-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
ETS-4 Is a Transcriptional Regulator of Life Span in
Aging is a complex phenotype responsive to a plethora of environmental inputs; yet only a limited number of transcriptional regulators are known to influence life span. How the downstream expression programs mediated by these factors (or others) are coordinated into common or distinct set of aging effectors is an addressable question in model organisms, such as C. elegans. Here, we establish the transcription factor ETS-4, an ortholog of vertebrate SPDEF, as a longevity determinant. Adult worms with ets-4 mutations had a significant extension of mean life span. Restoring ETS-4 activity in the intestine, but not neurons, of ets-4 mutant worms rescued life span to wild-type levels. Using RNAi, we demonstrated that ets-4 is required post-developmentally to regulate adult life span; thus uncoupling the role of ETS-4 in aging from potential functions in worm intestinal development. Seventy ETS-4-regulated genes, identified by gene expression profiling of two distinct ets-4 alleles and analyzed by bioinformatics, were enriched for known longevity effectors that function in lipid transport, lipid metabolism, and innate immunity. Putative target genes were enriched for ones that change expression during normal aging, the majority of which are controlled by the GATA factors. Also, some ETS-4-regulated genes function downstream of the FOXO factor, DAF-16 and the insulin/IGF-1 signaling pathway. However, epistasis and phenotypic analyses indicate that ets-4 functioned in parallel to the insulin/IGF-1 receptor, daf-2 and akt-1/2 kinases. Furthermore, ets-4 required daf-16 to modulate aging, suggesting overlap in function at the level of common targets that affect life span. In conclusion, ETS-4 is a new transcriptional regulator of aging, which shares transcriptional targets with GATA and FOXO factors, suggesting that overlapping pathways direct common sets of lifespan-related genes.
Published in the journal: . PLoS Genet 6(9): e32767. doi:10.1371/journal.pgen.1001125
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1001125Summary
Aging is a complex phenotype responsive to a plethora of environmental inputs; yet only a limited number of transcriptional regulators are known to influence life span. How the downstream expression programs mediated by these factors (or others) are coordinated into common or distinct set of aging effectors is an addressable question in model organisms, such as C. elegans. Here, we establish the transcription factor ETS-4, an ortholog of vertebrate SPDEF, as a longevity determinant. Adult worms with ets-4 mutations had a significant extension of mean life span. Restoring ETS-4 activity in the intestine, but not neurons, of ets-4 mutant worms rescued life span to wild-type levels. Using RNAi, we demonstrated that ets-4 is required post-developmentally to regulate adult life span; thus uncoupling the role of ETS-4 in aging from potential functions in worm intestinal development. Seventy ETS-4-regulated genes, identified by gene expression profiling of two distinct ets-4 alleles and analyzed by bioinformatics, were enriched for known longevity effectors that function in lipid transport, lipid metabolism, and innate immunity. Putative target genes were enriched for ones that change expression during normal aging, the majority of which are controlled by the GATA factors. Also, some ETS-4-regulated genes function downstream of the FOXO factor, DAF-16 and the insulin/IGF-1 signaling pathway. However, epistasis and phenotypic analyses indicate that ets-4 functioned in parallel to the insulin/IGF-1 receptor, daf-2 and akt-1/2 kinases. Furthermore, ets-4 required daf-16 to modulate aging, suggesting overlap in function at the level of common targets that affect life span. In conclusion, ETS-4 is a new transcriptional regulator of aging, which shares transcriptional targets with GATA and FOXO factors, suggesting that overlapping pathways direct common sets of lifespan-related genes.
Introduction
The emerging picture from studies with model organisms is that animal life span is regulated by coordination of gene regulatory networks in response to environmental inputs. In C. elegans a number of transcription factors function as genetic modifiers of aging, including Forkhead (DAF-16 and PHA-4) and GATA (ELT-3, ELT-5, and ELT-6) factors [1]–[3]. The transcription factors that function in life span determination often respond to evolutionarily conserved pathways and cellular processes, including insulin/IGF-1 signaling, c-Jun N-terminal kinase signaling (JNK), the Target of rapamycin pathway (TOR), caloric intake, mitochondrial respiration and signaling from the germ line [1], [2], [4]–[8]. Gene expression profiling has identified overlapping downstream targets for these transcription factors that regulate development, metabolism, reproduction, stress response and innate immunity [3], [9]–[11]. Consistent with this complexity, genetic tests show that disruption of individual downstream target genes has a modest impact on longevity. Therefore, the downstream effectors of each transcription factor are proposed to act collectively to mediate the significant impact of signaling pathways on life span [9].
In this study, we identify the ETS transcription factor, ETS-4, as a longevity determinant in C. elegans. Gene knock-out studies performed for 20 of the 26 ETS genes in mice have implicated ETS factors in diverse cellular processes such as proliferation, differentiation, migration, apoptosis, and cell-cell interactions [12]–[15]. However, discerning molecular mechanisms of ETS protein function in mice has been complicated by the large number of ETS paralogs expressed in any particular cell type [15], [16]. C. elegans, with only ten ets genes, provides a simpler and more genetically tractable model to investigate ETS factor function in vivo.
C. elegans ets-4 is the apparent ortholog of vertebrate SPDEF/SAM pointed domain containing ETS transcription factor (also known as PDEF/Prostate derived ETS factor) (Figure S1). The mRNA levels of SPDEF are altered in breast and prostate tumors [17], [18]. Studies using tumor cell lines show that SPDEF affects cell migration and invasion pathways [18]–[23]. A deletion allele in mice suggests a role in specialized intestinal epithelial cell differentiation [24]. Thus, SPDEF may have significant disease relevance, yet its physiological role in normal animal development and homeostasis is not completely understood.
To decipher ets-4 function, we undertook a reverse genetics approach in C. elegans and established ets-4 as a transcriptional regulator of longevity. Expression of ets-4 was observed in a number of tissues, however, transgenic rescue experiments implicated the intestine in the aging phenotype. Gene expression profiling identified ETS-4-regulated genes that function in life span determination. Strikingly, a significant number of these life span effectors have been previously shown to function downstream of the insulin/IGF-1 signaling pathway as well as the GATA factor, ELT-3. Genetic tests reveal that ets-4 functions in parallel to the insulin/IGF-1 signaling pathway, yet requires the FOXO transcription factor, daf-16, to modulate life span. Taken together, our findings identify the physiological role of ETS-4 in C. elegans and indicate transcriptional control of aging effectors by ETS-4.
Results
ETS-4 Is a Novel Longevity Determinant
To determine the function of ETS-4 in vivo, ets-4(ok165) worms carrying a deletion at the ets-4 locus were analyzed. Coding sequences for both the ETS DNA binding domain, and the PNT domain, which is a protein-protein interaction domain conserved in a subset of ETS proteins, were lacking in the ets-4(ok165) worms (Figure 1A and 1C). Animals carrying the ok165 allele lacked full-length ets-4 mRNA as confirmed by RT-PCR analysis (Figure 1B). Therefore, we propose that ets-4(ok165) is a null allele for ETS-4 function.
Fig. 1. Schematic of ets-4 gene structure and gene deletion analysis. 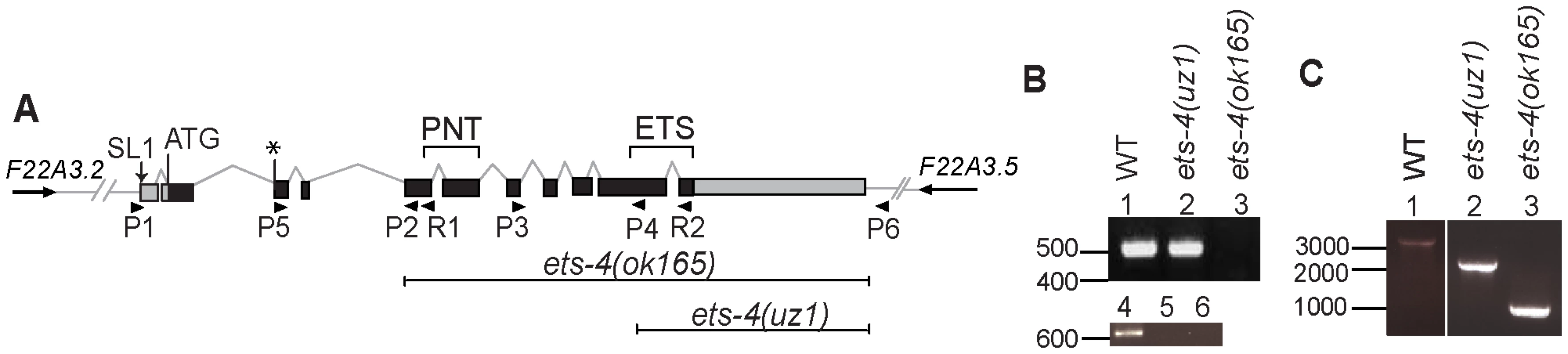
(A) Gene structure of ets-4. Black boxes represent exons linked by lines that indicate introns. The 3′UTR and 5′UTR are shaded gray. The proposed translation start site ATG, confirmed site of addition of the SL1 sequence and exons encoding the ETS and PNT domains are indicated. * denotes the start of an alternative isoform (WormBase web site, http://www.wormbase.org, release WS214). The orientations of neighboring genes are represented by arrows. Sequences deleted in the ok165 and uz1 alleles are marked. The position of primers used for reverse transcription (R1, R2) and PCR (P1, P2, P3, P4, P5, P6) analyses are indicated. (B) Agarose gel showing RT-PCR analysis of wild-type (WT), ets-4(uz1) and ets-4(ok165) worms with primers R1, P1, P2 (top panel) and R2, P3, P4 (bottom panel). DNA size indicated in bp. (C) Agarose gel showing PCR analysis of wild-type (WT), ets-4(uz1) and ets-4(ok165) worms with primers P5 and P6. DNA size indicated in bp. After outcrossing six times to the wild-type (N2) strain, the ets-4(ok165) worms were examined for phenotypes. The larval developmental time, measured as the time taken for L1 larvae to reach the young adult stage at 20°C, was 6–8 hr longer in ets-4(ok165) than wild-type worms (Figure 2A). Despite this 10–13% delay, ets-4(ok165) larvae exhibited apparently wild-type development and morphology. Neither arrests at particular larval stages nor molting defects were observed. The self brood size of ets-4(ok165) worms was similar to that of wild-type worms, suggesting normal fecundity (Figure 2B). However, a difference in the rate of egg-laying was observed (Figure 2C). During the peak egg-laying period (day 2 of egg-laying) the ets-4(ok165) worms laid significantly fewer eggs (109±5) than wild-type worms (142±6) (Figure 2C). In addition, ets-4(ok165) hermaphrodites produced significantly more progeny (39±6) later in life (day 4 of egg-laying) than wild-type worms (14±2) (Figure 2C). The ets-4(ok165) males exhibited mating efficiency comparable to wild-type males in crosses with temperature-sensitive fem-3(e2006) hermaphrodites [25] that are incapable of producing self-progeny at the restrictive temperature (data not shown). In summary, ets-4(ok165) worms exhibited a 10–13% delay in larval development and an altered egg-laying rate, but were wild-type in morphology, development and fecundity suggesting normal fitness.
