-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaA Viral microRNA Down-Regulates Multiple Cell Cycle Genes through mRNA 5′UTRs
Global gene expression data combined with bioinformatic analysis provides strong evidence that mammalian miRNAs mediate repression of gene expression primarily through binding sites within the 3′ untranslated region (UTR). Using RNA induced silencing complex immunoprecipitation (RISC-IP) techniques we have identified multiple cellular targets for a human cytomegalovirus (HCMV) miRNA, miR-US25-1. Strikingly, this miRNA binds target sites primarily within 5′UTRs, mediating significant reduction in gene expression. Intriguingly, many of the genes targeted by miR-US25-1 are associated with cell cycle control, including cyclin E2, BRCC3, EID1, MAPRE2, and CD147, suggesting that miR-US25-1 is targeting genes within a related pathway. Deletion of miR-US25-1 from HCMV results in over expression of cyclin E2 in the context of viral infection. Our studies demonstrate that a viral miRNA mediates translational repression of multiple cellular genes by targeting mRNA 5′UTRs.
Published in the journal: . PLoS Pathog 6(6): e32767. doi:10.1371/journal.ppat.1000967
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1000967Summary
Global gene expression data combined with bioinformatic analysis provides strong evidence that mammalian miRNAs mediate repression of gene expression primarily through binding sites within the 3′ untranslated region (UTR). Using RNA induced silencing complex immunoprecipitation (RISC-IP) techniques we have identified multiple cellular targets for a human cytomegalovirus (HCMV) miRNA, miR-US25-1. Strikingly, this miRNA binds target sites primarily within 5′UTRs, mediating significant reduction in gene expression. Intriguingly, many of the genes targeted by miR-US25-1 are associated with cell cycle control, including cyclin E2, BRCC3, EID1, MAPRE2, and CD147, suggesting that miR-US25-1 is targeting genes within a related pathway. Deletion of miR-US25-1 from HCMV results in over expression of cyclin E2 in the context of viral infection. Our studies demonstrate that a viral miRNA mediates translational repression of multiple cellular genes by targeting mRNA 5′UTRs.
Introduction
The recent discovery of a new class of regulatory genes known as microRNAs (miRNAs) has resulted in a paradigm shift in gene regulation research. miRNAs are small single-stranded RNA species of approximately 20–24 bases in length that regulate gene expression through post transcriptional mechanisms [1]. Expression of miRNAs is thought to be ubiquitous among multicellular eukaryotes [2]. In addition to eukaryotic miRNAs, more than 100 viral miRNAs have been identified, almost all of which are expressed by herpesviruses [3]. Targets for the majority of viral miRNAs are currently unknown due to the difficulty involved in identifying novel target transcripts. This remains one of the major challenges in elucidating the function of miRNAs. However, recent reports have begun to elucidate the various roles of viral miRNAs. These include blocking apoptosis, immune evasion and regulation of viral replication through targeting of both cellular and viral gene expression [4], [5].
HCMV, a member of the beta-herpesvirus sub family, encodes at least 11 miRNAs [6]–[8]. Previously, we demonstrated that the HCMV encoded miRNA, miR-UL112-1, targets a number of the virus's own genes, including the immediate early transactivator IE72 which is essential for driving acute replication of HCMV [9]. In addition, miR-UL112-1 targets the cellular gene MICB, resulting in protection against recognition by natural killer cells [10]. miR-UL112-1 may therefore play an important role in establishing and maintaining viral latency and persistence through regulation of viral gene expression and subversion of host antiviral pathways. Indeed, consensus is beginning to emerge that herpesvirus miRNAs may, in general, be important in establishing and maintaining viral persistence [9], [11]–[13].
Current studies indicate that miRNA targeting in mammalian cells occurs predominantly through binding to sequences within 3′UTRs [14]–[16]. The reason for this bias is unclear, although a recent study demonstrated that miRNA-mediated repression of a reporter construct was less efficient when the target site was placed in the ORF compared to the 3′UTR [17]. In contrast, inhibition of gene expression through targeting the 5′UTR has been demonstrated, at least in the context of an artificial reporter construct, indicating that miRNA targeting of 5′UTRs is possible [18]. However, statistical analysis of conserved miRNA target sequences and global biochemical screens have demonstrated that mammalian miRNA target sites rarely occur within 5′UTRs [14]–[16], [19]. The role of 5′UTRs in miRNA regulation is further complicated by a study that found miR-10a induces, rather than inhibits, protein expression through binding to 5′UTRs of cellular transcripts [20]. In addition, binding of the liver specific miRNA, miR-122 to the 5′UTR of Hepatitis C (HCV) genome is required for virus replication [21]. These studies suggest a model in which binding to the 5′UTR results in mechanistic effects divergent from 3′UTR binding.
Here, we identify cellular transcript targets of one of the most highly expressed HCMV encoded miRNAs, miR-US25-1, using a recently developed biochemical approach called RISC immunoprecipitation. Strikingly, the majority of identified transcripts contained miR-US25-1 target sites within the 5′UTR rather than the 3′UTR. The target transcripts include a number of genes associated with cell cycle control, including cyclin E2, as well as histone proteins, suggesting that miR-US25-1 is targeting functionally related genes. Crucially, we demonstrate that targeting of cyclin E2 by miR-US25-1 occurs in the context of HCMV infection and results in inhibition of cyclin E2 protein expression.
Results
Identification of cellular targets of miR-US25-1 through RISC immunoprecipitation
Identifying target transcripts is still one of the main challenges in determining the functional role of miRNAs. Although bioinformatic strategies have proven useful, they are limited by high false positive rates. Due to a lack of effective approaches, target transcripts for the majority of viral miRNAs, and miRNAs in general remain unknown. Alternative experimental approaches for the identification of miRNA targets that do not completely rely on bioinformatic predictions are therefore desirable. Recently, approaches for miRNA target identification have been devised that rely on immunoprecipitation of the RISC complex and the associated target transcripts [22], [23]. As part of the RISC complex, miRNAs bind to target transcripts and form stable interactions. Using a HEK293 cell line that expresses a tagged component of RISC (Argonaute 2 (Ago2) tagged with myc epitope) these complexes can be immunoprecipitated and targeted transcripts identified by microarray analysis (Figure 1a). This approach was used to identify cellular targets of one of the most highly expressed HCMV miRNAs, miR-US25-1. The Ago2 tagged cell line was transiently transfected with a construct encoding the full-length pre-miRNA of miR-US25-1, under the control of the human U6 polymerase III promoter. Three days post transfection cells were harvested and RISC complexes were immunoprecipitated with a myc-epitope specific antibody. mRNA levels within the immunoprecipitated complexes as well in whole cell lysates were quantified by microarray analysis. As previously described [22], [23], association of a specific mRNA with the RISC complex is represented by quantitative enrichment of the mRNA in the immunoprecipitated fraction relative to the total (whole cell) fraction. In order to determine which targets are specifically associated with miR-US25-1, fold enrichment of transcripts immunoprecipitated from miR-US25-1 transfected cells was compared to values from cells transfected with a vector expressing a negative control miRNA. miR-US25-1 specific targets are only expected to be enriched in cells expressing miR-US25-1. Transcripts were then ranked according to the level of enrichment with the highest enriched transcripts considered potential targets of miR-US25-1 (Table S1). Over all, the results indicate that miR-US25-1 RISC complexes associated with a relatively small population of transcripts (Figure 1b and c) and fold changes were skewed towards enrichment as would be expected if miR-US25-1 were targeting a specific population of genes. The majority of transcripts were not enriched, showing enrichment levels close to 1 (Figure 1b). Thirty-six transcripts showed greater than 2-fold enrichment, while only 1 transcript was reduced by more than 2-fold. The highest level of enrichment was 6.5 fold.
