-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
FSHD: A Repeat Contraction Disease Finally Ready to Expand (Our Understanding of Its Pathogenesis)
article has not abstract
Published in the journal: . PLoS Genet 6(10): e32767. doi:10.1371/journal.pgen.1001180
Category: Perspective
doi: https://doi.org/10.1371/journal.pgen.1001180Summary
article has not abstract
Facioscapulohumeral muscular dystrophy (FSHD), was one of the first diseases shown to be caused by an unstable repeat in the early 1990s along with spinal and bulbar muscular atrophy (SBMA), myotonic dystrophy (DM1), and fragile X mental retardation (FRAXA), where the latter three are caused by genetically expanding trinucleotide repeats [1]. However, FSHD differs considerably from the trinuclotide repeat diseases, as it is caused by a contraction of a macrosatellite (D4Z4 repeat, 3.3 kb/unit). Moreover, far less is understood about the pathogenic mechanism for FSHD, relative to SBMA, DM1, and FRAXA. This is not due to a shortage of experimental efforts, plausible hypotheses, or collaborative efforts towards understanding FSHD [2], [3]. The elucidation of FSHD is hampered by the size of the unstable repeat, its sequence complexity, the number of repeat units, and the presence of the repeat on Chromosomes 4 and 10, making analysis technically difficult. The difficulty is compounded further by the absence of an obvious gene, transcript, or protein in the unstable or proximal region; in fact, the D4Z4 repeats have been referred to as “junk” DNA or are thought to be a pseudogene, at best. As a result, FSHD has proved to be one of the most complex and challenging genetic diseases to even a glimpse an underlying pathogenic cause for FSHD. Several recent papers, including one in this issue of PLoS Genetics [4], have made significant advances that now permit us to expand our understanding of FSHD pathogenesis, a repeat contraction disease.
FSHD presents with weakness of facial muscles, stabilizers of the scapula, or dorsiflexors of the foot. The weakness is progressive with age. Disease severity is highly variable and shows some signs of anticipation, common to other repeat-associated diseases. FSHD is autosomal dominant, characterized by a deletion of D4Z4 repeat units, located in the subtelomere of chromosome 4q35 (Figure 1). Non-affected D4Z4 alleles are polymorphic having 11–100 repeat units; individuals affected with FSHD have 10 or fewer units, but must have at least one unit to show disease, which is now known to be the most telomeric unit. D4Z4 contractions can be inherited or occur as de novo mutations. The contracted D4Z4 repeat arrays show loss of DNA methylation and reduced histone 3 lysine 9 trimethylation, consistent with a more open chromatin structure [5]. The role of the altered chromatin in FSHD pathogenesis is controversial and has been suggested to enhance expression of adjacent genes like FRG1 or ANT1 [6]. More recently, FSHD2-affected individuals that display the altered chromatin but have non-contracted D4Z4 repeats have implicated the derepression of a DUX4 transcript encoded on the D4Z4 repeat units [7], [8]. However, the mechanism through which the altered chromatin at D4Z4 repeats contributes to FSHD remains unclear.
