-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaEmergence and Modular Evolution of a Novel Motility Machinery in Bacteria
Bacteria glide across solid surfaces by mechanisms that have remained largely mysterious despite decades of research. In the deltaproteobacterium Myxococcus xanthus, this locomotion allows the formation stress-resistant fruiting bodies where sporulation takes place. However, despite the large number of genes identified as important for gliding, no specific machinery has been identified so far, hampering in-depth investigations. Based on the premise that components of the gliding machinery must have co-evolved and encode both envelope-spanning proteins and a molecular motor, we re-annotated known gliding motility genes and examined their taxonomic distribution, genomic localization, and phylogeny. We successfully delineated three functionally related genetic clusters, which we proved experimentally carry genes encoding the basal gliding machinery in M. xanthus, using genetic and localization techniques. For the first time, this study identifies structural gliding motility genes in the Myxobacteria and opens new perspectives to study the motility mechanism. Furthermore, phylogenomics provide insight into how this machinery emerged from an ancestral conserved core of genes of unknown function that evolved to gliding by the recruitment of functional modules in Myxococcales. Surprisingly, this motility machinery appears to be highly related to a sporulation system, underscoring unsuspected common mechanisms in these apparently distinct morphogenic phenomena.
Published in the journal: . PLoS Genet 7(9): e32767. doi:10.1371/journal.pgen.1002268
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002268Summary
Bacteria glide across solid surfaces by mechanisms that have remained largely mysterious despite decades of research. In the deltaproteobacterium Myxococcus xanthus, this locomotion allows the formation stress-resistant fruiting bodies where sporulation takes place. However, despite the large number of genes identified as important for gliding, no specific machinery has been identified so far, hampering in-depth investigations. Based on the premise that components of the gliding machinery must have co-evolved and encode both envelope-spanning proteins and a molecular motor, we re-annotated known gliding motility genes and examined their taxonomic distribution, genomic localization, and phylogeny. We successfully delineated three functionally related genetic clusters, which we proved experimentally carry genes encoding the basal gliding machinery in M. xanthus, using genetic and localization techniques. For the first time, this study identifies structural gliding motility genes in the Myxobacteria and opens new perspectives to study the motility mechanism. Furthermore, phylogenomics provide insight into how this machinery emerged from an ancestral conserved core of genes of unknown function that evolved to gliding by the recruitment of functional modules in Myxococcales. Surprisingly, this motility machinery appears to be highly related to a sporulation system, underscoring unsuspected common mechanisms in these apparently distinct morphogenic phenomena.
Introduction
In Gram-negative bacteria, envelope machineries connecting the cell interior to the extracellular milieu must span all envelope layers, including the inner membrane, peptidoglycan and outer membrane. Despite these constraints, gram-negative bacteria have evolved sophisticated envelope nano-machines to interact with their environment. Conspicuous examples are bacterial organelles such as flagella, pili, and transport and secretion systems [1], [2]. In general, the structural genes encoding these systems are clustered within large transcriptional units allowing co-regulation of their expression. However, assembly also relies on additional complexity and must involve “just-in time” transcriptional regulations, specific targeting and protein self-assembly properties [3]. This raises the question of the evolutionary processes that led to the emergence of these macromolecular systems [4].
Non-homologous envelope macro-molecular structures mediate motility in bacteria. For example, bacteria swim in extremely viscous environments by means of a rotary flagellum, one of the most sophisticated known biological nano-machines [3]. Bacteria can also crawl across surfaces, for example, polymerization and de-polymerization of pilin fibers from the bacterial cell pole pull the cell forward, a “twitching” motility mechanism which also involves the coordinated assembly of many envelope proteins [5]–[7]. However, gram-negative rod-shaped bacteria are also able to move on surfaces by other means. For example, many bacteria move smoothly along their long axis in the absence of obvious extra-cellular organelles [8]. This gliding motility is associated with unusual flexibility of the cell body and can be found in very diverse bacterial phyla, such as, Bacteroidetes, Cyanobacteria and Deltaproteobacteria [8], [9]. In most species, the mechanism that drives gliding motility remains speculative. For example, in Flavobacterium johnsoniae (Bacteroidetes) gliding motility may be associated with a novel secretion apparatus. However, it is unclear whether this system is involved in assembly of the gliding machinery or constitutes the machinery itself [10]. Finally, gliding may be propelled differently in various species [8].
Despite decades of research, dedicated gliding motility machineries have not been identified unambiguously in any bacterial species, hampering detailed mechanistic studies and asking the question of the emergence of this process in bacteria. In Myxococcus xanthus, a gram negative deltaproteobacterium, surface motility allows the directed aggregation of thousands of cells into mounds that mature into fruiting bodies where the bacteria differentiate into spores [11]. Myxococcus cells can move by twitching motility, but in the absence of pili, the cells are still able to move, unmasking the activity of the gliding engine [11]. Recent cytological work suggested that motility is driven by protein complexes (Focal Adhesion Complexes, FAC) that push against the substratum as they accumulate periodically on the ventral side of the cell [12]–[14]. In a live cell assay, FACs can be observed as bright fluorescent fixed spots in cells expressing a fluorescent gliding motility protein (AglZ-YFP, [12]). The formation of AglZ-YFP foci requires the bacterial MreB-actin cytoskeleton [15] and a FACs-localized proton motive force-driven motor (AglRQS) was recently identified [13]. These observations suggest that AglRQS powers motility in concert with the MreB-cytoskeleton; however, how work from AglRQS is tranduced to the cell surface remains unknown and requires the identification of a motor-associated complex that spans the cell envelope.
In the past, 51 genes associated to defects in gliding motility were identified by transposon-based genetic screens, but the functional role of these genes in motility was not established [16], [17]. Recent work by Nan et al. [18] uncovered a new motility complex (AgmU, AglZ, AglT, AgmK, AgmX, AglW and CglB) and suggested that AgmU may be actively transported by PMF-utilizing motors [14]. However, the function of this complex and its direct link with the AglRQS motor remains to be established.
In this work, we aimed to identify the motility machinery conclusively. We re-investigated the 51 known M. xanthus gliding genes with the premise that the gliding machinery must have co-evolved with the AglRQS motor. This approach allowed us to identify a novel energy-driven protein complex, which we prove to be the basal gliding machinery. The results reveal the architecture of the gliding machinery and suggest a scenario of its emergence (and evolution) in bacteria.
Results/Discussion
Identification of candidate genes encoding the gliding machinery
Two independent transposon-based genetic screen studies [16], [17] identified 35 and 23 potential gliding motility genes, respectively (Table S1). Only seven genes overlapped in the two genetic studies, suggesting that the screens are not saturated and thus, the complete set of genes involved in gliding motility has likely not been identified. Nevertheless, these data constituted a good starting point and could indeed contain genes that encode the motility machinery. Irrespective of the exact motility mechanism, a number of cell envelope proteins should be part of the motility machinery. Therefore, we re-visited gene annotations specifically looking for genes encoding predicted membrane proteins, exported proteins and proteins containing motifs mediating protein-protein interaction, such as Tetratricopeptide repeat (TPR) and Coiled-coil domains (Table S1). A total of 28 genes were thus highlighted.
A careful survey of these 28 genes revealed that 13 gene hits were in fact clustered into four chromosomal regions of the M. xanthus DK 1622 genome. One region containing three hits, aglW (MXAN_5756, tolB), aglX (MXAN_5753, tolQ) and aglV (MXAN_5754, tolR), encoded together with MXAN_5755 (tolA) and MXAN_5757 (pal), bona fide components of a complete Tol-Pal system. Tol-Pal maintains envelope integrity and supports cell division in all bacteria where it has been studied [19], [20]. Thus, it is unlikely that Tol-Pal constitutes the motility machinery. Consistent with a general envelope function of the Myxococcus Tol-Pal, the aglV (tolR) mutant was also severely impaired in twitching motility [16]. We then focussed our analysis on the remaining three gene clusters (hereafter referred as Gliding1 (G1), Gliding 2 (G2) and Motor 1 (M1), Figure 1A). The G1 cluster contains eight genes, MXAN_4870-62, six of which have been hit by transposons: agmU (MXAN_4870), aglT (MXAN_4869), pglI (MXAN_4867), agmV (MXAN_4864/65, see below), agmK (MXAN_4863) and agmX (MXAN_4862) (Figure 1A). The G2 cluster contains four genes, MXAN_2538-41, two of them inactivated by transposons: agmO (MXAN_2538) and agnA (MXAN_2541). Finally, M1 contains the aglRQS genes themselves (MXAN_6862-60) and two hits by transposon insertions in aglR (MXAN_6862) and aglS (MXAN_6860). So overall, the G1, G2 and M1 clusters involve 15 genes, 10 of which have been previously hit by the transposon screens (Table 1).
