-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
DCAF26, an Adaptor Protein of Cul4-Based E3, Is Essential for DNA Methylation in
DNA methylation is involved in gene silencing and genome stability in organisms from fungi to mammals. Genetic studies in Neurospora crassa previously showed that the CUL4-DDB1 E3 ubiquitin ligase regulates DNA methylation via histone H3K9 trimethylation. However, the substrate-specific adaptors of this ligase that are involved in the process were not known. Here, we show that, among the 16 DDB1 - and Cul4-associated factors (DCAFs) encoded in the N. crassa genome, three interacted strongly with CUL4-DDB1 complexes. DNA methylation analyses of dcaf knockout mutants revealed that dcaf26 was required for all of the DNA methylation that we observed. In addition, histone H3K9 trimethylation was also eliminated in dcaf26KO mutants. Based on the finding that DCAF26 associates with DDB1 and the histone methyltransferase DIM-5, we propose that DCAF26 protein is the major adaptor subunit of the Cul4-DDB1-DCAF26 complex, which recruits DIM-5 to DNA regions to initiate H3K9 trimethylation and DNA methylation in N. crassa.
Published in the journal: . PLoS Genet 6(9): e32767. doi:10.1371/journal.pgen.1001132
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1001132Summary
DNA methylation is involved in gene silencing and genome stability in organisms from fungi to mammals. Genetic studies in Neurospora crassa previously showed that the CUL4-DDB1 E3 ubiquitin ligase regulates DNA methylation via histone H3K9 trimethylation. However, the substrate-specific adaptors of this ligase that are involved in the process were not known. Here, we show that, among the 16 DDB1 - and Cul4-associated factors (DCAFs) encoded in the N. crassa genome, three interacted strongly with CUL4-DDB1 complexes. DNA methylation analyses of dcaf knockout mutants revealed that dcaf26 was required for all of the DNA methylation that we observed. In addition, histone H3K9 trimethylation was also eliminated in dcaf26KO mutants. Based on the finding that DCAF26 associates with DDB1 and the histone methyltransferase DIM-5, we propose that DCAF26 protein is the major adaptor subunit of the Cul4-DDB1-DCAF26 complex, which recruits DIM-5 to DNA regions to initiate H3K9 trimethylation and DNA methylation in N. crassa.
Introduction
The Cul4-DDB1 complex, a major class of cullin-RING ubiquitin ligases (CRLs), is evolutionarily conserved from yeasts to humans. Previous studies have indicated that Cul4-DDB1-regulated ubiquitination is linked to multiple processes, such as cell cycle regulation, DNA replication licensing, DNA repair, and gene expression processes [1]–[3]. In the CRLs, cullin associates with substrates via adaptor molecules in the N terminus, and interacts with the E2 enzyme via the RING finger protein Hrt1/ROC1/Rbx1 in the C terminus [4]. Substrate-specific adaptors, such as the F-box-containing proteins in SCF complexes and the BTB domain-containing proteins in Cul3-based ubiquitin ligases, determine substrate-specific ubiquitination in many biological processes [5]. Although the different structural states of DDB1 may allow it to directly recruit substrates to the Cul4-based E3 platform, studies have demonstrated that ubiquitination of several characterized CUL4-DDB1 substrates requires additional substrate-specific adaptors [6], [7].
Recent studies showed that a class of adaptors called DCAFs (DDB1 - and Cul4-associated factors) [4] are employed by Cul4-based E3 ligases to identify specific proteins for ubiquitination. Most DCAFs are WD40-containing proteins with relatively conserved “WDXR” motifs that interact with DDB1 protein. However, several DCAF proteins lacking this conserved motif are still able to bind DDB1 in vivo [3], [4], [8], [9].
Among the well-characterized DCAF proteins, mammalian WDR5 and RBBP5 are essential components of the histone methyltransferase complex that methylates histone H3 on lysine 4 (H3K4) [10]–[12]. Further studies showed that Cul4-DDB1 can interact with WDR5 and RBBP5 and regulate histone H3K4 methylation. Down regulation of each of these genes by siRNA severely reduces the tri - and monomethylation of histone H3K4, but not H3K4 dimethylation [13]. Interestingly, inactivation of Cul4 or DDB1 also causes a significant inhibition of histone H3K9 and H3K27 trimethylation [13]. However, none of these DCAFs were shown to be involved directly in DNA methylation in eukaryotes.
In fission yeast, the Cul4-Rik1 E3 ubiquitin ligase associates with the histone methyltransferase Clr4 on heterochromatic regions to methylate histone H3K9, contributing to heterochromatin assembly and maintenance [14]. The catalytic activity of Cul4 is required for its proper function in heterochromatin formation. This study suggests that the activity of Cul4-based E3 ligase is required for histone H3K9 methylation. In addition, a WD-40-containing protein, Raf1/Dos1/Clr8/Cmc1, is required for histone H3K9 methylation and heterochromatin formation in S. pombe [15]–[18]. Thus, these studies imply that Raf1/Dos1/Clr8/Cmc1 functions as an adaptor protein associated with Cul4-Rik1 complex in S. pombe.
We previously demonstrated that Cul4-DDB1 E3 ligase is essential for DNA methylation in N. crassa by regulating histone H3K9 trimethylation [19]. These results suggest that Cul4-DDB1 ubiquitin ligase is required for epigenetic control in higher eukaryotes. However, the substrate-specific adaptors of Cul4-DDB1 E3 ligase and the requirement of DCAFs as the substrate adaptors in DNA methylation are unknown in N. crassa.
Here, we identified three DCAFs that could strongly interact with DDB1 protein in vivo, out of 16 candidates in the N. crassa genome. DNA methylation analysis in knockout strains of the 12 dcaf genes showed that dcaf26 was essential for DNA methylation in N. crassa. The dcaf26 deletion mutant also lost histone H3K9 trimethylation. Our protein interaction results suggest that DCAF26 functions as an adaptor subunit of the Cul4-DDB1-DCAF26 complex by recruiting histone methyltransferase DIM-5 to DNA methylation regions in N. crassa. In addition, we show that the interaction of DCAF26 with DDB1 was required to enable the Cul4-DDB1-DCAF26-DIM-5 complex to regulate H3K9 trimethylation and DNA methylation in this organism.
Results
Comparative analysis of N. crassa DCAF proteins
We searched the N. crassa genome for WD-40-containing proteins with WDXR and DxR motifs (also known as a DWD motif). The conserved 16 amino acid sequence [IFVL]-[IFVL]-[AGST]-[AGST]-[AGST]-x-[DE]-x(2)-[IFVL]-x-[IFVL] - [WY]-[DE]-[IFVL]-[RK] [20] was used as the seed to search for WD-40-containing proteins. Five putative DCAFs met the criteria (Table 1). Based on the sequences of hundreds of DCAF proteins from yeast to human that have been identified by protein interaction experiments and bioinformatics analysis, we identified another 11 putative DCAF proteins (Table 1). In total, we identified 16 putative DCAF proteins in the N. crassa genome (Figure 1A).
