-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Chromosome Movements Promoted by the Mitochondrial Protein SPD-3 Are Required for Homology Search during Meiosis
Pairing of homologous chromosomes during early meiosis is essential to prevent the formation of aneuploid gametes. Chromosome pairing includes a step of homology search followed by the stabilization of homolog interactions by the synaptonemal complex (SC). These events coincide with dramatic changes in nuclear organization and rapid chromosome movements that depend on cytoskeletal motors and are mediated by SUN-domain proteins on the nuclear envelope, but how chromosome mobility contributes to the pairing process remains poorly understood. We show that defects in the mitochondria-localizing protein SPD-3 cause a defect in homolog pairing without impairing nuclear reorganization or SC assembly, which results in promiscuous installation of the SC between non-homologous chromosomes. Preventing SC assembly in spd-3 mutants does not improve homolog pairing, demonstrating that SPD-3 is required for homology search at the start of meiosis. Pairing center regions localize to SUN-1 aggregates at meiosis onset in spd-3 mutants; and pairing-promoting proteins, including cytoskeletal motors and polo-like kinase 2, are normally recruited to the nuclear envelope. However, quantitative analysis of SUN-1 aggregate movement in spd-3 mutants demonstrates a clear reduction in mobility, although this defect is not as severe as that seen in sun-1(jf18) mutants, which also show a stronger pairing defect, suggesting a correlation between chromosome-end mobility and the efficiency of pairing. SUN-1 aggregate movement is also impaired following inhibition of mitochondrial respiration or dynein knockdown, suggesting that mitochondrial function is required for motor-driven SUN-1 movement. The reduced chromosome-end mobility of spd-3 mutants impairs coupling of SC assembly to homology recognition and causes a delay in meiotic progression mediated by HORMA-domain protein HTP-1. Our work reveals how chromosome mobility impacts the different early meiotic events that promote homolog pairing and suggests that efficient homology search at the onset of meiosis is largely dependent on motor-driven chromosome movement.
Published in the journal: . PLoS Genet 9(5): e32767. doi:10.1371/journal.pgen.1003497
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003497Summary
Pairing of homologous chromosomes during early meiosis is essential to prevent the formation of aneuploid gametes. Chromosome pairing includes a step of homology search followed by the stabilization of homolog interactions by the synaptonemal complex (SC). These events coincide with dramatic changes in nuclear organization and rapid chromosome movements that depend on cytoskeletal motors and are mediated by SUN-domain proteins on the nuclear envelope, but how chromosome mobility contributes to the pairing process remains poorly understood. We show that defects in the mitochondria-localizing protein SPD-3 cause a defect in homolog pairing without impairing nuclear reorganization or SC assembly, which results in promiscuous installation of the SC between non-homologous chromosomes. Preventing SC assembly in spd-3 mutants does not improve homolog pairing, demonstrating that SPD-3 is required for homology search at the start of meiosis. Pairing center regions localize to SUN-1 aggregates at meiosis onset in spd-3 mutants; and pairing-promoting proteins, including cytoskeletal motors and polo-like kinase 2, are normally recruited to the nuclear envelope. However, quantitative analysis of SUN-1 aggregate movement in spd-3 mutants demonstrates a clear reduction in mobility, although this defect is not as severe as that seen in sun-1(jf18) mutants, which also show a stronger pairing defect, suggesting a correlation between chromosome-end mobility and the efficiency of pairing. SUN-1 aggregate movement is also impaired following inhibition of mitochondrial respiration or dynein knockdown, suggesting that mitochondrial function is required for motor-driven SUN-1 movement. The reduced chromosome-end mobility of spd-3 mutants impairs coupling of SC assembly to homology recognition and causes a delay in meiotic progression mediated by HORMA-domain protein HTP-1. Our work reveals how chromosome mobility impacts the different early meiotic events that promote homolog pairing and suggests that efficient homology search at the onset of meiosis is largely dependent on motor-driven chromosome movement.
Introduction
Accurate chromosome segregation during meiosis requires the formation of physical attachments between homologous chromosomes (homologs). To achieve this, a series of events unfold during meiotic prophase to promote the formation of inter-homolog crossover events during meiotic recombination [1]. Crossovers, together with sister chromatid cohesion, provide the basis of stable mechanical connections between the homologs, and sites of crossing over are visualized as chiasmata in late meiotic prophase [2]. Crucially, before homologs can be tethered by crossover events, each chromosome must first recognize its correct pairing partner amongst all the chromosomes present in the nucleus.
In most organisms the pairing process can be divided into three phases that have distinctive genetic requirements: the initial encounters between the homologs, during which homology recognition must take place, the stabilization of these early interactions by recombination dependent or independent mechanisms, and the assembly of the synaptonemal complex (SC) in the interface between the homologs [3]. Although the SC is required for the full and intimate alignment of the homologs, it is clear that the SC per se has no role in discriminating between homologous and non-homologous chromosomes. In haploid plants or yeast the SC is promiscuously assembled between non-homologous chromosomes [4], [5], and yeast, Caenorhabditis elegans and mice mutants that lack components of the central region of the SC achieve high levels of pairing [6]–[8]. Furthermore, the uncontrolled assembly of the SC at the start of meiotic prophase has been shown to interfere with the pairing process [9]. Therefore, licensing of SC assembly needs to be carefully controlled during early prophase, and checkpoint-like mechanisms appear to be in place to ensure that SC assembly is coupled to successful homolog recognition [10]–[12]. The mechanisms that promote homolog pairing during early meiotic prophase remain one of the least understood aspects of meiosis.
The onset of homolog pairing coincides with a dramatic reorganization of chromosomes inside the nucleus that involves tethering of one or both chromosomal ends to the nuclear envelope (NE), which in most organisms leads to the clustering of all telomeres in a small area of the inner NE [13]. Chromosome attachment to the NE is mediated by SUN-domain proteins located on the inner NE, which interact with a transmembrane KASH-domain protein located on the outer NE [14], [15]. This SUN-KASH protein bridge spans across the NE, allowing the transmission of forces originated by the cytoskeleton to induce movement of meiotic chromosomes [11], [16], [17]. Rapid chromosome movements appear to be a conserved feature of meiotic prophase, and the parameters that define these movements have been investigated in fungi, plants, and C. elegans [18]–[23]. Preliminary studies suggest that chromosome movements also occur during mammalian meiosis [24], [25]. In fact, a SUN/KASH protein pair that interacts with cytoskeletal motors has recently been described in mammals [26], suggesting a high degree of conservation in the mechanism that mediates meiotic chromosome movement. Despite this conservation, there are important differences in the pattern of chromosomal movements observed in different species, and the function of meiotic chromosome motion remains under debate. This is exemplified by the fact that meiotic prophase chromosome movement has been implicated in many different events, including: promoting chromosomal encounters during early prophase [18], [22], [27], [28], [29], destabilizing inappropriate interactions between non-homologous chromosomes [11], [17], [21], [30], [31], licensing SC assembly [11], promoting meiotic progression [28], completion of biochemical steps during meiotic recombination [32], and regulation of crossover distribution [21], [30].
In the nematode C. elegans homolog pairing occurs in a region of the germ line known as transition zone, which corresponds to the leptotene and zygotene stages of meiosis. Transition zone nuclei display a polarized distribution of chromatin (referred to as chromosome clustering), and the acquisition of this conformation, which is thought to indicate active homology search, requires the CHK-2 kinase [33]. In transition zone nuclei the chromosomal end carrying the pairing center (PC) region, which is known to promote pairing and SC assembly, is attached to the inner NE [34]–[36]. PCs act by recruiting the polo-like kinase PLK-2 to the nuclear envelope, where PLK-2 induces the formation of dynamic SUN-1/ZYG-12 (the SUN/KASH pair in C. elegans) aggregates that are required to ensure faithful pairing [11], [37], [38], [39]. Chromosome movement mediated by the SUN-1/ZYG-12 bridge requires cytoplasmic dynein and microtubules [11], [23]. A second type of chromosome movement, characterized by increased diffusion of PCs on the NE, also starts at the onset of meiotic prophase, and it has been proposed that this diffusive movement could be a major contributor to pairing, while motor-driven movement may be required for licensing SC assembly [23].
Here we identify SPD-3, a mitochondrial protein, as a factor required to induce normal levels of motor-driven chromosome-end movement during early meiotic prophase in C. elegans. We show that the reduced movement of SUN-1 aggregates seen in spd-3(me85) mutants results in deficient homology recognition and improper SC assembly between non-homologous chromosomes, supporting a model in which cytoskeleton-dependent chromosome movements are required for the earliest steps of meiotic homology search.
Results
A Premature STOP codon in the spd-3 gene, which encodes a mitochondrial protein, impairs chiasma formation
In order to identify genes required for crossover formation, we performed a forward genetic screen to isolate mutants that displayed diakinesis oocytes with reduced numbers of chiasmata (Text S1). In wild-type oocytes, the six pairs of homologs are linked by chiasmata and therefore six DAPI-stained bodies are present (Figure 1A). In contrast, me85 mutants displayed diakinesis oocytes with up to twelve DAPI-stained bodies, demonstrating deficient chiasma formation (Figure 1A). Mapping of the me85 mutation using a comparative genome hybridization array [40, Text S1] suggested the presence of a mutation in the spd-3 gene, and sequencing of the spd-3 gene in me85 mutants confirmed the presence of an early STOP codon predicted to remove the last 61 amino acids of SPD-3 (Figure 1B). We next performed complementation tests between me85 mutants and two strains carrying spd-3 deletion alleles (tm2969 and ok1817, which are expected to be null spd-3 alleles), as well as a transgenic strain expressing a functional GFP-tagged version of SPD-3 [41]. Both spd-3 deletion alleles failed to complement the me85 mutation, and the spd-3::GFP transgene fully rescued the defects of me85 homozygous worms. Furthermore, western blot analysis demonstrated that protein extracts from me85 mutants lacked a band at the expected molecular weight for SPD-3, while SPD-3-positive bands were clearly visible in extracts from wild-type worms and from worms carrying the spd-3::GFP transgene (Figure 1C). These results confirm that the phenotypes seen in me85 mutants are due to the early STOP codon in spd-3.
