-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Uses a Unique Ligand-Binding Mode for Trapping Opines and Acquiring A Competitive Advantage in the Niche Construction on Plant Host
An ecological niche is defined, in a given environment, by the availability of nutritive resources, which can be specifically assimilated by certain living organisms to promote their proliferation. The bacterial pathogen Agrobacterium tumefaciens is able to engineer an ecological niche in the infected host via the transformation of the plant genome and diversion of the plant metabolism towards production of the opine nutrients. In this work, we quantified the selective advantage conferred to a member of the phytopathogenic species A. tumefaciens which is able to assimilate the opine nopaline. This opine is a condensate of arginine and α-ketoglurate that is produced both under linear and cyclic forms in the plant tumour environment. We further determined at the molecular and atomistic levels how A. tumefaciens is able to sense the nopaline molecules, and which metabolic pathways are activated in response. Overall, this work deciphered some key molecular events in the niche construction of the pathogen A. tumefaciens that is unique among living organisms and used to develop bioengineering tools.
Published in the journal: . PLoS Pathog 10(10): e32767. doi:10.1371/journal.ppat.1004444
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1004444Summary
An ecological niche is defined, in a given environment, by the availability of nutritive resources, which can be specifically assimilated by certain living organisms to promote their proliferation. The bacterial pathogen Agrobacterium tumefaciens is able to engineer an ecological niche in the infected host via the transformation of the plant genome and diversion of the plant metabolism towards production of the opine nutrients. In this work, we quantified the selective advantage conferred to a member of the phytopathogenic species A. tumefaciens which is able to assimilate the opine nopaline. This opine is a condensate of arginine and α-ketoglurate that is produced both under linear and cyclic forms in the plant tumour environment. We further determined at the molecular and atomistic levels how A. tumefaciens is able to sense the nopaline molecules, and which metabolic pathways are activated in response. Overall, this work deciphered some key molecular events in the niche construction of the pathogen A. tumefaciens that is unique among living organisms and used to develop bioengineering tools.
Introduction
The widespread pathogen Agrobacterium tumefaciens evolved a unique process of niche construction inside the host plant: it transfers a portion (T-DNA) of its tumour-inducing (Ti) plasmid into the nuclear genome of the infected plant cells [1], [2]. In the stably transformed host cells, the expression of the T-DNA genes drives synthesis of the plant growth factors auxin and cytokinins that enhance cell proliferation and vascular differentiation. In parallel, the surface and intercellular spaces of the neoplastic tissues of the tumour - that is a characteristic symptom of the crow-gall disease - are colonized by the bacterial pathogen. The carbon and nitrogen metabolisms of the transformed cells are also diverted. The T-DNA codes for the enzymatic synthesis of unusual metabolites. These diverse derivatives of sugars, phosphorylated or not, or amino, or organic acids are called opines [3]–[6]. As an example, the well-studied A. tumefaciens strain C58 induces accumulation of different opines such as nopaline that is formed from arginine and α-ketoglutarate (α-KG) and agrocinopines A and B that are non-nitrogenous phosphodiesters of sugars.
Besides the opine synthesis genes carried on the T-DNA, the Ti-plasmid also harbours genes involved in the uptake and catabolism of the opines by A. tumefaciens. In other words, the bacteria carrying a Ti-plasmid can benefit from the opines produced by the cells of the plant tumours that express the appropriate T-DNA genes. Aside, some opines, the so-called conjugative opines such as agrocinopine, trigger the quorum-sensing pathway which in turns activates the horizontal transfer (bacterial conjugation) of the Ti-plasmid [7]. As a consequence, the tumour niche was called the opine niche, and the niche construction process was theorized as the opine concept [3], [4]. This concept highlights the key-role of opines in the maintenance and dissemination of the Ti-plasmids among the agrobacteria colonizing the plant host. Experimental evaluation of the validity of the opine concept was performed by measuring the selective advantage conferred to opine-assimilating bacteria, including agrobacteria and other species, in opine-rich environments, such as a culture medium or the rhizosphere and phyllosphere of opine-producing plants [8]–[13]. However, to our knowledge, this hypothesis has not been tested in the natural opine niche, i.e. the plant tumour, making the opine concept in the Agrobacterium-plant host interaction formally still unproven.
In A. tumefaciens, the recognition and import of opines is conferred by periplasmic binding proteins (PBPs) and their associated ATP-binding cassette (ABC) transporters [14], [15]; but there is no known structure of the opine-PBP complex. In this work, we investigated the structural and biochemical properties of the PBP NocT of A. tumefaciens C58 which senses at least two opines: nopaline and pyronopaline, a lactam nopaline-derivative that can also be found in plant tumours [16], [17]. In addition, the complete set of A. tumefaciens genes transcriptionally responsive to these opines was identified and the fitness advantage conferred by opine assimilation upon A. tumefaciens was measured directly in the plant tumours. This integrative work highlights the structural and functional characteristics of opine binding and assimilation in the niche construction process evolved by the plant pathogen. It also definitely provides evidence to support the opine concept that directs the A. tumefaciens plant-host interaction.
Results
Structures and overall fold of NocT
The X-ray structures of the mature free-liganded NocT and the two liganded NocT with pyronopaline and nopaline were obtained at 1.89 Å, 1.55 Å and 2.29 Å resolutions, respectively (Table S1). Each crystal contained two very similar molecules in the asymmetric unit as indicated by the overall root mean square deviations (RMSD) for all Cα atoms of 0.6 Å, 0.46 Å, and 0.28 Å respectively. The two liganded forms adopted a similar closed conformation (RMSD of 0.3 Å for all Cα) while the unliganded form showed an open conformation (Figure 1) as commonly reported for the structures of PBPs solved with and without a ligand [18]–[20]. A 43° rotation around the hinge region of the C-terminal domain (residues 117–231) was observed once the N-terminal domains (residues 29–112 and 239–284) of the unliganded and liganded structures were superimposed leading to a movement of 16 Å for Thr168. NocT possesses a typical fold of cluster F within the PBP structural classification [21]. Indeed, a structural comparison of its closed form to all entries in the PDB using SSM-EBI (http://www.ebi.ac.uk/msd-srv/ssm [22]) showed that the most similar overall structures were PBPs from the same cluster F, i.e. the liganded structure of the histidine binding protein HisJ (PDB code 1HSL) of Escherichia coli and that of the lysine-arginine-ornithine binding protein LAO (PDB code 1LAF) of Salmonella enterica [23], [24]. RMSD values of NocT vs. HisJ and LAO ranged from 1.47 to 1.5 Å over 226/228 Cα atoms, corresponding to a sequence identity of 38 and 36% respectively.
Fig. 1. Ribbon representation of NocT in complex with nopaline and comparison with the unliganded form. 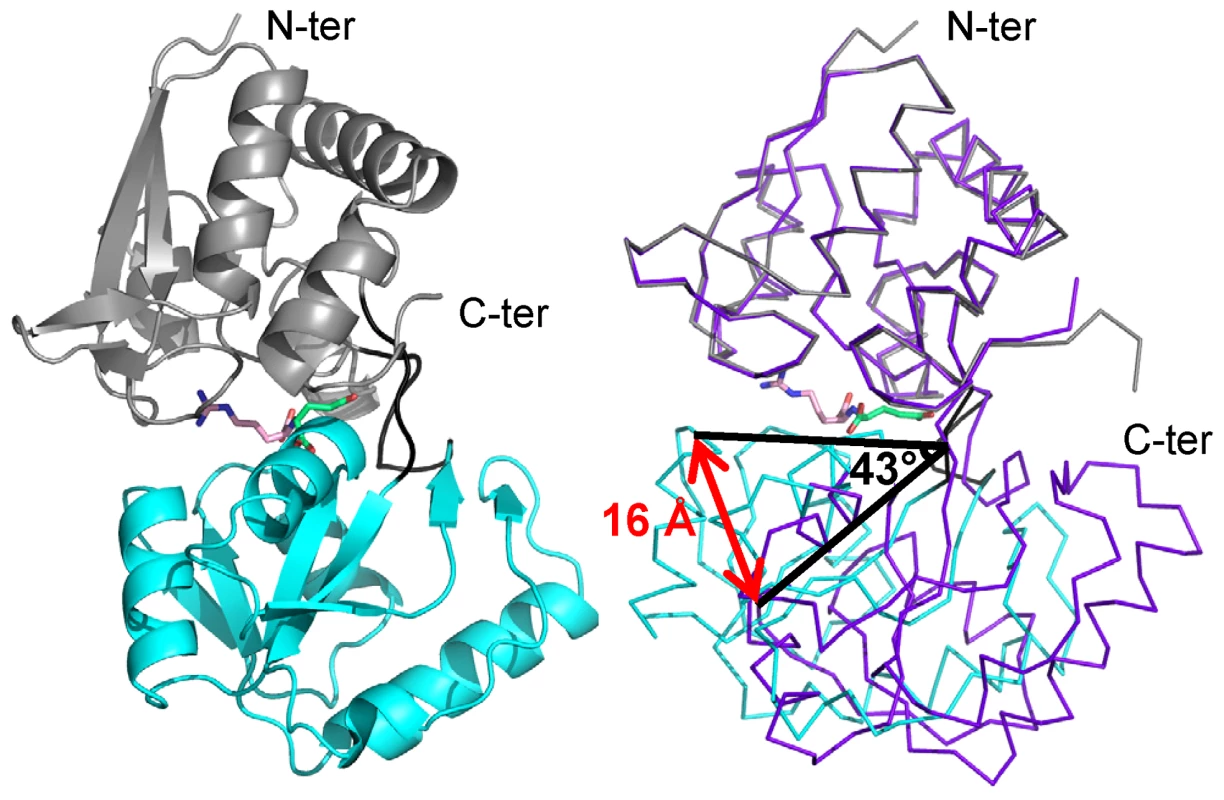
Left panel; Nopaline is located in the cleft between the two domains shown in grey and cyan. The short hinge region between the two domains is shown in black. The arginine part of the nopaline is shown in pink while its α-KG part in limegreen. Right panel, comparison of the unliganded structure of NocT in purple with the complexed nopaline in grey and cyan. NocT ligand binding site with nopaline and pyronopaline
The nopaline and pyronopaline bound between the two closed lobes were very well defined in their respective electron density maps (Figure 2). The ligand-binding site of NocT was defined by the Glu36, Tyr39, Tyr42, Trp77, Ala94, Ala95, Gly97, Arg102, Thr115, Met117, Gln165, Thr168, Ser169, His170, Ser207, Gly238 and Val239 residues. Overall both ligands shared the same protein binding mode for their arginine moiety. The side chain of this moiety was wedged between two aromatic residues (Trp77 and Tyr39) and pointed toward the opening of the cleft by making seven hydrogen bonds with the side chains of Gln165 and Glu36 and the carbonyl of Ala94 (Figure 2). Its carboxyl group made a salt-bridge with the side chain of Arg102 and interacted with both NH of Gly97 and Ser169 while its NH group formed a hydrogen bond with the carbonyl of Ala95. Remarkably this hydrogen bond cannot exist with pyronopaline due to the lactam structure of the molecule.
