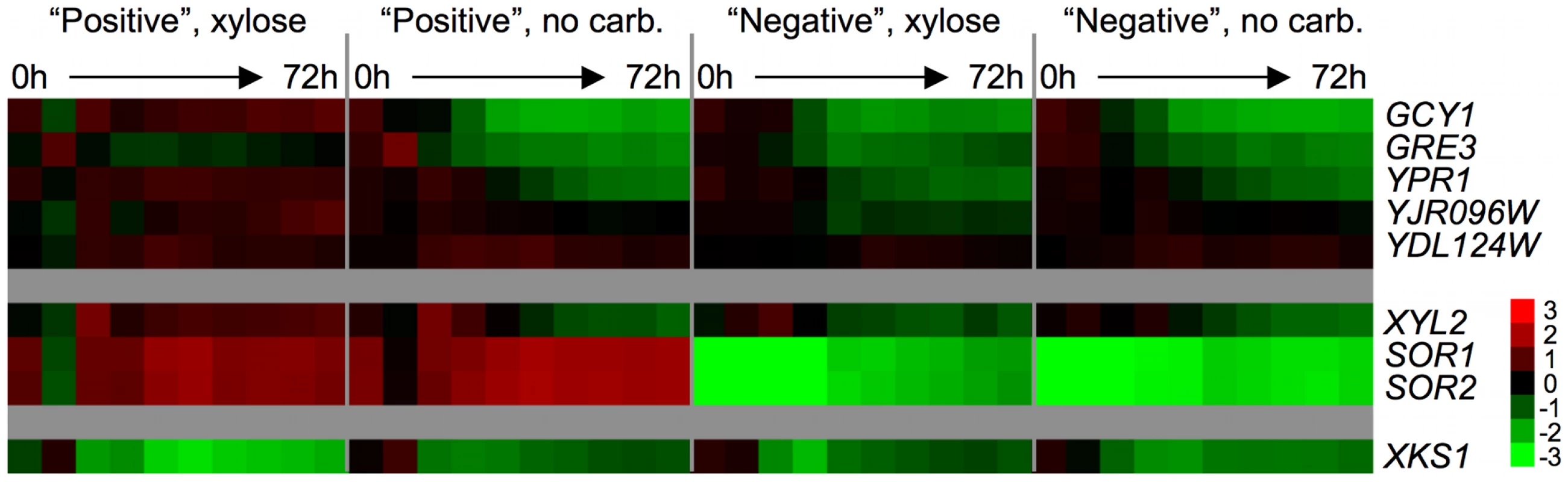
Relative mRNA abundance (compared to a pooled reference of all samples) for putative xylose pathway genes. Values are average Log2(sample/reference) ratios among 3 biological replicates for each time point. Time 0 is immediately following inoculation from a saturated YPD culture into “xylose” (2% xylose in minimal media) or “no carb.” (no carbon source in minimal media). Time points were taken every 8 hours for 72 hours. Interestingly, the sorbitol dehydrogenases SOR1 and SOR2, suggested to have the biochemical ability to oxidize xylitol, are highly expressed compared to the reference in the positive strain both in the presence and absence of xylose, and are strongly repressed vs. the pooled reference in the negative strains in both conditions across the time course. Because there is only one nucleotide difference between the coding sequences of SOR1 and SOR2, the probes on the array for these genes are only different by 1 base out of 60 and thus there is likely to be cross-hybridization of the mRNA's from the two SOR genes. It is also possible that there is hybridization of XDH1 mRNA to these probes, as there are only a few differences between XDH1 and the SOR1/2 probes on the microarray (6 for SOR1 and 7 for SOR2). Although we cannot determine which of the mRNA's (SOR1, SOR2 or XDH1) are hybridizing to the probes, it is nevertheless obvious that there is a distinct difference between the positive and negative spores in the expression levels of at least one of these putative dehydrogenase genes. No striking difference in the expression level of the xylulokinase, XKS1, was observed between any conditions or between any spores. The lack of change in the expression of XKS1 is somewhat unsurprising, as it has been previously reported that low levels of XKS1 are sufficient to allow xylose metabolism, while over-expression can enhance xylose fermentation in an engineered strain [29], [62]. Taken together, these data strongly suggest that the presence of XDH1 in the positive spores permits continued expression of some members of the endogenous xylose pathway when grown in xylose.
