-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaThe Secretory Pathway Calcium ATPase PMR-1/SPCA1 Has Essential Roles in Cell Migration during Embryonic Development
Maintaining levels of calcium in the cytosol is important for many cellular events, including cell migration, where localized regions of high calcium are required to regulate cytoskeletal dynamics, contractility, and adhesion. Studies show inositol-trisphosphate receptors (IP3R) and ryanodine receptors (RyR), which release calcium into the cytosol, are important regulators of cell migration. Similarly, proteins that return calcium to secretory stores are likely to be important for cell migration. The secretory protein calcium ATPase (SPCA) is a Golgi-localized protein that transports calcium from the cytosol into secretory stores. SPCA has established roles in protein processing, metal homeostasis, and inositol-trisphosphate signaling. Defects in the human SPCA1/ATP2C1 gene cause Hailey-Hailey disease (MIM# 169600), a genodermatosis characterized by cutaneous blisters and fissures as well as keratinocyte cell adhesion defects. We have determined that PMR-1, the Caenorhabditis elegans ortholog of SPCA1, plays an essential role in embryogenesis. Pmr-1 strains isolated from genetic screens show terminal phenotypes, such as ventral and anterior enclosure failures, body morphogenesis defects, and an unattached pharynx, which are caused by earlier defects during gastrulation. In Pmr-1 embryos, migration rates are significantly reduced for cells moving along the embryo surface, such as ventral neuroblasts, C-derived, and anterior-most blastomeres. Gene interaction experiments show changing the activity of itr-1/IP3R and unc-68/RyR modulates levels of embryonic lethality in Pmr-1 strains, indicating pmr-1 acts with these calcium channels to regulate cell migration. This analysis reveals novel genes involved in C. elegans cell migration, as well as a new role in cell migration for the highly conserved SPCA gene family.
Published in the journal: . PLoS Genet 9(5): e32767. doi:10.1371/journal.pgen.1003506
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003506Summary
Maintaining levels of calcium in the cytosol is important for many cellular events, including cell migration, where localized regions of high calcium are required to regulate cytoskeletal dynamics, contractility, and adhesion. Studies show inositol-trisphosphate receptors (IP3R) and ryanodine receptors (RyR), which release calcium into the cytosol, are important regulators of cell migration. Similarly, proteins that return calcium to secretory stores are likely to be important for cell migration. The secretory protein calcium ATPase (SPCA) is a Golgi-localized protein that transports calcium from the cytosol into secretory stores. SPCA has established roles in protein processing, metal homeostasis, and inositol-trisphosphate signaling. Defects in the human SPCA1/ATP2C1 gene cause Hailey-Hailey disease (MIM# 169600), a genodermatosis characterized by cutaneous blisters and fissures as well as keratinocyte cell adhesion defects. We have determined that PMR-1, the Caenorhabditis elegans ortholog of SPCA1, plays an essential role in embryogenesis. Pmr-1 strains isolated from genetic screens show terminal phenotypes, such as ventral and anterior enclosure failures, body morphogenesis defects, and an unattached pharynx, which are caused by earlier defects during gastrulation. In Pmr-1 embryos, migration rates are significantly reduced for cells moving along the embryo surface, such as ventral neuroblasts, C-derived, and anterior-most blastomeres. Gene interaction experiments show changing the activity of itr-1/IP3R and unc-68/RyR modulates levels of embryonic lethality in Pmr-1 strains, indicating pmr-1 acts with these calcium channels to regulate cell migration. This analysis reveals novel genes involved in C. elegans cell migration, as well as a new role in cell migration for the highly conserved SPCA gene family.
Introduction
Calcium is one of the most versatile and important small molecules in the cell: it is involved in cell processes starting at the beginning of new cell life during fertilization and ending with a role in cell death during apoptosis [1]. One developmental role for calcium is in cell migration. Studies have determined that migrating cells establish a relatively stable calcium gradient, with higher levels in the rear of the cell [2]. The increased calcium levels at the rear of the cell are likely important for regulating cell detachment and contraction [3], [4]. More recently, studies identified transient flickers of high calcium enriched at the leading edge of migrating cells and preceding directional changes. These calcium flickers may provide localized cues that help direct migration [4]–[6].
Calcium-sensitive molecules important for cell migration can be divided into two broad categories: those that sense and respond to differences in calcium levels and those that control levels of calcium by transporting it across membranes [1]. In this second class, channels that release calcium into the cytosol, such as the plasma-membrane-associated transient receptor potential (TRP) channels, partnering with the secretory pathway inositol-trisphosphate channels (IP3R) and ryanodine receptors (RyR), have been shown to play a key role in producing calcium flickers and in cell migration [1], [4], [7], [8]. The localized movement of calcium out of the cytosol must also be important for producing calcium flickers and gradients. Calcium uptake from the cytosol requires Ca2+ ATPase transporters found at three distinct subcellular locations: the plasma membrane (PMCA), the Sarco(endo)plasmic reticulum (SERCA) and the secretory pathway/Golgi (SPCA) [1], [9], [10]. SERCA, which localizes to both the endoplasmic reticulum (ER) and Golgi, helps to control cytosolic calcium levels in a variety of cells and is integral for proper IP3 signaling [1]. The Golgi-localized SPCA, which has not been as extensively characterized, transports both Ca2+ and Mn2+, playing an important role in homeostasis for both molecules. SPCA is also critical for protein processing in the Golgi, Ca2+ release in response to IP3 signaling, and stress tolerance [1], [9], [11]. While at least one of these Ca2+ ATPase proteins is likely to be involved in maintaining subcellular calcium gradients important for cell migration, this link has yet to be firmly established.
Genetic studies show the highly conserved SPCA1 protein has essential roles in embryonic development and disease. Loss of a single copy of the SPCA1 gene in humans causes Hailey-Hailey disease (MIM# 169600), a skin disorder characterized by recurrent lesions or blisters of the skin in areas subject to high stress [12]–[16]. SPCA1 is likely essential in humans, as more severe phenotypes are found in patients who suffer clonal loss of both copies of the gene [17], [18]. Mice embryos homozygous for null mutations in SPCA1 die with defects in neural tube closure, while heterozygotes show susceptibility to squamous cell tumors, a phenotype observed occasionally in humans with Hailey-Hailey [19]–[22]. The primary cellular defect in Hailey-Hailey disease patients is keratinocyte acantholysis or loss of cell adhesion, marked by aggregation of keratin intermediate filaments that have retracted from desmosomes [23], [24]. In mice, cell adhesion appears normal, but cells show signs of Golgi dysfunction, which likely induces the high levels of apoptosis observed in the neural tube and mesenchyme [19]. Based on this genetic analysis, it is not evident that SPCA, which has a conserved role at the molecular level, has a similarly conserved role in development, although each phenotype may be the result of secretory pathway stress [25].
Caenorhabditis elegans has served as a good model system for studying calcium signaling because it has the full complement of membrane-associated calcium channels. Research has revealed a role for the calcium channel ITR-1/IP3R in ovulation, gastrulation, pharyngeal pumping, defecation and other developmental processes [26]. It is expressed in the embryo and is the only channel shown to play a direct role in cell migration, as itr-1 mutants fail to complete ventral closure during C. elegans development [26]–[28]. The UNC-68/RyR localizes to both ER and Golgi and plays a key role in muscle function [7], [29]–[32]. The single SPCA1 gene, pmr-1, encodes a protein that, like its vertebrate counterpart, co-localizes with Golgi markers, when expressed in COS cells or in C. elegans [33]–[35]. Based on RNA in situ hybridization and microarray analysis, pmr-1 is expressed in the adult germ line and at moderate levels throughout embryogenesis in most blastomeres [36]–[38]. pmr-1 transcriptional and translational fusion constructs show expression starting from the 3-fold embryo and continuing until the adult stage in the nervous system, intestine, intestinal valve, hypodermis, spermatheca, and gonad [35], [39]. The absence of germ line and early embryo expression for these fusion constructs may indicate they are silenced [40] or contain incomplete regulatory sequences. PMR-1 protein also shows functional conservation, with important roles in Ca2+ spiking in response to IP3 signaling, stress response, thermotolerance, pathogen resistance, and metal homeostasis [26], [33]–[35], [41], [42].
Using alleles of pmr-1 obtained in forward genetic and deletion screens, we have identified a new, essential role for pmr-1 in embryonic development in C. elegans. Strains homozygous for mutations in pmr-1 show temperature-sensitive embryonic lethality caused by defects in ventral enclosure, apical enclosure, and morphogenesis. By identifying the temperature-sensitive period we have learned that pmr-1 plays an important role during the period preceding ventral enclosure. While pmr-1 mutants show normal cell fate specification, lineage, and cell ingressions during gastrulation, we see defects in the migration of cells along the surface of embryo following ingression in the C-lineage, ventral neuroblasts, and in anterior lineages, a cell migration process that appears similar to migrations during neurulation in vertebrates. Phenotypes in pmr-1 mutants can be suppressed or enhanced by changes in the activity of the RyR unc-68 and IP3R itr-1 genes, indicating that these cell migrations are sensitive to calcium levels. This study identifies new molecules important for embryonic cell migration in C. elegans and reveals a new role in cell migration for the calcium channel PMR-1/SPCA protein family.
Results
Isolation and genetic analysis of pmr-1 alleles
Using two independent forward genetic screens designed to identify conditional mutants essential for embryonic enclosure and morphogenesis, we obtained two alleles of the C. elegans SPCA1 homolog pmr-1, jc10 and ru5. Strains homozygous for either the jc10 or ru5 allele produce inviable progeny when grown at 25°C, although these strains show increased embryonic viability at lower temperatures (Table 1). The tm1840 and tm1750 alleles [43] show similar temperature sensitivity, although overall embryonic viability is significantly lower at both 15°C and 20°C. For all four alleles examined, Pmr-1 embryos that die during mid-embryogenesis display a range of terminal phenotypes, including ventral closure defects, head ruptures, morphogenesis defects, and pharynx unattached (Pun) phenotypes (Figure 1; Table 2).