Fig. 2. Nopaline (A) and pyronopaline (B) bound to the ligand binding site of NocT are shown as pink/limegreen and pink/blue stick in their simulated annealing Fo-Fc omit map contoured at 4 σ, respectively. 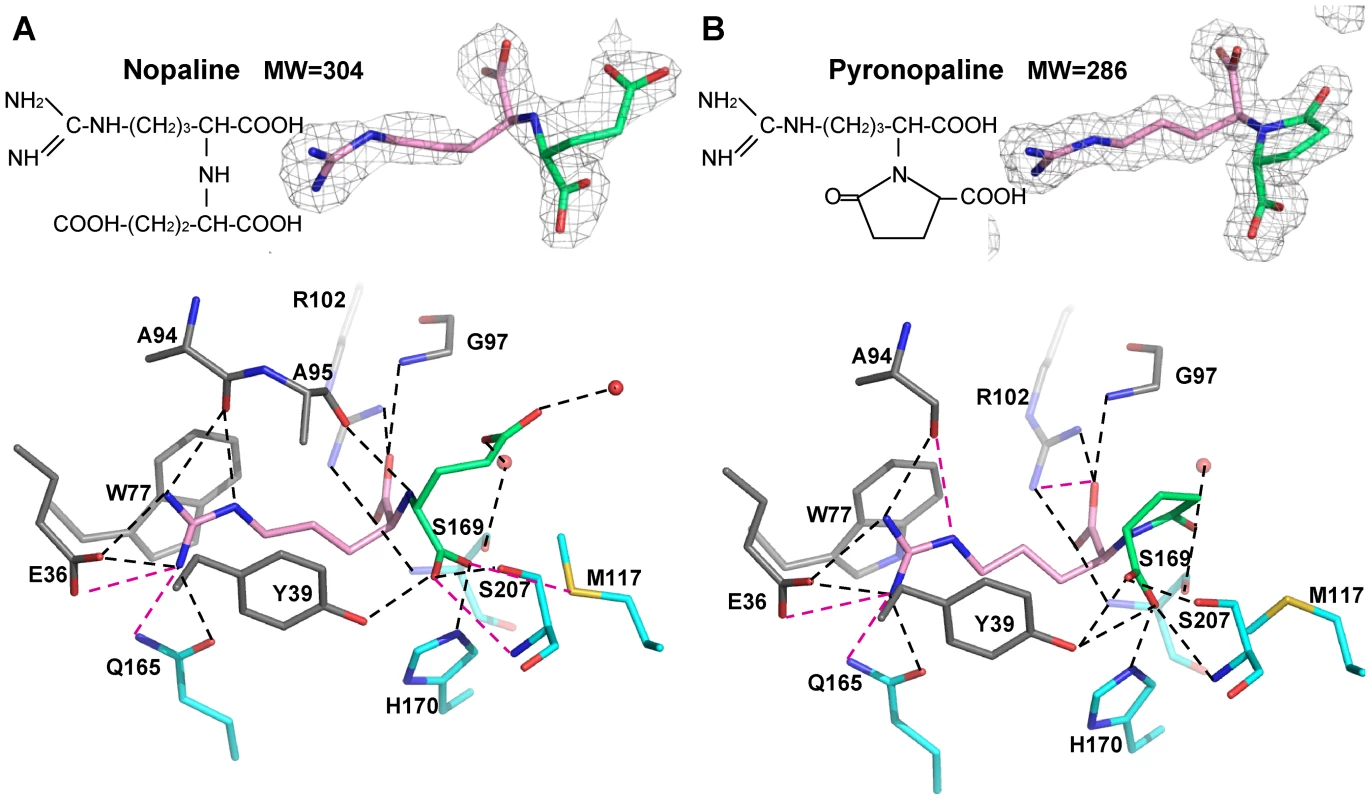
Hydrogen bonds between NocT and the ligand are shown as dashed lines in black (for distances below 3.2 Å) and magenta (for distances between 3.2 and 3.4 Å). Concerning the α-KG parts of both ligands, their first carboxyl group adopted a different position (shift around 1 Å of the two oxygen atoms), but made similar hydrogen bonds with NocT involving the side chains of Tyr39, His170 and Ser207 as well as the NH of Ser207. An additional bond was observed between the Met117 side chain and the nopaline. In the pyronopaline, the hydroxyl of the terminal carbon covalently linked to the N atom interacted with the side chain of Ser169 whereas in the nopaline, each oxygen from the terminal carboxylate group of the α-KG moiety formed two water-mediated interactions with the main chains of Thr115 for one water and with the CO of Phe235 and the S169 side chain for the other water (Figure S1). An analogous water molecule interacting with the CO main chain of Phe235 in the pyronopaline-liganded structure was shifted by 1.4 Å and bound the Gln99 side chain which appeared well ordered in this structure. The large mobile residue Met117 was forced to accommodate its side chain position according to the type of bound ligand and made Van der Walls contacts with the pyronopaline (Figure S1). The rearrangement of the side chain (shift of 1.9 Å) in the pyronopaline structure compared with the nopaline structure led to a rearrangement of the carboxyl moiety of the α-KG and of the H170 side chain (Figure S1).
Affinity of NocT to nopaline and pyronopaline
By intrinsic protein fluorescence titration, the dissociation constant (KD) values between NocT and nopaline and pyronopaline were 3.7±0.6 and 0.57±0.07 µM respectively (Figure S2). These apparent KD values were in the low micromolar range usually observed for PBP ligands [21]. Using isothermal titration calorimetry (ITC), the mean KD value between NocT and pyronopaline was determined as 0.58±0.05 µM (Figure S2), identical to that obtained by fluorescence measurement. The ITC data also confirmed the 1∶1 binding stoichiometry and demonstrated a positive enthalpy change upon pyronopaline binding. This binding reaction was entirely driven by a large favorable increase in entropy (TΔS = 12.6 kcal/mol). With both fluorescence titration and ITC techniques, no interaction could be measured between NocT and arginine; ITC also revealed no interaction between NocT and histidine or ornithine (Figure S2).
Comparison of the ligand binding site of NocT with other PBPs and nopaline-binding signature
Around the arginine moiety of the nopaline/pyronopaline ligand, the ligand binding site of NocT resembled those of the PBPs HisJ and LAO which have been shown to bind arginine and other amino acids (Figure 3). Two residues were conserved in NocT, HisJ and LAO: Tyr39 (Tyr14 in LAO) which stacked the arginine side chain ligand and Arg102 (Arg77 in LAO) which bound the carboxyl group of the arginine ligand. Glu36 was replaced by an equivalent Asp residue that preserved the interaction with the guanidinium group of the ligand. The position of the side chain arginine ligand and that of its carboxyl group were similar in the three PBPs leading to similar polar interactions. The amino group of the arginine ligand was tightly bound in HisJ and LOA with the side chains of a serine at position 72 (corresponding to Gly97 in NocT) and an aspartate at position 161 (Ser207 in NocT). Importantly, these two major interactions will be lost in NocT leading to a less efficient binding of an arginine ligand in line with the observed absence of detectable interaction between NocT and arginine by ITC and fluorescence titration. The presence of Gly97 in NocT seems especially essential for the spatial accommodation of the α-KG moiety of nopaline/pyronopaline whereas the equivalent residue (Ser72) in HisJ and LAO seems incompatible with nopaline or pyronopaline binding due to steric clash.
