-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaInterplay between Structure-Specific Endonucleases for Crossover Control during Meiosis
The number and distribution of crossover events are tightly regulated at prophase of meiosis I. The resolution of Holliday junctions by structure-specific endonucleases, including MUS-81, SLX-1, XPF-1 and GEN-1, is one of the main mechanisms proposed for crossover formation. However, how these nucleases coordinately resolve Holliday junctions is still unclear. Here we identify both the functional overlap and differences between these four nucleases regarding their roles in crossover formation and control in the Caenorhabditis elegans germline. We show that MUS-81, XPF-1 and SLX-1, but not GEN-1, can bind to HIM-18/SLX4, a key scaffold for nucleases. Analysis of synthetic mitotic defects revealed that MUS-81 and SLX-1, but not XPF-1 and GEN-1, have overlapping roles with the Bloom syndrome helicase ortholog, HIM-6, supporting their in vivo roles in processing recombination intermediates. Taking advantage of the ease of genetic analysis and high-resolution imaging afforded by C. elegans, we examined crossover designation, frequency, distribution and chromosomal morphology in single, double, triple and quadruple mutants of the structure-specific endonucleases. This revealed that XPF-1 functions redundantly with MUS-81 and SLX-1 in executing crossover formation during meiotic double-strand break repair. Analysis of crossover distribution revealed that SLX-1 is required for crossover suppression at the center region of the autosomes. Finally, analysis of chromosome morphology in oocytes at late meiosis I stages uncovered that SLX-1 and XPF-1 promote meiotic chromosomal stability by preventing formation of chromosomal abnormalities. We propose a model in which coordinate action between structure-specific nucleases at different chromosome domains, namely MUS-81, SLX-1 and XPF-1 at the arms and SLX-1 at the center region, exerts positive and negative regulatory roles, respectively, for crossover control during C. elegans meiosis.
Published in the journal: . PLoS Genet 9(7): e32767. doi:10.1371/journal.pgen.1003586
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003586Summary
The number and distribution of crossover events are tightly regulated at prophase of meiosis I. The resolution of Holliday junctions by structure-specific endonucleases, including MUS-81, SLX-1, XPF-1 and GEN-1, is one of the main mechanisms proposed for crossover formation. However, how these nucleases coordinately resolve Holliday junctions is still unclear. Here we identify both the functional overlap and differences between these four nucleases regarding their roles in crossover formation and control in the Caenorhabditis elegans germline. We show that MUS-81, XPF-1 and SLX-1, but not GEN-1, can bind to HIM-18/SLX4, a key scaffold for nucleases. Analysis of synthetic mitotic defects revealed that MUS-81 and SLX-1, but not XPF-1 and GEN-1, have overlapping roles with the Bloom syndrome helicase ortholog, HIM-6, supporting their in vivo roles in processing recombination intermediates. Taking advantage of the ease of genetic analysis and high-resolution imaging afforded by C. elegans, we examined crossover designation, frequency, distribution and chromosomal morphology in single, double, triple and quadruple mutants of the structure-specific endonucleases. This revealed that XPF-1 functions redundantly with MUS-81 and SLX-1 in executing crossover formation during meiotic double-strand break repair. Analysis of crossover distribution revealed that SLX-1 is required for crossover suppression at the center region of the autosomes. Finally, analysis of chromosome morphology in oocytes at late meiosis I stages uncovered that SLX-1 and XPF-1 promote meiotic chromosomal stability by preventing formation of chromosomal abnormalities. We propose a model in which coordinate action between structure-specific nucleases at different chromosome domains, namely MUS-81, SLX-1 and XPF-1 at the arms and SLX-1 at the center region, exerts positive and negative regulatory roles, respectively, for crossover control during C. elegans meiosis.
Introduction
Structure-specific endonucleases are required for several kinds of DNA repair processes such as nucleotide excision repair (NER), DNA interstrand crosslink repair (ICL) and double-strand break repair (DSBR). Homologous recombination is an error free repair pathway because the broken DNA ends are repaired from templates consisting of either homologous sequence at the sister chromatids or the homologous chromosomes. During meiotic recombination, at least one DNA double-strand break has to be repaired as a crossover (obligate crossover) by homologous recombination between non-sister chromatids of a homologous pair of chromosomes. Crossover formation is essential for generating genetic diversity and promoting accurate chromosome segregation. The double (or single) Holliday junction is believed to be the intermediate required to make a crossover product [1]. The opposite sense resolution of the double Holliday junction results in crossover products, while the same sense resolution results in non-crossover products [2]. Moreover, the convergent branch migration and decatenation of such intermediates, referred to as double Holliday junction dissolution, also results in non-crossover products.
Unprocessed double Holliday junctions are toxic for cycling cells. Usually, branch migration during Holliday junction dissolution depends on the Bloom syndrome helicase (BLM) and the decatenation process is catalyzed by topoisomerase III [3]. RMI1 and RMI2 are the essential cofactors of the dissolvasome, BTR (BLM-TOP3-RMI1-RMI2) complex [4], [5]. If double Holliday junctions are not processed by the BTR complex then Holliday junction resolvases play an essential role in avoiding breaks observed at anaphase. This outcome allowed for a synthetic lethal screen with sgs1, which encodes the BLM helicase homolog in yeast, and identified Mus81/Slx3-Mms4/Slx2 and Slx1–Slx4 [6]. Importantly, this screening strategy did not identify Rad1-Rad10, orthologs of the human XPF-ERCC1, and Yen1, the ortholog of human GEN1, because sgs1Δyen1Δ and sgs1Δrad1Δ are viable [7]. In vitro, MUS81-EME1, SLX1–SLX4, and GEN1 have Holliday junction resolvase activity [8]–[14]. While the 3′flap nuclease activity of Rad1 in yeast constitutes its main function during homologous recombination [15], the XPF homolog MEI-9 is required for the majority of the crossovers formed in Drosophila [16].
MUS81, SLX1 and XPF require EME1, SLX4 and ERCC1, respectively, for nuclease activity. SLX4 also acts as a scaffolding protein for several DNA repair proteins including MUS81 and XPF [10]–[12]. It is reported that the D. melanogaster and C. elegans orthologs of SLX4 (MUS312 and HIM-18, respectively) are required for crossover formation during meiosis [17], [18].
In C. elegans meiosis, it is proposed that between 5 and 12 DSBs are evenly distributed along each pair of chromosomes [19]–[21] and one of the DSB sites is designated as a future crossover site by COSA-1, MSH-5 and ZHP-3 [22], [23]. The number of crossovers is tightly regulated as only a single crossover occurs between each homologous chromosome pair. Crossover distribution is also regulated in many organisms. For example, crossover formation is suppressed at centromeres and telomeres in budding yeast [24]. It is also known that the single interhomolog crossover is frequently located at the terminal quarters of the autosomes and the terminal thirds of the X chromosome in C. elegans [25], [26]. Interestingly, crossover formation is suppressed at the center of the chromosomes compared to the arm regions. Recently three studies identified mutants that showed decreased levels of crossover suppression at the center of the autosomes [21], [27], [28]. The molecular mechanisms responsible for this suppression remain to be elucidated, and one of the factors required for this crossover suppression at the center region is SLX-1 [21].
Despite the importance of Holliday junction resolution, severe meiotic defects have not been reported among single mutants for most of the structure-specific endonucleases in yeast, flies, mice or worms, with notable exceptions including mus81 and eme1 mutants in fission yeast, and mei-9 in flies [9], [16]. Moreover, it is still not known whether these structure-specific endonucleases exhibit a Holliday junction resolution activity in vivo. To investigate whether these four structure-specific endonucleases coordinately function to form crossovers during meiotic prophase, likely as Holliday junction resolvases, we took advantage of the ease of genetic analysis and the power of high-resolution imaging in the well-defined spatial-temporal distribution of germline nuclei in C. elegans. We made single, double, triple and quadruple mutants of the structure-specific nucleases and analyzed phenotypes indicative of errors in chromosome segregation (decreased brood size, increased embryonic lethality, larval arrest and incidence of male offspring), as well as crossover designation, frequency and distribution, and bivalent morphology. Our studies demonstrate that: 1) HIM-18 interacts with MUS-81, SLX-1 and XPF-1; 2) XPF-1 acts redundantly with MUS-81 and SLX-1 for crossover formation; and 3) SLX-1 exhibits a region-specific crossover suppression activity. We propose that the structure-specific endonucleases coordinately function for both positive and negative control of a crossover. Moreover, this is the first report implicating the redundant actions of XPF-1 with both MUS-81 and SLX-1 in crossover formation.
Results
SLX-1, XPF-1 and MUS-81, but not GEN-1, interact with HIM-18/SLX-4
To understand the interaction networks between structure-specific endonucleases, we performed a matrix-based yeast two-hybrid assay by using full-length constructs generated from cDNA for him-18, slx-1, xpf-1, ercc-1, mus-81, eme-1 (F56A6.4), gen-1 and rad-54 (Figure 1). Although the ortholog of human EME1 (F56A6.4) was not previously known in C. elegans, we found it through a BLAST search. We detected seven protein-protein interactions, HIM-18-SLX-1, HIM-18-XPF-1, HIM-18-MUS-81, XPF-1-ERCC-1, MUS-81-EME-1, HIM-18-HIM-18 and ERCC-1-ERCC-1. We confirmed conserved interactions between each nuclease and its non-catalytic subunit, HIM-18-SLX-1, XPF-1-ERCC-1 and MUS-81-EME-1. Similar to mammals, HIM-18 interacts with three structure-specific endonucleases, namely SLX-1, XPF-1 and MUS-81. We detected two novel self-interactions: HIM-18-HIM-18 and ERCC-1-ERCC-1. These interactions support the suggestion that SLX4 and ERCC1 could make a very large (2M Dalton) protein complex in human HEK293 cells [10]. Although it is known that Mus81 interacts with Rad54 in S. cerevisiae [29], we did not detect this interaction in C. elegans. Interestingly, GEN-1 did not interact with any structure-specific endonucleases or their regulatory subunits. However, we cannot rule out the possibility that post-translational modification-dependent interactions might have been missed in a yeast two-hybrid assay. These data suggest that the structure-specific endonucleases identified thus far can be categorized into two classes, one consisting of HIM-18-associated nucleases (SLX-1, XPF-1 and MUS-81) and the second consisting of GEN-1.
