-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Inhibition of the Smc5/6 Complex during Meiosis Perturbs Joint Molecule Formation and Resolution without Significantly Changing Crossover or Non-crossover Levels
Meiosis is a specialized cell division used by diploid organisms to form haploid gametes for sexual reproduction. Central to this reductive division is repair of endogenous DNA double-strand breaks (DSBs) induced by the meiosis-specific enzyme Spo11. These DSBs are repaired in a process called homologous recombination using the sister chromatid or the homologous chromosome as a repair template, with the homolog being the preferred substrate during meiosis. Specific products of inter-homolog recombination, called crossovers, are essential for proper homolog segregation at the first meiotic nuclear division in budding yeast and mice. This study identifies an essential role for the conserved Structural Maintenance of Chromosomes (SMC) 5/6 protein complex during meiotic recombination in budding yeast. Meiosis-specific smc5/6 mutants experience a block in DNA segregation without hindering meiotic progression. Establishment and removal of meiotic sister chromatid cohesin are independent of functional Smc6 protein. smc6 mutants also have normal levels of DSB formation and repair. Eliminating DSBs rescues the segregation block in smc5/6 mutants, suggesting that the complex has a function during meiotic recombination. Accordingly, smc6 mutants accumulate high levels of recombination intermediates in the form of joint molecules. Many of these joint molecules are formed between sister chromatids, which is not normally observed in wild-type cells. The normal formation of crossovers in smc6 mutants supports the notion that mainly inter-sister joint molecule resolution is impaired. In addition, return-to-function studies indicate that the Smc5/6 complex performs its most important functions during joint molecule resolution without influencing crossover formation. These results suggest that the Smc5/6 complex aids primarily in the resolution of joint molecules formed outside of canonical inter-homolog pathways.
Published in the journal: . PLoS Genet 9(11): e32767. doi:10.1371/journal.pgen.1003898
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003898Summary
Meiosis is a specialized cell division used by diploid organisms to form haploid gametes for sexual reproduction. Central to this reductive division is repair of endogenous DNA double-strand breaks (DSBs) induced by the meiosis-specific enzyme Spo11. These DSBs are repaired in a process called homologous recombination using the sister chromatid or the homologous chromosome as a repair template, with the homolog being the preferred substrate during meiosis. Specific products of inter-homolog recombination, called crossovers, are essential for proper homolog segregation at the first meiotic nuclear division in budding yeast and mice. This study identifies an essential role for the conserved Structural Maintenance of Chromosomes (SMC) 5/6 protein complex during meiotic recombination in budding yeast. Meiosis-specific smc5/6 mutants experience a block in DNA segregation without hindering meiotic progression. Establishment and removal of meiotic sister chromatid cohesin are independent of functional Smc6 protein. smc6 mutants also have normal levels of DSB formation and repair. Eliminating DSBs rescues the segregation block in smc5/6 mutants, suggesting that the complex has a function during meiotic recombination. Accordingly, smc6 mutants accumulate high levels of recombination intermediates in the form of joint molecules. Many of these joint molecules are formed between sister chromatids, which is not normally observed in wild-type cells. The normal formation of crossovers in smc6 mutants supports the notion that mainly inter-sister joint molecule resolution is impaired. In addition, return-to-function studies indicate that the Smc5/6 complex performs its most important functions during joint molecule resolution without influencing crossover formation. These results suggest that the Smc5/6 complex aids primarily in the resolution of joint molecules formed outside of canonical inter-homolog pathways.
Introduction
Meiosis is the cell division by which haploid gametes are created in sexually reproducing organisms. It is specialized to preserve the chromosome number among generations and to create genetic diversity in a population. Meiosis begins with the replication of each homologous parental chromosome (homolog) into a pair of sister chromatids. Two sequential rounds of DNA segregation then follow. The first, MI, segregates the homologs away from each other, while the second, MII, separates the sister chromatids. This leads to the formation of four haploid cells from a single diploid parent. Prior to homolog segregation, programmed DNA double-strand breaks (DSBs) are induced that are repaired through a process called homologous recombination. In budding yeast and mice, recombination is essential for proper homolog segregation at MI. Together with sister chromatid cohesion, recombination facilitates segregation by creating stable attachments between the maternal and paternal homologs, thus ensuring their correct organization in preparation for anaphase I [1].
Meiotic DSBs are catalyzed by the enzyme Spo11 [2], [3]. After DSB induction, the ends of the DSB are resected to form single-stranded DNA overhangs that can invade a homologous sequence for repair. An initial DNA joint molecule (JM) is then formed following exchange of the broken end with a homologous sequence (Figure S1). The JM is further processed and enzymatically resolved according to its composition to generate two types of products: Those that mutually exchange DNA sequences between the homologs to physically attach them, called crossovers (COs), and those that repair without mutual exchange, called non-crossovers (NCOs) [4], [5]. Initial stabilization after invasion of the break end forms a transient JM called a single-end invasion (SEI) (Figure S1) [6]. Displacement of the invading strand of the SEI, such as in helicase-mediated unwinding by the BLM ortholog Sgs1, followed by ligation with the free DSB end, forms a NCO in a process called synthesis-dependent strand annealing (SDSA) (Figure S1A) [6]–[8]. Alternatively, the SEI can be stabilized and processed to form a stable JM intermediate known as a double-Holliday junction (dHJ) (Figure S1B) [9]. The dHJ must be cleaved by endonucleases or dissolved using a helicase in combination with a topoisomerase in order to be processed into its products [5], [10]. During meiosis, NCOs and dHJ-JMs form concurrently while COs form after dHJ disappearance, indicating that COs are the main products of dHJ resolution (Figure S1C) [11], [12]. NCOs, on the other hand, are primarily formed via SDSA [11]. Regulating the formation and resolution of dHJ-JMs is essential for homolog segregation at MI, and several factors have been identified that specifically promote CO formation without influencing overall DSB repair [10]. Most of these proteins belong to the meiosis-specific ZMM (Zip1-4, Mer3, Msh4, Msh5, Spo16) family, which stimulate COs by stabilizing dHJ formation [13]–[15]. The phosphatase PP4 (Pph3/Psy2) also promotes proper CO formation by stabilizing SEIs [16]. Moreover, recent evidence has implicated the mismatch repair components Exo1 and the MutLγ complex Mlh1–Mlh3 as crossover-specific JM resolution factors [17]. While the ZMM proteins regulate the majority of COs in budding yeast and mice, a subset is dependent on the endonuclease Mus81-Mms4 [18]–[20]. In fission yeast, however, all COs form via the Mus81-Eme1 (Mus81-Mms4 in budding yeast) pathway and are derived from single, rather than double, HJs [21]–[24].
Chromatin in budding yeast is organized in a loop-axis configuration [25]. Meiotic DSB hotspots are located in the DNA loops while recombination is carried out close to the meiotic axis [26], [27]. Normal DSB induction is dependent of the tethering of DSB hotspot sequences to accessory DSB proteins at the axis prior to break induction [28]–[30]. Hence, proper meiotic recombination relies on correct loop-axis configuration and events that change this architecture alter recombination events and outcomes [27]–[31]. Despite the presence of the sister chromatid, the homologous chromosome is preferred as a repair template during meiosis [32]. This inter-homolog (IH) bias is due to combined efforts of mechanisms that promote invasion of the homolog strand, and components of the meiotic axis that physically block sister invasion [32]–[37]. The meiotic axis includes the cohesin subunit Rec8, which is required for proper axis formation and loop organization [38]. In the absence of Rec8, the loop-axis configuration is perturbed and DSBs form at low levels with altered distribution as compared to wild-type cells [39],[40]. The axis-organizing function of Rec8 is also needed to maintain IH bias during the SEI-to-dHJ transition, even though Rec8 is actually a promoter of inter-sister (IS) recombination most likely due to its role in sister chromatid cohesion (see below) [40]. The presence of IH-promoting axis components antagonize the IS bias created by Rec8 to allow IH events to dominate [40]. To further promote IH-recombination, the ZMM proteins form a structure called the synaptonemal complex (SC) between the homologs. The SC holds the homologs close to one another during recombination, thereby facilitating homolog-directed strand invasion [14],[15]. Together these mechanisms establish a bias for IH recombination but do not eliminate IS recombination, with the possibility that up to one-third of all wild-type recombination events may be directed to the sister [40],[41]. These IS repair events rarely go via a detectable JM intermediate, most likely due to the decreased preference for recombination via a IS-JM in combination with fast turnover rates for IS-JMs that may arise [42]. If inefficiently resolved, an inter-sister DNA link on the telomere-proximal side of a CO will inhibit the segregation of homologs at MI, making it crucial for cells to properly process inter-sister recombination events.
As stated, the cohesin complex is a meiotic axis component required for proper recombination. It is also essential for sister chromatid cohesion during mitosis and meiosis [43]. Cohesin is a member of the evolutionarily conserved structural maintenance of chromosomes (SMC) family of proteins, which also includes the Smc5/6 complex. Components of the Smc5/6 complex were first identified as repair proteins working in the homologous recombination pathway [44]–[46]. The complex consists of eight subunits: Smc5, Smc6, Nse1, Mms21 (Nse2) and Nse3-6, and assists in the reduction of topological stress during replication as well as DSB repair in post-replicative vegetative cells [46]–[50]. Cells harboring mutations in SMC5, SMC6 or MMS21 accumulate recombination intermediates following DNA damage inflicted during mitotic S phase [51]–[53]. Mutating genes involved in the resolution of aberrant recombination structures at blocked replication forks, such as MUS81-MM4, SGS1 and TOP3, aggravates this phenotype [54]–[56]. Recent studies have pointed to a role for the Smc5/6 complex during meiotic recombination as well. A study in C. elegans showed that the Smc5 and Smc6 proteins are required to process recombination structures in germ line cells [57]. In fission yeast, nse1-3 are needed for proper meiotic chromosome segregation [58][59]. In addition, fission yeast cells harboring mutations in nse6 accumulate meiotic JMs in the form of single HJs that resemble those found in cells lacking the endonuclease Mus81 [59]. Although the HJs are DSB-dependent, the nse6 mutant used in this study was not meiosis-specific and accumulated recombination intermediates during mitosis and pre-meiotic S phase as well [59]. Thus, the meiotic intermediates observed may have been a consequence of lesions accumulated prior to meiotic induction. A study in budding yeast was also unable to isolate a meiosis-specific phenotype for mutants of the Smc5/6 protein complex. The segregation block in these smc6 mutants was not DSB-dependent and most likely caused by defects accumulated during mitosis or pre-meiotic S-phase [60]. Due to the discrepancies between these studies, the meiotic function of the Smc5/6 protein complex remains unclear.
