-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaChronobiomics: The Biological Clock as a New Principle in Host–Microbial Interactions
article has not abstract
Published in the journal: . PLoS Pathog 11(10): e32767. doi:10.1371/journal.ppat.1005113
Category: Pearls
doi: https://doi.org/10.1371/journal.ppat.1005113Summary
article has not abstract
How Do Mammals Organize Their Biological Clocks?
The rotation of the Earth around its own axis creates a fundamental challenge for life on this planet, related to the need of all living organisms to modify their physiology in accordance with daily variations in multiple geophysical properties, including light, temperature, and availability of nutrients. As a consequence, all domains of life have developed intrinsic timing systems (“biological clocks”) in order to efficiently adapt organismal activity to the fluctuating environmental conditions [1]. The mechanisms by which different organisms have solved this challenge are manifold. In mammals, the molecular clock is classically understood as a network of transcription factors that are rhythmically operational in virtually all cells of the body [2]. Specifically, the core circadian clock is constituted by the transcription factors Period/Cryptochrome and BMAL1/CLOCK, which not only control each other’s circadian expression pattern but also drive rhythmic oscillations in a large number of target genes, thereby orchestrating the daily activity profile of a cell [3]. Interestingly, mammalian molecular clocks are autonomous and self-sustained but can be entrained by external stimuli, such as light and nutrient availability, in order to adapt organism-intrinsic rhythmicity to fluctuating environmental conditions.
Are There Biological Clocks in Bacteria?
Until about 25 years ago, evidence for a biological clock had only been found in eukaryotes. Since then, however, bacterial circadian clocks have been well documented. Similar to eukaryotic clocks, they are endogenous, self-sustained, and can be entrained by environmental conditions. Of note, bacterial circadian rhythms have so far primarily been described in organisms that are light responsive, namely the cyanobacterium Synechococcus elongates [4]. In this bacterium, the molecular clock consists of three proteins (KaiA, KaiB, and KaiC). Remarkably, their oscillatory activity can be sustained in the absence of transcription and is characterized by rhythmic phosphorylation patterns [5]. In vitro reconstitution of this three-member clock leads to sustained rhythmic activity [6]. Of note, this rhythmic phosphorylation adapts to availability and intracellular storage of nutrients [7]. Although potential homologs in the genomic sequence of other bacteria have been found, their characterization awaits further studies. In addition, oxidation-reduction cycles of peroxiredoxin proteins have been found to be circadian across all domains of life [8]. Thus, circadian systems have putatively evolved in multiple distinct ways in different forms of life.
What Happens to Biological Clocks in Places Where Eukaryotes and Prokaryotes Share Their Habitats?
The study of diverse molecular clocks across the domains of life is especially interesting in scenarios in which prokaryotes and eukaryotes interact to form symbiotic communities. The first insight into such cross talk involving clock systems regulated across organisms came from a study of the squid Euprymna scolopes and its luminous endosymbiont Vibrio fischeri [9]. E. scolopes encodes genes for cryptochromes, whose oscillatory expression pattern is synchronized with and depends on symbiont luminescence in the light organ of the squid. Further evidence came from studies of the intestinal microbiota in mice, where it was observed that gut microbial colonization influences rhythmic signaling events in the ileal epithelium downstream of toll-like receptors (TLRs). This, in turn, regulates the organization of molecular clock activity and glucocorticoid production in the intestine [10]. The microbiota also impacts clock gene expression beyond the gastrointestinal track. Germ-free mice, which are born and raised under strictly sterile conditions in the absence of microorganisms, feature alterations in clock gene expression in the liver and the hypothalamus [11]. Together, these studies suggest that microbial colonization has a previously unappreciated function in the maintenance of circadian rhythms in the eukaryotic hosts. The underlying mechanisms for this activity remain obscure (Fig 1).
