-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
The HMW1C Protein Is a Glycosyltransferase That Transfers Hexose Residues to Asparagine Sites in the HMW1 Adhesin
The Haemophilus influenzae HMW1 adhesin is a high-molecular weight protein that is secreted by the bacterial two-partner secretion pathway and mediates adherence to respiratory epithelium, an essential early step in the pathogenesis of H. influenzae disease. In recent work, we discovered that HMW1 is a glycoprotein and undergoes N-linked glycosylation at multiple asparagine residues with simple hexose units rather than N-acetylated hexose units, revealing an unusual N-glycosidic linkage and suggesting a new glycosyltransferase activity. Glycosylation protects HMW1 against premature degradation during the process of secretion and facilitates HMW1 tethering to the bacterial surface, a prerequisite for HMW1-mediated adherence. In the current study, we establish that the enzyme responsible for glycosylation of HMW1 is a protein called HMW1C, which is encoded by the hmw1 gene cluster and shares homology with a group of bacterial proteins that are generally associated with two-partner secretion systems. In addition, we demonstrate that HMW1C is capable of transferring glucose and galactose to HMW1 and is also able to generate hexose-hexose bonds. Our results define a new family of bacterial glycosyltransferases.
Published in the journal: . PLoS Pathog 6(5): e32767. doi:10.1371/journal.ppat.1000919
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1000919Summary
The Haemophilus influenzae HMW1 adhesin is a high-molecular weight protein that is secreted by the bacterial two-partner secretion pathway and mediates adherence to respiratory epithelium, an essential early step in the pathogenesis of H. influenzae disease. In recent work, we discovered that HMW1 is a glycoprotein and undergoes N-linked glycosylation at multiple asparagine residues with simple hexose units rather than N-acetylated hexose units, revealing an unusual N-glycosidic linkage and suggesting a new glycosyltransferase activity. Glycosylation protects HMW1 against premature degradation during the process of secretion and facilitates HMW1 tethering to the bacterial surface, a prerequisite for HMW1-mediated adherence. In the current study, we establish that the enzyme responsible for glycosylation of HMW1 is a protein called HMW1C, which is encoded by the hmw1 gene cluster and shares homology with a group of bacterial proteins that are generally associated with two-partner secretion systems. In addition, we demonstrate that HMW1C is capable of transferring glucose and galactose to HMW1 and is also able to generate hexose-hexose bonds. Our results define a new family of bacterial glycosyltransferases.
Introduction
Glycosylation of proteins is an essential process that plays an important role in protein structure and function and represents a strategy to fine tune cell-cell recognition and signaling. For a long period of time, glycosylation of proteins was believed to be restricted to eukaryotes. However, in recent years glycoproteins have been identified increasingly in prokaryotes as well, including pathogenic bacteria such as Pseudomonas aeruginosa, Campylobacter spp., Neisseria spp., and E. coli, among others [1]–[10].
Nonencapsulated (nontypable) Haemophilus influenzae is a human specific pathogen that is a common cause of localized respiratory tract and invasive disease and initiates infection by colonizing the upper respiratory tract [11], [12]. Approximately 75–80% of isolates express two related high-molecular weight proteins called HMW1 and HMW2 that mediate high-level adherence to respiratory epithelial cells and facilitate the process of colonization [13], [14]. The HMW1 and HMW2 adhesins are encoded by homologous chromosomal loci that appear to represent a gene duplication event and contain 3 genes, designated hmw1A, hmw1B, and hmw1C and hmw2A, hmw2B, and hmw2C, respectively [15], [16].
HMW1 and HMW2 are synthesized as pre-pro-proteins (Figure 1A) and are secreted by the two-partner secretion system [17]–[19]. Amino acids 1–68 represent an atypical signal peptide and direct the pre-pro-proteins to the Sec apparatus, where they are cleaved by signal peptidase I [18]. The resulting pro-proteins are targeted to the HMW1B and HMW2B outer membrane translocators and undergo cleavage between amino acids 441 and 442, removing the pro-pieces and generating mature species that are 125 kDa and 120 kDa, respectively [18]–[21] (Figure 1A). Following translocation across the outer membrane, mature HMW1 and HMW2 remain non-covalently associated with the bacterial surface [18], [19].
Fig. 1. Purified proteins for examination of glycosylation of HMW1. 
Panel A shows a schematic of the HMW1 pre-pro-protein. The white bar represents the signal peptide, corresponding to amino acids 1–68. The gray bar represents the pro-piece, corresponding to amino acids 69–441. The black bar represents the mature protein, corresponding to amino acids 442–1536. The vertical ticks above the black bar represent sites of N-linked glycosylation. The portion of HMW1 that was used as the acceptor protein for in vitro glycosyltransferase assays is highlighted by the bracket and corresponds to amino acids 802–1406 and contains 18 sites of glycosylation. Panel B shows Coomassie blue-strained gels of purified strep-tagged HMW1802–1406 and purified HAT-tagged HMW1C. In recent work, we demonstrated that HMW1 is a glycoprotein and undergoes glycosylation in the cytoplasm in a process that is dependent upon HMW1C [22]. Functional analyses revealed that glycosylation of HMW1 protects against premature degradation, analogous to some eukaryotic proteins [22]. In addition, glycosylation appears to influence HMW1 tethering to the bacterial surface, a prerequisite for HMW1-mediated adherence [22]. Based on carbohydrate composition analysis of purified HMW1 using gas chromatography and combined gas chromatography-mass spectrometry, the modifying sugars include glucose, galactose, and possibly small amounts of mannose [22]. Analysis of HMW1 proteolytic fragments by mass spectrometry identified 31 sites of modification [23]. All of the modified sites were asparagine residues, in all except one case within the conventional sequence motif for eukaryotic N-linked glycosylation, namely NX(S/T) where X is any residue except for proline [23]. LC-MS/MS analysis, accurate mass measurement, and deuterium replacement studies established that the modifying glycan structures were mono-hexose or di-hexose units rather than N-acetylated hexosamine units that comprise the di-N-diacetyl chitobiose core of eukaryotic and many bacterial asparagine-linked glycans. These results suggested a novel N-linked carbohydrate-peptide transferase activity that does not require assembly of the monosaccharide units onto a lipid-linked intermediate [23].
In the present study, we studied the enzymatic mechanism responsible for the glycosylation of asparagine residues in HMW1. We found that the HMW1C protein encoded in the hmw1 gene cluster is capable of transferring glucose and galactose to the HMW1 adhesin. In addition, HMW1C is capable of generating hexose-hexose linkages.
Results
HMW1C is a glycosyltransferase
In earlier work we found that insertional inactivation of hmw1C in H. influenzae strain Rd-HMW1 resulted in a loss of glycosylation of HMW1 [22], suggesting that HMW1C participates in the process of glycosylation. Further analysis revealed that amino acids 386–439 in HMW1C share 40–41% identity and 51–65% similarity with a domain conserved in a family of eukaryotic O-GlcNAc transferases, including human O-GlcNAc transferase, rat O-GlcNAc transferase, and a plant protein called Spy [22], raising the possibility that HMW1C is a glycosyltransferase.
