-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
miR171-Targeted Scarecrow-Like Proteins Bind to GT -Elements and Mediate Gibberellin-Regulated Chlorophyll Biosynthesis under Light Conditions
Chlorophyll biosynthesis is essential for plant growth and development. To date, the regulatory mechanisms of chlorophyll biosynthesis have been well understood only in dark conditions. Previous reports showed that miR171-targeted SCL6/22/27 proteins were involved in chlorophyll biosynthesis. However, the molecular mechanism of SCL action remains unclear. In this study, we found that SCLs negatively regulated chlorophyll biosynthesis though suppressing the expression of the key gene PROTOCHLOROPHYLLIDE OXIDOREDUCTASE (POR). SCL27 is highly expressed at the basal cell proliferation region of young leaves, suggesting an important role of SCLs in inhibiting chloroplast development before cell expansion. In addition, GT-cis elements were required for SCL27 directly binding to the PORC promoter. Furthermore, we showed that SCLs mediated GA-regulated chlorophyll biosynthesis through direct interaction with DELLA proteins. The interaction between SCLs and DELLAs reduced the DNA binding activity of SCL27. Our uncovered GA-DELLA-SCL module and its DNA binding targets provide new insights into molecular mechanisms by which chlorophyll biosynthesis and cell proliferation are coordinately regulated during leaf development in response to developmental and environmental cues.
Published in the journal: . PLoS Genet 10(8): e32767. doi:10.1371/journal.pgen.1004519
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004519Summary
Chlorophyll biosynthesis is essential for plant growth and development. To date, the regulatory mechanisms of chlorophyll biosynthesis have been well understood only in dark conditions. Previous reports showed that miR171-targeted SCL6/22/27 proteins were involved in chlorophyll biosynthesis. However, the molecular mechanism of SCL action remains unclear. In this study, we found that SCLs negatively regulated chlorophyll biosynthesis though suppressing the expression of the key gene PROTOCHLOROPHYLLIDE OXIDOREDUCTASE (POR). SCL27 is highly expressed at the basal cell proliferation region of young leaves, suggesting an important role of SCLs in inhibiting chloroplast development before cell expansion. In addition, GT-cis elements were required for SCL27 directly binding to the PORC promoter. Furthermore, we showed that SCLs mediated GA-regulated chlorophyll biosynthesis through direct interaction with DELLA proteins. The interaction between SCLs and DELLAs reduced the DNA binding activity of SCL27. Our uncovered GA-DELLA-SCL module and its DNA binding targets provide new insights into molecular mechanisms by which chlorophyll biosynthesis and cell proliferation are coordinately regulated during leaf development in response to developmental and environmental cues.
Introduction
Chlorophylls are complexed with their binding proteins and serve two primary functions in photosynthesis: they trap light energy and transfer it to the reaction centers of photosystems [1], [2]. During light absorption and energy transfer, chlorophylls inevitably generate highly reactive singlet oxygen, particularly under strong light, leading to the inhibition of photosynthesis, plant growth and even to cell death [3], [4]. In addition, many chlorophyll precursors present in their free state are strong photosensitizers that produce reactive oxygen species upon light illumination. Therefore, the chlorophyll biosynthetic pathway is strictly regulated in response to developmental and environmental cues.
It has been well documented that chlorophyll biosynthesis is finely regulated at the multiple steps in the pathway and at both transcriptional and post-transcriptional levels [5]. For example, protochlorophyllide (Pchlide) levels of etiolated seedlings are negatively regulated by the phytochrome-interacting factors PIF1 and PIF3-5 [6]–[9], but positively regulated by two transposase-derived transcription factors, FAR1 (far-red impaired response 1) and FHY3 (far-red elongated hypocotyls 3) [10]; the activity of the key enzyme glutamyl-tRNA reductase (HEMA1) is inhibited directly by heme and the FLU protein via a feedback mechanism [2], [11]–[15], while Mg-chelatase is stimulated by the binding of genomes uncoupled 4 (GUN4) to the ChlH subunit (GUN5) of Mg-chelatase, protoporphyrin IX (PPIX) and Mg-PPIX [16]–[19]. It is worth emphasizing that the activity of the key enzyme Pchlide oxidoreductase (POR) is primarily subject to the transcriptional regulation [8], [20]–[21]. The Arabidopsis genome contains three differentially regulated POR genes. It has been shown that PORA is expressed in etiolated seedlings and its mRNA level drops sharply in light; PORB is expressed in both etiolated seedlings and light-grown plants; PORC expression is activated by light in a fluence rate-dependent manner [22]–[24]. Available evidence revealed that the expression of PORA and PORB is regulated by the transcription factors ethylene insensitive 3 (EIN3) and its homolog EIN3-like1 (EIL1) via directly binding to the EBS cis-elements in the promoter region [21]. Although PORC expression was reported to be directly induced by PIF1 [8], it remains unclear how PORC is regulated in light where PIF proteins are degraded.
Gibberellic acid (GA) is an important phytohormone that controls many aspects of plant development and growth via the GA-GID-DELLA signaling module in Arabidopsis [25]–[30]. With regard to the chlorophyll biosynthetic pathway, DELLA stabilization in the GA-deficient ga1-3 mutant leads to increased accumulation of Pchlide and PORs in etiolated seedlings, which are substantially more resistant to photo-oxidative damage after transferred from darkness to light [20]. DELLAs promote Pchlide biosynthesis by repressing the transcriptional activity of PIFs in the dark [20], [31], [32]. In contrast, the molecular mechanism underlying the DELLA-regulated POR expression is not fully understood. Recently, the miR171-targeted scarecrow-like (SCL) transcription factors SCL6/SCL6-IV, SCL22/SCL6-III and SCL27/SCL6-II (also known as hairy meristems [HAM] and lost meristems [LOM]) have been demonstrated to play an important role in the proliferation of meristematic cells, polar organization and chlorophyll synthesis [33]–[38]. However, it remains unknown how these SCL proteins control chlorophyll synthesis. Here, we provide convincing evidence that DELLA-regulated POR expression is, at least in part, mediated by miR171-targeted SCLs in light.
Results
miR171-targeted SCLs regulate chlorophyll biosynthesis via the key enzyme POR
As previously reported [38], both MIR171c over-expressors (MIR171c-OX) and scl6 scl22 scl27 triple mutants produce dark green leaves (Figure 1A and Figure S1A), which contain approximately 40% more chlorophyll than wild type (WT) leaves (Figure 1B). In contrast, the over-expression of miR171-resistant LUC-rSCL27 (fused to the luciferase gene) results in leaf yellowing (Figure 1A and Figure S1A) and a significant decrease in chlorophyll content (Figure 1B). These results indicate that miR171-targeted SCLs are negative regulators of chlorophyll biosynthesis.
Fig. 1. POR is critical for SCL-regulated chlorophyll biosynthesis in light. 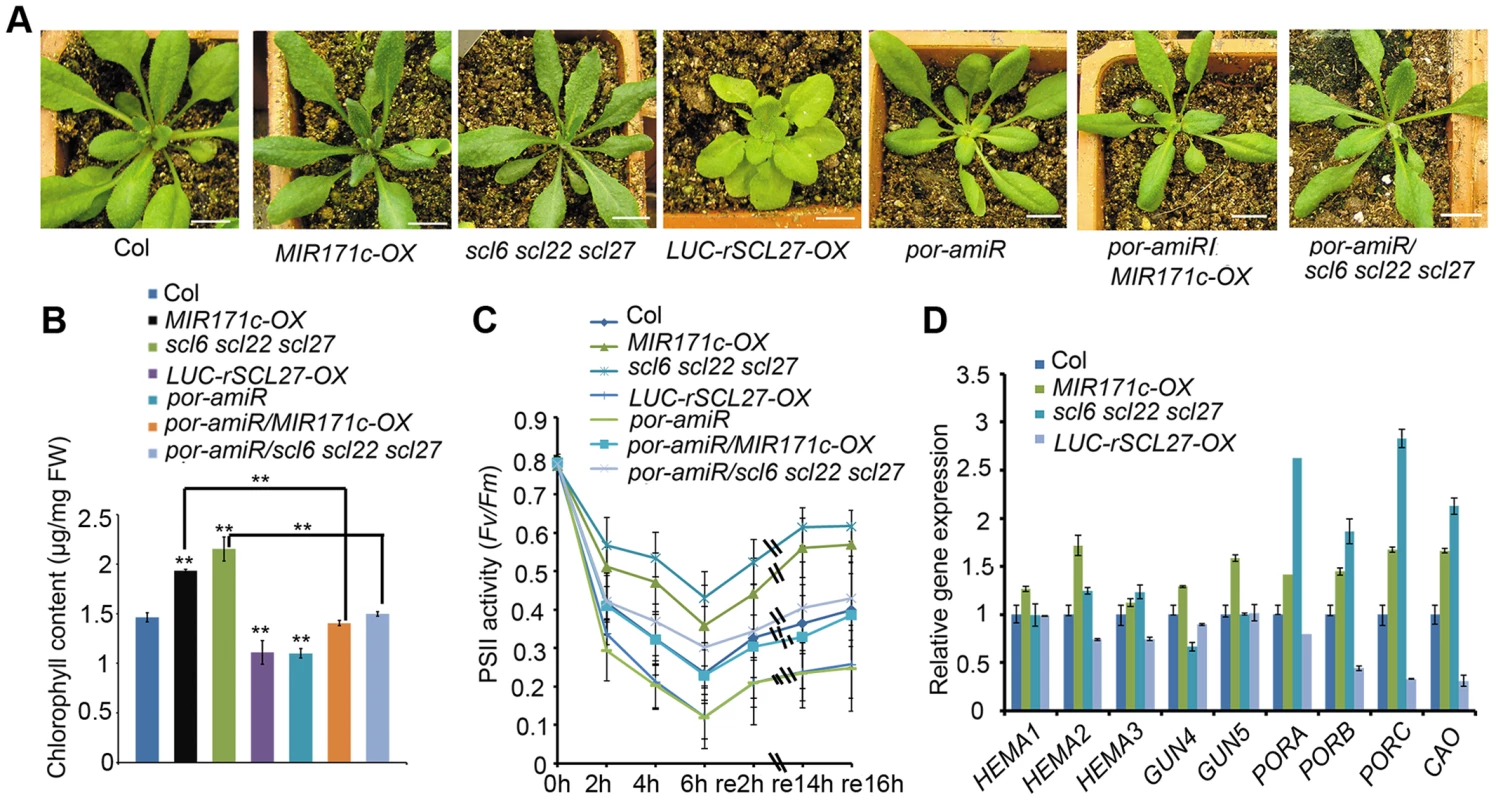
(A) Phenotypes of WT (Col), MIR171c-OX, scl6 scl22 scl27, 35S::LUC-rSCL27, por-amiR, por-amiR/MIR171c-OX and por-amiR/scl6 scl22 scl27 plants grown under a 16 h/8 h light/dark cycle. Bars = 1 cm. (B) Chlorophyll content of the genotypes shown in (A). FW, fresh weight. ** indicates p values (Student's t-test) <0.01 compared with WT or between the indicated two genotypes. Error bars indicate the s.d. (n = 4). (C) PSII activity (Fv/Fm) of the leaves described in (A) treated with excess light (800 µmol m−2 s−1) for the indicated times and then incubated in the dark. Error bars indicate the s.d. (n = 18). (D) qPCR analysis of HEMAs, GUN4, GUN5, PORs and CAO transcript levels using total RNA extracted from the leaves of the genotypes shown in (A). The relative expression levels were normalized to that of ACTIN2, and the relative expression in WT plants was set as 1. Error bars represent the s.d. (n = 3). Two biological replicates were performed and provided similar results. To explore the physiological role of SCL proteins in the regulation of chlorophyll biosynthesis, we constructed transgenic plants expressing rSCL27 fused to the β-glucuronidase (GUS) gene driven by the SCL27 native promoter, designated pSCL27::rSCL27-GUS. We examined the pattern of GUS expression in the 3 - to 11-day-old seedlings. The results of GUS staining clearly showed that the SCL27-GUS fusion protein started to accumulate in the newly developed leaves (Figure S2). In the first pair of leaves, the GUS signal was first observed in the 3-day-old seedlings through the whole leaves, and maintained at a relative stable level at the basal region until to the 7-day-old seedlings, and suddenly disappeared in the 8-day-old seedlings (Figure 2A–2B and Figure S2). Consistent with this observation, the scl6 scl22 scl27 mutant exhibited more intense chlorophyll fluorescence at the base of leaves than did the WT, whereas chlorophyll fluorescence intensity at the leaf apical region was identical to that in the WT (Figure 2C and 2D), suggesting that SCL proteins play an important role in inhibiting chloroplast development before cell expansion. This result is consistent with a previous report that leaf greening and cell expansion initiate at the leaf tip and proceed in a basipetal direction [39].