To further understand the transcriptome-wide response of these strains, we identified genes that changed significantly across the time course, compared these genes with other microarray datasets to identify any clear physiological responses, and looked for categories of functional enrichment within groups of up or down-regulated genes. Using Significance Analysis of Microarrays [SAM] [63] with a false discovery rate of 1%, we identified a list of 1266 genes whose expression levels were significantly changed over time. Specifically, we carried out a SAM analysis using the two-class (paired timecourse) option to identify genes whose expression changed over time within the positive spores, comparing the xylose to the no carbon condition. Next, we identified genes whose expression changed over time when comparing the positive to the negative spores in the presence of xylose, again using SAM with a two-class (paired timecourse) option. From the union of these two gene lists, we removed genes whose expression levels changed significantly over time within the negative strain, comparing the xylose to the no carbon condition (another two-class, paired timecourse analysis). Using this strategy, we generated an inclusive list of genes whose expression values change over time due to differences between the positive and negative strain, or due to differences between the presence and absence of xylose specifically in the positive strain. To identify the physiological responses that are associated with these gene expression differences, we retrieved data for these 1266 genes using HIDRA [64] from three other yeast microarray experiments [65]–[67] and organized the genes by K-means clustering with K = 10 [68] (Figure 8, Datasets S1, S2). For consistency with the other datasets, each of the four time-course experiments performed in this work were zero-transformed. To the right of the experiments from this paper are, respectively, a measure of how each gene's expression level correlates with increased growth rate [65], a gene expression time course over the diauxic shift [66], gene expression across a set of carbon sources (ethanol, sucrose, fructose, glucose, galactose, and raffinose) [67], and a series of time courses in various conditions including starvation, steady state growth, and other stresses [67]. We observed 5 groups (labeled on the right of the heat map) that appear to be strongly driven by similarity of the positive strain in 2% xylose to either growth rate or a stress response. For example, the genes in groups 1 and 4 (Figure 8) are more highly expressed over the time course in the positive strain in xylose when compared to the positive strain in no carbon source or the negative strain in either condition, and these genes also show a positive correlation with growth rate. As expected, when GO::TermFinder [69] is used on these groups to look for functional enrichment of biological processes, we observed processes known to be up-regulated in conjunction with a higher growth rate. Specifically, group 1 was significantly enriched for vesicle-mediated transport (GO:0016192, p = 2.11e-8) and cellular localization (GO:0051641, p = 3.13e-8) among others (Dataset S3) and group 4 is enriched for translation (GO:0006412, p = 2.26e-41) and ribosome biogenesis (GO:0042254, p = 5.87e-23) along with related processes (Dataset S4). Group 5 shows the same pattern, but largely with the opposite response, meaning that these are genes whose expression is negatively correlated with growth, and we observed that they are expressed at a lower relative level in the positive strain in xylose when compared to the no carbon condition or the negative strain in either condition; but we observed no functional enrichment in this group. Interestingly, within group 5 there is a small group of genes (labeled ‡) whose expression is induced over time relative to the reference in the positive strain in xylose, and repressed over time in the other conditions. This group includes SNO4, THI4, and HSP32, which are genes all at least putatively involved in thiamin biosynthesis. Thiamin biosynthesis is known to be important for sugar metabolism, and is a pathway in which higher expression of certain components has likely been selected for in a variety of industrial yeasts [7]. There is also a small group of genes within group 1 (labeled †) that behaves differently than the rest of the group, as it is strongly repressed relative to the reference in the positive strain in xylose. Within this group of seven genes, four of them could be involved in intracellular redox balancing as they all use NADP(H) as a cofactor (TRR1, OYE2, GDH1, ADH6). In general, these three groups suggest that XDH1 in the positive strain permits a “growth-like” transcriptional response in the presence of xylose, whereas in the absence of xylose or the absence of XDH1 the strains are exhibiting an expression pattern consistent with lack of growth and starvation (e.g. groups 4 and 5). We also observed two other groups that did not fit this pattern, but instead the positive strain in xylose exhibited a response more akin to various stresses. For example, in group 2 we observed lower relative expression in the positive strain in xylose compared to the other three conditions despite the fact that these genes are all strongly correlated with growth rate, and included functional enrichment for RNA metabolism (GO:0016070, p = 1.56e-6) and ribosome biogenesis (GO:0042254, p = 1.33e-5) (Dataset S5). Instead, they appear to be more similar to the expression patterns in strains experiencing nitrogen depletion, stationary phase, diamide, DTT, or hydrogen peroxide treatment, and 37°C heat shock. We observed a similar response in group 3, in which the expression level is opposite what we might expect if growth rate was the main cause of the expression differences but similar if the strains were exhibiting an environmental stress response. Interestingly, this group was enriched for pentose metabolic process (GO:0019321, p = 5.7e-3) and response to oxidative stress (GO:0006979, p = 7.88e-3) (Dataset S6). These data suggest that despite the fact that this set of genes is normally repressed in response to a higher growth rate, some of these genes may be responding to the presence of xylose.
Fig. 8. Gene expression timecourse. 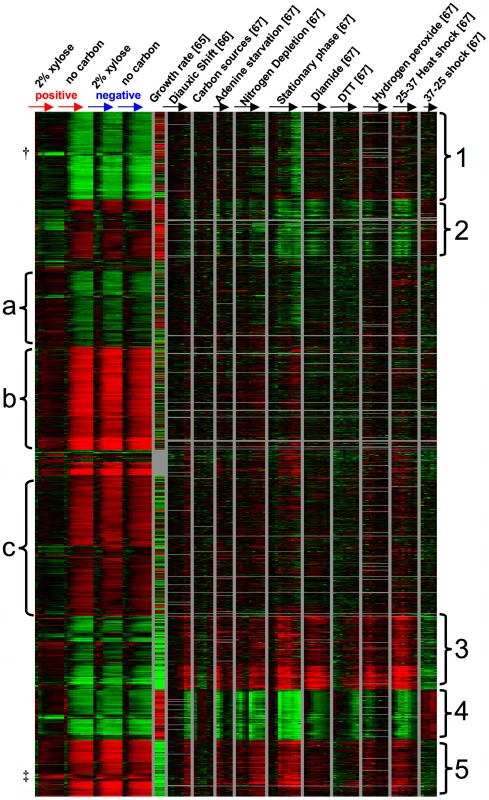
K-means (K = 10) clustering of gene expression values from this work and three other data sets. The 1,266 genes all changed significantly in this study in at least one two-class (paired timecourse) SAM analysis (see Results). Values from this study are time zero-transformed relative mRNA abundance (compared to a pooled reference of all samples from this work) and are averaged among 3 biological replicates at each time point. From left to right: xylose positive strain (2% xylose), xylose positive strain (no carbon source), xylose negative strain (2% xylose), xylose negative strain (no carbon source). “Growth rate” data were calculated by [65] and show the strength and direction of the transcriptional response of a given gene to a higher growth rate. “Diauxic shift” are zero-transformed data from [66], and all other data are from [67]; again all time-courses are zero-transformed. (†,‡ indicate small subgroups discussed in Results.) There were also three groups of genes that did not have an obvious visual relationship with either growth rate or stress response. Group (a) appears to be more highly expressed in the positive strain in xylose compared to no carbon or the negative strain in either condition. While this group contains no functional enrichment using GO::TermFinder, it does contain a number of genes related to carbon metabolism, including PFK1, PFK2, PGI1, GCR1, and GND1. The final two groups (b and c) both appear to be expressed at a lower level in the positive strain in xylose compared to the other three conditions. Both groups have functional enrichment for various processes related to transcription and its regulation (Datasets S7, S8). In general genes in these three groups (a–c) show larger magnitude expression changes (induction or repression relative to the reference) in the non-growth conditions than in the positive strain in the presence of xylose. These clusters could support the conclusion that in the absence of xylose or the absence of XDH1, strains are exhibiting a response (perhaps starvation) that is simply not induced in the presence of xylose in the positive strain. In summary, these microarray data suggest that the positive strain in the presence of xylose is capable of “growth” when compared to the negative strain or lack of xylose, but it is still exhibiting a less pronounced stress-like response. These data are not inconsistent with the positive strain recognizing and using xylose as a carbon source.
