-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
The Spalt Transcription Factors Generate the Transcriptional Landscape of the Wing Pouch Central Region
How signalling pathways regulate the formation of organs with a precise size and pattern of differentiation is a fundamental question in developmental genetics. One classical example of the link between signalling and organ development is the regulation of wing disc development by the Decapentaplegic/BMP (Dpp) signalling pathway in Drosophila. A key outcome of this pathway is the transcriptional activation of the spalt major (salm) and spalt related (salr) genes, both encoding transcription factors. In this manner, the identification of Salm/Salr target genes is a critical step towards the understanding of the mode of action of these proteins and the genetic logic underlying the regulation of wing development by the Dpp signalling pathway. In order to identify these target genes, we used expression microarrays, in situ hybridization and phenotypic analysis. We identified an unexpected complexity in the transcriptional landscape of the wing disc that includes genes positively and negatively regulated by Salm/Salr. These findings have major implications for the reconstruction of the genetic hierarchy initiated by the Dpp pathway and leading to the formation of a wing with a correct size and pattern, because some of the genes we identified could explain particular aspects of the sal mutant phenotype.
Published in the journal: . PLoS Genet 11(8): e32767. doi:10.1371/journal.pgen.1005370
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005370Summary
How signalling pathways regulate the formation of organs with a precise size and pattern of differentiation is a fundamental question in developmental genetics. One classical example of the link between signalling and organ development is the regulation of wing disc development by the Decapentaplegic/BMP (Dpp) signalling pathway in Drosophila. A key outcome of this pathway is the transcriptional activation of the spalt major (salm) and spalt related (salr) genes, both encoding transcription factors. In this manner, the identification of Salm/Salr target genes is a critical step towards the understanding of the mode of action of these proteins and the genetic logic underlying the regulation of wing development by the Dpp signalling pathway. In order to identify these target genes, we used expression microarrays, in situ hybridization and phenotypic analysis. We identified an unexpected complexity in the transcriptional landscape of the wing disc that includes genes positively and negatively regulated by Salm/Salr. These findings have major implications for the reconstruction of the genetic hierarchy initiated by the Dpp pathway and leading to the formation of a wing with a correct size and pattern, because some of the genes we identified could explain particular aspects of the sal mutant phenotype.
Introduction
The coordination of growth and patterning during the development of tissues and organs depends on the activity of signalling pathways acting in a context-dependent manner. For example the function of the Decapentaplegic (Dpp) signalling pathway is required to regulate cell viability and motility during dorsal closure [1], but the same pathway controls growth and patterning during imaginal disc development [2]. The developmental context is determined by the combinatory of transcription factors expressed in a given tissue, forming gene expression landscapes that influence cell behaviours and also control the response to universal signalling pathways.
The wing imaginal disc is an epithelial tissue that grows by cell proliferation during the larval development of the fly, and differentiates the wing and half of the thorax during pupal development [3]. The growth of the epithelium is accompanied by a progressive specification of spatial territories with different genetic identities. Several signalling pathways play a fundamental role during this process in part by regulating the expression of transcription factors. Among these pathways, the Dpp signalling pathway specifies the central region of the wing blade, its growth and patterning [2]. Several targets and additional components of the transcriptional regulation events triggered by Dpp signalling have being identified in Drosophila [2] including the T-box containing protein Bifid [4] and the Zn-fingers transcription factors Spalt major (Salm) and Spalt related (Salr) [5]. These proteins confer correct epithelial morphology and cell affinity to the central domain of the wing, and also regulate cell proliferation, viability and vein pattern formation [6–8].
Salm and Salr belong to a conserved family of transcriptional regulators that in vertebrates include four components (Spalt-like/Sall1-4) with important developmental roles during neural development and organogenesis [9]. In fact, two human Sall genes are related to the genetic diseases Townes Brocks Syndrome (SALL1) [10] and Okihiro Syndrome (SALL4) [11,12]. The Sal proteins can engage in a variety of interactions with other proteins and with DNA, and they can act as transcriptional repressors or activators [9,13–18]. Salm and Salr act as transcriptional repressors in Drosophila cultured cells, and the activity of at least Salr depends on the histone deacetylase complex NuRD [15]. However, the mechanisms by which Sal proteins regulate transcription are still not fully understood, although they include interaction with heterochromatic regions and recruitment of histone deacetylase complexes [13–15].
The spalt genes play a central role in mediating the effects of Dpp signalling during wing disc development [8], but the identity of Sal target genes is still unknown. Thus, only two gene complexes, the knirps and Iroquois gene complexes, have being identified as candidate downstream genes of Sal in the specification of vein territories [19,20]. However, Sal proteins are not only required for vein patterning, but also to promote cell division and survival in the central region of the wing [8], and they contribute to the maintenance of epithelial integrity in this territory [8,21]. The identity of the targets mediating these functional requirements is totally unknown. The identification of Salm/Salr target genes is of critical importance to understand the genetic hierarchy acting downstream of Dpp signalling in the wing disc.
Here we describe the global transcriptional changes that occur in salm/salr knockdown discs. These data, in combination with in situ hybridization assays and phenotypic analysis has allowed us to identify a collection of candidate genes that are regulated by the Sal proteins and might mediate their functions in the wing disc. Our work identifies an unsuspected transcriptional complexity occurring downstream of Spalt that involves repression as well as activation of gene expression.
Results
Genome-wide transcriptional changes observed in wing discs with reduced expression of salm and salr
In third instar wing discs salm and salr are expressed in a broad stripe of cells in the wing pouch (Fig 1A-1A’) that corresponds in the adult wing to a region including the vein L2 and extending to the L4/L5 intervein. This expression is directly regulated by Dpp signalling [22]. It is likely that Salm/Salr regulate the expression of a collection of target genes that in turn mediate the variety of functions assigned to them. To identify “Salm/Salr Target Genes” (STG), we compared the expression profile of wing discs in which the expression of both salm and salr genes was reduced by RNA interference with control wing discs. For these experiments, we used the salEPv-Gal4 driver, which reproduces the domain of salm/salr expression in the wing blade region of the imaginal disc [23] (Fig 1A). In addition, we took advantage of the temperature-sensitive Gal80 protein, which blocks the activity of Gal4 at 25°C but is inactive at 29°C [24]. Thus, in salEPv-Gal4 UAS-GFP; tub-Gal80ts/UAS-GFP discs grown at 25°C the expression of GFP was undetected in the wing blade (Fig 1B’), and the expression of Salm was unaffected (Fig 1B). This allowed us to precisely determine the time since when we started expressing the salm and salr RNA interferences. We chose to compare two time points, one 24 hours and other 48 hours after a 25°C to 29°C temperature shift, as this might allow to distinguish direct and indirect Salm/Salr transcriptional effects. We found that after 24 hours at 29°C (T24) the expression of Salm was already undetected in the wing blade (Fig 1C-1C’’; salEPv-Gal4 UAS-GFP /UAS-salm-i; tub-Gal80ts / UAS-salr-i), whereas the expression of GFP was strong in the central domain of the wing (Fig 1C-1C’). As expected, we also found that GFP expression was robust and Salm expression lost after 48 hours at 29°C (T48) (Fig 1D-1D’’). Wing discs observed at T48 differ from those analysed at T24 in that they displayed a severer phenotype of reduction in the size of the salEPv-Gal4 expression domain (compare Fig 1D and 1C). Older discs also showed a loss in FasIII expression (S1 Fig), indicating that the developmental defects characteristics of salm/salr loss develop progressively over time.
Fig. 1. Global results of the genome-wide transcriptional changes observed in wing discs with reduced expression of salm and salr. 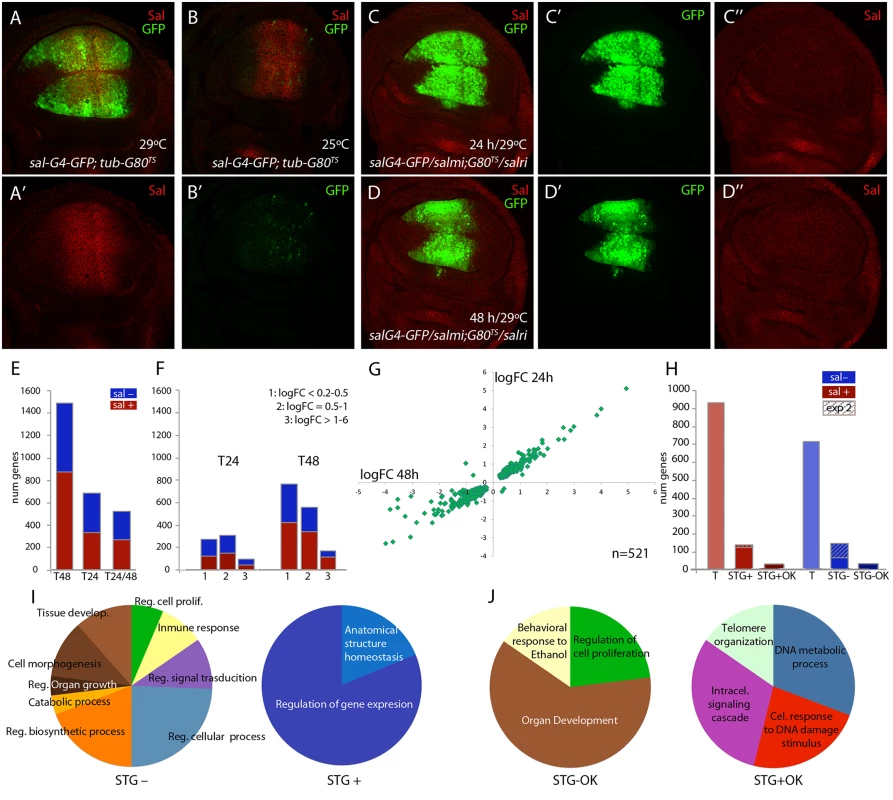
(A-A’) Expression of Salm (red in A and A’) and salEPv-Gal4 (green in A) in salEPv-Gal4 UAS-GFP; tub-Gal80ts/UAS-GFP wing discs raised at 29°C. (B-B’) Expression of Salm (red in B) and salEPv-Gal4 (green in B’) in salEPv-Gal4 UAS-GFP; tub-Gal80ts/UAS-GFP wing discs raised at 25°C. (C-C’’) Imaginal disc of salEPv-Gal4 UAS-GFP /UAS-salm-i; tub-Gal80ts /UAS-salr-i genotype (salm-i/salr-i 24h) raised at 29°C 24–28 hours before dissection, showing the expression of GFP (green in C and C’) and Salm (red in C and C’’). (D-D’’) Imaginal disc of salEPv-Gal4 UAS-GFP /UAS-salm-i; tub-Gal80ts /UAS-salr-i genotype (salm-i/salr-i 48h) raised at 29°C 44–48 hours before dissection showing the expression of GFP (green in D and D’) and Salm (red in D and D’’). (E) Number of genes which expression level changes with an adjusted p-value lower than 0.05 that were identified in the comparisons with controls of salm-i/salr-i at 48h (T48); salm-i/salr-i at 24h (T24) and at both time intervals (T24/T48). The blue and red sections of each column correspond to genes showing reduced (sal-; blue) and increassed (sal+; red) expression in salmi/salri compared to their corresponding controls. (F) Number of genes identified at T24 and T48 grouped by logFoldChange values. Blue represents genes which expression is reduced and red those genes which expression is increased. 1: logFC lower than 0.5; 2: logFC between 0.5 and 1 and 3: logFC major than 1. (G) Graphical representation of the logFoldChange values for those genes that were identified both in salm-i/salr-i 48h and salm-i/salr-i 24h. (H) Total number of identified genes from experiment 1 (T) with increased (sal+; red) and reduced (sal-; blue) expression in salm-i/salr-i discs compared to controls. Number of selected genes coming from experiments 1 and 2 (STG) with increased (STG+) and reduced (STG-) expression. Genes selected from experiment 2 are indicated by stripped columns. Number of best candidate sal repressed (STG+OK) and activated (STG-OK) genes. (I-J) Number of genes grouped in GO categories enriched for STG- and STG+ genes (I) and for STG-OK and STG+OK genes (J). In the case of control discs (salEPv-Gal4 UAS-GFP; tub-Gal80ts/UAS-GFP) we could not detect any difference between the expression profiles of discs grown 24 or 48 hours at 29°C. In contrast, in salEPv-Gal4 UAS-GFP/UAS-salm-i; tub-Gal80ts/UAS-salr-i (salm-i/salr-i) discs we detected 71 genes whose expression changes comparing discs at T48 and T24 (S1 Table). Most of these cases (69) correspond to genes whose expression was increased at T48 compared to T24. We also compared salm-i/salr-i and control discs at T24 and T48. In the first case (T24) we detected significant changes (adjusted p-value lower than 0.05) in the expression levels of 686 genes, of which 98 displayed a fold-change higher than 2 (Fig 1E and 1F and S2 Table). The number of changes was much higher at T48, where we found changes in the expression levels of 1488 genes, including 168 with a fold-change higher than 2 (Fig 1E and 1F; S2 Table). Most of the genes whose expression level changed at T24 were also detected at T48 hours (n = 521 out of 686). In these cases, there was a good correlation between the extent of change observed in the T24 and T48 classes (Fig 1G). Genes that were only detected at T24 (165) in general had low fold-change values, and only 10 had a fold-change higher than 2 (S2 Table). These lists of genes show a low overlap with other transcriptomic and genomic analyses carried out in the wing disc. For example, we only could find 13 coincidences with the list of 146 genes enriched in the notum or wing regions of the wing disc [25], 40 coincidences with the 1649 genes associated to Brinker binding peaks [26] and 10 coincidences with the 4954 genes identified as modified comparing sal mutant embryos with control ones [27]. These results indicate that the collection of genes identified in our microarrays corresponds to a novel set of genes that were not selected before in other genomic studies in the wing disc.