Fig. 1. pmr-1 mutant embryos show a range of terminal phenotypes at the temperatures tested. 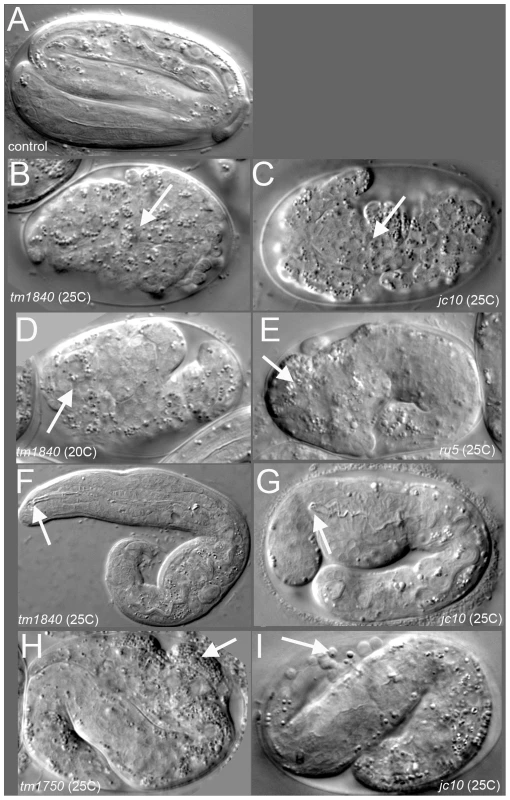
(A) Control embryos elongate into the vermiform shape just prior to hatching. In contrast, all four mutant alleles of pmr-1 produce the same range of terminal phenotypes that include B) and C) enclosure defects resulting in full body ruptures (arrow), D) and E) head ruptures (arrow), F) and G) a detached pharynx and body morphogenesis defects (arrow) or H) and I) cells outside the embryo (arrow). Embryos shown include deletion alleles pmr-1(tm1840) (B, D, F) and pmr-1(tm1750) (H), as well as hypomorphic alleles pmr-1(jc10) (C, G, I) and pmr-1(ru5) (E). See Table 2 for specific terminal phenotype frequencies for each allele. In all images, anterior is to the left. Tab. 1. The severity of the phenotypes in pmr-1 mutant strains is dependent on allele and temperature. 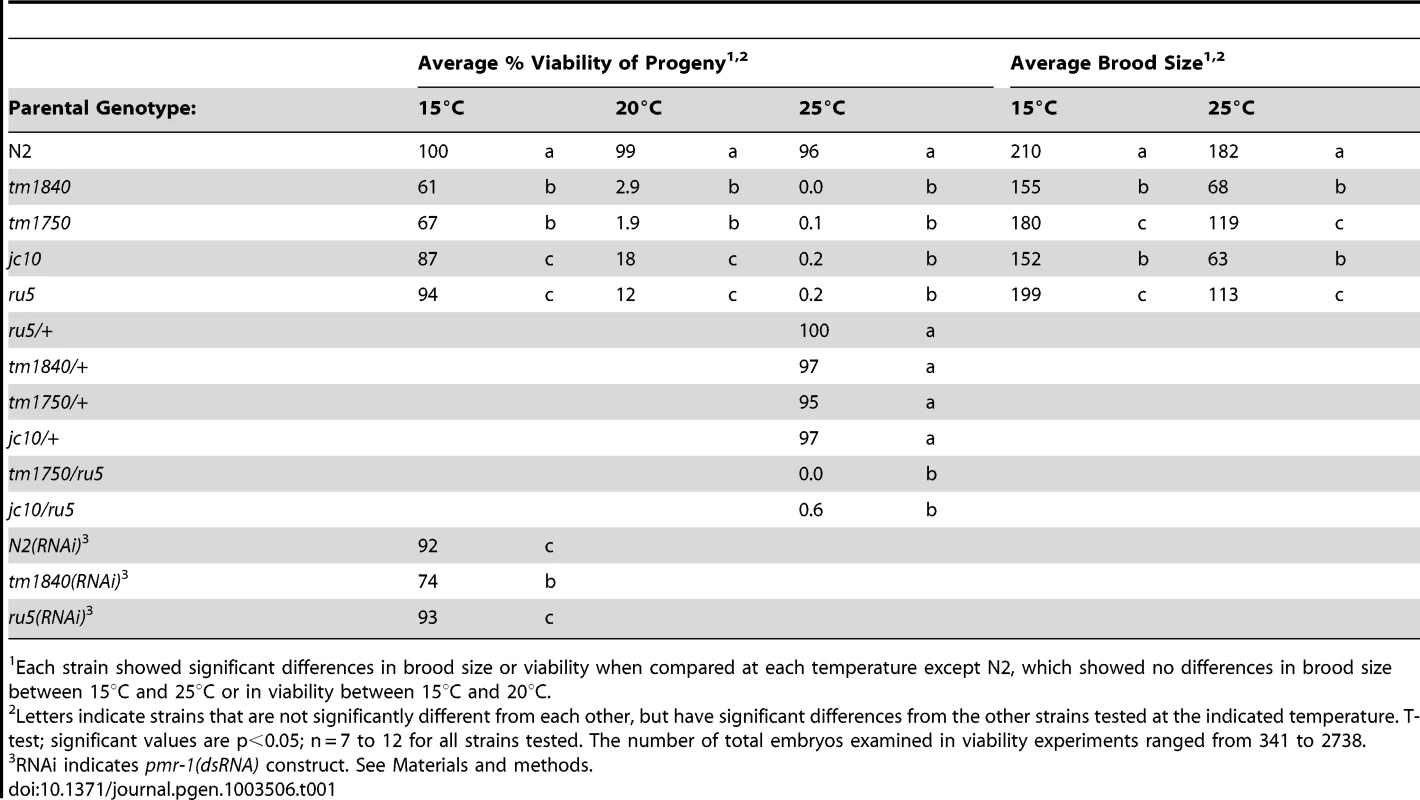
Each strain showed significant differences in brood size or viability when compared at each temperature except N2, which showed no differences in brood size between 15°C and 25°C or in viability between 15°C and 20°C. Tab. 2. pmr-1 alleles are pleiotropic at all temperatures. 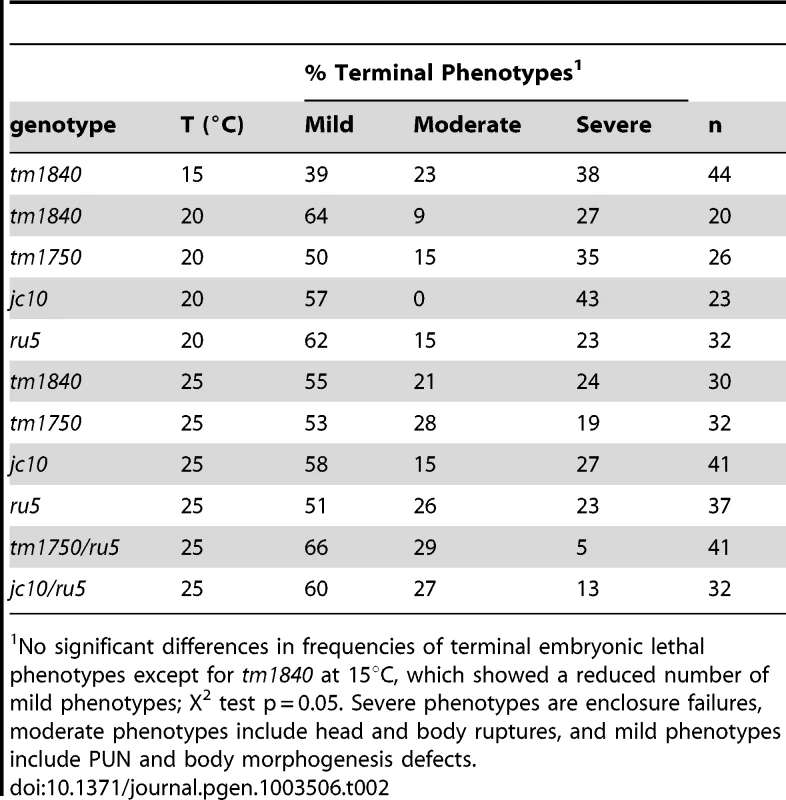
No significant differences in frequencies of terminal embryonic lethal phenotypes except for tm1840 at 15°C, which showed a reduced number of mild phenotypes; X2 test p = 0.05. Severe phenotypes are enclosure failures, moderate phenotypes include head and body ruptures, and mild phenotypes include PUN and body morphogenesis defects. We established that the embryonic lethal phenotypes observed in jc10 and ru5 strains are due to mutations in the pmr-1 gene by a series of criteria. Using conventional and snp-snp mapping strategies, the jc10 and ru5 alleles were mapped to a small interval on L.G. I (Figure S1A). Whole genome and conventional sequencing revealed that pmr-1 was the only gene in the mapped region that carried a mutation in both the jc10 and ru5 strains. Complementation analysis showed that jc10 and ru5 failed to complement each other and also failed to complement the pmr-1 deletions tm1750 and tm1840, consistent with the interpretation that all four mutations are allelic (Table 1; data not shown; Figure S1B). Transformation of ru5 strains with fosmids that contain the pmr-1 gene (Figure S1B, S1C) rescued the high temperature embryonic lethality. In contrast, we did not observe rescue in strains transformed with any other fosmids or cosmids from the mapped region (Figure S1B, S1C). Taken together, these experiments confirm that the defects in embryogenesis that we observe are due to disruption of pmr-1 gene activity.
The pmr-1(tm1840) deletion allele most likely represents a null, as the deletion eliminates an early exon, found in all of the major identified isoforms (Figure S1B) [33]. Based on strong sequence homology between PMR-1 and SERCA, whose crystal structure has been solved [44], the deletion would eliminate a transmembrane domain and a portion of the E1/E2 ATPase domain, thus rendering the expressed protein non-functional. However, we were unable to confirm whether the pmr-1(tm1840) allele is a null. Genetic tests with the dxDf2 deficiency strain were uninformative due to strong dominant phenotypes in the parental strain. Injection of dsRNA did not enhance or diminish the defects in the pmr-1(tm1840) strain (Table 1), which is consistent with expectations for a null mutation. However, injection of dsRNA into controls did not phenocopy the pmr-1(tm1840) allele, a result consistent with those reported by others (Table 1); [35], [45]. Alternative experiments to test for the null, such as use of the antisera generated against PMR-1 [34], were also ambiguous because of background activity of the antisera (data not shown).
While the tm1840 allele has the highest mortality at low temperatures, other alleles of pmr-1 show a range of phenotype severity. The pmr-1(tm1750) allele contains a deletion of coding sequence from just one isoform, but the severity of the phenotype, which is similar to pmr-1(tm1840), suggests either the isoform is critical or other isoforms are also disrupted (Table 1 and Table 2). The pmr-1(jc10) allele has a premature stop in an exon found in all isoforms (exon 4 of the pmr-1b isoform) that would eliminate transmembrane helices and the E1/E2 ATPase domain (Figure S1). The pmr-1(ru5) allele is a G142D substitution in a residue conserved from yeast to humans in PMR-1, but also conserved in SERCA (Figure S1) [46]. Based on homology with SERCA, this residue, in a beta strand that forms a portion of the A domain, would likely result in localized disruption of this domain, interfering with key interactions between the A and N domains of the E1/E2 ATPase [44]. Because all four alleles show the same embryonic lethal defects, varying only in the frequency of inviable embryos observed at a given temperature, the simplest interpretation is that they represent reduced or complete loss of function of pmr-1 gene activity, and are indicative of a key role for pmr-1 in embryogenesis.
In humans, Hailey-Hailey disease has a dominant inheritance pattern, likely due to haploinsufficiency of the pmr-1 homolog SPCA1 [10]. In C. elegans, analysis of dominance is complicated because we observe both maternal and zygotic rescue for pmr-1 alleles. pmr-1(ru5) hermaphrodites crossed to control males produce live progeny at 25°C, showing zygotic activity is sufficient to rescue the mutant phenotype. F1 hermaphrodites from a cross between pmr-1 and control strains produce between 96%–100% viable F2 embryos at 25°C, depending on the allele (Table 1). Since 25% of these F2 progeny should be homozygous for the pmr-1 loss-of-function alleles, these data suggest the maternally provided gene product is sufficient to rescue most of these embryos. However, for pmr-1(ru5), we were able to determine that the inheritance pattern is weakly semi-dominant. Of F2 progeny from a cross between pmr-1(ru5) and controls, 39% show a later developmental phenotype at 25°C, including larval lethality (3%), sterility (13%), small broods (3%), and inviable F3 progeny (20%; n = 236). These data show that some individuals heterozygous for pmr-1 loss-of-function alleles exhibit a phenotype. Although the literature reports that pmr-1 is expressed in most blastomeres throughout embryonic development [36]–[38], these genetic data suggest the wild type pmr-1 gene product is essential during a developmental period when both maternal and zygotic gene products are active.
pmr-1 is required for embryonic development
During C. elegans embryonic development the epidermal (hypodermal) cells that are born on the dorsal surface migrate to the ventral midline, enclosing the body of the embryo. The anterior-most epidermal cells also migrate to enclose the head, ultimately connecting epidermal tissue with the anterior foregut, creating the buccal cavity (Figure 1A) [47]–[49]. pmr-1 mutant embryos die with variable terminal phenotypes that include failure to enclose the body (Figure 1B, 1C) and head Figure 1D, 1E), as well as a detached pharynx (Pun) (Figure 1F, 1G). pmr-1 embryos that elongate also show defects in epidermal cell organization (1H, I). Additional phenotypes include reduced brood size and some larval lethality (Table 1). The terminal phenotypes due to pmr-1 disruption, such as ventral enclosure defects and head enclosure failures, are at least superficially similar to the neural tube closure failures in mice lacking the PMR-1 homolog SPCA, as well as the cell adhesion defects observed in Hailey-Hailey disease patients [19], [23], [24].
When pmr-1 is disrupted, the frequency of embryonic lethality varies depending on both allele and temperature. Given that all four alleles carry different types of genetic lesions, from point mutation to deletion, it is unlikely that they are temperature sensitive, per se, but rather there is a developmental process in which normal pmr-1 activity is increasingly important as temperature is increased. All alleles are also pleiotropic, showing a range of embryonic lethal phenotypes (Table 2). The frequency with which a specific terminal phenotype is observed is similar for all alleles, at all temperatures, and independent of whether the temperature shifts occur early or close to the temperature-sensitive period in development, as described below (Table 2). These data are consistent with the hypothesis that pmr-1 is playing a role in developmental events that affect cells positioned throughout the embryo, resulting in the variable effects.
PMR-1 is required during gastrulation
To better understand the defects leading to the Pmr-1 terminal phenotypes, we observed embryos during gastrulation using Nomarski optics. Using this approach, the first observable differences between control and pmr-1(ru5) embryos are in the position of the C blastomeres. In control embryos, the C-lineage blastomeres are positioned along the ventral surface at the posterior end of the embryo just prior to ingression (Figure 2A). In pmr-1(ru5) mutant embryos at the same developmental stage, these cells occupy a more dorsal position (Figure 2E). At the end of gastrulation, the gastrulation cleft closes in control embryos (Figure 2B). However, in pmr-1(ru5) embryos, the cleft remains open (Figure 2F), reflecting mispositioned ventral neuroblasts. We also saw later differences in cell position, with anterior blastomeres properly positioned in controls (Figure 2C), but altered in pmr-1(ru5) embryos (Figure 2G). Finally, we observed that the basement membrane, which initially surrounds the pharynx but is lost along the anterior most cells of the pharynx in control embryos (Figure 2D) [49], remains in place in pmr-1(ru5) embryos (Figure 2H). This analysis suggests the terminal phenotypes we observe are actually caused by problems in cell positioning earlier in embryogenesis.