Discussion
In this work we have shown that naturally occurring strains of Saccharomyces cerevisiae are capable of utilizing xylose without engineering or directed evolution, and have determined the genetic basis for this phenotype. While it has been known for many years that the xylose isomer xylulose is fermentable by S. cerevisiae, it has generally been thought that this species is incapable of metabolizing xylose. However, recent work has shown natural genetic variation for xylose utilization does exist, and that natural selection and breeding can improve xylose utilization in natural strains of S. cerevisiae [9]. By screening through many industrial and clinical isolates, we discovered variation within this species that permits utilization of this sugar, fermentation of which is an important prerequisite for the efficient generation of ethanol from lignocellulosic biomass sources. We have also shown that this ability to utilize xylose by Saccharomyces is conferred by the presence of a single gene, a novel putative xylitol dehydrogenase that we have named XDH1. This gene is both necessary and sufficient to permit xylose utilization in the normally non-xylose-utilizing S288c laboratory strain, and is absent from the reference genome sequence of S288c.
We also characterized the transcriptional response of one of our xylose-utilizing strains of S. cerevisiae to xylose in the presence and absence of XDH1. While these data do not allow us to draw conclusions as to whether or not this gene permits actual fermentation (rather than simply utilization) of xylose, we can make a number of observations. First, it is clear that the endogenous xylose pathway is capable of responding at the transcriptional level to the presence of xylose when this novel XDH is present. Secondly, we can infer that this sugar and its downstream metabolites are likely being funneled into central carbon metabolism via the pentose phosphate pathway as is consistent with what has previously been observed. This suggests that industrial or laboratory strains of S. cerevisiae may be more poised to ferment this pentose than previously thought, implying that we can better harness the standing genetic potential that already exists in nature and use it in combination with directed evolution and metabolic engineering to make an industrially applicable xylose fermentation strain.
The idea that Saccharomyces might be more “ready” to ferment xylose than previously thought is further supported by our genetic dissection of the xylose metabolic pathway endogenous to S. cerevisiae. We corroborated previous data that shows the xylulokinase encoded by XKS1 is functional and supports metabolism of xylose. We also demonstrated that GRE3 and YPR1, encoding two aldo-keto reductases, are each sufficient to allow xylose utilization in our strain background. The observation that a novel xylitol dehydrogenase is responsible for the xylose utilization phenotype, and the observation that the genes in the reference strain encoding enzymes putatively thought to oxidize xylitol (SOR1, SOR2, XYL2) are in fact detrimental to the phenotype, further support that the idea of a redox imbalance in S. cerevisiae favoring xylitol production over further metabolism is true [70], [71]. Finally, our results also suggest that some property of the XDH1 is able to reduce the cofactor imbalance and may be capable of pushing xylitol through the xylose metabolic pathway.
We also discovered Saccharomyces sensu stricto interspecific hybrids in our screen that appear to robustly utilize xylose by a mechanism independent of XDH1. Some of these strains are even more effective at utilizing xylose than the S. cerevisiae wine strains we have characterized here, and we are currently attempting to identify the locus (or loci) responsible for these other xylose phenotypes. Based upon the results in Table 2 that show S. bayanus xylose-positive strains that do not possess XDH1, it is likely that there is at least one other trait that is as yet unidentified. There may also be additional components of the xylose utilization pathway for which hypomorphic alleles exist in natural strains, as XDH1 is present in xylose-negative segregants of some xylose-positive strains we identified. We also suggest that the only other previously described [9], [72] xylose phenotype native to S. cerevisiae is likely to be XDH1-dependent, given that wine strains were included in the initial breeding. Because we and others have assayed strains that only contain a small sample of the variation that likely exists in the Saccharomyces gene pool, it is likely that there is additional variation present in nature that may be able to contribute to a xylose-positive phenotype.
Finally, we have developed a novel application of high-throughput sequencing for quickly mapping an unknown trait by BSA. Because we were able to identify a clear segregation pattern for our phenotype of interest, in this case a single locus, we were able to easily pool segregants and use sequencing to narrow down the genomic location using the high frequency of polymorphisms that segregated with our locus. Applying sequencing technology in addition to tiling arrays was critical as our phenotype resided in a region of the genome that is not present in the reference genome. Given that the number of genes responsible is small, we suggest that this application of high-throughput sequencing could be used broadly for associating other unknown genotypes to well-characterized phenotypes. It will be particularly applicable to other species that have small genomes and for which the genome sequence or tiling arrays are not readily available, or for such species that may contain variation not captured in their respective reference genomes.
While effective conversion of xylose to ethanol in an industrial setting by Saccharomyces yeasts has not yet reached its full potential, much progress has been made recently. We suggest that uncovering and studying the genes responsible for xylose utilization in wild strains of Saccharomyces may contribute directly to further improvements in lignocellulosic biomass fermentation. Additionally, the functions of these genes might continue to shed light on problematic areas in the metabolism of xylose, helping to inform directed evolution and metabolic engineering approaches.
