-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Abiotic Stresses Antagonize the Rice Defence Pathway through the Tyrosine-Dephosphorylation of OsMPK6
Chemical defence inducers make plants resistant to diseases such as rice blast. However, plants sometimes become more pathogen susceptible under abiotic stresses even in their presence. Because such regulation prioritizes the responses to the most life-threatening stress, it could be necessary for plants to survive in nature. However, it seems dispensable or even disadvantageous for crops cultivated under fertile conditions. Here, we show the molecular mechanism underlying one of such phenomena in rice. WRKY45 is a central transcription factor that regulates strong defence signalling mediated by salicylic acid. We found that WRKY45 is activated through phosphorylation by a protein kinase, OsMPK6, which is activated by dual phosphorylation in response to the defence signalling. We also found that OsMPK6 can be inactivated by tyrosine dephosphorylation in response to abiotic stresses such as low temperature and high salinity probably mediated by abscisic acid, leading to reduction of WRKY45-dependent disease resistance. Moreover, we found that specific tyrosine protein phosphatases dephosphorylate/inactivate OsMPK6 in response to abiotic stresses. Knockdown of their genes rendered rice plants resistant against blast disease even under the abiotic stresses, pointing to the way to further improve rice.
Published in the journal: . PLoS Pathog 11(10): e32767. doi:10.1371/journal.ppat.1005231
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1005231Summary
Chemical defence inducers make plants resistant to diseases such as rice blast. However, plants sometimes become more pathogen susceptible under abiotic stresses even in their presence. Because such regulation prioritizes the responses to the most life-threatening stress, it could be necessary for plants to survive in nature. However, it seems dispensable or even disadvantageous for crops cultivated under fertile conditions. Here, we show the molecular mechanism underlying one of such phenomena in rice. WRKY45 is a central transcription factor that regulates strong defence signalling mediated by salicylic acid. We found that WRKY45 is activated through phosphorylation by a protein kinase, OsMPK6, which is activated by dual phosphorylation in response to the defence signalling. We also found that OsMPK6 can be inactivated by tyrosine dephosphorylation in response to abiotic stresses such as low temperature and high salinity probably mediated by abscisic acid, leading to reduction of WRKY45-dependent disease resistance. Moreover, we found that specific tyrosine protein phosphatases dephosphorylate/inactivate OsMPK6 in response to abiotic stresses. Knockdown of their genes rendered rice plants resistant against blast disease even under the abiotic stresses, pointing to the way to further improve rice.
Introduction
Plants, as sessile organisms, are continuously exposed to various environmental stresses in nature. To cope with such conditions using limited resources, plants have evolved various mechanisms that enable resource allocation to the most life-threatening stress [1] [2]. Such tradeoffs between the responses to different stresses are often regulated by crosstalk between signalling pathways [3] [4] [5]. A number of studies have reported various signalling components that appear to influence signalling crosstalk. However, the precise molecular mechanisms that regulate the crosstalk remain poorly understood in most cases [6] [4] [7].
The salicylic acid (SA) signalling pathway plays a crucial role in pathogen defence. In Arabidopsis, NPR1, the transcriptional cofactor, plays a major role in the SA defence signalling pathway [8]. In rice (Oryzae sativa), in addition to OsNPR1/NH1, the rice ortholog of NPR1, the transcription factor (TF) WRKY45 plays a crucial role in the branched SA pathway [9] [10] [11] [12]. Up-regulation of WRKY45 by chemical defence inducers, such as benzothiadiazole (BTH), or its overexpression, renders rice plants resistant against several pathogens, including fungus, such as Magnaporthe oryzae causing blast disease, and bacterium, such as Xanthomonas oryzae pv. oryzae causing leaf blight disease [9] [13] [14], without major negative effects on plant growth. WRKY45 auto-regulates the transcription of its own gene [12] and is regulated by the ubiquitin-proteasome system [15].
Abscisic acid (ABA) signalling is mainly involved in plant responses to abiotic stresses, such as the cold, drought, and high salinity [16] [17]. However, ABA also acts as a modulator of defence responses against pathogens, both positively and negatively, with its negative role being more prevalent [18] [3] [19] [20] [4] [21] [5] [22]. Recent studies have shown that ABA antagonizes SA-signalling, thereby interfering with defence responses in tomato, Arabidopsis, and rice [23] [24] [25].
The WRKY TFs can be phosphorylated and activated by MAP kinases, as is the case with Arabidopsis WRKY33 [26] and Nicotiana benthamiana WRKY8 [27]. The negative regulation of MAP kinases through dephosphorylation by protein phosphatases, including Ser/Thr-specific phosphatases, dual-specificity phosphatases, and Tyr-specific phosphatases (PTPases), has been reported [28] [29]. We have previously reported that activated MAP kinases can phosphorylate WRKY45 in vitro, and MAP kinases can be activated in response to SA [30]. However, details of WRKY45 phosphorylation, and the biological significance of the phosphorylation have remained unknown.
Rice blast is one of the most serious crop diseases in the world. It has been reported that rice plants are more blast susceptible under abiotic stresses, such as low temperature and drought [31] [32] [33]. In rice, ABA treatment severely compromised M. oryzae resistance [34], which is mediated by suppression of WRKY45 and OsNPR1/NH1 genes via ABA signalling [25] [35]. These authors suggest that ABA signalling plays a role in increased blast susceptibility under low temperature.
In this report, we show the mechanism underlying blast resistance through the activation of WRKY45 by MAP kinase (MAPK)-dependent phosphorylation in the SA pathway. Moreover, we showed that the tyrosine dephosphorylation of the MAPK by PTPases, OsPTP1/2, is responsible for the MAPK inactivation under abiotic stresses or in the presence of exogenous ABA. Additionally, the knockdown of OsPTP1/2 uncoupled the induced blast resistance from the abiotic stresses. These findings should enable the development of technologies to protect rice from diseases even under the influence of environmental factors.
Results
Phosphorylation is required for full activation of WRKY45
WRKY45 was phosphorylated by OsMPK6 in vitro in the presence of a constitutively active form of OsMKK10-2 (MKK10-2D, Os03g0225100, LOC_Os03g12390.1), which is a rice MAPK kinase that phosphorylates and activates OsMPK6 in vitro [30]; however, its biological significance was unknown. To further analyse the WRKY45 phosphorylation, we therefore determined the sites in WRKY45 that are phosphorylated by OsMPK6. Incubation in vitro of fragmented WRKY45 polypeptides fused to a maltose binding protein (MBP) with OsMPK6 revealed that only two regions (amino acids 239–292 and 281–326) of WRKY45 were phosphorylated (S1 Fig). By a consequent substitution study on candidate phosphorylation sites (Ser or Thr), we found that Thr266, Ser294, and Ser299 were required for phosphorylation of WRKY45 by OsMPK6 in vitro (S2 Fig).
To assess the effects of the phosphorylation on transcriptional activity, we generated mutant forms of WRKY45 in which all the three amino acids were replaced by Asn (NNN) or Asp (DDD) to mimic dephosphorylation and phosphorylation of all three sites, respectively, and tested them by transient reporter assays (Fig 1A). The transactivation activity of DDD was 2–4-fold higher than that of NNN (Fig 1B), indicating that the phosphomimetic mutation elevated the transcriptional activity of WRKY45. These results suggest that WRKY45 phosphorylation results in its activation in vivo. The level of wild-type (WT) WRKY45 activity was between those of NNN and DDD (Fig 1B), consistent with its partial phosphorylation state. We attempted to generate transgenic plants overexpressing NNN and DDD mutants tagged with the myc sequence at their N-termini, but we obtained neither of them. Then, we focused on the carboxyl-terminal region that contained two of the three phosphorylation sites, the closely located Ser residues, Ser294 and Ser299, because the carboxyl-terminal region of WRKY45 is critical for transcriptional activity [15]. We generated mutants in which the two serines were substituted with Asp (TDD) or Asn (TNN), and attempted to overexpress the mutant cDNAs in rice transformants (Fig 1A). While we were unable to obtain the transformants for TDD, we obtained several for TNN. Whereas the TNN transformants accumulated the transgene-derived proteins to levels comparable, or even slightly higher, than those in WT WRKY45-overexpressing (ox) transformants, they showed no significant enhancement of blast resistance (Fig 1C), indicating that the substitution of these two serines compromised the function of WRKY45. To examine the contribution of the phosphorylation at these two Ser to transcriptional activity, we generated new mutants, DNN and NDD, and analysed them in the transient system (Fig 1A). The relative activity of NDD was as high as that of DDD, while that of DNN was as low as that of the NNN mutant (Fig 1D). These results suggest that the phosphorylation of Ser294 and/or Ser299, but not that of Thr266, is important for the transcriptional activity of WRKY45, consistent with the results of blast resistance test (Fig 1C).
