-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaMouse Pulmonary Adenoma Susceptibility 1 Locus Is an Expression QTL Modulating -4A
A person's risk of developing cancer depends on both genetic and environmental factors. To study the genetic predisposition to cancer without the influence of environmental variables, scientists study mice treated with urethane, a chemical carcinogen that induces lung tumors. By crossing inbred (genetically identical) strains of mice that are either resistant or susceptible to urethane-induced cancer, researchers can search for genes associated with tumor formation in the offspring. From previous work of this type using second-generation mice, it was already known that a region on chromosome 6 was associated with tumor formation. Now, a new study, carried out in a fourth-generation mouse population, focused to a single gene of chromosome 6 called Kras. This gene forms two different messenger RNA transcripts, called Kras-4A and Kras-4B, that produce two proteins with slightly different structure and, perhaps, function. The study found that mice susceptible to lung tumors have relatively more Kras-4A messenger RNA than resistant mice and that this difference may be due to small variations in the DNA near or within this gene.
Published in the journal: . PLoS Genet 10(4): e32767. doi:10.1371/journal.pgen.1004307
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004307Summary
A person's risk of developing cancer depends on both genetic and environmental factors. To study the genetic predisposition to cancer without the influence of environmental variables, scientists study mice treated with urethane, a chemical carcinogen that induces lung tumors. By crossing inbred (genetically identical) strains of mice that are either resistant or susceptible to urethane-induced cancer, researchers can search for genes associated with tumor formation in the offspring. From previous work of this type using second-generation mice, it was already known that a region on chromosome 6 was associated with tumor formation. Now, a new study, carried out in a fourth-generation mouse population, focused to a single gene of chromosome 6 called Kras. This gene forms two different messenger RNA transcripts, called Kras-4A and Kras-4B, that produce two proteins with slightly different structure and, perhaps, function. The study found that mice susceptible to lung tumors have relatively more Kras-4A messenger RNA than resistant mice and that this difference may be due to small variations in the DNA near or within this gene.
Introduction
The Pulmonary adenoma susceptibility 1 (Pas1) locus, mapping in the distal region of chromosome 6, is the major modulator of lung tumor susceptibility in mice [1], [2]. In inbred mouse strains, the Pas1 locus has a conserved haplotype consisting of six genes clustered in a ∼450-kb region [3]. From proximal to distal, these genes are branched chain aminotransferase 1 (Bcat1), lymphoid-restricted membrane protein (Lrmp), cancer susceptibility candidate 1 (Casc1), LYR motif containing 5 (Lyrm5), v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog (Kras) and intermediate filament tail domain containing 1 (Ifltd1). Four of these genes, namely Lrmp, Casc1, Kras and Ifltd1, have been singled out as candidate genes for Pas1 locus functions in fine-mapping studies [4]–[6]. Although for none of these genes is there a clear demonstration of a role in lung tumor susceptibility, there is substantial evidence supporting the involvement of Kras as the key effector of the Pas1 locus.
Kras encodes a small GTPase that functions as a molecular switch in signal transduction, influencing cell proliferation [7]. Permanently activating mutations, at codons 12, 13 and 61, are frequently found in both spontaneous and chemically induced lung tumors in mice [8], [9]. Moreover, in mouse models in which mutant Kras can be activated by somatic recombination in the lung, animals are highly susceptible to lung tumorigenesis and develop multiple lung tumors at 100% incidence with a short latency [10], [11]. Nonetheless, as heterozygous Kras knockout mice have higher susceptibility to chemically induced lung tumorigenesis than wild-type mice, the wild-type Kras allele may have a tumor suppression function [12]. The double role of Kras in lung carcinogenesis—tumor suppressor when wild-type and oncogene when mutated—led to the hypothesis that lung cancer susceptibility could result from the subtle balance between expression levels of wild-type and mutated Kras [13]. Given the frequent occurrence of activating Kras mutations in mouse lung tumors, an increase in Kras gene expression in lung tumor tissue could be expected to raise the level of active Kras protein, thereby providing the growth advantage characteristic of neoplasms. Evidence supporting this mechanism was provided by the observation that, in mice, Kras mRNA levels in normal lung tissue were ∼2-fold higher in strains susceptible to lung tumorigenesis (both highly susceptible A/J mice and intermediate-susceptible FVB/N and 129/Sv mice) than in a resistant strain (C57BL/6) [13].
Our understanding of the mode of action of Kras in lung tumorigenesis is further complicated by the existence of two main transcripts, namely Kras-4A and -4B, generated by alternative splicing of its fourth coding exon. The two transcripts differ in their 3′-termini and give rise to two proteins (of 189 and 188 residues, respectively) with different C-terminal sequences. Because the C-termini of Ras proteins function in plasma membrane binding (through both electrostatic binding of basic residues and hydrophobic binding of fatty acylated residues) [14], sequence variations in this region could affect the biological functions of the isoforms. Indeed, in transfected cells, human Kras-4A, but not Kras-4B, efficiently induced transformed foci and enabled anchorage-independent growth [15], [16]. Additional differences between the two isoforms regard their expression levels, which were found to be modulated during mouse embryogenesis and to vary in different tissues [17], [18]. Moreover, the ratio between Kras-4A and Kras-4B mRNA levels in lung tissue was higher in mouse strains susceptible to lung cancer than in resistant ones [17]. More recent evidence implicating the Kras-4A oncoprotein in lung tumorigenesis was provided by two studies in which mice expressing only the Kras-4B isoform had greater resistance to chemical carcinogenesis than did wild-type mice also expressing Kras-4A [19], [20].
Despite these observations, several aspects of the candidacy of Kras as the major effector of the Pas1 locus in lung tumor susceptibility remain to be clarified. Wild-type alleles from susceptible and resistant mice code for the same protein; consequently, the mechanism by which Kras determines genetic susceptibility to lung tumorigenesis could, instead, depend on its overall expression level or on the ratio of its isoforms in normal lung tissue.
To address these points and to clarify the involvement of the two Kras isoforms, we studied Kras mRNA levels and germline variants using an established model of urethane-induced lung tumorigenesis in an advanced intercross population between A/J mice (tumor susceptible) and C57BL/6 mice (resistant). This model was previously instrumental in defining the Pas1 locus based on the pattern of tumorigenesis in F2 intercross mice [1]. In the present study, we used F4 mice (of a new pedigree) in order to have a greater resolution power. After confirming Pas1 as a quantitative trait locus (QTL) modulating lung tumor multiplicity in this pedigree, we performed a genome-wide association study to look for expression QTL modulating the levels of Kras-4A and Kras-4B transcripts in lung tumors and normal lung tissue. Our findings indicate that Kras-4A and, to some extent, also Kras-4B are modulated by cis-acting elements within the Pas1 locus itself. These results underpin a possible causal relationship between germline variations and mRNA levels of the Kras-4A isoform, consequently influencing lung tumor susceptibility.
Results
A/J and C57BL/6 mice were mated and bred to the fourth generation (ABF4) to establish a pedigree for the current study. Male ABF4 mice (n = 183) were treated with a single injection of urethane at 4 weeks of age to induce lung tumor formation, sacrificed 36 weeks later and assessed for lung tumor multiplicity (Nlung). Values of Nlung ranged from 0 (in 27 animals) to a maximum of 33.
To assess the genetic control of susceptibility to lung tumorigenesis in this pedigree, we performed genome-wide linkage analysis on the 183 urethane-treated ABF4 mice using Illumina SNP-arrays, which permit the genotyping of up to 1449 single nucleotide polymorphisms (SNPs). After quality control filtering, genotype data were obtained for 548 informative (polymorphic) non-redundant SNPs dispersed over the whole genome. Simple interval mapping was performed to detect QTLs associated with squared root-transformed values of Nlung. This analysis identified a single major locus in the telomeric region of chromosome 6 (Figure 1A). The QTL peak had a LOD score of 48.2 and was centered around marker rs6265387 (Pthlh gene region) near the Pas1 locus; moreover, at the Kras 37-bp marker, located 2-Mb proximal to rs6265387 and distinguished from the SNP by 3 recombination events, the LOD score was almost identical (LOD = 48.0). The QTL accounted for 69% of the total phenotypic variance in the ABF4 population. When the mice were grouped according to genotype (Figure 1B), Nlung in the 59 animals homozygous for the A/J-derived allele (represented by a G at rs6265387) was 1.5-fold higher than that in the 82 heterozygous mice and ∼24-fold higher than that in the 42 mice homozygous for the C57BL/6-derived allele (represented by an A at rs6265387; P<1.0×10−6, ANOVA followed by Tukey's test for multiple comparisons).