From the total of genes that showed a significant change comparing salm-i/salr-i and control discs at T24 and T48, we decided to select those ones that changed with a logFC of at least 1. We distinguished between those that are present at both time points (n = 124) and those that changed only at T24 or T48 (n = 67). A total of 51 genes from these 191 selected ones (30%) were also identified in the microarrays described in experiment 2, in which we compared expression profiles from discs over-expressing sal (UAS-sal) and disc carrying salm/salr mutant clones with their respective controls (see material and methods; experiment 2). To generate a collection of candidates STG for subsequent analysis, we added a total of 95 genes that were only selected in experiment 2 with the best scores (Fig 1H). The joint list of STG (n = 286) includes 139 genes whose expression increases in salm/salr loss of function (STG+; Fig 1H and S3 Table) and 147 whose expression levels decreases in salm/salr loss of function conditions (STG-; Fig 1H and S3 Table). Based on subsequent in situ hybridization data and phenotypic analysis (see below), we further selected from the STG+ and STG - groups a collection of 32 and 34 genes, named STG+OK and STG–OK respectively, which were considered the best candidates to be regulated by Sal and to mediate the functions of Sal proteins (Fig 1H; see Table 1).
Tab. 1. Best candidates Salm/Salr Target Genes (STG OK). 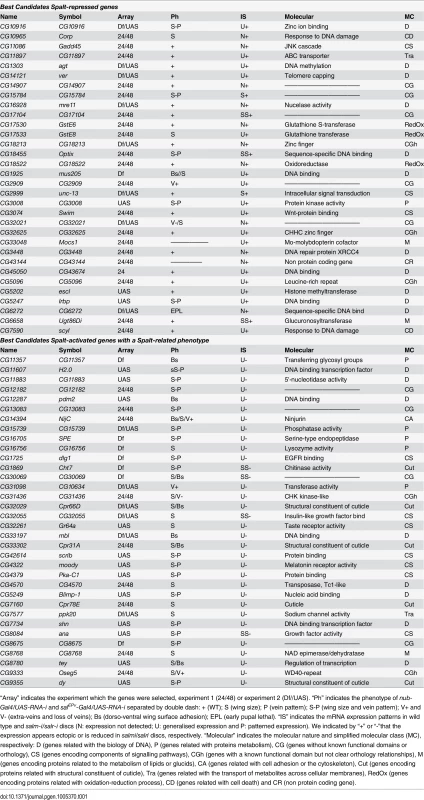
Best candidates Spalt-repressed genes and Best candidates Spalt-activated genes with a Spalt-related phenotype. Gene Ontology analysis
The STG+ and STG - (286) genes encode proteins belonging to a variety of molecular categories among which stand out those related to the biology of DNA (17%) and proteins (11%), and general metabolism of lipids or glucids (9%; S2 Fig). They also include a large collection of genes without known functional domains or orthology relationships (18%) as well as genes with some known functional domain but not clear orthology (8%) (S2 Fig). The class of genes related to the biology of DNA includes all sequence-specific transcription factors and other DNA modifying enzymes that impinge in the regulation of gene expression, and its prominence suggests that Salm/Salr proteins might regulate wing development in part by controlling the expression of other transcription factors acting as intermediaries. Some sets were differently represented in the STG+ and STG - groups, including protein metabolism (6% versus 16%), Red-Ox (9% vs 2%) and cell signalling (7% vs 14%). The biological significance of these variations is at present unknown.
To obtain information about possible enrichments of gene functions related to the known requirements of salm/salr, we inspected several collections of genes using the software DAVID [28,29] with the GO term “biological process” (S4 Table). For those genes whose RNA expression levels change with a adjusted p-value lower than 0.05 at 24h after salm/salr RNAi induction (686 genes), we only found a limited enrichment of GO terms related with the known functions of Sal (S4 Table). In contrast, the STG genes with reduced expression in salm/salr knockdown (STG-; 147) were enriched for the terms “regulation of cell proliferation” (5 genes), “tissue development” (9 genes), “cell morphogenesis” (9 genes), “regulation of signal transduction” (8 genes) and “regulation of biosynthetic process” (15 genes) (Fig 1I and S4 Table). The STG genes with augmented expression in salm/salr knockdown conditions (STG+; 139 genes) were only enriched in the descriptions “regulation of gene expression” (13 genes) and “anatomical structure homeostasis” (3 genes) Fig 1I and S4 Table).
mRNA expression in the wing disc of selected genes with increased expression in salm-i/salr-i compared to wild type wing discs
We studied the expression pattern of 100 of the 139 STG+ genes in wild type and in salm-i/salr-i backgrounds. Genes that were identified only as changing at T48 (39) were discarded for this analysis, as we assumed that, likely, this group included most genes whose expression changes indirectly in response to the loss of salm/salr function. Most of the analysed genes (n = 100) displayed a generalised pattern of expression in third instar wing disc (U, 69%; Fig 2A and examples in Fig 2D–2F) or their expression was not detected by in situ hybridization (N, 21%; Fig 2A and examples in Fig 2B and 2C). Only 10% of the genes analysed were expressed with a spatial pattern that, in general, was unrelated to the pattern of expression of salm/salr in the wing blade (P; Fig 2A and examples in Fig 2G–2I). The expression patterns of all genes included in these categories are shown in S3–S7 Figs, and selected representative examples are shown in Fig 2B–2I.
Fig. 2. Examples of mRNA expression of selected genes (STG+) with increased expression in salm-i/salr-i compared to wild type wing discs. 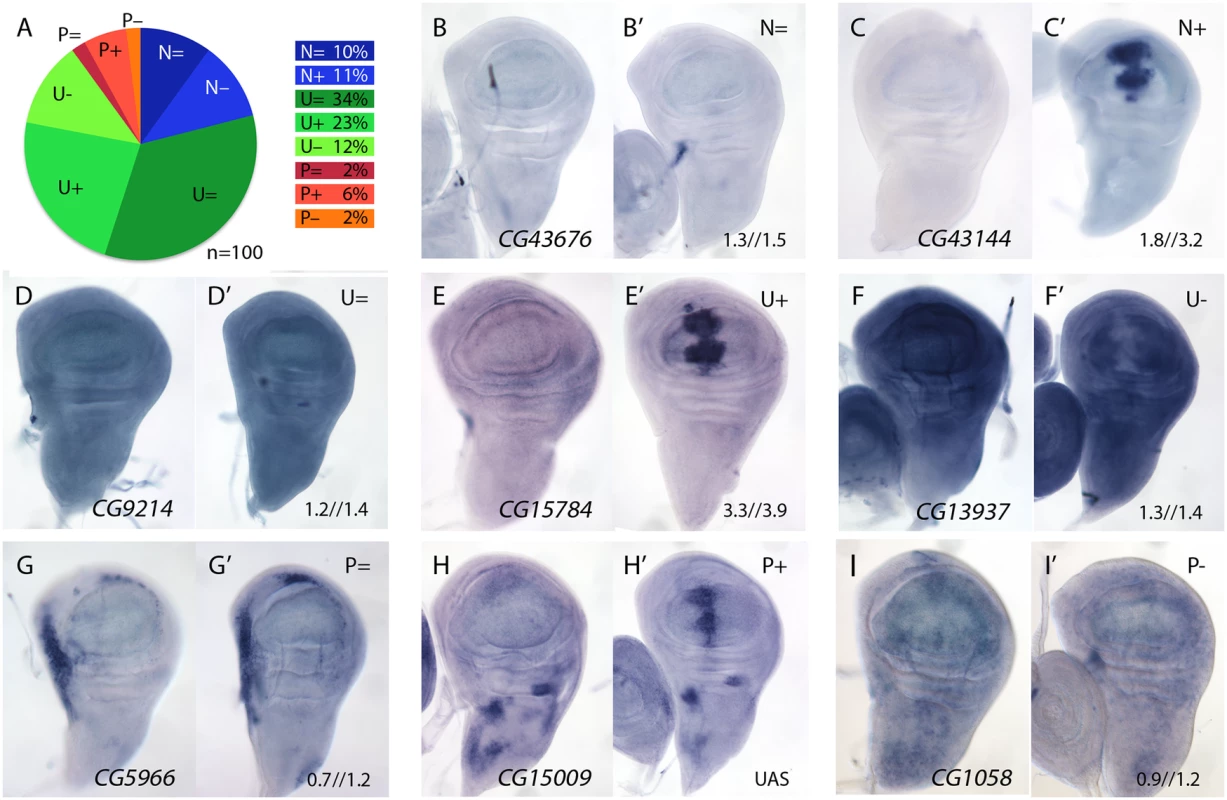
(A) Frequency of expression patterns encountered in wild type discs and observed changes in UAS-dicer2/+; salEPv-Gal4 UAS-GFP/UAS-salm-i; UAS-salr-i/+ wing discs. Expression not detected in the wing disc (N), generalised expression (U) and patterned expression (P). We indicated by “=“, “+” and “-”that the corresponding expression does not change, appears ectopic or is reduced in salm-i/salr-i wing discs, respectively. (B-I) Representative examples of in situ hybridizations in late third instar wing discs with probes against the genes CG43676 (N =; B-B’), CG43144 (N =; C-C’), CG9214 (U =; D-D’), CG15784 (U+; E-E’), CG13937 (U-; F-F’), CG5966 (P =; G-G’), CG15009 (P+; H-H’) and CG1058 (P-; I-I’). In each pair of panels, B-I corresponds to wild type discs and B’-I’ to UAS-dicer2/+; salEPv-Gal4 UAS-GFP/UAS-salm-i; UAS-salr-i/+ wing discs. The expression patterns class and logFC values at 24h and 48 h from experiment 1 or the microarray condition from experiment 2 are indicated at the top and to the bottom corner, respectively, of each panel in B’ to L’ images. The expression of all these genes was also studied in salm/salr loss of function conditions (UAS-dicer2/+; salEPv-Gal4 UAS-GFP/UAS-salm-i; UAS-salr-i/+). We found a significant change in the expression patterns detected by in situ hybridization in 40 cases (40%). In all these cases, and irrespectively of the normal pattern of expression of the gene, we detected a strong stripe of increased (for U and P genes) or ectopic (for N genes) expression localised in the central region of the wing disc (Fig 2C-2C’, 2E-2E’ and 2H-2H’; see also S3 and S4 Figs). For the remnant of genes (n = 60) either we could not detect any change in their spatial expression (46%; Fig 2A, 2B-2B’, 2D-2D’ and 2G-2G’) or their expressions were reduced (14%; Fig 2A, 2F-2F’ and 2I-2I’). We do not know the reasons underlying this discrepancy between the microarray data and the in situ hybridization analysis. However, as both techniques as subject to different sources of noise and experimental error, we considered that this discrepancy was acceptable, and focussed for any subsequent analysis in those genes that display a clear change by in situ hybridization in salm-i/salr-i knockdown discs. All cases in which we found an increase in expression in salm/salr knockdown discs are shown in S3 and S4 Figs, and those for which we could not detect a change in the in situ expression or their expressions were reduced are shown in S5–S7 Figs.
