-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
A Major Role of the RecFOR Pathway in DNA Double-Strand-Break Repair through ESDSA in
In Deinococcus radiodurans, the extreme resistance to DNA–shattering treatments such as ionizing radiation or desiccation is correlated with its ability to reconstruct a functional genome from hundreds of chromosomal fragments. The rapid reconstitution of an intact genome is thought to occur through an extended synthesis-dependent strand annealing process (ESDSA) followed by DNA recombination. Here, we investigated the role of key components of the RecF pathway in ESDSA in this organism naturally devoid of RecB and RecC proteins. We demonstrate that inactivation of RecJ exonuclease results in cell lethality, indicating that this protein plays a key role in genome maintenance. Cells devoid of RecF, RecO, or RecR proteins also display greatly impaired growth and an important lethal sectoring as bacteria devoid of RecA protein. Other aspects of the phenotype of recFOR knock-out mutants paralleled that of a ΔrecA mutant: ΔrecFOR mutants are extremely radiosensitive and show a slow assembly of radiation-induced chromosomal fragments, not accompanied by DNA synthesis, and reduced DNA degradation. Cells devoid of RecQ, the major helicase implicated in repair through the RecF pathway in E. coli, are resistant to γ-irradiation and have a wild-type DNA repair capacity as also shown for cells devoid of the RecD helicase; in contrast, ΔuvrD mutants show a markedly decreased radioresistance, an increased latent period in the kinetics of DNA double-strand-break repair, and a slow rate of fragment assembly correlated with a slow rate of DNA synthesis. Combining RecQ or RecD deficiency with UvrD deficiency did not significantly accentuate the phenotype of ΔuvrD mutants. In conclusion, RecFOR proteins are essential for DNA double-strand-break repair through ESDSA whereas RecJ protein is essential for cell viability and UvrD helicase might be involved in the processing of double stranded DNA ends and/or in the DNA synthesis step of ESDSA.
Published in the journal: . PLoS Genet 6(1): e32767. doi:10.1371/journal.pgen.1000774
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1000774Summary
In Deinococcus radiodurans, the extreme resistance to DNA–shattering treatments such as ionizing radiation or desiccation is correlated with its ability to reconstruct a functional genome from hundreds of chromosomal fragments. The rapid reconstitution of an intact genome is thought to occur through an extended synthesis-dependent strand annealing process (ESDSA) followed by DNA recombination. Here, we investigated the role of key components of the RecF pathway in ESDSA in this organism naturally devoid of RecB and RecC proteins. We demonstrate that inactivation of RecJ exonuclease results in cell lethality, indicating that this protein plays a key role in genome maintenance. Cells devoid of RecF, RecO, or RecR proteins also display greatly impaired growth and an important lethal sectoring as bacteria devoid of RecA protein. Other aspects of the phenotype of recFOR knock-out mutants paralleled that of a ΔrecA mutant: ΔrecFOR mutants are extremely radiosensitive and show a slow assembly of radiation-induced chromosomal fragments, not accompanied by DNA synthesis, and reduced DNA degradation. Cells devoid of RecQ, the major helicase implicated in repair through the RecF pathway in E. coli, are resistant to γ-irradiation and have a wild-type DNA repair capacity as also shown for cells devoid of the RecD helicase; in contrast, ΔuvrD mutants show a markedly decreased radioresistance, an increased latent period in the kinetics of DNA double-strand-break repair, and a slow rate of fragment assembly correlated with a slow rate of DNA synthesis. Combining RecQ or RecD deficiency with UvrD deficiency did not significantly accentuate the phenotype of ΔuvrD mutants. In conclusion, RecFOR proteins are essential for DNA double-strand-break repair through ESDSA whereas RecJ protein is essential for cell viability and UvrD helicase might be involved in the processing of double stranded DNA ends and/or in the DNA synthesis step of ESDSA.
Introduction
The bacterium Deinococcus radiodurans is extremely resistant to treatments such as ionizing radiation and desiccation. This resistance can be correlated with the ability of D. radiodurans to reconstruct a functional genome from hundreds of radiation or dessication-induced chromosomal fragments, while the genomes of most organisms are irreversibly shattered under the same conditions. The rapid reconstitution of an intact genome is dependent on extended synthesis-dependent strand annealing (ESDSA) and recombination [1],[2]. It was proposed that, following severe DNA damage, the fragmented DNA end is recessed in a 5′–3′ direction, liberating single stranded 3′ overhangs which, through RecA - and RadA-mediated strand invasion, prime DNA synthesis on overlapping fragments [2]. DNA synthesis is initiated by Pol III and elongated by Pol I or by Pol III and the newly synthesized single-strands anneal to complementary single stranded extensions forming long double stranded DNA intermediates which are assembled into intact circular chromosomes by RecA-mediated homologous recombination [2]. Though the dependence of ESDSA on RecA, Pol I, and Pol III activities is well documented [1],[2], little is known about the cellular factors required for the first steps of this process (i.e. the formation of the single stranded 3′ overhangs which promote RecA/RadA - dependent strand invasion to prime DNA synthesis).
Three enzymatic activities are required for presynaptic processing of double stranded DNA ends in the model bacterium Escherichia coli: a helicase, a 5′-3′exonuclease, and a mediator function for efficient RecA filament formation onto ssDNA (see for reviews [3]–[5]). All these activities are carried out by the RecBCD complex (or its functional homolog AddAB) which is the major component for initiation of recombinational repair of DNA double-strand-breaks (DSB) in wild-type cells. However, if RecBCD is inactivated, an alternate pathway, the RecF pathway, promotes recombinational DSB repair [6]–[10] in cells containing mutations in sbcB (suppressor of recBC), which encodes the 3′-5′ exonuclease I, and in sbcC (or sbcD) [11]. This pathway comprises the 5′-3′ single-strand DNA exonuclease RecJ, the RecQ helicase and the RecF, RecO and RecR proteins that act together to promote loading of RecA onto single stranded DNA.
Whereas examination of the phylogenetic distribution of RecBCD and AddAB complexes revealed that one or the other complex is present in most sequenced bacteria, D. radiodurans is naturally devoid of these two complexes but does encode a RecD homologue [12]. RecD protein was shown to be present in the absence of RecBC not only in D. radiodurans, but also in firmicutes and Streptomyces [13]. The deinococcal RecD protein is expressed and active as a DNA helicase [14]. Further work is required to assign RecD protein to a specific DNA repair pathway because conflicting data have been published concerning the in vivo role of RecD in radioresistance [15]–[16]. D. radiodurans possesses homologs of the key components of the RecF pathway: RecJ (DR1126), RecQ (DR1289), RecF (DR1089), RecO (DR0819), and RecR (DR0198) suggesting that the RecF pathway is the main recombinational repair pathway in this organism, as observed in other bacteria that lack RecBCD homologs [13]. D. radiodurans also lacks homologs of the SbcB nuclease, an inhibitor of the RecF pathway in E. coli. Moreover, it was shown that expression in trans of the SbcB protein from E. coli renders D. radiodurans cells radiation-sensitive [17].
In this paper, we investigate the role of the D. radiodurans proteins belonging to the RecF pathway in ESDSA and/or homologous recombination. We demonstrate that RecJ exonuclease is an essential protein for cell viability. We show that the RecF, RecO, RecR proteins as well as the RecA protein are absolutely required for massive DNA synthesis during DSB repair whereas RecQ appears to be substituted by the UvrD helicase to play a role in this process. We propose that RecJ, in conjunction with UvrD, could generate the single stranded tails on which RecFOR will stimulate RecA loading. Interestingly, an intact genome could be slowly reconstituted in the absence of RecA, RecF, RecO or RecR, suggesting alternate DSB repair through non-homologous end joining (NHEJ) and/or single-strand annealing (SSA).
Results
recJ is an essential gene in D. radiodurans
To determine the importance of the RecFOR pathway in DSB repair and radioresistance in D. radiodurans, we replaced the coding regions of key genes belonging to this pathway (recJ, recQ, recF, recO, and recR) with an antibiotic resistance cassette. The deletion-substitution alleles were constructed in vitro using the tripartite ligation method [18] and introduced by transformation into D. radiodurans to replace the corresponding wild-type alleles via homologous recombination. Because D. radiodurans contains from 4 to 10 genome equivalents [19],[20], the transformants were extensively purified on selective media in order to obtain the mutant homogenotes whose purity was verified by PCR. Whereas only few rounds of purification on selective antibiotic plates sufficed to obtain ΔrecQ, ΔrecF, ΔrecO and ΔrecR homogenotes (see Figure S1), in the case of recJ, the wild-type allele was present together with the ΔrecJ allele even after seven rounds of purification of three independent candidates (Figure 1), suggesting that RecJ protein is essential for cell viability.
