-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Cell-Autonomous Gβ Signaling Defines Neuron-Specific Steady State Serotonin Synthesis in
Levels of neurotransmitter serotonin synthesis shape disparate behaviors in evolutionary diverse organisms, but the mechanisms defining steady state serotonin synthesis in functionally distinct neuronal types remain unknown. A genetic screen for neuron-specific serotonin synthesis mutants in Caenorhabditis elegans revealed a unique Gβ GPB-1 signaling pathway operating in specific serotonergic neurons to define the baseline expression of serotonin synthesis rate-limiting enzyme tryptophan hydroxylase tph-1. Unlike in canonical heterotrimeric G protein signaling pathways where Gα subunits drive downstream effectors, we found that signaling through Gβ GPB-1 to the OCR-2 TRPV channel defines the baseline tph-1 expression. This Gβ signaling is not required for the establishment or maintenance of the serotonergic cell fates, but dedicated to set steady state 5-HT synthesis in mature neurons. Behavioral analyses showed that 5-HT synthesized in different neurons modulates distinct innate rhythmic behaviors. Our work identified a Gβ-mediated signaling pathway operating in differentiated neuronal cells to specify intrinsic functional diversities, and illuminate a mechanistic principle for genetic programming of neuron-specific steady state 5-HT synthesis in dedicated behavioral circuits.
Published in the journal: . PLoS Genet 11(9): e32767. doi:10.1371/journal.pgen.1005540
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005540Summary
Levels of neurotransmitter serotonin synthesis shape disparate behaviors in evolutionary diverse organisms, but the mechanisms defining steady state serotonin synthesis in functionally distinct neuronal types remain unknown. A genetic screen for neuron-specific serotonin synthesis mutants in Caenorhabditis elegans revealed a unique Gβ GPB-1 signaling pathway operating in specific serotonergic neurons to define the baseline expression of serotonin synthesis rate-limiting enzyme tryptophan hydroxylase tph-1. Unlike in canonical heterotrimeric G protein signaling pathways where Gα subunits drive downstream effectors, we found that signaling through Gβ GPB-1 to the OCR-2 TRPV channel defines the baseline tph-1 expression. This Gβ signaling is not required for the establishment or maintenance of the serotonergic cell fates, but dedicated to set steady state 5-HT synthesis in mature neurons. Behavioral analyses showed that 5-HT synthesized in different neurons modulates distinct innate rhythmic behaviors. Our work identified a Gβ-mediated signaling pathway operating in differentiated neuronal cells to specify intrinsic functional diversities, and illuminate a mechanistic principle for genetic programming of neuron-specific steady state 5-HT synthesis in dedicated behavioral circuits.
Introduction
Serotonin (5-HT) is a neuromodulator implicated in stress-triggered behaviors such as aggression, anxiety, as well as in diverse innate behaviors and physiological processes ranging from food intake to rhythmic motor acts and circadian cycles [1]. Increasing evidence suggests that too much or too little of 5-HT signals from particular neurons contribute to aspects of behavioral and physiological alterations [1,2]. Heterogeneity of 5-HT-producing neurons has been characterized in human and rodent central nervous system (CNS) based on anatomical distributions, axonal trajectories and electrophysiological properties [3]. While the signaling pathways specifying serotonergic cell fates have been studied extensively, little is known about the genetic program defining 5-HT production in mature nervous systems.
Studies in rodents suggest that transcriptional regulatory networks modulate CNS 5-HT synthesis throughout an animal’s life [2,4,5]. In the current paradigm, levels of 5-HT synthesis under favorite environment are considered the steady state or baseline 5-HT signals, and internal and external stressors may further enhance 5-HT synthesis for facilitating behavioral and physiological adaptation [4]. It is generally assumed that the baseline 5-HT synthesis is a feature of default serotonergic cell fates [6]. This view, however, does not address how basal levels of Tph2, encoding the CNS 5-HT synthesis rate-limiting enzyme tryptophan hydroxylase, display stereotyped spatiotemporal changes after the 5-HT cell fate established [7]. Further, early life experience can influence baseline Tph2 expression in the adulthood CNS [4]. Alternative theories propose that additional regulatory programs define 5-HT synthesis [5]. Indeed, the transcription factor PET-1 maintains Tph2 expression in 70% of CNS 5-HT neurons in adult mice to influence anxiety behavior [8]. Thus, steady state 5-HT synthesis could set the tone of innate neural circuitry and influence the sensitivity to stress-induced neural plasticity. However, the genetic programs that define steady state 5-HT synthesis in functionally distinct neurons and its mechanistic relation to stress-induced changes in 5-HT synthesis in an animal remain to be discovered.
We have focused on genetic dissection of serotonergic phenotypes in C. elegans. The small C. elegans nervous system has a classical 5-HT system with the characteristic diversities. Of 302 neurons in a hermaphrodite worm, three pairs, ADF chemosensory neurons, NSM pharyngeal secretory neurons and HSN motor neurons, produce 5-HT [9]. Each pair has unique morphological and functional features but expresses a common set of serotonergic phenotype genes: the sole tryptophan hydroxylase gene tph-1, vesicular monoamine transporter (VMAT) cat-1 for 5-HT release and 5-HT uptake transporter SERT mod-5. Using a green fluorescent protein reporter for tph-1 (tph-1::gfp), our laboratory and others have identified a variety of internal and external stresses via distinct signaling pathways upregulating tph-1 expression in specific neurons to influence behavior, emphasizing tph-1 transcription as a key to dynamic changes in 5-HT signaling [10–13].
The β subunits of the heterotrimeric guanine nucleotide-binding proteins (G proteins) have been implicated in multiple aspects of G protein signaling. Gβ proteins are members of a large family of proteins containing multiple copies of tryptophan-aspartic acid repeat (WD40) motifs that may interact with diverse partners [14]. In canonical G protein signaling pathways, Gβ serves as a negative regulator of Gα interaction with downstream effectors that drive cellular responses [15]. Evidence has been accumulating that Gβ proteins also directly signal to their own effectors, in both Gα-coupled receptor-dependent and receptor-independent mechanisms [16,17]. Co-crystal structure determination and NMR analysis revealed several effector-binding hotspots on the Gβ WD40 repeats domain, and biochemical studies demonstrated that purified Gβ may induce diverse cellular responses by directly interacting with ion channels, enzymes, and arrays of signaling molecules [16,17]. Indeed, pharmacological analyses and cell-based studies have implicated Gβ-mediated signaling in cell cycles, cellular differentiation and stress responses [16–18]. In general, however, the physiological roles of Gβ signaling in native cellular contexts remain to be elucidated.
In this paper, we report a cell-autonomous Gβ signaling defining baseline tph-1 expression. A genetic screen for 5-HT synthesis mutants identified a Gβ GPB-1 to TRPV channel OCR-2 signaling pathway that is necessary and sufficient to define baseline tph-1 expression, thus 5-HT synthesis, in ADF neurons. This GPB-1-mediated signaling is not required for establishment or maintenance of the cell fates, and is mechanistically separated from stress-induced tph-1 upregulation. Our data establish a role for Gβ-mediated signaling operating in mature neurons to specify the intrinsic functional properties. Gβ-mediated signaling could represent a genetic determinant underlying neuron-specific steady state 5-HT synthesis in functionally distinct neurons that regulate specific aspects of innate behavior.
Results
GPB-1 specifically regulates tph-1 expression in ADF neurons
We isolated the yz71 mutant through an unbiased forward genetic screen for mutations that specifically diminished tph-1::gfp expression in ADF serotonergic chemosensory neurons [12]. Under optimal growth conditions, yz71 mutants showed an ~90% reduction in ADF tph-1::gfp throughout the life, as compared to wild-type (WT) animals (Fig 1B and 1C). In contrast, tph-1::gfp in other neurons was not reduced in yz71 mutants (Fig 1B and 1C). 5-HT immunostaining of yz71 mutants showed dramatically diminished 5-HT levels in ADF (Fig 1B and 1D). We therefore concluded that yz71 impairs a gene function specifically required for tph-1 expression, thus 5-HT synthesis, in the ADF neurons.
Fig. 1. gpb-1 deficient mutants specifically diminish 5-HT synthesis in ADF neurons. 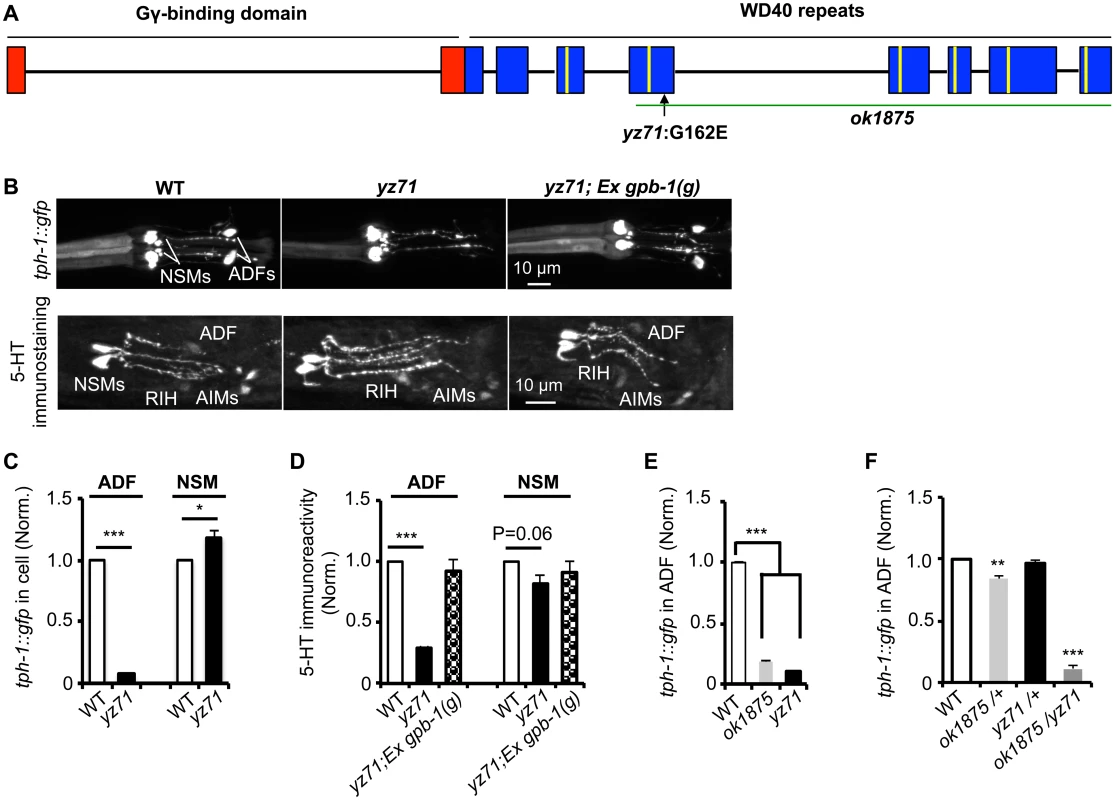
(A) gpb-1 mutant alleles. Lines denote introns and rectangles denote exons of gpb-1 gene. The predicted region binding to Gγ (red) and seven WD40 repeats of the propeller domain (blue) that interacts with Gα are in reference of previous work [28,53]. A green line indicates the deleted region in the ok1875 allele, and an arrow points to the location of the yz71 mutation. (B) Photomicrographs showing tph-1::gfp and 5-HT immunostaining were selectively diminished in ADF neurons in yz71 mutants and rescued by gpb-1(g) transgene. (C—D) Quantification of tph-1::gfp fluorescence (C) and 5-HT immunoreactivity (D) in ADF and NSM neurons in L4 stage animals. (E) Quantification of ADF tph-1::gfp in WT, gpb-1(ok1875)-null and yz71 at L1 stage. (F) Comparing ADF tph-1::gfp in WT with that in ok1875/yz71 hemizygotes, and ok1875/+ and yz71/+ heterozygotes. Data represent the average of 2 (5-HT staining) or 3 (GFP) repeats. N = 18–32/genotype/trial. The value of fluorescence of mutants and transgenic animals is normalized to that of WT animals, mean ± SEM, *p < 0.05, **p < 0.01, ***, p < 0.001, t-tests. Single nucleotide polymorphism (SNP)-based mapping narrowed the yz71 mutation to a 700 kb contig of LG II, where we identified a missense mutation causing a G162E substitution in the third WD40 repeat of the Gβ protein GPB-1 (Fig 1A). We confirmed that GPB-1(G162E) is responsible for diminished 5-HT in yz71 mutants, by determining that transgenic expressing WT genomic gpb-1 sequence restored ADF tph-1::gfp and 5-HT immunostaining (Fig 1B and 1D).