Fig. 2. ETS-4 affects larval developmental rate and egg-laying rate. 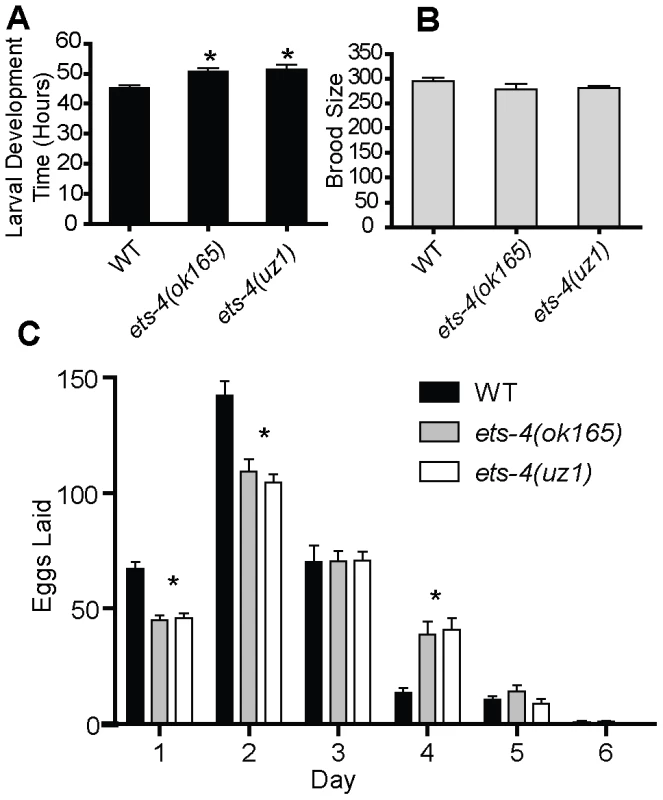
(A) The larval development time (hr) defined as the time from L1 to pre-fertile adult for wild-type (WT), ets-4(ok165) and ets-4(uz1) worms at 20°C. Values are mean ± SEM and * indicates p<0.05. (B) Total number of progeny (brood size, mean ± SEM) at 20°C was counted for individual WT (n = 11), ets-4(ok165) (n = 16) and ets-4(uz1) (n = 18) worms. (C) The number of eggs laid (mean ± SEM) during each day of the egg-laying period for WT, ets-4(ok165), and ets-4(uz1) worms at 20°C (* indicates p<0.001). Considering a post-developmental role for ets-4, we monitored the adult life span of ets-4(ok165) worms. Because different laboratory strains of C. elegans can have significantly different life spans [26], we compared the life spans of ets-4(ok165) animals with those of isogenic ets-4(+) controls obtained by outcrossing ets-4(ok165) to the wild-type (N2) strain. Age-matched wild-type or ets-4(ok165) L4 stage larvae were picked and the first day of adulthood counted as day one. At 25°C, the mean adult life span of ets-4(ok165) worms (18.0±0.4 days) was significantly longer than that of isogenic ets-4(+) wild-type worms (13.3±0.6 days) (Figure 3A and Table 1). Thus, loss of ets-4 led to a significantly longer mean adult life span compared to wild-type worms at 25°C (Figure 3A, Table 1 and Table S1). Since growth temperature strongly influences C. elegans longevity [27], the life span of ets-4(ok165) animals was also monitored at 20°C. The mean adult life span of ets-4(ok165) animals at 20°C (27.1±0.8 days) was significantly longer than that of isogenic ets-4(+) wild-type worms (15.4±0.6 days), confirming the extended life span phenotype of ets-4 null mutant animals (Figure 3B and Table 1). Because feeding defective (eat) mutant animals are also slow-growing and long-lived [6], [28], the feeding behavior of ets-4(ok165) was examined by recording the pharyngeal pumping and defecation rates. Under well-fed conditions, the feeding behavior of ets-4(ok165) worms was indistinguishable from that of wild-type animals (Figure S2). Thus, ETS-4 regulates life span without modifying feeding behavior.
Fig. 3. ETS-4 regulates C. elegans life span. 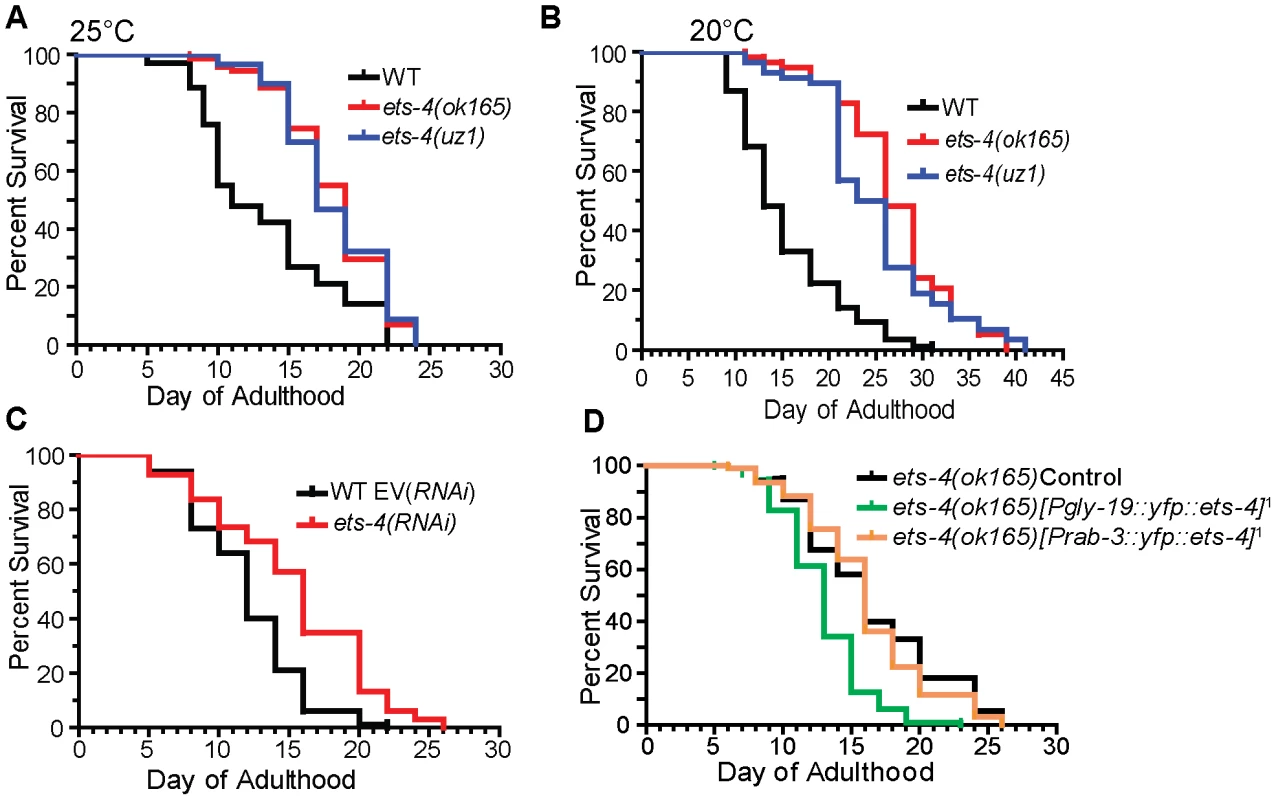
Survival curves from life span experiments. See Table 1 and Table S1 for data from additional trials, mean life span and statistical analyses. (A) Survival curves of wild-type (WT), ets-4(ok165) and ets-4(uz1) worms at 25°C. (B) Survival curves of wild-type (WT), ets-4(ok165) and ets-4(uz1) worms at 20°C. (C) Survival curves for wild-type (WT) worms grown at 25°C and subjected to ets-4(RNAi) or an empty vector (EV) control RNAi starting at the L4 stage. (D) Survival curves of ets-4(ok165) transgenic animals with tissue-specific expression of ets-4 in intestinal cells, ets-4(ok165);[Pgly-19::yfp::ets-4]1, or in neurons, ets-4(ok165);[Prab-3::yfp::ets-4]1. 1refers to one of the two independent transgenic lines generated. Injection control strain indicated as ets-4(ok165) Control. Tab. 1. Summary of Life Span Analysis for ets-4(ok165) and ets-4(uz1) Wormsa. 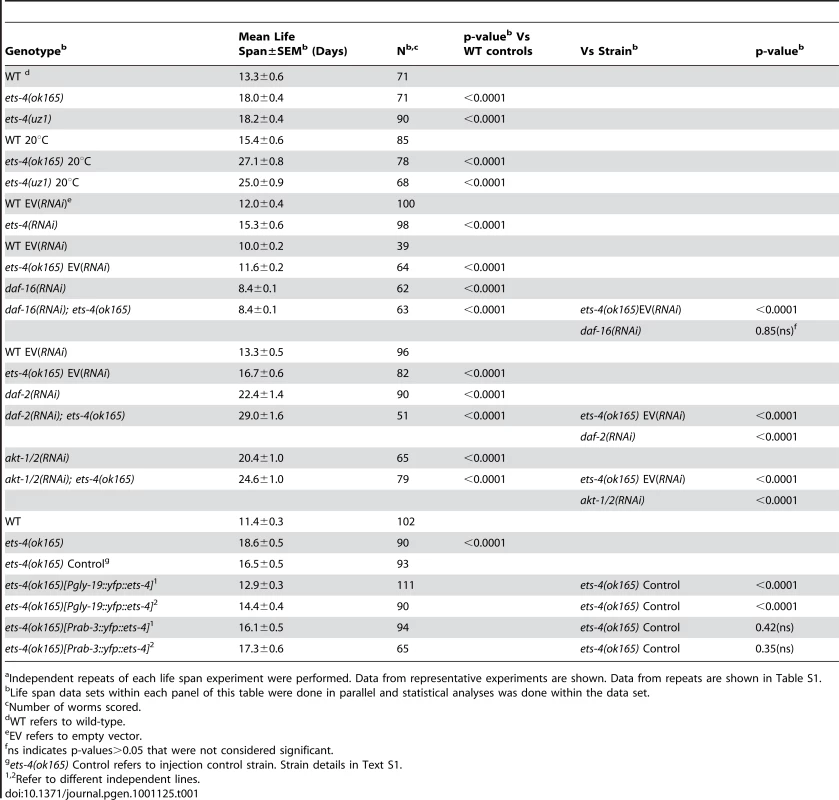
Independent repeats of each life span experiment were performed. Data from representative experiments are shown. Data from repeats are shown in Table S1. To further characterize the role of ets-4 in life span regulation, we initiated ets-4 inactivation by RNAi at the L4 larval stage to bypass potential earlier developmental roles [29]. Similar to the long-lived phenotype of ets-4(ok165) mutant animals, ets-4(RNAi) on wild-type worms resulted in significant extension of mean adult life span (Figure 3C and Table 1). These data suggest that ets-4 functions post-developmentally in regulation of adult life span.
We generated a second deletion allele of ets-4, uz1, which removes sequences coding for most of the ETS domain and the 3′UTR, but retains the PNT domain coding sequence (Figure 1A and 1C). An abridged ets-4 mRNA that could encode a truncated protein was detected in ets-4(uz1) worms (Figure 1B). Similar to the ets-4(ok165) worms, we observed slow larval growth, altered progeny production and extended life span phenotypes in ets-4(uz1) mutant animals (Figure 2, Figure 3A and 3B). Thus, examination of this second ets-4 deletion allele corroborated the loss-of-function phenotypes observed in ets-4(ok165) mutant animals and enabled further use of this allele. However, ets-4(uz1) worms exhibited several unique phenotypes not observed in ets-4(ok165) worms including ruptured vulva, distorted seam cell syncytia and broken alae (data not shown). We speculate that these added effects are likely due to the interfering activity of the truncated ETS-4 encoded by the uz1 allele that would lack the ability to bind to ETS-4 target genes and yet, may interact with various protein partners through the PNT domain. Therefore, the additional phenotypes associated with the ets-4(uz1) allele may be involved with, but not necessarily restricted to, normal ETS-4 function and were not studied further. Nevertheless, analyses of two distinct deletion alleles of ets-4 enabled the identification of ets-4 loss-of-function phenotypes.
Tissues That Control Life Span Express ets-4
To better understand how ETS-4 regulates life span, we sought to identify the tissues in which ETS-4 functions. GFP reporter constructs containing 5 kb of the ets-4 promoter alone (Pets-4::gfp) or including the genomic DNA coding for ETS-4 (ets-4::gfp) were used to generate several independent transgenic lines. Robust GFP expression for both constructs was observed in the intestinal cells of transgenic worms starting at the 3-fold embryonic stage and was maintained through larval development and in the adult (Figure 4A and 4B). GFP expression was also seen in several cells of the anterior and posterior bulbs of the pharynx and in seam cells (Figure 4B, 4C and 4D). Lastly, ets-4 expression was observed in a few unidentified cells of the vulva, hypodermal nuclei, several unidentified neurons, labial socket cells of the head, and a few cells of the rectum (data not shown). Thus, these results expanded the observations made in previous studies using shorter regions of the ets-4 promoter in GFP constructs [30]–[32] and demonstrated ets-4 expression in the intestine and neurons, key tissues known to regulate longevity in worms [33]–[35].