Fig. 1. Simplified schematic outlining the genetic requirements for FSHD and the current model for pathogenesis. 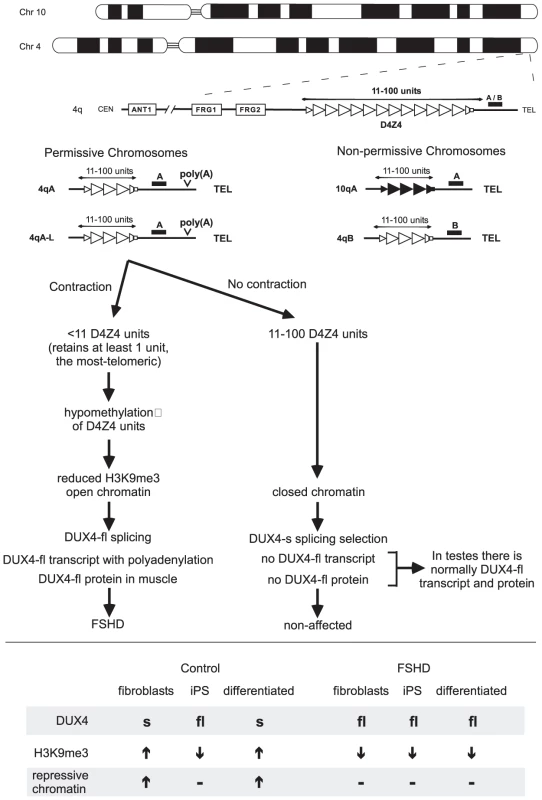
The Chromosome 4 D4Z4 repeats (open triangles) and its homolog on Chromosome 10 (closed triangles), indicating the 4qA/4qB polymorphisms that define the genetic background of the repeat. Individuals with FSHD have a D4Z4 repeat tract of <11 repeats, at least 1 unit on 4qA but not on 4qB or 10q chromosomes. All permissive chromosomes and FSHD individuals have a distal canonical highly efficient poly(A) motif ATTAAA. Non-permissive chromosomes have inefficient degenerate motifs. Both have alternative poly(A) motifs further downstream. Current model involving contraction, DUX4 transcription, polyadenylation, altered chromatin, regulated DUX4 splicing, tissue- and development-specific DUX-fl protein expression. See text for details. Lower, de-differentiation and differentiation affect DUX4-fl expression in control but not FSHD cells. See text for details. DUX4 Transcripts from the D4Z4 Repeats
The distal (most telomeric) unit of the D4Z4 repeat was recently shown to have a transcriptional profile that differs from internal units, and the transcript extended into telomeric regions [9], [10], [11]. This finding suggests that this very last, distal D4Z4 unit may be the key unit that must be retained after D4Z4 contractions to lead to disease (Figure 1). The DUX4 transcript from the distal D4Z4 unit is suggested to encode a double homeobox gene of unknown function, related to DUXC and Duxbl, which in mice is expressed in germ-line cells and in early phases of skeletal muscle development. Previously, Tapscott's group showed full-length RNA transcripts from the D4Z4 repeat spanning the DUX4 open reading frame, DUX4-fl, as well as a shorter transcript, DUX4-s, that utilized a cryptic splice donor, which retains the double-homeobox domains but loses the carboxyterminal end of DUX4 [11]. When expressed in cultured cells, the full length DUX4 caused reduced proliferation, induced morphological changes, increased sensitivity to oxidative damage, MyoD-repression, impaired myogenesis, and, at higher levels, led to cell death—all features compatible with observations in FSHD1 patient cells [11], [12]. This distal DUX4 transcript can be observed in FSHD1 patient myotubes but not in control myotubes [13]. Exactly what mediates the expression in FSHD has proved elusive.
Genetics Reveals a Role of a Polyadenylation Variant Downstream of the Most-Telomeric Chromomosome 4 D4Z4 Unit
Recent genetic advances made by a group of international labs, headed by Silvere van der Maarel, revealed the most-telomeric D4Z4 unit and its adjacent polyadenylation sequence to be crucial to FSHD pathogenesis [13] (Figure 1). Some Chromosome 4 backgrounds are categorized as permissive for FSHD disease or non-permissive to FSHD when D4Z4 contracts. Chromosome 10 repeats are typically non-permissive. Essentially, D4Z4 contractions to 1–10 units on permissive chromosomes are pathogenic, while contractions on non-permissive chromosomes are non-pathogenic. Sequence comparison of the permissive chromosomes with the common, non-permissive chromosomes failed to reveal a motif unique to the proximal D4Z4 units of the repeat array that might explain the permissiveness of the 4A161 chromosome [13]. Curiously, immediately distal to the most-telomeric D4Z4 unit, there was a polymorphism unique to the permissive 4A161 chromosome. This sequence polymorphism was subsequently shown to act as polyadenylation signal (ATTAAA) of the distal (most telomeric) DUX4 transcript. Notably, all permissive, but not non-permissive, chromosomes harbored the efficient poly(A) signal [9], [13]. This association should be investigated further.