To address the possibility that HMW1C is the glycosyltransferase responsible for N-linked glycosylation of HMW1, we purified HAT-tagged HMW1C and Strep-tagged HMW1802–1406 (Figure 1B). HMW1802–1406 corresponds to just over half of mature HMW1 (HMW1442–1536), contains 18 documented N-linked glycosylation sites, and was more amenable to purification than mature HMW1 (Figure 1A). Subsequently, we incubated approximately equimolar quantities of HAT-HMW1C and Strep-HMW1802–1406 with both UDP-α-D-glucose and UDP-α-D-galactose at room temperature for 60 minutes, then examined the reaction mixture for reactivity with the DIG-glycan reagents. As shown in Figure 2A, we observed efficient glycosylation of HMW1802–1406 that was dependent on both HMW1C and the UDP-hexoses. To extend this result, we performed the same experiment with UDP-α-D-glucose by itself, UDP-α-D-galactose by itself, GDP-α-D-mannose by itself, UDP-α-D-N-Acetylglucosamine by itself, and UPD-α-D-N-Acetylgalactosamine by itself. As shown in Figure 2B, we observed glycosylation with UDP-α-D-glucose alone and UDP-α-D-galactose alone but not with GDP-α-D-mannose, UDP-α-D-N-Acetylglucosamine, or UPD-α-D-N-Acetylgalactosamine alone. To determine whether smaller amounts of HMW1C are associated with appreciable glycosylation of HMW1802–1406, we repeated assays with a fixed amount of HMW1802–1406, fixed amounts of UDP-α-D-glucose and UDP-α-D-galactose, and dilutions of HMW1C. Based on analysis using DIG-glycan reagents, we observed efficient glycosylation with molar quantities of HMW1C that were less than one-tenth the molar quantity of HMW1802–1406 (data not shown).
Fig. 2. In vitro transferase assays, examining samples by SDS-PAGE and detecting glycosylation with DIG-glycan reagents. 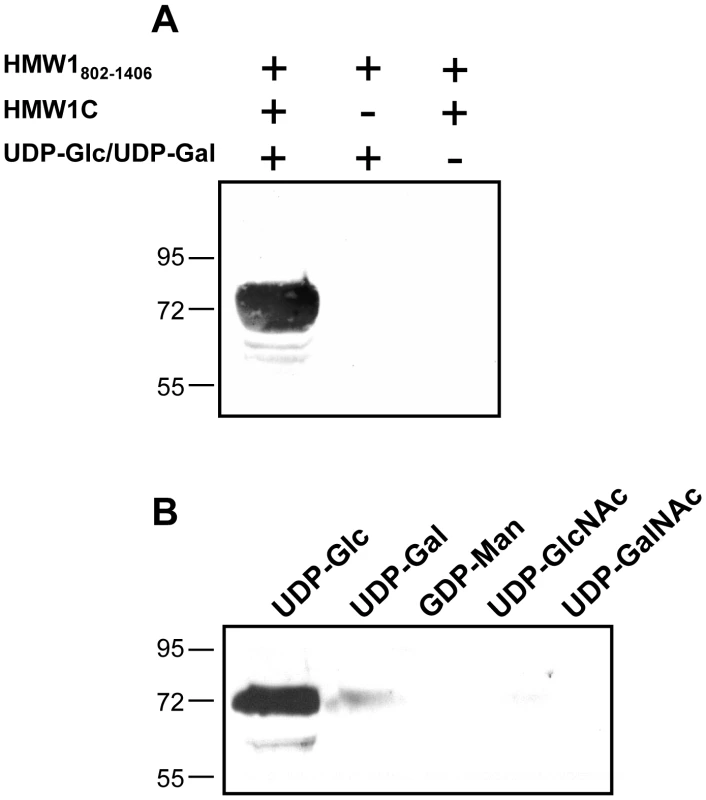
Panel A shows results with various combinations of purified HMW1802–1406, purified HMW1C, and UDP-α-D-glucose plus UDP-α-D-galactose. Panel B shows results with purified HMW1C, purified HMW1802–1406, and either UDP-α-D-glucose, UDP-α-D-galactose, GDP-α-D-mannose, UDP-α-D-N-Acetylglucosamine, or UPD-α-D-N-Acetylgalactosamine. LC-MS/MS analysis of HMW1 after in vitro glycosylation demonstrates specificity of glycosylation with glucose versus galactose
To address whether the glycosylation of HMW1802–1406 in in vitro reactions mimicked glycosylation of native HMW1 in whole bacteria and to gain further insight into which sugars modify which sites, we repeated reactions with purified Strep-tagged HMW1802–1406, purified HAT tagged HMW1C, and UDP-α-D-glucose alone, UDP-α-D-galactose alone, GDP-α-D-mannose alone, or UDP-α-D-glucose plus UDP-α-D-galactose plus GDP-α-D-mannose and then examined the reaction mixtures by LC-MS/MS. As a positive control we examined purified HMW1802–1406 recovered from DH5α/pASK-HMW1802–1406 + pHMW1C, and as a negative control we examined HMW1802–1406 recovered from DH5α/pASK-HMW1802–1406 (lacking pHMW1C).
As summarized in Table 1, we detected 10 of the 18 predicted sites of glycosylation and 11 distinct glycopeptides in HMW1802–1406, including 10 glycopeptides with a single site of glycosylation and one glycopeptide with two sites of glycosylation (KNITFEGGNITFGSR). Interestingly, of the 10 sites of glycosylation, all were modified in the in vitro reactions with UDP-α-D-glucose alone and with UDP-α-D-glucose plus UDP-α-D-galactose plus GDP-α-D-mannose. In contrast, only 6 of the 10 sites of glycosylation were modified in the in vitro reactions with UDP-α-D-galactose alone. Consistent with our observations using DIG-Glycan reagents, no sites were glycosylated in the in vitro reactions with GDP-α-D-mannose alone.
Tab. 1. Glycopeptides detected in in vitro glycosylation assays with HMW1802–1406. 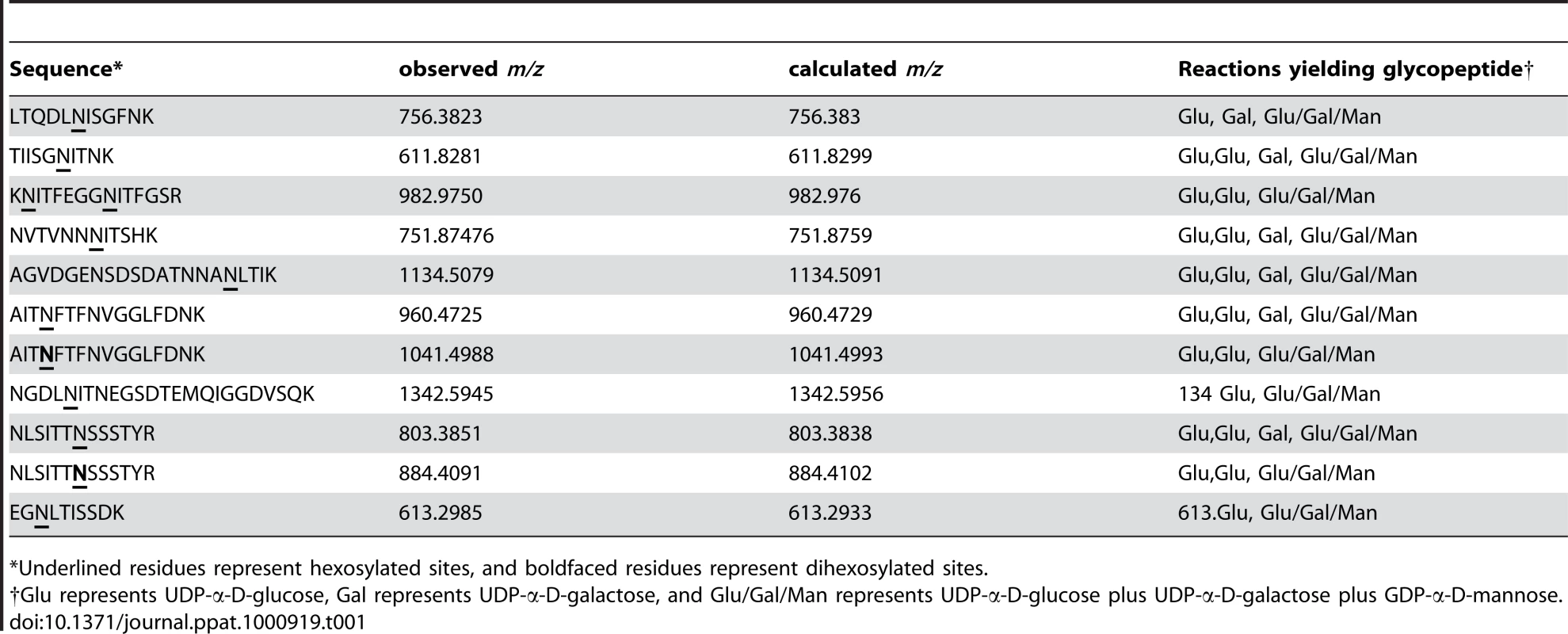
*Underlined residues represent hexosylated sites, and boldfaced residues represent dihexosylated sites. As demonstrated by the collision-induced fragmentation spectra shown in Figure 3 and Figure S1, the glycopeptide NLSITTNSSSTY (HMW1 amino acids 946–958, with glycosylation at N952) and the glycopeptide AITNFTFNVGGLFDNK (HMW1 amino acids 909–924, with glycosylation at N912) were present in two forms, including one with a mono-hexose at the predicted site of glycosylation and the other with a di-hexose at the predicted site of glycosylation. The forms containing a mono-hexose were detected in the in vitro reactions with UDP-α-D-glucose alone, UDP-α-D-galactose alone, and UDP-α-D-glucose plus UDP-α-D-galactose plus GDP-α-D-mannose, while the forms containing a di-hexose were detected only in the in vitro reactions with UDP-α-D-glucose alone and with UDP-α-D-glucose plus UDP-α-D-galactose plus GDP-α-D-mannose, suggesting that glucose must be the first hexose linked to asparagine in the glycopeptides containing di-hexose modification.
