-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
The Evolutionarily Conserved LIM Homeodomain Protein LIM-4/LHX6 Specifies the Terminal Identity of a Cholinergic and Peptidergic . Sensory/Inter/Motor Neuron-Type
The correct generation and maintenance of the nervous system is critical for the animal’s life. Dysregulation of these processes leads to multiple neurodevelopmental disorders. It has been a daunting challenge not only to identify the developmental mechanisms that determine neuronal cell fate, but also to understand the extent to which the mechanisms are evolutionarily conserved. Here, we describe a developmental mechanism that determines the fate of a specific cholinergic and peptidergic neuronal type in C. elegans. We show that the lim-4 LIM homeodomain transcription factor is necessary and sufficient to promote and maintain the specific cholinergic and peptidergic properties and functions via binding to unique DNA sequences. We also demonstrate that C. elegans lim-4 and human LHX6 show striking functional similarity; specifically, C. elegans LIM-4 or human LHX6 can induce cholinergic and peptidergic characteristics in human neuronal cell lines. Given the high conservation of these transcription factors, these developmental mechanisms are likely to be generally applicable in the nervous system of other organisms as well.
Published in the journal: . PLoS Genet 11(8): e32767. doi:10.1371/journal.pgen.1005480
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005480Summary
The correct generation and maintenance of the nervous system is critical for the animal’s life. Dysregulation of these processes leads to multiple neurodevelopmental disorders. It has been a daunting challenge not only to identify the developmental mechanisms that determine neuronal cell fate, but also to understand the extent to which the mechanisms are evolutionarily conserved. Here, we describe a developmental mechanism that determines the fate of a specific cholinergic and peptidergic neuronal type in C. elegans. We show that the lim-4 LIM homeodomain transcription factor is necessary and sufficient to promote and maintain the specific cholinergic and peptidergic properties and functions via binding to unique DNA sequences. We also demonstrate that C. elegans lim-4 and human LHX6 show striking functional similarity; specifically, C. elegans LIM-4 or human LHX6 can induce cholinergic and peptidergic characteristics in human neuronal cell lines. Given the high conservation of these transcription factors, these developmental mechanisms are likely to be generally applicable in the nervous system of other organisms as well.
Introduction
The proper generation and maintenance of cells in the nervous system is essential for multi-cellular organisms. Each neuron achieves its identity by the acquisition of many distinct features, including appropriate synaptic contacts and expression of distinct sets of neurotransmitters. Fate determination and specification of neuronal cells largely relies on interactions between trans-acting transcription factors and cis-regulatory elements of their target genes [1, 2]. The same transcription factors may be used again after neuronal cell fate determination to maintain the neuron’s integrity [3]. However, it has been challenging not only to discover the transcription factors, but also to identify their regulatory mechanisms and target genes critically associated with determination and maintenance of neuronal cell fate.
In the nematode Caenorhabditis elegans, about 40% of nervous system (~120 neurons) appear to be cholinergic, including a subset of motor neurons in the ventral nerve cord and several sensory, motor and interneurons in the head, and many of these cholinergic neurons co-express neuropeptides [4, 5]. Several genes that function in terminal differentiation and specification of cholinergic neuronal fate have been identified, including the Olf/EBF transcription factor unc-3 for the A-, B-, and AS-type ventral nerve cord and SAB motor neurons, LIM homeobox transcription factor ttx-3 (ortholog of mammalian Lhx2/9) for the AIY and AIA interneurons, Paired-like homeobox gene ceh-10 for the AIY interneurons, and POU homeobox gene unc-86 for the IL2 sensory neurons, URA motor neurons and URB interneurons [6, 7, 8, 9, 10]. These transcription factors act as terminal selectors to directly or indirectly regulate expression of most terminal differentiation genes, such as the cholinergic gene battery but not that of pan-neuronal genes, and broadly affect terminal differentiation of each cholinergic neuron types [2]. Although the terminal selector transcription factors that are required for terminal differentiation of half of the cholinergic neurons have been identified, mechanisms and genes that differentiate other morphologically and functionally different cholinergic neuron types remain to be elucidated.
The SMB multimodal sensory/inter/motor neurons consist of two pairs of neurons that are located in the head and innervate the head neck muscles (Fig 1A). Their processes, which run in ventral or dorsal sublateral cords to the tail and have electric and chemical synaptic contacts to other neurons in the head, were proposed to sense the stretch of body and regulate head locomotion [11]. In fact, laser ablation of the SMB neurons caused increased reversal frequency and wave amplitude of forward locomotion [12]. These neurons utilize at least two neurotransmitters, acetylcholine and a FMRFamide-related peptide, FLP-12 [5, 13]. Genes or molecules that are pivotal for the generation or differentiation of these SMB neurons have not been identified.
Fig. 1. Expression of a flp-12 neuropeptide reporter is abolished in the SMB neurons of lim-4 mutants. 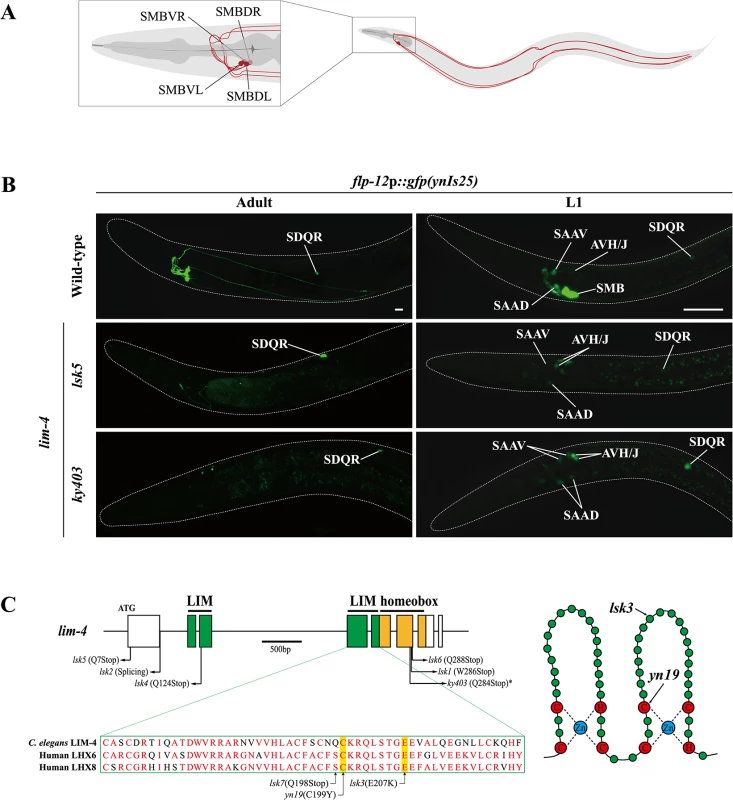
(A) Schematic drawing of the SMB neurons in C. elegans. Four cell bodies are located in the head (DL: dorsal left, DR: dorsal right, VL: ventral left, VR: ventral right) and their processes run in sublateral cords to the tail. (B) Expression of the flp-12 neuropeptide reporter is abolished specifically in the SMB neurons of lim-4 mutants. GFP expression in ynIs25 integrated strains is observed in the SMB and SDQ(L/R) neurons of wild-type adult (left column) or L1 larval (right column) stage animals while GFP expression in the SMB neurons is not detected in lim-4 null mutant alleles (lsk5, ky403). Faint expression of the flp-12p::gfp reporter in the SAA and AVH/J neurons of L1 larvae is not altered in lim-4 mutants. Anterior is at left in all images. Scale bars: 20 μm. (C) Genomic structure of lim-4 (left) and schematic structure of the LIM domain of LIM-4 (right). The sequence alignment of part of the LIM domain of C. elegans LIM-4 and human LHX6 and LHX8 is shown. Identical residues in at least two proteins are shown in red. Molecular lesions of lim-4 mutant alleles are indicated. *Mutation in ky403 was previously reported [14]. LIM domain and homeodomain are labeled in green and in yellow, respectively. Here, we show that the LIM homeodomain LIM-4 protein is necessary to drive expression of terminal differentiation genes, including the cholinergic gene battery and the flp-12 neuropeptide gene, but not pan-neuronal genes in the SMB neurons; consequently, in lim-4 mutants, the neuronal function of the SMB neurons is abolished. We find that LIM-4 maintains its own expression by autoregulation in the SMB neurons and ectopic expression of LIM-4 is sufficient to drive expression of the SMB marker in other cell types. Moreover, our promoter analyses and bioinformatic searches with the SMB marker genes identified a cis-regulatory motif that is necessary and sufficient to drive gene expression in the SMB neurons. We also show that two lim-4 human orthologs, LHX6 and LHX8, functionally substitute for lim-4 in C. elegans. Furthermore, expression of C. elegans LIM-4 or human LHX6 in the human neuroblastoma cell line induces cholinergic and peptidergic characteristics. We propose that there is an evolutionarily conserved role of lim-4/LHX6/LHX8 LIM homeobox genes as terminal selectors to differentiate cholinergic and peptidergic neuronal cells and provide insight into how neuronal characteristics such as neurotransmitter identity are acquired via trans-acting and cis-regulatory mechanisms.
Results
Expression of a neuropeptide gene in the SMB neurons is abolished in lim-4 mutants
To identify factors that specify the neuronal cell-fate of SMB, we performed a genetic screen to isolate animals in which the expression pattern of a terminal differentiation marker, flp-12p::gfp reporter, was disrupted exclusively in the SMB neurons. flp-12 encodes a FMRFamide-related neuropeptide and is expressed in a set of neurons that includes the SMB and SDQ neurons in adults [13]. Among mutants isolated from this screen, seven mutant alleles (named as lsk1,2,4,5,6,7, yn19) exhibited complete loss of flp-12 expression, while one mutant allele (lsk3) showed weak expression of flp-12 in all four SMB neurons at either adult (Fig 1B; S1 Fig; Table 1) or L1 larval developmental stage (Fig 1B; Table 1; S1 Table) animals. By contrast, expression of flp-12 in the SDQ and other neurons weakly expressing flp-12 was unaffected in all eight mutants (Fig 1B; S1 Fig), indicating that expression of flp-12 was specifically affected in the SMB neurons of these mutants.