Fig. 2. SCL-GUS accumulation and chlorophyll fluorescence intensity at the early stage of leaf growth. 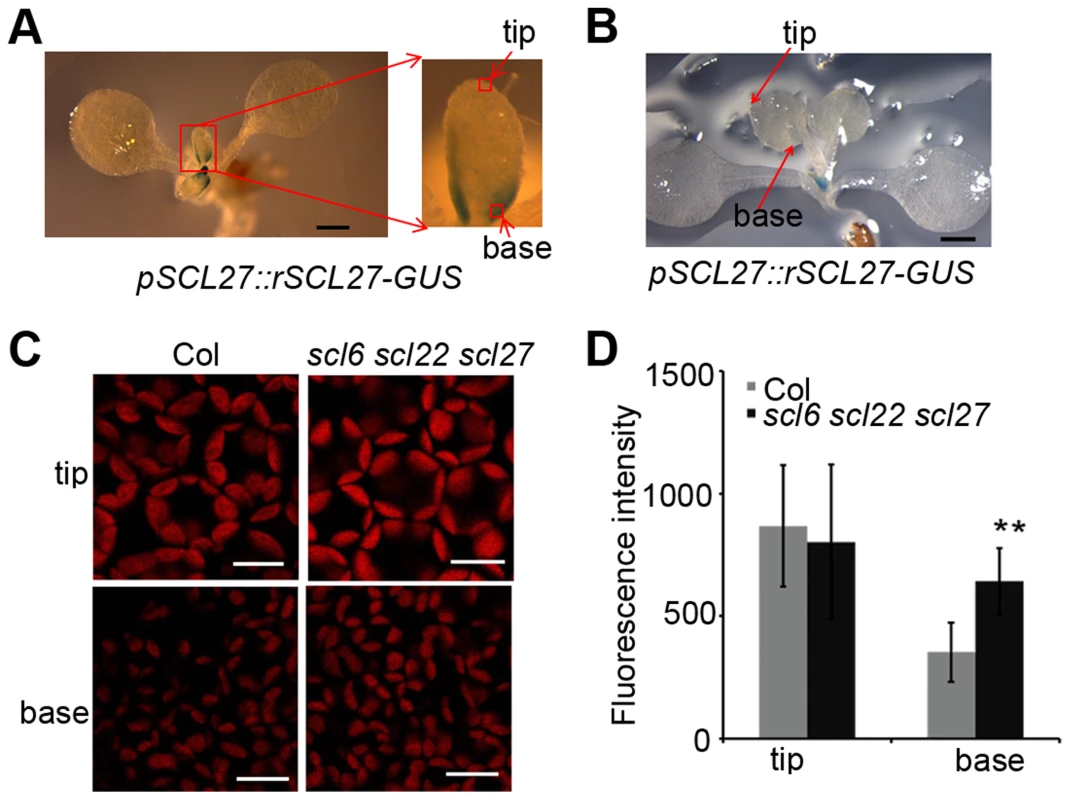
(A) The GUS activity of 6-day-old transgenic plants expressing pSCL27::rSCL27-GUS. Bar = 1 mm. (B) The GUS activity of 8-day-old transgenic plants expressing pSCL27::rSCL27-GUS. Bar = 1 mm. (C) Chlorophyll autofluorescence from the leaf tip and basal cells shown in (A). Bars = 20 µm. (D) Fluorescence intensity in the tip and basal cells of 6-day-old Col and scl6 scl22 scl27 seedlings. ** represents p values (Student's t-test) <0.01 relative to wild-type. Error bars indicate the s.d. (n = 18). We further evaluated the role of SCLs in plant adaptation to high light stress by measuring the ratio of variable fluorescence to maximum fluorescence (Fv/Fm), which reflects the maximal photochemical efficiency of photosystem II (PSII) photochemistry (PSII activity). Compared to WT plants, PSII activity decreased more slowly in MIR171c-OX and scl6 scl22 scl27 plants but decreased more rapidly in LUC-rSCL27-OX plants (Figure 1C) in light stress, indicating that miR171-targeted SCLs are also involved in plant adaptation to excess light. We also investigated the role of SCLs in the growth of etiolated seedlings and chloroplast development. As shown in Figure S3A–S3C, manipulation of SCL gene expression slightly but not significantly affected greening ratio, Pchlide content and etioplast ultrastructure of the 5-day-old dark-grown seedlings. However, stacked and stromal thylakoid membranes were thicker in chloroplasts from MIR171c-OX and scl6 scl22 scl27 mature leaves while was thinner in those from LUC-rSCL27-OX leaves, compared to WT (Figure S4A–S4B). Consistently, immunoblotting analysis showed that the levels of light-harvesting complex subunits including LHCB1, LHCB2, LHCB5, and LHCA1 were higher in MIR171c-OX and scl6 scl22 scl27 than in WT but lower in LUC-rSCL27 (Figure S4C). However, changes in SCL expression had no effect on the accumulation of PsaD (PSI subunit) and AtpB (ATP synthase beta subunit) in mature leaves (Figure S4C). Taken together, these results indicate that SCLs are involved in chlorophyll biosynthesis mainly in light but not in the dark.
To elucidate the molecular mechanism underlying SCL-regulated chlorophyll synthesis, we analyzed the transcriptional levels of several key genes in the pathway, including the genes encoding HEMA1, GUN4, GUN5, PORs and chlorophyll a oxygenase (CAO). Quantitative PCR (qPCR) and northern blotting assays showed that among the inspected genes the levels of PORs and CAO transcripts were higher in MIR171c-OX and scl6 scl22 scl27 while were lower in LUC-rSCL27-OX, compared to those in the WT (Figure 1D and Figure S1B). The expression levels of PORs and CAO were correlated well with chlorophyll content in the leaves of MIR171c-OX, scl triple mutant and LUC-rSCL27-OX plants, suggesting that the expression of PORs and CAO is regulated by SCLs. Immunoblotting analysis using a POR antibody that can recognize all three isoforms of POR showed that MIR171c-OX and scl6 scl22 scl27 plants accumulated higher levels of PORC and PORB proteins than did WT and LUC-rSCL27-OX plants (Figure S1C). Thus, the data obtained indicate that the expression of POR, the key enzyme in the chlorophyll biosynthetic pathway, is negatively regulated by SCLs.
To verify the role of POR in SCL-regulated chlorophyll synthesis, we down-regulated the expression of POR in WT and scl6 scl22 scl27 mutant plants using an artificial microRNA that was designed to specifically target the three POR genes. Transgenic plants (por-amiR) with substantially reduced levels of POR expression were identified using qPCR (Figure S1D and S1E). Knocking down POR expression in WT, MIR171c-OX and scl triple mutant plants led to a pale-green phenotype and a lower level of chlorophyll and PSII activity than in the corresponding controls (Figure 1A–1C). Taken together, these data indicate that miR171-targeted SCLs regulate chlorophyll biosynthesis via the key enzyme POR.
