-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Postnatal Survival of Mice with Maternal Duplication of Distal Chromosome 7 Induced by a / Imprinting Control Region Lacking Insulator Function
The misexpressed imprinted genes causing developmental failure of mouse parthenogenones are poorly defined. To obtain further insight, we investigated misexpressions that could cause the pronounced growth deficiency and death of fetuses with maternal duplication of distal chromosome (Chr) 7 (MatDup.dist7). Their small size could involve inactivity of Igf2, encoding a growth factor, with some contribution by over-expression of Cdkn1c, encoding a negative growth regulator. Mice lacking Igf2 expression are usually viable, and MatDup.dist7 death has been attributed to the misexpression of Cdkn1c or other imprinted genes. To examine the role of misexpressions determined by two maternal copies of the Igf2/H19 imprinting control region (ICR)—a chromatin insulator, we introduced a mutant ICR (ICRΔ) into MatDup.dist7 fetuses. This activated Igf2, with correction of H19 expression and other imprinted transcripts expected. Substantial growth enhancement and full postnatal viability was obtained, demonstrating that the aberrant MatDup.dist7 phenotype is highly dependent on the presence of two unmethylated maternal Igf2/H19 ICRs. Activation of Igf2 is likely the predominant correction that rescued growth and viability. Further experiments involved the introduction of a null allele of Cdkn1c to alleviate its over-expression. Results were not consistent with the possibility that this misexpression alone, or in combination with Igf2 inactivity, mediates MatDup.dist7 death. Rather, a network of misexpressions derived from dist7 is probably involved. Our results are consistent with the idea that reduced expression of IGF2 plays a role in the aetiology of the human imprinting-related growth-deficit disorder, Silver-Russell syndrome.
Published in the journal: . PLoS Genet 6(1): e32767. doi:10.1371/journal.pgen.1000803
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1000803Summary
The misexpressed imprinted genes causing developmental failure of mouse parthenogenones are poorly defined. To obtain further insight, we investigated misexpressions that could cause the pronounced growth deficiency and death of fetuses with maternal duplication of distal chromosome (Chr) 7 (MatDup.dist7). Their small size could involve inactivity of Igf2, encoding a growth factor, with some contribution by over-expression of Cdkn1c, encoding a negative growth regulator. Mice lacking Igf2 expression are usually viable, and MatDup.dist7 death has been attributed to the misexpression of Cdkn1c or other imprinted genes. To examine the role of misexpressions determined by two maternal copies of the Igf2/H19 imprinting control region (ICR)—a chromatin insulator, we introduced a mutant ICR (ICRΔ) into MatDup.dist7 fetuses. This activated Igf2, with correction of H19 expression and other imprinted transcripts expected. Substantial growth enhancement and full postnatal viability was obtained, demonstrating that the aberrant MatDup.dist7 phenotype is highly dependent on the presence of two unmethylated maternal Igf2/H19 ICRs. Activation of Igf2 is likely the predominant correction that rescued growth and viability. Further experiments involved the introduction of a null allele of Cdkn1c to alleviate its over-expression. Results were not consistent with the possibility that this misexpression alone, or in combination with Igf2 inactivity, mediates MatDup.dist7 death. Rather, a network of misexpressions derived from dist7 is probably involved. Our results are consistent with the idea that reduced expression of IGF2 plays a role in the aetiology of the human imprinting-related growth-deficit disorder, Silver-Russell syndrome.
Introduction
Parthenogenetic mouse embryos usually die before 6½ days post coitum (dpc). Occasionally they develop to the 25 somite forelimb bud stage or approximately 9½ dpc [1]–[5]. Parthenogenones possess two maternally-derived genomes and would be expected to possess abnormal levels of transcript of all known imprinted genes, that is, lack of expression of paternally expressed genes (two inactive copies), and over-expression of maternally expressed genes (two active copies). Their death is likely a composite effect of at least some of these misexpressions, although those involved are not well defined. Defining the causes is important for improving understanding of the aetiology of genomic imprinting [6]–[9] and the prevalence of sexual reproduction, which ‘has long been an evolutionary enigma’ [10].
Knowledge of the causes of parthenogenetic death has come from two sources. First, the union of unbalanced complementary gametes in intercrosses of mice carrying reciprocal or Robertsonian translocations yield, at low frequency, embryos with maternal duplication and paternal deficiency for particular Chr regions as defined by the translocation breakpoint [11]–[13]. Maternal duplication of twelve Chr regions results in developmental anomalies. Only three of these are associated with peri - or prenatal death, these being maternal duplication of proximal Chr 6 (MatDup.prox6)—prior to 11½ dpc [14], maternal duplication of distal Chr 7 (MatDup.dist7)—late fetal death [15], and maternal disomy of Chr 12—perinatal death, probably attributable to the distal region [16]. Second, knockouts of imprinted genes and imprinting control regions (ICRs) have provided information on the effects of disregulation of imprinted genes, for example, [17]–[21]. To better define the causes of failed parthenogenetic development, and learn more of how imprinted genes at dist7 work together to regulate normal development, we have examined some of the misexpressions of imprinted genes thought to contribute to the abnormal development of MatDup.dist7 conceptuses. These display a pronounced growth deficit of the fetus and placenta and die at the late fetal stage, or possibly at birth. Live MatDup.dist7 young have never been observed [13],[15] (J. Mann, unpublished data).
Dist7 is an important region in terms of genomic imprinting, containing over 20 imprinted genes [13],[22]. At least three of these are regulated by the Igf2/H19 imprinting control region (ICR), these being ‘insulin like growth factor 2’ (Igf2)—paternally expressed and encoding a mitogen important for embryonic growth [23],[24], ‘insulin II’ (Ins2)—paternally expressed in yolk sac [25], and the non-coding ‘H19 fetal liver mRNA’ (H19) gene—maternally expressed [26]. Other non-coding transcripts have been described, these being Mir483, contained within an intron of Igf2 [27] and for which imprinting status is unknown, Mir675, contained with an H19 exon and therefore likely to follow the imprinting pattern of the host gene [28],[29], and antisense transcripts within Igf2 [30]. The targets of the Mir483 and Mir675 miRNAs are unknown. The maternally-derived Igf2 allele is inactive due to the hypo-methylated maternal Igf2/H19 ICR functioning as a‘CCCTC-binding factor’ (CTCF)-based chromatin insulator. This lies between the Igf2 promoter and the shared Igf2-H19 enhancers, preventing their interaction. The maternal H19 promoter lies on the same side of the insulator as the enhancers, therefore interaction occurs. On the paternal Chr the ICR is hyper-methylated, preventing CTCF binding and insulator formation and allowing for paternal Igf2 promoter and enhancer interaction. The paternal H19 promoter, just distal to the methylated ICR, also becomes methylated, and is inactive. The Ins2 gene is located just distal to Igf2. The Ins2 parental alleles are affected in the same way as their Igf2 counterparts, but only in yolk sac. Ins2 is expressed biallelically in pancreas [25], [31]–[33].
Telomeric or distal to the Igf2/H19 ICR domain is a large cluster of imprinted genes under regulatory control of the Kv differentially methylated region (DMR)-1 (KvDMR1) ICR. The active state of maternally-derived genes within this cluster is coincident with maternal-specific ICR methylation and the inactive state of the promoter of the ‘KCNQ1 overlapping transcript 1’ (Kcnq1ot1) gene contained within the ICR. The paternal ICR is hypo-methylated, and paternal-specific elongation of the Kcnq1ot1 transcript is coincident with silencing in cis of genes within the cluster [17],[34],[35]. One of the genes regulated by this ICR is the ‘cyclin-dependent kinase inhibitor 1C (P57)’ (Cdkn1c) gene encoding a protein facilitating reduced cell proliferation, increased apoptosis and delayed cell differentiation [36],[37].