Tab. 1. Expression pattern of the SMB expressed markers in lim-4 mutant animals. 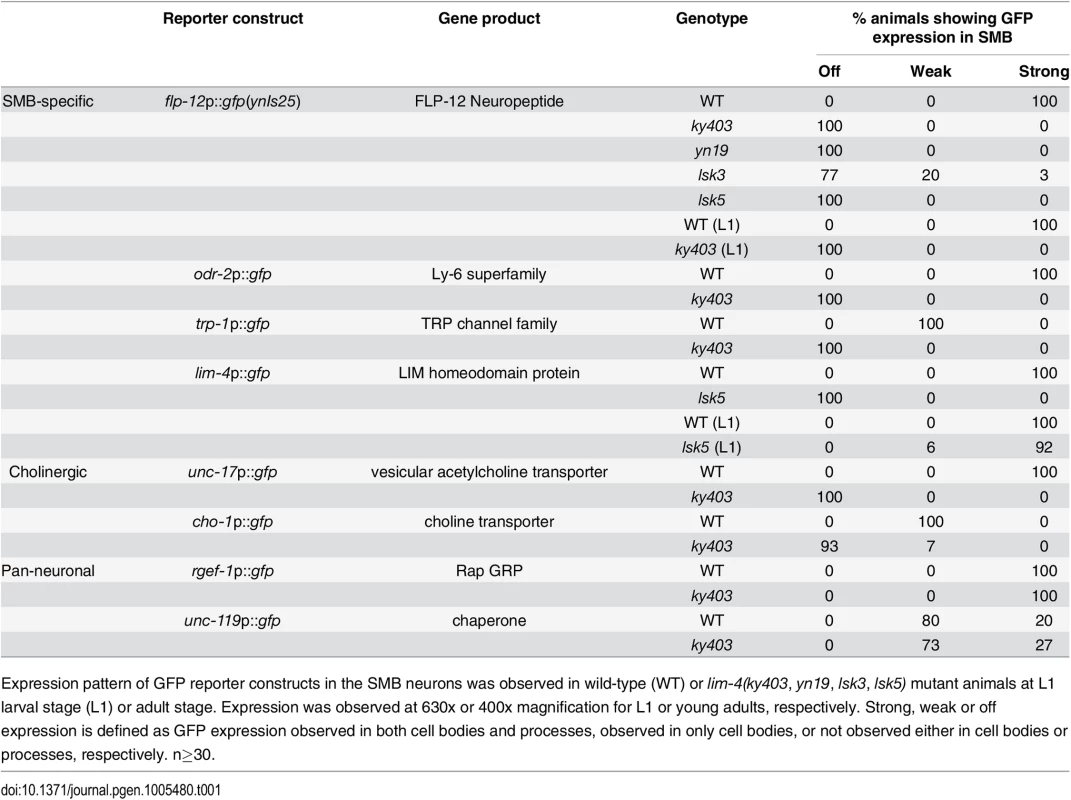
Expression pattern of GFP reporter constructs in the SMB neurons was observed in wild-type (WT) or lim-4(ky403, yn19, lsk3, lsk5) mutant animals at L1 larval stage (L1) or adult stage. Expression was observed at 630x or 400x magnification for L1 or young adults, respectively. Strong, weak or off expression is defined as GFP expression observed in both cell bodies and processes, observed in only cell bodies, or not observed either in cell bodies or processes, respectively. n≥30. From subsequent complementation test and three factor analysis, all mutations were found to be allelic to the previously identified lim-4(ky403) mutation. lim-4 encodes a LIM homeodomain protein that is required for specification of AWB and ADF chemosensory neuron identity [14, 15,16]. In lim-4(ky403) null mutants, AWB cell fate is changed to that of the AWC chemosensory neurons, thereby causing dye-filling defects in the AWB neurons [14]. Expression of flp-12 was also completely abolished in the SMB neurons of ky403 mutants (Fig 1B; Table 1). Like in the ky403 mutants, the AWB neurons failed to dye-fill in the lsk1-7 and yn19 mutants (S2 Fig; S2 Table). The molecular lesions of all eight mutants mapped to the coding region of the lim-4 gene (Fig 1C). Five mutant alleles (lsk1,4,5,6,7) had nonsense mutations that resulted in premature translation stop, suggesting that these mutations are null alleles. lsk2 had a mutation in the splice donor site after the 1st exon. yn19 and lsk3 had missense mutations within the coding region of the second LIM domain, resulting in C199Y and E207K substitutions, respectively. The cysteine residue (C199) is critical for forming a zinc finger motif in the LIM domain [15]. The glutamate residue (E207) resides in the LIM domain and is highly conserved through evolution (Fig 1C), suggesting that this residue is essential for LIM-4 function via protein-protein interactions. These findings indicate that LIM-4 has a role in regulating gene expression in the SMB neurons.
Expression of terminally differentiated SMB markers including cholinergic genes is abolished in lim-4 mutants
To determine the extent to which LIM-4 regulates gene expression in the SMB neurons, we examined additional SMB terminal differentiation genes, including odr-2 GPI-anchored cell surface protein [17], trp-1 TRPC channel [18], and cholinergic markers such as unc-17 vesicular acetylcholine transporter (VAChT) [19] and cho-1 choline transporter (ChT) (Fig 2A) [20]. In order to locate the SMB cell bodies, we used expression of ceh-17p::dsRed in the cell bodies of SIAV as a marker that is directly adjacent to the cell bodies of SMBD (S3 Fig) [21]. None of the SMB specific or cholinergic markers were expressed in the SMB neurons of lim-4 mutants while expression in other neuron types was generally not affected (Fig 2B and 2C; Table 1). We next tested expression of two well-characterized pan-neuronal gene markers, rgef-1 Ras guanine nucleotide releasing protein and unc-119 chaperone [6, 7]. Expression of these pan-neuronal genes was not altered in the SMB neurons of lim-4 mutants (Fig 2D; Table 1), indicating that the SMB cells may retain neuronal properties.
Fig. 2. LIM-4 regulates expression of the terminally differentiated markers in the SMB neurons. 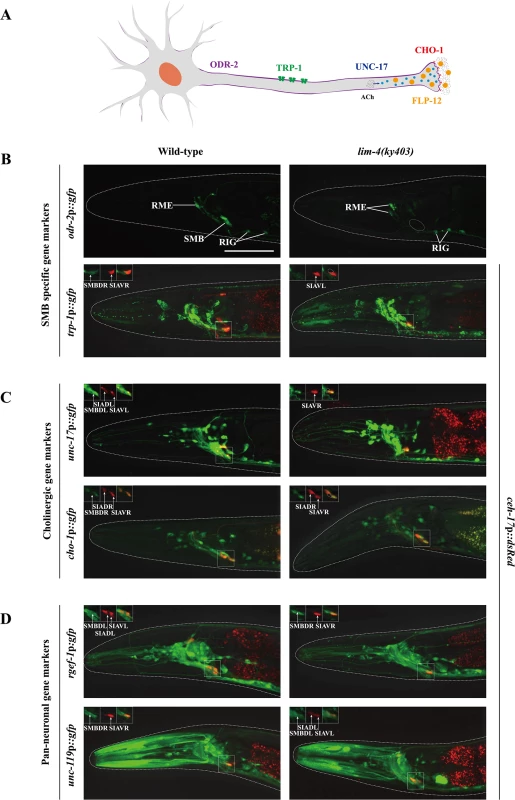
(A) Schematic drawing of expressed genes in the SMB neurons; odr-2 (GPI-anchored cell surface protein), trp-1 (TRPC channel), unc-17 (VAChT), cho-1 (ChT), and flp-12 (neuropeptide) (B-D) Expression of the indicated reporter constructs is shown in wild-type (left column) or lim-4(ky403) mutant (right column) animals. Merged images with the ceh-17p::mCherry reporter expression in the SIA neurons (shown in red) were shown for trp-1, unc-17, cho-1, rgef-1, or unc-119 promoter reporter for help in identification of the SMB neurons (see S3 Fig). Images are derived from z-stacks of confocal microscopy images while images in the upper-left boxed regions are single focal plane confocal microscopy images. Quantitative analysis of these phenotypes is shown in Table 1. Anterior is to the left. Scale bar: 50 μm. To determine whether the lim-4 SMB neurons adopted a different cell fate such as the structurally and/or functionally related sub-lateral nerve cord neurons including the SIA, SIB, and SMD neurons, we tested markers including ceh-17 for SIA [21], flp-22 for SMD [13] or ceh-24 for SIA, SIB, and SMD [22, 23]. None of these markers were ectopically expressed in lim-4 mutants (S4 Fig), suggesting that the cell fate of the SMB neurons is not transformed to that of the structurally and/or functionally related cell types.
The SMB neurons are generated from ABalpapap (SMBDL, SMBVL) or ABarappap (SMBDR, SMBVR) precursors, and three of their sister cells undergo programmed cell death before hatching (S5 Fig) [24]. We observed expression of pan-neuronal markers in SMB of adult lim-4 mutants (Fig 2D), suggesting that the SMB cells do not adopt the apoptotic fate of their sister cells. Based on these results, we conclude that LIM-4 activity does not initiate neuronal cell fate, but specifies SMB cell fate by regulating expression of terminal differentiation genes, thereby acting as a terminal selector transcription factor in the SMB neurons.
The function of the SMB neurons is compromised in lim-4 mutants
Wild-type animals move in sinusoidal waves of a consistent wave width and wavelength (Fig 3A) [25]. lim-4 mutants move in a coiled or loopy fashion (Fig 3A) [14]. To quantitate the loopy uncoordinated movement, the waveforms of these animals were measured by viewing tracks made in a bacterial lawn and compared to that of wild-type animals (Fig 3B). lim-4 mutants had significantly accentuated waveforms (Fig 3C). While the average wavelength for lim-4 null mutants (ky403 or lsk5) is similar or mildly decreased compared to that of wild-type animals, the average wave width for ky403 or lsk5 mutants (ky403: 359.46±13.51 μm, n = 30; lsk5: 374.27±16.47 μm, n = 30) is about 70% higher than that of wild-type animals (N2 : 194.54±4.28 μm, n = 30) (Fig 3A and 3C). yn19 and lsk3 missense mutants similarly exhibited significantly larger wave width (yn19: 312.31±7.75 μm, n = 30; lsk3: 365.43±11.97 μm, n = 30) (Fig 3A and 3C), suggesting that yn19 and lsk3 mutations also fully eliminate the contribution of LIM-4 to locomotion.
Fig. 3. lim-4 mutant animals moved in a coiled or loopy fashion due to the functional defects of the SMB neurons. 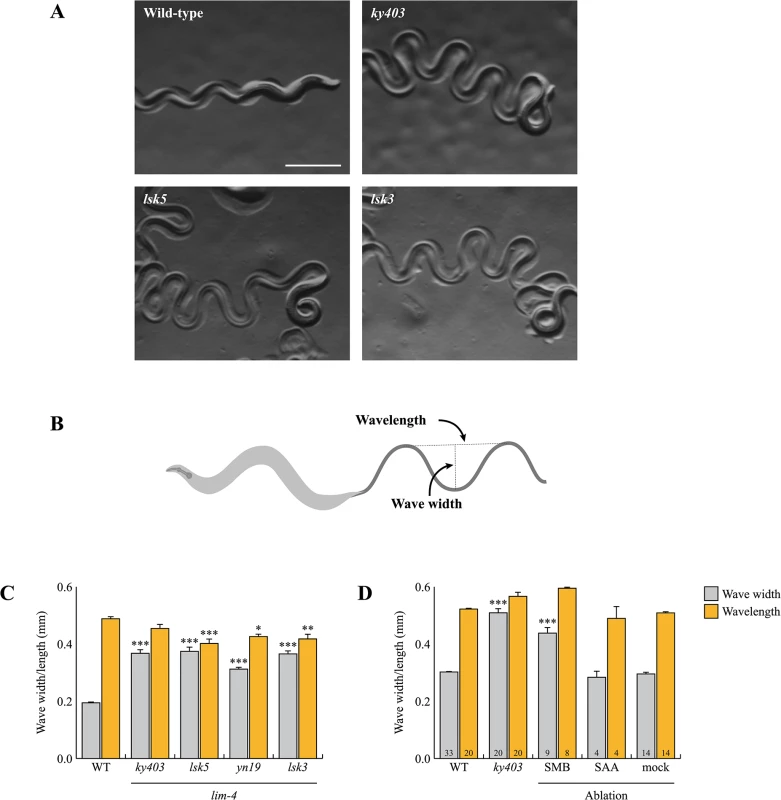
(A) Wild-type animals show a characteristic sinusoidal waveform whose tracks can also be observed in the bacterial lawn. lim-4(ky403, lsk5, lsk3) mutant animals showed an exaggerated waveform characterized by an increased wave width. Scale bar: 0.5 mm. (B) To quantitate locomotion, the waveforms of different animals were analyzed and compared to that of wild-type animals by viewing tracks made in a bacterial lawn under a microscope. Wave width and wavelength were measured and averaged as the distance from the peak to the trough of the sine wave and distance between one peak and the next corresponding peak, respectively. (C-D) Average of wave width and wavelength of the lim-4 mutant animals (C) or SMB/SAA-ablated animals (D). n≥30 for each. Error bars are the SEM. *, ** and *** indicate significantly different from wild-type at p<0.05, 0.01, and 0.001, respectively (one-way ANOVA test followed by the Tukey post-hoc test). The number of animals tested is indicated on the bars. Note that the size was measured by using the software (C) or a scale built into an eyepiece (D). To assess whether the loopy movement of lim-4 mutants is due to a functional defect of the SMB neurons, we ablated the SMB neurons by laser microsurgery. Consistent with a previous study [12], killing the SMB neurons resulted in a loopy or coiled movement phenotype; the average wave width was increased by over 50% compared to control animals and similar to that of lim-4 mutants (Fig 3D). We, however, noted that the SMB ablation did not result in as strong a loopy phenotype as the lim-4 mutations, suggesting that the mutations have additional effects on locomotion beyond elimination of SMB function. lim-4 is also expressed in the SAA neurons (see below) of which roles have been implicated in head locomotion [11]. Laser ablation of the SAA neurons did not cause a loopy or coiled movement, ruling out the possibility that defects of the SAA neurons result in movement defects of lim-4 mutants (Fig 3D). These results indicate that the SMB neurons function to regulate locomotion by modulating the wave width of the animal and that the loopy phenotype of lim-4 mutants is due to defects in the function of the SMB neurons.