Fig. 3. Collision-induced fragmentation spectra from glycosylated peptide NLSITTNSSSTYR (HMW1 amino acids 946–958). 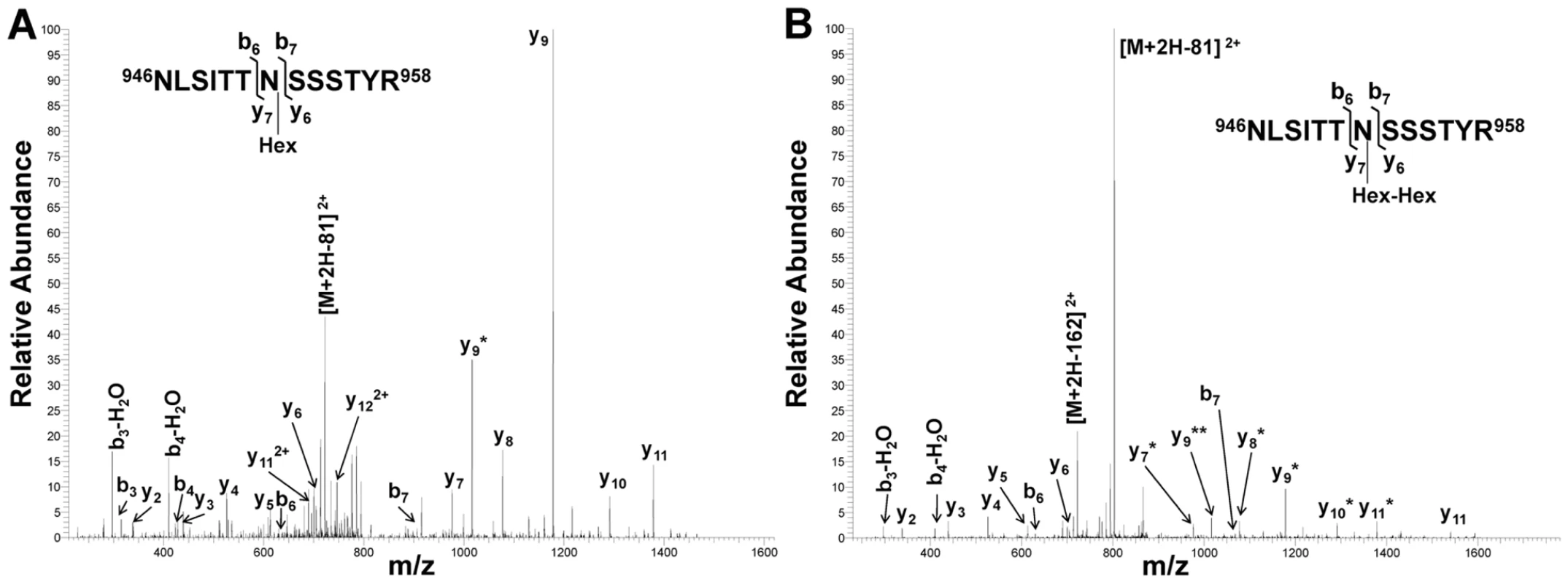
Panel A shows the CID spectrum of the glycopeptide that is modified with one hexose unit. Panel B shows the CID spectrum of the glycopeptide that is modified with a di-hexose. The asterisks indicate b and y fragmentation ions that underwent a neutral loss of one (*) or two (**) hexosyl residues. Together these findings demonstrate that the HMW1C protein is a glycosyltransferase and has a novel activity capable of transferring glucose and galactose to asparagine residues in HMW1 and creating hexose-hexose bonds. In addition, they demonstrate that the di-hexosylated sites at N951 and N912 are initially modified with a glucose monosaccharide.
Inactivation of galU eliminates glycosylation of HMW1
To extend our understanding of glycosylation of HMW1 and confirm our observation that HMW1 is modified with glucose and galactose in in vitro glycosylation assays, we examined the effect of insertional inactivation of galU (open reading frame HI0812 in strain Rd) on glycosylation of HMW1 in strain Rd-HMW1. The galU gene encodes glucose-1-phosphate uridyl transferase, which converts glucose-1-phosphate to UDP-glucose (Figure S2). UDP-glucose in turn can be converted directly to UDP-galactose by GalE (UDP Gal-4-epimerase) or can serve as the donor of UDP for conversion of galactose-1-phosphate to UDP-galactose. In assessing the effect of inactivation of galU, we incubated Rd-HMW1/galU in supplemented brain heart infusion broth [24], which contains glucose as the primary carbon source. Interestingly, inactivation of galU mimicked the effect of inactivation of hmw1C described in our earlier work [22], eliminating HMW1 glycosylation as assessed by DIG-glycan blots (Figure 4A), virtually eliminating HMW1 tethering to the bacterial surface (Figure 4B), and abolishing HMW1-mediated adherence (Figure 4C).