Materials and Methods
Strains
Strains used in this study are shown in Table S1 and Table S6. In order to cross diploid HO/HO wine strains to a haploid S288c strain, wine strains were transformed with either pGS35 (CEN/ARS, KanMX) or pGS36 (CEN/ARS, Hph) and the resulting transformants carrying the plasmid were sporulated (Hph is the gene that permits hygromycin B resistance). Spores were mixed with a haploid ho S288c strain carrying either pGS35 if the wine strain carried pGS36 or vice versa, and plated onto YPD plates supplemented with G418 (200µg/mL) and hygromycin B (150µg/mL).
Media, Growth Conditions, and Growth Quantification
To screen for xylose utilization, single colonies were pre-grown to saturation at 25°C in YP with 2% glucose and then diluted 1∶50 into YP with 2% xylose (Sigma) or no carbon source. 100µL cultures were grown for 5 days at 25°C in a sealed 96-well plate and absorbance was read at 595nm every 15 minutes in a TECAN Genios plate reader with orbital shaking. Xylose positives were identified by visual inspection of increasing OD in xylose compared to no carbon source, and were confirmed by retesting in both YP and Minimal [73] media.
Because growth on xylose is not exponential, we did not calculate a doubling time. Instead, to quantify xylose utilization we calculated a slope (change in OD over time) across the linear range of OD increase, from 20 to 80 hours in a typical TECAN growth experiment following the initial trehalose growth. Growth curves were done in at least triplicate (see Table S6 for all deletion strains), and a t-test was used to determine significant differences in rate of OD increase between deletion strains and “wild type” xylose positives.
Gene Expression Arrays
To analyze gene expression, cultures were pre-grown to saturation in YP with 2% glucose and diluted 1∶50 into a 1.1L culture of minimal medium [73] with 2% xylose or no carbon source. 100mL samples were collected starting immediately after inoculation (t = 0) and at subsequent 8 hour intervals for 72 hours by filtering with 0.45µm analytical test filter funnels (Nalgene) and were snap frozen in liquid nitrogen. RNA was extracted using a modified version of the hot phenol protocol, as described [74], [75]. A pooled reference sample was created by combining 350ng of each of the 120 RNA samples (10 time points for 6 strains in 2 conditions). 325ng of each total RNA sample and reference were labeled with Cy dyes (Amersham) using the Agilent Low RNA Input Linear Amplification Kit, and hybridized to Agilent Yeast Gene Expression Arrays (v2, 8x15K) for 17 hours at 65°C at 10rpm in a hybridization oven (Shel Lab). Arrays were scanned at 5µm resolution on an Agilent Scanner, and Agilent Feature Extraction v9.5.3.1 was used for extraction of data from the scanned images, and data normalization and calculation of log2 ratios. Gene expression data have been deposited in the GEO database with accession number GSE19121.
Bulk Segregant Analysis
Xylose-positive segregants of Simi White (Lallemand), Lalvin AC, and SIHA Activ-Hefe 4 were crossed once to S288c (GSY147), and the resulting diploids were then sporulated. F2 segregants were scored for xylose utilization in the TECAN plate reader as described above. 1.5mL of overnight YPD culture of each segregant grown was spun down, resuspended, and frozen in 300µL of sorbitol solution (0.9M sorbitol, 0.1M Tris pH 8, 0.1M EDTA). Samples were pooled by phenotype at this stage and genomic DNA was extracted as described [76]. The pools contained 39 positives and 39 negatives for Simi White, 19 positives and 16 negatives for Lalvin AC, and 16 positives and 16 negatives for SIHA.
Genomic DNA was labeled as described [77], and microarray-assisted BSA was done using Affymetrix GeneChip S. cerevisiae Tiling 1.0R Array basically as described [52], [78]. Briefly, a ratio of the log2 intensities for the perfect match probes was plotted across every chromosome for each nucleotide. The plots for each chromosome were scanned visually for local peaks in intensity. Tiling array data have been deposited in the GEO database with accession number GSE19121.
The same pools of genomic DNA were used for BSA by sequencing. 5µg of genomic DNA were prepared for sequencing using the Illumina Genomic DNA Sample Kit. Flow cells were prepared using the Illumina Standard Cluster Generation Kit v2, and samples were sequenced on the Illumina Genome Analyzer II. GAII data were analyzed with the Illumina 1.3.2 pipeline, and reads (with qualities) were aligned to the S288c genome with MAQ v0.7.1 [56] using default parameters. Reads from the positive pools that did not align to the reference genome were combined, and reads that contained any uncalled bases (“N”> = 1) were removed from further analysis. De novo assembly was performed on this filtered set of un-mapped reads using Velvet v0.7.55 [57] with default parameters and hash length = 13. All raw high throughput sequence data have been deposited in the SRA database with accession number SRP001391.
Cloning
The novel XDH was cloned into the NotI site of pRS316 [59] from GSY2469 (Simi White derivative) and GSY1362 (Lalvin AC derivative) by PCR using primers that contained NotI restriction sites. Primers are listed in Table S5. FY2 (S288c) was then transformed with the resulting plasmids (pGS104 and pGS105) via a slightly modified lithium acetate method [79]. Plasmids are listed in Table S6. Growth was assayed as described above in the TECAN plate reader.
Plasmid loss experiments were done as follows. The original transformant that was used to generate a TECAN growth curve was also streaked for single colonies on a YPD plate. These were grown and replica plated onto YPD and SC-URA plates, and colonies were picked from the YPD plate that either retained the plasmid (grew on the SC-URA replica plate) or lost the plasmid during mitosis (did not grow on the SC-URA replica plate) and were tested again in the TECAN.
Deletion Construction
Homologous recombination was used to create a disruption of the novel XDH. Primers are listed in Table S5. Briefly, KanMX6 was amplified from pFA6-KanMX6 [80], and approximately 400 bases up and downstream of the XDH homolog were amplified separately using primers that overlapped with the 5′ and 3′ primers used to amplify KanMX6. The three fragments were joined using Phusion DNA polymerase (Finnzymes) and the resulting deletion cassette was integrated into GSY2469 and GSY2468 (Simi White derivatives) by lithium acetate transformation. Correct integration of the deletion was confirmed by PCR and by showing opposing segregation of G418 resistance and the xylose trait in a cross to another xylose-positive haploid derivative.
