-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Rising from the Ashes: DNA Repair in
article has not abstract
Published in the journal: . PLoS Genet 6(1): e32767. doi:10.1371/journal.pgen.1000815
Category: Perspective
doi: https://doi.org/10.1371/journal.pgen.1000815Summary
article has not abstract
The extraordinary resistance of Deinococcus radiodurans to ionizing radiation (IR) and desiccation is slowly drawing more intense scrutiny. Relative to most other organisms, Deinococcus has a survival advantage measured in orders of magnitude. Exposure to 5 kGy of IR reduces the genome of any bacterium to hundreds of fragments. Deinococcus is no exception. However, Deinococcus seems to take this catastrophe in stride. Over a period of 3–4 hours, overlapping fragments are spliced together into complete chromosomes, and the cells soon resume normal growth. There is no measurable lethality. Attempts to understand the molecular basis of this phoenix-like capability has given rise to numerous hypotheses. Notable among them are proposals that the condensed nature of the Deinococcus genome [1] or an unusual capacity to avoid protein oxidation [2] are keys to radiation resistance.
Regardless of the physiological or metabolic adaptations that Deinococcus may employ to enhance survival, it is hard to explain extreme genome reconstitution without considering DNA repair. As originally defined by Daly and Minton [3], and reinforced more recently by Radman and colleagues [4],[5], genome reconstitution in Deinococcus proceeds in two phases. The first phase has been attributed to a process dubbed extended synthesis-dependent single-strand DNA annealing (ESDSA) [4],[5]. The second phase involves RecA protein-mediated double-strand break repair. Some initial studies suggested that the first phase of repair did not involve the Deinococcus RecA protein, but more recent work has documented a role for RecA in both phases [4],[5]. ESDSA involves considerable nuclease activity to generate single-stranded DNA, strand invasion mediated by the RecA and/or RadA proteins, and extensive DNA synthesis primed by the invading strands prior to the annealing steps [4],[5].
The emerging picture (Figure 1) provides a useful framework, but one with many questions. Most of these involve enzymes and their roles. The generation of single-stranded DNA in ESDSA requires the function of at least one nuclease and helicase and perhaps several of both. Proteins are also needed to load RecA protein onto single-strand DNA binding protein (SSB)-coated single-stranded DNA. The most important pathway for double-strand break repair in Escherichia coli utilizes the RecBCD enzyme for all of these roles. However, Deinococcus encodes no homologue of RecB or RecC. Deinococcus does encode homologues of every protein involved in what is considered (in E. coli) an auxiliary pathway for recombinational DNA repair—the RecFOR pathway. In the RecFOR pathway, the RecJ and RecQ proteins take up the nuclease and helicase roles, respectively, while the RecFOR proteins function to load RecA protein onto the DNA. The absence of RecBC homologues in Deinococcus seems to imply that the RecFOR path is particularly important.
Fig. 1. Two stages of genome reconstitution in Deinococcus radiodurans. 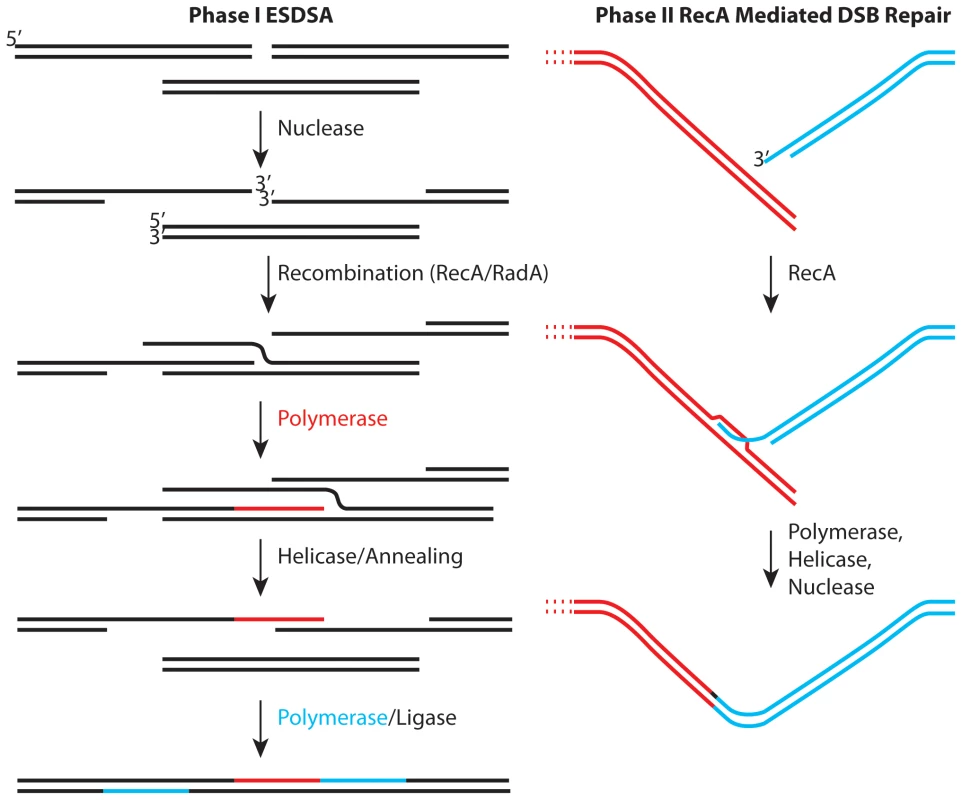
