-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Allele-Specific Induction of IL-1β Expression by C/EBPβ and PU.1 Contributes to Increased Tuberculosis Susceptibility
IL-1β is important for the initial establishment of antimicrobial adaptive immunity, but prolonged IL-1β expression can also cause progressive immunopathology during M. tuberculosis infection. The paradoxical activities of IL-1β in promoting both antimycobacterial immunity and chronic tissue damage have left the ultimate contribution of this cytokine to TB progression in human populations unclear. In this work, we address the role of IL-1β-mediated inflammation using a combination of human genetics and molecular biology, and suggest that exuberant IL-1β responses are causatively associated with TB progression and poor treatment outcome in humans. This work furthers our understanding of the immunological factors that underlie TB disease and provide a strong rationale for the development of specific anti-inflammatory adjunctive therapies that could improve the long-term outcome of TB treatment. In addition, these insights inform the design of future TB control efforts that include the rational design of disease-preventing vaccines and genotype-targeted delivery of TB chemotherapy.
Published in the journal: . PLoS Pathog 10(10): e32767. doi:10.1371/journal.ppat.1004426
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1004426Summary
IL-1β is important for the initial establishment of antimicrobial adaptive immunity, but prolonged IL-1β expression can also cause progressive immunopathology during M. tuberculosis infection. The paradoxical activities of IL-1β in promoting both antimycobacterial immunity and chronic tissue damage have left the ultimate contribution of this cytokine to TB progression in human populations unclear. In this work, we address the role of IL-1β-mediated inflammation using a combination of human genetics and molecular biology, and suggest that exuberant IL-1β responses are causatively associated with TB progression and poor treatment outcome in humans. This work furthers our understanding of the immunological factors that underlie TB disease and provide a strong rationale for the development of specific anti-inflammatory adjunctive therapies that could improve the long-term outcome of TB treatment. In addition, these insights inform the design of future TB control efforts that include the rational design of disease-preventing vaccines and genotype-targeted delivery of TB chemotherapy.
Introduction
Tuberculosis (TB), a chronic bacterial disease caused by Mycobacterium tuberculosis (Mtb), remains a major global health problem that claims 1.4 million lives annually. Natural infection with Mtb is initiated by the deposition of Mtb-containing aerosol droplets onto lung alveolar surfaces, and infection at this site can produce a wide spectrum of clinical outcomes. The vast majority of immunocompetent individuals contain the pathogen and remain indefinitely asymptomatic, a status defined as “latent” TB [1]. A smaller proportion of Mtb-infected people develop active disease, most often characterized by a progressive inflammatory pathology of the lung. The hematogenous dissemination of Mtb can also result in disease at a variety of extra pulmonary sites, commonly including bone and the central nervous system [2].
The mechanisms controlling TB progression remain elusive. Both Mendelian genetic susceptibilities in the IL-12 and IFN-γ axis [3], [4], and the association of TB with HIV-mediated lymphocyte depletion indicate that cell-mediated adaptive immunity is critical for controlling mycobacterial growth and containing disease. However, genetic association studies also commonly identify single nucleotide polymorphisms (SNPs) in pro-inflammatory innate immune mediators that are associated with TB susceptibility and regulate cellular function, such as TLRs, LTA4H, IL22, and IL6 [5], [6], [7], [8], [9], [10]. These associations suggest that disease progression may be determined at multiple levels in human populations and that the inflammatory response could play a decisive role.
As a potent proinflammatory cytokine, IL-1β plays an important role in many inflammation-related diseases as well as cancer. Accordingly, IL1B gene polymorphisms, especially functional SNPs -31 T>C (rs1143627) and -511 G>A (rs16944) are associated with susceptibility to a number of diseases [11], [12], [13], [14]. IL-1β is also important in the pathogenesis of TB in mice [15], and polymorphisms in the human IL1B gene have been suggested to have differing effects on TB susceptibility. Awomoyi reported the IL1B rs16944, but not +3953 T>C (rs1143634) SNP, was associated with susceptibility to TB in a Gambian population [16]. Kusuhara et al reported that three SNPs in IL1B (rs1143629, rs1143643 and rs3917368) are associated with TB susceptibility in a Japanese population [17]. Similarly, susceptibility to TB is associated with the polymorphic +3953 region in the IL1B gene [18]. Using an oligochip-based method, Sun et al genotyped two SNPs (rs1143627, rs16944) in a small cohort of the Chinese Han population (98 TB patients and 65 healthy controls), and found that the rs1143627T allele was associated with TB susceptibility [19]. On the contrary, a strong protection conferred by IL1B +3953 T-allele-carrying genotypes was observed in a Northwestern Colombian population [20]. While these studies implicated several SNPs in IL1B gene were associated with TB susceptibility, a conclusive role of IL1B SNPs has not yet been established due to the small sample sizes and different ethnic background in these studies. Furthermore, the mechanisms by which these polymorphisms might act remain unclear.
Recent mechanistic studies in the mouse model have implicated IL-1β mediated granulocytic inflammation as a potential driver of TB progression [21], [22], [23]. IL-1β is a prototypical proinflammatory cytokine that stimulates both local and systemic responses [24], and this cytokine plays a complex dual role in chronic infections. The production of IL-1β is important for the proper development of antimicrobial adaptive immunity during the initial stages of infection [25], [26]. However, prolonged IL-1β production promotes the continual recruitment of granulocytes to the lung, induces the expression of additional inflammatory mediators such as prostaglandin E2, and stimulates tissue-damaging metalloproteinases [27], [28]. As a result, the production of this cytokine must be repressed in chronically Mtb-infected animals to avoid progressive pathology [21], [29]. While, production of IL-1β correlates with the severity of human TB disease [30], [31], its paradoxical activities in promoting both antimycobacterial immunity and chronic tissue damage leave the ultimate contribution of this cytokine to TB progression in human populations unclear.
In this study, we identified a genetic polymorphism (rs1143627) in the promoter region of the IL1B gene that increases the C/EBPβ - and PU.1-dependent expression of the cytokine. The high-IL-1β-expressing genotype is associated with increased risk of active tuberculosis and poor clinical outcome. The observed correlation between IL-1β production, neutrophil recruitment, and TB susceptibility indicates a causative role for IL-1β-mediated granulocytic inflammation in TB progression.
Results
IL1B promoter polymorphism is associated with susceptibility to tuberculosis
Genetic variants in the IL1B gene, especially those in the promoter region, can affect cytokine expression and have been associated with susceptibility to inflammatory disorders [24], [32] and chronic infections [33], [34]. Since TB is a disease of chronic inflammation, we hypothesized that similar genetic variants might alter susceptibility to this disease. To test our hypothesis, the genotype distribution of 4 IL1B SNPs with potential regulatory effects was first determined in healthy controls and TB patients from a cohort in Shenzhen and then replicated in a Shanghai cohort (Table 1). Among them, the following three SNPs were located within the known promoter region, -31 T>C (rs1143627), -511 G>A (rs16944), and -1473 G>C (rs1143623). The final SNP, +7326 T>C (rs2853550), is located in the 3-UTR of IL1B gene. The minor allele frequencies of all 4 SNPs were>10% and were in Hardy-Weinberg equilibrium in both control groups (P>0.05). However, only the frequency of rs1143627 was significantly different between patients with active TB (TB, n = 1533) and healthy controls (HC, n = 1445) in the Shenzhen cohort. Specifically, a significantly higher frequency of the T allele at rs1143627 was observed among patients with active TB, compared with controls, indicating that this allele is associated with an increased risk of tuberculosis (odds ratio [OR] = 1.20; 95% confidence interval [CI], 1.09–1.33; P = 0.0004). At the genotype level, carriage of the IL1B rs1143627T allele increased the apparent risk of active TB (OR = 1.35; 95% CI, 1.14–1.59; P = 0.0005, dominant model). TT and TC genotypes also showed increased TB susceptibility compared to CC genotype using an additive model (OR = 1.44; 95%CI, 1.18–1.76; P = 0.0004; and OR = 1.30; 95%CI, 1.09–1.55; P = 0.004) (Table 2).
Tab. 1. Characteristics of patients with active TB and healthy controls in multiple cohorts. 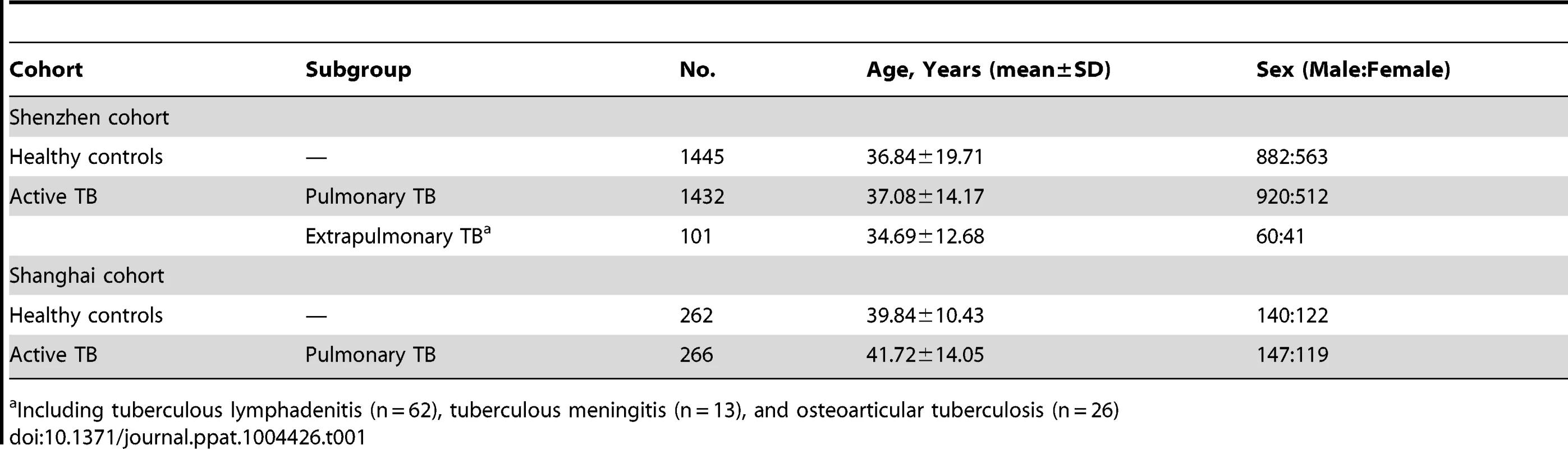
Including tuberculous lymphadenitis (n = 62), tuberculous meningitis (n = 13), and osteoarticular tuberculosis (n = 26) Tab. 2. Association between IL1B Gene SNPs and TB susceptibility in Shenzhen cohort. 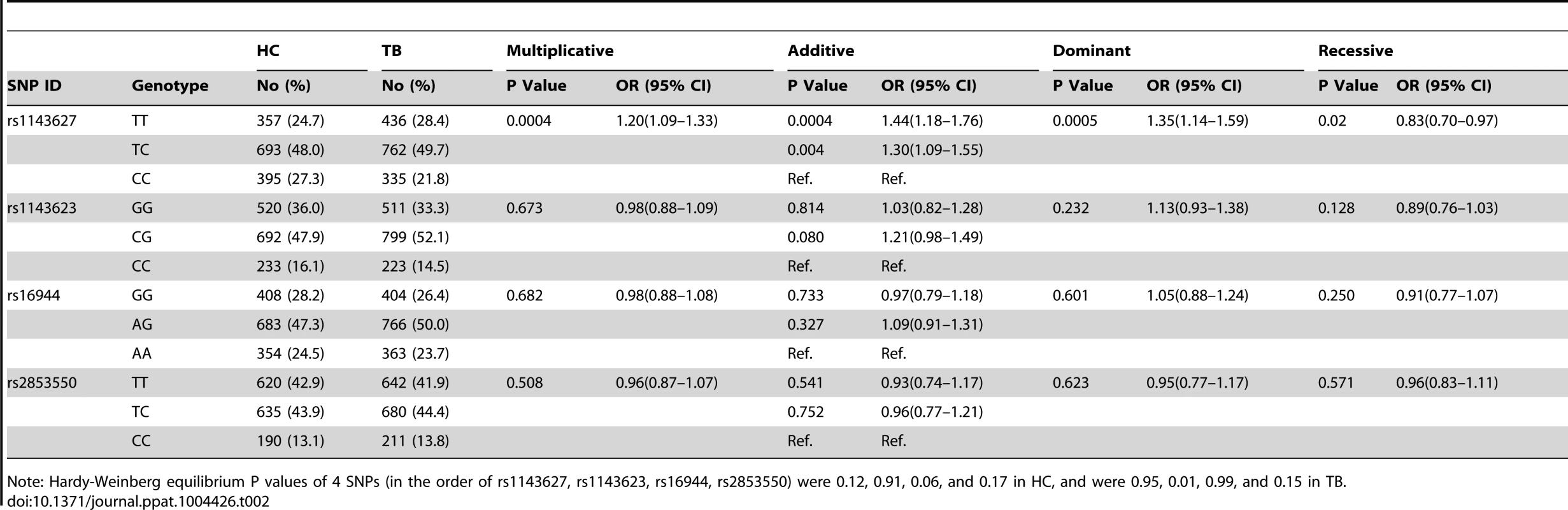
Note: Hardy-Weinberg equilibrium P values of 4 SNPs (in the order of rs1143627, rs1143623, rs16944, rs2853550) were 0.12, 0.91, 0.06, and 0.17 in HC, and were 0.95, 0.01, 0.99, and 0.15 in TB. The association of rs1143627 with tuberculosis was replicated in an independent Shanghai cohort. In this group the T allele was associated with TB susceptibility using a multiplicative model, with an adjusted OR of 1.40 (95% CI, 1.10–1.77, P = 0.007). Using an additive model the T allele was estimated to impart a 97% increase in risk (OR = 1.97; 95% CI, 1.20–3.23; P = 0.006) (Table 3). These observations from two large independent cohorts were similar and consistent with the reported distribution of rs1143627 and rs16944 alleles in separate small cohort of Chinese TB patients and healthy controls [35]. When we pooled the Shenzhen and shanghai data together (1799 cases and 1707 controls) for statistical analysis, the effect of rs1143627T was even stronger in all genetic models (multiplicative, additive, dominant and recessive, Table S1) than in either cohort alone. Thus, we conclude that the T allele of IL1B SNP rs1143627 is associated with susceptibility to active TB in the Chinese population.