Fig. 2. pmr-1 mutant embryos show differences in the positioning of cells during and after gastrulation. 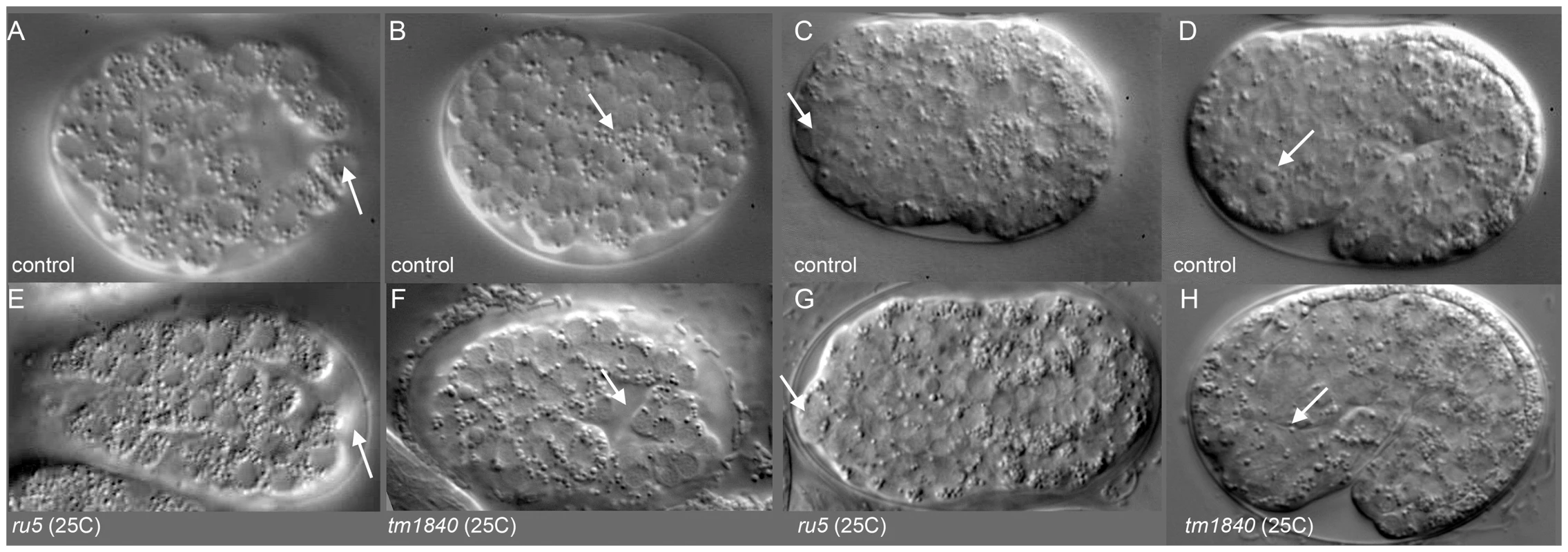
A) In control embryos, the C blastomeres are positioned at the posterior end of the embryo, on the ventral surface, surrounding the gastrulation cleft (arrow). E) In pmr-1 mutant embryos at the same developmental stage, the C blastomeres are shifted to a more dorsal position, out of the ventral focal plane (arrow). B) In control embryos, the gastrulation cleft closes as the last ventral cell ingresses (arrow). F) In pmr-1 mutant embryos at the same stage, the gastrulation cleft remains open (arrow). C) In control embryos, anterior cells form a smooth continuum (arrow), but are displaced in pmr-1 mutant embryos (G, arrow). D) In control embryos, the basement membrane that surrounds the anterior pharynx disappears as these cells migrate to the anterior (arrow, [49]. However, disruption in cell positioning disrupts this process in pmr-1 mutant embryos (H, arrow), preventing pharyngeal attachment. Genotypes are N2 control (A–E), pmr-1(tm1840) (F, H), or pmr-1(ru5) (E, G), all grown at 25°C. In all images, anterior is to the left. A–C and E–G are ventral views; D and H are lateral views. To better pinpoint the developmental time point when pmr-1 activity is required, we took advantage of the temperature sensitive phenotypes of pmr-1 alleles. Using the pmr-1(ru5) allele, which has viable progeny at 15°C but not at 25°C and is less fragile than the pmr-1 deletion alleles, we performed temperature shift experiments. In reciprocal temperature shifts, we found that embryos grown at the restrictive temperature 2 to 3 hours after the two-cell stage are not viable, while those grown at the permissive temperature during this time period survive (Figure 3). Temperature pulse experiments show a one-hour pulse from the restrictive to permissive temperature can significantly rescue viability only if the pulse occurs between 2 and 3 hours after two-cell stage, consistent with the hypothesis that pmr-1 is required only during this specific time period (). In contrast, one-hour pulses from 15°C to 25°C are insufficient to cause high lethality (). The temperature-sensitive period for pmr-1 is during gastrulation, during a period when anterior, c-lineage, and ventral blastomeres migrate (Figure 3; Figure S2), and significantly earlier than the events directly associated with the enclosure failure, Pun, and morphogenetic terminal phenotypes.
Fig. 3. The temperature-sensitive period for pmr-1(ru5) embryos is during anterior, C-lineage, and ventral cell migrations. 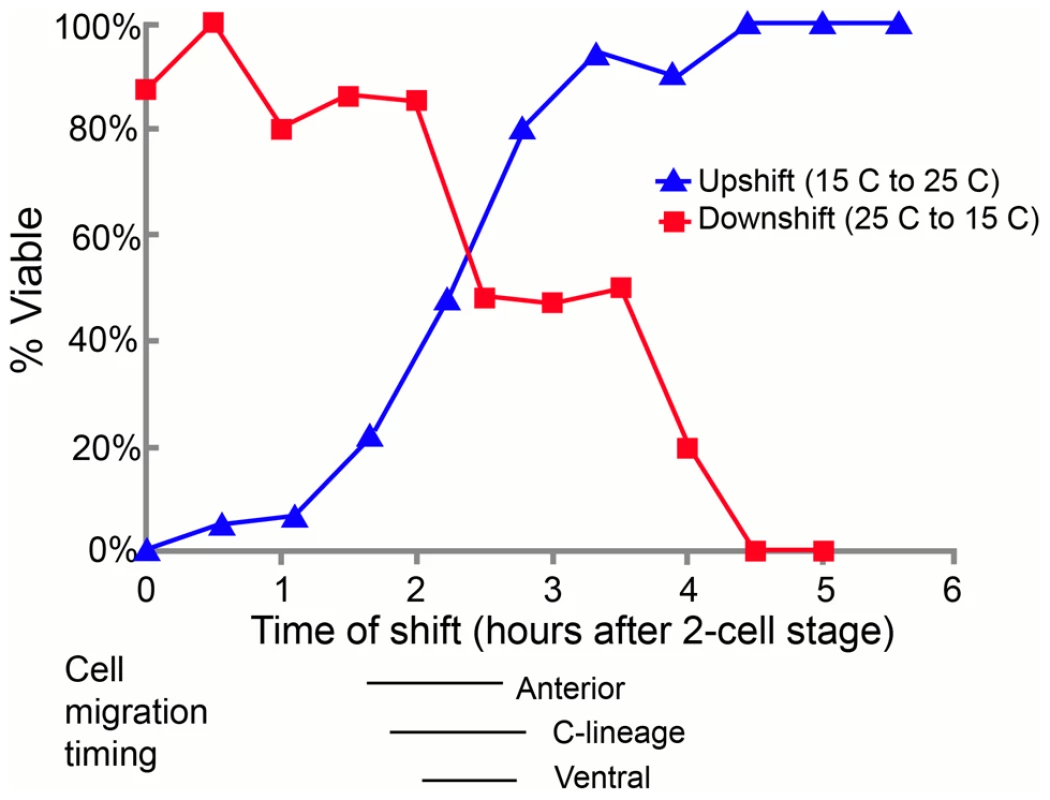
In the upshift experiments, embryos, extracted from gravid adults grown at the permissive temperature (15°C) from the mid-L4 stage, were switched to the restrictive temperature (25°C) at the indicated time. Embryos were then maintained at the restrictive temperature for the duration of embryogenesis and scored “viable” if they thrived past the L1 stage. In the downshift experiments, the same protocol was followed except that the embryos were switched from restrictive temperatures (25°C) to permissive temperatures (15°C) at the indicated times. The lines below the graph correspond to the times when anterior, C-derived, and ventral lineage cell migrations were assayed. All times were normalized to correspond to development at 25°C; n = 5 to 36 embryos at each time point. pmr-1 mutants do not have lineage or cell fate defects
Given that the temperature-sensitive period is during a stage of development when cells are rapidly dividing and differentiating, we wanted to determine if the defects observed in pmr-1 embryos were due to changes in cell lineage. In C. elegans, the lineage pattern is essentially invariant in control embryos [47]. We took advantage of this to compare the lineage of pmr-1(ru5) embryos to controls, from the 2-cell to ∼300-cell stage, using StarryNite software [50]–[53]. Our analysis of pmr-1(ru5) embryos indicates that the cell division timing and pattern for all lineages is indistinguishable from controls (Figure 4). We also found that the correct number of cells underwent programmed cell death with normal timing, beginning one to two divisions after the temperature-sensitive period (data not shown). These data indicate that pmr-1 embryos are not dying because of premature cell death or general defects in cell division timing or patterns.
Fig. 4. Cell division patterns and timing are normal in pmr-1(ru5) embryos. 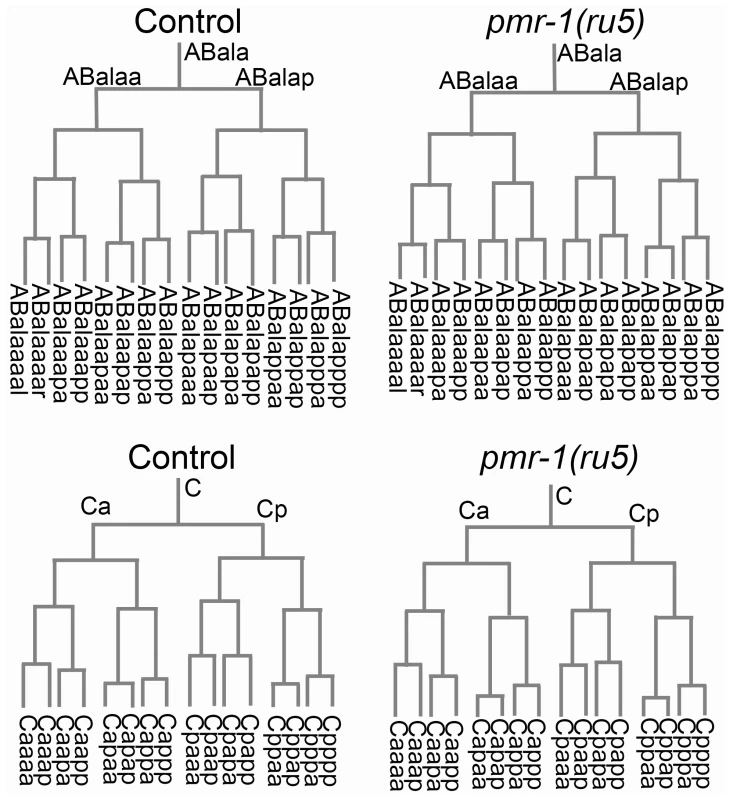
We determined the cell division patterns in control and pmr-1(ru5) embryos at 25°C to ∼300-cell stage using 4-D microscopy and cell lineage analysis software (Materials and Methods; n = 6 for each strain). Two representative lineages for control (left) and pmr-1(ru5) (right) embryos are shown. We did not observe any differences in cell lineage, cell deaths, or cell division timing in pmr-1(ru5) embryos throughout the temperature-sensitive period. Apoptotic cell deaths, which begin at least one cell division after the temperature sensitive period, occurred normally in all lineages examined (n = 2 controls; n = 4 pmr-1(ru5) strains). Although lineages appeared normal in pmr-1(ru5) embryos, we wanted to ascertain whether the cell fate was also normal. To address this question, we examined the expression patterns of GFP reporter fusion constructs that are expressed with specific timing, in a position-dependent manner within the embryo, or in specific tissues. Reporters included those expressed in the nervous system, hypodermis, muscle, intestine, and pharynx, some of which are derived from multiple lineages. We also included reporters, such as ceh-13, ceh-16 and vab-7, which are expressed in a position-dependent manner in multiple tissues and lineages [54]–[57]. In comparisons between control and pmr-1(ru5) embryos, the expression patterns of all of these reporter constructs were identical with only a few exceptions (Table 3; Figure S3, S4). These exceptions included differences in the position, but not in the number of cells expressing epidermal, neuroblast, and muscle markers. The other exception involved the expression of vab-7/even-skipped [54], which showed modest, but statistically significant delays in gene expression in C-lineage muscle and epidermal cells (Table 3; Figure S3). While we cannot rule out subtle effects on cell fate or changes in the fate of a small subset of cells, this analysis indicates that all major lineages and tissues are differentiating properly in pmr-1(ru5) embryos grown under restrictive conditions.