Fig. 4. Expression pattern of ets-4. 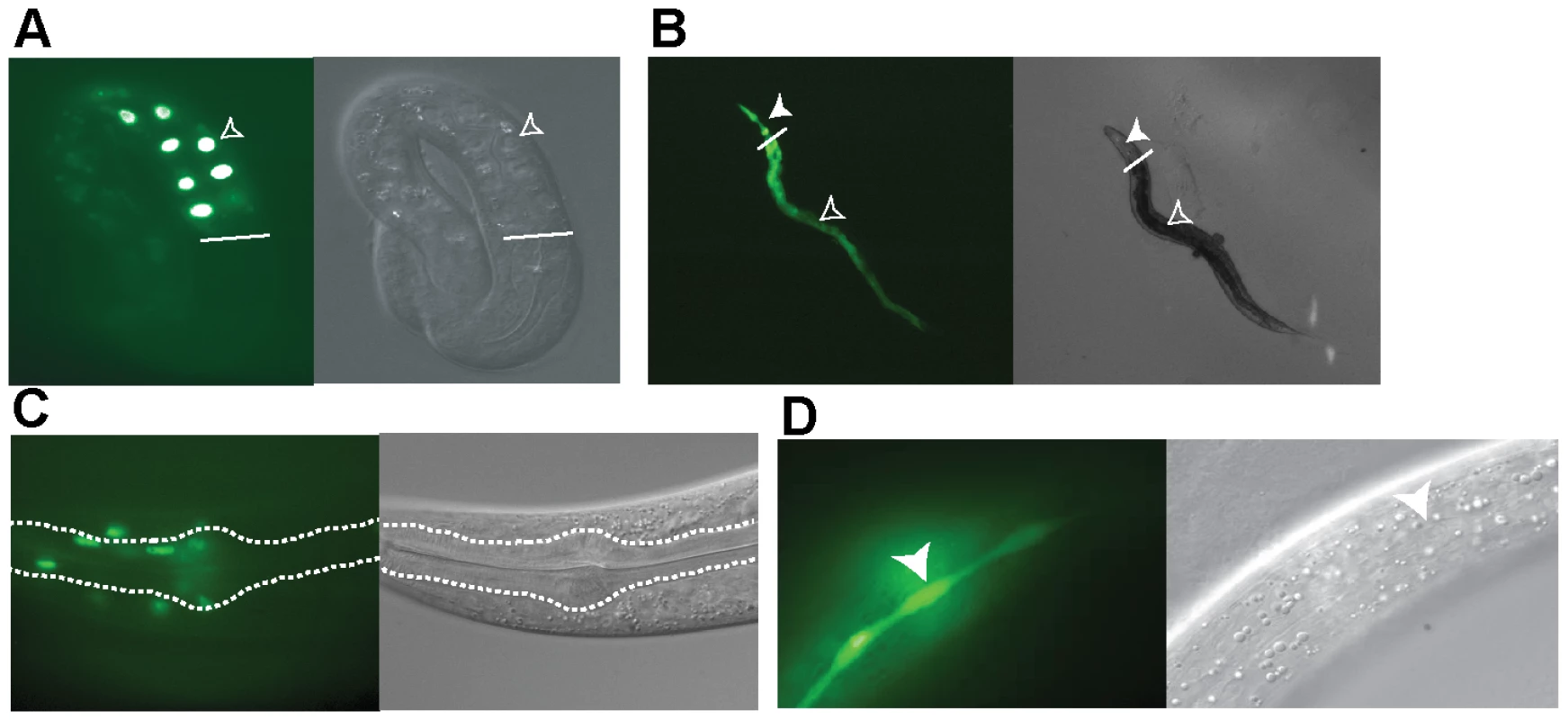
Transgenic worms expressing the Pets-4::gfp promoter fusion (B and D) or the ets-4::gfp promoter and open reading frame construct (A and C) are shown. Differential interference contrast (DIC) and fluorescence images were visualized using a Zeiss Axioskop 2 microscope. (A) GFP expression in the intestinal cells of a 3-fold stage embryo (open arrowhead). White line indicates start of the intestine. (B) GFP expression in the intestinal (open arrowhead) and pharyngeal (filled arrowhead) cells. White line indicates the demarcation between the pharynx and intestine. (C) GFP expression in the cells in and around the pharynx of an adult worm. The procorpus, metacarpus and part of the isthmus are outlined. (D) GFP expression in the seam cell syncytium (filled arrowhead) of an adult worm. ETS-4 Functions in the Intestine to Regulate Life Span
Studies in mammals, flies and worms have identified subsets of cells, including adipose, intestinal and neuronal tissues, that affect the rate of aging of the entire organism [33]–[40]. To identify cell types in which ETS-4 functions to modulate life span, a transgene encoding a YFP::ETS-4 fusion was expressed in intestinal cells or neurons of ets-4(ok165) worms by use of the gly-19 or rab-3 promoters, respectively [35], [41], [42]. Analysis of at least two independent transgenic lines showed that YFP::ETS-4 expressed in intestinal cells restored the life span of ets-4(ok165) worms to wild-type (Figure 3D, Table 1 and Table S1). In contrast, the extended life span phenotype of ets-4(ok165) worms was not affected by YFP::ETS-4 expression in neurons (Figure 3D, Table 1 and Table S1). Additionally, YFP::ETS-4 expression in either cell type did not rescue the altered egg-laying rate phenotype of ets-4(ok165) worms, indicating that the longevity and egg-laying phenotypes were separable (Figure S3). Animals expressing YFP::ETS-4 had wild-type brood sizes, suggesting normal fecundity and fitness (Figure S3). Moreover, because the life span of rescued lines was not shortened beyond wild-type controls, we concluded that YFP::ETS-4 fusion was not toxic in these tissues (Table 1). In summary, ETS-4 functions in the intestine to modulate life span.
ETS-4 Regulated Genes
Because ETS-4 is implicated to be a transcription factor, a role in life span determination predicts a set of ETS-4 target genes that function in aging. We took advantage of the two distinct strains with a disrupted ets-4 locus to investigate effects on gene expression. We chose late L4 stage larvae because of ease in staging and relevance to adult intestine function. Microarray-based expression profiling of wild-type and ets-4(ok165) larvae identified 145 genes whose expression was altered with 88 genes down-regulated 2.2 fold or more (Table S2). qRT-PCR analyses in age-matched, one-day old adults confirmed the differential expression observed for nine out of nine genes selected randomly from the top-thirty changed genes (Figure S4). As predicted by the broader phenotypic consequences observed in ets-4(uz1) worms, more genes (542) displayed altered expression in these animals than in ets-4(ok165) worms (Table S3). qRT-PCR controls in age-matched, 1-day old ets-4(uz1) and wild-type adult worms confirmed the expression changes observed for eight out of eight genes (Figure S4).
Because animals carrying either ets-4 deletion displayed similar life span extension, we predicted that the genes overlapping in these data sets would be enriched for aging effectors. A statistically significant overlap of 70 genes with altered expression in ets-4(ok165) and ets-4(uz1) worms was identified (p<0.0001) (Table 2 and Table S4). To determine, in an unbiased manner, whether these 70 ETS-4-regulated genes represented a particular biological pathway, we performed gene ontology analysis using GOstat [43]. The top five overrepresented categories yielded by this analysis include lipid transport (vit-2, vit-3, vit-4, and vit-5), multicellular organismal aging (vit-5, vit-2, thn-1, and lys-7) and fatty acid metabolic process (acdh-2, ech-9, and C48B4.1) (Table 3). Five control gene lists of the same size generated randomly from genes represented on the expression arrays did not show these classes to be overrepresented.
Tab. 2. Comparison of Genes Altered in ets-4 Mutant Worms to Other Published Gene Lists. 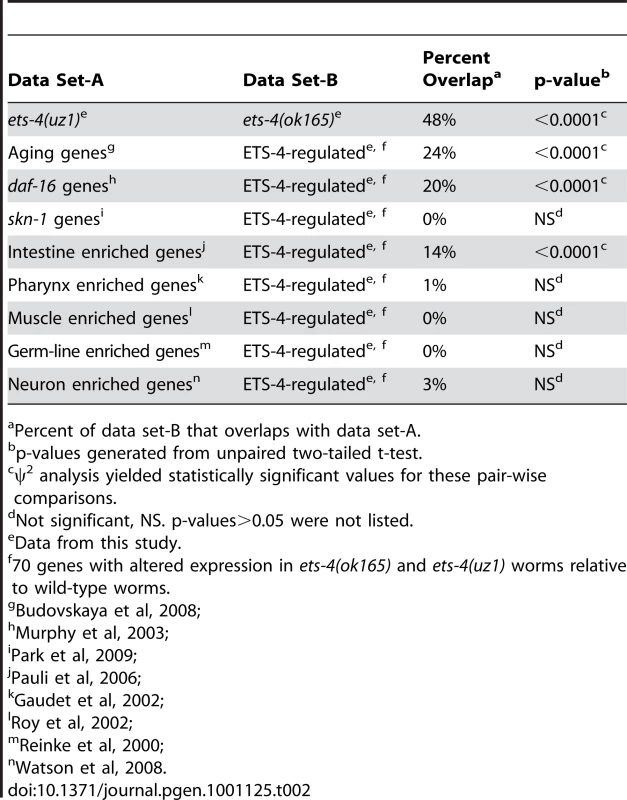
Percent of data set-B that overlaps with data set-A. Tab. 3. Top Five Overrepresented Ontology Terms of ETS-4-Regulated Genesa. 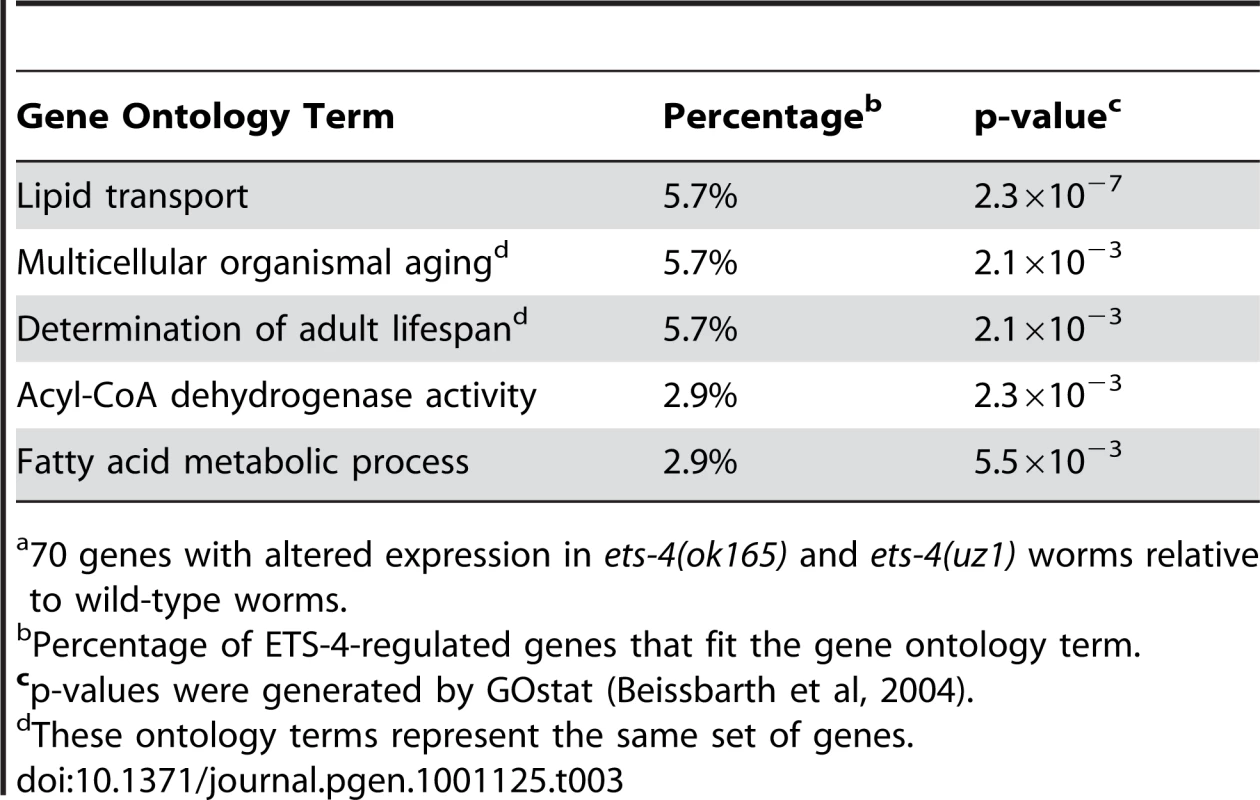
70 genes with altered expression in ets-4(ok165) and ets-4(uz1) worms relative to wild-type worms. Restoration of ETS-4 function in the intestine rescued life span of ets-4 null worms to wild-type (Figure 3D and Table 1). To test whether ETS-4-regulated genes were biased towards expression in a particular cell type, we compared our microarray data set to tissue-specific expression data from previous studies. Strikingly, the ETS-4-regulated gene list was significantly (p<0.0001) enriched only with intestinal genes [44], and not with germ-line [45], muscle [46], pharyngeal [47] or neuronal genes [48] (Table 2). This result did not vary when down - or up-regulated gene sets in the ets-4 mutant animals were analyzed separately for enrichment of cell-type specific gene expression (data not shown). Thus, the molecular signature of ETS-4, identified by gene expression profiling, substantiates a role for ets-4 in the intestine.