To probe G162E effects on GPB-1 functionality, we analyzed a gpb-1 deletion allele, ok1875. Homozygous gpb-1 null alleles are lethal [19], but those carrying maternal GPB-1 from heterozygous hermaphrodites can survive to larval stage 1 (L1) and displayed reduced ADF tph-1::gfp (Fig 1E). Importantly, a majority of yz71/ok1875 hemizygotes also died at L1, and the few that survived displayed reduced ADF tph-1::gfp (Fig 1F). In contrast, ADF tph-1::gfp levels in both yz71/+ and ok1875/+ heterozygotes were close to WT (Fig 1F). Taken together, these data identified that gpb-1 selectively regulates ADF tph-1 expression, and G162E is a recessive, reduction-of-function gpb-1 allele. Unlike gpb-1 null alleles, yz71 homozygotes are fertile, providing a genetic system for characterizing native GPB-1 function in an animal.
GPB-1(G162E) does not impair serotonergic cell fates
As GPB-1 functions early in embryogenesis [19,20], an immediate question was whether ADF neurons are present in yz71 mutants. In the mammalian CNS, postmitotic precursors of 5-HT neurons are born, migrate and acquire other neuronal features hours to days before committing to express 5-HT phenotype genes [2]. In C. elegans, ADFs are generated and migrate to the amphid sensilla during embryogenesis and switch on tph-1 expression after hatching [9,21]. We therefore investigated if gpb-1(yz71) would disrupt ADF formation, survival, or its serotonergic fate.
We first determined whether amphid sensory neurons were generated in gpb-1(yz71) mutants, using dye DiI filling. The pair of amphids in the head is the major C. elegans sensory organ, each comprising one ADF and eleven other classes of non-serotonergic sensory neurons with characteristic ciliated sensory endings sensing particular environmental cues [22]. We observed DiI filled into the typical six pairs of the amphid neurons in yz71 mutants as in WT animals (Fig 2A). Judged by DiI fluorescence, the overall neuronal organization and their axonal and dendritic morphology were preserved in yz71 mutants (Fig 2A). However, while DiI rarely filled into ADF neurons in WT animals (8%, N = 26), 31% of ADF were filled with DiI in yz71 mutants (N = 65). DiI also filled into a few non-amphid neurons in some yz71 mutants (Fig 2A).
Fig. 2. gpb-1(yz71) does not abrogate ADF cell fates. 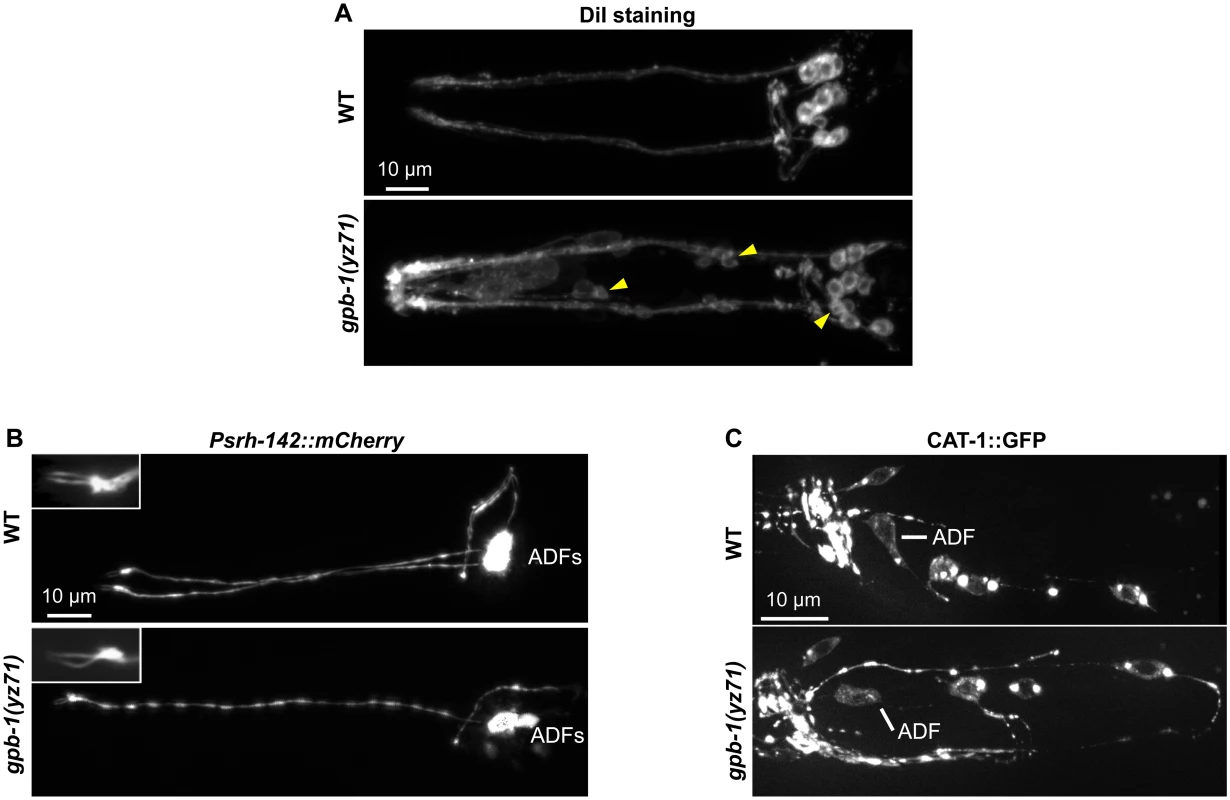
(A) Visualizing amphid neuronal architecture in living animals using DiI filling. In yz71 mutants, DiI filled into the typical 6 pairs of amphid neurons, although the dye could sometimes be detected in several additional neurons (arrowheads). (B) ADF-specific marker Psrh-142::mCherry was expressed and showed characteristic ADF cilia structure (insets) in yz71 mutants. (C) GFP-tagged VMAT CAT-1 protein was expressed in ADF of yz71 mutants. Animals at all developmental stages were examined, and images of L4 animals are shown. We next determined whether yz71 altered gross ADF cell fates, using a mCherry reporter for the ADF-specific marker gene srh-142. Fig 2B shows that Psrh-142::mCherry was expressed in yz71 mutants, and their ADFs displayed the characteristic axon, dendritic and cilia architectures.
To assess the general serotonergic cell fate, we used a functional GFP reporter for VMAT CAT-1 protein [23]. We did not detect an appreciable change in GFP-tagged CAT-1 expression or subcellular localization in yz71 mutants (Fig 2C). These data showed that ADF neurons were generated and maintained serotonergic fates in yz71 mutants, leading to our hypothesis that GPB-1 selectively mediates transcriptional control of 5-HT synthesis.
Deficits in baseline and stress-induced tph-1 expression are mechanistically separable
ADF tph-1 expression is highly sensitive to external and internal environment. Gβ of heterotrimeric G proteins could function in multiple signaling pathways in concert with different Gα subunits [16]. Therefore, GPB-1 function could define a mechanism setting baseline tph-1 expression, as well as be involved in signaling pathway(s) that mediate stress-induced tph-1 upregulation. To discern and define the role of GPB-1 in steady state and stress-induced 5-HT synthesis, we measured tph-1::gfp in yz71 mutants under optimal conditions and two well-established aversive conditions, dauer formation and pathogenic bacterial infection.
A stress-resistant state called dauer can be induced by a defined set of aversive growth conditions and is the most commonly used paradigm in C. elegans stress response studies [24]. Dauers enhance a battery of stress responsive genes to produce a series of physiological, morphological and behavioral remodeling [24]. Previously, we identified that alteration of ADF sensory cilia architecture caused either by dauer formation or genetic mutations in intraflagellar transport (IFT) triggers ADF tph-1 upregulation [10]. When yz71 mutants were induced to form dauers, ADF tph-1::gfp was significantly enhanced compared to their siblings under optimal growth conditions (Fig 3A). Likewise, we observed elevated ADF tph-1::gfp in yz71 mutants carrying defective IFT component che-2 (Fig 3B). However, ADF tph-1::gfp levels in yz71 dauers and yz71;che-2 double mutants were lower than WT dauers and che-2 single mutants, respectively (Fig 3A and 3B). These observations support the idea that dauer formation and GPB-1 regulate two mechanistically separable layers of tph-1 expression. yz71 mutants do not impair tph-1 upregulation induced by dauer formation or cilia structural alterations but have a lower baseline of tph-1 expression.
Fig. 3. tph-1::gfp expression in gpb-1(yz71) mutants under optimal and aversive conditions. 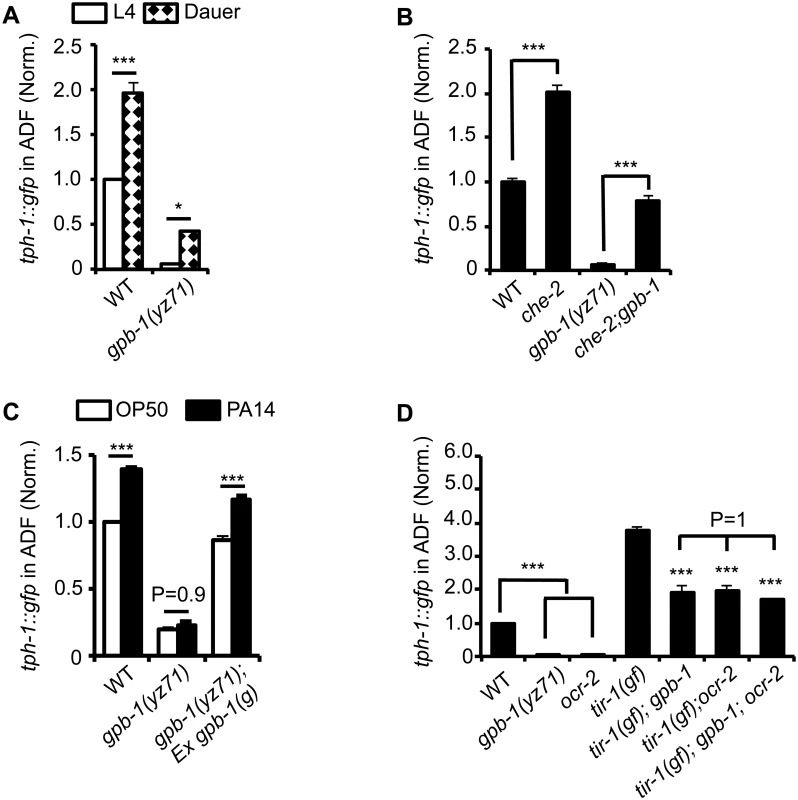
(A) Dauer formation induced by aversive growth conditions caused ADF tph-1::gfp upregulation in WT and yz71mutants, as compared to their respective L4 stage siblings. (B) Mutation in cilia structural gene che-2 was capable of triggering ADF tph-1::gfp upregulation in yz71 mutants. (C) Pathogen PA14 failed to induce ADF tph-1::gfp upregulation in yz71 mutants, as comparing tph-1::gfp between 1st day adults fed PA14 and non-pathogenic bacterial control OP50 for 6 hr. gpb-1(g) transgene restored the PA14 response in yz71 mutants. (D) Comparing ADF tph-1::gfp between gpb-1(yz71) and ocr-2 TRPV channel mutants. In both mutants, ADF tph-1::gfp was diminished under optimal growth conditions, but enhanced in tir-1(yz68gf) background as compared to the yz71 and ocr-2 single mutants. yz71 and ocr-2 did not produce an additive effect. For each assay, the value of ADF GFP fluorescence in WT animals under a stress paradigm and that of mutants is normalized to the value of WT animals under optimal conditions. Data represent the average of ≥ 3 trials ± SEM, * p < 0.05, *** p < 0.001, t-test for two group comparisons, and ANOVA for multi-group/condition comparisons. Pathogenic bacterial infection is another established aversive paradigm inducing ADF tph-1 upregulation. Feeding worms the human opportunistic pathogen Pseudomonas aeruginosa strain PA14, instead of nutritious E. coli OP50, specifically induces ADF tph-1 upregulation to facilitate behavioral avoidance of the pathogenic food [11]. PA14-induced ADF tph-1::gfp upregulation requires Gqα protein EGL-30 [25]. Consistent with Gβ as a regulator of Gα signaling, yz71 mutants failed to upregulate ADF tph-1::gfp during PA14 infection, and the gpb-1(g) transgene rescued the PA14 sensitivity (Fig 3C). However, whereas both egl-30(lf) and egl-30(gf) abrogated PA14-induced tph-1::gfp upregulation, neither the egl-30 mutation dramatically diminished ADF tph-1::gfp [10,25], suggesting that the baseline and pathogen-induced tph-1 expression are mediated by mechanistically separated GPB-1 functions.
