-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaBck2 Acts through the MADS Box Protein Mcm1 to Activate Cell-Cycle-Regulated Genes in Budding Yeast
The Bck2 protein is a potent genetic regulator of cell-cycle-dependent gene expression in budding yeast. To date, most experiments have focused on assessing a potential role for Bck2 in activation of the G1/S-specific transcription factors SBF (Swi4, Swi6) and MBF (Mbp1, Swi6), yet the mechanism of gene activation by Bck2 has remained obscure. We performed a yeast two-hybrid screen using a truncated version of Bck2 and discovered six novel Bck2-binding partners including Mcm1, an essential protein that binds to and activates M/G1 promoters through Early Cell cycle Box (ECB) elements as well as to G2/M promoters. At M/G1 promoters Mcm1 is inhibited by association with two repressors, Yox1 or Yhp1, and gene activation ensues once repression is relieved by an unknown activating signal. Here, we show that Bck2 interacts physically with Mcm1 to activate genes during G1 phase. We used chromatin immunoprecipitation (ChIP) experiments to show that Bck2 localizes to the promoters of M/G1-specific genes, in a manner dependent on functional ECB elements, as well as to the promoters of G1/S and G2/M genes. The Bck2-Mcm1 interaction requires valine 69 on Mcm1, a residue known to be required for interaction with Yox1. Overexpression of BCK2 decreases Yox1 localization to the early G1-specific CLN3 promoter and rescues the lethality caused by overexpression of YOX1. Our data suggest that Yox1 and Bck2 may compete for access to the Mcm1-ECB scaffold to ensure appropriate activation of the initial suite of genes required for cell cycle commitment.
Published in the journal: . PLoS Genet 9(5): e32767. doi:10.1371/journal.pgen.1003507
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003507Summary
The Bck2 protein is a potent genetic regulator of cell-cycle-dependent gene expression in budding yeast. To date, most experiments have focused on assessing a potential role for Bck2 in activation of the G1/S-specific transcription factors SBF (Swi4, Swi6) and MBF (Mbp1, Swi6), yet the mechanism of gene activation by Bck2 has remained obscure. We performed a yeast two-hybrid screen using a truncated version of Bck2 and discovered six novel Bck2-binding partners including Mcm1, an essential protein that binds to and activates M/G1 promoters through Early Cell cycle Box (ECB) elements as well as to G2/M promoters. At M/G1 promoters Mcm1 is inhibited by association with two repressors, Yox1 or Yhp1, and gene activation ensues once repression is relieved by an unknown activating signal. Here, we show that Bck2 interacts physically with Mcm1 to activate genes during G1 phase. We used chromatin immunoprecipitation (ChIP) experiments to show that Bck2 localizes to the promoters of M/G1-specific genes, in a manner dependent on functional ECB elements, as well as to the promoters of G1/S and G2/M genes. The Bck2-Mcm1 interaction requires valine 69 on Mcm1, a residue known to be required for interaction with Yox1. Overexpression of BCK2 decreases Yox1 localization to the early G1-specific CLN3 promoter and rescues the lethality caused by overexpression of YOX1. Our data suggest that Yox1 and Bck2 may compete for access to the Mcm1-ECB scaffold to ensure appropriate activation of the initial suite of genes required for cell cycle commitment.
Introduction
The temporal control of transcription is likely a universal feature of cell cycles, with clear transcriptional programs in yeast, bacteria and metazoans [1]–[4]. Bursts of gene expression in eukaryotes tend to be associated with major cell cycle transitions that are governed by cyclin-dependent kinases (Cdks) in association with regulatory subunits called cyclins [5]. Up-regulation of G1-specific forms of the Cdc28 Cdk is rate-limiting for cell cycle commitment. Control over Cdc28 activity is exerted at several levels, including transcriptional induction of cyclin gene expression; the G1/S phase transition features activation of a massive transcriptional program of ∼120 genes, including the G1 cyclin genes CLN1 and CLN2 [6]–[10]. Two heterodimeric transcription factors largely drive the G1/S cluster: SBF (Swi4,6-dependent cell cycle box binding factor, a heterodimer of Swi4 and Swi6) binds the so-called SCB promoter element (Swi4,6-dependent cell cycle box) found upstream of the cyclin genes and cell wall biosynthetic genes, while MBF (MluI cell cycle box binding factor, a heterodimer of Mbp1 and Swi6) preferentially acts through a distinct element called the MCB (MluI cell cycle box) found mostly upstream of DNA replication and repair genes.
Although the role of SBF and MBF at the G1/S transition is well established, deletion of Swi6, the common subunit of both complexes, does not cause cell cycle arrest, suggesting alternative pathways for activating G1 transcription. One of these pathways is defined by Bck2, a poorly understood cell cycle regulator whose deletion causes dramatic cell cycle phenotypes in certain genetic contexts. For example, deletion of both CLN3, which encodes a cyclin that activates Cdc28 at M/G1, and BCK2 causes synthetic lethality [11]–[13]. Furthermore, BCK2 is one of only two genes whose overexpression is known to bypass the lethality caused by mutation of all three G1-specific Cdc28 cyclins (CLN1, CLN2 and CLN3) [11]. Bck2 was discovered almost 20 years ago in screens for high copy suppressors of protein kinase C pathway mutants (including pkc1 itself) and G1 cyclin deficiencies [11], [14] and is thought to have an activating role in transcription of G1/S genes [12], [13]. Activation of G1/S genes by Bck2 depends partly, but not wholly, on SBF and MBF and Bck2 appears capable of activating G1/S transcription in the absence of Cdc28, indicating SBF - and MBF-independence [13]. Furthermore, a region in the promoter of CLN2, termed UAS2, which completely lacks SCB or MCB elements, is activated by BCK2 overexpression [12] and operates in a CDC28-independent manner [15]. Bck2 also has a role in regulating heat shock genes and adaptation of the protein kinase C pathway MAP kinase Slt2 [16], although it is not clear whether these roles depend on SBF. Collectively, these findings suggest that Bck2 may operate through an unidentified DNA-binding factor whose activity is Cdc28-independent.
The MADS box transcription factor Mcm1 has an important regulatory function at two points in the cell cycle – M/G1 and G2/M. During M/G1, Mcm1 functions as a critical constituent of a complex that forms on ECB (Early Cell cycle Box) elements in promoters of genes expressed at the M/G1 phase transition such as CLN3 and SWI4 [17]. Two related homeodomain proteins, Yox1 and Yhp1, act as repressors of Mcm1 on ECB elements by physically interacting with Mcm1 and with DNA binding sites next to the Mcm1 site in the ECB element [18]. Mcm1 is also a critical constituent of complexes that form during the G2/M phase transition to control the CLB2 cluster of genes, including the G2 cyclin gene CLB2 and CDC20, which encodes an activator of the anaphase promoting complex [19]. The CLB2 gene cluster is activated by the Clb-Cdc28 and Cdc5 kinases [20]–[23] and negatively regulated by protein kinase C, Pkc1 [24]; these kinases regulate a promoter-bound complex of Mcm1, Fkh2 and Ndd1 [25]–[29]. Some CLB2 cluster genes, but not CLB2 itself, contain hybrid elements composed of an Mcm1-binding site flanked by Yox1 - and Fkh2-binding sites [18]. In CLB2 cluster genes that contain such elements, Yox1 and Fkh2 compete for binding to DNA-bound Mcm1 despite the spatial separation of their DNA recognition elements [30]. Interestingly, no such juxtaposed binding motifs are obvious in the vicinity of ECB elements and no other binding partners for Mcm1 that positively regulate these genes have been identified. Mcm1 also has roles at the promoters of some other genes, including the G1/S gene CLN2, where it contributes to the presence of a nucleosome depleted region that is needed for reliable “on/off” expression once per cell cycle [31].
In this study, we show that Bck2 activates expression of the M/G1 genes CLN3 and SWI4, through an interaction with Mcm1 on ECB elements in the promoters of these genes. Moreover, increased Bck2 dosage leads to decreased Yox1 binding at M/G1 promoters and overproduction of BCK2 rescues the lethality caused by overexpression of YOX1, indicating that Bck2 and Yox1 may compete for access to Mcm1 on promoters. Consistent with this hypothesis, mutation of a key residue on Mcm1 known to prevent interaction with Yox1 also prevents interaction with Bck2. In addition, we show that Bck2 interacts with the promoters of the G1/S gene CLN2 and the G2/M gene CLB2. Our experiments reveal a previously unappreciated function for Bck2 as a cofactor for Mcm1 and suggest a more general role for Bck2 in regulating cell-cycle gene expression.
Results
Truncation Analysis of the BCK2 Gene
We reasoned that an exploration of Bck2-interacting proteins might illuminate the function of Bck2 in G1 progression. Bck2 is difficult to work with biochemically (for example endogenous Bck2 is undetectable by Western blot [16], [32, and data not shown], so we turned to the yeast two-hybrid (Y2H) system for our protein interaction screens. Bck2 has a potent transcriptional activation domain, and activates reporter gene expression when fused to the Gal4 DNA binding domain (DBD) in a Y2H reporter strain [33] (Figure 1), a property that has precluded identification of Bck2 binding partners using the Y2H screening method. To discover Bck2 derivatives that might be useful for two-hybrid screening, we created 20 truncations of the BCK2 gene and fused them to the GAL4 DBD (Figure 1). We first assessed the ability of each truncation construct to activate transcription of two reporter genes, in which the GAL4 UAS is upstream of either lacZ (Figure S1) or ADE2 (Figure S1B). Bck2 residues 662 to 851 were required for transcriptional activation (fragments 11–20), while the Bck2 N-terminal region had some apparent inhibitory activity (Figure S1A), as suggested by our finding that deletions of the N-terminus resulted in significant increases in reporter gene expression. A construct containing fragment 5, lacking the first 529 amino acids of Bck2, was the most potent Y2H auto-activator in the lacZ reporter assay (Figure S1A).