In this study, we employed meiosis-specific alleles of genes encoding for the Smc5/6 complex to investigate the meiotic role of the complex in the budding yeast Saccharomyces cerevisiae. Cells lacking components of the Smc5/6 complex during meiosis experience a segregation block that is dependent on DSB formation. Mutants are normal in meiotic prophase progression and DSB repair and have no significant defects in sister chromatid cohesion. Return-to-function studies indicate that the complex works at later stages of meiotic recombination. This function is most critical at times of JM resolution, and cells with non-functional Smc6 accumulate high levels of JMs in the form of both IS - and IH-JM intermediates. CO and NCO levels remain unchanged, indicating that the majority of IH-JMs are processed normally, and suggesting that most of the unresolved JM intermediates are derived from inter-sister recombination events. These findings demonstrate that the Smc5/6 protein complex is directly involved in meiotic recombination and suggest that Smc6 plays a key role in resolving recombination intermediates during meiosis, especially those that form between sister chromatids.
Results
The Smc5/6 protein complex is required for meiotic chromosome segregation
To initially address the meiosis-specific function of the Smc5/6 protein complex, the temperature-sensitive smc6-56 allele was utilized. This mutant has known mitotic recombination defects at high temperature [51],[61]. At permissive temperature, smc6-56 cells underwent normal meiotic divisions and formed viable spores (Figure S2A). When meiosis was carried out at non-permissive temperature from the time of meiotic induction, the smc6-56 mutant exhibited a mixture of two phenotypes: cells that did not appear to have entered the meiotic program and accumulated as mononucleates, and cells that failed to segregate chromosomes but formed spores (Figure S2B). A mixed cell population was also observed in a previous study when cells with the temperature-sensitive allele smc6-9 were grown at non-permissive temperature from the time of meiotic induction [60]. The authors of this paper concluded that the meiotic defects in smc6-9 cells were largely due to problems acquired during mitosis or pre-meiotic S phase [60]. The mononucleate population in the smc6-9 and smc6-56 mutants resembles that observed in mitotic cells harboring the smc6-56 allele, in which approximately half of the cells arrest in G2/M after replication at non-permissive temperature [49]. To focus on non-replicative meiotic functions, smc6-56 cells were allowed to complete pre-meiotic replication at permissive temperature before shifting to non-permissive temperature (Figure 1A, Figure S2C). Under such “soft-shift” conditions, the smc6-56 mutant only formed cells containing one unsegregated DNA mass outside four empty spores (Figure 1B). To confirm that this defect was not caused by high temperature and reflected the true meiotic phenotype of a smc5/6 mutant, meiotic-null (mn) alleles of SMC5, NSE4 and NSE2 were constructed by replacing their endogenous promoters with the mitosis-specific CLB2 promoter [62]. Because CLB2 is not down-regulated until after pre-meiotic S phase, replication defects were avoided using this system [63],[64]. As in the smc6-56 mutant, smc5-mn, nse4-mn and nse2-mn mutants were not able to segregate their chromosomes and instead formed cells with one DNA mass outside of four empty spores (Figure 1B). This demonstrates that the smc6-56 phenotype reflects a meiotic function of the Smc5/6 complex.
Fig. 1. Smc5/6 mutations cause a recombination-dependent segregation block without affecting meiotic progression. 
(A) Set-up of the soft-shift method. Cells were shifted to non-permissive temperature (33°C) upon completion of pre-meiotic S phase as judged from FACS profiles shown in Figure S2. (B) Meiotic time courses for wild-type (CB1017), smc6-56 (CB1032), smc5-mn (CB1872), nse4-mn (CB1511) and nse2-mn (CB2053) strains. At indicated times, cells were fixed and stained with DAPI to determine their nuclear content. Percent of MI+MII cells shown at left, percent of cells with one DNA mass outside four empty spores shown at right. Inset picture illustrates the “one DNA mass outside of four empty spores” phenotype, scale bar = 1 µm. Graphs represent a single synchronous meiotic time course. N = 200. (C) Meiotic spindle formation in wild type (CB1017) and smc6-56 (CB1032). Fixed whole cells were stained with an anti-α-tubulin antibody (green) and DAPI (red). The images represent spindle morphology in wild type and smc6-56 at prophase (pro), metaphase I (meta I), metaphase II (meta II), anaphase II (ana II) and after completing sporulation (spores). Meiotic progression demonstrated by plotting the fraction of cells with a single tubulin focus remaining at each time point on an inverted y-axis. N = 200. (D) Meiotic progression determined as percent of nuclei with full or partial Zip1 axes analyzed on meiotic spreads at indicated times. Picture demonstrates full Zip1 axes shown in green, scale bar = 2 µm. Dark blue line shows Zip1 axis formation when in the absence of NDT80 function. N = 100. (E) smc6-56 (CB1346) and wild-type (CB46) cells undergoing meiosis under soft-shift conditions were isolated and surface-spread to detect Zip1 (green) and epitope-tagged Rec8 (red). DNA was visualized with DAPI (gray). Scale bar = 1.8 µm. (F) Meiotic progression in spo11Δ (CB1302), spo11Δ smc6-56 (CB1301), spo11Δ sm5-mn (CB1754), spo11Δ nse4-mn (CB1510) and spo11Δ nse2-mn (CB2067) shown in percent of MI+MII cells. N = 200. Mutants of the Smc5/6 complex complete the meiotic program
In S. cerevisiae, spores form around duplicated spindle pole bodies regardless of DNA location [65]. Thus the “one DNA mass outside of four empty spores” phenotype of smc5/6 mutants suggests that they complete the meiotic program. To test this hypothesis, spindle morphology was monitored in the smc6-56 mutant. In line with the idea that smc5/6 mutants do not hinder meiotic progression, smc6-56 cells were able to duplicate their spindle-pole bodies and elongate their spindles despite abnormal spindle morphology due to failure to segregate the DNA (Figure 1C). To further challenge the assumption that smc5/6 cells complete the meiotic program, meiotic progression was analyzed by scoring the dynamics of Zip1 axes. Zip1 is a ZMM component of the SC [66]. Cells that are unable to complete recombination form incomplete Zip1 axes and do not progress past prophase [67]. Mutants lacking the transcription factor Ndt80 can initiate recombination but fail to signal downstream factors necessary to complete recombination and exit prophase and accumulate with full Zip1 axes [68]–[70]. Zip1 axes were formed and removed normally in smc6-56, smc5-mn and nse4-mn mutants (Figure 1D). The smc6-56 mutant was also normal in the timing and morphology of Zip1 and Rec8 axes (Figure 1E). Together, these data demonstrate that cells lacking Smc5/6 components fail to segregate their DNA but do not halt the meiotic cell cycle.
The segregation block in Smc5/6 mutants is dependent on meiotic recombination
To test if the segregation block in Smc5/6 complex mutants was due to meiotic recombination, nuclear divisions were monitored in a spo11Δ background. Cells devoid of SPO11 do not initiate meiotic recombination and improperly segregate their DNA since they lack attachments between the homologs [2]. Even though the resulting spores are unviable, DNA segregation can be monitored within the cells. Deletion of SPO11 in smc6-56, smc5-mn, nse4-mn and nse2-mn mutants abolished the segregation block (Figure 1F), indicating that the segregation defect in these cells is the result of problems during DSB repair.
To test whether the nuclear division failure was due to break-independent sister entanglements, segregation was examined in cells containing the smc6-56 mutation in a spo11Δ spo13Δ background. SPO13 is required to prevent biorientation of sister kinetochores at meiosis I, and, in the absence of recombination, spo13Δ cells undergo a single meiotic division, segregating sister chromatids to form cells with two viable, diploid spores called dyads [71],[72]. The spo11Δ spo13Δ smc6-56 mutant segregated its sisters efficiently and formed viable dyads under soft-shift conditions (Figure S3). These data confirm that the segregation block in smc6 mutants is not due to recombination-independent sister entanglements.
Sister chromatid cohesion and double-strand break repair are largely unaffected in the smc6-56 mutant
To further study meiosis in smc6-56 cells, sister chromatid separation was assessed at sites 35 kb from the centromere and 23 kb from the telomere of chromosome V. These regions were observed using the previously described GFP-tagged Tetracycline repressor/operator (TetR-GFP/Tet-O) system. This system is based on endogenously expressed TetR-GFP, which accumulates at multiple copies of Tet-Os inserted at the chromosomal region of interest, thereby allowing its visualization by fluorescence microscopy [73],[74]. Despite the full segregation block in smc6-56 cells, no major defect in sister chromatid cohesion or sister chromatid separation was observed at the centromere or telomere of chromosome V (Figure 2A). Final levels of sister chromatid separation did not reach those in wild-type cells, but the results showed that sister chromatids were able to separate within the unsegregated DNA masses in smc6-56 mutants. This suggests that the Smc5/6 complex has little influence on meiotic sister chromatid cohesion and implies that the chromosomes in sm6-56 cells are held together by cohesin-independent mechanisms. This notion is further supported by the finding that smc6-56 cells can separate their sister chromatids in a spo11Δ spo13Δ background (Figure S3).