Tab. 3. pmr-1(ru5ts) strains properly express cell fate and cell polarity markers. 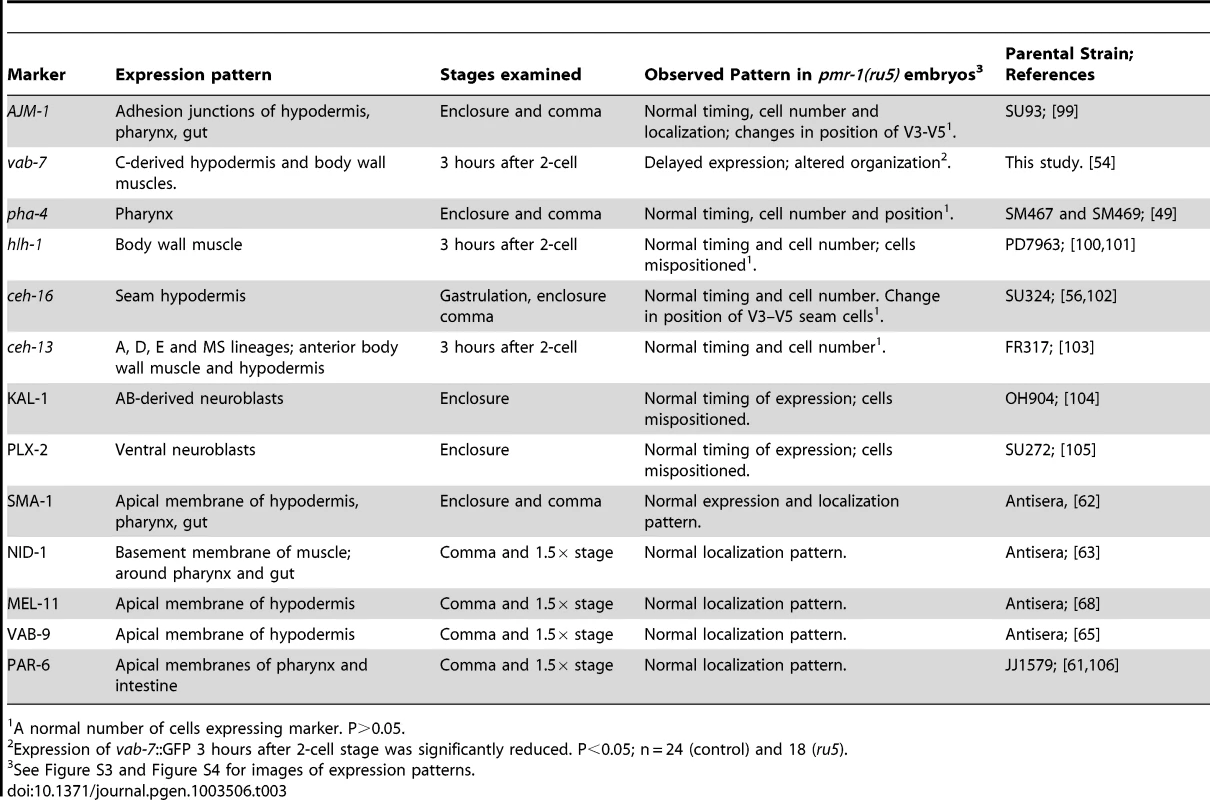
A normal number of cells expressing marker. P>0.05. Pmr-1 embryos have defects in cell migration
The observed differences in the position and timing of the vab-7 reporter, as well as the Nomarski imaging analysis (Figure 3; Table 3), suggested that pmr-1 mutants may have defects in cell migration. Since cells that express vab-7 are derived from the C lineage [54], we looked more closely at cell migration in C-derived blastomeres. In control embryos, the C-lineage blastomeres migrate from a dorsal position to a more ventral one along the posterior surface of the embryo. Muscle precursors, which have a more ventral starting position and migrate ahead of the hypodermal precursors, eventually ingress into the embryo at a posterior/ventral position (Figure 5) [58], [59]. When we compared control and pmr-1(ru5) embryos, we saw no differences in the timing of C-blastomere ingressions. Similarly, the timing of E, P4, MS, and D-derived blastomere ingressions were similar in control and pmr-1(ru5ts) embryos (n = 6 for each strain; t-test; p>0.05). This analysis shows that pmr-1 does not appear to be playing an essential role in cell ingression during C. elegans gastrulation.
Fig. 5. Cell migration defects in the C lineage cells of pmr-1(ru5) embryos. 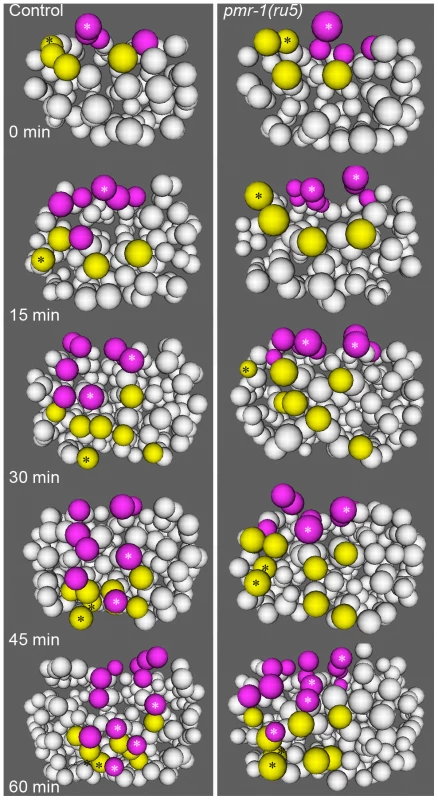
In control embryos (left panels), the C lineage muscle (yellow) and hypodermal (magenta) precursors migrate from dorsal to ventral positions. The migration rates of control embryos are significantly faster than in pmr-1(ru5) embryos (right panels). The muscle cell precursor Capa and its descendants (yellow; *) and the hypodermal cell precursor Cpap, and its descendants (magenta; *) show the migration and division of a single cell lineage in controls and pmr-1(ru5) mutant embryos. Average rates of migration of these cells for pmr-1(ru5) embryos are 55% of controls for Capa and 40% for Cpap (n = 6 for each strain; p<0.05). Each panel represents a 15-minute interval, starting at 1.8 hours (∼97 cells) after the 2-cell stage at 25°C, which corresponds with the temperature-sensitive period. Posterior view is shown, with dorsal at top. In contrast, pmr-1 does have a critical role in the migration of cells along the surface of the embryo. Using 4-dimensional representations of embryos created with Acetree, we measured the migration rates of C blastomeres along the posterior surface of the embryo (Materials and methods). In pmr-1(ru5) embryos, the rates of migration of both muscle and hypodermal cell precursors were significantly slower, at 55% and 40% of controls, respectively (Figure 5; t-test; p<0.05). Given that these cell migrations occur during the temperature sensitive period for pmr-1 gene activity, this analysis is consistent with the interpretation that pmr-1 plays an important role in cell migration during C. elegans embryonic development.
Given the variability of the pmr-1 terminal phenotypes, we examined several other lineages to determine if other cells exhibited cell migration defects. Terminal phenotype and gene expression analysis suggested that many cells are properly positioned in the embryo, so we focused our attention on cells that migrate during the temperature-sensitive period and whose migration defects might lead to enclosure failures and Pun phenotypes. In control embryos, the ventral neuroblasts derived from ABprp and ABplp lineages migrate from lateral to central positions along the ventral surface of the embryo, closing the gastrulation cleft (Figure 6A; [47], [58]. Hypodermal cells then crawl over these cells during enclosure [47], [48]. Analysis of pmr-1(ru5) embryos showed that these ventral cells are properly positioned at the beginning of the temperature-sensitive period, but many move a shorter distance than controls (Figure 6A), in some cases failing to close the gastrulation cleft, leading to later enclosure failures. The migration distances of lineages examined varied widely, and some cells migrated distances similar to controls. However, the majority of cells examined showed significant reduction in cell migration, on average 44% of controls in pmr-1(ru5) embryos (Figure 6B; T-test; p<0.05). These results show that pmr-1 is playing a key role in cell migration in cells that give rise to ventral neuroblasts, as well as epithelial cells.
Fig. 6. Cell migration defects in ventral cells of pmr-1(ru5) embryos. 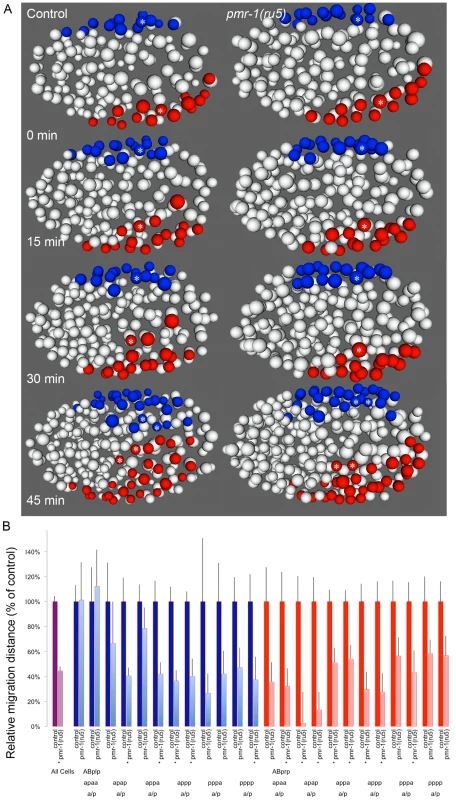
A. Ventral ABp-derived blastomeres migrate from the left (ABplp; blue) or right (ABprp; red) side of the embryo toward the ventral mid-line in control embryos (left panels). In pmr-1(ru5) embryos (right panels), the migration of these blastomeres is significantly reduced. For example, the rectal and interneuron precursor AB.plp appp (Blue; *) and the ring interneuron and ventral cord neuron precursor AB.prp appa (Red; *) migrate further in control than in pmr-1(ru5) embryos. Each panel represents a 15-minute interval, starting at 2 hours (∼162 cells) after the 2-cell stage at 25°C, which corresponds to the temperature-sensitive period. Ventral view is shown, with anterior to the left. B. In comparisons of average migration distance for ABprp (red) and ABplp (blue) blastomeres, cells migrated significantly farther in controls (dark boxes) than in pmr-1(ru5) (light boxes), with significant reductions in migration distance in at least one daughter in 8 of the 12 cells examined. The migration rates in pmr-1(ru5) embryos were 44% of those in controls, when all ventral blastomere migration values were compared (purple). T-test, * indicates p<0.05. Specific lineages tested are indicated. Measurements were taken starting at 2 hours after the 2-cell stage at 25°C (∼162 cells), and again 45 minutes later. In the anterior of control embryos, cells derived from the ABala and ABalp lineages, many of which ultimately differentiate into epithelial or nervous system cells, undergo dynamic migration patterns, with individual blastomeres crossing the dorsal-ventral or left-right midlines during embryogenesis. Some anterior blastomeres also make specific turns and ingress to form the anterior pharynx (Figure 7; [47], [58], [60]. In pmr-1(ru5) embryos, many anterior cells show migration defects. While some cells migrated normally in a subset of the pmr-1(ru5) embryos examined, many cells migrated shorter distances, in the wrong direction, or failed to make key turns, resulting in cells that were improperly positioned (Figure 7; t-test; p<0.05). Since these cells give rise to arcade cells, hypodermis, and ring ganglia, their failure to migrate properly during the temperature-sensitive period can readily account for the terminal phenotypes we observe in pmr-1(ru5) embryos. Taken together, these results show that pmr-1 plays a key role in cell migration and positioning during gastrulation.
