-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaSp6 and Sp8 Transcription Factors Control AER Formation and Dorsal-Ventral Patterning in Limb Development
In this report we examined the functional roles of Sp6 and Sp8 during limb development using compound loss-of-function mutants. Sp6 and Sp8, two members of the Sp gene family, are expressed in the limb bud ectoderm and function downstream of WNT/βcatenin signaling for Fgf8 induction. The analysis of the allelic series shows that the progressive reduction in the dose of Sp6 and Sp8 gene products leads to predictable morphology, from syndactyly, to split hand/foot malformation, oligodactyly, truncation and finally amelia, indicating that these two factors act in a complementary manner. The molecular characterization of the mutant limbs reveal that Sp6/Sp8 are required in a dose-dependent manner for Fgf8 and En1 induction, thereby placing them as an important link between the induction of the AER and the establishment of dorsal-ventral patterning during limb development.
Published in the journal: . PLoS Genet 10(8): e32767. doi:10.1371/journal.pgen.1004468
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004468Summary
In this report we examined the functional roles of Sp6 and Sp8 during limb development using compound loss-of-function mutants. Sp6 and Sp8, two members of the Sp gene family, are expressed in the limb bud ectoderm and function downstream of WNT/βcatenin signaling for Fgf8 induction. The analysis of the allelic series shows that the progressive reduction in the dose of Sp6 and Sp8 gene products leads to predictable morphology, from syndactyly, to split hand/foot malformation, oligodactyly, truncation and finally amelia, indicating that these two factors act in a complementary manner. The molecular characterization of the mutant limbs reveal that Sp6/Sp8 are required in a dose-dependent manner for Fgf8 and En1 induction, thereby placing them as an important link between the induction of the AER and the establishment of dorsal-ventral patterning during limb development.
Introduction
The apical ectodermal ridge (AER), a specialized thickened epithelium at the distal edge of the developing limb bud, is a major signaling center for limb development (for a review, see [1]). The AER, through the production of several members of the Fibroblast growth factor (Fgf) family, controls survival, proliferation and appropriate gene expression in the subjacent mesoderm [2]–[5].
The AER is formed through a complex and not completely understood process that starts with the induction of the AER precursor cells that are marked by their expression of Fgf8. In the mouse, these precursors are specified in the ventral ectoderm of the early limb bud to progressively compact at the tip of the bud to form the mature AER [6], [7]. The mature AER is a linear and regular band of polystratified (in mouse) and pseudostratified (in chick) epithelium rimming the distal dorsal-ventral boundary of the limb bud. Once the digit primordia have formed, the AER flattens and expression of Fgfs ceases, first over the interdigital spaces and later over the digit tips [8]. Cell lineage analysis has demonstrated that the AER is a transitory structure formed by a self-sustaining cell population that is exhausted before birth [9].
Initially, the expression of Fgf10 in the presumptive limb mesoderm activates the expression of Fgf8 in the overlying ectoderm through the induction of Wnt3a [10]–[13]. An ectodermally active Wnt/βcatenin pathway is required throughout limb development, first for AER induction and then for AER maintenance [14], [15]. The genetic removal of βcatenin from the limb ectoderm, before the initiation of Fgf8 expression, completely prevents limb development while its removal after Fgf8 expression leads to variable truncations [14], [15]. Another essential pathway involved in AER induction and maintenance is the Bone morphogenetic protein (Bmp) signaling pathway. Similar to Wnt/βcatenin signaling, Bmp signaling is required for AER induction, but paradoxically and in stark contrast to Wnt/βcatenin signaling, it exerts a negative influence on AER maintenance. Thus, when Bmp signaling is abolished from the limb ectoderm prior to AER induction, Fgf8 is never activated and the AER does not form resulting in amelic phenotypes [16], [17]. However, when Bmp signaling is removed from the limb ectoderm after Fgf8 and AER induction, the expression of Fgf8 is prolonged in the AER leading to syndactyly [16]. Bmp signaling has been proposed to act both upstream and downstream of Wnt/βcatenin signaling and, despite intensive study, the interactions between these pathways in the induction and maintenance of the AER remains only partially understood [14], [15].
Very interesting is the connection between the AER and the establishment of dorsal-ventral (DV) patterning [18]. During normal development the position of the mature AER always coincides with the DV boundary. However, it is well known that a normal functional AER can form in the absence of normal DV patterning. For instance, in the eudiplopodia chick mutant an extra AER appears within the dorsal ectoderm leading to extra double dorsal limb outgrowth [19]. Also, in the double Wnt7a;En1 (Engrailed1) mutant a virtually normal AER forms despite disturbed DV patterning [20], [21]. It has been suggested that the coordination between the position of the AER and the DV boundary depends on BMP signaling because, besides its above mentioned role on AER induction and maintenance, it also regulates DV patterning through the induction of En1, which in turn restricts Wnt7a to the dorsal ectoderm [17], [21]–[23].
Sp6 and Sp8, also known as epiprofin and buttonhead, respectively, are two members of the Sp transcription factor family that have been implicated in AER induction and maintenance [24]–[27]. Both share similar patterns of expression in the limb bud ectoderm and AER and function downstream of Wnt/βcatenin signaling and upstream of Fgf8. Based on their overlapping patterns of expression and on their individual loss-of function phenotypes, we suspected that these two factors act in a complementary manner in the induction and maintenance of the AER downstream of Wnt/βcatenin [26]. Therefore, in order to further elucidate the functions and potential redundancy of these two genes, we generated double Sp6;Sp8 null mutants. We also generated Sp6-null;Sp8-conditional mutants using an Sp8 floxed allele with both the AP2αCre and the Msx2Cre deleter lines. Interestingly, mutant embryos that lacked the four Sp6;Sp8 alleles or that retained a single Sp6 allele were tetra-amelic. Initial budding occurred, but Fgf8 was not activated in the limb ectoderm preventing further development. Mutants bearing a single functional copy of Sp8 displayed a split-hand/foot malformation phenotype (SHFM) with dorsalization of the digital tips. The phenotypic data together with the molecular defects identified in mutant limb buds indicate that Sp6 and Sp8 are together absolutely necessary for AER development and DV patterning.
Results
Both Sp6 and Sp8 are expressed in the entire prospective limb ectoderm and progressively become confined to the AER as the limb bud emerges. Loss of function of Sp6 [26], [28] results in soft tissue syndactyly in the forelimb and osseous syndactyly to complete phalangeal synostosis in the hindlimb, whereas the inactivation of Sp8 [24], [25] results in variable limb truncations most frequently at the level of the elbow/knee. Both mutations show a deficit in the maturation of the AER. Also, dorsalization of the ventral digit tips is a characteristic feature of Sp6 mutants and the molecular analysis of Sp8 mutants indicates that early limb buds become progressively dorsalized [24]–[26]. Although the individual inactivation of either Sp6 or Sp8 does not interfere with the initial activation of Fgf8 in the AER, several studies have demonstrated that both factors function downstream of Wnt/βcatenin signaling and that Sp8 is able to bind and activate the Fgf8 promoter [26], [27], [29], [30]. This, together with their similar expression patterns in the limb ectoderm, led us to propose that Sp6 and Sp8 transcription factors have a redundant function in the Wnt/βcatenin dependent induction of Fgf8 in the AER [26].
We have previously shown that Sp8 expression is maintained in the absence of Sp6 [26] and here we found that Sp6 is expressed in the absence of Sp8 although at a lower level than normal, and is progressively downregulated in concert with the downregulation of Fgf8 expression (Figure S1 and [24], [25]). Thus, Sp6 may directly or indirectly require Sp8 to maintain a normal level of expression. Nevertheless, the expression of Sp6 even at a reduced level in the Sp8 mutant could account for the induction and partial maintenance of Fgf8 supporting our hypothesis that both factors function in a redundant manner during limb development.
Limb phenotype of double Sp6;Sp8 mutants
To test our hypothesis we analyzed limb development in double Sp6;Sp8 mutants (Figure 1). For this genetic approach, we used the Sp6 (Sp6−; [28]) and the Sp8 (Sp8CreERT2, hereafter referred to as Sp8−; [24]) null alleles and we analyzed the progeny from crosses between Sp6+/−;Sp8+/− double heterozygous mice (Figure 1). Sp6+/−;Sp8+/− double heterozygous mice showed no obvious defect in either limb patterning or skeletogenesis, yet displayed subnormal fertility. Skeletal preparations of the neonates recovered from these crosses were used to characterize the limb phenotype; other phenotypic traits will be considered elsewhere.
Fig. 1. Effects of inactivating Sp6 and Sp8 in limb development. 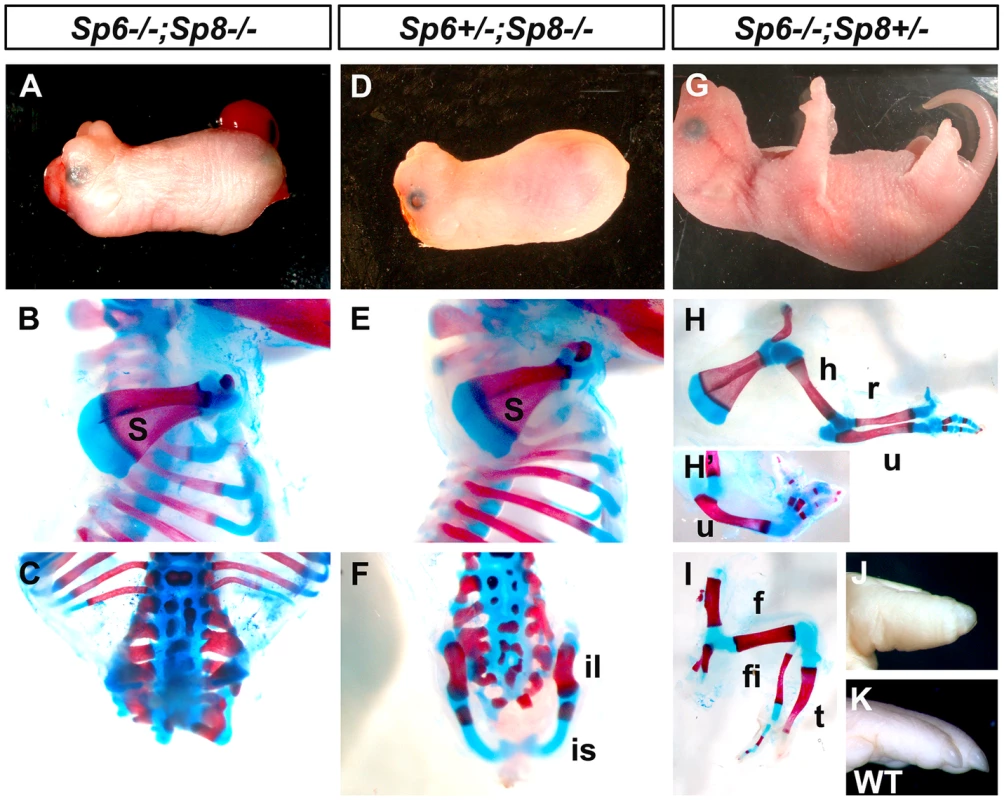
The external aspect (top row) and skeletal preparations of the forelimb (middle row) and hindlimb (bottom row) of newborns are shown for each genotype (genotypes indicated at the top). In the absence of the four functional alleles of Sp6 and Sp8 (A–C), or when only one functional allele of Sp6 remains (D–F) no limbs form. Underdeveloped hip bones with rudimentary ilium and ischium form when one functional allele of Sp6 is present (F). Animals with only one functional allele of Sp8 (G–I) display a split hand/foot malformation phenotype with occasional absence of the radius (H′) and more severe phenotype in the hindlimb (I). The digit tips in these limbs show conical nails (J), compare with normal digits (K). Abbreviations: s, scapula; h, humerus; r, radius; u, ulna, f, femur, t, tibia, fi, fibula, is: ischium il, ilium. Animals singly mutant for Sp6 or for Sp8 exhibited their previously described phenotypes including exencephaly and spina bifida in Sp8 mutants [24]–[26]. In our crosses, the majority of Sp8 mutant limbs were truncated at the level of the elbow/knee with the olecranon also present in half of the specimens. Remarkably, in 100% of newborn double mutants both forelimbs and hindlimbs were absent (Figure 1A–C; 3 out of 102). In these mutants, no skeletal elements formed distal to the scapula (Figure 1B). Caudal lumbar vertebrae were highly disorganized and the body appeared truncated caudal to the sacrum with only rudimentary cartilage contributing to the pelvis (Figure 1C and Figure S2). Also, animals in which both copies of the Sp8 gene and one copy of the Sp6 gene had been removed (Sp6+/−;Sp8−/−) were always tetra-amelic (Figure 1D–F; 10 out of 102). However, in contrast to double mutants, the pelvic girdles showed undeveloped iliac and ischial anlagen (Figure 1F and Figure S2). The effect of a single functional copy of Sp6 in the morphogenesis of the pelvic girdle is shown in detail in Figure 2.
