-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Novel Cyclic di-GMP Effectors of the YajQ Protein Family Control Bacterial Virulence
Cyclic di-GMP is a bacterial second messenger that acts to regulate a wide range of functions including those that contribute to the virulence of pathogens. Our knowledge of the different actions and receptors for this nucleotide is far from complete. An understanding of the action of these elements may be key to interference with the processes they control. Here we have used an affinity pull-down assay using cyclic di-GMP-coupled magnetic beads to identify cyclic di-GMP binding proteins in the plant pathogen Xanthomonas campestris. This analysis identified XC_3703, a protein of the YajQ family that was able to bind cyclic di-GMP with high affinity. Mutation of XC_3703 led to reduced virulence of X. campestris to plants and alteration in biofilm formation. Genetic and functional analysis of YajQ family members from the human pathogens Pseudomonas aeruginosa and Stenotrophomonas maltophilia showed that they also specifically bound cyclic di-GMP and contributed to virulence in model systems. The findings thus identify a new class of cyclic di-GMP effector that regulates bacterial virulence and raise the possibility that other members of the YajQ family, which occur widely in bacteria, also act in cyclic di-GMP signalling pathways.
Published in the journal: . PLoS Pathog 10(10): e32767. doi:10.1371/journal.ppat.1004429
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1004429Summary
Cyclic di-GMP is a bacterial second messenger that acts to regulate a wide range of functions including those that contribute to the virulence of pathogens. Our knowledge of the different actions and receptors for this nucleotide is far from complete. An understanding of the action of these elements may be key to interference with the processes they control. Here we have used an affinity pull-down assay using cyclic di-GMP-coupled magnetic beads to identify cyclic di-GMP binding proteins in the plant pathogen Xanthomonas campestris. This analysis identified XC_3703, a protein of the YajQ family that was able to bind cyclic di-GMP with high affinity. Mutation of XC_3703 led to reduced virulence of X. campestris to plants and alteration in biofilm formation. Genetic and functional analysis of YajQ family members from the human pathogens Pseudomonas aeruginosa and Stenotrophomonas maltophilia showed that they also specifically bound cyclic di-GMP and contributed to virulence in model systems. The findings thus identify a new class of cyclic di-GMP effector that regulates bacterial virulence and raise the possibility that other members of the YajQ family, which occur widely in bacteria, also act in cyclic di-GMP signalling pathways.
Introduction
Cyclic di-GMP (bis-(3′-5′) cyclic di-guanylate) is a second messenger in bacteria that acts to regulate a wide range of functions that include adhesion, biofilm formation, motility, synthesis of polysaccharides and synthesis of virulence factors in pathogens (recently reviewed by [1], [2], [3], [4]). The cellular level of cyclic di-GMP results from a balance between synthesis and degradation. Three protein domains are implicated in these processes: the GGDEF domain catalyzes synthesis of cyclic di-GMP from 2 molecules of GTP whereas EAL and HD-GYP domains catalyze hydrolysis of cyclic di-GMP, firstly to the linear nucleotide pGpG and then at different rates to GMP [1], [2], [3], [4]. All of these domains are named after conserved amino acid motifs. Most proteins with GGDEF/EAL/HD-GYP domains contain additional signal input domains, suggesting that their activities are responsive to signals or cues from the bacterial cell or its environment.
A number of cellular effectors or receptors for cyclic di-GMP have already been described in different bacteria [5], [6]. These include proteins with a PilZ domain, enzymatically inactive variants of GGDEF and EAL domains and a number of transcriptional regulators that do not possess a common domain organization [3], [5]. Furthermore, cyclic di-GMP is able to bind to untranslated regions of different mRNAs thereby affecting gene expression via riboswitches [3], [5]. Nevertheless, despite considerable progress, the mechanisms by which cyclic di-GMP exerts its action on diverse cellular processes remain incompletely understood, so that the discovery of further classes of cyclic di-GMP effectors is to be expected.
Here we have used an affinity pull down assay in order to identify potential cyclic di-GMP effectors in the phytopathogen Xanthomonas campestris pv. campestris (hereafter Xcc), the causal agent of black rot disease of cruciferous plants. As well as being a plant pathogen of global importance, Xcc is a model organism for molecular studies of plant-microbe interactions [7], [8]. Xcc has 37 proteins implicated in cyclic di-GMP synthesis and degradation and several of these are known to modulate synthesis of different virulence factors and virulence to plants [9], [10]. Thus far the only cyclic di-GMP effectors identified in Xanthomonas species are the transcription factor Clp, the enzymatically inactive GGDEF-EAL domain protein FimX and several PilZ domain proteins [5], [11], [12], [13], [14].
Our approach to reveal cyclic di-GMP binding proteins in Xcc described here identified XC_3703, a member of the YajQ family of proteins that is broadly distributed in bacteria. Mutational analysis showed that XC_3703 contributed to Xcc virulence to plants. Other members of the YajQ family from the human pathogens Stenotrophomonas maltophilia and Pseudomonas aeruginosa were also shown to preferentially bind cyclic di-GMP and to contribute to virulence in model systems. The findings thus identify a sub-group of the YajQ family of proteins as a new class of cyclic di-GMP effector.
Results
Identification of the cyclic di-GMP receptor protein XC_3703 in Xcc
To identify cyclic di-GMP receptor proteins, we performed an affinity pull-down assay using cyclic di-GMP–coupled magnetic beads and soluble protein extracts derived from the Xcc wild-type strain 8004. The selectively bound proteins were separated by SDS-polyacrylamide gel electrophoresis (Figure 1A) and were identified by peptide mass fingerprinting. Overall, 7 putative cyclic di-GMP binding proteins were identified from three cyclic di-GMP pull down experiments on Xcc 8004 lysates (Table S1). Three of these proteins were previously characterized cyclic di-GMP binding proteins from Xcc: the two PilZ domain-containing proteins XC_0965 and XC_3221 and the transcriptional regulator Clp, which is XC_0486. In addition, the analysis identified XC_1036, a protein containing a GGDEF domain with a predicted I-site (allosteric) cyclic di-GMP binding motif. Of particular interest to the work here was XC_3703 (MASCOT score 1156) a member of the highly conserved YajQ family of bacterial proteins. XC_3703 is related to YajQ of Escherichia coli (BLASTP E value is 10−20), a protein of unknown function that has motifs characteristic of nucleotide-binding proteins (Figure S1). YajQ family proteins are encoded by many bacterial genomes, to include both Gram-negative and Gram-positive bacteria. An amino acid sequence alignment of XC_3703 with sequences from a selected range of organisms is shown in Figure S1.
Fig. 1. Identification of Xanthomonas campestris XC_3703 as a cyclic di-GMP- binding protein. 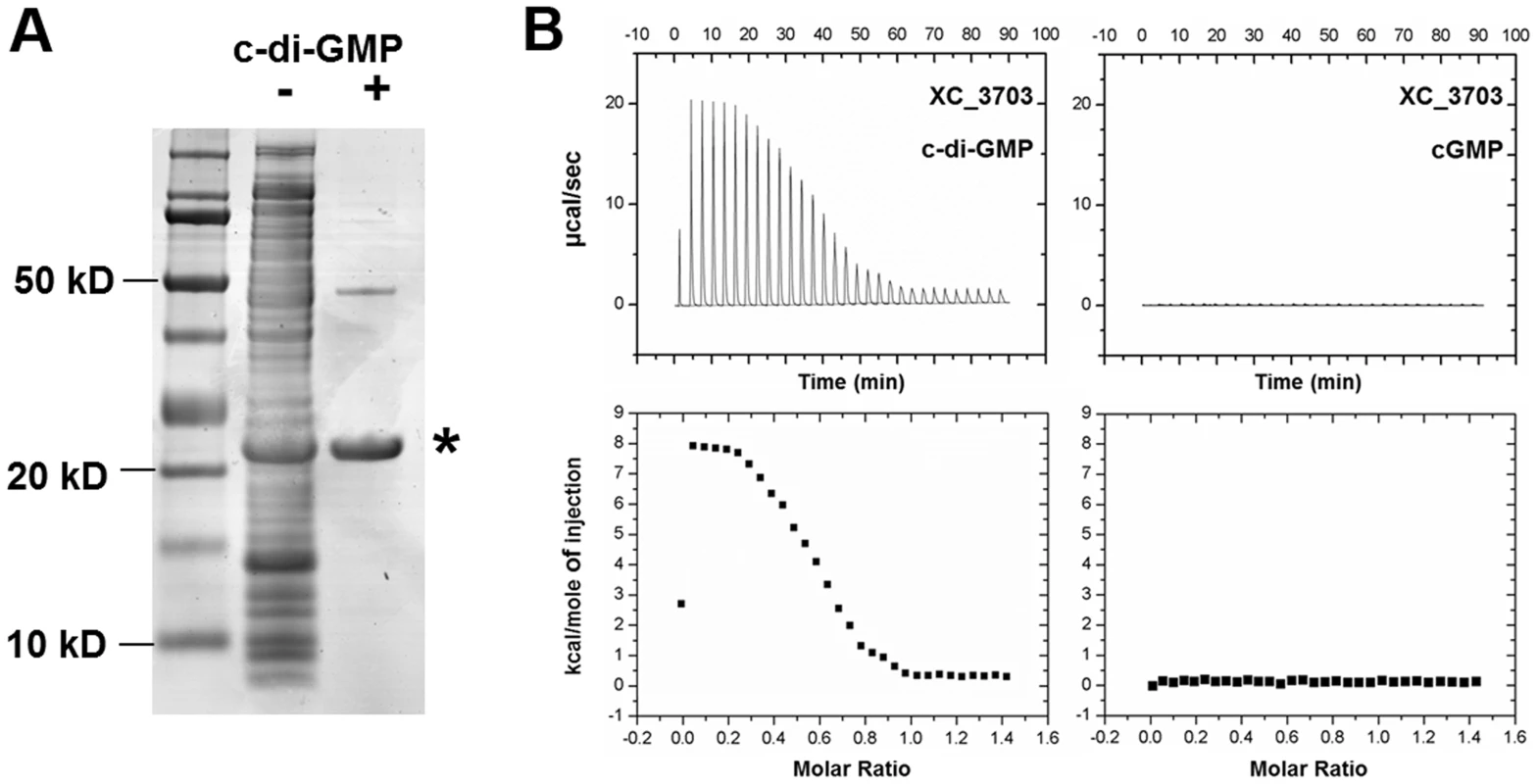
(A) SDS-Polyacrylamide gel separation of Xcc proteins retained on cyclic di-GMP-coupled beads. Lanes from left: Molecular weight markers; Total crude extract of Xcc soluble proteins; Proteins retained by the cyclic di-GMP-coupled beads. The protein band indicated by the asterisk was identified by mass spectrometry of tryptic peptides as XC_3703. (B) Isothermal titration calorimetry analysis of binding of cyclic di-GMP and cyclic GMP by XC_3703. Lower panels show the integrated data obtained from the raw data, after subtracting the heat of dilution. Injection of cyclic di-GMP yielded an endothermic binding isotherm. Experimental data were fitted using the MicroCal ORIGIN version 7.0 software and a Kd for cyclic di-GMP was calculated as 2 µM. No binding of cyclic GMP to XC_3703 was detected. XC_3703 binds cyclic di-GMP with high affinity
The ability of XC_3703 to bind different nucleotides including cyclic di-GMP was further examined using isothermal titration calorimetry (ITC) with the purified protein (see Materials and Methods). The full-length XC_3703 protein bound cyclic di-GMP with high affinity with a Kd of 2 µM (Figure 1B; Table S2). It appeared that binding was endothermic in nature, which is a characteristic of a binding mechanism that favours hydrogen bonding and hydrophobic interactions [15], [16]. The fitting of the binding isotherm data suggested a stoichiometry in which one molecule of cyclic di-GMP was bound by two molecules of the XC_3703 protein. The observed binding affinity is within the range of physiological cyclic di-GMP levels observed in Xcc and is similar to the binding affinity of a number of established cyclic di-GMP effector proteins. XC_3703 showed a much lower affinity for binding of ATP, GTP and cyclic di-AMP and no detectable binding of cyclic GMP or cyclic AMP (Figure 1B; Table S2). Taken together, these findings suggest that XC_3703 preferentially binds cyclic di-GMP.