Fig. 1. Putative DDB1- and Cul4-associated factors (DCAFs) in N. crassa. 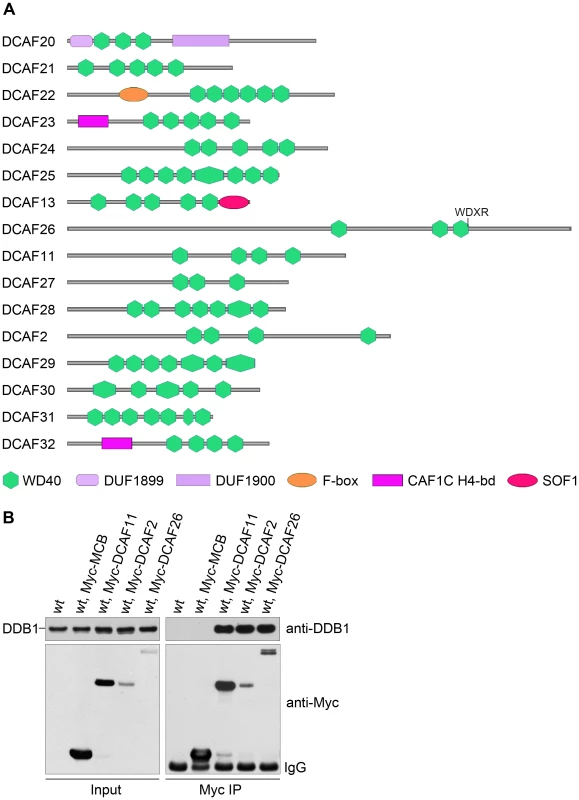
(A) Schematic representation of 16 putative DCAFs with predicted domains. (B) Interactions between Myc-tagged DCAFs and DDB1 protein. c-Myc antibody was used for immunoprecipitation, followed by western blot analysis using DDB1 or c-Myc antibodies. The wild-type strain and wild-type expressing Myc-tagged MCB (the regulatory subunit of cAMP-dependent protein kinase) strain were used as negative controls. Tab. 1. Putative DCAFs in <i>Neurospora crassa</i>. 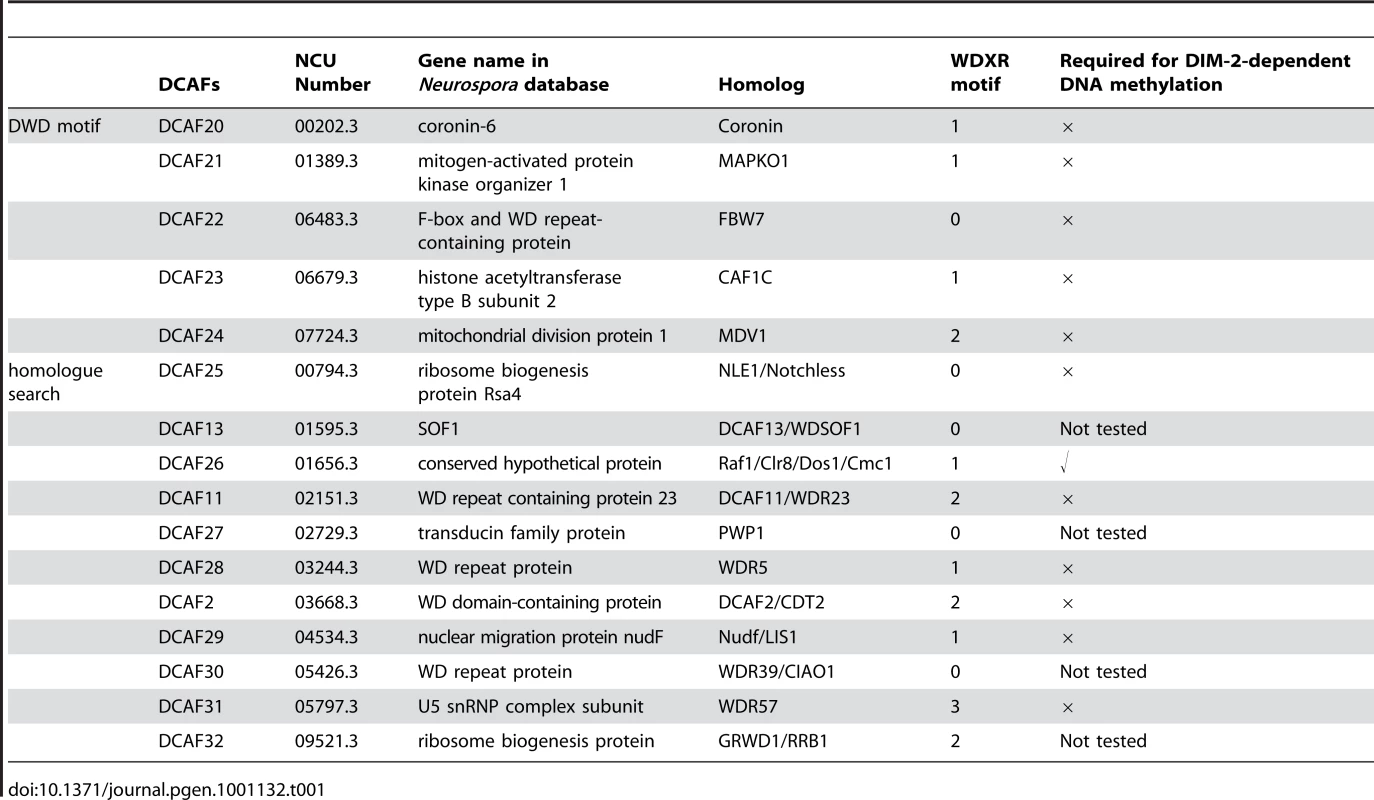
To identify the true DCAFs in N. crassa, we examined the interactions between DDB1 and the predicted DCAF proteins. We first made constructs in which the DCAF ORF was under the control of a quinic acid (QA)–inducible promoter [21]. To facilitate the detection of DCAF expression, five copies of the c-Myc epitope and six histidine residues [22] were inserted into the N terminus of the DCAF ORF. The constructs were then transformed into a wild-type strain and Myc-DCAF expression in the resulting transformants in the presence of QA was confirmed by western blot analysis using the c-Myc antibody. Afterwards, immunoprecipitation assays were used to examine the interactions between Myc-DCAF proteins and DDB1 in the transformants. As shown in Figure 1B, three Myc-tagged DCAFs, DCAF11, DCAF2, and DCAF26, interacted strongly with the DDB1 protein in vivo. Our DDB1 antibody depleted with tissue of the ddb1KO strain specifically recognized a 128-kDa band in the wild-type strain, but not in the ddb1 mutant (Figure S1). DCAF11 and DCAF2 had two conserved WDXR motifs, while DCAF26 had only one conserved WDXR motif (WNVR). In contrast, only weak interactions were detected between DDB1 and the other Myc-DCAFs (Figure S2). These results suggest that DCAF11, DCAF2, and/or DCAF26 could be the adaptor(s) in the Cul4-DDB1 E3 ligase complex in N. crassa.
Knockout of N. crassa dcaf genes
Our previous study showed that the Cul4-DDB1 E3 complex is required for DNA methylation in N. crassa [19]. To investigate the function of the putative DCAFs in DNA methylation, we tried to generate deletion strains of the 16 candidate dcaf genes by gene replacement in the ku70RIP strain. However, we could not obtain the homokaryotic deletion strains of four genes (NCU01595, NCU02729, NCU05426, and NCU09521), suggesting that they are important for cell viability in N. crassa. Several independent homokaryotic knockout strains of the other 12 dcaf genes were obtained by microconidia purification. PCR analysis confirmed the integration of the knockout cassette at the endogenous dcaf locus, and no dcaf ORF signals were detected in these knockout mutants.
DCAF26 is essential for DNA methylation
To identify the DCAF protein(s) required for DNA methylation, we measured the cytosine methylation states of all homokaryotic dcaf mutants. Genomic DNA of the wild-type strain and ku70RIP, dim-2KO, and dcaf mutants was digested with DpnII or BfuCI (Sau3AI). Undigested and digested DNA was then used as templates for PCR. Three representative methylated regions, the ku70RIP locus, ζ-η, and ψ63, were examined in the wild-type strain, ku70RIP, dim-2KO, and dcaf mutants. As shown in Figure 2, no PCR products were detected with DpnII - or BfuCI-digested genomic DNA as templates in the dcaf26KO mutant, the same as for the dim-2KO mutant. To further confirm these results, we tested the known methylation region on centromere VII of N. crassa. As expected, methylation on this DNA region was also lost in the dcaf26KO mutant (Figure 2), indicating that DCAF26 plays an essential role in DNA methylation. In contrast, the other 11 homokaryotic dcaf mutants exhibited normal cytosine methylation patterns at ζ-η and ψ63 regions, the same as those in the wild-type strain (Figure S3). This demonstrated that they were not essential for DIM-2–dependent DNA methylation (Table 1). Taken together, these results suggest that DCAF26 was a key regulator of DNA methylation in N. crassa.
Fig. 2. DCAF26 is essential for N. crassa DNA methylation. 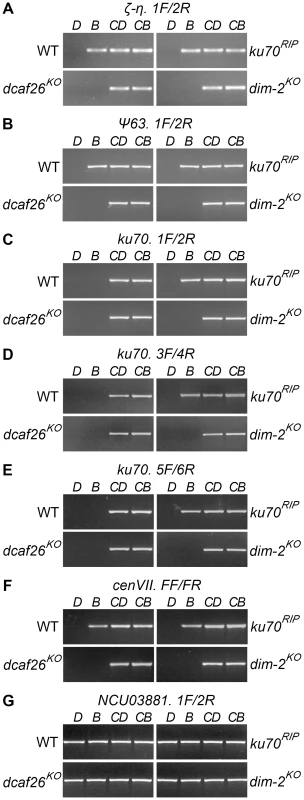
DNA methylation in the wild-type strain (WT) and ku70RIP, dcaf26KO, and dim-2KO strains on the (A) ζ-η region, (B) ψ63 region, (C–E) the ku70 and ku70RIP loci, and (F) the centromeric VII region detected by methylation-sensitive restriction digestion. (G) Digested genomic DNA served as a template for PCR of the NCU03381 region, which was covered by primer pairs and had no DpnII site. The knockout mutants were in a bd ku70RIP background. Genomic DNA digested by 5mC-sensitive BfuCI (B) or its 5mC-insensitive isoschizomer, DpnII (D), was amplified by PCR with the labeled primers. Untreated genomic DNA as template for PCR was used as the control (CD or CB). DCAF26 is required for H3K9 trimethylation
In N. crassa, proper DNA methylation depends on histone H3K9 trimethylation, which is controlled by the histone methyltransferase DIM-5 [23], [24]. When we compared the growth and developmental phenotypes of the dcaf knockout strains to those of the wild-type strain and cul4KO, ddb1KO, and dim-5KO mutants, we found that the dcaf26 mutant exhibited dense, cauliflower-like growth patterns with abnormal hyphae and asexual spores on plates (Figure 3A). This was similar to the phenotypes of the cul4, ddb1, and dim-5 deletion strains [19] (Figure 3A). The dcaf26KO mutant also exhibited slow growth rates on racetubes compared to that of the wild-type strain (Figure 3B); these rates were similar to those of the cul4KO, ddb1KO, and dim-5KO strains. These results suggest that DCAF26 affected DNA methylation by regulating H3K9 trimethylation in the same pathway as the Cul4-DDB1 complex. Therefore, we next examined H3K9 trimethylation levels by chromatin immunoprecipitation (ChIP) in the wild-type strain, the ku70RIP strain, and the dcaf26 mutant. Chromatin samples were immunoprecipitated with antibody against trimethylated Lys9 of histone H3 and analyzed by PCR with primers targeted to methylated DNA regions. As shown in Figure 3C, trimethylated H3K9 was associated with the methylated ku70RIP region in the ku70RIP strain, whereas trimethylated H3K9 at the ku70RIP region was abolished in the dcaf26 mutant. Similarly, trimethylated H3K9 at the ζ-η and ψ63 regions in the ku70RIP strain was observed at levels comparable to the wild-type strain (Figure 3C). In contrast, H3K9 trimethylation was lost in the dcaf26 mutant (Figure 3C). Taken together, these data indicate that N. crassa DCAF26 was required for trimethylation of histone H3K9 at methylated DNA regions.