Fig. 2. RT-qPCR quantification of Sp6 and Sp8 transcripts in the limb ectoderm of E10.5 control embryos. 
Histogram bars represent the average expression values after normalization to the ubiquitously expressed 18s-RNA (standard deviation shown as error bars). Sp8 (red) exhibits a higher level of expression than Sp6 (blue) both in forelimbs (FL) and in hindlimbs (HL) and both factors are expressed at higher level in the forelimb than in the hindlimb. Mutant mice in which both copies of the Sp6 gene and one copy of the Sp8 gene had been inactivated (Sp6−/−;Sp8+/−) had proximal-distal (PD) complete, but extremely malformed, limbs (Figure 1G–I, 11 out of 102). Consistently, the forelimb paw had the “claw-like” appearance typical of split-hand/foot malformation (SHFM) in which anterior digits were hypoplastic or missing and posterior digits were frequently fused (Figure 1G–H–H′). The radius was occasionally absent (Figure 1H′). Hindlimbs showed a more severe phenotype with the zeugopod constantly abnormal (Figure 1I). Although there was some variability, the majority of specimens displayed a misshaped and frequently truncated tibia and a thin fibula surmounted by one or two rows of small skeletal rods that we interpreted as digits (Figure 1I). The phenotype was variable among different animals and within individuals each paw showing specific deficiencies. No left or right severity preference was identified. This phenotype is comparable to the human SHFM, a highly variable malformation that has also been termed ectrodactyly, split hand, cleft hand and lobster claw hand [31]–[34]. Of most interest, the digits in both fore and hindlimbs of Sp6−/−;Sp8+/− mutants were bidorsal exhibiting circumferential nails and lacking ventral pads (Figure 1J–K).
In summary, our genetic analysis shows that Sp6/Sp8 transcription factors are together absolutely required for limb development. Furthermore, the data support our hypothesis that Sp6 and Sp8 perform complementary functions in the limb ectoderm. Interestingly, one single functional allele of Sp6 is insufficient, in the absence of an Sp8 allele, to support limb development. In contrast, one single functional allele of Sp8, in the absence of an Sp6 allele, permits development of all three PD segments, although displaying a SHFM phenotype.
Sp8 is expressed at higher level than Sp6 in the limb ectoderm
Since Sp6 and Sp8 display similar temporal and spatial patterns of expression in the limb ectoderm [24]–[26], one possible explanation for the difference in their functional capacity as described above is that Sp8 has specific functions that Sp6 cannot accomplish. However, it is also possible that these functional differences are due to differences in their levels of expression. Thus, to quantify the Sp8 and Sp6 levels of expression in the limb ectoderm we performed a quantitative RT-PCR assay in E10.5 control embryos. Our results showed that Sp8 was expressed more robustly than Sp6 during limb development (Figure 2). Expression of Sp8 was 3 fold higher than expression of Sp6 in the forelimb and 5 fold higher in the hindlimb. Our quantitative analysis also showed that the expression of both Sp6 and Sp8 was higher in the forelimb than in the hindlimb, although it should be noted that the development of the hindlimb is delayed compared to that of the forelimb at this stage, which could account for the forelimb/hindlimb disparity.
To investigate the basis for the differential level of expression of Sp6 and Sp8 in the limb, we performed an in silico analysis of their putative promoter regions (Figure S3). To enhance the identification of functionally relevant regulatory sequences, we limited our evaluation to regions 5′ of the coding sequences that were conserved across divergent species as determined by the mVista browser [35]. We further screened the conserved regions for potential transcription factor binding sites using Alibaba 2.1 and Sequencher 4.8 and then confirmed conservation between mouse and human. Our analysis identified 12 potential βcatenin/Lef1 binding sites 5′ to the Sp8 coding sequence, whereas Sp6 had only five. This finding provides a potential mechanism for the increased level of Sp8 transcription during limb development. In addition, the presence of 29 potential Sp binding sites in the region containing the putative Sp6 promoter and the 12 present in Sp8 supports a possible cross-regulation between Sp transcription factors as suggested by the lower Sp6 expression in absence of Sp8 (Figure S1). Based on our quantitative and in silico analysis we speculate that Sp8 makes a more substantial contribution to limb development than Sp6 because of a higher level of transcription, a speculation that requires further investigation.
Ap2αCre inactivation of Sp8 on an Sp6 deficient background
When performing the crosses between double heterozygous we found a reduced frequency of pregnancies in double heterozygous females and also that the fraction of double mutant offspring was significantly below the expected 1/16 Mendelian frequency. To circumvent these issues and avoid the neural phenotype from Sp8 null mutants, we used an Sp8 floxed conditional allele (Sp8f; [36]) to remove it specifically from the limb ectoderm. Among the available lines with Cre activity in the limb ectoderm (Msx2Cre [37]; Brn4Cre [17]; RARCre [38]; AP2αCre [39]; Mox2Cre [40]), we selected the AP2αCre line because it has been reported to drive very early Cre function in both fore and hindlimbs, at least before activation of Fgf8 [41]. Because Sp8 is already expressed at E7.5 in the embryonic ectoderm ([24], [25] and authors' personal observations), we decided to determine in more detail the activity of the AP2α;Cre transgenic line using the ROSA26 reporter strain (R26R; [42]). Our analysis showed AP2α;Cre activity in the early embryonic ectoderm at E8.5 indicating that the removal of the Sp8 floxed allele would occur before limb initiation (Figure S4).
Thus, we used the AP2α;Cre line, the conditional allele of Sp8 and the Sp6 null allele to generate the combined loss of function of Sp6 and Sp8 in the limb ectoderm (Figure S5). As to be expected, the double mutants (Sp6−/−;Sp8f/−;AP2αCre) and the mutants that retained a single allele of Sp6 (Sp6+/−;Sp8f/−;AP2αCre) were 100% tetra-amelic and showed similar phenotypes to those described above for the double ubiquitous deletions (Figure S5A–F). Also as expected, the Sp6−/−;Sp8f/+;AP2αCre genotype exhibited the SHFM phenotype with its typical variability (compare Figure 1H–I with Figure S5H–I). In sum, the limb phenotypes obtained using the Sp8 floxed allele and the AP2αCre line replicated exactly the phenotypes obtained with the constitutive deletions. Finally, it should be noted that the neural phenotype was not rescued in conditional mutants (Figure S5A, 5D) indicating an unanticipated wide overlap between the expression of AP2α and Sp8 in the neural tube (Figure S4).
Msx2Cre inactivation of Sp8 on an Sp6 deficient background
We also inactivated Sp8 from the limb ectoderm using the Msx2;Cre line simultaneously with the inactivation of Sp6. This Msx2;Cre transgenic line has been extensively monitored using the ROSA26 reporter strain [15], [37] and it is known that it drives Cre activity before Fgf8 activation of expression in the hindlimb but after Fgf8 expression and initiation of limb development in the forelimb. We reasoned that the use of this conditional mutant would provide information on the requirement of Sp8 once the early stages of limb initiation have occurred.
First of all we compared the phenotype of the limb conditional Sp8 mutant (Sp8f/−;Msx2Cre) with that of the Sp8 null mutant (Sp8−/−) in both forelimbs and hindlimbs (Figure 3A–F). Notwithstanding the variability, the phenotypes using the conditional allele were on average milder than those using the constitutive null allele [24], [25], [30] (Figure 3A–F). In the conditional Sp8f/−;Msx2Cre mutant, an initial burst of Sp8 expression permitted normal forelimb development up to the wrist and furthermore one or two incomplete posterior digits were formed (Figure 3E). In the hindlimbs, one posterior digit was always present although the tibia frequently appeared truncated (Figure 3F). This improvement in the phenotype (compare Figure 3A–C with Figure 3D–F) indicates that a transient early expression of Sp8 has a considerable impact on both fore and hind limb development.
Fig. 3. Msx2Cre removal of Sp8 on a Sp6 deficient background. 
The external aspect (top row) and skeletal preparations of the forelimb (middle row) and hindlimb (bottom row) of newborns are shown for each genotype (genotypes indicated at the top). Msx2Cre conditional removal allows transient expression of Sp8 in both forelimbs and hindlimbs which results in Sp8 conditional mutant (D–F) displaying a milder limb phenotype than ubiquitous mutants (A–C). One single conditional allele of Sp8 in the forelimb (G–H) seems to be equivalent to both functional alleles of Sp6 (A–B) while in the hindlimb is not sufficient for limb development (C, I). This conditional allele of Sp8 in addition to one single allele of Sp6 permits the formation of the three PD segments of the limb although with a single digit (J–L). Finally, one conditional allele of Sp8 plus a normal allele Sp8 results in SHFM (M–O). The conditional removal of Sp8 in the absence of Sp6 (Sp6−/−;Sp8f/−;Msx2Cre) resulted in a forelimb truncated at the elbow while the hindlimbs didn't develop (Figure 3G–I). When one copy of Sp6 remained (Sp6+/−;Sp8f/−;Msx2Cre) the phenotype notably improved with truncations at the level of the wrist/ankle associated with the formation of an incomplete digit (Figure 3K–L). Finally, when a functional copy of Sp8 remained besides the Sp8 floxed allele (Sp6−/−;Sp8+/f;Msx2Cre) the phenotype obtained was SHFM (Figure 3M–O).
In summary, when the phenotypes of our allelic series are classified according to severity, a clear correlation with the total dosage of Sp6 and Sp8 is observed (Figure 1, 3 and Figure S5). The more parsimonious explanation is that both transcription factors are functionally equivalent during limb development, although Sp8 makes a greater contribution than Sp6 presumably due to a higher level of expression (Figure 2). Our study also suggests that there is a threshold of expression below which no limb forms and that the level of Sp6 expression attained by a single allele of Sp6 is below this threshold.
A functional AER does not develop when the gene dosage of Sp6 and Sp8 is significantly reduced or completely eliminated
Since both Sp6 and Sp8 are involved in the Wnt/βcatenin dependent induction of Fgf8, it seems reasonable to presume that the amelic phenotype of double mutants may rely on a failure to induce a functional AER. Therefore we examined embryonic limbs at the stages when the limb bud is emerging and the AER is being induced. For this analysis we used the Sp6 and Sp8 constitutive null alleles. By E9.5, in the normal limb bud, several genes including Fgf8, Bmp4 and Msx2 are expressed in the ventral limb ectoderm forming the preAER [17], [22], [43]. These AER precursors will become progressively confined to the distal tip as the AER matures [7], [20] (Figure 4A, C, G, I, K, O).
Fig. 4. Fgf8 is not detected in double Sp6;Sp8 mutants. 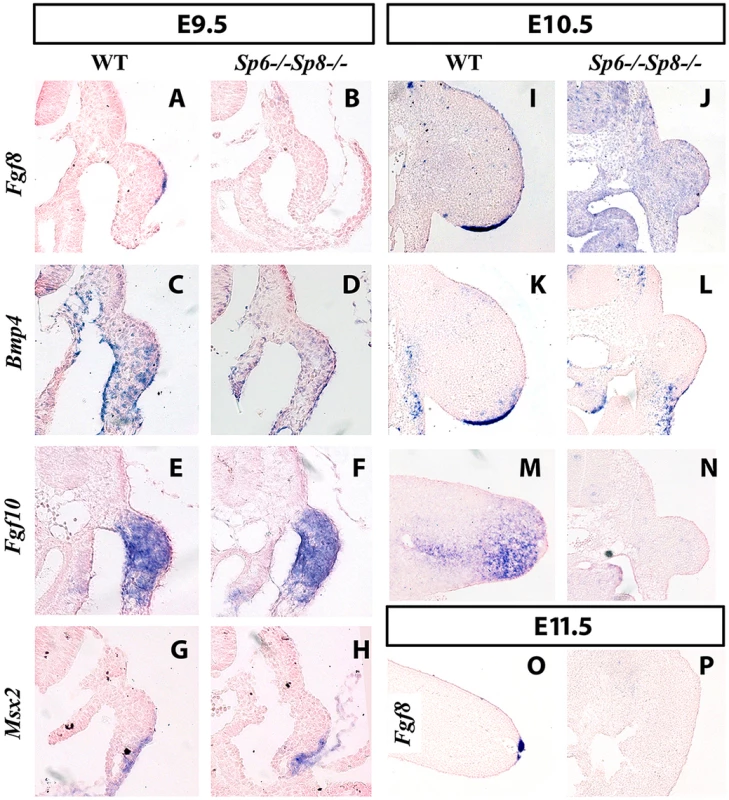
ISH to transverse sections through the level of the forelimbs at the stage indicated at the top and with the probe indicated on the left. Genotypes are also marked at the top of the figure. In the absence of Sp6 and Sp8, Fgf8 expression in the limb ectoderm is never detected as shown at E9.5 (A–B), E10.5 (I–J) and E11.5 (O–P). However, Bmp4 (C–D), Fgf10 (E–F) and Msx2 (G–H) are normally activated at E9.5 but not maintained at later stages (K–N). Note that the initial budding of the double mutant is similar to normal (A–H) but further growth is impaired (I–N) and complete regression has occurred by E11.5 (O–P). In all panels dorsal is up and distal to the right. However, in the absence of the four Sp6;Sp8 alleles (Sp6−/−;Sp8−/−) or when only one functional allele of Sp6 remained (Sp6+/−; Sp8−/−), Fgf8 was never detected in the limb ectoderm at any of the stages analyzed (Figure 4B for E9.5 (25–30 somites); Figure 4J for E10.5 (36–40 somites) and Figure 4P for E11.5). Because these two genotypes always showed identical expression patterns for all the genes analyzed, only the results of Sp6−/−;Sp8−/− mutants are shown in the Figures. In contrast to Fgf8, Bmp4 and Bmp2 expression was found to occur normally at E9.5 both in the limb ectoderm and limb mesoderm of double mutants and mutants with a single functional allele of Sp6 (Figure 4C–D). This was confirmed by the expression of Msx2, a bona fide target of Bmp signaling [44], [45] (Figure 4G–H). However, neither Bmp4 (Figure 4K–L) nor Msx2 were maintained in the limb ectoderm by E10.5. Disregarding the absence of Fgf8 expression, initiation of limb development was normal in Sp6−/−;Sp8−/− and Sp6+/−;Sp8−/− compound mutants with the formation of a small bulge; thus by E9.5 the phenotype was not yet evident (Figure 4A–H).