The first stage, extended synthesis-dependent single-strand annealing (ESDSA) is dominated by nuclease and DNA polymerase functions. The second stage is a more conventional RecA-mediated double-strand break repair process focused on the final splicing of large chromosomal segments. In two important reports in this issue of PLoS Genetics, Sommer and colleagues convert much recent speculation into substance. In the first report [6], the Chandler and Sommer laboratories collaborate to explore the mechanism of transposition of element ISDra2. This transposon is a member of a family of elements that transpose via single-stranded DNA intermediates. Transposition is activated by irradiation of Deinococcus. The work not only documents the transposition mechanism, it reinforces the proposition that extensive lengths of single-stranded genomic DNA are generated in the early stages of genome reconstitution in this bacterium. As a bonus, the work provides hope for the development of in vivo transposition as a tool for genetic manipulation of this genome.
The second report [7] clearly establishes the central role of the RecFOR pathway in genome reconstitution. Where the effects of RecFOR pathway gene deficiencies are generally modest in E. coli, they are dramatic in D. radiodurans. Sommer and colleagues demonstrate that cells lacking functional recF, recO, or recR genes are essentially as dysfunctional in genome reconstitution as recA mutants. Cells with these deficiencies are viable in the absence of extreme DNA damage, but grow much slower than wild type. Thus, the RecFOR pathway is important during normal replication, as well as during genome reconstitution. If the gene encoding the RecJ nuclease is inactivated, the cells are inviable. Surprisingly, cells lacking the RecQ helicase exhibit wild type resistance to IR. Instead, the helicase that appears to be critical is UvrD. In E. coli, the function of UvrD in recombinational DNA repair is to remove RecA filaments from the DNA [8],[9]. In Deinococcus, it almost certainly does more. In summary, Deinococcus now presents a unique opportunity to demonstrate what the RecFOR pathway can really do.
This RecFOR pathway may look a bit different from its cousin in E. coli. After irradiation, about 60 Deinococcus genes are induced, and quite a number of them have roles in genome reconstitution [10]. Many of these genes are novel. Knockouts do not have the dramatic effects of recAJFOR knockouts, but their cumulative effects can be significant. Some, like the DdrA protein (a distant homologue of the eukaryotic Rad52 protein [11]), and DdrB (a novel single-strand DNA binding protein [12]) must be worked into the schemes.
The work of Chandler and Sommer has a few more far-reaching implications. The spectacular feat of genome reconstitution after heavy irradiation does not require a completely new pathway for double-strand break repair, and no such pathway appears to be present. Instead, D. radiodurans relies heavily on a set of recombinational DNA repair functions that are recognizable in almost all species. In large measure, efficient genome reconstitution involves tweaking those repair proteins, providing a few novel augmentations, and perhaps modifying the environment in which all of these proteins function. However, the properties already noted that distinguish the RecFOR pathway in Deinococcus from the same process in E. coli bear reiteration. The roles that well-known repair proteins play in radioresistance are not perfectly predictable, based on what we understand about their function in E. coli. An orthologous relationship between proteins can inform speculation, but it must be subjected to experimental substantiation. Every DNA repair protein examined to date in D. radiodurans has provided one or more novel twists in our understanding of its function, structure, interaction with other proteins, and role in repair.