XC_3703 is required for virulence to plants and biofilm formation
Cyclic di-GMP is known to have a broad regulatory action in Xcc that includes influences on the synthesis of virulence factors and the formation of biofilms [7], [8]. The possible role of XC_3703 in these diverse regulatory actions was investigated by comparative phenotypic and transcriptomic analyses of the wild-type and XC_3703 deletion mutant. The XC_3703 mutant showed reduced virulence to Chinese Radish (Figure 2A) when tested by leaf clipping (see Materials and Methods) and reduced biofilm biomass when grown in complex medium (Figure 2B). Complementation restored these phenotypes towards wild-type.
Fig. 2. Transcriptome and phenotypic characterisation of an XC_3703 deletion mutant reveals roles in biofilm formation and virulence in Xcc. 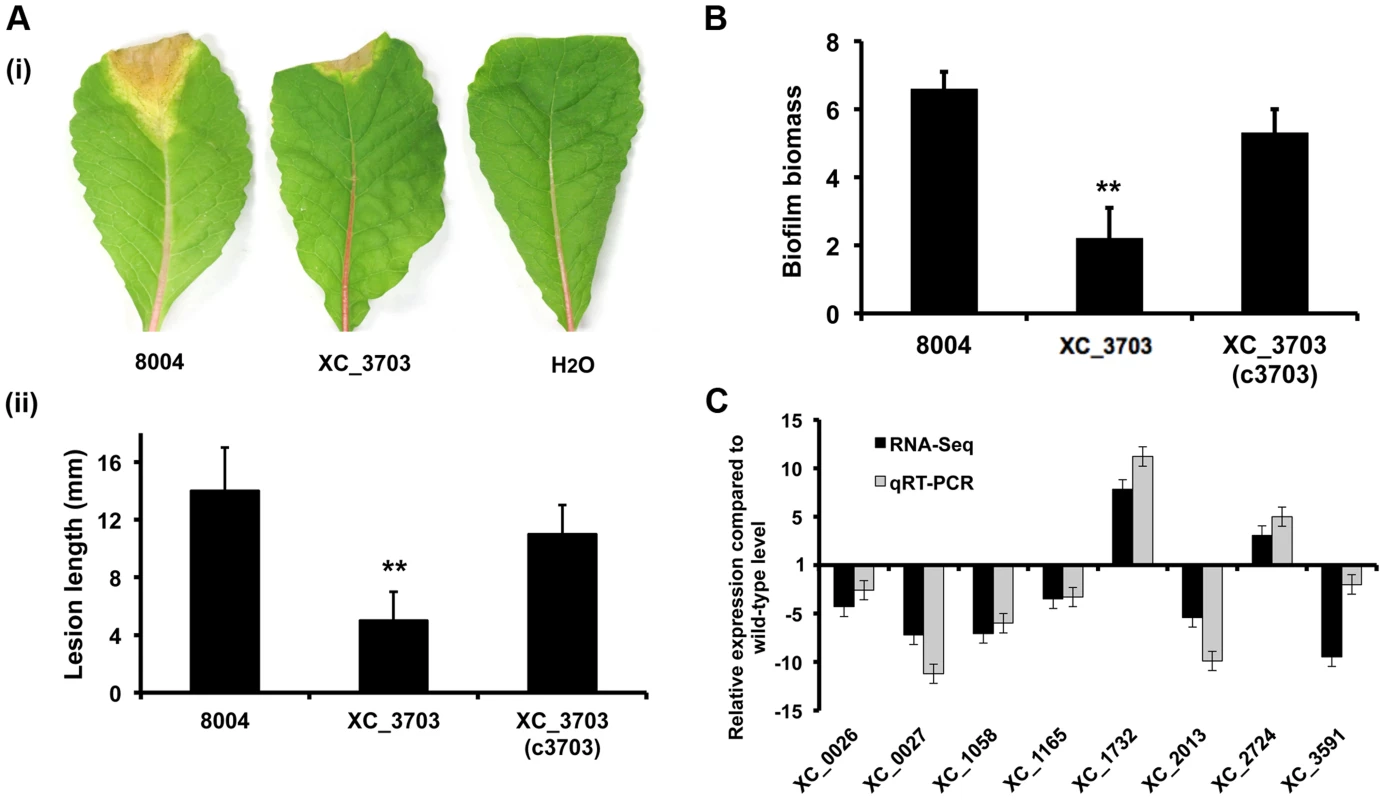
(A) The virulence of the Xcc strains was tested by measurement of the lesion length after bacteria were introduced into the vascular system of Chinese Radish by leaf clipping. (i) Representative virulence assays for (from left to right) Xcc wild-type (8004), XC_3703 deletion mutant and negative control (H2O). (ii) Re-integration of the gene encoding XC_3703 into a neutral site in the mutant chromosome (indicated as XC_3703 (cXC_3703)) restored the reduced virulence of the mutant towards wild-type. Values given are the mean and standard deviation of triplicate measurements each comprising of 30 leaves. Asterisks indicate values that are significantly different from the wild-type (p<0.01, two-tailed Student's t-test). (B) Deletion of XC_3703 also led to decreased biofilm and cell adhesion on a glass surface when assessed by crystal violet staining. Biofilm biomass is measured as a ratio of absorbance at 550 and 600 nm. Re-integration of the gene encoding XC_3703 into the chromosome (indicated as XC_3703 (c3703)) restores the phenotypes towards wild-type. Values given are the mean and standard deviation of triplicate measurements. Asterisks indicate values that are significantly different from the wild-type (p<0.01, two-tailed Student's t-test). (C) Differential expression of selected genes implicated in virulence or biofilm formation in the XC_3703 deletion mutant and wild-type as determined by RNA-Seq (dark grey) and qRT–PCR (light grey). Mutation of XC_3703 affected transcript levels of XC_0026 (cellulase), XC_0027 (endoglucanase), XC_1058 (pilin), XC_1165 (TonB receptor) XC_1732 (glycosyltransferase), XC_2013 (MASE1 domain-containing protein), XC_2724 (methyltransferase), and XC_3591 (pectate lyase). The qRT–PCR data were normalised to 16S rRNA and is presented as the fold change with respect to the wild-type for each gene. Data (means ± standard deviation) are representative of four independent biological experiments. The complete RNA-Seq data set is detailed in Table S4. Comparison of the transcriptome profiles of the wild-type and XC_3703 mutant by RNA-Seq showed that deletion of XC_3703 led to alteration in levels of transcript of a number of genes (Table S3, S4). These genes are associated with a range of biological functions that include bacterial motility and attachment, stress tolerance, virulence, regulation, transport, multidrug resistance, detoxification and signal transduction (Table S4).
The effect of XC_3703 mutation on the level of transcript was validated by quantitative reverse transcription polymerase chain reaction (qRT-PCR) (Figure 2C). The genes selected for these analyses represented those that have been previously implicated in virulence and biofilm regulation in Xcc but with a range of fold change of expression and of diverse functional classes. The relative expression levels of 12 genes measured using qRT-PCR reflected the differences in gene expression observed by transcriptome analysis (Figure 2C).
XC_3703 interacts with transcription regulator XC_2801
A number of cyclic di-GMP effector proteins exert their action through protein-protein interactions [6], [17]. To identify potential interacting proteins for XC_3703, we employed yeast two-hybrid (Y2H) analysis using the full-length XC_3703 protein as bait. Of the 36 preys isolated, the identities of 15 could be established by sequencing (Table S5). These represented partial or full-length proteins of diverse function including a putative octaprenyl-diphosphate synthase (XC_1377), a putative transcriptional regulator of the LysR family (XC_2801) and outer-membrane lipoprotein carrier protein (XC_2211). These particular protein interactions were confirmed when DNA fragments encoding the full length proteins were cloned and constructs were used individually as prey in Y2H analysis. The Y2H analysis was extended by Far-Western blotting experiments in which lysates of bacteria overexpressing the XC_3703 protein were separated by SDS polyacrylamide gel electrophoresis, transferred to nitrocellulose membranes and probed with the His6 tagged XC_1377, XC_2211 or XC_2801 which was then detected with an anti-His6 antiserum (Figure S2).
Since XC_2801 gave the strongest interaction signal we continued analysis with this transcriptional regulator. XC_2801 comprises a conserved N-terminal DNA binding domain and C-terminal LysR substrate binding domain (Figure 3A). Bacterial two-hybrid analysis using different truncated versions of XC_2801 showed that only full-length XC_2801 and the derivatives retaining the substrate binding domain interacted with XC_3703, while no interaction was seen with the isolated DNA binding domain (Figure 3A).