The current view considers that Fgf10 signaling from the limb mesoderm induces Wnt/βcatenin signaling in the ectoderm and this leads to Fgf8 activation and therefore AER induction in the ectoderm. Subsequently, Fgf8 from the ectoderm signals back to the mesoderm to maintain Fgf10, establishing an Fgf10-Fgf8 positive feedback loop necessary for further outgrowth [11]. Consistent with Sp6 and Sp8 acting downstream of Fgf10 and Wnt/βcatenin signaling, double mutant limb buds normally activated Fgf10 expression in the limb mesenchyme (Figure 4E–F at E9.5). However, due to the failure to activate Fgf8, the emergent limb buds cannot maintain Fgf10 in the limb mesoderm (Figure 4M–N) and regress so that by E11.5 no trace of the limb bud remained (Figure 4O–P).
These results demonstrate the absolute requirement of Sp6/Sp8 for Fgf8 activation in the limb ectoderm and are consistent with Sp6/Sp8 being necessary mediators of Wnt/βcatenin induction of Fgf8. Finally, our results also show that Bmp4 expression in the limb ectoderm, which requires βcatenin [14], [15], can occur in the total absence of Sp6 and Sp8 (this work, see Figure 4C–D) as well as in the absence of significant AER-related Fgf expression [46].
In the absence or significant reduction of Sp6/Sp8, limb development initiates, but later regresses by apoptosis
Next we investigated the reason of the regression of the emerging limb bud in double mutants. The phenotype of the double mutants is reminiscent of the chick mutant limbless. In limbless the limb bud arises normally, but due to the inability to form an AER, the entire bud undergoes cell death and disappears [47], [48]. Also, cell death is a constant feature after the surgical removal of the AER [3], [4] or genetic attenuation of Fgf signaling from the AER [38], [46], [49], [50]. Therefore, we analyzed cell death by TUNEL in our double mutant limb buds.
Abnormal cell death, compared with control littermates, was not detected at E9.5 in Sp6−/−;Sp8−/− and Sp6+/−;Sp8−/− compound mutants. However, extensive apoptosis was apparent both in the mesoderm and ectoderm of these mutant limb buds by E10.5 (Figure 5A–B). Cell death started and was most prominent in the central region of the bud but apoptotic cells were also observed in the ectoderm particularly at dorsal proximal and ventral level (Figure 5B). This extensive apoptosis can account for the regression of the limb bud and the amelic phenotype as in limbless [47], [48].
Fig. 5. Effects of inactivating Sp6 and Sp8 genes on cell survival and AER morphogenesis. 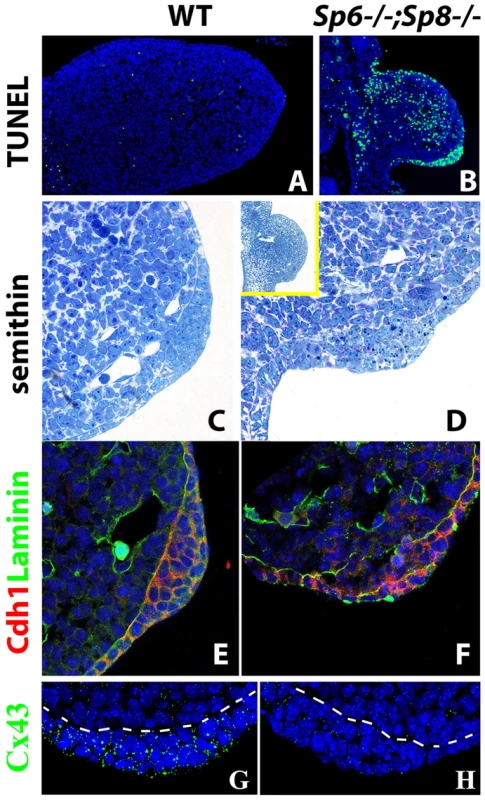
(A–B) TUNEL assay showing abundant apoptotic cells (green) both in the mesoderm and ectoderm of the E10.5 double mutant forelimb bud (B) compared to control (A). (C–D) semithin longitudinal section of araldite embedded control and double mutant limb buds showing the thickening in the ventral ectoderm of mutants. The insert in D shows a lower magnification to appreciate the ventral position of the ectoderm thickening in mutants. (E–F) Confocal images of double immunohistochemistry for Laminin-b, marking the basement membrane (green) and E-cadherin expressed specifically in the ectoderm (red) showing that the cells accumulated in the ventral limb ectoderm of mutant embryos are of ectodermal origin. (G–H) Confocal images of Connexin 43 immunostaining showing the enrichment of gap junctions in the control AER (green dots) but not in the double mutant AER. All the panels show forelimb buds at E10.5. In the immunostainings, the nuclei are counter stained with DAPI (blue). AER morphogenesis initiates even in the complete absence or significant reduction of Sp6 and Sp8
In the histological sections of double mutant limb buds we noticed a thickening of the ventral ectoderm that was particularly evident in the TUNEL assays because of the abundant cell death in this region (Figure 5A–B). To analyze this thickening with maximum detail, we performed semithin sections (1 micron thick) of araldite embedded embryos. Transverse sections through double mutant (Sp6−/−;Sp8−/−) and Sp6+/−;Sp8−/− embryos at the level of the forelimbs showed an irregular thickening of the ventral ectoderm by E10.5 (Figure 5C–D). The thickening didn't span the whole ventral ectoderm but was patchy and sometimes protruded into the mesoderm; it had the appearance of a ventrally positioned and immature AER, in which the apoptotic images were very abundant. To confirm that this thickening was of ectoderm origin, we used immunohistochemistry and confocal microscopy to localize E-Cadherin (Cdh1), which is an epithelial marker, and laminin, a major component of the basement membrane. The double immunohistochemistry demonstrated that the thickening was ectodermal as it expressed Cdh1 and was underlined by a laminin marked basement membrane (Figure 5E–F). To assess the functionality of this thickened ectoderm, we analyzed the expression of Connexin 43 (Cx43), a gap junction protein encoded by the Gja1 gene and considered a marker of the specialized AER ectoderm [51]. In contrast to the high expression present in the wild type AER, Cx43 was not detected above background in the thickened ectoderm of double mutants (Figure 5G–H).
Taken together our results indicate that Sp6/Sp8 factors are absolutely required for a functional AER, but dispensable for initial AER morphology confirming an independence between AER morphology and function.
Absence or significant reduction of Sp6/Sp8 activity in the limb ectoderm disrupts DV patterning
The known relationship between the specification of the AER and DV patterning together with the DV phenotypic alterations present in Sp6−/− and Sp6−/−;Sp8+/− mutants prompted us to analyze the state of DV patterning in our mutants. Furthermore, the ventral position of the mutant AER indicates a failure in the normal morphogenetic movements of the ectoderm that compact the AER, a process in which En1 has been implicated [6], [20]. Thus, we analyzed the expression of two genes relevant to DV patterning, Wnt7a and En1, in consecutive serial limb bud sections.
In the emerging limb bud (E9.25; 22–23 So), before En1 expression is detectable, Wnt7a is normally expressed in the dorsal ectoderm exceeding the mid-distal point of the bud and extending into the ventral ectoderm [7], [23] (author's personal observations). Shortly afterwards, the expression of the pre-AER markers Fgf8 and Bmp4 in the ventral ectoderm and of En1 in the more proximal ventral ectoderm progressively restricts Wnt7a to the dorsal ectoderm (Figure 6A, C).
Fig. 6. Effects of inactivating Sp6 and Sp8 genes on dorsal-ventral limb patterning. 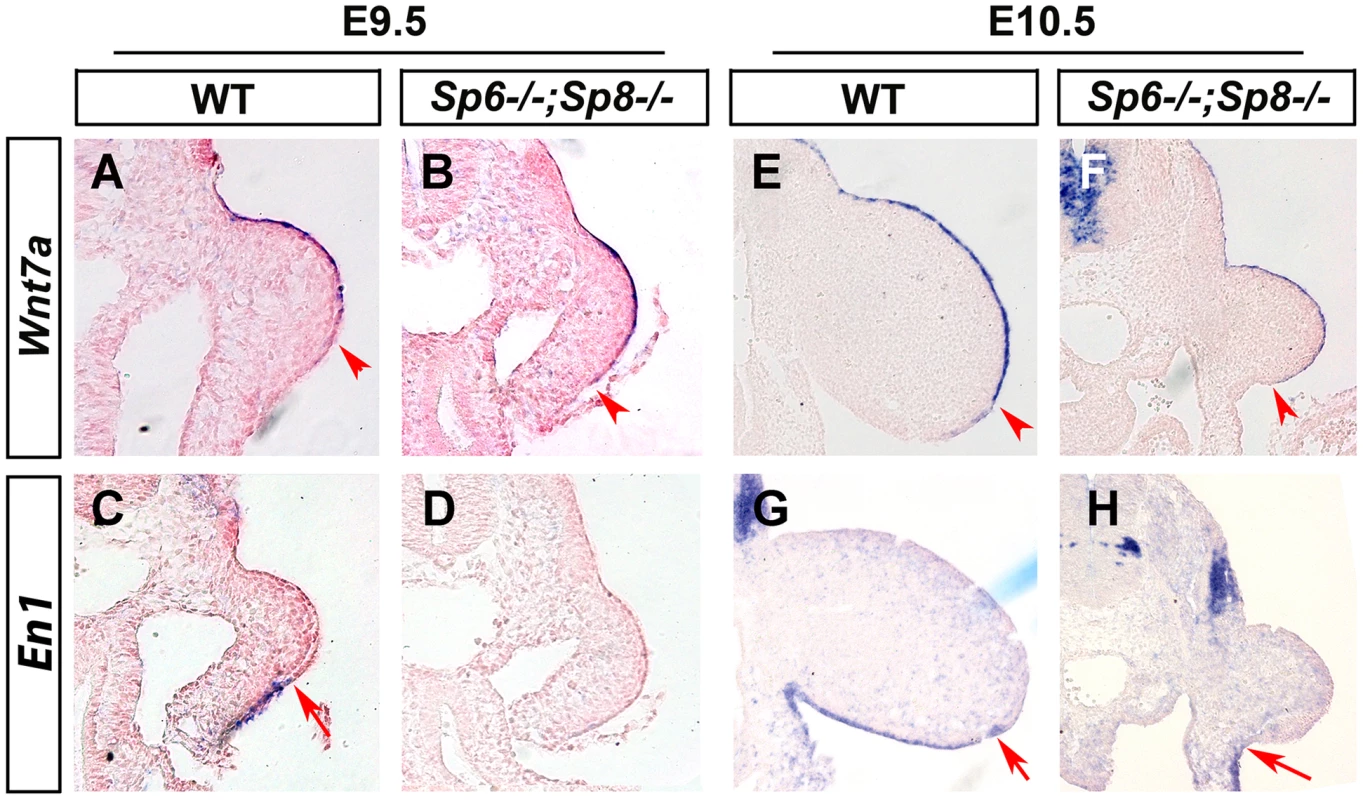
ISH to transverse sections through the level of the forelimbs at the stage indicated at the top and with the probe indicated on the left. Genotypes are also marked at the top. Note that, contrary to controls (A, E), Wnt7a is not restricted to the dorsal ectoderm in double mutant embryos (B, F). Accordingly, En1 expression is undetectable in the ventral limb ectoderm of mutant embryos (D, H) compared to controls (C, G). The arrowheads and arrows mark the distal limit of Wnt7a and En1 expression, respectively. In double mutant Sp6−/−; Sp8−/− and Sp6+/−;Sp8−/− embryos, the initial extended expression of Wnt7a was never restricted to the dorsal ectoderm and its expression persisted covering almost the entire limb ectoderm while En1 expression was not detected in the ventral ectoderm (Figure 6B–D, F–H). These results reveal that the absence or significant reduction of Sp6/Sp8 dosage interferes with the normal specification of DV patterning resulting in double dorsal distal limb buds. Our results also show that a virtually normal Bmp signaling in the early limb bud (Figure 4C–D and Figure 4G–H) is not sufficient for En1 expression in the absence of Sp6 and Sp8.