MatDup.dist7 fetuses are maternally duplicated for the hypo-methylated Igf2/H19 ICR and hyper-methylated KvDMR1 ICR regions, as well as for other imprinted transcripts at dist7. This epigenetic configuration is highly similar to that associated with the human imprinting-related growth deficit disorder, Silver-Russell syndrome (SRS) (OMIM 180860). More than half of cases are associated with hypo-methylation of the IGF2/H19 ICR, also known as ‘ICR1’. The disease is also associated with maternal duplication of the KvDMR1 ICR region, also known as ‘ICR2’, and maternal duplication of the 11p15.5 Chr region encompassing both ICRs. It is strongly suspected that SRS is caused by downregulation of IGF2, and, in a minority of cases, excess CDKN1C or other imprinted genes regulated by ICR2. However, empirical evidence is lacking [38]–[40].
The death of MatDup.dist7 fetuses has been difficult to decipher. Available evidence suggests that maternal duplication of the Igf2/H19 ICR regulatory domain alone is insufficient to explain the total phenotype observed. Mice with paternal inheritance of a tandem duplication of a chicken β-globin CTCF-based chromatin insulator, substituted for the endogenous Igf2/H19 ICR, are similar to MatDup.dist7 mice in having a fully functional hypo-methylated insulator on both parental Chrs. They lack Igf2 activity, have at least twofold over-expression of H19, with both parental alleles probably active, and would be expected to lack Ins2 activity in yolk sac. Nevertheless, their phenotype—dwarfism combined with postnatal viability—is essentially identical to Igf2 mutants [41]. Mice homozygous for this genetic modification, in a mix of strains 129S1/SvImJ and outbred Swiss CF-1, showed normal fecundity and were maintained as a random-bred line for several years (J. Mann, unpublished data). Further, lack of Igf2 activity is unlikely to be the sole cause of reduced growth in MatDup.dist7 fetuses. At 17½ dpc, their weight is approximately 40% of wild-type [42] (J. Mann and Walter Tsark, unpublished observations) compared to 50–60% of wild-type for Igf2 mutants and mice maternally inheriting the chicken insulator [41]. Overall, these observations indicate that the MatDup.dist7 phenotype of fetal growth deficit and death involves the misexpression of imprinted genes outside the influence of the Igf2/H19 ICR, and this has previously been suggested [42].
Available evidence also indicates that maternal duplication of the KvDMR1 ICR regulatory domain alone is insufficient to explain the total phenotype observed. Mice with paternal inheritance of a deletion of this element exhibit biallelic expression of adjacent imprinted genes. These mice, in a mix of mouse strains 129S4/SvJae and C57BL/6J, are postnatally viable. They show some reduction in size, and it has been indicated that this is caused by over-expression of Cdkn1c [35]. Reduced growth has also been observed in Cdkn1c-BAC transgenic mice. While these displayed high frequency perinatal mortality in strain 129/Sv, high postnatal viability was obtained in a mix of strains 129/Sv and outbred Swiss MF1 [43]. These observations indicate that MatDup.dist7 late fetal death, occurring in the context of mixed strains including outbred Swiss, involves the misexpression of imprinted genes outside the influence of the KvDMR1 ICR. Overall, these observations have led to suggestions that MatDup.dist7 death could be a composite effect of misexpressions derived from both imprinted domains, for example, Igf2 inactivity combined with Cdkn1c over-expression [43].
To define the role of imprinted genes regulated by the Igf2/H19 ICR in the MatDup.dist7 phenotype, we evaluated the effects of introducing a mutated Igf2/H19 ICR (ICRΔ) which cannot bind CTCF and form an insulator [44]. MatDup.dist7 fetuses carrying ICRΔ would be expected to be corrected in terms of the number of active alleles of Igf2—activation of one of two inactive alleles, H19—repression of one of two active alleles, and Ins2—activation of one of two inactive alleles in yolk sac. MatDup.dist7 fetuses carrying ICRΔ were significantly rescued in terms of growth and were able to survive to adulthood. These results demonstrate that the aberrant phenotype of MatDup.dist7 fetuses is highly dependent on the presence of two maternally-derived Igf2/H19 ICR chromatin insulators.
Results
Maternal Inheritance of ICRΔ Rescues Growth in Igf2 Null Mutants
Maternal inheritance of ICRΔ results in activation of Igf2 in cis such that total Igf2 RNA is 1.7 and 2.1 times the normal level in the liver and kidney of 17½ dpc fetuses, respectively, and also repression of H19 in cis, such that total H19 RNA is 0.2 and 0 times the normal level in these same tissues, respectively [44]. This configuration of expression—two active Igf2 and two inactive H19 alleles—is coincident with increased growth, an effect thought to be due to the former misexpression [18],[45],[46]. Lack of H19 RNA alone has no effect on Igf2 expression or imprinting and results in no discernible phenotype [47]. Maternal inheritance of ICRΔ would also be expected to result in activation of Ins2 in yolk sac.
To confirm that maternal inheritance of ICRΔ can mediate normal growth, we tested its function in mice paternally inheriting a null mutation of Igf2 (Igf2−). Mice of genotype (ICR+/+, Igf2+/−) are small due to lack of Igf2 activity, with the maternal allele inactive, and the paternal allele null [24]. Results are shown in Figure 1. Experimental young of genotype (ICRΔ/+, Igf2+/−), in which the maternally-derived Igf2 allele is activated in cis by ICRΔ, were not significantly different in weight to control (ICR+/+, Igf2+/+) mice at 6 weeks of age (females, P = 0.271; males, P = 0.035). Thus, a single maternal copy of ICRΔ induces sufficient Igf2 activity for achieving normal postnatal growth. We note that, in respect to growth with one versus two active Igf2 alleles, experimental (ICRΔ/+, Igf2+/−) animals with one active allele (maternal), were not significantly different in weight to (ICRΔ/+, Igf2+/+) animals with two active alleles (females, P = 0.378; males, P = 0.089). Further, (ICR+/+, Igf2+/+) females with one active allele (paternal), were not significantly different in weight to (ICRΔ/+, Igf2+/+) females with two active alleles (P = 0.04). However, in males, mice with one active allele (paternal) were lighter than mice with two active alleles, as expected (P = 0.002). Given the borderline probability values obtained, greater numbers of animals need to be analysed to accurately determine the relative growth rates of mice of the various genotypes.
Fig. 1. Weight gain in Igf2 mutants carrying ICRΔ. 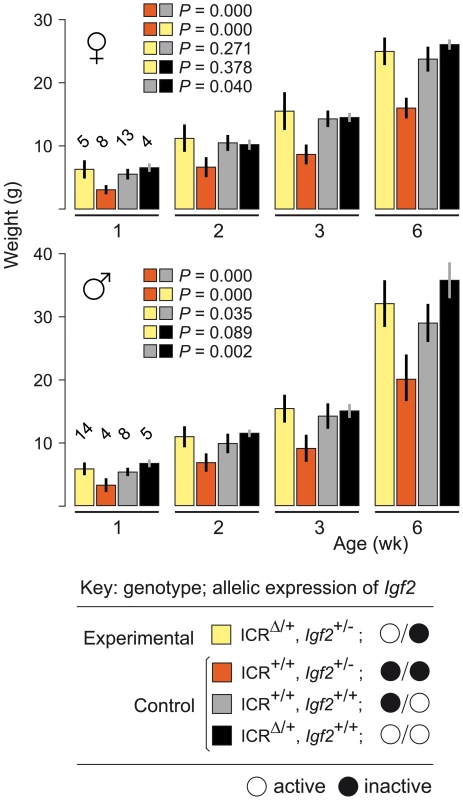
Young were obtained from (ICRΔ/+, Igf2+/+ ♀×ICR+/+, Igf2+/− ♂) matings. Bars are mean±s.d., with (n) given above the 1 wk bars. Key: alleles are maternal/paternal in derivation. The two-sample t-test was used to determine the probability that two 6 week samples (identified as paired squares) were equal. ICRΔ Rescues Growth and Viability in MatDup.dist7 Fetuses
MatDup.dist7 zygotes were produced in intercrosses of mice carrying the reciprocal translocation T(7;15)9H (T9H). Such intercrosses give rise to a high proportion of unbalanced zygotes, and litter size is small. Of balanced zygotes, only one in seven are expected to be MatDup.dist7, these identified by the dist7 marker, albino (c), a mutation of the ‘tyrosinase’ (Tyr) gene [15].
