-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Interaction between Two Timing MicroRNAs Controls Trichome Distribution in
MicroRNAs are important aging regulators in many organisms. In Arabidopsis the miR156-targeted SQUAMOSA PROMOTER BINDING PROTEIN LIKE (SPL) transcription factors play important roles as an endogenous age cue in programming phase transition and phase-dependent morphogenesis, including trichome patterning. However, how the timely increasing SPL output is modulated remains elusive. By dissecting the regulatory network controlling trichome formation on stem, we show that a group of GRAS family members, LOST MERISTEMS 1 (LOM1), LOM2 and LOM3, targeted by timing miR171, function in modulating the SPL activity through direct protein-protein interaction. LOMs promote trichome formation through attenuating the SPL (such as SPL9) activity of trichome repression. The LOM-SPL interaction affects many aspects of plant growth and development, including flowering, aging and chlorophyll biosynthesis. Interestingly, MIR171A gene expression is regulated by its own targets (LOMs), forming a feedback loop to program plant life. Our study establishes an age-dependent regulatory network composed of two timing miRNAs which act oppositely through direct interaction of their target proteins.
Published in the journal: . PLoS Genet 10(4): e32767. doi:10.1371/journal.pgen.1004266
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004266Summary
MicroRNAs are important aging regulators in many organisms. In Arabidopsis the miR156-targeted SQUAMOSA PROMOTER BINDING PROTEIN LIKE (SPL) transcription factors play important roles as an endogenous age cue in programming phase transition and phase-dependent morphogenesis, including trichome patterning. However, how the timely increasing SPL output is modulated remains elusive. By dissecting the regulatory network controlling trichome formation on stem, we show that a group of GRAS family members, LOST MERISTEMS 1 (LOM1), LOM2 and LOM3, targeted by timing miR171, function in modulating the SPL activity through direct protein-protein interaction. LOMs promote trichome formation through attenuating the SPL (such as SPL9) activity of trichome repression. The LOM-SPL interaction affects many aspects of plant growth and development, including flowering, aging and chlorophyll biosynthesis. Interestingly, MIR171A gene expression is regulated by its own targets (LOMs), forming a feedback loop to program plant life. Our study establishes an age-dependent regulatory network composed of two timing miRNAs which act oppositely through direct interaction of their target proteins.
Introduction
MicroRNA (miRNA) was first identified as the regulator of the juvenile-to-adult transition in Caenorhabditis elegans [1], and a similar function was later assigned to plant miRNA: miR156 and its target SQUAMOSA PROMOTER BINDING PROTEIN LIKE (SPL) genes define an endogenous aging and flowering pathway [2], [3]. The miR156 overexpression delays phase transition and startup of flowering. Similarly, accumulation of SPLs accelerates aging [2]–[4]. In Arabidopsis there are 17 SPL genes, 11 of which contain a miR156 target site. The miR156-targeted SPLs can be divided into four clades based on their protein structures, and the clades composed of SPL3/4/5 and SPL9/15 both promote phase transition [2]–[4]. Acting downstream of miR156-targeted SPLs, miR172 also plays roles in developmental timing of Arabidopsis [3]. In addition, there are other transcription regulators that affect SPL activities, such as DELLAs, the negative regulator of gibberellin signaling pathway, interact with SPLs to control flowering [5].
Trichomes are specialized epidermal cells, acting as barriers to protect plants from herbivores, UV irradiation, and excessive transpiration. In Arabidopsis, trichome distribution is spatially and temporally regulated, and the distribution pattern serves as a trait to distinguish between juvenile and adult leaves [6], [7]. During the early vegetative phase, trichomes are evenly distributed on the adaxial side of rosette leaves. Plants start transition from juvenile to adult phase when trichomes initiate on leaf abaxial side [7]. After entering into the reproductive stage, the number of trichomes is gradually reduced along the inflorescence stems. Floral organs are nearly glabrous except for a few trichomes on the abaxial surface of sepals. Genetic screens of Arabidopsis mutants have identified sets of regulators governing trichome formation. In brief, the trichome initiation complex of Arabidopsis comprises a WD40 protein TRANSPARENT TESTA GLABRA1 (TTG1), an R2R3 MYB protein GLABRA1 (GL1), and a basic helix-loop-helix transcription factor GL3 or its homolog, ENHANCER of GL3 (EGL3) [8]–[10]. This ternary complex initiates trichome cell development by activating GL2, which encodes a homeodomain leucine zipper transcription factor [11], [12]. A group of single-repeat R3 MYB factors, including TRIPTYCHON (TRY) [13], CAPRICE (CPC) [14], [15], ENHANCER OF TRY AND CPC1 (ETC1) [16], ETC2 [17], ETC3 [18], TRICHOMELESS1 (TCL1) [19] and TCL2 [20], [21], redundantly suppress trichome initiation by competing with GL1 for the binding site of GL3 to prevent the active complex formation [12]. In addition to these specific negative factors, hormone signaling components also affect the trichome regulatory complex [22].
We previously reported that the miR156-targeted SPLs temporally repress trichome distribution on stem and inflorescence through activating TCL1 and TRY [23]. Plants overexpressing miR156 developed ectopic trichomes on the stem and floral organs, whereas plants with elevated SPLs produced fewer trichomes after bolting [23]. Since miR156-SPLs define a major endogenous age cue, this provides a straightforward mechanism that connects plant phase transition with trichome development. Due to the easiness of observation, trichome formation on stem is an ideal system to investigate the interaction of miR156-SPL module with other developmental or environmental signaling pathways.
LOST MERISTEMS (LOMs), also known as AtHAMs because the first functionally characterized member of this subclade is HAIRY MERISTEM (HAM) from Petunia hybrid [24], are transcription factors belonging to the GRAS-domain containing family. There are three LOM genes in Arabidopsis, LOM1/2/3 (also known as AtHAM1/2/3 [25] or SCL6-2/3/4 [26]), which are targets of miR171 [27], [28]. Like other GRAS members, LOMs contain a highly conserved GRAS domain on C-terminus and a variable N-terminus. LOM1 and LOM2 are ∼65% identical at amino acid sequence level, and LOM3 has a shorter N-terminal domain. Previous studies revealed that LOMs function in diverse processes such as meristem maintenance, shoot branching, chlorophyll biosynthesis and root growth [25]–[27], [29]. Here, we report that miR171-targeted LOMs functionally interfere with selected SPLs through protein-protein interaction.
Results
miR171-LOM regulates trichome distribution
In Arabidopsis genome, there are four miR171 coding genes, MIR171A, B, C and MIR170. We found that overexpression (OE) of MIR171A/B/C under the 35S promoter reduced trichomes on stem very significantly (Figure 1A and 1B), which was not reported previously. To see if the reduction of trichomes was caused by decreased level of LOMs due to miR171 accumulation, we first checked the abundance of mature miR171 and the transcript levels of LOMs in these transgenic plants. Indeed, miR171 was over-accumulated (Figure 1C) and LOM expression declined drastically (Figure 1D). In addition, miR171 overexpression resulted in phenotypic changes similar to those of lom1 lom2 lom3 triple mutant (termed lomt hereinafter) as reported [26]: the narrower rosette leaves and the higher chlorophyll content (Figure 1E–1I). We then analyzed the effects of LOM overaccumulation by examining the 35S::LUC-rLOM1 plants, in which a miRNA-resistant LOM1 (rLOM1) was fused to firefly luciferase (LUC) gene and expressed constitutively. In addition to yellow-green leaves (Figure 1J) as reported [26], the 35S::LUC-rLOM1 plants produced supernumerary trichomes on stems, inflorescences and pedicels compared to the wild-type and lomt mutant plants (Figure 1K–1N), indicating that LOM1 accumulation induces ectopic trichomes after bolting.
