-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
A Quorum Sensing Regulated Small Volatile Molecule Reduces Acute Virulence and Promotes Chronic Infection Phenotypes
A significant number of environmental microorganisms can cause serious, even fatal, acute and chronic infections in humans. The severity and outcome of each type of infection depends on the expression of specific bacterial phenotypes controlled by complex regulatory networks that sense and respond to the host environment. Although bacterial signals that contribute to a successful acute infection have been identified in a number of pathogens, the signals that mediate the onset and establishment of chronic infections have yet to be discovered. We identified a volatile, low molecular weight molecule, 2-amino acetophenone (2-AA), produced by the opportunistic human pathogen Pseudomonas aeruginosa that reduces bacterial virulence in vivo in flies and in an acute mouse infection model. 2-AA modulates the activity of the virulence regulator MvfR (multiple virulence factor regulator) via a negative feedback loop and it promotes the emergence of P. aeruginosa phenotypes that likely promote chronic lung infections, including accumulation of lasR mutants, long-term survival at stationary phase, and persistence in a Drosophila infection model. We report for the first time the existence of a quorum sensing (QS) regulated volatile molecule that induces bistability phenotype by stochastically silencing acute virulence functions in P. aeruginosa. We propose that 2-AA mediates changes in a subpopulation of cells that facilitate the exploitation of dynamic host environments and promote gene expression changes that favor chronic infections.
Published in the journal: . PLoS Pathog 7(8): e32767. doi:10.1371/journal.ppat.1002192
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1002192Summary
A significant number of environmental microorganisms can cause serious, even fatal, acute and chronic infections in humans. The severity and outcome of each type of infection depends on the expression of specific bacterial phenotypes controlled by complex regulatory networks that sense and respond to the host environment. Although bacterial signals that contribute to a successful acute infection have been identified in a number of pathogens, the signals that mediate the onset and establishment of chronic infections have yet to be discovered. We identified a volatile, low molecular weight molecule, 2-amino acetophenone (2-AA), produced by the opportunistic human pathogen Pseudomonas aeruginosa that reduces bacterial virulence in vivo in flies and in an acute mouse infection model. 2-AA modulates the activity of the virulence regulator MvfR (multiple virulence factor regulator) via a negative feedback loop and it promotes the emergence of P. aeruginosa phenotypes that likely promote chronic lung infections, including accumulation of lasR mutants, long-term survival at stationary phase, and persistence in a Drosophila infection model. We report for the first time the existence of a quorum sensing (QS) regulated volatile molecule that induces bistability phenotype by stochastically silencing acute virulence functions in P. aeruginosa. We propose that 2-AA mediates changes in a subpopulation of cells that facilitate the exploitation of dynamic host environments and promote gene expression changes that favor chronic infections.
Introduction
Bacteria excrete small molecules that act as specific chemical signals to positively regulate specialized processes [1], including the production of virulence factors important for pathogenic infection, host colonization, and interspecies microbial interactions [2]. Using interconnected multi-layered regulatory networks, such as quorum sensing (QS) networks, bacteria sense and respond to external and internal bacterial cell signals as well as environmental cues, thereby adapting to exploit target hosts. Adaptation and coordination of gene expression is particularly important for pathogenic microorganisms that need to colonize changing host environments since their ability to sense and respond to host environmental cues is crucial for their survival.
Pseudomonas aeruginosa is an opportunistic human pathogen that causes chronic and acute infections, and is a major agent of morbidity and mortality in cystic fibrosis (CF) patients. Establishment of chronic P. aeruginosa respiratory or wound infections requires a complex adaptive process that mediates essential physiological changes allowing bacterial cells to survive and persist in a dynamic host environment. Although insights into chronic infection pathways have been reported [3]–[7], the specific bacterial signals that may promote the transition and/or adaptation of known pathogens from an acute to a chronic infection remain unknown.
Many P. aeruginosa virulence factors associated with acute infections are controlled by QS [8]. This pathogen has an extensively studied complex QS-communication network that facilitates cross-talk between organisms and impacts many P. aeruginosa group-related behaviors including virulence [8]. There are at least three known QS systems in P. aeruginosa: two are dependent on the acyl-homoserine-lactone (AHL) QS transcription factors LasR and RhlR [9], and a third is dependent on the 4-hydroxy-2-alkylquinolines (HAQs) LysR-type transcription factor MvfR (multiple virulence factor regulator) [10]. MvfR is critical for P. aeruginosa acute infection through its control of genes involved in the production of secreted virulence factors and in iron assimilation [10]–[13]. This regulator controls HAQ signaling and its own activity by positively regulating the expression of genes in the pqsABCDE [14] and phnAB [10] operons. These operons encode enzymes that catalyze the biosynthesis of at least 59 distinct low molecular weight compounds, most of which are structurally related to HAQs. While two of the most abundant HAQs, [4-hydroxy-2-heptylquinoline (HHQ) and 3,4-dihydroxy-2-heptylquinoline (Pseudomonas Quinolone Signal-PQS)] [14]–[16], function in vivo as ligands that bind and activate MvfR [14], [17], the biological functions of other PqsABCD/PhnAB biosynthetic products remain elusive.
In this study, we show that one of these abundant MvfR-regulated non-HAQ low molecular weight molecules, 2-aminoacetophenone (2-AA), that is used to diagnose P. aeruginosa infections in humans [18], reduces acute virulence by negatively fine-tuning the transcription and synthesis of the MvfR ligand HHQ, and promotes changes that are critical for pathogen adaptation and important for chronic infection.
Results/Discussion
2-AA Synthesis Is Controlled by MvfR but Is Not Required for the Activation of Its Regulon
To determine the functions of the abundant MvfR-regulated small molecules, we first compared the liquid chromatography/mass spectrometry (LC/MS) total ion chromatograms of culture-free supernatants from highly pathogenic wild-type P. aeruginosa (PA14) cells versus those of isogenic mvfR mutant cells. As shown in Figure 1A, the mvfR versus PA14 supernatant lacked HHQ and PQS, as well as three other abundant low molecular weight compounds: the HAQ molecule 4-hydroxy-2 heptylquinoline N-oxide (HQNO) [19], 2,4-dihydroxyquinoline (DHQ) [20], and the non-HAQ molecule 2-AA. 2-AA is a relatively simple, non-HAQ volatile molecule responsible for the grape-like odor of P. aeruginosa cultures as well as burn wounds infected with P. aeruginosa [18], [21]. Along with DHQ and HQNO, 2-AA is also produced and excreted by the HAQs-producing bacterium Burkholderia thailandensis [23] (Figure S1). Because HHQ and PQS both induce pqsABCDE expression [14], we investigated whether these three additional abundant molecules also induce expression of these genes. Maximum levels of 2-AA in the cell supernatant vary from micromolar to millimolar range depending on the growth medium used [18], [22]. We used Luria-Bertani (LB) broth media in experiments examining the effects of exogenous 2-AA supplementation on pqs operon gene transcription. We chose to use LB broth because it supports lower levels of 2-AA production (37.5 µM = 5 µg/ml Figure 1B) than other media [18]. Using a pqsA-green fluorescence protein (GFP)(ASV)-transcriptional reporter fusion in a pqsA::H double mutant background that does not produce any of these molecules (Figure S2), we exogenously added to LB media the above molecules. HQNO only modestly induced pqsABCDE expression, while DHQ and 2-AA did not induce pqsABCDE expression (Figure 1C). This finding indicates that unlike HHQ and PQS, 2-AA and DHQ molecules may have biological roles other than activating pqs operon transcription.
Fig. 1. 2-AA synthesis is controlled by MvfR. 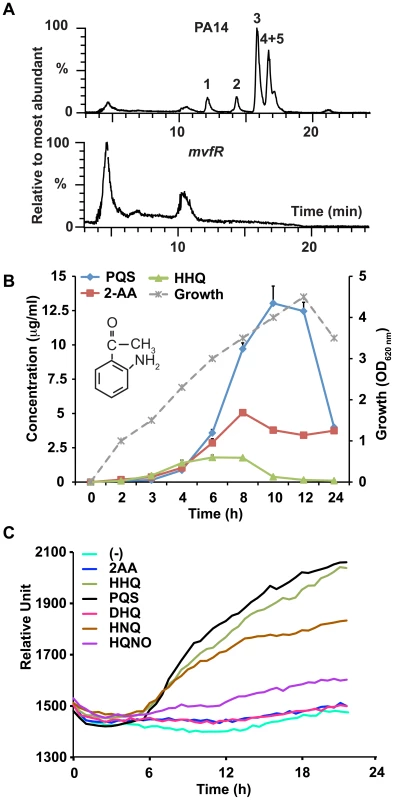
(A) LC/MS total ion chromatograms show the relative percentage of the most abundant small molecules in P. aeruginosa (PA14) (upper panel) and isogenic mvfR mutant (lower panel) supernatants after 9 h of growth. The most abundant molecules in PA14 cultures that were absent in mvfR mutant cultures were: DHQ (1); 2-AA (2); HQNO (3); and HHQ+PQS (4+5). The abundant peaks shown in mvfR are negligible compared to the amount of HHQ or PQS produced by PA14 since the percentage value on the Y axis is drawn with respect to the most abundant molecule detected. (B) Production kinetics of 2-AA in PA14 supernatants in LB. PA14 growth is shown as OD600 nm on the secondary Y axis versus time. The chemical structure of 2-AA is shown in the inset. (C) 2-AA is not a MvfR co-inducer. MvfR-dependent pqsA-GFP(ASV) expression in the pqsA::pqsH mutant with or without exogenous molecules (10 µg/ml), reported as relative fluorescence versus time, averaged for six replicates. 2-AA Down-regulates the Expression of MvfR-regulated Loci
The kinetics and dose-dependency effects of exogenously added 2-AA and DHQ on pqsA regulation were examined in greater detail using WT cells carrying the pqsA-GFP(ASV) transcriptional reporter fusion. Figure 2A shows that 2-AA, but not DHQ (Figure S3A), greatly reduced pqsA expression, in a dose-dependent manner, with 2-AA achieving the strongest inhibition at 200 µg/ml (1.5 mM). Bacterial growth was unaffected by either 2-AA or DHQ (Figure S3B and C). In light of these results, we focused our subsequent experimental efforts on 2-AA.