Interestingly, the genomic regions associated to the genes over-expressed in salm/salr knock-down discs (n = 40) were enriched in the H3K9me3 epigenetic mark (12,79% versus 3,09% in the genome, p-value 0.001), which is associated to heterochromatic regions [30]. These genes included an enriched set for the terms “DNA metabolic process”, “telomere organization” and “cellular response to DNA damage stimulus” formed by mre11, mus205, Irbp, agt, Dif and scyl (see Fig 1J and S4 Table, STG+OK). The functional relation between these genes and the Sal functions is unknown, but certainly deserves an in depth exploration.
A fraction of genes over-expressed in salm/salr knockdown background are targets of the JNK pathway
The JNK signalling pathway is activated in the Salm/Salr domain in salm/salr knockdown backgrounds [8]. This pathway can regulate gene expression through the action of the JUN and FOS transcription factors [31], and therefore, it is possible that some of the genes over-expressed in the central domain of salm/salr knockdown wing discs correspond to targets of the JNK pathway. To distinguish between genes repressed by Salm/Salr and genes activated by JNK signalling, we studied the expression of genes over-expressed in salm/salr knockdown background (n = 40) when JNK signalling is suppressed. To suppress this pathway, we over-expressed the JNK phosphatase puckered (puc) in wing discs where salm and salr expression was reduced by RNA interference. We found that the suppression of JNK by puc over-expression cancels the ectopic expression observed in salmi/salri background for 8 genes (Fig 3A, 3D–3F’ and S8 Fig). These genes were more frequently either not expressed in wild type discs (N+; 3 out of 11, see Fig 3B) or expressed in a generalised manner (U+; 4 out of 23, see Fig 3B). Only the ectopic expression of one gene that was normally expressed in a spatially restricted pattern turned out to be regulated by, or dependent on, JNK signalling (P+; 1 out of 6, see Fig 3B). Interestingly, most JNK-dependent genes were either undetected or detected with low logFC values after the 24h period at 29°C (Fig 3C), which is compatible with them being regulated by Salm/Salr indirectly. For 32 out of 40 genes analysed, we found that the over-expression of puc did not affect the characteristic ectopic expression caused by the loss of salm/salr (Fig 3A and 3G–3I’ and S8 Fig). Most genes whose expression behaves as independent of JNK displayed a significant logFC at both 24 and 48 hours intervals, which suggest that they might be direct targets of Salm/Salr regulation (Fig 3C). This set of 34 genes that show JNK-independent over-expression in salmi/salri discs was considered the collection of the best candidate Sal target genes regulated by repression (STG+OK; Fig 1G and 1J and Table 1).
Fig. 3. Analysis of JNK-dependent gene regulation for genes ectopically expressed in salm/salr knockdown discs. 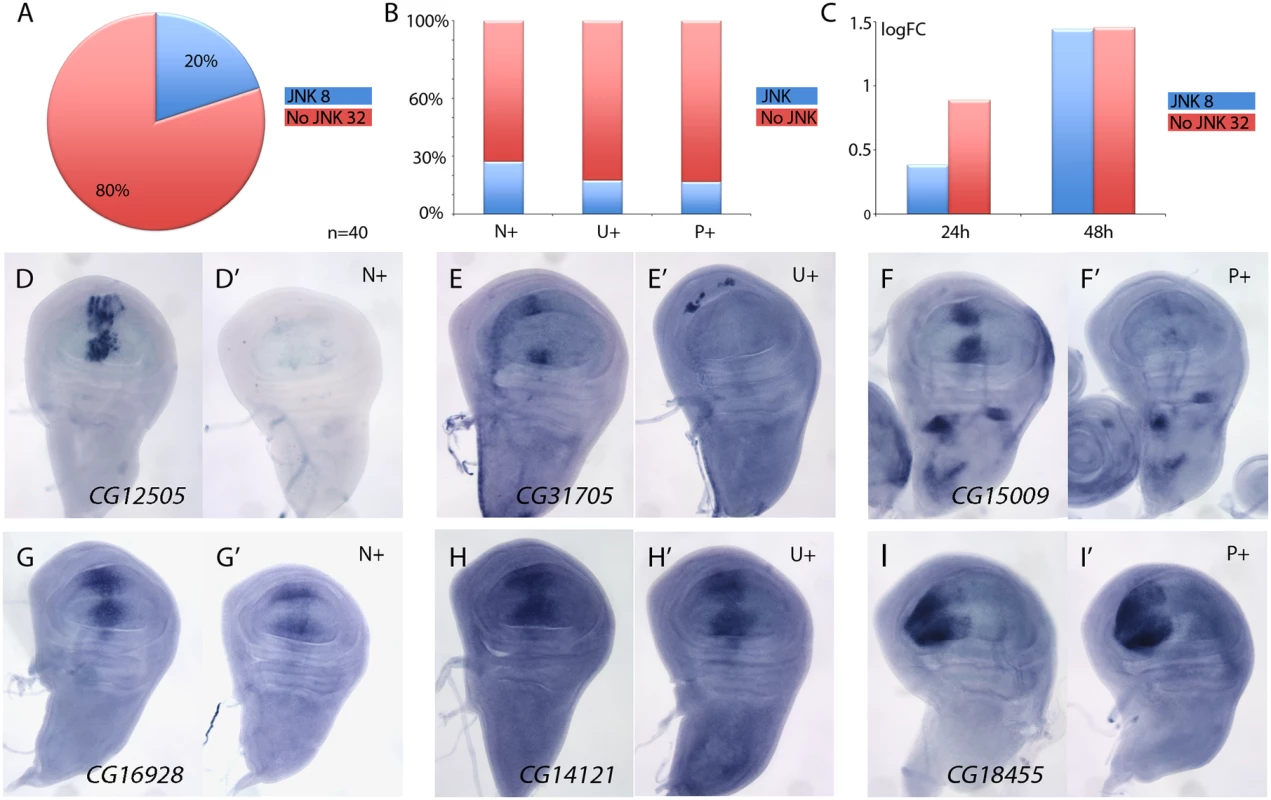
(A) Frequency of genes which ectopic expression in salm-i/salr-i discs is independent (80%, red; 32 genes) or dependent (20%, blue; 8 genes) of JNK signalling. (B) Percentage of JNK-dependent genes (blue) and JNK-independent genes (red) which are normally not expressed (N+), are expressed in a generalised manner (U+) or are expressed with a spatially restricted pattern (P+). (C) Average of logFC of JNK-dependent genes (blue columns) and JNK-independent genes (red columns) at 24h (left columns) and 48h (right columns) at 29°C. (D-F’) Representative examples of in situ hybridizations in late third instar wing discs with probes against the JNK-dependent genes CG12505 (D-D’), CG31705 (E-E’) and CG15009 (F-F’). The panels D-F correspond to salEPv-Gal4 UAS-GFP/UAS-salm-i; UAS-salr-i/UAS-GFP discs and D’-F’ to salEPv-Gal4 UAS-GFP/UAS-salm-i; UAS-salr-i/UAS-puc wing discs. (G-I’) Representative examples of in situ hybridizations in late third instar wing discs with probes against the JNK-independent genes CG16928 (G-G’), CG14121 (H-H’), and CG18455 (I-I’). The panels G-I correspond to salEPv-Gal4 UAS-GFP/UAS-salm-i; UAS-salr-i/UAS-GFP discs and G’-I’ to—salEPv-Gal4 UAS-GFP/UAS-salm-i; UAS-salr-i/UAS-puc wing discs. It is possible that some of the phenotypic aspects of loss of Sal function are related with the ectopic expression of genes showing a strong de-repression in the central domain of the wing pouch in salm/salr knockdown conditions. To identify whether this was the case, we introduced RNA interference for all 40 genes de-repressed in the salm-i/salr-i background. In none of these combinations (salEPv-Gal4 UAS-salm-i/UAS-RNA-i; UAS-salr-i/+) we could detect a significant change of the phenotype compared with control flies (salEPv-Gal4 UAS-salm-i/UAS-GFP; UAS-salr-i/+) indicating that the phenotype of salm/salr cannot be assigned to single genes being over-expressed.
mRNA expression in the wing disc of selected genes with decreased expression in salmi/salri compared to wild type wing discs
We also selected 147 genes whose expression was reduced in salm/salr knockdown discs (STG-; Fig 1H), and studied the expression pattern of 139 of them. Most of these genes are expressed in a generalised manner in the wing disc (84%; class U, Fig 4A), and only 14% displayed restricted expression patterns that relate to the formation of veins and interveins (Fig 4A; class P). For 2% of the selected genes we could not detect expression by in situ hybridization in the wing disc (Fig 4A–4C’; class N). The changes observed in the expression of genes detected as reduced in salmi/salri background were very coherent, and consisted in a strong reduction of their expression in the central region of the wing pouch (Fig 4E–4F’, 4H–4I’, S9–S12 Figs). This change was observed for genes that are normally expressed in all wing disc cells (class U; Fig 4E–4F’ and S9, S11 and S12 Figs), and for genes displaying restricted expression patterns (class P; Fig 4H–4I’ and S9 and S10 Figs).