SCL27 binds to the PORC promoter
The important role of PORs in SCL-regulated chlorophyll biosynthesis prompted us to investigate whether SCLs can directly control the promoter activity of POR genes. Because both PORC and MIR171 are regulated by light but not by the circadian clock [22]–[24], [40], we hypothesized that PORC was a direct target of SCLs. To test this hypothesis, we co-expressed the LUC reporter gene under the control of the PORC promoter (a 1685-bp genomic fragment upstream of the start codon) together with 6xMYC-rSCL27 in Nicotiana benthamiana leaves using a transient expression system. The expression of LUC was much lower in the leaves transformed with 6xMYC-rSCL27 than in leaves transformed with the empty vector and rSCL27-VP16 (a transcriptional activator) (Figure 3A), suggesting that the PORC promoter is a direct target of SCL27. To identify the PORC promoter region bound by SCL27, three fragments extending from the PORC start codon to −1685, −861 and −455 bp upstream were fused to the LUC reporter gene. LUC expression under the control of either pPORC-1685 or pPORC-861 was significantly reduced by 6xMYC-rSCL27 but not by rSCL27-VP16, whereas LUC expression driven by pPORC-455 was low and unaffected by 6xMYC-rSCL27 or rSCL27-VP16 (Figure 3A). Consistently, LUC expression under the control of pPORC-1685 or pPORC-861 was higher in MIR171c-OX and scl6 scl22 scl27 than in WT (Col), whereas LUC expression driven by pPORC-455 did not significantly differ between WT and MIR171c-OX or between WT and scl6 scl22 scl27 (Figure S5A). These data suggest that the promoter region between −861 bp and −455 bp is required for SCL27 binding to the PORC promoter.
Fig. 3. SCL27 binds to the PORC promoter. 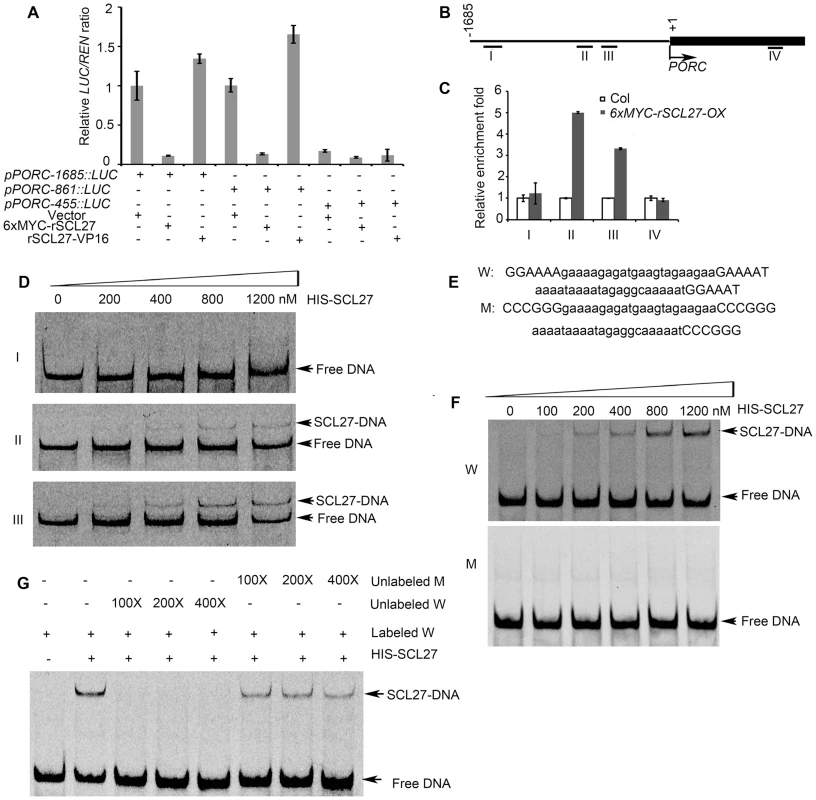
(A) Effect of SCL27 on the activity of three PORC promoter regions. The LUC reporter gene under the control of these promoter regions was transformed into N. benthamiana leaves, with or without 6xMYC-rSCL27 or rSCL27-VP16. The relative LUC activities were normalized to a 35S::REN internal control. Error bars indicate the s.d. (n = 4). Three biological replicates provided similar results. (B) Schematic diagram of the PORC promoter and the first exon region. Fragments Ι (−1524 bp to −1324 bp), ΙΙ (−778 bp to −598 bp), ΙΙΙ (−572 bp to −372 bp) and IV (1144 bp to 1246 bp) were used for ChIP. (C) ChIP-qPCR analysis of the relative enrichment of the DNA fragments mentioned in (B). The β-TUBULIN-2 promoter was used as a reference. Error bars indicate the s.d. (n = 3). Two biological replicates were performed and showed similar results. (D) EMSA analysis of SCL27 binding to fragments I, II and III. (E) DNA sequences. W and M contain GT and mutated-GT elements indicated by capital letters, respectively. (F) SCL27 binding to GT elements was analyzed using the indicated levels of purified SCL27 protein mixed with 1 nM of Cy5-fluorescently labeled 62-bp DNA fragments. (G) The specificity of the SCL27-DNA interaction was tested using a competition assay with 0.1, 0.2 and 0.4 µM of unlabeled W or unlabeled M fragments. We then performed chromatin immuno-precipitation (ChIP) and qPCR assays to further define the SCL27-binding region within the PORC promoter (Figure 3B). Our results showed that fragments II (−778 bp to −598 bp) and III (−572 bp to −372 bp) were enriched in immuno-precipitates from the transgenic plants over-expressing 6xMYC-rSCL27 but not in those from WT plants (Figure 3C), whereas fragments I (−1524 bp to −1324 bp) and IV (1144 bp to 1246 bp of the coding sequence, used as a negative control) were not enriched (Figure 3C), indicating that fragments II and III contain SCL27-binding cis-elements. We next performed electrophoretic mobility shift assays (EMSAs) to confirm whether SCL27 can directly bind to fragments II and III of the PORC promoter. Consistent with the ChIP-qPCR results, shifted bands were observed when purified recombinant SCL27 protein (Figure S5B) was incubated with DNA fragments II or III, and the intensity of the bands gradually increased with increasing concentrations of SCL27 (Figure 3D). However, no shifted band was detected when SCL27 was incubated with fragment I (Figure 3D). Taken together, our in vivo and in vitro data suggest that SCL27 inhibits PORC expression via directly binding to the PORC promoter.
GT elements have been reported to be important for light-regulated gene expression, and DNA fragments II and III contain these cis-elements [41]. To test whether GT-elements are important for SCL27 binding to the PORC promoter, we chose the 62-bp DNA fragment from −500 bp to −438 bp, which contains three G(A/G)(A/T)AA(A/T) GT element repeats [41] (Figure 3E). The EMSA results showed that purified recombinant SCL27 bound to the W fragment but not to the M fragment (Figure 3F). The formation of the SCL27-DNA complex was suppressed by a 100-, 200 - or 400-fold excess of unlabeled W fragment, but not by the unlabeled M fragment (Figure 3G). Thus, we conclude that GT elements are required for SCL27 to bind to the PORC promoter.
SCLs mediate DELLA-regulated POR expression in light
DELLA proteins up-regulate the expression of PORs either in a PIF-dependent or PIF-independent manner [20]. We therefore tested whether miR171-targeted SCLs mediated DELLA-regulated POR expression. For this purpose, we generated LUC-rSCL27-OX/ga1-3 or pSCL27::rSCL27/pRGA::RGAd17 (the GA-insensitive form of RGA) plants via sexual crossing. Over-expressing LUC-rSCL27 in the WT or ga1-3 genetic background led to pale-green phenotypes and significantly decreased chlorophyll content (Figure 4A, 4B and Figure S6A). Likewise, expressing pSCR27::rSCL27 in the pRGA::RGAd17 plants also resulted in a pale-green phenotype and decreased chlorophyll content (Figure S6B and S6C). qPCR analysis showed that over-expressing LUC-rSCL27 in WT and ga1-3 plants led to a dramatic decrease in PORC expression (Figure 4C). To confirm the epistasis of SCLs to DELLAs in the regulation of chlorophyll biosynthesis, we over-expressed MIR171c in WT (Ler) and della pentuple mutants. Indeed, over-expressing MIR171c in these plants resulted in dark green leaves and increased chlorophyll content (Figure 4D and 4E). Consistently, the level of PORC expression was higher in MIR171c over-expressors than in the corresponding control WT (Ler) and della pentuple plants (Figure 4F). These data indicate that DELLA-promoted chlorophyll biosynthesis and PORC expression are dependent on SCLs.