Fig. 1. SPD-3 is required for chiasma formation and localizes to mitochondria. 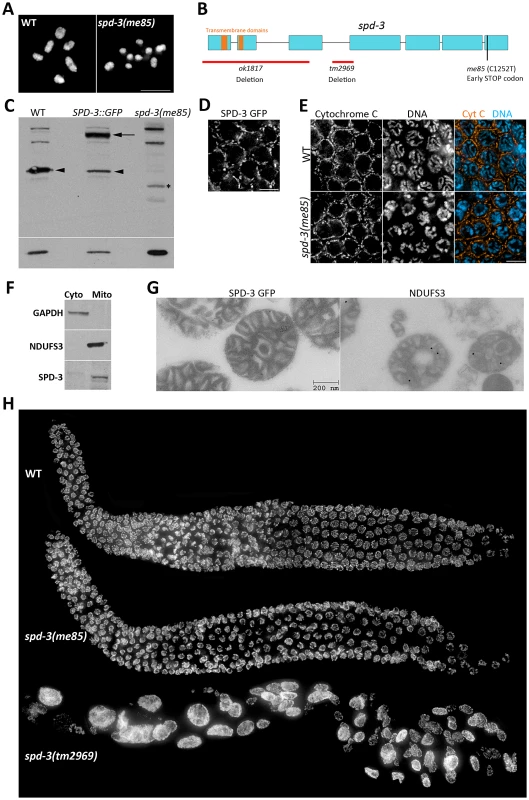
(A) Projection of diakinesis oocytes stained with DAPI. Note that while WT oocytes display 6 DAPI-stained bodies, corresponding to 6 bivalents, the spd-3(me85) mutant oocyte displays 9 DAPI-stained bodies, demonstrating a partial failure in chiasma formation. (B) Diagram of the spd-3 gene indicating the position of the me85 mutation and two deletion alleles. (C) Western blot probed with anti-SPD-3 antibodies, arrowheads point to the endogenous SPD-3 protein, arrow points the SDP-3::GFP fusion protein, and the asterisks labels a band that may correspond to a truncated SPD-3 protein made by spd-3(me85) mutants. The western blot shown at the bottom was probed with anti-tubulin antibodies and was used as a loading control. (D) Pachytene region of an ex-vivo germ line from a worm expressing SPD-3::GFP, which shows a localization pattern similar to that observed in (E) for immunolocalization of the mitochondrial protein cytochrome C in the pachytene region of fixed germ lines. (F) Western blots from cytosolic and mitochondrial extracts stained with the indicated antibodies. Note that SPD-3 is only found in the mitochondrial extract. (G) Electron micrographs of mitochondria purified from worms expressing SDP-3::GFP, and labeled with anti-GFP (left, 10 nm gold) and anti-NDUFS3 antibodies (right, 18 nm gold). (H) Whole germ lines from 16–18 hours post L4 worms stained with DAPI. BLAST searches of protein databases failed to identify clear SDP-3 homologs outside nematodes, but a search for conserved domains identified two putative transmembrane domains in the N terminus of SPD-3 (Figure 1B) and a previous study suggested that SPD-3 localizes to mitochondria in the early embryo [41]. We took three complementary approaches to verify that SDP-3 localizes to mitochondria. First, ex-vivo imaging of the pachytene region from germ lines carrying the spd-3::GFP transgene demonstrated that SDP-3::GFP localized to elongated structures outside of the nucleus that appeared very similar to the localization pattern of the mitochondrial protein cytochrome C in fixed germ lines (Figure 1D, 1E). SPD-3::GFP also displayed the same staining pattern in the mitotic and transition zones of the germ line (Figure S1). Cytochrome C localization in spd-3(me85) mutant germ lines did not reveal any obvious mitochondrial defects (Figure 1E). Second, using anti-NUO-2 (homologue of human NDUFS3) and anti-GAPDH antibodies as mitochondrial and cytosolic markers, we performed a western blot analysis of mitochondrial and cytosolic extracts purified from whole worm homogenates. NUO-2 and GAPDH were found in the mitochondrial and cytosolic fractions respectively, while SPD-3 was only present in the mitochondrial fraction (Figure 1F). Finally, we used immuno-electron microscopy to confirm the presence of SPD-3::GFP in purified mitochondria, using anti-NDUFS3 antibodies as a positive control (Figure 1G). These results confirm that SPD-3 localizes to mitochondria.
spd-3 deletion results in highly abnormal germ cells, but spd-3(me85) mutants display organized germ lines
During the complementation tests described above, we noticed that germ lines from mutants homozygous for either of the spd-3 deletions were highly disorganized, containing few nuclei that were of abnormal sizes (Figure 1H). This phenotype suggests that the mitotic divisions that precede meiosis were severely impaired in the absence of SDP-3. In contrast, young spd-3(me85) mutants (16–18 hours post L4) displayed well-organized germ lines that resembled those of age-matched wild-type controls in both size and appearance (Figure 1H). However, older spd-3(me85) mutants displayed enlarged nuclei in the mitotic region of the germ line, and the number of these abnormal nuclei increased with age (Figure S1). Although we did not detect enlarged nuclei in the meiotic region of young spd-3(me85) mutants and the analysis of diakinesis oocytes suggests a normal chromosome complement (see Figure 2C–2D), we can not completely rule out that some nuclei may enter meiosis with minor abnormalities. Similar to the defects observed in the mitotic region of the germ line in old (30 hours post L4 or older) spd-3(me85) mutants, embryos produced by spd-3(me85) homozygous mothers of any age displayed severe mitotic defects (Figure S1). This observation is in agreement with the previous finding that SPD-3 is required during the first mitotic division in the embryo [41]. These results show that in contrast to spd-3 deletion mutants, spd-3(me85) mutants have a late onset of mitotic defects in their germ lines. Since a defect in chiasma formation is already evident in young spd-3(me85) mutants, germ lines from these worms can be used to elucidate the meiosis-specific roles of SPD-3.
Fig. 2. SC assembly and recombination in spd-3(me85) mutants. 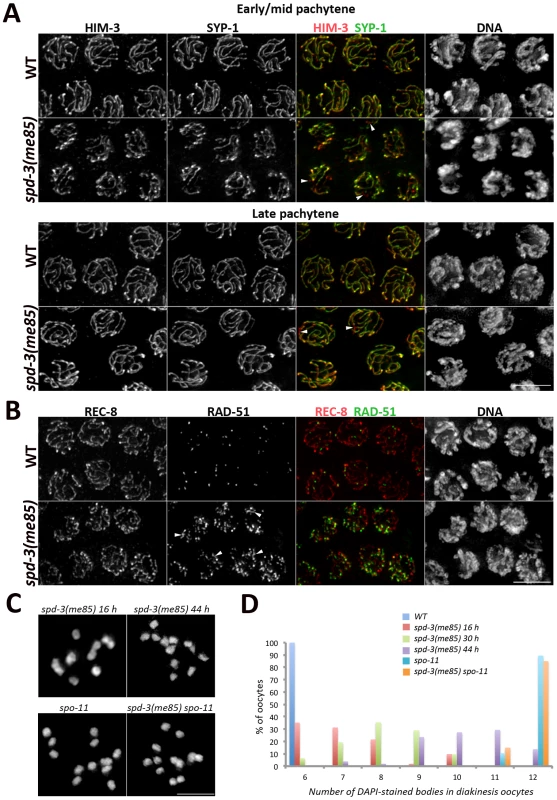
(A) Projections of pachytene nuclei stained with anti-HIM-3 and anti-SYP-1 antibodies and counterstained with DAPI. Arrowheads point to unsynapsed regions. (B) Projections of pachytene nuclei stained with anti-REC-8 and anti-RAD-51 antibodies and counterstained with DAPI. Arrowheads point to long stretches of RAD-51 signals present in the spd-3(me85) mutant. (C and D) Projections of individual diakinesis oocytes of the indicated genotype stained with DAPI (C) and quantification of the number of DAPI-stained bodies present in diakinesis oocytes of the indicated genotypes and ages (D). 6 DAPI-stained bodies corresponds to 6 bivalents (wild-type oocytes), while 12 corresponds to 12 univalents (no crossovers) and 7 to 11 indicate a mixture of bivalents and univalents. The number of diakinesis oocytes analyzed per genotype were: spd-3(me85) 16 hours (52 oocytes), spd-3(me85) 30 hours (31 oocytes), spd-3(me85) 44 hours (51 oocytes), wild-type control 16 hours (52 oocytes), spo-11 16 hours (30 oocytes), spd-3(me85) spo-11 16 hours (40 oocytes). A two-tailed Mann-Whitney test shows that the difference between any two of the analyzed genotypes is highly significant (p<0.001), apart from the comparison between spo-11 and spd-3(me85); spo-11 which is not different (p = 0.7). Scale bar = 5 µm in all panels. spd-3(me85) mutants are competent for SC assembly and crossover formation but defective in pairing
We set out to identify the primary defect that led to the chiasma deficit seen in spd-3(me85) mutant oocytes by investigating SC assembly, meiotic recombination and homolog pairing in germ lines from young spd-3(me85) mutants. We noticed that nuclei with variable degrees of chromosome clustering (polarized distribution of chromatin inside the nucleus as seen by DAPI staining) persisted well into the pachytene region in spd-3(me85) mutants (Figure S2). In wild-type germ lines chromosome clustering is only observed in transition zone nuclei (leptotene-zygotene), while in mutants that fail to assemble the SC chromosome clustering persists until late pachytene. Thus, we analyzed SC assembly in spd-3(me85) mutants using antibodies against HIM-3 (an axial element component) and SYP-1, a component of the central region of the SC [6], [42]. Loading of HIM-3 and SYP-1 started as nuclei entered meiosis and by early pachytene all nuclei displayed robust SYP-1 tracks covering most chromosomes, although some short chromosomal tracks remained unsynapsed (Figure 2A). By the mid to late pachytene transition, chromosome clustering was released and the overall level of synapsis was very similar to that seen in wild-type germ lines (Figure 2A). These results show that spd-3(me85) mutants are proficient in SC assembly.
We next monitored the dynamics of meiotic recombination intermediates by following the RAD-51 recombinase, which binds onto single stranded DNA following the formation of double strand breaks (DSBs). spd-3(me85) mutant germ lines displayed an extensive accumulation of RAD-51 foci throughout meiotic prophase (Figure 2B and Figure S2). Although this accumulation of recombination intermediates suggests that meiotic DSB repair is compromised in spd-3(me85) mutants, the analysis of diakinesis oocytes showed that spd-3(me85) mutants are competent in chiasma formation. Diakinesis oocytes from young spd-3(me85) mutants displayed on average 7.2 DAPI-stained bodies (corresponding to an average of 4.8 bivalents per oocyte), a significant decrease (p<2E−6 by the two-tailed Mann-Whitney test) compared with the 11.9 DAPI-stained bodies (0.1 bivalents) observed in chiasma-deficient spo-11 mutants, and a significant increase (p<2E-6) compared with the 6 DAPI-stained bodies (6 bivalents) observed in wild-type oocytes (Figure 2C–2D, Figure S2). Older spd-3(me85) mutants displayed a significant increase in the number of DAPI-stained bodies per oocyte, with an average of 8.2 (3.8 bivalents) at 30 hours post L4 (p<0.0002, compared to spd-3(me85) mutants at 16 hours post L4) and an average of 10.2 (1.8 bivalents) at 44 hours post L4 (p<2E-6) (Figure 2C–2D). These observations show that spd-3(me85) mutants are competent in chiasma formation, but that the number of chiasmata that they form decreases with age. Importantly, chiasmata were not observed in spd-3(me85); spo-11 double mutants, in which the number of DAPI-stained bodies (average = 11.8) is not significantly different (p = 0.7) from that seen in spo-11 controls (Figure 2C–2D). This analysis suggests that some DSBs are repaired as crossovers in spd-3(me85) mutants. In agreement with this interpretation, COSA-1 foci, which mark crossover sites in C. elegans [43], were observed in late pachytene nuclei of spd-3(me85) mutants (Figure S2). Therefore, the reduced number of chiasmata seen in spd-3(me85) mutant oocytes can not be explained solely by defects in the meiotic recombination machinery.