Fig. 4. Examples of mRNA expression of selected genes with reduced expression in salm-i/salr-i compared to wild type wing discs. 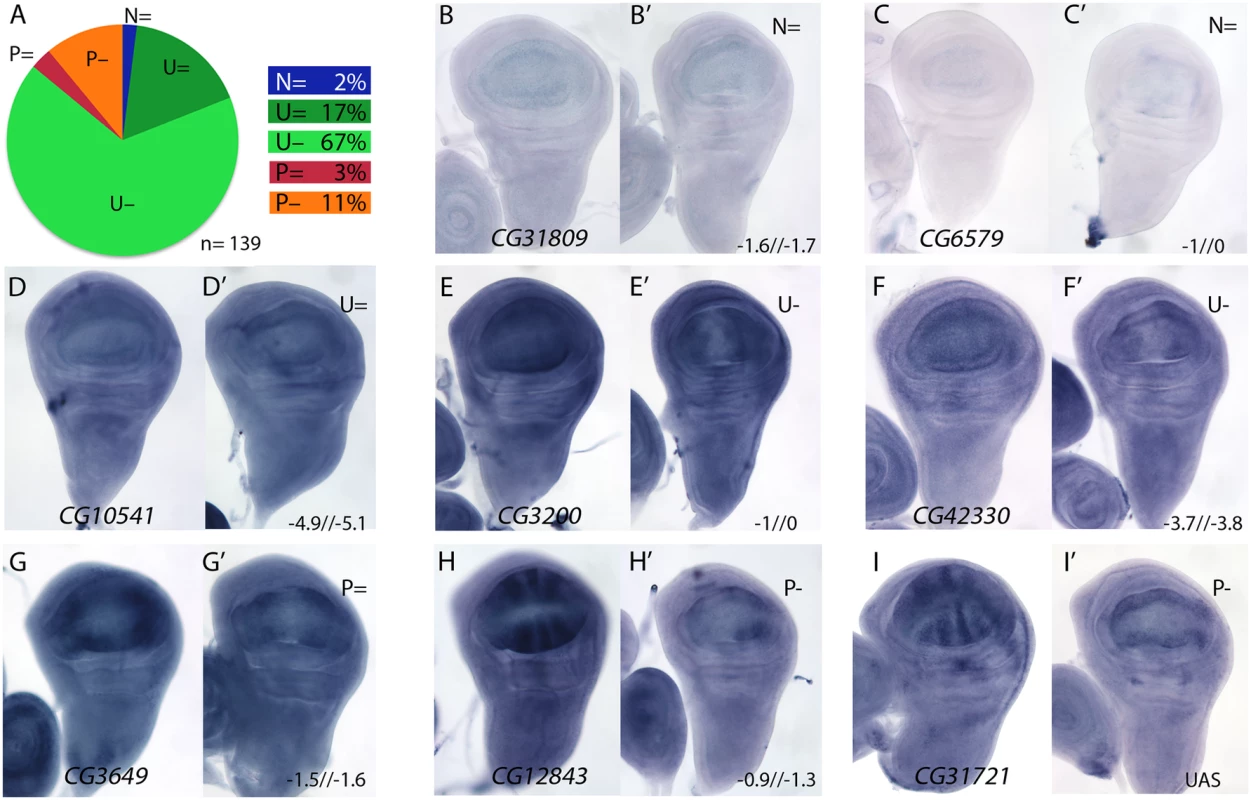
(A) Frequency of expression patterns in wild type discs and observed changes in UAS-dicer2/+; salEPv-Gal4 UAS-GFP/UAS-salm-i; UAS-salr-i/+ wing discs. Expression not detected in the wing disc (N), generalised expression (U) and patterned expression (P). We indicated by “=“, “+” and “-”that the expression does not change, appears ectopic or is reduced in salmi/salri discs, respectively. (B-I) Representative examples of in situ hybridizations in late third instar wing discs with probes against the genes CG31809 (N =; B-B’), CG6579 (N =; C-C’), CG10541 (U =; D-D’), CG3200 (U-; E-E’), CG42330 (U-; F-F’), CG3649 (P =; G-G’), CG12843 (P-; H-H’) and CG31721 (P-; I-I’). In each pair of panels, B-I corresponds to wild type discs and B’-I’ to UAS-dicer2/+; salEPv-Gal4 UAS-GFP/UAS-salm-i; UAS-salr-i/+ wing discs. The expression patterns class and logFC values at 24h and 48 h from experiment 1 or the microarray condition from experiment 2 are indicated at the top and to the bottom corner, respectively, of each B’ to L’ panel. For a 22% of the analysed cases we could not detect a change in the expression pattern comparing wild type and salm/salr knockdown discs (Fig 4A, Fig 4D-4D’, Fig 4G-4G’ and S13 Fig). We wondered whether some of these genes were regulated by the Dpp pathway, independently of Spalt, in the central region of the wing. To reduce Dpp activity we over-expressed Dad in this territory (salEPv-Gal4; UAS-dad; Fig 5A–5D). In this background the expression of Sal was strongly reduced (Fig 5C-5C’; compared with Fig 5B) and the resulting adult wing display a moderate to strong Dpp phenotype (Fig 5D), that could be partially rescued by the co-expression of either Salm (Fig 5E) or Salr (Fig 5F). Indeed, we found that the expression of CG32372 (Fig 5G-5G’’’) and CG17278 (Fig 5H-5H’’’) are not strongly affected in salm-i/salr-i discs (Fig 5G’, 5H’ and 5I’), but are reduced or absent in UAS-dad discs (Fig 5G’’ and 5H’’). Because the loss of CG32372 and CG17278 in UAS-dad discs cannot be rescued by the over-expression of Salm (Fig 5G’’’ and 5H’’’), we suggest that these genes are likely candidates for Dpp regulation independently of Salm/Salr. Indeed, CG32372 corresponds to larval translucida, a gene which expression was already shown to be regulated by Dpp [32]. We assume that genes expressed in the central domain of the wing disc and regulated by Dpp will be picked up in our microarray experiments as Sal-, because the extent of this territory is reduced in salm/salr knockdown discs.
Fig. 5. Genes regulated by Dpp signalling independently of Salm/Salr. 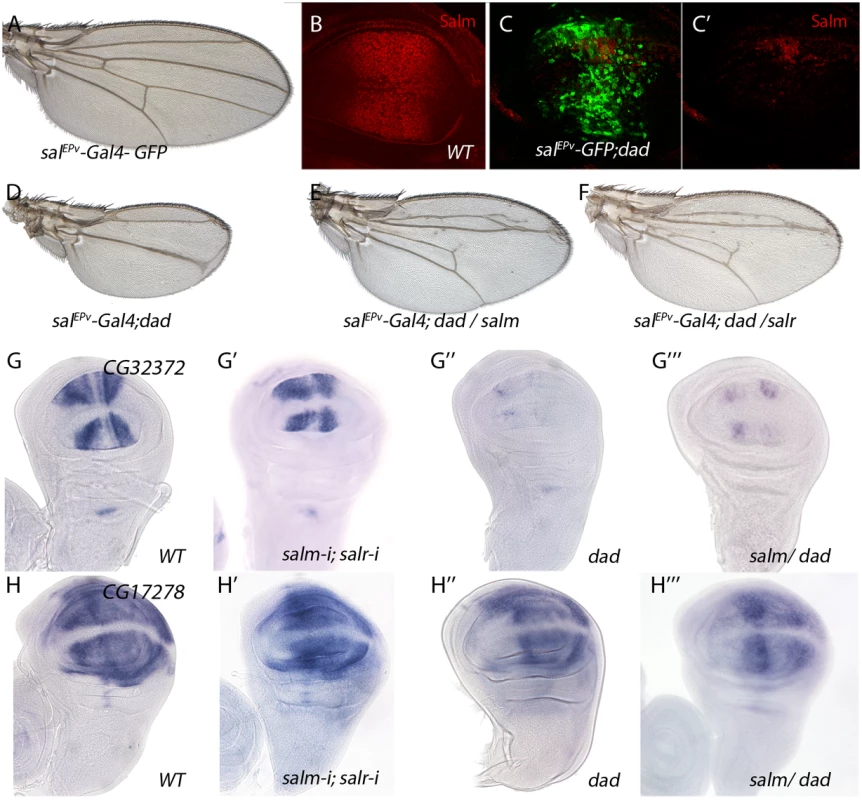
(A) Control salEPv-Gal4 UAS-GFP/+ wing. (B) Control third instar wing disc showing Spalt major expression (Red). (C-C’) salEPv-Gal4 UAS-GFP/+; UAS-dad/+ wing disc showing Salm (red in C and C’) and GFP expression (green in C). (D) salEPv-Gal4 UAS-GFP/+; UAS-dad/+ wing. (E) salEPv-Gal4 UAS-GFP/+; UAS-dad/UAS-salm wing. (F) salEPv-Gal4 UAS-GFP/+; UAS-dad/UAS-salr wing. Forced expression of Salm (E) or Salr (F) in the central region of salEPv-Gal4 UAS-GFP;UAS-dad (D) discs partially rescues the size and pattern defects caused by Dad over-expression. (G-G’’’) CG32372 in situ hybridization in third instar discs of salEPv-Gal4 UAS-GFP/+ (G), salEPv-Gal4 UAS-GFP/UAS-salm-i; UAS-salr-i/+ (G’), salEPv-Gal4 UAS-GFP/+; UAS-dad/+ (G’’) and salEPv-Gal4 UAS-GFP/+; UAS-dad/UAS-salm (G’’’). (H-H’’’) CG17278 in situ hybridization in third instar discs of salEPv-Gal4 UAS-GFP/+ (H), salEPv-Gal4 UAS-GFP/UAS-salm-i; UAS-salr-i/+ (H’), salEPv-Gal4 UAS-GFP/+; UAS-dad/+ (H’’) and salEPv-Gal4 UAS-GFP/+; UAS-dad/UAS-salm (H’’’). The over-expression of Salm does not rescue the loss of CG32372 or CG17278 expression caused by increased Dad expression. Phenotypic analysis of genes with modified level of expression in salm/salr knockdown discs
From the microarray and in situ hybridization data, it seems clear that the reduction of salm/salr function in the wing disc causes a profound alteration in the transcriptional landscape of the wing. Some of these changes might be direct, but it is likely that many others could be the consequence of inappropriate expression of Salm/Salr target genes encoding transcription factors, or, as we found for JNK target genes, the result of inappropriate activation of signalling pathways. For these reasons, it is unlikely that by interfering with the expression of each individual gene we were able to reproduce the effects of salm/salr loss during wing development. Despite of this, we wanted to make a preliminary identification of the functional requirements of the Salm/Salr candidate downstream genes by analysing the consequences of reducing their expression. To this end, we expressed in the wing disc (nub-Gal4/UAS-RNA-i and salEPv-Gal4/UAS-RNA-i) RNA interference directed against most genes belonging to the STG+ (126 out of 139) and STG - (139 out of 147) classes. As expected, we found that 66% of STG+ genes and 50% of STG - genes tested in Gal4/UAS-RNAi combinations resulted in wings that were normal in size and vein pattern (Figs 6A and 7A; class N and S5 Table). The remnant of the Gal4/UAS-RNA-i combinations resulted in wings with abnormal size, altered pattern of veins, or a collapse of the entire central region of the wing (Figs 6A and 6B, 7A and 7B; class Y and S5 Table).