Fig. 1. Phosphorylation at carboxyl-terminal serines activates WRKY45 in rice. 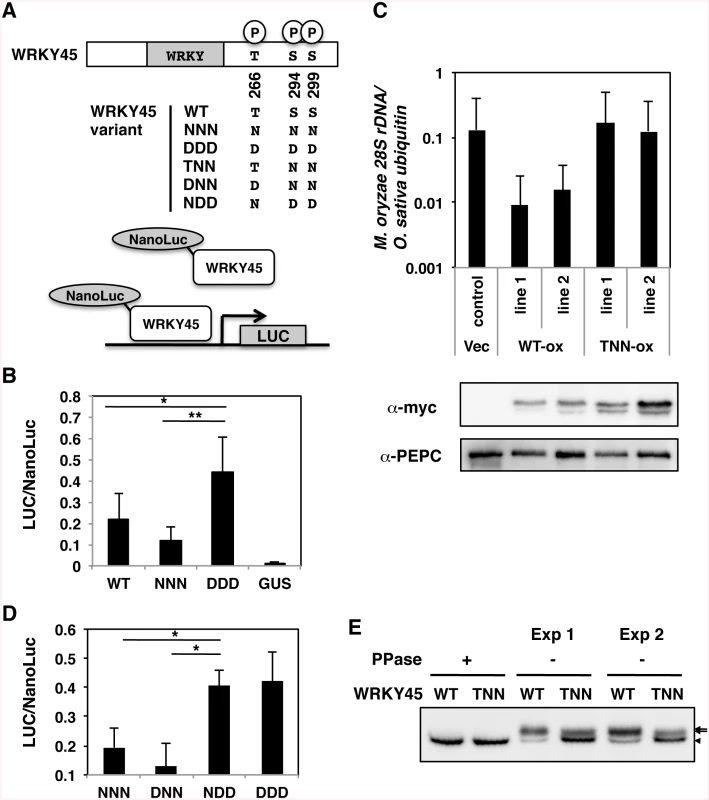
(A) Schematic representation of WRKY45 variants and the reporter assay system. Luciferase activities, based on the transactivation by NanoLuc-WRKY45 fusion proteins per total NanoLuc activity, were determined as specific transactivation activities. N and D represent dephosphomimetic (Asn) and phosphomimetic (Asp) substitutions at the Thr266, Ser294, and Ser299of the WRKY45 proteins. (B) Specific transactivation activities of wild-type (WT) and mutant WRKY45. The mutants in which the three amino acids were changed to N (NNN) or D (DDD) simultaneously were assayed as described in the Methods. Means of three or more biological replicates are shown with SD. *, P < 0.02; **, P<0.002 (Student’s t-test). (C) Blast resistance assay of myc-tagged WT and mutant WRKY45-overexpressing (ox) rice plants. Blast resistance was evaluated by quantifying M. oryzae 28S rDNA using genomic qPCR. Means of three or more biological replicates are shown with SD. Accumulation of the WT and mutant WRKY45 proteins, in which the two serines were substituted with Asn (TNN), were detected by anti-myc antibody. Phosphoenolpyruvate carboxylase (PEPC) was immunodetected as the loading control. (D) Specific transactivation activities of DNN and NDD mutants of WRKY45, in which the three amino acids were independently changed to N or D. The assays were performed similarly to those in (B). (E) Immunodetection of the WT and TNN mutant of WRKY45 proteins in plants was performed after treatment with (+) or without (-) lambda protein phosphatase (PPase) in vitro. Two independent sets of experiments were done (Exp 1 and -2). Arrows and an arrowhead indicate phosphorylated and dephosphorylated proteins, respectively. Numbers of bands appear to be different from those in 2C presumably because of different resolution of the gels. To examine whether these two serines in WRKY45 are phosphorylated in vivo, we treated the extracts from WT and TNN WRKY45-ox plants with lambda protein phosphatase (PPase). The electrophoretic mobility of the TNN form treated with the PPase was indistinguishable from that of WT WRKY45 (Fig 1E), indicating that the effect of Ser-to-Asn substitutions at the two sites on the electrophoretic mobility is negligible. Without the PPase treatment, two bands were seen in both WT and TNN extracts (Fig 1E), suggesting that both WT and the mutant WRKY45 proteins were phosphorylated in vivo. The slower mobility (upper) bands in WT extracts were broad and much more intense than the faster mobility (lower) band for the unphosphorylated WRKY45 protein. By contrast, the upper band in the TNN extract was less intense than the lower band and thinner. These reproducible results suggest that Ser294 and/or Ser299 are/is actually phosphorylated in plant cells. This idea was also supported by the results of Phos-Tag polyacrylamide gel electrophoresis [36] (S3 Fig). Taken together, the phosphorylation of Ser294 and/or Ser299 of WRKY45 and consequent activation of its transcriptional activity are required for the full functioning of WRKY45-dependent blast resistance.
OsMPK6 was dually phosphorylated in response to SA in rice plants
We have previously shown that OsMPK6 becomes active in response to SA [30]. Here, we monitored the activation state of OsMPK6 by examining the dual phosphorylation of MAPK at Thr and Tyr in the TEY-signature (position 225–227 in OsMPK6) [37] in calli. An immunoblot using anti-pTEpY antibody showed that a band, which is missing in osmpk6 mutant and corresponds to that for OsMPK6 in gel mobility, was intensified by the SA-treatment in WT calli (Fig 2). The increase of the band intensity in response to SA was rather weak, which we interpret to be due to sporadically elevated basal OsMPK6 phosphorylation level because of high SA levels in rice. In the osmpk6 mutant calli, a faster migrating band appeared, consistent with our previous observations in an in-gel kinase assay [30]. In leaves, OsMPK6 was also dually phosphorylated and another band appeared (Fig 2). These results suggest that OsMPK6 is a major MAPK activated by SA.
Fig. 2. Salicylic acid (SA) induces OsMPK6 phosphorylation. 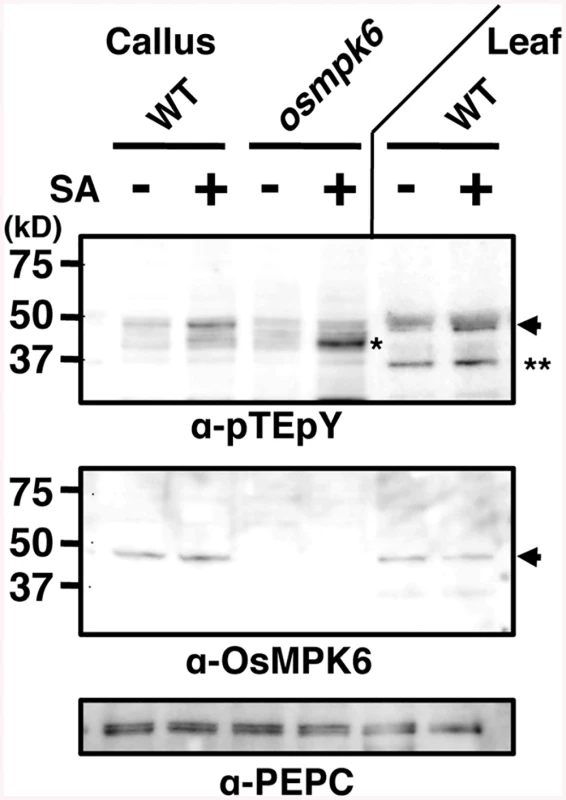
Rice calli or leaves were treated with 1 mM SA, and proteins with a doubly phosphorylated TEY motif were immunodetected with an anti-pTEpY antibody. NB was used as WT leaf source. OsMPK6 and PEPC were detected by specific antibodies. Arrows, OsMPK6; * and **, other putative MAPKs. Activation of OsMPK6 by OsMKK10-2 is sufficient for WRKY45 induction and blast resistance
To mimic the activation of the SA pathway that leads to the activation of OsMPK6 and then WRKY45, we performed in vitro kinase assays using MKK10-2D and mutant forms of OsMPK6 as substrates (Figs 3A and S4). A kinase-dead form of OsMPK6, K96R, was phosphorylated by MKK10-2D in vitro; however, another kinase-dead form, in which, in addition to the K96R mutation, the Thr and Tyr in the TEY-signature were replaced by Asp (K96R/T225D/Y227D), was not (Fig 3A). In addition, OsMPK6 mutants in which the Tyr and Thr were independently substituted, T225D and Y227A, were less phosphorylated than the WT OsMPK6 (S4 Fig). These results indicate that MKK10-2D phosphorylates the TEY-signature of OsMPK6 specifically and suggest that OsMPK6 is activated by OsMKK10-2 through the specific phosphorylation.
Fig. 3. Activation of OsMPK6 is sufficient for the WRKY45 induction and blast resistance. 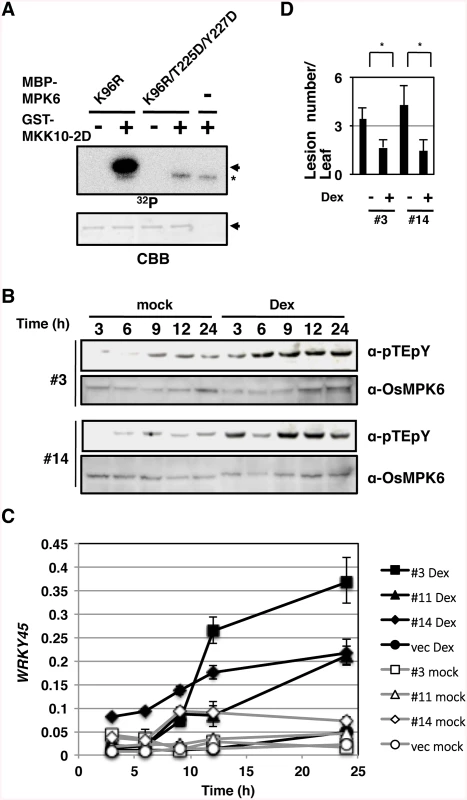
(A) MKK10-2D phosphorylates OsMPK6 in vitro. Glutathione-S-transferase (GST)- MKK10-2D exclusively phosphorylates Thr225 and Tyr227 of maltose binding protein (MBP)-MPK6 in vitro. K96R, a kinase-dead form of OsMPK6 in which Lys96 in the MBP-MPK6 was replaced by Arg; T225D and Y227D, OsMPK6 mutant in which Thr225 and Tyr227, respectively, were replaced by Asp; Arrows, MBP-MPK6; *, GST-MKK10-2D; CBB, Coomassie brilliant blue staining. (B) Dexamethasone (Dex)-induced MKK10-2D phosphorylates the TEY motif. Two independent GVG-MKK10-2D lines (#3 and #14) were treated with Dex, and proteins were detected with anti-pTEpY and anti-OsMPK6 antibodies. (C) Dex-induced MKK10-2D expression up-regulates WRKY45. GVG-MKK10-2D plants were Dex- or mock-treated, and the level of WRKY45 transcripts relative to that of rice ubiquitin transcripts was determined at indicated times by RT-qPCR. (D) MKK10-2D expression induces blast resistance. Dex-inducible MKK10-2D plants were inoculated with blast fungi with or without Dex pretreatment. Values are mean lesion number/leaf (n = 12 biological replicates) with SEM [58]. *, P < 0.1 (Student’s t-test). Then, we expressed MKK10-2D in rice plants using the dexamethasone (Dex)-inducible system (GVG-MKK10-2D) [38] and monitored the activation of OsMPK6 by pTEpY antibody. The dual phosphorylation was induced after the Dex-treatment in two independent GVG-MKK10-2D lines (Fig 3B, α-pTEpY). In these plants, WRKY45 expression (Fig 3C) and blast resistance (Fig 3D) were also induced after the Dex treatment. These results demonstrate that the activation of OsMPK6 by MKK10-2D is sufficient for the induction of WRKY45 expression and blast resistance without exogenous SA or BTH.