Fig. 1. Genetic clusters carrying gliding motility genes in M. xanthus. 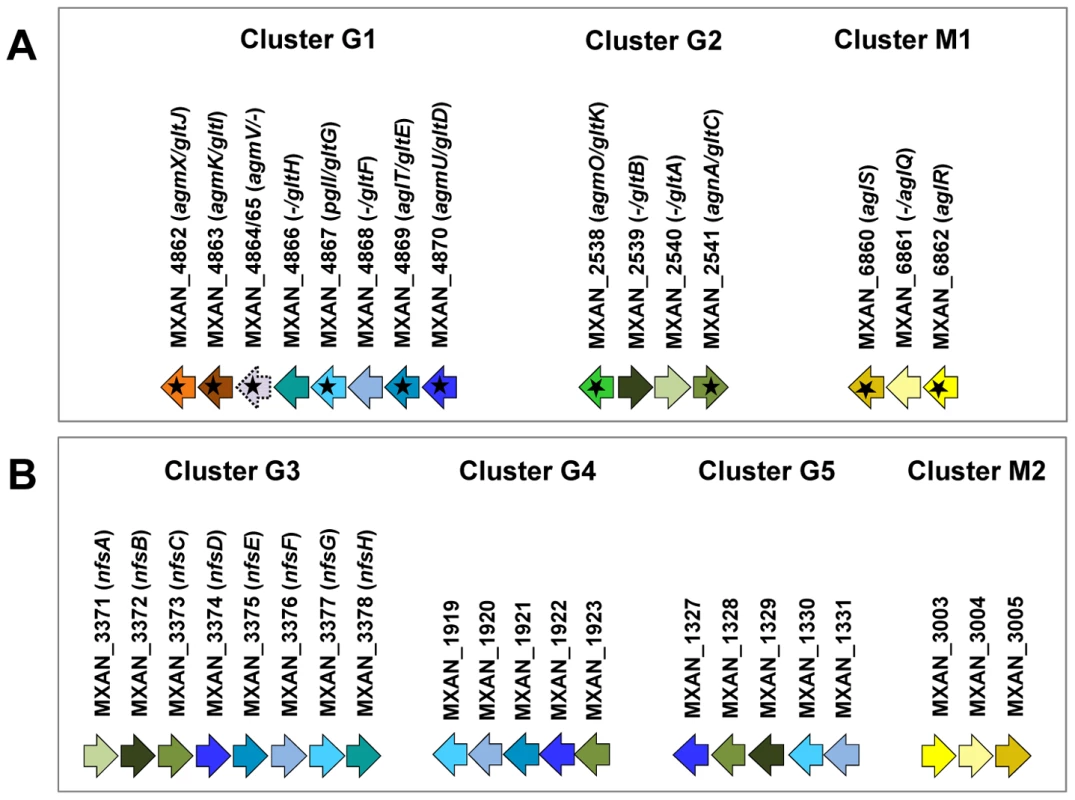
(A) Genetic organisation of the 15 genes composing the G1, G2 and M1 clusters encoding the putative components of the gliding machinery in Myxococcus xanthus DK 1622. Predicted genes are indicated with their locus_tag, and their former and new names. The arrow that represents the putative MXAN_4864 and MXAN_4865 genes is a dotted line because they are likely pseudogenes (see text for more details). Stars indicate genes that were hit by the transposon screens [16], [17]. (B) Genetic organization of the G3, G4, G5 and M2 homologue clusters in M. xanthus DK 1622. The colour code indicates homologous genes and will be used throughout the study. Tab. 1. Bioinformatic analysis of G1, G2, and M1 cluster genes. 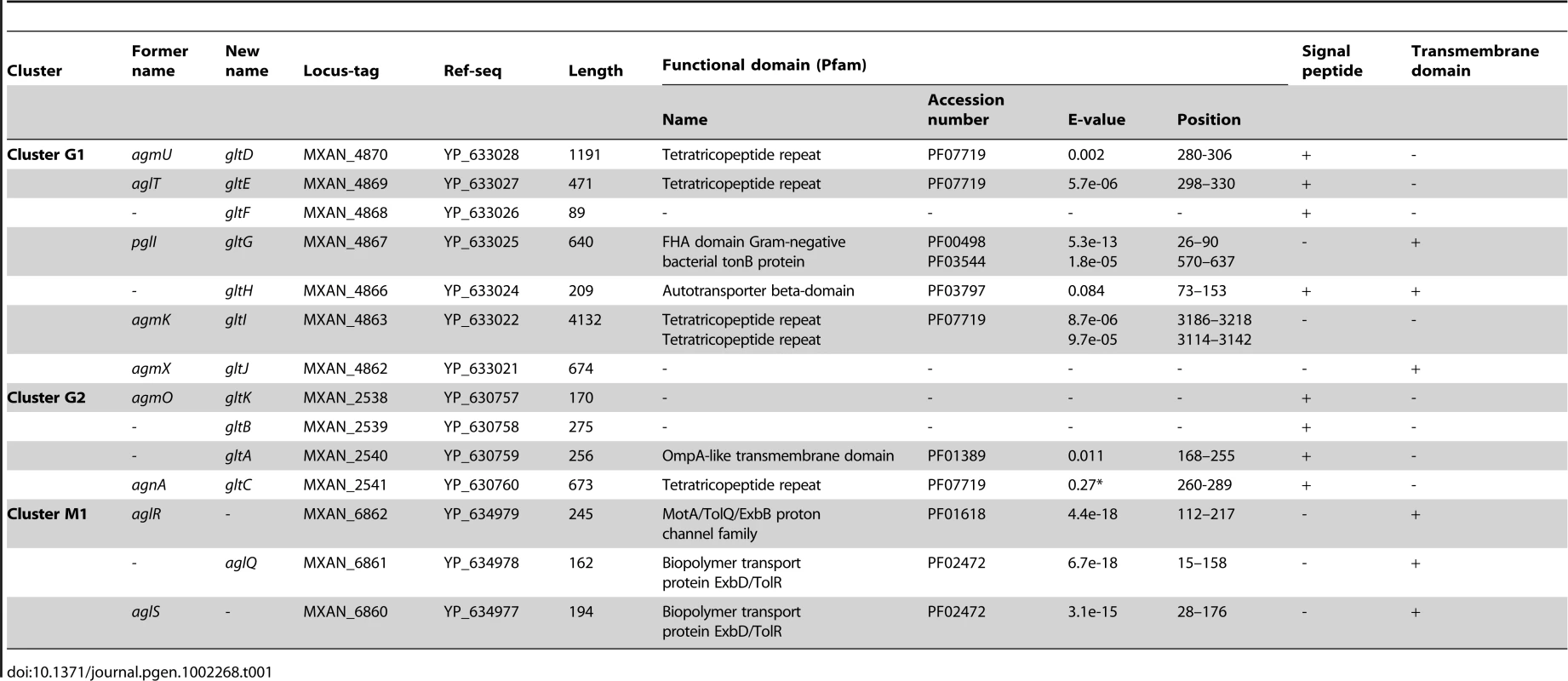
The M1 cluster encodes the component of a TolQR-like proton conducting motor, which has been characterized elsewhere [13]. The G1 and G2 cluster genes were analysed using public sequences and domain databases. The predicted MXAN_4866 (G1 region) and MXAN_2540 (G2 region) proteins are probably secreted and inserted in the outer membrane because they contain an Autotransporter ß-domain and adopt an OmpA-like fold, respectively (Table 1). AgmO may also be located in the outer-membrane because it carries a typical Outer-membrane Type-II signal sequence. TPR-repeats typically involved in multiprotein assemblies [21] are encoded by four G1 and G2 region genes: agmU, aglT, agmK, and agnA (Table 1). Among them, AgmU, AglT, and AgnA also carry signal peptides, suggesting that they are exported beyond the inner membrane. PglI (G1 region) is a predicted bi-topic transmembrane protein with a cytosolic ForkHead-Associated domain (FHA, [22]) and a periplasmic domain of unknown function. AgmX (G1 region) is also a potential integral membrane protein. MXAN_4868 and MXAN_2539 both carry N-terminal signal peptides but do not contain any conserved functional domains (Table 1). Finally, MXAN_4864 and MXAN_4865 are probably not actual genes and were discarded from this study (see Text S1 for justification) In the rest of this work, we tested if the G1 and G2 genes encode the AglRQS motor-associated gliding machinery. For clarity and to homogenize the nomenclature, we renamed all the G1 and G2 genes glt (gliding transducer, see below), with gltD, E, F, G, H, I and J corresponding respectively to agmU, aglT, MXAN_4868, pglI, MXAN_4866, agmK and agmX (G1) and gltC, A, B and K corfresponding to agnA, MXAN_2540, MXAN_ 2539 and agmO (G2, see below and Text S2 for justification).
The G1, G2, and M1 clusters may encode components of a single macro-molecular machinery
The G1, G2 and M1 clusters encode a majority of potential envelope proteins and a motor complex (see above). A tempting hypothesis would be therefore that all these components constitute the gliding machinery. We systematically investigated the taxonomic distribution of the 14 genes defining the G1, G2 and M1 clusters in the 1180 complete prokaryotic proteomes available at the beginning of this study (see methods). The 14 genes could be separated in two distinct groups based on taxonomic distribution: A first group (Group A) contained seven genes (gltF, gltH, gltI, gltJ, gltK, gltB and gltA) that were only present (and sometimes in several copies) in Myxococcales (i.e. Sorangium cellulosum, Plesiocystis pacifica, Haliangium ochraceum, Stigmatella aurantiaca, Myxococcus xanthus and the four Anaeromyxobacter sp.) and in Bdellovibrionales (Bdellovibrio bacteriovorus) (Figure 2A). Such restricted taxonomic distribution suggested that these genes appeared only recently during the evolution of the Deltaproteobacteria. By contrast, a second group (Group B) contains seven genes with a much broader taxonomic distribution (Figure 2A). Specifically, blastp and PSI-BLAST queries identified 142 GltD, 2545 GltE, 313 GltG, 83 GltC, 2677 AglR, 2348 AglQ and 2385 AglS homologues. The taxonomic distributions of all these homologues are very different, suggesting that the corresponding genes have undergone different evolutionary histories, which was confirmed by preliminary phylogenetic analyses (not shown). However, in all these phylogenetic trees, the M. xanthus sequences emerge within a monophyletic clade containing homologues from other Deltaproteobacteria but also from a set of unrelated bacteria (i.e. one Betaproteobacteria, several Gammaproteobacteria and one member of Fibrobacteres, Figure S4). This strongly suggests that, although these genes belong to large gene families of distinct evolutionary histories, the M. xanthus gltD, gltE, gltG, gltC, aglR, aglQ and aglS genes and their closest homologues share a similar evolutionary history. The presence of Group B genes (sometime in several copies per genomes) in a few distantly related bacteria (Figure S1) suggests a complex evolutionary history punctuated with horizontal gene transfer (HGT) and gene duplication events (see below). In all non-Deltaproteobacteria and in Geobacter, Group B genes clustered in a single genomic region, possibly an operon, arguing strongly that they encode a single functional unit (i.e. core complex, Figure 2B and Figure S1, Tables S2 and S3). In all these bacteria, the core complex contains an additional gene that has no homologues in Myxococcales and Bdellovibrionales (Figure S1). Remarkably, group B genes (and thus the core complex) group genes from the G1 (gltD, gltE and gltG), G2 (gltC) and M1 (aglQ, R and S) clusters (Figure 2A-2B). This suggests an evolutionary link between the G1, G2 and M1 gene clusters. Strengthening this prediction, homologues of G1 and G2 clusters are grouped on the chromosome of the four Anaeromyxobacter relatives (Figure S1). Then, we proceeded to test the functional relationships between the G1, G2 and M1 genes.
Fig. 2. Taxonomic distribution of the closest homologues of the 14 genes composing the G1, G2, and M1 clusters, and genetic organization of the core complex. 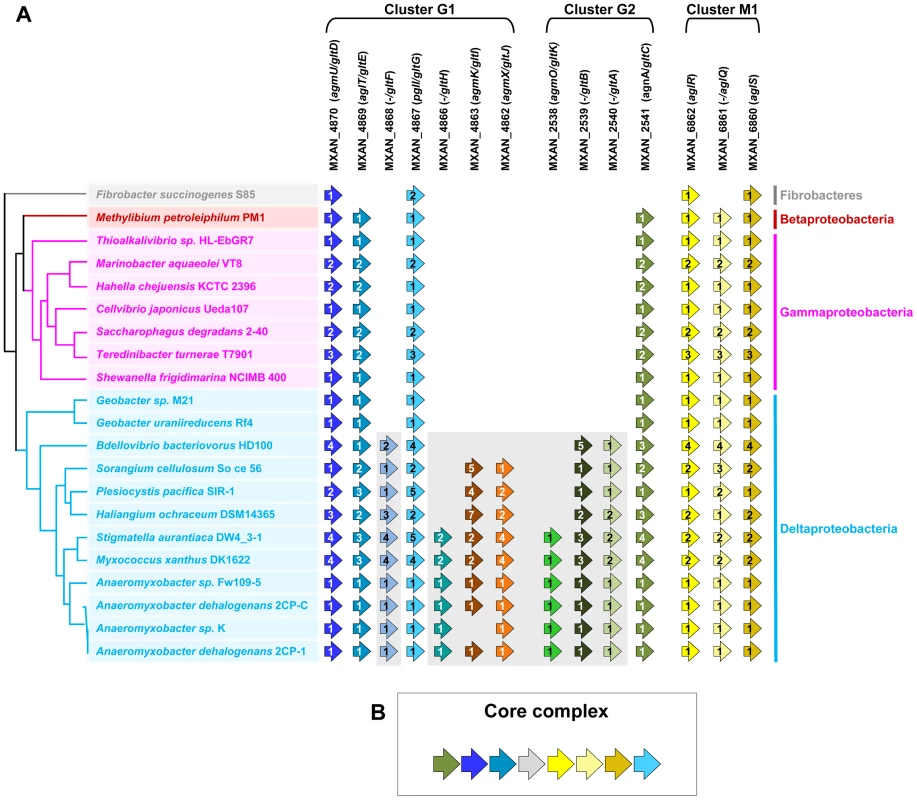
(A) For a given gene, the number of homologues in the corresponding genome is indicated by the numbers within arrows. The relationships between the species carrying the different homologues of the genes are indicated by the phylogeny on the left. Based on their taxonomic distribution, the 14 genes can be divided into Group A (grey background) and Group B (white background). (B) In all non Deltaproteobacteria and in Geobacter, the Group B genes clustered in a single genomic region. Genetic characterization of G1 and G2 gene clusters
In M. xanthus, many of the genes composing the G1 and G2 clusters were previously hit by genetic screens [16], [17]; however, the genes were only partially characterized, and it was not determined how they might be functionally related. More recently, Nan et al. [18] showed that individual deletions of the G1 genes gltD-J impair motility, but their analysis did not test whether these genes are structural. In fact, structural motility components cannot be simply discriminated from regulatory motility components solely based on mutational analysis and colony agar plate assays. First, the absence of motility at colony edges does not necessarily indicate that single cells are completely unable to move: for example, a class of directional mutants (FrzCDc, hyper active Frz-receptor mutants,[23]) forms smooth colony edges, yet, when observed under the microscope, individual cells glide but move back and forth at very high frequencies and thus show no net translocation (hyper-reversing cells, [23]). Thus, motility mutants must also be probed in single cell motility assays. Second, some mutations leading to complete motility defects can be suppressed by second-side mutations, showing that the mutated genes are not structural but regulatory. For example, the motility defect of the aglZ mutant is suppressed when frz, encoding a signal transduction system regulating the directionality of motility, is disrupted [24].