Fig. 1. Schematic showing diurnal cross talk between host and intestinal microbiota. 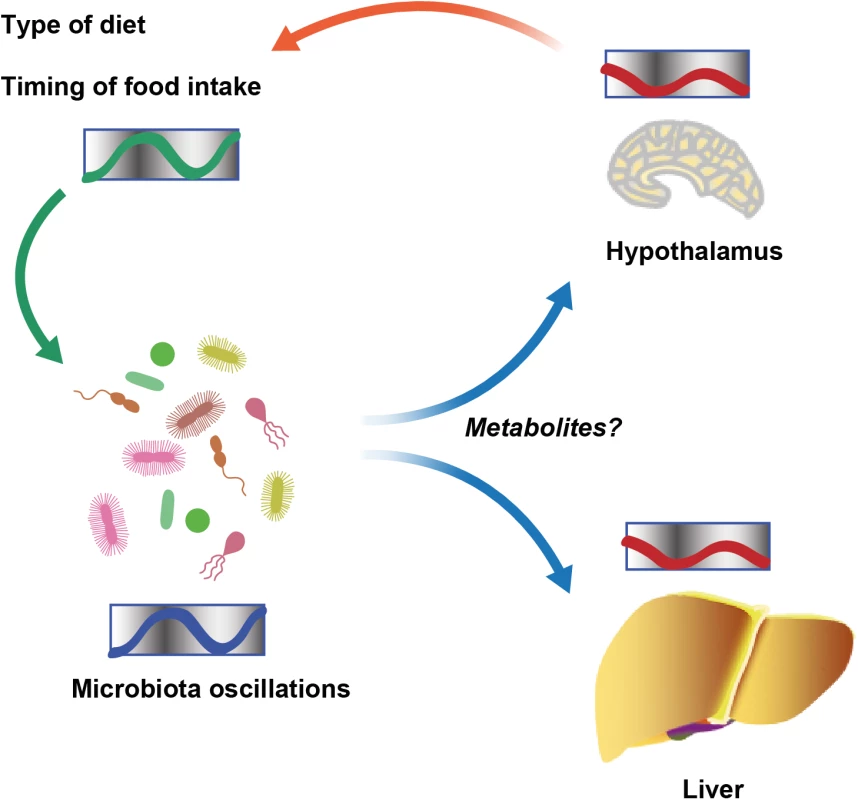
Recently, we uncovered that the circadian interactions between host and microbiota are not solely restricted to microbial control of host clock function but rather constitute a bidirectional cross talk [12]. We and others demonstrated that the intestinal microbiota undergoes rhythmic fluctuations in taxonomic composition and, consequently, genomic coding capacity [11–13]. As a result, different times of the day feature distinct microbial community configurations and microbiota functions. Thus, in addition to the intrinsic circadian timing systems identified in cyanobacteria, the intestinal microbiota undergoes community-scale rhythmicity on the level of metagenome composition [14]. These compositional fluctuations are controlled by the circadian clock of the host, since they cease in the absence of a functional host molecular clock [12]. Furthermore, the type of diet and the rhythmicity of food intake are major drivers of the oscillations in the intestinal microbial ecosystem [12,13]. In addition, recent insights using parenteral feeding raise the possibility that further, still-unidentified host factors are involved in regulating microbial oscillations [11]. Taken together, symbiotic microbial colonization is required for circadian homeostasis of the host. In turn, a functional host circadian clock ensures periodic oscillations of its symbiotic microbial ecosystem (Fig 1).
How Does the Biological Clock Influence the Antimicrobial Response?
While the above examples represent homeostatic diurnal cross talk between eukaryotes and prokaryotes in symbiotic communities, it is interesting to consider whether the underlying regulatory principles also apply to the specific cases in which the ecosystem is invaded by pathogens. Such situations are characterized by loss of mucosal homeostasis and the instigation of a rapid immune response. Indeed, many aspects of antimicrobial pathway and innate immune response regulation involve the circadian clock [15,16], as was first discovered in Drosophila [17]. In mice, the molecular clock regulates diurnal expression of proinflammatory genes in macrophages, thereby generating characteristic profiles of cytokine expression over the course of a day [18]. Furthermore, leukocyte abundance in the circulation and recruitment to peripheral tissues underlies strong circadian fluctuation [19]. The circadian clock also controls the expression of innate immune receptors, as has been described for TLR9 [20]. As a result, inflammatory responses, including susceptibility to sepsis, are strongly influenced by the time of day [19,20]. Major insights came from the study of cell-intrinsic clock functions in immune cells. Inflammatory monocytes with a cell type-specific deletion of Bmal1 feature a disrupted diurnal trafficking pattern, predisposing mice to inflammatory diseases [21].
In addition to the circadian regulation of innate immunity, the development of adaptive immune cells, in particular T helper (TH) 17 cells, has also been suggested to be regulated by the circadian clock. The clock transcription factor REV-ERBα controls the TH17-controlling transcription factor RORγt through NFIL3 [22], such that TH17 lineage specification is under circadian control. Recently, however, it has been found that mice with a T cell-specific deletion of Bmal1 do not feature defective adaptive immune responses [23], raising the possibility that the observed regulation of TH17 cells may involve cell-extrinsic factors.