OsMKK4 can also phosphorylate and activate OsMPK6 in vitro and in vivo [39] [30]. However, the induced expression of the OsMKK4 constitutively active form using the Dex induction system failed to induce WRKY45 expression, while phenylalanine ammonia lyase gene, as a positive control, was induced (S5 Fig). These results suggest that OsMKK10-2, but not OsMKK4, is involved in the MAPK cascade in the SA signalling pathway leading to WRKY45 up-regulation and blast resistance.
OsPTP1/2 dephosphorylates a Tyr in OsMPK6 in response to ABA
Then, we investigated the effects of ABA on the OsMPK6–WRKY45 pathway in GVG-MKK10-2D lines (Fig 4A and S6 Fig). We induced OsMKK10-2D in the GVG-MKK10-2D transformants by Dex treatment in the presence of ABA. Interestingly, the accumulation of WRKY45 transcripts was severely reduced by the ABA treatment, although the induction of the MKK10-2D transgene was unaffected by ABA (Fig 4A). These results imply a possibility that ABA-signalling interfered with the SA pathway by affecting some molecular event(s) between OsMKK10-2 activation and WRKY45 transcription. We examined whether ABA-signalling affects the phosphorylation status of OsMPK6. Strikingly, the ABA treatment significantly lowered the dual phosphorylation level of OsMPK6 in parallel with the repression of WRKY45 transcription (Fig 4A and S6 Fig). These results suggest that ABA-signalling dephosphorylates OsMPK6 or inhibits its phosphorylation by MKK10-2D.
Fig. 4. Tyr-specific dephosphorylation of OsMPK6 and suppression of WRKY45 expression in response to ABA. 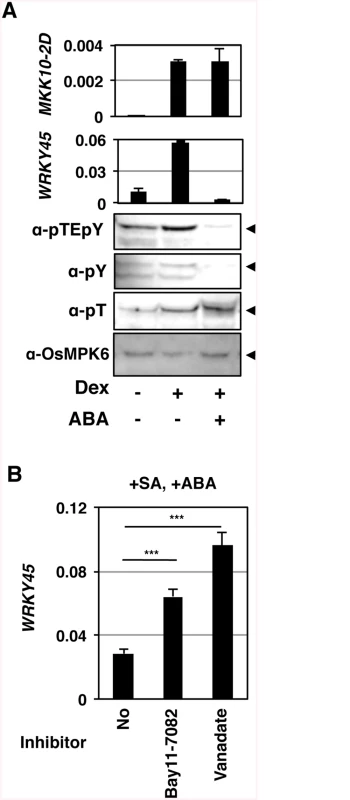
(A) GVG-MKK10-2D plants (line #3) were treated with 10 μM Dex and 10 μM ABA. Means of three technical repeats (RT-qPCR relative to ubiquitin) with pooled samples are shown with SD. pY, anti-phosho-Tyr; pT, anti-phospho-Thr; arrowheads, OsMPK6. Similar results were obtained with another transformant line (S6 Fig). (B) Tyr-specific phosphatase (PTPase) inhibitors inhibit the suppression of salicylic acid (SA)-induced WRKY45 transcription by ABA. WT rice plants (NB) were treated with 1 mM SA and 100 μM ABA, in the presence or absence of 50 μM Bay11-7082 or 2 mM vanadate. Means of six or more biological replicates (RT-qPCR relative to rice ubiquitin 1) are shown with SEM. ***, P < 0.001 (Student’s t-test). In Arabidopsis, MAPK can be dephosphorylated by Ser/Thr-phosphatases, dual-specificity phosphatases, and PTPases [28] [29]. This information, taken together with our results, led us to presume that OsMPK6 could be dephosphorylated in response to ABA, rather than ABA inhibiting the MAPK kinase. To further investigate the phosphorylation state of OsMPK6, we used anti-phospho-Tyr and anti-phospho-Thr specific antibodies in an immunoblot analysis. While the phospho-Thr signal was unaffected, the phospho-Tyr signal became faint in parallel with that of dual phosphorylation (Fig 4A and S6 Fig), suggesting that the Tyr, but not Thr, of OsMPK6 was dephosphorylated in response to ABA. Based on these results, we predicted the involvement of dephosphorylation of OsMPK6 by PTPases in the action of ABA. To assess the possible involvement of PTPases, we treated rice leaf segments with SA and ABA in the presence or absence of the PTPase inhibitors, Bay11-7082 [40] and vanadate. In the absence of these inhibitors, WRKY45 transcripts dramatically increased after the SA treatment; however, the co-treatment with ABA largely compromised the induction (S7 Fig) [25]. Meanwhile, the reduction of WRKY45 transcript levels by ABA was significantly less in the presence of these inhibitors (Fig 4B and S7 Fig). Taken together with the results of immunodetection described above, we postulated that ABA-responsive PTPase(s) dephosphorylated/inactivated OsMPK6, which in turn deactivated WRKY45 by under-phosphorylation, leading to the reduction of WRKY45 transcripts. The rice genome has two genes encoding putative Tyr-specific PTPases, OsPTP1 (Os12g0174800, LOC_Os12g07590) and OsPTP2 (Os11g0180200, LOC_Os11g07850.1) (S8 Fig), which have high homology to AtPTP1, a Tyr phosphatase that dephosphorylates MPK6 [41, 42]. Active site signature of Tyrosine phosphatase (S8 Fig), which is conserved in PTPs from all the organisms [43], is also present in OsPTP1 and -2, further lending support for these proteins being Tyr-specific phosphatases. To test this hypothesis, we generated rice transformants in which both PTPase genes were knocked down using the construct shown in Fig 5A (PTP-wkd). Then, we investigated the dual phosphorylation of OsMPK6 in WT [Nipponbare (NB)] and PTP-wkd rice plants after SA treatment in the absence or presence of ABA (Fig 5B). In NB, the dual phosphorylation level increased after the SA treatment, but the increase was completely cancelled in the presence of ABA. In PTP-wkd rice, ABA did not suppress the level of dual phosphorylation, which further increased after SA treatment. These results strongly suggest that OsPTP1/2 is involved in ABA-dependent dephosphorylation of OsMPK6. In the absence of ABA in PTP-wkd rice, the dual phosphorylation level was relatively high even without SA treatment; thus, OsPTP1/2 could also play a role in reducing the basal level of OsMPK6 phosphorylation.
Fig. 5. Dephosphorylation of OsMPK6 in response to ABA requires OsPTP1/2. 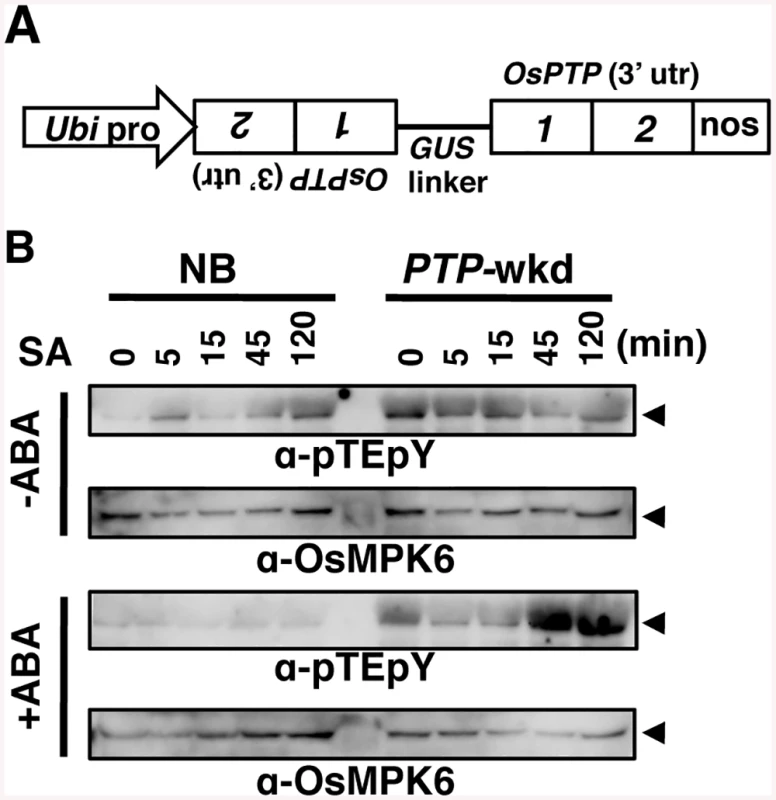
(A) Schematic representation of the double OsPTP1/2 knockdown (PTP-wkd) construct. (B) PTP knockdown antagonizes the TEY dephosphorylation by ABA. NB (WT) and PTP-wkd plants were treated for the periods indicated with 1 mM SA with or without 100 μM ABA, and proteins were detected using indicated antibodies. To test whether OsPTP1/2 directly dephosphorylate OsMPK6, we performed in vitro dephosphorylation assays (Fig 6). Phospho-Tyr, but not phosphor-Thr, of OsMPK6, due to phosphorylation by OsMKK10-2, decreased when incubated with WT OsPTP1 fused with MBP (Fig 6, upper panels). However, the decrease of signals was not significant when incubated with a mutant OsPTP1 in which catalytically essential cysteine [44] (positions 258, S8 Fig) was replaced with serine (Fig 6). We also monitored the ability of OsMPK6 to phosphorylate WRKY45 by the addition of recombinant WRKY45 and [γ-32P] ATP to the system preincubated with unlabelled ATP to activate OsMPK6. The WRKY45 phosphorylation was completely abolished in the presence of WT OsPTP1, whereas the phosphorylation level remained unchanged in the presence of the mutant PTPase (Fig 6, lower panels). These results indicate that OsPTP1 directly dephosphorylates OsMPK6 and inactivates its WRKY45 phosphorylation activity. These results support the notion that OsPTP1 mediates the suppression of the SA-OsMPK6-WRKY45 pathway via ABA-signalling. We did not detect evident activities for OsPTP2 using the same reaction mixtures, possibly because of suboptimal conditions.