Lipid storage and metabolism have been linked to longevity regulation [36], [39], [40], [49] Also, expression of yolk protein genes (vit genes) and genes regulating fatty acid β-oxidation, such as an acyl-CoA dehydrogenase (acdh-2), an acyl-CoA oxidase (C48B4.1) and an enoyl-CoA hydratase (ech-9), were down-regulated in ets-4 mutant animals (Table S4). Thus, to explore a possible mechanism for life span regulation by ETS-4, we examined lipid levels in ets-4 null mutant animals. Lipid extracts from synchronized, one-day old adult, wild-type and ets-4 null mutant animals were subjected to thin-layer chromatography (TLC). The phospholipid (PL) and triacyglyceride (TAG) fractions from ets-4(ok165) worms visualized on TLC plates were similar to wild-type (Figure S5). The relative levels of triacylglycerol stores, as well as the fatty acid composition of phospholipid and triacylglycerol fractions, quantified by gas chromatography in age-matched, one-day old ets-4(ok165) adult animals, were not altered compared to wild-type worms (Figure S5). Thus, the altered expression of genes involved in lipid metabolism did not affect total lipid levels in ets-4(ok165) worms. Therefore, lipid homeostasis in ets-4(ok165) animals could likely be maintained due to the compensatory activities of other enzymes regulating fatty acid metabolism. Alternatively, lipid uptake, transport or storage in different tissues of ets-4(ok165) worms may be affected without an alteration in the total lipid levels within the whole organism.
Discerning the transcriptional targets of ETS-4 enabled us to ask whether ETS-4-regulated genes change expression during the course of normal aging [3]. We found that 24% of the 70 ETS-4-regulated genes were previously identified age-regulated genes (Table 2 and Table S4). This is a significantly higher overlap than that expected by random chance (p<0.0001), corroborating a function for ETS-4 in normal aging (Table 2). Interestingly, the GATA transcription factors ELT-3, ELT-5, and ELT-6 were shown to direct the age-regulation of a large fraction of genes that change expression with age [3]. Our data suggested that ETS-4 participated in directing the expression of a significant proportion of age-regulated genes.
To decipher whether a common set of targets exists for transcription factors that function in life span determination, we compared ETS-4-regulated genes with those that function downstream of the well-characterized insulin/IGF-1 signaling pathway. A comparison to the genes that act downstream of two components of the insulin/IGF-1 signaling pathway (daf-2 and daf-16) [10] indicated a 20% overlap, which represents a significant enrichment (p<0.0001) (Table 2 and Table S4). The overlapping gene set included genes that were down-regulated as well as genes up-regulated in ets-4(ok165) animals (Table S4). Also, 50% of the overlapping genes were up-regulated in daf-2 pathway mutant animals and repressed in daf-16; daf-2 double mutant animals, while the other half displayed the opposite expression profile (Table S4) [10] Since ets-4 and daf-2 loss-of-function mutations cause an extended life span phenotype, we predicted that the direction of expression changes for genes involved in aging will be similar in these mutant animals. Indeed, the genes identified by our ontology analyses as functioning in multicellular organismal aging (Table 3) were either down-regulated (vit-5 and vit-2) or up-regulated (thn-1 and lys-7) in both ets-4(ok165) and daf-2(−) animals (Table S4) [10]. Taken together, our analyses revealed a shared pattern of gene expression changes between in ets-4 mutant animals, insulin/IGF-1 signaling pathway mutant animals, and aging wild-type worms. Thus, a common set of transcriptional targets for ETS-4, the GATA factors (ELT-3, ELT-5, and ELT-6), and DAF-16 exist.
ETS-4 Functions as a Sequence-specific DNA Binding Transcription Factor
Analyzing global transcriptional changes in ets-4 mutant animals identified a number of genes regulated by ETS-4. Direct transcriptional targets are predicted to have ETS-4 binding sites in the transcription start site (TSS)-proximal regions. As a first step towards testing this hypothesis, we characterized the DNA binding and transcriptional activity of ETS-4. We tested the binding of ETS-4 to ETS binding sites previously described, which display a 5′-GGAA/T-3′ core recognition sequence [13], [50], which is consistent with the reported motif for the vertebrate orthologue, SPDEF [51]. ETS-4, purified from a bacterial expression system, bound to ETS binding sites displaying either a GGAA or GGAT core motif with similar high affinity (KD∼10-9 M) (Figure 5A and Figure 5B). Furthermore, a wild-type, but not mutated, ETS binding site, competed with ETS-4 binding to the labeled DNA (Figure 5C). These results indicated that ETS-4 bound to consensus ETS sites in a sequence-specific manner.
Fig. 5. In vitro DNA binding activity of ETS-4. 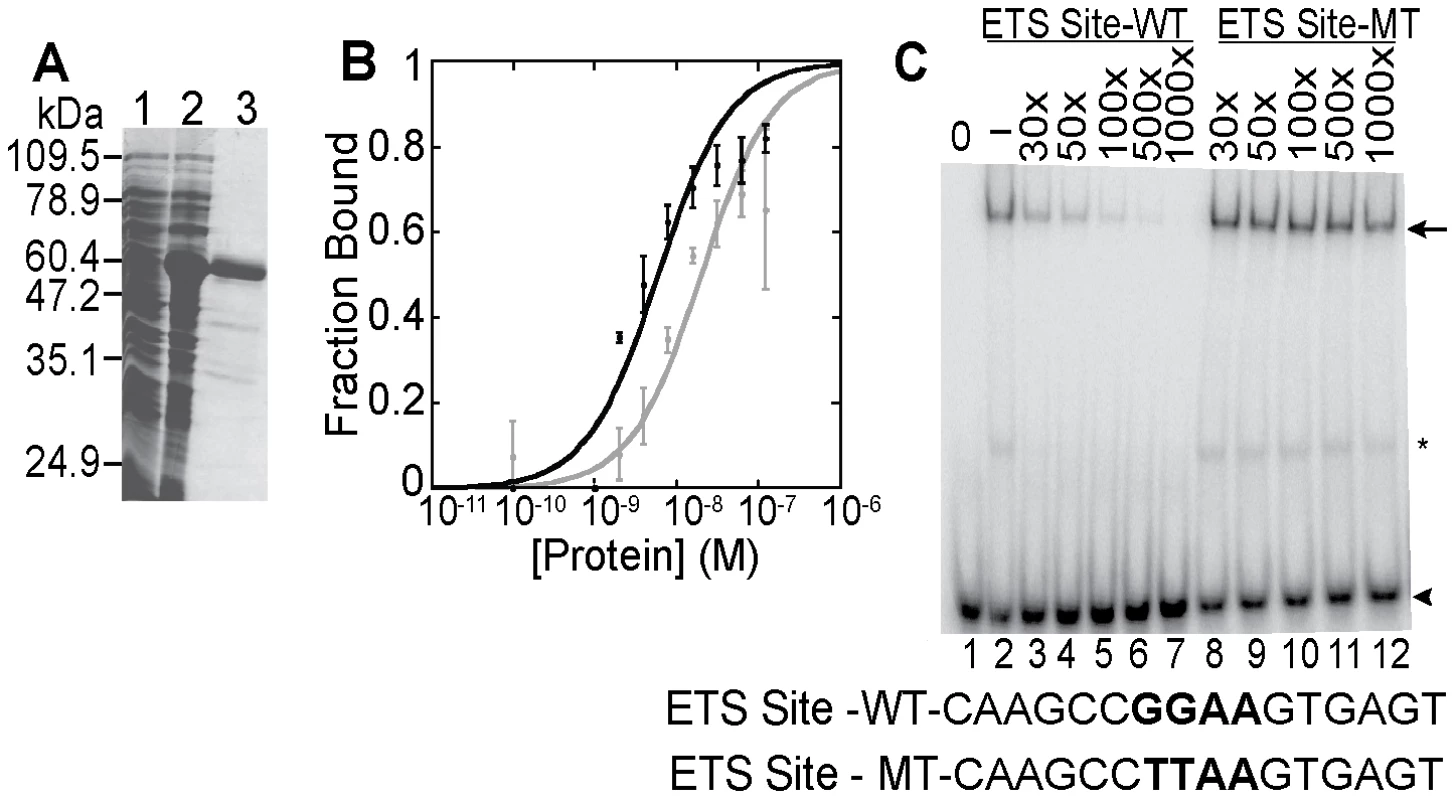
(A) Purification of recombinant ETS-4. SDS-PAGE gel, stained with coomassie blue, shows uninduced (lane 1), induced (lane 2) bacterial lysate and purified 6xHis tagged ETS-4 (lane 3). (B) DNA-binding isotherms derived from electrophoretic mobility shift assays (EMSAs) performed with increasing concentrations of recombinant ETS-4 (10−11 M to 10−7 M) incubated with radiolabeled DNA duplex containing a GGAA (black)- or GGAT (gray)-based ETS binding site. Binding curves represent the mean (±SD) from three independent EMSA experiments. (C) EMSA showing sequence specificity of ETS-4 binding. Radiolabeled DNA duplex (ETS Site-WT) was used at 10−10 M for all lanes. Unlabeled competitor DNA duplex was added in molar excess (X) (lanes 3–12). The competitor DNA contained either a GGAA sequence (ETS Site-WT) or a mutated sequence (TTAA) (ETS Site-MT). The arrowhead marks the mobility of unbound DNA. Binding of ETS-4 shifts the DNA to a slower migrating band (arrow). Band marked by asterisk is possibly due to an ETS-4 degradation product bound specifically to DNA. ETS proteins demonstrate functional diversity acting both as transcriptional activators and repressors [13],[52]. To determine the transcriptional activity of ETS-4, we tested ETS-4 in transcription assays in yeast and cultured mouse fibroblasts. In S. cerevisiae, LexA::ETS-4(1-345) activated transcription of a reporter gene via a promoter with LexA binding sites (Figure S6). ETS-4(1–345) also activated transcription in NIH3T3 cells (Figure S6). In yeast, the N-terminal ETS-4(1–125) fragment activated transcription of the reporter gene; however, this activity was lost with the inclusion of the PNT domain in ETS-4(1–200) (Figure S6). Indeed, we have mapped repressive function to the PNT domain including recruitment of repressive co-factors (data not shown). Thus, our data identify two regions of ETS-4 that activate transcription (amino acids 1–125 and 200–345) and suggest the ability of the PNT domain to modulate this activity towards a repressive function. This potential opposing transcriptional function is consistent with the hypothesis that both up - and down-regulated genes are direct ETS-4 targets.
To identify candidate direct targets of ETS-4, we looked for ETS binding sites conserved across six nematode species in the region from 1500 base pairs (bp) upstream to 500 bp downstream of the transcription start site (TSS) of ETS-4-regulated genes. The position specific weight matrix (PWM) used to search for ETS binding motifs encompasses the sites that ETS-4 bound in vitro (Figure 5 and Table S5). Using the MULTIZ alignment algorithm [53], at least one conserved ETS binding motif was identified in the transcriptional control regions of 54 of the 70 ETS-4 regulated genes (Table S5). Although this frequency did not represent an enrichment of conserved ETS sites compared to other randomly selected TSS-proximal regions (data not shown), we suspect this represents the general importance of ETS factors in many control regions and not the lack of biological relevance of these sites on ETS-4 regulated genes [54], [55]. It was striking that genes with conserved ETS binding motifs included not only 82% of the down-regulated genes, but also 72% of the up-regulated genes in ets-4 mutant animals (Table S5). This is consistent with the activating and repressive transcriptional functions of ETS-4 noted above. Also, several ETS proteins, in response to signaling pathways, fine-tune their transcriptional activity, functioning as activators or repressors [13], [52], [56]. In conclusion, we propose that ETS proteins may bind the transcriptional control regions and, thus, regulate directly the expression of 77% of the genes that were identified to function downstream of ETS-4.