Fig. 4. Effect of inactivation of the galU gene. 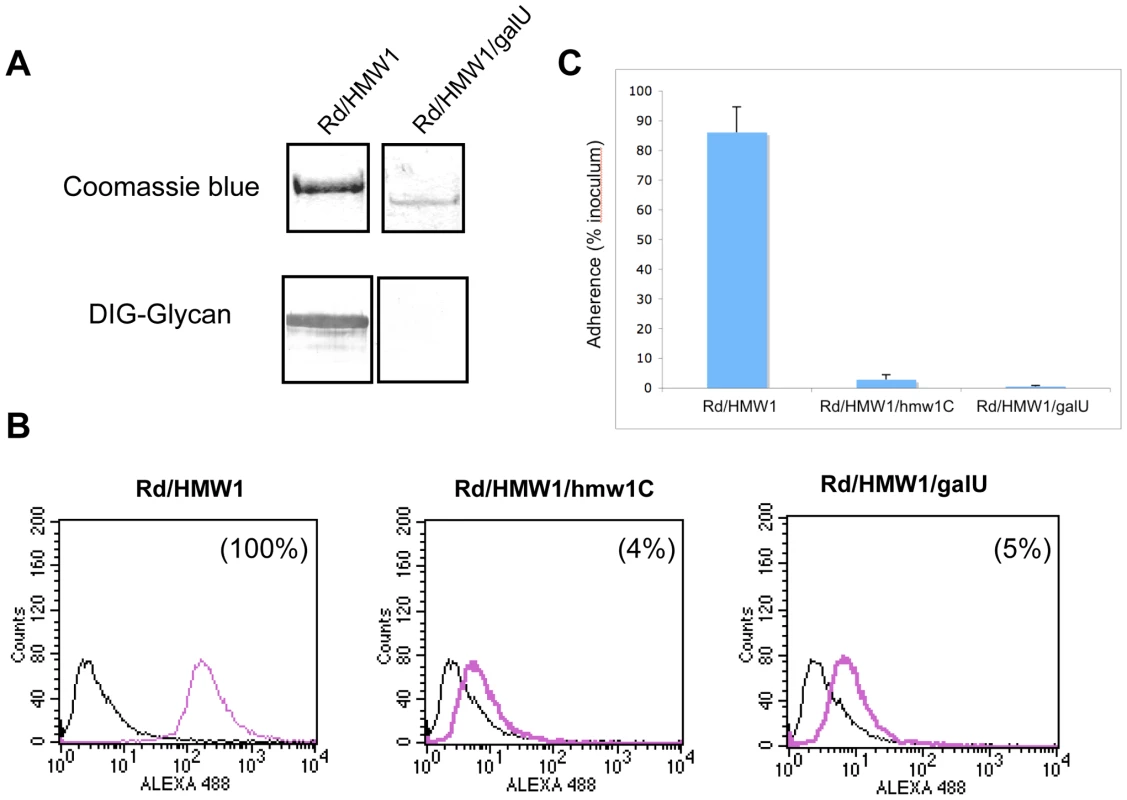
Panel A shows purified HMW1 recovered from strain Rd-HMW1 and Rd-HMW1/galU. With strain Rd-HMW1, HMW1 was recovered from the surface of whole bacteria. With strain Rd-HMW1/galU, HMW1 was recovered from the culture supernatant and concentrated, reflecting the fact that HMW1 is no longer surface associated in this strain. Purified protein was resolved by SDS-PAGE and was then either stained with Coomassie blue (upper gel) or examined for glycosylation using DIG-glycan reagents (lower gel). Panel B shows flow cytometry results with strains Rd-HMW1, Rd-HMW1/hmw1C, and Rd-HMW1/galU using antiserum GP85 against HMW1 to detect surface-associated HMW1. The number in the upper right corner of the plots of Rd-HMW1/hmw1C and Rd-HMW1/galU represents the percentage of surface-associated HMW1 in strain Rd-HMW1. Panel C shows in vitro adherence results comparing adherence by Rd-HMW1, Rd-HMW1/hmw1C, and Rd-HMW1/galU to Chang epithelial cells. Bars and error bars represent mean and standard deviations of measurements performed in triplicate. Consistent with our in vitro glycosyltransferase assays with purified HMW1802–1406 and HMW1C, these results indicate that UDP-glucose is required for glycosylation of HMW1 in H. influenzae under standard growth conditions in supplemented brain heart infusion broth.
Discussion
In this study, we found that the H. influenzae HMW1C protein encoded in the hmw1 gene cluster is a glycosyltransferase and is capable of transferring glucose and galactose to asparagine residues in the HMW1 adhesin, providing the first example of a glycosyltransferase that transfers hexose units rather than N-acetylated amino sugars to asparagine residues in protein targets. Further analysis revealed that HMW1C is capable of creating both hexose-asparagine and hexose-hexose linkages, suggesting multi-functionality as a glycosyltransferase.
All previously reported carbohydrate modification of asparagine residues in proteins in Eukarya and Bacteria involve the en bloc transfer of oligosaccharides from a lipid-linked intermediate by an oligosaccharyltransferase complex [25]. In Archaea, the mechanisms of N-glycosylation are less well understood. Glycosylation of asparagine residues with a trisaccharide moiety in the flagellin and S-layer proteins of Methanococcus voltae has been proposed to proceed via a lipid-linked intermediate [26]. More recently it has been shown that hexose units are attached directly to asparagine residues in an S layer glycoprotein of Haloferax volcanii [27]. A pentasaccharide with the structure Hex-X-hexuronic acid-HexA-HexA-Hex-peptide was identified at two glycosylation sites. Interestingly, these two sites were different from the conventional N-glycosylation sequence motif observed in eukaryotes and in HMW1. It is currently unclear whether the H. volcanii Hex-Asn linkage is formed from a lipid-linked intermediate or via activated monosaccharides as we have found with HMW1 and HMW1C.
In earlier work, we performed carbohydrate composition analysis on purified HMW1 and detected glucose, galactose, and small amounts of mannose [22]. Given the potential for contaminating sugars to be detected in this analysis, we were uncertain as to whether mannose was truly present as a modifying sugar in HMW1, especially given that it accounted for only 2.5–3% of the total carbohydrate [22]. Our analysis in the current study argues that mannose is not present in HMW1. In particular, in in vitro glycosyltransferase assays using purified HMW1802–1406, HMW1C, and GDP-α-D-mannose, we were unable to detect modification of HMW1802–1406 using either DIG-Glycan reagents or LC-MS/MS.
Based on assessment of the 10 glycopeptides that we detected in our in vitro glycosylation assays with HMW1802–1406, which corresponds to just over half of mature HMW1, we observed that HMW1C transfers glucose to all glyscosylated asparagines and transfers galactose to only a subset of glyscosylated asparagines. All of these glycosylation sites correspond to the conventional sequence motif of N-linked glycans, namely NX(S/T), with X being any amino acid except proline. Examination of the primary amino acid sequence of the sites that are modified only with glucose and the sites that are modified with either glucose or galactose in in vitro assays reveals no apparent distinction, suggesting that factors beyond the amino acid sequence influence the specificity or potentially the efficiency of glycosylation. This observation is consistent with the fact that only a fraction of conventional sequences motifs are glycosylated in HMW1 purified from H. influenzae [23].
Further analysis of the glycopeptides detected after in vitro glycosylation revealed two peptides that were modified with a di-hexose. Interestingly, in both cases the glycopeptides were detected only in the reactions performed with UDP-α-D-glucose alone and with UDP-α-D-glucose plus UDP-α-D-galactose plus GDP-α-D-mannose, indicating modification with UDP-α-D-glucose. In contrast, the corresponding glycopeptides containing a single hexose at the asparagines in question were detected in the reactions performed with UDP-α-D-glucose alone, with UDP-α-D-galactose alone, and with UDP-α-D-glucose plus UDP-α-D-galactose plus GDP-α-D-mannose, indicating modification with either glucose or galactose. Considered together, these results suggest that glucose must be linked to asparagine in the glycopeptides containing di-hexose modification. At this point, it is unclear whether the di-hexose is generated prior to modification of the acceptor asparagine residue or whether instead a single hexose is linked to the target asparagine and then a second hexose is linked to the first hexose, although the conventional interpretation is that the hexose is added to the protein and then the chain is extended. In either event, it appears that HMW1C is responsible for creating the hexose-hexose bond.
