-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaThe Microbiota Mediates Pathogen Clearance from the Gut Lumen after Non-Typhoidal Diarrhea
Many enteropathogenic bacteria target the mammalian gut. The mechanisms protecting the host from infection are poorly understood. We have studied the protective functions of secretory antibodies (sIgA) and the microbiota, using a mouse model for S. typhimurium diarrhea. This pathogen is a common cause of diarrhea in humans world-wide. S. typhimurium (S. tmatt, sseD) causes a self-limiting gut infection in streptomycin-treated mice. After 40 days, all animals had overcome the disease, developed a sIgA response, and most had cleared the pathogen from the gut lumen. sIgA limited pathogen access to the mucosal surface and protected from gut inflammation in challenge infections. This protection was O-antigen specific, as demonstrated with pathogens lacking the S. typhimurium O-antigen (wbaP, S. enteritidis) and sIgA-deficient mice (TCRβ−/−δ−/−, JH−/−, IgA−/−, pIgR−/−). Surprisingly, sIgA-deficiency did not affect the kinetics of pathogen clearance from the gut lumen. Instead, this was mediated by the microbiota. This was confirmed using ‘L-mice’ which harbor a low complexity gut flora, lack colonization resistance and develop a normal sIgA response, but fail to clear S. tmatt from the gut lumen. In these mice, pathogen clearance was achieved by transferring a normal complex microbiota. Thus, besides colonization resistance ( = pathogen blockage by an intact microbiota), the microbiota mediates a second, novel protective function, i.e. pathogen clearance. Here, the normal microbiota re-grows from a state of depletion and disturbed composition and gradually clears even very high pathogen loads from the gut lumen, a site inaccessible to most “classical” immune effector mechanisms. In conclusion, sIgA and microbiota serve complementary protective functions. The microbiota confers colonization resistance and mediates pathogen clearance in primary infections, while sIgA protects from disease if the host re-encounters the same pathogen. This has implications for curing S. typhimurium diarrhea and for preventing transmission.
Published in the journal: . PLoS Pathog 6(9): e32767. doi:10.1371/journal.ppat.1001097
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1001097Summary
Many enteropathogenic bacteria target the mammalian gut. The mechanisms protecting the host from infection are poorly understood. We have studied the protective functions of secretory antibodies (sIgA) and the microbiota, using a mouse model for S. typhimurium diarrhea. This pathogen is a common cause of diarrhea in humans world-wide. S. typhimurium (S. tmatt, sseD) causes a self-limiting gut infection in streptomycin-treated mice. After 40 days, all animals had overcome the disease, developed a sIgA response, and most had cleared the pathogen from the gut lumen. sIgA limited pathogen access to the mucosal surface and protected from gut inflammation in challenge infections. This protection was O-antigen specific, as demonstrated with pathogens lacking the S. typhimurium O-antigen (wbaP, S. enteritidis) and sIgA-deficient mice (TCRβ−/−δ−/−, JH−/−, IgA−/−, pIgR−/−). Surprisingly, sIgA-deficiency did not affect the kinetics of pathogen clearance from the gut lumen. Instead, this was mediated by the microbiota. This was confirmed using ‘L-mice’ which harbor a low complexity gut flora, lack colonization resistance and develop a normal sIgA response, but fail to clear S. tmatt from the gut lumen. In these mice, pathogen clearance was achieved by transferring a normal complex microbiota. Thus, besides colonization resistance ( = pathogen blockage by an intact microbiota), the microbiota mediates a second, novel protective function, i.e. pathogen clearance. Here, the normal microbiota re-grows from a state of depletion and disturbed composition and gradually clears even very high pathogen loads from the gut lumen, a site inaccessible to most “classical” immune effector mechanisms. In conclusion, sIgA and microbiota serve complementary protective functions. The microbiota confers colonization resistance and mediates pathogen clearance in primary infections, while sIgA protects from disease if the host re-encounters the same pathogen. This has implications for curing S. typhimurium diarrhea and for preventing transmission.
Introduction
Bacterial diarrhea is a global cause of morbidity and mortality. In most cases, the acute disease symptoms cease after a few days and the pathogen is eliminated from the gut. However, the mechanisms eliminating enteropathogenic bacteria from the gut lumen are poorly understood. Most “classical” effector mechanisms of the immune system are ineffective in the gut lumen (i.e. complement-mediated killing, opsonophagocytosis, T-cell mediated toxicity). In the gut, innate and adaptive immune responses such as antimicrobial peptides, natural and pathogen-specific mucosal secretory IgA (sIgA) antibodies are considered to be cardinal defense mechanisms. In addition to the host's immune system, the highly dense and diverse bacterial community in the gut (the microbiota; >500 different species [1], [2]) plays a key role by inhibiting pathogen growth in the gut lumen right from the beginning. This phenomenon is referred to as ‘colonization resistance’ and efficiently blocks infections by Clostridium difficile, Salmonella spp. and many other pathogenic bacteria [3]. Colonization resistance might be based on nutrient limitation, release of inhibitory metabolites, production of bactericidal compounds, the competition for binding sites and other, unidentified features of the dense microbial community [4], [5].
Much less is known about the mechanisms clearing enteropathogenic bacteria from the gut lumen once they have established an infection in this niche. ‘Pathogen clearance’ differs significantly from colonization resistance as both, the mucosa [6] and the microbiota, must recover from pathogen-inflicted disturbance while eliminating the pathogen [7]. Here, we have studied the mechanisms of pathogen clearance from the gut lumen using the example of non-typhoidal Salmonella (NTS) diarrhea.
NTS infections, including S. enterica spp. I serovar Typhimurium (S. tm), account for a significant share of food-borne diarrhea in Europe and Northern America. In sub-Saharan Africa, NTS are also an important cause of invasive disease with high mortality, particularly in HIV infected individuals [8]. In humans, colonization resistance confers partial protection, but antibiotic treatment increases the risk of Salmonella diarrhea [9], [10]. In the typical cases of NTS diarrhea, the pathogen begins to grow in the gut and disease symptoms manifest eight to 24h after consumption of contaminated food or water. Usually, the pathogen remains limited to the gastrointestinal tract and diarrhea subsides within several days. After cessation of symptoms, Salmonella remains detectable in the stool for weeks, several months or sometimes even longer [11], [12]. Pathogen clearance seems to fail in these long-term ‘asymptomatic excretors’. This is problematic, as ‘asymptomatic excretors’ pose a significant risk of transmission, in particular when food workers in restaurants or the food industry are affected [13].
So far, we can only speculate about mechanisms mediating pathogen clearance from the gut lumen. Antimicrobial peptides might be involved in some infections, but should not affect S. tm clearance, as this pathogen is particularly resistant against this type of compound [14], [15]. Antibody responses, i.e. pathogen-specific secretory IgA (sIgA), might also clear pathogens from the gut lumen. S. tm elicits profound antibody responses against LPS and protein antigens [16]. In systemic infection models antibody responses can confer some degree of protection [17], [18]. Previous work on the role of sIgA in intestinal S. tm infection yielded conflicting results. sIgA protected cultured epithelial cells from S. tm infection, but did not reduce intestinal pathogen densities [18]. Similar findings were made for the enteropathogenic bacterium Citrobacter rodentium [19]. However, the role of sIgA in pathogen clearance in models of acute Salmonella enterocolitis with high intestinal pathogen loads has not been addressed so far. Finally, we reasoned that the microbiota itself might contribute to pathogen clearance. It remained to be shown which mechanisms contribute to pathogen clearance.
We have used a Salmonella diarrhea mouse model to analyze the relative importance of sIgA and the intestinal microbiota in S. tm clearance after infection. In mice, the intestinal microbiota confers colonization resistance. Normally, <10% of mice permit pathogen growth and get mucosal inflammation upon oral S. tm infection [20]. Oral antibiotic-treatment alleviates colonization resistance and wild type S. tm grows up to very high densities in the intestinal lumen and induces mucosal inflammation (colitis) in 100% of the animals [6]. The gut inflammation allows S. tm to out-compete the microbiota thus promoting pathogen overgrowth [21]. Here, we have extended this mouse model to study pathogen clearance at later phases of the primary infection when acute mucosal inflammation has ceased. We analyzed the levels of pathogen shedding, sIgA responses and the role of the microbiota. This revealed that the microbiota plays an essential role in pathogen clearance. The implications for curing asymptomatic excretors and preventing S. tm diarrhea are discussed.
Results
Sm-treated mice recover from S. tmatt induced gut inflammation and display extensive differences in the kinetics of fecal pathogen clearance
In sm-treated mice, infection with an attenuated S. typhimurium strain (S. typhimurium SL1344 sseD; termed S. tmatt; Table S1) is known to recapitulate key aspects of the early stages of human NTS diarrhea, i.e. gut inflammation 8h after orogastric exposure with infection confined to the gastrointestinal tract [22]. Symptoms of the acute gut inflammation usually decline by 5–7 days after infection [23]. In order to assess, if this model may be useful to dissect the role of pathogen-specific sIgA and the intestinal microbiota in pathogen clearance at the final stage of a primary infection we analyzed the outcome of long-term S. tmatt infections [6], [24].
We monitored S. tmatt shedding for up to 60 days after infection. S. tmatt shedding in stool began to decrease after a few days, varied extensively between different animals and lasted for 2 to 8 weeks (Fig. 1A). At 60 day p.i., S. tmatt shedding was reduced below 105 cfu/g (p<0.05 day 1 vs. day 60 p.i.). At all stages, the infection remained largely confined to the gastrointestinal tract and draining mesenteric lymph nodes (MLN) and gut inflammation subsided after 7–44 days (Fig. 1B,C). Interestingly, we observed a high incidence of ‘asymptomatic excretors’ around day 44 post infection (p.i.). These mice were characterized by a low pathological score (≤3) and high cecum pathogen loads (≥105 cfu/g stool; Fig. 1C, right panel, green symbols). This may indicate that pathogen clearance from the gut lumen is not necessary in order to resolve gut inflammation. In fact, both might be independent from each other. We concluded that this model could be useful to analyze the mechanism of pathogen clearance from the gut lumen after infection.
Fig. 1. S. tmatt infection yields ‘asymptomatic excretors’ and elicits O-antigen specific immunity. 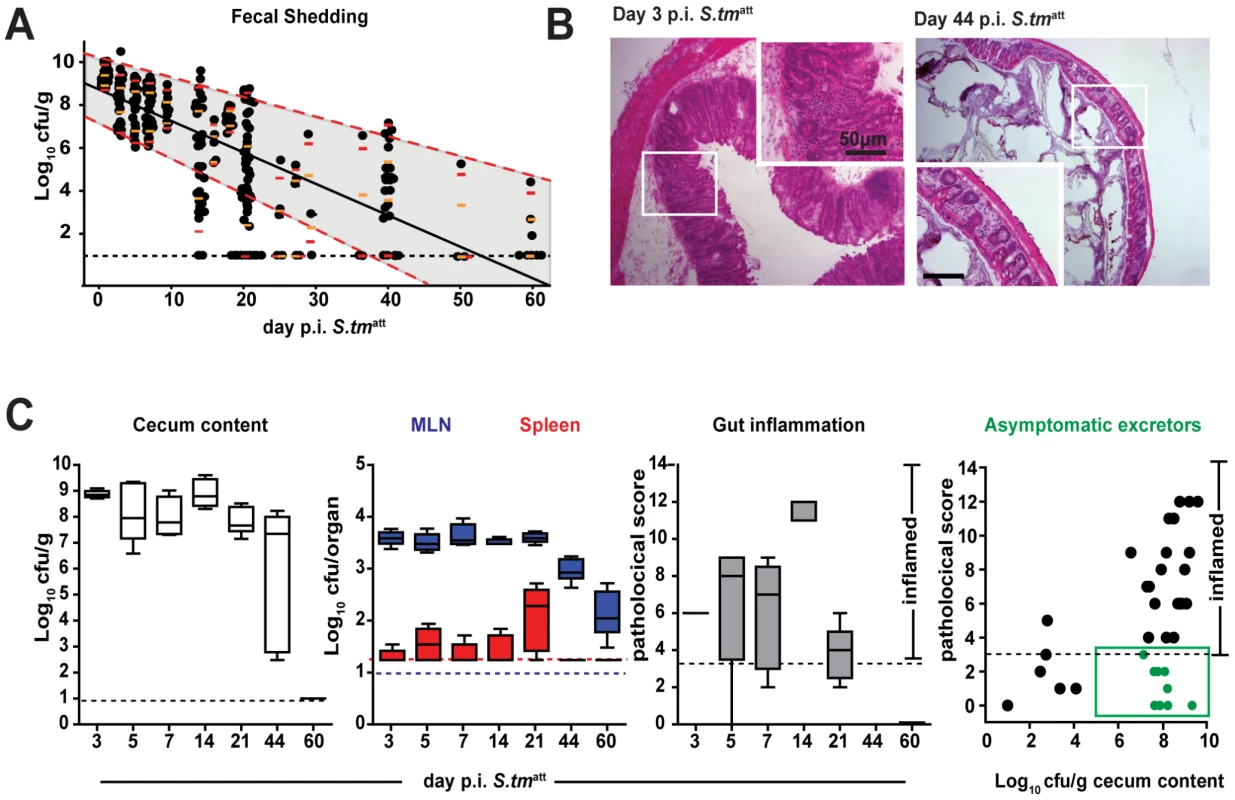
A. Time course of fecal S. tmatt shedding. Sm-treated mice were infected with S. tmatt and fecal Salmonella shedding was monitored (5<n<48 individual mice per time point). Medians are shown in black; orange and red: 5%, 25%, 75% and 95% quantiles. Dashed red lines: weighted linear regression on the 5% and 95% quantiles. B. H&E stained cross-section of the cecum at day 3 (left) and day 44 (right) post S. tmatt infection. Enlarged section (white box) is shown in inset. Scale bar: 50µm. C. S. tmatt loads in cecum (left panel; Log10 cfu/g), spleen (red) and MLN (blue; both Log10 cfu/organ; 2nd panel) and cecal mucosa inflammation (3rd panel) at the indicated times post S. tmatt infection. ‘Asymptomatic excretors’ (4th panel) are defined as showing a pathological score ≤3 while shedding ≥105 cfu/g S. tmatt.(green box). Each dot represents an individual mouse between day 3 and 60 S. tmatt immunization. Black dotted line: detection limit. S. tmatt induces O-antigen specific mucosal protection
Next, we wanted to address if mice that had experienced a primary S. tmatt infection in our model developed an adaptive immune response that would protect against gut inflammation upon re-infection with the same pathogen. This would be a pre-requisite for functional analysis of antibody responses in pathogen clearance. Therefore, we extended the infection model as depicted in Fig. 2A (‘immunization-challenge’ protocol). Sm-treated mice were infected with S. tmatt for 39 days as in the standard protocol ( = experimental group; mock-immunization = negative control). This allowed sufficient time for recovering from acute inflammation and the generation of a S. tm-specific adaptive immune response (Fig. 1C; see also Fig. 2C,D). At day 39, the mice were treated with ampicillin to transiently suppress the microbiota and eliminate any S. tmatt which may have persisted in the gut. We then challenged the animals with wild type S. typhimurium (wt; ampicillin resistant; 200 cfu by gavage). In the mock-immunized mice, wt S. typhimurium efficiently colonized the gut and elicited acute intestinal inflammation within two days after challenge (Fig. 2B). In contrast, the S. tmatt-immunized mice were generally protected against wt S. typhimurium-inflicted disease (8/10 mice with cecal pathology score ≤3; p = 0.0099; Fig. 2B).