Fig. 1. Lung tumor multiplicity in 183 urethane-treated male ABF4 mice is controlled by the Pas1 locus. 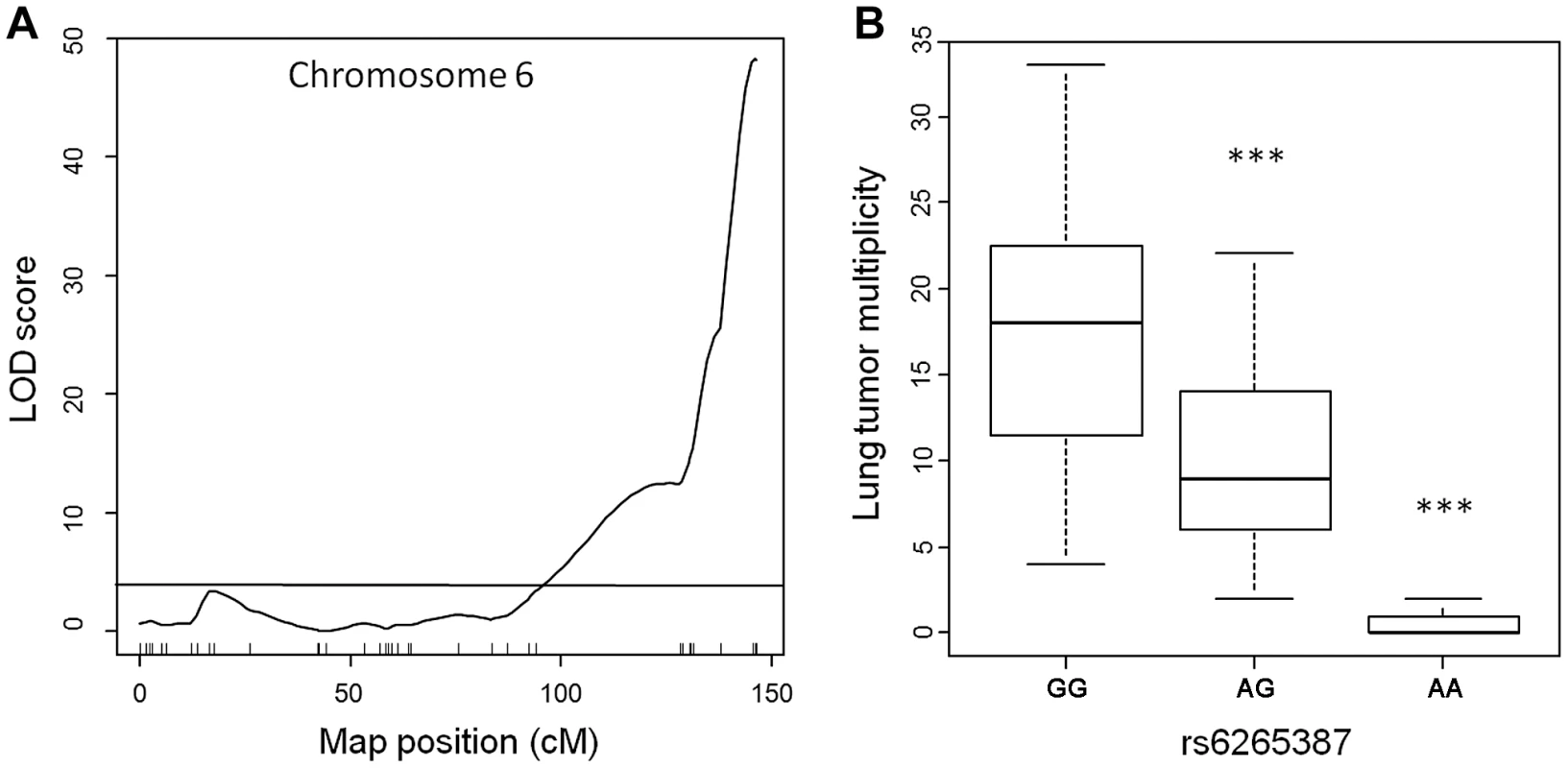
(a) LOD score plot for chromosome 6 on which a quantitative trait locus (QTL) for lung tumor multiplicity (square root transformed values) mapped to the telomeric region. The QTL peak (LOD score = 48, phenotypic variance explained = 69%) overlapped with the Pas1 locus. Tick marks show the position of 37 genotyped markers, including rs6265387 at the QTL peak. Horizontal line indicates the 95% LOD threshold. (b) Number of lung tumors per animal, grouped according to genotype at rs6265387. Mice homozygous for the A/J-derived allele (GG; n = 59) had more tumors than either heterozygous animals (n = 82) or mice homozygous for the C57BL/6-derived allele (AA; n = 42). ***P<1.0×10−6 versus the A/J-derived allele, ANOVA followed by Tukey's test for multiple comparisons. The line within each box represents the median; upper and lower edges of each box are 75th and 25th percentiles, respectively; upper and lower bars indicate the highest and lowest values less than one interquartile range from the extremes of the box. These results, which confirm the key role of the Pas1 locus in murine lung tumorigenesis, establish this new intercross population as suitable for studying Pas1 function. In this cross, lung carcinogenesis can be considered a monogenic trait due to the overwhelming genetic effect of the Pas1 locus. Therefore, we used this pedigree to test whether the Pas1 locus exerts its effect on lung tumorigenesis through a modulation of Kras mRNA levels. To this aim, we performed linkage analyses to identify expression QTLs associating with the levels of Kras-4A and Kras-4B mRNA.
Pas1 is an expression QTL controlling Kras-4A transcript levels in lung tumors of urethane-treated mice
From 80 of the 183 ABF4 urethane-treated mice used in the genetic linkage analysis, we were able to resect a tumor specimen from lung tissue. This subgroup included 37 mice homozygous at rs6265387 for the A/J-derived allele (associated with tumor susceptibility), 34 heterozygous mice, and 9 mice homozygous for the C57BL/6 resistant allele that nevertheless had developed a lung tumor. Indeed, the intrinsic low susceptibility to lung tumorigenesis of the 42 animals homozygous for the C57BL/6-derived allele (only 9 of these mice developed a lung tumor) did not allow us to analyze a subgroup of the original population with a genotype ratio typical of such intercrosses (i.e., 1∶2∶1).
RNA was extracted from each specimen and used in quantitative PCR to measure the levels of Kras-4A and -4B mRNA. Genome-wide linkage analysis for Kras-4A found a single expression QTL on chromosome 6 (peak LOD = 4.5), corresponding to the Pas1 locus (Figure 2A). In contrast, no expression QTL was found for Kras-4B on any chromosome, including chromosome 6 where LOD scores remained below the threshold for significance. When the 80 mice were grouped according to the genotype at rs6265387, we found that Kras-4A mRNA was highest in mice homozygous for the A/J-derived susceptible allele (GG), intermediate in heterozygous (AG) animals, and lowest in mice homozygous for the C57BL/6 resistant allele (AA); these differences were significant (GG vs. AG, P = 3.2×10−3; GG vs. AA, P = 8.9×10−5; ANOVA followed by Tukey's test for multiple comparisons, Figure 2B). These results implicate Kras-4A in the mechanism of Pas1-dependent tumor formation.