Since spd-3(me85) mutants are competent in SC assembly, and at least partly competent in crossover formation, we investigated if a defect in homolog pairing could account for the deficit in chiasmata observed in spd-3(me85) oocytes. We used FISH probes to monitor the pairing status of two autosomal regions (the PC region of chromosome III and an interstitial region on chromosome V carrying the 5S rDNA locus), as well as antibodies against HIM-8, a protein that binds specifically to the PC of the X chromosome [35]. All three loci displayed clearly reduced levels of pairing throughout meiotic prophase in spd-3(me85) mutant germ lines compared to wild-type controls (Figure 3B). These differences were significant (by a two-tailed Fisher's exact test) for the three loci in all the 5 zones in which germ lines were divided (Table S1), apart from the PC of chromosome III in zone 2, a region of the germ line that contains some pre-meiotic nuclei (Figure 3B). However, by zone 3 the PC of chromosome III was paired in 90% of wild-type nuclei, but only in 48% of spd-3(me85) mutant nuclei (Figure 3B), and this difference is highly significant (p = 7E-18). The 5S rDNA locus displayed the strongest pairing defect of the three loci, never reaching above 30% of pairing throughout meiotic prophase in spd-3(me85) mutants. These results confirm that SPD-3 is required for proper pairing during meiotic prophase.
Fig. 3. spd-3(me85) mutants display a pairing defect and non-homologous synapsis. 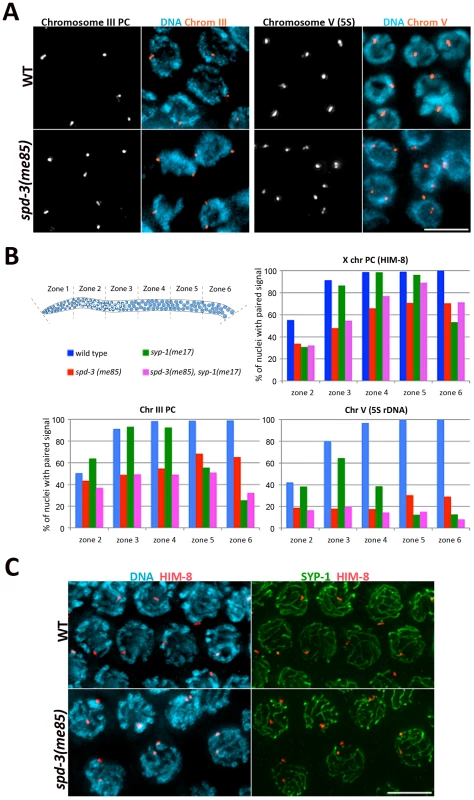
(A) Projections of pachytene nuclei following fluorescence in-situ hybridization (FISH) with probes against the PC region of chromosome III and the 5S rDNA locus on chromosome V and counterstained with DAPI. (B) Quantification of pairing in germ lines of the indicated phenotypes. The Y axis indicates the percentage of nuclei with paired signals (1 focus per nucleus) and the X axis indicates the region along the germ line: zone 2 premeiotic nuclei and start of transition zone, zone 3 transition zone and early pachytene, zones 4–6 early to late pachytene. Note the reduction of pairing of all three loci in spd-3(me85) mutants compared to wild-type controls, and that overall pairing is not improved in spd-3(me85); syp-1 double mutants compared to spd-3(me85) single mutants. Table S1 offers the statistical analysis of the differences in pairing levels between the different genotypes that were analyzed. (C) Projections of pachytene nuclei stained with anti-SYP-1 and anti-HIM-8 antibodies and counterstained with DAPI. spd-3(me85) mutants show nuclei with two HIM-8 signals, in which each HIM-8 spot is associated with a different track of SYP-1. Scale bar = 5 µm in all panels. Non-homologous synapsis is established in spd-3(me85) mutants
spd-3(me85) mutants show a pairing failure that is accompanied by nearly wild-type levels of SC assembly, suggesting that at least some stretches of SC must be assembled between non-homologous chromosomes. We investigated the fidelity of SC loading at the X chromosome PC by first identifying pachytene nuclei displaying two HIM-8 signals, which indicates a failure in X-chromosome pairing, and then determining the association between each HIM-8 signal and SC tracks labeled by SYP-1 (Figure 3C). Out of 40 nuclei with unpaired HIM-8 signals, 37 showed both HIM-8 signals associated with different SYP-1 stretches, and 3 nuclei showed only 1 of the two HIM-8 signals associated with a SYP-1 stretch. Out of 70 wild-type control nuclei, 69 showed paired HIM-8 signals associated with a single track of SC, while 1 nucleus showed paired HIM-8 signals that were not associated with the SC. Thus, X chromosomes are frequently involved in non-homologous synapsis, demonstrating that promiscuous SC loading occurs in spd-3(me85) mutants.
SPD-3 is required for the initial steps of homolog pairing
SC loading between non-homologous chromosomes demonstrates that pairing and SC assembly are not properly coordinated in spd-3(me85) mutants, raising the possibility that premature loading of the SC was interfering with homology search in spd-3(me85) mutants. If this was the case, inhibiting SC assembly in the germ lines of spd-3(me85) mutants should lead to improved pairing during early meiotic prophase, since pairing at this stage is independent of SC central region components [6]. Therefore, we quantified pairing levels of three loci (5S rDNA, and the pairing centers of chromosomes X and III) in spd-3(me85); syp-1 double mutants. In zones 2 and 3, corresponding to transition zone and early pachytene, the levels of pairing observed in spd-3(me85); syp-1 double mutants were not statistically different from the levels of pairing seen in spd-3(me85) mutants for any of the three loci (Figure 3B and Table S1). Furthermore, pairing in spd-3(me85); syp-1 mutants never arose above the levels seen in spd-3(me85) mutants at any stage for the 5S and chromosome III PC probes, while a small (but significant) increase in pairing was seen for the X chromosome PC only in zones 4 and 5. Therefore, preventing SC assembly does not alleviate the pairing defect present in spd-3(me85) mutants, suggesting that SPD-3 plays a role in the early steps of homolog pairing that precede SC assembly.
Pairing centers localize to SUN-1 aggregates on the nuclear envelope of spd-3(me85) mutants, but formation of large SUN-1 aggregates is impaired
Pairing at the onset of meiosis requires tethering of the chromosomal end carrying the PC to the inner NE, where PCs are seen localizing to aggregates formed by the SUN-1 protein [11], [39], [44]. To investigate if PC localization to SUN-1 aggregates on the NE was disrupted in spd-3(me85) mutants, we introduced a transgene expressing a functional SUN-1::GFP fusion protein [39] into the spd-3(me85) mutant background, and these germ lines were stained with anti-GFP (to visualize SUN-1) and anti-HIM-8 or anti-ZIM-3 antibodies to visualize PC regions. Transition zone nuclei from wild-type germ lines typically displayed between 2 and 4 large SUN-1 aggregates that colocalized with the PC-binding proteins. In spd-3(me85) mutants, both HIM-8 and ZIM-3 were always found associated with SUN-1 aggregates, showing that localization of PC regions to SUN-1 aggregates is not disrupted and suggesting normal tethering of PCs to the NE (Figure 4A and Videos S1, S2). However, transition zone nuclei of spd-3(me85) mutants displayed a clear increase in the overall number of SUN-1 aggregates and most aggregates were much smaller in size than those seen in wild-type controls. Small and round SUN-1 aggregates (foci) have been proposed to represent the attachment to the NE of a single chromosomal end, while larger SUN-1 aggregates (patches) are thought to include several chromosomal ends [39]. Quantification of SUN-1 aggregates during meiotic prophase revealed a strong increase of SUN-1 foci in spd-3(me85) mutant germ lines, where in zone three 50% of the nuclei displayed between 6 and 10 SUN-1 foci, while no nuclei with 6 or more foci were found in wild-type controls (Figure 4C). The extensive presence of SUN-1 foci in spd-3(me85) mutants suggests that many PC regions are attached to the NE in isolation, failing to be included in larger aggregates where PC regions from several chromosomes come together. Furthermore, SUN-1 aggregates persisted in substantial numbers until zone 5 of spd-3(me85) mutant germ lines, demonstrating a delay in the dissolution of SUN-1 aggregates.
Fig. 4. Localization of PC proteins to the NE in spd-3(me85) mutants. 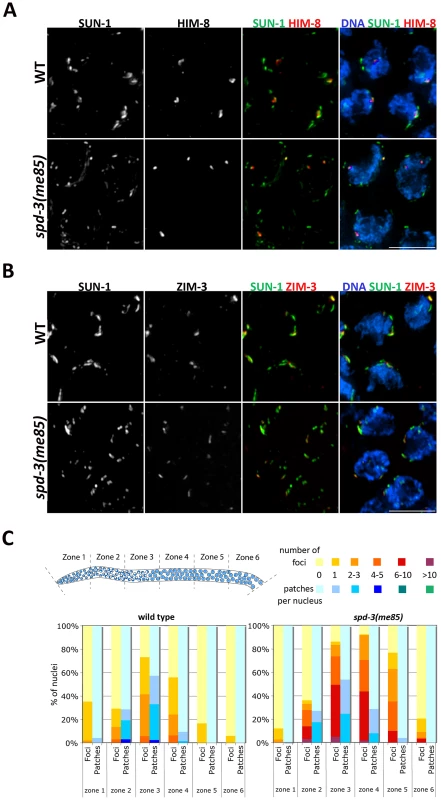
(A–B) Projections of transition zone nuclei from wild type and spd-3(me85) mutant worms expressing SUN-1::GFP and stained with anti-HIM-8 and anti-ZIM-3 antibodies and counterstained with DAPI. HIM-8 and ZIM-3 signals are associated with SUN-1 aggregates in both wild type and spd-3(me85) mutant, which display elevated numbers of SUN-1 aggregates. Videos S1 and S2 show three-dimensional reconstructions of nuclei from panel A. Scale bar = 5 µm in both panels. (C) Quantification of SUN-1 aggregates. Y axis indicates the percentage of nuclei with a given number of SUN-1 foci (aggregates smaller that 1.1 µm in diameter) or patches (aggregates larger than 1.1 µm in diameter), the X axis indicates regions along the germ line as indicated in the diagram. Note the big increase in SUN-1 foci in spd-3(me85) mutant and their persistence until mid/late pachytene. Molecular markers of the transition zone persist into pachytene in spd-3(me85) mutants, in a HTP-1-dependent manner
A key function of PCs is to recruit PLK-2 (Polo-Like Kinase 2) to the NE in transition zone nuclei, where PLK-2 induces dramatic changes in the organization of NE components, including the formation of large SUN-1 aggregates [37], [38]. These changes in NE organization coincide with the phosphorylation of SUN-1 at serine 8 (S8-Pi) and serine 12 (S12-Pi), both of which depend on CHK-2 [39], a kinase required for pairing [33], while S12-Pi also requires PLK-2 [37], [38]. Thus, we investigated if the failure to form large SUN-1 aggregates in spd-3(me85) mutants was due to defects in the recruitment of PLK-2 or SUN-1 phosphorylation. PLK-2 and SUN-1 phosphorylation appeared in a timely fashion on the NE of transition zone nuclei in spd-3(me85) mutants, where they formed numerous, and mostly small, aggregates that were similar in appearance to those observed when visualizing SUN-1::GFP (Figure 5). These results show that molecular markers of transition zone are normally recruited in spd-3(me85) mutants, suggesting normal CHK-2 activity. Indeed, the levels of SC assembly observed in spd-3(me85) mutants are largely dependent on CHK-2, since spd-3(me85); chk-2 double mutants displayed low levels of synapsis, as seen in chk-2 single mutants (Figure S3).