Fig. 2. S. tmatt induces O-antigen specific mucosal protection. 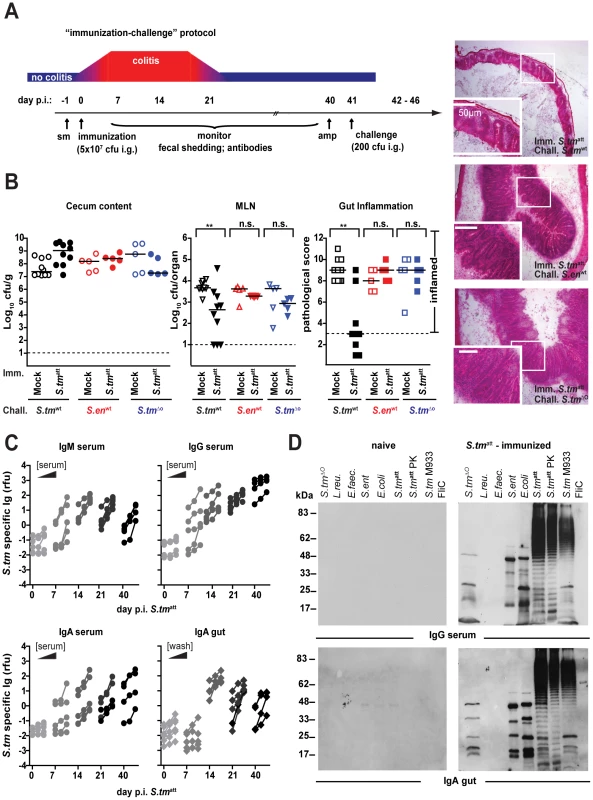
A. ‘Immunization-challenge’ model. At day 0, sm-treated mice are infected and S. tmatt (5×107cfu; i.g.). They develop enteric pathology (red) that usually declines by day 14–20 p.i. (blue). Afterwards, the mice appear healthy. At day 40 p.i., mice are treated with ampicillin (20mg i.g.) and challenged with S. tmwt (or other strains; 200cfu i.g.). The degree of Salmonella-induced gut inflammation and pathogen loads are determined at day 1–5 post challenge. B. S. tmatt immunized mice are protected in an O-antigen dependent fashion. S. tmatt (filled symbols) or mock (open symbols) immunized mice were challenged with S. tmwt (black; n = 10), S. enwt (red circles; n = 5) or S. tmΔO (blue; n = 5) at day 40 post immunization. At day 2 post challenge, challenge strain loads were assessed in the cecum content (left panel) and the MLN (middle panel). Dashed lines indicate detection limits. Right panels: inflammation of the cecal mucosa (score≤3 indicates no inflammation). Black bar: median; *p<0.05; **p<0.005; n.s. = not significant. H&E stained cross-sections of the cecum of S. tmatt-immunized mice at day 2 post challenge with S. tmwt (upper panel), S. enwt (middle panel) and S. tmΔO (lower panel). Enlarged section (white box) is shown in the lower panel. Scale bar: 50µm. C. Time course of the S. tmatt specific humoral immune response. Antibodies directed against the surface of S. tmatt in serum or gut wash of the S. tmatt -immunized mice from Fig. 1C were analyzed by bacterial FACS (Materials and Methods; Fig.S1). The Y-axis shows S. tmatt specifc Ig (relative fluorescence units; rfu), the X-axis different dilutions of serum or gut wash (1∶ 20, 1∶60, 1∶180) at different days post S. tmatt immunization. D. S. tmatt-immunized mice mount an O-antigen specific antibody response. Serum and gut wash Ig from naïve and S. tmatt infected mice (day 40 post infection) were analyzed by immunoblot against different bacterial lysates (S. tmΔO; L. reuteri; E. faecalis; S. entwt; E. coli; S. tmatt; S. tmatt digested with proteinase K; S. tm M933 [no flagella, no functional TTSS]; flagellin FliC). Ig was detected with the respective HRP-labeled secondary antibodies. The experiment is representative for 6 different animals. Bacterial surface structures and secreted proteins are dominant targets of adaptive immune responses [18], [25], [26], [27], [28], [29]. Therefore, we analyzed whether surface-protein or O-antigen specific immune responses might explain the protection of S. tmatt-immunized mice. No protection was observed against challenge with the NTS serotype S. enteritidis (S. enwt), harboring a different LPS-O-antigen, or an O-antigen deficient isogenic S. typhimurium mutant (S. tmΔO; ΔwbaP; Table S1; p>0.05 vs. colitis in mock immunized controls). Thus, S. tmatt-immunized mice mounted an adaptive immune response which protected from mucosal disease on re-infection with the pathogen in an O-antigen-dependent way.
S. tmatt induces pathogen-specific sIgA
The exquisite O-antigen specificity of protection from a second round of inflammation suggested that adaptive immunity and particularly sIgA may be the crucial mechanism not only for preventing inflammation on re-infection, but also for clearing pathogens from the gut. Therefore, we determined the kinetics of the Salmonella-specific humoral immune response by measuring specific Ig via surface staining of live, intact bacteria by flow cytometric analysis (Fig. 2C). This assay accurately differentiates S. tm specific antibodies from antibodies directed against closely related species, such as E. coli [30] (Fig.S1A–C). S. tm-specific IgM, IgG and IgA were detectable in the serum as early as 7 days post immunization. By day 14, all mice secreted S. tm -surface-specific sIgA into the gut lumen. Mucosal sIgA responses were confirmed by immunohistochemistry (Fig.S2). Salmonella antigens targeted by this strong, specific humoral immune response were analyzed by Western blotting. The antibody response was indeed pathogen-specific, as Lactobacillus reuteri RR and Enterococcus faecalis, two commensals isolated from our mouse colony, were not recognized (Fig. 2D; Fig.S3). In analogy to the human infection (Fig.S4), the antibody response included sIgA recognizing the O-antigen of S. tm (protease resistant ladder-like bands in the Western blot; Fig. 2D), a highly repetitive sugar structure of the lipopolysaccharide (LPS), coating the surface of the pathogen. In contrast, the O-antigens from S. enteritidis and E. coli, which have a different sugar structure or LPS from the O-antigen deficient mutant S. tmΔO were not recognized. In addition, antibodies to several prominent protein antigens were detected. Most of these protein antigens were conserved in different Salmonella and E. coli strains, but not in L. reuteri RR or E. faecalis.
It should be noted that acute mucosal inflammation seems necessary to elicit immune responses protecting from enterocolitis. It was also shown previously, that invasive Salmonella strains triggered more potent adaptive immune responses [31]. Mice not pretreated with sm before immunization (low antigen loads, no gut inflammation), sm-treated mice immunized with S. tmavir (high antigen loads, no gut inflammation) and parenterally immunized mice (S. tmatt i.v.; systemic antigen loads, no gut inflammation) did not mount detectable levels of O-antigen-specific sIgA. None of the mice were protected against wild type S. tm (S. tmwt) mediated enteropathogenesis (Fig.S5).
Overall, these data demonstrated that the LPS O-antigen was the dominant protective antigen and that mice mount a robust pathogen-specific sIgA response during the first round of infection. This is in line with earlier data from studies in the mouse typhoid fever model, in chicken and data from human patients [18], [25], [26], [27] (Fig.S4). However, from these first sets of experiments we could not conclude whether pathogen-specific sIgA was sufficient for S. tm clearance from the gut.
sIgA is dispensable for pathogen clearance from the gut lumen
In order to address sIgA functions in pathogen clearance, we analyzed the outcome of S. tmatt infection in different KO-mice lacking key mediators of functional adaptive immune responses. We determined whether T-cell dependent or -independent mucosal sIgA immune responses [30], [32], [33] were critical for termination of inflammation, pathogen clearance and protection from inflammation on re-infection. ‘Immunization-challenge’ experiments were performed on mice lacking the T-cell receptor (TCRβ−/−δ−/−; T-cell deficient), B-cells (JH−/−), IgA (IgA−/−) or sIgA and sIgM-transport into the gut lumen (pIgR−/−; Table S2). Two days after initial infection with S. tmatt, all knockout mice displayed pronounced gut inflammation (data not shown) and gut inflammation subsided by day 40 (Table 1). This demonstrated that the acute mucosal inflammation can be efficiently terminated in the absence of T-cells, B-cells, antibodies or sIgA. Furthermore, several IgA−/− (3/4) and pIgR−/− (2/5) animals managed to clear S. tmatt from the gut lumen by day 40 p.i. This indicated that pathogens can (at least in some cases) be cleared from the gut lumen, in the absence of pathogen-specific sIgA (and sIgM) in the gut lumen. In order to exclude differences attributable to alterations in microbiota composition between different mouse lines, we have compared the S. tmatt clearance kinetics between IgA−/− and wild type littermates (IgA+/−, IgA+/−, IgA+/+; Fig. 3). This verified that kinetics of pathogen clearance was not affected by presence or absence of sIgA.