Fig. 1. The interaction network between structure-specific endonucleases. 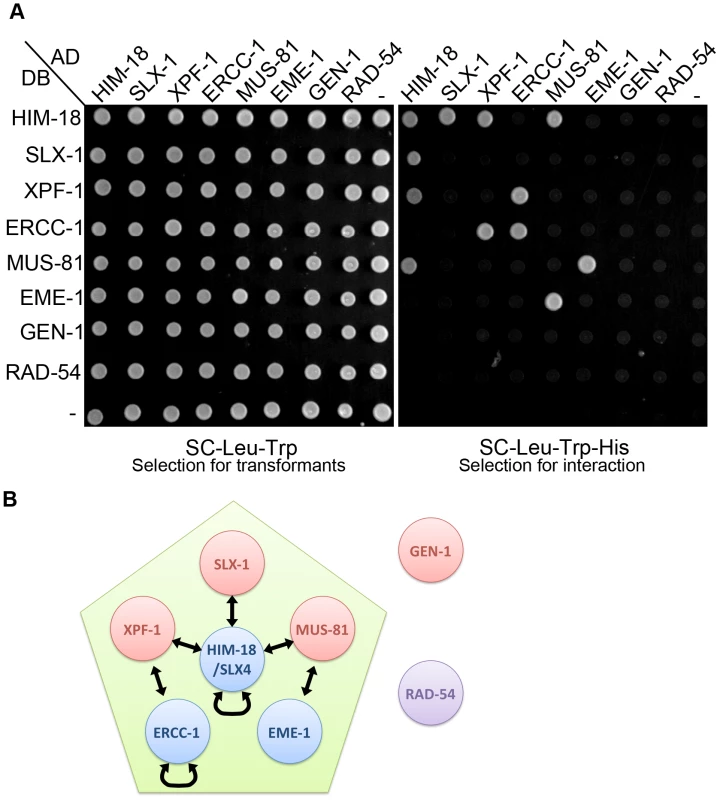
The yeast two-hybrid system was used to examine the protein interactions between HIM-18, SLX-1, MUS-81, EME-1, XPF-1, ERCC-1, GEN-1 and RAD-54. Proteins are fused to either the DNA binding domain (DB) or the activation domain (AD) of GAL4. (A) The diploid yeast strains containing plasmids pVV212 (GAL4 DNA binding domain (DB)+TRP1) and pVV213 (GAL4 activation domain (AD)+LEU2) can grow on SC-LEU-TRP plates. All pair-wise combinations of the interactions are assayed with vector alone as the negative control. Interactions were scored on selective medium lacking leucine, tryptophan and histidine (SC-LEU-TRP-HIS). (B) Schematic representation of the interaction network. Arrows indicate protein-protein interactions. mus-81 and slx-1 mutants show increased sterility
To investigate whether the structure-specific nucleases play a role in the germline, we measured the brood size in mus-81, slx-1, xpf-1 and gen-1 mutants (Figure 2A and Table S1). A decreased brood size is suggestive of increased sterility. The brood size was reduced to 60.5% and 68% of wild type levels in mus-81 and slx-1 single mutants, respectively, while there was no significant reduction in brood size for either xpf-1 (89%) or gen-1 (111.9%) single mutants compared to wild type. The increased sterility observed for mus-81 and slx-1 mutants suggests that MUS-81 and SLX-1 are required for normal germline function.
Fig. 2. Plate phenotypes of structure-specific endonuclease mutants. 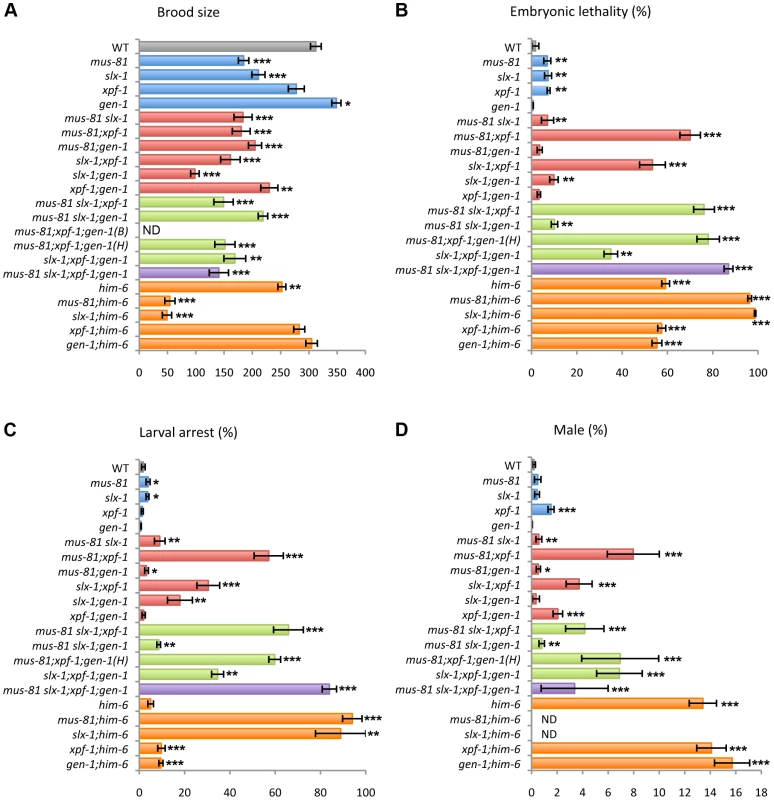
(A) Brood size. Entire brood sizes were scored for singled hermaphrodites of the indicated genotypes. (B) Embryonic lethality. Total numbers of dead eggs/total number of eggs laid were scored. (C) Larval arrest. Arrested L1–L4 worms/total number of hatched worms were scored. (D) Incidence of males. Total number of adult males/total number of adult worms were scored. ND, not determined due to low N values. B, Bristol and H, Hawaiian. Error bars indicate standard error of the mean. Asterisks indicate statistical difference compared to wild type (*P<0.05, **P<0.01, ***P<0.0001, by the two-tailed Mann-Whitney Test, 95% C.I.). The total N-values and the mean values for the plate phenotypes are summarized in Table S1. mus-81 and slx-1 exhibit synthetic mitotic defects with him-6, the C. elegans BLM homolog
It is known that mutants of Holliday junction resolvases show synthetic lethality with mutants of the Bloom syndrome helicase gene in yeast and mammals [6], [30]–[32]. To investigate whether mus-81, slx-1, xpf-1 and gen-1 genetically interact with him-6, we made double mutants of each nuclease with him-6 and examined the brood size, levels of embryonic lethality, larval arrest and the incidence of males detected among their progeny (Figure 2 and Table S1). Increases in either embryonic lethality or larval arrest are suggestive of mitotic defects. A high incidence of males (Him phenotype) is indicative of increased X chromosome nondisjunction and correlates with meiotic defects, whereas a combination of increased embryonic lethality accompanied by a high incidence of males is suggestive of increased aneuploidy resulting from meiotic missegregation of both autosomes and the X chromosome, respectively [33]. mus-81 and slx-1 show synthetic mitotic defects with him-6 while xpf-1 and gen-1 do not. However, due to the lack of viable adult progeny, which impedes assessing the frequency of males, we cannot rule out the possibility of synthetic meiotic defects as well. These results suggest that MUS-81 and SLX-1, but not XPF-1 and GEN-1, are essential in processing recombination intermediates in the absence of HIM-6.
xpf-1 exhibits synergistic effects with mus-81 and slx-1 for embryonic and larval development as well as meiotic X chromosome disjunction
To examine whether the structure-specific nucleases play either distinct or overlapping roles in the mitotic and/or meiotic programs, we measured embryonic lethality, larval arrest and the incidence of males observed among the progeny of single, double, triple and quadruple mutants of mus-81, slx-1, xpf-1 and gen-1 (Figure 2 and Table S1). While only an average of 7% and 7.6% embryonic lethality was observed respectively in mus-81 and xpf-1 single mutants, 70% embryonic lethality was observed in mus-81;xpf-1 double mutants (Figure 2B and Table S1). Synergistic effects were also observed regarding the phenotype of larval arrest in mus-81; xpf-1 double mutants, where 57.1% larval arrest was observed among the surviving progeny, compared to 3.9% and 1.3% in the mus-81 and xpf-1 single mutants, respectively (Figure 2C and Table S1). mus-81;xpf-1 double mutants also exhibited a higher incidence of males (8% males) among their progeny, indicative of X chromosome nondisjunction, compared to mus-81 and xpf-1 single mutants with 0.5% and 1.5%, respectively (xpf-1 vs. xpf-1;mus-81, P<0.0025) (Figure 2D and Table S1). These results suggest that compensating activities of MUS-81 and XPF-1 are required for embryonic viability, larval development and proper X chromosome segregation.
A similar outcome is observed when examining the same phenotypes described above in slx-1;xpf-1 double mutants compared to each single mutant (P<0.0001) (Figure 2B–2D and Table S1). These results suggest that SLX-1 and XPF-1 exhibit synergistic roles when it comes to embryonic, larval and meiotic development.
slx-1 and gen-1 show synthetic sterility and larval arrest
Unlike slx-1 single mutants, gen-1 single mutants exhibit neither increased sterility nor increased larval arrest compared to wild type (Figure 2A and Table S1). However, the brood size of slx-1;gen-1 double mutants decreased to 46% of slx-1 and 28% of gen-1 single mutants (P<0.0001) (Figure 2A and Table S1). The frequency of larval arrest observed among the progeny of slx-1;gen-1 double mutants increased 5.3-fold compared to slx-1 and 26-fold compared to gen-1 single mutants (P<0.0001) (Figure 2C and Table S1). However, there is no genetic interaction between slx-1 and gen-1 with regard to embryonic lethality or X chromosome nondisjunction (Figure 2B–2D and Table S1). Therefore, these results suggest that SLX-1 can fully compensate for absence of GEN-1 during gametogenesis and larval development, whereas GEN-1 can only partially compensate for loss of SLX-1.