To test the function of ETS-4 as a DNA binding transcription factor in a native context, we focused on vit-5, which is one of the genes down-regulated in ets-4 mutant worms that bears conserved ETS binding sites (Table S2, Table S4 and Table S5). vit-5 encodes a lipoprotein related to mammalian ApoB-100, a core LDL particle constituent [57]. Also, vit-5(RNAi) was shown previously to extend the mean life span of RNAi-sensitive wild-type worms [10]. Pvit-5::gfp expression was significantly reduced in the intestinal cells of ets-4(ok165) compared to wild-type worms suggesting that ETS-4 is necessary for inducing vit-5 expression (Figure S7). We conclude that ETS-4 is a transcription factor, fully competent to help orchestrate a transcriptional network involved in life span regulation.
ETS-4 Functions in Parallel to the Insulin/IGF-1 Receptor Signaling
To characterize how ets-4 modulates C. elegans lifespan, we tested whether ets-4 genetically interacted with other known longevity regulators. First, because ETS-4, ELT-3, and DAF-16 share common downstream targets, we asked if they function downstream of the same signaling pathway. The insulin/IGF-1 signaling pathway involves the activation of the insulin/IGF-1 receptor, DAF-2, which triggers a kinase cascade involving the serine/threonine kinases AKT-1 and AKT-2 culminating in the cytoplasmic sequestration and inhibition of the FOXO transcription factor DAF-16 [58], [59]. In addition to inhibiting DAF-16, the insulin/IGF-1 pathway also exerts a constant level of regulation on ELT-3 expression [3], [60]. We tested whether ets-4 modulated life span by acting downstream of the insulin/IGF-1 signaling pathway. To do this, we examined the genetic relationship between ets-4 and two major components of the insulin/IGF-1 signaling cascade - the receptor daf-2 and the kinases akt-1/2. For the life span comparisons, RNAi was used to inactivate insulin/IGF-1 signaling pathway genes in isogenic ets-4(ok165) and wild-type strains. If ets-4 regulates life span by acting downstream of the insulin/IGF-1 signaling pathway, then loss of ets-4 should not significantly extend the life span of long-lived worms lacking insulin/IGF-1 signaling. Alternatively, further extension of the life span of long-lived ets-4 null mutant worms upon inhibition of insulin/IGF-1 signaling would indicate that ets-4 functions in parallel to this pathway. As previously reported [1], [61], [62], daf-2(RNAi) animals were significantly long-lived (Table 1). Notably, daf-2(RNAi); ets-4(ok165) animals lived longer than either ets-4(ok165) or daf-2(RNAi) worms alone (Table 1 and Table S1). Similarly, RNAi against the kinases akt-1/akt-2 further extended the life span of the long-lived ets-4(ok165) worms (Figure 6A and Table 1). Thus, inhibition of two different components of the insulin/IGF-1 signaling pathway further increased the life span of long-lived ets-4(ok165) animals. Because RNAi, and not null mutations, was used to inactivate signaling pathway genes, the possibility that the insulin/IGF-1 receptor pathway partially contributes to the life span phenotypes of ets-4 null mutant animals cannot be completely eliminated. However, our data support a model whereby ets-4 functions in parallel to the insulin/IGF-1 signaling pathway to modulate life span (Figure 7).
Fig. 6. ets-4 acts in parallel to the insulin/IGF-1 signaling pathway converging onto daf-16. 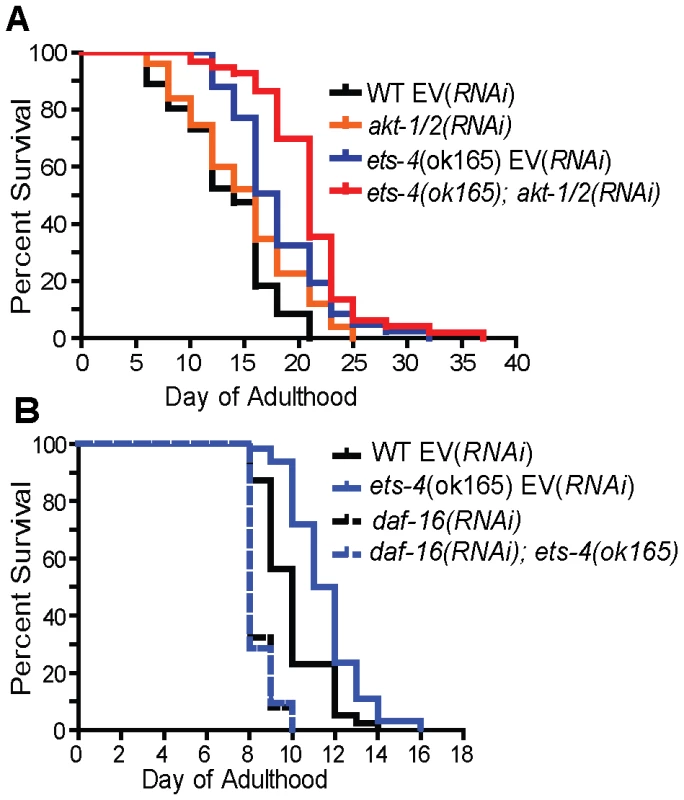
Survival curves from life span experiments conducted at 25°C. See Table 1 and Table S1 for data from additional trials, mean life span and statistical analysis. (A) Wild-type (WT) and ets-4(ok165) worms were grown at 25°C and subjected to akt-1/2(RNAi) or an empty vector (EV) control RNAi starting at the L4 stage. (B) Wild-type (WT) and ets-4(ok165) worms were grown at 25°C and subjected to daf-16(RNAi) or an empty vector (EV) control RNAi starting at the L4 stage. Fig. 7. Model for ETS-4 function as a transcriptional regulator. 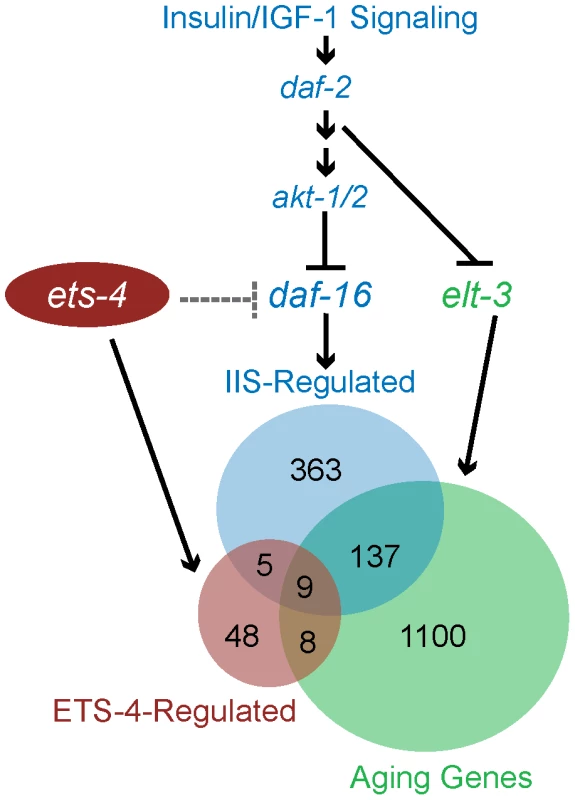
ETS-4 functions in parallel to the insulin/IGF-1 signaling pathway (blue) to modulate a set of downstream targets (red circle). ets-4 may function upstream of (dotted line) or in parallel (solid line) to antagonize daf-16 function in longevity regulation. ETS-4-regulated gene set overlaps with that of the insulin/IGF-1 signaling pathway acting through DAF-16 (blue circle) as well as that of ELT-3 (green circle), which directs the age-regulation of a majority of genes that change expression with age. We further explored the genetic relationship between ets-4 and the insulin/IGF-1 signaling pathway by investigating if ets-4 participated in other physiological processes regulated by the pathway. Signaling through DAF-2 is critical for dauer formation. Hence, the involvement of ets-4 in dauer formation was tested first. Assaying for dauer formation at 25°C and 27°C [63], [64], we found no defects in dauer formation in ets-4(ok165) mutant animals compared to wild-type worms (data not shown), suggesting that ets-4 does not play a significant role in this process. Next, we tested whether ets-4 null mutant animals exhibit altered response to environmental stress stimuli since insulin/IGF-1 signaling also regulates stress resistance. To assay for heat stress response, the survival of adult ets-4(ok165) worms was monitored after a shift to 35°C. As reported previously[65]-[68], daf-2(e1370) animals survived significantly longer than wild-type worms at 35°C, whereas daf-16(mgDf50) animals died faster (Figure S8). No significant differences in survival were seen between ets-4(ok165) and wild-type worms during the heat stress time-course (Figure S8), indicating that ets-4 is not required for response to heat stress.
Extension of life span is often associated with increased resistance to oxidative stress [69]. We determined whether ets-4 null mutant animals show altered response to oxidative stress by monitoring survival when exposed to a powerful oxidant, paraquat. No significant differences in survival were seen between ets-4(ok165) and wild-type worms during the majority of the oxidative stress time-course (Figure S8), indicating that ets-4(ok165) worms display wild-type response to oxidative stress. The participation of ETS-4 in the oxidative stress response pathway was also tested by assessing the genetic interaction of ets-4 with skn-1. Transcription factor SKN-1, the ortholog of mammalian Nrf proteins, is critical for oxidative stress resistance and acts in multiple longevity pathways, including the insulin/IGF-1 signaling cascade [70]. To test whether SKN-1 was required to mediate the life span extension observed in ets-4 null mutant animals, we inhibited skn-1 activity by RNAi and monitored the adult life span at 25°C. skn-1(RNAi) was previously reported to alter the expression of genes involved in oxidative stress response [71]. Also, inhibition of skn-1 by RNAi was shown to decrease the life span of daf-2 null mutant animals, but not control RNAi sensitive animals [72]. skn-1(RNAi) on ets-4(ok165) worms did not alter the extended life span phenotype of these worms (Figure S8), suggesting that the ets-4 null mutant animals are not susceptible to a partial loss of SKN-1 function. Additionally, to determine whether ETS-4 and SKN-1 shared downstream effectors, we compared a list of SKN-1-dependent target genes involved in oxidative stress response [71] to the 70 ETS-4-regulated genes. There was no significant enrichment for the stress responsive, SKN-1-dependent genes amongst genes that act downstream of ETS-4 (Table 2). Taken together, these data suggest that ETS-4 does not contribute to all physiological processes regulated by insulin/IGF-1 signaling and supports a model whereby ETS-4 functions, in part, independently of DAF-2 signaling to regulate life span.
ETS-4 Requires DAF-16 to Regulate Life Span
The FOXO transcription factor, DAF-16, is a well established regulator of life span that functions in the insulin/IGF-1 signaling pathway [1], [60], [73]–[75] and shares downstream longevity effectors with ETS-4 (Table 2 and Figure 7). We examined the genetic relationship between ets-4 and daf-16. Consistent with previous studies [1], [61], [75], inhibition of daf-16 activity by RNAi shortened life span (Figure 6B and Table 1). Further, ets-4(ok165) worms, when subjected to daf-16(RNAi), did not display an extended life span (Figure 6B and Table 1). Thus, daf-16 was required for the longevity phenotype of ets-4(ok165) worms suggesting that ets-4 functions upstream of, or in parallel to, daf-16 in lifespan regulation (Figure 7). Because the loss of ets-4 extends life span, whereas, inhibition of daf-16 activity shortens it, our genetic tests indicate that ets-4 antagonizes daf-16 function in longevity regulation by upstream effects, or in parallel.