Last but not least, we may soon see a first in vitro reconstitution of a complete DNA double-strand break repair reaction. In this arena, D. radiodurans is gradually eclipsing E. coli as the most pliable bacterial model system. The proteins, or at least most of them, are in hand. Fortuitously, the enzymes from Deinococcus appear to be more amenable to structural determination than their E. coli cousins. The only structural information currently available about RecF, RecO, and RecR come from the Deinococcus enzymes [13]–[17]. Activities are being characterized, and more surprises are anticipated. Deinococcus simply does it better.
Zdroje
1. Levin-ZaidmanS
EnglanderJ
ShimoniE
SharmaAK
MintonKW
2003 Ringlike structure of the Deinococcus radiodurans genome: a key to radioresistance? Science 299 254 256
2. DalyMJ
GaidamakovaEK
MatrosovaVY
VasilenkoA
ZhaiM
2007 Protein oxidation implicated as the primary determinant of bacterial radioresistance. PLoS Biol 5 e92 doi:10.1371/journal.pbio.0050092
3. DalyMJ
MintonKW
1996 An alternative pathway of recombination of chromosomal fragments precedes recA-dependent recombination in the radioresistant bacterium Deinococcus radiodurans. J Bacteriol 178 4461 4471
4. SladeD
LindnerAB
PaulG
RadmanM
2009 Recombination and replication in DNA repair of heavily irradiated Deinococcus radiodurans. Cell 136 1044 1055
5. ZahradkaK
SladeD
BailoneA
SommerS
AverbeckD
2006 Reassembly of shattered chromosomes in Deinococcus radiodurans. Nature 443 569 573
6. PasternakC
Ton-HoangB
CosteG
BailoneA
ChandlerM
2009 Irradiation-induced D. radiodurans genome fragmentation triggers transposition of a single resident insertion sequence. PLoS Genet 6 e1000799 doi:10.1371/journal.pgen.1000799
7. BentchikouE
ServantP
CosteG
SommerS
2009 A major role of the RecFOR pathway in DNA double-strand-break repair through ESDSA in Deinococcus radiodurans. PLoS Genet 6 e1000774 doi:10.1371/journal.pgen.1000774
8. CentoreRC
SandlerSJ
2007 UvrD limits the number and intensities of RecA-green fluorescent protein structures in Escherichia coli K-12. J Bacteriol 189 2915 2920
9. VeauteX
DelmasP
SelvaM
JeussetJ
Le CamE
2005 UvrD helicase, unlike Rep helicase, dismantles RecA nucleoprotein filaments in Escherichia coli. EMBO J 24 180 189
10. TanakaM
EarlAM
HowellHA
ParkMJ
EisenJA
2004 Analysis of Deinococcus radiodurans's transcriptional response to ionizing radiation and desiccation reveals novel proteins that contribute to extreme radioresistance. Genetics 168 21 33
11. HarrisDR
TanakaM
SavelievSV
JolivetE
EarlAM
2004 Preserving genome integrity: the DdrA protein of Deinococcus radiodurans R1. PLoS Biology 2 e304 doi:10.1371/journal.pbio.0040304
12. NoraisC
Chitteni-PattuS
WoodEA
InmanRB
CoxMM
2009 DdrB protein, an alternative Deinococcus radiodurans SSB induced by ionizing radiation. J Biol Chem 284 21402 21411
13. KorolevaO
MakharashviliN
CourcelleCT
CourcelleJ
KorolevS
2007 Structural conservation of RecF and Rad50: implications for DNA recognition and RecF function. EMBO J 26 867 877
14. LeirosI
TimminsJ
HallDR
McSweeneyS
2005 Crystal structure and DNA-binding analysis of RecO from Deinococcus radiodurans. EMBO J 24 906 918
15. MakharashviliN
KorolevaO
BeraS
GrandgenettDP
KorolevS
2004 A novel structure of DNA repair protein RecO from Deinococcus radiodurans. Structure 12 1881 1889
16. MakharashviliN
KorolevaO
KorolevS
2005 Structural studies of replication/recombination mediator proteins. Biophys J 88 14A 14A
17. TimminsJ
LeirosI
McSweeneyS
2007 Crystal structure and mutational study of RecOR provide insight into its mode of DNA binding. EMBO J 26 3260 3271
Štítky
Genetika Reprodukční medicína
Článek Activation of Mutant Enzyme Function by Proteasome Inhibitors and Treatments that Induce Hsp70Článek Maternal Ethanol Consumption Alters the Epigenotype and the Phenotype of Offspring in a Mouse ModelČlánek Genetic Dissection of Differential Signaling Threshold Requirements for the Wnt/β-Catenin PathwayČlánek Distinct Type of Transmission Barrier Revealed by Study of Multiple Prion Determinants of Rnq1
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2010 Číslo 1- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
