-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Formation of Complexes at Plasmodesmata for Potyvirus Intercellular Movement Is Mediated by the Viral Protein P3N-PIPO
Intercellular transport of viruses through cytoplasmic connections, termed plasmodesmata (PD), is essential for systemic infection in plants by viruses. Previous genetic and ultrastructural data revealed that the potyvirus cyclindrical inclusion (CI) protein is directly involved in cell-to-cell movement, likely through the formation of conical structures anchored to and extended through PD. In this study, we demonstrate that plasmodesmatal localization of CI in N. benthamiana leaf cells is modulated by the recently discovered potyviral protein, P3N-PIPO, in a CI:P3N-PIPO ratio-dependent manner. We show that P3N-PIPO is a PD-located protein that physically interacts with CI in planta. The early secretory pathway, rather than the actomyosin motility system, is required for the delivery of P3N-PIPO and CI to PD. Moreover, CI mutations that disrupt virus cell-to-cell movement compromise PD-localization capacity. These data suggest that the CI and P3N-PIPO complex coordinates the formation of PD-associated structures that facilitate the intercellular movement of potyviruses in infected plants.
Published in the journal: . PLoS Pathog 6(6): e32767. doi:10.1371/journal.ppat.1000962
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1000962Summary
Intercellular transport of viruses through cytoplasmic connections, termed plasmodesmata (PD), is essential for systemic infection in plants by viruses. Previous genetic and ultrastructural data revealed that the potyvirus cyclindrical inclusion (CI) protein is directly involved in cell-to-cell movement, likely through the formation of conical structures anchored to and extended through PD. In this study, we demonstrate that plasmodesmatal localization of CI in N. benthamiana leaf cells is modulated by the recently discovered potyviral protein, P3N-PIPO, in a CI:P3N-PIPO ratio-dependent manner. We show that P3N-PIPO is a PD-located protein that physically interacts with CI in planta. The early secretory pathway, rather than the actomyosin motility system, is required for the delivery of P3N-PIPO and CI to PD. Moreover, CI mutations that disrupt virus cell-to-cell movement compromise PD-localization capacity. These data suggest that the CI and P3N-PIPO complex coordinates the formation of PD-associated structures that facilitate the intercellular movement of potyviruses in infected plants.
Introduction
The ability of plant viruses to cross the cell wall barrier between an infected and adjacent healthy cell is a prerequisite to establish systemic infection [1]–[6]. Cell-to-cell movement of viruses occurs through plasmodesmata (PD), a specialized intercellular organelle, unique to the plant kingdom [1], [2]. PD are structurally complex microchannels that cross the cell wall and establish cytoplasmic and endomembrane continuity between neighboring cells [1], [2], [7], [8]. PD allow small molecules to diffuse between cells and regulate the intercellular trafficking of macromolecules or macromolecular complexes such as virions and ribonucleoprotein complexes [1], [2], [7], [8]. Viral cell-to-cell movement through PD is mediated by virus-encoded factors termed movement protein (MP) [9], [10]. Based on the characteristics of their intercellular transport, plant viruses can be classified into several groups. One group, which includes tobamoviruses, encodes a single dedicated MP that associates with, and increases the size exclusion limits of, PD to allow transport of virions or viral genomes (nucleic acids) through the modified channel [11], [12]. The second group includes many plant viruses with icosahedral particles, and requires both the MP and coat protein (CP) for cell-to-cell movement. This group, which includes nepo - and comoviruses, encodes an MP that forms PD-associated tubules that traverse the cell wall and through which virions move [13], [14]. The third group includes several viruses with filamentous particles, such as potexviruses, that contain a set of three movement genes called the triple gene block. These genes encode proteins that are assumed to function coordinately, but without forming the tubular structure, to transport viral particles or genomes through PD [3], [4], [15].
The family Potyviridae consists of approximately 30% of known plant viruses, including many agriculturally important viruses, e.g., Plum pox virus (PPV), Soybean mosaic virus (SMV), Turnip mosaic virus (TuMV), Tobacco etch virus (TEV) and Tobacco vein mottling virus (TVMV) [16], [17]. Potyviruses contain a flexuous filamentous virion morphology composed of approximately 2000 copies of CP, and possess dimensions of 680–900 nm by 11–15 nm [17]. The potyviral genome is a single-stranded, positive-sense RNA of approximately 10 kilobases (kb), containing a long open reading frame (ORF) encoding a polyprotein that is cleaved into ∼10 mature proteins [18], [19]. They are, from the N terminus of the polyprotein, P1, helper component proteinase (HC-Pro), P3, 6K1, cyclindrical inclusion (CI), 6K2, viral genome-linked protein of nuclear inclusion protein a (NIa-VPg), proteinase domain of NIa (NIa-Pro), nuclear inclusion protein b (NIb), and CP (Figure 1). In addition, a small ORF encoding a recently identified ∼25 kDa protein, P3N-PIPO, is embedded in the P3 coding region as a plus 2 frameshift sequence (Figure 1) [20]. Of these 11 potyviral proteins, CI, CP, HC-Pro and VPg have been suggested to be involved in viral cell-to-cell movement [9], [21], [22]. Accumulating evidence indicates that HC-Pro and VPg are essential in other aspects of the infection process such as viral genome replication or suppression on host defense (RNA silencing) [23], [24], where as CP and CI are more likely to be MPs [9], [25]–[29]. Mutations in the conserved core region of CP abolish virion assembly and cell-to-cell movement, suggesting potyviruses likely move as virions [25], [26]. High-resolution ultrastructural analyses indicate that CI forms the cone-shaped structures at the cell periphery adjacent to PD [27]–[29]. CP and viral RNA are present in these PD-associated structures in infected cells [27], [28]. Moreover, substitutions affecting the N terminus of CI can result in loss of cell-to-cell movement without compromising virus replication at the single cell level [9]. These findings support the idea that potyvirus intercellular transport involves interactions between virus particles, CI structures and PD.
Fig. 1. Schematic representation of the TuMV genome. 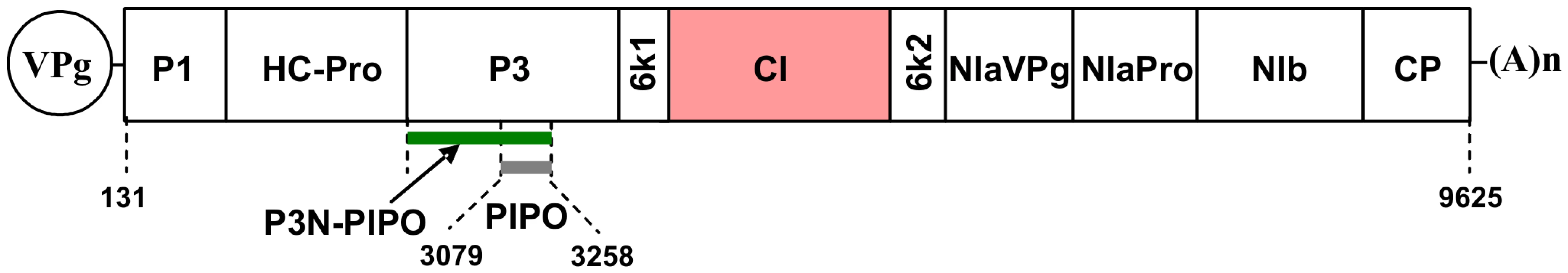
The circle represents the genome-linked viral protein, VPg. Two short horizontal lines represent 5′ and 3′ untranslational region, respectively. The large box represents the long open reading frame (from nucleotides 131 to 9625). The mature proteins resulting from processing the large polyprotein are indicated as smaller boxes. PIPO (from nucleotides 3079 to 3258) derived from a frameshif on the P3 cistron is indicated as a shot grey bar. P3N-PIPO is indicated in green and CI in red. The poly(A) tail is shown as (A)n. For clarity, the relative sizes of the mature proteins are not drawn to scale. In this work, we present evidence that localization of CI to PD was modulated by potyviral protein P3N-PIPO in a ratio dependent manner. Trafficking of P3N-PIPO and CI to PD was via the secretory pathway. In addition, CI mutations that impaired virus cell-to-cell movement lost the ability to associate with PD. In combination with previous data, these results suggest that CI and P3N-PIPO coordinate the formation of the PD-associated conical structures for intercellular transport.