Fig. 1. Schematic representation and test of deletion-substitution in the D. radiodurans recJ gene. 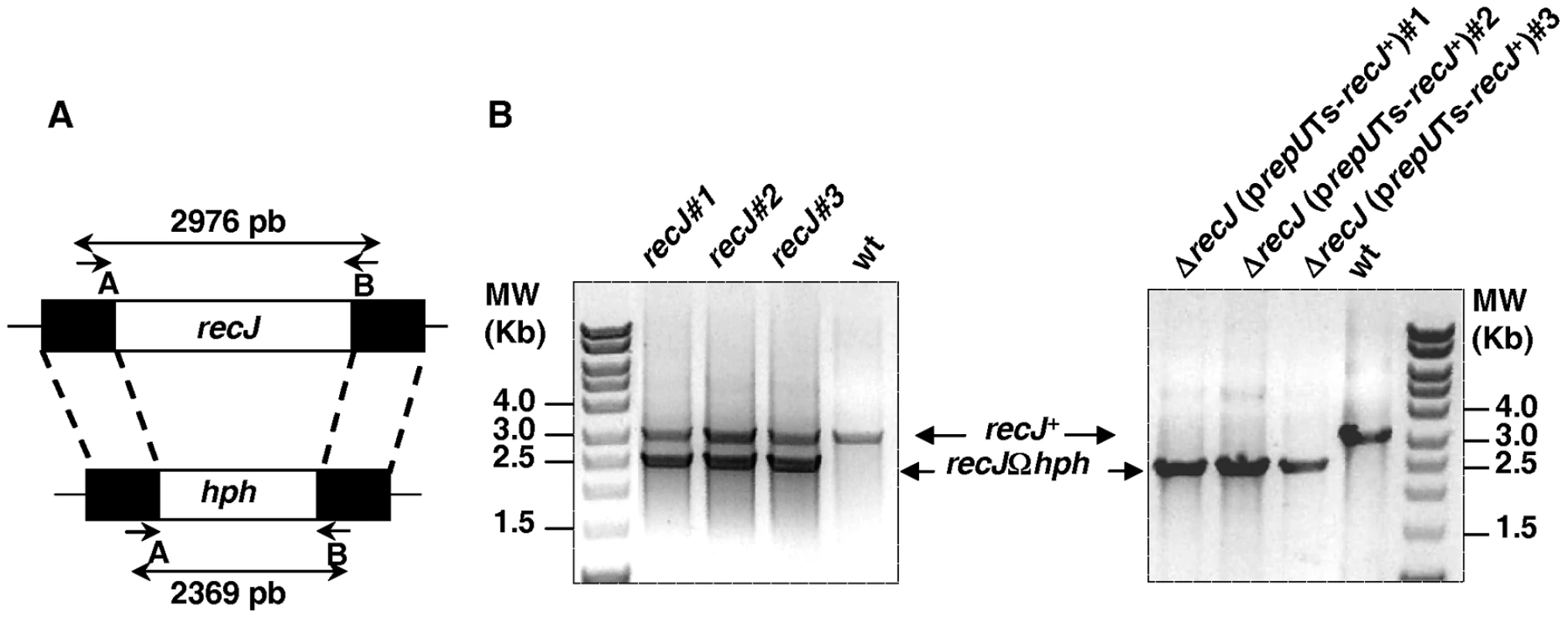
(A) Schematic representation of the allele replacement event in recJ gene. Short arrows indicate the position of specific primers used for diagnostic PCR. Primers are described in Table S1. (B) PCR analysis of three independent candidate recJ mutants and three independent ΔrecJ (prepUTS-recJ+) mutants. To obtain positive evidence for the essentiality of the recJ gene, we used the new diagnostic assay described by Nguyen et al [21]. For this purpose, the recJ gene was cloned onto a prepUTs vector thermosensitive for replication in D. radiodurans [21]. The sequence of DR1126 (recJ) in strain ATCC 13939 (GenBank, accession number QG856645) was found to differ from the DR1126 published sequence [22]. An additional G was found 7 nucleotides upstream the published putative GTG initiation codon and another additional G was found 58 nucleotides before the published putative TGA STOP codon giving rise to a RecJ protein containing 705 aa (versus 684 aa in the RecJ protein predicted from the previously published sequence) with 64 additional amino acids in the N-terminal domain and 43 aa missing in the C-terminal domain of the protein. The predicted sequence of the RecJ protein in strain ATCC 13939 displays a better alignment with the published protein sequences of the E. coli, Deinococcus geothermalis and Thermus thermophilus RecJ proteins (Figure S2). The recombinant plasmid was introduced into a recJ+ recipient and the chromosomal copy of recJ in the resulting merodiploid strain was inactivated (Figure 1). If recJ is an essential gene, the cells will die upon loss of the complementing plasmid at the non permissive temperature. As can be observed in Figure 2 (lanes 1–3), the ΔrecJ (prepUTs-recJ+) bacteria grew normally at 28° (the permissive temperature for the plasmid) but lose viability at 37° (the non-permissive temperature for the plasmid), demonstrating the essentiality of the recJ gene.
Fig. 2. recJ is essential for D. radiodurans viability. 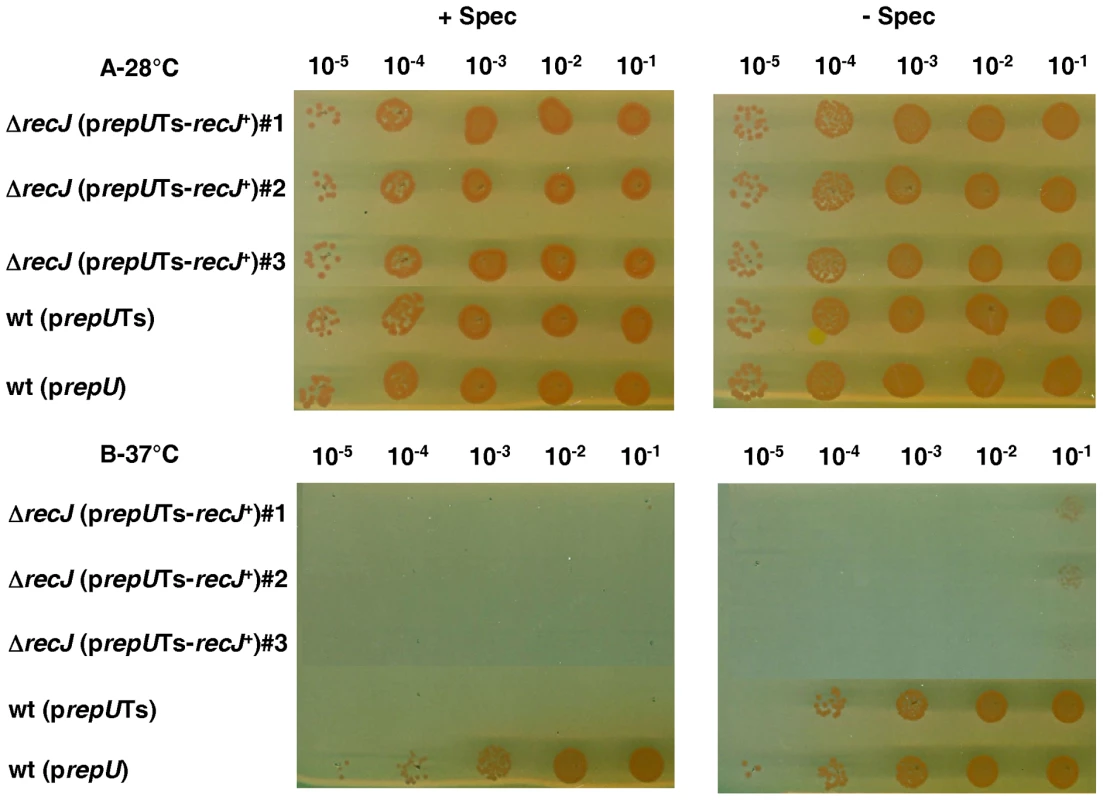
Strains were grown in liquid medium with spectinomycin at 28°C. The dilutions of cells were spotted on medium with or without spectinomycin at 28°C (A) or 37°C (B). Lane 1–3: strain GY14110 [ΔrecJ (prepUTs-recJ+)], lane 4: strain GY13781 containing thermosensitive plasmid p13840 (prepUTs), lane 5: strain GY13786 containing non-thermosensitive plasmid p11554 (prepU). Sub-lethal phenotype of ΔrecF, ΔrecO, and ΔrecR mutants
The ΔrecF, ΔrecO, and ΔrecR mutants, though viable, showed a greatly impaired growth. Indeed, the mutants had a generation time 4-fold longer than the wild-type (5 hours for the mutants versus 80 min for the wild-type) and comparable to that of a ΔrecA mutant. Furthermore, cells devoid of RecF, RecO or RecA proteins had a 10-fold reduced plating efficiency as compared to the wild-type strain and this defect was even more pronounced in the ΔrecR mutant, displaying a 30-fold reduced plating efficiency (Figure 3).