Fig. 3. Protein interaction, transcription and phenotypic profiling reveal an overlap in regulatory influence of XC_3703 and XC_2801. 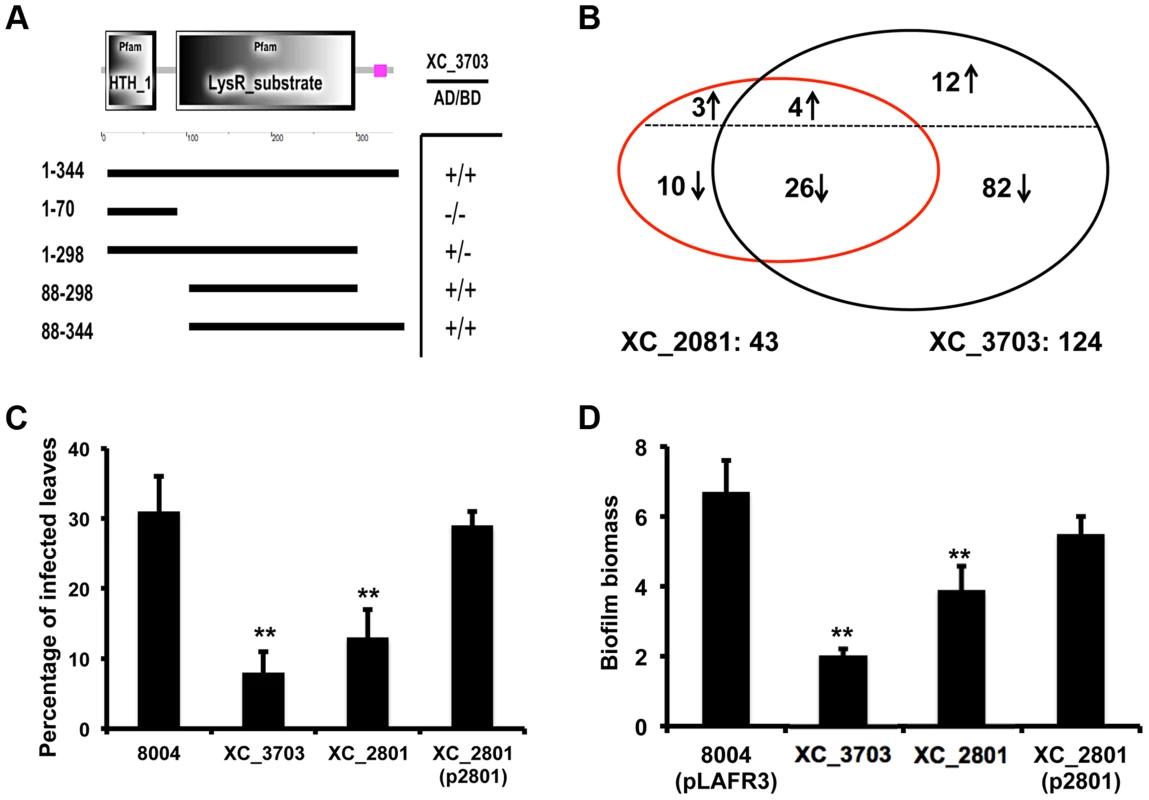
(A) Domains of XC_2801 were predicted by SMART with amino acid positions indicated. Domain abbreviations are; HTH_1: helix-turn-helix DNA binding domain; LysR_substrate: small molecule binding domain associated with LysR family transcriptional regulators. Black lines below figures represent constructs cloned into either pBT or pTRG vectors to assess potential protein-protein interactions with XC_3703 using the bacterial two-hybrid assay. (+) indicates that an interaction was detected. (B) Venn diagrams showing the overlap of genes whose expression is down-regulated or up-regulated in the XC_2801 or XC_3703 mutant strain. Changes in gene expression in XC_2801 and XC_3703 deletion mutants compared with the wild-type Xcc were measured by RNA-Seq. Further detail of data analysis is given in Materials and Methods section and the complete RNA Seq data sets are detailed in Table S4. (C) Virulence of different Xcc strains assessed after spray inoculation. The percentage of the total number of inoculated leaves that showed typical black rot disease symptoms at the leaf margin is given. Values are means and standard deviations of three replicates, each comprising 25 plants and approximately 100 leaves. Asterisks indicate values that are significantly different from the wild-type at p<0.01 by two-tailed Student's t-test. Complementation of the XC_2801 mutant by introduction of XC_2801 cloned into pLAFR3 (indicated as XC_2801(p2801)) restored the phenotype towards wild-type. (D) Biofilm and cell adhesion of the XC_2801 mutant to a glass surface was decreased compared to wild-type strain 8004. Complementation of the XC_2801 mutant by introduction of XC_2801 cloned into pLAFR3 (indicated as XC_2801(p2801)) restored the phenotype towards wild-type. Biofilm biomass was measured by crystal violet staining and expressed as a ratio of absorbance at 550/600 nm. Values given are the mean and standard deviation of triplicate measurements. Asterisks indicate values that are significantly different from the wild-type at p<0.01 by two-tailed Student's t-test. The regulatory action of XC_3703 overlaps with that of XC_2801
The findings outlined above indicate a potential interaction between XC_3703 and XC_2801. As a first step towards establishing a functional link between the two proteins, the phenotypic and transcriptional effects of mutation of XC_2801 were compared with mutation of XC_3703.
Mutation of XC_2801 significantly influenced the expression of 43 genes in Xcc; the regulated genes are associated with diverse functions that include extracellular enzyme production, attachment, biofilm formation and flagellar biosynthesis (Table S4). Quantitative RT-PCR methods were used to confirm alterations in expression of selected genes as revealed by RNA-Seq (Figure S3). Notably, comparison of the effects of mutation of XC_3703 and XC_2801 on the transcriptome revealed a significant overlap of regulatory influence (Figure 3B).
As outlined above, the XC_3703 mutant showed reduced virulence in Chinese Radish when plants are inoculated by leaf clipping. In contrast, the XC_2801 mutant showed no alteration in virulence when this method of inoculation was used. However, when plants were inoculated by spraying both XC_3703 and XC_2801 mutants showed reduced virulence, as indicated by a reduction in the percentage of inoculated leaves that develop black rot symptoms (Figure 3C). The effect of XC_2801 mutation on virulence was less pronounced than that of XC_3703 mutation however. Complementation of the XC_2801 mutant by introduction of XC_2801 cloned in pLAFR3 restored virulence to near wild-type level (Figure 3C).
Mutation of XC_2801 also had an effect on Xcc biofilm formation, although the effect was not as pronounced as that following mutation of XC_3703 (Figure 3D). Complementation with a clone expressing the full-length XC_2801 protein restored biofilm formation of the XC_2801 mutant to near wild-type levels. Taken together, these observations support the contention the effects of XC_3703 are exerted, at least in part, by interaction with XC_2801.
XC_2801 binds to target promoters only as a complex with XC_3703
The transcriptional analyses outlined above showed that levels of transcripts of the flhB, aaeA, fliL and flgG genes were decreased approximately fivefold in both the XC_3703 and XC_2801 mutant compared to the wild-type. The regulation of these genes by XC_2801 was also examined by the use of promoter fusions to gusA (see Materials and Methods). Differences in the level of GusA between the XC_2801 mutant and wild-type were seen with all four fusions (Figure 4A), demonstrating that XC_2801 (directly or indirectly) regulates the expression of flhB, aaeA, fliL and flgG.