Fig. 6. Phenotypic analysis of genes with increased expression in sal knockdown discs. 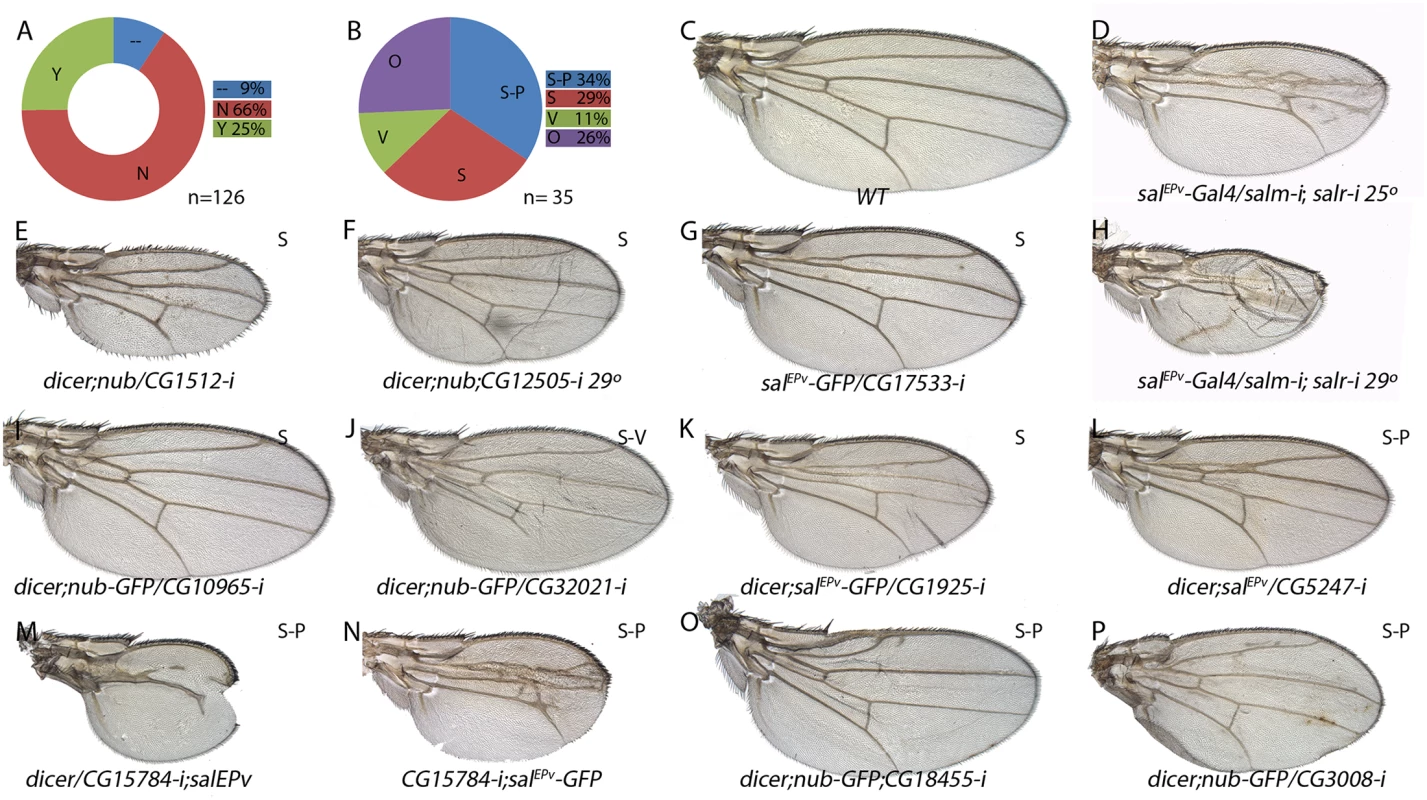
(A) Fraction of genes showing a wing mutant phenotype (Y; green) and no phenotype (N; red) from a total of 126 UAS-dicer2/+; nub-Gal4/ UAS-RNA-i or UAS-dicer2/+; salEPv-Gal4/ UAS-RNA-i combinations. Genes not analysed from the STG+ class are shown in blue (—) (B) Frequency of defects in both wing size and vein pattern (S-P; blue), changes in wing size (S; red), defects in vein formation with minor or no effect in wing size (V; green) and other wing morphology defects (O; purple) from a total of 35 UAS-RNA-i/Gal4 combinations. (C) Wild type wing. (E-F) Representative examples of adult wings knockdown for genes over-expressed in salm-i/salr-i mutant discs but activated in response to JNK. UAS-dicer2/+; nub-Gal4/UAS-CG1512-i (E) and UAS-dicer2/+; nub-Gal4/UAS-CG12505-i (F). (G-P) Representative examples of adult wings of UAS-RNA-i/Gal4 combinations for genes over-expressed in salm-i/salr-i knockdown discs independently of JNK activity: salEPv-Gal4 UAS-GFP/UAS-CG17533-i (G), UAS-dicer2/+; nub-Gal4/UAS-CG10965-i (I), UAS-dicer2/+; nub-Gal4/UAS-CG32021-i (J), UAS-dicer2/+; salEPv-Gal4/UAS-CG1925-i (K), /+;UAS-dicer2/+; salEPv-Gal4/ UAS-CG5247-i (L), UAS-dicer2/UAS-CG15784-i; salEPv-Gal4/+ (M), UAS-CG15784-i; salEPv-Gal4 UAS-GFP/+ (N), UAS-dicer2/+; salEPv-Gal4/UAS-CG18455-i (O) and UAS-dicer2/+; nub-Gal4/UAS-CG3008-i (P). Moderate and strong salm-i/salr-i knockdown phenotypes are shown for comparison in (D) and (H), in wings of salEPv-Gal4/ UAS-salm-i; UAS-salr-i/+ raised at 25°C and 29°C, respectively. Fig. 7. Phenotypic analysis of genes with reduced expression in sal knockdown discs. 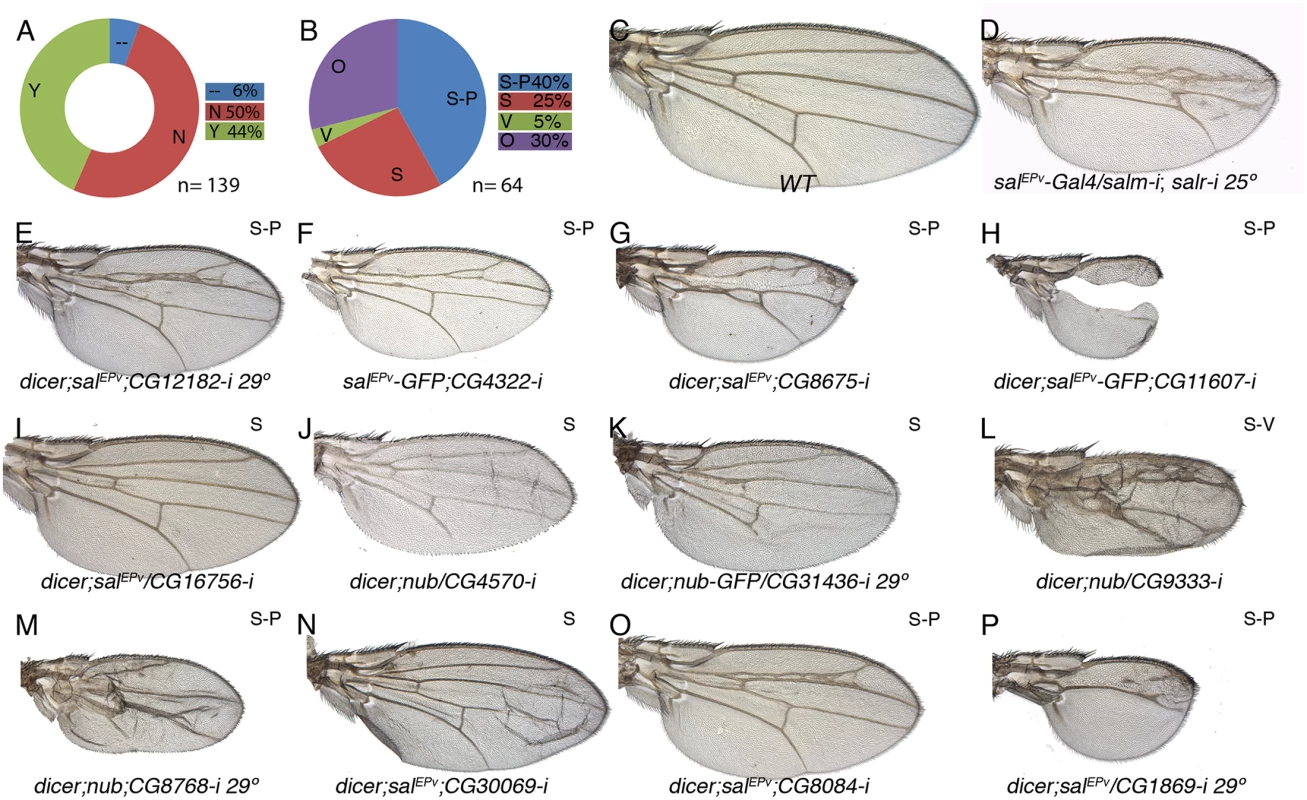
(A) Fraction of genes showing a wing mutant phenotype (Y; green) and no phenotype (N; red) from a total of 139 UAS-RNA-i/Gal4 combinations. Genes not analysed of the STG- class are shown in blue (—). (B) Frequency of mutant wings showing defects in both wing size and vein pattern (blue; S-P), changes in wing size (S; red), defects in vein formation with minor or no effect in wing size (V; green) and other morphological defects in the wing (O; purple) from a total of 64 UAS-RNA-i/Gal4 combinations. (C) Wild type wing. (D) salEPv-Gal4/UAS-salm-i; UAS-salr-i/+ wing. (E-P) Representative examples of adult wings knockdown for genes showing reduced expression by in situ hybridization in salm-i/salr-i wing discs. UAS-dicer2/+; salEPv-Gal4/UAS-CG12182-i grown at 29°C (E); salEPv-Gal4 UAS-GFP/UAS-CG4322-i (F), UAS-dicer2/+; salEPv-Gal4 / UAS-CG8675-i (G), UAS-dicer2/+; salEPv-Gal4/+; UAS-CG11607-i/+ (G), dicer2/+; salEPv-Gal4/ UAS-CG16756-i (I), UAS-dicer2/+; nub-Gal4/UAS-CG4570-i (J), UAS-dicer2/+; nub-Gal4/UAS-CG31436-i (K), UAS-dicer2/+; nub-Gal4/+; UAS-CG9333-i/+ (L), UAS-dicer2/+; nub-Gal4/UAS-CG8768-i (M), UAS-dicer2/+; salEPv-Gal4/UAS-CG30069-i (N), UAS-dicer2/+; salEPv-Gal4/+; UAS-CG8084-i/+ (O) and UAS-dicer2/+; salEPv-Gal4/ UAS-CG1869-i at 29°C (P). Some combinations between UAS-RNAi and nub-Gal4, which is expressed in the wing blade and hinge, resulted in pupal lethality (9 for STG+ and 14 for STG-; S5 Table). In these cases, and when we were able to inspect the pharate adults, the phenotype was the total absence of the wing (see for example S14B Fig and S15F Fig). A similar no-wing phenotype was also found in some combinations (8) that were able to produce adult escapers (S5 Table). The UAS-RNA-i for all these genes (pupal lethal and no-wing phenotype in S5 Table) were also combined with salEPv-Gal4, which is only expressed in the central region of the wing blade. In all cases, the resulting adult wings displayed a variety of phenotypes ranging from severe disruptions of the central wing territory (see for example Fig 6M) to subtle defects in the size and patterning of the wing (see for example Fig 6L). In addition to wings displaying both size and pattern defects (class S-P; 34% and 40% of viable combinations with a wing phenotype in the STG+ and STG - groups; Figs 6B, 6I–6P, 7B, 7E–7H, 7O and 7P), we also found a consistent phenotype in which the size of the wing was reduced, but the vein pattern remained intact (class S; 29% in the STG+ and 25% in the STG - groups; Figs 6B, 6E–6G, 6I–6K, 7B, 7I and 7J). With lesser frequencies we also found phenotypes in which the differentiation of the veins was disrupted, including cases with loss of veins or extra-veins (Figs 6B and 6J, 7B and 7L), and other phenotypes including failures in the adhesion of the dorsal and ventral wing surfaces (Fig 7N) and loss of wing margin structures. A representative collection of at least one genetic combination with the nub-Gal4 or salEPv-Gal4 drivers is shown in S14 and S15 Figs.
It is somehow surprising that the distribution of loss-of-function phenotypes is similar for genes that were identified as activated or repressed by Salm/Salr. However, the majority of genes displaying over-expression in salmi/salri discs independent of JNK display a wild type loss-of-function phenotype (21 out of 32), as expected if they contribute to the sal phenotype by over-expression. For the set of genes which expression in the central domain of the wing is activated by Salm/Salr, we focused in those showing phenotype related to Salm/Salr requirements, as they are the most attractive candidates to mediate the functions of Sal proteins during wing development (34 genes). This group was named STG-OK (see Fig 1G and Table 1), and the genes belonging to it display mostly size and pattern (16/34) and size (14/34) defects, and they are enriched for the GO terms “cell proliferation”, “organ development” and “behavioural response to ethanol” (see Fig 1J and S4 Table, STG-OK).
Discussion
The Spalt transcription factors occupy a central position in the genetic hierarchy linking Dpp signalling with the development of the Drosophila wing. Thus, this pathway directly regulates salm/salr expression in the central region of the wing blade, and these genes mediate most of the developmental roles of Dpp in this territory. In this manner, a necessary step to understand how Dpp signalling regulates wing development is to identify Spalt target genes that implement the variety of cellular responses elicited by the pathway. Here we found a complex transformation in the transcriptional landscape of salm/salr knockdown discs, and identify a collection of genes that might fulfil the criteria to be considered bona fide Spalt target genes. From these genes it should be possible to identify which of them are directly related with the known functions of the Dpp pathway in the wing.
The microarray experiments and the selection criteria
Our identification of candidate Spalt target genes is based on microarrays experiments in which we compared genome wide gene expression levels in genetic backgrounds with altered salm/salr function. The simplest assumption is that the expression of a gene that is activated or repressed by Salm/Salr would be reduced or increased, respectively, in a salm/salr knockdown background (Fig 8A and 8B). The ability to identify such changes in the wing disc is somehow complicated, because we are only modifying salm/salr expression in the central region of the wing blade, which is a small region of a complex structure such as the wing disc. In addition, the changes we identified in expression levels could be attributed to a variety of causes unrelated to direct regulation by Salm/Salr. For example, loss of salm/salr function interferes with the growth of the disc, changing the size-proportions of the wing versus the rest of the disc. For this reason, we expect that a fraction of genes we are selecting might correspond to those that are differentially expressed in the wing blade versus the rest of the disc [25]. Furthermore, we know that some genes regulated, directly or indirectly, by Salm/Salr are expressed only in a very restricted population of cells, which most likely would difficult their identification using mRNA extracted from the entire disc for the microarray experiments. This is the case of the Iroquois and knirps gene complexes, which are expressed only in the developing veins L3/L5 and L2, respectively, in the wing blade [20] and that were not identified in the microarray experiments. In addition, and inherent to all microarray-based experiments, changes in levels of expression as a consequence of the loss or gain of salm/salr function do not necessarily indicate a direct regulation of the affected gene. Despite these caveats, we expect that at least some direct targets of Salm/Salr would be included within the population of genes showing a significant change in expression levels in salm/salr knockdown discs.