Fig. 2. Kras-4A levels in lung tumors from 80 urethane-treated ABF4 mice are controlled by Pas1 locus. 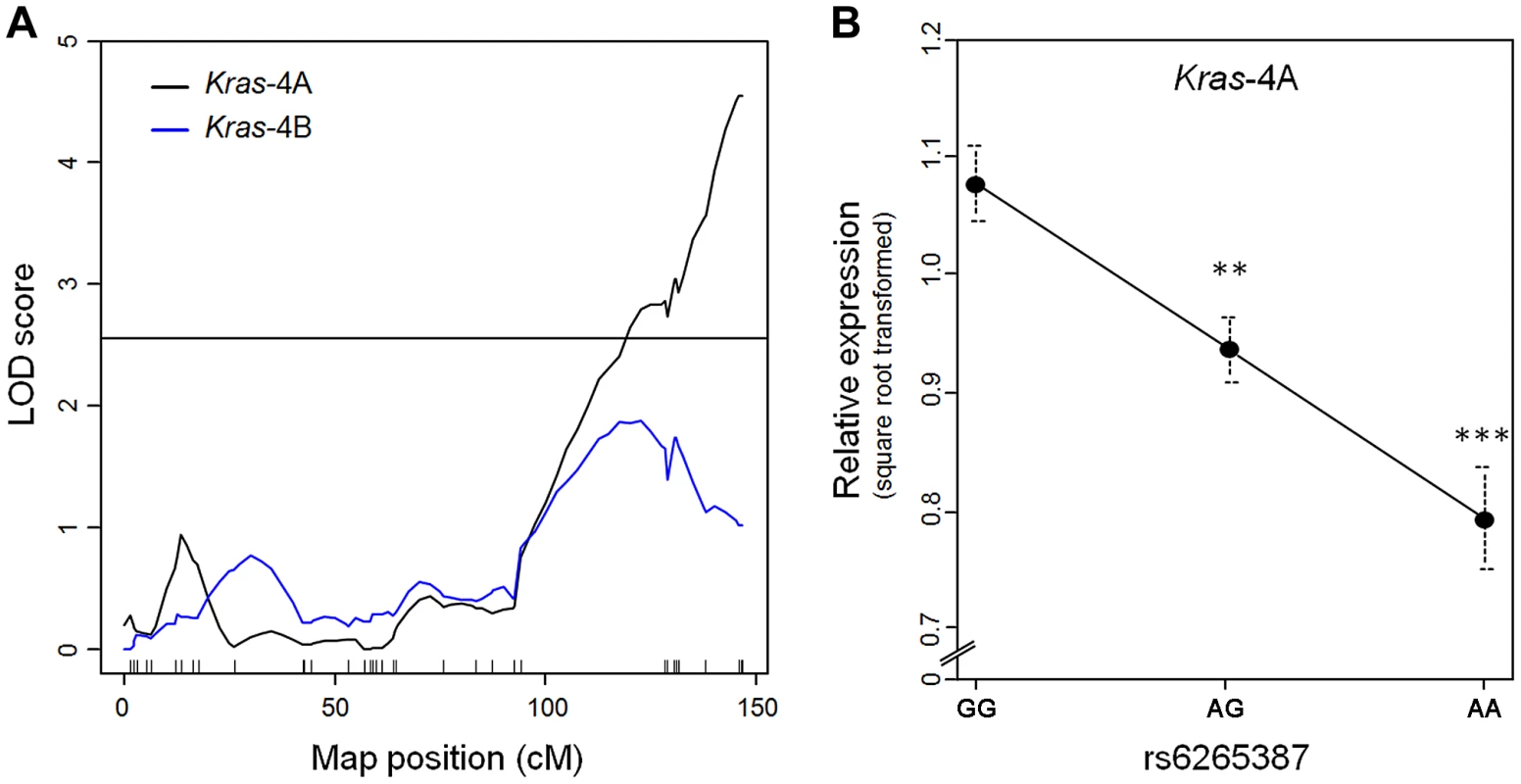
(a) Expression QTL analysis of Kras transcripts showed that square-root-transformed levels of Kras-4A linked to the Pas1 locus (LOD = 4.5). No significant linkage was observed for the Kras-4B mRNA isoform. Tick marks show the position of the genotyped markers in a recombinational map. Horizontal line at LOD = 2.55 marks the 95% threshold for significance. (b) Relative expression levels of the Kras-4A isoform according to rs6265387 genotype. Mice homozygous for the A/J-derived susceptible allele (GG, n = 37) had higher levels than either heterozygous animals (AG, n = 34) or mice homozygous for the C57BL/6 resistant allele (AA, n = 9). ***P<0.001, **P<0.01 vs. GG mice, ANOVA followed by Tukey's test for multiple comparisons. Values are means and SE. Pas1-dependent tumor susceptibility correlates with Kras-4A transcript levels in normal lung tissue
The genetic susceptibility to cancer is an intrinsic feature of normal tissue that, in urethane-treated mice, influences both the probability of tumor initiation and the number of tumors that develop. Consequently, it is likely that the fundamental elements underlying this phenomenon are present not only in tumors but also in normal tissue. To test this hypothesis, we attempted to validate in normal lung tissue the genetic linkage we observed in lung tumors. Therefore, we genotyped 111 untreated male ABF4 mice for nine markers on chromosome 6 spanning from 96.7 Mb to 148.3 Mb. These markers include 8 SNPs and one 37-bp sequence variation that in C57BL/6 mice occurs as a tandem repeat; this insertion mutation is located in the second intron of Kras [21]. Additionally, we assayed lung mRNA from these mice for the two Kras isoforms, and examined the association of mRNA levels with genotype.
A strong linkage was found between the level of Kras-4A mRNA and genotyped markers in the Pas1 locus, describing a LOD curve with a peak of 23.2 (Figure 3A); this expression QTL explained 62% of the phenotypic variance. A much weaker, yet statistically significant linkage for the Kras-4B transcript was observed (maximum LOD score = 4.1). These results confirm and strengthen the major role of Kras-4A isoform found in tumors (Figure 2). We then grouped the mice according to genotype at the Kras 37-bp insertion to examine the relationship between Kras mRNA levels and the number of inherited A/J-derived alleles of the Pas1 locus. A strong association was found for Kras-4A levels (P = 9.7×10−14, ANOVA; square-root-transformed values are shown in Figure 3B): mice with two A/J-derived alleles (i.e. no insertion, -/-) had on average 1.9-times more transcript than did mice receiving two C57BL/6-derived alleles (ins/ins), and heterozygous mice had intermediate levels. In contrast, the linkage between Pas1 genotype and Kras-4B transcript levels was weaker (P = 9.7×10−5, ANOVA, Figure 3C).