Fig. 2. 2-AA silences MvfR regulon in a subpopulation of cells and restricts HAQ mediated QS signaling relevant in acute infection. 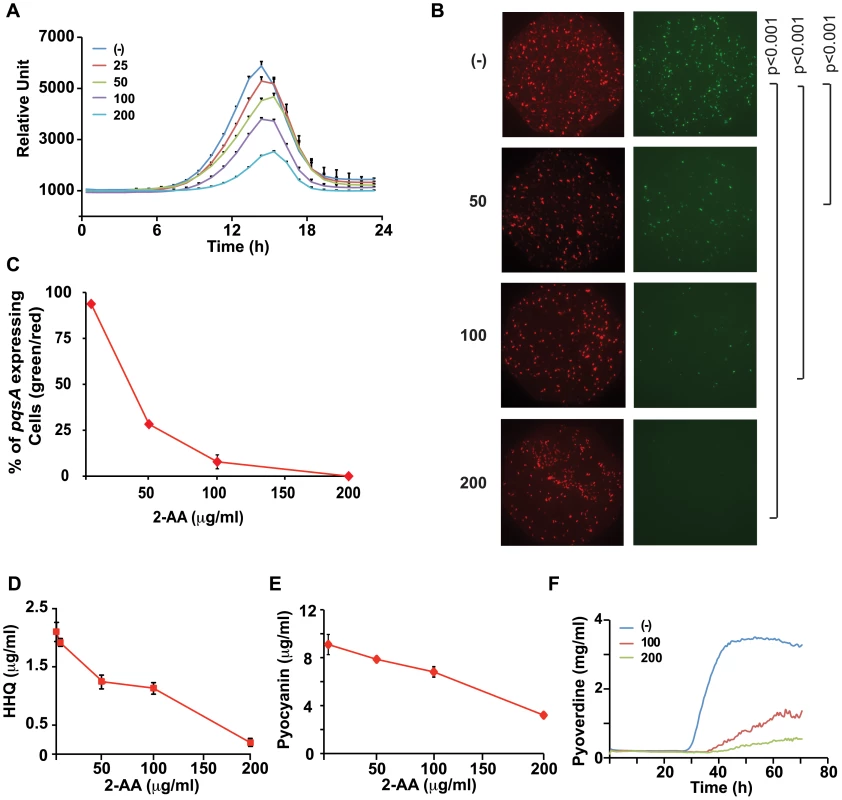
(A) Quantification of pqsA-GFP(ASV) expression in PA14 cells in response to increasing 2-AA concentration (µg/ml); the error bar represents the standard deviation of the 6 replicates. The difference in fluorescence intensity at the peak was statistically significant at all concentrations with p values <0.005. (B) Expression of pqsA-GFP(ASV) in response to 2-AA (µg/ml). Images of total number of cells in the optical field as assessed by membrane staining (red) and bacterial cells expressing pqsA-GFP(ASV)(green). (C) Quantification of pqsA expressing cells (percentage of green/red) in response to each 2-AA concentration. (D) Production of HHQ in supernatants collected from PA14 cells (OD 2.0) with exogenously added 2-AA at 0–200 µg/ml HHQ levels were quantified in triplicate by LC/MS. The error bars represent the standard deviation from triplicate samples. (E) Levels of pyocyanin in PA14 cultures grown (to OD 3.0) in LB and LB supplemented with varying (0–200 µg/ml) concentrations of 2-AA. Error bars represent standard deviation of the triplicate samples. These experiments were repeated at least three times with similar results. (F) Kinetics of pyoverdine production in the presence of 100 and 200 µg/ml of 2-AA. (-) indicates no exogenously added 2-AA. We visualized pqsA-GFP expression under a fluorescent microscope in the presence of a range of 2-AA concentrations. Surprisingly, but in accordance with the established dose-dependent effects of 2-AA, only a subpopulation of the cells showed a shut-down of pqsA-GFP activity in the presence of 50 µg/ml (0.375 mM) 2-AA (Figure 2B and C); a statistically significant number of cells had turned off the pqsA expression though some still fluoresced strongly. No fluorescing cells were seen at 200 µg/ml 2-AA (Figure 2B and C). Thus 2-AA appears to promote phenotypic heterogeneity in a genetically “homogenous” population and silence pqs operon expression in a fraction of the cells, suggesting that MvfR active and inactive cells co-exist to achieve a bistable phenotype [24]. The size of the 2-AA responsive population increased with higher 2-AA concentrations. The need for higher concentrations of 2-AA to produce significant detectable transcriptional changes may be due to the compound affecting only a subpopulation of cells.
2-AA inhibition of MvfR is likely mediated via negative feedback regulation given that MvfR controls 2-AA synthesis (Figure 1A). To corroborate our pqsA expression data, we measured levels of HHQ in 2-AA treated cells and found that HHQ production was also inhibited by 2-AA in a dose-dependent manner (Figure 2D). Consistent with the view that 2-AA down-regulates the MvfR regulon, we observed that 2-AA decreased production of pyocyanin (Figure 2E) and pyoverdine (Figure 2F), virulence factors whose synthesis depends on MvfR. A time course study of 2-AA production (Figure 1B) showed that 2-AA levels did not significantly decrease as occurs for the MvfR activators HHQ and PQS (Figure 1B), indicating that the production kinetics and stability of 2-AA are distinct from those of the MvfR activators. Together, this convergence of data strongly suggests that 2-AA has the novel biological activity of silencing the MvfR regulon.
2-AA Down-regulates pqsABCDE Operon Expression via MvfR, and by Targeting HAQ Biosynthesis Enzymes
To elucidate the mechanism by which 2-AA inhibits the MvfR regulon, we administered 2-AA together with HHQ to PA14 isogenic pqsA::pqsH double mutant cells, which do not produce any HAQs [14] or 2-AA (Figure S2) but have functional MvfR. The expression of the pqsA reporter in pqsA::pqsH cells requires activation of MvfR. While exogenous addition of HHQ to these cells induced pqsA reporter expression via activation of MvfR, co-addition of 2-AA, at 100 µg/ml (0.75 mM) and above, attenuated this expression in a dose-dependent manner, suggesting that 2-AA may negatively impact the MvfR-regulated operon pqsABCDE at the transcriptional level (Figure 3A) via MvfR. Unlike other anthranilic acid analogs, which act on PqsA activity and inhibit the synthesis of MvfR ligands [25], the apparent 2-AA inhibition described here can be regarded as independent of PqsA since the pqsA::pqsH mutant was used in these experiments. Moreover, 2-AA did not perturb MvfR protein levels or MvfR ligand stability (data not shown).