Fig. 1. Truncation analysis of the BCK2 gene. 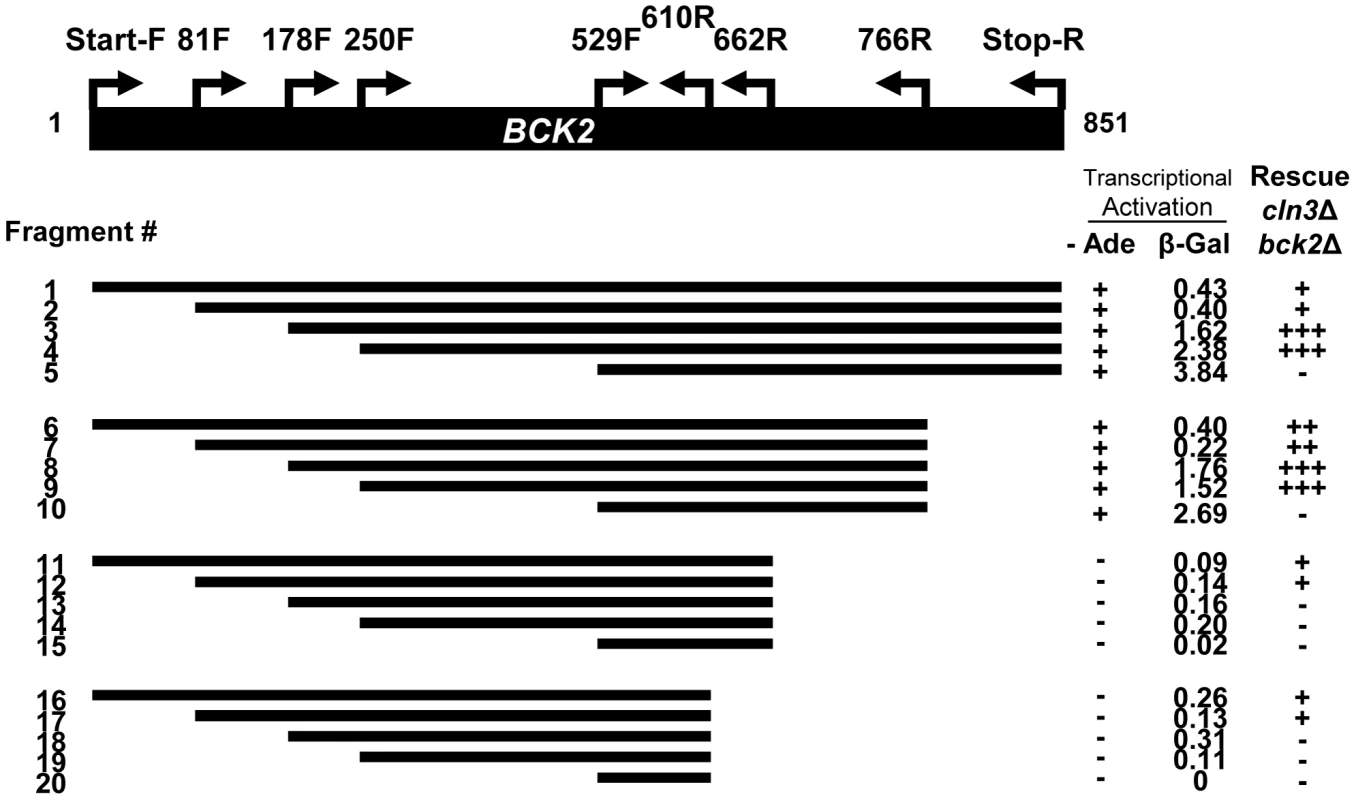
Fragments of the BCK2 gene (black bar) were amplified from genomic DNA using primer positions shown and designated as “amino acid+F (Forward), or R (reverse)”. PCR products were cloned into a yeast two-hybrid vector to create BCK2 fragments fused to the N-terminal GAL4 DBD (DNA Binding Domain). High density growth spots (in either the ADE2 transcription activation assay or the complementation assay) were called “+”, “++”, or “+++” depending on extent of growth. A complete absence of growth was called “−”. Numbers in the β-Gal column represent averaged quantities (per fusion protein) in Miller Units (U). Proteins that autoactivate in the Y2H system often have a role in transcription and this property may reflect the biological activity of the protein [33]. To explore the relationship between Y2H autoactivation and biological function, we assessed the ability of each Bck2 fragment to complement the synthetic lethal phenotype of a cln3Δbck2Δ strain (Figure S1C). We discovered that a fragment of Bck2 containing residues 250 to 766 was able to robustly complement the lethality of the cln3Δbck2Δ strain (Figure 1, Figure S1C). Consistent with the observation that the first 178 residues of Bck2 are not necessary for suppression of the pkc1 lysis phenotype by high-copy BCK2 [14], a derivative of Bck2 lacking the N-terminal 178 residues was also able to complement the inviability of the cln3Δbck2Δ strain (Figure S1C). Bck2 residues 529 to 851 alone failed to complement the cln3Δbck2Δ phenotype but were sufficient for Y2H auto-activation. This region is also insufficient to suppress the lysis phenotype of pkc1 mutants [14]. We conclude that the central region (250 to 766) of Bck2 lacking the N - and C-terminal ends is sufficient to complement essential in vivo functions of Bck2 and that this essential function is separable from the Y2H auto-activation activity of the Bck2 protein.
A Yeast-Two-Hybrid Screen Using Gal4 DBD-Bck2 as Bait Identifies Six Novel Interacting Proteins
Thus far, no known protein interaction partners of Bck2 easily explain the cell cycle transcription phenotypes associated with deletion of BCK2. To carry out a Y2H screen, we decided to use the largest Bck2 construct that did not auto-activate the ADE2 Y2H reporter gene, but complemented the inviability of the cln3Δbck2Δ strain (fragment 11; Figure 1). We chose this fragment for our screen since complementing regions often contain important protein-protein interaction domains. For example, the minimal region of the Ada2 protein required for complementation is the same region required for physical interaction with Gcn5 and Ada3 [34].
We used the ORFeome Y2H screening method [35] to discover potential Bck2-interacting proteins. We identified six proteins that interacted with Bck2: Mcm1, Yap6, Tpd3, Std1, Mth1, and Mot3 (Figure 2 and data not shown). With the exception of Tpd3, all of these proteins are transcriptional regulators that act proximal to DNA. As noted in the introduction, Mcm1 is a member of a class of MADS box transcription factors found in all eukaryotic organisms [36]–[38] and we explore the Mcm1-Bck2 interaction in detail below. Four of the other Bck2 partners we identified in our screen were also DNA binding proteins that had roles in ion homeostasis and nutrient sensing: (1) Yap6 is a basic leucine zipper (bZIP) transcription factor that activates a number of genes involved in sodium and lithium tolerance [39], [40]; (2) Std1 and Mth1 are both controllers of glucose-regulated gene expression [41] and are required for transcriptional repression of HXT (hexose transport) genes [42], [43]; (3) Mot3 is a Zn-finger transcription factor that activates a number of cell wall genes [44], and represses transcription of the DAN/TIR group of genes that encode cell wall mannoproteins during anaerobic growth [44]. The only protein that is not a transcription factor, Tpd3, is the scaffold subunit A of the heterotrimeric protein phosphatase 2A (PP2A) [45], which has roles at several points in the cell cycle and is involved in the TOR pathway for nutrient sensing [46]. We did not identify SWI4 or MBP1 in our Y2H screen, but a direct test revealed a weak interaction between Bck2 and Swi4 (Figure 2). The identification of genes with known roles in transcription and nutrient response is consistent with the apparent functions of Bck2, and validates our two-hybrid screen as a tool for identifying Bck2-interacting proteins.
Fig. 2. Bck2-interacting proteins identified in a genome-wide yeast two-hybrid screen. 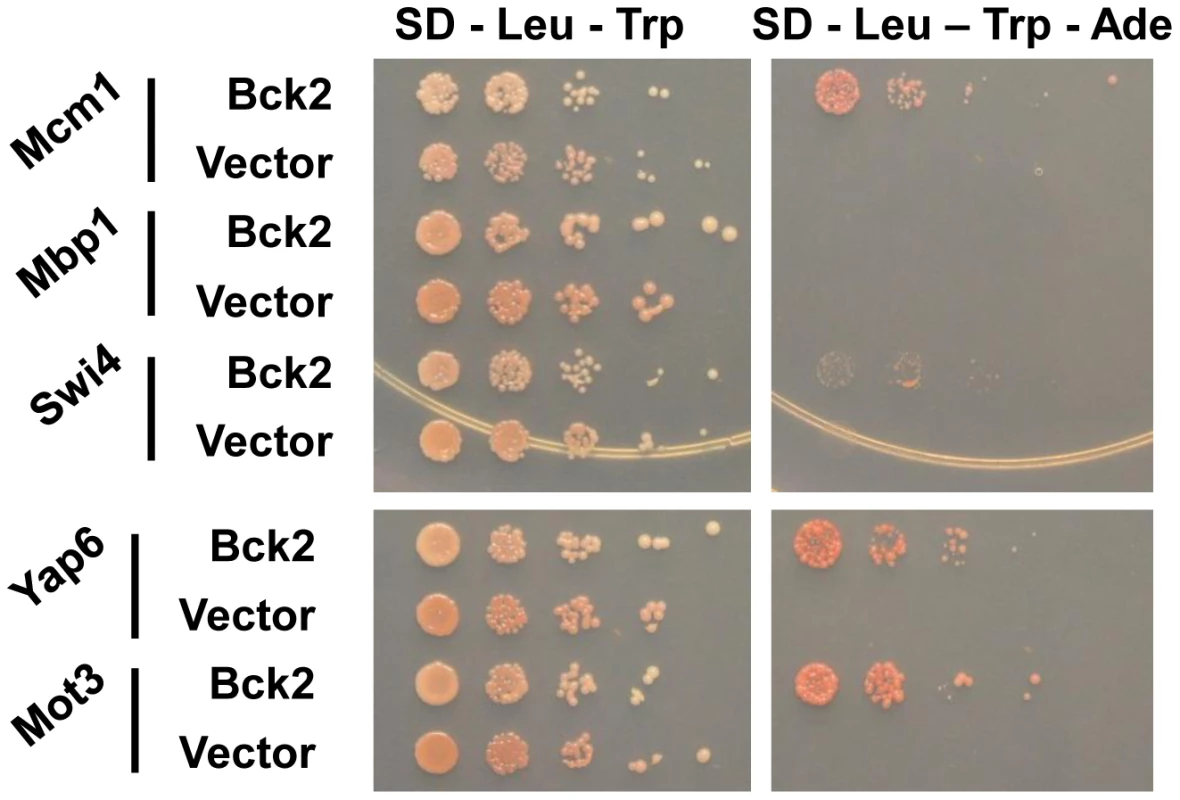
Yeast transformants carrying ADH1-GAL4 DBD (vector; LEU2) or ADH1-GAL4 DBD-BCK2 Fragment 11 (Bck2) in a two-hybrid bait strain (Y8930) were mated to yeast transformants of a two-hybrid prey strain (Y8800) bearing specific gene ORFs fused to the N-terminal GAL4 AD (activation domain; TRP1, i.e. ADH1-GAL4 AD-ORF plasmid). Diploids were selected by streaking on double plasmid selection medium (SD – Leu – Trp). Strains were grown to equivalent optical density, and spotted in serial 10-fold dilutions on double plasmid selection medium (SD – Leu – Trp) or medium where growth is proportional to transcription of the ADE2 gene (SD – Leu – Trp - Ade). Plates were incubated for 48 h at 30°C. Bck2 Activates Cell-Cycle-Regulated Genes that Contain Mcm1-Binding Sites in Their Promoters
Given the clear roles for Mcm1 in cell-cycle-dependent gene expression, we chose to focus our follow-up analysis on the Mcm1-Bck2 interaction. In addition to the Y2H interaction between Bck2 and Mcm1, several observations from earlier studies implicate Bck2 in the activation of Mcm1 target genes in M/G1 phase: (1) mRNA from the M/G1 gene SWI4 accumulates much more slowly in bck2Δ than WT cells in synchronized cultures, whereas SWI4 is upregulated in cells that overexpress BCK2 [12]; (2) high-copy BCK2 stimulates the expression of the Mcm1-dependent reporter gene, P-lacZ [47]; (3) overexpression of BCK2 causes increased transcription of CLN3 and SWI4 by microarray analysis [48]. However, up to now, no direct connection between BCK2 and M/G1 genes has been established. To evaluate the significance of the Mcm1-Bck2 physical interaction in vivo, we first assessed the effect of BCK2 deletion on expression of lacZ reporter genes whose expression was dependent on either multiple Mcm1-binding sites (4 x P-sites) [47] or the upstream activating sequences of four Mcm1-regulated genes expressed in M/G1 phase – CLN3, CDC6, CDC47, SWI4 [49] (Figure 3A). In these plasmid reporter assays, deletion of BCK2 had no effect on expression of a control ACT1-lacZ reporter gene. However, we saw a pronounced reduction in expression of the CLN3-lacZ, CDC6-lacZ, CDC47-lacZ, SWI4-lacZ, and P-lacZ reporter genes in the bck2Δ strain (Figure 3B). These results suggest a role for BCK2 in M/G1 gene expression. To verify the results of the reporter gene assays, we next examined endogenous levels of Mcm1 target gene expression in a bck2Δ strain (Figure 4). Since Mcm1 controls CLN3 and SWI4 transcript accumulation at a very early point in G1, we synchronized cultures in mitosis with a cdc20-3 temperature-sensitive allele and released them into the subsequent cell cycle. Cells in this experiment were slow growing and enriched in large-budded cells that precluded FACS analysis of cell cycle synchrony (data not shown). However, CLN2 transcription was highly periodic in both WT and bck2Δ cells, indicating that these cultures were synchronously released from the mitotic block. Consistent with previous reports, CLN2 transcript was reduced in the bck2Δ strain [11], [12] while levels of a control transcript (ALG9) were unaffected. Strikingly, the accumulation of CLN3 and SWI4 mRNAs was significantly reduced in bck2Δ cells, and peak expression was also delayed, at least for the CLN3 transcript. In wild-type cells, CLB2 mRNA peaked after G1 transcripts as expected. However, in bck2Δ cells CLB2 transcripts were delayed and only began to accumulate near the end of the time-course, after the peak of CLB2 expression seen in wild-type cells. Thus, BCK2 is required for the appropriate expression of the M/G1 genes CLN3 and SWI4, the G1/S gene CLN2, and the G2/M gene CLB2.