Fig. 3. D. radiodurans cells devoid of recA, recF, recO, or recR genes show reduced plating efficiency. 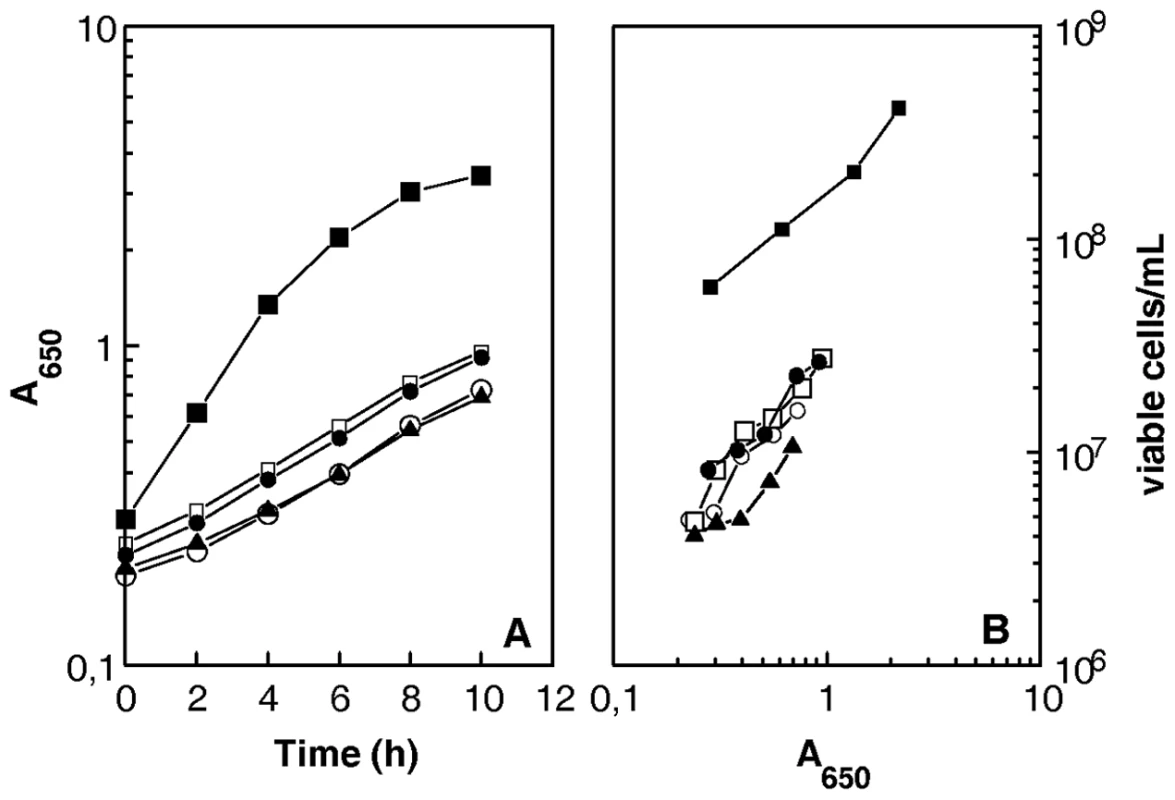
Bacterial strains GY12965 (ΔrecF, open circles), GY 12966 (ΔrecO, open squares), GY 12967 (ΔrecR, filled triangles), GY 12968 (ΔrecA, filled circles), and wild-type (closed squares) were incubated at 30°C. At different times, incubation samples were taken and the A650 values of the cultures and the numbers of viable cells/ml were measured. UvrD helicase is required for rapid repair of DNA double-strand breaks whereas RecQ and RecD helicases are dispensable
In E. coli, the RecQ helicase initiates DSB repair via the RecFOR pathway by unwinding duplex DNA in the 3′-5′ direction, while the single stranded DNA exonuclease RecJ hydrolyzes the 5′ strand to provide a DNA-substrate for RecA loading onto the 3′ strand [4],[23].
We found that inactivation of the RecQ helicase in D. radiodurans had no effect on radioresistance, because the knockout mutant was as resistant to γ-irradiation as the wild-type strain (Figure 4A). This result suggests that other helicase(s) might be involved in the initiation step of DSB repair in this organism. We tested the RecD and UvrD helicases for putative roles in DSB repair. We found that a ΔrecD deletion mutant was as radioresistant as the wild-type strain, whereas a ΔuvrD mutant showed a reduction in survival that ranged from 5-fold at 11.6 kGy to more than 100-fold at 17.8 kGy (Figure 4A). However, the mutant still retained significant radioresistance as compared to a repair-deficient ΔrecA strain (Figure 4B), suggesting that other helicase(s) may overlap in function with UvrD and thus lessen the effect of a ΔuvrD mutation. To test this hypothesis, we investigated whether the combined absence of UvrD and RecQ or UvrD and RecD proteins results in a more dramatic effect on radio-resistance. As seen in Figure 4A, the ΔuvrD ΔrecQ double mutant bacteria were not more sensitive to γ-rays than a ΔuvrD single mutant. In contrast the ΔuvrD ΔrecD double mutant bacteria were slightly more sensitive to γ-rays than a ΔuvrD single mutant, suggesting that the RecD helicase may have a partial back-up function in the absence of UvrD.
Fig. 4. Increased sensitivity to γ-irradiation of cells devoid of RecF, RecO, RecR, or UvrD. 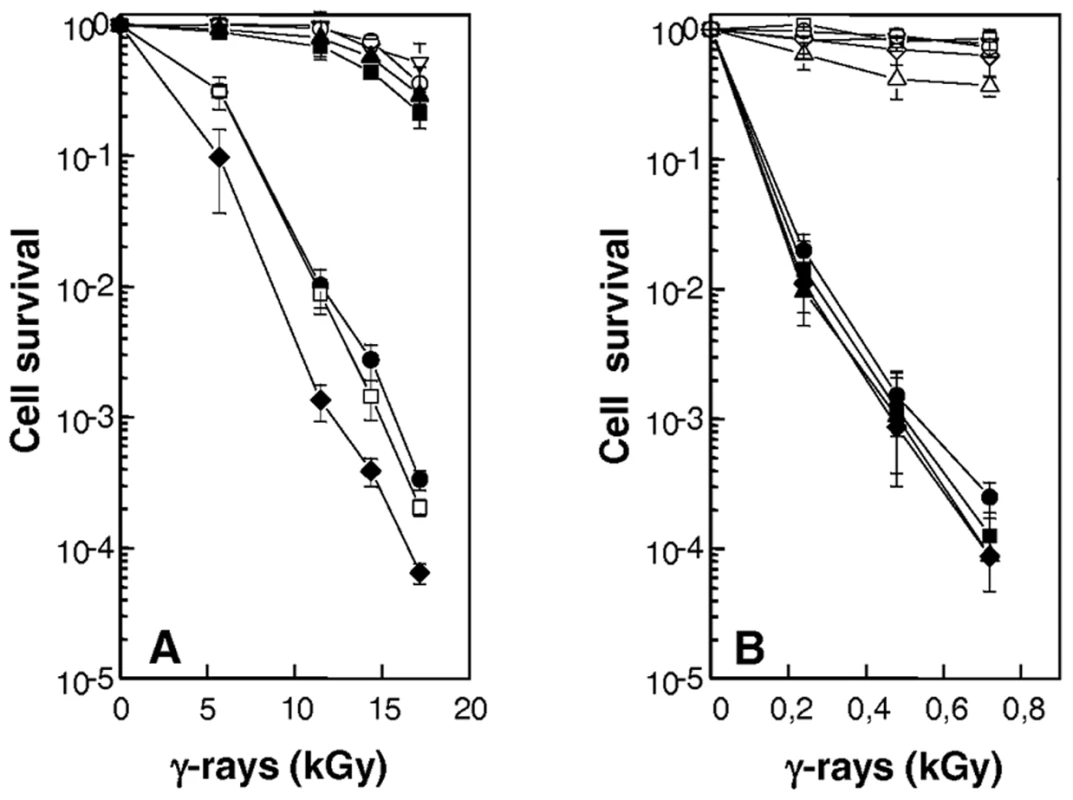
(A) Increased sensitivity of cells devoid of uvrD gene. R1 (wild-type, open inverted triangles), GY12957 (ΔrecQ, filled squares), GY12974 (ΔuvrD, filled circles), GY12975 (ΔrecQΔuvrD, open squares), GY12976 (ΔrecDΔuvrD, filled diamonds), GY12977 (ΔrecQΔrecD, filled triangles), GY13130 (ΔrecD, open circles) bacteria were exposed to γ-irradiation at doses indicated on the abscissa; and cell survival was measured as described in the Materials and Methods. (B) ΔrecFOR mutants are as sensitive as ΔrecA mutant to γ-irradiation. Bacterial strains GY12936 (wild-type/p11520, inverted triangles), GY14115 (ΔrecA/p11559, filled circles), GY14116 (ΔrecO/p11520, filled diamonds), GY14117 (ΔrecF/p11520, filled squares), GY14118 (ΔrecR/p11520, filled triangles), GY14111 (ΔrecA/p11562: recA+, open circles), GY14112 (ΔrecO/p11860: recO+, open diamonds), GY14113 (ΔrecF/p11862: recF+, open squares), GY14114 (ΔrecR/p11870: recR+, open triangles) were exposed to γ-irradiation at doses indicated on the abscissa, and cell survival was measured as described in the Materials and Methods. To investigate the possible role(s) of the UvrD helicase in DSB repair, we examined whether the ΔuvrD mutant was affected in two key steps of the ESDSA pathway: (i) the reassembly of broken DNA fragments and (ii) the associated massive DNA synthesis. Cells were exposed to 6.8 kGy γ-irradiation, a dose that introduces approximately 200 DSB per genome equivalent in a D. radiodurans cell [24]. Recovery from DNA damage was monitored by the appearance of the complete pattern of the 11 resolvable genomic DNA fragments generated by NotI digestion [25] and de novo DNA synthesis was measured by labelling DNA with a 15 min 3H-TdR pulse at different times post irradiation. As seen in Figure 4, ΔrecQ and ΔrecD cells repaired DSB with the same kinetics as the wild-type strain, reconstituting an intact genome within 3 h post-irradiation (Figure 5A). In contrast, in ΔuvrD bacteria, this process required approximately 8 h (Figure 5A), the kinetics of DSB repair had an increased latent phase (240 min in the mutant versus 90 min in the wild-type) during which DNA degradation took place and a slower rate of fragment assembly. Moreover, resumption of DNA synthesis was delayed in ΔuvrD mutant bacteria and its rate was 2-fold lower than that observed in wild-type bacteria (Figure 5B). These results suggest that UvrD plays a major role in DSB repair through ESDSA.