Fig. 4. miR171-targeted SCLs mediate DELLA-regulated POR expression in light. 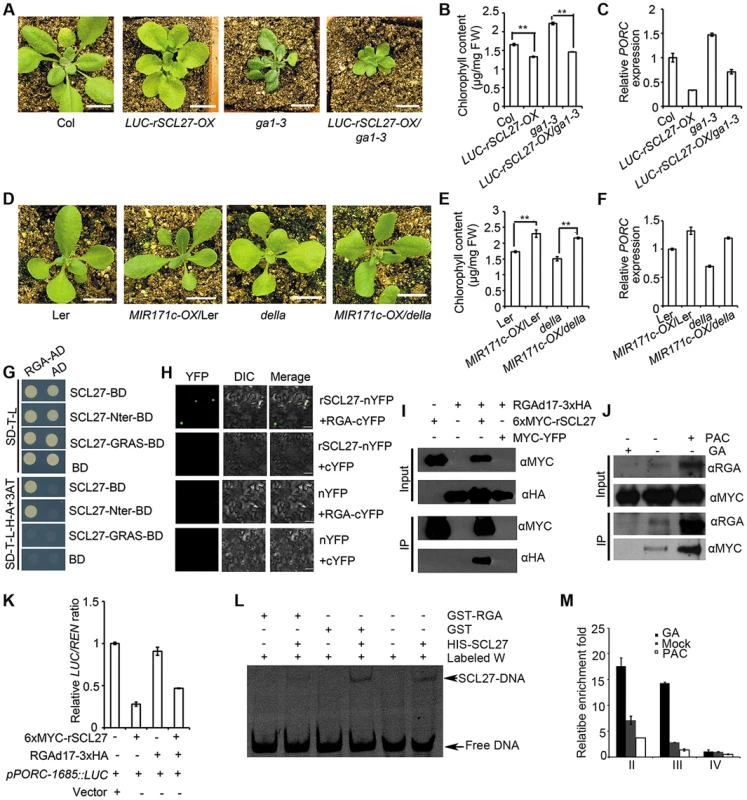
(A and D) Phenotypes of the indicated plants grown under long-day conditions. Bars = 1 cm. (B, C, E and F) Chlorophyll content (B and E) and relative PORC mRNA levels (C and F) of the plants shown in (A and D). ** indicates p values (Student's t-test) <0.01 between the indicated two genotypes; error bars indicate the s.d. (n = 4). PORC expression levels were normalized to that of ACTIN2, and the level of PORC expression in Col or Ler was set as 1. Error bars indicate the s.d. (n = 3). Two biological replicates were performed and showed similar results. FW, fresh weight. (G) Domain mapping of the interaction between SCL27 and RGA in yeast. (H) BiFC analysis of the interaction between SCL27 and RGA. Bars = 50 µm. (I) Co-IP assay of the interaction between SCL27 and RGA using a transient expression assay in N. benthamiana leaves. Fusion proteins were detected by immunoblotting with anti-MYC or anti-HA antibodies. (J) Transgenic plants over-expressing 6xMYC-rSCL27 were used for co-IP. Arabidopsis proteins were detected by anti-MYC or anti-RGA antibodies. (K) Effect of DELLA binding to SCL27 on PORC promoter activity. The pPORC-1685::LUC reporter gene was transformed with 6xMYC-rSCL27 and/or RGAd17-3xHA in N. benthamiana. Relative LUC activities were normalized to the 35S::REN internal control. The LUC/REN ratio in the leaves transformed with the vector was set as 1. Error bars indicate the s.d. (n = 4). Three biological replicates showed similar results. (L) The binding of SCL27 to DNA was analyzed using EMSA in the presence of RGA. GST was used as a control. (M) in vivo analysis of the binding of SCLs to PORC promoter regions in the presence of GA3 or PAC. Three-week-old 6xMYC-rSCL27-OX plants treated with GA or PAC for 2 days were used for ChIP and qPCR experiments. The β-TUBULIN-2 promoter was used as a reference. Error bars indicate the s.d. (n = 3). Two biological replicates showed similar results. To examine the role of the DELLA-SCL module in chlorophyll biosynthesis in the dark, we measured the greening ratio and Pchlide content of 5-day-old etiolated WT, MIR171c-OX, scl6 scl22 scl27 and LUC-rSCL27-OX seedlings in the presence of paclobutrazol (PAC), which increases the levels of DELLA proteins. Our results showed that changes in SCL expression did not affect the greening ratio and Pchlide content in the absence or presence of PAC (Figure S7A and S7B), indicating that SCLs are not involved in DELLA-promoted Pchlide biosynthesis in the dark.
We then tested whether DELLA proteins directly interact with SCLs in vitro and in vivo. Yeast two-hybrid assays showed that the DELLA protein RGA interacted with SCL27 via the N-terminal domain (Figure 4G). In addition, SCL22 bound to all DELLA proteins in yeast (Figure S8A), indicating that the interaction between SCLs and DELLAs is universal. The in vivo interaction between SCLs and DELLAs was examined by bimolecular fluorescence complementation (BiFC) and co-immunoprecipitation (Co-IP) assays using a transient expression system. A strong YFP signal was observed when either rSCL27-nYFP and RGA-cYFP or rSCL27-Nter-nYFP and RGA-cYFP were co-expressed in leaves (Figure 4H and Figure S8B). Co-IP results also showed that RGAd17-3xHA bound to 6xMYC-rSCL27 but not to the control MYC-YFP (Figure 4I). Furthermore, MYC-SCL27 was precipitated by the antibody against RGA in total proteins extracted from transgenic plants over-expressing 6XMYC-rSCL27 treated with PAC but not treated with GA3 (Figure 4J). These results demonstrate that RGA interacts directly with SCL27 both in vitro and in vivo. In addition, qPCR assays showed that the accumulation of SCL transcripts was not altered in plants treated with GA3 (Figure S9A) or PAC (Figure S9B), or in GA mutants, including ga1-3, gai-2 or rga gal1 rgl2 rgl3 plants (Figure S9C). Likewise, RGA and GAI expression was not apparently affected by SCL levels (Figure S9D). Thus, these results exclude the possibility that DELLAs and SCLs are mutually regulated at the transcriptional level.
DELLAs have been shown to regulate various biological processes by preventing transcription factors from binding to DNA [31]–[32], [42]–[46]. The antagonistic role of DELLAs and SCLs in the regulation of chlorophyll biosynthesis raises the possibility that DELLAs might inhibit SCL binding to DNA. To test this hypothesis, we analyzed the promoter activity of PORC using a dual-luciferase reporter assay by transforming RGAd17-3xHA and/or 6xMYC-rSCL27 into N. benthamiana leaves. The results showed that pPORC-1685::LUC reporter activity was significantly suppressed by 6xMYC-rSCL27 but was not affected by RGAd17-3xHA. The degree of inhibition of pPORC-1685::LUC reporter activity by SCL27 was partially mitigated by the co-expression of RGAd17-3xHA (Figure 4K). Consistent with these results, EMSA analysis showed that the interaction between RGA and SCL27 decreased the binding of SCL27 to DNA (Figure 4L and Figure S10A, S10B). Furthermore, ChIP-qPCR analysis also showed that the enrichment of fragments II and III containing GT elements in the PORC promoter (shown in Figure 2B) was higher in MYC antibody pulled-down precipitates from GA-treated 6xMYC-rSCL27-OX plants but lower in those from PAC-treated 6xMYC-rSCL27-OX plants than in those from plants given the mock treatment (Figure 4M and Figure S10C). Thus, our data demonstrate that the RGA-SCL27 interaction decreases SCL27 DNA-binding activity.
SCL27 activates MIR171 gene expression
In general, the level of miRNA expression is inversely correlated with the level of target gene expression. However, miR171 accumulation was reported to peak 6 hours earlier than that of SCL6 [40]. Recent studies have shown that the expression of miRNAs can be controlled by their target genes in a feedback manner [47]. Consistent with this idea, GT elements have been found in the promoters of MIR171s. qPCR assays showed that the expression levels of all MIR171 genes are much higher in LUC-rSCL27-OX plants than in WT plants, whereas the expression levels of these genes are lower in scl6 scl22 scl27 plants than in WT plants (Figure 5A), indicating that SCLs are positive regulators of MIR171 expression. However, the extent to which SCL27 regulated MIR171 expression differed among the MIR171 genes (Figure 5A). For example, SCL27 had the greatest effect on the level of MIR171a expression but had lower, similar effects on the expression levels of MIR171b and MIR171c (Figure 5A).
Fig. 5. SCL27 activates MIR171 gene expression in a feedback manner. 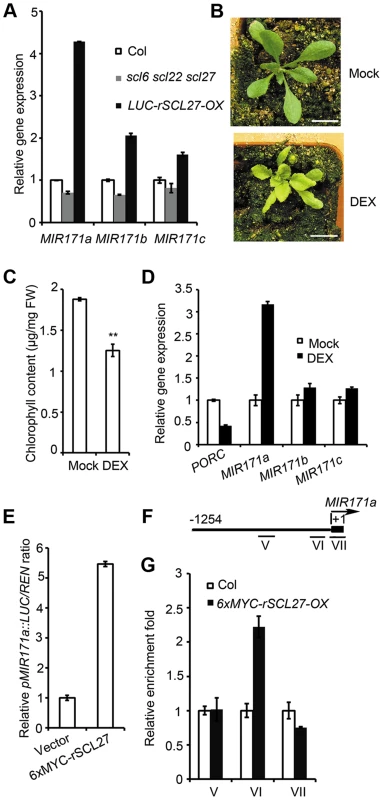
(A) qPCR analysis of MIR171a, MIR171b and MIR171c expression in Col, scl6 scl22 scl27 and LUC-rSCL27-OX plants. Relative expression levels of MIR171 genes were normalized to that of ACTIN2, and the relative expression in WT plants was set as 1. Error bars represent the s.d. (n = 3). Two biological replicates were performed with similar results. (B) 35S::6xMYC-rSCL27-GR/scl6 scl22 scl27 transgenic plants were treated with DEX (10 µM) or untreated (Mock) for 20 days. Bars = 1 cm. (C) Chlorophyll content of the plants shown in (B). ** indicates p value (Student's t-test) <0.01; Error bars indicate the s.d. (n = 4). (D) Relative expression of PORC, MIR171a, MIR171b and MIR171c in the plants shown in (B). Expression levels were normalized to that of ACTIN2. Expression levels in plants without DEX were set as 1 for each gene. Error bars represent the s.d. (n = 3). Two biological replicates were performed with similar results. (E) Relative activity of the MIR171a promoter. pMIR171a::LUC was transformed into N. benthamiana leaves with or without co-transformation of 6xMYC-rSCL27. Relative LUC activities were normalized to the 35S::REN internal control. Error bars indicate the s.d. (n = 4). Three biological replicates showed similar results. (F) Schematic diagram of MIR171a promoter regions V (−726 bp to −495 bp), VΙ (−260 bp to −71 bp) and VΙΙ (the precursor of MIR171a), which were used for ChIP experiments. (G) Relative enrichment of MIR171a promoter fragments in the immuno-precipitates. Leaves of 3-week-old Col and 6xMYC-rSCL27-OX plants were used for ChIP experiments. The enriched DNA fragments were quantified using qPCR. The β-TUBULIN-2 promoter was used as a reference. Error bars indicate the s.d. (n = 3). Similar results were obtained from three independent immuno-precipitation experiments. Additionally, we generated transgenic plants expressing 6xMYC-rSCL27 fused to the rat glucocorticoid receptor (GR) under the control of the 35S regulatory sequence in the scl triple mutant background; these plants were designated 35S::6xMYC-rSCL27-GR/scl6 scl22 scl27. Compared to mock (dimethyl sulfoxide, DMSO)-treated plants, transgenic plants treated with 10 µM dexamethasone (DEX) were pale green and accumulated less chlorophyll (Figure 5B and 5C). qPCR analysis showed that the level of PORC mRNA was rapidly decreased in the transgenic plants treated with DEX for 4 hours (Figure 5D). Using this inducible expression system, we found that MIR171a transcripts accumulated to levels more than 3-fold higher in DEX-treated plants than in the control, whereas two other MIR171 genes were only slightly up-regulated by SCL27 (Figure 5D). To confirm the observation that SCL27 activates MIR171 gene expression, the LUC reporter gene driven by the MIR171a promoter (pMIR171a::LUC) was co-transformed with or without 6xMYC-rSCL27 into N. benthamiana leaves. As shown in Figure 5E, pMIR171a::LUC activity was significantly increased by 6xMYC-rSCL27. These results indicate that SCLs can up-regulate MIR171 gene expression. To confirm that SCL27 directly regulates MIR171 gene expression, ChIP-qPCR was performed using three fragments: V (−726 bp to −495 bp, without GT elements), VI (−260 bp to −71 bp, containing GT elements) and VII (the precursor of MIR171a). Indeed, only fragment VI was enriched in MYC antibody pulled-down precipitates obtained from the 6xMYC-rSCL27 over-expressing plants but not in those obtained from the WT plants (Figure 5F and 5G). Taken together, these data clearly indicate that miR171 and its target SCLs form a feedback loop to finely regulate chlorophyll biosynthesis.