Fig. 3. Bck2 activates Mcm1-driven lacZ reporter constructs. 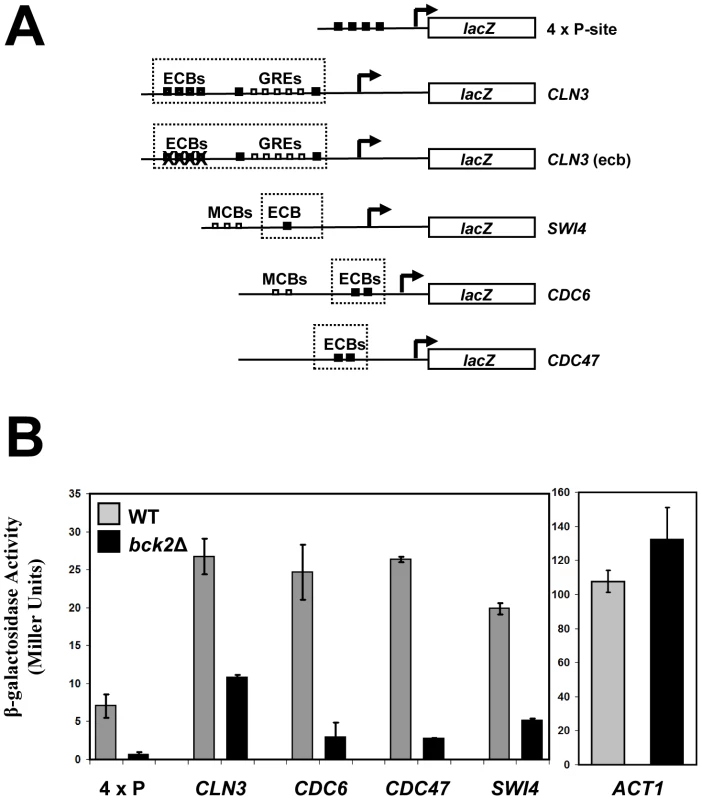
(A) Diagrams of plasmid reporter constructs used to assess the effect of BCK2 deletion or overexpression. Constructs containing either multiple synthetic Mcm1-binding sites upstream of the lacZ gene (4 x P-site, pCLM771) or endogenous promoters that contain ECB elements (CLN3 pBD1790, SWI4 pBD1577, CDC6 pBD1637, CDC47 pBD1951) are shown. Black boxes represent distinct Mcm1-binding sites such as Mcm1-binding P-site elements or ECB elements, whereas white boxes represent MCB elements or GRE elements. (B) WT (grey bars) or bck2Δ (black bars) yeast transformants carrying P-lacZ, CLN3-lacZ, SWI4-lacZ, CDC6-lacZ, CDC47-lacZ, and ACT1-lacZ were assessed for lacZ expression level. Asynchronous cells were grown to mid-log phase in selective medium and subjected to quantitative β-galactosidase assays to measure lacZ expression. Y-axis values are expressed in Miller units. Error bars reflect values obtained from 3 independent transformants in separate experiments. Fig. 4. Effect of BCK2 deletion on CLN2, ALG9, CLN3, SWI4, BCK2, and CLB2 mRNA accumulation during the cell cycle. 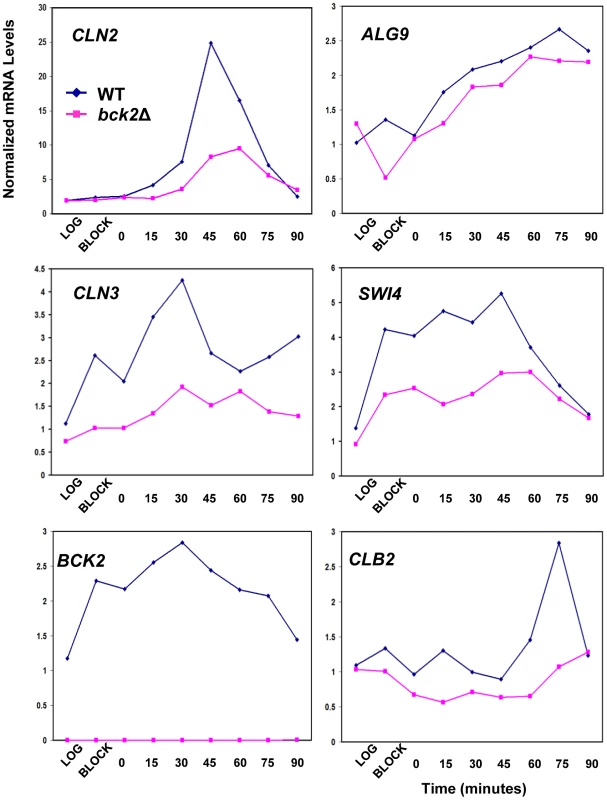
Y8890 (cdc20-3 WT, dark blue line with diamonds) and BY4897 cultures (cdc20-3 bck2Δ, dark pink line with squares) were grown to log-phase, arrested at M/G1 by incubating for 3.5 hours at 37°C (block), then released into the cell cycle by re-incubation at 21°C. Samples were harvested every 15 minutes and mRNA levels quantified by Q-PCR using ACT1 mRNA levels as a normalizing control. In WT cells, the peak of CLN2 transcription marks the G1/S transition, the peak of CLN3 and SWI4 marks M/G1, and the peak of CLB2 marks G2/M. Bck2 Requires ECB Elements for Transcriptional Activation of M/G1 Genes
The observation that Bck2 is required for proper transcription of M/G1 genes that contain Mcm1-binding sites suggested that Bck2 may function through ECB elements. To test this hypothesis, we assayed the effect of BCK2 deletion on expression of lacZ reporter plasmids containing either a WT CLN3 promoter that has intact ECB elements or a mutated CLN3 promoter that lacks functional ECB elements [50]. Deletion of BCK2 in combination with mutation of ECB elements caused a level of reporter gene expression similar to that seen with either perturbation alone (Figure 5A), indicating that Bck2 acts through ECB elements in order to control expression of M/G1 genes.
Fig. 5. Bck2 requires intact ECB elements for transcriptional activation. 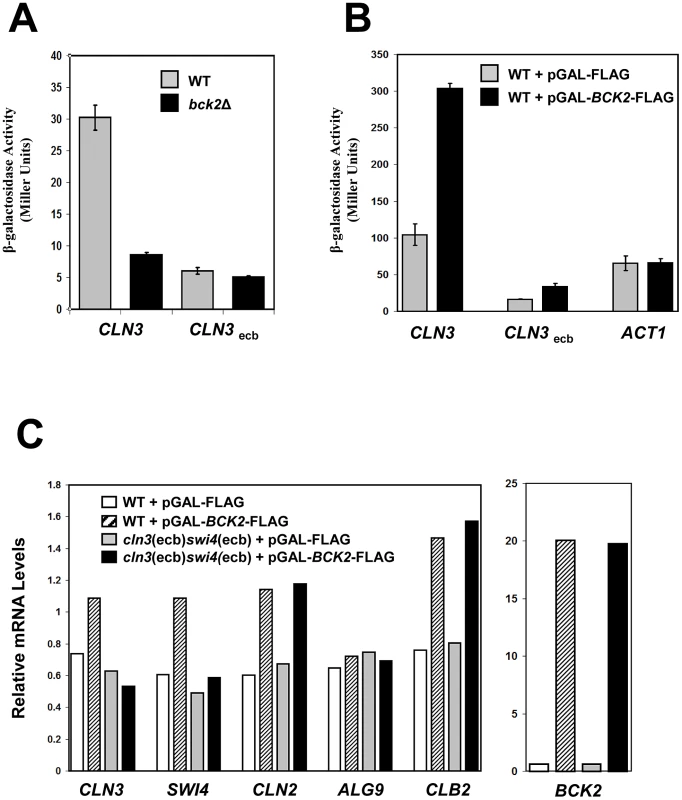
(A) WT (grey bars) or bck2Δ (black bars) yeast transformants harboring a CLN3-lacZ or CLN3ecb-lacZ reporter plasmid were grown to mid-log phase in selective medium and subjected to quantitative β-galactosidase assays. (B) WT yeast strains containing a CLN3-lacZ, CLN3ecb-lacZ or ACT1-lacZ reporter plasmid were co-transformed with pGAL-BCK2-FLAG (black bars) or vector (grey bars) and grown to mid-log phase in selective medium containing galactose (inducing conditions) and subjected to quantitative β-galactosidase assays to measure lacZ expression. Y-axis values are expressed in Miller units. Error bars reflect values obtained from 3 independent transformants in separate experiments. (C) A WT (BY2125) strain and a strain lacking functional ECB elements in the CLN3 and SWI4 promoters (BY2680; cln3(ecb)swi4(ecb)) were transformed with pGAL-BCK2-FLAG (hatched bars in WT; black bars in mutant) or vector (white bars in WT; grey bars in mutant). Transformants were grown to saturation in plasmid selective medium and subcultured in YPGal to mid-log phase before harvesting for quantification of mRNA levels by Q-PCR analysis using ACT1 mRNA levels as a normalizing control. Relative enrichment of CLN3, SWI4, CLN2, ALG9 and CLB2 mRNA normalized against ACT1 mRNA is shown in the left panel. Relative enrichment of BCK2 mRNA from the same samples normalized against ACT1 mRNA is shown in the right panel. To gather more evidence that Bck2 works through ECB elements, we next tested the effects of BCK2 overexpression on ECB-containing reporter gene expression. For this experiment, we used a lacZ reporter gene driven by a version of the CLN3 promoter in which ECB elements were mutated. As previously seen [50], when ECB elements were mutated, expression of CLN3pr-lacZ in wild-type cells was substantially reduced (Figure 5B). Overexpression of BCK2 induced expression of the wild-type CLN3pr-lacZ reporter gene approximately 3-fold, in a manner that was largely dependent on intact ECB elements (Figure 5B). We conclude that ECB elements are required for Bck2 to maximally activate transcription through the CLN3 promoter. To substantiate the requirement of ECB elements in transcriptional activation of M/G1-regulated genes by overproduced Bck2, we next assayed the effects of BCK2 overexpression on expression of the endogenous CLN3 and SWI4 genes. We compared the expression of CLN3 and SWI4 in a wild-type strain to a strain where ECB elements in the promoters of both CLN3 and SWI4 were mutated (cln3(ecb)swi4(ecb)) [49]. Consistent with our reporter gene assays, overproduction of Bck2 increased CLN3 and SWI4 transcript levels in a WT strain, but not the cln3(ecb)swi4(ecb) mutant strain (Figure 5C), indicating that Bck2 functions through ECB elements in endogenous M/G1 promoters.
Bck2 Also Activates Expression of G1/S and G2/M Genes
As previously seen, overproduction of Bck2 also increased CLN2 expression in WT cells [12], [51]; however, this induction was entirely independent of the ECB elements in the CLN3 and SWI4 gene promoters (Figure 5C). This result suggests that the induction of CLN2 transcription by overexpressed BCK2 is not an indirect consequence of increased CLN3 and SWI4 expression. Similarly, the induction of CLB2 expression by overexpressed BCK2 [12], [51] was also independent of the ECB elements in the CLN3 and SWI4 promoters (Figure 5C), again suggesting that the CLB2 induction is not an indirect effect of defects in M/G1 phase gene expression. Finally, overexpressed BCK2 did not alter expression of ALG9, a non-ECB containing gene, indicating that Bck2 is not an activator of global transcription. Together, our analyses of the effects of BCK2 deletion and overexpression show that Bck2 activates CLN3 and SWI4 transcription in an ECB-dependent manner, while also promoting expression of G1/S (CLN2) and G2/M-regulated (CLB2) genes by a mechanism that does not depend on its effect on early G1 genes.