Thus, structural machinery genes must minimally meet the following criteria: (i) gene deletion should result in complete loss of motility in mutants that also lack twitching motility both at colony and single cell scales and (ii), the motility defect should not be suppressed by a frz mutation [24]. Consequently, in this study, we systematically combined the deletions to pilA- or frzE-null mutation (encoding the major pilin sub-unit and the essential FrzE kinase, respectively). It is still possible that regulatory genes may work independently from Frz, but, altogether, the genetic, localization and interaction evidence strongly supports that the Glt proteins are structural (see below).
We therefore made markerless in frame deletions in all the G1 and G2 genes (except gltI and gltJ) and showed that the deletions did not create polar effects by quantitative RT-PCR (Table 2 and Table 3). Of note, the expression of gltH was up-regulated 4-5 folds when gltG was deleted, which may point to a regulatory function of GltG (Table 2).
Tab. 2. glt mRNA expression in cluster G1 deletion strains. 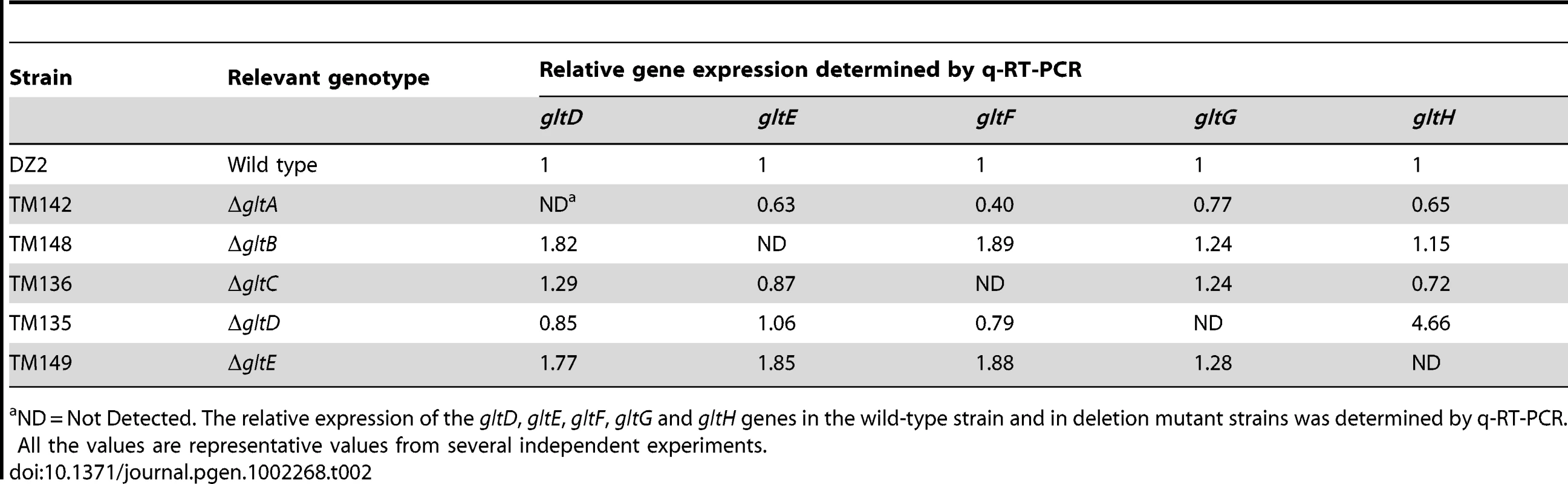
ND = Not Detected. The relative expression of the gltD, gltE, gltF, gltG and gltH genes in the wild-type strain and in deletion mutant strains was determined by q-RT-PCR. All the values are representative values from several independent experiments. Tab. 3. glt mRNA expression in the cluster G2 deletion strains. 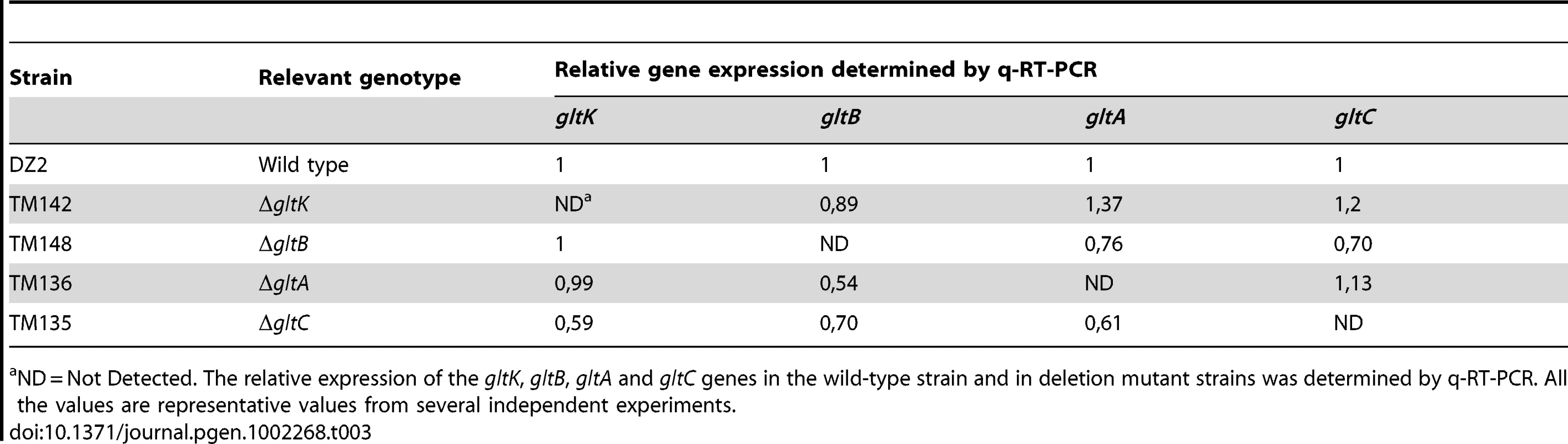
ND = Not Detected. The relative expression of the gltK, gltB, gltA and gltC genes in the wild-type strain and in deletion mutant strains was determined by q-RT-PCR. All the values are representative values from several independent experiments. On agar plate assays, the gltA-H and gltK mutants retained intact twitching motility but were completely deficient in single cell motility at the colony edges (Figure 3A). gltA-H and gltK pilA double mutants were all completely non-motile both at the colony and single cell levels (Figure 3A and data not shown), showing that the glt genes are specific and essential to gliding motility. In one exception, the gltH pilA mutant showed small scales “jerky” displacements on occasions, but the motility defect was still very severe (Figure 3A and Video S1).
Fig. 3. Group B genes encode structural components of the motility machinery. 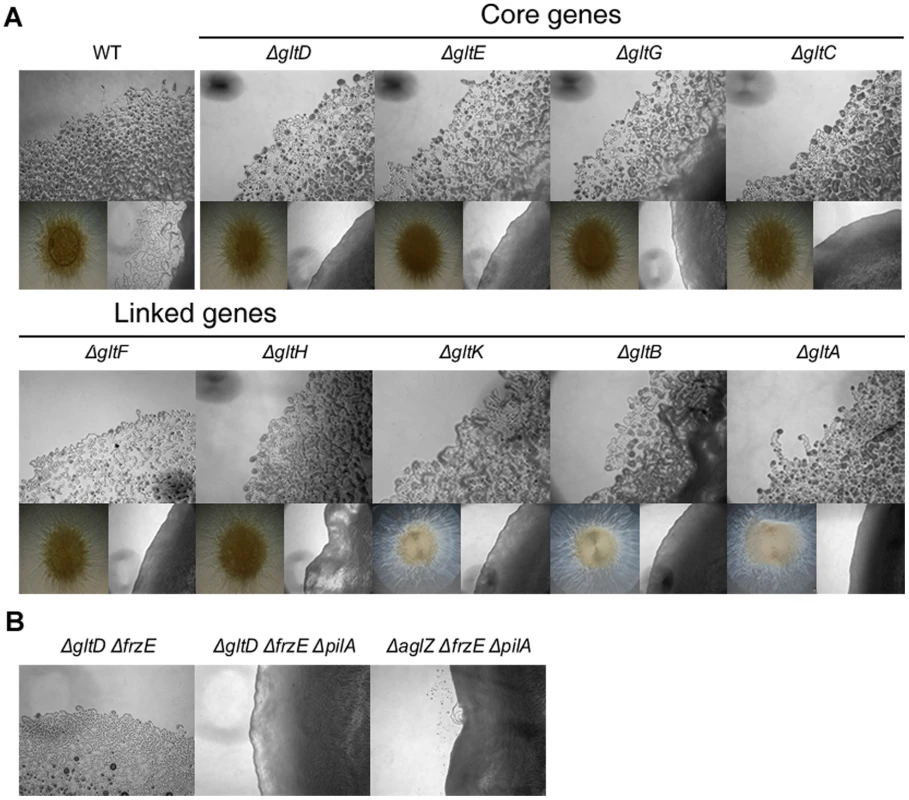
(A) Motility at the gltA-Hand gltK deletion mutants colony edges after 48h incubation at 32°C on hard (1.5%) (upper panel). Lower left panel: twitching motility is unaffected in the gltA-H and gltK deletion mutants and observed in the form of expanded colony swarms on soft (0.5%). Lower right panel: motility of double pilA gltA-H and pilA gltK deletion mutants showing the complete absence of motility in these mutants. (B) Hard agar colony edges of the gltD frzE, gltD frzE pilA and aglZ frzE pilA mutants showing the lack of motility restoration in the gltD mutant. Note that single cells are clearly visible in the aglZ frzE pilA mutant, consistent with previous literature [24]. In a second step, we observed that gltA-H and gltK frzE double mutants were also completely non-motile in the colony and single cell assays (Figure 3B and data not shown). As a control, we also tested the simultaneous deletion of frzE and aglZ and observed that colony and single motility were both restored, as previously described (Figure 3B, [24]). Interestingly, group swarming appeared enhanced on hard agar plates in all cases, suggesting that the frzE mutation enhanced twitching in those mutants (Figure S2). Nan et al. [18] reported that motility of a gltD mutant allele was restored when a frz mutation was introduced, however this conclusion was based on observation of colony edges. In fact, enhanced twitching motility in the double mutant may have been mis-interpreted for restored gliding motility. To test this, we further introduced a pilA mutation in the double gltD frzE mutant. Motility was completely abolished in the resulting triple mutant. In contrast, the triple aglZ frzE pilA mutant was motile under similar conditions, as expected (Figure 3B, compare middle and right panels). The enhanced twitching in the glt frzE double mutants points to intriguing couplings between gliding and twitching motility, which will need further investigation.
In conclusion, the glt genes are genetically separable from aglZ, and may thus encode structural components of the motility machinery. A comparable genetic analysis also suggested that aglR, Q and S are structural [13]. Thus, the genetic results are consistent with a functional link between aglRQS and the glt G1 and G2 group genes.