As a result of the circadian variation in immune system potency, the susceptibility of the host to pathogenic infection varies over the course of a day. For instance, the degree of the immune response to oral infection with Salmonella Typhimurium is more pronounced when the infection occurs during the active phase of the host [24], likely to anticipate a higher risk to acquire foreign microbial elements during the time of food intake. The time of day therefore also affects the ability of the host to clear infection. For both S. Typhimurium and Listeria monocytogenes, it has been found that pathogen clearance varies with circadian time, a phenomenon that is not apparent in mice with genetic deletion of clock components [21,24]. Whether the immune system is also involved in circadian variations in the antimicrobial response against commensals and whether such responses may regulate diurnal rhythms of the microbiome remain elusive.
What Are the Consequences of Interkingdom Diurnal Rhythmicity?
The findings described above establish diurnal activity as a new principle in host–microbial interactions. The microbiota has emerged as a major mediator in the interaction of the host with its environment, so rhythmic adaptation of the eukaryotic and prokaryotic part of the “metaorganism”to the time of day might have been an important selective feature during evolution. There are three important conclusions that can be drawn from the first studies in this emerging field. First, it seems that biological clock systems are at work in a large range of biological contexts, ranging from cell-intrinsic clocks consisting of only three proteins, to larger transcriptional networks, oscillations at the level of organ function, and whole-organism behavior, to even symbiotic community-wide cross regulation of diurnal activity between different domains of life. The full spectrum of rhythmicity levels that are featured by the microbiome remains to be investigated and might likewise encompass important cell-intrinsic, transcriptomic, and metabolomics oscillations. Such insights will be fundamental for the mechanistic understanding of daily rhythmicity and its consequences in host–microbial interactions.
Second, the diurnality in host–microbiota interactions might, at least in part, account for the critical role that the biological clock plays in regulation of the host’s metabolic homeostasis. Indeed, microbiota diurnal rhythms have been recently suggested to exist in humans, to be disturbed upon clock disruption of the host, and consequently to drive metabolic aberrations [12]. Thus, in addition to morbidity induced by pathogens, microbiome diurnal rhythmicity may also influence noninfectious disease pathogenesis, including common multifactorial diseases that have been linked to disruptions in the circadian clock [14].
Finally, the findings of interdependent diurnal behavior in symbiotic communities may have another important consequence. They suggest that a stable state in such an ecosystem is not characterized by the static maintenance of community structure and function, but rather by hour-scale fluctuations around a homeostatic set point. Such an oscillating system might be more capable of meeting the constraints imposed on the community by environmental and geophysical variations over the course of a day, since deviations from the normal state might be more rapidly achieved by modifying the amplitude or frequency of rhythmically occurring oscillations.
The field of circadian rhythms in host–microbial interactions is still in its infancy, and the major functional principles and mechanisms have yet to be understood. Nonetheless, the first studies have already shown the great promise that this field holds for our understanding of the factors regulating host–microbial interactions, ecosystem stability, susceptibility to infection, and metabolic disorders—a list that may be further expanded in the years to come.
Zdroje
1. Hardin PE, Panda S. Circadian timekeeping and output mechanisms in animals. Current opinion in neurobiology. 2013;23(5):724–31. doi: 10.1016/j.conb.2013.02.018 23731779
2. Dibner C, Schibler U, Albrecht U. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annual review of physiology. 2010;72 : 517–49. doi: 10.1146/annurev-physiol-021909-135821 20148687