Fig. 6. OsPTP1 directly dephosphorylates and inactivates OsMPK6 in vitro. 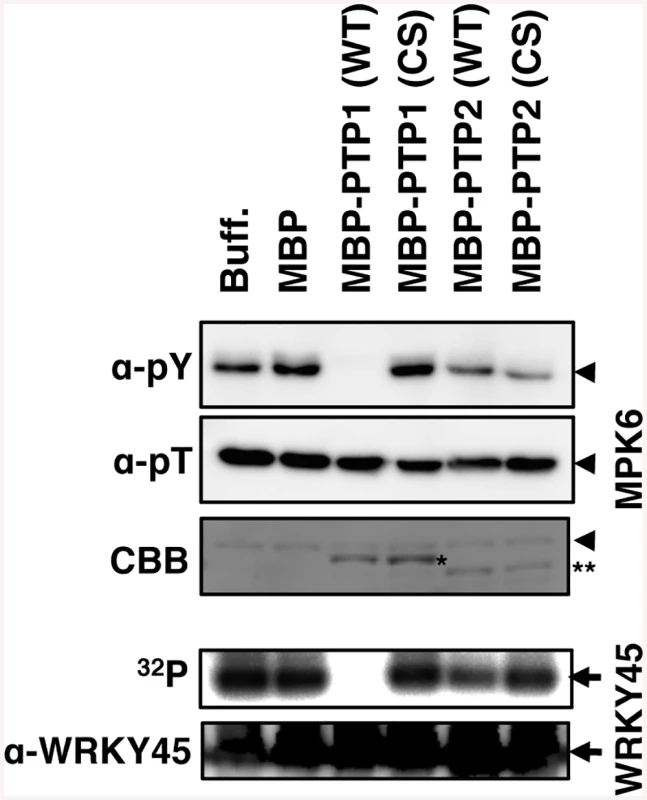
To assay OsMPK6 dephosphorylating activity of the PTPs (upper panels), after incubation of WT or mutant form MBP-PTP1/2 with GST-MKK10-2D for 2 h at room temperature, phospho-Tyr and phospho-Thr of MBP-MPK6 was monitored by immuno blot using anti-pY and anti-pT antibodies, respectively. To assay WRKY45 phosphorylating activity of OsMPK6 (lower panels), after incubation of WT or mutant form of MBP-PTP1/2 with GST-MKK10-2D and unlabelled ATP for 2 h at room temperature, the mixture was further incubated with [γ-32P]ATP and MBP-WRKY45 for 20 min and monitored for 32P-incorporation into MBP-WRKY45. The lanes MBP and Buff are the controls with or without maltose binding protein, respectively. CS represents mutant PTPases with Cys-to-Ser substitutions. Coomassie brilliant blue (CBB) staining and immunoblot with anti-WRKY45 antibody are shown as loading controls. Arrowheads, MBP-MPK6; arrows, MBP-WRKY45; *, MBP-PTP1; **, MBP-PTP2. Induced blast resistance was compromised by ABA or abiotic stresses
BTH, a chemical defence inducer, enhances disease resistance by acting on the SA signalling pathway in various plants, including rice [45] [46]. In Arabidopsis, the effect of BTH on defence responses is compromised by high salt conditions acting through ABA signalling [24]. To test the effects of abiotic stresses on BTH-induced blast resistance in rice, we pretreated WT rice plants, NB, with 10 μM ABA, low temperature (15°C/9°C, day/night), or high salinity (250 mM NaCl) in the presence or absence of 10 μM BTH, inoculated them with M. oryzae and monitored for fungal growth (Fig 7). BTH confers a strong resistance against blast disease in rice [9]. Co-treatment with ABA compromised the resistance as reported previously [25]. Interestingly, BTH-treated plants under the cold and high salinity conditions were more highly susceptible to blast than the control plants, similar to the ABA/BTH co-treated plants (Fig 7). These results imply a possibility that the abiotic stresses mediated by ABA signalling negatively affected the BTH-induced blast resistance in rice.
Fig. 7. Blast resistance of WT and PTP-wkd plants under abiotic stress conditions. 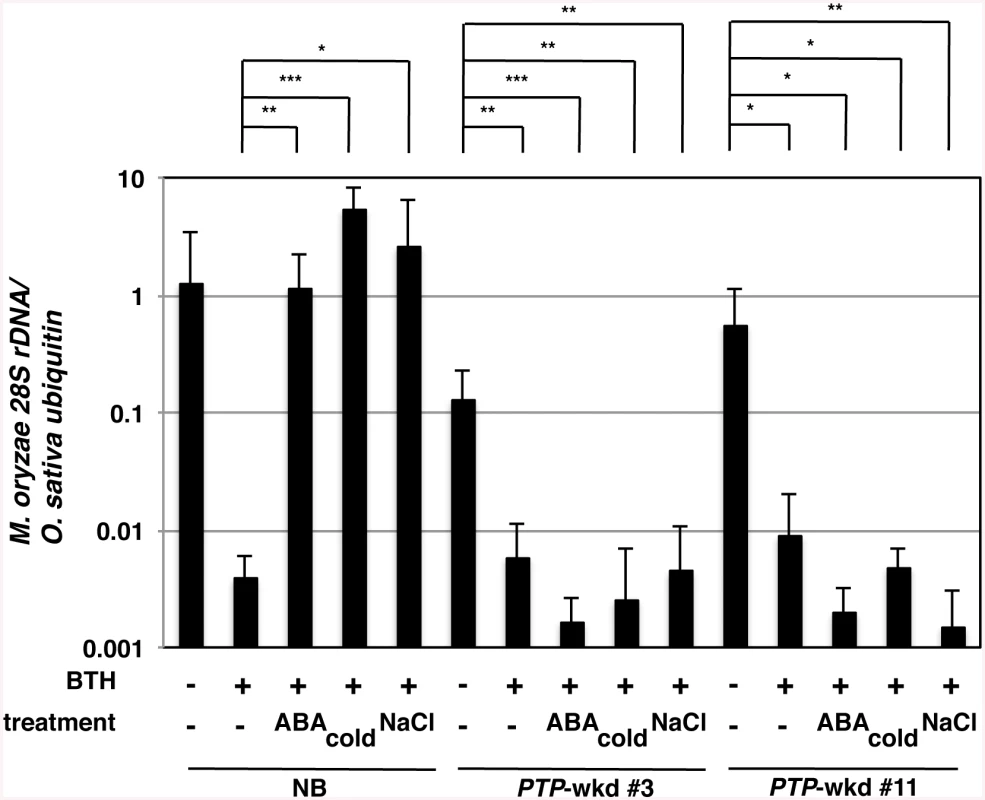
Detached leaf sheaths of wild-type Nipponbare [NB(WT)] or PTP-wkd (lines #3 and #11) plants treated with (+) or without (-) BTH (10 μM) and/or ABA (10 μM with 100 μM fluridone), or cold (15°C/9°C, day/night) were inoculated with Magnaporthe oryzae. Although we added fluridone with ABA to avoid feedback regulation of ABA biosynthesis, results were similar with or without fluridone. For the high salinity condition, intact plants were soaked in NaCl (250 mM) for 4 days prior to detaching leaves. Blast resistance was evaluated by quantifying M. oryzae 28S rDNA relative to that of rice ubiquitin 1 using genomic qPCR. Means of three or more biological replicates are shown with SD. *, P<0.1; **, P<0.02; ***, P<0.002 (Student’s t-test). Double knockdown of OsPTP1/2 desensitizes rice blast resistance to ABA-mediated abiotic stresses
Activation of OsMPK6 by MKK10-2D, which mimics the activation of the SA pathway by SA/BTH, conferred rice plants with blast resistance. OsPTP1/2 dephosphorylated and inactivated OsMPK6 probably in response to ABA. These results led us to examine whether PTP-wkd plants were less sensitive to the ABA-mediated abiotic stresses, in regard to BTH-induced disease resistance (Fig 7). Under normal conditions, BTH induced as strong a blast resistance in PTP-wkd plants as in NB (Fig 7). Unlike NB; however, PTP-wkd plants exhibited strong blast resistance even in the presence of ABA (Fig 7). Moreover, PTP-wkd plants showed strong blast resistance under abiotic stress conditions, low temperature (15/9°C, day/night), and high salinity (250 mM NaCl) (Fig 7). These results indicate the involvement of OsPTP1/2 in the suppression of SA/BTH-dependent blast resistance by ABA. Visible morphological and growth phenotypes were not observed in PTP-wkd under normal or stress conditions.
OsPTP1/2 is involved in the ABA-dependent repression of WRKY45 expression
SA/BTH-dependent defence system involves two independent sub-pathways, WRKY45 and OsNPR1 sub-pathways [9–12]. To investigate whether OsPTP1/2 act(s) on either sub-pathway specifically, we examined the gene expression patterns in PTP-wkd lines after the treatment with SA in the absence or presence of ABA (Fig 8 and S10 Fig). In the absence of ABA, WRKY45 transcript levels in NB and PTP-wkd lines were within a similar range (Fig 8A). In the presence of ABA, the transcript levels were significantly decreased in NB. By contrast, the transcript levels in PTP-wkd plants were increased to the levels without ABA (Fig 8A). WRKY62 is a direct target gene of WRKY45, and we have previously observed that this gene behaves similarly to WRKY45 in response to SA/BTH [9] [14]. In this experiment (Fig 8B), the expression pattern of WRKY62 paralleled that of WRKY45. We have previously reported that the expression of OsNPR1, as well as that of WRKY45, were suppressed by ABA [25]. Interestingly; however, no effect of the OsPTP1/2 double knockdown was observed on the suppression of OsNPR1 expression by ABA (Fig 8C). The SalT gene is an ABA-inducible gene [47] [25]; however, the effect of the OsPTP1/2 double knockdown was not observed on the expression of this gene either (Fig 8D). These results suggest that OsPTP1/2 acts specifically on the WRKY45 sub-pathway of SA-signalling under abiotic stress conditions (Fig 9). The expression of OsPTP1/2 genes was not positively affected by ABA (Fig 8E and 8F), high salinity, or low temperature condition (S9 Fig), suggesting that these genes are regulated post-transcriptionally by ABA and abiotic stresses.