Fig. 3. IgA deficiency does not affect kinetics of pathogen clearance. 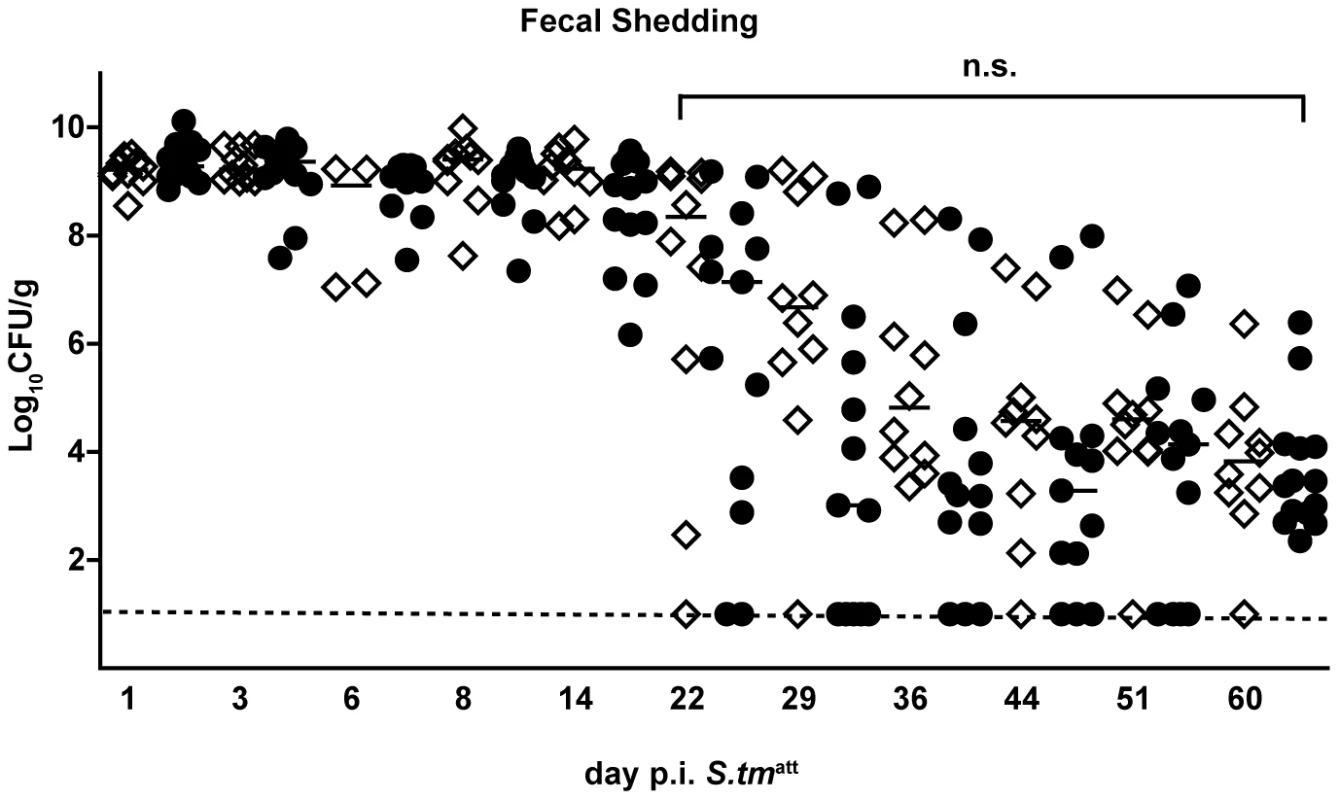
Time course of fecal S. tmatt shedding in IgA−/− and IgA-proficient littermates. IgA−/− and IgA-proficient littermates were generated by crossing IgA+/− mice in order to yield littermates with comparable gut flora. Sm-treated IgA−/− (n = 10; white diamonds), IgA+/− (n = 7; black circles) and wild type (n = 7; black circles) littermates were infected with S. tmatt and fecal Salmonella shedding was monitored until day 60 post infection. Black bar: medians. Dashed line: detection limit. Statistics: comparison of fecal shedding by IgA−/− vs. IgA+/− and IgA+/+ (n.s.: p>0.05). Tab. 1. Primary infection with S. tmatt. 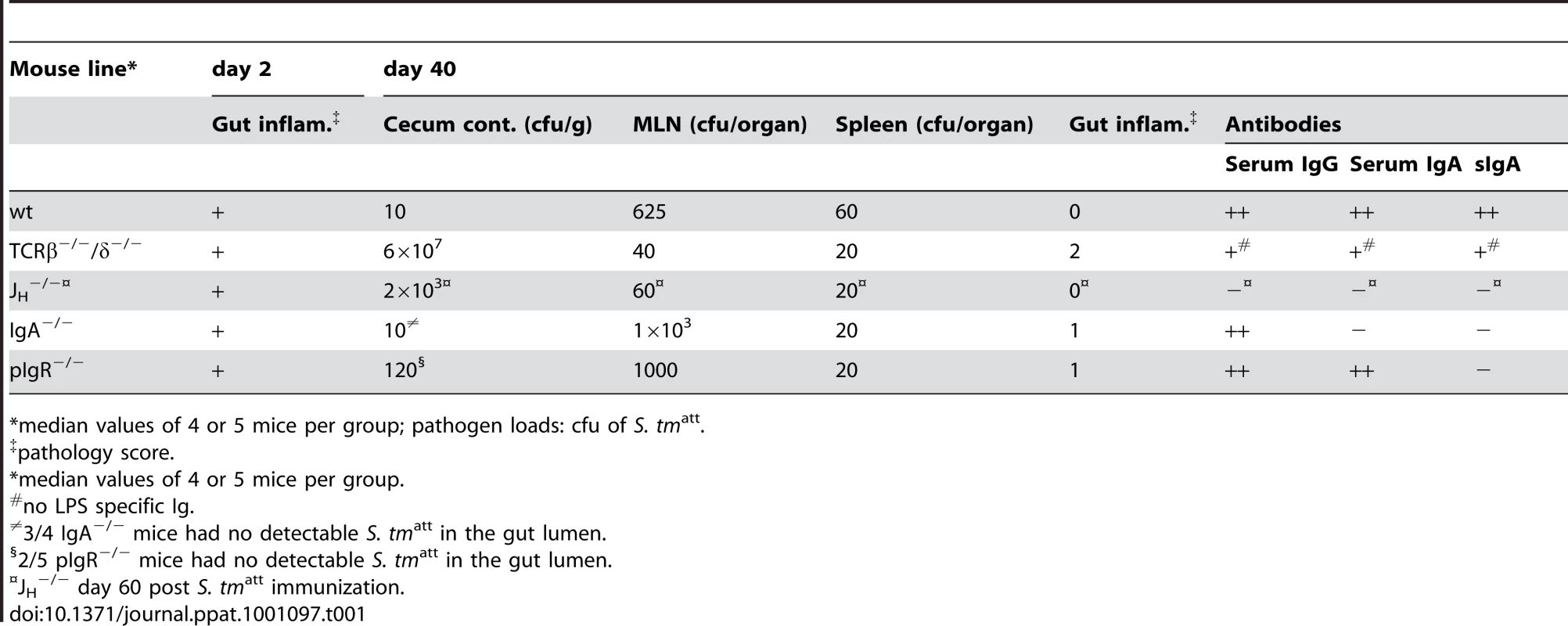
*median values of 4 or 5 mice per group; pathogen loads: cfu of S. tmatt. Strikingly, none of the S. tmatt -immunized knockout mice developed O-antigen specific antibodies and none were protected from intestinal inflammation upon challenge with S. tmwt (pathological score ≫3; Table 2 and Fig.S6). Thus, a T-cell dependent, adaptive mucosal sIgA response is essential for protection from secondary disease, but is dispensable for resolving the initial inflammatory response to S. tmatt and for clearing the pathogen from the intestinal lumen.
Tab. 2. Challenge infection (day 2 post S. tmwt challenge). 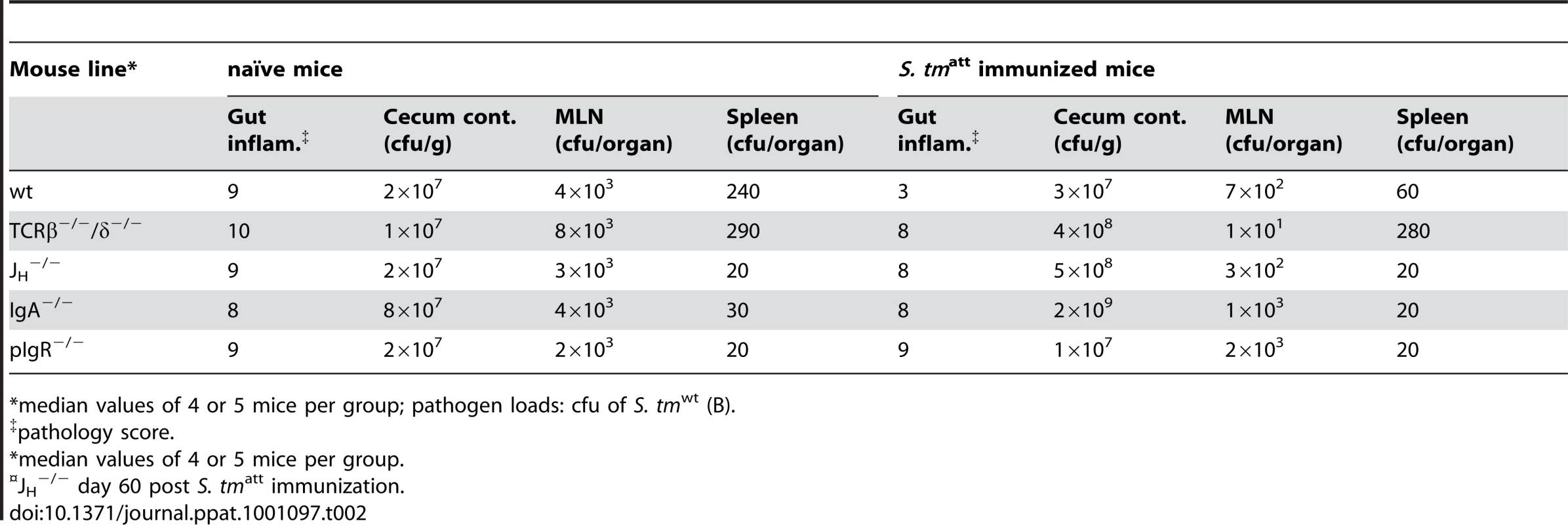
*median values of 4 or 5 mice per group; pathogen loads: cfu of S. tmwt (B). sIgA restricts intestinal pathogen growth and mucosal access
Though dispensable, our findings did not exclude that sIgA exerts an effector function [34] which contributes in some way to pathogen clearance. To identify such mechanisms, we analyzed the effects of sIgA on pathogen growth and its interaction with the host's intestinal mucosa in greater detail. First, we applied a modified ‘immunization-challenge’ protocol. Sm-treated mice were infected with S. tmatt, an equivalent S. enteritidis strain (S. enatt; S. enteritidis 125109 sseD; [35]) or mock. Antibody responses and S. tmatt/ S. enatt loads in the stool were monitored (Fig.S7 and data not shown). After 39 days, immunized mice were treated with ampicillin (elimination of microbiota and remaining S. tmatt or S. enatt) and challenged with a 1∶1 mixture of S. tmavir and S. enavir (ampicillin resistant; sseDinvG mutants; 200 cfu each by gavage; Table S1). These latter mutants can colonize the gut lumen of naïve mice for up to four days, remain confined to the gastrointestinal tract and they do not elicit enteropathogenesis, thus mimicking the situation in the intestines of ‘asymptomatic excretors’ [21], [24]. We decided not to use S. tmΔO for this type of competition experiments as it displays a pronounced competitive growth defect in mice when co-infections are performed with an isogenic wild type strain [36]. In the gut lumen of S. tmatt immunized mice, S. enavir out-competed S. tmavir (Fig. 4A; black symbols). In S. enatt immunized mice, S. tmavir out-competed S. enavir (red symbols), and in mock-immunized mice, both strains colonized with equal efficiency. Therefore, O-antigen specific sIgA may help controlling pathogen growth or survival in the gut lumen. Furthermore, S. tmavir (but not S. enavir) was aggregated in the gut lumen and occluded from the mucosal surface of S. tmatt immunized mice (Fig. 4A, right panels). Pathogen occlusion was confirmed by assessing pathogen loads in the gut tissue of challenged mice. In S. tmatt -immunized animals, S. tmwt tissue loads were 100-fold lower than in mock-immunized controls (Fig. 4B). In contrast, S. tmatt immunization did not prevent the invasion of S. enteritidis. Furthermore, S. tmatt immunized pIgR−/− mice, which cannot transport sIgA across the gut epithelium, failed to prevent gut tissue invasion by wt S. typhimurium into the mucosal tissue (Fig. 4B). Thus, the O-antigen-specific sIgA response conferred protection by restricting pathogen growth in the gut lumen and preventing the interaction of the pathogen with the intestinal mucosa. To some extent, this may also contribute to pathogen clearance from the gut lumen.
Fig. 4. Immunization elicits O-antigen specific sIgA which retards pathogen growth and restricts mucosal access. 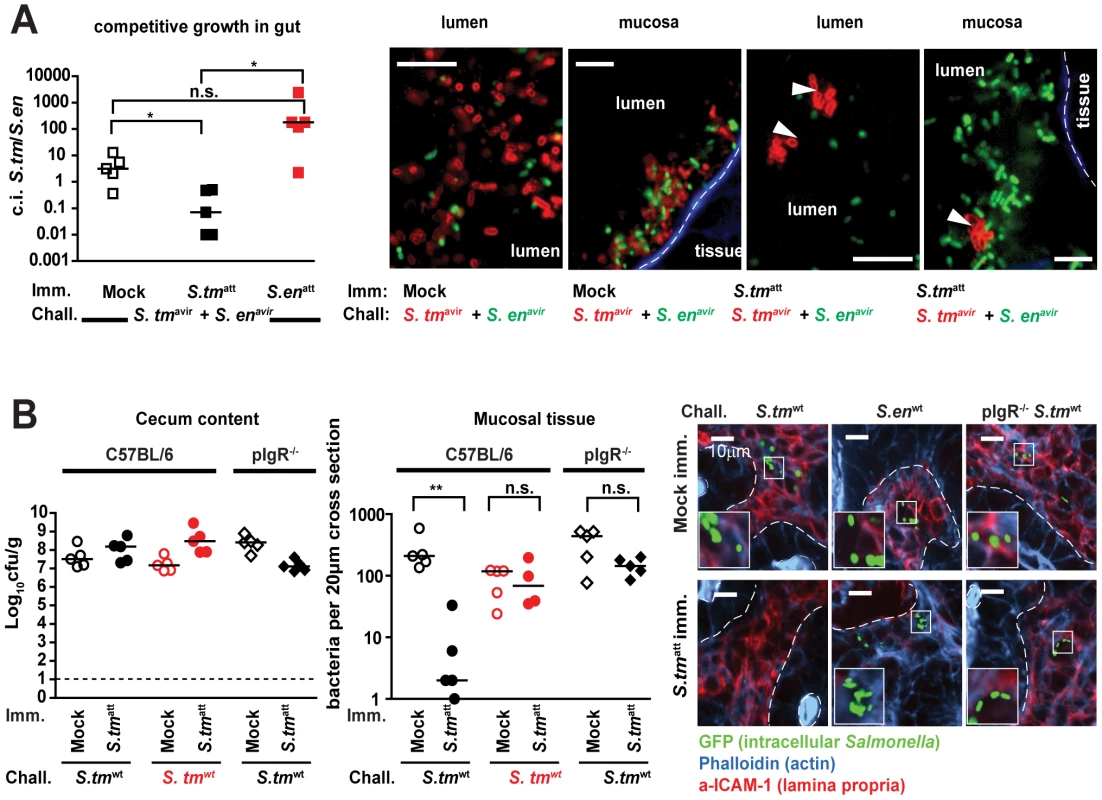
A. sIgA retards pathogen growth and occludes mucosal access. Mice were immunized with S. tmatt, S. enatt or mock (n = 5 per group), treated with ampicillin and challenged for 1 day with a 1∶1 mixture S. tmavir(ampR; pWKS30) and S. enavir (ampR; pM979-GFP+). None of the mice developed gut inflammation. Left panel: competitive index (c.i.) of S. tmavir(ampR; pWKS30) and S. enavir (ampR; pM979-GFP+) in the cecal lumen. Right panels: Immunofluorescence microscopy of the cecal lumen (left panels) and the mucosal surface (right panels; actin brush boarder = blue) to detect S. tmavir (red α-S. tm LPS stain) and S. enavir (green GFP+). Scale bar: 10µm. B. sIgA blocks mucosal invasion by S. tmwt. C57Bl/6 (circles) or pIgR−/− mice (diamonds) were immunized with mock (open symbols) or S. tmavir (closed symbols) and challenged with S. tmwt (black) or S. enwt (red). Bacterial loads in the cecal lumen (left) and Salmonella invasion into the mucosal tissue (middle) was determined at 2 days post challenge. Right panels: Immunofluorescence microscopy of GFP-expressing S. tmwt or S. enwt (green) in the cecal mucosa (red: ICAM-1, lamina propria; blue: Actin, epithelial brush border); White dotted lines mark epithelial-submucosal boarder; Scale bar: 10µm. Mice harboring a low-complexity microbiota become long-term asymptomatic excretors
While O-antigen-specific sIgA was indispensable to prevent disease, it did not seem to be a major determinant in pathogen clearance from the gut lumen. The onset of adaptive sIgA responses and cessation of symptoms seemed to occur well ahead of S. tmatt elimination from the intestines. Moreover, IgA deficiency did not affect pathogen clearance kinetics (Table 1; Fig. 3). This was different from most well studied paradigms of acute systemic infection where the onset of protective immunity coincides with declining pathogen loads. This strongly suggested that sIgA-independent mechanisms may underlie pathogen clearance from the gut.