Fig. 2. Sister chromatid cohesion, Rec8 dynamics and DSB repair are largely unchanged in smc6-56 mutants. 
(A) Sister chromatid separation of chromosome V. Percent of sister chromatid separation 35 Kb away from the centromere (CenV) in wild type (CB1197) and smc6-56 (CB1248) is shown in solid lines. Percent separation 50 Kb away from the right telomere (TelV) in wild type (CB1427) and smc6-56 (CB1426) indicated in dotted lines. N = 200. (B) Percent of cells with full Rec8 protein axes in wild type (CB46) and smc6-56 (CB1346). Rec8 axes were assessed by detecting epitope-tagged Rec8 (Rec8-3HA) on chromosome spreads using standard immunofluorescence techniques, picture illustrates a cell with full Rec8 axes shown in red, scale bar indicates 2 µm. N = 100. (C) DSB repair at the HIS4LEU2 hotspot on chromosome III in wild type (CB1183) and smc6-56 (CB1303). The curves represent mean break levels at the two DSB sites at the indicated time points. The Southern blot shown is representative for the three used for quantifications, DSB species were identified according to their size. (D) Cumulative DSB levels at the HIS4LEU2 hotspot in rad50S (CB2059) and rad50S smc6-56 (CB2060). The Southern blot shown is representative for those used to quantify DSBs in the rad50S background. Plots in (C) and (D) represent mean ± standard deviation from three independent experiments. All experiments were run under soft-shift conditions. To confirm that remaining cohesin was not the cause of the segregation block in smc6 mutants, cohesin dynamics were monitored on chromosome spreads using an epitope-tagged version of the meiosis-specific cohesin subunit, Rec8. After being loaded between sister chromatids following DNA replication, Rec8 is removed from chromosome arms at the first nuclear division but maintained at centromeres until MII [75]. If cohesin remains between sister chromatid arms at the first nuclear division, homolog segregation will be blocked due to the inability to resolve COs at the chromosomal level [76]. The smc6-56 mutant was able to properly localize and remove Rec8 from the chromosome axis (Figure 1E, Figure 2B), leading to the conclusion that the segregation block in this mutant is caused by cohesin-independent chromosome attachments.
To examine the role of the Smc5/6 complex during meiotic break repair, DSBs were monitored at the HIS4LEU2 hotspot on chromosome III [12][77][78]. In this assay, smc6-56 mutants were able to repair their DSBs efficiently at the two sites analyzed (Figure 2C). To investigate whether smc6-56 mutants have higher levels of break formation, DSB accumulation was investigated in a rad50S background. This mutant cannot resect the ends of the break and accumulates unprocessed DSBs [79]. The smc6-56 rad50S mutant had higher levels of breaks at one DSB site but normal levels at the other (Figure 2D). Whole-chromosome break patterns were similar in smc6-56 rad50S and rad50S on chromosomes III, IV and VI ( and data not shown). These data show that DSB repair and distribution are unchanged in smc6 mutants but that overall DSB levels may be higher, at least at specific sites.
Functional SMC6 is required during joint molecule resolution
The segregation block in the smc5/6 mutants is reminiscent of that observed in mutants that are unable to resolve JMs [8],[17],[80],[81]. Factors that promote JM resolution and subsequent prophase exit are activated by the transcription factor Ndt80. In the absence of NDT80, cells accumulate in late prophase with unresolved JMs [82],[82]. To initially assess if the Smc5/6 complex also plays a role during JM resolution, cells in which expression of NDT80 is controlled by an estradiol-inducible promoter (NDT80-IN) were utilized [83]. Combining the smc6-56 allele with NDT80-IN allowed the control of Smc6 activity by temperature shifts carried out concurrently to NDT80 induction. The smc6-56 cells were unable to segregate their DNA when taken into the ndt80 arrest at permissive temperature and released at non-permissive temperature (Figure 3A). This was not due to incomplete arrest at the time of temperature upshift and NDT80 induction, since smc6-56 cells kept in the ndt80 block three hours longer before shifting to non-permissive temperature showed the same segregation block (data not shown). At the final time point after release into non-permissive temperature after arrest at permissive temperature, smc6-56 cells were largely inviable (Figure 3B). This suggests that the structures which block segregation are also lethal to the cells. If the smc6-56 mutant instead underwent the ndt80 arrest under soft-shift conditions and was shifted to permissive temperature during release, meiotic divisions were restored and cells completed both MI and MII with wild-type kinetics (Figure 3C). These cells were also viable at the final time point (Figure 3D). These data imply that SMC6 is most critical during JM resolution, and suggest that unresolved JMs are the cause of the segregation block and inviablilty in smc6 mutants.
Fig. 3. The Smc5/6 complex performs its most critical functions at the time of joint molecule resolution. 
Meiotic progression and cell viability were determined following temperature shifts in strains carrying an inducible NDT80 allele (NDT80-IN) under the control of estradiol (ED). Meiotic progression given as percent of MI+MII cells in NDT80-IN SMC6 (CB1753) and NDT80-IN smc6-56 (CB1723) at the indicated time points. N = 200. For viability assessment, cells were collected at the indicated time points, sonicated briefly, diluted to the desired concentration, spread onto YPD plates and grown at permissive temperature for 3 days. Viability is given in percent as determined by the number of colony-forming units divided by the total number of cells plated. (A) Cells were accumulated in an ndt80 arrest at permissive temperature until 7 h when 1 µM β-estradiol (+ED, arrow) was added and cultures were shifted to non-permissive temperature. (B) Cell viability for cells undergoing meiosis under the same conditions described for (A) at 0 h and 24 h after meiotic induction. (C) Cells were accumulated in ndt80 arrest under soft-shift conditions at non-permissive temperature until 7 h when 1 µM β-estradiol (+ED, arrow) was added and cultures were shifted to permissive temperature. (D) Cell viability for cells undergoing meiosis under the same conditions described for (C) at 0 h and 24 h after meiotic induction. The smc6-56 mutant accumulates unresolved joint molecules
To assess whether JMs accumulate in smc6-56 mutants, recombination was examined at the molecular level at the ectopic URA3-ARG4 locus on chromosome III, which allows the detection of JMs in the form of dHJs using one-dimensional (1D) gel electrophoresis (Figure S5) [84]–[86]. This hotspot was used in combination with NDT80-IN under soft-shift conditions to test the hypothesis that the segregation block in smc6-56 cells is caused by the accumulation of unresolved JMs. The smc6-56 mutant accumulated a ≈3-fold higher number of JMs than the SMC6 strain (6.6% vs. 2.2% at 7 h) prior to NDT80 induction (Figure 4A–C). Following induction, approximately two-thirds of JMs in the smc6-56 mutant remained unresolved after 24 h. In spite of unresolved recombination intermediates, NCOs and COs accumulated at the same time and level in the smc6 mutant as compared to the SMC6 strain (Figure 4B,C). Recombination products were also observed at wild-type levels at the HIS4LEU2 hotspot in the smc6-56 mutant (data not shown).
Fig. 4. The smc6-56 mutant accumulates unresolved joint molecules but forms normal levels of recombination products. 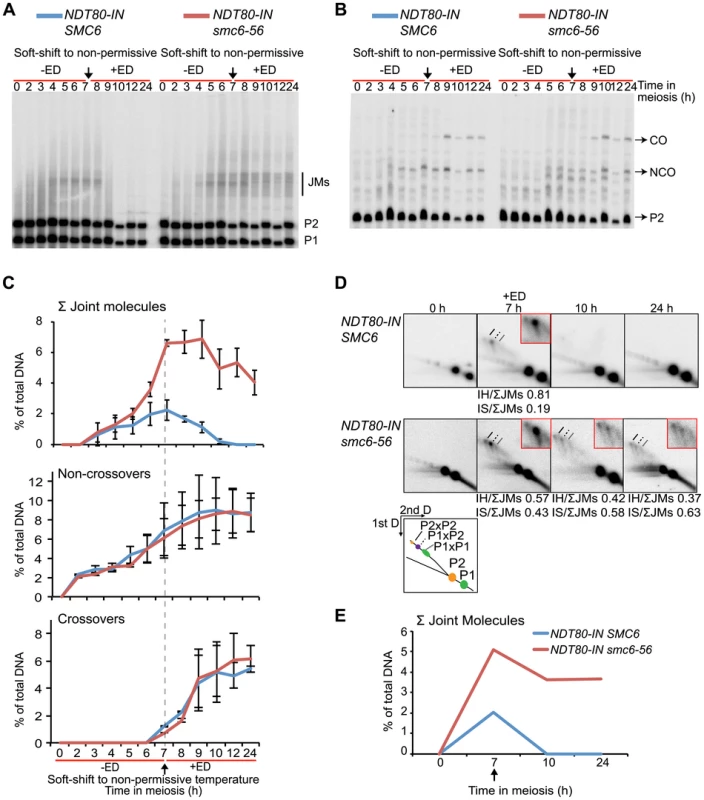
Analysis of recombination measured at the ectopic URA3-ARG4 interval on chromosome III (Figure S5) [84] in strains containing the estradiol-inducible NDT80 allele (NDT80-IN) under soft-shift conditions. NDT80-IN was induced at 7 h with 1 µM β-estradiol (+ED, arrow). Blue curves indicate NDT80-IN SMC6 (CB2096); red NDT80-IN smc6-56 (CB2097). (A) Representative Southern blot used to detect JMs after digesting with XmnI and probing for a region in ARG4. P1, P2 and JM regions based on expected sizes. (B) Representative Southern blot used to detect CO/NCO products at the same interval after digesting with XhoI/EcoRI and probing for HIS4 sequences. P2, CO and NCO regions based on expected sizes. (C) Quantifications of total JM levels, NCOs and COs in percent of total DNA from blots illustrated in (A) and (B). Dotted line and arrow indicate time of NDT80-IN induction. Plots represent mean ± standard deviation from three independent experiments. (D) Two-dimensional analysis of JM species. DNA from NDT80-IN SMC6 and NDT80-IN smc6-56 undergoing meiosis under soft-shift conditions was isolated and subjected to two-dimensional electrophoresis as described in Materials and Methods. The interpretive panel shows the assumed identity of JM species after probing for a region which recognizes both homologs. JMs between P1×P1 are the result of random breaks and are therefore expected to give a broader signal than P2×P2 and P2×P1. Dashed line, P2×P2 IS-JM; solid line, P2×P1 IH-JM; dotted line, P1×P1 IS-JM. The enlarged panels are enhanced images of the JM region from the designated time point. Ratio of IH-JMs/ΣJMs and IS-JMs/ΣJMs given below relevant images and were calculated as described in Figure S9. (E) Quantification of total JMs in percent of total DNA from the gels represented in (D). Arrow denotes NDT80 induction. Curves represent one experiment, with similar results from a second independent experiment presented in Figure S7A. To better identify JM species, native-native two-dimensional (2D) gel electrophoresis was utilized at the URA3-ARG4 locus in combination with the NDT80-IN system. This method of electrophoresis separates JMs by size in the first dimension and by shape plus size in the second dimension [87]. Prior to NDT80 induction, SMC6 cells accumulated a strong JM spot corresponding to the predicted size for IH-JMs (P1×P2) (Figure 4D, 7 h). This spot was flanked by two weaker regions: a slower-migrating spot predicted to be IS-JMs from P2 (P2×P2) and a faster-migrating, less defined spot corresponding to IS-JMs from P1 (P1×P1) (Figure 4D). To verify the identity of these flanking spots as IS-JMs, recombination was examined in cells lacking the axial element protein Hop1, in which the sister chromatid is preferred over the homolog as a repair template [42],[88]. As anticipated, the hop1Δ mutant lacked the middle spot corresponding to IH-JMs and acquired the two outer spots predicted for IS-JMs, with P2×P2 being the dominating IS species (Figure S6A). The indistinctness of the P1×P1 spot is due to the fact that the DSB hotspots at this locus are only located on the P2 homolog, directing the majority of inter-sister repair to this set of sister chromatids (Figure S5).