Tab. 3. Replication of association between rs1143627 SNP and TB susceptibility in Shanghai cohort. 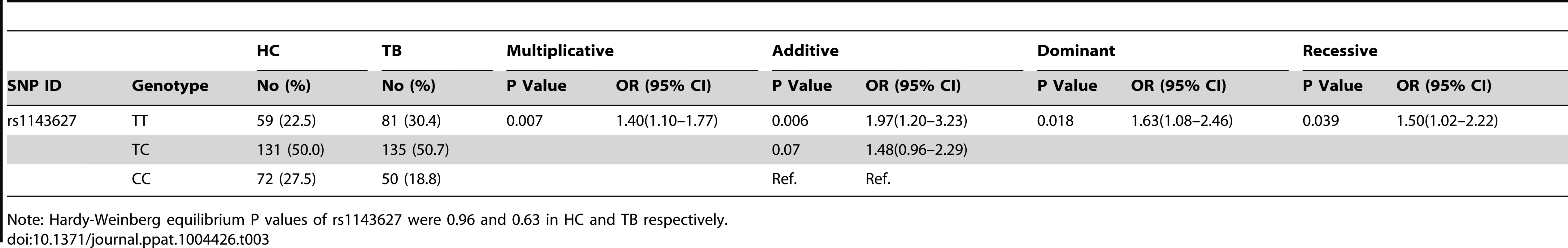
Note: Hardy-Weinberg equilibrium P values of rs1143627 were 0.96 and 0.63 in HC and TB respectively. While TB is primarily a disease of the lung, dissemination of the bacterium can also result in pathology at a variety of different tissue sites. To understand if the rs1143627 SNP differentially affected pulmonary versus extrapulmonary disease, we further investigated whether the distribution of rs1143627 alleles differed in patients suffering from distinct manifestations of TB. As shown in Table 4, the rs1143627T allele is significantly associated with both pulmonary TB (PTB, OR = 1.17; 95% CI, 1.06–1.30; P = 0.003) as well as extrapulmonary disease (ETB, OR = 1.78; 95% CI, 1.32–2.39; P<0.0001) using a multiplicative model. However, the frequency of the rs1143627T allele was significantly higher in patients with extrapulmonary TB than in those with pulmonary disease (P = 0.005). Thus, among TB patients, individuals carrying rs1143627T allele are more prone to develop extrapulmonary TB than those without (OR = 1.53, 95% CI, 1.14–2.05, multiplicative model).
Tab. 4. Association between rs1143627 SNP and susceptibility to various clinical phenotypes of TB in Shenzhen cohort. 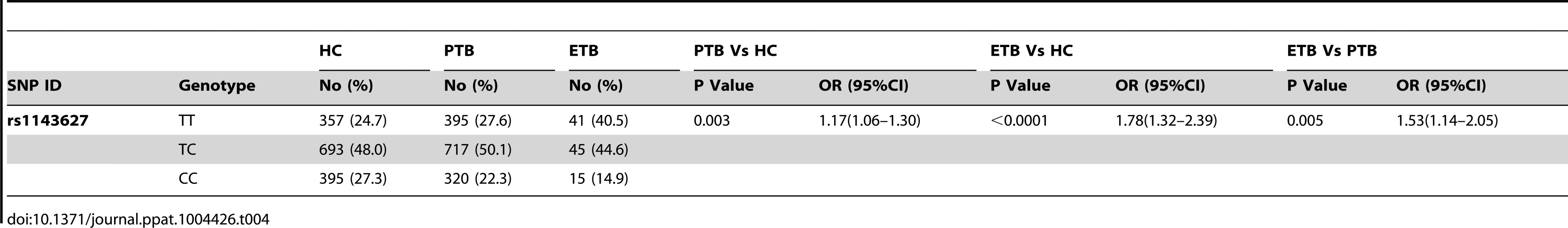
IL1B polymorphism is associated with more severe TB disease
The proinflammatory activity of IL-1β suggested that the rs1143627 polymorphism might affect the inflammatory response to Mtb, which could be exhibited at either systemic or local levels. No difference in erythrocyte sedimentation rate (ESR) or C-reactive protein (CRP) levels were apparent among pulmonary TB patients carrying different rs1143627 genotypes (TT, TC, CC) (Fig. S1). However, using high-resolution computed tomography (HRCT), we were able to directly quantify lung damage in these patients using a score based on radiographic manifestations including the presence of nodules, cavities, and bronchial lesions [36], [37]. A total of 453 out of 1432 patients with primary pulmonary TB received an HRCT score before anti-TB treatment. Among these individuals, the HRCT scores were significantly higher in patients carrying rs1143627TT genotype than those with rs1143627CC genotype (Fig. 1A). Even 2 years after the completion of anti-TB treatment, patients (n = 53) of the rs1143627TT genotype still displayed significantly higher HRCT scores than those carrying rs1143627CC, suggesting that the rs1143627 polymorphism is associated with long-term lung damage and disease outcome (Fig. 1C). In contrast, no difference of HRCT score was found in patients carrying different genotypes of rs16944 (Fig. 1B and D), an IL1B SNP that is associated with the risk of developing active TB in the Gambian population [16], but not in our current study. Taken together, these results indicated that the rs1143627T allele is associated with more severe pulmonary TB and the expression of extrapulmonary disease.
Fig. 1. SNP rs1143627 is associated with the severity of pulmonary TB disease. 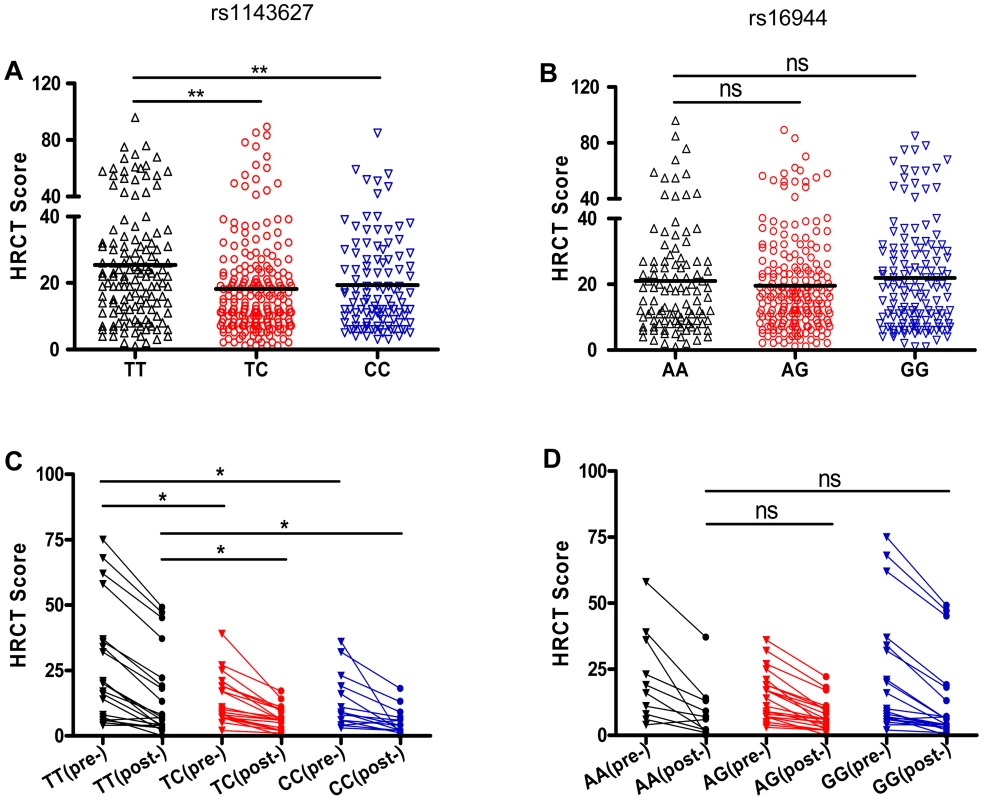
(A and B) The HRCT scores were determined in a total of 453 patients with pulmonary TB before initiation of anti-TB chemotherapy. The HRCT scores in patients carrying different rs1143627 genotypes (A) or rs16944 genotypes (B) were compared. (C and D) The HRCT scores were determined in 53 of 453 patients who have performed HRCT examinations before and 2 years after completion of anti-TB chemotherapy. The HRCT scores [before (pre-) and 2 years after (post-) initiation of treatment] in patients carrying different rs1143627 genotypes (C) or rs16944 genotypes (D) were compared. Differences between groups were compared with the ANOVA/Newman-Keuls multiple comparison test. *, p<0.05; **, p<0.01; ns, not significant. The rs1143627T allele increases IL-1β production, but not IL-Ra expression
Located at position -31 from the transcriptional start site, SNP rs1143627 is in the promoter region of IL1B gene. Previous reports indicated that rs1143627 affects transcription of IL-1β in response to lipopolysaccharide [38]. To determine whether this polymorphism affects gene expression in response to Mtb stimulation, we assessed IL-1β expression in CD14+ monocytes isolated from healthy controls carrying different rs1143627 genotypes in response to heat-killed Mtb or the 19 kDa lipoprotein derived from this bacillus. In response to both stimuli, monocytes isolated from individuals carrying rs1143627TT and TC genotypes produced significantly higher amounts of IL1B protein (Fig. 2A) and mRNA (Fig. 2C) than those carrying the CC genotype. Since the bioactivity of IL-1β also involves the antagonistic effects of IL-1Ra, we assessed the concentration of IL-1Ra in the culture supernatant and IL1RN mRNA cell lysates. As shown in Fig. 2B and D, no significant differences were observed in IL-1Ra expression upon Mtb lysate stimulation among different rs1143627 genotypes. Together, these results suggest that the rs1143627T allele specifically increased the amount of bioactive IL-1β produced by Mtb stimulation.