Thus, we hypothesized that the microbiota might play a crucial role in pathogen clearance. The microbiota is a dense bacterial community composed of approx. 500–1000 different species [9], [37]. It confers numerous beneficial effects to the host [38] including ‘colonization resistance’, i.e. a generalized interference with the growth of many pathogens in the gut of a naïve host [3]. Antibiotic treatment disrupts the normal microbiota, alleviates colonization resistance and constitutes a known risk factor for Salmonella infections in humans and mice [7], [9], [10], [21], [39]. Furthermore, the species composition of the microbiota - and by inference the degree of colonization resistance - can vary significantly between different individuals [40]. Therefore, the microbiota composition might explain why Salmonella shedding by ‘asymptomatic excretors’ can last for months or years.
In sm-treated mice, the microbiota is transiently reduced, but rapidly returns to pretreatment community composition, re-establishes ‘colonization resistance’ and ‘asymptomatic excretion’ occurs just transiently (Fig. 1C; [21], [41]). For this reason, our original infection model was not optimally suited for dissecting the differential role of the microbiota and sIgA in pathogen clearance.
To overcome this problem we used ‘L-mice’ which harbor a well defined, low complexity microbiota (L = ‘LCM mice’; [20]). L -mice are ex-germ free mice that are stably associated with the ‘Altered Schaedler Flora’ [42] comprising <20 species. The representatives with the highest abundance are ASF500 (Firmicutes; Clostridia; Clostridiales; Lachnospiraceae; unclassified_Lachnospiraceae) and ASF519 (Bacteroidetes; Bacteroidia; Bacteroidales; Porphyromonadaceae; Parabacteroides). Thus, the L microbiota resembles the conventional (C) microbiota of mice and men at broad lineages levels [43]. However, in spite of an equally high bacterial density as the C microbiota, the L microbiota does not confer colonization resistance [20]. Accordingly, S. tmatt efficiently colonized the gut lumen of L-mice at high levels (≥108cfu/g) and elicited pronounced enteropathogenesis by day 2 p.i. even without previous antibiotic treatment (Fig. 5A). After 40 days, all immunized L-mice had resolved acute inflammation, but kept on shedding S. tmatt at high levels for at least 83 days (Fig. 5A; see also below). This was not due to a defective O-antigen-specific sIgA response: sIgA responses in L-mice were as pronounced as in C-mice as indicated by the increased numbers of IgA+ cells in the cecal mucosa (Fig. 5B,C and Fig.S2) and by Western Blot analysis (Fig.S8 and Fig 2D). The strong adaptive mucosal immune response was also confirmed by gene expression profiling of the cecal mucosa (Fig. 5D, Fig.S9, Table S3). Furthermore, challenge experiments confirmed the O-antigen-specific protection from enteropathogenesis (Fig. 6). However, despite this O-antigen-specific sIgA response, high-level pathogen shedding persisted in all analyzed animals (Fig. 5A; see also below). Therefore, O-antigen-specific sIgA was insufficient for luminal S. tmatt clearance. This was in line with our hypothesis that elements of the normal, complex microbiota (which is lacking in L-mice) may play a key role in terminating fecal S. tmatt shedding.
Fig. 5. An O-antigen-specific sIgA response is insufficient to terminate S. tmatt shedding by L-mice. 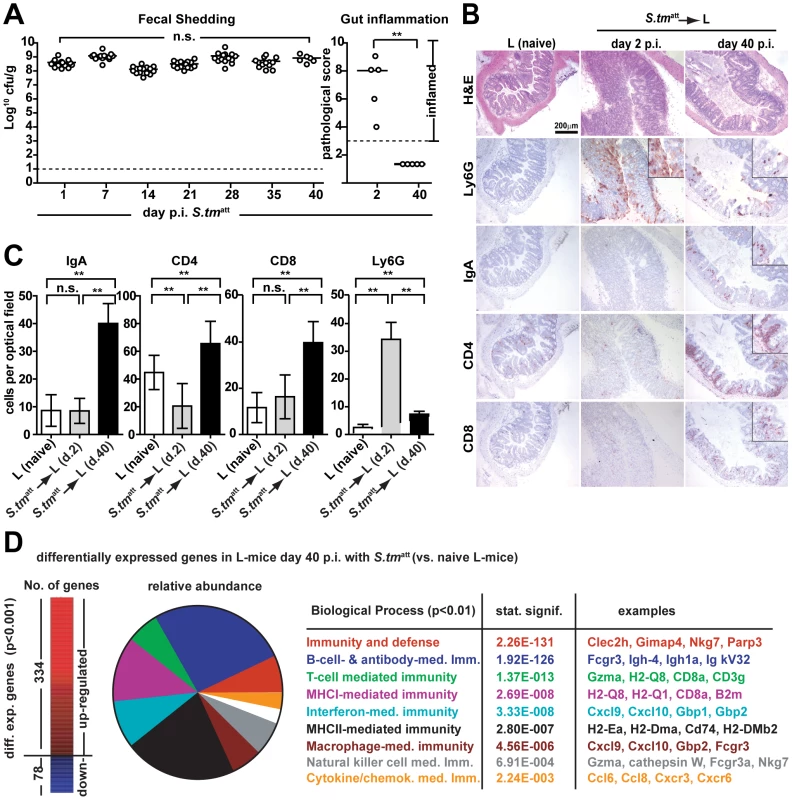
A. L-mice do not clear S. tmatt from the gut. L-mice (n = 15) were immunized with S. tmatt (no streptomycin-treatment) and fecal shedding was monitored for 40 days (left panel). Gut inflammation was analyzed at day 2 and day 40 p.i. (5 mice per time point; right panel). B. and C. L-mice mount a pronounced adaptive mucosal immune response. Cecal tissue of naïve L-mice and L-mice at days two and 40 p.i. was stained by immunohistochemistry for Ly6G (granulocytes), IgA, CD4 (T-cells) and CD8 (T-cells) cellular markers. Quantitative data were from 15 randomly selected 40× high power fields (hpf) from 3–5 mice per group. Y-axis: average cell number per 40× hpf (C). D. Gene expression analysis of naïve and L-mice at day 40 post S. tmatt immunization. Left: Numbers of differentially expressed genes in S. tmatt-immunized L-mice. Middle: relative abundance of biological pathways significantly upregulated in S. tmatt-immunized L-mice. Processes, p-values and characteristic examples are shown as a table (right). Fig. 6. O-antigen specific protection from enteropathogenesis of S. tmatt-immunized L-mice. 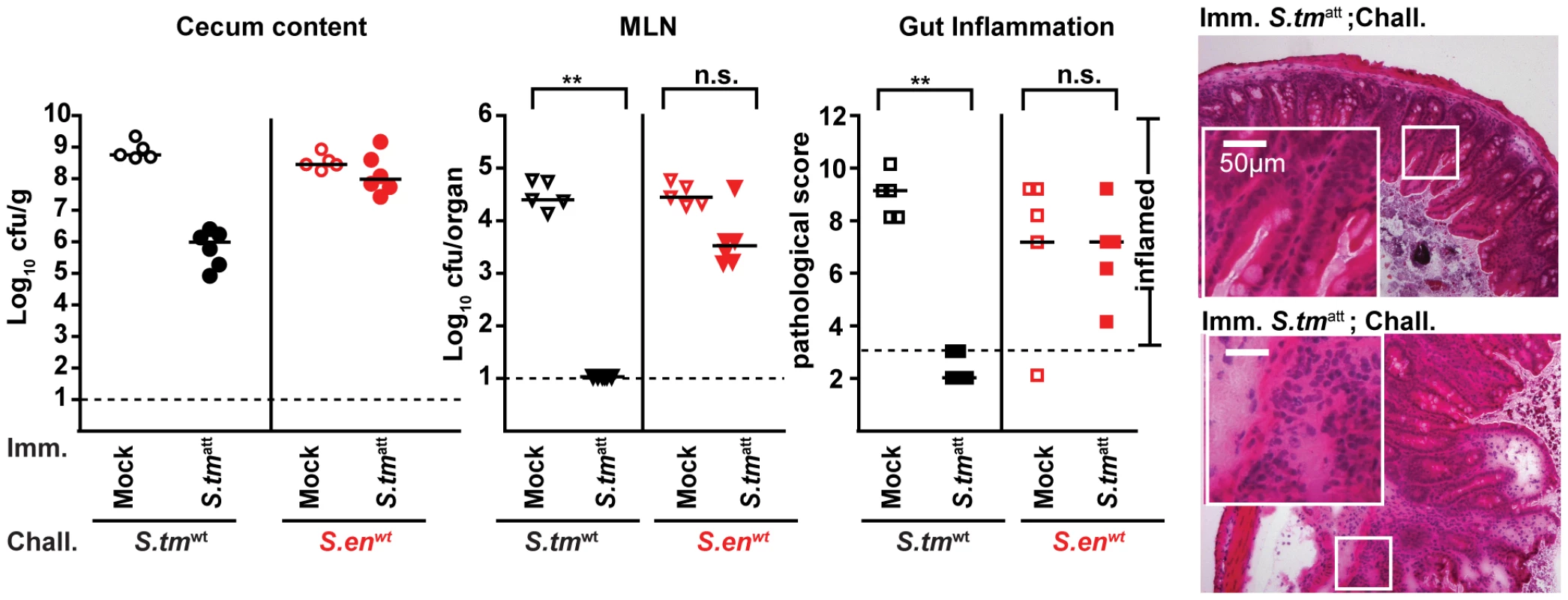
S. tmatt- (filled symbols) or mock- (open symbols) immunized mice were challenged for two days with S. tmwt (black; n = 10) or S. enwt (red circles; n = 5) at day 40 post immunization. Challenge-strain loads in the cecal content (left panel) and the MLN (middle panel) and cecal inflammation were assessed at day 2 post challenge. Dashed lines: detection limit or limit defining the absence of disease (pathological score ≤3). Black bars: median; *p<0.05; **p<0.005; n.s. = not significant. Right: H&E stained cross-sections of the cecum of S. tmatt-immunized L-mice at day two post challenge with S. tmwt (upper panel) or S. enwt (lower panel). Scale bar: 50µm. Transfer of a complex microbiota facilitates pathogen clearance
To formally define the importance of the commensal microbiota in pathogen clearance, two groups of L-mice were infected with S. tmatt for 83 days. The first group was kept under strict hygiene isolation and shed high loads of S. tmatt until the end of the experiment (S. tmatt→L; Fig. 7A; open symbols). The second group was exposed to C microbiota at day 40 by placing C donor mice into the same cage (S. tmatt→L/C; 6 independent cages). Both groups of mice developed the typical pathogen-specific, adaptive sIgA response by day 83 p.i. (Fig. 7B,C). Upon introduction of the C donor mice, fecal shedding decreased gradually and ceased in most of the S. tmatt→L/C mice by day 83 (<105cfu/g; Fig. 7A; black symbols) but not in the S. tmatt→L group. This suggested that pathogen clearance was mediated in some way by the complex microbiota.