Fig. 3. Structural comparison between the binding sites of NocT (shown in color domains as in Figure 1) in complex with nopaline and pyronopaline and the Salmonella PBP LAO in complex with arginine (PDB code 1LAF; in yellow). 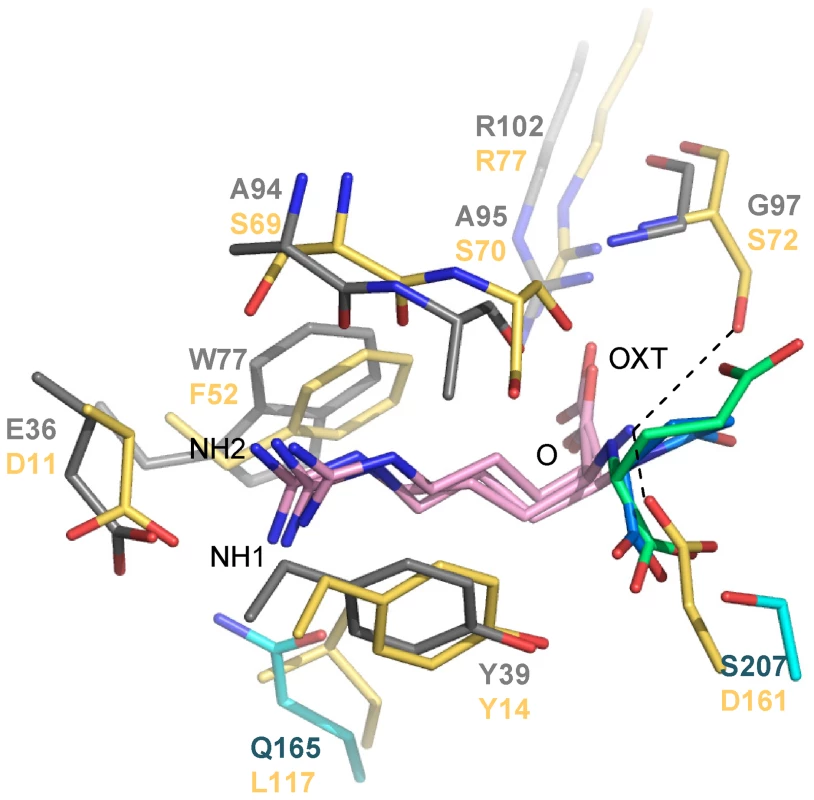
Nopaline, pyronopaline and arginine ligands are shown in pink/limegreen, pink/blue and pink respectively. In A. tumefaciens strains harbouring an octopine-type Ti plasmid, the PBP OccJ allows the uptake of the opine octopine which is a condensate of arginine and pyruvate [25]. Interestingly, NocT seems involved in the importation of nopaline and octopine (only in the presence of nopaline), whereas OccJ permits the importation of octopine but not that of nopaline ([25], [26]. The structure of OccJ is unknown. Sequence comparison between NocT and OccJ shows that among the 12 residues of NocT which directly interact with nopaline/pyronopaline, 4 are different in OccJ. These are G97, M117, H170 and S207 in NocT which correspond to serine, asparagine, alanine and asparagine residues in OccJ, respectively. As mentioned above in the NocT/LAO/HisJ comparison, the replacement of Gly97 in NocT by a serine at the equivalent position might be responsible for preventing OccJ from binding nopaline. Moreover M117, H170 and S207 interact only with the α-KG moiety of nopaline/pyronopaline. We therefore define these 4 amino acids G97M117H170S207 as the nopaline-binding signature.
To learn more on the role of Met117 residue in the interaction between NocT and its ligand, we constructed two NocT mutants with different short polar residues: NocT-M117N because an asparagine is found in the OccJ sequence and NocT-M117S because a serine is found in the LAO/HisJ sequences. Both mutants displayed a similar KD for pyronopaline that was 76-fold higher than that of the WT protein, proving that Met117 is a key component of the ligand affinity and belongs to the nopaline-binding signature. We also obtained the structure of NocT-M117N in complex with pyronopaline (Table S1). Prior to this, the stability of NocT-M117N was verified using differential scanning calorimetry (DSC) and, compared to the WT protein, the mutant presented similar Tm (60.12°C versus 60.25°C for the WT) and ΔH (1.59 105 cal/mol/°C versus 1.4 105 cal/mol/°C for the WT) (Figure S3), meaning that mutating M117 has no significant effect on protein stability. The structure of NocT-M117N in complex with pyronopaline confirmed that the replacement of Met117 with Asn led to a loss of hydrophobic interaction with the ligand and showed that His170 which is free to move towards Asn117 bound the carboxyl group of the α-KG of the pyronopaline in a position not observed in the WT complexes (Figure S4). A loss of the polar interaction between the side chain of Ser169 and the terminal OH of the pyronopaline ring was also observed due to steric hindrance.
The NocT phylogenetical cluster is branched to amino acid-binding PBPs
Five hundred bacterial NocT-homologous PBPs with a threshold set at at least 40% of identity were recovered using blastp at NCBI. To these were added 53 other homologous sequences found in the Agrobacterium genomes of the AgrobacterScope genome library (Genoscope, France). The relation tree constructed from these 553 sequences revealed different subgroups. The closest NocT relatives were used to build a novel relation tree rooted with the HisJ and LAO sequences (Figure 4). Members of the NocT subgroup were highly similar PBPs which belong to the A. tumefaciens strains C58, S56, Zutra3-1, Kerr14 (biovar 1), and A. radiobacter K84 (biovar 2). All of them are nopaline-assimilating agrobacteria. Their genomes exhibited a strong synteny with respect of the noc operon region of A. tumefaciens C58. Moreover, all these NocT proteins retained the nopaline-binding signature G97M117H170S207. Outside the NocT subgroup, the nopaline-binding signature was strongly degenerated, but the arginine-binding signature was conserved in some proteins, such as one PBP of the marine alpha-proteobacterium BAL199 (NCBI BioProject PRJNA54661). These features suggested that the ability to bind nopaline was highly specific to members of the NocT PBP-subgroup, and that NocT and some arginine-PBPs could have evolved from a common ancestor.
Fig. 4. NocT phylogeny and occurence of the nopaline-binding signature. 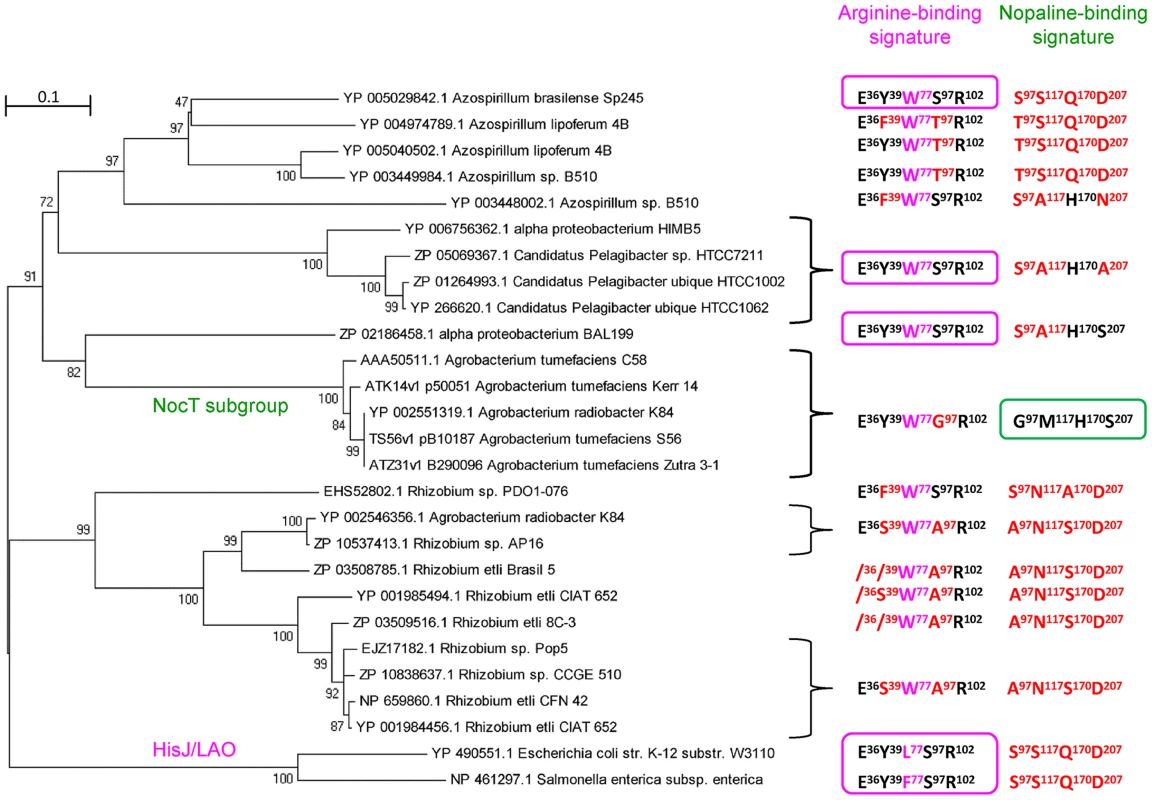
The displayed subtree was rooted with HisJ and LAO sequences. For each protein or protein cluster, the residues which are identical to (black) and different from (red) those involved in the binding of arginine (pink box) and nopaline (green box) are indicated. The W, F and L residues at position 77 (pink) may play a similar role in maintaining arginine. Nopaline/pyronopaline regulon in A. tumefaciens C58
The capability of NocT to bind pyronopaline suggested that A. tumefaciens C58 could use the same pathway for the assimilation of nopaline and pyronopaline. To test this hypothesis, we constructed two A. tumefaciens C58 KO-mutants that were affected in the binding and transport (nocT) and catabolism (ocd) of nopaline. When nopaline or pyronopaline were used as sole sources of carbon and nitrogen and when nopaline was used as a sole source of carbon in the presence of inorganic nitrogen, the nocT mutant could not grow while the growth of ocd mutant was strongly impaired (Figure 5A). These features established that nopaline and pyronopaline were assimilated by the same catabolic pathway Noc, which had been previously characterized for nopaline [27]. In this pathway (Figure 5B), the nopaline is cleaved into α-KG and arginine by the enzymatic complex NoxAB; then, the degradation of arginine into proline is completed by the enzymes Arc, ArcA and Ocd, and the conversion of proline into glutamate, a C and N-source, by the enzyme PutA ( = Atu4157) [27]–[30].