lim-4 is expressed and functions in the SMB neurons to regulate their terminal specification
lim-4 has previously been shown to be expressed in several neuronal types in the head of postembryonic animals; these neurons include the AWB, SIA, SAA, RID, RIV, and RMD neurons, but not the SMB neurons [14]. Like the SMB neurons, the SIA neurons project their processes into the sub-lateral nerve cords and their cell morphology and position are similar to those of the SMB neurons [11]. To determine whether lim-4 expression was mis-identified in the SIA neurons, we examined the expression pattern of lim-4p::gfp transgene (oyIs35) that includes 3.6 kb of upstream sequence [14, 26] and compared it to that of ceh-17p::dsRed, a SIA marker [21] or flp-12p::mCherry, a SMB marker [13], respectively (Fig 4A). We observed co-localization of lim-4 expression with that of flp-12 but not of ceh-17, indicating that lim-4 is expressed in the SMB neurons rather than the SIA neurons. In support of this re-assignment, the expression of other SMB markers such as odr-2p::gfp or trp-1p::gfp was completely abolished in the SMB neurons of lim-4 mutants, whereas expression of ceh-17 was not affected (Fig 2B; S4 Fig).
Fig. 4. lim-4 is expressed in and acts in the SMB neurons and its expression is autoregulated. 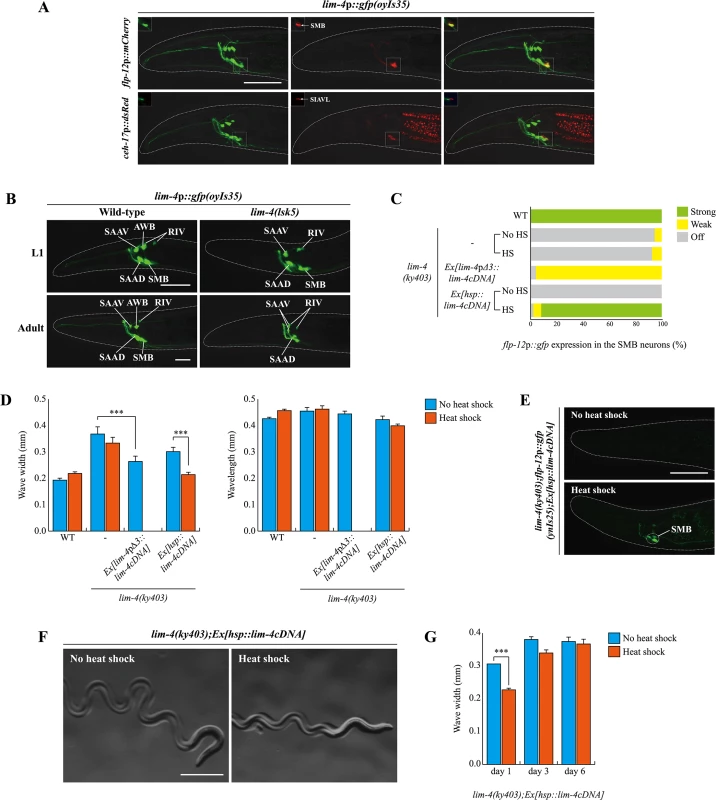
(A) GFP expression of oyIs35 animals carrying an integrated lim-4p::gfp reporter is overlapped with expression of the flp-12p::mCherry reporter (the SMB marker) but not with expression of the ceh-17p::dsRed reporter (the SIA marker). Images are derived from z-stacks of confocal microscopy images while images in the upper-left boxed regions are single focal plane confocal microscopy images. Anterior is to the left. Scale bar: 50 μm. (B) In lim-4 mutants, expression of the lim-4p::gfp reporter is abolished in the cell bodies and processes of the SMB neurons in adult but not L1 larval stage animals. GFP expression of the lim-4p::gfp reporter is shown in wild-type (left column) or lim-4(lsk5) mutant (right column) animals at L1 larval (top) or adult stage (bottom). Note that the lim-4p::gfp reporter is not expressed in the AWB neurons of lim-4 mutants [14]. Images are derived from z-stacks of confocal microscopy images. Quantitative analysis of these phenotypes is shown in Table 1. Anterior is to the left. Scale bar: 20 μm. (C) Percentage of animals of the indicated genotypes expressing stably integrated flp-12p:gfp reporter (ynIs25) is shown. Strong, weak or off expression is defined as GFP expression observed at 400x magnification in both cell bodies and processes, in only cell bodies, or not observed either in cell bodies or processes, respectively. Heat shocks were treated to L4 larval stage animals at 33°C twice for 30 minutes and after 14 hours, phenotypes were analyzed. Over two independent transgenic lines were tested. n≥50 for each. (D) The average of wave width or wavelength of the indicated genotypes. n≥30 for each. Error bars are the SEM. *** indicates significantly different between indicated animals at p<0.001 (one-way ANOVA test followed by the Tukey post-hoc test). Heat shocks were applied to L4 larval stage animals at 33°C twice for 30 minutes and phenotypes were analyzed after 14 hours. (E-F) Shown are representative pictures of lim-4 mutants containing the hsp::lim-4cDNA transgene with no heat shock or after heat shock treatment. Images are derived from z-stacks of confocal microscopy images (E: Scale bar: 50 μm) and derived from a light microscopy image (F: Scale bar: 0.5 mm). (G) lim-4 is required to maintain function of the SMB neurons. The average wave width was analyzed in lim-4(ky403);Ex[hsp::lim-4cDNA] at 1 day after, 3 day after or 6 day after heat shock treatment. n≥30 for each. Error bars are the SEM. *** Significantly different between no heat shock and heat shock treatment conditions at p<0.001(one-way ANOVA test followed by the Tukey post-hoc test). Expression of lim-4 was previously shown to be autoregulated in the AWB neurons but not in the other LIM-4-expressing neurons [14]. We confirmed that lim-4 expression in the AWB neurons was not seen in lim-4 mutants at L1 larval stage, whereas the expression of lim-4 in the other neurons including the SMB neurons, was detected (Fig 4B). However, lim-4 expression in the SMB neurons gradually decreased from the L1 larval stage until it became undetectable in the adult stage (Fig 4B; Table 1). Hence, lim-4 appears to be required to maintain its own expression in the SMB neurons but does not initiate its expression, further supporting its role as a terminal selector gene in the SMB neurons.
To determine whether lim-4 acts cell-autonomously within SMB, we tried to rescue lim-4 phenotypes by expressing a wild-type lim-4 cDNA driven under the control of lim-4pΔ3 promoter. The lim-4pΔ3 promoter includes minimal upstream regulatory sequences that drive transgene expression exclusively in the SMB neurons but not as strongly as the full promoter of lim-4 and more dominantly in the SMBD than SMBV neurons (see below), and was used to identify expression in SMB of genes tested in this study (S6 Fig). The gene expression and locomotion defects of lim-4 mutants were partially restored, while the dye-filling defects were still present and the normal average wavelength was not altered, indicating that LIM-4 acts in the SMB neurons to affect locomotion and transmitter specification (Fig 4C and 4D; S3 Table). Taken together, lim-4 is expressed and acts in the SMB neurons to specify the SMB cell-fate.
Postdevelopmental expression of LIM-4 is sufficient to restore the SMB-specific defects of lim-4 mutants
To determine when the activity of LIM-4 is required for the expression of the SMB markers and proper locomotive movement, we first expressed lim-4 with an inducible, ubiquitously expressed heat-shock promoter (hsp16.2) [27]. Upon transient supply of lim-4 gene activity at the fourth larval stage (i.e., long after the SMB neurons have differentiated in the embryo), expression of flp-12 was fully restored and the loopy phenotype of lim-4 mutants was rescued (Fig 4C–4F). These results demonstrate that post-developmental expression of LIM-4 is sufficient to restore the expression of the SMB markers and the function of the SMB neurons in lim-4 mutants. These data further indicate that the SMB neurons are not irreversibly switched to another cell-fate and demonstrate that loss of lim-4 does not result in irreversible developmental defects. The dye-filling defects of the AWB/ADF neurons in lim-4 mutants were partially rescued after multiple heat shocks (S3 Table) [14, 16].
We also used the inducible rescue assay to corroborate the prediction that lim-4 is continuously required to maintain the functional properties of the SMB neurons. To this end, we supplied lim-4 activity via the heat-shock promoter at L4 stage and then analyzed the animals after 14 hours at the young adult stage. In these animals, we found the locomotory defects to be partially rescued (Fig 4G). When assayed after a long time interval at 3 and 6 days (i.e., older adult stages), the animals again displayed a mutant phenotype indistinguishable from the control (Fig 4G), suggesting that the transient rescuing ability of the lim-4 gene activity has faded. These results demonstrate that lim-4 does not only initiate but also maintains the expression of the SMB terminal differentiation genes and, hence, the function of the SMB neurons.
Expression of lim-4 is sufficient to induce the SMB identity in other cell-types
To address whether expression of lim-4 is sufficient to induce the SMB identity in other cell-types, we first examined ectopic flp-12 expression upon transient supply of lim-4 gene activity via the heat-shock promoter at the embryonic stage. Although the heat-shock promoter should drive ubiquitous LIM-4 expression, ectopic flp-12 expression was not seen broadly elsewhere; interestingly, expression was limited in only one cell-type, the ALN neurons, in 25% of transgenic animals (n = 50) (Fig 5A). The ALN neurons are a pair of cholinergic oxygen-sensing neurons in the tail [5, 11, 28] that do not appear functionally or linearly related to the SMB neurons (S5 Fig).