Several rare, but highly informative FSHD individuals were identified that harbored unusual contracted hybrid D4Z4 repeats composed of D4Z4 units from Chromosome 4 and Chromosome 10—some hybrids resided on Chromosome 4, others on Chromosome 10 [13]. Two important conclusions can be made from these individuals. First, the cause of FSHD disease linked to Chromosome 10 excludes a previously suggested role for enhanced expression of the adjacent genes on 4q (FRG1, FRG2, ANT1, etc….) in the pathogenesis of FSHD [6], as these were not present on Chromosome 10. Secondly, in all affected individuals with the unusual hybrid repeats, the last D4Z4 unit of the contracted array originated from a permissive background and had the adjacent highly efficient polyadenylation motif, thereby strengthening the crucial role of this motif to FSHD pathogenesis.
In transfection experiments DUX4 transcripts derived from the permissive chromosome were stable and efficiently polyadenylated, whereas transcripts derived from non-permissive chromosomes were undetectable and polyadenylation inefficient [13]. Thus, a bona fide poly(A) signal unique to the permissive chromosomes produced stable transcripts with greater polyadenylation efficiency than non-permissive chromosomes, thereby providing strong evidence suggesting that increased polyadenylation, and hence stability, of the distal (most telomeric) DUX4 transcript may be causally implicated in FSHD pathogenesis (Figure 1). Despite lacking the polyadenylation motif that is present only on the FSHD permissive chromosome, testis (but not muscle cells) are able to express and stably retain the DUX4 transcript. Snider et al. [4] revealed that alternative polyadenylation arises at motifs more telomeric on the non-permissive and permissive chromosomes in testis, but not somatic cells. They suggest that this DUX4 alternative polyadenylation may be regulated in a tissue - and development-specific manner. This important advance revealed that DUX4 transcript and protein was naturally expressed and provided insight into how this expression was regulated, as well as avenues to how it could be misregulated in disease states. Analysis of a broader range of tissues and developmental stages will reveal insight into the function of DUX4.
Epigenetics May Regulate DUX4 Splicing
The full-length RNA transcripts from the D4Z4 repeat spanning the DUX4 open reading frame, DUX4-fl, and the shorter transcript, DUX4-s that utilized a cryptic splice donor produce full-length and truncated DUX4 proteins, respectively [11]. The presence of the full-length DUX4-fl mRNA in control human testes, but not in control muscle, which express the shorter DUX4-s mRNA, was shown by Snider et al. to be mediated by DUX4 splice site usage in this issue of PLoS Genetics [4]. Some FSHD samples also expressed DUX4-s transcript. Thus, both control and FSHD myoblasts and muscles transcribe DUX4, but the full-length DUXF-fl transcript is expressed only in FSHD cells and muscles. Expression of DUX4-fl, but not DUX4-s, can lead to both nuclear foci and increased apoptosis [4]. However, overexpression of either DUX4-fl or DUX4-s will suppress myogenesis [11], [12]. Understanding the natural function of the DUX4-fl protein and the DUX4-s protein, presuming its careful regulation supports a function, is now a pressing issue in FSHD research. Curiously, the relatively high levels of DUX4 expression in FSHD cells appears to be due to large numbers of transcripts produced in a small subset of cells, rather than a small number of transcripts produced in most cells. Understanding what regulates non-expression or expression in cells may provide insight into disease pathogenesis and possibly avenues for therapeutically down-regulating DUX4 expression in FSHD individuals. Similar insights may arise from understanding what regulates the splicing of the DUX4 transcript.
Snider et al. [4] suggest that DUX4 splice site usage may be regulated by epigenetic modifications of the D4Z4 region (Figure 1). Control fibroblasts expressed DUX4-s, but not DUX4-fl; however, when these cells were made into induced pluripotent stem cells (iPS) they switched to expression of DUX4-fl (Figure 1, lower portion). Upon differentiation of these control iPS cells, expression switched back from DUX4-fl to DUX4-s. This switch correlated with increased levels of repressive chromatin modification (H3K9me3) at the D4Z4 repeats in the differentiated cells. In contrast, in FSHD fibroblasts, their iPS derived cells, and their differentiated stages, DUX4-fl was consistently expressed. Thus, it appears that higher levels of repressive chromatin at the D4Z4 region in control cells may mediate the splicing that produces DUX4-s. Similarly, the reduced levels of repressive chromatin present in FSHD cells [5] may suppress the production of DUX4-s and allow expression of DUX4-fl.