We examined in more detail two possible ways by which ets-4 could function upstream of daf-16. First, we tested whether ETS-4 reduced DAF-16 levels through a transcriptional effect. Our gene expression profiling experiments indicated that the expression of daf-16 transcripts was not altered in ets-4 mutant worms (NCBI's Gene Expression Omnibus, accession number GSE17954). Further, upon crossing a daf-16::gfp reporter into ets-4(ok165) worms, we did not observe an alteration in the expression of DAF-16::GFP due to loss of ets-4 (data not shown). Second, we examined whether ETS-4 antagonized DAF-16 activity by promoting its cytoplasmic retention. The function of the transcription factor DAF-16 is modulated in response to stress conditions, such as heat-shock, by altering its nuclear localization [58], [76]. The intracellular localization of DAF-16::GFP was not affected by the loss of ets-4 under normal growth conditions and heat-shock (data not shown). These results suggest that ETS-4 does not regulate DAF-16 by altering its expression level or nuclear-cytoplasmic localization. Alternatively, ETS-4 and DAF-16 could function in parallel pathways to modulate distinct targets involved in longevity regulation (Figure 7). Or, an indirect modulation of DAF-16 activity by ETS-4 is possible, through the regulation of a required transcription co-factor. Taken together, we conclude that ETS-4 is a new life span determinant that functions in parallel to the insulin/IGF-1 signaling pathway, but requires the FOXO transcription factor, daf-16, to modulate life span.
Discussion
In this study, we describe a novel transcriptional regulator of longevity. Worms lacking ETS-4 exhibited a pronounced life span extension. Despite broad expression in multiple cell types, ETS-4 function in the adult intestine was important for longevity regulation. Notably, identification of an ETS-4-regulated gene set uncovered shared transcriptional targets with two other longevity modifiers, the FOXO and GATA factors.
ETS-4 Regulates Life Span
Loss of ets-4 function led to a substantial extension in mean adult life span. Additionally, ets-4 mutant worms exhibited an altered egg-laying rate. These data are consistent with the precedence of several life span altering mutations that affect multiple aspects of nematode biology, including reproduction and metabolism. This raises the question of whether these phenotypes are causative of increased longevity. Another key question with the identification of ETS-4 as a novel genetic modifier of aging with a broad expression pattern was whether a particular cell type was crucial for its function in longevity regulation. Our data showed that restoring ETS-4 function specifically in the intestine, but not neurons, rescued the extended life span of ets-4 null mutant animals back down to wild-type levels. Interestingly, other transcription factors that function as longevity determinants, including the FOXO protein DAF-16 and the GATA factors ELT-3, ELT-5, and ELT-6, regulate life span primarily through their function in the intestine [3], [34]. These data thus also support our model of shared transcriptional targets for these factors. Moreover, restoring expression of ETS-4 in the intestine of null mutant worms does not rescue the altered egg-laying phenotype. Thus, the longevity and altered egg-laying rate phenotypes are separable, implying a correlative rather than causative relationship between the two. Because relative to one another, the rates of aging of different tissues appear normal, it is proposed that a network of signaling and feedback regulation is involved in coordinating aging within an animal [33], [34]. Our data demonstrate that ETS-4 is a transcriptional regulator of this network that functions in the intestine to modulate the rate of aging in C. elegans.
Model for ETS-4 Function in Longevity Regulation
Gene expression profiling of long-lived worms carrying two different deletion alleles of ets-4, identified a robust set of ETS-4-regulated genes. Consistent with the longevity phenotype of ets-4 mutant worms, the ETS-4-regulated gene set was enriched for genes that modulate life span (Table 3). In addition to the regulation of these known life span determinants by ETS-4, 29 genes of unknown function were also misexpressed in ets-4 mutant animals and may contribute to the long-lived phenotype of these worms. DNA-binding studies and bioinformatics searches identified conserved ETS binding sites in promoter regions of 54 of the 70 ETS-4-regulated genes, suggesting that these were direct targets (Table S5). Notably, the ETS-4-regulated gene set was significantly enriched for genes that change expression during normal aging (Figure 7, Table 2 and Table S4), the majority of which were proposed targets of the GATA factor ELT-3 [3]. Consistent with the shared pattern of expression changes observed between aged worms and insulin/IGF-1 signaling pathway mutant animals [3], a significant proportion of ETS-4-regulated genes functioned downstream of the insulin/IGF-1 signaling pathway and the FOXO transcription factor DAF-16 [10], [11] (Figure 7, Table 2, and Table S4). RNAi against four of these common downstream effectors, vit-5, vit-2, thn-1, lys-7 alters worm life span [10]. These longevity effectors participate in lipid transport (vitellogenins/yolk proteins) and innate immune response (lysozymes and thaumatins). In addition to this set of overlapping targets for three transcriptional regulators of aging ELT-3, DAF-16 and ETS-4, notable targets were absent. For example, expression of genes mediating response to oxidative stress regulated by the transcription factor SKN-1 (superoxide dismutases) [71], heat-shock or toxicity (metallothioneins and xenobiotic metabolism genes) that are altered in daf-16 mutant worms remain unchanged in long-lived ets-4 mutant animals. We propose that ETS-4 participates in some, but not all biological processes regulated by FOXO and GATA factors. In summary, we introduce ETS-4 as a novel transcriptional regulator in the genetic network that modulates life span.
Insights into ETS function in Vertebrate Development and Disease
Our study is the first demonstration of an ETS factor modulating animal life span, thus illustrating the utility of C. elegans in identifying novel functions for ETS proteins not easily discerned in more complex systems. The mammalian ortholog of ets-4, SPDEF is expressed in the intestine and in tissues with high epithelial content, like breast and prostate [17], [77]. A recent study of a mouse mutant strain with a dysfunctional SPDEF allele showed impaired terminal differentiation of specialized intestinal secretory cells derived from the intestinal epithelium [24]. Markers for these secretory intestinal cells, the Paneth and goblet cells, were implicated as SPDEF targets [24]. In an interesting evolutionary convergence, our study illustrated ETS-4 function in the worm intestine, which is a tube comprised of 20 large epithelial cells [78]. The C. elegans intestine executes multiple functions carried out by distinct organs in higher eukaryotes, such as digestion and absorption of nutrients, synthesis and storage of fats, initiation of an innate immune response to pathogens and yolk production [44], [78]–[80]. Our work demonstrates the regulation of intestinal genes, such as lysozymes and vitellogenins by ETS-4. Additionally, restoring ETS-4 function in the worm intestine, but not neurons, reduced the life span of ets-4 null worms to wild-type, revealing a link between ETS function in the intestine and longevity regulation.
Our finding that ETS-4 has a role in C. elegans aging also provides a new connection between this ETS factor and cancer. Studies in human cell lines have implicated SPDEF function in tumorigenesis. Cell lines from breast and prostate tumors have altered SPDEF expression, although the significance of this misregulation remains controversial. Whereas some studies propose a putative role for SPDEF as a tumor suppressor, others suggest a prometastatic function [17], [18], [22], [23], [81], [82]. Given the strong correlation between physiological aging and tumor susceptibility, aging studies in C. elegans have been used to provide genetic insights into tumor biology [83]–[85]. Thus, the role of ETS-4 in aging raises new implications for the physiological role of mammalian ETS factors in development, homeostasis and disease.
Materials and Methods
C. elegans Strains and RNAi
C. elegans strains were maintained at 20°C as described previously [86] unless otherwise mentioned. The wild-type reference strain was N2 Bristol. The RB637: ets-4(ok165) X strain was obtained from the C. elegans Gene Knockout Project at OMRF (International C. elegans Gene Knockout Consortium). ets-4(uz1) was isolated by PCR-based screening of a library of worms mutated with EMS generated at the University of Utah. The deletion breakpoints of the uz1 on cosmid F22A3 are 14790 and 15861. Single-worm PCR [87] was used for screening and to determine the genotype of worms during crosses with ets-4(uz1) and ets-4(ok165) worms. For ets-4(ok165) and ets-4(uz1) identification, nested-PCR was carried out using the following primers: Forward primers: CAATGAACGGTACTGGCTCAG and Primer P5: TGCAATCTTCCAATCCAACCC; Reverse primers: ACTGCCGGAGGACAAATGTC and Primer P6: CATTGCGATTCCCATGTAACC; Primers for sequences deleted in the ok165 and uz1 alleles: GCTAGCCAGCACCAACAATCAA and ACACCAAACGCTGCTTCTTT. The ets-4(ok165) and ets-4(uz1) worms were outcrossed to the N2 strain six times before phenotypic analysis, including life span assays. We also used BC11290: dpy-5(e907) I; sEx11290[rCesC04F6.1::GFP + pCeh361] [31], TJ356: zIS356 IV [Pdaf-16::daf-16-gfp; rol-6(su1006)] [76], lin-15(n765ts) [88], daf-16(mgDf50) I, CF1041 daf-2(e1370) III [66].
RNAi was performed essentially as described previously [89] using HT115(DE3) E. coli expressing dsRNA from daf-16, daf-2, skn-1 (obtained from Julie Ahringer's RNAi library [90]), akt-1, akt-2 (obtained from the Marc Vidal's RNAi library [91]), ets-4 or carrying the L4440 control plasmid (EV for empty vector control). Each clone was sequenced to confirm its identity. Individual RNAi clones were grown overnight with Ampicillin (100 µg/ml) or Kanamycin (25 µg/ml) seeded onto NGM plates containing 1 mM IPTG (Sigma) and 25 µg/ml Carbenicillin (Sigma) and allowed to grow for 2 days at room temperature. For akt-1/2(RNAi), a 1 : 1 mix of akt-1 and akt-2 equal density overnight bacterial cultures was used to seed NGM plates containing 1 mM IPTG (Sigma) and 25 µg/ml Carbenicillin (Sigma). L4 stage larvae were placed on the RNAi plates and observed for phenotypes at 25°C.
RT-PCR and Microarray Analysis
Total RNA from mixed stage worms was isolated by phenol-chloroform extraction and subjected to DNaseI treatment using the RNeasy kit (Qiagen). 1–2 µg of total RNA was reverse-transcribed by SuperScript III (Invitrogen) according to manufacturer's protocol. Ten percent of the resultant cDNA was PCR-amplified by Taq DNA polymerase in a 50 µl reaction. Primer sequences: R1: GGCACAAGTTGTACTGATGTC, P1 (SL1 primer): GTTTAATTACCCAAG TTTGAG, P2: CAGATGACGGAGAATCAGGTC, R2: CTACAAGTTATAAGGAGGCAGG, P3: CTTCAGCCGCCTAGAAACTG and P4: CCAATATCTAGCCAGCAGGAG. The PCR product was sequenced to determine the position of SL1 attachment and splicing pattern.
To assess the expression of mRNA in synchronized populations of N2, ets-4(ok165) and ets-4(uz1) worms, levels of cDNA from the reverse transcription reaction were assessed by quantitative PCR according to manufacturer's protocol using the Roche LightCycler 480. Transcript levels were normalized to the averaged levels of cdc-42 and pmp-3 [92]. See Table S6 for primer sequences.
For microarray analysis, total RNA was isolated from L4 stage larvae as described above and reverse transcribed. Cy5 and Cy3 labeled cDNA was applied to C. elegans 22K gene expression arrays (Agilent). Data from three independent repeats of the experiment were analyzed. Lowess-normalized log (base 10) ratios of mutant/wild-type gene expression were obtained from the two-color Agilent C. elegans gene expression microarrays. The log ratios were analyzed using the Rank Products method [93] to identify consistently differentially expressed genes. Genes were selected using a probability of false prediction (PFP) cut off of 0.1, i.e. a false discovery rate of 10%. This technique corrects for multiple testing with repeated trials on random permutations of the data set.
The microarray data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series accession number GSE21851 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE21851).
Life Span Assays
Life span assays were conducted as described previously [94]. The ets-4(ok165) and ets-4(uz1) worms were outcrossed to the wild-type strain six times before the life span assays. Briefly, worms were grown for two or more generations at the assay temperature (25°C or 20°C) prior to the assay. Hermaphrodites were allowed to lay eggs for 6–8 hr on OP50 to obtain synchronous progeny for the experiment. L4 stage larvae were picked to plates spotted with RNAi bacteria containing 1 mM IPTG or OP50 bacteria and allowed to age at 25°C. The animals were moved to fresh plates daily during the reproductive period and every other day for the rest of the assay. The worms were scored for life by assessing movement to touch every 1–2 days [8]. Animals that bagged, exploded or crawled off the plate were excluded from the analysis. The first day of adulthood was counted as day one of the life span experiment. Life span curves and statistical data including p-values from Log-rank (Mantel-Cox) test were generated using GraphPad Prism version 5 software (GraphPad Software, San Diego, California USA).