Fig. 4. XC_2801 binds to specific promoters and only in the presence of XC_3703. 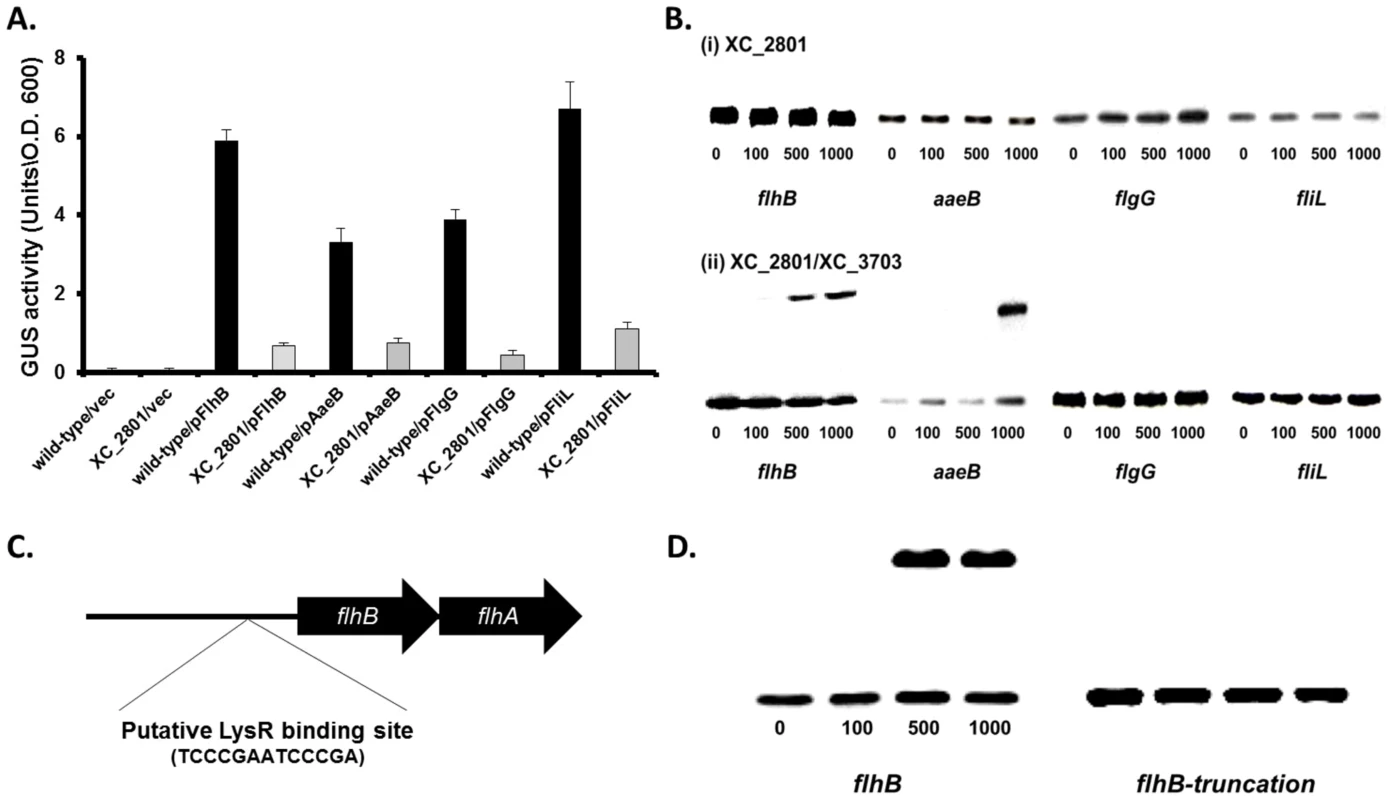
(A) Promoter-fusion analysis of the upstream regions flhB, aaeB, flgG and fliL revealed significantly decrease in promoter activity in the XC_2801 mutant strain. Values given are the mean and standard deviation of triplicate measurements. Values for promoter activity in the XC_2801 mutant strain are significantly different from the wild-type at p<0.01 by two-tailed Student's t-test. The decrease in promoter activity was consistently observed in three independent experiments each consisting of three biological replicates. (B) Binding of XC_2801 to the promoter regions of flhB, aaeB, flgG and fliL in the absence (i) or presence (ii) of XC_3703 assessed by the use of electromobility shift assay (EMSA). Each lane contained 1.5 nM DIG-labelled Probe DNA, and in addition nanomolar concentrations of purified His6-tag XC_2801 protein as indicated below each well. (C) Bioinformatics reveals a putative LysR family regulator-binding site (TCCCGAATCCCCGA) located 80-67 nucleotides upstream of the predicted translational start site of flhB. (D) Although a shift was observed with the flhB upstream (promoter) region, no shift was observed with an overlapping DNA fragment that lacks the putative XC_2801 binding site (indicated as truncated flhB). The nanomolar concentration of His-tagged XC_2801 used in each assay is indicated below each well. DIG-labelled DNA at 1.5 nM was used. To further investigate the role of XC_2801 in regulation of transcription and any potential interplay between XC_2801 and XC_3703, electrophoretic mobility shift assays (EMSA) were used to examine protein binding to target promoters. His-tagged expression constructs of XC_2801 and XC_3703 were generated and both were subsequently purified by nickel affinity column chromatography. Purified His-tagged XC_2801 fusion protein caused no mobility shift of DNA probes spanning the upstream region of flhB, aaeB, fliL and flgG (Figure 4B). Interestingly, incubation of His-tagged XC_2801 together with XC_3703 led to a mobility shift of a DNA probe spanning the upstream regions of flhB and aaeB but not fliC or flgG (Figure 4B) whereas XC_3703 alone had no effect (Figure S4). These observations, taken together with the two-hybrid and far-western analyses presented above, suggest that under the conditions used, XC_2801 only binds to the upstream region of flhB and aaeB as a complex with XC_3703.
As LysR-type transcriptional regulators (LTTRs) typically bind a specific motif, T-N11-A, the upstream region of flhBA was examined for such a motif. The sequence TCCCGAATCCCGA was identified 80–67 bp upstream of the putative translational start site of flhB (Figure 4C). To investigate if this motif was essential for protein binding, EMSA was performed using an overlapping DNA fragment that lacked the full putative binding site. The presence of both XC_2801 and XC_3703 proteins did not cause a mobility shift of this DNA fragment demonstrating that XC_2801//XC_3703 binding requires the complete TCCCGAATCCCGA motif (Figure 4D).
The binding of the XC_2801//XC_3703 complex to a target promoter is reversed by cyclic di-GMP
As outlined above, XC_3703 preferentially binds cyclic di-GMP with a high affinity. This raised the question as to whether cyclic di-GMP could influence the binding of the XC_2801//XC_3703 complex to target promoters. To investigate this, the EMSA assay for the binding of XC_3703//XC_2801 to the flhBA promoter was repeated in the presence of added cyclic di-GMP at different concentrations. Addition of cyclic di-GMP at 1 µM had no apparent effect, but at 10 µM completely prevented DNA binding (Figure 5A). Other nucleotides did not have the same effect (Figure S4). XC_2801 alone did not bind to the flhBA promoter (as shown above) and addition of cyclic di-GMP did not alter this outcome (Figure 5B). Furthermore, XC_2801 appeared to have no affinity for the nucleotide as measured by ITC (Figure S5). These results suggest that cyclic di-GMP inhibits the interaction of the XC_3703//XC_2801 complex with DNA by binding to XC_3703, thus preventing the transcription of flhBA.
Fig. 5. Binding of XC_2801//XC_3703 to the flhBA promoter is modulated by cyclic di-GMP. 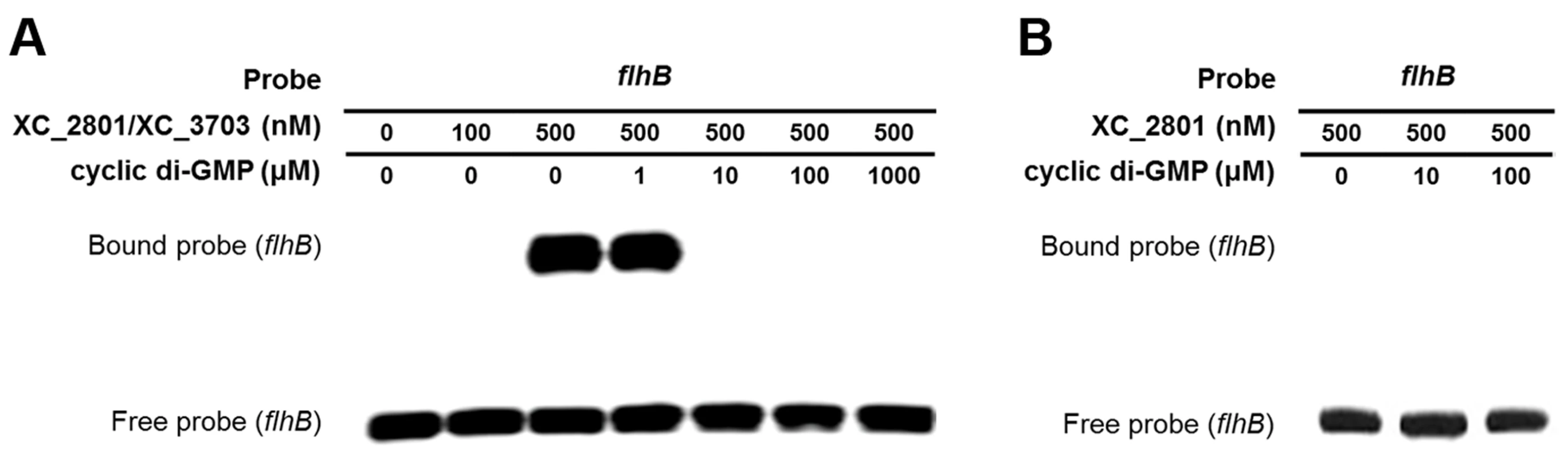
(A). The impact of various concentrations of cyclic di-GMP on the binding of XC_2801//XC_3703 to the flhBA promoter was assessed by the use of electromobility shift assay (EMSA). The DIG-labelled promoter fragment was incubated with purified XC_2801 and XC_3703 proteins and cyclic di-GMP. Concentrations of proteins and nucleotide are indicated above each well. (B) No binding of XC_2801 to the flhBA promoter region was seen in the presence of various concentrations of cyclic di-GMP as assessed by the use of EMSA. The DIG-labelled promoter fragment was incubated with purified XC_2801 and cyclic di-GMP. Concentrations of protein and nucleotide are indicated above each well. XC_3703 homologues that bind cyclic di-GMP have roles in virulence in several human pathogens
Bioinformatic analysis reveals that proteins of the YajQ family are widely conserved in bacteria (Figure S1). To examine whether cyclic di-GMP binding was a common feature of this family, recombinant YajQ-like proteins from various bacteria including Escherichia coli, Pseudomonas aeruginosa, Stenotrophomonas maltophilia, Bacillus cereus and Clostridium species were purified and their ability to bind different nucleotides was assessed. Interestingly, affinity for cyclic di-GMP binding was only seen in YajQ family proteins from P. aeruginosa (PA4395) and S. maltophilia (Smlt4090) (Table S1). YajQ family proteins from E. coli, Clostridium sp. and B. cereus by contrast exhibited a greater affinity for ATP and/or GTP than for any cyclic mono - or di-nucleotide.
The finding that XC_3703 can influence the virulence of Xcc prompted us to test whether PA4395 and Smlt4090 have a role in the virulence of P. aeruginosa and S. maltophilia respectively. Previous studies have established a correlation between the ability of strains of P. aeruginosa and S. maltophilia to attach and induce cytotoxicity in cultured mammalian cells and the virulence of these strains to mice [18], [19]. Mutation of either PA4395 or Smlt4090 led to reduced adhesion to monolayers of human bronchial epithelial cells compared to the wild-type (Figure 6 A, B). Importantly, the reduction in adhesion of these mutants could be restored to the level of the wild-type strain by complementation (Figure 6 A, B).
Fig. 6. Roles for XC_3703 homologues in virulence of the human pathogens Pseudomonas aeruginosa and Stenotrophomonas maltophilia. 