Fig. 2. The rs1143627 polymorphism affects IL-1β production by monocytes upon Mtb stimulation. 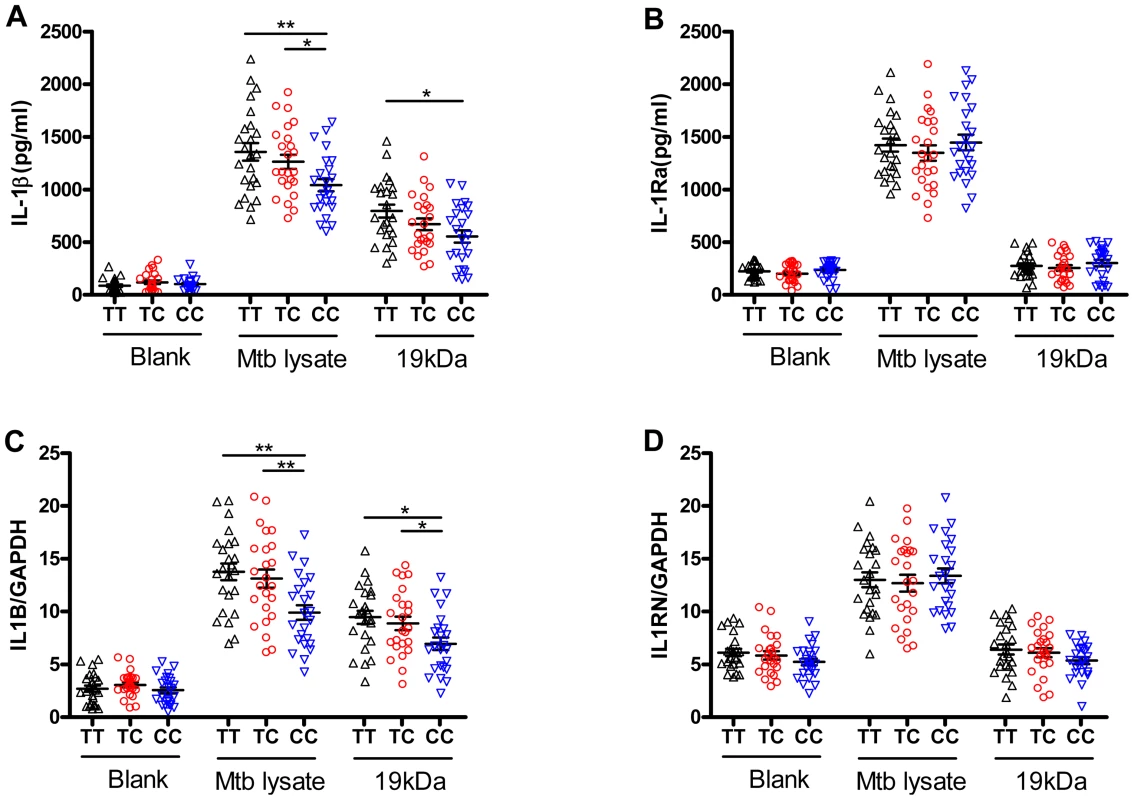
CD14+ monocytes were isolated from PBMCs of healthy controls carrying different rs1143627 genotypes (TT, n = 24; TC, n = 24; CC, n = 24). Purified CD14+ monocytes were cultured in the absence or presence of heat-killed Mtb lysate (20 µg/mL) or Mtb 19 kDa lipoprotein (0.5 µg/mL,). (A and B) The concentrations of IL-1β (A) and IL-Ra (B) in the culture supernatant were determined by ELISA after incubation for 24 h. (C and D) The IL1B (C) and IL1RN (D) mRNA levels were determined using SYBR Green-based real-time qPCR after incubation for 24 h, and the data was expressed as mRNA copy number relative to house keeping gene GAPDH. Differences between groups were compared with the ANOVA/Newman-Keuls multiple comparison test. *, p<0.05; **, p<0.01. The rs1143627T allele does not impair the anti-mycobacterial adaptive immune response, but promotes neutrophil infiltration to the lung
IL-1β is recognized to play an important role in shaping adaptive immunity, especially Th17 cell responses [39], [40]. To understand if the rs1143627T allele could promote TB disease by inhibiting the expression of adaptive immunity, we investigated whether this polymorphism altered the production of Mtb antigen-specific IFN-γ or IL-17A, the hallmark cytokines of Th1 and Th17 cells, respectively. PBMC isolated from healthy controls carrying different rs1143627 genotypes were cultured in the presence of heat-killed Mtb and the production of IFN-γ and IL-17A were compared. Consistent with our previous results with purified monocytes, we found that PBMC from individuals carrying rs1143627 TT and TC genotypes produced more IL-1β than those from individuals carrying the CC genotype (Fig. 3A). The rs1143627TT PBMC also produced slightly higher levels of IFN-γ and similar levels of IL-17A as rs1143627 CC cells (Fig. 3, B and C). The increased production of IFN-γ was a result of IL-1β secretion, as IFN-γ and IL-1β secretion were highly correlated in PBMC cultures and IFN-γ production could be inhibited by the addition of a blocking antibody to IL-1β (Fig. S2).
Fig. 3. rs1143627T-induced IL-1β has little effect on Mtb-specific lymphocyte responses, but IL-1β correlates with neutrophil recruitment to the lung. 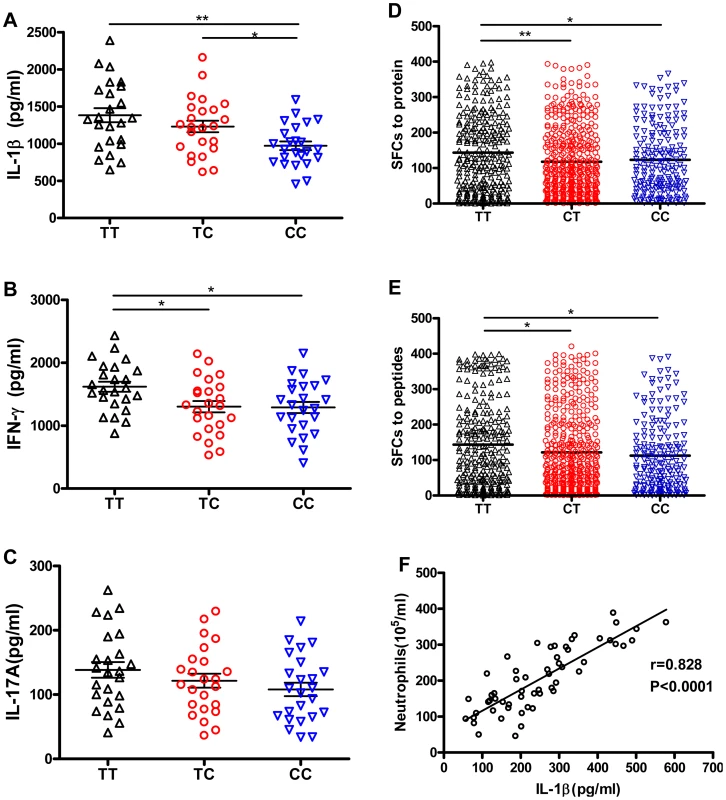
PBMCs from healthy controls carrying different rs1143627 genotypes (TT, n = 24; TC, n = 24; CC, n = 24) were cultured in the absence of Mtb lysate (20 µg/mL) for 48 h. (A-C) The concentrations of secreted IL-1β (A), IFN-γ (B) and IL-17A (C) were determined by ELISA. (D and E) Mtb antigen (ESAT-6 protein, or ESAT-6/CFP-10 peptide pool) specific IFN-γ production by PBMCs from patients with PTB carrying different rs1143627 genotypes was quantified by ELISPOT assay. Data were expressed as the number of IFN-γ SFCs per 2×105 PBMCs of each subjects. Differences between groups were compared with the ANOVA/Newman-Keuls multiple comparison test. (F) Correlation analysis between the levels of IL-1β and the number of neutrophils in the BALF. The coefficient r and P values are indicated. *, p<0.05; **, p<0.01. The effect of rs1143627 on the production of IFN-γ-producing cells was further investigated in a large cohort of pulmonary TB patients. Mtb antigen-specific IFN-γ production by PBMCs was detected by using a previously established IFN-γ Elispot assay that employs either ESAT-6 protein or an ESAT-6/CFP-10 peptide pool as a stimulant [41], [42]. Patients carrying the rs1143627TT genotype had significantly higher numbers of Mtb antigen-specific IFN-γ spot forming cells (SFCs) than those carrying the CC genotype (Fig. 3, D and E). In contrast, the numbers of IFN-γ SFCs were not different among patients carrying different rs16944 genotypes (Fig. S3). Thus, the TB-associated rs1143627T allele was associated with an augmentation of the canonical antimycobacterial response. We conclude that the susceptibility of individuals carrying the rs1143627T allele is unlikely to be the result of a failure to respond to mycobacterial antigen.
Another primary effect of IL-1β is the recruitment of neutrophils, a process that can exacerbate TB pathogenesis in animal models [21], [22], [23]. To investigate whether this cytokine could play a similar role in human TB, we simultaneously measured the level of IL-1β and the number of neutrophils in broncheoalveolar lavage fluid (BALF) from patients with active pulmonary TB. A significant correlation between the level of IL-1β and the number of recruited neutrophils was found (r = 0.828, P<0.0001) (Fig. 3F). Thus, the increased production of IL-1β associated with the rs1143627T allele could promote lung damage and TB progression by stimulating granulocytic inflammation.
SNP rs1143627 influences C/EBPβ and PU.1 binding to IL1B promoter
The rs1143627 defines a T>C mutation at the -31 position of the IL1Β promoter. We used electrophoretic mobility-shift analysis (EMSA) to determine if this polymorphism altered protein binding to this promoter region. Synthetic allele-specific oligonucleotides representing the polymorphic rs1143627 sites were incubated with nuclear protein extracts from human monocytic U937 cells stimulated without or with killed Mtb lysate. There was no difference in binding activity between rs1143627T oligonucleotide (T Probe) and rs1143627C oligonucleotide (C Probe) when U937 were not stimulated with Mtb lysate (Fig. 4A). Previous Mtb stimulation of these cells induced the formation of a DNA binding complex on the rs1143627T oligonucleotide (“complex 1”, Figure 4B). In contrast, Mtb exposure induced less complex 1 formation on the rs1143627C oligonucleotide (Fig. 4B, lane 2 and 6; Fig. 4C). Furthermore, complex 1 formation on radiolabelled rs1143627 was specifically blocked by competition with the unlabelled oligonucleotide (Fig. 4B, lanes 3, 4, 7, and 8). These results indicated that the binding of one or more proteins to the IL1B promoter region was altered by rs1143627 polymorphism, which could cause the observed difference in IL-1β expression that was associated with these genotypes.