Fig. 3. Kras-4A expression is highly controlled by Pas1 locus in normal lung. 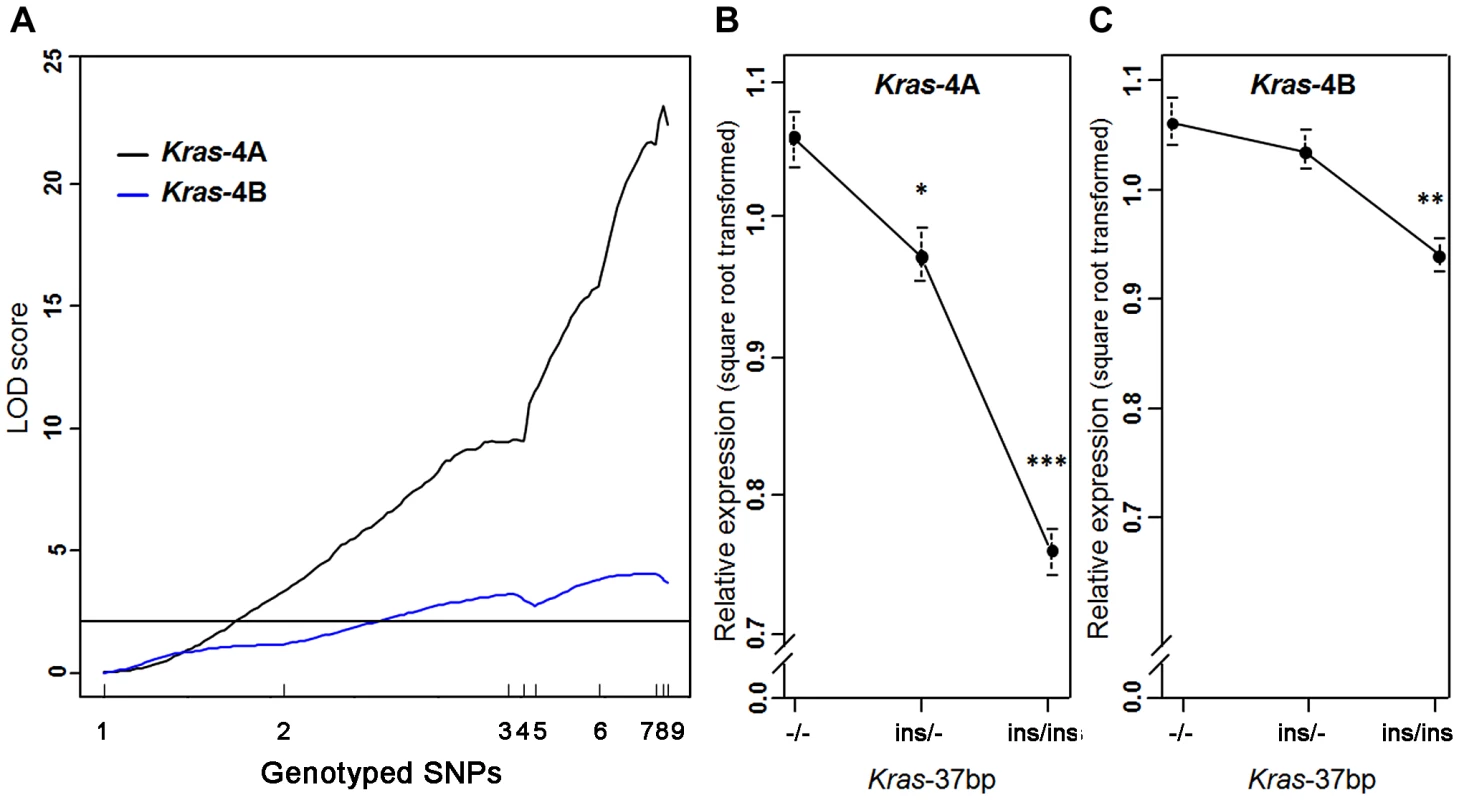
(a) Genetic linkage analysis of expression levels (square root transformed) of two Kras transcripts in 111 untreated ABF4 mice showed that the Kras-4A isoform was strongly linked to the Pas1 locus (LOD score = 23.2) whereas the Kras-4B isoform showed a weaker linkage (LOD score = 4.1). Horizontal line at LOD = 2.13 marks the threshold for significance. Tick marks show the position, in a recombinational map, of the nine genotyped markers spanning from chromosome 6 position 96.7 Mb to 148.3 Mb. (b, c) Expression levels of Kras-4A and -4B in normal lung tissue, by genotype for the 37-bp insertion mutation common to both isoforms. The A/J-derived susceptible allele is negative (-) for the insertion whereas the C57BL/6 allele is positive (ins); 30 mice were -/-, 52 ins/-, and 29 ins/ins. (b) For Kras-4A, P = 9.7×10−14, ANOVA. Tukey's test for multiple comparisons, *P = 0.01, ***P = 7.4×10−14 vs. -/-. (c) For Kras-4B, P = 9.7×10−5, ANOVA. Tukey's test for multiple comparisons, **P = 1.5×10−4 vs. -/-. Values are means and SE. We then examined whether this specific genetic control exerted by the Pas1 locus was also present in normal lung of the parental inbred strains A/J and C57BL/6 (Figure 4). The median level of the Kras-4A transcript in A/J (tumor-susceptible) mice was 2.3-fold higher than that of C57BL/6 (resistant) mice (P = 6.0×10−9, ANOVA; square-root-transformed values are shown in Figure 4A). In contrast, the Kras-4B levels were not significantly different between mouse strains (Figure 4B).
Fig. 4. Kras-4A is expressed at higher levels in susceptible (A/J) than resistant (C57BL/6) strains. 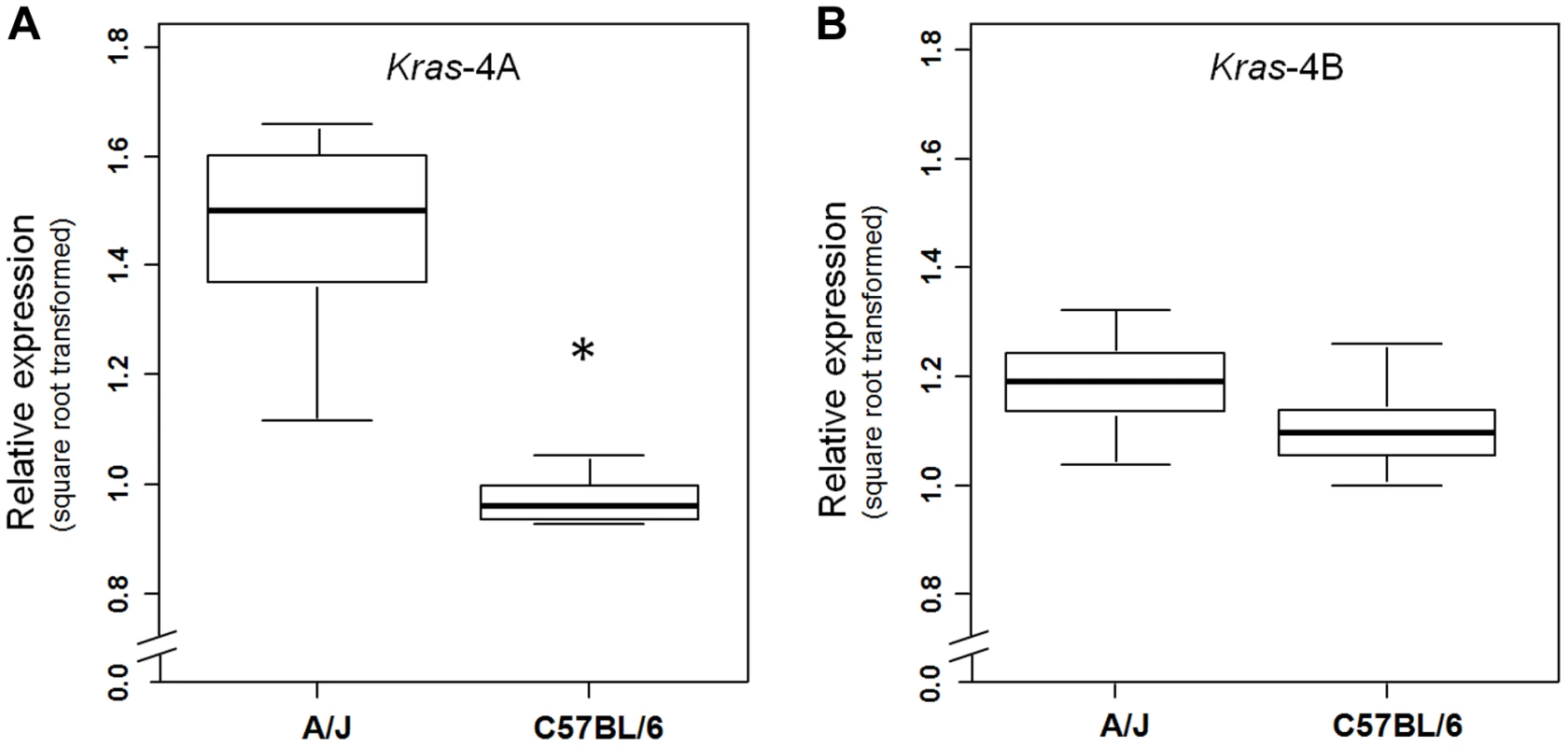
Expression levels of Kras-4A (a) and Kras-4B (b) isoforms were measured by qPCR in normal lung of 11 A/J and 11 C57BL/6 mice. * P = 7.1×10−5, ANOVA. The line within each box represents the median of square root transformed values; upper and lower edges of each box are 75th and 25th percentiles, respectively; upper and lower bars indicate the highest and lowest values less than one interquartile range from the extremes of the box. Altogether, these results indicate that the A/J-derived allele of the Pas1 locus, which confers susceptibility to lung tumorigenesis, is also associated with higher steady-state levels of the Kras-4A splice variant. These observations suggest that subtle modulation of Kras-4A mRNA production or stability may be the key effector of Pas1-controlled lung tumor susceptibility, possibly via a cis-acting element within Pas1 itself.