Fig. 7. Transfer of a complex microbiota curbs S. tmatt shedding by L-mice. 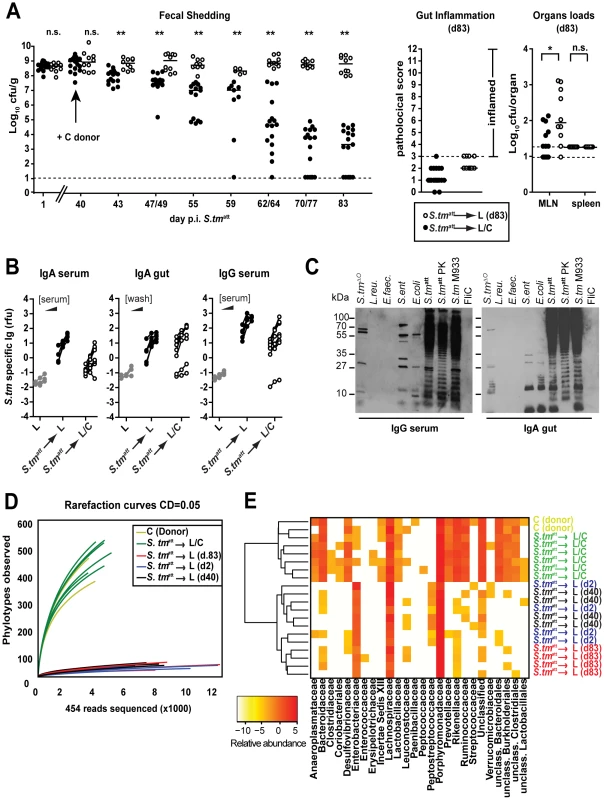
A. Exposure to conventional gut microbiota curbs pathogen shedding by ‘asymptomatic excretors’. L-mice were immunized with S. tmatt. At day 40 p.i., donor animals with a conventional microbiota (C-mice) were added to one group (black circles; n = 17; S. tmatt→L/C). The other group remained under hygienic isolation (open circles; n = 10; S. tmatt→L). Fecal S. tmatt shedding was monitored until day 83 p.i. Right panels show gut inflammation and organ loads at day 83 p.i.. B. and C. S. tm-specific Ig-response (serum IgA and IgG; IgA in gut wash; day 83 p.i.) in S. tmatt→L and S. tmatt→L/C mice was determined by bacterial FACS and by immunoblot as in Fig. 2A,B. D. A complex microbiota is transferred to L-mice by day 83 p. S. tmatt immunization. Collectors' curves (CD = 0.05) were created for each mouse from the total number of filtered sequences for representative mice: S. tmatt→L (d.2), S. tmatt→L (d.40), S. tmatt→L (d.83), S. tmatt→L/C and 2 C-mice (donor). E. Phylotypes of mice shown in (D) (Clustering distance CD = 0.05) were sorted according to their family taxon (family level; x-axis) and average clustering was performed on Euclidean distances calculated between abundance profiles for each mouse and every time-point sampled. Red color indicates high abundance (Log2), yellow color low abundance. In order to verify microbiota-transfer, we assessed microbiota composition using high-throughput 16S rRNA gene sequence analysis (Materials and Methods). S. tmatt→L/C mice displayed significantly higher diversity than S. tmatt→L mice as well as LCM mice at day 2 and 40 after S. tmatt immunization (Fig. 7D). The rarefaction curves indicated that S. tmatt→L/C mice had acquired a microbiota of the similar complexity as the C donor mice. This was confirmed by assessing the richness (actual diversity) of the samples by calculating the Shannon index (H) and species evenness (E) as well as the Chao1 diversity estimate (Table S4). Accordingly, the taxonomy assignment confirmed that the number of bacterial taxa in the stool increased significantly in S. tmatt→L/C mice. All S. tmatt→L mice carried high loads of Enterobacteriaceae (i.e. Salmonella spp., E. coli spp.; red colors) in their stools. In contrast, no Enterobacteriaceae were detected in the stools of 3 (out of 6) S. tmatt→L/C mice and the remaining 3 animals carried low levels of this family (yellow colors, Fig. 7E). In addition, the microbiota composition was similar between all S. tmatt→L/C animals as demonstrated by hierarchical cluster analysis of eubacterial family profiles (Fig. 7E). This indicated that pathogen displacement occurs in a reproducible, stereotypic fashion and may not result from random transfer of only few members of the conventional microbiota. Most importantly, these data demonstrate that members of the conventional microbiota can upon transfer lead to the termination of sustained pathogen shedding in L-mice.
It remained unclear whether pathogen clearance was mediated directly by the microbiota or by microbiota-induced mucosal responses. Recently, it has been shown that parts of the microbiota (i.e. segmented filamentous bacteria) induce mucosal TH-17 cell responses that can protect from pathogen infection [2]. However, we did not observe differences in IL-17A or IFN gamma-producing CD4 T-cells in the MLN of S. tmatt→L and S. tmatt→L/C animals by day 83 (Fig. 8). Furthermore, we tested if total MLN cells obtained from S. tmatt→L/C animals would, upon transfer into S. tmatt→L (d.40) induce clearance of intestinal S. tmatt. However, this was not the case (Fig.8). This was in line with the notion that the microbiota may directly mediate pathogen clearance.
Fig. 8. Adoptive transfer of MLN cells isolated form mice that cleared S. tmatt from the intestine into S. tmatt→L mice (day 40 p.i.) does not induce S. tmatt clearance. 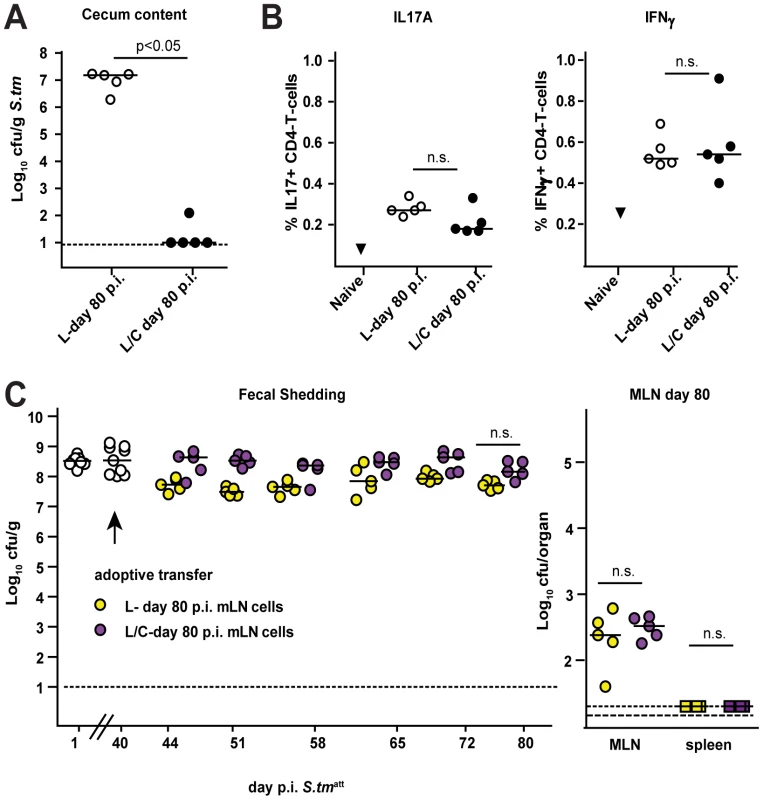
L-mice were immunized with S. tmatt. At day 40 p.i., animals with a conventional microbiota (C-mice) were added to one group (black circles; n = 5; S. tmatt→L/C). The other group remained under hygienic isolation (open circles; n = 5; S. tmatt→L). A. All mice were sacrificed at day 80 p.i. and S. tmatt levels in the cecal content were defined. B. Total MLN cells from the two groups of mice were isolated and analyzed with respect to the expression of intracellular cytokines IL-17A (left) and IFNγ (right) in CD4+ T-cells at day 80 post S. tmatt infection (% cytokine-expressing CD4+ T-cells). C. 3×107 MLN cells of ‘donor’ mice S. tmatt→L and S. tmatt→L/C described above were adoptively transferred into another cohort of S. tmatt→L mice (day 40 p.i.: = ‘aceptors’). Fecal S. tmatt shedding was followed in the ‘acceptors’ until day 80 post S. tmatt infection ( = day 40 post adoptive transfer, left). S. tmatt levels in the MLN of ‘acceptors’ at day 80 post S. tmatt infection ( = day 40 post adoptive transfer, left). Discussion
In this study, we have defined the contributions of sIgA and the microbiota in protecting the host from NTS infection. During the first encounter with the pathogen, the microbiota mediates at least two different protective functions, colonization resistance and pathogen clearance. The former is well established and prohibits the growth of diverse incoming pathogens, thus preventing colonization right on [3], [5], [20], [44]. Here, we identified pathogen clearance as a second protective function attributable to the microbiota. Pathogen clearance eliminates the pathogen from the gut lumen after an episode of acute infection, i.e. after Salmonella diarrhea. This differs from colonization resistance as the pathogen starts out at high density and the normal microbiota must re-grow from a state of depletion and disturbed composition (i.e. caused by the pathogen and the inflammatory response). Compared to the microbiota, an adaptive sIgA response mounted during the later stages of an acute infection contributes little to clearing the pathogen from the gut lumen. However, pathogen-specific sIgA protects from mucosal inflammation if the same pathogen is encountered for a second time. Thus, the microbiota and sIgA have complementary functions which jointly protect against enteropathogenic bacteria during the initial infection and subsequent exposure.
How does the microbiota mediate pathogen clearance? The lack of suitable assay systems has hampered addressing this question in the past. Clearly, S. tm clearance starts out in a situation where the pathogen has grown up in the gut lumen, inflicted disease and thereby slashed microbiota density, composition and/or function [45]. This situation is gradually reversed, involving the decrease of luminal pathogen loads as well as microbiota re-growth. Finally, normal microbiota composition, density and function are restored. Conceivably, some of the mechanisms conferring colonization resistance, i.e. bacteriocin production, inhibitory metabolites, oxygen depletion, receptor blocking, stimulating mucin - or antimicrobial peptide release, stabilization of the mucosal barrier, improvement of gut motility and/or nutrient limitation [5], might also contribute to different phases of pathogen clearance. Also, microbiota-mediated stimulation of the mucosal cellular immune system may be involved [2], [46] even though TH-17 mediated responses do not seem to contribute significantly to S. tm clearance, as indicated by adoptive transfer experiments (Fig. 8). The mechanisms mediating clearance and the relative importance of the microbiota, sIgA and other mucosal immune responses may differ between different pathogens or even between different strains of a given pathogen. Identifying the commensal species (or consortia) involved and the molecular mechanisms mediating pathogen clearance will be an important task for future research.
Does sIgA contribute to pathogen clearance? Pathogen specific sIgA is produced by wild type animals during the phase of pathogen clearance. O-antigen-specific sIgA led to aggregation of luminal pathogens, prevented access to the enterocyte surface and reduced net pathogen growth as indicated by a reduced competitive index. Surprisingly, this had little effect on S. tm clearance. Wild type mice and IgA−/− littermates displayed equivalent rates of pathogen clearance. Moreover, sIgA did not reduce pathogen loads in the stool, at least in the L-mice in the absence of a complex gut flora. Thus, for pathogen clearance at the end of a primary enteric S. tm infection, a pathogen-specific sIgA response is neither necessary nor sufficient.
Instead, sIgA protected from mucosal inflammation upon re-infection with the same pathogen. The LPS O-antigen was the key protective antigen of this adaptive immune response. Protection was attributable to pathogen-aggregation in the gut lumen, reduced net pathogen growth and pathogen-exclusion from the epithelial surface, thus inhibiting pathogen invasion into the gut tissue. This required sIgA transport into the gut lumen, as immunized pIgR−/− mice, which fail to transport sIgA across the intestinal epithelium, have equivalent pathogen loads in the gut mucosa as non-immunized littermates or wild type animals. Thus, pathogen specific antibodies do not seem to contribute much to protection, once S. tm has breached the mucosal barrier. This is in line with earlier work on the roles of antibody responses in systemic S. tm infection [17], [47]. In conclusion, our experiments show that O-antigen-specific sIgA responses protect against Salmonella-mediated gut inflammation upon re-infection.
It is interesting to consider the protective function of sIgA and the microbiota from an evolutionary perspective. The intestines of most animals are colonized by bacterial communities [43]. It seems safe to assume that microbiota have an evolutionary ancient function in protecting from infection. We speculate that this pertains to colonization resistance and to pathogen clearance and that both have evolved to provide protection against a broad range of pathogens. The elaborate adaptive immune system of modern mammals, including sIgA responses, evolved much later. It evolved in the presence of the protective functions provided by the microbiota, i.e. colonization resistance and pathogen clearance. The high efficiency of this microbiota-mediated protection may explain why sIgA responses have not evolved to affect the first round of infection with a given pathogen. This was simply not necessary. In contrast, evolving the sIgA response to protect in the case of repeated exposure to the same pathogen may have represented a strong benefit which cannot be accomplished by the microbiota. This evolutionary history may explain why the sIgA response contributes little during the primary infection. Anyhow, in modern mammals the microbiota and sIgA have quite different protective functions which complement each other during the initial - and subsequent encounters with a given pathogen.
An ‘unfavorable’ microbiota composition, e.g. in L-mice, can result in long term shedding. Asymptomatic NTS excretion is also observed in humans recovering from acute diarrhea. This period of asymptomatic excretion normally lasts for two to eight weeks, but may last for more than a year in a few patients. This poses a risk of transmission. In analogy to the long term shedding by L-mice, we propose that these individuals might lack some unidentified component of the normal microbiota. In L-mice, the pathogen is cleared upon transferring microbiota from a healthy donor. This may have implications for managing human long term asymptomatic excretors. Traditionally, patients are advised to adhere to strict personal hygiene and might even be isolated in order to reduce the risk of transmission. However, at the same time this deprives the patients from exposure to conventional microbiota from healthy individuals which might enhance pathogen clearance. So far, we do not know the species of the microbiota, the cellular interactions, and molecular mechanisms explaining pathogen clearance. However, the experimental systems presented in our study may provide the tools to address these important issues. Our findings provide a basis for future research on optimal management of ‘asymptomatic excretors’, NTS vaccine development and microbiota-directed therapy for acute diarrheal NTS infections.