In conclusion, D4Z4 repeats are not junk DNA: DUX4 encoded in the repeats is in fact a retrogene, most likely retrotransposed to a DNA gene from the DUXC mRNA [10]. Tapscott and colleagues have shown DUX4 is normally expressed in the male germline, and for the FSHD-permissive variant, the DUX4 transcript is efficiently polyadenylated in muscle, thus suggesting that DUX4 is not a “defective” pseudogene. Importantly, the aberrant suppression of DUX4-fl protein expression in muscle, both by transcript stabilization via disease-permissive polyadenylation and suppressed splicing to the shortened DUX4-s variant, seems to be important contributors to FSHD pathogenesis. The mechanisms through which the DUX4 transcripts and proteins lead to normal development or FSHD are future goals waiting to be expanded upon in this macrosatellite contracting disease.
Zdroje
1. PearsonCE
Nichol EdamuraK
ClearyJD
2005 Repeat instability: mechanisms of dynamic mutations. Nat Rev Genet 6 729 42
2. LemmersRJLF
van der MaarelSM
2009 Facioscapulohumeral Muscular Dystrophy.
PagonRA
BirdTC
DolanCR
StephensK
GeneReviews [Internet] Seattle (WA) University of Washington, Seattle; 1993 - [updated 2009 Jul 9]
3. DmitrievP
LipinskiM
VassetzkyYS
2009 Pearls in the junk: dissecting the molecular pathogenesis of facioscapulohumeral muscular dystrophy. Neuromuscul Disord 19 17 20
4. SniderL
GengLN
LemmersRJLF
KybaM
WareCB
2010 Facioscapulohumeral dystrophy: incomplete suppression of a retrotransposed gene. PLoS Genet 6 e1001181 doi:10.1371/journal.pgen.1001181
5. ZengW
de GreefJC
ChenYY
ChienR
KongX
2009 Specific loss of histone H3 lysine 9 trimethylation and HP1gamma/cohesin binding at D4Z4 repeats is associated with facioscapulohumeral dystrophy (FSHD). PLoS Genet 5 e1000559 doi:10.1371/journal.pgen.1000559
6. GabelliniD
GreenMR
TuplerR
2002 Inappropriate gene activation in FSHD: a repressor complex binds a chromosomal repeat deleted in dystrophic muscle. Cell 110 339 48
7. van OverveldPG
LemmersRJ
SandkuijlLA
EnthovenL
WinokurST
2003 Hypomethylation of D4Z4 in 4q-linked and non-4q-linked facioscapulohumeral muscular dystrophy. Nat Genet 35 315 317
8. de GreefJC
LemmersRJ
van EngelenBG
SacconiS
VenanceSL
2009 Common epigenetic changes of D4Z4 in contraction-dependent and contraction-independent FSHD. Hum Mutat 30 1449 1459
9. DixitM
AnsseauE
TassinA
WinokurS
ShiR
2007 DUX4, a candidate gene of facioscapulohumeral muscular dystrophy, encodes a transcriptional activator of PITX1. Proc Natl Acad Sci U S A 104 18157 18162
10. ClappJ
MitchellLM
BollandDJ
FantesJ
CorcoranAE
2007 Evolutionary conservation of a coding function for D4Z4, the tandem DNA repeat mutated in facioscapulohumeral muscular dystrophy. Am J Hum Genet 81 264 279
11. SniderL
AsawachaicharnA
TylerAE
GengLN
PetekLM
2009 RNA transcripts, miRNA-sized fragments and proteins produced from D4Z4 units: new candidates for the pathophysiology of facioscapulohumeral dystrophy. Hum Mol Genet 18 2414 2430
12. BosnakovskiD
XuZ
GangEJ
GalindoCL
LiuM
2008 An isogenetic myoblast expression screen identifies DUX4-mediated FSHD-associated molecular pathologies. EMBO J 27 2766 79
13. LemmersRJ
van der VlietPJ
KloosterR
SacconiS
CamañoP
2010 A Unifying genetic model for facioscapulohumeral muscular dystrophy. Science 329 1650 1653
Štítky
Genetika Reprodukční medicína
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2010 Číslo 10- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
- Délka menstruačního cyklu jako marker ženské plodnosti
- Intrauterinní inseminace a její úspěšnost
-
Všechny články tohoto čísla
- Common Genetic Variants and Modification of Penetrance of -Associated Breast Cancer
- FSHD: A Repeat Contraction Disease Finally Ready to Expand (Our Understanding of Its Pathogenesis)