Fig. 8. OsPTP1/2 act on the WRKY45 sub-pathway but not the OsNPR1 sub-pathway. 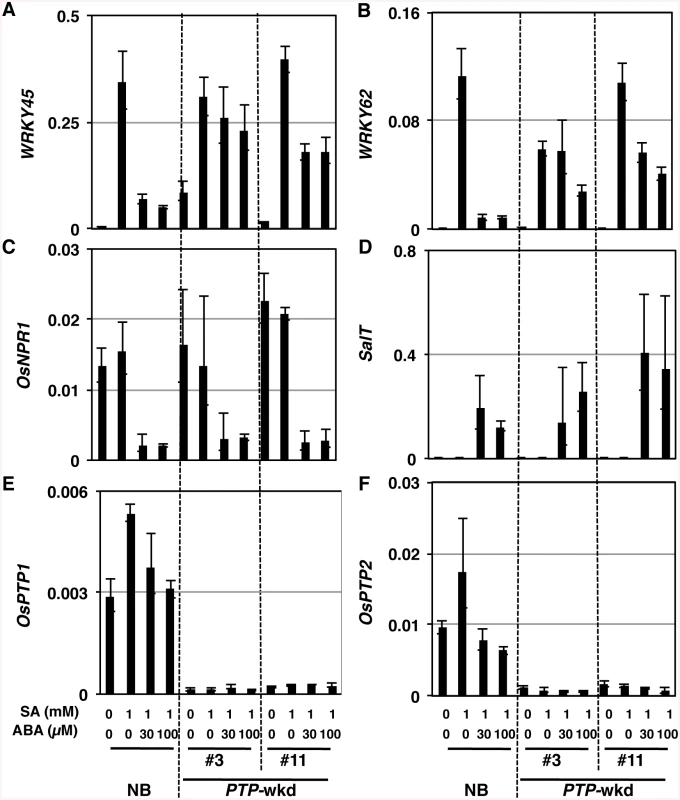
Transcript levels of each gene (relative to that of ubiquitin 1) were determined by RT-qPCR using RNA extracted from leaves treated with chemicals indicated. A. WRKY45, B. WRKY62, C. OsNPR1, D. SalT, E. OsPTP1, F. OsPTP2 Means of three technical repeats are shown with SD. Results of Student’s t-test are shown. *, P<0.1; **, P<0.05; ***, P<0.002; ****, P<0.001. Fig. 9. Proposed model for the mechanism underlying SA–ABA crosstalk through OsPTP1/2 against the OsMPK6–WRKY45-mediated SA pathway of the rice defence program. 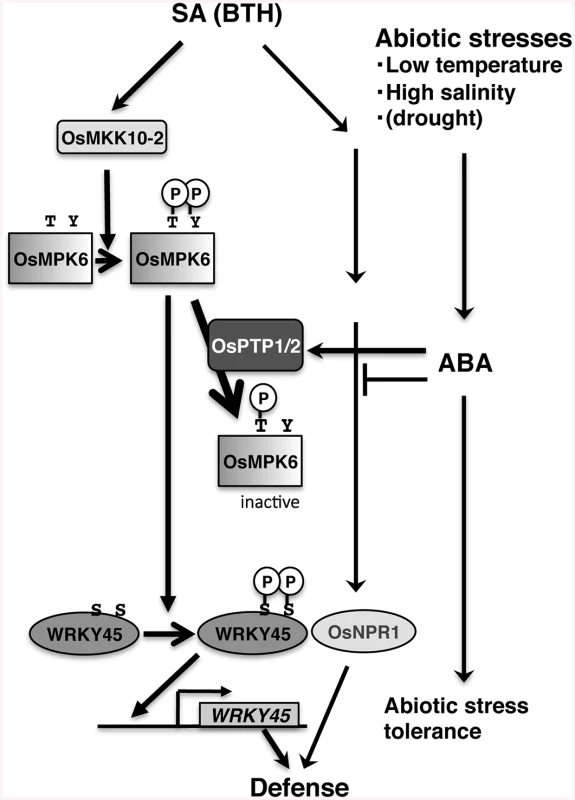
Discussion
Plants are constantly confronted to both abiotic and biotic stresses that seriously reduce their productivity. Plant responses to these stresses involve numerous complex physiological, molecular and cellular adaptations. When plants are simultaneously exposed to both abiotic and biotic stresses, plants respond in a specific manner depending on the interplays of signalling pathways that are invoked by respective stresses.
In this study, we illustrated the mechanisms underlying the WRKY45-dependent SA-signalling pathway for defence against pathogens in rice and how ABA-signalling antagonizes the WRKY45-dependent SA-signalling under abiotic stresses (Fig 9). In the absence of abiotic stresses, SA/BTH activates OsMPK6 by Thr/Tyr dual phosphorylation most probably through OsMKK10-2. The activated OsMPK6 phosphorylates one or two Ser residue(s) in the carboxyl-terminal region of WRKY45, thereby activating its transcriptional activity and conferring strong blast resistance to rice. ABA-signalling, which was activated by abiotic stresses, such as low temperature and high salinity, suppresses SA-signalling through the specific dephosphorylation of Tyr227 of OsMPK6 by OsPTP1/2. This results in compromised blast resistance in the presence of SA/BTH, likely because of reduced expression and activity of WRKY45.
Different role of tyrosine protein phosphatases in Arabidopsis and rice
Tyr-specific PTPases are also encoded in dicot genomes. Of these, only Arabidopsis AtPTP1 has been functionally characterized, and the tyrosine dephosphorylating activity of AtMPK6 (MPK6) has been reported [42] [41]. Elevated levels of SA, accompanied by PR-gene expression, have been reported in a double mutant of the AtPTP1 and MKP1 phosphatase genes [41]. However, no phenotype has been found in a single mutant of the AtPTP1 gene. Our finding for OsPTP1/2 is the first to report a function that is specific to plant PTPs; therefore, whether or not PTPs in other plant species have similar functions remains to be determined. In addition, this function is probably largely dependent on the pathosystem (combination of host and pathogen).
Managing signalling crosstalk for further improving crops
Why have (rice) plants developed this antagonistic signalling crosstalk? Presumably, this is the mechanism behind the trade-off between the responses to biotic and abiotic stresses, which prioritizes the most life-threatening stress through the allocation of limited resources under various situations. In nature, such a system is likely to help plants to survive changing environments in a cost-efficient manner. Serious rice losses due to blast disease occurred during the cold summers of 1993 and 2003 in Japan. Other research reported that low temperatures and drought render rice plants more susceptible to blast disease [32] [33]. Moreover, high salinity conditions can breakdown the disease resistance to Fusarium and Phytophthora in tomato [48]. In our experiments, the rice plants prioritized the responses to the cold or high salinity over disease resistance, which eventually compromised the BTH-induced blast resistance. Owing to this rice response under multiple stresses, most of the rice plants died of blast disease. Considering this consequence, this trade-off mechanism does not appear to have been beneficial to plants in our experiments. Crop cultivation is often conducted under resource-rich fertile conditions, and so were our experiments. Under such conditions, the trade-off seems to be unnecessary and even harmful for plants, as well as for farmers. Therefore, down-regulating such crosstalk seems to be favourable for agriculture as long as there is no unexpected adverse side effect. Theoretically, provided that the PTPases act specifically on the SA–ABA crosstalk, there is unlikely to be any such side effects. Indeed, we have not observed any significant adverse effects on the growth of PTP-wkd plants so far. The same could also hold for the trade-offs between particular stress responses and/or between a particular stress and plant growth/yield. On the basis of this speculation, down-regulating a particular signalling crosstalk could be an important goal of crop breeding. So far, little is known about the molecules, besides OsPTP1/2, that directly mediate the crosstalk. To date, several signaling components have been reported to mediate signalling crosstalk; however, most of them play indirect roles in the crosstalk. Modifications of such molecules would change the balance of responses to different stresses, but they are unlikely to eliminate the crosstalk, which would maximize the defence or tolerance to both stresses [22]. Currently, only a few molecules, such as BZR1, which mediates the crosstalk between brassinosteroid - and gibberellin-signaling, as well as innate immunity [49] [50], are known to play direct roles in the signalling crosstalk. Once researchers identify more of these molecules, it should become possible to develop multi-stress-tolerant crops without penalties on yields.
Methods
Plant materials and growth conditions
Rice plants (Oryza sativa subsp. japonica cv. Nipponbare) were grown in a greenhouse in soil (Bonsol No. 2; Sumitomo Chemical Co., Tokyo, Japan) at 30°C/26°C (day/night) with a relative humidity (RH) of approximately 60%.
Culturing and inoculation of M. oryzae
Culturing of the blast fungus M. oryzae (race 003) and fungal inoculations of rice plants were carried out essentially as described previously [25], with slight modifications. Briefly, M. oryzae conidia suspended in 0.02% Tween20 at a density of 150,000/ml were sprayed onto rice leaves, which had been pretreated with chemicals in 0.1 × MS in 50-ml tubes sealed with surgical tape (3M, St. Paul, MN, USA; cat# 3530–1). Detailed condition for each experiment is described below.
Normal-temperature conditions: after chemical pretreatments for 1 d in a growth chamber (30°C, 14-h day and 26°C, 10-h night; 60% RH), the solutions were removed, and the leaves were spray-inoculated with fungal conidia. The leaves were incubated in a dew chamber at 24°C for 24 h, and then for a further 5 d period in a growth chamber.
Low-temperature conditions: rice leaves were pretreated with chemicals for 2 d in a growth chamber (15°C, 14-h days and 9°C, 10-h nights; 60% RH). After fungal inoculations followed by incubation for 3 d under low-temperature conditions, solutions were removed, and leaves were further incubated for 5 d under normal-temperature conditions.
High-salinity conditions: before detaching leaves, whole plants were soaked in 250 mM NaCl for 4 d. Other procedures are the same as those of the normal condition.