G1 cluster proteins localize to the cell envelope
We next aimed to determine the subcellular localization of the suspected Glt protein complex. In absence of specific antibodies to detect all proteins, we only tested some proteins of the G1 cluster: GltD, E, F, G and H, all predicted to localize within the cell envelope (Table 1). We also tested the localization of a functional GltF-mCherry fusion with specific anti-mCherry antibodies. Cell fractionation experiments showed unambiguously that all five proteins localize in the cell envelopes (Figure 4). GltD was also present in the soluble fraction but to minor extents (Figure 4). GltF-mCherry was equally distributed in the soluble and membrane extracts (Figure 4). The GltF-mCherry fusion was functional (Figure S3 and data not shown), however it also seemed to be processed to some extent during the fractionation procedure (Figure 4), thus it cannot be excluded that its presence in the soluble fraction results from improper secretion.
Fig. 4. Envelope localization of the Glt proteins. 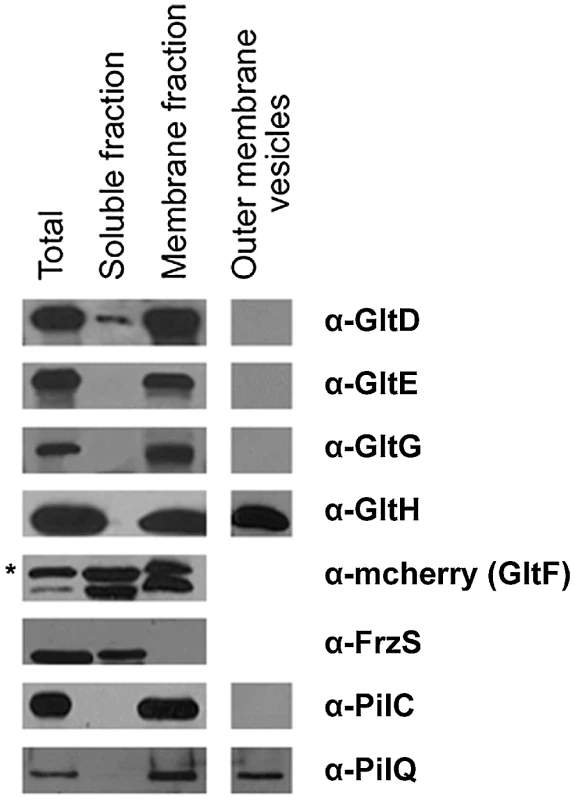
Envelope localization of the Glt proteins. GltF localization is determined by western detection of the GltF-mCherry fusion (as indicated by the asterisk). FrzS, PilC and PilQ were used as control markers of the soluble, inner membrane and outer membrane fractions, respectively. We next wanted to discriminate inner - and outer-membrane proteins. Separating the inner membrane from the outer membrane was difficult using standard sucrose density gradients or detergent-based methods (see Methods). We therefore decided to harvest outer-membrane-derived vesicles (see Methods). Extracted vesicles contained PilQ, the pilus Secretin but not PilC, localizing at the inner membrane, confirming that the vesicles were derived from the outer membrane. Only GltH was detected in the vesicle preparation, which is consistent with the presence of an auto-transporter ß-domain in this protein (Figure 4, Table 1). All together these results suggest that GltD-G form an inner membrane localized complex that extends through the periplasm and connect the cell surface via the outer-membrane protein GltH.
The Glt proteins form an AglRQS-associated dynamic motility complex
In a parallel study, we have demonstrated that the M1 cluster (aglRQS) encodes a proton-motive force-driven channel that produces motility traction forces at FACs [13]. The present study suggests that AglRQS and Glt proteins are functionally related, which needed to be proven experimentally. If the Glt proteins interact with the AglZ-AglRQS system, it would be expected that the Glt proteins also localize at FACs. A fluorescent functional GltD-mCherry fusion was already available [18]. We additionally obtained another functional fusion to GltF. In two other studies, GltD-mCherry was found to localize both in fixed clusters [18] and along a dynamic helix-like structure [14]. To rationalize this apparent dual localization pattern, it was proposed that GltD-mCherry molecules traffic along a helix and accumulate at FACs when they become engaged in propulsion [14]. In our hands, the pattern of GltD-, GltF-mCherry fluorescence in live cells was similar: fluorescence was mostly evident around the cell periphery; however, when we collected z-stacks of unprocessed images, fluorescent clusters became clearly apparent when the focal plane was focussed closer to the substratum (Figure 5A, 5B and Video S2). In moving cells, these clusters were fixed and largely co-localized with AglZ-YFP (Figure 5B and 5C). We were not able to resolve a helical pattern of GltD-mCherry around the cell periphery, but observing this structure may require mathematical image-deconvolution processing [14], which would explain this discrepancy. Nevertheless, these results show that GltD and GltF are recruited at FACs, and may be parts of a complex mediating contact between the exterior and the cell interior.
Fig. 5. The Glt proteins localize dynamically to the AglZ-YFP clusters in a AglQ-dependent manner. 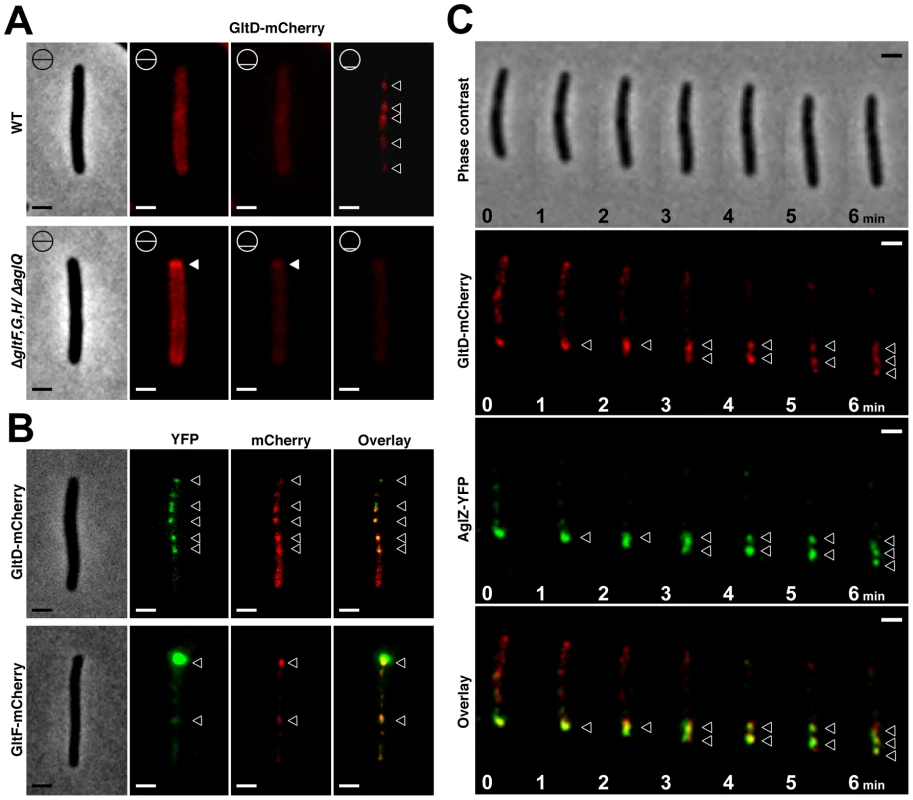
(A) Localization of GltD-mCherry in different z sections in WT (upper panel) and mutant backgrounds (lower panel). Shown are unprocessed fluorescent micrographs of the different sections (position of the section along the z axis is indicated by a barred circle). Open triangles indicate GltD clusters. Scale bar = 1 µm. (B) Co-localization of GltD- and GltF-mCherry with AglZ-YFP. Open triangles indicate clusters were the chimeric proteins co-localize. Scale bar = 1 µm. (C) dynamic localization of GltD-mCherry and AglZ-YFP during movement. Scale bar = 1 µm. GltD-mCherry dynamics are dependent on the PMF [14], suggesting that they result from the activity of a motor, possibly the AglRQS complex (M1). Indeed, in an aglQ mutant, GltD - and GltF-mCherry failed to accumulate at FACs and were only localized around the cell periphery and at the cell poles (Figure 5A. and data not shown). To prove that AglRQS directly fuels trafficking of the Glt proteins, we searched which Glt protein may interact with the motor. By analogy to the Tol/Exb system, the AglR protein would deliver motor work to an output protein through an H+-driven conformational change in its N-terminal transmembrane helix [25]. Thus, the best candidate for direct interaction with AglR is the GltG protein. Indeed, this predicted transmembrane protein has a proline-rich TonB-like motif, typically found in TolA and TonB, the effector transducers in the Tol/Exb systems [20]. Moreover, GltG is the only predicted transmembrane protein that belongs to the core complex together with AglRQS (Figure 2B). We tested a potential interaction between AglR and GltG in a bacterial two-hybrid assay [26] (Figure 6). Highly significant β-galactosidase activity was only obtained when AglR and GltG were expressed together, showing that these proteins interact specifically (Figure 6). Finally, GltD-mCherry cluster localization was also abolished in mutants lacking gltF, G and H, further suggesting that these proteins are parts of one motility complex within the focal adhesion clusters (Figure 5A). All together, the results suggest that the AglRQS-Glt proteins assemble a dynamic envelope spanning motility machinery at the focal adhesion sites.