Differential allelic expression of Kras-4A is due to sequence variants in cis-regulatory elements
To test whether the allele-specific levels of Kras isoforms are attributable to variations in cis-regulatory elements also mapping in the Pas1 locus, we examined the possibility of differential allelic expression. For this analysis, we considered two SNPs (rs29968550 and rs30022167, mapping in a region of Kras mRNA common to the two isoforms) whose genotypes permitted us to distinguish the A/J - from the C57BL/6J-derived allele. By pyrosequencing, we determined the frequencies of the two alleles in Kras-4A and -4B cDNA from normal lung tissue of 20 heterozygous untreated ABF4 mice and from lung tumor specimens of 15 heterozygous treated mice and in genomic DNA from the same animals, used as control for the amplification efficiency of the two alleles (Figure S1). Indeed, when allelic imbalance is tested, uneven amplification unrelated to differential transcription may occur in some of the assays. Therefore, the experimentally obtained allelic ratio for genomic DNA, which in principle is equal to 1 in heterozygotes, is used as the baseline to evaluate the ratios observed for cDNAs.
Based on the results for both SNPs, comparing the mean allelic ratios for Kras-4A cDNA with those of the corresponding genomic DNA, we observed a ∼2-fold higher expression of the A/J-derived allele in Kras-4A cDNA in both tumor and normal tissue indicating a higher steady state of this isoform associated with the A/J-derived allele, possibly due to either a higher production rate or a lower degradation rate (Figure 5). For Kras-4B, the mean allelic ratios for cDNA from tumors were similar to those for genomic DNA, meaning equal production from both alleles; in normal tissue, however, a slightly higher ratio for Kras-4B cDNA relative to genomic DNA was observed (Figure 5).
Fig. 5. Differential allelic expression of Kras-4A isoform indicates the existence of functional polymorphisms in Kras gene. 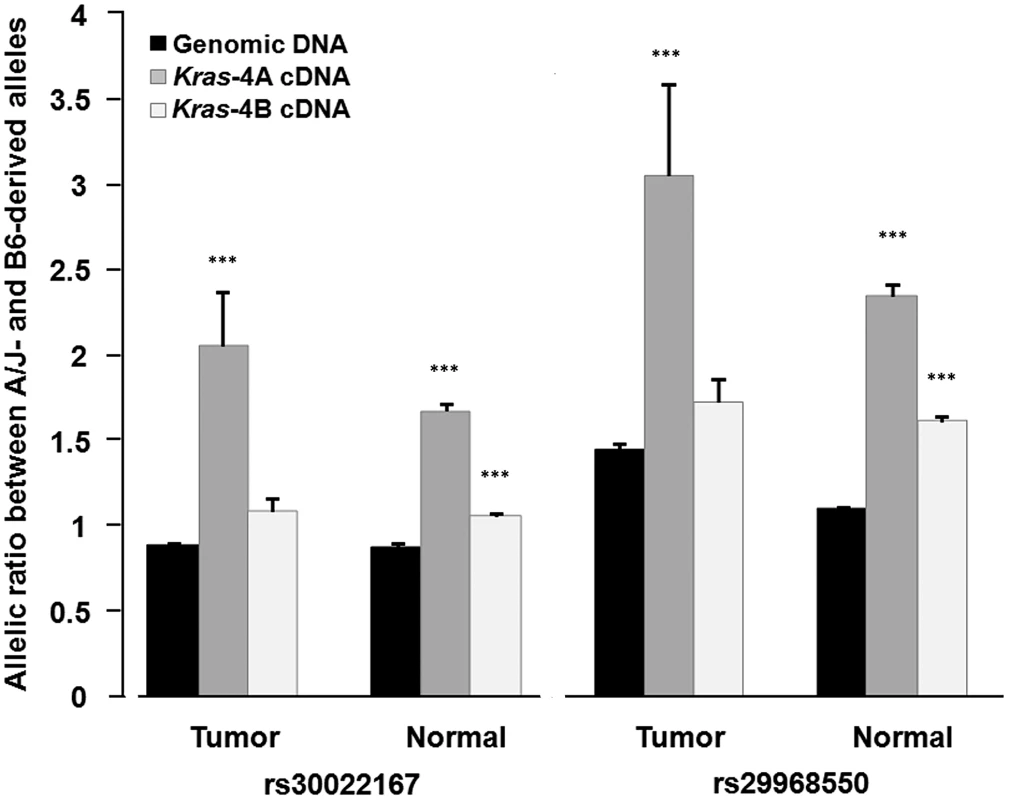
Allelic ratios were determined by pyrosequencing for rs30022167 and rs29968550, which map in a region of the Kras 3′-UTR common to both isoforms, on genomic DNA and cDNA from normal lung tissue (n = 20) and lung tumor specimens (n = 15) from ABF4 heterozygous mice. Values are mean and SE; *** P<0.001, two-sided Welch's t test (see Table 1 for complete data). In the statistical analysis, allelic ratios were log10-transformed to approximate a normal distribution before applying Welch's t test (Table 1). This analysis showed that the high allelic ratios for Kras-4A cDNA, observed at two SNPs and in both normal and tumoral lung tissue, are significantly different from those of genomic DNA (P<0.001, see Table 1). In the case of Kras-4B cDNA, the allelic ratios were significantly different from that of genomic DNA only for normal lung tissue (P<0.001). These results provide evidence for the differential allelic expression of Kras transcripts, with higher levels being produced from the A/J-derived allele in ABF4 intercross mice. These findings are compatible with the presence of allele-specific germline variations in cis-acting elements that mainly influence the selection or stability of the Kras-4A isoform.