Fig. 7. Cell migration defects in the anterior-most cells of pmr-1(ru5) embryos at 25°C. 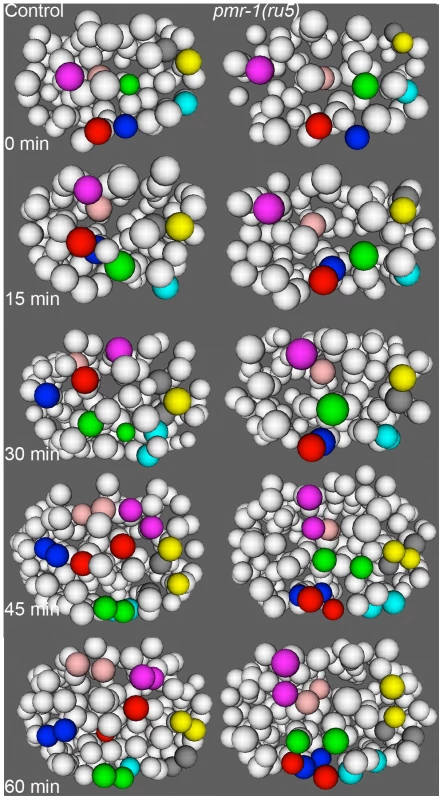
The anterior cells of control embryos (left panels) undergo distinct and reproducible cell rearrangements during gastrulation. CANL, labial, and ring ganglia precursors ABalapaa (magenta) and ABalaapa (blue) cross the left/right mid-line, labial and ring ganglia precursors ABalpppa (yellow) and ABalppap (gray) cross the dorsal/ventral mid-line and pharyngeal precursor ABalpapp (cyan) migrates from the left to a ventral position. Other cells, such as ring ganglia precursor ABalaaaa (red), migrate from peripheral to central positions or vice versa, as with pharyngeal precursor ABalpaaa (green) and labial/ring ganglia/CANL precursor ABalappa (pink). In a pmr-1(ru5) embryo (right panels), many of these anterior cells migrate shorter distances, migrate in the wrong direction, or fail to change directions, resulting in cells that are mis-positioned. While not every cell showed a migration defect in each embryo, measurements of the Cartesian coordinates for all of these 8 cells except ABalpppa (yellow), as well as three others not labeled above (labial and ring ganglia precursors ABalaaap, ABalpppp, and ABalppaa) showed significant positional differences in pmr-1(ru5) embryos (n = 6) compared to control embryos (n = 6; p<0.05). Each panel represents a 15-minute interval, starting at 1.6 hours (∼88 cells) after 2-cell stage at 25°C, which corresponds to the temperature-sensitive period. Anterior view is shown, with dorsal (top) and embryo right (left). Cell fate information from [97], [98]. pmr-1 mutants do not show disruptions in cell polarity, adhesion, or basement membrane structures
In Hailey-Hailey disease, cells show defects in cell attachment structures [23], [24]. The defects in cell migration we observe in pmr-1 mutants could be due to changes in cell adhesion, as well as in other cellular processes including cell polarity or establishment of the basement membrane. Our analysis of gene expression suggests that each of these processes is relatively normal in pmr-1(ru5ts) embryos. PAR-6 is an apically localized protein that plays a key role in cell polarity in a number of developmental processes [61]. SMA-1 is an actin-binding protein that localizes to the apical membrane of polarized epithelial cells [62]. NID-1 is a basement membrane protein expressed during embryonic development and nid-1(cg119) mutants have Pun phenotypes [63], [64]. AJM-1, MEL-11, and VAB-9 localize to apical adhesion complexes [65]–[68]. Each of these proteins is expressed in the correct cells and shows normal subcellular localization in pmr-1(ru5ts) embryos (Table 3; Figure S4). The localization is normal even in hypodermal cells that show positioning defects (Figure S4M-S4R). Similarly, KAL-1 and PLX-2, proteins that are expressed in ventral neuroblasts and play key roles in hypodermal cell migration and enclosure [69], [70], appear to be properly expressed in pmr-1(ru5) embryos (Figure S3I–S3L). While we cannot rule out subtle changes in protein localization and gene expression, these data indicate that the defects we observe in pmr-1 mutant embryos are not due to general defects in cell polarity, cell adhesion, or expression of basement membrane proteins.
Altering calcium channel activity can modulate pmr-1 mutant phenotypes
Given the conserved molecular roles of PMR-1/SPCA, we asked whether perturbations in Ca2+ signaling were chiefly responsible for the pmr-1 mutant phenotypes. To address this question, we altered the activity of either the ITR-1/IP3R or UNC-68/RyR calcium channels, reasoning that if calcium levels were critical for normal cell migration, changes in the activities of these receptors would suppress or enhance the pmr-1 embryonic lethal phenotypes. We made strains carrying pmr-1 mutations and itr-1(sy327), a gain-of-function allele that is thought to increase the affinity for IP3 [28], [71]. At 20C, the itr-1(sy327) strain has slightly reduced embryonic viability (91%) compared to controls while the pmr-1(tm1840) strain has 9% embryonic viability. The pmr-1(tm1840);itr-1(sy327) double mutant shows significant improvements in viability compared to the pmr-1(tm1840) single mutant (42%; Table 4). This affect is not allele specific, as we observed similar suppression of phenotypes for the pmr-1(jc10), pmr-1(ru5), and pmr-1(tm1750) lines when placed in an itr-1(sy327) background (Table 4). These data show itr-1 phenotypes are epistatic to pmr-1. We also made double mutants carrying pmr-1 alleles and itr-1(jc5), a loss-of-function allele [27]. While we were unable to generate double mutants with pmr-1(tm1840) or pmr-1(tm1750), suggesting the combination is lethal (data not shown, V.P., R.W. and R.M.), the pmr-1(ru5); itr-1(jc5) strain showed enhanced lethality compared to the pmr-1(ru5) strain alone (Table 4), suggesting the two genes act in parallel pathways during this stage of development. In contrast, depletion of unc-68 using RNAi significantly suppressed the embryonic lethality of pmr-1(ru5) strains (Table 4), indicating unc-68 acts in opposition to pmr-1 in a role very different from that of itr-1. Taken together, these data show that the embryonic lethality we observe in pmr-1 mutant embryos can be affected by changes in the activity of calcium channels. While this type of analysis does not allow us to rule out other possibilities, it provides strong support for the hypothesis that pmr-1 is acting through its role in Ca2+ signaling to affect cell migration.
Tab. 4. Genetic interactions with pmr-1. 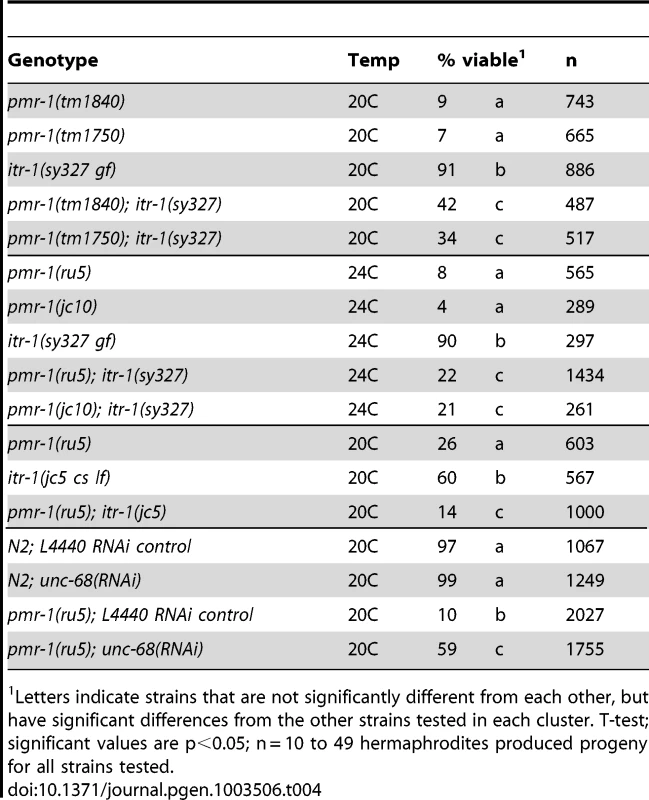
Letters indicate strains that are not significantly different from each other, but have significant differences from the other strains tested in each cluster. T-test; significant values are p<0.05; n = 10 to 49 hermaphrodites produced progeny for all strains tested. Discussion
Our analysis of PMR-1/SPCA1 in C. elegans has revealed a novel and essential role in cell migration for this highly conserved gene [1], [9]. Using a forward genetic screen designed to look for genes required for normal epidermal cell migration and adhesion, we identified mutations in the C. elegans SPCA1 gene pmr-1. Strains homozygous for pmr-1 show temperature-sensitive embryonic lethality, with terminal phenotypes that include ventral enclosure failures, head ruptures, and morphogenetic phenotypes including a detached pharynx (Pun) phenotype. Phenotype and temperature shift analysis indicates that the pmr-1 gene plays a crucial role in a specific period of embryonic development, during gastrulation. Cell fate specification, cell division patterns, apoptosis, and cell ingression during gastrulation are all similar to control embryos. However, the migrations of specific subsets of cells, along the posterior, anterior, and ventral surfaces of the embryo, are defective in pmr-1 mutant embryos. The specificity of the phenotype, coupled with the temperature shift analysis, indicate that PMR-1/SPCA plays an essential role in a specific set of cell migrations during gastrulation in the developing C. elegans embryo.
Given the broad expression pattern for pmr-1 in all cells, coupled with the proposed housekeeping functions one would expect for a calcium transporter [1], [9], the specific timing and proposed role for PMR-1 in ectodermal cell migration is unexpected. Studies examining the expression of pmr-1 indicate that PMR-1 protein localizes to the Golgi and that the gene product is present in the germ line and widely expressed in cells throughout embryogenesis, including during gastrulation [33]–[38]. This expression pattern, which is consistent with our genetic analysis that maternally or zygotically provided gene product is sufficient to rescue the Pmr-1 mutant phenotype, would suggest a broad role for the gene product. Yet our temperature shift and pulse analysis indicates requirements for PMR-1 can be bypassed during other stages of embryogenesis. The phenotype analysis identifies a very specific role in the migration of anterior, C-lineage, and ventral cells along the surface of the embryo during gastrulation, but with no discernable effects on other cell migration events such as ingression or ventral closure, nor on any other developmental pathway that might alter cell fate, division patterns, or cell polarity. The best explanation for these data is that PMR-1 may act redundantly in some aspects of embryogenesis, but plays a more critical or specific role in the cell migrations along the embryo surface that follow cell ingression, a markedly different developmental process [58], [60].
PMR-1/SPCA is part of a network of proteins that regulate calcium levels in the cell cytoplasm and calcium stores in the secretory pathway. Models for modulating cellular calcium levels start with a signal transduced through PLC and IP3 triggering release of Ca2+ into the cytoplasm from secretory stores through ITR-1/IP3 and UNC-68/RyR. The calcium ATPases SCA-1/SERCA and PMR-1/SPCA, located predominantly in the ER and Golgi, respectively, then pump Ca2+ back out of the cytoplasm into these organelles [7], [26]. A simple model would predict that reduced activity of a transporter acting in one direction is suppressed by reduced activity of channel acting in the opposite direction. This is the result we observe for interactions between pmr-1 and unc-68, where pmr-1(ru5) strains showed reduced lethality in an unc-68(RNAi) background (Table 4). In contrast, strains carrying pmr-1 and itr-1(jc5) loss-of-function alleles showed enhanced lethality at semi-permissive conditions while strains carrying the itr-1(sy327) gain-of-function allele showed significant suppression of pmr-1(ru5) embryonic lethality. This gene interaction analysis suggests that itr-1 and pmr-1 act in parallel pathways during embryonic development and that enhanced sensitivity of itr-1 to IP3 signaling in the itr-1(sy32y7) mutant [72] can help bypass the requirement for pmr-1. While these interpretations reflect the genetic analysis, they are puzzling because ITR-1 and PMR-1 act in opposite directions with respect to cytosolic calcium. Similarly, UNC-68 does not appear to have any developmental roles nor does it appear to be expressed at high levels in the embryo until later stages [7], [29]–[32].
A speculative model to explain the genetic interaction data we observe is that these proteins may play some redundant and some non-redundant roles in regulating calcium levels in the cell, due in part to the polarity of the secretory pathway. One would predict that when PMR-1 activity is reduced, the pool of calcium in secretory stores is also modestly reduced and the calcium levels in the cytoplasm might be elevated [34]. Reduction of UNC-68 or ITR-1 calcium release would, in theory, restore overall secretory pool levels. However, the calcium flicker model for cell migration predicts that calcium must be supplied in localized bursts at the leading edge of the cell [5], [6]. In a pmr-1 mutant, the pool of available calcium would be disproportionately reduced in the Golgi, the secretory organelle closest to the leading edge. If release is, as predicted, strongly dependent on ITR-1 [5], [6], an itr-1 loss-of-function mutant would further diminish calcium release, making the phenotype worse, while the itr-1(sy327) gain-of-function mutation would release calcium at lower levels of signal [28], [71], suppressing the phenotype. A second possibility to explain the suppressor data reflects the distinct mechanisms by which the channels are activated. While both the UNC-68/RYR and ITR-1/IP3R channels are sensitive to changes in cytosolic calcium, only the ITR-1/IP3R channels responds to an IP3 ligand [73], [74]. Reduced activity of the ITR-1/IP3R channel would make the secretory pathway insensitive to IP3 signaling, which when coupled with reduced stores would make the phenotype worse. Increasing IP3R sensitivity might compensate for altered cytosolic calcium, making the situation better, as observed. Finally, since the regulation of calcium signaling and transport is highly interdependent, with channels and targets acutely sensitive to minor changes in calcium concentration [1], [72]–[74], single loss-of-function mutations may result in some compensatory changes in the signaling system that cannot occur if two systems are defective.
While it is tempting to assume that disruption of pmr-1 has a direct effect on calcium-mediated cell signaling, disruptions in the gene might instead effect other calcium-dependent processes required for cell migration. The embryonic lethal phenotypes in Pmr-1 strains were observed at all the temperatures assayed, but they were more severe at high temperatures for all four alleles tested. Although our experiments were in the normal temperature range for C. elegans growth [75] and we saw no evidence of necrosis in pmr-1 mutant embryos, recent research indicates that pmr-1 is important for survival of heat shock, acting to prevent necrosis [42]. It is possible that the migrating cells of the embryo are particularly sensitive to changes in the activity of this stress resistance pathway. Alternatively, the cytoskeletal and membrane dynamics of migrating cells may simply be more sensitive to temperature than other developmental processes, or the timing is more precise, such that changes in pmr-1 activity have a stronger impact. Another possibility is that the changes in pmr-1 activity alter calcium-dependent processing of specific proteins in the Golgi. No matter the precise mechanism by which changes in calcium dynamics alter cell migration, our identification of a role for PMR-1, acting with ITR-1/IP3R and UNC-68/RyR, confirms earlier work that ITR-1 plays an important role in cell migration [27], [28] and identifies new genes important for normal cell migration in the C. elegans embryo.