Fig. 4. SNP rs1143627 variant affects C/EBPβ and PU.1 binding to the IL1B promoter. 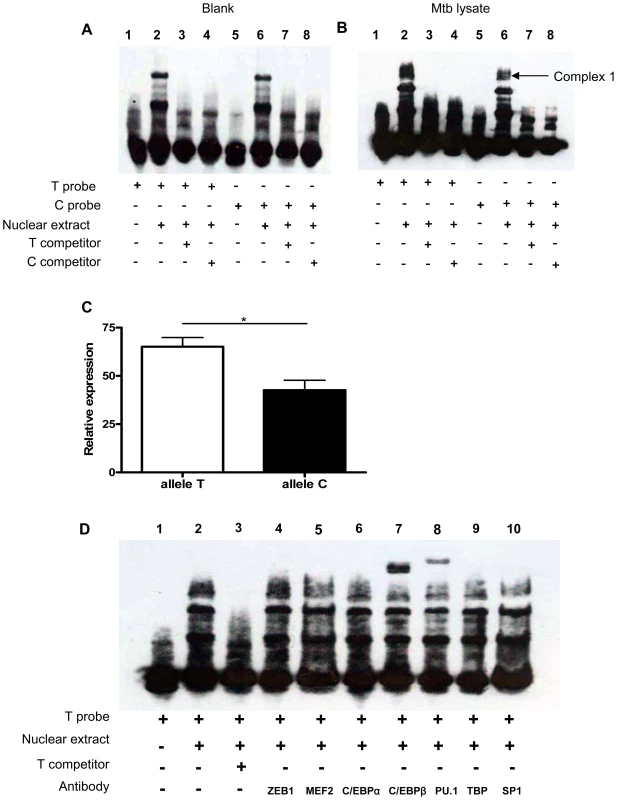
(A and B) Nuclear extracts were prepared PMA-differentiated U937 cells stimulated without (A) or with (B) Mtb lysate (20 µg/mL) for 24 h. Nuclear extracts were incubated with T-probe (lanes 1–4) or C-probe (lanes 5–8) in the absence or presence of unlabelled competitors and subjected to EMSA. (C) The relative abundance of complex 1 bound to T probe or C probe in (B) quantified by densitometry. Data are shown as the mean ± SEM, and differences between groups were compared with unpaired t tests. (D) Nuclear extracts prepared from (A) were incubated with T-probe in the absence (lane 2) or presence of antibodies against transcription factors including ZEB1, MEF2, C/EBPα, C/EBPβ, PU.1, TBP, and SP1 (lanes 4–10). Anti-C/EBPβ (lane 7) and anti-PU.1 (lane 8) supershift the DNA-protein complex 1. *, p<0.05. Bioinformatics-based prediction analysis using alibaba and match algorithm (http://www.gene-regulation.com) and previous studies of the IL1B promoter region indicated that transcription factors C/EBPa, C/EBPβ, PU.1, TBP and SP1 are involved in the regulation of IL1B gene expression and have the potential to bind in the polymorphic region [43], [44]. To determine if these proteins were part of the rs1143627-modulated complex 1, we conducted supershift experiments using antibodies against each transcription factor. Addition of antibodies against C/EBPβ and PU.1, but not antibodies against RSRFC4, C/EBPa, TBP or SP1, to the rs1143627T oligonucleotide binding reactions resulted in shift of complex 1 to a higher molecular weight species (Fig. 4D). Thus, the protein complex formed on the rs1143627-containing region includes PU.1 and C/EBPβ, suggesting that the T>C polymorphism could alter IL-1β expression by influencing the binding of transcription complexes that contain these factors.
Expression of PU.1 and C/EBPβ is induced by Mtb and temporally-correlated with IL-1β expression
To determine if C/EBPβ and PU.1 are produced in a manner that is consistent with the proposed role in IL-1β regulation, we quantified the expression of all three genes in the monocyte-like U937 cells after stimulation with heat killed Mtb or infection with the live attenuated strain of Mtb, H37Ra. Consistent with previous reports, both stimuli induced IL1B mRNA expression (Fig. 5A). More importantly, we found Mtb exposure also induced the expression of C/EBPB (Fig. 5B) and PU.1 (Fig. 5C), but no significant difference was observed in TBP expression (Fig. 5D). Furthermore, the mRNA level of IL1B was significantly correlated with that of both C/EBPB (r = 0.887, P<0.0001, Fig. 5E) and PU.1 (r = 0.811, P = 0.0001, Fig. 5F), and not correlated with TBP expression (r = 0.314, P = 0.236, Fig. 5G). The induction of PU.1 and C/EBPB mRNA expression occurred earlier after stimulation than IL1B (Fig. 5H-K), which is consistent with a role for PU.1 and C/EBPβ in regulating the IL1B promoter in response to Mtb infection.
Fig. 5. C/EBPB and PU.1 expression induced by Mtb temporally correlates with IL1B expression. 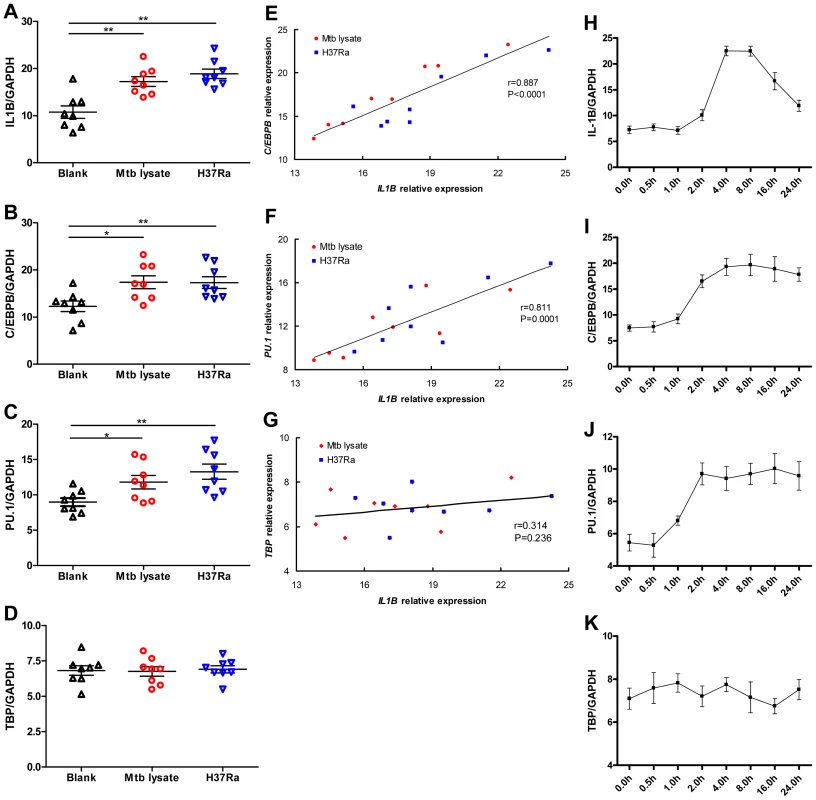
(A-D) SYBR Green-based real-time qPCR assay for the gene expression of IL1B (A), C/EBPB (B), PU.1 (C), and TBP (D) by differentiated U937 cells in response to Mtb lysate (20 µg/mL) or H37Ra infection (MOI = 10). The data are expressed as mRNA copy number relative to the house keeping gene GAPDH. Differences between groups were compared with the ANOVA/Newman-Keuls multiple comparison test. (E-G) Correlation analysis between the C/EBPB (E), PU.1 (F), or TBP (G) expression and IL1B expression. The coefficient r and P values are indicated. (H-K) Dynamic change of IL1B (H), C/EBPB (I), PU.1 (J) and TBP (K) expression by differentiated U937 upon stimulation with Mtb lysate (20 µg/mL). *, p<0.05; **, p<0.01. PU.1 and C/EBPβ differentially transactivate IL1B promoter variants determined by the rs1143627 polymorphism
To assess the role of PU.1 and C/EBPβ in Mtb-induced IL1B transcription, we investigated whether ectopic PU.1 and C/EBPβ expression affects IL1B promoter activity in HeLa cells. As shown in Fig. 6A, transfection of a reporter plasmid containing a 1371 bp fragment of the human IL1B promoter (−1292 to +79) produced very low luciferase levels. However, cotransfection of the IL1B promoter plasmid with PU.1 or C/EBPβ expression plasmids significantly enhanced luciferase levels, and cotransfection of all three plasmids further increased promoter activity (Fig. 6A). Expression levels of PU.1 and C/EBPβ were similar when transfected either individually or in combination (Fig. 6C).
Fig. 6. Allele-specific effects of rs1143627 are mediated through PU.1 and C/EBPβ. 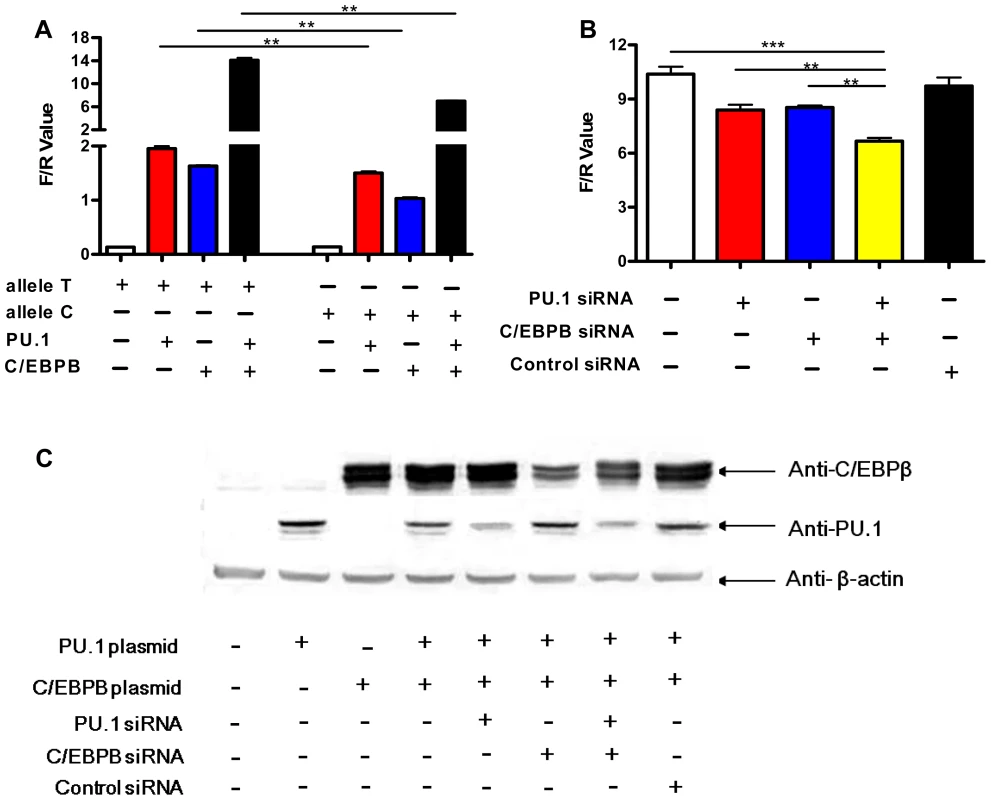
(A) IL1B promoter luciferase reporter plasmids carrying rs1143627 T or C allele were transfected into HeLa cells, with or without co-transfecting PU.1- or C/EBPβ-expressing plasmids. Luciferase activities of HeLa cells were determined and normalized to Renillla luciferase activities. (B) HeLa cells cotransfected with the T-allele IL1B promoter luciferase reporter plasmids, PU.1- and C/EBP-β- expressing plasmids were infected with PU.1, C/EBPβ, or control siRNA-carrying lentiviral vector constructs. Luciferase activities of HeLa cells were determined and normalized to Renillla luciferase activities. Differences between groups were compared with the ANOVA/Newman-Keuls multiple comparison test. (C) Western-blotting assay shows the expression of PU.1 or C/EBPβ after transfection or siRNA knockdown using monoclonal antibodies against PU.1 and C/EBPβ. **, p<0.01, ***, p<0.001. siRNA knockdown studies verified the contribution PU.1 and C/EBPβ in Mtb-induced IL1B transcriptional activity in this transfection system. The expression of both transcription factors could be partially inhibited by siRNA transfection (Fig. 6C). Knockdown of either protein significantly inhibited the Mtb-induced IL1B promoter activity (Fig. 6B). Simultaneous knockdown had an additive effect (Fig. 6B), consistent with the collaborative activity observed in co-transfection experiments (Fig. 6A). Thus, these transcription factors simultaneously contribute to IL1B promoter activity in HeLa cells.