Mutants retaining a single functional allele of Sp8 exhibit a split-hand/foot malformation
The presence of a single allele of Sp8 (Sp6−/−;Sp8+/− or Sp6−/−;Sp8f/+;AP2αCre or Sp6−/−;Sp8f/+;Msx2Cre), was sufficient to allow the elaboration of all three segments along the PD axis, although the autopod was characterized by the loss or malformation of central elements creating a SHFM.
To understand the molecular basis of this phenotype, we analyzed the expression of Fgf8 during limb development in Sp6−/−;Sp8+/− mutants. This analysis showed that the AER precursors were irregularly specified in the ventral ectoderm. The whole mount in situ hybridization at E10 showed obvious gaps and irregularities in the area in which Fgf8 should be uniformly expressed (Figure 7A). During further development, the expression of Fgf8 became robust in the posterior AER, but was absent in the central-anterior areas except for a typical spot of residual anterior expression (Figure 7B–C). The expression of Bmp4 in the ectoderm always replicated the same abnormal pattern as Fgf8 (Figure 7A–C). Furthermore, the compaction and maturation of the AER was defective as it remained flat and broad with occasional extensions into the ventral ectoderm (arrow in Figure 7C and 7E′). Thus, in harmony with previous reports [32], [52], [53], the SHFM phenotype in our Sp6−/−;Sp8+/− mutants derives from a failure to properly establish and maintain the AER, preferentially in the central to anterior limb region.
Fig. 7. Molecular and morphological analysis of Sp6−/−;Sp8+/− mutant limb buds. 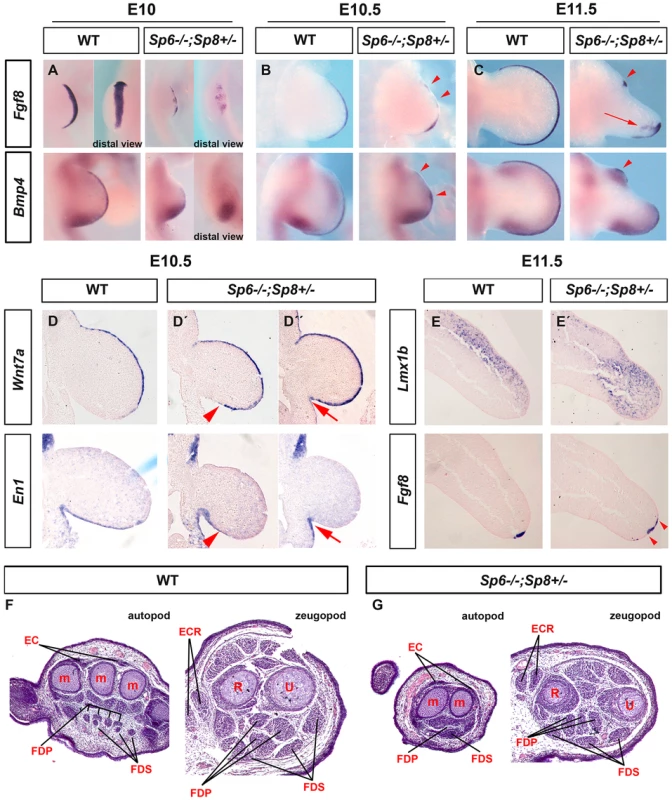
(A–C) WMISH for Fgf8 and Bmp4 showing irregular activation in the limb bud ectoderm of Sp6−/−;Sp8+/− E10 (A), E10.5 (B) and E11.5 (C) forelimb buds compared to control littermates. Note the irregular early activation and predominant posterior maintenance of Fgf8 and Bmp4 expression, except for a residual focus of anterior expression (red arrowheads). (D, D′, D″) ISH for Wnt7a and En1 to consecutive (7 microns apart) sections of control and mutant E10.5 forelimb buds (D, D′ and D″). Note the variable expansion of Wnt7a into the ventral ectoderm always associated with a corresponding proximal restriction of En1 (D′, D″) indicated by red arrowheads (D′) and red arrows (D″). (E–E′) ISH for Lmx1b and Fgf8 in consecutive sections of control and Sp6−/−;Sp8+/− E11.5 forelimb buds. The Lmx1b expression invades the ventral mesoderm distally under the broad and flat AER. (F–G) Hematoxylin-Eosin stained transverse histological sections at the autopod and zeugopod level of E15.5 control (F) and Sp6−/−;Sp8+/− (G) limbs. Some of the individual muscles and tendons are labeled. Abbreviations: EC, extensor digitorium communis; FDS, flexor digitorium sublimis; FDP, Flexor digitorium Profundus; ECR, extensor carpi radiallis; m, metacarpal; R, radius; U, ulna. In Sp6−/−;Sp8+/− mutants the expression of Wnt7a and En1 was consistently abnormal but highly variable even within a single limb bud. Wnt7a was always found to abnormally extend into the ventral ectoderm to a variable degree that always correlated with a complementary ventral expression of En1 (Figure 7D, D′, D″). This was easily appreciated when consecutive sections of the same limb bud were hybridized for Wnt7a and En1 as shown in Figure 7D′ and 7D″. Accordingly, the expression of Lmx1b, the downstream target of Wnt7a responsible for the dorsalization of the dorsal mesoderm [54], was found to variably extend under the flattened and broad AER into the ventral mesoderm distally (Fig. 7E–E′). These molecular alterations explain the bidorsal tips of Sp6−/−;Sp8+/− mutants. To ascertain possible DV defects at more proximal levels we performed a histological analysis on transversal sections of E15.5 mutant and control limbs. Our results showed that DV patterning of muscles and tendons were preserved for the most part in the stylopod and zeugopod, but were less well defined in the autopod (Figure 7F–G).
In humans, isolated or non-syndromic SHFM is a genetically heterogeneous developmental disorder of which six loci have been identified [31]–[34]. SHFM Type I, the most frequent variety, is due to a mutation on chromosome 7, in a region that contains the two homeobox genes DLX5 and DLX6 [55]–[57]. SHFM Type IV maps to chromosome 3 and it has been shown that TP63 is the gene involved [31], [58]. Furthermore, it has been shown that Dlx5 and Dlx6 are transcriptional targets of Tp63 [59], [60]. Tp63 is a member of the p53 family of transcription factors crucial for stratified epithelial differentiation [61], [62] and Dlx5 and Dlx6 are members of the family of distalless-related homeodomain transcription factors (Dlx1–Dlx6) that play key roles in limb development. Therefore, we analyzed the expression of Tp63 and Dlx5 and Dlx6 in our SHFM mutants, to determine whether the Tp63 pathway was involved. Our analysis showed that Tp63 and Dlx5 and Dlx6 were normally expressed in the Sp6−/−;Sp8+/− mutant except for the flattened AER morphology (Figure 8A–D). Finally, the analysis of double mutants showed that Tp63, Dlx5 and Dlx6 were initially expressed normally in the complete absence of Sp6 and Sp8 (Figure 8E–F) suggesting that if Sp6/Sp8 are components of the Tp63 network, they act downstream of Dlx5 and Dlx6. The expression of Tp63 in Sp6−/−;Sp8−/− mutants was further confirmed by immunohistochemistry (Figure 8G–H).
Fig. 8. Tp63 and Dlx5 expression in mutant limb buds. 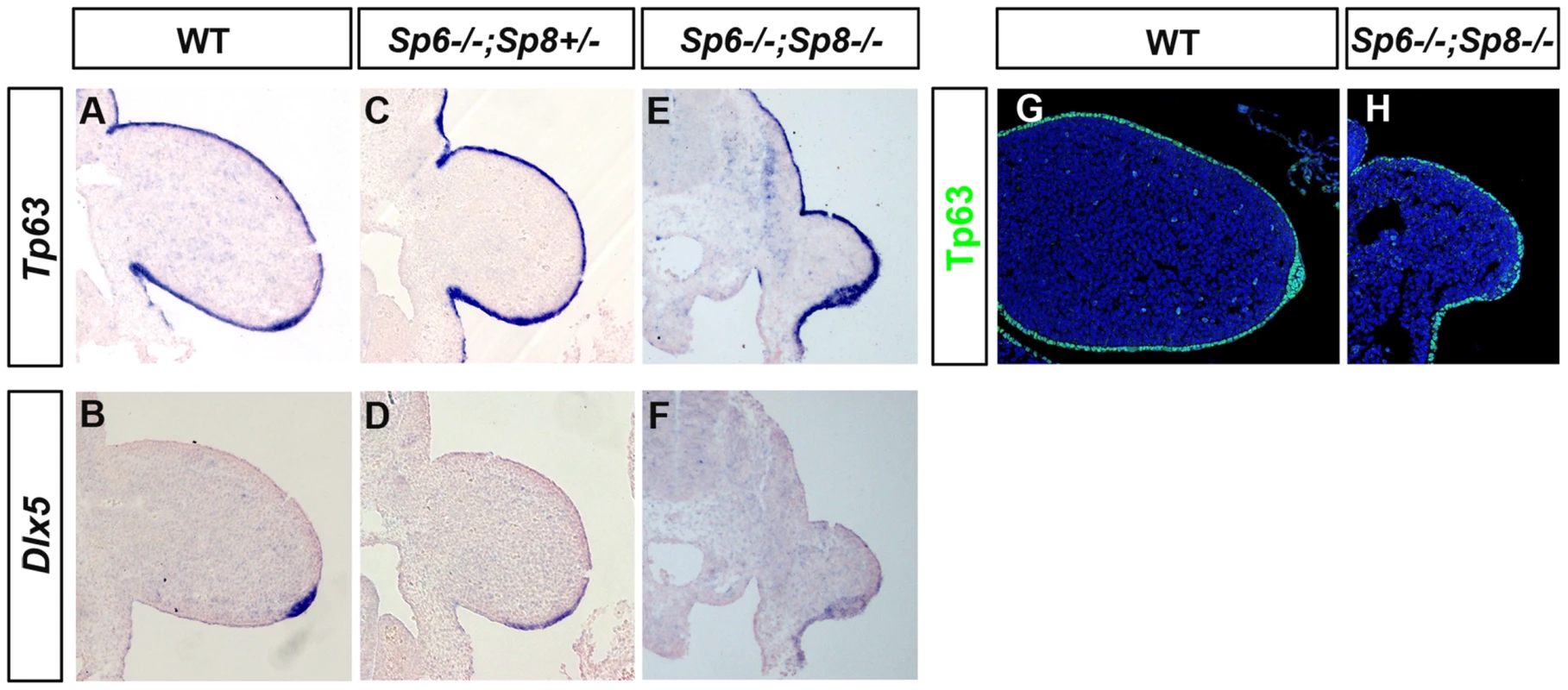
(A–F) Tp63 and Dlx5 expression is normally detected in the limb ectoderm of control (A, B), Sp6−/−;Sp8+/− (C, D) and Sp6−/−;Sp8−/− (E, F) mutants although Dlx5 is downregulated. (G–H) Immunostaining for Tp63 (green) showing expression in the Sp6−/−;Sp8−/− double mutant limb bud similar to wild type littermate. All the panels show longitudinal sections of E10.5 forelimb buds. Discussion
Sp6 and Sp8 play complimentary functions in limb development
There are numerous examples in limb development of related genes with similar patterns of expression playing redundant functions and therefore providing robustness to the system. Among these are members of the Fgf, Bmp, and Hox gene families in which the overall final gene dosage is the key parameter for normal morphology [49], [63], [64]. Here, by using a variety of loss-of-function alleles we have identified that Sp6 and Sp8 control AER development and DV patterning in a redundant and dose-dependent manner. However, both genes do not contribute equally which may in part be due to their differential levels of transcription.