Fig. 3. Negative regulation of the MvfR regulon by 2-AA is a result of down-regulation of pqsABCDE expression and interference with MvfR activity via inhibition of HHQ biosynthesis. 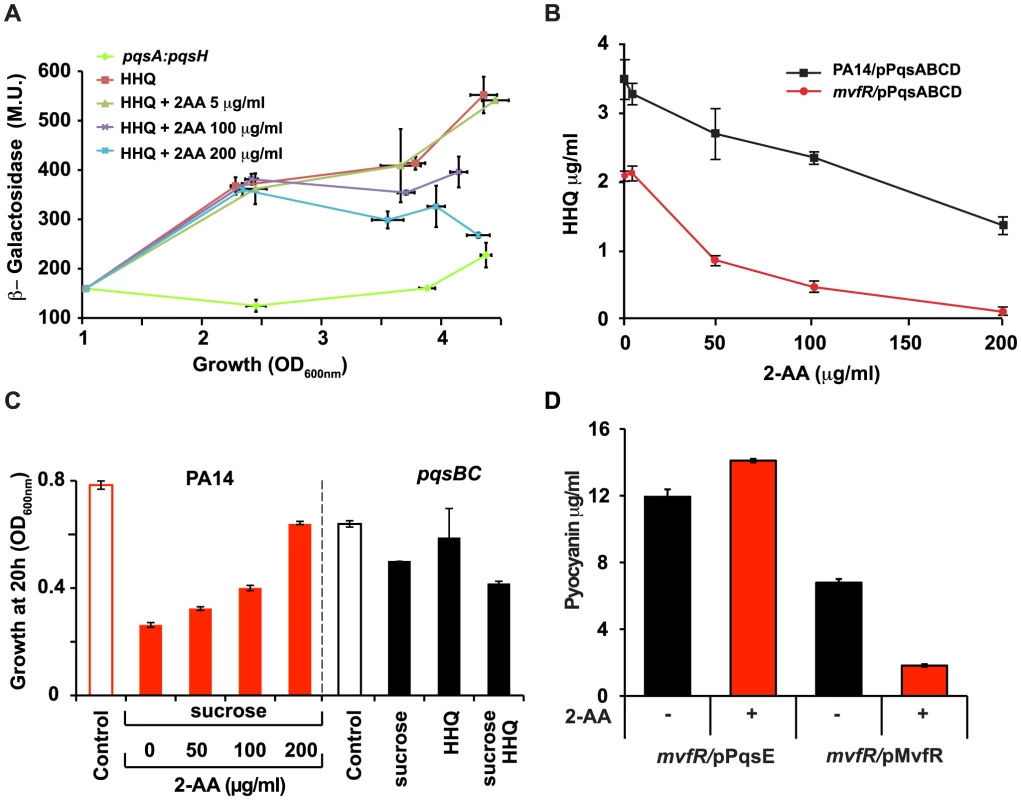
(A) pqsA promoter response was assessed by measuring pqsA-LacZ expression in the PA14 isogenic pqsA::pqsH double mutant in the presence of HHQ (10 µg/ml) or HHQ together with 2-AA at the indicated concentrations. β-galactosidase activity is given in Miller Units, and plotted against growth assessed at OD600 nm. The experiment was repeated three times with similar results. (B) Levels of HHQ in the supernatant (OD 2.0) of PA14 and mvfR mutant cells constitutively expressing pqsABCD on a plasmid. HHQ levels were quantified in triplicate by LC/MS. The error bars represent the standard deviation of triplicate samples. (C) Growth of PA14 and pqsB::C mutant cells carrying pqsA-SacB after 20 h of inoculation in the presence or absence of sucrose (10%), and/or with exogenously added various concentrations of 2-AA. The pqsA promoter of pqsA-SacB in the pqsB::C mutant was induced by exogenous addition of 10 µg/ml HHQ where indicated. (D) Concentration of pyocyanin in mvfR mutant cells (OD 3.0) over-expressing MvfR or PqsE under a constitutive promoter in the presence or absence of 2-AA (200 µg/ml). Error bars represent standard deviations of triplicate samples. These experiments were repeated at least three times with similar results. To assess whether down-regulation of MvfR activity by 2-AA is due to reduced ligand levels, we engineered mvfR mutant cells to synthesize HHQ independently of MvfR by constitutively expressing pqsABCD. These cells produced HHQ in the absence of 2-AA whereas HHQ production decreased in the presence of 2-AA in a dose-dependent manner (Figure 3B), indicating that inhibition of the MvfR regulon can also occur independently of MvfR. This post-transcriptional inhibition could be mediated through interference with ligand biosynthesis.
The pqsABBCDE operon gene pqsA is required for the synthesis of 2-AA and HHQ (Figure S2). In our effort to understand 2-AA biosynthesis, we created various mutants in the PQS operon. Surprisingly, the pqsB::C mutant produced 2-AA in the absence of HHQ (Figure S2), demonstrating that neither pqsB and pqsC is required for 2-AA synthesis. Addition of PQS to pqsB::C mutant cultures induced pqs operon transcription, resulting in WT levels of 2-AA (Figure S2). However, as expected the addition of PQS did not result in production of HHQ (data not shown).
We used pqsB::C mutant cells with an additional, more sensitive reporter system to further assess the effects of endogenous 2-AA produced by the pqsB::C mutant on transcription of the mvfR regulon and its biological relevance in the absence of HHQ, as well as to quantify 2-AA effects on the pqs operon. We fused the pqsA promoter to the Bacillus subtilis sacB gene that codes for the levansucrase product, which is toxic when cells are grown in the presence of sucrose. The sacB gene has previously been incorporated into allelic exchange vectors as a means of counter-selection [23]. As shown in Figures 3C and S4, PA14 cells with the pqsA-sacB fusion gene incorporated stably into their chromosome did not grow in the presence of sucrose, while the corresponding isogenic pqsB::C mutant cells did grow significantly, corroborating the findings that 2-AA suppresses pqsA promoter activation and indicating that this effect is stronger in the absence of HHQ. Bacterial cell proliferation occurred even following exogenous addition of HHQ, which promotes further induction of the pqs operon (Figure 3C and S4). Importantly, the endogenous level of 2-AA in the pqsB::C mutant cells in presence of the inducer, had a suppression efficacy comparable to 100 µg/ml of exogenously added 2-AA Figure 3C. These results are physiologically important and consistent with the putative role of 2-AA in down-regulation of MvfR regulon. These results also suggest there may be limited uptake of exogenously added 2-AA thereby requiring addition of higher concentrations of 2-AA to generate a physiological response by exogenously added 2-AA.
The last gene in the pqs operon, pqsE, is essential for pyocyanin production [16], [26]–[27], but dispensable for HAQ synthesis [12], [15]. Importantly, 2-AA did not reduce pyocyanin in an mvfR mutant that constitutively expressed pqsE, even though it efficiently inhibited pyocyanin production when MvfR was constitutively expressed (Figure 3D). Hence we can deduce that 2-AA negative regulation of the MvfR regulon is a result of the down-regulation of pqsABCDE operon expression and interference with MvfR activity upstream of PqsE, via inhibition of HHQ biosynthesis.
2-AA Treatment Reduces the Mortality of PA14-infected Flies and Mice
We have shown previously that P. aeruginosa pathogenesis can be studied in Drosophila melanogaster [28]–[30]. Drosophila shares striking similarities with mammals in terms of its overall physiology and innate immunity signal transduction pathway components [28], [31]. P. aeruginosa strain PA14 is highly virulent in flies, causing high mortality; meanwhile, mvfR mutants exhibit reduced virulence, causing reduced mortality [30]. Therefore, since 2-AA negatively regulates MvfR, we first tested whether it reduces P. aeruginosa virulence in Drosophila. As shown in the Figure 4A, flies co-injected with P. aeruginosa and 2-AA succumb to infection significantly later than flies injected with only P. aeruginosa.
Fig. 4. 2-AA reduces P. aeruginosa virulence. 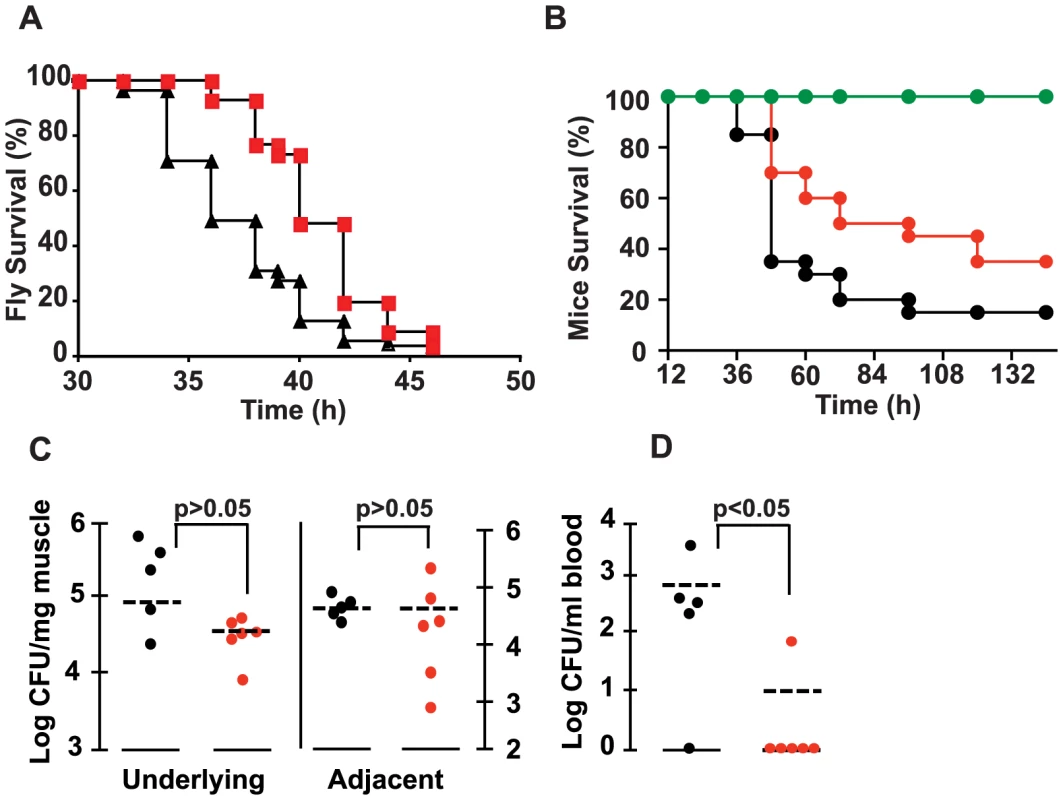
(A) Survival kinetics of flies injected with a PA14 bacterial suspension with (∼1.6 ng/fly) and without 2-AA. Data were averaged from two experiments with n(PA14) = 55 and n(PA14+2-AA) = 56. Significance of difference of survival rate was calculated using the log-rank test of the Kaplan Meier survival estimate (p = 0.0056). (B) Survival rates of mice infected with PA14 versus PA14 + 67 µg of 2-AA/mouse. The data correspond to the averages of two independent experiments, with n(PA14) = 20 and n(PA14+2-AA) = 20. Significance of difference of survival rates was calculated using the Kaplan-Meier method (p = 0.03), with a hazard ratio of 1.8932 (95% CI, 1.0664 to 6.0718). (C and D) Bacterial loads in the local and adjacent muscle and in blood were determined for control and 2-AA treated mice, at 20 h post-burn and infection. The statistical significance of the CFU/mg muscle tissue differences between the control and experimental mice were determined using the Mann-Whitney test for independent samples, with p = 0.43 for the difference in the underlying and adjacent muscle in response to 2-AA, and p = 0.045 for the difference in the blood in response to 2-AA. CFU data are presented as log 10. Black PA14, red PA14+2-AA, green 2-AA only. To confirm the in vivo efficacy of 2-AA in attenuating the virulence of PA14 cells and validate the above findings in a mammalian model of infection, we used the well-studied acute mouse burn and infection model [32], in which we have shown previously that mvfR mutant cells cause attenuated virulence [10], [25]. Indeed, as shown in Figure 4B, 2-AA also attenuated the virulence of PA14 cells in mice. Mice inoculated with PA14 and injected once with 2-AA survived significantly longer than control mice inoculated with PA14 alone. Importantly, this effect could not have been due to reduced PA14 proliferation since 2-AA did not affect the bacterial colony forming units (CFUs) in the muscle tissue underlying the burn injury and infection site (Figure 4C). Importantly, 2-AA greatly hampered the systemic spread of bacteria in the blood of the infected mice (Figure 4D), which is a significant problem in humans with P. aeruginosa infections. Silencing of MvfR regulon activity, and thus of acute virulence factor gene expression, is likely responsible in large part for the reduced systemic dissemination of PA14 cells and attenuated PA14 virulence observed.