We then utilized this reporter assay to determine if altered PU.1 and/or C/EBPβ transactivation could account for rs1143627 allele-specific regulation of IL1B gene expression. When -31 T>C alterations were introduced into the IL1B promoter we observed no difference in basal transcriptional activity in HeLa cells. However, upon co-transfection with PU.1, C/EBPβ, or both, the T allele produced significantly higher luciferase activity than the C variant (Fig. 6A). The degree of increased activity observed for the rs1143627 T allele was similar to that observed in primary monocytes and PBMC (Fig. 2 and Fig. 3A). Taken together, these results indicate that the SNP rs1143627 alters a cis-regulatory element in the IL1B gene that alters C/EBPβ - and PU.1-dependent expression of IL1B and susceptibility to TB.
Discussion
Proinflammatory cytokines, such as IL-1β can play complex and dichotomous roles during chronic infections. This cytokine is initially required to prime the anti-mycobacterial immune response [25], [26], [45], and may also promote resistance to initial infection through the induction of epithelial antimicrobial peptides [46]. However, the prolonged production of this cytokine must be restrained to prevent chronic tissue damage [21]. Similarly complex roles for proinflammatory pathways have been described during a variety of viral and bacterial infections [47], [48], [49]. As a result, the production of these cytokines often correlates with the severity of human disease, but their complex biological activities make it difficult to determine if this expression is causing pathology or limiting it.
In this study, we define a mechanistic link between a polymorphism in the promoter of the IL1B gene and the severity of TB disease, which implies a causal relationship between the expression of this cytokine and the progression of pathology. A similar association between high IL1B expressing genotypes and inflammatory diseases has been observed in a number of previous studies. Associations between TB and IL1B polymorphisms at -511G>A, +3953T>C, and +3962T>C have been reported in a variety of populations [16], [20], [50], [51], [52], although our study is the first to describe a link between high Mtb-induced IL-1β production, inflammation, and TB disease. The specific IL1B promoter allele (rs1143627T) that was associated with TB in our large cohort was previously reported to be enriched in a smaller study of Chinese TB patients [35]. Indeed, this allele is also associated with other diseases that involve IL-1β-dependent inflammation or cell death, such as influenza [53], keratoconus [54], [55], and coronary artery disease [56]. Thus, it appears that TB susceptibility can be determined by similar mechanisms to those that underlie other inflammatory diseases of diverse etiology.
We found that a single base change in the IL1B promoter increased the synergistic activity of the C/EBPβ and PU.1 transcription factors leading to increased IL-1β expression in monocytes and PBMC. These two transcription factors are known to act in a coordinated manner to drive macrophage differentiation [57], and our studies suggest they also promote the effector functions of these cells. Individuals carrying the high-IL-1β-producing rs1143627T allele were more prone to develop active TB, to have severe pathology in the lung, and to harbor extrapulmonary lesions. The association between IL-1β production and severe TB disease is consistent with our recent finding that NLRP3 inflammasome dependent IL-1β enhances neutrophil recruitment and exacerbates pulmonary pathology in mice infected with Mtb [21]. In the chronically-infected mouse, IL-1β activity is restrained at a posttranslational level through the nitric oxide (NO)-dependent nitrosylation of NLRP3 [21], an essential component of the inflammasome complex that processes pro-IL-1β into its active form. As this regulatory pathway has also been described in human cells [58], the processing of rs1143627T-induced pro-IL-1β into its bioactive form is likely mediated either by residual NLRP3 activity or through inflammasome-independent processes.
Two general mechanisms could account for the TB susceptibility that is associated with increased IL-1β expression; inhibition of specific antimicrobial immunity or exacerbated inflammatory tissue damage. The former hypothesis is unlikely to explain our current observations, as IFN-γ and IL-17 responses were either unchanged or enhanced in the TB susceptible individuals. The correlation that we observed between IL-1β, IFN-γ and TB disease supports the emerging view of IFN-γ as a poor correlate of protective immunity [59]. Mtb antigen-specific IFN-γ production by Th1, multifunctional Th1, or Th1/Th17 cells, is associated with clinical severity and bacterial load in TB patients, and not protective immunity [36], [60], [61]. Instead, the significant correlation we observed between IL-1β levels and the number of neutrophils in BALF of patients with active TB supports a specific role for granulocytic inflammation in promoting the progression of TB disease. This model is consistent with the pathological role played by neutrophil recruitment in a number of TB-susceptible mouse strains [22], [62], [63], and the predominance of this cell type in the BALF of humans with active TB [29], [64].
The high IL-1β expressing rs1143627TT genotype was associated with the severity of lung pathology in TB patients both before and after anti-tuberculosis treatment. This finding is consistent with the poor treatment response of individuals with a high frequency of IL-17/IFN-γ double-producing cells [61], which our studies suggest as a possible marker of IL-1β production (Fig. 3). These observations suggest that sustained IL-1β production causes persistent lung damage that could contribute to the permanently decreased lung function observed in patients with advanced and/or recurrent TB [65]. Thus, anti-inflammatory treatments targeted to the IL-1 pathway could be a useful adjunct therapy to mitigate the long-term pulmonary impairment caused by Mtb infection, particularly for individuals of high IL-1β-expressing genotypes.
Materials and Methods
Ethics statement
All protocols for this study were reviewed and approved by the Research Ethics Committee of Shenzhen Third People's Hospital (No. 2012–003), and conducted according to the Declaration of Helsinki. The use, for research purposes, of excess BALF leftover from clinically indicated bronchoscopies was deemed exempt from a requirement for informed consent beyond the consent normally obtained for this clinical procedure. The Research Ethics Committee approved the collection of peripheral blood exclusively for research purposes with the written informed consent of all participants.
Subjects and samples
Three case-control cohorts were used in this study to investigate the association between IL1B gene polymorphisms and susceptibility to TB. All subjects were genetically unrelated members of the Chinese Han population. The Shenzhen experimental cohort involved 1533 patients with active TB and 1445 healthy controls, which have been used in our previous IL6 polymorphism study[10]. Of the 1533 patients, 1432 were diagnosed with pulmonary TB (PTB), and 101 were with extrapulmonanry TB (ETB) including tuberculous lymphadenitis (n = 62), tuberculous meningitis (n = 13), and osteoarticular TB (n = 26). The Shanghai cohort was used for validation and consisted of 266 PTB patients and 262 controls, which has been used for a previous genetic study on IL-17F polymorphisms and TB susceptibility [66]. The diagnosis of tuberculosis was based on clinical symptoms, radiological evidence, and findings from Mtb examination as described previously [10], [67]. Healthy controls with normal chest radiograph findings and without a clinical history of TB were recruited. The characteristics of the study population are shown in Table 1. The whole blood samples were store at −80°C after collection for DNA extraction. Peripheral blood mononuclear cells (PBMCs) were isolated from whole blood through density gradient centrifugation over Ficoll-Hypaque as described elsewhere [68] and stored in nitrogen. The broncho-alveolar lavage fluid (BALF) was collected from Mtb culture confirmed pulmonary TB patients (n = 57) before initiation of anti-TB chemotherapy.
SNP selection and genotyping
Genomic DNA was prepared from peripheral whole blood according to the standard protocols of QIAamp DNA Blood Mini kit (Qiagen, Hilden, Germany) as described previously[10]. Since we were particularly interested in SNPs with regulatory activity, we focused on SNPs located in putative transcription factor binding sites and microRNA target sites. To search for functional SNPs, we referred to Jaspar, UniPROBE, TRANSFAC, and PITA databases, then calculated the binding score of alleles to transcription factor, or the minimum hybridization energy and thermodynamics to microRNA as described [9]. Three SNPs in the promoter (rs1143627 T>C, rs16944 G>A, rs1143623 G>C) and one in 3-UTR (rs2853550 T>C) of the IL1B gene were genotyped using the MassARRAY system (Sequenom, San Diego, CA) as described elsewhere [9], [10]. The relative height (intensity) of the peaks and the signal-to-noise (SNR) ratio were analyzed using Caller software to call genotypes in real-time. Typer software can apply cluster analysis to the genotype calls assigned by the Caller software. After cluster analysis, manual curation of spectra was performed to further validate the outcome. Assays with low call rates (<90%) were discarded or redesigned, i.e., all assays shown in this study have a call rate of>90%.
HRCT examination and scoring
HRCT were performed at 10 mm section interval (120 kV, 50–450 mAs), (1 mm slice thickness, 1.5s scanning time) with a window level between 2550 and 40 Hounsfield Units (HU) and window width between 300 and 1600 HU using the Toshiba Aquilion 64 CT Scanner (Toshiba, Tokyo, Japan). HRCT scans were analyzed two independent chest radiologists and final conclusions on the findings were reached by consensus. The arbitrary scores were based on the percentage of lung parenchyma abnormality as previously described [36], [37].
Cell preparation and cultures
Human monocytic cell lines U937 (ATCC CRL 1593) and HeLa cells (strain S3) were cultured in antibiotic-free Dulbecco's modified Eagle medium (DMEM) containing 10% fetal bovine serum (FBS) (Hyclone, Logan, UT, USA). After differentiation in the presence of phorbol 12-myristate 13-acetate (PMA, 20 ng/ml) for 24 hours, U937 cells were rested in fresh complete medium for 24 h before further stimulation. The differentiated U937 cells were exposed to heat-killed M. tuberculosis lysate (20 µg/mL), or live Mtb H37Ra strain at a multiplicity of infection of 10∶1. The cells were harvested after additional 24 hours and used for gene expression assays or nuclear protein binding activity assays.
Primary monocytes were isolated from PBMCs of 72 healthy controls (n = 24 for each rs1143627 genotype) by positive selection using magnetic CD14 MicroBeads (Miltenyi Biotech, Germany) according to the manufacturer's instructions. Purified CD14+ monocytes and PBMCs were transferred into a 96-well plate in serum-free AIM-V medium (Gibco, Carlsbad, CA, USA), with 2×105 cells/well in the absence or presence of heat killed Mtb lysate (20 µg/mL) or Mtb 19 kDa lipoprotein (0.5 µg/mL; Lionex GmbH, Braunschweig, Germany). The cells were harvested after 24 h or 48 h and used for gene expression assays, and the supernatant was collected to determine cytokine (IL-1β, IL-1Ra, IFN-γ, and IL-17A) production using ELISA.