Fig. 5. Markers of the transition zone persist in spd-3(me85) mutants. 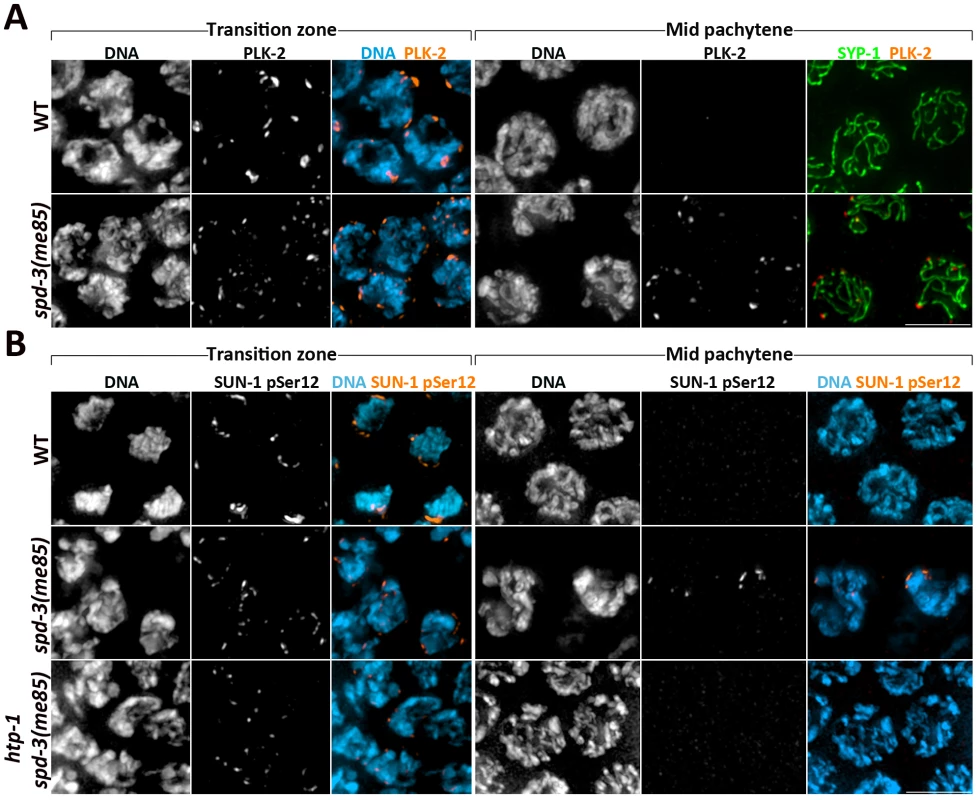
(A) Projections of transition zone and pachytene nuclei from wild type and spd-3(me85) mutants stained with anti-PLK-2 and anti-SYP-1 antibodies, and counterstained with DAPI. Note that both PLK-2 signals and chromosome clustering (seen by DAPI staining) persist until mid pachytene in spd-3(me85) mutants. The WT mid-pachytene nuclei correspond to the region of the germ line in which PLK-2 is no longer seen forming aggregates on the NE, and in which PLK-2 had not started to accumulate on the SC, which occurred in late pachytene nuclei. (B) Projections of transition zone and pachytene nuclei from the indicated genotypes stained with antibodies specific for SUN-1 phosphorylation at Serine 12 and counterstained with DAPI. Note that both SUN-1 pSer12 staining and chromosome clustering persist in mid-pachytene nuclei of spd-3(me85) mutants, and that this persistence is HTP-1 dependent. Scale bar = 5 µm in all panels. Interestingly, all three markers (PLK-2, S8-Pi and S12-Pi) persisted on the NE until the mid pachytene region of the germ line, suggesting that exit from early meiotic prophase is delayed in spd-3(me85) mutants (Figure 5 and Figure S3). The axial element component HTP-1 coordinates pairing and SC assembly and has been proposed to prevent exit from transition zone until homolog pairs are stabilized by the SC [10]; therefore, we tested if persistence of transition zone markers in spd-3(me85) mutants required HTP-1. spd-3(me85); htp-1 double mutant germ lines displayed few nuclei labeled by SUN-1 S12-Pi as well as very few nuclei with chromosome clustering (Figure 5B). This shows that exit from transition zone is actively delayed in spd-3(me85) mutants, in a HTP-1-dependent fashion. Overall, these observations demonstrate that known molecular regulators of pairing and synapsis, including CHK-2, PLK-2 and HTP-1, are active in spd-3(me85) mutants.
Movement of SUN-1 aggregates is impaired in spd-3(me85) mutants
The formation of large SUN-1 aggregates requires chromosome-end movement on the NE at the start of meiotic prophase [39]. We investigated the dynamics of SUN-1 aggregates in transition zone nuclei by live imaging of strains expressing a sun-1::GFP transgene. Filming was performed over a period of 15 minutes using the parameters described in [22]. SUN-1 aggregates were highly dynamic in wild-type controls, in which fusion and splitting events between SUN-1 aggregates were frequently observed (Video S3). In contrast, filming of SUN-1 aggregates in spd-3(me85) mutants revealed severely reduced movement, smaller SUN-1 aggregates and reduced instances of fusion events (Video S4). The reduced mobility of SUN-1 aggregates in spd-3(me85) mutants was reminiscent of the limited movement observed in strains expressing a sun-1::GFP transgene carrying the G311V mutation (sun-1(jf18) allele), which is known to impair the movement of SUN-1 aggregates [39] (Video S5).
Using the plotting tools developed in [22], we tracked the movement of individual SUN-1 aggregates and calculated the projected speed and the area explored by the aggregates in spd-3(me85), sun-1(jf18) and wild-type controls. Tracking in wild-type nuclei showed extensive overlap of individual SUN-1 tracks, while the displacement tracks in spd-3(me85) mutants showed little overlap, with most tracks covering a small area around a fixed position. Similar displacement tracks were observed in sun-1(jf18) mutants (Figure 6). Quantification of the area covered by SUN-1 aggregates in spd-3(me85) mutants showed an average arc of 39° (maximum 103°, minimum 11°), a substantial reduction compared to wild-type controls (average arc 90°, maximum 164°, minimum 21°). Interestingly, the area covered by SUN-1 aggregates in sun-1(jf18) mutants (average arc 26°, maximum 50°, minimum 11°) was reduced compared to spd-3(me85) mutants. Finally, the analysis of the distribution of projected speeds demonstrated a clear reduction in the speed of SUN-1 aggregates in spd-3(me85) mutants, with just 40% of aggregates moving at speeds above 40 nm/s, compared to 65% in wild-type controls (Figure 6). Furthermore, aggregates moving at high speed (160 nm/s and higher) represented 10% in wild-type nuclei, but only 0.82% in spd-3(me85) mutants, and 0.29% in sun-1(jf18) mutants. The decrease in high-speed moving aggregates suggests a defect in motor-driven motion, since dynein-dependent movement of SUN-1/ZYG-12 aggregates is characterized by an average speed of 190 nm/s [23]. This quantitative analysis demonstrates that movement of SUN-1 aggregates is reduced in spd-3(me85) mutants, although not as much as in sun-1(jf18) mutants, which are completely deficient in homolog pairing [44].
Fig. 6. spd-3(me85) mutants display reduced mobility of SUN-1 aggregates. 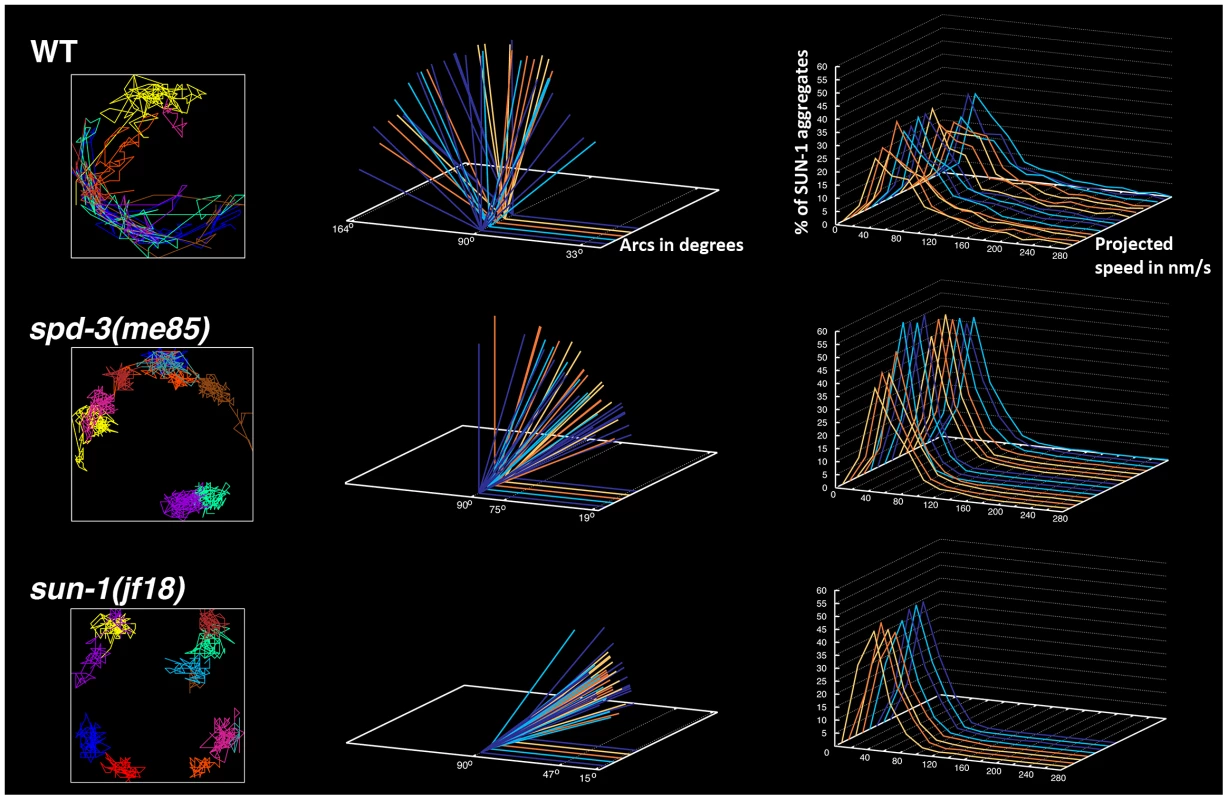
Left-hand side column: examples of the displacement tracks of all SUN-1::GFP aggregates within a nucleus over a period of 15 minutes. spd-3(me85) and sun-1(jf18) mutants show obvious reductions in both the area explored by SUN-1::GFP aggregates and in the overlap of different tracks. Middle column: each arc represents the distance traveled by a SUN-1::GFP aggregate inside a nucleus, with larger angles indicating larger distance traveled. Arcs corresponding to all SUN-1 aggregates from 5 nuclei are shown per genotype, arcs are severely reduced in spd-3(me85) mutants, although not as much as in sun-1(jf18) mutants. Right-hand column: Each line represents the distribution of the projected speeds of all SUN-1 aggregates inside a nucleus over a period of 15 minutes. spd-3(me85) and sun-1(jf18) mutants show a strong reduction in the percentage of SUN-1 aggregates moving at high speeds. Knockdown of cytoskeletal motors phenocopies the defect in the movement of SUN-1 aggregates seen in spd-3(me85) mutants
The experiments described above show that spd-3(me85) mutants are proficient in the molecular events that promote pairing, such as recruitment of PLK-2 to PCs, but deficient in forming dynamic SUN-1 aggregates. Movement of SUN-1 aggregates requires the activity of cytoskeletal motors that interact with ZYG-12, the KASH domain partner of SUN-1, on the outside of the NE [11], [23]. Thus, we investigated if knocking down dynein impaired the formation of functional SUN-1 aggregates in a similar fashion to that observed in spd-3(me85) mutants. Since dynein is also required for the mitotic divisions, we used a strain expressing DHC-1::GFP fusion to test RNAi conditions that would induce dynein knockdown in transition zone nuclei, while avoiding the presence of aneuploid nuclei in the meiotic region of the germ line. After 48 hours of RNAi treatment at 20°C, the DHC-1::GFP signal was not present in transition zone nuclei, confirming dynein knockdown (Figure S4). Next, we performed dhc-1 RNAi on a strain expressing a SUN-1::GFP fusion protein, and confirmed that dynein knockdown impaired the formation of large SUN-1 aggregates (Figure 7A). Similar results were observed following RNAi of dynein light chain or dynactin (Figure 7A). Quantification of SUN-1 movement following dhc-1 RNAi showed defects highly reminiscent to those seen in spd-3(me85) mutants, with SUN-1 tracks covering small areas and moving at slower speeds than controls (Figure 7B and Videos S6, S7).