Fig. 3. DCAF26 is essential for histone H3K9 trimethylation. 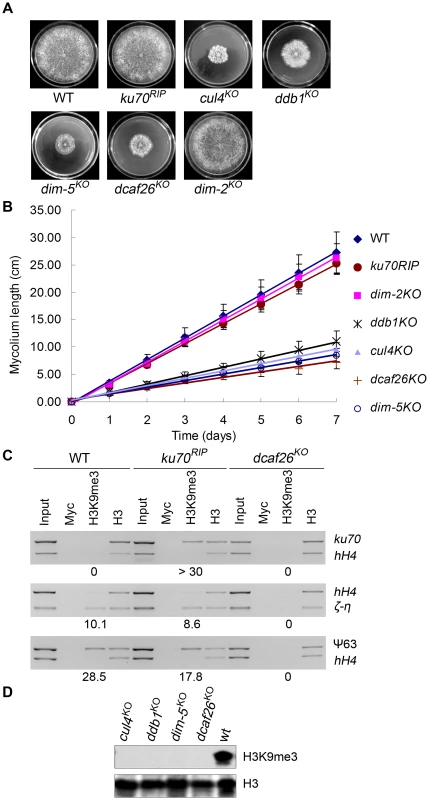
(A) Dense, cauliflower-like growth patterns of dcaf26KO, cul4KO, ddb1KO, and dim-5 KO strains on plates with minimal media (30°C, 32h). (B) Growth rate of the wild-type, ku70RIP, dcaf26KO, cul4KO, ddb1KO, dim-5KO, and dim-2KO strains, measured at 25°C using the racetube assay in constant darkness. (C) Loss of histone H3K9 trimethylation at ku70RIP, ζ-η, and ψ63 regions in the dcaf26KO strain. Levels of H3K9 trimethylation at the indicated locations were determined by ChIP assay. Myc antibody was used as the negative control, and H3 antibody was used as the positive control and as the control for integrity of the nucleosome structure. (D) Western blot analysis of global H3 and H3K9 trimethylation in the wild-type strain and mutants. To investigate the effect of DCAF26 protein on global trimethylated H3K9 in N. crassa, we tested the levels of histone H3 or trimethylated H3K9 using western blot analysis in wild-type, dcaf26KO, cul4KO, ddb1KO, and dim-5KO strains. As shown in Figure 3D, all of the strains had similar levels of histone H3. In contrast to the robust H3K9 trimethylation in the wild-type strain, very little trimethylated H3K9 was detected in dcaf26KO, cul4KO, ddb1KO, dim-5KO strains. Together, these results strongly suggest that DCAF26 protein was required for histone H3K9 trimethylation.
DCAF26 is the key component for recruiting DIM-5 to the Cul4-DDB1 complex
Having identified DCAF26 as a DCAF protein that interacted strongly with DDB1 protein, we next tested whether DCAF26 was a key component in the Cul4-DDB1-DIM-5 complex for DNA and histone H3K9 methylation. We did so by performing an immunoprecipitation assay to detect interactions between Myc-DCAF26 and Flag-Cul4 and between Myc-DCAF26 and Flag-DIM-5. As shown in Figure 4A, Myc-tagged DCAF26 specifically interacted with one of two Flag-Cul4 species (neddylated/unneddylated Cul4). To determine whether DCAF26 preferentially interacts with neddylated or unneddylated Cul4, we loaded protein extract from a csn-2KO strain, in which Cul4 remained in a hyperneddylated state, side by side with Myc-DCAF26/Flag-Cul4 to show that the upper band in the input and the Myc-DCAF26-interacting Cul4 band were neddylated Cul4 (Figure 4A), similar to the association between DIM-5 and neddylated Cul4 species in N. crassa [19]. We then examined the interactions between DCAF26 protein and DIM-5. As expected, the Flag antibody pulled down the Myc-tagged DCAF26 in the strain that coexpresses Flag-DIM-5 and Myc-DCAF26 (Figure 4B), indicating that DCAF26 was a component of the Cul4-DDB1-DIM-5 complex. These data explain the similar phenotypes in dcaf26KO, cul4KO, ddb1KO, and dim-5KO mutants.
Fig. 4. DCAF26 is the key component for recruiting DIM-5 to the Cul4-DDB1 complex. 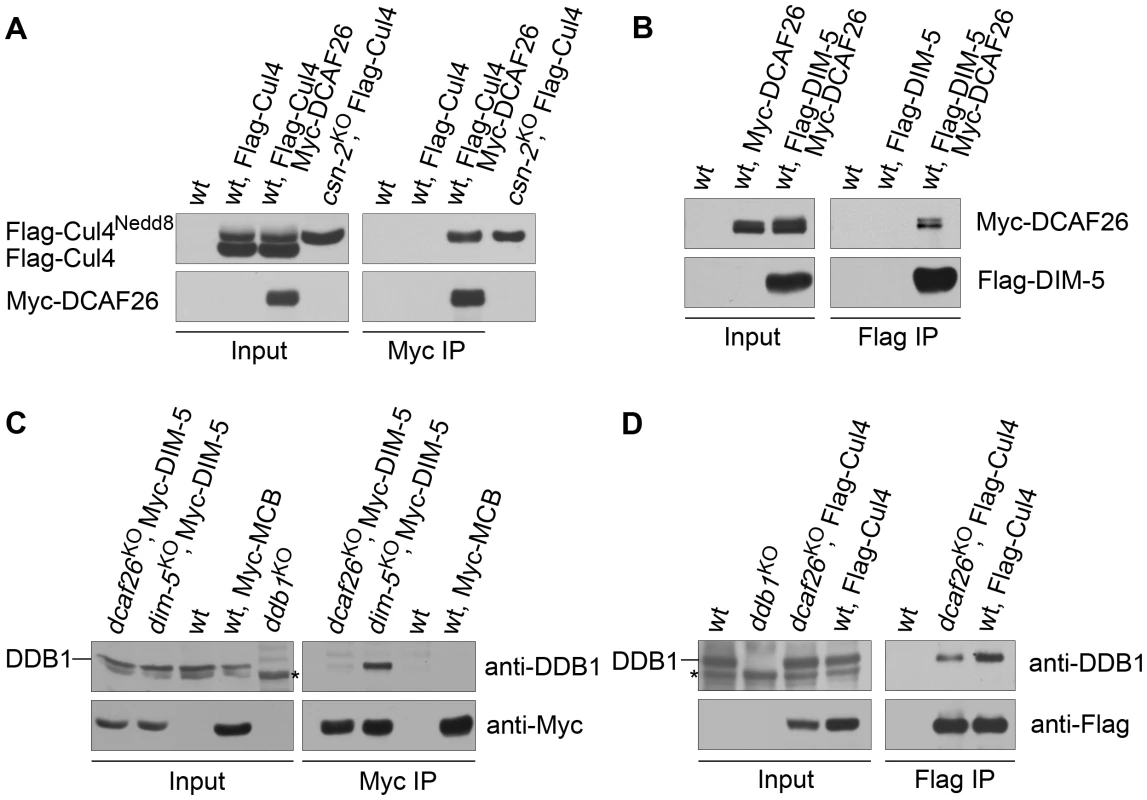
(A) Immunoprecipitation assay using Myc antibody showing the association between Myc-DCAF26 and Flag-Cul4. Myc-DCAF26 interacted with neddylated rather than unneddylated Flag-Cul4. The wild-type strain and the wild-type strain expressing Flag-Cul4 only were used as negative controls. Constitutively hyperneddylated Flag-Cul4 expressed in the csn-2KO strain was used as the positive control for neddylated Flag-Cul4 (lanes 4 and lane 8). (B) The interaction between Myc-DCAF26 and Flag-DIM-5. (C) Immunoprecipitation assay with Myc antibody showing that DDB1 formed a complex with Myc-DIM-5 in the dim-5KO complemented strain with expressing Myc-DIM-5, but not in the dcaf26KO strain. (D) Interactions of Flag-Cul4 and DDB1 independent of DCAF26 protein. Asterisks indicate nonspecific bands detected by anti-DDB1 serum. We next investigated the function of DCAF26 in this complex. We checked the interactions of DDB1-DIM-5 in the dcaf26KO strain and in a dim-5KO, qa-Myc-DIM-5 transformant. As shown in Figure 4C, the interactions between DDB1 and DIM-5 were severely impaired in the dcaf26KO strain, while DIM-5 strongly interacted with DDB1 in the presence of DCAF26. This indicates that DCAF26 was required for recruiting DIM-5 to the Cul4-DDB1 complex. Furthermore, the interaction between DDB1 and Cul4 was not affected in the dcaf26KO mutant (Figure 4D), confirming that DCAF26 was an adaptor protein in the Cul4-DDB1-DIM-5 complex.