Bck2 Localizes to M/G1, G1/S, and G2/M Phase Promoters
Since Bck2 functions through ECB elements (Figure 5) and physically interacts with Mcm1 (Figure 2), we next asked if Bck2 localized to the promoter regions of M/G1-phase genes. We first used chromatin immunoprecipitation (ChIP) with a strain carrying a TAP-tagged allele of Bck2 to assess association of Bck2 with various promoters. We detected a reproducible enrichment of promoter DNA in the Bck2 ChIP, but the signal was very low relative to Swi4 or Mcm1 ChIPs (data not shown). To improve our assay, we repeated the ChIP experiment using a strain in which a FLAG-tagged derivative of Bck2 was conditionally overproduced (Figure 6A). Under inducing conditions (galactose), Bck2-FLAG IPs were enriched in CLN2, CLN3 and SWI4 promoter DNA relative to non-inducing conditions (raffinose) (Figure 6A) or vector control (data not shown). The enhanced enrichment of CLN3 promoter DNA compared to SWI4 promoter DNA in Bck2 IPs likely reflects the presence of more ECB elements in the CLN3 promoter (6 versus 1). Association of Bck2 with the CLN3 and SWI4 promoters was entirely dependent on the presence of ECB elements, while association with the CLN2 promoter was unaffected, consistent with our gene expression analysis (Figure 5). Our findings are supported by a recent study that identified Bck2 as a constituent of DNA-bound complexes containing Mcm1 [52]. We conclude that Bck2 localizes to the promoters of CLN3 and SWI4 in a manner that depends on ECB elements.
Fig. 6. Bck2 localizes to the promoters of M/G1, G1/S and G2/M genes. 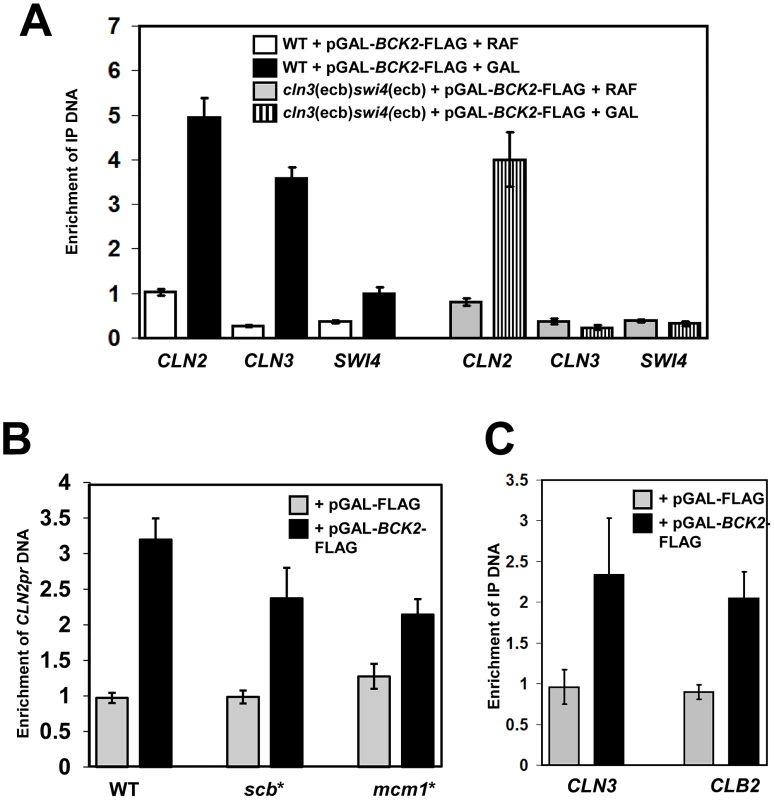
(A) Bck2 localization to M/G1 genes depends on ECBs. WT strain (BY2125; W303) or a strain containing mutated ECB elements in the CLN3 and SWI4 promoters (BY2680; cln3(ecb)swi4(ecb)) was transformed with a pGAL-BCK2-FLAG plasmid and grown separately in raffinose- (non-inducing conditions) or galactose-containing medium (inducing conditions) to mid-log phase. Cultures were harvested and anti-FLAG ChIPs were analyzed for CLN2, CLN3 and SWI4 promoter DNA by Q-PCR. The Y-axis measures enrichment of promoter DNA for the target gene indicated relative to enrichment of non-promoter DNA from an untranscribed region of chromosome II. (B) Bck2 localization to the CLN2 promoter is reduced when SCBs or Mcm1-binding sites are mutated. Vector and pGAL-BCK2-FLAG were transformed into WT (GC46), and strains with 3 SCBs mutated (yLB76-scb*) and 2 Mcm1-binding sites mutated (yLB76-mcm1*), cells were grown in inducing conditions, and Bck2-Flag localization to the CLN2 promoter was analyzed by Q-PCR. (C) Bck2 localizes to the CLB2 promoter. WT cells containing vector or pGAL-BCK2-FLAG were grown in inducing conditions and Bck2-Flag localization to the CLN3 and CLB2 promoter was analyzed by ChIP. Bck2 also localized to the CLN2 promoter in our ChIP assays. The CLN2 promoter contains three SCB elements, which are required for cell-cycle-specific gene expression, and two Mcm1 binding sites, which contribute to the presence of a nucleosome depleted region but have no effect on average expression levels or cell-cycle-specific expression [31]. We assayed the ability of Bck2 to localize to the CLN2 promoter in strains that contained mutations in either set of elements [31]. Mutation of either the SCBs (scb*) or the Mcm1-binding sites (mcm1*) led to a modest reduction in Bck2 localization to the CLN2 promoter (Figure 6B), suggesting that Bck2 may be recruited to CLN2 by either SBF or Mcm1. The SBF-dependence of Bck2 recruitment to the CLN2 promoter was further supported by a reduced Bck2 ChIP in the absence of SWI4 or SWI6 (Figure S2).
Since we saw that BCK2 was required for proper expression of CLB2 (Figure 4), and that overproduced Bck2 led to increased CLB2 RNA levels (Figure 5C), we next asked whether Bck2 could also localize to the CLB2 promoter. Mcm1 binds to the CLB2 promoter in cooperation with another DNA-binding protein Fkh2, which is activated by Ndd1 [25]–[29]. Our ChIP experiments revealed that Bck2 localized to the CLB2 promoter with an efficiency comparable to that seen with the CLN3 promoter (Figure 6C), and the binding was partially dependent on FKH2 (Figure S3).
Bck2 May Compete with Yox1 for Access to Mcm1 on ECB Elements
As noted earlier, the closely related homeodomain proteins Yox1 and Yhp1 act as repressors of Mcm1 by interacting directly with Mcm1 at DNA binding sites adjacent to the actual Mcm1 DNA binding site within the ECB [18]. Activation of Mcm1 on ECB elements correlates with removal of Yox1 from ECB elements, while deletion of YHP1 has little effect on the level or periodicity of gene expression, and Yhp1 is not part of the predominant complex at ECB elements [18]. These observations suggested that Bck2 may activate CLN3 and SWI4 transcription through ECB elements by promoting the removal of Yox1. To test this hypothesis, we first assessed Yox1 binding to ECB elements within the CLN3 promoter when Bck2 levels were elevated. Overexpression of BCK2 significantly reduced the amount of Yox1 associated with the CLN3 promoter (Figure 7A; left panel), implicating Bck2 in Yox1 removal. Yhp1 was not localized to the CLN3 promoter to the same extent as Yox1, nor was the association affected by BCK2 overproduction, consistent with a secondary role for Yhp1 [18]. Consistent with previous reports that Mcm1 remains localized to promoters throughout the cell cycle [18], we observed that Mcm1 localization was not significantly affected by BCK2 dosage (Figure 7A, right panel). These experiments suggest that Bck2 overproduction may lead to the displacement of Yox1 repressor from the CLN3 promoter, which is correlated with activation of M/G1-regulated genes [18].
Fig. 7. Bck2 may compete with Yox1 for interaction with Mcm1. 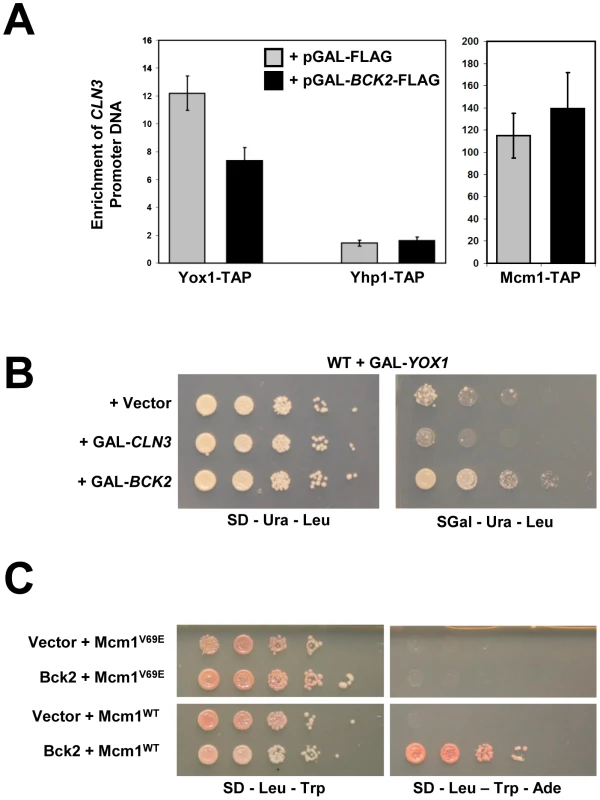
(A) YOX1-TAP, YHP1-TAP and MCM1-TAP strains carrying a pGAL-BCK2-FLAG plasmid (black bars) or vector (grey bars), were individually grown in plasmid selective medium containing galactose (inducing conditions) to mid-log phase. Cultures were harvested and anti-TAP ChIPs were performed using IgG sepharose resin and analyzed for CLN3 promoter DNA using Q-PCR. Error bars reflect values obtained after multiple Q-PCR runs from the same experiment. (B) A WT strain (Y7092) was co-transformed with a GAL-YOX1 (URA3) plasmid and one of Vector, GAL-CLN3, or GAL-BCK2 (LEU2) plasmid. Transformant cultures of equivalent optical density were spotted in serial 5-fold dilutions on selective medium that was either non-inducing (glucose) or inducing (galactose), and incubated for 72 h at 30°C. (C) A yeast two-hybrid bait strain (Y8930) carrying either plasmid ADH1-GAL4 DB (vector) or plasmid ADH1-GAL4 DB-BCK2 Fragment 11 (Bck2) were mated to yeast two-hybrid prey strain (Y8800) carrying either ADH1-GAL4 AD-MCM1WT or ADH1-GAL4-MCM1V69E to create diploid yeast strains. Diploids were spotted in serial 10-fold dilutions on double plasmid selection medium, or medium where growth is proportional to transcription of the ADE2 gene, and incubated for 48–72 h at 30°C. Overexpression of YOX1 is toxic, likely because high levels of Yox1 cause constitutive repression of its target genes [18]. To test the model that Bck2 may compete with Yox1 binding to Mcm1 at ECB elements, we asked if the toxicity caused by YOX1 overexpression was suppressed by concurrent overexpression of BCK2. Previous work on the CLB2 gene cluster, which is expressed at the G2/M phase transition, has shown that Mcm1 acts as a common scaffold for recruitment of Yox1 and the forkhead protein Fkh2 [30]. These physical interactions with Mcm1 are mutually exclusive and are mediated by distinct Yox1 and Fkh2 DNA binding elements that flank a central Mcm1 DNA binding site. When bound to Mcm1 on promoters, Fkh2 recruits the positively acting co-regulator Ndd1 in order to activate the CLB2 cluster genes [26]. Constitutive overexpression of YOX1 inhibits Ndd1 binding to the Yox1-regulated SPO12 promoter [30], consistent with a mechanism based on mutual exclusivity between an activator and repressor. Our observation that overproduction of Bck2 leads to reduced Yox1 on the CLN3 promoter (Figure 7A) suggested that the YOX1 dose-lethality phenotype might be suppressed by concurrently overproducing BCK2. Indeed, we observed that overexpression of BCK2 was able to significantly suppress the lethality caused by overexpressing YOX1 (Figure 7B). Overexpression of CLN3 failed to rescue the YOX1 overexpression phenotype, suggesting that the BCK2 rescue does not simply reflect an indirect effect of reduced G1 phase. Although the suppression of YOX1 toxicity by overexpressed BCK2 does not show that the effects are direct, it is consistent with a competitive relationship between Bck2 and Yox1 for interaction with the Mcm1 scaffold.