Fig. 5. The nopaline/pyronopaline regulon in A. tumefaciens C58. 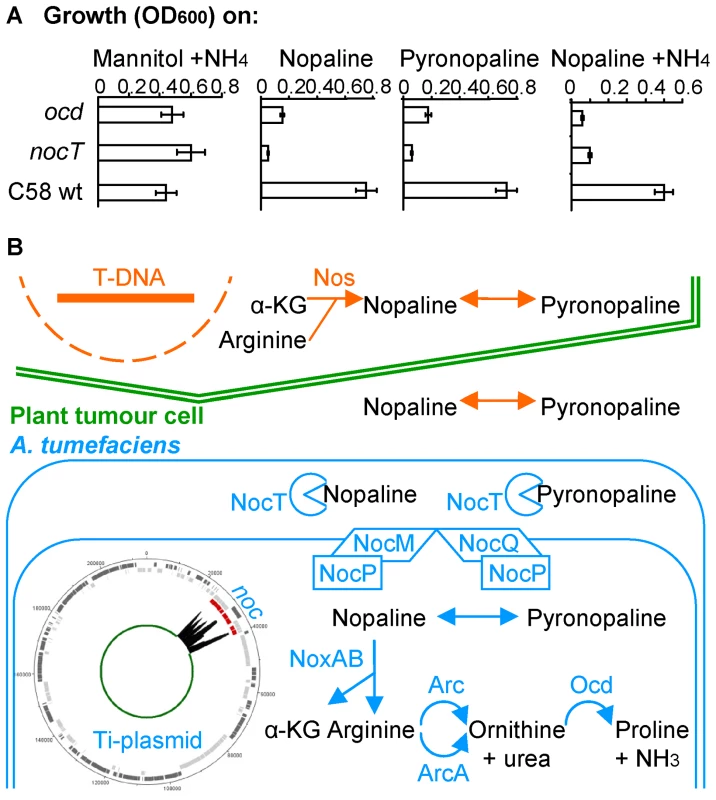
A, growth of the A. tumefaciens C58 wt and nocT and ocd mutants on AB medium supplemented with mannitol and NH4, or nopaline or pyronopaline as sole sources of carbon and nitrogen. The OD600 values were taken after 24 hours; B, schematic representation of the nopaline/pyronopaline synthesis and degradation pathways in the plant tumour. The NocPTQM (ABC transporter), NoxAB (nopaline oxidase A and B), Arc (arginase) and Ocd (ornithine cyclodeaminase) proteins are encoded in the noc region of the Ti-plasmid. A transcriptome graph indicates the position and expression fold change of the noc genes on the Ti-plasmid in presence of nopaline/pyronopaline. The ArcA protein is encoded in the linear chromosome. Spontaneous conversion between nopaline and pyronopaline is represented by double arrows. Furthermore, we delineated the nopaline and pyronopaline regulon by transcriptomics. In this experiment, an accR mutant of A. tumefaciens C58 was used because this mutant expresses the transcriptional regulator NocR at a higher level than does the wild type strain [31]. In presence of nopaline NocR induces the expression of the noc genes [32]. Consequently, an optimal expression of the nopaline regulon was expected in the accR genetic background. Noticeably, the accR mutant also mimics the condition of an exposure to the opine agrocinopine that accumulates in A. tumefaciens C58-induced plant tumours [33]. Comparative transcriptomics in cell culture of an accR mutant grown in the presence or absence of nopaline and pyronopaline should therefore reveal nopaline/pyronoplaine responsive genes in a context reminiscent of the plant tumour environment. With this approach, 32 differentially (Fch>3; P<0.05) expressed genes of A. tumefaciens C58 were identified in the presence of nopaline/pyronopaline, 8 of them being downregulated and 24 upregulated (Table S2). Subsequent RT-qPCR experiments on a selection of genes (cysJ, arcA, nocT, nocP and noxB) confirmed the micro-array results (Table S2). The highest upregulated genes (Fch>20) included the noc operons of the Ti-plasmid [14] and an arginase-encoding gene (arcA = atu4007) located on the linear chromosome. As no other catabolic functions were affected, we suggest to refer to these highly-expressed genes as the nopaline/pyronopaline core-regulon (Table S2).
NocT-mediated selective advantages in the plant-tumour niche
In the tumours induced on tomato plants, we measured the level of nopaline and pyronopaline. Nopaline but not pyronopaline could be detected by mass spectrometry. Moreover, nopaline accumulated at significant and equivalent levels in tumour tissues induced by the C58-control and by the nocT strains (Figure 6A). We therefore confirmed that the tumour niche is a natural nopaline-rich environment. However, as the quantification of nopaline was performed on whole plant tumour extracts, it remains possible that only a small fraction of this nopaline is available to the A. tumefaciens cells colonizing the exterior surface of the tumour.
Fig. 6. Involvement of NocT in the fitness of A. tumefaciens in the plant tumour. 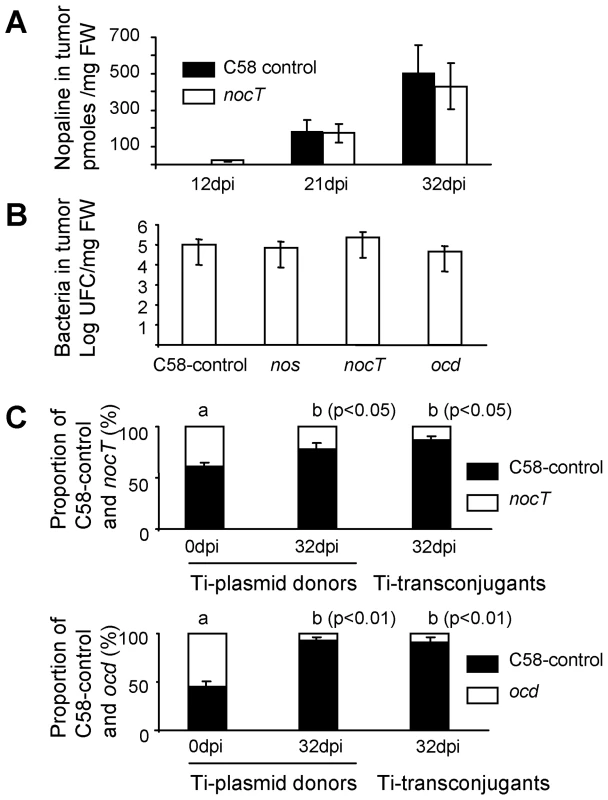
A, nopaline level in plant tumours induced by A. tumefaciens C58-control and nocT derivative; B, cell number of A. tumefaciens C58-control and its nos, nocT and ocd derivatives when they individually colonize plant tumour; C, competitive infection between the A. tumefaciens C58-control strain and nocT or ocd derivatives, all acting as Ti-plasmid donors in the presence of the recipient A. tumefaciens C58.00. Proportion (%) of the Ti-plasmid genotypes among donor strains in the inoculum (0 dpi) and the mature tumours (32 dpi) and among transconjugants (32 dpi) is indicated. Exact Fischer test was used for statistical analysis (p-value thresholds are indicated on the graph). Thereafter, we investigated the selective advantage conferred upon A. tumefaciens C58 by the nopaline pathway in plant tumours. In addition to the nocT and ocd mutants that were impaired in the assimilation of nopaline, we constructed a nos mutant that was defective for the synthesis of the nopaline by the tumour cells (see nopaline pathway in the Figure 5). When these mutants were individually used to inoculate tomato plants, they colonized the tumour tissues at population sizes (105 CFU/mg FW) similar to that reached by the C58-control strain (Figure 6B). Hence, the tumour environment provides enough nutrients to support the growth of the pathogens whatever their capacities to produce, transport or catabolize the nopaline and pyronopaline.
In a second experiment, the nos, nocT and ocd KO-mutants were co-inoculated with the C58-control strain in presence of the recipient strain C58.00 (free of Ti and At plasmids). For each of these conditions the mixed populations colonized the plant tumours at levels (total cell number at 105 CFU/mg FW) that were similar to those reached in the single infections. The frequency of Ti-plasmid donors was determined at the infection time and in the plant tumours to allow the calculation of competitive indexes (CI, see materials and methods). As expected, the CI value reached 1.0 in the competition between the nos mutant and C58-control. In this case, nopaline that is synthesized via the expression of the wild-type T-DNA gene, benefited equally to the two co-inoculated nopaline-utilizing populations (nos mutant and C58-control). In contrast, CI values were only 0.35 and 0.07 in the competitions that involved nocT or ocd mutants and C58-control, respectively. These low CI values revealed that the C58-control population significantly outcompeted nocT and ocd populations which were unable to assimilate nopaline (Figure 6C).