Interactions between DCAF26 and DDB1 are essential for DNA and H3K9 methylation
The finding that N. crassa DCAF26 interacts with DDB1 and neddylated Cul4 to form a complex prompted us to investigate the functional importance of the interaction between DCAF26 and DDB1. As shown in Figure 5A, DCAF26 and Cul4 protein interactions were totally abolished in the ddb1KO mutant, suggesting that DDB1 served as a bridge between Cul4 and DCAF26 to form the complex. This result suggests that DDB1, DCAF26, and their interactions contributed the H3K9 and DNA methylation functions of the Cul4-DDB1-DCAF26-DIM-5 complex.
Fig. 5. WD3 region of DCAF26 is necessary for binding between DCAF26 and DDB1. 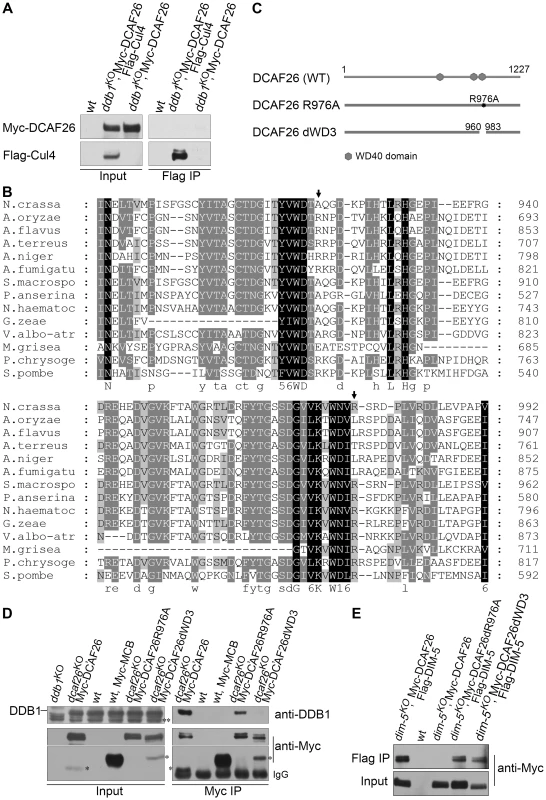
(A) Immunoprecipitation assay with Flag antibody showing that Flag-Cul4 failed to form a complex with Myc-DCAF26 in the ddb1KO strain. (B) Amino acid alignment of the conserved WD40 regions and WDXR motifs of different fungi DCAF26 homologs. The identical residues among different homologs are shaded. Arrows point out the arginine in the WDXR motif. (C) Graphic diagrams showing the domain structure of DCAF26 and different DCAF26 internal point mutations or deletion mutants. (D) Immunoprecipitation assay with c-Myc antibody showing that Myc-DCAF26 failed to interact with DDB1 in the DCAF26dWD3 mutant, but not in the DCAF26R976A mutant, compared to the strong interaction between DDB1 and wild-type DCAF26. Double asterisks indicate nonspecific bands detected by anti-DDB1 serum. Asterisks indicate degraded species of Myc-DCAF26. (E) Immunoprecipitation assay with Flag antibody showing that the mutant Myc-DCAF26 in the DCAF26R976A and DCAF26dWD3 strains still interacted with Flag-DIM-5. DCAF26 (NCU01656.3) contains some interesting sequence features, including one WDXR motif (WNVR) and WD-40 repeat regions. When the DCAF26 protein sequence was queried in a blast search against protein databases, DCAF26 was found to be similar to various fungal homologs. Many DCAF26 homologs contain one WDXR motif and one WDTA, or another WDXR motif located separately between consecutive “propeller blade” folds of the protein (Figure 5B). N. crassa DCAF26 protein contains one WDTA (aa 917–920) motif and one conserved WDXR (WNVR973–976) motif (Figure 5B).
To determine which domains of DCAF26 are involved in the DCAF26-DDB1 interaction, we mutated arginine 976 to alanine in the WNVR motif (Figure 5C) in qa-Myc-His-DCAF26 construct. As shown in Figure 5D, this mutation of DCAF26 reduced binding with DDB1 compared to the binding between wild-type DCAF26 and DDB1 in dcaf26KO transformants. However, this DCAF26 mutation did not affect the interactions between DCAF26R(976)A and DIM-5 (Figure 5E). We then deleted 24 amino acids (960–983) from DCAF26; this amino acid stretch included the WNVR motif and the third WD-40 domain (Figure 5C). As shown in Figure 5D, the interaction of DCAF26dWD3 with DDB1 was totally abolished in dcaf26KO qa-Myc-DCAF26dWD3 transformants. However, this DCAF26 deletion did not affect interactions between DCAF26dWD3 and DIM-5 (Figure 5E). These results indicate that the WNVR motif and adjacent amino acids were important for interactions with DDB1, not interactions with DIM-5.
To further examine the function of the interaction between DCAF26 and DDB1, we investigated the function of these mutated DCAF26 in dcaf26KO transformants. As shown in Figure 6A, the expression of Myc-tagged wild-type DCAF26 fully rescued the growth and developmental defects of the dcaf26KO mutant, resulting in a similar growth rate as the wild-type strain on plates. Importantly, the DNA methylation (Figure 6B) and the H3K9 trimethylation (Figure 6C and 6D) in the dcaf26KO, qa-Myc-His-DCAF26 transformant were also restored. The expression of Myc-DCAF26R(976)A mutant protein can restore the growth and developmental phenotypes (Figure 6A), as well as DNA methylation (Figure 6B) and H3K9 trimethylation (Figure 6C and 6D) of dcaf26KO mutant. These results indicated that the weak interaction of DCAF26-DDB1 is sufficient for the formation of Cul4-DDB1-DCAF26-DIM-5 complex. In contrast, the expression of Myc-DCAF26dWD3 mutant protein failed to rescue the growth and developmental phenotypes (Figure 6A) and the defects of DNA methylation (Figure 6B) and H3K9 trimethylation (Figure 6C and 6D) of dcaf26KO mutant, indicating that the interaction of DCAF26-DDB1 was required for the proper function of Cul4-DDB1-DCAF26-DIM-5 complex. Taken together, these results further demonstrate that DCAF26 is a critical component in the Cul4-DDB1 ubiquitin ligase in N. crassa.
Fig. 6. Interactions between DCAF26 and DDB1 are required for proper functioning of the Cul4-DDB1-DCAF26-DIM-5 complex. 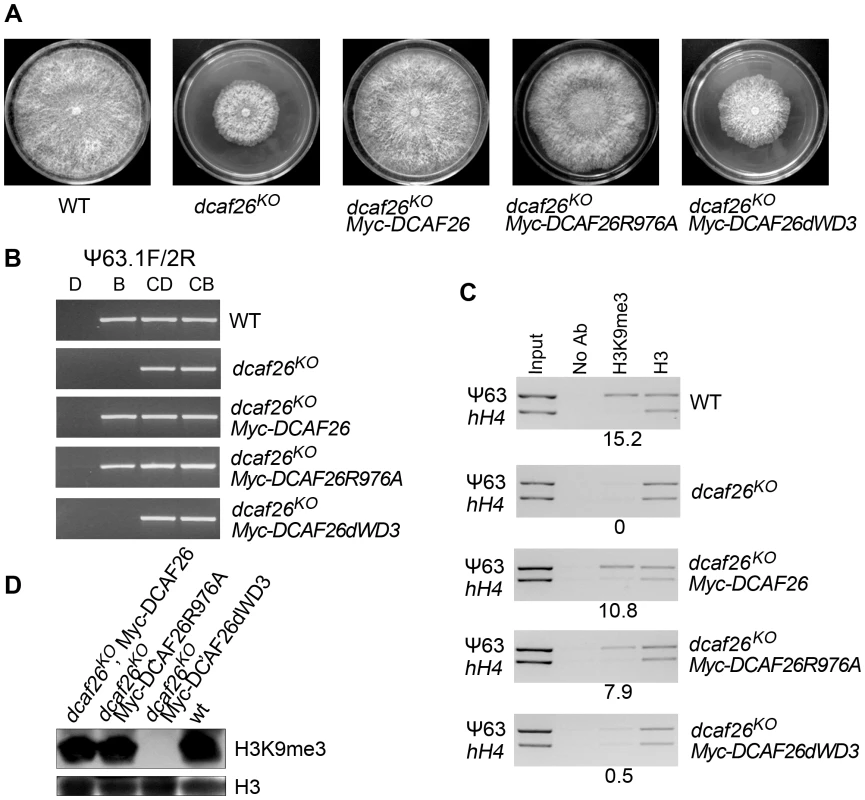
(A) Growth and developmental defects of the dcaf26KO strain were complemented by wild-type Myc-DCAF26 or mutated Myc-DCAF26R976A, but not Myc-DCAF26dWD3. (B) Loss of DNA methylation in dcaf26KO was rescued by expressing Myc-DCAF26 or Myc-DCAF26R976A, but not Myc-DCAF26dWD3. (C) Loss of H3K9 trimethylation at the ψ63 region in the dcaf26KO strain was rescued by expressing Myc-DCAF26 or Myc-DCAF26R976A, but not Myc-DCAF26dWD3. (D) Western blot analysis showing that the loss of global H3K9 trimethylation in dcaf26KO was restored by Myc-DCAF26 or Myc-DCAF26R976A, but not Myc-DCAF26dWD3. H3 antibody was used as the positive control and as the control for integrity of the nucleosome structure. Purification of DCAF26 and identification of DCAF26-associated proteins
Recent studies demonstrate the biochemical function of the DCAF protein DDB2 as a substrate receptor for XPC ubiquitination in DDB1-DDB2-CUL4A-Rbx1 complex in human cell lines [25]. Since we have demonstrated that DCAF26 is required for recruiting DIM-5 to the Cul4-DDB1 complex, we wondered whether there are other substrates associated with the complex.