- Genome-Wide Identification of Targets and Function of Individual MicroRNAs in Mouse Embryonic Stem Cells
- Allele-Specific Down-Regulation of Expression Induced by Retinoids Contributes to Climate Adaptations
- The Meiotic Recombination Checkpoint Suppresses NHK-1 Kinase to Prevent Reorganisation of the Oocyte Nucleus in
- Actin Depolymerizing Factors Cofilin1 and Destrin Are Required for Ureteric Bud Branching Morphogenesis
- DSIF and RNA Polymerase II CTD Phosphorylation Coordinate the Recruitment of Rpd3S to Actively Transcribed Genes
- Continuous Requirement for the Clr4 Complex But Not RNAi for Centromeric Heterochromatin Assembly in Fission Yeast Harboring a Disrupted RITS Complex
- Genome-Wide Association Study of Blood Pressure Extremes Identifies Variant near Associated with Hypertension
- The Cytosine Methyltransferase DRM2 Requires Intact UBA Domains and a Catalytically Mutated Paralog DRM3 during RNA–Directed DNA Methylation in
- β-Actin and γ-Actin Are Each Dispensable for Auditory Hair Cell Development But Required for Stereocilia Maintenance
- Genetic Association Study Identifies as a Risk Gene for Idiopathic Dilated Cardiomyopathy
- Evidence for a Xer/ System for Chromosome Resolution in Archaea
- Four Novel Loci (19q13, 6q24, 12q24, and 5q14) Influence the Microcirculation
- Lifespan Extension by Preserving Proliferative Homeostasis in
- Ancient and Recent Adaptive Evolution of Primate Non-Homologous End Joining Genes
- Loss of the p53/p63 Regulated Desmosomal Protein Perp Promotes Tumorigenesis
- Altering a Histone H3K4 Methylation Pathway in Glomerular Podocytes Promotes a Chronic Disease Phenotype
- Characterization of LINE-1 Ribonucleoprotein Particles
- Conserved Genes Act as Modifiers of Invertebrate SMN Loss of Function Defects
- Alternative Splicing at a NAGNAG Acceptor Site as a Novel Phenotype Modifier
- Tight Regulation of the Gene of the KplE1 Prophage: A New Paradigm for Integrase Gene Regulation
- Conjugative DNA Transfer Induces the Bacterial SOS Response and Promotes Antibiotic Resistance Development through Integron Activation
- Nasty Viruses, Costly Plasmids, Population Dynamics, and the Conditions for Establishing and Maintaining CRISPR-Mediated Adaptive Immunity in Bacteria
- Stress-Induced Activation of Heterochromatic Transcription
- H3K27me3 Profiling of the Endosperm Implies Exclusion of Polycomb Group Protein Targeting by DNA Methylation
- Simultaneous Disruption of Two DNA Polymerases, Polη and Polζ, in Avian DT40 Cells Unmasks the Role of Polη in Cellular Response to Various DNA Lesions
- Characterising and Predicting Haploinsufficiency in the Human Genome
- Dual Functions of ASCIZ in the DNA Base Damage Response and Pulmonary Organogenesis
- Pervasive Cryptic Epistasis in Molecular Evolution
- Transition from Positive to Neutral in Mutation Fixation along with Continuing Rising Fitness in Thermal Adaptive Evolution
- Comprehensive Analysis Reveals Dynamic and Evolutionary Plasticity of Rab GTPases and Membrane Traffic in
- Regulates Tissue-Specific Mitochondrial DNA Segregation
- Role for the Mammalian Swi5-Sfr1 Complex in DNA Strand Break Repair through Homologous Recombination
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Genome-Wide Identification of Targets and Function of Individual MicroRNAs in Mouse Embryonic Stem Cells
- Common Genetic Variants and Modification of Penetrance of -Associated Breast Cancer
- Allele-Specific Down-Regulation of Expression Induced by Retinoids Contributes to Climate Adaptations
- Simultaneous Disruption of Two DNA Polymerases, Polη and Polζ, in Avian DT40 Cells Unmasks the Role of Polη in Cellular Response to Various DNA Lesions
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