Discussion
miR171 and its target SCL proteins have been reported to play an important role in plant development and growth [33]–[38]. However, little is known about the molecular mechanisms by which the miR171-SCL module functions. In this study, we found compelling evidence showing that SCLs are GT element-binding transcriptional factors that can suppress or promote gene expression in Arabidopsis. Given that GT elements are widely distributed in tandem repeats within the promoter regions of many photosynthetic and plastid ribosomal genes [41], it is reasonable to assume that the miR171-SCL module can regulate the expression of other genes in a manner similar to that used for the PORC gene.
In higher plants, light and GA are important signals that antagonistically regulate chloroplast biogenesis, which is a complicated process including chloroplast division and the formation of the photosynthetically active chloroplast [31], [32]. It is well established that PIFs, which are negative regulators of chlorophyll biosynthesis, are critical downstream effectors in light and GA signal transduction pathways [20], [31], [32]. PIFs bind directly to the conserved DNA G-box motif of gene promoters and regulate the chlorophyll biosynthetic pathway by inhibiting Pchlide accumulation and inducing POR gene expression in an additive, redundant or specific manner [6]–[9], [20]. This regulatory mechanism involving PIFs is apparently important for the prevention of free Pchlide accumulation and the subsequent greening of etiolated seedlings upon light exposure [20], [31], [32]. Based on the results derived from this study, we suggest that SCLs play an important role in regulating chlorophyll biosynthesis under light conditions (Figure 6), in which PIFs are rapidly degraded. In addition, PIF proteins can be sequestered by DELLAs, the levels of which are elevated in light and decreased in the dark, blocking the ability of PIFs to bind to their target gene promoters [20]. Thus, the SCLs and PIFs control chlorophyll biosynthesis in different yet cooperative manners, and PIFs are replaced by miR171-targeted SCLs to inhibit chlorophyll biosynthesis in light. Since both the levels of DELLA proteins and miR171 expression are elevated in light, the inhibition of SCLs on chlorophyll biosynthesis is coordinately relieved at transcriptional and post-translational levels, while the positive feedback regulation pathway in which SCLs activate miR171 expression might be important for auto-regulating the homeostasis of SCL proteins in light.
Fig. 6. A working model of GA-regulated chlorophyll biosynthesis under the light condition. 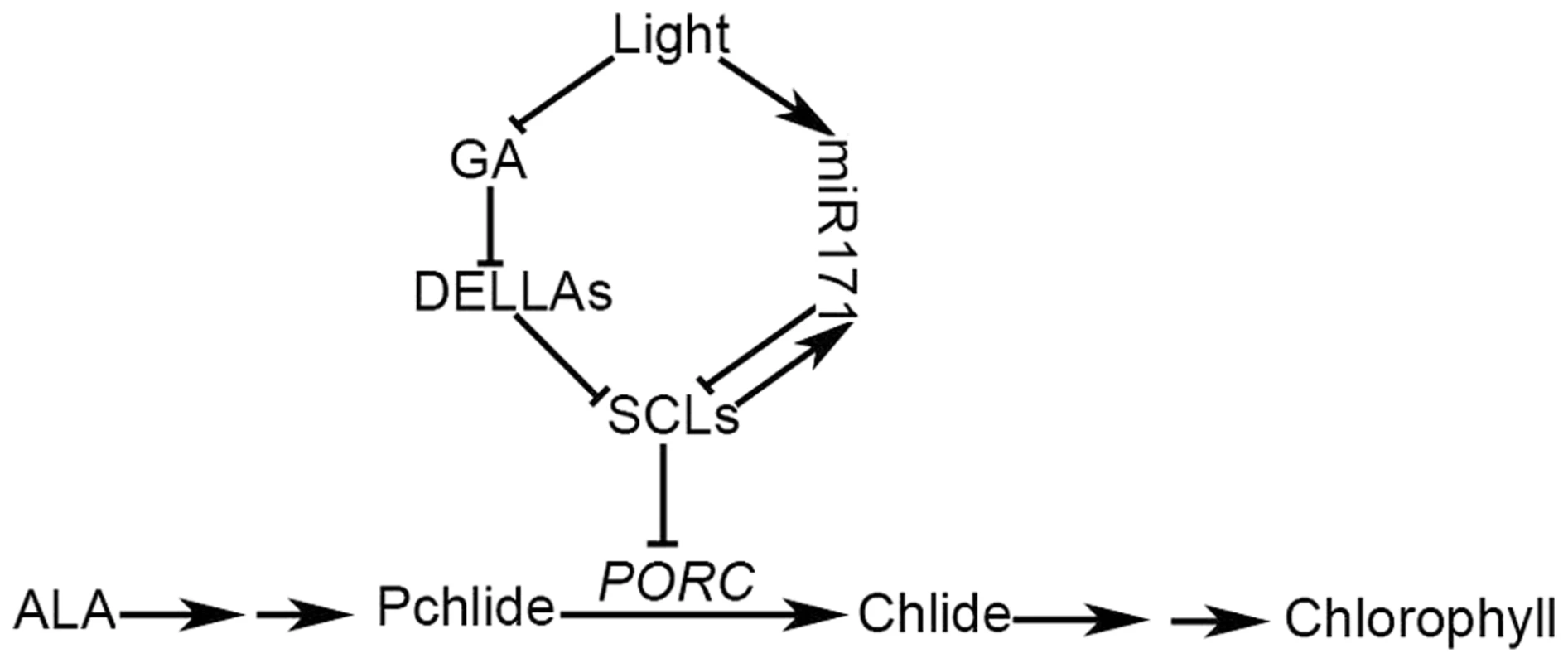
In light, GA biosynthesis is inhibited, resulting in an increase in DELLAs. Accumulated DELLAs bind to SCLs and subsequently terminate the SCL suppression of PORC expression. On the other hand, light promotes miR171 expression, leading to a decrease in SCL expression. Thus, the inhibition of SCLs on PORC expression is maximally relieved in light. miR171 and its target SCLs form a feedback regulatory loop that maintains the light-dependent diurnal oscillation of miR171 and SCL expression. Arrows indicate verified positive regulation and the chlorophyll biosynthesis pathway; bars indicate verified negative regulation. In addition to environmental cues, chloroplast development is regulated by developmental signals. Early leaf growth is divided into two sequential cellular processes after primordium initiation: cell proliferation and cell expansion [39]. Usually, chloroplast development is suppressed in the cell proliferation region at the leaf base, then remains relatively stable over a certain period, and finally is abolished abruptly. Once a cell has stopped proliferating, it enters the stage of cell expansion, which is triggered by chloroplast differentiation [39]. Thus, chloroplast differentiation plays an important role in the timing of the transition from cell proliferation to cell expansion. However, blocking chloroplast differentiation and retrograde signaling from chloroplasts to the nuclei using norflurazon cannot completely stop cell expansion, suggesting that other mechanisms are also involved in the phase shift [39]. Our data showed that a negative regulator of chloroplast development, SCL27, is highly expressed at the base of growing leaves, and chloroplast development proceeds more rapidly in a scl triple mutant than in WT. These results suggest that miR171-targeted SCLs play an important role in suppressing chloroplast development in dividing cells during early leaf growth. Interestingly, leaf size is apparently altered in SCL27 over-expressors and the scl triple mutant compared to that in WT (Figure 1A). One explanation is that SCLs function as coordinators that simultaneously regulate chloroplast development and cell proliferation; another possibility is that the onset of SCL-regulated chloroplast development leads to a change in the timing of cell proliferation exit. Further investigation is required to elucidate the molecular mechanism by which SCLs coordinately regulate leaf size and chloroplast development.
Chloroplast biogenesis is also coordinated with cell expansion during leaf growth to achieve optimal photosynthesis rates. For example, leaf greening accompanies cell expansion, which initiates at the leaf tip and proceeds in a basipetal direction in Arabidopsis [39]. It has been demonstrated that GA plays a critical role in controlling cell expansion and chloroplast biogenesis through DELLA proteins in both dicot and monocot plant species [48]. The number of thylakoid membranes per granum and the chloroplast density per cell are increased in the ga1-3 mutant, indicating that more chlorophyll is synthesized in the mutant chloroplasts. It is likely that DELLA proteins, which are stabilized in the ga1-3 mutant, promote chlorophyll biosynthesis by suppressing the inhibitory transcriptional activity of SCLs. Thus, it appears that the DELLA-SCL module functions to balance chloroplast development and cell expansion, which is accompanied by a dramatic increase in photosynthesis.
Furthermore, we observed another complex phenomenon: over-expression of SCLs did not completely rescue the dark-green phenotype of the ga1-3 mutant, indicating that DELLAs transmit signals that affect chlorophyll biosynthesis by regulating other interacting proteins. A number of DELLA-interacting transcriptional factors have been identified thus far [31]–[32], [42]–[46], including EIN3 and EIL1, which are downstream effectors of ethylene signaling. DELLAs de-repress EIN3 and EIL1 function during apical hook formation in etiolated seedlings [43]. Interestingly, EIN3 and EIL1 were also shown to regulate chlorophyll biosynthesis by repressing the accumulation of Pchlide and by activating the expression of POR genes (PORA and PORB) [21]. It is likely that DELLAs regulate PORA and PORB expression directly via EIN3 and EIL1. DELLAs might also indirectly regulate chlorophyll biosynthesis through other interacting transcriptional factors, including brassinosteroid-resistant 1 (BZR1) and the jasmonic acid ZIM-domain proteins (JAZs) [42], [49], [50]. Taken together, the findings described here indicate that DELLAs are critical factors integrating various signaling pathways to dynamically regulate chlorophyll biosynthesis.