Mcm1 contains a hydrophobic pocket found on the surface of the MADS DNA-binding domain, and mutation of this pocket by the introduction of a V69E mutation disrupts interaction with Fkh2 [53]–[55], which prevents binding of both Yox1 and Fkh2 to Mcm1 [30]. We hypothesized that the competitive binding mechanism that allows Mcm1 to activate genes transcribed at the G2/M transition may also function for genes transcribed at the M/G1 transition. Specifically, we wondered whether Bck2 might activate Mcm1 at ECB elements through competition with Yox1 for binding to Mcm1. Consistent with our hypothesis, we found that the Y2H-based interaction between Bck2 and Mcm1 was abolished in the Mcm1V69E mutant (Figure 7C). We conclude that Bck2 may act to remove Yox1 by a competitive binding mechanism similar to that seen at G2/M phase promoters [30].
Discussion
Bck2 Activates Early G1 Genes by Interacting with Mcm1 at ECB Elements
Our work identifies a previously unappreciated role for Bck2 in the regulation of M/G1-specific transcription in budding yeast, and also reveals functions for Bck2 in regulating late G1 and G2/M genes (Figure 8). In contrast to the CLB2 gene cluster, no positively acting partner protein had yet been found that cooperates with Mcm1 to regulate M/G1-expressed genes. We describe several observations showing that Bck2 functions to control Mcm1 activity on promoter elements to ensure the proper regulation of early G1 events. First, Bck2 is required to activate expression of the M/G1 genes SWI4, CLN3, CDC6, and CDC47. Second, we show a requirement for intact Mcm1-binding ECB elements within the promoters of CLN3 and SWI4, consistent with the physical interaction between Bck2 and Mcm1 that we see by Y2H. Third, Bck2 localizes to early G1 promoters in an ECB-dependent manner.
Fig. 8. Summary of Bck2-dependent regulation of cell cycle gene expression. 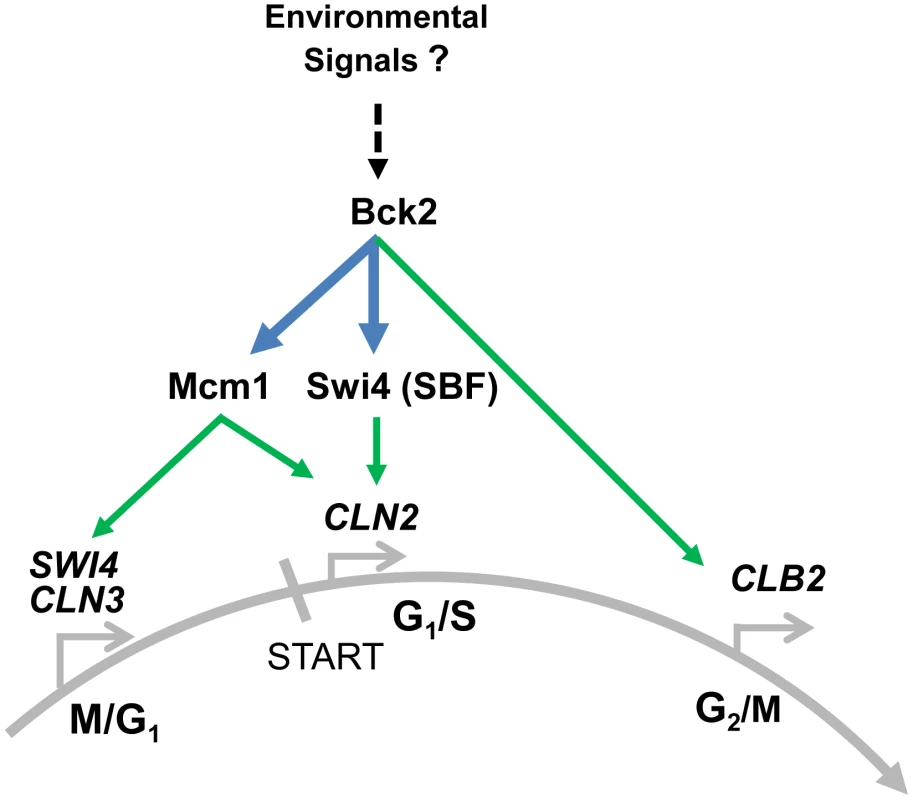
In pre-START cells Bck2 binds to Mcm1 and promotes expression of M/G1 genes such as SWI4 and CLN3, which encode important constituents of the transcriptional ‘switch’ for START. Cln3 then contributes to activation of Swi4, a component of SBF, which induces expression of G1/S genes such as CLN2. Bck2 also induces CLN2 expression directly through physical association with Swi4 and with Mcm1. In G2/M Bck2 promotes expression of CLB2, possibly through interaction with Mcm1. We speculate that Bck2 may be responding to environmental signals such as nutrients in its role of activating cell cycle gene expression (see text for details). Bold lines indicate interactions, both protein-protein (in blue) and protein-DNA (in green) described in this work. Our finding that Bck2 is needed for expression of Mcm1-regulated genes in M/G1 phase may explain previous observations that Bck2 activates G1/S gene expression in a pathway distinct from that involving Cln-Cdc28 activation of promoter-bound SBF/MBF. For example, high-copy BCK2 activates SBF/MBF target genes in a cdc28-4 mutant [13] and suppresses the G1-arrest of a cln1Δcln2Δcln3Δ strain [11]. These activities likely reflect increased SWI4 expression, because Bck2 activates SWI4 transcription [12], [48], and high-copy suppression by BCK2 of the G1-arrest of a cln1Δcln2Δcln3Δ strain requires SWI4 [11]. Importantly, SWI4 is induced in a cdc28-13 mutant [7]. There is no evidence that Cdk activity is required for activation of M/G1-expressed genes.
Bck2-Dependent Activation of G1/S Genes
Bck2 has also been shown to activate G1/S genes, in a manner that is both partially dependent and partially independent of SBF/MBF activity. For example, overproduction of Bck2 stimulates expression of an SCB-lacZ reporter gene, activation of which is strictly dependent on SBF [12]. Large-scale mass spectrometry experiments reveal an interaction between Bck2 and Swi4 [56] and we see a weak Y2H interaction between BCK2 and SWI4 (Figure 2). In our experiments, localization of Bck2 to the CLN2 promoter was modestly reduced when the SCB sites were mutated (Figure 6B) and Bck2 localization was partially dependent on SWI4 and SWI6 (Figure S2), suggesting that SBF has a role in recruiting Bck2. However, consistent with an SBF-independent activity, Bck2 can activate several natural SBF/MBF target gene promoters in the absence of either SWI4 or MBP1 [13], or the elements SBF/MBF bind [12]. In our experiments, mutation of the Mcm1-binding sites in the CLN2 promoter also modestly reduced Bck2 binding (Figure 6B), suggesting that Mcm1 also contributes to Bck2 recruitment to the CLN2 promoter. Since SCB elements are much more important for proper CLN2 expression than Mcm1-binding sites [31], it is likely that SBF has a more important role than Mcm1 in recruiting Bck2 to the CLN2 promoter.
Bck2-Dependent Activation of G2/M Genes
We present several lines of evidence that suggest a role for Bck2 in activation of G2/M genes. First, a bck2Δ strain has delayed CLB2 expression (Figure 4). Second,overexpression of BCK2 leads to increased expression of CLB2 [12] (Figure 5C), and other G2/M genes [51]. Third, we show that Bck2 localizes to the CLB2 promoter (Figure 6C), in a manner dependent on FKH2 (Figure S3). Activation of G2/M genes is controlled by Ndd1, which is recruited to promoters by binding Fkh2 [25]–[29]; it is not clear why FKH2 is involved in Bck2 localization to the CLB2 promoter. One possibility is that Bck2 binds to both Fkh2 and Mcm1 at G2 promoters, similar to its binding to SBF and Mcm1 at G1/S promoters; however, we did not detect an interaction between BCK2 and FKH2 in a pairwise Y2H assay (data not shown). Regardless, the mechanism of action of Bck2 at G2/M promoters must differ from that at M/G1 promoters where Bck2 appears to act on Mcm1 alone. Unlike the G2/M promoters, M/G1 promoters [6] do not contain DNA binding sites for Fkh2 [29] or other positive regulators, indicating that induction of M/G1 [18] genes depends on a positively-acting protein that presumably acts through Mcm1.
Bck2 as an Activator of Other Mcm1-Regulated Genes
Mcm1, together with class-specific regulators, controls the expression of several different groups of genes in addition to the cell-cycle-regulated genes discussed above: (1) activation of genes involved in mating together with Ste12; (2) regulation of cell-type-specific genes together with α1 or α2; and (3) control of genes involved in arginine metabolism together with Arg80 (for review see [57]). Other observations suggest that Bck2 may be involved in regulating expression of other classes of Mcm1-dependent genes. First, BCK2 was identified as a gene whose overexpression strongly induced the expression of FUS1 reporter genes [58]. Second, a more recent study examining transcriptional profiles found that overexpression of BCK2 led to expression of cell-cycle-regulated genes from multiple cell-cycle stages, consistent with our findings [51]. Furthermore, overexpression of BCK2 induced a large number of genes involved in mating, but not genes involved in cell type or nitrogen metabolism [51]. Indeed these authors suggested that Bck2 may elicit gene expression via Ste12-Mcm1. We have shown that Bck2 acts through Mcm1 to promote expression of three classes of cell cycle genes; we suggest that Bck2 likely regulates mating genes in a similar manner.
Regulation of Bck2
We have shown in this work that Bck2 regulates cell cycle gene expression. We suggest that the role of Bck2 may be to fine-tune expression of different classes of Mcm1-regulated genes. How might Bck2 itself be regulated? Bck2 is rich in serine and threonine residues and has been shown to be phosphorylated at multiple sites at different stages of the cell cycle [59], [60]. Bck2 has been linked to several cell-cycle-regulating kinases and phosphatases in genetic studies (the Protein Kinase C pathway [16], [48], Sit4 [12], [61], Cdc28 [13], [48], Cbk1 [16]) and in mass spectrometry and phosphopeptide analyses (Cdc15 [62], Cdc28 [60], Fus3 and Kss1 [56]). Thus Bck2 may play a role in linking detection of nutrients or other conditions that affect cell cycle progression to Mcm1-dependent gene expression (Figure 8). For example, during early G1 phase, yeast cells assess nutrient status, in part through the Tor pathway and the phosphatase Sit4 [63]. BCK2 has been proposed to function in the SIT4 pathway of CLN activation [12], [61]. Both Tor and Sit4 signaling are also required for proper M/G1 gene expression [63], [64], and like tor1Δ cells and sit4Δ cells, bck2Δ cells are rapamycin sensitive [16], [63], [65]. Thus Bck2 may be part of the signal transduction pathway linking nutrient status to passage through M/G1 of the cell cycle. Consistent with a potential role for Bck2 in integrating environmental and cell cycle signals, our Y2H screen identified several transcription factors and regulators with established functions in regulating gene expression in response to various nutrients or stresses, including nutrient sensing through the TOR pathway (see Results and Figure 2). How Bck2 may work together with various proteins to regulate nutrient and stress responses remains to be determined.
Bck2 Mode of Action
Based on the high level of similarity between budding yeast Mcm1 and human Serum Response Factor (SRF) [30], [54], [55], [66]–[68], the Bck2 protein might represent a budding yeast analog of a specific SRF co-activator in mammalian cells. For example, the myocardin/MKL family members are SRF co-activators which are enriched at target promoters in serum-rich medium [69]. Our discovery of Bck2 as a cofactor for Mcm1, coupled with accumulating evidence for a role for Bck2 in nutrient sensing, suggests that Bck2 binding may also increase under nutrient-rich conditions. Further studies will be required to firmly test the analogy between the Mcm1-Bck2 and SRF-MKL pathways and to illuminate how Bck2 may link gene expression, the cell cycle machinery and environmental signals to ensure appropriate cell proliferation.
Materials and Methods
Yeast Strains and Plasmids
Yeast strains used in this study (Table 1) were derivatives of either S288C or W303, with the exception of the Y2H strains. Plasmids are described in Table 2. Yeast cultures were grown in YEP (1% yeast extract, 2% bactopeptone) supplemented with 2% glucose. Synthetic minimal medium supplemented with the appropriate nutrients was used to select for plasmid maintenance and gene replacements. Yeast transformation and general manipulation of yeast cells were performed using standard techniques.