Disease development was evaluated by quantifying M. oryzae 28S rDNA by qPCR [51]. Three or more biological repeats were performed for each disease resistance assay.
Plasmid construction
Site-directed mutagenesis was performed with a QuickChange Multi Site-directed mutagenesis kit (Stratagene, La Jolla, CA, USA) according to the manufacturer’s instructions. For constructs using NanoLuc, cDNA encoding NanoLuc was subcloned from the pNL vector (Promega, Madison, WI, USA). For overexpression of WT and mutant WRKY45, cDNA amino-terminally fused with 3X myc tag was cloned into the pZH vector under the control of the maize ubiquitin promoter. For Dex-inducible MKK10-2D, OsMKK10-2D cDNA [30] was subcloned into the pINDEX vector [52]. For PTP-wkd, the 3′-untranslated regions of OsPTP1 and -2 genes were amplified using u7 (5′ - CACCCGGGTATCCCTAAGGCAGGA-3′) and u8 (5′ - AAATGATTCAGTTTAAACCTACTAACTCTCTTTAATTCCGT-3′), and u9 (5′ - TTAGTAGGTTTAAACTGAATCATTTCTATGGAACAATCAGT-3′) and u10 (5′ - AGGCCTGGGTGGGCAGGAGAAGCG-3′) primers, respectively. Then, we performed an overlapping second PCR using the u7 and u10 primers, and the products of the first reactions. Each amplified fusion gene was cloned into pENTR/D-TOPO (Invitrogen, Carlsbad, CA, USA) and subsequently transferred into the pANDA vector [53].
Reporter assay
A reporter assay in the rice leaf sheath was essentially performed as described previously [14]. The mixture of plasmids for the expression of effectors consisted of the plasmids encoding wild-type or mutant proteins amino-terminally fused with and without NanoLuc. The effectors with or without the N-terminal NanoLuc were mixed at the ratio of 1 : 10. A total of 3 μg of the LUC reporter plasmid and 0.5 μg of the effecter plasmid mixture were used per assay. LUC and Nanoluc activities were determined using luciferin and furimazine as substrates, respectively, and their ratios (LUC/Nanoluc) were compared.
Rice transformation
Agrobacterium-mediated transformations of rice calli were performed as described previously [54] [55]. Plants were regenerated from transformed calli by selecting for hygromycin resistance.
Chemical treatments of leaves for protein or RNA analyses
Chemical treatments of leaves were conducted essentially as described previously [25]. Leaf blades from rice plants at the four-leaf stage were cut into segments approximately 0.5 cm long and submerged in a solution containing the chemicals prepared in 0.002% Silwet L-77. The leaf segments were incubated in the light at 30°C for the periods indicated. We applied ABA 1 h before SA or Dex treatments. The PTPase inhibitors were applied 1 h before the ABA treatment.
Immunoblot assays
Immunoblot assays were carried out essentially as described previously [15]. Proteins were extracted with 50 mM Hepes-KOH, pH 7.5, containing Complete Protease Inhibitor Cocktail (Roche Diagnostics, Mannheim, Germany), 1 mM PMSF, and protein phosphatase inhibitor cocktails (phosphatase inhibitor cocktail 1 and 2; Sigma, St Louis, MO, USA) or alternatively PhosTop (Roche Diagnostics). For the treatment with PPase, the protein phosphatase inhibitor cocktails were excluded. After centrifugation, supernatants (containing 6–20 μg protein) were subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE; 10% 29 : 1acrylamide:bis-acrylamide), followed by electroblotting. In the case of the Phos-Tag SDS-PAGE, 10 μM Phos-Tag acrylamide (Wako, Tokyo, Japan) and 20 μM Zn(NO3)2 were included in the 7.5% polyacrylamide gel (29 : 1 acrylamide:bis-acrylamide). Other procedures were performed as described previously [36].
Immunodetection was performed using SNAPid (Millipore, Billerica, MA, USA) according to the manufacturer’s instructions. The antibodies used were as follows: anti-pTEpY (Promega, cat# V8031) at a 1/1,500 dilution; anti-OsMPK6 [39] at a 1/1,500 dilution; anti-phosphoenolpyruvate carboxylase [56] at a 1/30,000 dilution; anti-pY (Millipore, clone 4G10 platinum) at a 1/1,500 dilution; anti-pT [Promega, anti-pT183 MAPK pAb (rabbit)] at a 1/4,000 dilution; and anti-WRKY45 [15] at a 1/300 dilution.
RT-qPCR
Total RNA was isolated from rice leaves treated with chemicals as described above using Trizol reagent (Invitrogen). cDNA was synthesized using ReverTraAce (Toyobo, Tokyo, Japan). Quantitative PCR was run on a Thermal Cycler Dice TP800 system (Takara Bio, Tokyo, Japan) using the SYBR premix ExTaq mixture (Takara Bio) as described previously [9]. Sequences of primers used for RT-qPCR are listed in S1 Table.
Phosphorylation and dephosphorylation assay in vitro
Phosphorylation and dephosphorylation assays were carried out as described previously [57] [39], with modifications. GST-MKK10-2D and MBP-MPK6 (WT or mutant) were incubated in reaction buffer (10 mM Hepes-KOH, pH 7.5, 5 mM EGTA, 20 mM MgCl2, 1 mM DTT) containing 0.5 mM ATP at 25°C for 20 min. For dephosphorylation assays, MBP-PTP1 or -2 was added and the reaction mixture was incubated for an additional 20 min. For WRKY45 phosphorylation activity assays, the same reaction mixtures were pre-incubated for 20 min, and reactions were initiated by adding MBP-WRKY45 and 37 kBq [γ-32P]ATP. The mixtures were incubated for an additional 20 min, and then terminated by adding Laemmle’s sample buffer and boiling. Labelled proteins were analysed by SDS-PAGE. Coomassie brilliant blue staining was performed as loading controls.
Accession numbers
WRKY45: Os05g0322900, LOC_Os05g25770
OsMKK10-2: Os03g0225100, LOC_Os03g12390.1
OsMPK6: Os06g0154500, LOC_Os06g06090
OsPTP1: Os12g0174800, LOC_Os12g07590
OsPTP2: Os11g0180200, LOC_Os11g07850.1
Supporting Information
Zdroje
1. Matyssek R, Agerer R, Ernst D, Munch JC, Osswald W, Pretzsch H, et al. The plant's capacity in regulating resource demand. Plant biology. 2005;7(6):560–80. doi: 10.1055/s-2005-872981 16388460.
2. Tian D, Traw MB, Chen JQ, Kreitman M, Bergelson J. Fitness costs of R-gene-mediated resistance in Arabidopsis thaliana. Nature. 2003;423(6935):74–7. doi: 10.1038/nature01588 12721627.
3. Fujita M, Fujita Y, Noutoshi Y, Takahashi F, Narusaka Y, Yamaguchi-Shinozaki K, et al. Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Curr Opin Plant Biol. 2006;9(4):436–42. doi: 10.1016/j.pbi.2006.05.014 16759898.
4. Pieterse CM, Van der Does D, Zamioudis C, Leon-Reyes A, Van Wees SC. Hormonal modulation of plant immunity. Annu Rev Cell Dev Biol. 2012;28 : 489–521. doi: 10.1146/annurev-cellbio-092910-154055 22559264.
5. Sharma R, De Vleesschauwer D, Sharma MK, Ronald PC. Recent advances in dissecting stress-regulatory crosstalk in rice. Mol Plant. 2013;6(2):250–60. doi: 10.1093/mp/sss147 23292878.
6. Vert G, Chory J. Crosstalk in cellular signaling: background noise or the real thing? Dev Cell. 2011;21(6):985–91. doi: 10.1016/j.devcel.2011.11.006 22172668; PubMed Central PMCID: PMC3281494.
7. DenancÈ N, S·nchez-Vallet A, Goffner D, Molina A. Disease resistance or growth: the role of plant hormones in balancing immune responses and fitness costs. Frontiers in Plant Science. 2013;4. doi: 10.3389/fpls.2013.00155
8. Cao H, Glazebrook J, Clarke JD, Volko S, Dong X. The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell. 1997;88(1):57–63. 9019406.
9. Shimono M, Sugano S, Nakayama A, Jiang CJ, Ono K, Toki S, et al. Rice WRKY45 plays a crucial role in benzothiadiazole-inducible blast resistance. Plant Cell. 2007;19(6):2064–76. doi: 10.1105/tpc.106.046250 17601827; PubMed Central PMCID: PMC1955718.
10. Sugano S, Jiang C-J, Miyazawa S-I, Masumoto C, Yazawa K, Hayashi N, et al. Role of OsNPR1 in rice defense program as revealed by genome-wide expression analysis. Plant Molecular Biology. 2010;74(6):549–62. Epub 2010/10/07. doi: 10.1007/s11103-010-9695-3 20924648.
11. Takatsuji H, Jiang C-J, Sugano S. Salicylic acid signaling pathway in rice and the potential applications of its regulators. JARQ. 2010;44(3):217–23. ISI:000284669300001.
12. Nakayama A, Fukushima S, Goto S, Matsushita A, Shimono M, Sugano S, et al. Genome-wide identification of WRKY45-regulated genes that mediate benzothiadiazole-induced defense responses in rice. BMC Plant Biology. 2013;13(1):150. doi: 10.1186/1471-2229-13-150
13. Shimono M, Koga H, Akagi AYA, Hayashi N, Goto S, Sawada M, et al. Rice WRKY45 plays important roles in fungal and bacterial disease resistance. Molecular Plant Pathology. 2012;13(1):83–94. doi: 10.1111/j.1364-3703.2011.00732.x 21726399
14. Inoue H, Hayashi N, Matsushita A, Xinqiong L, Nakayama A, Sugano S, et al. Blast resistance of CC-NB-LRR protein Pb1 is mediated by WRKY45 through protein-protein interaction. Proc Natl Acad Sci U S A. 2013;110(23):9577–82. doi: 10.1073/pnas.1222155110 23696671; PubMed Central PMCID: PMC3677490.