To better understand the substrate adaptor role of DCAF26 for the Cul4-DDB1 E3 ligase, we attempted to purify this protein in N. crassa. To do so, we expressed Myc-His-DCAF26 protein by inoculating the dcaf26KO, qa-Myc-His-DCAF26 strain in liquid media containing quinic acid. Then Myc-His-DCAF26 protein was purified by nickel-column followed by immunoprecipitation using the c-Myc monoclonal antibody. As shown in Figure 7A, several major protein bands were specifically observed in the Myc-His-DCAF26 sample but not in the negative control (WT lane). The LC-MS/MS analysis of excised gel bands led to the identification of five well-known proteins, DCAF26, Cul4, DDB1, Nedd8, DIM-5 (the histone H3K9 methyltransferase in N. crassa) (Figure 7A and 7B). All proteins were represented with three or more peptides. The coexistence of Nedd8 peptides with Cul4 was consistent with previous results showing that DCAF26 preferentially interacted with neddylated Cul4 proteins in vivo, further confirming that DCAF26 is a key component of Cul4-DDB1 E3 ligase. In addition, we also identified other four DCAF26 co-purified proteins (Figure 7A and 7B) encoded by NCU07855, NCU04152, NCU06123, and NCU11350, respectively. Among these proteins, NCU04152 (DIM-7) is also required for DNA methylation and H3K9 trimethylation (Figure S4). Since the submission of this paper, similar results were recently shown by Lewis et al. [26]. Furthermore, the absence of ubiquitin peptides in the purification products suggests that either the ubiquitinated substrate is released immediately from Cul4-DDB1 complex or its ubiquitination level is too low to be detected.
Fig. 7. Identification of DCAF26-associated proteins. 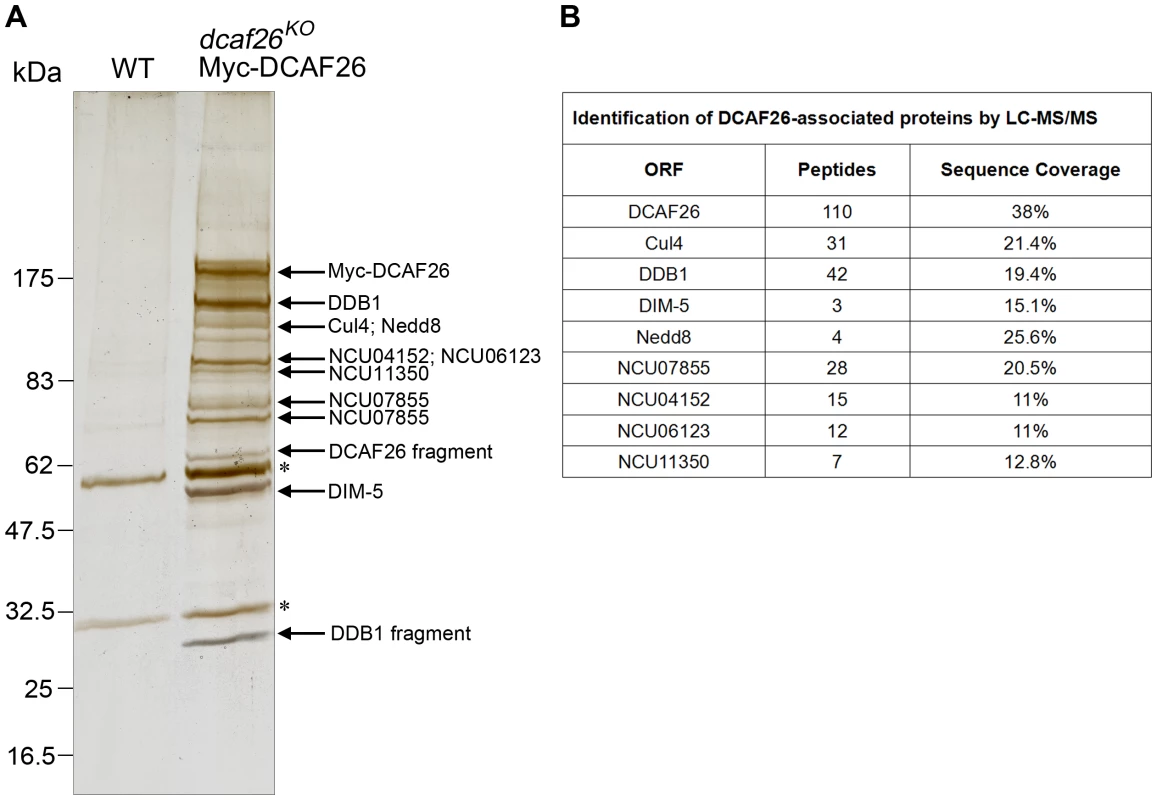
(A) Silver-stained SDS-PAGE showing the two-step purification of N. crassa DCAF26. Wild-type strain was used as the negative control. Asterisks indicate the two IgG bands. (B) Eight DCAF26-associated proteins were identified by LC-MS/MS. Discussion
Cul4-based E3 ubiquitin ligases are evolutionarily conserved multifunctional complexes in eukaryotes. In this study, using a genetic screen to find genes that encode proteins interacting with DDB1 and Cul4 in N. crassa, we identified DCAF26 as a new component required for DNA methylation. dcaf26KO mutants exhibited the loss of all known DNA methylation and H3K9 trimethylation. Our data demonstrate that DCAF26 interacts with DDB1, which in turn recruits DIM-5 via the Cul4-DDB1-DCAF26 complex to regulate H3K9 trimethylation and DNA methylation.
DCAF26 is an essential component in the Cul4-DDB1 complex for recruiting DIM-5 to regulate H3K9 trimethylation
Recent studies demonstrated that three proteins, DIM-5 [23], HP1 [27], and DIM-2 [28], are essential for DNA methylation in N. crassa, which function in histone H3K9 trimethylation and DNA methylation. To understand the regulation of DNA methylation in N. crassa, we sought to identify the proteins operating upstream of DIM-5. Previously, we showed that DDB1 and Cul4 function in an early step of DNA methylation by forming a complex with DIM-5 to regulate H3K9 trimethylation in N. crassa [19]. To determine the precise function of this complex, we sought to identify the DDB1 - and Cul4-associated factors (DCAFs) involved in this pathway. In mammalian cells, the DCAF protein WDR5 is a core component of the histone methylation complex essential for H3K4 methylation [12], and loss of WDR5 specifically affects tri - and monomethylated H3K4 [13]. However, there is limited evidence to suggest that histone H3 methylation is directed by DCAF protein, which recruits a specific histone methyltransferase. In fission yeast, a putative DCAF protein, Dos1/Raf1/Clr8/Cmc1 15–18, interacts with Cul4, Rik1, and Clr4 (histone H3K9 methyltransferase) to regulate the heterochromatin formation. It was reported that Dos1, which is associated with Rik1 and important for the function of the Clr4-Rik1 complex, is essential for the recruitment of Clr4 in the RNAi-dependent heterochromatin pathway [17].
To find out how N. crassa DCAF proteins participate in histone H3K9 methylation and DNA methylation, we explored potential DCAF proteins that might interact with Cul4-DDB1 E3 ubiquitin ligase. We found that DCAF26 was associated with DDB1 and was essential for H3K9 trimethylation and DNA methylation. Protein interaction experiments revealed that, as with DIM-5 [19], DCAF26 preferentially interacted with neddylated Cul4 species. Interactions between DDB1 and DIM-5 were dependent on DCAF26; this indicates that DCAF26 served as a link between Cul4-DDB1 and DIM-5 in this pathway, thus regulating H3K9 trimethylation. Interestingly, in fission yeast, the loss of H3K9 methylation in dos1 deletion mutants is similar to that in rik1 and clr4 mutants, and more severe than in RNAi mutants [17], suggesting that Dos1 is required for histone H3 modification in the same pathway as Rik1 and Clr4.