Similar to the results obtained from the 1D gels, levels of total JMs from the 2D gels were approximately 2.5-fold higher in the smc6-56 mutant than in the SMC6 strain prior to NDT80 induction (Figure 4D–E). A homolog-specific probe identified many of these JMs as IS-JMs in the smc6-56 mutant (Figure S6B). Though some JMs in the smc6-56 mutant were resolved following release, about two-thirds of the total persisted at the final time point (Figure 4E). Further examination of JM composition revealed that smc6-56 cells formed JMs composed of a higher ratio of IS-JMs compared to SMC6 cells in the ndt80 arrest (Figure 4D 7 h, 0.43 vs. 0.19 of total). After NDT80 induction, the ratio of IS-JMs to total JMs increased to over 0.6 in smc6-56 cells, though some IH-JMs persisted as well. Similar results were obtained in an independent experiment (Figure S7A). These data indicate that SMC6 prevents the formation of excess JMs and facilitates the resolution of JMs; especially those formed between sister chromatids.
SMC6 is required for the resolution of joint molecules
To test if SMC6 is needed for JM resolution, JMs were examined under conditions when the Smc6 protein was functional during the ndt80-mediated arrest and then made non-functional during release. In this situation, the smc6-56 mutant was inviable and unable to segregate its DNA (Figure 3A–B). JM formation was normal when smc6-56 cells were arrested at permissive temperature (Figure 5A–E). When shifted to non-permissive temperature at the time of NDT80 induction, JMs were not fully resolved, and no significant decrease in CO or NCO levels was detected (Figure 5A–E). Possible reasons for the counter-intuitive finding that IH-JMs persist without a detectable decrease in CO formation are considered in the discussion. Upon closer examination, the ratio of IS-JMs out of total JMs at the final time point is increased to 0.67 from its ratio of 0.23 at the time of ndt80 release (Figure 5D). IH-JMs remained as well, but some were apparently resolved, as reflected in the decreased IH-JM ratio and formation of COs at later time points. Similar results were found in an independent experiment (Figure S7B). Together these results implicate a role for SMC6 in the resolution of IS-JMs and, to a lesser extent, IH-JMs that form under normal conditions.
Fig. 5. Smc6 protein function is required to resolve a subset of joint molecules. 
Cells were accumulated in an ndt80 arrest at permissive temperature (green lines, −ED) until NDT80 induction when cultures were shifted to non-permissive temperature (red lines, +ED) at 7 h. Arrows indicate addition of 1 µM β-estradiol. Blue curves indicate NDT80-IN SMC6 (CB2096) and red curves indicate NDT80-IN smc6-56 (CB2097). One-dimensional JM and CO/NCO detection and two-dimensional JM analysis were performed using the same locus and techniques described in Figure 4. (A) Representative Southern blot used to detect joint molecules. (B) Representative Southern blot used to detect CO/NCO species. (C) Quantifications of total JM levels, NCO and CO products from blots in (A) and (B), respectively. Dotted line and arrow indicate time of NDT80 induction. Plots represent mean ± standard deviation from three independent experiments. (D) Identification of JM species using two-dimensional electrophoresis on NDT80-IN SMC6 and NDT80-IN smc6-56 undergoing NDT80 arrest at permissive temperature and release at non-permissive temperature. The enlarged panels are enhanced images of the JM region from the designated time point. Ratio of IH-JMs/ΣJMs and IS-JMs/ΣJMs given below relevant images and were calculated as described in Figure S9. (E) Quantification of total JMs in percent of total DNA from the gels represented in (D). Arrow denotes NDT80-IN induction. Curves represent a single experiment, with similar results from a second independent experiment shown in Figure S7B. Joint molecules accumulated in the absence of SMC6 function are resolved after restoration of Smc6 activity
When Smc6 is non-functional during ndt80 arrest but functional during release, cells successively completed nuclear divisions (Figure 3C) and formed normal levels of CO and NCO products (Figure 6A–C). Prior to NDT80 induction, the smc6-56 mutant formed ≈3-fold higher total JM levels (Figure 6C, E). At this time point, the ratio of IS-JMs to the total on the 2D gels was 0.62 in the smc6 mutant, compared to 0.20 in the SMC6 strain. When Smc6 function was restored at the time of NDT80 induction, all JMs were resolved (Figure 6A,C,D,E). Similar results were obtained from 2D gels from an independent experiment (Figure S7C). This supports the notion that the rescue in nuclear divisions is due to the restoration of JM resolution. The JMs that accumulated in the absence of SMC6 function were not lethal, as viability was also restored when the cells were shifted to permissive temperature (Figure 3D). These results show that all JMs formed without functional Smc6 can properly be resolved if Smc6 function is restored during JM resolution.
Fig. 6. Joint molecules accumulated without functional Smc6 protein can be resolved when its function is restored during resolution. 
Cells were accumulated in an ndt80 arrest with soft-shift to non-permissive temperature (red lines, −ED). NDT80 was induced concurrently with the shift to permissive temperature (green lines, +ED) at 7 h. Arrows indicate addition of 1 µM β-estradiol. Blue curves indicate NDT80-IN SMC6 (CB2096) and red curves indicate NDT80-IN smc6-56 (CB2097). One-dimensional JM and CO/NCO detection and two-dimensional JM analysis were done using the same locus and techniques described in Figure 4. (A) Representative Southern blot used to detect joint molecules. (B) Representative Southern blot used to detect CO/NCO species. (C) Quantifications of total JM levels, NCO and CO products from blots in (A) and (B), respectively. Dotted line and arrow indicate time of NDT80 induction. Plots represent mean ± standard deviation from three independent experiments. (D) Identification of JM species using two-dimensional electrophoresis on NDT80-IN SMC6 and NDT80-IN smc6-56 undergoing NDT80 arrest at non-permissive temperature and release at permissive temperature. The enlarged panels are enhanced images of the JM region from the designated time point. Ratio of IH-JMs/ΣJMs and IS-JMs/ΣJMs given below relevant images and were calculated as described in Figure S9. (E) Quantification of total JMs in percent of total DNA from the gels represented in (D). Arrow denotes NDT80-IN induction. Curves represent one experiment, with similar results from a second independent experiment shown in Figure S7C. Smc6 localization on meiotic chromosomes depends on cohesin
To gain additional insights into the function of the Smc5/6 complex during meiotic recombination, an N-terminal epitope-tagged version of Smc6 was used to analyze the complex's binding on chromosome spreads using immunofluorescence. The tagged version of SMC6 was fully functional and neither impeded events during meiotic prophase nor delayed meiotic segregation (data not shown). Smc6-Myc appeared on chromosomes during early prophase around the time of Rec8 foci formation (Figure 7A). When the Rec8 axis began to organize, Smc6's binding became more profuse and formed an axis-like structure. On full-length axes, Smc6 localized at regions with weaker Rec8 signals as well as at sites with more profuse Rec8 signals (Figure 7C, solid and dashed arrows, respectively). Consistent with results from a previous study [60], Smc6 bound abundantly to the rDNA, seen by the brightly staining Smc6 region (Figure 7A, green) corresponding to weak DAPI staining. After late prophase, the Smc6-Myc signal became diffuse and disappeared prior to MI. Removing cohesin seemed to reduce the amount of Smc6 foci and eliminated the axis-like pattern of Smc6 (Figure 7B). Western blotting revealed that Smc6 protein levels were similar in the wild type and in the rec8Δ mutant, indicating that the diminished levels of Smc6 binding were not due to decreased Smc6 protein levels in this cohesin mutant (Figure S8). This demonstrates that cohesin is required for the proper organization of Smc6 foci on chromosomes and suggests that localization of the Smc5/6 complex is influenced by meiotic axis structure and/or the presence of sister chromatid cohesion.