Mutation of the genes encoding homologs of XC_3703 in P. aeruginosa (PA4395) and S. maltophilia (Smlt4090) resulted in strains with reduced capability to adhere to host cells and with reduced virulence in a murine model of infection. (A, B) Adhesion to human cells and internalization of strains of P. aeruginosa and S. maltophilia. Bronchial epithelial monolayers were co-cultured with ∼100 bacteria per cell for 2 hours. The bronchial epithelial cells were then washed and bacteria from cell lysates were plated on LB agar. Colonies were counted after 48 h. Values given are means and standard deviations of triplicate measurements. Asterisks indicate values that are significantly different from the appropriate wild-type (p<0.05, Student's t test). Complementation restored adhesion towards wild-type levels. (C, D). Mutation of PA4395 in P. aeruginosa or Smlt4090 in S. maltophilia leads to reduced colonization of mice compared to wild-type. C57BL/6 mice were intra-tracheally infected with 1×108 CFU of wild-type P. aeruginosa PA14 or the PA4395 mutant (C); wild-type S. maltophilia K279a or the Smlt4090 mutant (D). Animals were sacrificed 1, 3 and 5 days after infection and 10-fold serial dilutions of lung homogenates were plated to determine the bacterial load. The means and standard deviations of triplicate measurements are shown. Asterisks indicate values that are significantly different from the appropriate wild-type (p<0.05, Student's t test). On the basis on these findings, we extended these studies to utilize the persistence mouse model of pulmonary infection. C57Bl/6 mice were intra-tracheally infected with wild-type P. aeruginosa, the PA4395 mutant, wild-type S. maltophilia or the Smlt4090 mutant and clearance of strains from the lung was examined at 1, 3 and 5 days post infection (see Materials and Methods). Both the PA4395 and Smlt4090 mutants had reduced persistence in C57BL/6 mice compared to their respective wild-type strains (Figure 6 C, D).
The findings from these infection models indicate that YajQ family proteins of P. aeruginosa and S. maltophilia significantly contribute to virulence of these opportunistic pathogens.
Discussion
The YajQ family of proteins is broadly distributed in bacteria, with typically one member of this family in each species. YajQ family proteins have motifs characteristic of nucleotide or nucleic acid-binding proteins [16], [20]. Accordingly, YajQ from Escherichia coli has been shown to bind GTP and tRNA [16]. Here we show that some members of the YajQ family are novel cyclic di-GMP effectors that selectively bind this second messenger and act to regulate bacterial virulence. In particular we show that XC_3703 from Xanthomonas campestris influences the transcription of genes that contribute to virulence in plants and biofilm formation. We provide evidence that this action of XC_3703 is exerted, at least in part, through protein-protein interactions with the LysR family regulator XC_2801 in a manner that is negatively regulated by cyclic di-GMP. XC_3703 must exert a regulatory effect via additional pathways not involving XC_2801, but these currently remain unidentified.
We further identified the YajQ protein PA4395 of P. aeruginosa as a cyclic di-GMP binding protein. Previous studies to identify cyclic di-GMP-binding proteins from P. aeruginosa in a global fashion have used pull-downs with a cyclic di-GMP analog (2′-aminohexylcarbamoyl-cyclic di-GMP) covalently coupled to Sepharose beads [21] or a tri-functional capture compound incorporating cyclic di-GMP, biotin and a photo-activated reactive group [22]. Both of these studies identified a range of proteins, including six common proteins with an established function in cyclic di-GMP binding, but neither identified PA4395. Interestingly, of the novel proteins described in these two studies, only five were identified by both approaches, leading to the suggestion that the different compounds used for the experiments might be specific for a certain subset of proteins [22].
Knowledge of the cellular function of proteins of the YajQ family has been thus far restricted to the protein of Pseudomonas syringae that is implicated as a host factor controlling transcription in the bacteriophage Φ6 [23]. YajQ of P. syringae exerts an effect on phage RNA polymerase by protein-protein interactions involving the major protein (P1) of the phage core. Intriguingly this action is modulated in later phases of viral infection, even though YajQ is still present [23]. The underlying mechanism is unknown, although it has been suggested that a change in the chemical environment of the host cell between early and late phases of infection is responsible [23]. Our findings raise the possibility that one controlling factor could be the level of cyclic di-GMP in the bacterial cell.
In this study, XC_3703 was observed to bind cyclic di-GMP with an affinity of ∼2 µM. Measurements of cyclic di-GMP levels in wild-type Xcc strains under various conditions indicate that this value is within the physiological range of the concentration of the nucleotide [24], [25], [26]. The affinities of cyclic di-GMP effectors from a variety of organisms have been found to vary considerably, ranging from 10–15 µM down to ∼100 nM [27]. In Xcc, the transcription factor Clp binds cyclic di-GMP with an affinity of 3.5 µM, whereas the EAL domain of FimX has an affinity of 0.4 µM [11], [12]. The difference in binding affinities of these effectors has been suggested to represent a method whereby different responsive elements are selectively activated (or inactivated) as the cellular concentration of cyclic di-GMP changes [27].
Our experiments also show that not all members of the YajQ family preferentially bind cyclic di-GMP, suggesting that they have other cellular roles. This contention is supported by the observation that YajQ family members are found in bacteria that do not apparently have cyclic di-GMP signaling, such as Haemophilus influenzae [20], [28]. Proteins of the YajQ family show high level of amino acid sequence similarity but with some sequence divergence in the central part of the protein. Further work including structural studies will be required to identify residues that are critical for cyclic di-GMP binding, which may then allow bioinformatic discrimination of those family members that have this capability.
XC_3703 represents the first member of a new class of cyclic di-GMP binding protein and our findings suggest that the regulatory action of XC_3703 is exerted in part through protein-protein interaction with XC_2801, a protein of the LysR family that is the most abundant type of transcriptional regulator in bacteria [29]. Studies of members of the LysR family of transcriptional regulators indicate considerable structural flexibility, which may account for their variety of regulatory modes and versatility [30]. In general, the binding of small molecular weight molecules to the C-terminal co-inducer-binding domain modulates the activity of this family of regulators. XC_3703 also interacts with the C-terminal domain of XC_2801, suggesting a variation on the co-inducer mode of regulation (Figure S6). It appears that cyclic di-GMP specifically binds to XC_3703 with high affinity and that binding prevents the interaction of XC_3703//XC_2801 with a target promoter sequence (flhBA). In this manner, although XC_2801 alone does not bind cyclic di-GMP, transcriptional activation by XC_2801 is prevented when the concentration of cyclic di-GMP is high. Consistent with this view, the levels of expression of genes that we show here to be regulated by XC_2801 are reduced under conditions in which the cellular levels of cyclic di-GMP are high [30]. The absence of binding of XC_2801 or XC_2801//XC_3703 to the promoters of the fliL and flgG genes in EMSAs may indicate that the expression of these genes is indirectly regulated by XC_2801. We cannot however exclude that XC_2801 may also respond to a small-molecular weight co-inducer, which would be missing from these assays.
Notably, mutation of XC_2801 affects expression of only a subset of the genes that are regulated by XC_3703. Furthermore, loss of XC_3703 influences virulence of Xcc when the bacteria are introduced into the leaf vascular system by leaf clipping, but loss of XC_2801 has no effect. From these observations we surmised that XC_3703 must exert additional regulatory effects, independently of XC_2801, that contribute to bacterial virulence. The Y2H analysis of proteins that interact with XC_3703 may give clues to the identity of such additional pathways. Intriguingly, interaction is seen between XC_3703 and HpaR1 (XC_2736), a GntR family transcriptional regulator that has been previously shown to control the hypersensitive response and virulence in Xcc [25]. Further work is required to determine the importance of this interaction to the virulence of Xcc.
Finally, we determined that YajQ proteins from the opportunistic human pathogens P. aeruginosa and S. maltophilia are capable of binding cyclic di-GMP and showed by mutational analysis that these proteins significantly influence virulence, biofilm formation and persistence in models of human infection. S. maltophilia is a xanthomonad, related to Xcc whereas P. aeruginosa is more distantly related. One challenge is to establish if regulatory interactions between proteins of the YajQ and LysR families have a role in cyclic di-GMP signaling in these other bacteria. Interference with cyclic di-GMP signaling has emerged as a promising approach towards treatment of bacterial biofilm formation and the control of virulence and disease. Potential targets include the enzymes involved in cyclic di-GMP synthesis or degradation and the effectors by which cyclic di-GMP exerts its action. Our knowledge of the mechanisms of cyclic di-GMP signaling and associated effectors is however incomplete, and further understanding will be key to the progress of a promising approach for disease control into effective therapeutic strategies.
Materials and Methods
Bacterial strains, plasmids, and culture conditions
Xanthomonas campestris pv campestris (Xcc) strains and culture conditions have been described previously [31], [32], [33]. Most experiments were carried out in NYGB medium, which comprises 5 g liter−1 bacteriological peptone (Oxoid, Basingstoke, U.K.), 3 g liter−1 yeast extract (Difco), and 20 g liter−1 glycerol. For biofilm formation, Xcc was grown in L medium, which comprises 10 g liter−1 bactotryptone (Difco), 5 g liter−1 yeast extract, 5 g liter−1 sodium chloride, and 1 g liter−1 D-glucose. E. coli strains were grown in LB medium at 37°C. Other plasmids and strains used are shown in Table S6. Where required antibiotics were used at concentrations of 100 µg ml−1 for ampicillin, 50 µg ml−1 for rifampicin, 20 µg ml−1 kanamycin and 15 µg ml−1 tetracycline.
General molecular biology methods
Common molecular biological methods such as isolation of plasmid and chromosomal DNA, polymerase chain reaction (PCR), plasmid transformation as well as restriction digestion were carried out using standard protocols [34]. PCR products were cleaned using the Qiaquick PCR purification kit (Qiagen) and DNA fragments were recovered from agarose gels using Qiaquick minielute gel purification kit (Qiagen). Oligonucleotide primers were purchased from Sigma-Genosys.
Construction of insertion and deletion mutant strains
In-frame deletion of selected genes was carried out using pK18mobsac as described previously for XC_3703 and XC_2801 [31], [32]. Mutants were also created by the disruption of genes with the use of the plasmid pK18mob as described previously for Xcc [31].
Construction of protein expression vectors and protein purification
The DNA fragments encoding the proteins of interest were synthesized by Gene Oracle in pGOv4 and sub-cloned into pET47b or pLAFR3 before transformation into E. coli BL21 (DE3). Genomic regions are described in Table S7. BL21 (DE3) cells were grown in LB media and induced with 0.25 mM IPTG; protein overexpression was carried out at 37°C for 1 h. Purification was achieved by Ni2+ affinity chromatography using the N-terminal His6 tag followed by tag cleavage using recombinant 3C protease.