Fig. 1. MiR171-LOMs regulate trichome initiation. 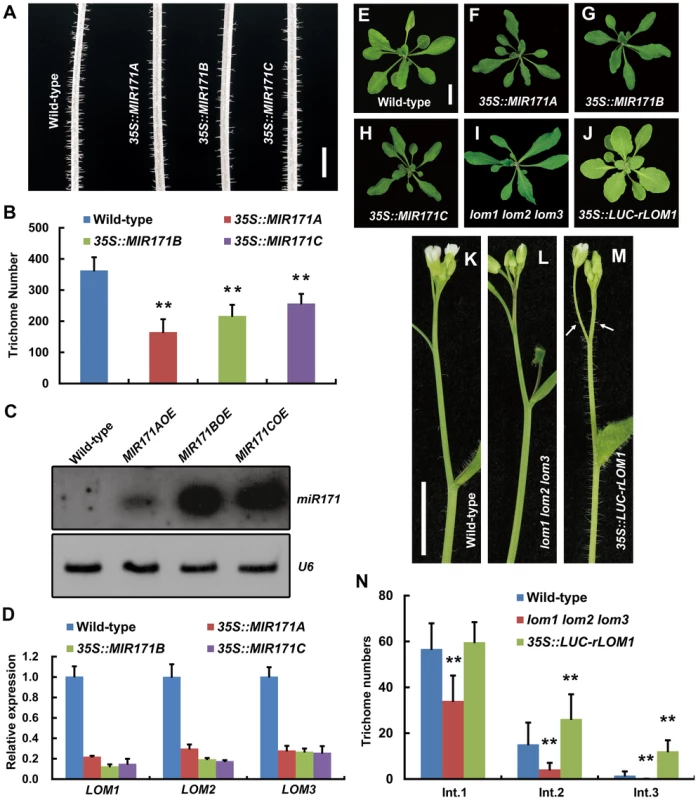
(A) Trichome distribution on main stem of wild-type (Col-0) and MIR171OE (35S::MIR171A/B/C) plants; MIR171 overexpression decreased trichome density. Scale bar represents 5 mm. (B) Overexpression of MIR171 reduced the total number of trichomes on stems. Trichome numbers on main stems (0 to 5 cm from bottom) were counted and data were given as mean s.d. (n = 18) and analyzed by t test. **P<0.01. (C) RNA blots showing high accumulation of mature miR171 in 10-day-old MIR171OE plants. U6 was used as an internal reference. (D) qRT-PCR showing repressed expression of LOM1, LOM2 and LOM3 in MIR171OE plants. Expression level in the wild-type was set to 1. Errors bars indicate s.d. (n = 3). Three biological replicates were analyzed, with consistent results. (E–J) View of 15-day-old plants of indicated genotypes. Overexpression of MIR171 resulted in dark-green and narrower leaves. MIR171OE transgenic plants showed similar phenotypes as lomt (lom1 lom2 lom3) mutant (I). On the contrary, 35S::LUC-rLOM1 transgenic plants (J) showed yellow-green leaves. Scale bar in (E) represents 1 cm for (E–J). (K–M) Spatial distribution of trichomes on main stem of wild-type, lomt and 35S::LUC-rLOM1 plants. Arrows in (M) indicate the ectopic trichomes on the main inflorescence stems and pedicels. Scale bar in (K) represents 1 cm for (K–M). (N) Trichome density on main stem of wild-type, lomt and 35S::LUC-rLOM1 plants. The Int.1 to Int.3 represents the main stem internodes from bottom to top. The y axis indicates trichome number per centimeter of each internode. Data are given as mean s.d. (n>16) and analyzed by t test. **P<0.01. To further examine the role of LOMs in trichome production, we analyzed transgenic Arabidopsis expressing Myc-tagged rLOM1, 2 and 3, under the 35S or their native promoters, respectively. LOM1::Myc-rLOM1 and LOM2::Myc-rLOM2 plants produced supernumerary or ectopic trichomes on stems, inflorescences, and even on pedicels that are glabrous in wild-type plants (Figure 2A–2C); and 35S::Myc-rLOM1/2/3 plants exhibited a similar phenotype as that of 35S::LUC-rLOM1 with higher but varied degrees of trichome enrichment (Figure 2D–2F). To exclude the effect of Myc tag on LOM activities, we generated 35S::rLOM1 transgenic Arabidopsis, which exhibited the same phenotypic change as 35S::Myc-rLOM1 and 35S::LUC-rLOM1. Clearly, the three miR171-regulated LOMs have the ability to promote trichome initiation at flowering stage.
Fig. 2. Elevated LOMs trigger ectopic trichomes and down-regulate TRY and TCL1. 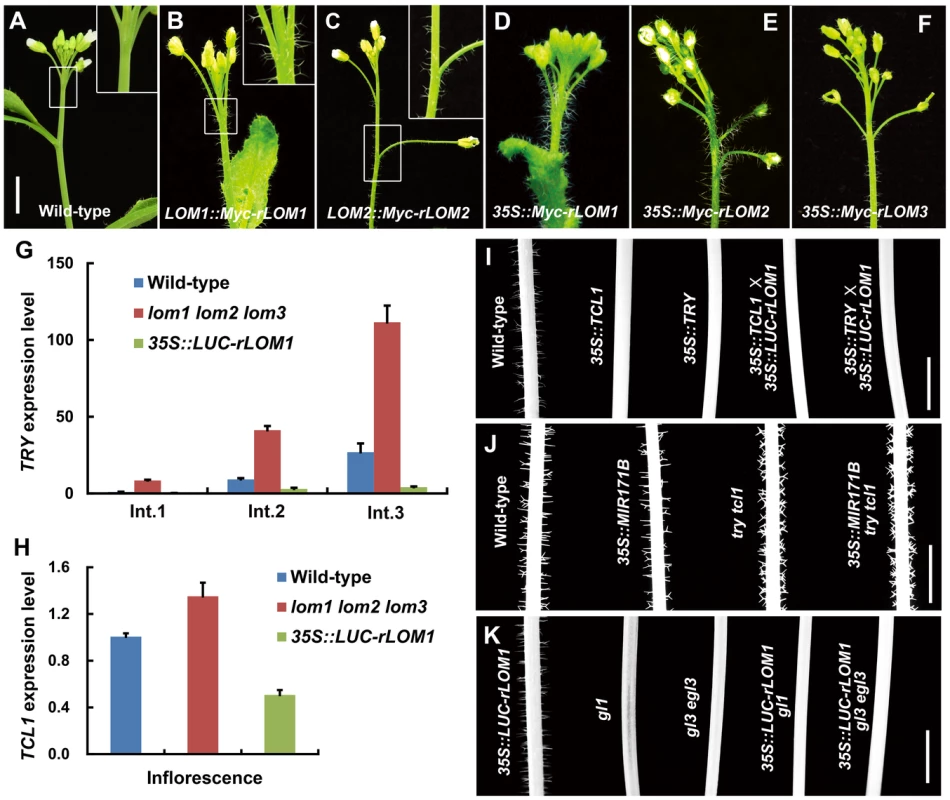
(A–F) Trichomes on main stems and inflorescences of different plants as indicated. The insertion panel in the top right corner in (A–C) represents magnified image of the area marked by square; note that ectopic trichomes appeared on inflorescence stems and pedicels in (B) and (C). Scale bar shared in (A–F), 1 cm. (G) Relative expression level of the trichome repressor gene TRY in main stem of wild-type, lomt and LUC-rLOM1OE plants. The Int.1 to Int.3 represent the stem internodes from bottom to top. Expression level in Int.1 of wild-type was set to 1. Error bars indicate s.d. (n = 3). (H) Relative expression level of TCL1 in inflorescences of wild-type, lomt and LUC-rLOM1OE plants. Errors bars indicate s.d. (n = 3). For (G) and (H) three biological replicates were analyzed with similar results. (I–K) Trichome distribution on main stems (Int.1) of different genetic background plants as indicated. Note that TCL1 and TRY are epistatic to LOM1, as LUC-rLOM1OE did not induce trichome production in TCL1OE and TRYOE backgrounds (I). MIR171BOE did not reduce trichome density in try tcl1 double mutant (J). 35S::LUC-rLOM1 failed to rescue the glabrous phenotype of gl1 and gl3 egl3 mutants (K). Scale bars in (I–K) represent 1 cm. Stem trichome repressors act at downstream of miR171-LOMs
Based on the Arabidopsis trichome model [12], ectopic trichomes could be triggered by either the increase of trichome promoting factors or the reduction of single-repeat R3 MYB repressors. For example, gibberellin and cytokinin promote trichome formation by activating the expression of C2H2 transcription factor genes GIS, GIS2 and ZFP8, which successively promote the transcription of GL1 and GL3 [30]–[32]. However, analysis of the lomt and the 35S::LUC-rLOM1 plants by quantitative RT-PCR (qRT-PCR) did not show an evident change of transcript levels of GIS, GIS2, ZFP8 and GL3 (Figure S1A). We then examined the expression of the R3 MYB repressor genes and found that transcript changes of CPC, ETC1 and TCL2 were uncoupled from trichome production in the lomt and the 35S::LUC-rLOM1 plants (Figure S1B and S1C), whereas the mRNA levels of TRY and TCL1 matched the phenotypes (Figures 2G and 2H). Indeed, among these single MYB-domain factors, TRY and TCL1 are the major negative regulators of stem and inflorescence trichomes [13], [19], [23]. TRY expression in the main stems was up-regulated in lomt and repressed in 35S::LUC-rLOM1 plants (Figure 2G). And similarly, TCL1 expression was also down-regulated in 35S::LUC-rLOM1 inflorescences (Figure 2H), consistent with the ectopic trichomes on inflorescence stems and pedicels. These data indicate that transcriptional repression of TRY and TCL1 occurred in LOM-overexpressors that produced ectopic trichomes.
To dissect genetic interaction between LOMs and the trichome repressor genes, we crossed 35S::LUC-rLOM1 to 35S::TRY and 35S::TCL1 plants, respectively. Overexpression of LOM1 failed to induce trichome production as both hybrid progeny exhibited the glabrous phenotype like 35S::TRY or 35S::TCL1 (Figure 2I). On the other hand, down-regulation of LOMs in try tcl1 double mutant by MIR171B overexpression did not reduce trichome production as it worked in wild-type background (Figure 2J). These data indicate that miR171-LOMs act upstream of TRY and TCL1 in promoting trichome formation. We also introduced 35S::LUC-rLOM1 into gl1 and gl3 egl3 backgrounds respectively, and the resultant plants barely developed trichomes (Figure 2K), demonstrating that, whether promoted by LOMs or not, trichome initiation in Arabidopsis requires the GL1-GL3/EGL3-TTG1 ternary complex.