MUS-81 and SLX-1 have redundant or compensatory roles with XPF-1 in crossover formation
It is known that a single crossover occurs at the terminal quarters along autosomes and at the terminal thirds along the X chromosome in C. elegans meiosis [25], [26]. Recently, the boundaries between different chromosome domains (tips, arms and center regions) have been reported using high-density single-nucleotide polymorphism (SNP) genotyping on a large panel of recombinant inbred advanced intercross lines (RIAILs) in C. elegans [26] (Figure 3A). Recombination was not observed at either the left or the right tips (∼0.5 Mb from telomeric ends) of each chromosome [26]. To examine whether the four structure-specific nucleases show either distinct or overlapping roles in meiotic crossover formation we compared crossover frequencies and distribution as in [34] along both chromosomes V and X between wild type and all single, double, triple and quadruple mutants (Figure 3). Specifically, we monitored four SNP sites located near the ends of chromosomes V and X (positions a and d), and at the boundaries between the arms and the center (positions b and c) of these two chromosomes (Figure 3A). The SNP sites selected were closely juxtaposed to the boundaries defined in the Rockman and Kruglyak study [26]. This analysis allowed us to compare the crossover frequency and distribution on the left arm, center and right arm of these chromosomes.
Fig. 3. Roles for the structure-specific nucleases in regulating crossover frequency and distribution. 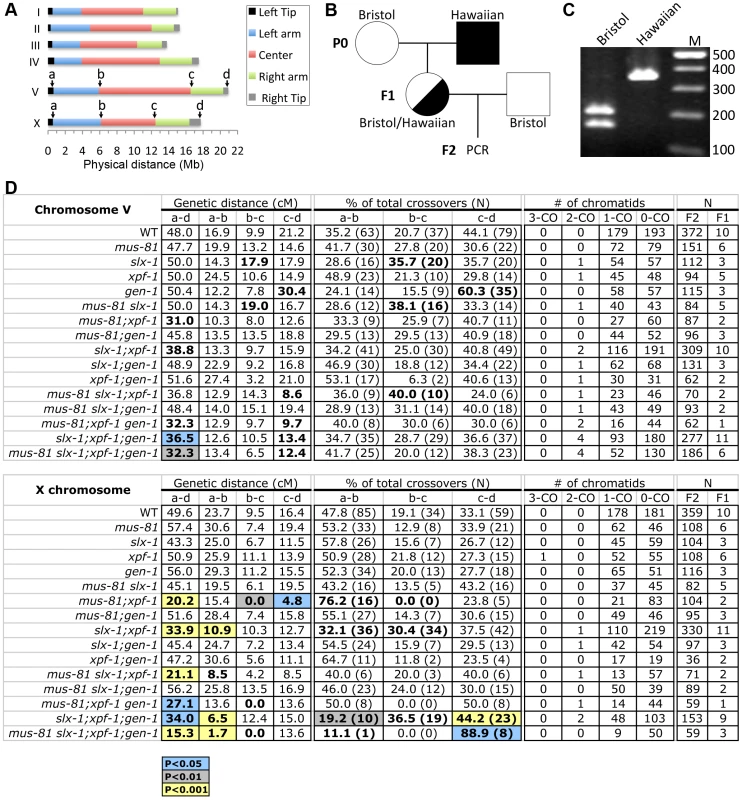
(A) Chromosome domains. Left tip, left arm, center, right arm and right tip are indicated according to [26]. SNPs that alter the restriction length and are located closest to the boundary of each chromosome domain were chosen for chromosomes V and X. (B) Schematic representation of the strategy for obtaining the F2 progeny for snip-SNP analysis. Circles indicate hermaphrodites. Squares indicate males. White, Bristol. Black, Hawaiian. (C) The representative band pattern of PCR products at position “a” on the X chromosome. The restriction enzyme (BspHI) treatment was performed before loading. (D) Crossover frequencies and distributions observed in the entire a–d interval, left arm (a–b), center (b–c), and right arm (c–d) on chromosomes V and X. Highlighted cells indicate statistical difference compared to wild type (blue; P<0.05, grey; P<0.01, and yellow; P<0.001 by the Fisher's Exact Test and corrected by Sidak adjustment for multiple comparisons). Bold type indicates statistical significance (P<0.05) before Sidak correction. We assayed 48 cM and 49.6 cM intervals corresponding to 96.7% and 96.9% of the whole lengths of chromosomes V and X, respectively (Figure 3A to 3D). None of the single mutants tested exhibited a statistically significant change in the crossover frequency in this interval (interval a–d) for either chromosome compared to wild type (Figure 3D). However, we observed a significant reduction of crossover frequency in all mus-81; xpf-1 and slx-1; xpf-1 backgrounds (Figure 3D). Specifically, we observed the following frequencies compared to wild type on chromosomes V and X, respectively: mus-81;xpf-1 (65% and 41% of wild type), mus-81;xpf-1;gen-1 (67% and 55%), mus-81slx-1;xpf-1;gen-1 (67% and 31%), slx-1;xpf-1 (81% and 68%), and slx-1;xpf-1;gen-1 (76% and 69%) (Figure 3D). Notably, while the following mutants exhibited a significant decrease (P<0.05) in crossover frequency when compared to wild type for the a–d interval on chromosome V: mus-81; xpf-1 (P = 0.0041), slx-1;xpf-1(P = 0.0133), mus-81 slx-1; xpf-1 (P = 0.0870, borderline significance likely due to the low N-value), and mus-81;xpf-1;gen-1 (P = 0.0142) (Table S2), we no longer observed this significance after Sidak correction for multiple comparisons (Figure 3D and Table S3). However, this may be an artifact of the stringency of the Sidak correction as suggested by our observations of increased embryonic lethality and frequency of males in these mutants accompanied by chromosomal abnormalities in late meiotic prophase I (Figure 2 and see below). These data therefore suggest that MUS-81, XPF-1 and SLX-1 all contribute to crossover formation, with XPF-1 functioning redundantly with MUS-81 and SLX-1, respectively. This is further supported by the high embryonic lethality observed in all the mus-81; xpf-1 and slx-1; xpf-1, but not mus-81slx-1, backgrounds (Figure 2B and Table S1). Interestingly, no further significant reduction was observed when a gen-1 mutation was introduced into the mus-81;xpf-1 or slx-1; xpf-1 backgrounds. Therefore, C. elegans GEN-1 does not seem to be involved in crossover formation, although the accompanying study by O'Neil et al. [35] observed that microinjection of human GEN1 rescues the accumulation of joint molecules in mus-81;xpf-1 double mutants.
Finally, crossover frequency on the X chromosome of mus-81;xpf-1 double mutants is 60% of the level observed in slx-1;xpf-1 double mutants (P = 0.0101). Furthermore, introduction of an slx-1 mutation did not further affect the crossover frequency observed in mus-81;xpf-1 double mutants. Thus, a mus-81 mutation causes a more severe effect than an slx-1 mutation in the xpf-1 background. Taken together, these data suggest that both MUS-81 and SLX-1 have either redundant or compensatory roles with XPF-1 in regulating crossovers on both autosomes and the X chromosome.
Regulation of crossover interference is impaired in multi-nuclease mutants
In general, only a single crossover occurs between each pair of homologous chromosomes during prophase I of C. elegans meiosis [36]. Therefore, crossover interference, by which the formation of an interhomolog crossover in a given chromosome region discourages formation of additional crossovers nearby, is strictly enforced in C. elegans [37]. Thus, the occurrence of multiple crossover events between homologous chromosomes indicates misregulation of crossover interference in this system. While we did not observe multiple crossovers in wild type for either chromosomes V or X, these were observed in multi-nuclease mutants (Figure 3D). For example, 4.1% and 7.1% of total crossover events, calculated as in [34], were double crossovers on chromosome V in slx-1;xpf-1;gen-1 triple and mus-81slx-1;xpf-1;gen-1 quadruple mutants, respectively. These results are consistent with the observations by Agostinho et al. [38] and suggest that structure-specific nucleases may be involved in the regulation of crossover interference.
SLX-1 is required for regulation of crossover distribution
As we previously reported, crossover distribution in slx-1 mutants shifts to the center of chromosome V (35.7% of total crossovers) where crossover formation is tightly suppressed in wild type (20.7% of total crossovers) (P = 0.0312; Fisher's Exact test) (Table S2). Notably, this statistical significance is no longer observed following Sidak correction for multiple comparisons (P = 0.3784) (Figure 3D and Table S3). However, there are additional phenotypes that can be explained, at least in part, by a deregulation in crossover distribution, such as the increased embryonic and larval lethality, decreased brood size, and increased chromosome abnormalities (see below) observed in slx-1 single mutants. In addition, a similar shift was observed in Agostinho et al. [38]. These results suggest that SLX-1 exhibits either anti-crossover activity or pro-non-crossover activity at the center region of the autosomes.
The increased crossover levels previously detected on the center of the X chromosome in slx-1 mutants compared to wild type [21] were not recapitulated here. This is due to the interval previously used in this analysis, which included regions of both the center and the right arm of this chromosome. In our current study, the more strictly defined SNP sites at the boundaries of the arms and the center, allow us to observe a more precise crossover distribution in each chromosome domain and to correlate our findings to the global analysis presented for wild type in Rockman and Kruglyak [26]. Therefore, we conclude that SLX-1 does not inhibit interhomolog crossover formation at the center region of the X chromosome.