Notwithstanding the phenotypic variation associated with each particular genotype, when the predominant phenotypes obtained from the allelic series of compound Sp6 and Sp8 mutants are categorized in order of increasing severity, a strong correlation with gene dosage is observed (schematically shown for the forelimb phenotypes in Figure 9). A progressive reduction in the dose of Sp6 and Sp8 gene products leads to predictable morphology, from syndactyly, to SHFM, oligodactyly, truncation and finally amelia. This comparative analysis shows that the amount of gene product provided by a single functional allele of Sp8 permits the complete development of the PD axis while one functional allele of Sp6 does not, most likely because the gene product provided is below the critical threshold required for AER induction. Both alleles of Sp6 provide less gene product than a single allele of Sp8 and equivalent to a transient expression of one copy of Sp8, as occurs in the forelimb when the Msx2;Cre deleter line is used. Collectively, the data from our allelic series indicate that Sp6 and Sp8 are, for the most part, functionally equivalent and work in concert during limb development.
Fig. 9. Illustration showing the correlation between the Sp6/Sp8 gene dose and the severity of the limb phenotype. 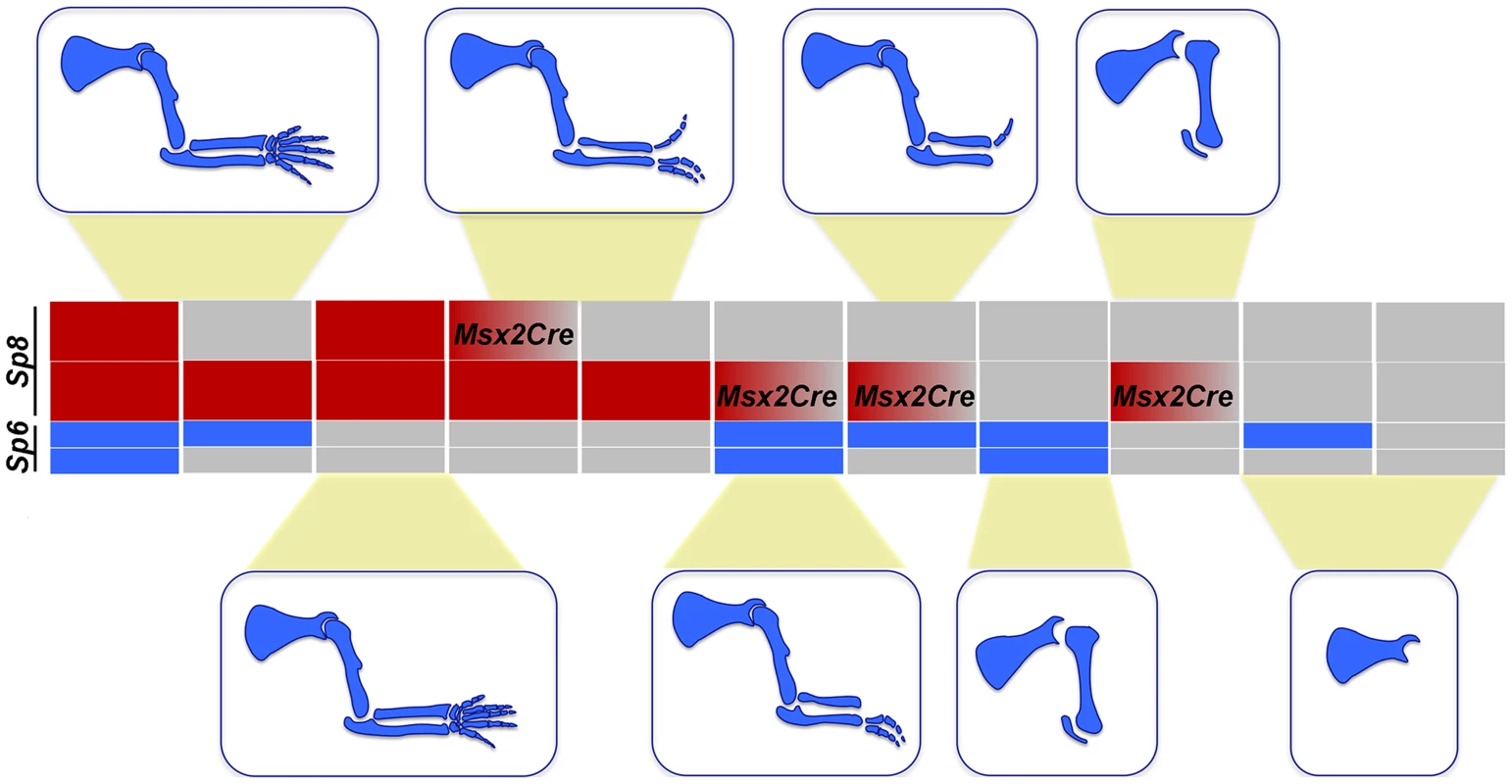
Blue boxes represent the Sp6 alelles and red boxes the Sp8 alelles. Grey boxes represent null alleles and boxes with a red to grey graduation represent conditionally removal with the Msx2Cre allele. We found that the putative Sp8 promoter has an increased number of potential βcatenin/Lef1 binding sites compared to Sp6, which might account for the higher levels of Sp8 expression. Interestingly, another member of the Sp family also expressed in the limb ectoderm, Sp9 [27], is unable to promote limb development in the absence of Sp6/Sp8 possibly because of its low level of expression [27]. Supporting this notion, there is a decreased number of βcatenin/Lef1 binding sites within the Sp9 putative promoter region when compared to Sp6.
We also considered whether the differences in Sp functional capacity could be due to structural differences. Comparative analysis of known protein domains and multiple alignment of Sp6, Sp8 and Sp9 revealed variability in the amino ends with the only common domains shared by these transcription factors being the zinc finger domains located in the carboxy ends. No structure-function correlation in the variable amino terminal domains was evident. For example, Sp6 and Sp8 are structurally disparate, but function in a complementary fashion. In contrast, Sp9 is structurally more similar to Sp8, than Sp6 is to Sp8 and yet does not show a complementary function in the limb. Therefore, even though these factors differ in their amino terminal domains, which may be functional in a different context [65], it is reasonable to speculate that in the limb, their functional capacity relies on their level of expression; this remains to be demonstrated.
Sp6 and Sp8 are absolutely necessary for Fgf8 induction and maintenance
Two of the main phenotypic features in our allelic series are truncations and SHFM. Studies in different mouse models and experimental manipulations in chick have established that these phenotypes can result from perturbations in AER functioning [1] and, accordingly, our analysis showed that Sp6/Sp8 are required for the formation and maintenance of a functional AER.
The first phase in the formation of the AER is the induction of AER precursor cells in the limb ectoderm characterized by the expression of Fgf8. This depends on at least three important signaling inputs: i) Fgf10 produced in the limb mesoderm and signaling through the Fgf receptor 2b (Fgfr2b) expressed within the ectoderm [12], [13], [66]–[70], ii) Wnt/βcatenin signaling produced in the limb ectoderm and signaling preferentially to the ventral limb ectoderm [14], [15] and iii) Bmp signaling, mainly from the limb ectoderm, but also possibly from the limb mesoderm that signals through the Bmpr1a receptor in the limb ectoderm [16], [17], [22], [71], [72]. Although the crosstalk between these three inputs is complex and not completely understood, both the Fgf10 and the Bmp signaling pathways have been shown to act upstream of Wnt/βcatenin signaling in the induction of the AER [14], [16].
The analysis we have performed shows that when the dose of Sp6/Sp8 is significantly reduced, Fgf8 is not activated, disregarding initial normal Fgf10 expression and Bmp signaling. Because both Sp6 and Sp8 have been shown to function downstream of Wnt/βcatenin signaling [26], [27], [30], and Sp8 has been shown to bind and activate the Fgf8 promoter [29], our results fit with a model in which Sp6 and Sp8 function as transcriptional activators of Fgf8 downstream of Wnt/βcatenin signaling in the limb ectoderm (Figure 10). Sp6 and Sp8 function together and in a dose-dependent manner as necessary mediators of the Wnt/βcatenin-Fgf8 regulatory loop. Our phenotypic and molecular studies indicate that the level of gene product produced by a single Sp8 allele is around the minimum dose required for the activation and maintenance of Fgf8 expression while that produced by a single Sp6 allele does not reach this minimum.
Fig. 10. Regulatory pathways mediated by Sp6 and Sp8. 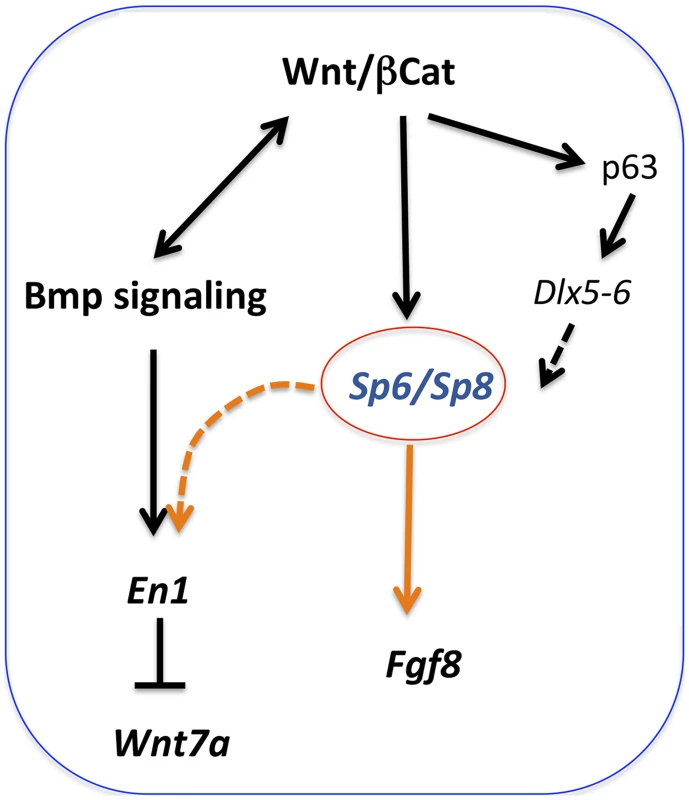
Sp6 and Sp8 are necessary mediators of the Wnt/βcatenin-dependent induction of Fgf8 in the limb ectoderm. In addition, these two factors also collaborate with BMP signaling in the induction of En1 in the ventral limb ectoderm. Finally, Sp6 and Sp8 may also act downstream of Tp63 and Dlx genes. It is known that the Wnt/βcatenin signaling pathway is not only required for AER induction, but also for its maintenance. The limb truncations observed when, in the absence of Sp6, Sp8 is removed from the forelimb ectoderm after the AER has been induced (Sp6−/−;Sp8f/−;Msx2Cre), indicate an ongoing role for Sp8 in AER maintenance, further supporting our model.
Most interestingly, our analysis shows that the complete absence of Sp6 and Sp8 transcription factors does not prevent the initiation of AER morphology confirming the independence between AER function and morphology. This is in high contrast to βcatenin loss-of-function mutants in the limb ectoderm that completely lack any evidence of a morphological AER or ectoderm thickening [14], [15]. This difference may reflect the requirement of βcatenin for a proper AER morphology as has already been suggested [15], [30], [49] and corroborates that the Wnt3/βcatenin-Sp6/Sp8-Fgf8 regulatory loop is not a simple lineal one. Tp63, a crucial factor for AER morphology and Fgf8 maintenance of expression [61], [62], and a well-established target of Wnt/βcatenin in the ectoderm [73] may be at the root of this difference. Characterization of separate Sp6/Sp8 and Tp63 mediated pathways may help to uncouple βcatenin's multiple roles in AER formation and function.
βcatenin is also necessary for the expression of other AER markers (i.e. Bmp4, Msx2 [14], [15]) in addition to Fgf8. Sp6 and Sp8 are necessary for the expression of Fgf8, but Bmp ligands and Msx2 are normally activated in the total absence of Sp6/Sp8. Collectively, these data demonstrate that Sp6 and Sp8 mediate only part of the βcatenin functions in the limb ectoderm, principally the induction of Fgf8.
Recently, it has been shown that a conserved Wnt-Sp8-Fgf8 genetic cassette is also used to regulate the outgrowth of other body appendages such as the genital tubercle [30]. This work identified Sp8 as partially mediating the regulation of Fgf8 by the canonical Wnt/βcatenin pathway, a function that we demonstrate here that is fully accomplished by Sp6 and Sp8 together. Their result showing the failure of forced expression of Sp8 in the AER (R26Sp8;Msx2) to rescue the phenotype of βcatenin loss-of-function in the limb ectoderm is very likely due to Sp8 not reaching, in these experiments, the minimum level of expression required for Fgf8 induction.