Fig. 5. DNA repair and DNA synthesis in ΔrecQ, ΔrecD, and ΔuvrD mutants. 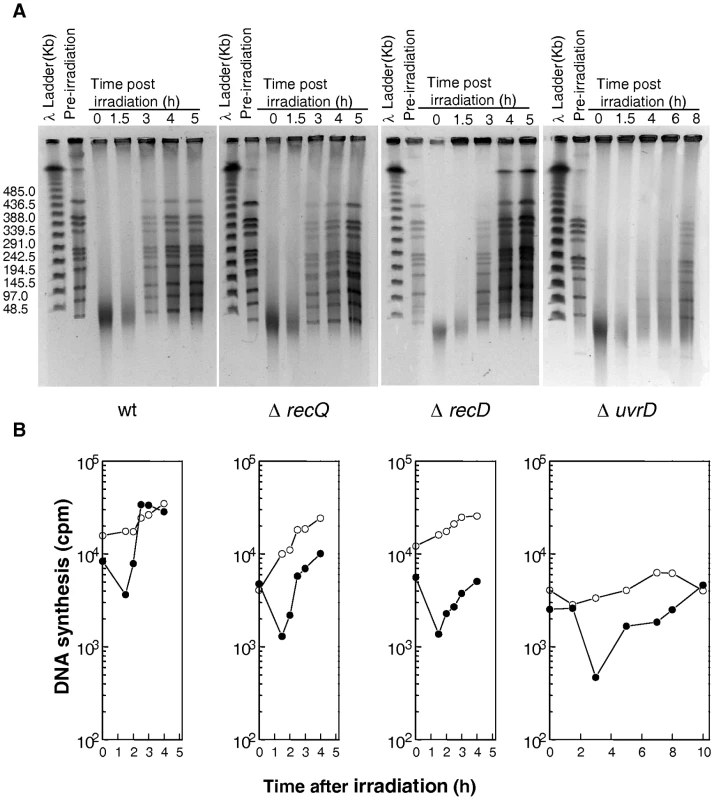
(A) Kinetics of DSB repair in wild-type, ΔrecQ, ΔrecD, and ΔuvrD mutants followed by pulsed-field gel electrophoresis (PFGE). PFGE shows NotI treated DNA from unirradiated cells (lane pre-irradiation) and from irradiated cells (6,8 kGy) immediately after irradiation (0) and at the indicated incubation times (hours). (B) Rate of DNA synthesis in wild-type, ΔrecQ, ΔrecD, and ΔuvrD mutants. Incorporation of [3H]thymidine during 15-min pulse labelling measures the global rate of DNA synthesis in 6.8 kGy-irradiated (filled circles) and unirradiated (open circles) bacteria. A major role of RecFOR in D. radiodurans radioresistance
The ΔrecF, ΔrecO and ΔrecR mutants were as radiosensitive as a ΔrecA mutant (Figure 4B). The radiosensitivity of the ΔrecF and ΔrecO mutants was fully complemented by a plasmid expressing RecF or RecO proteins in trans, whereas, in the case of the ΔrecR mutant, bacteria expressing recR+ in trans only recovered 90% of wild-type survival after γ-irradiation (Figure 4B). Because recR belongs to a putative operon, the radiosensitivity of the knock-out mutant may be due in part to a polar effect of our construct on a downstream gene or to a sub - or overoptimal plasmid-based expression of the RecR protein.
The kinetics of DNA double-strand-break repair in the three mutants was very similar to that observed in a ΔrecA mutant (Figure 6A). There was a slight and progressive reassembly of the radiation-induced DNA fragments that culminates at 24h post-irradiation incubation in the restitution of a complete pattern of the 11 NotI resolvable fragments (Figure 6A). However, only very faint bands of reconstituted chromosome were observed 24h post-irradiation incubation suggesting that a complete genome was only present in a small subpopulation of the mutant cells. The initial degradation of the damaged DNA that can be seen in the wild-type during the first hour of post-irradiation incubation (Figure 5A) was also markedly reduced in the three recFOR mutants (Figure 6A), as was previously observed for a ΔrecA mutant [2]; Figure 6A).
Fig. 6. DNA repair and DNA synthesis in ΔrecA, ΔrecF, ΔrecO, and ΔrecR mutants. 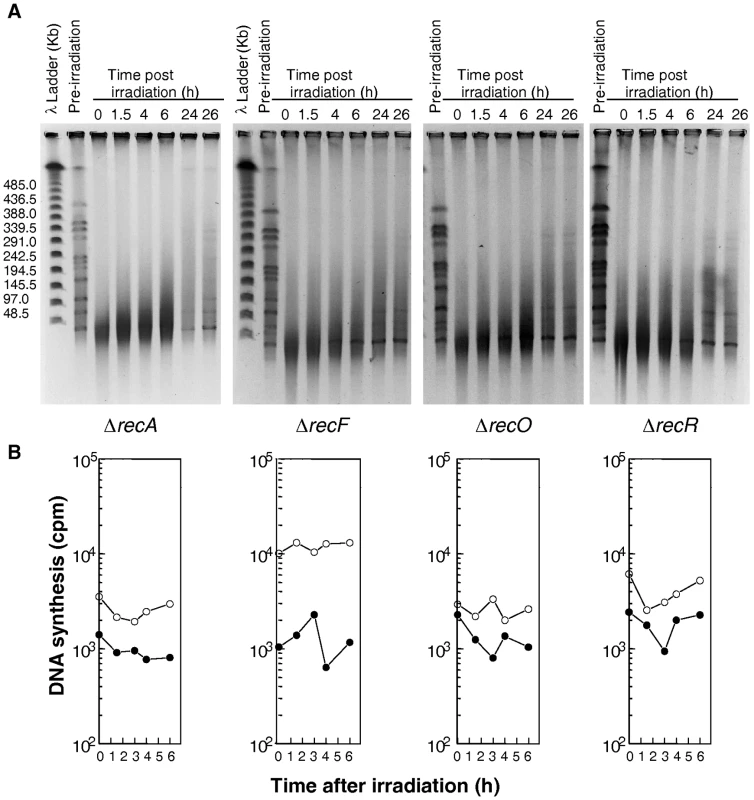
(A) Kinetics of DSB repair in wild-type, ΔrecA, ΔrecF, ΔrecO, and ΔrecR mutants. PFGE shows NotI treated DNA from unirradiated cells (lane pre-irradiation) and from irradiated cells (6,8kGy) immediately after irradiation (0) and at the indicated incubation times (hours). (B) Rate of DNA synthesis in wild-type, ΔrecA, ΔrecF, ΔrecO, and ΔrecR mutants. Incorporation of [3H]thymidine during 15-min pulse labelling measures the global rate of DNA synthesis in irradiated (filled circles) and unirradiated (circles) bacteria. The reconstitution of the complete genomic NotI pattern in the irradiated recFOR mutants did not result from the multiplication of rare survivors, because there was no observable increase in the number of CFU during 24 hours of incubation of irradiated cells (data not shown). Pulses of 3H-TdR showed that no DNA synthesis was observed during the 6 hours following γ-irradiation (Figure 6B) nor during the late fragment assembly in ΔrecF, ΔrecO and ΔrecR bacteria (data not shown), as observed previously in ΔrecA bacteria [2]. Moreover, the late genome reconstitution in these mutants is not sufficient to ensure cell survival.
In conclusion, our results suggest that RecF, RecO and RecR proteins, like RecA protein, play a central role in Deinococcal radioresistance, probably because they are absolutely required for loading RecA onto its DNA substrate to perform efficient double-strand-break repair via ESDSA and recombinational repair pathways.
Discussion
Recent studies have shown that the ability of D. radiodurans to cope with the DNA-shattering effect of elevated doses of γ-irradiation or dessication involves a robust DNA repair process called extended synthesis-dependent strand annealing (ESDSA) in which long tracts of newly synthesized DNA are made [1],[2]. Whereas the dependence of massive DNA synthesis on Pol I, Pol III and RecA (or its homolog RadA) in ESDSA is well documented [2], little is known about the cellular factors required for the initial steps of this process: (i) the formation of the single stranded 3′ overhangs and (ii) the loading of RecA on this recombinogenic substrate to prime DNA synthesis.
Major role of RecFOR proteins in DNA double-strand-break repair in Deinococcus radiodurans
D. radiodurans is naturally devoid of RecB and RecC proteins but contains homologs of key proteins of the E. coli RecF pathway: RecJ, RecQ, RecF, RecO and RecR. We found that cells devoid of RecF, RecO or RecR proteins were as radiosensitive as cells devoid of RecA. The ΔrecF, ΔrecO and ΔrecR mutants, as previously shown for a ΔrecA mutant [2], supported a slow and progressive reassembly of the radiation-induced DNA fragments. As in ΔrecA cells, genome reassembly was not accompanied by significant DNA synthesis, suggesting that cells devoid of RecF, RecO or RecR proteins are deficient for ESDSA, with repair of DSB probably mediated by RecA-independent pathways, such as single-strand annealing (SSA) or non-homologous end joining (NHEJ). The mutants also showed an important lethal sectoring during normal growth, similar to that observed in a ΔrecA mutant, in which about 90% of the visible cells failed to give rise to colonies [26]. The similarity of the ΔrecFOR and ΔrecA phenotypes supports the hypothesis that RecA activity in D. radiodurans is totally dependent on a functional RecF pathway.