3. Mohawk JA, Green CB, Takahashi JS. Central and peripheral circadian clocks in mammals. Annual review of neuroscience. 2012;35 : 445–62. doi: 10.1146/annurev-neuro-060909-153128 22483041
4. Kondo T, Strayer CA, Kulkarni RD, Taylor W, Ishiura M, Golden SS, et al. Circadian rhythms in prokaryotes: luciferase as a reporter of circadian gene expression in cyanobacteria. Proceedings of the National Academy of Sciences of the United States of America. 1993;90(12):5672–6. 8516317
5. Tomita J, Nakajima M, Kondo T, Iwasaki H. No transcription-translation feedback in circadian rhythm of KaiC phosphorylation. Science. 2005;307(5707):251–4. 15550625
6. Nakajima M, Imai K, Ito H, Nishiwaki T, Murayama Y, Iwasaki H, et al. Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science. 2005;308(5720):414–5. 15831759
7. Pattanayak GK, Phong C, Rust MJ. Rhythms in energy storage control the ability of the cyanobacterial circadian clock to reset. Current biology: CB. 2014;24(16):1934–8. doi: 10.1016/j.cub.2014.07.022 25127221
8. Edgar RS, Green EW, Zhao Y, van Ooijen G, Olmedo M, Qin X, et al. Peroxiredoxins are conserved markers of circadian rhythms. Nature. 2012;485(7399):459–64. doi: 10.1038/nature11088 22622569
9. Heath-Heckman EA, Peyer SM, Whistler CA, Apicella MA, Goldman WE, McFall-Ngai MJ. Bacterial bioluminescence regulates expression of a host cryptochrome gene in the squid-Vibrio symbiosis. mBio. 2013;4(2). doi: 10.1128/mBio.00167-13 23549919
10. Mukherji A, Kobiita A, Ye T, Chambon P. Homeostasis in intestinal epithelium is orchestrated by the circadian clock and microbiota cues transduced by TLRs. Cell. 2013;153(4):812–27. doi: 10.1016/j.cell.2013.04.020 23663780
11. Leone V, Gibbons SM, Martinez K, Hutchison AL, Huang EY, Cham CM, et al. Effects of Diurnal Variation of Gut Microbes and High-Fat Feeding on Host Circadian Clock Function and Metabolism. Cell host & microbe. 2015;17(5):681–689. doi: 10.1016/j.chom.2015.03.006 25891358
12. Thaiss CA, Zeevi D, Levy M, Zilberman-Schapira G, Suez J, Tengeler AC, et al. Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostasis. Cell. 2014;159(3):514–29. doi: 10.1016/j.cell.2014.09.048 25417104
13. Zarrinpar A, Chaix A, Yooseph S, Panda S. Diet and feeding pattern affect the diurnal dynamics of the gut microbiome. Cell metabolism. 2014;20(6):1006–17. doi: 10.1016/j.cmet.2014.11.008 25470548
14. Thaiss CA, Zeevi D, Levy M, Segal E, Elinav E. A day in the life of the meta-organism: diurnal rhythms of the intestinal microbiome and its host. Gut microbes. 2015;6(2):137–42. doi: 10.1080/19490976.2015.1016690 25901892
15. Curtis AM, Bellet MM, Sassone-Corsi P, O'Neill LA. Circadian clock proteins and immunity. Immunity. 2014;40(2):178–86. doi: 10.1016/j.immuni.2014.02.002 24560196
16. Scheiermann C, Kunisaki Y, Frenette PS. Circadian control of the immune system. Nature reviews Immunology. 2013;13(3):190–8. doi: 10.1038/nri3386 23391992
17. Shirasu-Hiza MM, Dionne MS, Pham LN, Ayres JS, Schneider DS. Interactions between circadian rhythm and immunity in Drosophila melanogaster. Current biology: CB. 2007;17(10):R353–5. 17502084
18. Keller M, Mazuch J, Abraham U, Eom GD, Herzog ED, Volk HD, et al. A circadian clock in macrophages controls inflammatory immune responses. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(50):21407–12. doi: 10.1073/pnas.0906361106 19955445
19. Scheiermann C, Kunisaki Y, Lucas D, Chow A, Jang JE, Zhang D, et al. Adrenergic nerves govern circadian leukocyte recruitment to tissues. Immunity. 2012;37(2):290–301. doi: 10.1016/j.immuni.2012.05.021 22863835
20. Silver AC, Arjona A, Walker WE, Fikrig E. The circadian clock controls toll-like receptor 9-mediated innate and adaptive immunity. Immunity. 2012;36(2):251–61. doi: 10.1016/j.immuni.2011.12.017 22342842
21. Nguyen KD, Fentress SJ, Qiu Y, Yun K, Cox JS, Chawla A. Circadian gene Bmal1 regulates diurnal oscillations of Ly6C(hi) inflammatory monocytes. Science. 2013;341(6153):1483–8. doi: 10.1126/science.1240636 23970558
22. Yu X, Rollins D, Ruhn KA, Stubblefield JJ, Green CB, Kashiwada M, et al. TH17 cell differentiation is regulated by the circadian clock. Science. 2013;342(6159):727–30. doi: 10.1126/science.1243884 24202171