Interestingly, homology analysis reveals 42–68% identity and 58–83% similarity between the full-length HMW1C sequence and proteins in a number of other gram-negative bacterial pathogens, including the enterotoxigenic E. coli (ETEC) EtpC protein and predicted proteins in Yersinia pseudotuberculosis, Y. enterocolitica, Y. pestis, H. ducreyi, Actinobacillus pleuropneumoniae, Mannheimia spp., Xanthomonas spp., and Burkholderia spp, among others (Table S1). In ETEC, Y. pseudotuberculosis, Y. enterocolitica, and Y. pestis, these homologs are encoded by genes that are adjacent to known or predicted two-partner secretion loci. The H. ducreyi, Mannheimia succiniciproducens, and Burkholderia xenovorans genomes contain genes that encode predicted two-partner secretion proteins as potential targets for the HMW1C homologs, although these genes are in unlinked locations. The ETEC EtpC protein is encoded by a two-partner secretion locus called etpBAC and has been shown to be required for glycosylation of the EtpA adhesin, a high-molecular weight protein that has a predicted molecular mass of ∼177 kDa and promotes adherence to intestinal epithelial cells and colonization of the intestine in mice [28], [29]. These observations suggest that that there is a family of bacterial HMW1C-like proteins with glycosyltransferase activity.
To summarize, in eukaryotes N-linked glycosylation occurs in the endoplasmic reticulum and involves an oligosaccharyltransferase that catalyzes the transfer of the oligosaccharide from the lipid donor dolichylpyrophosphate to the acceptor protein. Similarly, in bacteria, N-glycosylation generally occurs in the periplasm and involves an oligosaccharyltransferase that transfers the glycan structure from a lipid donor to the acceptor protein. In contrast, in the case of the H. influenzae HMW1 adhesin, N-linked glycosylation occurs in the cytoplasm and involves direct transfer of hexose units to the acceptor protein by HMW1C, with no requirement for a lipid donor. In this study, we have established that the H. influenzae HMW1C protein is a multi-functional enzyme that is capable of transferring glucose and galactose to asparagine residues in selected conventional N-linked sequence motifs in HMW1 and is also capable of creating hexose-hexose linkages. Based on homology analysis, it is likely that a variety of other bacteria possess HMW1C-like proteins with similar enzymatic activity. In future work, we will examine whether these HMW1C-like proteins are identical to HMW1C in terms of the glycan units that they transfer and the acceptor protein sequence motifs that they recognize.
Materials and Methods
Bacterial strains and plasmids
The strains and plasmids used in this study are listed in Table 2. H. influenzae strain Rd-HMW1 is a derivative of strain Rd that contains the intact hmw1 locus and expresses fully functional HMW1 [22]. H. influenzae strain Rd-HMW1/hmw1C is a derivative of strain Rd-HMW1 that contains an insertionally inactivated hmw1C gene [22]. The H. influenzae Rd-HMW1 derivative harboring a kanamycin cassette in galU was constructed by transforming competent Rd-HMW1 with genomic DNA recovered from RdgalU and selecting for kanamycin resistance [30].
Tab. 2. Strains and plasmids used in this study. 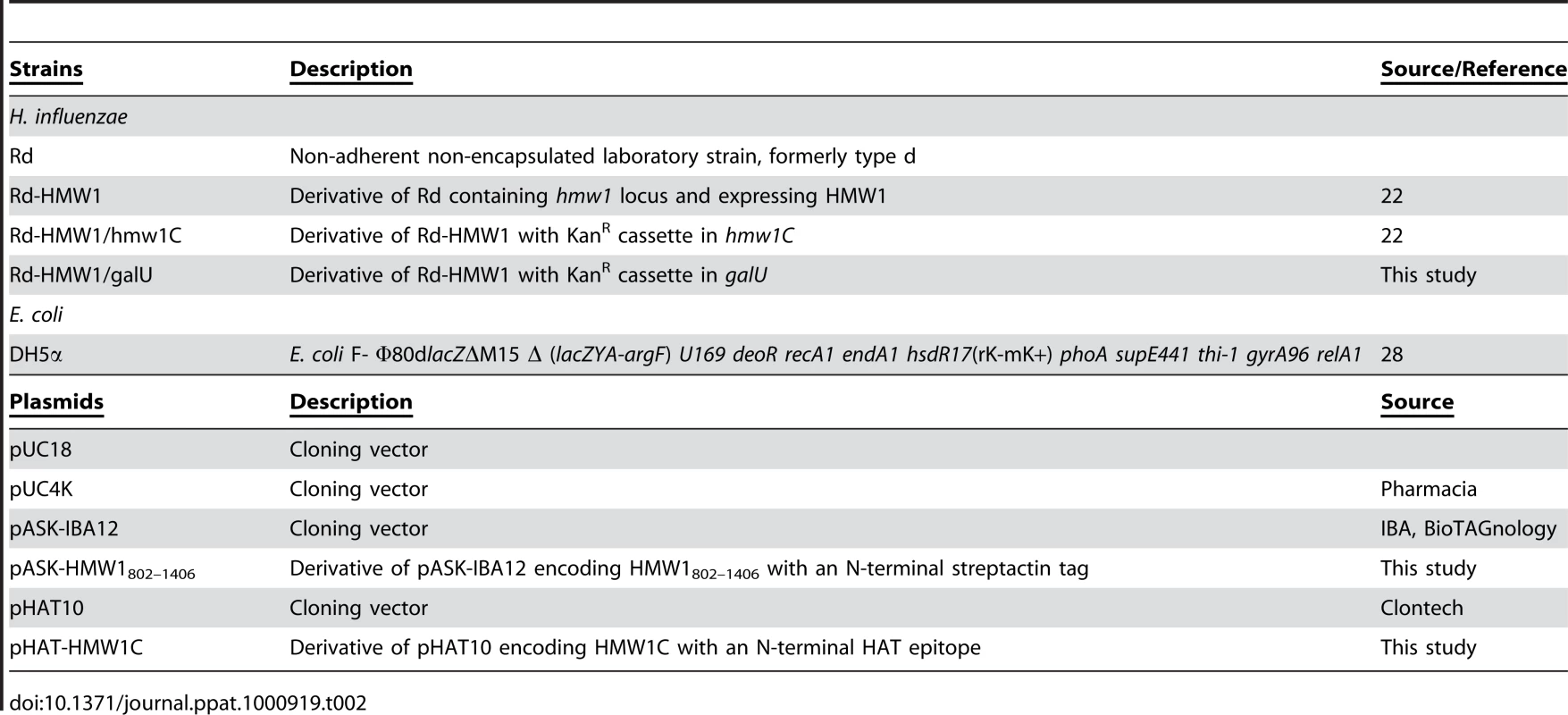
In order to overexpress HMW1802–1406 with a Strep tag at the N terminus, the fragment encoding HMW1802–1406 was amplified by PCR from pHMW1-14 using a 5′ primer that incorporated a BamHI site and a 3′ primer that incorporated a SalI site. The PCR amplicon was digested with BamI and SalI and then ligated into BamHI-SalI-digested pASK-IBA12 (IBA, BioTAGnology), creating pASK-HMW1802–1406.
In order to overexpress the HMW1C protein with a HAT epitope at the N terminus, the hmw1C gene was amplified by PCR from pHMW1-14 using a 5′ primer that incorporated a BamHI site and a 3′ primer that incorporated an EcoRI site. The PCR amplicon was digested with BamHI and EcoRI and then ligated into BamHI-EcoRI-digested pHAT10 (Clontech), creating pHAT-HMW1C.