Fig. 8. Gene regulatory network triggered by Dpp. 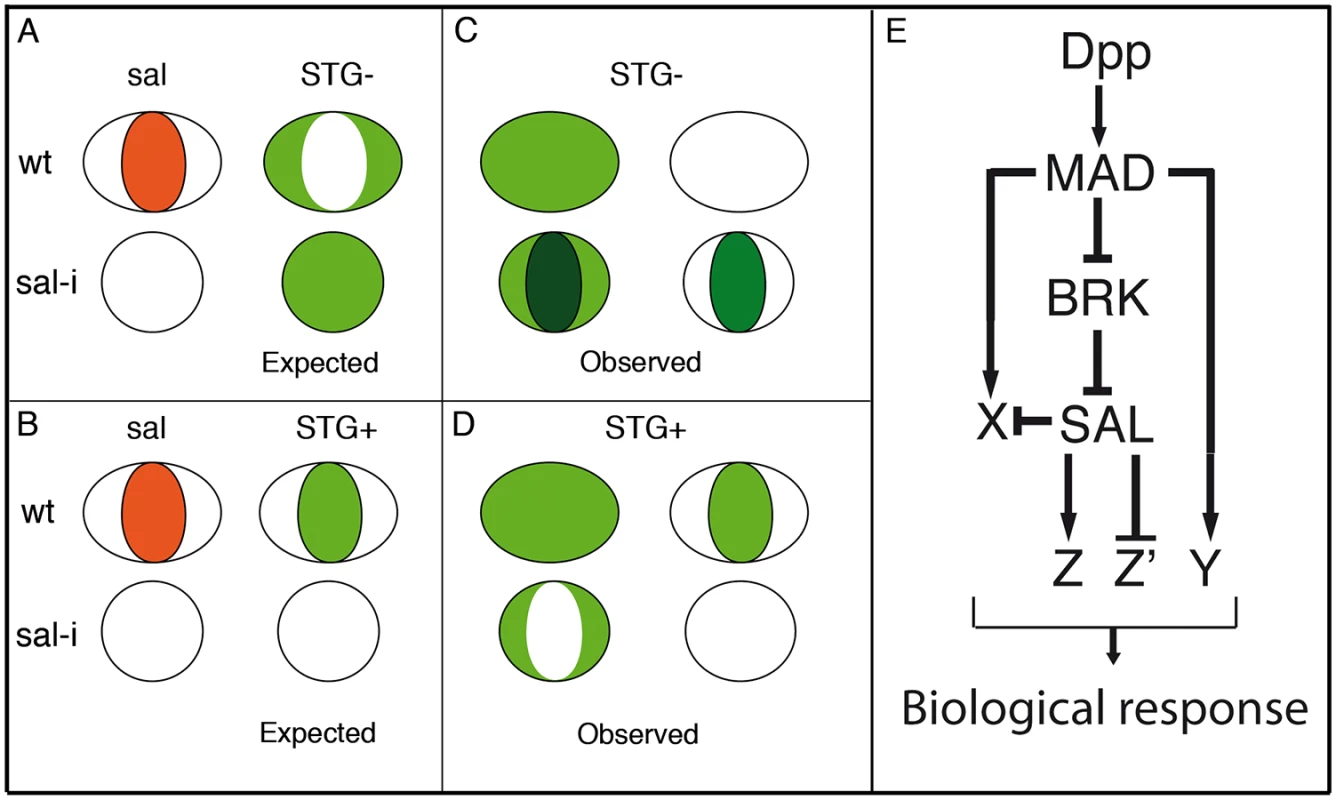
(A-D) Schematic representation of the wing pouch (ovals and circles) showing the expected (green fill in A-B) and observed (green fill in C-D) expression in wild type discs (ovals) and in salmi/salri mutant discs (circles). The behaviour of genes repressed by Sal is shown in A (expected) and C (observed), and for genes activated by Sal in B (expected) and D (observed). (E) The expression of Salm/Salr (SAL) is regulated by the Dpp pathway components Brinker (BRK) and Mad (MAD). Dpp, using Mad or Mad and Brk, also activate the expression of additional genes (X and Y) which in turn can be subject to further regulation by Salm/Salr (X) or independent of them (Y). Finally, the Salm/Salr transcription factors regulate by activation (Z) or repression (Z’) the expression of their direct target genes, contributing to the set of active genes in the centre of the wing blade that confer this territory its particular characteristics. The salm/salr genes are expressed in a restricted region of the wing blade, and for this reason we decided to visualize directly the expression pattern of the genes identified in the microarrays by in situ hybridization. Since we identified a large number of genes in the microarrays (1653), we needed to restrict the expression analysis only to a fraction of this gene pool. The criteria we used to shortlist the genes for in situ hybridization experiments was based in the logFC values, prioritizing in some cases those genes that change early in response to the reduction in salm/salr expression. Setting a logFC value of 1 as threshold (2x fold change difference), we ended up with a short list of 286 candidate genes (STG), and analysed most of them by in situ hybridization in wild type and in salm/salr knockdown discs.
Expression patterns of candidate Salm/Salr target genes
The expression patterns of the selected genes fall into three classes: first, a fraction of genes are expressed in a restricted pattern, which in some cases is related to the expression of Salm/Salr. Some of the genes expressed in the central region of the wing pouch, such as CG1342 and CG7201 behave as positive targets of Salm/Salr (Fig 4H and 4I’; Z in Fig 8E), whereas genes expressed in a complementary pattern, for example CG2999 (S4O-S4O’ Fig), behave as negative targets of Salm/Salr (Z’ in Fig 8). In other cases (CG32372 and CG17278) we were able to assign them to direct positive regulation by Dpp without a requirement for Salm/Salr function (Y in Fig 8E). The category of genes with restricted expression in the wing disc includes several whose expressions are related to the subdivision of the wing blade into pro-vein and inter-vein territories. The expression of these genes (CG1273 and CG33970 in the veins (S10A-S10A’ and S10H-S10H’ Fig) and CG12843, CG18657 and CG30069 in the interveins (Fig 4H-4H’ and S10B-S10B’ Fig and S10D-S10D’ Fig) are regulated by Salm/Salr only in the central region of the wing pouch, suggesting that they contain complex regulatory regions conferring them independent regulation in the central versus the most anterior and posterior regions of the developing wing blade.
A second class includes all the genes that are expressed in a generalised manner in the wing pouch or in the entire wing disc (Fig 8C and 8D). For many of these genes we found that Salm/Salr positively regulate their expression in the central region of the wing pouch (Fig 8D). Finally, we found a third class of genes that display a robust ectopic expression in the central region of the wing in response to the loss of salm/salr function (Fig 8C). Most of these genes are either not expressed at all in the wing disc, or are expressed in a generalised manner. All 40 genes included in this group show a consistent behaviour as negative targets of Salm/Salr, and they uncover an unexpected aspect of the genetic regulatory structure of the wing disc. Thus, it seems that one key contributions of Salm/Salr function to wing development is to prevent the deployment of a particular program of gene activation that is operative in the central region of the wing disc (X in Fig 8E). Because this territory coincides with the domain of Dpp signalling, it is likely that the pathway activates a battery of genes in the central domain of the wing disc whose expression is repressed by Salm/Salr, which are also Dpp targets (X in Fig 8E). This double regulatory mechanism consisting in direct activation and indirect repression triggered by the Dpp pathway might be very important to ensure that only a selected fraction of possible target genes are expressed in a particular tissue in response to the pathway. One important result of such dual positive/negative mode of regulation is to confer versatility to the genetic response to the pathway, refining the inventory of target genes that are activated in each developmental context.
We were able to further dissect the group of genes behaving as negative targets of Salm/Salr into two classes based on their dependence on JNK activity. Thus, we found that some of these genes are expressed in salm-i/salr-i background as a consequence of ectopic JNK activity, whereas the expression of the majority of them behaves as independent of JNK.
Functional requirements of candidate Salm/Salr target genes
The double objective of our approach was to identify the genes that are regulated by Salm/Salr and to assign them biological functions that might account for the known Salm/Salr requirements during wing blade development. These requirements include cell division and viability, pattern formation and a variety of epithelial characteristics including cell affinity. Despite the broad requirements of Salm/Salr functions, it was unexpected to find such a large number of gene expression changes in salm/salr backgrounds. The in-depth analysis of each gene is beyond the scope of this work, but we identified candidates that likely play a direct role downstream of Salm/Salr in some of the processes affected by these proteins.
In the case of genes repressed by Salm/Salr, we found that most of them do not show a loss of function phenotype, as they are not normally expressed in the wing disc. We attempted to assign a requirement for these genes in the development of the salm/salr mutant phenotype using an RNAi approach, for which we knocked down individually all genes that are ectopically expressed in salm/salr discs, but we could not identify a significant individual contribution for any of the 37 genes analysed. Although we do not know to what extent inappropriate ectopic expression of genes that are negatively regulated by Salm/Salr contribute to the spalt loss of function phenotype in the wing, it is likely that it is their combined over-expression what contributes to the cell division and cell viability defects characteristics of salm/salr mutant cells. The molecular functions of these genes falls into a variety of categories, but it is remarkable that 11 of them are related to DNA metabolism, including putative transcription factors (CG6794 and CG6272), DNA repair (CG16928, CG1925, CG3448 and CG5247) and other putative DNA binding or modifying proteins (CG1303, CG14121, CG45050, CG5096 and CG5202). Genes repressed by Salm/Salr also include 7 proteins that function in general metabolism (CG17533, CG18522, CG17530, CG1851, CG33048, CG2999 and CG6658) and two gene pairs formed by a non-coding RNA and a coding gene (CG1512/CG43144 and CG12505/CG10102). Intriguingly, these groups of genes are enriched in a chromatin mark (H3K9me3) related to the formation of heterochromatin, suggesting that Salm/Salr in flies may, as proposed for human Sall1 [13], participate in gene repression through its association with heterochromatin regions. Only one gene repressed by Salm/Salr, CG18455 (optix), displayed a strong vein pattern phenotype that can be related to the function of Sal in defining the position of the vein L2, and our preliminary work confirms that the homeobox DNA-binding domain protein Optix is a component of the genetic network regulated by Salm/Salr in the positioning of the vein L2.
In the case of genes detected as activated by Salm/Salr, we further refined the initial list of 139 candidates (S5 Table) to a group of 34 genes which expression changed in salm/salr knockdown conditions and also displayed a loss-of-function phenotype (STG-OK; Table 1). The most frequent phenotypes include defects in the size of the wing (14 cases) and alterations in both the size and pattern of the wing (16 cases). These phenotypes are similar to the salm/salr loss-of-function conditions, suggesting that these genes might be functionally related to Sal functions. At this stage, it is tempting to speculate that genes affecting only the size of the wing may be related to the regulation of cell proliferation or cell viability, two process that are influenced by Salm/Salr in the central domain of the disc. Also, genes whose loss of function phenotypes resemble the loss of salm/salr function and affect simultaneously the size and pattern of the wing are of singular interest, and the corresponding genes are obvious candidates to implement the characteristics imposed by Salm/Salr to the central region of the wing blade. The group of best candidates Sal-activated genes includes three cases particularly interesting such as CG14394 (Ninjurin, NijC), CG1725 (disclarge, dlg1), CG42614 (scribble, scrib). The corresponding proteins encode components related to epithelial architecture, a process that is severely compromised in salm/salr mutations, and consequently they are bona fide candidates to regulate this aspect of Sal function. Additional components regulating the cytoskeleton or cell adhesion were also identified in the microarrays, but they were not further considered because they did not show a loss-of-function phenotype, such as CG7478 (Actin 79B) and CG42330 (Dscam4), or because we did not detect a change in their expression by in situ hybridization (CG43079 (nrm), CG10541 (Tektin C) and CG7607).