LOMs attenuate SPL activities of trichome repression
Our previous report showed that SPLs repress stem and inflorescence trichome production through activating TCL1 and TRY gene expression [23]. Because up-regulation of LOMs (35S::LUC-rLOM1) and down-regulation of SPLs (35S::MIR156F) both reduced TCL1 and TRY expression and triggered ectopic trichomes, we wondered if miR171-LOMs induced trichome formation through affecting the miR156-targeted SPLs. To test this hypothesis, we crossed 35S::MIR171B to 35S::MIR156F plants. Although 35S::MIR171B repressed trichome formation on stem in wild-type plants, it did not change the ectopic trichome distribution induced by 35S::MIR156F (Figure 3A–3D), suggesting a requirement of miR156-targeted SPLs in miR171-mediated trichome suppression. Correspondently, mutation of LOM genes caused a further trichome reduction in 35S::MIM156 plants (Figure 3E and 3F), suggesting an enhancement of SPL activity in the absence of LOMs. High level of LOM1 (LOM1::Myc-rLOM1) stimulated while high level of SPL9 (SPL9::GFP-rSPL9) repressed stem trichome formation (Figure 3G and 3H). The plants expressing both (LOM1::Myc-rLOM1×SPL9::GFP-rSPL9) showed an intermediate phenotype in terms of stem trichomes, though less than the wild-type (Figure 3G and 3H). Together, these data suggest an antagonistic effect of LOMs on the SPLs. Although the intermediate phenotype mentioned above, at this stage it does not rule out the possibility of independent effects of these proteins on common downstream targets, such as TRY and TCL1.
Fig. 3. LOMs and SPLs regulate trichome formation antagonistically. 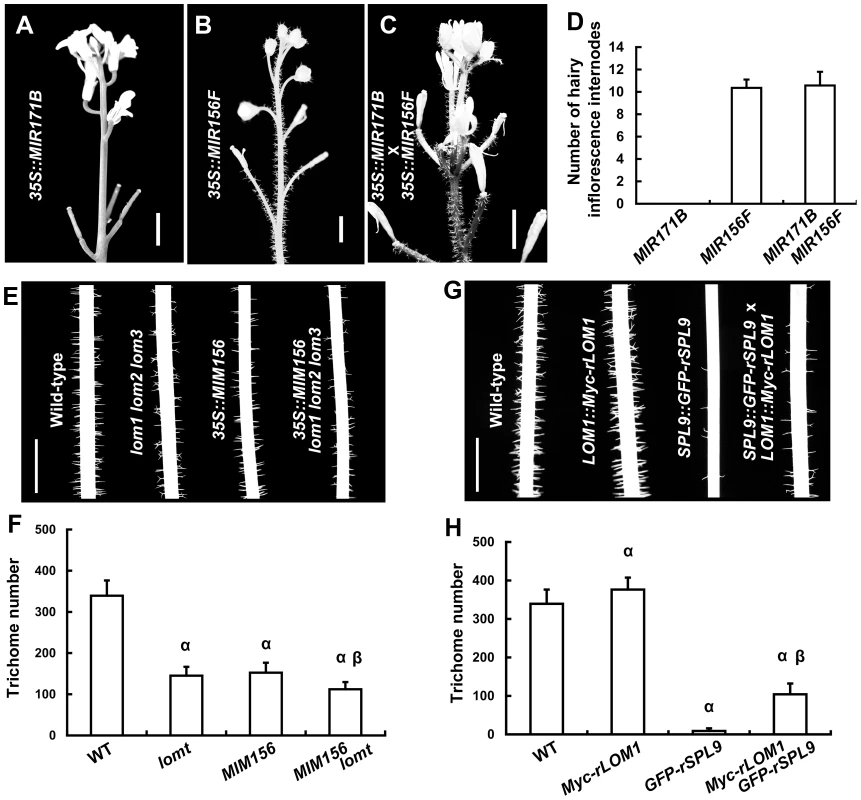
(A–C) Trichome distribution on stems and inflorescences of 35S::MIR171B (A), 35S::MIR156F (B) and the progeny of 35S::MIR171B×35S::MIR156F plants (C). Bars = 5 mm in (A–C). (D) Number of main inflorescence stem internodes with trichomes, counting from the first pedicel. Data are given as mean s.d. (n = 14). (E) Trichomes on stems (Int.1) of the indicated genotypes; note that trichome production was further reduced in 35S::MIM156 lomt plants. (F) Trichome numbers of plants in (E). The y axis indicates trichome number on main stem (0 to 5 cm from the bottom). Data are given as mean s.d. (n = 20); α represents P values (t-test)<0.01 relative to wild-type, and β represents P values (t-test)<0.01 relative to lomt and MIM156, respectively. Note that the trichome number of MIM156 lomt was very significantly less than that in MIM156 or lomt plants. (G) Trichomes on the stems (Int.1) of the indicated genotypes. SPL9::GFP-rSPL9 transgenic plants were almost glabrous, and the LOM1::Myc-rLOM1 plants produced more trichomes than the wild-type. (H) Trichome numbers (0 to 5 cm from the basal of main stems) of plants in (G). Data are given as mean s.d. (n = 20). α represents P values (t-test)<0.01 relative to wild-type, and β represents P values (t-test)<0.01 relative to Myc-rLOM1 and GFP-rSPL9, respectively. LOMs interact with SPLs
In order to further elucidate the correlation between LOMs and SPLs in trichome regulation, we analyzed TCL1 promoter activities in plants of different backgrounds. GUS activity conferred by TCL1::GUS was decreased in 35S::LUC-rLOM1 and increased in 35S::MIR171B plants (Figure 4A–4C), consistent with qRT-PCR results (Figure 2H). The increase of GUS activity in 35S::MIR171B was abolished when the mutant promoters of TCL1mu3::GUS and TCL1mu4::GUS (see [23]) were used, in which the SPL binding sites in TCL1 promoter were disrupted (Figures 4D–4G). Clearly, the SPL binding motifs were involved in regulation of TCL1 by LOMs, further supporting that LOMs modulate trichome formation via SPLs. 35S::LUC-rLOM1 and 35S::MIR156F plants exhibited other phenotypic similarities in addition to ectopic trichome production, such as short plastochron, delayed flowering time, yellow-green leaves, enhanced shoot branching and limp stem (see [26] and Figure S2), suggesting that the antagonistic effects between LOMs and SPLs could be general rather than limited to trichome regulation. It was reported that DELLAs physically interact with SPLs and gibberellin promotes flowering partially through releasing this interaction [5]. Since both LOM and DELLA proteins belong to two close subclades of the GRAS family [33], we wondered if LOMs also interacted with SPLs. We first examined the subcellular localization of SPL9 and LOM1. 35S::GFP-rSPL9 and 35S::mCherry-rLOM1 were transiently expressed in Nicotiana benthamiana leaves, and the fusion proteins of GFP-SPL9 and mCherry-LOM1 were found co-localized in speckles in the nucleus (Figure 4H). We then performed a biomolecular fluorescence complementation (BiFC) assay by fusing SPL9 to the C-terminal half of LUC (cLUC-rSPL9) and LOMs to the N-terminal half (LOMs-nLUC). A strong LUC activity was detected in leaves co-infiltrated with the respective chimerical constructs (Figure 4I). The interaction between SPL9 and LOMs was further confirmed in a yeast two-hybrid assay (Figure 4J). Domain deletion revealed that the N-terminal domain of LOM1 was responsible for its interaction with SPL9 (Figure 4K). In addition, LOMs were able to bind to SPL2 (Figure S3A), which is remarkably also involved in trichome development [34]. Taken together, these results demonstrate that the miR171-targeted LOMs physically interact with the miR156-targeted SPL9 and SPL2 and may result in attenuation of the SPL function such as regulating trichome patterning. Based on the facts that miR156 and miR171 are conserved in plant kingdom and excessive miRNAs cause opposite phenotypes, the interaction between the two miRNA targets coordinate many developmental and morphogenesis events beyond trichome formation.