23. Hemmers S, Rudensky AY. The Cell-Intrinsic Circadian Clock Is Dispensable for Lymphocyte Differentiation and Function. Cell reports. 2015;11(9):1339–49. doi: 10.1016/j.celrep.2015.04.058 26004187
24. Bellet MM, Deriu E, Liu JZ, Grimaldi B, Blaschitz C, Zeller M, et al. Circadian clock regulates the host response to Salmonella. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(24):9897–902. doi: 10.1073/pnas.1120636110 23716692
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek TRIM21 Promotes cGAS and RIG-I Sensing of Viral Genomes during Infection by Antibody-Opsonized VirusČlánek Effector OspB Activates mTORC1 in a Manner That Depends on IQGAP1 and Promotes Cell ProliferationČlánek Fundamental Roles of the Golgi-Associated Aspartyl Protease, ASP5, at the Host-Parasite InterfaceČlánek Modulation of the Surface Proteome through Multiple Ubiquitylation Pathways in African Trypanosomes
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2015 Číslo 10- Stillova choroba: vzácné a závažné systémové onemocnění
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Choroby jater v ordinaci praktického lékaře – význam jaterních testů
- Diagnostický algoritmus při podezření na syndrom periodické horečky
-
Všechny články tohoto čísla
- Expression of Concern: Misregulation of Underlies the Developmental Abnormalities Caused by Three Distinct Viral Silencing Suppressors in Arabidopsis
- Preparing for the Next Epidemic with Basic Virology
- Effectively Communicating the Uncertainties Surrounding Ebola Virus Transmission
- Translating Basic Research into Clinical Applications: Malaria Research at an NIH Lab
- A Gut Odyssey: The Impact of the Microbiota on Spore Formation and Germination
- Papillomavirus E6 Oncoproteins Take Common Structural Approaches to Solve Different Biological Problems
- Chronobiomics: The Biological Clock as a New Principle in Host–Microbial Interactions
- Dimensions of Horizontal Gene Transfer in Eukaryotic Microbial Pathogens
- Addressing the Complications of Ebola and Other Viral Hemorrhagic Fever Infections: Using Insights from Bacterial and Fungal Sepsis
- Time for Chocolate: Current Understanding and New Perspectives on Cacao Witches’ Broom Disease Research
- Ganglioside and Non-ganglioside Mediated Host Responses to the Mouse Polyomavirus
- Crosslinking of a Peritrophic Matrix Protein Protects Gut Epithelia from Bacterial Exotoxins
- Structure Elucidation of Coxsackievirus A16 in Complex with GPP3 Informs a Systematic Review of Highly Potent Capsid Binders to Enteroviruses
- CD39 Expression Identifies Terminally Exhausted CD8 T Cells
- Abiotic Stresses Antagonize the Rice Defence Pathway through the Tyrosine-Dephosphorylation of OsMPK6
- Dissociation of Tissue Destruction and Bacterial Expansion during Bubonic Plague
- Interferon-γ: The Jekyll and Hyde of Malaria
- CCR2 Inflammatory Dendritic Cells and Translocation of Antigen by Type III Secretion Are Required for the Exceptionally Large CD8 T Cell Response to the Protective YopE Epitope during Infection
- A New Glycan-Dependent CD4-Binding Site Neutralizing Antibody Exerts Pressure on HIV-1
- The Suramin Derivative NF449 Interacts with the 5-fold Vertex of the Enterovirus A71 Capsid to Prevent Virus Attachment to PSGL-1 and Heparan Sulfate
- Trans-generational Immune Priming Protects the Eggs Only against Gram-Positive Bacteria in the Mealworm Beetle
- Peripheral Vγ9Vδ2 T Cells Are a Novel Reservoir of Latent HIV Infection
- Respiratory Syncytial Virus Disease Is Mediated by Age-Variable IL-33
- TRIM21 Promotes cGAS and RIG-I Sensing of Viral Genomes during Infection by Antibody-Opsonized Virus
- Modeling the Effects of Vorinostat Reveals both Transient and Delayed HIV Transcriptional Activation and Minimal Killing of Latently Infected Cells
- Identification of a Novel Lipoprotein Regulator of Spore Germination
- Calcium Regulation of Hemorrhagic Fever Virus Budding: Mechanistic Implications for Host-Oriented Therapeutic Intervention
- Antigen-Specific Th17 Cells Are Primed by Distinct and Complementary Dendritic Cell Subsets in Oropharyngeal Candidiasis
- Comparative Life Cycle Transcriptomics Revises Genome Annotation and Links a Chromosome Duplication with Parasitism of Vertebrates