The ICRΔ mutation was introduced into female T9H/+ parents and was inherited by MatDup.dist7 zygotes (Figure 2). Expected allelic activity of Igf2 and Cdkn1c in the three possible MatDup.dist7 genotypes is shown (Figure 2B). ICRΔ-induced activation of Igf2 was confirmed in 13½ dpc MatDup.dist7 fetuses obtained in (T9H/+, Tyrc/c, ICRΔ/+ ♀×T9H/+, Tyr+/+, ICR+/+ ♂) intercrosses. The level of Igf2 transcript in MatDup.dist7 ICRΔ.+ fetuses was the same as in control ICR+/+ fetuses with one active allele, while it was almost double the normal amount in MatDup.dist7 ICRΔ.Δ fetuses with probably two active alleles (Figure 3A and 3B). Increased total Igf2 RNA was also seen in mice which maternally inherit ICRΔ and have an active maternal and paternal allele of Igf2 (Figure 3A and 3B). Also, MatDup.dist7 fetuses of all genotypes contained at least double the amount of Cdkn1c RNA relative to controls, probably because of two active alleles (Figure 3A and 3B). These intercross matings were allowed to proceed to term and we immediately began to observe viable albino or MatDup.dist7 young which were of overtly similar size to agouti littermates. A MatDup.dist7 animal and its two littermates at 10 days post-partum is shown (Figure 4). All MatDup.dist7 young obtained were of genotype ICRΔ.+ or recombinant ICRΔ.Δ. Seven of 52 mice born were MatDup.dist7 which is similar to the expected frequency, indicating that ICRΔ was always able to increase growth and rescue viability. In age - and litter-matched animals, a significant weight deficit of approximately 17% in MatDup.dist7 animals became apparent at 6 weeks of age when compared with controls carrying an equivalent number of active Igf2 alleles, that is, MatDup.dist7 ICRΔ.+ with control ICR+/+ (one active allele each) and MatDup.dist7 ICRΔ.Δ recombinant with control ICRΔ/+ (probably two active alleles each) (Figure 5A).
Fig. 2. Production of MatDup.dist7 fetuses. 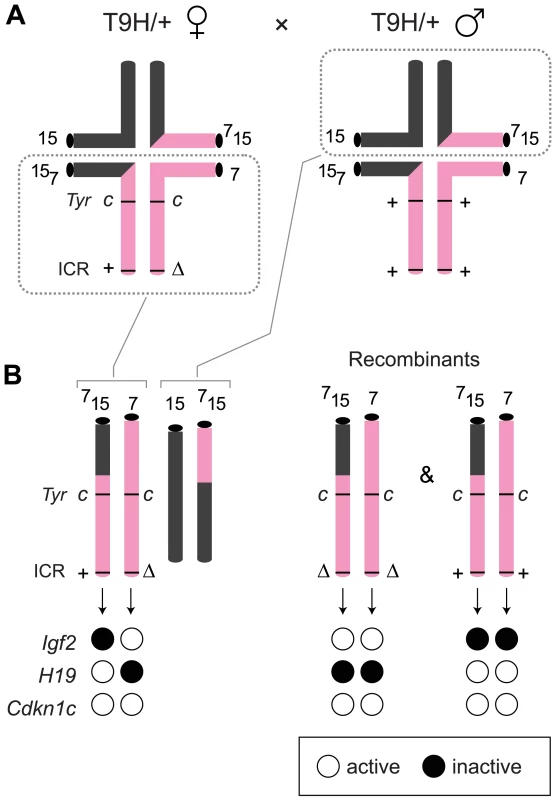
(A) Quadrivalents at meiosis I occurring in (T9H/+, Tyrc/c, ICRΔ/+ ♀×T9H/+, Tyr+/+, ICR+/+ ♂) matings. Female reciprocal translocation heterozygote (T9H/+) parent is homozygous for the dist7 marker albino (Tyrc/c) and carries ICRΔ (ICRΔ/+), while the male T9H/+ parent is wild-type at both of these loci (Tyr+/+, ICR+/+). (B) Genotypes of MatDup.dist7 fetuses obtained from the union of unbalanced complementary gametes. MatDup.dist7 individuals are readily identified as albinos from 12½ dpc by lack of eye or fur pigmentation. Many of these are ICRΔ.+ although a high frequency of homozygous recombinants, ICRΔ.Δ and ICR+.+, are also obtained. Chrs depicted are actually paired chromatids. Underneath is depicted allele-specific expression—MatDup.dist7 ICRΔ.+ and ICRΔ.Δ fetuses have one and probably two activated Igf2 alleles, and should have one and two repressed H19 alleles, respectively. Fig. 3. Expression of imprinted genes in 13½ dpc MatDup.dist7 fetuses. 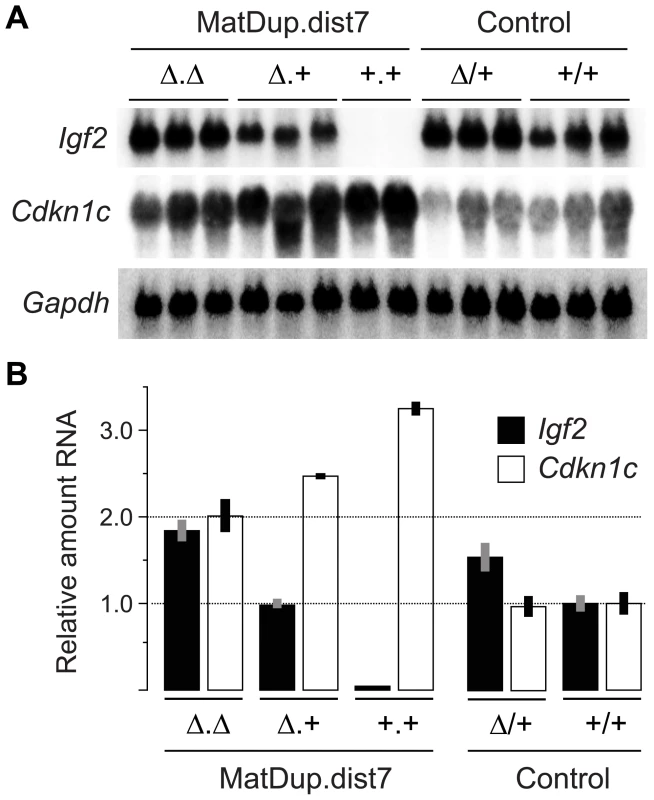
(A) Northern blots for the imprinted genes Igf2 and Cdkn1c, and for the housekeeping Gapdh gene for normalization. Each lane is an individual fetus. ICR genotype is given immediately above the lanes. (B) Northern blots in (A) were quantitated to show relative RNA levels. Values for Igf2 and Cdkn1c were normalized to Gapdh RNA, calibrated to control ICR+/+ values, and adjusted to a mean of 1.0. Values are mean±s.d. with (n) as shown in (A). Mating scheme to breed these animals was as described in the legend to Figure 2. Fig. 4. Neonatal MatDup.dist7 mouse rescued by ICRΔ. 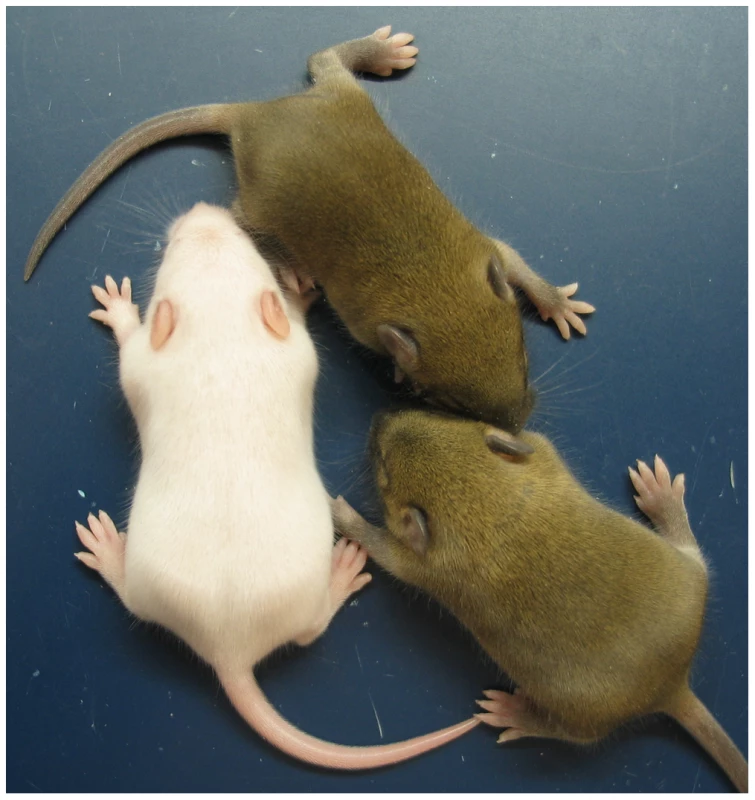
MatDup.dist7 (ICRΔ.+, Cdkn1c+.+) albino neonate with two agouti littermates at 10 days post-partum. Mating scheme to breed these animals was as described in the legend to Figure 2. Fig. 5. Weight gain in post-partum MatDup.dist7 mice rescued by ICRΔ. 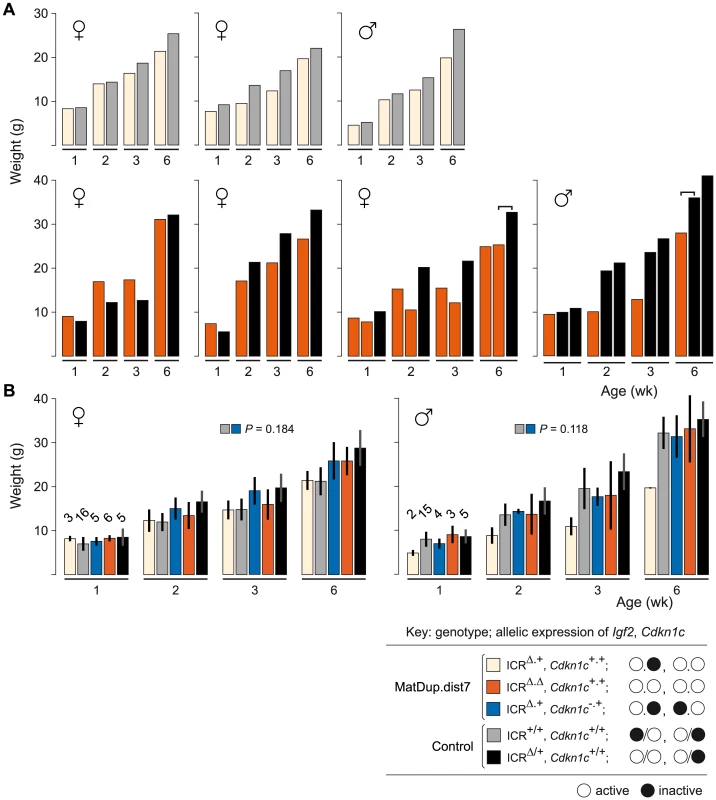