Brood Size, Egg-Laying Rate and Development Time Assays
Single L4 stage larvae were allowed to lay eggs at 20°C and transferred to a fresh NGM plate every day till the end of the reproductive period. The number of eggs laid and the number of hatched progeny were counted. The average number of eggs laid each day during the egg-laying period and the total number of progeny per worm (brood size) were plotted. Development time assays were done as described previously [95]. Briefly, synchronized L1 larvae of each genotype were grown at 20°C. The animals were monitored every 3 hr after they had reached L4 stage till they were pre-fertile adults. Each experiment with at least ten worms per genotype was repeated twice, independently. Unpaired t-test analyses were performed to calculate p-values.
Supporting Information
Zdroje
1. KenyonC
ChangJ
GenschE
RudnerA
TabtiangR
1993 A C. elegans mutant that lives twice as long as wild type. Nature 366 461 464
2. PanowskiSH
WolffS
AguilaniuH
DurieuxJ
DillinA
2007 PHA-4/Foxa mediates diet-restriction-induced longevity of C. elegans. Nature 447 550 555
3. BudovskayaYV
WuK
SouthworthLK
JiangM
TedescoP
2008 An elt-3/elt-5/elt-6 GATA transcription circuit guides aging in C. elegans. Cell 134 291 303
4. OhSW
MukhopadhyayA
SvrzikapaN
JiangF
DavisRJ
2005 JNK regulates lifespan in Caenorhabditis elegans by modulating nuclear translocation of forkhead transcription factor/DAF-16. Proc Natl Acad Sci U S A 102 4494 4499
5. SheafferKL
UpdikeDL
MangoSE
2008 The Target of Rapamycin pathway antagonizes pha-4/FoxA to control development and aging. Curr Biol 18 1355 1364
6. LakowskiB
HekimiS
1998 The genetics of caloric restriction in Caenorhabditis elegans. Proc Natl Acad Sci U S A 95 13091 13096
7. LeeSS
LeeRY
FraserAG
KamathRS
AhringerJ
2003 A systematic RNAi screen identifies a critical role for mitochondria in C. elegans longevity. Nat Genet 33 40 48
8. HsinH
KenyonC
1999 Signals from the reproductive system regulate the lifespan of C. elegans. Nature 399 362 366
9. HouthoofdK
VanfleterenJR
2007 Public and private mechanisms of life extension in Caenorhabditis elegans. Mol Genet Genomics 277 601 617
10. MurphyCT
McCarrollSA
BargmannCI
FraserA
KamathRS
2003 Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature 424 277 283
11. MurphyCT
2006 The search for DAF-16/FOXO transcriptional targets: approaches and discoveries. Exp Gerontol 41 910 921
12. SharrocksAD
2001 The ETS-domain transcription factor family. Nat Rev Mol Cell Biol 2 827 837
13. GravesBJ
PetersenJM
1998 Specificity within the ets family of transcription factors.
WoudeGV
KleinG
Advances in Cancer Research San Diego Academic Press 1 55
14. MaroulakouIG
BoweDB
2000 Expression and function of Ets transcription factors in mammalian development: a regulatory network. Oncogene 19 6432 6442
15. HollenhorstPC
JonesDA
GravesBJ
2004 Expression profiles frame the promoter specificity dilemma of the ETS family of transcription factors. Nucleic Acids Res 32 5693 5702
16. GalangCK
MullerWJ
FoosG
OshimaRG
HauserCA
2004 Changes in the expression of many Ets family transcription factors and of potential target genes in normal mammary tissue and tumors. J Biol Chem 279 11281 11292
17. FeldmanRJ
SementchenkoVI
GayedM
FraigMM
WatsonDK
2003 Pdef expression in human breast cancer is correlated with invasive potential and altered gene expression. Cancer Res 63 4626 4631
18. GuX
ZerbiniLF
OtuHH
BhasinM
YangQ
2007 Reduced PDEF expression increases invasion and expression of mesenchymal genes in prostate cancer cells. Cancer Res 67 4219 4226
19. OettgenP
FingerE
SunZ
AkbaraliY
ThamrongsakU
2000 PDEF, a novel prostate epithelium-specific ets transcription factor, interacts with the androgen receptor and activates prostate-specific antigen gene expression. J Biol Chem 275 1216 1225
20. ChenH
NandiAK
LiX
BieberichCJ
2002 NKX-3.1 interacts with prostate-derived Ets factor and regulates the activity of the PSA promoter. Cancer Res 62 338 340
21. GunawardaneRN
SgroiDC
WrobelCN
KohE
DaleyGQ
2005 Novel role for PDEF in epithelial cell migration and invasion. Cancer Res 65 11572 11580
22. TurnerDP
MoussaO
SauaneM
FisherPB
WatsonDK
2007 Prostate-derived ETS factor is a mediator of metastatic potential through the inhibition of migration and invasion in breast cancer. Cancer Res 67 1618 1625
23. TurnerDP
FindlayVJ
KirvenAD
MoussaO
WatsonDK
2008 Global gene expression analysis identifies PDEF transcriptional networks regulating cell migration during cancer progression. Mol Biol Cell 19 3745 3757
24. GregorieffA
StangeDE
KujalaP
BegthelH
van den BornM
2009 The Ets-Domain Transcription Factor Spdef Promotes Maturation of Goblet and Paneth Cells in the Intestinal Epithelium. Gastroenterology
25. HodgkinJ
1986 Sex determination in the nematode C. elegans: analysis of tra-3 suppressors and characterization of fem genes. Genetics 114 15 52
26. GemsD
RiddleDL
2000 Defining wild-type life span in Caenorhabditis elegans. J Gerontol A Biol Sci Med Sci 55 B215 219
27. Van VoorhiesWA
WardS
1999 Genetic and environmental conditions that increase longevity in Caenorhabditis elegans decrease metabolic rate. Proc Natl Acad Sci U S A 96 11399 11403
28. AveryL
1993 The genetics of feeding in Caenorhabditis elegans. Genetics 133 897 917
29. CurranSP
RuvkunG
2007 Lifespan regulation by evolutionarily conserved genes essential for viability. PLoS Genet 3 e56
30. HartAH
ReventarR
BernsteinA
2000 Genetic analysis of ETS genes in C. elegans. Oncogene 19 6400 6408
31. McKaySJ
JohnsenR
KhattraJ
AsanoJ
BaillieDL
2003 Gene expression profiling of cells, tissues, and developmental stages of the nematode C. elegans. Cold Spring Harb Symp Quant Biol 68 159 169
32. Reece-HoyesJS
ShinglesJ
DupuyD
GroveCA
WalhoutAJ
2007 Insight into transcription factor gene duplication from Caenorhabditis elegans Promoterome-driven expression patterns. BMC Genomics 8 27
33. MurphyCT
LeeSJ
KenyonC
2007 Tissue entrainment by feedback regulation of insulin gene expression in the endoderm of Caenorhabditis elegans. Proc Natl Acad Sci U S A 104 19046 19050
34. LibinaN
BermanJR
KenyonC
2003 Tissue-specific activities of C. elegans DAF-16 in the regulation of lifespan. Cell 115 489 502
35. IserWB
GamiMS
WolkowCA
2007 Insulin signaling in Caenorhabditis elegans regulates both endocrine-like and cell-autonomous outputs. Dev Biol 303 434 447
36. HwangboDS
GershmanB
TuMP
PalmerM
TatarM
2004 Drosophila dFOXO controls lifespan and regulates insulin signalling in brain and fat body. Nature 429 562 566
37. BroughtonS
PartridgeL
2009 Insulin/IGF-like signalling, the central nervous system and aging. Biochem J 418 1 12
38. WolkowCA
KimuraKD
LeeMS
RuvkunG
2000 Regulation of C. elegans life-span by insulinlike signaling in the nervous system. Science 290 147 150
39. BluherM
KahnBB
KahnCR
2003 Extended longevity in mice lacking the insulin receptor in adipose tissue. Science 299 572 574
40. GiannakouME
GossM
JungerMA
HafenE
LeeversSJ
2004 Long-lived Drosophila with overexpressed dFOXO in adult fat body. Science 305 361
41. NonetML
StauntonJE
KilgardMP
FergestadT
HartwiegE
1997 Caenorhabditis elegans rab-3 mutant synapses exhibit impaired function and are partially depleted of vesicles. J Neurosci 17 8061 8073
42. SchluterOM
SchmitzF
JahnR
RosenmundC
SudhofTC
2004 A complete genetic analysis of neuronal Rab3 function. J Neurosci 24 6629 6637
43. BeissbarthT
SpeedTP
2004 GOstat: find statistically overrepresented Gene Ontologies within a group of genes. Bioinformatics 20 1464 1465
44. PauliF
LiuY
KimYA
ChenPJ
KimSK
2006 Chromosomal clustering and GATA transcriptional regulation of intestine-expressed genes in C. elegans. Development 133 287 295
45. ReinkeV
SmithHE
NanceJ
WangJ
Van DorenC
2000 A global profile of germline gene expression in C. elegans. Mol Cell 6 605 616
46. RoyPJ
StuartJM
LundJ
KimSK
2002 Chromosomal clustering of muscle-expressed genes in Caenorhabditis elegans. Nature 418 975 979
47. GaudetJ
MangoSE
2002 Regulation of organogenesis by the Caenorhabditis elegans FoxA protein PHA-4. Science 295 821 825
48. WatsonJD
WangS
Von StetinaSE
SpencerWC
LevyS
2008 Complementary RNA amplification methods enhance microarray identification of transcripts expressed in the C. elegans nervous system. BMC Genomics 9 84
49. SchonesDE
CuiK
CuddapahS
RohTY
BarskiA
2008 Dynamic regulation of nucleosome positioning in the human genome. Cell 132 887 898
50. NyeJA
PetersenJM
GuntherCV
JonsenMD
GravesBJ
1992 Interaction of murine Ets-1 with GGA-binding sites establishes the ETS domain as a new DNA-binding motif. Genes Dev 6 975 990
51. BadisG
BergerMF
PhilippakisAA
TalukderS
GehrkeAR
2009 Diversity and complexity in DNA recognition by transcription factors. Science 324 1720 1723
52. KoppJL
WilderPJ
DeslerM
KimJH
HouJ
2004 Unique and selective effects of five Ets family members, Elf3, Ets1, Ets2, PEA3, and PU.1, on the promoter of the type II transforming growth factor-beta receptor gene. J Biol Chem 279 19407 19420
53. BlanchetteM
KentWJ
RiemerC
ElnitskiL
SmitAF
2004 Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res 14 708 715
54. HollenhorstPC
ShahAA
HopkinsC
GravesBJ
2007 Genome-wide analyses reveal properties of redundant and specific promoter occupancy within the ETS gene family. Genes Dev 21 1882 1894
55. HollenhorstPC
ChandlerKJ
PoulsenRL
JohnsonWE
SpeckNA
2009 DNA specificity determinants associate with distinct transcription factor functions. PLoS Genetics 5 e1000778
56. YangSH
JaffrayE
HayRT
SharrocksAD
2003 Dynamic interplay of the SUMO and ERK pathways in regulating Elk-1 transcriptional activity. Mol Cell 12 63 74
57. SpiethJ
BlumenthalT
1985 The Caenorhabditis elegans vitellogenin gene family includes a gene encoding a distantly related protein. Mol Cell Biol 5 2495 2501
58. LeeRY
HenchJ
RuvkunG
2001 Regulation of C. elegans DAF-16 and its human ortholog FKHRL1 by the daf-2 insulin-like signaling pathway. Curr Biol 11 1950 1957
59. ParadisS
RuvkunG
1998 Caenorhabditis elegans Akt/PKB transduces insulin receptor-like signals from AGE-1 PI3 kinase to the DAF-16 transcription factor. Genes Dev 12 2488 2498
60. KenyonC
2005 The plasticity of aging: insights from long-lived mutants. Cell 120 449 460
61. BoehmM
SlackF
2005 A developmental timing microRNA and its target regulate life span in C. elegans. Science 310 1954 1957
62. HsuAL
MurphyCT
KenyonC
2003 Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science 300 1142 1145
63. LeeSS
KennedyS
TolonenAC
RuvkunG
2003 DAF-16 target genes that control C. elegans life-span and metabolism. Science 300 644 647
64. LiJ
EbataA
DongY
RizkiG
IwataT
2008 Caenorhabditis elegans HCF-1 functions in longevity maintenance as a DAF-16 regulator. PLoS Biol 6 e233
65. LithgowGJ
WhiteTM
MelovS
JohnsonTE
1995 Thermotolerance and extended life-span conferred by single-gene mutations and induced by thermal stress. Proc Natl Acad Sci U S A 92 7540 7544
66. GemsD
SuttonAJ
SundermeyerML
AlbertPS
KingKV
1998 Two pleiotropic classes of daf-2 mutation affect larval arrest, adult behavior, reproduction and longevity in Caenorhabditis elegans. Genetics 150 129 155