2-AA Promotes the Emergence of P. aeruginosa Phenotypes that likely Promote Chronic Lung Infections
One may wonder why a pathogen such as P. aeruginosa would produce a molecule that decreases its own virulence. However, given that successful adaptation of an organism depends on its ability to regulate gene expression in response to its changing environment and thereby maximize its long-term survival, the existence of such a mechanism should not be surprising. There is ongoing debate regarding the role of QS in chronic infection due to inactivation of LasR in P. aeruginosa isolates from CF sputum, while a lack of QS has been proposed to facilitate adaptation during a chronic infection [33]. Since 2-AA down-regulates the expression of QS-related acute virulence functions, we questioned if the same molecule could promote other functions that support adaptation during chronic infections. We therefore examined a variety of phenotypes associated with chronic infection by P. aeruginosa.
A significant fraction of P. aeruginosa cells isolated from chronically infected CF patients accumulate multiple mutations in genes affecting acute virulence functions, including mucA, the regulator of alginate production, the QS regulator LasR, type III secretion system, multidrug efflux pumps, genes involved in motility, and DNA repair genes such as mutS [2], [34]–[38]. Mutations in the lasR gene are particularly notable, not only because they accumulate in bacteria colonizing the lungs of chronically infected CF patients, but also because they have been shown to promote long-term growth and survival of CF isolates in vitro [33]. Therefore, we first examined the frequency of occurrence of lasR mutations in populations of P. aeruginosa grown in the presence of increasing concentrations of 2-AA. We found that PA14 cells exposed to increasing 2-AA concentrations for 10 d accumulated significantly more lasR mutations than untreated cells (Figure 5A). Sequence analysis showed that these lasR mutant lines harbor simple deletions or single nucleotide non-synonymous mutations in the lasR gene that produce inactive LasR protein (Table 1). One of the lasR mutant lines also carried mutations in the intergenic region of the fleQ flagellar gene regulator, which is important for motility, and five clones carried mutations in the intergenic region of mexT, a regulator of multidrug efflux. Mutations in these genes lead to loss of flagellar motility and antibiotic resistance, respectively [39]–[40]. However, no mutations in the DNA repair gene mutS, in type III secretion system genes (exsA, pscQ and popD), or in the virulence factor vfR or rpoN, which are also associated with chronic infections [2], were identified in any of the lasR mutant lines sequenced. Additionally, 2-AA does not appear to be mutagenic, as it did not stimulate mutagenesis in a standard Ames test (data not shown). We did not identify any mutation in lasR within a day of incubation when silencing of mvfR was observed, suggesting that it's silencing is not due to the loss of lasR. Nevertheless, the ability of 2-AA to promote lasR mutations was particularly pronounced after 10 d of incubation in isogenic mutS cells (Figure 5A), which are defective in DNA repair function. In contrast to the 2-AA dependent non-linear accumulation of lasR mutants observed in the WT background, a more linear accumulation of lasR mutants in the mutS mutant background is observed (Figure 5A). These data suggest that once a DNA repair mechanism is compromised, as in mutS mutants commonly found in P. aeruginosa cultures from CF sputum [37]–[38], 2-AA effect is more prominent.
Fig. 5. 2-AA promotes phenotypes associated with pathogenic bacteria at chronic infection sites. 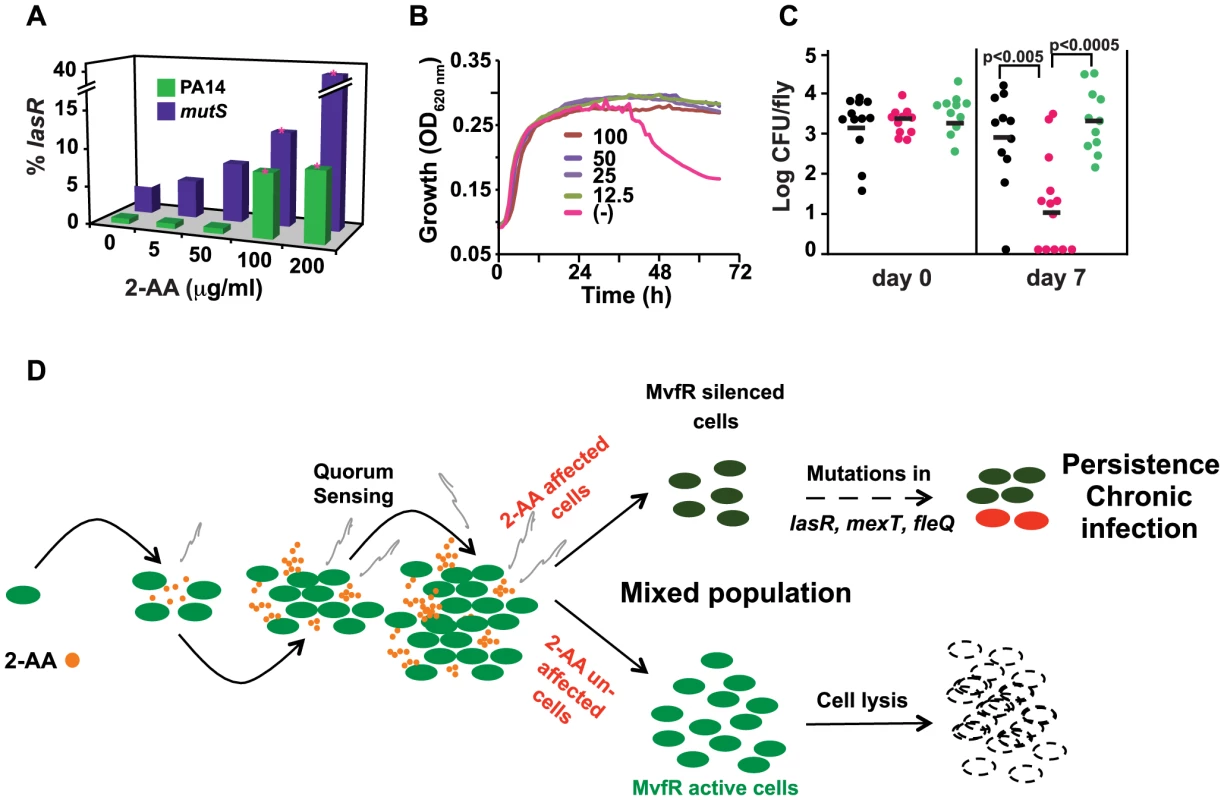
(A) 2-AA promotes increased accumulation of lasR mutations in PA14 and mutS cells in a concentration-dependent manner. Data are averages of two independent experiments, performed in triplicate. Significance of difference in number of lasR mutants was calculated using Student's t-test, assuming equal variance. Asterisks represent significant differences relative to the untreated control group. For both PA14 and mutS mutant cells, lasR mutant accumulation was significantly increased with 100 µg/ml and 200 µg/ml 2-AA (p's = 0.02 and 0.0001 for PA14, and p's = 0.05 and 0.01 for mutS, respectively). (B) 2-AA promotes prolonged stationary phase growth. Untreated PA14 cells began to lyse by 40 h, whereas PA14 cells treated with 2-AA at all tested concentrations did not. (C) 2-AA promotes persistent infection in flies. Bacterial CFUs per fly were assessed at 7 d post-P. aeruginosa feeding (6–9 flies per time point). Significant differences in CFUs after 7 d were seen for pqsA (pink) versus PA14 (black) or pqsB::C (green); Student's t-test p values are indicated in the figure. The data presented were combined from two independent experiments. (D) A model for 2-AA function. As the volatile (squiggly grey lines) 2-AA accumulates overtime in a microenvironment (i.e. biofilms, wounds or CF lungs) it creates a heterogeneous population of cells that consists of WT and physiologically and genetically different cells. The unaltered WT cells undergo lysis. On the other hand, the subpopulation of cells with silenced MvfR can continue to accumulate mutations that are beneficial for adaption to chronic infection. Tab. 1. List of clones bearing lasR, mexT, and fleQ mutations. 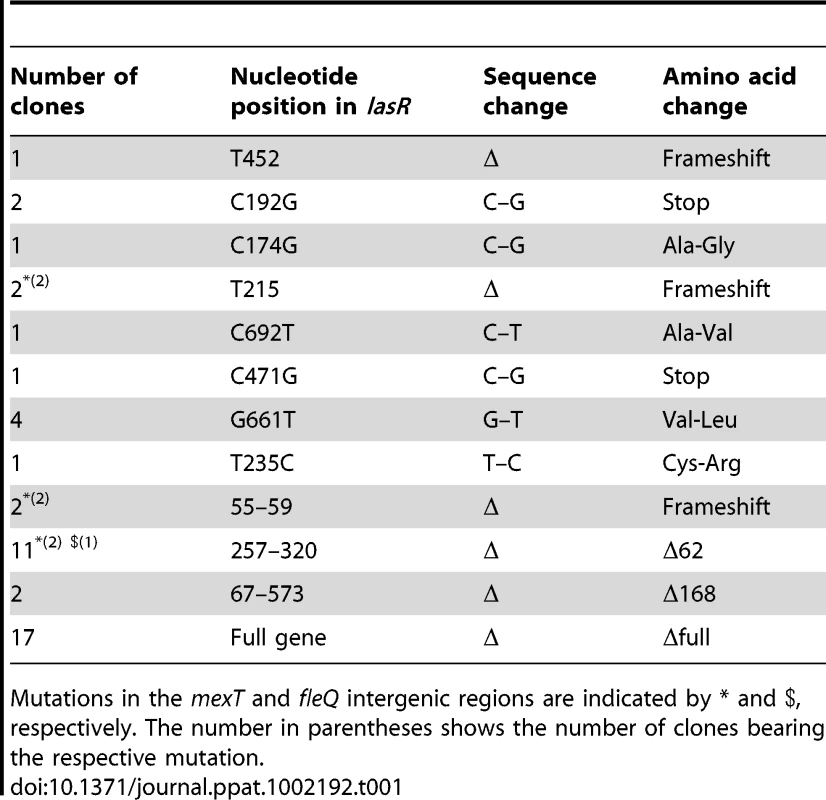
Mutations in the mexT and fleQ intergenic regions are indicated by * and $, respectively. The number in parentheses shows the number of clones bearing the respective mutation. Although the mechanisms and advantages of lasR mutations are not clearly understood, it has been proposed that lasR mutation may provide a growth advantage [33]. We further tested the effects of 2-AA on long-term survival of P. aeruginosa. When grown under the condition of limited iron and aeration, PA14 cells reach stationary phase within 10–12 h and undergo significant lysis after 40 h (Figure 5B). However, addition of 2-AA inhibited entry into the lytic phase and the treated cells remained in stationary phase, promoting bacterial cell long-term survival (Figure 5B). To investigate whether 2-AA can also promote long-term survival in vivo, we developed a non-vertebrate, whole animal persistence infection assay in Drosophila melanogaster. Using the fly-P. aeruginosa feeding assay [28], flies were fed for 2 d with P. aeruginosa WT (PA14), 2-AA non-producer PA14 isogenic mutant strain pqsA, and the 2-AA producer strain pqsB::C. Flies were then transferred onto bacteria-free food, and the CFUs were quantified at 7 d post-feeding. Bacterial load decreased over time in the pqsA-infected flies, while flies infected with the 2-AA producing strains PA14 and pqsB::C sustained ∼3 log greater CFUs (Figure 5C), indicating that 2-AA helps bacteria to survive and persist, and thus promotes better fitness in a dynamic host environment. Due to the longer incubation duration, we cannot rule out the possibility that the persistence seen here may be due to lasR mutation induced by 2-AA. Together with the above in vitro studies, these in vivo results corroborate the view that 2-AA plays a key role in switching cells to a phase that promotes concomitant adaptations that enable P. aeruginosa to persist in a chronic infection.