To genetically dissect the endogenous xylose pathway, deletions of the xylose pathway genes were crossed into a haploid Simi White derivative that was previously backcrossed twice to S288c (GSY2469). Diploid strains heterozygous for deletions of GCY1, GRE3, YPR1, YJR096W, XYL2, and XKS1 were purchased from Invitrogen. Deletions of SOR1 and SOR2 were not available from the deletion collection as they are in large genomic regions of essentially 100% identity. Deletions were constructed as described [81], except with approximately 80 bases of homology to the regions immediately up and downstream of the SOR1/2 open reading frames rather than 40. Transformants were crossed to pgu1Δ and lrg1Δ to differentiate between sor1Δ and sor2Δ. Segregation of deletions was tracked by colony PCR when creating strains with more than two deletions, as the deletions are all marked with G418R. Primers are listed in Table S5, and strains are listed in Table S6.
Supporting Information
Zdroje
1. SomervilleC
2007 Biofuels. Curr Biol 17 115 119
2. BassoLC
de AmorimHV
de OliveiraAJ
LopesML
2008 Yeast selection for fuel ethanol production in Brazil. FEMS Yeast Res 8 1155 1163
3. MatsushikaA
InoueH
KodakiT
SawayamaS
2009 Ethanol production from xylose in engineered Saccharomyces cerevisiae strains: current state and perspectives. Appl Microbiol Biotechnol 84 37 53
4. Hahn-HägerdalB
GalbeM
Gorwa-GrauslundMF
LidenG
ZacchiG
2006 Bio-ethanol–the fuel of tomorrow from the residues of today. Trends Biotechnol 24 549 556
5. FarrellAE
PlevinRJ
TurnerBT
JonesAD
O'HareM
2006 Ethanol can contribute to energy and environmental goals. Science 311 506 508
6. ArguesoJL
CarazzolleMF
MieczkowskiPA
DuarteFM
NettoOV
2009 Genome structure of a Saccharomyces cerevisiae strain widely used in bioethanol production. Genome Res 19 2258 2270
7. StambukB
DunnB
Alves-JrS
DuvalE
SherlockG
2009 Industrial Fuel Ethanol Yeasts Contain Adaptive Copy Number Changes in Genes Involved in Vitamin B1 and B6 Biosynthesis. Genome Res 19 2271 2278
8. SahaBC
2003 Hemicellulose bioconversion. J Ind Microbiol Biotechnol 30 279 291
9. AttfieldPV
BellPJL
2006 Use of population genetics to derive nonrecombinant Saccharomyces cerevisiae strains that grow using xylose as a sole carbon source. FEMS Yeast Res 6 862 868
10. ChiangLC
GongCS
ChenLF
TsaoGT
1981 d-Xylulose Fermentation to Ethanol by Saccharomyces cerevisiae. Appl Environ Microbiol 42 284 289
11. WangPY
ShopsisC
SchneiderH
1980 Fermentation of a pentose by yeasts. Biochemical and Biophysical Research Communications 94 248 254
12. GongCS
ClaypoolTA
McCrackenLD
MaunCM
UengPP
1983 Conversion of pentoses by yeasts. Biotechnol Bioeng 25 85 102
13. ChangQ
GriestT
HarterT
PetrashJ
2007 Functional studies of aldo-keto reductases in Saccharomyces cerevisiae. BBA-Molecular Cell Research 1773 321 329
14. TraffKL
JonssonLJ
Hahn-HägerdalB
2002 Putative xylose and arabinose reductases in Saccharomyces cerevisiae. Yeast 19 1233 1241
15. ToivariMH
SalusjarviL
RuohonenL
PenttilaM
2004 Endogenous xylose pathway in Saccharomyces cerevisiae. Appl Environ Microbiol 70 3681 3686
16. JeffriesTW
2006 Engineering yeasts for xylose metabolism. Curr Opin Biotechnol 17 320 326
17. KötterP
CiriacyM
1993 Xylose fermentation by Saccharomyces cerevisiae. Appl Microbiol Biotechnol 38 776 783
18. KötterP
AmoreR
HollenbergCP
CiriacyM
1990 Isolation and characterization of the Pichia stipitis xylitol dehydrogenase gene, XYL2, and construction of a xylose-utilizing Saccharomyces cerevisiae transformant. Curr Genet 18 493 500
19. HallbornJ
WalfridssonM
AiraksinenU
OjamoH
Hahn-HägerdalB
1991 Xylitol production by recombinant Saccharomyces cerevisiae. Nat Biotechnol 9 1090 1095
20. TantirungkijM
NakashimaN
SekiT
YoshidaT
1993 Construction of xylose-assimilating Saccharomyces cerevisiae. Journal of Fermentation and Bioengineering 75 83 88
21. HoN
ChenZ
BrainardA
1998 Genetically Engineered Saccharomyces Yeast Capable of Effective Cofermentation of Glucose and Xylose. Appl Environ Microbiol 64 1852 1859
22. JinYS
JonesS
ShiNQ
JeffriesTW
2002 Molecular cloning of XYL3 (D-xylulokinase) from Pichia stipitis and characterization of its physiological function. Appl Environ Microbiol 68 1232 1239
23. WalfridssonM
BaoX
AnderlundM
LiliusG
BulowL
1996 Ethanolic fermentation of xylose with Saccharomyces cerevisiae harboring the Thermus thermophilus xylA gene, which expresses an active xylose (glucose) isomerase. Appl Environ Microbiol 62 4648 4651
24. AmoreR
WilhelmM
HollenbergC
1989 The fermentation of xylose – an analysis of the expression of Bacillus and Actinoplanes xylose isomerase genes in yeast. Appl Microbiol Biotechnol 30 351 357
25. KuyperM
HarhangiH
StaveA
WinklerA
JettenM