The phenotypes we observe in C. elegans pmr-1 mutants have at least some superficial similarities to those observed in vertebrates with defects in the SPCA1 gene, including humans with Hailey-Hailey disease. In both C. elegans and in vertebrates, we observe semi-dominant phenotypes and the severity of the observed phenotypes are influenced by environmental stresses. The terminal phenotypes, including apparent loss of cell adhesion in the epidermis of humans with Hailey-Hailey or failures to close the rostral neural tube in mice embryos homozygous for SPCA loss-of-function alleles [12]–[16], [19], [23], [24] also look similar to the enclosure failures and head ruptures we observe in C. elegans embryos.
One major difference between the worm pmr-1 mutant phenotype and Hailey-Hailey disease patients is the nature of defects in cell adhesion. In Hailey-Hailey disease, symptoms include acantholysis of keratinocytes, marked by keratin dissociation from desmosomes [23], [24]. Our analysis of pmr-1 mutants suggests that the gene does not play a direct role in cell adhesion. Proteins that localize to adhesion complexes, such as MEL-11, AJM-1, and VAB-9, look normal in pmr-1 mutant embryos and epistasis analysis indicates that vab-9 and pmr-1 are in different pathways (unpublished data, J. Simske). While intermediate filament and desmosomal structures have not yet been carefully examined in pmr-1 mutants, we do not suspect an association. While pmr-1 is essential during gastrulation, analysis by others indicates genes that encode intermediate filament and attachment structures in C. elegans, such as IFA-1, IFA-2/MUA-6, IFA-3, IFA-4, IFB-1, VAB-10, MUP-4, and LET-805, assemble later in development, playing key roles in the embryonic elongation that follows ventral closure [76]–[83]. This analysis strongly suggests intermediate filaments are unlikely to be directly involved in the PMR-1-mediated cell migration, although it is possible PMR-1 may play a later, non-essential role in maintaining these attachment structures. Similarly, we did not see elevated levels of cell death in pmr-1 mutants, as reported for the mice knockouts [19]. These data suggest that there may be differences between SPCA function in C. elegans and in vertebrate systems, perhaps due to fundamental differences in ectodermal cell structure. Alternatively, the terminal phenotypes observed in vertebrates could reflect earlier defects.
Given our results in C. elegans, as well as the strong sequence conservation of the gene, it is tempting to speculate that the SPCA's have cell migration roles in vertebrates. The relative simplicity of C. elegans as model system, as well as powerful new lineaging tools [50]–[53], has allowed us to distinguish between the terminal phenotype and the actual defects in cell migration found in pmr-1 mutants. In humans, keratinocytes differentiate in response to changes in extracellular calcium, migrating from the stratum basale into upper layers of the epidermis as they differentiate [84]. It is conceivable that in Hailey-Hailey patients, stress-damaged cells are replaced inefficiently because of a defect in cell migration, caused by altered calcium levels, leading to the observed lesions and keratinocyte defects [23], [24]. Similarly, the rostral neural tube closure failings in SPCA mouse knockouts [19] could be due to defects in migration of specific subsets of ectodermal cells during this stage of development.
In summary, we have shown that the PMR-1/SPCA1 Ca2+/Mn2+ ATPase plays a key role in cell migration during C. elegans embryonic development. Strains carrying mutant pmr-1 show defects in migration of cells along the surface of the embryo following the ingression of cells during gastrulation. The cell migration defects are likely caused by changes in intracellular calcium levels, as they can be enhanced or suppressed by changes in activity of the ITR-1/IP3R or UNC-68/RyR calcium channels. This analysis reveals a new role for calcium signaling in the migration of specific blastomeres during C. elegans development. It also reveals a new role in cell migration for the SPCA1 family of genes.
Materials and Methods
Growth conditions, strains, and alleles
Strains were grown and maintained under standard conditions [85]. Wild type strain N2 was used as a control. The ru5 and jc10 alleles of pmr-1 were identified in two independent embryonic conditional embryonic lethal screens using the mutagen EMS [85]; J. Simske). The tm1840 and tm1750 alleles were produced in the National Bioresource Project (kindly provided by S. Mitani; [43]. All pmr-1 mutant strains, which were maintained at 15°C, were backcrossed to the N2 strain at least 3 times. The deletions in tm1840 and tm1750 were confirmed using PCR analysis. PCR mapping lines CB4856, RW700, deficiency lines CB2775 eDf9/eDf24 I, CB2770 eDf4/eDf24 I, CB2769 eDf3/eDf24 I, KR2838 hDf17/hIn1[unc-54(h1040)], JK1542 ces-1(n703) qDf7/dpy-5(e61) srf-2(yj262) unc-75(e950) I, SL536 dxDf2/spe-9(eb19) unc-101(m1) I, and conventional mapping lines MT465 dpy-5 (e61)I, bli-2(e768)II, unc-32(e189)III, MT464 unc-5(e53)IV, dpy-11(e224)V, lon-2(e678)X; DR210 dpy-5(e61), daf-16(m26), unc-75(e950) I, CB2010 unc-54(e675), dpy-5(e61)I, JK228 glp-4(bn2), unc-54(e675) I were used in mapping experiments. SU180 itr-1(jc5) jcIs1 IV, PS2368 itr-1(sy327) unc-24(e138) IV and QQ101 vab-9(ju6) II strains were used for double mutant analysis. For analysis using GFP fusions, F2 progeny exhibiting the Pmr-1 phenotype and carrying the appropriate markers were selected from crosses between pmr-1(ru5) males and the following strains: RW10026 unc-119(ed3); stIs10026, SM467 pIs7; rol-6(su1006), SM469 pIs6, SU93 jcIs1, PD7963 ccIs7963, SU324 jcIs26, FR317 swIs1, OH904 otIs33, SU272 jcIs1, evIs136, and JJ1579 zuIs77. Strains expressing vab-7::GFP were generated by microparticle bombardment of pJA64 and pDPMM016b into a pmr-1(ru5)I; unc-119(ed3)III strain [86] and backcrossed to wild type to generate controls.
Mapping, complementation, and genome-sequencing experiments
The ru5 and jc10 alleles were mapped using conventional and deficiency mapping strategies [87]. The position of the ru5 allele was further refined using PCR-based and snp-snp mapping techniques [88]–[90]. Genomic DNA was extracted from the ru5 strain using standard methods and the whole genome was sequenced using Illumina sequencing and MAQGene, as described [91]. Alterations in candidate genes were confirmed in ru5 using PCR amplification followed by subcloning (Strataclone, Agilent Technologies) and conventional sequencing (U. of Iowa Carver Center, U of Wisconsin, and Fred Hutchinson DNA sequencing facilities). The coding regions of these candidate genes were then sequenced in jc10. In complementation analysis, ru5 males were crossed to jc10, tm1840, or tm1750 hermaphrodites and the F1 progeny scored for embryonic lethality at 25°C. For complementation rescue analysis, ru5 young adults were transformed by injection of 25–100 µg/ml of a fosmid or cosmid covering each of the candidate genes, along with the co-transformation marker pRF4 [92], [93]. Rol Progeny were assayed for survivorship at 25°C for each injected construct.
Mutant and RNAi viability and brood size analysis
To determine rates of viability, L4 hermaphrodites were shifted from 15°C to the indicated temperature. Eggs from single hermaphrodites were counted and the number of resultant late larva or adult progeny scored. In brood size experiments, single L4 hermaphrodites were shifted to the indicated temperature and total broods quantified as previously described [94]. Both brood size and viability data were compared using t-tests. Terminal phenotypes were scored during mid-to-late embryogenesis. Embryos that failed to enclose were classified as “severe”, those with head or later body ruptures as “moderate”, and those with cell position or pharynx unattached (Pun) phenotypes as “mild”. We used chi-square analysis to compare the frequency of each class for a given strain or condition. For RNA interference experiments with pmr-1, young adult hermaphrodites were injected with a pmr-1 dsRNA construct mv ZK256.1a at 0.9 mg/ml using standard methods [95]. Injected worms were allowed to recover and lay eggs. The embryos of injected progeny were examined for levels of embryonic lethality and terminal phenotypes. For RNAi with unc-68, we used an RNAi feeding protocol [95].
Temperature shift experiments
Embryos were extracted from gravid pmr-1(ru5) adults grown at the indicated temperature from the early-to-mid-L4 stage, shifted to the second temperature at the indicated time point, and then maintained at that second temperature for the duration of embryogenesis. Each embryo was scored for hatching and terminal phenotypes. Experiments at the two temperatures were calibrated based on comparisons of the timing of key developmental events.
Lineaging and cell migration analysis
For lineaging and cell migration analysis, pmr-1(ru5) embryos carrying nuclear-localized GFP were imaged at 25°C according to the protocol outlined in [50], [52], [53]. Four-dimensional representations of embryos were generated using the Acetree program [51]. pmr-1(ru5) lineages (n = 6) were compared to controls (n = 6 to 8 ) (kindly provided by M. Boeck, P. Weisdepp, and R. Waterston). Files for each embryo were synchronized to the same developmental time points based on cell division patterns in several lineages. Because of the different migration patterns for the lineages tested, several techniques were used for measuring cell movement. To analyze ingression, the timing of the ingressions of P4, D, MS, and C-lineage daughters in N2 was compared to pmr-1(ru5) embryos. C-lineage cell migration was calculated by measuring the distance of the migrating cell nucleus relative to the ventral-most cell in the embryo. The average slopes of distance over time were used for comparisons. Ventral cell migration was calculated by measuring the distance of the migrating cell nucleus relative to the left or right edge of the embryo and total migration distances over time were compared. The position of the anterior–most cells was measured using polar or Cartesian coordinates, the positions plotted over time, and positions compared in each dimension. In all cases, t-tests were used to determine if the differences between the controls and pmr-1(ru5) embryos were statistically significant.
Gene expression analysis
For analysis of the expression patterns of GFP-fusion proteins, control and pmr-1(ru5) embryos were grown at 25°C and compared at specific developmental stages. In some experiments, the position and number of nuclei expressing markers were analyzed using t-tests. For analysis using in situ immunofluorescence, control and pmr-1 strains were collected and fixed using a freeze cracking technique and stained with anti-SMA-1, anti-NID-1, anti-AJM-1, anti-SQV-8, or anti-PMR-1 antisera [34], [62], [63], [66], [96]. Nomarski and fluorescent images of embryos were collected using an Olympus spinning disc microscope using Slidebook, Nikon E600 microscopes using a coolsnap CCD camera controlled by Metaview software, and Olympus DeltaVision deconvolution microscope (Applied Precision, Issaquah, WA); projected images were created using 4D macros within NIH Image and Image J.
Supporting Information
Zdroje
1. BriniM, CarafoliE (2009) Calcium pumps in health and disease. Phys Rev 89 : 1341–1378.
2. BrundageRA, FogartyKE, TuftRA, FayFS (1991) Calcium gradients underlying polarization and chemotaxis of eosinophils. Science 254 : 703–706.
3. RidleyAJ, SchwartzMA, BurridgeK, FirtelRA, GinsbergMH, et al. (2003) Cell Migration: Integrating signals from front to back. Science 302 : 1704–1709.
4. WeiC, WangX, ZhengM, ChengH (2012) Calcium gradients underlying cell migration. Curr Opin Cell Biol 24 : 254–261.
5. WeiC, WangX, ChenM, OuyangK, ZhengM, et al. (2010) Flickering calcium microdomains signal turning of migrating cells. Can J Physiol Pharmacol 88 : 105–110.
6. WeiC, WangX, ChenM, OuyangK, SongLS, et al. (2009) Calcium flickers steer cell migration. Nature 457 : 901–906.
7. ZampeseE, PizzoP (2012) Intracellular organelles in the saga of Ca2+ homeostasis: different molecules for different purposes? Cell Mol Life Sci 69 : 1077–1104.
8. HoweAK (2011) Cross-talk between calcium and protein kinase A in the regulation of cell migration. Curr Opin Cell Biology 23 : 554–561.
9. VandecaetsbeekI, VangheluweP, RaeymaekersL, WuytackF, VanoevelenJ (2011) The Ca2+ Pumps of the endoplasmic reticulum and Golgi apparatus. Cold Spring Harbor Perspectives in Biology 3: a004184.
10. MissiaenL, DodeL, VanoevelenJ, RaeymaekersL, WuytackF (2007) Calcium in the Golgi apparatus. Cell Calcium 41 : 405–416.
11. HeW, HuZ (2012) The role of the Golgi-resident SPCA Ca2+ Mn2+ pump in ionic homeostasis and neural function. Neurochem Res 37 : 455–468.
12. HaileyH, HaileyH (1939) Familial benign chronic pemphigus. Arch Dermatol Syphilol 39 : 679–685.
13. HuZ, BonifasJM, BeechJ, BenchG, ShigiharaT, et al. (2000) Mutations in ATP2C1, encoding a calcium pump, cause Hailey-Hailey disease. Nature Genet 24 : 61–65.