(A) Weight gain in litter matched MatDup.dist7 (ICRΔ.+, Cdkn1c+.+) or (ICRΔ.Δ, Cdkn1c+.+) young obtained from matings of (T9H/+, Tyrc/c, ICRΔ/+ ♀×T9H/+, Tyr+/+, ICR+/+ ♂) and (T9H/+, Tyrc/c, ICRΔ/+, Cdkn1c+/− ♀×T9H/+, Tyr+/+, ICR+/+, Cdkn1c+/+ ♂) mice. Each graph represents a single litter where at least one control with the same number of active Igf2 alleles was obtained. Each bar represents a single animal. The paired-sample t-test was used to determine the probability that the weight of MatDup.dist7 and control mice was equal. For three-animal litters, the pairs used in the statistical test are indicated by the bracket above the bars. The P value was <0.01 for 6 week old mice, indicating a significant difference, while there was no significant difference in weight at 1, 2, and 3 weeks of age. (B) All weight gain data collected from the two sets of matings as described in (A) regardless of littermate matching. Bars are mean±s.d., with (n) given above the 1 wk bars. The two-sample t-test was used to determine the probability that two 6 week samples (identified as paired squares) were equal. Key: alleles are maternal.maternal in derivation for MatDup.dist7 and maternal/paternal in derivation for controls. Involvement of Cdkn1c Expression in MatDup.dist7 Death
CDKN1C may antagonize the growth promoting effects of IGF2 [17],[48], and it has been suggested that excess CDKN1C may combine with lack of IGF2 to cause MatDup.dist7 death [43]. To test this possibility, we introduced a null allele of Cdkn1c (Cdkn1c−) into MatDup.dist7 fetuses to enforce its monoallelic expression. In (T9H/+, Tyrc/c, ICR+/+ Cdkn1c+/− ♀×T9H/+, Tyr+/+, ICR+/+ Cdkn1c+/+ ♂) matings, all of 55 young obtained were agouti controls, that is, at least six albino MatDup.dist7 (ICR+.+, Cdkn1c+.−) pups were expected, but none were observed. This result is not consistent with the idea that MatDup.dist7 death results only from the combined action of the Cdkn1c and Igf2 misexpressions.
Involvement of Cdkn1c Expression in MatDup.dist7 ICRΔ.+ Postnatal Growth Deficit
To test for a role of Cdkn1c over-expresssion in the growth deficit at 6 weeks of age of rescued postnatal MatDup.dist7 ICRΔ.+ animals, we introduced Cdkn1c− into MatDup.dist7 fetuses such that they were of genotype (ICRΔ.+, Cdkn1c+.−). This genotype should be normalized for the number of active alleles of imprinted genes regulated by the Igf2/H19 ICR, and also be normalized for Cdkn1c expression, that is, all of these imprinted genes should be monoallelically expressed. In (T9H/+, Tyrc/c, ICRΔ/+, Cdkn1c+/− ♀×T9H/+, Tyr+/+, ICR+/+, Cdkn1c+/+ ♂) matings, viable MatDup.dist7 ICRΔ.+, Cdkn1c+.− young were obtained and these did not display a significant weight deficit at 6 weeks of age—with the caveat that the weight measurements are relative to control young obtained in the previous matings (Figure 5B). Their weights could not be compared to littermates as, given the mating scheme, agouti littermates were always positive for ICRΔ—inheritance of Cdkn1c− being lethal—and therefore possessed two active copies of Igf2. In any event, these results are consistent with the possibility that biallelic expression of Cdkn1c does contribute to a reduction in postnatal growth in MatDup.dist7 ICRΔ.+ or ICRΔ.Δ, Cdkn1c+.+ animals.
Discussion
We have shown that maternal introduction of a mutant Igf2/H19 ICR, which lacks chromatin insulator activity, into MatDup.dist7 fetuses substantially alters their abnormal phenotype—small size and death at the late fetal stage—to one of near normal growth rate and survival to adulthood. This result clearly demonstrates the dependence of this phenotype on a misexpression of imprinted genes caused by the presence of two active maternally-derived Igf2/H19 ICR chromatin insulators. As this ICR is known to regulate the expression of at least three dist7 imprinted genes—H19, Ins2, Igf2, and a number of non-coding transcripts—correction in the misexpression of one or more of these was probably responsible for the result obtained. Activation of Igf2 was likely an important correction, this being the only alteration in expression induced by ICRΔ expected to affect growth.