Results
Formation of TuMV CI punctate spots along the cell wall is mediated by P3N-PIPO in a CI:P3N-PIPO ratio-dependent manner
Previous high-resolution ultrastructural studies with several potyviruses have revealed that CI forms conical structures in close proximity to PD [27]–[29]. To study CI localization, Agrobacterium-mediated transient expression was used in this study. In Nicotiana benthamiana leaf cells, TuMV CI tagged with the momomeric red fluorescent protein (mRFP) formed thread-like structures that aggregated in the cytoplasm 48 hrs post-infiltration (Figure 2A, panel I). In contrast, CI-mRFP formed punctate spots along the cell wall in most cells (approximately 80%) when coexpressed with a recombinant infectious clone of TuMV tagged with the green fluorescent protein (GFP) (Figure 2A, panel II). These punctate bodies of CI-mRFP spanned walls of the two adjoining cells, appearing in patterns as PD [30]. At higher magnifications, the CI-mRFP fluorescence labeled thread-like structures of variable lengths (Figure 2B, panels I, II). The CI-mRFP structures resembled single or paired elongated bars that extended from the plasma membrane, which was labeled by REM-GFP (Figure 2B, panels I, II) [31]. Interestingly, by 72 hrs post-infiltration, many thread-like structures of CI-mRFP retracted from the cell wall and aggregated in the cytoplasm in the majority of infected cells (approximately 95%) (Figure 2A, panel III). Thus, association of the CI-mRFP with the cell wall appeared to be dependent on one or more virus-encoded factors in a transient fashion.
Fig. 2. Subcellular localization of TuMV CI in N. benthamiana leaf cells. 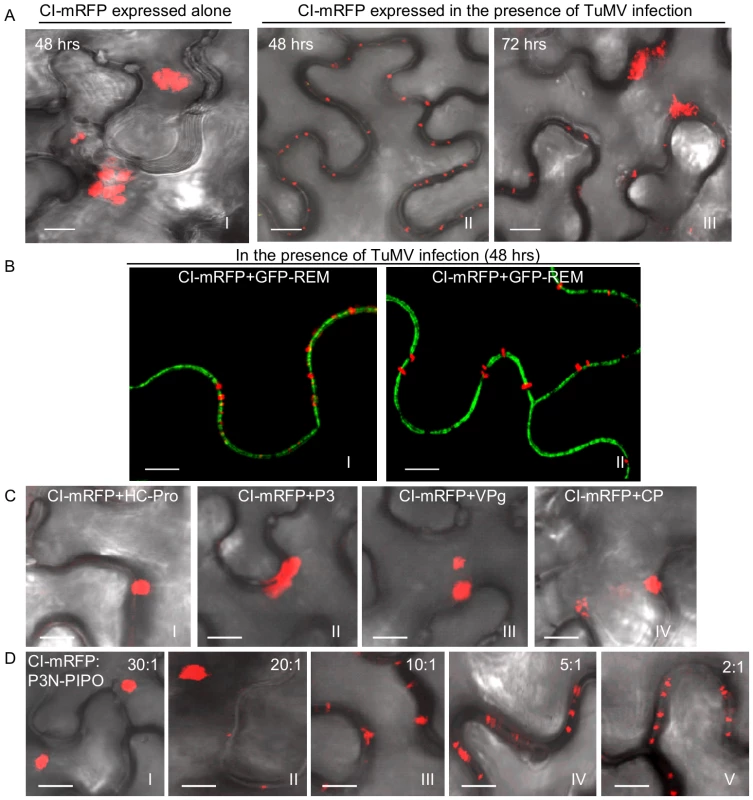
(A) Localization of CI-mRFP expressed alone in the leaf cells 48 hrs post-agroinfiltration (panel I) or in TuMV-infected leaf tissues 48 hrs (panel II) or 72 hrs (panel III) post-agroinfiltration. (B) Localization of CI-mRFP coexpressing with the plasma membrane marker GFP-REM (panels I, II) 48 hrs post-agroinfiltration. (C) Localization of CI-mRFP in the leaf cells coexpressing other viral proteins, i.e., HC-Pro (panel I), P3 (panel II), VPg (panel III) or CP (panel IV). Images were taken 48 hrs post-agroinfiltration. (D) Localization of CI-mRFP in the cells coexpressing P3N-PIPO. The ratio of agrobacterial culture mixtures containing plasmid CI to plasmid P3N-PIPO is indicated. Bars, 8 µm. To determine if any TuMV protein facilitated targeting of the CI-mRFP to the cell wall, each viral protein (P1, HC-Pro, P3, P3N-PIPO, 6K1, 6K2, NIa, NIb and CP) was coexpressed individually with CI-mRFP. As illustrated using HC-Pro, P3, VPg and CP, coexpression with most TuMV proteins had no effect on the cytoplasmic aggregation of CI-mRFP (Figure 2C). In contrast, coexpression with P3N-PIPO resulted in the cell wall-associated punctate CI-mRFP (Figure 2D). Cell wall association was dependent on the CI-mRFP:P3N-PIPO ratio, with at least some CI-mRFP forming cytoplasmic aggregates at ratios of 10∶1 and higher (Figure 2D, Figure S1). These data suggest that CI is directed to cell walls through a dose-dependent interaction with, or is influenced by, P3N-PIPO.
TuMV P3N-PIPO is a PD-located protein
To examine the intracellular distribution of P3N-PIPO in planta, P3N-PIPO-GFP was transiently expressed in N. benthamiana leaf cells via agroinfiltration. P3N-PIPO-GFP localized at the cell wall as punctate spots 48 hrs post-infiltration (Figure 3A, panel I). Under higher magnifications, paired punctate structures that spanned the adjoining cell walls were clearly evident (Figure 3A, panel II). P3N-PIPO-GFP was coexpressed with PDLP1-mRFP, a type I membrane plasmodesmatal protein [32]. P3N-PIPO-GFP and PDLP1-mRFP colabeled the punctate structures on the wall (Figure 3B), suggesting that P3N-PIPO is a PD-localized protein. To further confirm the localization of P3N-PIPO, P3N-PIPO tagged with the yellow fluorescent protein (YFP) and the actin marker mTalin tagged with the cyan fluorescent protein (CFP) were coexpressed in N. benthamiana leaf cells. In the unplasmolyzed cells, P3N-PIPO-YFP formed punctate spots on the cell wall, while mTalin-CFP fluorescence was evenly distributed throughout the cytoplasm compartment (Figure 3C, control). After plasmolysis treatment, P3N-PIPO-YFP remained within the cell wall, whereas the mTalin-CFP-labeled cytoplasm detached from the cell wall (Figure 3C, plasmolyzed).
Fig. 3. TuMV P3N-PIPO is a PD-localized protein and mediates the targeting of CI to PD in N. benthamiana. 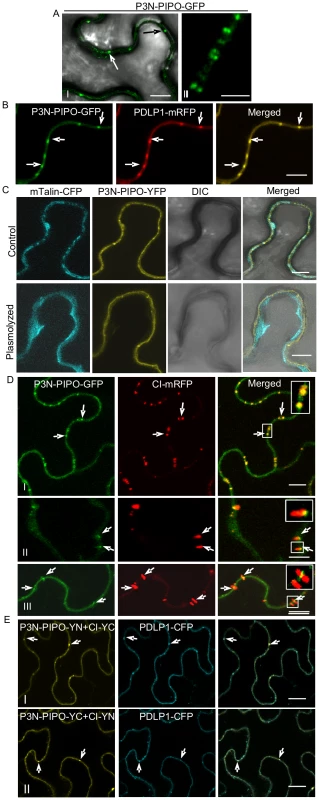
(A, panels I, II) Localization of P3N-PIPO-GFP transiently expressed in the cell treated 48 hrs post-agroinfiltration. Paired P3N-PIPO structures under a higher magnification (panel II). (B) Colocalization of P3N-PIPO-GFP with the PD marker PDLP1-mRFP. Arrows point to PD costained by P3N-PIPO-GFP and PDLP1-mRFP. (C) Cells coexpressing P3N-PIPO-YFP and mTalin-CFP as a cell membrane marker (control). Fluorescence of P3N-PIPO-YFP in plasmolyzed leaf tissue containing mTalin-CFP (plasmolyzed). DIC, Differential interference contrast. (D) Colocalization of P3N-PIPO with CI-mRFP 48 hrs (panel I) and 72 hrs (panels II, III) post-agroinfiltration. Arrows point to the PD-localized P3N-PIPO-GFP and CI-mRFP. Insets are the enlarged images of the areas in white boxes in the corresponding panels. (E) Interactions of TuMV CI and P3N-PIPO proteins in vivo. BiFC analysis (48 hrs post-agroinfiltration) was used to assess interactions in cells coexpressing CI-YC and P3N-PIPO-YN (panel I), CI-YN and P3N-PIPO-YC (panel II). Arrows indicate the strong BiFC fluorescence at PD costained by the PD marker, PDLP1-CFP. Bars, 8 µm. To determine if P3N-PIPO colocalizes with the CI protein at PD, P3N-PIPO-GFP was coexpressed with CI-mRFP. At 48 hrs post-infiltration, the CI-mRFP punctate fluorescence predominantly overlapped the P3N-PIPO fluorescence at PD (Figure 3D, panel I). At 72 hrs, the size of P3N-PIPO-derived structures remained the same, whereas the PD-localized fluorescence of CI-mRFP expanded as thin thread-like structures into the cytoplasm (Figure 3D, panels II, III). The majority of the red fluorescent threads of CI-mRFP were seen to extend from P3N-PIPO-labeled spots (Figure 3D, panels II, III). These results suggest that CI is targeted to PD in the presence of P3N-PIPO, colocalizes with P3N-PIPO at PD, and subsequently forms PD-rooted thread-like structures.