Fig. 7. The Smc6 protein localizes to meiotic chromosomes during prophase in a cohesin- dependent manner. 
Cells were isolated at the indicated time points and surface-spread to detect epitope-tagged proteins Smc6 (Smc6-13Myc, green) and Rec8 (Rec8-3HA, red) using standard immunofluorescence techniques. DNA visualized with DAPI shown in grey. (A) Wild type (CB1361). (B) rec8Δ (CB1411). (C) Enlarged image of a representative cell from the wild type (CB1361) at the 4 h time point. Solid arrows indicate an Smc6 site at a weaker-staining Rec8 site. Dashed arrows indicate a strong Smc6 site on top of a strong Rec8 site. Discussion
The results presented here suggest that the Smc5/6 complex prevents excessive JM formation and aids in JM resolution during meiosis. This resolution function is particularly critical for JM intermediates formed between sister chromatids. Similar to other mutants defective in JM processing, smc5/6 mutants experience a recombination-dependent segregation block without halting meiotic progression [8],[17],[80],[81]. This could indicate that Smc6 works together with established resolution pathways, such as those mediated by the ZMMs or Sgs1/Mus81-Mms4. Unlike those mutants, however, the smc6-56 mutant does not lead to any detectable decrease in CO or NCO levels, suggesting that it works predominately outside of canonical meiotic recombination pathways. Given the normal levels of COs and NCOs, and the nature of the persisting JMs, we propose that in the absence of Smc6, cells accumulate primarily IS-JMs, but also a subset of IH-JMs (Figure 8).
Fig. 8. Model for the role of the Smc5/6 complex during meiosis. 
Schematic diagram depicting the result of having non-functional Smc6 protein before and/or after NDT80 induction as indicated (red = non-permissive/non-functional, green = permissive/functional) and described in the text. When JM formation occurs without Smc6 function, overall JM levels are 2.5–3-fold higher than in cells with functional Smc6 (Figure 4C,E, Figure 6C,E). The ratio of IS-JMs to total JMs in the smc6-56 mutant is twice that in SMC6 cells (Figure 4D, Figure 6D). Although IH-JMs are still the dominating species, the absence of Smc6 diminishes the IH-bias slightly. This may be due to higher levels if IS recombination in the smc6-56 mutant or due to an accumulation of normally transient IS-JM intermediates that cannot be resolved without functional Smc6. When NDT80 is induced in the continued absence of Smc6 function, about three-fourths of total JMs persist and the IS-JM ratio increases further, though some IH-JMs also persist (Figure 4C,D). Surprisingly, CO formation is normal in spite of persisting IH-JMs. One explanation for this could be that these lingering IH-JMs are not significant enough in number to cause a detectable decrease in CO levels (Figure 8). As an alternative, extra IH-JMs could come from additional recombination-initiating events, which has been shown to occur in some mutants [11]. The smc6-56 mutant does form slightly higher numbers of DSBs at one break site in the rad50S background (Figure 2D, DSBII). If this also occurs at other sites, it may account for some of the increase in the levels of recombination in the smc6 mutant.
More recombination-initiating events would also explain the IH-JMs that are never resolved, despite normal CO levels, when Smc6 is non-functional from the time of meiotic induction (Figure 4A–D). This suggests that cells lacking functional Smc6 accumulate recombination intermediates that will require Smc6 for their resolution. In line with this hypothesis, when cells lacking Smc6 function during JM formation are released from the ndt80 arrest in the presence of functional Smc6, all JMs are resolved, despite the higher ratio of IS-JMs at the time of release and higher overall JM levels (Figure 6). COs and NCOs are formed efficiently (Figure 6B,C), and DNA segregation and viability is rescued (Figure 3C,D). This indicates that the JMs formed in the mutant can be properly resolved when Smc6 function is restored (Figure 8). This reversible phenotype is similar to what has been observed for the inter-sister recombination intermediates which accumulate in mitotic cells lacking Smc6 function [89].
Finally, when Smc6 is functional during JM formation in an ndt80-mediated arrest, both the levels and ratios of JMs are normal (Figure 5A,C,D,E). Upon release into conditions that render Smc6 non-functional, however, three-fourths of total JMs persist and cells are unable to segregate their DNA (Figure 5C, Figure 3A). The remaining JMs are composed of both IS-JMs and IH-JMs, but the ratio of IS-JMs increases upon shift to non-permissive temperature (Figure 5D). A fraction of IH-JMs is resolved, as reflected in the efficient formation of COs and decrease in the IH-JM ratio (Figure 5A–D). One explanation for why these unresolved IH-JMs do not lead to a detectable decrease in CO formation is that they contribute to a very small portion of total CO levels not distinguishable in the assays used here (Figure 8). NCO formation is also normal, indicating that the remaining JMs are not caused by converted SDSA events (Figure 5B,C). These data suggest that a subset of IH-JMs that form under normal conditions require Smc6 for their resolution, while nearly all IS-JMs seem to rely on Smc6 for their resolution (Figure 8). This finding correlates with the role of the Smc5/6 complex in the resolution of sister chromatid intermediates during homologous recombination during mitosis in yeast [53] and in germ line cells in C. elegans [57].
Even though the presented evidence suggests a role for the Smc5/6 complex outside of canonical recombination pathways, the possibility that it works together with other recombination pathways cannot be ruled out (Eva Hoffman, personal communication; Franz Klein, personal communication). In a wild-type meiosis, almost all NCOs are derived from the SDSA pathway and do not resolve via a JM intermediate [11],[12]. In contrast, in cells lacking SGS1, what should have been SDSA events are instead stabilized and transformed into dHJs [8]. These are later resolved into both NCO and CO products, thereby delaying the timing of NCO formation until JM resolution is induced [8]. With that in mind, the additional JMs in the smc6-56 mutant presented here could come from the conversion of NCO-forming SDSA events into JM intermediates. However, the smc6-56 mutant is not defective in the timing of NCO formation under non-permissive conditions, and forms most of its NCOs prior to NDT80 induction (Figure 4B,C, Figure 6B,C). Final NCO levels are also normal, indicating that the JMs that remain in the smc6-56 mutant are not derived from the conversion of SDSA events into unresolvable JMs.
Here we show that the smc6-56 mutant is able to establish sister chromatid cohesion and efficiently localize and remove cohesin from chromosomes (Figure 2B, Figure 1C). Mutants in other subunits of the Smc5/6 complex have been reported to inhibit full removal of meiotic cohesin (Eva Hoffman, personal communication). One explanation for this discrepancy could be that some components of the Smc5/6 complex work in different pathways. The segregation block in an smc5-mn mutant is also reported to be partially rescued when Rec8 is artificially removed from chromosomes (Eva Hoffman, personal communication). While this result could point towards a function for Smc5 in cohesin removal, it could also support the notion that the Smc5/6 complex is needed to resolve IS recombination intermediates. Removing sister chromatid cohesion reduces the likelihood of IS repair, thereby decreasing the level of IS recombination and allowing some smc5-mn cells to segregate their chromosomes. While the smc6-56 mutant can separate some of its sister chromatids at the telomere and centromere on chromosome V, this segregation is not complete (Figure 2A). This is not due to an abnormal version of cohesion, as cells forced to undergo a mitosis-like division in the absence of recombination can separate their sister chromatids (Figure S3). Instead, the sisters are most likely held together by DNA attachments and not cohesin, as illustrated by data showing that the smc6-56 mutant accumulates unresolved JMs between sister chromatids. In addition, the centromeric and telomeric regions have been suggested to be hotspots for meiotic inter-sister repair [90], which could explain why less than half of the sister chromatids are able to separate at these regions despite cohesin removal in smc6-56 cells.
Smc6 localizes to meiotic chromosomes as well as to the rDNA (Figure 7A). Preliminary evidence suggests that its binding pattern may reveal its precise localization on the meiotic axis, i.e. whether it sits at or between Rec8 sites (Figure 7C). Indeed, abolishing sister chromatid cohesion by removing Rec8 reduces the binding of Smc6 to chromosomes, suggesting that cohesin may guide Smc6 localization (Figure 7B). It is possible that the absence of sister chromatid cohesion reduces the likelihood of inter-sister recombination, which in turn diminishes the loading of Smc6. Alternatively, or in addition, the stable association of Smc6 to chromosomes may require proper axis formation and/or cohesion as such. Deletion of REC8 only diminishes the binding of Smc6, indicating that other factors dictate the loading of Smc6 to meiotic chromosomes. It will be interesting to learn what role the Smc5/6 complex plays in meiotic chromatin organization in order to gain further insights into its role during recombination.
In conclusion, this study identifies a crucial role for the Smc5/6 complex in processing of recombination intermediates during meiotic recombination. Mutants in the Smc5/6 complex acquire high levels of recombination intermediates between homologs and sister chromatids. The majority of IH-JMs are resolved, as reflected by the decreased ratio of IH-JMs at the final time points and the normal level of COs. IS-JMs, on the other hand, seem to depend on the function of Smc6 for their resolution. We therefore propose that the main impediment to homolog segregation in smc5/6 mutants is unresolved linkages between sister chromatids, though some homolog attachments contribute to the segregation defect as well.
Materials and Methods
Yeast strains and methods
Strains used for this investigation are derivatives of SK1 [91] and are shown in Table S1. Gene deletions and C-terminal epitope tags were introduced using standard methods [92]. The smc6-56 allele was integrated at its endogenous locus and contains three point mutations in the coil-coil region of the protein which render it temperature-sensitive [61]. Meiotic nulls for SMC5, NSE4 and NSE2 were made by replacing the endogenous promoters with the CLB2 promoter using one-step gene replacement [62]. The NDT80-IN strains have been described [82],[83].