Fig. 6. The AglRQS motor interacts directly with the gliding motility machinery. 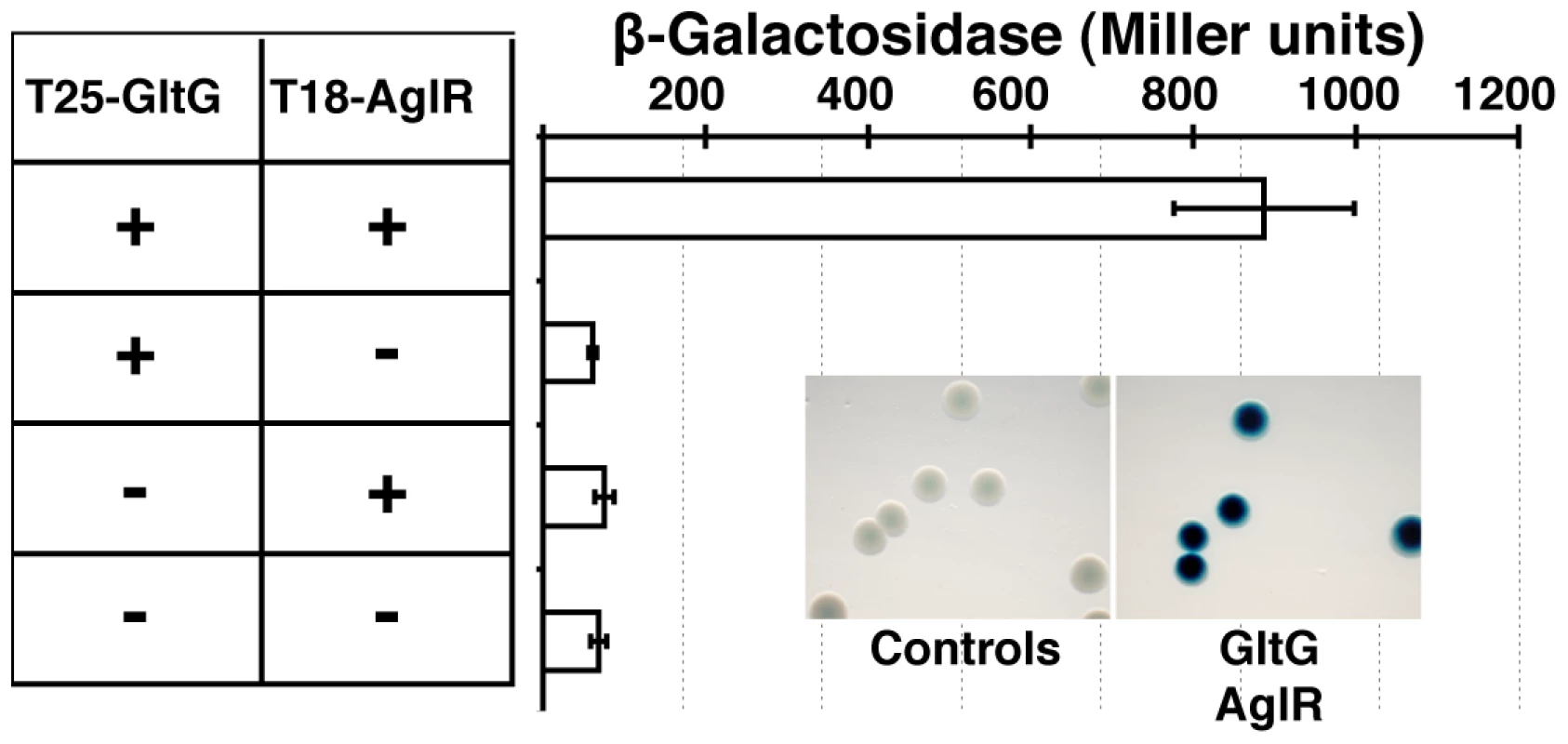
AglR interacts with GltG in a bacterial two-hybrid assay. Emergence and evolutionary history of the gliding machinery
Taken together, the computational and experimental results strongly suggest that the G1, G2 and M1 clusters contain genes encoding the major components of the gliding motility machinery. The most striking result of our in silico analysis is the discovery of a conserved core of genes (Group B) coding for several homologues of the gliding machinery components in non-gliding bacteria. To obtain further insights on the evolutionary mechanisms underlying the emergence of the gliding apparatus, we conducted an in-depth phylogenomic analysis (see Text S3). The phylogenies of the closest homologues of the seven genes defining the conserved core of genes (i.e. Group B) showed similar topologies (Figure S4). However, these analyses were based on a fairly small number of unambiguously aligned positions and as a result most of the nodes of the inferred trees were weakly supported (Bootstrap Values (BV) <90% and Posterior Probabilities (PP) <0.95, Figure S4). This caveat precluded the precise elucidation of the evolutionary histories of the components. To improve the resolution of the phylogenetic trees, we combined the group B genes gltD, E , G and AglR,Q,S in two distinct supermatrices (See Methods).
As expected, the trees based on each supermatrix showed better resolutions than the individual gene trees (compare PP and BV in Figure 6 and Figure S4). Consistent with the single phylogenies (Figure S4), two separate clades (at odds with the species phylogeny) were observed in the resulting phylogenetic trees (PP = 1.00 and BV = 100%, Figure 7): More precisely, the three Geobacter representatives (Deltaproteobacteria) emerged within the Gammaproteobacteria, whereas F. succinogenes and the other Deltaproteobacteria, belonging to distinct phyla [27], emerged together in the glt and agl phylogenetic trees (Figure 7). Moreover, the relationships among the gammaproteobacterial sequences were mostly incongruent with the species phylogeny (Figure 2 and Figure 7). The discrepancy between the organism and gene trees precluded the clear identification of the precise bacterial lineage where the core complex originated, possibly the Gamma - or Deltaproteobacteria (Figure 8A). Nevertheless, HGT of the core complex is apparent: first, between Gammaproteobacteria and Deltaproteobacteria, then among Gammaproteobacteria and from Deltaproteobacteria to Fibrobacter, and last, from Gammaproteobacteria or Betaproteobacteria to Geobacter (Figure 8A, circles 1 to 4). In contrast, the restricted taxonomic distribution of the Group A genes indicates that they appeared and were recruited more recently during differentiation of the Deltaproteobacteria. An evolutionary scenario may thus be suggested: gltA, B and F likely appeared in the common ancestor of the Myxococcales and Bdellovibrionales, whereas gltI and gltJ (MXAN_4863-62) probably appeared in the ancestor of the Myxococcales, while gltK and H may have been acquired more recently (Figure 8A).
Fig. 7. Co-evolution of gltD-E-G and aglR-Q-S. 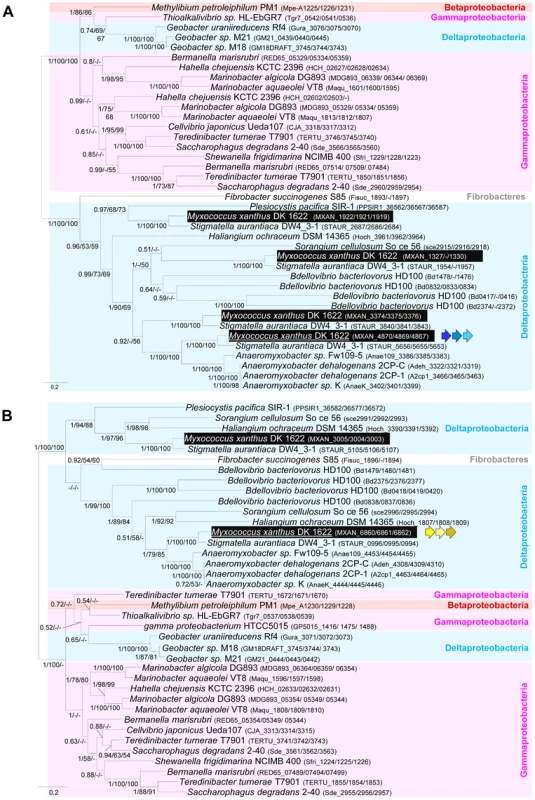
Rooted Bayesian phylogenetic trees of concatenated alignments of (A) GltD, GltE and GltG (39 sequences, 586 positions) and (B) AglR, AglQ and AglS (38 sequences, 376 positions). The root has been placed according to the phylogenies of the individual proteins. Numbers at nodes indicate posterior probabilities (PP) computed by MrBayes and bootstrap values (BV) computed by Treefinder and PhyML. Only PP and BV above 0.5 and 50% are shown. The scale bars represent the average number of substitutions per site. In each phylogenetic tree the putative M. xanthus gliding motility proteins are underlined and are illustrated with colour-coded gene symbols. For each species the individual locus_tags of the concatenated proteins are indicated in brackets. The position of multiple duplications of the concatenated proteins in M. xanthus are highlighted by black rectangles. Fig. 8. Evolution and structure of the Myxococcus gliding motility machinery. 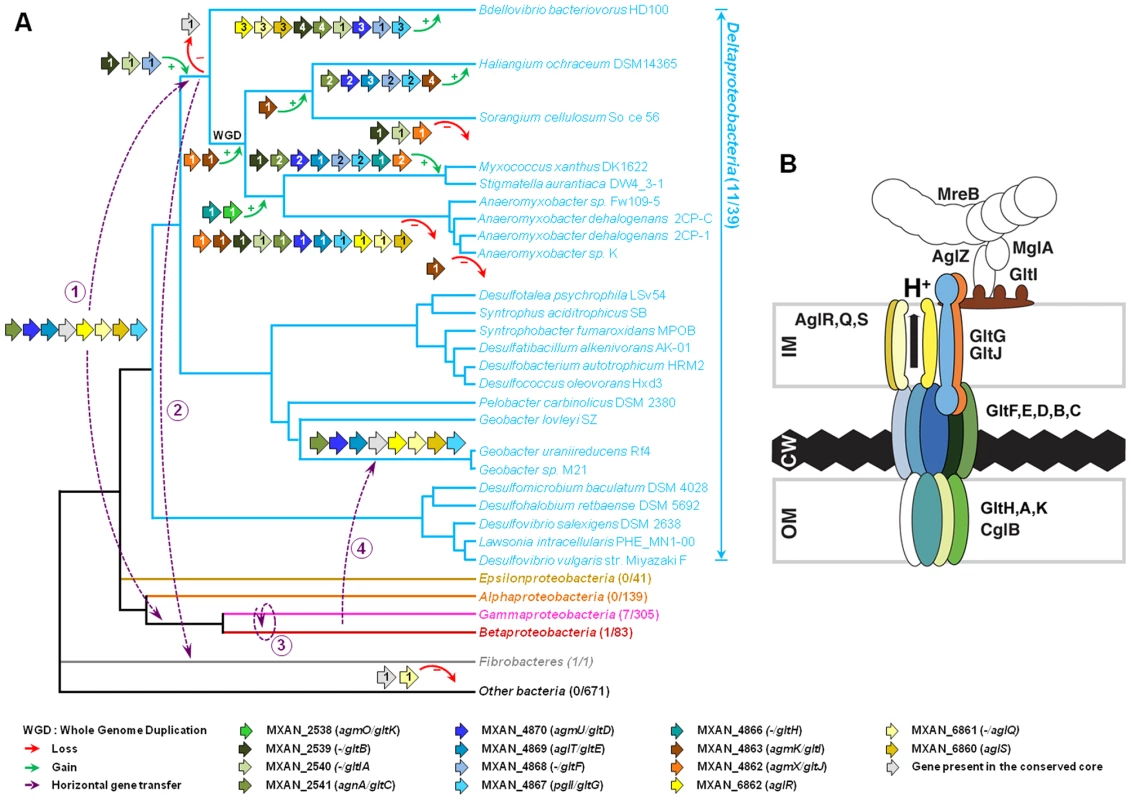
(A) Evolutionary scenario describing the emergence and evolution of the gliding motility machinery in M. xanthus. The relationships between organisms carrying close homologues of the 14 genes encoding putative components of the gliding machinery in M. xanthus are represented by the phylogeny. Green and red arrows respectively indicate gene acquisition and gene loss. The number of gene copies that were acquired or lost is indicated within arrows. The purple dotted arrows represent horizontal gene transfer events of one or several components. WGD marks the putative whole genome duplication event that occurred in the ancestor of Myxococcales. For each gene, locus_tag, former (agm/agl/agn) and new (glt and agl) names are provided. The number of complete genomes that contain homologues of glt and agl genes compared to the total number of complete genomes available at the beginning of this study are indicated in brackets. (B) The Myxococcus gliding machinery. The diagram compiles data from this work and published literature. Components were added based on bioinformatic predictions, mutagenesis, interaction and localization studies. Exhaustive information is not available for all proteins and thus the diagram largely is subject to modifications once more data will be available. Known interactions within the complex from experimental evidence are AglR-GltG, AglZ-MglA and interactions within the AglRQS molecular motor [13], [15]. For clarity, the proteins were colour-coded as in the rest of the manuscript. The evolutionary history of the genes involved in the gliding machinery is complicated by multiple duplication events, sometime followed by gene losses, which occurred in Myxococcales and Bdellovibrionales but also in Gammaproteobacteria (e.g. two Marinobacter, Teredinibacter turnerae and Saccharophagus degradans) and Fibrobacteres (Figure 8A and Figure S1). As a result, the gene clusters are sometimes present in several copies in the genomes of some species (i.e. the G3, G4, G5 and M2 clusters in M. xanthus, Figure 1B). Interestingly, none of these copies can substitute for the motility functions of the G1, G2 and M1 genes suggesting that duplications were associated with the emergence of novel functions (see below).