Following exposure of D. radiodurans to ionizing radiation, there is a rapid and extensive degradation of chromosomal DNA that plays an important role in the repair process in this species (reviewed by [24]). The initial degradation of damaged DNA can be observed using pulsed-field electrophoresis as a reduction of the amount of the double stranded DNA fragments during the first 90 min of post-irradiation incubation in the wild-type cells, prior to the onset of fragment assembly. Slade et al observed that DNA degradation is markedly reduced in a ΔrecA mutant, leading the authors to propose that RecA itself regulates maturation of double-strand ends by controlling both DNA degradation and DNA synthesis [2]. We found that DNA degradation was also reduced in ΔrecF, ΔrecO or ΔrecR mutants, as well as in a ΔrecA mutant. RecA may play a regulatory role in the control of expression of nuclease-like activities in response to DNA damage, while RecFOR proteins may be indirectly involved in DNA degradation by facilitating the formation of the RecA filament on single stranded DNA. It would be interesting to analyse DNA degradation in the Deinococcal recA424 mutant, which retains the RecA coprotease activity while remaining deficient in recombination activity [27].
Biochemical studies using RecFOR proteins from E. coli indicate that these proteins act together as mediators of the formation of the pre-synaptic RecA filament onto single stranded DNA. Current models agree on the formation of two complexes, RecFR and RecOR. RecOR is generally thought to be responsible for rendering SSB-coated ssDNA accessible to RecA. RecFR targets dsDNA or dsDNA-ssDNA junctions and is responsible for the targeting of RecA to the ssDNA region of gaps [28]–[31]. More recently, it was proposed that RecR is the key component with which RecA interacts, whereas the RecO protein can displace SSB and bind to single stranded DNA independently of RecR, yet does not load RecA until RecR is added [32],[33]. When RecF is present, a RecFOR loading pathway, independent of RecO-SSB interactions, is preferred [33].
Recent X-ray structural analysis of RecO and RecR proteins from D. radiodurans confirms the existence of a RecOR complex in this organism. RecR molecules form a ring structure that can encircle both dsDNA and ssDNA [34],[35]. The structure of the RecF protein from D. radiodurans has also recently been elucidated, showing that the RecF protein exhibits extensive structural similarity with the head domain of the eukaryotic Rad50 protein [36]. More recently, a model of recognition of the ds-DNA ss-DNA junction in D. radiodurans through a DNA-protein and protein-protein interaction was proposed: RecR interacts with ssDNA coated by RecO-SSB, which leads to the elevation of the local concentration of RecR and stimulates RecF binding in the adjacent ds-DNA [37].
Essentiality of RecJ protein
While inactivation of RecA or RecFOR proteins in D. radiodurans reduced cell viability, inactivation of RecJ resulted in a fully lethal phenotype. In other bacterial species, mutations in recJ are highly synergistic with those in recBCD. In E. coli, recBC recJ mutants are recombination deficient, extremely UV-sensitive and highly growth disrupted [11],[38]. In Salmonella typhimurium, recB recJ mutants also display a similar phenotype [39]. More recently, it was shown that a recJ knock-out is colethal with recBCD or recD deletions in Acinetobacter baylyi [40]. The strongly reduced viability (or lethality) of recBC recJ bacteria was attributed to severe deficiencies in repair of spontaneous DNA damage and inactivated replication forks [39],[40]. It should be noted that, whereas E. coli and S. typhimurium contain at least three 5′-3′ exonucleases [RecJ, Exo V (RecBCD), Exo VII (XseAB)], the genome of A. baylyi encodes only Exo V and RecJ, and that of D. radiodurans encodes only RecJ and one of the two subunits of Exo VII. We propose that Exo VII has some back-up activity in E. coli or S. typhimurium when RecJ and ExoV are inactivated, an activity that is missing in A. baylyi and D. radiodurans.
In E. coli, RecJ and RecFOR were proposed to be required to restore DNA synthesis after UV-induced damage [41],[42]. The mechanism by which lesion-blocked replication forks recover in E. coli is thought to involve the formation of reverse replication fork intermediate stabilized by RecA and RecF and degraded by the RecQ-RecJ helicase-nuclease when RecA or RecF are absent [41]. The fork regression allows DNA repair enzymes to remove the blocking lesion, thus restoring processive replication. In the absence of RecJ, the recovery of replication is significantly delayed and both replication recovery and cell survival become dependent on translesion synthesis by DNA polymerase V [42]. D. radiodurans does not encode a bypass DNA polymerase belonging to the Y family, and under these conditions RecJ may be essential for restoration of replication forks after arrest, even in cells not treated by DNA damaging agents. Frequent DNA double-strand-breaks were thought to arise spontaneously ranging from 0.2–1 per genome replication in E. coli [4],[43]. However, a more direct quantification of DNA double-strand-breaks indicated that the rate of spontaneous breakage is 20 to 100-fold lower than predicted, only one percent of the cells having one or more DNA double-strand-breaks per genome replication [44].
Because cells devoid of RecA or RecFOR are viable, the ΔrecJ lethal phenotype cannot be only due to a possible deficiency in DSB repair, leading us to postulate that RecJ is required in D. radiodurans for more than one cellular process and that inactivation of all of these processes (DSB repair, fork reversion, restoration of a fork structure after regression …) may be lethal for the cell.
Involvement of UvrD in DNA double-strand-break repair
In E. coli, the RecJ exonuclease has been mainly associated with the RecQ helicase in recombination and repair (see, for review, [4]). The RecQ protein from D. radiodurans shows unusual domain architecture with three tandem HRDC (Helicase RNase D C-terminal) domains in addition to the conserved helicase and RQC (RecQ C-terminal) domains. The tandem arrangement of HRDC domains regulates the specificity of the binding of RecQ to DNA substrates [45],[46]. Here, we found that ΔrecQ mutants displayed a wild-type level of resistance to γ-irradiation, exhibiting the same kinetics as the wild-type strain for fragment reassembly and DNA synthesis after irradiation. In another report, a recQ knock-out mutant was shown to be highly sensitive to H2O2 and slightly more sensitive than the wild-type strain to elevated γ-irradiation doses [46]. It was recently proposed that recQ deletion, by causing transcriptome alteration, would generate ROS accumulation and Fe and Mn alterations [47]. Our findings suggest that the RecQ helicase in D. radiodurans plays only a minor role in DSB repair, probably as consequence of redundant functions provided by other helicase(s). Mutants devoid of RecD behave like ΔrecQ mutants in that they show wild-type radioresistance and repair capacity. The Deinococcal RecD protein has been characterized in vitro as a helicase with 5′-3′ polarity (opposite to that of RecQ) and low processivity [14].
In contrast, we found that inactivation of UvrD (helicase II) markedly reduced Deinococcal radioresistance and severely delayed the kinetics of DSB repair. UvrD has been largely characterized for its role in nucleotide excision repair (NER) and mismatch repair (MMR) in E. coli (reviewed by [48]). However, the altered kinetics of repair and the radiosensitivity of ΔuvrD bacteria are unlikely to result from a deficiency in the NER pathway because uvrA deficient mutant bacteria display a wild-type survival pattern following exposure to ionizing-radiation (Figure S3). The ΔmutS bacteria deficient for MMR were also shown to be as radioresistant as wild-type bacteria [18]. Interestingly, the delayed kinetics of DSB repair in cells devoid of UvrD coincided with DNA synthesis (albeit significantly less extensive than that observed in the wild-type cells) suggesting that ESDSA repair could take place but only inefficiently in this mutant. We propose that UvrD is involved in ESDSA and that the redundant activity of other helicase(s) is responsible for the residual DNA repair capacity observed in the ΔuvrD mutant. The fact that the ΔrecQΔuvrD and the ΔrecDΔuvrD double mutant bacteria were not as radiosensitive as ΔrecA bacteria suggests that neither RecQ nor RecD can solely fulfil this role, and that other helicase(s) may be involved. Helicase IV (HelD) has been implicated as partner of the RecJ exonuclease in the RecF pathway in E. coli, together with Helicase II and RecQ [49]. Mutational inactivation of Helicase IV has no effect on the radioresistance of D. radiodurans [50]. Alternatively, RecA itself, by binding to double stranded DNA ends, could unwind DNA and provide a DNA substrate for RecJ or another 5′-3′ exonuclease. Indeed, in vitro, the D. radiodurans RecA protein binds preferentially to double stranded DNA [51].
In E. coli, the UvrD protein was not shown to be required for DNA double-strand-break repair. In contrast, it was shown to possess an anti-recombination activity, which has been related to its capacity to disrupt the RecA nucleoprotein filament [52],[53]. This activity is conserved among many species [54]. Thus, as in other species, the D. radiodurans UvrD protein might not be involved directly in the maturation of DNA double-strand ends. Several observations suggest that E. coli UvrD may be involved in DNA replication [55]–[58] and it was shown to be required for DNA replication of several different rolling-circle plasmids in E. coli [59]. Thus, the D. radiodurans UvrD protein might also act in the DNA synthesis step of ESDSA.