67. HorikawaM
SakamotoK
2009 Fatty-acid metabolism is involved in stress-resistance mechanisms of Caenorhabditis elegans. Biochem Biophys Res Commun 390 1402 1407
68. MabonME
ScottBA
CrowderCM
2009 Divergent mechanisms controlling hypoxic sensitivity and lifespan by the DAF-2/insulin/IGF-receptor pathway. PLoS One 4 e7937
69. FinkelT
HolbrookNJ
2000 Oxidants, oxidative stress and the biology of ageing. Nature 408 239 247
70. OliveiraRP
Porter AbateJ
DilksK
LandisJ
AshrafJ
2009 Condition-adapted stress and longevity gene regulation by Caenorhabditis elegans SKN-1/Nrf. Aging Cell 8 524 541
71. ParkSK
TedescoPM
JohnsonTE
2009 Oxidative stress and longevity in Caenorhabditis elegans as mediated by SKN-1. Aging Cell 8 258 269
72. TulletJM
HertweckM
AnJH
BakerJ
HwangJY
2008 Direct inhibition of the longevity-promoting factor SKN-1 by insulin-like signaling in C. elegans. Cell 132 1025 1038
73. LarsenPL
AlbertPS
RiddleDL
1995 Genes that regulate both development and longevity in Caenorhabditis elegans. Genetics 139 1567 1583
74. OggS
ParadisS
GottliebS
PattersonGI
LeeL
1997 The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature 389 994 999
75. LinK
DormanJB
RodanA
KenyonC
1997 daf-16: An HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science 278 1319 1322
76. HendersonST
JohnsonTE
2001 daf-16 integrates developmental and environmental inputs to mediate aging in the nematode Caenorhabditis elegans. Curr Biol 11 1975 1980
77. JedlickaP
Gutierrez-HartmannA
2008 Ets transcription factors in intestinal morphogenesis, homeostasis and disease. Histol Histopathol 23 1417 1424
78. McGheeJD
2007 The C. elegans intestine. WormBook 1 36
79. KimbleJ
SharrockWJ
1983 Tissue-specific synthesis of yolk proteins in Caenorhabditis elegans. Dev Biol 96 189 196
80. SchulenburgH
KurzCL
EwbankJJ
2004 Evolution of the innate immune system: the worm perspective. Immunol Rev 198 36 58
81. GhadersohiA
PanD
FayaziZ
HicksDG
WinstonJS
2007 Prostate-derived Ets transcription factor (PDEF) downregulates survivin expression and inhibits breast cancer cell growth in vitro and xenograft tumor formation in vivo. Breast Cancer Res Treat 102 19 30
82. SchaeferJS
SabherwalY
ShiHY
SriramanV
RichardsJ
2010 Transcriptional regulation of p21/CIP1 cell cycle inhibitor by PDEF controls cell proliferation and mammary tumor progression. J Biol Chem 285 11258 11269
83. PinkstonJM
GariganD
HansenM
KenyonC
2006 Mutations that increase the life span of C. elegans inhibit tumor growth. Science 313 971 975
84. Pinkston-GosseJ
KenyonC
2007 DAF-16/FOXO targets genes that regulate tumor growth in Caenorhabditis elegans. Nat Genet 39 1403 1409
85. BrunetA
2007 Aging and cancer: killing two birds with one worm. Nat Genet 39 1306 1307
86. BrennerS
1974 The genetics of Caenorhabditis elegans. Genetics 77 71 94
87. WicksSR
YehRT
GishWR
WaterstonRH
PlasterkRH
2001 Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nat Genet 28 160 164
88. FergusonEL
HorvitzHR
1989 The multivulva phenotype of certain Caenorhabditis elegans mutants results from defects in two functionally redundant pathways. Genetics 123 109 121
89. KamathRS
Martinez-CamposM
ZipperlenP
FraserAG
AhringerJ
2001 Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol 2 RESEARCH0002
90. KamathRS
FraserAG
DongY
PoulinG
DurbinR
2003 Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421 231 237
91. RualJF
CeronJ
KorethJ
HaoT
NicotAS
2004 Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res 14 2162 2168
92. HoogewijsD
De HenauS
DewildeS
MoensL
CouvreurM
2008 The Caenorhabditis globin gene family reveals extensive nematode-specific radiation and diversification. BMC Evol Biol 8 279
93. BreitlingR
ArmengaudP
AmtmannA
HerzykP
2004 Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett 573 83 92
94. HansenM
HsuAL
DillinA
KenyonC
2005 New genes tied to endocrine, metabolic, and dietary regulation of lifespan from a Caenorhabditis elegans genomic RNAi screen. PLoS Genet 1 119 128
95. LeeSJ
KenyonC
2009 Regulation of the longevity response to temperature by thermosensory neurons in Caenorhabditis elegans. Curr Biol 19 715 722
Štítky
Genetika Reprodukční medicína
Článek Allelic Variation at the 8q23.3 Colorectal Cancer Risk Locus Functions as a Cis-Acting Regulator ofČlánek Allelic Selection of Amplicons in Glioblastoma Revealed by Combining Somatic and Germline AnalysisČlánek Lactic Acidosis Triggers Starvation Response with Paradoxical Induction of TXNIP through MondoAČlánek Rice a Cinnamoyl-CoA Reductase-Like Gene Family Member, Is Required for NH1-Mediated Immunity to pv.Článek Differentiation of Zebrafish Melanophores Depends on Transcription Factors AP2 Alpha and AP2 Epsilon
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2010 Číslo 9- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
- Intrauterinní inseminace a její úspěšnost
- Délka menstruačního cyklu jako marker ženské plodnosti
-
Všechny články tohoto čísla
- Optimal Strategy for Competence Differentiation in Bacteria
- Mutational Patterns Cannot Explain Genome Composition: Are There Any Neutral Sites in the Genomes of Bacteria?
- Frail Hypotheses in Evolutionary Biology
- Genetic Architecture of Complex Traits and Accuracy of Genomic Prediction: Coat Colour, Milk-Fat Percentage, and Type in Holstein Cattle as Contrasting Model Traits
- Allelic Variation at the 8q23.3 Colorectal Cancer Risk Locus Functions as a Cis-Acting Regulator of
- Allelic Selection of Amplicons in Glioblastoma Revealed by Combining Somatic and Germline Analysis
- Germline Variation Controls the Architecture of Somatic Alterations in Tumors
- Mice Doubly-Deficient in Lysosomal Hexosaminidase A and Neuraminidase 4 Show Epileptic Crises and Rapid Neuronal Loss
- Analysis of Population Structure: A Unifying Framework and Novel Methods Based on Sparse Factor Analysis
- FliO Regulation of FliP in the Formation of the Flagellum
- Cdc20 Is Critical for Meiosis I and Fertility of Female Mice
- dMyc Functions Downstream of Yorkie to Promote the Supercompetitive Behavior of Hippo Pathway Mutant Cells
- DCAF26, an Adaptor Protein of Cul4-Based E3, Is Essential for DNA Methylation in
- Genome-Wide Double-Stranded RNA Sequencing Reveals the Functional Significance of Base-Paired RNAs in
- An Immune Response Network Associated with Blood Lipid Levels
- Genetic Variants and Their Interactions in the Prediction of Increased Pre-Clinical Carotid Atherosclerosis: The Cardiovascular Risk in Young Finns Study
- The Histone H3K36 Methyltransferase MES-4 Acts Epigenetically to Transmit the Memory of Germline Gene Expression to Progeny
- Long- and Short-Term Selective Forces on Malaria Parasite Genomes
- Lactic Acidosis Triggers Starvation Response with Paradoxical Induction of TXNIP through MondoA
- Identification of Early Requirements for Preplacodal Ectoderm and Sensory Organ Development
- Orphan CpG Islands Identify Numerous Conserved Promoters in the Mammalian Genome
- Analysis of the Basidiomycete Reveals Conservation of the Core Meiotic Expression Program over Half a Billion Years of Evolution
- ETS-4 Is a Transcriptional Regulator of Life Span in
- The SR Protein B52/SRp55 Is Required for DNA Topoisomerase I Recruitment to Chromatin, mRNA Release and Transcription Shutdown
- The Baker's Yeast Diploid Genome Is Remarkably Stable in Vegetative Growth and Meiosis
- Chromatin Landscape Dictates HSF Binding to Target DNA Elements
- The APETALA-2-Like Transcription Factor OsAP2-39 Controls Key Interactions between Abscisic Acid and Gibberellin in Rice
- Accurately Assessing the Risk of Schizophrenia Conferred by Rare Copy-Number Variation Affecting Genes with Brain Function
- Widespread Over-Expression of the X Chromosome in Sterile F Hybrid Mice
- The Characterization of Twenty Sequenced Human Genomes
- The Genome of a Pathogenic : Cooptive Virulence Underpinned by Key Gene Acquisitions
- A Single Element Maintains Repression of the Key Developmental Regulator
- Identification of New Genetic Risk Variants for Type 2 Diabetes
- Effect of Correlated tRNA Abundances on Translation Errors and Evolution of Codon Usage Bias
- Evidence of Selection upon Genomic GC-Content in Bacteria
- Proteomic Changes Resulting from Gene Copy Number Variations in Cancer Cells
- Rice a Cinnamoyl-CoA Reductase-Like Gene Family Member, Is Required for NH1-Mediated Immunity to pv.
- Longitudinal Genome-Wide Association of Cardiovascular Disease Risk Factors in the Bogalusa Heart Study
- Response to Mechanical Stress Is Mediated by the TRPA Channel Painless in the Heart
- DNMT3L Modulates Significant and Distinct Flanking Sequence Preference for DNA Methylation by DNMT3A and DNMT3B
- Identifying Signatures of Natural Selection in Tibetan and Andean Populations Using Dense Genome Scan Data
- Incremental Genetic Perturbations to MCM2-7 Expression and Subcellular Distribution Reveal Exquisite Sensitivity of Mice to DNA Replication Stress
- Loss of Maternal ATRX Results in Centromere Instability and Aneuploidy in the Mammalian Oocyte and Pre-Implantation Embryo
- Comparative Genomic Hybridization (CGH) Reveals a Neo-X Chromosome and Biased Gene Movement in Stalk-Eyed Flies (Genus )
- Differentiation of Zebrafish Melanophores Depends on Transcription Factors AP2 Alpha and AP2 Epsilon
- Gene–Environment Interactions at Nucleotide Resolution
- Dementia Revealed: Novel Chromosome 6 Locus for Late-Onset Alzheimer Disease Provides Genetic Evidence for Folate-Pathway Abnormalities
- Critical Functions of Rpa3/Ssb3 in S-Phase DNA Damage Responses in Fission Yeast
- Preferential Re-Replication of Heterochromatin in the Absence of Geminin
- The Potential for Enhancing the Power of Genetic Association Studies in African Americans through the Reuse of Existing Genotype Data
- Evidence That Mutation Is Universally Biased towards AT in Bacteria
- Perturbation Analysis of Heterochromatin-Mediated Gene Silencing and Somatic Inheritance
- Diversity of Eukaryotic DNA Replication Origins Revealed by Genome-Wide Analysis of Chromatin Structure
- Genetic Deletion of the Desmosomal Component Promotes Tumor Microinvasion in a Mouse Model of Pancreatic Neuroendocrine Carcinogenesis
- The Metabolic Enzyme ManA Reveals a Link between Cell Wall Integrity and Chromosome Morphology
- SNPs Associated with Cerebrospinal Fluid Phospho-Tau Levels Influence Rate of Decline in Alzheimer's Disease
- Synthesizing and Salvaging NAD: Lessons Learned from
- A Central Regulatory System Largely Controls Transcriptional Activation and Repression Responses to Phosphate Starvation in Arabidopsis
- An Insect Herbivore Microbiome with High Plant Biomass-Degrading Capacity
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Synthesizing and Salvaging NAD: Lessons Learned from
- Optimal Strategy for Competence Differentiation in Bacteria
- Long- and Short-Term Selective Forces on Malaria Parasite Genomes
- Identifying Signatures of Natural Selection in Tibetan and Andean Populations Using Dense Genome Scan Data
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