The relocalization of CI-mRFP to PD in the presence of P3N-PIPO might suggest an interaction between CI and P3N-PIPO. To test this hypothesis, a bimolecular fluorescence complementation (BiFC) assay was carried out. CI-YN and P3N-PIPO-YC hybrid proteins, as well as the reverse hybrid combination, were coexpressed in N. benthamiana leaf cells. Strong BiFC fluorescence from both hybrid combinations was detected along the cell wall, highlighted by PD fluorescence that coincided with PD-associated PDLP1-CFP 48 hrs post-infiltration (Figure 3E, panels I, II). No BiFC fluorescence was detected in the negative control samples expressing combinations with non-hybrid YC or YN (data not shown). Taken together these data suggest that P3N-PIPO is a PD-located protein and may direct targeting of CI to PD through protein-protein interaction.
PD targeting of P3N-PIPO and CI requires a functional secretory pathway and is independent of the actomyosin motility system
Trafficking of plasmodesmatal proteins, such as PDLP1 (a type I membrane receptor-like protein) and C1RGP (a class 1 reversibly glycosylated polypeptide) to PD has been shown to exploit the secretory pathway [32], [33]. Chemical or protein inhibitors were used to investigate the role of the secretory pathway in P3N-PIPO trafficking to PD. Both Brefeldin A (BFA) and a GTP-restricted mutant of the Sar1 protein [Sar1(H74L)] can block the ER-Golgi vesicular transport pathway [34]. In BFA-treated plant leaf cells, targeting of PDLP1-CFP or P3N-PIPO-YFP to PD was inhibited, with both PDLP1-CFP and P3N-PIPO-YFP accumulating in large compartments that are typical of BFA-treated cells (Figure 4A, B, panels I, II; Figure S2) [32]. Inhibition of PD localization of both proteins was incomplete (Figure S2), possibly due to only partial disruption of trafficking or to residual localization and accumulation prior to drug treatment. Consistent with recent findings [32], coexpression of non-tagged Sar1(H74L) caused the retention of PDLP1-CFP in the ER network (web-like fluorescence pattern) and inhibited the trafficking of PDLP1-CFP to PD (Figure 4A, C, panels I; Figure S2). In contrast, when coexpressed with Sar1(H74L), P3N-PIPO trafficking to PD was also inhibited and P3N-PIPO-YFP was cytosolic in the cells (Figure 4A, C, panels II; Figure S2). This is in agreement with the result that no typical transmembrane domains were found in TuMV P3N-PIPO using computer-assisted prediction tools described in Material and Methods. Of 100 cells observed under the confocal microscope, approximately 20% of punctate structures of PDLP1-CFP and P3N-PIPO-YFP were PD-located after BFA treatment or coexpression with untagged Sar1(H74L) (Figure S2). It is therefore concluded that, like some other PD-localized proteins, P3N-PIPO utilizes the secretory pathway for delivery to PD.
Fig. 4. Targeting of P3N-PIPO and CI to PD requires the BFA-sensitive secretory pathway and is independent of the acto-myosin motility system. 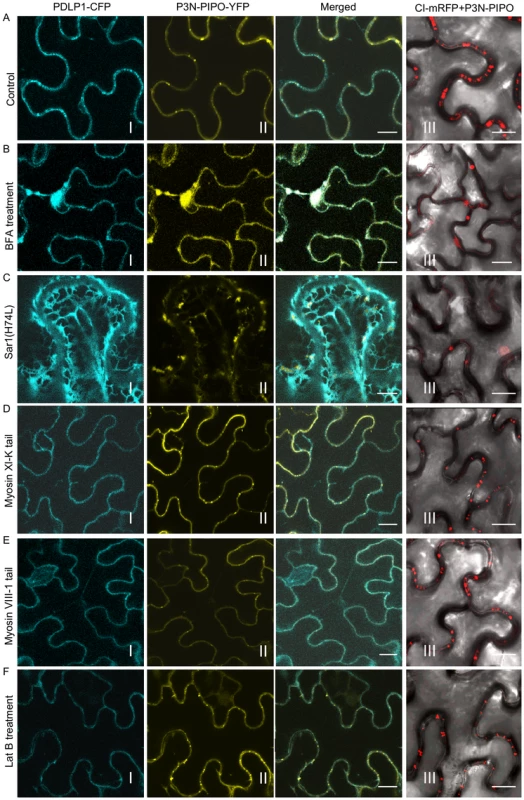
PD marker PDLP-1-CFP (panels I), P3N-PIPO-YFP (panels II) and CI-mRFP in the presence of untagged P3N-PIPO (panels III) were transiently expressed in leaf cells treated with water (Control, A), 50 µg/mL BFA (BFA, B), co-agroinfiltrated with the untagged COPII mutant Sar1(H74L) [Sar1(H74L), C], co-agroinfiltrated with the untagged myosin XI-K tail (Myosin XI-K tail, D), co-agroinfiltrated with the untagged myosin VIII-1 tail (Myosin VIII-1 tail, E), and 25 µM Lat B (Lat B, F). Images were taken 48 hrs post-agroinfiltration. N, nucleus. Bars, 10 µm. The actomyosin motility system, empowered by myosin motors, has been implicated in the targeting of viral MP proteins to PD [30] and intercellular spread of some plant viruses [35]. Higher plants possess two groups of myosins, class VIII and class XI [27]. The tails of myosins XI-K and VIII-1, which behave as dominant-negative antagonists to myosin function, were used to examine the role of myosins in the targeting of P3N-PIPO to PD. Interestingly, while overexpression of the tails inhibited targeting of PDLP1-CFP to PD, they had no apparent effects on P3N-PIPO-YFP steady-state accumulation patterns (Figure 4A, D, E, panels I, II; Figure S2). As the requirement of myosin motors for intracellular trafficking implies the utilization of the actin microfilaments, we further tested the effect of Latrunculin B (Lat B), an inhibitor of actin polymerization, on the targeting of P3N-PIPO to PD. Lat B treatment did not affect the targeting of P3N-PIPO to PD (Figure 4A, F, panel II; Figure S2). These data suggest that the actomyosin motility system, which is involved in the trafficking of plasmodesmatal proteins PDLP1 and some other viral MPs to PD, is not involved in PD-localization of P3N-PIPO.
The effect of secretory pathway inhibitors and dominant-negative mutants on the localization of CI was investigated. In the absence of P3N-PIPO or TuMV infection, none of these inhibitors interfered with the formation of CI-mRFP aggregates in the cytoplasm (data not shown). Consistently, analysis of the TuMV CI sequence with several computer-assisted prediction tools described in Material and Methods did not reveal any typical membrane domains. However, both BFA treatment and overexpression of Sar1(H4L) inhibited PD-localization of CI-mRFP in the presence of P3N-PIPO (Figure 4A, B, C, panels III; Figure S2). The targeting of CI-mRFP to PD was not obviously affected in leaf cells expressing the myosins tails or treated with LatB (Figure 4D, E, F, panels III; Figure S2). Thus, PD targeting of P3N-PIPO or CI (in the presence of P3N-PIPO) requires a functional secretory pathway and is independent of the actomyosin motility system.
TuMV CP associates with PD-located CI structures during virus infection
The assembled virion is likely the form in which potyviruses move through PD [25]–[28]. To assess the subcellular distribution of virions relative to the CI protein during virus infection, YFP-CP and CI-mRFP were coexpressed in N. benthamiana leaf cells infected with a recombinant TuMV-GFP infectious clone, or a TuMV infectious clone expressing a 6K-GFP fusion protein (TuMV::6K-GFP) [36], [37]. As described earlier, CI trafficked to PD and formed aggregates in the cytoplasm at 48 to 72 hrs post-agroinfiltration (Figure 2A). The fibrillar structures of YFP-CP were also observed either in the cytoplasm or along the cell wall (Figure 5). In the cytoplasm, CI aggregates are often associated with 6K vesicles where virus replication takes place (Figure 5, panel III) [37]. The fibrillar structures of CP represent virions or CP-containing nucleoprotein (NP) complexes [28]. In the cytoplasm, the fibrillar structures of CP were in close proximity to chloroplasts during virus infection (Figure 5, panel II). This was in contrast to CP punctate structures forming away from chloroplasts when expressed alone (Figure 5, panel I). Moreover, the fibrillar structures of YFP-CP were associated with PD-localized structures of mRFP-CI in the presence of P3N-PIPO during virus infection (Figure 5, panel IV), consistent with published results that the CP of Pea seed-borne mosaic virus (PSbMV) is present in the CI conical structures adjacent to PD [27], [28] and that TEV CP is required for cell-to-cell movement [25], [26].