Crossover designation is not altered in most endonuclease-defective mutants
ZHP-3, the C. elegans homolog of S. cerevisiae Zip3, is a crossover-promoting factor [23], [39]. ZHP-3 initially localizes along the length of chromosomes, but becomes restricted to six distinct foci per nucleus in late pachytene, marking the six crossover precursor sites (one per homolog pair) [23], [39]. Since SNP analysis revealed that crossover frequency was reduced in mus-81;xpf-1 and slx-1;xpf-1 double mutants, we assessed the number of ZHP-3 foci per nucleus in the nuclease-defective mutants to determine if a similar reduction in ZHP-3 foci could be observed (Table 1). All single mutants, as well as all double, triple, and quadruple nuclease mutant combinations did not result in changes in the number of ZHP-3 foci (∼6 per nucleus). This is consistent with the observed number of ZHP-3 foci in the single nuclease-defective mutants and the double nuclease-defective mutants by Agostinho et al. and O'Neil et al. [35], [38]. Thus, the reduction in crossover frequency observed for the mus-81;xpf-1 and slx-1;xpf-1 backgrounds by SNP analysis could not be detected by cytological crossover analysis based on scoring the number of ZHP-3 foci per nucleus. This outcome suggests that crossover designation is not drastically affected in the nuclease-defective mutants, but that subsequent resolution of those events into crossovers is impaired.
Tab. 1. ZHP-3 foci in late pachytene and diplotene nuclei of the nuclease-deficient mutants. 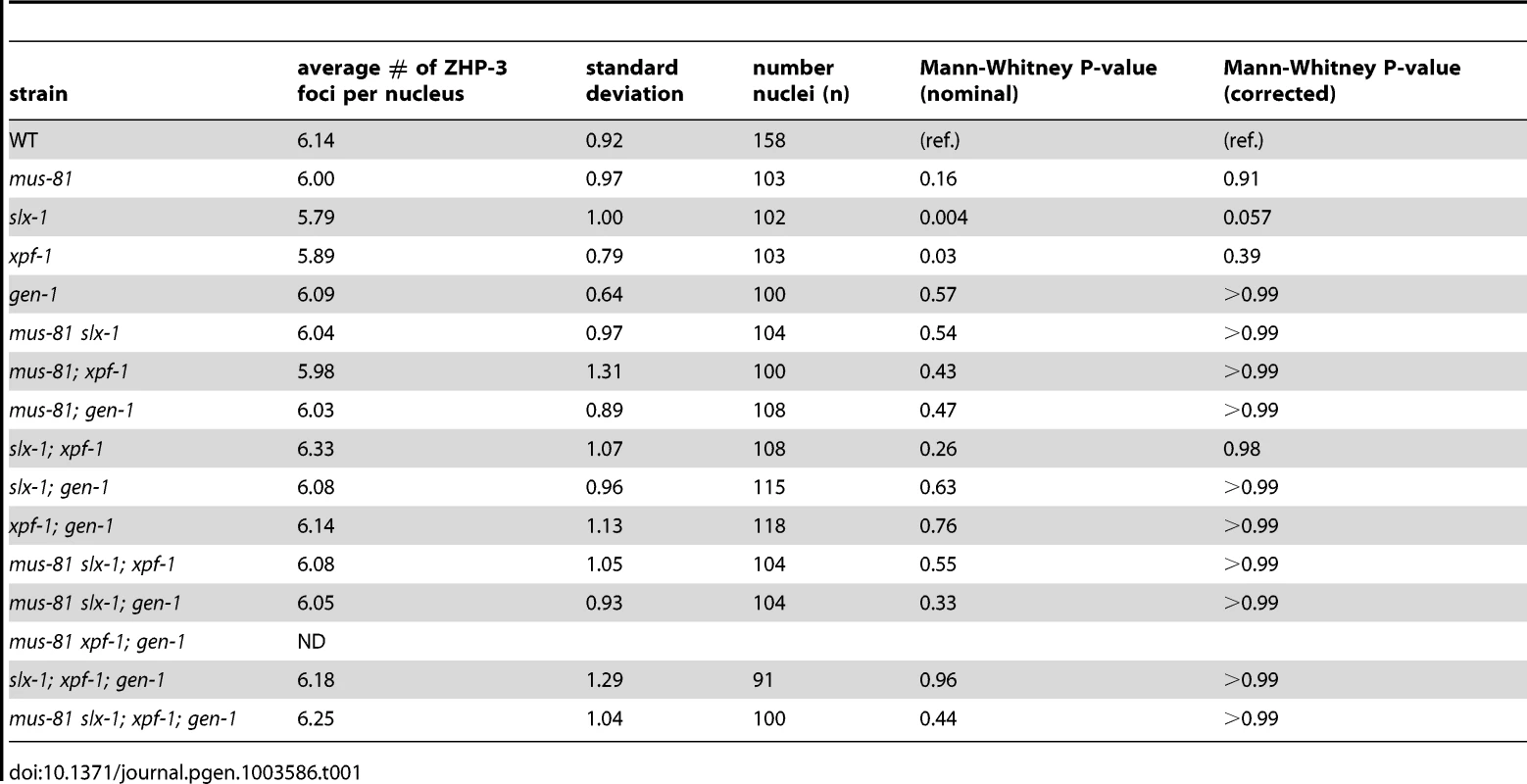
The absence of structure-specific endonucleases results in severe chromosomal abnormalities
Increased sterility, embryonic lethality, and X chromosome nondisjunction in the absence of the MUS-81, XPF-1, SLX-1, and GEN-1 endonucleases indicate defects in meiotic chromosome segregation. To observe the meiotic chromosomal defects, we examined chromosome morphology in −1 oocytes at diakinesis and +1 oocytes with chromosomes at anywhere from prometaphase I to anaphase I in the endonuclease mutants (Figure 4A–H). Following formation of the single off-centered crossover between homologous chromosomes, the bivalents remodel around the crossover site adopting a cruciform-structure comprised of a short and long arms (Figure 4C) [40]. To monitor bivalent morphology, and precisely examine homolog attachment, we stained oocytes with DAPI and antibodies recognizing the meiosis-specific α-kleisin REC-8 and the aurora B kinase AIR-2 [41], [42]. REC-8 localizes along both the long and short arms during diakinesis, and is removed at the short arm at anaphase I, thereby allowing homologs to segregate to opposite poles of the spindle [41]. AIR-2 localizes as two rings only along the short arm of the bivalents until the metaphase to anaphase I transition and it has been proposed to phosphorylate REC-8 along the short arm, thus promoting its turnover [18], [43]. The bivalents in the endonuclease-defective mutants exhibited a range of chromosome defects suggestive of impaired DSBR and/or lack of mature interhomolog crossover formation, which included chromatin bridges, premature homolog separation, DNA fragments, and a frayed appearance (Figure 4) [18], [44]–[48]. The xpf-1 and slx-1 single mutants exhibited elevated numbers of oocytes carrying chromosomal aberrations (38% (17/45) and 43% (10/23); P<0.0001, respectively; Figure 4A), whereas these were not drastically increased in the mus-81 and gen-1 single mutants (0% (0/21), P = 1, and 4% (1/26), P = 0.23, respectively). However, all double, triple, and quadruple nuclease mutant combinations showed an increase in the frequency of oocytes with chromosomal abnormalities compared to wild type (Figure 4A). Thus, the absence of either SLX-1 or XPF-1 individually increases the frequency of oocytes with aberrations and any combination of mutations in slx-1, xpf-1, mus-81, or gen-1 also results in defective bivalent morphology.
Fig. 4. Oocytes in structure-specific endonuclease mutants exhibit chromosomal abnormalities. 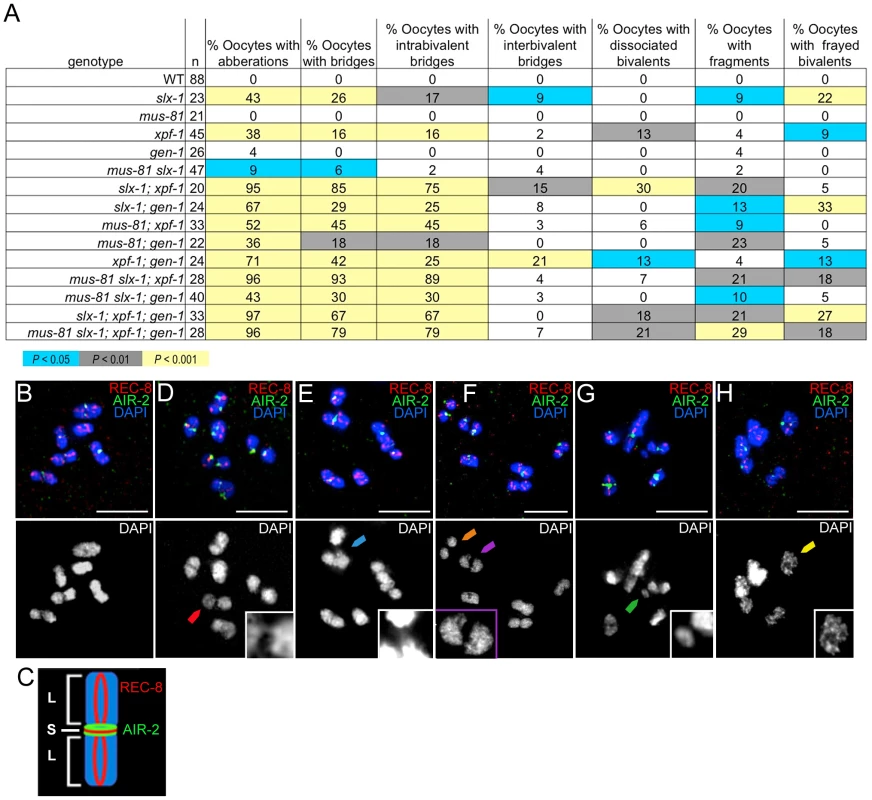
(A) Percentage of oocytes carrying chromosomal abnormalities in the nuclease-deficient backgrounds. Each oocyte is scored for the absence of defects or the presence of either a single or multiple chromosomal defects. Shading within the chart indicates level of statistical significance compared to wild type by the Fisher's exact test. (B) A wild type −1 oocyte at diakinesis has no observable chromosomal defects by analysis of REC-8 (red) and AIR-2 (green) immunolocalization and DAPI-stained chromatin (blue). (C) The illustration shows a single cruciform bivalent comprised of a short and long arms and depicts the normal localization of REC-8 and AIR-2. (D) A chromatin bridge between homologs (red arrowhead) is observed in a −1 oocyte from an slx-1; xpf-1 double mutant germline. Inset shows magnified image of the intrabivalent chromatin bridge. (E) An example of an interbivalent chromatin bridge (blue arrowhead) in a −1 oocyte from an slx-1;xpf-1 double mutant. The interbivalent chromatin bridge is shown at a higher magnification in the inset. (F) Two pairs of homologs prematurely separated at their short arms (orange and purple arrowheads) observed in a −1 oocyte from an slx-1;xpf-1 double mutant. The separated homologs (dissociated bivalents) are identified by the uncoupling of the two AIR-2 rings (green), normally observed at the region of contact between the short arms, and the lack of observable DAPI-staining chromatin between the separated short arms. Purple arrowhead points to the dissociated bivalent magnified in the inset. (G) One DNA fragment (green arrowhead) has separated from chromosomes in a −1 oocyte from an slx-1 mutant germline. Inset depicts a higher magnification of the DNA fragment indicated by the green arrowhead. (H) A −1 oocyte from an slx-1;gen-1 germline with frayed bivalents. The inset shows a magnified single frayed bivalent from the oocyte. Scale bars, 5 µm. Merged images are shown with separated channels in Figure S1. Analysis of chromosomes in the oocytes in the nuclease-defective mutants revealed chromatin bridges within bivalents (intrabivalent) and between bivalents (interbivalent; Figure 4). With the exception of the mus-81 single mutant (0%; 0/21) and the gen-1 single mutant (0%; 0/26), all single, double, triple, and quadruple nuclease mutants had higher than wild type levels (0%; 0/88) of chromatin bridges (intrabivalent and/or interbivalent), observed in anywhere from 6% to 93% of oocytes. Among the other single nuclease mutants (xpf-1 (16%; 7/45) and slx-1 (26%; 6/23)), the slx-1 mutant had the highest frequency of oocytes with chromatin bridges within bivalents and/or between bivalents. The double mutant combinations with the highest frequency of oocytes with chromatin bridges included: the slx-1;xpf-1 (85%; 17/20) and the mus-81; xpf-1 mutants (45%; 15/33). Interbivalent chromatin bridges were observed in the slx-1 single mutant (9%; 2/23), slx-1;xpf-1 double mutant (15%; 3/20), and the xpf-1;gen-1 double mutant (21%; 5/24). We hypothesize that the chromatin bridges between bivalents arise from multichromatid strand invasions that can be removed by nucleases or are typically prevented by helicases. High levels of oocytes with chromatin bridges have also been observed for these double mutants in the accompanying studies by Agostinho et al. and O'Neil et al [35], [38].