The Myxococcus gliding machinery and its distribution in bacteria
The data strongly suggest that an ancestral cluster of genes containing GltD, E, G, C and AglR, Q, S (Group B genes) evolved into a motility machinery after sequential recruitment of new components, namely GltF, H, I, J, A B and K (Group A genes). Obviously, ancient genetic linkages were lost during the evolution of the gliding machinery, explaining why it has not been previously identified and precluding the rapid identification of all the motility genes. The Agl/Glt complex is likely the gliding machinery because: (i), individual mutations of all the aglRQS [13] and glt genes resulted in complete motility defects and were not suppressed by a second-site frz mutation. (ii), GltD - and GltF-mCherry fusions showed similar localization patterns and localized to fixed FACs like the AglRQS proteins [13] (iii) GltD localization depended on GltF, G and H and, (iv) AglR interacted with GltG in a bacterial two-hybrid study and the localization of GltD-mCherry depended on AglQ. We thus propose that mechanical work from the motor is transduced to the cell surface by the Glt complex through a specific interaction with GltG. Here, we have identified a minimal motility machinery gene set, and it cannot be excluded that more Glt proteins may emerge as functional and phylogenomic approaches will be continued. For example, interaction studies identified GltD, E, I, J to be part of a complex also containing CglB and AglW [18]. The functional relevance of these interactions still needs to be demonstrated. CglB is an outer membrane motility lipoprotein [28] that harbours a very restricted taxonomy distribution, being present only in Myxococcales genomes (except the four Anaeromyxobacter). CglB may be a surface-exposed components of the complex. However, AglW may not be specifically linked to motility because its genomic localization and its amino acid sequence indicate that it is a bona fide structural component of the Tol/Pal system (TolB).
The discovery of the Agl/Glt machinery now provides a new framework to elucidate the gliding motility mechanism (Figure 8B). A low-resolution architecture of the apparatus may be suggested by the genetic/localization/interaction and bioinformatic results (Figure 8B). AglRQS and GltG may constitute an inner membrane platform (this study) linked to the MreB cytoskeleton via proteins such as AglZ and MglA [15] on the cytosolic side and anchored to the substratum by a GltA-K complex in the cell envelope (Figure 8B). Nan et al. [14] proposed that the motility complex does not traverse the peptidoglycan layer but rather deforms it, generating transverse waves propagating down the axis of the cell. While this is plausible, the finding that GltH (and also potentially GltK and GltA based on bioinformatics predictions) is a critical outer-membrane component of the machinery rather argues that the motility complex is continuous through the cell envelope and contacts the cell exterior directly. More work will be required to understand the individual functions of the Glt proteins but the identification of the machinery gene set now opens investigations to understand the motility mechanism as a whole.
Elucidating the motility mechanism may be greatly facilitated by functional studies of the core complex (aglR, Q, S and gltD, E, G, C). The conservation and genetic linkage of these genes in gammaproteobacterial genomes suggest that they encode a functional protein complex of unknown function in these bacteria. It is unlikely that the core complex drives motility on its own because gliding is not documented in most bacteria where it is found and our study shows that the corresponding genes are not sufficient to drive motility. Based on this later observation, we propose that additional functional blocks (such as Group A genes) have been added sequentially to the original protein complex to convert it into a motility machinery (Figure 8A). What are these building blocks and how many of them remain to be discovered?
A recent study unambiguously showed that Bdellovibrio bacteriovorus is a bona fide glider [29]. While we cannot definitively rule out the independent emergence of gliding motility in this bacterium, we consider it unlikely: the Bdellovibrio genome contains four sets of expanded core complex suggesting that the Bdellovibrionales and Myxococcales gliding apparati are linked evolutionarily. Gliding motility may thus have emerged quite early in the ancestor of the Myxococcales and the Bdellovibrionales. The absence of homologues of GltH, I, J and K, all essential gliding proteins in Myxococcus, in the Bdellovibrio genome suggests that there are species specific requirements for gliding motility (Figure S1). Bdellovibrio cells are unusually small (less than 1 µm in length and 0.5 in diameter vs > µm in length and 1 µm in diameter for Myxococcus), which could explain some structural differences between gliding apparati. Based on the phylogenetic analysis of the agl components, the genes composing the Bd0828-0838 locus appear more closely related to aglRQS and may therefore constitute the best candidate to encode the Bdellovibrio gliding apparatus (Figure 7B). The Bd0828-0838 cluster also contains many homologues of M. xanthus gliding genes (with the exception of gltH, I, J and K, Figure 7A and Figure S1). Based on the Bdellovibrio example, it is tempting to speculate that any bacterium containing AglR,Q,S, GltD, E, F, G, C, A and B is a potential glider. This is for instance the case of Myxococcus close relatives, Stigmatella aurantiaca and the four Anaeromyxobacter species (Figure S1).
Finally, the M. xanthus gliding machinery is not conserved in bacteria belonging in other phyla (e.g. Bacteroidetes or Cyanobacteria), confirming that gliding motility evolved several times independently in Bacteria, as suggested by Jarrell and McBride [8].
AglRQS/Glt-like machineries are exquisitely specialized
The presence of multiple copies of the G1, G2 and M1 clusters in Myxococcales (e.g. G3, G4, G5 and M2, in M. xanthus Figure 1B) likely results from duplication events. These duplications may be linked to the whole genome duplication event that occurred in the ancestor of the Myxococcales [30] and/or resulted from punctual gene-duplications during differentiation of the terminal branch of the Deltaproteobacteria. Duplications provide the raw material for the evolution of new gene functions [31], [32] and, for example, several regulation networks may have emerged this way in Myxococcus [30]. This study shows that the G3, G4, G5 and M2 clusters cannot compensate disruptions in the glt and aglRQS genes, already suggesting that they encode distinct functions. To further test this, we generated polar mutations in all the gltD homologues (MXAN_1922, G4; MXAN_1327, G5 and MXAN_3374, G3) and a deletion in MXAN_3004 (M2), the aglQ homologue [13]. None of these mutations impacted motility at any appreciable level, (Figure S5 and [13]). If the function of the G4, G5 and M2 regions is unknown, the G3 region was recently shown to be critical for sporulation and named nfs (necessary for sporulation, [33]). As expected, our nfsD (MXAN_3374) mutant failed to mature spores (data not shown). The nfsA-H genes are clustered in a single genomic region containing close homologues of G1 and G2 region genes (with the exception of GltI, J and K). Strikingly, the short evolutionary distances separating the nfs and glt genes in individual gene trees and in the glt supermatrix indicate that the nfsA-H genes are in fact the closest homologues of the glt genes (Figure 7 and Figure S4). Thus, the Glt and Nfs systems are a clear example of exquisite machinery specialization: in these cases, the ancestral core machinery has terminally differentiated to drive sporulation or gliding motility. In absence of more mechanistic insights, it is not clear which of the two processes is the most recent but this finding points to unsuspected similarities in these two distinct morphological processes. Comparative molecular analysis of the nsf and glt systems should be powerful to understand how these machineries function and how they can be specialized to enforce sporulation or gliding.
Conclusions
In summary, the mechanism of gliding motility has remained mysterious despite three decades of research. A converging array of evidence now shows that motility is not propelled by slime secretion but results from PMF-energized trans-envelope complexes periodically distributed along the cell body (this study and [12], [13], [18]). However, how force is transduced from the AglRQS motor to the Glt proteins through the entire cell envelope and ultimately how that translates into motion, remains to be elucidated. The identification of the components of the gliding machinery now paves the way to address these questions. An immediate goal will be to characterize the motility machinery in an exhaustive manner, which we should be able to resolve combining bioinformatics, genetics and cell biology. In addition, the M. xanthus genome contains several gene clusters deriving from the ancestral core complex, but these copies are not functionally redundant and even specify non-motility related functions (i.e. the sporulation nfs system). Thus, Glt-like systems are remarkably linked to two fundamental processes of the Myxococcus life cycle and their acquisition may thus have been critical to the recent diversification of the Deltaproteobacteria. In the future, comparative analysis in M. xanthus, but also in the Delta and Gammaproteobacteria should be a powerful approach to elucidate the pathways that led to the evolution and diversification of complex bacterial envelope machineries.
Materials and Methods
Bacterial strains, plasmids, and growth
Primers and plasmids are listed in Tables S4 and S5. See Tables S6 and S7 for strains and their mode of construction. M. xanthus strains were grown at 32°C in CYE rich media as previously described [23]. Plasmids were introduced in M. xanthus by electroporation. Mutants and transformants were obtained by homologous recombination based on a previously reported method. Complementation of gltG and expression of GltF-mCherry were obtained after ectopic integration of the genes of interest at the Mx8-phage attachment site in appropriate deletion backgrounds (Table S6).
For phenotypic assays, cells (10 µl), at a concentration of 4×109 cfu ml−1, were spotted on CYE plates containing agar concentrations of 0.5% or 1.5%, incubated at 32°C and photographed after 48 h with an Olympus SZ61 binocular or a Nikon Eclipse (model TE2000E) microscope (4x objective).
mRNA extraction and QT-Reverse Transcription PCR
RNA from appropriate strains was extracted using a standard RNA purification kit (Promega). One microgram of total RNA was reverse-transcribed following the recommendations of the iScript cDNA Synthesis Kit (Bio-Rad). The resulting cDNA was then diluted (1/16), and 5 µl were used for the quantitative reverse transcription-PCR (q-RT-PCR) reaction. This step was performed on a Mastercycler ep realplex instrument (Eppendorf), using the SYBR Premix Ex Taq (Perfect Real Time) PCR kit (Takara Bio Group, Japan) according to manufacturer instructions in a final volume of 20 µl. Specific primers used for the reactions are described in Table S4. Melting curves were systematically analyzed to control for the specificity of the PCR reactions. The relative units were calculated from a standard curve plotting four different dilutions (1/80, 1/400, 1/2,000, and 1/10,000) against the PCR cycle number at which the measured fluorescence intensity reached the threshold (CT), corresponding to ∼10 times the standard deviation and thus significantly above the noise band of the baseline.
Western blotting
Western blotting was performed as previously described [34] with 1/1000-1/5000 dilutions of polyclonal α-GltD, α-GltE, α-GltG, α-GltH (all raised for this study) and α-mCherry[13], α-PilC, α-PilQ [35] and α-FrzS [36].
Preparation of cell membrane fractions and OMVs
Membrane Fractions and OMVs were purified from exponentially-growing cell cultures. Vegetative cells of M. xanthus were grown in CYE medium to an OD600 nm = 0.7. For membrane fractions, cells were harvested by centrifugation at 8.000 rpm for 10 min at RT, resuspended in 50 mM Tris-HCl pH 7.6 and lysed by sonication. Cell debris were removed by low speed centrifugation (14000 rpm). The supernatants were then centrifuged at 45,000 g for 1 hr at 4°C. The resulting supernatants are enriched in soluble proteins. Pellets containing the crude envelope fractions (Inner and outer membrane) were resuspended in 50 mM Tris-HCl pH 7.6 and homogenized. The quality of the fractionation procedure was tested with antibodies to FrzS (soluble protein [36]) and PilC (inner membrane protein [35]).
Standard procedures to separate the inner membrane from the outer membrane using sucrose density gradients [37] did not successfully separate the two membranes. Detergent-based methods have been used successfully in Myxococcus, however in our case we could not prevent rapid degradation of the Glt proteins during the separation process [35]. OMVs are largely derived from the outer membranes, which was recently confirmed by proteomic analysis of the Myxococcus outer-membranes [38]. Thus, to test which Glt proteins are in the outer membranes we tested their presence in purified vesicules. For OMVs purification, cells and were discarded by centrifugation (8.000 rpm for 10 min at RT) and the culture supernatant was used for the isolation of vesicles. Culture supernatants (1 L) were passed through a 0.2 µm vacuum filter (Millipore). The resulting filtrate was centrifuged at 125 000× g for 2 h at 4°C to recover membrane vesicles. The supernatant was carefully removed and the vesicle pellet was resuspended in 50 mM Tris-HCl pH 7.6 and centrifuged at 180 000× g for 2 h at 4°C to concentrate and wash vesicles. The quality of the purification procedure was tested by electron microscopy (not shown) and antibodies to PilQ (outer membrane protein [35]) and PilC (inner membrane protein [35]).
Bacterial two-hybrid experiments
Bacterial two-hybrid experiments, plate and ß-Galactosidase assays were performed as previously described [26] and as recommended by the manufacturer (Euromedex).
Time lapse video-microscopy
Time lapse experiments were performed as previously described [39]. Microscopic analysis was performed using an automated and inverted epifluorescence microscope TE2000-E-PFS (Nikon, France). The microscope is equipped with “The Perfect Focus System” (PFS) that automatically maintains focus so that the point of interest within a specimen is always kept in sharp focus at all times, in spite of any mechanical or thermal perturbations. Images were recorded with a CoolSNAP HQ 2 (Roper Scientific, Roper Scientific SARL, France) and a 40x/0.75 DLL “Plan-Apochromat” or a 100x/1.4 DLL objective. All fluorescence images were acquired with appropriate filters with a minimal exposure time to minimize bleaching and phototoxicity effects.
Cell tracking was performed automatically using a previously described macro under the METAMORPH software (Molecular devices), when appropriate, manual measurements were also performed to correct tracking errors with tools built into the software. Typically, the images were equalized, straightened and overlaid under both ImageJ 1.40 g (National Institute of Health, USA) and METAMORPH.
Annotation and mapping of gliding motility genes
The genetic screens of Youderian et al. ([16]) and Yu and Kaiser ([17]) allowed the identification of 35 and 23 potential gliding motility genes, respectively (Table S1). The function of the corresponding proteins was investigated using sequence similarity based approaches against the non-redundant (nr) database at the National Center for Biotechnology Information (NCBI, http://www.ncbi.nlm.nih.gov/) and Pfam (release 24.0) databases ([40]). The presence and location of signal peptide signal cleavage sites and of transmembrane helix were the predicted using the signalP 3.0 server ([41]) and TMHMM server v.2.0 ([42]). Finally, the location and the neighbourhood of each gene in the chromosome of M. xanthus DK 1622 were investigated using the complete genome sequence available at the NCBI ([30]; CP000113).
Datasets construction
Homologues of each M. xanthus candidate protein were retrieved from a local database containing all complete prokaryotic proteomes available at the NCBI (April 8, 2010) using blastp with default parameters [43]. We also include in our analyses the genome sequences of Stigmatella auriantiaca DW4/3-1 and of Plesiocystis pacifica SIR-1 that came out in November 2010 and whose assembly is ongoing, respectively, both genomes being available at the NCBI. Importantly, the distinction between homologous and non-homologous sequences was assessed by visual inspection of each blastp outputs (no arbitrary cut-off on the E-value or score). To ensure the exhaustive sampling of homologues, iterative blastp queries were performed using homologues identified at each step as new seeds. PSI-BLAST queries were also used in order to recover very divergent homologues [43]. The absence of homologue in any complete proteome was systematically verified by tblastn queries against the nucleotide sequence of the corresponding genome. For each candidate protein, the retrieved homologues were gathered in a dataset. The corresponding sequences were aligned using the ClustalW2 program (Default parameters, [44]). Each alignment was visually inspected and manually refined when necessary using the ED program from the MUST package [45]. Regions where the homology between amino acid positions was doubtful were manually removed using NET from the MUST package.
Working on complete genomes may introduce major biases due to the taxonomic sampling of available complete genomes. Accordingly, for each candidate protein a second set of datasets based on homologues retrieved from the non-redundant (nr) protein database (the most exhaustive public database) at the NCBI was assembled. The taxonomic distribution and the phylogeny of homologues retrieved from either the nr database or from complete genomes showed similar patterns (data not shown). Thus, our analyses based on complete genomes are representative and reflect the taxonomic distribution of known homologues. Accordingly, only the results based on complete genomes will be presented in the results section.
The preliminary phylogenetic analyses of the candidate proteins allowed the identification of closest relatives of M. xanthus sequences. For each protein these homologues were gathered in a second dataset, the sequences were aligned and the resulting alignment was manually refined and cleaned like previously described. For the phylogenetic analyses of some of these datasets, we were removed some divergent sequences that can bias the phylogenetic reconstruction.
One approach to improve the resolution of the phylogenetic trees is to combine the genes that share a common evolutionary history in a single large alignment (also called supermatrix), [46]–[48]. Among the seven genes composing the Group B, gltD, E and G homologues are always clustered together in genomes and their individual phylogenies are very similar. Thus, these genes likely share a similar evolutionary history and can be used to build a supermatrix. For similar reasons, we combined the aglR, Q and S alignments in a second supermatrix. In contrast, gltC could neither be included in the glt nor in the agl supermatrix because it does not cluster physically with the corresponding genes in most Deltaproteobacteria. The Glt and Agl supermatrices were manually constructed by combining the cleaned alignments of GltDEG and AglQRS, respectively. When more than one homologue of these genes were present in a given genome, the genes were combined according to their physical linkage on the chromosome. For instance in the case of AglQRS, in M. xanthus the genes were combined as following: (i) MXAN_6860, _6861, _6862 and (ii) MXAN_3005, _3004, _3003.
Phylogenetic analyses
For each individual and concatenated alignment, both Maximum likelihood (ML) and Bayesian phylogenetic trees were computed. ML analyses were run using PHYML version 3.0 with the Le and Gascuel (LG) model (amino acid frequencies estimated from the dataset) and a gamma distribution (4 discrete categories of sites and an estimated alpha parameter) to take into account evolutionary rate variations across sites [49]. The robustness of each branch was estimated by the non-parametric bootstrap procedure implemented in PhyML (100 replicates of the original dataset with the same parameters). Additional ML analyses were performed using TreeFinder with the same parameters [50]. Bayesian analyses were performed using MrBayes [51] with a mixed model of amino acid substitution including a gamma distribution (4 discrete categories) and an estimated proportion of invariant sites. MrBayes was run with four chains for 1 million generations and trees were sampled every 100 generations. To construct the consensus tree, the first 1500 trees were discarded as “burnin”.
Supporting Information
Zdroje
1. RegoATFronzesRWaksmanG 2010 SnapShot: Bacterial Appendages I. Cell 140 162 162.e1 doi:10.1016/j.cell.2009.12.047
2. RegoATFronzesRWaksmanG 2010 SnapShot: Bacterial appendages II. Cell 140 294 294.e1 doi:10.1016/j.cell.2010.01.006
3. ErhardtMNambaKHughesKT 2010 Bacterial nanomachines: the flagellum and type III injectisome. Cold Spring Harb Perspect Biol 2 a000299 doi:10.1101/cshperspect.a000299
4. PallenMJMatzkeNJ 2006 From The Origin of Species to the origin of bacterial flagella. Nat. Rev. Microbiol 4 784 790 doi:10.1038/nrmicro1493
5. MerzAJSoMSheetzMP 2000 Pilus retraction powers bacterial twitching motility. Nature 407 98 102 doi:10.1038/35024105
6. SkerkerJMPrasolMSPerchukBSBiondiEGLaubMT 2005 Two-component signal transduction pathways regulating growth and cell cycle progression in a bacterium: a system-level analysis. PLoS Biol 3 e334 doi:10.1371/journal.pbio.0030334
7. PelicicV 2008 Type IV pili: e pluribus unum? Mol. Microbiol 68 827 837 doi:10.1111/j.1365-2958.2008.06197.x
8. JarrellKFMcBrideMJ 2008 The surprisingly diverse ways that prokaryotes move. Nat. Rev. Microbiol 6 466 476 doi:10.1038/nrmicro1900
9. MignotT 2007 The elusive engine in Myxococcus xanthus gliding motility. Cell. Mol. Life Sci 64 2733 2745 doi:10.1007/s00018-007-7176-x
10. SatoKNaitoMYukitakeHHirakawaHShojiM 2010 A protein secretion system linked to bacteroidete gliding motility and pathogenesis. Proc. Natl Acad Sci U S A 107 276 281 doi:10.1073/pnas.0912010107
11. MaurielloEMFMignotTYangZZusmanDR 2010 Gliding motility revisited: how do the myxobacteria move without flagella? Microbiol. Mol. Biol. Rev 74 229 249 doi:10.1128/MMBR.00043-09
12. MignotTShaevitzJWHartzellPLZusmanDR 2007 Evidence that focal adhesion complexes power bacterial gliding motility. Science 315 853 856 doi:10.1126/science.1137223
13. SunMWartelMCascalesEShaevitzJWMignotT 2011 Motor-driven intracellular transport powers bacterial gliding motility. Proc. Natl Acad Sci 108 U.S.A 7559 7564 doi:10.1073/pnas.1101101108
14. NanBChenJNeuJCBerryRMOsterG 2011 Myxobacteria gliding motility requires cytoskeleton rotation powered by proton motive force. Proc Natl Acad Sci 108 U.S.A 2498 2503 doi:10.1073/pnas.1018556108
15. MaurielloEMFMouhamarFNanBDucretADaiD 2010 Bacterial motility complexes require the actin-like protein, MreB and the Ras homologue, MglA. EMBO J 29 315 326 doi:10.1038/emboj.2009.356
16. YouderianPBurkeNWhiteDJHartzellPL 2003 Identification of genes required for adventurous gliding motility in Myxococcus xanthus with the transposable element mariner. Mol. Microbiol 49 555 570
17. YuRKaiserD 2007 Gliding motility and polarized slime secretion. Mol. Microbiol 63 454 467 doi:10.1111/j.1365-2958.2006.05536.x
18. NanBMaurielloEMFSunI-HWongAZusmanDR 2010 A multi-protein complex from Myxococcus xanthus required for bacterial gliding motility. Mol. Microbiol 76 1539 1554 doi:10.1111/j.1365-2958.2010.07184.x
19. YehY-CComolliLRDowningKHShapiroLMcAdamsHH 2010 The caulobacter Tol-Pal complex is essential for outer membrane integrity and the positioning of a polar localization factor. J Bacteriol 192 4847 4858 doi:10.1128/JB.00607-10
20. LloubèsRCascalesEWalburgerABouveretELazdunskiC 2001 The Tol-Pal proteins of the Escherichia coli cell envelope: an energized system required for outer membrane integrity? Res. Microbiol 152 523 529
21. BlatchGLLässleM 1999 The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. Bioessays 21 932 939 doi:10.1002/(SICI)1521-1878(199911)21 : 11<932::AID-BIES5>3.0.CO;2-N
22. HammetAPikeBLMcNeesCJConlanLATenisN 2003 FHA domains as phospho-threonine binding modules in cell signaling. IUBMB Life 55 23 27 doi:10.1080/1521654031000070636
23. BustamanteVHMartínez-FloresIVlamakisHCZusmanDR 2004 Analysis of the Frz signal transduction system of Myxococcus xanthus shows the importance of the conserved C-terminal region of the cytoplasmic chemoreceptor FrzCD in sensing signals. Mol. Microbiol 53 1501 1513 doi:10.1111/j.1365-2958.2004.04221.x
24. MaurielloEMFNanBZusmanDR 2009 AglZ regulates adventurous (A-) motility in Myxococcus xanthus through its interaction with the cytoplasmic receptor, FrzCD. Mol. Microbiol 72 964 977 doi:10.1111/j.1365-2958.2009.06697.x