Role of Sp6 and Sp8 in dorsal-ventral patterning
During normal development the AER forms at the DV boundary of the limb bud reflecting a tight link between AER formation and DV patterning. Based on the analysis of the limbless, En1 mutants and on misexpression experiments in chick, it was hypothesized that the expression of En1 in the ventral ectoderm might function to establish a DV interface as a prerequisite for AER induction [74], [75]. However, there are several examples of normal AERs forming in the absence of a DV boundary, such as eudiplopodia, the double Wnt7a;En1 mutant and experiments in chick creating bidorsal limbs [19]–[21], [76].
Here we report that DV patterning is also disrupted when the Sp6/Sp8 gene dose is perturbed. In the amelic phenotypes, even if the limb does not form, the molecular analysis of the emerging limb buds indicates that they are bi-dorsal as Wnt7a expression is extended along most of the limb ectoderm while En1 is not detected. Interestingly, the failure to activate En1 occurs despite normal expression of Bmp ligands in the limb ectoderm and mesoderm. In the SHFM phenotypes the digital tips display conical nails. In these limb buds the AER is irregularly induced and where maintained it remains flat, broad and immature. This correlates with an extension of Wnt7a expression into the ventral ectoderm and a proximally restricted expression of En1 [20], [21], [23], [77]. Lmx1b expands into the ventral mesoderm distally explaining the bi-dorsal phenotypic traits in the digits of Sp6−/−;Sp8+/− mutants, while DV patterning is largely preserved at more proximal levels.
We found that in the absence of a sufficient amount of Sp6/Sp8 gene products Bmp signaling is not sufficient to induce En1. Sp family members are known to bind and interact with other transcription factors, including Smads. Thus, we hypothesized that Sp6/Sp8 transcription factors interact/cooperate with Smad proteins downstream of Bmp signaling to mediate En1 activation [78], [79] (Figure 10). This interaction could occur at the protein level or by summative or synergistic effects on the En1 promoter. Interestingly, the putative En1 promoter exhibits 25 potential Sp binding sites and 12 Smad binding sites that are conserved between human and mouse. Further investigation will be required to clarify this relationship (Figure S6).
The Sp6;Sp8 double mutant limbs are reminiscent of those of the limbless mutation in chicken. Limbless is a simple Mendelian autosomal recessive mutation characterized by tetra-amelia in the homozygous condition [80]. The mutation causes defects in no other organs, although it is effectively lethal because the chicks are unable to hatch without legs [47], [48]. Limb development initiates in limbless embryos and the early limb buds are morphologically indistinguishable from normal embryos until stage 19. However, the early limb buds are bidorsal and don't form an apical ridge [74], [75], [81]. The limb bud mesoderm undergoes cell death beginning in the mid-distal mesoderm at stage 19–20 so that by stage 24, no signs of limb buds remain [47], [75]. The similarities between our mutant and the limbless mutation may indicate a common target gene. After chromosomal mapping of the limbless mutation, Robb and coworkers [82] suggested Sp8 as a priority candidate. This is reinforced by the fact that Sp6 seems to be absent in chickens (Figure S3). However, further studies to validate this suspicion have not been done. Interestingly, limbless does not display the neural phenotype characteristic of Sp8 mutants, i.e., except for the limb phenotype the embryo is normal. This could be explained by a defect in a limb specific Sp8 regulatory element in limbless. However, the lack of any AER morphology in limbless, in contrast to the double Sp6;Sp8 mutants, decreases the likelihood that Sp8 is the gene targeted.
Sp6 and Sp8 and split hand/foot malformation
In humans, the SHFM is a genetically heterogeneous congenital malformation characterized by a deficit in the formation of the central elements of the hands and feet that results in a central cleft associated with fusion and malformations of the remaining digits. The phenotype is highly variable, even between the limbs of a single affected individual, and ranges from a mild central syndactyly to severe loss of elements with oligodactyly and sometimes even affecting the zeugopod. It is currently accepted that this phenotype is the result of a premature regression of the central part of the AER [52], [53], [59]. Remarkably, the limb phenotype of the embryos that develop with a single copy of Sp8 reproduces the human SHFM condition. The molecular analysis of these mutant limb buds indicates that the product obtained from one allele of Sp8, in the absence of Sp6, barely reaches the threshold required for Fgf8 induction. This is based on the low levels of Fgf8 transcription achieved and also on the irregular expression domain that likely results from a cell autonomous effect of the mutation. Due to normal biologic variation, the level of Sp8 attained may reach the threshold required for Fgf8 induction in some cells, but not in others. Interestingly, at later stages Fgf8 expression is not maintained in central regions suggesting that this later deficit in Fgf8 expression is the cause of the SHFM phenotype in Sp6;Sp8 compound mutants. Since the irregular early activation of Fgf8 has not been observed in other models of SHFM, its possible contribution to the phenotype remains to be investigated [52], [53], [59].
Removal of all known AER-related Bmp ligands (Bmp2, Bmp4 and Bmp7) from the AER using Msx2Cre also results in SHFM [71]. However, in Sp6−/−;Sp8+/− mutants, Bmp4 is still expressed in the remaining AER suggesting that this SHFM phenotype is not caused by the loss of Bmp expression in the AER. In fact, since Bmp signalling is required for the induction of Fgf8, the SHFM phenotype following AER-related Bmp removal can also be explained by an irregular induction of Fgf8.
Of great interest is the recent genetic analysis of Fgf8 regulation that has identified nearly 50 Fgf8-regulatory modules in a 220 Kb region centromeric to the gene [83]. All the AER-specific enhancers, many of them embedded in the FBXW4 gene, drive expression all along the AP extension of the AER. Interestingly, SHFM type III [84], [85] is caused by duplications of this genomic region that disrupt the normal architecture of the multiple enhancers likely affecting Fgf8 expression [83]. Therefore, SHFM type III is likely the result of Fgf8 misregulation [83].
As previously mentioned, despite the identification of 6 loci involved in SHFM, only TP63 (SHFM type IV) and DLX5 and DLX6 (SHFM type I) have been unequivocally associated with this malformation [34]. Mutations in WNT10B (SHFM type VI) were also identified to be causative for SHFM, although there is some doubt on whether these mutations are sufficient for the phenotype [86]–[88]. Since similar phenotypes are frequently caused by disruption of different components of a regulatory network, we have considered the possibility that Sp6 and Sp8 genes might be part of the Tp63 network. Indeed, the phenotypes of our mutants are identical, including the DV component, to those recently reported in a new identified human mutation in DLX5 [57]. However, the fact that Tp63, Dlx5 and Dlx6 have essentially normal expression patterns in the early Sp6/Sp8 mutant limb bud indicates that, if Sp6/Sp8 transcription factors act within the Tp63 network, they function downstream of Tp63 and Dlx factors. Tp63 is necessary for the formation and maintenance of a normal epidermal layer [61], [62]. In mouse, removal of Tp63 results in several abnormalities including limb truncations that are most similar to the Sp8-null phenotype [24], [25], [61], [62] suggesting that Tp63 may preferentially control Sp8, but not Sp6 in mice. In any case, the relationship between the Tp63-Dlx and the Sp-Fgf8 regulatory modules, both downstream of Wnt/βcatenin, add an extra level of complexity to limb development that requires further investigation.
Conclusions
This study provides compelling evidence for the absolute requirement of Sp6 and Sp8 for limb development as in their complete absence, or substantial reduction, no limbs form. By using a variety of loss-of-function alleles to remove the activity of Sp6 and Sp8 genes, we reveal that these two factors work together and in a dose-dependent manner as necessary mediators for AER development and DV patterning.
Our study supports a model in which these two factors work together downstream of Wnt/βcatenin signaling in the induction of Fgf8 and also downstream of Bmp signaling in the induction of En1 establishing a link between proximal-distal and dorsal-ventral patterning.
Materials and Methods
Ethics statement and mouse strains
All animal procedures were conducted accordingly to the EU regulations and 3R principles and reviewed and approved by the Bioethics Committee of the University of Cantabria. Mutant mouse lines were described previously: Sp6 null allele [28]; Sp8 null allele [24]; Sp8 floxed allele [36]; AP2αCre [39] and Msx2Cre lines [37]; R26R [42]. Mice and embryos were genotyped by PCR, using genomic DNA extracted from tail biopsies and yolk sacs, respectively.
Skeletal preparation
After removing skin and viscera, mouse embryos were fixed in 95% ethanol. Alizarin Red and Alcian blue skeletal staining was performed according to standard protocols, cleared by KOH treatment and stored in glycerol.
In situ hybridization
In situ hybridization (ISH) was performed in whole-mount and in sections following standard procedures using the previously described Bmp4 [64], Dlx5 and Dlx6 [53], En1 [23] Fgf8 [43], Fgf10 [89], Lmx1b [90], Msx2 [91], Tp63 [62], Sp6 [28] and Wnt7a [92] antisense riboprobes.
RNA quantification by real-time PCR
Embryonic fore and hind - limb buds were dissected in cold RNAse-free PBS from E10.5 wild type embryos. Total RNA was isolated separately from 3 pools of 8 forelimbs or 8 hindlimbs each. cDNA synthesis was done using standard conditions.
Real-time RT-PCR was carried out on an Mx3005P cycler, using the SYBRGreen PCR Master Mix (Invitrogen) and the data were analyzed using the MxPro software (Stratagene). Results were tested statistically performing ANOVA and Student-T test, being statistically significant when p<0.05.
Expression of Sp6 and Sp8 was normalized to that of housekeeping gene 18sRNA. The primers used (5′ to 3′ orientation) were: Sp6-F: tgctaaccgctgtctgtgg; Sp6-R:ctggtatgtctggagaggttgc; Sp8-F: ttatctccaaggtgcacacg; Sp8-R:gcttgaaccaggactcatacg; 18sRNA-R: ttggcaatgtttcgctc;18sRNA-F: cgccgctagaggtgaaattt.
Cell death assay
Detection of cell death was performed in sections of paraffin embedded tissue using terminal deoxynucleotidyl transferase mediated dUTP nick-end labelling (TUNEL) with the Apoptag Fluorescein Direct In Situ Apoptosis Detection Kit (Intergen) following the manufacturer's instructions.
β-gal reporter analysis
For detection of β-galactosidase activity, R26R;Ap2αCre double transgenic embryos were fixed for 30 min, rinsed in PBS and incubated in the presence of X-gal as described [93].
Immunohistochemistry
Immunohistochemistry was performed in paraffin sections using the anti E-cadherin (Byoscience, # 610182), anti Laminin (Abcam, # ab11575), anti Tp63 (Abcam, # Ab53039) and anti Connexin43 (Abcam, # ab11370) primary antibodies. Antigen retrieval was performed by incubation with proteinase K (10 µg/ml) for E-cadherin and laminin or with citrate buffer in pressure cooker for Tp63 and Connexin43. Alexa®488 and TexasRED fluorescently tagged secondary antibodies were used. Vecthasield containing DAPI for nuclear counter staining was used as mounting medium. Confocal images were acquired in a SP-5 laser-scan confocal microscope (Leica Microsystems).
In silico analysis
Conservation of En1, Sp6 and Sp8 loci between mouse, human, opossum, chicken and zebrafish was determined using pairwise alignment software (mVista browser, http://genome.lbl.gov/vista/). Conserved noncoding regions were further analyzed for potential transcription factor binding sites using AliBaba 2.1 (http://www.generegulation.com/pub/programs/alibaba2/index.html) and Sequencher 4.8 (Gene Codes Inc.) informatic software.
Supporting Information
Zdroje
1. Fernandez-TeranM, RosMA (2008) The Apical Ectodermal Ridge: morphological aspects and signaling pathways. Int J Dev Biol 52 : 857–871.
2. NiswanderL, JeffreyS, MartinGR, TickleC (1994) A positive feedback loop coordinates growth and patterning in the vertebrate limb. Nature 371 : 609–612.
3. DudleyAT, RosMA, TabinCJ (2002) A re-examination of proximodistal patterning during vertebrate limb development. Nature 418 : 539–544.
4. RoweDA, CairnsJM, FallonJF (1982) Spatial and temporal patterns of cell death in limb bud mesoderm after apical ectodermal ridge removal. Dev Biol 93 : 83–91.
5. FallonJF, LopezA, RosMA, SavageMP, OlwinBB, et al. (1994) FGF-2: apical ectodermal ridge growth signal for chick limb development. Science 264 : 104–107.
6. KimmelRA, TurnbullDH, BlanquetV, WurstW, LoomisCA, et al. (2000) Two lineage boundaries coordinate vertebrate apical ectodermal ridge formation. Genes Dev 14 : 1377–1389.