Fig. 5. Association of TuMV CP with CI in TuMV-infected N. benthamiana leaf cells. 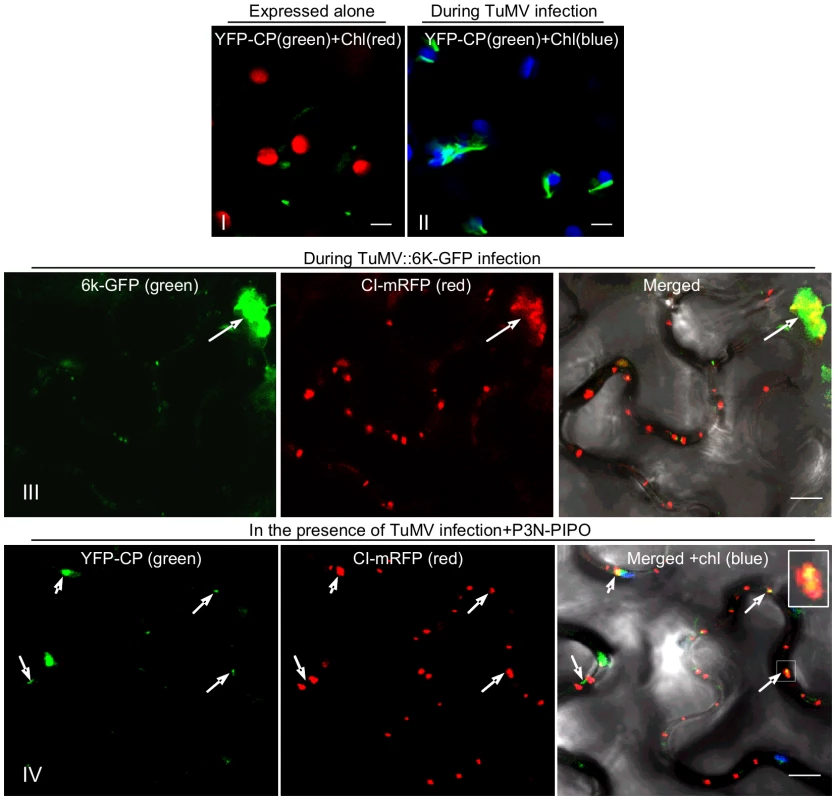
(Panels I, II) TuMV CP tagged by YFP (YFP-CP) in the cytoplasm when expressed alone (panel I) or during virus infection (panel II). (Panel III) When coexpressed with the recombinant TuMV::6K-GFP infectious clone, some mRFP-CP is also present in proximity to the 6K-GFP-labeled replication complex (arrow). (Panel IV) TuMV YFP-CP attachment to PD-associated CI structures.(arrows; Inset) in the cell periphery in the presence of P3N-PIPO during virus infection. All images are taken 48 hrs post-agroinfiltration. Chl, chloroplasts. Bars, 8 µm. PD targeting of CI is essential for potyvirus cell-to-cell movement
To determine if the P3N-PIPO protein of potyviruses other than TuMV direct targeting of their respective CI to PD, P3N-PIPO and CI of TEV were analyzed in N. benthamiana cells. Consistent with observations described above for TuMV, TEV P3N-PIPO colocalized with PDLP1 at PD (Figure 6A). TEV CI-mRFP formed aggregates in the cytoplasm when expressed alone (Figure 7A, panel I) and was targeted to PD with associated thread-like structures when coexpressed with TEV P3N-PIPO-GFP (Figure 7A, panel II).
Fig. 6. Subcellular localization of TEV P3N-PIPO and its interaction with TEV CI and the intercellular movement-defective mutant CI(AA3,4). 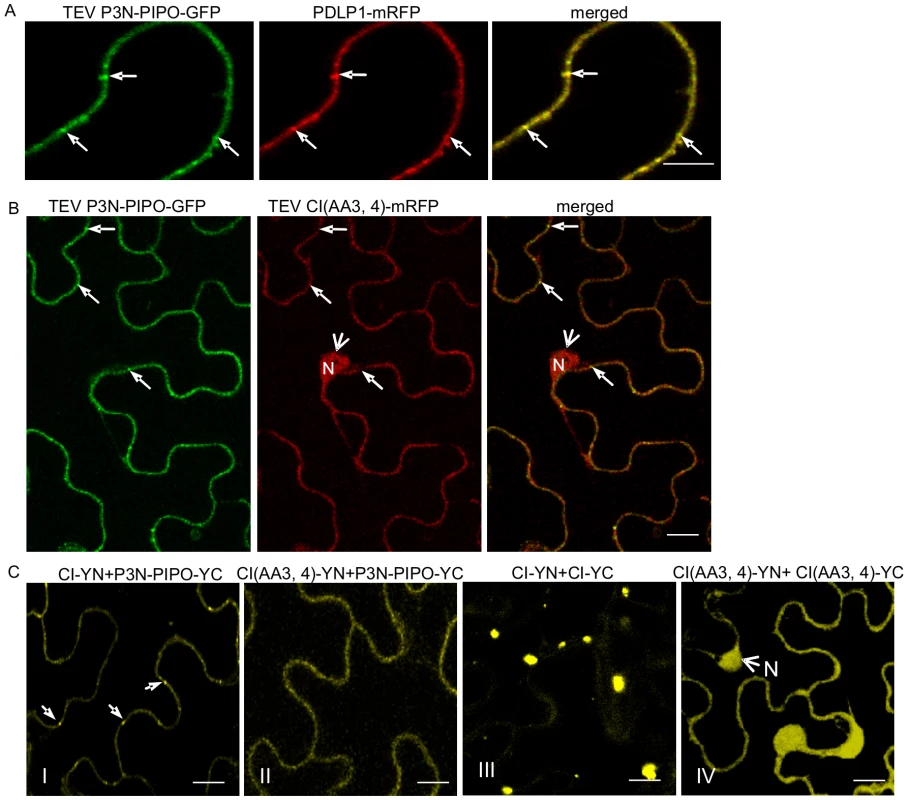
(A) Colocalization of TEV P3N-PIPO-GFP with the PD marker PDLP1-mRFP. Arrows point to the PD-located P3N-PIPO-GFP and PDLP1-mRFP. (B) Coexpression of TEV CI(AA3,4)-mRFP does not change PD-localization of TEV P3N-PIPO-GFP. Arrows indicate PD-located P3N-PIPO-GFP. Arrowheads indicate nucleus-localization of TEV CI(AA3,4)-mRFP. (C) BiFC analysis of interactions of TEV CI-YN and TEV P3N-PIPO-YC (panel I), TEV CI (AA3, 4)-YN and TEV P3N-PIPO-YC(panel II), TEV CI-YN and TEV CI-YC (panel III), and TEV CI (AA3, 4)-YN and TEV CI (AA 3, 4)-YC. N, nucleus. Bars, 10 µm. Fig. 7. Two TEV CI mutants defective in cell-to-cell movement fail to form either cytoplasmic inclusions (when expressed alone) or PD-associated structures (in the presence of P3N-PIPO). 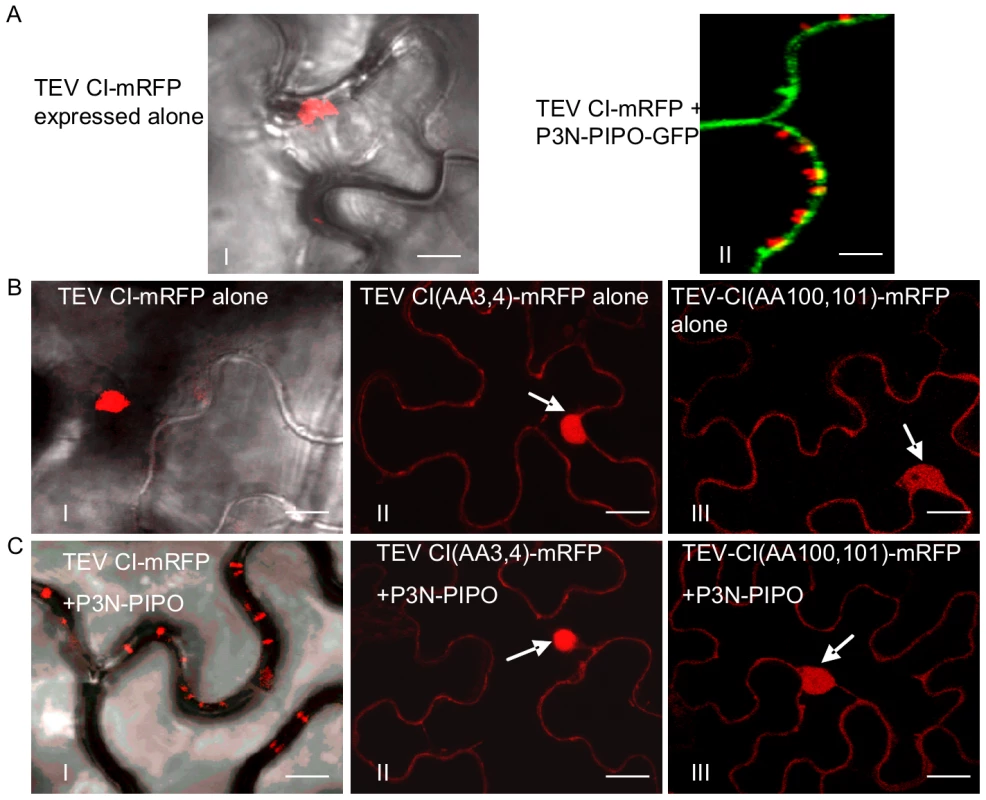
(A) TEV CI-mRFP forms aggregates in the cytoplasm when expressed alone (panel I) and punctate spots along the cell walls when coexpressed with P3N-PIPO-GFP (panel II). (B) When expressed alone, TEV CI(AA3, 4)-mRFP (panel II) and TEV CI(AA100,101)-mRFP (panel III) are distributed in the nucleus and in periphery rather than forming typical inclusions in the cytoplasm (panel I). (C) In the presence of P3N-PIPO, TEV CI(AA3, 4)-mRFP (panel II) and TEV CI(AA100,101)-mRFP (panel III) are distributed in the nucleus and cell periphery rather than targeting PD (panel I). All images are taken 48 hrs post-agroinfiltration. Bars, 8 µm. Previous genetic analysis of TEV identified CI mutants with substitutions affecting two aspartic residues at positions 3 and 4, or two positively charged amino acids (lysine and argentine) at positions 100 and 101, with defects in cell-to-cell movement [9]. These mutants amplified to levels equivalent to that of the parental virus in isolated cells. When expressed alone or coexpressed with TEV P3N-PIPO, the two TEV CI mutant proteins, CI(AA3, 4)-mRFP and CI(AA100,101)-mRFP, were observed in the nucleus and cell periphery, but not in punctate PD or late-forming cytoplasmic aggregate patterns typical for the parental CI-mRFP protein (Figure 7B, C). Furthermore, coexpression of CI(AA3,4)-mRFP with P3N-PIPO-GFP did not affect the targeting of P3N-PIPO-GFP to PD (Figure 6B). BiFC experiments were performed to examine if TEV P3N-PIPO interacted with CI(AA3,4). As expected, the BiFC fluorescence resulting from parental CI-YN and P3N-PIPO-YC, or their reverse hybrid combination, was localized to PD (Figure 6C, panel I). Such targeting was compromised in cells with CI(AA3, 4)-YN and P3N-PIPO-YC or their reverse hybrid combination (Figure 6C, panel II). Though the mutated CI did interact with P3N-PIPO, the BiFC signal was evident in the cell periphery but not within distinct, punctate foci indicative of PD (Figure 6C, panel II). BiFC experiments also localized the self-interacting TEV CI mutant CI(AA3, 4) to the nucleus and cell periphery (Figure 6C, panel IV) rather than to typical inclusions in the cytoplasm (Figure 6B, panel III). BiFC fluorescence was not detected in cells expressing non-hybrid YN and YC proteins (data not shown). The lack of PD-localization of the mutant TuMV and TEV CI proteins may explain the cell-to-cell movement defects reported previously [9].
Discussion