Synthesis of HAQs Analogs of Burkholderia are also Inhibited by 2-AA
Production of the 2-AA has been reported in plants, invertebrate animals, and vertebrate animals, including humans [41]. Moreover, several plant and human eubacterial pathogens, including Pseudomonas, Bordetella, Burkholderia, Ralstonia, Streptomyces, and Mycobacteria, as well as the Archeae Sulfolobus, encode putative PqsA and MvfR homologues, and in some cases, pqsBCD-related loci [42]. As such, these species might also produce 2-AA and use this molecule to down-regulate their respective virulence functions. Indeed, Burkholderia thailandensis, highly related and often used as a non-pathogenic surrogate for the level 3 pathogen B. pseudomallei, was confirmed to produce 2-AA (Figure S1). We assessed whether this 2-AA could inhibit HAQ biosynthesis in B. thailandensis, and found that exogenous 2-AA inhibited production of the HHQ and HNQ methylated analogs HMHQ and HMNQ (Figure S5A and B) [23], suggesting that 2-AA may perform analogous functions in B. thailandensis as observed in P. aeruginosa. 2-AA producing species may have a selective advantage in mixed microbial communities by adversely influencing the expression or activities of fitness traits of their neighbors.
Here we provide evidence that 2-AA, a small volatile molecule produced by P. aeruginosa (and B. thailandensis), is a QS regulated molecule and an important modulator of acute and chronic virulence functions. Although much work has focused on how QS cell-cell signaling leads to the activation of a plethora of virulence factors, little is known about the silencing of these factors. Degradation of AHL by an AHL acylase [43] and interference of LasR and RhlR binding to their promoter by the transcription factors QscR [44] and RsaL [45] have been shown to down-regulate AHL mediated QS. We recently demonstrated the existence of an interplay between QS systems, their components, and environmental factors that negatively impact HAQs [27]. However, no signaling molecule has been identified to date that negatively regulates acute virulence QS related functions or promotes bacterial adaptation and long-term persistence. Furthermore, no volatile compounds have been reported to be involved in QS regulation and virulence. Our results demonstrate that the low molecular weight molecule 2-AA silences the MvfR regulon via a negative feedback mechanism, most likely via repressed synthesis of MvfR ligands transcriptionally and post-transcriptionally, which restricts HAQ-mediated QS signaling and thus acute virulence functions, thereby enabling chronic infection. We propose the idea that there may be QS regulated aerial communication among bacteria, as 2-AA is a QS-regulated molecule and a volatile signal [46]. Such aerial communication may occur and be critical in a microenvironment such as the one found in biofilms and in infected wounds or CF lungs. It is important to note that suboptimal inhibitory concentration of 2-AA affects only a subpopulation of cells, suggesting that MvfR active and inactive cells co-exist causing a bistable phenotype. Similar bi/multistability phenotypes have been described as naturally occurring for several well known systems regulated by feedback mechanism, such as regulation of the Lac operon in E. coli, mucoidy in P. aeruginosa, lysogeny of the lambda phage, and transformation competency and sporulation in B. subtilis [24], [47]. In our model (Figure 5D), we propose that 2-AA inhibits MvfR via feedback regulation and promotes longer cell survival in vitro and in vivo in flies, presumably by interfering with cell lysis and inducing mutations in lasR. Additionally, the accumulation of lasR mutations that allow adaptation to low oxygen conditions [48] suggest that this mechanism contributes to overall pathogen fitness in chronic infections. Whether, lasR mutations arise in WT cells, MvfR silenced cells, and/or metabolically altered cells, remains to be determined. In corroboration, the Drosophila persistence assay showed that 2-AA promotes bacterial long-term survival and as such fitness.
Assessment of the potential role of 2-AA in the establishment and persistence of chronic infections has been hindered by a lack of clinically relevant chronic infection models. Nevertheless, the Drosophila persistence assay, using 2-AA producing and non-producing isogenic strains, permits us to at least determine the impact of this small molecule in a dynamic host environment that shares many immune functions with the human. Our identification of a P. aeruginosa QS regulated volatile molecule that limits virulence and invasive functions associated with acute infections while promoting phenotypic and genetic changes associated with chronic infection elucidates novel avenues for combating bacterial adaptation and survival in the chronic infection environment. These results also suggest that interfering with the MvfR pathway could prevent both acute and chronic infections and that 2-AA synthesis provides a potential target for the development of new anti-virulence drugs in the combatance of P. aeruginosa, and possibly of other pathogenic bacteria such as Burkholderia. This study uncovers insights that paradigmatically pave the way for the search for 2-AA-like volatile small molecules that promote pathogen adaptation and establishment of chronic infections caused by foreboding human pathogens.
Materials and Methods
Ethics Statement
This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved by the Committee on the Ethics of Animal Experiments of the Massachusetts General Hospital (Permit Number: 2006N000093/2). All Procedures were performed under sodium pentobarbital anesthesia, and all efforts were made to minimize suffering.
Bacterial Strains and Growth Conditions
A P. aeruginosa strain known as RifR human clinical isolate UCBPP-PA14 (PA14) was used in the present experiments [11]. All of the PA14 mutants described in this paper are isogenic to UCBPP-PA14. The bacteria were grown at 37°C on LB broth or on plates of LB agar containing appropriate antibiotics unless otherwise indicated. The overnight PA14 cultures were grown in LB and diluted the following day in fresh media with or without 2-AA. Bacterial growth kinetics was determined by taking OD600 nm measurements.