Fig. 5. lim-4 is sufficient to induce cholinergic or peptidergic marker expression in other cell-types. 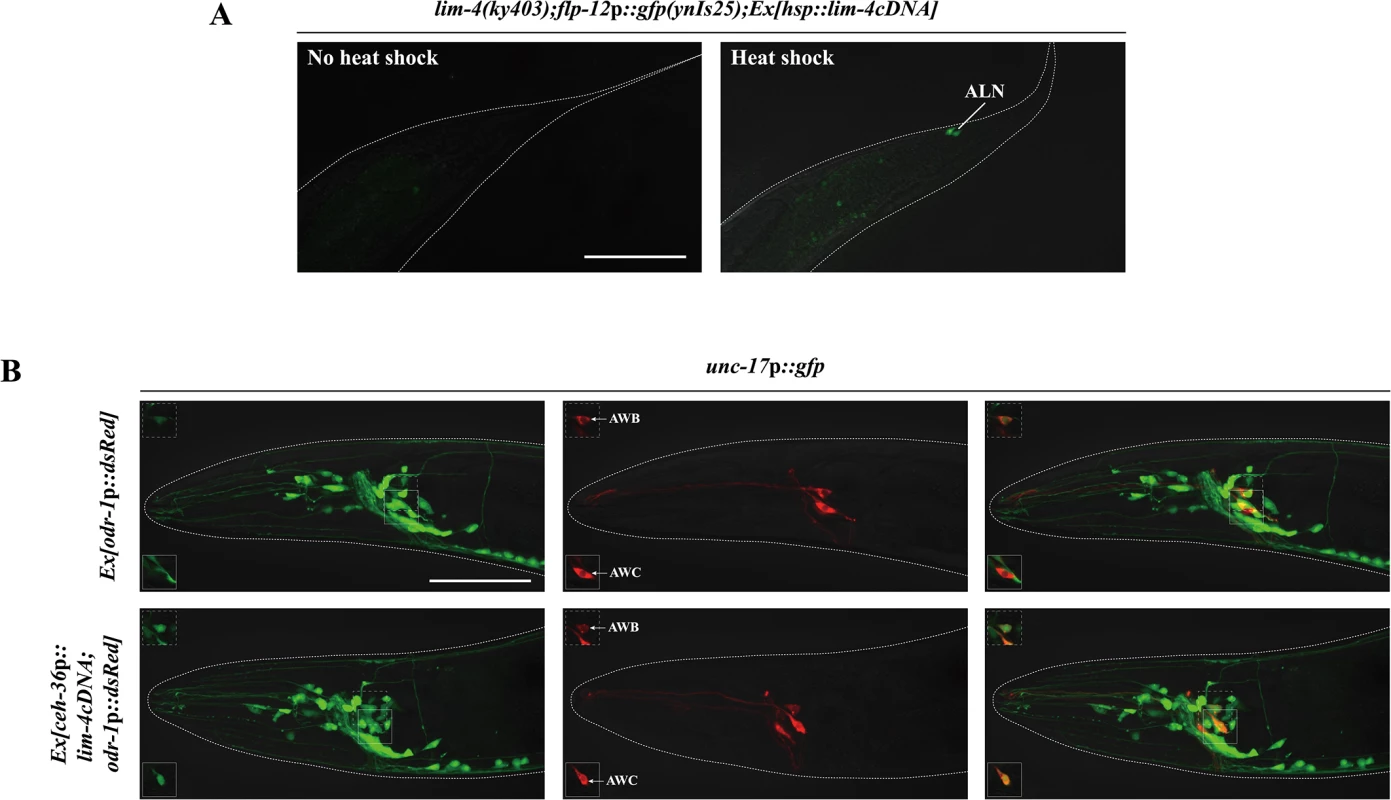
(A) Ectopic expression of LIM-4 is sufficient to drive flp-12 expression in the ALN cholinergic neurons. Heat shocks were applied to 2 or 3 fold stage of embryos at the 37°C twice for 30 minutes and phenotypes of adults were analyzed. Images are derived from z-stacks of confocal microscopy images. Posterior is at right. Scale bar: 50 μm. (B) Ectopic expression of LIM-4 in AWC induces unc-17 expression. Ex[odr-1p::dsRed] transgenic animals express dsRed in AWC and AWB [59]. Note that we observed unc-17 expression in the AWB neurons of wild-type animals. Images are derived from z-stacks of confocal microscopy images while images in the upper or bottom-left boxed regions are single focal plane confocal microscopy images. Anterior is at left. Scale bar: 50 μm. We next attempted to express LIM-4 in the glutamatergic chemosensory neurons AWC using the promoter of the ceh-36 homeobox gene [26, 29]. Ectopic expression of unc-17 VAChT was detected in AWC (Fig 5B) while the flp-12 was not ectopically expressed in AWC (S7 Fig). We further induced broader ectopic expression of LIM-4 in the subset of glutamatergic neurons under a specific eat-4 glutamate transporter gene promoter that drives reporter expression in 11 (but not in AWC) out of 38 glutamatergic neuron classes in the hermaphrodites [30]. Expression of LIM-4 in these cells did not drive ectopic expression of cho-1 ChT or affect expression of eat-4 (S8 Fig), suggesting that expression of LIM-4 alone is not sufficient to generally induce cholinergic cell fate in a subset of glutamatergic neurons. These results suggest that lim-4 is partially sufficient to drive expression of the SMB markers in a context-dependent manner.
LIM-4 regulates gene expression via a cis-regulatory motif in the SMB markers
Homeodomain transcription factors generally bind well-defined DNA sequences to control transcription of target genes [31]. Systemic analysis of homeodomain DNA-binding specificities allowed prediction of the recognition motif of each homeodomain protein, and a cis-regulatory motif containing the consensus TAAT core DNA sequences was predicted to be the binding site for the homeodomain of LIM-4 and its mammalian and Drosophila homologs (LHX6/8 and Arrowhead, respectively) (Fig 6A; S9 Fig) [32, 33]. Indeed, LHX6 and LHX8 have been shown to directly bind to the predicted DNA sequences (ATAATCA) in the promoter regions of the Shh gene [34].
Fig. 6. A cis-regulatory motif is necessary and sufficient to drive expression of terminally differentiated SMB markers. 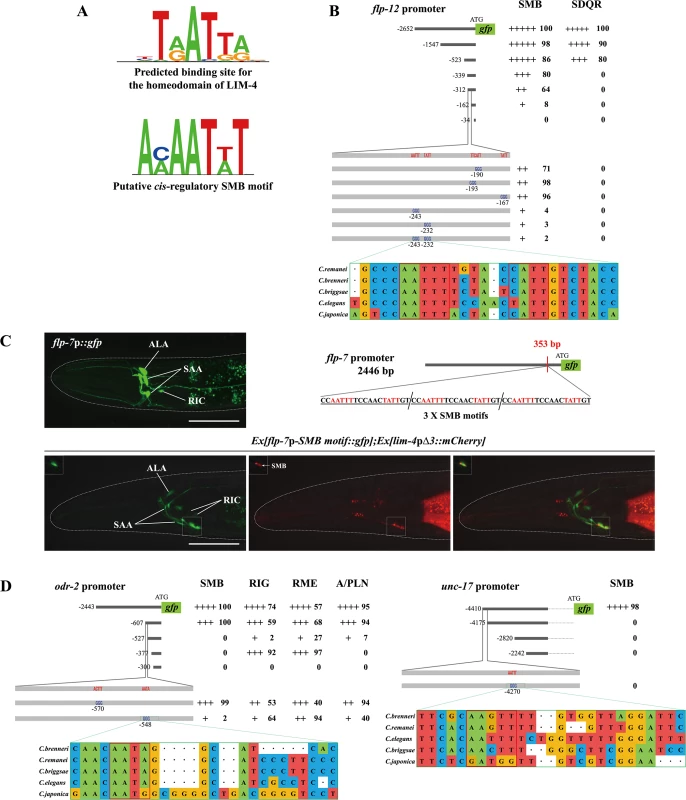
(A) Shown are the predicted binding site (top) for the homeodomain of LIM-4 from a web based tool, PreMoTF (http://stormo.wustl.edu/PreMoTF) [33] and the putative cis-regulatory SMB motif (bottom) identified from promoter analysis of flp-12, odr-2, and unc-17 genes. (B) The percentage of transgenic animals expressing flp-12p:: gfp reporter construct in the indicated neurons is shown. Strength of GFP expression is indicated by the number of + symbols. Wild-type nucleotides are indicated in red, mutated nucleotides in blue. At least two independent extrachromosomal lines for each construct were examined except a flp-12 promoter (-2652) construct of which number was derived from one integrated line. n≥50 for each. Identified cis-regulatory sequences found in other Caenorhabditis species are shown below. Analysis between -523 bp and -339 bp upstream of flp-12 promoter is shown in S10 Fig. (C) Insertion of the SMB motif into the non-SMB expressed flp-7 gene promoter induces flp-7 expression in the SMB neurons. The inserted cis-regulatory sequences identified from the flp-12 promoter analysis and the inserted site in the flp-7 promoter are indicated. GFP expression is overlapped with expression of the lim-4pΔ3::mCherry reporter in the SMB neurons of wild-type animals. Images are derived from z-stacks of confocal microscopy images while images in the upper-left boxed regions are single focal plane confocal microscopy images. Anterior is to the left. Scale bars: 50 μm. (D) The percentage of transgenic animals expressing odr-2p::gfp or unc-17p::gfp reporter construct in the indicated neurons is shown. Strength of GFP expression is indicated by the number of + symbols. Wild-type nucleotides are indicated in red, mutated nucleotides in blue. At least two independent extrachromosomal lines for each construct were examined. n≥50 for each. Identified cis-regulatory sequences found in other Caenorhabditis species are shown below. To identify cis-regulatory motifs required to drive expression of the SMB marker genes in the SMB neurons, DNA sequences within the promoters of the SMB markers were serially deleted and the resultant transgenic animals were examined for altered expression patterns. From these analyses, we first determined a minimal region within the flp-12 promoter for flp-12 expression. Deletion of a 150 bp sequence located ~162 bp upstream of the translation start sequence caused decreased gfp expression in the SMB neurons (Fig 6B; S10 Fig). Within the 150 bp region, we next found four AT - rich DNA sequences that are fully conserved in the promoters of the flp-12 orthologs in the related Caenorhabditis species (Fig 6B). Mutations of two AT-rich DNA sequences resulted in an almost complete loss of gfp expression, while mutations of the other two sequences did not affect the gfp expression (Fig 6B), indicating that the former two motifs are necessary for the expression of flp-12 in the SMB neurons. DNA sequences of these motifs (AAAATTG and ACAATAG) share limited sequence conservation with putative LIM-4 binding sequences, and will be referred to as SMB motifs (Fig 6A). To test whether these SMB motifs are sufficient to drive gene expression in the SMB neurons, we inserted three copies of the SMB motifs in the promoter of flp-7, which is normally expressed in the several head neurons, but not in the SMB neurons (Fig 6C) [13]. Transgenic animals expressing a flp-7p-SMB motif::gfp reporter construct still exhibited gfp expression in flp-7 expressing neurons, indicating that insertion of the SMB motifs within the regulatory region of flp-7 does not alter the flp-7 expression pattern. In addition, we observed consistent expression of flp-7 in the SMB neurons in 100% transgenic animals (n = 50) (Fig 6C), suggesting that these SMB motifs are necessary and sufficient to drive gene expression in the SMB neurons. These results are consistent with the hypothesis that LIM-4 directly binds the SMB motifs to regulate expression of flp-12 gene in the SMB neurons.
To define additional regulatory motifs for expression of SMB markers, we examined the promoter regions of two additional SMB markers, odr-2 and unc-17, and defined the regions essential for SMB expression. These regions in the odr-2 or unc-17 promoters contained the SMB motifs found in the flp-12 promoter (Fig 6D). We mutated these motifs in the context of the odr-2 and unc-17 reporter genes and found that these mutations reduced expression of the reporter genes (Fig 6D). These results demonstrate that distinct cis-regulatory motifs can determine cell-specific expression or something of this sort.
A cis-regulatory region in the lim-4 promoter that is required for lim-4 expression in the AWB neurons was previously identified [35]. We performed analogous deletion analysis experiments in transgenic animals to dissect the lim-4 promoter to identify motifs required for lim-4 expression in the SMB neurons (S11 Fig). As proof-of-principle, we also identified the lim-4 regulatory sequences for the AWB expression (S11 Fig). However, we could not identify simple cis-regulatory motifs in the lim-4 promoter required for the SMB expression (S11 Fig). Instead, multiple regions in the lim-4 promoter act in concert to regulate LIM-4 expression in the SMB neurons, suggesting a complexity of cis-regulatory motifs in the lim-4 promoter to ensure proper LIM-4 expression in the SMB neurons.
The function of lim-4 is conserved in human
The mammalian genome contains two LIM-4 orthologs, LHX6 and LHX8. In mice, these genes are largely expressed in the developing and adult striatum and orchestrate specification of interneuron identities; specifically, LHX6 and LHX8 are required to determine GABAergic/peptidergic and cholinergic interneuronal cell fate, respectively. In addition, these genes have redundant function to regulate expression of shh in MGE neurons [34]. LIM-4 exhibits a high degree of protein sequence homology to LHX6 and LHX8 (in particular, 60% identical in its homeodomain) (Fig 7A) [14], suggesting a functional conservation of these proteins.