Fig. 4. LOMs repress SPL function through direct protein-protein interaction. 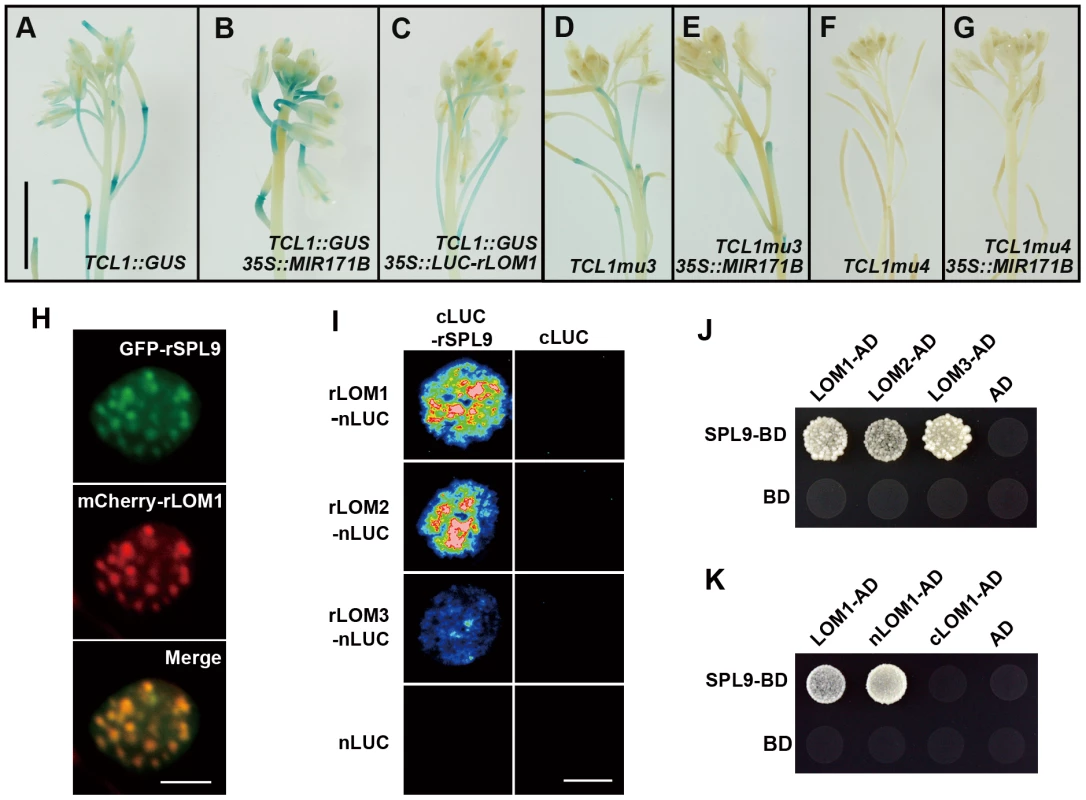
(A–C) TCL1::GUS reporter in indicated backgrounds. GUS activity was increased in MIR171BOE (B) and decreased in LUC-rLOM1OE (C) compared to wild-type (A) inflorescences. (D–G) The increase of GUS activity in MIR171BOE plants was abolished when the SPL binding sites in TCL1 promoter was mutated (see [23]). MIR171BOE did not increase the activity of TCL1mu3::GUS (E) and TCL1mu4::GUS (G), compared to that shown in (D) and (F), respectively. Scale bar in (A) represents 1 cm for (A–G). (H) SPL9 and LOM1 were co-localized in nucleus. 35S::GFP-rSPL9 and 35S::mCherry-rLOM1 were transiently expressed in N. benthamiana leaves and the fluorescence merged in nucleus. Scale bar, 10 µm. (I) BiFC assay of the interaction between LOMs and SPL9. rLOMs-nLUC or nLUC was transiently co-expressed with cLUC-rSPL9 or cLUC in N. benthamiana leaves; only the combination of rLOMs-nLUC and cLUC-rSPL9 triggered the luciferase activity when luciferin was infiltrated. Scale bar, 1 cm. (J) Yeast two-hybrid assay of protein-protein interaction. SPL9-BD combined with LOM1/2/3-AD conferred yeast growth in -Leu-Trp-His medium. (K) SPL9 interacts with the N-terminal (1–250) of LOM1 (nLOM1) in yeast two-hybrid system. LOMs repress SPL activities of gene regulation
To substantiate the potentially wide implication of LOM-SPL interaction, we examined the effect of LOMs on the flowering pathway. In long-day conditions, LOM1 overexpression delayed flowering time (Figure 5A and Figure S3B–S3E) and consistently down-regulated the expression of the MADS-box gene, SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1), which was under the direct control of miR156-tageted SPLs [2]; by contrast, SOC1 was up-regulated in lomt mutant along with earlier flowering (Figure 5A and 5B). As the N-terminal domain of LOM1 (nLOM1) was responsible for interaction with SPL9 (Figure 4K), we overexpressed nLOM1 fused to the nuclear localization signal (NLS). The 35S::nLOM1-NLS-YFP plants showed normal development (Figures 5C and 5D) except for late flowering and more trichomes on stems (Figures 5E–5G). A drastic drop of the SOC1 transcript levels (Figure 5F) was accompanied by severe delay of flowering (Figure 5E) in 35S::nLOM1-NLS-YFP plants. It is the same to rLOM1OE plants that nLOM1-NLS-YFP triggered more stem trichomes (Figure 5G). However, remarkable flowering delay and ectopic trichomes were not observed in 35S::nLOM1-YFP plants, in which the NLS was removed and the nLOM1-YFP signal was diffused in cytoplasm (Figure 5H). Because nLOM1 does not contain the DNA binding domain, these data further support that it is the LOM-SPL interaction that caused, or at least contributed to, the flowering delay and trichome increase.
Fig. 5. Overexpression of N-terminal of LOM1 in nucleus triggers a flowering delay. 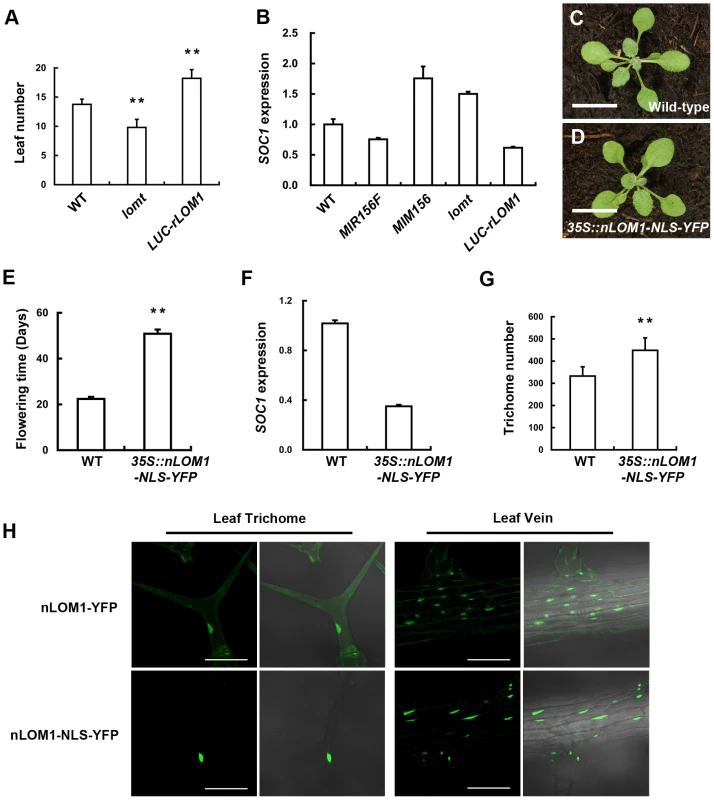
(A) The number of leaves at flowering of wild-type, lomt mutant and LUC-rLOM1OE plants under long-day condition. Data are given as mean s.d (n>61), and analyzed by t test. **P<0.01. (B) qRT-PCR analysis of expression of SOC1 in 7-day-old long-day-grown seedlings. SOC1 was repressed in 35S::MIR156F and increased in 35S::MIM156 as reported [2]. (C and D) View of 10-day-old wild-type (C) and 35S::nLOM1-NLS-YFP (D) plants. 35S::nLOM1-NLS-YFP plants look normal. Scale bars, 1 cm. (E) 35S::nLOM1-NLS-YFP delayed flowering very significantly under long-day condition. The number of days to flowering was counted when the floral buds were visible. Data are given as mean s.d. (n = 18) and analyzed by t test, **P<0.01. (F) Expression of SOC1. SOC1 was down-regulated in 35S::nLOM1-NLS-YFP plants, consistent with delayed flowering. (G) Trichome numbers of wild-type and 35S::nLOM1-NLS-YFP plants. The y axis indicates the total trichome number on main stems. Data are given as mean s.d. (n = 32) and analyzed by t test, **P<0.01. (H) Subcellular localization of nLOM1-YFP and nLOM1-NLS-YFP in leaf trichome (left panel) and leaf vein (right panel). Scale bars, 100 µm. We noted that, among the three LOMs, LOM3 has a shorter and more diversified N-terminal region (Figure S4). Interestingly, in comparison with 35S::Myc-rLOM1 and 35S::Myc-rLOM2 plants, overexpression of rLOM3 only led to a mild increase of trichome production (Figure 2F) and less serious developmental abnormalities. This result again points to the importance of the N-terminal domains of LOM proteins.
LOMs induce miR171 accumulation
To see whether LOMs affected the SPL output through mediating miR156 accumulation, we compared the miR156 amount in wild-type, lomt and 35S::LUC-rLOM1 plants by RNA blots. Under long-day conditions, the miR156 accumulation was similar in 10-day-old seedlings (Figure 6A). Although slightly higher in 20-day-old 35S::LUC-rLOM1 plants, the miR156 level was not altered in the stem of lomt or LUC-rLOM1OE plants (Figure 6B), neither was SPL9 expression (Figure S5). Thus it is unlikely that LOMs have a significant effect on miR156 abundance.
Fig. 6. LOMs regulate MIR171 expression. 
(A) RNA blot analysis of mature miR171 and miR156 in wild-type, lomt and LUC-rLOM1OE plants. Seedlings (10-day-old) and rosette leaves (20-day-old plants) were harvested for total RNA isolation. (B) RNA blot analysis of mature miR171 and miR156 in stem (Int.2 from the bottom) of 35-day-old plants. (C) RNA blots of miR171 in 10-day-old LOM overexpression plants; 35S::Myc-rLOM1/2/3 promoted miR171 accumulation. (D) qRT-PCR analysis of the pri-miRNA transcripts of MIR171A, B and C in 10-day-old plants of indicated genotypes. All three pri-miRNA transcripts were up-regulated in LUC-rLOM1OE and reduced in lomt mutant plants. Error bars indicate s.d. (n = 3). Three biological replicates were analyzed, with similar results. (E) Expression of three LOM genes in 10-day-old plants of indicated genotypes. LOM2 and LOM3 were slightly reduced in LUC-rLOM1OE plants probably due to elevated miR171 accumulation. Error bars indicate s.d. (n = 3). Three biological replicates were analyzed, with similar results. (F) GUS staining of MIR171A/B/C::GUS in different backgrounds. The GUS activity was stronger in LUC-rLOM1OE and weaker in MIR171BOE plants than that in wild-type. Interestingly, the miR171 level was clearly increased in 35S::LUC-rLOM1 but decreased in lomt mutant plants (Figure 6A), and overexpression of LOM2 or LOM3 also promoted miR171 accumulation (Figure 6C). qRT-PCR showed that the MIR171A transcript was much higher (up to 3-fold) in 35S::LUC-rLOM1 but lower in lomt mutant than in wild-type plants, whereas the expression of MIR171B and C showed a less degree response to LOM1 (Figure 6D). LOM2 and LOM3 expression was slightly reduced in 35S::LUC-rLOM1 plants (Figure 6E), possibly due to the elevated miR171 levels. We also generated the MIR171A/B/C::GUS reporter lines and crossed them to 35S::LUC-rLOM1 and 35S::MIR171B plants, respectively. Compared to wild-type, GUS activities of MIR171A::GUS and MIR171B::GUS were enhanced in 35S::LUC-rLOM1 and weakened in 35S::MIR171B plants, respectively (Figure 6F).