LC/MS
Quantification of 2-AA and HAQs in bacterial culture supernatants was performed as described previously [14], [49]. The HAQs were separated on a C18 reverse-phase column connected to a triple quadrupole mass spectrometer, using a water/acetonitrile gradient [49]. Positive electrospray in MRM mode with 2×10−3 mTorr argon and 30 V as the collision gas and energy was employed to quantify 2-AA and HAQs, using the following ion transitions: 2-AA 136>91, HHQ 244>159, HHQ-D4 248>163, PQS 260>175, and PQS-D4 264>179. To quantify HAQs, Pseudomonas PA14 cells were grown in LB supplemented with different concentrations of 2-AA (Sigma, USA), with untreated LB being used as a negative control. The culture supernatant was collected at different stages of growth mixed with an equal volume of methanol, and the compounds were analyzed by LC/MS. All samples were analyzed in triplicate.
Bacterial Growth Curves
Bacterial cells were diluted 1/100 from overnight cultures and grown in six replicates in 20 µl in a 96-well plate (Corning, Inc., Corning, NY). Bacterial growth was measured as a function of optical density at OD600 nm using Tecan F200 automated plate reader (Infinite F200, Tecan Group Ltd, Männedorf, Switzerland).
For the pqsA-sucB assay, the bacteria were diluted from overnight cultures to OD600 nm 0.1 in NaCl-free LB with tetracycline (50 µg/ml) and grown (Sigma, USA), with or without 10% sucrose (200 µl). Growth was measured in triplicate every 30 min for up to 25 h, using a Sunrise plate reader (Tecan Group Ltd, Männedorf, Switzerland).
For lysis experiments, the LB medium was treated with Chelex 100 resin 100–200 mesh, sodium glutamate (50 g/l)(BioRad, Hercules, CA) for 1 h followed by filtration with 0.2 µm filters (Corning, NY). Bacterial cells were diluted 1/100 in presence of various concentrations of 2AA. Triplicates (200 µl volumes) were inoculated in 96 wells plates. The plates were incubated and growth was recorded every 30 min for 3 d using a Sunrise plate reader (Tecan Group Ltd, Männedorf, Switzerland).
pqsA-GFP(ASV) Fluorescence Measurement and pqsA-LacZ, ß-galactosidase Assay
PA14 or pqsA::pqsH cells carrying pAC37 plasmids with pqsA promoter fused to short-lived GFP {pqsA-GFP(ASV)} [50] were grown overnight in LB supplemented with gentamycin (50 µg/ml). The cultures were diluted in the morning with the test compound at desired concentrations and aliquoted (200 µl) into 6 replicates in a 96-well assay plate (Corning, Inc. Corning, NY). Green fluorescent protein (GFP) fluorescence (excitation at 485 nm, emission at 535 nm) and OD600 nm for growth was measured every 30 min using a Tecan F200 automated plate reader (Infinite F200, Tecan Group Ltd, Männedorf, Switzerland). Values presented were above the background fluorescence from the empty strain. For measurement of ß-galactosidase activity, pqsA::pqsH cells harboring pGX5, which carries the pqsA-lacZ reporter gene [51], were diluted to OD600 nm = 0.05; and OD600 nm and ß-galactosidase activity (expressed as Miller Units) were measured at the indicated optical densities. Assays were performed in triplicate. All of the experiments were repeated at least three times.
Construction of pqsB::C
The mutant was constructed as described by Lesic et al [52] such that most of the pqsB and pqsC genes were replaced with a Kanamycin resistance marker. The deletion was made from nt 151 in pqsB to nt 456 in pqsC (within the coding regions), leaving 150 bp of the 5′ end of the pqsB coding region and 149 bp of the 3′ end of the pqsC coding region. The mutants were selected on LB plates containing Kanamycin (200 µg/ml).
Visualization of 2-AA Affected Cells
PA14 cells harboring the plasmid pAC37, which contained the pqsA-GFP(ASV) reporter fusion, were grown to mid-logarithmic phase in the presence of various concentrations of 2-AA. The cells were washed in phosphate buffered saline (PBS) and their membranes were stained with FM-64 (Invitrogen) according to the manufacturer's instructions. Five-microliter aliquots of bacterial cells were spotted onto slides and covered with Poly-L-lysine coated coverslips (Sigma Aldrich, US). The bacteria were visualized using an Eclipse E800 (Nikon) microscope with FM-64-stained membranes visualized as red and pqsA-GFP visualized as green. The pictures were processed using Spot V4.0.9 (Diagnostic Instruments) software.
Plasmid Construction
To constitutively express pqsABCD, a genomic fragment containing pqsABCD was amplified by PCR using pqsABamHI 5′ CATGGATCCAACGTTCTGTCATGTCCACG3′ and PqsDPst1 5′CGACTGCAGTCAACATGGCCGGTTCAC3′ primers from an H44 cosmid [53] as template. The BamH1 and Pst1 digested PCR product was ligated to a pDN18 vector digested with BamH1 and Pst1. The construct was introduced into E. coli Top10 (Invitrogen) cells and P. aeruginosa mvfR mutant cells by electroporation.
To construct pqsA-sucB reporter fusion, the pqsA promoter was amplified using the primers 5′GACTAGTCGAGCAAGGGTTGTAACGGTTTTTG3′ and 5′GAAGATCTGACAGAACGTTCCCTCTTCAGCGA3′ and the PA14 chromosome was used as template. The sacB gene was amplified using the primer pairs 5′GAAGATCTATGAACATCAAAAAGTTTGCA3′ and 5′AAACTGCAGGTTGATAAGAAATAAAAGAAAATGCC3′ from pKOBEG-sacB plasmid [54]. PqsA promoter (PpqsA) was then digested with SpeI/BglII and the fragment containing sacB was digested with BglII/PstI. The two fragments were ligated to the CTX (TetR) plasmid digested with SpeI/BglII. The ligated vector was eletroporated into E. coli SM10 lambda pir and used to integrate the CTX-PpqsA-sacB to PA14 chromosome. Rif and TetR plates were used to select for the PA14 CTX-PpqsA-sacB clones and the presence of CTX-PpqsA-sacB in the PA14 chromosome was further confirmed by PCR.
Pyocyanin Quantification
Overnight PA14 cultures were diluted to OD600 nm = 0.05 in 5 ml LB or LB +200 µg/ml 2-AA. The bacteria were grown in triplicate at OD 3.0. Pyocyanin was extracted with chloroform from 5 ml cell culture supernatant and then extracted with an equal volume of HCl (0.2 N); optical density was measured at OD520 nm. The amount of pyocyanin was quantified by multiplying the OD520 nm value by 17.072 to obtain values in µg/ml [55].
Pyoverdine Quantification
PA14 bacterial cells (200 µl) were grown in 96-well plates in D-TSB medium with a range of 2-AA concentrations. The plates were incubated and pyoverdine production and growth were measured every 30 min by a Tecan F200 automated plate reader (Infinite F200, Tecan Group Ltd, Männedorf, Switzerland). Pyoverdine levels were measured using excitation at 400 nm and emission at 460 nm. The values were normalized to cell growth (OD600 nm). Pyoverdine concentrations were calculated using a calibration curve of fluorescence for a range of pyoverdine concentrations (Sigma Aldrich, USA).
Mortality Studies
Fly mortality
Fly maintenance and the infection assays were performed as described previously [29]. For the survival assays, 50–60 flies were infected with a bacterial suspension of 5×107 cells/ml (grown in LB to OD600 nm = 3.0) in the presence (1.6 ng of 2-AA /fly) and absence of 2AA. The bacteria were mixed with 2-AA immediately before injection, and the flies were incubated at 21°C following infection. Fly mortality was assessed over time as described previously [29]. The experiment was performed twice with qualitatively similar results between the replicates. Bacterial inoculations were accomplished via pricking the middle dorsolateral thorax of the fly with a needle previously dipped into the bacterial suspension. The number of inoculated cells was ∼100 (2 nl) on average per fly. Statistical significance of survival kinetics was assessed by the Kaplan Meier method.
Mouse mortality
A thermal injury mouse model [32] was used, as described previously [11], to assess bacterial pathogenicity in 6-week-old CD1 mice (Charles River, Boston, MA, USA). The animal protocol was approved by the Massachusetts General Hospital Institutional Animal Care and Use Committee. Following administration of anesthesia, a full-thickness thermal burn injury involving 5–8% of the total body surface area was produced on the dermis of the shaved mouse abdomen, and an inoculum of 2.5×105 PA14 cells in 100 µl of saline or 2-AA solution (67 µg of 2-AA in 100 µl saline/mouse, mixture prepared immediately before injection) was injected intradermaly into the burn eschar. Mortality was recorded for 1 week. CFU counts were performed for 5–6 mice per group. Local muscle biopsies underneath the eschar, and from adjacent muscle on either side of the burn eschar, 20 h post-burn and infection were collected and homogenized in 1 ml of 1× PBS. The samples were diluted and plated on LB-agar plates containing rifampicin (50 mg/L). The systemic spread of the bacteria was assessed by counting the CFUs in the blood at 20 h post-burn and infection.