Construction of gusA reporter plasmids
Reporter plasmids pG2277 (flhB), pG3487 (aaeB), pG2239 (flgG) and pG2262 (fliL) were constructed by cloning the putative promoter region and ribosome binding site (∼500-bp region upstream of the start codon) of XC_2277, XC_3487, XC_2239 and XC_2262 respectively into the broad-host-range cloning vector pLAFRJ, which harbors the coding region (without promoter and RBS) of β-glucuronidase (gusA) gene in its MCS (multiple cloning site). The putative promoter region and RBS of XC_2277, XC_3487, XC_2239 and XC_2262 were amplified from the chromosomal DNA of Xcc8004 using the primer pairs detailed in Table S7. The amplified DNA fragment, confirmed by sequencing, was inserted 9 bp upstream of the promoterless gusA ATG start codon in the vector pLAFRJ to create the recombinant plasmid. The recombinant plasmid obtained was further confirmed by restriction analysis and PCR. All reporter plasmids were introduced into the Xcc strains of interest through conjugation as previously described [25].
Affinity pull-down assay
A volume of 50 ml of an Xcc culture with an OD600 of 1 was harvested and suspended in 1 ml 10 mM Tris·HCl (pH 7.5), 50 mM NaCl buffer containing EDTA-free complete protease inhibitor (Roche). Cells were mixed with 0.1-mm glass beads and lysed in a Fast-Prep machine twice for 45 seconds. Samples were centrifuged for 5 min at 17,000× g and subsequently for 1 h at 100,000× g to obtain cytoplasmic protein extracts. A volume of 50 ml streptavidin dynabeads (Invitrogen) coupled with 2.4 µM biotinylated cyclic-di-GMP (BioLog) were incubated with 1.2 mg cytoplasmic proteins in 1.5 mL 10% (vol/vol) glycerol, 1 mM MgCl2, 5 mM Tris (pH 7.5), 230 mM NaCl, 0.5 mM DTT, and 4 mM EDTA containing 50 µg/ml BSA for 30 min at room temperature. Samples were washed four times with the same buffer lacking BSA and suspended in 50 µl protein sample buffer. Samples were boiled for 5 min, beads removed, and 18 µl run on 12% (wt/vol) SDS/PAGE gels. Protein identification was carried out by in-gel proteolysis and the resultant peptides were analysed by LC-MS/MS on the LTQ Orbitrap Classic (FingersPrints, UK).
Isothermal titration calorimetry (ITC) experiments
The dissociation constant (Kd) between wild-type XC_3703 (or selected homolog) with cyclic di-GMP, cyclic di-AMP, ATP, cyclic GMP or cyclic AMP was measured by using an VP-ITC MicroCalorimeter. Titrations were carried out at 25°C in an assay buffer containing 20 mM Tris-Cl (pH 8.0), 80 mM NaCl. Samples of XC_3703 for ITC measurements were dialyzed extensively against the assay buffer overnight. The concentrations of protein in the cell ranged between 10 to 300 µM and that of the nucleotide in the syringe was in the range of 1–4 mM. A volume of 8 µl of nucleotide was injected into the cell with a time lag of 180 s between each injection for a total of 30 times. ITC data were analyzed by integrating heat exchange amount after subtracting background dilution heat from the apparent values. Data fitting was based on a one-site binding model using the using the MicroCal ORIGIN version 7.0 software.
RNA extraction and preparation for expression profiling
Three independent cultures of each selected Xanthomonas strain were sub-cultured and grown to logarithmic phase (0.7–0.8 OD600) at 30°C in NYGB broth without selection. 800 µl of RNA protect (Qiagen) was added to 400 µl culture and incubated at room temperature for 5 min. Cell suspensions were centrifuged, the supernatant was discarded, and pellets were stored at −80°C. After thawing, 100 µl TE-lysozyme (400 µg/ml) was added and samples were incubated at room temperature. Total RNA was isolated using the RNeasy Mini Kit (Qiagen) whereby cells were homogenised utilising a 20-gauge needle and syringe. Samples were treated with DNase (Ambion) according to manufacturer's instructions and the removal of DNA contamination was confirmed by PCR.
Library construction and cDNA sequencing
RNA quality was assessed on a Bioanalyser PicoChip (Agilent) and RNA quantity was measured using the RNA assay on QuBit fluorometer (Life Technologies). Ribosomal RNA was depleted with Ribo-Zero rRNA Removal Kits for Gram-Negative Bacteria (Epicentre). The percentage of rRNA contamination was checked on a Bioanalyser PicoChip (Agilent). The rRNA-depleted sample was processed using the Illumina TruSeq RNA v2 sample preparation kit. In brief, the sample was chemically fragmented to ∼200 nt in length and the cleaved RNA fragments were primed with random hexamers into first strand cDNA using reverse transcriptase and random primers. The RNA template was removed and a replacement strand was synthesised to generate ds cDNA. The ds cDNA was end repaired to remove the overhangs from the fragmentation into blunt ends. A single ‘A’ nucleotide was added to the 3′ ends on the blunt fragments, which is complementary to a ‘T’ nucleotide on the 3′ end of the Illumina adapters. At this stage, adapters containing 6 nt barcodes were used for different samples to allow the pooling of multiple samples together. The resulted barcoded samples were enriched by 10 cycles of PCR to amplify the amount of DNA in the library. The final cDNA libraries were sequenced on an Illumina HiSeq2000 as per manufacturer's instructions. The RNA-Seq raw data files are accessible through XanthomonasGbrowse: http://browser.tgac.bbsrc.ac.uk/cgi-bin/gb2/gbrowse/Xanthomonas_8004/.
Computational analysis
Cluster generation was performed using the Illumina cBot and the cDNA fragments were sequenced on the Illumina HiSeq2000 following a standard protocol. The fluorescent images were processed to sequences using the Real-Time Analysis (RTA) software v1.8 (Illumina). Raw sequence data were demultiplexed by assigning the sequenced reads of each sample with their corresponding indexed reads. The reads for each sample were then processed through the primary analysis pipeline to generate FASTQ files.
Read mapping, annotation and quantification of transcript levels
Reads for each sample were aligned to the Xanthomonas campestris pv campestris 8004 NC_007086 assembly (Integrated Microbial Genomes (IMG) database, taxon object ID 637000343) using Bowtie version 0.12.7 with default parameters. Transcript abundance was determined for the gene models annotated in the IMG Xanthomonas campestris pv campestris 8004 genome release IMG/W 2.0 using the Bowtie RNA-Seq BAM alignments for each of the samples. To estimate the level of transcription for each gene, the number of reads that mapped within each annotated coding sequence (CDS) was determined.
Analysis of differential expression
Differential expression was assessed using Cufflinks (Cufflinks reports an expression value for each transcript and gene in Fragments Per Kilobase of exon model per Million mapped fragments (FPKM). A test statistic is also calculated which, after Benjamini-Hochberg correction for multiple-testing was used to determine significant changes in expression between each pair of samples (false discovery rate 0.05).
Quantitative Real-time PCR
Quantitative RT-PCRs were used to validate RNA-Seq data. Reverse transcription PCR was achieved using a cDNA synthesis kit (Promega) according to the manufacturer's instructions. Specific RT-PCR primers were used to amplify central fragments of approximately 200 bp in length from different genes. For qRT-PCRs, quantification of gene expression and melting curve analysis were completed using a LightCycler (Roche) and Platinum SYBR Green qPCR Supermix-UGD (Invitrogen) was used according to manufacturer's instructions. The constitutively expressed housing keeping gene, 16S rRNA was used as a reference to standardize all samples and replicates.
Yeast two-hybrid screen
The methods used for construction and analysis of the Xcc genomic DNA prey library in the plasmid vector pOAD were as described previously [17]. Xcc DNA sequence coding for the XC_3703 used as bait was amplified by PCR using Xcc genomic DNA as template and primers designed based on the Xcc genome sequence. These primers were designed to contain unique restriction sites to facilitate cloning into the pOBD vector downstream of the Gal4 DNA-binding domain. After transformation into E. coli DH5α cells, individual colonies were picked for plasmid isolation and confirmation by DNA sequencing. The bait construct was transformed into Saccharomyces cerevisiae, and this strain was used to screen the Xcc genomic DNA prey library. Approximately 2×108 yeast transformants (carrying both and prey vectors) were plated on selective media that lacked adenine, histidine, tryptophan, and leucine. Fifteen hundred transformants displaying prototrophy were transferred to plates with adenine and histidine but lacking tryptophan and leucine. Preys were identified by isolation of plasmids and sequencing.
For measurement of interactions of the segments of XC_2801 with XC_3703, doubly transformed yeast strains were grown in YPD medium (10 g liter−1 Bacto-yeast extract (Difco), 20 g liter−1 Bactopeptone (Difco), and 2 g liter−1 D-glucose) were performed as described [17].
Far-western assays
In vitro binding assays were performed as described previously [34]. Briefly, ≈20 µg of purified recombinant proteins or total lysates of E. coli cultures expressing recombinant proteins were resolved by SDS/PAGE (12% acrylamide) and transferred to nitro - cellulose membranes. The membranes were blocked with TBS-TTmilk [140 mM NaCl, 20 mM TrisHCl (pH 7.4), 0.1% Triton X-100, 0.1% Tween-20, 5% powdered milk] for 1 h and then probed with anti-His6 antiserum as described previously [35].
Electrophoretic gel mobility shift assay
The DNA probes used for EMSA were prepared by PCR amplification of the desired upstream region of flhB, using oligonucleotides as the primers (primers and amplified regions are described in Table S7). The purified PCR products were 3′-end-labelled with digoxigenin following the manufacturer's instruction (Roche). The EMSA was carried out using the DIG Gel Shift Kit 2nd Generation (Roche) as recommended by the manufacturer with some modifications. Ten fmoles of the DIG-labelled fragment and a range of 0–1000 nM of XC_2801 protein were added to the binding reaction. The mixture was allowed to proceed at room temperature for 45 min. The samples were separated by electrophoresis on 6% native polyacrylamide gels and transferred to Hybond-N blotting membrane (Amersham). Protein–DNA complexes were visualized by NBT/BCIP according to the manufacturer's instructions (Roche).
Plant virulence assays
The virulence of Xcc to Chinese Radish was estimated after bacteria were introduced into the leaves by leaf clipping and spraying as previously detailed [10], [31], [36]. For leaf clipping assay, bacteria grown overnight in NYGB medium were washed and re-suspended in water to an OD at 600 nm of 0.001. For leaf clipping the last completely expanded leaf was cut with scissors dipped in the bacterial suspensions. Thirty leaves were inoculated for each strain tested. Lesion length was measured 14 days after inoculation. Each strain was tested in at least four separate experiments. For spraying assays, five-week old seedlings with fully expanded leaves were used. Bacteria were grown overnight in NYGB medium, centrifuged and re-suspended in water to a cell density OD of 0.01 at 600 nm for spraying. A volume of 50 ml of the bacterial suspension was inoculated onto the leaves of 25 plants (approximately 100 leaves) by spraying. Three replicates of each independent experiment were carried out. Ten days after inoculation the relative virulence was determined as the percentage of the total inoculated leaves that showed the typical black rot disease symptoms at the leaf margin. The experiment was repeated three times.