15. Matsushita A, Inoue H, Goto S, Nakayama A, Sugano S, Hayashi N, et al. Nuclear ubiquitin proteasome degradation affects WRKY45 function in the rice defense program. The Plant Journal. 2013;73(2):302–13. doi: 10.1111/tpj.12035 23013464
16. Vlot AC, Dempsey DA, Klessig DF. Salicylic Acid, a multifaceted hormone to combat disease. Annu Rev Phytopathol. 2009;47 : 177–206. doi: 10.1146/annurev.phyto.050908.135202 19400653.
17. Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol. 2010;61 : 651–79. doi: 10.1146/annurev-arplant-042809-112122 20192755.
18. Mauch-Mani B, Mauch F. The role of abscisic acid in plant-pathogen interactions. Curr Opin Plant Biol. 2005;8(4):409–14. doi: 10.1016/j.pbi.2005.05.015 15939661.
19. Asselbergh B, De Vleesschauwer D, Hofte M. Global switches and fine-tuning-ABA modulates plant pathogen defense. Mol Plant Microbe Interact. 2008;21(6):709–19. doi: 10.1094/MPMI-21-6-0709 18624635.
20. Cao FY, Yoshioka K, Desveaux D. The roles of ABA in plant-pathogen interactions. J Plant Res. 2011;124(4):489–99. doi: 10.1007/s10265-011-0409-y 21380629.
21. De Vleesschauwer D, Gheysen G, Höfte M. Hormone defense networking in rice: tales from a different world. Trends in Plant Science. 2013;18(10):555–65. doi: 10.1016/j.tplants.2013.07.002 23910453.
22. Takatsuji H, Jiang C-J. Plant hormone crosstalks under biotic stresses. In: Tran L-SP, Pal S, editors. Phytohormones: a window to metabolism, signaling and biotechnological applications: Springer New York; 2014. p. 323–50.
23. Audenaert K, De Meyer GB, Hofte MM. Abscisic acid determines basal susceptibility of tomato to Botrytis cinerea and suppresses salicylic acid-dependent signaling mechanisms. Plant Physiol. 2002;128(2):491–501. doi: 10.1104/pp.010605 11842153; PubMed Central PMCID: PMC148912.
24. Yasuda M, Ishikawa A, Jikumaru Y, Seki M, Umezawa T, Asami T, et al. Antagonistic interaction between systemic acquired resistance and the abscisic acid-mediated abiotic stress response in Arabidopsis. Plant Cell. 2008;20(6):1678–92. Epub 2008/07/01. tpc.107.054296 [pii] doi: 10.1105/tpc.107.054296 18586869.
25. Jiang C-J, Shimono M, Sugano S, Kojima M, Yazawa K, Yoshida R, et al. Abscisic acid interacts antagonistically with salicylic acid signaling pathway in rice–Magnaporthe grisea Interaction. Molecular Plant-Microbe Interactions. 2010;23(6):791–8. doi: 10.1094/MPMI-23-6-0791 20459318
26. Mao G, Meng X, Liu Y, Zheng Z, Chen Z, Zhang S. Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. Plant Cell. 2011;23(4):1639–53. Epub 2011/04/19. doi: 10.1105/tpc.111.084996 21498677; PubMed Central PMCID: PMC3101563.
27. Ishihama N, Yamada R, Yoshioka M, Katou S, Yoshioka H. Phosphorylation of the Nicotiana benthamiana WRKY8 Transcription Factor by MAPK Functions in the Defense Response. Plant Cell. 2011. Epub 2011/03/10. tpc.110.081794 [pii] doi: 10.1105/tpc.110.081794 21386030.
28. Bartels S, Gonzalez Besteiro MA, Lang D, Ulm R. Emerging functions for plant MAP kinase phosphatases. Trends Plant Sci. 2010;15(6):322–9. Epub 2010/05/11. doi: 10.1016/j.tplants.2010.04.003 20452268.
29. Rodriguez MC, Petersen M, Mundy J. Mitogen-activated protein kinase signaling in plants. Annu Rev Plant Biol. 2010;61 : 621–49. doi: 10.1146/annurev-arplant-042809-112252 20441529.
30. Ueno Y, Yoshida R, Kishi-Kaboshi M, Matsushita A, Jiang C-J, Goto S, et al. MAP kinases phosphorylate rice WRKY45. Plant Signaling & Behavior. 2013;8(6):e24510.
31. Kahn RP, Libby JL. The effect of environmental factors and plant ages on the infection of rice by the blast fungus, Pyricularia oryzae. Phytopathology. 1958;48 : 25–30.
32. Bonman JM, Sanchez LM, Mackill AO. Effects of water deficit on rice blast. II. Disease-development in the field. J Plant Prot Trop. 1988;5 : 67–73.
33. Gill MA, Bonman JM. Effects of water deficit on rice blast. I. Influence of water deficit on components of resistance. J Plant Prot Trop. 1988;5 : 61–6.
34. Koga H, Dohi K, Mori M. Abscisic acid and low temperatures suppress the whole plant-specific resistance reaction of rice plants to the infection of Magnaporthe grisea. Physiological and Molecular Plant Pathology. 2004;65(1):3–9. doi: 10.1016/j.pmpp.2004.11.002
35. Yazawa K, Jiang C-J, Kojima M, Sakakibara H, Takatsuji H. Reduction of abscisic acid levels or inhibition of abscisic acid signaling in rice during the early phase of Magnaporthe oryzae infection decreases its susceptibility to the fungus. Physiological and Molecular Plant Pathology. 2012;78 : 1–7. doi: 10.1016/j.pmpp.2011.12.003
36. Kinoshita E, Kinoshita-Kikuta E. Improved Phos-tag SDS-PAGE under neutral pH conditions for advanced protein phosphorylation profiling. Proteomics. 2011;11(2):319–23. doi: 10.1002/pmic.201000472 21204258.
37. Anderson NG, Maller JL, Tonks NK, Sturgill TW. Requirement for integration of signals from two distinct phosphorylation pathways for activation of MAP kinase. Nature. 1990;343(6259):651–3. 2154696
38. Aoyama T, Chua NH. A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 1997;11(3):605–12. Epub 1997/03/01. 9107046.
39. Kishi-Kaboshi M, Okada K, Kurimoto L, Murakami S, Umezawa T, Shibuya N, et al. A rice fungal MAMP-responsive MAPK cascade regulates metabolic flow to antimicrobial metabolite synthesis. Plant J. 2010;63(4):599–612. doi: 10.1111/j.1365-313X.2010.04264.x 20525005; PubMed Central PMCID: PMC2988419.
40. Krishnan N, Bencze G, Cohen P, Tonks NK. The anti-inflammatory compound BAY 11–7082 is a potent inhibitor of Protein Tyrosine Phosphatases. FEBS J. 2013. doi: 10.1111/febs.12283 23578302.
41. Bartels S, Anderson JC, Gonzalez Besteiro MA, Carreri A, Hirt H, Buchala A, et al. MAP kinase phosphatase1 and protein tyrosine phosphatase1 are repressors of salicylic acid synthesis and SNC1-mediated responses in Arabidopsis. Plant Cell. 2009;21(9):2884–97. doi: 10.1105/tpc.109.067678 19789277; PubMed Central PMCID: PMC2768924.
42. Gupta R. Redox control of protein tyrosine phosphatases and mitogen-activated protein kinases in plants. Plant Physiology. 2003;132(3):1149–52. doi: 10.1104/pp.103.020792 12857797
43. Guan KL. The mitogen activated protein kinase signal transduction pathway: from cell surface to the nucleus. Cell Signal. 1994;6 : 581–9. 7857762
44. Xu Q, Fu HH, Gupta R, Luan S. Molecular characterization of a tyrosine-specific protein phosphatase encoded by a stress-responsive gene in Arabidopsis. The Plant Cell. 1998;10(5):849–57. 9596642
45. Görlach J, Volrath S, Knauf-Beiter G, Hengy G, Beckhove U, Kogel KH, et al. Benzothiadiazole, a novel class of inducers of systemic acquired resistance, activates gene expression and disease resistance in wheat. Plant Cell. 1996;8(4):629–43. doi: 10.1105/tpc.8.4.629 8624439
46. Lawton KA, Friedrich L, Hunt M, Weymann K, Delaney T, Kessmann H, et al. Benzothiadiazole induces disease resistance in Arabidopsis by activation of the systemic acquired resistance signal transduction pathway. The Plant Journal. 1996;10(1):71–82. doi: 10.1046/j.1365-313X.1996.10010071.x 8758979
47. Rabbani MA, Maruyama K, Abe H, Khan MA, Katsura K, Ito Y, et al. Monitoring expression profiles of rice genes under cold, drought, and high-salinity stresses and abscisic acid application using cDNA microarray and RNA gel-blot analyses. Plant Physiol. 2003;133(4):1755–67. doi: 10.1104/pp.103.025742 14645724; PubMed Central PMCID: PMC300730.
48. Besri M. Effects of salinity on plant diseases development. In: Lieth H, Al Masoom A, editors. Towards the rational use of high salinity tolerant plants. Tasks for vegetation science. 28: Springer Netherlands; 1993. p. 67–74.
49. Li QF, Wang C, Jiang L, Li S, Sun SS, He JX. An interaction between BZR1 and DELLAs mediates direct signaling crosstalk between brassinosteroids and gibberellins in Arabidopsis. Sci Signal. 2012;5(244):ra72. doi: 10.1126/scisignal.2002908 23033541.
50. Lozano-Duran R, Macho AP, Boutrot F, Segonzac C, Somssich IE, Zipfel C. The transcriptional regulator BZR1 mediates trade-off between plant innate immunity and growth. Elife. 2013;2:e00983. doi: 10.7554/eLife.00983 24381244; PubMed Central PMCID: PMC3875382.