A scenario for DNA double-strand-break repair through ESDSA in D. radiodurans
Taking into account our results and those of others [2],[60],[61], we propose a model for the role of the proteins of the RecF pathway in ESDSA (Figure 7). In this model, RecJ or an as-yet unidentified exonuclease associated with the UvrD helicase, could generate 3′ single stranded DNA ends required for priming of massive DNA synthesis. Alternatively, RecA itself, by binding to double stranded DNA ends, could unwind DNA and provide a DNA substrate for RecJ or another exonuclease.
Fig. 7. Model of initiation of DNA double-strand-break repair through ESDSA in <i>D. radiodurans</i>. 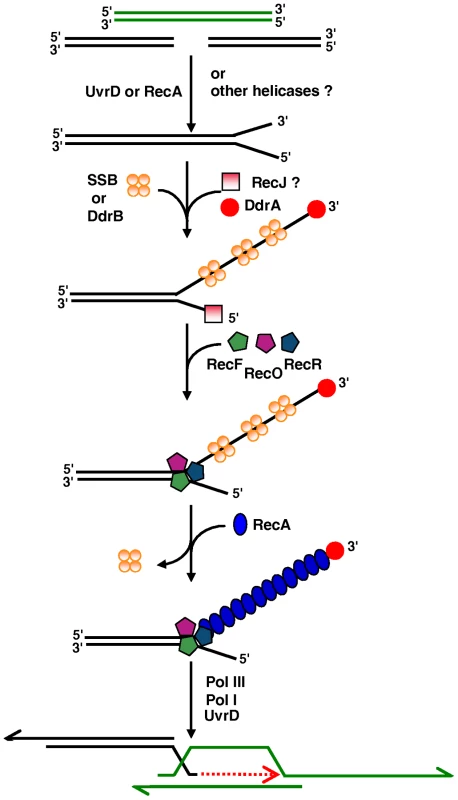
Analysis of the transcriptome of D. radiodurans revealed a large group of genes that are up-regulated in response to either desiccation or ionizing radiation [62]. The deinococcal specific ddrA (DR0423) and ddrB (DR0070) genes were found among the most highly induced in response to each stress and their inactivation promotes sensitization of the mutant cells to ionizing radiation [62]. The DdrA protein is involved in protection of 3′ DNA single stranded ends [60] and presumably ensures long-lived recombinational substrates [63]. The DdrB protein binds single stranded DNA but not duplex DNA and is the prototype of a new bacterial SSB family [61]. The induction of an alternative SSB following irradiation has potentially broad significance for efficient genome reconstitution. We propose that during initiation of ESDSA, DdrA protects the 3′ DNA ends whereas SSB or the SSB-like DdrB binds to single stranded DNA.
Our results supporting the idea that RecA activity in D. radiodurans is totally dependant on a functional RecF pathway, lead us to propose that RecFOR renders SSB or DdrB - coated single stranded DNA accessible to RecA and favors formation of a RecA nucleoprotein filament required for invasion of a double stranded homologous DNA. Finally, as described previously [2], Pol III and Pol I can promote DNA synthesis, eventually with the help of the UvrD helicase.
Moreover, the compact D. radiodurans nucleoid structure that remains unaltered after high-dose γ-irradiation may passively contribute to radioresistance by preventing the dispersion of free DNA ends [64],[65]. Such a condensed genome may provide suitable scaffolds for DNA repair through ESDSA, recombinational and/or DNA end joining processes.
In conclusion, we demonstrate the essential role of key components of the D. radiodurans RecF pathway in ESDSA. We show for the first time that (i) inactivation of only one exonuclease, RecJ, results in cell lethality (ii) cells devoid of RecF, RecO or RecR display greatly impaired growth (iii) RecF, RecO or RecR proteins are essential for radioresistance through ESDSA (iv) UvrD helicase has an unexpected crucial function in DNA double-strand-break repair through ESDSA.
Materials and Methods
Bacterial strains, cultures, media, and transformation
Bacterial strains and plasmids are listed in Table 1 and Table 2, respectively. The Escherichia coli strain DH5α was used as the general cloning host and strain SCS110 was used to propagate plasmids prior to introduction into D. radiodurans via transformation [66]. All D. radiodurans strains were derivatives of strain R1 (ATCC 13939). D. radiodurans was grown in TGY2X (1% tryptone, 0.2% dextrose, 0.6% yeast extract) or in TGYA (0.5% tryptone, 0.2% dextrose, 0.15% yeast extract) at 30°C with aeration or on TGY1X plates solidified with 1.5% agar. E. coli strains were grown in Luria-Bertani (LB) broth (Gibco Laboratories). When necessary, media were supplemented with the appropriate antibiotics used at the following final concentrations: chloramphenicol 3 µg/mL for D. radiodurans; kanamycin 6 µg/mL for D. radiodurans; tetracycline 2.5 µg/mL for D. radiodurans; hygromycin 50 µg/mL; spectinomycin 40 µg/mL for E. coli and 75 µg/mL for D. radiodurans. Transformation of D. radiodurans with PCR products, genomic DNA, or plasmids was performed as previously described [26].
DNA manipulations
Plasmid DNA was extracted from E. coli using the QIAprep spin miniprep kit (Qiagen). Chromosomal DNA of D. radiodurans was isolated as previously described [67]. Amplification of plasmid or genomic DNA by PCR was performed with DyNAzyme EXT DNA polymerase (Finnzyme) or Extensor Hi-Fidelity PCR enzyme Mix (ABgene). Oligonucleotides used are listed in Table S1.
Deletion of genes in D. radiodurans
The recF, recO, recR, recA, uvrD, recD, recQ, recJ disruption mutants were constructed by the tripartite ligation method [18]. The mutated alleles constructed in vitro were then used to transform D. radiodurans to replace their wild-type counterpart by homologous recombination. The genetic structure and the purity of the mutants were checked by PCR using primers described in Table S1.
Construction of plasmids
Plasmid p11869 is a derivative of the thermosensitive plasmid p13840 [21]. To construct p11869, the recJ gene was amplified by PCR using the primer pair (PS441/PS442) and the product was cloned into plasmid p13840 between the NdeI/XhoI sites.
Plasmids p11862, p11860 and p11870 carrying the recF, recO, recR genes, respectively, under the control of their natural promoter were used to express the recF, recO, recR genes in a ΔrecF, ΔrecO, ΔrecR background. To construct plasmid p11860, the recO gene was amplified by PCR using the primer pair (PS402/PS403) and the resultant product was cloned into plasmid p11520 [68] between the SacI/BamHI sites. Plasmid p11870, containing the recR gene, was constructed in a similar manner using the primer pairs PS414/PS415. The recF gene was cloned into plasmid p11520 between the SpeI/BglII sites in a similar manner using the primers PS410/PS411 to obtain p11862. All constructions were verified by DNA sequencing.
Plasmid p11562 [63], expressing recA from a PSpac promoter, was used to transform GY12968: ΔrecAΩkan giving rise to strain GY14111. The expression of recA was induced by adding 10 mM IPTG to the media.
Treatment of D. radiodurans with γ-irradiation
Exponential cultures, grown in TGY2X (supplemented with spectinomycin when necessary), were concentrated to an A650 = 10 in TGY2X and irradiated on ice with a 137Cs irradiation system (Institut Curie, Orsay, France) at a dose rate of 44.7 Gy/min. Following irradiation, diluted samples were plated on TGY plates. Colonies were counted after 3–4 days incubation at 30°C.
Assay of recJ gene essentiality
The essentiality of genes was evaluated in a growth experiment in which the strains grown at 28°C in liquid medium with spectinomycin, were serially diluted, plated on TGY agar and incubated at 28°C or 37°C in the presence or absence of spectinomycin [21].
Kinetics of DNA repair measured by pulse-field gel electrophoresis
Non-irradiated or irradiated (6.8 kGy) cultures were diluted in TGY2X to an A650 = 0.2 and incubated at 30°C. At different post-irradiation recovery times, culture aliquots (5mL) were removed to prepare DNA plugs as described previously [60]. The agarose embedded DNA plugs were digested for 16 h at 37°C with 10 units of NotI restriction enzyme. After digestion, the plugs were subjected to pulsed field gel electrophoresis as described previously [69].
Rate of DNA synthesis measured by DNA pulse labelling
The rate of DNA synthesis was measured according to a modified protocol from Zahradka et al [1]. Exponential cultures, grown in TGYA, were concentrated to an A650 = 20 in TGYA and irradiated as described previously. Non-irradiated or irradiated cultures (6.8 kGy) were diluted in TGYA to an A650 = 0.2 and incubated at 30°C. At different time 0.5mL samples were taken and mixed with 0.1mL pre-warmed TGYA containing 4.8 µCi [methyl-3H]thymidine (PerkinElmer, specific activity 70–90 Ci/mmol). Radioactive pulses of 15 min were terminated by addition of 2 mL ice-cold 10% TCA. Samples were kept on ice for at least 1 h, and then collected by vacuum filtration onto Whatman GF/C filters followed by washing twice with 5mL 5% TCA and twice with 5mL 96% ethanol. Filters were dried for 10 min under a heat source and placed in 4 mL scintillation liquid. The precipitated counts were measured in a liquid scintillation counter (Packard, TRI - carb 1600 TR).