- The Autophagy Receptor TAX1BP1 and the Molecular Motor Myosin VI Are Required for Clearance of Salmonella Typhimurium by Autophagy
- Carcinogenic Parasite Secretes Growth Factor That Accelerates Wound Healing and Potentially Promotes Neoplasia
- Effector OspB Activates mTORC1 in a Manner That Depends on IQGAP1 and Promotes Cell Proliferation
- Dengue Virus Infection of Requires a Putative Cysteine Rich Venom Protein
- Distinct Viral and Mutational Spectrum of Endemic Burkitt Lymphoma
- Fundamental Roles of the Golgi-Associated Aspartyl Protease, ASP5, at the Host-Parasite Interface
- Phenotypic and Functional Alterations in Circulating Memory CD8 T Cells with Time after Primary Infection
- Systematic Identification of Cyclic-di-GMP Binding Proteins in Reveals a Novel Class of Cyclic-di-GMP-Binding ATPases Associated with Type II Secretion Systems
- Influenza Transmission in the Mother-Infant Dyad Leads to Severe Disease, Mammary Gland Infection, and Pathogenesis by Regulating Host Responses
- Myeloid Cell Arg1 Inhibits Control of Arthritogenic Alphavirus Infection by Suppressing Antiviral T Cells
- The White-Nose Syndrome Transcriptome: Activation of Anti-fungal Host Responses in Wing Tissue of Hibernating Little Brown Myotis
- Influenza Virus Reassortment Is Enhanced by Semi-infectious Particles but Can Be Suppressed by Defective Interfering Particles
- Identification of the Mechanisms Causing Reversion to Virulence in an Attenuated SARS-CoV for the Design of a Genetically Stable Vaccine
- Differentiation-Dependent KLF4 Expression Promotes Lytic Epstein-Barr Virus Infection in Epithelial Cells
- The Histone Acetyltransferase Hat1 Regulates Stress Resistance and Virulence via Distinct Chromatin Assembly Pathways
- C-di-GMP Regulates Motile to Sessile Transition by Modulating MshA Pili Biogenesis and Near-Surface Motility Behavior in
- Modulation of the Surface Proteome through Multiple Ubiquitylation Pathways in African Trypanosomes
- Crystal Structure of the Human Cytomegalovirus Glycoprotein B
- Depletion of . GlmU from Infected Murine Lungs Effects the Clearance of the Pathogen
- Immunologic Control of Papillomavirus Type 1
- Requires Host Rab1b for Survival in Macrophages
- Structure Analysis Uncovers a Highly Diverse but Structurally Conserved Effector Family in Phytopathogenic Fungi
- PD-L1 Expression on Retrovirus-Infected Cells Mediates Immune Escape from CD8 T Cell Killing
- Phospho-dependent Regulation of SAMHD1 Oligomerisation Couples Catalysis and Restriction
- IL-4 Induced Innate CD8 T Cells Control Persistent Viral Infection
- Crystal Structures of a Piscine Betanodavirus: Mechanisms of Capsid Assembly and Viral Infection
- BCG Skin Infection Triggers IL-1R-MyD88-Dependent Migration of EpCAM CD11b Skin Dendritic cells to Draining Lymph Node During CD4+ T-Cell Priming
- Antigenic Characterization of the HCMV gH/gL/gO and Pentamer Cell Entry Complexes Reveals Binding Sites for Potently Neutralizing Human Antibodies
- Rescue of a Plant Negative-Strand RNA Virus from Cloned cDNA: Insights into Enveloped Plant Virus Movement and Morphogenesis
- Geminivirus Activates to Accelerate Cytoplasmic DCP2-Mediated mRNA Turnover and Weakens RNA Silencing in
- Disruption of Sphingolipid Biosynthesis Blocks Phagocytosis of
- The Fungal Exopolysaccharide Galactosaminogalactan Mediates Virulence by Enhancing Resistance to Neutrophil Extracellular Traps
- The Timing of Stimulation and IL-2 Signaling Regulate Secondary CD8 T Cell Responses
- Structural and Functional Analysis of Murine Polyomavirus Capsid Proteins Establish the Determinants of Ligand Recognition and Pathogenicity
- The Dual Role of an ESCRT-0 Component HGS in HBV Transcription and Naked Capsid Secretion
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Chronobiomics: The Biological Clock as a New Principle in Host–Microbial Interactions
- Interferon-γ: The Jekyll and Hyde of Malaria
- Crosslinking of a Peritrophic Matrix Protein Protects Gut Epithelia from Bacterial Exotoxins
- Antigen-Specific Th17 Cells Are Primed by Distinct and Complementary Dendritic Cell Subsets in Oropharyngeal Candidiasis
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