Tab. 1. Saccharomyces cerevisiae strains used in this study. 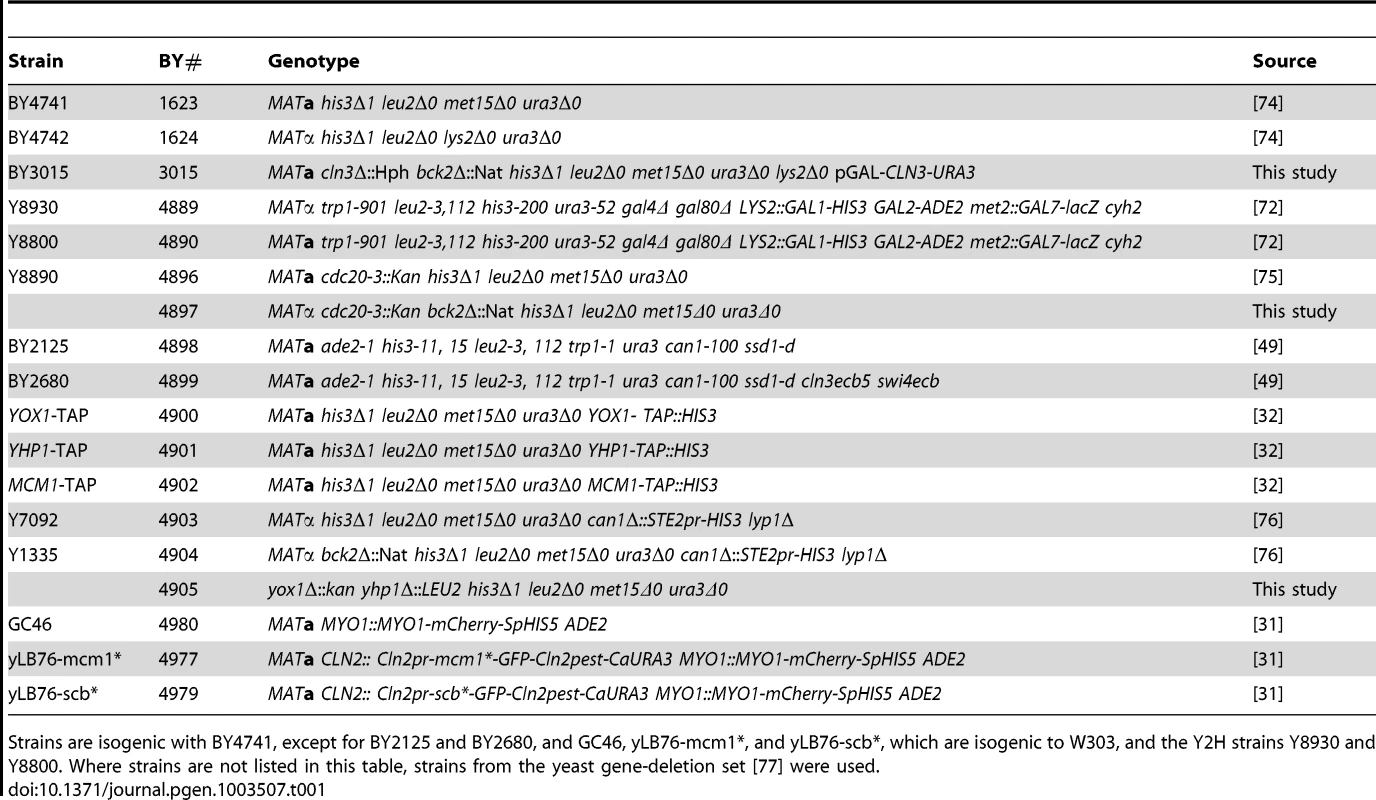
Strains are isogenic with BY4741, except for BY2125 and BY2680, and GC46, yLB76-mcm1*, and yLB76-scb*, which are isogenic to W303, and the Y2H strains Y8930 and Y8800. Where strains are not listed in this table, strains from the yeast gene-deletion set [77] were used. Tab. 2. Plasmids used in this study. 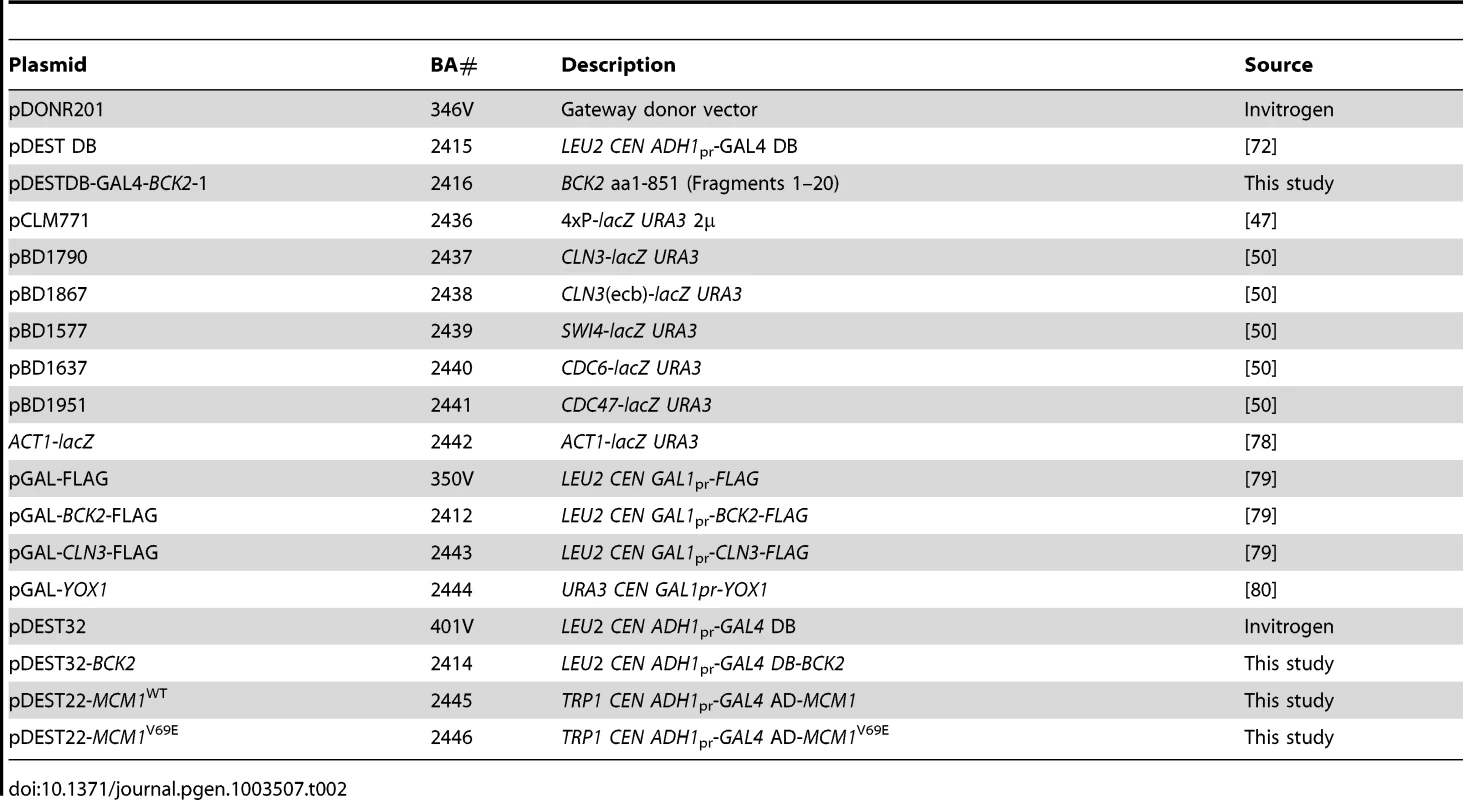
Cloning and Construction of BCK2 Truncations
Fragments of the BCK2 gene were amplified from genomic DNA using PCR primers designed to be compatible with the Gateway system of recombinational cloning (Table S1). All PCR products were recombined into the donor vector pDONR201 using the BP clonase II system (Invitrogen) and positive clones were fully sequence-confirmed. BCK2 truncations within pDONR201 were recombined into the destination vector pDEST-DB using the LR clonase II system.
β-Galactosidase Assays
Quantitative β-galactosidase assays were performed as follows. Exponentially growing cells at an optical density at 600 nm of 0.2 to 0.25 were harvested. Extracts were prepared by vortexing the cells in 1 ml Z-buffer (0.1 M NaPO4 [pH 7.0], 0.01 M KCl, 1 mM MgSO4, 4 mM 2-mercaptoethanol)+20 µl 0.1% SDS+40 µl chloroform for 45 seconds. After 5 min, 0.2 ml of o-nitrophenylgalactoside (Sigma; at 4 mg/ml in Z buffer) solution was added and the reaction was incubated at 30°C until a slight yellowing was observed. The reactions were stopped by the addition of 0.5 ml of 1 M Na2CO3, and the samples were centrifuged for 3 minutes at 13,000× g. The A420 of the supernatant was determined. β-Galactosidase units were calculated using the following formula: units = (A420)*(1000)/(time of reaction, in minutes)*(volume of extract in assay = 1 ml)*(cellular concentration in OD600 values). For each time point, the assays were performed on three separate cultures, and the average is reported. Qualitative β-galactosidase overlay assays were performed as described [70].
Complementation Analysis
Transformants of a cln3Δbck2Δ pGAL-CLN3 -URA3 strain (BY3015) bearing ADH1-GAL4 DBD-BCK2-LEU2 (1–20) plasmids were grown in plasmid selective medium, prepared at equivalent optical density and spotted in serial 10-fold dilutions onto plasmid selective medium containing either galactose lacking 5′FOA or glucose+5′FOA, and incubated for 48 h at 30°C.
Genome-Wide Y2H Screen
The Y2H ORFeome method [71] was used to screen for Bck2-interacting proteins by mating a prey strain with a bait strain. The AD ORFeome (prey strain) is a pooled collection of AD-ORF plasmids [72] in a 96-well format. The pooled AD ORFeome [72] was grown in a 96-well culture block containing 600 µl per well of SD – Trp media at 30°C for 48 hours. Five µl of AD ORFeome culture were spotted from the 96-well culture block onto large YPD plates and allowed to dry. Next, 5 µl of a DBD ORF strain culture (bait strain transformed with a single DBD-ORF plasmid) was dispensed directly onto the ORFeome strain spots. Five diploid control strains were spotted onto the same plate at an empty location in order to ensure the quality of selection plates and to help evaluate the phenotype of interactions. The plate was incubated at 30°C until growth of spots was apparent before replica plating onto SD – Leu – Trp and grown for 3 days at 30°C in order to select only diploid yeast. This plate was replica plated onto a final selection plate containing SD – Leu – Trp – Ade and grown for 5 days until foci were observed. At least 3 foci per spot were picked, and subjected to colony PCR using primers homologous to the sequences flanking the ORF within the AD plasmid. Yeast cells were scraped from plates into 30 µl of lysis solution (2.5 mg zymolyase in 1 ml 1 M sorbitol), and incubated at 37°C for 15 minutes then 95°C for 5 minutes before addition of 120 µl of ddH2O. PCR reactions were performed in 25 µl volumes containing 5 µl of the yeast cell preparation described. PCR reactions were performed using 5 minute extension times in order to ensure that large ORF inserts were isolated. PCR reactions were electrophoresed on agarose gels, purified using the PureLink kit (Invitrogen) and sent for sequencing analysis.
Direct Pair-Wise Y2H Assays
Yeast transformants carrying ADH1-GAL4 DBD (vector; LEU2) or ADH1-GAL4 DBD-BCK2 Fragment 11 (Bck2) in a two-hybrid bait strain (Y8930) were mated to yeast transformants of a two-hybrid prey strain (Y8800) bearing specific ADH1-GAL4-AD-ORF-TRP plasmids. Diploids were selected by streaking cells onto double plasmid selection medium (SD – Leu – Trp). Diploids of equivalent optical density were spotted in serial 10-fold dilutions on double plasmid selection medium (SD – Leu – Trp), or medium where growth was proportional to transcription of the ADE2 gene (SD – Leu – Trp - Ade), and incubated for 48 h at 30°C. Six diploid strains carrying different combinations of AD and DBD ORF-fusions [72], [73] were used as a spectrum of positive and negative controls.