Measurement of genes expression by qRT-PCR
Total RNA extraction was performed with the RNAeasy Mini kit (Qiagen, Valencia, CA), and residual DNA was digested using RNAse-free DNAse (Qiagen). cDNA was synthesized using an oligo-dT primer and SuperScript II reverse transcriptase (Invitrogen, Carlsbad, CA). Gene expression was measured using a previously described SYBR Green-based real-time quantitative PCR [69]. For all assays, target genes were normalized against the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) level. The qPCR primers were designed as follows: for the IL1B gene:, 5'-TTCTTCGACACATGGGATAACG-3' (forward) and 5'-TGGAGAACACCACTTGTTGCT-3' (reverse); for the IL1RN gene: 5'-GGAAGATGTGCCTGTCCTGT-3' (forward) and 5'-TCTCGCTCAGGTCAGTGATG-3' (reverse); for the PU.1 gene: 5'-CAGCTCTACCGCCACATGGA-3' (forward) and 5'-TAGGAGACCTGGTGGCCAAGA-3' (reverse); for the C/EBPB gene: 5'-AACTCTCTGCTTCTCCCTCTG-3' (forward) and 5'-AAGCCCGTAGGAACATCTTT-3' (reverse); for the TBP gene: 5'-TCTGGGATTGTACCGCAGC-3' (forward) and 5'-CGAAGTGCAATGGTCTTTAGG-3' (reverse); for the GAPDH gene: 5'-GCACCGTCAAGGCTGAGAAC-3' (forward) and 5'-TGGTGAAGACGCCAGTGGA-3' (reverse).
Measurement of secreted cytokine by ELISA
The levels of IL-1β, IL-1Ra, IFN-γ, IL-17A in the supernatants of stimulated PBMCs or CD14+ monocytes were determined using commercially available ELISA kits (R&D, Minneapolis, MN), following the manufacturer's instructions.
Plasmid constructs
The human IL1B promoter (−1292 to +79) was amplified by PCR and inserted into pGL3-Basic vector (Promega) upstream of the firefly luciferase coding region at XhoI and NcoI sites. Positive clones were subjected to site-directed mutagenesis using Quick Change Site-Directed mutagenesis Kit (Stratagene, La Jolla, CA) to obtain the desired alleles. Expression vectors for the full-length PU.1 and C/EBPB were constructed by inserting the respective coding regions into pcDNA3.1 expression vector (life technology, Carlsbad, CA) at XhoI and EcoRI sites. The primer used for cloning were as follows: for PU.1, 5'-TATCTCGAGAACTTGTGCTGGCCCTGCAATG-3' (forward) and 5'-TATGAATTCTGTGGGGCGGGTGGCGCCGCT-3' (reverse); for C/EBPB, 5'-TATCTCGAGGAGTCAGAGCCGCGCACGGGACT-3' (forward) and 5'-TATGAATTCTGCAGTGGCCGGAGGAGGCGAGCAGGGGCT-3' (reverse).
Transfection and dual-luciferase assay
HeLa cells (2×105) were plated in 24-well plates 24 h before transfection. A total of 0.8 ug plasmid DNA including 0.3 ug of either rs1143627 T or rs1143627 C reporter vector, 0.2 mg of pcDNA3.1-PU.1 or pcDNA3.1-C/EBPB expression vector, and 0.1 mg of pRL-TK control vector, were co-transfected into the cells using Lipofectamine 2000 reagent (life technology). Cells were stimulated by PMA (50 ng/ml) for 20 h, following 24 h of transfection. Cells were then harvested and lysed in 150 uL passive lysis buffer (Promega), the lysates were assayed for both the firefly and Renilla luciferase activities using the dual-luciferase reporter assay system (Promega). Promoter activity was measured as the ratio between firefly and Renilla luciferase. The transfection for each construct was performed three times and each construct was assayed for promoter activity in duplicates.
Western blotting
HeLa cells transfected with plasmid expressing PU.1 or C/EBPβ were collected 16–20 h after transfection. The cells pellet was lysed using lysis buffer (10 mM Tris, pH 7.5, 150 mM NaCl, 1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride, 0.2 mM sodium orthovanadate, 0.5% Nonidet P-40) supplemented with a cocktail of protease inhibitors (Sigma-Aldrich, Steinheim, Germany). The lysates were separated by SDS-PAGE on a 12% polyacrylamide gel and then transferred onto a polyvinylidene difluoride membrane. The membranes were sequentially probed with the respective primary antibodies, followed by appropriate HRP-conjugated secondary antibodies (Promega, Madison, WI) and then visualized by exposure to X-ray films.
Electrophoretic mobility shift assay (EMSA)
Nuclear extracts were prepared from U937 monocytes as reported previously [70]. Two sets of complementary DNA-oligonucleotide sequences containing IL1B rs1143627 C or T allele were designed and biotin-labelled or left unlabelled (Life technology). The oligonucleotides used were as follows: rs1143627C variant, 5'-CCTACTTCTGCTTTTGAAAGCCATAAAAACAGCGA GGGAGAAA-3'; and rs1143627T variant, 5'-CCTACTTCTGCTTTTGA AAGCTATAAAAACAGCGAGGAGAAA-3'. Equimolar amounts of each strand were combined in annealing buffer (10 mM Tris, 1 mM EDTA and 50 mM NaCl) by heating to 95°C for 2 min, and cooling slowly to room temperature over 2 h. EMSA assays were performed by using the LightShift chemiluminescent EMSA kit (Pierce, Rockford, IL). Binding reactions contained 20fmol biotin-labelled double - stranded probe, 2.0 uL nuclear extract and 1.0 ug poly (dI:dC) in a total volume of 20 uL binding buffer (10 mM Tris, 50 mM KCl, 3 mM MgCl2, 0.1 mM EDTA and 1.0 mM DTT). After incubation for 20 min at room temperature, complexes were separated on a 6% native polyacrylamide gel, and blotted onto a positively charged nylon membrane (Millipore, Billerica, MA), and visualized by exposure to X-ray films. EMSA images were quantified by densitometry using Quantity One, version 4.5, software (Bio-Rad). For relative quantification, the integrated optical density value was determined with background values taken below each band of interest to account for non-specific antibody staining in the lane.
For competition experiments, a 200-fold molar excess of unlabelled probe was added prior to addition of labeled probe. To assess the putative binding sites within the IL1B promoter in the region encompassing the SNP rs1143627, we used two bioinformatic tools, AliBaba2 and TFSearch, to predict the interaction of the different probes with proteins and set the cutoff for the dissimilarity matrix at 15%. In antibody supershift assays, anti-ZEB1, MEF2, C/EBPα, C/EBPβ, PU.1, TBP, and SP1 (each at the final concentration of 1 ug/mL, Santa Cruz Biotechnology, Santa Cruz, CA) were added after binding of nuclear extracts to labeled probe and incubated for 20 min at room temperature.
IFN-γ ELISPOT assay
A previously established in-house IFN-γ ELISPOT assay was used to measure Mtb antigen specific IFN-γ spot forming cells (SFCs) in peripheral blood samples from patients with TB [41], [42]. Briefly, a total of 2×105 cells/well of PBMCs were cultured in duplicate in 96-well plates in the presence of ESAT6 protein (protein) or peptide pools derived from ESAT6/CFP10 (peptides) for 24 h. PBMCs cultured in medium alone or in the presence of phytohemagglutinin (Sigma) at 2.5 ug/ml were used as negative or positive controls, respectively. IFN-γ producing cells were visualized as spot forming cells after incubation with biotinylated anti–IFN-γ monoclonal antibody (eBioscience) for 4 h, followed by streptavidin-alkaline phosphatase conjugate for 2 h and then substrate solution. The number of SFCs was counted using an automated ELISPOT reader (BioReader 4000 Pro-X; Biosys, Germany). The number of SFCs to protein and peptides was expressed as the number of SFCs per 0.2 million PBMCs. The background number of SFCs in the negative control well was subtracted.
Lentiviral-mediated RNA interference
For knockdown of PU.1, or C/EBPB, we used lentiviral vectors expressing gene-specific small interfering RNA (siRNA) to specifically block expression. Oligonucleotide sequences of PU.1 and C/EBPB specific siRNAs were as follows: PU.1 siRNA, 5'-GCTTCGCCGAGAACAACTTCA-3', C/EBPB siRNA, 5'-TTCTCCGAACGTGTCACGTTTC-3'. The stem-loop oligonucleotides were synthesized and cloned into a lentivirus-based vector carrying the green fluorescent protein (GFP) gene (pGCSIL-GFP, Genechem, Shanghai, China). A universal sequence (LV1-NC: 5'-TTCTCCGAACGT GTCACGT-3') was used as a negative control for RNA interference. Individual human shRNA lentiviral clones were prepared and isolated as previously described [71]. For infection with siRNA-carrying lentiviral vector constructs, the viruses were diluted in serum-free OptiMEM (life technonogy) and cells were infected at a multiplicity of infection of 10 for 3 h in the presence of 5 ug/mL polybrene. After 48 h of infection in serum-containing medium, cells were harvested and tested for PU.1 or C/EBPβ expression by Western-blot.
Statistical analysis
The Hardy-Weinberg Equilibrium (HWE) for IL1B polymorphisms distribution was analyzed in healthy controls and cases. The allelic and genotypic frequencies of SNPs between cases and controls were compared using the Pearson X2 test. The unconditional logistic regression adjusted by gender and age were performed to calculate the Odd ratios (ORs), 95% confidence intervals (CIs) and corresponding P values under four alternative models (multiplicative, additive, dominant and recessive). The one-way analysis of variance (ANOVA)/Newman-Keuls multiple comparison test was used for statistical analyses to compare the differences among multiple groups. The paired t test was used to compare the IFN-γ production with or without treatment of anti-IL-1β. Correlations were assessed using Spearman's rank correlation, and Pearson's t test was used to analyze the correlation. We used GraphPad Prism software (version 4.0) for all the statistic analysis. Two-tailed statistical tests were conducted with a significance level of 0.05.
Accession numbers of genes mentioned in the manuscript
IL1B: [Homo sapiens (human)]. NCBI Gene ID: 3553.
C/EBPB: [Homo sapiens (human)]. NCBI Gene ID: 1051.
PU.1: [Homo sapiens (human)]. NCBI Gene ID: 6688.
TBP: [Homo sapiens (human)]. NCBI Gene ID: 6908.
IL1RN: [Homo sapiens (human)]. NCBI Gene ID: 3557.
Supporting Information
Zdroje
1. AhmadS (2011) Pathogenesis, immunology, and diagnosis of latent Mycobacterium tuberculosis infection. Clin Dev Immunol 2011 : 814943.
2. SiaIG, WielandML (2011) Current concepts in the management of tuberculosis. Mayo Clin Proc 86 : 348–361.
3. El BaghdadiJ, GrantAV, SabriA, El AzbaouiS, ZaidiH, et al. (2013) [Human genetics of tuberculosis]. Pathol Biol (Paris) 61 : 11–16.
4. CottleLE (2011) Mendelian susceptibility to mycobacterial disease. Clin Genet 79 : 17–22.
5. MischEA, MacdonaldM, RanjitC, SapkotaBR, WellsRD, et al. (2008) Human TLR1 deficiency is associated with impaired mycobacterial signaling and protection from leprosy reversal reaction. PLoS Negl Trop Dis 2: e231.
6. CawsM, ThwaitesG, DunstanS, HawnTR, LanNT, et al. (2008) The influence of host and bacterial genotype on the development of disseminated disease with Mycobacterium tuberculosis. PLoS Pathog 4: e1000034.
7. ThuongNT, HawnTR, ThwaitesGE, ChauTT, LanNT, et al. (2007) A polymorphism in human TLR2 is associated with increased susceptibility to tuberculous meningitis. Genes Immun 8 : 422–428.
8. TobinDM, VaryJCJr, RayJP, WalshGS, DunstanSJ, et al. (2010) The lta4h locus modulates susceptibility to mycobacterial infection in zebrafish and humans. Cell 140 : 717–730.
9. ZhangG, ChenX, ChanL, ZhangM, ZhuB, et al. (2011) An SNP selection strategy identified IL-22 associating with susceptibility to tuberculosis in Chinese. Sci Rep 1 : 20.
10. ZhangG, ZhouB, WangW, ZhangM, ZhaoY, et al. (2012) A functional single-nucleotide polymorphism in the promoter of the gene encoding interleukin 6 is associated with susceptibility to tuberculosis. J Infect Dis 205 : 1697–1704.