Fig. 7. Depletion of cytoskeletal motors impairs SUN-1::GFP mobility. 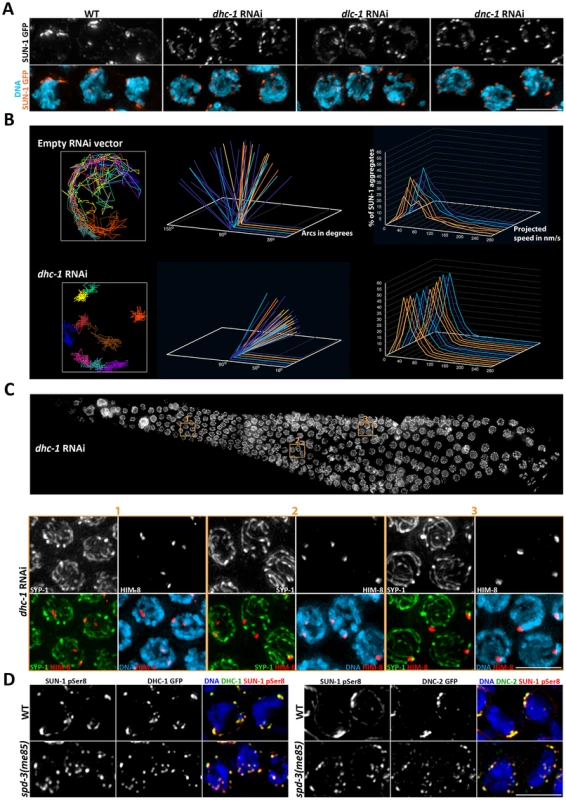
(A) Projections of transition zone nuclei from worms expressing SUN-1::GFP and subjected to RNAi of the indicated genes encoding components of the dynein motor complex, and showing the formation of numerous small SUN-1 aggregates that contrast to the larger -and fewer- aggregates seen in wild-type nuclei. (B) Plotting of SUN-1 movement following dynein knockdown. dhc-1 RNAi results in a reduction of the distance covered and speeds of SUN-1 aggregates. See legend of Figure 6 for a description of the graphs. (C) Top panel: whole germ line following dhc-1 RNAi stained with DAPI. Bottom panel: projection of the nuclei included in the three regions outlined on the whole-germ line panel. Nuclei are stained with anti-SYP-1 and anti-HIM-8 antibodies and counterstained with DAPI. Some nuclei from zones 1 and 2 show separated HIM-8 signals associated with different tracks of SYP-1. (D) Projection of transition zone nuclei from wild type and spd-3(me85) mutant worms expressing DHC-1::GFP or DNC-2::GFP and stained with anti-GFP antibodies and antibodies specific for serine 8 phosphorylation on SUN-1, and counterstained with DAPI. DHC-1 and DNC-2 localize to SUN-1 aggregates marked by SUN-1 S8Pi in the NE of both wild type and spd-3(me85) mutant nuclei. Scale bar = 5 µm in all panels. Given that dhc-1 RNAi elicited defects in SUN-1 clustering and movement similar to those seen in spd-3(me85) mutants, we investigated if pairing and SC assembly were also affected by dynein knockdown by staining dhc-1 RNAi germ lines with anti-SYP-1 and anti-HIM-8 antibodies. The presence of SYP-1 throughout the germ line demonstrated proficient SC assembly, while some nuclei in transition zone and early pachytene showed two separated HIM-8 signals, demonstrating a pairing defect (Figure 7C). Nuclei from mid/late pachytene, which presumably had undergone pairing before dynein knockdown, displayed paired HIM-8 signals. Using more severe dhc-1 RNAi conditions (55 hours at 25°C) we observed defects in SC assembly, as previously reported [11]. However, under the dynein RNAi conditions used here, defects in SUN-1 clustering and homolog pairing became visible before SC assembly was impaired. We also noted the presence of enlarged nuclei in the premeiotic region of dhc-1 RNAi germ lines, which is similar to the mitotic defects seen in older spd-3(me85) mutant germ lines. These observations suggest that dynein function may be affected in spd-3(me85) mutants.
The similarities between the defects seen in spd-3(me85) mutants and worms subjected to dhc-1 RNAi led us to ask if recruitment of cytoskeletal motors to the outer NE was impaired in spd-3(me85) mutant germ lines. Both dynein and dynactin were recruited to the NE in transition zone nuclei of spd-3(me85) mutants, where they fully colocalized with SUN-1 aggregates marked by SUN-1 S8-Pi antibodies (Figure 7D). We also observed that ZYG-12, the KASH domain partner of SUN-1 that is required to link chromosomes to the cytoskeleton in transition zone nuclei [11], was recruited to SUN-1 aggregates (Figure S4). Thus, spd-3(me85) mutants are proficient in recruiting cytoskeletal motors to the NE of transition zone nuclei.
Inhibition of mitochondrial respiration impairs movement of SUN-1 aggregates
As dynein is an ATP-driven molecular motor and we have shown that SPD-3 localizes to mitochondria, we investigated whether decreasing ATP levels by interfering with mitochondrial function would also impair the movement of SUN-1 aggregates. We filmed the movement of SUN-1 aggregates following exposure to different concentrations (and times) of sodium azide, which causes a reversible inhibition of the cytochrome C oxidase that induces ATP depletion [45]. We started by testing azide doses lower than 15 mM, which were previously shown to be sublethal in C. elegans [46]. Remarkably, worms exposed to 5 mM sodium azide for 10 minutes displayed severely reduced SUN-1 aggregate movement in transition zone nuclei, as well as reduced intensity of the GFP signal per aggregate (Videos S8, S9). We next tried lower concentrations of azide with longer exposure times (0.1 mM for 5 hours) and also observed clearly impaired SUN-1 movement, but stronger GFP signal (Video S10). Importantly, 100% of the worms recovered from exposure to these azide treatments. Furthermore, following removal from the azide solution, SUN-1 aggregates regained movement. Thus, movement of SUN-1 aggregates is highly sensitive to mitochondrial disruption.
Given the above observations, we investigated whether mitochondrial function was impaired in spd-3(me85) mutants by direct measurement of oxygen consumption using a Seahorse metabolic analyzer [47, see Material and Methods and legend of Figure S4]. The basal rate of oxygen consumption was significantly lower in spd-3(me85) mutants than in wild-type controls (Figure S4). Together, these observations suggest that deficient mitochondrial function may be responsible for the defect in SUN-1 aggregate movement observed in spd-3(me85) mutants.
Discussion
We have shown that defects in the mitochondria-localizing protein SPD-3 cause a severe reduction in the mobility of SUN-1 aggregates at the start of meiotic prophase. Since all analyzed PC-binding proteins localize to SUN-1 aggregates in spd-3(me85) mutants, we infer that movement of PCs tethered to the NE is also reduced in spd-3(me85) mutants. This reduced motility is accompanied by a failure in homolog pairing and extensive SC assembly between non-homologous chromosomes, supporting a requirement for chromosome-end movement in homology search. All tested components of meiotic chromosomes and of the machinery required to move meiotic chromosomes, including the SUN/KASH pair, PLK-2, and cytoskeletal motors, are normally recruited at the start of meiotic prophase in spd-3(me85) mutants. But despite normal recruitment of pairing-promoting proteins, movement of SUN-1 aggregates is nevertheless greatly impaired in spd-3(me85) mutants. We propose that this defect is due to the reduced function of cytoskeletal motors in spd-3(me85) mutants, which is likely caused by a defect in mitochondrial function. Several observations support this possibility. First, we have shown that dynein depletion mimics the SUN-1 aggregate movement defects seen in spd-3(me85) mutants. Second, the mitotic divisions in spd-3(me85) mutant embryos and in the premeiotic region of older spd-3(me85) mutant germ lines show defects that also mimic those seen when dynein function is compromised, such as centrosome detachment from the nucleus (Figure S1) and formation of large aneuploid nuclei. Third, dynein is a motor that uses ATP as a source of energy to generate force, and we have shown that impairing mitochondrial function leads to a rapid inhibition of SUN-1 aggregate movement in transition zone nuclei. Although we have not evaluated the possible contribution of other ATP-consuming motors to the mobility of SUN-1 aggregates, if motors such as kinesins are also involved in this process, their function is likely to be affected in spd-3(me85) mutants. In addition, SPD-3 was not identified as an interactor of dynein or dynactin in immunoprecipitation experiments of these motors (data not shown), suggesting that SPD-3 could affect these cytoskeletal motors by means that do not involve a direct physical interaction. Thus, the spd-3(me85) mutant offers a unique opportunity to investigate the impact of reduced SUN-1/PC mobility on meiotic prophase events without detectably impairing the integrity of any of the proteins known to promote pairing directly by localizing to the NE or chromosomes.