Tab. 1. Differential allelic expression of Kras isoforms in lung tumor specimens and normal lung tissues from ABF4 intercross mice. 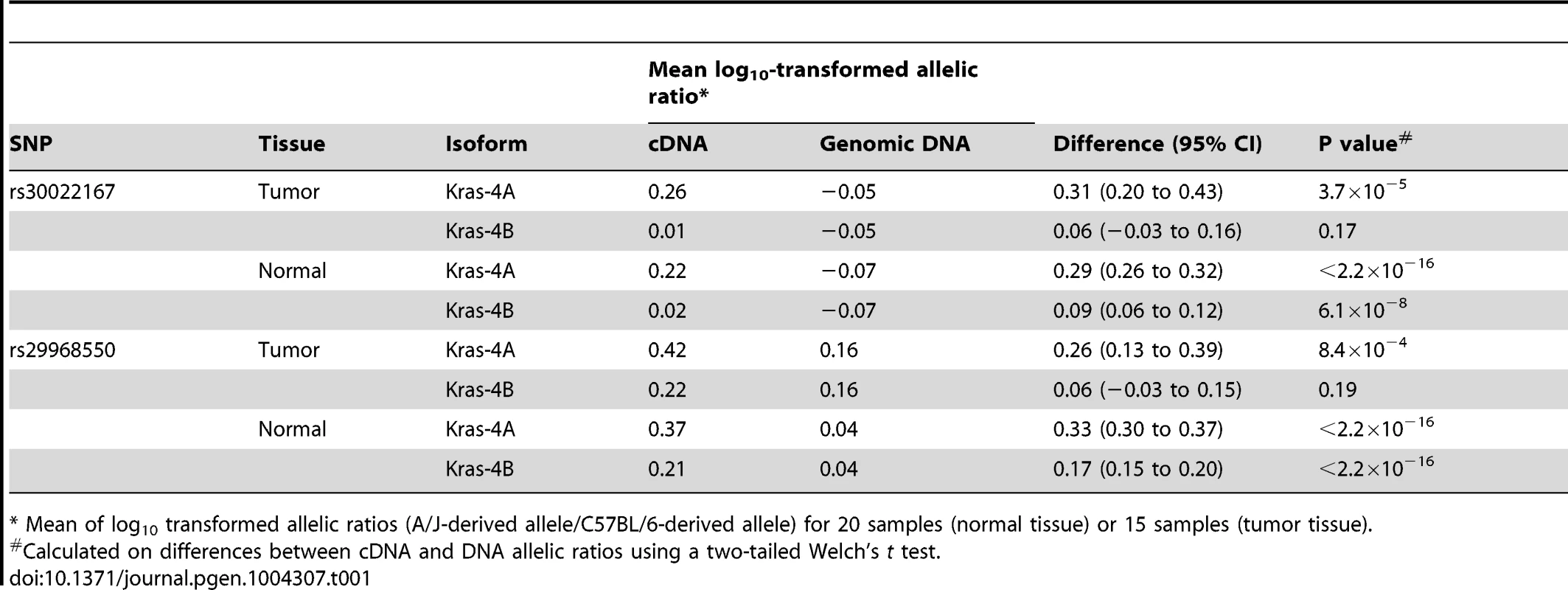
* Mean of log10 transformed allelic ratios (A/J-derived allele/C57BL/6-derived allele) for 20 samples (normal tissue) or 15 samples (tumor tissue). Discussion
In this study, we verified the major role of the Pas1 locus in modulating lung tumorigenesis in an ABF4 advanced intercross population, where the lung tumor multiplicity (Nlung) phenotype behaved as a monogenic trait under the control of the Pas1 locus, explaining almost 70% of the phenotypic variance. In addition, we found that Pas1 is an expression QTL because it controls the level of the Kras-4A isoform in urethane-induced lung tumors. This genetic control is an inherited trait, in that it was already present in normal lung tissue of untreated mice. Finally, we observed the preferential expression of Kras-4A from the A/J-derived allele in heterozygous mice, suggesting the existence of allele-specific germline variations in cis-acting elements that influence splicing bias, selection or stability of this isoform.
We carried out this genetic study in an ABF4 intercross since it was an effective way to accumulate, in a population of a relatively small size, 3-fold more recombination events than those possible in a conventional F2 intercross population of the same size [22]. Indeed, each ABF4 mouse underwent six informative meioses compared to the two informative meioses of an F2 mouse, thus providing improved resolution for mapping loci affecting strain-related phenotypes [22]. This methodological choice allowed us to obtain a sharp peak of linkage between Pas1 and Nlung, with very high association (LOD score = 48 in 183 ABF4 mice). The high rate of recombination in this intercross was also useful for testing our hypothesis that Kras isoform expression is modulated by Pas1.
Animals homozygous for the C57BL/6-derived allele had low susceptibility to lung tumorigenesis and, therefore, rarely developed lung tumors after urethane treatment. Hence, the genetic class with the resistant phenotype was under-represented in the experiment designed to identify expression QTLs in tumors. Another difficulty that presented in our research was the known dysregulation of gene expression architecture in cancer, including mouse lung tumors [23], [24], which could have reduced the detectability of expression QTLs. Despite these limitations, we observed a significant linkage of the Pas1 locus with mRNA levels of Kras-4A (LOD score = 4.5 in 80 mice) but not of Kras-4B.
In normal lung tissues from untreated mice, where all three genotypes were present at roughly the expected ratio and where no pathologic alterations could have played a confounding role, we observed that the Kras-4A isoform levels were again significantly linked with the Pas1 locus (LOD score = 23.2 in 111 mice). In these untreated animals, the Kras-4B isoform also had significant linkage with Pas1, although the LOD score of 4.1 was only ∼17% that obtained for Kras-4A.
The differential allelic expression of Kras-4A and, to some extent, Kras-4B, in normal lung tissue and in lung tumor specimens, indicated the existence of regulatory polymorphisms located within the Pas1 locus. These cis-acting elements have not yet been identified. However, since the Kras gene promoter has been shown to have similar activity in susceptible and resistant mice [25], it is conceivable that these cis-acting elements are located in other regions such as introns or the 3′-untranslated region of the Kras transcript. Regulatory elements in these regions can be expected to impact more on mRNA splicing and stability than on promoter activity. Several polymorphisms in non-coding regions of Kras have already been found in various mouse inbred strains and the functional roles of some of them have been investigated [21], [25], [26]; these studies, however, reached contrasting conclusions. Future investigations could focus on the targeted re-sequencing of the A/J-derived Kras gene and flanking regions, to search for not yet identified polymorphisms that affect splicing selection or mRNA stability of Kras isoforms. The candidate functional polymorphisms could then be tested by cloning into reporter vectors for transfection and assay of cell lines.
Overall, these results shed new light on the role of expression QTLs in mouse lung tumorigenesis, since they indicate that the genetic control exerted by Pas1 mostly affects the expression of the Kras-4A transcript. These results point to the 4A isoform as the functional element of Pas1, the major locus modulating lung tumorigenesis in mice.
Materials and Methods
Ethics statement
All animals received humane care according to the criteria outlined in a protocol approved during the meeting board of December 21, 2006, by the institutional ethical committee for animal use (CESA) at the Fondazione IRCCS Istituto Nazionale dei Tumori.
Mouse crosses, tissues, DNA and RNA
A pedigree of intercross mice was generated by mating lung tumor-susceptible A/J mice (A; 5 females) with lung tumor-resistant C57BL/6 mice (B; 5 males). From generation ABF2 to ABF4, male and female mice were individually labeled, randomly selected using a random number generator and bred, avoiding the pairing of siblings (20 female and 20 male ABF2 mice, and 55 female and 55 male ABF3 mice were mated). After weaning, ABF4 mice were sexed and ear-tagged, and a section of tail was collected for DNA extraction. Male ABF4 mice were then randomly assigned to two groups: untreated mice (n = 131) were raised under standard conditions, while 188 male mice were treated with a single intraperitoneal injection of urethane (1 g/kg body weight) at 4 weeks of age to induce the development of lung tumors [27].
To obtain normal lung tissue, 111 untreated AFB4 mice, 11 A/J and 11 C57BL/6 mice (all males) were anesthetized and killed at 16 weeks of age; lung lobes were isolated and frozen in liquid nitrogen. To obtain lung tumor tissue, the 183 urethane-treated mice were killed at 40 weeks of age; the chest was opened, the trachea was dissected to permit needle access, and the lungs were filled with 0.5 ml RNALater solution. Then, the lung lobes were removed, placed in RNALater, and kept overnight at 4°C degrees. The next day, lungs were examined for surface tumors. For each mouse, we recorded the total number of tumors (Nlung) and, whenever possible, resected one tumor of about 1–1.5 mm under a stereo-microscope. These tumor specimens were frozen and stored at −80°C degrees.
Genomic DNA was extracted from tail samples using the DNeasy Blood & Tissue Kit (Qiagen, Valencia, CA, USA) and quantified using Picogreen dsDNA Quantitation Kit (Invitrogen, Life Technologies, Paisley, UK). Total RNA was extracted from normal lung tissue and from lung tumor specimens using RNeasy Midi Kit (Qiagen), purified with RNeasy MinElute Cleanup (Qiagen), and quantified by spectrophotometry (ND-1000 spectrophotometer, NanoDrop, Wilmington, DE, USA). RNA integrity was verified using the RNA 6000 Nano Assay Kit (Agilent Technologies, Palo Alto, CA, USA); the mean RIN value was 8.9 (SD = 0.8; range, 6.7 to 9.9).