To investigate if MIR171 genes were directly regulated by their targets, we used an inducible system to test the activity of LOM1 in activating MIR171 expression. The 10-day-old 35S::rLOM1-GR lomt seedlings were sprayed with 10 mM dexamethasone (DEX) to allow the translocation of LOM1-GR fusion protein into the nucleus. An obvious transcript increase of MIR171A and, to a less extent, of MIR171B was observed after 4 hours, whereas MIR171C was not induced during this period (Figure 7A). Furthermore, in transient assays using N. benthamiana leaves, the level of luciferase activity controlled by the MIR171A promoter was elevated significantly when LOM1 was co-expressed, while the MIR171B and MIR171C promoters exhibited a weak or marginal response (Figure 7B). To identify the LOM1 binding regions, we dissected the truncated promoters of the three MIR171 genes (Figure 7C). Successive deletions from the 5′-end revealed a 143-bp promoter fragment of MIR171A (−201 to −343) which conferred the LOM1 induction (Figure 7D and Figure S6). However, the response of MIR171B and MIR171C promoters to LOM1 was negligible in N. benthamiana leaves (Figure 7E, 7F and Figure S6). Finally, we performed a chromatin immunoprecipitation (ChIP) assay using the 10-day-old 35S::Myc-rLOM1 plants, which showed LOM1 bound strongly to A4 region (Figure 7G), corresponding to the fragments identified in promoter deletion assays (Figure 7D and Figure S6). LOM1 weakly bound to promoters of MIR171B and MIR171C directly (Figure 7H and 7I). Together, these data reveal a regulatory feedback loop between LOMs and MIR171 genes, particularly MIR171A (Figure 8), which explains the seemingly contradictory phenomenon that miR171 and its target LOMs show similar expression patterns and both are mounting to a high level in inflorescence (Figure S7).
Fig. 7. LOM1 directly binds MIR171A promoter. 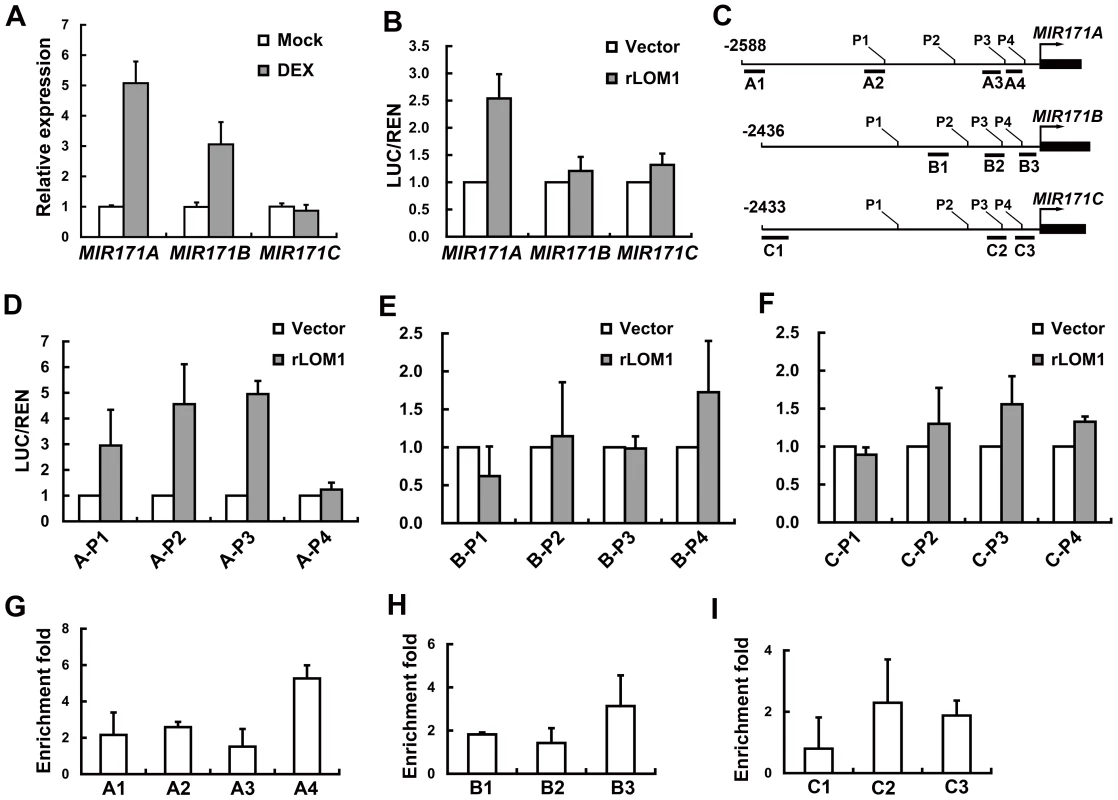
(A) Expression of MIR171A, B and C in rosette leaves of 10-day-old 35S::rLOM1-GR lomt seedlings after a 4-h DEX or a mock treatment. Error bars indicate s.d. of three technical replicates, and the results were consistent in three biological replicates. (B) Relative reporter activities showing the induction of MIR171 transcription by LOM1. N. benthamiana leaves were used for transient expression of the reporter (MIR171A/B/C::LUC) in combination with the effector (empty vector or 35S::rLOM1). The relative LUC activities normalized to the REN activity are shown (LUC/REN, n = 3). (C) Schematic diagram of MIR171A (top), MIR171B (middle) and MIR171C (bottom) upstream genomic regions. Black box represents stem-loop of the pri-miR171; P1–P4 represent the start sites of truncated promoters used in dual-LUC assays in (D–F); A1–A4, B1–B3 and C1–C3 represent the DNA amplicons for ChIP assays in (G–I). (D–F) Relative activities of truncated promoters of MIR171A (D), MIR171B (E) and MIR171C (F) in response to LOM1 induction. (G–I) ChIP enrichment of MIR171 promoter regions bound by Myc-LOM1. DNA fragments, isolated from rosette leaves of 10-day-old 35S::Myc-LOM1 and wild-type plants, were quantitatively analyzed by PCR, and β-TUBULIN-2 promoter was used as a reference. Error bars indicate s.d. of three quantitative PCR replicates. Fig. 8. A model for miR171-LOM and miR156-SPL interaction in regulating trichome formation and other biological events. 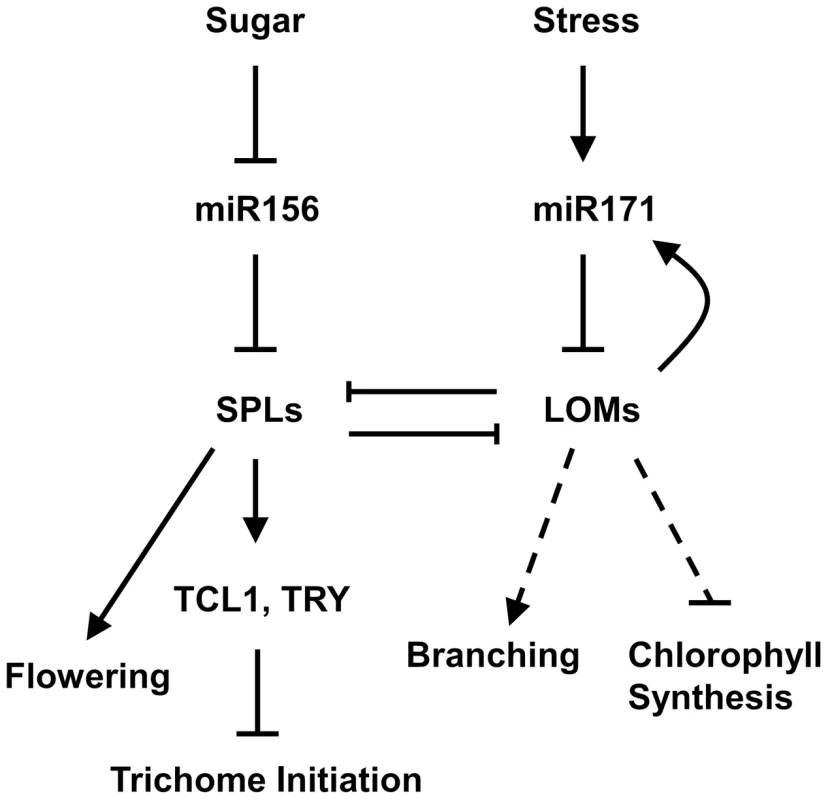
Trichomes are distributed on stem acropetally, with a high density at the basal part and sparse to nearly glabrous at apical part and inflorescence. The miR156-targeted SPLs up-regulate trichome negative factors TRY and TCL1 to repress trichome production on stem and inflorescence, whereas miR171-targeted LOMs counteract the SPLs through protein-protein interaction. LOMs positively regulate MIR171, forming a feedback loop to maintain their homeostasis, which influences the SPL output. Arrows indicate positive regulation, blunt ends indicate negative regulation, and the dashed line indicates the unidentified pathway. Discussion
By dissecting molecular mechanisms that control Arabidopsis trichome distribution, we found that the miR171-targeted LOMs directly interact with the miR156-targeted SPL9 and SPL2, leading to inhibition of the SPL activities. Remarkably, miR156 and miR171 function antagonistically in regulating many aspects of plant growth and development, including but far beyond the age-dependent trichome formation. Because both miR156 and miR171 are timing regulators, the interaction between their targets shed a new light on the endogenous network of plant aging. A similar but mechanically distinct scenario has been elucidated in C. elegans, where lin-4 and miR-239 oppositely regulate lifespan by common downstream genes [35], [36].