To a large extent, development can be contemplated as the deployment of gene regulatory hierarchies and their implementation in particular cellular behaviours. The analysis of wing disc development has provided numerous examples of how signalling pathways are engaged in these processes, in part though the regulation of gene expression. However, we still had a poor understanding of the regulatory events triggered by signalling pathways or transcription factors. This work aimed to reconstruct the genetic cascade initiated by Dpp signalling that acting through the Spalt transcription factors controls the growth and patterning of the central region of the wing disc. Our analysis uncovered an unsuspected complexity in the transcriptional landscape regulated by Salm/Salr, consisting in batteries of activated and repressed genes (Fig 8E). The identification of Salm/Salr candidate target genes also opens the possibility of searching for the DNA regulatory regions that confer response to Salm/Salr, and to solve the mechanisms of Salm/Salr transcriptional regulation.
Material and Methods
Genetic strains
We used the following Gal4 lines: salEPv-Gal4 [23], nub-Gal4, sd-Gal4, 756-Gal4 [5] and tub-Gal80ts [24], the UAS lines UAS-GFP [33], UAS-salm-i (ID 3029 VDRC), UAS-salr-i (ID 28386 VDRC), UAS-puc [34], UAS-dicer2 [35], UAS-sal [5], UAS-dad and the sal/salr deficiency Df(2L)32FP5 [36]. The expression of salEPv-Gal4 is restricted to the central region of the wing imaginal disc between the vein L2 and intervein L4–L5, and the expression of nub-Gal4, sd-Gal4 and 756-Gal4 occurs in the entire wing pouch (sd-Gal4) and hinge (756-Gal4 and nub-Gal4). The UAS lines used to express RNA interference for the genes selected in microarray experiments are listed in Table 2 and were obtained from Bloomington Stock Center, Vienna Drosophila RNAi Center (VDCR) and NIG-FLY RNAi. Unless otherwise stated, crosses were done at 25°C. Lines not described in the text can be found in FlyBase [37].
Microarray experiments
We did two sets of microarray experiments. In one set (Experiment 1), we compared the expression profiles of 2 genetic conditions at different times:
Experiment 1:
salEPv-Gal4 UAS-GFP; tub-Gal80ts/ UAS-GFP (control discs).
salEPv-Gal4 UAS-GFP / UAS-salm-i; tub-Gal80ts / UAS-salr-i (experimental discs).
In all experiments the larvae were raised at 25°C and then transferred to 29°C for a period of 24h (T24) or 48h (T48) before the dissection of the imaginal discs. At 25°C the Gal80ts protein blocks Gal4 activity, whereas at 29°C Gal4 is active and drives the expression of the selected UAS constructs. We extracted mRNA from 4 biological replicas for each experimental genotype and temperature conditions: salEPv-Gal4 UAS-GFP/UAS-salm-i; tub-Gal80ts/UAS-salr-i 24h and 48h at 29°C and salEPv-Gal4 UAS-GFP; tub-Gal80ts/UAS-GFP 24h and 48h at 29°C. The total amount of mRNA varied in each replica from 2.4 to 5.1 μg (S6 Table). RNA was extracted following a TRIzol protocol (TRIzol Reagent Ambion) and posterior DNase treatment (DNA-freeTM kit Ambion) from groups of 40–80 discs previously stored at -80°C in RNA stabilization solution (RNAlater Ambion) until homogenisation and mRNA extraction. The RNA samples were sent to BIOARRAY (http://www.bioarray.es) for further processing and hybridization in Agilent arrays specifically designed for Drosophila melanogaster (ID 043135). In this work we describe exhaustively the result of these experiments.
Experiment 2:
In a second set of experiments, we compared the expression profiles of 4 genetic conditions:
sd-Gal4/+; FRT40/ FRT40 M(2)z; UAS-FLP/+
sd-Gal4/+; Df(2L)salFP5 FRT40/ FRT40 M(2)z; UAS-FLP/+
756-Gal4/ UAS-sal
756-Gal4 / UAS-GFP
We collected imaginal discs, extracted mRNA, and compared the expression profile of genotypes 1 with 2 (Df) and 3 with 4 (UAS). For these experiments we used Affymetrix arrays, and the genes which expression varied more consistently were selected.
Experiments 1 and 2 differs in several fundamental aspects; first, in experiment 1 all cells included in the domain of salEPv-Gal4 expression display a similar and simultaneous reduction in salm and salr expression, whereas in experiment 2 (Df) the wing is a mosaic of Df(2L)salFP5 homozygous and heterozygous cells. Furthermore, the over-expression of salm in the entire wing (UAS) causes a strong reduction in wing size [5], and many 756-Gal4/UAS-sal wings develop only as stumps of wing tissue. Despite the differences between these experiments, we found that 10% of the genes identified in experiment 1 were also identified in experiment 2 (14% at 24h and 8% at 48h after temperature change). As this coincidence was low, we decided to apply more stringent criteria to restrict the number of genes selected from experiment 1 (see results). From the resulting genes, a fraction of 30% was also identified in experiment 2. To construct our final list of candidate genes to be further analysed, we finally added a set of genes displaying very good scores from experiment 2 that were not identified or selected in experiment 1 (see S3 Table). We reasoned that combining the results from both experiments, which are very different in many respects, and applying stringent selection criteria contributed to add some robustness to the final list of genes that we were able to analyse.
Immunocytochemistry
We used rabbit anti-Salm and rat anti-Salm [36], mouse anti-FasIII (Hybridoma bank at Iowa University). Secondary antibodies were from Jackson Immunological Laboratories (used at 1/200 dilution). Imaginal wing discs were dissected, fixed and stained as described in [38]. Confocal images were captured using a LSM510 confocal microscope. All images were processed with the program ImageJ 1.45s (NIH, USA) and Adobe Photoshop CS6.
In situ hybridization
Imaginal discs were dissected and fixed in 4% formaldehyde for 20 min at room temperature, washed in PBS-0.1% Tween (PBT), and re-fixed for 20 min at room temperature with 4% formaldehyde, 0.1% Tween. After three washes in PBT, discs were stored at -20°C in hybridization solution (SH; 50% formamide, SSC 5X, 100 μg/ml salmon sperm DNA, 50 μg/ml heparin and 0.1% Tween). Disc were pre-hybridized for 2 hours at 55°C in SH, and hybridized with digoxigenin-labelled RNA probes at 55°C. The probes were previously denaturalized at 80°C for 10 min. After hybridization, discs were washed in SH and PBT and incubated for 2 hours at room temperature in a 1 : 4000 dilution of anti-DIG antibody (Roche). After incubation, the discs were washed in PBT and the detection of probes was done with NBT and BCIP solution (Roche). The discs were mounted in 70% glycerol. Pictures were taken using a Spot digital camera coupled to a Zeiss Axioplam microscope using the 20X objectives. All images were processed with Adobe Photoshop CS6.
The probes were generated using cDNA from the collections of Expression Sequence Tags (EST) of Berkeley Drosophila Genome Project, or by PCR from genomic DNA (S3 and S7 Tables), using primers with RNA polymerase T7 and T3 sequences in their ends described in S7 Table. The transcription was done using the RNA polymerase T7, T3 or SP6 (Roche) during 2 hours and the probes were precipitated and suspended in H2O DEPC.
Functional annotation: Gene Ontology (GO) analysis
Functional annotation of Gene Ontology terms was performed using the software DAVID [28–29]. We searched for the GO term “Biological process” (GOTERM_BP_4) in a list of FlyBase Gene ID for the collections of genes identified in the microarray experiments against the Drosophila melanogaster genome as background. We accepted as a significant enrichment those GO terms who have a p-value lower than 0.05.
Supporting Information
Zdroje
1. Beira JV, Springhorn A, Gunther S, Hufnagel L, Pyrowolakis G, Vincent JP (2014) The Dpp/TGFbeta-Dependent Corepressor Schnurri Protects Epithelial Cells from JNK-Induced Apoptosis in Drosophila Embryos. Developmental Cell 31 : 240–247. doi: 10.1016/j.devcel.2014.08.015 25307481
2. Affolter M, Basler K (2007) The Decapentaplegic morphogen gradient: from pattern formation to growth regulation. Nature Rew. Genetics, 8 : 663–674. 17703237
3. de Celis J (2003) Pattern formation in the Drosophila wing: The development of the veins. BioEssays 25 : 443–451. 12717815
4. Grimm S, Pflugfelder GO (1996) Control of the gene optomotor-blind in Drosophila wing development by decapentaplegic and wingless. Science 271 : 1601–1604.
5. de Celis JF, Barrio R, Kafatos FC (1996) A gene complex acting downstream of dpp in Drosophila wing morphogenesis. Nature 381 : 421–424. 8632798
6. Umemori M, Takemura M, Maeda K, Ohba K, Adachi-Yamada T (2007) Drosophila T-box transcription factor Optomotor-blind prevents pathological folding and local overgrowth in wing epithelium through confining Hh signal. Dev Biol 308 : 68–81. 17573067
7. Shen J, Dahmann C, Pflugfelder G (2010) Spatial discontinuity of Optomotor-blind expression in the Drosophila wing imaginal disc disrupts epithelial architecture and promotes cell sorting. BMC Dev Biol 10 : 23.
8. Organista MF, De Celis JF (2013) The Spalt transcription factors regulate cell proliferation, survival and epithelial integrity downstream of the Decapentaplegic signalling pathway. Biology Open 2 : 37–48.
9. de Celis JF, Barrio R (2009) Regulation and function of Spalt proteins during animal development. The International Journal of Developmental Biology 53 : 1385–1398. doi: 10.1387/ijdb.072408jd 19247946
10. Kohlhase J, Wischermann A, Reichenbach H, Froster U, Engel W (1998) Mutations in the SALL1 putative transcription factor gene cause Townes - Brocks syndrome. Nat Genet 18 : 81–83. 9425907
11. Kohlhase J, Heinrich M, Liebers M, Frohlich Archangelo L, Reardon W, Kispert A (2002) Cloning and expression analysis of SALL4, the murine homologue of the gene mutated in Okihiro syndrome. Cytogenetic and Genome Research 98 : 274–277. 12826753
12. Al-Baradie R, Yamada K, St Hilaire C, Chan WM, Andrews C, McIntosh N, Nakano M, Martonyi EJ, Raymond WR, Okumura S, et al (2002) Duane radial ray syndrome (Okihiro syndrome) maps to 20q13 and results from mutations in SALL4, a new member of the SAL family. American Journal of Human Genetics 71 : 1195–1199. 12395297
13. Netzer C, Rieger L, Brero A, Zhang C, Hinzke M, et al. (2001) SALL1, the gene mutated in Townes-Brocks syndrome, encodes a transcriptional repressor which interacts with TRF1/PIN2 and localizes to pericentromeric heterochromatin. Hum Mol Genet 10 : 3017–3024. 11751684
14. Netzer C, Bohlander S, Hinzke M, Chen Y, L K (2006) Defining the heterochromatin localization and repression domains of SALL1. Biochim Biophys Acta 1762 : 386–391. 16443351
15. Sánchez J, Talamillo A, González M, Sánchez-Pulido L, Jiménez S, Pirone L, Sutherland J, Barrio R (2011) Drosophila Sal and Salr are transcriptional repressors. Biochem J 438 : 437–445. doi: 10.1042/BJ20110229 21689070
16. Wu Q, Chen X, Zhang J, Loh Y, Low T, et al. (2006) Sall4 Interacts with Nanog and Co-occupies Nanog Genomic Sites in Embryonic Stem Cells. J Biol Chem 281 : 24090–24094. 16840789
17. Yamashita K, Sato A, Asashima M, PC W, Nishinakamura R (2007) Mouse homolog of SALL1, a causative gene for Townes-Brocks syndrome, binds to A/T-rich sequences in pericentric heterochromatin via its Cterminal zinc finger domains. Genes Cells 12 : 171–182. 17295837
18. Zhang J, Tam W, Tong G, Wu Q, Chan H, et al (2006) Sall4 modulates embryonic stem cell pluripotency and early embryonic development by the transcriptional regulation of Pou5f1. Nat Cell Biol 8 : 1114–1123. 16980957
19. Lunde K, Biehs B, Neuber U, Bier E (1998) The knirps and knirps-related genes organize development of the second wing vein in Drosphila. Development 125 : 4145–4154. 9753669
20. de Celis J, Barrio R (2000) Function of the spalt/spalt-related gene complex in positioning the veins in the Drosophila wing. Mech Dev 91 : 31–41. 10704828
21. Milan M, Perez L, Cohen SM (2002) Short-range cell interactions and cell survival in the Drosophila wing. Developmental Cell 2 : 797–805. 12062091
22. Barrio R, de Celis JF (2004) Regulation of spalt expression in the Drosophila wing blade in response to the Decapentaplegic signalling pathway. PNAS 101 : 6021–6026.