Genome-wide SNP and Pas1 locus genotyping
Genomic DNA from urethane-treated ABF4 mice was used for genome-wide SNP genotyping carried out with the GoldenGate Genotyping Assay according to the manufacturer's protocol (Illumina, San Diego, CA, USA), using the Mouse MD Linkage Panel representing 1449 mouse loci.
Additionally, the Pas1 locus in untreated ABF4 mice was genotyped for nine genetic markers, including eight SNPs (rs6349084, rs33863668, rs31000839, rs31005929, rs13479063, rs33893742, rs13479082, rs3711088) and a 37-bp tandem repeat in Kras [21]. Briefly, a genomic fragment surrounding each marker was PCR-amplified in reactions containing 30 ng genomic DNA, 1 U AmpliTaq Gold (Applied Biosystems, Life Technologies), 1× AmpliTaq Gold buffer (Applied Biosystems, Life Technologies), 1.5 mM MgCl2, 200 µM dNTPs and 5 pmol of each of a pair of specific primers (Table S1) in a total volume of 25 µl. To genotype the eight SNPs, PCR products were pyrosequenced on a PyroMark Q96 ID system running PyroMark Q96 ID Software (Qiagen). To genotype the Kras 37-bp repeat, PCR amplicons were analyzed by 3% agarose gel electrophoresis for fragment size.
Quantitative real-time PCR
RNA from normal lung (1 µg) or from lung tumor specimens (0.5 µg) was used to synthesize cDNA by reverse transcription using the Transcriptor First Strand cDNA Synthesis Kit (Roche, Basel, Switzerland). Kras-4A and -4B transcript levels in the cDNA were measured in quantitative PCR (qPCR) assays using Fast SYBR Green PCR Master Mix (Applied Biosystems) and 300 nM intron-spanning primers (Table S1). Hypoxanthine phosphoribosyltransferase 1 (Hprt1, Table S1) was used as reference gene. Relative expression levels were calculated using the comparative Ct method using one of the synthesized cDNA samples as calibrator.
Detection of differential allelic expression of Kras-4A and -4B transcripts
The allelic expression of Kras-4A and Kras-4B isoforms was analyzed taking into consideration two SNPs in the 3′-UTR region of the Kras gene (Figure S1): rs30022167 (A or C, for the A/J - or C57BL/6-derived allele, respectively) and rs29968550 (A or G, for the A/J - or C57BL/6-derived allele, respectively). Among the animals heterozygous for markers at the Pas1 locus, we selected 20 mice from the untreated group and 15 from the urethane-treated group, from which we already had genomic DNA. In addition, from these animals, we also had cDNA from normal lung (n = 20) or from tumor specimens (n = 15), having been reverse-transcribed for the qPCR experiments. For each Kras isoform, we carried out a first amplification step on the cDNA samples using a forward primer located in exon 4A (Kras-4A) or in the junction between exon 3 and exon 4B (Kras-4B) and a common reverse primer located in the 3′-UTR region of Kras gene downstream of rs30022167 and rs29968550 (Table S1 and Figure S1). The resulting PCR product as well as a sample of genomic DNA from the same animal were PCR-amplified using SNP-specific primers. These PCR products were pyrosequenced on a PyroMark Q96 ID system running PyroMark Q96 ID Software (Qiagen). For each SNP, the proportions of the two alleles present in each sample (genomic DNA, Kras-4A cDNA and Kras-4B cDNA from untreated and tumor-bearing mice) were determined from pyrogram peak heights, and allelic ratios (A/J-derived allele/C57BL/6-derived allele) were calculated.
Statistical analyses
Nlung values and mRNA levels were square-root transformed to improve the normality of distribution. The transformed values and genotype data were analyzed by simple interval mapping using R/qtl [28] to identify QTLs. LOD scores were considered significant if greater than the 95% LOD threshold (α = 0.05), calculated by 10,000 permutations. The percent phenotypic variance explained by a given QTL was calculated from the LOD score by the following formula: R2 = [1−10(−2LOD/n)], where n is the sample size [28].
Differences between genotype groups were analyzed by one-way ANOVA followed by Tukey's test for multiple comparisons. Log10-transformed allelic ratios were compared between cDNA and genomic DNA using a two-tailed Welch's t test. Data were considered significant when P<0.01.
Supporting Information
Zdroje
1. GariboldiM, ManentiG, CanzianF, FalvellaFS, RadiceMT, et al. (1993) A major susceptibility locus to murine lung carcinogenesis maps on chromosome 6. Nature Genet 3 : 132–136.
2. ManentiG, DraganiTA (2005) Pas1 haplotype-dependent genetic predisposition to lung tumorigenesis in rodents: A meta-analysis. Carcinogenesis 26 : 875–882.
3. ManentiG, GalbiatiF, Giannì BarreraR, PettinicchioA, AcevedoA, et al. (2004) Haplotype sharing suggests that a genomic segment containing six genes accounts for the pulmonary adenoma susceptibility 1 (Pas1) locus activity in mice. Oncogene 23 : 4495–4504.
4. WangM, LemonWJ, LiuG, WangY, IraqiFA, et al. (2003) Fine mapping and identification of candidate pulmonary adenoma susceptibility 1 genes using advanced intercross lines. Cancer Res 63 : 3317–3324.
5. WangM, FutamuraM, WangY, YouM (2005) Pas1c1 is a candidate for the mouse pulmonary adenoma susceptibility 1 locus. Oncogene 24 : 1958–1963.
6. LiuP, WangY, VikisH, MaciagA, WangD, et al. (2006) Candidate lung tumor susceptibility genes identified through whole-genome association analyses in inbred mice. Nat Genet 38 : 888–895.
7. KingPD, LubeckBA, LapinskiPE (2013) Nonredundant functions for ras GTPase-activating proteins in tissue homeostasis. Sci Signal 6: re1.
8. YouM, CandrianU, MaronpotRR, StonerGD, AndersonMW (1989) Activation of the ki-ras protooncogene in spontaneously occurring and chemically induced lung tumors of the strain A mouse. Proc Natl Acad Sci USA 86 : 3070–3074.
9. ReFC, ManentiG, BorrelloMG, ColomboMP, FisherJH, et al. (1992) Multiple molecular alterations in mouse lung tumors. Mol Carcinog 5 : 155–160.
10. JohnsonL, MercerK, GreenbaumD, BronsonRT, CrowleyD, et al. (2001) Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature 410 : 1111–1116.
11. MeuwissenR, LinnSC, van dV, MooiWJ, BernsA (2001) Mouse model for lung tumorigenesis through cre/lox controlled sporadic activation of the K-ras oncogene. Oncogene 20 : 6551–6558.
12. ZhangZ, WangY, VikisHG, JohnsonL, LiuG, et al. (2001) Wildtype Kras2 can inhibit lung carcinogenesis in mice. Nat Genet 29 : 25–33.
13. ToMD, Perez-LosadaJ, MaoJH, HsuJ, JacksT, et al. (2006) A functional switch from lung cancer resistance to susceptibility at the Pas1 locus in Kras2LA2 mice. Nat Genet 38 : 926–930.
14. PriorIA, HancockJF (2012) Ras trafficking, localization and compartmentalized signalling. Semin Cell Dev Biol 23 : 145–153.
15. VoiceJK, KlemkeRL, LeA, JacksonJH (1999) Four human ras homologs differ in their abilities to activate raf-1, induce transformation, and stimulate cell motility. J Biol Chem 274 : 17164–17170.
16. PlowmanSJ, ArendsMJ, BrownsteinDG, LuoF, DevenneyPS, et al. (2006) The K-ras 4A isoform promotes apoptosis but does not affect either lifespan or spontaneous tumor incidence in aging mice. Exp Cell Res 312 : 16–26.
17. WangY, YouM, WangY (2001) Alternative splicing of the K-ras gene in mouse tissues and cell lines. Exp Lung Res 27 : 255–267.
18. PellsS, DivjakM, RomanowskiP, ImpeyH, HawkinsNJ, et al. (1997) Developmentally-regulated expression of murine K-ras isoforms. Oncogene 15 : 1781–1786.