7. BellSM, SchreinerCM, ScottWJ (1998) The loss of ventral ectoderm identity correlates with the inability to form an AER in the legless hindlimb bud. Mech Dev 74 : 41–50.
8. Salas-VidalE, ValenciaC, CovarrubiasL (2001) Differential tissue growth and patterns of cell death in mouse limb autopod morphogenesis. Dev Dyn 220 : 295–306.
9. GuoQ, LoomisC, JoynerAL (2003) Fate map of mouse ventral limb ectoderm and the apical ectodermal ridge. Dev Biol 264 : 166–178.
10. KengakuM, CapdevilaJ, Rodriguez-EstebanC, De La PenaJ, JohnsonRL, et al. (1998) Distinct WNT pathways regulating AER formation and dorsoventral polarity in the chick limb bud. Science 280 : 1274–1277.
11. OhuchiH, NakagawaT, YamamotoA, AragaA, OhataT, et al. (1997) The mesenchymal factor, FGF10, initiates and maintains the outgrowth of the chick limb bud through interaction with FGF8, an apical ectodermal factor. Development 124 : 2235–2244.
12. MinH, DanilenkoDM, ScullySA, BolonB, RingBD, et al. (1998) Fgf-10 is required for both limb and lung development and exhibits striking functional similarity to Drosophila branchless. Genes Dev 12 : 3156–3161.
13. SekineK, OhuchiH, FujiwaraM, YamasakiM, YoshizawaT, et al. (1999) Fgf10 is essential for limb and lung formation. Nat Genet 21 : 138–141.
14. SoshnikovaN, ZechnerD, HuelskenJ, MishinaY, BehringerRR, et al. (2003) Genetic interaction between Wnt/beta-catenin and BMP receptor signaling during formation of the AER and the dorsal-ventral axis in the limb. Genes Dev 17 : 1963–1968.
15. BarrowJR, ThomasKR, Boussadia-ZahuiO, MooreR, KemlerR, et al. (2003) Ectodermal Wnt3/beta-catenin signaling is required for the establishment and maintenance of the apical ectodermal ridge. Genes Dev 17 : 394–409.
16. Pajni-UnderwoodS, WilsonCP, ElderC, MishinaY, LewandoskiM (2007) BMP signals control limb bud interdigital programmed cell death by regulating FGF signaling. Development 134 : 2359–2368.
17. AhnK, MishinaY, HanksMC, BehringerRR, CrenshawEB3rd (2001) BMPR-IA signaling is required for the formation of the apical ectodermal ridge and dorsal-ventral patterning of the limb. Development 128 : 4449–4461.
18. ZellerR, DubouleD (1997) Dorso-ventral limb polarity and origin of the ridge: on the fringe of independence? Bioessays 19 : 541–546.
19. GoetinckPF (1964) Studies on Limb Morphogenesis. Ii. Experiments with the Polydactylous Mutant Eudiplopodia. Dev Biol 10 : 71–91.
20. LoomisCA, KimmelRA, TongCX, MichaudJ, JoynerAL (1998) Analysis of the genetic pathway leading to formation of ectopic apical ectodermal ridges in mouse Engrailed-1 mutant limbs. Development 125 : 1137–1148.
21. CyganJA, JohnsonRL, McMahonAP (1997) Novel regulatory interactions revealed by studies of murine limb pattern in Wnt-7a and En-1 mutants. Development 124 : 5021–5032.
22. PizetteS, Abate-ShenC, NiswanderL (2001) BMP controls proximodistal outgrowth, via induction of the apical ectodermal ridge, and dorsoventral patterning in the vertebrate limb. Development 128 : 4463–4474.
23. LoomisCA, HarrisE, MichaudJ, WurstW, HanksM, et al. (1996) The mouse Engrailed-1 gene and ventral limb patterning. Nature 382 : 360–363.
24. TreichelD, SchockF, JackleH, GrussP, MansouriA (2003) mBtd is required to maintain signaling during murine limb development. Genes Dev 17 : 2630–2635.
25. BellSM, SchreinerCM, WaclawRR, CampbellK, PotterSS, et al. (2003) Sp8 is crucial for limb outgrowth and neuropore closure. Proc Natl Acad Sci U S A 100 : 12195–12200.
26. TalamilloA, DelgadoI, NakamuraT, de-VegaS, YoshitomiY, et al. (2010) Role of Epiprofin, a zinc-finger transcription factor, in limb development. Dev Biol 337 : 363–374.
27. KawakamiY, EstebanCR, MatsuiT, Rodriguez-LeonJ, KatoS, et al. (2004) Sp8 and Sp9, two closely related buttonhead-like transcription factors, regulate Fgf8 expression and limb outgrowth in vertebrate embryos. Development 131 : 4763–4774.
28. NakamuraT, UndaF, de-VegaS, VilaxaA, FukumotoS, et al. (2004) The Kruppel-like factor epiprofin is expressed by epithelium of developing teeth, hair follicles, and limb buds and promotes cell proliferation. J Biol Chem 279 : 626–634.
29. SaharaS, KawakamiY, Izpisua BelmonteJC, O'LearyDD (2007) Sp8 exhibits reciprocal induction with Fgf8 but has an opposing effect on anterior-posterior cortical area patterning. Neural Dev 2 : 10.
30. LinC, YinY, BellSM, VeithGM, ChenH, et al. (2013) Delineating a conserved genetic cassette promoting outgrowth of body appendages. PLoS Genet 9: e1003231.
31. GuerriniL, CostanzoA, MerloGR (2011) A symphony of regulations centered on p63 to control development of ectoderm-derived structures. J Biomed Biotechnol 2011 : 864904.
32. DuijfPH, van BokhovenH, BrunnerHG (2003) Pathogenesis of split-hand/split-foot malformation. Hum Mol Genet 12 Spec No 1: R51–60.
33. ElliottAM, EvansJA (2006) Genotype-phenotype correlations in mapped split hand foot malformation (SHFM) patients. Am J Med Genet A 140 : 1419–1427.
34. GurrieriF, EvermanDB (2013) Clinical, genetic, and molecular aspects of split-hand/foot malformation: An update. Am J Med Genet A 161 : 2860–2872.
35. FrazerKA, PachterL, PoliakovA, RubinEM, DubchakI (2004) VISTA: computational tools for comparative genomics. Nucleic Acids Res 32: W273–279.
36. ZembrzyckiA, GrieselG, StoykovaA, MansouriA (2007) Genetic interplay between the transcription factors Sp8 and Emx2 in the patterning of the forebrain. Neural Dev 2 : 8.
37. SunX, LewandoskiM, MeyersEN, LiuYH, MaxsonREJr, et al. (2000) Conditional inactivation of Fgf4 reveals complexity of signalling during limb bud development. Nat Genet 25 : 83–86.
38. MoonAM, CapecchiMR (2000) Fgf8 is required for outgrowth and patterning of the limbs. Nat Genet 26 : 455–459.
39. MacateeTL, HammondBP, ArenkielBR, FrancisL, FrankDU, et al. (2003) Ablation of specific expression domains reveals discrete functions of ectoderm - and endoderm-derived FGF8 during cardiovascular and pharyngeal development. Development 130 : 6361–6374.
40. DelgadoI, Dominguez-FrutosE, SchimmangT, RosMA (2008) The incomplete inactivation of Fgf8 in the limb ectoderm affects the morphogenesis of the anterior autopod through BMP-mediated cell death. Dev Dyn 237 : 649–658.
41. BouletAM, MoonAM, ArenkielBR, CapecchiMR (2004) The roles of Fgf4 and Fgf8 in limb bud initiation and outgrowth. Dev Biol 273 : 361–372.
42. SorianoP (1999) Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet 21 : 70–71.
43. CrossleyPH, MartinGR (1995) The mouse Fgf8 gene encodes a family of polypeptides and is expressed in regions that direct outgrowth and patterning in the developing embryo. Development 121 : 439–451.
44. GrahamA, Francis-WestP, BrickellP, LumsdenA (1994) The signalling molecule BMP4 mediates apoptosis in the rhombencephalic neural crest. Nature 372 : 684–686.
45. TumpelS, Sanz-EzquerroJJ, IsaacA, EblaghieMC, DobsonJ, et al. (2002) Regulation of Tbx3 expression by anteroposterior signalling in vertebrate limb development. Dev Biol 250 : 251–262.
46. SunX, MarianiFV, MartinGR (2002) Functions of FGF signalling from the apical ectodermal ridge in limb development. Nature 418 : 501–508.
47. FallonJF, FrederickJM, CarringtonJL, LanserME, SimandlBK (1983) Studies on a limbless mutant in the chick embryo. Prog Clin Biol Res 110 Pt A: 33–43.
48. CarringtonJL, FallonJF (1988) Initial limb budding is independent of apical ectodermal ridge activity; evidence from a limbless mutant. Development 104 : 361–367.
49. MarianiFV, AhnCP, MartinGR (2008) Genetic evidence that FGFs have an instructive role in limb proximal-distal patterning. Nature 453 : 401–405.
50. Fernandez-TeranM, RosMA, MarianiFV (2013) Evidence that the limb bud ectoderm is required for survival of the underlying mesoderm. Dev Biol 381 : 341–352.
51. GreenCR, BowlesL, CrawleyA, TickleC (1994) Expression of the connexin43 gap junctional protein in tissues at the tip of the chick limb bud is related to the epithelial-mesenchymal interactions that mediate morphogenesis. Dev Biol 161 : 12–21.
52. IanakievP, KilpatrickMW, ToudjarskaI, BaselD, BeightonP, et al. (2000) Split-hand/split-foot malformation is caused by mutations in the p63 gene on 3q27. Am J Hum Genet 67 : 59–66.
53. RobledoRF, RajanL, LiX, LufkinT (2002) The Dlx5 and Dlx6 homeobox genes are essential for craniofacial, axial, and appendicular skeletal development. Genes Dev 16 : 1089–1101.
54. RiddleRD, EnsiniM, NelsonC, TsuchidaT, JessellTM, et al. (1995) Induction of the LIM homeobox gene Lmx1 by WNT7a establishes dorsoventral pattern in the vertebrate limb. Cell 83 : 631–640.
55. MarinoniJC, StevensonRE, EvansJP, GeshuriD, PhelanMC, et al. (1995) Split foot and developmental retardation associated with a deletion of three microsatellite markers in 7q21.2-q22.1. Clin Genet 47 : 90–95.
56. SchererSW, CheungJ, MacDonaldJR, OsborneLR, NakabayashiK, et al. (2003) Human chromosome 7: DNA sequence and biology. Science 300 : 767–772.
57. ShamseldinHE, FadenMA, AlashramW, AlkurayaFS (2012) Identification of a novel DLX5 mutation in a family with autosomal recessive split hand and foot malformation. J Med Genet 49 : 16–20.
58. BrunnerHG, HamelBC, Van BokhovenH (2002) The p63 gene in EEC and other syndromes. J Med Genet 39 : 377–381.
59. Lo IaconoN, ManteroS, ChiarelliA, GarciaE, MillsAA, et al. (2008) Regulation of Dlx5 and Dlx6 gene expression by p63 is involved in EEC and SHFM congenital limb defects. Development 135 : 1377–1388.
60. KouwenhovenEN, van HeeringenSJ, TenaJJ, OtiM, DutilhBE, et al. (2010) Genome-wide profiling of p63 DNA-binding sites identifies an element that regulates gene expression during limb development in the 7q21 SHFM1 locus. PLoS Genet 6: e1001065.
61. YangA, SchweitzerR, SunD, KaghadM, WalkerN, et al. (1999) p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature 398 : 714–718.
62. MillsAA, ZhengB, WangXJ, VogelH, RoopDR, et al. (1999) p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature 398 : 708–713.
63. ShethR, GregoireD, DumouchelA, ScottiM, PhamJM, et al. (2013) Decoupling the function of Hox and Shh in developing limb reveals multiple inputs of Hox genes on limb growth. Development 140 : 2130–2138.
64. BandyopadhyayA, TsujiK, CoxK, HarfeBD, RosenV, et al. (2006) Genetic analysis of the roles of BMP2, BMP4, and BMP7 in limb patterning and skeletogenesis. PLoS Genet 2: e216.
65. ZhaoC, MengA (2005) Sp1-like transcription factors are regulators of embryonic development in vertebrates. Dev Growth Differ 47 : 201–211.
66. OhuchiH, HoriY, YamasakiM, HaradaH, SekineK, et al. (2000) FGF10 acts as a major ligand for FGF receptor 2 IIIb in mouse multi-organ development. Biochem Biophys Res Commun 277 : 643–649.
67. XuX, WeinsteinM, LiC, NaskiM, CohenRI, et al. (1998) Fibroblast growth factor receptor 2 (FGFR2)-mediated reciprocal regulation loop between FGF8 and FGF10 is essential for limb induction. Development 125 : 753–765.