Transformation and mutagenesis
Plasmids were introduced into E. coli by chemical transformation [31]. DNA was introduced into H. influenzae using the MIV method of transformation described by Herriott et al. [32]. Transformants were selected by plating on agar containing kanamycin, and mutations were confirmed by PCR analysis using primers that anneal to regions flanking the target gene.
Protein purification
To purify HMW1802–1406, E. coli strain DH5α/pASK-HMW1802–1406 was grown at 37°C to an OD600 of 0.7, then induced for 2 hrs with the addition of 100 µg/ml of anhydro-tetracycline (Sigma). Cells were harvested, resuspended in 100 mM Tris pH 8.0, 150 mM NaCl with Complete Mini protease inhibitor (Roche), and lysed by sonication. Insoluble material was removed by centrifugation at 12,500 × g for 30 min. The supernatant was loaded onto a Strep-Tactin Superflow cartridge and eluted according to the manufacturer's instructions (IBA, BioTAGnology). Eluted fractions were analyzed for purity by SDS-PAGE and were pooled. To purify HMW1C, E. coli strain DH5α/pHAT-HMW1C was grown at 37°C overnight. Cells were recovered, resuspended in 50 mM sodium phosphate buffer pH 7.0, 300 mM NaCl (bufferA), and lysed by sonication. Insoluble material was removed by centrifugation at 12,500 × g for 30 min. The supernatant was loaded onto a 1 ml Talon column (Clontech) and eluted with a gradient of 0 to 300 mM imidazole in Buffer A. Fractions were analyzed for purity by SDS-PAGE and were pooled.
Glycosyltransferase assay
In standard in vitro glycosyltransferase assays, 1.5 µg (23 pmole) of purified HMW1802–1406 was combined with a mixture containing 20 µl of 50 mM UDP-α-D-glucose, 50 mM UDP-α-D-galactose, 50 mM GDP-α-D-mannose, 50 mM UDP-α-D-N-Acetylglucosamine, or 50 mM UDP-α-D-N-acetylgalactosamine (Calbiochem) either as individual sugars or as mixtures. The reactions were initiated with addition of 1.5 µg (21 pmole) of purified HMW1C in a final volume of 150 µl in 25 mM Tris pH 7.2, 150 mM NaCl. Samples were incubated for 60 minutes at room temperature and then further incubated at 4°C overnight.
Carbohydrate detection
To detect protein glycosylation, DIG Glycan reagents (Roche) were employed. Use of these reagents is based on the oxidation of hydroxyl groups in carbohydrates to aldehydes either in solution or bound to nitrocellulose membranes. Digoxigenin is then covalently linked to the aldehyde groups, and an anti-digoxigenin alkaline-phosphatase conjugated agent is used for detection of labeled carbohydrates.
Flow cytometry
FACS analysis was performed by the Duke University Medical Center Cancer Research Center Flow Cytometry Shared Resource Center using a Becton Dickinson FACS Calibur instrument at a wavelength of 488 nm. Bacterial suspensions were fixed with 1% formaldehyde in PBS at room temperature for 30 min. After washing once with Tris buffered saline (TBS), bacteria were resuspended in 1 ml of TBS, 50 mM EDTA, 0.1% bovine serum albumin, and a 1∶1000 dilution of guinea pig antiserum GP85 directed against HMW1 [33] and were incubated with gentle rocking at room temperature for 1 hr. Samples were then centrifuged, washed twice with PBS, and resuspended in 200 µl of PBS, 0.1% bovine serum albumin, and a 1∶200 dilution of Alexa Fluor488 anti-guinea pig antibody (Molecular Probes). Samples were incubated with gentle rocking at room temperature for 1 hr. After two additional washes with PBS, bacterial pellets were re-suspended in 1 ml of PBS and were then analyzed. Data were analyzed with CELLQUEST software (Becton Dickinson). To quantify histograms, markers were drawn on plots, and positive events within the markers were determined as a percentage of the positive control (set at 100%).
Adherence assays
Adherence assays were performed with Chang epithelial cells (human conjunctiva; ATCC CCL 20.2) (Wong-Kilbourne derivative clone 1-5c-4) as described previously [16]. Percent adherence was calculated by dividing the number of adherent colony-forming units by the number of inoculated colony-forming units. All strains were examined in triplicate, and each assay was repeated at least two times.
Protein analysis
Whole cell sonicates were prepared by suspending bacterial pellets in 10 mM HEPES, pH 7.4 and sonicating to clarity. Proteins were resolved by SDS-PAGE using 10% polyacrylamide gels. Western blots were performed using guinea pig antiserum GP85 against the HMW1 protein [30].
Protein digestion and peptide preparation
Samples were precipitated using the 2D protein clean up kit (GE Healthcare) according to the manufacturer's instructions. Bovine serum albumin (100 ng) was added to each sample as an internal standard. Pellets were dissolved in 40 µl 9 M urea and aliquoted into 0.5 ml microfuge tubes. Samples (20 µl in 9 M urea) were reduced with 5 mM TCEP at pH 8.0 at room temperature for 30 min and were alkylated with 10 mM iodoacetamide (Bio-Rad) in the dark at room temperature for 30 min. TCEP and iodoacetamide were quenched with 5 mM dithiothreitol (DTT) at room temperature for 10 min. The reduced and alkylated proteins were digested with 1 µg of endoproteinase Lys-C (Roche) at 37°C overnight. Samples were diluted with 64 µl H2O to reduce the concentration of urea to 2 M and were then digested with 4 µg trypsin (Sigma) at 37°C overnight. Peptides were acidified with 5.5 µl formic acid (Sigma) and extracted 6 times with 10–200 µl NuTip porous graphite carbon wedge tips (Glygen) according to the manufacturer's directions and were then eluted into 1.5 ml autosampler vials with 60% acetonitrile (Burdick & Jackson) in 0.1% formic acid. The peptide digests were evaluated for quality and detergent contaminants using MALDI-TOF/TOF [34] prior to LC-MS analysis. For MALDI-TOF/TOF analysis, the peptide sample (0.5 µl) was mixed with an equal volume of MALDI matrix solution (Agilent Technologies) prior to spotting. For nano-LC-FTICR-MS analysis, the peptide sample was dried and immediately dissolved in 10 µl aqueous acetonitrile/formic acid (1%/1%).
Mass spectrometry
The complex mixtures of peptides and glycopeptides from HMW1802–1406 were analyzed using high-resolution nano-LC-MS on a hybrid mass spectrometer consisting of a linear quadrupole ion-trap and an Orbitrap (LTQ-Orbitrap XL, Thermo-Fisher). The liquid chromatographs were nanoflow HPLC systems (NanoLC-1Dplus™ and NanoLC-Ultra™) that were interfaced to the mass spectrometer with a nanospray source (PicoView PV550; New Objective). The in-house packed LC column (Jupiter C12 Proteo, 4 µm particle size, 90 Å pore size [Phenomenex]) was equilibrated in 98% solvent A (aqueous 0.1% formic acid) and 2% solvent B (acetonitrile containing 0.1% formic acid). The samples (10 µL) were injected from autosampler vials using the LC-systems autosamplers at a flow rate of 1.0 µL/min and were eluted using a segmented linear gradient (250 nL/min) with solvent B: isocratic at 2% B, 0–2 min; 2% B to 40% B, 2–65 min; 40% B to 80% B, 65–70 min; isocratic at 80% B, 70–72 min; 80% B to 2% B, 72–77 min; and isocratic at 2% B, 77–82 min. The survey scans (m/z 350–2000) (MS1) were acquired at high resolution (60,000 at m/z = 400) in the Orbitrap, and the MS/MS spectra (MS2) were acquired in the linear ion trap at low resolution, both in profile mode. The maximum injection times for the MS1 scan in the Orbitrap and the LTQ were 50 ms and 100 ms, respectively. The automatic gain control targets for the Orbitrap and the LTQ were 2×105 and 3×104, respectively. The MS1 scans were followed by six MS2 events in the linear ion trap with wideband collision activation in the ion trap (parent threshold = 1000; isolation width = 2.0 Da; normalized collision energy = 30%; activation Q = 0.250; activation time = 30 ms). Dynamic exclusion was used to remove selected precursor ions (−0.25/+1.5 Da) after MS2 acquisition with a repeat count of 2, a repeat duration of 30 s, and a maximum exclusion list size of 200. The following ion source parameters were used: capillary temperature 200 °C, source voltage 2.5 kV, source current 100 µA, and the tube lens at 79 V. The data were acquired using Xcalibur, version 2.0.7 (Thermo-Fisher).