Previous work established that PA14 triggers ADF tph-1 upregulation via a cell-autonomous Toll-interlukin-1 receptor domain adaptor protein TIR-1 activated MAPK signaling pathway [12,26]. As another method to distinguish GPB-1 function between the baseline and pathogen-induced tph-1 expression, we generated double mutants of yz71 and tir-1(yz68gf). tir-1(gf) mutants constitutively enhance ADF tph-1::gfp by activating the MAPK signaling [12]. If GPB-1 functions purely in the pathogen-responsive pathway, tir-1(gf);yz71 double mutants would display ADF tph-1::gfp levels similar to one of the single mutants. In contrary, tir-1(gf);yz71 double mutants displayed an intermediate ADF tph-1::gfp level between the two single mutants (Fig 3D). This result suggests that GPB-1 functions upstream of TIR-1 in the pathogen responsive pathway, while a TIR-1-independent GPB-1 function defines baseline tph-1 expression.
We further tested this hypothesis by generating double mutants of tir-1(gf) and ocr-2. Disruption of cell-autonomous OCR-2 TRPV channel specifically diminishes ADF baseline tph-1 expression [12,27]. If yz71 disrupts the OCR-2 pathway, ADF tph-1::gfp levels in tir-1(gf);ocr-2 and tir-1(gf);yz71 should be similar. This was what we observed (Fig 3D). Further, tir-1(gf);gpb-1(yz71);ocr-2 triple mutants did not display a lower ADF tph-1::gfp level compared to the double mutants (P = 1, ANOVA) (Fig 3D). Combined, these data suggest that GPB-1 function is essential for EGL-30 Gqα signaling during pathogen infection, and an EGL-30 - and TIR-1-independent GPB-1 function in the OCR-2 signaling pathway directs baseline ADF tph-1 expression.
Cell-autonomous GPB-1 directs baseline tph-1 expression
GPB-1 is expressed broadly in neurons and non-neuronal tissues [19]. To identify and discern the GPB-1 action sites for basal and pathogen-induced tph-1 expression, we generated transgenic lines expressing gpb-1 cDNA in defined cells and tested their ability to rescue ADF tph-1::gfp expression in yz71 mutants under optimal conditions and during PA14 infection.
We first expressed GPB-1 in all neurons, or in hypodermal and glial cells surrounding amphid sensory cilia, or in the gut. Fig 4A shows that expressing GPB-1 in neurons, but not in the other tissues, rescued both the basal and PA14-induced tph-1::gfp expression.
Fig. 4. Baseline and pathogen-induced ADF tph-1::gfp expression involve separated GPB-1 functions. 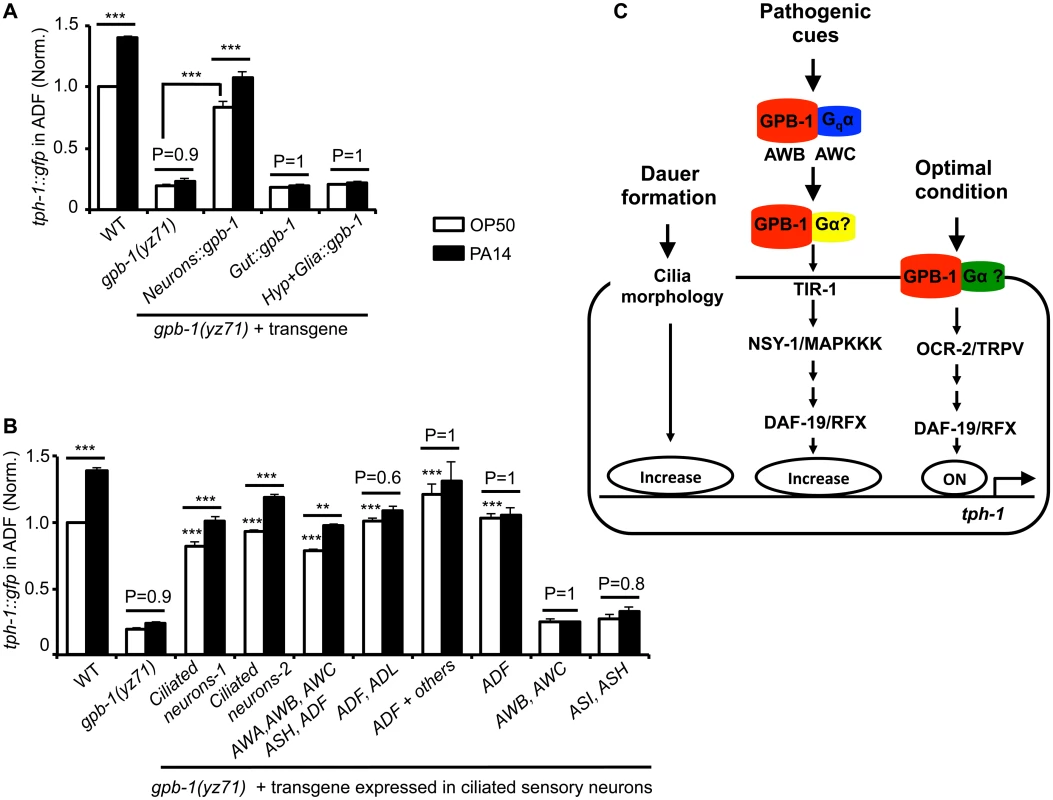
(A) Transgenic expressing gpb-1 cDNA in all neurons rescued both baseline and pathogen PA14-induced ADF tph-1::gfp expression in yz71 mutants. (B) Expressing gpb-1 cDNA specifically in ADF restored baseline tph-1::gfp but not the response to PA14. ADF is the only neuronal type expressed in common by the three transgenes that selectively rescued the baseline ADF tph-1::gfp (the ADF and ADL expression was driven by a lin-11 promoter, and the ADF + others expression was driven by the cat-1 promoter). The transgenes expressed in additional amphid neurons including ADF rescued both the baseline and PA14-induced tph-1::gfp expression, but the transgenes that are not expressed in ADF failed to rescue under either condition. For each assay, the value of ADF GFP fluorescence in WT animals fed PA14 and that of mutants and transgenic animals is normalized to the value of WT animals under optimal conditions. Data represent the average of ≥ 2 trials ± SEM. The differences between yz71 mutants and the transgenic animals under optimal growth conditions are marked on the top of white bars, and the differences between the same strains fed OP50 and PA14 are indicated, ** p < 0.01, *** p < 0.001, ANOVA followed by Tukey tests. (C) Model for GPB-1 regulation of the baseline and pathogen-induced tph-1 expression in ADF. Cell-autonomous GPB-1 is necessary and sufficient for the baseline tph-1 expression. GPB-1 regulates PA14-induced tph-1 upregulation by controlling Gqα EGL-30, which acts in AWB and AWC neurons in the pathogen-induced signaling pathway [25], and other Gα signaling in additional amphid neurons. As Gqα EGL-30 functions in AWB and AWC amphid neurons to mediate PA14-induced tph-1::gfp upregulation in ADF [25], we tested whether GPB-1 acts in AWB and AWC to direct both the basal and PA14-induced tph-1 expression in ADF. Surprisingly, expressing GPB-1 in AWB and AWC failed to rescue tph-1::gfp under either condition (Fig 4B). In contrast, expressing GPB-1 in all the ciliated neurons or selectively in AWA, AWB, AWC, ASH and ADF restored ADF tph-1::gfp under both the conditions (Fig 4B). To probe a cell-autonomous role for GPB-1 in ADF, we generated two transgenes that share in common only in expressing GPB-1 in ADF and a third transgene expressing GPB-1 specifically in ADF. All the three transgenes robustly rescued the basal ADF tph-1::gfp expression, but none responded to PA14 (Fig 4B). We concluded that pathogen-induced tph-1 upregulation requires GPB-1 in AWB, AWC and additional amphid sensory neurons. In contrast, cell autonomous GPB-1 function is necessary and sufficient to define the baseline tph-1 expression in the ADF neurons (Fig 4C).
G162E alters a conserved residue at the interface between Gβ and switch II helix of Gα
How might GPB-1(G162E) disrupt G protein functionalities? GPB-1 shares 86% amino acid identity with human and bovine Gβ1 (Fig 5A). To gain insights into GPB-1 functional mechanisms regulating tph-1 expression, we modeled GPB-1(G162E) over the crystallographic structure of rat Giα1 and bovine β1γ2 heterotrimer [28]. The seven WD40 repeats of Gβ fold into a circular β-bladed propeller which makes two major contacts with Gα: the side of the propeller interacts with the Gα N-terminal helix, and the narrow propeller region forms an interface with the Gα switch II helix [28] (Fig 5Bi and 5Bii). Formation of Gβ/Gα switch II interface favors the inactive GDP-binding state, whereas GTP binding of Gα triggers the switch II helix rotating 120° thereby permitting both Gβ and Gα proteins to interact with their effectors [28,29]. G162E occurs right at the Gβ/Gα switch II interface (Fig 5Biii and 5Biv).
Fig. 5. G162E occurs at the Gβ surface that binds to Gα switch II helix and Gβ effectors. 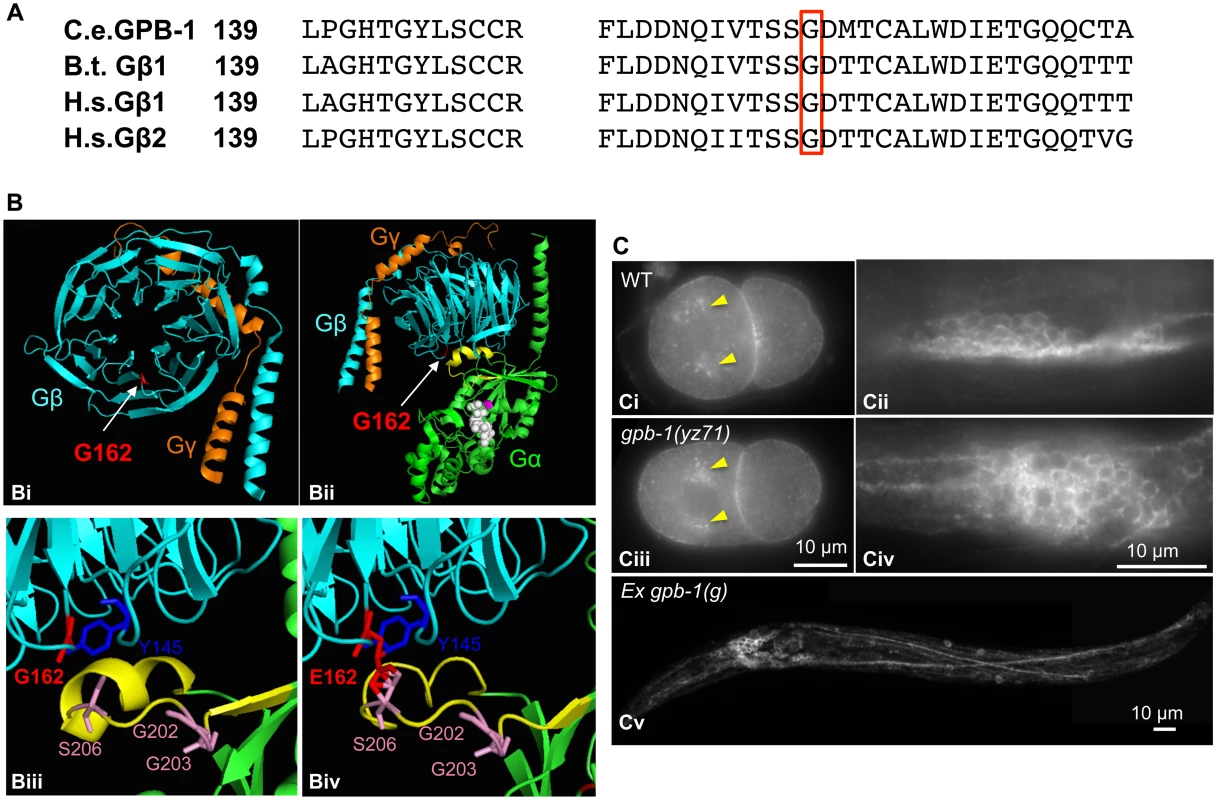
(A) Comparing amino acid sequence surrounding G162 (red boxed) between C. elegans GPB-1, bovine (B.t.) Gβ1, and human (H.s.) Gβ1 and Gβ2, all of which contain 340 amino acid residues and the numbers refer to residue positions in the corresponding proteins. (B) Modeling G162E substitution over bovine β1. (Bi) Cartoon presentation of the crystallographic structure of rat Giα1 and bovine β1γ2 heterotrimer rendered with PyMOL software. β subunit is colored cyan, and γ subunit colored orange. G162 (the side chain colored red) is located on the surface of the narrow side of the propeller architecture of Gβ. (Bii) Another view of the heterotrimer showing the interactions between Gβ and GDP-bound Gα (green). G162 is located on the surface that binds to the Gα switch II helix (yellow). GDP is in white, and the nearby Gα-S47 that is critical for the GTP binding is colored magenta. (Biii) An enlarged view showing the interface between Gβ and Gα switch II helix. Gβ-G162 sits nearby Gβ-Y145, a critical residue in an effector binding hotspot [17]. Gβ-G162 is in close proximity to Gα-S206 in the switch II helix. The conformation of Gα-G202 and G203 is critical for GTP binding [32]. (Biv) An enlarged view showing the Gβ and Gα switch II interface, with G162 changed to E in the Gβ sequence. (C) GPB-1 immunostaining. Ci and Ciii. In both WT and yz71 two-cell embryos, GPB-1 was enriched in the asters (arrowheads) and in the region between cells. Cii and Civ. In WT and yz71 animals, GPB-1 can be detected in the cell membrane in neurons. Cv. gpb-1(g) overexpression transgenic animals showing GPB-1 localized to axons and dendrites. Because gpb-1(yz71) is a reduction-of-function allele, G162E is likely to disrupt Gβ gene function. Whole mount GPB-1 immunostaining indicated that G162E does not grossly impair GPB-1 expression or subcellular localization. For example, both yz71 and WT 2-cell-stage embryos showed GPB-1 enriched in the aster and cell membranes between cells (Fig 5Ci and 5Ciii). In yz71 animals, as in WT and GPB-1-overexpressing animals, GPB-1 was detected in neuronal and non-neuronal tissues, and localized to the cell membrane as well as in axons and dendrites in neurons (Fig 5Cii, 5Civ and 5Cv), as seen in published GPB-1 expression patterns using different GPB-1 antibodies [19]. Because heterotrimeric complex formation is required for Gβ membrane localization [28], our data suggest that GPB-1(G162E) is likely capable of forming G protein complexes but disrupts the signaling.
Diminished baseline tph-1 in gpb-1 mutants is not caused by reduced Gα signaling
Gβ could serve as a regulator of Gα signaling that drives tph-1 expression. Alternatively, Gβ itself could function as the signaling molecule to define tph-1 expression in ADF neurons. We first considered the possibility that altered Gα-mediated signaling underscores diminished tph-1 expression in gpb-1 mutants. ADF expresses seven Gα genes. Loss-of-function mutants of egl-30, gpa-3, gpa-10, gpa-13 and odr-3 did not dramatically diminish ADF tph-1::gfp [10,25,27]. gsa-1(lf) is lethal, but RNAi of gsa-1 also did not diminish ADF tph-1::gfp (134±14%, N = 50) compared to empty vector control (N = 28). Further, two deletion alleles of Goα goa-1, n363 and sa734, both markedly elevated ADF tph-1::gfp [10,25] (Fig 6A). Repeated attempts failed to generate a goa-1(n363);gpb-1(yz71) double mutant. As both GOA-1 and GPB-1 function in early embryogenesis [19,20,30], eliminating GOA-1 in gpb-1(yz71) background could be lethal. We therefore generated a double mutant of ocr-2 and goa-1(n363), and found that ocr-2;goa-1(n363) double mutants displayed diminished ADF tph-1::gfp as in ocr-2 single mutants (Fig 6A), suggesting that the absence of GOA-1 protein elevated tph-1 expression by activating the OCR-2 TRPV pathway.
Fig. 6. GPB-1 regulates baseline ADF tph-1::gfp expression in a GOA-1-dependent manner. 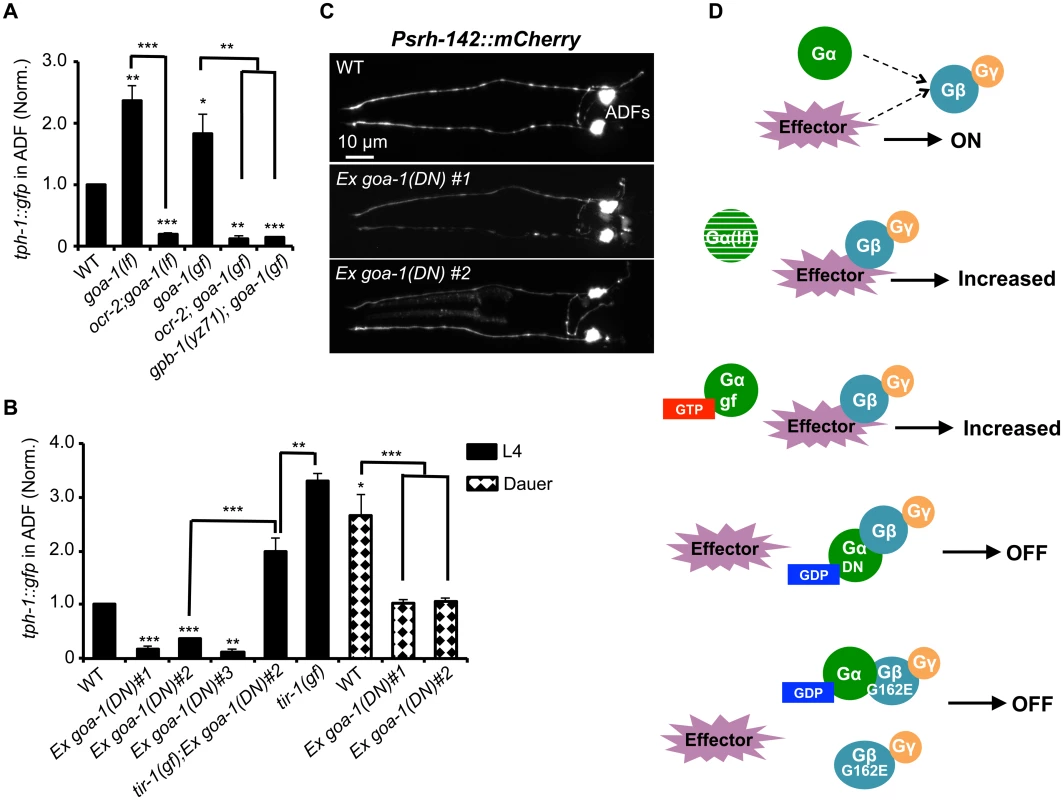
(A) Both GOA-1 deletion (goa-1lf) and GTP-bound hyperactive GOA-1(Q205L) (goa-1gf) mutations elevated ADF tph-1::gfp compared to WT. gpb-1(yz71) and ocr-2 TRPV mutations suppressed tph-1::gfp in goa-1(lf) and goa-1(gf) backgrounds. (B) Dominant negative GDP-bound GOA-1(S47C) transgene (goa-1DN) diminished ADF tph-1 expression in three independent transgenic lines. goa-1(DN) transgenic animals remained capable of enhancing ADF tph-1::gfp when induced to form dauers or in tir-1(gf) background. For each assay, the value of ADF GFP fluorescence in dauers and that of mutants and transgenic animals is normalized to the value of WT animals under optimal conditions. Data represent the average of ≥ 2 trials ± SEM. The differences between WT and mutants are marked on the top of bar, and the differences between comparison groups are indicated, *p < 0.05, ** p < 0.01, *** p < 0.001, t-test for two group comparisons, and ANOVA for multi-group/condition comparisons. (C) ADF-specific marker Psrh-142::mCherry was expressed and showed characteristic ADF axon and dendritic morphology in goa-1(DN) transgenic animals. Animals at all developmental stages were analyzed, and images of L4 animals are shown. (D) A schematic of GPB-1 and GOA-1 interaction on tph-1 expression. Effector molecules compete with Goα for Gβ binding to drive tph-1 expression. In goa-1(lf) an goa-1(gf) mutants, the effector constitutively binds to Gβ. GDP-bound conformation of the GOA-1(DN) protein blocks the effector binding. GPB-1(G162E) obstructs the interactions with both GOA-1 and the effector, leading to constitutively diminished tph-1 regardless of the presence of GDP- or GTP-bound Goα. We then tested a scenario in which GPB-1(G162E) inhibits tph-1 expression by inducing constitutive GOA-1 activation. We obtained a transgenic strain overexpressing gain-of-function GOA-1(Q205L) protein [31]. GOA-1(Q205L) is thought to constitutively activate GOA-1 signaling by locking the protein in the GTP-bound conformation [31]. Indeed, GOA-1(Q205L) retarded locomotion and egg laying, opposite to that seen with goa-1(lf) mutants [31]. However, contradicting to our hypothesis, GOA-1(Q205L) conferred an OCR-2-dependent increase in ADF tph-1::gfp as the goa-1 deletion alleles (Fig 6A). Further, GOA-1(Q205L) failed to induce tph-1::gfp in yz71 mutant background (Fig 6A), indicating that GOA-1(Q205L)-induced tph-1 expression requires GPB-1 at work.
These results prompted us to hypothesize that both lacking GOA-1 and GTP-bound GOA-1 constitutively activate GPB-1-mediated signaling that sets baseline tph-1 expression. To test this possibility, we constructed a dominant negative (DN) mutation in GOA-1 analogous to the mammalian Goα(S47C) mutation, which is locked in the GDP-bound conformation, and therefore constitutively tethers βγ and simultaneously shuts off both Goα and βγ mediated signaling [32]. We were unable to generate transgenic line expressing GOA-1(S47C) under the goa-1 promoter, perhaps also because goa-1 plays a critical role in early embryogenesis [19,20]. Therefore, we expressed GOA-1(S47C) under the VMAT cat-1 promoter. Three independent transgenic lines all dramatically reduced ADF tph-1::gfp (Fig 6B). Like gpb-1(yz71) mutants (Fig 2B), the GOA-1(S47C) transgene did not reduce the expression levels of ADF-specific marker gene Psrh-142::mCherry or alter ADF morphology (Fig 6C). Further, when GOA-1(S47C) transgenic lines were induced to form dauers or crossed into tir-1(gf) background, ADF tph-1::gfp expression levels were enhanced as compared to non-dauer GOA-1(S47C) transgenic animals (Fig 6B). Like gpb-1(yz71) dauers and tir-1(gf);gpb-1(yz71) double mutant (Fig 3A and 3D), ADF tph-1::gfp levels in GOA-1(S47C) transgenic dauers and tir-1(gf);GOA-1(S47C) animals were lower than WT dauers and tir-1(gf) single mutants (Fig 6B). Combined, these data support our hypothesis that GOA-1(S47C) inactivated GPB-1 in ADF leading to diminished baseline tph-1::gfp expression. Interestingly, the Pcat-1::goa-1(S47C) transgene also reduced tph-1::gfp in NSM. We speculate that NSM tph-1 expression might also be sensitive to G protein signaling, which was blocked by GOA-1(S47C). These data together endorse the model in which GPB-1-mediated Gβ signaling sets the baseline tph-1 expression in ADF, and this GPB-1 signaling is controlled by its interaction with GOA-1 (Fig 6D).
tph-1 expression in ADF neurons controls specific innate behaviors
Studies of tph-1 mutants have demonstrated a requirement for 5-HT in a variety of innate and stress-induced behaviors [9,11]. As the first step toward understanding the physiological importance of cell-specific regulation of 5-HT synthesis, we investigated the role of ADF-produced 5-HT in three well-characterized 5-HT-regulated innate behaviors: pharyngeal pumping, locomotion and egg laying. Because of the broad expression and multiple functional roles, disruption of gpb-1 and ocr-2 could impair multiple sites in the neural circuits. To identify the role of 5-HT produced in specific neurons, we therefore utilized established transgenic lines expressing tph-1 either in ADF (ADF::tph-1) or in NSM (NSM::tph-1)[11]. We integrated these transgenes onto the chromosomes of tph-1 deletion mutants and compared them with tph-1 mutants.
C. elegans feeds continuously on a E. coli bacterial lawn, by rhythmic contractions of the pharyngeal muscles [33]. The rate of pharyngeal pumping is a measure of C. elegans feeding behavior and is modulated by food availability and 5-HT [33]. Abundant food or applying exogenous 5-HT stimulates the pumping rates. In the presence of food, tph-1 null mutants displayed reduced pumping rates compared to WT animals, as previously observed [9]. We observed that the ADF::tph-1 transgene robustly rescued the pumping rate (Fig 7A). In contrast, the NSM::tph-1 transgene did not produce a significant effect (Fig 7A), consistent with the report that laser ablation of NSMs did not reduce the pumping rate [33]. Because ADF senses salts, biotin and nucleotide derivatives [22], ADF-produced 5-HT may modulate the neural circuit that relays attractive signals of nutritious compounds to feeding behavior.
Fig. 7. ADF-produced 5-HT modulates specific 5-HT-regulated innate behaviors. 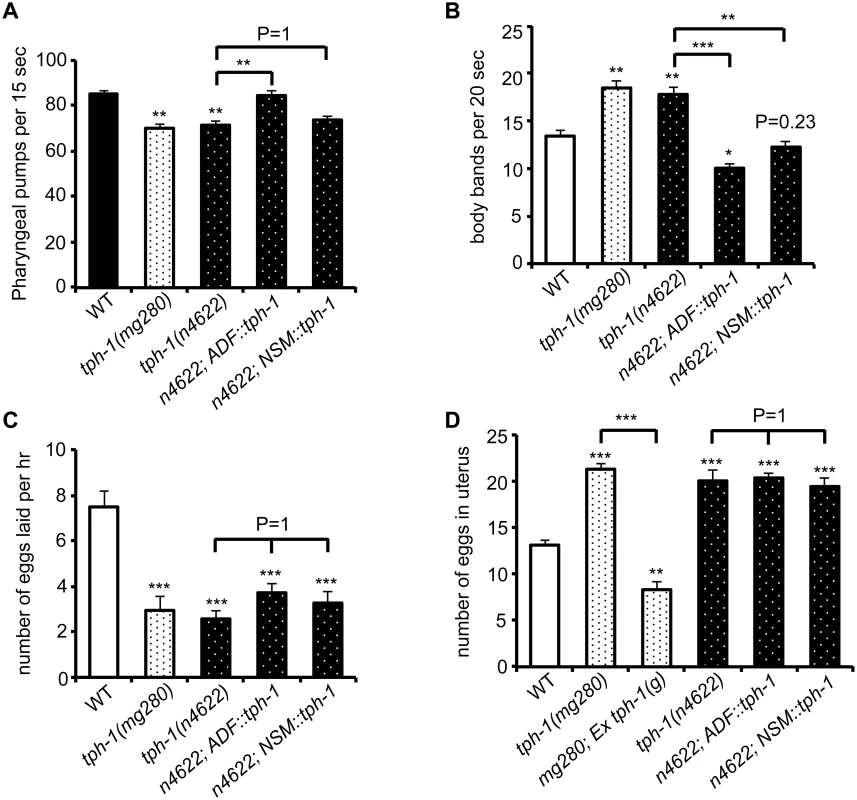
(A) tph-1 expressed in ADF not in NSM rescued pumping rates of tph-1 mutants. (B) tph-1 expressed in ADF and in NSM concerted to reduce locomotory rates. (C—D) tph-1 expressed in ADF and NSM is not required for 5-HT-regulation of egg-laying rates or egg accumulation in the uterus. Data represent the summary of 4–9 trials. Differences between the groups are indicated, and the differences to WT are marked above the bars, ** p < 0.01, *** p < 0.001 (ANOVA followed by Bonferroni test). Another C. elegans rhythmic behavior controlled by both food availability and 5-HT is locomotory rates. C. elegans slows down on a bacterial lawn, and applying 5-HT inhibits locomotion of WT animals [34,35]. tph-1 null alleles displayed increased locomotory rates on a bacterial lawn (Fig 7B). Interestingly, both ADF::tph-1 and NSM::tph-1 robustly rescued hyperactive locomotion of tph-1 mutants (Fig 7B). The pharyngeal NSMs are thought to sense bacteria passing through the pharynx [33], while ADF might sense food signals in the environment. Thus our observations could reflect how 5-HT from distinct neuronal types may converge distinct sensory perceptions to a particular behavioral output.
Because such transgenic arrays are typically overexpressed, we asked whether ADF::tph-1 would alter every 5-HT-regulated behavior, by testing egg-laying behavior. Exogenous 5-HT stimulates the rate of eggs released from the uterus [36]. Conversely, tph-1 mutants reduce the rate of egg laying and accumulate fertilized eggs in the uterus [9]. Neither ADF::tph-1 nor NSM::tph-1 significantly improved egg-laying behavior, although transgenic expressing genomic tph-1 sequence fully rescued (Fig 7C and 7D), supporting the notion that the HSN 5-HT-producing motor neurons are connected to the vulva muscles and control egg laying [37]. These results together support a model in which the steady state 5-HT synthesis in ADF tunes the innate behavioral circuits coordinating food intake and food searching movement.
Discussion
In this study, we identified a cell-autonomous requirement of GPB-1 Gβ signaling for baseline tph-1 expression in a pair of C. elegans ADF chemosensory neurons. Genetic analysis suggests that this GPB-1 function is negatively regulated by GOA-1 and acts upstream of the OCR-2 TRPV channel. Behavioral analysis showed that 5-HT produced in specific neuronal types controls defined innate behaviors. We therefore propose that regulation of baseline Tph gene expression under favorite environment represents another layer of transcriptional network that shapes and maintains the serotonergic systems, and could potentially be re-programmed in a cell-specific manner in the mature nervous systems to influence behavior.
Dedicated transcriptional program defines baseline 5-HT synthesis
Several lines of our data suggest that GPB-1-OCR-2-mediated tph-1 expression is mechanistically separated from serotonergic fate establishment in ADF neurons. First, gpb-1(yz71) mutants did not diminish VMAT CAT-1. Second, the Pcat-1::gpb-1 transgene fully restored the baseline tph-1::gfp in yz71 mutants, showing that GPB-1 directs 5-HT synthesis after the serotonergic identity established. Consistently, OCR-2 and its downstream RFX transcription factor DAF-19 are essential for ADF tph-1 but not cat-1 expression [12,27]. In mammals, the baseline Tph2 expression in raphe 5-HT-producing neurons displays a circadian rhythmicity according to glucocorticoid oscillation [38,39]. Conditional knocking out PET-1 in adult mice abrogated Tph2 but not VMAT expression in specific sets of raphe 5-HT-producing neurons, leading to enhanced anxiety behavior [8]. These findings emphasize cell-specific regulation of Tph gene expression in mature neurons as a key to maintain 5-HT signaling in evolutionary diverse organisms.
Our data indicated that cell-autonomous GPB-1 sets the ADF tph-1 expression baseline. At present, it is unclear how this signaling pathway is activated. This signaling is unlikely to be activated by environmental cues, because cilia mutants that cannot detect external cues do not diminish ADF tph-1 expression [10]. Accumulating evidence indicates that G proteins can be activated by receptor-independent cell intrinsic cues [16,17]. While further studies are required to define the GPB-1 activation mechanism, GPB-1-regulation of tph-1 differs from the mechanisms that maintain the terminally differentiated neuronal cell identity [5], as GPB-1 is not essential for preserving ADF identity or axon/dendritic architecture. Thus, the GPB-1-OCR-2 signaling pathway in ADF is likely to represent another layer of transcriptional network that is dedicated to direct baseline neurotransmitter synthesis in specific neuronal types. Although genetic dissection of 5-HT production in mammalian CNS remains not feasible, evidence has been mounting that genetic variants that reduce Tph2 expression attend quantitative effects on 5-HT-modulated behavior in rodents and humans [2].
Gβ GPB-1, not Gα, is a key regulator of tph-1 baseline expression in ADF neurons
Genetic dissection of transcriptional regulation of 5-HT synthesis in this work took advantage of the viable GPB-1(G162E) reduction-of-function allele. Analysis of the baseline and pathogen-induced tph-1::gfp expression has revealed two mechanistically separable mechanisms of GPB-1. Our cell-specific rescue experiments, combined with the report that Gqα EGL-30 function in AWC and AWB neurons is necessary and sufficient for PA14-induced ADF tph-1 upregulation [25], argue that GPB-1 is a regulator for EGL-30 signaling in AWC and AWB that relay infection cues to ADF. In contrast, GPB-1 regulation of the tph-1 baseline expression depends on Goα GOA-1.
Like GPB-1, GOA-1 is broadly expressed and has been shown to regulate mitotic spindle positions in embryogenesis as well as a variety of behaviors [40]. The role of GOA-1 in regulating baseline tph-1 expression, however, differs from all these examples in which GOA-1 signaling is essential. In contrast, both goa-1 deletion and hyperactive GOA-1(Q205L) conferred elevated ADF tph-1::gfp. These data, together with the results that ocr-2 and GPB-1(G162E) diminished ADF tph-1::gfp in the goa-1 deletion and GOA-1(Q205L) mutants, point to GOA-1 as a negative regulator of the GPB-1-OCR-2 signaling pathway that drives baseline tph-1 expression (Fig 6D). However, we do not eliminate the possibility that GOA-1 signaling can transduce other sensory cues to influence 5-HT synthesis.
How might G162E disrupt Gβ function? We found that, similar to GPB-1(G162E), expressing GOA-1(S47C), analogous to the mammalian Goα(S47C) that is locked in the GDP-bound conformation, diminished ADF tph-1::gfp. Thus one possibility is that GPB-1(G162E) promotes the heterotrimeric complex in the inactive conformation. Modeling GPB-1 sequence over the Giα1β1γ2 heterotrimer crystallographic structure revealed that G162 is located at the Gβ surface that binds to Gα switch II helix. GTP-binding of Gα triggers an 120° rotation of the switch II helix, thereby switching on the G protein signaling [28,41]. Thus G162E could block switch II helix rotation. Alternatively, G162E might alter the conformation of nearby G202 and G203 in the switch II helix critical for GTP binding [32], reducing the GTP affinity. These models are also consistent with GPB-1(G162E) abrogation of EGL-30-dependent pathogen-induced tph-1 upregulation, indicating that G162E obstructs a key point of GPB-1/Gα interaction. However, these models do not address how GPB-1(G162E) diminished tph-1::gfp induced by GOA-1(Q205L), which is locked in the GTP-bound conformation. As the transgenes form multiple copies of gene arrays, expressing GOA-1(Q205L) driven by the goa-1 promoter could reduce endogenous goa-1 transcription due to promoter titration. In addition, GOA-1(Q205L) protein could compete with endogenous GOA-1 for the trafficking machinery and functional sites to liberate GPB-1 as well as GPB-1(G162E). Thus, another possibility would be that G162E also impairs effector signaling that regulates tph-1 expression.
There is increasing evidence that Gβ interacts directly with a wide range of effectors to modulate diverse downstream cellular responses [16,17]. It is interesting to note that the first example was the activation of a cardiac potassium channel by purified Gβ [16]. A common lesson learnt from X-ray crystal structure determination, NMR analysis and biochemical analysis is that Gβ effector-binding sites are masked by Gβ/Gα switch II binding [17]. In this regard, Gβ-Y145 is a key residue in one such effector-binding hotspot and sits closely to G162 in the crystal structure [17] (Fig 5Biii). Possibly, GDP-bound GOA-1(S47C) locks the switch II into the conformation unable for effector binding to GPB-1, whereas GPB-1(G162E) produces a dual disruption on Gα switch II conformational changes and Gβ effector binding that defines the tph-1 baseline, thus steady state 5-HT synthesis, in the ADF neurons (Fig 6D).
Methods
Strains
C. elegans strains were maintained at 20°C on NGM agar plates seeded with a lawn of E. coli OP50 as a food source [42]. WT animals were Bristol strain N2. The Hawaiian isolate CB4856 was used in genetic mapping of gpb-1(yz71) mutation. Mutant strains used in this study were: che-2(e1033), eri-1(mg366);lin-15B(n744), gpb-1(ok1875)/mIn1 [mIs14 dpy-10(e128)], goa-1(n363), ocr-2(yz5), tir-1(yz68), MT15434 tph-1(mg280), and tph-1(n4622). Transgenic strains were: CX6741: tph-1(mg280);Ex[ADF::tph-1][11], CX7749: tph-1(mg280);Ex[NSM::tph-1][11], tph-1(mg280); yzEx126[tph-1(g); Rol-6] [9], GR1333: yzIs71[tph-1::gfp; Rol - 6(d)][9], Is[cat-1::gfp][23], PS1493: dpy-20(e1362);syIs9[pJMGoQL; Dpy-20(+)] [31], and yzEx010[Psrh-142::mCherry;elt-2::gfp][43].
Identification of gpb-1(yz71) mutation
yz71 was isolated from a genetic screen for mutants with diminished tph-1::gfp in the ADF neurons after ethyl methane sulfonate mutagenesis of GR1333 animals as described previously [12]. Genetic mapping using single-nucleotide polymorphisms (SNP) of CB4856 localized yz71 to a 700 kb contig on LG II. Sequencing yz71 genomic DNA revealed two mutations in this contig. After the two mutations separated by backcrossing with N2, diminished ADF tph-1::gfp segregated only with the G-to-A transition causing a G162E substitution in the third predicted WD40 repeat of the Gβ protein GPB-1. The G-to-A transition abolished a restriction enzyme BspE1 cutting site, which was used as another method to validate the yz71 mutation.
Generation of transgenic animals
All constructs were generated by PCR. gpb-1(g) was a genomic DNA fragment encompassing 2,027 bp 5’-noncoding sequence, exons, introns and 933 bp 3’-UTR of the gpb-1 gene, similar to previously reported [19]. To express gpb-1 in specific cells, gpb-1 cDNA sequence was inserted between a heterologous promoter and unc-54 3’-UTR. gpb-1 cDNA was amplified from cDNA mixture prepared from total RNA of WT animals. The following published promoter sequences were used: 1.3 kb rab-3 promoter expressed in all neurons [44], 2.9 kb ges-1 promoter expressed in the intestine [45], 3.7 kb daf-6 promoter expressed in hypodermis and glia [46], 2.7 kb che-2 [47] and 1.8 kb dyf-1 [10] promoters both expressed in all ciliated neurons, 2.7 kb odr-3 promoter expressed in ciliated sensory neurons AWA, AWB, AWC, ADF, ASH, PHA and PHB [48], 4.1 kb lin-11 genomic fragment encompassing 623 bp promoter to 14 bp of exon 5 expressed in ADF and ADL [49], with a stop codon and SL2 sequence inserted between the lin-11 and gpb-1 cDNA sequences, 4.6 kb cat-1 promoter expressed in monoaminergic neurons [27], 3.4 kb srh-142 promoter expressed in ADF [43], 2.4 kb odr-1 promoter expressed in AWB and AWC neurons [50], and 2.6 kb sra-6 promoter expressed in ASH, ASI plus a few unidentified neurons and intestine [51].
To overexpress GOA-1(S47C) in serotonergic neurons, goa-1 cDNA was amplified from WT cDNA mixture, and the codon Ser47 (TCG) was changed to Cys47 (TGT), using PCR primer ggagaatcaggaaaatgtactattg, and the mutagenized sequence was fused to the cat-1 promoter sequence and unc-54 3’-UTR.
For generating transgenic worm lines, individual constructs from three independent PCR reactions were pooled to reduce potential PCR errors, and the pooled PCR products were purified (Qiagen) and microinjected into WT or yz71 mutants carrying an integrated tph-1::gfp. For several transgenic lines, a construct was first injected into WT animals and then crossed into yz71 mutant background. The plasmids containing either elt-2::gfp or unc-122::rfp were co-injected as a transgenic marker. Typically, two transgenic lines from one injection were analyzed, and data from one representative line are presented. For overexpressing GOA-1(S47C), DNA from two independent preparations were individually injected, and transgenic lines from both injections are presented.
RNAi
RNA-interference (RNAi) experiments were performed in the background of eri-1;lin-15B to enhance RNAi efficiency in neurons, as previously described [12]. RNAi assays were carried out by feeding worms an E. coli HT115 clone expressing dsRNA of a target gene or the control empty L4440 vector (Ahringer RNAi library, University of Cambridge, England). RNAi clones were individually cultured overnight in Luria broth containing 100 μg/ml ampicillin. 600 μl of bacterial culture were spread evenly to cover the surface of assay plates containing NGM medium supplemented with 6mM IPTG and 25μg/ml carbenicillin, and incubated overnight at room temperature. Eggs from eri-1;lin-15B animals carrying tph-1::gfp were placed onto each plate, incubated at 20°C, and GFP levels in ADF of resultant L4 and second day adults were quantified.
Quantification of tph-1::gfp expression
The expression of a chromosomally integrated tph-1::gfp reporter in ADF or NSM neurons in living worms under optimal growth conditions, during pathogen PA14 infection, or in the dauer stage was evaluated by measuring GFP fluorescence intensity, as we have done previously [10,12]. Briefly, images of individual neurons were captured under a 40x objective lens at a fixed exposure time, using an AxioImager Z1 microscope equipped with proper filters and AxioCam MR digital camera (Zeiss, Northwood, NY). The external contour of the cell body in the images was delineated, and fluorescence intensity within the entire cell body was measured using the ImageJ software (National Institute of Health, Maryland).
For quantifying tph-1::gfp intensity during pathogen infection, first day young adult worms were transferred to standard slow killing assay plates seeded with either PA14 culture [52] or control plates seeded with OP50, incubated at 25°C for 6 hr, images of the ADF neurons were captured and GFP intensity quantified.
For quantifying tph-1::gfp intensity in dauers, WT and mutants were induced to form dauers by dauer pheromones and high growth temperature at 25°C as previously described [10,12]. Gravid adults from each strain were transferred to NGM plates supplied with dauer pheromone, allowed to lay eggs in a 25°C incubator, the adults were then removed from the plates, and dauers developed from hatched eggs on the plates were analyzed 3 days later. For each strain the value of dauers was compared to that of L4 sibling grown on NGM plates without pheromone and assayed on the same day. For some experiments, starvation was used as a second method to induce dauer formation. Data from dauers induced by the two conditions are comparable.
Data represent the average of at least three trials unless specified otherwise. For each trial, 15–25 animals per genotype per condition and treatment were analyzed and compared to the controls assayed on the same day. WT animals under the same conditions and treatments were analyzed for every experiment.
Indirect immunofluorescence microscopy and dye DiI filling
5-HT immunostaining and quantification of 5-HT immunoreactivity in individual neurons were performed with whole mount worms as we described previously [9], using rabbit antibody against 5-HT (purchased from Dr. H.W.M. Steinbusch, Maastricht University, Masstricht, The Netherlands). The staining patterns were visualized via Alexa Fluor 594 conjugated goat anti-rabbit antibodies (Invitrogen) under an AxioImager Z1 microscope equipped with proper filters. To quantify the intensity of 5-HT immunoreactivity, images of ADF or NSM neurons in individual worms were captured under a 40x objective lens at a fixed exposure time with 100% UV exposure level. For each image, fluorescence intensity of a 10x10 pixels area within a cell body was quantified using the ImageJ software. To exclude staining background, fluorescence intensity over a 10x10 pixels area posterior to the cell body in the same image was quantified, and the value of the background was subtracted from the value of the corresponding neuronal area.
GPB-1 immunostaining was performed according to a published protocol [19] with modifications. Briefly, well-fed mixed stages of worms and embryos were washed off culture plates, rinsed with water, transferred to 0.01% poly-L-lysine-coated slide, crushed by pressing the sample with a coverslip, and frozen on dry ice. The coverslip was then removed and the samples were treated with methanol and acetone. Following serial rehydration, the preparations were incubated with a blotting mix of 0.1% Tween 20 and 5% non-fat milk overnight at 4°C, and stained with rabbit antibody against GPB-1 [20]. The staining patterns were visualized via Alexa Fluor 594 conjugated goat anti-rabbit antibodies.
DiI staining was performed as we previously described [10,12], by soaking well-fed living worms in M9 buffer containing 10 μg/ml DiI for 2 hr, and visualization of the staining pattern under a fluorescence microscope with a proper filter.
Structural modeling of Gβ(G162E) substitution
WD40 repeats are in reference of published predicted GPB-1 sequence [53]. For homology comparisons, the amino acid sequence of C. elegans GPB-1, human Gβ1 GNB1 (GenBank: CAG33065.1), human Gβ2 GNB2 (GenBank: CAG46530.1) and bovine β1 GNB1 (NCBI reference sequence: NP_786971.2) were aligned using the ClustalW2 software [54]. The bovine Gβ1 protein shares 100% and 86% amino acid identity to human Gβ1 and C. elegans GPB-1, respectively.
Crystal structure of the rat Giα and bovine β1γ2 heterotrimer (PDB entry: 1GP2)[28] was used to model the G162E substitution with a focus on the interface between Gβ and Gα switch II helix. The public crystal structural data were downloaded from the protein data bank (http://www.rcsb.org/pdb/explore/explore.do?structureId=1GP2, PDB entry: 1GP2), and displayed as cartoon structures using the PyMOL software (http://www.pymol.org). To model Gβ/Gα switch II helix contacting interface, Gβ-G162 and other contacting residues in the Gβ and Gα identified by the crystallography were manually selected, colored and their side chains displayed. To model the G162E substitution, G162 was manually replaced by E in the Gβ sequence, using the Crystallographic Object-Oriented Toolkit (COOT) [55], and modeled again using PyMOL.
Behavior assays
All behavioral tests were performed with first day young adults, using protocols as we have previously described [9,56], with modifications. Well-fed L4 of WT, mutant and transgenic animals were picked onto fresh plates seeded with a lawn of OP50 as food source, incubated at 20°C, and resultant adults were assayed about 24 hr later. Prior to the assays, animals were allowed to stay at room temperature for 3–5 hr. The rate of pharyngeal pumping was assayed using a stereo dissecting microscope at an 80x magnification. The number of pumps of individual animals within a 15 sec interval was recorded by Moticam 2300 digital camera using the Motic Images Plus 2.0 software (http://www.motic.com/en/index), and the pumping rate in the movie was manually counted. For each assay, 5–12 animals of each genotype were analyzed.
The locomotion rates were evaluated by two methods. The first method measures the locomotory activities by video recording the movement of individual worms on the center of an OP50 lawn on an agar plate in a 30 sec interval, using the Moticam connected to a dissecting microscope. Locomotory rates were determined by counting the number of bends of the anterior body region of individual animals in the movie. Animals that linger around the edge of the bacterial lawn for more than 5 sec were not analyzed. The second method counted the number of the body bends manually under a dissecting scope. 5–15 animals of each genotype were analyzed for every trial.
Egg-laying behavior was evaluated by two assays. First assay measures the number of eggs laid by a worm within a one-hour interval. For each strain, 10 animals were transferred onto a fresh plate, and the number of eggs laid on the plates was scored one hour later. The second assay determines eggs carried in the uterus of animals that were used for the egg-laying count assay, determining that the reduced number of eggs laid was not simply due to reduced number of eggs in the uterus. To score the number of eggs in the uterus, individual worms were placed into a drop of solution containing 3% of commercial bleach and 1N NaOH on an agar pad on a glass slide. The bleach solution dissolved the body of animals, and eggs, which were protected by their eggshells, were scored immediately. For each behavioral test, the assays were repeated at least 4 times, and representative data pooled from 4–9 trials are presented.
Statistics
Unpaired Student’s t-test was used for comparisons between a mutant and WT, or between two mutants or two treatments. For comparison between multiple groups, one-way ANOVA followed by Bonferroni test were performed. For comparison between multiple groups with different treatments, two-way ANOVA followed by Tukey’s test were performed.
Zdroje
1. Müller CP, Jacobs BL (2010) Handbook of the behavioral neurobiology of serotonin. Handbook of behavioral neuroscience v 18. 1st ed. Amsterdam: Elsevier/Academic Press.
2. Deneris ES, Wyler SC (2012) Serotonergic transcriptional networks and potential importance to mental health. Nature neuroscience 15 : 519–527. doi: 10.1038/nn.3039 22366757
3. Smidt MP, van Hooft JA (2013) Subset specification of central serotonergic neurons. Frontiers in cellular neuroscience 7 : 200. doi: 10.3389/fncel.2013.00200 24198761
4. Hale MW, Shekhar A, Lowry CA (2011) Development by environment interactions controlling tryptophan hydroxylase expression. Journal of chemical neuroanatomy 41 : 219–226. doi: 10.1016/j.jchemneu.2011.05.002 21640184
5. Deneris ES, Hobert O (2014) Maintenance of postmitotic neuronal cell identity. Nature neuroscience 17 : 899–907. doi: 10.1038/nn.3731 24929660
6. Goridis C, Rohrer H (2002) Specification of catecholaminergic and serotonergic neurons. Nature reviews Neuroscience 3 : 531–541. 12094209
7. Gutknecht L, Kriegebaum C, Waider J, Schmitt A, Lesch KP (2009) Spatio-temporal expression of tryptophan hydroxylase isoforms in murine and human brain: convergent data from Tph2 knockout mice. European neuropsychopharmacology: the journal of the European College of Neuropsychopharmacology 19 : 266–282.
8. Liu C, Maejima T, Wyler SC, Casadesus G, Herlitze S, et al. (2010) Pet-1 is required across different stages of life to regulate serotonergic function. Nature neuroscience 13 : 1190–1198. doi: 10.1038/nn.2623 20818386
9. Sze JY, Victor M, Loer C, Shi Y, Ruvkun G (2000) Food and metabolic signalling defects in a Caenorhabditis elegans serotonin-synthesis mutant. Nature 403 : 560–564. 10676966
10. Moussaif M, Sze JY (2009) Intraflagellar transport/Hedgehog-related signaling components couple sensory cilium morphology and serotonin biosynthesis in Caenorhabditis elegans. Journal of Neuroscience 29 : 4065–4075. doi: 10.1523/JNEUROSCI.0044-09.2009 19339602
11. Zhang Y, Lu H, Bargmann CI (2005) Pathogenic bacteria induce aversive olfactory learning in Caenorhabditis elegans. Nature 438 : 179–184. 16281027
12. Xie Y, Moussaif M, Choi S, Xu L, Sze JY (2013) RFX transcription factor DAF-19 regulates 5-HT and innate immune responses to pathogenic bacteria in Caenorhabditis elegans. PLoS genetics 9: e1003324. doi: 10.1371/journal.pgen.1003324 23505381
13. Pocock R, Hobert O (2010) Hypoxia activates a latent circuit for processing gustatory information in C. elegans. Nature neuroscience 13 : 610–614. doi: 10.1038/nn.2537 20400959
14. Neer EJ, Schmidt CJ, Nambudripad R, Smith TF (1994) The ancient regulatory-protein family of WD-repeat proteins. Nature 371 : 297–300. 8090199
15. Gilman AG (1987) G proteins: transducers of receptor-generated signals. Annual review of biochemistry 56 : 615–649. 3113327
16. Khan SM, Sleno R, Gora S, Zylbergold P, Laverdure JP, et al. (2013) The expanding roles of Gbetagamma subunits in G protein-coupled receptor signaling and drug action. Pharmacol Rev 65 : 545–577. doi: 10.1124/pr.111.005603 23406670
17. Lin Y, Smrcka AV (2011) Understanding molecular recognition by G protein betagamma subunits on the path to pharmacological targeting. Mol Pharmacol 80 : 551–557. doi: 10.1124/mol.111.073072 21737569
18. Ferreira T, Wilson SR, Choi YG, Risso D, Dudoit S, et al. (2014) Silencing of odorant receptor genes by G protein betagamma signaling ensures the expression of one odorant receptor per olfactory sensory neuron. Neuron 81 : 847–859. doi: 10.1016/j.neuron.2014.01.001 24559675
19. Zwaal RR, Ahringer J, van Luenen HG, Rushforth A, Anderson P, et al. (1996) G proteins are required for spatial orientation of early cell cleavages in C. elegans embryos. Cell 86 : 619–629. 8752216
20. Thyagarajan K, Afshar K, Gonczy P (2011) Polarity mediates asymmetric trafficking of the Gbeta heterotrimeric G-protein subunit GPB-1 in C. elegans embryos. Development 138 : 2773–2782. doi: 10.1242/dev.063354 21652650
21. Sulston JE, Schierenberg E, White JG, Thomson JN (1983) The embryonic cell lineage of the nematode Caenorhabditis elegans. Developmental biology 100 : 64–119. 6684600
22. Bargmann CI, Horvitz HR (1991) Chemosensory neurons with overlapping functions direct chemotaxis to multiple chemicals in C. elegans. Neuron 7 : 729–742. 1660283
23. Sze JY, Zhang S, Li J, Ruvkun G (2002) The C. elegans POU-domain transcription factor UNC-86 regulates the tph-1 tryptophan hydroxylase gene and neurite outgrowth in specific serotonergic neurons. Development 129 : 3901–3911. 12135927
24. Hu PJ (2007) Dauer. WormBook: the online review of C elegans biology: 1–19.
25. Qin Y, Zhang X, Zhang Y (2013) A neuronal signaling pathway of CaMKII and Gqalpha regulates experience-dependent transcription of tph-1. The Journal of neuroscience: the official journal of the Society for Neuroscience 33 : 925–935.
26. Shivers RP, Kooistra T, Chu SW, Pagano DJ, Kim DH (2009) Tissue-specific activities of an immune signaling module regulate physiological responses to pathogenic and nutritional bacteria in C. elegans. Cell Host Microbe 6 : 321–330. doi: 10.1016/j.chom.2009.09.001 19837372
27. Zhang S, Sokolchik I, Blanco G, Sze JY (2004) Caenorhabditis elegans TRPV ion channel regulates 5HT biosynthesis in chemosensory neurons. Development 131 : 1629–1638. 14998926
28. Wall MA, Coleman DE, Lee E, Iniguez-Lluhi JA, Posner BA, et al. (1995) The structure of the G protein heterotrimer Gi alpha 1 beta 1 gamma 2. Cell 83 : 1047–1058. 8521505
29. Mixon MB, Lee E, Coleman DE, Berghuis AM, Gilman AG, et al. (1995) Tertiary and quaternary structural changes in Gi alpha 1 induced by GTP hydrolysis. Science 270 : 954–960. 7481799
30. Tsou MF, Hayashi A, Rose LS (2003) LET-99 opposes Galpha/GPR signaling to generate asymmetry for spindle positioning in response to PAR and MES-1/SRC-1 signaling. Development 130 : 5717–5730. 14534135
31. Mendel JE, Korswagen HC, Liu KS, Hajdu-Cronin YM, Simon MI, et al. (1995) Participation of the protein Go in multiple aspects of behavior in C. elegans. Science 267 : 1652–1655. 7886455
32. Slepak VZ, Quick MW, Aragay AM, Davidson N, Lester HA, et al. (1993) Random mutagenesis of G protein alpha subunit G(o)alpha. Mutations altering nucleotide binding. The Journal of biological chemistry 268 : 21889–21894. 8408043
33. Avery L, Horvitz HR (1989) Pharyngeal pumping continues after laser killing of the pharyngeal nervous system of C. elegans. Neuron 3 : 473–485. 2642006
34. Sawin ER (1996) Genetic and cellular analysis of modulated behaviors in Caenorhabditis elegans. PhD Thesis, Massachusetts Institute of Technology, Cambridge, Massachusetts.
35. Gurel G, Gustafson MA, Pepper JS, Horvitz HR, Koelle MR (2012) Receptors and other signaling proteins required for serotonin control of locomotion in Caenorhabditis elegans. Genetics 192 : 1359–1371. doi: 10.1534/genetics.112.142125 23023001
36. Horvitz HR, Chalfie M, Trent C, Sulston JE, Evans PD (1982) Serotonin and octopamine in the nematode Caenorhabditis elegans. Science 216 : 1012–1014. 6805073
37. Desai C, Garriga G, McIntire SL, Horvitz HR (1988) A genetic pathway for the development of the Caenorhabditis elegans HSN motor neurons. Nature 336 : 638–646. 3200316
38. Malek ZS, Sage D, Pevet P, Raison S (2007) Daily rhythm of tryptophan hydroxylase-2 messenger ribonucleic acid within raphe neurons is induced by corticoid daily surge and modulated by enhanced locomotor activity. Endocrinology 148 : 5165–5172. 17595225
39. Nexon L, Poirel VJ, Clesse D, Pevet P, Raison S (2009) Complex regional influence of photoperiod on the nycthemeral functioning of the dorsal and median raphe serotoninergic system in the Syrian hamster. Eur J Neurosci 30 : 1790–1801. doi: 10.1111/j.1460-9568.2009.06986.x 19863652
40. Bastiani C, Mendel J (2006) Heterotrimeric G proteins in C. elegans. WormBook: the online review of C elegans biology: 1–25.
41. Lambright DG, Noel JP, Hamm HE, Sigler PB (1994) Structural determinants for activation of the alpha-subunit of a heterotrimeric G protein. Nature 369 : 621–628. 8208289
42. Brenner S (1973) The genetics of behaviour. Br Med Bull 29 : 269–271. 4807330
43. Zheng X, Chung S, Tanabe T, Sze JY (2005) Cell-type specific regulation of serotonergic identity by the C. elegans LIM-homeodomain factor LIM-4. Dev Biol 286 : 618–628. 16168406
44. Nonet ML, Staunton JE, Kilgard MP, Fergestad T, Hartwieg E, et al. (1997) Caenorhabditis elegans rab-3 mutant synapses exhibit impaired function and are partially depleted of vesicles. J Neurosci 17 : 8061–8073. 9334382
45. McGhee JD, Birchall JC, Chung MA, Cottrell DA, Edgar LG, et al. (1990) Production of null mutants in the major intestinal esterase gene (ges-1) of the nematode Caenorhabditis elegans. Genetics 125 : 505–514. 2379823
46. Perens EA, Shaham S (2005) C. elegans daf-6 encodes a patched-related protein required for lumen formation. Dev Cell 8 : 893–906. 15935778
47. Fujiwara M, Ishihara T, Katsura I (1999) A novel WD40 protein, CHE-2, acts cell-autonomously in the formation of C. elegans sensory cilia. Development 126 : 4839–4848. 10518500
48. Roayaie K, Crump JG, Sagasti A, Bargmann CI (1998) The G alpha protein ODR-3 mediates olfactory and nociceptive function and controls cilium morphogenesis in C. elegans olfactory neurons. Neuron 20 : 55–67. 9459442
49. Hobert O, D'Alberti T, Liu Y, Ruvkun G (1998) Control of neural development and function in a thermoregulatory network by the LIM homeobox gene lin-11. J Neurosci 18 : 2084–2096. 9482795
50. Yu S, Avery L, Baude E, Garbers DL (1997) Guanylyl cyclase expression in specific sensory neurons: a new family of chemosensory receptors. Proc Natl Acad Sci U S A 94 : 3384–3387. 9096403
51. Troemel ER, Chou JH, Dwyer ND, Colbert HA, Bargmann CI (1995) Divergent seven transmembrane receptors are candidate chemosensory receptors in C. elegans. Cell 83 : 207–218. 7585938
52. Tan MW, Mahajan-Miklos S, Ausubel FM (1999) Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proc Natl Acad Sci U S A 96 : 715–720. 9892699
53. van der Voorn L, Gebbink M, Plasterk RH, Ploegh HL (1990) Characterization of a G-protein beta-subunit gene from the nematode Caenorhabditis elegans. J Mol Biol 213 : 17–26. 2110981
54. Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, et al. (2003) Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res 31 : 3497–3500. 12824352
55. Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60 : 2126–2132. 15572765
56. Jafari G, Xie Y, Kullyev A, Liang B, Sze JY (2011) Regulation of extrasynaptic 5-HT by serotonin reuptake transporter function in 5-HT-absorbing neurons underscores adaptation behavior in Caenorhabditis elegans. The Journal of neuroscience: the official journal of the Society for Neuroscience 31 : 8948–8957.
Štítky
Genetika Reprodukční medicína
Článek The Chromatin Protein DUET/MMD1 Controls Expression of the Meiotic Gene during Male Meiosis inČlánek Tissue-Specific Gain of RTK Signalling Uncovers Selective Cell Vulnerability during Embryogenesis
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 9- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
- Intrauterinní inseminace a její úspěšnost
- Délka menstruačního cyklu jako marker ženské plodnosti
-
Všechny články tohoto čísla
- Retraction: RNAi-Dependent and Independent Control of LINE1 Accumulation and Mobility in Mouse Embryonic Stem Cells
- Signaling from Within: Endocytic Trafficking of the Robo Receptor Is Required for Midline Axon Repulsion
- A Splice Region Variant in Lowers Non-high Density Lipoprotein Cholesterol and Protects against Coronary Artery Disease
- The Chromatin Protein DUET/MMD1 Controls Expression of the Meiotic Gene during Male Meiosis in
- A NIMA-Related Kinase Suppresses the Flagellar Instability Associated with the Loss of Multiple Axonemal Structures
- Slit-Dependent Endocytic Trafficking of the Robo Receptor Is Required for Son of Sevenless Recruitment and Midline Axon Repulsion
- Expression of Concern: Protein Under-Wrapping Causes Dosage Sensitivity and Decreases Gene Duplicability
- Mutagenesis by AID: Being in the Right Place at the Right Time
- Identification of as a Genetic Modifier That Regulates the Global Orientation of Mammalian Hair Follicles
- Bridges Meristem and Organ Primordia Boundaries through , , and during Flower Development in
- Evaluating the Performance of Fine-Mapping Strategies at Common Variant GWAS Loci
- KLK5 Inactivation Reverses Cutaneous Hallmarks of Netherton Syndrome
- Differential Expression of Ecdysone Receptor Leads to Variation in Phenotypic Plasticity across Serial Homologs
- Receptor Polymorphism and Genomic Structure Interact to Shape Bitter Taste Perception
- Cognitive Function Related to the Gene Acquired from an LTR Retrotransposon in Eutherians
- Critical Function of γH2A in S-Phase
- Arabidopsis AtPLC2 Is a Primary Phosphoinositide-Specific Phospholipase C in Phosphoinositide Metabolism and the Endoplasmic Reticulum Stress Response
- XBP1-Independent UPR Pathways Suppress C/EBP-β Mediated Chondrocyte Differentiation in ER-Stress Related Skeletal Disease
- Integration of Genome-Wide SNP Data and Gene-Expression Profiles Reveals Six Novel Loci and Regulatory Mechanisms for Amino Acids and Acylcarnitines in Whole Blood
- A Genome-Wide Association Study of a Biomarker of Nicotine Metabolism
- Cell Cycle Regulates Nuclear Stability of AID and Determines the Cellular Response to AID
- A Genome-Wide Association Analysis Reveals Epistatic Cancellation of Additive Genetic Variance for Root Length in
- Tissue-Specific Gain of RTK Signalling Uncovers Selective Cell Vulnerability during Embryogenesis
- RAB-10-Dependent Membrane Transport Is Required for Dendrite Arborization
- Basolateral Endocytic Recycling Requires RAB-10 and AMPH-1 Mediated Recruitment of RAB-5 GAP TBC-2 to Endosomes
- Dynamic Contacts of U2, RES, Cwc25, Prp8 and Prp45 Proteins with the Pre-mRNA Branch-Site and 3' Splice Site during Catalytic Activation and Step 1 Catalysis in Yeast Spliceosomes
- ARID1A Is Essential for Endometrial Function during Early Pregnancy
- Predicting Carriers of Ongoing Selective Sweeps without Knowledge of the Favored Allele
- An Interaction between RRP6 and SU(VAR)3-9 Targets RRP6 to Heterochromatin and Contributes to Heterochromatin Maintenance in
- Photoreceptor Specificity in the Light-Induced and COP1-Mediated Rapid Degradation of the Repressor of Photomorphogenesis SPA2 in Arabidopsis
- Autophosphorylation of the Bacterial Tyrosine-Kinase CpsD Connects Capsule Synthesis with the Cell Cycle in
- Multimer Formation Explains Allelic Suppression of PRDM9 Recombination Hotspots
- Rescheduling Behavioral Subunits of a Fixed Action Pattern by Genetic Manipulation of Peptidergic Signaling
- A Gene Regulatory Program for Meiotic Prophase in the Fetal Ovary
- Cell-Autonomous Gβ Signaling Defines Neuron-Specific Steady State Serotonin Synthesis in
- Discovering Genetic Interactions in Large-Scale Association Studies by Stage-wise Likelihood Ratio Tests
- The RCC1 Family Protein TCF1 Regulates Freezing Tolerance and Cold Acclimation through Modulating Lignin Biosynthesis
- The AMPK, Snf1, Negatively Regulates the Hog1 MAPK Pathway in ER Stress Response
- The Parkinson’s Disease-Associated Protein Kinase LRRK2 Modulates Notch Signaling through the Endosomal Pathway
- Multicopy Single-Stranded DNA Directs Intestinal Colonization of Enteric Pathogens
- Recurrent Domestication by Lepidoptera of Genes from Their Parasites Mediated by Bracoviruses
- Three Different Pathways Prevent Chromosome Segregation in the Presence of DNA Damage or Replication Stress in Budding Yeast
- Identification of Four Mouse Diabetes Candidate Genes Altering β-Cell Proliferation
- The Intolerance of Regulatory Sequence to Genetic Variation Predicts Gene Dosage Sensitivity
- Synergistic and Dose-Controlled Regulation of Cellulase Gene Expression in
- Genome Sequence and Transcriptome Analyses of : Metabolic Tools for Enhanced Algal Fitness in the Prominent Order Prymnesiales (Haptophyceae)
- Ty3 Retrotransposon Hijacks Mating Yeast RNA Processing Bodies to Infect New Genomes
- FUS Interacts with HSP60 to Promote Mitochondrial Damage
- Point Mutations in Centromeric Histone Induce Post-zygotic Incompatibility and Uniparental Inheritance
- Genome-Wide Association Study with Targeted and Non-targeted NMR Metabolomics Identifies 15 Novel Loci of Urinary Human Metabolic Individuality
- Outer Hair Cell Lateral Wall Structure Constrains the Mobility of Plasma Membrane Proteins
- A Large-Scale Functional Analysis of Putative Target Genes of Mating-Type Loci Provides Insight into the Regulation of Sexual Development of the Cereal Pathogen
- A Genetic Selection for Mutants Reveals an Interaction between DNA Polymerase IV and the Replicative Polymerase That Is Required for Translesion Synthesis
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Arabidopsis AtPLC2 Is a Primary Phosphoinositide-Specific Phospholipase C in Phosphoinositide Metabolism and the Endoplasmic Reticulum Stress Response
- Bridges Meristem and Organ Primordia Boundaries through , , and during Flower Development in
- KLK5 Inactivation Reverses Cutaneous Hallmarks of Netherton Syndrome
- XBP1-Independent UPR Pathways Suppress C/EBP-β Mediated Chondrocyte Differentiation in ER-Stress Related Skeletal Disease
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