19. ToMD, WongCE, KarnezisAN, Del RosarioR, Di LauroR, et al. (2008) Kras regulatory elements and exon 4A determine mutation specificity in lung cancer. Nat Genet 40 : 1240–1244.
20. PatekCE, ArendsMJ, WallaceWA, LuoF, HaganS, et al. (2008) Mutationally activated K-ras 4A and 4B both mediate lung carcinogenesis. Exp Cell Res 314 : 1105–1114.
21. ChenB, JohansonL, WiestJS, AndersonMW, YouM (1994) The second intron of the K-ras gene contains regulatory elements associated with mouse lung tumor susceptibility. Proc Natl Acad Sci USA 91 : 1589–1593.
22. DarvasiA, SollerM (1995) Advanced intercross lines, an experimental population for fine genetic mapping. Genetics 141 : 1199–1207.
23. BonnerAE, LemonWJ, DevereuxTR, LubetRA, YouM (2004) Molecular profiling of mouse lung tumors: Association with tumor progression, lung development, and human lung adenocarcinomas. Oncogene 23 : 1166–1176.
24. MelkamuT, ZhangX, TanJ, ZengY, KassieF (2010) Alteration of microRNA expression in vinyl carbamate-induced mouse lung tumors and modulation by the chemopreventive agent indole-3-carbinol. Carcinogenesis 31 : 252–258.
25. Jones-BolinSE, JohanssonE, PalmisanoWA, AndersonMW, WiestJS, et al. (1998) Effect of promoter and intron 2 polymorphisms on murine lung K-ras gene expression. Carcinogenesis 19 : 1503–1508.
26. TimofeevaOA, GorshkovaEV, LevashovaZB, KobzevVF, FilipenkoML, et al. (2002) Pulmonary carcinogenesis susceptibility-associated single-nucleotide polymorphisms in K-ras intron 2 affect the binding of factor gata-6 but not gene expression. Mol Biol (Mosk) 36 : 817–824.
27. ShimkinMB, StonerGD (1975) Lung tumors in mice: Application to carcinogenesis bioassay. Adv Cancer Res 21 : 1–58.
28. BromanKW, WuH, SenS, ChurchillGA (2003) R/qtl: QTL mapping in experimental crosses. Bioinformatics 19 : 889–890.
Štítky
Genetika Reprodukční medicína
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 4- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- IVF a rakovina prsu – zvyšují hormony riziko vzniku rakoviny?
-
Všechny články tohoto čísla
- The Challenges of Mitochondrial Replacement
- Concocting Cholinergy
- Genome-Wide Diet-Gene Interaction Analyses for Risk of Colorectal Cancer
- Statistical Power to Detect Genetic (Co)Variance of Complex Traits Using SNP Data in Unrelated Samples
- Mouse Pulmonary Adenoma Susceptibility 1 Locus Is an Expression QTL Modulating -4A
- Transcription-Associated R-Loop Formation across the Human CGG-Repeat Region
- Epigenetic Regulation by Heritable RNA
- Protein Quantitative Trait Loci Identify Novel Candidates Modulating Cellular Response to Chemotherapy
- Genome-Wide Profiling of Yeast DNA:RNA Hybrid Prone Sites with DRIP-Chip
- The Mechanism of Gene Targeting in Human Somatic Cells
- A LINE-1 Insertion in DLX6 Is Responsible for Cleft Palate and Mandibular Abnormalities in a Canine Model of Pierre Robin Sequence
- Interaction between Two Timing MicroRNAs Controls Trichome Distribution in
- DNA Glycosylases Involved in Base Excision Repair May Be Associated with Cancer Risk in and Mutation Carriers
- The Myc-Mondo/Mad Complexes Integrate Diverse Longevity Signals
- Evolutionarily Diverged Regulation of X-chromosomal Genes as a Primal Event in Mouse Reproductive Isolation
- Mutations in Conserved Residues of the microRNA Argonaute ALG-1 Identify Separable Functions in ALG-1 miRISC Loading and Target Repression
- Genetic Predisposition to In Situ and Invasive Lobular Carcinoma of the Breast
- Isl1 Directly Controls a Cholinergic Neuronal Identity in the Developing Forebrain and Spinal Cord by Forming Cell Type-Specific Complexes
- A Synthetic Community Approach Reveals Plant Genotypes Affecting the Phyllosphere Microbiota
- The Sequence-Specific Transcription Factor c-Jun Targets Cockayne Syndrome Protein B to Regulate Transcription and Chromatin Structure
- Determining the Control Circuitry of Redox Metabolism at the Genome-Scale
- DNA Repair Pathway Selection Caused by Defects in , , and Telomere Addition Generates Specific Chromosomal Rearrangement Signatures
- Methylome Diversification through Changes in DNA Methyltransferase Sequence Specificity
- Folliculin Regulates Ampk-Dependent Autophagy and Metabolic Stress Survival
- Fine Mapping of Dominant -Linked Incompatibility Alleles in Hybrids
- Unexpected Role of the Steroid-Deficiency Protein Ecdysoneless in Pre-mRNA Splicing
- Three Groups of Transposable Elements with Contrasting Copy Number Dynamics and Host Responses in the Maize ( ssp. ) Genome
- Sox5 Functions as a Fate Switch in Medaka Pigment Cell Development
- Synergistic Interactions between the Molecular and Neuronal Circadian Networks Drive Robust Behavioral Circadian Rhythms in
- Chromatin Landscapes of Retroviral and Transposon Integration Profiles
- Widespread Use of Non-productive Alternative Splice Sites in
- Ras GTPase-Like Protein MglA, a Controller of Bacterial Social-Motility in Myxobacteria, Has Evolved to Control Bacterial Predation by
- Cell Type-Specific Functions of Genes Revealed by Novel Adipocyte and Hepatocyte Circadian Clock Models
- Phenotype Ontologies and Cross-Species Analysis for Translational Research
- Embryogenesis Scales Uniformly across Temperature in Developmentally Diverse Species
- In Pursuit of the Gene: An Interview with James Schwartz
- Molecular Mechanisms of Hypoxic Responses via Unique Roles of Ras1, Cdc24 and Ptp3 in a Human Fungal Pathogen
- Analysis of the Genome and Transcriptome of var. Reveals Complex RNA Expression and Microevolution Leading to Virulence Attenuation
- Genotypic and Functional Impact of HIV-1 Adaptation to Its Host Population during the North American Epidemic
- RNA Editome in Rhesus Macaque Shaped by Purifying Selection
- Proper Actin Ring Formation and Septum Constriction Requires Coordinated Regulation of SIN and MOR Pathways through the Germinal Centre Kinase MST-1
- Interplay of the Serine/Threonine-Kinase StkP and the Paralogs DivIVA and GpsB in Pneumococcal Cell Elongation and Division
- A Quality Control Mechanism Coordinates Meiotic Prophase Events to Promote Crossover Assurance
- CNNM2 Mutations Cause Impaired Brain Development and Seizures in Patients with Hypomagnesemia
- The RNA-Binding Protein QKI Suppresses Cancer-Associated Aberrant Splicing
- Uncoupling Transcription from Covalent Histone Modification
- Rad51–Rad52 Mediated Maintenance of Centromeric Chromatin in
- FRA2A Is a CGG Repeat Expansion Associated with Silencing of
- A General Approach for Haplotype Phasing across the Full Spectrum of Relatedness
- A Novel Highly Divergent Protein Family Identified from a Viviparous Insect by RNA-seq Analysis: A Potential Target for Tsetse Fly-Specific Abortifacients
- A Central Role for in Regulation of Islet Function in Man
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- The Sequence-Specific Transcription Factor c-Jun Targets Cockayne Syndrome Protein B to Regulate Transcription and Chromatin Structure
- The Mechanism of Gene Targeting in Human Somatic Cells
- Genetic Predisposition to In Situ and Invasive Lobular Carcinoma of the Breast
- Widespread Use of Non-productive Alternative Splice Sites in
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