In the present study, TuMV CI was localized to the cell wall at the early stages of infection (Figure 2B, panel I; Figure 1A, panel II), and to thread-like structures emanating from punctate bodies (Figure 2B, panel II). CI structures disassociated from the cell wall and accumulated as aggregates in the cytoplasm at later time points (Figure 2A, panel III). By contrast, when expressed alone, CI-mRFP only aggregated in the cytoplasm (Figure 2A, panel I). P3N-PIPO was found to direct the CI fusion protein to the cell wall-associated punctate bodies, which colocalized with PD protein marker PDLP1 (Figure 2C, D; Figure 3). Significantly, the CI protein from multiple potyviruses was shown to associate with P3N-PIPO, and target to PD in a P3N-PIPO-dependent manner (Figure 2; Figure 7). This is consistent with several ultrastructural studies showing that the CI protein of Sorghum mosaic virus (Figure S3), TVMV [27] and Wheat spindle streak mosaic virus [38] form conical structures adjacent to PD, and possibly traverse the PD in a fine thread-like structure. It is worth noting that polyclonal antibodies against TVMV P3 specifically labeled cell wall-bound deposits of CI in TVMV-infected cells [27], [39]. Since TVMV P3N-PIPO and P3 share an N-terminal region of approximately 20 kDa [20], the P3 polyclonal antibodies may actually have recognized P3N-PIPO. This speculation is also supported by the finding that potyviral P3 protein does not interact with CI [40]. These data point to a role for P3N-PIPO directing CI to PD, anchoring the CI proteins therein and/or facilitating the deposition of CI through PD.
We show that P3N-PIPO is a PD-located protein and interacts with CI (Figure 3). Unlike PDLP1, both P3N-PIPO and CI lack a typical transmembrane domain. Disrupting the ER-Golgi secretory pathway obstructed the targeting of P3N-PIPO and CI to PD, whereas impairing the actomyosin motility system had little effect on their PD localization (Figure 4). Thus, targeting of P3N-PIPO and CI to PD requires a functional secretory pathway but not the actomyosin motility system. Interestingly, targeting of PDLP1 to PD is mediated by a transmembrane domain and requires both the ER-Golgi secretory pathway and the actomyosin motility system (Figure 4) [32]. It appears that some viral MPs, i.e., Beet yellow virus MP, require the actomyosin motility system for their PD targeting, while others, such as Tobacco mosaic virus MP, do not [30]. Since targeting of P3N-PIPO and PDLP1 to PD shares the BFA-sensitive secretory pathway and differs in the requirement of the actomyosin motility system (Figure 4), exactly how the secretion route branches from the ER-Golgi pathway to PD is yet to be determined.
The delivery of CI-mRFP to PD was mediated by P3N-PIPO in a ratio dependent manner. As P3N-PIPO is translated via ribosomal frameshifting [20], the ratio of CI to P3N-PIPO biosynthesis depends on the frameshift efficiency. In PSbMV–infected pea cotyledons, cell wall-associated CI conical structures were found only in the infection front where active genome translation and replication was occurring [28]. In cells behind this zone, CI was no longer associated with the cell wall, which is consistent with the temporal series of events documented in the current study. The transient association of CI structures with PD could be conditional upon the level of viral genome translation, frameshifting and the subsequent CI:P3N-PIPO ratio.
Targeting of CI to PD by P3N-PIPO may also be affected by CI self-interactions. Two movement-defective CI mutants (DD3, 4AA and KK101,102AA), previously shown to have reduced self-interacting strength [41], lost ability to form cytoplasmic structures (Figure 6C, panel III; Figure 7B). When these mutants were either expressed alone or in the presence of P3N-PIPO, they were distributed to the cell periphery and the nucleus (Figure 7B,C). Though these mutants could still interact with P3N-PIPO (Figure 6C), they were unable to accumulate at PD to form the thread-like structure (Figure 7C). Therefore, the proper self-assembly of CI is necessary for its interaction with P3N-PIPO to form the cone-shape structure at PD for viral cell-to-cell movement.
During the reviewing process of this paper, Wen and Hajimorad reported that mutation of the putative SMV PIPO impedes cell-to-cell movement [42], providing genetic evidence that P3N-PIPO is a potyviral MP. Thus, our data reveals the mechanism by which P3N-PIPO functions as an MP. Taking into account these new findings, we propose a model for the formation of movement complexes that facilitate intercellular transport of potyviruses (Figure 8). Initial events likely involve the recruitment of nascent virus particles by CI or self-interacting CI structures at membrane-bound sites of replication adjacent to chloroplasts. Next, CI-virion complexes may associate with either pre-targeted P3N-PIPO followed by trafficking to PD, or with PD-associated P3N-PIPO. CI structures are then proposed to grow from P3N-PIPO-anchored sites at PD, forming thread-like structures that might recruit additional virus particles for transport. Actual movement of virus particles through PD may be facilitated by PD-traversing CI complexes, although the details of this proposed event are not known. It is difficult to imagine a mechanistic role for the late-forming cytoplasmic aggregates in virus movement, as these form during a post-movement phase of infection. This model bears some similarity to that proposed by Jackson and colleagues for cell-to-cell movement of hordeiviruses [3], [4], which encode three MPs (TGB1, TGB2 and TGB3). The hordeivirus model states that TGB1 binds the viral RNAs to form movement complexes. Interaction with TGB3 targets the TGB1-RNA movement complex to PD. TGB2 then interacts with TGB3 to stabilize the movement complex at PD and facilitate PD gating. The key deficits of the proposed models center around the mechanisms underlying gating, the mechanisms to generate force, and the impact of these virus-associated events on the normal functions of PD in intercellular trafficking and defense. These represent critical areas of future study.