The xpf-1 mutation tended to increase the frequency of oocytes where homologs have dissociated prematurely, as indicated by bivalents that have been separated at the short arm and confirmed by the presence of uncoupled AIR-2 rings (6%–30%; Figure 4). Separation at the short arm likely indicates that fragile chromatin connections between homologs have been broken. With the exception of the mus-81; xpf-1 double mutant and mus-81 slx-1; xpf-1 triple mutant, all strains lacking XPF-1 had a statistically significant increased level of dissociated bivalents compared to wild type (Figure 4A). The mus-81; xpf-1 and mus-81 slx-1; xpf-1 mutants had a borderline increase in prematurely dissociated homologs (P = 0.079 and P = 0.063, respectively).
The slx-1 single mutant (9%; 2/23), most double mutant combinations, all triple mutants, and the quadruple mutant had DNA fragments in 9% to 29% of oocytes (Figure 4). The mus-81 slx-1 and xpf-1; gen-1 double mutants were the only mutant combinations that did not exhibit increased DNA fragments compared to wild type (P = 0.35 and P = 0.22, respectively).
Thus, absence of XPF-1, SLX-1, or combinations of any two of the four nucleases generally increased the occurrence of chromosomal abnormalities, which is consistent with the genome instability revealed by their concomitant increase in sterility, embryonic lethality, and male progeny. These plate phenotypes likely resulted from failure to properly process recombination intermediates without those endonucleases as apparent by the aberrant bivalent morphology. The nuclease-defective mutants were prone to contain oocytes with chromatin bridges, which supports the role of these nucleases in joint molecule resolution.
Discussion
Our goal was to uncover the functional overlap and differences between the structure-specific nucleases MUS-81, SLX-1, XPF-1 and GEN-1 in promoting genomic stability in the C. elegans germline. We found that HIM-18/SLX-4 interacts with SLX-1, XPF-1 and MUS-81 by yeast two-hybrid. Thus, HIM-18/SLX-4 in C. elegans may act as a platform for the three different nucleases to regulate their activity, as previously shown in vitro for the human homologs [10], but it is unknown if their interactions with HIM-18/SLX-4 are mutually exclusive. We show that xpf-1 is synergistic with either mus-81 or slx-1 for chromosome nondisjunction as detected by plate phenotyping (embryonic lethality, sterility, larval arrest and frequency of males), SNP mapping of crossover frequency and distribution, and the assessment of bivalent abnormalities such as chromatin bridges. This is evidence that MUS-81 and SLX-1 may act in the same pathway, which is parallel to that of XPF-1, to form crossovers and resolve chromatin bridges. Specifically, MUS-81 and SLX-1 may cleave the same recombination intermediates but their nuclease activity cannot be replaced by XPF-1 because of their potentially distinct substrate preferences. From plate phenotyping, we found that mus-81 and slx-1, but not xpf-1, exhibit synthetic mitotic defects with him-6, albeit we cannot rule out the possibility of synthetic meiotic defects as well; we infer that MUS-81 and SLX-1 may cleave recombination intermediates if they are not dissolved by HIM-6. Similar to yeast Yen1, C. elegans GEN-1 appears to have a minor role in promoting genomic stability that is revealed only in the absence of other nucleases; the gen-1 mutation can enhance sterility, larval arrest, and bivalent morphology defects. Here we find additional evidence supporting our previous study that SLX-1 reduces crossovers in the center of chromosomes. Although XPF-1 with MUS-81 or SLX-1 are nearly essential for progeny viability, we found that the quadruple nuclease mutant still retains some crossovers, a subset of which are no longer subject to interference. Thus, crossovers resolved by other nucleases may not be regulated by the mechanism that reinforces crossover distribution and positioning. We propose a model in which coordinate action between MUS-81, SLX-1 and XPF-1 promotes crossovers at the arms and presence of SLX-1 inhibits crossovers at the center region of chromosomes.
In addition to MUS-81, SLX-1 and XPF-1 other candidate nucleases are involved in crossover formation
Based on our crossover analysis, MUS-81, SLX-1 and XPF-1 are factors contributing to obligate crossover formation (Figure 5A). However, because mus-81slx-1; xpf-1; gen-1 quadruple mutants still show 67% and 31% of crossovers on chromosomes V and X, respectively, this suggests that additional nucleases involved in meiotic crossover formation may exist in C. elegans. Recently, it has been shown that the biochemically characterized resolvases, Yen1, Mus81-Mms4, Slx1–Slx4, the Bloom syndrome helicase homolog Sgs1 and a mismatch repair complex, Exo1-Mlh1-Mlh3, are required for joint molecule resolution in yeast [49]. Although exo-1 and mlh-1 single mutants are viable in C. elegans (T. Saito et al., unpublished results and [35], [50], [51]), we cannot eliminate the possibility that EXO-1 and MLH-1 may act in a redundant manner during crossover formation in this organism. There is no MLH-3 ortholog in C. elegans; however, we have found that its potential nuclease motif, DQHAX2EX4E, is conserved in PMS-2 [52]. Furthermore, FAN-1, which interacts with MLH-1 and is required for interstrand crosslink repair, may also act as a Holliday junction resolvase because the VRR_NUC domain, which is conserved in the FAN-1 homolog of the archaeon Sulfolobus solfataricus, can cleave Holliday junctions [53].
Fig. 5. Interplay between structure-specific endonucleases for crossover control. 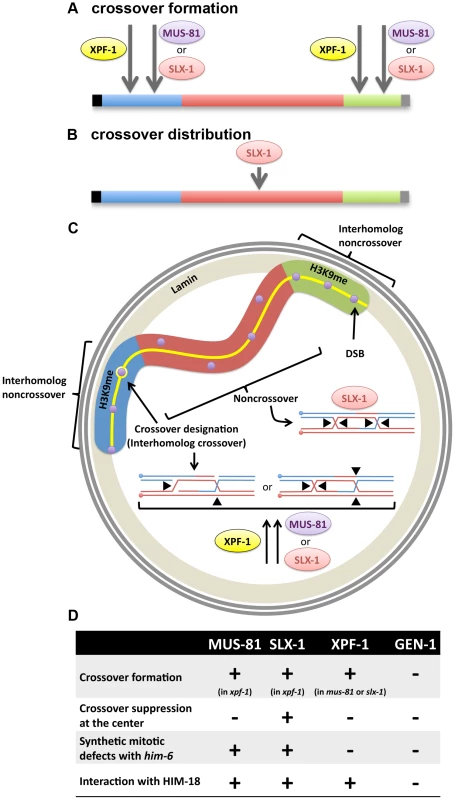
(A) Obligate crossover regulation (assurance). MUS-81, SLX-1 and XPF-1 are factors contributing to obligate crossover formation at the arm region of the chromosome. (B) Regulation of crossover (CO) distribution. SLX-1 is important for proper crossover distribution. (C) Hypothetical spatial control of meiotic recombination. H3K9me enriched arm regions (blue and green) of the chromosome are tethered to the nuclear membrane by the transmembrane protein LEM-2. There is supposed to be an interhomolog bias at the arm regions, given that DSBs are more prone to result in interhomolog crossover formation on these regions. One of the DSBs (purple circles) at one of the arm regions is designated as the future crossover site by pro-crossover factors ZHP-3, MSH-4, MSH-5 and COSA-1 (yellow circle). At the crossover-designated site, either (1) cooperative nicking of the D-loop and the half junction or (2) opposite sense resolution of the double Holliday junction, may result in a crossover product. We propose that XPF-1 redundantly functions with MUS-81 and SLX-1, respectively, to resolve those joint molecules. The other DSBs at the arm region may be repaired by SDSA or dHJ dissolution to make noncrossovers. At the center region (red), where there is less interhomolog bias than on the arm regions, there may be a higher incidence of intersister repair (indicated by purple circles not superimposed onto the synaptonemal complex, represented by the yellow line). If double Holliday junctions are formed between homologous chromosomes, SLX-1 may promote noncrossover formation by same sense resolution of the double Holliday junction in that chromosome region. (D) Crossover phenotypes (crossover formation and distribution), synthetic mitotic defects with him-6 and physical interaction with SLX4/HIM-18 are summarized for each structure-specific endonuclease. Another aspect to consider, regarding Holliday junction resolution, is that it is reminiscent of the decatenation activity exhibited by the type I topoisomerase. This is supported by reports that the vaccinia virus topoisomerase and the human topoisomerase I can resolve synthetic Holliday junctions [54], [55]. Thus, it will be interesting to investigate whether the C. elegans topoisomerase family of proteins can cleave artificial Holliday junctions in vitro and whether they are required for crossover formation in vivo. In addition, the SLX4/HIM-18 complex remains a possible candidate because recently another nuclease, the human SNM1B/Apollo, was found to interact with SLX4 [56]. The SNM1B homolog in C. elegans is MRT-1, which is required for DNA crosslink repair and telomerase activity [57]. Finally, LEM-3/Ankle1, a protein that contains an Ankyrin repeat, LEM domain (for lamina-associated polypeptide, emerin, MAN1 domain) and a GIY-YIG type nuclease domain that is also found in SLX-1, has been recently identified. LEM domain-containing proteins connect the nuclear membrane and chromatin. Chromatin immunoprecipitation analysis (ChIP-on-chip and ChIP-seq) revealed that histone H3K9me and LEM-2 are enriched at the arm regions of the chromosomes, where crossovers are frequently observed [58]–[60]. Further studies will therefore aim to identify the additional crossover-specific Holliday junction resolvases operating during meiosis.