Fig. 7. C. elegans LIM-4 or human LHX6 induces expression of cholinergic makers and neuronal characteristics in human neuroblastoma cells. 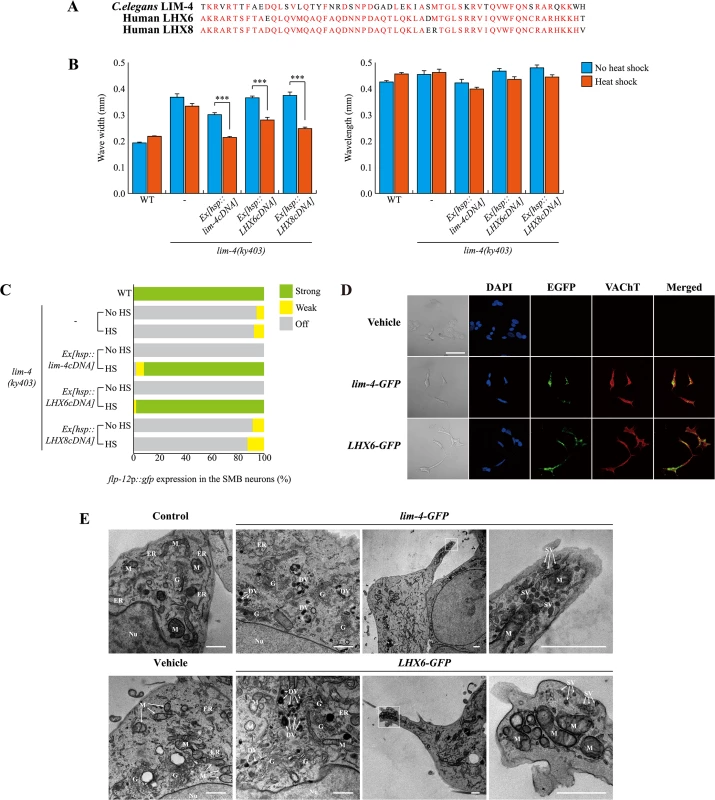
(A) Shown is the sequence alignment of the homeodomain of C. elegans LIM-4 and human LHX6 and LHX8. Identical residues in at least two proteins are shown in red. (B) Average of wave width or wavelength of the indicated genotypes. n≥30 for each. Error bars are the SEM. *** indicates different between heat shock and no heat shock conditions at p<0.001 (student t-test). Heat shocks were applied to L4 larval stage animals at 33°C twice for 30 minutes and phenotypes were analyzed after 14 hours. Data of WT,-, or Ex[hsp::lim-4cDNA] are from Fig 4D. (C) Percentage of animals of the indicated genotypes expressing stably integrated flp-12p::gfp reporter (ynIs25) is shown. Strong, weak or off expression is defined as GFP expression observed at 400x magnification in both cell bodies and processes, in only cell bodies, or not observed either in cell bodies or processes, respectively. Over two independent transgenic lines were tested. n≥50 for each. Data of WT,-, or Ex[hsp::lim-4cDNA] are from Fig 4C. (D) Confocal images of SH-SY5Y human neuroblastoma cell line transfected by empty vector, C. elegans lim-4 or human LHX6 and immunostained with VAChT antibodies. Scale bar: 50 μm. Note that cells expressing either LHX6-GFP or lim-4-GFP exhibit spiky protrusions. (E) Ultrastructural analysis of LIM-4 or LHX6 transfected SH-SY5Y cell lines. Images from transmission electron microscope for untransfected (control), empty vector, lim-4-GFP, or LHX6-GFP transfected cells are shown. Left two columns are from peri-nuclear region and right two columns are from protruded region (far right images are higher magnification of the boxed area). Nu: nucleus, G: Golgi apparatus, M: mitochondria, ER: endoplasmic reticulum, DV: dense core vesicle, SV: synaptic vesicle. Scale bars: 1 μm. Confocal images of these control and transfected SH-SY5Y cells grown on the MatTek culture dish are shown in S14 Fig. To test for functional homology, we first tried to rescue C. elegans lim-4 mutants by expressing human LHX6 or LHX8 cDNA under the control of the heat shock promoter. Similar to C. elegans lim-4 cDNA, human LHX6 or LHX8 cDNA fully restored altered locomotion of lim-4 mutants (Fig 7B; S12A Fig). Moreover, LHX6 also fully rescued the defect of flp-12 expression in lim-4 mutants while LHX8 did not rescue (Fig 7C; S12B Fig), indicating that LHX6 may have a higher degree of functional conservation to LIM-4 than LHX8.
LHX8 has been shown to be required for the development and maintenance of cholinergic neurons in mouse basal forebrain [36, 37]. Overexpression of LHX8 was sufficient to differentiate rat hippocampal neural stem cells or newborn neurons into cholinergic neuron types [38, 39] and induced expression of cholinergic markers in a human neuroblastoma cell line [40]. Whether LHX6 has a similar role in specification of cholinergic cell fate has not been explored. Thus, we tested whether overexpression of human LHX6 or C. elegans LIM-4 could promote expression of cholinergic markers in human neuroblastoma SH-SY5Ycells. These cells appear to mimic immature catecholaminergic neurons when untreated [41, 42], but can differentiate into various mature neuron-like phenotypes depending on the addition of differentiation-inducing agents [43]. We generated stable SH-SY5Y cell lines expressing either LHX6-GFP, lim-4-GFP, or empty vehicle and asked whether transfected cell lines express cholinergic markers. Expression of either LHX6-GFP or lim-4-GFP was detected predominantly in cell nuclei, supporting the action of LHX-6 and LIM-4 as transcription factors (Fig 7D; S13A Fig). Transfected cells were immunoreactive to VAChT or choline acetyltransferase (ChAT) antibodies and exhibited higher endogenous ChAT message levels, as assayed by quantitative reverse transcription polymerase chain reaction (RT-PCR), compared to that in cells transfected with an empty vehicle (Fig 7D; S13B Fig). Thus, human LHX6 and even C. elegans lim-4 are sufficient to promote expression of cholinergic markers in human cells. We also noted that cells expressing either LHX6-GFP or lim-4-GFP were morphologically different to cells bearing empty vehicle: empty vehicle bearing cells tended to grow in clusters and were round shape; by contrast, LHX6-GFP or lim-4-GFP expressing cells formed less clusters and appeared as spiky neuronal cells (Fig 7D; S13B Fig). These results, therefore, indicate that LHX6 or LIM-4 can induce differentiation of SH-SY5Y cells into cholinergic as well as neuronal phenotypes.
We further examined the morphology of transfected cells by electron microscopy. Untransfected or empty vesicle transfected SH-SY5Y cells exhibited typical shapes of mitochondria, endoplasmic reticulum (ER), and Golgi apparatus in the peri-nuclear region (Fig 7E). In either lim-4 or LHX6 transfected cells, however, we observed additional ultrastructural components, such as 100nm large-dense core vesicles, which may contain neuropeptides, near the Golgi apparatus (Fig 7E) [44]. Furthermore, in the spiky protruded region of lim-4 or LHX6 transfected cells, we also identified mitochondria and small synaptic vesicles that may represent axon terminals of neurons (Fig 7E). These results further support that expression of LIM-4 or LHX6 may produce synaptic vesicles containing large neuropeptides and small molecule neurotransmitters in human cell lines.
Discussion
In these studies, we have identified an important regulator that controls terminal differentiation of a distinct neuronal cell-type in C. elegans. The SMB neurons appear to have mixed neuronal functions. Their long, unbranched and synapse-free processes along the body may serve as proprioreceptors to sense body stretch. In addition, they contact over 20 sensory, inter, or motor neurons via chemical or electrical synapses and may integrate additional extrinsic or intrinsic cues to regulate head muscle contraction [11]. The SMB neurons co-express a unique combination of neurotransmitters, acetylcholine and FLP-12 FMRFamide-like neuropeptides. Thus, an intriguing question is how the SMB sensory/inter/motor neurons acquire their unique characteristics. Our experiments show that the LIM-4 LIM homeodomain transcription factor is necessary and sufficient to promote and probably maintain SMB-specific properties and functions. In lim-4 null mutants, the neurotransmitter identity and neuronal function of the SMB neurons are completely lost but pan-neuronal features are not affected. Transient LIM-4 expression in lim-4 mutants not only restores the SMB characteristics and functions, but also induces ectopic expression of a SMB-expressed neurotransmitter in another cell-type. Our promoter analysis suggests that LIM-4 directly regulates expression of the SMB terminal differentiation marker via well conserved homeodomain binding sequences and also controls its own expression. Hence, we propose that lim-4 acts as a terminal selector gene to broadly specify the SMB neuronal identity [2, 45].
A few terminal selector genes that determine cholinergic cell-fate in C. elegans have been identified. For example, the Olf/EBF gene unc-3, the heterodimer of LIM homeobox gene ttx-3 and Paired-like homeobox gene ceh-10, and POU homeobox gene unc-86 regulate terminal differentiation of the A-, B-, and AS-type ventral nerve cord or SAB motor neurons, the AIY interneurons, and the IL2 sensory neurons, URA motor neurons and URB interneurons, respectively [6, 7, 8, 9, 10]. ttx-3 also acts as a terminal selector in the AIA interneurons [10]. These genes regulate expression of not only the cholinergic gene battery, including unc-17 (VAChT), but also other terminally differentiated cell-specific markers. Furthermore, these trans-acting factors appear to directly bind to the evolutionarily conserved cis-regulatory elements of most, if not all, their target genes. In the case of the unc-17 promoter region, distinct cis-regulatory target sites, such as the COE motif for UNC-3 and the AIY motif for TTX-3/CHE-10, are systemically organized (S15A Fig). In this study, we identified an additional cis-regulatory element, called the SMB motif, in the unc-17 gene (S15A Fig), suggesting that the elaborate cis-regulatory architecture ensures expression of cell-specific characteristics. Since additional terminal selector genes required for specification of over 50 uncharacterized cholinergic cell-types need to be identified, additional motifs must exist in the cholinergic gene battery such as unc-17 (S15A Fig).
Recent work has shown that distinct combination of 13 different terminal selector genes defines identity of 25 different glutamatergic cell-types in C. elegans, suggesting that the combinatorial codes of terminal selector transcription factors are a general theme for determining cell-type specificity [30]. In fact, in the AIY cholinergic neurons, two terminal selector transcription factors, ttx-3 and ceh-10, form a heterodimer that directly regulates expression of their target genes via a common cis-regulatory bipartite motif; mutations of each gene lead to complete loss of the AIY specific neuronal identity [6, 9]. We have tried to identify the putative binding partner(s) of LIM-4 in the SMB neurons by analyzing the expression pattern of the flp-12 reporter construct in mutants for which genes were previously reported to be expressed in the SMB neurons such as the fax-1 nuclear receptor and cog-1 Nkx6-type homeobox transcription factor [46, 47]. None of these mutations affects flp-12 expression in the SMB neurons (S4 Table). Therefore, it is not yet clear which transcription factors work in combination with lim-4 to control the terminal differentiation of the SMB neurons. Because our results demonstrate that lim-4 expression is initiated at an early developmental stage and then autoregulated afterward, we also tested the possibility that expression of fax-1 or cog-1 in the SMB neurons may regulate lim-4 expression in SMB. In chemosensory neuron types, the lin-11 LIM homeobox, ceh-37 Otx, mls-2 HMX/NKX homeobox and nhr-67 Tailless/TLX genes control expression of terminal selector genes in the AWA, AWB, AWC, and ASE neurons, respectively [26, 29, 48, 49]. However, lim-4 expression is not altered in fax-1 or cog-1 mutants (S4 Table), indicating that these SMB-expressed transcription factors may have more specific roles in development or differentiation of the SMB neurons. We will continue searching for the transcription factors that partner with LIM-4 or act upstream or downstream of LIM-4 to control terminal differentiation of the SMB neurons.