The MS2 spectra were analyzed both by searching a customized protein database that contained the sequences of HMW802–1406 and by expert manual interpretation. The exact masses of the glycopeptides and fragmentation ions were calculated using the Molecular Weight Calculator, version 6.45 (http://ncrr.pnl.gov/software/). For database searches, the LC-MS files were processed using MASCOT Distiller (Matrix Science, version 2.3.0.0) with the settings previously described [35]. The resulting MS2 centroided files were used for database searching with MASCOT, version 2.1.6, and the following parameters: enzyme, trypsin; MS tolerance = 10 ppm; MS/MS tolerance = 0.8 Da with a fixed carbamidomethylation of Cys residues and the following variable modifications: Methionine, oxidation; Pyro-glu (N-term); Maximum Missed Cleavages = 5; and 1+, 2+, and 3+ charge states.
Supporting Information
Zdroje
1. BenzI
SchmidtMA
2001 Glycosylation with heptose residues mediated by the aah gene product is essential for adherence of the AIDA-1 adhesin. Mol Microbiol 40 1403 1413
2. BrimerC
MontieT
1998 Cloning and comparison of fliC genes and identification of glycosylation in the flagellin of Pseudomonas aeruginosa a-type strains. J Bacteriol 180 3209 3217
3. CastricP
1995 pilO, a gene required for glycosylation of Pseudomonas aeruginosa 1244 pilin. Microbiology 141 1247 1254
4. DoigP
KinsellaN
GuerryP
TrustTJ
1996 Characterization of a post-translational modification of Campylobacter flagellin: identification of a sero-specific glycosyl moiety. Mol Microbiol 19 379 387
5. LindenthalC
ElsinghorstEA
1999 Identification of a glycoprotein produced by enterotoxigenic Escherichia coli. Infect Immun 67 4084 4091
6. SchmidtMA
RileyLW
BenzI
2003 Sweet new world: glycoproteins in bacterial pathogens. Trends Microbiol 11 554 561
7. SzymanskiCM
YaoR
EwingCP
TrustTJ
GuerryP
1999 Evidence for a system of general protein glycosylation in Campylobacter jejuni. Mol Microbiol 32 1022 1030
8. SzymanskiCM
WrenBW
2005 Protein glycosylation in bacterial mucosal pathogens. Nature Rev Microbiol 3 225 237
9. VirjiM
1997 Post-translational modifications of meningococcal pili. Identification of common substituents: glycans and α-glycerophosphate – a review. Gene 192 141 147
10. WackerM
LintonD
HitchenPG
Nita-LazarM
HaslamSM
2002 N-linked glycosylation in Campylobacter jejuni and its functional transfer into E. coli. Science 298 1790 1793
11. TurkDC
1984 The pathogenicity of Haemophilus influenzae. J Med Microbiol 18 1 16
12. St GemeJWIII
1993 Nontypeable Haemophilus influenzae disease: epidemiology, pathogenesis, and prospects for prevention. Infect Agents Dis 2 1 16
13. St GemeJWIII
FalkowS
BarenkampSJ
1993 High-molecular-weight proteins of nontypeable Haemophilus influenzae mediate attachment to human epithelial cells. Proc Natl Acad Sci, USA 90 2875 2879
14. St GemeJWIII
GrassS
1998 Secretion of the Haemophilus influenzae HMW1 and HMW2 adhesins involves a periplasmic intermediate and requires the HMWB and HMWC proteins. Mol Microbiol 27 617 630
15. BarenkampSJ
St GemeJWIII
1994 Genes encoding high molecular weight adhesion proteins of nontypeable Haemophilus influenzae are part of gene clusters. Infect Immun 62 3320 3328
16. BuscherAZ
BurmeisterK
BarenkampSJ
St GemeJWIII
2004 Evolutionary and functional relationships among the nontypeable Haemophilus influenzae HMW family of adhesins. J Bacteriol 186 4209 4217
17. Jacob-DubuissonF
LochtC
AntoineR
2001 Two-partner secretion in Gram-negative bacteria: a thrifty, specific pathway for large virulence proteins. Mol Microbiol 40 306 313
18. GrassS
St GemeJWIII
2000 Maturation and secretion of the non-typable Haemophilus influenzae HMW1 adhesin: roles of the N-terminal and C-terminal domains. Mol Microbiol 36 55 67
19. St GemeJWIII
KumarVV
CutterD
BarenkampSJ
1998 Prevalence and distribution of the hmw and hia genes and the HMW and Hia adhesins among genetically diverse strains of nontypeable Haemophilus influenzae. Infect Immun 66 364 368
20. BarenkampSJ
LeiningerE
1992 Cloning, expression, and DNA sequence analysis of genes encoding nontypeable Haemophilus influenzae high-molecular-weight proteins related to filamentous hemagglutinin of Bordetella pertussis. Infect Immun 60 1302 1313
21. SuranaNK
GrassS
HardyGG
HuilinL
ThanassiDG
St. GemeJWIII
2004 Evidence for conservation of architecture and physical properties of Omp85-like proteins throughout evolution. Proc Natl Acad Sci USA 101 14497 14502
22. GrassS
2003 The Haemophilus influenzae HMW1 adhesin is glycosylated in a process that requires HMW1C and phosphoglucomutase, an enzyme involved in lipooligosaccharide biosynthesis. Mol Microbiol 48 737 751
23. GrossJ
BuscherAZ
SwordsWE
ApicellaMA
BarenkampSJ
2008 The Haemophilus influenzae HMW1 adhesin is glycosylated with an unusual N-linked carbohydrate modification. J Biol Chem 283 26010 26015
24. AndersonP
JohnstonRBJr
SmithDH
1972 Human serum activity against Haemophilus influenzae type b. J Clin Invest 51 31 38
25. KelleherDJ
GilmoreR
2006 An evolving view of the eukaryotic oligosaccharyltransferase. Glycobiology 16 47R 62R
26. ChabanB
VoisinS
KellyJ
LoganSM
JarrellKF
2006 Identification of genes involved in the biosynthesis and attachment of Methanococcus voltae N-linked glycans: insight into N-linked glycosylation pathways in Archaea. Mol Microbiol 61 259 268
27. Abu-QarnM
Yurist-DoutschS
GiordanoA
TraunerA
MorrisHR
2007 Haloferax volcanii AglB and AglD are involved in N-glycosylation of the S-layer glycoprotein and proper assembly of the surface layer. J Mol Biol 374 1224 1236
28. FleckensteinJM
RoyK
FischerJF
BurkittM
2006 Identification of a two-partner secretion locus. Infect Immun 74 2245 2258
29. RoyK
HamiltonD
AllenKP
RandolphMP
FleckensteinJM
2008 The EtpA exoprotein of enterotoxigenic Escherichia coli promotes intestinal colonization and is a protective antigen in an experimental model of murine infection. Infect Immun 76 2106 2112
30. HoodDW
DeadmanME
AllenT
MasoudH
MartinA
2006 Use of the complete genome sequence information of Haemophilus influenzae strain Rd to investigate lipopolysaccharide biosynthesis. Mol Microbiol 22 951 965
31. SambrookJ
RusselDW
2001 Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Press, New York
32. HerriottRM
MeyerEM
VogtM
1970 Defined nongrowth media for stage II development of competence in Haemophilus influenzae. J Bacteriol 101 517 524
33. BuscherAZ
GrassS
HeuserJ
RothR
St GemeJWIII
2006 Surface anchoring of a bacterial adhesin secreted by the two-partner secretion pathway. Mol Microbiol 61 470 83
34. MedzihradszkyKF
CampbellJM
BaldwinMA
FalickAM
JuhaszP
2000 The characteristics of peptide collision-induced dissociation using a high-performance MALDI-TOF/TOF tandem mass spectrometer. Anal Chem 72 552 558
35. NittisT
GuittatL
LeDucRD
DaoB
DuxinJB
2010 Revealing novel telomere proteins using in vivo crosslinking, tandem affinity purification and label-free quantitative LC-FTICR-MS. Mol Cell Proteomics Jan 22, 2010 (Epub ahead of print)
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Mouse Senile Amyloid Fibrils Deposited in Skeletal Muscle Exhibit Amyloidosis-Enhancing ActivityČlánek The Role of Intestinal Microbiota in the Development and Severity of Chemotherapy-Induced MucositisČlánek Crystal Structure of HIV-1 gp41 Including Both Fusion Peptide and Membrane Proximal External RegionsČlánek Demonstration of Cross-Protective Vaccine Immunity against an Emerging Pathogenic Ebolavirus Species
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2010 Číslo 5- Stillova choroba: vzácné a závažné systémové onemocnění
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Jak souvisí postcovidový syndrom s poškozením mozku?