Fig. 1. RISC-IP analysis of HCMV encoded miRNA, miR-US25-1. 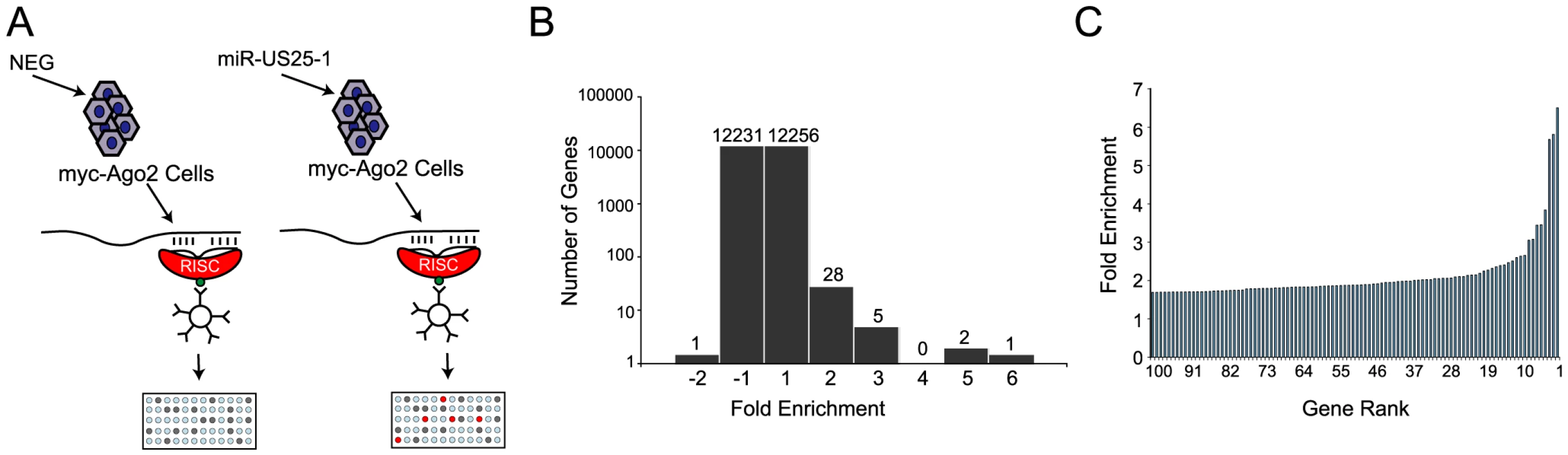
(a) Schematic representation of c-myc tagged Ago2 pull downs. (b) Distribution of fold enrichment and (c) enrichment levels of top 100 transcripts following RISC-IP analysis using c-myc tagged Ago2 approach. To increase confidence in target identification, a second, parallel approach was used in which a biotinylated synthetic mimic of miR-US25-1 was transfected into HEK293 cells and miR-US25-1 specific miRNA protein complexes (miRNPs) were isolated using streptavidin bead pull downs (Figure 2a and Table S2). In contrast to the previous approach this should only pull down direct targets of miR-US25-1. Again, most genes showed little or no enrichment with a relatively small population of transcripts showing exponential increase in enrichment (Figure 2b and c). However, the levels of enrichment were much higher following biotin isolation, reaching a maximum of 23.6 fold. In this case fold changes did not skew towards enrichment as miR-US25-1 was compared to a second HCMV miRNA, miR-US5-2, rather than a negative control. Transcripts showing a negative enrichment ratio, likely represent targets of miR-US5-2.
Fig. 2. RISC-IP analysis of HCMV encoded miRNA, miR-US25-1. 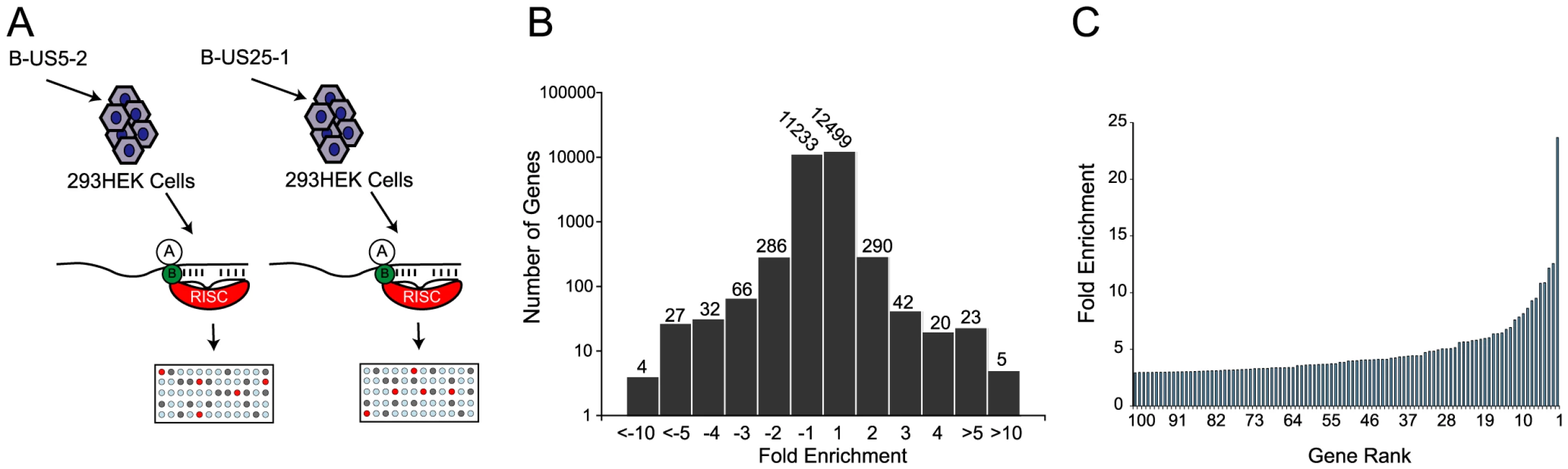
(a) Schematic representation of biotin pull downs. (b) Distribution of fold enrichment and (c) enrichment levels of top 100 transcripts following RISC-IP analysis using biotin approach. The two data sets were compared to determine whether the same genes were identified by both RISC pull down approaches. As the enrichment levels in the biotin approach were higher than those found with the myc-Ago2 approach, averaging the enrichment levels would result in bias towards the biotin data set. To avoid this bias, the data sets were combined using rank sum analysis. Transcripts were assigned a rank based on the comparative level of enrichment (highest enriched = rank 1, lowest = rank 24526) then the average rank between the myc-Ago2 approach and the biotin approach was calculated for each gene. Although differences existed in the rankings of the two data sets (for example TRIM28 was ranked 1st in the c-myc approach, but 188th in the biotin approach) a population of transcripts were enriched by both approaches. Fifteen of the top 20 genes showed greater than 2 fold enrichment by both approaches, giving high confidence that these transcripts were likely targets of miR-US25-1. Table 1 shows the top 20 ranked genes by rank sum analysis including a summary description of their function and the enrichment levels by each approach. A number of these targets are involved in cell cycle control (cyclin E2, BRCC3, PSMA4 and EID1 [24]–[27]) and tumor progression (ASRGL1, CD147, MAPRE2 and ASPSCR1 [28]–[31]), while three of the targets encode histone genes (LOC440093, H3F3B, HIST2H4A), indicating that miR-US25-1 targets functionally related genes.