2003 High-level functional expression of a fungal xylose isomerase: the key to efficient ethanolic fermentation of xylose by Saccharomyces cerevisiae? FEMS Yeast Research 4 69 78
26. KuyperM
WinklerA
DijkenJ
PronkJ
2004 Minimal metabolic engineering of Saccharomyces cerevisiae for efficient anaerobic xylose fermentation: a proof of principle. FEMS Yeast Research 4 655 664
27. KuyperM
HartogMMP
ToirkensMJ
AlmeringMJH
WinklerAA
2005 Metabolic engineering of a xylose-isomerase-expressing Saccharomyces cerevisiae strain for rapid anaerobic xylose fermentation. FEMS Yeast Res 5 399 409
28. MadhavanA
TamalampudiS
UshidaK
KanaiD
KatahiraS
2009 Xylose isomerase from polycentric fungus Orpinomyces: gene sequencing, cloning, and expression in Saccharomyces cerevisiae for bioconversion of xylose to ethanol. Appl Microbiol Biotechnol 82 1067 1078
29. JohanssonB
ChristenssonC
HobleyT
Hahn-HägerdalB
2001 Xylulokinase overexpression in two strains of Saccharomyces cerevisiae also expressing xylose reductase and xylitol dehydrogenase and its effect on fermentation of xylose and lignocellulosic hydrolysate. Appl Environ Microbiol 67 4249 4255
30. ToivariMH
AristidouA
RuohonenL
PenttilaM
2001 Conversion of xylose to ethanol by recombinant Saccharomyces cerevisiae: importance of xylulokinase (XKS1) and oxygen availability. Metab Eng 3 236 249
31. JinYS
JeffriesTW
2003 Changing flux of xylose metabolites by altering expression of xylose reductase and xylitol dehydrogenase in recombinant Saccharomyces cerevisiae. Appl Biochem Biotechnol 106 277 286
32. Träff-BjerreKL
JeppssonM
Hahn-HägerdalB
Gorwa-GrauslundM-F
2004 Endogenous NADPH-dependent aldose reductase activity influences product formation during xylose consumption in recombinant Saccharomyces cerevisiae. Yeast 21 141 150
33. JeppssonM
TraffK
JohanssonBr
Hahn-HägerdalB
Gorwa-GrauslundM
2003 Efect of enhanced xylose reductase activity on xylose consumption and product distribution in xylose-fermenting recombinant Saccharomyces cerevisiae. FEMS Yeast Res 3 167 175
34. WalfridssonM
HallbornJ
PenttiläM
KeränenS
Hahn-HägerdalB
1995 Xylose-metabolizing Saccharomyces cerevisiae strains overexpressing the TKL1 and TAL1 genes encoding the pentose phosphate pathway enzymes transketolase and transaldolase. Appl Environ Microbiol 61 4184 4190
35. JeppssonM
JohanssonB
Hahn-HägerdalB
Gorwa-GrauslundMF
2002 Reduced Oxidative Pentose Phosphate Pathway Flux in Recombinant Xylose-Utilizing Saccharomyces cerevisiae Strains Improves the Ethanol Yield from Xylose. Appl Environ Microbiol 68 1604 1609
36. TraffKL
Otero CorderoRR
van ZylWH
Hahn-HägerdalB
2001 Deletion of the GRE3 aldose reductase gene and its influence on xylose metabolism in recombinant strains of Saccharomyces cerevisiae expressing the xylA and XKS1 genes. Appl Environ Microbiol 67 5668 5674
37. WalfridssonM
AnderlundM
BaoX
Hahn-HägerdalB
1997 Expression of different levels of enzymes from the Pichia stipitis XYL1 and XYL2 genes in Saccharomyces cerevisiae and its effects on product formation during xylose utilisation. Appl Microbiol Biotechnol 48 218 224
38. VerhoR
LondesboroughJ
PenttiläM
RichardP
2003 Engineering redox cofactor regeneration for improved pentose fermentation in Saccharomyces cerevisiae. Appl Environ Microbiol 69 5892 5897
39. PetschacherB
NidetzkyB
2008 Altering the coenzyme preference of xylose reductase to favor utilization of NADH enhances ethanol yield from xylose in a metabolically engineered strain of Saccharomyces cerevisiae. Microb Cell Fact 7 9
40. Van VleetJH
JeffriesTW
OlssonL
2008 Deleting the para-nitrophenyl phosphatase (pNPPase), PHO13, in recombinant Saccharomyces cerevisiae improves growth and ethanol production on D-xylose. Metabolic Engineering 10 360 369
41. BengtssonO
Hahn-HägerdalB
Gorwa-GrauslundMF
2009 Xylose reductase from Pichia stipitis with altered coenzyme preference improves ethanolic xylose fermentation by recombinant Saccharomyces cerevisiae. Biotechnol Biofuels 2 9
42. SondereggerM
SauerU
2003 Evolutionary engineering of Saccharomyces cerevisiae for anaerobic growth on xylose. Appl Environ Microbiol 69 1990 1998
43. PitkänenJ-P
RintalaE
AristidouA
RuohonenL
PenttiläM
2005 Xylose chemostat isolates of Saccharomyces cerevisiae show altered metabolite and enzyme levels compared with xylose, glucose, and ethanol metabolism of the original strain. Appl Microbiol Biotechnol 67 827 837
44. WahlbomCF
van ZylWH
JönssonLJ
Hahn-HägerdalB
OteroRRC
2003 Generation of the improved recombinant xylose-utilizing Saccharomyces cerevisiae TMB 3400 by random mutagenesis and physiological comparison with Pichia stipitis CBS 6054. FEMS Yeast Res 3 319 326
45. NiH
LaplazaJM
JeffriesTW