Roles of SLX-1 in regulating crossover positioning
It has been reported that XND-1, HIM-5 and SLX-1 are required for the suppression of crossovers at the center of the autosomes [21], [27], [28]. However, the molecular mechanism for this chromosome region-specific crossover suppression is unknown. A possible mechanism for crossover suppression may involve same sense resolution of double Holliday junctions at the center of chromosomes by SLX-1 (Figure 5B and 5C). In addition, it is known that there are epigenetic differences between the arms and the center region of chromosomes in C. elegans (Figure 5C) [60]. Specifically, histone H3K9 me1/2/3 is enriched at the arm regions and H3K4me3 is enriched at the center in embryonic and larval stages. Whether this kind of epigenetic mark is maintained during meiotic recombination in the mature germline (adult worms) remains to be determined. Given the presence of a PHD-finger in SLX-1, it remains to be tested whether SLX-1 may act as a region specific crossover suppressor in part by recognizing these or other epigenetic marks defining chromosome domains or boundaries.
A model for biased crossover resolution of Holliday junctions during meiosis
How is crossover formation regulated in C. elegans meiosis? It has been previously estimated that the number of DSBs is around 5–12/homologous chromosome pair during meiotic prophase in C. elegans [19]–[21]. Therefore, it remains unclear how only one of these DSBs is destined for repair as an interhomolog crossover while all other DSBs are repaired as noncrossovers including interhomolog noncrossovers, intersister crossovers and intersister noncrossovers.
If there is no bias in how either a single or double Holliday junction is resolved, the expectation is that the crossover/noncrossover ratio should be 1∶1. However, only opposite-sense resolution of a double Holliday junction allows for crossover formation. Since crossovers are essential during meiosis, biasing factors that reinforce opposite-sense resolution must exist to ensure crossover formation. COSA-1, MSH-5 and ZHP-3 are factors that promote crossover formation but it is not known if their biochemical activities directly promote opposite-sense resolution. Based on our observations, Holliday junction resolvases do not play a role in designating a single DSB as the site destined for repair as an interhomolog crossover (Table 1).
One possible hypothesis to explain the single crossover at one of the arm regions is that there are both interhomolog and noncrossover biases operating at the arm regions (Figure 5C). Once one of the DSBs at the arm region is marked by pro-crossover factors, such as ZHP-3, MSH-4, MSH-5 and COSA-1, the designated DSB site may undergo resolution by the redundant activities of HIM-18-binding nucleases, SLX-1, MUS-81 and XPF-1. A double nicked Holliday junction cleaved by Mus81-Eme1 in yeast [61] has been suggested to only result in a crossover. In C. elegans, SLX-1, XPF-1 or MUS-81 may act on a recombination intermediate consisting of a D-loop and a half junction, which resembles two nicked Holliday junctions, to form a crossover (Figure 5C). Further studies will reveal the biochemical activities of these proteins in more detail. Given the role of RTEL-1 in catalyzing D-loop disruption in vitro [48], we propose that all undesignated DSBs at the arm regions are converted into noncrossover products via an RTEL-1-dependent SDSA pathway.
In budding yeast, plants and mice, the MSH4-MSH5-dependent crossover pathway (class I crossovers) is thought to be MUS81-independent [62], [63]. However, our study suggests that MUS-81 may be able to make a portion of the MSH-4 - and MSH-5-dependent crossovers in certain mutant situations in C. elegans.
In summary, we have addressed the functional overlaps and differences between these four structure-specific endonucleases regarding their regulatory roles in crossover control during meiosis. This has revealed novel roles for MUS-81, SLX-1 and XPF-1 in promoting the obligate crossover, and SLX-1 in regulating crossover distribution.
Materials and Methods
C. elegans genetics
C. elegans strains were cultured at 20°C under standard conditions [64]. The strains used in this study are listed in Table S4.
Determining crossover frequencies and distribution
Meiotic crossover frequencies and distribution were assayed utilizing single-nucleotide polymorphism (SNP) markers as in [34]. The SNP markers located at the boundaries of the chromosome domains were chosen based on data from WormBase (WS231) and [26]. The SNP markers and primers used are listed in Table S5. PCR and restriction digests of single worm lysates were performed as described in [65]. Statistical analysis was performed using the two-tailed Fisher's Exact test and Chi square test, 95% C.I., as in [28], [66], and corrected by Sidak adjustment for multiple comparisons.
Yeast two-hybrid analysis
The yeast two-hybrid assay was performed according to [67]. Full-length cDNAs for HIM-18, SLX-1, XPF-1, ERCC-1, MUS-81, EME-1, GEN-1, and RAD-54 were cloned into a Gateway donor vector (pDONR223). Each construct was then subcloned into 2μ Gateway destination vectors pVV213 (activation domain (AD), LEU2+) and pVV212 (Gal4 DNA binding domain (DB), TRP1+). AD-Y and DB-X fusions were transformed into MATa Y8800 and MATα Y8930 yeast strains, respectively. MATa Y8800 and MATα Y8930 were mated on YPD plates and diploids carrying both plasmids were selected on SC-Leu-Trp plates. The interactions were assessed by growth on SC-Leu-Trp-His plates at 30°C.
Immunofluorescence and imaging
Whole-mounted dissected germ lines from adult hermaphrodites 21–24 h post-L4 larval stage were subjected to 1% formaldehyde fixation, as described in [18], with the exception of the addition of cold water fish skin gelatin (0.1%; Sigma) in the 1% BSA blocking solution. The following primary antibodies were used at the indicated dilutions: α-REC-8 (1∶100; Abcam), α-AIR-2 (1∶100; [68]), and α-ZHP-3 (1∶500; [23]).
Images were acquired at 100× magnification with or without 1.5× auxiliary magnification as stacks of optical sections at 0.2-µm intervals using an IX-70 microscope (Olympus) and a cooled CCD camera (model CH350; Roper Scientific) controlled by the DeltaVision system (Applied Precision). Images were subjected to deconvolution analysis using the SoftWorx Suite 3.0 program (Applied Precision) using an enhanced ratio algorithm with 15 iterations. Statistical analysis of ZHP-3 foci was performed by the two-tailed Mann-Whitney Test, 95% C.I., and corrected by Sidak adjustment for multiple comparisons.
Supporting Information
Zdroje
1. HollidayR (1964) A mechanism for gene conversion in fungi. Genetical Research 5 : 282–304.
2. SzostakJW, Orr-WeaverTL, RothsteinRJ, StahlFW (1983) The double-strand-break repair model for recombination. Cell 33 : 25–35.
3. WuL, HicksonID (2003) The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature 426 : 870–874.
4. SinghTR, AliAM, BusyginaV, RaynardS, FanQ, et al. (2008) BLAP18/RMI2, a novel OB-fold-containing protein, is an essential component of the Bloom helicase-double Holliday junction dissolvasome. Genes Dev 22 : 2856–2868.
5. XuD, GuoR, SobeckA, BachratiCZ, YangJ, et al. (2008) RMI, a new OB-fold complex essential for Bloom syndrome protein to maintain genome stability. Genes Dev 22 : 2843–2855.
6. MullenJR, KaliramanV, IbrahimSS, BrillSJ (2001) Requirement for three novel protein complexes in the absence of the Sgs1 DNA helicase in Saccharomyces cerevisiae. Genetics 157 : 103–118.
7. BlancoMG, MatosJ, RassU, IpSC, WestSC (2010) Functional overlap between the structure-specific nucleases Yen1 and Mus81-Mms4 for DNA-damage repair in S. cerevisiae. DNA Repair (Amst) 9 : 394–402.
8. ChenXB, MelchionnaR, DenisCM, GaillardPH, BlasinaA, et al. (2001) Human Mus81-associated endonuclease cleaves Holliday junctions in vitro. Mol Cell 8 : 1117–1127.
9. BoddyMN, GaillardPH, McDonaldWH, ShanahanP, YatesJR3rd, et al. (2001) Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell 107 : 537–548.
10. MunozIM, HainK, DeclaisAC, GardinerM, TohGW, et al. (2009) Coordination of structure-specific nucleases by human SLX4/BTBD12 is required for DNA repair. Mol Cell 35 : 116–127.
11. FekairiS, ScaglioneS, ChahwanC, TaylorER, TissierA, et al. (2009) Human SLX4 is a Holliday junction resolvase subunit that binds multiple DNA repair/recombination endonucleases. Cell 138 : 78–89.