Although chromosome movements induced by cytoskeletal forces appear to be a conserved feature of meiotic prophase, there is little agreement about the contribution of this chromosome end-led motion to the pairing process [31]. Particularly unclear is the extent to which cytoskeletal forces contribute to homology search at the start of meiotic prophase. For example, S. cerevisiae mutants deficient in telomere-led movement show delayed -but eventually highly successful - pairing, which may be promoted by chromosome movements induced by processes such as thermal motion, chromatin remodeling, or DNA and RNA metabolism [27]. On the other hand, homolog associations during meiotic prophase of S. pombe, which lack a conventional SC, are largely dependent on dynein-driven movement [29]. A recent report shows that the earliest homolog associations detected in mouse meiosis, which precede initiation of meiotic recombination, are dependent on Sun1 [48], suggesting that telemore-led movement may be required for early homology search in mammals. In C. elegans, movement of PCs attached to SUN-1 aggregates has been proposed to induce chromosome encounters [22], but the onset of meiotic prophase is also marked by a CHK-2-dependent increase in the diffusion of PCs on the NE, and this dynein-independent movement has been proposed to be an important contributor to homolog pairing [23]. Our analysis of spd-3(me85) mutants provides substantial evidence that motor-driven chromosome motion is required to ensure homolog encounters at the start of meiotic prophase. We have shown that although CHK-2 is active in spd-3(me85) mutants, suggesting that increased PC diffusion on the NE upon meiotic entrance occurs normally, spd-3(me85) mutants show a severe pairing defect. Furthermore, our quantitative analysis shows that the area explored on the NE by SUN-1 aggregates is severely reduced in spd-3(me85) mutants compared to wild-type controls, but that this reduction is not as severe as that seen in sun-1(jf18) mutants, which show a stronger pairing defect [11], [44]. An important difference between these two SUN-1 aggregate movement-deficient mutants is that while sun-1(jf18) mutants fail to recruit dynein to the outer NE [11], dynein is normally recruited to the NE in spd-3(me85) mutants. Thus, residual dynein-driven chromosome movement may be sufficient to induce a significant increase in the pairing levels of spd-3(me85) mutants compared to sun-1(jf18) mutants, although not enough to achieve wild-type levels of pairing. Our analysis suggests a correlation between the extent of motor-driven chromosome movement and the success of homolog pairing.
Further support for cytoskeletal forces playing a key role in homology search comes from the analysis of pairing in the absence of SC, which prevents “trapping” of improper chromosomal interactions in mutants that display non-homologous synapsis, such as spd-3(me85) or sun-1(jf18). SC assembly has been shown to interfere with pairing of the X chromosome PC in sun-1(jf18) mutants [11], [44], however, preventing SC assembly in spd-3(me85) mutants only induces a limited improvement in the pairing levels of the X chromosome PC (seen only in mid-pachytene nuclei) and no improvement at any stage in the pairing levels of chromosome III PC or the 5S rDNA locus on chromosome V. Since sun-1(jf18) mutants display increased diffusion of PCs upon meiosis entrance [23], this diffusive movement may account for the increased pairing of the X chromosome PC observed when the SC is removed in sun-1(jf18) mutants. In this scenario, SC removal may allow the extended time required for the encounter of homologous PCs under reduced movement conditions. However, we have observed that although spd-3(me85) mutants display an extended transition zone, the overall impact of SC removal on pairing is minimal. This suggests that the amount of PC movement present in spd-3(me85) mutants, which is likely to include diffusive motion plus a residual component of dynein-driven movement, is below the threshold required to ensure full pairing of PC regions. Reduced and delayed pairing of PC regions has also been reported following dynein depletion, with chromosome V PC only reaching 60% of pairing by the mid pachytene region [11]. Furthermore, reduced movement of SUN-1 aggregates has a dramatic impact on the pairing levels of chromosomal regions located away from PC regions, and pairing in these regions does not improve following SC removal [This work, 44]. Since full homolog alignment can sometimes be achieved in the absence of SC [49], and non-PC regions have been proposed to display an SC-independent pairing capability [50], our observations suggest that reduced movement of PC regions on the NE has a negative impact on the pairing ability of all chromosomal regions, not just PCs. During wild-type meiosis pairing is rapidly achieved at the start of meiotic prophase, and we propose that this is an important feature of meiosis and that motor-driven movement is essential to achieve timely and robust homolog pairing during early prophase, which is a prerequisite to complete later meiotic events and ensure successful chiasma formation.
Movement-deficient mutants such as sun-1(jf18), chk-2, and worms lacking all PC-binding proteins or PLK activity show defects in pairing and an absence of chromosome clustering in transition zone nuclei, suggesting that these two events are mechanistically linked and require PC-led chromosome movement [33], [37], [38], [44]. However, the reduced movement of PCs attached to SUN-1 aggregates in spd-3(me85) mutants causes a severe decrease in pairing levels without preventing chromosome clustering. Furthermore, the molecular markers associated with chromosome clustering (PLK-2 and SUN-1 phosphorylation) persist in spd-3(me85) mutants, demonstrating an extension of transition zone and delayed meiotic progression. Exit from transition zone marks the end of the homology search-competent phase of meiotic prophase, and is a regulated process under the control of the HORMA-domain protein HTP-1, which prevents release of chromosome clustering until homolog interactions are stabilized by loading of the SC [9], [10], [51]. Since the accumulation of transition zone nuclei in spd-3(me85) mutants is HTP-1 dependent, this suggests that the mechanism that prevents dispersal of chromosome clustering until homologous pairs are linked by the SC remains functional under reduced movement conditions. HTP-1 is also part of a checkpoint-like coupling mechanism that prevents SC assembly when pairing fails [10]–[12], however, this regulatory function of HTP-1 appears to be impaired by reduced movement, since extensive non-homologous synapsis occurs in spd-3(me85) mutants. The failure of this coupling in spd-3(me85) mutants may appear surprising given that HTP-1 has been proposed to prevent SC assembly when dynein is depleted [11]. However, dynein localizes to the NE in spd-3(me85) mutants, and our quantitative analysis shows that a residual component of motor-driven motion likely persists in spd-3(me85) mutants. Under these conditions, coupling of SC assembly to successful pairing fails and SC assembly ensues despite pairing failure. Our observations suggest that the early meiotic events that lead to proper pairing show a different degree of dependency on chromosome mobility: timely homolog encounters and coupling of SC assembly to homology verification are highly susceptible to reduced chromosome mobility, while acquisition of chromosome clustering and HTP-1-dependent persistence of the molecular markers of the transition zone can occur under reduced movement conditions.
The formation of stable inter-homolog interactions is the hallmark of meiosis, and we favor a model in which the homolog encounters required to begin the pairing process are actively promoted by the chromosomal movements that occur at the start of meiotic prophase. Increased chromosome mobility is also observed following DNA damage in mitotic yeast cells [52]. This damage-induced movement facilitates pairing of homologous loci, allowing proper repair of the broken chromosome with its homologous partner. Thus, increased chromosome movement may be the primary response to start the search for a homologous chromosome, and this response may be conserved beyond meiosis.
Materials and Methods
Genetics
All C. elegans strains were cultured using standard conditions and, unless otherwise noted, grown at 20°C. The wild-type Bristol N2 strain was used as a control. The genetic screen and mapping of the spd-3(me85) mutation are described in Text S1. Two deletion alleles of spd-3 were generated by the National Bioresource Project (tm2969) and by the C. elegans knockout consortium (ok1817). The following mutations were used in this study: spd-3(me85), spd-3(tm2969), spd-3(ok1817), spo-11(me44), syp-1(me17), htp-1(gk174), sun-1(jf18), sun-1(ok1282), chk-2(me64). The following transgenes were used: JfSi1 [Psun-1::GFP, cb unc-119(+)], ojIs31[pie-1::spd-3::GFP, unc-119(+)], ojIs57 [pie-1::dnc-2::gfp unc-119(+)], orIs17 [Pdhc-1::GFP::dhc-1 unc-119(+)], ojls9 [zyg-12ABC::GFP unc-119(ed3)], ojIs5 [pie-1::dnc-1::GFP, unc-119(+)], meIs8[unc-119(+) pie-1::gfp::cosa-1] II.
Plotting of SUN-1 aggregate movement
Plotting was performed following the method described in [22]. Briefly, 16 hours post L4 hermaphrodites were placed in a drop of M9 containing 10 mM levamisol on a 2% agarose pad, a coverslip was placed on top and sealed with melted vaseline. Data collection was performed using a Delta Vision Deconvolution system (Applied Precision) equipped with an Olympus 1×70 microscope and a CoolSNAPHQ2 Monochrome camera. Images from transition zone nuclei were acquired in a series of 1 µm-spaced Z-stacks, with a time lapse of 5 sec over 15 min (181 frames), and with the following parameters: 30% light intensity, 200 msec exposure, 60× objective and image size of 512×512 pixels.
For plotting, maximum intensity projections of the Z-stacks for each time point were created using softWoRx to create movies of SUN-1 movement. Autoquant X2 was used to reduce the background of the movies, and then Metamorph Offline was used to align the projections and track the SUN-1 aggregates. The position of SUN-1 aggregates was plotted using Gnuplot and XTerminal. For a detailed description of the plotting tools see [22]. The total number of nuclei and SUN-1::GFP aggregates that were plotted per genotype was: for spd-3(me85) mutants 20 nuclei containing 215 tracks, for sun-1(jf18) 9 nuclei containing 108 tracks, and for wild-type controls 15 nuclei containing 119 tracks.
Western blots
Fifty worms of the desired genotype were picked into 30 µl of 1× TE containing a protease inhibitor cocktail (Roche). The worm suspension was frozen in liquid N2 and then thawed before adding 5 µl of 6× loading buffer and boiling the samples for 10 min. Samples were run in 6% acrylamide gels and blotted at 4°C, then the membranes were probed with rabbit α-SPD-3 antibodies (1∶2000), followed by goat α-rabbit HRP-conjugated (Upstate). α-SPD-3 antibodies were generated by SDIX using Genomic Antibody Technology against a 100 amino acid peptide that included residues 373–472 of the SPD-3 protein.
Mitochondria were purified using a Q proteome Mitochondria isolation kit (Qiagen, 37612) with modifications (see supplemental information). High purity mitochondrial extracts were mixed with loading buffer, boiled for 10 minutes, run in acrylamide gels and blotted as described above. The following primary antibodies and dilutions were used: mouse α-GFP (Roche, 11814460001) 1∶500, mouse α-GAPDH (Applied Biosciences, AM4300) 1∶2000, mouse α-NDUFS3 (Abcam, ab 14711) 1∶2000, and rabbit α-SPD-3 (1∶2000).
Immunofluorescence
Immunostaining was performed as described in [10]. Briefly, 20 age-matched hermaphrodites were dissected in 15 µl of 1× egg buffer containing 0.1% TWEEN 20, fixed for 5 min in 1% paraformaldehyde and immersed in liquid N2 before the coverslip was removed using a razor blade. Slides were then placed in −20°C methanol for at least 2 min. Following a wash in 1× PBS (0.1% TWEEN 20), slides were blocked for 1 hour in 1× PBS containing 0.1% TWEEN 20 and 0.7% BSA. The following primary antibodies and dilutions were used: mouse α-REC-8 (Abcam 38372) 1∶50; rabbit α-HTP-1 (this study) 1∶200; rabbit α-HIM-3 [42] 1∶200; guinea pig α-SYP-1 [6] 1∶200; rabbit α-HIM-8 (Novus biologicals) 1∶500; rabbit α-RAD-51 [53] 1∶200 ; rabbit α-ZIM-3 (gift from Verena Jantsch) 1∶300; rabbit α-SUN-1 S8-Pi [39] 1∶600; rabbit α-SUN-1 S12-Pi [39] 1∶200; rabbit α-PLK-2 [54] 1∶500; mouse α-Cytochrome C (Abcam, ab110325) 1∶200; mouse α-GFP (Roche, 11814460001) 1∶300; rabbit α-GFP-Alexa fluor 488 conjugated (Invitrogen, A-21311) 1∶300.