14. KellermayerR (2005) Hailey-Hailey disease is an orthodisease of PMR-1 deficiency in Saccharomyces cerevisiae. FEBS Letters 579 : 2021–2025.
15. Missiaenv, RaeymaekersL, DodeL, VanoevelenJ, Van BaelenK, et al. (2004) SPCA1 pumps and Hailey-Hailey disease. Biochem Biophys Res Comm 322 : 1204–1213.
16. SudbrakR, BrownJ, Dobson-StoneC, CarterS, RamserJ, et al. (2000) Hailey-Hailey disease is caused by mutations in ATP2C1 encoding a novel Ca2+ pump. Hum Mol Genet 12 : 1131–1140.
17. HwangL, LeeJ, RichardG, UittoJ, HsuS (2004) Type 1 segmental manifestation of Hailey-Hailey disease. J Am Acad Dermatol 49 : 712–714.
18. Poblete-GutierrezP, WiederholtT, KonigA, JugertF, MarquardtY, et al. (2004) Allelic loss underlies type 2 segmental Hailey-Hailey disease, providing molecular confirmation of a novel genetic concept. J Clin Invest 114 : 1407–1409.
19. OkunadeGW, MillerML, AzharM, AndringaA, SanfordL, et al. (2007) Loss of the Atp2c1 Secretory Pathway Ca2+-ATPase (SPCA1) in mice causes Golgi stress, apoptosis, and midgestational death in homozygous embryos and squamous cell tumors in adult heterozygotes. J Biol Chem 282 : 26517–26527.
20. ChunSI, WhangKC, SuWP (1988) Squamous cell carcinoma arising in Hailey-Hailey disease. J Cutan Pathol 15 : 234–237.
21. CockayneSE, RasslDM, ThomasSE (2000) Squamous cell carcinoma arising in Hailey-Hailey disease of the vulva. Br J Dermatol 142 : 540–542.
22. HolstVA, FairKP, WilsonBB, PattersonJW (2000) Squamous cell carcinoma arising in Hailey-Hailey disease. J Am Acad Dermatol 43 : 368–371.
23. MetzeD, HammH, SchoratA, LugerT (1995) Involvement of the adherens junction-actin filament system in acantholytic dyskeratosis of Hailey-Hailey disease. A histological, ultrastructural, and histochemical study of lesional and non-lesional skin. J Cutan Pathol 23 : 211–222.
24. HashimotoK, FujiwaraK, TadaJ, HaradaM, SetoyamaM, et al. (1995) Desmosomal dissolution in Grover's disease, Hailey-Hailey's disease and Darier's disease. J Cutan Pathol 1995 : 488–501.
25. ShullG, MillerML, PrasadV (2011) Secretory pathway stress responses as possible mechanisms of disease involving Golgi Ca2+ pump dysfunction. Biofactors 37 : 150–158.
26. BaylisH, Vazquez-ManriqueR (2012) Genetic analysis of IP3 and calcium signaling pathways in C. elegans. Biochimica et Biophysica Acta General subjects 1280 : 1253–1268.
27. Thomas-VirnigCL, SimsP, SimskeJS, HardinJ (2004) The Inositol 1,4,5-Trisphosphate Receptor Regulates Epidermal Cell Migration in Caenorhabditis elegans. Curr Biol 14 : 1882–1887.
28. WalkerDS, GowerNJ, LyS, BradleyGL, BaylisHA (2002) Regulated disruption of inositol 1,4,5-trisphosphate signaling in Caenorhabditis elegans reveals new functions in feeding and embryogenesis. Mol Biol Cell 1329–1337.
29. MaryonEB, SaariB, AndersonP (1998) Muscle-specific functions of ryanodine receptor channels in Caenorhabditis elegans. J Cell Sci 111 : 2885–2895.
30. SakubeY, AndoH, KagawaH (1997) An abnormal ketamine response in mutants defective in the ryanodine receptor gene ryr-1 (unc-68) of Caenorhabditis elegans. J of Mol Biol 267 : 849–864.
31. LiuQ, ChenB, YankovaM, MorestK, MaryonEB, et al. (2005) Presynaptic Ryanodine Receptors are required for normal quantal size at the Caenorhabditis elegans neuromuscular junction. J Neuroscience 25 : 6745–6754.
32. ZalkR, LehnartSE, MarksAR (2007) Modulation of the Ryanodine Receptor and intracellular calcium. Ann Rev Biochem 76 : 367–385.
33. Van BaelenK, VanoevelenJ, MissiaenL, WuytackF (2001) The Golgi pmr1 p-type ATPase of Caenorhabditis elegans. J Biol Chem 276 : 10683–10691.
34. MissiaenL, Van AckerK, ParysJB, De SmedtH, Van BaelenK, et al. (2001) Baseline cytosolic Ca2+ oscillations derived from a non-endoplasmic reticulum Ca2+ store. J Biol Chem 276 : 39161–39170.
35. ChoJ, KoK, GunasekaranS, AhnnJ (2005) Caenorhabditis elegans PMR1, a P-type Calcium ATPase is important for calcium/manganese homeostasis and oxidative stress response. FEBS Letters 578 : 778–782.
36. CelnikerSE, DillonLAL, GersteinMB, GunsalusKC, HenikoffS, et al. (2009) Unlocking the secrets of the genome. Nature 459 : 927–930.
37. TabaraH, MotohashiT, KoharaY (1996) A multi-well version of in situ hybridization on whole mount embryos of Caenorhabditis elegans. Nucl Acids Res 24 : 2119–2124.
38. LevinM, HashimshonyT, WagnerF, YanaiI (2012) Developmental milestones punctuate gene expression in the Caenorhabditis embryo. Dev Cell 22 : 1101–1108.
39. Hunt-NewburyR, ViveirosR, JohnsenR, MahA, AnastasD, et al. (2007) High-throughput in vivo analysis of gene expression in Caenorhabditis elegans. PLoS Biol 5: e237 doi:10.1371/journal.pbio.0050237.
40. KellyWG, XuS, MontgomeryMK, FireA (1997) Distinct requirements for somatic and germline expression of a generally expressed Caenorhabditis elegans gene. Genetics 146 : 227–238.
41. BandyopadhyayJ, SongH, ParkB, SingaraveluG, SunJ, et al. (2009) Functional assessment of Nramp-like metal transporters and manganese in Caenorhabditis elegans. Biochem Biophys Res Commun 390 : 136–141.
42. KourtisN, NikoletopoulouV, TavernarakisN (2012) Small heat-shock proteins protect from heat-stroke-associated neurodegeneration. Nature 490 : 213–218.
43. Gengyo-AndoK, MitaniS (2000) Characterization of mutations induced by ethyl methanesulfonate, UV, and trimethylpsoralen in the nematode Caenorhabditis elegans. Biochem Biophysica Res Comm 269 : 64–69.
44. ToyoshimaC, MizutaniT (2004) Crystal structure of the calcium pump with a bound ATP analogue. Nature 430 : 529–535.
45. SonnichsenB, KoskiLB, WalshA, MarschallP, NeumannB, et al. (2005) Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature 434 : 462–469.
46. Wormbase (2012) http://www.wormbase.org/db/get?name=WBGene00004063class=Gene.
47. SulstonJE, SchierenbergE, WhiteJG, ThomsonJN (1983) The embryonic cell lineage of the nematode Caenorhabditis elegans. Developmental Biology 100 : 64–119.
48. Williams-MassonEM, HeidPJ, LavinCA, HardinJ (1998) The cellular mechanism of epithelial rearrangement during morphogenesis of the Caenorhabditis elegans dorsal hypodermis. Developmental Biology 204 : 263–276.
49. PortereikoM, MangoS (2001) Early morphogenesis of the Caenorhabditis elegans pharynx. Dev Biol 233 : 482–494.
50. BaoZ, MurrayJI, BoyleT, OoiSL, SandelMJ, et al. (2006) Automated cell lineage tracing in Caenorhabditis elegans. PNAS 2707–2712.
51. BoyleTJ, BaoZ, MurrayJI, ArayaCL, WaterstonRH (2006) AceTree: a tool for visual analysis of Caenorhabditis elegans embryogenesis. BMC Bioinformatics 7 : 275.
52. MurrayJI, BaoZ, BoyleTJ, BoeckME, MericleBL, NicholasTJ, et al. (2008) Automated analysis of embryonic gene expression with cellular resolution in C. elegans. Nature Methods 5 : 703–709.
53. MurrayJI, BaoZ, BoyleT, WaterstonRH (2006) The lineaging of fluorescently-labeled Caenorhabditis elegans embryos with StarryNite and AceTree. Nature Protocols 1 : 1468–1476.
54. AhringerJ (1996) Posterior patterning by the Caenorhabditis elegans even-skipped homolog vab-7. Genes Dev 10 : 1120–1130.
55. WittmannC, BossingerO, GoldsteinB, FleischmannM, KohlerR, et al. (1997) The expression of the C. elegans labial-like Hox gene ceh-13 during early embryogenesis relies on cell fate and on anteroposterior cell polarity. Development 124 : 4193–4200.
56. CassataG, ShemerG, MorandiP, DonhauserR, PodbilewiczB (2005) ceh-16/engrailed patterns the embryonic epidermis of Caenorhabditis elegans. Development 132 : 739–749.
57. BrunschwigK, WittmanC, SchnabelR, BurglinT, ToblerH, et al. (1999) Anterior organization of the C. elegans embryo by the labial-like Hox gene ceh-13. Development 126 : 1537–1546.
58. Nance, J. et al.. Gastrulation in C. elegans (September 26, 2005), WormBook, ed. The C. elegans Research Community, WormBook, doi/10.1895/wormbook.1.23.1, http://www.wormbook.org.
59. RohrschneiderM, NanceJ (2009) Polarity and cell fate specification in the control of Caenorhabditis elegans gastrulation. Dev Dynamics 238 : 789–796.
60. HarrellJB, GoldsteinB (2011) Internalization of multiple cells during C. elegans gastrulation depends on common cytoskeletal mechanisms but different cell polarity and cell fate regulators. Dev Biol 350 : 1–12.
61. NanceJ, MunroEM, PriessJ (2003) C. elegans PAR-3 and PAR-6 are required for apicobasal asymmetries associated with cell adhesion and gastrulation. Development 130 : 5339–5350.
62. PraitisV, CicconeE, AustinJ (2005) SMA-1 spectrin has essential roles in epithelial cell sheet morphogenesis in C. elegans. Dev l Biol 283 : 157–170.
63. KangS, KramerJ (2000) Nidogen is nonessential and not required for normal type IV collagen localization in Caenorhabditis elegans. Mol Biol Cell 3911–3923.
64. TrzebiatowskaA, TopfU, SauderU, DrabikowskiK, Chiquet-EhrismannR (2008) Caenorhabditis elegans teneurin, ten-1, is required for gonadal and pharyngeal basement membrane integrity and acts redundantly with integrin ina-1 and dystroglycan dgn-1. Mol Biol Cell 19 : 3898–3908.
65. SimskeJS, KoppenM, SimsP, HodgkinJ, YonkofA, et al. (2003) The cell junction protein VAB-9 regulates adhesion and epidermal morphology in C. elegans. Nat Cell Biol 5 : 619–625.
66. KoppenM, SimskeJS, SimsPA, FiresteinBL, HallDH, et al. (2001) Cooperative regulation of AJM-1 controls junctional integrity in Caenorhabditis elegans epithelia. Nat Cell Biol 3 : 983–991.
67. PieknyAJ, MainsPE (2002) Rho-binding kinase (LET-502) and myosin phosphatase (MEL-11) regulate cytokinesis in the early Caenorhabditis elegans embryo. J Cell Sci 115 : 2271–2282.
68. PieknyAJ, JohnsonJL, ChamGD, MainsPE (2003) The Caenorhabditis elegans nonmuscle myosin genes nmy-1 and nmy-2 function as redundant components of the let-502/Rho-binding kinase and mel-11/myosin phosphatase pathway during embryonic morphogenesis. Development 130 : 5695–5704.
69. NakaoF, HudsonM, SuzukiM, PecklerZ, KurokawaR, et al. (2007) The plexin PLX-2 and the ephrin EFN-4 have distinct roles in MAB-20/Semaphorin 2A signaling in C. elegans morphogenesis. Genetics 176 : 1591–1607.
70. RugarliE, Di SchiaviE, HilliardM, ArbucciS, GhezziC, et al. (2002) The Kallmann syndrome gene homolog in C. elegans is involved in epidermal morphogenesis and neurite branching. Development 129 : 1283–1294.
71. ClandininT, DeModenaJ, SternbergP (1998) Inositol trisphosphate mediates a RAS-independent response to LET-23 receptor tyrosine kinase activation in C. elegans. Cell 92 : 523–533.
72. EspeltMV, EstevezAY, YinX, StrangeK (2005) Oscillatory calcium signaling in the isolated Caenorhabditis elegans intestine: Role of the Inositol-1,4,5-trisphosphate Receptor and Phospholipases C beta and gamma. J Gen Physiol 126 : 379–392.