Fig. 8. Model for potyvirus intercellular transport through PD. 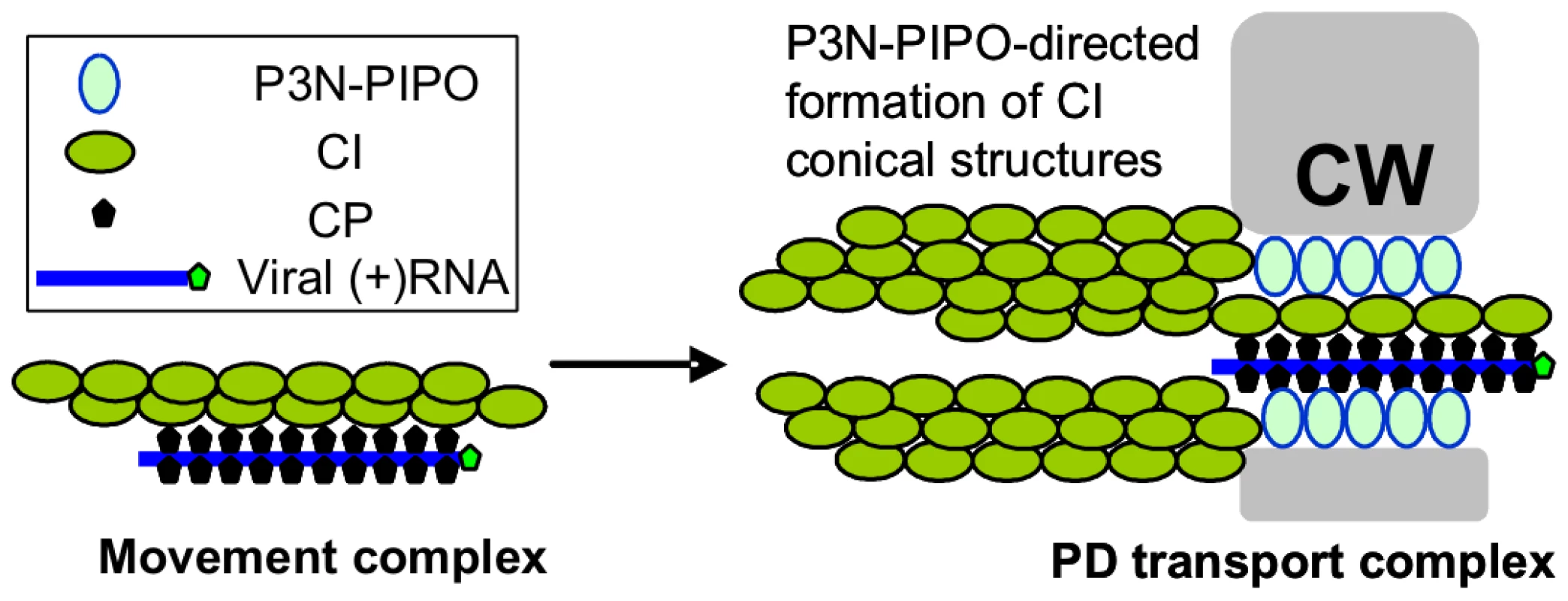
The virion-CI movement complex is intracellularly transported to the modified PD where CI forms conical structures anchored by the PD-located P3N-PIPO. The virion is then fed through the CI structures and PD to enter the adjacent cell. CW, cell wall. Materials and Methods
Plasmid construction
Gateway technology (Invitrogen, Burlington, Ontario, Canada) was used to generate all plasmid clones used in this work. Gene sequences were amplified by PCR using Phusion DNA polymerase (NEB). The resulting DNA fragments were purified and transferred by recombination into the entry vector pDONR221 (Invitrogen) using BP clonase II (Invitrogen) following the supplier's recommendations. Insertions in the resulting pDONR clones were verified by DNA sequencing. Destination vectors pEarleygate100, pEarleygate102, pMDC83, pMDC43, pGWB454 and pGWB455 were used to express untagged protein, C-terminal CFP-HA fusion protein, C-terminal GFP-6xHis fusion protein, N-terminal GFP fusion protein, C-terminal mRFP fusion protein, and N-terminal mRFP fusion protein, respectively [43]–[46]. The vectors used in the BiFC assay were modified from pEarlyGate vectors 201 and 202, where the YFP N-terminus comprises amino acids 1–174 and the C-terminus comprises amino acids 174–239 [36]. The recombinant TuMV infectious clones containing 6K-GFP (TuMV:6K-GFP) and GFP (TuMV:GFP) were as described previously [37], [47]. Plasmid pTEV7DA-GFP was described [48]. The P1, HC-Pro, P3, P3N-PIPO, 6K1, CI, 6K2, NIa, NIb and CP coding regions of TuMV were amplified by PCR, recombined into pDONR221, and then into the binary destination vector pEarleygate100 for expressing nontagged proteins. The entry vector pDONR221 containing TuMV P3N-PIPO or CI were transferred by recombination into the binary destination vectors pEarleygate102, pMDC83 and pGWB454 to produce plasmids TuMV P3N-PIPO-CFP, TuMV P3N-PIPO-GFP, TuMV P3N-PIPO-mRFP, TuMV CI-GFP, and TuMV P3N-PIPO-mRFP. The entry vector pDONR221 containing TuMV CP was transferred by recombination into the binary destination vectors pMDC43 and pGWB455 to produce plasmids TuMV GFP-CP and TuMV mRFP-CP, respectively. The TEV P3N-PIPO, wild-type CI, and two CI mutants (DD3, 4AA and KR100,101AA) were amplified by PCR, recombined into pDONR221, and then into the binary destination vectors pEarleygate100, pMDC43 or pGWB455 to generate plasmids TEV P3N-PIPO (nontagged), TEV P3N-PIPO-GFP, TEV-CI-mRFP, TEV CI(AA3, 4)-mRFP and TEV CI(AA100,101)-mRFP. For BiFC experiments, the pDONR221vectors containing TuMV P3N-PIPO and CI were recombined into BiFC vectors YN and YC to generate P3N-PIPO-YN (YC) and CI-YN (YC), respectively. The pDONR221 vectors containing TEV P3N-PIPO, CI, CI(AA3, 4) and CI(AA100,101) were recombined into BiFC vectors YN and YC to generate TEV P3N-PIPO-YN (YC), CI-YN (YC), CI(AA3, 4)-YN (YC) and CI(AA100,101)-YN (YC), respectively. The plasmid containing the PDLP1:GFP was a kind gift from Andrew J. Maule (John Innes Centre, UK). The DNA fragment for PDLP1 was amplified and recombined into pDONR221 and then into the binary destination vector pGWB454 to give PDLP1-mRFP. The construct for expressing GFP-REM was kindly provided by Sébastien Mongrand (Centre National de la Recherche Scientifique-University of Bordeaux, France) [23]. The construct for expressing the myosins XI-K and VIII-1 tails were kindly provided by Valerian V. Dolja (Oregon State University) [30], [49]. The construct for expressing the actin filament marker mTalin-CFP was previously described [37].
Transient expression in N. benthamiana
Binary vectors were transformed into Agrobacterium tumefaciens GV3101. For agroinfiltration, agrobacterial cultures were grown overnight in LB containing appropriate antibiotics. The agrobacteria were collected by centrifugation, and then resuspended in 10 mM MgCl2 containing 100 µM acetosyringone. After a minimum of 2 h incubation at room temperature, the culture was diluted to an optical density of 0.2–0.5 at 600 nm (OD600). N. benthamiana plants were agroinfiltrated with appropriate agrobacterial cultures and the agroinfiltrated plants were maintained under normal growth conditions for 2 to 4 days. For plasmolysis, plant tissue was infiltrated with 30% glycerol and viewed immediately.
For co-agroinfiltration assays with different ratio of CI and P3N-PIPO expression plasmids, different volumes of agrobacterial cells (OD600 = 0.6) containing expression plasmids untagged TuMV P3N-PIPO, TuMV P3N-PIPO-6xHis, TuMV CI-mRFP or TuMV CI-YFP-HA were mixed to achieve desired ratios of CI to PIPO. The total cell population of each mixture was kept constant for each infiltration experiment. Co-agroinfiltration of different plasmid combinations, i.e., TuMV CI-mRFP and TuMV P3N-PIPO or TuMV CI-YFP-HA and TuMV P3N-PIPO-6xHis was repeated at least three times. For western blot analyses, leaf samples were examined under the confocal microscope to confirm the subcellular localization of the fluorescent fusion proteins. Total proteins were extracted from N. benthamiana leaves agroinfiltrated with mixtures of agrobacterial cells containing TuMV CI-YFP-HA and TuMV P3N-PIPO-6xHis. Immunoblot was conducted with anti-HA IgG (Sigma, St. Lous, MO) and anti-His IgG (Abcam, Cambridge, MA) following the protocols recommended by the suppliers.