Materials and Methods
Plant materials and growth conditions
MIR171c-OX, scl6 scl22 scl27 triple mutants, 35S::LUC-rSCL27, ga1-3, gai-2 (SAIL_587_CO2), rga rgl1 rgl2 rgl3 mutants, and pRGA::RGAd17 are in the Arabidopsis thaliana Columbia ecotype (Col) background [27], [38], [44]; the ga1-3 mutant was backcrossed with the wild type (Col) for six generations; the por-amiR and pSCL27::SCL27-GUS were transformed in Col background; the por-amiR/MIR171c-OX, LUC-rSCL27-OX/ga1-3, and rSCL27/RGAd17 plants were generated by crossing; the della pentuple is in the Ler ecotype [44]. Seeds were germinated and grown on the half Murashige and Skoog (MS) media containing 1% sucrose and 0.7% phytoagar. All plants were grown at 21°C under light (110 µmol. m−2. s−1) in long days (16-h light/8-h dark).
Plasmid construction and plant transformation
About 1.7 - and 1.2-kb promoter fragments at the upstream of the start codon were amplified from PORC and MIR171a genes in the Col genome, respectively, with primers listed in Table S1. The amplified fragments were inserted in the XhoΙ/BamHI sites of the pGREEN0800LUC vector [51], [52] to produce pPORC-1685::LUC and pMIR171a::LUC vectors. The pPORC-861::LUC and pPORC-455::LUC vectors were constructed in a similar way. To make the POR amiRNA vector, the amiRNA target sequences for POR genes and primers including POR I miR-s, POR II miR-a, POR III miR*s and POR IV miR*a were designed using the WMD3 Web microRNA Designer (http://wmd3.weigelworld.org/cgi-bin/webapp.cgi) and listed in Table S1. The amiRNA precursor was amplified by overlapping PCR from the pRS300 template to produce the fragment containing the POR target amiRNA foldback. DNA fragments were gel-purified and cloned into the Gateway cloning vector pENTR-SD/D/TOPO (Invitrogen) according to the manufacturer's instructions. After sequencing confirmation, the cloned DNA fragments were transferred to the 35S over-expression vector (pGWB2) (Invitrogen) using LR clonase (Invitrogen).
For yeast two-hybrid analysis, SCL22 cDNA was cloned into the pGBKT7 vector (Clontech). RGA, GAI, RGL1, RGL2 and RGL3 cDNAs were cloned into the pGADT7 vector (Clontech). SCL27 and RGA cDNAs were cloned into pDEST22 (Invitrogen); cDNAs encoding SCL27 and its N-terminal (1–267 amino acids) and GRAS domain (268–640 amino acids) were cloned into pDEST32 (Invitrogen). The primers used for these constructs are given in Table S1. For BiFC analysis, SCL27, SCL27 N-terminal, and SCL27-GRAS sequences were cloned into pCAMBIA1300 (nYFP), whereas RGA was cloned into pCAMBIA1300 (cYFP).
For in vitro protein-DNA binding analysis, SCL27 and RGA was cloned into the pET28b and pGEX6p-3 vectors, respectively. The constructs were transformed into the expression strain BL21 for protein expression. For co-IP analysis, RGAd17 and miR171-resistent SCL27 (rSCL27) were cloned into the binary vector with 3xHA or 6xMYC. Transgenic plants were generated by the floral dipping method [53] and were screened with 50 mg/mL of kanamycin sulfate or 50 mg/mL of hygromycin.
Physiological and transmission electron microscopy assays
Seedling greening was analyzed by exposing 5-day-old dark-grown seedlings to white light (16 h-light/8 h-dark) for 2 days. Chlorophyll autofluorescence was analyzed using a confocal laser scanning microscope (Olympus, FV10-ASW). In the PAC-treated etiolated seedlings, 0.01 µM of PAC was used. Greening ratio was determined by counting the percentage of green cotyledons of each genotype. Pchlide was extracted from 5-day-old etiolated seedlings with 1 mL of ice-cold 80% acetone in the dark. The samples were centrifuged at 13000 rpm for 10 min, and fluorescence was excited by the wavelength of 440 nm and scanned from 600 nm to 700 nm using a fluorescence spectrophotometer (Hitachi) at room temperature [54]. The results were presented by relative fluorescence per seedling. Chlorophyll was measured as described previously [55]. The Fv/Fm parameter was measured using light-stressed leaf discs after 15-min adaptation to darkness [56]. For electron microscopy observation, cotyledons of 5-day-old etiolated and 25-day-old seedlings were fixed and processed as previously described [57], and examined with an H-7650 transmission electron microscope (Hitachi).
Yeast two-hybrid assay
Plasmids were transformed into yeast strain AH109 by the lithium chloride–polyethyleneglycol method according to the manufacturer's manual (Clontech). The transformants were selected on SD-Leu-Trp plates. The protein-protein interactions were tested on SD-Trp-Leu -His-Ade plates with or without 3-amino-1, 2, 4-triazole.
BiFC analysis
The A. tumefaciens strain GV3101 transformed with each of the two constructs for BiFC analysis was cultured in the solution containing 10 mM MES, 10 mM MgCl2, and 100 µM acetosyringone to an optical density (OD600) of 0.6 to 0.8. Then, two strains were mixed and incubated at the room temperature for at least 2 h. The YFP fluorescence was analyzed using a confocal laser scanning microscope (Olympus, FV10-ASW) 48 to 96 h after N. benthamiana leaves were infiltrated with the mixture.
GUS staining
Plant materials were submerged in 90% acetone for 15 min, and then transferred into 0.5 mg/mL X-Gluc solution (0.1 M sodium phosphate buffer, pH 7.0, 10 mM EDTA, 0.1% Triton X-100, 0.5 mM potassium ferrocyanide, 0.5 mM potassium ferricyanide). Plant materials were vacuumized, kept at 37°C and decolorized in 70% ethanol.
Co-immunoprecipitation and immunoblot assays
Agrobacteria-infiltrated N. benthamiana leaves and transgenic plants over-expressing 6xMYC-rSCL27 were used for Co-IP analyses. The soluble proteins were extracted with the extraction buffer (50 mM Heps [pH 7.5], 150 mM NaCl, 10 mM EDTA [pH 8.0], 0.2% Nonidet P-40, 10% glycerol, 1% PVPP, 2 mM DTT, 1× Complete Protease Inhibitor Cocktail [Sigma]). The beads were washed with the buffer (50 mM Heps [pH 7.5], 200 mM NaCl, 10 mM EDTA [pH 8.0], 0.1% Nonidet P-40, 10% glycerol). Immunoprecipitation was performed with the anti-MYC antibody using N. benthamiana leaves. For Arabidopsis samples, immunoprecipitation was performed with the anti-RGA antibody. RGAd17-3xHA and 6xMYC-SCL27 fusion proteins were detected by immunoblotting with anti-HA (Sigma) and anti-MYC antibodies (Santa Cruz). To analyze POR, LHCB1, LHCB2, LHCB5, LHCA1, PsaD, and AtpB protein levels in vivo, samples (0.1 g) were ground in liquid nitrogen and suspended with 200 µL extraction buffer (125 mM Tris [pH 8.8], 4% SDS, 20% glycerol, 5% β-Me). Total protein was extracted by incubating the samples in boiled water for 5 min, and then centrifuged at 13 000 rpm for 10 min. Proteins were detected with the anti-POR, LHCB1, LHCB2, LHCB5, LHCA1, PsaD, and AtpB antibodies (Agrisera) after total proteins were separated onto a SDS-PAGE gel and transferred to Hybond-ECL Nitrocellulose membrane (Amersham Biosciences).
Transient transcription dual-luciferase (Dual-LUC) assays
The transient expression assay (Dual-LUC) was carried out as described previously [52]. Agrobacteria-infiltrated N. benthamiana leaves were used for LUC/REN analyses. Leaf samples were collected for the transient expression assay using commercial Dual-LUC reaction (DLR) reagents, according to the manufacturer's instruction (Promega).
Quantitative PCR and Northern blot analysis
One µg of total RNAs was used for reverse transcription in a 20 µL reaction system using the RNA PCR (AMV) kit (Promega). Quantitative PCR was performed with SYBR-Green PCR Mastermix (Takara), and amplification was real-time monitored on stepone and steponeplus real-time PCR system (Applied Biosystems). ACTIN2 was used as an internal control for normalization. The primers are listed in Table S1. Northern blot analysis was carried out as described [58].
ChIP analysis
ChIP experiments were performed according to published protocols [59]. Briefly, about 3 g tissues of 3-week-old 6xMYC-rSCL27-OX transgenic plants were harvested. For GA3 or PAC treatment, samples were harvested from the plants treated with 10 µM GA3 and 0.1 µM PAC for 2 day. After fixation, the materials were resuspended in extraction buffer followed by sonification. One third of the solution was saved as input total DNA without precipitation; another one-third was mixed with the MYC-fused agarose (Sigma); and the remaining one-third was precipitated in parallel with HA-fused agarose as a negative control. The resulting DNA samples were purified using a PCR purification kit (Qiagen). The relative concentrations of the DNA fragments were analyzed by qPCR, using the β-TUBULIN2 gene promoter as the reference.
EMSA
The EMSA was performed as reported previously [60]. The primers used were shown in Table S1. The Cy5 fluorescence-labeled DNA (1 nM) was incubated with the indicated amount of the purified His-SCL27 protein in 20 µL of the binding buffer. The concentration of the proteins used for the competitive assay in Figure 4L was 1000 nM. After incubation at 30°C for 20 min, the reaction mixture was electrophoresed at 4°C on a 6% native polyacrylamide gel in 0.5×Tris-borate-EDTA for 2 h (about 200-bp) or 1 h (62-bp) at 100 V. Fluorescence-labeled DNA on the gel was then detected with the Starion FLA-9000 (FujiFilm, Japan).