The survival of MatDup.dist7 mice with ICRΔ is more difficult to decipher. As discussed in the Introduction section, it is unlikely that the Igf2/H19 ICR-derived misexpressions are solely responsible for their death, as mice with two functional chromatin insulators—a maternally-derived Igf2/H19 ICR, and a paternally-derived chicken insulator substituted for the Igf2/H19 ICR, possess the same combination of misexpressions as MatDup.dist7 mice in respect to this region, yet these animals have normal postnatal viability [41]. Further evidence is provided by observations of the effects of misexpression of each imprinted gene alone. First, for H19, no overt effect on phenotype is observed in transgenic mice with ectopic over-expression [49]–[52]. Biallelic or over-expression of H19 has been suggested to cause perinatal death of mice produced by combining a non-growing oocyte genome (ng), carrying a deletion of the distal Chr 12 IG-DMR ICR (Δ12), with a fully grown oocyte genome (fg)—ngΔ12/fg mice [53]. However, these mice would be predicted to have the equivalent expression profile of imprinted genes as mice with maternal inheritance of the chicken insulator substitution. The latter mice are viable, despite twofold over-expression of H19 [41]. Therefore, the perinatal death of ngΔ12/fg mice may result from the combined action of H19 RNA excess—or possibly Igf2 RNA absence—and small imperfections in expression derived from the non-growing oocyte genome, for example, as related to the IG-DMR ICR deletion. Second, for Ins2, mice lacking in expression of this gene are viable [54]. Third, for Igf2, mice lacking expression are dwarfed and have impaired lung development [55], but are usually viable. High postnatal survival frequency of Igf2 mutants is seen in inbred strain 129/SvEv [23],[24] although in this strain we have observed a low level of perinatal death (J. Mann, unpublished observations). In the present study, in a mix of strains 129/SvEv and outbred Swiss CF-1, we observed high frequency survival. Also, in this same strain mix, we maintained a Igf2−/− random-bred line for a number of years which had normal fecundity (J. Mann, unpublished data). On the other hand, use of a second Igf2 null mutation [56] revealed that lack of IGF2 in strain C57BL/6J results in death at birth. This effect was not peculiar to this second knockout allele as homozygous mutants can be obtained in strain 129 (M. Constancia, personal communication). In the present study, MatDup.dist7 young were a mix of strains 129S1/SvImJ, CF-1, C57BL/6J and CBA/Ca. In this mix, lack of Igf2 activity is highly likely to be compatible with survival. Given these various lines of evidence, the present experiments strongly suggest that misexpressed imprinted genes, as regulated by the Igf2/H19 ICR, work in combination with misexpressions derived outside of this region of influence in causing the total MatDup.dist7 phenotype.
The significant rescue in growth probably mediated by Igf2 activation may also be directly related to MatDup.dist7 survival in that it could compensate for negative effects derived from outside the Igf2/H19 ICR region. Nevertheless, we cannot rule out the possibility that Ins2 inactivity in yolk sac, excess H19 RNA, or the misexpression of non-coding RNAs regulated by the Igf2/H19 ICR make a contribution to the lethal effect. These possibilities could be investigated through correction of their misexpression in MatDup.dist7 fetuses, then determining growth and survival. For example, correction of H19 over-expression could be achieved by introducing a deletion of the transcript region only.
The imprinted genes operating outside the influence of the Igf2/H19 ICR that contribute to MatDup.dist7 death would be expected to require maternal-, rather than paternal-specific imprinting or methylation for attaining differential expression in the normal context. This is because for full-term development, there is apparently no other requirement, aside from Igf2/H19 ICR methylation, for paternal imprinting at dist7 [57]. The cluster of genes requiring maternal-specific methylation of the KvDMR1 ICR for activity fulfills this criterion. While the introduction of a null mutation of Cdkn1c, and hence enforced monoallelic expression of this gene, did not rescue MatDup.dist7 fetuses, this does not rule out the possibility that CDKN1C excess has a role in causing MatDup.dist7 death. In MatDup.dist7 (ICR+.+, Cdkn1c+.+) fetuses, Cdkn1c RNA levels were found to be more than three times that of controls, suggesting that each maternally-derived Cdkn1c allele was upregulated 1.5-fold. Therefore, CDKN1C could still be in excess in MatDup.dist7 (ICR+.+, Cdkn1c+.−) animals. Also, there remains the possibility that excess Cdkn1c RNA may contribute as part of a network of misexpressions derived from the cluster regulated by the KvDMR1 ICR. For example, biallelic expression of the ‘pleckstrin homology-like domain, familiy A, member 2’ (Phlda2) gene results in placental growth retardation and marginal fetal growth restriction [58], and upregulation of PHLDA2 is correlated with growth retardation in humans [59],[60]. Also, it has been suggested that excess expression of the ‘achaete-scute complex homolog 2 (Drosophila) (Ascl2) gene could cause the MatDup.dist7 lethal effect [42]. The phenotype of MatDup.dist7 fetuses could also involve misexpressions of dist7 imprinted genes lying outside of the influence of the two known ICRs. For example, ‘adenosine monophosphate deaminase 3’ (Ampd3)—maternally expressed in placenta, and identified in a transcriptome analysis of MatDup.dist7 conceptuses [22], ‘inositol polyphosphate-5-phosphatase F’ (Innp5f)—an isoform paternally expressed in brain [61], and ‘cathepsin D’ (Ctsd)—possible paternal-specific expression [62].
The postnatal weight deficit of approximately 17% in MatDup.dist7 young at 6 weeks of age was similar to that in mice paternally inheriting a deleted KvDMR1 ICR. This deletion results in biallelic expression of imprinted genes regulated by this ICR, including Cdkn1c [17],[34]. Indeed, excess CDKN1C has been indicated as the cause of the weight deficit [35]. Consistent with this possibility is that the weight of MatDup.dist7 ICRΔ.+, Cdkn1c+.− young was normal at 6 weeks of age. However, we note that MatDup.dist7 neonates displayed no significant weight deficit until reaching adulthood, while in mice paternally inheriting the deleted KvDMR1 ICR, the weight deficit is present in fetuses and persists throughout postnatal development [17]. More data regarding weight gain in relation to the inheritance of ICRΔ, in MatDup.dist7 young and otherwise, is required to confirm these observations.
In terms of MatDup.dist7 death, additional experiments are required to determine exactly which combination of misexpressions are involved. The total MatDup.dist7 phenotype has been ascribed to the very distal portion of Chr 7 as defined by the reciprocal translocation T(7;11)65H (T65H) [42]. This translocation has a breakpoint far more distal on Chr 7 relative to the T9H translocation used in this study, although is still proximal to the two clusters of imprinted genes regulated by the Igf2/H19 and KvDMR1 ICRs. However, some caution should be exercised in ascribing the total effect to this region. While it was shown that T65H - and T9H-MatDup.dist7 fetuses are of similar morphology [42], the postnatal viability of the former was not investigated. If T65H-MatDup.dist7 fetuses are also inviable, then the composite lethal effect is likely to be contained within the two aforementioned clusters of imprinted genes. Evidence that the KvDMR1 cluster contributes to the effect could be obtained by determining the viability of MatDup.dist7 fetuses carrying a deletion of this whole cluster. This would result in enforced monoallelic expression of all genes under regulation of the KvDMR1 ICR, including Cdkn1c, and these mice and would be expected to be postnatally viable, although small because of Igf2 inactivity. Such a deletion, made through truncation of Chr 7 at a point distal to the Ins2 gene, has been described [63]. A complication with this possible experiment is the existence of imprinted genes at dist7 which are not regulated by either ICR. Another experiment could be to breed mice with paternal inheritance of the chicken β-globin insulator substituted for the Igf2/H19 ICR [41] combined with paternal inheritance of the KvDMR1 ICR deletion [17]. These would misexpress all imprinted genes under regulatory control of both ICRs. If these were the only misexpressions involved in the MatDup.dist7 phenotype, then the phenotype should be reproduced.
MatDup.dist7 fetuses provide an epigenetic model of a subtype of human Silver-Russell syndrome (SRS) involving maternal duplication of the orthologous Chr region, 11p15.5, which encompasses ICR1 and ICR2. In these fetuses, we have shown that abrogation of ICR1 insulator function was able to restore Igf2 expression, concomitant with restoration of growth and survival. The most common subtype of SRS, that involving hypo-methylation of ICR1, is perhaps better modelled in mice maternally inheriting the chicken insulator in place of ICR1. These animals provide information on the effects of the presence of two functional insulators at the Igf2/H19 region as the only epigenetic lesion. In these fetuses, we previously showed that DNA methylation was abrogated while insulator function remained intact. This resulted in reduced Igf2 activity and growth retardation [41]. Both of these findings support the idea that reduced expression of IGF2 during fetal development is causal in the development of SRS. They also support the suggestion that the failure to detect low concentrations of serum IGF2 in SRS patients is related to downregulation of IGF2 by this stage [38]. Further genetic manipulation in these mouse models should provide additional implications for the human disease.