Inhibitor studies
Inhibitor studies were done following transient coexpression of PDLP1-CFP and P3N-PIPO-YFP or coexpression of CI-mRFP and untagged P3N-PIPO in 4-week-old N. benthamiana plants. The infiltrated plant leaves were infused with BFA (50 µg/ml in water) at 40 h post-infiltration with the expression constructs and viewed 12 h post-BFA treatment. For the disruption of actin, leaf tissues were treated with 25 µM Lat B (Sigma-Aldrich, Oakville, Ontario, Canada) for 2 h. Negative controls were run in parallel.
Confocal microscopy
Plant tissue was imaged at room temperature using a Leica TCS SP2 inverted confocal microscope with an Argon ion laser. GFP was excited at 488 nm and the emitted light was captured at 505 to 555 nm. Light emitted at 630–680 nm was used to record chlorophyll autofluorescence. YFP was excited at 514 nm and captured at 525-650 nm. mRFP was excited at 543 nm and captured at 590–630 nm. Images were captured digitally and handled using the Leica LCS software.
Sequence sources and analyses
The TEV and TuMV genome sequences used in this study were retrieved from GenBank (http://www.ncbi.nlm.nih.gov/Genbank/) with accession numbers DQ986288.1 and EF028235.1, respectively. Transmembrane helices in the P3, P3N-PIPO and CI proteins of TuMV were predicted as previously described [50].
Supporting Information
Zdroje
1. Maule AJ
2008 Plasmodesmata: structure, function and biogenesis. Curr Opin Plant Biol 11 680 686
2. LucasWJ
HamBK
KimJ-K
2009 Plasmodesmata-bridging the gap between neiboring cells. Trend Cell Biol 19 495 503
3. JacksonAO
LimHS
BraggJ
GanesanU
LeeMY
2009 Hordeivirus replication, movement, and pathogenesis. Annu Rev Phytopathol 47 385 422
4. Verchot-LubiczJ
2005 A new cell-to-cell transport model for Potexviruses. Mol Plant-Microbe Interact 18 283 290
5. NelsonRS
CitovskyV
2005 Plant viruses: invaders of cells and pirates of cellular pathways. Plant Physiol 138 1809 1814
6. BoevinkP
OparkaKJ
2005 Virus-host interactions during movement processes. Plant Physiol 138 1815 1821
7. LeeJ-Y
TaokaK
YooB-C
Ben-NissahG
KimD-J
2005 Plasmodesmal-associated protein kinase in tobacco and Arabidopsis recognizes a subset of non-cell-autonomous proteins. Plant Cell 17 2817 2831
8. OparkaKJ
2004 Getting the message across: how do plant cells exchange macromolecular complexes? Trend Plant Sci 9 33 41
9. CarringtonJC
JensenPE
SchaadMC
1998 Genetic evidence for an essential role for potyvirus CI protein in cell-to-cell movement. Plant J 14 393 400
10. LucasWJ
2006 Plant viral movement proteins: agent for cell-to-cell trafficking of viral genomes. Virology 344 169 184
11. KawakamiS
WatanabeY
BeachyRN
2004 Tobacco mosaic virus infection spreads cell to cell as intact replication complexes. Proc Natl Acad Sci U S A 101 6291 6296
12. WrightKM
WoodNT
RobertsAG
ChapmanS
BoevinkP
2007 Targeting of TMV movement protein to plasmodesmata requires the actin/ER network: evidence from FRAP. Traffic 8 21 31
13. LaporteC
VetterG
LoudesAM
RobinsonDG
HillmerS
2003 Involvement of the secretory pathway and the cytoskeleton in intracellular targeting and tubule assembly of Grapevine fanleaf virus movement protein in tobacco BY-2 cells. Plant Cell 15 2058 2075
14. PouwelsJ
van der VeldenT
WillemseJ
BorstJW
van LentJ
2004 Studies on the origin and structure of tubules made by the movement protein of Cowpea mosaic virus. J Gen Virol 85 3787 3796
15. LimHS
BraggJN
GanesanU
RuzinS
SchichnesD
2009 Subcellular localization of the barley stripe mosaic virus triple gene block proteins. J Virol 83 9432 9448
16. BergerPH
AdamsMJ
BarnettOW
BruntAA
HammondJ
2005 Family Potyviridae.
FauquetCM
MayoMA
ManiloffJ
DesselbergerU
BallLA
Virus taxonomy, Classification, Nomenclature of Viruses Eighth Report of the International Committee on Taxonomy of Viruses. Academic Press 819 841
17. KooninEV
WolfYI
NagasakiK
DoljaVV
2008 The Big Bang of picorna-like virus evolution antedates the radiation of eukaryotic supergroups. Nat Rev Microbiol 6 925 939
18. RajamäkiM-L
Mäki-ValkamaT
MäkinenK
ValkonenJPT
2004 Infection with potyviruses.
TalbotN
Plant-Pathogen Interactions Oxford, UK Blackwell Publishing 68 91
19. Urcuqui-InchimaS
HaenniAL
BernardiF
2001 Potyvirus proteins: a wealth of functions. Virus Res 74 157 175
20. ChungBY
MillerWA
AtkinsJF
FirthAE
2008 An overlapping essential gene in the Potyviridae. Proc Natl Acad Sci U S A 105 5897 5902
21. RojasMR
ZerbiniFM
AllisonRF
GilbertsonRL
LucasWJ
1997 Capsid protein and helper component-proteinase function as potyvirus cell-to-cell movement proteins. Virology 237 283 295
22. NicolasO
DunningtonSW
GotowLF
PironeTP
HellmannGM
1997 Variations in the VPg protein allow a potyvirus to overcome va gene resistance in tobacco. Virology 237 452 459
23. KasschauKD
CarringtonJC
1998 A counterdefensive strategy of plant viruses: suppression of posttranslational gene silencing. Cell 95 461 470
24. PuustinenP
MäkinenK
2004 Uridylylation of the potyvirus VPg by viral replicase NIb correlates with the nucleotide binding capacity of VPg. J Biol Chem 279 38103 38110
25. DoljaVV
HaldemanR
RobertsonNL
DoughertyWG
CarringtonJC
1994 Distinct functions of capsid protein in assembly and movement of tobacco etch potyvirus in plants. EMBO J 13 1482 1491
26. DoljaVV
Haldeman-CahillR
MontgomeryAE
VandenBoschKA
CarringtonJC
1995 Capsid protein determinants involved in cell-to-cell and long distance movement of tobacco etch potyvirus. Virology 207 1007 1016
27. Rodríguez-CerezoE
FindlayK
ShawJG
LomonossoffGP
QiuSG
LinsteadP
1997 The coat and cylindrical inclusion proteins of a potyvirus are associated with connections between plant cells. Virology 236 296 306
28. RobertsIM
WangD
FindlayK
MauleAJ
1998 Ultrastructural and temporal observations of the potyvirus cylindrical inclusions (Cls) show that the Cl protein acts transiently in aiding virus movement. Virology 245 173 181
29. RobertsIM
WangD
ThomasCL
MauleAJ
2003 Seed transmission of Pea seed borne mosaic virus in pea exploits novel symplastic pathways and is, in part, dependent upon chance. Protoplasma 222 31 43
30. AvisarD
ProkhnevskyAI
DoljaVV
2008 Class VIII myosins are required for plasmodesmatal localization of a closterovirus Hsp70 homolog. J Virol 82 2836 2843
31. RaffaeleS
BayerE
LafargeD
CluzetS
German RetanaS
2009 Remorin, a solanaceae protein resident in membrane rafts and plasmodesmata, impairs potato virus X movement. Plant Cell 21 1541 1555
32. ThomasCL
BayerEM
RitzenthalerC
Fernandez-CalvinoL
MauleAJ
2008 Specific targeting of a plasmodesmal protein affecting cell-to-cell communication. PLoS Biol 6 e7
33. SagiG
KatzA
Guenoune-GelbartD
EpelBL
2005 Class 1 reversibly glycosylated polypeptides are plasmodesmal-associated proteins delivered to plasmodesmata via the Golgi apparatus. Plant Cell 17 1788 1800
34. daSilvaLL
SnappEL
DeneckeJ
Lippincott-SchwartzJ
HawesC
BrandizziF
2004 Endoplasmic reticulum export sites and Golgi bodies behave as single mobile secretory units in plant cells. Plant Cell 16 1753 1771
35. HarriesPA
ParkJW
SasakiN
BallardKD
MauleAJ
2009 Differing requirements for actin and myosin by plant viruses for sustained intercellular movement. Proc Natl Acad Sci U S A 106 17594 17599
36. HuangTS
WeiT
LaliberteJF
WangA
2010 A host RNA helicase-like protein, AtRH8, interacts with the Potyviral genome-linked protein, VPg, associates with the virus accumulation complex, and is essential for infection. Plant Physiol 152 255 266
37. WeiT
HuangTS
McNeilJ
LalibertéJF
HongJ
2010 Sequential recruitment of the endoplasmic reticulum and chloroplasts for plant potyvirus replication. J Virol 84 799 809
38. LangenbergWG