Unlike agrocinopines A and B, nopaline is not a conjugative opine in A. tumefaciens C58, hence the regulation of the plasmid Ti-transfer genes is independent of the presence and concentration of nopaline. In the above plant tumour assays, we were therefore also able to evaluate the capacity of each of the competitors to disseminate its Ti-plasmid into the recipient strain C58.00. In plant tumours, Ti-plasmid transconjugants reached up to 102 CFU/mg FW. The proportion of transconjugants which acquired the Ti-plasmid from each of the two donors in competition was measured (Figure 6C). We observed that, among transconjugants, the relative abundance of the Ti-plasmid that conferred nopaline-assimilation was higher than that of the Ti-plasmid that did not confer nopaline-assimilation. Preferential accumulation of the nopaline-using transconjugants we observed could simply mirror the relative abundance of Ti-plasmid donors in the tumour niche. The competitive advantage which is conferred by opine assimilation could therefore also contribute to dominance of opine-using transconjugants over non-users.
Discussion
This work revealed the structural basis of the PBP NocT that is required for the binding and assimilation of the opines nopaline and pyronopaline in A. tumefaciens C58. Structural and amino-acid sequence analyses showed that NocT was unique among more than 150 PBPs that are encoded by the A. tumefaciens C58 genome [34]. Genome data-base analysis also showed that close homologues of NocT can be only found in the nopaline-assimilating strains of the genus Agrobacterium: either in the pathogenic strains C58, S56, Zutra 3-1, and Kerr 14 belonging to the A. tumefaciens species complex, or in non-pathogenic ones such as the biocontrol strain A. radiobacter K84 which carries the nopaline binding nocT gene on a 185-kbp plasmid [35]. In combination with toxin production [36], [37], nopaline assimilation might contribute to the capacity of A. radiobacter K84 to compete with A. tumefaciens pathogens in the plant environment. Based on our data we cannot rule out the possibility that other nopaline-assimilating isolates of different Agrobacterium species for whom genome sequence is still unknown could also possess a NocT PBP [38], [39].
Previous reports established that the two PBPs LAO (Salmonella) and HisJ (Escherichia), structurally close to NocT, can bind basic amino acids such as arginine [23], [24]. The resemblance between NocT and LAO/HisJ structures and ligand binding sites is not surprising because nopaline and pyronopaline are L-arginine derivatives. However, we showed here that NocT cannot bind arginine (nor histidine and ornithine) probably due to the absence of two essential side chains (Ser72 and Asp161 in LAO corresponding to Gly97 and Ser207 in NocT), both locking the α-amine. The absence of binding between arginine and NocT might confer two advantages. First it might prevent arginine from competing with nopaline for uptake by the Noc system and second it might prevent NocT from becoming saturated with arginine if it cannot pass on to the nopaline transporter. In parallel the presence of a Ser in LAO and HisJ at the equivalent position of Gly97 in NocT which creates a steric hindrance for the accommodation of nopaline and pyronopaline might prevent these molecules from competing with arginine, ornithine or histidine for being bound by their respective PBPs.
The NocT structure is proposed as a reference for further structural comparisons with PBPs involved in the sensing and uptake of other opines, such as agrocinopine [15] and octopine [35]. The opine octopine is a condensate of arginine and pyruvate, therefore structurally close to nopaline. In A. tumefaciens strains harbouring an octopine-type Ti plasmid, octopine is recognized by the PBP OccJ and then transported into the cytoplasm where it acts both as a nutrient and a conjugative signal [25]. Interestingly, NocT is involved in the uptake of nopaline and that of octopine in the presence of nopaline; in contrast, OccJ is involved in the uptake of octopine but not nopaline [25], [26]. Although OccJ and NocT share little homology (48% of identity) 8 of the 12 residues interacting with nopaline in NocT are conserved in OccJ. Based on the 4 differences we defined a nopaline-binding signature and highlighted the importance of Gly97 in NocT for spatial accommodation of nopaline/pyronopaline. This signature notably explains why among LAO, HisJ, NocT and OccJ - which all share strong similarities for the arginine moiety of their ligand - NocT is unique in its ability to bind nopaline/pyronopaline. Further elucidation of the octopine-liganded structures of the PBPs OccJ and NocT would be informative in order to understand the binding properties of octopine and compare it with those of nopaline. Moreover, as enzymes encoded by the noc genes are able, once activated, to catabolize octopine [25], [26], it would be of great interest to investigate the mechanisms which drive the specific assimilation of nopaline and octopine in A. tumefaciens C58, and notably the role of the transcriptional regulator NocR which controls the expression of the noc genes. The question about how co-existing A. tumefaciens populations evolved a specialized or generalist capacity in the synthesis and degradation pathways of opines remains to be explored.
Some opines, i.e. nopaline, succinanopine and leucinopine can be spontaneously converted into cyclic derivatives under acidic conditions [4], [16], [17]. Pyronopaline was detected in tumours induced by A. tumefaciens C58 on the monocot Asparagus officinalis [17], but its occurrence was not explored further in other plant hosts. In this work, pyronopaline was not detected in the tomato plant-tumours induced by the same pathogen (i.e. A. tumefaciens strain C58), suggesting that plant genotype and/or growth conditions may influence the nopaline/pyronopaline conversion in vivo. Even though the formation of the gamma-lactam ring strongly modifies the α-KG part of nopaline, we showed that NocT bound pyronopaline with an affinity higher than that it exhibited for nopaline. Interestingly, nopaline formed with NocT one more polar protein interactions than pyronopaline. However, these interactions were shorter for the pyronopaline. In terms of affinity, the present study highlighted the important contribution of hydrophobic contacts as the positive enthalpy of 2.1 kcal/mol indicated. The tight contacts of the Met117 side chain with the pyronopaline probably made this ligand conformation preferential for NocT. A combination of transcriptomics and genetics established that A. tumefaciens C58 assimilated nopaline and pyronopaline via the same Ti-plasmid encoded pathway. Hence, regardless of the nopaline/pyronopaline equilibrium in the plant tumours, A. tumefaciens appears to mobilize a unique binding, transport, and assimilation system to use them as nutrients.
With respect to the T-DNA/opine-mediated niche construction process, this work experimentally evidenced the validity of the opine concept [3], [4] within the natural agrobacterial environment that the plant tumour is. Indeed, we demonstrated that binding and assimilation of the opine nopaline contributed to the fitness of A. tumefaciens strain C58 in the plant tumour when nopaline assimilating and non-assimilating bacteria were co-infected. On the opposite, when both bacterial types were infected separately, they multiplied to reach a similar level in the plant tumour. The plant tumour is a nutrient-rich environment in which sugars, amino acids, phosphate and sulfate accumulate [40]. This observation supports the notion that the presence of nopaline, made available for bacteria in tumours, does not increase the carrying capacity of plant-tumour habitat for Agrobacterium pathogens, but selects those able to assimilate it. Using transgenic plants producing opines in all plant tissues but free of Agrobacterium-induced tumour, several studies investigated colonization and fitness of opine assimilating and non-assimilating Pseudomonas in rhizosphere and phyllosphere [10], [12], [13]. In all the opine-rich compartments, opine assimilating Pseudomonas outcompeted non-assimilating Pseudomonas. In the same reports, the carrying capacity of the plant tissues was only increased in the carbon-poor phyllosphere compartment, but was not affected in the carbon-rich rhizosphere compartment. The magnitude of the nutrient bias induced by opines seems therefore to directly influence the dynamics and fitness of the bacterial populations. However, it was previously reported that opine accumulation in plant tumour may favor its translocation to other parts of the host plant [41]. This suggests that opines which are stocked in plant tissues could benefit to the pathogen at different times of the infection cycle even after the death of the host plant, by allowing the maintenance of virulent populations (carrying a Ti-plasmid) in soil until the infection of a new host.
Materials and Methods
Purification of NocT and its constructed mutants
Using 5′-GGAATTCCATATGAAGGACTACAAAAGCATT and 5′-TTTGCGGCCGCTTAATGGTGATGGTGATGGTGCTGCTTGGGGGAGGCGTC primers, nocT gene of A. tumefaciens C58 was amplified and then cloned into pET-9aSN1 expression vector (a gift from S. Chéruel, IBBMC, University Paris Sud, Orsay, France). The pET-9aSN1-nocT was used as template to generate directed mutations with QuikChange II XL directed mutagenesis kit (Stratagene). For mutations Met117 to Ser (M117S) and Asn (M117N), the synthetic forward primers 5′–TATCTCCTCAGCCCGAGTACGTTCTTG and 5′-TATCTCCTCACGCCGAATACGTTCTTG and their reverse complemented primers were designed. The nucleotide sequence of all alleles was confirmed by DNA-sequence analysis (GATC, France).