Interaction between DDB1 and DCAF26 is essential for H3K9 trimethylation and DNA methylation
Immunoprecipitation experiments revealed that DCAF26 bridges the Cul4-DDB1 complex to DIM-5 and functions in the same DNA methylation pathway in N. crassa. If DCAF26 protein interacts with DDB1, Cul4, and DIM-5 to form a complex, we would expect the association of DCAF26-DDB1 to play an essential role in the function of the Cul4-DDB1-DCAF26-DIM-5 complex. Recent studies showed that many DCAF proteins contain two conserved WDXR motifs that are the key interacting modules with DDB1 in Cul4-DDB1 E3 ubiquitin ligases [3], [4], [13], [29], [30]. In this study, we showed that deletion of a region with a WDXR motif in DCAF26, but not the single-amino acid substitution of WNVR976A, eliminated DCAF26-DDB1 interactions and H3K9 trimethylation and DNA methylation in vivo, indicating that the interaction between DCAF26 and DDB1 is required for H3K9 trimethylation and DNA methylation. Although several studies showed that the arginines in conserved WDXR motifs in DCAF proteins are required for the association between DCAFs and DDB1, some DCAFs that lack the conserved motifs can still interact with DDB1 and Cul4 proteins [8], [9]. Interestingly, S. pombe Dos1/Raf1/Clr8/Cmc1 contains two conserved WDXR motifs [4]; however, mutagenesis studies of these motifs to examine the interactions with Rik1 protein have not yet been performed. Alignment of these two motifs and adjacent regions in S. pombe Dos1 with the corresponding region in N. crassa showed that they share a high similarity, suggesting that this region is important for interactions with DDB1. Indeed, the results of our deletion and mutagenesis studies revealed that the WDXR-containing WD3 region, but not the arginine residue, was necessary for productive assembly with DDB1 into a functional H3K9 methyltransferase complex under physiological conditions. In addition, we found that the WDXR-containing WD3 region was not required for interactions between DCAF26 and DIM-5, and confirmed that the association of DCAF26-DDB1 was essential for recruiting DIM-5 to regulate H3K9me3. Thus, the Cul4-DDB1-DCAF26 complex serves to recruit the histone methyltransferase DIM-5 for H3K9 trimethylation, which serves as a mark for DNA methylation.
The substrate(s) of the Cul4-DDB1-DCAF26 ubiquitin ligase
Our results indicate that DCAF26 protein was necessary for recruiting DIM-5 into the Cul4-DDB1 complex. In addition, both DCAF26 and DIM-5 preferentially interacted with neddylated Cul4 species. These data suggest that the Cul4-DDB1-DCAF26 complex is required to recruit DIM-5 or to ubiquitinate a specific substrate(s) to perform its epigenetic functions. In fission yeast, in vitro data show that H2B can be polyubiquitinated by a purified complex containing Cul4-Rik1-Raf1-Raf2-Clr4 [16]. In mammalian cells, the putative targeting substrate of Cul4-DDB1-WDR5 is histone H3 [13]. In human cells, Cul4-DDB1DDB2 promotes the ubiquitination of histones H3 and H4 [31]. However, under normal growth conditions, the Cul4-DDB1-DCAF26 complex mainly recruits DIM-5 to regulate H3K9 trimethylation and DNA methylation. In contrast, histones H2B and H4 were copurified with ubiquitin, Cmc1, and Clr4 in Rik1 purification products and were suggested as the likely targets of Cul4-mediated ubiquitination in fission yeast [15]. In mammalian cells, XPC was identified as a substrate directly targeted by the DCAF protein DDB2 to the DDB1-DDB2-CUL4A-Rbx1 E3 complex [25]. In S. pombe, the DCAF protein Cdt2 is required for the degradation of Spd1 through ubiquitination by the Cul4-DDB1 E3 ligase [7]. However, no direct binding is observed between Cdt2 and its target protein Cdt1 in mammalian cells [29]. These results suggest that it may be difficult to identify the substrate of a specific DCAF by copurifying its interacting proteins. We tried to identify the substrate of the Cul4-DDB1-DCAF26 complex by purification of Myc-His-DCAF26 in N. crassa. In Myc-His-DCAF26 affinity purification products, Cul4, DDB1, DIM-5, and Nedd8 were copurified, strongly suggesting that they form an E3 ubiquitin ligase. Unlike the Rik1 purification in fission yeast, no ubiquitinated protein was detected in the purification. This result suggests that the substrate is ubiquitinated at low levels, such as mono-ubiquitination, by the Cul4-DDB1-DCAF26 complex, or that the ubiquitinated substrate is released immediately from its E3 complex. Alternatively, it is also possible that ubiquitinated protein(s) do not associate tightly with this complex during H3K9 methylation and DNA methylation. The activating signals of DNA methylation can trigger assembly of the Cul4-DDB1-DCAF26 complex, in which neddylated Cul4 promotes the complex formation with DDB1 and DCAF26, and then recruits DIM-5 at specific DNA regions for H3K9 trimethylation. This functional complex would link the initial signals of DNA methylation, and downstream epigenetic modifications on histones and DNA.
Together, our results suggest that in the DNA methylation process, the biochemical function of the Cul4-DDB1-DCAF26 complex is to recruit DIM-5 to control H3K9 methylation and DNA methylation in N. crassa. Because Cul4-DDB1 complexes are conserved in higher eukaryotes, we propose that analogous Cul4-DDB1-DCAF complexes may have a similar role in these organisms.
Materials and Methods
Strains and culture conditions
In this study, 87-3 (bd a) was used as the wild-type strain. All of the dcaf gene knockout mutants were newly generated from the bd ku70RIP genetic background strain and are listed in Table 1. The ddb1KO, cul4KO, dim-5KO, and dim-2KO strains generated previously [19] were also included in this study. The 301-6 (bd, his-3, A), ddb1KO (bd, his-3), and dim-5KO (bd, his-3) strains were used as the host strain for the his-3 targeting constructs.
Liquid cultures were grown in minimal medium (1× Vogel's, 2% glucose). For QA-induced gene expression, 0.01 M QA (pH 5.8) was added into liquid medium containing 1× Vogel's, 0.1% glucose, and 0.17% arginine [32]. For the racetube assay, the medium contained 1× Vogel's, 0.1% glucose, 0.17% arginine, 50 ng/mL biotin, and 1.5% agar.
Plasmid construction and creation of cotransformation strains
Full-length open reading frames (ORFs) and the 3′-UTR of all putative DCAF proteins were PCR amplified from genomic DNA and cloned into pqa-5Myc-6His. To study the interaction of Myc-tagged DCAF26 with Flag-tagged Cul4 or Flag-DIM-5, the Myc-tagged construct was introduced into transformants expressing Flag-Cul4 or Flag-DIM-5 by cotransformation with pBT6 (containing the benomyl resistance gene, obtained from the Fungal Genetics Stock Center). Western blot analyses using a monoclonal c-Myc antibody (9E10, Santa Cruz Biotechnology) were performed to identify the positive cotransformants. Immunoprecipitation assays using c-Myc antibody or Flag antibody (F3165-5MG, Sigma) were performed to test the interactions between Myc-tagged DCAF26 and Flag-Cul4 or Flag-DIM-5 in positive cotransformants. Protein extraction, quantification, western blot analysis, and immunoprecipitation assays were performed as described previously [32], [33].
Generation of dcaf knockout mutants by gene replacement
Sixteen dcaf gene knockout mutants were generated following the knockout procedures described previously [19]. Briefly, the entire ORF knockout cassette of each gene was created by PCR. The hph gene replacement cassette was introduced into the ku70RIP strain by electroporation. The transformants with hph at the ORF of the targeted dacf gene locus were passaged on minimal slants with hygromycin for five generations. The homokaryotic knockout strains of these dacf genes were then obtained by conidia purification and confirmed by PCR analysis.
DNA methylation analysis
The protocol of DNA methylation assay was the same as described previously [19]. Briefly, genomic DNA (200 ng) was digested with DpnII or BfuCI (Sau3AI) in 50 µL reaction system. No enzyme was added to the control samples. The PCR primers for ku70, ζ-η, Ψ63, and cen VII regions are listed in Table 2. PCR was performed using 1 µL of the digested DNA as a template in a 50 µL reaction system, with a program of 5 min at 94°C, followed by 31 cycles at 94°C (30 sec), 53°C (30 sec), and 72°C (1 min). The PCR products were resolved by electrophoresis on 2% agarose gels. Each experiment was performed independently at least three times.