- Délka menstruačního cyklu jako marker ženské plodnosti
- Intrauterinní inseminace a její úspěšnost
-
Všechny články tohoto čísla
- Irradiation-Induced Genome Fragmentation Triggers Transposition of a Single Resident Insertion Sequence
- A Major Role of the RecFOR Pathway in DNA Double-Strand-Break Repair through ESDSA in
- Kidney Development in the Absence of and Requires
- Modeling of Environmental Effects in Genome-Wide Association Studies Identifies and as Novel Loci Influencing Serum Cholesterol Levels
- Inverse Correlation between Promoter Strength and Excision Activity in Class 1 Integrons
- Activation of Mutant Enzyme Function by Proteasome Inhibitors and Treatments that Induce Hsp70
- Postnatal Survival of Mice with Maternal Duplication of Distal Chromosome 7 Induced by a / Imprinting Control Region Lacking Insulator Function
- The Werner Syndrome Protein Functions Upstream of ATR and ATM in Response to DNA Replication Inhibition and Double-Strand DNA Breaks
- Maternal Ethanol Consumption Alters the Epigenotype and the Phenotype of Offspring in a Mouse Model
- Understanding Gene Sequence Variation in the Context of Transcription Regulation in Yeast
- miR-30 Regulates Mitochondrial Fission through Targeting p53 and the Dynamin-Related Protein-1 Pathway
- Elevated Levels of the Polo Kinase Cdc5 Override the Mec1/ATR Checkpoint in Budding Yeast by Acting at Different Steps of the Signaling Pathway
- Alternative Epigenetic Chromatin States of Polycomb Target Genes
- Co-Orientation of Replication and Transcription Preserves Genome Integrity
- A Comprehensive Map of Insulator Elements for the Genome
- Environmental and Genetic Determinants of Colony Morphology in Yeast
- U87MG Decoded: The Genomic Sequence of a Cytogenetically Aberrant Human Cancer Cell Line
- The MCM-Binding Protein ETG1 Aids Sister Chromatid Cohesion Required for Postreplicative Homologous Recombination Repair
- Genetic Dissection of Differential Signaling Threshold Requirements for the Wnt/β-Catenin Pathway
- Differential Localization and Independent Acquisition of the H3K9me2 and H3K9me3 Chromatin Modifications in the Adult Germ Line
- Genetic Crossovers Are Predicted Accurately by the Computed Human Recombination Map
- Collaborative Action of Brca1 and CtIP in Elimination of Covalent Modifications from Double-Strand Breaks to Facilitate Subsequent Break Repair
- Distinct Type of Transmission Barrier Revealed by Study of Multiple Prion Determinants of Rnq1
- Genome-Wide Association Study Identifies as a Novel Susceptibility Gene for Osteoporosis
- and Regulate Reproductive Habit in Rice
- Nonsense-Mediated Decay Enables Intron Gain in
- Altered Gene Expression and DNA Damage in Peripheral Blood Cells from Friedreich's Ataxia Patients: Cellular Model of Pathology
- The Systemic Imprint of Growth and Its Uses in Ecological (Meta)Genomics
- The Gift of Observation: An Interview with Mary Lyon
- Genotype and Gene Expression Associations with Immune Function in
- The Elongator Complex Regulates Neuronal α-tubulin Acetylation
- Rising from the Ashes: DNA Repair in
- Mis-Spliced Transcripts of Nicotinic Acetylcholine Receptor α6 Are Associated with Field Evolved Spinosad Resistance in (L.)
- BRIT1/MCPH1 Is Essential for Mitotic and Meiotic Recombination DNA Repair and Maintaining Genomic Stability in Mice
- Non-Coding Changes Cause Sex-Specific Wing Size Differences between Closely Related Species of
- Evidence for Pervasive Adaptive Protein Evolution in Wild Mice
- Evolutionary Mirages: Selection on Binding Site Composition Creates the Illusion of Conserved Grammars in Enhancers
- VEZF1 Elements Mediate Protection from DNA Methylation
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- A Major Role of the RecFOR Pathway in DNA Double-Strand-Break Repair through ESDSA in
- Kidney Development in the Absence of and Requires
- The Werner Syndrome Protein Functions Upstream of ATR and ATM in Response to DNA Replication Inhibition and Double-Strand DNA Breaks
- Alternative Epigenetic Chromatin States of Polycomb Target Genes
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