E. coli Rosetta pLysS (Merck) was transformed by the recombinant plasmid. Cells were grown at 37°C in tryptone-yeast extract (TY) broth supplemented with 0.5 mM IPTG to induce NocT production, then centrifuged, resuspended in a buffer that consisted in 50 mM Tris–HCl, pH 8.0, 20 mM imidazole and 500 mM NaCl and disrupted by sonication. After centrifugation at 20 000 g for 30 min at 4°C, the supernatant was loaded onto a 5 mL Ni-NTA agarose column (GE Healthcare). Elution of NocT was performed with the following buffer: 50 mM Tris-HCl pH 8.0, 300 mM imidazole and 500 mM NaCl. NocT containing fractions were loaded onto a gel filtration column (HiLoad 26/60 Superdex 200 prep grade, GE Healthcare) equilibrated with 50 mM Tris-HCl pH 8.0 and 150 mM NaCl. NocT (29 221.6 Da including His tag without the signal peptide) was concentrated to 80 mg/mL (2.73 mM).
Purification, synthesis and quantification of opines
Nopaline was extracted from crushed tomato plant tumour tissues and purified according to the procedure described by Tempé [42]. Pyronopaline was obtained by synthesis using arginine and α-KG as precursors in the presence of sodium cyanoborohydride as described by Tempé [42]. This synthesis resulted in a mix of nopaline and pyronopaline. The complete conversion of this mix into pyronopaline was obtained as described previously [17]. The quantification of nopaline was performed as previously described [31] from macerates of whole tomato tumours.
All solutions of nopaline, pyronopaline and nopaline/pyronopaline were checked by mass spectrometry. The mass spectrometry measurements were performed in negative mode with an electrospray Q/TOF mass spectrometer (Q/TOF Premier, Waters) equipped with the Nanomate device (Advion) with compounds diluted in 50% acetonitrile and 1% formic acid. The Mass Lynx 4.1 software was used for acquisition and data processing. The external calibration was performed with NaI clusters (2 µg/µL, isopropanol/H2O 50/50, Waters) in the acquisition m/z mass range and the estimated mass accuracy is ±0.01 Da (at 300 Da). The spectra of nopaline and pyronopaline are shown in the figure S5.
Crystallization and structure determination
Crystallization conditions for unliganded (40 mg/mL or 1.37 mM) and liganded NocT (solution of 2.73 mM protein and 8 mM ligand) as well as for the M117N mutant were screened in sitting-drop vapour-diffusion experiments using the Classics and PEG II kits from Qiagen on a nanodrop robot (Cartesian, Proteomic Solution). Condition 34 from Classics was manually optimized at 293 K with home made solution in hanging drops composed of a 1∶1 volume ratio of crystallization solution (2 M Ammonium Sulfate (AS), 0.1 M Na Citrate pH 5.6, 0.2 M K tartrate and 5% PEG 400). Similar manually optimization of condition 31 from PEG II (30% PEG 4000, 0.1 M Tris pH 8 and 0.1M LiSO4) for both WT and mutant liganded proteins led to plate-shaped crystals. Crystals were transferred to a cryo-protectant solution (paraffin oil for AS precipitant or 20% (w/v) PEG 400 for PEG precipitant) and flash-frozen in liquid nitrogen. Diffraction data were collected at 100 K on the PROXIMA I beamline at SOLEIL synchrotron (Saint-Aubin, France). Data collection and processing statistics are given in Table S1. Structure determination of all crystals were performed by molecular replacement with PHASER [43] using first the coordinates of the N-terminal (residues 1–86 and 197–238) and the C-terminal (residues 93–185) of the E.coli histidine binding protein HisJ (1HSL) as two search models for the free-liganded structure and next our NocT model for the wild-type or mutant liganded-NocT complexes. Refinement was performed with BUSTER-2.10 [44] with NCS restraints as all asymmetric units contain two protein molecules. One TLS group was assigned for each structure. Electron density maps were evaluated using COOT [45]. Refinement details are shown in Table S1. Molecular graphics images were generated using PYMOL (http://www.pymol.org).
KD measurements by fluorescence titration and microcalorimetry
Ligand-NocT interaction was monitored by autofluorescence by exciting the protein at a wavelength of 295 nm and monitoring the quenching of fluorescence emission of tryptophan residues at 340 nm. All experiments were performed at 25°C in 25 mM Tris-HCl pH 8.0 and 150 mM NaCl with a fixed amount of proteins (5 µM) and increasing concentrations of ligand using a SpectraMax M5 microplate reader (Molecular Devices). Each ligand exhibited no emission signal at 340 nm. Fluorescence measurements were done in triplicates. The data were analysed using Origin 7 software and fitted to the equation f = ΔFluorescencemax * abs(x)/(KD+abs(x)).
ITC experiments were performed with an ITC200 isothermal titration calorimeter from MicroCal Llc (Northampton, MA). The experiments were carried out at 20°C. Protein concentration in the microcalorimeter cell (0.2 ml) varied from 100 to 150 µM. Nineteen injections of 2 µl of the ligand solution concentration from 1.5 to 1.6 mM were performed at intervals of 180 s while stirring at 1000 rpm. The experimental data were fitted to theoretical titration curves with software supplied by MicroCal (ORIGIN). This software uses the relationship between the heat generated by each injection and ΔH (enthalpy change in Kcal.mol−1), Ka (the association binding constant in M−1), n (the number of binding sites), total protein concentration and free and total ligand concentrations.
Differential scanning calorimetry
Thermal stability of wild type and M117N-NocT (20.5 µM) was studied by differential scanning calorimetry (DSC) on a MicroCal model VP-DSC in a standard buffer. Each measurement was preceded by a baseline scan with the standard buffer. All solutions were degassed just before loading into the calorimeter. Scans were performed at 1K.min-1 between 20 and 90°C. The heat capacity of the buffer was subtracted from that of the protein sample before analysis. Thermodynamic parameters were determined by fitting the data to the following equation ΔCp(T) = (Kd(T) ΔHcal ΔHvH)/((1+Kd(T))2 RT2) where Kd is the equilibrium constant for a two-state process, ΔHvH is the enthalpy calculated on the basis of a two-state process and ΔHcal is the measured enthalpy.
Growth conditions and construction of the A. tumefaciens C58 mutants
The A. tumefaciens C58 derivatives carrying pTi-accR::Gm and pTi::Gm (used as C58-control in plant assay) in which the genes accR (atu6138) and atu6147 were disrupted by inserting a gentamicin resistance cassette were already constructed [46]. The A. tumefaciens C58 derivatives harbouring the Ti-plasmid pTi-nos::Km, pTi-ocd::Gm, pTi-nocT::Gm were obtained by insertion of a gentamicin (Gm) or kanamycin (Km) resistance cassette into the genes nos ( = atu6015), ocd ( = atu6016), and nocT ( = atu6027) as described previously [46].
A. tumefaciens was cultivated at 30°C in Agrobacterium broth (AB) minimal medium supplemented with ammonium chloride (1 g/L) and mannitol (2 g/L) except when an alternative source of carbon and nitrogen is indicated, or in Luria-Bertani modified medium (LBm; NaCl 5 g/L). In growth assay nopaline and pyronopaline were added as a sole carbon and nitrogen source at 3 mM (ca 1 g/L). The antibiotics gentamycin and kanamycin were added at 25 µg/mL and 50 µg/mL, respectively.
Transcriptomic analysis
Overnight cultures of the A. tumefaciens accR mutant were sub-cultured at an initial OD600 of 0.05 in 50 ml of AB medium containing ammonium chloride (1 g/L) and mannitol (2 g/L) which was supplemented or not with the mix nopaline/pyronopaline at 1 mM. Cells were grown at 28°C for approximately 9 hours until early exponential phase (OD600 = 0.4). RNA extraction was performed using a phenol-based procedure, according to Planamente et al. [19]. Construction of cDNA libraries, hybridization and signal quantification were performed by the PartnerChip platform (Génopole Evry, France). Experiments were performed in triplicates. Normalized data of samples were pairwise compared and P values corresponding to statistical t-test were attributed for each gene.
Plant infection and numeration of bacterial populations
Tomato plants (F1 hybrid Dona, Vilmorin, France) were grown in greenhouse under long day conditions and controlled temperature (24–26°C). One-month old plants were scalpel wounded between first and second stem nodes and inoculated with the agrobacteria as described previously [20]. For each independent experiment, five to seven plant tumours (32 dpi) were crushed into NaCl 0.8% to recover the bacteria which were then spotted onto selective agar media to enumerate colony forming units (CFU). In the case of mixed infections, the proportions of the genotypes (C58-control and nos, nocT and ocd KO-mutants) were measured in the inoculum (Pi) and the plant tumour (Pt) using antibiotic resistances and PCR with appropriate primers. This allowed calculation of the competitive index CI = (Ptmutant/Ptcontrol)/(Pimutant/Picontrol) as described by Macho et al. [47]. The A. tumefaciens derivative C58.00 that contains no plasmid but harbours a chromosomal resistance to rifampicin, was used as a recipient strain. The transconjugants that acquired a Ti-plasmid from the donor strains were enumerated using antibiotic resistances and PCR with appropriate primers.
Phylogenetic analysis
Sequences were analyzed using blastP from NCBI (http://blast.ncbi.nlm.nih.gov/) and MicroScope (https://www.genoscope.cns.fr/agc/microscope/about/collabprojects.php). Alignements of NocT and related sequences were conducted using the ClustalW software. Relationship tree was build using the MEGA software, Version 5. The phylogeny was inferred using the neighbor-joining method. The bootstrap consensus tree inferred from 1000 replicates was taken to represent the evolutionary history of the taxa analyzed. The evolutionary distances are in units of the number of amino acid substitutions per site.