Sporulation
Liquid media, pre-sporulation and sporulation conditions were done using SPS media according to previously described methods [93]. Cultures were grown with vigorous shaking in baffled flasks at least ten times larger than the culture volume to achieve optimum synchrony. Permissive temperature was defined as 25°C, non-permissive temperature was 33°C. For the soft-shift setup, pre-sporulation plates and cultures were grown at 25°C. Once shifted to sporulation media, the cells were grown for 2.5 hours at 25°C before raising the temperature to 33°C. All experiments were performed at least twice with results similar to those presented in the figures here. Expression of NDT80-IN was induced by the addition of β-estradiol at a final concentration of 1 µM 7 h after meiotic induction.
Molecular analyses
The HIS4LEU2 locus used for DSB analysis has been described [6],[78]. The ectopic locus on chromosome III used for 1D and 2D JM analyses as well as CO/NCO detection is illustrated in Figure S5 and has been described [11][11]. For native-native two-dimensional gel electrophoresis, psoralen cross-linked DNA was extracted from meiotic cultures as described in [94] and references therein. After digesting with XmnI, DNA samples were run on a 0.4% SeaKem GTG agarose gel (Lonza) lacking ethidium bromide in 1X TBE (90 mM Tris-borate, 2 mM EDTA pH 8) at 1 V/cm for 24 hours at room temperature. Gels were stained for 10 minutes in 1X TBE containing 0.3 µg/mL ethidium bromide, and lanes were excised and laid perpendicular to the direction of current for the second dimension. The gel for the second dimension, 0.8% SeaKem GTG agarose (Lonza) in 1X TBE plus 0.3 µg/mL ethidium bromide, was cast around the gel slices and allowed to harden. Electrophoresis in the second dimension was carried out in 4°C for 6 hours at 4 V/cm in 1X TBE containing 0.3 µg/mL ethidium bromide with pumping from the cathode to the anode. Gels were subjected to Southern blot analysis and probed with ARG4 coding sequences (+165 to +1413). DNA preparation and one-dimensional electrophoresis for JM assays were done as described using conditions that stabilize JM intermediates [95]. JMs were analyzed using XmnI digests probed with ARG4 coding sequences (+165 to +1413, argD), COs/NCOs were analyzed using XhoI/EcoRI double digests probed with HIS4 coding sequences (+538 to +718, hisU). DNA was transferred to nylon membranes via downward capillary transfer using standard techniques. After cross-linking the DNA, the membranes were pre-hybridized in Church buffer (1% w/v BSA, 1 mM EDTA, 0.5 M phosphate buffer, 7% w/v SDS) for approximately 4 hours at 65°C and hybridized with the radioactively labeled probe overnight at 65°C. After washing, signals were detected on an imaging plate with a FLA-7000 image reader and quantified using Multi Gauge software, all from Fujifilm.
Quantifications of 1D and 2D gels were done using Multi Gauge software by selecting equivalent regions of interest, including one to measure the background of the region. Sizes of expected products were determined using molecular weight standards. The signal was corrected for background and divided by the sum of the measured region and the standard region (in all cases, the parental region). Similar blots were treated equally; for instance, regions of interest used to measure CO/NCO species were the same size for each blot. Further details regarding quantifications of JM species from 2D gels are given in Figure S9. All experiments were performed at least twice with results similar to those presented.
Cytology and immunofluorescence
Nuclear morphology was scored by DAPI (4′,6-diamidino-2-phenylindole) staining of ethanol-fixed cells using standard protocols. In situ immuno-staining of fixed whole cells for microtubule detection was performed using conventional techniques with a monoclonal mouse anti-alpha Tubulin antibody (DM1A, Abcam) at a 1∶1000 dilution. Stained slides were mounted and DAPI-stained using ProLong Gold (Invitrogen). Meiotic spreading was done on SuperFrost Plus slides according to the protocol previously described [96] with the exception that 5% Lipsol was used as a detergent. A 1∶500 dilution of rabbit-anti-Zip1 (gift from K. Schmekel) was used to detect Zip1. Smc6-13Myc was detected using 1∶200 mouse-anti-Myc (Invitrogen) and Rec8-3HA was detected using 1∶200 rat-anti-HA (Roche). Stained slides were mounted and DAPI-stained using ProLong Gold (Invitrogen). Image acquisition of a single focal plane was done in Volocity (Perkin Elmer) with a Leica confocal microscope. Image processing and analysis was carried out in Volocity.
Additional methods describing results shown in supporting figures can be found in Text S1.
Supporting Information
Zdroje
1. KlecknerN (1996) Meiosis: how could it work? Proc Natl Acad Sci U S A 93 : 8167–8174.
2. KeeneyS (2008) Spo11 and the Formation of DNA Double-Strand Breaks in Meiosis. Genome Dyn Stab 2 : 81–123.
3. KeeneyS, GirouxCN, KlecknerN (1997) Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88 : 375–384.
4. MimitouEP, SymingtonLS (2008) Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature 455 : 770–774.
5. SchwartzEK, HeyerWD (2011) Processing of joint molecule intermediates by structure-selective endonucleases during homologous recombination in eukaryotes. Chromosoma 120 : 109–127.
6. HunterN, KlecknerN (2001) The single-end invasion: an asymmetric intermediate at the double-strand break to double-holliday junction transition of meiotic recombination. Cell 106 : 59–70.
7. PaquesF, HaberJE (1999) Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev 63 : 349–404.
8. De MuytA, JessopL, KolarE, SourirajanA, ChenJ, et al. (2012) BLM helicase ortholog Sgs1 is a central regulator of meiotic recombination intermediate metabolism. Mol Cell 46 : 43–53.
9. SchwachaA, KlecknerN (1995) Identification of double Holliday junctions as intermediates in meiotic recombination. Cell 83 : 783–791.
10. YoudsJL, BoultonSJ (2011) The choice in meiosis - defining the factors that influence crossover or non-crossover formation. J Cell Sci 124 : 501–513.
11. AllersT, LichtenM (2001) Differential timing and control of noncrossover and crossover recombination during meiosis. Cell 106 : 47–57.
12. StorlazziA, XuL, CaoL, KlecknerN (1995) Crossover and noncrossover recombination during meiosis: timing and pathway relationships. Proc Natl Acad Sci U S A 92 : 8512–8516.
13. BornerGV, KlecknerN, HunterN (2004) Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell 117 : 29–45.
14. LynnA, SoucekR, BornerGV (2007) ZMM proteins during meiosis: crossover artists at work. Chromosome Res 15 : 591–605.
15. ShinoharaM, OhSD, HunterN, ShinoharaA (2008) Crossover assurance and crossover interference are distinctly regulated by the ZMM proteins during yeast meiosis. Nat Genet 40 : 299–309.
16. FalkJE, ChanAC, HoffmannE, HochwagenA (2010) A Mec1 - and PP4-dependent checkpoint couples centromere pairing to meiotic recombination. Dev Cell 19 : 599–611.
17. ZakharyevichK, TangS, MaY, HunterN (2012) Delineation of joint molecule resolution pathways in meiosis identifies a crossover-specific resolvase. Cell 149 : 334–347.
18. de los SantosT, HunterN, LeeC, LarkinB, LoidlJ, et al. (2003) The Mus81/Mms4 endonuclease acts independently of double-Holliday junction resolution to promote a distinct subset of crossovers during meiosis in budding yeast. Genetics 164 : 81–94.
19. HollowayJK, BoothJ, EdelmannW, McGowanCH, CohenPE (2008) MUS81 generates a subset of MLH1–MLH3-independent crossovers in mammalian meiosis. PLoS Genet 4: e1000186.
20. OsmanF, DixonJ, DoeCL, WhitbyMC (2003) Generating crossovers by resolution of nicked Holliday junctions: a role for Mus81-Eme1 in meiosis. Mol Cell 12 : 761–774.
21. SmithGR, BoddyMN, ShanahanP, RussellP (2003) Fission yeast Mus81.Eme1 Holliday junction resolvase is required for meiotic crossing over but not for gene conversion. Genetics 165 : 2289–2293.
22. CromieG, SmithGR (2008) Meiotic Recombination in Schizosaccharomyces pombe: A Paradigm for Genetic and Molecular Analysis. Genome Dyn Stab 3 : 195.
23. CromieGA, HyppaRW, SmithGR (2008) The fission yeast BLM homolog Rqh1 promotes meiotic recombination. Genetics 179 : 1157–1167.
24. CromieGA, HyppaRW, TaylorAF, ZakharyevichK, HunterN, et al. (2006) Single Holliday junctions are intermediates of meiotic recombination. Cell 127 : 1167–1178.
25. FilipskiJ, MuchaM (2002) Structure, function and DNA composition of Saccharomyces cerevisiae chromatin loops. Gene 300 : 63–68.
26. BlatY, ProtacioRU, HunterN, KlecknerN (2002) Physical and functional interactions among basic chromosome organizational features govern early steps of meiotic chiasma formation. Cell 111 : 791–802.
27. KlecknerN (2006) Chiasma formation: chromatin/axis interplay and the role(s) of the synaptonemal complex. Chromosoma 115 : 175–194.
28. AcquavivaL, SzekvolgyiL, DichtlB, DichtlBS, de La Roche Saint AndreC, et al. (2013) The COMPASS subunit Spp1 links histone methylation to initiation of meiotic recombination. Science 339 : 215–218.
29. SommermeyerV, BeneutC, ChaplaisE, SerrentinoME, BordeV (2013) Spp1, a member of the Set1 Complex, promotes meiotic DSB formation in promoters by tethering histone H3K4 methylation sites to chromosome axes. Mol Cell 49 : 43–54.