Our experiments suggest that misexpression of imprinted genes caused by two maternal copies of the Igf2/H19 ICR constitute one component of a composite barrier to parthenogenetic development that was not previously predicted. The lethal effect in MatDup.dist7 fetuses may be specific to later stages of development, and may not normally occur in parthenogenones given their peri-implantation death. Nevertheless, high-level paternal - and maternal-specific expression of Igf2 and H19, respectively, is present shortly after implantation, at least by 6½ dpc [64]. Therefore, it cannot be ruled out that these misexpressions, and others regulated by the Igf2/H19 ICR, play a role in what probably is a complex composite lethal effect involving a network of misexpressed imprinted genes. Indeed, the fact that parthenogenones fail earlier in development than embryos with maternal duplication of any single Chr region, indicates that misexpressions of imprinted genes from different regions are cumulative or synergistic in their deleterious effects. Further, at the molecular level, it has been shown that disregulation of the imprinted genes ‘pleiomorphic adenoma gene-like 1’ (Plagl1) and H19 can affect the expression of other imprinted genes in an imprinted gene expression network [65],[66].
Previous observations have shown that the normal activity of imprinted genes regulated by the Igf2/H19 ICR are one of a small number of developmentally critical expression profiles provided exclusively by imprinting through the male germ line, provided that most if not all other imprinted genes are not misexpressed [57]. The present results raise the possibility that full-term parthenogenetic development could be achieved by correcting the misexpressions of only a few imprinted genes in order to repair the total expression network. One necessary correction would be to activate the ‘paternally expressed 10’ (Peg10) gene. Lack of expression of this gene results in death by 10½ dpc, and this misexpression alone would be expected to present a barrier to parthenogenesis. It would be expected to contribute to, or could be solely responsible for, the embryonic death of MatDup.prox6 mice, which occurs prior to 11½ dpc [20].
Materials and Methods
Mouse Lines
Line no.; genotype; strain; source, how produced, or reference: Line-1; 129S1/SvImJ (129S1); Tyr+/+; The Jackson Laboratory, stock no. 002448. Line-2; outbred Swiss CF-1; Tyrc/c; Charles River Laboratories. Line-3; T9H/T9H, Tyr+/+; mix of C57BL/6J (B6) and CBA/Ca (CB); The Jackson Laboratory, stock no. 001752. Line-4; T9H/T9H, Tyrc/c; mix of B6, CB and CF-1; made by mating line-2 with -3, then intercrossing. Line-5; Tyrc/c, ICRΔ/Δ; mix of CF-1 and 129S1; made by mating ICRΔ/+ mice [44] with line-2, then intercrossing. Line-6; Tyrc/c, Cdkn1c+/−; mix of 129S7/SvEvBrd (129S7), B6 and CF-1; made by mating Cdkn1c+/− mice [37] with line-2, then intercrossing. Line-7; T9H/T9H, Tyrc/c, Cdkn1c+/−; mix of strains B6, CB, CF-1, and 129S7; made by mating line-4 with -6, then intercrossing. Line-8; Igf2+/−; 129/SvEv [23].
Matings
Production of experimental (ICRΔ/+, Igf2+/−) mice (Figure 1): Female parents (ICRΔ/+, Igf2+/+) were bred in (line-5 ♀×line-1 ♂) matings. Male parents (ICR+/+, Igf2+/−) were of line-8. Young were a mix of strains 129 and CF-1. Production of MatDup.dist7 ICRΔ.+ and ICRΔ.Δ mice (Figure 2): Female parents (T9H/+, Tyrc/c, ICRΔ/+) were bred in (line-5 ♀×line-4 ♂) matings. Male parents (T9H/+, Tyr+/+, ICR+/+) were bred in (line-3 ♀×line-1 ♂) matings. Young were a mix of strains 129S1, CF-1, B6, and CB. Production of MatDup.dist7 Cdkn1c−.+ young, attempted: Female parents (T9H/+, Tyrc/c, Cdkn1c+/−) were bred in (line-4 ♀×line-6 ♂) matings. Male parents (T9H/+, Tyr+/+, ICR+/+) were bred in (line-3 ♀×line-1 ♂) matings. Young were a mix of strains 129, B6, CB, and CF-1. Production of MatDup.dist7 (ICRΔ.+, Cdkn1c+.−) young (Figure 5B): Female parents (T9H/+, Tyrc/c, ICRΔ/+, Cdkn1c+/−) were bred in (line-5 ♀×line-7 ♂) matings. Male parents (T9H/+, Tyr+/+, ICR+/+) were bred in (line-3 ♀×line-1 ♂) matings. Young were a mix of strains 129S1, 129S7, B6, CB, and CF-1.
Genotyping
For the ICR, two pairs of primers were used. The first pair was specific for the mutant ICR, identical to a pair previously described [41]: 5′ - GCCC ACCA GCTG CTAG CCATC -3′ and 5′ - CCTA GAGA ATTC GAGG GACC TAAT AAC -3′, 240 bp amplicon identified ICRΔ.+ and ICRΔ.Δ animals. The second pair was specific for ICR+, with primers binding to sequence positions that were modified in ICRΔ [44]: 5′ - AACA AGGG AACG GATG CTAC CG -3′ and 5′ - GCAA TATG TAGT ATTG TACT GCCA CCAC -3′, lack of a 506 bp amplicon identified ICRΔ.Δ animals. For Cdkn1c, the null allele was identified using primers specific for the selection cassette using in gene targeting: 5′ - CTCA GAGG CTGG GAAG GGGT GGGT C -3′, within the mouse ‘phosphoglycerate kinase 1’ (Pgk1) promoter, and 5′ - ATAC TTTC TCGG CAGG AGCA AGGT G -3′, within the neo coding sequence, 520 bp amplicon.
Northern Blots
Fetuses were genotyped, and total RNA recovered using RNAzol (Tel-Test) after homogenization of the total fetus minus the head. Probes for Igf2 and ‘glyceraldehyde-3-phosphate dehydrogenase’ (Gapdh) RNA were as previously described [67]. The Cdkn1c probe was made by RT-PCR using primers; 5′ - GCCG GGTG ATGA GCTG GGAA -3′ and 5′ - AGAG AGGC TGGT CCTT CAGC -3′, 221 bp amplicon. Northern blots were performed with 32P radiolabelled probes as described previously [68]. The three probes were hybridized independently to the same blots after stripping. Radioactivity of bands was quantitated using a Typhoon PhosphorImager (Molecular Dynamics). For each lane, values for Igf2 and Cdkn1c were normalized to the Gapdh value.
Zdroje
1. KaufmanMH
BartonSC
SuraniMA
1977 Normal postimplantation development of mouse parthenogenetic embryos to the forelimb bud stage. Nature 265 53 55
2. BartonSC
SuraniMA
NorrisML
1984 Role of paternal and maternal genomes in mouse development. Nature 311 374 376
3. MannJR
Lovell-BadgeRH
1984 Inviability of parthenogenones is determined by pronuclei, not egg cytoplasm. Nature 310 66 67
4. McGrathJ
SolterD
1984 Completion of mouse embryogenesis requires both the maternal and paternal genomes. Cell 37 179 183
5. MannJR
Lovell-BadgeRH
1988 Two maternally derived X chromosomes contribute to parthenogenetic inviability. Development 104 129 136
6. HurstLD
McVeanGT
1998 Do we understand the evolution of genomic imprinting? Curr Opin Genet Dev 8 701 708
7. WilkinsJF
HaigD
2003 What good is genomic imprinting: the function of parent-specific gene expression. Nat Rev Genet 4 359 368
8. CoanPM
BurtonGJ
Ferguson-SmithAC
2005 Imprinted genes in the placenta: a review. Placenta 26 Suppl A S10 20
9. WoodAJ
OakeyRJ
2006 Genomic imprinting in mammals: emerging themes and established theories. PLoS Genet 2 e147
10. AviseJC
2008 Clonality: The genetics, ecology, and evolution of sexual abstinence in vertebrate animals. New York Oxford University Press