Accession codes
The atomic coordinates and structure factors of the unliganded NocT and the complexes with nopaline and pyronopaline as well as the M117N-NocT in complex with pyronopaline have been deposited in the protein data bank (http://www.rcsb.org) under accession codes 4P0I, 4POX, 4POW and 4PP0 respectively.
Supporting Information
Zdroje
1. GelvinSB (2003) Agrobacterium-mediated plant transformation: the biology behind the “gene-jockeying” tool. Microbiol Mol Biol Rev 67 : 16–37.
2. PitzschkeA, HirtH (2010) New insights into an old story: Agrobacterium-induced tumour formation in plants by plant transformation. 29 : 1021–1032.
3. SchellJ, Van MontaguM, De BeuckeleerM, De BlockM, DepickerA, et al. (1979) Interactions and DNA transfer between Agrobacterium tumefaciens, the Ti-plasmid and the plant host. Proc R Soc Lond B Biol Sci 204 : 251–266.
4. Tempé J, Petit A (1983) La piste des opines. In: Pühler A, editor. Molecular genetics of the bacteria-plant interaction. Berlin-Heidelberg: Springer-Verlag. pp. 14–32.
5. DessauxY, PetitA, TempéJ (1993) Chemistry and biochemistry of opines, chemical mediators of parasitism. Phytochem 34 : 31–38.
6. Flores-MirelesAL, EberhardA, WinansSC (2012) Agrobacterium tumefaciens can obtain sulphur from an opine that is synthesized by octopine synthase using S-methylmethionine as a substrate. Mol Microbiol 84 : 845–856.
7. PiperKR, Beck von BodmanS, FarrandSK (1993) Conjugation factor of Agrobacterium tumefaciens regulates Ti plasmid transfer by autoinduction. Nature 362 : 448–450.
8. GuyonP, ChiltonMD, PetitA, TempéJ (1980) Agropine in “null-type” crown gall tumours: Evidence for generality of the opine concept. Proc Natl Acad Sci USA 77 : 2693–2697.
9. BellCR, CummingsNE, CanfieldML, MooreLW (1990) Competition of octopine-catabolizing Pseudomonas spp. and octopine-type Agrobacterium tumefaciens for octopine in chemostats. Appl Environ Microbiol 56 : 2840–2846.
10. WilsonM, SavkaMA, HwangI, FarrandSK, LindowSE (1995) Altered Epiphytic Colonization of Mannityl Opine-Producing Transgenic Tobacco Plants by a Mannityl Opine-Catabolizing Strain of Pseudomonas syringae. Appl Environ Microbiol 61 : 2151–2158.
11. GuyonP, PetitA, TempéJ, DessauxY (1993) Transformed plants producing opines specifically promote growth of opine-degrading agrobacteria. Mol Plant-Microbe Interact 6 : 92–98.
12. OgerP, PetitA, DessauxY (1997) Genetically engineered plants producing opines alter their biological environment. Nat Biotechnol 15 : 369–372.
13. SavkaMA, FarrandSK (1997) Modification of rhizobacterial populations by engineering bacterium utilization of a novel plant-produced resource. Nat Biotechnol 15 : 363–368.
14. SchardlCL, KadoCI (1983) A functional map of the nopaline catabolism genes on the Ti plasmid of Agrobacterium tumefaciens C58. Mol Gen Genet 191 : 10–16.
15. KimH, FarrandSK (1997) Characterization of the acc operon from the nopaline-type Ti plasmid pTiC58, which encodes utilization of agrocinopines A and B and susceptibility to agrocin 84. J Bacteriol 179 : 7559–7572.
16. HallLM, SchrimsherJL, TaylorKB (1983) A new opine derived from nopaline. J Biol Chem 258 : 7276–7279.
17. HernalsteensJP, Thia-ToongL, SchellJ, Van MontaguM (1984) An Agrobacterium-transformed cell culture from the monocot Asparagus officinalis. EMBO J 3 : 3039–3041.
18. OhBH, PanditJ, KangCH, NikaidoK, GokcenS, et al. (1993) Three-dimensional structures of the periplasmic lysine/arginine/ornithine-binding protein with and without a ligand. J Biol Chem 268 : 11348–11355.
19. PlanamenteS, MondyS, HommaisF, VigourouxA, MoréraS, et al. (2013) Structural basis for selective GABA binding in bacterial pathogens. Mol Microbiol 86 : 1085–1099.
20. PlanamenteS, VigourouxA, MondyS, NicaiseM, FaureD, et al. (2010) A conserved mechanism of GABA binding and antagonism is revealed by structure-function analysis of the periplasmic binding protein Atu2422 in Agrobacterium tumefaciens. J Biol Chem 24 : 30294–30303.
21. BerntssonRP, SmitsSH, SchmittL, SlotboomDJ, PoolmanB (2010) A structural classification of substrate-binding proteins. FEBS Lett 584 : 2606–2617.
22. KrissinelE, HenrickK (2004) Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr 60 : 2256–2268.
23. YaoN, TrakhanovS, QuiochoFA (1994) Refined 1.89-A structure of the histidine-binding protein complexed with histidine and its relationship with many other active transport/chemosensory proteins. Biochem 33 : 4769–4779.
24. OhBH, AmesGF, KimSH (1994) Structural basis for multiple ligand specificity of the periplasmic lysine-, arginine-, ornithine-binding protein. J Biol Chem 269 : 26323–26330.
25. ZankerH, von LintigJ, SchröderJ (1992) Opine transport genes in the octopine (occ) and nopaline (noc) catabolic regions in Ti plasmids of Agrobacterium tumefaciens. J Bacteriol 174 : 841–849.
26. KlapwijkPM, OudshoornM, SchilperoortRA (1977) Inducible permease involved in the uptake of octopine, lysopine and octopinic acid by Agrobacterium tumefaciens strains carrying virulence-associated plasmids. Microbiology 102 : 1–11.
27. SansN, SchröderG, SchröderJ (1987) The Noc region of Ti plasmid C58 codes for arginase and ornithine cyclodeaminase. Eur J Biochem 167 : 81–87.
28. FarrandSK, DessauxY (1986) Proline biosynthesis encoded by the noc and occ loci of Agrobacterium Ti plasmids. J Bacteriol 167 : 732–734.
29. ChoK, FuquaC, WinansSC (1997) Transcriptional regulation and locations of Agrobacterium tumefaciens genes required for complete catabolism of octopine. J Bacteriol 179 : 1–8.
30. SchardlCL, KadoCI (1983) Ti plasmid and chromosomal ornithine catabolism genes of Agrobacterium tumefaciens C58. J Bacteriol 155 : 196–202.
31. LangJ, PlanamenteS, MondyS, DessauxY, MoréraS, et al. (2013) Concerted transfer of the virulence Ti plasmid and companion At plasmid in the Agrobacterium tumefaciens-induced plant tumour. Mol Microbiol 90 : 1178–1189.
32. Von LintigJ, KreuschD, SchroderJ (1994) Opine-regulated promoters and LysR-type regulators in the nopaline (noc) and octopine (occ) catabolic regions of Ti plasmids of Agrobacterium tumefaciens. J Bacteriol 176 : 495–503.
33. Beck von BodmanS, HaymanGT, FarrandSK (1992) Opine catabolism and conjugal transfer of the nopaline Ti plasmid pTiC58 are coordinately regulated by a single repressor. Proc Natl Acad Sci USA 89 : 643–647.
34. WoodDW, SetubalJC, KaulR, MonksDE, KitajimaJP, et al. (2001) The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science 294 : 2317–2323.
35. SlaterSC, GoldmanBS, GoodnerB, SetubalJC, FarrandSK, et al. (2009) Genome sequences of three agrobacterium biovars help elucidate the evolution of multichromosome genomes in bacteria. J Bacteriol 191 : 2501–2511.
36. RobertsWP, TateME, KerrA (1977) Agrocin 84 is a 6-N-phosphoramidate of an adenine nucleotide analogue. Nature 265 : 379–381.
37. KimJG, ParkBK, KimSU, ChoiD, NahmBH, et al. (2006) Bases of biocontrol: sequence predicts synthesis and mode of action of agrocin 84, the Trojan horse antibiotic that controls crown gall. Proc Natl Acad Sci USA 103 : 8846–8851.
38. BouzarH, MooreLW (1987) Isolation of different agrobacterium biovars from a natural oak savanna and tallgrass prairie. Appl Environ Microbiol 53 : 717–721.
39. BouzarH, OuadahD, KrimiZ, JonesJB, TrovatoM, et al. (1993) Correlative association between resident plasmids and the host chromosome in a diverse Agrobacterium soil population. Appl Environ Microbiol 59 : 1310–1317.
40. DeekenR, EngelmannJC, EfetovaM, CzirjakT, MüllerT, et al. (2006) An integrated view of gene expression and solute profiles of Arabidopsis tumours: a genome-wide approach. Plant Cell 18 : 3617–3634.
41. SavkaMA, BlackRC, BinnsAN, FarrandSK (1996) Translocation and exudation of tumour metabolites in crown galled plants. Mol Plant Microbe Interact 9 : 310–313.
42. Tempé J (1983) Chemistry and biochemistry of open-chain imino-acids. In: Weistein B, editor. Chemistry and biochemistry of amino acids, peptides and proteins. New York: Marcel Dekker Inc. pp. 113–203.