- Infekční komplikace virových respiračních infekcí – sekundární bakteriální a aspergilové pneumonie
-
Všechny články tohoto čísla
- Quorum Sensing Inhibition Selects for Virulence and Cooperation in
- The HMW1C Protein Is a Glycosyltransferase That Transfers Hexose Residues to Asparagine Sites in the HMW1 Adhesin
- Analysis of Virion Structural Components Reveals Vestiges of the Ancestral Ichnovirus Genome
- Mouse Senile Amyloid Fibrils Deposited in Skeletal Muscle Exhibit Amyloidosis-Enhancing Activity
- Global Migration Dynamics Underlie Evolution and Persistence of Human Influenza A (H3N2)
- The Type III Effectors NleE and NleB from Enteropathogenic and OspZ from Block Nuclear Translocation of NF-κB p65
- VEGF Promotes Malaria-Associated Acute Lung Injury in Mice
- Identification of a Mutant PfCRT-Mediated Chloroquine Tolerance Phenotype in
- The Early Stage of Bacterial Genome-Reductive Evolution in the Host
- Host-Detrimental Role of Esx-1-Mediated Inflammasome Activation in Mycobacterial Infection
- Elevation of Intact and Proteolytic Fragments of Acute Phase Proteins Constitutes the Earliest Systemic Antiviral Response in HIV-1 Infection
- The Pleiotropic CymR Regulator of Plays an Important Role in Virulence and Stress Response
- Alternative Sigma Factor σ Modulates Prophage Integration and Excision in
- Effect of Neuraminidase Inhibitor–Resistant Mutations on Pathogenicity of Clade 2.2 A/Turkey/15/06 (H5N1) Influenza Virus in Ferrets
- Massive APOBEC3 Editing of Hepatitis B Viral DNA in Cirrhosis
- NK Cells and γδ T Cells Mediate Resistance to Polyomavirus–Induced Tumors
- Is Genetically Diverse in Animals and Appears to Have Crossed the Host Barrier to Humans on (At Least) Two Occasions
- Adenylate Cyclase Toxin Mobilizes Its β Integrin Receptor into Lipid Rafts to Accomplish Translocation across Target Cell Membrane in Two Steps
- The Role of Intestinal Microbiota in the Development and Severity of Chemotherapy-Induced Mucositis
- HIV-1 Transmitting Couples Have Similar Viral Load Set-Points in Rakai, Uganda
- Few and Far Between: How HIV May Be Evading Antibody Avidity
- Galectin-9/TIM-3 Interaction Regulates Virus-Specific Primary and Memory CD8 T Cell Response
- Perforin Expression Directly by HIV-Specific CD8 T-Cells Is a Correlate of HIV Elite Control
- The Set3/Hos2 Histone Deacetylase Complex Attenuates cAMP/PKA Signaling to Regulate Morphogenesis and Virulence of
- Infidelity of SARS-CoV Nsp14-Exonuclease Mutant Virus Replication Is Revealed by Complete Genome Sequencing
- Combining ChIP-chip and Expression Profiling to Model the MoCRZ1 Mediated Circuit for Ca/Calcineurin Signaling in the Rice Blast Fungus
- Internalin B Activates Junctional Endocytosis to Accelerate Intestinal Invasion
- A Complex Small RNA Repertoire Is Generated by a Plant/Fungal-Like Machinery and Effected by a Metazoan-Like Argonaute in the Single-Cell Human Parasite
- Opc Invasin Binds to the Sulphated Tyrosines of Activated Vitronectin to Attach to and Invade Human Brain Endothelial Cells
- Muc2 Protects against Lethal Infectious Colitis by Disassociating Pathogenic and Commensal Bacteria from the Colonic Mucosa
- PdeH, a High-Affinity cAMP Phosphodiesterase, Is a Key Regulator of Asexual and Pathogenic Differentiation in
- Isolates with Antimony-Resistant but Not -Sensitive Phenotype Inhibit Sodium Antimony Gluconate-Induced Dendritic Cell Activation
- The Microbiota and Allergies/Asthma
- Environmental Factors Determining the Epidemiology and Population Genetic Structure of the Group in the Field
- Prolonged Antigen Presentation Is Required for Optimal CD8+ T Cell Responses against Malaria Liver Stage Parasites
- Crystal Structure of HIV-1 gp41 Including Both Fusion Peptide and Membrane Proximal External Regions
- Susceptibility to Anthrax Lethal Toxin-Induced Rat Death Is Controlled by a Single Chromosome 10 Locus That Includes
- Demonstration of Cross-Protective Vaccine Immunity against an Emerging Pathogenic Ebolavirus Species
- Effective, Broad Spectrum Control of Virulent Bacterial Infections Using Cationic DNA Liposome Complexes Combined with Bacterial Antigens
- High Multiplicity Infection by HIV-1 in Men Who Have Sex with Men
- The -Specific Human Memory B Cell Compartment Expands Gradually with Repeated Malaria Infections
- EBV Promotes Human CD8 NKT Cell Development
- Persistent Growth of a Human Plasma-Derived Hepatitis C Virus Genotype 1b Isolate in Cell Culture
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Quorum Sensing Inhibition Selects for Virulence and Cooperation in
- The Role of Intestinal Microbiota in the Development and Severity of Chemotherapy-Induced Mucositis
- Crystal Structure of HIV-1 gp41 Including Both Fusion Peptide and Membrane Proximal External Regions
- Susceptibility to Anthrax Lethal Toxin-Induced Rat Death Is Controlled by a Single Chromosome 10 Locus That Includes
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