51. Qi M, Yang Y. Quantification of magnaporthe grisea during infection of rice plants using real-time polymerase chain reaction and northern blot/phosphoimaging analyses. Phytopathology. 2002;92(8):870–6. doi: 10.1094/PHYTO.2002.92.8.870 18942966
52. Ouwerkerk P, de Kam R, Hoge J, Meijer A. Glucocorticoid-inducible gene expression in rice. Planta. 2001;213 : 370–8. doi: 10.1007/s004250100583 11506359
53. Miki D, Shimamoto K. Simple RNAi vectors for stable and transient suppression of gene function in rice. Plant and Cell Physiology. 2004;45(4):490–5. doi: 10.1093/pcp/pch048 15111724
54. Hiei Y, Ohta S, Komari T, Kumashiro T. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. The Plant Journal. 1994;6(2):271–82. doi: 10.1046/j.1365-313X.1994.6020271.x 7920717
55. Fuse T, Sasaki T, Yano M. Ti-plasmid vectors useful for functional analysis of rice genes. Plant BioTechology. 2001;18 : 219–22.
56. Ueno Y, Imanari E, Emura J, Yoshizawa-Kumagaye K, Nakajima K, Inami K, et al. Immunological analysis of the phosphorylation state of maize C4-form phosphoenolpyruvate carboxylase with specific antibodies raised against a synthetic phosphorylated peptide. Plant J. 2000;21(1):17–26. 10652147.
57. Huang Y, Li H, Gupta R, Morris PC, Luan S, Kieber JJ. ATMPK4, an Arabidopsis homolog of mitogen-activated protein kinase, is activated in vitro by AtMEK1 through threonine phosphorylation. Plant Physiology. 2000;122(4):1301–10. doi: 10.1104/pp.122.4.1301 10759527
58. Akagi A, Jiang C-J, Takatsuji H. Magnaporthe oryzae Inoculation of rice seedlings by spraying with a spore suspension. Bioprotocol. 2015;5(11):e1486.
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek TRIM21 Promotes cGAS and RIG-I Sensing of Viral Genomes during Infection by Antibody-Opsonized VirusČlánek Effector OspB Activates mTORC1 in a Manner That Depends on IQGAP1 and Promotes Cell ProliferationČlánek Fundamental Roles of the Golgi-Associated Aspartyl Protease, ASP5, at the Host-Parasite InterfaceČlánek Modulation of the Surface Proteome through Multiple Ubiquitylation Pathways in African Trypanosomes
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2015 Číslo 10- Stillova choroba: vzácné a závažné systémové onemocnění
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Jak souvisí postcovidový syndrom s poškozením mozku?
- Familiární středomořská horečka
-
Všechny články tohoto čísla
- Expression of Concern: Misregulation of Underlies the Developmental Abnormalities Caused by Three Distinct Viral Silencing Suppressors in Arabidopsis
- Preparing for the Next Epidemic with Basic Virology
- Effectively Communicating the Uncertainties Surrounding Ebola Virus Transmission
- Translating Basic Research into Clinical Applications: Malaria Research at an NIH Lab
- A Gut Odyssey: The Impact of the Microbiota on Spore Formation and Germination
- Papillomavirus E6 Oncoproteins Take Common Structural Approaches to Solve Different Biological Problems
- Chronobiomics: The Biological Clock as a New Principle in Host–Microbial Interactions
- Dimensions of Horizontal Gene Transfer in Eukaryotic Microbial Pathogens
- Addressing the Complications of Ebola and Other Viral Hemorrhagic Fever Infections: Using Insights from Bacterial and Fungal Sepsis
- Time for Chocolate: Current Understanding and New Perspectives on Cacao Witches’ Broom Disease Research
- Ganglioside and Non-ganglioside Mediated Host Responses to the Mouse Polyomavirus
- Crosslinking of a Peritrophic Matrix Protein Protects Gut Epithelia from Bacterial Exotoxins
- Structure Elucidation of Coxsackievirus A16 in Complex with GPP3 Informs a Systematic Review of Highly Potent Capsid Binders to Enteroviruses
- CD39 Expression Identifies Terminally Exhausted CD8 T Cells
- Abiotic Stresses Antagonize the Rice Defence Pathway through the Tyrosine-Dephosphorylation of OsMPK6
- Dissociation of Tissue Destruction and Bacterial Expansion during Bubonic Plague
- Interferon-γ: The Jekyll and Hyde of Malaria
- CCR2 Inflammatory Dendritic Cells and Translocation of Antigen by Type III Secretion Are Required for the Exceptionally Large CD8 T Cell Response to the Protective YopE Epitope during Infection
- A New Glycan-Dependent CD4-Binding Site Neutralizing Antibody Exerts Pressure on HIV-1
- The Suramin Derivative NF449 Interacts with the 5-fold Vertex of the Enterovirus A71 Capsid to Prevent Virus Attachment to PSGL-1 and Heparan Sulfate
- Trans-generational Immune Priming Protects the Eggs Only against Gram-Positive Bacteria in the Mealworm Beetle
- Peripheral Vγ9Vδ2 T Cells Are a Novel Reservoir of Latent HIV Infection
- Respiratory Syncytial Virus Disease Is Mediated by Age-Variable IL-33
- TRIM21 Promotes cGAS and RIG-I Sensing of Viral Genomes during Infection by Antibody-Opsonized Virus
- Modeling the Effects of Vorinostat Reveals both Transient and Delayed HIV Transcriptional Activation and Minimal Killing of Latently Infected Cells
- Identification of a Novel Lipoprotein Regulator of Spore Germination
- Calcium Regulation of Hemorrhagic Fever Virus Budding: Mechanistic Implications for Host-Oriented Therapeutic Intervention
- Antigen-Specific Th17 Cells Are Primed by Distinct and Complementary Dendritic Cell Subsets in Oropharyngeal Candidiasis
- Comparative Life Cycle Transcriptomics Revises Genome Annotation and Links a Chromosome Duplication with Parasitism of Vertebrates
- The Autophagy Receptor TAX1BP1 and the Molecular Motor Myosin VI Are Required for Clearance of Salmonella Typhimurium by Autophagy
- Carcinogenic Parasite Secretes Growth Factor That Accelerates Wound Healing and Potentially Promotes Neoplasia
- Effector OspB Activates mTORC1 in a Manner That Depends on IQGAP1 and Promotes Cell Proliferation
- Dengue Virus Infection of Requires a Putative Cysteine Rich Venom Protein
- Distinct Viral and Mutational Spectrum of Endemic Burkitt Lymphoma
- Fundamental Roles of the Golgi-Associated Aspartyl Protease, ASP5, at the Host-Parasite Interface
- Phenotypic and Functional Alterations in Circulating Memory CD8 T Cells with Time after Primary Infection
- Systematic Identification of Cyclic-di-GMP Binding Proteins in Reveals a Novel Class of Cyclic-di-GMP-Binding ATPases Associated with Type II Secretion Systems
- Influenza Transmission in the Mother-Infant Dyad Leads to Severe Disease, Mammary Gland Infection, and Pathogenesis by Regulating Host Responses
- Myeloid Cell Arg1 Inhibits Control of Arthritogenic Alphavirus Infection by Suppressing Antiviral T Cells
- The White-Nose Syndrome Transcriptome: Activation of Anti-fungal Host Responses in Wing Tissue of Hibernating Little Brown Myotis
- Influenza Virus Reassortment Is Enhanced by Semi-infectious Particles but Can Be Suppressed by Defective Interfering Particles
- Identification of the Mechanisms Causing Reversion to Virulence in an Attenuated SARS-CoV for the Design of a Genetically Stable Vaccine
- Differentiation-Dependent KLF4 Expression Promotes Lytic Epstein-Barr Virus Infection in Epithelial Cells
- The Histone Acetyltransferase Hat1 Regulates Stress Resistance and Virulence via Distinct Chromatin Assembly Pathways
- C-di-GMP Regulates Motile to Sessile Transition by Modulating MshA Pili Biogenesis and Near-Surface Motility Behavior in
- Modulation of the Surface Proteome through Multiple Ubiquitylation Pathways in African Trypanosomes
- Crystal Structure of the Human Cytomegalovirus Glycoprotein B
- Depletion of . GlmU from Infected Murine Lungs Effects the Clearance of the Pathogen
- Immunologic Control of Papillomavirus Type 1
- Requires Host Rab1b for Survival in Macrophages
- Structure Analysis Uncovers a Highly Diverse but Structurally Conserved Effector Family in Phytopathogenic Fungi
- PD-L1 Expression on Retrovirus-Infected Cells Mediates Immune Escape from CD8 T Cell Killing
- Phospho-dependent Regulation of SAMHD1 Oligomerisation Couples Catalysis and Restriction
- IL-4 Induced Innate CD8 T Cells Control Persistent Viral Infection
- Crystal Structures of a Piscine Betanodavirus: Mechanisms of Capsid Assembly and Viral Infection
- BCG Skin Infection Triggers IL-1R-MyD88-Dependent Migration of EpCAM CD11b Skin Dendritic cells to Draining Lymph Node During CD4+ T-Cell Priming
- Antigenic Characterization of the HCMV gH/gL/gO and Pentamer Cell Entry Complexes Reveals Binding Sites for Potently Neutralizing Human Antibodies
- Rescue of a Plant Negative-Strand RNA Virus from Cloned cDNA: Insights into Enveloped Plant Virus Movement and Morphogenesis
- Geminivirus Activates to Accelerate Cytoplasmic DCP2-Mediated mRNA Turnover and Weakens RNA Silencing in
- Disruption of Sphingolipid Biosynthesis Blocks Phagocytosis of
- The Fungal Exopolysaccharide Galactosaminogalactan Mediates Virulence by Enhancing Resistance to Neutrophil Extracellular Traps
- The Timing of Stimulation and IL-2 Signaling Regulate Secondary CD8 T Cell Responses
- Structural and Functional Analysis of Murine Polyomavirus Capsid Proteins Establish the Determinants of Ligand Recognition and Pathogenicity
- The Dual Role of an ESCRT-0 Component HGS in HBV Transcription and Naked Capsid Secretion
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Chronobiomics: The Biological Clock as a New Principle in Host–Microbial Interactions
- Interferon-γ: The Jekyll and Hyde of Malaria
- Crosslinking of a Peritrophic Matrix Protein Protects Gut Epithelia from Bacterial Exotoxins
- Antigen-Specific Th17 Cells Are Primed by Distinct and Complementary Dendritic Cell Subsets in Oropharyngeal Candidiasis
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