30. PanizzaS, MendozaMA, BerlingerM, HuangL, NicolasA, et al. (2011) Spo11-accessory proteins link double-strand break sites to the chromosome axis in early meiotic recombination. Cell 146 : 372–383.
31. StorlazziA, GarganoS, Ruprich-RobertG, FalqueM, DavidM, et al. (2010) Recombination proteins mediate meiotic spatial chromosome organization and pairing. Cell 141 : 94–106.
32. SchwachaA, KlecknerN (1997) Interhomolog bias during meiotic recombination: meiotic functions promote a highly differentiated interhomolog-only pathway. Cell 90 : 1123–1135.
33. BishopDK, ParkD, XuL, KlecknerN (1992) DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell 69 : 439–456.
34. ShinoharaA, GasiorS, OgawaT, KlecknerN, BishopDK (1997) Saccharomyces cerevisiae recA homologues RAD51 and DMC1 have both distinct and overlapping roles in meiotic recombination. Genes Cells 2 : 615–629.
35. NiuH, WanL, BaumgartnerB, SchaeferD, LoidlJ, et al. (2005) Partner choice during meiosis is regulated by Hop1-promoted dimerization of Mek1. Mol Biol Cell 16 : 5804–5818.
36. HollingsworthNM, PonteL (1997) Genetic interactions between HOP1, RED1 and MEK1 suggest that MEK1 regulates assembly of axial element components during meiosis in the yeast Saccharomyces cerevisiae. Genetics 147 : 33–42.
37. CarballoJA, JohnsonAL, SedgwickSG, ChaRS (2008) Phosphorylation of the axial element protein Hop1 by Mec1/Tel1 ensures meiotic interhomolog recombination. Cell 132 : 758–770.
38. RevenkovaE, JessbergerR (2006) Shaping meiotic prophase chromosomes: cohesins and synaptonemal complex proteins. Chromosoma 115 : 235–240.
39. KugouK, FukudaT, YamadaS, ItoM, SasanumaH, et al. (2009) Rec8 guides canonical Spo11 distribution along yeast meiotic chromosomes. Mol Biol Cell 20 : 3064–3076.
40. KimKP, WeinerBM, ZhangL, JordanA, DekkerJ, et al. (2010) Sister cohesion and structural axis components mediate homolog bias of meiotic recombination. Cell 143 : 924–937.
41. GoldfarbT, LichtenM (2010) Frequent and efficient use of the sister chromatid for DNA double-strand break repair during budding yeast meiosis. PLoS Biol 8: e1000520.
42. SchwachaA, KlecknerN (1994) Identification of joint molecules that form frequently between homologs but rarely between sister chromatids during yeast meiosis. Cell 76 : 51–63.
43. PetersJM, NishiyamaT (2012) Sister chromatid cohesion. Cold Spring Harb Perspect Biol 4: a011130.
44. FousteriMI, LehmannAR (2000) A novel SMC protein complex in Schizosaccharomyces pombe contains the Rad18 DNA repair protein. EMBO J 19 : 1691–1702.
45. AndrewsEA, PalecekJ, SergeantJ, TaylorE, LehmannAR, et al. (2005) Nse2, a component of the Smc5-6 complex, is a SUMO ligase required for the response to DNA damage. Mol Cell Biol 25 : 185–196.
46. LehmannAR, WalickaM, GriffithsDJ, MurrayJM, WattsFZ, et al. (1995) The rad18 gene of Schizosaccharomyces pombe defines a new subgroup of the SMC superfamily involved in DNA repair. Mol Cell Biol 15 : 7067–7080.
47. KegelA, Betts-LindroosH, KannoT, JeppssonK, StromL, et al. (2011) Chromosome length influences replication-induced topological stress. Nature 471 : 392–396.
48. StromL, LindroosHB, ShirahigeK, SjogrenC (2004) Postreplicative recruitment of cohesin to double-strand breaks is required for DNA repair. Mol Cell 16 : 1003–1015.
49. LindroosHB, StromL, ItohT, KatouY, ShirahigeK, et al. (2006) Chromosomal association of the Smc5/6 complex reveals that it functions in differently regulated pathways. Mol Cell 22 : 755–767.
50. De PiccoliG, Cortes-LedesmaF, IraG, Torres-RosellJ, UhleS, et al. (2006) Smc5-Smc6 mediate DNA double-strand-break repair by promoting sister-chromatid recombination. Nat Cell Biol 8 : 1032–1034.
51. BranzeiD, SollierJ, LiberiG, ZhaoX, MaedaD, et al. (2006) Ubc9 - and mms21-mediated sumoylation counteracts recombinogenic events at damaged replication forks. Cell 127 : 509–522.
52. SollierJ, DriscollR, CastellucciF, FoianiM, JacksonSP, et al. (2009) The Saccharomyces cerevisiae Esc2 and Smc5-6 proteins promote sister chromatid junction-mediated intra-S repair. Mol Biol Cell 20 : 1671–1682.
53. ChavezA, GeorgeV, AgrawalV, JohnsonFB (2010) Sumoylation and the structural maintenance of chromosomes (Smc) 5/6 complex slow senescence through recombination intermediate resolution. J Biol Chem 285 : 11922–11930.
54. MorikawaH, MorishitaT, KawaneS, IwasakiH, CarrAM, et al. (2004) Rad62 protein functionally and physically associates with the smc5/smc6 protein complex and is required for chromosome integrity and recombination repair in fission yeast. Mol Cell Biol 24 : 9401–9413.
55. HwangJY, SmithS, CeschiaA, Torres-RosellJ, AragonL, et al. (2008) Smc5-Smc6 complex suppresses gross chromosomal rearrangements mediated by break-induced replications. DNA Repair (Amst) 7 : 1426–1436.
56. ChenYH, ChoiK, SzakalB, ArenzJ, DuanX, et al. (2009) Interplay between the Smc5/6 complex and the Mph1 helicase in recombinational repair. Proc Natl Acad Sci U S A 106 : 21252–21257.
57. BickelJS, ChenL, HaywardJ, YeapSL, AlkersAE, et al. (2010) Structural maintenance of chromosomes (SMC) proteins promote homolog-independent recombination repair in meiosis crucial for germ cell genomic stability. PLoS Genet 6: e1001028.
58. PebernardS, McDonaldWH, PavlovaY, YatesJR3rd, BoddyMN (2004) Nse1, Nse2, and a novel subunit of the Smc5-Smc6 complex, Nse3, play a crucial role in meiosis. Mol Biol Cell 15 : 4866–4876.
59. Wehrkamp-RichterS, HyppaRW, PruddenJ, SmithGR, BoddyMN (2012) Meiotic DNA joint molecule resolution depends on Nse5-Nse6 of the Smc5-Smc6 holocomplex. Nucleic Acids Res 40 : 9633–9646.
60. FarmerS, San-SegundoPA, AragonL (2011) The Smc5-Smc6 complex is required to remove chromosome junctions in meiosis. PLoS One 6: e20948.
61. OnodaF, TakedaM, SekiM, MaedaD, TajimaJ, et al. (2004) SMC6 is required for MMS-induced interchromosomal and sister chromatid recombinations in Saccharomyces cerevisiae. DNA Repair (Amst) 3 : 429–439.
62. LeeBH, AmonA (2003) Role of Polo-like kinase CDC5 in programming meiosis I chromosome segregation. Science 300 : 482–486.
63. GrandinN, ReedSI (1993) Differential function and expression of Saccharomyces cerevisiae B-type cyclins in mitosis and meiosis. Mol Cell Biol 13 : 2113–2125.
64. DahmannC, FutcherB (1995) Specialization of B-type cyclins for mitosis or meiosis in S. cerevisiae. Genetics 140 : 957–963.
65. NeimanAM (2011) Sporulation in the budding yeast Saccharomyces cerevisiae. Genetics 189 : 737–765.
66. SymM, EngebrechtJA, RoederGS (1993) ZIP1 is a synaptonemal complex protein required for meiotic chromosome synapsis. Cell 72 : 365–378.
67. RoederGS, BailisJM (2000) The pachytene checkpoint. Trends Genet 16 : 395–403.
68. TungKS, HongEJ, RoederGS (2000) The pachytene checkpoint prevents accumulation and phosphorylation of the meiosis-specific transcription factor Ndt80. Proc Natl Acad Sci U S A 97 : 12187–12192.
69. ChuS, HerskowitzI (1998) Gametogenesis in yeast is regulated by a transcriptional cascade dependent on Ndt80. Mol Cell 1 : 685–696.
70. XuL, AjimuraM, PadmoreR, KleinC, KlecknerN (1995) NDT80, a meiosis-specific gene required for exit from pachytene in Saccharomyces cerevisiae. Mol Cell Biol 15 : 6572–6581.
71. KatisVL, MatosJ, MoriS, ShirahigeK, ZachariaeW, et al. (2004) Spo13 facilitates monopolin recruitment to kinetochores and regulates maintenance of centromeric cohesion during yeast meiosis. Curr Biol 14 : 2183–2196.
72. KlapholzS, EspositoRE (1980) Isolation of SPO12-1 and SPO13-1 from a natural variant of yeast that undergoes a single meiotic division. Genetics 96 : 567–588.
73. MichaelisC, CioskR, NasmythK (1997) Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell 91 : 35–45.
74. TanakaT, CosmaMP, WirthK, NasmythK (1999) Identification of cohesin association sites at centromeres and along chromosome arms. Cell 98 : 847–858.
75. KleinF, MahrP, GalovaM, BuonomoSB, MichaelisC, et al. (1999) A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell 98 : 91–103.
76. BuonomoSB, ClyneRK, FuchsJ, LoidlJ, UhlmannF, et al. (2000) Disjunction of homologous chromosomes in meiosis I depends on proteolytic cleavage of the meiotic cohesin Rec8 by separin. Cell 103 : 387–398.