Biofilm assays
Biofilm was assessed by attachment to glass and was determined by crystal violet staining. Log-phase-grown bacteria were diluted to OD600 nm = 0.02 in L media broth and 5 ml was incubated at 30°C for 24 h in 14-ml glass tubes. After gently pouring off the media, bacterial pellicles were washed twice with water and were then stained with 0.1% crystal violet. Tubes were washed and rinsed with water until all unbound dye was removed [25], [31]. Bound crystal violet was eluted in ethanol and measured at OD595. Three independent assays were carried out for each strain.
Ethics statement
Animal experiments were conducted in compliance with a protocol (# 16-051-11) approved by The Animal Experimentation Ethics Committee (AEEC) of University College Cork or Cork University Hospital. The Minister for Health accredits the University College Cork for conducting animal experiments. AEEC is guided by legislative requirements, in particular the Cruelty to Animals Act as amended and supplemented by the European Communities.
Infection and treatment of animals
Mouse infection was performed using a variation on the mouse model described by [35], [37]. Briefly, P. aeruginosa or S. maltophilia strains were grown in Luria broth at 37°C overnight with shaking, after which bacteria were collected by centrifugation and resuspended in PBS. The exact number of bacteria was determined by plating serial dilutions of each inoculum on Luria broth agar plates. Female C57BL/6 mice (approximately 8-weeks old) were anesthetized and infected by the intratracheal route with 20 µl of culture of wild-type strains or mutants at a final inoculum of 5×106 colony forming unit (CFU) per mouse. Mice were killed at 1, 3 and 5 days post-infection by intraperitoneal injection of 0.3 ml of 30% pentobarbital. Lungs and spleens were harvested aseptically and homogenized in sterile PBS. A 10-fold serial dilution of lung homogenates was plated on Pseudomonas isolation agar (for P. aeruginosa strains) or NGBA media (for S. maltophilia strains). The results (means ± standard deviation) are expressed as CFU/organ.
Epithelial cell co-culture
P. aeruginosa or S. maltophilia strains were grown on bronchial cells using a co-culture model system as previously described [37], [38], [39]. Briefly, bronchial epithelial monolayers were co-cultured with 100 bacteria per cell for 2 hours. The bronchial epithelial cells were then washed and bacteria from cell lysates were plated on LB agar. Colonies were counted after 48 hours.
Supporting Information
Zdroje
1. BoydCD, O'TooleGA (2012) Second messenger regulation of biofilm formation: Breakthroughs in understanding c-di-GMP effector systems. Annu Rev Cell Dev Biol 28 : 439–462.
2. HenggeR (2009) Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol 7 : 263–273.
3. RomlingU, GalperinMY, GomelskyM (2013) Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev 77 : 1–52.
4. SchirmerT, JenalU (2009) Structural and mechanistic determinants of c-di-GMP signalling. Nat Rev Microbiol 7 : 724–735.
5. RyanRP, Tolker-NielsenT, DowJM (2012) When the PilZ don't work: effectors for cyclic di-GMP action in bacteria. Trends Microbiol 20 : 235–242.
6. SondermannH, ShikumaNJ, YildizFH (2012) You've come a long way: c-di-GMP signaling. Curr Opin Microbiol 15 : 140–146.
7. MansfieldJ, GeninS, MagoriS, CitovskyV, SriariyanumM, RonaldP, DowM, VerdierV, BeerSV, MachadoMA, et al. (2012) Top 10 plant pathogenic bacteria in molecular plant pathology. Mol Plant Pathol 13 : 614–629.
8. RyanRP (2013) Cyclic di-GMP signalling and the regulation of bacterial virulence. Microbiol-SGM 159 : 1286–1297.
9. RyanRP, FouhyY, LuceyJF, CrossmanLC, SpiroS, HeYW, ZhangLH, HeebS, CamaraM, WilliamsP, et al. (2006) Cell-cell signaling in Xanthomonas campestris involves an HD-GYP domain protein that functions in cyclic di-GMP turnover. Proc Natl Acad Sci USA 103 : 6712–6717.
10. RyanRP, FouhyY, LuceyJF, JiangB-L, HeY-Q, FengJ-X, TangJ-L, DowJM (2007) Cyclic di-GMP signalling in the virulence and environmental adaptation of Xanthomonas campestris. Mol Microbiol 63 : 429–442.
11. ChinK-H, LeeY-C, TuZ-L, ChenC-H, TsengY-H, YangJ-M, RyanRP, McCarthyY, DowJM, WangAHJ, et al. (2010) The cAMP receptor-like protein CLP is a novel c-di-GMP receptor linking cell-cell signaling to virulence gene expression in Xanthomonas campestris. J Mol Biol 396 : 646–662.
12. GuzzoCR, DungerG, SalinasRK, FarahCS (2013) Structure of the PilZ-FimX(EAL)-c-di-GMP complex responsible for the regulation of bacterial type IV pilus biogenesis. J Mol Biol 425 : 2174–2197.
13. McCarthyY, RyanRP, O'DonovanK, HeY-Q, JiangB-L, FengJ-X, TangJ-L, DowJM (2008) The role of PilZ domain proteins in the virulence of Xanthomonas campestris pv. campestris. Mol Plant Pathol 9 : 819–824.
14. TaoF, HeY-W, WuD-H, SwarupS, ZhangL-H (2010) The cyclic nucleotide monophosphate domain of Xanthomonas campestris global regulator Clp defines a new class of cyclic-di-GMP effectors. J Bacteriol 192 : 1020–1029.
15. LundbackT, HardT (1996) Sequence-specific DNA-binding dominated by dehydration. Proc Natl Acad Sci USA 93 : 4754–4759.
16. SaveanuC, MironS, BorzaT, CraescuCT, LabesseG, GagyiC, PopescuA, SchaefferF, NamaneA, Laurent-WinterC, et al. (2002) Structural and nucleotide-binding properties of YajQ and YnaF, two Escherichia coli proteins of unknown function. Protein Sci 11 : 2551–2560.
17. RyanRP, McCarthyY, KielyPA, O'ConnorR, FarahCS, ArmitageJP, DowJM (2012) Dynamic complex formation between HD-GYP, GGDEF and PilZ domain proteins regulates motility in Xanthomonas campestris. Mol Microbiol 86 : 557–567.
18. FouhyY, ScanlonK, SchouestK, SpillaneC, CrossmanL, AvisonMB, RyanRP, DowJM (2007) Diffusible signal factor-dependent cell-cell signaling and virulence in the nosocomial pathogen Stenotrophomonas maltophilia. J Bacteriol 189 : 4964–4968.
19. MulcahyH, O'CallaghanJ, O'GradyEP, AdamsC, O'GaraF (2006) The posttranscriptional regulator RsmA plays a role in the interaction between Pseudomonas aeruginosa and human airway epithelial cells by positively regulating the type III secretion system. Infect Immun 74 : 3012–3015.
20. TeplyakovA, ObmolovaG, BirN, ReddyP, HowardAJ, GillilandGL (2003) Crystal structure of the YajQ protein from Haemophilus influenzae reveals a tandem of RNP-like domains. J Struct Funct Genomics 4 : 1–9.
21. DuevelJ, BertinettiD, MoellerS, SchwedeF, MorrM, et al. (2012) A chemical proteomics approach to identify c-di-GMP binding proteins in Pseudomonas aeruginosa. J Microbiol Methods 88 : 229–236.
22. NesperJ, ReindersA, GlatterT, SchmidtA, JenalU (2012) A novel capture compound for the identification and analysis of cyclic di-GMP binding proteins. J Proteomics 75 : 4874–4878.
23. QiaoX, SunY, QiaoJ, MindichL (2008) The role of host protein YajQ in the temporal control of transcription in bacteriophage Phi 6. Proc Natl Acad Sci USA 105 : 15956–15960.
24. HaD-G, MerrittJH, HamptonTH, HodgkinsonJT, JanecekM, SpringDR, WelchM, O'TooleGA (2011) 2-Heptyl-4-Quinolone, a precursor of the Pseudomonas quinolone signal molecule, modulates swarming motility in Pseudomonas aeruginosa. J Bacteriol 193 : 6770–6780.
25. LuX-H, AnS-Q, TangD-J, McCarthyY, TangJ-L, DowJM, RyanRP (2012) RsmA regulates biofilm formation in Xanthomonas campestris through a regulatory network involving cyclic di-GMP and the Clp transcription factor. PLoS One 7 : 12.
26. RyanRP, LuceyJ, O'DonovanK, McCarthyY, YangL, Tolker-NielsenT, DowJM (2009) HD-GYP domain proteins regulate biofilm formation and virulence in Pseudomonas aeruginosa. Environ Microbiol 11 : 1126–1136.
27. PultzIS, ChristenM, KulasekaraHD, KennardA, KulasekaraB, MillerSI (2012) The response threshold of Salmonella PilZ domain proteins is determined by their binding affinities for c-di-GMP. Mol Microbiol 86 : 1424–1440.
28. GalperinMY (2010) Diversity of structure and function of response regulator output domains. Curr Opin Microbiol 13 : 150–159.
29. MaddocksSE, OystonPCF (2008) Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins. Microbiol-SGM 154 : 3609–3623.
30. MomanyC, NeidleEL (2012) Defying stereotypes: the elusive search for a universal model of LysR-type regulation. Mol Microbiol 83 : 453–456.
31. AnS-Q, FebrerM, McCarthyY, TangD-J, ClissoldL, KaithakottilG, SwarbreckD, TangJ-L, RogersJ, DowJM, et al. (2013) High-resolution transcriptional analysis of the regulatory influence of cell-to-cell signalling reveals novel genes that contribute to Xanthomonas phytopathogenesis. Mol Microbiol 88 : 1058–1069.