43. McCoyAJ, Grosse-KunstleveRW, AdamsPD, WinnMD, StoroniLC, et al. (2007) Phaser crystallographic software. J Appl Crystallogr 40 : 658–674.
44. BlancE, RoversiP, VonrheinC, FlensburgC, LeaSM, et al. (2004) Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr D Biol Crystallogr 60 : 2210–2221.
45. EmsleyP1, CowtanK (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60 : 2126–2132.
46. HaudecoeurE, TannièresM, CirouA, RaffouxA, DessauxY, et al. (2009) Different regulation and roles of lactonases AiiB and AttM in Agrobacterium tumefaciens C58. Mol Plant Microbe Interact 22 : 529–537.
47. MachoAP, GuidotA, BarberisP, BeuzónCR, GeninS (2010) A competitive index assay identifies several Ralstonia solanacearum type III effector mutant strains with reduced fitness in host plants. Mol Plant Microbe Interact 23 : 1197–1205.
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Identification of the Microsporidian as a New Target of the IFNγ-Inducible IRG Resistance SystemČlánek Human Cytomegalovirus Drives Epigenetic Imprinting of the Locus in NKG2C Natural Killer CellsČlánek APOBEC3D and APOBEC3F Potently Promote HIV-1 Diversification and Evolution in Humanized Mouse ModelČlánek Role of Non-conventional T Lymphocytes in Respiratory Infections: The Case of the Pneumococcus
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2014 Číslo 10- Stillova choroba: vzácné a závažné systémové onemocnění
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Jak souvisí postcovidový syndrom s poškozením mozku?
- Infekční komplikace virových respiračních infekcí – sekundární bakteriální a aspergilové pneumonie
-
Všechny články tohoto čísla
- Theory and Empiricism in Virulence Evolution
- -Related Fungi and Reptiles: A Fatal Attraction
- Adaptive Prediction As a Strategy in Microbial Infections
- Antimicrobials, Stress and Mutagenesis
- A Novel Function of Human Pumilio Proteins in Cytoplasmic Sensing of Viral Infection
- Social Motility of African Trypanosomes Is a Property of a Distinct Life-Cycle Stage That Occurs Early in Tsetse Fly Transmission
- Autophagy Controls BCG-Induced Trained Immunity and the Response to Intravesical BCG Therapy for Bladder Cancer
- Identification of the Microsporidian as a New Target of the IFNγ-Inducible IRG Resistance System
- mRNA Structural Constraints on EBNA1 Synthesis Impact on Antigen Presentation and Early Priming of CD8 T Cells
- Infection Causes Distinct Epigenetic DNA Methylation Changes in Host Macrophages
- Neutrophil Crawling in Capillaries; A Novel Immune Response to
- Live Attenuated Vaccine Protects against Pulmonary Challenge in Rats and Non-human Primates
- The ESAT-6 Protein of Interacts with Beta-2-Microglobulin (β2M) Affecting Antigen Presentation Function of Macrophage
- Characterization of Uncultivable Bat Influenza Virus Using a Replicative Synthetic Virus
- HIV Acquisition Is Associated with Increased Antimicrobial Peptides and Reduced HIV Neutralizing IgA in the Foreskin Prepuce of Uncircumcised Men
- Uses a Unique Ligand-Binding Mode for Trapping Opines and Acquiring A Competitive Advantage in the Niche Construction on Plant Host
- Involvement of a 1-Cys Peroxiredoxin in Bacterial Virulence
- Ethanol Stimulates WspR-Controlled Biofilm Formation as Part of a Cyclic Relationship Involving Phenazines
- Densovirus Is a Mutualistic Symbiont of a Global Crop Pest () and Protects against a Baculovirus and Bt Biopesticide
- Insights into Intestinal Colonization from Monitoring Fluorescently Labeled Bacteria
- Mycobacterial Antigen Driven Activation of CD14CD16 Monocytes Is a Predictor of Tuberculosis-Associated Immune Reconstitution Inflammatory Syndrome
- Lipoprotein LprG Binds Lipoarabinomannan and Determines Its Cell Envelope Localization to Control Phagolysosomal Fusion
- Dampens the DNA Damage Response
- MicroRNAs Suppress NB Domain Genes in Tomato That Confer Resistance to
- Novel Cyclic di-GMP Effectors of the YajQ Protein Family Control Bacterial Virulence
- Vaginal Challenge with an SIV-Based Dual Reporter System Reveals That Infection Can Occur throughout the Upper and Lower Female Reproductive Tract
- Detecting Differential Transmissibilities That Affect the Size of Self-Limited Outbreaks
- One Small Step for a Yeast - Microevolution within Macrophages Renders Hypervirulent Due to a Single Point Mutation
- Expression Profiling during Arabidopsis/Downy Mildew Interaction Reveals a Highly-Expressed Effector That Attenuates Responses to Salicylic Acid
- Human Cytomegalovirus Drives Epigenetic Imprinting of the Locus in NKG2C Natural Killer Cells
- Interaction with Tsg101 Is Necessary for the Efficient Transport and Release of Nucleocapsids in Marburg Virus-Infected Cells
- The N-Terminus of Murine Leukaemia Virus p12 Protein Is Required for Mature Core Stability
- Sterol Biosynthesis Is Required for Heat Resistance but Not Extracellular Survival in
- Allele-Specific Induction of IL-1β Expression by C/EBPβ and PU.1 Contributes to Increased Tuberculosis Susceptibility
- Host Cofactors and Pharmacologic Ligands Share an Essential Interface in HIV-1 Capsid That Is Lost upon Disassembly
- APOBEC3D and APOBEC3F Potently Promote HIV-1 Diversification and Evolution in Humanized Mouse Model
- Structural Basis for the Recognition of Human Cytomegalovirus Glycoprotein B by a Neutralizing Human Antibody
- Systematic Analysis of ZnCys Transcription Factors Required for Development and Pathogenicity by High-Throughput Gene Knockout in the Rice Blast Fungus
- Epstein-Barr Virus Nuclear Antigen 3A Promotes Cellular Proliferation by Repression of the Cyclin-Dependent Kinase Inhibitor p21WAF1/CIP1
- The Host Protein Calprotectin Modulates the Type IV Secretion System via Zinc Sequestration
- Cyclophilin A Associates with Enterovirus-71 Virus Capsid and Plays an Essential Role in Viral Infection as an Uncoating Regulator
- A Novel Alpha Kinase EhAK1 Phosphorylates Actin and Regulates Phagocytosis in
- The pH-Responsive PacC Transcription Factor of Governs Epithelial Entry and Tissue Invasion during Pulmonary Aspergillosis
- Sensing of Immature Particles Produced by Dengue Virus Infected Cells Induces an Antiviral Response by Plasmacytoid Dendritic Cells
- Co-opted Oxysterol-Binding ORP and VAP Proteins Channel Sterols to RNA Virus Replication Sites via Membrane Contact Sites
- Characteristics of Memory B Cells Elicited by a Highly Efficacious HPV Vaccine in Subjects with No Pre-existing Immunity
- HPV16-E7 Expression in Squamous Epithelium Creates a Local Immune Suppressive Environment via CCL2- and CCL5- Mediated Recruitment of Mast Cells
- Dengue Viruses Are Enhanced by Distinct Populations of Serotype Cross-Reactive Antibodies in Human Immune Sera
- CD4 Depletion in SIV-Infected Macaques Results in Macrophage and Microglia Infection with Rapid Turnover of Infected Cells
- A Sialic Acid Binding Site in a Human Picornavirus
- Contact Heterogeneity, Rather Than Transmission Efficiency, Limits the Emergence and Spread of Canine Influenza Virus
- Myosins VIII and XI Play Distinct Roles in Reproduction and Transport of
- HTLV-1 Tax Stabilizes MCL-1 via TRAF6-Dependent K63-Linked Polyubiquitination to Promote Cell Survival and Transformation
- Species Complex: Ecology, Phylogeny, Sexual Reproduction, and Virulence
- A Critical Role for IL-17RB Signaling in HTLV-1 Tax-Induced NF-κB Activation and T-Cell Transformation
- Exosomes from Hepatitis C Infected Patients Transmit HCV Infection and Contain Replication Competent Viral RNA in Complex with Ago2-miR122-HSP90
- Role of Non-conventional T Lymphocytes in Respiratory Infections: The Case of the Pneumococcus
- Kaposi's Sarcoma-Associated Herpesvirus Induces Nrf2 during Infection of Endothelial Cells to Create a Microenvironment Conducive to Infection
- A Relay Network of Extracellular Heme-Binding Proteins Drives Iron Acquisition from Hemoglobin
- Glutamate Secretion and Metabotropic Glutamate Receptor 1 Expression during Kaposi's Sarcoma-Associated Herpesvirus Infection Promotes Cell Proliferation
- Reduces Malaria and Dengue Infection in Vector Mosquitoes and Has Entomopathogenic and Anti-pathogen Activities
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Novel Cyclic di-GMP Effectors of the YajQ Protein Family Control Bacterial Virulence
- MicroRNAs Suppress NB Domain Genes in Tomato That Confer Resistance to
- The ESAT-6 Protein of Interacts with Beta-2-Microglobulin (β2M) Affecting Antigen Presentation Function of Macrophage
- Characterization of Uncultivable Bat Influenza Virus Using a Replicative Synthetic Virus
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