68. LuP, EwaldAJ, MartinGR, WerbZ (2008) Genetic mosaic analysis reveals FGF receptor 2 function in terminal end buds during mammary gland branching morphogenesis. Dev Biol 321 : 77–87.
69. De MoerloozeL, Spencer-DeneB, RevestJM, HajihosseiniM, RosewellI, et al. (2000) An important role for the IIIb isoform of fibroblast growth factor receptor 2 (FGFR2) in mesenchymal-epithelial signalling during mouse organogenesis. Development 127 : 483–492.
70. YuK, OrnitzDM (2008) FGF signaling regulates mesenchymal differentiation and skeletal patterning along the limb bud proximodistal axis. Development 135 : 483–491.
71. ChoiKS, LeeC, MaatoukDM, HarfeBD (2012) Bmp2, Bmp4 and Bmp7 are co-required in the mouse AER for normal digit patterning but not limb outgrowth. PLoS One 7: e37826.
72. MaatoukDM, ChoiKS, BouldinCM, HarfeBD (2009) In the limb AER Bmp2 and Bmp4 are required for dorsal-ventral patterning and interdigital cell death but not limb outgrowth. Dev Biol 327 : 516–523.
73. FerrettiE, LiB, ZewduR, WellsV, HebertJM, et al. (2011) A conserved Pbx-Wnt-p63-Irf6 regulatory module controls face morphogenesis by promoting epithelial apoptosis. Dev Cell 21 : 627–641.
74. GrieshammerU, MinowadaG, PisentiJM, AbbottUK, MartinGR (1996) The chick limbless mutation causes abnormalities in limb bud dorsal-ventral patterning: implications for the mechanism of apical ridge formation. Development 122 : 3851–3861.
75. RosMA, Lopez-MartinezA, SimandlBK, RodriguezC, Izpisua BelmonteJC, et al. (1996) The limb field mesoderm determines initial limb bud anteroposterior asymmetry and budding independent of sonic hedgehog or apical ectodermal gene expressions. Development 122 : 2319–2330.
76. MichaudJL, LapointeF, Le DouarinNM (1997) The dorsoventral polarity of the presumptive limb is determined by signals produced by the somites and by the lateral somatopleure. Development 124 : 1453–1463.
77. LoganC, HornbruchA, CampbellI, LumsdenA (1997) The role of Engrailed in establishing the dorsoventral axis of the chick limb. Development 124 : 2317–2324.
78. PardaliK, KurisakiA, MorenA, ten DijkeP, KardassisD, et al. (2000) Role of Smad proteins and transcription factor Sp1 in p21(Waf1/Cip1) regulation by transforming growth factor-beta. J Biol Chem 275 : 29244–29256.
79. Safe S, Kim K (2004) Progress in Nucleic Acid Research and Molecular Biology. Elsevier Academic Press.
80. PrahladKV, SkalaG, JonesDG, BrilesWE (1979) Limbless: a new genetic mutant in the chick. J Exp Zool 209 : 427–434.
81. NoramlyS, PisentiJ, AbbottU, MorganB (1996) Gene expression in the limbless mutant: polarized gene expression in the absence of Shh and an AER. Dev Biol 179 : 339–346.
82. RobbEA, GitterCL, ChengHH, DelanyME (2011) Chromosomal mapping and candidate gene discovery of chicken developmental mutants and genome-wide variation analysis of MHC congenics. J Hered 102 : 141–156.
83. MarinicM, AktasT, RufS, SpitzF (2013) An integrated holo-enhancer unit defines tissue and gene specificity of the Fgf8 regulatory landscape. Dev Cell 24 : 530–542.
84. SidowA, BulotskyMS, KerrebrockAW, BirrenBW, AltshulerD, et al. (1999) A novel member of the F-box/WD40 gene family, encoding dactylin, is disrupted in the mouse dactylaplasia mutant. Nat Genet 23 : 104–107.
85. FriedliM, NikolaevS, LyleR, ArcangeliM, DubouleD, et al. (2008) Characterization of mouse Dactylaplasia mutations: a model for human ectrodactyly SHFM3. Mamm Genome 19 : 272–278.
86. UgurSA, TolunA (2008) Homozygous WNT10b mutation and complex inheritance in Split-Hand/Foot Malformation. Hum Mol Genet 17 : 2644–2653.
87. BlattnerA, HuberAR, RothlisbergerB (2010) Homozygous nonsense mutation in WNT10B and sporadic split-hand/foot malformation (SHFM) with autosomal recessive inheritance. Am J Med Genet A 152A: 2053–2056.
88. AzizA, Irfanullah, KhanS, ZimriFK, MuhammadN, et al. (2014) Novel homozygous mutations in the WNT10B gene underlying autosomal recessive split hand/foot malformation in three consanguineous families. Gene 534 : 265–271.
89. HaradaH, ToyonoT, ToyoshimaK, YamasakiM, ItohN, et al. (2002) FGF10 maintains stem cell compartment in developing mouse incisors. Development 129 : 1533–1541.
90. ChenH, LunY, OvchinnikovD, KokuboH, ObergKC, et al. (1998) Limb and kidney defects in Lmx1b mutant mice suggest an involvement of LMX1B in human nail patella syndrome. Nat Genet 19 : 51–55.
91. RobertB, LyonsG, SimandlBK, KuroiwaA, BuckinghamM (1991) The apical ectodermal ridge regulates Hox-7 and Hox-8 gene expression in developing chick limb buds. Genes Dev 5 : 2363–2374.
92. ParrBA, McMahonAP (1995) Dorsalizing signal Wnt-7a required for normal polarity of D-V and A-P axes of mouse limb. Nature 374 : 350–353.
93. WhitingJ, MarshallH, CookM, KrumlaufR, RigbyPW, et al. (1991) Multiple spatially specific enhancers are required to reconstruct the pattern of Hox-2.6 gene expression. Genes Dev 5 : 2048–2059.
94. MatysV, Kel-MargoulisOV, FrickeE, LiebichI, LandS, et al. (2006) TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res 34: D108–110.
Štítky
Genetika Reprodukční medicína
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 8- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- IVF a rakovina prsu – zvyšují hormony riziko vzniku rakoviny?
-
Všechny články tohoto čísla
- DNA Double Strand Break Repair in Mitosis Is Suppressed by Phosphorylation of XRCC4
- Inference of Transposable Element Ancestry
- The Population Genetics of Evolutionary Rescue
- Retinoic Acid Activates Two Pathways Required for Meiosis in Mice
- Pooled Segregant Sequencing Reveals Genetic Determinants of Yeast Pseudohyphal Growth
- Comprehensive Identification of Single Nucleotide Polymorphisms Associated with Beta-lactam Resistance within Pneumococcal Mosaic Genes
- SMA-Causing Missense Mutations in Display a Wide Range of Phenotypes When Modeled in
- Branch Migration Prevents DNA Loss during Double-Strand Break Repair
- Transcriptome Sequencing from Diverse Human Populations Reveals Differentiated Regulatory Architecture
- Genetic Deletion of SEPT7 Reveals a Cell Type-Specific Role of Septins in Microtubule Destabilization for the Completion of Cytokinesis
- Tethering Sister Centromeres to Each Other Suggests the Spindle Checkpoint Detects Stretch within the Kinetochore
- Global Genetic Variations Predict Brain Response to Faces
- Demography and the Age of Rare Variants
- The Response to High CO Levels Requires the Neuropeptide Secretion Component HID-1 to Promote Pumping Inhibition
- Sp6 and Sp8 Transcription Factors Control AER Formation and Dorsal-Ventral Patterning in Limb Development
- The Groucho Co-repressor Is Primarily Recruited to Local Target Sites in Active Chromatin to Attenuate Transcription
- A Transposable Element Insertion Confers Xenobiotic Resistance in Drosophila
- The Genomic Architecture of Population Divergence between Subspecies of the European Rabbit
- Human Social Genomics
- Gene Expansion Shapes Genome Architecture in the Human Pathogen : An Evolutionary Genomics Analysis in the Ancient Terrestrial Mucorales (Mucoromycotina)
- Canonical Non-Homologous End Joining in Mitosis Induces Genome Instability and Is Suppressed by M-phase-Specific Phosphorylation of XRCC4
- Roles of Type 1A Topoisomerases in Genome Maintenance in
- The TRIM-NHL Protein LIN-41 Controls the Onset of Developmental Plasticity in
- Wnt-Mediated Repression via Bipartite DNA Recognition by TCF in the Hematopoietic System
- KDM6 Demethylase Independent Loss of Histone H3 Lysine 27 Trimethylation during Early Embryonic Development
- Integration of UPR and Oxidative Stress Signaling in the Control of Intestinal Stem Cell Proliferation
- miR171-Targeted Scarecrow-Like Proteins Bind to GT -Elements and Mediate Gibberellin-Regulated Chlorophyll Biosynthesis under Light Conditions
- Syndecan-1 Is Required to Maintain Intradermal Fat and Prevent Cold Stress
- LIN-3/EGF Promotes the Programmed Cell Death of Specific Cells in by Transcriptional Activation of the Pro-apoptotic Gene
- A System for Genome-Wide Histone Variant Dynamics In ES Cells Reveals Dynamic MacroH2A2 Replacement at Promoters
- Multiple Regulation of Rad51-Mediated Homologous Recombination by Fission Yeast Fbh1
- A Genome-Wide Association Study of the Maize Hypersensitive Defense Response Identifies Genes That Cluster in Related Pathways
- The RNA Helicases AtMTR4 and HEN2 Target Specific Subsets of Nuclear Transcripts for Degradation by the Nuclear Exosome in
- Asymmetric Division and Differential Gene Expression during a Bacterial Developmental Program Requires DivIVA
- A Model-Based Approach for Identifying Signatures of Ancient Balancing Selection in Genetic Data
- Chromatin Insulator Factors Involved in Long-Range DNA Interactions and Their Role in the Folding of the Drosophila Genome
- Conditional Inactivation of Upstream Binding Factor Reveals Its Epigenetic Functions and the Existence of a Somatic Nucleolar Precursor Body
- Evidence for Divisome Localization Mechanisms Independent of the Min System and SlmA in
- Patterns of Admixture and Population Structure in Native Populations of Northwest North America
- Response Regulator Heterodimer Formation Controls a Key Stage in S Development
- A Genetic Strategy to Measure Circulating Insulin Reveals Genes Regulating Insulin Production and Secretion
- EVA-1 Functions as an UNC-40 Co-receptor to Enhance Attraction to the MADD-4 Guidance Cue in
- Dysfunction of the CNS-Heart Axis in Mouse Models of Huntington's Disease
- An Otx/Nodal Regulatory Signature for Posterior Neural Development in Ascidians
- Phosphorylation of a Central Clock Transcription Factor Is Required for Thermal but Not Photic Entrainment
- Genome-Wide Patterns of Genetic Variation within and among Alternative Selective Regimes
- EF-P Dependent Pauses Integrate Proximal and Distal Signals during Translation
- Ku-Mediated Coupling of DNA Cleavage and Repair during Programmed Genome Rearrangements in the Ciliate
- Functional Specialization Among Members Of Knickkopf Family Of Proteins In Insect Cuticle Organization
- Playing RNase P Evolution: Swapping the RNA Catalyst for a Protein Reveals Functional Uniformity of Highly Divergent Enzyme Forms
- The Translational Regulators GCN-1 and ABCF-3 Act Together to Promote Apoptosis in
- Meta-Analysis of Genome-Wide Association Studies in African Americans Provides Insights into the Genetic Architecture of Type 2 Diabetes
- A -Regulatory Mutation of Causes Silky-Feather in Chickens
- VIB1, a Link between Glucose Signaling and Carbon Catabolite Repression, Is Essential for Plant Cell Wall Degradation by
- A Population Genetic Signal of Polygenic Adaptation
- A Conserved Dopamine-Cholecystokinin Signaling Pathway Shapes Context–Dependent Behavior
- The MAP Kinase p38 Is Part of Circadian Clock
- The Cohesin Subunit Rad21 Is Required for Synaptonemal Complex Maintenance, but Not Sister Chromatid Cohesion, during Drosophila Female Meiosis
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Meta-Analysis of Genome-Wide Association Studies in African Americans Provides Insights into the Genetic Architecture of Type 2 Diabetes
- KDM6 Demethylase Independent Loss of Histone H3 Lysine 27 Trimethylation during Early Embryonic Development
- The RNA Helicases AtMTR4 and HEN2 Target Specific Subsets of Nuclear Transcripts for Degradation by the Nuclear Exosome in
- EF-P Dependent Pauses Integrate Proximal and Distal Signals during Translation
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