Fly Feeding Infection and Bacterial Persistence
Fly feeding on a P. aeruginosa-containing solution was performed as described previously [28]. Briefly, 5–7-day-old female Oregon-R flies (N = 26 per group) were fed for 1 d with a mixture of 0.05 ml of LB bacterial culture at OD600 nm = 1.8, 1 ml of 20% sucrose, and 4 ml of water. Thus, the feeding mix contained a final concentration of 1% LB, ∼3×107 bacteria/ml, and 4% sucrose. A sterile cotton ball was placed at the bottom of each fly vial and was impregnated with 5 ml of the feeding mix. The flies in each treatment group were sub-divided into three fly vials (13 flies per vial), sealed with a clean cotton ball, and incubated at 25°C. A day later, the flies were transferred to 50 ml plastic screw-cap tubes (10 flies per tube). The tubes were perforated with a heated 0.9-mm needle to enable aeration, while the caps were perforated with a heated 1.2-mm needle and covered with a 2.3 cm Whatman disc (Fisher scientific, USA). A 0.2-ml volume of 4% sucrose solution was dropped onto the Whatman disc and covered with parafilm to provide food for the flies with minimal contamination. New fly tubes were prepared daily and the flies were incubated at 25°C. CFU counts per fly were measured at the indicated time points by dipping 6–9 flies per time point in 95% ethanol for 3 s and letting them dry out on paper tissues (Kimwipes). Then each fly was ground up using a 1.5-ml tube pestle in 0.1 ml of LB, and 1 : 10 dilutions were plated on LB plates. Statistical analysis of the CFU data was performed using the Student's t-test.
Assessment of Mutations
PA14 and mutS mutant were grown overnight in LB. The saturated culture was diluted 1 : 10 in triplicate in 4% sucrose with varying concentrations of 2-AA, or without 2-AA. The diluted cultures were incubated statically at 25°C for 10 d. The cells were then diluted and plated to allow the numbers of total colonies to be counted the following day. After 2 d of plating, the lasR cells were identified by their excessive pyocyanin production. The percentage of lasR mutants in each sample was calculated and statistical significance was determined by Student's t-test, assuming equal variance. The lasR mutant colonies were further confirmed by sequencing of the complete lasR gene. In addition, the complete sequences of mexA, mexE, mexS, mexT, mexZ, wspF, fleQ, exsA, accC, vfR, popD, mutS, nfxB, rpoN, rhlR, pqsA, pqsb, pqsE, pqsH, and mvfR were also determined in these lasR mutant colonies.
Supporting Information
Zdroje
1. BasslerBLLosickR 2006 Bacterially speaking. Cell 125 237 246
2. SmithEEBuckleyDGWuZSaenphimmachakCHoffmanLR 2006 Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc Natl Acad Sci U S A 103 8487 8492
3. GoodmanALKulasekaraBRietschABoydDSmithRS 2004 A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev Cell 7 745 754
4. HassettDJKorfhagenTRIrvinRTSchurrMJSauerK 2010 Pseudomonas aeruginosa biofilm infections in cystic fibrosis: insights into pathogenic processes and treatment strategies. Expert Opin Ther Targets 14 117 130
5. HausslerS 2010 Multicellular signalling and growth of Pseudomonas aeruginosa. Int J Med Microbiol 300 544 548
6. MougousJDCuffMERaunserSShenAZhouM 2006 A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 312 1526 1530
7. SchobertMJahnD 2010 Anaerobic physiology of Pseudomonas aeruginosa in the cystic fibrosis lung. Int J Med Microbiol 300 549 556
8. WilliamsPCamaraM 2009 Quorum sensing and environmental adaptation in Pseudomonas aeruginosa: a tale of regulatory networks and multifunctional signal molecules. Curr Opin Microbiol 12 182 191
9. ShinerEKRumbaughKPWilliamsSC 2005 Inter-kingdom signaling: deciphering the language of acyl homoserine lactones. FEMS Microbiol Rev 29 935 947
10. CaoHKrishnanGGoumnerovBTsongalisJTompkinsR 2001 A quorum sensing-associated virulence gene of Pseudomonas aeruginosa encodes a LysR-like transcription regulator with a unique self-regulatory mechanism. Proc Natl Acad Sci U S A 98 14613 14618
11. RahmeLGStevensEJWolfortSFShaoJTompkinsRG 1995 Common virulence factors for bacterial pathogenicity in plants and animals. Science 268 1899 1902
12. GallagherLAMcKnightSLKuznetsovaMSPesciECManoilC 2002 Functions required for extracellular quinolone signaling by Pseudomonas aeruginosa. J Bacteriol 184 6472 6480
13. OglesbyAGFarrow3rdJMLeeJHTomarasAPGreenbergEP 2008 The influence of iron on Pseudomonas aeruginosa physiology: a regulatory link between iron and quorum sensing. J Biol Chem 283 15558 15567
14. XiaoGDezielEHeJLepineFLesicB 2006 MvfR, a key Pseudomonas aeruginosa pathogenicity LTTR-class regulatory protein, has dual ligands. Mol Microbiol 62 1689 1699
15. DezielELepineFMilotSHeJMindrinosMN 2004 Analysis of Pseudomonas aeruginosa 4-hydroxy-2-alkylquinolines (HAQs) reveals a role for 4-hydroxy-2-heptylquinoline in cell-to-cell communication. Proc Natl Acad Sci U S A 101 1339 1344
16. DezielEGopalanSTampakakiAPLepineFPadfieldKE 2005 The contribution of MvfR to Pseudomonas aeruginosa pathogenesis and quorum sensing circuitry regulation: multiple quorum sensing-regulated genes are modulated without affecting lasRI, rhlRI or the production of N-acyl-L-homoserine lactones. Mol Microbiol 55 998 1014
17. WadeDSCalfeeMWRochaERLingEAEngstromE 2005 Regulation of Pseudomonas quinolone signal synthesis in Pseudomonas aeruginosa. J Bacteriol 187 4372 4380
18. CoxCDParkerJ 1979 Use of 2-aminoacetophenone production in identification of Pseudomonas aeruginosa. J Clin Microbiol 9 479 484
19. LepineFMilotSDezielEHeJRahmeLG 2004 Electrospray/mass spectrometric identification and analysis of 4-hydroxy-2-alkylquinolines (HAQs) produced by Pseudomonas aeruginosa. J Am Soc Mass Spectrom 15 862 869
20. LepineFDekimpeVLesicBMilotSLesimpleA 2007 PqsA is required for the biosynthesis of 2,4-dihydroxyquinoline (DHQ), a newly identified metabolite produced by Pseudomonas aeruginosa and Burkholderia thailandensis. Biol Chem 388 839 845
21. LabowsJNMcGinleyKJWebsterGFLeydenJJ 1980 Headspace analysis of volatile metabolites of Pseudomonas aeruginosa and related species by gas chromatography-mass spectrometry. J Clin Microbiol 12 521 526
22. Scott-ThomasAJSyhreMPattemorePKEptonMLaingR 2010 2-Aminoacetophenone as a potential breath biomarker for Pseudomonas aeruginosa in the cystic fibrosis lung. BMC Pulm Med 10 56
23. VialLLepineFMilotSGroleauMCDekimpeV 2008 Burkholderia pseudomallei, B. thailandensis, and B. ambifaria produce 4-hydroxy-2-alkylquinoline analogues with a methyl group at the 3 position that is required for quorum-sensing regulation. J Bacteriol 190 5339 5352
24. ThattaiMvan OudenaardenA 2004 Stochastic gene expression in fluctuating environments. Genetics 167 523 530
25. LesicBLepineFDezielEZhangJZhangQ 2007 Inhibitors of pathogen intercellular signals as selective anti-infective compounds. PLoS Pathog 3 1229 1239
26. Farrow3rdJMSundZMEllisonMLWadeDSColemanJP 2008 PqsE functions independently of PqsR-Pseudomonas quinolone signal and enhances the rhl quorum-sensing system. J Bacteriol 190 7043 7051
27. HazanRHeJXiaoGDekimpeVApidianakisY 2010 Homeostatic Interplay between Bacterial Cell-Cell Signaling and Iron in Virulence. PLoS Pathog 6 e1000810
28. ApidianakisYPitsouliCPerrimonNRahmeL 2009 Synergy between bacterial infection and genetic predisposition in intestinal dysplasia. Proc Natl Acad Sci U S A
29. ApidianakisYRahmeLG 2009 Drosophila melanogaster as a model host for studying Pseudomonas aeruginosa infection. Nat Protoc 4 1285 1294
30. LauGWGoumnerovBCWalendziewiczCLHewitsonJXiaoW 2003 The Drosophila melanogaster toll pathway participates in resistance to infection by the gram-negative human pathogen Pseudomonas aeruginosa. Infect Immun 71 4059 4066
31. FerrandonDImlerJLHetruCHoffmannJA 2007 The Drosophila systemic immune response: sensing and signalling during bacterial and fungal infections. Nat Rev Immunol 7 862 874
32. StevensEJRyanCMFriedbergJSBarnhillRLYarmushML 1994 A quantitative model of invasive Pseudomonas infection in burn injury. J Burn Care Rehabil 15 232 235
33. D'ArgenioDAWuMHoffmanLRKulasekaraHDDezielE 2007 Growth phenotypes of Pseudomonas aeruginosa lasR mutants adapted to the airways of cystic fibrosis patients. Mol Microbiol 64 512 533
34. NguyenDSinghPK 2006 Evolving stealth: genetic adaptation of Pseudomonas aeruginosa during cystic fibrosis infections. Proc Natl Acad Sci U S A 103 8305 8306
35. MatheeKNarasimhanGValdesCQiuXMatewishJM 2008 Dynamics of Pseudomonas aeruginosa genome evolution. Proc Natl Acad Sci U S A 105 3100 3105
36. JainMRamirezDSeshadriRCullinaJFPowersCA 2004 Type III secretion phenotypes of Pseudomonas aeruginosa strains change during infection of individuals with cystic fibrosis. J Clin Microbiol 42 5229 5237