FISH
20 hermaphrodite worms were dissected and processed as described in the immunostaining protocol, with the difference that they were fixed for 2 min in 7.4% paraformaldehyde. Once the cover slip was freeze cracked, the slides were placed in −20°C methanol and then allowed to warm up to room temperature in methanol. Slides were then washed in 50% methanol, 1× SSC containing 0.1% TWEEN-20 and then in 2× SSC (0.1% TWEEN-20), before germ lines were dehydrated by incubating the slides in 70%, 90% and 100% ethanol (3 minutes each). Slides were air dried before adding the hybridization mix containing labeled FISH probes in 2× SSC, 50% formamide and 10% dextran sulfate. Probe labeling, hybridization, and post-hybridization washes were performed as described in [10]. Probes were made against the 5S rDNA on chromosome V, and the cosmid T17A3, which labels the PC end of chromosome III.
Image acquisition and processing
All images from FISH and immunostaining experiments were acquired with a Delta Vision system using a 100× lens. Projections from three-dimensional Z-stacks were made using softWoRx and images were mounted in Photoshop. Rotation of three-dimensional reconstructions were made using Volocity.
RNA interference
All RNAi experiments were performed by feeding worms with HT115 bacteria transformed with a vector for IPTG-inducible expression of dsRNA. Bacteria containing the desired vector, as well as empty vector (L4440) controls, were grown overnight at 37°C in 20 ml of LB containing 50 µg/ml ampicillin. Cultures were then spun down and resuspended in 1 ml LB, before 100 µl of bacteria were seeded onto NGM agar plates containing 1 mM IPTG and 25 µg/ml ampicillin. Expression of dsRNA was induced by incubating the plates at 37°C overnight. L4 worms were added to the RNAi plates and incubated at 20°C unless otherwise noted. Worms were transferred to freshly seeded RNAi plates every 20 hours. RNAi clones are described in Text S1.
Immuno-electron microscopy
For immunolabelling experiments, isolated mitochondria were fixed with 0.1% glutaraldehyde, dehydrated through a graded ethanol series and embedded in LR White resin. Thin sections (70 nm) were labeled using either goat α-GFP (Rockland Immunochemicals) or mouse α-NDUFS3 diluted in blocking buffer containing 0.8% bovine serum albumin, 0.01% Tween20, and 0.1% fish scale gelatin (Nycomed, Amersham) in PBS. The secondary IgG antibodies against either GFP or NDUFS3 were coupled to 10 nm (goat anti-rabbit: British Biocell) or 18 nm (goat anti-mouse: Dianova) colloidal gold, respectively. The antibody complexes were stabilized with 1% glutaraldehyde in PBS and the labeled sections were poststained with 2% uranyl acetate followed by lead citrate and viewed in an FEI TECNAI 12 transmission electron microscope operated at 100 kV.
Azide treatment
Synchronized young adult worms expressing SUN-1::GFP were incubated in S medium containing OP-50 bacteria in 1.5 ml tubes. Sodium azide was added to a final concentration of 5 mM or 0.1 mM and the tubes were incubated with shaking at 20°C for the desired time. Following completion of the incubation time, worms were transferred to seeded NGM plates and immediately used for live imaging of SUN-1 aggregates as described above.
Measurement of oxygen consumption
Basal levels of oxygen consumption were measured based on the methods described in [47], and using a Seahorse XF24 analyzer. Worms were picked as L4 and allowed to grow for 36 hours on seeded NGM plates, at which point they were picked in groups of 50–60 worms to 2 ml tubes containing M9 buffer. Worms were then washed 3 times with M9 to eliminate residual bacteria and finally resuspended in 500 µl of M9. Each group of 50–60 worms was transferred to an individual well of a 24-well standard Seahorse plate (100777-004) and oxygen consumption was measured 7 times per well. In total 10 wells were used per genotype (500–600 worms). At the end of the measurements the exact number of worms present in each well was counted using a dissecting microscope and the respiration rates were normalized to the number of worms in each individual well.
Supporting Information
Zdroje
1. Martinez-PerezE, ColaiacovoMP (2009) Distribution of meiotic recombination events: talking to your neighbors. Curr Opin Gen Dev 19 : 105–112.
2. PetronczkiM, SiomosMF, NasmythK (2003) Un menage a quatre: the molecular biology of chromosome segregation in meiosis. Cell 112 : 423–440.
3. BhallaN, DernburgAF (2008) Prelude to a division. Annu Rev Cell Dev Biol 24 : 397–424.
4. SantosJL, JimenezMM, DiezM (1994) Meiosis in haploid rye: extensive synapsis and low chiasma frequency. Heredity 73 : 580–588.
5. LoidlJ, NairzK, KleinF (1991) Meiotic chromosome synapsis in a haploid yeast. Chromosoma 100 : 221–228.
6. MacQueenAJ, ColaiacovoMP, McDonaldK, VilleneuveAM (2002) Synapsis-dependent and -independent mechanisms stabilize homolog pairing during meiotic prophase in C. elegans. Genes Dev 16 : 2428–2442.
7. SymM, RoederGS (1994) Crossover interference is abolished in the absence of a synaptonemal complex protein. Cell 79 : 283–292.
8. de VriesFA, de BoerE, van den BoschM, BaarendsWM, OomsM, et al. (2005) Mouse Sycp1 functions in synaptonemal complex assembly, meiotic recombination, and XY body formation. Genes Dev 19 : 1376–1389.
9. ZhangW, MileyN, ZastrowMS, MacQueenAJ, SatoA, et al. (2012) HAL-2 promotes homologous pairing during Caenorhabditis elegans meiosis by antagonizing inhibitory effects of synaptonemal complex precursors. PLoS Genet 8: e1002880 doi:10.1371/journal.pgen.1002880.
10. Martinez-PerezE, VilleneuveAM (2005) HTP-1-dependent constraints coordinate homolog pairing and synapsis and promote chiasma formation during C. elegans meiosis. Genes Dev 19 : 2727–2743.
11. SatoA, IsaacB, PhillipsCM, RilloR, CarltonPM, et al. (2009) Cytoskeletal forces span the nuclear envelope to coordinate meiotic chromosome pairing and synapsis. Cell 139 : 907–919.
12. CouteauF, ZetkaM (2005) HTP-1 coordinates synaptonemal complex assembly with homolog alignment during meiosis in C. elegans. Genes Dev 19 : 2744–2756.
13. ScherthanH (2007) Telomere attachment and clustering during meiosis. Cell Mol Life Sci 64 : 117–124.
14. FridkinA, PenknerA, JantschV, GruenbaumY (2009) SUN-domain and KASH-domain proteins during development, meiosis and disease. Cell Mol Life Sci 66 : 1518–1533.
15. HiraokaY, DernburgAF (2009) The SUN rises on meiotic chromosome dynamics. Dev Cell 17 : 598–605.
16. ChikashigeY, HaraguchiT, HiraokaY (2007) Another way to move chromosomes. Chromosoma 116 : 497–505.
17. KoszulR, KimKP, PrentissM, KlecknerN, KameokaS (2008) Meiotic chromosomes move by linkage to dynamic actin cables with transduction of force through the nuclear envelope. Cell 133 : 1188–1201.
18. SheehanMJ, PawlowskiWP (2009) Live imaging of rapid chromosome movements in meiotic prophase I in maize. Proc Natl Acad Sci USA 106 : 20989–20994.
19. ChikashigeY, DingDQ, FunabikiH, HaraguchiT, MashikoS, et al. (1994) Telomere-led premeiotic chromosome movement in fission yeast. Science 264 : 270–273.
20. ScherthanH, WangH, AdelfalkC, WhiteEJ, CowanC, et al. (2007) Chromosome mobility during meiotic prophase in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 104 : 16934–16939.
21. ConradMN, LeeCY, ChaoG, ShinoharaM, KosakaH, et al. (2008) Rapid telomere movement in meiotic prophase is promoted by NDJ1, MPS3, and CSM4 and is modulated by recombination. Cell 133 : 1175–1187.
22. BaudrimontA, PenknerA, WoglarA, MachacekT, WegrostekC, et al. (2010) Leptotene/zygotene chromosome movement via the SUN/KASH protein bridge in Caenorhabditis elegans. PLoS Genet 6: e1001219 doi:10.1371/journal.pgen.1001219.
23. WynneDJ, RogO, CarltonPM, DernburgAF (2012) Dynein-dependent processive chromosome motions promote homologous pairing in C. elegans meiosis. J Cell Biol 196 : 47–64.
24. MorelliMA, WerlingU, EdelmannW, RobersonMS, CohenPE (2008) Analysis of meiotic prophase I in live mouse spermatocytes. Chromosome Res 16 : 743–760.
25. ParvinenM, SoderstromKO (1976) Chromosome rotation and formation of synapsis. Nature 260 : 534–535.
26. MorimotoA, ShibuyaH, ZhuX, KimJ, IshiguroK, et al. (2012) A conserved KASH domain protein associates with telomeres, SUN1, and dynactin during mammalian meiosis. J Cell Biol 198 : 165–172.
27. LeeCY, ConradMN, DresserME (2012) Meiotic chromosome pairing is promoted by telomere-led chromosome movements independent of bouquet formation. PLoS Genet 8: e1002730 doi:10.1371/journal.pgen.1002730.
28. Sonntag BrownM, ZandersS, AlaniE (2011) Sustained and rapid chromosome movements are critical for chromosome pairing and meiotic progression in budding yeast. Genetics 188 : 21–32.
29. DingDQ, YamamotoA, HaraguchiT, HiraokaY (2004) Dynamics of homologous chromosome pairing during meiotic prophase in fission yeast. Dev Cell 6 : 329–341.
30. WanatJJ, KimKP, KoszulR, ZandersS, WeinerB, et al. (2008) Csm4, in collaboration with Ndj1, mediates telomere-led chromosome dynamics and recombination during yeast meiosis. PLoS Genet 4: e1000188 doi:10.1371/journal.pgen.1000188.
31. KoszulR, KlecknerN (2009) Dynamic chromosome movements during meiosis: a way to eliminate unwanted connections? Trends Cell Biol 19 : 716–724.
32. KosakaH, ShinoharaM, ShinoharaA (2008) Csm4-dependent telomere movement on nuclear envelope promotes meiotic recombination. PLoS Genet 4: e1000196 doi:10.1371/journal.pgen.1000196.
33. MacQueenAJ, VilleneuveAM (2001) Nuclear reorganization and homologous chromosome pairing during meiotic prophase require C. elegans chk-2. Genes Dev 15 : 1674–1687.