Tab. 2. List of primers used in this study. 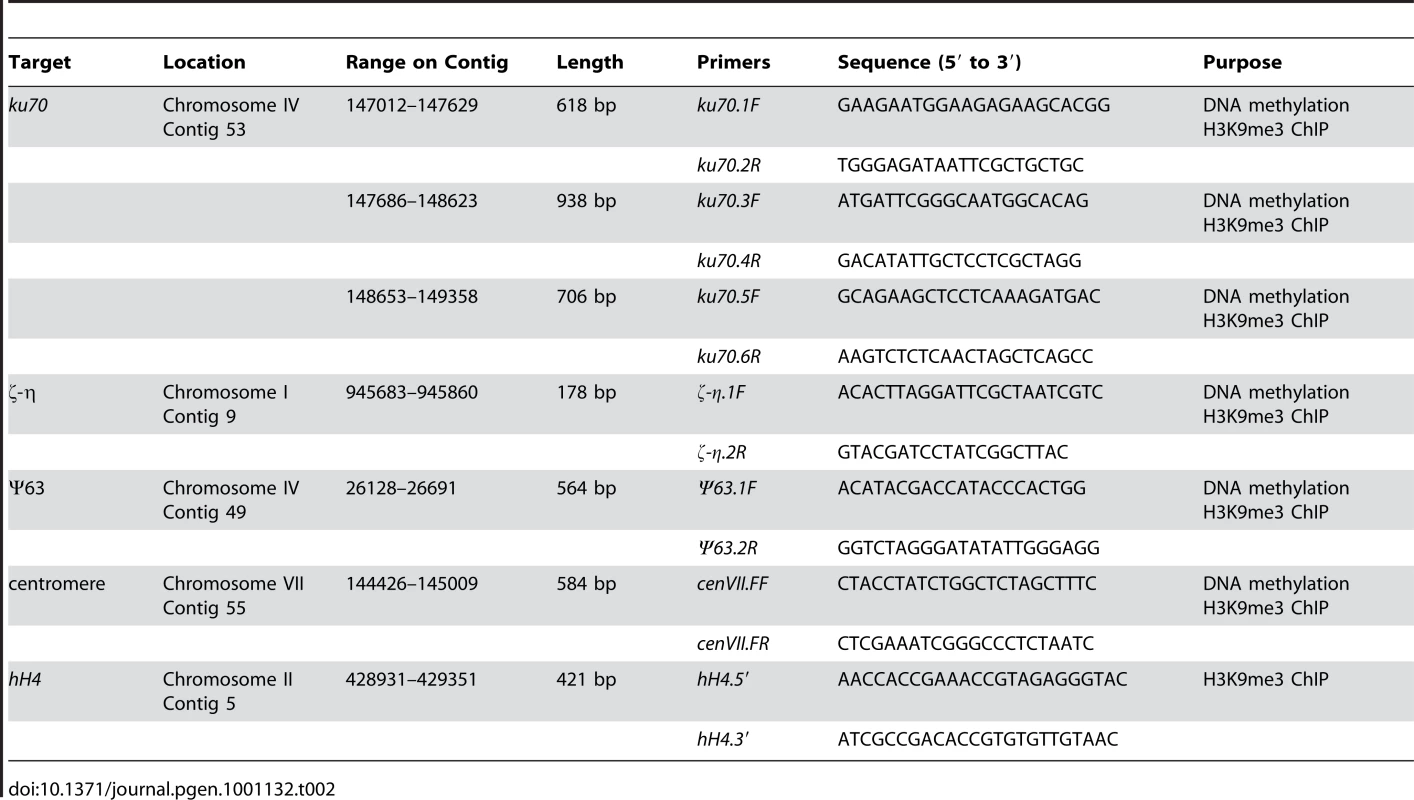
ChIP analysis
For the ChIP assay, tissues were fixed in minimal media containing 1% formaldehyde for 10 min at 25°C. ChIP was performed using 8 µL of antibody to H3K9me3 (07-442, Upstate Biotechnology) or 10µL of antibody to H3 (06-755, Upstate Biotechnology) for 6 mg/ml protein. After washing with 70% ethanol, extracted DNA pellets were resuspended in 30 µL of double-distilled water, and 0.5 µL of the DNA solution was used for PCR. The primers for ku70, ζ-η, Ψ63, and hH4 regions are listed in Table 2. PCR conditions were as follows: 5 min at 94°C, 26–30 cycles at 94°C (30 sec), 53°C (30 sec), and 72°C (1 min). Different PCR cycles were tested to ensure that DNA amplification was within the exponential amplification range. PCR products were resolved by electrophoresis on 2% agarose gels. ChIP assays with anti-Myc antibody or no antibody were used as negative controls. Each experiment was independently performed at least three times.
Western blot analysis of histone H3 and trimethylated H3K9
N. crassa histone proteins were extracted from the wild-type strain, dcaf26KO, ddb1KO, cul4KO, and dim-5KO strains, as described previously [34]. Equal amounts of histone protein extracts were loaded onto 15% SDS-PAGE gels. Western blot analysis was performed using antibodies against trimethylated H3 Lys9 (07-442 Upstate Biotechnology) or H3 (06-755 Upstate Biotechnology).
Purification of Myc-His-DCAF26 from N. crassa
dcaf26KO Myc-His-DCAF26 strain and the wild-type strain (as the negative control) were cultured for approximately 20 hr in constant light (LL) in liquid medium containing QA (0.01 M QA, 1× Vogel's, 0.1% glucose, and 0.17% arginine). Approximately 10 g of tissue from each strain grown in LL was harvested. The purification procedure was the same as described previously [22]. Fractions containing purified Myc-His-DCAF26 were immunoprecipitated by adding 20 µL of c-Myc monoclonal antibody-coupled agarose beads (9E10AC, Santa Cruz Biotechnology). The precipitates were analyzed by SDS-PAGE (2%–20%), which was subsequently silver stained following the manufacturer's instructions (ProteoSilver Plus, Sigma). The specific bands were excised and subjected to tryptic digestion and liquid chromatography–mass spectrometry/mass spectrometry analysis (LC-MS/MS).
Supporting Information
Zdroje
1. O'ConnellBC
HarperJW
2007 Ubiquitin proteasome system (UPS): what can chromatin do for you? Curr Opin Cell Biol 19 206 214
2. HigaLA
ZhangH
2007 Stealing the spotlight: CUL4-DDB1 ubiquitin ligase docks WD40-repeat proteins to destroy. Cell Div 2 5
3. LeeJ
ZhouP
2007 DCAFs, the missing link of the CUL4-DDB1 ubiquitin ligase. Mol Cell 26 775 780
4. AngersS
LiT
YiX
MacCossMJ
MoonRT
2006 Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery. Nature 443 590 593
5. PetroskiMD
DeshaiesRJ
2005 Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol 6 9 20
6. GroismanR
KuraokaI
ChevallierO
GayeN
MagnaldoT
2006 CSA-dependent degradation of CSB by the ubiquitin-proteasome pathway establishes a link between complementation factors of the Cockayne syndrome. Genes Dev 20 1429 1434
7. LiuC
PoiteleaM
WatsonA
YoshidaSH
ShimodaC
2005 Transactivation of Schizosaccharomyces pombe cdt2+ stimulates a Pcu4-Ddb1-CSN ubiquitin ligase. Embo J 24 3940 3951
8. LiT
ChenX
GarbuttKC
ZhouP
ZhengN
2006 Structure of DDB1 in complex with a paramyxovirus V protein: viral hijack of a propeller cluster in ubiquitin ligase. Cell 124 105 117
9. FukumotoY
DohmaeN
HanaokaF
2008 Schizosaccharomyces pombe Ddb1 recruits substrate-specific adaptor proteins through a novel protein motif, the DDB-box. Mol Cell Biol 28 6746 6756
10. DouY
MilneTA
TackettAJ
SmithER
FukudaA
2005 Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell 121 873 885
11. RuthenburgAJ
WangW
GrayboschDM
LiH
AllisCD
2006 Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex. Nat Struct Mol Biol 13 704 712
12. WysockaJ
SwigutT
MilneTA
DouY
ZhangX
2005 WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell 121 859 872
13. HigaLA
WuM
YeT
KobayashiR
SunH
2006 CUL4-DDB1 ubiquitin ligase interacts with multiple WD40-repeat proteins and regulates histone methylation. Nat Cell Biol 8 1277 1283
14. JiaS
KobayashiR
GrewalSI
2005 Ubiquitin ligase component Cul4 associates with Clr4 histone methyltransferase to assemble heterochromatin. Nat Cell Biol 7 1007 1013
15. HongEJ
VillenJ
GeraceEL
GygiSP
MoazedD
2005 A cullin E3 ubiquitin ligase complex associates with Rik1 and the Clr4 histone H3-K9 methyltransferase and is required for RNAi-mediated heterochromatin formation. RNA Biol 2 106 111
16. HornPJ
BastieJN
PetersonCL
2005 A Rik1-associated, cullin-dependent E3 ubiquitin ligase is essential for heterochromatin formation. Genes Dev 19 1705 1714
17. LiF
GotoDB
ZaratieguiM
TangX
MartienssenR
2005 Two novel proteins, dos1 and dos2, interact with rik1 to regulate heterochromatic RNA interference and histone modification. Curr Biol 15 1448 1457
18. ThonG
HansenKR
AltesSP
SidhuD
SinghG
2005 The Clr7 and Clr8 directionality factors and the Pcu4 cullin mediate heterochromatin formation in the fission yeast Schizosaccharomyces pombe. Genetics 171 1583 1595
19. ZhaoY
ShenY
YangS
WangJ
HuQ
2010 Ubiquitin ligase components Cullin4 and DDB1 are essential for DNA methylation in Neurospora crassa. J Biol Chem 285 4355 4365
20. LeeJH
TerzaghiW
GusmaroliG
CharronJB
YoonHJ
2008 Characterization of Arabidopsis and rice DWD proteins and their roles as substrate receptors for CUL4-RING E3 ubiquitin ligases. Plant Cell 20 152 167
21. AronsonBD
JohnsonKA
LorosJJ
DunlapJC
1994 Negative feedback defining a circadian clock: autoregulation of the clock gene frequency. Science 263 1578 1584
22. HeQ
ChengP
HeQ
LiuY
2005 The COP9 signalosome regulates the Neurospora circadian clock by controlling the stability of the SCFFWD-1 complex. Genes Dev 19 1518 1531
23. TamaruH
SelkerEU
2001 A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature 414 277 283
24. TamaruH
ZhangX
McMillenD
SinghPB
NakayamaJ
2003 Trimethylated lysine 9 of histone H3 is a mark for DNA methylation in Neurospora crassa. Nat Genet 34 75 79
25. SugasawaK
OkudaY
SaijoM
NishiR
MatsudaN
2005 UV-induced ubiquitylation of XPC protein mediated by UV-DDB-ubiquitin ligase complex. Cell 121 387 400
26. LewisZA
AdhvaryuKK
HondaS
ShiverAL
SelkerEU
2010 Identification of DIM-7, a protein required to target the DIM-5 H3 methyltransferase to chromatin. Proc Natl Acad Sci U S A 107 8310 8315
27. FreitagM
HickeyPC
KhlafallahTK
ReadND
SelkerEU
2004 HP1 is essential for DNA methylation in neurospora. Mol Cell 13 427 434
28. KouzminovaE
SelkerEU
2001 dim-2 encodes a DNA methyltransferase responsible for all known cytosine methylation in Neurospora. Embo J 20 4309 4323
29. JinJ
AriasEE
ChenJ
HarperJW
WalterJC
2006 A family of diverse Cul4-Ddb1-interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1. Mol Cell 23 709 721
30. HeYJ
McCallCM
HuJ
ZengY
XiongY
2006 DDB1 functions as a linker to recruit receptor WD40 proteins to CUL4-ROC1 ubiquitin ligases. Genes Dev 20 2949 2954
31. WangH
ZhaiL
XuJ
JooHY
JacksonS
2006 Histone H3 and H4 ubiquitylation by the CUL4-DDB-ROC1 ubiquitin ligase facilitates cellular response to DNA damage. Mol Cell 22 383 394
32. ChengP
YangY
HeintzenC
LiuY
2001 Coiled-coil domain-mediated FRQ-FRQ interaction is essential for its circadian clock function in Neurospora. Embo J 20 101 108
33. GarceauNY
LiuY
LorosJJ
DunlapJC
1997 Alternative initiation of translation and time-specific phosphorylation yield multiple forms of the essential clock protein FREQUENCY. Cell 89 469 476
34. BraunsteinM
RoseAB
HolmesSG
AllisCD
BroachJR
1993 Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes Dev 7 592 604
Štítky
Genetika Reprodukční medicína
Článek Allelic Variation at the 8q23.3 Colorectal Cancer Risk Locus Functions as a Cis-Acting Regulator ofČlánek Allelic Selection of Amplicons in Glioblastoma Revealed by Combining Somatic and Germline AnalysisČlánek Lactic Acidosis Triggers Starvation Response with Paradoxical Induction of TXNIP through MondoAČlánek Rice a Cinnamoyl-CoA Reductase-Like Gene Family Member, Is Required for NH1-Mediated Immunity to pv.Článek Differentiation of Zebrafish Melanophores Depends on Transcription Factors AP2 Alpha and AP2 Epsilon
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2010 Číslo 9- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- IVF a rakovina prsu – zvyšují hormony riziko vzniku rakoviny?