Previous studies show that lim-4 determines the proper cell-type specification of the AWB and serotonergic ADF neuron types [14, 16]. The AWB and ADF neurons are two classes of amphidial chemosensory neurons in the head of worms that detect volatile chemical repellants and putative food signals, respectively [50]. In lim-4 mutants, the AWB neurons lack AWB-specific characteristics and functions, such as expression of putative 7-TM receptor str-1, AWB-specific cilia and axon morphology, and abilities to take up lipophilic dyes, and instead acquire features and functions of the AWC olfactory neurons, such as expression of the putative 7-TM receptor str-2 and AWC-specific cilia and axon structures, suggesting that lim-4 acts as a cell fate switch between the AWB and AWC neurons [14]. In case of ADF cell-fate specification, lim-4 acts transiently in the precursor cells of ADF and regulates part of the terminal differentiation process; lim-4 mutants lack expression of a set of serotonergic markers, including tryptophan hydroxylase tph-1, but do not affect expression of a putative 7-TM receptor srh-142 [16]. The ceh-37 Otx gene is required for expression of lim-4 in AWB and srh-142 in ADF, suggesting that ceh-37 also differentially affects the AWB and ADF cell fates [26]. In addition, lim-4 has a role in regulating axon morphology of the SAA neurons but not expression of terminal differentiation markers [14]. We propose that the SMB neurons in lim-4 mutants do not adopt a functionally or lineage-related cell fate, but remained undifferentiated because they lose expression of most, if not all, terminal differentiation genes. Thus, lim-4 plays distinct roles in neuronal development in a context-dependent manner and acts as a bona fide terminal selector for the differentiation of SMB (S15B Fig).
The LIM homeobox gene family has a high degree of structural conservation through evolution amongst the homeobox gene superfamily [51–53]. However, their functional conservation among distantly related species is relatively unexplored. We demonstrate that C. elegans lim-4 and human LHX6 and LHX8 (also referred as L3 or LHX7) show striking functional similarity; LHX6 completely rescues locomotive defects and flp-12 expression phenotypes of lim-4 mutants, whereas human LHX8 restores only locomotion but not flp-12 expression in lim-4 mutants. Furthermore, expression of either LHX6 or lim-4 is sufficient to drive cholinergic differentiation in human neuroblastoma cells. The role of LHX8 in cholinergic cell-fate determination in the mammalian nervous system has been well characterized [36, 54, 55]. Deletion of the murine LHX8 (LHX7), causes a subtype of cholinergic interneurons to convert into another subtype of GABAergic interneurons [55]. However, the study of LHX6 function has focused on GABAergic fate specification [56, 57]. We have uncovered that LHX6 also plays a role in cholinergic cell fate determination in C. elegans or human neuroblastoma cells and acts as a terminal selector to control the differentiation of neuronal subtypes. Cholinergic neurons in the mammalian forebrain have crucial roles in locomotive and cognitive functions and thus, understanding and manipulation of cholinergic cell fate specification may be beneficial to identify therapeutic targets and methods for neurodiseases resulting from cholinergic neuronal dysfunction.
Materials and Methods
Strains
N2 Bristol strain was used as wild-type strain. Mutant strains and transgenic strains used in this study are listed in S5 Table. All strains were maintained at 20°C.
Isolation of lim-4 mutants
The flp-12p::gfp(ynIs25) integrated strain was used to performed EMS mutagenesis according to Sulston and Hodgkin (1988). Eight alleles (yn19, lsk1, lsk2, lsk3, lsk4, lsk5, lsk6, lsk7) in which GFP expression was completely abolished, were isolated from screening ~15,000 haploid genomes, found to be allelic to each other, and mapped on LG X. Based on three factor crosses using the double mutants unc-6 dpy-6, dpy-8 unc-6, and unc-2 dpy-8, yn19 was located approximately 2.4 MU downstream of unc-2 and 4.3 MU upstream of dpy-8. In this region, we did complementation tests with a lim-4(ky403) mutant and found that they were allelic. The molecular lesions were identified by sequencing amplification products of lim-4. All lim-4 alleles were outcrossed with N2 at least five times before phenotypic analysis. To observe locomotion phenotypes of lim-4 mutants (ky403, yn19, lsk3, lsk5), the integrated flp-12p::gfp array was removed by mating with N2 males.
Molecular biology and transgenic worms
For promoter analysis, promoter regions of odr-2 and unc-17 were amplified by PCR from N2 genomic DNA and were inserted into the pPD95.77 vector [27]. lim-4p::gfp [14] was gifted from Piali Sengupta. Promoter regions of each reporter construct were deleted with various digestion enzymes or PCR fusions. Mutagenesis was performed using QuikChange II XL Site-Directed Mutagenesis Kit (Stratagene) according to the manufacturer’s protocol.
For the constructs used to test for rescue, the hsp16.2 promoter was fused with the following cDNAs: lim-4 cDNA (kind gift form Oliver Hobert), human LHX6 cDNA (BC103937) and human LHX8 cDNA (BC040321) (Thermo Fisher Scientific). The lim-4p∆3 or odr-1 promoter were used to generate the lim-4p∆3::lim-4cDNA or odr-1p::lim-4cDNA constructs, respectively.
To generate the flp-7p-SMBmotif::gfp construct, three copies of SMB motif oligomers that have SacI enzyme site at 5’ and 3’ ends were synthesized and inserted into SacI site at -353 bp region of the flp-7 promoter.
otIs518; otIs534 (cho-1fosmid::yfp; eat-4fosmid::mChOpti) and the eat-4p∆5::gfp construct were kind gifts from Oliver Hobert. To express lim-4 cDNA in the subset of glutamatergic neurons, lim-4 cDNA was replaced with gfp to generate eat-4p∆5::lim-4cDNA. Then, 2.5 ng of eat-4p∆5::lim-4cDNA was injected into the otIs518; otIs534 strain with 50 ng rol-6 as an injection marker.
To express lim-4 cDNA in the AWC neurons during development stage, ceh-36p was fused with lim-4 cDNA and 5 ng was injected into vsIs48 (unc-17p::gfp) and ynIs82 (flp-12p::gfp) with 50ng of odr-1p::dsRed as an injection marker. As the control, 50 ng odr-1p::dsRed was injected into vsIs48 (unc-17p::gfp).
ceh-17p::dsRed was kindly gifted from Satoshi Suo.
Heat shock treatment and phenotype analysis
Heat shocks were administered to fourth larva stage (L4) of the transgenic worms at 33°C twice for 30 minutes with an hour incubation at 20°C between heat shocks for recovery modified from [7]. After heat shocks, worms were incubated at 20°C for 14 hours to reach the young adult stage when lim-4 phenotypes were assayed. To observe ectopic expression of flp-12p::gfp in other cell types, heat shocks were administered at the embryo 2 - or 3-fold stage two times at 37°C for 30 minutes with an hour incubation at 20°C between heat shocks. Heat shocks were administered at L4 animals to observe locomotion at days 1, 3, and 6. The L4 stage was counted as day 0.
The level of flp-12p::gfp expression in the SMB neurons was quantified as strong, weak, off. Strong was determined as robust expression in the SMB neuronal cell bodies and processes. Weak was defined as faint expression in cell bodies and no expression in the processes. Off was defined as no flp-12p::gfp expression in either cell bodies and processes. To measure wave width and wavelength of the worm tracks, Leica microscope software (Leica Application Suite Advanced Fluorescence Lite 3.5. Ink) was used. Wavelength was defined as distance between one peak and the next corresponding peak, and wave width was distance from the peak to the trough of the sine wave. The average of six consecutive wave width and wavelength from each worm track was quantified as the individual data.
Laser ablation
Laser ablation experiments were performed as previously described [58]. L1 larvae of the integrated lim-4p::gfp (oyIs35) strain were anesthetized with 10 mM sodium azide and all four SMB or SAA neurons were killed by a nitrogen dye-pulsed laser (Photonic Instruments, St. Charles, IL). Animals were recovered for 3 days at 20°C. After performing locomotion assays, GFP expression of lim-4p::gfp reporter was observed to confirm laser ablations in the SMB or SAA neurons.
Bioinformatics analysis
The conservation of the cis-regulatory motif from the promoter analysis of the flp-12, lim-4, odr-2, and unc-17 were examined by using the USCS genome browser (http://genome.ucsc.edu/). DNA sequences from the five different Caenorhabditis species were obtained from the UCSC website, and aligned using the ClustalW2 in EBI (European Bioinformatics Institutes; http://www.ebi.ac.uk/Tools/msa/clustalw2/) to identify cis-regulatory regions including SMB motifs. The position frequency matrix (PFM) of LIM-4, LHX6, and LHX8 predicted binding sites were derived from a web based tool, PreMoTF (http://stormo.wustl.edu/PreMoTF). Predicted conserved motif sequence logo was obtained from the Seq2Logo website (http://www.cbs.dk/biotools/Seq2Logo/).
Microscopy
Fluorescent microscopic images were taken with a Zeiss LSM700 Confocal microscope and were obtained using ZEN 2009 Light Edition software. For light microscopic images of worms, Leica High-performance Fluorescence Stereomicroscopy M205FA was used and Leica Application Suite Advanced Fluorescence Lite 3.5 software was used to measure the phenotype.
Cell culture
The SH-SY5Y human neuroblastoma cell line (ATCC) was cultured with 10% complete medium (1 : 1 mixture of DMEM and Ham's F12 medium and 10% supplemental fetal bovine serum, 100 U/ml penicillin, and 100 μg/mL streptomycin) in a humidified, 5% CO2-95% air, 37°C incubator.
Construction recombinant lentiviruses and transfection into the SH-SY5Y cells
HEK293T cells were purchased from American Type culture Collection (ATCC) and were cultured in DMEM with 10% FBS. cDNA for lim-4 or LHX6 was cloned into the lentiviral vector, pRetroX-IRES-ZsGreen1 (Clontech). Transformation was performed with lentivirus constructs and packaging vector into 293T cells using a lipofectamine 2000 reagent (Invitrogen). At 72 hours post transfection, the viral particles were harvested by filtration using a 0.45mm syringe filter.
Wells of glass bottom dishes (MatTek) were coated 0.1μM fibronectin (Sigma) for 1 h at 37°C. After removing the fibronectin solution, SH-SY5Y cells were seeded at density of 5×104 cells per well and grown to confluence in 50%. Viruses were added to cells at 37°C for 120 min followed by addition of an equal volume of DMEM and Ham's F12 medium with 10% fetal bovine serum, and incubation for a further 24 h. Cells then were washed with phosphate-buffered saline, and DMEM and Ham's F12 medium with 1% fetal bovine serum. After an additional 24 h in growth medium, cells were washed in 1 : 1 DMEM and Ham's F12 medium with 1% fetal bovine serum. Under these conditions, up to 90% of cells were infected.
Immunocytochemistry
Cells were fixed using 4% paraformaldehyde, followed by 0.1% triton-X permeabilization and incubation with antibodies. Fixed cells were incubated at 4°C overnight with the primary antibodies, including human-cross reactive rabbit anti-ChAT (Millipore) and human-cross reactive rabbit anti-VAChT (Synaptic Systems). Bound antibodies were visualized with Alexa Fluor 594 FluoroNanogold-anti-rabbit–conjugated secondary antibodies (Nanoprobes). Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI, Sigma).
qRT-PCR
Total RNA was obtained from the cells by using a Trizol Reagent (Invitrogen) according to the manufacturer’s instructions. cDNAs were synthesized using High-Capacity cDNA reverse transcription kits (Applied Biosystems). Quantitative real-time RT PCR was performed using the SYBR Green PCR master mix kit (Applied Biosystems) on the ABI 7500 Real Time PCR System under the following conditions: Cycling conditions were 2 min at 50°C, 10 min at 95°C, followed by 40 cycles of 95°C for 15s and 60°C for 1 min. Primer sets were designed using primer express 3.0 software based on the human gene sequences from GenBank and are as follows: ChAT (sense: 5'-GGCTCAGAACAGCAGCATCA -3' and antisense: 5'-GAGACGGCGGAAATTAATGACA -3'); GAPDH (sense: 5'-ACCCACTCCTCCACCTTT GA-3' and antisense: 5'-TGTTGCTGTAGCCAAATTCGTT-3'). The housekeeping gene GAPDH was used as an internal standard. Reaction specificity was confirmed by melting curve analysis.