2007 Transposon mutagenesis to improve the growth of recombinant Saccharomyces cerevisiae on D-xylose. Appl Environ Microbiol 73 2061 2066
46. CarretoL
EirizMF
GomesAC
PereiraPM
SchullerD
2008 Comparative genomics of wild type yeast strains unveils important genome diversity. BMC Genomics 9 524
47. DunnB
LevineRP
SherlockG
2005 Microarray karyotyping of commercial wine yeast strains reveals shared, as well as unique, genomic signatures. BMC Genomics 6 53
48. KvitekDJ
WillJL
GaschAP
2008 Variations in stress sensitivity and genomic expression in diverse S. cerevisiae isolates. PLoS Genet 4 e1000223 doi:10.1371/journal.pgen.1000223
49. FayJC
BenavidesJA
2005 Evidence for domesticated and wild populations of Saccharomyces cerevisiae. PLoS Genet 1 e5 doi:10.1371/journal.pgen.0010005
50. LitiG
CarterDM
MosesAM
WarringerJ
PartsL
2009 Population genomics of domestic and wild yeasts. Nature 458 337 341
51. SchachererJ
ShapiroJA
RuderferDM
KruglyakL
2009 Comprehensive polymorphism survey elucidates population structure of Saccharomyces cerevisiae. Nature 458 342 345
52. BrauerMJ
ChristiansonCM
PaiDA
DunhamMJ
2006 Mapping novel traits by array-assisted bulk segregant analysis in Saccharomyces cerevisiae. Genetics 173 1813 1816
53. QuarrieS
Lazic-JancicV
KovacevicD
SteedA
PekicS
1999 Bulk segregant analysis with molecular markers and its use for improving drought resistance in maize. Journal of Experimental Botany 50 1299 1306
54. BornemanAR
ForganAH
PretoriusIS
ChambersPJ
2008 Comparative genome analysis of a Saccharomyces cerevisiae wine strain. FEMS Yeast Res 8 1185 1195
55. LouisEJ
1995 The chromosome ends of Saccharomyces cerevisiae. Yeast 11 1553 1573
56. LiH
RuanJ
DurbinR
2008 Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res 18 1851 1858
57. ZerbinoDR
BirneyE
2008 Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18 821 829
58. NovoM
BigeyF
BeyneE
GaleoteV
GavoryF
2009 Eukaryote-to-eukaryote gene transfer events revealed by the genome sequence of the wine yeast Saccharomyces cerevisiae EC1118. Proc Natl Acad Sci U S A 106 16333 16338
59. SikorskiR
HieterP
1989 A System of Shuttle Vectors and Yeast Host Strains Designed for Efficient Manipulation of DNA in Saccharomyces cerevisiae. Genetics 122 19 27
60. Rodriguez-PeñaJ
CidV
ArroyoJ
1998 The YGR194c (XKS1) gene encodes the xylulokinase from the budding yeast Saccharomyces cerevisiae. FEMS Microbiology Letters 162 155 160
61. HoN
ChangS
1989 Cloning of yeast xylulokinase gene by complementation of E. coli and yeast mutations. Enzyme Microb Technol 11 417 421
62. JinYS
NiH
LaplazaJM
JeffriesTW
2003 Optimal growth and ethanol production from xylose by recombinant Saccharomyces cerevisiae require moderate D-xylulokinase activity. Appl Environ Microbiol 69 495 503
63. TusherVG
TibshiraniR
ChuG
2001 Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A 98 5116 5121
64. HibbsM
WallaceG
DunhamMJ
KaiL
TroyanskayaO
2007 Viewing the Larger Context of Genomic Data through Horizontal Integration. Information Visualization, 2007 IV '07 11th International Conference
65. BrauerMJ
HuttenhowerC
AiroldiEM
RosensteinR
MateseJC
2008 Coordination of growth rate, cell cycle, stress response, and metabolic activity in yeast. Mol Biol Cell 19 352 367
66. DeRisiJL
IyerVR
BrownPO
1997 Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278 680 686
67. GaschAP
SpellmanPT
KaoCM
Carmel-HarelO
EisenMB
2000 Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell 11 4241 4257
68. EverittBS
1974 Cluster analysis London Heinemann Educational [for] the Social Science Research Council
69. BoyleEI
WengS
GollubJ
JinH
BotsteinD
2004 GO::TermFinder–open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics 20 3710 3715
70. BruinenbergP
BotP
DijkenJ
ScheffersW
1983 The role of redox balances in the anaerobic fermentation of xylose by yeasts. Appl Microbiol Biotechnol 18 287 292
71. JeffriesT
1983 Utilization of xylose by bacteria, yeasts, and fungi.
FiechterA
JeffriesT
Pentoses and Lignin Berlin/Heidelberg Springer 1 32
72. BellPJ
HigginsVJ
AttfieldPV
2001 Comparison of fermentative capacities of industrial baking and wild-type yeasts of the species Saccharomyces cerevisiae in different sugar media. Lett Appl Microbiol 32 224 229
73. AdamsJ
HanschePE
1974 Population studies in microorganisms. I. Evolution of diploidy in Saccharomyces cerevisiae. Genetics 76 327 338
74. SchmittME
BrownTA
TrumpowerBL
1990 A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res 18 3091 3092
75. LeeA
HansenKD
BullardJ
DudoitS
SherlockG
2008 Novel low abundance and transient RNAs in yeast revealed by tiling microarrays and ultra high-throughput sequencing are not conserved across closely related yeast species. PLoS Genet 4 e1000299 10.1371/journal.pgen.1000299