-
Všechny články tohoto čísla
- Optimal Strategy for Competence Differentiation in Bacteria
- Mutational Patterns Cannot Explain Genome Composition: Are There Any Neutral Sites in the Genomes of Bacteria?
- Frail Hypotheses in Evolutionary Biology
- Genetic Architecture of Complex Traits and Accuracy of Genomic Prediction: Coat Colour, Milk-Fat Percentage, and Type in Holstein Cattle as Contrasting Model Traits
- Allelic Variation at the 8q23.3 Colorectal Cancer Risk Locus Functions as a Cis-Acting Regulator of
- Allelic Selection of Amplicons in Glioblastoma Revealed by Combining Somatic and Germline Analysis
- Germline Variation Controls the Architecture of Somatic Alterations in Tumors
- Mice Doubly-Deficient in Lysosomal Hexosaminidase A and Neuraminidase 4 Show Epileptic Crises and Rapid Neuronal Loss
- Analysis of Population Structure: A Unifying Framework and Novel Methods Based on Sparse Factor Analysis
- FliO Regulation of FliP in the Formation of the Flagellum
- Cdc20 Is Critical for Meiosis I and Fertility of Female Mice
- dMyc Functions Downstream of Yorkie to Promote the Supercompetitive Behavior of Hippo Pathway Mutant Cells
- DCAF26, an Adaptor Protein of Cul4-Based E3, Is Essential for DNA Methylation in
- Genome-Wide Double-Stranded RNA Sequencing Reveals the Functional Significance of Base-Paired RNAs in
- An Immune Response Network Associated with Blood Lipid Levels
- Genetic Variants and Their Interactions in the Prediction of Increased Pre-Clinical Carotid Atherosclerosis: The Cardiovascular Risk in Young Finns Study
- The Histone H3K36 Methyltransferase MES-4 Acts Epigenetically to Transmit the Memory of Germline Gene Expression to Progeny
- Long- and Short-Term Selective Forces on Malaria Parasite Genomes
- Lactic Acidosis Triggers Starvation Response with Paradoxical Induction of TXNIP through MondoA
- Identification of Early Requirements for Preplacodal Ectoderm and Sensory Organ Development
- Orphan CpG Islands Identify Numerous Conserved Promoters in the Mammalian Genome
- Analysis of the Basidiomycete Reveals Conservation of the Core Meiotic Expression Program over Half a Billion Years of Evolution
- ETS-4 Is a Transcriptional Regulator of Life Span in
- The SR Protein B52/SRp55 Is Required for DNA Topoisomerase I Recruitment to Chromatin, mRNA Release and Transcription Shutdown
- The Baker's Yeast Diploid Genome Is Remarkably Stable in Vegetative Growth and Meiosis
- Chromatin Landscape Dictates HSF Binding to Target DNA Elements
- The APETALA-2-Like Transcription Factor OsAP2-39 Controls Key Interactions between Abscisic Acid and Gibberellin in Rice
- Accurately Assessing the Risk of Schizophrenia Conferred by Rare Copy-Number Variation Affecting Genes with Brain Function
- Widespread Over-Expression of the X Chromosome in Sterile F Hybrid Mice
- The Characterization of Twenty Sequenced Human Genomes
- The Genome of a Pathogenic : Cooptive Virulence Underpinned by Key Gene Acquisitions
- A Single Element Maintains Repression of the Key Developmental Regulator
- Identification of New Genetic Risk Variants for Type 2 Diabetes
- Effect of Correlated tRNA Abundances on Translation Errors and Evolution of Codon Usage Bias
- Evidence of Selection upon Genomic GC-Content in Bacteria
- Proteomic Changes Resulting from Gene Copy Number Variations in Cancer Cells
- Rice a Cinnamoyl-CoA Reductase-Like Gene Family Member, Is Required for NH1-Mediated Immunity to pv.
- Longitudinal Genome-Wide Association of Cardiovascular Disease Risk Factors in the Bogalusa Heart Study
- Response to Mechanical Stress Is Mediated by the TRPA Channel Painless in the Heart
- DNMT3L Modulates Significant and Distinct Flanking Sequence Preference for DNA Methylation by DNMT3A and DNMT3B
- Identifying Signatures of Natural Selection in Tibetan and Andean Populations Using Dense Genome Scan Data
- Incremental Genetic Perturbations to MCM2-7 Expression and Subcellular Distribution Reveal Exquisite Sensitivity of Mice to DNA Replication Stress
- Loss of Maternal ATRX Results in Centromere Instability and Aneuploidy in the Mammalian Oocyte and Pre-Implantation Embryo
- Comparative Genomic Hybridization (CGH) Reveals a Neo-X Chromosome and Biased Gene Movement in Stalk-Eyed Flies (Genus )
- Differentiation of Zebrafish Melanophores Depends on Transcription Factors AP2 Alpha and AP2 Epsilon
- Gene–Environment Interactions at Nucleotide Resolution
- Dementia Revealed: Novel Chromosome 6 Locus for Late-Onset Alzheimer Disease Provides Genetic Evidence for Folate-Pathway Abnormalities
- Critical Functions of Rpa3/Ssb3 in S-Phase DNA Damage Responses in Fission Yeast
- Preferential Re-Replication of Heterochromatin in the Absence of Geminin
- The Potential for Enhancing the Power of Genetic Association Studies in African Americans through the Reuse of Existing Genotype Data
- Evidence That Mutation Is Universally Biased towards AT in Bacteria
- Perturbation Analysis of Heterochromatin-Mediated Gene Silencing and Somatic Inheritance
- Diversity of Eukaryotic DNA Replication Origins Revealed by Genome-Wide Analysis of Chromatin Structure
- Genetic Deletion of the Desmosomal Component Promotes Tumor Microinvasion in a Mouse Model of Pancreatic Neuroendocrine Carcinogenesis
- The Metabolic Enzyme ManA Reveals a Link between Cell Wall Integrity and Chromosome Morphology
- SNPs Associated with Cerebrospinal Fluid Phospho-Tau Levels Influence Rate of Decline in Alzheimer's Disease
- Synthesizing and Salvaging NAD: Lessons Learned from
- A Central Regulatory System Largely Controls Transcriptional Activation and Repression Responses to Phosphate Starvation in Arabidopsis
- An Insect Herbivore Microbiome with High Plant Biomass-Degrading Capacity
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Synthesizing and Salvaging NAD: Lessons Learned from
- Optimal Strategy for Competence Differentiation in Bacteria
- Long- and Short-Term Selective Forces on Malaria Parasite Genomes
- Identifying Signatures of Natural Selection in Tibetan and Andean Populations Using Dense Genome Scan Data
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