Tab. 1. Top 20 ranked cellular targets of miR-US25-1. 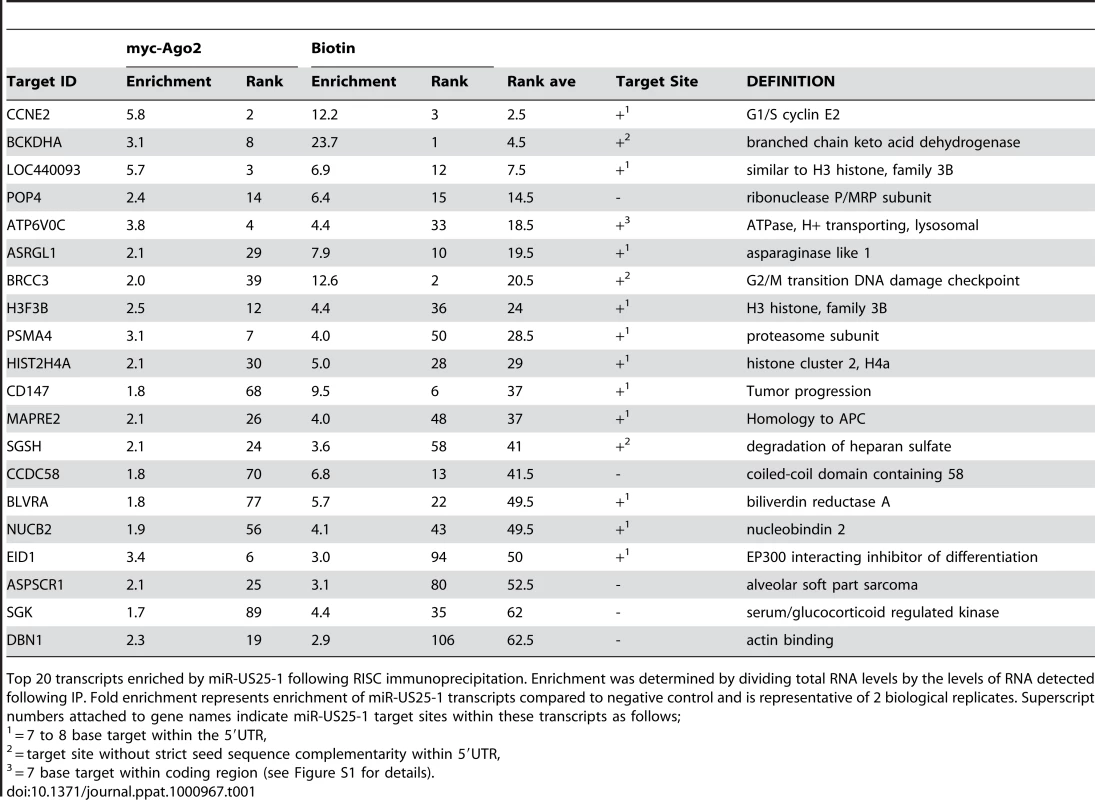
Top 20 transcripts enriched by miR-US25-1 following RISC immunoprecipitation. Enrichment was determined by dividing total RNA levels by the levels of RNA detected following IP. Fold enrichment represents enrichment of miR-US25-1 transcripts compared to negative control and is representative of 2 biological replicates. Superscript numbers attached to gene names indicate miR-US25-1 target sites within these transcripts as follows; miR-US25-1 targets sites within the 5′UTR of transcripts
If the observed enrichment were due to direct targeting by miR-US25-1, the identified population of transcripts would be expected to contain binding sites for the miRNA. Binding of the 5′ end of the miRNA, specifically nucleotides 1–8, known as the seed sequence, are thought to be particularly important [1], [32], [33]. Therefore, the transcripts in the database were searched for seed sequence matches complimentary to nucleotides, 1–7, 2–8 and 1–8 of miR-US25-1. The number of seed matches within the top 50 enriched transcripts from Table S3 was then compared to the number of matches within the rest of the gene list, thereby determining whether genes highly enriched, were more likely to contain predicted miR-US25-1 target sites. Strikingly, miR-US25-1 seed matches were significantly overrepresented within the 5′UTRs of enriched genes (Figure 3 and Table 2). Twelve of the top 20 genes shown in Table 1 contained at least one 7 base seed match within the 5′UTR. In addition, a further 3 genes contain target sites within the 5′UTRs that do not strictly adhere to Watson-Crick base pairing within the seed region. One gene, ATP6V0C, contained a 7 base target within the coding region, 100 bases down stream of the 5′UTR (Table 1 and Figure S1). Crucially the number of target sites within the top 50 genes increased from 19 and 14 with the biotin and c-myc approach individually, to 24 target sites in the combined data set, providing additional evidence that the combined approach provides a more robust method of identifying target transcripts.
Fig. 3. Majority of miR-US25-1 target sites reside within 5′UTRs. 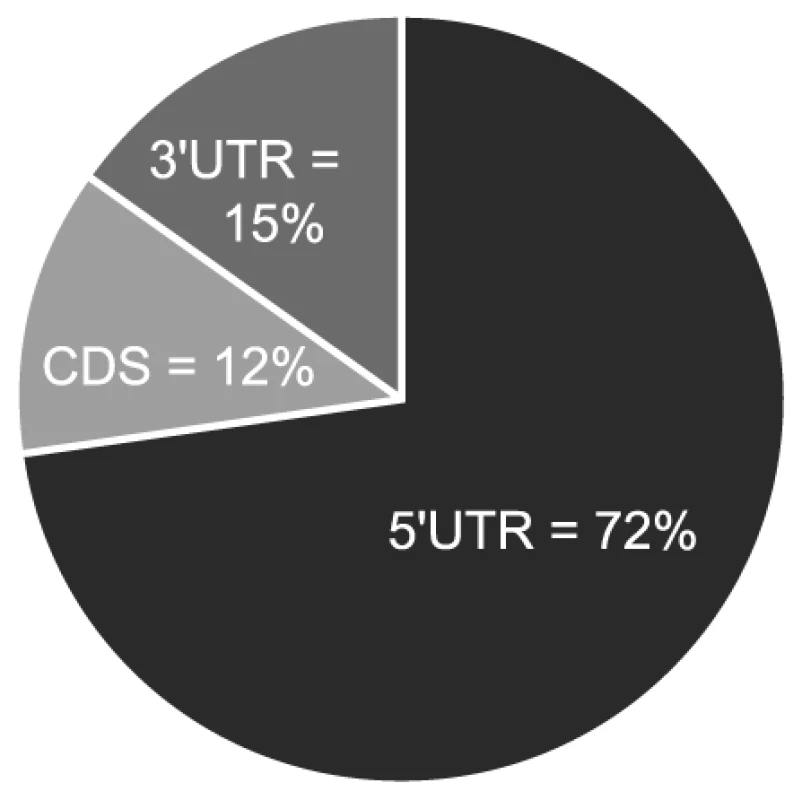
Pie chart shows the percentage of miR-US25-1 seed matches within the 5′UTR, ORF or 3′UTR of top 50 enriched transcripts. Tab. 2. Over representation of seed sequence target sites within top 50 enriched transcripts. 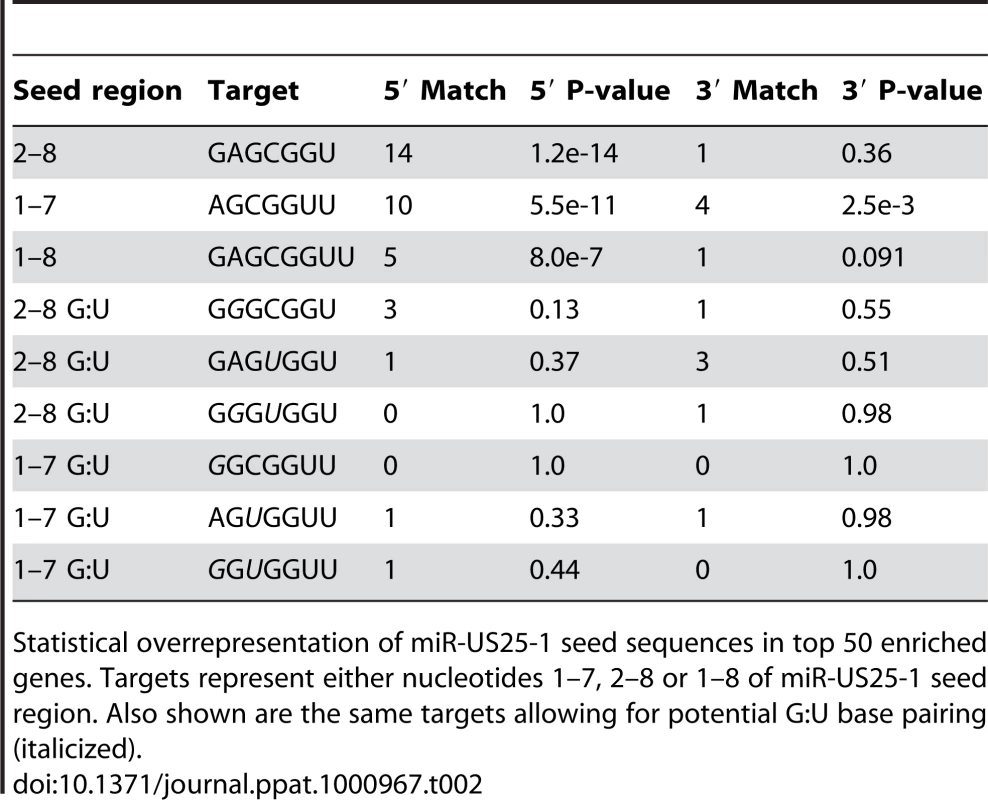
Statistical overrepresentation of miR-US25-1 seed sequences in top 50 enriched genes. Targets represent either nucleotides 1–7, 2–8 or 1–8 of miR-US25-1 seed region. Also shown are the same targets allowing for potential G:U base pairing (italicized). To confirm that enrichment of identified transcripts was due to binding of miR-US25-1 to the 5′UTR, the 5′UTRs and ∼500 bases of upstream genomic sequence of two of the top target genes, cyclin E2 and H3F3B, were cloned in front of a luciferase reporter construct (Figure 4a and b). These constructs were cotransfected into c-myc tagged Ago2 expressing 293 