Materials and Methods
Animals
Specified pathogen-free (SPF) wild type C57BL/6 mice, JH−/− [48] and IgA−/− [49], pIgR−/− [50] and TCRβ−/−δ−/− mice [51] (7–10 weeks old; all C57BL/6 background) were bred at the Rodent center HCI (RCHCI) under barrier conditions in individually ventilated cages (Ehret). IgA−/−, IgA+/− and IgA+/+ littermates were generated by crossing IgA−/− with C57BL/6 mice and breeding IgA+/−×IgA+/− animals. L-mice were generated by colonizing germfree C57BL/6 mice with the Altered Schaedler flora (ASF). Mice, housed in a bubble isolator, were inoculated at eight weeks of age by intra-gastric and intra-rectal administration of 107–108 cfu of ASF bacteria on consecutive days (www.taconic.com/library). Later, L-mice (C57BL/6 background) were maintained under barrier conditions in IVCs with autoclaved chow and autoclaved, acidified water. Mice with complex microbiota were never housed together with these in the same room to prevent contamination with additional commensal bacteria.
Ethics statement
All animal experiments were approved (license 201/2004 and 201/2007 Kantonales Veterinäramt Zürich) and performed according to local guidelines (TschV, Zurich) and the Swiss animal protection law (TschG).
Infection experiments
Salmonella infections were performed in individually ventilated cages at the RCHCI, Zurich, as previously described [52]. In brief, wild type C57BL/6 mice, JH−/−, IgA−/−, pIgR−/− and TCRβ−/−δ−/− mice were pretreated with 20mg of streptomycin (sm) by gavage. 24h later, the mice were inoculated with 5×107 CFU of S. tmatt, PBS (mock) or as indicated. ‘Challenge infections’ were performed 40 days later (or as indicated). Mice were treated with ampicillin (20mg; by gavage) and 24h later infected with a dose of 200 CFU of the respective ampicillin-resistant (pM973) challenge strain. Samples of cecal tissue were cryo-embedded, and inflammation was quantified on cryosections (5 µm, cross-sectional) stained with hematoxylin and eosin (H&E). Pathogen-colonization was assessed as described, below.
Histology
H&E-stained cecum cryosections were scored as described, evaluating submucosal edema, PMN infiltration, goblet cells and epithelial damage yielding a total score of 0–13 points [53].
Analysis of Salmonella loads in cecal content, mesenteric lymph nodes, and spleen
Mesenteric lymph nodes (MLN), spleen and liver were removed aseptically and homogenized in cold PBS (0.5% tergitol, 0.5% BSA). The cecum content was suspended in 500ìl cold PBS and bacterial loads were determined by plating on MacConkey agar plates (50 ìg ml−1 streptomycin) as described [21]. Colonization levels of the challenge strain (carrying pM973 with an antibiotic marker) and immunization strain (S. tmatt; kmR) were determined by selective plating (100ìg ml−1 ampicillin or 30 ìg ml−1 kanamycin; levels of challenge strain: ampR-kmR Salmonella). For co-infection experiments shown in Fig. 2C, competitive indices were calculated according to the formula CI = ratio S. tm:S. encecal content/ratio S. tm:S. eninoculum.
Adoptive transfer of MLN cells
MLN were harvested from S. tmatt→L (total of 5 mice) and S. tmatt→L/C (total of 5 mice) at day 80 post S. tmatt infection. Single cell suspensions were prepared using 100µm cell strainers and 40µg/ml DNAse (Roche). S. tmatt→L mice (at day 40 post S. tmatt infection) were injected intravenously with 3×107 cells (pooled for each of the two groups) in 200µl PBS. Fecal S. tmatt shedding was monitored for another 40 days.
Intracellular cytokine staining
Single-MLN-cell suspensions were prepared as described above. For intracellular staining of IFN-γ and IL-17A, 1×107 nucleated MLN cells were cultured for 3 h in 1 ml of RPMI 1680 supplemented with 10% heat-inactivated FCS and stimulated with PMA (5pg/ml) /ionomycin (500pg/ml). After adding 20 µg/ml brefeldin A, the cells were incubated for another 3h at 37°C. Cells were harvested and washed in ice-cold FACS buffer (PBS, 2% heat-inactivated FCS, 5 mM EDTA, and 0.02% sodium azide). Cells were resuspended in FACS buffer and stained on the surface with fluorescently-labeled antibodies for 30 min on ice. For intracellular staining of IL-17A and IFN-γ, cells were washed once and fixed/permeabilized for 10 min at room temperature using 500 µl of FIX/perm solution (FACSLyse; BD Biosciences; diluted to 2× concentration in distilled water and 0.05% Tween 20). Cells were washed once and stained with directly conjugated Abs against IFN-γ-APC (BD) and IL-17A-PE (Biolegend). Cells were then washed again and resuspended in PBS. Data were collected on a LSRII flow cytometer (BD Biosciences) and analyzed using FlowJo software (Tree Star).
Gut wash
The intestine was flushed with 2 ml of a washing buffer containing PBS, 0.05M EDTA pH8.0 and 66µM PMSF. Intestinal wash was briefly vortexed and centrifuged at 4°C, 30 min, 40.000 rpm (Eppendorf centrifuge). Aliquots of supernatants were stored at −80°C.
Immunofluorescence microscopy
Bacteria harboring pM973 (GFP expression after tissue entry) in the lamina propria and epithelium were enumerated by fluorescence microscopy as described [22] using cryo-sections of PFA-fixed cecal tissue stained with Armenian hamster anti-CD54 (clone 3E2; stains lamina propria) antibody (Becton Dickinson), Cy3-conjugated goat anti–Armenian hamster Ig (Jackson ImmunoResearch Laboratories), DAPI (stains DNA; Sigma-Aldrich), and Alexa647-conjugated phalloidin (stains polymerized actin; Fluoprobes). We evaluated three 20 µm thick sections of the cecum per mouse and plotted for each mouse the average of the three values.
For detecting S. tm and S. en pM979 in the gut lumen in situ, cecal tissues were recovered and treated as described recently [54]. Briefly, the tissues were fixed in paraformaldehyde (4% in PBS, pH 7.4 over night, 4°C), washed with PBS, equilibrated in PBS (20% sucrose, 0.1% NaN3 over night, 4°C), embedded in O.C.T. (Sakura, Torrance, CA), snap-frozen in liquid nitrogen and stored at −80°C. Cryosections (7ìm) were air-dried for 2 h at room temperature, fixed in 4% paraformaldehyde (5 min), washed and blocked in 10% (w/v) normal goat serum in PBS for 1h. S. tm was detected by staining for 1h with a polyclonal rabbit á-Salmonella-O-antigen group B serum (factors 1, 4, 5 and 12, Difco; 1∶500 in PBS, 10% (w/v) goat serum) and Cy3-conjugated secondary goat-α-rabbit antibody. S. en pM979 expresses gfp under the control of a constitutive promoter and bacteria were detected in the green channel. F-Actin (epithelial brush border) was visualized by staining with Alexa-647-conjugated phalloidin, as indicated (Molecular Probes). Sections were mounted with Vectashield hard set (Vector laboratories) and sealed with nail polish. Images were recorded with a microscope (Axiovert 200; Carl Zeiss, Inc.), an Ultraview confocal head (PerkinElmer), and a krypton argon laser (643-RYB-A01; Melles Griot). Infrared, red, and green fluorescence was recorded confocally, and blue fluorescence was recorded by epifluorescence microscopy.
Immunohistochemistry
Frozen consecutive sections of spleen, liver, cecum, colon and small intestine (7ìm thick) were briefly fixed (10 min) in acetone and blocked for 30 min with phosphate buffered saline (PBS) containing 0.5% bovine serum albumin (BSA). Sections were then incubated with the primary antibody for 1 h at room temperature. Primary antibodies included: B220/CD45R (Pharmingen 553084; 1∶200) , CD4 (clone YTS191; 1∶200) and CD8 (clone YTS169; 1∶200) for T-cells (kindly provided by Rolf Zinkernagel; 1∶50), Ly-6G (Gr-1) for neutrophils (clone RB6-8C5; 1∶600), F4/80 for macrophages (Serotec MCAP 497; 1∶50), CD11c for dendritic cells (BD Biosciences 553800; 1∶100) and IgA (rat-anti-mouse IgA; Pharmingen 556969; clone C10-3; 1∶4000). Secondary antibodies and detection chromagens were applied and visualized using standard methods (see also [55]).
Statistical analysis
Statistical analysis was performed using the exact Mann-Whitney U test (Prism 4.0c). A P value of <0.05 (two tailed) was considered to be statistically significant. In mouse experiments, values were set to the minimal detectable value (10 cfu for cecum; 10 CFU for MLNs; 20 CFU for the spleen) for samples harboring “no bacteria.” Two figures (Fig. 1A and 4F) were generated using the statistical software package R. To assess the distribution of Salmonella loads in mice during the 60 day infection experiments, median and quantiles (corresponding to 0.05, 0.25, 0.75 and 0.95 probabilities) were plotted for each day or group of days. We performed a linear regression on medians and both 0.05 and 0.95 quantiles, weighted by the number of data points sampled for each day.
The OTUs abundance heatmap represents the mouse normalized OTU abundances (log2) clustered by average linkage clustering on Euclidean distances. This was generated using the function ‘heatmap.2’ from the ‘gplots’ R library.
Immunoblot analysis
The equivalent of 1 OD600 units/ml (where OD600 is the optical density at 600nm) of liquid o.n. cultures of S. tmΔO, Lactobacillus reuteri RR, Enterococcus faecalis, S. enwt, E.coli, S. tmwt, S. tmwt proteinase K treated (Gibco/Life Technologies; 0.4 mg/ml; 1h 57°C) or S. tm M933 was pelleted by centrifugation at 14,000×g for 2 min, and the supernatant was discarded. Cells were resuspended in Laemmli sample buffer (0.065 M Tris-HCl [pH 6.8], 2% [wt/vol] sodium dodecyl sulfate [SDS], 5% [vol/vol] β-mercaptoethanol, 10% [vol/vol] glycerol, 0.05% [wt/vol] bromophenol blue) and lysed for 5 min at 95°C. Equal amounts of the different strains and purified S. tm flagellin FliC were loaded on a 12% SDS-polyacrylamide gel and proteins wer separated by electrophoresis. Immunoblots were stained with mouse serum (diluted 1∶200 in PBS) or intestinal lavages (diluted 1∶20 in PBS) from naïve or immunized mice, goat-α-mouse-IgA HRP (Southern Biotech), goat-anti-mouse-IgG HRP (Bethyl Laboratories) and developed using an ECL kit (Amersham). The same protocol was used for the analysis of the human patient serum (dilution 1∶20 in PBS), where goat-anti-human-IgA-HRP (2050-05, NEB) and goat-anti-human-IgG-HRP (2040-05, NEB) were used as secondary antibodies.
Bacterial FACS
Analysis was performed as described in [30]. 3ml LB cultures were inoculated from single colonies of plated bacteria and cultured overnight at 37°C without shaking. 1ml of culture was gently pelleted for 4min at 7,000 rpm in an Eppendorf minifuge and washed 3× with sterile-filtered PBS (1% BSA, 0.05% sodium azide) before resuspending to yield a final density of 107 bacteria per ml. Mouse serum was diluted 1∶20 in PBS (1% BSA, 0.05% sodium azide) and heat-inactivated at 60°C for 30min. The serum solution was then spun at 13,000 rpm in an Eppendorf minifuge for 10min to remove any bacteria-sized contaminants and the supernatant was used to perform serial dilutions (1∶20, 1∶60, 1∶180). 25µl serum solution and 25µl bacterial suspension were mixed and incubated at 4°C for 1h. Bacteria were washed twice before resuspending in monoclonal FITC-anti-mouse IgA (559354; BD Pharmingen), PE-anti-mouse total IgG (715-116-151; Jackson Immunoresearch Europe) and APC-anti-mouse IgM (550676; BD Pharmingen). After a further hour of incubation bacteria were washed once with PBS (1% BSA, 0.05% sodium azide) and then resuspended in PBS (2% PFA) for acquisition on a FACSCalibur using FSC and SSC parameters in logarithmic mode. Data were analysed using FlowJo software (Treestar, USA). Analysis of specific IgA in intestinal lavages was achieved using an identical protocol, using a dilution of 1∶2, 1∶6 and 1∶18 of gut wash.
Microarray-Analysis
The cecum tissue was excised (3 biological replicates per group), washed in cold PBS, placed in 300µl RLT-buffer (RNeasy Mini Kit, Qiagen; 1% β-Mercaptoethanol) and snap-frozen in liquid nitrogen. Total RNA was extracted with the Nucleospin RNA II kit (Macherey Nagel, Germany) and prepared for hybridization as recommended by the manufacturer (Applied biosystems, USA). Briefly, 2µg of total RNA and a T7-oligo(dT) primer were used for reverse transcription. The double-stranded cDNA was purified and converted to DIG labeled-cRNA by in vitro transcription using DIG-UTP (Roche, Germany). The cRNA was purified, fragmented and hybridized on ABI Mouse Genome Survey v2.0 microarrays for 16h. The microarray was washed and incubated with anti-DIG antibodies conjugated to alkaline phosphatase and a chemiluminescent substrate. The microarrays were scanned with the Applied Biosystems 1700 chemiluminescent microarray analyzer [56]. Normalization was achieved using the NeONORM method [57]. Significance of log2 fold changes (log2Q) were determined based on a double-log normal distribution hypothesis of signal intensities using mixture ANOVA methodology [56]. A change in the gene expression profiles was considered as significant if p<0.001. Heat maps were created according to standard methods [56]. Gene Ontology (GO) annotations were analyzed using the Panther Protein Classification System (http://www.pantherdb.org). Microarray data were deposited in the publicly available database: http://mace.ihes.fr with accession number: 2947924142.