Cell Synchronization
A cdc20-3 temperature-sensitive strain (BY4896, Table 1) was grown in YPD medium at 21°C, arrested in M phase by incubation at 37°C for 3.5 hours, and released into the cell cycle by transferring the culture back to 21°C. Arrest was determined by visualization of large budded cells under a light microscope.
mRNA Purification and RT–qPCR
Total RNA was isolated by phenol-chloroform extraction and further purified using the RNeasy kit (Qiagen). RNA was transcribed into cDNA using the Superscript II Reverse Transcriptase kit (Invitrogen) and RNA was then removed by addition of NaOH. Reactions were run on the ABI 7500 system (Applied Biosystems) using standard Q-PCR conditions. Data were analyzed using ABI7500 system software. VIC and FAM labelled fluorogenic primers (ABI), used to detect CLN2, ALG9, CLN3, SWI4, BCK2, CLB2 and ACT1 cDNA, are described in Table S1.
Mutagenesis
To construct the Mcm1V69E yeast two-hybrid prey plasmid, the Mcm1WT prey plasmid was subject to in vitro mutagenesis using the QuikChange site-directed mutagenesis kit (Stratagene).
ChIP Assays
Yeast strains transformed with pGAL-FLAG plasmids were grown in raffinose-containing minimal media overnight, and then grown separately in raffinose - (non-inducing conditions) or galactose-containing medium (inducing conditions) to mid-log phase. Cultures were harvested and anti-FLAG ChIPs were analyzed for CLN2, CLN3, CLB2 and SWI4 promoter DNA by TaqMan Q-PCR (Applied Biosystems) using primers with homology to target gene promoter DNA (Table S1). Enrichment of promoter DNA was determined relative to non-promoter DNA from an untranscribed region of chromosome II (Table S1).
Supporting Information
Zdroje
1. LaubMT, McAdamsHH, FeldblyumT, FraserCM, ShapiroL (2000) Global analysis of the genetic network controlling a bacterial cell cycle. Science 290 : 2144–2148.
2. ChoRJ, HuangM, CampbellMJ, DongH, SteinmetzL, et al. (2001) Transcriptional regulation and function during the human cell cycle. Nat Genet 27 : 48–54.
3. OlivaA, RosebrockA, FerrezueloF, PyneS, ChenH, et al. (2005) The cell cycle-regulated genes of Schizosaccharomyces pombe. PLoS Biol 3: e225 doi:10.1371/journal.pbio.0030225.
4. LuY, MahonyS, BenosPV, RosenfeldR, SimonI, et al. (2007) Combined analysis reveals a core set of cycling genes. Genome Biol 8: R146.
5. MorganDO (1997) Cyclin-dependent kinases: engines, clocks, and microprocessors. Annu Rev Cell Dev Biol 13 : 261–291.
6. SpellmanPT, SherlockG, ZhangMQ, IyerVR, AndersK, et al. (1998) Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell 9 : 3273–3297.
7. ChoRJ, CampbellMJ, WinzelerEA, SteinmetzL, ConwayA, et al. (1998) A genome-wide transcriptional analysis of the mitotic cell cycle. Mol Cell 2 : 65–73.
8. WittenbergC, ReedSI (2005) Cell cycle-dependent transcription in yeast: promoters, transcription factors, and transcriptomes. Oncogene 24 : 2746–2755.
9. OrlandoDA, LinCY, BernardA, WangJY, SocolarJE, et al. (2008) Global control of cell-cycle transcription by coupled CDK and network oscillators. Nature 453 : 944–947.
10. GranovskaiaMV, JensenLJ, RitchieME, ToedlingJ, NingY, et al. (2010) High-resolution transcription atlas of the mitotic cell cycle in budding yeast. Genome Biol 11: R24.
11. EpsteinCB, CrossFR (1994) Genes that can bypass the CLN requirement for Saccharomyces cerevisiae cell cycle START. Mol Cell Biol 14 : 2041–2047.
12. Di ComoCJ, ChangH, ArndtKT (1995) Activation of CLN1 and CLN2 G1 cyclin gene expression by BCK2. Mol Cell Biol 15 : 1835–1846.
13. WijnenH, FutcherB (1999) Genetic analysis of the shared role of CLN3 and BCK2 at the G(1)-S transition in Saccharomyces cerevisiae. Genetics 153 : 1131–1143.
14. LeeKS, HinesLK, LevinDE (1993) A pair of functionally redundant yeast genes (PPZ1 and PPZ2) encoding type 1-related protein phosphatases function within the PKC1-mediated pathway. Mol Cell Biol 13 : 5843–5853.
15. StuartD, WittenbergC (1994) Cell cycle-dependent transcription of CLN2 is conferred by multiple distinct cis-acting regulatory elements. Mol Cell Biol 14 : 4788–4801.
16. KuraviVK, KurischkoC, PuriM, LucaFC (2011) Cbk1 kinase and Bck2 control MAP kinase activation and inactivation during heat shock. Mol Biol Cell 22 : 4892–4907.
17. MaiB, MilesS, BreedenLL (2002) Characterization of the ECB binding complex responsible for the M/G(1)-specific transcription of CLN3 and SWI4. Mol Cell Biol 22 : 430–441.
18. PramilaT, MilesS, GuhaThakurtaD, JemioloD, BreedenLL (2002) Conserved homeodomain proteins interact with MADS box protein Mcm1 to restrict ECB-dependent transcription to the M/G1 phase of the cell cycle. Genes Dev 16 : 3034–3045.
19. AlthoeferH, SchleifferA, WassmannK, NordheimA, AmmererG (1995) Mcm1 is required to coordinate G2-specific transcription in Saccharomyces cerevisiae. Mol Cell Biol 15 : 5917–5928.
20. DarievaZ, Pic-TaylorA, BorosJ, SpanosA, GeymonatM, et al. (2003) Cell cycle-regulated transcription through the FHA domain of Fkh2p and the coactivator Ndd1p. Curr Biol 13 : 1740–1745.
21. ReynoldsD, ShiBJ, McLeanC, KatsisF, KempB, et al. (2003) Recruitment of Thr 319-phosphorylated Ndd1p to the FHA domain of Fkh2p requires Clb kinase activity: a mechanism for CLB cluster gene activation. Genes Dev 17 : 1789–1802.
22. Pic-TaylorA, DarievaZ, MorganBA, SharrocksAD (2004) Regulation of cell cycle-specific gene expression through cyclin-dependent kinase-mediated phosphorylation of the forkhead transcription factor Fkh2p. Mol Cell Biol 24 : 10036–10046.
23. DarievaZ, BulmerR, Pic-TaylorA, DorisKS, GeymonatM, et al. (2006) Polo kinase controls cell-cycle-dependent transcription by targeting a coactivator protein. Nature 444 : 494–498.
24. DarievaZ, HanN, WarwoodS, DorisKS, MorganBA, SharrocksAD (2012) Protein kinase C regulates late cell cycle-dependent gene expression. Mol Cell Biol 32 : 4651–61.
25. LoyCJ, LydallD, SuranaU (1999) NDD1, a high-dosage suppressor of cdc28-1N, is essential for expression of a subset of late-S-phase-specific genes in Saccharomyces cerevisiae. Mol Cell Biol 19 : 3312–3327.
26. KorandaM, SchleifferA, EndlerL, AmmererG (2000) Forkhead-like transcription factors recruit Ndd1 to the chromatin of G2/M-specific promoters. Nature 406 : 94–98.
27. KumarR, ReynoldsDM, ShevchenkoA, GoldstoneSD, DaltonS (2000) Forkhead transcription factors, Fkh1p and Fkh2p, collaborate with Mcm1p to control transcription required for M-phase. Curr Biol 10 : 896–906.
28. PicA, LimFL, RossSJ, VealEA, JohnsonAL, et al. (2000) The forkhead protein Fkh2 is a component of the yeast cell cycle transcription factor SFF. EMBO J 19 : 3750–3761.
29. ZhuG, SpellmanPT, VolpeT, BrownPO, BotsteinD, et al. (2000) Two yeast forkhead genes regulate the cell cycle and pseudohyphal growth. Nature 406 : 90–94.
30. DarievaZ, ClancyA, BulmerR, WilliamsE, Pic-TaylorA, et al. (2010) A competitive transcription factor binding mechanism determines the timing of late cell cycle-dependent gene expression. Mol Cell 38 : 29–40.
31. BaiL, OndrackaA, CrossFR (2011) Multiple sequence-specific factors generate the nucleosome-depleted region on CLN2 promoter. Mol Cell 42 : 465–476.
32. GhaemmaghamiS, HuhWK, BowerK, HowsonRW, BelleA, et al. (2003) Global analysis of protein expression in yeast. Nature 425 : 737–741.
33. TitzB, ThomasS, RajagopalaSV, ChibaT, ItoT, et al. (2006) Transcriptional activators in yeast. Nucleic Acids Res 34 : 955–967.
34. CandauR, BergerSL (1996) Structural and functional analysis of yeast putative adaptors. Evidence for an adaptor complex in vivo. J Biol Chem 271 : 5237–5245.
35. RualJF, VenkatesanK, HaoT, Hirozane-KishikawaT, DricotA, et al. (2005) Towards a proteome-scale map of the human protein-protein interaction network. Nature 437 : 1173–1178.
36. WynneJ, TreismanR (1992) SRF and MCM1 have related but distinct DNA binding specificities. Nucleic Acids Res 20 : 3297–3303.
37. TreismanR (1994) Ternary complex factors: growth factor regulated transcriptional activators. Curr Opin Genet Dev 4 : 96–101.
38. ShoreP, SharrocksAD (1995) The MADS-box family of transcription factors. Eur J Biochem 229 : 1–13.
39. FernandesL, Rodrigues-PousadaC, StruhlK (1997) Yap, a novel family of eight bZIP proteins in Saccharomyces cerevisiae with distinct biological functions. Mol Cell Biol 17 : 6982–6993.
40. MendizabalI, RiosG, MuletJM, SerranoR, de LarrinoaIF (1998) Yeast putative transcription factors involved in salt tolerance. FEBS Lett 425 : 323–328.
41. SchmidtMC, McCartneyRR, ZhangX, TillmanTS, SolimeoH, et al. (1999) Std1 and Mth1 proteins interact with the glucose sensors to control glucose-regulated gene expression in Saccharomyces cerevisiae. Mol Cell Biol 19 : 4561–4571.
42. LakshmananJ, MosleyAL, OzcanS (2003) Repression of transcription by Rgt1 in the absence of glucose requires Std1 and Mth1. Curr Genet 44 : 19–25.
43. KimJH, JohnstonM (2006) Two glucose-sensing pathways converge on Rgt1 to regulate expression of glucose transporter genes in Saccharomyces cerevisiae. J Biol Chem 281 : 26144–26149.
44. AbramovaN, SertilO, MehtaS, LowryCV (2001) Reciprocal regulation of anaerobic and aerobic cell wall mannoprotein gene expression in Saccharomyces cerevisiae. J Bacteriol 183 : 2881–2887.
45. van ZylW, HuangW, SneddonAA, StarkM, CamierS, et al. (1992) Inactivation of the protein phosphatase 2A regulatory subunit A results in morphological and transcriptional defects in Saccharomyces cerevisiae. Mol Cell Biol 12 : 4946–4959.
46. JiangY, BroachJR (1999) Tor proteins and protein phosphatase 2A reciprocally regulate Tap42 in controlling cell growth in yeast. EMBO J 18 : 2782–2792.
47. LiS, AultA, MaloneCL, RaittD, DeanS, et al. (1998) The yeast histidine protein kinase, Sln1p, mediates phosphotransfer to two response regulators, Ssk1p and Skn7p. EMBO J 17 : 6952–6962.