25. GoemaereELCascalesELloubèsR 2007 Mutational analyses define helix organization and key residues of a bacterial membrane energy-transducing complex. J. Mol. Biol 366 1424 1436 doi:10.1016/j.jmb.2006.12.020
26. KarimovaGPidouxJUllmannALadantD 1998 A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc Natl Acad Sci 95 U.S.A 5752 5756
27. KriegNRLudwigWWhitmanWBHedlung BP PasterBJ 2011 Bergey's Manual of Systematic Bacteriology. Dans 948
28. NudlemanEWallDKaiserD 2005 Cell-to-cell transfer of bacterial outer membrane lipoproteins. Science 309 125 127 doi:10.1126/science.1112440
29. LambertCFentonAKHobleyLSockettRE 2011 Predatory bdellovibrio bacteria use gliding motility to scout for prey on surfaces. J Bacteriol 193 3139 3141 doi:10.1128/JB.00224-11
30. GoldmanBSNiermanWCKaiserDSlaterSCDurkinAS 2006 Evolution of sensory complexity recorded in a myxobacterial genome. Proc Natl Acad Sci 103 U.S.A 15200 15205 doi:10.1073/pnas.0607335103
31. SémonMWolfeKH 2007 Consequences of genome duplication. Curr. Opin. Genet. Dev 17 505 512 doi:10.1016/j.gde.2007.09.007
32. Van de PeerYMaereSMeyerA 2009 The evolutionary significance of ancient genome duplications. Nat. Rev. Genet 10 725 732 doi:10.1038/nrg2600
33. MüllerF-DTreuner-LangeAHeiderJHuntleySMHiggsPI 2010 Global transcriptome analysis of spore formation in Myxococcus xanthus reveals a locus necessary for cell differentiation. BMC Genomics 11 264 doi:10.1186/1471-2164-11-264
34. ZhangYFrancoMDucretAMignotT 2010 A bacterial Ras-like small GTP-binding protein and its cognate GAP establish a dynamic spatial polarity axis to control directed motility. PLoS Biol 8 e1000430 doi:10.1371/journal.pbio.1000430
35. BulyhaISchmidtCLenzPJakovljevicVHöneA 2009 Regulation of the type IV pili molecular machine by dynamic localization of two motor proteins. Mol. Microbiol 74 691 706 doi:10.1111/j.1365-2958.2009.06891.x
36. MignotTMerlieJPJrZusmanDR 2005 Regulated pole-to-pole oscillations of a bacterial gliding motility protein. Science 310 855 857 doi:10.1126/science.1119052
37. ViarreVCascalesEBallGMichelGPFFillouxA 2009 HxcQ liposecretin is self-piloted to the outer membrane by its N-terminal lipid anchor. J. Biol. Chem 284 33815 33823 doi:10.1074/jbc.M109.065938
38. KahntJAguiluzKKochJTreuner-LangeAKonovalovaA 2010 Profiling the outer membrane proteome during growth and development of the social bacterium Myxococcus xanthus by selective biotinylation and analyses of outer membrane vesicles. J Proteome Res 9 5197 5208 doi:10.1021/pr1004983
39. DucretAMaisonneuveENotareschiPGrossiAMignotT 2009 A microscope automated fluidic system to study bacterial processes in real time. PLoS ONE 4 e7282 doi:10.1371/journal.pone.0007282
40. FinnRDMistryJTateJCoggillPHegerA 2010 The Pfam protein families database. Nucleic Acids Res 38 D211 222 doi:10.1093/nar/gkp985
41. BendtsenJDNielsenHvon HeijneGBrunakS 2004 Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol 340 783 795 doi:10.1016/j.jmb.2004.05.028
42. KroghALarssonBvon HeijneGSonnhammerEL 2001 Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305 567 580 doi:10.1006/jmbi.2000.4315
43. AltschulSFMaddenTLSchäfferAAZhangJZhangZ 1997 Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25 3389 3402
44. LarkinMABlackshieldsGBrownNPChennaRMcGettiganPA 2007 Clustal W and Clustal X version 2.0. Bioinformatics 23 2947 2948 doi:10.1093/bioinformatics/btm404
45. PhilippeH 1993 MUST, a computer package of Management Utilities for Sequences and Trees. Nucleic Acids Res 21 5264 5272
46. BrownJRDouadyCJItaliaMJMarshallWEStanhopeMJ 2001 Universal trees based on large combined protein sequence data sets. Nat. Genet 28 281 285 doi:10.1038/90129
47. BrochierCBaptesteEMoreiraDPhilippeH 2002 Eubacterial phylogeny based on translational apparatus proteins. Trends Genet 18 1 5
48. DelsucFBrinkmannHPhilippeH 2005 Phylogenomics and the reconstruction of the tree of life. Nat. Rev. Genet 6 361 375 doi:10.1038/nrg1603
49. GuindonSGascuelO 2003 A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol 52 696 704
50. JobbGvon HaeselerAStrimmerK 2004 TREEFINDER: a powerful graphical analysis environment for molecular phylogenetics. BMC Evol. Biol 4 18 doi:10.1186/1471-2148-4-18
51. HuelsenbeckJPRonquistF 2001 MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17 754 755
Štítky
Genetika Reprodukční medicína
Článek Produces a Long Noncoding RNA and Has General Functions in the Germline, Stem Cells, and BrainČlánek Inference of Relationships in Population Data Using Identity-by-Descent and Identity-by-StateČlánek A Genome-Wide Meta-Analysis of Six Type 1 Diabetes Cohorts Identifies Multiple Associated LociČlánek Retrotransposon-Induced Heterochromatin Spreading in the Mouse Revealed by Insertional PolymorphismsČlánek Essential Roles of BCCIP in Mouse Embryonic Development and Structural Stability of ChromosomesČlánek The Receptor Tyrosine Kinase Alk Controls Neurofibromin Functions in Drosophila Growth and LearningČlánek PCNA Ubiquitination Is Important, But Not Essential for Translesion DNA Synthesis in Mammalian CellsČlánek Genome-Wide Association Study Identifies Four Loci Associated with Eruption of Permanent Teeth
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2011 Číslo 9- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- IVF a rakovina prsu – zvyšují hormony riziko vzniku rakoviny?
-
Všechny články tohoto čísla
- Temporal Trends in Results Availability from Genome-Wide Association Studies
- Association of eGFR-Related Loci Identified by GWAS with Incident CKD and ESRD
- Genetic Variants at Chromosomes 2q35, 5p12, 6q25.1, 10q26.13, and 16q12.1 Influence the Risk of Breast Cancer in Men
- Large-Scale Gene-Centric Analysis Identifies Novel Variants for Coronary Artery Disease
- Genetic Association for Renal Traits among Participants of African Ancestry Reveals New Loci for Renal Function
- Transcriptome Kinetics Is Governed by a Genome-Wide Coupling of mRNA Production and Degradation: A Role for RNA Pol II
- Conserved Regulation of p53 Network Dosage by MicroRNA–125b Occurs through Evolving miRNA–Target Gene Pairs
- Heterozygous Mutations of Are Associated with an Increased Risk of Isolated Metopic Craniosynostosis in Humans and Mice
- Study of FoxA Pioneer Factor at Silent Genes Reveals Rfx-Repressed Enhancer at and a Potential Indicator of Esophageal Adenocarcinoma Development
- Cholesterol Metabolism Is Required for Intracellular Hedgehog Signal Transduction
- Produces a Long Noncoding RNA and Has General Functions in the Germline, Stem Cells, and Brain
- Age-Dependent Recombination Rates in Human Pedigrees
- Sequence Conservation and Functional Constraint on Intergenic Spacers in Reduced Genomes of the Obligate Symbiont
- Sex Chromosome Mosaicism and Hybrid Speciation among Tiger Swallowtail Butterflies
- A Negative Feedback Loop That Limits the Ectopic Activation of a Cell Type–Specific Sporulation Sigma Factor of
- Phased Whole-Genome Genetic Risk in a Family Quartet Using a Major Allele Reference Sequence
- Mutations in or near the Transmembrane Domain Alter PMEL Amyloid Formation from Functional to Pathogenic
- Inactivation of Alters Melanosome Shape But Has Only a Subtle Effect on Visible Pigmentation
- Novel Interactions between Actin and the Proteasome Revealed by Complex Haploinsufficiency
- Germline Genetic Variants Disturbing the /LIN28 Double-Negative Feedback Loop Alter Breast Cancer Susceptibility
- Separation of Recombination and SOS Response in RecA Suggests LexA Interaction Sites
- Inference of Relationships in Population Data Using Identity-by-Descent and Identity-by-State
- Misregulation of Scm3p/HJURP Causes Chromosome Instability in and Human Cells
- A Noncoding Point Mutation of Causes Multiple Developmental Malformations and Obesity in Twirler Mice
- The Evolutionarily Conserved Longevity Determinants HCF-1 and SIR-2.1/SIRT1 Collaborate to Regulate DAF-16/FOXO
- A Genome-Wide Metabolic QTL Analysis in Europeans Implicates Two Loci Shaped by Recent Positive Selection
- Bacterial Communities of Diverse Species: Ecological Context of a Host–Microbe Model System
- A Genome-Wide Meta-Analysis of Six Type 1 Diabetes Cohorts Identifies Multiple Associated Loci
- Elongator Complex Influences Telomeric Gene Silencing and DNA Damage Response by Its Role in Wobble Uridine tRNA Modification
- Elevated Proteasome Capacity Extends Replicative Lifespan in
- Genome-Wide Analysis of Heteroduplex DNA in Mismatch Repair–Deficient Yeast Cells Reveals Novel Properties of Meiotic Recombination Pathways
- MicroRNA Predictors of Longevity in
- An Iterative Genetic and Dynamical Modelling Approach Identifies Novel Features of the Gene Regulatory Network Underlying Melanocyte Development
- Atypical AT Skew in Firmicute Genomes Results from Selection and Not from Mutation
- Retrotransposon-Induced Heterochromatin Spreading in the Mouse Revealed by Insertional Polymorphisms
- Genomic Analysis of QTLs and Genes Altering Natural Variation in Stochastic Noise
- The Abnormal Phenotypes of Cartilage and Bone in Calcium-Sensing Receptor Deficient Mice Are Dependent on the Actions of Calcium, Phosphorus, and PTH
- Cell Type–Specific Transcriptome Analysis Reveals a Major Role for and miR-200b in Mouse Inner Ear Morphogenesis
- Essential Roles of BCCIP in Mouse Embryonic Development and Structural Stability of Chromosomes
- IAP1-Mediated Ubiquitylation Controls Activation of the Initiator Caspase DRONC Independent of Protein Degradation
- VANG-1 and PRKL-1 Cooperate to Negatively Regulate Neurite Formation in
- The Receptor Tyrosine Kinase Alk Controls Neurofibromin Functions in Drosophila Growth and Learning
- Comparative and Functional Genomics of PD630 for Biofuels Development
- Identification of Type 1 Diabetes–Associated DNA Methylation Variable Positions That Precede Disease Diagnosis
- PCNA Ubiquitination Is Important, But Not Essential for Translesion DNA Synthesis in Mammalian Cells
- Genetic Effects at Pleiotropic Loci Are Context-Dependent with Consequences for the Maintenance of Genetic Variation in Populations
- Genome-Wide Association Study Identifies Four Loci Associated with Eruption of Permanent Teeth
- Bmp and Nodal Independently Regulate Expression to Maintain Unilateral Nodal Activity during Left-Right Axis Specification in Zebrafish
- Inter-Allelic Prion Propagation Reveals Conformational Relationships among a Multitude of [] Strains
- Emergence and Modular Evolution of a Novel Motility Machinery in Bacteria
- Histone Methyltransferase MET-2 Shields the Male X Chromosome from Checkpoint Machinery and Mediates Meiotic Sex Chromosome Inactivation
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Retrotransposon-Induced Heterochromatin Spreading in the Mouse Revealed by Insertional Polymorphisms
- The Evolutionarily Conserved Longevity Determinants HCF-1 and SIR-2.1/SIRT1 Collaborate to Regulate DAF-16/FOXO
- Genome-Wide Analysis of Heteroduplex DNA in Mismatch Repair–Deficient Yeast Cells Reveals Novel Properties of Meiotic Recombination Pathways
- Association of eGFR-Related Loci Identified by GWAS with Incident CKD and ESRD
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