Supporting Information
Zdroje
1. ZahradkaK
SladeD
BailoneA
SommerS
AverbeckD
2006 Reassembly of shattered chromosomes in Deinococcus radiodurans. Nature 443 569 573
2. SladeD
LindnerAB
PaulG
RadmanM
2009 Recombination and replication in DNA repair of heavily irradiated Deinococcus radiodurans. Cell 136 1044 1055
3. SmithGR
1989 Homologous recombination in prokaryotes: enzymes and controlling sites. Genome 31 520 527
4. KuzminovA
1999 Recombinational repair of DNA damage in Escherichia coli and bacteriophage lambda. Microbiol Mol Biol Rev 63 751 813
5. KowalczykowskiSC
2000 Initiation of genetic recombination and recombination-dependent replication. Trends Biochem Sci 25 156 165
6. HoriiZ
ClarkAJ
1973 Genetic analysis of the recF pathway to genetic recombination in Escherichia coli K12: isolation and characterization of mutants. J Mol Biol 80 327 344
7. ClarkAJ
SandlerSJ
WillisDK
ChuCC
BlanarMA
1984 Genes of the RecE and RecF pathways of conjugational recombination in Escherichia coli. Cold Spring Harb Symp Quant Biol 49 453 462
8. NakayamaH
NakayamaK
NakayamaR
IrinoN
NakayamaY
1984 Isolation and genetic characterization of a thymineless death-resistant mutant of Escherichia coli K12: identification of a new mutation (recQ1) that blocks the RecF recombination pathway. Mol Gen Genet 195 474 480
9. KolodnerR
FishelRA
HowardM
1985 Genetic recombination of bacterial plasmid DNA: effect of RecF pathway mutations on plasmid recombination in Escherichia coli. J Bacteriol 163 1060 1066
10. MahdiAA
LloydRG
1989 Identification of the recR locus of Escherichia coli K-12 and analysis of its role in recombination and DNA repair. Mol Gen Genet 216 503 510
11. LloydRG
EvansNP
BuckmanC
1987 Formation of recombinant lacZ+ DNA in conjugational crosses with a recB mutant of Escherichia coli K12 depends on recF, recJ, and recO. Mol Gen Genet 209 135 141
12. CromieGA
2009 Phylogenetic ubiquity and shuffling of the bacterial RecBCD and AddAB recombination complexes. J Bacteriol 191 5076 5084
13. RochaEP
CornetE
MichelB
2005 Comparative and evolutionary analysis of the bacterial homologous recombination systems. PLoS Genet 1 e15 doi:10.1371/journal.pgen.0010015
14. WangJ
JulinDA
2004 DNA helicase activity of the RecD protein from Deinococcus radiodurans. J Biol Chem 279 52024 52032
15. ZhouQ
ZhangX
XuH
XuB
HuaY
2007 A new role of Deinococcus radiodurans RecD in antioxidant pathway. FEMS Microbiol Lett 271 118 125
16. ServinskyMD
JulinDA
2007 Effect of a recD mutation on DNA damage resistance and transformation in Deinococcus radiodurans. J Bacteriol 189 5101 5107
17. MisraHS
KhairnarNP
KotaS
ShrivastavaS
JoshiVP
2006 An exonuclease I-sensitive DNA repair pathway in Deinococcus radiodurans: a major determinant of radiation resistance. Mol Microbiol 59 1308 1316
18. MennecierS
CosteG
ServantP
BailoneA
SommerS
2004 Mismatch repair ensures fidelity of replication and recombination in the radioresistant organism Deinococcus radiodurans. Mol Genet Genomics 272 460 469
19. HansenMT
1978 Multiplicity of genome equivalents in the radiation-resistant bacterium Micrococcus radiodurans. J Bacteriol 134 71 75
20. Harsojo
KitayamaS
MatsuyamaA
1981 Genome multiplicity and radiation resistance in Micrococcus radiodurans. J Biochem (Tokyo) 90 877 880
21. NguyenHH
Bouthier de la TourC
ToueilleM
VannierF
SommerS
2009 The essential histone-like protein HU plays a major role in Deinococcus radiodurans nucleoid compaction. Mol Microbiol 73 240 252
22. WhiteO
EisenJA
HeidelbergJF
HickeyEK
PetersonJD
1999 Genome sequence of the radioresistant bacterium Deinococcus radiodurans R1. Science 286 1571 1577
23. MendoncaVM
KlepinHD
MatsonSW
1995 DNA helicases in recombination and repair: construction of a delta uvrD delta helD delta recQ mutant deficient in recombination and repair. J Bacteriol 177 1326 1335
24. BattistaJR
1997 Against all odds: the survival strategies of Deinococcus radiodurans. Annu Rev Microbiol 51 203 224
25. KikuchiMNI
KitayamaS
WatanabeH
YamamotoK
1999 Genomic organization of the radioresistant bacterium Deinococcus radiodurans : physical map and evidence for multiple replicons. FEMS Microbiol Lett 174 151 157
26. Bonacossa de AlmeidaC
CosteG
SommerS
BailoneA
2002 Quantification of RecA protein in Deinococcus radiodurans reveals involvement of RecA, but not LexA, in its regulation. Mol Genet Genomics 268 28 41
27. SatohK
NarumiI
KikuchiM
KitayamaS
YanagisawaT
2002 Characterization of RecA424 and RecA670 proteins from Deinococcus radiodurans. J Biochem (Tokyo) 131 121 129
28. ShanQ
BorkJM
WebbBL
InmanRB
CoxMM
1997 RecA protein filaments: end-dependent dissociation from ssDNA and stabilization by RecO and RecR proteins. J Mol Biol 265 519 540
29. WebbBL
CoxMM
InmanRB
1997 Recombinational DNA repair: the RecF and RecR proteins limit the extension of RecA filaments beyond single-strand DNA gaps. Cell 91 347 356
30. BorkJM
CoxMM
InmanRB
2001 The RecOR proteins modulate RecA protein function at 5′ ends of single-stranded DNA. EMBO J 20 7313 7322
31. MorimatsuK
KowalczykowskiSC
2003 RecFOR proteins load RecA protein onto gapped DNA to accelerate DNA strand exchange: a universal step of recombinational repair. Mol Cell 11 1337 1347
32. InoueJ
HondaM
IkawaS
ShibataT
MikawaT
2008 The process of displacing the single-stranded DNA-binding protein from single-stranded DNA by RecO and RecR proteins. Nucleic Acids Res 36 94 109
33. SakaiA
CoxMM
2009 RecFOR and RecOR as distinct RecA loading pathways. J Biol Chem 284 3264 3272
34. LeeBI
KimKH
ShimSM
HaKS
YangJK
2004 Crystallization and preliminary X-ray crystallographic analysis of the RecR protein from Deinococcus radiodurans, a member of the RecFOR DNA-repair pathway. Acta Crystallogr D Biol Crystallogr 60 379 381
35. LeirosI
TimminsJ
HallDR
McSweeneyS
2005 Crystal structure and DNA-binding analysis of RecO from Deinococcus radiodurans. EMBO J 24 906 918
36. KorolevaO
MakharashviliN
CourcelleCT
CourcelleJ
KorolevS
2007 Structural conservation of RecF and Rad50: implications for DNA recognition and RecF function. EMBO J 26 867 877
37. MakharashviliN
MiT
KorolevaO
KorolevS
2009 RecR-mediated modulation of RecF dimer specificity for single - and double-stranded DNA. J Biol Chem 284 1425 1434
38. LovettST
ClarkAJ
1984 Genetic analysis of the recJ gene of Escherichia coli K-12. J Bacteriol 157 190 196
39. GarzonA
BeuzonCR
MahanMJ
CasadesusJ
1996 recB recJ mutants of Salmonella typhimurium are deficient in transductional recombination, DNA repair and plasmid maintenance. Mol Gen Genet 250 570 580
40. KicksteinE
HarmsK
WackernagelW
2007 Deletions of recBCD or recD influence genetic transformation differently and are lethal together with a recJ deletion in Acinetobacter baylyi. Microbiology 153 2259 2270
41. CourcelleJ
DonaldsonJR
ChowKH
CourcelleCT
2003 DNA damage-induced replication fork regression and processing in Escherichia coli. Science 299 1064 1067
42. CourcelleCT
ChowKH
CaseyA
CourcelleJ
2006 Nascent DNA processing by RecJ favors lesion repair over translesion synthesis at arrested replication forks in Escherichia coli. Proc Natl Acad Sci U S A 103 9154 9159
43. CoxMM
GoodmanMF
KreuzerKN
SherrattDJ
SandlerSJ
2000 The importance of repairing stalled replication forks. Nature 404 37 41
44. PenningtonJM
RosenbergSM
2007 Spontaneous DNA breakage in single living Escherichia coli cells. Nat Genet 39 797 802
45. KilloranMP
KeckJL
2006 Three HRDC domains differentially modulate Deinococcus radiodurans RecQ DNA helicase biochemical activity. J Biol Chem 281 12849 12857
46. HuangL
HuaX
LuH
GaoG
TianB
2007 Three tandem HRDC domains have synergistic effect on the RecQ functions in Deinococcus radiodurans. DNA Repair (Amst) 6 167 176
47. ChenH
HuangL
HuaX
YingL
HuY
2009 Pleiotropic Effects of RecQ in Deinococcus radiodurans. Genomics
48. MatsonSW
RobertsonAB
2006 The UvrD helicase and its modulation by the mismatch repair protein MutL. Nucleic Acids Res 34 4089 4097
49. MendoncaVM
Kaiser-RogersK
MatsonSW
1993 Double helicase II (uvrD)-helicase IV (helD) deletion mutants are defective in the recombination pathways of Escherichia coli. J Bacteriol 175 4641 4651
50. CaoZ
JulinDA
2009 Characterization in vitro and in vivo of the DNA helicase encoded by Deinococcus radiodurans locus DR1572. DNA Repair (Amst) 8 612 619