Bacterial DNA extraction and 16SrRNA gene specific PCR
Total DNA was extracted from cecal contents using a QIAmp DNA stool mini kit (Qiagen). Bacterial lysis was enhanced using 0.1mm glass beads in buffer ASF and a Tissuelyzer device (5 minutes, 30Hz; Qiagen). V5-V6 regions of bacterial 16S rRNA were amplified using primers B-V5 (5′ GCCTTGCCAGCCCGCTCAG ATT AGA TAC CCY GGT AGT CC 3′) and A-V6-TAGC (5′GCCTCCCTCGCGCCATCAG [TAGC] ACGAGCTGACGACARCCATG 3′). The brackets contain one of the 20 different 4-mer tag identifiers [TAGC, TCGA, TCGC, TAGA, TGCA, ATCG, AGCT, AGCG, ATCT, ACGT, GATC, GCTA, GCTC, GATA, GTCA, CAGT, CTGA, CAGA, CTGT, CGTA]. Cycling condition were as follows: 95°C, 10min; 22 cycles of (94°C, 30s; 57°C, 30s; 72°C, 30s); 72°C, 8min; 4°C, ∞; Reaction conditions (50µl) were as follows: 50ng template DNA; 50 mM KCl, 10 mM Tris-HCl pH 8.3, 1.5 mM Mg2+, 0.2mM dNTPs; 40pmol of each primer, 5U of Taq DNA polymerase (Mastertaq; Eppendorf).
PCR products of different reactions were pooled, ethanol-precipitated and fragments of ∼300bp were purified by gel electrophoresis, excised and recovered using a gel-extraction kit (Machery-Nagel). Amplicon sequencing of the PCR products was performed using a 454 FLX instrument (70×70 Picotitre plate) according to the protocol recommended by the supplier (www.454.com). PCR to detect ASF bacteria in the feces was done as described in [42].
Quality filtering and reads sorting
We applied quality control to 454 reads in order to avoid artificial inflation of ecosystem diversity estimates [58]. Reads containing the consensus sequence (‘ACGAGCTGACGACA[AG]CCATG’) of the V6 reverse primer were filtered with respect to their length (200nt≤length≤300nt). Quality filtering was then applied to include only sequences containing one of the exact 4nt tag sequences and displaying at maximum one ambiguous nucleotide ‘N’. The latter criterion has been reported as a good indicator of sequence quality for a single read [41]. We identified 6,754 reads with an incorrect primer sequence, 1,155 reads shorter than 200nt, 8 reads longer than 300nt and 119 reads containing more than one ‘N’. After filtering, 140,237 reads remained (out of an initial total of 149,786 raw reads) and were processed as described below for OTU definition and chimera filtering. To estimate the reliability of sample discrimination using our primer-tagging approach, we assessed the number of reads observed to have an illegitimate 4-mer tag (i.e., different from our set of 20 tags). The sequencing plate produced a total of 141,784 quality-filtered reads from which 1,547 contained an incorrect tag (1.09%). Given that 256 distinct 4-mer tags are possible and that we used only 20 of these, the majority of sequencing or primer errors in this region are detectable. Correcting for the small fraction of undetectable errors (20/256) and division by four yields an error rate of 0.296% per single nucleotide - at the position of the tag in the primer (this includes errors during primer synthesis as well as sequencing). Since most errors are actually visible as errors, the rate of unintentional ‘miscall’ of sample identity is 0.092%.
Definition of OTUs
To reduce computational time and complexity, we built OTUs using the complete filtered dataset covering all non-redundant reads from the 20 samples. Exactly identical sequences were represented by one representative only; after OTU computation, redundant sequences were taken into account for OTU abundance analysis. For subsequent taxonomy classification, we included additional quality-filtered 16S rRNA reference sequences, selected from the Greengenes database (http://greengenes.lbl.gov/Download/Sequence_Data/Greengenes_format/greengenes16SrRNAgenes.txt.gz, release 01-28-2009 [59]). This reference database is based on full-length non-chimerical sequences with a minimum length of 1100nt (in order to fully cover the V6 region of all entries). No archaeal sequences were included in the analysis.
The alignment of non-redundant reads from all mice with the reference database was performed using the secondary-structure aware Infernal aligner (http://infernal.janelia.org/, release 1.0, [60]) and based on the 16S rRNA bacterial covariance model of the RDP database (http://rdp.cme.msu.edu/; [61]). Before defining OTUs, we first removed reference sequences for which the alignment was not successful (Infernal bit-score<0). The alignment was then processed to include an equivalent amount of information from every read. To do so, we identified the consensus reverse primer sequence of the V6 region within the aligned sequence of Escherichia coli K12, as a reference. The full alignment was then trimmed from the start position (defined by the E. coli V6 reverse primer) and ended after 200nt. This also ensured a further limitation of the effect of pyrosequencing errors by trimming the 3′ end of each read, a region which is more sequencing-error prone (the trimmed and aligned reads length ranged from 152 to 231nt) [58]. Using this alignment, OTUs were built by hierarchical cluster analysis at various distances (0.01, 0.03, 0.05 and 0.10) using the ‘complete linkage clustering’ tool of the RDP pyrosequencing pipeline http://pyro.cme.msu.edu/; [61]).
Taxonomy assignment
In a first step, taxonomy was inferred for all reads using the stand-alone version of the RDP classifier (http://sourceforge.net/projects/rdp-classifier, revision 2.0, [62]). Taxon-level predictions were considered reliable when supported by a minimum bootstrap value of 80%. In order to predict taxonomy for each OTU, we either used any reference sequences present within a cluster, or the taxonomy of the reads present in the cluster, as predicted by the RDP classifier. To increase the resolution of the prediction, we privileged any reference sequences over the reads. For each OTU, taxonomy was inferred by a simple majority vote: if more than half of the reference sequences (or reads) present within a cluster agreed on a taxon, the OTU was annotated according to this taxon. In case of conflicts, we assigned a consensus taxon to a higher phylogenetic level for which the majority vote condition was met.
Chimera estimation
Deep pyrosequencing on the 454 platform has revealed extensive microbial diversity that was previously undetected with culture-dependent methods [63]. Nevertheless, sequencing data generated from pools of PCR products have to be interpreted carefully; limitations and biases of the PCR technique have to be taken into account. This can lead to over-estimations of microbial diversity as has been recently reported [58], [64]. Moreover, during amplification, chimerical sequences can be generated.
On such short sequences, recombination points (recombination can occur from an incompletely extended primer or by template-switching; [65]) are extremely difficult to detect. Recently, a new tool to filter noise and remove chimera in 454 pyrosequencing data has been published [64]. There, the authors suggest that because of sequencing errors, diversity estimates may be at least an order of magnitude too high. To our best knowledge, at the time of our analysis, there were no available tools to detect chimera within libraries of short 454 reads. Therefore, in order to detect chimeras we decided to compare taxonomies assigned to N-terminal and C-terminal read fragments using BLASTn. In order to ensure a reasonable alignment length and a relatively high identity to the matching reference sequences, we only analyzed reads for which both fragments had a minimum identity of 95% and a minimum bit-score of 150 (these cutoffs were selected heuristically). A given read was deemed chimeric when the taxonomies of the best hits of each half were clearly not congruent (i.e., differing at the phylum level). Our simple chimera detection method resulted in a slightly higher rate of detected chimera compared to the method of Quince et al., 2009 (∼4.5% compare to ∼3% in their example), suggesting that our approach is at least of comparable stringency [64].
Supporting Information
Zdroje
1. BackhedF
LeyRE
SonnenburgJL
PetersonDA
GordonJI
2005 Host-bacterial mutualism in the human intestine. Science 307 1915 1920
2. IvanovII
AtarashiK
ManelN
BrodieEL
ShimaT
2009 Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139 485 498
3. van der WaaijD
Berghuis-de VriesJM
LekkerkerkL-v
1971 Colonization resistance of the digestive tract in conventional and antibiotic-treated mice. J Hyg (Lond) 69 405 411
4. VollaardEJ
ClasenerHA
1994 Colonization resistance. Antimicrob Agents Chemother 38 409 414
5. StecherB
HardtWD
2008 The role of microbiota in infectious disease. Trends Microbiol 16 107 114
6. BarthelM
HapfelmeierS
Quintanilla-MartinezL
KremerM
RohdeM
2003 Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect Immun 71 2839 2858
7. BarmanM
UnoldD
ShifleyK
AmirE
HungK
2008 Enteric salmonellosis disrupts the microbial ecology of the murine gastrointestinal tract. Infect Immun 76 907 915
8. HohmannEL
2001 Nontyphoidal salmonellosis. Clin Infect Dis 32 263 269
9. DethlefsenL
HuseS
SoginML
RelmanDA
2008 The Pervasive Effects of an Antibiotic on the Human Gut Microbiota, as Revealed by Deep 16S rRNA Sequencing. PLoS Biol 6 e280
10. DoreK
BuxtonJ
HenryB
PollariF
MiddletonD
2004 Risk factors for Salmonella typhimurium DT104 and non-DT104 infection: a Canadian multi-provincial case-control study. Epidemiol Infect 132 485 493
11. BuchwaldDS
BlaserMJ
1984 A review of human salmonellosis: II. Duration of excretion following infection with nontyphi Salmonella. Rev Infect Dis 6 345 356
12. BalfourAE
LewisR
AhmedS
1999 Convalescent excretion of Salmonella enteritidis in infants. J Infect 38 24 25
13. GreigJD
ToddEC
BartlesonCA
MichaelsBS
2007 Outbreaks where food workers have been implicated in the spread of foodborne disease. Part 1. Description of the problem, methods, and agents involved. J Food Prot 70 1752 1761
14. BaderMW
SanowarS
DaleyME
SchneiderAR
ChoU
2005 Recognition of antimicrobial peptides by a bacterial sensor kinase. Cell 122 461 472
15. GunnJS
LimKB
KruegerJ
KimK
GuoL
1998 PmrA-PmrB-regulated genes necessary for 4-aminoarabinose lipid A modification and polymyxin resistance. Mol Microbiol 27 1171 1182
16. BrownA
HormaecheCE
1989 The antibody response to salmonellae in mice and humans studied by immunoblots and ELISA. Microb Pathog 6 445 454
17. MittruckerHW
RaupachB
KohlerA
KaufmannSH
2000 Cutting edge: role of B lymphocytes in protective immunity against Salmonella typhimurium infection. J Immunol 164 1648 1652
18. UrenTK
WijburgOL
SimmonsC
JohansenFE
BrandtzaegP
2005 Vaccine-induced protection against gastrointestinal bacterial infections in the absence of secretory antibodies. Eur J Immunol 35 180 188
19. MaaserC
HousleyMP
IimuraM
SmithJR
VallanceBA
2004 Clearance of Citrobacter rodentium requires B cells but not secretory immunoglobulin A (IgA) or IgM antibodies. Infect Immun 72 3315 3324
20. StecherB
ChaffronS
KappeliR
HapfelmeierS
FreedrichS
2010 Like will to like: abundances of closely related species can predict susceptibility to intestinal colonization by pathogenic and commensal bacteria. PLoS Pathog 6 e1000711
21. StecherB
RobbianiR
WalkerAW
WestendorfAM
BarthelM
2007 Salmonella enterica Serovar Typhimurium Exploits Inflammation to Compete with the Intestinal Microbiota. PLoS Biol 5 e244
22. HapfelmeierS
StecherB
BarthelM
KremerM
MüllerA
2005 The Salmonella Pathogenicity Island (SPI)-1 and SPI-2 Type III Secretion Systems Allow Salmonella Serovar Typhimurium to trigger Colitis via MyD88-Dependent and MyD88-Independent Mechanisms. J Immunol 174 1675 1685
23. CoombesBK
CoburnBA
PotterAA
GomisS
MirakhurK
2005 Analysis of the contribution of Salmonella pathogenicity islands 1 and 2 to enteric disease progression using a novel bovine ileal loop model and a murine model of infectious enterocolitis. Infect Immun 73 7161 7169
24. HapfelmeierS
HardtWD
2005 A mouse model for S. typhimurium-induced enterocolitis. Trends Microbiol
25. CurtissR3rd
KellySM
HassanJO
1993 Live oral avirulent Salmonella vaccines. Vet Microbiol 37 397 405
26. SimonsenJ
MolbakK
FalkenhorstG
KrogfeltKA
LinnebergA
2009 Estimation of incidences of infectious diseases based on antibody measurements. Stat Med 28 1882 1895
27. AngelakopoulosH
HohmannEL
2000 Pilot study of phoP/phoQ-deleted Salmonella enterica serovar typhimurium expressing Helicobacter pylori urease in adult volunteers. Infect Immun 68 2135 2141
28. KangHY
SrinivasanJ
CurtissR3rd