EM
Control and transfected SH-SY5Y cells grown on the MatTek culture dish were fixed for 2 h at 4°C in PBS containing 2.5% glutaraldehyde. After three washes in PBS, the cells were postfixed with 1% osmium tetroxide on ice for 2 h and washed three times again in PBS. The cells were then embedded in Epon 812 mixture and polymerized in an oven at 60°C for 24 hours after dehydration in increasing concentrations of ethanol (50, 70, 80, 90, 95 and 100%) and propylene oxide series (20 min each). The embedded blocks were trimmed and sectioned on an ultramicrotome with a diamond knife and ultrathin sections were collected on Formvar-coated copper grids. The grids were stained with 2.5% uranyl acetate (7 min) and Reynolds lead citrate (2 min), and were viewed with a transmission electron microscope (Technai G2 Spirit Twin, FEI, USA) at 120 kV.
Supporting Information
Zdroje
1. Guillemot F. Spatial and temporal specification of neural fates by transcription factor codes. Development 2007;134 : 3771–80. 17898002
2. Hobert O. Regulation of terminal differentiation programs in the nervous system. Annu Rev Cell Dev Biol 2011;27 : 681–96. doi: 10.1146/annurev-cellbio-092910-154226 21985672
3. Deneris ES, Hobert O. Maintenance of postmitotic neuronal cell identity. Nat Neurosci 2014;17 : 899–907. doi: 10.1038/nn.3731 24929660
4. Li C, Kim K. Neuropeptides. WormBook 2008;1–36. doi: 10.1895/wormbook.1.142.1
5. Duerr JS, Han HP, Fields SD, Rand JB. Identification of Major Classes of Cholinergic Neurons in the Nematode Caenorhabditis elegans. J Comp Neurol 2008;506 : 398–408. 18041778
6. Altun-Gultekin Z, Andachi Y, Tsalik EL, Pilgrim D, Kohara Y, Hobert O. A regulatory cascade of three homeobox genes, ceh-10, ttx-3 and ceh-23, controls cell fate specification of a defined interneuron class in C. elegans. Development 2001;128 : 1951–69. 11493519
7. Kratsios P, Stolfi A, Levine M, Hobert O. Coordinated regulation of cholinergic motor neuron traits through a conserved terminal selector gene. Nat Neurosci 2011;15 : 205–14. doi: 10.1038/nn.2989 22119902
8. Kratsios P, Pinan-Lucarré B, Kerk SY, Weinreb A, Bessereau JL, Hobert O. Transcriptional coordination of synaptogenesis and neurotransmitter signaling. Curr Biol 2015;25 : 1282–95. doi: 10.1016/j.cub.2015.03.028 25913400
9. Wenick AS, Hobert O. Genomic cis-regulatory architecture and trans-acting regulators of a single interneuron-specific gene battery in C. elegans. Dev Cell 2004;6 : 757–70. 15177025
10. Zhang F, Bhattacharya A, Nelson JC, Abe N, Gordon P, Lloret-Fernandez C, et al. The LIM and POU homeobox genes ttx-3 and unc-86 act as terminal selectors in distinct cholinergic and serotonergic neuron types. Development 2014;141 : 422–35. doi: 10.1242/dev.099721 24353061
11. White JG, Southgate E, Thomson JN, Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci 1986;314 : 1–340. 22462104
12. Gray JM, Hill JJ, Bargmann CI. A circuit for navigation in Caenorhabditis elegans. Proc Natl Acad Sci U S A 2005;102 : 3184–91. 15689400
13. Kim K, Li C. Expression and regulation of an FMRFamide-related neuropeptide gene family in Caenorhabditis elegans. J Comp Neurol 2004;475 : 540–50. 15236235
14. Sagasti A, Hobert O, Troemel ER, Ruvkun G, Bargmann CI. Alternative olfactory neuron fates are specified by the LIM homeobox gene lim-4. Genes Dev 1999;13 : 1794–806. 10421632
15. Dawid IB1, Toyama R, Taira M. LIM domain proteins. C R Acad Sci III. 1995 Mar;318(3):295–306. 7788499
16. Zheng X, Chung S, Tanabe T, Sze JY. Cell-type specific regulation of serotonergic identity by the C. elegans LIM-homeodomain factor LIM-4. Dev Biol 2005;286 : 618–28. 16168406
17. Chou JH, Bargmann CI, Sengupta P. The Caenorhabditis elegans odr-2 gene encodes a novel Ly-6-related protein required for olfaction. Genetics 2001;157 : 211–24. 11139503
18. Colbert HA, Smith TL, Bargmann CI. OSM-9, a novel protein with structural similarity to channels, is required for olfaction, mechanosensation, and olfactory adaptation in Caenorhabditis elegans. J Neurosci 1997;17 : 8259–69. 9334401
19. Alfonso A, Grundahl K, Duerr JS, Han HP, Rand JB. The Caenorhabditis elegans unc-17 gene: a putative vesicular acetylcholine transporter. Science 1993;261 : 617–9. 8342028
20. Okuda T, Haga T, Kanai Y, Endou H, Ishihara T, Katsura I. Identification and characterization of the high-affinity choline transporter. Nat Neurosci 2000;3 : 120–5. 10649566
21. Pujol N, Torregrossa P, Ewbank JJ, Brunet JF. The homeodomain protein CePHOX2/CEH-17 controls antero-posterior axonal growth in C. elegans. Development 2000;127 : 3361–71. 10887091
22. Kennerdell JR, Fetter RD, Bargmann CI. Wnt-Ror signaling to SIA and SIB neurons directs anterior axon guidance and nerve ring placement in C. elegans. Development 2009;136 : 3801–10. doi: 10.1242/dev.038109 19855022
23. Harfe BD, Fire A. Muscle and nerve-specific regulation of a novel NK-2 class homeodomain factor in Caenorhabditis elegans. Development 1998;125 : 421–9. 9425137
24. Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol 1983;100 : 64–119. 6684600
25. Brenner S. The genetics of Caenorhabditis elegans. Genetics 1974;77 : 71–94. 4366476
26. Lanjuin A, VanHoven MK, Bargmann CI, Thompson JK, Sengupta P. Otx/otd homeobox genes specify distinct sensory neuron identities in C. elegans. Dev Cell 2003;5 : 621–33. 14536063
27. Fire A, Harrison SW, Dixon D. A modular set of lacZ fusion vectors for studying gene expression in Caenorhabditis elegans. Gene 1990;93 : 189–198. 2121610
28. Chang AJ, Chronis N, Karow DS, Marletta MA, Bargmann CI. A distributed chemosensory circuit for oxygen preference in C. elegans. PLoS Biol 2006;4: e274. 16903785
29. Kim K, Kim R, Sengupta P. The HMX/NKX homeodomain protein MLS-2 specifies the identity of the AWC sensory neuron type via regulation of the ceh-36 Otx gene in C. elegans. Development 2010;137 : 963–74. doi: 10.1242/dev.044719 20150279
30. Serrano-Saiz E, Poole RJ, Felton T, Zhang F, De La Cruz ED, Hobert O. Modular control of glutamatergic neuronal identity in C. elegans by distinct homeodomain proteins. Cell 2013;155 : 659–73. doi: 10.1016/j.cell.2013.09.052 24243022
31. Bürglin TR. Homeodomain Sybtypes and Functional Diversity. A Handbook of Transcription Factors (ed. Hughes T.R..) Subcellular Biochemistry 2011;52. doi: 10.1007/978-90-481-9069-0_5
32. Noyes MB, Christensen RG, Wakabayashi A, Stormo GD, Brodsky MH, Wolfe SA. Analysis of homeodomain specificities allows the family-wide prediction of preferred recognition sites. Cell 2008;133 : 1277–89. doi: 10.1016/j.cell.2008.05.023 18585360
33. Christensen RG, Enuameh MS, Noyes MB, Brodsky MH, Wolfe SA, Stormo GD. Recognition models to predict DNA-binding specificities of homeodomain proteins. Bioinformatics 2012;28: i84–9. doi: 10.1093/bioinformatics/bts202 22689783
34. Flandin P, Zhao Y, Vogt D, Jeong J, Long J, Potter G, et al. Lhx6 and Lhx8 coordinately induce neuronal expression of Shh that controls the generation of interneuron progenitors. Neuron 2011;70 : 939–50. doi: 10.1016/j.neuron.2011.04.020 21658586
35. Nokes EB, Van Der Linden AM, Winslow C, Mukhopadhyay S, Ma K, Sengupta P. Cis-regulatory mechanisms of gene expression in an olfactory neuron type in Caenorhabditis elegans. Dev Dyn 2009;238 : 3080–92. doi: 10.1002/dvdy.22147 19924784
36. Zhao Y, Marín O, Hermesz E, Powell A, Flames N, Palkovits M, et al. The LIM-homeobox gene Lhx8 is required for the development of many cholinergic neurons in the mouse forebrain. Proc Natl Acad Sci U S A 2003;100 : 9005–10. 12855770
37. Mori T, Yuxing Z, Takaki H, Takeuchi M, Iseki K, Hagino S, et al. The LIM homeobox gene, L3/Lhx8, is necessary for proper development of basal forebrain cholinergic neurons. Eur J Neurosci 2004;19 : 3129–41. 15217369
38. Zhu P, Li H, Jin G, Tian M, Tan X, Shi J, et al. LIM-homeobox gene Lhx8 promote the differentiation of hippocampal newborn neurons into cholinergic neurons in vitro. In Vitro Cell Dev Biol Anim 2013;49 : 103–7. doi: 10.1007/s11626-013-9582-8 23385486
39. Shi J, Li H, Jin G, Zhu P, Tian M, Qin J, et al. Lhx8 promote differentiation of hippocampal neural stem/progenitor cells into cholinergic neurons in vitro. In Vitro Cell Dev Biol Anim 2012;48 : 603–9. doi: 10.1007/s11626-012-9562-4 23150137
40. Li H, Jin G, Zhu P, Zou L, Shi J, Yi X, et al. Upregulation of Lhx8 increase VAChT expression and ACh release in neuronal cell line SHSY5Y. Neurosci Lett 2014;559 : 184–8. doi: 10.1016/j.neulet.2013.11.047 24316404
41. Lopes FM, Schröder R, da Frota ML Jr, Zanotto-Filho A, Müller CB, Pires AS, et al. Comparison between proliferative and neuron-like SH-SY5Y cells as an in vitro model for Parkinson disease studies. Brain Res 2010;1337 : 85–94. doi: 10.1016/j.brainres.2010.03.102 20380819
42. Biedler JL, Roffler-Tarlov S, Schachner M, Freedman LS. Multiple neurotransmitter synthesis by human neuroblastoma cell lines and clones. Cancer Res 1978;38 : 3751–7. 29704
43. Kovalevich J, Langford D. Considerations for the use of SH-SY5Y neuroblastoma cells in neurobiology. Methods Mol Biol 2013;1078 : 9–21. doi: 10.1007/978-1-62703-640-5_2 23975817
44. Danks K, Wade JA, Batten TF, Walker JH, Ball SG, Vaughan PF. Redistribution of F-actin and large dense-cored vesicles in the human neuroblastoma SH-SY5Y in response to secretagogues and protein kinase Calpha activation. Brain Res Mol Brain Res. 1999 Feb 5;64(2):236–45. 9931495
45. Hobert O. Regulatory logic of neuronal diversity: terminal selector genes and selector motifs. Proc Natl Acad Sci U S A 2008;105 : 20067–71. doi: 10.1073/pnas.0806070105 19104055
46. Wightman B, Ebert B, Carmean N, Weber K, Clever S. The C. elegans nuclear receptor gene fax-1 and homeobox gene unc-42 coordinate interneuron identity by regulating the expression of glutamate receptor subunits and other neuron-specific genes. Dev Biol 2005;287 : 74–85. 16183052
47. Palmer R, Inoue T, Sherwood DR, Jiang LI, Sternberg PW. Caenorhabditis elegans cog-1 locus encodes GTX/Nkx6.1 homeodomain proteins and regulates multiple aspects of reproductive system development. Dev Biol 2002;252 : 202–13. 12482710
48. Sarafi-Reinach TR, Melkman T, Hobert O, Sengupta P. The lin-11 LIM homeobox gene specifies olfactory and chemosensory neuron fates in C. elegans. Development 2001;128 : 3269–81. 11546744