cells with plasmids expressing either miR-US25-1 or the negative control plasmid. RISC-IP analysis was conducted as described above, with levels of luciferase transcript measured using specific RT-PCR primers to the coding region of the reporter gene. Figure 4c shows miR-US25-1 expression resulted in enrichment of luciferase transcript, indicating that the 5′UTRs are indeed sufficient for miR-US25-1 binding. Deletion of the identified seed sequence targets from the 5′UTRs resulted in a loss of enrichment, confirming that the 5′UTR sequences are sufficient and that the target sites are necessary for miR-US25-1 binding.
Fig. 4. miR-US25-1 represses gene expression. 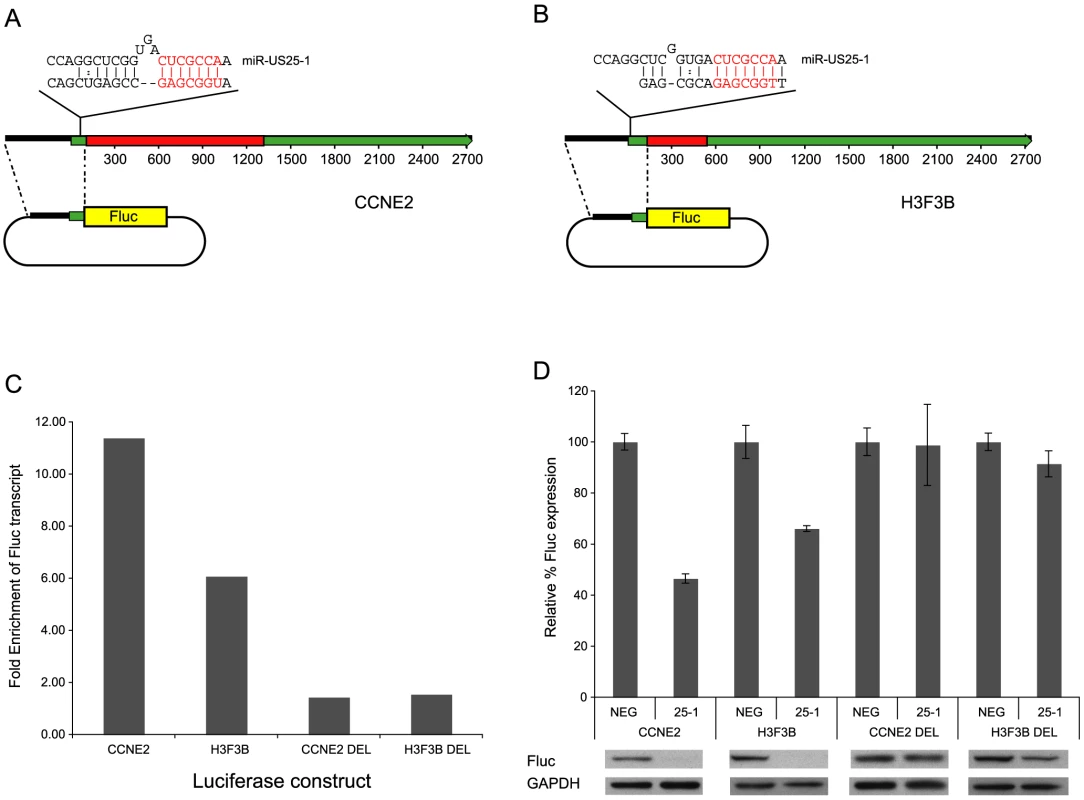
(a) Schematic representation of miR-US25-1 target transcripts, cyclin E2 and (b) H3F3B. Red boxes represent the open reading frames, green boxes the UTRs. The position of the target site within the 5UTR is indicated as well as the predicted binding between miR-US25-1 and target sites. The seed region is highlighted in red. (c) 5′UTR and promoter region of cyclin E2 (CCNE2) and H3F3B were cloned upstream of a luciferase reporter construct. Cells were cotransfected with firefly luciferase (Fluc) constructs and either miR-US25-1 expression plasmid or a negative control plasmid. Following RISC-IP analysis, levels of luciferase transcript were analyzed by RT-PCR and enrichment determined by comparing IP levels with total levels. Target sites were replaced with an EcoRI site to create deletion constructs. Bars represent the fold increase in enrichment following miR-US25-1 expression compared to control. Transcript levels were normalized to GAPDH. (d) Luciferase constructs described above were cotransfected with renillin luciferase plasmid and either miR-US25-1 expression plasmid (US25-1) or the control plasmid (NEG). Luciferase activity was normalized to renillin levels then calculated as percentage of the negative control, which was set to 100%. Error bars indicate s.d. from 3 independent experiments. Equivalent western blots for each transfection is shown below luciferase graph indicating Fluc protein levels. Targeting of 5′UTRs by miR-US25-1 results in decreased gene expression
To determine the effect on gene expression of miR-US25-1 binding on the identified 5′UTR targets, luciferase assays were conducted using the 5′UTR constructs described above. In each case expression of miR-US25-1 resulted in a significant reduction in luciferase activity and protein levels (Figure 4d). miR-US25-1 regulation was dependent on the cloned 5′UTRs and the seed target sites as deletion of these target sites rescued expression of luciferase. Expression of miR-US25-1 appears to have resulted in a greater reduction of luciferase protein as determined by western blot, compared to luciferase activity. We speculate that the reduction in luciferase activity is not linearly reflecting the reduction in actually protein levels, possible due to enzymatic nature of the luciferase assay. Direct measurement of luciferase protein may therefore be a more sensitive measure of miRNA regulation.