11. CattanachBM
BeecheyCV
1997 Genomic imprinting in the mouse: possible final analysis.
ReikW
SuraniA
Genomic Imprinting Oxford IRL Press 118 145
12. CattanachBM
BeecheyCV
PetersJ
2004 Interactions between imprinting effects in the mouse. Genetics 168 397 413
13. WilliamsonCM
BlakeA
ThomasS
BeecheyCV
HancockJ
MRC Harwell, Oxfordshire. World Wide Web Site - Mouse Imprinting Data and References - http://har.mrc.ac.uk/research/genomic_imprinting/
14. BeecheyCV
2000 Peg1/Mest1 locates distal to the currently defined imprinting region on mouse proximal chromosome 6 and identifies a new imprinting region affecting growth. Cytogenet Cell Genet 90 309 314
15. SearleAG
BeecheyCV
1990 Genome imprinting phenomena on mouse chromosome 7. Genet Res 56 237 244
16. TevendaleM
WatkinsM
RasberryC
CattanachB
Ferguson-SmithAC
2006 Analysis of mouse conceptuses with uniparental duplication/deficiency for distal chromosome 12: comparison with chromosome 12 uniparental disomy and implications for genomic imprinting. Cytogenet Genome Res 113 215 222
17. FitzpatrickGV
SolowayPD
HigginsMJ
2002 Regional loss of imprinting and growth deficiency in mice with a targeted deletion of KvDMR1. Nat Genet 32 426 431
18. LeightonPA
IngramRS
EggenschwilerJ
EfstratiadisA
TilghmanSM
1995 Disruption of imprinting caused by deletion of the H19 gene region in mice. Nature 375 34 39
19. LinSP
YoungsonN
TakadaS
SeitzH
ReikW
2003 Asymmetric regulation of imprinting on the maternal and paternal chromosomes at the Dlk1-Gtl2 imprinted cluster on mouse chromosome 12. Nat Genet 35 97 102
20. OnoR
NakamuraK
InoueK
NaruseM
UsamiT
2006 Deletion of Peg10, an imprinted gene acquired from a retrotransposon, causes early embryonic lethality. Nat Genet 38 101 106
21. PetersJ
WilliamsonCM
2008 Control of imprinting at the Gnas cluster. Adv Exp Med Biol 626 16 26
22. SchulzR
MenheniottTR
WoodfineK
WoodAJ
ChoiJD
2006 Chromosome-wide identification of novel imprinted genes using microarrays and uniparental disomies. Nucleic Acids Res 34 e88
23. DeChiaraTM
EfstratiadisA
RobertsonEJ
1990 A growth-deficiency phenotype in heterozygous mice carrying an insulin-like growth factor II gene disrupted by targeting. Nature 345 78 80
24. DeChiaraTM
RobertsonEJ
EfstratiadisA
1991 Parental imprinting of the mouse insulin-like growth factor 2 gene. Cell 64 849 859
25. GiddingsSJ
KingCD
HarmanKW
FloodJF
CarnaghiLR
1994 Allele specific inactivation of insulin 1 and 2, in the mouse yolk sac, indicates imprinting. Nat Genet 6 310 313
26. BartolomeiMS
ZemelS
TilghmanSM
1991 Parental imprinting of the mouse H19 gene. Nature 351 153 155
27. LandgrafP
RusuM
SheridanR
SewerA
IovinoN
2007 A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129 1401 1414
28. MinenoJ
OkamotoS
AndoT
SatoM
ChonoH
2006 The expression profile of microRNAs in mouse embryos. Nucleic Acids Res 34 1765 1771
29. CaiX
CullenBR
2007 The imprinted H19 noncoding RNA is a primary microRNA precursor. RNA 13 313 316
30. MooreT
ConstanciaM
ZubairM
BailleulB
FeilR
1997 Multiple imprinted sense and antisense transcripts, differential methylation and tandem repeats in a putative imprinting control region upstream of mouse Igf2. Proc Natl Acad Sci U S A 94 12509 12514
31. BellAC
FelsenfeldG
2000 Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405 482 485
32. HarkAT
SchoenherrCJ
KatzDJ
IngramRS
LevorseJM
2000 CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature 405 486 489
33. SzabóP
TangSH
RentsendorjA
PfeiferGP
MannJR
2000 Maternal-specific footprints at putative CTCF sites in the H19 imprinting control region give evidence for insulator function. Curr Biol 10 607 610
34. Mancini-DinardoD
SteeleSJ
LevorseJM
IngramRS
TilghmanSM
2006 Elongation of the Kcnq1ot1 transcript is required for genomic imprinting of neighboring genes. Genes Dev 20 1268 1282
35. ShinJY
FitzpatrickGV
HigginsMJ
2008 Two distinct mechanisms of silencing by the KvDMR1 imprinting control region. EMBO J 27 168 178
36. YanY
FrisenJ
LeeMH
MassagueJ
BarbacidM
1997 Ablation of the CDK inhibitor P57KIP2 results in increased apoptosis and delayed differentiation during mouse development. Genes Dev 11 973 983
37. ZhangP
LiegeoisNJ
WongC
FinegoldM
HouH
1997 Altered cell differentiation and proliferation in mice lacking p57Kip2 indicates a role in Beckwith-Wiedemann syndrome. Nature 387 151 158
38. BinderG
SeidelAK
WeberK
HaaseM
WollmannHA
2006 IGF-II serum levels are normal in children with Silver-Russell syndrome who frequently carry epimutations at the IGF2 locus. J Clin Endocrinol Metab 91 4709 4712
39. Abu-AmeroS
MonkD
FrostJ
PreeceM
StanierP
2008 The genetic aetiology of Silver-Russell syndrome. J Med Genet 45 193 199
40. EggermannT
2009 Epigenetic regulation of growth: lessons from Silver-Russell syndrome. Endocr Dev 14 10 19
41. SzabóPE
TangSH
ReedMR
SilvaFJ
TsarkWM
2002 The chicken β-globin insulator element conveys chromatin boundary activity but not imprinting at the mouse Igf2/H19 domain. Development 129 897 904
42. BeecheyCV
BallST
TownsendKM
JonesJ
1997 The mouse chromosome 7 distal imprinting domain maps to G-bands F4/F5. Mamm Genome 8 236 240
43. AndrewsSC
WoodMD
TunsterSJ
BartonSC
SuraniMA
2007 Cdkn1c (p57Kip2) is the major regulator of embryonic growth within its imprinted domain on mouse distal chromosome 7. BMC Dev Biol 7 53
44. SzabóPE
TangSH
SilvaFJ
TsarkWM
MannJR
2004 Role of CTCF binding sites in the Igf2/H19 imprinting control region. Mol Cell Biol 24 4791 4800
45. RipocheMA
KressC
PoirierF
DandoloL
1997 Deletion of the H19 transcription unit reveals the existence of a putative imprinting control element. Genes Dev 11 1596 1604
46. EggenschwilerJ
LudwigT
FisherP
LeightonPA
TilghmanSM
1997 Mouse mutant embryos overexpressing IGF-II exhibit phenotypic features of the Beckwith-Wiedemann and Simpson-Golabi-Behmel syndromes. Genes Dev 11 3128 3142
47. JonesBK
LevorseJM
TilghmanSM
1998 Igf2 imprinting does not require its own DNA methylation or H19 RNA. Genes Dev 12 2200 2207
48. CasparyT
ClearyMA
PerlmanEJ
ZhangP
ElledgeSJ
1999 Oppositely imprinted genes p57Kip2 and Igf2 interact in a mouse model for Beckwith-Wiedemann syndrome. Genes Dev 13 3115 3124
49. PfeiferK
LeightonPA
TilghmanSM
1996 The structural H19 gene is required for transgene imprinting. Proc Natl Acad Sci U S A 93 13876 13883
50. AinscoughJF
KoideT
TadaM
BartonS
SuraniMA
1997 Imprinting of Igf2 and H19 from a 130 kb YAC transgene. Development 124 3621 3632
51. ElsonDA
BartolomeiMS
1997 A 5′ differentially methylated sequence and the 3′-flanking region are necessary for H19 transgene imprinting. Mol Cell Biol 17 309 317
52. CarrMS
GetekKA
LevorseJM
SchmidtJV
2006 Expression of a modified H19 RNA does not cause embryonic lethality in mice. Mamm Genome 17 5 13
53. KawaharaM
WuQ
Ferguson-SmithAC
KonoT
2007 Appropriate expression of imprinted genes on mouse chromosome 12 extends development of bi-maternal embryos to term. FEBS Lett 581 5178 5184
54. DuvillieB
CordonnierN
DeltourL
Dandoy-DronF
ItierJM
1997 Phenotypic alterations in insulin-deficient mutant mice. Proc Natl Acad Sci U S A 94 5137 5140
55. SilvaD