37. OliverACantonRCampoPBaqueroFBlazquezJ 2000 High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science 288 1251 1254
38. MaciaMDBlanquerDTogoresBSauledaJPerezJL 2005 Hypermutation is a key factor in development of multiple-antimicrobial resistance in Pseudomonas aeruginosa strains causing chronic lung infections. Antimicrob Agents Chemother 49 3382 3386
39. KohlerTEppSFCurtyLKPechereJC 1999 Characterization of MexT, the regulator of the MexE-MexF-OprN multidrug efflux system of Pseudomonas aeruginosa. J Bacteriol 181 6300 6305
40. JyotJDasguptaNRamphalR 2002 FleQ, the major flagellar gene regulator in Pseudomonas aeruginosa, binds to enhancer sites located either upstream or atypically downstream of the RpoN binding site. J Bacteriol 184 5251 5260
41. TulpMBohlinL 2005 Rediscovery of known natural compounds: nuisance or goldmine? Bioorg Med Chem 13 5274 5282
42. DiggleSPLumjiaktasePDipilatoFWinzerKKunakornM 2006 Functional genetic analysis reveals a 2-Alkyl-4-quinolone signaling system in the human pathogen Burkholderia pseudomallei and related bacteria. Chem Biol 13 701 710
43. SioCFOttenLGCoolRHDiggleSPBraunPG 2006 Quorum quenching by an N-acyl-homoserine lactone acylase from Pseudomonas aeruginosa PAO1. Infect Immun 74 1673 1682
44. ChuganiSAWhiteleyMLeeKMD'ArgenioDManoilC 2001 QscR, a modulator of quorum-sensing signal synthesis and virulence in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 98 2752 2757
45. de KievitTSeedPCNezezonJPassadorLIglewskiBH 1999 RsaL, a novel repressor of virulence gene expression in Pseudomonas aeruginosa. J Bacteriol 181 2175 2184
46. SchulzSDickschatJS 2007 Bacterial volatiles: the smell of small organisms. Nat Prod Rep 24 814 842
47. BalabanNQMerrinJChaitRKowalikLLeiblerS 2004 Bacterial persistence as a phenotypic switch. Science 305 1622 1625
48. HoffmanLRRichardsonARHoustonLSKulasekaraHDMartens-HabbenaW 2010 Nutrient availability as a mechanism for selection of antibiotic tolerant Pseudomonas aeruginosa within the CF airway. PLoS Pathog 6 e1000712
49. LepineFDezielEMilotSRahmeLG 2003 A stable isotope dilution assay for the quantification of the Pseudomonas quinolone signal in Pseudomonas aeruginosa cultures. Biochim Biophys Acta 1622 36 41
50. YangLBarkenKBSkindersoeMEChristensenABGivskovM 2007 Effects of iron on DNA release and biofilm development by Pseudomonas aeruginosa. Microbiology 153 1318 1328
51. XiaoGHeJRahmeLG 2006 Mutation analysis of the Pseudomonas aeruginosa mvfR and pqsABCDE gene promoters demonstrates complex quorum-sensing circuitry. Microbiology 152 1679 1686
52. LesicBRahmeLG 2008 Use of the lambda Red recombinase system to rapidly generate mutants in Pseudomonas aeruginosa. BMC Mol Biol 9 20
53. HeJBaldiniRLDezielESaucierMZhangQ 2004 The broad host range pathogen Pseudomonas aeruginosa strain PA14 carries two pathogenicity islands harboring plant and animal virulence genes. Proc Natl Acad Sci U S A 101 2530 2535
54. DerbiseALesicBDacheuxDGhigoJMCarnielE 2003 A rapid and simple method for inactivating chromosomal genes in Yersinia. FEMS Immunol Med Microbiol 38 113 116
55. KurachiM 1958 Studies of the biosynthesis of pyocyanine. II. Isolation and determination of pyocyanine. Bull Inst Chem Res 36 174 187
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Crystal Structure of Reovirus Attachment Protein σ1 in Complex with Sialylated OligosaccharidesČlánek A Protein Thermometer Controls Temperature-Dependent Transcription of Flagellar Motility Genes inČlánek Modulation of NKp30- and NKp46-Mediated Natural Killer Cell Responses by Poxviral Hemagglutinin
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2011 Číslo 8- Stillova choroba: vzácné a závažné systémové onemocnění
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Choroby jater v ordinaci praktického lékaře – význam jaterních testů
- Diagnostický algoritmus při podezření na syndrom periodické horečky
-
Všechny články tohoto čísla
- Phenotypic Screens, Chemical Genomics, and Antimalarial Lead Discovery
- Characterisation of Regulatory T Cells in Nasal Associated Lymphoid Tissue in Children: Relationships with Pneumococcal Colonization
- Crystal Structure of Reovirus Attachment Protein σ1 in Complex with Sialylated Oligosaccharides
- Absence of Cross-Presenting Cells in the Salivary Gland and Viral Immune Evasion Confine Cytomegalovirus Immune Control to Effector CD4 T Cells
- Transcriptomic Analysis of Host Immune and Cell Death Responses Associated with the Influenza A Virus PB1-F2 Protein
- A Quorum Sensing Regulated Small Volatile Molecule Reduces Acute Virulence and Promotes Chronic Infection Phenotypes
- Autocrine Regulation of Pulmonary Inflammation by Effector T-Cell Derived IL-10 during Infection with Respiratory Syncytial Virus
- A Protein Thermometer Controls Temperature-Dependent Transcription of Flagellar Motility Genes in
- Association of Human TLR1 and TLR6 Deficiency with Altered Immune Responses to BCG Vaccination in South African Infants
- Histo-Blood Group Antigens Act as Attachment Factors of Rabbit Hemorrhagic Disease Virus Infection in a Virus Strain-Dependent Manner
- MrkH, a Novel c-di-GMP-Dependent Transcriptional Activator, Controls Biofilm Formation by Regulating Type 3 Fimbriae Expression
- Beta-HPV 5 and 8 E6 Promote p300 Degradation by Blocking AKT/p300 Association
- Modulation of NKp30- and NKp46-Mediated Natural Killer Cell Responses by Poxviral Hemagglutinin
- Transportin 3 Promotes a Nuclear Maturation Step Required for Efficient HIV-1 Integration
- Coordination of KSHV Latent and Lytic Gene Control by CTCF-Cohesin Mediated Chromosome Conformation
- A Novel Persistence Associated EBV miRNA Expression Profile Is Disrupted in Neoplasia
- The Plant Pathogen pv. Is Genetically Monomorphic and under Strong Selection to Evade Tomato Immunity
- IL-10 Blocks the Development of Resistance to Re-Infection with
- Anti-Apoptotic Machinery Protects the Necrotrophic Fungus from Host-Induced Apoptotic-Like Cell Death during Plant Infection
- Crystal Structure of PrgI-SipD: Insight into a Secretion Competent State of the Type Three Secretion System Needle Tip and its Interaction with Host Ligands
- Evades Immune Recognition of Flagellin in Both Mammals and Plants
- Tumor Cell Marker PVRL4 (Nectin 4) Is an Epithelial Cell Receptor for Measles Virus
- Provides Insights into the Evolution of the Salmonellae
- B Cell Repertoire Analysis Identifies New Antigenic Domains on Glycoprotein B of Human Cytomegalovirus which Are Target of Neutralizing Antibodies
- Thy1 Nk Cells from Vaccinia Virus-Primed Mice Confer Protection against Vaccinia Virus Challenge in the Absence of Adaptive Lymphocytes
- The Cytokine Network of Acute HIV Infection: A Promising Target for Vaccines and Therapy to Reduce Viral Set-Point?
- Dendritic Cell Status Modulates the Outcome of HIV-Related B Cell Disease Progression
- Differential Contribution of PB1-F2 to the Virulence of Highly Pathogenic H5N1 Influenza A Virus in Mammalian and Avian Species
- A Communal Bacterial Adhesin Anchors Biofilm and Bystander Cells to Surfaces
- Two Group A Streptococcal Peptide Pheromones Act through Opposing Rgg Regulators to Control Biofilm Development
- Activation of HIV Transcription by the Viral Tat Protein Requires a Demethylation Step Mediated by Lysine-specific Demethylase 1 (LSD1/KDM1)
- Unique Evolution of the UPR Pathway with a Novel bZIP Transcription Factor, Hxl1, for Controlling Pathogenicity of
- Disruption of PML Nuclear Bodies Is Mediated by ORF61 SUMO-Interacting Motifs and Required for Varicella-Zoster Virus Pathogenesis in Skin
- Flagellar Motility Is Not Directly Required to Maintain Attachment to Surfaces
- Viral Infection Induces Expression of Novel Phased MicroRNAs from Conserved Cellular MicroRNA Precursors
- Functional Cure of SIVagm Infection in Rhesus Macaques Results in Complete Recovery of CD4 T Cells and Is Reverted by CD8 Cell Depletion
- Recruitment of the Major Vault Protein by InlK: A Strategy to Avoid Autophagy
- The Steroid Catabolic Pathway of the Intracellular Pathogen Is Important for Pathogenesis and a Target for Vaccine Development
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Tumor Cell Marker PVRL4 (Nectin 4) Is an Epithelial Cell Receptor for Measles Virus
- Two Group A Streptococcal Peptide Pheromones Act through Opposing Rgg Regulators to Control Biofilm Development
- Differential Contribution of PB1-F2 to the Virulence of Highly Pathogenic H5N1 Influenza A Virus in Mammalian and Avian Species
- Recruitment of the Major Vault Protein by InlK: A Strategy to Avoid Autophagy
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