23. Cruz C, Glavic A, Casado M, de Celis JF (2009) A gain-of-function screen identifying genes required for growth and pattern formation of the Drosophila melanogaster wing. Genetics 183 : 1005–1026. doi: 10.1534/genetics.109.107748 19737745
24. McGuire S, Le P, Osborn A, Matsumoto K, Davis R (2003) Spatiotemporal rescue of memory dysfunction in Drosophila. Science 302 : 1765–1768. 14657498
25. Butler MJ, Jacobsen TL, Cain DM, Jarman MG, Hubank M, Whittle JS, Phillips R, Simcox A (2003) Discovery of genes with highly restricted expression patterns in the Drosophila wing disc using DNA oligonucleotide microarrays. Development 130 : 659–670. 12505997
26. Doumpas N, Ruiz‐Romero M, Blanco E, Edgar B, Corominas M, Teleman AA (2013) Brk regulates wing disc growth in part via repression of Myc expression. EMBO reports 14, 261–268. doi: 10.1038/embor.2013.1 23337628
27. Ferreiro MJ, Rodríguez-Ezpeleta N, Pérez C, Hackenberg M, Aransay AM, Barrio R, Cantera R. (2012) Whole transcriptome analysis of a reversible neurodegenerative process in Drosophila reveals potential neuroprotective genes. BMC Genomics 13 : 483. doi: 10.1186/1471-2164-13-483 22978642
28. Huang D, Sherman B, Lempicki R (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols 4 : 44–57. doi: 10.1038/nprot.2008.211 19131956
29. Huang D, Sherman B, Lempicki R (2009) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37 : 1–13. doi: 10.1093/nar/gkn923 19033363
30. modMine: flexible access to modENCODE data. Nucleic Acids Res. 2012 January; 40(D1): D1082–D1088
31. Hibi M, Lin A, Smeal T, Minden A, Karin M (1993) Identification of an oncoprotein - and UV-responsive protein kinase that binds and potentiates the c-Jun activation domain. Genes Dev 7 : 2135–2148. 8224842
32. Szuperák M, Salah S, Meyer EJ, Nagarajan U, Ikmi A, Gibson MC (2011) Feedback regulation of Drosophila BMP signaling by the novel extracellular protein larval translucida. Development 138 : 715–24. doi: 10.1242/dev.059477 21266407
33. Ito K, Awano W, Suzuki K, Hiromi Y, Yamamoto D (1997) The Drosophils mushroom body is a quadruple structure of clonal units each of which contains a virtually identical set of neurones and glial cells. Development 124 : 761–771. 9043058
34. Martin-Blanco E, Gampel A, Ring J, Virdee K, Kirov N, Tolkovsky AM, Martinez-Arias A (1998) puckered encodes a phosphatase that mediates a feedback loop regulating JNK activity during dorsal closure in Drosophila. Genes & Dev 12 : 557–570.
35. Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, et al (2007) A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature, 448 : 151–156. 17625558
36. Barrio R, de Celis JF, Bolshakov S, Kafatos FC (1999) Identification of regulatory regions driving the expression of the Drosophila spalt complex at different developmental stages. Dev Biol 215 : 33–47. 10525348
37. Pierre S St., Ponting L, Stefancsik R, McQuilton P, Consortium F (2014) FlyBase 102—advanced approaches to interrogating FlyBase. Nucleic Acids Res 42 : 80–88.
38. de Celis JF (1997) Expression and function of decapentaplegic and thick veins during the differentiation of the veins in the Drosophila wing. Development 124 : 1007–1018. 9056776
Štítky
Genetika Reprodukční medicína
Článek Loss and Gain of Natural Killer Cell Receptor Function in an African Hunter-Gatherer PopulationČlánek Let-7 Represses Carcinogenesis and a Stem Cell Phenotype in the Intestine via Regulation of Hmga2Článek Binding of Multiple Rap1 Proteins Stimulates Chromosome Breakage Induction during DNA ReplicationČlánek SLIRP Regulates the Rate of Mitochondrial Protein Synthesis and Protects LRPPRC from DegradationČlánek Protein Composition of Infectious Spores Reveals Novel Sexual Development and Germination Factors inČlánek The Formin Diaphanous Regulates Myoblast Fusion through Actin Polymerization and Arp2/3 RegulationČlánek Runx1 Transcription Factor Is Required for Myoblasts Proliferation during Muscle Regeneration
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 8- Růst a vývoj dětí narozených pomocí IVF
- Délka menstruačního cyklu jako marker ženské plodnosti
- Vztah užívání alkoholu a mužské fertility
- Intrauterinní inseminace a její úspěšnost
- Šanci na úspěšný průběh těhotenství snižují nevhodné hladiny progesteronu vznikající při umělém oplodnění
-
Všechny články tohoto čísla
- Putting the Brakes on Huntington Disease in a Mouse Experimental Model
- Identification of Driving Fusion Genes and Genomic Landscape of Medullary Thyroid Cancer
- Evidence for Retromutagenesis as a Mechanism for Adaptive Mutation in
- TSPO, a Mitochondrial Outer Membrane Protein, Controls Ethanol-Related Behaviors in
- Evidence for Lysosome Depletion and Impaired Autophagic Clearance in Hereditary Spastic Paraplegia Type SPG11
- Loss and Gain of Natural Killer Cell Receptor Function in an African Hunter-Gatherer Population
- Trans-Reactivation: A New Epigenetic Phenomenon Underlying Transcriptional Reactivation of Silenced Genes
- Early Developmental and Evolutionary Origins of Gene Body DNA Methylation Patterns in Mammalian Placentas
- Strong Selective Sweeps on the X Chromosome in the Human-Chimpanzee Ancestor Explain Its Low Divergence
- Dominance of Deleterious Alleles Controls the Response to a Population Bottleneck
- Transient 1a Induction Defines the Wound Epidermis during Zebrafish Fin Regeneration
- Systems Genetics Reveals the Functional Context of PCOS Loci and Identifies Genetic and Molecular Mechanisms of Disease Heterogeneity
- A Genome Scale Screen for Mutants with Delayed Exit from Mitosis: Ire1-Independent Induction of Autophagy Integrates ER Homeostasis into Mitotic Lifespan
- Non-synonymous FGD3 Variant as Positional Candidate for Disproportional Tall Stature Accounting for a Carcass Weight QTL () and Skeletal Dysplasia in Japanese Black Cattle
- The Relationship between Gene Network Structure and Expression Variation among Individuals and Species
- Calmodulin Methyltransferase Is Required for Growth, Muscle Strength, Somatosensory Development and Brain Function
- The Wnt Frizzled Receptor MOM-5 Regulates the UNC-5 Netrin Receptor through Small GTPase-Dependent Signaling to Determine the Polarity of Migrating Cells
- Nbs1 ChIP-Seq Identifies Off-Target DNA Double-Strand Breaks Induced by AID in Activated Splenic B Cells
- CCNYL1, but Not CCNY, Cooperates with CDK16 to Regulate Spermatogenesis in Mouse
- Evidence for a Common Origin of Blacksmiths and Cultivators in the Ethiopian Ari within the Last 4500 Years: Lessons for Clustering-Based Inference
- Of Fighting Flies, Mice, and Men: Are Some of the Molecular and Neuronal Mechanisms of Aggression Universal in the Animal Kingdom?
- Hypoxia and Temperature Regulated Morphogenesis in
- The Homeodomain Iroquois Proteins Control Cell Cycle Progression and Regulate the Size of Developmental Fields
- Evolution and Design Governing Signal Precision and Amplification in a Bacterial Chemosensory Pathway
- Rac1 Regulates Endometrial Secretory Function to Control Placental Development
- Let-7 Represses Carcinogenesis and a Stem Cell Phenotype in the Intestine via Regulation of Hmga2
- Functions as a Positive Regulator of Growth and Metabolism in
- The Nucleosome Acidic Patch Regulates the H2B K123 Monoubiquitylation Cascade and Transcription Elongation in
- Rhoptry Proteins ROP5 and ROP18 Are Major Murine Virulence Factors in Genetically Divergent South American Strains of
- Exon 7 Contributes to the Stable Localization of Xist RNA on the Inactive X-Chromosome
- Regulates Refractive Error and Myopia Development in Mice and Humans
- mTORC1 Prevents Preosteoblast Differentiation through the Notch Signaling Pathway
- Regulation of Gene Expression Patterns in Mosquito Reproduction
- Molecular Basis of Gene-Gene Interaction: Cyclic Cross-Regulation of Gene Expression and Post-GWAS Gene-Gene Interaction Involved in Atrial Fibrillation
- The Spalt Transcription Factors Generate the Transcriptional Landscape of the Wing Pouch Central Region
- Binding of Multiple Rap1 Proteins Stimulates Chromosome Breakage Induction during DNA Replication
- Functional Divergence in the Role of N-Linked Glycosylation in Smoothened Signaling
- YAP1 Exerts Its Transcriptional Control via TEAD-Mediated Activation of Enhancers
- Coordinated Evolution of Influenza A Surface Proteins
- The Evolutionary Potential of Phenotypic Mutations
- Genome-Wide Association and Trans-ethnic Meta-Analysis for Advanced Diabetic Kidney Disease: Family Investigation of Nephropathy and Diabetes (FIND)
- New Routes to Phylogeography: A Bayesian Structured Coalescent Approximation
- SLIRP Regulates the Rate of Mitochondrial Protein Synthesis and Protects LRPPRC from Degradation
- Satellite DNA Modulates Gene Expression in the Beetle after Heat Stress
- SHOEBOX Modulates Root Meristem Size in Rice through Dose-Dependent Effects of Gibberellins on Cell Elongation and Proliferation
- Reduced Crossover Interference and Increased ZMM-Independent Recombination in the Absence of Tel1/ATM
- Suppression of Somatic Expansion Delays the Onset of Pathophysiology in a Mouse Model of Huntington’s Disease
- Protein Composition of Infectious Spores Reveals Novel Sexual Development and Germination Factors in
- The Evolutionarily Conserved LIM Homeodomain Protein LIM-4/LHX6 Specifies the Terminal Identity of a Cholinergic and Peptidergic . Sensory/Inter/Motor Neuron-Type
- SmD1 Modulates the miRNA Pathway Independently of Its Pre-mRNA Splicing Function
- piRNAs Are Associated with Diverse Transgenerational Effects on Gene and Transposon Expression in a Hybrid Dysgenic Syndrome of .
- Retinoic Acid Signaling Regulates Differential Expression of the Tandemly-Duplicated Long Wavelength-Sensitive Cone Opsin Genes in Zebrafish
- The Formin Diaphanous Regulates Myoblast Fusion through Actin Polymerization and Arp2/3 Regulation
- Genome-Wide Analysis of PAPS1-Dependent Polyadenylation Identifies Novel Roles for Functionally Specialized Poly(A) Polymerases in
- Runx1 Transcription Factor Is Required for Myoblasts Proliferation during Muscle Regeneration
- Regulation of Mutagenic DNA Polymerase V Activation in Space and Time
- Variability of Gene Expression Identifies Transcriptional Regulators of Early Human Embryonic Development
- The Drosophila Gene Interacts Genetically with and Shows Female-Specific Effects of Divergence
- Functional Activation of the Flagellar Type III Secretion Export Apparatus
- Retrohoming of a Mobile Group II Intron in Human Cells Suggests How Eukaryotes Limit Group II Intron Proliferation
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Exon 7 Contributes to the Stable Localization of Xist RNA on the Inactive X-Chromosome
- YAP1 Exerts Its Transcriptional Control via TEAD-Mediated Activation of Enhancers
- SmD1 Modulates the miRNA Pathway Independently of Its Pre-mRNA Splicing Function
- Molecular Basis of Gene-Gene Interaction: Cyclic Cross-Regulation of Gene Expression and Post-GWAS Gene-Gene Interaction Involved in Atrial Fibrillation
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