VenihakiM
GuoWH
LopezMF
2006 IGF2 deficiency results in delayed lung development at the end of gestation. Endocrinology 147 5584 5591
56. MurrellA
HeesonS
BowdenL
ConstanciaM
DeanW
2001 An intragenic methylated region in the imprinted Igf2 gene augments transcription. EMBO Rep 2 1101 1106
57. KawaharaM
WuQ
TakahashiN
MoritaS
YamadaK
2007 High-frequency generation of viable mice from engineered bi-maternal embryos. Nat Biotechnol 25 1045 1050
58. SalasM
JohnR
SaxenaA
BartonS
FrankD
2004 Placental growth retardation due to loss of imprinting of Phlda2. Mech Dev 121 1199 1210
59. McMinnJ
WeiM
SchupfN
CusmaiJ
JohnsonEB
2006 Unbalanced placental expression of imprinted genes in human intrauterine growth restriction. Placenta 27 540 549
60. ApostolidouS
Abu-AmeroS
O'DonoghueK
FrostJ
OlafsdottirO
2007 Elevated placental expression of the imprinted PHLDA2 gene is associated with low birth weight. J Mol Med 85 379 387
61. ChoiJD
UnderkofflerLA
WoodAJ
CollinsJN
WilliamsPT
2005 A novel variant of Inpp5f is imprinted in brain, and its expression is correlated with differential methylation of an internal CpG island. Mol Cell Biol 25 5514 5522
62. LuediPP
HarteminkAJ
JirtleRL
2005 Genome-wide prediction of imprinted murine genes. Genome Res 15 875 884
63. OhR
HoR
MarL
GertsensteinM
PaderovaJ
2008 Epigenetic and phenotypic consequences of a truncation disrupting the imprinted domain on distal mouse chromosome 7. Mol Cell Biol 28 1092 1103
64. SzabóPE
MannJR
1995 Allele-specific expression and total expression levels of imprinted genes during early mouse development: implications for imprinting mechanisms. Genes Dev 9 3097 3108
65. VarraultA
GueydanC
DelalbreA
BellmannA
HoussamiS
2006 Zac1 regulates an imprinted gene network critically involved in the control of embryonic growth. Dev Cell 11 711 722
66. GaboryA
RipocheMA
Le DigarcherA
WatrinF
ZiyyatA
2009 H19 acts as a trans regulator of the imprinted gene network controlling growth in mice. Development 136 3413 3421
67. McLaughlinKJ
SzabóP
HaegelH
MannJR
1996 Mouse embryos with paternal duplication of an imprinted chromosome 7 region die at midgestation and lack placental spongiotrophoblast. Development 122 265 270
68. SzabóP
MannJR
1994 Expression and methylation of imprinted genes during in vitro differentiation of mouse parthenogenetic and androgenetic embryonic stem cell lines. Development 120 1651 1660
Štítky
Genetika Reprodukční medicína
Článek Activation of Mutant Enzyme Function by Proteasome Inhibitors and Treatments that Induce Hsp70Článek Maternal Ethanol Consumption Alters the Epigenotype and the Phenotype of Offspring in a Mouse ModelČlánek Genetic Dissection of Differential Signaling Threshold Requirements for the Wnt/β-Catenin PathwayČlánek Distinct Type of Transmission Barrier Revealed by Study of Multiple Prion Determinants of Rnq1
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2010 Číslo 1- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
- Délka menstruačního cyklu jako marker ženské plodnosti
- Intrauterinní inseminace a její úspěšnost
-
Všechny články tohoto čísla
- Irradiation-Induced Genome Fragmentation Triggers Transposition of a Single Resident Insertion Sequence
- A Major Role of the RecFOR Pathway in DNA Double-Strand-Break Repair through ESDSA in
- Kidney Development in the Absence of and Requires
- Modeling of Environmental Effects in Genome-Wide Association Studies Identifies and as Novel Loci Influencing Serum Cholesterol Levels
- Inverse Correlation between Promoter Strength and Excision Activity in Class 1 Integrons
- Activation of Mutant Enzyme Function by Proteasome Inhibitors and Treatments that Induce Hsp70
- Postnatal Survival of Mice with Maternal Duplication of Distal Chromosome 7 Induced by a / Imprinting Control Region Lacking Insulator Function
- The Werner Syndrome Protein Functions Upstream of ATR and ATM in Response to DNA Replication Inhibition and Double-Strand DNA Breaks
- Maternal Ethanol Consumption Alters the Epigenotype and the Phenotype of Offspring in a Mouse Model
- Understanding Gene Sequence Variation in the Context of Transcription Regulation in Yeast
- miR-30 Regulates Mitochondrial Fission through Targeting p53 and the Dynamin-Related Protein-1 Pathway
- Elevated Levels of the Polo Kinase Cdc5 Override the Mec1/ATR Checkpoint in Budding Yeast by Acting at Different Steps of the Signaling Pathway
- Alternative Epigenetic Chromatin States of Polycomb Target Genes
- Co-Orientation of Replication and Transcription Preserves Genome Integrity
- A Comprehensive Map of Insulator Elements for the Genome
- Environmental and Genetic Determinants of Colony Morphology in Yeast
- U87MG Decoded: The Genomic Sequence of a Cytogenetically Aberrant Human Cancer Cell Line
- The MCM-Binding Protein ETG1 Aids Sister Chromatid Cohesion Required for Postreplicative Homologous Recombination Repair
- Genetic Dissection of Differential Signaling Threshold Requirements for the Wnt/β-Catenin Pathway
- Differential Localization and Independent Acquisition of the H3K9me2 and H3K9me3 Chromatin Modifications in the Adult Germ Line
- Genetic Crossovers Are Predicted Accurately by the Computed Human Recombination Map
- Collaborative Action of Brca1 and CtIP in Elimination of Covalent Modifications from Double-Strand Breaks to Facilitate Subsequent Break Repair
- Distinct Type of Transmission Barrier Revealed by Study of Multiple Prion Determinants of Rnq1
- Genome-Wide Association Study Identifies as a Novel Susceptibility Gene for Osteoporosis
- and Regulate Reproductive Habit in Rice
- Nonsense-Mediated Decay Enables Intron Gain in
- Altered Gene Expression and DNA Damage in Peripheral Blood Cells from Friedreich's Ataxia Patients: Cellular Model of Pathology
- The Systemic Imprint of Growth and Its Uses in Ecological (Meta)Genomics
- The Gift of Observation: An Interview with Mary Lyon
- Genotype and Gene Expression Associations with Immune Function in
- The Elongator Complex Regulates Neuronal α-tubulin Acetylation
- Rising from the Ashes: DNA Repair in
- Mis-Spliced Transcripts of Nicotinic Acetylcholine Receptor α6 Are Associated with Field Evolved Spinosad Resistance in (L.)
- BRIT1/MCPH1 Is Essential for Mitotic and Meiotic Recombination DNA Repair and Maintaining Genomic Stability in Mice
- Non-Coding Changes Cause Sex-Specific Wing Size Differences between Closely Related Species of
- Evidence for Pervasive Adaptive Protein Evolution in Wild Mice
- Evolutionary Mirages: Selection on Binding Site Composition Creates the Illusion of Conserved Grammars in Enhancers
- VEZF1 Elements Mediate Protection from DNA Methylation
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- A Major Role of the RecFOR Pathway in DNA Double-Strand-Break Repair through ESDSA in
- Kidney Development in the Absence of and Requires
- The Werner Syndrome Protein Functions Upstream of ATR and ATM in Response to DNA Replication Inhibition and Double-Strand DNA Breaks
- Alternative Epigenetic Chromatin States of Polycomb Target Genes
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