51. KimJI
SharmaAK
AbbottSN
WoodEA
DwyerDW
2002 RecA Protein from the extremely radioresistant bacterium Deinococcus radiodurans: expression, purification, and characterization. J Bacteriol 184 1649 1660
52. MorelP
HejnaJA
EhrlichSD
CassutoE
1993 Antipairing and strand transferase activities of E. coli helicase II (UvrD). Nucleic Acids Res 21 3205 3209
53. VeauteX
DelmasS
SelvaM
JeussetJ
Le CamE
2005 UvrD helicase, unlike Rep helicase, dismantles RecA nucleoprotein filaments in Escherichia coli. EMBO J 24 180 189
54. LestiniR
MichelB
2007 UvrD controls the access of recombination proteins to blocked replication forks. EMBO J 26 3804 3814
55. OssannaN
MountDW
1989 Mutations in uvrD induce the SOS response in Escherichia coli. J Bacteriol 171 303 307
56. KlinkertMQ
KleinA
Abdel-MonemM
1980 Studies on the functions of DNA helicase I and DNA helicase II of Escherichia coli. J Biol Chem 255 9746 9752
57. KuhnB
Abdel-MonemM
1982 DNA synthesis at a fork in the presence of DNA helicases. Eur J Biochem 125 63 68
58. LahueRS
AuKG
ModrichP
1989 DNA mismatch correction in a defined system. Science 245 160 164
59. BruandC
EhrlichSD
2000 UvrD-dependent replication of rolling-circle plasmids in Escherichia coli. Mol Microbiol 35 204 210
60. HarrisDR
TanakaM
SavelievSV
JolivetE
EarlAM
2004 Preserving genome integrity: the DdrA protein of Deinococcus radiodurans R1. PLoS Biol 2 e304 doi:10.1371/journal.pbio.0020304
61. NoraisCA
Chitteni-PattuS
WoodEA
InmanRB
CoxMM
2009 DdrB protein, an alternative Deinococcus radiodurans SSB induced by ionizing radiation. J Biol Chem 284 21402 21411
62. TanakaM
EarlAM
HowellHA
ParkMJ
EisenJA
2004 Analysis of Deinococcus radiodurans's transcriptional response to ionizing radiation and desiccation reveals novel proteins that contribute to extreme radioresistance. Genetics 168 21 33
63. JolivetE
LecointeF
CosteG
SatohK
NarumiI
2006 Limited concentration of RecA delays DNA double-strand break repair in Deinococcus radiodurans R1. Mol Microbiol 59 338 349
64. Levin-ZaidmanS
EnglanderJ
ShimoniE
SharmaAK
MintonKW
2003 Ringlike structure of the Deinococcus radiodurans genome: a key to radioresistance? Science 299 254 256
65. ZimmermanJM
BattistaJR
2005 A ring-like nucleoid is not necessary for radioresistance in the Deinococcaceae. BMC Microbiol 5 17
66. MeimaR
RothfussHM
GewinL
LidstromME
2001 Promoter cloning in the radioresistant bacterium Deinococcus radiodurans. J Bacteriol 183 3169 3175
67. MennecierS
ServantP
CosteG
BailoneA
SommerS
2006 Mutagenesis via IS transposition in Deinococcus radiodurans. Mol Microbiol 59 317 325
68. BentchikouE
ServantP
CosteG
SommerS
2007 Additive effects of SbcCD and PolX deficiencies in the in vivo repair of DNA double-strand breaks in Deinococcus radiodurans. J Bacteriol 189 4784 4790
69. LecointeF
ShevelevIV
BailoneA
SommerS
HubscherU
2004 Involvement of an X family DNA polymerase in double-stranded break repair in the radioresistant organism Deinococcus radiodurans. Mol Microbiol 53 1721 1730
70. MoseleyBE
CoplandHF
1978 Four mutants of Micrococcus radiodurans defective in the ability to repair DNA damaged by mitomycin-C, two of which have wild-type resistance to ultraviolet radiation. Mol Gen Genet 160 331 337
71. EarlAM
RankinSK
KimKP
LamendolaON
BattistaJR
2002 Genetic evidence that the uvsE gene product of Deinococcus radiodurans R1 is a UV damage endonuclease. J Bacteriol 184 1003 1009
Štítky
Genetika Reprodukční medicína
Článek Activation of Mutant Enzyme Function by Proteasome Inhibitors and Treatments that Induce Hsp70Článek Maternal Ethanol Consumption Alters the Epigenotype and the Phenotype of Offspring in a Mouse ModelČlánek Genetic Dissection of Differential Signaling Threshold Requirements for the Wnt/β-Catenin PathwayČlánek Distinct Type of Transmission Barrier Revealed by Study of Multiple Prion Determinants of Rnq1
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2010 Číslo 1- Růst a vývoj dětí narozených pomocí IVF
- Délka menstruačního cyklu jako marker ženské plodnosti
- Vztah užívání alkoholu a mužské fertility
- Šanci na úspěšný průběh těhotenství snižují nevhodné hladiny progesteronu vznikající při umělém oplodnění
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
-
Všechny články tohoto čísla
- Irradiation-Induced Genome Fragmentation Triggers Transposition of a Single Resident Insertion Sequence
- A Major Role of the RecFOR Pathway in DNA Double-Strand-Break Repair through ESDSA in
- Kidney Development in the Absence of and Requires
- Modeling of Environmental Effects in Genome-Wide Association Studies Identifies and as Novel Loci Influencing Serum Cholesterol Levels
- Inverse Correlation between Promoter Strength and Excision Activity in Class 1 Integrons
- Activation of Mutant Enzyme Function by Proteasome Inhibitors and Treatments that Induce Hsp70
- Postnatal Survival of Mice with Maternal Duplication of Distal Chromosome 7 Induced by a / Imprinting Control Region Lacking Insulator Function
- The Werner Syndrome Protein Functions Upstream of ATR and ATM in Response to DNA Replication Inhibition and Double-Strand DNA Breaks
- Maternal Ethanol Consumption Alters the Epigenotype and the Phenotype of Offspring in a Mouse Model
- Understanding Gene Sequence Variation in the Context of Transcription Regulation in Yeast
- miR-30 Regulates Mitochondrial Fission through Targeting p53 and the Dynamin-Related Protein-1 Pathway
- Elevated Levels of the Polo Kinase Cdc5 Override the Mec1/ATR Checkpoint in Budding Yeast by Acting at Different Steps of the Signaling Pathway
- Alternative Epigenetic Chromatin States of Polycomb Target Genes
- Co-Orientation of Replication and Transcription Preserves Genome Integrity
- A Comprehensive Map of Insulator Elements for the Genome
- Environmental and Genetic Determinants of Colony Morphology in Yeast
- U87MG Decoded: The Genomic Sequence of a Cytogenetically Aberrant Human Cancer Cell Line
- The MCM-Binding Protein ETG1 Aids Sister Chromatid Cohesion Required for Postreplicative Homologous Recombination Repair
- Genetic Dissection of Differential Signaling Threshold Requirements for the Wnt/β-Catenin Pathway
- Differential Localization and Independent Acquisition of the H3K9me2 and H3K9me3 Chromatin Modifications in the Adult Germ Line
- Genetic Crossovers Are Predicted Accurately by the Computed Human Recombination Map
- Collaborative Action of Brca1 and CtIP in Elimination of Covalent Modifications from Double-Strand Breaks to Facilitate Subsequent Break Repair
- Distinct Type of Transmission Barrier Revealed by Study of Multiple Prion Determinants of Rnq1
- Genome-Wide Association Study Identifies as a Novel Susceptibility Gene for Osteoporosis
- and Regulate Reproductive Habit in Rice
- Nonsense-Mediated Decay Enables Intron Gain in
- Altered Gene Expression and DNA Damage in Peripheral Blood Cells from Friedreich's Ataxia Patients: Cellular Model of Pathology
- The Systemic Imprint of Growth and Its Uses in Ecological (Meta)Genomics
- The Gift of Observation: An Interview with Mary Lyon
- Genotype and Gene Expression Associations with Immune Function in
- The Elongator Complex Regulates Neuronal α-tubulin Acetylation
- Rising from the Ashes: DNA Repair in
- Mis-Spliced Transcripts of Nicotinic Acetylcholine Receptor α6 Are Associated with Field Evolved Spinosad Resistance in (L.)
- BRIT1/MCPH1 Is Essential for Mitotic and Meiotic Recombination DNA Repair and Maintaining Genomic Stability in Mice
- Non-Coding Changes Cause Sex-Specific Wing Size Differences between Closely Related Species of
- Evidence for Pervasive Adaptive Protein Evolution in Wild Mice
- Evolutionary Mirages: Selection on Binding Site Composition Creates the Illusion of Conserved Grammars in Enhancers
- VEZF1 Elements Mediate Protection from DNA Methylation
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- A Major Role of the RecFOR Pathway in DNA Double-Strand-Break Repair through ESDSA in
- Kidney Development in the Absence of and Requires
- The Werner Syndrome Protein Functions Upstream of ATR and ATM in Response to DNA Replication Inhibition and Double-Strand DNA Breaks
- Alternative Epigenetic Chromatin States of Polycomb Target Genes
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání