-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Recurrent Tissue-Specific mtDNA Mutations Are Common in Humans
Mitochondrial DNA (mtDNA) variation can affect phenotypic variation; therefore, knowing its distribution within and among individuals is of importance to understanding many human diseases. Intra-individual mtDNA variation (heteroplasmy) has been generally assumed to be random. We used massively parallel sequencing to assess heteroplasmy across ten tissues and demonstrate that in unrelated individuals there are tissue-specific, recurrent mutations. Certain tissues, notably kidney, liver and skeletal muscle, displayed the identical recurrent mutations that were undetectable in other tissues in the same individuals. Using RFLP analyses we validated one of the tissue-specific mutations in the two sequenced individuals and replicated the patterns in two additional individuals. These recurrent mutations all occur within or in very close proximity to sites that regulate mtDNA replication, strongly implying that these variations alter the replication dynamics of the mutated mtDNA genome. These recurrent variants are all independent of each other and do not occur in the mtDNA coding regions. The most parsimonious explanation of the data is that these frequently repeated mutations experience tissue-specific positive selection, probably through replication advantage.
Published in the journal: . PLoS Genet 9(11): e32767. doi:10.1371/journal.pgen.1003929
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003929Summary
Mitochondrial DNA (mtDNA) variation can affect phenotypic variation; therefore, knowing its distribution within and among individuals is of importance to understanding many human diseases. Intra-individual mtDNA variation (heteroplasmy) has been generally assumed to be random. We used massively parallel sequencing to assess heteroplasmy across ten tissues and demonstrate that in unrelated individuals there are tissue-specific, recurrent mutations. Certain tissues, notably kidney, liver and skeletal muscle, displayed the identical recurrent mutations that were undetectable in other tissues in the same individuals. Using RFLP analyses we validated one of the tissue-specific mutations in the two sequenced individuals and replicated the patterns in two additional individuals. These recurrent mutations all occur within or in very close proximity to sites that regulate mtDNA replication, strongly implying that these variations alter the replication dynamics of the mutated mtDNA genome. These recurrent variants are all independent of each other and do not occur in the mtDNA coding regions. The most parsimonious explanation of the data is that these frequently repeated mutations experience tissue-specific positive selection, probably through replication advantage.
Introduction
Mitochondrial DNA (mtDNA) heteroplasmy is commonly thought to be the product of either maternal inheritance [1] or rare, random somatic mutations that undergo subsequent expansion within an individual via genetic drift [2], [3]. Inherited heteroplasmy should be present in many, but perhaps not all, tissues while somatic mutations spread only as a result of cell division subsequent to the mutation event. Somatic mutations will be restricted to those cells or tissues derived from a common progenitor, and should follow patterns of development. Such intra-individual patterns should therefore differ among individuals as a function of where and when the initial mutation occurred.
Under these standard models, both inherited and somatic heteroplasmies should differ among unrelated individuals. However, until recent advances in sequencing technology it was impossible to assay low levels of heteroplasmy across the entire mitochondrial genome. We applied massively parallel sequencing technology to test these models by deeply sequencing the same ten tissues from two unrelated individuals. Unexpectedly we found that certain tissues, notably kidney, liver and skeletal muscle, have recurrent mtDNA mutations that were undetectable in other tissues in the same individuals. These mutations were found across unrelated individuals in these same tissues. Neither the maternal inheritance nor the random somatic mutation models explain the observed patterns of recurrent mtDNA heteroplasmy. The common recurrence of these tissue-specific mutations indicates a completely different model of mtDNA heteroplasmy, namely a decidedly non-random process that results in particular mutations, but only in specific tissues.
Results
Sequencing, Coverage and Copy Number
Next generation sequencing provides several advantages over previous methods in that it allows detection of very low heteroplasmy levels across the entire mtDNA genome without having to target specific sites. We sequenced mtDNA from 10 tissues (kidney, lung, liver, small bowel, large bowel, skeletal muscle, spleen, brain white matter, skin above belt and skin below belt) obtained at autopsy from two cancer-free individuals (Text S1). In brief, mtDNA sequences were generated as 100 nt paired-end reads on Illumina HiSeq 2000 machines, were aligned to the human reference genome rCRS [4] using BWA [5], and then were locally realigned and recalibrated using GATK [6]. Variants were reported as heteroplasmic if their frequencies were ≥1% on both strands from reads with a mapping quality score ≥30. We further eliminated variants with any of the following artifacts: strand bias, low average base quality score, or clustering at read ends. Strand bias was evaluated at both base pair and motif levels. We identified 20 heteroplasmic sites with mutation levels >1% in our two subjects (Table 1).
Tab. 1. Complete list of heteroplasmic sites. 
The numbers are the percentages of non-reference alleles as defined by the Cambridge Reference Sequence, NC_012920.1. The “-” denotes no heteroplasmy detected above the threshold of 1%. The coverage of mtDNA sequencing varied across tissues, ranging from over 7,000 to almost 80,000 (Figure 1A). By comparing mtDNA coverage to that for autosomal chromosomes, we estimated the mtDNA copy number per cell for each sample (Figure 1B). The two subjects had similar estimated mtDNA copy numbers in each tissue with values consistent with expectations based on previous data, ranging from a few hundred mtDNA per cell in spleen to a few thousand in the skeletal muscle [7], liver, and kidney.
Fig. 1. Coverage data. 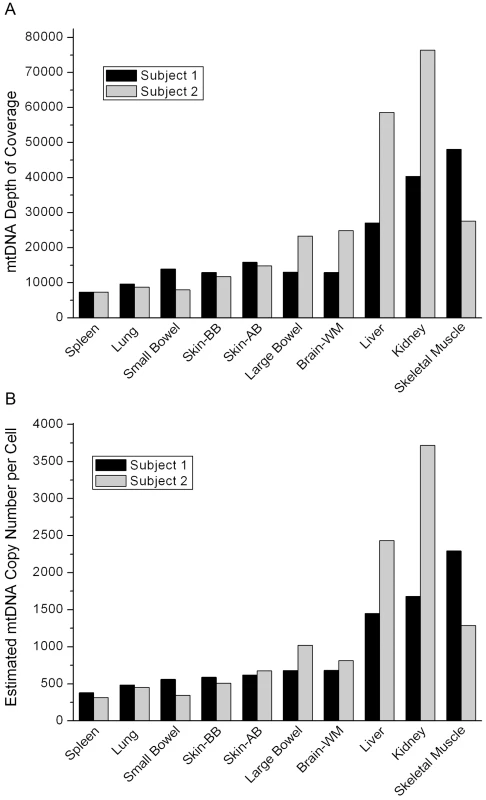
(A) The coverage of the mtDNA in each tissue in the two individuals sequenced in our study. (B) The mtDNA copy number per cell for each tissue estimated from the coverage data. Both subjects died of myocardial infarction and had no evidence of cancer or occult cancer. Subject 1 was a male, age 57 years, while subject 2 was a female, age 71 years (Table 2). This age and gender difference may explain the difference in mtDNA copy number in skeletal muscle between the two subjects.
Tab. 2. Demographics for our subjects. 
MI: myocardial infarction. Recurrence of Site-Specific Heteroplasmy
Unexpectedly, we found eight sites to be heteroplasmic in the same tissues in both of our subjects (Figure 2). Contamination was unlikely since the pattern of these sites fit no known haplogroup, and the samples from the two subjects were collected at different times and sequenced at different facilities (see Materials and Methods). Most of these variants exist in the general population but are rare (Table S2) [8]. Some of these variants have been previously reported as being heteroplasmic in these same tissues but the recurrence and tissue-specificity of these mtDNA variations was not discussed [9]. In the four unrelated individuals combined from our study and that of He et al [9] there were 10 recurrent mutations. All of the recurrent mutations lay within the mtDNA control region, and nine of the ten recurrent mutations occurred in multiple individuals but only in specific tissues (site 16093 was the only exception and this mutation was found in a wide range of tissues). Six of the recurrent mutation sites were observed in both studies (Figure 2). Surprisingly, these recurrent tissue-specific mutations are all close to regulatory sites for mtDNA replication, indicating that these variations are likely to alter the replication dynamics of the mutated mtDNA molecules.
Fig. 2. Graphical representation of the human mtDNA control region (sites 576-1 and 16569-16024). 
All sites found to be heteroplasmic in two or more subjects are listed, with measured percent heteroplasmy given after each tissue name. SM – skeletal muscle. H – heavy strand of mtDNA, L – light strand, OH1 and OH2 – origins of replication of the heavy strand. Site numbering is from rCRS. Three of the ten tissues studied (liver, kidney and skeletal muscle) harbored multiple mtDNA mutations that were shared across two or more of the four individuals combined from both studies (Figure 2). Mutations at sites 60 and 72 occurred in liver and kidney in both studies, while mutations at sites 94 and 203 were repeatedly detected in liver and/or kidney only in our subjects. Of these mutations, three sites (60, 72 and 94) when present in an individual, always occurred in both liver and kidney. Another three occurred in skeletal muscle of all four patients (sites 64, 189 and 408). Heteroplasmy at positions 189 and 408 has been found in skeletal muscle of the elderly [10]. A mutation at site 67 was found only in skeletal muscle in our two subjects. There was no observable linkage disequilibrium among the sites (all pairwise r2<0.007), indicating that these are independent mutations and are not due to contamination. Heteroplasmy for these tissue-specific recurrent mutations ranged from 1–21%, but was not detected in the other tissues. Strikingly, the level of heteroplasmy was similar across individuals in the same tissues (Figure 2). The depth of sequence coverage in these tissues (skeletal muscle, liver and kidney) ranged from 27,000–76,000× (Figure 1A), providing high confidence in our observation of heteroplasmy in these samples. Of the tissues we examined (with the notable exception of brain white matter), skeletal muscle, liver and kidney are the ones most often affected by mitochondrial disease [11].
Although the entire mtDNA genome was sequenced to high depth, we found no single base pair substitutions outside the control region that were repeated between individuals. The common 4977 deletion [12], [13], [14] was found in many tissues as expected. Here we have focused only on those heteroplasmic sites common to multiple subjects; the full list of identified heteroplasmic sites is given in Table 1. These data clearly indicate a non-random distribution (6.0e−4<p<6.0e−6) of recurrent heteroplasmic mutations that flank important regulatory elements for mtDNA replication (Figure 2). Several of the repeated variants were clustered (sites 60–72) near a recently reported origin of replication for the H strand [15], [16] at sites 54–57. Three of the other repeated mutations (189, 203 and 16093) occur very near the boundaries of the displacement loop [15]. Finally, site 408, which was heteroplasmic in the skeletal muscle of all four individuals, lies within the L strand promoter that initiates the RNA primer for mtDNA replication.
One variant, 16093, was unusual in that it was observed in all tested tissues in one subject from each study, with tissue-specific heteroplasmy levels that were strikingly similar across these two individuals (Figure 3; r = 0.93, p<0.003). These subjects were of similar age (59 and 57 years), so it is possible that the 16093 heteroplasmy increases with age at different rates in different tissues, leading to the similar heteroplasmy levels across tissues in these subjects. The 16093 site lies within a loop of a predicted large secondary structure of the mtDNA and is known to be hyper-mutable [17]. Individuals with the 16093C variant in blood tend to have C/T heteroplasmy in buccal cells [18]. In one of our subjects and in patient 11 from He et al. [9], the 16093C variant is the major allele in most tissues (except muscle), and these are the same two individuals who have widespread 16093T/C heteroplasmy across all tested tissues (Figure 2). The similar, tissue-dependent heteroplasmy levels at 16093 reinforce the observation that heteroplasmy levels at other sites are also comparable across individuals (Figures 2 and 3).
Fig. 3. Tissue dependent heteroplasmy levels of the 16093 T/C variant across two subjects. 
The heteroplasmy levels in these two subjects have a correlation of 0.93 (p = 0.003). The diagonal line shows equal values in the two subjects. Patient 11 is from He et al. [9]. Repeated Indel Mutations
We also found two insertion/deletion (indel) somatic mutations that were repeated across the two sequenced subjects in a tissue-specific pattern. Both are length variations in polynucleotide tracts. The human mtDNA reference sequence (rCRS) has a stretch of six guanines, denoted by G6, from sites 66–71. Both of our subjects had measurable heteroplasmy for the G5 variant, decreasing the length of this poly-G tract by one nucleotide. This variant was found in the same four tissues in both subjects: kidney (0.9% and 3.6% in subjects 1 and 2 respectively), large bowel (0.7% and 2.4%), small bowel (1.0% and 4.3%) and the white matter of the brain (1.9% and 3.6%). This poly-G tract is located adjacent to one of the recurrent heteroplasmic SNPs (Figure 2). In kidney, the G5 variant did not occur on the same reads as the heteroplasmic variant at site 72, demonstrating that this is not a sequencing artifact and that the variants are on different mtDNA molecules. Considering its location, it is reasonable to hypothesize that this poly-G length variant also affects mtDNA replication.