Except promoting the expression of miR172, SPLs positively regulate miR156 expression as well [3]. However, the miR171-LOM module is different from the miR156-SPL. The miR156 and its targets, such as SPL9, show reverse expression patterns [2]–[4], whereas miR171 and LOMs have a congruous temporal expression pattern (Figure S7). The level of LOMs elevates with age, leading to progressively activation of MIR171 genes, which in turn keep the LOM transcripts under the fine control. This regulatory feedback loop ensures the homeostasis of miR171 and its targets. Furthermore, since the LOMs themselves are transcription factors, the effect of LOM-SPL interaction can be bilateral. Taken chlorophyll content as an example, miR156 overexpression suppresses chlorophyll biosynthesis and this suppression is LOM-dependent (Figure S8A and S8B). Several transgenic lines of 35S::rLOM1-YFP and 35S::Myc-rLOM2 produced yellowish leaves during early vegetative stage due to decreased chlorophyll content as reported [26]. But this phenotype became less evident with plant aging (Figure S8C–S8E) when the SPL level was increasing, suggesting a possibility that the miR156-targeted SPLs enhance chlorophyll biosynthesis at least partially through negatively regulating the LOM activity. Finally, although overexpression of LOM1 and SPL9 simultaneously resulted in an intermediate phenotype, such as the trichome number shown in Figure 3, the two transcription factors may act independently on downstream genes. Identification of the targets of LOM factors will further our understanding of the biological significance of LOM-SPL interaction.
In addition to LOMs, DELLAs could bind to SPLs as well. DELLAs are degraded in response to gibberellin [37]–[39], resulting in release of the factors they bind, and the SPL-DELLA interaction integrates the hormone signals to the miR156-SPL pathway in regulating plant flowering [5]. MiR171 is not only regulated by endogenous cues, but also responds to environmental stress, such as cold, high salt and hydration [40]–[43]. Thus the LOM-SPL interaction may transduce environmental stimuli into endogenous signaling to adjust plant phase transition and development. Because both LOMs and SPLs mount with plant age, and their over-accumulation have adverse effects on plant growth and development [2], [3], [26], [44], we propose that plants have employed LOMs as a damper to measure the increasingly higher SPL output at protein level in aged plants when the miR156 level is low, and likewise SPLs may temporally restrict the activity of LOMs. Since the miR156-targeted SPLs function as a key endogenous age cue and SPL9 is one of the highly active SPL members, the LOM-SPL interaction has a profound contribution to programming plant life.
Mining of genome data revealed that both miR156 and miR171 are highly conserved in land plants from moss (Physcomitrella patens) to flowering plants of both monocots and dicots [45], and in crop plants they control important agronomic traits [46], [47]. In the diploid cotton species of Gossypium raimondii there are 15 MIR171 and seven putative LOM genes, of which five LOMs contain the miR171 recognition sites (see [48] and Figure S9). It would be interesting to examine if LOMs are involved in regulating cotton fiber (seed trichome) development. Notably miR171a* is also functional in gene silencing and the miR171a*-SU(VAR)3-9 HOMOLOG8 pair was proposed to have evolved very recently in the Arabidopsis lineage [44]. A recent report showed that overexpression of miR171 (Hvu-pri-miR171a) in barley up-regulated miR156 and repressed vegetative phase transitions [49], which is in contrast with the opposite effects of the two miRNAs in Arabidopsis described herein and reported by others [26], [29], [50]. Whether the interplay between the two conserved aging miRNAs varies with plant taxa is a subject of further study.
Materials and Methods
Plant materials and constructs
Plants of Arabidopsis thaliana, ecotype Columbia (Col-0), and Nicotiana benthamiana were grown at 22°C in long days (16 h light/8 h dark). The lom triple mutant [26], 35S::LUC-rLOM1 [26], 35S::MIR156F [23], 35S::MIM156 [23], SPL9::GFP-rSPL9 [23], TCL1::GUS [23], TCL1mu3/4::GUS [23] have been described previously. The gl1 (SALK_039478), try (SALK_029760), tcl1 (SALK_055460) and gl3 egl3 (CS66490) mutants were obtained from Arabidopsis Biological Resource Center (ABRC).
For MIR171A/B/C::GUS constructs, the promoters of MIR171A/B/C (∼2 kb) were PCR amplified using PrimeSTAR HS DNA polymerase (TaKaRa) and individually fused to the GUS coding region. For LOM constructs, the miRNA-resistant versions were created by two-round PCR. The resultant fragment was inserted into a vector which harbors a 35S::6×Myc cassette to generate 35S::Myc-rLOM1/2/3. Then the 35S promoter was replaced by a native promoter to generate LOM1::Myc-rLOM1 and LOM12::Myc-rLOM2. At least 30 T1 seedlings were analyzed for each construct. For BiFC constructs, coding regions of the miRNA-resistant LOMs and SPL9 were PCR amplified and cloned into JW771 and JW772 [51], respectively. Primers are listed in Supplementary Table S1.
Trichome number and flowering time measurement
The trichome numbers were counted on each internode of stem, and the number was divided by the length of each internode to calculate the trichome density (number per centimeter). Flowering time was measured by counting the total number of leaves (rosette and cauline leaves) and the number of days to flowering under long-day condition. The data were given as mean s.d. and analyzed by t test.
Gene expression analyses
RNAs were extracted with Trizol reagent (Invitrogen) following the manufacturer's instructions. Total RNAs of 1 µg were used for reverse transcription in a 20 µL reaction system with M-MLV Reverse Transcriptase kit (Invitrogen). The fragments of interest were amplified by RT-PCR using sequence-specific primers (see Supplementary Table S1). Real-time PCR was performed with SYBR Premix Ex Taq II (Takara), and amplification was monitored on the Mastercycler ep RealPlex2 (Eppendorf). The gene expression level was normalized to reference gene β-TUBULIN2 (At5g62690). For DEX induction, 10-day-old 35S::rLOM1-GR lomt seedlings were sprayed with 10 mM DEX (Sigma-Aldrich) or alcohol (mock control). After 4 hours, rosette leaves were harvested.
Protein–protein interaction assays
For subcellular localization assay, 35S::GFP-rSPL9 and 35S::mCherry-rLOM1 were transiently expressed in N. benthamiana leaves. After 3 days, the materials were observed using confocal microscope OLYMPUS FV1000.
Yeast two-hybrid assay was performed using the Matchmaker GAL4 Two-Hybrid System according to the manufacturer's manual (Clontech). Full-length or truncated cDNAs of LOM1 were inserted into pGBKT7 and those of SPLs into pGADT7, respectively. Plasmids were transferred into yeast strain AH109 (Clontech) by the LiCl-PEG method. The interactions were tested on SD/-Leu/-Trp/-His plates supplemented with 15 mM 3-amino-1,2,4,-triazole. Three independent clones for each transformation were tested.
BiFC assays were performed as described [51], [52]. Briefly, four chimerical constructs were used. SPL9 was fused to C-terminal half of luciferase (cLUC-rSPL9) and three LOMs to the N-terminal half (LOMs-nLUC); cLUC and nLUC alone were used as controls. Agrobacterium tumefaciens cells were re-suspended in infiltration buffer (10 mM MgCl2, 10 mM MES pH 5.7, 150 µm acetosyringone) at OD600 = 0.8. 35S::P19-HA and the suspension was co-infiltrated to inhibit gene silencing [53]. After 3 days, a total of 0.8 mM luciferin was infiltrated and the LUC activity was monitored. The following pairs of constructs were used for co-infiltration: cLUC-rSPL9 and LOMs-nLUC, cLUC and LOMs-nLUC, cLUC-rSPL9 and nLUC, as well as cLUC and nLUC.
Small RNA blot
Total RNAs were extracted using Trizol reagent (Invitrogen), and 5–20 µg of the total RNA were resolved on 17% polyacrylamide gels under denaturing conditions (7 M urea). RNAs were then transferred to HyBond-N+ membranes (GE Healthcare) by semidry blotting, and membranes were hybridized with oligonucleotide DNA probes labeled with digoxigenin using the DIG Oligonucleotide 3-End Labeling Kit, Second Generation (Roche). Oligonucleotide sequences are listed in Supplementary Table S1 online.
ChIP assays
Chromatin immunoprecipitation (ChIP) experiments were performed as described [54]. Tissues (∼2 g) of 10-day-old 35S::Myc-rLOM1 or wild-type seedlings were harvested and then cross-linked in formaldehyde solution (1%) under a vacuum. The material was washed and ground in liquid nitrogen, the resultant powder was re-suspended in extraction buffer (0.4 M sucrose, 10 mM Tris-HCl, pH 8.0, 10 mM MgCl2, 5 mM mercaptoethanol, 0.1 mM PMSF, and 1× protease inhibitor [Roche]) and lysis buffer (50 mM HEPES, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% deoxycholate sodium, and 0.1% SDS), successively, followed by sonification (output 3, 6×10 s). An anti-Myc antibody (Abmart) was added for precipitation. After several washes, DNA samples were reversely cross-linked and then purified using a PCR purification kit (Qiagen). The relative amounts of the DNA amplicons were analyzed by quantitative PCR using the β-TUBULIN2 gene promoter as a reference. Relative enrichment was calculated by normalizing the value in 35S::Myc-rLOM1 against the value in wild-type.
Dual-LUC assays
A dual-luc method using N. benthamiana plants was used [55]. Briefly, the effector plasmid is 35S::rLOM1 or empty vector, and the reporter plasmid, pGreen-0800-LUC, harbors two luciferases: the firefly luciferase (LUC) controlled by the MIR171 promoter, and the Renilla (REN) luciferase controlled by the constitutive 35S promoter. The Agrobacterium strain containing the reporter was mixed with the effector strain (at the reporter:effector ratio of 1∶3). The mixture was infiltrated into leaves of N. benthamiana. Three days later, leaf samples were collected for the dual-luc assay using commercial Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer's instruction. The LUC activity was normalized to REN. Three biological repeats were measured for each sample.
GUS staining
GUS activity was assayed by staining. Plant materials were submerged in 0.5 mg/mL X-Gluc solution (0.1 M sodium phosphate buffer, pH 7.0, 10 mM EDTA, 0.1% Triton X-100, 0.5 mM potassium ferrocyanide, 0.5 mM potassium ferricyanide), vacuumized and kept at 37°C. Subsequent materials were decolorized in 70% ethanol.
Accession numbers
LOM1 (At2G45160), LOM2 (At3G60630), LOM3 (At4G00150), MIR171A (At3G51375), MIR171B (At1G11735), MIR171C (At1G62035), SPL9 (At2g42200), SPL2 (At5G43270), MIR156F (At5G26147), SOC1 (At2g45660), GL1 (At3G27920), GL3 (At5G41315), EGL3 (At1G63650), TCL1 (At2g30432), TRY (At5G53200), GIS (At3g58070), GIS2 (At5g06650 ), ZFP8 (At2g41940), β-TUBULIN-2 (At5g62690) and Ph-HAM (AY112704).
Supporting Information
Zdroje
1. LeeRC, FeinbaumRL, AmbrosV (1993) The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75 : 843–854.
2. WangJW, CzechB, WeigelD (2009) miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138 : 738–749.
3. WuG, ParkMY, ConwaySR, WangJW, WeigelD, et al. (2009) The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138 : 750–759.
4. WuG, PoethigRS (2006) Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development 133 : 3539–3547.
5. YuS, GalvaoVC, ZhangYC, HorrerD, ZhangTQ, et al. (2012) Gibberellin regulates the Arabidopsis floral transition through miR156-targeted SQUAMOSA PROMOTER BINDING-LIKE transcription factors. Plant Cell 24 : 3320–3332.
6. LawsonEJ, PoethigRS (1995) Shoot development in plants: time for a change. Trends Genet 11 : 263–268.
7. TelferA, BollmanKM, PoethigRS (1997) Phase change and the regulation of trichome distribution in Arabidopsis thaliana. Development 124 : 645–654.
8. PayneCT, ZhangF, LloydAM (2000) GL3 encodes a bHLH protein that regulates trichome development in Arabidopsis through interaction with GL1 and TTG1. Genetics 156 : 1349–1362.
9. PeschM, HulskampM (2004) Creating a two-dimensional pattern de novo during Arabidopsis trichome and root hair initiation. Curr Opin Genet Dev 14 : 422–427.
10. RamsayNA, GloverBJ (2005) MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci 10 : 63–70.
11. RerieWG, FeldmannKA, MarksMD (1994) The GLABRA2 gene encodes a homeo domain protein required for normal trichome development in Arabidopsis. Genes Dev 8 : 1388–1399.
12. IshidaT, KurataT, OkadaK, WadaT (2008) A genetic regulatory network in the development of trichomes and root hairs. Annu Rev Plant Biol 59 : 365–386.
13. EschJJ, ChenMA, HillestadM, MarksMD (2004) Comparison of TRY and the closely related At1g01380 gene in controlling Arabidopsis trichome patterning. Plant J 40 : 860–869.
14. WadaT, TachibanaT, ShimuraY, OkadaK (1997) Epidermal cell differentiation in Arabidopsis determined by a Myb homolog, CPC. Science 277 : 1113–1116.
15. SchellmannS, SchnittgerA, KirikV, WadaT, OkadaK, et al. (2002) TRIPTYCHON and CAPRICE mediate lateral inhibition during trichome and root hair patterning in Arabidopsis. EMBO J 21 : 5036–5046.
16. KirikV, SimonM, HuelskampM (2004) Schiefelbein J (2004) The ENHANCER OF TRY AND CPC1 gene acts redundantly with TRIPTYCHON and CAPRICE in trichome and root hair cell patterning in Arabidopsis. Dev Biol 268 : 506–513.
17. KirikV, SimonM, WesterK, SchiefelbeinJ, HulskampM (2004) ENHANCER of TRY and CPC 2 (ETC2) reveals redundancy in the region-specific control of trichome development of Arabidopsis. Plant Mol Biol 55 : 389–398.
18. SimonM, LeeMM, LinY, GishL, SchiefelbeinJ (2007) Distinct and overlapping roles of single-repeat MYB genes in root epidermal patterning. Dev Biol 311 : 566–578.
19. WangS, KwakSH, ZengQ, EllisBE, ChenXY, et al. (2007) TRICHOMELESS1 regulates trichome patterning by suppressing GLABRA1 in Arabidopsis. Development 134 : 3873–3882.
20. GanL, XiaK, ChenJG, WangS (2011) Functional characterization of TRICHOMELESS2, a new single-repeat R3 MYB transcription factor in the regulation of trichome patterning in Arabidopsis. BMC Plant Biol 11 : 176.
21. Tominaga-WadaR, NukumizuY (2012) Expression Analysis of an R3-Type MYB Transcription Factor CPC-LIKE MYB4 (TRICHOMELESS2) and CPL4-Related Transcripts in Arabidopsis. Int J Mol Sci 13 : 3478–3491.
22. QiT, SongS, RenQ, WuD, HuangH, et al. (2011) The Jasmonate-ZIM-domain proteins interact with the WD-Repeat/bHLH/MYB complexes to regulate Jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell 23 : 1795–1814.
23. YuN, CaiWJ, WangS, ShanCM, WangLJ, et al. (2010) Temporal control of trichome distribution by microRNA156-targeted SPL genes in Arabidopsis thaliana. Plant Cell 22 : 2322–2335.
24. StuurmanJ, JaggiF, KuhlemeierC (2002) Shoot meristem maintenance is controlled by a GRAS-gene mediated signal from differentiating cells. Genes Dev 16 : 2213–2218.
25. EngstromEM, AndersenCM, Gumulak-SmithJ, HuJ, OrlovaE, et al. (2011) Arabidopsis homologs of the Petunia HAIRY MERISTEM gene are required for maintenance of shoot and root indeterminacy. Plant Physiol 155 : 735–750.
26. WangL, MaiYX, ZhangYC, LuoQ, YangHQ (2010) MicroRNA171c-targeted SCL6-II, SCL6-III, and SCL6-IV genes regulate shoot branching in Arabidopsis. Mol Plant 3 : 794–806.
27. LlaveC, XieZ, KasschauKD, CarringtonJC (2002) Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297 : 2053–2056.
28. RhoadesMW, ReinhartBJ, LimLP, BurgeCB, BartelB, et al. (2002) Prediction of plant microRNA targets. Cell 110 : 513–520.
29. SchulzeS, SchaferBN, ParizottoEA, VoinnetO, TheresK (2010) LOST MERISTEMS genes regulate cell differentiation of central zone descendants in Arabidopsis shoot meristems. Plant J 64 : 668–678.
30. GanY, KumimotoR, LiuC, RatcliffeO, YuH, et al. (2006) GLABROUS INFLORESCENCE STEMS modulates the regulation by gibberellins of epidermal differentiation and shoot maturation in Arabidopsis. Plant Cell 18 : 1383–1395.
31. ZhouZ, AnL, SunL, GanY (2012) ZFP5 encodes a functionally equivalent GIS protein to control trichome initiation. Plant Signal Behav 7 : 28–30.
32. ZhouZ, SunL, ZhaoY, AnL, YanA, et al. (2013) Zinc Finger Protein 6 (ZFP6) regulates trichome initiation by integrating gibberellin and cytokinin signaling in Arabidopsis thaliana. New Phytol 198 : 699–708.
33. EngstromEM (2011) Phylogenetic analysis of GRAS proteins from moss, lycophyte and vascular plant lineages reveals that GRAS genes arose and underwent substantial diversification in the ancestral lineage common to bryophytes and vascular plants. Plant Signal Behav 6 : 850–854.
34. ShikataM, KoyamaT, MitsudaN, Ohme-TakagiM (2009) Arabidopsis SBP-box genes SPL10, SPL11 and SPL2 control morphological change in association with shoot maturation in the reproductive phase. Plant Cell Physiol 50 : 2133–2145.
35. Smith-VikosT, SlackFJ (2012) MicroRNAs and their roles in aging. Journal of Cell Science 125 : 7–17.
36. de LencastreA, PincusZ, ZhouK, KatoM, LeeSS, et al. (2010) MicroRNAs both promote and antagonize longevity in C. elegans. Curr Biol 20 : 2159–2168.
37. MuraseK, HiranoY, SunTP, HakoshimaT (2008) Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature 456 : 459–463.
38. GriffithsJ, MuraseK, RieuI, ZentellaR, ZhangZL, et al. (2006) Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell 18 : 3399–3414.
39. SunTP (2011) The molecular mechanism and evolution of the GA-GID1-DELLA signaling module in plants. Curr Biol 21: R338–345.
40. LiuHH, TianX, LiYJ, WuCA, ZhengCC (2008) Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. RNA 14 : 836–843.
41. ZhouL, LiuY, LiuZ, KongD, DuanM, et al. (2010) Genome-wide identification and analysis of drought-responsive microRNAs in Oryza sativa. J Exp Bot 61 : 4157–4168.
42. ZhaoM, TaiH, SunS, ZhangF, XuY, et al. (2012) Cloning and characterization of maize miRNAs involved in responses to nitrogen deficiency. PLoS One 7: e29669.
43. LiangG, HeH, YuD (2012) Identification of nitrogen starvation-responsive microRNAs in Arabidopsis thaliana. PLoS One 7: e48951.
44. ManavellaPA, KoenigD, Rubio-SomozaI, BurbanoHA, BeckerC, et al. (2013) Tissue-specific silencing of Arabidopsis SU(VAR)3-9 HOMOLOG8 by miR171a. Plant Physiol 161 : 805–812.
45. AxtellMJ, SnyderJA, BartelDP (2007) Common functions for diverse small RNAs of land plants. Plant Cell 19 : 1750–1769.
46. MiuraK, IkedaM, MatsubaraA, SongXJ, ItoM, et al. (2010) OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat Genet 42 : 545–549.
47. JiaoY, WangY, XueD, WangJ, YanM, et al. (2010) Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat Genet 42 : 541–544.
48. WangK, WangZ, LiF, YeW, WangJ, et al. (2012) The draft genome of a diploid cotton Gossypium raimondii. Nat Genet 44 : 1098–1103.
49. CurabaJ, TalbotM, LiZ, HelliwellC (2013) Over-expression of microRNA171 affects phase transitions and floral meristem determinancy in barley. BMC Plant Biol 13 : 6.
50. WangJW, SchwabR, CzechB, MicaE, WeigelD (2008) Dual effects of miR156-targeted SPL genes and CYP78A5/KLUH on plastochron length and organ size in Arabidopsis thaliana. Plant Cell 20 : 1231–1243.
51. GouJY, FelippesFF, LiuCJ, WeigelD, WangJW (2011) Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant Cell 23 : 1512–1522.
52. ChenH, ZouY, ShangY, LinH, WangY, et al. (2008) Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol 146 : 368–376.
53. PappI, MetteMF, AufsatzW, DaxingerL, SchauerSE, et al. (2003) Evidence for nuclear processing of plant micro RNA and short interfering RNA precursors. Plant Physiol 132 : 1382–1390.
54. HongGJ, XueXY, MaoYB, WangLJ, ChenXY (2012) Arabidopsis MYC2 interacts with DELLA proteins in regulating sesquiterpene synthase gene expression. Plant Cell 24 : 2635–2648.
55. BanerjeeR, SchleicherE, MeierS, VianaRM, PokornyR, et al. (2007) The signaling state of Arabidopsis cryptochrome 2 contains flavin semiquinone. J Biol Chem 282 : 14916–14922.
56. ArnonDI (1949) Copper enzymes in isolated chloroplasts: Polyphenol oxidase in Beta vulgaris. Plant Physiol 24 : 1–15.
Štítky
Genetika Reprodukční medicína
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 4- Akutní intermitentní porfyrie
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Intrauterinní inseminace a její úspěšnost
- Růst a vývoj dětí narozených pomocí IVF
- Pánevní endometrióza spojená s volnou tekutinou v peritoneální dutině snižuje úspěšnost otěhotnění po intrauterinní inseminaci
-
Všechny články tohoto čísla
- The Challenges of Mitochondrial Replacement
- Concocting Cholinergy
- Genome-Wide Diet-Gene Interaction Analyses for Risk of Colorectal Cancer
- Statistical Power to Detect Genetic (Co)Variance of Complex Traits Using SNP Data in Unrelated Samples
- Mouse Pulmonary Adenoma Susceptibility 1 Locus Is an Expression QTL Modulating -4A
- Transcription-Associated R-Loop Formation across the Human CGG-Repeat Region
- Epigenetic Regulation by Heritable RNA
- Protein Quantitative Trait Loci Identify Novel Candidates Modulating Cellular Response to Chemotherapy
- Genome-Wide Profiling of Yeast DNA:RNA Hybrid Prone Sites with DRIP-Chip
- The Mechanism of Gene Targeting in Human Somatic Cells
- A LINE-1 Insertion in DLX6 Is Responsible for Cleft Palate and Mandibular Abnormalities in a Canine Model of Pierre Robin Sequence
- Interaction between Two Timing MicroRNAs Controls Trichome Distribution in
- DNA Glycosylases Involved in Base Excision Repair May Be Associated with Cancer Risk in and Mutation Carriers
- The Myc-Mondo/Mad Complexes Integrate Diverse Longevity Signals
- Evolutionarily Diverged Regulation of X-chromosomal Genes as a Primal Event in Mouse Reproductive Isolation
- Mutations in Conserved Residues of the microRNA Argonaute ALG-1 Identify Separable Functions in ALG-1 miRISC Loading and Target Repression
- Genetic Predisposition to In Situ and Invasive Lobular Carcinoma of the Breast
- Isl1 Directly Controls a Cholinergic Neuronal Identity in the Developing Forebrain and Spinal Cord by Forming Cell Type-Specific Complexes
- A Synthetic Community Approach Reveals Plant Genotypes Affecting the Phyllosphere Microbiota
- The Sequence-Specific Transcription Factor c-Jun Targets Cockayne Syndrome Protein B to Regulate Transcription and Chromatin Structure
- Determining the Control Circuitry of Redox Metabolism at the Genome-Scale
- DNA Repair Pathway Selection Caused by Defects in , , and Telomere Addition Generates Specific Chromosomal Rearrangement Signatures
- Methylome Diversification through Changes in DNA Methyltransferase Sequence Specificity
- Folliculin Regulates Ampk-Dependent Autophagy and Metabolic Stress Survival
- Fine Mapping of Dominant -Linked Incompatibility Alleles in Hybrids
- Unexpected Role of the Steroid-Deficiency Protein Ecdysoneless in Pre-mRNA Splicing
- Three Groups of Transposable Elements with Contrasting Copy Number Dynamics and Host Responses in the Maize ( ssp. ) Genome
- Sox5 Functions as a Fate Switch in Medaka Pigment Cell Development
- Synergistic Interactions between the Molecular and Neuronal Circadian Networks Drive Robust Behavioral Circadian Rhythms in
- Chromatin Landscapes of Retroviral and Transposon Integration Profiles
- Widespread Use of Non-productive Alternative Splice Sites in
- Ras GTPase-Like Protein MglA, a Controller of Bacterial Social-Motility in Myxobacteria, Has Evolved to Control Bacterial Predation by
- Cell Type-Specific Functions of Genes Revealed by Novel Adipocyte and Hepatocyte Circadian Clock Models
- Phenotype Ontologies and Cross-Species Analysis for Translational Research
- Embryogenesis Scales Uniformly across Temperature in Developmentally Diverse Species
- In Pursuit of the Gene: An Interview with James Schwartz
- Molecular Mechanisms of Hypoxic Responses via Unique Roles of Ras1, Cdc24 and Ptp3 in a Human Fungal Pathogen
- Analysis of the Genome and Transcriptome of var. Reveals Complex RNA Expression and Microevolution Leading to Virulence Attenuation
- Genotypic and Functional Impact of HIV-1 Adaptation to Its Host Population during the North American Epidemic
- RNA Editome in Rhesus Macaque Shaped by Purifying Selection
- Proper Actin Ring Formation and Septum Constriction Requires Coordinated Regulation of SIN and MOR Pathways through the Germinal Centre Kinase MST-1
- Interplay of the Serine/Threonine-Kinase StkP and the Paralogs DivIVA and GpsB in Pneumococcal Cell Elongation and Division
- A Quality Control Mechanism Coordinates Meiotic Prophase Events to Promote Crossover Assurance
- CNNM2 Mutations Cause Impaired Brain Development and Seizures in Patients with Hypomagnesemia
- The RNA-Binding Protein QKI Suppresses Cancer-Associated Aberrant Splicing
- Uncoupling Transcription from Covalent Histone Modification
- Rad51–Rad52 Mediated Maintenance of Centromeric Chromatin in
- FRA2A Is a CGG Repeat Expansion Associated with Silencing of
- A General Approach for Haplotype Phasing across the Full Spectrum of Relatedness
- A Novel Highly Divergent Protein Family Identified from a Viviparous Insect by RNA-seq Analysis: A Potential Target for Tsetse Fly-Specific Abortifacients
- A Central Role for in Regulation of Islet Function in Man
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- The Sequence-Specific Transcription Factor c-Jun Targets Cockayne Syndrome Protein B to Regulate Transcription and Chromatin Structure
- The Mechanism of Gene Targeting in Human Somatic Cells
- Genetic Predisposition to In Situ and Invasive Lobular Carcinoma of the Breast
- Widespread Use of Non-productive Alternative Splice Sites in
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