77. XuL, KlecknerN (1995) Sequence non-specific double-strand breaks and interhomolog interactions prior to double-strand break formation at a meiotic recombination hot spot in yeast. EMBO J 14 : 5115–5128.
78. CaoL, AlaniE, KlecknerN (1990) A pathway for generation and processing of double-strand breaks during meiotic recombination in S. cerevisiae. Cell 61 : 1089–1101.
79. AlaniE, PadmoreR, KlecknerN (1990) Analysis of wild-type and rad50 mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell 61 : 419–436.
80. JessopL, LichtenM (2008) Mus81/Mms4 endonuclease and Sgs1 helicase collaborate to ensure proper recombination intermediate metabolism during meiosis. Mol Cell 31 : 313–323.
81. OhSD, LaoJP, TaylorAF, SmithGR, HunterN (2008) RecQ helicase, Sgs1, and XPF family endonuclease, Mus81-Mms4, resolve aberrant joint molecules during meiotic recombination. Mol Cell 31 : 324–336.
82. CarlileTM, AmonA (2008) Meiosis I is established through division-specific translational control of a cyclin. Cell 133 : 280–291.
83. BenjaminKR, ZhangC, ShokatKM, HerskowitzI (2003) Control of landmark events in meiosis by the CDK Cdc28 and the meiosis-specific kinase Ime2. Genes Dev 17 : 1524–1539.
84. SourirajanA, LichtenM (2008) Polo-like kinase Cdc5 drives exit from pachytene during budding yeast meiosis. Genes Dev 22 : 2627–2632.
85. AllersT, LichtenM (2001) Intermediates of yeast meiotic recombination contain heteroduplex DNA. Mol Cell 8 : 225–231.
86. JessopL, AllersT, LichtenM (2005) Infrequent co-conversion of markers flanking a meiotic recombination initiation site in Saccharomyces cerevisiae. Genetics 169 : 1353–1367.
87. BellL, ByersB (1983) Separation of Branched from Linear DNA by Two-Dimensional Gel-Electrophoresis. Analytical Biochemistry 130 : 527–535.
88. HollingsworthNM, GoetschL, ByersB (1990) The HOP1 gene encodes a meiosis-specific component of yeast chromosomes. Cell 61 : 73–84.
89. Bermudez-LopezM, CeschiaA, de PiccoliG, ColominaN, PaseroP, et al. (2010) The Smc5/6 complex is required for dissolution of DNA-mediated sister chromatid linkages. Nucleic Acids Res 38 : 6502–6512.
90. BlitzblauHG, BellGW, RodriguezJ, BellSP, HochwagenA (2007) Mapping of meiotic single-stranded DNA reveals double-stranded-break hotspots near centromeres and telomeres. Curr Biol 17 : 2003–2012.
91. KaneSM, RothR (1974) Carbohydrate metabolism during ascospore development in yeast. J Bacteriol 118 : 8–14.
92. LongtineMS, McKenzieA3rd, DemariniDJ, ShahNG, WachA, et al. (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14 : 953–961.
93. ElrodSL, ChenSM, SchwartzK, ShusterEO (2009) Optimizing sporulation conditions for different Saccharomyces cerevisiae strain backgrounds. Methods Mol Biol 557 : 21–26.
94. OhSD, JessopL, LaoJP, AllersT, LichtenM, et al. (2009) Stabilization and electrophoretic analysis of meiotic recombination intermediates in Saccharomyces cerevisiae. Methods Mol Biol 557 : 209–234.
95. AllersT, LichtenM (2000) A method for preparing genomic DNA that restrains branch migration of Holliday junctions. Nucleic Acids Res 28: e6.
96. NairzK, KleinF (1997) mre11S–a yeast mutation that blocks double-strand-break processing and permits nonhomologous synapsis in meiosis. Genes Dev 11 : 2272–2290.
Štítky
Genetika Reprodukční medicína
Článek Ribosome Synthesis and MAPK Activity Modulate Ionizing Radiation-Induced Germ Cell Apoptosis inČlánek Fission Yeast Shelterin Regulates DNA Polymerases and Rad3 Kinase to Limit Telomere Extension
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 11- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
- Délka menstruačního cyklu jako marker ženské plodnosti
- Intrauterinní inseminace a její úspěšnost
-
Všechny články tohoto čísla
- Molecular Recognition by a Polymorphic Cell Surface Receptor Governs Cooperative Behaviors in Bacteria
- The Light Skin Allele of in South Asians and Europeans Shares Identity by Descent
- Ribosome Synthesis and MAPK Activity Modulate Ionizing Radiation-Induced Germ Cell Apoptosis in
- Retrotransposon Silencing During Embryogenesis: Cuts in LINE
- Roles of XRCC2, RAD51B and RAD51D in RAD51-Independent SSA Recombination
- Parallel Evolution of Chordate Regulatory Code for Development
- A Genetic Approach to the Recruitment of PRC2 at the Locus
- Deletion of the Murine Cytochrome P450 Locus by Fused BAC-Mediated Recombination Identifies a Role for in the Pulmonary Vascular Response to Hypoxia
- Elevated Mutagenesis Does Not Explain the Increased Frequency of Antibiotic Resistant Mutants in Starved Aging Colonies
- Deletion of an X-Inactivation Boundary Disrupts Adjacent Gene Silencing
- Interplay between Active Chromatin Marks and RNA-Directed DNA Methylation in
- Recombinogenic Conditions Influence Partner Choice in Spontaneous Mitotic Recombination
- Crosstalk between NSL Histone Acetyltransferase and MLL/SET Complexes: NSL Complex Functions in Promoting Histone H3K4 Di-Methylation Activity by MLL/SET Complexes
- A New Role for the GARP Complex in MicroRNA-Mediated Gene Regulation
- RNAi-Dependent and Independent Control of LINE1 Accumulation and Mobility in Mouse Embryonic Stem Cells
- Loss of DNMT1o Disrupts Imprinted X Chromosome Inactivation and Accentuates Placental Defects in Females
- Inhibition of the Smc5/6 Complex during Meiosis Perturbs Joint Molecule Formation and Resolution without Significantly Changing Crossover or Non-crossover Levels
- Disruption of Lipid Metabolism Genes Causes Tissue Overgrowth Associated with Altered Developmental Signaling
- Translation Initiation Factors eIF3 and HCR1 Control Translation Termination and Stop Codon Read-Through in Yeast Cells
- Recruitment of TREX to the Transcription Machinery by Its Direct Binding to the Phospho-CTD of RNA Polymerase II
- MYB97, MYB101 and MYB120 Function as Male Factors That Control Pollen Tube-Synergid Interaction in Fertilization
- Oct4 Is Required ∼E7.5 for Proliferation in the Primitive Streak
- Contrasted Patterns of Crossover and Non-crossover at Meiotic Recombination Hotspots
- Transposable Prophage Mu Is Organized as a Stable Chromosomal Domain of
- Ash1l Methylates Lys36 of Histone H3 Independently of Transcriptional Elongation to Counteract Polycomb Silencing
- Fine-Mapping the Genetic Association of the Major Histocompatibility Complex in Multiple Sclerosis: HLA and Non-HLA Effects
- Genomic Mechanisms Accounting for the Adaptation to Parasitism in Nematode-Trapping Fungi
- Decoding a Signature-Based Model of Transcription Cofactor Recruitment Dictated by Cardinal Cis-Regulatory Elements in Proximal Promoter Regions
- Removal of Misincorporated Ribonucleotides from Prokaryotic Genomes: An Unexpected Role for Nucleotide Excision Repair
- Fission Yeast Shelterin Regulates DNA Polymerases and Rad3 Kinase to Limit Telomere Extension
- Activin Signaling Targeted by Insulin/dFOXO Regulates Aging and Muscle Proteostasis in
- Activin-Like Kinase 2 Functions in Peri-implantation Uterine Signaling in Mice and Humans
- Demographic Divergence History of Pied Flycatcher and Collared Flycatcher Inferred from Whole-Genome Re-sequencing Data
- Recurrent Tissue-Specific mtDNA Mutations Are Common in Humans
- The Histone Variant His2Av is Required for Adult Stem Cell Maintenance in the Testis
- The Maternal-to-Zygotic Transition Targets Actin to Promote Robustness during Morphogenesis
- Reconstructing the Population Genetic History of the Caribbean
- and Are Required for Growth under Iron-Limiting Conditions
- Whole Genome, Whole Population Sequencing Reveals That Loss of Signaling Networks Is the Major Adaptive Strategy in a Constant Environment
- Neuron-Specific Feeding RNAi in and Its Use in a Screen for Essential Genes Required for GABA Neuron Function
- RNA∶DNA Hybrids Initiate Quasi-Palindrome-Associated Mutations in Highly Transcribed Yeast DNA
- Mouse BAZ1A (ACF1) Is Dispensable for Double-Strand Break Repair but Is Essential for Averting Improper Gene Expression during Spermatogenesis
- Genetic and Functional Studies Implicate Synaptic Overgrowth and Ring Gland cAMP/PKA Signaling Defects in the Neurofibromatosis-1 Growth Deficiency
- DUX4 Binding to Retroelements Creates Promoters That Are Active in FSHD Muscle and Testis
- Pathways-Driven Sparse Regression Identifies Pathways and Genes Associated with High-Density Lipoprotein Cholesterol in Two Asian Cohorts
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- and Are Required for Growth under Iron-Limiting Conditions
- Genetic and Functional Studies Implicate Synaptic Overgrowth and Ring Gland cAMP/PKA Signaling Defects in the Neurofibromatosis-1 Growth Deficiency
- The Light Skin Allele of in South Asians and Europeans Shares Identity by Descent
- RNA∶DNA Hybrids Initiate Quasi-Palindrome-Associated Mutations in Highly Transcribed Yeast DNA
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