48. Martin-YkenH, DagkessamanskaiaA, TalibiD, FrancoisJ (2002) KNR4 is a member of the PKC1 signalling pathway and genetically interacts with BCK2, a gene involved in cell cycle progression in Saccharomyces cerevisiae. Curr Genet 41 : 323–332.
49. MacKayVL, MaiB, WatersL, BreedenLL (2001) Early cell cycle box-mediated transcription of CLN3 and SWI4 contributes to the proper timing of the G(1)-to-S transition in budding yeast. Mol Cell Biol 21 : 4140–4148.
50. McInernyCJ, PartridgeJF, MikesellGE, CreemerDP, BreedenLL (1997) A novel Mcm1-dependent element in the SWI4, CLN3, CDC6, and CDC47 promoters activates M/G1-specific transcription. Genes Dev 11 : 1277–1288.
51. FerrezueloF, AldeaM, FutcherB (2009) Bck2 is a phase-independent activator of cell cycle-regulated genes in yeast. Cell Cycle 8 : 239–252.
52. LambertJP, FillinghamJ, SiahbaziM, GreenblattJ, BaetzK, et al. (2010) Defining the budding yeast chromatin-associated interactome. Mol Syst Biol 6 : 448.
53. MeadJ, BruningAR, GillMK, SteinerAM, ActonTB, VershonAK (2002) Interactions of the Mcm1 MADS box protein with cofactors that regulate mating in yeast. Mol Cell Biol 22 : 4607–4621.
54. TanS, RichmondTJ (1998) Crystal structure of the yeast MATalpha2/MCM1/DNA ternary complex. Nature 391 : 660–666.
55. BorosJ, LimFL, DarievaZ, Pic-TaylorA, HarmanR, et al. (2003) Molecular determinants of the cell-cycle regulated Mcm1p-Fkh2p transcription factor complex. Nucleic Acids Res 31 : 2279–2288.
56. BreitkreutzA, ChoiH, SharomJR, BoucherL, NeduvaV, et al. (2010) A global protein kinase and phosphatase interaction network in yeast. Science 328 : 1043–1046.
57. ShoreP, SharrocksAD (1995) The MADS-box family of transcription factors. Eur J Biochem 229 : 1–13.
58. EdwardsMC, LiegeoisN, HoreckaJ, DePinhoRA, SpragueGFJr, et al. (1997) Human CPR (cell cycle progression restoration) genes impart a Far - phenotype on yeast cells. Genetics 147 : 1063–1076.
59. ChiA, HuttenhowerC, GeerLY, CoonJJ, SykaJE, et al. (2007) Analysis of phosphorylation sites on proteins from Saccharomyces cerevisiae by electron transfer dissociation (ETD) mass spectrometry. Proc Natl Acad Sci U S A 104 : 2193–8.
60. HoltLJ, TuchBB, VillénJ, JohnsonAD, GygiSP, et al. (2009) Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science 325 : 1682–1686.
61. MunozI, SimonE, CasalsN, ClotetJ, ArinoJ (2003) Identification of multicopy suppressors of cell cycle arrest at the G1-S transition in Saccharomyces cerevisiae. Yeast 20 : 157–169.
62. BodenmillerB, WankaS, KraftC, UrbanJ, CampbellD, et al. (2010) Phosphoproteomic analysis reveals interconnected system-wide responses to perturbations of kinases and phosphatases in yeast. Sci Signal 3: rs4.
63. BarbetNC, SchneiderU, HelliwellSB, StansfieldI, TuiteMF, et al. (1996) TOR controls translation initiation and early G1 progression in yeast. Mol Biol Cell 7 : 25–42.
64. Fernandez-SarabiaMJ, SuttonA, ZhongT, ArndtKT (1992) SIT4 protein phosphatase is required for the normal accumulation of SWI4, CLN1, CLN2, and HCS26 RNAs during late G1. Genes Dev 6 : 2417–2428.
65. CutlerNS, PanX, HeitmanJ, CardenasME (2001) The TOR signal transduction cascade controls cellular differentiation in response to nutrients. Mol Biol Cell 12 : 4103–4113.
66. PassmoreS, ElbleR, TyeBK (1989) A protein involved in minichromosome maintenance in yeast binds a transcriptional enhancer conserved in eukaryotes. Genes Dev 3 : 921–935.
67. ChristC, TyeBK (1991) Functional domains of the yeast transcription/replication factor MCM1. Genes Dev 5 : 751–763.
68. TreismanR, AmmererG (1992) The SRF and MCM1 transcription factors. Curr Opin Genet Dev 2 : 221–226.
69. PipesGC, CreemersEE, OlsonEN (2006) The myocardin family of transcriptional coactivators: versatile regulators of cell growth, migration, and myogenesis. Genes Dev 20 : 1545–1556.
70. BarralY, JentschS, MannC (1995) G1 cyclin turnover and nutrient uptake are controlled by a common pathway in yeast. Genes Dev 9 : 399–409.
71. XinX, RualJF, Hirozane-KishikawaT, HillDE, VidalM, et al. (2009) Shifted Transversal Design smart-pooling for high coverage interactome mapping. Genome Res 19 : 1262–1269.
72. YuH, BraunP, YildirimMA, LemmensI, VenkatesanK, et al. (2008) High-quality binary protein interaction map of the yeast interactome network. Science 322 : 104–110.
73. GelperinDM, WhiteMA, WilkinsonML, KonY, KungLA, et al. (2005) Biochemical and genetic analysis of the yeast proteome with a movable ORF collection. Genes Dev 19 : 2816–2826.
74. BrachmannCB, DaviesA, CostGJ, CaputoE, LiJ, et al. (1998) Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14 : 115–132.
75. LiZ, VizeacoumarFJ, BahrS, LiJ, WarringerJ, et al. (2011) Systematic exploration of essential yeast gene function with temperature-sensitive mutants. Nat Biotechnol 29 : 361–367.
76. CostanzoM, BaryshnikovaA, BellayJ, KimY, SpearED, et al. (2010) The genetic landscape of a cell. Science 327 : 425–431.
77. WinzelerEA, ShoemakerDD, AstromoffA, LiangH, AndersonK, et al. (1999) Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285 : 901–906.
78. HughesRE, LoRS, DavisC, StrandAD, NealCL, et al. (2001) Altered transcription in yeast expressing expanded polyglutamine. Proc Natl Acad Sci U S A 98 : 13201–13206.
79. HoY, GruhlerA, HeilbutA, BaderGD, MooreL, et al. (2002) Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415 : 180–183.
80. HuY, RolfsA, BhullarB, MurthyTV, ZhuC, et al. (2007) Approaching a complete repository of sequence-verified protein-encoding clones for Saccharomyces cerevisiae. Genome Res 17 : 536–543.
Štítky
Genetika Reprodukční medicína
Článek Attachment Site Selection and Identity in Bxb1 Serine Integrase-Mediated Site-Specific RecombinationČlánek High-Resolution Transcriptome Maps Reveal Strain-Specific Regulatory Features of Multiple IsolatesČlánek Neuropeptides Function in a Homeostatic Manner to Modulate Excitation-Inhibition Imbalance inČlánek Implicates Tyrosine-Sulfated Peptide Signaling in Susceptibility and Resistance to Root Infection
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 5- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- IVF a rakovina prsu – zvyšují hormony riziko vzniku rakoviny?
-
Všechny články tohoto čísla
- Functional Elements Are Embedded in Structurally Constrained Sequences
- RNA–Mediated Epigenetic Heredity Requires the Cytosine Methyltransferase Dnmt2
- Loss of Expression and Promoter Methylation of SLIT2 Are Associated with Sessile Serrated Adenoma Formation
- Attachment Site Selection and Identity in Bxb1 Serine Integrase-Mediated Site-Specific Recombination
- Human Genetics in Rheumatoid Arthritis Guides a High-Throughput Drug Screen of the CD40 Signaling Pathway
- Genome-Wide Analysis in German Shepherd Dogs Reveals Association of a Locus on CFA 27 with Atopic Dermatitis
- Liver X Receptors Protect from Development of Prostatic Intra-Epithelial Neoplasia in Mice
- Chromosomal Organization and Segregation in
- A Statistical Framework for Joint eQTL Analysis in Multiple Tissues
- Cell Polarity and Patterning by PIN Trafficking through Early Endosomal Compartments in
- Bck2 Acts through the MADS Box Protein Mcm1 to Activate Cell-Cycle-Regulated Genes in Budding Yeast
- High-Resolution Transcriptome Maps Reveal Strain-Specific Regulatory Features of Multiple Isolates
- Neuropeptides Function in a Homeostatic Manner to Modulate Excitation-Inhibition Imbalance in
- A Compendium of Nucleosome and Transcript Profiles Reveals Determinants of Chromatin Architecture and Transcription
- Wnt Signaling Regulates the Lineage Differentiation Potential of Mouse Embryonic Stem Cells through Tcf3 Down-Regulation
- Filamin and Phospholipase C-ε Are Required for Calcium Signaling in the Spermatheca
- The Specificity and Flexibility of L1 Reverse Transcription Priming at Imperfect T-Tracts
- Imputation-Based Meta-Analysis of Severe Malaria in Three African Populations
- Implicates Tyrosine-Sulfated Peptide Signaling in Susceptibility and Resistance to Root Infection
- Clathrin and AP2 Are Required for Phagocytic Receptor-Mediated Apoptotic Cell Clearance in
- Encodes CDF Transporters That Excrete Zinc from Intestinal Cells of and Act in a Parallel Negative Feedback Circuit That Promotes Homeostasis
- Global Properties and Functional Complexity of Human Gene Regulatory Variation
- DNA Binding of the Cell Cycle Transcriptional Regulator GcrA Depends on N6-Adenosine Methylation in and Other
- Side Effects: Substantial Non-Neutral Evolution Flanking Regulatory Sites
- From Paramutation to Paradigm
- From Mouse to Human: Evolutionary Genomics Analysis of Human Orthologs of Essential Genes
- Distinct Translational Control in CD4 T Cell Subsets
- Female Bias in and Regulation by the Histone Demethylase KDM6A
- ATM–Dependent MiR-335 Targets CtIP and Modulates the DNA Damage Response
- HDAC7 Is a Repressor of Myeloid Genes Whose Downregulation Is Required for Transdifferentiation of Pre-B Cells into Macrophages
- The Majority of Primate-Specific Regulatory Sequences Are Derived from Transposable Elements
- Identification of Meiotic Cyclins Reveals Functional Diversification among Plant Cyclin Genes
- EGL-13/SoxD Specifies Distinct O and CO Sensory Neuron Fates in
- Congruence of Additive and Non-Additive Effects on Gene Expression Estimated from Pedigree and SNP Data
- Using Extended Genealogy to Estimate Components of Heritability for 23 Quantitative and Dichotomous Traits
- Ikbkap/Elp1 Deficiency Causes Male Infertility by Disrupting Meiotic Progression
- Analysis of the Genetic Basis of Disease in the Context of Worldwide Human Relationships and Migration
- Duplication and Retention Biases of Essential and Non-Essential Genes Revealed by Systematic Knockdown Analyses
- Strong Purifying Selection at Synonymous Sites in
- , a Susceptibility Gene for Type 1 and Type 2 Diabetes, Modulates Pancreatic Beta Cell Apoptosis via Regulation of a Splice Variant of the BH3-Only Protein
- Chromosome Movements Promoted by the Mitochondrial Protein SPD-3 Are Required for Homology Search during Meiosis
- The Secretory Pathway Calcium ATPase PMR-1/SPCA1 Has Essential Roles in Cell Migration during Embryonic Development
- The Genomic Signature of Crop-Wild Introgression in Maize
- CDK4 T172 Phosphorylation Is Central in a CDK7-Dependent Bidirectional CDK4/CDK2 Interplay Mediated by p21 Phosphorylation at the Restriction Point
- Genome-Wide Identification of Regulatory RNAs in the Human Pathogen
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Using Extended Genealogy to Estimate Components of Heritability for 23 Quantitative and Dichotomous Traits
- HDAC7 Is a Repressor of Myeloid Genes Whose Downregulation Is Required for Transdifferentiation of Pre-B Cells into Macrophages
- Female Bias in and Regulation by the Histone Demethylase KDM6A
- ATM–Dependent MiR-335 Targets CtIP and Modulates the DNA Damage Response
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