miR-US25-1 regulates expression of cyclin E2 and TRIM28 during viral infection
Although these results confirm that miR-US25-1 can bind to a specific population of cellular transcripts, it is important to determine whether these genes are targeted in the context of a viral infection. HEK293 cells are not permissive to HCMV and have a different gene expression profile than cells that are HCMV permissive. To enable RISC-IP analysis of permissive cell lines, a direct antibody to Ago2 was generated using a peptide from the N-terminus of the protein. This antibody was shown to efficiently recognize endogenous Ago2 (Figure S2a and b). RISC complexes were immunoprecipitated from either uninfected human primary fibroblast cells or cells infected with HCMV. The associated RNA was isolated and subjected to RT-PCR analysis using primers specific to the top target cyclin E2 and TRIM28. Although TRIM28 was not in the top 20 targets shown in Table 2, it was the top target following immunoprecipitation of tagged Ago2 from cells transfected with the pSIREN construct (Table S1). As shown in Figure 5a, cyclin E2 and TRIM28 were effectively enriched from cells infected with HCMV compared to the uninfected control cells. In addition, immunoprecipitation using a pre-bleed control serum, which is not expected to pull down Ago2, did not result in enrichment, indicating that the effect was specifically due to association with RISC complexes.
Fig. 5. miR-US25-1 targets 5′UTR's in context of viral infection. 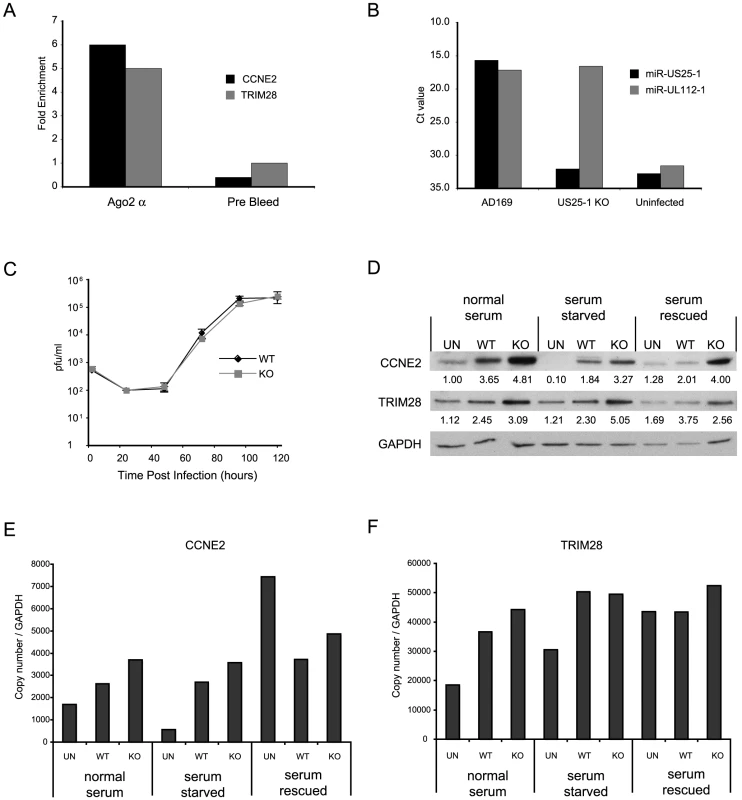
(a) RISC-IP analysis was conducted on uninfected human primary fibroblast cells or cells infected with HCMV using a direct Ago2 antibody. Results show levels of enrichment of cyclin E2 or TRIM28 transcript from infected cells compared to uninfected cells. RISC-IP was also conducted using pre-bleed antibody derived from rabbits before antigen inoculation. (b) miR-US25-1 was deleted from HCMV. Levels of miR-US25-1 and miR-UL112-1 were determined by RT-PCR analysis following infection of human primary fibroblast cells with either wild type HCMV or the knock out virus. RNA from uninfected cells is used as a negative control. (c) Viral growth of miR-US25-1 knock out virus was compared to wild type HCMV following low (MOI of 0.5) or high (MOI of 10) multiplicity infection of human primary fibroblast cells. Cells plus supernatant were collected at indicated times and assayed on primary human fibroblast cells by limiting dilution (d) Levels of cyclin E2 and TRIM28 protein were determined following high multiplicity infection (MOI of 10) of human primary fibroblast cells with either wild type virus or miR-US25-1 knock out virus. Cells were either grown in normal serum conditions, serum starved conditions or serum starved cells with serum replaced 10 hours prior to harvest. Cells were harvested 72 hours post infection. Relative densities of bands normalized to GAPDH are shown below each lane. Total RNA was also isolated and transcript levels for cyclin E2 (e) and TRIM28 (f) determined by RT-PCR. Transcript copy number was normalized to GAPDH levels. To determine the specific effects of miR-US25-1 on the expression of target proteins in the context of viral infection, miR-US25-1 pre-miRNA coding region was deleted from the virus. As shown in Figure 5b, successful disruption of miR-US25-1 expression was confirmed by RT-PCR analysis. Cells infected with wild type HCMV express high levels of both miR-US25-1 and miR-UL112-1, whereas miR-US25-1 levels were below background in cells infected with the knockout virus. miR-UL112-1 levels were equivalent to wild type levels indicating efficient infection with the miR-US25-1 knockout virus. Low (MOI 0.5 – Figure 5c) and high (MOI 10 – supplementary Figure S2c) multiplicity growth curve analysis show the knockout virus was able to replicate with wild type kinetics in human primary fibroblast cells. The effects of virally expressed miR-US25-1 on two top targets, cyclin E2 and TRIM28, were determined by western blot analysis. As cyclin E protein levels are regulated throughout the cell cycle, various serum conditions were used to look at the effects of virus infection during normal cycling populations, populations arrested by serum starvation and cells induced from a resting state using replacement of serum. Western blot analysis indicates that serum starvation effectively repressed cyclin E2 expression as expected and serum rescue resulted in resumption of cyclin E2 expression. Furthermore, as is the case with cyclin E1, cyclin E2 levels were induced by HCMV infection in all serum conditions. Figure 5d also shows a clear increase in expression of cyclin E2, and to a lesser extent TRIM28, in cells infected with the miR-US25-1 knock-out virus compared to wild type infected cells, demonstrating that miR-US25-1 reduces the expression of these target genes. Time course experiments show that expression of cyclin E2 was equivalent between wild type and KO virus infection 24 hours post infection, and regulation by miR-US25-1 does not occur until approximately 48 hours post infection (supplementary Figure S2d). This concurs with previous studies showing levels of miR-US25-1 increase during the progression of the viral infection and suggests that miR-US25-1 levels at 24 hours post infection are not high enough to produce measurable effects on cyclin E2 protein levels [7]. A slight increase (approximately 1.5 fold) was observed in RNA levels of cyclin E2 and TRIM28, consistent with previous reports that miRNA targeting can cause moderate decreases in transcript levels (Figure 5e and f). Following serum rescue, levels of cyclin E2 RNA increase in mock infected cells. The fact that cyclin E2 protein levels do not show a similar increase at this time likely reflects the delay between transcriptional activation and protein translation.
Discussion
These observations provide the first comprehensive identification of multiple cellular targets of a viral miRNA using RISC-IP analysis. Strikingly, the study demonstrates that miR-US25-1 mediates inhibition of gene expression through the novel mechanism of targeting 5′UTR sequences. Furthermore, we demonstrate that miR-US25-1 targets multiple cellular genes related to cell cycle control. To our knowledge this is the first example of a viral miRNA targeting 5′UTRs and is the first demonstration of an endogenous miRNA repressing protein expression through targeting sequences within the 5′UTR.
These results are in contrast to previous studies demonstrating miRNA targeting of 5′UTRs. Targeting of cellular 5′UTRs by miR-10a moderately increased gene expression while targeting of the HCV genome by miR-122 was shown to be required for viral replication [20], [21]. Clearly, targeting of 5′UTRs by miRNAs can mediate distinct positive or negative regulatory effects depending on the context. It will be interesting to determine how miRNAs mediate distinct regulatory effects and whether inhibition of protein expression through miRNA targeting of 5′UTRs is more common than previously thought, or whether this mechanism is specific to miR-US25-1 or viral miRNAs.
The functions of a number of cellular genes identified in this study have important implications for the biology of HCMV and viruses in general. Infection with HCMV has long been known to manipulate the cell cycle by altering the expression of cyclin dependent kinases (CDKs) and their associated cyclin subunits [27]. Cyclin E proteins are expressed early in G1 phase where they bind to and activate CDK2, resulting in progression into S phase. Previous studies have demonstrated that HCMV induces resting G0 cells to enter the cell cycle whereupon the virus blocks further progression at the G1/S boundary [34]. By blocking the cell cycle at the G1/S phase the virus creates a cellular environment conducive for DNA replication. HCMV induced expression of cyclin E1 is thought to play an important role in driving cells into the G1/S phase [35].
Here, we show the virus also induces expression of cyclin E2 early in infection, then moderates cyclin E2 protein levels through targeting by miR-US25-1. miR-US25-1 may therefore function as a rheostat regulator, modulating expression of cyclin E2 to generate the correct balance in protein induction. This may contribute to the viruses ability to block cell cycle progression at the G1/S phase, or to protect the infected cell against toxicity. Over-expression of cyclin E has been linked to sensitivity to apoptosis and unchecked induction of cyclin E2 may be detrimental to the virus [36], [37].
Alternatively, miR-US25-1 function may be unrelated to cell cycle control. Recent studies have suggested that herpesvirus miRNAs may be important during persistent or latent infection [9], [11]–[13]. HCMV is thought to reside within haematopoietic stem cell populations that give rise to latently infected monocyte and macrophage cells [38], [39]. By targeting genes involved in cell cycle progression and differentiation, the virus could manipulate the production of cells generated by latently infected progenitors to favor certain cell types such as monocytes and macrophages. Although deletion of US25-1 did not result in a phenotypic effect on the replication following infection of primary human fibroblast cells, regulation of the target genes identified may be important in other cell types, such as endothelial cells or macrophage cells, or during the latent or persistent phase of the virus life cycle.
Finally, the study of viral miRNAs may provide a powerful method for identifying novel cellular regulatory and antiviral pathways. This study suggests that viral miRNAs, like cellular miRNAs, may function through targeting multiple genes within related pathways. Many of target genes identified in this study are functionally related and contain the same 5′UTR sequence motif. It is likely that miR-US25-1 has evolved to target this 5′UTR motif, thereby subverting the regulatory pathway for the benefit of the virus. Investigation of viral miRNAs may therefore lead to discovery of additional novel cellular pathways.
Materials and Methods
RISC-IP analysis
RISC-IP analysis was carried as out previously described [22], [23]. In brief HEK293 cells stably transfected with c-myc tagged Ago2 were transfected with the pSIREN expression plasmid or a synthetic biotinylated siRNA using Fugene (Roche) or RNAimax (Invitrogen) according to manufacturers specifications. Three days post infection cells were lysed, samples taken for total RNA levels and miRNP complexes immunoprecipitated using anti-c-myc antibody beads (Sigma) or streptavidin beads. RNA was isolated using Trizol and analyzed for quality using an Agilent Bioanalyzer and transcript levels determined on the Illumina HumanRef-8 platform. Microarray data was analyzed using Gene sifter software. Enrichment of specific transcripts, through association with miRNP complexes was determined by dividing the immunoprecipitated levels of transcripts by the total levels, thereby taking into consideration any direct effects of miR-US25-1 on transcript levels. This approach initially identifies any transcript associated with any miRNP complex. To specifically identify those transcripts targeted by miR-US25-1, the enrichment profile was compared to cells transfected with a negative control vector, resulting in exclusion of transcripts enriched through association with cellular miRNAs. Transcripts were then ranked according to the level of enrichment with the highest enriched transcripts considered potential targets of miR-US25-1. For example, in cells transfected with the negative control plasmid, cyclin E2 levels were 1958 in the total sample and 2219 in the IP sample, giving an enrichment value of 1.1. In cells transfected with miR-US25-1 expression plasmid, cyclin E2 levels were 2718 in the total sample compared to 16744 in the IP sample, giving an enrichment value of 6.1. By dividing the enrichment value from cells transfected with US25-1 compared to the control cells (6.1/1.1) the overall enrichment ratio is calculated as 5.4.
Argonaute specific antibody was generated by immunization of rabbits with a peptide corresponding to the N terminal region of Argonaute 2 (5-MYSGAGPALAPPAPPPPIQGYAFKPPPRPD3′). For virus infections, primary human fibroblast cells (Clontech) were infected at high multiplicity (MOI of 10) with the laboratory lab strain AD169. RISC-IP analysis was conducted as above, except antibody to endogenous Ago2 was used to immunoprecipitated miRNP complexes and transcript levels determined using direct RT-PCR primer probe sets (ABI) for CCNE2, TRIM28, and GAPDH.
Sequence analysis
Transcript sequences were down loaded from NCBI using RefSeq ID's. Transcript data sets were searched for seed sequence matches using a Java based script program. Statistical overrepresentation of seed matches within the top 50 transcripts from Table S3 was determined by Fisher exact test. Predicted binding between miR-US25-1 and target sites within the 10 most highly enriched transcripts were determined using the online RNA folding algorithm, mfold (http://mfold.bioinfo.rpi.edu/cgi-bin/rna-form1.cgi).
Luciferase assay
5′UTRs and approximately 500 bases upstream sequence of target genes were PCR amplified from human fibroblast DNA and cloned upstream of the pGL4 luc2 (Promega) firefly luciferase construct. 5′UTR luciferase constructs were cotransfected with a renillin control construct and miRNA mimics using Lipofectamine 2000 (Invitrogen) into HEK293 cells according to manufacturers instructions. Cells were harvested 18 hours post transfection and luciferase levels measured using Promega's dual reporter kit. Protein levels were determined using an anti-firefly luciferase antibody (Sigma).
Western blot analysis
Human primary fibroblast cells were grown in either 10% serum or 0.01% serum for 18 hours before infection at high multiplicity (MOI of 10) with either wild type AD169 or miR-US25-1 knock out virus. Serum rescued cells were recovered with 10% serum 48 hours post infection. Seventy-two hours post infection, cells were harvested using RIPA buffer and total protein levels determined by BCA analysis. Thirty micrograms total protein was loaded and proteins detected using primary antibodies to cyclin E2 (Abcam), TRIM28 (Cellsignal), and GAPDH (Abcam) and secondary HRP-conjugated antibodies (Jackson labs) with ECL reagent (GE bioscience).
miRNA expression cassettes
The predicted pre-miRNA region of miR-US25-1 plus approximately 100 additional bases were PCR amplified (primers shown in supplementary Table S4) and cloned into the pSIREN expression plasmid (Clontech). pSIREN NEG was supplied by Clontech. Synthetic miRNA mimics were purchased from Dharmacon.
miR-US25-1 KO virus
miR-US25-1 pre-miRNA coding region was deleted from AD169 BAC clone using BAC technology as previously described [40]. Briefly a PCR amplified cassette containing FRT flanked Kanamycin was recombined into AD169 BAC genome replacing the miR-US25-1 coding region using primers listed in Table S4. The Kanamycin cassette was then removed by recombining the FRT sites through inducible FLIP recombinase. The resulting BAC was isolated and electroporated into human primary fibroblast cells to produce infectious virus.
RT-PCR analysis
Total RNA was harvested using Trizol and concentrations determined on a nano-drop spectrophotometer. 100 ng of total RNA was then reverse transcribed using either random hexemers or specific RT primers for miRNA RT-PCR. Specific primer probe sets were then used for real time amplification using TAQMAN probes. All primers and probes shown in Table S4. Gene specific primer probe sets were from ABI.
Supporting Information
Zdroje
1. BartelDP
2004 MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116 281 297
2. BartelDP
2009 MicroRNAs: target recognition and regulatory functions. Cell 136 215 233
3. Griffiths-JonesS
GrocockRJ
van DongenS
BatemanA
EnrightAJ
2006 miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res 34 D140 144
4. GottweinE
CullenBR
2008 Viral and cellular microRNAs as determinants of viral pathogenesis and immunity. Cell Host Microbe 3 375 387
5. GreyF
HookL
NelsonJ
2008 The functions of herpesvirus-encoded microRNAs. Med Microbiol Immunol 197 261 267
6. DunnW
TrangP
ZhongQ
YangE
van BelleC
2005 Human cytomegalovirus expresses novel microRNAs during productive viral infection. Cell Microbiol 7 1684 1695
7. GreyF
AntoniewiczA
AllenE
SaugstadJ
McSheaA
2005 Identification and characterization of human cytomegalovirus-encoded microRNAs. J Virol 79 12095 12099
8. PfefferS
SewerA
Lagos-QuintanaM
SheridanR
SanderC
2005 Identification of microRNAs of the herpesvirus family. Nat Methods 2 269 276
9. GreyF
MeyersH
WhiteEA
SpectorDH
NelsonJ
2007 A Human Cytomegalovirus-Encoded microRNA Regulates Expression of Multiple Viral Genes Involved in Replication. PLoS Pathog 3 e163
10. Stern-GinossarN
ElefantN
ZimmermannA
WolfDG
SalehN
2007 Host immune system gene targeting by a viral miRNA. Science 317 376 381
11. MurphyE
VanicekJ
RobinsH
ShenkT
LevineAJ
2008 Suppression of immediate-early viral gene expression by herpesvirus-coded microRNAs: implications for latency. Proc Natl Acad Sci U S A 105 5453 5458
12. UmbachJL
KramerMF
JurakI
KarnowskiHW
CoenDM
2008 MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature 454 780 783
13. ZiegelbauerJM
SullivanCS
GanemD
2009 Tandem array-based expression screens identify host mRNA targets of virus-encoded microRNAs. Nat Genet 41 130 134
14. FarhKK
GrimsonA
JanC
LewisBP
JohnstonWK
2005 The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science 310 1817 1821
15. LewisBP
BurgeCB
BartelDP
2005 Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120 15 20
16. LimLP
LauNC
Garrett-EngeleP
GrimsonA
SchelterJM
2005 Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433 769 773
17. GuS
JinL
ZhangF
SarnowP
KayMA
2009 Biological basis for restriction of microRNA targets to the 3′ untranslated region in mammalian mRNAs. Nat Struct Mol Biol 16 144 150
18. LeeI
AjaySS
YookJI
KimHS
HongSH
2009 New class of microRNA targets containing simultaneous 5′-UTR and 3′-UTR interaction sites. Genome Res 19 1175 1183
19. ChiSW
ZangJB
MeleA
DarnellRB
2009 Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature 460 479 486
20. OromUA
NielsenFC
LundAH
2008 MicroRNA-10a binds the 5′UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell 30 460 471
21. JoplingCL
YiM
LancasterAM
LemonSM
SarnowP
2005 Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science 309 1577 1581
22. EasowG
TelemanAA
CohenSM
2007 Isolation of microRNA targets by miRNP immunopurification. Rna 13 1198 1204
23. KarginovFV
ConacoC
XuanZ
SchmidtBH
ParkerJS
2007 A biochemical approach to identifying microRNA targets. Proc Natl Acad Sci U S A 104 19291 19296
24. BoudreauHE
BroustasCG
GokhalePC
KumarD
MewaniRR
2007 Expression of BRCC3, a novel cell cycle regulated molecule, is associated with increased phospho-ERK and cell proliferation. Int J Mol Med 19 29 39
25. JungT
CatalgolB
GruneT
2009 The proteasomal system. Mol Aspects Med 30 191 296
26. MiyakeS
SellersWR
SafranM
LiX
ZhaoW
2000 Cells degrade a novel inhibitor of differentiation with E1A-like properties upon exiting the cell cycle. Mol Cell Biol 20 8889 8902
27. PaytonM
CoatsS
2002 Cyclin E2, the cycle continues. Int J Biochem Cell Biol 34 315 320
28. AvramisVI
TiwariPN
2006 Asparaginase (native ASNase or pegylated ASNase) in the treatment of acute lymphoblastic leukemia. Int J Nanomedicine 1 241 254
29. IaconoKT
BrownAL
GreeneMI
SaouafSJ
2007 CD147 immunoglobulin superfamily receptor function and role in pathology. Exp Mol Pathol 83 283 295
30. LadanyiM
LuiMY
AntonescuCR
Krause-BoehmA
MeindlA
2001 The der(17)t(X;17)(p11;q25) of human alveolar soft part sarcoma fuses the TFE3 transcription factor gene to ASPL, a novel gene at 17q25. Oncogene 20 48 57
31. WangY
ZhouX
ZhuH
LiuS
ZhouC
2005 Overexpression of EB1 in human esophageal squamous cell carcinoma (ESCC) may promote cellular growth by activating beta-catenin/TCF pathway. Oncogene 24 6637 6645
32. BrenneckeJ
StarkA
RussellRB
CohenSM
2005 Principles of microRNA-target recognition. PLoS Biol 3 e85
33. DoenchJG
SharpPA
2004 Specificity of microRNA target selection in translational repression. Genes Dev 18 504 511
34. KalejtaRF
ShenkT
2002 Manipulation of the cell cycle by human cytomegalovirus. Front Biosci 7 d295 306
35. JaultFM
JaultJM
RuchtiF
FortunatoEA
ClarkC
1995 Cytomegalovirus infection induces high levels of cyclins, phosphorylated Rb, and p53, leading to cell cycle arrest. J Virol 69 6697 6704
36. MazumderS
GongB
AlmasanA
2000 Cyclin E induction by genotoxic stress leads to apoptosis of hematopoietic cells. Oncogene 19 2828 2835
37. UglandH
BoquestAC
NaderiS
CollasP
BlomhoffHK
2008 cAMP-mediated induction of cyclin E sensitizes growth-arrested adipose stem cells to DNA damage-induced apoptosis. Mol Biol Cell 19 5082 5092
38. SinclairJ
SissonsP
2006 Latency and reactivation of human cytomegalovirus. J Gen Virol 87 1763 1779
39. Soderberg-NauclerC
FishKN
NelsonJA
1997 Reactivation of latent human cytomegalovirus by allogeneic stimulation of blood cells from healthy donors. Cell 91 119 126
40. RueCA
JarvisMA
KnocheAJ
MeyersHL
DeFilippisVR
2004 A cyclooxygenase-2 homologue encoded by rhesus cytomegalovirus is a determinant for endothelial cell tropism. J Virol 78 12529 12536
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Insight into the Mechanisms of Adenovirus Capsid Disassembly from Studies of Defensin NeutralizationČlánek Serum-Dependent Selective Expression of EhTMKB1-9, a Member of B1 Family of Transmembrane KinasesČlánek Host Cell Invasion and Virulence in Sepsis Is Facilitated by the Multiple Repeats within FnBPA
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2010 Číslo 6- Stillova choroba: vzácné a závažné systémové onemocnění
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Jak souvisí postcovidový syndrom s poškozením mozku?
- Parazitičtí červi v terapii Crohnovy choroby a dalších zánětlivých autoimunitních onemocnění
-
Všechny články tohoto čísla
- Cognitive Dysfunction Is Sustained after Rescue Therapy in Experimental Cerebral Malaria, and Is Reduced by Additive Antioxidant Therapy
- DNA Watermarking of Infectious Agents: Progress and Prospects
- Self-Protection against Gliotoxin—A Component of the Gliotoxin Biosynthetic Cluster, GliT, Completely Protects Against Exogenous Gliotoxin
- Modifies the Tsetse Salivary Composition, Altering the Fly Feeding Behavior That Favors Parasite Transmission
- Requirement of NOX2 and Reactive Oxygen Species for Efficient RIG-I-Mediated Antiviral Response through Regulation of MAVS Expression
- The Enteropathogenic Effector EspF Targets and Disrupts the Nucleolus by a Process Regulated by Mitochondrial Dysfunction
- Epithelial p38α Controls Immune Cell Recruitment in the Colonic Mucosa
- Coexpression of PD-1, 2B4, CD160 and KLRG1 on Exhausted HCV-Specific CD8+ T Cells Is Linked to Antigen Recognition and T Cell Differentiation
- Insight into the Mechanisms of Adenovirus Capsid Disassembly from Studies of Defensin Neutralization
- EspA Acts as a Critical Mediator of ESX1-Dependent Virulence in by Affecting Bacterial Cell Wall Integrity
- An RNA Element at the 5′-End of the Poliovirus Genome Functions as a General Promoter for RNA Synthesis
- Tetherin Restricts Productive HIV-1 Cell-to-Cell Transmission
- Epstein-Barr Virus-Encoded LMP2A Induces an Epithelial–Mesenchymal Transition and Increases the Number of Side Population Stem-like Cancer Cells in Nasopharyngeal Carcinoma
- Protein Kinase A Dependent Phosphorylation of Apical Membrane Antigen 1 Plays an Important Role in Erythrocyte Invasion by the Malaria Parasite
- Requirement for Ergosterol in V-ATPase Function Underlies Antifungal Activity of Azole Drugs
- The SOCS-Box of HIV-1 Vif Interacts with ElonginBC by Induced-Folding to Recruit Its Cul5-Containing Ubiquitin Ligase Complex
- A Crucial Role for Infected-Cell/Antibody Immune Complexes in the Enhancement of Endogenous Antiviral Immunity by Short Passive Immunotherapy
- Modulation of the Arginase Pathway in the Context of Microbial Pathogenesis: A Metabolic Enzyme Moonlighting as an Immune Modulator
- Role of Abl Kinase and the Wave2 Signaling Complex in HIV-1 Entry at a Post-Hemifusion Step
- Serum-Dependent Selective Expression of EhTMKB1-9, a Member of B1 Family of Transmembrane Kinases
- Epigenetic Repression of by Latent Epstein-Barr Virus Requires the Interaction of EBNA3A and EBNA3C with CtBP
- Host Cell Invasion and Virulence in Sepsis Is Facilitated by the Multiple Repeats within FnBPA
- Role of PKR and Type I IFNs in Viral Control during Primary and Secondary Infection
- A Kinome RNAi Screen Identified AMPK as Promoting Poxvirus Entry through the Control of Actin Dynamics
- Cryptococcal Cell Morphology Affects Host Cell Interactions and Pathogenicity
- NleG Type 3 Effectors from Enterohaemorrhagic Are U-Box E3 Ubiquitin Ligases
- Human Cytomegalovirus UL29/28 Protein Interacts with Components of the NuRD Complex Which Promote Accumulation of Immediate-Early RNA
- Complement Receptor 1 Is a Sialic Acid-Independent Erythrocyte Receptor of
- A Viral microRNA Down-Regulates Multiple Cell Cycle Genes through mRNA 5′UTRs
- Immunotoxin Complementation of HAART to Deplete Persisting HIV-Infected Cell Reservoirs
- Protein Expression Redirects Vesicular Stomatitis Virus RNA Synthesis to Cytoplasmic Inclusions
- Entry and Fusion of Emerging Paramyxoviruses
- Paramyxovirus Entry and Targeted Vectors for Cancer Therapy
- Suppressing Glucose Transporter Gene Expression in Schistosomes Impairs Parasite Feeding and Decreases Survival in the Mammalian Host
- Formation of Complexes at Plasmodesmata for Potyvirus Intercellular Movement Is Mediated by the Viral Protein P3N-PIPO
- Fungal Cell Gigantism during Mammalian Infection
- Two Novel Point Mutations in Clinical Reduce Linezolid Susceptibility and Switch on the Stringent Response to Promote Persistent Infection
- Rotavirus Structural Proteins and dsRNA Are Required for the Human Primary Plasmacytoid Dendritic Cell IFNα Response
- The Epigenetic Landscape of Latent Kaposi Sarcoma-Associated Herpesvirus Genomes
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Requirement of NOX2 and Reactive Oxygen Species for Efficient RIG-I-Mediated Antiviral Response through Regulation of MAVS Expression
- Formation of Complexes at Plasmodesmata for Potyvirus Intercellular Movement Is Mediated by the Viral Protein P3N-PIPO
- Insight into the Mechanisms of Adenovirus Capsid Disassembly from Studies of Defensin Neutralization
- Two Novel Point Mutations in Clinical Reduce Linezolid Susceptibility and Switch on the Stringent Response to Promote Persistent Infection
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