1993 Structural proteins of three viruses in the Potyviridae adhere only to their homologous cylindrical inclusions in mixed infections. J Struct Biol 110 188 195
39. Rodríguez-CerezoE
AmmarED
PironeTP
ShawJG
1993 Association of the non-structural P3 viral protein with cylindrical inclusions in potyvirus-infected cells. J Gen Virol 74 1945 1949
40. GuoD
RajamäkiM-.L
SaarmaM
ValkonenJPT
2001 Towards a protein interaction map of potyviruses: Protein interaction matrixes of two potyviruses based on the yeast two-hybrid system. J Gen Virol 82 935 939
41. Gómez de CedrónM
OsabaL
LópezL
GarcíaJA
2006 Genetic analysis of the function of the plum pox virus CI RNA helicase in virus movement. Virus Res 116 136 145
42. WenRH
HajimoradMR
2010 Mutational analysis of the putative pipo of soybean mosaic virus suggests disruption of PIPO protein impedes movement. Virology 400 1 7
43. CurtisMD
GrossniklausU
2003 A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133 462 469
44. EarleyKW
HaagJR
PontesO
OpperK
JuehneT
2006 Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45 616 629
45. KarimiM
De MeyerB
HilsonP
2005 Modular cloning in plant cells. Trends Plant Sci 10 103 105
46. NakagawaT
SuzukiT
MurataS
NakamuraS
HinoT
2007 Improved Gateway binary vectors: high-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci Biotechnol Biochem 71 2095 2100
47. WeiT
WangA
2008 Biogenesis of cytoplasmic membranous vesicles for plant potyvirus replication occurs at the endoplasmic reticulum exit sites in a COPI - and COPII-dependent manner. J Virol 82 12252 12264
48. SchaadMC
JensenPE
CarringtonJC
1997 Formation of plant RNA virus replication complexes on membranes: role of an endoplasmic reticulum-targeted viral protein. EMBO J 16 4049 4059
49. AvisarD
ProkhnevskyAI
MakarovaKS
KooninEV
DoljaVV
2008 Myosin XI-K is required for rapid trafficking of Golgi stacks, peroxisomes, and mitochondria in leaf cells of Nicotiana benthamiana. Plant Physiol 146 1098 1108
50. CuiX
WeiT
Chowda-ReddyRV
SunGY
WangA
2010 The Tobacco etch virus P3 protein forms mobile inclusions via the early secretory pathway and traffic along actin microfilaments. Virology 397 56 63
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Insight into the Mechanisms of Adenovirus Capsid Disassembly from Studies of Defensin NeutralizationČlánek Serum-Dependent Selective Expression of EhTMKB1-9, a Member of B1 Family of Transmembrane KinasesČlánek Host Cell Invasion and Virulence in Sepsis Is Facilitated by the Multiple Repeats within FnBPA
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2010 Číslo 6- Stillova choroba: vzácné a závažné systémové onemocnění
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Jak souvisí postcovidový syndrom s poškozením mozku?
- Infekční komplikace virových respiračních infekcí – sekundární bakteriální a aspergilové pneumonie
-
Všechny články tohoto čísla
- Cognitive Dysfunction Is Sustained after Rescue Therapy in Experimental Cerebral Malaria, and Is Reduced by Additive Antioxidant Therapy
- DNA Watermarking of Infectious Agents: Progress and Prospects
- Self-Protection against Gliotoxin—A Component of the Gliotoxin Biosynthetic Cluster, GliT, Completely Protects Against Exogenous Gliotoxin
- Modifies the Tsetse Salivary Composition, Altering the Fly Feeding Behavior That Favors Parasite Transmission
- Requirement of NOX2 and Reactive Oxygen Species for Efficient RIG-I-Mediated Antiviral Response through Regulation of MAVS Expression
- The Enteropathogenic Effector EspF Targets and Disrupts the Nucleolus by a Process Regulated by Mitochondrial Dysfunction
- Epithelial p38α Controls Immune Cell Recruitment in the Colonic Mucosa
- Coexpression of PD-1, 2B4, CD160 and KLRG1 on Exhausted HCV-Specific CD8+ T Cells Is Linked to Antigen Recognition and T Cell Differentiation
- Insight into the Mechanisms of Adenovirus Capsid Disassembly from Studies of Defensin Neutralization
- EspA Acts as a Critical Mediator of ESX1-Dependent Virulence in by Affecting Bacterial Cell Wall Integrity
- An RNA Element at the 5′-End of the Poliovirus Genome Functions as a General Promoter for RNA Synthesis
- Tetherin Restricts Productive HIV-1 Cell-to-Cell Transmission
- Epstein-Barr Virus-Encoded LMP2A Induces an Epithelial–Mesenchymal Transition and Increases the Number of Side Population Stem-like Cancer Cells in Nasopharyngeal Carcinoma
- Protein Kinase A Dependent Phosphorylation of Apical Membrane Antigen 1 Plays an Important Role in Erythrocyte Invasion by the Malaria Parasite
- Requirement for Ergosterol in V-ATPase Function Underlies Antifungal Activity of Azole Drugs
- The SOCS-Box of HIV-1 Vif Interacts with ElonginBC by Induced-Folding to Recruit Its Cul5-Containing Ubiquitin Ligase Complex
- A Crucial Role for Infected-Cell/Antibody Immune Complexes in the Enhancement of Endogenous Antiviral Immunity by Short Passive Immunotherapy
- Modulation of the Arginase Pathway in the Context of Microbial Pathogenesis: A Metabolic Enzyme Moonlighting as an Immune Modulator
- Role of Abl Kinase and the Wave2 Signaling Complex in HIV-1 Entry at a Post-Hemifusion Step
- Serum-Dependent Selective Expression of EhTMKB1-9, a Member of B1 Family of Transmembrane Kinases
- Epigenetic Repression of by Latent Epstein-Barr Virus Requires the Interaction of EBNA3A and EBNA3C with CtBP
- Host Cell Invasion and Virulence in Sepsis Is Facilitated by the Multiple Repeats within FnBPA
- Role of PKR and Type I IFNs in Viral Control during Primary and Secondary Infection
- A Kinome RNAi Screen Identified AMPK as Promoting Poxvirus Entry through the Control of Actin Dynamics
- Cryptococcal Cell Morphology Affects Host Cell Interactions and Pathogenicity
- NleG Type 3 Effectors from Enterohaemorrhagic Are U-Box E3 Ubiquitin Ligases
- Human Cytomegalovirus UL29/28 Protein Interacts with Components of the NuRD Complex Which Promote Accumulation of Immediate-Early RNA
- Complement Receptor 1 Is a Sialic Acid-Independent Erythrocyte Receptor of
- A Viral microRNA Down-Regulates Multiple Cell Cycle Genes through mRNA 5′UTRs
- Immunotoxin Complementation of HAART to Deplete Persisting HIV-Infected Cell Reservoirs
- Protein Expression Redirects Vesicular Stomatitis Virus RNA Synthesis to Cytoplasmic Inclusions
- Entry and Fusion of Emerging Paramyxoviruses
- Paramyxovirus Entry and Targeted Vectors for Cancer Therapy
- Suppressing Glucose Transporter Gene Expression in Schistosomes Impairs Parasite Feeding and Decreases Survival in the Mammalian Host
- Formation of Complexes at Plasmodesmata for Potyvirus Intercellular Movement Is Mediated by the Viral Protein P3N-PIPO
- Fungal Cell Gigantism during Mammalian Infection
- Two Novel Point Mutations in Clinical Reduce Linezolid Susceptibility and Switch on the Stringent Response to Promote Persistent Infection
- Rotavirus Structural Proteins and dsRNA Are Required for the Human Primary Plasmacytoid Dendritic Cell IFNα Response
- The Epigenetic Landscape of Latent Kaposi Sarcoma-Associated Herpesvirus Genomes
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Requirement of NOX2 and Reactive Oxygen Species for Efficient RIG-I-Mediated Antiviral Response through Regulation of MAVS Expression
- Formation of Complexes at Plasmodesmata for Potyvirus Intercellular Movement Is Mediated by the Viral Protein P3N-PIPO
- Insight into the Mechanisms of Adenovirus Capsid Disassembly from Studies of Defensin Neutralization
- Two Novel Point Mutations in Clinical Reduce Linezolid Susceptibility and Switch on the Stringent Response to Promote Persistent Infection
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