73. FoskettJK, WhiteC, CheungK-H, MakD-OD (2007) Inositol Trisphosphate Receptor Ca2 Release Channels. Physiol Rev 87 : 593–658.
74. Van PetegemF (2012) Ryanodine Receptors: Structure and function. J Biol Chem 287 : 31624–31632.
75. Stiernagle, T. Maintenance of C. elegans (February 11, 2006), WormBook, ed. The C. elegans Research Community, WormBook, doi/10.1895/wormbook.1.101.1, http://www.wormbook.org.
76. KarabinosA, SchmidtH, HarborthJ, SchnabelR, WeberK (2001) Essential roles for four cytoplasmic intermediate filament proteins in Caenorhabditis elegans development. Proc Natl Acad Sci U S A 98 : 7863–7868.
77. HapiakV, HreskoMC, SchrieferLA, SaiyasisongkhramK, BercherM, et al. (2003) mua-6, a gene required for tissue integrity in Caenorhabditis elegans, encodes a cytoplasmic intermediate filament. Developmental Biology 263 : 330–342.
78. WooWM, GoncharovA, JinY, ChisholmAD (2004) Intermediate filaments are required for C. elegans epidermal elongation. Dev Biol 267 : 216–229.
79. CarberryK, WiesenfahrtT, WindofferR, BossingerO, LeubeRE (2009) Intermediate Filaments in Caenorhabditis elegans. Cell Motility and the Cytoskeleton 66 : 852–864.
80. DingM, GoncharovA, JinY, ChisholmAD (2003) C. elegans ankyrin repeat protein VAB-19 is a component of epidermal attachment structures and is essential for epidermal morphogenesis. Development 130 : 5791–5801.
81. BosherJM, HahnBS, LegouisR, SookhareeaS, WeimerRM, et al. (2003) The Caenorhabditis elegans vab-10 spectraplakin isoforms protect the epidermis against internal and external forces. J Cell Biol 161 : 757–768.
82. HreskoMC, SchrieferLA, ShrimankarP, WaterstonRH (1999) Myotactin, a novel hypodermal protein involved in muscle-cell adhesion in Caenorhabditis elegans. J Cell Biol 146 : 659–672.
83. HongL, ElblT, WardJ, Franzini-ArmstrongC, RybickaKK, et al. (2001) MUP-4 is a novel transmembrane protein with functions in epithelial cell adhesion in Caenorhabditis elegans. J Cell Biol 154 : 403–414.
84. FuchsE, RaghavanS (2002) Getting under the skin of epidermal morphogenesis. Nature Reviews, Genetics 3 : 199–209.
85. BrennerS (1974) The genetics of Caenorhabditis elegans. Genetics 77 : 71–94.
86. PraitisV, CaseyE, CollarD, AustinJ (2001) Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics 157 : 1217–1226.
87. Fay, D. Genetic mapping and manipulation: Chapter 5-SNPs: Three-point mapping (February 17, 2006), WormBook, ed. The C. elegans Research Community, WormBook, doi/10.1895/wormbook.1.94.1, http://www.wormbook.org.
88. WilliamsBD, SchrankB, HuynhC, ShownkeenR, WaterstonRH (1992) A genetic mapping system in C. elegans based on polymorphic sequence-tagged sites. Genetics 131 : 609–624.
89. WicksS, YehR, GishW, WaterstonR, PlasterkR (2001) Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nature Genetics 28 : 160–164.
90. Fay, D. Genetic mapping and manipulation: Chapter 5-SNPs: Three-point mapping (February 17, 2006), WormBook, ed. The C. elegans Research Community, WormBook, doi/10.1895/wormbook.1.94.1, http://www.wormbook.org.
91. BigelowH, DoitsidouM, SarinS, HobertO (2009) MAQGene: Software to facilitate C. elegans mutant genome sequence analysis. Nature Methods 6 : 549.
92. MelloCC, KramerJM, StinchcombD, AmbrosV (1991) Efficient gene transfer in C. elegans : extrachromosomal maintanance and integration of transforming sequences. EMBO Journal 10 : 3959–3970.
93. EvansTC, CrittendenSL, KoyoyianniV, KimbleJ (1994) Translational control of maternal glp-1 mRNA establishes an asymmetry in the C. elegans embryo. Cell 77 : 183–194.
94. McKeownC, PraitisV, AustinJ (1998) sma-1 encodes a BH-spectrin homolog required for Caenorhabditis elegans morphogenesis. Development 125 : 2087–2098.
95. Ahringer, J., ed. Reverse genetics (April 6, 2006), WormBook, ed. The C. elegans Research Community, WormBook, doi/10.1895/wormbook.1.47.1, http://www.wormbook.org.
96. HadwigerG, DourS, ArurS, FoxP, NonetM (2010) A monoclonal antibody toolkit for C. elegans. PLoS ONE 5: e10161 doi:10.1371/journal.pone.0010161.
97. Sulston, J E and White, J G (1988) “Parts list”. In: Wood, W. B., Wood, W. B.s. The Nematode Caenorhabditis elegans. Cold Spring Harbor, New York, USA: Cold Spring Harbor Laboratory Press. pp. 415–431.
98. WormAtlas, Altun, Z.F., Herndon, L.A., Crocker, C., Lints, R.and Hall, D.H.(eds); 2002–22012. http://www.wormatlas.org
99. MohlerWA, SimskeJS, Williams-MassonEM, HardinJD, WhiteJG (1998) Dynamics and ultrastructure of developmental cell fusions in the Caenorhabditis elegans hypodermis. Curr Biol 8 : 1087–1090.
100. KrauseM, HarrisonSW, XuSQ, ChenL, FireA (1994) Elements regulating cell - and stage-specific expression of the C. elegans MyoD family homolog hlh-1. Developmental Biology 166 : 133–148.
101. ChenLM, KrauseM, Sepanski, FireA (1994) The Caenorhabditis elegans MYOD homologue HLH-1 is essential for proper muscle function and complete morphogenesis. Development 120 : 1631–1640.
102. KingRS, MaidenSL, HawkinsNC, KiddAR, KimbleJ, et al. (2009) The N - or C-terminal domains of DSH-2 can activate the C. elegans Wnt/beta-catenin asymmetry pathway. Developmental Biology 238 : 234–244.
103. ChalfieM, TuY, EuskirchenG, WardWW, PrasherDC (1994) Green fluorescent protein as a marker for gene expression. Science 263 : 802–805.
104. Altun-GultekinZF, AndachiY, TsalikEL, PilgrimD, KoharaY, et al. (2001) A regulatory cascade of three homeobox genes, ceh-10, ttx-3 and ceh-23, controls cell fate specification of a defined interneuron class in C. elegans. Development 128 : 1951–1969.
105. IkegamiR, SimokatK, ZhengH, BrownL, GarrigaG, et al. (2012) Semaphorin and Eph receptor signaling guide a series of cell movements for ventral enclosure in C. elegans. Current Biology 22 : 1–11.
106. GranaTM, CoxEA, LynchAM, HardinJ (2010) SAX-7/L1CAM and HMR-1/cadherin function redundantly in blastomere compaction and non-muscle myosin accumulation during Caenorhabditis elegans gastrulation. Developmental Biology 344 : 731–744.
Štítky
Genetika Reprodukční medicína
Článek Attachment Site Selection and Identity in Bxb1 Serine Integrase-Mediated Site-Specific RecombinationČlánek Bck2 Acts through the MADS Box Protein Mcm1 to Activate Cell-Cycle-Regulated Genes in Budding YeastČlánek High-Resolution Transcriptome Maps Reveal Strain-Specific Regulatory Features of Multiple IsolatesČlánek Neuropeptides Function in a Homeostatic Manner to Modulate Excitation-Inhibition Imbalance inČlánek Implicates Tyrosine-Sulfated Peptide Signaling in Susceptibility and Resistance to Root Infection
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 5- Akutní intermitentní porfyrie
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Růst a vývoj dětí narozených pomocí IVF
- IVF a rakovina prsu – zvyšují hormony riziko vzniku rakoviny?
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
-
Všechny články tohoto čísla
- Functional Elements Are Embedded in Structurally Constrained Sequences
- RNA–Mediated Epigenetic Heredity Requires the Cytosine Methyltransferase Dnmt2
- Loss of Expression and Promoter Methylation of SLIT2 Are Associated with Sessile Serrated Adenoma Formation
- Attachment Site Selection and Identity in Bxb1 Serine Integrase-Mediated Site-Specific Recombination
- Human Genetics in Rheumatoid Arthritis Guides a High-Throughput Drug Screen of the CD40 Signaling Pathway
- Genome-Wide Analysis in German Shepherd Dogs Reveals Association of a Locus on CFA 27 with Atopic Dermatitis
- Liver X Receptors Protect from Development of Prostatic Intra-Epithelial Neoplasia in Mice
- Chromosomal Organization and Segregation in
- A Statistical Framework for Joint eQTL Analysis in Multiple Tissues
- Cell Polarity and Patterning by PIN Trafficking through Early Endosomal Compartments in
- Bck2 Acts through the MADS Box Protein Mcm1 to Activate Cell-Cycle-Regulated Genes in Budding Yeast
- High-Resolution Transcriptome Maps Reveal Strain-Specific Regulatory Features of Multiple Isolates
- Neuropeptides Function in a Homeostatic Manner to Modulate Excitation-Inhibition Imbalance in
- A Compendium of Nucleosome and Transcript Profiles Reveals Determinants of Chromatin Architecture and Transcription
- Wnt Signaling Regulates the Lineage Differentiation Potential of Mouse Embryonic Stem Cells through Tcf3 Down-Regulation
- Filamin and Phospholipase C-ε Are Required for Calcium Signaling in the Spermatheca
- The Specificity and Flexibility of L1 Reverse Transcription Priming at Imperfect T-Tracts
- Imputation-Based Meta-Analysis of Severe Malaria in Three African Populations
- Implicates Tyrosine-Sulfated Peptide Signaling in Susceptibility and Resistance to Root Infection
- Clathrin and AP2 Are Required for Phagocytic Receptor-Mediated Apoptotic Cell Clearance in
- Encodes CDF Transporters That Excrete Zinc from Intestinal Cells of and Act in a Parallel Negative Feedback Circuit That Promotes Homeostasis
- Global Properties and Functional Complexity of Human Gene Regulatory Variation
- DNA Binding of the Cell Cycle Transcriptional Regulator GcrA Depends on N6-Adenosine Methylation in and Other
- Side Effects: Substantial Non-Neutral Evolution Flanking Regulatory Sites
- From Paramutation to Paradigm
- From Mouse to Human: Evolutionary Genomics Analysis of Human Orthologs of Essential Genes
- Distinct Translational Control in CD4 T Cell Subsets
- Female Bias in and Regulation by the Histone Demethylase KDM6A
- ATM–Dependent MiR-335 Targets CtIP and Modulates the DNA Damage Response
- HDAC7 Is a Repressor of Myeloid Genes Whose Downregulation Is Required for Transdifferentiation of Pre-B Cells into Macrophages
- The Majority of Primate-Specific Regulatory Sequences Are Derived from Transposable Elements
- Identification of Meiotic Cyclins Reveals Functional Diversification among Plant Cyclin Genes
- EGL-13/SoxD Specifies Distinct O and CO Sensory Neuron Fates in
- Congruence of Additive and Non-Additive Effects on Gene Expression Estimated from Pedigree and SNP Data
- Using Extended Genealogy to Estimate Components of Heritability for 23 Quantitative and Dichotomous Traits
- Ikbkap/Elp1 Deficiency Causes Male Infertility by Disrupting Meiotic Progression
- Analysis of the Genetic Basis of Disease in the Context of Worldwide Human Relationships and Migration
- Duplication and Retention Biases of Essential and Non-Essential Genes Revealed by Systematic Knockdown Analyses
- Strong Purifying Selection at Synonymous Sites in
- , a Susceptibility Gene for Type 1 and Type 2 Diabetes, Modulates Pancreatic Beta Cell Apoptosis via Regulation of a Splice Variant of the BH3-Only Protein
- Chromosome Movements Promoted by the Mitochondrial Protein SPD-3 Are Required for Homology Search during Meiosis
- The Secretory Pathway Calcium ATPase PMR-1/SPCA1 Has Essential Roles in Cell Migration during Embryonic Development
- The Genomic Signature of Crop-Wild Introgression in Maize
- CDK4 T172 Phosphorylation Is Central in a CDK7-Dependent Bidirectional CDK4/CDK2 Interplay Mediated by p21 Phosphorylation at the Restriction Point
- Genome-Wide Identification of Regulatory RNAs in the Human Pathogen
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Using Extended Genealogy to Estimate Components of Heritability for 23 Quantitative and Dichotomous Traits
- HDAC7 Is a Repressor of Myeloid Genes Whose Downregulation Is Required for Transdifferentiation of Pre-B Cells into Macrophages
- Female Bias in and Regulation by the Histone Demethylase KDM6A
- ATM–Dependent MiR-335 Targets CtIP and Modulates the DNA Damage Response
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