2002 Immune responses to recombinant pneumococcal PspA antigen delivered by live attenuated Salmonella enterica serovar typhimurium vaccine. Infect Immun 70 1739 1749
29. BergmanMA
CummingsLA
BarrettSL
SmithKD
LaraJC
2005 CD4+ T cells and toll-like receptors recognize Salmonella antigens expressed in bacterial surface organelles. Infect Immun 73 1350 1356
30. SlackE
HapfelmeierS
StecherB
VelykoredkoY
StoelM
2009 Innate and adaptive immunity cooperate flexibly to maintain host-microbiota mutualism. Science 325 617 620
31. MartinoliC
ChiavelliA
RescignoM
2007 Entry route of Salmonella typhimurium directs the type of induced immune response. Immunity 27 975 984
32. WijburgOL
UrenTK
SimpfendorferK
JohansenFE
BrandtzaegP
2006 Innate secretory antibodies protect against natural Salmonella typhimurium infection. J Exp Med 203 21 26
33. MacphersonAJ
GattoD
SainsburyE
HarrimanGR
HengartnerH
2000 A primitive T cell-independent mechanism of intestinal mucosal IgA responses to commensal bacteria. Science 288 2222 2226
34. BrandtzaegP
2009 Mucosal immunity: induction, dissemination, and effector functions. Scand J Immunol 70 505 515
35. SuarM
PeriaswamyB
SonghetP
MisselwitzB
MullerA
2009 Accelerated type III secretion system 2-dependent enteropathogenesis by a Salmonella enterica serovar enteritidis PT4/6 strain. Infect Immun 77 3569 3577
36. IlgK
EndtK
MisselwitzB
StecherB
AebiM
2009 O-antigen-negative Salmonella enterica serovar Typhimurium is attenuated in intestinal colonization but elicits colitis in streptomycin-treated mice. Infect Immun 77 2568 2575
37. TurnbaughPJ
HamadyM
YatsunenkoT
CantarelBL
DuncanA
2008 A core gut microbiome in obese and lean twins. Nature
38. DuerkopBA
VaishnavaS
HooperLV
2009 Immune responses to the microbiota at the intestinal mucosal surface. Immunity 31 368 376
39. BohnhoffM
DrakeBL
MillerCP
1954 Effect of streptomycin on susceptibility of intestinal tract to experimental Salmonella infection. Proc Soc exp Biol, NY 86 132 137
40. TurnbaughPJ
HamadyM
YatsunenkoT
CantarelBL
DuncanA
2009 A core gut microbiome in obese and lean twins. Nature 457 480 484
41. HuseSM
DethlefsenL
HuberJA
WelchDM
RelmanDA
2008 Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. PLoS Genet 4 e1000255
42. Sarma-RupavtarmRB
GeZ
SchauerDB
FoxJG
PolzMF
2004 Spatial distribution and stability of the eight microbial species of the altered schaedler flora in the mouse gastrointestinal tract. Appl Environ Microbiol 70 2791 2800
43. LeyRE
HamadyM
LozuponeC
TurnbaughPJ
RameyRR
2008 Evolution of mammals and their gut microbes. Science 320 1647 1651
44. NardiRM
SilvaME
VieiraEC
BambirraEA
NicoliJR
1989 Intragastric infection of germfree and conventional mice with Salmonella typhimurium. Braz J Med Biol Res 22 1389 1392
45. RawlsJ
2007 Enteric infection and inflammation alter gut microbial ecology. Cell host and microbe 2 73 74
46. Gaboriau-RouthiauV
RakotobeS
LecuyerE
MulderI
LanA
2009 The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity 31 677 689
47. MittruckerHW
KaufmannSH
2000 Immune response to infection with Salmonella typhimurium in mice. J Leukoc Biol 67 457 463
48. ChenJ
TrounstineM
AltFW
YoungF
KuraharaC
1993 Immunoglobulin gene rearrangement in B cell deficient mice generated by targeted deletion of the JH locus. Int Immunol 5 647 656
49. HarrimanGR
BogueM
RogersP
FinegoldM
PachecoS
1999 Targeted deletion of the IgA constant region in mice leads to IgA deficiency with alterations in expression of other Ig isotypes. J Immunol 162 2521 2529
50. UrenTK
JohansenFE
WijburgOL
KoentgenF
BrandtzaegP
2003 Role of the polymeric Ig receptor in mucosal B cell homeostasis. J Immunol 170 2531 2539
51. MombaertsP
MizoguchiE
LjunggrenHG
IacominiJ
IshikawaH
1994 Peripheral lymphoid development and function in TCR mutant mice. Int Immunol 6 1061 1070
52. StecherB
BarthelM
SchlumbergerMC
HaberliL
RabschW
2008 Motility allows S. Typhimurium to benefit from the mucosal defence. Cell Microbiol 10 1166 1180
53. HapfelmeierS
MullerAJ
StecherB
KaiserP
BarthelM
2008 Microbe sampling by mucosal dendritic cells is a discrete, MyD88-independent step in DeltainvG S. Typhimurium colitis. J Exp Med 205 437 450
54. StecherB
HapfelmeierS
MullerC
KremerM
StallmachT
2004 Flagella and Chemotaxis Are Required for Efficient Induction of Salmonella enterica Serovar Typhimurium Colitis in Streptomycin-Pretreated Mice. Infect Immun 72 4138 4150
55. MuellerC
2002 Tumour necrosis factor in mouse models of chronic intestinal inflammation. Immunology 105 1 8
56. EilebrechtS
PellayFX
OdenwalderP
BrysbaertG
BeneckeBJ
2008 EBER2 RNA-induced transcriptome changes identify cellular processes likely targeted during Epstein Barr Virus infection. BMC Res Notes 1 100
57. NothS
BrysbaertG
BeneckeA
2006 Normalization using weighted negative second order exponential error functions (NeONORM) provides robustness against asymmetries in comparative transcriptome profiles and avoids false calls. Genomics Proteomics Bioinformatics 4 90 109
58. KuninV
EngelbrektsonA
OchmanH
HugenholtzP
2009 Wrinkles in the rare biosphere: pyrosequencing errors lead to artificial inflation of diversity estimates. Environ Microbiol
59. DeSantisTZ
HugenholtzP
LarsenN
RojasM
BrodieEL
2006 Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72 5069 5072
60. EddySR
2002 A memory-efficient dynamic programming algorithm for optimal alignment of a sequence to an RNA secondary structure. BMC Bioinformatics 3 18
61. ColeJR
WangQ
CardenasE
FishJ
ChaiB
2009 The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 37 D141 145
62. WangQ
GarrityGM
TiedjeJM
ColeJR
2007 Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73 5261 5267
63. SoginML
MorrisonHG
HuberJA
WelchDM
HuseSM
2006 Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc Natl Acad Sci U S A 103 12115 12120
64. QuinceC
LanzenA
CurtisTP
DavenportRJ
HallN
2009 Accurate determination of microbial diversity from 454 pyrosequencing data. Nat Methods 6 639 641
65. KanagawaT
2003 Bias and artifacts in multitemplate polymerase chain reactions (PCR). J Biosci Bioeng 96 317 323
66. JohansenFE
PeknaM
NorderhaugIN
HanebergB
HietalaMA
1999 Absence of epithelial immunoglobulin A transport, with increased mucosal leakiness, in polymeric immunoglobulin receptor/secretory component-deficient mice. J Exp Med 190 915 922
67. JensenLJ
KuhnM
StarkM
ChaffronS
CreeveyC
2009 STRING 8–a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res 37 D412 416
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek SRFR1 Negatively Regulates Plant NB-LRR Resistance Protein Accumulation to Prevent AutoimmunityČlánek Inhibition of TIR Domain Signaling by TcpC: MyD88-Dependent and Independent Effects on VirulenceČlánek Phylogenetic Approach Reveals That Virus Genotype Largely Determines HIV Set-Point Viral LoadČlánek A Family of Plasmodesmal Proteins with Receptor-Like Properties for Plant Viral Movement Proteins
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2010 Číslo 9- Stillova choroba: vzácné a závažné systémové onemocnění
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Jak souvisí postcovidový syndrom s poškozením mozku?
- Familiární středomořská horečka
-
Všechny články tohoto čísla
- Azole Drugs Are Imported By Facilitated Diffusion in and Other Pathogenic Fungi
- Two Genes on A/J Chromosome 18 Are Associated with Susceptibility to Infection by Combined Microarray and QTL Analyses
- Impact of Simian Immunodeficiency Virus Infection on Chimpanzee Population Dynamics
- Breaking the Stereotype: Virulence Factor–Mediated Protection of Host Cells in Bacterial Pathogenesis
- The Canine Papillomavirus and Gamma HPV E7 Proteins Use an Alternative Domain to Bind and Destabilize the Retinoblastoma Protein
- Rescue of HIV-1 Release by Targeting Widely Divergent NEDD4-Type Ubiquitin Ligases and Isolated Catalytic HECT Domains to Gag
- Steric Shielding of Surface Epitopes and Impaired Immune Recognition Induced by the Ebola Virus Glycoprotein
- Dynamics of the Multiplicity of Cellular Infection in a Plant Virus
- HLA Class I Binding of HBZ Determines Outcome in HTLV-1 Infection
- Pathogenic Bacteria Target NEDD8-Conjugated Cullins to Hijack Host-Cell Signaling Pathways
- The HA and NS Genes of Human H5N1 Influenza A Virus Contribute to High Virulence in Ferrets
- SRFR1 Negatively Regulates Plant NB-LRR Resistance Protein Accumulation to Prevent Autoimmunity
- Cyclin-Dependent Kinase Activity Controls the Onset of the HCMV Lytic Cycle
- The N-Terminal Domain of the Arenavirus L Protein Is an RNA Endonuclease Essential in mRNA Transcription
- Generation of Neutralizing Antibodies and Divergence of SIVmac239 in Cynomolgus Macaques Following Short-Term Early Antiretroviral Therapy
- Inhibition of TIR Domain Signaling by TcpC: MyD88-Dependent and Independent Effects on Virulence
- Intracellular Proton Conductance of the Hepatitis C Virus p7 Protein and Its Contribution to Infectious Virus Production
- The Transcriptome of the Human Pathogen at Single-Nucleotide Resolution
- The Epidermal Growth Factor Receptor (EGFR) Promotes Uptake of Influenza A Viruses (IAV) into Host Cells
- Surface Co-Expression of Two Different PfEMP1 Antigens on Single -Infected Erythrocytes Facilitates Binding to ICAM1 and PECAM1
- Sequestration and Tissue Accumulation of Human Malaria Parasites: Can We Learn Anything from Rodent Models of Malaria?
- Phylogenomics of Ligand-Gated Ion Channels Predicts Monepantel Effect
- Generation of Covalently Closed Circular DNA of Hepatitis B Viruses via Intracellular Recycling Is Regulated in a Virus Specific Manner
- CpG-Methylation Regulates a Class of Epstein-Barr Virus Promoters
- Molecular and Evolutionary Bases of Within-Patient Genotypic and Phenotypic Diversity in Extraintestinal Infections
- A Bistable Switch and Anatomical Site Control Virulence Gene Expression in the Intestine
- Are Members of the Fungal Genus (a) Commensals; (b) Opportunists; (c) Pathogens; or (d) All of the Above?
- Structures of Receptor Complexes of a North American H7N2 Influenza Hemagglutinin with a Loop Deletion in the Receptor Binding Site
- Phylogenetic Approach Reveals That Virus Genotype Largely Determines HIV Set-Point Viral Load
- The Coevolution of Virulence: Tolerance in Perspective
- Involvement of the Cytokine MIF in the Snail Host Immune Response to the Parasite
- Structure of the Extracellular Portion of CD46 Provides Insights into Its Interactions with Complement Proteins and Pathogens
- A Family of Plasmodesmal Proteins with Receptor-Like Properties for Plant Viral Movement Proteins
- High Content Phenotypic Cell-Based Visual Screen Identifies Acyltrehalose-Containing Glycolipids Involved in Phagosome Remodeling
- A Novel Small Molecule Inhibitor of Hepatitis C Virus Entry
- The Microbiota Mediates Pathogen Clearance from the Gut Lumen after Non-Typhoidal Diarrhea
- RNA Polymerases (L-Protein) Have an N-Terminal, Influenza-Like Endonuclease Domain, Essential for Viral Cap-Dependent Transcription
- Pathogen Specific, IRF3-Dependent Signaling and Innate Resistance to Human Kidney Infection
- Cellular Entry of Ebola Virus Involves Uptake by a Macropinocytosis-Like Mechanism and Subsequent Trafficking through Early and Late Endosomes
- The Length of Vesicular Stomatitis Virus Particles Dictates a Need for Actin Assembly during Clathrin-Dependent Endocytosis
- Formation of Mobile Chromatin-Associated Nuclear Foci Containing HIV-1 Vpr and VPRBP Is Critical for the Induction of G2 Cell Cycle Arrest
- Association of Tat with Promoters of PTEN and PP2A Subunits Is Key to Transcriptional Activation of Apoptotic Pathways in HIV-Infected CD4+ T Cells
- Metal Hyperaccumulation Armors Plants against Disease
- Cyclin-Dependent Kinase-Like Function Is Shared by the Beta- and Gamma- Subset of the Conserved Herpesvirus Protein Kinases
- Role of Acetyl-Phosphate in Activation of the Rrp2-RpoN-RpoS Pathway in
- Ebolavirus Is Internalized into Host Cells Macropinocytosis in a Viral Glycoprotein-Dependent Manner
- A Novel Family of IMC Proteins Displays a Hierarchical Organization and Functions in Coordinating Parasite Division
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Structure of the Extracellular Portion of CD46 Provides Insights into Its Interactions with Complement Proteins and Pathogens
- The Length of Vesicular Stomatitis Virus Particles Dictates a Need for Actin Assembly during Clathrin-Dependent Endocytosis
- Inhibition of TIR Domain Signaling by TcpC: MyD88-Dependent and Independent Effects on Virulence
- The Coevolution of Virulence: Tolerance in Perspective
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