12. SvendsenJM, SmogorzewskaA, SowaME, O'ConnellBC, GygiSP, et al. (2009) Mammalian BTBD12/SLX4 assembles a Holliday junction resolvase and is required for DNA repair. Cell 138 : 63–77.
13. IpSC, RassU, BlancoMG, FlynnHR, SkehelJM, et al. (2008) Identification of Holliday junction resolvases from humans and yeast. Nature 456 : 357–361.
14. HabrakenY, SungP, PrakashL, PrakashS (1994) Holliday junction cleavage by yeast Rad1 protein. Nature 371 : 531–534.
15. DaviesAA, FriedbergEC, TomkinsonAE, WoodRD, WestSC (1995) Role of the Rad1 and Rad10 proteins in nucleotide excision repair and recombination. J Biol Chem 270 : 24638–24641.
16. SekelskyJJ, McKimKS, ChinGM, HawleyRS (1995) The Drosophila meiotic recombination gene mei-9 encodes a homologue of the yeast excision repair protein Rad1. Genetics 141 : 619–627.
17. YildizO, MajumderS, KramerB, SekelskyJJ (2002) Drosophila MUS312 interacts with the nucleotide excision repair endonuclease MEI-9 to generate meiotic crossovers. Mol Cell 10 : 1503–1509.
18. SaitoTT, YoudsJL, BoultonSJ, ColaiacovoMP (2009) Caenorhabditis elegans HIM-18/SLX-4 interacts with SLX-1 and XPF-1 and maintains genomic integrity in the germline by processing recombination intermediates. PLoS Genet 5: e1000735.
19. NottkeAC, Beese-SimsSE, PantalenaLF, ReinkeV, ShiY, et al. (2011) SPR-5 is a histone H3K4 demethylase with a role in meiotic double-strand break repair. Proc Natl Acad Sci U S A 108 : 12805–12810.
20. RosuS, LibudaDE, VilleneuveAM (2011) Robust crossover assurance and regulated interhomolog access maintain meiotic crossover number. Science 334 : 1286–1289.
21. SaitoTT, MohideenF, MeyerK, HarperJW, ColaiacovoMP (2012) SLX-1 Is Required for Maintaining Genomic Integrity and Promoting Meiotic Noncrossovers in the Caenorhabditis elegans Germline. PLoS Genet 8: e1002888.
22. YokooR, ZawadzkiKA, NabeshimaK, DrakeM, ArurS, et al. (2012) COSA-1 reveals robust homeostasis and separable licensing and reinforcement steps governing meiotic crossovers. Cell 149 : 75–87.
23. BhallaN, WynneDJ, JantschV, DernburgAF (2008) ZHP-3 acts at crossovers to couple meiotic recombination with synaptonemal complex disassembly and bivalent formation in C. elegans. PLoS Genet 4: e1000235.
24. PanJ, KeeneyS (2007) Molecular cartography: mapping the landscape of meiotic recombination. PLoS Biol 5: e333.
25. BarnesTM, KoharaY, CoulsonA, HekimiS (1995) Meiotic recombination, noncoding DNA and genomic organization in Caenorhabditis elegans. Genetics 141 : 159–179.
26. RockmanMV, KruglyakL (2009) Recombinational landscape and population genomics of Caenorhabditis elegans. PLoS Genet 5: e1000419.
27. WagnerCR, KuerversL, BaillieDL, YanowitzJL (2010) xnd-1 regulates the global recombination landscape in Caenorhabditis elegans. Nature 467 : 839–843.
28. MeneelyPM, McGovernOL, HeinisFI, YanowitzJL (2012) Crossover distribution and frequency are regulated by him-5 in Caenorhabditis elegans. Genetics 190 : 1251–1266.
29. InterthalH, HeyerWD (2000) MUS81 encodes a novel helix-hairpin-helix protein involved in the response to UV - and methylation-induced DNA damage in Saccharomyces cerevisiae. Mol Gen Genet 263 : 812–827.
30. AndersenSL, KuoHK, SavukoskiD, BrodskyMH, SekelskyJ (2011) Three structure-selective endonucleases are essential in the absence of BLM helicase in Drosophila. PLoS Genet 7: e1002315.
31. CoulonS, GaillardPH, ChahwanC, McDonaldWH, YatesJR3rd, et al. (2004) Slx1–Slx4 are subunits of a structure-specific endonuclease that maintains ribosomal DNA in fission yeast. Mol Biol Cell 15 : 71–80.
32. WechslerT, NewmanS, WestSC (2011) Aberrant chromosome morphology in human cells defective for Holliday junction resolution. Nature 471 : 642–646.
33. HodgkinJ, HorvitzHR, BrennerS (1979) Nondisjunction mutants of the nematode Caenorhabditis elegans. Genetics 91 : 67–94.
34. NabeshimaK, VilleneuveAM, HillersKJ (2004) Chromosome-wide regulation of meiotic crossover formation in Caenorhabditis elegans requires properly assembled chromosome axes. Genetics 168 : 1275–1292.
35. O'NeilNJ, MartinJS, YoudsJL, WardJD, PetalcorinMI, et al. (2013) Joint molecule resolution requires the redundant activities of MUS-81 and XPF-1 during C. elegans meiosis. PLoS Genet 9: e1003582.
36. MeneelyPM, FaragoAF, KauffmanTM (2002) Crossover distribution and high interference for both the X chromosome and an autosome during oogenesis and spermatogenesis in Caenorhabditis elegans. Genetics 162 : 1169–1177.
37. Martinez-PerezE, ColaiacovoMP (2009) Distribution of meiotic recombination events: talking to your neighbors. Curr Opin Genet Dev 19 : 105–112.
38. AgostinhoA, MeierB, SonnevilleR, JagutM, WoglarA, et al. (2013) Combinatorial regulation of meiotic Holliday junction resolution in C. elegans by HIM-6/(BLM) helicase, SLX-4, and SLX-1, MUS-81 and XPF-1 nucleases. PLoS Genet 9: e1003591.
39. JantschV, PasierbekP, MuellerMM, SchweizerD, JantschM, et al. (2004) Targeted gene knockout reveals a role in meiotic recombination for ZHP-3, a Zip3-related protein in Caenorhabditis elegans. Mol Cell Biol 24 : 7998–8006.
40. NabeshimaK, VilleneuveAM, ColaiacovoMP (2005) Crossing over is coupled to late meiotic prophase bivalent differentiation through asymmetric disassembly of the SC. J Cell Biol 168 : 683–689.
41. PasierbekP, JantschM, MelcherM, SchleifferA, SchweizerD, et al. (2001) A Caenorhabditis elegans cohesion protein with functions in meiotic chromosome pairing and disjunction. Genes Dev 15 : 1349–1360.
42. SchumacherJM, GoldenA, DonovanPJ (1998) AIR-2: An Aurora/Ipl1-related protein kinase associated with chromosomes and midbody microtubules is required for polar body extrusion and cytokinesis in Caenorhabditis elegans embryos. J Cell Biol 143 : 1635–1646.
43. RogersE, BishopJD, WaddleJA, SchumacherJM, LinR (2002) The aurora kinase AIR-2 functions in the release of chromosome cohesion in Caenorhabditis elegans meiosis. J Cell Biol 157 : 219–229.
44. AdamoA, CollisSJ, AdelmanCA, SilvaN, HorejsiZ, et al. (2010) Preventing nonhomologous end joining suppresses DNA repair defects of Fanconi anemia. Mol Cell 39 : 25–35.
45. AllardP, ColaiacovoMP (2010) Bisphenol A impairs the double-strand break repair machinery in the germline and causes chromosome abnormalities. Proc Natl Acad Sci U S A 107 : 20405–20410.
46. MartinJS, WinkelmannN, PetalcorinMI, McIlwraithMJ, BoultonSJ (2005) RAD-51-dependent and -independent roles of a Caenorhabditis elegans BRCA2-related protein during DNA double-strand break repair. Mol Cell Biol 25 : 3127–3139.
47. RinaldoC, BazzicalupoP, EderleS, HilliardM, La VolpeA (2002) Roles for Caenorhabditis elegans rad-51 in meiosis and in resistance to ionizing radiation during development. Genetics 160 : 471–479.
48. YoudsJL, MetsDG, McIlwraithMJ, MartinJS, WardJD, et al. (2010) RTEL-1 enforces meiotic crossover interference and homeostasis. Science 327 : 1254–1258.
49. ZakharyevichK, TangS, MaY, HunterN (2012) Delineation of joint molecule resolution pathways in meiosis identifies a crossover-specific resolvase. Cell 149 : 334–347.
50. LemmensBB, TijstermanM (2011) DNA double-strand break repair in Caenorhabditis elegans. Chromosoma 120 : 1–21.
51. LemmensBB, JohnsonNM, TijstermanM (2013) COM-1 Promotes Homologous Recombination during Caenorhabditis elegans Meiosis by Antagonizing Ku-Mediated Non-Homologous End Joining. PLoS Genet 9: e1003276.
52. NishantKT, PlysAJ, AlaniE (2008) A mutation in the putative MLH3 endonuclease domain confers a defect in both mismatch repair and meiosis in Saccharomyces cerevisiae. Genetics 179 : 747–755.
53. BondCS, KvaratskheliaM, RichardD, WhiteMF, HunterWN (2001) Structure of Hjc, a Holliday junction resolvase, from Sulfolobus solfataricus. Proc Natl Acad Sci U S A 98 : 5509–5514.
54. HedeMS, PetersenRL, FrohlichRF, KrugerD, AndersenFF, et al. (2007) Resolution of Holliday junction substrates by human topoisomerase I. J Mol Biol 365 : 1076–1092.
55. SekiguchiJ, SeemanNC, ShumanS (1996) Resolution of Holliday junctions by eukaryotic DNA topoisomerase I. Proc Natl Acad Sci U S A 93 : 785–789.
56. SalewskyB, SchmiesterM, SchindlerD, DigweedM, DemuthI (2012) The nuclease hSNM1B/Apollo is linked to the Fanconi anemia pathway via its interaction with FANCP/SLX4. Hum Mol Genet 21 : 4948–56.