11. HayashiF, WatanabeM, NanbaT, InoueN, AkamizuT, et al. (2009) Association of the -31C/T functional polymorphism in the interleukin-1beta gene with the intractability of Graves' disease and the proportion of T helper type 17 cells. Clin Exp Immunol 158 : 281–286.
12. LiC, WangC (2013) Current evidences on IL1B polymorphisms and lung cancer susceptibility: a meta-analysis. Tumour Biol 34 : 3477–3482.
13. ChenML, LiaoN, ZhaoH, HuangJ, XieZF (2014) Association between the IL1B (−511), IL1B (+3954), IL1RN (VNTR) polymorphisms and Graves' disease risk: a meta-analysis of 11 case-control studies. PLoS One 9: e86077.
14. El-OmarEM, CarringtonM, ChowWH, McCollKE, BreamJH, et al. (2000) Interleukin-1 polymorphisms associated with increased risk of gastric cancer. Nature 404 : 398–402.
15. MishraBB, RathinamVA, MartensGW, MartinotAJ, KornfeldH, et al. (2013) Nitric oxide controls the immunopathology of tuberculosis by inhibiting NLRP3 inflammasome-dependent processing of IL-1beta. Nat Immunol 14 : 52–60.
16. AwomoyiAA, CharuratM, MarchantA, MillerEN, BlackwellJM, et al. (2005) Polymorphism in IL1B: IL1B-511 association with tuberculosis and decreased lipopolysaccharide-induced IL-1beta in IFN-gamma primed ex-vivo whole blood assay. J Endotoxin Res 11 : 281–286.
17. KusuharaK, YamamotoK, OkadaK, MizunoY, HaraT (2007) Association of IL12RB1 polymorphisms with susceptibility to and severity of tuberculosis in Japanese: a gene-based association analysis of 21 candidate genes. Int J Immunogenet 34 : 35–44.
18. NaslednikovaIO, UrazovaOI, VoronkovaOV, StrelisAK, NovitskyVV, et al. (2009) Allelic polymorphism of cytokine genes during pulmonary tuberculosis. Bull Exp Biol Med 148 : 175–180.
19. SunH, WangY, MaX, PeiF, ZhangY, et al. (2007) A method of oligochip for single nucleotide polymorphism genotyping in the promoter region of the interleukin-1 beta gene and its clinical application. Oligonucleotides 17 : 336–344.
20. GomezLM, CamargoJF, CastiblancoJ, Ruiz-NarvaezEA, CadenaJ, et al. (2006) Analysis of IL1B, TAP1, TAP2 and IKBL polymorphisms on susceptibility to tuberculosis. Tissue Antigens 67 : 290–296.
21. MishraBB, RathinamVA, MartensGW, MartinotAJ, KornfeldH, et al. (2012) Nitric oxide controls the immunopathology of tuberculosis by inhibiting NLRP3 inflammasome-dependent processing of IL-1beta. Nat Immunol 14 : 52–60.
22. DorhoiA, DeselC, YeremeevV, PradlL, BrinkmannV, et al. (2010) The adaptor molecule CARD9 is essential for tuberculosis control. J Exp Med 207 : 777–792.
23. DorhoiA, IannacconeM, FarinacciM, FaeKC, SchreiberJ, et al. (2013) MicroRNA-223 controls susceptibility to tuberculosis by regulating lung neutrophil recruitment. J Clin Invest.
24. GabayC, LamacchiaC, PalmerG (2010) IL-1 pathways in inflammation and human diseases. Nat Rev Rheumatol 6 : 232–241.
25. JuffermansNP, FlorquinS, CamoglioL, VerbonA, KolkAH, et al. (2000) Interleukin-1 signaling is essential for host defense during murine pulmonary tuberculosis. J Infect Dis 182 : 902–908.
26. YamadaH, MizumoS, HoraiR, IwakuraY, SugawaraI (2000) Protective role of interleukin-1 in mycobacterial infection in IL-1 alpha/beta double-knockout mice. Lab Invest 80 : 759–767.
27. O'KaneCM, ElkingtonPT, JonesMD, CaviedesL, TovarM, et al. (2010) STAT3, p38 MAPK, and NF-kappaB drive unopposed monocyte-dependent fibroblast MMP-1 secretion in tuberculosis. Am J Respir Cell Mol Biol 43 : 465–474.
28. HarrisJE, Fernandez-VilasecaM, ElkingtonPT, HorncastleDE, GraeberMB, et al. (2007) IFNgamma synergizes with IL-1beta to up-regulate MMP-9 secretion in a cellular model of central nervous system tuberculosis. Faseb J 21 : 356–365.
29. LyadovaIV, TsiganovEN, KapinaMA, ShepelkovaGS, SosunovVV, et al. (2010) In mice, tuberculosis progression is associated with intensive inflammatory response and the accumulation of Gr-1 cells in the lungs. PLoS One 5: e10469.
30. ChensueSW, DaveyMP, RemickDG, KunkelSL (1986) Release of interleukin-1 by peripheral blood mononuclear cells in patients with tuberculosis and active inflammation. Infect Immun 52 : 341–343.
31. TsaoTC, LiL, HsiehM, LiaoS, ChangKS (1999) Soluble TNF-alpha receptor and IL-1 receptor antagonist elevation in BAL in active pulmonary TB. Eur Respir J 14 : 490–495.
32. HarrisonP, PointonJJ, ChapmanK, RoddamA, WordsworthBP (2008) Interleukin-1 promoter region polymorphism role in rheumatoid arthritis: a meta-analysis of IL-1B-511A/G variant reveals association with rheumatoid arthritis. Rheumatology (Oxford) 47 : 1768–1770.
33. MigitaK, MaedaY, AbiruS, NakamuraM, KomoriA, et al. (2007) Polymorphisms of interleukin-1beta in Japanese patients with hepatitis B virus infection. J Hepatol 46 : 381–386.
34. Martinez-CarrilloDN, Garza-GonzalezE, Betancourt-LinaresR, Monico-ManzanoT, Antunez-RiveraC, et al. (2010) Association of IL1B -511C/-31T haplotype and Helicobacter pylori vacA genotypes with gastric ulcer and chronic gastritis. BMC Gastroenterol 10 : 126.
35. SunH, WangY, MaX, PeiF, SunH, et al. (2007) A method of oligochip for single nucleotide polymorphism genotyping in the promoter region of the interleukin-1 beta gene and its clinical application. Oligonucleotides 17 : 336–344.
36. QiuZ, ZhangM, ZhuY, ZhengF, LuP, et al. (2012) Multifunctional CD4 T cell responses in patients with active tuberculosis. Sci Rep 2 : 216.
37. OrsF, DenizO, BozlarU, GumusS, TasarM, et al. (2007) High-resolution CT findings in patients with pulmonary tuberculosis: correlation with the degree of smear positivity. J Thorac Imaging 22 : 154–159.
38. WenAQ, WangJ, FengK, ZhuPF, WangZG, et al. (2006) Effects of haplotypes in the interleukin 1beta promoter on lipopolysaccharide-induced interleukin 1beta expression. Shock 26 : 25–30.
39. ZielinskiCE, MeleF, AschenbrennerD, JarrossayD, RonchiF, et al. (2012) Pathogen-induced human TH17 cells produce IFN-gamma or IL-10 and are regulated by IL-1beta. Nature 484 : 514–518.
40. LasiglieD, TraggiaiE, FedericiS, AlessioM, BuoncompagniA, et al. (2012) Role of IL-1 beta in the development of human T(H)17 cells: lesson from NLPR3 mutated patients. PLoS One 6: e20014.
41. ChenX, YangQ, ZhangM, GranerM, ZhuX, et al. (2009) Diagnosis of active tuberculosis in China using an in-house gamma interferon enzyme-linked immunospot assay. Clin Vaccine Immunol 16 : 879–884.
42. ZhangM, WangH, LiaoM, ChenX, GranerM, et al. (2010) Diagnosis of latent tuberculosis infection in bacille Calmette-Guerin vaccinated subjects in China by interferon-gamma ELISpot assay. Int J Tuberc Lung Dis 14 : 1556–1563.
43. LindH, HaugenA, ZienolddinyS (2007) Differential binding of proteins to the IL1B -31 T/C polymorphism in lung epithelial cells. Cytokine 38 : 43–48.
44. KominatoY, GalsonD, WatermanWR, WebbAC, AuronPE (1995) Monocyte expression of the human prointerleukin 1 beta gene (IL1B) is dependent on promoter sequences which bind the hematopoietic transcription factor Spi-1/PU.1. Mol Cell Biol 15 : 58–68.
45. FremondCM, TogbeD, DozE, RoseS, VasseurV, et al. (2007) IL-1 receptor-mediated signal is an essential component of MyD88-dependent innate response to Mycobacterium tuberculosis infection. J Immunol 179 : 1178–1189.
46. VerwayM, BouttierM, WangTT, CarrierM, CalderonM, et al. (2013) Vitamin D Induces Interleukin-1beta Expression: Paracrine Macrophage Epithelial Signaling Controls M. tuberculosis Infection. PLoS Pathog 9: e1003407.
47. BhowmickR, Tin MaungNH, HurleyBP, GhanemEB, GronertK, et al. (2013) Systemic Disease during Streptococcus pneumoniae Acute Lung Infection Requires 12-Lipoxygenase-Dependent Inflammation. J Immunol.
48. LitvakV, RatushnyAV, LampanoAE, SchmitzF, HuangAC, et al. (2012) A FOXO3-IRF7 gene regulatory circuit limits inflammatory sequelae of antiviral responses. Nature 490 : 421–425.
49. CillonizC, ShinyaK, PengX, KorthMJ, ProllSC, et al. (2009) Lethal influenza virus infection in macaques is associated with early dysregulation of inflammatory related genes. PLoS Pathog 5: e1000604.
50. TrajkovD, TrajchevskaM, ArsovT, PetlichkovskiA, StrezovaA, et al. (2009) Association of 22 cytokine gene polymorphisms with tuberculosis in Macedonians. Indian J Tuberc 56 : 117–131.
51. Motsinger-ReifAA, AntasPR, OkiNO, LevyS, HollandSM, et al. (2010) Polymorphisms in IL-1beta, vitamin D receptor Fok1, and Toll-like receptor 2 are associated with extrapulmonary tuberculosis. BMC Med Genet 11 : 37.
52. WilkinsonRJ, PatelP, LlewelynM, HirschCS, PasvolG, et al. (1999) Influence of polymorphism in the genes for the interleukin (IL)-1 receptor antagonist and IL-1beta on tuberculosis. J Exp Med 189 : 1863–1874.
53. LiuY, LiS, ZhangG, NieG, MengZ, et al. (2013) Genetic variants in IL1A and IL1B contribute to the susceptibility to 2009 pandemic H1N1 influenza A virus. BMC Immunol 14 : 37.
54. MikamiT, MeguroA, TeshigawaraT, TakeuchiM, UemotoR, et al. (2013) Interleukin 1 beta promoter polymorphism is associated with keratoconus in a Japanese population. Mol Vis 19 : 845–851.
55. KimSH, MokJW, KimHS, JooCK (2008) Association of -31T>C and -511 C>T polymorphisms in the interleukin 1 beta (IL1B) promoter in Korean keratoconus patients. Mol Vis 14 : 2109–2116.
56. RechcinskiT, GrebowskaA, KurpesaM, SztybrychM, PerugaJZ, et al. (2009) Interleukin-1b and interleukin-1 receptor inhibitor gene cluster polymorphisms in patients with coronary artery disease after percutaneous angioplasty or coronary artery bypass grafting. Kardiol Pol 67 : 601–610.