32. AnS-Q, LuG-T, SuH-Z, LiR-F, HeY-Q, JiangB-L, TangD-J, TangJ-L (2011) Systematic mutagenesis of all predicted gntR genes in Xanthomonas campestris pv. campestris reveals a GntR family transcriptional regulator controlling hypersensitive response and virulence. Mol Plant Microbe Interact 24 : 1027–1039.
33. SlaterH, Alvarez-MoralesA, BarberCE, DanielsMJ, DowJM (2000) A two-component system involving an HD-GYP domain protein links cell-cell signalling to pathogenicity gene expression in Xanthomonas campestris. Mol Microbiol 38 : 986–1003.
34. Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual. 2nd Edition Volumes 1, 2 and 3.
35. TwomeyKB, O'ConnellOJ, McCarthyY, DowJM, O'TooleGA, PlantBJ, RyanRP (2012) Bacterial cis-2-unsaturated fatty acids found in the cystic fibrosis airway modulate virulence and persistence of Pseudomonas aeruginosa. ISME J 26 : 939–950.
36. AnS-Q, AllanJH, McCarthyY, FebrerM, DowJM, RyanRP (2014) The PAS domain-containing histidine kinase RpfS is a second sensor for the diffusible signal factor of Xanthomonas campestris. Mol Microbiol 92 : 586–597.
37. McCarthyY, YangL, TwomeyKB, SassA, Tolker-NielsenT, MahenthiralingamE, DowJM, RyanRP (2010) A sensor kinase recognizing the cell-cell signal BDSF (cis-2-dodecenoic acid) regulates virulence in Burkholderia cenocepacia. Mol Microbiol 77 : 1220–1236.
38. AndersonGG, Moreau-MarquisS, StantonBA, O'TooleGA (2008) In vitro analysis of tobramycin-treated Pseudomonas aeruginosa biofilms on cystic fibrosis-derived airway epithelial cells. Infect Immun 76 : 1423–1433.
39. Moreau-MarquisS, StantonBA, O'TooleGA (2008) Pseudomonas aeruginosa biofilm formation in the cystic fibrosis airway. Pulm Pharmacol Therap 21 : 595–599.
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Identification of the Microsporidian as a New Target of the IFNγ-Inducible IRG Resistance SystemČlánek Human Cytomegalovirus Drives Epigenetic Imprinting of the Locus in NKG2C Natural Killer CellsČlánek APOBEC3D and APOBEC3F Potently Promote HIV-1 Diversification and Evolution in Humanized Mouse ModelČlánek Role of Non-conventional T Lymphocytes in Respiratory Infections: The Case of the Pneumococcus
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2014 Číslo 10- Stillova choroba: vzácné a závažné systémové onemocnění
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Jak souvisí postcovidový syndrom s poškozením mozku?
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Infekční komplikace virových respiračních infekcí – sekundární bakteriální a aspergilové pneumonie
-
Všechny články tohoto čísla
- Theory and Empiricism in Virulence Evolution
- -Related Fungi and Reptiles: A Fatal Attraction
- Adaptive Prediction As a Strategy in Microbial Infections
- Antimicrobials, Stress and Mutagenesis
- A Novel Function of Human Pumilio Proteins in Cytoplasmic Sensing of Viral Infection
- Social Motility of African Trypanosomes Is a Property of a Distinct Life-Cycle Stage That Occurs Early in Tsetse Fly Transmission
- Autophagy Controls BCG-Induced Trained Immunity and the Response to Intravesical BCG Therapy for Bladder Cancer
- Identification of the Microsporidian as a New Target of the IFNγ-Inducible IRG Resistance System
- mRNA Structural Constraints on EBNA1 Synthesis Impact on Antigen Presentation and Early Priming of CD8 T Cells
- Infection Causes Distinct Epigenetic DNA Methylation Changes in Host Macrophages
- Neutrophil Crawling in Capillaries; A Novel Immune Response to
- Live Attenuated Vaccine Protects against Pulmonary Challenge in Rats and Non-human Primates
- The ESAT-6 Protein of Interacts with Beta-2-Microglobulin (β2M) Affecting Antigen Presentation Function of Macrophage
- Characterization of Uncultivable Bat Influenza Virus Using a Replicative Synthetic Virus
- HIV Acquisition Is Associated with Increased Antimicrobial Peptides and Reduced HIV Neutralizing IgA in the Foreskin Prepuce of Uncircumcised Men
- Uses a Unique Ligand-Binding Mode for Trapping Opines and Acquiring A Competitive Advantage in the Niche Construction on Plant Host
- Involvement of a 1-Cys Peroxiredoxin in Bacterial Virulence
- Ethanol Stimulates WspR-Controlled Biofilm Formation as Part of a Cyclic Relationship Involving Phenazines
- Densovirus Is a Mutualistic Symbiont of a Global Crop Pest () and Protects against a Baculovirus and Bt Biopesticide
- Insights into Intestinal Colonization from Monitoring Fluorescently Labeled Bacteria
- Mycobacterial Antigen Driven Activation of CD14CD16 Monocytes Is a Predictor of Tuberculosis-Associated Immune Reconstitution Inflammatory Syndrome
- Lipoprotein LprG Binds Lipoarabinomannan and Determines Its Cell Envelope Localization to Control Phagolysosomal Fusion
- Dampens the DNA Damage Response
- MicroRNAs Suppress NB Domain Genes in Tomato That Confer Resistance to
- Novel Cyclic di-GMP Effectors of the YajQ Protein Family Control Bacterial Virulence
- Vaginal Challenge with an SIV-Based Dual Reporter System Reveals That Infection Can Occur throughout the Upper and Lower Female Reproductive Tract
- Detecting Differential Transmissibilities That Affect the Size of Self-Limited Outbreaks
- One Small Step for a Yeast - Microevolution within Macrophages Renders Hypervirulent Due to a Single Point Mutation
- Expression Profiling during Arabidopsis/Downy Mildew Interaction Reveals a Highly-Expressed Effector That Attenuates Responses to Salicylic Acid
- Human Cytomegalovirus Drives Epigenetic Imprinting of the Locus in NKG2C Natural Killer Cells
- Interaction with Tsg101 Is Necessary for the Efficient Transport and Release of Nucleocapsids in Marburg Virus-Infected Cells
- The N-Terminus of Murine Leukaemia Virus p12 Protein Is Required for Mature Core Stability
- Sterol Biosynthesis Is Required for Heat Resistance but Not Extracellular Survival in
- Allele-Specific Induction of IL-1β Expression by C/EBPβ and PU.1 Contributes to Increased Tuberculosis Susceptibility
- Host Cofactors and Pharmacologic Ligands Share an Essential Interface in HIV-1 Capsid That Is Lost upon Disassembly
- APOBEC3D and APOBEC3F Potently Promote HIV-1 Diversification and Evolution in Humanized Mouse Model
- Structural Basis for the Recognition of Human Cytomegalovirus Glycoprotein B by a Neutralizing Human Antibody
- Systematic Analysis of ZnCys Transcription Factors Required for Development and Pathogenicity by High-Throughput Gene Knockout in the Rice Blast Fungus
- Epstein-Barr Virus Nuclear Antigen 3A Promotes Cellular Proliferation by Repression of the Cyclin-Dependent Kinase Inhibitor p21WAF1/CIP1
- The Host Protein Calprotectin Modulates the Type IV Secretion System via Zinc Sequestration
- Cyclophilin A Associates with Enterovirus-71 Virus Capsid and Plays an Essential Role in Viral Infection as an Uncoating Regulator
- A Novel Alpha Kinase EhAK1 Phosphorylates Actin and Regulates Phagocytosis in
- The pH-Responsive PacC Transcription Factor of Governs Epithelial Entry and Tissue Invasion during Pulmonary Aspergillosis
- Sensing of Immature Particles Produced by Dengue Virus Infected Cells Induces an Antiviral Response by Plasmacytoid Dendritic Cells
- Co-opted Oxysterol-Binding ORP and VAP Proteins Channel Sterols to RNA Virus Replication Sites via Membrane Contact Sites
- Characteristics of Memory B Cells Elicited by a Highly Efficacious HPV Vaccine in Subjects with No Pre-existing Immunity
- HPV16-E7 Expression in Squamous Epithelium Creates a Local Immune Suppressive Environment via CCL2- and CCL5- Mediated Recruitment of Mast Cells
- Dengue Viruses Are Enhanced by Distinct Populations of Serotype Cross-Reactive Antibodies in Human Immune Sera
- CD4 Depletion in SIV-Infected Macaques Results in Macrophage and Microglia Infection with Rapid Turnover of Infected Cells
- A Sialic Acid Binding Site in a Human Picornavirus
- Contact Heterogeneity, Rather Than Transmission Efficiency, Limits the Emergence and Spread of Canine Influenza Virus
- Myosins VIII and XI Play Distinct Roles in Reproduction and Transport of
- HTLV-1 Tax Stabilizes MCL-1 via TRAF6-Dependent K63-Linked Polyubiquitination to Promote Cell Survival and Transformation
- Species Complex: Ecology, Phylogeny, Sexual Reproduction, and Virulence
- A Critical Role for IL-17RB Signaling in HTLV-1 Tax-Induced NF-κB Activation and T-Cell Transformation
- Exosomes from Hepatitis C Infected Patients Transmit HCV Infection and Contain Replication Competent Viral RNA in Complex with Ago2-miR122-HSP90
- Role of Non-conventional T Lymphocytes in Respiratory Infections: The Case of the Pneumococcus
- Kaposi's Sarcoma-Associated Herpesvirus Induces Nrf2 during Infection of Endothelial Cells to Create a Microenvironment Conducive to Infection
- A Relay Network of Extracellular Heme-Binding Proteins Drives Iron Acquisition from Hemoglobin
- Glutamate Secretion and Metabotropic Glutamate Receptor 1 Expression during Kaposi's Sarcoma-Associated Herpesvirus Infection Promotes Cell Proliferation
- Reduces Malaria and Dengue Infection in Vector Mosquitoes and Has Entomopathogenic and Anti-pathogen Activities
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Novel Cyclic di-GMP Effectors of the YajQ Protein Family Control Bacterial Virulence
- MicroRNAs Suppress NB Domain Genes in Tomato That Confer Resistance to
- The ESAT-6 Protein of Interacts with Beta-2-Microglobulin (β2M) Affecting Antigen Presentation Function of Macrophage
- Characterization of Uncultivable Bat Influenza Virus Using a Replicative Synthetic Virus
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