57. MeierB, BarberLJ, LiuY, ShtesselL, BoultonSJ, et al. (2009) The MRT-1 nuclease is required for DNA crosslink repair and telomerase activity in vivo in Caenorhabditis elegans. EMBO J 28 : 3549–3563.
58. GersteinMB, LuZJ, Van NostrandEL, ChengC, ArshinoffBI, et al. (2010) Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science 330 : 1775–1787.
59. IkegamiK, EgelhoferTA, StromeS, LiebJD (2010) Caenorhabditis elegans chromosome arms are anchored to the nuclear membrane via discontinuous association with LEM-2. Genome Biol 11: R120.
60. LiuT, RechtsteinerA, EgelhoferTA, VielleA, LatorreI, et al. (2011) Broad chromosomal domains of histone modification patterns in C. elegans. Genome Res 21 : 227–236.
61. OsmanF, DixonJ, DoeCL, WhitbyMC (2003) Generating crossovers by resolution of nicked Holliday junctions: a role for Mus81-Eme1 in meiosis. Mol Cell 12 : 761–774.
62. HollingsworthNM, BrillSJ (2004) The Mus81 solution to resolution: generating meiotic crossovers without Holliday junctions. Genes Dev 18 : 117–125.
63. SchwartzEK, HeyerWD (2011) Processing of joint molecule intermediates by structure-selective endonucleases during homologous recombination in eukaryotes. Chromosoma 120 : 109–127.
64. BrennerS (1974) The genetics of Caenorhabditis elegans. Genetics 77 : 71–94.
65. DavisMW, HammarlundM, HarrachT, HullettP, OlsenS, et al. (2005) Rapid single nucleotide polymorphism mapping in C. elegans. BMC Genomics 6 : 118.
66. MetsDG, MeyerBJ (2009) Condensins regulate meiotic DNA break distribution, thus crossover frequency, by controlling chromosome structure. Cell 139 : 73–86.
67. WalhoutAJ, VidalM (2001) High-throughput yeast two-hybrid assays for large-scale protein interaction mapping. Methods 24 : 297–306.
68. de CarvalhoCE, ZaaijerS, SmolikovS, GuY, SchumacherJM, et al. (2008) LAB-1 antagonizes the Aurora B kinase in C. elegans. Genes Dev 22 : 2869–2885.
Štítky
Genetika Reprodukční medicína
Článek Independent Evolution of Transcriptional Inactivation on Sex Chromosomes in Birds and MammalsČlánek The bHLH Subgroup IIId Factors Negatively Regulate Jasmonate-Mediated Plant Defense and DevelopmentČlánek Selective Pressures to Maintain Attachment Site Specificity of Integrative and Conjugative ElementsČlánek Reassembly of Nucleosomes at the Promoter Initiates Resilencing Following Decitabine ExposureČlánek Hepatocyte Growth Factor Signaling in Intrapancreatic Ductal Cells Drives Pancreatic Morphogenesis
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 7- Akutní intermitentní porfyrie
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Růst a vývoj dětí narozených pomocí IVF
- IVF a rakovina prsu – zvyšují hormony riziko vzniku rakoviny?
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
-
Všechny články tohoto čísla
- An Solution for Crossover Formation
- Genome-Wide Association Mapping in Dogs Enables Identification of the Homeobox Gene, , as a Genetic Component of Neural Tube Defects in Humans
- Independent Evolution of Transcriptional Inactivation on Sex Chromosomes in Birds and Mammals
- Stepwise Activation of the ATR Signaling Pathway upon Increasing Replication Stress Impacts Fragile Site Integrity
- Genomic Analysis of Natural Selection and Phenotypic Variation in High-Altitude Mongolians
- Modification of tRNA by Elongator Is Essential for Efficient Translation of Stress mRNAs
- Role of CTCF Protein in Regulating Locus Transcription
- Gene Set Signature of Reversal Reaction Type I in Leprosy Patients
- Mapping of PARK2 and PACRG Overlapping Regulatory Region Reveals LD Structure and Functional Variants in Association with Leprosy in Unrelated Indian Population Groups
- Is Required for Formation of the Genital Ridge in Mice
- Monopolin Subunit Csm1 Associates with MIND Complex to Establish Monopolar Attachment of Sister Kinetochores at Meiosis I
- Recombination Dynamics of a Human Y-Chromosomal Palindrome: Rapid GC-Biased Gene Conversion, Multi-kilobase Conversion Tracts, and Rare Inversions
- Mechanisms of Protein Sequence Divergence and Incompatibility
- Histone Methyltransferase DOT1L Drives Recovery of Gene Expression after a Genotoxic Attack
- Female Behaviour Drives Expression and Evolution of Gustatory Receptors in Butterflies
- Combinatorial Regulation of Meiotic Holliday Junction Resolution in by HIM-6 (BLM) Helicase, SLX-4, and the SLX-1, MUS-81 and XPF-1 Nucleases
- The bHLH Subgroup IIId Factors Negatively Regulate Jasmonate-Mediated Plant Defense and Development
- The Role of Interruptions in polyQ in the Pathology of SCA1
- Dietary Restriction Induced Longevity Is Mediated by Nuclear Receptor NHR-62 in
- Fine Time Course Expression Analysis Identifies Cascades of Activation and Repression and Maps a Putative Regulator of Mammalian Sex Determination
- Genome-scale Co-evolutionary Inference Identifies Functions and Clients of Bacterial Hsp90
- Oxidative Stress and Replication-Independent DNA Breakage Induced by Arsenic in
- A Moonlighting Enzyme Links Cell Size with Central Metabolism
- Budding Yeast Greatwall and Endosulfines Control Activity and Spatial Regulation of PP2A for Timely Mitotic Progression
- The Conserved Intronic Cleavage and Polyadenylation Site of CstF-77 Gene Imparts Control of 3′ End Processing Activity through Feedback Autoregulation and by U1 snRNP
- The BTB-zinc Finger Transcription Factor Abrupt Acts as an Epithelial Oncogene in through Maintaining a Progenitor-like Cell State
- The Cohesion Protein SOLO Associates with SMC1 and Is Required for Synapsis, Recombination, Homolog Bias and Cohesion and Pairing of Centromeres in Drosophila Meiosis
- The RNA-binding Proteins FMR1, Rasputin and Caprin Act Together with the UBA Protein Lingerer to Restrict Tissue Growth in
- Pattern Dynamics in Adaxial-Abaxial Specific Gene Expression Are Modulated by a Plastid Retrograde Signal during Leaf Development
- A Network of HMG-box Transcription Factors Regulates Sexual Cycle in the Fungus
- Bacterial Adaptation through Loss of Function
- ENU-induced Mutation in the DNA-binding Domain of KLF3 Reveals Important Roles for KLF3 in Cardiovascular Development and Function in Mice
- Interplay between Structure-Specific Endonucleases for Crossover Control during Meiosis
- FGF Signalling Regulates Chromatin Organisation during Neural Differentiation via Mechanisms that Can Be Uncoupled from Transcription
- The Arabidopsis RNA Binding Protein with K Homology Motifs, SHINY1, Interacts with the C-terminal Domain Phosphatase-like 1 (CPL1) to Repress Stress-Inducible Gene Expression
- Selective Pressures to Maintain Attachment Site Specificity of Integrative and Conjugative Elements
- The Conserved ADAMTS-like Protein Lonely heart Mediates Matrix Formation and Cardiac Tissue Integrity
- The cGMP-Dependent Protein Kinase EGL-4 Regulates Nociceptive Behavioral Sensitivity
- RBM5 Is a Male Germ Cell Splicing Factor and Is Required for Spermatid Differentiation and Male Fertility
- Disease-Related Growth Factor and Embryonic Signaling Pathways Modulate an Enhancer of Expression at the 6q23.2 Coronary Heart Disease Locus
- Yeast Pol4 Promotes Tel1-Regulated Chromosomal Translocations
- A Dual Role for SOX10 in the Maintenance of the Postnatal Melanocyte Lineage and the Differentiation of Melanocyte Stem Cell Progenitors
- SLC26A4 Targeted to the Endolymphatic Sac Rescues Hearing and Balance in Mutant Mice
- Odoriferous Defensive Stink Gland Transcriptome to Identify Novel Genes Necessary for Quinone Synthesis in the Red Flour Beetle,
- Prediction of Complex Human Traits Using the Genomic Best Linear Unbiased Predictor
- Gene × Physical Activity Interactions in Obesity: Combined Analysis of 111,421 Individuals of European Ancestry
- Reassembly of Nucleosomes at the Promoter Initiates Resilencing Following Decitabine Exposure
- Exquisite Light Sensitivity of Cryptochrome
- miR-133a Regulates Adipocyte Browning In Vivo
- Strabismus Promotes Recruitment and Degradation of Farnesylated Prickle in Planar Polarity Specification
- Hepatocyte Growth Factor Signaling in Intrapancreatic Ductal Cells Drives Pancreatic Morphogenesis
- Is a Potential Tumor Suppressor Gene Commonly Inactivated by Epigenetic Mechanisms in Colorectal Cancer
- Joint Molecule Resolution Requires the Redundant Activities of MUS-81 and XPF-1 during Meiosis
- The Mating Competence of Geographically Diverse Strains in Their Natural and Unnatural Sand Fly Vectors
- Defective Repair of Oxidative Base Lesions by the DNA Glycosylase Nth1 Associates with Multiple Telomere Defects
- Effective Blocking of the Enhancer Requires Cooperation between Two Main Mechanisms Suggested for the Insulator Function
- Trans-Ancestral Studies Fine Map the SLE-Susceptibility Locus
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- SLC26A4 Targeted to the Endolymphatic Sac Rescues Hearing and Balance in Mutant Mice
- Bacterial Adaptation through Loss of Function
- The Cohesion Protein SOLO Associates with SMC1 and Is Required for Synapsis, Recombination, Homolog Bias and Cohesion and Pairing of Centromeres in Drosophila Meiosis
- Gene × Physical Activity Interactions in Obesity: Combined Analysis of 111,421 Individuals of European Ancestry
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