76. TrecoDA
1987 Preparation of Yeast DNA.
AusubelF
BrentR
KingstonR
MooreD
SeidmanJ
Curr Protoc Mol Biol New York John Wiley and Sons, Inc 13.11.11 13.11.12
77. KaoKC
SherlockG
2008 Molecular characterization of clonal interference during adaptive evolution in asexual populations of Saccharomyces cerevisiae. Nat Genet 40 1499 1504
78. GreshamD
RuderferDM
PrattSC
SchachererJ
DunhamMJ
2006 Genome-wide detection of polymorphisms at nucleotide resolution with a single DNA microarray. Science 311 1932 1936
79. SchiestlRH
GietzRD
1989 High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet 16 339 346
80. WachA
BrachatA
PohlmannR
PhilippsenP
1994 New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10 1793 1808
81. LongtineMS
McKenzieA3rd
DemariniDJ
ShahNG
WachA
1998 Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14 953 961
Štítky
Genetika Reprodukční medicína
Článek Epistasis of Transcriptomes Reveals Synergism between Transcriptional Activators Hnf1α and Hnf4αČlánek SMA-10/LRIG Is a Conserved Transmembrane Protein that Enhances Bone Morphogenetic Protein SignalingČlánek B1 SOX Coordinate Cell Specification with Patterning and Morphogenesis in the Early Zebrafish EmbryoČlánek Abundant Quantitative Trait Loci Exist for DNA Methylation and Gene Expression in Human Brain
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2010 Číslo 5- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- IVF a rakovina prsu – zvyšují hormony riziko vzniku rakoviny?
-
Všechny články tohoto čísla
- Digital Quantification of Human Eye Color Highlights Genetic Association of Three New Loci
- Common Genetic Variants near the Brittle Cornea Syndrome Locus Influence the Blinding Disease Risk Factor Central Corneal Thickness
- Age- and Temperature-Dependent Somatic Mutation Accumulation in
- All About Mitochondrial Eve: An Interview with Rebecca Cann
- Aging and Chronic Sun Exposure Cause Distinct Epigenetic Changes in Human Skin
- Transposed Genes in Arabidopsis Are Often Associated with Flanking Repeats
- A Survey of Genomic Traces Reveals a Common Sequencing Error, RNA Editing, and DNA Editing
- Gene Transposition Causing Natural Variation for Growth in
- The Polyproline Site in Hinge 2 Influences the Functional Capacity of Truncated Dystrophins
- Epistasis of Transcriptomes Reveals Synergism between Transcriptional Activators Hnf1α and Hnf4α
- Integration of Light Signals by the Retinoblastoma Pathway in the Control of S Phase Entry in the Picophytoplanktonic Cell
- The Proprotein Convertase Encoded by () Is Required in Corpora Cardiaca Endocrine Cells Producing the Glucose Regulatory Hormone AKH
- Sgs1 and Exo1 Redundantly Inhibit Break-Induced Replication and Telomere Addition at Broken Chromosome Ends
- A MATE-Family Efflux Pump Rescues the 8-Oxoguanine-Repair-Deficient Mutator Phenotype and Protects Against HO Killing
- The Relationship among Gene Expression, the Evolution of Gene Dosage, and the Rate of Protein Evolution
- SMA-10/LRIG Is a Conserved Transmembrane Protein that Enhances Bone Morphogenetic Protein Signaling
- The Nuclear Receptor DHR3 Modulates dS6 Kinase–Dependent Growth in
- Involvement of Global Genome Repair, Transcription Coupled Repair, and Chromatin Remodeling in UV DNA Damage Response Changes during Development
- B1 SOX Coordinate Cell Specification with Patterning and Morphogenesis in the Early Zebrafish Embryo
- Linkage and Association Mapping of Flowering Time in Nature
- Bulk Segregant Analysis by High-Throughput Sequencing Reveals a Novel Xylose Utilization Gene from
- Abundant Quantitative Trait Loci Exist for DNA Methylation and Gene Expression in Human Brain
- Ablation of Whirlin Long Isoform Disrupts the USH2 Protein Complex and Causes Vision and Hearing Loss
- Characterization of Oxidative Guanine Damage and Repair in Mammalian Telomeres
- DNA Adenine Methylation Is Required to Replicate Both Chromosomes Once per Cell Cycle
- Genome-Wide Copy Number Variation in Epilepsy: Novel Susceptibility Loci in Idiopathic Generalized and Focal Epilepsies
- FACT Prevents the Accumulation of Free Histones Evicted from Transcribed Chromatin and a Subsequent Cell Cycle Delay in G1
- GC-Biased Evolution Near Human Accelerated Regions
- Liver and Adipose Expression Associated SNPs Are Enriched for Association to Type 2 Diabetes
- Myeloid Cell-Restricted Insulin Receptor Deficiency Protects Against Obesity-Induced Inflammation and Systemic Insulin Resistance
- The Mating Type Locus () and Sexual Reproduction of : Insights into the Evolution of Sex and Sex-Determining Chromosomal Regions in Fungi
- B-Cyclin/CDKs Regulate Mitotic Spindle Assembly by Phosphorylating Kinesins-5 in Budding Yeast
- Post-Replication Repair Suppresses Duplication-Mediated Genome Instability
- Genome Sequence of the Plant Growth Promoting Endophytic Bacterium sp. 638
- Transcription Factors Mat2 and Znf2 Operate Cellular Circuits Orchestrating Opposite- and Same-Sex Mating in
- The Use of Orthologous Sequences to Predict the Impact of Amino Acid Substitutions on Protein Function
- Mutation in Archain 1, a Subunit of COPI Coatomer Complex, Causes Diluted Coat Color and Purkinje Cell Degeneration
- Shelterin-Like Proteins and Yku Inhibit Nucleolytic Processing of Telomeres
- Affecting Function Causes a Dilated Heart in Adult
- Manipulation of Behavioral Decline in with the Rag GTPase
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Common Genetic Variants near the Brittle Cornea Syndrome Locus Influence the Blinding Disease Risk Factor Central Corneal Thickness
- All About Mitochondrial Eve: An Interview with Rebecca Cann
- The Relationship among Gene Expression, the Evolution of Gene Dosage, and the Rate of Protein Evolution
- SMA-10/LRIG Is a Conserved Transmembrane Protein that Enhances Bone Morphogenetic Protein Signaling
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