49. Sarin S, Antonio C, Tursun B, Hobert O. The C. elegans Tailless/TLX transcription factor nhr-67 controls neuronal identity and left/right asymmetric fate diversification. Development 2009;136 : 2933–44. doi: 10.1242/dev.040204 19641012
50. Bargmann CI, Horvitz HR. Chemosensory neurons with overlapping functions direct chemotaxis to multiple chemicals in C. elegans. Neuron 1991;7 : 729–42. 1660283
51. Holland PW, Takahashi T. The evolution of homeobox genes: Implications for the study of brain development. Brain Res Bull 2005;66 : 484–90. 16144637
52. Hobert O, Westphal H. Functions of LIM-homeobox genes. Trends Genet 2000;16 : 75–83. 10652534
53. Srivastava M, Larroux C, Lu DR, Mohanty K, Chapman J, Degnan BM, et al. Early evolution of the LIM homeobox gene family. BMC Biol 2010;8 : 4. doi: 10.1186/1741-7007-8-4 20082688
54. Cho HH, Cargnin F, Kim Y, Lee B, Kwon RJ, Nam H, et al. Isl1 directly controls a cholinergic neuronal identity in the developing forebrain and spinal cord by forming cell type-specific complexes. PLoS Genet 2014;10: e1004280. doi: 10.1371/journal.pgen.1004280 24763339
55. Lopes R, Verhey van Wijk N, Neves G, Pachnis V. Transcription factor LIM homeobox 7 (Lhx7) maintains subtype identity of cholinergic interneurons in the mammalian striatum. Proc Natl Acad Sci U S A 2012;109 : 3119–24. doi: 10.1073/pnas.1109251109 22315402
56. Fragkouli A, van Wijk NV, Lopes R, Kessaris N, Pachnis V. LIM homeodomain transcription factor-dependent specification of bipotential MGE progenitors into cholinergic and GABAergic striatal interneurons. Development 2009;136 : 3841–51. doi: 10.1242/dev.038083 19855026
57. Vogt D, Hunt RF, Mandal S, Sandberg M, Silberberg SN, Nagasawa T, et al. Lhx6 directly regulates Arx and CXCR7 to determine cortical interneuron fate and laminar position. Neuron 2014;82 : 350–64. doi: 10.1016/j.neuron.2014.02.030 24742460
58. Chao MY, Komatsu H, Fukuto HS, Dionne HM, Hart AC. Feeding status and serotonin rapidly and reversibly modulate a Caenorhabditis elegans chemosensory circuit. Proc Natl Acad Sci U S A 2004;101 : 15512–7. 15492222
59. L’Etoile ND, Bargmann CI. Olfaction and odor discrimination are mediated by the C. elegans guanylyl cyclase ODR-1. Neuron 2000;25 : 575–86. 10774726
60. Hedgecock EM, White JG. Polyploid tissues in the nematode Caenorhabditis elegans. Dev Biol 1985;107 : 128–33. 2578115
Štítky
Genetika Reprodukční medicína
Článek Loss and Gain of Natural Killer Cell Receptor Function in an African Hunter-Gatherer PopulationČlánek Let-7 Represses Carcinogenesis and a Stem Cell Phenotype in the Intestine via Regulation of Hmga2Článek Binding of Multiple Rap1 Proteins Stimulates Chromosome Breakage Induction during DNA ReplicationČlánek SLIRP Regulates the Rate of Mitochondrial Protein Synthesis and Protects LRPPRC from DegradationČlánek Protein Composition of Infectious Spores Reveals Novel Sexual Development and Germination Factors inČlánek The Formin Diaphanous Regulates Myoblast Fusion through Actin Polymerization and Arp2/3 RegulationČlánek Runx1 Transcription Factor Is Required for Myoblasts Proliferation during Muscle Regeneration
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 8- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
- Intrauterinní inseminace a její úspěšnost
- Délka menstruačního cyklu jako marker ženské plodnosti
-
Všechny články tohoto čísla
- Putting the Brakes on Huntington Disease in a Mouse Experimental Model
- Identification of Driving Fusion Genes and Genomic Landscape of Medullary Thyroid Cancer
- Evidence for Retromutagenesis as a Mechanism for Adaptive Mutation in
- TSPO, a Mitochondrial Outer Membrane Protein, Controls Ethanol-Related Behaviors in
- Evidence for Lysosome Depletion and Impaired Autophagic Clearance in Hereditary Spastic Paraplegia Type SPG11
- Loss and Gain of Natural Killer Cell Receptor Function in an African Hunter-Gatherer Population
- Trans-Reactivation: A New Epigenetic Phenomenon Underlying Transcriptional Reactivation of Silenced Genes
- Early Developmental and Evolutionary Origins of Gene Body DNA Methylation Patterns in Mammalian Placentas
- Strong Selective Sweeps on the X Chromosome in the Human-Chimpanzee Ancestor Explain Its Low Divergence
- Dominance of Deleterious Alleles Controls the Response to a Population Bottleneck
- Transient 1a Induction Defines the Wound Epidermis during Zebrafish Fin Regeneration
- Systems Genetics Reveals the Functional Context of PCOS Loci and Identifies Genetic and Molecular Mechanisms of Disease Heterogeneity
- A Genome Scale Screen for Mutants with Delayed Exit from Mitosis: Ire1-Independent Induction of Autophagy Integrates ER Homeostasis into Mitotic Lifespan
- Non-synonymous FGD3 Variant as Positional Candidate for Disproportional Tall Stature Accounting for a Carcass Weight QTL () and Skeletal Dysplasia in Japanese Black Cattle
- The Relationship between Gene Network Structure and Expression Variation among Individuals and Species
- Calmodulin Methyltransferase Is Required for Growth, Muscle Strength, Somatosensory Development and Brain Function
- The Wnt Frizzled Receptor MOM-5 Regulates the UNC-5 Netrin Receptor through Small GTPase-Dependent Signaling to Determine the Polarity of Migrating Cells
- Nbs1 ChIP-Seq Identifies Off-Target DNA Double-Strand Breaks Induced by AID in Activated Splenic B Cells
- CCNYL1, but Not CCNY, Cooperates with CDK16 to Regulate Spermatogenesis in Mouse
- Evidence for a Common Origin of Blacksmiths and Cultivators in the Ethiopian Ari within the Last 4500 Years: Lessons for Clustering-Based Inference
- Of Fighting Flies, Mice, and Men: Are Some of the Molecular and Neuronal Mechanisms of Aggression Universal in the Animal Kingdom?
- Hypoxia and Temperature Regulated Morphogenesis in
- The Homeodomain Iroquois Proteins Control Cell Cycle Progression and Regulate the Size of Developmental Fields
- Evolution and Design Governing Signal Precision and Amplification in a Bacterial Chemosensory Pathway
- Rac1 Regulates Endometrial Secretory Function to Control Placental Development
- Let-7 Represses Carcinogenesis and a Stem Cell Phenotype in the Intestine via Regulation of Hmga2
- Functions as a Positive Regulator of Growth and Metabolism in
- The Nucleosome Acidic Patch Regulates the H2B K123 Monoubiquitylation Cascade and Transcription Elongation in
- Rhoptry Proteins ROP5 and ROP18 Are Major Murine Virulence Factors in Genetically Divergent South American Strains of
- Exon 7 Contributes to the Stable Localization of Xist RNA on the Inactive X-Chromosome
- Regulates Refractive Error and Myopia Development in Mice and Humans
- mTORC1 Prevents Preosteoblast Differentiation through the Notch Signaling Pathway
- Regulation of Gene Expression Patterns in Mosquito Reproduction
- Molecular Basis of Gene-Gene Interaction: Cyclic Cross-Regulation of Gene Expression and Post-GWAS Gene-Gene Interaction Involved in Atrial Fibrillation
- The Spalt Transcription Factors Generate the Transcriptional Landscape of the Wing Pouch Central Region
- Binding of Multiple Rap1 Proteins Stimulates Chromosome Breakage Induction during DNA Replication
- Functional Divergence in the Role of N-Linked Glycosylation in Smoothened Signaling
- YAP1 Exerts Its Transcriptional Control via TEAD-Mediated Activation of Enhancers
- Coordinated Evolution of Influenza A Surface Proteins
- The Evolutionary Potential of Phenotypic Mutations
- Genome-Wide Association and Trans-ethnic Meta-Analysis for Advanced Diabetic Kidney Disease: Family Investigation of Nephropathy and Diabetes (FIND)
- New Routes to Phylogeography: A Bayesian Structured Coalescent Approximation
- SLIRP Regulates the Rate of Mitochondrial Protein Synthesis and Protects LRPPRC from Degradation
- Satellite DNA Modulates Gene Expression in the Beetle after Heat Stress
- SHOEBOX Modulates Root Meristem Size in Rice through Dose-Dependent Effects of Gibberellins on Cell Elongation and Proliferation
- Reduced Crossover Interference and Increased ZMM-Independent Recombination in the Absence of Tel1/ATM
- Suppression of Somatic Expansion Delays the Onset of Pathophysiology in a Mouse Model of Huntington’s Disease
- Protein Composition of Infectious Spores Reveals Novel Sexual Development and Germination Factors in
- The Evolutionarily Conserved LIM Homeodomain Protein LIM-4/LHX6 Specifies the Terminal Identity of a Cholinergic and Peptidergic . Sensory/Inter/Motor Neuron-Type
- SmD1 Modulates the miRNA Pathway Independently of Its Pre-mRNA Splicing Function
- piRNAs Are Associated with Diverse Transgenerational Effects on Gene and Transposon Expression in a Hybrid Dysgenic Syndrome of .
- Retinoic Acid Signaling Regulates Differential Expression of the Tandemly-Duplicated Long Wavelength-Sensitive Cone Opsin Genes in Zebrafish
- The Formin Diaphanous Regulates Myoblast Fusion through Actin Polymerization and Arp2/3 Regulation
- Genome-Wide Analysis of PAPS1-Dependent Polyadenylation Identifies Novel Roles for Functionally Specialized Poly(A) Polymerases in
- Runx1 Transcription Factor Is Required for Myoblasts Proliferation during Muscle Regeneration
- Regulation of Mutagenic DNA Polymerase V Activation in Space and Time
- Variability of Gene Expression Identifies Transcriptional Regulators of Early Human Embryonic Development
- The Drosophila Gene Interacts Genetically with and Shows Female-Specific Effects of Divergence
- Functional Activation of the Flagellar Type III Secretion Export Apparatus
- Retrohoming of a Mobile Group II Intron in Human Cells Suggests How Eukaryotes Limit Group II Intron Proliferation
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Exon 7 Contributes to the Stable Localization of Xist RNA on the Inactive X-Chromosome
- YAP1 Exerts Its Transcriptional Control via TEAD-Mediated Activation of Enhancers
- SmD1 Modulates the miRNA Pathway Independently of Its Pre-mRNA Splicing Function
- Molecular Basis of Gene-Gene Interaction: Cyclic Cross-Regulation of Gene Expression and Post-GWAS Gene-Gene Interaction Involved in Atrial Fibrillation
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