Accession numbers
SCL27 (At2G45160), SCL22 (At3G60630), SCL6 (At4G00150), MIR171A (At3G51375), MIR171B (At1G11735), MIR171C (At1G62035), β-TUBULIN-2 (At5G62690), HEMA1 (AT1G58290), GUN4 (AT3G59400), GUN5 (AT5G13630), PORA (AT5G54190), PORB (AT4G27440), PORC (AT1G03630), GAI (AT1G14920), RGA (AT2G01570), RGL1 (AT1G66350), RGL2 (AT3G03450), and RGL3 (AT5G17490).
Supporting Information
Zdroje
1. MeskauskieneR, NaterM, GoslingsD, KesslerF, op den CampR, ApelK (2001) FLU: A negative regulator of chlorophyll biosynthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA 98 : 12826–12831.
2. GoslingsD, MeskauskieneR, KimC, LeeKP, NaterM, et al. (2004) Concurrent interactions of heme and FLU with Glu tRNA reductase (HEMA1), the target of metabolic feedback inhibition of tetrapyrrole biosynthesis, in dark - and light-grown Arabidopsis plants. Plant J 40 : 957–967.
3. Krieger-LiszkayA (2005) Singlet oxygen production in photosynthesis. J Exp Bot 56 : 337–346.
4. ReinbotheC, El BakkouriM, BuhrF, MurakiN, NomataJ, et al. (2010) Chlorophyll biosynthesis: spotlight on protochlorophyllide reduction. Trends Plant Sci 15 : 614–624.
5. MochizukiN, TanakaR, GrimmB, MasudaT, MoulinM, et al. (2010) The cell biology of tetrapyrroles: a life and death struggle. Trends Plant Sci 15 : 488–498.
6. HuqE, Al-SadyB, HudsonM, KimC, ApelK, et al. (2004) Phytochrome-interacting factor 1 is a critical bHLH regulator of chlorophyll biosynthesis. Science 305 : 1937–1941.
7. MonteE, Al-SadyB, LeivarP, QuailPH (2007) Out of the dark: how the PIFs are unmasking a dual temporal mechanism of phytochrome signalling. J Exp Bot 58 : 3125–3133.
8. MoonJ, ZhuL, ShenH, HuqE (2008) PIF1 directly and indirectly regulates chlorophyll biosynthesis to optimize the greening process in Arabidopsis. Proc Natl Acad Sci USA 105 : 9433–9438.
9. StephensonPG, FankhauserC, TerryMJ (2009) PIF3 is a repressor of chloroplast development. Proc Nat. Acad Sci USA 106 : 7654–7659.
10. TangW, WangW, ChenD, JiQ, JingY, et al. (2012) Transposase-derived proteins FHY3/FAR1 interact with PHYTOCHROME-INTERACTING FACTOR1 to regulate chlorophyll biosynthesis by modulating HEMB1 during deetiolation in Arabidopsis. Plant Cell 25 : 1984–2000.
11. TerryMJ, KendrickRE (1999) Feedback inhibition of chlorophyll synthesis in the phytochrome-deficient aurea and yellowgreen-2 mutants of tomato. Plant Physiol 119 : 143–152.
12. CornahJE, TerryMJ, SmithAG (2003) Green or red: what stops the traffic in the tetrapyrrole pathway? Trends Plant Sci 8 : 224–230.
13. VothknechtUC, KannangaraCG, von WettsteinD (1998) Barley glutamyl tRNAGlu reductase: mutations affecting haem inhibition and enzyme activity. Phytochemistry 47 : 513–519.
14. SrivastavaA, BealeSI (2005) Glutamyl-tRNA reductase of Chlorobium vibrioforme is a dissociable homodimer that contains one tightly bound heme per subunit. J Bacteriol 187 : 4444–4450.
15. MeskauskieneR, ApelK (2002) Interaction of FLU, a negative regulator of tetrapyrrole biosynthesis, with the glutamyl-tRNA reductase requires the tetratricopeptide repeat domain of FLU. FEBS Lett 532 : 27–30.
16. LarkinRM, AlonsoJM, EckerJR, ChoryJ (2003) GUN4, a regulator of chlorophyll synthesis and intracellular signaling. Science 299 : 902–906.
17. DavisonPA, SchubertHL, ReidJD, IorgCD, HerouxA, et al. (2005) Structural and biochemical characterization of Gun4 suggests a mechanism for its role in chlorophyll biosynthesis. Biochemistry 44 : 7603–7612.
18. VerdeciaMA, LarkinRM, FerrerJL, RiekR, ChoryJ, et al. (2005) Structure of the Mg-chelatase cofactor GUN4 reveals a novel hand-shaped fold for porphyrin binding. PLoS Biol 3: e151.
19. AdhikariND, FroehlichJE, StrandDD, BuckSM, KramerDM, et al. (2011) GUN4-porphyrin complexes bind the ChlH/GUN5 subunit of Mg-Chelatase and promote chlorophyll biosynthesis in Arabidopsis. Plant Cell 23 : 1449–1467.
20. CheminantS, WildM, BouvierF, PelletierS, RenouJP, et al. (2011) DELLAs regulate chlorophyll and carotenoid biosynthesis to prevent photooxidative damage during seedling deetiolation in Arabidopsis. Plant Cell 5 : 1849–1860.
21. ZhongS, ZhaoM, ShiT, ShiH, AnF, et al. (2009) EIN3/EIL1 cooperate with PIF1 to prevent photo-oxidation and to promote greening of Arabidopsis seedlings. Proc Natl Acad Sci USA 106 : 21431–21436.
22. OosawaN, MasudaT, AwaiK, FusadaN, ShimadaH, et al. (2000) Identification and light-induced expression of a novel gene of NADPH-protochlorophyllide oxidoreductase isoform in Arabidopsis thaliana. FEBS Lett 474 : 133–136.
23. SuQ, FrickG, ArmstrongG, ApelK (2001) POR C of Arabidopsis thaliana: A third light - and NADPH-dependent protochlorophyllide oxidoreductase that is differentially regulated by light. Plant Mol Biol 47 : 805–813.
24. MasudaT, TakamiyaK (2004) Novel Insights into the Enzymology, Regulation and Physiological Functions of Light-dependent Protochlorophyllide Oxidoreductase in Angiosperms. Photosynth Res 81 : 1–29.
25. DillA, SunTP (2001) Synergistic de-repression of gibberellins signaling by removing RGA and GAI function in Arabidopsis thaliana. Genetics 159 : 777–785.
26. LeeS, ChengH, KingKE, WangW, HeY, et al. (2002) Gibberellin regulates Arabidopsis seed germination via RGL2, a GAI/RGA-like gene whose expression is up-regulated following imbibition. Genes Dev 16 : 646–658.
27. TylerL, ThomasSG, HuJ, DillA, AlonsoJM, et al. (2004) DELLA proteins and gibberellin-regulated seed germination and floral development in Arabidopsis. Plant Physiol 135 : 1008–1019.
28. AlabadíD, GilJ, BlázquezMA, García-MartínezJL (2004) Gibberellins repress photomorphogenesis in darkness. Plant Physiol 134 : 1050–1057.
29. AlabadíD, Gallego-BartoloméJ, OrlandoL, García-CárcelL, RubioV, et al. (2008) Gibberellins modulate light signaling pathways to prevent Arabidopsis seedling de-etiolation in darkness. Plant J 53 : 324–335.
30. SunX, XueB, JonesWT, RikkerinkE, DunkerAK, et al. (2011) A functionally required unfoldome from the plant kingdom: intrinsically disordered N-terminal domains of GRAS proteins are involved in molecular recognition during plant development. Plant Mol Biol 77 : 205–223.
31. de LucasM, DavièreJM, Rodríguez-FalcónM, PontinM, Iglesias-PedrazJM, et al. (2008) A molecular framework for light and gibberellin control of cell elongation. Nature 451 : 480–484.
32. FengS, MartinezC, GusmaroliG, WangY, ZhouJ, et al. (2008) Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451 : 475–479.
33. LlaveC, KasschauKD, RectorMA, CarringtonJC (2002) Endogenous and silencing-associated small RNAs in plants. Plant Cell 14 : 1605–1619.
34. RhoadesMW, ReinhartBJ, LimLP, BurgeCB, BartelB, et al. (2002) Prediction of plant microRNA targets. Cell 110 : 513–520.
35. BolleC (2004) The role of GRAS proteins in plant signal transduction and development. Planta 218 : 683–692.
36. SchulzeS, SchäferBN, ParizottoEA, VoinnetO, TheresK (2010) LOST MERISTEMS genes regulate cell differentiation of central zone descendants in Arabidopsis shoot meristems. Plant J 64 : 668–678.
37. CurabaJ, TalbotM, LiZ, HelliwellC (2013) Over-expression of microRNA171 affects phase transitions and floral meristem determinancy in barley. BMC Plant Biol 13 : 6.
38. WangL, MaiYX, ZhangYC, LuoQ, YangHQ (2010) MicroRNA171c-targeted SCL6-II, SCL6-III, and SCL6-IV genes regulate shoot branching in Arabidopsis. Mol Plant 3 : 794–806.
39. AndriankajaM, DhondtS, De BodtS, VanhaerenH, CoppensF, et al. (2012) Exit from proliferation during leaf development in Arabidopsis thaliana: a not-so-gradual process. Dev Cell 22 : 64–78.
40. SiréC, MorenoAB, Garcia-ChapaM, López-MoyaJJ, San SegundoB (2009) Diurnal oscillation in the accumulation of Arabidopsis microRNAs, miR167, miR168, miR171 and miR398. FEBS Lett 583 : 1039–1044.
41. ZhouDX (1999) Regulatory mechanism of plant gene transcription by GT-elements and GT-factors. Trends Plant Sci 4 : 210–214.
42. HouX, LeeLY, XiaK, YanY, YuH (2010) DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev Cell 19 : 884–894.
43. ZhangZL, OgawaM, FleetCM, ZentellaR, HuJ, et al. (2011) Scarecrow-like 3 promotes gibberellin signaling by antagonizing master growth repressor DELLA in Arabidopsis. Proc Natl Acad Sci USA 108 : 2160–2165.
44. YuS, GalvãoVC, ZhangYC, HorrerD, ZhangTQ, et al. (2012) Gibberellin regulates the Arabidopsis floral transition through miR156-targeted SQUAMOSA promoter binding-like transcription factors. Plant Cell 24 : 3320–3332.
45. AnF, ZhangX, ZhuZ, JiY, HeW, et al. (2012) Coordinated regulation of apical hook development by gibberellins and ethylene in etiolated Arabidopsis seedlings. Cell Research 22 : 915–927.
46. LiQF, WangC, JiangL, LiS, SunSS, et al. (2012) An interaction between BZR1 and DELLAs mediates direct signaling crosstalk between brassinosteroids and gibberellins in Arabidopsis. Science Signaling 5: ra72.
47. WuG, ParkMY, ConwaySR, WangJW, WeigelD, et al. (2009) The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138 : 750–759.
48. JiangX, LiH, WangT, PengC, WangH, et al. (2012) Gibberellin indirectly promotes chloroplast biogenesis as a means to maintain the chloroplast population of expanded cells. Plant J 72 : 768–780.
49. Gallego-BartoloméJ, MinguetEG, Grau-EnguixF, AbbasM, LocascioA, et al. (2012) Molecular mechanism for the interaction between gibberellin and brassinosteroid signaling pathways in Arabidopsis. Proc Natl Acad Sci USA 109 : 13446–13451.
50. BaiMY, ShangJX, OhE, FanM, BaiY, et al. (2012) Brassinosteroid, gibberellin and phytochrome impinge on a common transcription module in Arabidopsis. Nat Cell Biol 14 : 810–817.
51. HellensRP, AllanAC, FrielEN, BolithoK, GraftonK, et al. (2005) Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 18 : 1–13.
52. LiuH, YuX, LiK, KlejnotJ, YangH, et al. (2008) Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science 322 : 1535–1539.
53. CloughSJ, BentAF (1998) Floral dip: a simplified method for Agrobacterium - mediated transformation of Arabidopsis thaliana. Plant J 16 : 735–743.
54. AronssonH, SchöttlerMA, KellyAA, SundqvistC, DörmannP, et al. (2008) Monogalactosyldiacylglycerol deficiency in Arabidopsis affects pigment composition in the prolamellar body and impairs thylakoid membrane energization and photoprotection in leaves. Plant Physiol 148 : 580–592.
55. ArnonDI (1949) Copper enzymes in isolated chloroplasts: Polyphenol oxidase in Beta vulgaris. Plant Physiol 24 : 1–15.
56. HavauxM, Dall'ostoL, BassiR (2007) Zeaxanthin has enhanced antioxidant capacity with respect to all other xanthophylls in Arabidopsis leaves and functions independent of binding to PSII antennae. Plant Physiol 145 : 1506–1520.
57. Harris N, Spence J, Oparka KJ (1994) A practical approach. In: Harris N, Oparka KJ, eds. Plant Cell Biology. United Kingdom: Oxford University Press. pp: 51–68.
58. HuangJ, TakanoT, AkitaS (2000) Expression of α-expansin genes in young seedlings of rice (oryza sativa L.). Planta 211 : 467–473.
59. YuN, CaiWJ, WangS, ShanCM, WangLJ, et al. (2010) Temporal control of trichome distribution by microRNA156-targeted SPL genes in Arabidopsis thaliana. Plant Cell 22 : 2322–2335.
60. ZhangL, LeynSA, GuY, JiangW, RodionovDA, et al. (2012) Ribulokinase and transcriptional regulation of arabinose metabolism in Clostridium acetobutylicum. J Bacteriol 194 : 1055–1064.
Štítky
Genetika Reprodukční medicína
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 8- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- IVF a rakovina prsu – zvyšují hormony riziko vzniku rakoviny?
-
Všechny články tohoto čísla
- DNA Double Strand Break Repair in Mitosis Is Suppressed by Phosphorylation of XRCC4
- Inference of Transposable Element Ancestry
- The Population Genetics of Evolutionary Rescue
- Retinoic Acid Activates Two Pathways Required for Meiosis in Mice
- Pooled Segregant Sequencing Reveals Genetic Determinants of Yeast Pseudohyphal Growth
- Comprehensive Identification of Single Nucleotide Polymorphisms Associated with Beta-lactam Resistance within Pneumococcal Mosaic Genes
- SMA-Causing Missense Mutations in Display a Wide Range of Phenotypes When Modeled in
- Branch Migration Prevents DNA Loss during Double-Strand Break Repair
- Transcriptome Sequencing from Diverse Human Populations Reveals Differentiated Regulatory Architecture
- Genetic Deletion of SEPT7 Reveals a Cell Type-Specific Role of Septins in Microtubule Destabilization for the Completion of Cytokinesis
- Tethering Sister Centromeres to Each Other Suggests the Spindle Checkpoint Detects Stretch within the Kinetochore
- Global Genetic Variations Predict Brain Response to Faces
- Demography and the Age of Rare Variants
- The Response to High CO Levels Requires the Neuropeptide Secretion Component HID-1 to Promote Pumping Inhibition
- Sp6 and Sp8 Transcription Factors Control AER Formation and Dorsal-Ventral Patterning in Limb Development
- The Groucho Co-repressor Is Primarily Recruited to Local Target Sites in Active Chromatin to Attenuate Transcription
- A Transposable Element Insertion Confers Xenobiotic Resistance in Drosophila
- The Genomic Architecture of Population Divergence between Subspecies of the European Rabbit
- Human Social Genomics
- Gene Expansion Shapes Genome Architecture in the Human Pathogen : An Evolutionary Genomics Analysis in the Ancient Terrestrial Mucorales (Mucoromycotina)
- Canonical Non-Homologous End Joining in Mitosis Induces Genome Instability and Is Suppressed by M-phase-Specific Phosphorylation of XRCC4
- Roles of Type 1A Topoisomerases in Genome Maintenance in
- The TRIM-NHL Protein LIN-41 Controls the Onset of Developmental Plasticity in
- Wnt-Mediated Repression via Bipartite DNA Recognition by TCF in the Hematopoietic System
- KDM6 Demethylase Independent Loss of Histone H3 Lysine 27 Trimethylation during Early Embryonic Development
- Integration of UPR and Oxidative Stress Signaling in the Control of Intestinal Stem Cell Proliferation
- miR171-Targeted Scarecrow-Like Proteins Bind to GT -Elements and Mediate Gibberellin-Regulated Chlorophyll Biosynthesis under Light Conditions
- Syndecan-1 Is Required to Maintain Intradermal Fat and Prevent Cold Stress
- LIN-3/EGF Promotes the Programmed Cell Death of Specific Cells in by Transcriptional Activation of the Pro-apoptotic Gene
- A System for Genome-Wide Histone Variant Dynamics In ES Cells Reveals Dynamic MacroH2A2 Replacement at Promoters
- Multiple Regulation of Rad51-Mediated Homologous Recombination by Fission Yeast Fbh1
- A Genome-Wide Association Study of the Maize Hypersensitive Defense Response Identifies Genes That Cluster in Related Pathways
- The RNA Helicases AtMTR4 and HEN2 Target Specific Subsets of Nuclear Transcripts for Degradation by the Nuclear Exosome in
- Asymmetric Division and Differential Gene Expression during a Bacterial Developmental Program Requires DivIVA
- A Model-Based Approach for Identifying Signatures of Ancient Balancing Selection in Genetic Data
- Chromatin Insulator Factors Involved in Long-Range DNA Interactions and Their Role in the Folding of the Drosophila Genome
- Conditional Inactivation of Upstream Binding Factor Reveals Its Epigenetic Functions and the Existence of a Somatic Nucleolar Precursor Body
- Evidence for Divisome Localization Mechanisms Independent of the Min System and SlmA in
- Patterns of Admixture and Population Structure in Native Populations of Northwest North America
- Response Regulator Heterodimer Formation Controls a Key Stage in S Development
- A Genetic Strategy to Measure Circulating Insulin Reveals Genes Regulating Insulin Production and Secretion
- EVA-1 Functions as an UNC-40 Co-receptor to Enhance Attraction to the MADD-4 Guidance Cue in
- Dysfunction of the CNS-Heart Axis in Mouse Models of Huntington's Disease
- An Otx/Nodal Regulatory Signature for Posterior Neural Development in Ascidians
- Phosphorylation of a Central Clock Transcription Factor Is Required for Thermal but Not Photic Entrainment
- Genome-Wide Patterns of Genetic Variation within and among Alternative Selective Regimes
- EF-P Dependent Pauses Integrate Proximal and Distal Signals during Translation
- Ku-Mediated Coupling of DNA Cleavage and Repair during Programmed Genome Rearrangements in the Ciliate
- Functional Specialization Among Members Of Knickkopf Family Of Proteins In Insect Cuticle Organization
- Playing RNase P Evolution: Swapping the RNA Catalyst for a Protein Reveals Functional Uniformity of Highly Divergent Enzyme Forms
- The Translational Regulators GCN-1 and ABCF-3 Act Together to Promote Apoptosis in
- Meta-Analysis of Genome-Wide Association Studies in African Americans Provides Insights into the Genetic Architecture of Type 2 Diabetes
- A -Regulatory Mutation of Causes Silky-Feather in Chickens
- VIB1, a Link between Glucose Signaling and Carbon Catabolite Repression, Is Essential for Plant Cell Wall Degradation by
- A Population Genetic Signal of Polygenic Adaptation
- A Conserved Dopamine-Cholecystokinin Signaling Pathway Shapes Context–Dependent Behavior
- The MAP Kinase p38 Is Part of Circadian Clock
- The Cohesin Subunit Rad21 Is Required for Synaptonemal Complex Maintenance, but Not Sister Chromatid Cohesion, during Drosophila Female Meiosis
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Meta-Analysis of Genome-Wide Association Studies in African Americans Provides Insights into the Genetic Architecture of Type 2 Diabetes
- KDM6 Demethylase Independent Loss of Histone H3 Lysine 27 Trimethylation during Early Embryonic Development
- The RNA Helicases AtMTR4 and HEN2 Target Specific Subsets of Nuclear Transcripts for Degradation by the Nuclear Exosome in
- EF-P Dependent Pauses Integrate Proximal and Distal Signals during Translation
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