57. LaiosaCV, StadtfeldM, XieH, de Andres-AguayoL, GrafT (2006) Reprogramming of committed T cell progenitors to macrophages and dendritic cells by C/EBP alpha and PU.1 transcription factors. Immunity 25 : 731–744.
58. MaoK, ChenS, ChenM, MaY, WangY, et al. (2013) Nitric oxide suppresses NLRP3 inflammasome activation and protects against LPS-induced septic shock. Cell Res 23 : 201–212.
59. GallegosAM, van HeijstJW, SamsteinM, SuX, PamerEG, et al. (2011) A gamma interferon independent mechanism of CD4 T cell mediated control of M. tuberculosis infection in vivo. PLoS Pathog 7: e1002052.
60. CaccamoN, GugginoG, JoostenSA, GelsominoG, Di CarloP, et al. (2010) Multifunctional CD4(+) T cells correlate with active Mycobacterium tuberculosis infection. Eur J Immunol 40 : 2211–2220.
61. JuradoJO, PasquinelliV, AlvarezIB, PenaD, RovettaAI, et al. (2012) IL-17 and IFN-gamma expression in lymphocytes from patients with active tuberculosis correlates with the severity of the disease. J Leukoc Biol 91 : 991–1002.
62. NandiB, BeharSM (2011) Regulation of neutrophils by interferon-gamma limits lung inflammation during tuberculosis infection. J Exp Med 208 : 2251–2262.
63. KellerC, HoffmannR, LangR, BrandauS, HermannC, et al. (2006) Genetically determined susceptibility to tuberculosis in mice causally involves accelerated and enhanced recruitment of granulocytes. Infect Immun 74 : 4295–4309.
64. EumSY, KongJH, HongMS, LeeYJ, KimJH, et al. (2010) Neutrophils are the predominant infected phagocytic cells in the airways of patients with active pulmonary TB. Chest 137 : 122–128.
65. HnizdoE, SinghT, ChurchyardG (2000) Chronic pulmonary function impairment caused by initial and recurrent pulmonary tuberculosis following treatment. Thorax 55 : 32–38.
66. PengR, YueJ, HanM, ZhaoY, LiuL, et al. (2013) The IL-17F sequence variant is associated with susceptibility to tuberculosis. Gene 515 : 229–232.
67. ChenX, ZhangM, LiaoM, GranerMW, WuC, et al. (2010) Reduced Th17 response in patients with tuberculosis correlates with IL-6R expression on CD4+ T Cells. Am J Respir Crit Care Med 181 : 734–742.
68. ChenX, ZhouB, LiM, DengQ, WuX, et al. (2007) CD4(+)CD25(+)FoxP3(+) regulatory T cells suppress Mycobacterium tuberculosis immunity in patients with active disease. Clin Immunol 123 : 50–59.
69. NovikovA, CardoneM, ThompsonR, ShenderovK, KirschmanKD, et al. (2011) Mycobacterium tuberculosis triggers host type I IFN signaling to regulate IL-1beta production in human macrophages. J Immunol 187 : 2540–2547.
70. TodaY, TsukadaJ, MisagoM, KominatoY, AuronPE, et al. (2002) Autocrine induction of the human pro-IL-1beta gene promoter by IL-1beta in monocytes. J Immunol 168 : 1984–1991.
71. YuLL, ChangK, LuLS, ZhaoD, HanJ, et al. (2013) Lentivirus-mediated RNA interference targeting the H19 gene inhibits cell proliferation and apoptosis in human choriocarcinoma cell line JAR. BMC Cell Biol 14 : 26.
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Identification of the Microsporidian as a New Target of the IFNγ-Inducible IRG Resistance SystemČlánek Human Cytomegalovirus Drives Epigenetic Imprinting of the Locus in NKG2C Natural Killer CellsČlánek APOBEC3D and APOBEC3F Potently Promote HIV-1 Diversification and Evolution in Humanized Mouse ModelČlánek Role of Non-conventional T Lymphocytes in Respiratory Infections: The Case of the Pneumococcus
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2014 Číslo 10- Stillova choroba: vzácné a závažné systémové onemocnění
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Jak souvisí postcovidový syndrom s poškozením mozku?
- Familiární středomořská horečka
-
Všechny články tohoto čísla
- Theory and Empiricism in Virulence Evolution
- -Related Fungi and Reptiles: A Fatal Attraction
- Adaptive Prediction As a Strategy in Microbial Infections
- Antimicrobials, Stress and Mutagenesis
- A Novel Function of Human Pumilio Proteins in Cytoplasmic Sensing of Viral Infection
- Social Motility of African Trypanosomes Is a Property of a Distinct Life-Cycle Stage That Occurs Early in Tsetse Fly Transmission
- Autophagy Controls BCG-Induced Trained Immunity and the Response to Intravesical BCG Therapy for Bladder Cancer
- Identification of the Microsporidian as a New Target of the IFNγ-Inducible IRG Resistance System
- mRNA Structural Constraints on EBNA1 Synthesis Impact on Antigen Presentation and Early Priming of CD8 T Cells
- Infection Causes Distinct Epigenetic DNA Methylation Changes in Host Macrophages
- Neutrophil Crawling in Capillaries; A Novel Immune Response to
- Live Attenuated Vaccine Protects against Pulmonary Challenge in Rats and Non-human Primates
- The ESAT-6 Protein of Interacts with Beta-2-Microglobulin (β2M) Affecting Antigen Presentation Function of Macrophage
- Characterization of Uncultivable Bat Influenza Virus Using a Replicative Synthetic Virus
- HIV Acquisition Is Associated with Increased Antimicrobial Peptides and Reduced HIV Neutralizing IgA in the Foreskin Prepuce of Uncircumcised Men
- Uses a Unique Ligand-Binding Mode for Trapping Opines and Acquiring A Competitive Advantage in the Niche Construction on Plant Host
- Involvement of a 1-Cys Peroxiredoxin in Bacterial Virulence
- Ethanol Stimulates WspR-Controlled Biofilm Formation as Part of a Cyclic Relationship Involving Phenazines
- Densovirus Is a Mutualistic Symbiont of a Global Crop Pest () and Protects against a Baculovirus and Bt Biopesticide
- Insights into Intestinal Colonization from Monitoring Fluorescently Labeled Bacteria
- Mycobacterial Antigen Driven Activation of CD14CD16 Monocytes Is a Predictor of Tuberculosis-Associated Immune Reconstitution Inflammatory Syndrome
- Lipoprotein LprG Binds Lipoarabinomannan and Determines Its Cell Envelope Localization to Control Phagolysosomal Fusion
- Dampens the DNA Damage Response
- MicroRNAs Suppress NB Domain Genes in Tomato That Confer Resistance to
- Novel Cyclic di-GMP Effectors of the YajQ Protein Family Control Bacterial Virulence
- Vaginal Challenge with an SIV-Based Dual Reporter System Reveals That Infection Can Occur throughout the Upper and Lower Female Reproductive Tract
- Detecting Differential Transmissibilities That Affect the Size of Self-Limited Outbreaks
- One Small Step for a Yeast - Microevolution within Macrophages Renders Hypervirulent Due to a Single Point Mutation
- Expression Profiling during Arabidopsis/Downy Mildew Interaction Reveals a Highly-Expressed Effector That Attenuates Responses to Salicylic Acid
- Human Cytomegalovirus Drives Epigenetic Imprinting of the Locus in NKG2C Natural Killer Cells
- Interaction with Tsg101 Is Necessary for the Efficient Transport and Release of Nucleocapsids in Marburg Virus-Infected Cells
- The N-Terminus of Murine Leukaemia Virus p12 Protein Is Required for Mature Core Stability
- Sterol Biosynthesis Is Required for Heat Resistance but Not Extracellular Survival in
- Allele-Specific Induction of IL-1β Expression by C/EBPβ and PU.1 Contributes to Increased Tuberculosis Susceptibility
- Host Cofactors and Pharmacologic Ligands Share an Essential Interface in HIV-1 Capsid That Is Lost upon Disassembly
- APOBEC3D and APOBEC3F Potently Promote HIV-1 Diversification and Evolution in Humanized Mouse Model
- Structural Basis for the Recognition of Human Cytomegalovirus Glycoprotein B by a Neutralizing Human Antibody
- Systematic Analysis of ZnCys Transcription Factors Required for Development and Pathogenicity by High-Throughput Gene Knockout in the Rice Blast Fungus
- Epstein-Barr Virus Nuclear Antigen 3A Promotes Cellular Proliferation by Repression of the Cyclin-Dependent Kinase Inhibitor p21WAF1/CIP1
- The Host Protein Calprotectin Modulates the Type IV Secretion System via Zinc Sequestration
- Cyclophilin A Associates with Enterovirus-71 Virus Capsid and Plays an Essential Role in Viral Infection as an Uncoating Regulator
- A Novel Alpha Kinase EhAK1 Phosphorylates Actin and Regulates Phagocytosis in
- The pH-Responsive PacC Transcription Factor of Governs Epithelial Entry and Tissue Invasion during Pulmonary Aspergillosis
- Sensing of Immature Particles Produced by Dengue Virus Infected Cells Induces an Antiviral Response by Plasmacytoid Dendritic Cells
- Co-opted Oxysterol-Binding ORP and VAP Proteins Channel Sterols to RNA Virus Replication Sites via Membrane Contact Sites
- Characteristics of Memory B Cells Elicited by a Highly Efficacious HPV Vaccine in Subjects with No Pre-existing Immunity
- HPV16-E7 Expression in Squamous Epithelium Creates a Local Immune Suppressive Environment via CCL2- and CCL5- Mediated Recruitment of Mast Cells
- Dengue Viruses Are Enhanced by Distinct Populations of Serotype Cross-Reactive Antibodies in Human Immune Sera
- CD4 Depletion in SIV-Infected Macaques Results in Macrophage and Microglia Infection with Rapid Turnover of Infected Cells
- A Sialic Acid Binding Site in a Human Picornavirus
- Contact Heterogeneity, Rather Than Transmission Efficiency, Limits the Emergence and Spread of Canine Influenza Virus
- Myosins VIII and XI Play Distinct Roles in Reproduction and Transport of
- HTLV-1 Tax Stabilizes MCL-1 via TRAF6-Dependent K63-Linked Polyubiquitination to Promote Cell Survival and Transformation
- Species Complex: Ecology, Phylogeny, Sexual Reproduction, and Virulence
- A Critical Role for IL-17RB Signaling in HTLV-1 Tax-Induced NF-κB Activation and T-Cell Transformation
- Exosomes from Hepatitis C Infected Patients Transmit HCV Infection and Contain Replication Competent Viral RNA in Complex with Ago2-miR122-HSP90
- Role of Non-conventional T Lymphocytes in Respiratory Infections: The Case of the Pneumococcus
- Kaposi's Sarcoma-Associated Herpesvirus Induces Nrf2 during Infection of Endothelial Cells to Create a Microenvironment Conducive to Infection
- A Relay Network of Extracellular Heme-Binding Proteins Drives Iron Acquisition from Hemoglobin
- Glutamate Secretion and Metabotropic Glutamate Receptor 1 Expression during Kaposi's Sarcoma-Associated Herpesvirus Infection Promotes Cell Proliferation
- Reduces Malaria and Dengue Infection in Vector Mosquitoes and Has Entomopathogenic and Anti-pathogen Activities
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Novel Cyclic di-GMP Effectors of the YajQ Protein Family Control Bacterial Virulence
- MicroRNAs Suppress NB Domain Genes in Tomato That Confer Resistance to
- The ESAT-6 Protein of Interacts with Beta-2-Microglobulin (β2M) Affecting Antigen Presentation Function of Macrophage
- Characterization of Uncultivable Bat Influenza Virus Using a Replicative Synthetic Virus
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