34. MacQueenAJ, PhillipsCM, BhallaN, WeiserP, VilleneuveAM, et al. (2005) Chromosome sites play dual roles to establish homologous synapsis during meiosis in C. elegans. Cell 123 : 1037–1050.
35. PhillipsCM, WongC, BhallaN, CarltonPM, WeiserP, et al. (2005) HIM-8 binds to the X chromosome pairing center and mediates chromosome-specific meiotic synapsis. Cell 123 : 1051–1063.
36. PhillipsCM, DernburgAF (2006) A family of zinc-finger proteins is required for chromosome-specific pairing and synapsis during meiosis in C. elegans. Dev Cell 11 : 817–829.
37. HarperNC, RilloR, Jover-GilS, AssafZJ, BhallaN, et al. (2011) Pairing centers recruit a Polo-like kinase to orchestrate meiotic chromosome dynamics in C. elegans. Dev Cell 21 : 934–947.
38. LabellaS, WoglarA, JantschV, ZetkaM (2011) Polo kinases establish links between meiotic chromosomes and cytoskeletal forces essential for homolog pairing. Dev Cell 21 : 948–958.
39. PenknerAM, FridkinA, GloggnitzerJ, BaudrimontA, MachacekT, et al. (2009) Meiotic chromosome homology search involves modifications of the nuclear envelope protein Matefin/SUN-1. Cell 139 : 920–933.
40. MaydanJS, OkadaHM, FlibotteS, EdgleyML, MoermanDG (2009) De Novo identification of single nucleotide mutations in Caenorhabditis elegans using array comparative genomic hybridization. Genetics 181 : 1673–1677.
41. DinkelmannMV, ZhangH, SkopAR, WhiteJG (2007) SPD-3 is required for spindle alignment in Caenorhabditis elegans embryos and localizes to mitochondria. Genetics 177 : 1609–1620.
42. ZetkaMC, KawasakiI, StromeS, MullerF (1999) Synapsis and chiasma formation in Caenorhabditis elegans require HIM-3, a meiotic chromosome core component that functions in chromosome segregation. Genes Dev 13 : 2258–2270.
43. YokooR, ZawadzkiKA, NabeshimaK, DrakeM, ArurS, et al. (2012) COSA-1 reveals robust homeostasis and separable licensing and reinforcement steps governing meiotic crossovers. Cell 149 : 75–87.
44. PenknerA, TangL, NovatchkovaM, LadurnerM, FridkinA, et al. (2007) The nuclear envelope protein Matefin/SUN-1 is required for homologous pairing in C. elegans meiosis. Dev Cell 12 : 873–885.
45. WallaceKB, StarkovAA (2000) Mitochondrial targets of drug toxicity. Annu Rev Pharmacol 40 : 353–388.
46. LagidoC, PettittJ, FlettA, GloverLA (2008) Bridging the phenotypic gap: real-time assessment of mitochondrial function and metabolism of the nematode Caenorhabditis elegans. BMC Physiol 8 : 7 doi:10.1186/1472-6793-8-7.
47. YamamotoH, WilliamsEG, MouchiroudL, CantoC, FanW, et al. (2011) NCoR1 is a conserved physiological modulator of muscle mass and oxidative function. Cell 147 : 827–839.
48. BoatengKA, BellaniMA, GregorettiIV, PrattoF, Camerini-OteroRD (2013) Homologous pairing preceding SPO11-mediated double-strand breaks in mice. Dev Cell 24 : 196–205.
49. NabeshimaK, Mlynarczyk-EvansS, VilleneuveAM (2011) Chromosome painting reveals asynaptic full alignment of homologs and HIM-8-dependent remodeling of X chromosome territories during Caenorhabditis elegans meiosis. PLoS Genet 7: e1002231 doi:10.1371/journal.pgen.1002231.
50. DombeckiCR, ChiangAC, KangHJ, BilgirC, StefanskiNA, et al. (2011) The chromodomain protein MRG-1 facilitates SC-independent homologous pairing during meiosis in Caenorhabditis elegans. Dev Cell 21 : 1092–1103.
51. SmolikovS, EizingerA, Schild-PrufertK, HurlburtA, McDonaldK, et al. (2007) SYP-3 restricts synaptonemal complex assembly to bridge paired chromosome axes during meiosis in Caenorhabditis elegans. Genetics 176 : 2015–2025.
52. Mine-HattabJ, RothsteinR (2012) Increased chromosome mobility facilitates homology search during recombination. Nat Cell Biol 14 : 510–517.
53. ColaiacovoMP, MacQueenAJ, Martinez-PerezE, McDonaldK, AdamoA, et al. (2003) Synaptonemal complex assembly in C. elegans is dispensable for loading strand-exchange proteins but critical for proper completion of recombination. Dev Cell 5 : 463–474.
54. NishiY, RogersE, RobertsonSM, LinR (2008) Polo kinases regulate C. elegans embryonic polarity via binding to DYRK2-primed MEX-5 and MEX-6. Development 135 : 687–697.
55. HamillDR, SeversonAF, CarterJC, BowermanB (2002) Centrosome maturation and mitotic spindle assembly in C. elegans require SPD-5, a protein with multiple coiled-coil domains. Dev Cell 3 : 673–684.
Štítky
Genetika Reprodukční medicína
Článek Attachment Site Selection and Identity in Bxb1 Serine Integrase-Mediated Site-Specific RecombinationČlánek Bck2 Acts through the MADS Box Protein Mcm1 to Activate Cell-Cycle-Regulated Genes in Budding YeastČlánek High-Resolution Transcriptome Maps Reveal Strain-Specific Regulatory Features of Multiple IsolatesČlánek Neuropeptides Function in a Homeostatic Manner to Modulate Excitation-Inhibition Imbalance inČlánek Implicates Tyrosine-Sulfated Peptide Signaling in Susceptibility and Resistance to Root Infection
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 5- Růst a vývoj dětí narozených pomocí IVF
- Délka menstruačního cyklu jako marker ženské plodnosti
- Vztah užívání alkoholu a mužské fertility
- Intrauterinní inseminace a její úspěšnost
- Šanci na úspěšný průběh těhotenství snižují nevhodné hladiny progesteronu vznikající při umělém oplodnění
-
Všechny články tohoto čísla
- Functional Elements Are Embedded in Structurally Constrained Sequences
- RNA–Mediated Epigenetic Heredity Requires the Cytosine Methyltransferase Dnmt2
- Loss of Expression and Promoter Methylation of SLIT2 Are Associated with Sessile Serrated Adenoma Formation
- Attachment Site Selection and Identity in Bxb1 Serine Integrase-Mediated Site-Specific Recombination
- Human Genetics in Rheumatoid Arthritis Guides a High-Throughput Drug Screen of the CD40 Signaling Pathway
- Genome-Wide Analysis in German Shepherd Dogs Reveals Association of a Locus on CFA 27 with Atopic Dermatitis
- Liver X Receptors Protect from Development of Prostatic Intra-Epithelial Neoplasia in Mice
- Chromosomal Organization and Segregation in
- A Statistical Framework for Joint eQTL Analysis in Multiple Tissues
- Cell Polarity and Patterning by PIN Trafficking through Early Endosomal Compartments in
- Bck2 Acts through the MADS Box Protein Mcm1 to Activate Cell-Cycle-Regulated Genes in Budding Yeast
- High-Resolution Transcriptome Maps Reveal Strain-Specific Regulatory Features of Multiple Isolates
- Neuropeptides Function in a Homeostatic Manner to Modulate Excitation-Inhibition Imbalance in
- A Compendium of Nucleosome and Transcript Profiles Reveals Determinants of Chromatin Architecture and Transcription
- Wnt Signaling Regulates the Lineage Differentiation Potential of Mouse Embryonic Stem Cells through Tcf3 Down-Regulation
- Filamin and Phospholipase C-ε Are Required for Calcium Signaling in the Spermatheca
- The Specificity and Flexibility of L1 Reverse Transcription Priming at Imperfect T-Tracts
- Imputation-Based Meta-Analysis of Severe Malaria in Three African Populations
- Implicates Tyrosine-Sulfated Peptide Signaling in Susceptibility and Resistance to Root Infection
- Clathrin and AP2 Are Required for Phagocytic Receptor-Mediated Apoptotic Cell Clearance in
- Encodes CDF Transporters That Excrete Zinc from Intestinal Cells of and Act in a Parallel Negative Feedback Circuit That Promotes Homeostasis
- Global Properties and Functional Complexity of Human Gene Regulatory Variation
- DNA Binding of the Cell Cycle Transcriptional Regulator GcrA Depends on N6-Adenosine Methylation in and Other
- Side Effects: Substantial Non-Neutral Evolution Flanking Regulatory Sites
- From Paramutation to Paradigm
- From Mouse to Human: Evolutionary Genomics Analysis of Human Orthologs of Essential Genes
- Distinct Translational Control in CD4 T Cell Subsets
- Female Bias in and Regulation by the Histone Demethylase KDM6A
- ATM–Dependent MiR-335 Targets CtIP and Modulates the DNA Damage Response
- HDAC7 Is a Repressor of Myeloid Genes Whose Downregulation Is Required for Transdifferentiation of Pre-B Cells into Macrophages
- The Majority of Primate-Specific Regulatory Sequences Are Derived from Transposable Elements
- Identification of Meiotic Cyclins Reveals Functional Diversification among Plant Cyclin Genes
- EGL-13/SoxD Specifies Distinct O and CO Sensory Neuron Fates in
- Congruence of Additive and Non-Additive Effects on Gene Expression Estimated from Pedigree and SNP Data
- Using Extended Genealogy to Estimate Components of Heritability for 23 Quantitative and Dichotomous Traits
- Ikbkap/Elp1 Deficiency Causes Male Infertility by Disrupting Meiotic Progression
- Analysis of the Genetic Basis of Disease in the Context of Worldwide Human Relationships and Migration
- Duplication and Retention Biases of Essential and Non-Essential Genes Revealed by Systematic Knockdown Analyses
- Strong Purifying Selection at Synonymous Sites in
- , a Susceptibility Gene for Type 1 and Type 2 Diabetes, Modulates Pancreatic Beta Cell Apoptosis via Regulation of a Splice Variant of the BH3-Only Protein
- Chromosome Movements Promoted by the Mitochondrial Protein SPD-3 Are Required for Homology Search during Meiosis
- The Secretory Pathway Calcium ATPase PMR-1/SPCA1 Has Essential Roles in Cell Migration during Embryonic Development
- The Genomic Signature of Crop-Wild Introgression in Maize
- CDK4 T172 Phosphorylation Is Central in a CDK7-Dependent Bidirectional CDK4/CDK2 Interplay Mediated by p21 Phosphorylation at the Restriction Point
- Genome-Wide Identification of Regulatory RNAs in the Human Pathogen
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Using Extended Genealogy to Estimate Components of Heritability for 23 Quantitative and Dichotomous Traits
- HDAC7 Is a Repressor of Myeloid Genes Whose Downregulation Is Required for Transdifferentiation of Pre-B Cells into Macrophages
- Female Bias in and Regulation by the Histone Demethylase KDM6A
- ATM–Dependent MiR-335 Targets CtIP and Modulates the DNA Damage Response
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