The second repeated heteroplasmic indel was in an 8-nucleotide poly-A tract at positions 12418–12425. This is the longest poly-A tract in the rCRS. In both subjects, we found the shorter A7 variant in the kidney samples only (1.0% heteroplasmy in subject 1 and 1.6% in subject 2). We also found the longer A9 variant in both kidney samples, but at very low levels (<1%). This indel is the only repeated mutation that we found in the coding region of the mitochondrial genome. It is located near the start of the MT-ND5 gene (12337–14148) and causes a frameshift mutation, severely altering almost the entire length of the ND5 protein, an essential component of complex I of the electron transfer chain.
Molecular Validation and Replication
To confirm that the observed heteroplasmy was not due to sequencing artifact, we performed an alternative analysis for site G94A because it could be assayed using RFLP analysis. Specifically, the G allele at G94A permits digestion with the restriction enzyme BcoDI. Sensitivity of this assay was determined using a titration of plasmid constructs with and without the restriction site. This assay was specific to mtDNA because the primers did not amplify nuclear DNA (Figure S1A). We detected as low as 2.5% of the undigested variant (Figure S1B). RFLP analysis of all samples from the two sequenced individuals demonstrated that the tissues shown to be heteroplasmic from the sequencing analyses had both digested and undigested bands and were therefore heteroplasmic (Figure 4). As a negative control, we also analyzed this site in spleen DNA and found it to be consistent with the sequencing results (Figure 4). This result was statistically significant (p<0.001).
Fig. 4. Mutation at position 94 is verified by RFLP analysis. 
DNA isolated from kidney (K), liver (L), and spleen (S) from subjects 1, 2, 3 and 4 were subjected to PCR and RFLP analysis. The presence of the mutation at position 94 is seen as the presence of the upper (uncut) band by gel electrophoresis. RFLP analysis of PCR products from wild type (94G), mutant (94A), and mixed (2.5% 94A) plasmid DNA are also loaded for comparison. To test the generality of this heteroplasmy we examined two additional cancer-free individuals (Subjects 3 and 4 in Table 2) using the same RFLP assay. This analysis replicated heteroplasmy in liver in both additional subjects and in kidney in one individual. No significant heteroplasmy was detected in spleen in either subject, providing additional support for the tissue-specificity of this SNP (Figure 4). The result was also statistically significant (p<0.001).
Discussion
Previous Literature on the Recurrent Mutation Sites
Several of our observed heteroplasmic sites have been identified in studies of human disease. The T408A mutation, which was present in the muscle of all four sequenced individuals (two from the present study and two from He et al.), has been reported as an age-related somatic mutation in muscle [10], [19], [20], [21]. It has also been associated with disease in an investigation of a patient with a mitochondrial depletion syndrome [22] that was fatal at a young age (14 years), where the T408A mutation exhibited heteroplasmy at high levels (>70%) in all investigated maternal relatives, but was not detectible in the patient. The authors speculated that the T408A mutation interacted with a hypothesized nuclear DNA factor that affected mtDNA replication, thus leading to the mtDNA depletion in this patient. The A189G mutation, which was also present in the muscle of all four sequenced individuals, has also been reported in studies of aging muscle [10], [19], [20], [21], [23], [24], [25], and is often reported together with T408A. Both of these mutations increase in heteroplasmy level in muscle slowly with increasing age [19].
Heteroplasmy at site 16093 has often been reported in a range of tissues [18], [26], [27], [28], [29], [30], [31], [32], consistent with the observation of this variant being in all tissues in one of our two sequenced subjects and one of the subjects from He et al. The T414G mutation has been reported to accumulate with age in fibroblasts and skeletal muscle [33], [34] and we detected this mutation in one individual in a skin sample (Table 1). Our observation of heteroplasmy at 189, 408, 414, and 16093 and the previous studies reporting these same variants provide support for the validity of the next-generation sequencing data.
We found that four heteroplasmic somatic mutations (T60C, T72C, G94A, and G203A) recur, but only in liver and/or kidney. Given that liver and kidney arise from endoderm and mesoderm respectively, it is unlikely these mutations share a common developmental origin. These four sites are also global population polymorphisms, though at low frequencies [8] (Table S2). Some of these have been previously reported in the context of human disease. The T72C variant has been reported as a somatic mutation in the brain tissue of both Alzheimer's cases and controls [35], although it was not detected in any sequenced brain samples in this study. G94A has recently been reported in two Chinese pedigrees transmitting Lebers Hereditary Optic Neuropathy but in these families this variant was an inherited fixed polymorphism, not a heteroplasmic somatic mutation [36]. Despite the rarity of G203A in the global population (estimated as 0.3% in a survey of human mtDNA sequences deposited in GenBank) [8] it has been identified as a fixed variant in patients with deafness in two independent studies in different ethnicities [37], [38].
Implications of the Recurrent Mutations
Our results indicate that mtDNA heteroplasmy due to somatic mutation is unexpectedly recurrent and tissue specific. By using a sensitive deep-sequencing technique across a wide range of tissues in multiple subjects we were able to test the hypothesis that specific mtDNA variations preferentially accumulate in particular tissues [34], [39]. One possible explanation for observing the same mtDNA heteroplasmic variants in two or more tissue types within the same person is that a mutation occurred early in embryonic development, before the tissues differentiated from their common progenitor. However, this hypothesis cannot explain the repeated observation of the same mutations in the same tissues in unrelated individuals. The occurrence of repeated mutations in the same tissues at sites that closely correspond to regulatory elements for mtDNA replication indicates somatic selection as the most likely mechanism driving the increase and maintenance of these heteroplasmic mutations. This inference is further supported by independent evidence that liver and kidney exhibit positive selection for mtDNA variants in a mouse model formed by artificially mixing the mtDNA of two different mouse strains [40], [41], [42]. In-vivo BrdU labeling in these mice over a time course of 50 hours did not detect any difference in labeling in liver samples between the two mtDNA haplotypes [41], leading the authors to conclude that replicative advantage was not the driving force for the segregation in these mice. However, we would argue that a 50 hour window is not comparable to the decades of replication advantage that would need to occur in our subjects. Recently, Sharpley et al [43] also generated a separate mouse model of heteroplasmy by mixing the naturally occurring NZB and 129S6 mtDNA sequences. This mouse model also showed that the segregation of the two mtDNA genomes varied in a tissue-specific manner, with liver and kidney having the strongest selection for the NZB version of the genome. Finally, liver has been argued to be under selection for nuclear aneuploidy and polyploidy, indicating that selection may have a special role in this tissue [44]. It is reasonable that the tissue specific selection of these mtDNA variants is due to regulation by nuclear-encoded mitochondrial genes with tissue-dependent expression, as has been shown in one of the mouse models in spleen [40]. In contrast to previous work documenting a wide variety of heteroplasmic sites and their functional implications, the unique value of this study is the comparison of mtDNA heteroplasmy across multiple tissues in several individuals and the demonstration that several somatic variants recur in a tissue-specific pattern.
The pattern of tissue-specific mutations we have found across multiple individuals could be explained by a few alternatives, including positive or negative selection. Under positive selection, mutations in certain tissues would increase in frequency due to their advantage. Under negative selection, mutations could occur at a high rate but would be removed from all tissues except for those where the recurrence is observed, where negative selection is presumably relaxed. Of these two alternatives, positive selection is the most likely explanation because under negative selection mutations should be scattered widely across the mtDNA control region, not just the recurrent ones at specific sites related to replication (Figure 2). In contrast, positive selection could simply be explained by a replication or other functional advantage in high copy number tissues due to increased mtDNA replication. It is important to note than any functional difference among the variant mtDNA molecules, if any, are due only to the sites we describe because all of our observed heteroplasmic sites are independent of each other, and there were no recurrent heteroplasmic sites in the coding regions.
Another alternative is that there are tissue-specific mutational hotspots within the mtDNA. For example, interferon-induced cytidine deaminases are capable of generating somatic mtDNA mutation in a tissue-specific fashion [45]. Although this alternative is not mutually exclusive to the selection argument, we still favor differential selection based on its simplicity, and on prior data suggesting that two of the tissues in which we observed recurrent mtDNA mutations, liver and kidney, also undergo selection in two separate heteroplasmic mouse models [40], [41], [42], [43]. The mouse models provide evidence suggesting that selection in the absence of any de-novo mutation generation can cause tissue specific heteroplasmy patterns because in both mouse models the two mtDNA haplotypes were artificially introduced through cell fusion and were not generated via a mutation process. Furthermore, even if the heteroplasmic sites we observed are mutational hotspots, their locations in the mtDNA genome are highly suggestive of functional roles (Figure 2). Homoplasmy in tumors has also been shown to possibly derive from random processes alone, but this computer simulation study is not directly analogous to recurrent heteroplasmy in normal tissues of multiple individuals [46]. Therefore, the most parsimonious and reasonable explanation for our data is positive selection in liver, kidney and skeletal muscle for certain mutations in and around the regions controlling mtDNA replication.
In a very different model system (i.e. a mouse strain with an abnormally high mutation rates due to a defective mtDNA polymerase) evidence for lower mutation load in the D loop was described [47]. Specifically, in this mouse model the accumulation of point mutations in the mtDNA was lower in the D loop region than the rest of the mtDNA. It is impossible to determine conclusively from these data whether this pattern is due to selection or to a variation in mutation rate, but it does demonstrate a non-random pattern in this part of the mtDNA, something we also observed but in a different direction.
Our data provide strong support for the conclusion that the current models of mtDNA variation are inadequate to explain what we now call “recurrent heteroplasmy”. The pattern of common, recurrent mutations we observed provides strong evidence that mtDNA heteroplasmy at several sites is non-random and is most likely the result of tissue-specific positive selection acting on the replication of mtDNA. The restriction of these mutations to liver, kidney and skeletal muscle indicates that the mtDNA replication process may vary across tissues, leading to tissue-specific selective forces, which correspond with high copy number tissues.
Materials and Methods
Tissue Collection
The protocols were approved by the Vanderbilt University Institutional Review Board. Samples were collected at autopsy within 48 hours post-mortem by a trained pathologist (RDH). Tissue samples for DNA extraction were collected using clean and sterile scalpels, placed in petri dishes, and transferred to 50 ml tubes containing ice-cold Dulbecco's phosphate-buffered saline (DPBS), rinsed again with DPBS and stored at −80°C until DNA extraction with exceptions as described below. Separate portions of the tissue sections were preserved in 10% formalin. Skin samples were collected from the ventral torso, from both above-belt (Skin-AB) and below-belt (Skin-BB) (e.g. above or below the waistline). Skeletal muscle was obtained from the diaphragm. The small and large bowel samples consisted of mucosal tissue that was collected by carefully scraping the loose mucosal layer from the internal surface of bowel sections. Bone marrow tissue was collected by flushing rib or vertebral body sections with DPBS and collecting the flushed material in a 50 ml tube on ice, which was then centrifuged to collect the cellular material. Splenocytes were isolated as previously described [48]. Gray and white brain samples were separated manually in a petri dish using a sterile scalpel. Demographic information for the four subjects is in Table 2.
DNA Extraction
For each tissue two DNA extractions were performed. The tissue was lysed using the DNeasy Blood and Tissue Kit (Qiagen 69504). Once the tissue was lysed and incubated at 56°C overnight, one set of DNA extractions was transferred into a 2.0 ml tube (Sarstedt 72.694.406) and put on the QIAsymphony for automated extraction (Qiagen). The protocols used on the QIAsymphony were Tissue_LC_200_V5 and Tissue_HC_200_V5 depending on the tissue type. The second set of DNA extractions was transferred to Autopure Qubes D (Qiagen 949022) and 3 ml of Cell Lysis Solution (Qiagen 949006) was added to each tissue sample. These samples were then placed on the Autopure LS (Qiagen) for automated extraction. The protocol used on the Autopure was Cell Lysate. The resulting DNA was stored in Nunc Cryotubes (Nunc 377267). DNA from each protocol was calibrated and samples combined prior to sequencing.
Sequencing and Data Processing
Sequences were generated as 100 nt paired-end reads on Illumina HiSeq 2000 machines. The two subjects were sequenced at different locations (subject 1 at Macrogen in South Korea and subject 2 at Illumina in California). Each sample was sequenced on 3–5 lanes, yielding 1.14–1.99 billion reads, which were aligned to the human reference genome hg19+rCRS (revised Cambridge Reference Sequence, NC_012920.1) using BWA [5] (ver. 0.5.9-r16). We performed local realignment and base quality score recalibration using GATK [6] (ver. 1.0.5974). The number of mapped reads ranged from 1.09–1.84 billion, with more than 90% of all reads being mapped except skeletal muscle from subject 2 (86.2%) (Table S1). Other programs were also used at various steps: samtools (ver. 0.1.16) for sorting and indexing bam files, bamtools (ver. 0.8.1025) for splitting and merging bam files, and picard (version 1.48) for marking duplicates and fixing mates after local realignment.
All samples were also genotyped on the Illumina Human Omni1 Quad chip with approximately one million SNPs in the nuclear genome. The consistency rate between the sequence - and chip-based SNP calls was >99.86% for all samples after standard quality control filtering. This indicates that the sequencing data were of high quality.
Identification of Heteroplasmy and Testing for Artifacts Due to Non-Circularity of the Reference Genome
We screened for heteroplasmy in mtDNA using reads with MAPQ≥30. For each site, we calculated the fraction of bases A, C, G, T on the forward and reverse strands. A site was called heteroplasmic if it had ≥1% frequency for two or more bases on both strands and the variant did not have any of the following alignment artifacts: 1) strand bias, 2) clustering at read ends, and 3) low average base quality score. Due to the high read depth (Figure 1, Table S1) all duplicate reads were retained. Heteroplasmy estimates were assessed with and without the duplicate reads and heteroplasmy levels were not influenced by the duplicates.
The linear mtDNA reference genome (rCRS) was created by cutting the circular mtDNA at a fixed position. This may generate alignment artifacts near the linearization site (i.e., the ends of the mtDNA reference): 1) a read overlapping the linearization site may be unmapped or require heavy clipping to be aligned, 2) a read may be aligned but its paired-end mate may not be, and 3) a read may be aligned to one end of the reference but its mate to the other end. As a result, reads close to the linearization site may have low mapping quality scores and may be disproportionately filtered out. As our data are 100 nt reads with insert sizes mostly between 250 bp and 400 bp (Figure S2), these artifacts may influence the results hundreds of bases away from the mtDNA “ends”. To prevent artifacts due to mtDNA circularity we also created a new mtDNA reference genome by shifting the rCRS starting point to position 7002, and repeated the whole data processing steps as described above. Heteroplasmy was virtually identical between the two alignments, with less than 0.1% difference in heteroplasmy estimates.
In addition, the artificial N base at 3107 of the rCRS reference can lead to alignment artifacts. This N was removed before the alignment was made.
Tests for Other Potential Sequencing Artifacts
For all observed heteroplasmic sites, we checked for various sequencing artifacts. The mtDNA control region harbors multiple poly-nucleotide tracts that could lead to sequencing artifacts. Since these artifacts often have strand bias, we filtered out all sites with strand bias. In addition, for each heteroplasmic site we performed a motif analysis similar to that described in detail for site 310 in the supplementary material, to identify artifacts (Table S4). We also checked for the presence of artifacts due to sequence similarities between the nuclear and mtDNA genomes (NUMT), and none could be detected (see Text S1). The reported sites are free of any artifacts.
Distributions of Cycle and Strand for the Mutant Alleles
Alignment errors are known to cause artifacts that often show strand bias and excessive occurrence of mutant alleles at read ends. We found no cycle (i.e. position on the read) or strand bias for the mutant alleles at the heteroplasmic sites we identified. We extracted all bases with mapping quality score ≥30 and base quality score ≥20 at each heteroplasmic site and compared the distributions of cycle and strand for the major and mutant alleles. Figure S3 shows the distributions of C and A alleles for site 64 in the skeletal muscle of Subject 1. The mutant allele was uniformly distributed across the read length on both strands, showing no strand or cycle bias. Other heteroplasmic sites we identified had similar patterns.
Levels of Linkage Disequilibrium between Heteroplasmic Sites
For heteroplasmic sites close enough to be on the same DNA read or read pair, we assessed whether the minor alleles are on the same haplotype background, or in other words, if they are in linkage disequilibrium (LD). LD between the variants could be a sign of either contamination or sequencing artifacts. Specifically, for every pair of sites ≤100 bp apart (e.g., 60-72-94 in liver and kidney tissues and 64–67 in skeletal muscles), we extracted reads that covered both positions and had MAPQ≥30. We further required that the reads had all bases matched (i.e., CIGAR string “100M”) or had clipping at one end (i.e., CIGAR string matching the regular expression pattern “[0–9]*S[0–9]*M” or “[0–9]*M[0–9]*S”), and the bases at the two sites had base quality score ≥20. We then tallied haplotypes and calculated r2 between the two sites. All r2 values were very close to zero (<0.004), indicating no LD between any heteroplasmic sites.
For every pair of sites >100 bp apart (e.g. between 60-72-94 and 203 in liver tissues; among 64–67, 189, and 408 in skeletal muscles), we extracted read pairs that covered both positions and had MAPQ≥30 and then followed the above procedure. Again, all r2 values were very close to zero (<0.007), indicating no LD in the mtDNA.
Sequencing Error Rate
The sequencing error rate is reflected in the recalibrated base quality scores. For example, a base quality (BQ) score of 25 means the error rate for that base is 0.32%, BQ = 27 means 0.2%, and BQ = 30 means 0.1%. These error rates are much lower than the 1% detection cutoff we used for the determination of heteroplasmic sites. Figure S4 shows the distribution of recalibrated base quality scores. For all our samples, 82.4% bases had recalibrated base quality score ≥30, 91.4% had scores ≥27, and 94.4% had scores ≥25. These results provide assurance that the bases we used for our inferences had high quality and an error rate much lower than our heteroplasmy detection threshold.
Estimation of mtDNA Copy Number
We calculated the depth of coverage for autosomes and mtDNA as:
The multiplier 100 was used because we had 100 nt reads. The mtDNA depth ranged from 5651×–119203×, and the autosome depth ranged 37×–61× (Table S1). Assuming each cell carries a diploid (2×) nuclear genome, the mtDNA copy number was estimated as: The estimated mtDNA copy number ranged from 315 to 5880 (Table S1).Statistical Tests
To test for non-randomness of the recurrent mutations, we calculated the probability that a mutation occurred at these 10 sites under two extreme scenarios. Using a model of constant mutation rate, c, along the whole mtDNA genome, the probability for a mutation to occur anywhere would be 16569c and the probability for it to occur at these 10 sites would be 10c. Thus the probability for an observed mutation in a DNA sample to occur only at any of these 10 sites is 10/16569 = 6.0e−4. Now suppose a mutation has been observed in a specific tissue of an individual. The probability to observe the same mutation in another individual only in the same tissue (out of 10 tissues) and on the same site (out of 10 sites) is further reduced to 6.0e−6. The recurrence patterns of the reported mutations fall between these two extreme scenarios, and therefore their p-values are between 6.0e−4 and 6.0e−6.
To test for correlation of 16093 heteroplasmy levels between the two individuals (Figure 3), we calculated the Pearson correlation coefficient and its associated p-value. The correlation was 0.93 and the p-value was 0.0028.
We calculated the p-value to evaluate the significance of our RFLP validation and replication results. For the validation part, we performed RFLP on six tissues (kidney, liver, spleen from Subjects 1–2). Let a = P(detect 94A|94A is absent), the probability of falsely detecting 94A in RFLP analysis while it was absent. Then the probability of seeing 94A in two kidneys, two livers but not the two spleens is a4(1−a)2. The value of a is probably lower than the false positive rate for sequencing analysis, which would be at most 0.2 (4 out of 20 tissues when the sequencing results were assumed to be false). Even at a = 0.2, the p-value will be a4(1−a)2 = 0.001. The p-value will be much smaller at a lower value of a; for example, p = 5.6e−6 if a = 0.05, and p = 9.8e−9 if a = 0.01. The p-value for the replication part can be similarly evaluated.
Haplogroup Determination
We determined the mtDNA haplogroups for our subjects: T2a1 for subject 1 and H1a1 for subject 2 (Table S3). Haplogrouping was performed using the H-Mito program (http://www.phylotree.org) supplied by Mannis van Oven [49].
RFLP Analysis
To provide molecular validation of sequencing results, we performed RFLP analysis using control plasmids and patient DNA from suspected heteroplasmic and homoplasmic tissues. We focused on position 94, which sequencing results identified as heteroplasmic in kidney and liver. Control samples consisted of 100% wild-type plasmids at position 94 (G), 100% mutant plasmids (A), or 97.5% wild-type and 2.5% mutant plasmids. DNA samples from suspected heteroplasmic kidney and liver tissues, and from suspected homoplasmic spleen tissue were analyzed for subjects 1–4. Fifteen nanograms of the control plasmid or patient DNA was amplified with 10 µM D-loop-targeted forward (5′-GATCACAGGTCTATCACCCTATTAAC-3′) and reverse (5′-CAGATACTGCGACATAGGGTGCT-3′) primers (Operon) and Platinum PCR Supermix (Invitrogen) according to manufacturer's directions. Following amplification, PCR products were digested for 8 h at 37°C with the restriction enzyme BcoDI, which cuts the wild-type but not mutant PCR product at position 94. Successful digestion resulted in cutting of the 130-bp PCR product into 90 - and 40-bp fragments. We added 5 µL 5× gel-loading dye (KD Medical/MediaTech) to each 20-uL reaction after restriction digest, and loaded 12 µL of digest products and gel-loading dye into each well of a 3% agarose (Sigma) gel in 1× TBE (Cellgro) with 0.0125% ethidium bromide (Bio-Rad). The gel was run at 150 V for 2 h, then UV imaged for 200 ms in a Syngene G:Box imager.
Supporting Information
Zdroje
1. CreeLM, SamuelsDC, ChinneryPF (2009) The inheritance of pathogenic mitochondrial DNA mutations. Biochimica Et Biophysica Acta-Molecular Basis of Disease 1792 : 1097–1102.
2. ElsonJL, SamuelsDC, TurnbullDM, ChinneryPF (2001) Random intracellular drift explains the clonal expansion of mitochondrial DNA mutations with age. American Journal of Human Genetics 68 : 802–806.
3. LarssonNG (2010) Somatic Mitochondrial DNA Mutations in Mammalian Aging. Annual Review of Biochemistry 79 : 683–706.
4. AndrewsRM, KubackaI, ChinneryPF, LightowlersRN, TurnbullDM, et al. (1999) Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nature Genetics 23 : 147–147.
5. LiH, DurbinR (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25 : 1754–1760.
6. McKennaA, HannaM, BanksE, SivachenkoA, CibulskisK, et al. (2010) The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Research 20 : 1297–1303.
7. MillerFJ, RosenfeldtFL, ZhangCF, LinnaneAW, NagleyP (2003) Precise determination of mitochondrial DNA copy number in human skeletal and cardiac muscle by a PCR-based assay: lack of change of copy number with age. Nucleic Acids Research 31 ((11)): e61.
8. PereiraL, FreitasF, FernandesV, PereiraJB, CostaMD, et al. (2009) The Diversity Present in 5140 Human Mitochondrial Genomes. American Journal of Human Genetics 84 : 628–640.
9. HeYP, WuJ, DressmanDC, Iacobuzio-DonahueC, MarkowitzSD, et al. (2010) Heteroplasmic mitochondrial DNA mutations in normal and tumour cells. Nature 464 : 610–614.
10. WangY, MichikawaY, MallidisC, BaiY, WoodhouseL, et al. (2001) Muscle-specific mutations accumulate with aging in critical human mtDNA control sites for replication. Proceedings of the National Academy of Sciences of the United States of America 98 : 4022–4027.
11. DimauroS, DavidzonG (2005) Mitochondrial DNA and disease. Annals of Medicine 37 : 222–232.
12. Corral-DebrinskiM, HortonT, LottMT, ShoffnerJM, BealMF, et al. (1992) Mitochondrial-DNA Deletions in Human Brain - Regional Variability and Increase with Advanced Age. Nature Genetics 2 : 324–329.
13. CortopassiGA, ShibataD, SoongNW, ArnheimN (1992) A Pattern of Accumulation of a Somatic Deletion of Mitochondrial-DNA in Aging Human Tissues. Proceedings of the National Academy of Sciences of the United States of America 89 : 7370–7374.
14. KrishnanKJ, ReeveAK, SamuelsDC, ChinneryPF, BlackwoodJK, et al. (2008) What causes mitochondrial DNA deletions in human cells? Nature Genetics 40 : 275–279.
15. FishJ, RauleN, AttardiG (2004) Discovery of a major D-loop replication origin reveals two modes of human mtDNA synthesis. Science 306 : 2098–2101.
16. YasukawaT, YangMY, JacobsHT, HoltIJ (2005) A bidirectional origin of replication maps to the major noncoding region of human mitochondrial DNA. Molecular Cell 18 : 651–662.
17. PereiraF, SoaresP, CarneiroJ, PereiraL, RichardsMB, et al. (2008) Evidence for Variable Selective Pressures at a Large Secondary Structure of the Human Mitochondrial DNA Control Region. Molecular Biology and Evolution 25 : 2759–2770.
18. AndrewT, CallowayCD, StuartS, LeeSH, GillR, et al. (2011) A Twin Study of Mitochondrial DNA Polymorphisms Shows that Heteroplasmy at Multiple Sites Is Associated with mtDNA Variant 16093 but Not with Zygosity. Plos One 6 ((8)) e22332 doi:10.1371/journal.pone.0022332
19. Del BoR, BordoniA, BoneschiFM, CrimiM, SciaccoM, et al. (2002) Evidence and age-related distribution of mtDNA D-loop point mutations in skeletal muscle from healthy subjects and mitochondrial patients. Journal of the Neurological Sciences 202 : 85–91.
20. Del BoR, CrimiM, SciaccoM, MalferrariG, BordoniA, et al. (2003) High mutational burden in the mtDNA control region from aged muscles: a single-fiber study. Neurobiology of Aging 24 : 829–838.
21. AvitalG, BuchshtavM, ZhidkovI, TuvalJ, DadonS, et al. (2012) Mitochondrial DNA heteroplasmy in diabetes and normal adults: role of acquired and inherited mutational patterns in twins. Human Molecular Genetics 21 : 4214–4224.
22. BarthelemyC, de BaulnyHO, LombesA (2002) D-loop mutations in mitochondrial DNA: link with mitochondrial DNA depletion? Human Genetics 110 : 479–487.
23. CormioA, MilellaF, VecchietJ, FelzaniG, GadaletaMN, et al. (2005) Mitochondrial DNA mutations in RRF of healthy subjects of different age. Neurobiology of Aging 26 : 655–664.
24. da CostaCK, KiyomotoBH, SchmidtB, OliveiraASB, GabbaiAA, et al. (2007) Age-related mitochondrial DNA point mutations in patients with mitochondrial myopathy. Journal of the Neurological Sciences 263 : 139–144.
25. PayneBAI, WilsonIJ, Yu-Wai-ManP, CoxheadJ, DeehanD, et al. (2013) Universal heteroplasmy of human mitochondrial DNA. Human Molecular Genetics 22 : 384–390.
26. AllardMW, PolanskeyD, MillerK, WilsonMR, MonsonKL, et al. (2005) Characterization of human control region sequences of the African American SWGDAM forensic mtDNA data set. Forensic Science International 148 : 169–179.
27. AllardMW, PolanskeyD, WilsonMR, MonsonKL, BudowleB (2006) Evaluation of variation in control region sequences for Hispanic individuals in the SWGDAM mtDNA data set. Journal of Forensic Sciences 51 : 566–573.
28. MalyarchukBA, DerenkoMV (2001) Variation of human mitochondrial DNA: Distribution of hot spots in hypervariable segment I of the major noncoding region. Russian Journal of Genetics 37 : 823–832.
29. PaiCY, HsiehLL, LeeTC, YangSB, LinvilleJ, et al. (2006) Mitochondrial DNA sequence alterations observed between blood and buccal cells within the same individuals having betel quid (BQ)-chewing habit. Forensic Science International 156 : 124–130.
30. PicornellA, GimenezP, CastroJA, RamonMM (2006) Mitochondrial DNA sequence variation in Jewish populations. International Journal of Legal Medicine 120 : 271–281.
31. TullyG, BarrittSM, BenderK, BrignonE, CapelliC, et al. (2004) Results of a collaborative study of the EDNAP group regarding mitochondrial DNA heteroplasmy and segregation in hair shafts. Forensic Science International 140 : 1–11.
32. TullyLA, ParsonsTJ, SteighnerRJ, HollandMM, MarinoMA, et al. (2000) A sensitive denaturing gradient-gel electrophoresis assay reveals a high frequency of heteroplasmy in hypervariable region 1 of the human mtDNA control region. American Journal of Human Genetics 67 : 432–443.
33. MichikawaY, MazzucchelliF, BresolinN, ScarlatoG, AttardiG (1999) Aging-dependent large accumulation of point mutations in the human mtDNA control region for replication. Science 286 : 774–779.
34. MurdockDG, ChristacosNC, WallaceDC (2000) The age-related accumulation of a mitochondrial DNA control region mutation in muscle, but not brain, detected by a sensitive PNA-directed PCR clamping based method. Nucleic Acids Research 28 : 4350–4355.
35. CoskunPE, BealMF, WallaceDC (2004) Alzheimer's brains harbor somatic mtDNA control-region mutations that suppress mitochondrial transcription and replication. Proceedings of the National Academy of Sciences of the United States of America 101 : 10726–10731.
36. ShuL, ZhangYM, HuangXX, ChenCY, ZhangXN (2012) Complete mitochondrial DNA sequence analysis in two southern Chinese pedigrees with Leber hereditary optic neuropathy revealed secondary mutations along with the primary mutation. International Journal of Ophthalmology 5 : 28–31.
37. JanssenGMC, NeuA, t'HartLM, van de SandeCMT, MaassenJA (2006) Novel mitochondrial DNA length variants and genetic instability in a family with diabetes and deafness. Experimental and Clinical Endocrinology & Diabetes 114 : 168–174.
38. YamasobaT, GotoY, OkaY, NishinoI, TsukudaK, et al. (2002) Atypical muscle pathology and a survey of cis-mutations in deaf patients harboring a 1555 A-to-G point mutation in the mitochondrial ribosomal RNA gene. Neuromuscular Disorders 12 : 506–512.
39. CoskunPE, Ruiz-PesiniE, WallaceDC (2003) Control region mtDNA variants: Longevity, climatic adaptation, and a forensic conundrum. Proceedings of the National Academy of Sciences of the United States of America 100 : 2174–2176.
40. BattersbyBJ, Loredo-OstiJC, ShoubridgeEA (2003) Nuclear genetic control of mitochondrial DNA segregation. Nature Genetics 33 : 183–186.
41. BattersbyBJ, ShoubridgeEA (2001) Selection of a mtDNA sequence variant in hepatocytes of heteroplasmic mice is not due to differences in respiratory chain function or efficiency of replication. Human Molecular Genetics 10 : 2469–2479.
42. JenuthJP, PetersonAC, ShoubridgeEA (1997) Tissue-specific selection for different mtDNA genotypes in heteroplasmic mice. Nature Genetics 16 : 93–95.
43. SharpleyMS, MarciniakC, Eckel-MahanK, McManusM, CrimiM, et al. (2012) Heteroplasmy of Mouse mtDNA Is Genetically Unstable and Results in Altered Behavior and Cognition. Cell 151 : 333–343.
44. DuncanAW, TaylorMH, HickeyRD, NewellAEH, LenziML, et al. (2010) The ploidy conveyor of mature hepatocytes as a source of genetic variation. Nature 467 : 707–710.
45. SuspeneR, AynaudMM, GuetardD, HenryM, EckhoffG, et al. (2011) Somatic hypermutation of human mitochondrial and nuclear DNA by APOBEC3 cytidine deaminases, a pathway for DNA catabolism. Proceedings of the National Academy of Sciences of the United States of America 108 : 4858–4863.
46. CollerHA, KhrapkoK, BodyakND, NekhaevaE, Herrero-JimenezP, et al. (2001) High frequency of homoplasmic mitochondrial DNA mutations in human tumors can be explained without selection. Nature Genetics 28 : 147–150.
47. StewartJB, FreyerC, ElsonJL, WredenbergA, CansuZ, et al. (2008) Strong purifying selection in transmission of mammalian mitochondrial DNA. Plos Biology 6 : 63–71.
48. ChandyKC, BeetonC (2007) Preparing T cell growth factor from rat splenocytes. Journal of Visualized Experiments 10: e402.
49. van OvenM, KayserM (2009) Updated Comprehensive Phylogenetic Tree of Global Human Mitochondrial DNA Variation. Human Mutation 30: E386–E394.
Štítky
Genetika Reprodukční medicína
Článek Ribosome Synthesis and MAPK Activity Modulate Ionizing Radiation-Induced Germ Cell Apoptosis inČlánek Fission Yeast Shelterin Regulates DNA Polymerases and Rad3 Kinase to Limit Telomere Extension
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 11- Akutní intermitentní porfyrie
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Růst a vývoj dětí narozených pomocí IVF
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
-
Všechny články tohoto čísla
- Molecular Recognition by a Polymorphic Cell Surface Receptor Governs Cooperative Behaviors in Bacteria
- The Light Skin Allele of in South Asians and Europeans Shares Identity by Descent
- Ribosome Synthesis and MAPK Activity Modulate Ionizing Radiation-Induced Germ Cell Apoptosis in
- Retrotransposon Silencing During Embryogenesis: Cuts in LINE
- Roles of XRCC2, RAD51B and RAD51D in RAD51-Independent SSA Recombination
- Parallel Evolution of Chordate Regulatory Code for Development
- A Genetic Approach to the Recruitment of PRC2 at the Locus
- Deletion of the Murine Cytochrome P450 Locus by Fused BAC-Mediated Recombination Identifies a Role for in the Pulmonary Vascular Response to Hypoxia
- Elevated Mutagenesis Does Not Explain the Increased Frequency of Antibiotic Resistant Mutants in Starved Aging Colonies
- Deletion of an X-Inactivation Boundary Disrupts Adjacent Gene Silencing
- Interplay between Active Chromatin Marks and RNA-Directed DNA Methylation in
- Recombinogenic Conditions Influence Partner Choice in Spontaneous Mitotic Recombination
- Crosstalk between NSL Histone Acetyltransferase and MLL/SET Complexes: NSL Complex Functions in Promoting Histone H3K4 Di-Methylation Activity by MLL/SET Complexes
- A New Role for the GARP Complex in MicroRNA-Mediated Gene Regulation
- RNAi-Dependent and Independent Control of LINE1 Accumulation and Mobility in Mouse Embryonic Stem Cells
- Loss of DNMT1o Disrupts Imprinted X Chromosome Inactivation and Accentuates Placental Defects in Females
- Inhibition of the Smc5/6 Complex during Meiosis Perturbs Joint Molecule Formation and Resolution without Significantly Changing Crossover or Non-crossover Levels
- Disruption of Lipid Metabolism Genes Causes Tissue Overgrowth Associated with Altered Developmental Signaling
- Translation Initiation Factors eIF3 and HCR1 Control Translation Termination and Stop Codon Read-Through in Yeast Cells
- Recruitment of TREX to the Transcription Machinery by Its Direct Binding to the Phospho-CTD of RNA Polymerase II
- MYB97, MYB101 and MYB120 Function as Male Factors That Control Pollen Tube-Synergid Interaction in Fertilization
- Oct4 Is Required ∼E7.5 for Proliferation in the Primitive Streak
- Contrasted Patterns of Crossover and Non-crossover at Meiotic Recombination Hotspots
- Transposable Prophage Mu Is Organized as a Stable Chromosomal Domain of
- Ash1l Methylates Lys36 of Histone H3 Independently of Transcriptional Elongation to Counteract Polycomb Silencing
- Fine-Mapping the Genetic Association of the Major Histocompatibility Complex in Multiple Sclerosis: HLA and Non-HLA Effects
- Genomic Mechanisms Accounting for the Adaptation to Parasitism in Nematode-Trapping Fungi
- Decoding a Signature-Based Model of Transcription Cofactor Recruitment Dictated by Cardinal Cis-Regulatory Elements in Proximal Promoter Regions
- Removal of Misincorporated Ribonucleotides from Prokaryotic Genomes: An Unexpected Role for Nucleotide Excision Repair
- Fission Yeast Shelterin Regulates DNA Polymerases and Rad3 Kinase to Limit Telomere Extension
- Activin Signaling Targeted by Insulin/dFOXO Regulates Aging and Muscle Proteostasis in
- Activin-Like Kinase 2 Functions in Peri-implantation Uterine Signaling in Mice and Humans
- Demographic Divergence History of Pied Flycatcher and Collared Flycatcher Inferred from Whole-Genome Re-sequencing Data
- Recurrent Tissue-Specific mtDNA Mutations Are Common in Humans
- The Histone Variant His2Av is Required for Adult Stem Cell Maintenance in the Testis
- The Maternal-to-Zygotic Transition Targets Actin to Promote Robustness during Morphogenesis
- Reconstructing the Population Genetic History of the Caribbean
- and Are Required for Growth under Iron-Limiting Conditions
- Whole Genome, Whole Population Sequencing Reveals That Loss of Signaling Networks Is the Major Adaptive Strategy in a Constant Environment
- Neuron-Specific Feeding RNAi in and Its Use in a Screen for Essential Genes Required for GABA Neuron Function
- RNA∶DNA Hybrids Initiate Quasi-Palindrome-Associated Mutations in Highly Transcribed Yeast DNA
- Mouse BAZ1A (ACF1) Is Dispensable for Double-Strand Break Repair but Is Essential for Averting Improper Gene Expression during Spermatogenesis
- Genetic and Functional Studies Implicate Synaptic Overgrowth and Ring Gland cAMP/PKA Signaling Defects in the Neurofibromatosis-1 Growth Deficiency
- DUX4 Binding to Retroelements Creates Promoters That Are Active in FSHD Muscle and Testis
- Pathways-Driven Sparse Regression Identifies Pathways and Genes Associated with High-Density Lipoprotein Cholesterol in Two Asian Cohorts
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- and Are Required for Growth under Iron-Limiting Conditions
- Genetic and Functional Studies Implicate Synaptic Overgrowth and Ring Gland cAMP/PKA Signaling Defects in the Neurofibromatosis-1 Growth Deficiency
- The Light Skin Allele of in South Asians and Europeans Shares Identity by Descent
- RNA∶DNA Hybrids Initiate Quasi-Palindrome-Associated Mutations in Highly Transcribed Yeast DNA
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání





