-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
DNA Double Strand Break Repair in Mitosis Is Suppressed by Phosphorylation of XRCC4
article has not abstract
Published in the journal: . PLoS Genet 10(8): e32767. doi:10.1371/journal.pgen.1004598
Category: Perspective
doi: https://doi.org/10.1371/journal.pgen.1004598Summary
article has not abstract
Cells are continuously subjected to DNA damage, the most cytotoxic form of which is the DNA double-strand break (DSB). DSBs arise from endogenous sources, such as collapsed replication forks, or can be caused by exogenous agents that include ionizing radiation, reactive oxygen species, and chemotherapeutic drugs such as topoisomerase II poisons [1]. During interphase, DSBs are repaired by one of two main pathways: classical non-homologous end joining (C-NHEJ) or homologous recombination repair (HRR) [2]. The fate of DSBs that arise during mitosis, however, has been poorly characterized. In this issue of PLOS Genetics, Shinohara and colleagues demonstrate that DSB induction in mitosis leads to the formation of anaphase bridges, a hallmark of genomic instability, and demonstrate that their formation requires primarily the C-NHEJ pathway [3].
C-NHEJ, which is active throughout interphase, is considered a rapid but error-prone pathway that rejoins DSB ends with minimal end processing. The main steps in C-NHEJ are detection of the DSB by the Ku heterodimer, followed by recruitment of end-processing factors and the DNA ligase IV-XRCC4-XLF complex. Regulation of C-NHEJ end processing is controlled by the DNA-dependent protein kinase catalytic subunit (DNA-PKcs) [4], [5]. HRR, on the other hand, occurs only in S and G2 and is initiated by the binding of the MRN complex (composed of Mre11, Rad50, and Nbs1), which is followed by MRN - and CtIP-mediated resection to create long 3′ overhanging ends required for Rad51-dependent strand invasion. Unlike C-NHEJ, HRR requires an undamaged DNA template for repair, usually the sister chromatid, and results in slower, but more precise, repair [6]. In addition to the two main repair pathways, DSBs can also be repaired by an error-prone, alternative end-joining pathway (Alt-NHEJ). Alt-NHEJ requires end-resection and involves CtIP, XRCC1, and DNA ligase III, but not the C-NHEJ factors [7].
DSBs initiate an elaborate signaling cascade that involves protein phosphorylation and ubiquitylation. This results in accumulation of proteins on chromatin surrounding the DSB to form DNA damage-induced foci and is required for regulation of repair and for cell cycle checkpoint arrest. Checkpoint signaling is initiated primarily by ataxia telangiectasia mutated (ATM)-dependent phosphorylation of histone H2AX, which in turn leads to recruitment of the checkpoint mediator MDC1 and the E3 ubiquitin ligases RNF8 and RNF168, as well as 53BP1 and BRCA1 [2], [8].
While repair of DSBs and cell cycle checkpoint arrest in interphase cells is relatively well understood, less is known regarding how cells respond to DSBs that are incurred when cells are in mitosis. This is of particular importance as errors in mitosis can lead to chromosome aberrations, genomic instability, polyploidy, or mitotic catastrophe [9], [10]. Early studies indicated that DSB repair pathways are suppressed in mitosis [11], and that DSBs originating in mitosis are not repaired until the subsequent G1 phase (reviewed in [12], [13]). In 2010, Jackson and colleagues reported that DSBs formed in mitosis initiate a partial DNA damage response in which ATM is activated, H2AX is phosphorylated, and MDC1 is recruited to foci [14]. Downstream recruitment of RNF8, RNF168, 53BP1, and BRCA1 fails to occur, however [14], [15]. Because 53BP1 promotes NHEJ [2], one mechanism for suppression of NHEJ in mitosis may be loss of 53BP1 from foci. Indeed, Durocher and colleagues recently showed that phosphorylation of RNF8 and 53BP1 by cyclin dependent kinase (CDK1) and the mitotic polo-like kinase 1 (PLK1) prevents their recruitment to DNA damage foci, thereby inactivating repair and minimizing opportunities to produce deleterious telomeric fusions [16].
What then of other repair processes in mitosis? Shinohara and colleagues report that anaphase bridge formation requires primarily the C-NHEJ pathway, as the number of bridges was decreased in cells depleted for the essential C-NHEJ protein, XRCC4. By contrast, depletion of CtIP resulted in increased anaphase bridge formation, suggesting that Alt-NHEJ or HRR suppresses anaphase bridge formation. Moreover, they show that both CDK1 and PLK1 protein kinases contribute to phosphorylation of XRCC4 in mitosis and conclude that phosphorylation of serine 326 in the extreme C-terminus of XRCC4 reduces anaphase bridge formation and inhibits DSB repair (Figure 1). The authors propose that CDK1/PLK1-dependent phosphorylation of XRCC4 serves as a switch to inhibit C-NHEJ in mitosis, preventing anaphase bridges and genomic instability [3]. These results are reminiscent of a previous study by this group in which they showed that CDK-dependent phosphorylation of the budding yeast homologue of XRCC4 (Lif1) promotes resection-mediated alternative end joining pathways, thus, in effect, suppressing NHEJ [17].
Fig. 1. Model for suppression of NHEJ in mitosis by phosphorylated XRCC4. 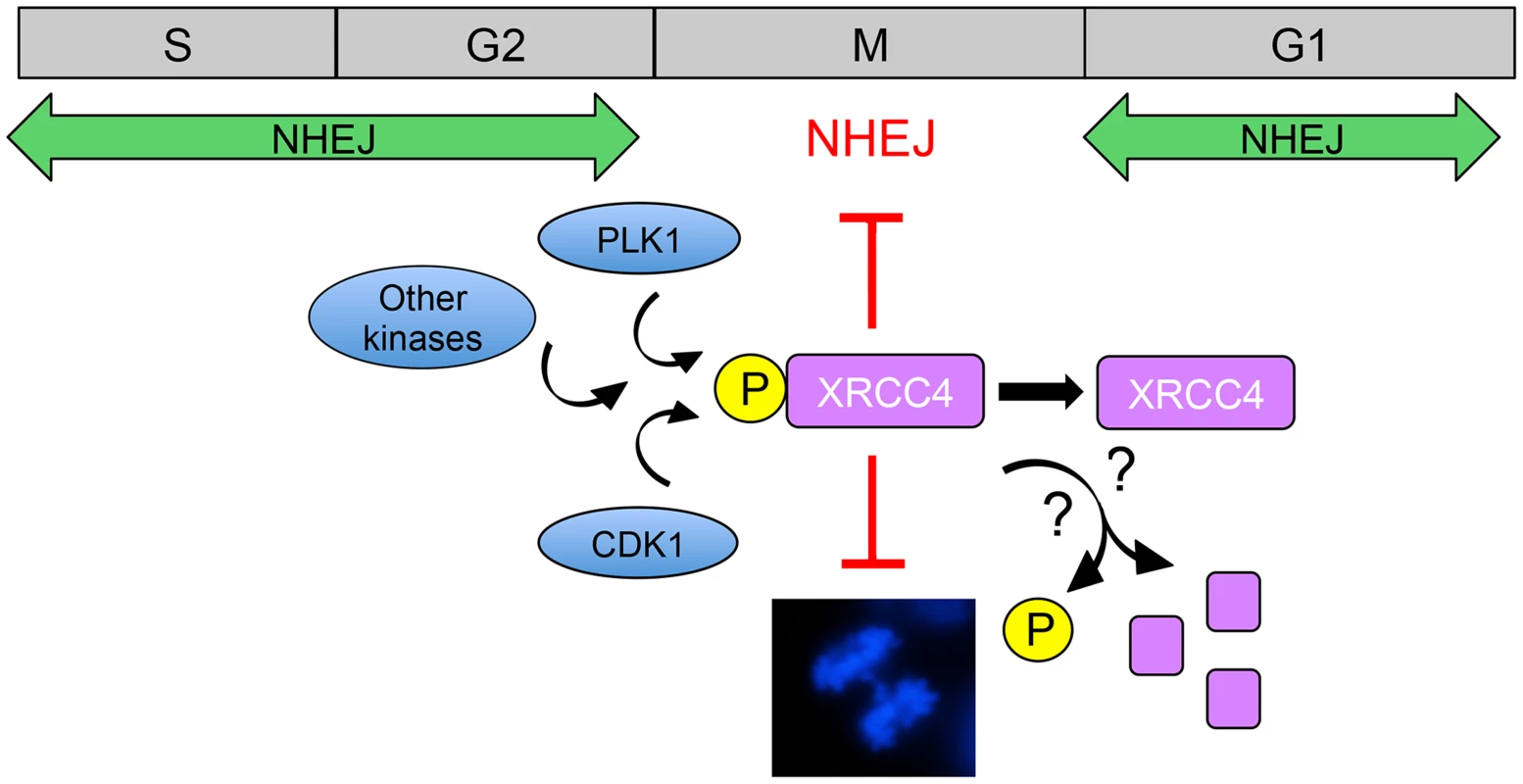
NHEJ is the major pathway for the repair of radiation and topoisomerase II poison-induced DSBs in interphase mammalian cells. CDK1 and PLK1 are activated as cells enter mitosis and contribute to phosphorylation of XRCC4 on serine 326 in the unstructured, C-terminal tail [3]. It is also likely that other protein kinases are involved in phosphorylation of XRCC4 in mitosis. In ways that are yet to be determined, phosphorylated XRCC4 suppresses C-NHEJ in mitosis, preventing formation of anaphase bridges (shown in the DAPI-stained mitotic cell in the lower panel, from [22]). Serine 326 phosphorylation of XRCC4 in mitosis is transient [3], suggesting that phosphorylated XRCC4 is either dephosphorylated or degraded as cells exit mitosis, allowing C-NHEJ to resume in the subsequent G1 phase. The study of Shinohara and colleagues raises interesting questions regarding the mechanism of XRCC4-mediated suppression of C-NHEJ. The authors show that DNA ligase IV, but not XRCC4, localizes to mitotic chromosomes and that failure of XRCC4 localization suppresses NHEJ. This is unexpected because in interphase, XRCC4 binds with high affinity to DNA ligase IV and, indeed, stabilizes the DNA ligase IV protein [18], [19]. Phosphorylation of XRCC4 in mitosis was not responsible for its failure to localize to mitotic chromosomes and phosphorylation did not affect the interaction of XRCC4 with DNA ligase IV or XLF. The mechanism underlying inactivation of XRCC4 function in mitosis, therefore, remains to be determined. While clearly implicating both CDK1 and PLK1 in phosphorylation of XRCC4 in mitosis, the results suggest that other protein kinases may also be involved. For example, phosphorylated XRCC4 runs as several bands on denaturing polyacrylamide gels. Though serine 326 is phosphorylated in at least two bands, inhibition of either CDK1 or PLK1 blocked serine 326 phosphorylation in only one band, suggesting that phosphorylation of XRCC4 in mitosis may be more complex than described to date.
The findings of Shinohara and colleagues also raise the question of what happens to XRCC4 at the end of mitosis. Is it dephosphorylated by protein phosphatases to allow NHEJ to proceed in the next G1 or is the phosphorylated form of XRCC4 targeted for degradation by the proteasome? Indeed, there are ample precedents for both. Protein phosphatases such as protein phosphatase 2A (PP2A) dephosphorylate multiple proteins required for entry into or exit from mitosis [20] and multiple proteins are degraded by the anaphase-promoting complex/cyclosome (APC/C) to allow mitotic exit [21]. It is also interesting to note that while XRCC4 is phosphorylated to inhibit C-NHEJ, another component of the pathway, DNA-PKcs, is required for accurate mitosis, as its down-regulation or inhibition leads to misaligned chromosomes and mitotic defects [22], [23]. DNA-PKcs is phosphorylated by PLK1 early in mitosis and is dephosphorylated by protein phosphatase 6 (PP6) at mitotic exit [22]. It will be interesting to determine how phosphorylation of DNA-PKcs and possibly other components of the C-NHEJ pathway contribute to maintenance of genome stability during mitosis. Together, these studies shed light on how DSB repair processes are regulated to prevent genomic instability during mitosis.
Zdroje
1. CicciaA, ElledgeSJ (2010) The DNA damage response: making it safe to play with knives. Mol Cell 40 : 179–204.
2. PanierS, BoultonSJ (2014) Double-strand break repair: 53BP1 comes into focus. Nat Rev Mol Cell Biol 15 : 7–18.
3. TerasawaM, ShinoharaA, ShinoharaM (2014) Canonical Non-homologous End Joining in Mitosis Induces Genome Instability and Is Suppressed by M-phase-specific Phosphorylation of XRCC4 via CDKs. PLoS Genet 10: e1004563.
4. LieberMR (2010) The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem 79 : 181–211.
5. WangC, Lees-MillerSP (2013) Detection and Repair of Ionizing Radiation-Induced DNA Double Strand Breaks: New Developments in Nonhomologous End Joining. Int J Radiat Oncol Biol Phys 86 : 440–449.
6. San FilippoJ, SungP, KleinH (2008) Mechanism of eukaryotic homologous recombination. Annu Rev Biochem 77 : 229–257.
7. ZhangY, JasinM (2011) An essential role for CtIP in chromosomal translocation formation through an alternative end-joining pathway. Nat Struct Mol Biol 18 : 80–84.
8. van AttikumH, GasserSM (2009) Crosstalk between histone modifications during the DNA damage response. Trends Cell Biol 19 : 207–217.
9. GanemNJ, PellmanD (2012) Linking abnormal mitosis to the acquisition of DNA damage. J Cell Biol 199 : 871–881.
10. GordonDJ, ResioB, PellmanD (2012) Causes and consequences of aneuploidy in cancer. Nat Rev Genet 13 : 189–203.
11. ZirkleRE, BloomW (1953) Irradiation of parts of individual cells. Science 117 : 487–493.
12. HeijinkAM, KrajewskaM, van VugtMA (2013) The DNA damage response during mitosis. Mutat Res 750 : 45–55.
13. MorrisonC, RiederCL (2004) Chromosome damage and progression into and through mitosis in vertebrates. DNA Repair (Amst) 3 : 1133–1139.
14. GiuntaS, BelotserkovskayaR, JacksonSP (2010) DNA damage signaling in response to double-strand breaks during mitosis. J Cell Biol 190 : 197–207.
15. GiuntaS, JacksonSP (2011) Give me a break, but not in mitosis: the mitotic DNA damage response marks DNA double-strand breaks with early signaling events. Cell Cycle 10 : 1215–1221.
16. OrthweinA, Fradet-TurcotteA, NoordermeerSM, CannyMD, BrunCM, et al. (2014) Mitosis inhibits DNA double-strand break repair to guard against telomere fusions. Science 344 : 189–193.
17. MatsuzakiK, TerasawaM, IwasakiD, HigashideM, ShinoharaM (2012) Cyclin-dependent kinase-dependent phosphorylation of Lif1 and Sae2 controls imprecise nonhomologous end joining accompanied by double-strand break resection. Genes Cells 17 : 473–493.
18. SibandaBL, CritchlowSE, BegunJ, PeiXY, JacksonSP, et al. (2001) Crystal structure of an Xrcc4-DNA ligase IV complex. Nat Struct Biol 8 : 1015–1019.
19. CritchlowSE, BowaterRP, JacksonSP (1997) Mammalian DNA double-strand break repair protein XRCC4 interacts with DNA ligase IV. Curr Biol 7 : 588–598.
20. HuntT (2013) On the regulation of protein phosphatase 2A and its role in controlling entry into and exit from mitosis. Adv Biol Regul 53 : 173–178.
21. LindonC (2008) Control of mitotic exit and cytokinesis by the APC/C. Biochem Soc Trans 36 : 405–410.
22. DouglasP, YeR, Trinkle-MulcahyL, NealJA, De WeverV, et al. (2014) Polo-like kinase 1 (PLK1) and protein phosphatase 6 (PP6) regulate DNA-dependent protein kinase catalytic subunit (DNA-PKcs) phosphorylation in mitosis. Biosci Rep 34: e00113 doi:10.1042/BSR20140051
23. LeeKJ, LinYF, ChouHY, YajimaH, FattahKR, et al. (2011) Involvement of DNA-dependent protein kinase in normal cell cycle progression through mitosis. J Biol Chem 286 : 12796–12802.
Štítky
Genetika Reprodukční medicína
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 8- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
- Délka menstruačního cyklu jako marker ženské plodnosti
- Intrauterinní inseminace a její úspěšnost
-
Všechny články tohoto čísla
- DNA Double Strand Break Repair in Mitosis Is Suppressed by Phosphorylation of XRCC4
- Inference of Transposable Element Ancestry
- The Population Genetics of Evolutionary Rescue
- Retinoic Acid Activates Two Pathways Required for Meiosis in Mice
- Pooled Segregant Sequencing Reveals Genetic Determinants of Yeast Pseudohyphal Growth
- Comprehensive Identification of Single Nucleotide Polymorphisms Associated with Beta-lactam Resistance within Pneumococcal Mosaic Genes
- SMA-Causing Missense Mutations in Display a Wide Range of Phenotypes When Modeled in
- Branch Migration Prevents DNA Loss during Double-Strand Break Repair
- Transcriptome Sequencing from Diverse Human Populations Reveals Differentiated Regulatory Architecture
- Genetic Deletion of SEPT7 Reveals a Cell Type-Specific Role of Septins in Microtubule Destabilization for the Completion of Cytokinesis
- Tethering Sister Centromeres to Each Other Suggests the Spindle Checkpoint Detects Stretch within the Kinetochore
- Global Genetic Variations Predict Brain Response to Faces
- Demography and the Age of Rare Variants
- The Response to High CO Levels Requires the Neuropeptide Secretion Component HID-1 to Promote Pumping Inhibition
- Sp6 and Sp8 Transcription Factors Control AER Formation and Dorsal-Ventral Patterning in Limb Development
- The Groucho Co-repressor Is Primarily Recruited to Local Target Sites in Active Chromatin to Attenuate Transcription
- A Transposable Element Insertion Confers Xenobiotic Resistance in Drosophila
- The Genomic Architecture of Population Divergence between Subspecies of the European Rabbit
- Human Social Genomics
- Gene Expansion Shapes Genome Architecture in the Human Pathogen : An Evolutionary Genomics Analysis in the Ancient Terrestrial Mucorales (Mucoromycotina)
- Canonical Non-Homologous End Joining in Mitosis Induces Genome Instability and Is Suppressed by M-phase-Specific Phosphorylation of XRCC4
- Roles of Type 1A Topoisomerases in Genome Maintenance in
- The TRIM-NHL Protein LIN-41 Controls the Onset of Developmental Plasticity in
- Wnt-Mediated Repression via Bipartite DNA Recognition by TCF in the Hematopoietic System
- KDM6 Demethylase Independent Loss of Histone H3 Lysine 27 Trimethylation during Early Embryonic Development
- Integration of UPR and Oxidative Stress Signaling in the Control of Intestinal Stem Cell Proliferation
- miR171-Targeted Scarecrow-Like Proteins Bind to GT -Elements and Mediate Gibberellin-Regulated Chlorophyll Biosynthesis under Light Conditions
- Syndecan-1 Is Required to Maintain Intradermal Fat and Prevent Cold Stress
- LIN-3/EGF Promotes the Programmed Cell Death of Specific Cells in by Transcriptional Activation of the Pro-apoptotic Gene
- A System for Genome-Wide Histone Variant Dynamics In ES Cells Reveals Dynamic MacroH2A2 Replacement at Promoters
- Multiple Regulation of Rad51-Mediated Homologous Recombination by Fission Yeast Fbh1
- A Genome-Wide Association Study of the Maize Hypersensitive Defense Response Identifies Genes That Cluster in Related Pathways
- The RNA Helicases AtMTR4 and HEN2 Target Specific Subsets of Nuclear Transcripts for Degradation by the Nuclear Exosome in
- Asymmetric Division and Differential Gene Expression during a Bacterial Developmental Program Requires DivIVA
- A Model-Based Approach for Identifying Signatures of Ancient Balancing Selection in Genetic Data
- Chromatin Insulator Factors Involved in Long-Range DNA Interactions and Their Role in the Folding of the Drosophila Genome
- Conditional Inactivation of Upstream Binding Factor Reveals Its Epigenetic Functions and the Existence of a Somatic Nucleolar Precursor Body
- Evidence for Divisome Localization Mechanisms Independent of the Min System and SlmA in
- Patterns of Admixture and Population Structure in Native Populations of Northwest North America
- Response Regulator Heterodimer Formation Controls a Key Stage in S Development
- A Genetic Strategy to Measure Circulating Insulin Reveals Genes Regulating Insulin Production and Secretion
- EVA-1 Functions as an UNC-40 Co-receptor to Enhance Attraction to the MADD-4 Guidance Cue in
- Dysfunction of the CNS-Heart Axis in Mouse Models of Huntington's Disease
- An Otx/Nodal Regulatory Signature for Posterior Neural Development in Ascidians
- Phosphorylation of a Central Clock Transcription Factor Is Required for Thermal but Not Photic Entrainment
- Genome-Wide Patterns of Genetic Variation within and among Alternative Selective Regimes
- EF-P Dependent Pauses Integrate Proximal and Distal Signals during Translation
- Ku-Mediated Coupling of DNA Cleavage and Repair during Programmed Genome Rearrangements in the Ciliate
- Functional Specialization Among Members Of Knickkopf Family Of Proteins In Insect Cuticle Organization
- Playing RNase P Evolution: Swapping the RNA Catalyst for a Protein Reveals Functional Uniformity of Highly Divergent Enzyme Forms
- The Translational Regulators GCN-1 and ABCF-3 Act Together to Promote Apoptosis in
- Meta-Analysis of Genome-Wide Association Studies in African Americans Provides Insights into the Genetic Architecture of Type 2 Diabetes
- A -Regulatory Mutation of Causes Silky-Feather in Chickens
- VIB1, a Link between Glucose Signaling and Carbon Catabolite Repression, Is Essential for Plant Cell Wall Degradation by
- A Population Genetic Signal of Polygenic Adaptation
- A Conserved Dopamine-Cholecystokinin Signaling Pathway Shapes Context–Dependent Behavior
- The MAP Kinase p38 Is Part of Circadian Clock
- The Cohesin Subunit Rad21 Is Required for Synaptonemal Complex Maintenance, but Not Sister Chromatid Cohesion, during Drosophila Female Meiosis
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Meta-Analysis of Genome-Wide Association Studies in African Americans Provides Insights into the Genetic Architecture of Type 2 Diabetes
- KDM6 Demethylase Independent Loss of Histone H3 Lysine 27 Trimethylation during Early Embryonic Development
- The RNA Helicases AtMTR4 and HEN2 Target Specific Subsets of Nuclear Transcripts for Degradation by the Nuclear Exosome in
- EF-P Dependent Pauses Integrate Proximal and Distal Signals during Translation
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



